- 1Department of Bioenergy Science and Technology, Chonnam National University, Gwangju, South Korea
- 2Department of Life Science, Sogang University, Seoul, South Korea
Drought is a critical environmental stress that limits growth and development of plants and reduces crop productivity. The aerial part of land plants is covered with cuticular waxes to minimize water loss. To understand the regulatory mechanisms underlying cuticular wax biosynthesis in Arabidopsis under drought stress conditions, we characterized the role of an AP2/DREB type transcription factor, RAP2.4. RAP2.4 expression was detected in one-week-old seedlings and rosette leaves, stems, stem epidermis, cauline leaves, buds, flowers, and siliques of 6-week-old Arabidopsis. The levels of RAP2.4 transcripts increased with treatments of abscisic acid (ABA), mannitol, NaCl, and drought stress. Under drought, total wax loads decreased by approximately 11% and 10%, and in particular, the levels of alkanes, which are a major wax component, decreased by approximately 11% and 12% in rap2.4-1 and rap2.4-2 leaves, respectively, compared with wild type (WT) leaves. Moreover, the transcript levels of cuticular wax biosynthetic genes, KCS2 and CER1, decreased by approximately 15–23% and 32–40% in rap2.4-1 and rap2.4-2 leaves, respectively, relative to WT 4 h after drought treatment, but increased by 2- to 12-fold and 3- to 70-fold, respectively, in three independent RAP2.4 OX leaves relative to WT. Epicuticular wax crystals were observed on the leaves of RAP2.4 OX plants, but not on the leaves of WT. Total wax loads increased by 1.5- to 3.3-fold in leaves of RAP2.4 OX plants relative to WT. Cuticular transpiration and chlorophyll leaching occurred slowly in the leaves of RAP2.4 OX plants relative to WT. Transcriptional activation assay in tobacco protoplasts showed that RAP2.4 activates the expression of KCS2 and CER1 through the involvement of the consensus CCGAC or GCC motifs present in the KCS2 and CER1 promoter regions. Overall, our results revealed that RAP2.4 is a transcription factor that activates cuticular wax biosynthesis in Arabidopsis leaves under drought stress conditions.
Introduction
Drought is a severe environmental stress implicated in the reduction of plant growth and crop productivity. As plants are sessile, they must cope with drought stress for their optimal growth and development. During the transition of land plants from aquatic to terrestrial environments, they developed a key surface structure, the outermost cuticle layer covering their aerial tissues, to protect them from terrestrial stresses such as drought, excess light, and UV light (Shepherd and Griffiths, 2006; Bernard and Joubès, 2013; Yeats and Rose, 2013). Therefore, the primary function of the cuticle layer is to reduce water-loss through the epidermis, except through the stomata (Yeats and Rose, 2013; Lee and Suh, 2015a). The cuticle, which is the first physical barrier between plants and the environment, plays a role in the protection of plants from excess UV light, pathogenic spores, and insect attack (Eigenbrode and Espelie, 1995; Sieber et al., 2000; Holmes and Keiller, 2002; Yeats and Rose, 2013) and also in the prevention of organ fusion during plant development (Ingram and Nawrath, 2017). The hydrophobic cuticle layer mainly comprises the cutin polyester matrix, intracuticular waxes embedded in the cutin matrix, and epicuticular waxes. Cutin polyester is mainly formed by interlinking through ester bonds of ω-hydroxy C16 or C18 fatty acids and their derivatives (Beisson et al., 2012; Bakan and Marion, 2017). Major components of cuticular waxes are very long chain fatty acids (VLCFA, longer than C20) and their derivatives including alkanes, aldehydes, primary and secondary alcohols, and wax esters (Lee and Suh, 2013; Hegebarth and Jetter, 2017).
Cuticular wax biosynthesis mainly occurs in the epidermal cells of plants (Suh et al., 2005). The C16 and C18 fatty acids synthesized in the plastids are further elongated to VLCFAs by the fatty acid elongase (FAE) complex in the endoplasmic reticulum (ER) (Millar et al., 1999; Zheng et al., 2005; Bach et al., 2008; Beaudoin et al., 2009; Kunst and Samuels, 2009; Haslam and Kunst, 2013; Kim et al., 2013; Morineau et al., 2016). The VLCFAs are modified to VLC aliphatic compounds, which are aldehydes, alkenes, secondary alcohols, and ketones via the alkane-forming pathway and primary alcohols and wax-esters via the alcohol-forming pathway (Rowland et al., 2006; Greer et al., 2007; Li et al., 2008; Bourdenx et al., 2011; Li-Beisson et al., 2013; Bernard and Joubès, 2013; Lee and Suh, 2015a). The wax molecules synthesized in the ER are secreted through the ATP-binding cassette transporter in the plasma membrane and with the help of glycosylphosphatidyl-anchored lipid transfer proteins to extracellular space, and then deposited on the surface of epidermal cells (DeBono et al., 2009; Lee et al., 2009; Kim et al., 2012). The total wax load and composition varied greatly among plant species, in an organ-specific manner, or by environmental factors (Holmes and Keiller, 2002; Shepherd and Griffiths, 2006; Kosma et al., 2009; Seo et al., 2011; Go et al., 2014; Lee et al., 2016; Kim et al., 2018). In particular, the total wax load increased by approximately 2-fold in Arabidopsis and tree tobacco (Nicotiana glauca) leaves under drought stress conditions (Kosma et al., 2009; Seo et al., 2011; Lee et al., 2016). Increased total wax loads in leaves reduced cuticular transpiration rate and conferred resistance to drought (Seo et al., 2011; Lee et al., 2016). Noticeably, the increased levels of VLC-alkane components in Arabidopsis are prominent under drought stress conditions, indicating that the molecular regulatory mechanism underlying the alkane-forming pathway may be important to understand one of the plant strategies to cope with drought (Kosma et al., 2009; Xue et al., 2017).
It has long been questioned how land plants precisely regulate total wax loads in response to drought stress. Several transcription factors have been reported to be implicated in the upregulation of wax biosynthesis in plants under drought (Seo et al., 2011; Lee et al., 2016). WIN1/SHN1, which belongs to the AP2/ERF transcription factor family, is a transcriptional activator that induces the expression of KCS1, ECERIFERUM1 (CER1), and CER2 genes and its overexpression in Arabidopsis and N. tabacum caused increased resistance to drought (Aharoni et al., 2004; Broun et al., 2004; Kannangara et al., 2007; Djemal and Khoudi, 2019). Similarly, ectopic expression of Medicago truncatula WXP1 and WXP2 transcription factors containing the AP2 domain led to increased wax load and enhanced drought tolerance (Zhang et al., 2005; Zhang et al., 2007). In addition, the Arabidopsis R2R3-type transcription factor MYB96, which is induced by drought and ABA treatment, directly activates the expression of KCS1, KCS2, KCS6, KCR1, and CER3 genes involved in wax biosynthesis (Seo et al., 2011). Its functional homolog, MYB94, is a transcription activator that increases the levels of WSD1, KCS2, CER2, FAR3, and ECR transcripts in Arabidopsis (Lee and Suh, 2015b). MYB94 and MYB96 additively activate cuticular wax biosynthesis by binding to the same MYB consensus motifs in wax biosynthetic gene promoters (Lee et al., 2016). Interestingly, total wax loads of double knockout myb94 myb96 leaves decreased by approximately 44% and 52% relative to the wild type under well-watered and drought stress conditions, respectively, indicating that about 50% of total wax biosynthesis in Arabidopsis leaves is dependent on both MYB94 and MYB96 (Lee et al., 2016). However, it is still unclear how the biosynthesis of the remaining wax in myb94 myb96 is regulated in Arabidopsis under both conditions.
Arabidopsis transcriptome analysis showed that the expression of an AP2/DREB transcription factor, RELATED TO APETALA 2.4 (RAP2.4, At1g78080) was induced by drought and salt stress treatments (Feng et al., 2005). M. truncatula WXP1, which shares the highest sequence homology with Arabidopsis RAP2.4, displayed increased total wax loads and enhanced drought tolerance in transgenic alfalfa (Medicago sativa) (Zhang et al., 2005). Lin et al. (2008) observed enhanced drought tolerance in Arabidopsis overexpressing RAP2.4 (RAP2.4 OX), but no significant differences were detected in water loss between wild type and RAP2.4 OX lines. These results suggest that regulation of cuticular wax biosynthesis may be involved in the drought stress response mediated by Arabidopsis RAP2.4.
In this study, we investigated the role of an AP2/DREB-type transcription factor, RAP2.4 in cuticular wax biosynthesis in Arabidopsis leaves under drought stress condition. Quantitative Real-Time-PCR (qRT-PCR) analysis showed that RAP2.4 expression is induced by treatment with ABA, and drought, salt, and osmotic stress. Under drought stress condition, total wax loads significantly decreased in Arabidopsis rap2.4-1 and rap2.4-2 leaves relative to the wild type under drought stress conditions, but no remarkable differences between the wild type and rap2.4 mutants were observed under well-watered conditions. In addition, we observed that ectopic expression of RAP2.4 increased total wax loads in Arabidopsis leaves and delayed water-loss through the cuticular layer of leaves. Transactivation assay of RAP2.4 in N. benthamiana protoplasts revealed that the expression of KCS2 and CER1 involved in VLCFA and alkane biosynthesis, respectively, is induced by RAP2.4 expression and in particular, the CCGAC or GCC consensus motifs present in the KCS2 and CER1 promoter regions are required for RAP2.4-mediated elevation of KCS2 and CER1 expression. These results demonstrate that an AP2/DREB-type transcription factor, RAP2.4 activates cuticular wax biosynthesis by increasing the expression of KCS2 and CER1 in Arabidopsis leaves under drought. This study suggests that RAP2.4, in addition to MYB96 and MYB94, is also an important component of the transcriptional gene regulatory network involved in drought-induced wax biosynthesis in Arabidopsis.
Materials and Methods
Plant Materials and Growth Conditions
The Arabidopsis T-DNA insertion mutants rap2.4-1 (SALK_020767) and rap2.4-2 (SALK_093377) were obtained from the Arabidopsis Biological Resource Center (ABRC1). To isolate rap2.4 mutants, the genomic DNA was isolated from rosette leaves of 2-week-old Arabidopsis using DNA extraction buffer [200 mM Tris-Cl (pH 8.0), 250 mM NaCl, 25 mM EDTA (pH 8.0), 0.5% SDS]. Genomic DNA and gene-specific primers (Supplementary Table 1) were used for DNA-based PCR. The seeds of Arabidopsis wild type (Col-0), transgenic Arabidopsis lines overexpressing RAP2.4, and rap2.4 Arabidopsis mutants were surface-sterilized with 75% EtOH solution containing 0.05% Triton X-100 and 100% EtOH. The Arabidopsis seeds were germinated in 1/2 MS medium (0.22% Murashige and Skoog media, 1% sucrose, 0.7% phytoagar, pH 5.7). Transgenic Arabidopsis seeds were germinated in 1/2 MS medium containing kanamycin (25 μg/ml). Seven-day-old Arabidopsis seedlings were transferred to sterile soil (soil:vermiculite:perlite, 3:2:1). All growth conditions were maintained at 24 ± 2°C under long-day conditions (16 h/8 h).
For treatment of drought stress, 7-day-old seedlings of wild type (Col-0) and rap2.4 mutants were transferred to plastic pots [350 (W) × 270 (D) × 130 (H)] filled with 500 g of soil. The transferred seedlings were covered with plastic wrap to maintain humidity for 5 days, The plastic wrap was punched with a razor blade to maintain ambient humidity; the plastic wrap was removed after 2 days. The soil was first soaked with water and then the plants were exposed to drought stress for 2 weeks. Next, the soil was re-soaked with water and then, the plants were subjected to a second drought treatment for 2 weeks.
Construction of Binary Vectors and Arabidopsis Transformation
To generate transgenic Arabidopsis overexpressing RAP2.4, RAP2.4 cDNA was amplified using At1g78080 F1/At1g78080 R1 primers (Supplementary Table 1) and Arabidopsis seedling cDNA. The amplified products were translationally fused with the gene encoding enhanced yellow fluorescent protein (eYFP) between the CaMV35S promoter and the terminator of ribulose 1,5-biphosphate carboxylase/oxygenase small subunit (rbc-T) from Pisum sativum in the modified pPZP212 binary vector2 (Hajdukiewicz et al., 1994). The recombinant vector was introduced into Arabidopsis (ecotype, Col-0) using Agrobacterium-mediated transformation (Clough and Bent, 1998). Transgenic Arabidopsis seedlings (RAP2.4 OX) were selected in 1/2 MS medium containing carbenicillin (100 μg/ml) and kanamycin (25 μg/ml). The selected transgenic plants (T2 or T3 plants) were used for further experiments.
Isolation of Total RNA and Gene Expression Analysis
To analyze the expression of RAP2.4 in various organs or tissues of Arabidopsis, we collected 1-week-old seedlings and rosette leaves, stems, flowers, buds, and siliques of 6-week-old Arabidopsis. To investigate the expression patterns after treatment of osmotic stress, salt stress, or exogenous ABA, 10-day-old seedlings were floated and incubated in 1/2 MS liquid medium containing 200 mM mannitol, 200 mM NaCl, or 100 μM ABA for 1, 2, and 6 h at 25]°C with shaking at 40 rpm. For exposure to drought stress, 10-day-old seedlings were transferred to cellulose paper (Whatman) and air-dried for 1 and 2 h. To investigate the expression patterns of cuticular wax biosynthetic genes in 4-week-old Arabidopsis wild type and rap2.4 mutants after the drying event, the aerial parts of wild type and rap2.4 mutants were cut and placed on the cellulose paper and air-dried for 1, 2, and 4 h. To analyze the expression patterns of cuticular wax biosynthetic genes in the wild type and transgenic Arabidopsis overexpressing RAP2.4, the rosette leaves were collected from 4-week-old Arabidopsis. Total RNA was isolated using the Nucleospin RNA Plant Extraction Kit (MACHEREY-NAGEL) following the manufacturer’s protocols. Total RNA was used for cDNA synthesis using GostripTM Reverse Transcriptase (Promega) following the manufacturer’s protocols. The cDNA and gene-specific primers (Supplementary Table 1) were used for both the semi-RT-PCR and the quantitative RT-PCR (CFX96 thermal cycler, Bio-Rad). The KAPA SYBR FAST qRT-PCR kit (KAPA Biosystems) was used for quantitative RT-PCR. Quantification of the PP2AA3 (At1g13320) transcripts was used for normalization of the RAP2.4 transcripts and cuticular wax biosynthetic genes.
Scanning Electron Microscope (SEM) Analysis
To observe the cuticular wax crystal on the surface of leaves, rosette leaves from 4-week-old wild type and transgenic Arabidopsis overexpressing RAP2.4 were used according to methods described in a previous study (Lee and Suh, 2015b).
Cuticular Wax Analysis Using Gas Chromatography (GC)
The cuticular wax were extracted from rosette leaves of 5- to 6-week-old wild type, rap2.4 mutants, and three independent transgenic Arabidopsis overexpressing RAP2.4 (OX20, OX26, and OX31) by shaking in chloroform for 30 s. Extracts were evaporated under nitrogen gas after adding the internal standard (1-tricosanol, docosanoic acid, and n-octacosane). Then, concentrated wax was dissolved in a solution of pyridine and bis-N,O-trimethylsilyl trifluoroacetamide (BSTFA, Sigma) (1:1, v/v) and incubated at 90°C for 30 min. The silylated waxes were concentrated under nitrogen gas and they were re-dissolved with premixed heptane-toluene solution (1:1, v/v). The conditions of the GC analysis were described in a previous study (Lee and Suh, 2015b).
Transpiration and Chlorophyll Leaching Assays
For the cuticular transpiration assay, well-watered 3-week-old Arabidopsis plants were incubated in the dark for 12 h, and the shoot was carefully separated from the root. Subsequently the shoot was fully watered for 1 h by floating on water. After removal of water from the shoot surface, the weights of the shoots were measured for 2.5 h with 15 min. intervals while maintaining the dark condition.
For the chlorophyll leaching assay, the shoots without roots of well-watered 3-week-old Arabidopsis were exposed to the dark for 12 h, and then immediately immersed in 80% ethanol with shaking at 40 rpm. The extracted chlorophyll content was measured for 3 h with 15 min. intervals on a spectrophotometer (Ultrasec 3100 pro; Amersham Biosciences) at wavelengths of 647 and 664 nm. The chlorophyll content was calculated as μmol Chlorophyll/fresh weight (FW, g) = [7.93 (A664) + 19.53 × (A647)]/FW (g).
Transcriptional Activation Assay in N. benthamiana Protoplasts
The transcriptional activation assays were performed as described in Lee and Suh (2015b). For making reporter constructs, the minimal CaMV 35S promoter and LUC gene of Gal4(3X):LUC plasmid (Tiwari et al., 2003) were amplified with Min35S pro_PstI-F/Min35S pro_XbaI-R primer set and Luc XmaI F/Luc SacI R primer set, respectively, and then the CaMV 35S promoter and GUS gene of pBI221 were replaced with the amplified DNA fragments, minimal CaMV 35S promoter and LUC gene, respectively (Supplementary Table 1). The generated vector named as pBI:LUC. Next, the oligonucleotides (KCS2 BS1, KCS2 BS2, CER1 BS1, and CER1 BS2) containing the consensus CCGAC or GCC motifs, which are present in the promoter regions of KCS2 and CER1 and their mutated oligonucleotides (KCS2 mBS1, KCS2 mBS2, CER1 mBS1, and CER1 mBS2), where the consensus CCGAC and GCC sequences were changed to TTTTT and TTT sequences, were designed. The HindIII and SalI restriction enzyme sites were included in the designed oligonucleotides at 5’- and 3’-end, respectively. The synthesized oligonucleotides were annealed, and then the DNA fragments were cloned in HindIII and SalI sites of the pBI:LUC vector. The pPZP212 and pRAP2.4 binary vectors were used as the effector. The pPZP212 or pRAP2.4. binary vector was co-transfected with each reporter vector into N. benthamiana leaf protoplast using polyethylene glycol (PEG)-mediated transfection (Yoo et al., 2007). The pBI221 expression vector was used as the normalization factor. Twenty hours after transfection, total protein extracted from the protoplasts was subjected to enzymatic assays to quantifyluciferase (LUC) activity using a dual-luciferase assay system (Promega). Quantification of the GUS level was used for normalization of quantification of LUC activity. The fluorescence of LUC and GUS were detected under GLOMAX (Promega).
Results
Expression of RAP2.4 in Arabidopsis
To investigate the expression levels of RAP2.4 gene in various Arabidopsis organs, we isolated total RNAs from 7-day-old seedlings and rosette leaves, stem, cauline leaves, floral buds, flowers, and siliques of 6-week-old plants, and then subjected them to qRT-PCR. When the transcript level of RAP2.4 was compared with that of PP2AA3, which is used as a reference gene to assess the quantity and quality of RNA samples (Czechowski et al., 2005), the level of RAP2.4 transcripts was approximately 1.6- to 3.4-fold higher in the various Arabidopsis organs tested (Figure 1A). The highest expression of RAP2.4 was observed in cauline leaves. To measure the increased levels of RAP2.4 transcripts in Arabidopsis after drought, osmotic, and salt stress treatments or ABA applications, 10-day-old seedlings were air-dried or incubated on 1/2 MS media supplemented with 200 mM mannitol, 200 mM NaCl, or 100 μM ABA for 0, 1, 2, or 6 h. The transcript levels of RAP2.4 increased by approximately 2.9- to 3.1-fold with drought treatment, 2.0- to 3.2-fold with 200 mM mannitol treatment, 1.8- to 2.9-fold with salt stress, and 2.1- to 3.1-fold with ABA applications (Figure 1B).
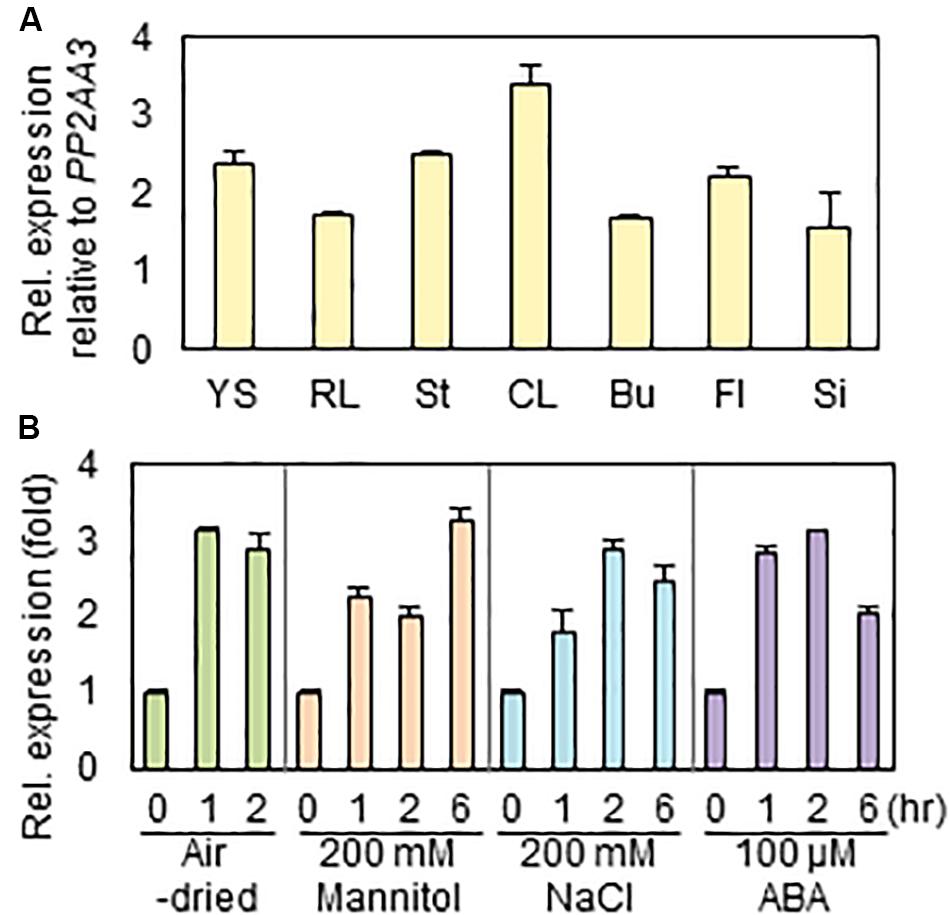
Figure 1. Expression of RAP2.4 in Arabidopsis. (A) The transcript level of RAP2.4 in various organs of Arabidopsis. Total RNA was isolated from 1-week-old seedlings (YS), rosette leaves (RL), stems (St), cauline leaves (CL), buds (Bu), flowers (Fl), and silique (Si) of 6-week-old Arabidopsis. Transcript levels were determined by quantitative Real Time-Polymerase Chain Reaction (qRT-PCR) and relative expression was determined using the ΔΔCt method. PP2AA3 (At1g13320) was used to determine the quantity and quality of the cDNAs. Each value is the mean ± SD of three independent measurements. (B) Expression of RAP2.4 transcripts after application of exogenous ABA and treatment of salt and osmotic stress, and drought. One-week-old seedlings were transferred to the 1/2 MS liquid medium supplemented with 200 mM mannitol, 200 mM NaCl, or 100 μM ABA for the indicated times. For drought stress, the transferred seedlings were air-dried for the indicated times. Transcript levels were determined by qRT-PCR and relative expression was determined using the ΔΔCt method. PP2AA3 (At1g13320) was used to determine the quantity and quality of the cDNAs. Each value is the mean ± SD of three independent measurements.
Isolation of rap2.4 Knock-Out Mutants and Generation of Transgenic Arabidopsis Plants Overexpressing rap2.4
To investigate the role of the RAP2.4 gene in cuticular wax biosynthesis, T-DNA-inserted rap2.4 mutant seeds (SALK_020767C and SALK_093377) were obtained from the Arabidopsis Biological Resource Center (www.arabidopsis.org). The T-DNA insertions were designated between 710 and 711 bp from initiation of transcription in rap2.4-1 (SALK_020767C) and 1504 and 1505 bp from initiation of transcription in rap2.4-2 (SALK_093377) in the TAIR site (see footnote 1) (Figure 2A). To check the T-DNA insertions from rap2.4-1 and rap2.4-2 mutants, genomic DNA was extracted from leaves of rap2.4-1 and rap2.4-2, and then genomic DNA PCR was performed. PCR bands, which were amplified using At1g78080-NF1 and At1g78080-CR1 primers, were detected in wild type (Col-0) but not in rap2.4-1 and rap2.4-2 mutants. However, PCR bands, which were amplified using LBa1 and At1g78080-CR1 or At1g78080-NF1 and LBa1 primers, were detected in rap2.4-1 and rap2.4-2 mutants but not in the wild type (Col-0) (Figure 2B). In RT-PCR analysis of 4-week-old wild type rap2.4-1, and rap2.4-2 leaves, RAP2.4 transcripts were detected in the wild type but not in rap2.4-1 and rap2.4-2, indicating that rap2.4-1 and rap2.4-2 are knockout mutants (Figure 2C).
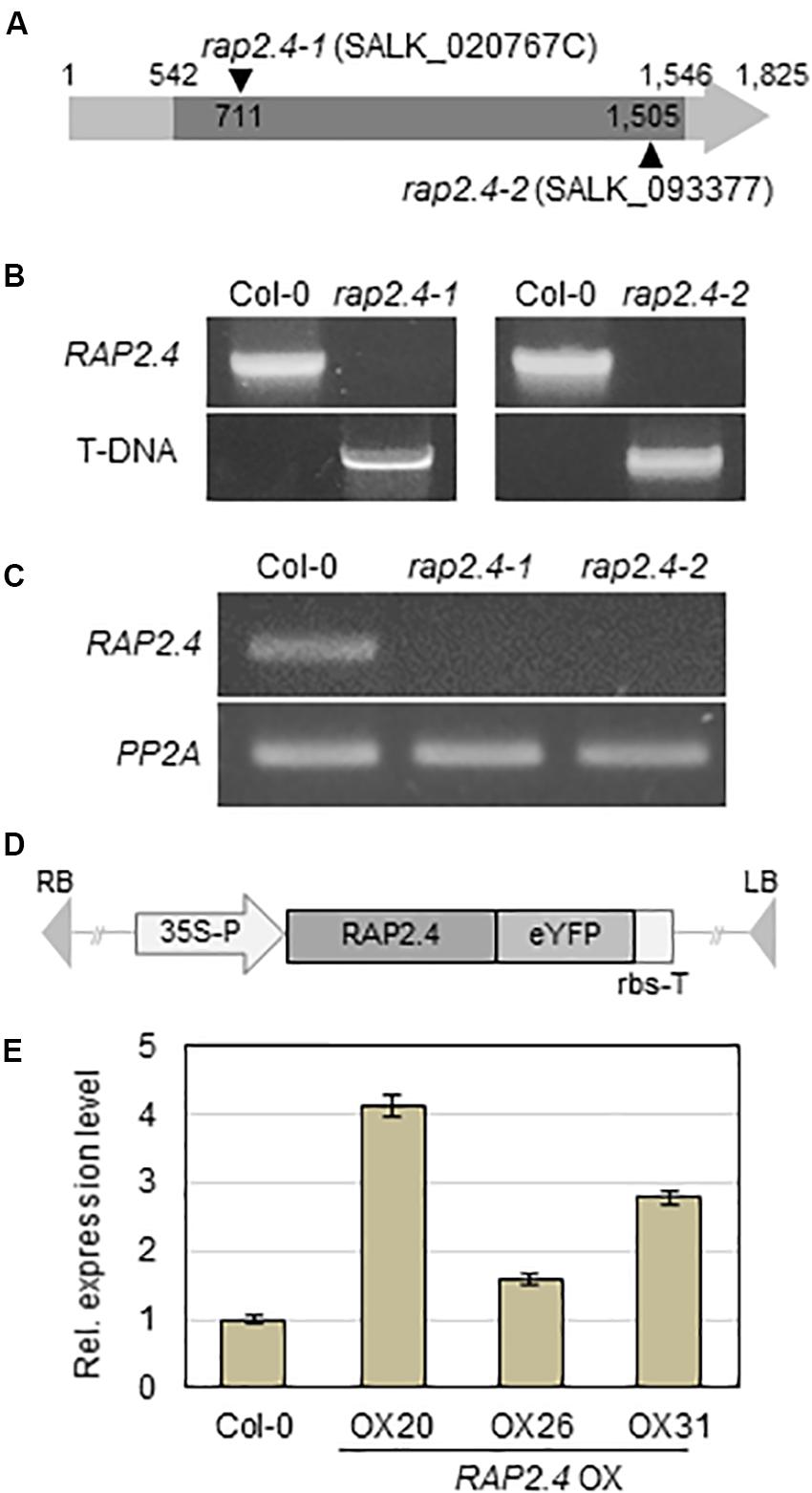
Figure 2. Isolation of Arabidopsis RAP2.4 knock-out mutant and generation of transgenic Arabidopsis overexpressing RAP2.4. (A) Schematic diagram of T-DNA-inserted Arabidopsis rap2.4 mutants. The rap2.4-1 and rap2.4-2 are RAP2.4 knock-out mutant alleles. Genomic DNA-PCR (B) and reverse transcription (RT)-PCR (C) analyses of RAP2.4 in Arabidopsis wild type (Col-0) and rap2.4 mutants. Total RNA was extracted from 4-week-old rosette leaves and subjected to RT-PCR. PP2AA3 (At1g13320) was used to determine the quantity and quality of the cDNAs. (D) Schematic diagram of RAP2.4 overexpression construct under the control of the CaMV 35S promoter (35S-P). rbs-T, terminator of ribulose 1,5-biphosphate carboxylase/oxygenase small subunit from Pisum sativum. eYFP, enhanced yellow fluorescent protein. RB, right border. LB, left border. (E) Quantitative real-time (RT)-PCR analysis of three independent transgenic Arabidopsis plants overexpressing RAP2.4 (OX20, OX26, and OX31). Total RNA was extracted from 4-week-old rosette leaves. PP2AA3 (At1g13320) was used to determine the quantity and quality of the cDNAs. Transcript levels were determined by qRT-PCR and relative expression was determined using the ΔΔCt method. Each value is the mean of three independent measurements. Bars indicate standard deviation.
To generate transgenic Arabidopsis plants overexpressing RAP2.4, RAP2.4 was inserted between the CaMV 35S promoter and the terminator of ribulose 1,5-biphosphate carboxylase/oxygenase small subunit (rbs-T) (Figure 2D). The generated binary vector was transformed into Arabidopsis plants via the Agrobacterium-mediated transformation method. qRT-PCR showed that the levels of RAP2.4 transcripts in RAP2.4 overexpression lines, OX20, OX26, and OX31 increased by 4.7-fold, 1.6-fold, and 2.8-fold, respectively, compared to the wild type (Figure 2E). During growth and development, we observed that growth of RAP2.4 OX lines was smaller than that of the wild type and the growth-retarded phenotype was prominent in RAP2.4 OX20. However, no noticeable differences in growth and development were observed between wild type and rap2.4 mutants (Supplementary Figure 1).
Cuticular Wax Content and Composition in Arabidopsis Wild Type and rap2.4 Mutants Under Well-Watered and Drought Stress Conditions
To investigate the effect of loss-of-function of RAP2.4 on cuticular wax composition and amount, cuticular waxes were extracted from leaves and stems of wild type, rap2.4-1, and rap2.4-2 mutants which were grown under well-watered or drought stress conditions, and measured by gas chromatography (GC) with a flame ionization detector (FID). Wild type and rap2.4 mutants showed no significant differences in leaf wax content and composition under sufficiently watered conditions (Figures 3A,C). Under the drought stress condition, however, total wax loads decreased by approximately 10% in both rap2.4-1 and rap2.4-2 leaves relative to the wild type (Figures 3B,D). Approximately 13–19%, 11–14%, 11–12%, and 12% reduction in the levels of each VLC-alkane, C27, C29, C31, and C33, respectively, were observed in rap2.4 mutants relative to the wild type under drought (Figures 3B,D). However, there were no significant differences in the levels of other wax components even though plants were grown under drought (Figures 3B,D).
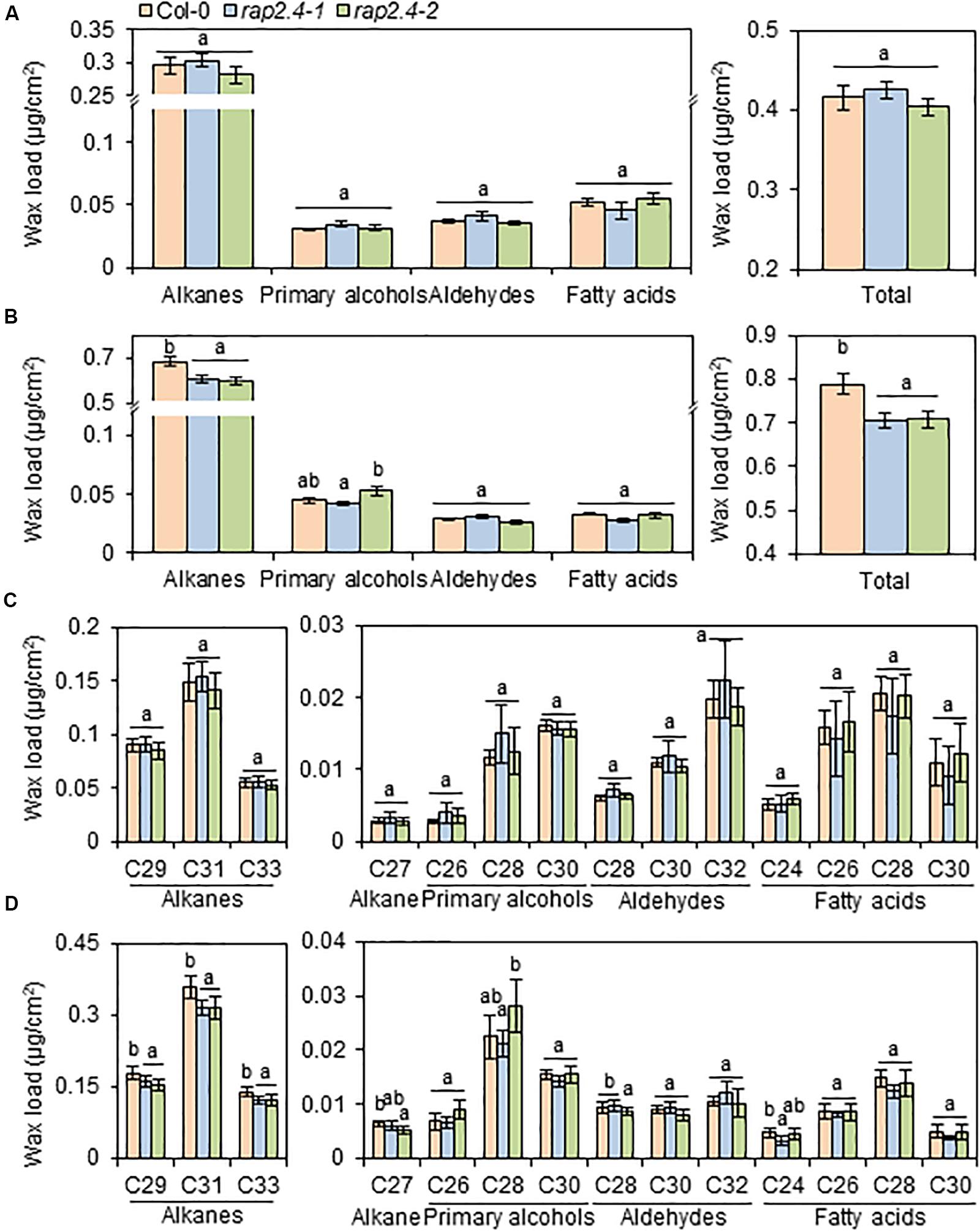
Figure 3. Cuticular wax amount and composition in the leaves of Arabidopsis wild type (Col-0) and rap2.4 mutants under normal and drought conditions. (A) Cuticular wax amount in leaves of plants grown under the normal growth conditions (temperature, 24 ± 2°C; humidity, approximately 50%; light condition, 100–120 μmol m–2s–1). (B) Cuticular wax amount in leaves of plants grown under the drought stress conditions. (C) Cuticular wax composition in leaves of plants grown under normal growth conditions. (D) Cuticular wax composition in leaves of plants grown under drought stress conditions. Cuticular waxes were extracted from leaves of 5- to 6-week-old Arabidopsis grown under normal growth and drought stress conditions. Each value is the mean of three independent measurements. Bars indicate standard deviation. Different letters denote statistically significant differences at P < 0.05 (Tukey’s test), following a one-way ANOVA test with treatment as the variable factor.
Epicuticular Wax Crystals and Cuticular Wax Content and Composition in Arabidopsis Wild Type and RAP2.4 Overexpression Lines
Because the wax-deficient phenotype of rap2.4 mutants was observed under drought, we next examined the formation of epicuticular wax crystals in leaves of wild type and RAP2.4 overexpression lines using SEM. SEM analysis showed that epicuticular wax crystals were clearly observed in leaves of RAP2.4 OX20, OX26, and OX31 lines, but not in leaves of the wild type (Figure 4A).
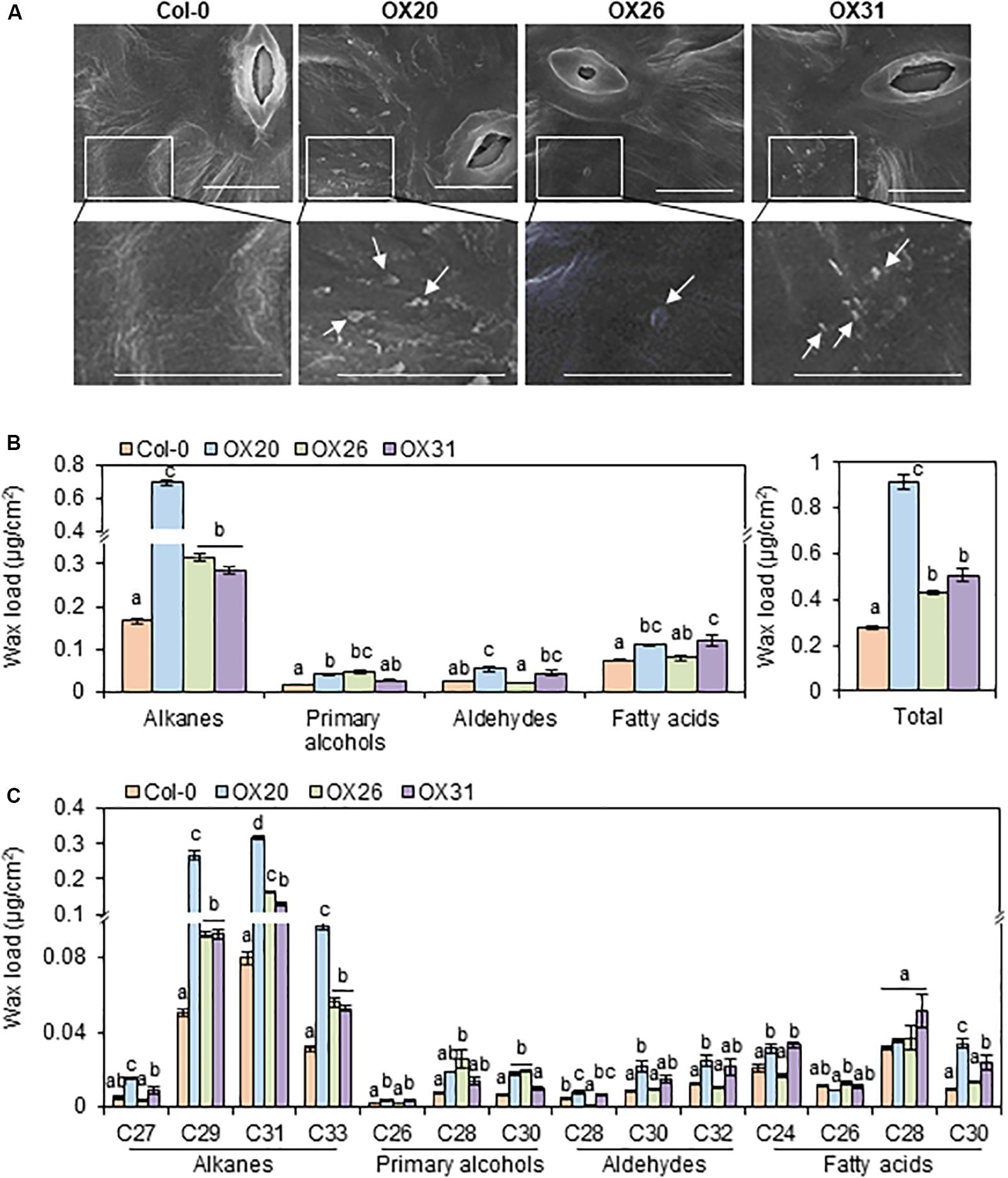
Figure 4. Epicuticular wax crystals (A) and cuticular wax amount and composition (B) in the leaves of Arabidopsis wild type (Col-0) and RAP2.4 OX lines. (A) Scanning electron microscopy analysis of epicuticular wax crystals on leaves of 3- to 4-week-old Arabidopsis wild type (Col-0) and RAP2.4 OX lines. The images (inset) in the upper panel were magnified and shown in the lower panel. Bars = 10 μm. White arrows indicate wax crystals. (B) Cuticular wax amount in leaves of 5-week-old Arabidopsis wild type (Col-0) and RAP2.4 OX lines. (C) Cuticular wax composition in leaves of 5-week-old Arabidopsis wild type (Col-0) and RAP2.4 OX lines. Each value is the mean of three independent measurements. Bars indicate standard error. Different letters denote statistically significant differences at P < 0.05 (Tukey’s test), following a one-way ANOVA test with treatment as the variable factor.
Subsequently, the content and composition of cuticular waxes were measured from leaves and stems of wild type and RAP2.4 overexpression lines using GC-FID (Supplementary Figure 2). Total wax content increased by approximately 3.3-, 1.5-, and 1.8-fold in RAP2.4 OX20, OX26, and OX31, respectively, compared with to that in the wild type (Figure 4B). The largest increase (1.7- to 4.2-fold) in leaves of all three RAP2.4 OX lines relative to that in the wild type was observed in the levels of VLC-alkanes (C27, C29, C31, and C33), which are major wax components (more than 70% of total wax loads) in Arabidopsis leaves (Figure 4C). In particular, approximately 3.2-, 5.3-, 4.0-, and 3.1-fold increase in the levels of C27, C29, C31, and C33, respectively, was observed in leaves of RAP2.4 OX20 compared with that of the wild type (Figure 4C). The significant increase in the levels of C29, C31, and C33 VLC-alkanes was also observed in leaves of RAP2.4 OX26, and OX31 relative to wild type (Figure 4C). An increase in the levels of other components, VLC-primary alcohols (C26, and C28), VLC-aldehydes (C28, C30, and C32), and VLC-fatty acids (C24 and C30) was also detected in leaves of RAP2.4 OX20 and OX31 relative to the levels in the wild type, except for C30 VLC-PA and C28 VLCFAs (Figure 4C). The levels of C28 and C30 VLC-PA were increased in leaves of RAP2.4 OX26 relative to wild type (Figure 4C).
Cuticular Transpiration and Chlorophyll Leaching Assays in Arabidopsis Wild Type and RAP2.4 Overexpression Lines
An increase in total wax loads in leaves of RAP2.4 OX lines prompted us to measure cuticular transpiration and chlorophyll leaching assays in the Arabidopsis wild type and RAP2.4 overexpression lines. For the cuticular transpiration assay, 3-week-old wild type and RAP2.4 OX lines were dark-treated for 12 h to completely close stomata in leaves, whole aerial parts of plants were floated on water under dark for 1 h, and then, cuticular transpiration rate was measured. As shown in Figure 5A, the water loss through the cuticle was slower in all three RAP2.4 OX lines than in the wild type.
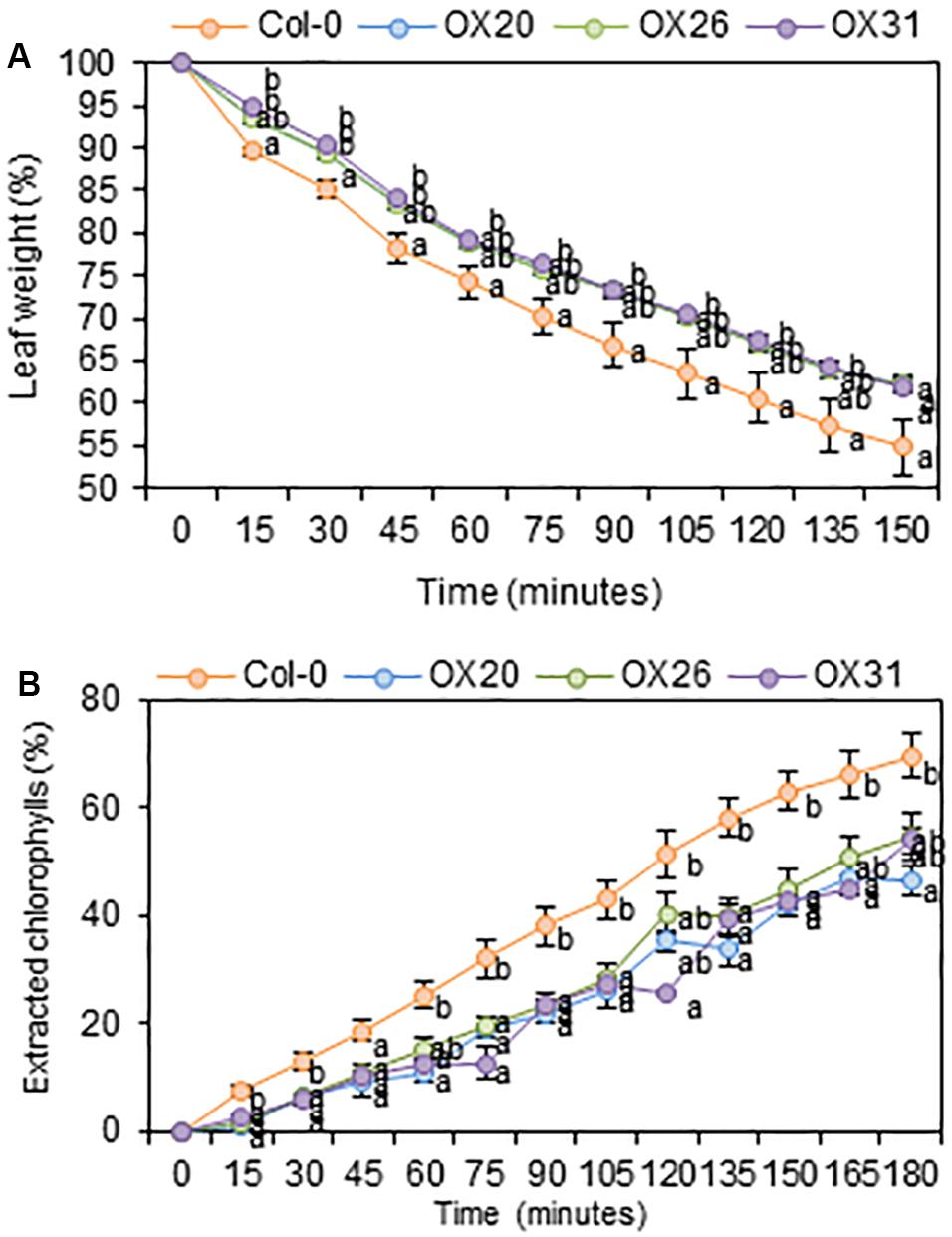
Figure 5. Cuticular transpiration (A) and chlorophyll leaching (B) assays of Arabidopsis wild type (Col-0) and RAP2.4 OX lines. (A) Cuticular transpiration assay in leaves of 3-week-old Arabidopsis wild type (Col-0) and RAP2.4 OX lines. (B) Chlorophyll leaching in leaves of 3-week-old Arabidopsis wild type (Col-0) and RAP2.4 OX lines. Each value represents the mean of three independent measurements. Bars indicate the standard error of the mean. Different letters denote statistically significant differences at P < 0.05 (Tukey’s test), following a one-way ANOVA test with treatment as the variable factor.
For the chlorophyll leaching assay, 3-week-old wild type and RAP2.4 OX lines were incubated for 12 h under dark, whole aerial parts of the plants were immersed in 80% ethanol, and then, the chlorophyll content leached from the leaves was measured. As shown in Figure 5B, the rate of chlorophyll leaching was slower in the RAP2.4 OX lines than in the wild type.
Expression of Genes Involved in Cuticular Wax Biosynthesis and Its Regulation in Arabidopsis Wild Type, rap2.4 Mutants, and RAP2.4 OX Leaves Under Well-Watered and/or Drought
Next, we examined whether the significant decrease in total wax loads and levels of alkanes observed in rap2.4-1 and rap2.4-2 leaves relative to that in the wild type under drought stress conditions correlates with the reduced expression of genes involved in cuticular wax biosynthesis and its regulation. Total RNA was isolated from wild type and rap2.4 mutant leaves, which were air-dried for 0, 1, 2, and 4 h and then subjected to qRT-PCR analysis. The RAP2.4 gene displayed the highest expression in wild type leaves 1 to 2 h after drought treatment. The expression of RD29A (At5g52310), which is known as a drought-inducible gene (Shinozaki and Yamaguchi-Shinozaki, 2007), was upregulated in both wild type and rap2.4 mutant leaves 1 h after drought treatment and continuously increased in the wild type up to 4 h after drought treatment. However, no further increase in the levels of RD29A was detected in rap2.4 mutant leaves 2 and 4 h after drought treatment. Increased levels of KCS2 (At1g04220) and CER1 (At1g02205) transcripts were approximately 20% and 40% lower, respectively, in rap2.4 leaves than in wild type leaves 4 h after drought treatment (Figure 6A). However, no significant differences were observed in the levels of KCR1 (At1g67730), PAS2 (At5g10480), KCS6 (At1g68530), CER3 (At5g57800), MYB94 (At3g47600), and MYB96 (At5g62470) expression between wild type and rap2.4 leaves after drought treatment (Supplementary Figure 3).
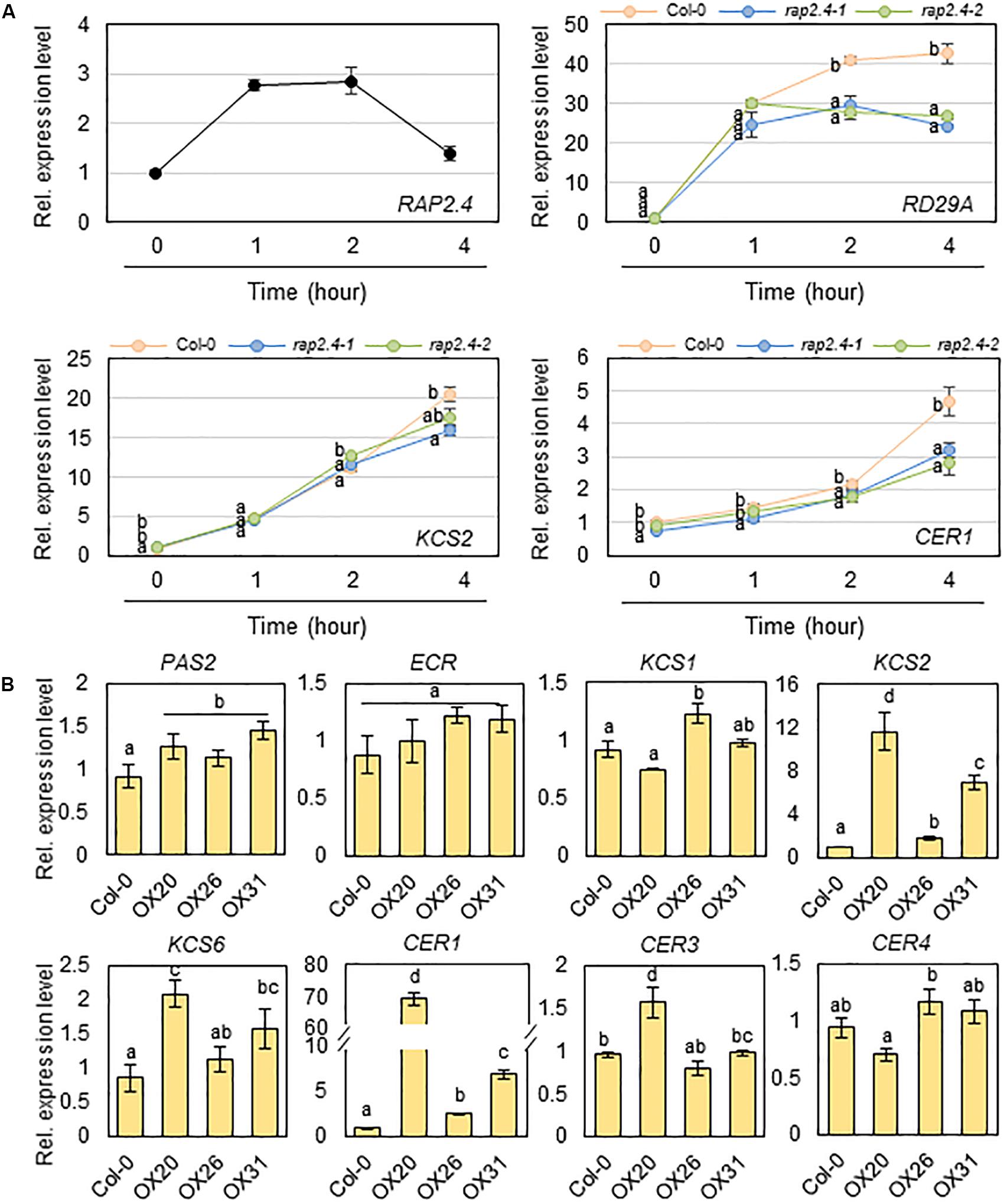
Figure 6. Quantitative RT-PCR analysis of wax biosynthetic genes in 4-week-old wild type (Col-0), rap2.4 mutants, and RAP2.4 OX lines. (A) The transcript level of RAP2.4 in 4-week-old Arabidopsis wild type (Col-0) and the transcript level of RD29A, KCS2, and CER1 in 4-week-old Arabidopsis wild type (Col-0) and rap2.4 mutants. Rosette leaves from 4-week-old Arabidopsis grown in soil were air-dried for 0, 1, 2, and 4 h. Each value represents the mean of three independent measurements. Bars indicate the standard deviation of the mean. Different letters denote statistically significant differences at P < 0.05 (Tukey’s test), following a one-way ANOVA test with treatment as the variable factor. (B) Quantitative RT-PCR analysis of wax biosynthetic genes in 4-week-old Arabidopsis wild type (Col-0) and RAP2.4 OX lines. The RNAs were extracted from rosette leaves of Arabidopsis grown in soil for three weeks. Each value represents the mean of three independent measurements. Bars indicate the standard deviation of the mean. Different letters denote statistically significant differences at P < 0.05 (Tukey’s test), following a one-way ANOVA test with treatment as the variable factor.
In addition, the expression of KCS2 and CER1 was markedly upregulated in all three RAP2.4 OX lines relative to that in the wild type (Figure 6B). The levels of KCS1 (At1g01120), KCS6, CER3, and CER4/FAR3 (At4g33790) were increased by 1.2- to 2.0-fold in 2 or 3 RAP2.4 OX lines compared with that in the wild type, but no significant change in the levels of PAS2 and ECR (At3g55360) expression was observed between the wild type and RAP2.4 OX lines (Figure 6B). These results indicate that KCS2 and CER1 are directly or indirectly regulated by the RAP2.4 transcription factor under drought.
Transcriptional Activation Assay of RAP2.4 in N. benthamiana Protoplasts
Based on the observation that the expression of KCS2 and CER1 was altered in rap2.4 mutants and RAP2.4 OX lines compared with that in the wild type, we further examined whether RAP2.4 directly regulate the expression of KCS2 and CER1. According to the previous report that RAP2.4 specifically recognizes the CCGAC core sequences of the drought response element (DRE) and GCC box (Rae et al., 2011), the consensus CCGAC and GCC motifs were searched in the promoter regions of KCS2 and CER1. For reporter constructs, the selected DNA fragments (KCS2 BS1, KCS2 BS2, CER1 BS1, and CER1 BS2] containing the consensus CCGAC or GCC motifs and their mutated DNA fragments (KCS2 mBS1, KCS2 mBS2, CER1 mBS1, and CER1 mBS2), where the consensus CCGAC and GCC sequences were converted to TTTTT and TTT sequences, respectively, were transcriptionally ligated to the minimal CaMV 35S promoter in the pBI:LUC harboring the luciferase reporter gene (Figures 7A,B). For effector constructs, the coding region of RAP2.4 was inserted between the CaMV 35S promoter and nopaline synthase (Nos) terminator in the pPZP212 vector and named pRAP2.4 (Figure 7B). The reporter and effector vectors were co-transfected into N. benthamiana protoplasts with a vector harboring GUS for internal control to monitor transformation efficiencies and luciferase and GUS activities were measured. The ratio of LUC activity to GUS activity in protoplasts expressing LUC driven by the KCS2-P1, KCS2-P2, and CER1-P1 promoters was approximately 14.5-, 3.5-, and 2.8-fold elevated, respectively, but no significant increase in LUC gene expression driven by the CER1-P2 promoter was observed, upon co-expression with RAP2.4 compared with the pPZP212 control. By contrast, the co-transformation of pRAP2.4 with KCS2-mP1, KCS2-mP2, CER1-mP1, or CER1-mP2 did not elevate the LUC gene expression (Figure 7C), indicating that the CCGAC or GCC consensus motifs present in the KCS2 (BS1 and BS2) and CER1 (BS1) promoter regions are essential for the expression of KCS2 and CER1 by RAP2.4 and RAP2.4 may be directly involved in the transcriptional activation of KCS2 and CER1 genes.
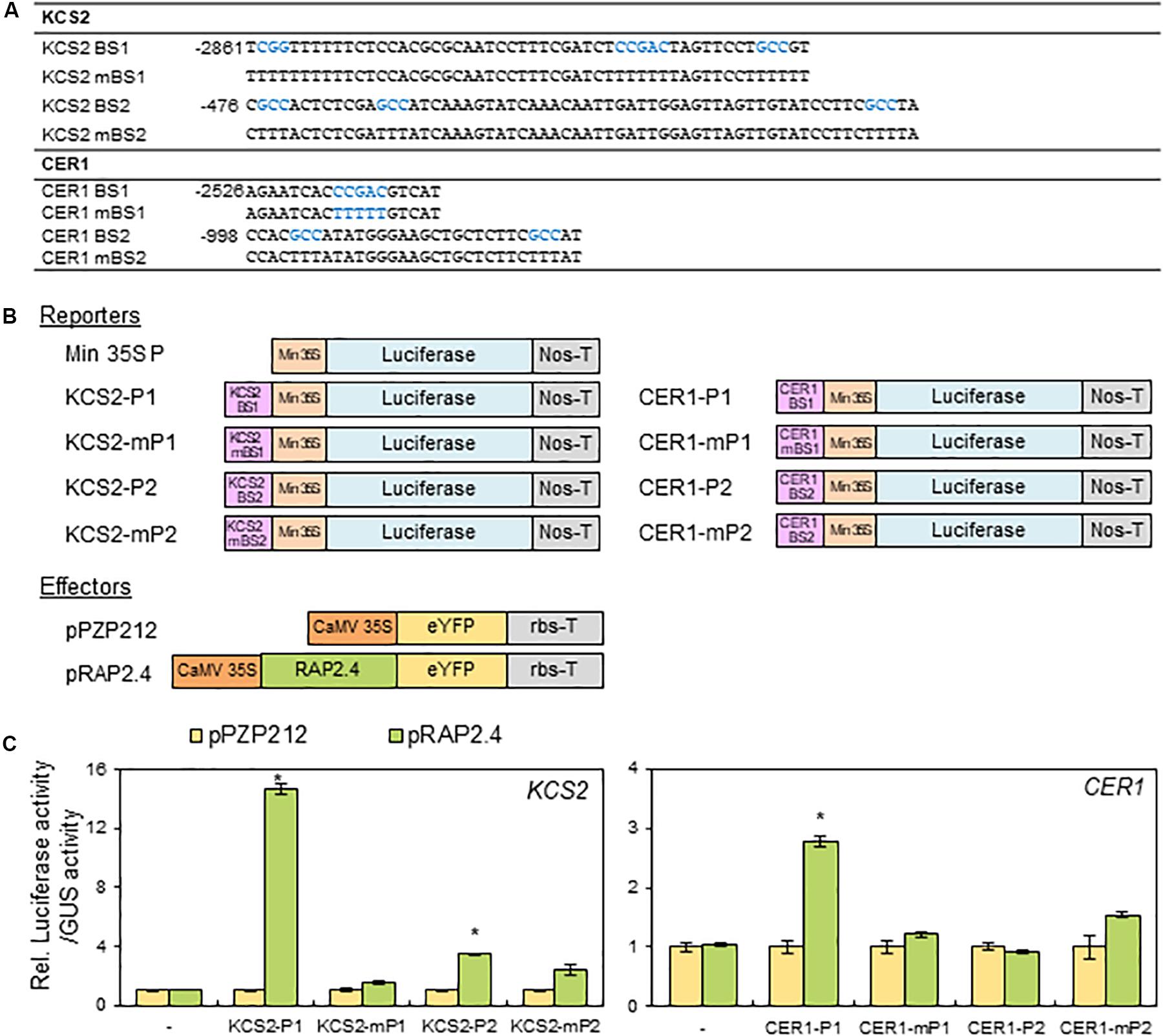
Figure 7. Transactivation assay of RAP2.4 in N. benthamiana. (A) The oligonucleotide sequences (BS) including RAP2.4 binding consensus sequences, which are present in the promoter regions of KCS2 and CER1 genes. Core binding sequences shown in blue were mutated, resulting in mBSs, to verify specific binding. (B) Schematic diagrams of reporter and effector constructs for transcriptional activation assay. For the reporter constructs, the synthesized oligonucleotides were annealed, and then the DNA fragments were ligated between the CaMV 35S minimal promoter (Min 35S) and the luciferase gene. For the effector construct, RAP2.4 was translationally fused with the gene encoding eYFP between the CaMV 35S promoter (CaMV35SP) and the terminator of ribulose 1,5-biphosphate carboxylase/oxygenase small subunit from Pisum sativum (rbs-T). Nos-T, The terminator of the nopaline synthase gene. (C) Transcriptional activation assay in N. benthamiana leaf protoplasts. N. benthamiana leaf protoplasts were co-transformed with reporter and effector constructs, and luciferase and GUS activities were determined fluorometrically. Luciferase activity was normalized by dividing this value by GUS activity. Each value is the mean of four independent measurements (t test, *P < 0.01). Bars indicate the standard error of the mean.
Discussion
When land plants face water deficit stress, they close the stomata (Brodribb and McAdam, 2017) and the aerial organs of land plants increase total loads of cuticular waxes, which are present in the outermost layer of the cuticle, to reduce non-stomatal water loss (Riederer and Schreiber, 2001; Domínguez et al., 2017). In particular, an increase in the levels of alkanes among the diverse wax components is prominent in Arabidopsis under drought (Kosma et al., 2009; Lee and Suh, 2015a). Although involvement of MYB96 and MYB94 transcription factors has been reported in the upregulation of cuticular wax biosynthesis in Arabidopsis (Seo et al., 2011; Lee and Suh, 2015b; Lee et al., 2016), increased total wax loads were still observed in myb96 myb94 mutants relative to the wild type under drought. This observation prompted us to find another transcription factor, which upregulates cuticular wax biosynthesis in Arabidopsis under drought. In this study, we show that ABA-, drought-, osmotic-, and salt-stress inducible RAP2.4 activates cuticular wax biosynthesis in Arabidopsis leaves under drought stress conditions. qRT-PCR and transcriptional activation assays revealed that RAP2.4 upregulates the expression of KCS2 and CER1, which are required for the production of alkanes, and contributes to an increase in total wax loads which is critical for drought resistance in Arabidopsis leaves.
The aerial parts of land plants increase or decrease total wax loads by the up- or down-regulation of genes involved in cuticular wax biosynthesis under different environmental conditions such as drought, dark, or pathogen infection or in an organ-specific manner (Raffaele et al., 2008; Buschhaus and Jetter, 2011; Seo et al., 2011; Go et al., 2014; Park et al., 2016). Therefore, it has been speculated that cuticular wax biosynthesis is mainly regulated by transcriptional regulatory mechanism, which is supported by the identification of several transcription factors, WIN1/SHN1, MYB30, MYB96, MYB94, DEWAX, DEWAX2, and WRI4 involved in the regulation of wax biosynthesis (Aharoni et al., 2004; Raffaele et al., 2008; Seo et al., 2011; Go et al., 2014; Lee and Suh, 2015b; Park et al., 2016; Kim et al., 2018). Among the several transcription factors, ABA- and drought-inducible MYB96 and MYB94 transcription factors were reported to additively function in the upregulation of cuticular wax biosynthesis under both well-watered and water-deficit conditions (Lee et al., 2016). However, the wax-deficient phenotype was observed in the leaves of rap2.4-1 and rap2.4-2 mutants under only drought stress conditions (Figure 3), suggesting that RAP2.4 plays a role in the upregulation of cuticular wax biosynthesis under drought. The hypothesis is consistent with the enhanced expression of RAP2.4 by ABA, salt, drought, and osmotic stress treatments (Figure 1B). In addition, in microarray analyses of 2-week-old Arabidopsis WT versus myb96-D (MYB96 OX) leaves and 3- to 4-week-old Arabidopsis WT versus MYB94 OX leaves, the levels of RAP2.4 expression was only approximately 1.4- and 1.2-fold elevated in myb96-D (MYB96 OX) and MYB94 OX leaves relative to WT, respectively (Seo et al., 2011; Lee et al., 2016). No significant differences were observed in the levels of MYB94 and MYB96 expression between Arabidopsis WT and rap2.4 leaves 1 h and 2 h after drought treatment (Supplementary Figure 2). Therefore, these results suggest that a DERB-type RAP2.4 transcription factor and MYB96/MYB94 transcription factors might independently regulate cuticular wax biosynthesis under drought stress conditions.
In wax chemical analysis in the leaves of rap2.4-1 and rap2.4-2 relative to that of the wild type under drought, a significant reduction was observed in the levels of alkanes, C29, C31, and C33, which comprise approximately 85% of total wax loads (Figure 3). Whereas a remarkable increase in the alkanes loads was observed in the leaves of RAP2.4 OX lines, although an elevation in the content of other components was detected (Figure 4). These results are consistent with the upregulation of KCS2 and CER1, which function in VLCFA and alkane synthesis, respectively (Figure 6). Transcriptional activation assay (Figure 7) showed that the expression KCS2 and CER1 is increased by the expression of RAP2.4 in tobacco protoplasts, indicating that KCS2 and CER1 are the target genes of RAP2.4. Among wax biosynthetic genes, the expression of KCS2 and CER1 genes are the most highly induced (approximately 10- and 5-fold, respectively) in Arabidopsis in response to drought (Seo et al., 2011). In myb96 myb94 seedlings, approximately 2- or 10-fold increase in the levels of KCS2 and CER1 transcripts were still detected under drought-treated conditions compared with that in the well-watered conditions (Lee et al., 2016). In addition to MYB96 and MYB94, therefore, RAP2.4 functions in the upregulation of KCS2 and CER1 under drought. In particular, electrophoretic mobility shift assay (EMSA) and/or chromatin immunoprecipitation (ChIP) assay showed that MYB96 and MYB94 transcription factors directly interact with the same cis-element (TAACTACTAACTA) in the promoter region of KCS2 gene, suggesting that it is possible that ABA- and drought-induced expression of KCS2 is synergistically regulated by MYB96 and MYB94 transcription factors (Seo et al., 2011; Lee et al., 2016). However, neither MYB96 nor MYB94 transcription factors were able to specifically bind to the putative consensus sequences in the CER1 promoter region (Seo et al., 2011; Lee et al., 2016). In this study, transcriptional activation assay of RAP2.4 in N. benthamiana protoplasts revealed that the CCGAC or GCC consensus motifs in the promoter regions of KCS2 and CER1 are required for the expression of KCS2 and CER1 driven by RAP2.4 (Figure 7), indicating that ABA- and drought-inducible RAP2.4 may be directly involved in the transcriptional activation of KCS2 and CER1 genes under drought.
The stability of the MYB96 and MYB30 proteins was reported to be controlled by MYB30-INTERACTING E3 LIGASE 1 (MIEL1) E3 ubiquitin ligase, which controls protein turnover (Lee et al., 2017). In particular, Lee et al. (2017) reported that wax accumulation is balanced in Arabidopsis stems by preventing the adverse effects of excess MYB96 protein through degradation of MYB96. Based on the evidence that the transcript levels of MIEL1 decreased significantly in 18-day-old Arabidopsis under osmotic (300 mM mannitol), salt (150 mM NaCl), and drought stress conditions3, MYB96 could be stabilized to play a role in the activation of wax biosynthesis under drought (Seo et al., 2011). It was reported that BTB/POZ and MATH domain (BPM) proteins, which are substrate adaptors for Cullin 3-based E3 ubiquitin ligase, are negative regulators to control the stability of AP2/ERF transcription factors, DREB2A and WRI1, which are critical in the modulation of heat stress response and fatty acid metabolism, respectively (Chen et al., 2013; Morimoto et al., 2017). As Weber and Hellmann (2009) reported that RAP2.4 interacts with BPM proteins, it will be interesting to investigate whether the BPM and RAP2.4 interaction is involved in the molecular mechanisms underlying the regulation of drought-induced wax biosynthesis. The recent evidence that the Kelch-domain containing F-box protein, Small and Glossy Leaves 1(SAGL1) regulates cuticular wax biosynthesis by modulating the stability of CER3 in response to changes in humidity (Kim et al., 2019) supports that post-translational regulatory mechanism as well as transcriptional and post-transcriptional control mechanisms play important roles in the regulation of cuticular wax biosynthesis (Aharoni et al., 2004; Hooker et al., 2007; Raffaele et al., 2008; Seo et al., 2011; Lam et al., 2012, 2015; Go et al., 2014; Lee and Suh, 2015b; Park et al., 2016; Kim et al., 2018, 2019).
RAP2.4/WOUND INDUCED DEDIFFERENTIATION 1 (WIND1) has been reported to be implicated in plant development regulated by light and ethylene (Lin et al., 2008) and promotion of somatic cell dedifferentiation in response to wounding (Iwase et al., 2011). In particular, Lin et al. (2008) reported that RAP2.4 OX seedlings or plants displayed shorter hypocotyls, reduced apical hook curvature in darkness, increased and longer root hairs, reduced size of cotyledons, and early flowering. In addition to the developmental defects in RAP2.4 OX plants, overexpression of RAP2.4 confers an enhanced tolerance to drought stress (Lin et al., 2008), but little was known about the molecular mechanism underlying drought tolerance in RAP2.4 OX plants. In this study, we observed a wax-deficient phenotype in drought-treated rap2.4-1 and rap2.4-2 mutants, even though no noticeable phenotype was observed in the rap2.4 mutants (Lin et al., 2008). In conclusion, an AP2/DREB-type transcription factor, RAP2.4, is a transcriptional activator which upregulates KCS2 and CER1 genes and thereby, the total wax loads increased in Arabidopsis leaves under drought. This study also suggests that RAP2.4 might be useful in the production of alkanes and the development of crops with enhanced drought tolerance.
Data Availability Statement
All datasets for this study are included in the article/Supplementary Material.
Author Contributions
SY, HK, and MS conceived and designed the research and wrote the manuscript. SY, HK, and RK conducted the experiments. SY, HK, RK, JK, and MS analyzed the data. JK edited the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by grants from the National Research Foundation (NRF-2019R1A2B5B02070204) of South Korea and the Next-Generation BioGreen 21 Program (No. PJ013422) of the Rural Development Administration, South Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Young Sam Go and Juyoung Kim for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00895/full#supplementary-material
Footnotes
- ^ https://www.arabidopsis.org/
- ^ https://www.novoprolabs.com/vector/V10826, pRAP2.4.
- ^ http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi
References
Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., and Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16, 2463–2480. doi: 10.1105/tpc.104.022897
Bach, L., Michaelson, L. V., Haslam, R., Bellec, Y., Gissot, L., and Marion, J. (2008). The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. U.S.A. 105, 14727–14731. doi: 10.1073/pnas.0805089105
Bakan, B., and Marion, D. (2017). Assembly of the cutin polyester: from cells to extracellular cell walls. Plants 6:E57. doi: 10.3390/plants6040057
Beaudoin, F., Wu, X., Li, F., Haslam, R. P., Markham, J. E., Zheng, H., et al. (2009). Functional characterization of the Arabidopsis β-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol. 150, 1174–1191. doi: 10.1104/pp.109.137497
Beisson, F., Li-Beisson, Y., and Pollard, M. (2012). Solving the puzzles of cutin and suberin polymer biosynthesis. Curr. Opin. Plant Biol. 15, 329–337. doi: 10.1016/j.pbi.2012.03.003
Bernard, A., and Joubès, J. (2013). Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog. Lipid Res. 52, 110–129. doi: 10.1016/j.plipres.2012.10.002Get
Bourdenx, B., Bernard, A., Domergue, F., Pascal, S., Léger, A., Roby, D., et al. (2011). Overexpression of Arabidopsis ECERIFEFUM1 promotes wax VLC-alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156, 29–45. doi: 10.1104/pp.111.172320
Brodribb, T. J., and McAdam, S. A. M. (2017). Evolution of the stomatal regulation of plant water content. Plant Physiol. 174, 639–649. doi: 10.1104/pp.17.00078
Broun, P., Poindexter, P., Osborne, E., Jiang, C. Z., and Riechmann, J. L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 4706–4711. doi: 10.1073/pnas.0305574101
Buschhaus, C., and Jetter, R. (2011). Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces? J. Exp. Bot. 62, 841–853. doi: 10.1093/jxb/erq366
Chen, L., Lee, J. H., Weber, H., Tohge, T., Witt, S., Roje, S., et al. (2013). Arabidopsis BPM proteins function as substrate adaptors to a cullin3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 25, 2253–2264. doi: 10.1105/tpc.112.107292
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. doi: 10.1104/pp.105.063743
DeBono, A., Yeats, T. H., Rose, J. K., Bird, D., Jetter, R., Kunst, L., et al. (2009). Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21, 1230–1238. doi: 10.1105/tpc.108.064451
Djemal, R., and Khoudi, H. (2019). Combination of the endogenous promoter-intron significantly improves salt and drought tolerance conferred by TdSHN1 transcription factor in transgenic tobacco. Plant Physiol. Biochem. 139, 435–445. doi: 10.1016/j.plaphy.2019.04.009
Domínguez, E., Heredia-Guerrero, J. A., and Heredia, A. (2017). The plant cuticle: old challenges, new perspectives. J. Exp. Bot. 68, 5251–5255. doi: 10.1093/jxb/erx389
Eigenbrode, S. D., and Espelie, K. E. (1995). Effects of plant epicuticular lipids on insect herbivores. Annu. Rev. Entomol. 40, 171–194. doi: 10.1146/annurev.en.40.010195.001131
Feng, J. X., Liu, D., Pan, Y., Gong, W., Ma, L. G., Luo, J. C., et al. (2005). An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol. 59, 853–868. doi: 10.1007/s11103-005-1511-0
Go, Y. S., Kim, H., Kim, H. J., and Suh, M. C. (2014). Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell 26, 1666–1680. doi: 10.1105/tpc.114.123307
Greer, S., Wen, M., Bird, D., Wu, X., Samuels, L., Kunst, L., et al. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 145, 653–667. doi: 10.1104/pp.107.107300
Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. doi: 10.1007/bf00014672
Haslam, T. M., and Kunst, L. (2013). Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210, 93–107. doi: 10.1016/j.plantsci.2013.05.008
Hegebarth, D., and Jetter, R. (2017). Cuticular waxes of Arabidopsis thaliana shoots: cell-type-specific composition and biosynthesis. Plants 6:27. doi: 10.3390/plants6030027
Holmes, M. G., and Keiller, D. R. (2002). Effects of pubescence and waxes on the reflectance of leaves in the ultraviolet and photosynthetic wavebands: a comparison of a range of species. Plant Cell Environ. 25, 85–93. doi: 10.1046/j.1365-3040.2002.00779.x
Hooker, T. S., Lam, P., Zheng, H., and Kunst, L. (2007). A core subunit of the RNA processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 19, 904–913. doi: 10.1105/tpc.106.049304
Ingram, G., and Nawrath, C. (2017). The roles of the cuticle in plant development: organ adhesions and beyond. J. Exp. Bot. 68, 5307–5321. doi: 10.1093/jxb/erx313
Iwase, A., Mitsuda, N., Koyama, T., Hiratsu, K., Kojima, M., Arai, T., et al. (2011). The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Cur. Biol. 21, 508–514. doi: 10.1016/j.cub.2011.02.020
Kannangara, R., Branigan, C., Liu, Y., Penfield, T., Rao, V., Mouille, G., et al. (2007). The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19, 1278–1294. doi: 10.1105/tpc.106.047076
Kim, H., Go, Y. S., and Suh, M. C. (2018). DEWAX2 transcription factor negatively regulates cuticular wax biosynthesis in Arabidopsis leaves. Plant Cell Physiol. 59, 966–977. doi: 10.1093/pcp/pcy033
Kim, H., Lee, S. B., Kim, H. J., Min, M. K., Hwang, I., and Suh, M. C. (2012). Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 53, 1391–1403. doi: 10.1093/pcp/pcs083
Kim, H., Yu, S.-I., Jung, S. H., Lee, B.-H., and Suh, M. C. (2019). The F-box protein SAGL1 and ECERIFERUM3 regulate cuticular wax biosynthesis in response to changes in humidity in Arabidopsis. Science 31, 2223–2240. doi: 10.1105/tpc.19.00152
Kim, J., Jung, J. H., Lee, S. B., Go, Y. S., Kim, H. J., Cahoon, R., et al. (2013). Arabidopsis 3-ketoacyl-coenzyme a synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 162, 567–580. doi: 10.1104/pp.112.210450
Kosma, D. K., Bourdenx, B., Bernard, A., Parsons, E. P., Lü, S., Joubès, J., et al. (2009). The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 151, 1918–1929. doi: 10.1104/pp.109.141911
Kunst, L., and Samuels, L. (2009). Plant cuticules shine: advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 12, 721–727. doi: 10.1016/j.pbi.2009.09.009
Lam, P., Zhao, L., Eveleigh, N., Yu, Y., Chen, X., and Kunst, L. (2015). The exosome and trans-acting small interfering RNAs regulate cuticular wax biosynthesis during Arabidopsis inflorescence stem development. Plant Physiol. 167, 323–336. doi: 10.1104/pp.114.252825
Lam, P., Zhao, L., McFarlane, H. E., Aiga, M., Lam, V., Hooker, T. S., et al. (2012). RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis. Plant Physiol. 159, 1385–1395. doi: 10.1104/pp.112.199646
Lee, H. G., Kim, J., Suh, M. C., and Seo, P. J. (2017). The MIEL1 E3 ubiquitin ligase negatively regulates cuticular wax biosynthesis in Arabidopsis stems. Plant Cell Physiol. 58, 1249–1259. doi: 10.1093/pcp/pcx065
Lee, S. B., Go, Y. S., Bae, H. J., Park, J. H., Cho, S. H., Cho, H. J., et al. (2009). Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola. Plant Physiol. 150, 42–54. doi: 10.1104/pp.109.137745
Lee, S. B., Kim, H. U., and Suh, M. C. (2016). MYB94 and MYB96 additively activate cuticular wax biosynthesis in Arabidopsis. Plant Cell Physiol. 57, 2300–2311. doi: 10.1093/pcp/pcw147
Lee, S. B., and Suh, M. C. (2013). Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Mol. Plant 6, 246–249. doi: 10.1093/mp/sss159
Lee, S. B., and Suh, M. C. (2015a). Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 34, 557–572. doi: 10.1007/s00299-015-1772-2
Lee, S. B., and Suh, M. C. (2015b). Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 56, 48–60. doi: 10.1093/pcp/pcu142
Li, F., Wu, X., Lam, P., Bird, D., Zheng, H., Samuels, L., et al. (2008). Identification of the wax ester synthase/acyl-coenzyme A: diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physio. 148, 97–107. doi: 10.1104/pp.108.123471
Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M. X., Arondel, V., Bates, P. D., et al. (2013). Acyl-lipid metabolism. Arabidopsis Book 11:e0161. doi: 10.1199/tab.0161
Lin, R. C., Park, H. J., and Wang, H. Y. (2008). Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol. Plant 1, 42–57. doi: 10.1093/mp/ssm004
Millar, A. A., Clemens, S., Zachgo, S., Giblin, E. M., Taylor, D. C., and Kunst, L. (1999). CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11, 825–838. doi: 10.1105/tpc.11.5.825
Morimoto, K., Ohama, N., Kidokoro, S., Mizoi, J., Takahashi, F., Todaka, D., et al. (2017). BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, E8528–E8536. doi: 10.1073/pnas.1704189114
Morineau, C., Gissot, L., Bellec, Y., Hematy, K., Tellier, F., Renne, C., et al. (2016). Dual fatty acid elongase complex interactions in Arabidopsis. PLoS One 11:e0160631. doi: 10.1371/journal.pone.0160631
Park, C. S., Go, Y. S., and Suh, M. C. (2016). Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 88, 257–270. doi: 10.1111/tpj.13248
Rae, L., Lao, N. T., and Kavanagh, T. A. (2011). Regulation of multiple aquaporin genes in Arabidopsis by a pair of recently duplicated DREB transcription factors. Planta 234, 429–444. doi: 10.1007/s00425-011-1414-z
Raffaele, S., Vailleau, F., Léger, A., Joubès, J., Miersch, O., Huard, C., et al. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20, 752–767. doi: 10.1105/tpc.107.054858
Riederer, M., and Schreiber, L. (2001). Protecting against water loss: analysis of the barrier properties of plant cuticles. J. Exp. Bot. 52, 2023–2032. doi: 10.1093/jexbot/52.363.2023
Rowland, O., Zheng, H., Hepworth, S. R., Lam, P., Jetter, R., and Kunst, L. (2006). CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 142, 866–877. doi: 10.1104/pp.106.086785
Seo, P. J., Lee, S. B., Suh, M. C., Park, M. J., Go, Y. S., and Park, C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23, 1138–1152. doi: 10.1105/tpc.111.083485
Shepherd, T., and Griffiths, W. D. (2006). The effects of stress on plant cuticular waxes. New Phytol. 171, 469–499. doi: 10.1111/j.1469-8137.2006.01826.x
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Sieber, P., Schorderet, M., Ryser, U., Buchala, A., Kolattukudy, P., Métraux, J. P., et al. (2000). Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12, 721–738. doi: 10.1105/tpc.12.5.721
Suh, M. C., Samuels, A. L., Jetter, R., Kunst, L., Pollard, M., Ohlrogge, J., et al. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139, 1649–1665. doi: 10.1104/pp.105.070805
Tiwari, S. B., Hagen, G., and Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543. doi: 10.1105/tpc.008417
Weber, H., and Hellmann, H. (2009). Arabidopsis thaliana BTB/POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family. FEBS J. 276, 6624–6635. doi: 10.1111/j.1742-4658.2009.07373.x
Xue, D., Zhang, X., Lu, X., Chen, G., and Chen, Z. H. (2017). Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front. Plant Sci. 8:621. doi: 10.3389/fpls.2017.00621
Yeats, T. H., and Rose, J. K. (2013). The formation and function of plant cuticles. Plant Physiol. 163, 5–20. doi: 10.1104/pp.113.222737
Yoo, S., Cho, Y., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, J. Y., Broeckling, C. D., Blancaflor, E. B., Sledge, M. K., Sumner, L. W., and Wang, Z. Y. (2005). Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 42, 689–707. doi: 10.1111/j.1365-313X.2005.02405.x
Zhang, J. Y., Broeckling, C. D., Sumner, L. W., and Wang, Z. Y. (2007). Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol. Biol. 64, 265–278. doi: 10.1007/s11103-007-9150-2
Keywords: AP2/DREB-type, Arabidopsis, cuticular wax, drought, RAP2.4, transcription factor
Citation: Yang SU, Kim H, Kim RJ, Kim J and Suh MC (2020) AP2/DREB Transcription Factor RAP2.4 Activates Cuticular Wax Biosynthesis in Arabidopsis Leaves Under Drought. Front. Plant Sci. 11:895. doi: 10.3389/fpls.2020.00895
Received: 02 October 2019; Accepted: 02 June 2020;
Published: 03 July 2020.
Edited by:
Guodong Wang, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, ChinaReviewed by:
Rongfeng Huang, Institute of Biotechnology (CAAS), ChinaQiao Zhao, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, China
Copyright © 2020 Yang, Kim, Kim, Kim and Suh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mi Chung Suh, bWNzdWhAc29nYW5nLmFjLmty
†These authors have contributed equally to this work
 Sun Ui Yang
Sun Ui Yang Hyojin Kim
Hyojin Kim Ryeo Jin Kim2
Ryeo Jin Kim2 Jungmook Kim
Jungmook Kim Mi Chung Suh
Mi Chung Suh