
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 June 2020
Sec. Plant Membrane Traffic and Transport
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00892
 Bingzhu Hou1,2
Bingzhu Hou1,2 Yuanyue Shen1*
Yuanyue Shen1*Abscisic acid (ABA) plays important roles in many aspects of plant growth and development, and responses to diverse stresses. Although much progress has been made in understanding the molecular mechanisms of ABA homoeostasis and signaling, the mechanism by which plant cells integrate ABA trafficking and signaling to regulate plant developmental processes is poorly understood. In this study, we used Arabidopsis STOMATAL CYTOKINESIS DEFECTIVE 2/RIPENING-REGULATED PROTEIN 1 (SCD2/RRP1) mutants and overexpression plants, in combination with transcriptome and protein-interaction assays, to investigate SCD2/RRP1 involvement in the integration of ABA trafficking and signaling in seed germination and seedling growth. Manipulation of SCD2/RRP1 expression affected ABA sensitivity in seed germination and seedling growth, as well as transcription of several ABA transporter genes and ABA content. RNA-sequencing analysis of Arabidopsis transgenic mutants suggested that SCD2/RRP1 was associated with ABA signaling via a type 2C protein phosphatase (PP2C) protein. The N- and C-terminal regions of SCD2/RRP1 separately interacted with both PYRABACTIN RESISTANCE 1 (PYR1) and ABA INSENSITIVE 1 (ABI1) on the plasma membrane, and SCD2/RRP1 acted genetically upstream of ABI1. Interestingly, ABA inhibited the interaction of SCD2/RRP1 with ABI1, but did not affect the interaction of SCD2/RRP1 with PYR1. These results suggested that in Arabidopsis SCD2/RRP1participates in early seed development and growth potentially through clathrin-mediated endocytosis- and clathrin-coated vesicle-mediated ABA trafficking and signaling. These findings provide insight into the mechanism by which cells regulate plant developmental processes through ABA.
During the lifetime of higher plants, the primary nutrient organs such as roots, stems, and leaves grow first for survival, followed by reproductive organs such as flowers, fruits, and seeds for sexual reproduction in response to diverse internal and external cues. At the cellular level, plant developmental processes essentially involve continuous cell division, differentiation, enlargement, and senescence. Each of these cellular processes is under the strict control of phytohormones.
Abscisic acid (ABA), a growth inhibitor identified in the early 1960s (Liu and Carnsdagger, 1961; Ohkuma et al., 1963), plays important roles not only in a variety of plant growth and developmental processes, including seed maturation and dormancy, seed germination, seedling and root growth, floral transition, fruit ripening, and stomatal movement, but also participates in the adaptive responses of plants to biotic and abiotic stresses, including drought, high salinity, chilling, and pathogen attack (Leung and Giraudat, 1998; Finkelstein et al., 2002; Himmelbach et al., 2003; Cutler et al., 2010; Zhang, 2014). These physiological responses are triggered by endogenous ABA contents, which are tightly controlled by ABA biosynthesis, catabolism, and transport (Nambara and Marion-Poll, 2005; Umezawa et al., 2006; Boursiac et al., 2013; Kuromori et al., 2018). In plants, ABA biosynthesis is synthesized from carotenoids, which are derived from the precursor of geranylgeranyl diphosphate by the methylerythritol phosphate pathway in plastids (Lichtenthaler, 1999). Geranylgeranyl diphosphate is also used for the biosynthesis of chlorophyll and gibberellins (Zi et al., 2014). To a large extent, free ABA homoeostasis is synergistically regulated at the synthesis–degradation and conjugation–hydroxylation levels by a set of critical enzymes, including 9-cis-epoxycarotenoid dioxygenase (NCED) for synthesis, ABA 8′-hydroxylases (CYP707A) for degradation, ABA glucosyltransferase for conjugation, and ABA β-glucosidase for hydroxylation (Schwartz et al., 2003; Lee et al., 2006). Several G-subfamily members of ATP-binding cassette (ABC) transporters, including ABCG 22/25/30/40, function in ABA trafficking (Boursiac et al., 2013; Merilo et al., 2015; Kuromori et al., 2018). Notably, AtNPF4.6/AtNRT1.2/AtAIT1 (a low-affinity nitrate transporter) and AtDTX50 (a member of the DTX/multidrug-toxic compound extrusion family) are ABA importers (Kanno et al., 2012; Léran et al., 2014). These diverse subcellular enzymes associated with ABA homoeostasis suggest the presence of ABA in different subcellular compartments, which is consistent with the multiple functions of ABA in plants in response to developmental and environmental cues (Xu et al., 2013).
Previous pharmacological and biochemical studies suggested the presence of both transmembrane and cytosolic ABA receptors in plants (Hornberg and Weiler, 1984; Allan et al., 1994; Assmann, 1994). Subsequently, multiple ABA receptors have been identified, including the plasma-membrane-localized GTG1/GTG2 (Pandey et al., 2009), a class of cytosolic PYR/PYL/RCAR (Ma et al., 2009; Park et al., 2009), and a plastid/chloroplast magnesium-chelatase H subunit ABAR/CHLH (Shen et al., 2006). Two core ABA signaling pathways in Arabidopsis(Arabidopsis thaliana) have been proposed, comprising the “ABA–PYR/PYL/RCAR–PP2C–SnRK2” pathway (Fujii et al., 2009), and the “ABA–ABAR–WRKY40–ABI5” pathway (Shang et al., 2010). Genetics and structural biology have confirmed the signaling transduction mechanism in the ABA–PYR/PYL/RCAR–PP2C–SnRK2 cascade: ABA binds to PYR/PYL/RCARs, which bind to and inhibit PP2C activity, and, as a result, the activated SnRK2s phosphorylate downstream targets to trigger ABA physiological responses (Fujii et al., 2009; Melcher et al., 2009; Miyazono et al., 2009; Park et al., 2009; Santiago et al., 2009; Soon et al., 2012). The ABA–PYR1–PP2C1–SnRK2 core signaling pathway also functions in strawberry fruit ripening (Chai et al., 2011; Jia et al., 2013).
It is known that ABA participates in complex biological processes and plays important roles in plant growth and development, as well as numerous stress responses. At the cellular level, plant growth and development requires continual cell division, enlargement, and adaptation in response to developmental and environmental cues. The multiple ABA receptors in different subcellular compartments contribute to the rapid response to physiological reactions. Although much progress has been made in elucidating the molecular mechanisms of ABA homoeostasis, transport, and signaling, the mechanism by which plant cells integrate ABA trafficking and signaling at different subcellular levels to regulate organ developmental processes is poorly understood.
STOMATAL CYTOKINESIS DEFECTIVE 2 (SCD2) functions in Arabidopsis cytokinesis and cell expansion through clathrin-mediated plasma membrane (PM) endocytosis and clathrin-coated vesicles (CCVs; McMichael et al., 2013). Interestingly, strawberry RIPENING-REGULATED PROTEIN 1 (FaRRP1), a ripening-regulated protein involved in fruit ripening (Shen et al., 2012, GenBank: JQ619656.1), is a homolog of SCD2. Given that ABA is a crucial regulator of ripening (Jia et al., 2011), clarification of the relationship between SCD2/RRP1 and ABA is important to understand ABA trafficking and signaling. In the present study, we used a series of Arabidopsis SCD2/RRP1 mutants and overexpression plants, in combination with transcriptome and protein-interaction analyses involving yeast two-hybrid (Y2H), co-immunoprecipitation (CoIP), bimolecular fluorescence complementation (BiFC), and luciferase (LUC) complementation assays, to investigate SCD2/RRP1 involvement in the integration of ABA trafficking and signaling in seed germination and seedling growth. The findings provide insight into the molecular mechanisms of endocytosis and CCVs mediated by the SCD2/RRP1 protein.
Arabidopsis accession Columbia (Col-0) wild-type (WT) plants were used in this study. The RRP1 T-DNA insertion line SALK_063106, hereafter termed the rrp1-1 mutant, was ordered from the Arabidopsis Biological Resource Center. The rrp1-2 and rrp1-3 mutants were generated using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) technology. The RRP1-overexpression lines (Super : RRP1-1 and Super : RRP1-3) were generated by cloning the coding sequence of RRP1 into the pBIB vector under the control of the Super promoter. Seeds were surface sterilized with 70% (v/v) ethanol, sown on 0.8% (w/v) agar plates containing full-strength Murashige and Skoog (MS) medium supplemented with 1% or 3% (w/v) sucrose, stratified for three days at 4°C, and then grown under continuous light at 22°C.
For analysis of Arabidopsis gene expression, quantitative real-time PCR (qPCR) was performed with SYBR Premix Ex Taq (Takara) on a 7500 Real Time PCR System (Life Technologies, USA). The cDNAs were diluted 10 times with ddH2O, and 2 μl was used for PCR. The reaction conditions comprised 40 cycles at 95°C for 15 s and 60°C for 1 min. Relative gene expression was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Arabidopsis ACTIN2/8 was used as an internal standard. The primers used are listed in Supplemental Table 1. The experiments were repeated three times.
Three plants were used for each analysis. Gas chromatography–mass spectrometry (GC-MS) was used to analyze ABA content based on 3H-ABA (3,000 cpm) as an internal standard. Frozen receptacles (1 g) were mixed with sodium diethyldithiocarbamatetrihydrate, quartz sand, and 80% methanol containing radioactiveABA. The mixture and extracts were prepared in accordance with the method of Jia et al. (2011). The experiment was repeated three times.
For germination assays, seeds were surface sterilized and stratified at 4°C for 72 h in the dark before germination. More than 300 seeds of each genotype were sown on the same plate containing MS medium supplemented with 1% (w/v) sucrose and 0, 0.5, or 1 μM ABA, and incubated at 22°C under constant illumination of 60 μmol·m−2·s−1. Germination was defined as initial emergence of the radicle through the seed coat. The percentage seed germination was recorded daily during the germination test. For seedling growth experiments, seeds were germinated after stratification on standard MS medium. About 72 h after stratification, the seeds were transferred to MS medium supplemented with different concentrations of ABA and incubated in the vertical position. Seedling growth was assessed at the indicated times after transfer, and the length of the primary root was measured using a ruler.
Six Arabidopsis plants were randomly selected from the frozen samples for total RNA isolation and cDNA synthesis. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Dusseldorf, Germany). DNase digestion was performed to remove contaminating DNA using the RNase-Free DNase Set (Qiagen). The RNA samples were processed using the RNA Library Prep Kit (New England BioLabs, Ipswich, MA, USA) and sequenced using an Illumina HiSeq2000 platform. The raw reads were filtered with the FASTQ_Quality_Filter tool from the FASTX-Toolkit. Reads longer than 35 bp and Q score > 20 were selected. All valid reads were combined to perform de novo splicing using the paired-end method with Trinity software (Wang et al., 2010). Data analysis was conducted in accordance with the method of Wang et al. (2017). The experiments were repeated three times.
To determine whether the interaction between ABI1 or PYR1 and RRP1 takes place in planta, the coding sequences of Arabidopsis ABI1 or PYR1 and RRP1 were cloned into the pSPYNE and pSPYCE vectors, respectively. The primer sequences used are listed in Supplemental Table 1. Plasmids containing YFPN-ABI1or YFPN-PYR1 and YFPC-RRP1 were introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Nicotiana benthamiana leaves. Three days after infiltration, the yellow fluorescent protein (YFP) fluorescence signal was detected using a Zeiss LSM 710 META confocal microscope.
Arabidopsis protoplasts transformed with the p35S:RRP1-Flag and pSuper : ABI1-GFP or pSuper : PYR1-GFP constructs were incubated in W5 buffer (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES, pH 5.7) for 14–16 h. The pSuper : GFP construct was used as a negative control. The primers used to construct the vectors are listed in Supplemental Table 1. Total proteins were extracted from transformed protoplasts with 1-ml extraction buffer (10 mM HEPEs, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100, protease inhibitor cocktail, and 1 mM PMSF). The supernatant was immunoprecipitated with anti-Flag agarose (Sigma-Aldrich) for 2 h at 4°C. The immunoprecipitates were separated in a 12% SDS-PAGE gel and detected with anti-Flag and anti-GFP antibodies (Abmart).
Y2H assays were performed using the Matchmaker GAL4-based two-hybrid system (Clontech, Palo Alto, CA, USA). Full-length or truncated cDNA of the corresponding genes was inserted into the pGADT7 and pGBKT7 vectors at the EcoRI/BamHI restriction sites, respectively. The primer sequences used are listed in Supplemental Table 1. The constructs were transformed into Saccharomyces cerevisiaestrain AH109 using a lithium acetate method. Yeast cells were cultured on selective medium −Leu/−Trp in accordance with the manufacturer’s instructions. Transformed colonies were plated onto selective medium −Leu/−Trp/−His/−Ade/+X-α-gal supplemented with 0 or 40 μM ABA to test for possible interactions.
For the Luc assays, the Arabidopsis RRP1 and ABI1 coding regions were amplified and cloned into the pCM1300-nLUC and pCM1300-cLUC vectors, respectively. The primers used to construct the vectors are listed in Supplemental Table 1. The plasmids were introduced into Agrobacterium tumefaciens strain GV3101 and co-infiltrated into the leaves of N. benthamiana. After incubation for 2 days, 0 or 100 μM ABA was sprayed onto the infected surface of infiltrated leaves for 6 h. The abaxial leaf surface was sprayed with 1 mM luciferin and then incubated in the dark for 10 min. The LUC signals were captured using a cooled CCD imaging camera (1300B, Roper) at −110°C. Relative LUC activity per cm2 infiltrated leaf area was calculated using Winview32 software. Three independent experiments were performed.
To identify characteristics of the Arabidopsis SCD2/RRP1 protein, a Protein Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) search using the 578-amino-acid sequence (Arabidopsis AT3G48860) determined that it is a coiled-coil domain-containing protein that belongs to a conserved superfamily containing a SMC (structural maintenance of chromosomes) domain, which have ATP-binding domains at the N- and C-termini and two extended coiled-coil domains separated by a hinge in the middle (Supplemental Figure 1A). Among the hits showing the highest homology, only RRP1 (Shen et al., 2012, GenBank: JQ619656.1, unpublished) and SCD2 (McMichael et al., 2013) have been annotated (Supplemental Figure 1B).
In the scd2-1 mutant, the guanine residue at position 1372 of the SCD2 open reading frame is deleted, leading to inactivation of the SCD2 domain, and plants show dwarfism and infertility (McMichael et al., 2013). Therefore, we screened for mutants in which gene expression, but not growth and development, were affected for further analysis. A set of transgenic Arabidopsis plants, comprising T-DNA insertion mutants, CRISPR/Cas9-edited mutants, and overexpression lines, was constructed. The T-DNA insertion line rrp1-1 (SALK_063106) was ordered from the Arabidopsis Biological Resource Center. The CRISPR/Cas 9-generated rrp1 mutants were produced using a pair of closely located sgRNA targets (T1 and T2) in RRP1 (Figure 1A). In addition, two T1 lines of CRISPR/Cas9-generated rrp1 mutants, named rrp1-2 and rrp1-3, were screened and confirmed by sequencing, which demonstrated that the rrp1-2 mutant was a homozygous mutant with a 68-bp deletion without a frame shift, whereas rrp1-3/+ was a heterozygous mutant with a 56-bp deletion with a frame shift inducing a premature stop codon (Figure 1B). No off-target mutations were detected (Supplemental Table 2). The heterozygous rrp1-3/+ plants grown on MS medium supplemented with 3% sucrose segregated on the basis of defects in plant growth and development (Figure 1C). Consistent with previous reports, homozygous rrp1-3 plants grown in soil were dwarfed and infertile in comparison with the WT (Figure 1D). Real-time PCR analysis showed that RRP1 expression was slightly repressed in the T-DNA mutants (rrp1-1) and elevated in the overexpression lines (Super : RRP1-1 and Super : RRP1-3) in comparison with the WT (Figure 1E). These results demonstrated that alteration of Arabidopsis SCD2/RRP1 expression affected plant growth.
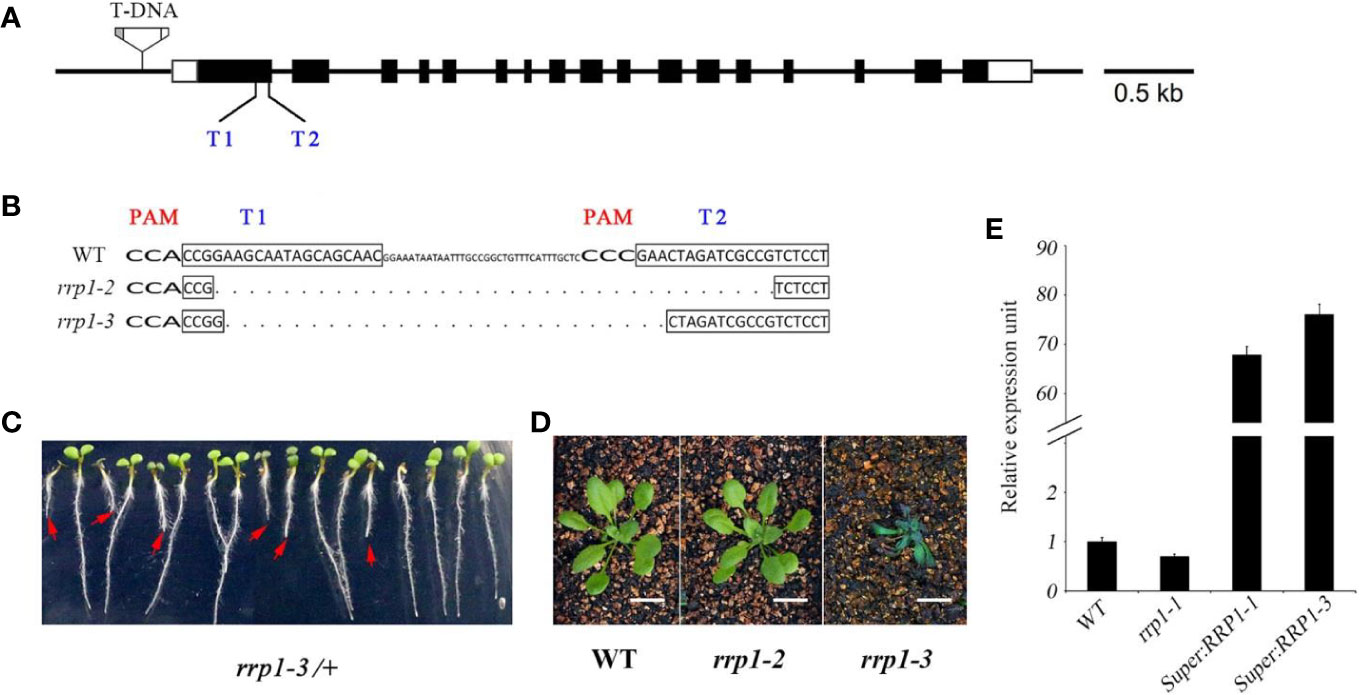
Figure 1 Manipulation of Arabidopsis SCD2/RRP1 expression affects Arabidopsis seedling growth. (A) Schematic illustration of RRP1 genomic regions with the rrp1-1 T-DNA insertion and CRISPR/Cas9 targets. Empty boxes represent 5′ and 3′ untranslated regions, black boxes represent exons, and black lines represent introns. T1 and T2 represent two target sites for CRISPR/Cas9 editing. The T-DNA insertion is not drawn to scale. Bar = 0.5 kb. (B) Homozygous rrp1 mutants of the T3 generation were generated by CRISPR/Cas9 editing. Mutation of RRP1 was evaluated by sequencing. The protospacer adjacent motifs are marked with wide bold letters, the black boxes indicate targets, and the dashed line indicates regions of deletion. (C) Heterozygous rrp1-3/+ mutants of the T2 generation grown vertically on Murashige and Skoog medium supplemented with 3% sucrose. The red arrows indicate homozygotes. (D) Phenotypic comparison between rrp1-2, rrp1-3, and wild type (WT) plants 4 weeks after germination. (E) Transcripts of the rrp1-1 mutant and a RRP1-overexpression line (Super : RRP1-3) were tested by quantitative real-time PCR. The data represent the mean ± SE (n = 3).
To explore the role of SCD2/RRP1 in ABA-responsive physiological processes in Arabidopsis, owing to the infertility of homozygous rrp1-3 plants, we used the rrp1-1, rrp1-2, Super : RRP1-1, and Super : RRP1-3 transgenic plants to investigate seed germination and seedling growth on MS medium supplemented with 0, 0.5, or 1 μM ABA. All lines exhibited a similar phenotype (Figure 2A, left panel). The rrp1-1 and rrp1-2 mutants showed insensitivity to ABA on medium supplemented with 1 μM ABA, whereas the Super : RRP1-1 and Super : RRP1-3 lines were hypersensitive to 0.5 μM ABA compared with the WT (Figure 2A, right panel). No significant differences in percentage seed germination among the genotypes was observed on ABA-free MS medium. On medium supplemented with 0.5 or 1 μM ABA, the rrp1-1 and rrp1-2 mutants showed a significant increase in percentage seed germination compared with the WT (Figure 2B). In contrast, the seed germination percentages of the Super : RRP1-1 and Super : RRP1-3 lines were similar to that of the WT on medium containing 0.5 μM ABA, whereas germination decreased significantly on medium supplemented with 1 μM ABA (Figure 2B). The cotyledon-greening percentage was similar among the transgenic and WT plants on ABA-free medium (Figure 2C); in contrast, the rrp1-1 and rrp1-2 mutants showed higher percentages, whereas the Super : RRP1-1 and Super : RRP1-3 lines showed lower percentages, of cotyledon greening compared with that of the WT (Figure 2C).
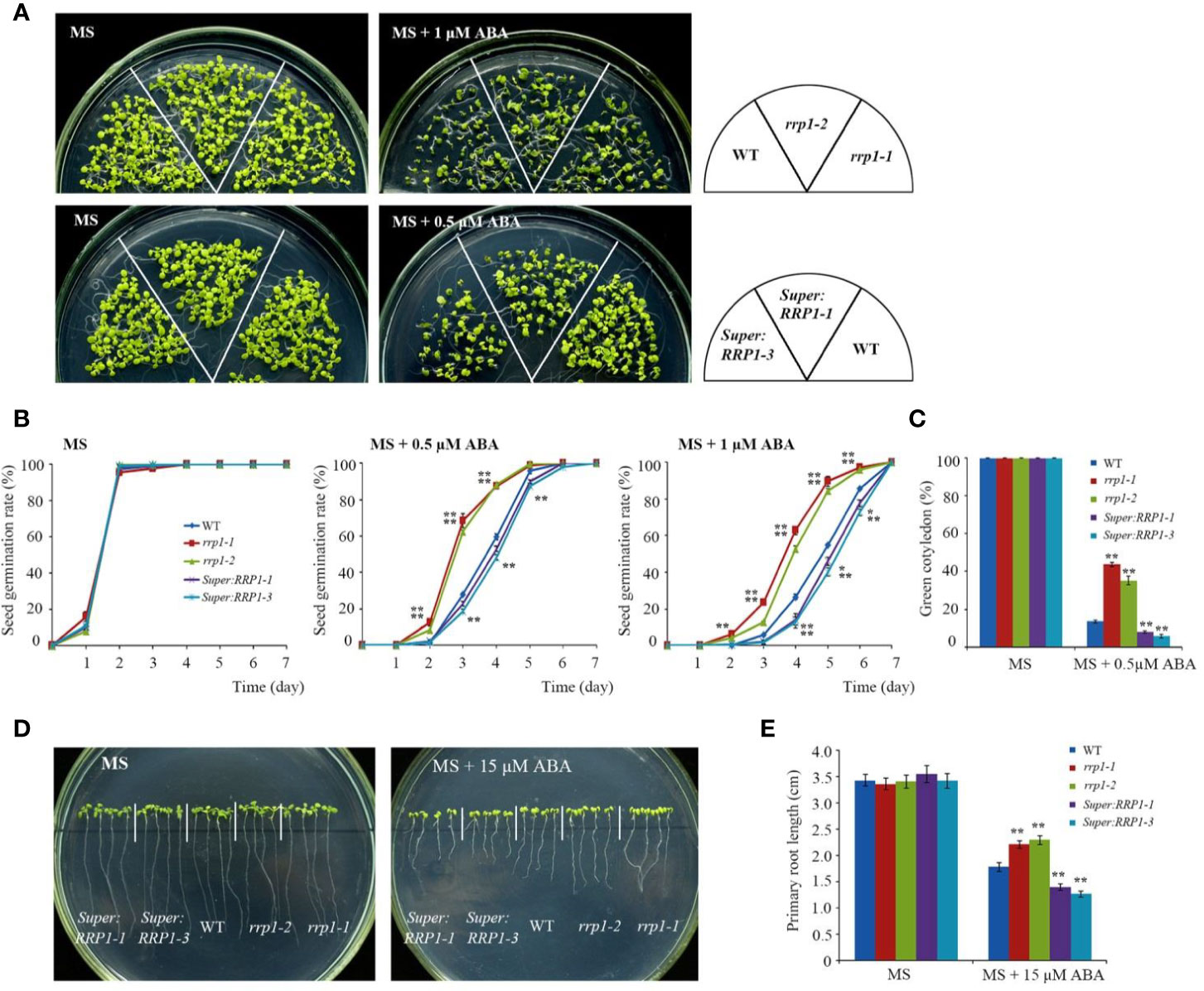
Figure 2 ABA sensitivity in the rrp1 mutants and the RRP1-overexpression Arabidopsis lines. (A) Phenotypic comparison analysis. Imbibed seeds were transferred to Murashige and Skoog (MS), MS + 0.5 μM ABA, and MS + 1 μM ABA medium and grown for 14 days. (B) Seed germination assay. Imbibed seeds were transferred to MS and MS medium supplemented with 0.5 or 1 μM ABA. Seed germination frequency was calculated at a given time. Data are the mean ± SE (n = 3). More than 300 seeds were measured in each replicate. (C) Cotyledon-greening analysis. Imbibed seeds were transferred to MS or MS + 0.5 μM ABA medium for 6 days before determination of cotyledon-greening percentage. Data are the mean ± SE (n = 3). More than 300 seeds were measured in each replicate. (D, E) Primary root length in the presence or absence of exogenous ABA. The 3-day-old seedlings were transferred to MS or MS + 15 μM ABA medium for 7 days, and then photographs were taken and the primary root length was measured. Asterisks in (B, C, E) indicate statistically significant differences compared with wild-type plants: *, P < 0.05; **, P < 0.01.
To examine whether SCD2/RRP1 was involved in ABA-mediated root growth, 3-day-old seedlings of rrp1 mutants, RRP1-OE lines, and the WT were transferred to MS medium supplemented with 0 or 15 μM ABA for 7 days. When grown on MS medium lacking ABA, the primary root length of Super : RRP1-3 was similar, whereas that of the rrp1-3homozygotes was shorter, compared with the primary root of WT seedlings (Figures 2D, E). When grown on the medium supplemented with 15 μM ABA, the primary root lengths of the Super : RRP1-1 and Super : RRP1-3 lines were shorter, whereas those of the rrp1-1and rrp1-2 mutants were longer, compared with the primary root of the WT (Figures 2D, E). Taken together, these data demonstrated that SCD2/RRP1 served as a positive regulator of ABA-mediated Arabidopsis seed germination and seedling growth.
To identify candidate SCD2/RRP1-regulated genes in the transgenic plants, three groups comprising the rrp1-3 mutants (homozygotes gained by CRISPR), the RRP1-overexpression line (Super : RRP1-3), and the WT were used for RNA-sequencing (RNA-seq) in accordance with a previous report on differentially expressed genes (DEGs) at the log2 (2 RPKM) level (Wang et al., 2017). Comparing the rrp1-3 mutants with the WT, 341 DEGs were annotated with 156 upregulated and 185 downregulated among the mapped 19,130 genes in the two libraries; comparing Super : RRP1-3 with the WT, 96 DEGs were annotated with 23 upregulated and 73 downregulated among the mapped 19,102 genes in the two libraries; and comparing Super : RRP1-3 with the rrp1-3 mutants, 331 DEGs were annotated with 171 upregulated and 160 downregulated among the mapped 19,105 genes in the two libraries (Table 1). It was notable that among the three-group comparisons, the majority of DEG-mapped pathways were detected in the comparison of Super : RRP1-3 with the rrp1-3 mutants; nine pathways were detected, among which the top-five pathways comprised α-linolenic acid metabolism, glucosinolate biosynthesis, 2-oxocarboxylic acid metabolism, degradation of valine, leucine and isoleucine, and plant hormone signal transduction.
To further investigate whether SCD2/RRP1 is involved in phytohormone signaling pathways, the DEGs mapped to this pathway were analyzed in relation to gibberellin, ABA, ethylene, brassinolide, jasmonic acid (JA), salicylic acid, cytokinin, and auxin. Five DEG families, consisting of PP2C, JAZ, MYC2, A-ARR, and PR1, were annotated to phytohormone pathways (Figure 3): ABA (ABI1: ABA-insensitive 1; HAI1: highly ABA-induced 1), JA (JAZ1, 2, 3, 5, 6, 9, and 10: Jasmonate ZIM-domain 1, 2, 3, 5, 6, 9, and 10; MYC2: myelocytomatosis protein 2, a basic helix-loop-helix Leu zipper transcription factor), cytokinin (ARR15: two-component type-A response regulator), and salicylic acid (PR1: pathogenesis-related protein 1; Figure 4). Interestingly, the majority of these DEGs were upregulated in the OE plants and downregulated in the CRISPR/Cas9 plants in comparison with the WT (Figure 4). Given that these transgenic plants showed no response to JA in seed germination and seedling growth (data not shown), these results suggested that SCD2/RRP1 might be involved in ABA signaling through the ABI1 protein, which is a crucial negative regulator of ABA signaling.
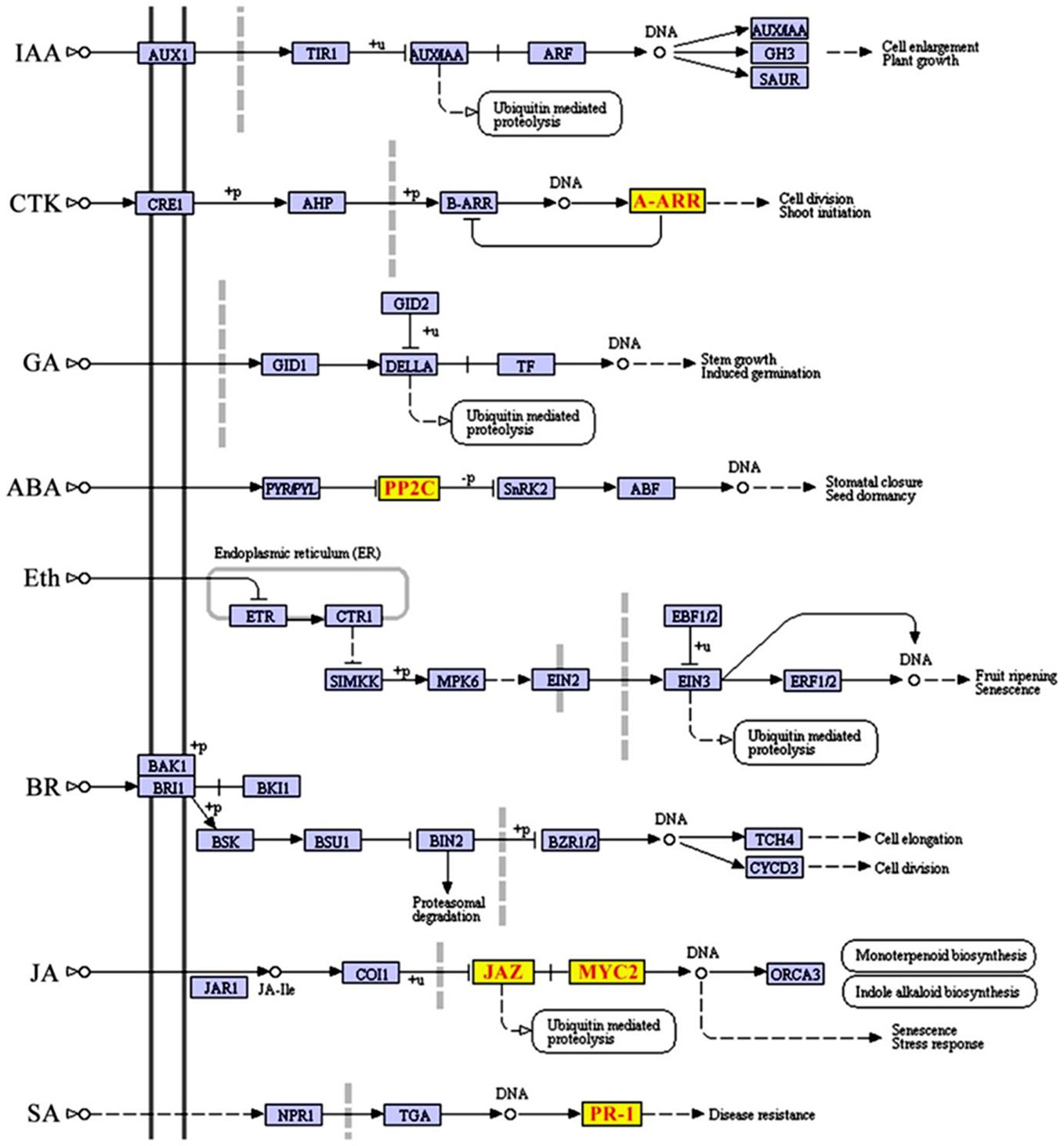
Figure 3 Differentially expressed genes (DEGs) mapped to the plant hormone signal transduction pathway according to a Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. KEGG terms with a corrected P-value less than 0.05 were considered to be significantly enriched by DEGs. Five DEGs, comprising JAZ, MYC2, ARR-A, PR1, and PP2C, were screened and transcript levels were upregulated, which are shown in red letters on yellow blocks in the plant hormone signal transduction pathway.
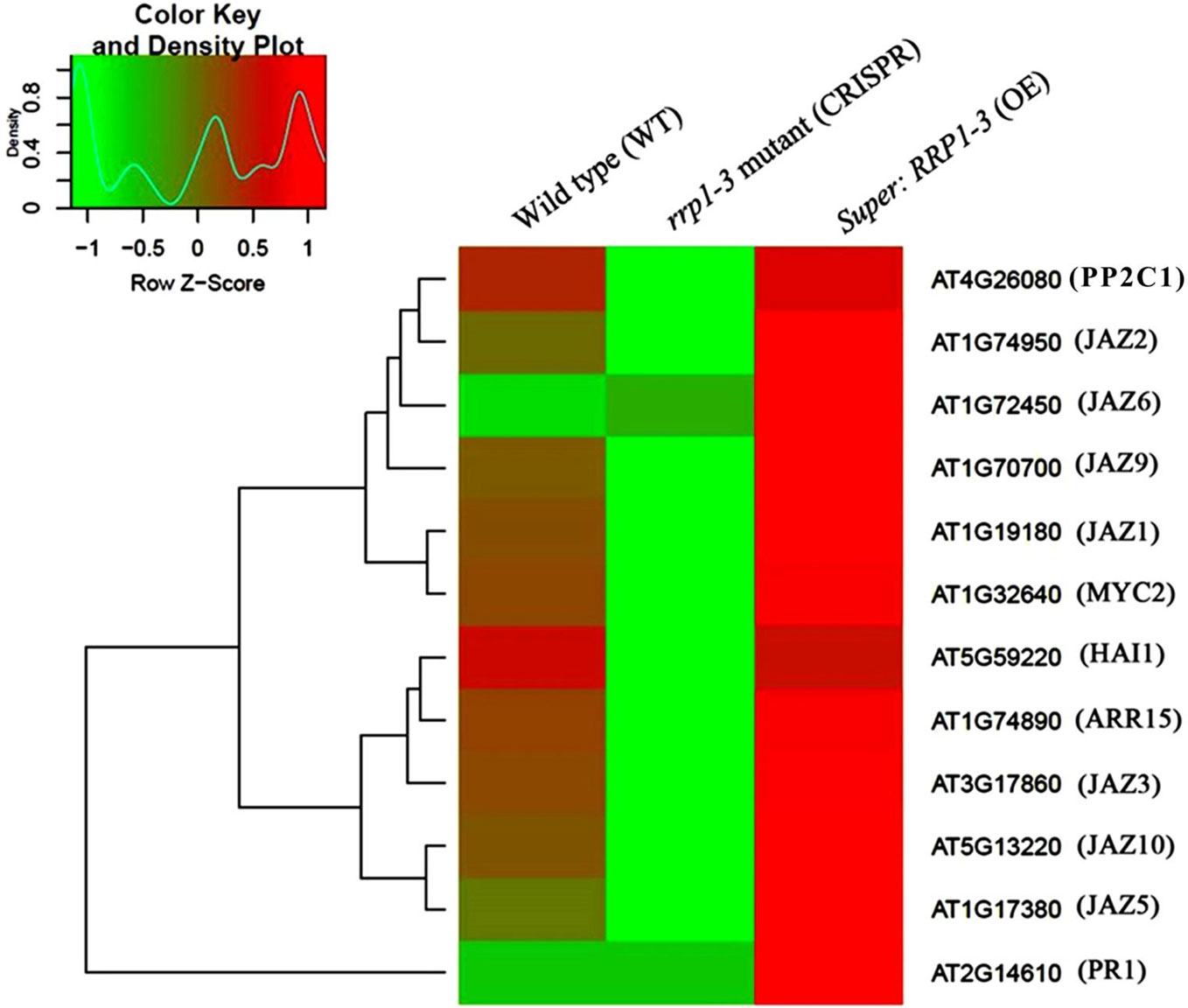
Figure 4 Heatmap and cluster analysis of transcripts for the important genes associated with RRP1 in Arabidopsis. Data for gene expression levels were normalized to the z-score with the formula log10 (FPKM + 1) and are indicated by a color key and density plot; from green to red represents a relative gene expression level from low to high, respectively. ABI1, ABA-insensitive 1; HAI1, highly ABA-Induced1; JAZ1, 2, 3, 5, 6, 9, and 10, Jasmonate ZIM-domain1, 2, 3, 5, 6, 9, and 10; MYC2, myelocytomatosis protein 2 (a basic helix-loop-helix Leu zipper transcription factor); ARR15, two-component type-A response regulator; and PR1 (pathogenesis-related protein 1).
To explore whether SCD2/RRP1 has a direct relationship with ABI1, BiFC and a N. benthamiana transient expression analysis were performed by co-infiltration of Agrobacterium tumefaciens harboring ABI1-YNE and RRP1-YCE into N. benthamianaleaf epidermal cells in vivo. After cultivation for 2.5 days, the fluorescence signal at 488 nm was observed. The YFP fluorescence signal was detected from the two-gene, co-infected leaves, whereas the control infected with the empty vector showed no YFP signal; notably, the interaction signals may be entirely cytosolic (Figure 5A).
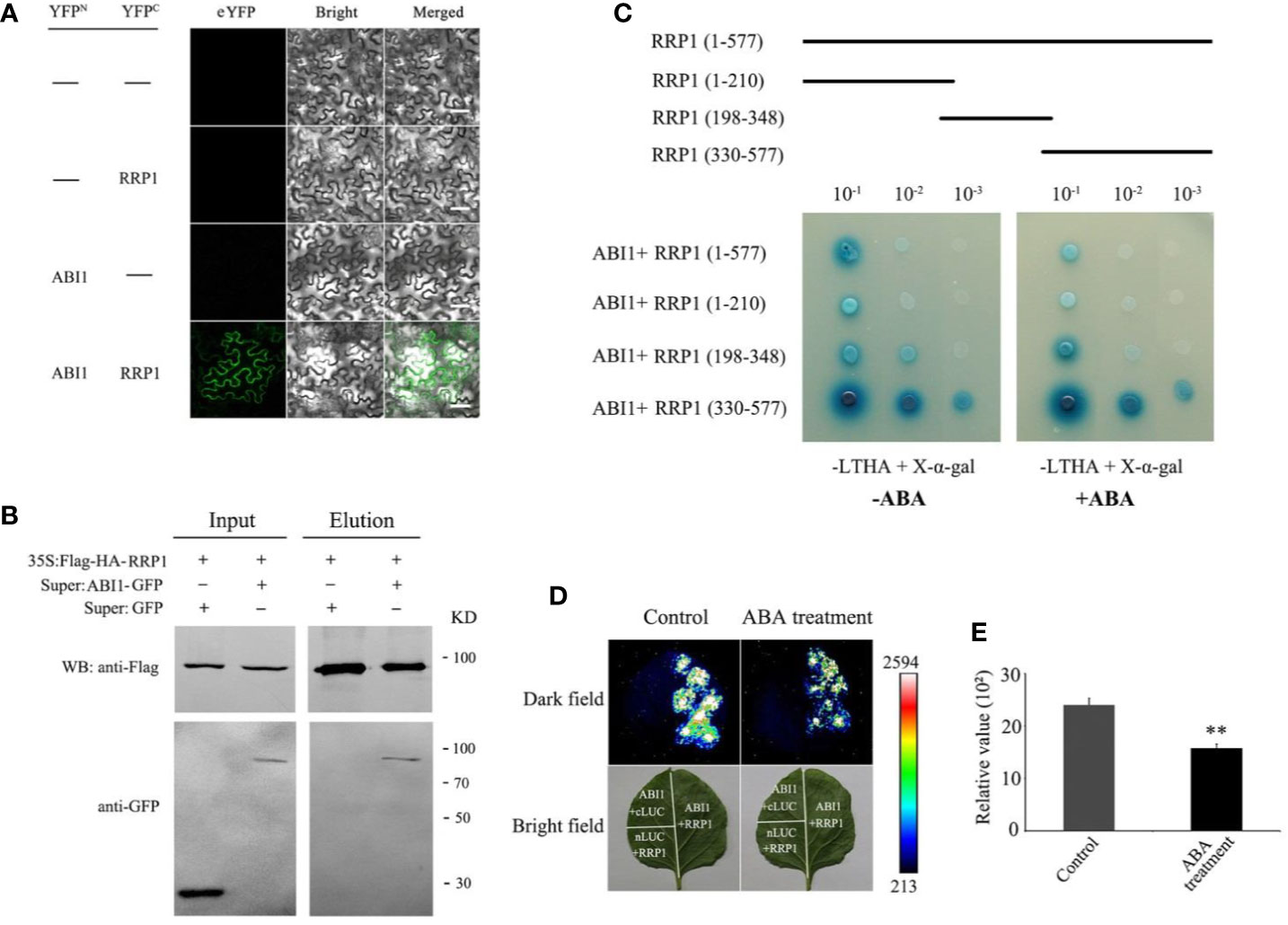
Figure 5 Interaction of RRP1 with ABI1 in Arabidopsis and the influence of ABA. (A) Bimolecular fluorescence complementation (BiFC) assay showing the interaction of RRP1 and ABI1. eYFP, enhanced yellow fluorescent protein. YFPN and YFPC are the N terminus and C terminus of eYFP, respectively. Scale bars, 20 μm. (B) Co-immunoprecipitation (Co-IP) assay of RRP1 with ABI1. Super : RRP1-GFP and 35S:Flag-HA-ABI1, or Super : GFP and 35S:Flag-HA-ABI1 were co-transfected into Arabidopsis protoplasts. Co-IP was conducted with anti-Flag beads from total isolated proteins, and immunoblotting analysis was performed with anti-GFP and anti-Flag antibodies. (C) The C-terminus of RRP1 primarily interacts with ABI1, and ABA weakens RRP1–ABI1 interaction in a Y2H assay. Top panels show a schematic representation of the different RRP1 deletions in the yeast vectors. Yeast transformants were spotted onto the selective medium SD/−Leu/−Trp/−His/−Ade (−LTHA)/+X-α-gal supplemented with 0 or 40 μM ABA. (D) ABA weakens RRP1–ABI1 interaction in a firefly luciferase (LUC) complementation imaging system. Nicotiana benthamiana leaves were transformed with the construct pairs ABI1-nLUC/RRP1-cLUC and negative controls (ABI1-nLUC/cLUC and nLUC/RRP1-cLUC). The leaves were observed for fluorescence imaging 6 h after spraying 0 or 100 μM ABA on the infected leaf surface. (E) Quantitative analysis of luminescence intensity. Each value is the mean ± SE of three independent experiments. Asterisks indicate statistically significant differences compared with the control: **, P < 0.01.
To further confirm this interaction, CoIP was used based on construction of RRP1-Flag and ABI1-GFP fusion protein vectors, which were co-transformed into Arabidopsis protoplasts and cultured for 16 h. The total protoplast proteins were extracted for CoIP western blotting, which revealed that the ABI1-GFP protein could be co-precipitated by anti-Flag beads and then detected by the anti-GFP antibody, and vice versa, whereas this was not observed for the control infected with the empty vector (Figure 5B). These results further demonstrated that RRP1 interacts with ABI1 in vivo.
To identify the crucial regions in SCD2/RRP1 for interaction with ABI1, the coding sequence of RRP1 was truncated into three sections (1–210, 198–348, and 330–577 bp). These cDNA fragments were ligated into the pGADT7 vector, co-transformed into yeast AH109 cells with the pGBKT7-RRP1 construct, then plated separately on SD/−Trp/−Leu/−His/−Ade/+X-α-Gal medium supplemented with 0 or 40 μM ABA, and cultured for 2–5 days. ABI1 interacted strongly with the polypeptide encoded by the 330–577 fragments, but interacted only weakly with the polypeptides encoded by the 1–210 and 198–348 fragments, and these interactions may be affected by ABA (Figure 5C).
To verify the effect of ABA on the interaction between RRP1 and ABI1, a LUC complementation assay in vivo was conducted. The coding sequences of RRP1 and ABI1 were separately ligated to generate the ABI1-nLUC and RRP1-cLUC constructs. Agrobacterium tumefaciens harboring the ABI1-nLUC and RRP1-cLUC fusion plasmids were co-infected into the lower epidermal cells of N. benthamiana leaves. After cultivation for 2.5 days, the infected leaf surfaces were sprayed with 0 μM (control) or 100 μM ABA solution. After culture for 6 h, the leaf surfaces were sprayed with LUC fluorescent substrate and reacted for 10 min, then the fluorescence signal was detected. The N. benthamianaleaves treated with 100 μM ABA showed weaker fluorescence compared with those treated with 0 μM ABA (Figures 5D, E). These results confirmed that SCD2/RRP1 is capable of interacting with ABI1, and indicated that interaction occurs at the C-terminus of SCD2/RRP1, which is affected by ABA.
To explore the genetic relationship between SCD2/RRP1 and ABI1, the ABA-insensitive rrp1-2 mutant and ABA-sensitive Super : RRP1-3 plants (Figure 1) as well as the ABA-sensitive abi1-cas9 mutants (our previously constructed knockout mutant at nucleotide position 536; data not shown) were used to generate F2 plants comprising abi1-cas9/rrp1-2 and abi1-cas9/Super : RRP1-3 by hybridization, screening, and identification (data not shown).
First, various phenotypes were observed using the abi1-cas9, rrp1-2, abi1-cas9/rrp1-2, and WT plants. Seed germination and growth analysis showed that on standard MS medium, all plant materials germinated and grew with no significant differences observed (Figure 6A, left, and Figure 6B). However, on medium supplemented with 0.5 μM ABA, the rrp1-2 mutant displayed an ABA-insensitive phenotype, whereas the abi1-cas9/rrp1-2double mutant showed an ABA-sensitive phenotype, similar to that of the abi1-cas9 mutant (Figures 6A, right, C). Analysis of the seedling cotyledon-greening percentage showed that on standard MS medium, all transgenic plants showed similar growth trends. On medium supplemented with 0.5 μM ABA, the rrp1-2 mutant showed a higher cotyledon-greening percentage, whereas the abi1-cas9/rrp1-2 double mutant showed a lower percentage, compared with that of the WT, similar tothe abi1-cas9 mutant (Figure 6D).
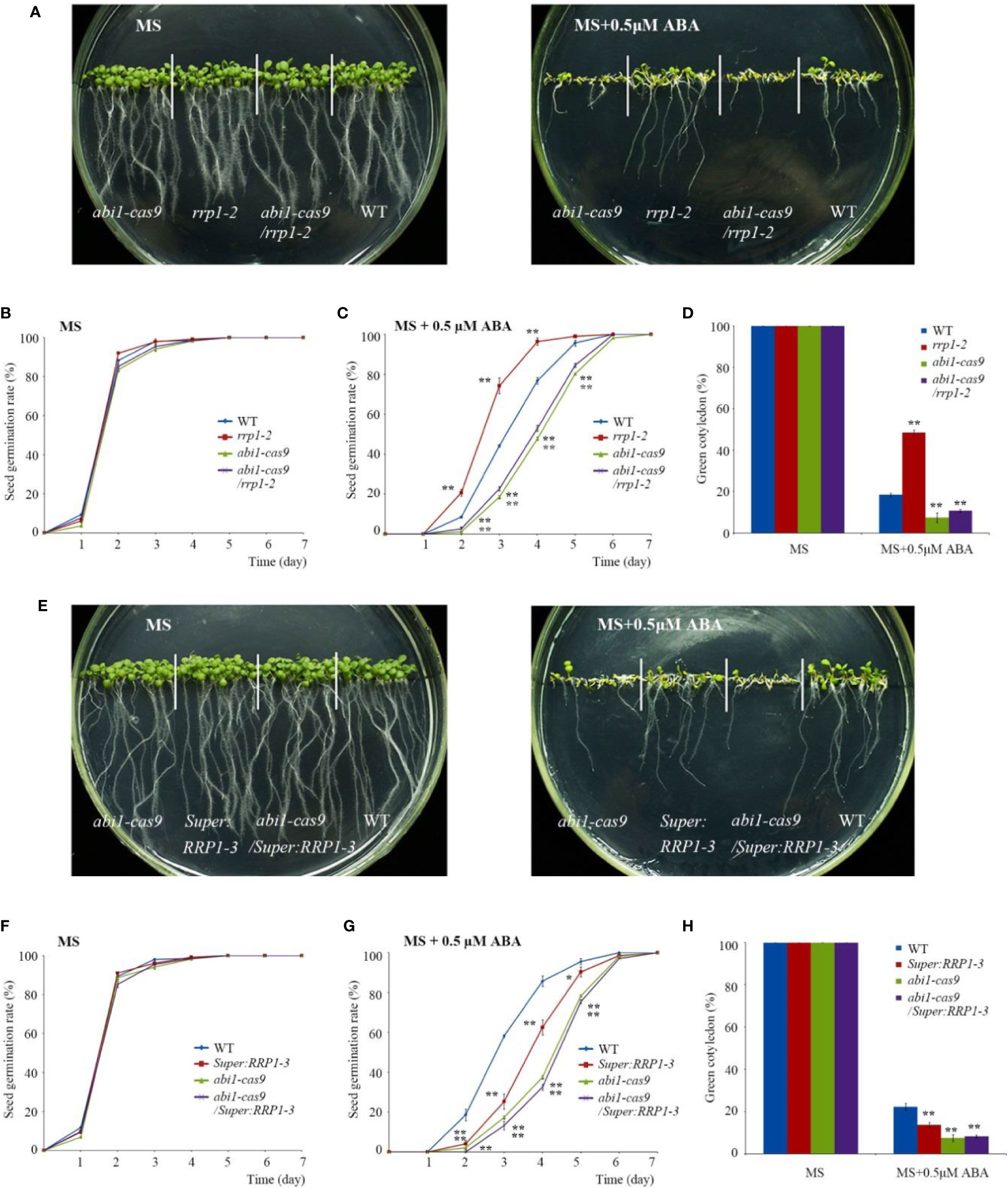
Figure 6 Function of RRP1 upstream of ABI1. (A) Phenotypic comparison analysis. Imbibed seeds were transferred to Murashige and Skoog (MS) and MS + 0.5 μM abscisic acid (ABA) medium and grown for 9 days. (B, C) Seed germination assay. Imbibed seeds were transferred to MS (B) and MS medium supplemented with 0.5 μM ABA (C). The seed germination frequency was calculated at a given time. Data are the mean ± SE (n = 3). More than 300 seeds were measured in each replicate. (D) Cotyledon-greening analysis. Imbibed seeds were transferred to MS or MS + 0.5 μM ABA medium for 7 days before determination of cotyledon-greening percentage. Data are the mean ± SE (n = 3). More than 300 seeds were measured in each replicate. (E) Phenotypic comparison analysis. Imbibed seeds were transferred to MS and MS + 0.5 μM ABA medium and grown for 11 days. (F, G) Seed germination assay. Imbibed seeds were transferred to MS (B) and MS medium containing 0.5 μM ABA (C). The seed germination frequency was calculated at a given time. Data are the mean ± SE (n = 3). More than 300 seeds were measured in each replicate. (H) Cotyledon-greening analysis. Imbibed seeds were transferred to MS or MS + 0.5 μM ABA medium for 7 days before determination of cotyledon-greening percentage. Data are the mean ± SE (n = 3). More than 300 seeds were measured in each replicate. Asterisks indicate statistically significant differences compared with the wild-type plants: *, P < 0.05; **, P < 0.01.
Second, various phenotypes were observed using the abi1-cas9, Super : RRP1-3, abi1-cas9/Super : RRP1-3, and the WT plants. On standard MS medium, all plant materials germinated and grew with no significant differences observed (Figures 6E, left, F). On medium supplemented with 0.5 μM ABA, germination and growth of Super : RRP1-3 and abi1-cas9/Super : RRP1-3 plants displayed an ABA-sensitive phenotype, and the degree of sensitivity of the abi1-cas9/Super : RRP1-3 plants was similar to that of the abi1-cas9 mutant (Figures 6E, right, G). Analysis of the seedling cotyledon-greening percentage showed that on standard MS medium, all transgenic plants showed similar growth trends. However, on medium supplemented with 0.5 μM ABA, Super : RRP1-3 and abi1-cas9/Super : RRP1-3 plants showed a lower cotyledon-greening percentage compared with that of the WT, but the percentage of the abi1-cas9/Super : RRP1-3 plants was similar to that of the abi1-cas9 mutant (Figure 6H).
Taken together, these results indicated that ABI1 knockout impaired not only the ABA-insensitive phenotypes of the rrp1-2 mutant, but also the ABA-sensitive phenotype of the RRP1-overexpressing lines in seed germination and seedling growth, which suggested that SCD2/RRP1 is genetically upstream of AtABI1 in the response to ABA.
Interaction of SCD2/RRP1 with ABI1 prompted us to explore the relationship between SCD2/RRP1 and PYR1. Therefore, Y2H, BiFC, and CoIP assays were conducted as described for the assays with ABI1. In the BiFC and CoIP interaction systems, RRP1 interacted directly with PYR1 in vivo, and interaction in the cytoplasm was observed (Figures 7A, B). The Y2H assay showed that PYR1 interacted with the polypeptide encoded by the 1–210 and 198–348 fragments of RRP1, and the interactions were not affected by ABA (Figure 7C). These results confirmed that SCD2/RRP1 is capable of interacting with PYR1 and the interaction can occur in the cytoplasm, and that interaction occurs in the N-terminus of RRP1 in an ABA-independent manner.
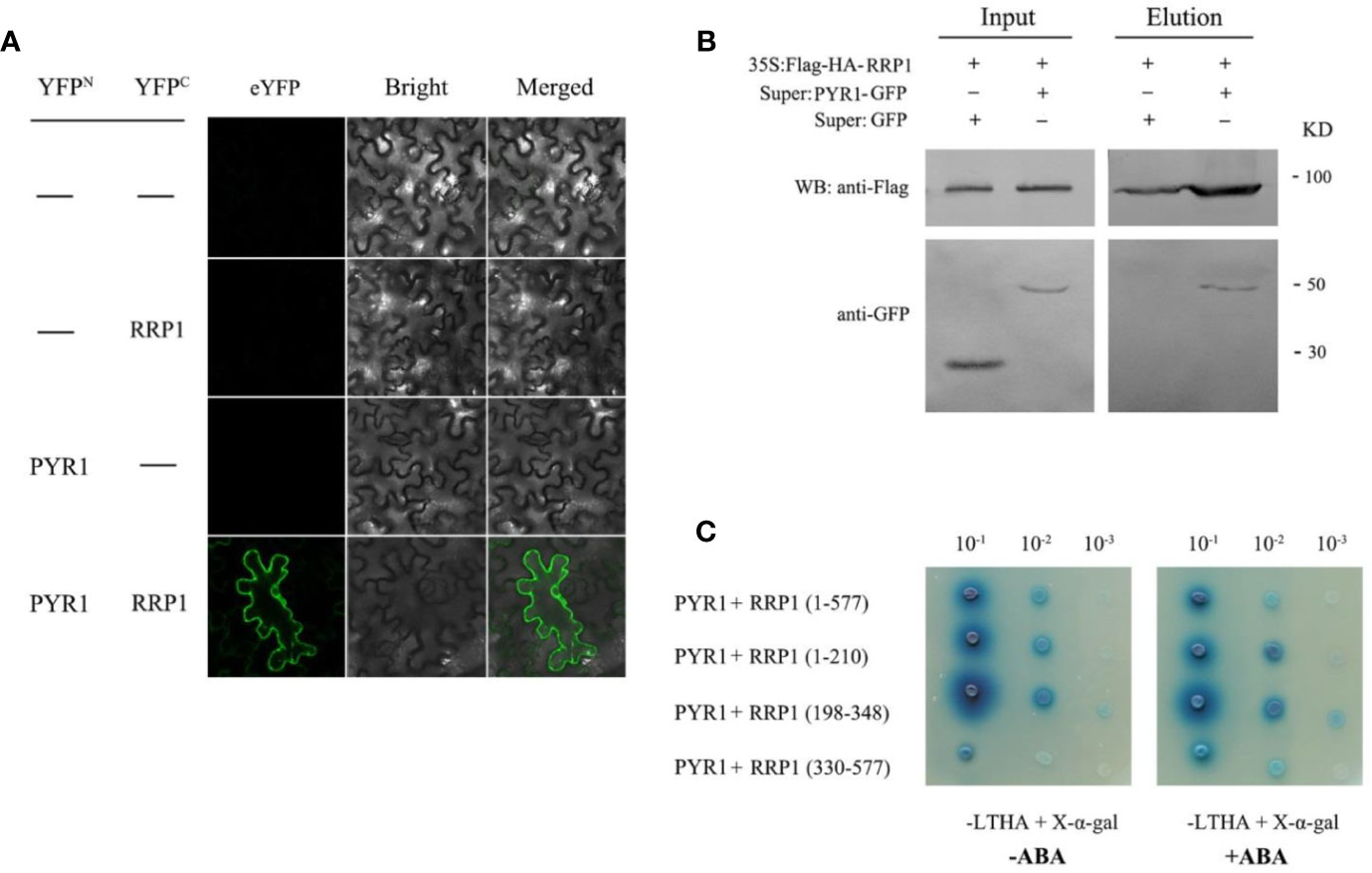
Figure 7 Interaction of RRP1 with PYR1 in Arabidopsis. (A) BiFC assay showing interaction of RRP1 and PYR1. eYFP, enhanced yellow fluorescent protein. YFPN and YFPC are the N terminus and C terminus of eYFP, respectively. Scale bars, 20 μm. (B) Co-IP assay of RRP1 with PYR1. Super : PYR1-GFP and 35S:Flag-HA-RRP1, or Super : GFP and 35S:Flag-HA-RRP1 were co-transfected into Arabidopsis protoplasts. Co-IP was conducted with anti-Flag beads from total isolated proteins, and immunoblotting analysis was performed with anti-GFP and anti-Flag antibodies. (C) The N-terminus of RRP1 primarily interacts with PYR1. Yeast transformants were spotted onto the selective medium SD/−Leu/−Trp/−His/−Ade (−LTHA)/+X-α-gal supplemented with 0 or 40 μM ABA.
Transport of ABA in Arabidopsis cells is regulated by ABCG40, NPF4.6, and ABCG30, which are involved in ABA uptake, and DTX50, ABCG25, and ABCG31, which participate in ABA efflux (Dong et al., 2015; Merilo et al., 2015; Kuromori et al., 2018). To investigate whether manipulation of SCD2/RRP1 gene expression influences overall transport capacity of ABA in WT, rrp1-3, and Super : RRP1-3 cells, the ABA contents and mRNA levels of several ABA transporter genes were analyzed on the basis of the RNA-seq data. The qPCR analysis showed that with regard to ABA uptake and transport, ABCG40 transcripts were upregulated by sixty-fold in the rrp1-3 mutant, whereas the transcripts of NPF4.6 and ABCG30 were significantly downregulated in overexpression lines. With regard to ABA efflux, the DTX50 transcript level was significantly downregulated by four-fold in the rrp1-3 mutant, whereas the transcript levels of ABCG25 and ABCG31 were not significantly affected in the overexpression lines (Figure 8A). The ABA contents were reduced in the rrp1-3 mutant, but no significant effect was observed in the Super-RRP1-3 line in comparison with the control (Figure 8B). These results further indicated that SCD2/RRP1 was associated with ABA trafficking.
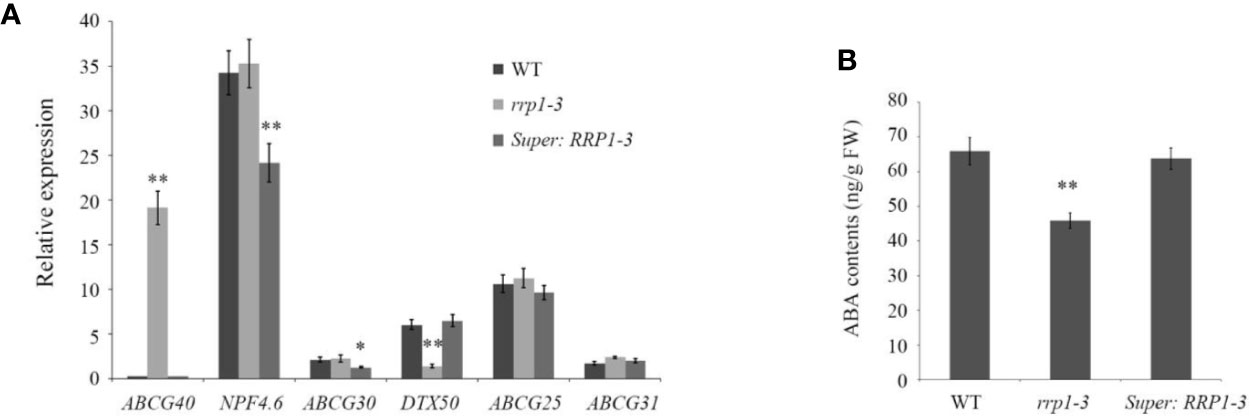
Figure 8 Analysis of relative expression levels of ABA transporter genes and ABA contents intransgenic Arabidopsis plants. (A) Transcripts of a set of ABA transporter genes were analyzed by quantitative real-time PCR in the wild-type, rrp1-3, and Super : RRP1-3 plants. (B) ABA contents in the wild-type, rrp1-3, and Super : RRP1-3 plants. The data represent the mean ± SE (n = 3). Asterisks indicate statistically significant differences compared with wild-type plants: *, P < 0.05; **, P < 0.01.
The Arabidopsis SCD1 protein, a Rab guanine nucleotide exchange factor, functions in cytokinesis and cell expansion by membrane trafficking (Falbel et al., 2003). Similarly, SCD2 is co-localized with SCD1 in the clathrin light chain at sites of endocytosis in the PM, functions at hypostasis to SCD1, and both proteins are required for clathrin-mediated plasma-membrane endocytosis and CCVs, which are crucial to cytokinesis and cell expansion (McMichael et al., 2013).
Whether SCD2/RRP1functions in relation to phytohormones is yet unknown. In the present study, we provide evidence to demonstrate that SCD2/RRP1 regulates Arabidopsis seed germination and seedling growth via ABA: (1) manipulation of SCD2/RRP1 expression in the mutants affects ABA sensitivity in seed germination and seedling growth (Figure 2); (2) SCD2/RRP1 can interact with ABI1, which is a core component of ABA core signaling (Fujii et al., 2009; Ma et al., 2009), and the interaction is inhibited by ABA (Figure 5); (3) SCD2/RRP1 can interact with PYR1, which is an ABA receptor (Fujii et al., 2009; Ma et al., 2009; Figure 7); and (4) manipulation of SCD2/RRP1 expression affects ABA contents and transcription of several ABA transporter genes (Figure 8).
Sessile plants have developed a series of mechanisms for growth and survival. Endocytosis is a mechanism by which certain molecules are transported into cells across the PM (Zwiewka et al., 2015; Mettlen et al., 2018). Clathrin-mediated endocytosis, which is the best-characterized endocytic pathway, is a major regulator of cell-surface protein internalization in response to extracellular and intracellular cues (Kitakura et al., 2011; Irani et al., 2012; Zwiewka et al., 2015; Belda-Palazon et al., 2016; Mettlen et al., 2018). Interestingly, clathrin-mediated endocytosis is involved in auxin transport (Kitakura et al., 2011), brassinosteroid signaling (Irani et al., 2012), and ABA signaling (Belda-Palazon et al., 2016). However, mechanisms mediated by clathrin-mediated endocytosis for cell-surface protein internalization and trafficking are not fully understood in response to phytohormones.
Arabidopsis SCD2 functions in clathrin-mediated membrane transport, including PM endocytosis, which is required for cytokinesis and cell expansion (McMichael et al., 2013). Manipulation of Arabidopsis SCD2/RRP1expression affects transcription of several ABA transporter genes and ABA contents (Figure 8), thus RRP1 may potentially be involved in ABA trafficking through clathrin-mediated membrane transport and endocytosis. Interestingly, interaction of SCD2/RRP1 with PYR1 and ABI1 was confirmed (Figures 5, 6, 7). Furthermore, ABA did not affect interaction of the N-terminus of SCD2/RRP1 with PYR1, but inhibited interaction of the C-terminus of SCD2/RRP1 with ABI1 (Figures 5 and 7), which suggested that SCD2/RRP1 may recruit PYR1 and ABI1 onto the PM and CCVs. The ABA–PYR1–ABI1–SnRK2.2 cascade is a core signaling mechanism, by which ABA regulates plant growth and development as well as stress responses (Fujii et al., 2009; Ma et al., 2009; Jia et al., 2013). Given that SCD2 is a peripheral membrane protein (McMichael et al., 2013), we thus speculate on the function of SCD2/RRP1 in Arabidopsis growth and development (Figure 9). The SCD2/RRP1 protein may recruit PYR/PYLs/RCARs and PP2Cs to form a three-protein complex near the PM, and then may initiate clathrin-mediated endocytosis and CCVs. ABA can inhibit interaction of RRP1 with ABI1 and, as a result, promotes ABA responsiveness and the ABA–PYR/PYLs/RCARs–PP2Cs–SnRK2 cascade, ultimately regulating plant growth and development. Further exploration is a prerequisite for visualization of the cellular processes in SCD2/RRP1-mediated ABA trafficking and signaling by endocytosis and vesicle transport.
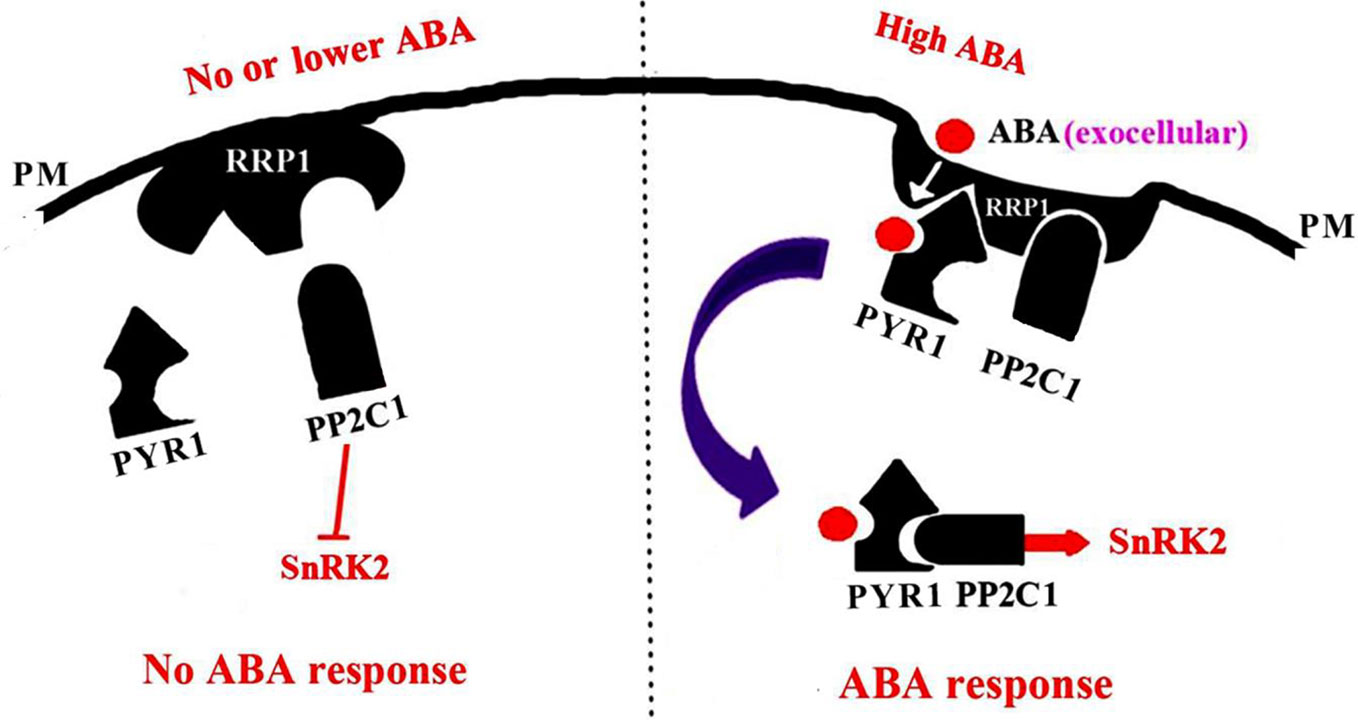
Figure 9 A model for SCD2/RRP1-mediated endocytosis of ABA and ABA signaling. When the ABA content is low or absent, RRP1 may bind to and activate ABI1 activity and, as a result, promote inhibition of SnRK2 activity and ABA response. When the ABA content is high, ABA binds to RRP1 to promote the RRP1 recruitment of PYR1 and ABI1 to the plasma membrane (PM) to form a three-protein complex, which then further facilitates SCD2/RRP1-mediated endocytosis of ABA and ABA signaling.
The datasets generated for this study can be found in the NCBI BioProject ID PRJNA635263.
BH performed the experiments. YS designed the research and wrote the article.
the National Natural Science Foundation of China (Project 31672125).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Qijun Chen for providing CRISPR/Cas9 vectors in Arabidopsis. This work was supported by the China National Science Foundation (31672125), the National Key Research and Development Program (2018YFD1000200), Beijing Natural Science Foundation (6171001), Sichuan Lomon Biotechnology Co. Ltd (2018001). We thank Robert McKenzie, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00892/full#supplementary-material
Allan, A. C., Fricker, M. D., Ward, J. L., Beale, M. H., Trewavas, A. J. (1994). Two transduction pathways mediate rapid effects of abscisic acid in Commelina guard cells. Plant Cell 6, 1319–1328. doi: 10.2307/3869829
Assmann, S. M. (1994). Ins and outs of guard cell ABA receptors. Plant Cell 6, 1187–1190. doi: 10.1105/tpc.6.9.1187
Belda-Palazon, B., Rodriguez, L., Fernandez, M. A., Castillo, M. C., Anderson, E. A., Gao, C., et al. (2016). FYVE1/FREE1 Interacts with the PYL4 ABA Receptor and Mediates its Delivery to the Vacuolar Degradation Pathway. Plant Cell 28, 2291–2311. doi: 10.1105/tpc.16.00178
Boursiac, Y., Léran, S., Corratgé-Faillie, C., Gojon, A., Krouk, G., Lacombe, B. (2013). ABA transport and transporters. Trends Plant Sci. 8, 325–333. doi: 10.1016/j.tplants.2013.01.007
Chai, Y. M., Jia, H. F., Li, C. L., Dong, Q. H., Shen, Y. Y. (2011). FaPYR1 is involved in strawberry fruit ripening. J. Exp. Bot. 62, 5079–5089. doi: 10.1093/jxb/err207
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Dong, T., Park, Y., Hwang, I. (2015). Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 58, 29–48. doi: 10.1042/bse0580029
Falbel, T. G., Koch, L. M., Nadeau, J. A., Seguí-Simarro, J. M., Sack, F. D., Bednarek, S. Y. (2003). SCD1 is required for cytokinesis and polarized cell expansion in Arabidopsis thaliana. Development 130, 4011–4024. doi: 10.1242/dev.00619
Finkelstein, R. R., Gampala, S. S., Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14, S15–S45. doi: 10.1105/tpc.010441
Fujii, H., Chinnusamy, V., Rodrigues, A., Rubio, S., Aatoni, R., Park, S. Y., et al. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. doi: 10.1038/nature08599
Himmelbach, A., Yang, Y., Grill, E. (2003). Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 6, 470–479. doi: 10.1016/S1369-5266(03)00090-6
Hornberg, C., Weiler, E. W. (1984). High-affinity binding sites for abscisic acid on the plasmalemma of Vicia faba guard cells. Nature 310, 321–324. doi: 10.1038/310321a0
Irani, N. G., Di Rubbo, S., Mylle, E., Van den Begin, J., Schneider-Pizoń, J., Hniliková, J., et al. (2012). Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8, 583–589. doi: 10.1038/nchembio.958
Jia, H. F., Chai, Y. M., Li, C. L., Lu, D., Luo, J. J., Qin, L., et al. (2011). Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 157, 188–199. doi: 10.1104/pp.111.177311
Jia, H. F., Lu, D., Sun, J. H., Li, C. L., Xing, Y., Qin, L., et al. (2013). Type 2C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening. J. Exp. Bot. 64, 1677–1687. doi: 10.1093/jxb/ert028
Kanno, Y., Hanada, A., Chiba, Y., Ichikawa, T., Nakazawa, M., Matsui, M., et al. (2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. United States America 109, 9653–9658. doi: 10.1073/pnas.1203567109
Kitakura, S., Vanneste, S., Robert, S., Löfke, C., Teichmann, T., Tanaka, H., et al. (2011). Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23, 1920–1931. doi: 10.1105/tpc.111.083030
Kuromori, T., Seo, M., Shinozaki, K. (2018). ABA Transport and Plant Water Stress Responses. Trends Plant Sci. Trends Plant Sci. 23 (6), 513–522. doi: 10.1016/j.tplants.2018.04.001
Léran, S., Varala, K., Boyer, J. C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Lee, K. H., Piao, H. L., Kim, H. Y., Choi, S. M., Jiang, F., Hartung, W., et al. (2006). Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126, 1109–1120. doi: 10.1016/j.cell.2006.07.034
Leung, J., Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. doi: 10.1146/annurev.arplant.49.1.199
Lichtenthaler, H. K. (1999). The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Biol. 50, 47–65. doi: 10.1146/annurev.arplant.50.1.47
Liu, W. C., Carnsdagger, H. R. (1961). Isolation of abscisin, an abscission accelerating substance. Science 134, 384–385. doi: 10.1126/science.134.3476.384
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
McMichael, C. M., Reynolds, G. D., Koch, L. M., Wang, C., Jiang, N., Nadeau, J., et al. (2013). Mediation of clathrin-dependent trafficking during cytokinesis and cell expansion by Arabidopsis stomatal cytokinesis defective proteins. Plant Cell 25, 3910–3925. doi: 10.1105/tpc.113.115162
Melcher, K., Ng, L. M., Zhou, X. E., Soon, F.-F., Xu, Y., Suino-Powell, K. M., et al. (2009). A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462 (7273), 602–608. doi: 10.1038/nature08613
Merilo, E., Jalakas, P., Kollist, H., Brosché, M. (2015). The role of ABA recycling and transporter proteins in rapid stomatal responses to reduced air humidity, elevated CO2 and exogenous ABA. Mol. Plant 8, 657–659. doi: 10.1016/j.molp.2015.01.014
Mettlen, M., Chen, P. H., Srinivasan, S., Danuser, G., Schmid, S. L. (2018). Regulation of clathrin-mediated endocytosis. Annu. Rev. Biochem 87, 871–896. doi: 10.1146/annurev-biochem-062917-012644
Miyazono, K., Miyakawa, T., Sawano, Y., Kubota, K., Kang, H. J., Asano, A., et al. (2009). Structural basis of abscisic acid signalling. Nature 462 (7273), 609–614. doi: 10.1038/nature08583
Nambara, E., Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185. doi: 10.1146/annurev.arplant.56.032604.144046
Ohkuma, K., Lyon, J. L., Addicott, F. T., Smith, O. E. (1963). Abscisin II, an abscission-accelerating substance from young cotton fruit. Science 142, 1592–1593. doi: 10.1126/science.142.3599.1592
Pandey, S., Nelson, D. C., Assmann, S. M. (2009). Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136, 136–148. doi: 10.1016/j.cell.2008.12.026
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Santiago, J., Dupeux, F., Round, A., Antoni, R., Park, S. Y., Jamin, M., et al. (2009). The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462 (7273), 665–668. doi: 10.1038/nature08591
Schwartz, S. H., Qin, X., Zeevaart, J. A. (2003). Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131, 1591–1601. doi: 10.1104/pp.102.017921
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Shen, Y. Y., Wang, X. F., Wu, F. Q., Du, S. Y., Cao, Z., Shang, Y., et al. (2006). The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443, 823–826. doi: 10.1038/nature05176
Shen, Y. Y., Luo, J. J., Liu, J. (2012). Fragaria x ananassa ripening-regulated protein (RRP1) mRNA, complete cds. Available at: https://www.ncbi.nlm.nih.gov/nuccore/395335145
Soon, F. F., Ng, L. M., Zhou, X. E., West, G. M., Kovach, A., Tan, M. H., et al. (2012). Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335 (6064), 85–88. doi: 10.1126/science.1215106
Umezawa, T., Okamoto, M., Kushiro, T., Nambara, E., Oono, Y., Seki, M., et al. (2006). CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 46, 171–182. doi: 10.1111/j.1365-313X.2006.02683.x
Wang, Z. Y., Fang, B. P., Chen, J. Y., Zhang, X. Z., Luo, Z. X., Huang, L. F., et al. (2010). De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweet potato (Ipomoea batatas). BMC Genomics 11, 726. doi: 10.1186/1471-2164-11-726
Wang, Q. H., Zhao, C., Zhang, M., Li, Y. Z., Shen, Y. Y., Guo, J. X. (2017). Transcriptome analysis around the onset of strawberry fruit ripening uncovers an important role of oxidative phosphorylation in ripening. Sci. Rep. 7, 41477. doi: 10.1038/srep41477
Xu, Z. Y., Kim, D. H., Hwang, I. (2013). ABA homeostasis and signaling involving multiple subcellular compartments and multiple receptors. Plant Cell Rep. 32, 807–813. doi: 10.1007/s00299-013-1396-3
Zi, J., Mafu, S., Peters, R. J. (2014). To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu. Rev. Plant Biol. 65, 259–286. doi: 10.1146/annurev-arplant-050213-035705
Keywords: Arabidopsis, seed germination, seedling growth, abscisic acid, SCD2/RRP1, PYR1, ABI1
Citation: Hou B and Shen Y (2020) A Clathrin-Related Protein, SCD2/RRP1, Participates in Abscisic Acid Signaling in Arabidopsis. Front. Plant Sci. 11:892. doi: 10.3389/fpls.2020.00892
Received: 30 March 2020; Accepted: 01 June 2020;
Published: 18 June 2020.
Edited by:
Kendal Hirschi, Baylor College of Medicine, United StatesReviewed by:
Toshiro Shigaki, The University of Tokyo, JapanCopyright © 2020 Hou and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanyue Shen, c2ZtbkB0b20uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.