- 1Key Laboratory of Sweetpotato Biology and Biotechnology, Ministry of Agriculture and Rural Affairs/Beijing Key Laboratory of Crop Genetic Improvement/Laboratory of Crop Heterosis and Utilization, Ministry of Education, College of Agronomy & Biotechnology, China Agricultural University, Beijing, China
- 2College of Agronomy, Qingdao Agricultural University, Qingdao, China
BTB-TAZ (BT)-domain proteins regulate plant development and pathogen defense. However, their roles in resistance to abiotic stresses remain largely unknown. In this study, we found that the sweetpotato BT protein-encoding gene IbBT4 significantly enhanced the drought tolerance of Arabidopsis. IbBT4 expression was induced by PEG6000, H2O2 and brassinosteroids (BRs). The IbBT4-overexpressing Arabidopsis seeds presented higher germination rates and longer roots in comparison with those of WT under 200 mM mannitol stress. Under drought stress the transgenic Arabidopsis plants exhibited significantly increased survival rates and BR and proline contents and decreased water loss rates, MDA content and reactive oxygen species (ROS) levels. IbBT4 overexpression upregulated the BR signaling pathway and proline biosynthesis genes and activated the ROS-scavenging system under drought stress. Yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays revealed that the IbBT4 protein interacts with BR-ENHANCED EXPRESSION 2 (BEE2). Taken together, these results indicate that the IbBT4 gene provides drought tolerance by enhancing both the BR signaling pathway and proline biosynthesis and further activating the ROS-scavenging system in transgenic Arabidopsis.
Introduction
The effects of drought on agricultural production are increasing worldwide (Zhu, 2002; Yang et al., 2010; Boyer et al., 2013; Chiogna et al., 2018). Phytohormones are essential for plant growth, development and protective responses against severe environmental stresses (Dong et al., 2014; Vargas et al., 2014; Jin et al., 2016). Brassinosteroids (BRs) compose a group of naturally- occurring steroidal phytohormones that are involved in the regulation of adaptation to environmental stresses, such as salinity, drought, heat and cold, as well as multiple aspects of growth and development in plants (Nolan et al., 2017; Planas-Riverola et al., 2019).
In the BR biosynthesis pathway, the BR-specific biosynthetic precursor campesterol (CR) is first converted to campestanol (CN) and then to brassinolide (BL) via the early and late C-6 oxidation pathways. In addition, early C-22 and C-23 hydroxylation branches are the dominant upstream BR biosynthesis pathways (Ohnishi et al., 2006a, b). Several key enzymes, including DWARF4 (DWF4), constitutive photomorphogenesis and dwarfism (CPD), De-etiolated-2(DET2), C-6 oxidase 1, 2 (BR6ox1, 2), and rotundifolia 3 (ROT3), regulate BR biosynthesis (Kim et al., 1999; Nomura et al., 2001; Shimada et al., 2001; Ohnishi et al., 2012). The BR signaling pathway starts with the membrane-localized receptor brassinosteroid-insensitive 1 (BRI1) and co-receptor BRI1-associated kinase 1 (BAK1) (Li and Nam, 2002; Kinoshita et al., 2005). Via inactivation of brassinosteroid-insensitive 2 (BIN2), a negative regulator of BR signaling, BR signaling ultimately culminates in the activation of BRI1-EMS suppressor 1 (BES1)/brassinazole-resistant 1 (BZR1) family transcription factors (Wang et al., 2002; Yin et al., 2002). Thus, the transcription factors BZR1 and BES1 directly regulate the transcription of BR-responsive genes in plants (Wang et al., 2002; Nolan et al., 2017; Planas-Riverola et al., 2019).
The plant-specific BTB-TAZ (BT)-domain protein subfamily, including five members, BT1 to BT5, belong to the bric-a-brac/tramtrack/broad complex (BTB) protein family. These members contain an N-terminal BTB domain, a transcriptional adapter zinc finger (TAZ) domain and a C-terminal calmodulin-binding (CaMB) domain (Du and Poovaiah, 2004). Ren et al. (2007) found that AtBT2 interacts with AtTAC1 and regulates primary root growth in Arabidopsis. In addition, AtBT2 and AtBT3 were found to be essential for gametophyte development in Arabidopsis (Robert et al., 2009). By interacting with AtGTE9 and AtGTE11, AtBT2 imparted resistance to abscisic acid (ABA) and glucose in Arabidopsis (Mandadi et al., 2009; Misra et al., 2018). Overexpression of AtBT2 partially rescued growth inhibition caused by reductions in AtNLP activity and affected the development of nitrate-dependent lateral roots in Arabidopsis (Sato et al., 2017). MdBT2 delayed leaf senescence by inducing the ubiquitination and degradation of MdbHLH93 in transgenic Arabidopsis and apple (An et al., 2019). AtBT4 was reported to regulate resistance to Botrytis cinerea and Pseudomonas syringae in Arabidopsis (Hao et al., 2013; Zheng et al., 2019). However, the roles of BTs in the resistance to abiotic stresses remain largely unknown in plants.
In this study, we cloned a BTB-TAZ domain protein-encoding gene IbBT4 from sweetpotato (Ipomoea batatas (L.) Lam.). It was found that this gene provided drought tolerance by enhancing both the BR signaling pathway and proline biosynthesis and further activating the ROS-scavenging system in transgenic Arabidopsis.
Materials and Methods
Plant Materials
The sweetpotato line Xushu55-2, which is tolerant to drought, was used for isolating the IbBT4 gene and analysing its expression profile. The plants were cultured on Murashige and Skoog (MS) media at 27 ± 1 °C under 13 h of daylight and 54 μM m–2 s–1. Nicotiana benthamiana was used for the subcellular localization of the IbBT4 protein. The function of this gene was analyzed in Arabidopsis thaliana Columbia-0 (WT). N. benthamiana and A. thaliana plants were grown in an artificial climate chamber at 22 ± 1°C and 30% humidity under 16 h of daylight and 54 μM m–2s–1.
Cloning and Sequence Analysis of IbBT4 and Its Promoter
Total complete RNA was isolated from in vitro-grown Xushu55-2 plants and then reverse transcribed to generate cDNA (Zhou et al., 2019). Based on the expressed sequence tag (EST) selected from the transcriptome sequencing data of Xushu55-2 (Zhu et al., 2018), the 5′-untranslated region (UTR) and 3′-UTR of IbBT4 were amplified using rapid amplification of cDNA ends (RACE) procedure using 5′- Full RACE Kit and 3′-Full RACE Core Set Ver.2.0 Kit (TaKaRa, Beijing, China). The cDNA sequence was analyzed by NCBI1. The coding sequence (CDS) was cloned by PCR with specific primers. Its genomic sequence was cloned from Xushu55-2 genomic DNA via PCR in conjunction with specific primers. The promoter region was cloned with Universal Genome Walker 2.0 Kit (TaKaRa, Dalian, China). All of the specific primers are listed in Supplementary Table S1. IbBT4 was annotated in the NCBI databases2. Amino acid sequence alignments, phylogenetic relationships and exon-intron structure were analyzed with DNAMAN software, MEGA 7.0 software and the Splign tool, respectively, and the molecular weight and theoretical isoelectric point (pI) were calculated via online software (Kang et al., 2019). The cis-acting regulatory elements in the promoter region were analyzed via the PlantCARE database.
Subcellular Localization
The coding sequence of IbBT4 was amplified with specific primers (Supplementary Table S1) and inserted into the pMDC83 vector together with green fluorescent protein (GFP). The fusion construct (35S:IbBT4:GFP) and the empty vector (35S:GFP, control) were subsequently transferred into Agrobacterium tumefaciens strain EHA105, respectively, which was then infiltrated into the leaf epidermal cells of 4-week-old N. benthamiana plants with a needleless syringe. At 48 h after agroinfiltration, the infiltrated leaf sections were observed at room temperature using a laser scanning confocal microscope with an argon laser (Olympus, Tokyo, Japan). The cells containing GFP were excited at 488 nm and emissions were recorded at 505–555 nm.
Expression Analysis of IbBT4
Total RNA from the root, stem and leaf tissues of the in vitro-grown Xushu55-2 plants and the leaf, stem, hair root, fibrous root and storage root tissues of 80 days old field-grown plants was extracted for analysing the expression of IbBT4 using quantitative real-time PCR (qRT-PCR), and the specific primers used are listed in Supplementary Table S1 (Zhou et al., 2019). The expression levels of IbBT4 in different tissues were normalized to those of Ibactin (AY905538), and the relative expression levels were calibrated using leaf tissue. Four-week-old in vitro-grown plants of Xushu55-2 were treated with Hoagland solution that comprised H2O (control), 30% PEG6000, 100 μM H2O2 and 100 nM BR, respectively, and after 0, 1, 3, 6, 12, and 24 h of treatment, whole plants were sampled for analysing the IbBT4 expression profiles. The expression levels of IbBT4 in the different treatments were normalized to those of Ibactin (AY905538) and calibrated using the plants sampled at 0 h after treatment. Three plants were used per treatment, and 3 biological replications were included in each treatment.
Arabidopsis Transformation
The coding sequence of IbBT4 was inserted into a pCAMBIA1300 vector to generate a pCAMBIA1300-35S-IbBT4 overexpression vector construct using specific primers (Supplementary Table S1), after which the construct was then introduced into A. tumefaciens strain GV3101. The floral-dip method was used to produce transgenic Arabidopsis plants, which were subsequently grown in pots to produce T3 seeds by screening with 50 mg/L hygromycin. The expression of IbBT4 in the transgenic Arabidopsis plants was analyzed according to the method of Zhou et al. (2019).
Drought Tolerance Assay of Transgenic Arabidopsis
To identify the drought tolerance of transgenic Arabidopsis, T3 and wild-type (WT) seeds were sown in the same plates with 1/2- strength MS media with or without (control) 200 mM mannitol, and after 5 days, their germination rates were measured. Approximately 50 seeds of each line were investigated. Five-day-old seedlings were cultured on 1/2 MS media with or without (control) 200 mM mannitol and after 2 weeks the primary root length was measured. Each material was treated with 5 seedlings per treatment and 3 biological replications were included. Furthermore, one-week-old seedlings growing on 1/2 MS media were planted in pots (7 cm × 7 cm) filled with a soil:vermiculite:humus mixture (1:1:1, v/v/v) for 2 weeks and then stressed by a 4-week-long drought followed by 2-days of re-watering to observe the phenotypes; the seedlings were grown in an artificial climate chamber at 22 ± 1°C and 30% humidity under 16 h of daylight and 54 μM m–2 s–1. Plants grown in pots under normal conditions for 6 weeks were used as controls. The experiments were set up with 3 biological replications and 5 plants per treatment.
Measurements of Components and Expression Analysis of Genes Related to Drought Tolerance
The leaves were detached from the T3 and WT plants grown for 4 weeks under normal conditions and their water loss rates were analyzed as described by Li et al. (2019). The leaves of the T3 and WT plants grown for 2 weeks under normal conditions followed by 2 weeks of drought stress and for 4 weeks under normal conditions (control) were used to measure the proline and MDA contents and SOD activity with assay kits (Comin Biotechnology Co., Ltd., Suzhou, China). Their BR and reactive oxygen species (ROS) contents were determined using an indirect enzyme-linked immune sorbent assay (ELISA) kit (Comin Biotechnology Co., Ltd., Suzhou, China) and nitro blue tetrazolium (NBT) staining (Song et al., 2015), respectively. The leaves of these plants were also used to analyze the transcript levels of the genes involved in the BR biosynthesis and signaling pathways, proline biosynthesis and the ROS-scavenging system using qRT-PCR and Atactin (NM112764); the with specific primers used are listed in Supplementary Table S1. The relative expression levels in T3 were calibrated using those of WT. Three plants were evaluated per sample, and 3 biological replications were included.
Yeast Two-Hybrid (Y2H) Assays
The coding sequence of IbBT4 was fused to the yeast expression vector pGBKT7 (pBD) for transactivation activity assays (Zhou et al., 2019). The pBD-IbBT4 bait plasmid and prey plasmid library components were co-transformed into Y2H Gold yeast according to the MatchmakerTM Gold Yeast Two-Hybrid System User Manual protocol (Clontech). The coding sequences of IbBEE2 and AtBEE2 were cloned into a pGADT7 (pAD) vector, after which the bait and prey plasmids were transformed into Y2H Gold yeast. The pAD/pBD-IbBT4 and pBD/pAD-BEE2 were used as negative controls and the pBD-53/pAD was used as a positive control. The transformed yeasts were spread on SD/-Trp/-His/-Leu/-Ade/X-a-Gal plates to test protein-protein interactions at 30°C for 5–8 days. The specific primers used are shown in Supplementary Table S1.
Bimolecular Fluorescence Complementation (BiFC) Assays
The coding sequence of IbBT4 was inserted into pSPYNE-35S, which was then fused to the N-terminus of yellow fluorescence protein (YFP); moreover, the coding sequence of IbBEE2 or AtBEE2 was inserted into pSPYCE-35S, which was then fused to the C-terminus of YFP (Supplementary Table S1). The plasmids were introduced into A. tumefaciens strain GV3101, the cells of which were then infiltrated into N. benthamiana leaves. After 2 days, YFP signals were observed under a confocal laser scanning microscope (FV-1000, Olympus, Japan) equipped with an argon laser. The excitation wavelength was 488 nm and recording wavelength ranged from 505 to 555 nm.
Statistical Analysis
The data are presented as the means ± SEs of three biological replicates and were analyzed with Student’s t-tests (two-tailed analyses). Significance levels at P < 0.05 and P < 0.01 are indicated with ∗ and ∗∗, respectively.
Results
Sequence Analysis of IbBT4 and Its Promoter
The IbBT4 gene was cloned from the drought-tolerant sweetpotato line Xushu55-2 by RACE and submitted to GenBank (accession no. MT387197). The 1792 bp cDNA of IbBT4 contains a 323 bp 5′-UTR and a 350 bp 3′-UTR. Its CDS was 1119 bp and encoded for a polypeptide of 372 aa with a molecular weight of 43.13 kDa and a predicted pI of 9.46. This gene contains a BTB_POZ_BT domain, a ZnF_TAZ domain and a BACK domain (Figure 1A). The IbBT4 protein was most closely related to that of Ipomoea nil (XP_019195495.1, 86%) and shared high sequence identity with BT4 proteins from Capsicum baccatum (PHT56768.1, 68.38%), Solanum pennellii (XP_015064154.1, 67.52%), Nicotiana attenuata (XP_019266630.1, 68.41%), Glycine max (XP_003516617.1, 52.08%), and Arabidopsis thaliana (AT5G67480.2, 49.48%) (Figure 1B). The 2804 bp genomic DNA of IbBT4 consisted of 5 exons and 4 introns, similar to that of InBT4, CbBT4, SpBT4, NaBT4, and GmBT4, but not AtBT4 (Figure 1C). The 1910 bp promoter region of IbBT4 contains several stress-responsive cis-acting regulatory elements, including GARE- motif, O2-site, LTR, TC-rich repeats, TCA-element, MBS, ARE, and MBSI (Supplementary Figure S1). The presence of these cis-acting elements in the promoter regions suggests that the expression of IbBT4 might be induced by abiotic stresses.
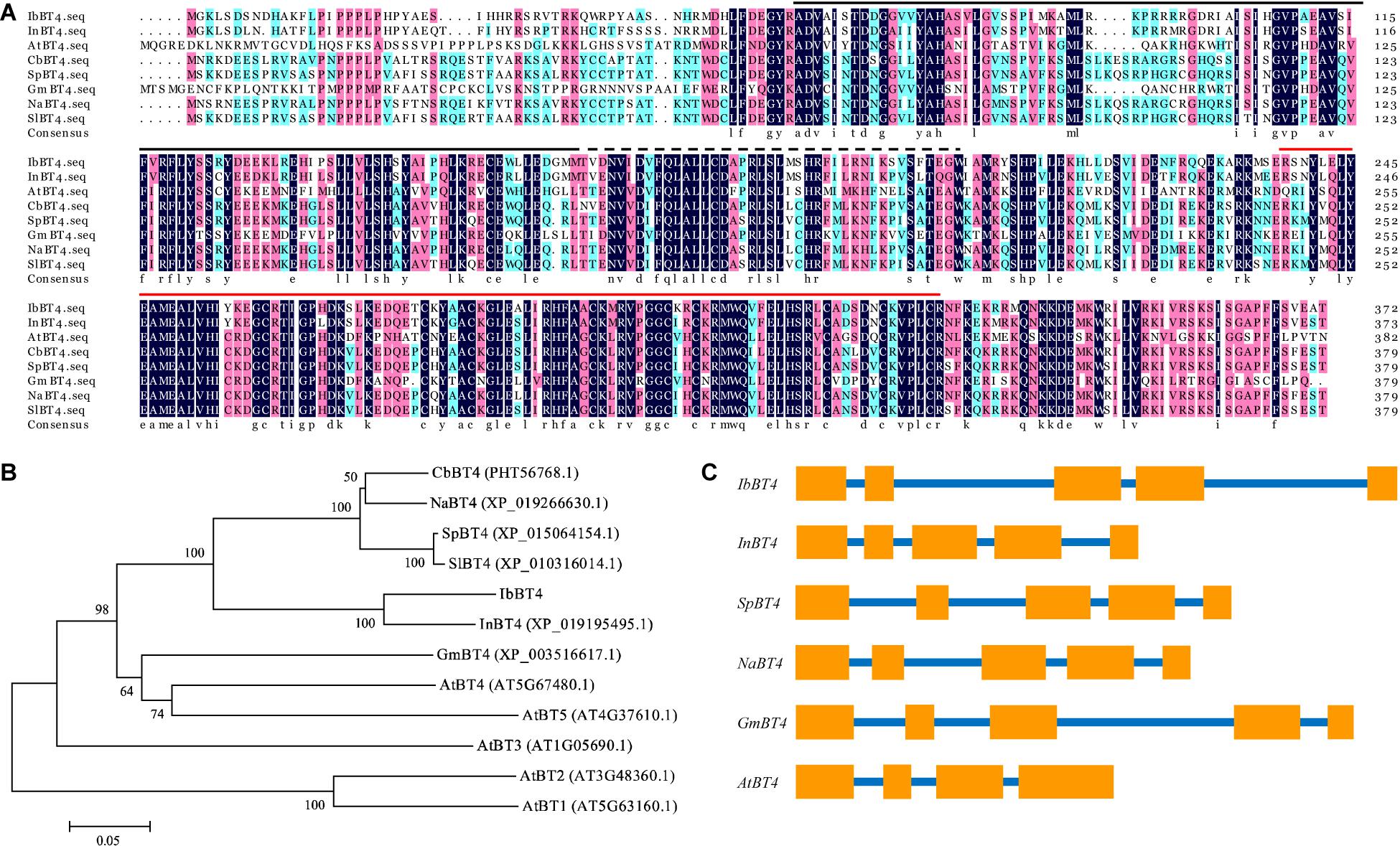
Figure 1. Sequence analysis of IbBT4. (A) Sequence alignment of IbBT4 and its homologs from other plant species. The conserved amino acids are indicated by a dark background.  , BTB_POZ_BT domain;
, BTB_POZ_BT domain;  , ZnF_TAZ domain;
, ZnF_TAZ domain;  , BACK domain. (B) Phylogenetic analysis of IbBT4 and BT proteins from other plant species. (C) Exon and intron constituents of BT4 genes. The exons are represented by boxes; the introns, by lines.
, BACK domain. (B) Phylogenetic analysis of IbBT4 and BT proteins from other plant species. (C) Exon and intron constituents of BT4 genes. The exons are represented by boxes; the introns, by lines.
Subcellular Location of IbBT4
To analyze the protein subcellular localization, the CDS of IbBT4 (no stop codon) was fused with GFP and transiently expressed in the leaf epidermal cells of N. benthamiana. The images from the leaf epidermal cells showed IbBT4-GFP fluorescence in the nuclei, while GFP fluorescence of the control was observed in the entire cell (Figure 2). Thus, IbBT4 was localized in the nucleus.
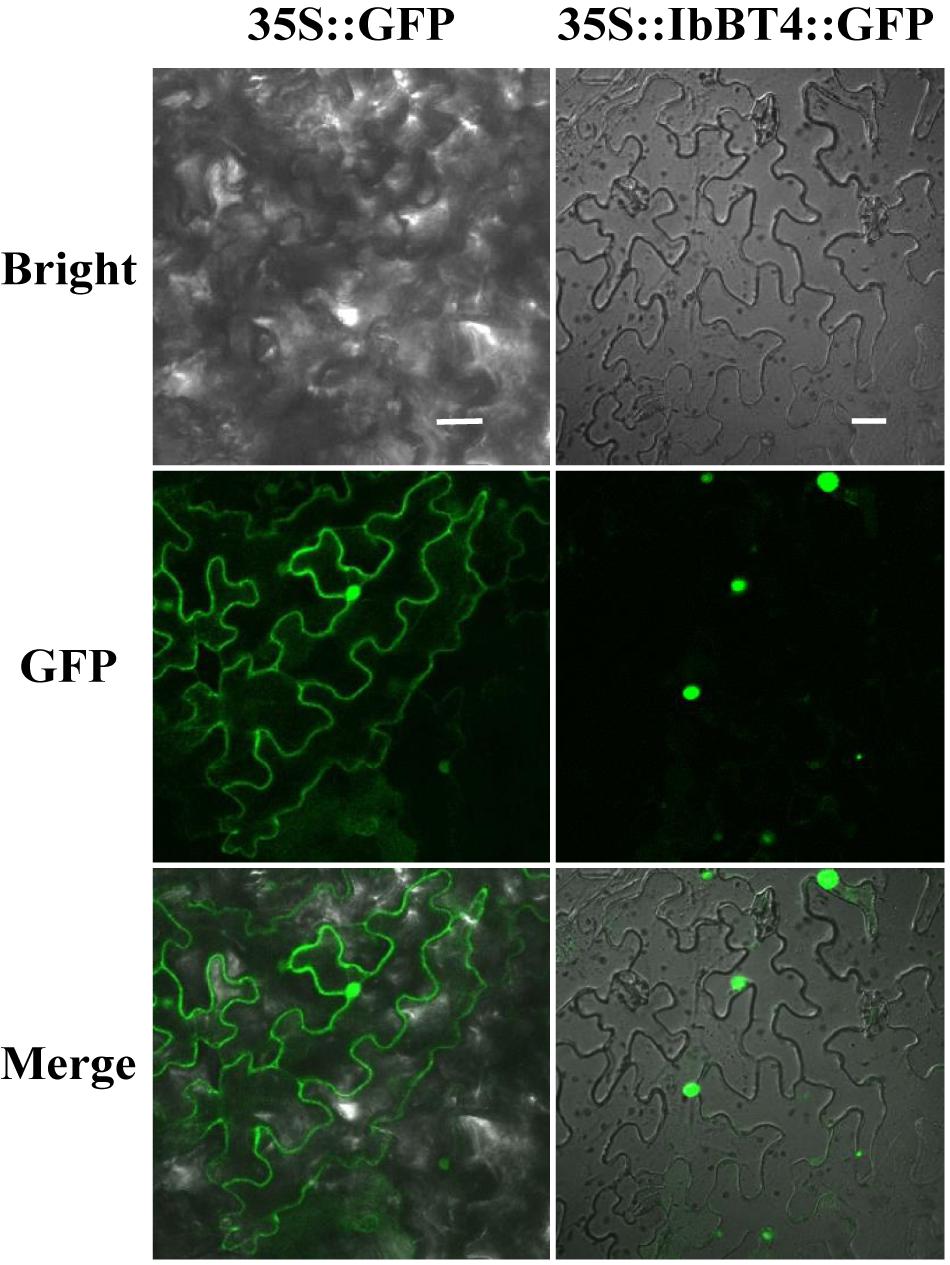
Figure 2. Subcellular localization of IbBT4 in tobacco leaf hypodermal cells. Confocal scanning microscopy images showing the localization of IbBT4-GFP fusion proteins to nuclei in the right column vs. GFP as the control in the left column. Bars = 20 μm.
Expression Profiles of IbBT4 in Sweetpotato
To investigate the potential working site of IbBT4 in sweetpotato, we analyzed its expression level in different tissues of Xushu55-2. The highest expression level was found in the leaves (Supplementary Figure S2). To further analyze its potential function in response to abiotic stresses, the expression of IbBT4 was checked using the 4-week-old in vitro-grown plants of Xushu55-2. It was found that IbBT4 was strongly induced by PEG6000, H2O2 and BR, and peaked (2.67-fold) at 6 h, (3.54-fold) at 6 h, and (130.18-fold) at 3 h (Figure 3). These results indicate that IbBT4 might be involved in drought, H2O2 and BR response pathways.
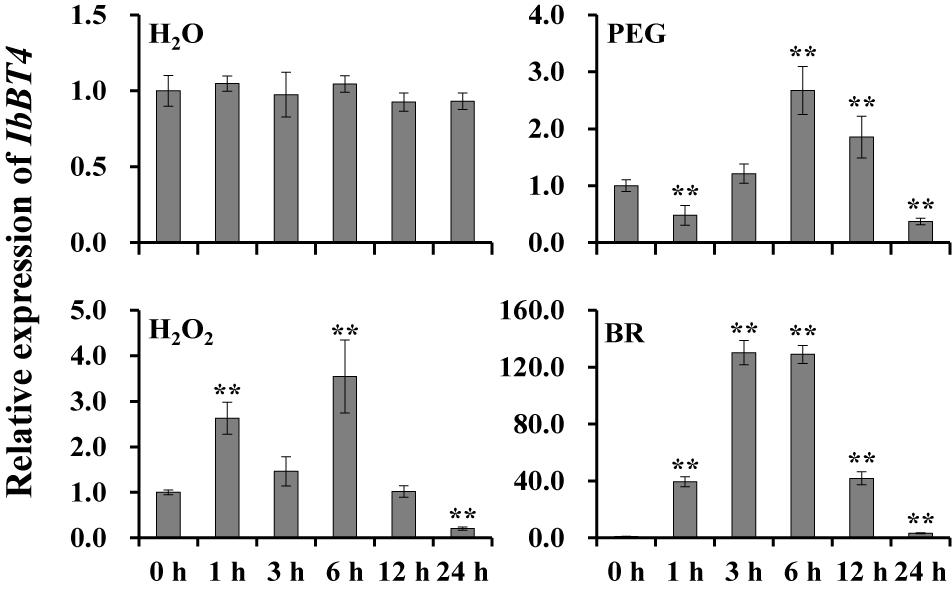
Figure 3. Expression analysis of IbBT4 in in vitro-grown Xushu55-2 plants after different time points (h) in response to H2O (control), 30% PEG6000, 100 μM H2O2 and 100 nM BR, respectively. The data are presented as the means ± SEs (n = 3). **Indicates a significant difference at P < 0.01 according to Student’s t-test.
Enhanced Drought Tolerance of Transgenic Arabidopsis
To identify the function of IbBT4, this gene was transferred to Arabidopsis. Seventeen putatively transgenic Arabidopsis plants were produced and 8 of them, named L1, L2, …, L8, were confirmed to be transgenic by PCR analysis. T3 lines were generated from these 8 transgenic plants by screening with hygromycin. qRT-PCR analysis indicated that 3 (L2, L3, and L7) of them had relatively high expression levels of IbBT4 (Supplementary Figure S3). These 3 transgenic lines were selected, and their drought tolerance was evaluated.
To investigate whether IbBT4 provides plant drought tolerance, transgenic Arabidopsis plants were evaluated in vitro and in vivo for their drought tolerance. No differences in seed germination rates between the transgenic lines and WT were observed under normal conditions, but all of the transgenic lines showed significantly higher germination rates than did the WT under 200 mM mannitol treatment (Figures 4A,B). Five-day-old transgenic and WT seedlings were used for drought stress tests. The seedlings were grown on 1/2 MS media with or without 200 mM mannitol for 2 weeks. The primary root length was measured as an indicator of the stress tolerance of plants. Similar growth between the transgenic lines and WT was observed on 1/2 MS media without mannitol, but the transgenic plants produced significantly longer primary roots than did the WT on 1/2 MS media with 200 mM mannitol (Figures 5A,B). To further evaluate the drought tolerance of the transgenic lines, one-week-old seedlings grown on 1/2 MS media were potted for 2 weeks and then stressed by a 4-week drought in an artificial climate chamber. Under normal conditions, the transgenic lines and WT exhibited similar phenotypes (Figure 6A), but lower water loss rates were found in the transgenic lines than in the WT (Figure 6B). Under drought stress, compared with WT, the transgenic lines showed better growth, increased survival rates, increased BR and proline contents and increased SOD activity but decreased MDA content and decreased ROS accumulation (Figures 6A–H). Taken together, these results demonstrated that overexpression of IbBT4 enhanced drought tolerance in Arabidopsis.
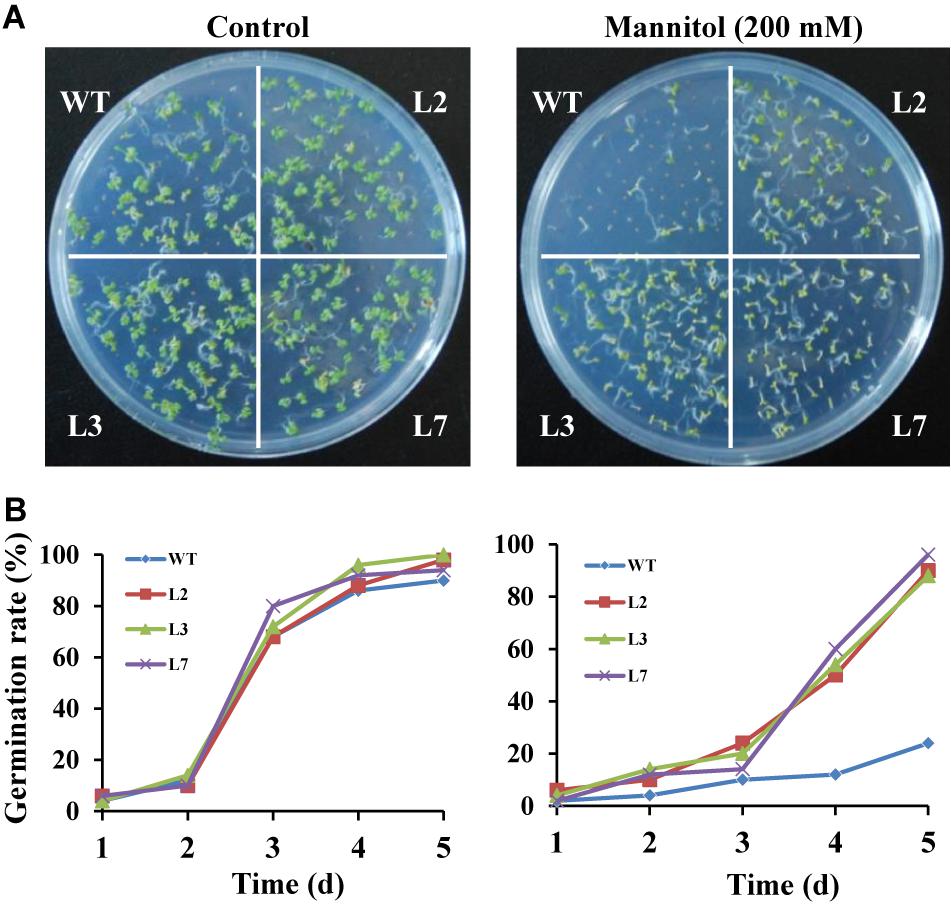
Figure 4. Germination assays of transgenic Arabidopsis and WT seeds sown for 5 days on 1/2 MS media without (control) or with 200 mM mannitol. (A) Growth vigor of the transgenic and WT seedlings. (B) Germination rates of the transgenic and WT seeds.
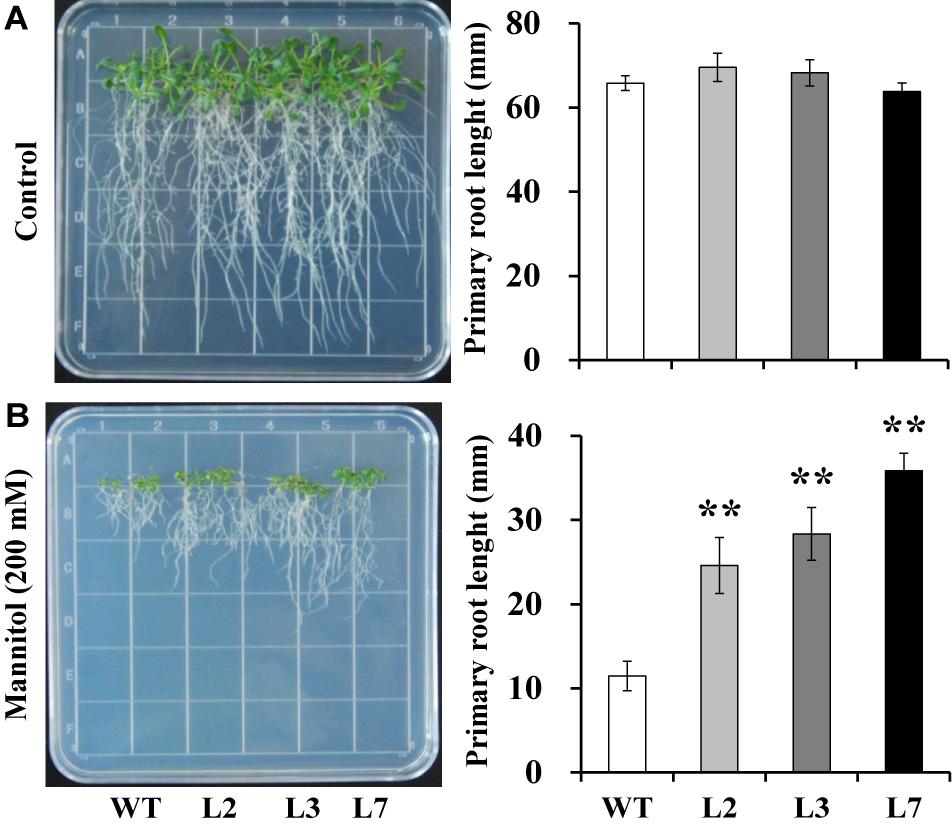
Figure 5. Responses of transgenic Arabidopsis and WT seedlings cultured for 2 weeks on 1/2 MS media without (control, A) or with 200 mM mannitol (B). The data are presented as the mean ± SEs (n = 3). **Indicates a significant difference from that of WT at P < 0.01 by Student’s t-test.
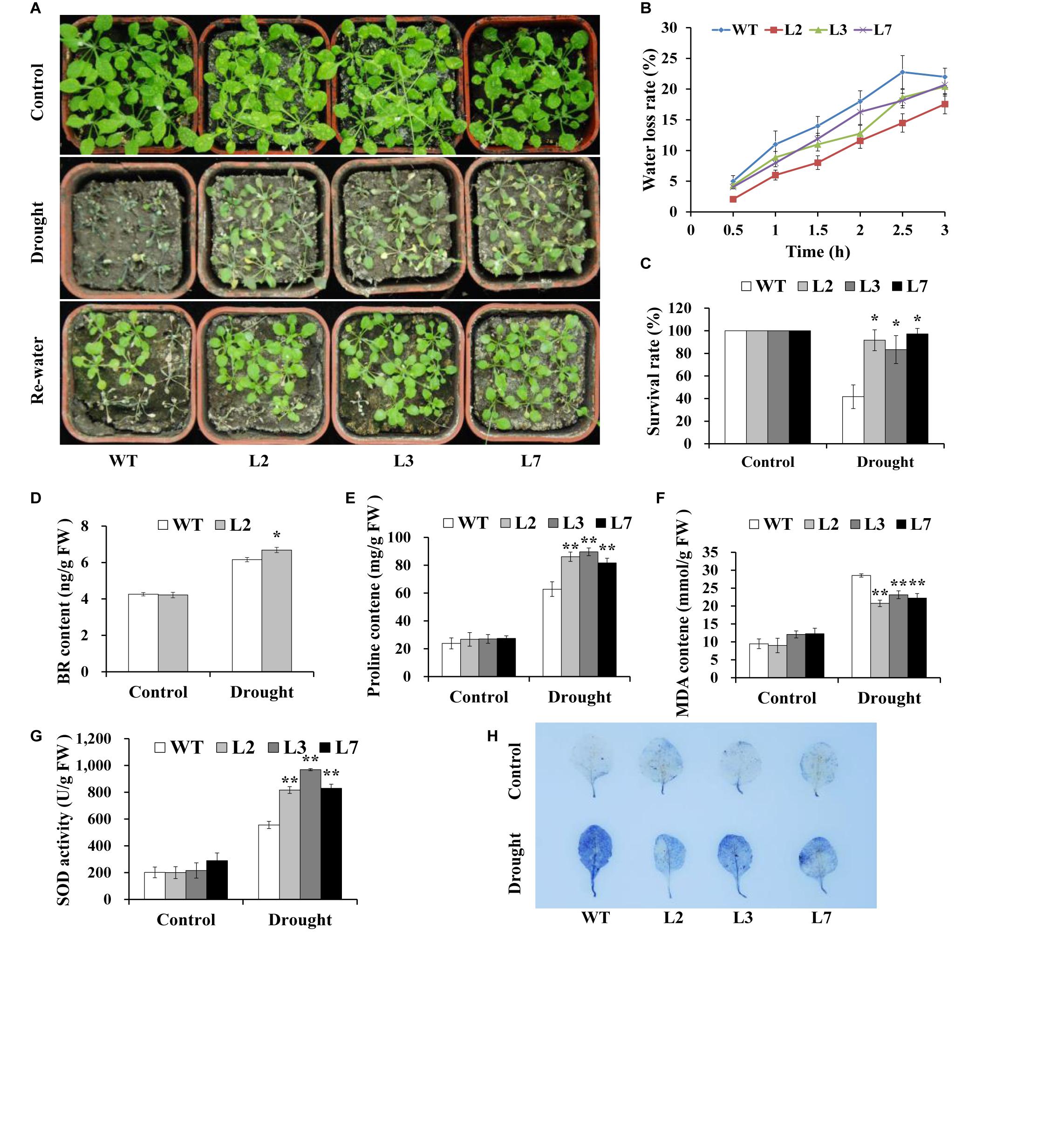
Figure 6. Responses of transgenic Arabidopsis and WT plants to drought stress. (A) Phenotypes of the transgenic plants vs. WT stressed by a 4-week-long drought period followed by 2 days of re-watering after 2 weeks of normal treatment. A 6-week-long normal treatment was used as a control. (B) Water loss rate of detached leaves from transgenic Arabidopsis and WT plants grown for 4 weeks under normal conditions. (C) Survival rate of transgenic and WT plants after 2 days of re-watering. (D–G) BR, proline and MDA contents and the SOD activity in the leaves of the transgenic and WT plants grown for 2 weeks under normal conditions followed by 2 weeks of drought stress and for 4 weeks under normal conditions (control), respectively. (H) NBT staining. * and ** indicate significant differences from those of WT at P < 0.05 and P < 0.01, respectively, according to Student’s t-test.
Underlying Mechanism by Which IbBT4 Provides Drought Tolerance
To explore the underlying mechanism of the involvement of IbBT4 in drought tolerance, the genes involved in the BR signaling pathway, proline biosynthesis and the ROS-scavenging system were analyzed by qRT-PCR using the leaves of the T3 and WT plants grown for 2 weeks under normal conditions followed by 2-week-long drought stress and for 4 weeks under normal conditions. Under drought stress, the genes involved in BR biosynthesis showed different expression patterns. AtROT3 and AtBR6ox1 were upregulated, and AtCPD was downregulated in the transgenic Arabidopsis lines compared with the WT; AtDWF4, AtDET2, AtCYP90D1, and AtBR6ox2 showed no differences (Figure 7). In the BR signaling pathway, AtBES1 was upregulated, AtBIN and AtBRI1 were downregulated, and AtBZR1 exhibited no differences under drought stress (Figure 7). Two key enzyme genes, AtP5CR and AtP5CS, which are involved in proline biosynthesis were upregulated under drought stress (Figure 7). The ROS-scavenging genes encoding superoxide dismutase (SOD), glutathione peroxidase (GPX), peroxidase (APX) and catalase (CAT), but not peroxidase (POD), were systematically upregulated under drought stress (Figure 7).
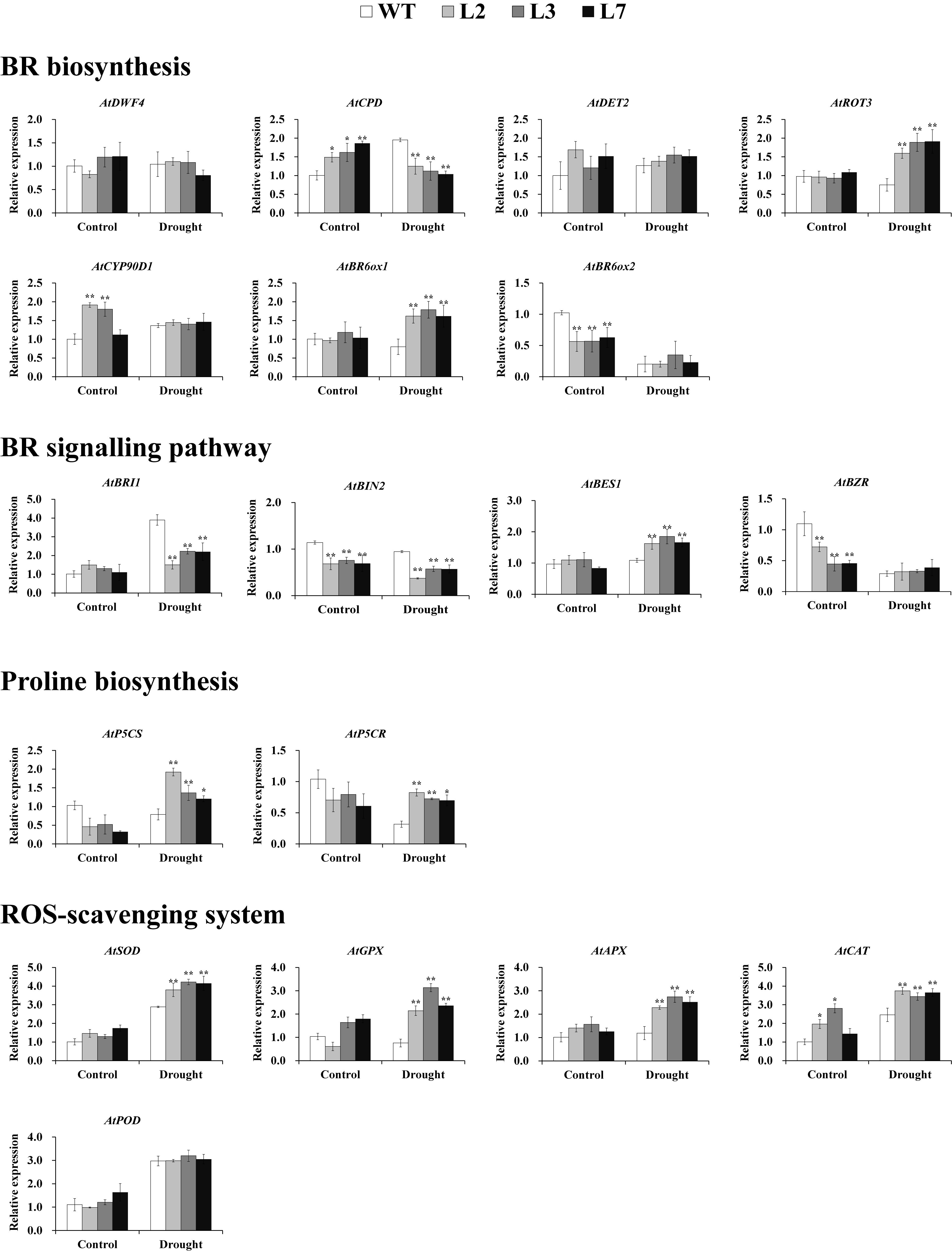
Figure 7. Transcript levels of relevant genes in the leaves of transgenic Arabidopsis and WT plants grown for 2 weeks under normal conditions followed by 2 weeks of drought stress and for 4 weeks under normal conditions (control), respectively. * and ** indicate significant differences from those of WT at P < 0.05 and P < 0.01, respectively, according to Student’s t-test.
The yeast cells harboring pGAL4 (+) grew well and turned blue, but the cells carrying pBD-IbBT4 and pBD (−) failed to grow on media containing X-α-Gal (SD/-Trp/-His/X), indicating that IbBT4 had no transactivation activity (Supplementary Figure S4). To further explore the functional mechanism of IbBT4, we performed Y2H assays. The yeast cells carrying negative controls failed to grow on media containing X-α-Gal (SD/-Leu/-Trp/-His/-Ade), and the positive control as well as yeast cells with pBD-BT4/pBEE2 were grew well and turned blue. The results showed that IbBT4 interacted with IbBEE2/AtBEE2 in cells of the yeast strain Y2H Gold (Figure 8A). The BiFC assay also showed that IbBT4 interacted with IbBEE2/AtBEE2 in the tobacco nucleus (Figure 8B). Friedrichsen et al. (2002) found that BEE2 positively regulated BR signaling in Arabidopsis. Therefore, it is inferred that IbBT4 might enhance drought tolerance by interacting with AtBEE2 in transgenic Arabidopsis.
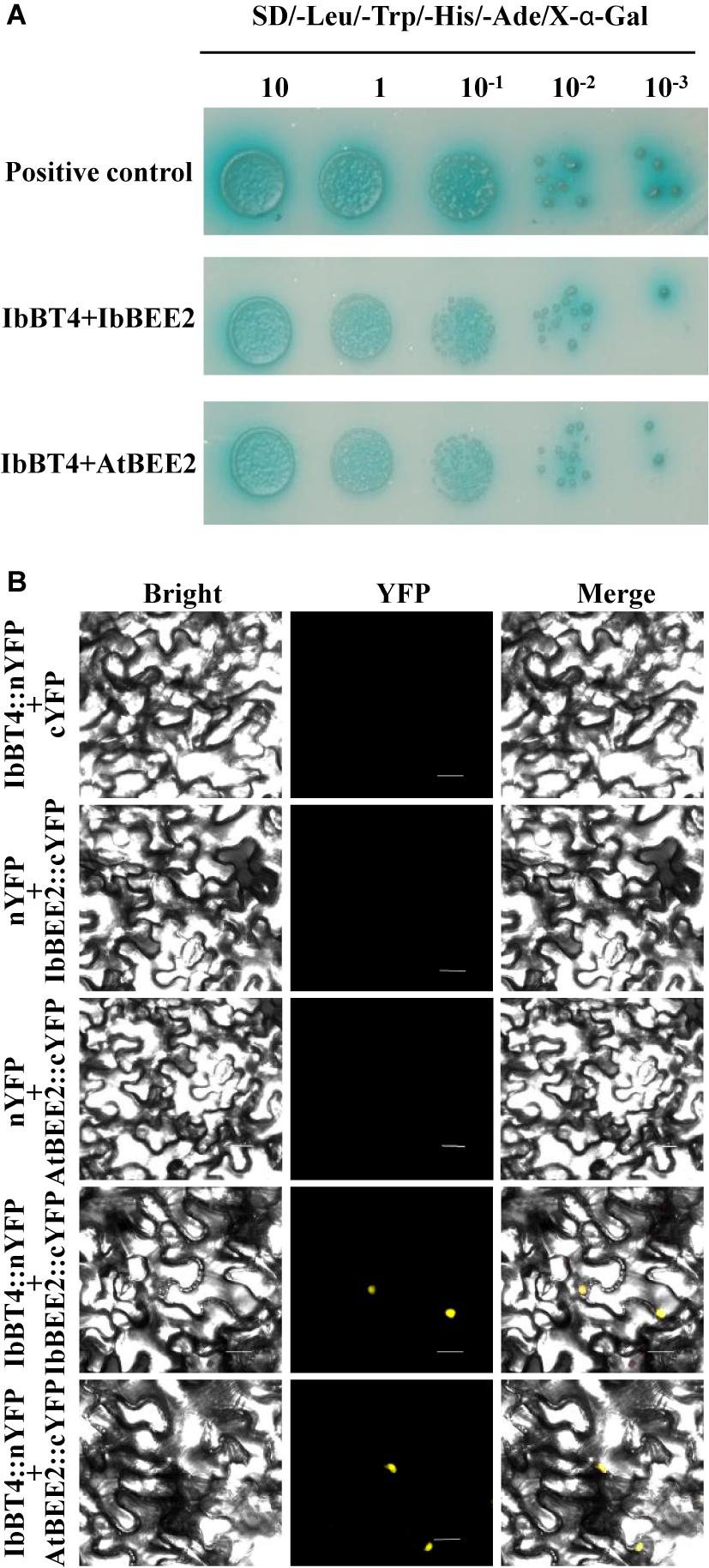
Figure 8. IbBT4 interacts with BEE2 in the nucleus. (A) Y2H assay revealing that IbBT4 interacts with BEE2 in Y2HGold cells. (B) BiFC assay showing that IbBT4 interacts with BEE2 in the tobacco nucleus. The yellow fluorescent protein (YFP) signals were predominantly localized in the nucleus.
Discussion
IbBT4 Enhances Drought Tolerance
The BTs have been reported to regulate plant development and pathogen defense (Du and Poovaiah, 2004; Ren et al., 2007; Robert et al., 2009; Sato et al., 2017; Zheng et al., 2019). However, their roles in the regulation of responses to abiotic stresses are still unclear. In our study, we cloned the IbBT4 gene from sweetpotato and found that the IbBT4 protein shared the highest identity with that of AtBT4 from Arabidopsis (Figure 1B). In Arabidopsis, BT1 and BT2 are localized in the nucleus and the cytoplasm, whereas BT3, BT4 and BT5 were determined to be as cytosolic proteins (Robert et al., 2009). This study showed that IbBT4 was localized in the nucleus (Figure 2). Thus, it is possible that the function of IbBT4 is functionally homologous to that of AtBT1 and AtBT2. The expression of IbBT4 was induced by PEG6000, H2O2 and BR (Figure 3) and compared with the WT plants, the IbBT4-overexpressing Arabidopsis plants displayed significantly enhanced drought tolerance (Figures 4–6). These results suggest that IbBT4 positively regulates plant drought tolerance.
IbBT4 Positively Regulates the BR Signaling Pathway
BRs regulate multiple aspects of plant growth and adaptations to cope with abiotic stresses, such as drought, cold, heat, and salinity stress (Zhu et al., 2013; Singh and Savaldi-Goldstein, 2015; Nolan et al., 2017; Planas-Riverola et al., 2019). The Arabidopsis BR biosynthetic gene AtDWARF4 conferred drought tolerance to Brassica napus (Sahni et al., 2016). Morevoer, the Arabidopsis BR-deficient mutant det2-9 accumulates relatively high levels of O2– in the roots (Lv et al., 2018). In the BR signaling pathway, AtBRI1 positively regulates Arabidopsis responses to cold and drought stresses (Kim et al., 2010; Ye et al., 2017). As a repressor of BR signaling, BIN2 modulates the degradation of BES1 (Nolan et al., 2017). Gain-of-function Arabidopsis BR mutants (bes1-D) showed decreased drought tolerance (Ye et al., 2017), and overexpression of TaBZR2 led to enhanced drought tolerance in wheat (Cui et al., 2019).
In our study, an increased BR level was detected in the IbBT4-overexpressing Arabidopsis plants under drought stress (Figure 6D). Overexpression of IbBT4 also led to upregulation of AtROT3, AtBR6ox1 and AtBES1 and downregulation of AtBIN2 under drought stress (Figure 7). These results indicated that IbBT4 positively regulates the BR signaling pathway in transgenic Arabidopsis (Figure 9). Furthermore, we found that IbBT4 interacts with BEE2 through Y2H and BiFC assays (Figure 8). Friedrichsen et al. (2002) identified three closely related basic helix-loop-helix (bHLH) transcription factors, BEE1, BEE2, and BEE3, as products of early response genes required for the full BR response and suggested that they are functionally redundant positive regulators of BR signaling in Arabidopsis. Similarly, Zhao et al. (2016) showed that MdBT1 and MdBT2 from apple interact with MdbHLH104, and MdBT2 regulates MdbHLH104 degradation via ubiquitination and the 26S proteasome pathway, thereby controlling the activity of plasma membrane H (+)-ATPases and the acquisition of iron. Therefore, it is thought that IbBT4 might regulate the BR signaling pathway by interacting with BEE2, which results in enhanced drought tolerance in transgenic Arabidopsis.
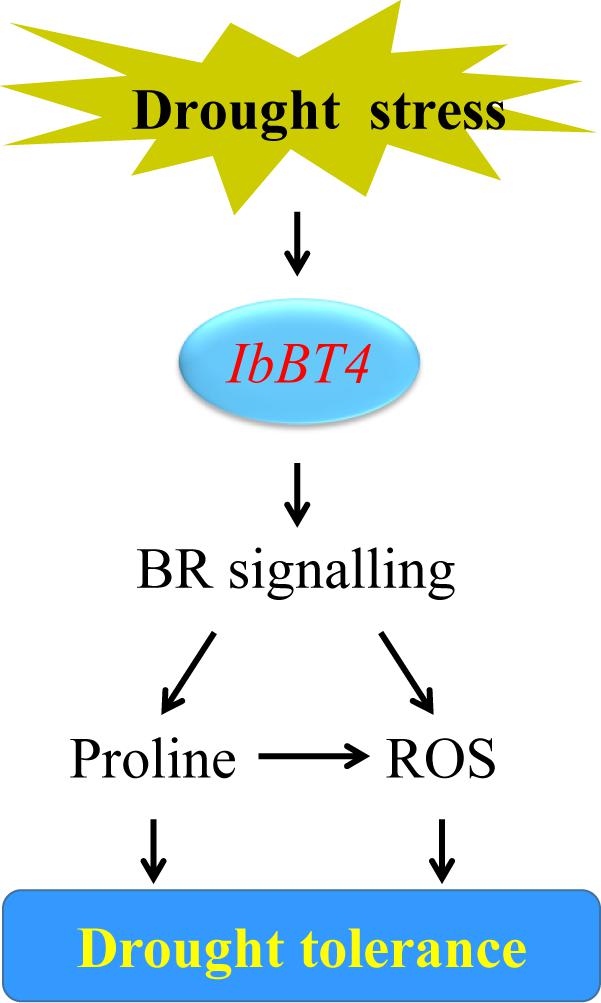
Figure 9. Diagram showing a proposed model for the regulation of IbBT4 in the drought tolerance of transgenic Arabidopsis.
The ABA signaling pathway plays important roles in plant responses to drought stress. Overexpression of IbARF5 and IbbZIP1 from sweetpotato enhanced drought tolerance in transgenic Arabidopsis through the ABA signaling pathway (Kang et al., 2018, 2019). It has also been reported that H2O2 plays a role in upstream signaling molecules of the ABA signaling pathway (Leon et al., 2014; Saxena et al., 2016; Phillips and Ludidi, 2017). We also investigated the function of IbBT4 in the ABA signaling pathway and found that the expression of IbBT4 was induced by ABA and H2O2 (Figure 3 and Supplementary Figure S5). However, no difference in seed germination or plant growth on MS media supplemented with ABA was observed between transgenic Arabidopsis and WT. Under drought stress, there was no difference in the content of ABA or transcript levels of the ABA biosynthesis related 9-cis-epoxycarotenoid dioxygenase (NCED) and ABA DEFICIENT 1 (ABA1) genes in the leaves of the transgenic Arabidopsis and WT plants (Supplementary Figure S5). These results suggest that IbBT4 might not be involved in ABA signaling pathway in transgenic Arabidopsis.
IbBT4 Increases Proline Accumulation and Activates the ROS-Scavenging System
It has been shown that BRs mediate adaptation to abiotic stresses by activating antioxidant machinery, promoting the production of osmoprotectants and fine-tuning stress-responsive transcript machinery (Planas-Riverola et al., 2019). Increases in SOD, CAT, APX, and POD of plants treated with BR indicate that BR act on the antioxidant system and reduce ROS levels, which increase tolerance to the water deficit in cowpea plants (Lima and Lobato, 2017). TaBZR2 and TaGST1 modulate drought tolerance in wheat by mediating the crosstalk between BR and ROS-scavenging system (Cui et al., 2019). Overexpression of BRL3, a vascular-enriched member of the BR receptor family, triggered proline accumulation and conferred drought stress tolerance to Arabidopsis (Fàbregas et al., 2018). Proline is essential for plants responses to abiotic stresses, and proline accumulation can protect plants against ROS damage by altering the activity of antioxidant enzymes (Alia et al., 2001; Khedr et al., 2003; Ozden et al., 2009; Lehmann et al., 2010; Carvalho et al., 2013; Liu et al., 2014). The ROS-scavenging system can protect the structure and function of biomolecules by detoxifying ROS to reduce oxidative damage in plant cells (Melchiorre et al., 2009; Gill and Tuteja, 2010; Liu et al., 2014; Wang et al., 2017).
In this study, we found that the proline content increased and that its key biosynthesis enzyme-encoding genes AtP5CR and AtP5CS were upregulated (Figures 6E, 7). In addition, in transgenic Arabidopsis under drought stress, the SOD activity increased; the ROS-scavenging system genes, including AtSOD, AtGPX, AtAPX, and AtCAT, were upregulated; and ROS levels decreased (Figures 6G, 7). These results suggest that overexpression of IbBT4 increases proline accumulation and activates the ROS-scavenging system, which leads to enhanced drought tolerance in transgenic Arabidopsis (Figure 9).
Conclusion
This study revealed that IbBT4 positively regulates plant drought tolerance. Under drought stress, IbBT4 overexpression in Arabidopsis significantly enhanced drought tolerance; increased BR and proline contents and SOD activity; decreased ROS and MDA levels; and upregulated genes involved in the BR signaling pathway, proline biosynthesis and the ROS-scavenging system. These results suggest that the IbBT4 gene provides drought tolerance by enhancing both the BR signaling pathway and proline biosynthesis and further activating the ROS-scavenging system in transgenic Arabidopsis.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
QL and YZ conceived and designed the experiments. YZ and HZu performed the experiments. YZ, HZa, SH, and SG analysed the data. QL, NZ, SX, and ZW contributed to the reagents, materials, and analysis tools. QL and YZ wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key R&D Program of China (2019YFD1001303/2019YFD1001300) and the China Agriculture Research System (CARS-10, Sweetpotato).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00877/full#supplementary-material
FIGURE S1 | Promoter of IbBT4 showing different cis-acting regulatory elements associated with abiotic stress responses.
FIGURE S2 | Expression analysis of IbBT4 indifferent tissues of Xushu55-2. The data are presented as the means ± SEs (n = 3). The different capital letters indicate a significant difference at P < 0.01 according to Student’s t-test. L: Leaf; S: Stem; HR: Hairy root; FR: Fibrous root; SR: Storage root.
FIGURE S3 | Expression analysis of IbBT4 intransgenic Arabidopsis plants. Atactin was used as an internal control. The data are presented as the means ± SEs (n = 3). ∗∗ Indicates a significant difference from L1 at P < 0.01 according to Student’s t-test.
FIGURE S4 | Transactivation activity assay of IbBT4 inyeast. (A) Transformed yeast cells harbouring different expression vectors were drawn onto SD/-Trp media. pBD (−) and pGAL4 (+) were used as negative and positive controls, respectively. (B) Transformed yeast cells harbouring different expression vectors were drawn onto SD/-Trp/-His media supplemented with X-α-Gal. pBD (−) and pGAL4 (+) were used as negative and positive controls, respectively.
FIGURE S5 | Analysis of the function of IbBT4 inthe ABA signalling pathway. (A) Expression analysis of IbBT4 in in vitro-grown Xushu55-2 plants after different time points (h) in response to 100 μM ABA. (B) Responses of transgenic Arabidopsis and WT seeds sown on 1/2 MS media with 0, 0.1, 0.2 and 0.3 μM ABA for 1 week. (C) Responses of transgenic Arabidopsis and WT seedlings cultured for 2 weeks on 1/2 MS media supplemented with 1 μM ABA. (D) ABA content in the leaves of transgenic Arabidopsis and WT plants grown for 2 weeks under normal conditions followed by 2 weeks of drought stress and for 4 weeks under normal conditions (control), respectively. (E–F) Transcript levels of ABA-related genes in the leaves of transgenic Arabidopsis and WT plants grown for 2 weeks under normal conditions followed by 2 weeks of drought stress and for 4 weeks under normal conditions (control), respectively. The data are presented as the means ± SEs (n = 3). ∗∗ Indicates a significant difference at P < 0.01 according to Student’s t-test.
TABLE S1 | Primers used in this study.
Footnotes
References
Alia, Mohanty, P., and Matysik, J. (2001). Effect of proline on the production of singlet oxygen. Amino Acids 21, 195–200. doi: 10.1007/s007260170026
An, J. P., Zhang, X. W., Bi, S. Q., You, C. X., Wang, X. F., and Hao, Y. J. (2019). MdbHLH93, an apple activator regulating leaf senescence, is regulated by ABA and MdBT2 in antagonistic ways. New Phytol. 222, 735–751. doi: 10.1111/nph.15628
Boyer, J. S., Byrne, P., Cassman, K. G., Cooper, M., Delmer, D., Greene, T., et al. (2013). The US drought of 2012 in perspective: a call to action. Glob. Food Secur. Agr. 2, 139–143. doi: 10.1016/j.gfs.2013.08.002
Carvalho, D. K., Campos, M. K. F. D., Domingues, D. S., Pereira, L. F. P., and Vieira, L. G. E. (2013). The accumulation of endogenous proline induces changes in gene expression of several antioxidant enzymes in leaves of transgenic Swingle citrumelo. Mol. Biol. Rep. 40, 3269–3279. doi: 10.1007/s11033-012-2402-5
Chiogna, G., Skrobanek, P., Narany, T. S., Ludwig, R., and Stumpp, C. (2018). Effects of the 2017 drought on isotopic and geochemical gradients in the Adige catchment. Italy. Sci. Total Environ. 645, 924–936. doi: 10.1016/j.scitotenv.2018.07.176
Cui, X. Y., Gao, Y., Gou, J., Yu, T. F., Zheng, W. J., Lui, Y. W., et al. (2019). BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 180, 605–620. doi: 10.1104/pp.19.00100
Dong, Y. P., Fan, G. Q., Zhao, Z. L., and Deng, M. J. (2014). Compatible solute, transporter protein, transcription factor, and hormone-related gene expression provides an indicator of drought stress in Paulownia fortunei. Funct. Integr. Genomic. 14, 479–491. doi: 10.1007/s10142-014-0373-4
Du, L. Q., and Poovaiah, B. W. (2004). A novel family of Ca2+/calmodulin-binding proteins involved in transcriptional regulation: interaction with fsh/Ring3 class transcription activators. Plant Mol. Biol. 54, 549–569. doi: 10.1023/B:PLAN.0000038269.98972.bb
Fàbregas, N., Lozano-Elena, F., Blasco-Escamez, D., Tohge, T., Martinez-Andujar, C., Albacete, A., et al. (2018). Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 9:4680. doi: 10.1038/s41467-018-06861-3
Friedrichsen, D. M., Nemhauser, J., Muramitsu, T., Maloof, J. N., Alonso, J., Ecker, J. R., et al. (2002). Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162, 1445–1456. doi: 10.1023/A:1021280325474
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Hao, C. C., Jia, J., Chen, Z., Xing, J. H., Weng, Q. Y., Wang, F. R., et al. (2013). Functional analysis of BT4 of Arabidopsis thaliana in resistance against Botrytis cinerea. Austr. Plant Pathol. 42, 393–401. doi: 10.1007/s13313-013-0202-6
Jin, Y., Zheng, H. Y., Jin, X. J., Zhang, Y. X., and Ren, C. Y. (2016). Effect of ABA, SA and JA on soybean growth under drought stress and re-watering. Soybean Sci. s565:1. doi: 10.11861/j.issn.1000-9841
Kang, C., He, S. Z., Zhai, H., Li, R. J., Zhao, N., and Liu, Q. C. (2018). A sweetpotato auxin response factor gene (IbARF5) is involved in carotenoid biosynthesis and salt and drought tolerance in transgenic Arabidopsis. Front. Plant Sci. 9:1307. doi: 10.3389/fpls.2018.01307
Kang, C., Zhai, H., He, S. Z., Zhao, N., and Liu, Q. C. (2019). A novel sweetpotato bZIP transcription factor gene, IbbZIP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Cell Rep. 38, 1373–1382. doi: 10.1007/s00299-019-02441-x
Khedr, A. H. A., Abbas, M. A., Wahid, A. A. A., Quick, W. P., and Abogadallah, G. M. (2003). Proline induces the expression of salt stress responsive proteins and may improve the adaptation of Pancratium maritimum L. to salt stress. J. Exp. Bot. 54, 2553–2562. doi: 10.1093/jxb/erg277
Kim, G. T., Tsukaya, H., Saito, Y., and Uchimiya, H. (1999). Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96, 9433–9437. doi: 10.1073/pnas.96.16.9433
Kim, S. Y., Kim, B. H., Lim, C. J., Lim, C. O., and Nam, K. H. (2010). Constitutive activation of stress-inducible genes in a brassinosteroid-insensitive 1 (bri1) mutant results in higher tolerance to cold. Physiol. Plant. 138, 191–204. doi: 10.1111/j.1399-3054.2009.01304.x
Kinoshita, T., Cano-Delgado, A., Seto, H., Hiranuma, S., Fujioka, S., Yoshida, S., et al. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433, 167–171. doi: 10.1038/nature03227
Lehmann, S., Funck, D., Szabados, L., and Rentsch, D. (2010). Proline metabolism and transport in plant development. Amino Acids 39, 949–962. doi: 10.1007/s00726-010-0525-3
Leon, J., Castillo, M. C., Coego, A., Lozano-Juste, J., and Mir, R. (2014). Diverse functional interactions between nitric oxide and abscisic acid in plant development and responses to stress. J. Exp. Bot. 65, 907–921. doi: 10.1093/jxb/ert454
Li, J., and Nam, K. H. (2002). Regulation of brassinosteroid signalling by a GSK3/SHAGGY-like kinase. Science 295, 1299–1301. doi: 10.1126/science.1065769
Li, Y., Zhang, H., Zhang, Q., Liu, Q. C., Zhai, H., Zhao, N., et al. (2019). An AP2/ERF gene, IbRAP2-12, from sweetpotato is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Sci. 281, 19–30. doi: 10.1016/j.plantsci.2019.01.009
Lima, J. V., and Lobato, A. K. S. (2017). Brassinosteroids improve photosystem II efficiency, gas exchange, antioxidant enzymes and growth of cowpea plants exposed to water deficit. Physiol. Mol. Biol. Plants 23, 59–72. doi: 10.1007/s12298-016-0410-y
Liu, D. G., He, S. Z., Zhai, H., Wang, L. J., Zhao, Y., Wang, B., et al. (2014). Overexpression of IbP5CR enhances salt tolerance in transgenic sweetpotato. Plant Cell Tissue Organ. Cult. 117, 1–16. doi: 10.1007/s11240-013-0415-y
Lv, B., Tian, H., Zhang, F., Liu, J., Lu, S., Bai, M., et al. (2018). Brassinosteroids regulate root growth by controlling reactive oxygen species homeostasis and dual effect on ethylene synthesis in Arabidopsis. PLoS Genet. 14:e1007144. doi: 10.1371/journal.pgen.1007144
Mandadi, K. K., Misra, A., Ren, S., and McKnight, T. D. (2009). BT2, a BTB protein, mediates multiple responses to nutrients, stresses, and hormones in Arabidopsis. Plant Physiol. 150, 1930–1939. doi: 10.1104/pp.109.139220
Melchiorre, M., Robert, G., Trippi, V., Racca, R., and Lascano, H. R. (2009). Superoxide dismutase and glutathione reductase overexpression in wheat protoplast: photooxidative stress tolerance and changes in cellular redox state. Plant Growth Regul. 57, 57–68. doi: 10.1007/s10725-008-9322-3
Misra, A., McKnight, T. D., and Mandadi, K. K. (2018). Bromodomain proteins GTE9 and GTE11 are essential for specific BT2-mediated sugar and ABA responses in Arabidopsis thaliana. Plant Mol. Biol. 96, 393–402. doi: 10.1007/s11103-018-0704-2
Nolan, T., Chen, J., and Yin, Y. (2017). Cross-talk of brassinosteroid signaling in controlling growth and stress responses. Biochem. J. 474, 2641–2661. doi: 10.1042/BCJ20160633
Nomura, T., Sato, T., Bishop, G. J., Kamiya, Y., Takatsuto, S., and Yokota, T. (2001). Accumulation of 6-deoxocathasterone and 6-deoxocastasterone in Arabidopsis, pea and tomato is suggestive of common rate-limiting steps in brassinosteroid biosynthesis. Phytochemistry 57, 171–178. doi: 10.1016/s0031-9422(00)00440-4
Ohnishi, T., Godza, B., Watanabe, B., Fujioka, S., Hategan, L., Ide, K., et al. (2012). CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J. Bio. Chem. 287, 31551–31560. doi: 10.1074/jbc.M112.392720
Ohnishi, T., Szatmari, A. M., Watanabe, B., Fujita, S., Bancos, S., Koncz, C., et al. (2006a). C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18, 3275–3288. doi: 10.1105/tpc.106.045443
Ohnishi, T., Watanabe, B., Sakata, K., and Mizutani, M. (2006b). CYP724B2 and CYP90B3 function in the early C-22 hydroxylation steps of brassinosteroid biosynthetic pathway in tomato. Biosci. Biotechnol. Biochem. 70, 2071–2080. doi: 10.1271/bbb.60034
Ozden, M., Demirel, U., and Kahraman, A. (2009). Effects of proline on antioxidant system in leaves of grapevine (Vitis vinifera L.) exposed to oxidative stress by H2O2. Sci. Hortic. 119, 163–168. doi: 10.1016/j.scienta.2008.07.031
Phillips, K., and Ludidi, N. (2017). Drought and exogenous abscisic acid alter hydrogen peroxide accumulation and differentially regulate the expression of two maize RD22-like genes. Sci. Rep. 7:8821. doi: 10.1038/s41598-017-08976-x
Planas-Riverola, A., Gupta, A., Betegon-Putze, I., Bosch, N., Ibanes, M., and Cano-Delgado, A. I. (2019). Brassinosteroid signaling in plant development and adaptation to stress. Development 146:dev151894. doi: 10.1242/dev.151894
Ren, S., Mandadi, K. K., Boedeker, A. L., Rathore, K. S., and McKnight, T. D. (2007). Regulation of telomerase in Arabidopsis by BT2, an apparent target of TELOMERASE ACTIVATOR1. Plant Cell 19, 23–31. doi: 10.1105/tpc.106.044321
Robert, H. S., Quint, A., Brand, D., Vivian-Smith, A., and Offringa, R. (2009). BTB and TAZ domain scaffold proteins perform a crucial function in Arabidopsis development. Plant J. 58, 109–121. doi: 10.1111/j.1365-313X.2008.03764.x
Sahni, S., Prasad, B. D., Liu, Q., Grbic, V., Sharpe, A., Singh, S. P., et al. (2016). Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci. Rep. 6:28298. doi: 10.1038/srep28298
Sato, T., Maekawa, S., Konishi, M., Yoshioka, N., Sasaki, Y., Maeda, H., et al. (2017). Direct transcriptional activation of BT genes by NLP transcription factors is a key component of the nitrate response in Arabidopsis. Biochem. Biophys. Res. Commun. 483, 380–386. doi: 10.1016/j.bbrc.2016.12.135
Saxena, I., Srikanth, S., and Chen, Z. (2016). Cross talk between H2O2 and interacting signal molecules under plant stress response. Front. Plant Sci. 7:570. doi: 10.3389/fpls.2016.00570
Shimada, Y., Fujioka, S., Miyauchi, N., Kushiro, M., Takatsuto, S., Nomura, T., et al. (2001). Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 126, 770–779. doi: 10.1104/pp.126.2.770
Singh, A. P., and Savaldi-Goldstein, S. (2015). Growth control: brassinosteroid activity gets context. J. Exp. Bot. 66, 1123–1132. doi: 10.1093/jxb/erv026
Song, X. X., Fang, J., Han, X. J., He, X. L., Liu, M. Y., Hu, J. J., et al. (2015). Overexpression of quinone reductasefrom Salix matsudana Koidz enhances salt tolerance in transgenic Arabidopsis thaliana. Gene 576, 520–527. doi: 10.1016/j.gene.2015.10.069
Vargas, L., Santa, B. A., Mota, F. J., Carvalho, T. G. D., Rojas, C. A., Vaneechoutte, D., et al. (2014). Drought tolerance conferred to sugarcane by association with Gluconacetobacter diazotrophicus: a transcriptomic view of hormone pathways. PLoS One 9:e114744. doi: 10.1371/journal.pone.0114744
Wang, L. Q., Li, Z., Lu, M. Z., and Wang, Y. C. (2017). ThNAC13, a NAC transcription factor from Tamarix hispida, confers salt and Osmotic stress tolerance to transgenic Tamarix and Arabidopsis. Front. Plant Sci. 8:635. doi: 10.3389/fpls.2017.00635
Wang, Z. Y., Nakano, T., Gendron, J., He, J. X., Chen, M., Vafedos, D., et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–515. doi: 10.1016/s1534-5807(02)00153-3
Yang, S. J., Vanderbeld, B., Wan, J. X., and Huang, Y. F. (2010). Narrowing down the targets: towards successful genetic engineering of drought-tolerant crop. Mol. Plant 3, 469–490. doi: 10.1093/mp/ssq016
Ye, H. X., Liu, S. Z., Tang, B. Y., Chen, J. N., Xie, Z. L., Nolan, T. M., et al. (2017). RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 8:14573. doi: 10.1038/ncomms14573
Yin, Y. H., Wang, Z. Y., Mora-Garcia, S., Li, J. M., Yoshida, S., Asami, T., et al. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. doi: 10.1016/s0092-8674(02)00721-3
Zhao, Q., Ren, Y. R., Wang, Q. J., Wang, X. F., You, C. X., Hao, Y.-J., et al. (2016). Ubiquitination-Related MdBT scaffold proteins target a bHLH transcription factor for iron homeostasis. Plant Physiol. 172, 1973–1988. doi: 10.1104/pp.16.01323
Zheng, X., Xing, J. H., Zhang, K., Pang, X., Zhao, Y. T., Wang, G. Y., et al. (2019). Ethylene response factor ERF11 activates BT4 transcription to regulate immunity to Pseudomonas syringae. Plant Physiol. 182, 1132–1151. doi: 10.1104/pp.18.01209
Zhou, Y. Y., Zhu, H., He, S. Z., Zhai, H., Zhao, N., Xing, S. H., et al. (2019). A novel sweetpotato transcription factor gene IbMYB116 enhances drought tolerance in transgenic Arabidopsis. Front. Plant Sci. 10:1025. doi: 10.3389/fpls.2019.01025
Zhu, H., Zhou, Y. Y., Zhai, H., He, S. Z., Zhao, N., and Liu, Q. C. (2018). Transcriptome profiling reveals insights into the molecular mechanism of drought tolerance in sweetpotato. J. Inter. Agri. 18, 9–23. doi: 10.1016/S2095-3119(18)61934-3
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Keywords: sweetpotato, IbBT4, drought tolerance, BR signaling pathway, Arabidopsis
Citation: Zhou Y, Zhai H, He S, Zhu H, Gao S, Xing S, Wei Z, Zhao N and Liu Q (2020) The Sweetpotato BTB-TAZ Protein Gene, IbBT4, Enhances Drought Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 11:877. doi: 10.3389/fpls.2020.00877
Received: 24 February 2020; Accepted: 28 May 2020;
Published: 23 June 2020.
Edited by:
Omar Borsani, Universidad de la República, UruguayReviewed by:
Sabina Vidal, Universidad de la República, UruguayYong Hwa Cheong, Sunchon National University, South Korea
Copyright © 2020 Zhou, Zhai, He, Zhu, Gao, Xing, Wei, Zhao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingchang Liu, bGl1cWNAY2F1LmVkdS5jbg==
 Yuanyuan Zhou1
Yuanyuan Zhou1 Hong Zhai
Hong Zhai Shaozhen He
Shaozhen He Hong Zhu
Hong Zhu Qingchang Liu
Qingchang Liu