- 1College of Agriculture, Jilin Agricultural University, Changchun, China
- 2Crop Science Post-doctoral Station, Jilin Agricultural University, Changchun, China
The ethylene response factor (ERF) plays a crucial role in plant innate immunity. However, the molecular function of ERF in response to Exserohilum turcicum (E. turcicum) remains unknown in maize. In this study, a novel ERF gene, designated as ZmERF105, was firstly isolated and characterized. The ZmERF105 protein contains an APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) domain and a conserved LSPLSPHP motif in its C-terminal region. ZmERF105 protein was exclusively localized to the nucleus. ZmERF105 expression responded to E. turcicum treatment. Yeast one-hybrid and transcription activity assays revealed that ZmERF105 is an activator of transcription and binds to GCC-box elements. Over-expression of ZmERF105 was shown to increase maize resistance against E. turcicum, and erf105 mutant lines displayed opposite phenotype. Moreover, the activities of superoxide dismutase (SOD) and peroxidase (POD) in the ZmERF105 over-expression lines were markedly higher than in the wild-type maize lines (WT) after infection with E. turcicum, and were compromised in the erf105 mutant lines. Simultaneously, ZmERF105 over-expression lines enhanced the expression of several pathogenesis-related (PR) genes, including ZmPR1a, ZmPR2, ZmPR5, ZmPR10.1, and ZmPR10.2 after infection with E. turcicum. In contrast, the expression of PR genes was reduced in erf105 mutant lines. Our work reveals that ZmERF105 as a novel player of the ERF network and positively regulates the maize resistance response to E. turcicum.
Introduction
Maize (Zea mays L.) is one of the most important food crops in the world. Its productivity is frequently hampered by pathogens and so improved disease resistance is an important goal in many breeding projects (Galiano-Carneiro and Miedaner, 2017; Weems and Bradley, 2018). Northern corn leaf blight (NCLB), caused by Exserohilum turcicum (E. turcicum), is a destructive disease of maize worldwide (Chang and Fan, 1986). It can reduce crop yields approximately 50%, severe infection even results in a total yield loss (Raymundo and Hooker, 1981; Perkins and Pedersen, 1987). Resistant cultivars are the most effective and economical way of controlling NCLB. Nine qualitative Ht genes which confer resistance to specific races of E. turcicum have already been identified in maize, including Ht1, Ht2, Ht3, HtN, Htm1, Htn1, HtP, ht4, and rt (Carson, 1995; Welz and Geiger, 2000; Hurni et al., 2015). Qualitative resistance often results in breakdown of disease resistance because of a change in the pathogen population; quantitative resistance breeding has become the primary method for NCLB control (Carson, 1995; Welz and Geiger, 2000). Thus, it is necessary to isolate resistance genes and analyze their funtions in order to improve maize resistance to E. turcicum.
Plants have evolved an array of defense mechanisms that enable them to protect themselves from pathogen infection (Boller and He, 2009; Cui H. T. et al., 2009; Bigeard et al., 2015; Birkenbihl et al., 2017). APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) are thought to be the major transcription factors (TFs) that regulate the plant defense system (Nakano et al., 2006; Li et al., 2011; Pieterse et al., 2012; Lai et al., 2013; Lu et al., 2013; Sherif et al., 2013). They were usually classified to three families, including AP2, RAV, and ERF family (Licausi et al., 2013). The ERF family is sometimes further divided into two major subfamilies, the ERF subfamily and the DREB subfamily (Sakuma et al., 2002). The ERFs contain a highly conserved AP2/ERF domain consisting of 57 or 59 amino acid residues, and the AP2/ERF domain binds to the cis-acting element AGCCGCC (GCC-box) presents in the promoters of pathogenesis-related (PR) genes (Ohme-Takagi and Shinshi, 1995; Zarei et al., 2011; Cheng et al., 2013; Franco-Zorrilla et al., 2014). The ERF gene was firstly isolated in tobacco (Ohme-Takagi and Shinshi, 1995), and successively identified in other plant species, such as wheat (Xu et al., 2007), tomato (Pan et al., 2010), soybean (Zhai et al., 2013) and rice (Zhang et al., 2013).
ERF genes acting as transcription activators or repressors involved in plant defense reactions (McGrath et al., 2005; Sherif et al., 2013; Huang et al., 2016; Li et al., 2018; Huang et al., 2019). Over-expression of AtERF1 activated the expression of PLANT DEFENSINS (PDF1.2) directly, and increased the plants resistance against a range of pathogens (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003). A T-DNA insertion mutant of AtERF14 increased susceptibility to infection by Fusarium oxysporum (F. oxysporum) (Oñate-Sánchez et al., 2007). AtERF96 positively regulated the Arabidopsis resistance to necrotrophic pathogen through enhancing the expression of PDF1.2a, PR-3, PR-4, and ORA59 (Catinot et al., 2015). The transcription activator, AtERF15 could positively regulate the Arabidopsis resistance against Pseudomonas syringae pv. tomato (Pst) DC3000 and Botrytis cinerea (B. cinerea) (Zhang et al., 2015). Although many ERF TFs identified so far are transcription activators, several ERFs were shown to act as negatively regulators of plant defense mechanisms (McGrath et al., 2005; Maruyama et al., 2013; Tian et al., 2015). Over-expression of AtERF4 was more susceptible to F. oxysporum, and knockout mutants of AtERF9 showed enhanced resistance to B. cinerea by suppressing the expression of the PDF1.2 (McGrath et al., 2005; Maruyama et al., 2013). StERF3, which contains an EAR motif, negatively regulates resistance to Phytophthora infestans (P. infestans) (Tian et al., 2015).
Previous reports have demonstrated that many ERF genes respond to various stimulus, such as ethylene (ET), salicylic acid (SA), and jasmonic acid (MeJA) (Grant and Jones, 2009; Cheng et al., 2013; Müller and Munné-Bosch, 2015; Yang et al., 2015; Zhang et al., 2017; Xie et al., 2019). Transcription of the TaERF1 is induced by ABA, ET, and SA (Xu et al., 2007). Expression of GmERF7 is induced by treatment with MeJA, ET, and ABA (Zhai et al., 2013). Transcription of the GmERF113 can be induced by ABA, ET, and SA (Zhao et al., 2017). These stimulus have been associated with resistance against different types of pathogens, such as (hemi) biotrophic pathogens are modulated via SA signaling pathway while necrotrophic pathogens are mediated through MeJA/ET signaling pathway (Ward et al., 1991; Farmer et al., 2003; Yang et al., 2005; Vlot et al., 2009; Verhage et al., 2010; Caarls et al., 2017). For example, AtERF1, AtERF5, AtERF6, and ORA59 are suggested to participate in JA/ET-regulated defense responses and increased plants resistance against B. cinerea by promoting the expression of AtPDF1.2 and ChiB (Lorenzo et al., 2003; Pre et al., 2008; Leon-Reyes et al., 2010; Zarei et al., 2011; Son et al., 2012; Moffat et al., 2012; Cheng et al., 2013; Van der Does et al., 2013; He et al., 2017; Kim et al., 2018). ERF11 acted as a regulator of the SA-mediated signaling pathway to enhance the Arabidopsis defense response against Pst DC3000 by directly regulating the expression of BT4 (Zheng et al., 2019).
Although the regulatory functions of the ERF genes have been widely explored in plants, the role of ERF genes in response to E. turcicum remain unknown. In a previous study, a novel ERF gene was significantly induced after infection with E. turcicum in the resistant inbred line “Mo17” than in the susceptible inbred line “Huobai”. In this work, we firstly isolated and characterized this ERF gene in maize, designated as ZmERF105 (GenBank No. NM001177195). ZmERF105 protein was exclusively localized to the nucleus. ZmERF105 acts as a transcriptional activator that binds to GCC-box elements. ZmERF105 expression responded to E. turcicum treatment. ZmERF105 positively regulated plant resistance against E. turcicum via regulating the expression of defense-related genes and the activities of antioxidant enzymes. These data suggested that ZmERF105 plays an important role in plant defense reactions, and may be useful in molecular breeding to improve the defensive capacity of maize against E. turcicum.
Materials and Methods
Plant Materials and Stress Treatments
E. turcicum and maize inbred lines were provided by Maize Breeding Team in Jilin Agricultural University. The seeds of the maize inbred line B73 were germinated in a greenhouse under a long-day photoperiod (16-h light, 8-h dark), and watered once every 3 days. Plant stress treatments were performed at the three-leaf stage. For hormone treatments, maize seedlings were sprayed with 0.1 mM MeJA, 0.1 mM ABA, or 0.5 mM SA. ET treatment was performed in sealed plexiglass chamber by dissolving 2 ml of 40% ethephon and 1 g of NaHCO3 in 200 ml of H2O.
E. turcicum (mixed races) was inoculated into the maize inbred line Mo17 (resistant inbred line) and Huobai (susceptible inbred line) according to the methods described by Wu et al. (2014), The plants were inoculated with two to three drops of conidial suspension at six-leaf stage, and the concentration of conidia was estimated to be about 1 × 105 spores ml−1. After inoculation, the plants were kept at 100% relative humidity to ensure spore germination. The leaves were collected at 6, 12, 24, 48, and 72 h after the treatment, and were frozen in liquid nitrogen for RNA extraction.
Cloning of ZmERF105 and Phylogenetic Analysis
The full-length cDNA sequence of ZmERF105 was amplified from the leaves of the maize inbred line B73 using reverse transcription PCR (RT-PCR). The amplified products were cloned into the pMD-18T vector (Takara) for sequencing (Sangon, Shanghai, China). The amino acid sequences of ZmERF105 and other AP2/ERF members were obtained from NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignment was analyzed using DNAMAN software, and the phylogenetic tree was generated with the neighbor-joining method using MEGA 5.0 software. The primers used for ZmERF105 cloning were provided in Supplementary Table S1.
Quantitative Real-Time PCR Analysis
Total RNA was extracted from maize leaves using Trizol reagent (Invitrogen, China) and reverse transcription was performed by using 1 µg of total RNA. Quantitative real-time PCR (qRT-PCR) was performed on a QuantStudio 3 instrument (Thermo, USA) using a SYBR Mixture system (TOYOBO, Japan). The relative expression levels were calculated by the 2−ΔΔCt method with the maize housekeeping gene ZmTub (GRMZM2G066191). The expression analysis was performed using three biological and three technical replicates, and statistically analyzed using Student’s t-test (*P < 0.05, **P < 0.01). The primers used for expression analysis were provided in Supplementary Table S1.
Maize Transformation and Assessment of Plant Disease Resistance
The coding sequence of ZmERF105 was ligated into the NcoI and PmlI sites of pCAMBIA3301 vector to construct the CaMV 35s: ZmERF105 plasmid, and the recombinant plasmid was introduced into Agrobacterium tumefaciens strain LBA4404 using the freeze-thaw method (Holsters et al., 1978). Germinating embryos of the maize inbred line H99 (susceptible inbred line) were used as explants transformed with the Agrobacterium-mediated method (Zhou et al., 2011). T2 transgenic maize lines were further confirmed using PCR and qRT-PCR. Primers used for the vector construction and detection were presented in Supplementary Table S1.
erf105 UFMu mutant lines (W22, mu2041934) were obtained from the Maize Genetics Cooperation Stock Center. The homozygous mutant lines were identified using PCR. The transcription levels were detected using qRT-PCR as described previously. Primers used for detection were presented in Supplementary Table S1.
To investigate the transgenic and mutant maize lines resistance against E. turcicum, artificial inoculation procedures were performed as described by Wu et al. (2014). The wild-type maize lines (WT) were used as the control. The living ear leaves of T2 transgenic lines and mutant lines were infected with E. turcicum. Disease symptoms on infected leaves were observed with a Nikon D7000 camera at 5 days post-inoculation (dpi). The relative lesion area was measured as described by Cui H. W. et al. (2009) using Photoshop CS3.
Subcellular Localization of ZmERF105
The full-length ZmERF105 sequence was cloned and ligated into the KpnI and XbaI sites of pCAMBIA1300 vector to construct the CaMV 35s: ZmERF105-GFP plasmid. After sequencing confirmation, CaMV 35s: ZmERF105-GFP plasmid and empty vector were transiently expressed in N. benthamiana (Liu et al., 2010). The GFP fluorescence signal was visualized and photographed with a laser scanning confocal microscope (Leica TCS SP5, Germany). The primers used for expression analysis were provided in Supplementary Table S1.
Yeast One-Hybrid Assay
The coding sequence of ZmERF105 was amplified and ligated into the NcoI and EcoRI sites of the pGADT7 vector. The specific DNA fragments, GCC (ATCCATAAGAGCCGCCACTAAAATAAGACCGATCAA) and mutated GCC (mGCC) (ATCCATAAGATCCTCCACTAAAATAAGACCGATCAA) were inserted into the pHIS2 vector. The plasmids were transformed into yeast strain Y187 and the transformants were selected on SD (-Trp/-Leu). Transformed colonies were subsequently grown on SD (-Trp/-His/Leu) medium with 100 mM 3-amino-1, 2, 4-triazole (3-AT), and were cultured at 28°C for 3 days. Primers used for the vector construction were presented in Supplementary Table S1.
Transcription Activity Assay
For transcription activity assay, the coding sequence of ZmERF105 was inserted into NcoI and PmlI sites of pCAMBIA3301 vector to construct the CaMV 35s: ZmERF105 effector plasmid. The reporter plasmid was constructed according to the method described by Dong et al. (2015). The combined reporter and effector plasmids were cotransformed into the Arabidopsis protoplasts according to a protocol described by Yoo et al. (2007). The relative GUS activity was determined as described by Chen et al. (2006). Primers used for the vector construction were presented in Supplementary Table S1.
Enzyme Activity Assay
The fresh leaves (approximately, 0.1 g) of seedlings were harvested and homogenized in 1 ml of 50 mM ice-cold phosphate buffer (pH 7.8). The extract was centrifuged at 12,000 rpm for 20 min at 4°C. Superoxide dismutase (SOD) and Peroxidase (POD) activities were measured following the method that described by Li et al. (2015).
Results
Isolation and Sequence Analysis of ZmERF105
The full-length cDNA sequence of ZmERF105 (GenBank No. NM001177195) was isolated from the leaves of maize inbred line B73 using RT-PCR. ZmERF105 has an open reading frame (ORF) of 990 bp, encoding a polypeptide of 329 amino acids with a predicted molecular mass of 34.5 kDa and a theoretical PI of 5.77. The deduced ZmERF105 protein contains a conserved AP2/ERF domain consisting of 58 amino acids, with conserved alanine (A) and aspartic acid (D) in it, suggesting that it is a member of the ERF family (Figure 1). The AP2/ERF domain contains conserved YRG and RAYD elements, which may play a key role in DNA binding and protein interaction, respectively (Figure 1). In addition, the ZmERF105 protein contains a conserved LSPLSPHP motif in its C-terminal region which is a mitogen-activated protein kinase (MAPK) phosphorylation site (Figure 1). The results from searching the Phytozome database (http://www.phytozome.net/maize) showed that ZmERF105 did not have intron and is located on chromosome 5. The phylogenetic tree analysis indicated that ZmERF105 protein shows highly similarity to SiERF105 (GenBank No. XP004953326), SbERF5 (GenBank No. XP021315889), and OsERF5 (GenBank No. XP015624058), respectively (Figure 2A). According to Fujimoto et al. (2000), ZmERF105 protein belongs to class III ERF group. The results from searching the SWISS-MODEL database (https://swissmodel.expasy.org/) showed that the 3D structure of ZmERF105 has a long C-terminal α-helix (α) wrapped in a three-stranded anti-parallel β-sheet (β1–β3) (Figure 2B).
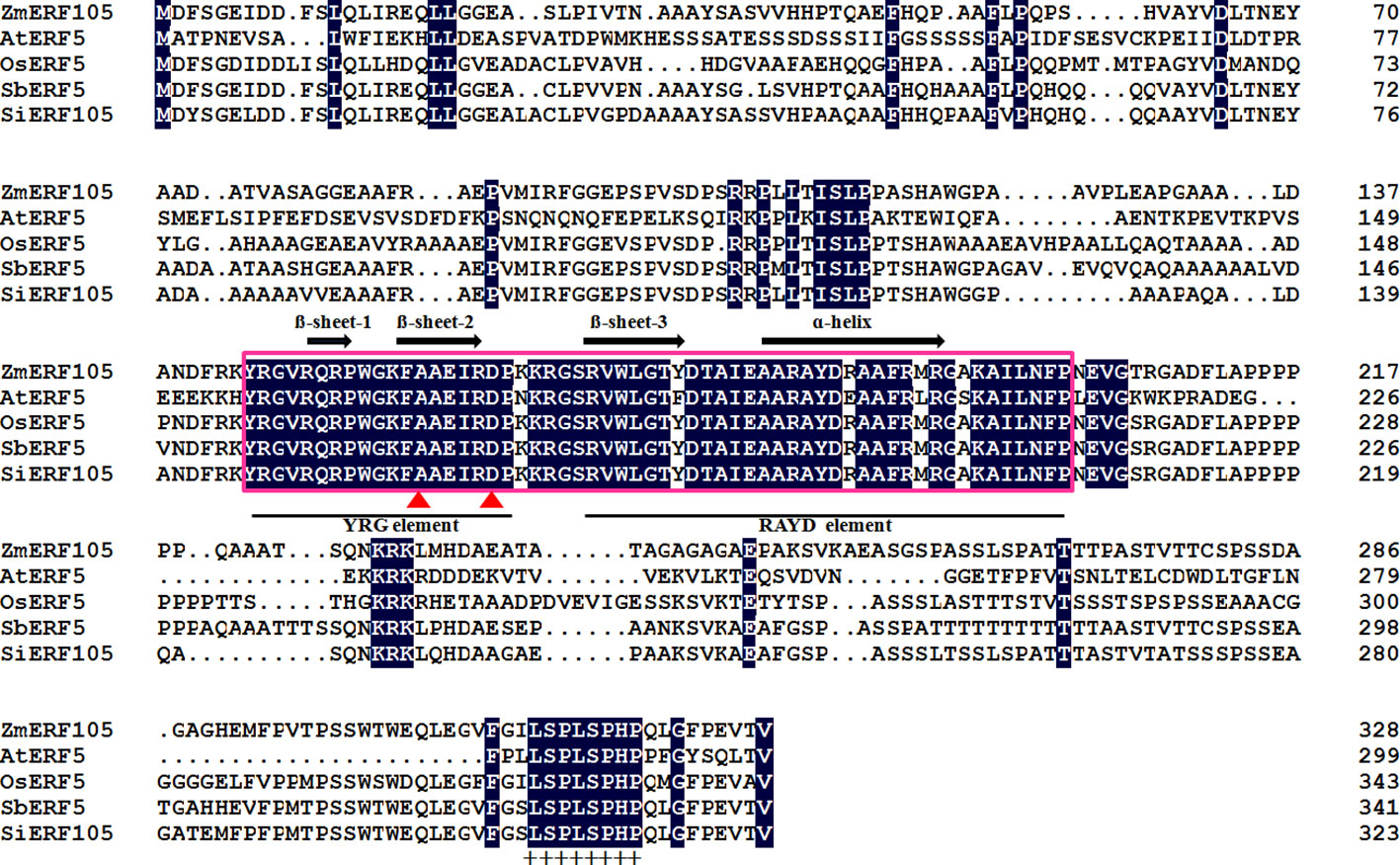
Figure 1 Conserved motifs of the ZmERF105 protein sequence in alignment with other ERF transcription factors. The sequence alignment was performed using DNAMAN software. The AP2 DNA binding domain is indicated by pink box. The one α-helix and three β-sheets are marked above the corresponding sequences. The YRG and RAYD elements are indicated below the alignment. The conserved alanine and aspartic acid residues in the AP2 domain are marked by red triangles. A conserved LSPLSPHP motif is marked by ‘‘+’’. AtERF5 (NP568679) is derived from Arabidopsis thaliana, OsERF5 (XP015624058) is derived from Oryza sativa, SbERF5 (XP021315889) is derived from Sorghum bicolor, and SiERF105 (XP004953326) is derived from Setaria italica.
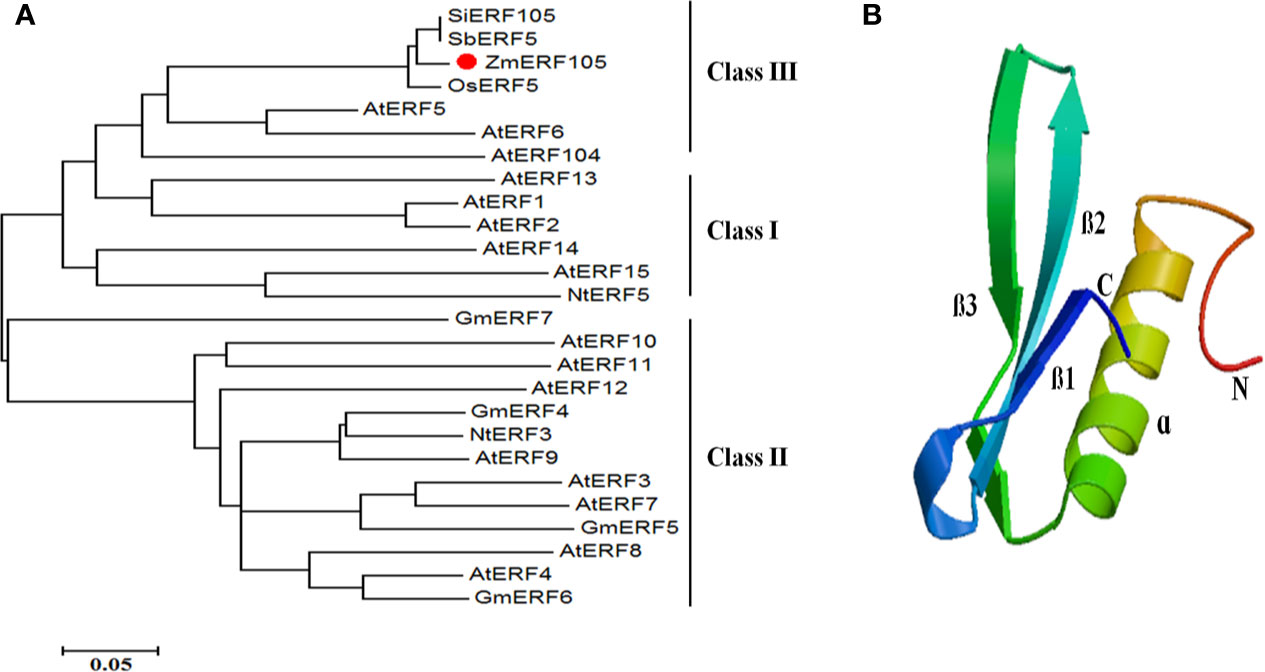
Figure 2 The phylogenetic tree and predicted three-dimensional structure of ZmERF105 protein. (A) The phylogenetic tree was generated with the neighbor-joining method using MEGA 5.0 software. ZmERF105 is indicated by the red dot. The accession numbers are as follows: AtERF1 (NP188965), AtERF2 (NP199533), AtERF3 (NP175479), AtERF4 (NP188139), AtERF5 (NP568679), AtERF6 (NP567529), AtERF7 (NP188666), AtERF8 (NP175725), AtERF9 (NP199234), AtERF10 (NP171876), AtERF11 (NP174159), AtERF12 (NP174158), AtERF13 (NP182011), AtERF14 (NP171932), AtERF15 (NP9850162), AtERF104 (NP_200968), GmERF4 (ACE76905), GmERF5 (AEX25891), GmERF6 (AEQ55267), GmERF7 (AEQ55266), NtERF3 (BAJ72664), NtERF5 (AAU81956), SbERF5 (XP021315889), SiERF105 (XP004953326), and OsERF5 (XP015624058). (B) Predicted three-dimensional structure of ZmERF105 protein.
Expression Profiles of ZmERF105 Under Various Stresses
In order to better understand the function of ZmERF105, the expression levels of ZmERF105 were investigated using qRT-PCR. The results demonstrated that ZmERF105 was expressed, with the highest expression in stems, followed by the leaves and the roots (Figure 3A). Under SA and ET treatments, the expression levels of ZmERF105 peaked at 5 and 10 h, respectively (Figures 3C, F). MeJA treatment induced a down-regulation of ZmERF105 expression at 2 h, followed by a slow increase, with maximum at 24 h (Figure 3D). In contrast, ZmERF105 expression rapidly reached the maximum at 2 h after ABA treatment, followed by a rapid decline in 5–10 h, and increased at 24 h (Figure 3E).
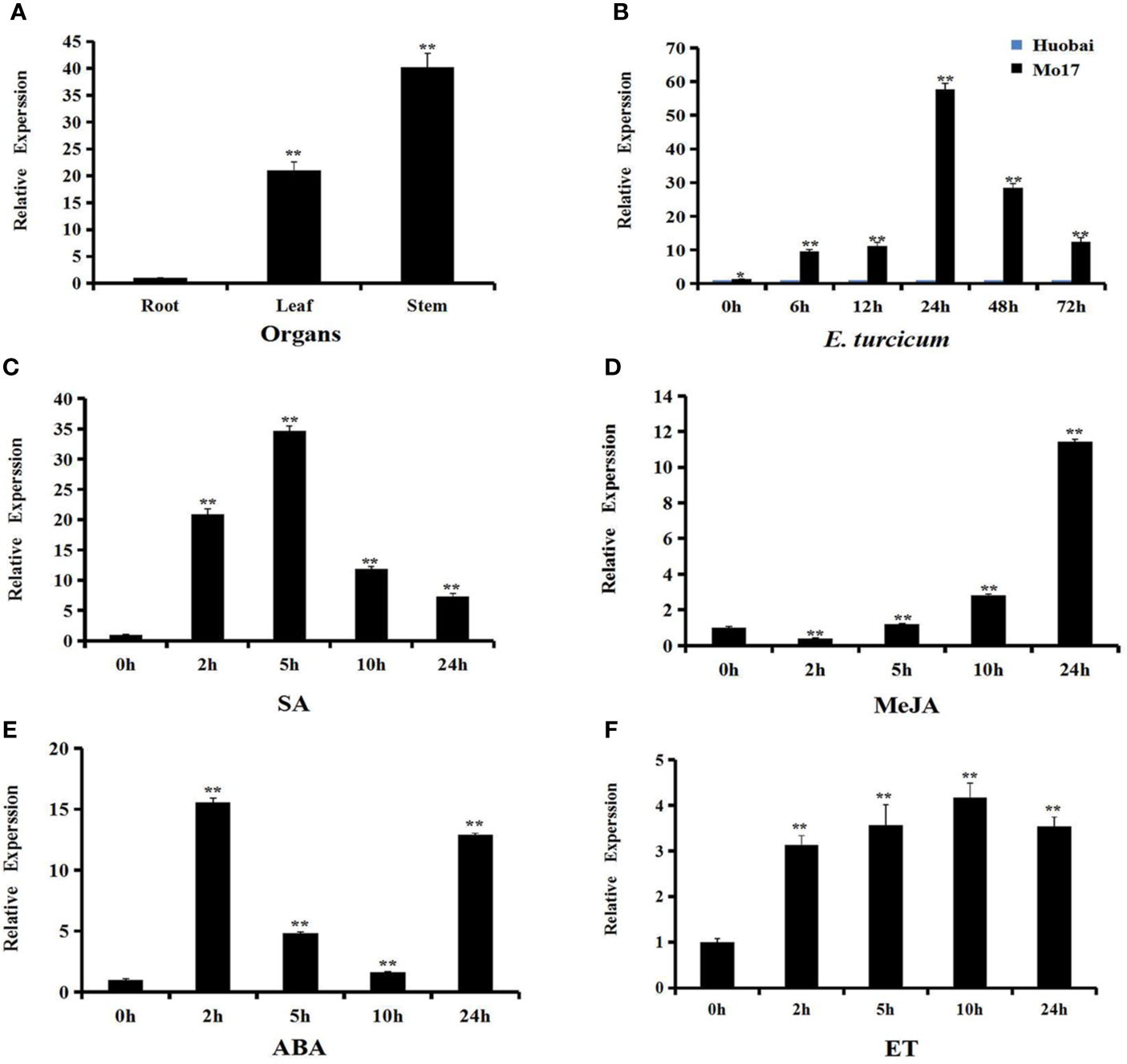
Figure 3 Expression profiles of ZmERF105 in maize. (A) Expression analysis of ZmERF105 in different organs. (B) Relative expression levels of ZmERF105 in Mo17 (resistant inbred line) and Huobai (susceptible inbred line) plants infection with E. turcicum. The samples were collected at 0, 6, 12, 24, 48, and 72 h after E. turcicum infection. The relative expression levels were compared with susceptible inbred line Huobai at same points. (C–F) Expression analysis of ZmERF105 under hormone treatments, including 0.5mM SA, 0.1mM MeJA, 0.1mM ABA or ET. The relative expression levels were calculated by the 2-ΔΔCt method with the maize housekeeping gene ZmTub (GRMZM2G066191) as an internal control. The experiment was performed using three biological and technical replicates each, and analyzed using Student' t-tests (*P < 0.05, **P < 0.01). Bars indicate standard error of the mean (SE).
ZmERF105 expression was detected in Mo17 (resistant inbred line) and Huobai (susceptible inbred line) plants after infection with E. turcicum. As shown in Figure 3B, the expression level of ZmERF105 was higher in Mo17 leaves than that in Huobai leaves during E. turcicum infection. A significant up-regulation of ZmERF105 expression was detected at 6 h and reached a maximum level at 24 h. These results demonstrated that ZmERF105 may involve in response to pathogen and multiple stimulus.
Subcellular Localization of the ZmERF105 Protein
To determine the subcellular location of ZmERF105 protein, a ZmERF105-GFP fusion protein under the control of the 35S promoter was obtained and introduced into the epidermal cells of N. benthamiana. Consistent with its function as a TF, the ZmERF105-GFP fusion protein was exclusively localized to the nucleus, and the fluorescent signals of the control were observed in whole cells (Figure 4).
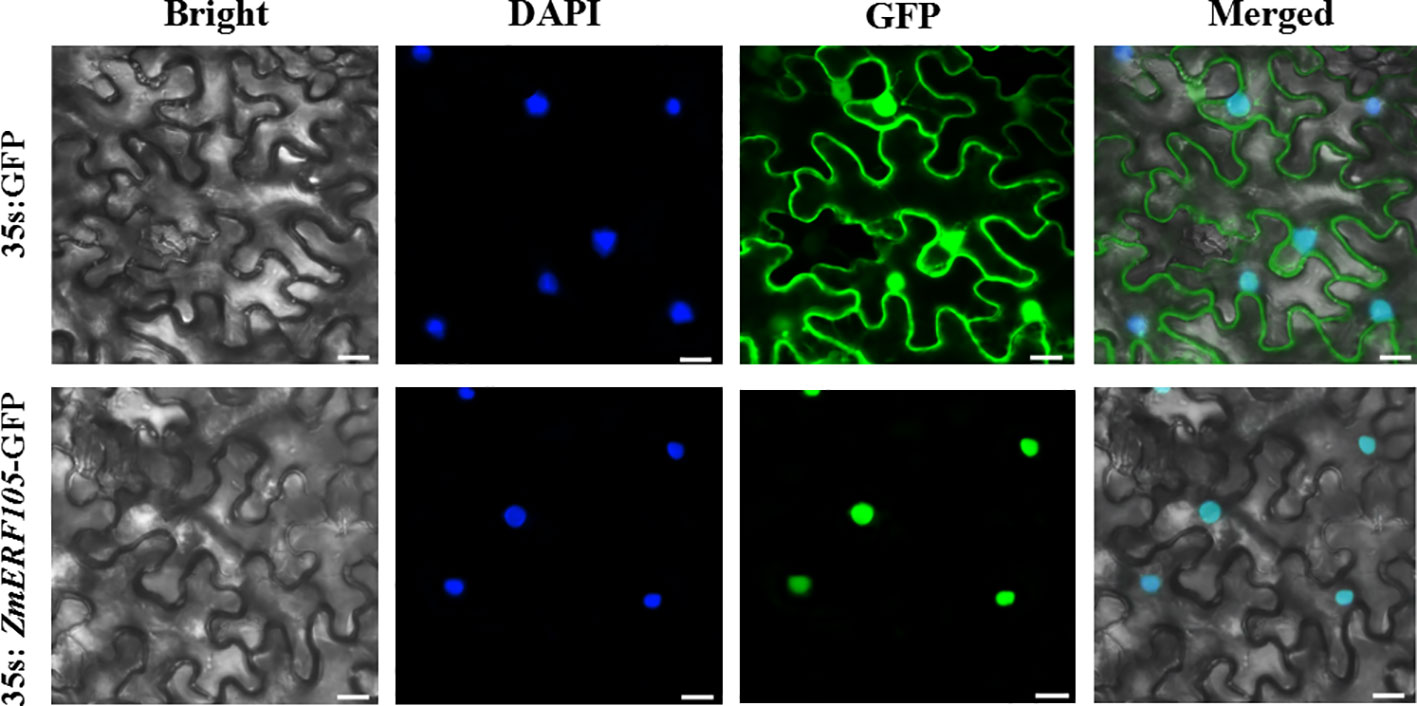
Figure 4 Subcellular localization of ZmERF105-GFP fusion protein in the epidermal cells of N. benthamiana. N. benthamiana leaves transiently expressing GFP alone (upper) and ZmERF105-GFP (bottom) proteins were observed with a confocal microscope. Bars = 25 μm.
ZmERF105 Binds to GCC-Box Elements and Functions as a Transcription Activator
Previous studies have shown that ERF genes contain a conserved DNA-binding domain which can bind to GCC-box elements (Zhang et al., 2004; Tang et al., 2007). To further investigate whether ZmERF105 specifically binds to GCC-box elements, a yeast one-hybrid assay was performed. As shown in Figure 5A, the bait stain (pHIS2-GCC) transfected with prey vector (pGADT7-ZmERF105) could grow on SD (-Leu/-Trp/-His) medium containing 100 mM 3-AT, while mutant bait stain (pHIS2-mGCC) could not. The result indicated that ZmERF105 can bind specifically to GCC-box elements
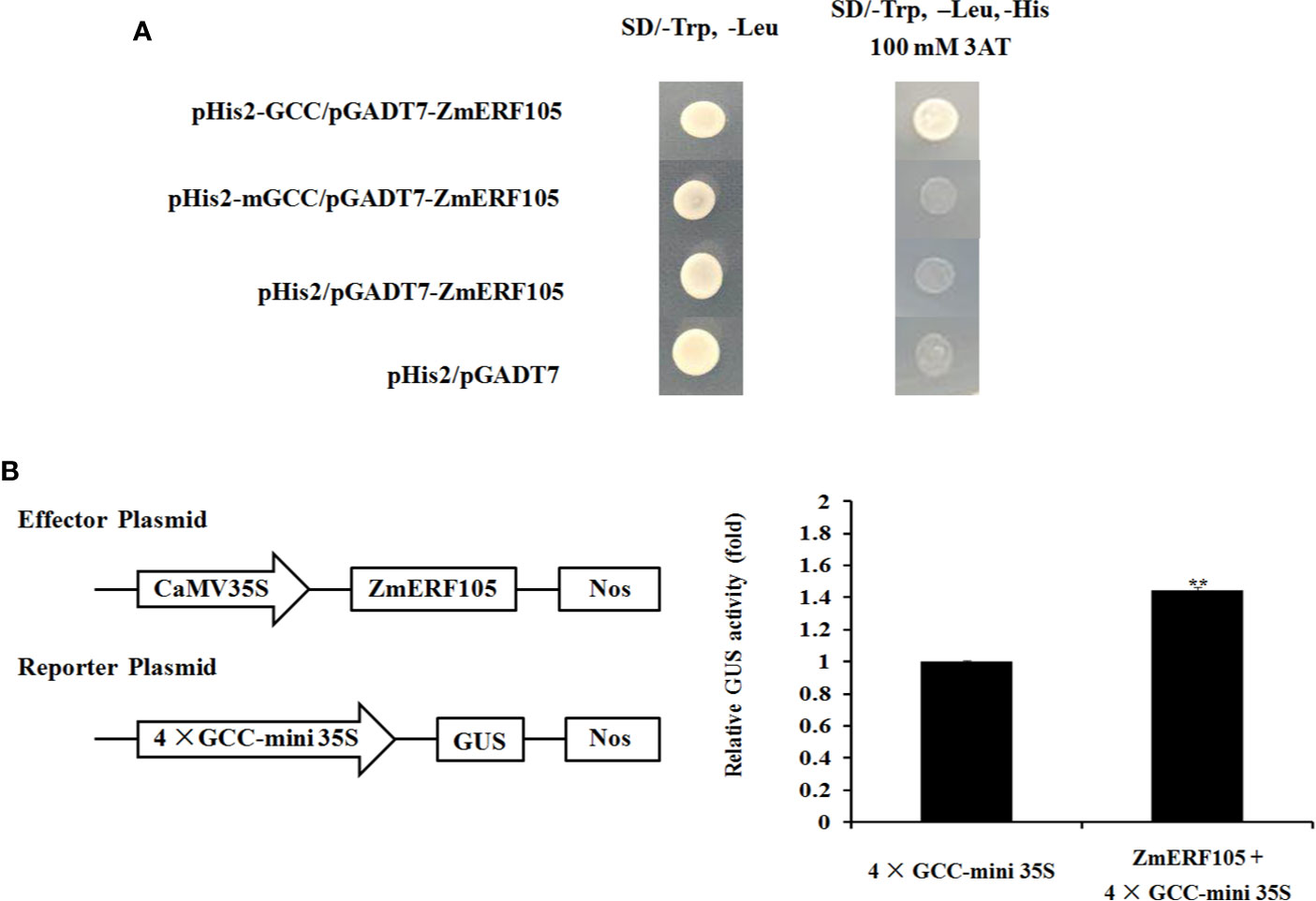
Figure 5 Binding to the GCC-box elements and transcription activity analysis of ZmERF105. (A) The binding activity of ZmERF105 to the GCC-box elements in yeast one-hybrid assay. Yeast cells were selected on SD (-Trp/-Leu) and SD (-Trp/-Leu/-His) media supplemented with 100 mM 3-AT. (B) Transcription activity analysis of ZmERF105 in Arabidopsis protoplasts. The numbers show the fold increase in GUS activity compared with the reporter vector driven by 4× GCC 35S mini promoter alone. The experiment was performed using three biological and technical replicates each and analyzed using Student’s t-tests (**P < 0.01). Bars indicate standard error of the mean (SE).
Transient transcription activity assay was used to determine whether ZmERF105 functions as a transcription activator. Expression of ZmERF105 was driven by the CaMV 35S promoter, and the GUS reporter gene was driven by 4× GCC 35S mini promoter. As shown in Figure 5B, the relative GUS activity of the Arabidopsis protoplasts transformed with was the CaMV 35S: ZmERF105 and 4× GCC 35S: GUS was approximately 1.4 times as high as the control. These results demonstrated that ZmERF105 functions as a transcriptional activator and binds to GCC-box elements.
ZmERF105 Positively Regulates Maize Resistance Against E. turcicum
To investigate whether ZmERF105 can improve resistance to E. turcicum in transgenic maize plants, T2 transgenic plants, constituting three independent ZmERF105 over-expressing transgenic lines (OE3, OE4, and OE7) was used to investigate the response to E. turcicum. The expression of ZmERF105 in the representative transgenic lines (OE3, OE4, and OE7) was further detected by qRT-PCR (Figure 6B). As illustrated in Figures 6A, C, the sizes of the lesions in over-expressing lines were significantly smaller than in the wild-type maize lines (WT) at 5 dpi. In addition, the relative expression of ZmERF105 had about 0.2-fold and 0.3-fold declines in erf105 mutant lines, respectively (Figure 6E). After inoculation with E. turcicum, the erf105 mutant lines displayed significantly susceptibility compared with the WT at 5 dpi (Figures 6D, F). These results demonstrated that ZmERF105 positively regulates the maize resistance against E. turcicum.
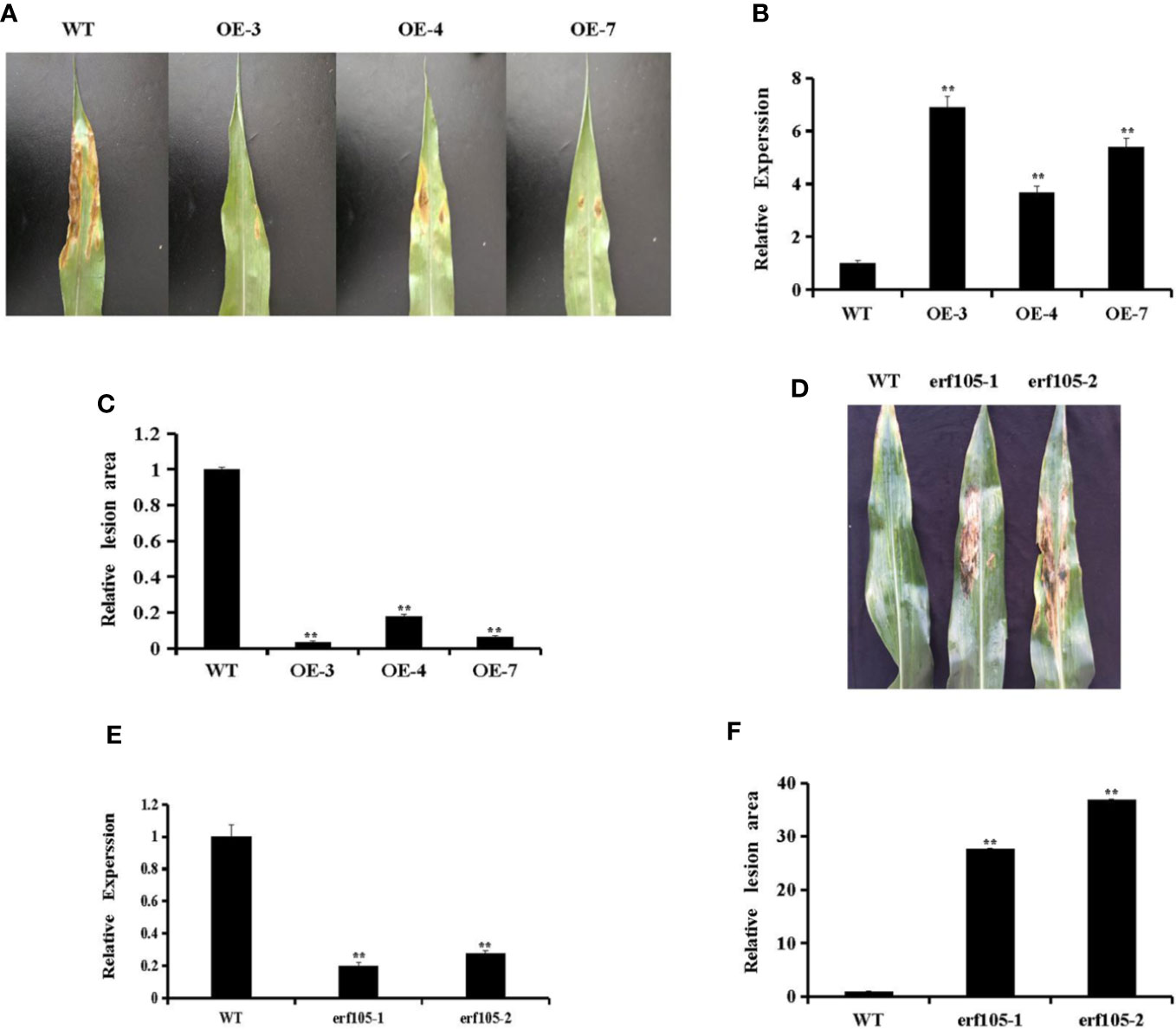
Figure 6 Responses of ZmERF105 over-expression and erf105 mutant lines to E. turcicum. (A) Disease symptoms on the leaves of T2 transgenic lines at 5 dpi. (B) Relative expression analysis of ZmERF105 in T2 transgenic lines. (C) The relative lesion areas of the transgenic lines at 5 dpi. (D) Disease symptoms on the leaves of erf105 mutant lines at 5 dpi. (E) Relative expression analysis of ZmERF105 in erf105 mutant lines. (F) The relative lesion areas of the erf105 mutant lines at 5 dpi. The experiment was performed using three biological and technical replicates each and analyzed using Student’s t-tests (**P < 0.01). Bars indicate standard error of the mean (SE).
ZmERF105 Affects Antioxidant Enzyme Activity
SOD and POD are the most important antioxidant enzyme in the process of ROS scavenging reaction, and it was suggested to participate in various defense mechanisms (Liu et al., 2016). To further confirm whether the role of ZmERF105 in resistance against E. turcicum is related to SOD and PDD activities, we examined the activities of SOD and POD in ZmERF105 over-expression and erf105 mutant lines. After inoculation E. turcicum, both SOD and POD activities were significantly high in ZmERF105 over-expression lines compared with those in the mock-inoculated lines at 24 h, and were remarkably decreased in the erf105 mutant lines (Figure 7). These results suggested that ZmERF105 increases the activities of the antioxidant enzymes in maize resistance to E. turcicum.
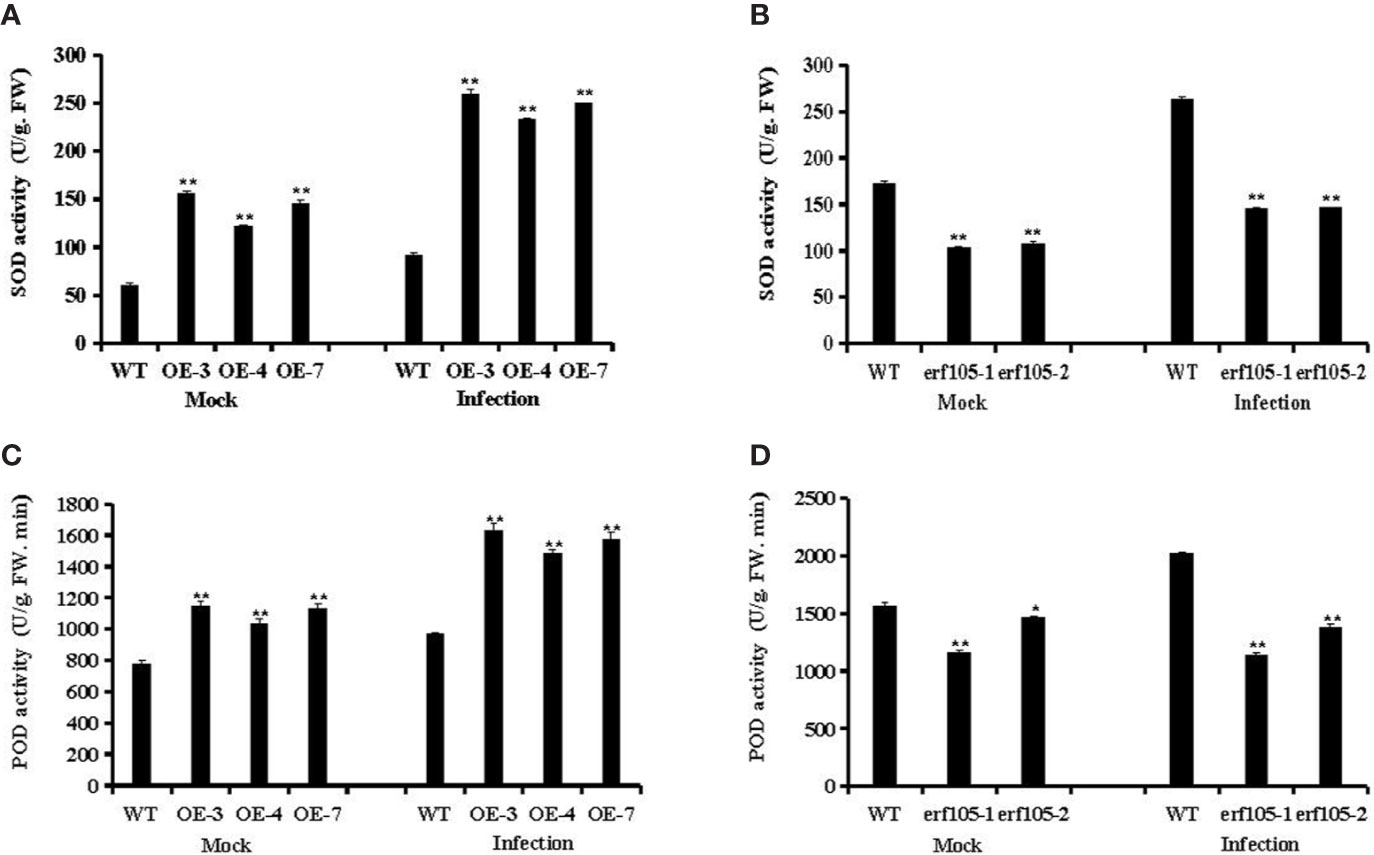
Figure 7 Analysis of antioxidant enzyme activity under mock treatment and infected by E. turcicum at 24-h post-inoculation (hpi). (A, B) The activity of the SOD in ZmERF105 over-expression and erf105 mutant lines, respectively. (C, D) The activity of the POD in ZmERF105 over-expression and erf105 mutant lines, respectively. The activity of the WT sample [mock-treated wild-type (WT) plants] was set to unity. The experiment was performed on three biological replicates, each with three technical replicates, and statistically analyzed using Student’s t-test (*P < 0.05, **P < 0.01). Bars indicate standard error of the mean (SE).
ZmERF105 Involves in Various Plant Defense Responses
ERF genes are suggested to participate in plant defense responses through regulating the expression of PR genes (Lorenzo et al., 2003; Pre et al., 2008; Moffat et al., 2012; Son et al., 2012). To further analyze the role of ZmERF105 in resistance against E. turcicum in maize, we examined the expression of PR genes that including ZmPR1a (GRMZM2G465226), ZmPR2 (GRMZM5G456997), ZmPR5 (GRMZM2G402631), ZmPR10.1 (GRMZM2G112488), and ZmPR10.2 (GRMZM2G112538). As shown in Figure 8, after 24h incubation with E. turcicum, the expression levels of these PR genes were highly induced in ZmERF105 over-expressing lines compared with those in the mock-inoculated lines, and were remarkably reduced in the erf105 mutant lines. These results indicated that ZmERF105 may enhance maize defense against E. turcicum by directly or indirectly regulating these PR genes.
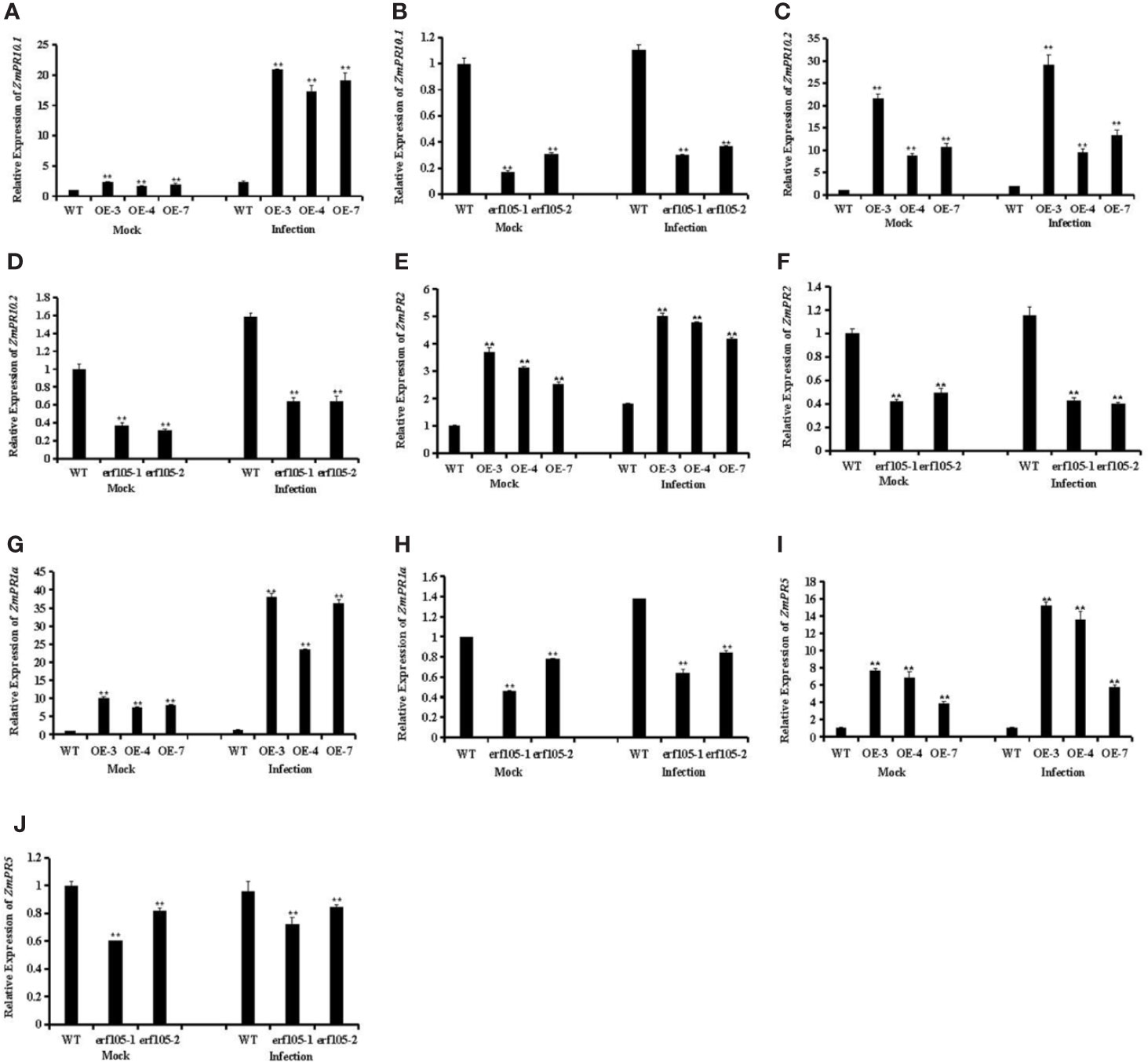
Figure 8 Relative expression levels of defense-related genes under mock treatment and infected by E. turcicum at 24 hpi in ZmERF105 over-expression and erf105 mutant lines, respectively. (A, B) The expression of the ZmPR10.1. (C, D) The expression of the ZmPR10.2. (E, F) The expression of the ZmPR2. (G, F) The expression of the ZmPR1a. (H, I) The expression of the ZmPR5. The relative expression levels were calculated by the 2−ΔΔCt method with the maize housekeeping gene ZmTub (GRMZM2G066191) as an internal control. The expression of the WT sample [mock-treated wild-type (WT) plants] was set to unity. The experiment was performed using three biological and technical replicates each and analyzed using Student’s t-tests (**P < 0.01). Bars indicate standard error of the mean (SE).
Due to the expression levels of SA-associated genes ZmPR1a and ZmPR5 exhibited significant increase in the ZmERF105 over-expression lines and were decreased in the erf105 mutant lines after E. turcicum infection, the expression level of ZmERF105 was induced by SA treatment, we speculated whether ZmERF105 is required for SA induced defense response. To test this hypothesis, we further assessed SA induces resistance to E. turcicum infection in erf105 mutant lines. As shown in Supplementary Figure S1, erf105 mutant lines showed less severe disease symptom by application of exogenous 10 mM SA than the water-treated lines at 5 dpi (Supplementary Figure S1). These results demonstrated that ZmERF105 may involve in SA-induced defense response. To further confirm whether ZmERF105 involved in MeJA, ET, and ABA induced defense responses, we detected the expression of MeJA-associated gene ZmLox1 (GRMZM2G156861), ET-associated gene ZmACS6 (GRMZM2G04361), and ABA-associated gene ZmRD20 (GRMZM2G342685), respectively. The results showed that the expression levels of ZmLox1 and ZmACS6 significantly increased in the ZmERF105 over-expression lines and were decreased in the erf105 mutant lines after E. turcicum infection, while ZmRD20 showed opposite results (Supplementary Figure S2). These results demonstrated that ZmERF105 may act positively as regulators of MeJA and ET signaling pathways in resistance to E. turcicum, and negatively in ABA signaling pathway.
Discussion
Plants have evolved an array of defense mechanisms to protect themselves when they are exposed to a wide variety of pathogens (Schwessinger and Ronald, 2012; Fu and Dong, 2013; Birkenbihl et al., 2017). ERFs are one of the most important TFs in the plant defense syetem (McGrath et al., 2005; Pre et al., 2008; Maruyama et al., 2013). In maize, a total of 98 predicted ERF members have been identified (Liu et al., 2013). In this study, a novel maize ERF gene, ZmERF105, was isolated and identified. ZmERF105 is involved in the pathogen response pathway and positively regulates the maize resistance against E. turcicum.
To our knowledge, ERF families were divided into ERF subfamily and DREB subfamily according to the amino acids difference at position 14th and 19th in AP2/ERF domain (Sakuma et al., 2002). Similar to other ERF proteins, ZmERF105 protein contains a conserved AP2/ERF domain consisting of 58 amino acids, with conserved alanine (A) and aspartic acid (D) in it (Figure 1), suggesting that it is a member of the ERF subfamily. Sequence analysis showed that ZmERF105 protein contains a LSPLSPHP motif in its C-terminal region, which maybe indicate the role of ZmERF105 in MAPK signaling pathway (Figure 1).
ERF genes have been shown to act as transcription activators or repressors and are able to bind to GCC-box elements (Fujimoto et al., 2000). For example, AtERF1, ORA59, AtERF6, and AtERF96 serve as transcriptional activators while GmERF5 and StERF3 serve as transcriptional repressors, and they can bind to GCC-box elements (Zarei et al., 2011; Wang et al., 2013; Catinot et al., 2015; Dong et al., 2015; Tian et al., 2015). In this study, we found that nuclear loci of ZmERF105 binds specifically to the GCC-box elements by yeast one-hybrid assay and shows transactivation activity in Arabidopsis protoplast (Figures 4 and 5). Our results suggested that ZmERF105 may function as a transcription activator and regulate defense-related genes expression by binding to GCC-box elements present in their promoters.
Most of ERFs act as activators and positively regulate the plant resistance against pathogens (Berrocal-Lobo et al., 2002; Son et al., 2012). Over-expression of AtERF1, ORA59, AtERF6, AtERF96, and AtERF104 were shown to increase plant resistance against B. cinerea (Berrocal-Lobo et al., 2002; Pre et al., 2008; Bethke et al., 2009; Zarei et al., 2011; Meng et al., 2013; Wang et al., 2013; Catinot et al., 2015). The transcription activator, AtERF15 could positively regulate the Arabidopsis resistance against Pst DC3000 and B. cinerea (Zhang et al., 2015). Over-expression of GmERF113 in transgenic soybean led to increased resistance to Phytophthora Sojae (P.sojae) (Zhao et al., 2017). In our research, the expression of ZmERF105 was significantly induced after infection with E. turcicum in the resistant inbred line “Mo17” than in the susceptible inbred line “Huobai” (Figure 3B). Therefore, we speculate that ZmERF105 may play important role in response to E. turcicum. As a result, we further analyzed the function of ZmERF105 in resistance to E. turcicum. Over-expression of ZmERF105 was shown to increase maize resistance against E. turcicum, and erf105 mutant lines displayed opposite phenotype (Figure 6). These results demonstrated that ZmERF105 positively regulates the maize resistance response to E. turcicum.
ERF genes are mainly involved in plant defense response by directly regulating the expression of defense-related genes (Meng et al., 2013; Wang et al., 2013). AtERF96 positively regulates the Arabidopsis resistance to B. cinerea by enhancing the expression of PDF1.2a, PR-3, and PR-4 (Catinot et al., 2015). Over-expression of AtERF1, AtERF5, AtERF6, and ORA59 increased Arabidopsis resistance against B. cinerea by promoting the expression of AtPDF1.2 (Lorenzo et al., 2003; Zarei et al., 2011; Son et al., 2012; Moffat et al., 2012; Cheng et al., 2013). Over-expression of GmERF3 in transgenic tobacco enhanced resistance against tobacco mosaic virus (TMV) and activating the expression of PR1, PR2, and PR4 (Zhang et al., 2009). Over-expression of GmERF113 increased soybean resistance to P. sojae and increased transcript levels of GmPR1 and GmPR10-1 (Zhao et al., 2017). Our previous research demonstrated that ZmERF105 over-expression lines enhanced the expression of several PR genes, including ZmPR1a, ZmPR2, ZmPR5, ZmPR10.1 and ZmPR10.2 after infection with E. turcicum, while the expression levels of these PR genes were reduced in erf105 mutant lines (Figure 8). We speculated that ZmERF105 may enhance maize defense against E. turcicum by directly or indirectly regulating these PR genes.
POD and SOD are the most important antioxidant enzymes to help eliminate the excessive accumulation of ROS in plants, to induce resistance or repair the damage (Liu et al., 2016). In the present study, the activities of SOD and POD in the ZmERF105 over-expression lines were markedly higher than in the WT after infection with E. turcicum, and were compromised in the erf105 mutant lines (Figure 7). These results demonstrated that ZmERF105 can strengthen ROS scavenging capability to provide sufficient protection against oxidative damage.
The phytohormones ET, JA, SA, and ABA are important for plants to distinguish and resist distinctive pathogens (Pieterse et al., 2012; Van der Does et al., 2013). Over-expression of AtERF5 resulted in increased resistance to B. cinerea through MeJA/ET signaling pathway, while showed increased susceptibility to the hemibiotroph Pst DC3000 via suppressing SA-mediated signaling pathway (Son et al., 2012; Moffat et al., 2012). AtERF1 and ORA59 are more resistant to B. cinerea via MeJA/ET-mediated signaling pathway (Berrocal-Lobo et al., 2002; Pre et al., 2008). AtERF15 involved in defense against B. cinerea and Pst DC3000 through SA signaling pathway (Zhang et al., 2015). ERF11 acted as a regulator of the SA-mediated signaling pathway to enhance the Arabidopsis defense response against Pst DC3000. The expression levels of SA-associated genes ZmPR1a and ZmPR5 exhibited significant increases in the ZmERF105 over-expression lines but were decreased in the erf105 mutant lines after E. turcicum infection (Figure 8). Furthermore, the expression of ZmERF105 was induced by SA treatment (Figure 3C) and SA induces resistance to E. turcicum infection in erf105 mutant lines (Supplementary Figure S1). These results demonstrated that ZmERF105 may involve in defense response to E. turcicum in the regulation of SA signaling pathways. In addition, we further demonstrated that ZmERF105 may positively regulate the expression of MeJA-associated gene and ET-associated gene in response to E. turcicum, and negatively regulate the expression of ABA-associated gene Supplementary Figure S2). The results showed that ZmERF105 may act positively as regulators of MeJA and ET signaling pathways in resistance to E. turcicum, and negatively in ABA signaling pathway.
In conclusion, ZmERF105 is a novel ERF gene and plays an important role in the plant defense system. ZmERF105 protein was exclusively localized to the nucleus. ZmERF105 acts as a transcriptional activator that binds to GCC-box elements. ZmERF105 expression responded to E. turcicum treatment. ZmERF105 positively regulated plant resistance against E. turcicum via regulating the expression of defense-related genes and the activities of antioxidant enzymes. Overall, our study provides new information to dissect the poorly understood mechanism of ZmERF105 in plant immune pathways.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
Conceived and designed the experiments: LJ and WGY. Performed the experiments and drafted the manuscript: ZZ, YL, SL, HW, and WM. Analyzed the data: ZW, WY, JC, and XR. Contributed reagents/materials/analysis tools: LJ and WGY.
Funding
This research was supported by Science and Technology Development Project of Jilin Province (20190301013NY), Doctor Foundation of Jilin Agricultural University (201801) and “13th Five-Year” Science and Technology Project of Education Department of Jilin Province (JJKH20180660KJ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Maize Genetics Cooperation Stock Center for the maize mutants.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00850/full#supplementary-material
References
Berrocal-Lobo, M., Molina, A., Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. doi: 10.1046/j.1365-313x.2002.01191.x
Bethke, G., Unthan, T., Uhrig, J. F., Pöschl, Y., Gust, A. A., Scheel, D., et al. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. U. S. A. 106, 8067–8072. doi: 10.1073/pnas.0810206106
Bigeard, J., Colcombet, J., Hirt, H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539. doi: 10.1016/j.molp.2014.12.022
Birkenbihl, R. P., Liu, S., Somssich, I. E. (2017). Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 38, 1–9. doi: 10.1016/j.pbi.201-7.04.004
Boller, T., He, S. Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324, 742–744. doi: 10.1126/science.1171647
Caarls, L., Van der Does, D., Hickman, R., Jansen, W., Verk, M. C., Proietti, S., et al. (2017). Assessing the role of ETHYLENE RESPONSE FACTOR transcriptional repressors in salicylic acid-mediated suppression of jasmonic acid-responsive genes. Plant Cell Physiol. 58, 266–278. doi: 10.1093/pcp/pcw187
Carson, M. L. (1995). A new gene in maize conferring the “Chlorotic Halo” reaction to infection by Exserohilum turcicum. Plant Dis. 79, 717–720. doi: 10.1094/PD-7-9-0717
Catinot, J., Huang, J. B., Huang, P. Y., Tseng, M. Y., Chen, Y. L., Gu, S. Y., et al. (2015). ETHYLENE RESPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate- and ethylene-responsive defence genes. Plant Cell Environ. 38, 2721–2734. doi: 10.1111/pce.12583
Chang, H. S., Fan, K. C. (1986). Comparative studies on some biology and pathology of corn and broom corn isolates of Exserohilum turcicum (Pass) Leonard & Suggs. Bot. Bull. Acad. Sinica 27, 209–218.
Chen, S. B., Tao, L. Z., Zeng, L. R., Vega-Sanchez, M. E., Umemura, K., Wang, G. L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7, 417–427. doi: 10.1111/J.1364-3703.2006.00346.x
Cheng, M. C., Liao, P. M., Kuo, W. W., Lin, T. P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Cui, H. T., Xiang, T. T., Zhou, J. M. (2009). Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell. Microbiol. 11, 1453–1461. doi: 10.1111/j.1462-5822.2009.01359.x
Cui, H. W., Yang, Y. L., Li, J. T., Luo, W. F., Miao, A. M., Hu, Z. X., et al. (2009). A faster method for measuring relative lesion area on leaves based on software photoshop. J. Anhui Agric. Sci. 37, 10760–10762, 10805. doi: 10.3969/j.issn.0517-6611.2009.22.192
Dong, L. D., Cheng, Y. X., Wu, J. J., Cheng, Q., Li, W. B., Fan, S. J., et al. (2015). Overexpression of GmERF5, a new member of the soybean EAR motif- containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 66, 2635–2647. doi: 10.1093/jxb/erv078
Farmer, E. E., Alméras, E., Krishnamurthy, V. (2003). Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol. 6, 372–378. doi: 10.1016/s1369-5266(03)00045-1
Franco-Zorrilla, J. M., Lopez-Vidriero, I., Carrasco, J. L., Godoy, M., Vera, P., Solano, R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. U. S. A. 111, 2367–2372. doi: 10.1073/pnas.1316278111
Fu, Z. Q., Dong, X. N. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Galiano-Carneiro, A. L., Miedaner, T. (2017). Genetics of Resistance and Pathogenicity in the Maize/Setosphaeria turcica Pathosystem and Implications for Breeding. Front. Plant Sci. 8, 1490. doi: 10.3389/fpls.2017.01490
Grant, M. R., Jones, J. D. (2009). Hormone (dis) harmony moulds plant health and disease. Science 324, 750–752. doi: 10.1126/science.1173771
He, X., Jiang, J. S., Wang, C. Q., Dehesh, K. (2017). ORA59 and EIN3 interaction couples jasmonate-ethylene synergistic action to antagonistic salicylic acid regulation of PDF expression. J. Integr. Plant Biol. 59, 275–287. doi: 10.1111/jipb.12524
Holsters, M., De, W. D., Depicker, A., Messens, E., Van, M. M., Schell, J. (1978). Transfection and of Agrobacterium tumefaciens. Mol. Genet. Genomics 163, 181–187. doi: 10.1007/BF00267408
Huang, P. Y., Catinot, J., Zimmerli, L. (2016). Ethylene response factors in Arabidopsis immunity. J. Exp. Bot. 67, 1231–1241. doi: 10.1093/jxb/erv518
Huang, P. Y., Zhang, J. S., Jiang, B., Chan, C., Yu, J. H., Liu, Y. P., et al. (2019). NINJA-associated ERF19 negatively regulates Arabidopsis pattern-triggered immunity. J. Exp. Bot. 70, 1033–1047. doi: 10.1093/jxb/ery414
Hurni, S., Scheuermann, D., Krattinger, S. G., Kessel, B., Wicker, T., Herren, G., et al. (2015). The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. U.S.A. 112, 8780–8785. doi: 10.1073/pnas.1502522112
Kim, N. Y., Jang, Y. J., Park, O. K. (2018). AP2/ERF Family Transcription Factors ORA59 and RAP2.3 Interact in the Nucleus and Function Together in Ethylene Responses. Front. Plant Sci. 9, 167. doi: 10.3389/fpls.2018.01675
Lai, Y., Dang, F. F., Lin, J., Yu, L., Shi, Y. L., Xiao, Y. H., et al. (2013). Overexpression of a Chinese cabbage BrERF11 transcription factor enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Physiol. Biochem. 62, 70–78. doi: 10.1016/j.plaphy.2012.10.010
Leon-Reyes, A., Du, Y. J., Koornneef, A., Proietti, S., Körbes, A. P., Memelink, J., et al. (2010). Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol. Plant Microbe Interact. 23, 187–197. doi: 10.1094/MPMI-23-2-0187
Li, Z. F., Zhang, L. X., Yu, Y. W., Quan, R. D., Zhang, Z. J., Zhang, H. W., et al. (2011). The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in. Arabidopsis Plant J. 68, 88–99. doi: 10.1111/j.1365-313X.2011.04670.x
Li, Z. J., Zhu, B., Wang, B., Gao, J. J., Fu, X. Y., Yao, Q. H. (2015). Stress responses to trichlorophenol in Arabidopsis and integrative analysis of alteration in transcriptional profiling from microarray. Gene 555, 159–168. doi: 10.1016/j.gene.2014.10.059
Li, Z. J., Tian, Y. S., Xu, J., Fu, X. Y., Gao, J. J., Wang, B., et al. (2018). A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against Pseudomonas syringae pv. tomato DC3000. Plant Physiol. Biochem. 132, 683–695. doi: 10.1016/j.plaphy.2-018.08.022
Licausi, F., Ohme-Takagi, M., Perata, P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Liu, L. J., Zhang, Y. Y., Tang, S. Y., Zhao, Q. Z., Zhang, Z. H., Zhang, H. W., et al. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61, 893–903. doi: 10.1111/j.1365-313X.2009.0-4109
Liu, S. X., W, X. L., Wang, H. W., X, H. B., Y, X. H., Y, J. B., et al. (2013). Genome-wide analysis of ZmDREB Genes and their association with natural variation in drought tolerance at seedling stage of Zea Mays L. PloS Genet. 9, e1003790. doi: 10.1371/journal.pgen.1003790
Liu, N. N., Ma, X. W., Zhou, S. H., Wang, P., Sun, Y., Li, X. C., et al. (2016). Molecu-lar and functional characterization of a polygalacturonase-inhibiting protein from Cynanchum komarovii that confers fungal resistance in Arabidopsis. PloS One 11, e146959. doi: 10.1371/journal.pone.014695
Lorenzo, O., Piqueras, R., Sánchez-Serrano, J. J., Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Lu, X., Jiang, W. M., Zhang, L., Zhang, F., Zhang, F. Y., Shen, Q., et al. (2013). AaERF1 positively regulates the resistance to Botrytis cinerea in Artemisia annua. PloS One 8, e57657. doi: 10.1371/journal.pone.0057657
Müller, M., Munné-Bosch, S. (2015). Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. doi: 10.1104/pp.15.00677
Maruyama, Y., Yamoto, N., Suzuki, Y., Chiba, Y., Yamazaki, K. I., Sato, T., et al. (2013). The Arabidopsis transcriptional repressor ERF9 participates in resistance against necrotrophic fungi. Plant Sci. 213, 79–87. doi: 10.1016/j.plantsci.2013.0-8.008
McGrath, K. C., Dombrecht, B., Manners, J. M., Schenk, P. M., Edgar, C. I., Maclean, D. J., et al. (2005). Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via agenome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139, 949–959. doi: 10.1104/pp.105.068544
Meng, X. Z., Xu, J., He, Y. X., Yang, K. Y., Mordorski, B., Liu, Y. D., et al. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25, 1126–1142. doi: 10.1105/tpc.112.109074
Moffat, C. S., Ingle, R. A., Wathugala, D. L., Saunders, N. J., Knight, H., Knight, M. R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PloS One 7, e35995. doi: 10.1371/journal.pone.0035995
Nakano, T., Suzuki, K., Fujimura, T., Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Oñate-Sánchez, L., Anderson, J. P., Young, J., Singh, K. B. (2007). AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 143, 400–409. doi: 10.1104/pp.106.086637
Ohme-Takagi, M., Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. doi: 10.1105/tpc.7.2.173
Pan, I. C., Li, C. W., Su, R. C., Cheng, C. P., Lin, C. S., Chan, M. T. (2010). Ectopic expression of an EAR motif deletion mutant of SlERF3 enhances tolerance to salt stress and Ralstonia solanacearum in tomato. Planta 232, 1075–1086. doi: 10.1007/s00425-010-1235-5
Perkins, J. M., Pedersen, W. L. (1987). Disease development and yield losses associated with northern leaf blight on corn. Plant Dis. 71, 940–943. doi: 10.1094/PD-71-0940
Pieterse, C. M., Van der Does, D., Zamioudis, C., Leon-Reyes, A., Van Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Pré, M., Atallah, M., Champion, A., De Vos, M., Pieterse, C. M. J., Memelink, J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357. doi: 10.1104/pp.108.117523
Raymundo, A. D., Hooker, A. L. (1981). Measuring the relationship between northern corn leaf blight and yield losses. Plant Dis. 65, 325–327. doi: 10.1094/PD-65-325
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., Yamaguchi-Shinoz- aki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.-2001.6299
Schwessinger, B., Ronald, P. C. (2012). Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63, 451–482. doi: 10.114-6/annurev-arplant-042811-105518
Sherif, S., El-Sharkawy, I., Paliyath, G., Jayasankar, S. (2013). PpERF3b, a transcriptional repressor from peach, contributes to disease susceptibility and side branching in EAR-dependent and -independent fashions. Plant Cell Rep. 32, 1111–1124. doi: 10.1007/s00299-013-1405-6
Son, G. H., Wan, J., Kim, H. J., Nguyen, X. C., Chung, W. S., Hong, J. C., et al. (2012). Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol. Plant Microbe Interact. 25, 48–60. doi: 10.1094/MPMI-06-11-0165
Tang, M. J., Sun, J. W., Liu, Y., Chen, F., Shen, S. H. (2007). Isolation and functional characterization of the JcERF gene, a putative AP2/EREBP domain-containing transcription factor, in the woody oil plant Jatropha curcas. Plant Mol. Biol. 63, 419–428. doi: 10.1007/s11103-006-9098-7
Tian, Z. D., He, Q., Wang, H. X., Liu, Y., Zhang, Y., Shao, F., et al. (2015). The potato ERF transcription factor StERF3 negatively regulates resistance to Phytophthora infestans and salt tolerance in potato. Plant Cell Physiol. 56, 992–1005. doi: 10.1093/pcp/pcv025
Van der Does, D., Leon-Reyes, A., Koornneef, A., Van Verk, M. C., Rodenburg, N., Pauwels, L., et al. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcripti-on factor ORA59. Plant Cell 25, 744–761. doi: 10.1105/tpc.112.108548
Verhage, A., Van Wees, S. C., Pieterse, C. M. (2010). Plant immunity: it’s the hormones talking, but what do they say? Plant Physiol. 154, 536–540. doi: 10.1104/pp.110.161570
Vlot, A. C., Dempsey, D. A., Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, P. C., Du, Y. Y., Zhao, X. L., Miao, Y. C., Song, C. P. (2013). The MPK6-ERF6-ROS- responsive cis-acting Element7/GCC box complex modulat-es oxidative gene transcription and the oxidative response in Arabidopsis. Plant Physiol. 161, 1392–1408. doi: 10.1104/pp.112.210724
Ward, E. R., Uknes, S. J., Williams, S. C., Dincher, S. S., Wiederhold, D. L., Alexander, D. C., et al. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085–1094. doi: 10.1105/tpc.3.10.1085
Weems, J. D., Bradley, C. A. (2018). Exserohilum Turcicum Race Population Distribution in the North Central United States. Plant Dis. 102, 292–299. doi: 10.1094/PDIS-01-17-0128-RE
Welz, H. G., Geiger, H. H. (2000). Genes for resistance to northern corn leaf blig-ht in diverse maize populations. Plant Breed. 119, 1–14. doi: 10.1046/j.1439-0523.2000.00462.x
Wu, F. L., Shu, J. H., Jin, W. B. (2014). Identification and Validation of miRNAs Associated with the Resistance of Maize (Zea mays L.) to Exserohilum turcicum. PloS One 9, e87251. doi: 10.1371/journal.pone.0087251
Xie, Z. L., Nolan, T. M., Jiang, H., Yin, Y. H. (2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 10, 228. doi: 10.3389/fpls.2019.00228
Xu, Z. S., Xia, L. Q., Chen, M., Cheng, X. G., Zhang, R. Y., Li, L. C., et al. (2007). Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 65, 719–732. doi: 10.1007/s11103-007-9237-9
Yang, Z., Tian, L. N., Latoszek-Green, M., Brown, D., Wu, K. Q. (2005). Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 58, 585–596. doi: 10.1007/s11103-005-7294-5
Yang, L., Li, B., Zheng, X. Y., Li, J., Yang, M., Dong, X., et al. (2015). Salicylic acid biosynthesis is enhanced and contributes to increased biotrophic pathogen resistance in Arabidopsis hybrids. Nat. Commun. 6, 7309. doi: 10.1038/ncomms-8309
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zarei, A., Körbes, A. P., Younessi, P., Montiel, G., Champion, A., Memelink, J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 75, 321–331. doi: 10.1007/s11103-010-9728-y
Zhai, Y., Wang, Y., Li, Y. J., Lei, T. T., Yan, F., Su, L. T., et al. (2013). Isolation and molecular characterization of GmERF7, a soybean ethylene-response factor thatincreases salt stress tolerance in tobacco. Gene 513, 174–183. doi: 10.1016/j.gene.2012.10.018
Zhang, H. W., Huang, Z. J., Xie, B. Y., Chen, Q., Tian, X., Zhang, X. L., et al. (2004). The ethylene-, jasmonate-, abscisic acid-, and NaCl-responsive tomato transcrip-tion factor JERF1 modulates expression of GCC box-containing genes and salt tolerance intobacco. Planta 220, 262–270. doi: 10.1007/s00425-004-1347-x
Zhang, G. Y., Chen, M., Li, L. C., Xu, Z. S., Chen, X. P., Guo, J. M., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60, 3781–3796. doi: 10.1093/jxb/erp214
Zhang, H. W., Zhang, J. F., Quan, R. D., Pan, X. W., Wan, L. Y., Huang, R. F. (2013). EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta 237, 1443–1451. doi: 10.1007/s004-25-013-1852-x
Zhang, H. J., Huang, L., Dai, Y., Liu, S. X., Hong, Y. B., Tian, L. M., et al. (2015). Arabidopsis AtERF15 positively regulates immunity against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea. Front. Plant Sci. 6, 686. doi: 10.3389/fpls.2015.00686
Zhang, W., Corwin, J. A., Copeland, D., Feusier, J., Eshbaugh, R., Chen, F., et al. (2017). Plastic transcriptomes stabilize immunity to pathogen diversity: the jasmonic acid and salicylic acid networks within the Arabidopsis/Botrytis pathosystem. Plant Cell 29, 2727–2752. doi: 10.1105/tpc.17.00348
Zhao, Y. L., Chang, X., Qi, D. Y., Dong, L. D., Wang, G. J., Fan, S. J., et al. (2017). A novel soybean ERF transcription factor, GmERF113, increases resistance to Phytophthora sojae infection in soybean. Front. Plant Sci. 8, 299. doi: 10.3389/fpls.2017.00299
Zheng, X., Xing, J. H., Zhang, K., Pang, X., Zhao, Y. T., Wang, G. Y., et al. (2019). Ethylene response factor ERF11 activates BT4 transcription to regulate immunity to Pseudomonas syringae. Plant Physiol. 180, 1132–1151. doi: 10.1104/pp.18.01209
Keywords: maize, E. turcicum, ethylene response factor, ZmERF105, transcription factor
Citation: Zang Z, Lv Y, Liu S, Yang W, Ci J, Ren X, Wang Z, Wu H, Ma W, Jiang L and Yang W (2020) A Novel ERF Transcription Factor, ZmERF105, Positively Regulates Maize Resistance to Exserohilum turcicum. Front. Plant Sci. 11:850. doi: 10.3389/fpls.2020.00850
Received: 06 February 2020; Accepted: 27 May 2020;
Published: 16 June 2020.
Edited by:
Víctor Flors, University of Jaume I, SpainReviewed by:
Jorge Vicente, University of Nottingham, United KingdomM. Teresa Sanchez-Ballesta, Instituto de Ciencia y Tecnología de Alimentos y Nutrición (ICTAN), Spain
Copyright © 2020 Zang, Lv, Liu, Yang, Ci, Ren, Wang, Wu, Ma, Jiang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangyu Jiang, TGlhbmd5dTAxMTNAMTYzLmNvbQ==; Weiguang Yang, WXdnNzg5QDEyNi5jb20=
†These authors have contributed equally to this work
 Zhenyuan Zang1†
Zhenyuan Zang1† Liangyu Jiang
Liangyu Jiang