
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 19 June 2020
Sec. Plant Biotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00843
This article is part of the Research TopicBiofuels and BioenergyView all 14 articles
 Mitra Mazarei1,2,3
Mitra Mazarei1,2,3 Holly L. Baxter1,2
Holly L. Baxter1,2 Avinash Srivastava2,4
Avinash Srivastava2,4 Guifen Li2,4
Guifen Li2,4 Hongli Xie2,4
Hongli Xie2,4 Alexandru Dumitrache2,5
Alexandru Dumitrache2,5 Miguel Rodriguez Jr.2,3,5
Miguel Rodriguez Jr.2,3,5 Jace M. Natzke2,5
Jace M. Natzke2,5 Ji-Yi Zhang2,4
Ji-Yi Zhang2,4 Geoffrey B. Turner2,6
Geoffrey B. Turner2,6 Robert W. Sykes2,6
Robert W. Sykes2,6 Mark F. Davis2,3,6
Mark F. Davis2,3,6 Michael K. Udvardi2,3,4
Michael K. Udvardi2,3,4 Zeng-Yu Wang2,4
Zeng-Yu Wang2,4 Brian H. Davison2,3,5
Brian H. Davison2,3,5 Elison B. Blancaflor2,4
Elison B. Blancaflor2,4 Yuhong Tang2,4*
Yuhong Tang2,4* Charles Neal Stewart Jr.1,2,3*
Charles Neal Stewart Jr.1,2,3*Switchgrass (Panicum virgatum L.) is a lignocellulosic perennial grass with great potential in bioenergy field. Lignocellulosic bioenergy crops are mostly resistant to cell wall deconstruction, and therefore yield suboptimal levels of biofuel. The one-carbon pathway (also known as C1 metabolism) is critical for polymer methylation, including that of lignin and hemicelluloses in cell walls. Folylpolyglutamate synthetase (FPGS) catalyzes a biochemical reaction that leads to the formation of folylpolyglutamate, an important cofactor for many enzymes in the C1 pathway. In this study, the putatively novel switchgrass PvFPGS1 gene was identified and its functional role in cell wall composition and biofuel production was examined by RNAi knockdown analysis. The PvFPGS1-downregulated plants were analyzed in the field over three growing seasons. Transgenic plants with the highest reduction in PvFPGS1 expression grew slower and produced lower end-of-season biomass. Transgenic plants with low-to-moderate reduction in PvFPGS1 transcript levels produced equivalent biomass as controls. There were no significant differences observed for lignin content and syringyl/guaiacyl lignin monomer ratio in the low-to-moderately reduced PvFPGS1 transgenic lines compared with the controls. Similarly, sugar release efficiency was also not significantly different in these transgenic lines compared with the control lines. However, transgenic plants produced up to 18% more ethanol while maintaining congruent growth and biomass as non-transgenic controls. Severity of rust disease among transgenic and control lines were not different during the time course of the field experiments. Altogether, the unchanged lignin content and composition in the low-to-moderate PvFPGS1-downregulated lines may suggest that partial downregulation of PvFPGS1 expression did not impact lignin biosynthesis in switchgrass. In conclusion, the manipulation of PvFPGS1 expression in bioenergy crops may be useful to increase biofuel potential with no growth penalty or increased susceptibility to rust in feedstock.
The addition or removal of one-carbon units (C1 metabolism) is required for the synthesis and regulation of many biological compounds and metabolic processes. The synthesis of basic compounds and a range of methylated molecules in all organisms requires C1 metabolism (Appling, 1991). In plants, a balanced supply of C1 units is required for the synthesis of numerous plant secondary metabolites (e.g., lignin and phytohormones) and hemicellulose (Hanson and Roje, 2001; Eckardt, 2007; Urbanowicz et al., 2012). As the major sink of methyl units, lignin biosynthesis is affected by changes of enzymes in the C1 pathway (Shen et al., 2002; Tang et al., 2014; Li et al., 2015). Folylpolyglutamate synthetase (FPGS) catalyzes a biochemical reaction that leads to the formation of folylpolyglutamate, an important cofactor for many enzymes in the C1 pathway (Shane, 1989). In plant cells, folates, mainly as polyglutamylated byproducts, are found in chloroplasts, mitochondria, and cytosol (Rébeillé et al., 2006; Hanson and Gregory, 2011). In Arabidopsis, there are three genes each encoding FPGS isoforms that are localized to chloroplasts (FPGS1), mitochondria (FPGS2), and cytosol (FPGS3) (Ravanel et al., 2001).
Switchgrass (Panicum virgatum L.) is a lignocellulosic perennial grass known for its high yield of biomass, wide adaptability, and ability to grow on marginal soil conditions. These characteristics have made switchgrass a promising bioenergy feedstock (van der Weijde et al., 2013). One of the major problems with lignocellulosic crops is the resistance of the cell wall to deconstruction for efficient conversion into biofuels (known as biomass recalcitrance) (Himmel and Bayer, 2009). Cell wall lignin is one of the main causes of recalcitrance, which limits efficient conversion of biomass into biofuels (Chen and Dixon, 2007; David and Ragauskas, 2010). Cell wall components have been key targets to reduce feedstock recalcitrance; manipulating cell wall biosynthesis gene expression has been the primary strategy (Nelson et al., 2017; Biswal et al., 2018; Brandon and Scheller, 2020). Several studies with transgenic plants have been conducted in greenhouses under tightly controlled environmental conditions and the actively growing green tissue has been analyzed most often (Fu et al., 2011a, b, 2012; Xu et al., 2011; Shen et al., 2012, 2013; Liu et al., 2018). In contrast, end-of-season senesced tissue is most often used for biofuel production. Thus, greenhouse experiments may not be predictive of transgenic plants performance and recalcitrance of field-grown plants. Field-grown plants are exposed to a wider range of biotic and abiotic stresses not present in the greenhouse. Therefore, field experiments are especially important for modified plants to better predict agronomic performance across multiple growing seasons.
We previously showed that reduced lignin content and improved cell wall digestibility was observed in an Arabidopsis mutant with a disrupted FPGS1 gene (Srivastava et al., 2015). We also showed that disruption in both FPGS1 and caffeoyl-CoA-3-O-methyltransferase (CCoAOMT), a lignin biosynthetic enzyme, resulted in further reduction in lignin content and improvement in cell wall digestibility in Arabidopsis (Xie et al., 2019). These studies prompted us to examine the possible role of FPGS in improving biofuel production for switchgrass.
In the present study, a novel switchgrass PvFPGS1 gene was identified and its functional role was examined by downregulation using RNAi technology in switchgrass. A field experiment using multiple transgenic switchgrass lines downregulating PvFPGS1 was conducted for three field growing seasons (2014–2016) to evaluate (i) PvFPGS1 transcript levels, (ii) growth traits and biomass production, (iii) cell wall composition, sugar release, and conversion into biofuel, and (iv) susceptibility to rust disease.
Using the FPGS cDNA sequence of Arabidopsis AtFPGS1 (At5g05980), TBLASTN was used to identify the homologous gene sequences from switchgrass EST databases (Zhang et al., 2013) as well as from the draft switchgrass genome (Panicum virgatum v1.1 DOE-JGI) at Phytozome. A FPGS family gene tree was originally constructed using neighbor-joining in the software MEGA6 (Tamura et al., 2013) and a potential isolog of AtFPGS1 was identified from switchgrass. A phylogeny tree for FPGS protein family was constructed by neighbor-joining using Geneious Prime 2019 software (www.geneious.com).
The RNAi construct was made using the switchgrass PvFPGS1 gene sequence (Pavir.Ib00114). A 462 bp sequence (Supplementary Figure S1) was amplified by PCR from switchgrass cultivar ‘Alamo’ using primer pair PvDFB-RNAi-F: 5′-AAGCAGGGGCATAAGGACA-3′ and PvDFB-RNAi-R: 5′-ATCGATTTGTTCAGGCTCAGC-3′. The target fragment was cloned into pCR8 entry vector and confirmed by sequencing. The target fragment was then sub-cloned into pANIC-8A RNAi-vector (Supplementary Figure S2) (Mann et al., 2012) to be driven under the maize ubiquitin 1 (ZmUbi1) promoter. Transgenic plants were produced using NFCX01 clonal of switchgrass ‘Alamo’ via Agrobacterium-mediated transformation (Xi et al., 2009).
Plants were grown in greenhouse under the 16-h day/8-h night light at 28°C day/22°C night temperature.
The T0 generation of PvFPGS1-downregulated plants were used in field experiments. The plants included six independent transgenic lines and one non-transgenic control (wild type). The plants were transplanted onto a field on June 05, 2014. The field was located at the University of Tennessee Plant Sciences Unit of the East Tennessee Research and Education Center (ETREC). The field site was 24.2 m × 15.1 m. Three replicate plots for each transgenic and control lines were distributed throughout the field in a randomized complete block design (RCBD). Each transgenic and control replicate plot contained four vegetatively propagated clones of each line. Replicate lines were spaced 152 cm apart with 76 cm between the four clonal plants within every single replicate. The experimental plots were surrounded by non-transgenic border plants (Supplementary Figure S3).
The field trial was conducted for three consecutive growing seasons, in which no environmental anomalies were observed. The soil fertility was in the range of switchgrass recommendations and no soil amendments were added. The plants were irrigated only for the first 2 months as needed after transplantation for establishment. Weeds were removed by tilling or hand. No herbicides were applied for the duration of the study. Following USDA APHIS BRS guidelines, plants were observed daily during the reproductive stage and emerging panicles were removed from all plants (transgenic, non-transgenic control and border plants) at R0-R1 developmental stage (Moore et al., 1991) by cutting the plant below the top node containing the inflorescence.
Baseline expression of FPGS1 in different tissues of switchgrass at the R1 growth stage was tested in greenhouse-grown plants using quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Total RNA was extracted from 1st, 2nd, 3rd, and 4th internodes; 1st, 2nd, 3rd, and 4th nodes; leaf blade; leaf sheath; crown; inflorescence; and root of non-trangenic lines or from the leaf blade, leaf sheath, and 2nd internode of transgenic lines at the R1 growth stage. For field-grown plants, samples were collected from green plants in August of each growing season. All samples were collected at the same date and time of each year analyzed. Tillers at the R0 developmental stage were chosen at random from two plants within each replicate. Each tiller was excised below the top internode. The resulting top portion of the tiller with the two intact top leaves was flash-frozen in liquid nitrogen and stored at −80°C for qRT-PCR analysis. Total RNA was isolated from the frozen tissues using SpectrumTM Plant Total RNA Kits (Sigma-Aldrich, St. Louis, MO, United States) following the manufacturer’s instructions. RNA quality was checked with Agilent Bioanalyzer 2100 (Agilent, Palo Alto, CA, United States) and quantified using QubitTMRNA BR Assay Kit (Fisher Scientific, Santa Clara, CA, United States). Five micrograms of total RNA was treated with TURBO DNA-freeTM Kit (Invitrogen, Carlsbad, CA, United States) to remove any potential genomic DNA contaminants. Two micrograms of DNA-free total RNA was used for first-strand cDNA synthesis using SuperScriptTM III First-Strand Synthesis System (Invitrogen). qRT-PCR was performed with Power SYBRTM Green PCR Master Mix (Applied Biosystems, Foster City, CA, United States) using ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). The primer pair used for the PvFPGS1 transcript analysis was PvDFB-RNAi-qRT-F1: 5′-CAAAGAGCTTCGGAGTTGG-3′ and PvDFB-RNAi-qRT-R1: 5′-GGTAGGGGATCAGTACGATTGA-3′. Data were collected and analyzed using the SDS 2.2.1 software (Applied Biosystems). The relative transcript quantification was normalized by the levels of switchgrass ubiquitin 1 (PvUbi1) transcripts (Shen et al., 2009) using primer pair SWUbi_F304: 5′-TTCGTGGTGGCCAGTAAGC-3′ and SWUbi_R367: 5′-AGAGACCAGAAGACCCAGGTACAG-3′.
Growth measurements were recorded each December (end-of-season) of the three growing seasons in the field after all aboveground biomass was completely senesced. For tiller height, the tallest tiller from each individual plant was measured from soil level to the tip of the top leaf. For plant width, the circumference at the mid-section of each whole plant was measured. Tiller numbers were tallied for each plant. For biomass yield, whole aboveground senesced biomass was harvested. The biomass was oven-dried at 43°C for 96 h and weighed to determine total dry aboveground biomass. The dried biomass samples were chipped into 5–8 pieces prior to milling. The chipped samples were milled with a Wiley mill (Thomas Scientific, Model 4, Swedesboro, NJ, United States) through a 20-mesh screen (1.0 mm particle size). This milled biomass was used for cell wall characterization and bioconversion analyses.
Lignin content and composition were determined by pyrolysis molecular beam mass spectrometry (py-MBMS) using the NREL high-throughput method wherein soluble extractive and starch were removed from the biomass samples (Sykes et al., 2009; Decker et al., 2012). Lignin content was estimated from the relative intensities of the lignin precursor peaks. S/G lignin monomer ratio was determined by dividing the sum of the intensity of syringyl peaks by the sum of the intensity of guaiacyl peaks. Sugar release by enzymatic hydrolysis was determined using the NREL high-throughput method as previously described (Selig et al., 2010). Cell wall residues prepared by removing soluble extractives and starch were subjected to a hot water pretreatment (180°C for 17.5 min) followed by a 72-h incubation at 40°C with hydrolyzing enzymes. Glucose and xylose release were determined by colorimetric assays, and total sugar release is the sum of glucose + xylose released (Studer et al., 2009).
Ethanol yield was determined by separate hydrolysis and fermentation (SHF) as described previously (Dumitrache et al., 2017). Biomass samples were incubated at 50°C for 5 days with hydrolyzing enzymes. The resulting sugars were fermented at 35°C for 72 h with Saccharomyces cerevisiae D5α (ATCC 200062). Ethanol yield was determined at the endpoint by HPLC quantification (Bio-Rad, Hercules, CA, United States).
The whole tillers and internode of the greenhouse-grown plants at the R1 developmental stage were harvested for RNA-seq analysis as described previously (Rao et al., 2019). RNA-seq was conducted at Joint Genome Institute (JGI) using Illumina TruSeq technology. Four biological replicates for each sample group were included. For each sample of the PvFPGS1-downregulated and control lines, a total of 40–50 million paired-end (PE) reads of 150 bp was generated. Paired-end Illumina reads after filtering and trimming treatment were mapped to the Switchgrass genome Panicum virgatum v3.1 using HISAT2 (Kim et al., 2015) with default parameters. The selected genes were annotated with switchgrass genome v5.11. Genes whose expression was different from the control were selected through comparison between each transgenic line and its control, using differential analysis software such as DESeq (Anders and Huber, 2010) with default settings of adjusted P-value < 0.05. Genes were annotated against Arabidopsis, rice and other model species using blast search.
Plants were evaluated for rust disease caused by Puccinia novopanici (formerly known as Puccinia emaculata) infection. Disease severity was assessed at weekly time points during the second (2015) and third (2016) growing seasons between July and August as described by Baxter et al. (2018). Two plants within each replicate were selected at random. A single tiller from each plant was tagged and all leaves on the selected tillers were examined for rust severity. The coverage of the top leaf surface with rust uredia was visually evaluated using the following scale: 0 = 0%, 1 ≤ 5%, 2 ≤ 10%, 3 ≤ 25%, 4 ≤ 40%, 5 ≤ 55%, 6 ≤ 70%, and 7 ≤ 100%. Because of the severity of the rust, the whole field was treated with fungicide in late August of each growing season. All data reported were collected before fungicide treatments. Fungicides used included “Quilt” (Syngenta Canada Inc., Guelph, ON, Canada) at a rate of 0.21 ml/m2, and “Heritage” (Syngenta Crop Protection, Greensboro, NC, United States) at a rate of 20 ml/m2.
Means were analyzed with one-way ANOVA using Fisher’s least significant difference method in SAS version 9.4 (SAS Institute Inc., Cary, NC, United States). Differences were considered statistically significant where P-values were less than 0.05.
Switchgrass FPGS (PvFPGS) was initially identified using the FPGS amino acid sequences from Arabidopsis thaliana (At5g05980). Analysis of the sequences showed that switchgrass assembly v1.1 has only three isoforms of FPGS: Pavir.Ib00114.1, Pavir.Ib03621.1, and Pavir.Ia04781.1. A phylogenetic tree was constructed using all three isoforms of switchgrass and Arabidopsis as well as FPGS variants from several other species. These species, Populus trichocarpa, Medicago truncatula, Oryza sativa, Zea mays, and Panicum hallii from Phytozome 12, served as reference to elucidate homologous relationship among different members of FPGSs. Based on homology analysis, Pavir.Ib00114 was identified and named PvFPGS1 (Figure 1). There are two other isoforms of FPGS in switchgrass genome: PvFPGS2 (Pavir.Ia04781.1) and PvFPGS3 (Pavir.Ib03621.1). Based on this whole genome level phylogeny analysis of FPGSs, the relationship of FPGSs between monocot and dicot is not as straightforward as one to one, especially AtFPGS3 had no isologs in other species. However, among all three FPGSs between Arabidopsis and switchgrass, AtFPGS1 is still closest to PvFPGS1 with an identity of 59.4%. Moreover, PvFPGS1 is isolog of maize brown midrib4 (bm4) which encode a functional FPGS and its loss-of-function leads to lower lignin content (Li et al., 2015).
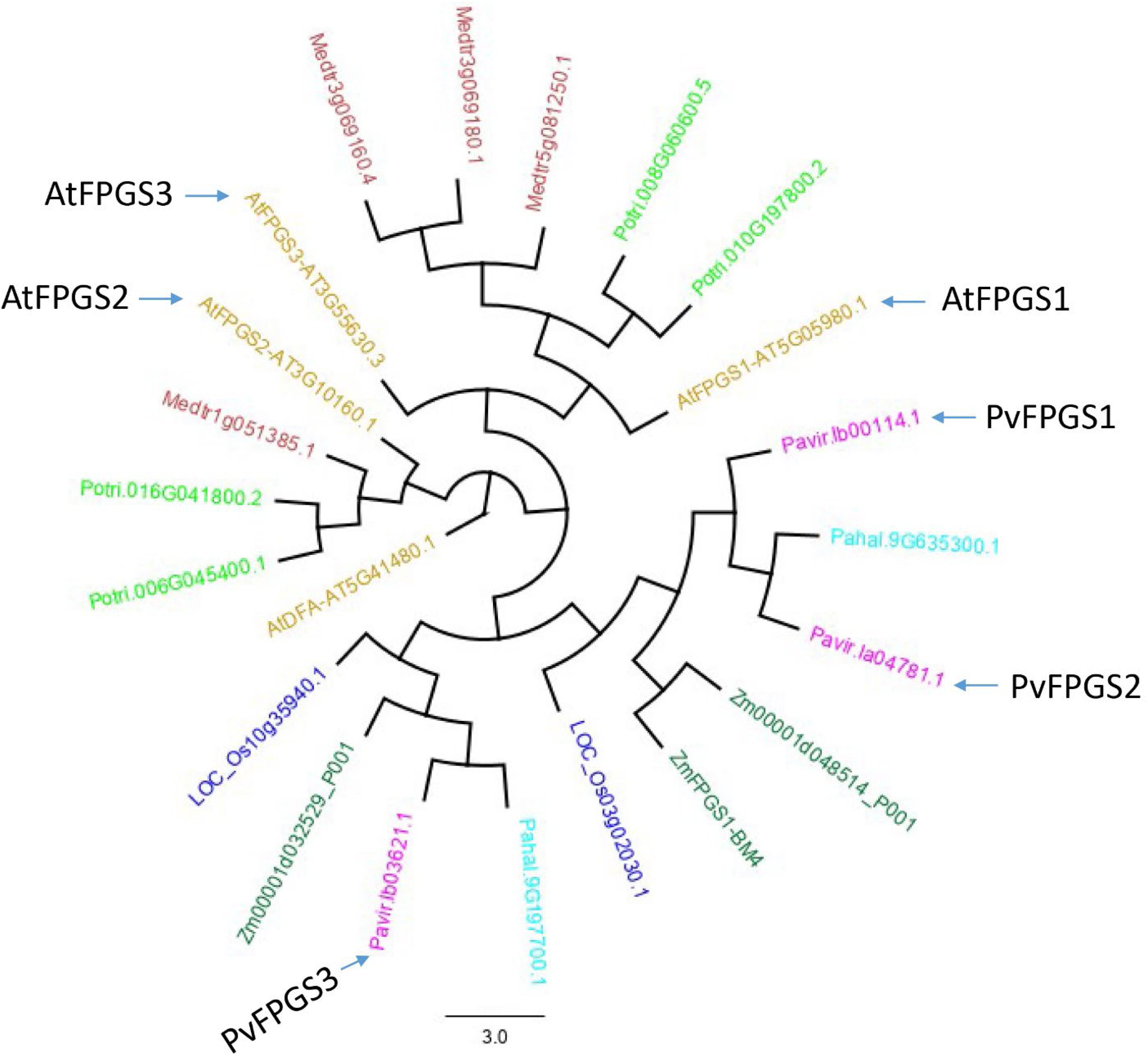
Figure 1. The gene tree of the FPGS protein family. Various members are shown by plant species: Arabidopsis thaliana (gold), Populus trichocarpa (light green), Medicago truncatula (brown), Oryza sativa (blue), Zea mays (green), Panicum hallii (light blue), and Panicum virgatum (pink) from Phytozome 12 (https://phytozome.jgi.doe.gov/) showing relationship based on amino acid sequences. The phylogenetic analysis shows that switchgrass genome has three isoforms of FPGS: PvFPGS1 (Pavir.Ib00114.1), PvFPGS2 (Pavir.Ia04781.1), and PvFPGS3 (Pavir.Ib03621.1). Based on this whole genome level comparison, the relationship of FPGSs between monocot and dicot is not as straightforward. However, among all three FPGSs in Arabidopsis and switchgrass, AtFPGS1 is still closest to PvFPGS1. In this graph, AtDFA encodes a dihydrofolate synthetase, homologous to FPGS but with different functions and serve as outgroup for this tree.
Transcript abundance via qRT-PCR analysis indicated that PvFPGS1 in non-transgenic plants is expressed in stems, leaves, crown, inflorescences, and roots at the R1 developmental stage. The level of PvFPGS1 expression was highest in the crown and leaves, and lowest in the root (Figure 2). PvFPGS1 transcripts were detected in all the tissue types tested. In the transgenic plants, depending on the transgenic event, PvFPGS1 transcript abundance was reduced by 23–82% in the joint leaf blade, leaf sheath, and 2nd internode tissues of the RNAi-transgenic lines (Supplementary Figure S4). Regardless of the level of reduction in PvFPGS1 expression, the growth and phenotype of transgenic lines was not apparently different from non-transgenic controls under greenhouse conditions (Figure 3).
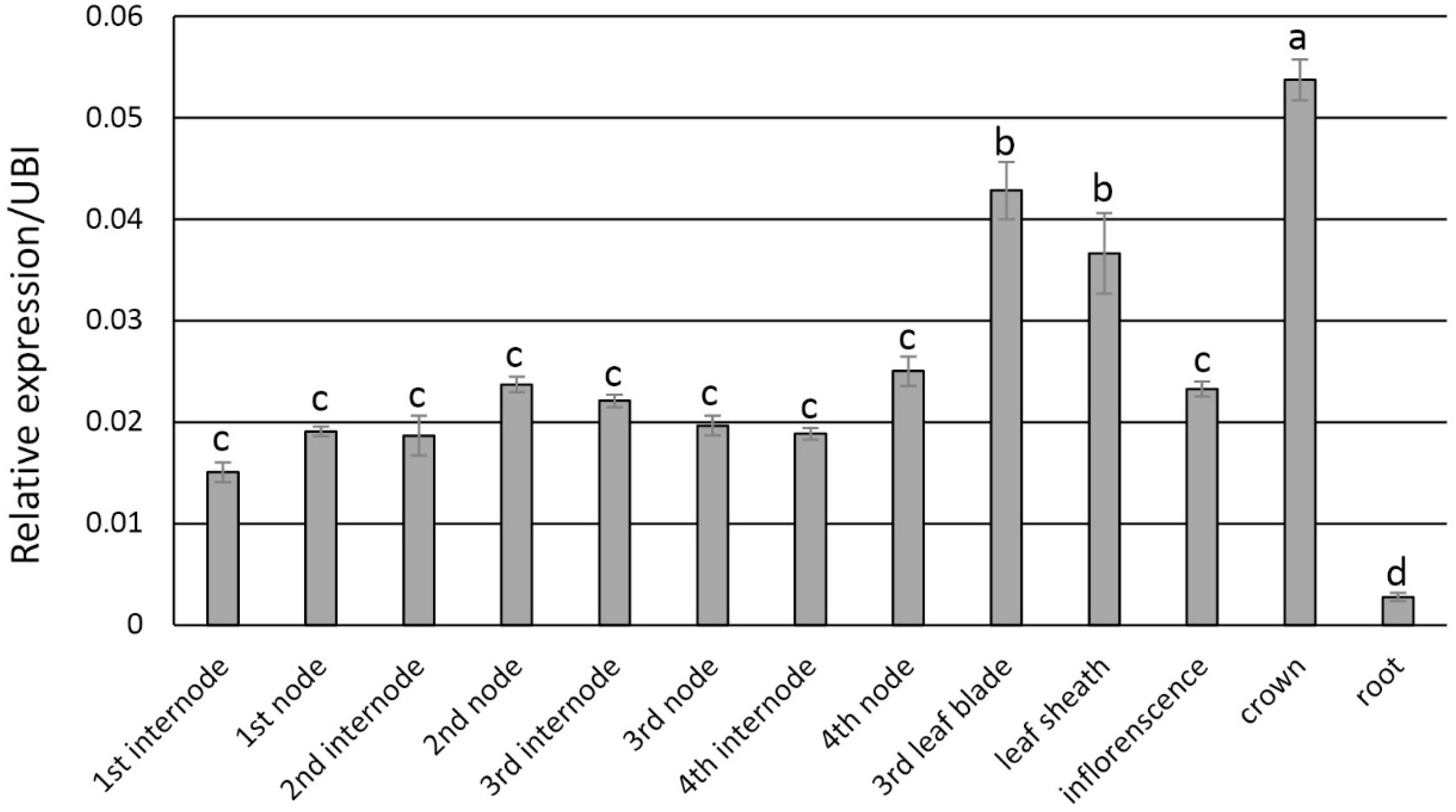
Figure 2. Expression patterns of PvFPGS1 in different plant tissues as determined by qRT-PCR. Plant samples for RNA extraction used in the qRT-PCR experiments were collected at R1 (reproductive stage 1) developmental stage. The relative levels of transcripts were normalized to the switchgrass ubiquitin 1 gene expression (UBI). Bars represent mean values of three biological replicates ± standard error. Means were compared by a one-way ANOVA and letter groupings were obtained using Fisher’s least significant difference method. Bars with different letters are significantly different at the 5% level.

Figure 3. Representative PvFPGS1-RNAi transgenic and non-transgenic control lines grown under greenhouse conditions at 3 months old.
From greenhouse studies, we selected six independent transgenic lines for the field experiment, which had a range of decreased PvFPGS1 expression. The plants included the transgenic lines T2 (decreased expression by 71%), T8 (decreased expression by 72%), T10 (decreased expression by 73%), T12 (decreased expression by 82%), T32 (decreased expression by 67%), T115 (decreased expression by 63%), and one non-transgenic control. The field study was conducted for three consecutive growing seasons (2014–2016) (Figure 4).

Figure 4. Photos of FPGS1-downregulated switchgrass in the field. (a) October 01, 2014; (b) November 21, 2014; (c) August 26, 2015; (d) December 08, 2015; (e) August 16, 2016; (f) November 17, 2016.
Transgene expression in the field-grown PvFPGS1-downregulated switchgrass lines was studied by qRT-PCR on sequentially harvested tissue over the course of the field study. PvFPGS1 transcript levels were reduced in the transgenic lines for field year one (2014), year two (2015), and year three (2016) compared to the control. The PvFPGS1 expression was lowest in transgenic line T8 with 80–89% decrease in transcript level compared to the control in years one, two, and three. The decrease in PvFPGS1 transcript level was followed by transgenic lines T2 (67–83%), T10 (76–81%), T12 (78–81%), T32 (72–77%), and T115 (by 58–86%) relative to the control during the three growing seasons (Figure 5).
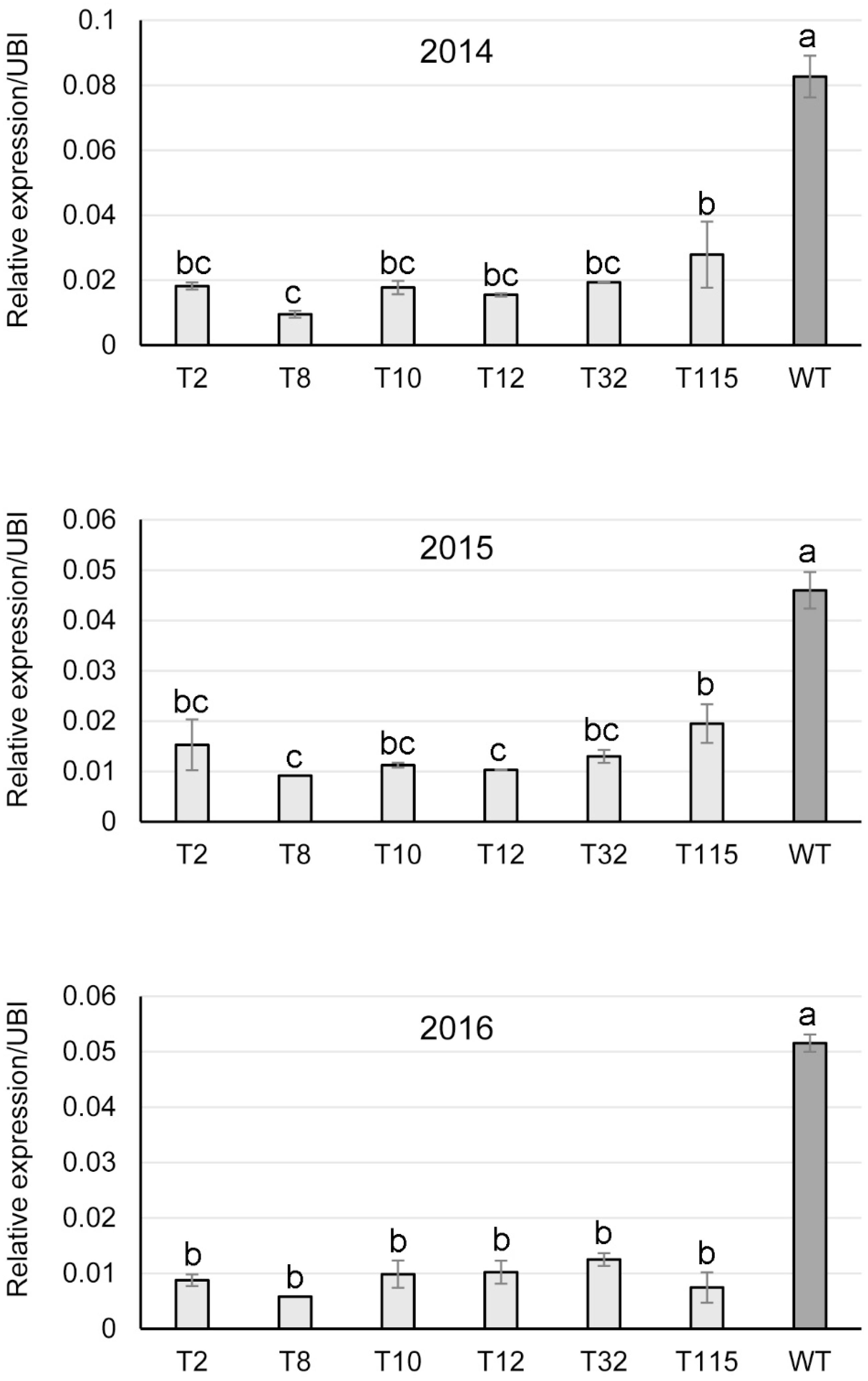
Figure 5. Relative transcript levels of PvFPGS1 in RNAi-transgenic lines as determined by qRT-PCR. Samples were collected in year one (2014), year two (2015) and year 3 (2016) of the field trial. WT: non-transgenic control. The relative levels of transcripts were normalized to the switchgrass ubiquitin 1 gene expression (UBI). Bars represent mean values of three biological replicates ± standard error, with the exception of line T8 in 2015 and 2016, which is from one surviving plant during the two growing seasons. Means within each year were compared by a one-way ANOVA and letter groupings were obtained using Fisher’s least significant difference method. Bars with different letters are significantly different at the 5% level.
For each growing season, the following end-of-year growth characteristics were assessed: tiller height, plant width, tiller number, and aboveground dry biomass. Biomass production of transgenic lines T10, T12, and T32 was comparable to that of the control in the first year (2014), while lines T2 (decreased biomass by 45%), T8 (decreased biomass by 81%), and T115 (decreased biomass by 62%) exhibited yield reduction. The yield reductions in these lines were congruent with decreased tiller height, plant width, and tiller number by up to 56%. In the second year (2015), the transgenic lines showed similar biomass yield and growth traits to that of the control with the exception of line T8, in which only one plant (out of 12 plants) survived the first field winter. The sole T8 survival had decreased biomass by 95% accompanied by up to 69% decrease in tiller height, plant width, and tiller number compared to the control. Similar to the second year, transgenic lines did not differ from the control in biomass production in the third year (2016), except for the T8 surviving plant which had a 99% decrease in biomass yield accompanied by up to 94% reduction in tiller height, plant width, and tiller number relative to the control (Table 1).
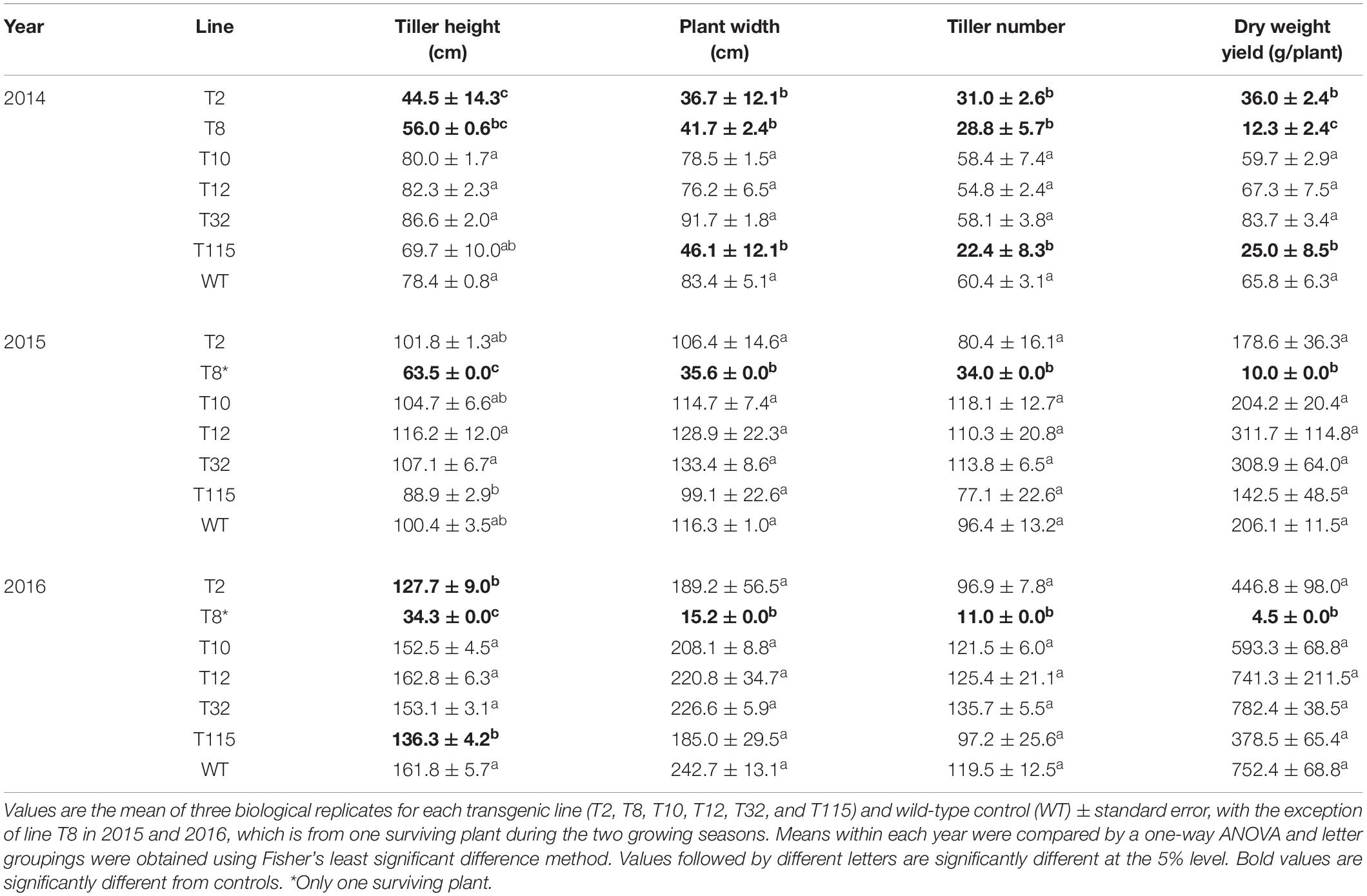
Table 1. Morphology and dry weight yield of PvFPGS1-downregulated lines at the first (2014), second (2015), and third (2016) growing seasons per plant.
Cell wall lignin content and the S/G ratio were measured for aboveground biomass harvested at end-of-season of each year by pyrolysis molecular beam mass spectrometry (py-MBMS). Decrease in lignin content (14% reduction) and S/G ratio (18% reduction) were only observed in line T8 relative to the control in year one (2014). There were no significant differences in lignin content or S/G ratio between all the other transgenic lines and the control lines at the first (2014), second (2015), and third (2016) growing seasons (Table 2).
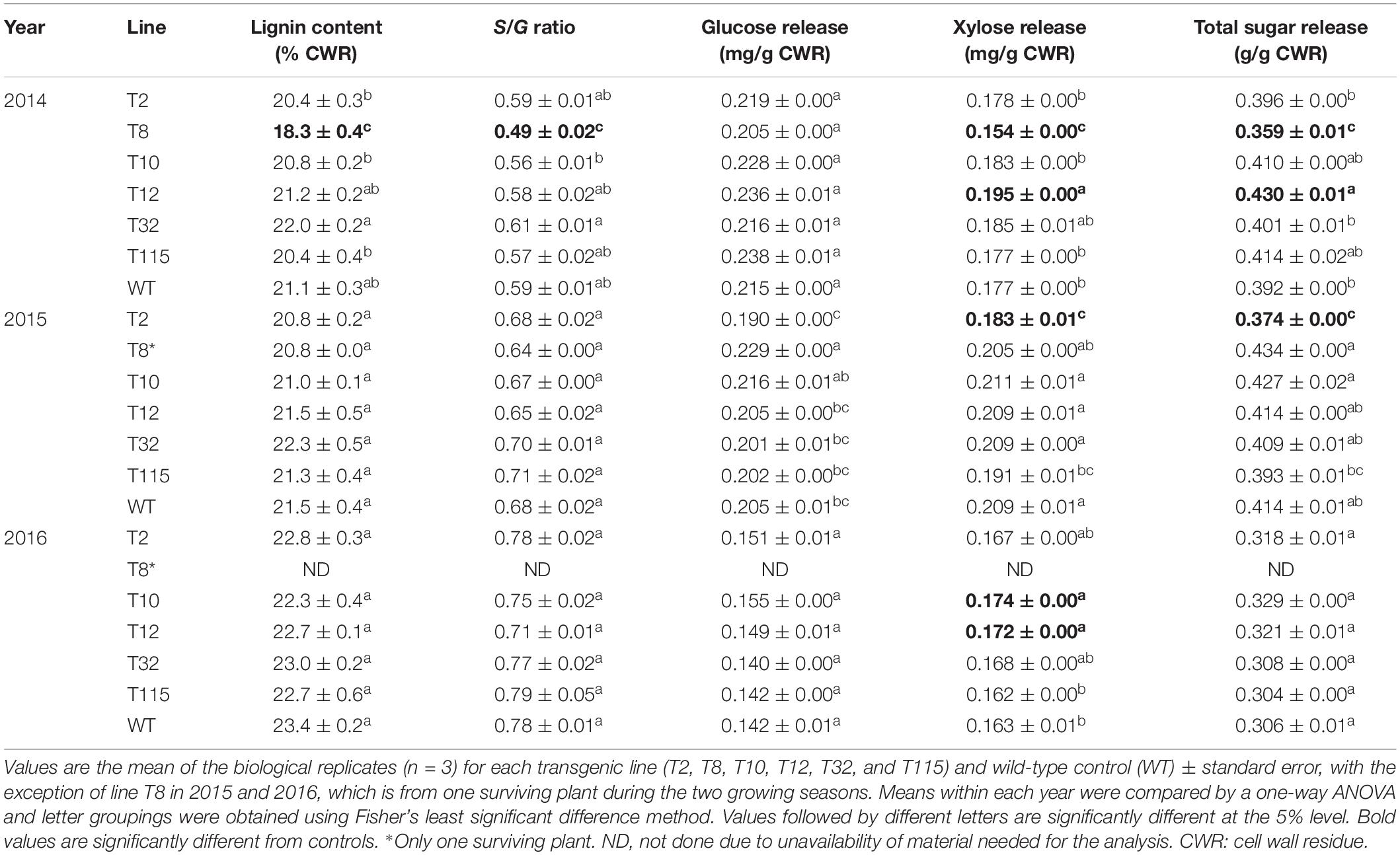
Table 2. Lignin content, S/G ratios, and sugar release of PvFPGS1-downregulated lines at the first (2014), second (2015), and third (2016) growing seasons.
Enzymatic hydrolysis was used to determine the sugar release for aboveground biomass harvested at end of each season. In year one (2014), line T8 exhibited a decrease in xylose (13%) and total sugar (8%) release relative to the control, whereas line T12 had a 10% higher xylose and total sugar release than control. In year two (2015), a decrease in xylose (12%) and total sugar (10%) release was only observed in line T2. In year three (2016), there was an increase in xylose release in lines T10 (7%) and T12 (5%). There were no significant differences in sugar release between the other transgenic and the control lines at the first (2014), second (2015), and third (2016) growing seasons (Table 2).
The fermentation potential of aboveground biomass harvested at end-of-season of each year was determined by separate hydrolysis and fermentation (SHF). Lines T2 and T10 with moderate levels of the decreased PvFPGS1 expression were selected to evaluate bioconversion efficiency. Line T10 showed an increase in ethanol yield in year one (2014) by 7%, year two (2015) by 18%, and year three (2016) by 9% relative to the control. There were no differences in ethanol yield between line T2 and the control at the three growing seasons (Figure 6).
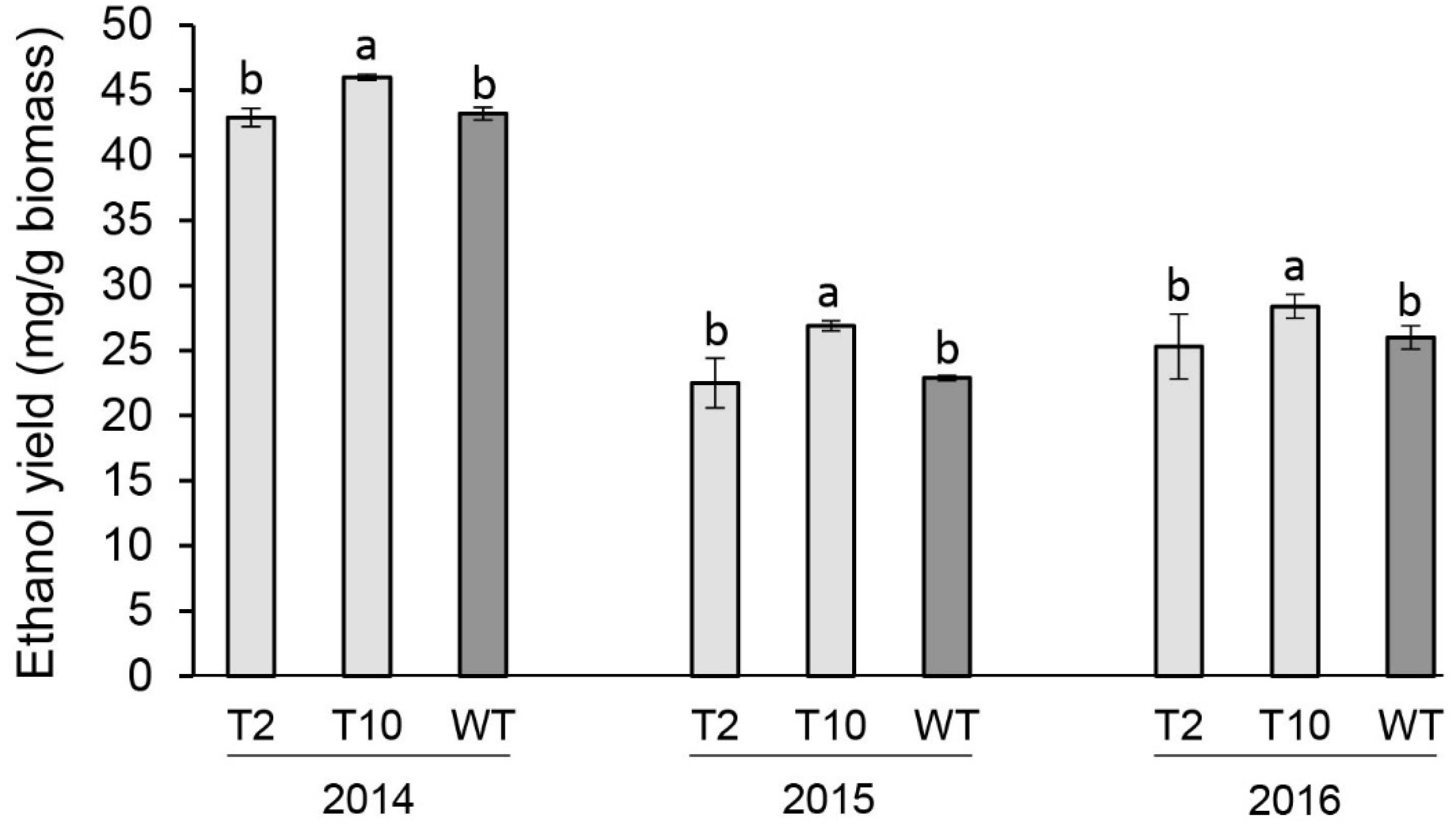
Figure 6. Ethanol yields of FPGS1-downregulated switchgrass in the first (2014), second (2015), and third (2016) growing seasons. Bars are the mean value of three biological replicates for each transgenic line (T2 and T10) and wild-type control (WT) ± standard error. Means within each year were compared by a one-way ANOVA and letter groupings were obtained using Fisher’s least significant difference method. Bars with different letters are significantly different at the 5% level.
Transcriptome analysis by RNA-seq was conducted to identify underlying pathways through which modification may further improve biofuel production. In both tiller and internode, the number of putative genes of different expression in the PvFPGS1-downregulated line T10 compared with control was similar, with 148 higher and 145 lower in tiller while 134 higher and 167 lower in the internode (Supplementary Table S1). FPGS1 has two subtypes presented in the annotated switchgrass genome, one in K genome and one in the N genome, each was named based on their subgenome, namely FPGS1K (Pavir.9KG559800 V5.1, originally Pavir.Ib00114.1 in V1.1), and FPGS1N (Pavir.9NG789900 V5.1, originally Pavir.Ia04781.1 in V1.1). Sequence identity between these two subtypes is 96.7%, and both were significantly reduced to 20–30% of the control levels, indicating these target genes were successfully downregulated in line T10. Reads for switchgrass FPGS2 is not well mapped in this RNA-seq data and its expression pattern will be worth looking at in future study. In total, there were 565 genes that were differentially expressed in line T10. Based on gene annotation, 38 differentially expressed genes were putatively involved in activities related to plant cell wall (Supplementary Table S2). Among them, in addition to the FPGS1 target genes, seven genes encode enzymes in the flavonoid and aromatic amino acids synthesis pathways which directly interact with the phenylpropanoid pathway, the upstream of lignin biosynthesis pathway. These genes include chalcone synthase (CHS) (Pavir.8NG107400 and Pavir.8KG261900), chalcone isomerase (Pavir.9NG037000), chorismate mutase (Pavir.1KG086400), isochorismate synthase (Pavir.2KG294000), anthocyanidin reductase (Pavir.7NG368500), and isoflavone 7-O-glucosyltransferase (Pavir.9NG095000). Peroxidase and laccase are enzymes involved in lignin polymerization and deposition (Tobimatsu and Schuetz, 2019). Three peroxidase (Pavir.2KG513700, Pavir.2NG638600, and Pavir.7NG321900) and three laccase genes (Pavir.5KG623300, Pavir.5NG585100, and Pavir.5KG613400) are among the selected genes. One gene potentially involved in monolignol metabolism, monolignol beta-glucoside homolog without catalytic acid/base, also present in the selected gene list (Pavir.7NG350700). Ten genes involved in different steps of some type of sugar and wall polysaccharide synthesis and modification are also among the selected genes. At the upstream of the pathway, two genes involved in myo-inositol metabolism have reduced expression in line T10, myo-inositol-1-phosphate synthase (Pavir.9KG643000) and myo-inositol 2-dehydrogenase (Pavir.2NG256100). Following these steps, four UDP nucleotide sugar metabolism enzymes are also among the mostly downregulated gene list: UDP-arabinose 4-epimerase (Pavir.9KG554500), UDP-arabinopyranose mutase 1 related (Pavir.9NG214400), UDP-glycosyltransferase 73B4 (Pavir.7NG278900), UDP-glucuronic acid decarboxylase (Pavir.9KG487800). Genes involved in pectin and hemicellulose also are among genes of changed expression in line T10. Among them are polygalacturonate 4-alpha-galacturonosyltransferase (GAUT4) (Pavir.2KG409000), cellulose synthase (CESA8) (Pavir.2KG167600), licheninase/mixed linkage beta-glucanase (Pavir.3NG120500); three glycotransferase (Pavir.5NG133600, Pavir.1KG262700, and Pavir.4KG337800) are all downregulated in line T10; the last group of genes on the list that are directly involved in cell wall structure are five wall associated proteins: four hydroxyproline-rich glycoprotein family protein (Pavir.2KG047700, Pavir.2KG047500, Pavir.3KG332300, and Pavir.9NG522300); the other one is wall-associated kinase 2 (Pavir.7NG318000). One interesting gene that is downregulated in the internode of line T10 encodes an EamA-like transporter family member (Pavir.5KG129300), homology to nodulin (MtN21) and walls are thin 1 (WAT1), which is a tonoplast-localized protein required for secondary wall formation in fibers (Ranocha et al., 2010).
All plants were evaluated for rust disease severity during the second (2015) and third (2016) growing seasons. Rust disease was detected in late July and infection was advanced largely through late August for both growing seasons. In order to maintain the field for downstream analyses, the field was sprayed with fungicides. Disease severity of rust infection was rated weekly prior to fungicide treatments based on the percentage of the leaf area coverage with rust uredia. The disease severity ranged from 0 to 9% during the second (2015) and 4–18% during the third (2016) growing seasons. There were no significant differences in rust susceptibility between transgenic and control lines for both growing seasons (Supplementary Figures S5, S6).
A clear understanding of cell wall enzymology is needed to engineer reduced recalcitrance in bioenergy crops. We previously reported that the loss of function of Arabidopsis FPGS1 resulted in lignin reduction and improved saccharification efficiency in Arabidopsis (Srivastava et al., 2015; Xie et al., 2019). Given the link between the FPGS and lignin biosynthetic pathways, our findings gave rise to the hypothesis that FPGS plays a functional role in reducing recalcitrance in switchgrass, a leading lignocellulosic bioenergy feedstock. The present study describes identification of switchgrass FPGS gene (PvFPGS1). Our study strengthens the FPGS-recalcitrance hypothesis.
In Arabidopsis, there are three genes that each encode FPGS isoforms: FPGS1, FPGS2, and FPGS3 (Ravanel et al., 2001). The homologous sequence in switchgrass discovered using AtFPGS1 was named PvFPGS1. At whole genome level, PvFPGS1 has two subtypes, each belong to the N and K subgenome, they show similarity closer to AtFPGS1 and AtFPGS3, farther from AtFPGS2.
Endogenous expression of PvFPGS1 transcripts was highest in the crown of plants. Transcript abundance decreased sequentially in leaves, internodes, inflorescences, and roots. The expression profile of PvFPGS1 is slightly different from Arabidopsis as the expression profile of Arabidopsis AtFPGS1 showed the highest expression in stems compared to other tissues (Srivastava et al., 2015). Secondary cell walls consisted of high amount of lignin provide much of the rigidity in stem tissue, in contrast to the flexible organs such as roots. Consistently, expression of secondary cell wall-related genes has been shown to be higher in stems (Zhao and Bartley, 2014; Mazarei et al., 2018; Rao et al., 2019). Noteworthy, other than inflorescence, the tissues of high PvFPGS1 expression in switchgrass are all significantly lignified (Crowe et al., 2017).
All transgenic PvFPGS1-downregulated lines had reduced targeted transcript levels over the three growing seasons of the field trial. These results confirm that the PvFPGS1-downregulated lines grown under field conditions sustained the reduction in PvFPGS1 transcript levels compared to the control. For most of transgenic lines, plant growth was comparable to the control plants. However, in the field, the transgenic PvFPGS1-downregulating line T8, which had the highest decrease in PvFPGS1 transcript levels (up to 89%) among lines either did not survive or had up to 99% reduction in biomass production over the control. The reduced biomass yield was accompanied with significant decrease in tiller height, plant width, and tiller number relative to the control. Yet, the transgenic line T8, which had the highest reduction in PvFPGS1 expression had normal growth under greenhouse conditions. These observations that the transgenic lines with normal plant growth and development under greenhouse conditions could not survive or had substantial biomass loss under field conditions emphasize the importance of performing field studies. Likewise, an association between levels of transgene expression and biomass production for switchgrass grown under field conditions has been shown for transgenic PvMYB4-overexpressing lines, where the transgenic lines with higher expression levels of PvMYB4 did not survive and/or had substantial reduction in biomass relative to control when grown under field conditions (Baxter et al., 2015). Yet, transgenic lines with low to moderate decreases in PvFPGS1 expression levels produced biomass yield comparable to that of the controls. These observations highlight the significance of an optimized level of gene expression in transgenic plants.
Lignin content and S/G ratios were not significantly different in low-to-moderate downregulated PvFPGS1 transgenic lines. Similarly, these transgenic lines showed similar sugar release efficiency compared with the control lines. Although our results are in contrast to Arabidopsis FPGS1 findings in which a loss of function was associated with reduced lignin content, coupled with an increase in sugar release efficiency (Srivastava et al., 2015; Xie et al., 2019), they used homozygous null-mutants in their studies. Similar observations were shown in maize where disruption in a gene encoding the isolog of PvFPGS1 resulted in low lignin (Li et al., 2015). We observed similar reductions in lignin in the T8 line where expression of FPGS1 was reduced to 80%. Cell wall lignin is one of the barriers to lignocellulosic biofuel production limiting digestion of cellulose into fermentable sugars. The manipulation of cell walls to decrease lignin content has been shown to improve bioconversion efficiency (Chen and Dixon, 2007; Baxter et al., 2014, 2015; Bonawitz et al., 2014; Eudes et al., 2014; Wilkerson et al., 2014; Hu et al., 2018). Since we did not find any noticeable difference in lignin content and composition in the selected PvFPGS1-downregulated lines, it is possible that the difference is very subtle and it is noticeable when we significantly disrupt the FPGS gene and associated supplies to the methylation pathway. This may also explain why there were not significant differences in sugar release efficiency between the selected PvFPGS1-downregulated lines and the controls.
Nevertheless, the PvFPGS1-downregulated line T10 produced up to 18% more ethanol than controls over the course of three field seasons. These observations may suggest that PvFPGS1 expression is associated with improved biofuel production. Transcriptome data provides some clues to the increased ethanol production in the transgenic line T10. In grass plant, lignin can be synthesized from both phenylalanine and tyrosine and it is tightly linked to other secondary metabolite pathways, and tightly regulated through metabolites flux (Recent review by Vanholme et al., 2019). The secondary metabolites that derived from the same upper biochemical pathways but branched off to produce different type of flavonoids including anthocyanidin2,3. Genes in several steps leading to the synthesis of anthocyanidin show affected expression in line T10 indicating that this metabolite pathway could be affected. Work has shown that disturbance in flavone synthase II in rice (Lam et al., 2017) and CHS in maize (Eloy et al., 2017) have changed lignin content and digestibility. Increased level of CHS in line T10 with increased ethanol production is in agreement with this evidence. Even though the lignin content was not significantly different in line T10, decreased transcript levels in several laccases and peroxidases hint that the level of polymerization of lignin could be reduced, which could contribute to its increased ethanol production.
Furthermore, among the enzymes involved in wall simple sugar to polysaccharide synthesis and wall modifications, a number of the genes show affected expression levels in line T10. Wall polysaccharide formed a network of cellulosic, hemicellulosic, and pectic polysaccharides and protein (Bashline et al., 2014) and changes are dynamic. Although not all expression changes will have consequential effects as the plants maintain normal growth and have similar sugar release, however, changes in some steps have led accumulated changes that have contributed to the increased ethanol productions in line T10. In rice, mutants of a putative glycosyltransferase in a grass-specific subfamily of GT61, are deficient in ferulic acid, coumaric acid, and aromatic compounds and exhibit an increased saccharification efficiency (Chiniquy et al., 2012). It is worth noting that one of the genes annotated as glycosyltransferase family 61 protein among the selected gene list. Lower expression of this gene could indirectly affect the level of these small metabolites and hence could contribute to the higher level of ethanol in line T10. The other gene that could be of significant effect is gene encoding licheninase/mixed linkage beta-glucanase. This enzyme could cause release of smaller oligosaccharides (DP < 6) from graminaceous hemicelluloses (Yoshida and Komae, 2006). Some smaller oligosaccharides were shown to be recalcitrant to fermentation (Jonathan et al., 2017). In the line T10, expression level of licheninase is reduced and thus could add another factor that potentially contribute to its higher ethanol yield.
Besides, phenolic composition of switchgrass has been shown to be another important factor affecting recalcitrance (Tschaplinski et al., 2012; Yee et al., 2012; Shen et al., 2013; Baxter et al., 2015). Studies with PvCOMT-downregulated switchgrass plants had higher levels of certain phenolic compounds which inhibited the microbial fermentation (Tschaplinski et al., 2012; Yee et al., 2012). In contrast, PvMYB4-overexpressing switchgrass plants grown in the greenhouse and field had lower amounts of phenolic compounds that inhibit microbial fermentation (Shen et al., 2013; Baxter et al., 2015). Interestingly, one PvMYB4-overexpressing line showed no increase in sugar release efficiency but still resulted in higher levels of ethanol production when grown in the field (Baxter et al., 2015). These studies suggest that there are factors other than lignin, e.g., phenolic compounds, that play roles in improved biofuel production observed in transgenic switchgrass overexpressing PvMYB4 (Shen et al., 2013; Baxter et al., 2015). Given that C1 metabolism pathway is involved in synthesis of variety of polymers and plant secondary metabolites, it is tempting to speculate that the increased ethanol production in the PvFPGS1-downregulated line may be caused by changes in phenolic fermentation inhibitors. Elucidating of this mechanism will be the subject of future study.
Successful establishment and sustainability of bioenergy feedstocks are key factors for production of fuel from biomass (Stewart and Cromey, 2011). Performing field studies of transgenic plants is crucial to examine consequences of their genetic modifications on plant defenses. Of particular significance is the rust disease caused by fungal pathogen P. novopanici that is identified as potentially damaging to switchgrass fields (Uppalapati et al., 2013). During the last two growing seasons, rust severity in PvFPGS1-downregulated lines was not different from that of control plants. Levels of PvFPGS1 expression appears to not be a factor in switchgrass rust disease.
In conclusion, we have shown that genetic manipulation of PvFPGS1 could lead to improved biofuel production without negatively impacting plant growth and biomass yield. These results provide further insights into the effect of knockdown expression of PvFPGS1 on improving biofuel production in switchgrass. As of interest is to use PvFPGS1 in complementation studies of the Arabidopsis and maize mutants with loss of function of native FPGS orthologs. This could further elucidate functionality of FPGS and provide information on potential strategies to enhance productivity in bioenergy crops. Further research on the FPGS genes enhance understanding of the factors associated with reducing recalcitrance. Our study provides a starting point for a more rigorous exploration of the role of PvFPGS1 in the bioenergy field.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
MM designed the experiments, participated in characterization of field-grown plants and preparation of plant samples for cell wall analysis, analyzed the data, and wrote the manuscript. HB performed rust disease phenotyping and statistical analysis, and participated in preparation of plant samples for cell wall analysis. AS performed transgenic lines generation and phylogenetic tree work. GL and HX performed gene expression analysis. AD, MR, JN, and BD performed ethanol yield analysis. J-YZ and MU performed cloning of the target gene. GT, RS, and MD performed lignin and sugar release analyses. Z-YW produced the transgenic plants. YT performed RNA-seq analysis. EB and YT conceived of the experimental approach and made significant intellectual contributions about the target gene and cell wall biology. CS conceived of the field study and its design and coordination, and assisted with interpretation of results and revisions to the manuscript. All authors contributed to text and data analysis. All authors read and approved the final manuscript.
This work was primarily supported by BioEnergy Science Center, and secondarily by the Center for Bioenergy Innovation. The BioEnergy Science Center and the Center for Bioenergy Innovation are United States Department of Energy Bioenergy Research Centers, supported by the Office of Biological and Environmental Research in the Department of Energy’s Office of Science. This manuscript has been partially authored by UT-Battelle, LLC, under contract DE-AC05-00OR22725 with the DOE. Funding was also provided by the Ivan Racheff Endowment and a USDA Hatch grant to CS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We greatly thank Joint Genome Institute (JGI) for RNA-seq data generation and Xin Chen and Chunman Zuo for assistance with RNA-seq data analysis. We thank Crissa Doeppke and Melvin Tucker for assistance with cell wall characterization. We also thank Ben Wolfe, Marcus Laxton, and the UT field staff for assistance with data collection and general field maintenance, and Reggie Millwood for assistance with the USDA APHIS BRS permit regulations. We also thank two reviewers for helpful comments that greatly improved the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00843/full#supplementary-material
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106.
Appling, D. R. (1991). Compartmentation of folate-mediated one-carbon metabolism in eukaryotes. FASEB J. 5, 2645–2651. doi: 10.1096/fasebj.5.12.1916088
Bashline, L., Lei, L., Li, S., and Gu, Y. (2014). Cell wall, cytoskeleton, and cell expansion in higher plants. Mol. Plant 7, 586–600. doi: 10.1093/mp/ssu018
Baxter, H. L., Mazarei, M., Dumitrache, A., Natzke, J., Rodriguez, M., Gou, J., et al. (2018). Transgenic miR156 switchgrass in the field: growth, recalcitrance and rust susceptibility. Plant Biotechnol. J. 6, 39–49. doi: 10.1111/pbi.12747
Baxter, H. L., Mazarei, M., Labbe, N., Kline, L. M., Cheng, Q., Windham, M. T., et al. (2014). Two-year field analysis of reduced recalcitrance transgenic switchgrass. Plant Biotechnol. J. 12, 914–924. doi: 10.1111/pbi.12195
Baxter, H. L., Poovaiah, C. R., Yee, K. L., Mazarei, M., Rodriguez, M. Jr., Thompson, O. A., et al. (2015). Field evaluation of transgenic switchgrass plants overexpressing PvMYB4 for reduced biomass recalcitrance. BioEnergy Res. 8, 910–921. doi: 10.1007/s12155-014-9570-1
Biswal, A. K., Atmodjo, M. A., Li, M., Baxter, H. L., Yoo, C. G., and Pu, Y. (2018). Sugar release and growth of biofuel crops are improved by downregulation of pectin biosynthesis. Nat. Biotechnol. 36, 249–257. doi: 10.1038/nbt.4067
Bonawitz, N. D., Kim, J. I., Tobimatsu, Y., Ciesielski, P. N., Anderson, N. A., Ximenes, E., et al. (2014). Disruption of mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509, 376–380. doi: 10.1038/nature13084
Brandon, A. G., and Scheller, H. V. (2020). Engineering of bioenergy crops: dominant genetic approaches to improve polysaccharide properties and composition in biomass. Front. Plant Sci. 11:282. doi: 10.3389/fpls.2020.00282
Chen, F., and Dixon, R. A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761. doi: 10.1038/nbt1316
Chiniquy, D., Sharma, V., Schultink, A., Baidoo, E. E., Rautengarten, C., Cheng, K., et al. (2012). XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc. Natl. Acad. Sci. U.S.A. 109, 17117–17122. doi: 10.1073/pnas.1202079109
Crowe, J. D., Feringa, N., Pattathil, S., Merritt, B., Foster, C., and Dines, D. (2017). Identification of developmental stage and anatomical fraction contributions to cell wall recalcitrance in switchgrass. Biotechnol. Biofuels 10:184. doi: 10.1186/s13068-017-0870-5
David, K., and Ragauskas, A. J. (2010). Switchgrass as an energy crop for biofuel production: a review of its ligno-cellulosic chemical properties. Energy Environ. Sci. 3, 1182–1190. doi: 10.1039/B926617H
Decker, S. R., Carlile, M., Selig, M. J., Doeppke, C., Davis, M., Sykes, R., et al. (2012). Reducing the effect of variable starch levels in biomass recalcitrance screening. Methods Mol. Biol. 908, 181–195. doi: 10.1007/978-1-61779-956-3_17
Dumitrache, A., Natzke, J., Rodriguez, M. Jr., Yee, K. L., Thompson, O. A., Poovaiah, C. R., et al. (2017). Transgenic switchgrass (Panicum virgatum L.) targeted for reduced recalcitrance to bioconversion: a two-year comparative analysis of field-grown lines modified for target gene or genetic element expression. Plant Biotechnol. J. 15, 688–697. doi: 10.1111/pbi.12666
Eckardt, N. A. (2007). Gibberellins are modified by methylation in planta. Plant Cell 19, 3–6. doi: 10.1105/tpc.107.050955
Eloy, N. B., Voorend, W., Lan, W., Saleme, M. L., Cesarino, I., Vanholme, R., et al. (2017). Silencing CHALCONE SYNTHASE in maize impedes the incorporation of tricin into lignin and increases lignin Content. Plant Physiol. 173, 998–1016. doi: 10.1104/pp.16.01108
Eudes, A., Liang, Y., Mitra, P., and Loqué, D. (2014). Lignin bioengineering. Curr. Opin. Biotechnol. 26, 189–198. doi: 10.1016/j.copbio.2014.01.002
Fu, C., Mielenz, J. R., Xiao, X., Ge, Y., Hamilton, C. Y., Rodriguez, M., et al. (2011a). Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl. Acad. Sci. U.S.A. 108, 3803–3808. doi: 10.1073/pnas.1100310108
Fu, C., Sunkar, R., Zhou, C., Shen, H., Zhang, J. Y., Matts, J., et al. (2012). Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol. J. 10, 443–452. doi: 10.1111/j.1467-7652.2011.00677.x
Fu, C., Xiao, X., Xi, Y., Ge, Y., Chen, F., Bouton, J., et al. (2011b). Downregulation of cinnamyl alcohol dehydrogenase (CAD) leads to improved saccharification efficiency in switchgrass. BioEnergy Res. 4, 153–164. doi: 10.1007/s12155-010-9109-z
Hanson, A. D., and Gregory, J. F. (2011). Folate biosynthesis, turnover, and transport in plants. Annu. Rev. Plant Biol. 62, 105–125. doi: 10.1146/annurev-arplant-042110-103819
Hanson, A. D., and Roje, S. (2001). One-carbon metabolism in higher plants. Annu. Rev. Plant Biol. 52, 119–137. doi: 10.1146/annurev.arplant.52.1.119
Himmel, M. E., and Bayer, E. A. (2009). Lignocellulose conversion to biofuels: current challenges, global perspectives. Curr. Opin. Biotechnol. 20, 316–317. doi: 10.1016/j.copbio.2009.05.005
Hu, Z., Zhang, G., Muhammad, A., Abdul Samad, R., Wang, Y., Walton, J. D., et al. (2018). Genetic loci simultaneously controlling lignin monomers and biomass digestibility of rice straw. Sci. Rep. 8:3636. doi: 10.1038/s41598-018-21741-y
Jonathan, M. C., DeMartini, J., Van Stigt Thans, S., Hommes, R., and Kabel, M. A. (2017). Characterisation of non-degraded oligosaccharides in enzymatically hydrolysed and fermented, dilute ammonia-pretreated corn stover for ethanol production. Biotechnol. Biofuels 10:112. doi: 10.1186/s13068-017-0803-3
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Lam, P. Y., Tobimatsu, Y., Takeda, Y., Suzuki, S., Yamamura, M., Umezawa, T., et al. (2017). Disrupting flavone synthase II alters lignin and improves biomass digestibility. Plant Physiol. 174, 972–985. doi: 10.1104/pp.16.01973
Li, L., Skinner, S. H., Liu, S., Beuchle, D., Tang, H. M., Yeh, C.-T., et al. (2015). The maize brown midrib4 (bm4) gene encodes a functional folylpolyglutamate synthase. Plant J. 81, 493–504. doi: 10.1111/tpj.12745
Liu, W., Mazarei, M., Ye, R., Peng, Y., Shao, Y., Baxter, H. L., et al. (2018). Switchgrass (Panicum virgatum L.) promoters for green tissue-specific expression of the MYB4 transcription factor for reduced-recalcitrance transgenic switchgrass. Biotechnol. Biofuels 11:122. doi: 10.1186/s13068-018-1119-7
Mann, D. G. J., LaFayette, P. R., Abercrombie, L. L., King, Z. R., Mazarei, M., Halter, M. C., et al. (2012). Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol. J. 10, 226–236. doi: 10.1111/j.1467-7652.2011.00658.x
Mazarei, M., Baxter, H. L., Li, M., Biswal, A. K., Kim, K., Meng, X., et al. (2018). Functional analysis of cellulose synthase CesA4 and CesA6 genes in switchgrass (Panicum virgatum) by overexpression or RNAi-mediated gene silencing. Front. Plant Sci. 9:1114. doi: 10.3389/fpls.2018.01114
Moore, K. J., Moser, L. E., Vogel, K. P., Waller, S. S., Johnson, B. E., and Pedersen, J. F. (1991). Describing and quantifying growth stages of perennial forage grasses. Agron. J. 83, 1073–1077. doi: 10.2134/agronj1991.00021962008300060027x
Nelson, R. S., Stewart, C. N. Jr., Gou, J., Holladay, S., Gallego-Giraldo, L., Flanagan, A., et al. (2017). Development and use of a switchgrass (Panicum virgatum L.) transformation pipeline by the BioEnergy Science Center to evaluate plants for reduced cell wall recalcitrance. Biotechnol. Biofuels 10:309. doi: 10.1186/s13068-017-0991-x
Ranocha, P., Denancé, N., Vanholme, R., Freydier, A., Martinez, Y., Hoffmann, L., et al. (2010). Walls are thin 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 63, 469–483. doi: 10.1111/j.1365-313X.2010.04256.x
Rao, X., Chen, X., Shen, H., Ma, Q., Li, G., Tang, Y., et al. (2019). Gene regulatory networks for lignin biosynthesis in switchgrass (Panicum virgatum). Plant Biotechnol. J. 17, 580–593. doi: 10.1111/pbi.13000
Ravanel, S., Cherest, H., Jabrin, S., Grunwald, D., Surdin-Kerjan, Y., Douce, R., et al. (2001). Tetrahydrofolate biosynthesis in plants: molecular and functional characterization of dihydrofolate synthetase and three isoforms of folylpolyglutamate synthetase in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 15360–15365. doi: 10.1073/pnas.261585098
Rébeillé, F., Ravanel, S., Jabrin, S., Douce, R., Storozhenko, S., and Van Der Straeten, D. (2006). Folates in plants: biosynthesis, distribution, and enhancement. Physiol. Plant. 126, 330–342. doi: 10.1111/j.1399-3054.2006.00587.x
Selig, M. J., Tucker, M. P., Sykes, R. W., Reichel, K. L., Brunecky, R., Himmel, M. E., et al. (2010). Lignocellulose recalcitrance screening by integrated high-throughput hydrothermal pretreatment and enzymatic saccharification. Ind. Biotechnol. 6, 104–111. doi: 10.1089/ind.2010.0009
Shane, B. (1989). Folylpolyglutamate synthesis and role in the regulation of one-carbon metabolism. Vitam. Horm. 45, 263–335. doi: 10.1016/S0083-6729(08)60397-0
Shen, B., Li, C., and Tarczynski, M. C. (2002). High free-methionine and decreased lignin content result from a mutation in the Arabidopsis S-adenosyl-l-methionine synthetase 3 gene. Plant J. 29, 371–380. doi: 10.1046/j.1365-313X.2002.01221.x
Shen, H., Fu, C., Xiao, X., Ray, T., Tang, Y., Wang, Z., et al. (2009). Developmental control of lignification in stems of lowland switchgrass variety Alamo and the effects on saccharification efficiency. BioEnerg Res. 2, 233–245. doi: 10.1007/s12155-009-9058-6
Shen, H., He, X., Poovaiah, C. R., Wuddineh, W. A., Ma, J., Mann, D. G., et al. (2012). Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 193, 121–136. doi: 10.1111/j.1469-8137.2011.03922.x
Shen, H., Poovaiah, C. R., Ziebell, A., Tschaplinski, T. J., Pattathil, S., Gjersing, E., et al. (2013). Enhanced characteristics of genetically modified switchgrass (Panicum virgatum L.) for high biofuel production. Biotechnol. Biofuels 6:71. doi: 10.1186/1754-6834-6-71
Srivastava, A. C., Chen, F., Ray, T., Pattathil, S., Peña, M. J., Avci, U., et al. (2015). Loss of function of folylpolyglutamate synthetase 1 reduces lignin content and improves cell wall digestibility in Arabidopsis. Biotechnol. Biofuels 8:224. doi: 10.1186/s13068-015-0403-z
Stewart, A., and Cromey, M. (2011). Identifying disease threats and management practices for bio-energy crops. Curr. Opin. Environ. Sustain. 3, 75–80. doi: 10.1016/j.cosust.2010.10.008
Studer, M. H., DeMartini, J. D., Brethauer, S., McKenzie, H. L., and Wyman, C. E. (2009). Engineering of a high-throughput screening system to identify cellulosic biomass, pretreatments, and enzyme formulations that enhance sugar release. Biotechnol. Bioeng. 105, 231–238. doi: 10.1002/bit.22527
Sykes, R., Yung, M., Novaes, E., Kirst, M., Peter, G., and Davis, M. (2009). High-throughput screening of plant cell-wall composition using pyrolysis molecular beam mass spectroscopy. Methods Mol. Biol. 581, 169–183. doi: 10.1007/978-1-60761-214-8_12
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 302725–2729. doi: 10.1093/molbev/mst197
Tang, H. M., Liu, S., Hill-Skinner, S., Wu, W., Reed, D., Yeh, C. T., et al. (2014). The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. Plant J. 77, 380–392. doi: 10.1111/tpj.12394
Tobimatsu, Y., and Schuetz, M. (2019). Lignin polymerization: how do plants manage the chemistry so well? Curr. Opin. Biotechnol. 56, 75–81. doi: 10.1016/j.copbio.2018.10.001
Tschaplinski, T. J., Standaert, R. F., Engle, N. L., Martin, M. Z., Sangha, A. K., Parks, J. M., et al. (2012). Down-regulation of the caffeic acid O-methyltransferase gene in switchgrass reveals a novel monolignol analog. Biotechnol. Biofuels 5:71. doi: 10.1186/1754-6834-5-71
Uppalapati, S. R., Serba, D. D., Ishiga, Y., Szabo, L. J., Mittal, S., Bhandari, H. S., et al. (2013). Characterization of the rust fungus, Puccinia emaculata, and evaluation of genetic variability for rust resistance in switchgrass populations. BioEnergy Res. 6, 458–468. doi: 10.1007/s12155-012-9263-6
Urbanowicz, B. R., Pena, M. J., Ratnaparkhe, S., Avci, U., Backe, J., Steet, H. F., et al. (2012). 4-O-methylation of glucuronic acid in Arabidopsis glucuronoxylan is catalyzed by a domain of unknown function family 579 protein. Proc. Natl. Acad. Sci. U.S.A. 109, 14253–14258. doi: 10.1073/pnas.1208097109
van der Weijde, T., Alvim Kamei, C. L., Torres, A. F., Vermerris, W., Dolstra, O., Visser, R. G., et al. (2013). The potential of C4 grasses for cellulosic biofuel production. Front. Plant Sci. 4:1. doi: 10.3389/fpls.2013.00107
Vanholme, R., De Meester, B., Ralph, J., and Boerjan, W. (2019). Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 56, 230–239. doi: 10.1016/j.copbio.2019.02.018
Wilkerson, C. G., Mansfield, S. D., Lu, F., Withers, S., Park, J. Y., Karlen, S. D., et al. (2014). Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344, 90–93. doi: 10.1126/science.1250161
Xi, Y., Fu, C., Ge, Y., Nandakumar, R., Hisano, H., Bouton, J., et al. (2009). Agrobacterium-mediated transformation of switchgrass and inheritance of the transgenes. Bioenerg. Res. 2, 275–283. doi: 10.1007/s12155-009-9049-7
Xie, H., Engle, N. L., Venketachalam, S., Yoo, C. G., Barros, J., Lecoultre, M., et al. (2019). Combining loss of function of FOLYLPOLYGLUTAMATE SYNTHETASE1 and CAFFEOYL-COA 3-O-METHYLTRANSFERASE1 for lignin reduction and improved saccharification efficiency in Arabidopsis thaliana. Biotechnol. Biofuels 12:108. doi: 10.1186/s13068-019-1446-3
Xu, B., Escamilla-Treviño, L. L., Sathitsuksanoh, N., Shen, Z., Shen, H., Zhang, Y. H., et al. (2011). Silencing of 4-coumarate:coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol. 192, 611–625. doi: 10.1111/j.1469-8137.2011.03830.x
Yee, K. L., Rodriguez, M. Jr., Tschaplinski, T. J., Engle, N. L., Martin, M. Z., Fu, C., et al. (2012). Evaluation of the bioconversion of genetically modified switchgrass using simultaneous saccharification and fermentation and a consolidated bioprocessing approach. Biotechnol. Biofuels 5:81. doi: 10.1186/1754-6834-5-81
Yoshida, K., and Komae, K. (2006). A rice family 9 glycoside hydrolase isozyme with broad substrate specificity for hemicelluloses in type II cell walls. Plant Cell Physiol. 47, 1541–1554. doi: 10.1093/pcp/pcl020
Zhang, J. Y., Lee, Y. C., Torres-Jerez, I., Wang, M., Yin, Y., Chou, W. C., et al. (2013). Development of an integrated transcript sequence database and a gene expression atlas for gene discovery and analysis in switchgrass (Panicum virgatum L.). Plant J. 74, 160–173. doi: 10.1111/tpj.12104
Keywords: folylpolyglutamate synthetase, switchgrass, RNAi-gene silencing, PvFPGS1, lignocellulosic, biofuel
Citation: Mazarei M, Baxter HL, Srivastava A, Li G, Xie H, Dumitrache A, Rodriguez M Jr, Natzke JM, Zhang J-Y, Turner GB, Sykes RW, Davis MF, Udvardi MK, Wang Z-Y, Davison BH, Blancaflor EB, Tang Y and Stewart CN Jr (2020) Silencing Folylpolyglutamate Synthetase1 (FPGS1) in Switchgrass (Panicum virgatum L.) Improves Lignocellulosic Biofuel Production. Front. Plant Sci. 11:843. doi: 10.3389/fpls.2020.00843
Received: 26 March 2020; Accepted: 26 May 2020;
Published: 19 June 2020.
Edited by:
Mengzhu Lu, Chinese Academy of Forestry, ChinaReviewed by:
Mingyang Quan, Beijing Forestry University, ChinaCopyright © 2020 Mazarei, Baxter, Srivastava, Li, Xie, Dumitrache, Rodriguez, Natzke, Zhang, Turner, Sykes, Davis, Udvardi, Wang, Davison, Blancaflor, Tang and Stewart. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuhong Tang, eXRhbmdAbm9ibGUub3Jn; Charles Neal Stewart Jr., bmVhbHN0ZXdhcnRAdXRrLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.