
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 19 June 2020
Sec. Plant Biotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00842
Feed supplementation with β-arginine-aspartate dipeptides (β-Asp-Arg DP) shows growth promoting effects in feeding trials with fish and might also be beneficial for pig and poultry farming. Currently, these DPs are generated from purified cyanophycin (CGP), with the help of the CGP-degrading enzyme cyanophycinase (CGPase). As alternative to an in vitro production, the DPs might be directly produced in feed crops. We already demonstrated that CGP can be produced in plastids of tobacco and potato, yielding up to 9.4% of the dry weight (DW). We also transiently co-expressed CGPases in the cytosol without degrading CGP in intact cells, while degradation occurs in the homogenized plant tissue. However, transient co-expression is not feasible for field-grown CGP plants, which is necessary for bulk production. In the present study, we proved that stable co-expression of the CGPase CphE241 in CGP-producing tobacco is sufficient to degrade 2.0% CGP/DW nearly completely within 3 h after homogenization of the leaves. In intact senescing leaves, CGP is partially released to the cytosol and degraded into DPs which limits the overall accumulation of CGP but not the level of the stable DPs. Even after 48 h, 54 μmol β-Asp-Arg DP/g DW could be detected in the extract, which correspond to 1.5% DP/DW and represents 84% of the expected amount. Thus, we developed a system for the production of β-Asp-Arg DP in field-grown plants.
The supply of feed with sufficient amounts of (conditionally) essential amino acids (AA) such as lysine (Lys), arginine (Arg), tryptophan (Trp) or methionine (Met) is one of the major challenges in livestock production (Ufaz and Galili, 2008). Since they are underrepresented in plant-based animal diets, feed is supplemented with AA that are chemically synthesized or produced via microbial fermentation (FAO, 2004; BMBF, 2007). Beyond their nutritional value as building blocks for proteins, many AAs provide also physiological benefits, e.g. immunomodulatory effects, which promote health and wellbeing (Wu et al., 2009, 2014). In this respect, Arg proved to be unique and even though not essential in adult animals such as pigs, poultry, and fish, dietary deficiency of Arg can result in metabolic, neurological or reproductive dysfunctions (Wu, 2009; Wu et al., 2009). However, free AAs inhibit each other’s uptake, which confines their use as feed additive1. In contrast, di-, tri- and oligopeptides do not undergo such restrictions. In addition, dipeptides (DP) are resorbed more efficiently compared to free mono AA due to the presence of a specific di- and tripeptide specific transporters in the enterocytes of the small intestine that show a higher substrate affinity compared to transporters for free AA (Webb, 1986; Chen et al., 1999; Klang et al., 2005; Lu and Klaassen, 2006; Broer, 2008; Sallam and Steinbuchel, 2010; Santos et al., 2012; Terova et al., 2013; Hou et al., 2017). The beneficial effects of arginine-aspartate (β-Asp-Arg) and lysine-aspartate (β-Asp-Lys) DP were demonstrated in a long-term feeding trial with fish in which an increased body weight gain has been observed (Cysal, personal communication). Currently, both types of DPs are derived from the polypeptide Cyanophycin (CGP), produced in transgenic Escherichia coli (E. coli).
Cyanophycin is naturally produced in cyanobacterial and non-cyanobacterial prokaryotes as transient storage for carbon, nitrogen and energy (Frommeyer et al., 2014). It is synthesized by the CGP-synthetases (CphAs) via non-ribosomal protein biosynthesis in two steps: (1) the α-amino group of arginine is linked to the β-carboxylate group of aspartate via a β-iso-peptide bond to a β-arginine-aspartate dipeptides (β-Asp-Arg DP) with a molecular weight of 0.2 kDa, (2) the DP is subsequently connected to the nascent CGP polypeptide by an α-peptide bond. CGP aggregates to intracellular membraneless granules (Law et al., 2009). In addition to Arg, other AAs such as Lys are incorporated as well in lower amounts (Frommeyer et al., 2014). The molecular weight of the polymer is polydisperse and ranges in cyanobacteria between 25 and 100 kDa, but when expressed in E. coli only between 20 and 40 kDa. Due the β-peptide bond, which does not occur in proteins, CGP can only be degraded by so-called cyanophycinases (CGPase), but is resistant to common proteases (Law et al., 2009). CGPases are divided into CphB- and CphE-type enzymes, which both degrade CGP into DP monomers. While CphBs are expressed by CGP-producing prokaryotes in the cytosol and remobilize CGP in the event of starvation, CphEs are secreted by non-CGP producing prokaryotes in order to exploit CGP from other bacteria. Interestingly, CphE-expressing bacteria were found in the colon of animals as part of the natural microbiota (Sallam and Steinbuchel, 2009a). Since AA and DP uptake occurs in the small intestine (Broer, 2008; Sallam and Steinbuchel, 2009a), the produced DP cannot be absorbed by the animal.
The recombinant production of CGP has already been established in yeast, fungi or E. coli and cultivation conditions were optimized, increasing the accumulation level up to 40% per dry weight (DW) and promoting the incorporation of Lys (Frommeyer et al., 2014). Likewise, the CphE-type CGPase from Pseudomonas alcaligenes DIP-1 (CphEal) was expressed in E. coli (Sallam and Steinbuchel, 2009b; Sallam et al., 2011). This research resulted in the establishment of the Cysal GmbH (see text footnote 1). Cysal produces both CGP and the CphE-type CGPase in individual E. coli strains, and incubates the purified substrate with the enzyme to generate the DPs that are sold as feed additives (Sallam and Steinbuchel, 2011). However, the bioreactor-based production necessitates a complex infrastructure and is limited in terms of scalability. Hence, this system is commonly used for the synthesis of high-value, but not for cost-sensitive products such as supplements for animal diets (Merlin et al., 2014; Sabalza et al., 2014; Walwyn et al., 2015).
Alternatively, recombinant low-value products can be produced in plants in an economical manner using existing agricultural infrastructure and farming practices as modeled in case studies (Tusé et al., 2014; Tschofen et al., 2016). Studies estimated that the production costs are on average less than a tenth compared to conventional microbial and mammalian cell cultures (Broz et al., 2013; Tusé et al., 2014; Chen, 2015; Gecchele et al., 2015; Walwyn et al., 2015; Dirisala et al., 2016; Nandi et al., 2016; Buyel et al., 2017). The synthesis of CGP in plants was established via expression of the CGP-synthetase gene from Thermosynechococcus elongatus BP-1 (cphATe) in plastids of stably transformed tobacco (Nicotiana tabacum) and potato (Solanum tuberosum) plants, yielding up to 9.4% leaf DW in commercial tobacco cultivars (Neumann et al., 2005; Hühns et al., 2008, 2009; Neubauer et al., 2012; Nausch et al., 2016). Plastid targeting was achieved via fusion of the coding region of cphATe to the transit peptide of the integral protein of photosystem II PsbY (Hühns et al., 2008). Plastidic CGP is almost exclusively composed of β-Asp-Arg DPs and has a molecular weight of 15–20 kDa. Noteworthy, the cytosolic expression yielded only low amounts of CGP and induced severe phenotypic disorders (Neumann et al., 2005).
Even though CGP-degrading prokaryotes were found in the digestive tract of animals (Sallam and Steinbuchel, 2009a), feeding of CGP-producing plants as diet might not lead to an absorption of β-Asp-Arg DPs, since the bacteria where only found in the colon, while DP uptake occurs in the small intestine (Chen et al., 1999; Klang et al., 2005; Lu and Klaassen, 2006; Broer, 2008; Terova et al., 2013). Hence, co-delivery of a CGPases would be necessary. Ideally, both substrate and degrading enzymes are produced by the same plant. To prevent premature CGP degradation and subsequent metabolizing of the resulting DPs, CGP, and CGPase need to be spatially separated. The production of both a CphB-type CGPase from T. elongatus BP-1 (CphBTe, 30.7 kDa) and a mutant of the CphE-type CGPase CphEal, termed as CphE241 (44.7 kDa), in the cytosol of plants was already shown via transient expression in tobacco (Ponndorf et al., 2016; Nausch and Broer, 2017). In addition, transient expression of both cytosolic enzymes in plastidic CGP-producing tobacco demonstrated that CGP was not degraded until the plant material was homogenized. Complete CGP degradation was only observed for CphE241, but not for CphBTe, which was assigned to a drastically higher enzyme activity of CphE241 (Ponndorf et al., 2017). In a feeding study with mice, in which purified plant-produced CGP and CphBTe were added to the diet, it could be shown that the CGPase is capable to degrade CGP in the digestive tract and that the β-Asp-Arg DPs were transferred to the blood (Ponndorf et al., 2016). This should also be true for CphEal, since this CGPase type naturally occurs in the colon (Sallam and Steinbuchel, 2009a) and since it has been demonstrated that CphEal is active in a pH range from 5 to 9 (Sallam and Steinbuchel, 2011).
Forage should be produced at large scale in the field. Hence the CGPase has to be stably expressed in the CGP-producing plants. Since yields achieved by transient expression are commonly drastically higher compared to the accumulation in stably nuclear transformed plants (Xu et al., 2012; Kopertekh and Schiemann, 2017; Schillberg et al., 2019) stable expression of CphE241 might not be sufficient to degrade CGP completely in an acceptable time frame. While expression levels up to 80% of the total soluble protein (TSP) were reported for the transient assay, corresponding to 5 mg/g fresh weight (FW), recombinant proteins do not exceed 1% of TSP in stably transformed plants, rarely exceeding 100 μg/g FW and reaching up to a maximum of 500 μg/g FW (Schillberg et al., 2019).
Here we show that the CGPase CphE241 can be stably expressed in high amounts in CGP-producing plants and that the CphE241 activity is sufficient to degrade the 2.0% CGP/DW present in the plant almost completely in 3 h. The premature release of CGP in intact older leaves into the cytosol limits the overall CGP accumulation during plant growth. However, the premature degradation does not affect the DP production, since DPs are not metabolized in plants. Conversion of 2.0% CGP/DW resulted in ∼54 μmol DP/g DW which corresponds to 14.7 mg DP/g DW or 1.5% DP/DW.
CphE241, a mutant of the CphE-type CGPase CphEal (Sallam et al., 2011) encoded by cphE241syn (Nausch and Broer, 2017), was selected for stable transformation of the commercial N. tabacum cultivar Badischer Geudertheimer (BG) event BG 176 which produces 2–2.5% CGP/DW. CphE241syn was transferred from the plasmid pEX-K4-CphE241syn (Nausch and Broer, 2017) into the vector pLH-IL6ER (Nausch et al., 2012). Via BamHI/NruI restriction/ligation, the coding region of IL6ER was replaced between 5′ region [consisting of the Cauliflower Mosaic Virus (CaMV) 35S promoter with double-enhancer (Odell et al., 1985), the tobacco mosaic virus (TMV) Ω translational enhancer (Gallie et al., 1987)] and the 3′ region [containing the CaMV 35S terminator (Odell et al., 1985)]. The entire gene was subsequently cloned via SfiI restriction/ligation into the plant transformation vector pLH7000 [Acc. No. AY234330 (Hausmann and Toepfer, 1999)], carrying the bialaphos-resistance gene (bar) from Streptomyces hygroscopicus (Thompson et al., 1987) as selection marker, forming pLH7000-35s-cphE241syn (Figure 1) for constitutive expression. The final plasmid was verified by restriction and sequencing (Eurofins Genomics GmbH). All vectors were transferred into the Rhizobium radiobacter (formerly Agrobacterium tumefaciens) strain C58C1 for stable transformation of the N. tabacum event BG 176, which was already transformed with the plastid-targeted cyanophycin synthetase gene from Thermosynecchococcus elongatus BP-1 [psby-cphATe (Hühns et al., 2008)]. This second transformation was termed as super-transformation.

Figure 1. The constitutive 35S promoter is used for the expression of cphE241syn and the bar coding region. Schematic representation of the T-DNA region of the pLH7000-35s-cphE241syn construct used for cytosolic expression of the cyanophycinase CphE241. LB, left border; RB, right border; En, Cauliflower Mosaic Virus (CaMV) 35S enhancer; p35S, CaMV 35S core promoter; t35S, CaMV 35S terminator; Ω, Ω translational enhancer of the TMV; bar, coding region for the bialaphos-resistance protein from Streptomyces hygroscopicus; cphE241syn, codon-optimized, synthetic coding region for the CphE241 protein.
For super-transformation with the CphE241 encoding T-DNA, the T2 individual BG 176-4-3 of the one-copy event BG 176 (Nausch et al., 2016) was used. Tobacco seeds were surface sterilized in a saturated calcium hypochlorite solution, germinated on Linsmaier and Skoog (LS) medium (4.4 g/L of LS medium including vitamins (Duchefa, Haarlem, Netherlands), 30 g/L sucrose, 6.5 g/L plant agar (Duchefa), [pH 5.7]. In vitro grown plants were maintained at 24/22°C day/night temperature with a 16 h photoperiod. Approximately 1-month old tobacco leaves were used for Agrobacterium-mediated transformation as described in Wohlleben et al. (1988), using 100 μg/mL kanamycin and 20 μ/mL D/L-phosphinothricin for selection of transgenic cells. Calli/Explant and Shoot/Explant ratios were determined 6 weeks after transformation. Regenerated shoots were selected on LS medium containing 20 μg/mL D/L-phosphinothricin, 100 μg/mL kanamycin, and 500 μg/mL cefotaxim. Shoots were transferred for rooting to LS medium with herbicide and antibiotics. Total DNA was isolated from regenerated plants as described by Nausch et al. (2012) and transgene integration confirmed by PCR using CphEsyn-fw5/CphEsyn-rv1 for the coding region of cphE241syn, barPP-fw/-rv for the coding region of the selection marker, and PsbY-fw/CphA-rv1 for the cphA gene (Table 1).
At least 100 seeds from self-fertilized transgenic plants were germinated on LS-medium containing 100 μg/mL kanamycin and 20 μg/mL D/L-phosphinothricin. Randomly selected seedlings were cultivated for 6 weeks on selective medium, and 4 weeks after the last subculture 48 plants carrying the transgenes were transferred to the greenhouse.
Transgenic individuals, both T0 super-transformants and T1 descendants, were transferred from tissue culture 4 weeks after the last subculture, directly into 5 L pots containing peat soil (Stender AG, Schramberg, Germany). Plants were fertilized once a week using 0.5% Wuxal Super (Hermann Meyer KG, Rellingen, Germany).
Dry weight was determined by oven drying (105°C, 3 h) followed by ashing in a muffle furnace (600°C, 5 h). Crude protein was determined by the Dumas combustion method using a Vario Max C/N/S-Analyzer (Elementar Analysensysteme GmbH, Hanau, Germany). Cyanophycin was measured as described in the section “CGP quantification”. AA (without Trp) were determined by HPLC (Shimadzu, Kyoto, Japan) using a cation column (LC K06, Alltech-Grom GmbH, Rottenburg-Hailfingen, Germany). AA (without Trp) were determined by HPLC (Shimadzu, Kyoto, Japan) using a cation column (LC K06, Alltech-Grom GmbH, Rottenburg-Hailfingen, Germany) according to Hackl et al. (2010). The temperature program of the column was set between 57°C and 74°C, and the pH gradient from 3.45 to 10.85. The buffer flow rate was 0.45 mL/min. AA were mixed with ninhydrin at a flow rate of 0.25 mL/min for tinting at 128°C and determined with an UV-detector at 570 nm (proline at 440 nm).”
Cyanophycin was quantified as described by Nausch et al. (2016) which bases upon the method of Simon (1973). Three samples per individual plant of 30–35 mg of freeze-dried leaf material were homogenized with ceramic pills in a Precellys 24 homogenisator (VWR International GmbH, Erlangen, Germany) and incubated in 1 mL 50 mM Tris [pH 8.0] for 30 min. After centrifugation, the pellet was resuspended in 1 mL of 0.1 M HCL and incubated for 1 h. After another centrifugation step, 800 μL of the supernatant was used for CGP analysis. 1–10 μL of sample were filled up with 0.1 M HCL to a final volume of 800 μL and 200 μL of 5x RotiQuant Bradford reagent (Carl Roth GmbH + Co. KG, Karlsruhe, Germany) added. After 5 min incubation, samples were measured at 595 nm. A calibration curve was prepared with purified CGP from potato tubers, isolated as described by Neubauer et al. (2012). OD values of leaf samples from non-transgenic plants were subtracted from OD values of samples of non-transgenic near-isogenic control plants.
In our previous study, we demonstrated that when CphE241syn is present in the plant sample, CGP is degraded during CGP quantification in the incubation step with Tris buffer and before measuring the OD at 595 nm (Ponndorf et al., 2017). Thus, we defined this protocol as protocol with active CphE enzyme. By omitting the Tris extraction step and direct incubation in 1 mL HCL, we could prevent the CGP degradation, hence in this protocol the CphE enzyme is inactivated. Comparing the results of both methods served as in vitro activity assay. For each individual plant, the in vitro activity assays was repeated 3–5 times.
Leaves were collected and cut along the leaf midrib into two halves in order to prepare two pooled samples from each plant. One pool was frozen immediately without homogenization to analyze the CGP content in the intact leaves at the time point of sampling (T0 sample), while the other pool was homogenized using a PT2100-Homogenizer (Kinematica AG, Littau-Lucerne, Switzerland; 30,000 rpm; 30–45 s) and incubated overnight at 22–24°C (T24 sample). Samples were freeze-dried and CGP quantified. The pools were freeze-dried and CGP quantified with at least 3–5 analysis per pool.
HCL-extracts from the CGP quantification were used for Coomassie-staining. 88 μL of a 72% trichloroacetic acid (TCA) solution were added to the 800 μL HCL-extracts in order to precipitate acid soluble proteins and CGP. After incubating for 2 h at 4°C and centrifugation (16,100 g, 15 min, 4°C), the pellet was resuspended in 100 μL loading buffer containing 10% glycerin, 150 mM Tris [pH 6.8], 3% SDS, 1% β-mercaptoethanol and 2.5% bromophenol blue. The pH was adjusted to 6.8 with a saturated Tris solution. After denaturation at 95°C for 5 min, 1–10 μL of the samples were separated by a 12% SDS-PAGE and Coomassie-stained. PageRulerPlus Prestained Protein Ladder Mix (Thermo Fisher Scientific, Brunswick, Germany) served as marker and 5 μg CGP, purified from potato tubers, as positive control.
Dipeptides were analyzed in five samples per plant. Approximately 200 mg of dry plant material were supplemented with 1 mL of 5 mmol/L HCl, the suspensions were vortexed for 30 s and centrifuged (15,000 g, 10 min, 4°C). The supernatants were transferred to new 1.5 mL polypropylene tubes and lyophilized. The residues were reconstituted in 50 μL of 20% (v/v) aqueous acetonitrile and derivatized with N2-(5-fluoro-2,4-dinitrophenyl)-L-valine amide (L-FDVA) as described by Ehrlich and co-workers (Ehrlich et al., 2009) with modifications. In detail, aliquots of the reconstituted extracts (20 μL) were supplemented with 7 μL of water and pH was adjusted to 8.0 with 1 mol/L NaHCO3 using indicator paper (typically 8 μL of NaHCO3 were required). Afterwards, 32 μL of 36.7 mmol/L L-FDVA acetone solution were added, and derivatization of amines was accomplished during 90 min at 40°C under continuous shaking (350 rpm), before the reactions were stopped by addition of 25 μL of 1 mol/L HCl. After the change of solution color to yellow, 50 μL of acetonitrile and 114 μL of water were added, and the samples were intensively vortexed. Finally, 500 μL 0.1% (v/v) formic acid were added, the samples were centrifuged (15,000 g, 10 min, 24°C), and 200 μL of the supernatants were transferred to the inserts of the vials for HPLC.
Analysis of the L-FDVA DP derivates relied on the method of Antonova et al. (2019) with some modifications. In detail, 4–100 μL of each sample were loaded on a reversed phase Hypersil GOLD aQ column (100 × 1 mm, 1.9 μm particle size), installed on a Dionex Ultimate 3000 UHPLC System (Thermo Fisher Scientific, Bremen, Germany). The separations were performed at the flow rate of 150 μL/min, at 40°C in a linear gradient mode, with eluents A and B being water and acetonitrile, both containing 0.1% (v/v) formic acid. After a 2-min isocratic step at 5% eluent B, L-FDVA derivatives of the dipeptide were separated in the sequential gradients to 20, 33, and 42% eluent B in one, seven, and 4 min, respectively. After a second isocratic segment (4 min at 42% eluent B) a further gradient to 70% eluent B was run in 3 min. The column effluents were transferred on-line into a hybrid LTQ-Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, Bremen, Germany), equipped with a heated electrospray ionization (HESI) source and operated in the positive ion mode. The analysis was performed under ion spray (IS) voltage of 4.0 kV, with nebulizer and auxiliary gases set to 35 and 30 arbitrary units, respectively. The capillary temperature was set to 275°C. The spectra were acquired at the mass to charge ratio (m/z) range of 400–2,000 and resolution of 30,000. The DP ß-L-aspartyl-L-arginine was identified in chromatograms by m/z, tR and co-elution with the authentic standard. Quantitative analysis relied on the standard addition approach as described elsewhere (Bilova et al., 2017). Thereby, analyte abundances were obtained by integration of corresponding extracted ion chromatograms (XICs) at specific tR. Peak integration was performed in Xcalibur 2.2 software.
Purified β-Asp-Arg DPs, which were used for the calibration curve, were kindly provided by the Cysal GmbH (Münster, Germany; Dr. M. Krehenbrink)1.
cDNA was prepared and PCR analysis conducted as described by Nausch and Broer (2017), except that an Oligo-dT-primer was used instead of an random hexamer primer. Total RNA was isolated from 100 mg tobacco leaf tissue using Trizol reagent according to the manufacturer’s instructions (Thermo Fisher Scientific, Brunswick, Germany). RNA integrity was assessed by visualizing the 28S and 18S rRNA bands under UV light in a denaturing 0.8% MOPS-agarose gel containing ethidium bromide. For reverse transcription (RT) into cDNA, total RNA samples were incubated with DNase I at 37°C for 1 h, followed by cDNA synthesis using a commercial kit [RevertAidTM H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific)]. 200 units of reverse transcriptase were added to 5 μg of DNase-treated RNA, 10 mM dNTP mix, 5 μM Oligo-dT-primer and incubated at 42°C for 1 h. The reaction was stopped for 10 min at 70°C. PCR was conducted with the DreamTaq DNA polymerase (Thermo Fisher Scientific). Undiluted cDNA (2 μL) were amplified in a 50 μL PCR reaction with following parameters: After an initial denaturation step (95°C for 3 min), 35 amplification cycles (95°C for 30 s, 58°C for 30 s, 72°C for 1.5 min) were carried out and completed by a final elongation step (72°C for 10 min). The PCR was performed as multiplex assay with construct-specific and actin-specific primers. The cphE241syn mRNA was detected using the primer pair CphEsyn-fw5/rv1, for psby-cphATe mRNA the primer pair PsbY-fw/CphA-rv1 was used (Table 1). Actin, as a housekeeping gene was used to compare the quantity of target mRNA/cDNA between samples and to detect contamination of samples with genomic DNA.
For Western blot, crude plant extracts were prepared either from freeze-dried leaf tissue. Approximately, 200 mg plant material were added to 1 mL NPI-lysis buffer (50 mM NaH2PO4 [pH = 8.0], 300 mM NaCl, 10 mM imidazole), homogenized with ceramic pills using a Precellys 24 homogenisator (VWR International GmbH, Erlangen, Germany) and incubated for 30 min at 4°C in a shaker. After centrifugation (16,100 g, 4°C, 15 min) the supernatant was transferred to another reaction tube and centrifuged again. Alternatively, the Tris extract from the CGP-quantification assay was used for Western blot. The TSP concentration was quantified via PierceTM Coomassie Plus Assay Kit (Thermo Fisher Scientific, Brunswick, Germany) according to the manufacturer’s instructions. For the detection of CphE241, the Western blot was conducted as described by Nausch and Broer (2017), using an anti-His antibody as primary antibody (Nausch and Broer, 2017). TSP samples were freeze-dried and resuspended in 25 μL loading buffer containing 10% glycerin, 150 mM Tris [pH 6.8], 3% SDS, 1% β-mercaptoethanol, and 2.5% bromophenol blue. After denaturing at 95°C for 5 min, samples were separated by a 12% SDS-PAGE and electrophoretically transferred to a 0.45 μm Hybond ECL nitrocellulose membrane (GE Healthcare Europe GmbH, Freiburg im Breisgau, Germany), using a Bio-Rad Trans-Blot semi-dry transfer cell. 2 mA/cm2 were applied for 1 h using 50 mM Tris, 40 mM glycine, 0.01% SDS, and 20% methanol as the transfer buffer [pH 8.5]. Membranes were blocked with TBSTween20 (20 mM Tris [pH 7.6], 150 mM NaCl, 0.05% Tween20) and 5% non-fat milk powder for 2 h at 24°C. After three washes with TBSTween20, membranes were probed at 24°C for 2 h with a mouse monoclonal anti-His antibody (cat.: DIA 900; Dianova, Hamburg Germany) at 1:1000 dilution in Signal Boost ImmunoReaction Enhancer solution I (Merck KGaA, Darmstadt, Germany). Following another washing step, the membranes were further probed at 24°C for 1 h with a horseradish peroxidase (HRP)-conjugated donkey anti-mouse antibody (cat. no. 715-035-151; Dianova) at 1:10,000 dilution in ImmunoReaction Enhancer Solution II. Again, three washes were done with TBS without Tween20. The signals were detected by incubation in ECL chemiluminescence reagent (ECL reagent I: 1 M Tris [pH = 8.5]; 250 mM Luminol, 90 mM p-Coumaric Acid; ECL reagent II: 1 M Tris [pH 8.5], 30% H2O2) and subsequent exposure of membranes to a Kodak Biomax light X-ray film (VWR; Darmstadt, Germany) for 1–15 min. For the verification of observed differences in the CphE241 expression level, Western blots were repeated 2–3 times.
CphE241syn, produced in E. coli and isolated via Ni-NTA purification as described by Nausch and Broer (2017) served as positive control.
Southern blot analysis was conducted according to Hühns et al. (2008). Chromosomal DNA was digested with BamHI and HindIII. The probe was amplified from the plasmid pLH7000-35s-CphE241syn using the primers cphEsyn-fw and cphEsyn-rv1 (Table 1).
Statistical analysis was performed with SPSS, using either the non-parametric (Mann–Whitney-U or unpaired T-test) or univariate ANOVA (including the post hoc Bonferroni, Duncan and Tukey test). A P-value P ≤ 0.05 (two-sided) was considered to be significant.
Overexpression of CGP can be used to enrich plants with Asp and Arg. In two different cultivars of tobacco, 2.5% CGP/DW result in an increase of Asp and Arg of 1% DW each (Supplementary Table SM1). In order to make the AA stored in CGP bioavailable, we aimed to co-express a CGPase. For co-expression of the CGPase CphE241 in tobacco with plastidic CGP-production, we selected T2 plants of BG transformant (event) 176, described in our previous study (Nausch et al., 2016), because (1) this line carries one-copy of CGP-synthetase encoding transgene, (2) was bred to homozygosity, and (3) has a homogenous and reliable CGP-accumulation of ∼2.5% CGP/leaf DW. These T2 plants were super-transformed with the gene for CphE241syn under the control of the constitutive CaMV 35s (pLH7000-35s-CphE241syn, Figure 1), using the bar gene and the herbicide phosphinothricin as selection system. Putative events, transformed with the empty vector pLH7000 were named BG 176 + EV and putative events from the transformation with pLH7000-35s-CphE241syn, named BG 176 + CphE241. The presence of the transgene was verified in regenerated plants via PCR, using CphEsyn-fw5/CphEsyn-rv1 for the coding region of cphE241syn, and barPP-fw/-rv for the coding region of the selection marker (Table 1). However, via Western blot the presence of the CphE241 protein was only detected in two out of 90 super-transformants (event 120 and 121; Figure 2 and Supplementary Table SM2). In three independent clones each, the signal of CphE241 was reproducibly 2–3 times higher in event 120 compared to 121 (data not shown). All 90 events were also analyzed for the CphE241 enzyme activity, using the in vitro activity assay. Only in the two events 120 and 121, in which the CphE241 protein could be detected (Figure 2) was CGP degradation observed (data not shown). In order to exclude gene silencing as a reason for the absence of Cph241 in the other events, the presence of the cphE241syn mRNA was analyzed in 15 super-transformants, including event 120 and 121 as control. The mRNA could be detected in event 120 and 121 and 4 other events (data not shown).
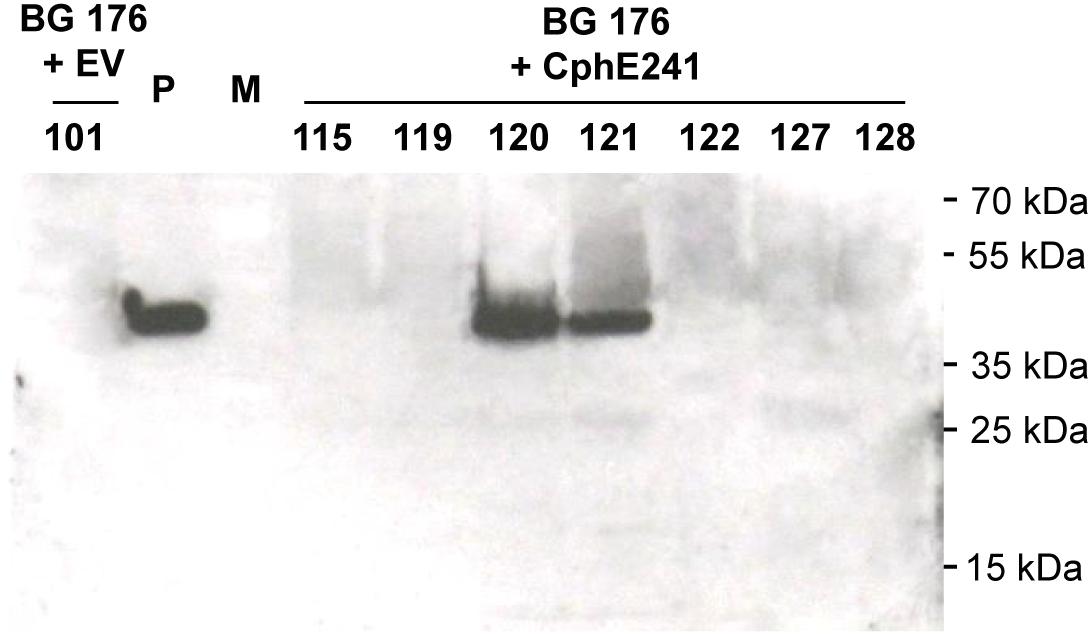
Figure 2. CphE241 could be detected in leaves of the in vitro grown events 120 and 121 by Western blot. Samples from 7 super-transformants were taken from leaves of 4 week old in vitro plants. 100 μg TSP per sample were analyzed in the Western blot analysis. BG 176 + EV: super-transformants, carrying the empty vector, BG 176 + CphE241: super-transformants, carrying the CphE241 encoding T-DNA. Numbers: secondary transformation events. P: 5 ng of purified CphE241syn, produced in Escherichia coli. M: PageRulerPlus Prestained Protein Ladder Mix (Thermo Fisher Scientific). Since the marker proteins are not conjugated with the HRP enzyme, they cannot be detected by ECL. The corresponding apparent molecular weights of the protein standards are given on the right side of the image.
To analyze whether the individual variability between plants affects the expression of CphE241 and the capability to degrade CGP, the assay was conducted with five clones each of both events. In all clones CGP degradation occurred in a similar degree. CGP degradation was always nearly complete in all clones of event 120 in contrast to the clones of 121 (Figure 3A). CGP degradation was verified by Coomassie-staining of the samples from the in vitro CphE241 activity assay (Figure 3B). In Western blots, the amount of enzyme was similar in all clones of event 120 and 121, respectively (Figure 3C).
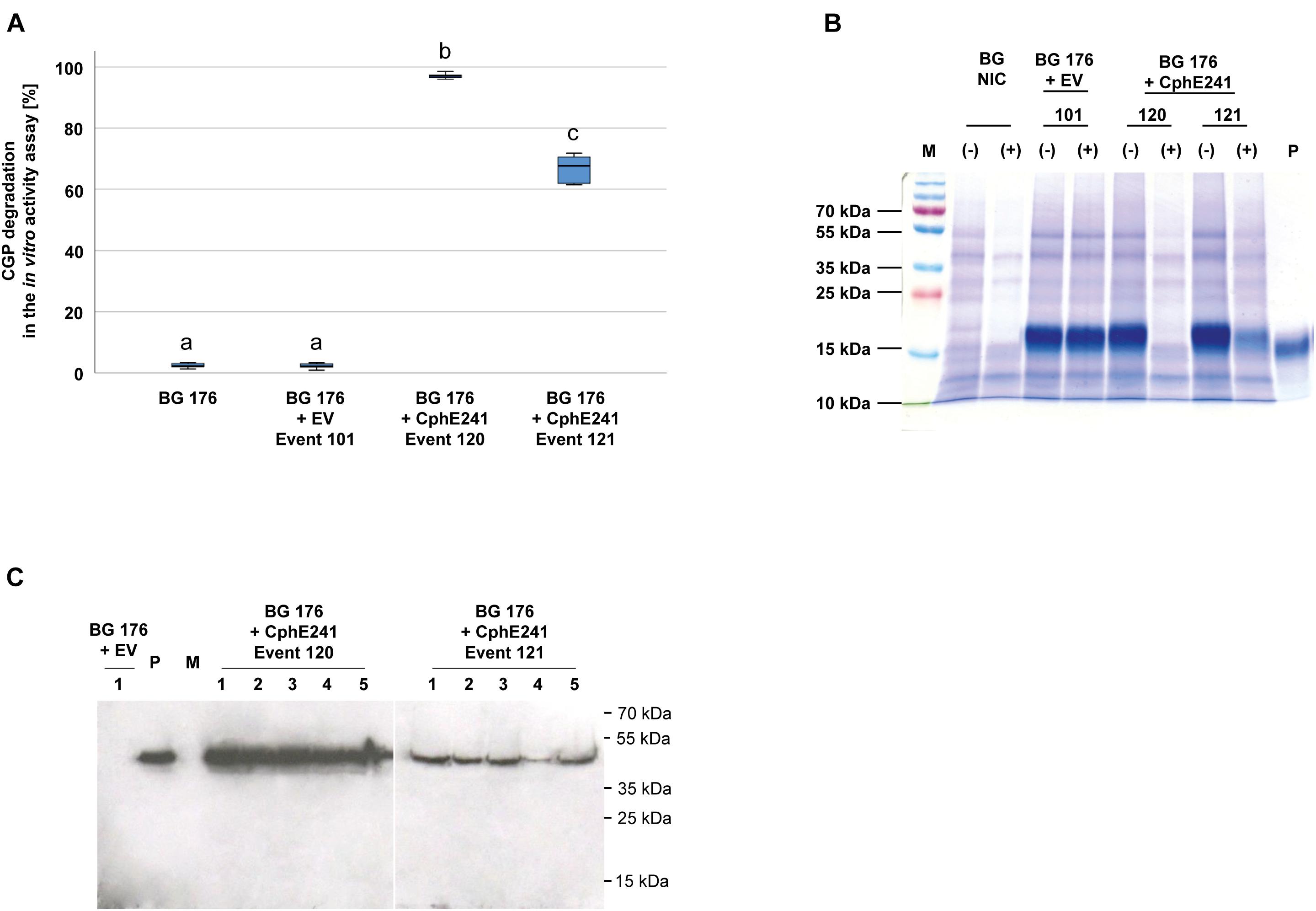
Figure 3. CGP degradation is almost complete in homogenized leaves from in vitro plants of event 120 but not in event 121. Five clones for each event were analyzed. Three fully developed, green leaves each were taken from 4 week old in vitro grown plants. (A) Box plot showing the CGP degradation in% in vitro in the presence of CphE241. The differences between a, b, and c are significant (ANOVA including post hoc Bonferroni, p < 0.05). Error bars represent the standard deviation; (B) CGP degradation visualized in a Coomassie-stained polyacrylamide gel. (–): inactivated CphE241, (+) active CphE241; P: purified CGP from transgenic potato tubers; M: marker as in (C); BG NIC: near-isogenic control plants, BG 176 + EV: super-transformant, carrying the empty vector. BG 176 + CphE241: super-transformants, carrying the CphE241 encoding T-DNA; (C) Western blot analysis: 100 μg TSP per sample were analyzed, P: 5 ng of purified CphE241syn, produced in Escherichia coli. M: PageRulerPlus Prestained Protein Ladder Mix (Thermo Fisher Scientific). Since the marker proteins are not conjugated with the HRP enzyme, they cannot be detected by ECL. The corresponding apparent molecular weights of the protein standards are given on the right side of the image; 1–5 different clones of the respective events 120 or 121.
The elite event BG 176 + CphE241 event 120 was inbred in order to investigate whether the CphE241 expression will increase or decrease in the offspring. Seeds of the self-pollinated T0 super-transformants were grown on regeneration medium with phosphinothricin for the CphE241 expression cassette, kanamycin for the CphA expression cassette and cefotaxim to inhibit Agrobacteria growth. Out of the T1 offspring, 48 plants were randomly selected and the presence of the transgenes psby-cphATe and cphE241syn was verified via PCR (data not shown). All T1 siblings were transferred to the greenhouse and analyzed via the in vitro CphE241 activity assay in the age of 6 weeks. All plants of BG 176 + CphE241 event 120 were capable to degrade CGP (Figure 4A). In 54.2% of the individuals the enzyme degraded more than 90% of the CGP, 35.4% showed a moderate CphE241 activity with a CGP degradation ranging from 50 to 90% and only 10.4% a low CphE241 activity with less than 50% CGP degradation. The 4 T1 individuals with the highest CphE241 activity (120-32, 120-42, 120-46, and 120-47) that was similar to the T0 parent, showed a reproducibly higher CphE241 steady-state protein level in the Western blot compared to the T0 parent (Figure 4B).
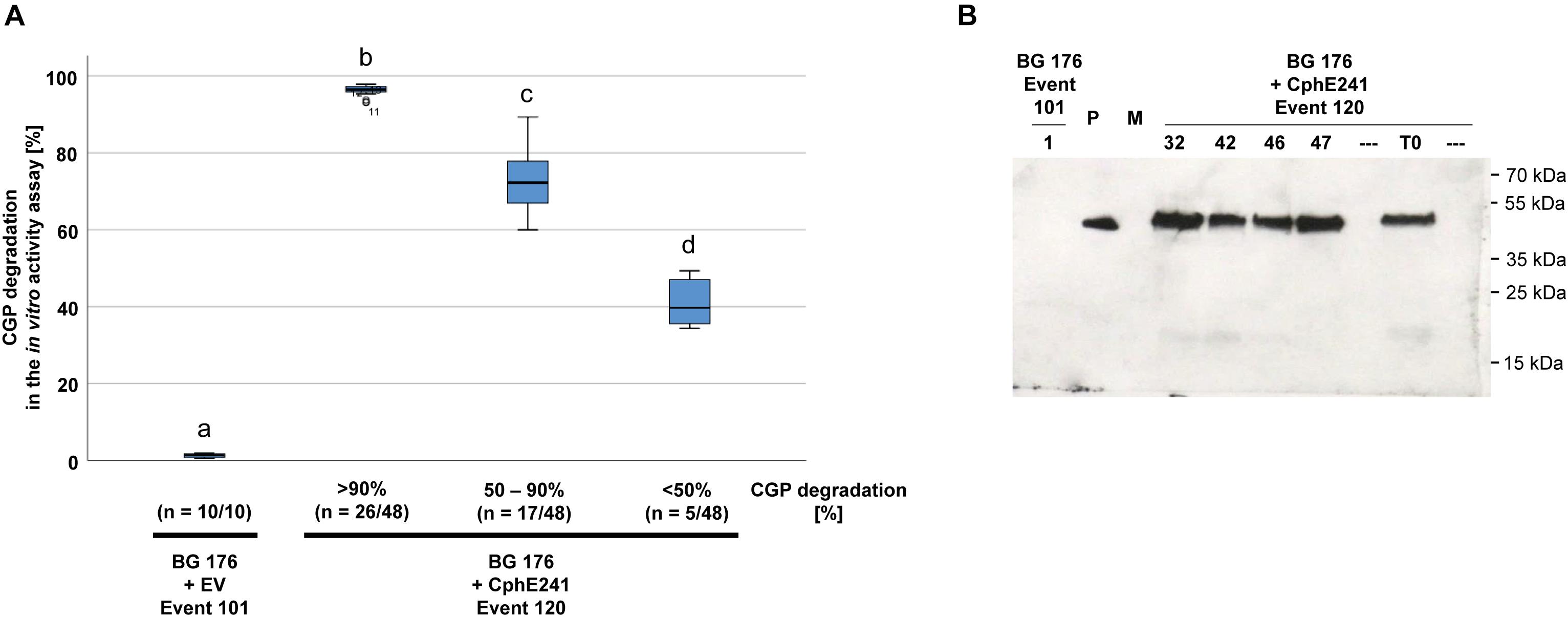
Figure 4. CGP degradation in the T1 offspring of event 120 can be similar to the one of the T0 parent. Forty-eight siblings were analyzed. Three samples each were taken from fully developed, green leaves of 6 week old, greenhouse grown plants. (A) Box plot showing the in vitro CphE241 enzyme activity assay. The differences between a, b, c, and d are significant (ANOVA including post hoc Bonferroni, p < 0.05). Error bars represent the standard deviation; (B) Western blot analysis of CphE241: 100 μg TSP per sample were analyzed, P: 5 ng of purified CphE241, produced in Escherichia coli. M: PageRulerPlus Prestained Protein Ladder Mix (Thermo Fisher Scientific); BG 176: primary transformant, BG 176 + EV: super-transformants, carrying the empty vector, BG 176 + CphE241: super-transformants. Since the marker proteins are not conjugated with the HRP enzyme, they cannot be detected by ECL. The corresponding apparent molecular weights of the protein standards are given on the right side of the image.
The elite event BG 176 + CphE241 event 120 and the four T1 individuals 120-32, 120-42, 120-46, and 120-47, which were selected for breeding of the T2 generation were analyzed for the copy number of the cphE241syn transgene (Figure 5A). The T0 super-transformant 120 carries at least 4 copies of cphE241 which are all present in the T1 individual 120-32 (Figure 5B). In the descendants 120-42 and 120-46, 3 copies were detected and in 120-47 only 1 copy was found (Figure 5B). Since all 4 T1 descendants did not differ in their CphE241 expression level, the high expression in this transgenic line might not be due to the complex integration pattern in event 120, but to the one integration locus, present in the T1 individual 120-47. This may also explain the variability in the CphE expression and CGP degradation activity observed in the T1 generation.
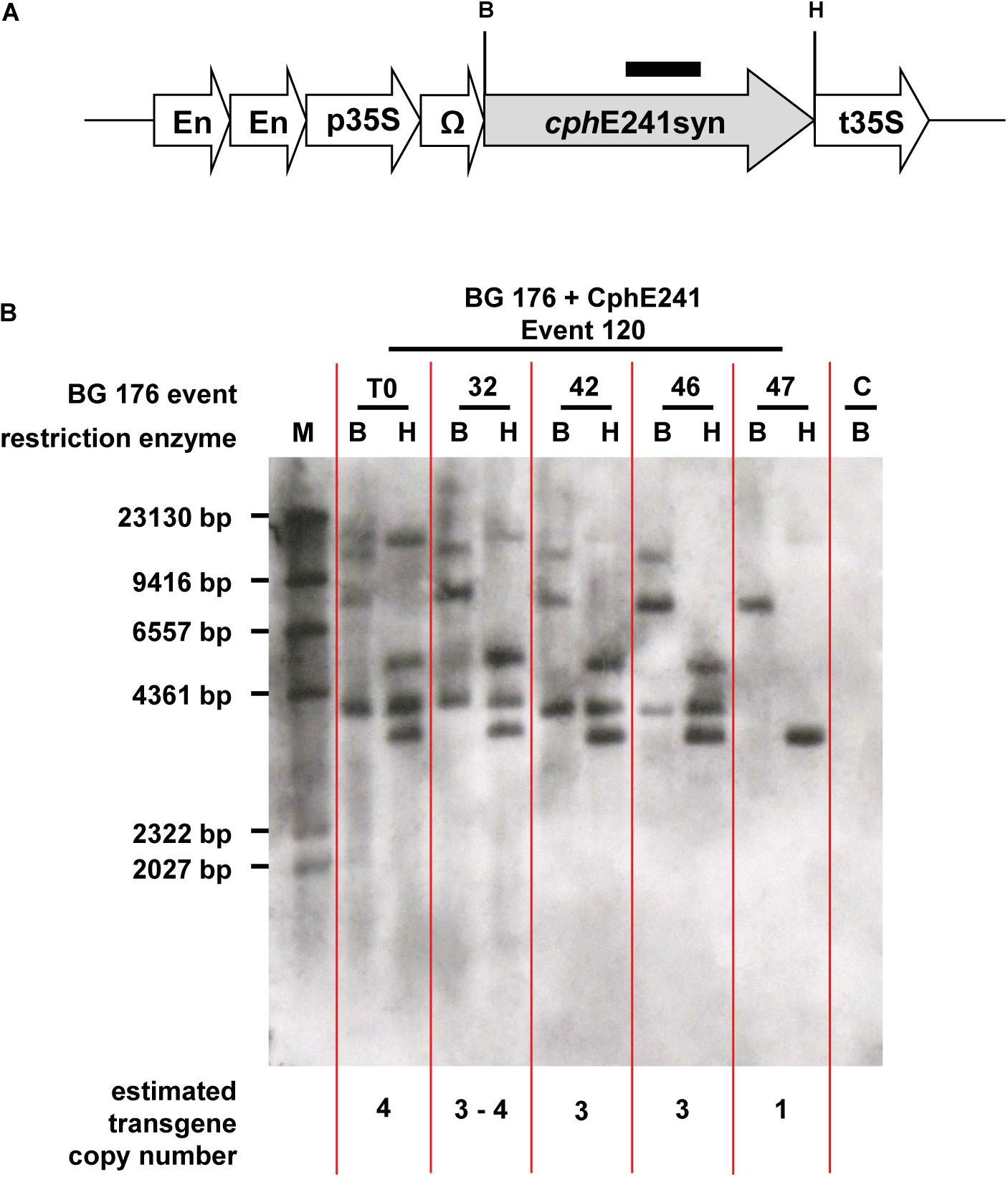
Figure 5. According to Southern blot analysis one transgene integration locus in event 120 seems to be decisive for high CphE241 expression. (A) transgene: black bar: cphE241syn hybridization probe; B/H: restriction enzymes sites of BamHI and HindIII; cphE241syn: synthetic codon-optimized coding region of the cyanophycinase CphE from Pseudomonas alcaligenes DIP-1, including a point mutation at the amino acid position 241 of the mature protein from glycine to aspartic acid, En, Cauliflower Mosaic Virus (CaMV) 35S enhancer; p35S, CaMV 35S core promoter; t35S, CaMV 35S terminator; Ω, 5′Ω translational enhancer of the TMV. (B) Chromosomal DNA digested with BamHI and HindIII and hybridized with the cphE241syn probe. M: DIG-labeled DNA Molecular Weight Marker II (Roche, Mannheim, Germany); C: BG 176 + EV event 101, T0: super-transformant of BG 176 + CphE241 event 120; 32, 42, 46, and 47: T1 descendants 120-32, 120-42, 120-46, and 120-47.
Based on the CGP degradation in in vitro grown plants we focused on event 120 for further analysis. In order to identify the optimal harvest time (highest amount of CGP/time in the greenhouse), three independent clones each were harvested in 1 week intervals over a period of 12 weeks. In the control plant BG 176, relative CGP levels per DW and total CGP amount per plant continuously increased until week 11 (Figures 6A,B). In contrast, in the event 120, both relative (Figure 6A) and total (Figure 6B) CGP levels increased only during the first 6 weeks and remained constant from then on in the intact cells. In the same plant material, we observed a reduced CGP degradation starting from week 6 in the in vitro CphE241 activity assay (Figure 6C) which complies to a reduced CphE241 signal strength as shown in Western blots (Figure 6D).
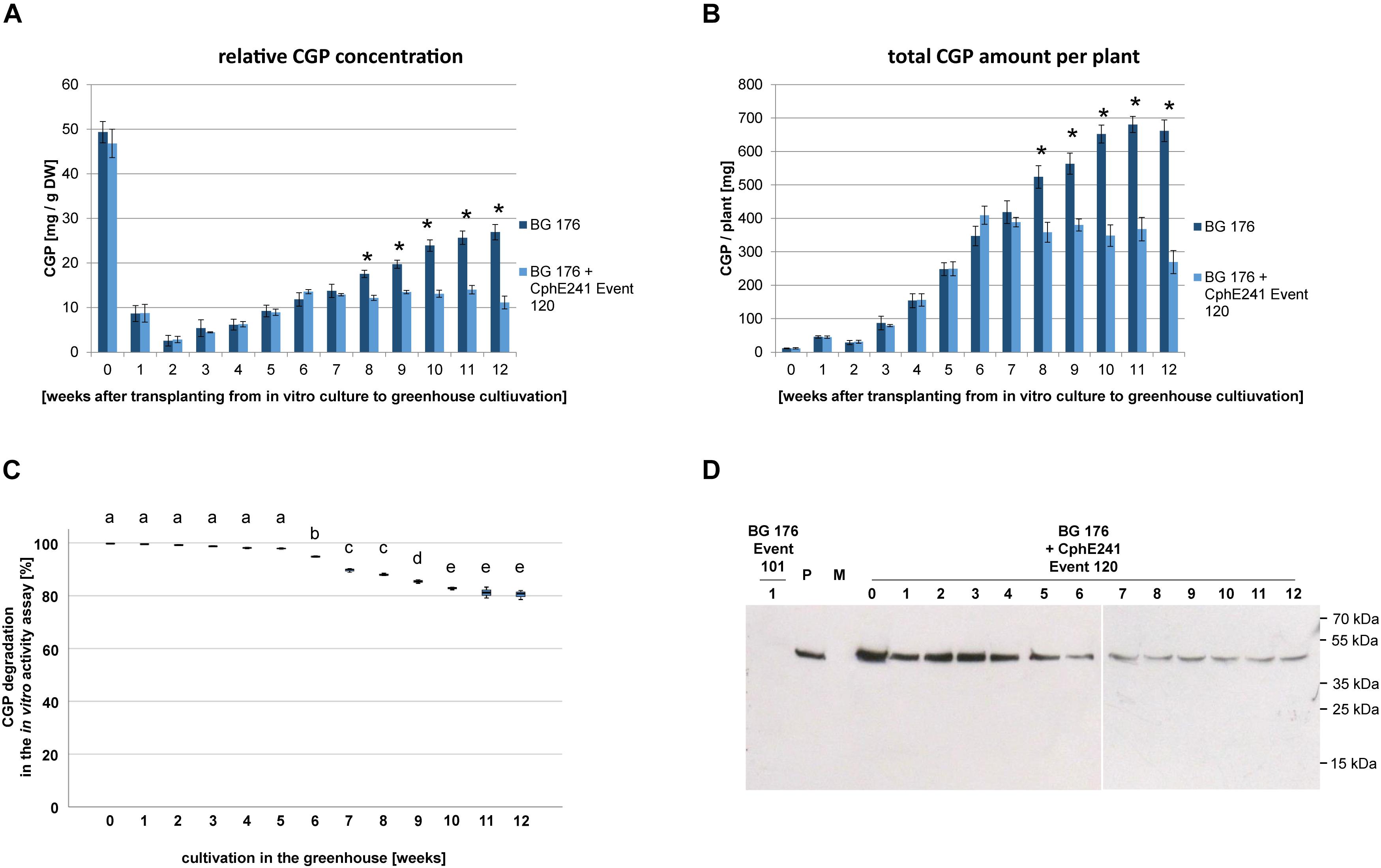
Figure 6. CGP accumulates in greenhouse grown clones of event 120 up to the age of 6 weeks. For each time point, the leaves of each three clones were harvested and pooled. Three to five samples per pool were analyzed. (A) Bar chart showing the relative CGP accumulation per DW; (B) Bar chart showing the absolute CGP accumulation per plant; (C) Box plot showing the CphE241 in vitro activity assay; The asterisk in (A) and (B) indicates a significant difference between BG 176 and BG 176 + CphE241 event 120. The differences between a–e in (C) are significant. For (A,B) ANOVA including post hoc Bonferroni (p < 0.05) was applied. Error bars represent the standard deviation; (D) Western blot analysis of CphE241: 100 μg TSP per sample were analyzed. BG 176: primary transformant. P: 5 ng of purified CphE241, produced in Escherichia coli. M: PageRulerPlus Prestained Protein Ladder Mix (Thermo Fisher Scientific). Since the marker proteins are not conjugated with the HRP enzyme, they cannot be detected by ECL. The corresponding apparent molecular weights of the protein standards are given on the right side of the image; BG 176 + CphE241: super-transformants, carrying the CphE241 encoding T-DNA. Numbers: 0: day of transfer of in vitro grown plants to the greenhouse; 1–12: weeks of cultivation in the greenhouse, n = 3.
To analyze green, vital and senescing leaves separately, we grew clones of BG 176 + 7000 event 101 and BG 176 + 35-CphE241 event 120 for 8 weeks in the greenhouse and sampled vital, green and senescing, yellow leaves separately (Supplementary Figure SM1). As shown in Figure 7A, in clones of the event 101 producing only CGP, CGP levels were similar in vital and senescing leaves with 19.4 (±1.6) and 18.7 (±1.3) mg CGP/g DW, while in event 120 in old leaves CGP was significantly decreased with 15.2 (±3.1) mg CGP/g DW compared to 20.2 (±1.1) mg CGP/g DW in young leaves. The CphE241 activity assay demonstrated that the CGP degradation was reduced in older leaves (Figure 7B) in line with a reduced CphE241 signal strength (Figure 7C). β-Asp-Arg DP were significantly increased in senescing leaves of event 120 with 1445.3 (±407.8) nmol DP/g DW compared to 868.5 (±105.0) in young leaves, respectively (Table 2).
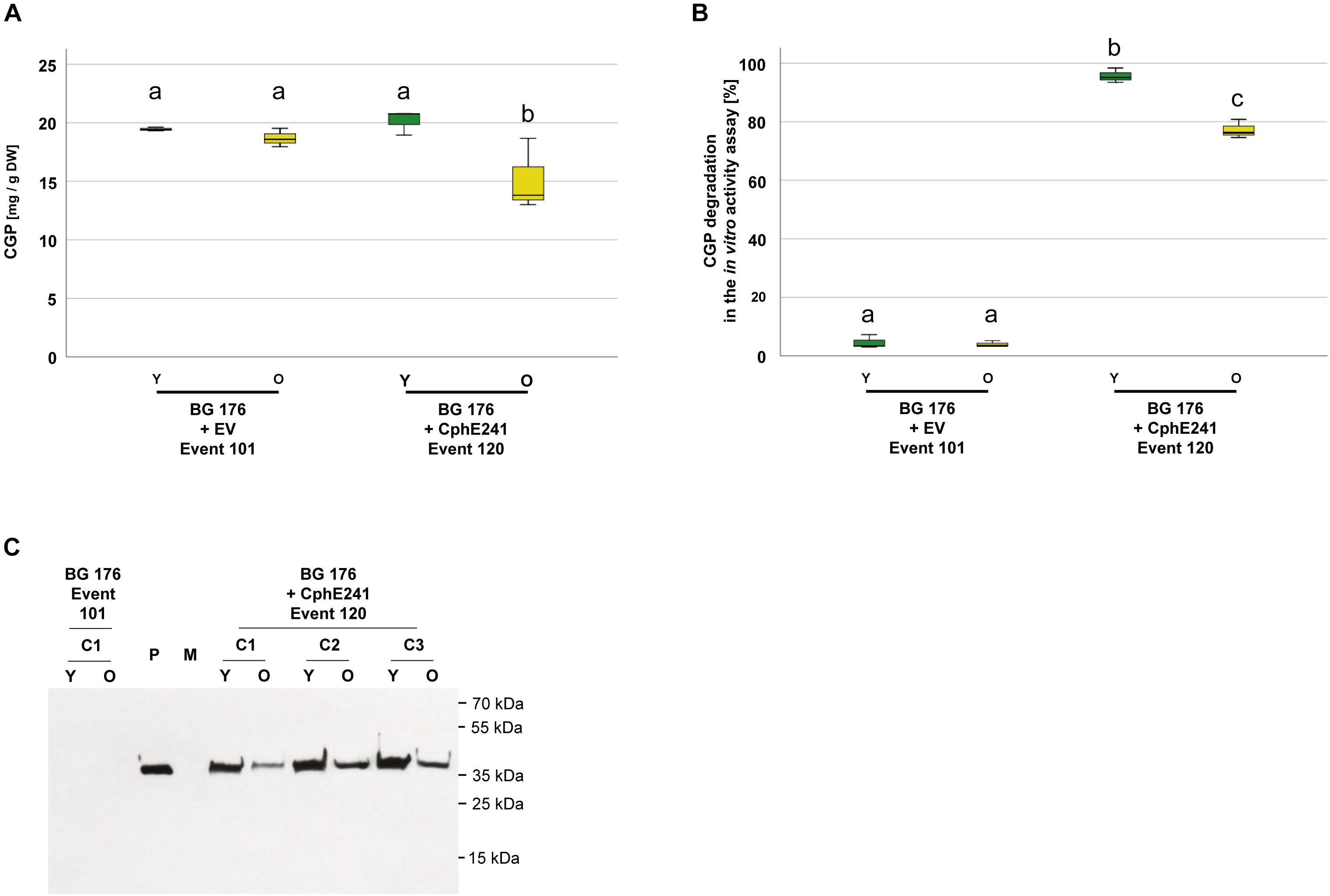
Figure 7. CGP degradation occurs both in green, fully developed and yellow, senescing leaves of event 120. Three clones were grown for 8 weeks in the greenhouse and fully developed, green leaves and yellow, senescing leaves sampled separately. The assay was repeated 3–5 times for each clone. (A) Box plot showing the CGP content, (B) Box plot showing the in vitro CphE241 activity assay; The differences in (A) and (B) between a, b, and c are significant (ANOVA including post hoc Bonferroni, p < 0.05). Error bars represent the standard deviation; (C) Western blot analysis of CphE241: 100 μg TSP per sample were analyzed. P: 5 ng of purified CphE241, produced in Escherichia coli. M: PageRulerPlus Prestained Protein Ladder Mix (Thermo Fisher Scientific). Since the marker proteins are not conjugated with the HRP enzyme, they cannot be detected by ECL. The corresponding apparent molecular weights of the protein standards are given on the right side of the image; c: clone, Y green young leaves, O: yellow old, senescing leaves, n = 3.

Table 2. Content of β-Asp-Arg DPs in different leaf types of the super-transformants BG 176 + 7000 event 101 and BG 176 + CphE241 event 120.
To assess the CphE241 activity in homogenized leaves (in planta), we analyzed the CGP degradation in leaf macerates. Twenty clones each of BG 176 + 7000 event 101 and BG 176 + CphE241 event 120 were grown for 6 weeks in the greenhouse and all leaves per plant were harvested. 10 of the 20 clones per plant line were directly shock frozen in liquid nitrogen (0 h). The leaves of the other 10 clones were homogenized in a commercial blender, incubated for 24 h at 24°C. Samples were taken at different time points. The amount of CGP at 0 h was set as 100%. In the BG 176 primary transformant, the CGP content was similar at 0 h and 24 h with 100% and 99% (Figure 8A), which corresponds to 18.12 (±2.17) mg/DW and 18.04 (±1.75) mg/DW, respectively. Neither obvious CGP degradation (Figure 8A) nor significant accumulation of β-Asp-Arg DPs occurred (data not shown). In event 120, CGP was only found at significant levels at 0 h with 19.34 (±2.91) mg/g DW (set as 100%), 0.75 h (73.2%), and 1.5 h (49.7%), but was almost completely degraded within 3 h with 2.16 mg (±0.74) mg/g DW (11.9%) left (Figure 8A). From then on the residual CGP level remained nearly constant and averaged at 24 h 1.32 (±0.68) mg CGP/g DW (6.8%) (Figure 8A). In parallel to the degradation of CGP, β-Asp-Arg, DPs accumulated in the homogenized tissue (24 h) with 54.05 (±7.32) μmol/g DW (Table 2), which was drastically higher than the 1.5 μmol/g DW, found in the intact senescing leaves. For complete CGP degradation 71.27 (±10.74) μmol DP/g DW are expected, while 90% degradation should result in 64.1 (±9.7) μmol DP/g DW, respectively. Thus, we measured 76.8% (±11.8) of the maximal possible amount of DP in case of complete CGP degradation and 84.4% (±9.4) of the apparently degraded CGP. The CphE241 enzyme could be detected by Western blotting in both sample types, but the level tended to be lower in the homogenized 24 h samples (Figure 8B). We could show that the DPs were stable after complete degradation of CGP at least for 48 h after homogenization (Figure 8C). The stability of DP was also demonstrated in vitro after addition of low amounts of purified DP (10, 50, and 250 pmol/μL which corresponded the column loads of 0.2, 1, and 5 pmol) to extraction buffer and plant extract of BG NIC plants for 1, 4, 8, and 24 h (Supplementary Figure SM2).
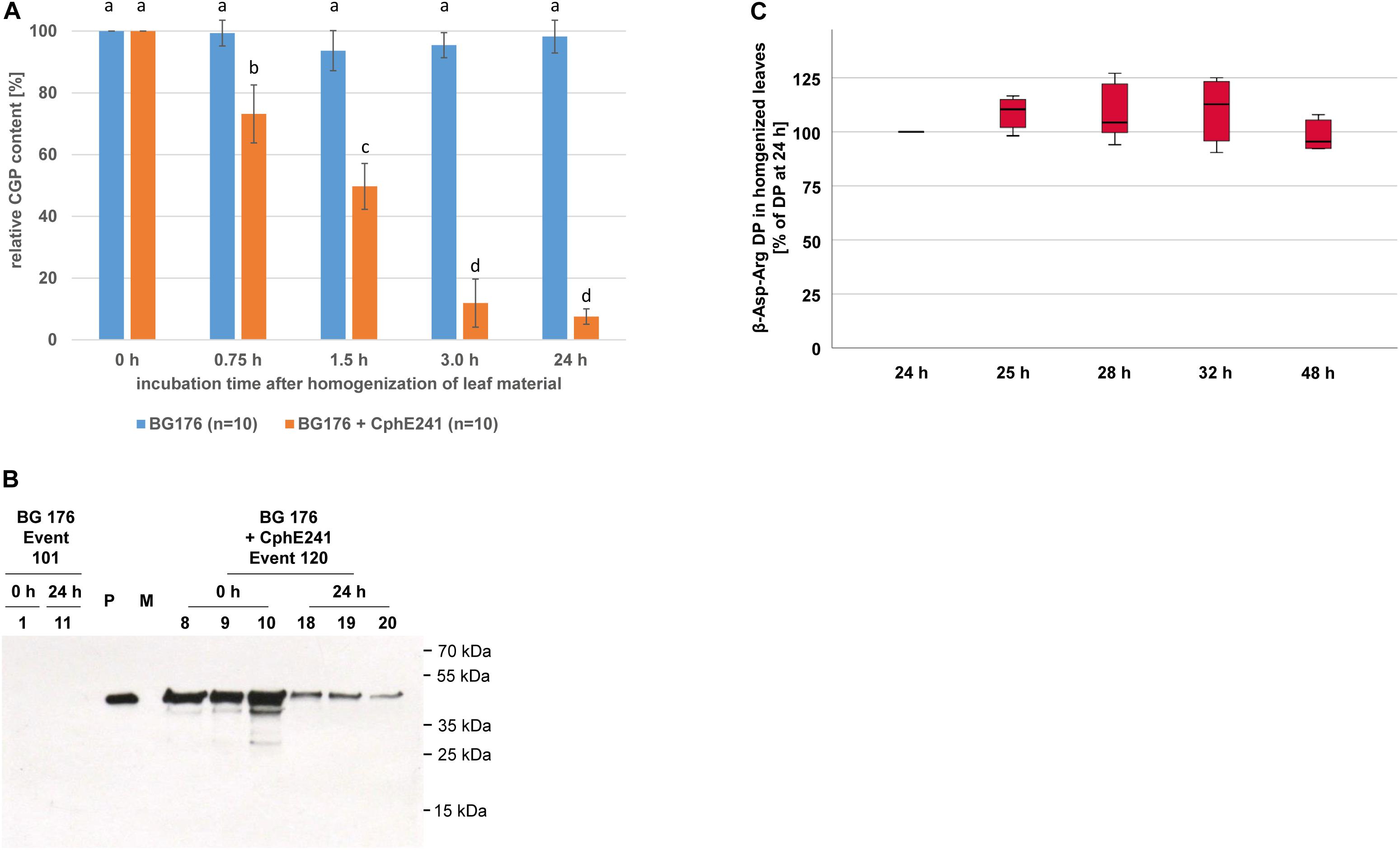
Figure 8. CGP degradation is almost complete 3 h after homogenization of leaves of event 120 and the β-Asp-Arg DP produced seems to be stable in the macerate. Ten clones of 6 week old, greenhouse grown plants were used for the in planta activity assay. The assay was 3–5 times repeated for each clone. (A) Bar chart showing the CGP degradation within 24 h; The differences between a, b, c, and d are significant (ANOVA including post hoc Bonferroni, p < 0.05). Error bars represent the standard deviation; (B) Western blot analysis of the CphE241 enzyme: 100 μg of TSP per sample were analyzed. P: 5 ng of purified CphE241, produced in Escherichia coli. M: PageRulerPlus Prestained Protein Ladder Mix (Thermo Fisher Scientific). Since the marker proteins are not conjugated with the HRP enzyme, they cannot be detected by ECL. The corresponding apparent molecular weights of the protein standards are given on the right side of the image; 0 h: leaf material directly after harvest, 24 h: leaf material after homogenization and incubation for 24 h at 24°C. (C) Box plot showing the stability of β-Asp-Arg DP in harvested, homogenized leaf material. The amount of β-Asp-Arg DPs at 24 h was set as 100%, (n = 6); BG 176 + EV: super-transformants, carrying the empty vector, BG 176 + CphE241: super-transformants, carrying the CphE241 encoding T-DNA, Numbers: clones. Differences between the samples were not significant (ANOVA including post hoc Bonferroni, p < 0.05). Error bars represent the standard deviation.
In this study, we demonstrated that the stable expression of CGPase CphE241 is sufficient to degrade 2.0% CGP/leaf DW in CGP-producing tobacco almost completely within 3 h after harvest. Although the premature degradation of CGP in intact but senescent CGPase producing plants slightly limits the CGP accumulation, this does not affect the production of β-Asp-Arg DP, since these are stable in the plants. The produced 54 μmol DP/g DW correspond to 14.7 mg DP/g DW or 1.5% DP/DW, respectively. Since Arg represents 49% of the mass of the DP, this amount is equal to 0.7% Arg/g DW. Feed for pigs and poultry, supplemented with Arg up to 1.6% DW, showed substantial effects on growth and well-being of the animals (Waseem et al., 2019). This concentration might be achieved as well when the CGPase is co-expressed in events with double of the amount of CGP. Since expression up to 9.4% CGP/DW could already be achieved (Nausch et al., 2016), this might be realistic, in case the CGPase production is sufficient.
However, under 90 events tested, there were only 2 (event 120 and 121) that produced CphE241 in a detectable amount. Based on the fact that the coding regions of both the psby-cphATe and the cphE241 coding region as well as the selection markers nptII and bar were expressed via the 35s promotor, the homologous sequences might induce a transgene silencing, which has been observed in multi-copy events (Rajeevkumar et al., 2015). Nevertheless, all 90 super-transformants produced CGP and were phosphinothricin- and kanamycin-resistant. That the cphE241 but neither the psby-cphATe gene nor the nptII and bar gene are silenced seems to be unlikely, if the 35S promoter present in all four genes causes the silencing. In addition, cphE241 mRNA was present in 6 out of 17 events that were analyzed via RT-PCR and event 120 with the high expression of CphE241 carries at least 4 copies of the cphE241 gene. It is remarkable that four events did not produce detectable levels of the protein despite the presence of the specific mRNA. Since the protein is stable for at least 24 h and the general translatability of the mRNA was proven in event 120 and 121, we can only assume that either the mRNA in the four events is somehow different from the mRNA in events 120 and 121 or the amount of cphE241syn RNA was too low to lead to detectable amounts of protein in the other events.
Analyzing the CphE241 expression under different conditions showed that the enzyme seems to be stable and active both in intact green, intact senescing and in homogenized leaves. This corresponds to our previous results where the CphE241 was as stable as the cyanobacterial CphEal (Nausch and Broer, 2017). Hence there is no obvious reason for the low number of events expressing detectable amounts of CphE241. The fact, that the expression in event 121 is not sufficient to degrade the CGP in event 176 as efficient as event 120 clearly shows that a high expression is necessary for complete degradation of CGP. The high expression of CphE241 in the T0 event 120 was also transferred to its T1 offspring and seems to be associated with one integration locus, since the offspring 120-47 has only one integration locus, but a similar CphE241 expression level as in the other offspring and the T0 parent that shows a complex integration pattern. Breeding the event 120 to homozygosity of this locus might provide a pool for large-scale production of the CGP/CGPase-tobacco plants as feed.
It is remarkable that the amount of CGP did not increase in plants older than 6 weeks, if the CGPase is present in the cytosol. Under greenhouse conditions, BG tobacco shifts from vegetative growth to seed production in the age of 6 weeks (Nausch et al., 2016), and leaf senescence is expected to start (Biswal et al., 2013). Flowering and leaf senescence are associated with endoproteolytic processes in leaves and nutrient translocation from leaves to seeds (Masclaux et al., 2000; Li et al., 2016). Thus, we assume that CGP is released into the cytosol due to the decomposition of plastids in senescent tissue and degraded, if a CGPase is present. The very low amounts of DP, that were observed in intact tissue, can be explained by the occasional degradation of plastids that also occurs in green, photosynthetic active leaves, releasing plastidic CGP into the cytosol (Biswal et al., 2013). The CGP conversion in old leaves might be compensated by CGP production in young leaves, leading to constant steady state level in the whole plant.
The usage of feed that contains CGP and CGPase to provide sufficient amounts of β-Asp-Arg DPs in the digestive tract requires a very active enzyme that is able to digest CGP in a short period. We could show in this publication that the main CGP degradation occurs already within 3 h in the extract. Nevertheless, in case the plants are feed without prior extraction, the enzyme has to be stable and effective in the digestive tract, and the persistence of the feed should be sufficient to provide high amounts the CGP. Ponndorf et al. (2016) could show that CphBTe, is stable enough to degrade CGP in the digestive tract of mice (Ponndorf et al., 2016). We could also demonstrate that there are no detectable differences in protein stability between CphBTe and CphE241 and that CphE241 showed a drastically higher enzyme activity, both as purified proteins under in vitro conditions and in crude leaf extracts (Ponndorf et al., 2017). Since the stability and activity of CphE241 is equal to that of CphEal (Nausch and Broer, 2017), CphEal is stable in a wide range of conditions (Sallam and Steinbuchel, 2011), and CphEal was found in the colon of animals (Sallam and Steinbuchel, 2009a), it might be assumed that CphE241 is also active in the intestine. Hence, it might be concluded that CGP degradation will also take place in the intestine of animals when CGP/CphE241 co-expressing fresh plants are directly fed and when the plant material is digested in the gut.
Nevertheless, tobacco is an uncommon feed. But comparing the production of CGP in tobacco and potato (Hühns et al., 2008, 2009), tobacco has the highest capacity to produce CGP without penalty on biomass (yield) and phenotype. In addition, tobacco produces high yields of leaf biomass without special demands on soil and fertilization (Billenkamp, 2010a, b, 2013), yielding up to 140–165 t leaf FW or 16–32 t leaf DW per year and hectare in a moderate climate when using mechanized farming practices (Wildman, 1979; Long, 1984; van Beilen et al., 2007). The tobacco leaves contain 20–30% protein/DW (Kung et al., 1980; Fantozzi and Sensidoni, 1983; De Jong, 1991), thus making it to an interesting feed crop for livestock. The low amounts of Arg in leaves of common tobacco cultivars (Kung and Tso, 1978; Wilkinson et al., 1981; Moldoveanu, 2005; Zeng et al., 2015) can be substituted by CGP and CGPase cooexpression as shown in this manuscript. The event 120 descendant 47 which produces sufficient amounts of CGPase carrying only one copy of the transgene can be used to transfer CGP production and degradation to nicotine-free cultivars like SL6322 or to the leaf-rich, gene edited “Virginia Smoking Tobacco” variety (Schachtsiek and Stehle, 2019). Since we could also show that CGP-producing BG tobacco can be ensiled and that CGP is not degraded during this process (Nausch et al., 2016), tobacco might become a valuable feed plant if β-Asp-Arg DP is also stable in the silage.
Since DPs have the potential to promote animal welfare in livestock, this approach might also be suitable for other feed crops such as cereals like barley (Hordeum vulgare) and maize (Zea mays), legumes like soybean (Glycine max) and pea (Pisum sativum), or crucifers like rapeseed (Brassica napus) or false flax (Camelina sativa). Moreover, whereas both conventional and previous biotechnological breeding efforts failed to increase (conditionally) essential AA such as Lys, Arg, Trp or Met in these plants (Ufaz and Galili, 2008), we could show in this study that the level of Arg was substantially increased via CGP/DP production. If other AA such as Lys would also be incorporated in plant-produced CGP/DP as it is the case in microorganisms (Frommeyer et al., 2014), DP would be a suitable tool to increase the underrepresented AA in these crops.
The datasets generated for this study are available on request to the corresponding author.
HN, AF, PW, and IB conceived the research and analyzed the data. HN, MD, and SH conducted the experiments. HN and IB wrote the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
This study was funded by the University of Rostock and the Leibniz Institute of Plant Biochemistry.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We want to express our warmest thanks to Martin Bachmann and Annette Zeyner from the Martin-Luther-University Halle-Wittenberg, Institute of Agricultural and Nutritional Sciences, Chair of Animal Nutrition [Halle (Saale), Germany] for their support in establishing the project. We are also grateful to Sonja Konopka and Kerstin Thoss for the passionate care of the transgenic BG plants. Purified β-Asp-Arg dipeptides were kindly provided by the Cysal GmbH [Münster, Germany; Dr. M. Krehenbrink (http://www.cysal.de/)].
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00842/full#supplementary-material
Antonova, K., Vikhnina, M., Soboleva, A., Mehmood, T., Heymich, M., Leonova, T., et al. (2019). Analysis of chemically labile glycation adducts in 3 seed proteins: case study of methylglyoxal-derived 4 hydroimidazolone 1 (MG-H1). Int. J. Mol. Sci. 20:3659. doi: 10.3390/ijms20153659
Billenkamp, N. (2010a). [Cultivar Description Burley 2010]. Sortenbeschreibung Burley 2010. Karlsruhe: Agricultural Technology Centre Augustenberg.
Billenkamp, N. (2010b). [Cultivar Description Virgin 2010]. Sortenbeschreibung Virgin 2010. Karlsruhe: Agricultural Technology Centre Augustenberg.
Billenkamp, N. (2013). [Guidepost for Tobacco Growing 2013]. Wegweiser Durch die Tabakanzucht 2013. Karlsruhe: Agricultural Technology Centre Augustenberg.
Bilova, T., Paudel, G., Shilyaev, N., Schmidt, R., Brauch, D., Tarakhovskaya, E., et al. (2017). Global proteomic analysis of advanced glycation end products in the Arabidopsis proteome provides evidence for age-related glycation hot spots. J. Biol. Chem. 292, 15758–15776. doi: 10.1074/jbc.M117.794537
Biswal, B., Krupinska, K., and Biswal, U. C. (2013). Plastid Development in Leaves during Growth and Senescence. Dirdrecht: Springer.
BMBF (2007). Weiße Biotechnologie – Chancen für Neue Produkte und Umweltschonende Prozesse [White Biotechnology – Prospekts for New Products and Environmentally Sound Processes]. Available online at: https://www.bmbf.de/ (accessed May 27, 2020).
Broer, S. (2008). Amino acid transport across mammalian intestinal and renal epithelia. Physiol. Rev. 88, 249–286. doi: 10.1152/physrev.00018.2006
Broz, A., Huang, N., and Unruh, G. (2013). Plant-based protein biomanufacturing. Genet. Eng. Biotechnol. News 33, 32–33. doi: 10.1089/gen.33.4.15
Buyel, J. F., Twyman, R. M., and Fischer, R. (2017). Very-large-scale production of antibodies in plants: the biologization of manufacturing. Biotechnol. Adv. 35, 458–465. doi: 10.1016/j.biotechadv.2017.03.011
Chen, H., Wong, E. A., and Webb, K. E. (1999). Tissue distribution of a peptide transporter mRNA in sheep, dairy cows, pigs, and chickens. J. Anim. Sci. 77, 1277–1283.
Chen, Q. (2015). Plant-made vaccines against West Nile virus are potent, safe, and economically feasible. Biotechnol. J. 10, 671–680. doi: 10.1002/biot.201400428
De Jong, D. W. (1991). Tobacco leaf protein: an evaluation of the use of putative chemical growth enhancers for tobacco leaf protein production. Beitr. Tabakforsch. Int 15, 33–41. doi: 10.2478/cttr-2013-0619
Dirisala, V. R., Nair, R. R., Srirama, K., Reddy, P. N., Rao, K. R. S. S., Satya Sampath Kumar, N., et al. (2016). Recombinant pharmaceutical protein production in plants: unraveling the therapeutic potential of molecular pharming. Acta Physiol. Plant. 39:18. doi: 10.1007/s11738-016-2315-3
Ehrlich, H., Hanke, T., Born, R., Fischer, C., Frolov, A., Langrock, T., et al. (2009). Mineralization of biomimetically carboxymethylated collagen fibrils in a model dual membrane diffusion system. J. Membrane Sci. 326, 254–259. doi: 10.1016/j.memsci.2008.10.003
Fantozzi, P., and Sensidoni, A. (1983). Protein extraction from tobacco leaves: technological, nutritional and agronomical aspects. Plant Foods Human Nutr. 32, 351–368. doi: 10.1007/bf01091194
Frommeyer, M., Wiefel, L., and Steinbüchel, A. (2014). Features of the biotechnologically relevant polyamide family “cyanophycins” and their biosynthesis in prokaryotes and eukaryotes. Crit. Rev. Biotechnol. 36, 153–164. doi: 10.3109/07388551.2014.946467
Gallie, D. R., Sleat, D. E., Watts, J. W., Turner, P. C., and Wilson, T. M. (1987). The 5’-leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo. Nucleic Acids Res. 15, 3257–3273. doi: 10.1093/nar/15.8.3257
Gecchele, E., Merlin, M., Brozzetti, A., Falorni, A., Pezzotti, M., and Avesani, L. (2015). A comparative analysis of recombinant protein expression in different biofactories: bacteria, insect cells and plant systems. J. Vis. Exp. 97:e52459. doi: 10.3791/52459
Hackl, W., Pieper, B., Pieper, R., Korn, U., and Zeyner, A. (2010). Effects of ensiling cereal grains (barley, wheat, triticale and rye) on total and pre-caecal digestibility of proximate nutrients and amino acids in pigs. J. Anim. Physiol. Anim. Nutr. 94, 729–735. doi: 10.1111/j.1439-0396.2010.01032.x
Hausmann, L., and Toepfer, R. (1999). “[Development of plasmid vectors]. Entwicklung von Plasmid-vektoren,” in [Lectures for Plant Breeding: Tailored Bioengineering of Canola Cultivars]. Vorträge für Pflanzenzüchtung: Bioengineering für Rapssorten Nach Maß, eds D. Brauer, G. Röbbelen, and R. Töpfer (Quedlinburg: Vorträge für Pflanzenzüchtung), 153–171.
Hou, Y., Wu, Z., Dai, Z., Wang, G., and Wu, G. (2017). Protein hydrolysates in animal nutrition: industrial production, bioactive peptides, and functional significance. J. Anim. Sci. Biotechnol. 8:24. doi: 10.1186/s40104-017-0153-9
Hühns, M., Neumann, K., Hausmann, T., Klemke, F., Lockau, W., Kahmann, U., et al. (2009). Tuber-specific cphA expression to enhance cyanophycin production in potatoes. Plant Biotechnol. J. 7, 883–898. doi: 10.1111/j.1467-7652.2009.00451.x
Hühns, M., Neumann, K., Hausmann, T., Ziegler, K., Klemke, F., Kahmann, U., et al. (2008). Plastid targeting strategies for cyanophycin synthetase to achieve high-level polymer accumulation in Nicotiana tabacum. Plant Biotechnol. J. 6, 321–336. doi: 10.1111/j.1467-7652.2007.00320.x
Klang, J. E., Burnworth, L. A., Pan, Y. X., Webb, K. E. Jr., and Wong, E. A. (2005). Functional characterization of a cloned pig intestinal peptide transporter (pPepT1). J. Anim. Sci. 83, 172–181. doi: 10.2527/2005.831172x
Kopertekh, L., and Schiemann, J. (2017). Transient production of recombinant pharmaceutical proteins in plants: evolution and perspectives. Curr. Med. Chem. 24, 365–380. doi: 10.2174/0929867324666170718114724
Kung, S. D., Saunder, J. A., Tso, T. C., Vaughan, D. A., Womack, M., Staples, R. C., et al. (1980). Tobacco as a potential food source and smoke material: nutritional evaluation of tobacco leaf protein. J. Food Sci. 45, 320–322. doi: 10.1111/j.1365-2621.1980.tb02605.x
Kung, S. D., and Tso, T. C. (1978). Tobacco as potential food source and smoke material: soluble protein content, extraction and amino acid composition. J. Food Sci. 43, 1844–1847. doi: 10.1111/j.1365-2621.1978.tb07428.x
Law, A. M., Lai, S. W.S., Tavares, J., and Kimber, M. S. (2009). The structural basis of beta-peptide-specific cleavage by the serine protease cyanophycinase. J. Mol. Biol. 392, 393–404. doi: 10.1016/j.jmb.2009.07.001
Li, L., Zhao, J., Zhao, Y., Lu, X., Zhou, Z., Zhao, C., et al. (2016). Comprehensive investigation of tobacco leaves during natural early senescence via multi-platform metabolomics analyses. Sci. Rep. 6:37976. doi: 10.1038/srep37976
Lu, H., and Klaassen, C. (2006). Tissue distribution and thyroid hormone regulation of Pept1 and Pept2 mRNA in rodents. Peptides 27, 850–857. doi: 10.1016/j.peptides.2005.08.012
Masclaux, C., Valadier, M.-H., Brugière, N., Morot-Gaudry, J.-F., and Hirel, B. (2000). Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211, 510–518. doi: 10.1007/s004250000310
Merlin, M., Gecchele, E., Capaldi, S., Pezzotti, M., and Avesani, L. (2014). Comparative evaluation of recombinant protein production in different biofactories: the green perspective. Biomed Res. Int. 2014:136419. doi: 10.1155/2014/136419
Moldoveanu, S. C. (2005). Analysis of protein amino acids in tobacco using microwave digestion of plant material. Beitr. Tabakforsch. Int. 21, 451–465. doi: 10.2478/cttr-2013-0813
Nandi, S., Kwong, A. T., Holtz, B. R., Erwin, R. L., Marcel, S., and McDonald, K. A. (2016). Techno-economic analysis of a transient plant-based platform for monoclonal antibody production. mAbs 8, 1456–1466. doi: 10.1080/19420862.2016.1227901
Nausch, H., and Broer, I. (2017). Cyanophycinase CphE from P. alcaligenes produced in different compartments of N. benthamiana degrades high amounts of cyanophycin in plant extracts. Appl. Microbiol. Biotechnol. 101, 2397–2413. doi: 10.1007/s00253-016-8020-8
Nausch, H., Hausmann, T., Ponndorf, D., Huhns, M., Hoedtke, S., Wolf, P., et al. (2016). Tobacco as platform for a commercial production of cyanophycin. N. Biotechnol. 33, 842–851. doi: 10.1016/j.nbt.2016.08.001
Nausch, H., Mikschofsky, H., Koslowski, R., Meyer, U., Broer, I., and Huckauf, J. (2012). High-level transient expression of ER-targeted human interleukin 6 in Nicotiana benthamiana. PLoS One 7:e48938. doi: 10.1371/journal.pone.0048938
Neubauer, K., Hühns, M., Hausmann, T., Klemke, F., Lockau, W., Kahmann, U., et al. (2012). Isolation of cyanophycin from tobacco and potato plants with constitutive plastidic cphATe gene expression. J. Biotechnol. 158, 50–58. doi: 10.1016/j.jbiotec.2011.12.008
Neumann, K., Stephan, D. P., Ziegler, K., Huhns, M., Broer, I., Lockau, W., et al. (2005). Production of cyanophycin, a suitable source for the biodegradable polymer polyaspartate, in transgenic plants. Plant Biotechnol. J. 3, 249–258. doi: 10.1111/j.1467-7652.2005.00122.x
Odell, J. T., Nagy, F., and Chua, N. H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313, 810–812. doi: 10.1038/313810a0
Ponndorf, D., Broer, I., and Nausch, H. (2017). Expression of CphB- and CphE-type cyanophycinases in cyanophycin-producing tobacco and comparison of their ability to degrade cyanophycin in plant and plant extracts. Transgenic Res. 26, 491–499. doi: 10.1007/s11248-017-0019-0
Ponndorf, D., Ehmke, S., Walliser, B., Thoss, K., Unger, C., Görs, S., et al. (2016). Stable production of cyanophycinase in Nicotiana benthamiana and its functionality to hydrolyse cyanophycin in the murine intestine. Plant Biotechnol. J. 15, 605–613. doi: 10.1111/pbi.12658
Rajeevkumar, S., Anunanthini, P., and Sathishkumar, R. (2015). Epigenetic silencing in transgenic plants. Front. Plant Sci. 6:693. doi: 10.3389/fpls.2015.00693
Sabalza, M., Christou, P., and Capell, T. (2014). Recombinant plant-derived pharmaceutical proteins: current technical and economic bottlenecks. Biotechnol. Lett. 36, 2367–2379. doi: 10.1007/s10529-014-1621-3
Sallam, A., Kalkandzhiev, D., and Steinbüchel, A. (2011). Production optimization of cyanophycinase ChpEal from Pseudomonas alcaligenes DIP1. AMB Express 1, 1–9. doi: 10.1186/2191-0855-1-38
Sallam, A., and Steinbuchel, A. (2009a). Cyanophycin-degrading bacteria in digestive tracts of mammals, birds and fish and consequences for possible applications of cyanophycin and its dipeptides in nutrition and therapy. J. Appl. Microbiol. 107, 474–484. doi: 10.1111/j.1365-2672.2009.04221.x
Sallam, A., and Steinbuchel, A. (2009b). Process for the Preparation of Dipeptides from Cyanophycin Employing the Isolated Pseudomonas Alcaligenes DIP1 CGPase CphEal. Patent EP 2133419A1.
Sallam, A., and Steinbuchel, A. (2010). Dipeptides in nutrition and therapy: cyanophycin-derived dipeptides as natural alternatives and their biotechnological production. Appl. Microbiol. Biotechnol. 87, 815–828. doi: 10.1007/s00253-010-2641-0
Sallam, A., and Steinbuchel, A. (2011). Biotechnological Production of Cyanophycin Dipeptides. Patent US 8980845 B2.
Santos, S., Torcato, I., and Castanho, M. A. (2012). Biomedical applications of dipeptides and tripeptides. Biopolymers 98, 288–293. doi: 10.1002/bip.22067
Schachtsiek, J., and Stehle, F. (2019). Nicotine-free, nontransgenic tobacco (Nicotiana tabacum l.) edited by CRISPR-Cas9. Plant Biotechnol. J. 17, 2228–2230. doi: 10.1111/pbi.13193
Schillberg, S., Raven, N., Spiegel, H., Rasche, S., and Buntru, M. (2019). Critical analysis of the commercial potential of plants for the production of recombinant proteins. Front. Plant Sci. 10:720. doi: 10.3389/fpls.2019.00720
Simon, R. D. (1973). Measurement of the cyanophycin granule polypeptide contained in the blue-green alga anabaena cylindrica. J. Bacteriol. 114, 1213–1216. doi: 10.1128/jb.114.3.1213-1216.1973
Terova, G., Robaina, L., Izquierdo, M., Cattaneo, A., Molinari, S., Bernardini, G., et al. (2013). PepT1 mRNA expression levels in sea bream (Sparus aurata) fed different plant protein sources. Springerplus 2:17. doi: 10.1186/2193-1801-2-17
Thompson, C. J., Movva, N. R., Tizard, R., Crameri, R., Davies, J. E., Lauwereys, M., et al. (1987). Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 6, 2519–2523. doi: 10.1002/j.1460-2075.1987.tb02538.x
Tschofen, M., Knopp, D., Hood, E., and Stöger, E. (2016). Plant molecular farming: much more than medicines. Annu. Rev. Anal. Chem. 9, 271–294. doi: 10.1146/annurev-anchem-071015-041706
Tusé, D., Tu, T., and McDonald, K. A. (2014). Manufacturing economics of plant-made biologics: case studies in therapeutic and industrial enzymes. BioMed Res. Int. 2014:256135. doi: 10.1155/2014/256135
Ufaz, S., and Galili, G. (2008). Improving the content of essential amino acids in crop plants: goals and opportunities. Plant Physiol. 147:954. doi: 10.1104/pp.108.118091
van Beilen, J. B., Möller, R., Toonen, M., Salentijn, E., and Clayton, D. (2007). Industrial Crop Platforms for the Production of Chemicals and Biopolymers. Newbury: CPL Press.
Walwyn, D. R., Huddy, S. M., and Rybicki, E. P. (2015). Techno-economic analysis of horseradish peroxidase production using a transient expression system in Nicotiana benthamiana. Appl. Biochem. Biotechnol. 175, 841–854. doi: 10.1007/s12010-014-1320-5
Waseem, M., Raza, A., Nawab, A., Tang, S., Waseem Ghani, M., Li, G., et al. (2019). Importance of arginine as immune regulator in animal nutrition. Int. J. Vet. Sci. Res. 5, 1–10. doi: 10.18488/journal.110.2019.51.1.10
Webb, K. E. (1986). Amino acid and peptide absorption from the gastrointestinal tract. Fed. Proc. 45, 2268–2271.
Wilkinson, R. E., Kasperbauer, M. J., and Young, C. T. (1981). Free amino acid content of Burley tobacco leaves developed under different light and temperature conditions. J. Agric. Food Chem. 29, 658–660. doi: 10.1021/jf00105a056
Wohlleben, W., Arnold, W., Broer, I., Hillemann, D., Strauch, E., and Punier, A. (1988). Nucleotide sequence of the phosphinothricin N-acetyltransferase gene from Streptomyces viridochromogenes Tü494 and its expression in Nicotiana tabacum. Gene 70, 25–37. doi: 10.1016/0378-1119(88)90101-1
Wu, G. (2009). Amino acids: metabolism, functions, and nutrition. Amino Acids 37, 1–17. doi: 10.1007/s00726-009-0269-0
Wu, G., Bazer, F. W., Dai, Z., Li, D., Wang, J., and Wu, Z. (2014). Amino acid nutrition in animals: protein synthesis and beyond. Annu. Rev. Anim. Biosci. 2, 387–417. doi: 10.1146/annurev-animal-022513-114113
Wu, G., Bazer, F. W., Davis, T. A., Kim, S. W., Li, P., Marc Rhoads, J., et al. (2009). Arginine metabolism and nutrition in growth, health and disease. Amino Acids 37, 153–168. doi: 10.1007/s00726-008-0210-y
Xu, J., Dolan, M. C., Medrano, G., Cramer, C. L., and Weathers, P. J. (2012). Green factory: plants as bioproduction platforms for recombinant proteins. Biotechnol. Adv. 30, 1171–1184. doi: 10.1016/j.biotechadv.2011.08.020
Keywords: Nicotiana tabacum, cyanophycin, co-expression, cyanophycinase CphE, β-Asp-Arg dipeptides
Citation: Nausch H, Dorn M, Frolov A, Hoedtke S, Wolf P and Broer I (2020) Direct Delivery of Health Promoting β-Asp-Arg Dipeptides via Stable Co-expression of Cyanophycin and the Cyanophycinase CphE241 in Tobacco Plants. Front. Plant Sci. 11:842. doi: 10.3389/fpls.2020.00842
Received: 22 February 2020; Accepted: 26 May 2020;
Published: 19 June 2020.
Edited by:
Joachim Hermann Schiemann, Julius Kühn-Institut, GermanyReviewed by:
Rima Menassa, Agriculture and Agri-Food Canada, CanadaCopyright © 2020 Nausch, Dorn, Frolov, Hoedtke, Wolf and Broer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Henrik Nausch, aGVucmlrLm5hdXNjaEB1bmktcm9zdG9jay5kZQ==; aGVucmlrLm5hdXNjaEBpbWUuZnJhdW5ob2Zlci5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.