
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 11 June 2020
Sec. Plant Breeding
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00810
 Zongxiang Zhan1†
Zongxiang Zhan1† Yingfen Jiang2,3†
Yingfen Jiang2,3† Nadil Shah2
Nadil Shah2 Zhaoke Hou2
Zhaoke Hou2 Yuanwei Zhou4
Yuanwei Zhou4 Bicheng Dun2
Bicheng Dun2 Shisheng Li5
Shisheng Li5 Li Zhu5
Li Zhu5 Zaiyun Li2
Zaiyun Li2 Zhongyun Piao1*
Zhongyun Piao1* Chunyu Zhang2*
Chunyu Zhang2*Clubroot caused by Plasmodiophora brassicae is a severe threat to the production of Brassica napus, worldwide. The cultivation of resistant varieties is the most efficient and environmentally friendly way to limit disease spread. We developed a highly resistant B. napus line, ZHE226, containing the resistance locus PbBa8.1. However, ZHE226 seeds contain high erucic acid content, which limits its cultivation owing to its low edible oil quality. A segregation population of BC3F2 was developed by crossing ECD04, a resistant European turnip donor, with Huangshuang5, an elite variety with no erucic acid in its seeds, as a recurrent plant. Fine mapping using the bulk segregation analysis sequencing (BSA-Seq) approach detected PbBa8.1 within a 2.9 MB region on chromosome A08. Interestingly, the previously reported resistance gene Crr1a was found in the same region. Genetic analysis revealed that the CAP-134 marker for Crr1a was closely linked with clubroot resistance (CR). Thus, PbBa8.1 and Crr1a might be allelic for CR. Moreover, comparative and genetic analysis showed that high erucic acid in the seeds of ZHE226 was due to linkage drag of fatty acid elongase 1 (FAE1) in the ECD04 line, which was located in the interval of PbBa8.1 with a physical and genetic distance of 729 Kb and 1.86 cm, respectively. Finally, a clubroot-resistant line with a low erucic acid content was successfully developed through gene-specific molecular marker assistant selection from BC4F4. These results will accelerate CR breeding programs in B. napus.
Clubroot is caused by a soil-borne, biotrophic obligate pathogen, Plasmodiophora brassicae (Woronin) and is a severe disease of rapeseed (Brassica napus) and other cruciferous crop species worldwide (Dixon, 2009). The life cycle of clubroot pathogen is comprised of two stages: root hair infection stage and cortex infection stage. Gall formation in infected roots is the most characteristic symptom of the disease, causing obstruction in nutrient uptake and resulting in premature death of the plant (Johnson and Karling, 1944; Suwabe et al., 2003; Hwang et al., 2011). The resting spores of clubroot pathogen can survive in the soil for over 20 years, making it difficult to control infection with cultural or chemical measures (Voorrips, 1995). Clubroot severely impairs the sustainable and healthy development of the rapeseed industry, especially in China and Canada. Several strategies have been developed for controlling clubroot, among which breeding of resistant cultivars is considered to be the most economical and environmentally friendly method (Donald et al., 2001; Diederichsen et al., 2009; Shigeru et al., 2010). The majority of clubroot-resistant germplasm with resistance genes/quantitative trait loci (QTL) were derived from European fodder turnips and were successfully used in CR breeding programs in Chinese cabbage and canola (Diederichsen et al., 2006; Rahman et al., 2011). The first winter canola clubroot resistant line, cv. “Mendel,” originated from a resynthesized B. napus (AACC, 2n = 38) line and was developed through the crossing of Brassica oleracea “ECD15” (CC, 2n = 18) and Brassica rapa “ECD04” (AA, 2n = 20) (Diederichsen and Sacristan, 2010). “Mendel” was successfully crossed with the Canadian spring canola and several lines were developed with high resistance to a number of P. brassicae pathotypes in Canada (Rahman et al., 2014). To date, more than 13 resistance genes/QTLs have been mapped in five chromosomes of the A-genome of B. rapa (Huang et al., 2019). Crr1 and PbBa8.1 resistance genes are located in the same locus of the A08 chromosome (Chen et al., 2013). Crr1 contains Crr1a and Crr1b, and Crr1a was successfully cloned by Katsunori et al. (2013).
The ZHE226 line (B. napus, AACC, 2n = 38), containing the resistance gene PbBa8.1, was derived by backcrossing with an elite rapeseed conventional variety, Huashuang5, while ECD04 was used as a CR donor plant (Zhan et al., 2015). ECD04 is an elite plant that contains 5 CR loci and is resistant to four different isolates of P. brassicae (Pb2, Pb4, Pb7, and Pb10) (Chen et al., 2013). The PbBa8.1 gene provides resistance to a number of clubroot-causing field isolates. The improved CR homozygous line, ZHE226, containing this locus exhibited strong resistance to pathotype 4 of P. brassicae in severely infested fields in Anhui, Hubei, and Sichuan provinces of China. However, after several backcrosses with Huashuang5, ZHE226 seeds and their offspring developed an increased and unstable content of erucic acid (C22:1) ranging from 1.39 to 18.95% in different growing seasons, as measured using near-infrared ray (NIR) techniques, while the glucosinolate content in the seeds was similar to that of Huashuang5. Erucic acid is a long-chain monounsaturated fatty acid that accumulates in the seeds of cruciferous plants and reduces the quality of oil the plants produce. It is predominantly oxidized in peroxisomes rather than in mitochondria, and promotes the production of reactive oxygen species and various cytosolic lipid metabolites. In some cases, it causes cardiotoxicity in animals, such as rodents and pigs (Fumiaki et al., 2013). High levels of erucic acid ingested through cooking with rapeseed oil is associated with ocular and respiratory tract diseases (Vles et al., 1978; Wang et al., 2010). Intake of high levels of erucic acid may also affect fertility and prenatal development, and cause damage to the human myocardial fibers (Zhang et al., 2008). The maximum level of erucic acid is found in canola seeds where it makes up 5% of the total fatty acids present, and this is considered to be an acceptable amount (Singh et al., 2011; Tian et al., 2018). However, reducing the content of erucic acid in oil to an acceptable level, or close to negligible amounts, is one of the most significant breeding goals for improving the canola oil quality (Vesna et al., 2010). Therefore, selecting clubroot-resistant materials with low erucic acid content (CR-LEA) is crucial for rapeseed breeding programs.
The fatty acid elongase 1 (FAE1) gene encodes a seed-specific enzyme of β-ketoacyl-CoA synthase that catalyzes the first condensation step in the elongation of very-long-chain fatty acids and limits erucic acid biosynthesis (Roscoe et al., 2001; Sun et al., 2013). The FAE1enzyme extends the fatty acid chain length from C18 to C20 and C22 (Lassner et al., 1996; Han et al., 2001). It was initially cloned in Arabidopsis by directed transposon tagging with the maize element activator (Ac) (James et al., 1995). Dorrell and Downey (1964) and Karim et al. (2016) indicated that erucic acid synthesis was determined by a single non-dominant gene, in B. rapa (Brassica campestris), whereas, Siebel and Pauls (1989) reported that erucic acid is simply inherited and controlled by two additive genes in B. napus. The two additive alleles are located on the A08 and C03 chromosomes in B. napus, accounting for ∼71% of the genetic variation in erucic acid synthesis (Qiu et al., 2006). The first oilseed cultivar with low erucic acid content (LEA) oilseed cultivar, Oro, was bred using the LEA Liho crop as parental material (Gang et al., 2008). The use of Oro as a donor plant has become a worldwide standard for the breeding of cultivars with LEA in seeds. Erucic acid content in ZHE226 was modestly high, although it was backcrossed with the LEA variety Huashuang5 for several generations. However, it is unclear whether FAE1 is a linkage drag of PbBa8.1, considering both genes are located on the A08 chromosome (Fourmann et al., 1998; Chen et al., 2013). Aside from the high erucic acid content in the seeds, ZHE226 is a CR line with high seed yield performance, with a similar genetic background to the recurrent parental variety Huashuang5. Therefore, in this study, a relationship between the genetically close linkage of the CR locus PbBa8.1 and the high erucic acid determining gene FAE1 was revealed, and a valuable CR line with LEA content was developed. Overall, this study will accelerate CR breeding programs in B. napus.
The segregation population of BC3F2 with CR was developed by crossing/backcrossing of ECD04 (European turnip, donor plant) containing the CR gene PbBa8.1 with Huangshuang5 (CR susceptible, recurrent plant), which is an elite conventional rapeseed variety with zero erucic acid and low glucosinolate content in the seeds. This population was used for bulked segregation analysis coupled with whole genome sequencing (BSA-Seq). Another segregation population, BC4F3, was developed by backcrossing BC3F2 homozygous for the PbBa8.1 gene with Huashuang5 (Figure 1). Linkage analyses of PbBa8.1 and FAE1 were carried out for the generation of this line. European turnip ECD04, a CR donor plant containing PbBa8.1with a high concentration of erucic acid content in seed, was used as a control for FAE1 gene cloning.
Plasmodiophora brassicae samples with severe galling were collected from the Huangshan locality, amplified on B. napus-susceptible material, and stored at −20°C. The resistance test in the control condition was carried out as previously described by Chen et al. (2013). Briefly, frozen galls were thawed at room temperature, homogenized, and resting spores were extracted. The spore concentration was adjusted to107 resting spores per milliliter in sterile distilled water and this spore suspension was used for host root inoculation. One-week-old seedlings were inoculated with resting spores by injecting 1 mL of the spore suspension into the root zone around the plant (Chen et al., 2013; Pang et al., 2018). The greenhouse day/night temperature was maintained at 25/20°C. The plants were watered regularly. Forty days post-inoculation (dpi), the plants were carefully dug out, their roots were washed, and roots were examined for resistance (Zhan et al., 2017).
Genomic DNA from young leaves was extracted using the DNeasy Plant Kit (TIANGEN, Beijing, China). DNA quality and quantity were measured using a Nano-Drop 2000 (Thermo Fisher Scientific, Waltham, MA, United States). An equal amount of DNA (2.0 μg/sample) from 30 BC3F2-resistant plants were mixed to construct the resistant bulk sample (R-pool) and a similar amount of DNA from 30 BC3F2-susceptible plants with severe clubs were mixed to form susceptible bulk samples (S-pool). The DNA samples were sequenced using the Illumina HiSeq 2000 platform (Illumina, San Diego, CA, United States) with 20× read depth. Quality control (QC) parameters were set as follows: QC procedures were reads with ≥10% unidentified nucleotides, reads with >50% bases having phred quality <5, reads with >10 nt aligned to the adapter, and putative PCR duplicates generated by PCR amplification in the library construction process were removed, and ≤10% mismatches were allowed. The clean reads were mapped to the reference genome of B. napus1 using the Burrows-Wheeler Aligner (BWA).
SAMtools and Varscan2 were employed to screen the InDel markers (Koboldt et al., 2012). The flanking sequence of the InDel site was extracted for primer development using Primer3 software (Untergasser et al., 2012). The parameters were set as follows: ≤3 nucleotide mismatch for the primers were allowed, ≤2 gaps in the PCR production were allowed, and PCR production size was set to 50–250 base pairs (bp). All primers were screened with the template genomic sequences from B. rapa and B. napus by e-PCR. The markers with only one amplification product in B. napus that also anchored on the same chromosome of B. rapa were allowed.
SNP/InDel detection and variant annotation were performed for the two pools (R and S-pool) using the Unified Genotyper function (GATK software). The ANNOVAR tool was used to annotate SNPs or InDels based on the reference genome (Mckenna et al., 2010). The read depth information for homozygous SNPs in the offspring pools was obtained to calculate the SNP index (frequency). The SNP indices in both pools that were under 0.3 were filtered out. The sliding window method was employed to present the SNP index of the whole genome. The average SNP index in each window was used as the SNP index. Default settings included a window size of 100 kb at a step size of 20 kb. A 95% interval of the permutation test was selected as a candidate locus from the delta of all indices (Takagi et al., 2013).
Genomic DNA from young leaves of ZHE226, ECD04, and Huashuang5 was extracted using the CTAB method (Murray and Thompson, 1980). Primers were designed according to the gene sequence of FAE1 (GU325717.1) published by NCBI3 The purified PCR products were amplified with Fast-Pfu Polymerase, ligated to the pEASY-Blunt vector (TransGen Biotech Co., Beijing, China) and transformed into Escherichia coli DH5α competent cells (Takara). At least four positive clones with a correct insertion size were selected and sequenced (AuGCT, Beijing, China). The gene-specific markers, AW and AM (Supplementary Table S1), were designed based on the SNP of FAE1 that differed between Huashuang5 and ECD04 lines using the amplification refractory mutation system (ARMS) method (Newton et al., 1989).
To determine the erucic acid content and genotypes in the segregating BC4F3 population at the earliest stage, the half-seed technique was employed (Anand and Downey, 1981). Briefly, a portion of the seed (0.5 mg) was cut carefully without damaging the embryo and transferred to a 5 mL glass tube. Then, 1.5 mL of 2.5% methanol sulfate solution and 350 μL of methylbenzene was added. The glass tubes were carefully sealed and kept at 90°C for 30–45 min. After cooling to room temperature, 1 mL of double-distilled H2O (ddH2O) and 1 mL n-hexane were added to each tube, blended, and tubes were centrifuged at 1000 rpm for 5 min using a Heraeus Fresco 21 microcentrifuge (Thermo Fisher Scientific, Germany). The supernatant containing the fatty acid methylated ester was transferred to autosampler vials and 0.5 μL samples were injected and analyzed by gas chromatography (GC) (HP7890A, Agilent) at a nitrogen carrier gas flow rate of 30 mL/min. The initial oven temperature was 180°C for 2 min, followed by 10°C/min to a final temperature of 220°C, which was held for 12 min.
Fatty acid standards [palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2), linolenic acid (C18:3), eicosenoic acid (C20:1), and erucic acid (C22:1) were purchased from Sigma-Aldrich China (Shanghai)]. The mean values and standard deviations from three biological replicates were calculated. Quantification of each fatty acid was carried out by the percentage of peak values using corresponding standard samples (Zhang et al., 2013).
A total of 933 BC3F2 individuals derived from ECD04 and Huashuang5 were inoculated with P. brassicae pathotype 4 collected from Huangshan, China. Of these individuals, 720 were resistant (R) to clubroot and 213 were susceptible (S), with a segregation ratio of 3:1, indicating a dominant pattern for inheritance of resistance [N = 933, χ2 = 2.49 < χ20.05(1) = 3.84] (Figure 2). The BSA-resequencing method was employed for fine-mapping of the resistance locus. A total of 137,056,905 and 138,277,214 clean reads were obtained from R- and S-pools after filtering with the next generation sequencing (NGS) tool kit, respectively (Supplementary Table S2, SAR accession: PRJNA605484). Over 95% of the clean reads were mapped onto the genome of B. napus (Brassica_napus_v4.1.chromosomes) (Supplementary Table S2), and a total of 303,345 SNPs and 18,818 InDels were detected between the two pools.

Figure 2. Clubroot resistant tests with pathogen of Huangshan in the BC3F2 generation. Root symptom evaluation of resistant BC3F2 individuals, susceptible individuals, and Huashuang5 (left to right).
The CR locus, PbBa8.1, was mapped to a physical region between 8.15 and 11.22 Mb on chromosome A08 of B. napus based on the ΔSNP index (Figure 3A). Approximately 57.32% of the SNPs were mapped to the A08 chromosome, and more than 12,000 SNPs were located in the candidate region. The large number of SNPs on the A08 chromosome may be due to the advanced population used in the experiment. A high-density physical map was then constructed (Figure 3A). A total of 121 indel markers were screened in the candidate region (Supplementary Table S3) with a density of 33.41 markers per Mb. Molecular markers were selected for synteny analysis, and the results revealed that the previously reported resistance gene, Crr1, overlapped with PbBa8.1 on the A08 chromosome between the markers of BSA1 and BRMS-088 in the physical map of the present study, with an interval of 1.3 Mb (Figure 3B and Supplementary Table S1).
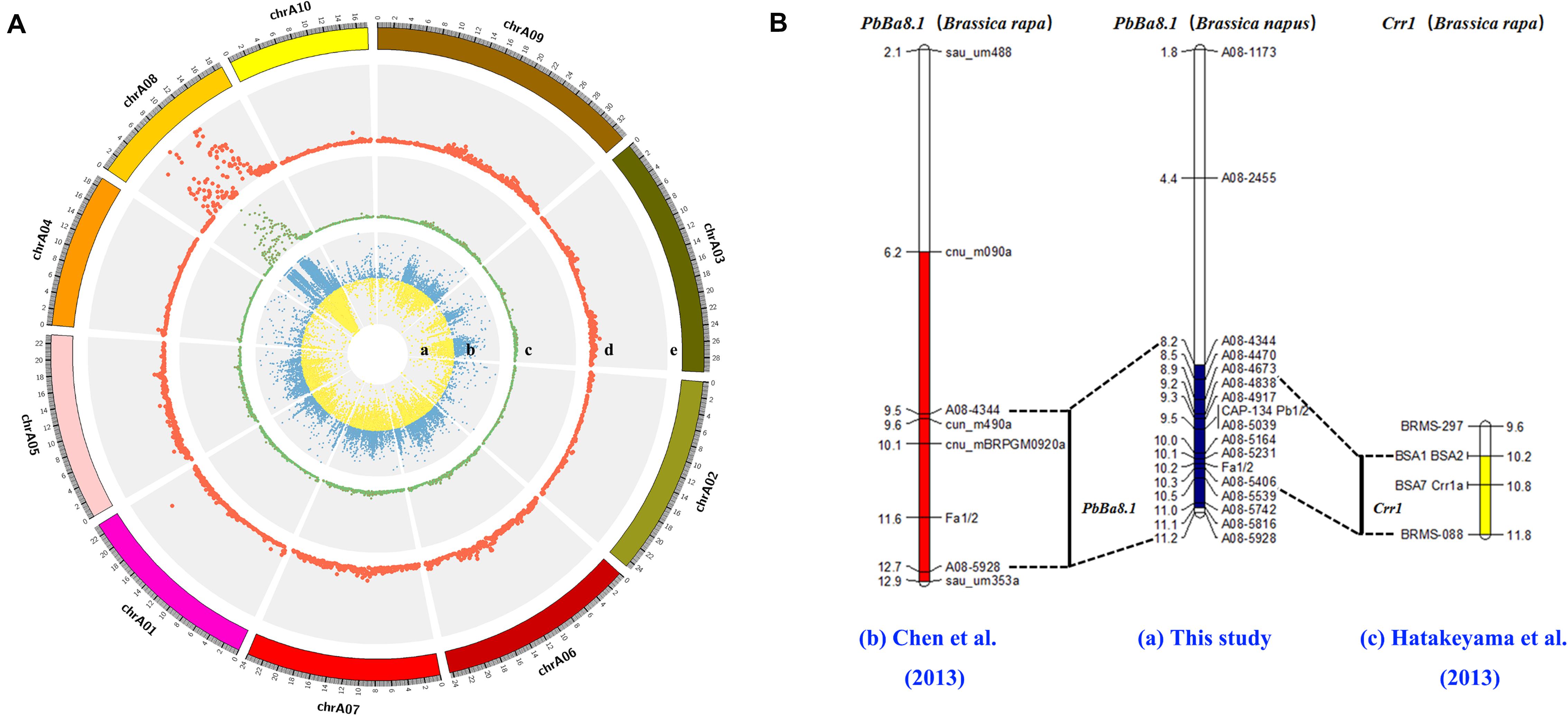
Figure 3. Fine mapping of PbBa8.1 in B. napus and synteny analysis with other resistant loci. (A) SNP distribution in the A genome and candidate region prediction. (B) Physical mapping of PbBa8.1 with InDel markers and synteny analysis with other resistant loci. The blue bar represents the candidate region of PbBa8.1 in B. napus. The red bar represents the candidate region of PbBa8.1 in B. rapa. The yellow bar represents the candidate region of Crr1 in B. rapa.
Although Crr1a was derived from “Siloga S2” and exhibits minor CR alone, while PbBa8.1 is a dominant locus that is likely controlled by at least one of the divergent functional alleles of Crr1a. To prove this hypothesis, a pair of gene-specific primers were designed based on the Crr1a sequence and used to amplify a 380 bp fragment from the total genomic DNA of ECD04 and Huashuang5 as templates. Sequencing results revealed that the 380 bp sequence from Huashuang5 is exactly the same as the reported Crr1a wild type sequence, indicating this gene does not significantly impact CR in Huashuang5. Fortunately, variations were observed in the corresponding sequence of ECD04 (Figure 4A), and this region was sufficient to generate a cleaved amplified polymorphic sequences (CAPS) marker (CAP-134) based on the sequence differences (249–251 sequence position), resulting in an extra restriction enzyme cutting site for TaqI in the ECD04 genome (Figure 4A). Using the CAP-134 marker, 200 randomly selected lines from the S-pool of the above BC3F2 generation were genotyped and 196 were found to be homozygous for the marker, while four were heterozygous (Figure 4B). To confirm whether the above four heterozygous plants were infected by other mixed pathotypes in the field, the self-pollinated seeds from these four heterozygous individuals were planted in the same infested field as Huangshan5 in the next growing season. The results showed a 3:1 segregation ratio for resistance and susceptibility indicating that, rather than recombination, these four plants were randomly infected by other pathotypes in the field with mixed features. Further the co-segregation characteristics of CAP-134 with PbBa8.1 was proven based on a large population.
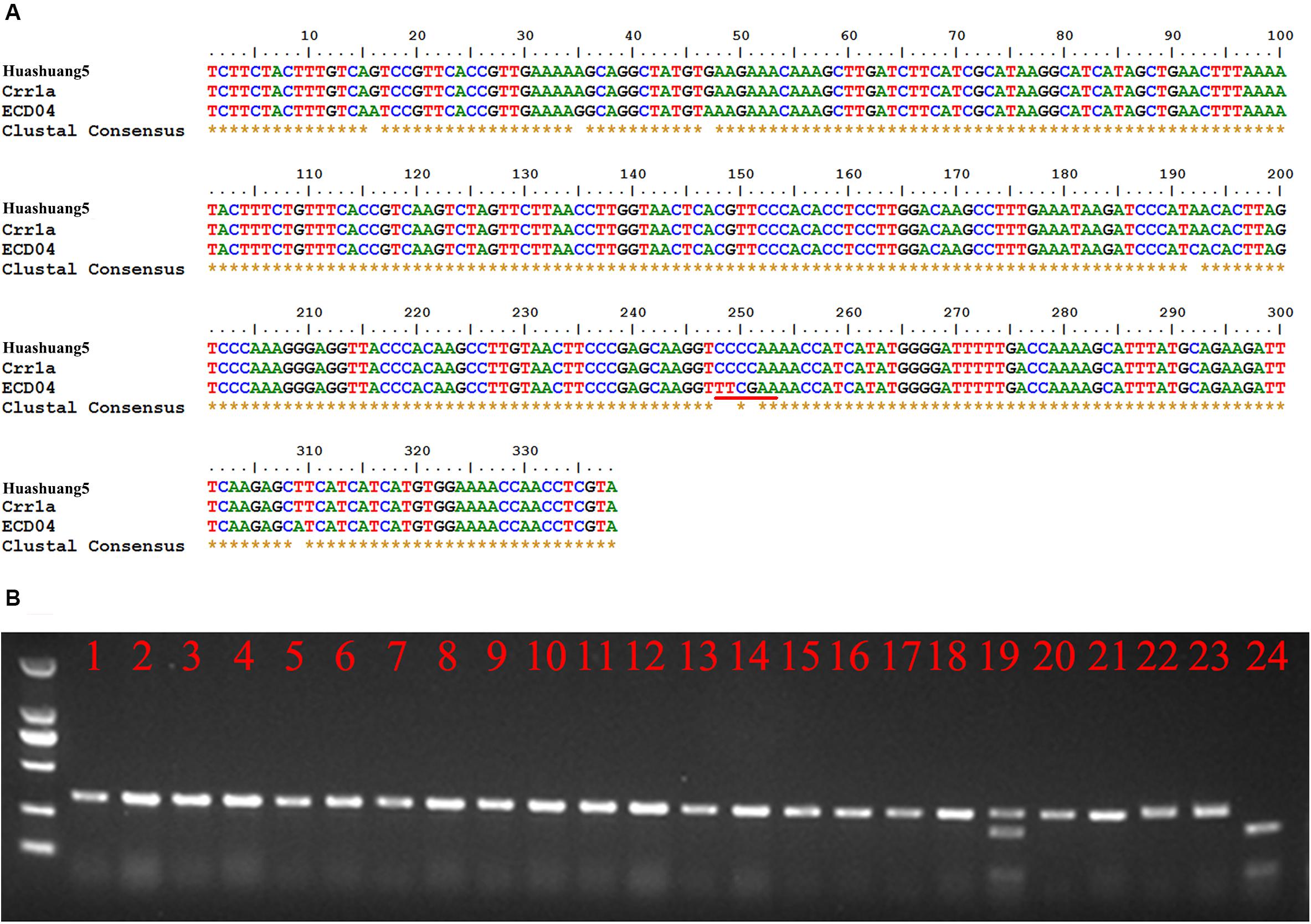
Figure 4. Gene-specific marker, CAP-134, designed based on sequence differences between Huashuang5 and ECD04. (A) Homologous nucleotide sequence alignment of Crr1a of ECD04 and Huashuang5. The red line indicates the restriction endonuclease sites of TaqI. (B) Genotype analysis of randomly selected susceptible individuals of the BC3F2 generation. Lines 1–22 show the susceptible plants. Lines 23 and 24 show Huangshuang5 and ECD04, respectively.
The FAE1 enzyme catalyzes the first condensation step in the elongation of very longchain monosaturated fatty acids (VLCMFA). There are two copies of FAE1 on the C3 and A08 chromosomes in B. napus (BnC3FAE1 and BnA8FAE1). The FAE1 gene was mapped to chromosome A08 to a 10.18 MB region (Brassica_napus_v4.1.chromosomes), as shown in Figure 3B. The physical distance between CAP-134 and BnA8FAE1was approximately 729 Kb, indicating a linkage drag of the genes causing high erucic acid content and CR. In order to confirm this hypothesis, the oil composition of mature seeds from ZHE226, Huashuang5, and ECD04 were measured using GC. The erucic acid composition in ZHE226 and ECD04 seeds was 24.88 and 44.58%, respectively, (Supplementary Table S4). However, erucic acid was not detected in Huashuang5, which confirmed that the high erucic acid haplotype of FAE1 in ZHE226 was inherited from ECD04 and not Huashuang5 (Figure 5). To further elucidate this result, genomic DNA from BnA8FAE1 was isolated by PCR and then sequenced and aligned (Supplementary Figure S1). As expected, the sequences of ECD04 and ZHE226 were identical to GU325717.1 (BnA8FAE1); however, there was a single transition of cytosine (C) to thymine (T) at position of 845of the FAE1 coding sequence in Huashuang5, which causes the serine (Ser) residue to transform to a phenylalanine (Phe) residue at amino acid position 282 (Table 1 and Supplementary Figure S1). These results indicate that this single C to T transition causes a loss of function mutation in BnA8fae1 in Huashuang5.
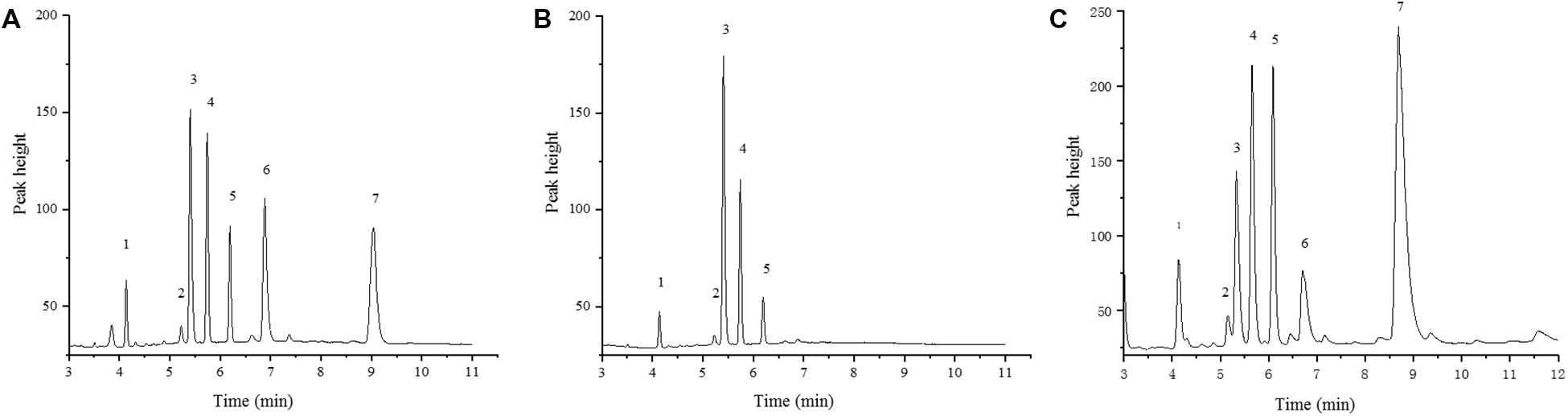
Figure 5. Gas Chromatograms of ZHE226, Huashuang5, and ECD04 seed fatty acid composition. (A) ZHE226; (B) Huashuang5; (C) ECD04. No. 1–7 represents palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2), linolenic acid (C18:3), eicosenoic acid (C20:1), and erucic acid (C22:1), respectively.
In order to screen CR lines with LEA content, 296 individual seeds of the BC4F3 segregation population were used for erucic acid quantification, and genotyping of BnA8FAE1/BnA8fae1 was performed using gene-specific marker pairs AW/AM. Phenotyping and genotyping results revealed that the average erucic acid content in the seeds of BnA8fae1/BnA8fae1, BnA8FAE1/BnA8fae1, and BnA8FAE1/BnA8FAE1 were 0, 9.04, and 19.26%, respectively, indicating an additive effect of BnA8FAE1 on erucic acid content. Using the gene-specific marker pair of Pb1/Pb2, converted from CAP-134 with ARMS method, the average erucic acid content in the seeds of pbBa8.1/pbBa8.1, PbBa8.1/pbBa8.1 andPbBa8.1/PbBa8.1 was 0.24, 8.83, and 18.95%, respectively (Figure 6). There were no significant differences in the average content of erucic acid between genotypes of PbBa8.1/PbBa8.1 and BnA8FAE1/BnA8FAE1, BnA8FAE1/BnA8fae1 and PbBa8.1/pbBa8.1, and BnA8fae1/BnA8fae1 and pbBa8.1/pbBa8.1. This confirmed that BnA8FAE1 is the linkage drag of PbBa8.1, and that FAE1 derived from ECD04 has an additive effect on erucic acid accumulation. A total of 11 individuals with recombination occurring between the two loci were identified and 4 of them were heterozygous for the PbBa8.1 locus with zero erucic acid in the seeds (Supplementary Table S5), because they carried the BnA8fae1gene. The genetic distance between PbBa8.1 and BnA8FAE1 was calculated to be approximately 1.86 cm based on the recombination rate in B. napus (Figure 7).
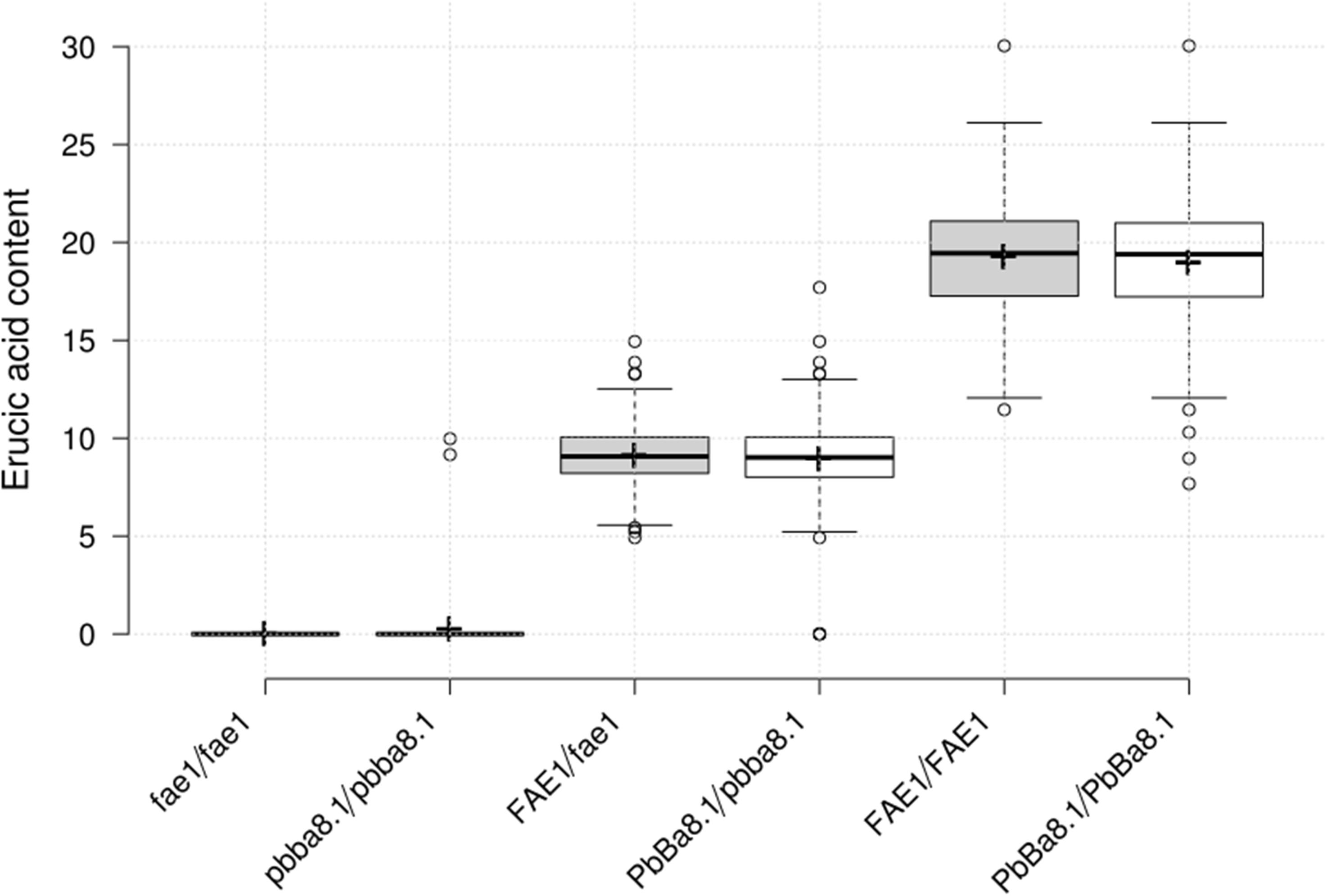
Figure 6. Erucic acid content of the different genotypes in the BC4F3 generation. Centerline represents the median; upper and lower boxes indicate upper and lower quartiles (Q3 and Q1) calculated with R software; the dotted line extends 1.5 times the upper and lower quartiles; circles indicate outliers; crosses represent sample means; n = 82, 80, 141, 141, 74, 75.
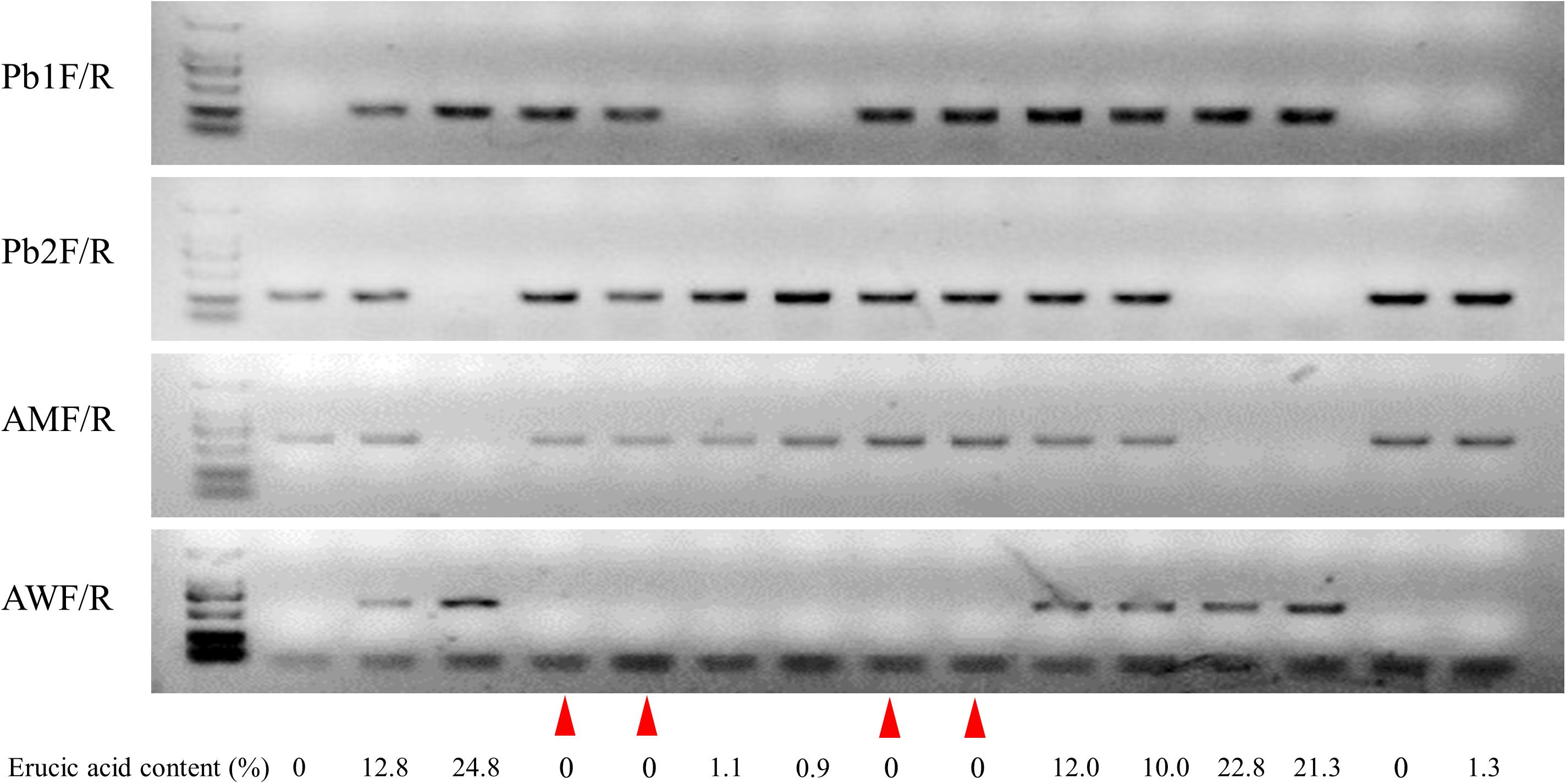
Figure 7. Genotypes and their corresponding erucic acid content in the BC4F3 generation. Lines 1–3 show Huashuang5, BC3F1 generation, and ZHE226, respectively. The remaining lines show heterozygous individuals selected from the BC4F3 generation. The red triangles represent the exchange of individuals with low erucic acid content.
The above four recombinant lines (Supplementary Table S4) heterozygous for PbBa8.1 as confirmed by Pb1/Pb2,with no erucic acid content as confirmed by GC analysis, were selected and self-pollinated to obtain homozygous CR lines. To confirm the erucic acid content of the self-pollinated seeds of these four recombinant lines, the NIRS technique was used to measure fatty acid composition. Very LEA contents of 2.8, 1.0, 1.0, and 1.4%were found in each of the four lines. Subsequently, the four different self-pollinated populations were tested for CR in the Huangshan region. A total of 173 homozygous and stable CR individuals were obtained by associated marker selection of the CR lines. Further, 16 randomly selected self-pollinated homozygous CR plants from the four different BC4F4 populations were and selected for quality analysis by NIRS. The erucic acid content of all these plants was less than 1.5% (Supplementary Table S6). Additionally, four lines were randomly selected for CR tests in the greenhouse. In contrast to the control plants, these lines exhibited durable resistance to the clubroot pathogen collected at the Huangshan locality (Figure 8).

Figure 8. Clubroot resistance of four independent CR-LEA line. Panels (A–D) show Huashuang5 and the others show homozygous disease-resistant lines without erucic acid.
Clubroot is a serious threat to rapeseed production globally, and particularly in China. Breeding of highly resistant varieties is the most effective way to control this disease (Diederichsen et al., 2009). CR turnips are widely used for CR breeding and resistance gene mapping. Most CR loci are race-specific. Previously, we developed a CR line, ZHE226, that contains the PbBa8.1 gene, which confers strong resistance against P. brassicae pathotype 4 (Zhan et al., 2015). In the early generation of this elite line, the erucic acid content was not determined because it was expected that the offspring should have characteristic LEA and glucosinolate contents. However, in the BC3F2 generation, a high erucic acid content in the seed oil was observed in all CR lines, although glucosinolate content was the same as that of the parental Huangshuang5 line. In the present study, we predicted that the PbBa8.1 locus might be closely linked with the FAE1 gene that is located in the same chromosome, and could increase erucic acid levels. Fine mapping revealed that a linkage drag of high erucic acid determined by the haplotype BnA8FAE1 is associated with the CR locus PbBa8.1. The recombination rate between the candidate genes, PbBa8.1 and BnA8FAE1, was approximately 1.86%. In this study, a novel line, Huashuang 5R, with a very low seed erucic acid content and high resistance to clubroot was developed successfully. PbBa8.1 exhibited dominant resistance against pathogen type 4 collected from the Huangshan region (Anhui Province). These results are consistent with the fact that PbBa8.1 revealed a high resistance against a number of P. brassicae field isolates from Anhui, Sichuan, and Hubei provinces (Shah et al., 2019).
Our fine-mapping data revealed that Crr1a and PbBa8.1 might be allelic and present in the same region in the genetic linkage map. However, Crr1a was cloned from the resistant line G004, encoding a Toll-Interleukin-1 receptor/nucleotide-binding site/leucine-rich repeat (TIR-NB-LRR) protein. Genetic analysis of the F2 population showed that the heterozygous Crr1 locus demonstrates a minor resistance against Ano-01 (Katsunori et al., 2013). Together with Crr2, Crr1 synergistically enhanced resistance (Suwabe et al., 2003).Thus, we can conclude that PbBa8.1 and Crr1 may be allelic, but differ in function. However, further study is required to explore whether other resistance genes are present in this region.
Interspecific hybridization breeding is aimed at introgression of a desirable trait from a wild donor line into an elite receptor line. However, undesirable and desirable traits are sometimes closely linked to each other in the alien segment from the donor that integrates into the genome of the receptor; this phenomenon is called linkage drag. Linkage drag can be removed by backcrossing. The probability of linkage drag depends on homology and the distance between two loci; the closer the linkage is, the more difficult it is to separate it, as it may require a large population and many generations of backcrossing. For example, linkage drag between high glucosinolate content in seeds and the restoration of Ogr-INGA cytoplasmic male sterility from the radish into B. napus took more than 10 years to relieve (Primard-Brisset et al., 2005). In tomato, the linkage breakdown of nematode resistance and undesirable fruit characteristics took 12 years to separate (Acquaah, 2012). However, in rice blast resistance, linkage drag was effectively removed in two cycles of recombinant selection (Feng et al., 2019). Cao et al. (2010) reported that backcross breeding and marker-assisted selection are effective tools to break linkage drag between low oil content and erucic acid in canola cultivars. Interestingly, in the current study, the linkage drag between the CR resistance locus and high erucic acid was relieved in 3 years, indicating a significantly high homology in the region of PbBa8.1 of the turnip ECD04 to that of B. napus Huashuang5. Linkage drag could have been disrupted earlier if a large segregation population was used in the early segregated generations.
The partial allelic sequence results showed that Crr1a is identical to Huashuang5 (Figure 4). However, no genes homologous to Crr1a were predicted after Blast against the whole genome of B. napus. This is reasonable since Huashuang5 is completely susceptible to P. brassicae. Further, the marker, CAP-134, derived from the different sequences between ECD04 and Huashuang5, co-segregated with CR and could be very useful for promoting CR breeding for canola.
Minor galls were observed (approximately 8%) in heterozygous lines harboring PbBa8.1. However, a 3:1 segregation ratio for CR was repeatedly obtained when self-pollinated plants from heterozygous lines were tested in the field or in greenhouse conditions. On the other hand, the mixture of the clubroot pathogens with variance in virulence and uneven distribution in the field could be the cause of the discrepancy. Similar results were reported by Strelkov et al. (2016). Therefore, we suggest that successful fine mapping of CR genes, single spore isolation, and genetic recombinant studies should be employed in future programs. It is also crucial that the desired lines carefully self-pollinate first and then be examined for inoculation tests for the next generation.
All datasets generated for this study are included in the article/Supplementary Material.
ZZ analyzed the data, performed the experiments, and drafted the manuscript. YJ performed the experiments and helped to draft the manuscript. NS helped to analyze the data. ZH and BD contributed to perform the experiments. SL and LZ helped to analyze the data. ZL and ZP helped to draft the manuscript. CZ conceived the study, participated in its coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
This project was supported by grants from the National Natural Science Foundation of China (grant No. 31871659) and the China Agriculture Research System (CARS-12).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00810/full#supplementary-material
FIGURE S1 | Nucleotide sequence alignment of FAE1 between ECD04 and Huasuang 5. Red frame indicated the Nucleotide mutation site.
TABLE S1 | All the primer sequences used in this study.
TABLE S2 | The statistics of the R- and S-pools sequence alignment to the reference genome.
TABLE S3 | List of the InDel markers designed in the candidate region.
TABLE S4 | Fatty acid composition in seeds of ZHE226, Huashuang5 and ECD04.
TABLE S5 | Recombinant plants and their erucic acid content detected in BC3F2 generation.
TABLE S6 | Seed oil quality detection of 16 CR-LEA lines selected in BC3F4 generation and Huashuang5.
Acquaah, G. (2012). Principles of Plant Genetics And Breeding, 2nd Edn, Hoboken, NJ: John Wiley & Sons, Ltd.
Anand, I. J., and Downey, R. K. (1981). A study of erucic acid alleles in digenomic rapeseed (Brassica napus L.). Can. J. Plant Sci. 61, 198–202. doi: 10.4141/cjps81-030
Cao, Z., Tian, F., Wang, N., Jiang, C., Lin, B., Xia, W., et al. (2010). Analysis of QTLs for erucic acid and oil content in seeds on A8 chromosome and the linkage drag between the alleles for the two traits in Brassica napus. J. Genet. Genomics 37, 231–240. doi: 10.1016/S1673-8527(09)60041-2
Chen, J., Jing, J., Zhan, Z., Zhang, T., Zhang, C., and Piao, Z. (2013). Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS One 8:e85307. doi: 10.1371/journal.pone.0085307
Diederichsen, E., Beckmann, J., Schondelmeier, J., and Dreyer, F. (2006). Genetics of clubroot resistance in Brassica napus ‘Mendel’. Acta Hortic. 706, 307–312. doi: 10.17660/ActaHortic.2006.706.35
Diederichsen, E., Frauen, M., Linders, E. G. A., Hatakeyama, K., and Hirai, M. (2009). Status and perspectives of clubroot resistance breeding in crucifer crops. J. Plant Growth Regul. 28, 265–281. doi: 10.1007/s00344-009-9100-0
Diederichsen, E., and Sacristan, M. D. (2010). Disease response of resynthesized Brassica napus L. lines carrying different combinations of resistance to Plasmodiophora brassicae Wor. Plant Breed. 115, 5–10. doi: 10.1111/j.1439-0523.1996.tb00862.x
Dixon, G. R. (2009). The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 28, 194–202. doi: 10.1007/s00344-009-9090-y
Donald, E. C., Porter, I. J., and Lancaster, R. A. (2001). Band incorporation of fluazinam (Shirlan) into soil to control clubroot of vegetable brassica crops. Aust. J. Exp. Agric. 41, 1223–1226. doi: 10.1071/ea00135
Dorrell, D. G., and Downey, R. K. (1964). The inheritance of erucic acid content in rapeseed (Brassica campestris). Can. J. Plant Sci. 44, 104–111. doi: 10.4141/cjps64-019
Feng, X., Lin, K., Zhang, W., Nan, J., Zhang, X., Wang, C., et al. (2019). Improving the blast resistance of the elite rice variety Kongyu-131 by updating the pi21 locus. BMC Plant Biol. 19:249. doi: 10.1186/s12870-019-1868-x
Fourmann, M., Barret, P., Renard, M., Pelletier, G., Delourme, R., and Brunel, D. (1998). The two genes homologous to Arabidopsis FAE1 co-segregate with the two loci governing erucic acid content in Brassica napus. Theor. Appl. Genet. 96, 852–858. doi: 10.1007/s001220050812
Fumiaki, I., Lemaitre, R. N., King, I. B., Xiaoling, S., Steffen, L. M., Folsom, A. R., et al. (2013). Long-chain monounsaturated Fatty acids and incidence of congestive heart failure in 2 prospective cohorts. Circulation 127:1512. doi: 10.1161/CIRCULATIONAHA.112.001197
Gang, W., Wu, Y., Ling, X., Li, X., and Lu, C. (2008). Zero erucic trait of rapeseed (Brassica napus L.) results from a deletion of four base pairs in the fatty acid elongase 1 gene. Theor. Appl. Genet. 116, 491–499. doi: 10.1007/s00122-007-0685-z
Han, J., Lühs, W., Sonntag, K., Zähringer, U., Borchardt, D. S., Wolter, F. P., et al. (2001). Functional characterization of β-ketoacyl-CoA synthase genes from Brassica napus L. Plant Mol. Biol. 46, 229–239. doi: 10.1023/A:1010665121980
Huang, Z., Peng, G., Gossen, B. D., and Yu, F. (2019). Fine mapping of a clubroot resistance gene from turnip using SNP markers identified from bulked segregant RNA-Seq. Mol. Breed. 39:1038. doi: 10.1007/s11032-019-1038-8
Hwang, S. F., Ahmed, H. U., Zhou, Q., Strelkov, S. E., Gossen, B. D., Peng, G., et al. (2011). Influence of cultivar resistance and inoculum density on root hair infection of canola (Brassica napus) by Plasmodiophora brassicae. Plant Pathol. 60, 820–829. doi: 10.1111/j.1365-3059.2011.02457.x
James, D. W., Lim, E., Keller, J., Plooy, I., Ralston, E., and Dooner, H. K. (1995). Directed tagging of the Arabidopsis Fatty Acid Elongation1 (FAE1) gene with the maize transposon activator. Plant Cell 7, 309–319. doi: 10.1105/tpc.7.3.309
Johnson, T. W., and Karling, J. S. (1944). The plasmodiophorales. Science 99, 299–301. doi: 10.2307/1295321
Karim, M. M., Tonu, N. N., Hossain, M. S., Funaki, T., Meah, M. B., Hossain, D. M., et al. (2016). Marker-assisted selection of low erucic acid quantity in short duration Brassica rapa. Euphytica 208, 535–544. doi: 10.1007/s10681-015-1596-8
Katsunori, H., Keita, S., Norio, T. R., Takeyuki, K., Tsukasa, N., Hiroyuki, F., et al. (2013). Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS One 8:e54745. doi: 10.1371/journal.pone.0054745
Koboldt, D. C., Zhang, Q., Larson, D. E., Shen, D., McLellan, M. D., Lin, L., et al. (2012). VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576. doi: 10.1101/gr.129684.111
Lassner, M. W., Lardizabal, K., and Metz, J. G. (1996). A jojoba β-Ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell 8, 281–292. doi: 10.2307/3870271
Mckenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110.20
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Newton, C. R., Graham, A., Heptinstall, L. E., Powell, S. J., Summers, C., Kalsheker, N., et al. (1989). Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17, 2503–2516. doi: 10.1093/nar/17.7.2503
Pang, W., Fu, P., Li, X., Zhan, Z., Yu, S., and Piao, Z. (2018). Identification and mapping of the clubroot resistance gene CRd in chinese cabbage (Brassica rapa ssp. pekinensis). Front Plant Sci. 9:653. doi: 10.3389/fpls.2018.00653
Primard-Brisset, C., Poupard, J. P., Horvais, R., Eber, F., Pelletier, G., Renard, M., et al. (2005). A new recombined double low restorer line for the Ogu -INRA cms in rapeseed (Brassica napus L.). Theor. Appl. Genet. 111, 736–746. doi: 10.1007/s00122-005-2059-8
Qiu, D., Morgan, C., Shi, J., Long, Y., Liu, J., Li, R., et al. (2006). A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor. Appl. Genet. 114, 67–80. doi: 10.1007/s00122-006-0411-2
Rahman, H., Peng, G., Yu, F., Falk, K. C., Kulkarni, M., and Selvaraj, G. (2014). Genetics and breeding for clubroot resistance in Canadian spring canola (Brassica napus L.). Can. J. Plant Pathol. 36, 122–134. doi: 10.1080/07060661.2013.862571
Rahman, H., Shakir, A., and Hasan, M. J. (2011). Breeding for clubroot resistant spring canola (Brassica napus L.) for the Canadian prairies: can the european winter canola cv. mendel be used as a source of resistance? Can. J. Plant Sci. 91, 447–458. doi: 10.4141/cjps10073
Roscoe, T. J., Lessire, R., Puyaubert, J., Renard, M., and Delseny, M. (2001). Mutations in the fatty acid elongation 1 gene are associated with a loss of β-ketoacyl-CoA synthase activity in low erucic acid rapeseed. FEBS Lett. 492, 107–111. doi: 10.1016/S0014-5793(01)02243-8
Shah, N., Sun, J., Yu, S., Yang, Z., Wang, Z., Huang, F., et al. (2019). Genetic variation analysis of field isolates of clubroot and their responses to Brassica napus lines containing resistant gene CRb, PbBa8.1 and their combination in homozygous and heterozygous state. Mol. Breed. 39:153. doi: 10.1007/s11032-019-1075-3
Shigeru, M., Koji, S., Hiroyuki, H., Yasuko, T., Takeshi, O., and Norifusa, M. (2010). Effects of cyazofamid against Plasmodiophora brassicae Woronin on Chinese cabbage. Pest Manag. Sci. 59, 287–293. doi: 10.1002/ps.627
Siebel, J., and Pauls, K. P. (1989). Inheritance patterns of erucic acid content in populations of Brassica napus microspore-derived spontaneous diploids. Theor. Appl. Genet. 77, 489–494. doi: 10.1007/BF00274268
Singh, L. L., Sudarsan, S. D., Jetley, R. P., Fitzgerald, B., and Milanova, M. (2011). U.S. Food and drug administration. J. Am. Pharm. Assoc. 12, 29–32.
Strelkov, S. E., Hwang, S.-F., Manolii, V. P., Cao, T., and Feindel, D. (2016). Emergence of new virulence phenotypes of Plasmodiophora brassicae on canola (Brassica napus) in Alberta, Canada. Eur. J. Plant. Pathol. 145, 517–529. doi: 10.1007/s10658-016-0888-8
Sun, X., Pang, H., Li, M., Peng, B., Guo, H., Yan, Q., et al. (2013). Evolutionary pattern of the FAE1 gene in Brassicaceae and its correlation with the erucic acid trait. PLoS One 8:e83535. doi: 10.1371/journal.pone.0083535
Suwabe, K., Tsukazaki, H., Iketani, H., Hatakeyama, K., Fujimura, M., Nunome, T., et al. (2003). Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor. Appl. Genet. 107, 997–1002. doi: 10.1007/s00122-003-1309-x
Takagi, H., Abe, A., Yoshida, K., Kosugi, S., Natsume, S., Mitsuoka, C., et al. (2013). QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74, 174–183. doi: 10.1111/tpj.12105
Tian, E., Liang, H., Wang, J., Guo, J., Cao, H., and Lin, S. (2018). Variation and correlation of erucic acid, oleic acid and glucosinolate contents in Brassica rapa Seeds. Asian Agric. Res. 10, 57–60.
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Vesna, K., Elzbieta, M., Barton, D. L., Michael, E., Reed, D. W., and Taylor, D. C. (2010). Restoring enzyme activity in nonfunctional low erucic acid Brassica napus fatty acid elongase 1 by a single amino acid substitution. FEBS J. 269, 5625–5631. doi: 10.1046/j.1432-1033.2002.03270.x
Vles, R. O., Bijster, G. M., and Timmer, W. G. (1978). Nutritional evaluation of low-erucic-acid rapeseed oils. Toxicol. Aspects Food Saf. 1, 23–32. doi: 10.1007/978-3-642-66896-8_3
Voorrips, R. E. (1995). Plasmodiophora brassicae: aspects of pathogenesis and resistance in Brassica oleracea. Euphytica 83, 139–146. doi: 10.1007/BF01678041
Wang, N., Shi, L., Tian, F., Ning, H., Wu, X., Long, Y., et al. (2010). Assessment of FAE1 polymorphisms in three Brassica species using EcoTILLING and their association with differences in seed erucic acid contents. BMC Plant Biol. 10:137. doi: 10.1186/1471-2229-10-137
Zhan, Z., Nwafor, C. C., Hou, Z., Gong, J., Zhu, B., Jiang, Y., et al. (2017). Cytological and morphological analysis of hybrids between Brassicoraphanus, and Brassica napus for introgression of clubroot resistant trait into Brassica napus L. PLoS One 12:e0177470. doi: 10.1371/journal.pone.0177470
Zhan, Z. X., Jiang, Y. F., Zhu, Z. Y., Zhang, C. S., Yang, Q. Y., Qian, L. I., et al. (2015). Development of close linked marker to PbBa8.1 conferring canola resistance to Plasmodiophora brassicae. Chin. J. Oil Crop Sci. 6, 766–771.
Zhang, S. F., Fu, T. D., and Zhu, J. C. (2008). Genetic analysis of erucic acid in Brassica napus L. using mixed major gene and polygene inheritance model. Sci. Agric. Sin. 41, 3343–3349.
Keywords: clubroot, Brassica napus, erucic acid, disease resistance, genetic improvement
Citation: Zhan Z, Jiang Y, Shah N, Hou Z, Zhou Y, Dun B, Li S, Zhu L, Li Z, Piao Z and Zhang C (2020) Association of Clubroot Resistance Locus PbBa8.1 With a Linkage Drag of High Erucic Acid Content in the Seed of the European Turnip. Front. Plant Sci. 11:810. doi: 10.3389/fpls.2020.00810
Received: 05 February 2020; Accepted: 19 May 2020;
Published: 11 June 2020.
Edited by:
Jacqueline Batley, The University of Western Australia, AustraliaReviewed by:
Rudolph Fredua-Agyeman, University of Alberta, CanadaCopyright © 2020 Zhan, Jiang, Shah, Hou, Zhou, Dun, Li, Zhu, Li, Piao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongyun Piao, enlwaWFvQHN5YXUuZWR1LmNu; Chunyu Zhang, emhjaHlAbWFpbC5oemF1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.