- Université Clermont Auvergne, INRAE, UMR 1095 Genetics, Diversity and Ecophysiology of Cereals, Clermont-Ferrand, France
Fusarium head blight (FHB), primarily caused by Fusarium graminearum, is one of the most devastating fungal wheat diseases. During the past decades, many efforts have been deployed to dissect FHB resistance, investigating both the wheat responses to infection and, more recently, the fungal determinants of pathogenicity. Although no total resistance has been identified so far, they demonstrated that some plant functions and the expression of specific genes are needed to promote FHB. Associated with the increasing list of F. graminearum effectors able to divert plant molecular processes, this fact strongly argues for a functional link between susceptibility-related factors and the fate of this disease in wheat. In this review, we gather more recent data concerning the involvement of plant and fungal genes and the functions and mechanisms in the development of FHB susceptibility, and we discuss the possibility to use them to diversify the current sources of FHB resistance.
Introduction
Fusarium head blight (FHB) is a cereal fungal disease primarily induced by Fusarium graminearum (Goswami and Kistler, 2004; Xu and Nicholson, 2009). In wheat, FHB has a direct impact on yield and grain quality, reducing grain weight as well as changing protein accumulation. FHB also causes serious health concerns through the contamination of grains by mycotoxins (e.g., deoxynivalenol, DON, a group 3 carcinogenic toxin), which are resilient to most transformation processes (Li et al., 2014). FHB has become a major threat for wheat crops since the early 1990s, especially in the main producing areas, such as North America, Europe, and China (Zhang et al., 2012). For example, economic losses have been estimated to a total of $1.176 billion over 2015 and 2016 in the United States (Wilson et al., 2018). Such losses are expected to increase as a result of an amplification of the frequency and the intensity of FHB outbreaks due to rises in temperatures and occasional increases in air humidity expected with the climate change (Luck et al., 2011; Shah et al., 2014).
Although the combined use of tolerant wheat cultivars, fungicides, and specific management practices (e.g., tillage and crop rotation) can reduce part of the losses due to the disease (Haidukowski et al., 2005; Hollingsworth et al., 2008; Salgado et al., 2014; Dahl and Wilson, 2018), no efficient strategy can fully control FHB epidemics so far (Mesterházy et al., 2005; Tóth et al., 2008). Primarily addressed through the search for genetic resistance, the last two decades of prolific FHB researches turn out with more than 550 quantitative trait loci (QTLs) (Steiner et al., 2017; Venske et al., 2019), covering the whole genome of wheat but with little effect on resistance improvement and failing in identifying regular resistant genes. Twenty years after the identification of Fhb1, the most stable and efficient locus for wheat resistance to FHB (Bai et al., 1999), it was recently shown that a deletion spanning the start codon or an N-terminal mutation of the TaHRC gene encoding a putative histidine-rich calcium-binding protein explains part of the Fhb1-mediated resistance (Li et al., 2019; Su et al., 2019). Although still in dispute, TaHRC constitutes the first susceptibility (S) gene to FHB in wheat and directly questions the involvement of susceptibility factors in the disease progress. With this in mind, the purpose of this review is to discuss the growing interest about the determinism of susceptibility to FHB by gathering information from both interacting partners and by emphasizing on its benefits in diversifying the current sources of FHB resistance.
Fhb Infection Process: Is Susceptibility Behind the Mirror?
Although the recessive nature of some plant resistances has been established for decades, the concept of susceptibility factors, encoded by the so-called susceptibility genes (S genes), has been clearly defined in 2002 by describing the function of the pmr6 plant gene that promotes the infection process and supports the pathogen’s growth and development (Vogel et al., 2002). Many S genes are now described in plants [reviewed in van Schie and Takken (2014)]. Albeit relatively few, those controlling the wheat/pathogens interactions fit well with this model. For example, the monodehydroascorbate reductase gene, TaMDHAR4, has been demonstrated to promote wheat stripe rust infection (Feng et al., 2014). Its mutation results in reducing the hyphae growth of the biotrophic pathogen Puccinia striiformis, thus inhibiting its sporulation and enhancing necrosis at the infection site. Further studies in the same interaction evidenced two other S genes, TaMDAR6 and TaSTP13 (Abou-Attia et al., 2016; Huai et al., 2020), emphasizing the existence of several S genes in the wheat genome and suggesting that some of them might be implicated in wheat susceptibility-related mechanisms to FHB. If most studies have focused on the genetic determinants of wheat resistance to FHB so far, an interesting alternative is to consider the molecular and the physiological processes that make the host plant susceptible to F. graminearum. An extensive literature dealing with large-scale analyses has already shown that, compared to resistant cultivars, the most susceptible ones are characterized by a specific deregulation of genes involved in a wide range of molecular processes (e.g., transcription factors, enzymes involved in primary and secondary metabolism, and defense-related genes), suggesting the intricate participation of a wealth of potential susceptibility factors (Ding et al., 2011; Gottwald et al., 2012; Erayman et al., 2015; Pan et al., 2018; Wang et al., 2018; Brauer et al., 2019).
Genetics Demonstration of the Existence of Wheat Susceptibility Factors to FHB
The involvement of putative susceptibility determinants during FHB development has been primarily suggested by studies using wheat aneuploïd lines (Figure 1). Ma et al. (2006) first evidenced that ditelosomic lines lacking in specific chromosome arms displayed an enhanced resistance to F. graminearum infection, suggesting the removal of pivotal susceptibility factors along with chromosome fragment deletion. Likewise, Garvin et al. (2015), in an attempt to introgress a new FHB resistance locus from the cultivar (cv.) “Freedom” into the susceptible cv. “Apogee,” have shown that the most resistant line was characterized by the deletion of a chromosome segment of about 19% of the length of the 3DL arm in comparison with the cv. “Apogee” (Figure 1). The wheat line missing this genomic interval resulted in up to 59% decrease of FHB severity as compared to cv. “Apogee” and displayed a significant reduction of DON accumulation (Garvin et al., 2015). Similarly, another chromosomal fragment of 31.7 Mbp on the short arm of chromosome 4D was demonstrated to contain potential wheat susceptibility factors to FHB (Hales et al., 2020). Its deletion leads to a significant decrease of F. graminearum spreading in wheat spikes. Evidence of susceptibility factors to FHB has also been provided through allele mining studies. The dwarfing allele at the locus Rht-D1 (Rht-D1b, formerly termed Rht2) has not been associated to FHB susceptibility by a direct effect of the plant height but rather through a pleiotropic or linkage effect (Draeger et al., 2007). Further experiments demonstrated that, in the cv. “Spark,” FHB resistance was largely conferred by the wild allele of the Rht-D1 gene, while the Rht-D1b allele found in the susceptible lines was responsible for approximately 50% of the phenotypic variance associated with the magnitude of initial infection (Srinivasachary et al., 2009). A similar increase of FHB infection has been described for the two particular alleles of the vernalization-related genes Vrn-A1 and Vrn-B1 (Xu et al., 2020). An FHB susceptibility source has also been identified in the “Sumai 3” Qfhs.kibr-2DS QTL (Figure 1), in which a specific allele encoding a multidrug resistance-associated protein was identified in the susceptible “Sumai 3”-derived cv. named “Gamenya,” unveiling that the FHB susceptibility determinants could be fortuitously inherited from resistant cultivars (Handa et al., 2008; Basnet et al., 2012; Niwa et al., 2014). Such examples suggest that a substantial subset of S genes/factors could be present in the wheat genome and highly conserved among the wheat cultivars. Although most of these studies provide only indirect evidences about the molecular determinism of FHB susceptibility, these results emphasize the relevance of considering the diversity of S genes as a complementary and promising approach to improve wheat resistance to FHB.
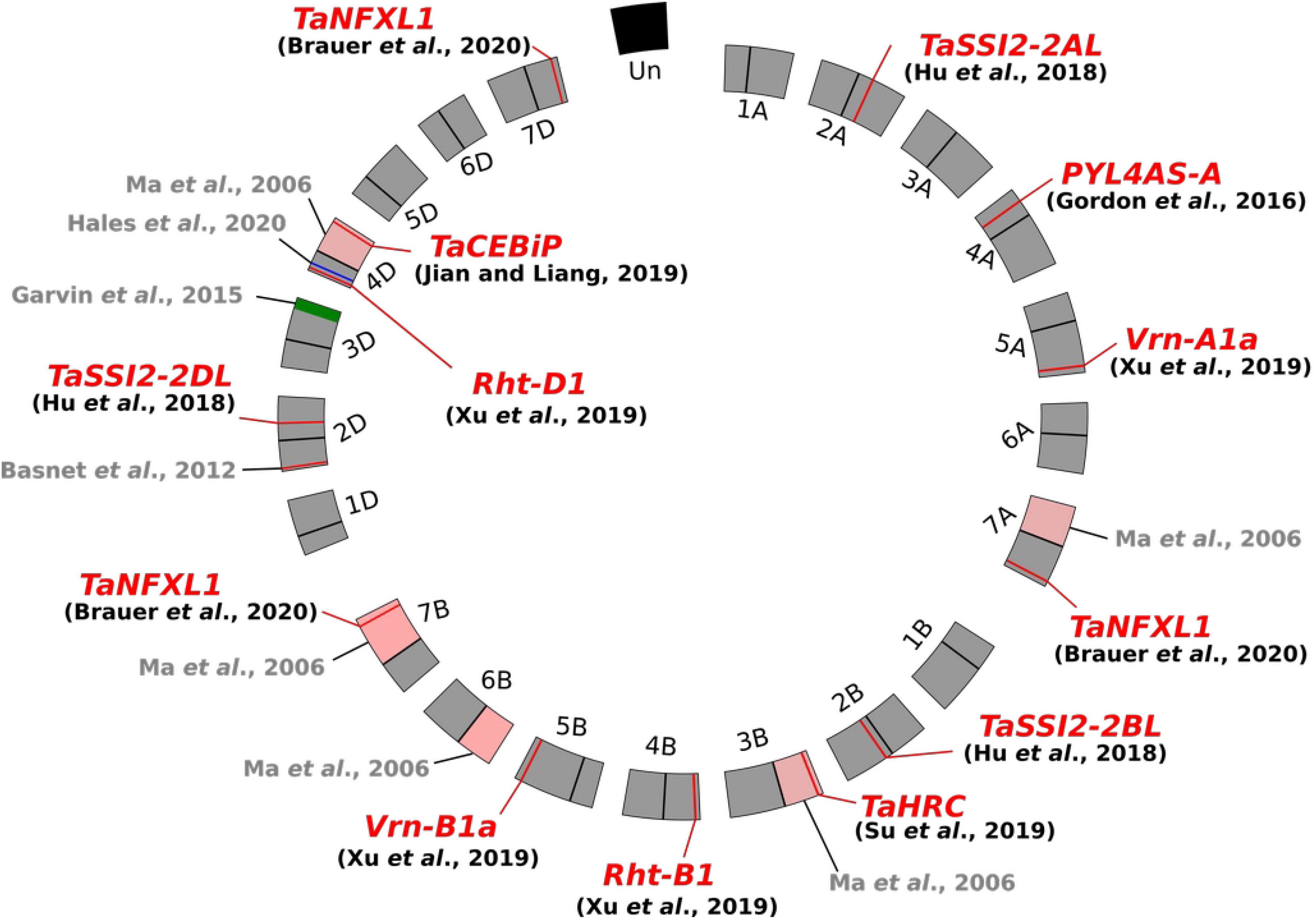
Figure 1. Circos plot of the wheat genome (Triticum aestivum) exemplifying Fusarium head blight susceptibility determinants. The three-component genome is represented as a circle including the A, B, and D genomes and their respective chromosomes. The pink areas refer to the deleted chromosome arms in the ditelosomic lines (Ma et al., 2006), the green and blue zones refer to the deleted genomic regions, and the red lines indicate the gene position.
Role of Phytohormones in FHB Development
Several works have already suggested that wheat hormonal pathways play a favorable role in FHB development. For instance, reducing EIN2 expression in wheat, one of the major components of ethylene signaling, decreased the disease symptoms and DON accumulation in the grains (Chen et al., 2009). Further large-scale transcriptomics showed that the ethylene pathway was specifically induced in the FHB-susceptible NAUH117 line, as compared to the resistant Wangshuibai landrace (Xiao et al., 2013). The abscisic acid (ABA) signaling pathway has also been demonstrated to favor F. graminearum infection in wheat spikes (Gordon et al., 2016; Wang et al., 2018). A virus-induced gene silencing approach demonstrated a role of the wheat ABA receptor Ta_PYL4AS_A (Figure 1), and its close homologs, in mediating FHB susceptibility and in decreasing mycotoxin accumulation (Gordon et al., 2016). Likewise, transcriptional and hormonal profiling showed that wheat genes involved in auxin biosynthesis were highly up-regulated, along with auxin accumulation, in the susceptible cultivars during the F. graminearum infection process as compared to the resistant ones (Biselli et al., 2018; Wang et al., 2018; Brauer et al., 2019). Salicylic and jasmonic acids are also widely described for their role in modulating FHB responses and in discriminating resistant vs. susceptible cultivars as well (Ding et al., 2011; Gottwald et al., 2012; Sun et al., 2016; Wang et al., 2018). Upon F. graminearum infection, their respective actions occur in two phases, an initial induction of salicylic acid happens at the early stages followed by the synthesis of jasmonic acid at the later stages (Ding et al., 2011). In addition, the silencing of the wheat TaSSI2 gene has been shown to increase FHB resistance, promoting salicylic acid signaling (Hu et al., 2018) and potentially altering the jasmonic acid pathway as demonstrated in Arabidopsis ssi2 mutant lines (Kachroo et al., 2004). This illustrates further the involvement of these two antagonist hormones in the FHB progress and suggests that susceptibility may involve systemic signals capable of deeply reshaping the plant physiology.
Shaping FHB Susceptibility in the Course of Grain Development
With a period of maximal susceptibility occurring within 3 days after anthesis (Beccari et al., 2019), FHB develops concomitantly with the grain filling period, during which a large number of plant physiological processes allow a massive accumulation of sugars, lipids, and proteins (Nadaud et al., 2010), resulting in a possible nutrient reservoir in the infection area. Spike ontogeny could thus indirectly and sequentially set up a range of susceptibility factors that can partly explain the dynamics of fungal development during the infection course (Chetouhi et al., 2015, 2016). Extensively boosted during the endosperm expansion, in planta F. graminearum growth is associated with massive protein abundance adjustments detectable simultaneously in both plant and fungal proteomes at 48–72 h post-inoculation at anthesis transition (Fabre et al., 2019b). At this stage, extensive metabolic changes in wheat rachis nodes have been reported, including a strong increase of gibberellic acid amount as well as glycolysis intermediates, suggesting that a release of wheat storage carbohydrates could possibly be used for fungal metabolic requirements (Bönnighausen et al., 2018). This is also supported by the converging evidences of large decreases in the expression of genes involved in sucrose and starch metabolism (Erayman et al., 2015; Chetouhi et al., 2016). Starch components, such as amylopectin and amylose, are known to be difficult for the fungus to recycle as a carbon source and for DON production (Oh et al., 2016), unlike sucrose (Jiao et al., 2008; Kawakami et al., 2014), suggesting that the deregulation of the host energy processes at the early stage of the disease could be one of the key factors that determine wheat susceptibility. However, the links are not obvious since many other studies have shown that primary metabolism and especially photosynthesis are extensively rearranged (Long et al., 2015; Biselli et al., 2018; Li et al., 2018; Fabre et al., 2019b). Although this can substantially limit the accumulation of sugars, this could also be seen as a means of constraining the energy requirements necessary to trigger defense mechanisms (Bolton, 2009). In agreement with many previous studies suggesting a key role of chloroplast in plant susceptibility (Lo Presti et al., 2015; Sowden et al., 2018; Han and Kahmann, 2019; Kretschmer et al., 2020), the remodeling of its functioning in wheat spikes during FHB is suspected to be a link between the plant defense responses and the adjustments of primary metabolism. This raises direct questions about the mechanisms used by the fungus to achieve such effects (Fabre et al., 2019b).
Fusarium graminearum Effectors: Knocking at the Wheat Cell Door to Trigger Susceptibility?
Several studies have already demonstrated that plant susceptibility factors could be diverted by a range of pathogen effectors. Consisting of proteins, RNA, and metabolites, effectors are molecules synthesized by the pathogen, delivered in host tissues, and able to alter the structure and the function of the host cell (Hogenhout et al., 2009; Lo Presti et al., 2015). Compared to bacteria, knowledge about fungal effectors remains relatively limited (Niu et al., 2013). However, a number of studies have provided essential information on the ability of F. graminearum in interfering with wheat molecular processes. Identifying F. graminearum effectors and understanding their roles in the infectious process could be a relevant strategy for identifying wheat susceptibility factors.
Breaking Wheat’s Defenses
One of the first characterized effectors of F. graminearum is the secreted DON mycotoxin (Miller and Young, 1985). Although its synthesis is not necessary for the penetration phase, its role in fungal spreading within the spike has been reported (Bai et al., 2001). DON acts as an inhibitor of protein and nucleotide synthesis in the host cell (Audenaert et al., 2013). Through such an effect, DON is supposed to alter the mitochondrial functions of many eukaryotes, and its role in inhibiting programmed cell death as well as in the expression of defense compounds (chitinases, peroxidases, and pathogen-related proteins) has already been described (Brown et al., 2011; Audenaert et al., 2013; Diamond et al., 2013). A recent report has shown that DON promotes the TaNFXL1 transcription factor in wheat, leading to FHB susceptibility through uncharacterized mechanisms (Brauer et al., 2020). Other F. graminearum effectors have been reported so far, revealing that proteins belonging to the cell-wall-degrading enzymes (CWDEs) are important promoters of wheat susceptibility to FHB (Quarantin et al., 2016, 2019; Paccanaro et al., 2017; Lu and Faris, 2019). For instance, several studies identified F. graminearum xylanases with a direct impact on cell wall weakening and an indirect role in enhancing hypersensitive-like symptoms in plant tissues (Paper et al., 2007; Pollet et al., 2009; Sella et al., 2013; Tundo et al., 2015). The FGL1 lipase, another CWDE effector (Voigt et al., 2005), was shown to physically interact with the wheat immunophilin protein FKBP12, altering the establishment of the FKBP12/ERG complex, which finally triggers cell death (Niu et al., 2013). In addition, by degrading the plant cell wall, the FGL1 effector promotes the release of free fatty acids that inhibit the callose deposits associated with the immune responses (Blümke et al., 2014). Similarly, the arabinanase Arb93b, induced during the early stage of FHB, was shown to suppress ROS-activated defense along with its arabinan-degrading activity (Hao et al., 2019). Besides protein effectors, sRNA products have also been reported to control plant responses. The 18-nt-length sRNA (Fg-sRNA1) targets a wheat chitin elicitor-binding protein, which is likely to function in wheat disease resistance signaling pathways (Jian and Liang, 2019). The identification of non-targeted allelic variants could thus guide future research toward “loss-of-susceptibility” forms of resistances.
Predicted Effector Searches Reveal an Increasingly Complex Arsenal
Although the catalog of characterized F. graminearum effectors remains limited, substantial efforts using genomics approaches have provided a large set of new candidates. Using the reference genome sequence, Brown et al. (2012) established a predicted secretome of 574 proteins sharing the structural features of secreted proteins (small size, cysteine-rich proteins, and signal peptides). This revealed a diverse hydrolytic arsenal and a range of putative effectors that could be potentially delivered in the wheat tissues. Secretome was further investigated by focusing on the 190 small secreted cystein-rich proteins (SS): the extracellular localization was confirmed for 25 of them, and the expression of 34 of them was demonstrated as regulated during the FHB progress (Lu and Edwards, 2015). The sequence analysis suggested that 17 SS harbor conserved functional domains such as glycosyl-hydrolase or pathogenesis-related domains, and two of them were homologous to Ecp2, a well-known effector produced by the tomato pathogen Cladosporium fulvum (Van den Ackerveken et al., 1993). Other studies dealing with in vitro or in planta approaches have also been successful in enlarging the list of candidate effectors (Paper et al., 2007; Yang et al., 2012; Ji et al., 2013). By evidencing the protein repertoire found specifically in the extracellular part of the plant tissues or identified in liquid culture and confirmed in planta using qRT-PCR, these extended the range of putative function including a number of proteases, esterase, and nucleases. Based on the few structural information available about fungal effectors (Sperschneider et al., 2018), several exploratory reports provided novel insights in their diversity and dynamics (Lysøe et al., 2011; Brown et al., 2017; Fabre et al., 2019a, b). By dissecting the asymptomatic and the symptomatic stages of the FHB infection, Brown et al. (2017) revealed particular gene groups with specific abundance patterns illustrating the early expression of genes involved in the transport of amino acids, in polyamine synthesis and ABC transporters, while hydrolytic carbohydrate-active enzymes and lipases were found at later stages. The delivery of putative effectors by waves at specific stages of the infection has also been confirmed at the protein level using an in planta dual-proteome approach (Fabre et al., 2019b). This study further demonstrated that putative effectors could be already accumulated in spores or synthesized within hours, and extensive co-variations were evidenced between abundance changes of effectors and the regulation of plant chloroplast proteins, especially at the beginning of grain cellularization. In addition, strong links were evidenced between the abundance of candidate effectors and strain aggressiveness (Fabre et al., 2019a), emphasizing that increased knowledge of the fungal component could lead to a better understanding of the processes involved in host susceptibility.
Concluding Remarks
The past decades of researches on FHB in wheat have provided a wealth of information on the genetic and the molecular determinants of the disease progress in spikes, mostly focused on resistance mechanisms. Although still marginally investigated, wheat susceptibility factors to FHB are emerging as key components that determine the fate of the disease, involving a complex molecular dialogue based on the interplay of fungal effectors and their plant targets. Understanding wheat susceptibility still requires many efforts on both partners and needs to fill the gap between wheat and fungal studies. This knowledge will open new strategies in order to control this complex plant/fungus interaction, providing alternative forms of resistance that are potentially more sustainable. While still a challenge, such loss-of-susceptibility forms have already demonstrated their potential to provide efficient and durable sources of disease resistance in crops (Pavan et al., 2010). They represent a promising strategy to control FHB epidemics and may provide a complementary approach to the introgression of gain-of-function resistance genes.
Author Contributions
FF, FR, and TA organized, prepared, and drafted the manuscript. TL and LB designed, reviewed, and finalized the manuscript.
Funding
This work was supported by the Region Auvergne-Rhône-Alpes, the Fonds Européen de Développement Régional (FEDER support), and by the Agence Nationale de la Recherche (NewMyco project, ANR-15-CE21-0010). The present work falls within the thematic area of the French Government IDEX-ISITE initiative 16-IDEX-0001 (CAP 20-25).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. C. Saintenac for his critical comments on the manuscript.
References
Abou-Attia, M. A., Wang, X., Nashaat Al-Attala, M., Xu, Q., Zhan, G., and Kang, Z. (2016). TaMDAR6 acts as a negative regulator of plant cell death and participates indirectly in stomatal regulation during the wheat stripe rust-fungus interaction. Physiol. Plant. 156, 262–277. doi: 10.1111/ppl.12355
Audenaert, K., Vanheule, A., Höfte, M., and Haesaert, G. (2013). Deoxynivalenol: a major player in the multifaceted response of Fusarium to its environment. Toxins 6, 1–19. doi: 10.3390/toxins6010001
Bai, G., Kolb, F. L., Shaner, G., and Domier, L. L. (1999). Amplified fragment length polymorphism markers linked to a major quantitative trait locus controlling scab resistance in wheat. Phytopathology 89, 343–348. doi: 10.1094/PHYTO.1999.89.4.343
Bai, G. H., Plattner, R., Desjardins, A., and Kolb, F. (2001). Resistance to Fusarium head blight and deoxynivalenol accumulation in wheat. Plant Breed. 120, 1–6.
Basnet, B. R., Glover, K. D., Ibrahim, A. M. H., Yen, Y., and Chao, S. (2012). A QTL on chromosome 2DS of “Sumai 3” increases susceptibility to Fusarium head blight in wheat. Euphytica 186, 91–101. doi: 10.1007/s10681-011-0495-x
Beccari, G., Arellano, C., Covarelli, L., Tini, F., Sulyok, M., and Cowger, C. (2019). Effect of wheat infection timing on Fusarium head blight causal agents and secondary metabolites in grain. Int. J. Food Microbiol. 290, 214–225. doi: 10.1016/j.ijfoodmicro.2018.10.014
Biselli, C., Bagnaresi, P., Faccioli, P., Hu, X., Balcerzak, M., Mattera, M. G., et al. (2018). Comparative transcriptome profiles of near-isogenic hexaploid wheat lines differing for effective alleles at the 2DL FHB resistance QTL. Front. Plant Sci. 9:37. doi: 10.3389/fpls.2018.00037
Blümke, A., Falter, C., Herrfurth, C., Sode, B., Bode, R., Schäfer, W., et al. (2014). Secreted fungal effector lipase releases free fatty acids to inhibit innate immunity-related callose formation during wheat head infection. Plant Physiol. 165, 346–358. doi: 10.1104/pp.114.236737
Bolton, M. (2009). Primary metabolism and plant defense – Fuel for the fire. Mol. Plant-Microbe Interact. 22, 487–497. doi: 10.1094/MPMI-22-5-0487
Bönnighausen, J., Schauer, N., Schäfer, W., and Bormann, J. (2018). Metabolic profiling of wheat rachis node infection by Fusarium graminearum – decoding deoxynivalenol-dependent susceptibility. New Phytol. 221, 459–469. doi: 10.1111/nph.15377
Brauer, E. K., Balcerzak, M., Rocheleau, H., Leung, W., Schernthaner, J., Subramaniam, R., et al. (2020). Genome editing of a deoxynivalenol-induced transcription factor confers resistance to Fusarium graminearum in wheat. Mol. Plant-Microbe Interact. 33, 553–560. doi: 10.1094/MPMI-11-19-0332-R
Brauer, E. K., Rocheleau, H., Balcerzak, M., Pan, Y., Fauteux, F., Liu, Z., et al. (2019). Transcriptional and hormonal profiling of Fusarium graminearum-infected wheat reveals an association between auxin and susceptibility. Physiol. Mol. Plant Pathol. 107, 33–39. doi: 10.1016/j.pmpp.2019.04.006
Brown, N. A., Antoniw, J., and Hammond-Kosack, K. E. (2012). The predicted secretome of the plant pathogenic fungus Fusarium graminearum: a refined comparative analysis. PLoS One 7:e33731. doi: 10.1371/journal.pone.0033731
Brown, N. A., Bass, C., Baldwin, T. K., Chen, H., Massot, F., Carion, P. W. C., et al. (2011). Characterisation of the Fusarium graminearum-Wheat Floral Interaction. J. Pathog. 2011:e626345. doi: 10.4061/2011/626345
Brown, N. A., Evans, J., Mead, A., and Hammond-Kosack, K. E. (2017). A spatial temporal analysis of the Fusarium graminearum transcriptome during symptomless and symptomatic wheat infection. Mol. Plant Pathol. 18, 1295–1312. doi: 10.1111/mpp.12564
Chen, X., Steed, A., Travella, S., Keller, B., and Nicholson, P. (2009). Fusarium graminearum exploits ethylene signalling to colonize dicotyledonous and monocotyledonous plants. New Phytol. 182, 975–983. doi: 10.1111/j.1469-8137.2009.02821.x
Chetouhi, C., Bonhomme, L., Lasserre-Zuber, P., Cambon, F., Pelletier, S., Renou, J.-P., et al. (2016). Transcriptome dynamics of a susceptible wheat upon Fusarium head blight reveals that molecular responses to Fusarium graminearum infection fit over the grain development processes. Funct. Integr. Genomics 16, 183–201. doi: 10.1007/s10142-016-0476-471
Chetouhi, C., Bonhomme, L., Lecomte, P., Cambon, F., Merlino, M., Biron, D. G., et al. (2015). A proteomics survey on wheat susceptibility to Fusarium head blight during grain development. Eur. J. plant Pathol. 141, 407–418. doi: 10.1007/s10658-014-0552-550
Dahl, B., and Wilson, W. W. (2018). Risk premiums due to Fusarium Head Blight (FHB) in wheat and barley. Agric. Syst. 162, 145–153. doi: 10.1016/j.agsy.2018.01.025
Diamond, M., Reape, T. J., Rocha, O., Doyle, S. M., Kacprzyk, J., Doohan, F. M., et al. (2013). The Fusarium mycotoxin deoxynivalenol can inhibit plant apoptosis-like programmed cell death. PLoS One 8:e69542. doi: 10.1371/journal.pone.0069542
Ding, L., Xu, H., Yi, H., Yang, L., Kong, Z., Zhang, L., et al. (2011). Resistance to hemi-biotrophic F. graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLoS One 6:e19008. doi: 10.1371/journal.pone.0019008
Draeger, R., Gosman, N., Steed, A., Chandler, E., Thomsett, M., Srinivasachary, A., et al. (2007). Identification of QTLs for resistance to Fusarium head blight, DON accumulation and associated traits in the winter wheat variety Arina. Theor. Appl. Genet. 115, 617–625. doi: 10.1007/s00122-007-0592-593
Erayman, M., Turktas, M., Akdogan, G., Gurkok, T., Inal, B., Ishakoglu, E., et al. (2015). Transcriptome analysis of wheat inoculated with Fusarium graminearum. Front. Plant Sci. 6:867. doi: 10.3389/fpls.2015.00867
Fabre, F., Bormann, J., Urbach, S., Roche, S., and Langin, T. (2019a). Unbalanced roles of fungal aggressiveness and host cultivars in the establishment of the Fusarium head blight in bread wheat. Front. Microbiol. 10:2857. doi: 10.3389/fmicb.2019.02857
Fabre, F., Vignassa, M., Urbach, S., Langin, T., and Bonhomme, L. (2019b). Time-resolved dissection of the molecular crosstalk driving Fusarium head blight in wheat provides new insights into host susceptibility determinism. Plant Cell Environ. 42, 2291–2308. doi: 10.1111/pce.13549
Feng, H., Liu, W., Zhang, Q., Wang, X., Wang, X., Duan, X., et al. (2014). TaMDHAR4, a monodehydroascorbate reductase gene participates in the interactions between wheat and Puccinia striiformis f. sp. tritici. Plant Physiol. Biochem. 76, 7–16. doi: 10.1016/j.plaphy.2013.12.015
Garvin, D. F., Porter, H., Blankenheim, Z. J., Chao, S., Dill-Macky, R., and Gustafson, J. P. (2015). A spontaneous segmental deletion from chromosome arm 3DL enhances Fusarium head blight resistance in wheat. Genome 58, 479–488. doi: 10.1139/gen-2015-2088
Gordon, C. S., Rajagopalan, N., Risseeuw, E. P., Surpin, M., Ball, F. J., Barber, C. J., et al. (2016). Characterization of Triticum aestivum abscisic acid receptors and a possible role for these in mediating Fusairum head blight susceptibility in wheat. PLoS One 11:e164996. doi: 10.1371/journal.pone.0164996
Goswami, R. S., and Kistler, H. C. (2004). Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. doi: 10.1111/J.1364-3703.2004.00252.X
Gottwald, S., Samans, B., Lück, S., and Friedt, W. (2012). Jasmonate and ethylene dependent defence gene expression and suppression of fungal virulence factors: two essential mechanisms of Fusarium head blight resistance in wheat? BMC Genomics 13:369. doi: 10.1186/1471-2164-13-369
Haidukowski, M., Pascale, M., Perrone, G., Pancaldi, D., Campagna, C., and Visconti, A. (2005). Effect of fungicides on the development of Fusarium head blight, yield and deoxynivalenol accumulation in wheat inoculated under field conditions with Fusarium graminearum and Fusarium culmorum. J. Sci. Food Agric. 85, 191–198. doi: 10.1002/jsfa.1965
Hales, B., Steed, A., Giovannelli, V., Burt, C., Lemmens, M., Molnar-Lang, M., et al. (2020). Type II Fusarium head blight susceptibility factor identified in wheat. bioRxiv [Preprint] doi: 10.1101/2020.02.06.937425
Han, X., and Kahmann, R. (2019). Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front. Plant Sci. 10:822. doi: 10.3389/fpls.2019.00822
Handa, H., Namiki, N., Xu, D., and Ban, T. (2008). Dissecting of the FHB resistance QTL on the short arm of wheat chromosome 2D using a comparative genomic approach: from QTL to candidate gene. Mol. Breed. 22, 71–84. doi: 10.1007/s11032-008-9157-7
Hao, G., McCormick, S., Vaughan, M., Naumann, T., Kim, H.-S., Proctor, R. H., et al. (2019). Fusarium graminearum arabinanase (Arb93B) enhances wheat head blight susceptibility by suppressing plant immunity. Mol. Plant-Microbe Interact. 32, 888–898. doi: 10.1094/mpmi-06-18-0170-r
Hogenhout, S. A., Van der Hoorn, R. A. L., Terauchi, R., and Kamoun, S. (2009). Emerging concepts in effector biology of plant-associated organisms. Mol. Plant-Microbe Interact. 22, 115–122. doi: 10.1094/MPMI-22-2-0115
Hollingsworth, C. R., Motteberg, C. D., Wiersma, J. V., and Atkinson, L. M. (2008). Agronomic and economic responses of spring wheat to management of Fusarium head blight. Plant Dis. 92, 1339–1348. doi: 10.1094/PDIS-92-9-1339
Hu, L., Mu, J., Su, P., Wu, H., Yu, G., Wang, G., et al. (2018). Multi-functional roles of TaSSI2 involved in Fusarium head blight and powdery mildew resistance and drought tolerance. J. Integr. Agric. 17, 368–380. doi: 10.1016/S2095-3119(17)61680-61680
Huai, B., Yang, Q., Wei, X., Pan, Q., Kang, Z., and Liu, J. (2020). TaSTP13 contributes to wheat susceptibility to stripe rust possibly by increasing cytoplasmic hexose concentration. BMC Plant Biol. 20:49. doi: 10.1186/s12870-020-2248-2242
Ji, X. L., Yan, M., Yang, Z. D., Li, A. F., and Kong, L. R. (2013). Shotgun analysis of the secretome of Fusarium graminearum. Indian J. Microbiol. 53, 400–409. doi: 10.1007/s12088-013-0392-391
Jian, J., and Liang, X. (2019). One small RNA of Fusarium graminearum targets and silences CEBiP gene in common wheat. Microorganisms 7:425. doi: 10.3390/microorganisms7100425
Jiao, F., Kawakami, A., and Nakajima, T. (2008). Effects of different carbon sources on trichothecene production and Tri gene expression by Fusarium graminearum in liquid culture. FEMS Microbiol. Lett. 285, 212–219. doi: 10.1111/j.1574-6968.2008.01235.x
Kachroo, A., Venugopal, S. C., Lapchyk, L., Falcone, D., Hildebrand, D., and Kachroo, P. (2004). Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 5152–5157. doi: 10.1073/pnas.0401315101
Kawakami, A., Nakajima, T., and Hirayae, K. (2014). Effects of carbon sources and amines on induction of trichothecene production by Fusarium asiaticum in liquid culture. FEMS Microbiol. Lett. 352, 204–212. doi: 10.1111/1574-6968.12386
Kretschmer, M., Damoo, D., Djamei, A., and Kronstad, J. (2020). Chloroplasts and plant immunity: where are the fungal effectors? Pathogens 9, 1–16. doi: 10.3390/pathogens9010019
Li, G., Zhou, J., Jia, H., Gao, Z., Fan, M., Luo, Y., et al. (2019). Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat. Genet. 51, 1106–1112. doi: 10.1038/s41588-019-0426-427
Li, X., Zhao, L., Fan, Y., Jia, Y., Sun, L., Ma, S., et al. (2014). Occurrence of mycotoxins in feed ingredients and complete feeds obtained from the Beijing region of China. J. Anim. Sci. Biotechnol. 5, 1–8. doi: 10.1186/2049-1891-5-37
Li, X., Zhong, S., Chen, W., Fatima, S. A., Huang, Q., Li, Q., et al. (2018). Transcriptome analysis identifies a 140 kb region of chromosome 3B containing genes specific to Fusarium head blight resistance in wheat. Int. J. Mol. Sci. 19:852. doi: 10.3390/ijms19030852
Lo Presti, L., Lanver, D., Schweizer, G., Tanaka, S., Liang, L., Tollot, M., et al. (2015). Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545. doi: 10.1146/annurev-arplant-043014-114623
Long, X. Y., Balcerzak, M., Gulden, S., Cao, W., Fedak, G., Wei, Y. M., et al. (2015). Expression profiling identifies differentially expressed genes associated with the Fusarium head blight resistance QTL 2DL from the wheat variety Wuhan-1. Physiol. Mol. Plant Pathol. 90, 1–11. doi: 10.1016/j.pmpp.2015.02.002
Lu, S., and Edwards, M. C. (2015). Genome-wide analysis of small secreted cysteine-rich proteins identifies candidate effector proteins potentially involved in Fusarium graminearum-wheat interactions. Phytopathology 106, 166–176. doi: 10.1094/PHYTO-09-15-0215-R
Lu, S., and Faris, J. D. (2019). Fusarium graminearum KP4-like proteins possess root growth-inhibiting activity against wheat and potentially contribute to fungal virulence in seedling rot. Fungal Genet. Biol. 123, 1–13. doi: 10.1016/j.fgb.2018.11.002
Luck, J., Spackman, M., Freeman, A., TreBicki, P., Griffiths, W., Finlay, K., et al. (2011). Climate change and diseases of food crops. Plant Pathol. 60, 113–121. doi: 10.1111/j.1365-3059.2010.02414.x
Lysøe, E., Seong, K.-Y., and Kistler, H. C. (2011). The transcriptome of Fusarium graminearum during the infection of wheat. Mol. Plant-Microbe Interact. MPMI 24, 995–1000. doi: 10.1094/MPMI-02-11-0038
Ma, H. X., Bai, G. H., Gill, B. S., and Hart, L. P. (2006). Deletion of a chromosome arm altered wheat resistance to Fusarium head blight and deoxynivalenol accumulation in Chinese spring. Plant Dis. 90, 1545–1549. doi: 10.1094/PD-90-1545
Mesterházy, Á, Bartók, T., Kászonyi, G., Varga, M., Tóth, B., and Varga, J. (2005). Common resistance to different Fusarium spp. causing Fusarium head blight in wheat. Eur. J. Plant Pathol. 112, 267–281. doi: 10.1007/s10658-005-2853-2859
Miller, J. D., and Young, J. C. (1985). Deoxynivalenol in an experimental Fusarium graminearum infection of wheat. Can. J. Plant. Pathol. 7, 132–134.
Nadaud, I., Girousse, C., Debiton, C., Chambon, C., Bouzidi, M. F., Martre, P., et al. (2010). Proteomic and morphological analysis of early stages of wheat grain development. Proteomics 10, 2901–2910. doi: 10.1002/pmic.200900792
Niu, X.-W., Zheng, Z.-Y., Feng, Y.-G., Guo, W.-Z., and Wang, X.-Y. (2013). The Fusarium graminearum virulence factor FGL targets an FKBP12 immunophilin of wheat. Gene 525, 77–83. doi: 10.1016/j.gene.2013.04.052
Niwa, S., Kubo, K., Lewis, J., Kikuchi, R., Alagu, M., and Ban, T. (2014). Variations for Fusarium head blight resistance associated with genomic diversity in different sources of the resistant wheat cultivar “Sumai 3.”. Breed. Sci. 64, 90–96. doi: 10.1270/jsbbs.64.90
Oh, M., Son, H., Choi, G. J., Lee, C., Kim, J. C., Kim, H., et al. (2016). Transcription factor ART1 mediates starch hydrolysis and mycotoxin production in Fusarium graminearum and F.verticillioides. Mol. Plant Pathol. 17, 755–768. doi: 10.1111/mpp.12328
Paccanaro, M. C., Sella, L., Castiglioni, C., Giacomello, F., Martínez-Rocha, A. L., D’Ovidio, R., et al. (2017). Synergistic effect of different plant cell wall-degrading enzymes is important for virulence of Fusarium graminearum. Mol. Plant-Microbe Interact. 30, 886–895. doi: 10.1094/MPMI-07-17-0179-R
Pan, Y., Liu, Z., Rocheleau, H., Fauteux, F., Wang, Y., and McCartney, C. (2018). Transcriptome dynamics associated with resistance and susceptibility against fusarium head blight in four wheat genotypes. BMC Genomics 19:642. doi: 10.1186/s12864-018-5012-5013
Paper, J. M., Scott-Craig, J. S., Adhikari, N. D., Cuomo, C. A., and Walton, J. D. (2007). Comparative proteomics of extracellular proteins in vitro and in planta from the pathogenic fungus Fusarium graminearum. Proteomics 7, 3171–3183. doi: 10.1002/pmic.200700184
Pavan, S., Jacobsen, E., Visser, R. G. F., and Bai, Y. (2010). Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 25, 1–12. doi: 10.1007/s11032-009-9323-6
Pollet, A., Sansen, S., Raedschelders, G., Gebruers, K., Rabijns, A., Delcour, J. A., et al. (2009). Identification of structural determinants for inhibition strength and specificity of wheat xylanase inhibitors TAXI-IA and TAXI-IIA. FEBS J. 276, 3916–3927. doi: 10.1111/j.1742-4658.2009.07105.x
Quarantin, A., Castiglioni, C., Schäfer, W., Favaron, F., and Sella, L. (2019). The Fusarium graminearum cerato-platanins loosen cellulose substrates enhancing fungal cellulase activity as expansin-like proteins. Plant Physiol. Biochem. 139, 229–238. doi: 10.1016/j.plaphy.2019.03.025
Quarantin, A., Glasenapp, A., Schäfer, W., Favaron, F., and Sella, L. (2016). Involvement of the Fusarium graminearum cerato-platanin proteins in fungal growth and plant infection. Plant Physiol. Biochem. 109, 220–229. doi: 10.1016/j.plaphy.2016.10.001
Salgado, J. D., Madden, L. V., Paul, P. A., Pathology, P., and Ohio, T. (2014). Efficacy and economics of integrating in-field and harvesting strategies to manage Fusarium head blight of wheat. Plant Dis. 98, 1407–1421. doi: 10.1094/PDIS-01-14-0093-RE
Sella, L., Gazzetti, K., Faoro, F., Odorizzi, S., D’Ovidio, R., Schäfer, W., et al. (2013). A Fusarium graminearum xylanase expressed during wheat infection is a necrotizing factor but is not essential for virulence. Plant Physiol. Biochem. 64, 1–10. doi: 10.1016/j.plaphy.2012.12.008
Shah, D. A., De Wolf, E. D., Paul, P. A., and Madden, L. V. (2014). Predicting Fusarium head blight epidemics with boosted regression trees. Phytopathology 104, 702–714. doi: 10.1094/PHYTO-10-13-0273-R
Sowden, R. G., Watson, S. J., and Jarvis, P. (2018). The role of chloroplasts in plant pathology. Essays Biochem. 62, 21–39. doi: 10.1042/EBC20170020
Sperschneider, J., Dodds, P. N., Gardiner, D. M., Singh, K. B., and Taylor, J. M. (2018). Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 19, 2094–2110. doi: 10.1111/mpp.12682
Srinivasachary, A., Gosman, N., Steed, A., Hollins, T. W., Bayles, R., Jennings, P., et al. (2009). Semi-dwarfing Rht-B1 and Rht-D1 loci of wheat differ significantly in their influence on resistance to Fusarium head blight. Theor. Appl. Genet. 118, 695–702. doi: 10.1007/s00122-008-0930-930
Steiner, B., Buerstmayr, M., Michel, S., Schweiger, W., Lemmens, M., and Buerstmayr, H. (2017). Breeding strategies and advances in line selection for Fusarium head blight resistance in wheat. Trop. Plant Pathol. 42, 165–174. doi: 10.1007/s40858-017-0127-127
Su, Z., Bernardo, A., Tian, B., Chen, H., Wang, S., Ma, H., et al. (2019). A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat. Genet. 51, 1099–1105. doi: 10.1038/s41588-019-0425-428
Sun, Y., Xiao, J., Jia, X., Ke, P., He, L., Cao, A., et al. (2016). The role of wheat jasmonic acid and ethylene pathways in response to Fusarium graminearum infection. Plant Growth Regul. 80, 69–77. doi: 10.1007/s10725-016-0147-141
Tóth, B., Kászonyi, G., Bartók, T., Varga, J., and Mesterházy, Á (2008). Common resistance of wheat to members of the Fusarium graminearum species complex and F. culmorum. Plant Breed. 127, 1–8. doi: 10.1111/j.1439-0523.2008.01412.x
Tundo, S., Moscetti, I., Faoro, F., Lafond, M., Giardina, T., Favaron, F., et al. (2015). Fusarium graminearum produces different xylanases causing host cell death that is prevented by the xylanase inhibitors XIP-I and TAXI-III in wheat. Plant Sci. 240, 161–169. doi: 10.1016/j.plantsci.2015.09.002
Van den Ackerveken, G. F., Van Kan, J. A., Joosten, M. H., Muisers, J. M., Verbakel, H. M., and De Wit, P. J. (1993). Characterization of two putative pathogenicity genes of the fungal tomato pathogen Cladosporium fulvum. Mol. Plant-Microbe Interact. 6, 210–215. doi: 10.1094/MPMI-6-210
van Schie, C. C. N., and Takken, F. L. W. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-145854
Venske, E., dos Santos, R. S., Farias, D., da, R., Rother, V., da Maia, L. C., et al. (2019). Meta-analysis of the QTLome of Fusarium head blight resistance in bread wheat: refining the current puzzle. Front. Plant Sci. 10:727. doi: 10.3389/fpls.2019.00727
Vogel, J. P., Raab, T. K., Schiff, C., and Somerville, S. C. (2002). PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14, 2095–2106. doi: 10.1105/tpc.003509
Voigt, C. A., Schäfer, W., and Salomon, S. (2005). A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J. 42, 364–375. doi: 10.1111/j.1365-313X.2005.02377.x
Wang, L., Li, Q., Liu, Z., Surendra, A., Pan, Y., Li, Y., et al. (2018). Integrated transcriptome and hormone profiling highlight the role of multiple phytohormone pathways in wheat resistance against Fusarium head blight. PLoS One 13:e207036. doi: 10.1371/journal.pone.0207036
Wilson, W., Dahl, B., and Nganje, W. (2018). Economic costs of Fusarium head blight, scab and deoxynivalenol. World Mycotoxin J. 11, 291–302. doi: 10.3920/WMJ2017.2204
Xiao, J., Jin, X., Jia, X., Wang, H., Cao, A., Zhao, W., et al. (2013). Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genomics 14:197. doi: 10.1186/1471-2164-14-197
Xu, K., He, X., Dreisigacker, S., He, Z., and Singh, P. K. (2020). Anther extrusion and its association with Fusarium head blight in CIMMYT wheat germplasm. Agronomy 10, 1–16. doi: 10.3390/agronomy10010047
Xu, X., and Nicholson, P. (2009). Community ecology of fungal pathogens causing Wheat head blight. Annu. Rev. Phytopathol. 47, 83–103. doi: 10.1146/annurev-phyto-080508-081737
Yang, F., Jensen, J. D., Svensson, B., Jørgensen, H. J. L., Collinge, D. B., and Finnie, C. (2012). Secretomics identifies Fusarium graminearum proteins involved in the interaction with barley and wheat. Mol. Plant Pathol. 13, 445–453. doi: 10.1111/j.1364-3703.2011.00759.x
Keywords: Triticum aestivum, Fusarium graminearum, scab, susceptibility factors, S genes, fungal effectors, new resistance sources
Citation: Fabre F, Rocher F, Alouane T, Langin T and Bonhomme L (2020) Searching for FHB Resistances in Bread Wheat: Susceptibility at the Crossroad. Front. Plant Sci. 11:731. doi: 10.3389/fpls.2020.00731
Received: 29 March 2020; Accepted: 07 May 2020;
Published: 11 June 2020.
Edited by:
Paul Christiaan Struik, Wageningen University and Research, NetherlandsReviewed by:
Wei-Hua Tang, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences (CAS), ChinaMoez Hanin, University of Sfax, Tunisia
Copyright © 2020 Fabre, Rocher, Alouane, Langin and Bonhomme. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ludovic Bonhomme, bHVkb3ZpYy5ib25ob21tZUBpbnJhZS5mcg==
†These authors have contributed equally to this work
 Francis Fabre
Francis Fabre Florian Rocher
Florian Rocher Tarek Alouane
Tarek Alouane Thierry Langin
Thierry Langin Ludovic Bonhomme
Ludovic Bonhomme