
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 27 May 2020
Sec. Plant Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00691
 Chunlei Zhou1†
Chunlei Zhou1† Qibing Lin2†
Qibing Lin2† Jie Lan1†
Jie Lan1† Tianyu Zhang1
Tianyu Zhang1 Xi Liu1
Xi Liu1 Rong Miao1
Rong Miao1 Changling Mou1
Changling Mou1 Thanhliem Nguyen1,3
Thanhliem Nguyen1,3 Jiachang Wang1
Jiachang Wang1 Xiao Zhang1
Xiao Zhang1 Liang Zhou1
Liang Zhou1 Xingjie Zhu1
Xingjie Zhu1 Qian Wang1
Qian Wang1 Xin Zhang2
Xin Zhang2 Xiuping Guo2
Xiuping Guo2 Shijia Liu1
Shijia Liu1 Ling Jiang1*
Ling Jiang1* Jianmin Wan1,2*
Jianmin Wan1,2*For efficient plant reproduction, seed dormancy delays seed germination until the environment is suitable for the next generation growth and development. The phytohormone abscisic acid (ABA) plays important role in the induction and maintenance of seed dormancy. Previous studies have identified that WRKY transcription factors can regulate ABA signaling pathway. Here, we identified an Oswrky29 mutant with enhanced dormancy in a screen of T-DNA insertion population. OsWRKY29 is a member of WRKY transcription factor family which located in the nuclear. The genetic analyses showed that both knockout and RNAi lines of OsWRKY29 had enhanced seed dormancy whereas its overexpression lines displayed reduced seed dormancy. When treated with ABA, OsWRKY29 knockout and RNAi lines showed greater sensitivity than its overexpression lines. In addition, the expression levels of ABA positive response factors OsVP1 and OsABF1 were higher in the OsWRKY29 mutants but were lower in its overexpression lines. Further assays showed that OsWRKY29 could bind to the promoters of OsABF1 and OsVP1 to inhibit their expression. In summary, we identified a new ABA signaling repressor OsWRKY29 that represses seed dormancy by directly downregulating the expression of OsABF1 and OsVP1.
The core role of seeds is to reproduce offspring. Seed plants have evolved diverse strategies to ensure survival of their offspring. Among those strategies, seed dormancy prevents immediate germination until appropriate seasonal conditions favor their survival and proliferation (Koornneef et al., 2002; Shu et al., 2016; Penfield, 2017). Pre-harvest sprouting is one of the major problems in cereal production, damaging about 20% of the acreage of hybrid rice in southern China (Hu et al., 2003). By preventing pre-harvest sprouting, seed dormancy can maintain crop yield and quality (Liu et al., 2019). Therefore, it is important to understand and exploit the underlying molecular mechanisms.
The phytohormones abscisic acid (ABA) play major roles in regulating seed dormancy: ABA accumulates in the developing embryo, inducing and maintaining seed dormancy (Finkelstein et al., 2008; Nambara et al., 2010). ABA works through a complex signaling pathway, most components of which are identified. The core components include ABA receptors PYR/PYL/RCAR, negative regulators (protein phosphatase PP2Cs), positive effectors (protein kinase SnRK2s), and various downstream transcription factors such as ABF1, ABF2, ABF3, ABF4, and ABI5 (Finkelstein and Lynch, 2000; Kang et al., 2002; Kim et al., 2004; Fujii et al., 2007; Ma et al., 2009; Park et al., 2009; Yoshida et al., 2010; Fuchs et al., 2014; Yu et al., 2015). In the absence of ABA, PP2Cs dephosphorylate SnRK2s to inhibit their kinase activity. In the presence of ABA, PYR/PYL/RCAR receptors bind to ABA and PP2C, thus unleashing the repression of PP2Cs on the activity of SnRK2s (Hauser et al., 2011). Activated SnRK2s subsequently phosphorylated downstream ABRE-binding protein/ABRE-binding (AREB/ABF) transcription factors to activate ABA responsive genes. Among AREB/ABF transcription factors, ABA insensitive 5 (ABI5), a member of the basic leucine zipper transcription (bZIP) factor family, plays a central role in regulating ABA-responsive genes in seeds of Arabidopsis (Finkelstein and Lynch, 2000). In addition, ABI3, a B3-type APETALA2 domain transcription factor, have been reported to interact with ABI5 and acts upstream of ABI5 to induce the expression of ABA-responsive genes, thus repressing seed germination in Arabidopsis (Lopez-Molina et al., 2002). The function of ABI3 in inducing seed dormancy was likely conserved in plants because CnABI3 in Chamaecyparis nootkatensis (Zeng et al., 2003), VIVIPAROUA 1 (VP1) in maize (Robichaud et al., 1979) and OsVP1 in rice (Gao et al., 2010) all can induce seed dormancy. A bZIP transcription factor, ABF1, is also an important downstream gene in ABA signaling. In Arabidopsis, the abf1 mutant seed exhibited less dormancy and hyposensitive to ABA (Sharma et al., 2011). In rice, the upregulated expression of some ABA-responsive genes was suppressed in Osabf1 mutants (Hossain et al., 2010). Overexpression lines of OsABF1 (OsbZIP12) were hypersensitive to ABA (Joo et al., 2014).
The WRKY family, one of the largest transcription factor families in plants, is involved in regulating many plant processes such as pathogen defense, stress responses and plant development (Wang et al., 2010; Chen et al., 2013; Hu et al., 2013; Li et al., 2016). In Oryza sativa spp. japonica, the WRKY transcription factor superfamily consists of an estimated 98 members that fall into four major structural groups based on number of WRKY domains and specific features of their zinc finger-like motifs (Ross et al., 2007). Most WRKY proteins have a high binding affinity to the DNA sequence (T)(T)TGAC(C/T), which is known as W-box (Ross et al., 2007). Although WRKY transcription factors can bind to the same W-box they still have functional differences in regulating seed dormancy by modulating the ABA signaling pathway. Some WRKY proteins promote seed dormancy by positively modulating ABA-responsive genes. For example, AtWRKY2 promotes expression of ABI3 and ABI5; meanwhile, ABA-induced AtWRKY2 transcription requires the participation of ABI3 and ABI5 (Jiang and Yu, 2009), suggesting a positive feedback loop between AtWRKY2 and ABI3 or ABI5 in regulating ABA response. AtWRKY63 activates ABA-responsive genes such as ABF2, RD29A, and COR47 (Ren et al., 2010). AtWRKY41 protein is reported to positively regulate ABI3 expression (Ding et al., 2014). AtWRKY6 promotes ABA signaling by direct downregulating the expression of RAV1, which is a repressor of ABI3, ABI4 and ABI5 expression (Huang et al., 2016). OsWRKY72 and OsWRKY77 activate promoter activity of ABA-induced gene HVA22 in barley aleurone cells (Xie et al., 2005). Some WRKY proteins repress seed dormancy by negatively modulating ABA-responsive genes. For example, AtWRKY18, AtWRKY40 and AtWRKY60 negatively regulate ABA signaling by directly repressing the expression of ABA-responsive genes, MYB2, ABI4, ABF4, ABI5, DREB1A, and RAB18 (Shang et al., 2010). OsWRKY24 and OsWRKY45 repress ABA-induced HVA22 expression in barley aleurone cells (Xie et al., 2005). However, the exact role of WRKY transcription factors in ABA response is obscure and their possible roles are often overlooked in studies of ABA signaling (Rushton et al., 2012). Thus, more WRKY genes need to be studied for better understood of their exact role in regulating ABA signaling.
In this study, we identified a new WRKY transcription factor, OsWRKY29, which negatively regulates seed dormancy in rice. Knockout and RNA interference of OsWRKY29 enhanced seed dormancy whereas its overexpression reduced seed dormancy. The germinating seeds of OsWRKY29 knockout and RNAi lines displayed hypersensitivity whereas these of OsWRKY29-overexpression lines displayed hyposensitivity to ABA. Compared with wild type, the expression of ABA positive response genes OsABF1 and OsVP1 was increased in OsWRKY29 mutants but reduced in OsWRKY29-overexpression lines. Further analyses showed that OsWRKY29 directly binds to W-boxes in the promoters of OsABF1 and OsVP1 to repress their expression. These results suggest that OsWRKY29 represses seed dormancy by weakening ABA response.
Oswrky29 mutant (2D-41043L) screened from a set of activation-tagged T-DNA insertion lines [in cultivar (cv.) Hwayoung (Hy)] (Jeong et al., 2002) was kindly provided by Prof. Gynheung An (Department of Plant Systems Biotech, Kyung Hee University, South Korea). All plants were grown at two field sites, Tuqiao in Nanjing and Sanya in Hainan province.
Dormancy levels were assessed according to the method of Wan et al. (2005), but the date of seeds harvest and the number of experimental samples were modified. Filled intact grains for evaluation of dormancy were collected at 45 days post heading. Freshly harvested seeds were placed on double sheets of filter paper in a 9 cm diameter Petri dish, moistened with distilled water, and maintained at 30°C and 100% relative humidity for 5 days. Each plant was tested in three replicates. 30 seeds were measured in each replicate. These seeds in three replicates are from the same harvest of the same plant. A germinated seed was declared when the plumule or radicle reached half the seed length. The percentage of germinated seeds on the 5th day was used as a measure of dormancy. For detection of sensitivity to ABA the harvested seeds were exposed to 50°C for 72 h to break dormancy (Lu et al., 2011). The heat-treated seeds (dormancy released stage) were used for ABA sensitivity tests. Seeds were germinated in 1, 3, and 5μM ABA solutions and the germination percentage was counted on the 5th or 7th day. Each plant was tested in three replicates. Thirty seeds were measured in each replicate.
Primer Premier 5.0 software was used to design primers, and the primers used in confirming T-DNA insertion sites are listed in Supplementary Table S1. Total RNA was extracted from tissues using a RNAprep Pure Plant Kit (Tiangen Biotech Co. Ltd., Beijing, China) according to the manufacturer’s instructions. First-strand cDNA was synthesized using a PrimeScriptTM II 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian). Synthesized cDNAs were used for RT-qPCR with a SYBR Premix Ex Taq kit (TaKaRa, Dalian, China) on an ABI 7500 real-time PCR system (Thermo Fisher Scientific, Waltham, MA, United States). Determination of relative changes in gene expression levels was based on three biological replicates. Primers were designed using the GenScript real-time PCR (TaqMan) primer design tool1 (Primers are listed in Supplementary Table S1). The rice UBQ5 gene (LOC_Os03g13170) was used to normalize cDNA quantity.
In order to knock out the OsWRKY29 gene, we cloned 20 bp gene-specific spacer sequences into the sgRNA-Cas9 vector and then introduced it into calli of cv. Hwayoung (primer pair OsWRKY29-CRISPR in Supplementary Table S1) by Agrobacterium-mediated transformation. We identified positive transgenic individuals by sequencing.
OsWRKY29 RNA interference plants were generated by amplification of 228 bp cDNA of OsWRKY29 using primer pair OsWRKY29-RNAi-SacI and OsWRKY29-RNAi-SnaBI (Supplementary Table S1). We fused OsWRKY29-RNAi-SacI into the SacI restriction site of the LH-FAD1390RNAi binary vector to construct an OsWRKY29-RNAi-SacI vector, then fused OsWRKY29-RNAi-SnaBI into the SnaBI restriction site of the OsWRKY29-RNAi-SacI vector to construct an OsWRKY29-RNAi vector, which was introduced into calli of Nipponbare by Agrobacterium-mediated transformation.
OsWRKY29-overexpressing plants were generated by amplification of full-length CDS of OsWRKY29 using primer pair OsWRKY29-OX (Supplementary Table S1) and fused into the KpnI/BamHI site of the pCUBi1390 binary vector; and the resulting UBi: OsWRKY29-overexpression construct was inserted into calli of Nipponbare by Agrobacterium-mediated transformation.
Full-length CDS of OsWRKY29 for subcellular localization of OsWRKY29 protein was inserted into the XbaI/BamHI restriction site upstream of GFP in expression vector pAN580 driven by a double CaMV35S promoter to generate the OsWRKY29-GFP construct (primer pair OsWRKY29-GFP in Supplementary Table S1). The full-length CDS was also inserted into the BglII/PstI restriction site downstream of GFP in expression vector pAN580 driven by a double CaMV35S promoter to generate a GFP-OsWRKY29 construct (primer pair GFP-OsWRKY29 in Supplementary Table S1). The OsWRKY29-GFP and GFP-OsWRKY29 plasmids were introduced into rice protoplasts according to a published protocol (Zhang et al., 2011). Fluorescence of GFP was observed using a laser scanning confocal microscope (LSM 700; Zeiss).
Two reporters were used in Transcription activity assays. One contained the firefly luciferase (LUC) gene fused with 5 × GAL4 binding site, and the other was a renilla luciferase (REN) gene under control of CaMV35S promoter as the internal control. Full-length of OsWRKY29 was fused with the GAL4-BD and driven by a CaMV35S promoter to generate effector. The plasmids with effector (GAL4-BD-WRKY29) and two reporters were co-transformed into rice protoplasts as described (Zhang et al., 2011). LUC activity was quantified with a Dual-Luciferase Assay Kit (Promega, Beijing, China) according to the manufacturer’s recommendations and relative LUC activity was calculated by the ratio of LUC/Ren. Each sample was tested with three replicates.
Yeast one-hybrid analysis was performed as described in a previous report (Lin et al., 2007). We cloned the full-length coding region of OsWRKY29 into the pB42AD vector at the EcoRI restriction site to produce a pB42AD-OsWRKY29 construct (primer pair pB42AD-OsWRKY29 in Supplementary Table S1). The promoter regions or mutant promoter regions of OsVP1 and OsABF1 were cloned into the pLacZi reporter vector (primer pairs listed in Supplementary Table S1) at the XhoI restriction site to generate pLacZi-OsVP1, pLacZi-OsABF1, mpLacZi-OsVP1, and mpLacZi-OsABF1 constructs. The pB42AD-OsWRKY29 construct was co-transformed with pLacZi-OsVP1, pLacZi-OsABF1, mpLacZi-OsVP1, and mpLacZi-OsABF1 constructs into yeast strain EGY48, respectively. Transformants were cultured on SD-Trp/-Ura plates at 30°C for 3 days, followed by transfer into 5-bromo-4-chloro-3-indolyl-b-d-galactopyranoside plates for blue color development. Yeast cultures with the empty pB42AD combination with the pLacZi reporter constructs were used as negative controls.
LUC activity assays were performed according to a previously described method (Duan et al., 2019). In order to generate ProOsABF1:LUC, mProOsABF1:LUC, ProOsVP1:LUC and mProOsVP1:LUC reporter constructs, we amplified ∼2 kb promoter regions of OsABF1 and OsVP1 using primer pairs ProOsABF1:LUC, mProOsABF1:LUC, ProOsVP1:LUC and mProOsVP1:LUC (primer pairs listed in Supplementary Table S1), and then fused them into the NcoI restriction site of the pGreenII 0800-LUC vector. The luciferase gene from Renilla reniformis (Ren) under control of the CaMV35S promoter was used as the internal control. The cDNA of OsWRKY29 was amplified (primer pair listed in Supplementary Table S1) and inserted into the XbaI/BamHI restriction sites of the pAN580 vector to generate a d35S:OsWRKY29 effector construct. The d35S:OsWRKY29 effector combined with four reporters, respectively, and the corresponding combinations were co-transformed into rice protoplasts as described (Zhang et al., 2011). Empty pAN580 vector co-transformed with the reporter constructs were used as vector controls. LUC activity was quantified with a Dual-Luciferase Assay Kit (Promega, Beijing, China) according to the manufacturer’s recommendations and relative LUC activity was calculated by the ratio of LUC/Ren. Each sample was tested in three replicates.
In vitro ChIP assays were performed according to a published method (Li et al., 2017). Total DNA of cv. Hwayoung and purified OsWRKY29-MBP protein were used for ChIP assays. Total DNA from two-week-old seedlings was sheared into 100-500 bp fragments using an ultrasonic crusher. Full-length coding sequence of OsWRKY29 was cloned into the expression vector pMAL-c2x (primer pairs listed in Supplementary Table S1) to generate MBP fusion protein. Expression of MBP and OsWRKY29-MBP in Escherichia coli strain BL21 (DE3) cells (TransGen, Beijing, China) was induced by 0.5 mM isopropylb-D-thiogalactoside at 16°C for 16 h. Fusion proteins were purified using Amylose Resin (New England BioLabs, Beijing, China) according to the manufacturer’s protocol, and protein concentrations were determined by the BSA quantitative assay. MBP and OsWRKY29-MBP were co-incubated with DNA fragments in the incubation buffer for 2 h (50 mM Tris, 1 mM EDTA, 100 mM KCl, adjusted to pH 7.0 by HCl, 5% glycerol, 0.1% Triton X-100, with freshly made 100 mM DTT added to the reaction solution to make a final concentration of 1 mM DTT). Afterward, the Amylose Resin was washed three times using the incubation buffer; 4 mL 5 M NaCl was added into for each 100 mL volume and the sample was incubated for 4 h to break down cross-linked OsWRKY29-MBP and DNA fragments. DNA fragments were subsequently collected with a DNA Recovery Kit (Zymo, Beijing, China) following the manufacturer’s recommendations. DNA fragments between 100 and 500 bp were used for ChIP-qPCR.
The prepared DNA for in vitro ChIP assays was also used for qPCR using relevant primer pairs (Supplementary Table S1) in a SYBR Premix Ex Taq (TaKaRa) with an ABI7500 Real-Time PCR instrument. PCR were performed in triplicate for each sample, and fold enrichment was calculated against the UBQ5 gene. MBP served as a negative control.
OsWRKY29 proteins (fused with maltose-binding protein [MBP] tags) were expressed in E. coli strain BL21 (DE3) (TransGen, Beijing, China) and affinity-purified. The probes containing the putative OsABF1 and OsVP1 binding sites were synthesized by Invitrogen (Shanghai) (primer pairs listed in Supplementary Table S2). The probes were labeled with biotin at the 5′ end and the competitive probes lacked the label. electrophoretic mobility shift assay (EMSA) was performed with the LightShift Chemiluminescent EMSA Kit (Thermo, No. 20148) following the manufacturer’s protocol. Briefly, equal amounts of complementary oligonucleotides were incubated at 95°C for 10 min, cooled slowly to 15°C (0.1°C/1s), and diluted to a 20 fmol final concentration. The DNA binding reaction was performed with 2 μl of probe, 2 μl Binding Buffer, 1 μg purified MBP or OsWRKY29-MBP protein, with or without competitive probes, and brought to 20 μl with added water and incubated at room temperature for 30 min. Samples were immediately applied to pre-run native polyacrylamide gel containing 6.5% acrylamide in 0.5 X Tris-Borate-EDTA buffer. After electro-blotting onto a nylon membrane (Millipore) the oligonucleotides were cross-linked using UV-light. The membrane was incubated in a blocking solution for 15 min, followed by incubation in a conjugate/blocking buffer solution for another 15 min. After intensive washing with a washing buffer, Substrate Equilibration Buffer was applied to the membrane for 5 min. Finally, Substrate Working Solution was added to visualize the signal.
A search of the T-DNA insertion database OryGenesDB2, showed that pGA2772 (activation-tagged T-DNA vector) was inverse-integrated in the second intron of LOC–Os07g02060 in the T-DNA insertion line (Figure 1A). To confirm the T-DNA insertion site in the Oswrky29 mutant, three primers, P1, P2, and P3 (P1 and P2 from the OsWRKY29 genome sequence and P3 from the right border (RB) of the inserted T-DNA), were designed for PCR analysis. When P1 and P2 were used for PCR, a single band was detected only in the wild type and when P3 and P2 were used for PCR, a single band was detected in the Oswrky29 mutant (Figure 1B). Identification of the latter sequence revealed two parts: one from the right border of the T-DNA, the other from the OsWRKY29 genome sequence (Supplementary Figure S1). These results confirmed that the mutant carried a T-DNA insertion in the second intron of OsWRKY29. To test if the T-DNA insertion affected the transcription of OsWRKY29, we performed RT-qPCR and found that the expression of OsWRKY29 was reduced in the mutant relative to the wild type Hwayoung (Hy) (Figure 1C).
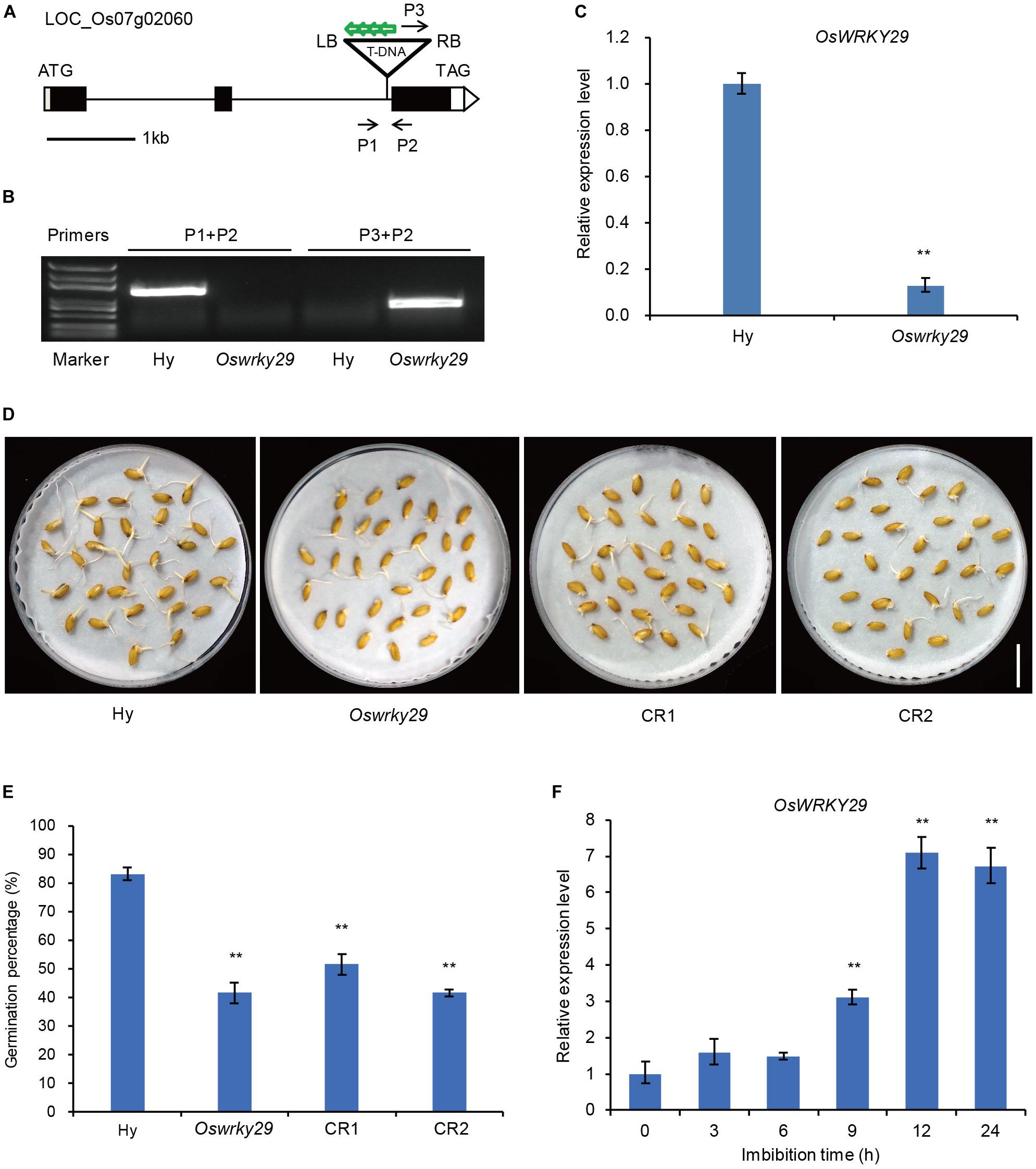
Figure 1. Seed dormancy phenotypes of Oswrky29 T-DNA insertion mutant. (A) T-DNA insertion site in Oswrky29 (LOC_Os07g02060) mutant is indicated by a triangle. Black boxes indicate exons, lines indicate introns, and white boxes indicate the untranslated regions. P1, P2, and P3 are the primers used to identify the T-DNA insertion site. Green arrows indicate four copies of 35S enhancers in the T-DNA insertion sequence. RB and LB indicate the right and left borders of the T-DNA insertion. (B) Identification of the T-DNA insertion by PCR. The primers are labeled in panel (A). (C) RT-qPCR of the OsWRKY29 transcript in Hwayoung (Hy) and Oswrky29 seeds. Fresh seeds were collected at 45 days post heading for RT-qPCR analysis. Values are means ± SD (n = 3). The Student’s t-test analysis indicates a significant difference (**P < 0.01). (D) Seed germination phenotypes of Hy, Oswrky29 mutant and OsWRKY29-CRISPR transgenic lines CR1 and CR2. Fresh seeds were collected at 45 days post heading for germination test. Scale bar, 2 cm. (E) Germination percentages for Hy, OsWRKY29 mutant, CR1 and CR2. Fresh seeds were collected at 45 days post heading for germination test. Values are means ± SD (n = 3), 30 seeds were measured in each replicate. The Student’s t-test analysis indicates a significant difference (**P < 0.01). (F) Analysis of OsWRKY29 expression levels during seed imbibition in water determined by RT-qPCR. Fresh seeds were collected at 45 days post heading and imbibition in water. RT-qPCR was performed at the indicated times. Values are means ± SD (n = 3). The Student’s t-test analysis indicates a significant difference (**P < 0.01).
Some WRKY proteins were reported to be involved in the regulation of seed dormancy (Xie et al., 2005; Ding et al., 2014; Huang et al., 2016). To test whether OsWRKY29 also regulates seed dormancy, the fresh harvested seeds of Oswrky29 mutant and OsWRKY29 knockout transgenic lines (CR1 and CR2) (Supplementary Figure S2) were used to perform seed germination assays. The results showed that germination percentage of Oswrky29 mutant was 41% compared to 83% for the wild type Hy (Figures 1D,E), suggesting that Oswrky29 mutant had stronger seed dormancy than its wild type Hy. Consistent with this, OsWRKY29 knockout transgenic lines (CR1 and CR2) also displayed enhanced seed dormancy (Figures 1D,E). These results showed that both the Oswrky29 mutant and OsWRKY29 knockout lines had stronger seed dormancy than Hy. Further, OsWRKY29 expression was evidently decreased with the grain development (Supplementary Figure S3), but during seed imbibition, OsWRKY29 expression was gradually upregulated (Figure 1F), indicating that the expression level of OsWRKY29 is negatively related to seed dormancy. Taken together, our results suggest that the absence of OsWRKY29 enhances seed dormancy.
To further confirm that knockdown of OsWRKY29 was responsible for the Oswrky29 mutant phenotype, two independent OsWRKY29 RNAi lines in Nipponbare (NIP) background, Ri-1 and Ri-2, in which OsWRKY29 transcript levels were evidently reduced, were chosen to test seed dormancy (Figure 2A). Seed germination of Ri-1 and Ri-2 was significantly reduced compared with NIP (Figures 2B,C). In contrast, the overexpression lines of OsWRKY29, OE-1 and OE-2, whose OsWRKY29 transcript levels were evidently increased (Figure 2A), showed significantly higher germination percentage (almost 100%) compared to their receptor NIP (Figures 2B,C). Thus, knockdown of OsWRKY29 apparently enhanced seed dormancy whereas its overexpression had an opposite effect, further supporting the notion that OsWRKY29 functions as a negative regulator of seed dormancy.
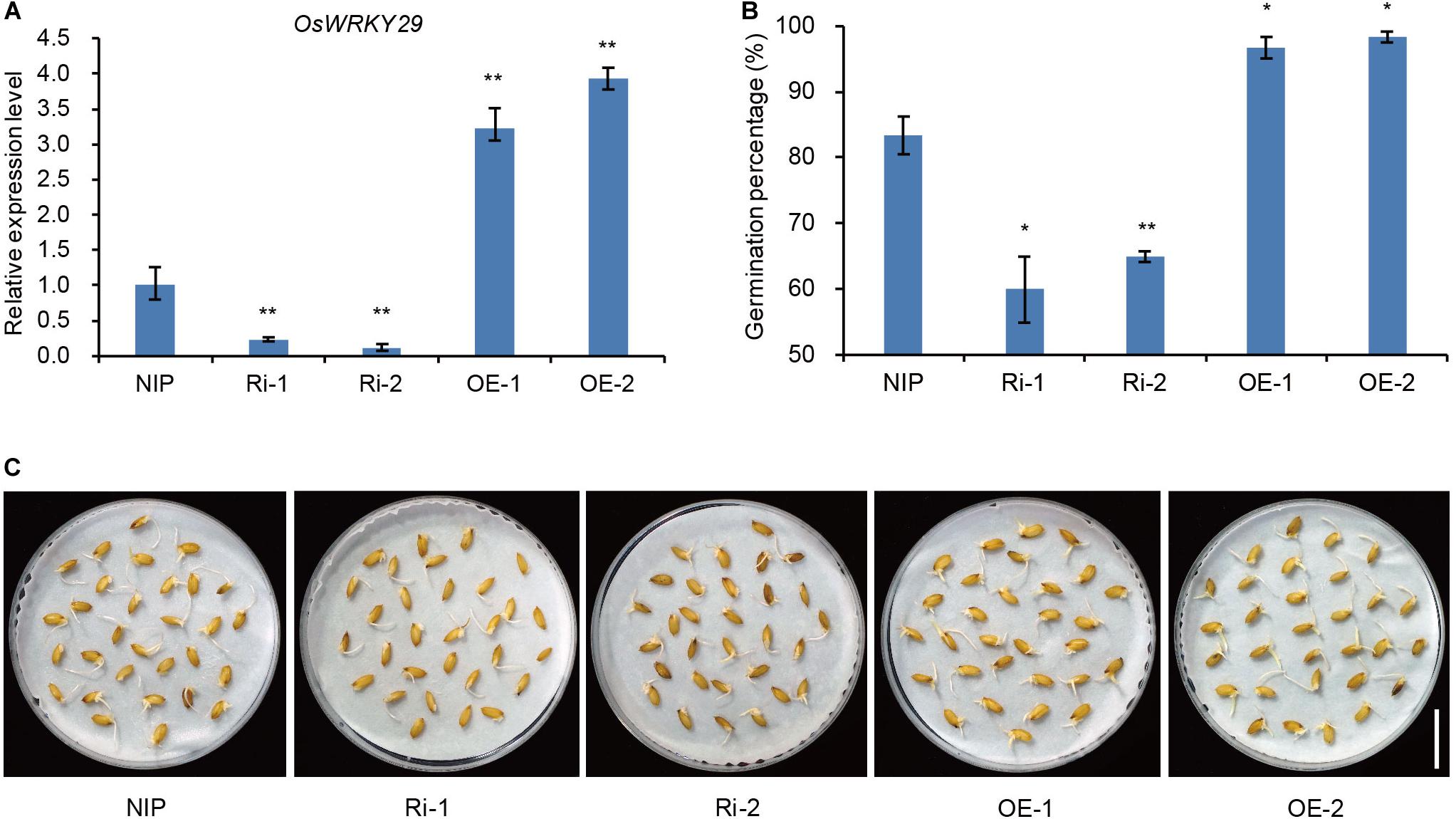
Figure 2. Seed dormancy phenotypes of OsWRKY29 transgenic plants. (A) OsWRKY29 expression levels in seeds of the NIP, OsWRKY29 Ri and OE lines determined by RT-qPCR. Fresh seeds were collected at 45 days post heading for RT-qPCR analysis. Values are means ± SD (n = 3). The Student’s t-test analysis indicates a significant difference (**P < 0.01). (B) Seed germination percentages of NIP, OsWRKY29 Ri and OE lines after five days of imbibition. Fresh seeds were collected at 45 days post heading for germination test. Values are means ± SD (n = 3), 30 seeds were measured in each replicate. The Student’s t-test analysis indicates a significant difference (*P < 0.05, **P < 0.01). (C) Seed dormancy phenotypes of NIP, OsWRKY29 Ri and OE lines after five days of imbibition. Fresh seeds were collected at 45 days post heading for germination test. Scale bar, 2 cm.
OsWRKY29, with three exons and two introns, encodes a 290 amino acid protein. By BLAST search in the NCBI database, we identified its homologs (including most reported homologs) in Arabidopsis and rice (Supplementary Figure S4A). Alignment some of these homologs using Clustal X revealed a conserved WRKYGQK motif (WRKY domain) and a C2H2 zinc-finger motif in OsWRKY29 and its homologs, suggesting that OsWRKY29 and its homologs would bind similar DNA motifs to regulate transcription (Supplementary Figure S4B).
The subcellular localization analyses showed that fusion proteins OsWRKY29-GFP and GFP-OsWRKY29 were uniformly localized in the nucleus (Figure 3A), consistent with function as a transcription factor. A RT-qPCR analysis revealed that OsWRKY29 was widely expressed in rice tissues, with the highest expression level of OsWRKY29 transcript in leaves and lowest expression in stems (Figure 3B). To test the transcription activity of OsWRKY29, we performed transient dual-luciferase (LUC) assays in rice protoplasts. The result revealed that OsWRKY29 led to an obvious downregulation of relative luciferase activity compared to the control (Figure 3C). These results indicated that OsWRKY29 acts as a transcriptional repressor.
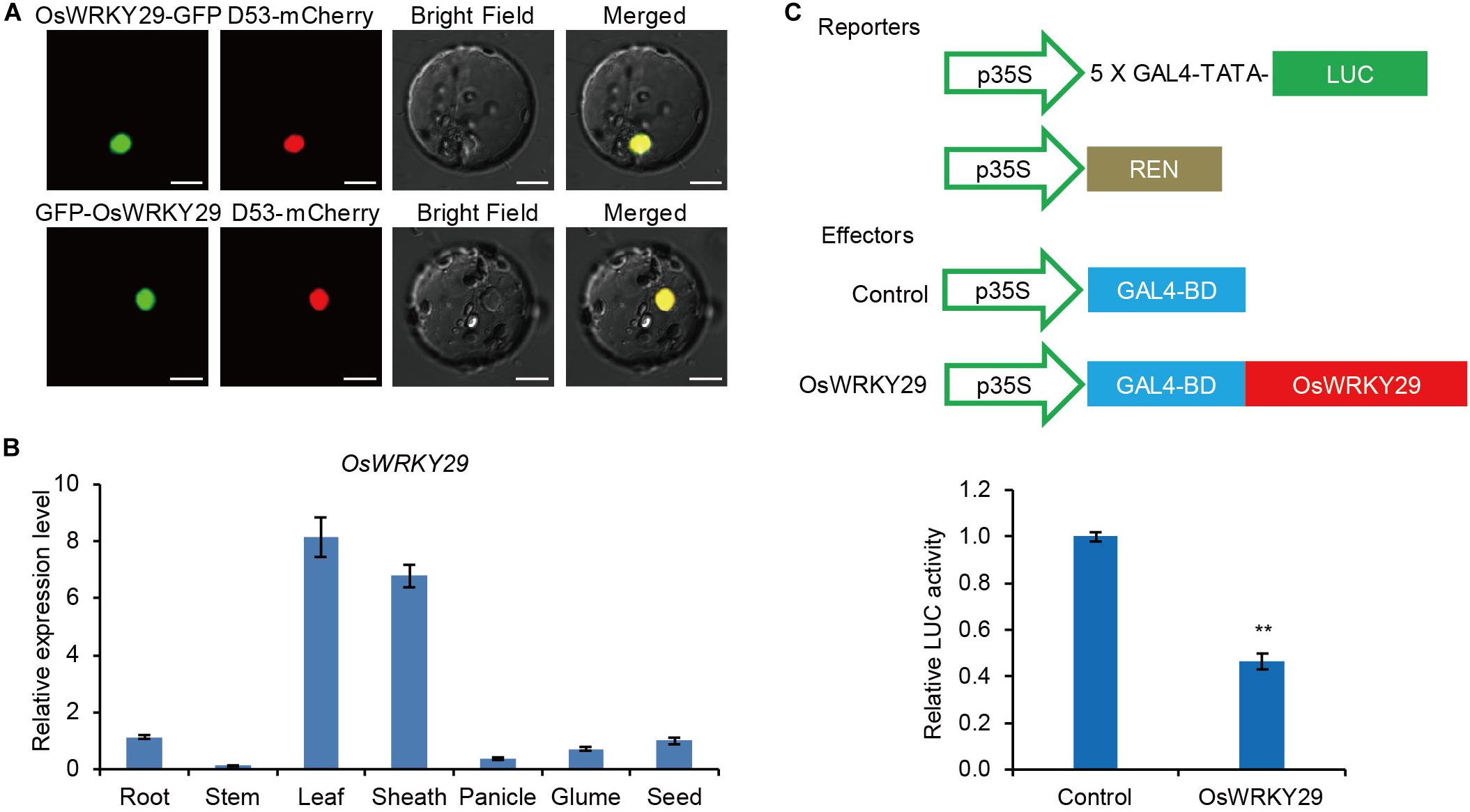
Figure 3. Subcellular-localization, expression pattern, and transcription activity of the OsWRKY29 gene. (A) Subcellular localizations of the OsWRKY29-GFP fusion protein and GFP-OsWRKY29 fusion protein in rice protoplasts. D53-mCherry was used as the nuclear marker. Scale bar, 10 μm. (B) Relative expression of OsWRKY29 in various tissues including root (2 weeks old), stems (heading stage), leaf (heading stage), leaf sheath (heading stage), panicle (2 cm), Glume (in blossom), and seed (30 DPA). Values are means ± SD (n = 3). (C) Transcription activity assay of OsWRKY29 in rice protoplasts. The reporters and effectors used in the assay were generated as shown. Firefly luciferase (LUC) and renilla luciferase (REN) activities were determined 16 h post-transformation. The relative luciferase activities in control and OsWRKY29-expressed samples were calculated by normalizing the LUC values against REN. Values are means ± SD (n = 3). The Student’s t-test analysis indicates a significant difference (**P < 0.01).
Previous studies showed that WRKY transcription factors can regulate many stress responses through modulating the ABA signaling pathway. Moreover, ABA signaling was considered to be essential for regulating seed dormancy and germination (Shang et al., 2010; Tao et al., 2011). To investigate whether OsWRKY29 regulates seed dormancy by modulating the ABA signaling pathway, we broke seed dormancy by drying seeds at 50°C for 72 h and treated seeds (dormancy released stage) of Hy, Oswrky29 mutant and OsWRKY29 knockout lines (CR1 and CR2) with or without ABA. Under treatment with ABA, Oswrky29 mutant and OsWRKY29 knockout lines showed reduced seed germination percentage relative to wild-type Hy (Supplementary Figures S5A–E), showing that Oswrky29 mutant and OsWRKY29 knockout lines had enhanced sensitivity to ABA. With the germination of Hy seeds, the increase in OsWRKY29 transcript level was clearly repressed by the treatment with 3 μM ABA compared to the treatment without ABA (-ABA) (Supplementary Figure S5F). Similarly, the OsWRKY29 transcript level was also significantly repressed by 100 μM ABA applied to Hy seedlings (Supplementary Figure S6). These observations showed that ABA can inhibit the expression of OsWRKY29 and the lack of OsWRKY29 enhanced sensitivity to ABA-mediated inhibition of seed germination.
To further confirm the function of OsWRKY29 in the ABA signaling pathway, OsWRKY29-overexpressing lines (OE-1, OE-2) and OsWRKY29 RNAi lines (Ri-1, Ri-2) were also used to study the physiological role of OsWRKY29 in ABA response. Heat-treated seeds (dormancy released stage) of OsWRKY29-overexpression lines and OsWRKY29 RNAi lines were treated with or without ABA. Without the ABA treatment (0 μM ABA), seeds of OsWRKY29-overexpression lines displayed faster germination (Figures 4A,B) and OsWRKY29 RNAi seeds displayed slower germination compared to NIP (Figures 4A,B). And with the ABA treatment, the germination of OsWRKY29-overexpression lines showed a slower decline trend in germination whereas the germination of OsWRKY29 RNAi lines showed a faster decline trend relative to NIP (Figures 4C–E). Knockdown of OsWRKY29 enhances while the overexpression of OsWRKY29 reduces ABA sensitivity during seed germination especially by 3 μM ABA treatment (Supplementary Figure S7). These data indicated that OsWRKY29 is a negative regulator in ABA-inhibited seed germination.
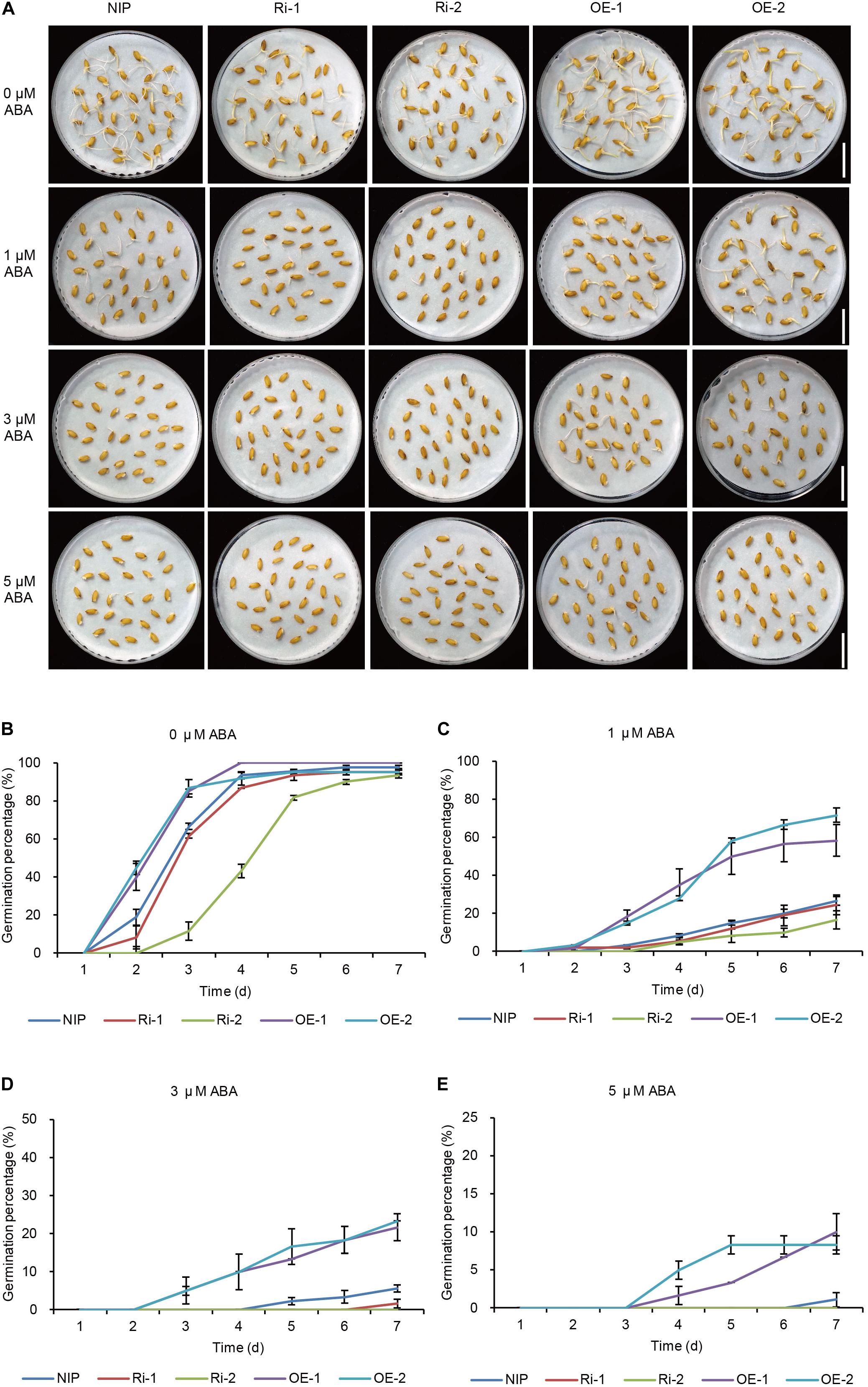
Figure 4. ABA responses of OsWRKY29 RNA interference (Ri) and overexpression (OE) lines. (A) Seed germination phenotypes of Nipponbare (NIP), OsWRKY29 Ri and OE lines treated with ABA at 7th day. Scale bar, 2 cm. (B–E) Germination time courses in water containing 0 μM ABA (B), 1 μM ABA (C), 3 μM ABA (D), and 5 μM ABA (E). Seed dormancy was broken by drying at 50°C for 72 h. The heat-treated seeds (dormancy released stage) were used for ABA treatment. Values are means ± SD (n = 3).
To understand the role of OsWRKY29 in ABA-mediated seed dormancy, expression levels of several ABA-related genes were analyzed in seeds of WT, Oswrky29 mutant and CR2, including ABA response genes, OsABI-LIKE1 (OsABIL1) (Li et al., 2015), OsABI-LIKE2 (OsABIL2) (Li et al., 2015), OsABI5 (Zou et al., 2008), OsVP1 (Hobo et al., 1999; Miyoshi et al., 2002), TRAB1 (Hobo et al., 1999; Kobayashi et al., 2005), OsABF1 (Hossain et al., 2010), OsABF2 (Yang et al., 2011; Tang et al., 2012), OsbZIP23 (Xiang et al., 2008), and OsbZIP72 (Lu et al., 2009), and ABA synthesis and metabolism genes, OsNCED1, OsNCED2, OsNCED3, OsNCED4, OsNCED5, OsABA8ox1, OsABA8ox2, and OsABA8ox3 (Zhu et al., 2009). Expression levels of positive ABA responsive factors, OsVP1 and OsABF1, and ABA synthesis gene OsNCED3, were higher in both Oswrky29 and CR2, and ABA metabolism gene OsABA8ox3, was only higher in Oswrky29 (Figures 5A,B). However, expression levels of other ABA-related genes, including OsABIL1, OsABIL2, OsABI5, TRAB1, OsABF1, OsABF2, OsbZIP23, OsbZIP72, OsNCED1, OsNCED2, OsNCED4, OsNCED5, OsABA8ox1, and OsABA8ox2 showed no evident difference among WT, Oswrky29 and CR2 (Figures 5A,B). We further verified that the transcript levels of OsVP1, OsABF1 and OsNCED3 were upregulated in OsWRKY29 Ri lines but downregulated in OsWRKY29 OE lines compared with their receptor NIP (Figure 5C). Interestingly, the expression level of OsABA8ox3 showed no difference among NIP, Ri and OE lines. In addition, the expression levels of some late embryogenesis abundant protein (LEA) genes (OsLEA3-1, OsLEA3-2, OsLEA5 and OsEm1) were higher in Ri lines and lower in OE lines, compared to NIP (Figure 5C). These results suggest that OsWRKY29 negatively regulates several ABA-related genes.
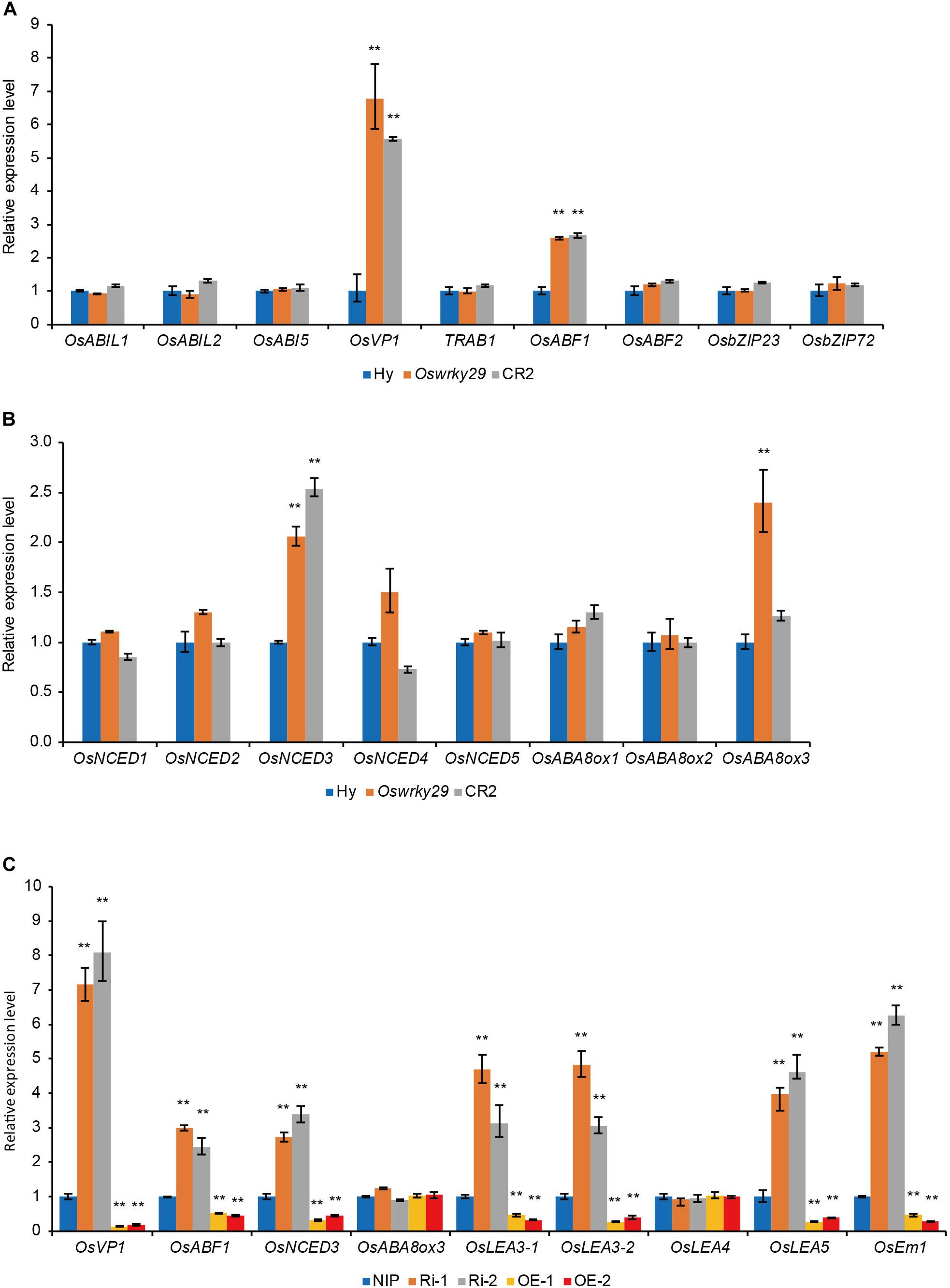
Figure 5. The expression levels of ABA-related genes. (A) Relative transcript levels of ABA response genes in seeds of Hy, Oswrky29 and CR2. Fresh seeds were collected at 45 days post heading for RT-qPCR analysis. Values are means ± SD (n = 3). The Student’s t-test analysis indicates a significant difference (**P < 0.01). (B) Relative transcript levels of ABA synthesis and metabolism genes in seeds of Hy, Oswrky29 and CR2. Fresh seeds were collected at 45 days post heading for RT-qPCR analysis. Values are means ± SD (n = 3). The Student’s t-test analysis indicates a significant difference (**P < 0.01). (C) Relative transcript levels of several ABA-related genes in seeds of NIP, OsWRKY29 Ri lines and OsWRKY29 OE lines. Fresh seeds were collected at 45 days post heading for RT-qPCR analysis. Values are means ± SD (n = 3). The Student’s t-test analysis indicates a significant difference (**P < 0.01).
Previous studies demonstrated that WRKY proteins regulate expression of their target genes by binding to W-boxes in their promoters. Analyses of 2 kb promoter sequences identified two W-boxes in the OsABF1 promoter, four W-boxes in OsVP1 promoter and one W-box in the OsNCED3 promoter (Figure 6A and Supplementary Figures S8, S9A). We hypothesized that OsWRKY29 binds to the W-boxes of the promoters of OsABF1, OsVP1, and OsNCED3 to regulate their expression.
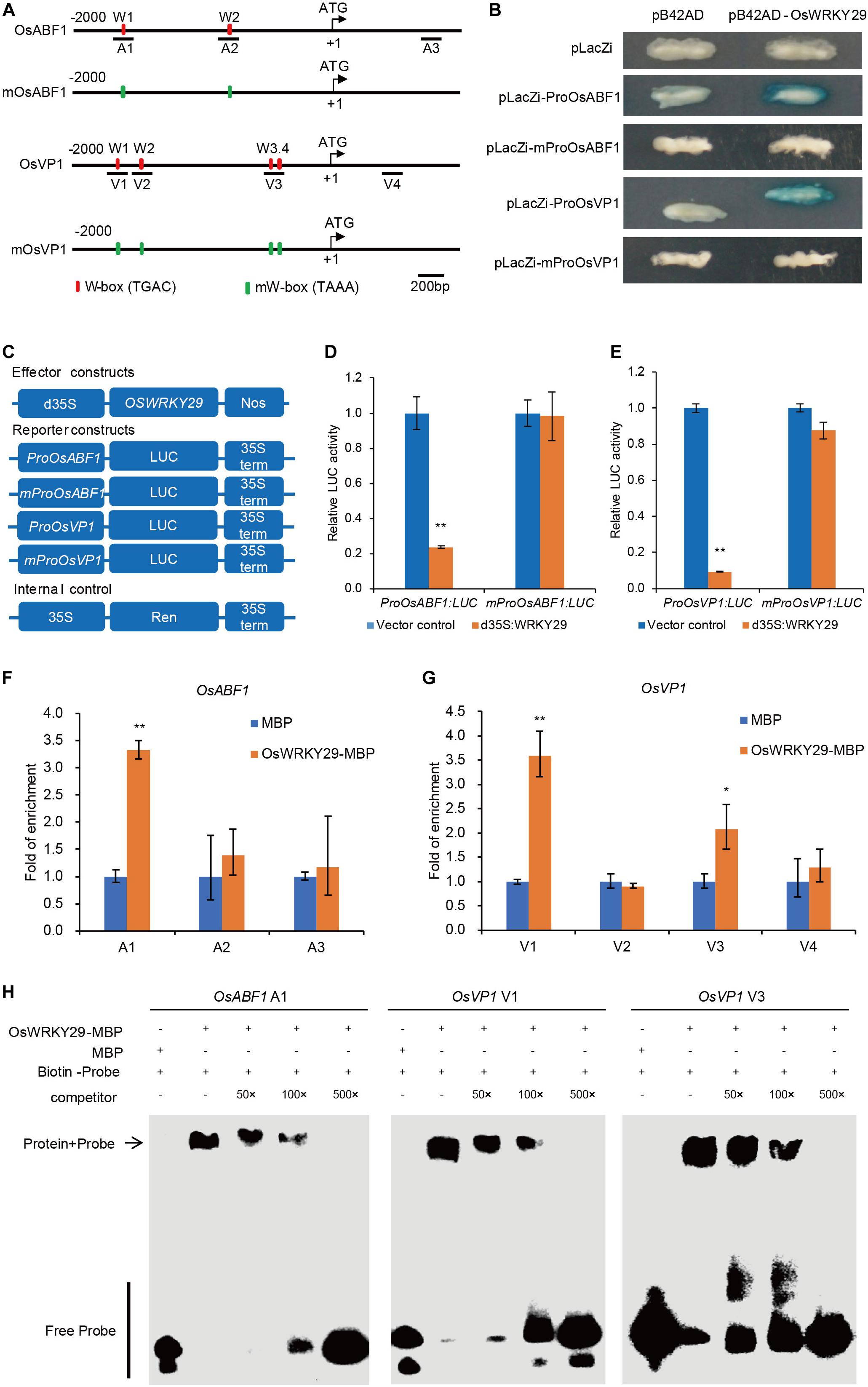
Figure 6. OsWRKY29 represses OsABF1 and OsVP1 expression by binding to their promoters. (A) Schematic indicating sequences 2 kb upstream of the start sites and parts of the coding sequences of OsABF1 and OsVP1. The translational start sites (ATG) shown at position + 1. Red rectangles represent W-boxes. Green rectangles represent mW-boxes (TGAC mutated to TAAA). The numbers (A1–A3 and V1–V4) indicate the tested regions. (B) Yeast one-hybrid assays showing OsWRKY29 binding to the promoters of OsABF1 and OsVP1. (C) Schematic diagram of the effector and reporter constructs. Full-length coding region of OsWRKY29 under control of a double 35S promoter was used as the effector. The firefly luciferase gene LUC driven by the OsABF1 and OsVP1 promoters and mutant promoters. The Renilla luciferase gene Ren driven by the 35S promoter were used as the reporter and internal control, respectively. (D,E) Relative LUC activities of ProOsABF1:LUC and mProOsABF1:LUC reporters (D) and ProOsVP1:LUC and mProOsVP1:LUC reporters (E) after co-expression with 35S:OsWRKY29 in rice protoplasts. 35S empty vector was used as control. Relative LUC activity was calculated by LUC/Ren. Values are means ± SD (n = 3). The Student’s t-test analysis indicates a significant difference (**P < 0.01). (F,G) ChIP-qPCR assays shows the target DNA fragments bound by OsWRKY29-MBP. OsWRKY29-MBP and MBP alone were incubated with total rice DNA for 4 h, pulled down, washed, and subjected to qPCR for OsABF1 (F) and OsVP1 (G). Fold enrichment was normalized against the ubiquitin promoter. Values are means ± SD (n = 3). The Student’s t-test analysis indicates a significant difference (*P < 0.05, **P < 0.01). (H) EMSA analysis showing the binding of recombinant OsWRKY29 protein to the promoters of OsABF1 (A1) and OsVP1 (V1 and V3).
To test the above inference, we first performed yeast one-hybrid (Y1H) assays to evaluate if OsWRKY29 directly binds to the promoters of OsABF1, OsVP1, and OsNCED3. As shown in Figure 6B and Supplementary Figure S9, the color of the control was unchanged whereas all experimental groups except OsNCED3 turned blue, suggesting that OsWRKY29 directly binds to the promoters of OsABF1 and OsVP1 but not OsNCED3. Then, we mutated these W-boxes from TGAC to TAAA in this experiment, and found that OsWRKY29 can’t bind to the mutated promoters of OsABF1 and OsVP1 (Figure 6B), suggesting W-boxes are necessary for OsWRKY29 to bind the promoter of OsABF1 or OsVP1. We further performed transient dual-luciferase (LUC) assays in rice protoplasts to test the transcriptional regulatory relationship between OsWRKY29 and OsABF1 or OsVP1. As shown in Figures 6C–E, OsWRKY29 greatly repressed the expression of the luciferase (LUC) reporter gene driven by the OsABF1 or OsVP1 promoter; but when the W-boxes were changed from TGAC to TAAA, the inhibition activity of OsWRKY29 on the mutated promoter of OsABF1 and OsVP1 was disappeared, indicating that OsWRKY29 inhibits the transcription activity of the OsABF1 and OsVP1 dependent on W-boxes.
We also performed an in vitro qPCR assay after chromatin immunoprecipitation (ChIP-qPCR). OsWRKY29 enriched on A1 promoter region of OsABF1 as well as the V1 and V3 promoter regions of OsVP1 rather than other promoter regions of OsABF1 and OsVP1 (Figures 6F,G), suggesting that OsWRKY29 could specifically bind to the A1 region of OsABF1 promoter or V1 and V3 regions of OsVP1 promoter. Furthermore, the EMSA showed that OsWRKY29-MBP recombinant proteins bound to the DNA probes in the A1 region of OsABF1 promoter or both the V1 and V3 regions of OsVP1 promoter. Moreover, unlabeled competing probes could effectively reduce the binding ability of OsWRKY29 in a dose-dependent manner (Figure 6H). Taken together, these results support the view that OsWRKY29 represses OsABF1 and OsVP1 expression by binding to the W-box region in the promoter of OsABF1 or OsVP1 in rice.
In this study, we identified a dormancy enhancement T-DNA insertion mutant Oswrky29 in rice (Figures 1A–E). The T-DNA insertion within the second intron of OsWRKY29 can reduced its expression level, which was responsible for the enhanced dormancy of Oswrky29 mutant (Figure 1). In addition, OsWRKY29 expression is negatively related to seed dormancy during seed development or seed germination (Figure 1F and Supplementary Figures S3, S5F) and its expression was inhibited by ABA in germinating seeds and early seedlings (Supplementary Figures S5F, S6). As a member of the WRKY transcription factor family, OsWRKY29, including a WRKY domain and a C2H2 type zinc-finger motif (Supplementary Figure S4B), is localized in the nucleus and acts as a transcriptional repressor (Figures 3A,C). ABA sensitivity assays indicated that OsWRKY29 likely represses seed dormancy through regulation of ABA signaling pathway (Figure 4 and Supplementary Figures S5, S7). Further assays showed that OsWRKY29 inhibits seed dormancy likely by repressing the expression of ABA positive response genes such as OsABF1 and OsVP1 by binding to their promoters (Figures 5, 6).
In summary, these results suggest that, in the absence of ABA, OsWRKY29 binds to the promoters of OsVP1 and OsABF1 (positive ABA response factors) to repress their expression, thus weakening ABA-mediated repression of seed germination. The presence of ABA represses OsWRKY29 expression, then releasing its repression on the expression of OsVP1 and OsABF1, thus permitting higher expression of OsVP1 and OsABF1 to enhance seed dormancy (Figure 7).
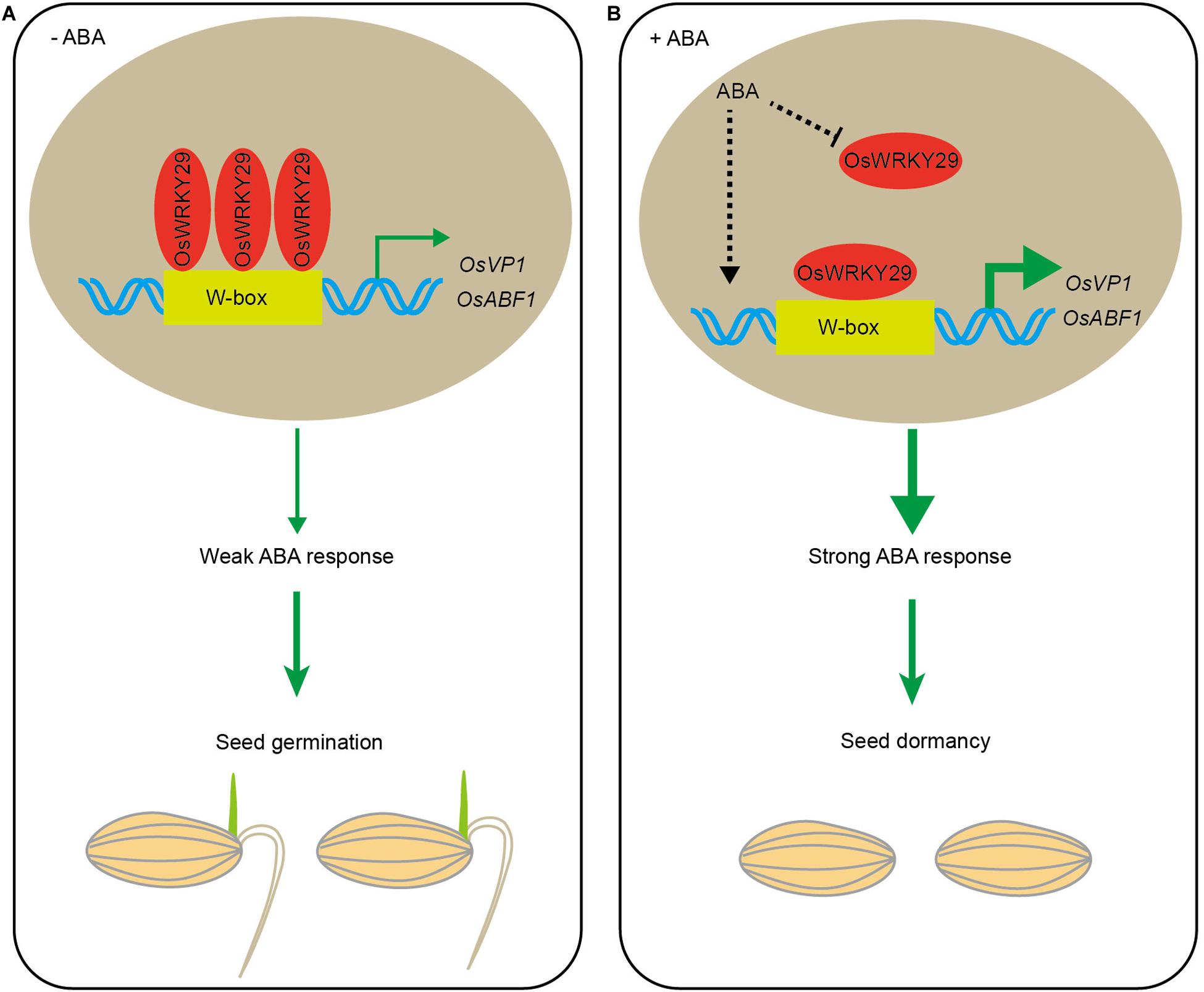
Figure 7. Hypothetical model for regulation of seed dormancy by OsWRKY29. (A) In the absence of ABA, OsWRKY29 binds to the OsVP1 and OsABF1 promoters to repress their expression, thus weakening the ABA response and permitting seed germination. (B) The presence of ABA represses OsWRKY29 expression to relieve its repression on the expression of OsVP1 and OsABF1, thus resulting in the high expression of OsVP1 and OsABF1 to strengthen ABA response and promote seed dormancy.
WRKY transcription factors are reported to be involved in regulation of stress response through ABA signaling. In Arabidopsis, abo3, a T−DNA insertion mutant of AtWRKY63, is hypersensitive to ABA, impairs ABA-induced stomatal closure, and has lower drought tolerance during both seedling establishment and growth (Ren et al., 2010). In rice, OsWRKY45 alleles play different roles in regulating ABA signaling and salt stress but have similar roles in response to drought and cold: OsWRKY45-1 negatively and OsWRKY45-2 positively regulate ABA signaling; however, OsWRKY45-2 but not OsWRKY45-1 negatively regulates the response of rice to salt stress (Tao et al., 2011). It was shown here that OsWRKY29 is also involved in ABA signaling by directly regulating expression of OsVP1 and OsABF1, suggesting that OsWRKY29 is probably involved in ABA-mediated regulation of stress response. Previous studies indicated that OsABF1 is a universally positive regulator of drought tolerance; abf1 mutants are more sensitive to salinity and drought treatments (Zhang et al., 2017). OsABF1 also regulates transition to floral development in rice in response to drought stress by directly activating gene OsWRKY104 (Zhang et al., 2016). As an upstream regulator of OsABF1, OsWRKY29 might also be involved in regulation of drought tolerance and flowering.
Some WRKY transcription factors function as promoters of ABA signaling by transcriptional activation and inhibition activities. For example, Atwrky2 knockout mutants reduced the expression of some ABA-responsive genes ABI3, ABI5, Em1, and Em6; conversely, ABA-induced AtWRKY2 accumulation requires ABI5, ABI3, ABA2 and ABA3 during seed germination, indicating a positive feedback loop between AtWRKY2 and ABI3 or ABI5 in regulating ABA response (Jiang and Yu, 2009). Induction of AtWRKY63 transcription by ABA was impaired in abi1, abi2, and abi5 lines and the abo3 mutation reduced the expression of ABF2, RD29A and COR47 (Ren et al., 2010). AtWRKY41 was reported to promote ABA signaling via direct regulation of ABI3 transcript levels (Ding et al., 2014). The transcript of AtWRKY6 was repressed during seed germination but induced by exogenous ABA; and vice versa, AtWRKY6 promoted ABA signaling by directly downregulating expression of RAV1 (Huang et al., 2016). OsWRKY72 and OsWRKY77 promote ABA-induced HVA22 expression in barley aleurone cells (Xie et al., 2005). Some WRKY proteins also act as repressors of ABA signaling. For example, AtWRKY18, AtWRKY40 and AtWRKY60 regulate ABA response by a de-repression mechanism by which the Mg-chelatase H subunit / putative ABA receptor (ABAR) recruits the WRKY proteins (which function as negative regulators of ABA signaling) from the nucleus to chloroplast and releases downstream ABA response genes, such as MYB2, ABI4, ABF4, ABI5, DREB1A, and RAB18 (Shang et al., 2010). OsWRKY24 and OsWRKY45 repress the induction of ABA to HVA22 in barley aleurone cells (Xie et al., 2005).
Differing from the positive feedback loop between AtWRKY2 and ABA signaling, OsWRKY29 was inhibited by ABA in germinating seeds and early seedlings (Supplementary Figures S5F, S6), suggesting a negative feedback loop between OsWRKY29 and the ABA signaling pathway. To verify if the molecular mechanism of OsWRKY29 was similar to those of AtWRKY18, AtWRKY40 and AtWRKY60, we investigated the subcellular localization of OsWRKY29 when treated with ABA and found that it was still localized in the nucleus (Supplementary Figure S10). Therefore, the repressive mechanism of OsWRKY29 on ABA signaling is probably different from that of AtWRKY18, AtWRKY40, and AtWRKY60.
Previous studies showed that WRKY proteins are key nodes in ABA-responsive networks: some WRKY proteins are negative regulators and others act as positive regulators of ABA response by promoting or inhibiting downstream ABA response genes. Other studies have shown that excessively strong or weak ABA responses can disrupt the balance between plant growth and stress response (Ding and De Smet, 2013; Skubacz et al., 2016; Yang et al., 2019). Taken together, these findings suggest that plants can recruit ABA response promoters such as OsWRKY72 and OsWRKY77 or repressors such as OsWRKY29 from an array of WRKY transcription factors to balance ABA signal responses for optimal growth and development.
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession numbers: OsWRKY29, Os07g0111400; OsABIL1, Os01g0583100; OsABIL2, Os05g0592800; OsABI5, Os01g0859300; OsVP1, Os01g0911700; TRAB1, Os08g0472000; OsABF1, Os01g0867300; OsABF2, Os06g0211200; OsbZIP23, Os02g0766700; OsbZIP72, Os09g0456200; OsNCED1, Os02g0704000; OsNCED2, Os12g04- 35200; OsNCED3, Os03g0645900; OsNCED4, Os07g0154100; OsNCED5, Os12g0617400; OsABA8ox1, Os02g0703600; OsAB- A8ox2, Os08g0472800; OsABA8ox3, Os09g0457100; OsLEA- 3-1, Os05g0542500; OsLEA3-2, Os03g0322900; OsLEA4, Os06g0110200; OsLEA5, Os05g0584200; OsEm1, Os05g0584200; OsWRKY16, Os01g0665500; OsWRKY49, Os05g0565900; OsW- RKY11, Os01g0626400; OsWRKY8, Os05g0583000; AtWRKY28, At4g18170; AtWRKY8, At5g46350; AtWRKY48, At5g49520; AtWRKY43, At2g46130; AtWRKY13, At4g39410; OsWRKY34, Os02g0652100; OsWRKY7, Os05g0537100; OsWRKY77, Os01- g0584900; AtWRKY2, At5g56270; OsWRKY24, Os01g0826400; AtWRKY63, At1g66600; OsWRKY45, Os05g0322900; AtWRK- Y41, At4g11070; AtWRKY6, At1g62300; AtWRKY40, At1g8- 0840; AtWRKY18, At4g31800; AtWRKY60, At2g25000; OsW- RKY72, Os11g0490900; AtWRKY45, At3g01970; UBQ5, and Os03g0234200.
All datasets generated for this study are included in the article/Supplementary Material.
JW supervised the project. CZ, LJ, and JW designed the research. CZ, JL, TZ, XL RM, CM, and TN performed the experiments. JW, XiaoZ, LZ, XingZ, and QW provided the technical assistance. CZ, QL, and LJ analyzed the data and wrote the manuscript. XinZ and XG generated the transgenic plants. SL cultivated the transgenic plants in the field. QL, LJ, and JW revised the manuscript. All authors have read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31871712 and 91735304), the National Key Research and Development Program of China (2016YFD0100901), and the Jiangsu Science and Technology Development Program (BE2018388).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Gynheung An (Department of Plant Systems Biotech, Kyung Hee University, Yongin, South Korea) for providing the Oswrky29 mutant seeds. We also acknowledge support from the Key Laboratory of Biology, Genetics and Breeding of Japonica Rice in the Mid-lower Yangtze River, Ministry of Agriculture, China and Jiangsu Collaborative Innovation Center for Modern Crop Production.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00691/full#supplementary-material
Chen, L. G., Zhang, L. P., Li, D. B., Wang, F., and Yu, D. Q. (2013). WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, E1963–E1971. doi: 10.1073/pnas.1221347110
Ding, Z., and De Smet, I. (2013). Localised ABA signalling mediates root growth plasticity. Trends Plant Sci. 18, 533–535. doi: 10.1016/j.tplants.2013.08.009
Ding, Z. J., Yan, J. Y., Li, G. X., Wu, Z. C., Zhang, S. Q., and Zheng, S. J. (2014). WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 79, 810–823. doi: 10.1111/tpj.12597
Duan, E. C., Wang, Y. H., Li, X. H., Lin, Q. B., Zhang, T., Wang, Y. P., et al. (2019). OsSHI1 regulates plant architecture through modulating the transcriptional activity of IPA1 in rice. Plant Cell 31, 1026–1042. doi: 10.1105/tpc.19.00023
Finkelstein, R., Reeves, W., Ariizumi, T., and Steber, C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. doi: 10.1146/annurev.arplant.59.032607.092740
Finkelstein, R. R., and Lynch, T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. doi: 10.1105/tpc.12.4.599
Fuchs, S., Tischer, S. V., Wunschel, C., Christmann, A., and Grill, E. (2014). Abscisic acid sensor RCAR7/PYL13, specific regulator of protein phosphatase coreceptors. Proc. Natl. Acad. Sci. U.S.A. 111, 5741–5746. doi: 10.1073/pnas.1322085111
Fujii, H., Verslues, P. E., and Zhu, J. K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19, 485–494. doi: 10.1105/tpc.106.048538
Gao, Y. H., Liu, J. K., Fan, J., Peng, Y., Huang, K., Zhang, D. F., et al. (2010). Construction and transformation of RNAi vector of OsVP1 for a regulatory gene of pre-harvest sprouting in Oryza sativa. Sci. Agric. Sinica 43, 1321–1327. doi: 10.4028/www.scientific.net/AMM.37-38.1549
Hauser, F., Waadt, R., and Schroeder, J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–R355. doi: 10.1016/j.cub.2011.03.015
Hobo, T., Kowyama, Y., and Hattori, T. (1999). A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. U.S.A. 96, 15348–15353. doi: 10.1073/pnas.96.26.15348
Hossain, M. A., Lee, Y., Cho, J. I., Ahn, C. H., Lee, S. K., Jeon, J. S., et al. (2010). The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 72, 557–566. doi: 10.1007/s11103-009-9592-9
Hu, W. M., Ma, H. S., Fan, L. J., and Ruan, S. L. (2003). Characteristics of preharvest sprouting in sterile lines in hybrid rice seeds production. Acta Agron. Sin. 29, 441–446. doi: 10.3321/j.issn:0496-3490.2003.03.022
Hu, Y., Chen, L., Wang, H., Zhang, L., Wang, F., and Yu, D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745. doi: 10.1111/tpj.12159
Huang, Y., Feng, C. Z., Ye, Q., Wu, W. H., and Chen, Y. F. (2016). Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development. PLoS Genet. 12:e1005833. doi: 10.1371/journal.pgen.1005833
Jeong, D. H., An, S., Kang, H. G., Moon, S., Han, J. J., Park, S., et al. (2002). T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 130, 1636–1644. doi: 10.1104/pp.014357
Jiang, W., and Yu, D. (2009). Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 9:96. doi: 10.1186/1471-2229-9-96
Joo, J., Lee, Y. H., and Song, S. I. (2014). Overexpression of the rice basic leucine zipper transcription factor OsbZIP12 confers drought tolerance to rice and makes seedlings hypersensitive to ABA. Plant Biotechnol. Rep. 8, 431–441. doi: 10.1007/s11816-014-0335-2
Kang, J. Y., Choi, H. I., Im, M. Y., and Kim, S. Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. doi: 10.1105/tpc.010362
Kim, S., Kang, J. Y., Cho, D. I., Park, J. H., and Kim, S. Y. (2004). ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40, 75–87. doi: 10.1111/j.1365-313X.2004.02192.x
Kobayashi, Y., Murata, M., Minami, H., Yamamoto, S., Kagaya, Y., Hobo, T., et al. (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44, 939–949. doi: 10.1111/j.1365-313X.2005.02583.x
Koornneef, M., Bentsink, L., and Hilhorst, H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5, 33–36. doi: 10.1016/S1369-5266(01)00219-9
Li, C., Shen, H., Wang, T., and Wang, X. (2015). ABA regulates subcellular redistribution of OsABI-LIKE2, a negative regulator in ABA signaling, to control root architecture and drought resistance in Oryza sativa. Plant Cell Physiol. 56, 2396–2408. doi: 10.1093/pcp/pcv154
Li, W., Wang, H. P., and Yu, D. Q. (2016). Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol. Plant 9, 1492–1503. doi: 10.1016/j.molp.2016.08.003
Li, W., Zhu, Z., Chern, M., Yin, J., Yang, C., Ran, L., et al. (2017). A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170:e115. doi: 10.1016/j.cell.2017.06.008
Lin, R., Ding, L., Casola, C., Ripoll, D. R., Feschotte, C., and Wang, H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318, 1302–1305. doi: 10.1126/science.1146281
Liu, X., Wang, J., Yu, Y., Kong, L., Liu, Y., Liu, Z., et al. (2019). Identification and characterization of the rice pre-harvest sprouting mutants involved in molybdenum cofactor biosynthesis. New Phytol. 222, 275–285. doi: 10.1111/nph.15607
Lopez-Molina, L., Mongrand, B., Mclachlin, D. T., Chait, B. T., and Chua, N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328. doi: 10.1046/j.1365-313X.2002.01430.x
Lu, B. Y., Xie, K., Yang, C. Y., Wang, S. F., Liu, X., Zhang, L., et al. (2011). Mapping two major effect grain dormancy QTL in rice. Mol. Breed. 28, 453–462. doi: 10.1007/s11032-010-9495-0
Lu, G., Gao, C. X., Zheng, X. N., and Han, B. (2009). Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229, 605–615. doi: 10.1007/s00425-008-0857-3
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Miyoshi, K., Kagaya, Y., Ogawa, Y., Nagato, Y., and Hattori, T. (2002). Temporal and spatial expression pattern of the OSVP1 and OSEM genes during seed development in rice. Plant Cell Physiol. 43, 307–313. doi: 10.1093/pcp/pcf040
Nambara, E., Okamoto, M., Tatematsu, K., Yano, R., Seo, M., and Kamiya, Y. (2010). Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 20, 55–67. doi: 10.1017/S0960258510000012
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Penfield, S. (2017). Seed dormancy and germination. Curr. Biol. 27, R874–R878. doi: 10.1016/j.cub.2017.05.050
Ren, X., Chen, Z., Liu, Y., Zhang, H., Zhang, M., Liu, Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. doi: 10.1111/j.1365-313X.2010.04248.x
Robichaud, C., Wong, J., and Sussex, I. (1979). Control of in vitro growth of viviparous embryo mutants of maize by abscisic acid. Dev. Genet. 1, 325–330. doi: 10.1002/dvg.1020010405
Ross, C. A., Liu, Y., and Shen, Q. X. J. (2007). The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 49, 827–842. doi: 10.1111/j.1744-7909.2007.00504.x
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Sharma, P. D., Singh, N., Ahuja, P. S., and Reddy, T. V. (2011). Abscisic acid response element binding factor 1 is required for establishment of Arabidopsis seedlings during winter. Mol. Biol. Rep. 38, 5147–5159. doi: 10.1007/s11033-010-0664-3
Shu, K., Liu, X. D., Xie, Q., and He, Z. H. (2016). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9, 34–45. doi: 10.1016/j.molp.2015.08.010
Skubacz, A., Daszkowska-Golec, A., and Szarejko, I. (2016). The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7:1884. doi: 10.3389/fpls.2016.01884
Tang, N., Zhang, H., Li, X., Xiao, J., and Xiong, L. (2012). Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158, 1755–1768. doi: 10.1104/pp.111.190389
Tao, Z., Kou, Y., Liu, H., Li, X., Xiao, J., and Wang, S. (2011). OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot. 62, 4863–4874. doi: 10.1093/jxb/err144
Wan, J. M., Cao, Y. J., Wang, C. M., and Ikehashi, H. (2005). Quantitative trait loci associated with seed dormancy in rice. Crop Sci. 45, 712–716. doi: 10.2135/cropsci2005.0712
Wang, H., Avci, U., Nakashima, J., Hahn, M. G., Chen, F., and Dixon, R. A. (2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. U.S.A. 107, 22338–22343. doi: 10.1073/pnas.1016436107
Xiang, Y., Tang, N., Du, H., Ye, H. Y., and Xiong, L. Z. (2008). Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 148, 1938–1952. doi: 10.1104/pp.108.128199
Xie, Z., Zhang, Z. L., Zou, X., Huang, J., Ruas, P., Thompson, D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. doi: 10.1104/pp.104.054312
Yang, G. D., Yu, Z. P., Gao, L., and Zheng, C. C. (2019). SnRK2s at the crossroads of growth and stress responses. Trends Plant Sci. 24, 672–676. doi: 10.1016/j.tplants.2019.05.010
Yang, X., Yang, Y. N., Xue, L. J., Zou, M. J., Liu, J. Y., Chen, F., et al. (2011). Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol. 156, 1397–1409. doi: 10.1104/pp.111.173427
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Yu, F. F., Wu, Y. R., and Xie, Q. (2015). Precise protein post-translational modifications modulate ABI5 activity. Trends Plant Sci. 20, 569–575. doi: 10.1016/j.tplants.2015.05.004
Zeng, Y., Raimondi, N., and Kermode, A. R. (2003). Role of an ABI3 homologue in dormancy maintenance of yellow-cedar seeds and in the activation of storage protein and Em gene promoters. Plant Mol. Biol. 51, 39–49. doi: 10.1023/a:1020762304937
Zhang, C., Li, C., Liu, J., Lv, Y., Yu, C., Li, H., et al. (2017). The OsABF1 transcription factor improves drought tolerance by activating the transcription of COR413-TM1 in rice. J. Exp. Bot. 68, 4695–4707. doi: 10.1093/jxb/erx260
Zhang, C., Liu, J., Zhao, T., Gomez, A., Li, C., Yu, C., et al. (2016). A drought-inducible transcription factor delays reproductive timing in rice. Plant Physiol. 171, 334–343. doi: 10.1104/pp.16.01691
Zhang, Y., Su, J., Duan, S., Ao, Y., Dai, J., Liu, J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7:30. doi: 10.1186/1746-4811-7-30
Zhu, G. H., Ye, N. H., and Zhang, J. H. (2009). Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol. 50, 644–651. doi: 10.1093/pcp/pcp022
Keywords: ABA signaling, OsABF1, Oryza sativa, seed dormancy, OsVP1
Citation: Zhou C, Lin Q, Lan J, Zhang T, Liu X, Miao R, Mou C, Nguyen T, Wang J, Zhang X, Zhou L, Zhu X, Wang Q, Zhang X, Guo X, Liu S, Jiang L and Wan J (2020) WRKY Transcription Factor OsWRKY29 Represses Seed Dormancy in Rice by Weakening Abscisic Acid Response. Front. Plant Sci. 11:691. doi: 10.3389/fpls.2020.00691
Received: 03 December 2019; Accepted: 01 May 2020;
Published: 27 May 2020.
Edited by:
Rujin Chen, Lanzhou University, ChinaReviewed by:
Jong-Seong Jeon, Kyung Hee University, South KoreaCopyright © 2020 Zhou, Lin, Lan, Zhang, Liu, Miao, Mou, Nguyen, Wang, Zhang, Zhou, Zhu, Wang, Zhang, Guo, Liu, Jiang and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Jiang, amlhbmdsaW5nQG5qYXUuZWR1LmNu; Jianmin Wan, d2Fuam1AbmphdS5lZHUuY24=; d2Fuamlhbm1pbkBjYWFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.