
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 03 June 2020
Sec. Plant Membrane Traffic and Transport
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00610
This article is part of the Research Topic Metal Transport in Plants View all 11 articles
 Sho Nishida1*
Sho Nishida1* Ryoji Tanikawa2
Ryoji Tanikawa2 Shota Ishida2
Shota Ishida2 Junko Yoshida3
Junko Yoshida3 Takafumi Mizuno3
Takafumi Mizuno3 Hiromi Nakanishi4
Hiromi Nakanishi4 Naoki Furuta2*
Naoki Furuta2*A number of metal hyperaccumulator plants, including nickel (Ni) hyperaccumulators, have been identified in the genus Noccaea. The ability to accumulate Ni in shoots varies widely among species and ecotypes in this genus; however, little is known about the molecular mechanisms underlying this intra- and inter-specific variation. Here, in hydroponic culture, we compared Ni accumulation patterns between Noccaea japonica, which originated in Ni-enriched serpentine soils in Mt. Yubari (Hokkaido, Japan), and Noccaea caerulescens ecotype Ganges, which originated in zinc/lead-mine soils in Southern France. Both Noccaea species showed extremely high Ni tolerance compared with that of the non-accumulator Arabidopsis thaliana. But, following treatment with 200 μM Ni, N. caerulescens showed leaf chlorosis, whereas N. japonica did not show any stress symptoms. Shoot Ni concentration was higher in N. caerulescens than in N. japonica; this difference was due to higher efficiency of root-to-shoot Ni translocation in N. caerulescens than N. japonica. It is known that the vacuole Ni transporter IREG2 suppresses Ni translocation from roots to shoots by sequestering Ni in the root vacuoles. The expression level of the IREG2 gene in the roots of N. japonica was 10-fold that in the roots of N. caerulescens. Moreover, the copy number of IREG2 per genome was higher in N. japonica than in N. caerulescens, suggesting that IREG2 expression is elevated by gene multiplication in N. japonica. The heterologous expression of IREG2 of N. japonica and N. caerulescens in yeast and A. thaliana confirmed that both IREG2 genes encode functional vacuole Ni transporters. Taking these results together, we hypothesize that the elevation of IREG2 expression by gene multiplication causes the lower root-to-shoot Ni translocation in N. japonica.
In plants, heavy metals absorbed from the soils by the roots accumulate in roots and shoots. Some heavy metals, such as zinc (Zn), manganese (Mn), and copper (Cu) are essential nutrients for plants, but excess accumulation of these metals can be toxic. Thus, plants found in natural environments contain heavy metals within certain ranges of concentrations, which can be defined as the “normal” ranges (Marschner, 1995). Some plant species, however, accumulate heavy metals in their aerial parts at unusually high levels without any stress symptoms. These species are called “metal hyperaccumulators” (Brooks et al., 1977). The threshold that defines metal hyperaccumulation depends on the element; for example, Zn/Mn hyperaccumulators contain >10,000 mg kg–1, and nickel (Ni)/Cu hyperaccumulators contain >1,000 mg kg–1 on a dry weight basis (Reeves et al., 2018). More than 700 species have been identified as metal hyperaccumulators, and most of these have been discovered in metal-contaminated soils (Reeves et al., 2018). Metal hyperaccumulators have been studied both as rare species with unique molecular mechanisms for abnormal metal accumulation, and as model organisms for adaptation to extreme environments (Hanikenne and Nouet, 2011). Metal hyperaccumulators have also received attention from researchers developing technologies for decontaminating metal-polluted soils using plants, so-called “phytoremediation,” or for harvesting rare metals from soils, so-called “phytomining” (Salt et al., 1998; Chaney et al., 2007).
Metal hyperaccumulators have several characteristic traits: enhanced metal absorption by roots, efficient metal translocation from roots to shoots, and elevated metal tolerance (Lasat et al., 1996; Kramer et al., 1997; Zhao et al., 2002; Caille et al., 2005). In recent years, the molecular mechanisms involved in those traits have been uncovered by molecular biological approaches. Zn/cadmium (Cd) transporter Heavy Metal ATPase 4 (HMA4), which plays a role in Zn/Cd loading into the xylem in roots, was shown to be involved in efficient Zn/Cd translocation from roots to shoots in the Zn/Cd hyperaccumulator Arabidopsis halleri using HMA4-knockdown plants (Hanikenne et al., 2008). HMA3 is thought to play a role in sequestering excess Cd in leaf vacuoles and contribute to high Cd tolerance and accumulation in shoots of the Zn/Cd/Ni hyperaccumulator Noccaea caerulescens (formerly Thlaspi caerulescens) ecotype Ganges (Ueno et al., 2011). Natural resistance-associated macrophage protein 1 (NRAMP1) is thought to mediate Cd influx across the endodermal plasma membrane in root vascular tissues and contribute to efficient root-to-shoot Cd translocation in N. caerulescens ecotype Ganges (Milner et al., 2014). The expression levels of HMA4, HMA3 and NRAMP1 are much higher in the metal hyperaccumulators than in the non-accumulators, and the elevated expression levels are required for metal hyperaccumulating abilities (Hammond et al., 2006; Hanikenne et al., 2008; Ueno et al., 2011; Milner et al., 2014). Hanikenne et al. (2008) attributed the elevated expression of HMA4 in A. halleri to the triplication of HMA4 in the genome and the mutation of cis-regulatory sequences. Elevated expression of HMA4 caused by gene multiplication is also found in the Zn/Cd hyperaccumulating ecotypes of N. caerulescens (Lochlainn et al., 2011). Elevated HMA3 and NRAMP1 expression levels in N. caerulescens ecotype Ganges are also likely caused by gene multiplication (Ueno et al., 2011; Milner et al., 2014), indicating that the elevated expression of metal transporter genes caused by gene multiplication are key evolutionary events for metal hyperaccumulators. High expression of many other genes involved in metal transport and metal homeostasis is also thought to be implicated in metal hyperaccumulation (Hanikenne and Nouet, 2011).
Most metal hyperaccumulators (more than 500 species) are Ni hyperaccumulators (Reeves et al., 2018). Ni hyperaccumulators are found on serpentine soils, which contain Ni at a level excessive for normal plants. In the Ni hyperaccumulating ecotype of N. caerulescens, nicotianamine in root cells and xylem forms a complex with Ni and is involved in the long-distance translocation of Ni from roots to shoots (Vacchina et al., 2003; Mari et al., 2006). Expression of Yellow-stripe like 3 (YSL3), encoding the Ni–nicotianamine complex transporter, is higher in the roots and shoots of N. caerulescens than in those of the non-accumulator Arabidopsis thaliana (Gendre et al., 2007), suggesting that elevated YSL3 expression increases Ni–nicotianamine mobility and contributes to efficient root-to-shoot Ni translocation. In another Noccaea Ni hyperaccumulator, N. goesingense, elevated serine acetyltransferase activity and elevated glutathione concentration are essential for enhanced Ni tolerance (Freeman et al., 2004, 2005; Na and Salt, 2011).
Assunção et al. (2003) showed that accumulation patterns of Ni, Zn, and Cd vary among N. caerulescens ecotypes originating from different soil types, and that an ecotype growing in serpentine soils is superior in Ni tolerance and accumulation to ecotypes from other metalliferous or non-metalliferous soils. Peer et al. (2003, 2006) showed that there is a large variation in Ni accumulation patterns among the species or accessions of Brassicaceae metal hyperaccumulators including a number of Noccaea species. In their results, shoot Ni concentration was not always correlated with the shoot concentrations of other metals, suggesting that there is a genetic factor specifically determining Ni hyperaccumulation.
The molecular mechanism of Ni accumulation has been well studied in A. thaliana. Ni is absorbed by roots via iron (Fe) uptake transporter Iron-regulated transporter 1 (IRT1) and unknown transporters (Nishida et al., 2011, 2015), and then translocated to shoots via the xylem. Excessively absorbed Ni is sequestered in root vacuoles by Ni transporter Iron-regulated 2 (IREG2) (also known as Ferroportin 2; Schaaf et al., 2006; Morrissey et al., 2009). Excess Ni induces Fe deficiency due to competition between Ni and Fe (Nishida et al., 2012). Then, IRT1 expression is induced by the Ni-induced Fe deficiency, resulting in accelerated Ni absorption via IRT1 (Nishida et al., 2012). IREG2 is also induced by Ni-induced Fe deficiency and detoxifies excess Ni (Schaaf et al., 2006). This synchronized induction of IREG2 and IRT1, which is regulated by FER-like Fe deficiency-induced transcription factor (FIT), is essential for the limited Ni tolerance shown by A. thaliana (Schaaf et al., 2006). Knockdown of IREG2 decreases root Ni and increases shoot Ni, indicating that IREG2 can determine the efficiency of Ni translocation from roots to shoots (Schaaf et al., 2006; Merlot et al., 2014). IREG1, the ortholog of IREG2, plays a role in Fe/cobalt (Co) translocation from roots to shoots by loading Fe/Co into the xylem in roots, but the contribution of IREG1 to Ni translocation was not confirmed (Morrissey et al., 2009). Merlot et al. (2014) reported that the expression level of a IREG homolog is higher in the Ni hyperaccumulator Psychotria gabriellae than in the related non-accumulator Psychotria semperflorens and the overexpression of P. gabriellae IREG increases Ni tolerance in yeast and A. thaliana, suggesting that IREG contributes to high Ni tolerance in P. gabriellae.
The objective of the present study was to reveal a genetic factor associated with variation in shoot Ni accumulation in Ni hyperaccumulators. We assessed Ni accumulation patterns in roots and shoots in two related Ni hyperaccumulators: Noccaea japonica, which originated from a serpentine soil, and N. caerulescens ecotype Ganges, which originated from a Zn/Pb-mine soil. We also investigated the involvement of IREG2 in the differential Ni accumulation patterns between the two Noccaea plant types.
Noccaea japonica growing in a serpentine soil in Mt. Yubari (Hokkaido, Japan), N. caerulescens ecotype Ganges growing in a Zn/Pb-mine soil in Southern France, and A. thaliana accession Col-0 were used. The ireg2-1 T-DNA insertion line (SALK_74442; Schaaf et al., 2006) was obtained from the Arabidopsis Biological Resource Center (Ohio State University, United States). An ireg2-1 homozygous line was identified using polymerase chain reaction (PCR) with primers flanking the insertion site and the left-border primer of T-DNA (Supplementary Table S1).
Half-strength Molecular Genetics Research Laboratory (MGRL) medium containing 8.6 μM Fe-EDTA at pH 5.5 was used as the hydroponic culture solution (Nishida et al., 2011). The procedure used for hydroponic culture was described previously (Nishida et al., 2011). Since N. japonica and N. caerulescens grow slower than A. thaliana, the preculture periods prior to metal exposure were 6 weeks for the Noccaea species and 4 weeks for A. thaliana. Two plants were cultured in every pot and pooled as one biological replicate. Plants were grown in a growth chamber at 22°C under a 10-h light/14-h dark cycle.
In agar-plate tests, half-strength MGRL medium containing 1.2% agar (agar type A, Sigma-Aldrich Co), 1% sucrose, 8.6 μM Fe-EDTA, and 5 mM 2-morpholinoethanesulfonic acid (MES) at pH 5.5 was used. Seeds were surface sterilized and sown on agar plates. Fifteen seedlings were pooled as one biological replicate. Plants were cultured in a growth chamber at 22°C under a 16-h light/8-h dark cycle.
Plants were exposed to hydroponic culture solutions supplemented with 0–200 NiCl2 or 0.5–1000 μM ZnCl2 for a week. Hydroponic solutions were buffered with 5 mM MES (pH 5.5). After exposure, plants were separated into roots and shoots. Roots were washed with deionized water and 50 mM EDTA-Na2 to remove metals bound to the cell walls, and then blotted. Harvested samples were weighed to determine the fresh weight, dried at 70°C, and weighted to determine the dry weight. Dried samples were digested with HNO3 and H2O2 in a heat block and diluted with ultra-pure water. Elemental analysis was performed using inductively coupled plasma optical emission spectrometers (ICPS-7500, Shimadzu Co.; SPS5100, Hitachi High-Tech Science Co).
Noccaea japonica and N. caerulescens plants were cultured hydroponically as described above. Total RNA was extracted from their roots following the method of Suzuki et al. (2004). Residual DNA was digested by DNase I (Takara Bio Inc), and first-strand cDNA was synthesized using a PrimeScript RT-PCR kit (Takara Bio Inc) with oligo (dT) primers. The resulting cDNA was used for gene cloning as follows. The mRNA sequence of NcIREG2, which was previously obtained by the Transcriptome Shotgun Assembly project for N. caerulescens, is available at DDBJ/EMBL/GenBank as “Nc_isotig06993_isogroup03561” (Lin et al., 2014; Supplementary Figure S3). Full-length IREG2 fragments from N. japonica and N. caerulescens were amplified using forward and reverse primers designed on regions corresponding to the 5’ UTR and 3’ UTR of NcIREG2, respectively, and a high-fidelity DNA polymerase (PrimeSTAR HS DNA Polymerase, Takara Bio Inc). PCR products were sequenced using primers designed on the internal regions of IREG2 (Supplementary Table S1). Since single nucleotide polymorphisms were detected in NjIREG2, NjIREG2 fragments were cloned separately in an Escherichia coli plasmid vector and sequenced as described later. IREG2 sequences were aligned by using Clustal W1. IREG2 sequences are deposited in the DNA Data Bank of Japan under the accession numbers LC522035 (NcIREG2) and LC522036 (NjIREG2).
Plants pre-cultured in hydroponic solution were exposed to 200 μM NiCl2 for a week. Total RNA was then extracted from the roots and treated with DNase I. cDNA was synthesized using a ReverTra Ace qPCR RT kit (Toyobo Co., Ltd), and quantitative RT-PCR (qRT-PCR) was performed using THUNDERBIRD SYBR qPCR Mix (Toyobo Co., Ltd). qPCR was performed in a real-time PCR system (LightCycler 96 System, Roche Diagnostics KK). The relative transcript level of IREG2 was calculated using a calibration curve and normalized with the EF1α transcript level. The IREG2-specific primers were designed on sequences that are completely identical between NcIREG2 and NjIREG2. To design EF1α-specific primers, a portion of the EF1α sequence was amplified from genomic DNA of N. japonica and N. caerulescens by PCR with EF1α common primers, which were designed on a region conserved in higher plants. EF1α fragments were sequenced, and a forward primer and a reverse primer, spanning an intron, were designed on the sequences. The specificities of qPCR primers were confirmed by dissociation curve analysis and gel electrophoresis of PCR products. Primer information is summarized in Supplementary Table S1.
Genomic DNA was extracted from young leaves of plants grown in a soil culture, and then IREG2 copy number was estimated by qPCR as described previously (Ueno et al., 2011; Milner et al., 2014). The qPCR protocol was as described in the section “Quantitative Reverse Transcription (RT)-PCR Assay,” except that the genomic DNA was used as the template instead of cDNA. Relative abundance of the IREG2 gene was normalized with that of the SHR gene (AT4G37650), which is a putative single copy gene, or the expressed sequence tag–based indel marker RR11nr025, of N. caerulescens (Ueno et al., 2011; Milner et al., 2014). Primer sequences are summarized in Supplementary Table S1.
The open-reading frames of NcIREG2 and NjIREG2 were amplified from cDNA by PCR with PrimeSTAR HS DNA Polymerase using a 5′ primer with Kpn1 site and 3′ primer with Xho1 site (Supplementary Table S1). PCR products were cloned into the pMD20-T vector by TA cloning (Takara Bio. Inc). The resulting IREG2-pMD-T plasmids were extracted from three independent colonies of E. coli and then sequenced. IREG2 fragments were subcloned into pENTR4 (Invitrogen) at the Kpn1–Xho1 sites, and subsequently subcloned into pGWB2 plant overexpression vector carrying the CaMV 35S promoter (Nakagawa et al., 2007) using Gateway LR Clonase II Enzyme mix (Invitrogen). The plasmid constructs were transferred into Agrobacterium tumefaciens strain GV3101 (pMP90), and the obtained transformants were used to transform the ireg2-1 homozygous line using the standard floral dip method. T1 transformants were selected on agar plates containing 10 μg mL–1 hygromycin, and then transferred to a soil culture. T2 progenies of four independent T1 lines were used for the following experiments. Fifteen seedlings of each T2 line grown on agar plates for 2 weeks (without hygromycin) were pooled and subjected to qRT-PCR. The relative expression level of IREG2 was calculated using EF1α as the internal reference gene.
Seeds were surface-sterilized and sown on agar plates supplemented with 0–70 μM NiCl2 (without hygromycin) and cultured for 14 days. After exposure, seedlings were pooled for each line and growth and Ni concentrations in roots and shoots were determined as described above.
The IREG2-pMD-T plasmids were digested with KpnI, and the plasmid ends were blunted by T4 DNA polymerase. The resulting linear plasmids were digested with XhoI, and IREG2 fragments were isolated by gel electrophoresis. The pKT10-Gal-HA-BS yeast expression vector was digested with EcoRI, and the plasmid ends were blunted by a Klenow fragment; the plasmid was then digested by SalI. IREG2 fragments were ligated to the blunt end and SalI-digested end of pKT10-Gal-HA-BS. The obtained plasmids were transformed into Saccharomyces cerevisiae strain BY4741 (MATa his3 leu2 met15 ura3) by the standard method. The transformants were selected on a yeast nitrogen base (YNB) uracil minus medium, which consisted of 0.67% YNB, 2% glucose, and appropriate amino acids. The assay for Ni tolerance was performed as described by Nishida et al. (2011) using YNB uracil minus media containing 0 or 750 μM NiCl2, and 2% galactose (pH 5.5).
Plants cultured hydroponically were exposed to 0 or 200 NiCl2 for a week. After exposure, roots were harvested and subjected to nicotianamine extraction following Nozoye et al. (2014). Nicotianamine was measured using high-performance liquid chromatography (HPLC) as described previously (Mori and Nishizawa, 1987).
Noccaea japonica, N. caerulescens (ecotype Ganges), and A. thaliana were exposed to 0, 25, or 200 μM Ni for a week in hydroponic culture, and Ni accumulation and symptoms were compared; A. thaliana was not tested at 200 μM Ni because this high concentration was lethal for this species. Following 25 μM Ni treatment, A. thaliana showed severe chlorosis, a typical symptom of Ni-induced Fe deficiency, over the whole shoot, and the growth in roots and shoots was significantly reduced compared with the un-treated controls (Figures 1A,B). In contrast, both N. japonica and N. caerulescens showed no visible symptoms or growth reduction, confirming that these Noccaea plant types have extremely high tolerance to Ni. Following 200 μM Ni treatment, N. caerulescens but not N. japonica exhibited chlorosis between the veins of young leaves; neither plant types showed significant growth reduction (Figures 1A,B and Supplementary Figure S1). Ni-induced Fe deficiency induces IRT1 expression in roots and accelerates Fe absorption. This means that Fe concentration in roots increases with Ni concentration in the medium (Nishida et al., 2011, 2012). Here, Fe concentration in roots was significantly increased by treatment with 25 μM Ni in N. caerulescens (Supplementary Figure S1B) but not N. japonica. Taken together, the data for chlorosis symptoms and root Fe concentration indicate that N. caerulescens shows higher Ni sensitivity than N. japonica.
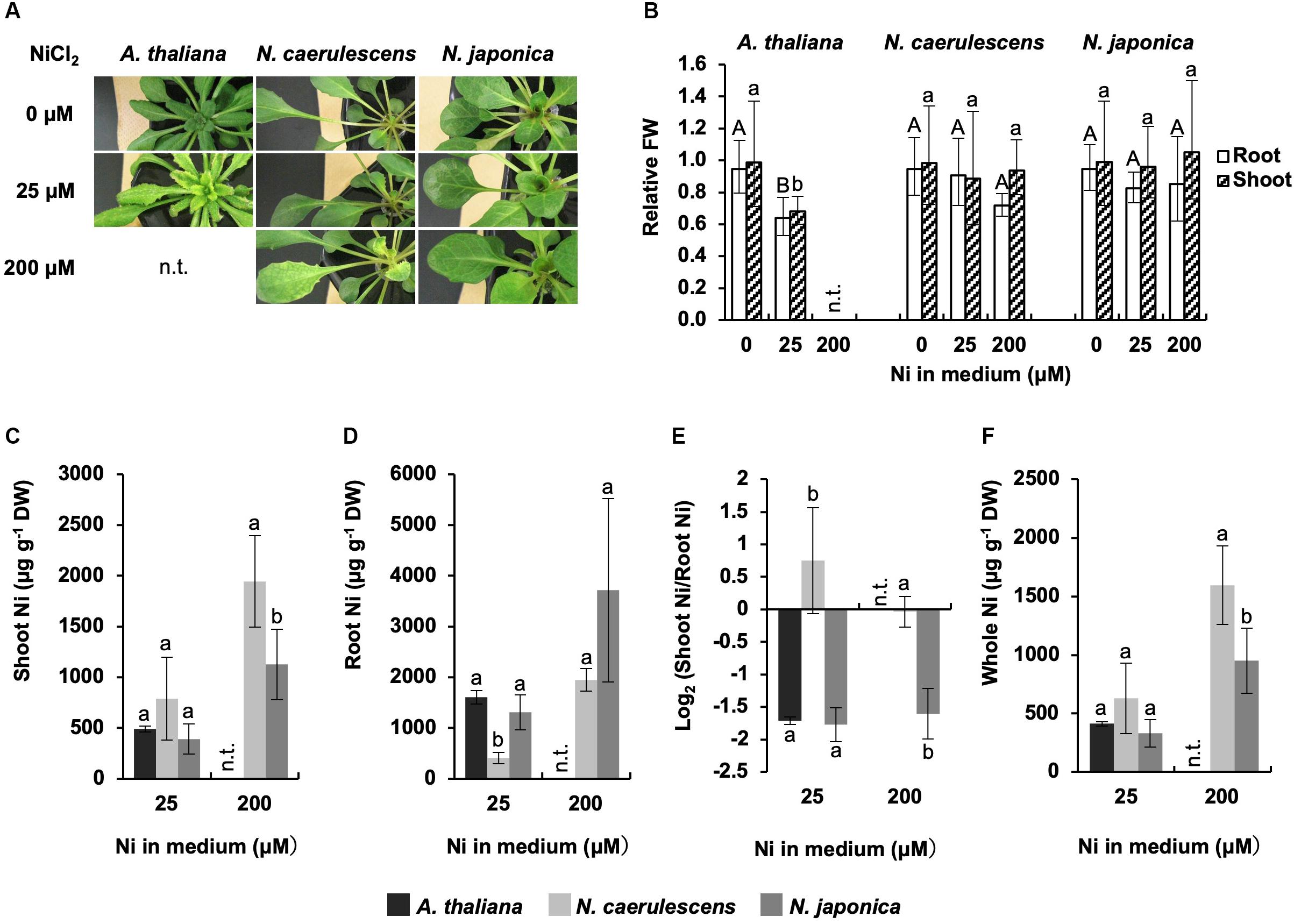
Figure 1. Ni tolerance and accumulation. Plants were exposed to the indicated concentrations of NiCl2 for 7 days in hydroponic culture. (A) Plants after Ni treatment. (B) Fresh weight (FW) relative to un-treated control. (C,D) Ni concentration expressed relative to dry weight (DW) in shoots (C) and roots (D). (E) Ratio of shoot Ni to root Ni concentration. (F) Ni concentration in whole plants. Values are geometric (B,E) or arithmetic (C,D,F) means ± SD of four biological replicates. Different letters indicate a statistically significant difference (P < 0.05; ANOVA, Tukey–Kramer) between different Ni treatments in each part (B) or different species in each treatment (C–F). n.t., not tested. Mean values of shoot FW/root FW under 0 μM Ni: 2.64 g/1.14 g (A. thaliana); 2. 02 g/1.01 g (N. caerulescens); 1.87 g/0.78 g (N. japonica).
Ni concentration in shoots was significantly higher in N. caerulescens than N. japonica following treatment with 200 μM Ni; the same tendency was observed for 25 μM Ni but the difference was not significant (Figure 1C). In contrast, Ni concentration in roots was significantly lower in N. caerulescens than N. japonica following treatment with either 25 or 200 μM Ni (Figure 1D). When the Ni levels in shoots and roots were compared, shoot Ni concentration was similar to or higher than root Ni concentration in N. caerulescens individuals, whereas shoot Ni concentration was lower than root Ni concentration in all N. japonica individuals tested. The ratio of shoot Ni to root Ni in N. caerulescens was significantly higher than that in N. japonica (Figure 1E). These results indicate that the activity of Ni translocation from roots to shoots was higher in N. caerulescens than in N. japonica. Following treatment with 200 μM Ni, the Ni concentration in whole plants was significantly higher in N. caerulescens than in N. japonica (Figure 1F) indicating that Ni absorption activity was higher in N. caerulescens.
We also investigated Zn accumulation and symptoms in N. japonica, N. caerulescens, and A. thaliana following treatment with a normal Zn concentration(0.5 μM Zn) or excess Zn (150 or 1000 μM Zn). Growth ofA. thaliana was greatly inhibited by 150 μMZn treatment compared with 0.5 μM Zn treatment, and serious necrosis developed in leaves following 1000 μM Zn treatment (Figure 2A). In contrast, both N. japonica and N. caerulescens showed high Zn tolerance, though there was a tendency for a slight decrease in growth under excess Zn conditions (Figures 2A,B and Supplementary Figure S2); this decrease was significant for 1000 μM Zn in N. caerulescens only. Shoot Zn concentrations in the Noccaea species were over twice that in A. thaliana under the normal Zn condition, while root Zn concentrations were lower in the Noccaea species than in A. thaliana (Figure 2C). These differences were statistically significant, confirming higher activity of root-to-shoot Zn translocation in the Noccaea species. In the comparison between Noccaea species, Zn concentrations in shoots and whole plants were significantly higher in N. japonica than in N. caerulescens under the normal Zn condition (Figure 2C), but no significant difference in Zn accumulation was observed under excess Zn conditions (Figures 2D,E,G). There was no difference in the ratio of shoot Zn to root Zn between N. japonica and N. caerulescens under excess Zn conditions (Figure 2F).
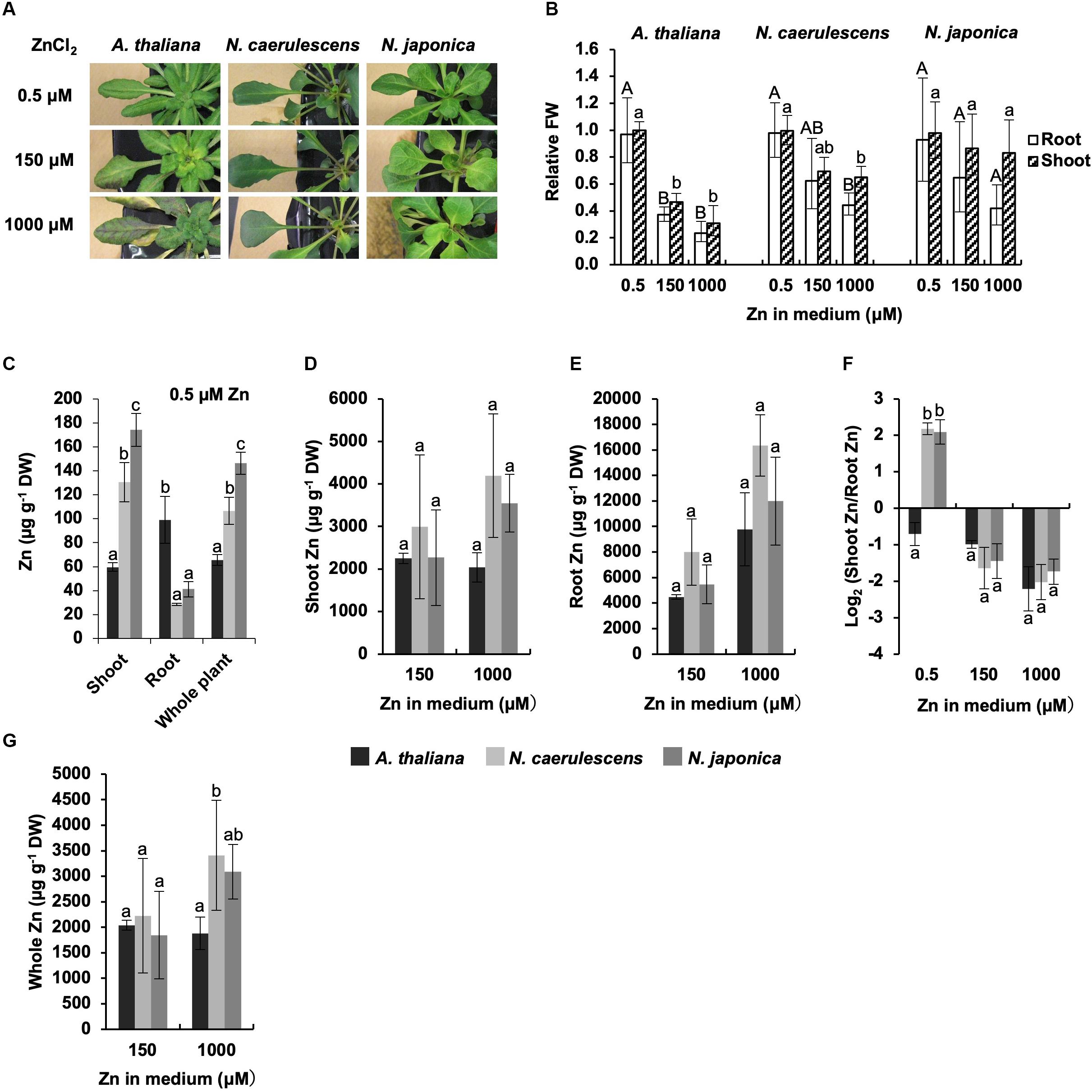
Figure 2. Zn tolerance and accumulation. Plants were exposed to the indicated concentrations of ZnCl2 for 7 days in hydroponic culture. (A) Plants after Zn treatments. (B) Fresh weight (FW) relative to un-treated control. (C) Zn concentration expressed relative to dry weight (DW) in shoots, roots, and whole plants under normal Zn condition (0.5 μM ZnCl2). (D,E) Zn concentrations in shoots (D) and root (E) under excess Zn conditions (150 and 1000 μM ZnCl2). (F) Ratio of shoot Zn to root Zn. (G) Zn concentration in whole plants. Values are geometric (B,F) or arithmetic (C–E,G) means ± SD of four biological replicates. Different letters indicate a statistically significant difference (P < 0.05; ANOVA, Tukey–Kramer) between different Zn treatments in each part (B), different species in each part (C), or different species in each treatment (D–G). n.t., not tested. Mean values of shoot FW/root FW under 0.5 μM Zn: 3.04 g/1.08 g (A. thaliana); 2. 72 g/1.67 g (N. caerulescens); 1.36 g/0.67 g (N. japonica).
The observed significant difference between N. japonica and N. caerulescens in terms of Ni, but not Zn, translocation from roots to shoots, suggests that the inter-species difference in Ni translocation is due to a Ni-specific mechanism. The efficiency of Ni translocation from roots to shoots depends on the abundances of vacuolar Ni2+ transporter IREG2 and metal chelator nicotianamine in roots. Nicotianamine accumulation in roots affects translocation of both Ni and Zn (Mari et al., 2006; Uraguchi et al., 2019), whereas IREG2 transports Ni2+ but not Zn2+. Therefore, we considered IREG2 to be the more likely candidate for the Ni-specific mechanism. We cloned the cDNAs of IREG2 from the roots of N. japonica and N. caerulescens (termed NjIREG2 and NcIREG2, respectively), and determined their sequences (Supplementary Figure S3). From N. japonica, three different IREG2 sequences were obtained (Supplementary Figure S4). These sequences were almost identical (>99.5% identity), and therefore one of them was used for further analysis. A single IREG2 sequence was obtained from N. caerulescens. The polypeptides encoded by NjIREG2 and NcIREG2 were almost identical to each other (99.4% identity) and consisted of 501 aa residues (Supplementary Figure S5). NjIREG2 and NcIREG2 shared considerable sequence identity (79%) with AtIREG2 (512 aa), suggesting that the functional role of IREG2 is conserved between the two Noccaea plant types.
We compared the expression level of IREG2 in roots between N. caerulescens and N. japonica using qRT-PCR. The expression level in N. japonica was over 10-fold that in N. caerulescens following treatment with 200 μM Ni (Figure 3A). This suggests that the IREG2 activity that sequesters Ni in root vacuoles is higher and Ni translocation to shoots is lower in N. japonica than in N. caerulescens. We also compared the genomic copy number of IREG2 between the two Noccaea plant types. The abundances of IREG2 gene relative to the abundance of the putative single copy gene SHR or the indel marker site RR11nr025 were compared between the two plant types. The copy number of IREG2 per genome in N. japonica was estimated to be 6–8 times that in N. caerulescens (Figure 3B). This suggests that the higher expression of IREG2 in N. japonica than in N. caerulescens is due to a higher copy number of IREG2 in N. japonica.
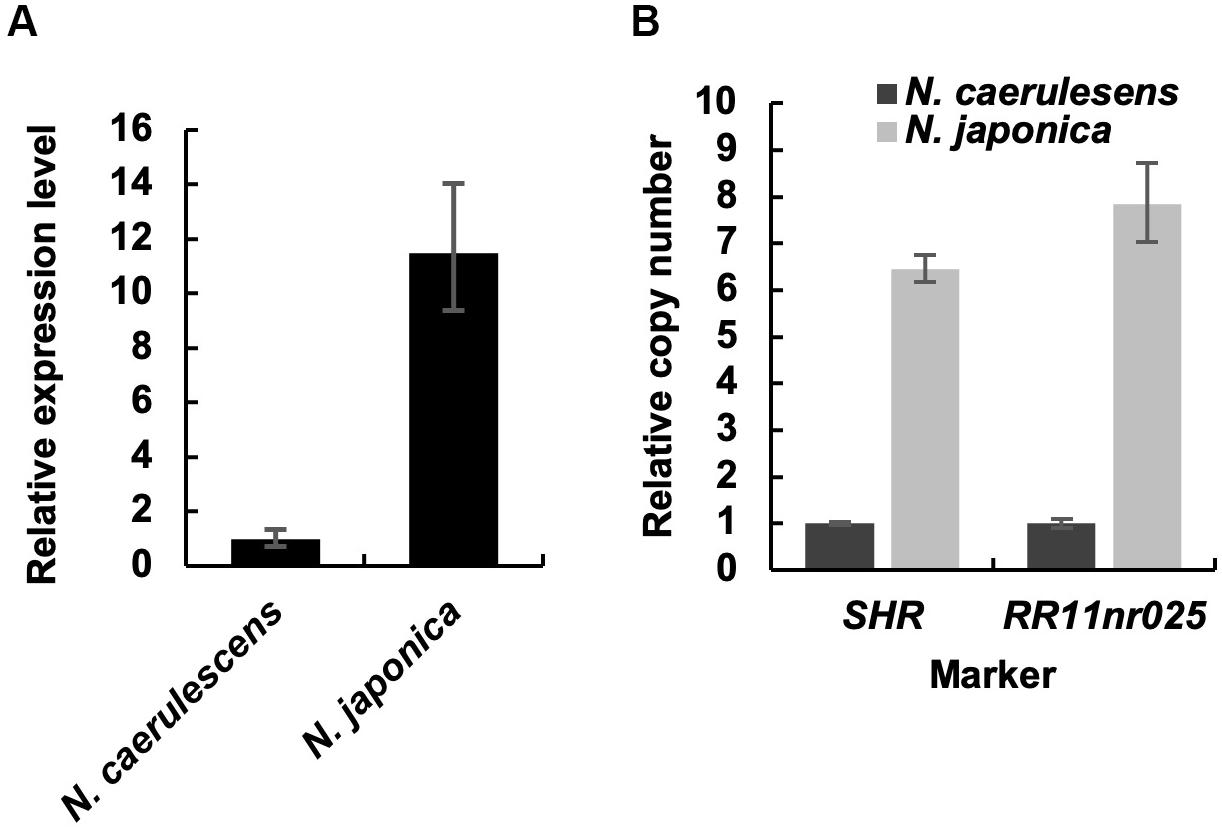
Figure 3. Root expression level and genomic copy number of IREG2 in the two Noccaea plant types. (A) Plants were exposed to 200 μM NiCl2 for 7 days in hydroponic culture, and the roots were subjected to qRT-PCR. The relative expression levels of IREG2 were calculated using EF1α as the internal reference gene. (B) The copy number of IREG2 per genome was quantified by qPCR and normalized with the expressed sequence tag–based indel marker RR11nr025 and the gene SHR. Values are means ± SD of three biological replicates.
According to Schaaf et al. (2006), heterologous expression of AtIREG2 increases the Ni tolerances of S. cerevisiae, and also overexpression of AtIREG2 increases the Ni tolerance of A. thaliana. Therefore, to determine whether NjIREG2 and NcIREG2 are functional homologs of AtIREG2, we first established S. cerevisiae strains expressing NjIREG2 or NcIREG2 and assayed their Ni tolerance. Both strains showed increased Ni tolerance compared with the vector control strain (Figure 4A). We then evaluated the Ni tolerance of A. thaliana transgenic lines expressing NcIREG2 or NjIREG2. cDNA fragments of NjIREG2 and NcIREG2 driven by the 35S promoter were separately introduced into the ireg2-1 homozygous line, and four independent lines constitutively expressing NcIREG2 or NjIREG2 were selected (Figure 4B). The Col-0 and ireg2-1 lines showed severe growth inhibition under 70 μM Ni compared with un-treated control, whereas the transgenic lines expressing NjIREG2 or NcIREG2 showed greatly increased Ni tolerance (Figures 4C–E). These results confirmed that NjIREG2 and NcIREG2 encode functional IREG2 proteins. Moreover, line NcIREG2#5, in which IREG2 expression level was considerably lower than in the other NcIREG2- or NjIREG2-expressing lines (Figure 4B), showed significantly lower root Ni accumulation compared with the others (Figure 4G). In addition, the ratio of shoot Ni to root Ni in NcIREG2#5 tended to be higher than that in the other lines (Figures 4F–H). These results indicate that the ability to sequester Ni in root vacuoles is lower in NcIREG2#5 than in the other lines, and supports the notion that the expression level of IREG2 determines the efficiency of root-to-shoot Ni translocation. Except for NcIREG2#5, there was no significant difference in Ni concentrations in roots and shoots between the NjIREG2-expressing lines and NcIREG2-expressing lines, suggesting that Ni sequestration activity is comparable between NcIREG2 and NjIREG2.
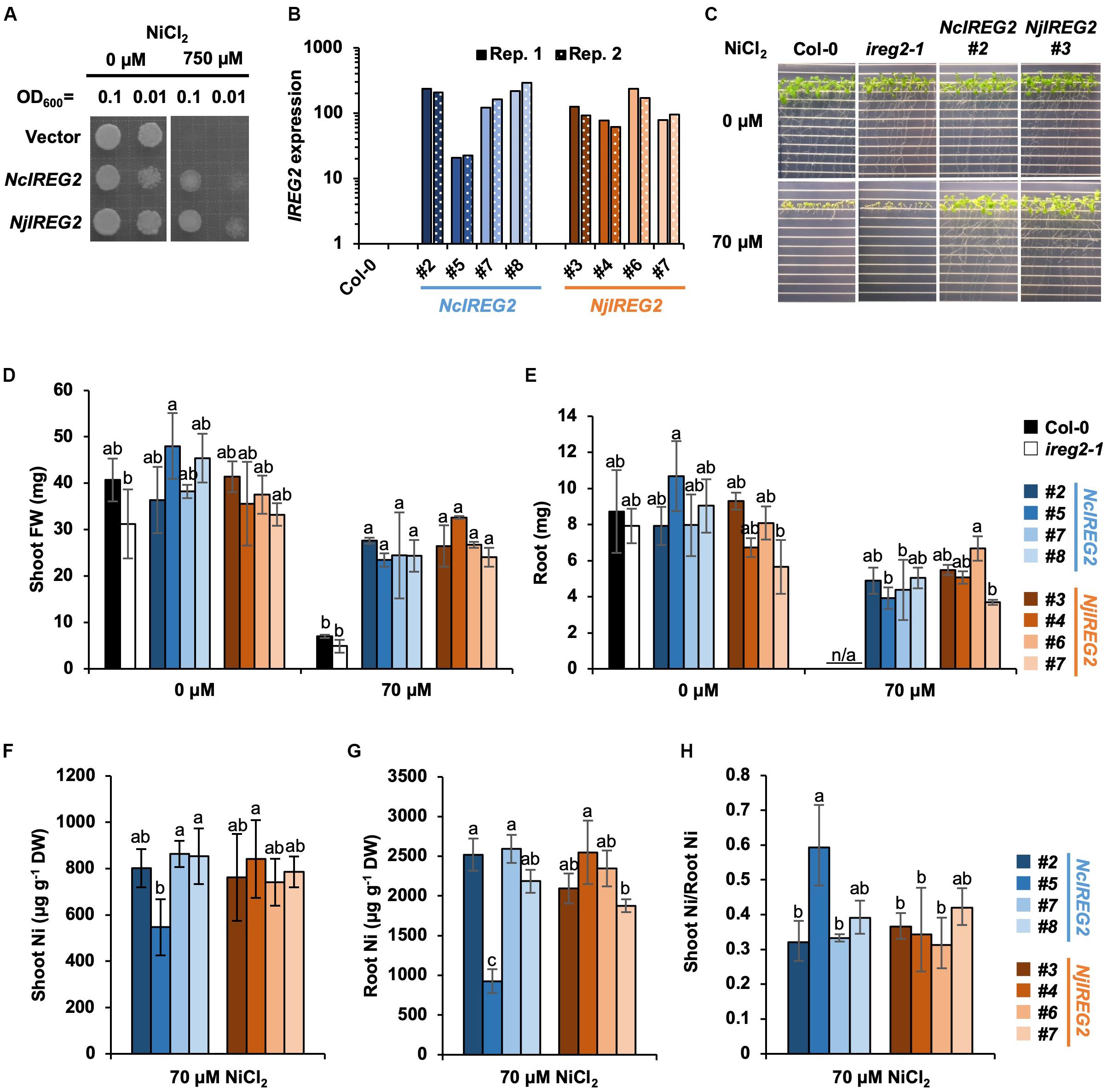
Figure 4. Heterologous expression of NcIREG2 and NjIREG2 in yeast and A. thaliana. (A) Saccharomyces cerevisiae cells were transformed with NcIREG2, NjIREG2, or empty vector and then grown on YNB media containing 0 or 750 μM NiCl2 at 30°C for 4 days. (B) Expression level of IREG2 in the transgenic lines overexpressing NcIREG2 or NjIREG2. The ireg2-1 homozygous line was transformed with 35S:NcIREG2 or 35S:NjIREG2, and four independent lines were selected for each construct. Relative expression levels of IREG2 (Col-0 = 1) were determined by qRT-PCR. Data of each biological replicate (Rep. 1 and Rep. 2) are shown. (C–E) Growth of 14-day-old plants after the treatments. (F,G) Ni concentration expressed relative to dry weight (DW) in roots and shoots. (H) Ratio of shoot Ni to root Ni concentration. Values are means ± SD of four biological replicates. Different letters indicate a statistically significant difference (P < 0.05) among lines in each Ni treatment (ANOVA, Tukey–Kramer).
Here, we showed that the efficiency of root-to-shoot Ni translocation is significantly lower in N. japonica than in N. caerulescens (ecotype Ganges), and that this is possibly due to higher IREG2 expression in N. japonica than in N. caerulescens. Moreover, we showed that the genomic copy number of IREG2 is much higher in N. japonica than in N. caerulescens, which likely causes the higher expression of IREG2 in N. japonica. The multiple IREG2 transcripts obtained from N. japonica (Supplementary Figure S4) could be derived from the multiplicated IREG2. These findings indicate that IREG2 could be a genetic factor determining variation in shoot Ni hyperaccumulation. A hypothetical model of the IREG2-dependent mechanism causing differential shoot Ni accumulation in the two Noccaea plant types is shown in Figure 5: Ni is absorbed by IRT1 (or unknown Ni transporters) into root cells; a part of the Ni absorbed is sequestered in root vacuoles by IREG2, and the other part is loaded into the xylem and translocated from roots to shoots; in N. japonica, the elevated expression of IREG2 in roots increases Ni accumulation in root vacuoles and decreases the amount of Ni to be loaded into xylem, thereby reducing the amount of Ni translocated from roots to shoots. A similar mechanism was shown for Cd accumulation in rice and A. thaliana: HMA3 suppresses Cd translocation from roots to shoots by sequestering Cd in the root vacuoles and determines variation in shoot Cd accumulation among cultivars or accessions (Ueno et al., 2010; Chao et al., 2012). Metal sequestration in root vacuoles is probably a major trait determining the efficiency of metal translocation from roots to shoots. We observed that the Ni concentration in the whole plant was higher in N. caerulescens than N. japonica following 200 μM Ni treatment (Figure 1F), indicating that Ni absorption activity was higher in N. caerulescens. Both the higher Ni absorption activity and the lower IREG2 expression in N. caerulescens likely contribute to its higher sensitivity to Ni compared with N. japonica (Figure 1A and Supplementary Figure S1). IRT1 expression is known to accelerate Ni absorption in A. thaliana (Nishida et al., 2012). We therefore hypothesize that more Ni is absorbed under excess Ni conditions in N. caerulescens than N. japonica because the induction of IRT1 by Ni-induced Fe deficiency is higher in N. caerulescens. This hypothesis assumes that IRT1 is predominantly responsible for Ni absorption from soils. Further analyses are necessary to confirm the involvement of IRT1 in Ni hyperaccumulation in N. caerulescens and N. japonica.
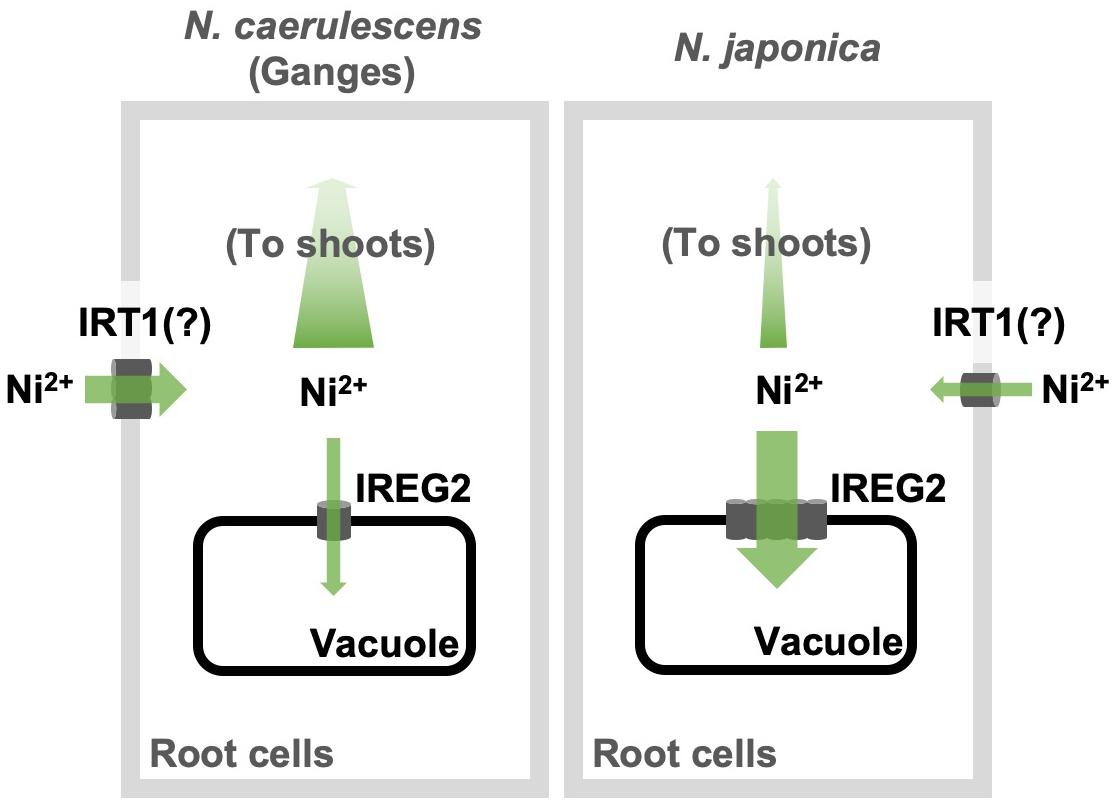
Figure 5. Schematic model of IREG2-dependent mechanism causing differential Ni accumulation in N. japonica and N. caerulescens. The gray rectangles and black rectangles denote the root cell membranes and the vacuole membranes, respectively.
In Ni hyperaccumulators, shoot Ni accumulation depends in part on nicotianamine accumulation in the roots: i.e., more nicotianamine accumulated in the roots increases the mobility of Ni in root cells and facilitates Ni translocation from roots to shoots (Curie et al., 2008). However, although the ratio of shoot Ni to root Ni concentration (an indicator of root-to-shoot translocation) was higher in N. caerulescens than N. japonica, we observed no significant difference in the ratio of shoot Zn to root Zn concentration between the two Noccaea plant types. If root nicotianamine were responsible for this difference, one might expect the root nicotianamine concentration to be higher in N. caerulescens than N. japonica, but this was not the case (Supplementary Figure S6). This finding provides further evidence that nicotianamine, which affects both Ni and Zn translocation, is not responsible for the difference in root-to-shoot Ni translocation between the two plant types. Richau et al. (2009) reported that shoot Ni accumulation in a Ni-hyperaccumulating N. caerulescens accession depends on the enhanced accumulation of free histidine in the roots: i.e., chelation of Ni by free histidine accumulated in the roots inhibits Ni sequestration into the root vacuoles and promotes Ni translocation from roots to shoots. However, Persans et al. (1999) denied the implication of root histidine accumulation in shoot Ni hyperaccumulation in the Ni hyperaccumulator N. goesingenese. Although the role of free histidine in Ni hyperaccumulation is still controversial, the possibility that free histidine is another factor determining variation in Ni hyperaccumulation in Noccaea cannot be ruled out.
It has been reported that N. caerulescens shows more efficient root-to-shoot Ni translocation compared with metal non-accumulator plants such as A. thaliana (Assunção et al., 2003; Mari et al., 2006). Here, we showed that the ratio of shoot Ni to root Ni concentration was significantly higher in N. caerulescens than in A. thaliana (Figure 1), confirming a higher Ni translocation in N. caerulescens. This difference is probably due to elevated nicotianamine accumulation in the roots of N. caerulescens as reported previously (Mari et al., 2006). A transcriptome analysis showing that IREG2 is not differentially expressed between N. caerulescens (a Zn/Pb-mine ecotype) and A. thaliana Col-0 (van de Mortel et al., 2006) suggests that IREG2 expression is not involved in the difference in Ni translocation. In contrast, our data suggest that the efficiency of Ni translocation from roots to shoots in N. japonica is similar to that of A. thaliana following treatment with 25 μM Ni (Figure 1E). Root nicotianamine accumulation in N. japonica was comparable with that in N. caerulescens, but the potential for efficient Ni translocation caused by elevated nicotianamine accumulation in roots might be offset by the elevated IREG2 expression. Efficient translocation of metals from roots to shoots is generally found in metal hyperaccumulators. However, our results imply that efficient Ni translocation is not necessary for Ni hyperaccumulation. Shoot Ni concentrations could reach the hyperaccumulation level in N. japonica growing in serpentine soils for a long time (Mizuno et al., 2003).
The expression levels of metal transporter genes (e.g., HMA3, HMA4, and NRAMP1) involved in shoot metal hyperaccumulation are elevated by gene duplication in metal hyperaccumulator species, and gene duplication is recognized as a mechanism facilitating evolution of metal hyperaccumulators (Hanikenne et al., 2008; Ueno et al., 2011; Milner et al., 2014). IREG2 is multiplicated in N. japonica and accordingly IREG2 expression is elevated. But, the elevated IREG2 reduces root-to-shoot Ni translocation by sequestering Ni in the root vacuoles (Schaaf et al., 2006; Morrissey et al., 2009) and so does not contribute to the trait of efficient Ni translocation from roots to shoots. N. japonica grows in serpentine soil areas, whereas N. caerulescens Ganges, the ecotype studied here, grows in Zn/Pb mine soil areas. IREG2 expression in N. caerulescens is higher in an ecotype from serpentine soils than in ecotypes from Zn/Pb mine soils or non-metalliferous soil (Halimaa et al., 2014). We assume that elevation of IREG2 expression was required to survive under excess Ni conditions. Whole genome sequence analysis of natural populations of the non-accumulator Arabidopsis lyrata revealed that a non-synonymous substitution in IREG2 is strongly linked to adaptation to serpentine soils (Turner et al., 2010). IREG2 might be a major target to be duplicated or modified for adaptation to Ni-rich soils in wild plants. For future study, it would be important to investigate the involvement of IREG2 in intra- and inter-specific variations in Ni hyperaccumulation pattern and Ni tolerance using a wide variety of species and accessions of Brassicaceae metal hyperaccumulators.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
SN designed the study and wrote the manuscript. SN, RT, SI, JY, and HN contributed to analyses. HN and NF assisted in the preparation of the manuscript. All authors contributed to data interpretation and approved the final version of the manuscript.
This work was supported, in part, by a grant from the Institute of Science and Engineering of Chuo University (NF and SN) and JSPS KAKENHI Grant Numbers JP16K18667, JP19K05756, and JP18KK0426 (SN).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared past co-authorship, with one of the authors, HN.
We thank the ABRC Stock Center for providing seeds of T-DNA insertion lines.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00610/full#supplementary-material
Assunção, A. G., Bookum, W. M., Nelissen, H. J., Vooijs, R., Schat, H., and Ernst, W. H. (2003). Differential metal-specific tolerance and accumulation patterns among Thlaspi caerulescens populations originating from different soil types. New Phytol. 159, 411–419. doi: 10.1046/j.1469-8137.2003.00819.x
Brooks, R. R., Lee, J., Reeves, R. D., and Jaffré, T. (1977). Detection of nickeliferous rocks by analysis of herbarium specimens of indicator plants. J. Geochem. Explorat. 7, 49–57. doi: 10.1016/0375-6742(77)90074-7
Caille, N., Zhao, F. J., and McGrath, S. P. (2005). Comparison of root absorption, translocation and tolerance of arsenic in the hyperaccumulator Pteris vittata and the nonhyperaccumulator Pteris tremula. New Phytol. 165, 755–761. doi: 10.1111/j.1469-8137.2004.01239.x
Chaney, R. L., Angle, J. S., Broadhurst, C. L., Peters, C. A., Tappero, R. V., and Sparks, D. L. (2007). Improved understanding of hyperaccumulation yields commercial phytoextraction and phytomining technologies. J. Environ. Qual. 36, 1429–1443. doi: 10.2134/jeq2006.0514
Chao, D. Y., Silva, A., Baxter, I., Huang, Y. S., Nordborg, M., Danku, J., et al. (2012). Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana. PLoS Genet. 8:e1002923. doi: 10.1371/journal.pgen.1002923
Curie, C., Cassin, G., Couch, D., Divol, F., Higuchi, K., Le Jean, M., et al. (2008). Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann. Bot. 103, 1–11. doi: 10.1093/aob/mcn207
Freeman, J. L., Garcia, D., Kim, D., Hopf, A., and Salt, D. E. (2005). Constitutively elevated salicylic acid signals glutathione-mediated nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Physiol. 137, 1082–1091. doi: 10.1104/pp.104.055293
Freeman, J. L., Persans, M. W., Nieman, K., Albrecht, C., Peer, W., Pickering, I. J., et al. (2004). Increased glutathione biosynthesis plays a role in nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Cell 16, 2176–2191. doi: 10.1105/tpc.104.023036
Gendre, D., Czernic, P., Conéjéro, G., Pianelli, K., Briat, J. F., Lebrun, M., et al. (2007). TcYSL3, a member of the YSL gene family from the hyper-accumulator Thlaspi caerulescens, encodes a nicotianamine-Ni/Fe transporter. Plant J. 49, 1–15. doi: 10.1111/j.1365-313x.2006.02937.x
Halimaa, P., Lin, Y. F., Ahonen, V. H., Blande, D., Clemens, S., Gyenesei, A., et al. (2014). Gene expression differences between Noccaea caerulescens ecotypes help to identify candidate genes for metal phytoremediation. Environ. Sci. Technol. 48, 3344–3353. doi: 10.1021/es4042995
Hammond, J. P., Bowen, H. C., White, P. J., Mills, V., Pyke, K. A., Baker, A. J., et al. (2006). A comparison of the Thlaspi caerulescens and Thlaspi arvense shoot transcriptomes. New Phytol. 170, 239–260. doi: 10.1111/j.1469-8137.2006.01662.x
Hanikenne, M., and Nouet, C. (2011). Metal hyperaccumulation and hypertolerance: a model for plant evolutionary genomics. Curr. Opin. Plant Biol. 14, 252–259. doi: 10.1016/j.pbi.2011.04.003
Hanikenne, M., Talke, I. N., Haydon, M. J., Lanz, C., Nolte, A., Motte, P., et al. (2008). Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453:391. doi: 10.1038/nature06877
Kramer, U., Smith, R. D., Wenzel, W. W., Raskin, I., and Salt, D. E. (1997). The role of metal transport and tolerance in nickel hyperaccumulation by Thlaspi goesingense halacsy. Plant Physiol. 115, 1641–1650. doi: 10.1104/pp.115.4.1641
Lasat, M. M., Baker, A. J., and Kochian, L. V. (1996). Physiological characterization of root Zn2+ absorption and translocation to shoots in Zn hyperaccumulator and nonaccumulator species of Thlaspi. Plant Physiol. 112, 1715–1722. doi: 10.1104/pp.112.4.1715
Lin, Y. F., Severing, E. I, te Lintel Hekkert, B., Schijlen, E., and Aarts, M. G. (2014). A comprehensive set of transcript sequences of the heavy metal hyperaccumulator Noccaea caerulescens. Front. Plant Sci. 5:261. doi: 10.3389/fpls.2014.00261
Lochlainn, S. Ó, Bowen, H. C., Fray, R. G., Hammond, J. P., King, G. J., White, P. J., et al. (2011). Tandem quadruplication of HMA4 in the zinc (Zn) and cadmium (Cd) hyperaccumulator Noccaea caerulescens. PLoS One 6:e17814. doi: 10.1371/journal.pone.0017814
Mari, S., Gendre, D., Pianelli, K., Ouerdane, L., Lobinski, R., Briat, J. F., et al. (2006). Root-to-shoot long-distance circulation of nicotianamine and nicotianamine–nickel chelates in the metal hyperaccumulator Thlaspi caerulescens. J. Exp. Bot. 57, 4111–4122. doi: 10.1093/jxb/erl184
Merlot, S., Hannibal, L., Martins, S., Martinelli, L., Amir, H., Lebrun, M., et al. (2014). The metal transporter PgIREG1 from the hyperaccumulator Psychotria gabriellae is a candidate gene for nickel tolerance and accumulation. J. Exp. Bot. 65, 1551–1564. doi: 10.1093/jxb/eru025
Milner, M. J., Mitani-Ueno, N., Yamaji, N., Yokosho, K., Craft, E., Fei, Z., et al. (2014). Root and shoot transcriptome analysis of two ecotypes of Noccaea caerulescens uncovers the role of NcNramp1 in Cd hyperaccumulation. Plant J. 78, 398–410. doi: 10.1111/tpj.12480
Mizuno, N., Nosaka, S., Mizuno, T., Horie, K., and Obata, H. (2003). Distribution of Ni and Zn in the leaves of Thlaspi japonicum growing on ultramafic soil. Soil Sci. Plant Nutr. 49, 93–97. doi: 10.1080/00380768.2003.10409984
Mori, S., and Nishizawa, N. (1987). Methionine as a dominant precursor of phytosiderophores in Graminaceae plants. Plant Cell Physiol. 28, 1081–1092.
Morrissey, J., Baxter, I. R., Lee, J., Li, L., Lahner, B., Grotz, N., et al. (2009). The ferroportin metal efflux proteins function in iron and cobalt homeostasis in Arabidopsis. Plant Cell 21, 3326–3338. doi: 10.1105/tpc.109.069401
Na, G., and Salt, D. E. (2011). Differential regulation of serine acetyltransferase is involved in nickel hyperaccumulation in Thlaspi goesingense. J. Biol. Chem. 286, 40423–40432. doi: 10.1074/jbc.m111.247411
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. doi: 10.1263/jbb.104.34
Nishida, S., Aisu, A., and Mizuno, T. (2012). Induction of IRT1 by the nickel-induced iron-deficient response in Arabidopsis. Plant Signal. Behav. 7, 329–331. doi: 10.4161/psb.19263
Nishida, S., Kato, A., Tsuzuki, C., Yoshida, J., and Mizuno, T. (2015). Induction of nickel accumulation in response to zinc deficiency in Arabidopsis thaliana. Int. J. Mol. Sci. 16, 9420–9430. doi: 10.3390/ijms16059420
Nishida, S., Tsuzuki, C., Kato, A., Aisu, A., Yoshida, J., and Mizuno, T. (2011). AtIRT1, the primary iron uptake transporter in the root, mediates excess nickel accumulation in Arabidopsis thaliana. Plant Cell Physiol. 52, 1433–1442. doi: 10.1093/pcp/pcr089
Nozoye, T., Kim, S., Kakei, Y., Takahashi, M., Nakanishi, H., and Nishizawa, N. K. (2014). Enhanced levels of nicotianamine promote iron accumulation and tolerance to calcareous soil in soybean. Biosci. Biotechnol. Biochem. 78, 1677–1684. doi: 10.1080/09168451.2014.936350
Peer, W. A., Mahmoudian, M., Freeman, J. L., Lahner, B., Richards, E. L., Reeves, R. D., et al. (2006). Assessment of plants from the Brassicaceae family as genetic models for the study of nickel and zinc hyperaccumulation. New Phytol. 172, 248–260. doi: 10.1111/j.1469-8137.2006.01820.x
Peer, W. A., Mamoudian, M., Lahner, B., Reeves, R. D., Murphy, A. S., and Salt, D. E. (2003). Identifying model metal hyperaccumulating plants: germplasm analysis of 20 Brassicaceae accessions from a wide geographical area. New Phytol. 159, 421–430. doi: 10.1046/j.1469-8137.2003.00822.x
Persans, M. W., Yan, X., Patnoe, J. M. M., Krämer, U., and Salt, D. E. (1999). Molecular dissection of the role of histidine in nickel hyperaccumulation in Thlaspi goesingense (Hálácsy). Plant Physiol. 121, 1117–1126. doi: 10.1104/pp.121.4.1117
Reeves, R. D., Baker, A. J., Jaffré, T., Erskine, P. D., Echevarria, G., and van der Ent, A. (2018). A global database for plants that hyperaccumulate metal and metalloid trace elements. New Phytol. 218, 407–411. doi: 10.1111/nph.14907
Richau, K. H., Kozhevnikova, A. D., Seregin, I. V., Vooijs, R., Koevoets, P. L., Smith, J. A. C., et al. (2009). Chelation by histidine inhibits the vacuolar sequestration of nickel in roots of the hyperaccumulator Thlaspi caerulescens. New Phytol. 183, 106–116. doi: 10.1111/j.1469-8137.2009.02826.x
Salt, D. E., Smith, R. D., and Raskin, I. (1998). Phytoremediation. Annu. Rev. Plant Biol. 49, 643–668.
Schaaf, G., Honsbein, A., Meda, A. R., Kirchner, S., Wipf, D., and von Wirén, N. (2006). AtIREG2 encodes a tonoplast transport protein involved in iron-dependent nickel detoxification in Arabidopsis thaliana roots. J. Biol. Chem. 281, 25532–25540. doi: 10.1074/jbc.m601062200
Suzuki, Y., Kawazu, T., and Koyama, H. (2004). RNA isolation from siliques, dry seeds, and other tissues of Arabidopsis thaliana. Biotechniques 37, 542–544. doi: 10.2144/04374bm03
Turner, T. L., Bourne, E. C., Von Wettberg, E. J., Hu, T. T., and Nuzhdin, S. V. (2010). Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat. Genet. 42:260. doi: 10.1038/ng.515
Ueno, D., Milner, M. J., Yamaji, N., Yokosho, K., Koyama, E., Clemencia Zambrano, M., et al. (2011). Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Plant J. 66, 852–862. doi: 10.1111/j.1365-313x.2011.04548.x
Ueno, D., Yamaji, N., Kono, I., Huang, C. F., Ando, T., Yano, M., et al. (2010). Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. 107, 16500–16505. doi: 10.1073/pnas.1005396107
Uraguchi, S., Weber, M., and Clemens, S. (2019). Elevated root nicotianamine concentrations are critical for Zn hyperaccumulation across diverse edaphic environments. Plant Cell Environ. 42, 2003–2014. doi: 10.1111/pce.13541
Vacchina, V., Mari, S., Czernic, P., Marquès, L., Pianelli, K., Schaumlöffel, D., et al. (2003). Speciation of nickel in a hyperaccumulating plant by high-performance liquid chromatography- inductively coupled plasma mass spectrometry and electrospray MS/MS assisted by cloning using yeast complementation. Anal. Chem. 75, 2740–2745. doi: 10.1021/ac020704m
van de Mortel, J. E., Villanueva, L. A., Schat, H., Kwekkeboom, J., Coughlan, S., Moerland, P. D., et al. (2006). Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 142, 1127–1147. doi: 10.1104/pp.106.082073
Keywords: iron regulated 2, metal hyperaccumulator, nickel, nicotianamine, Noccaea, phytoremediation, transporter, zinc
Citation: Nishida S, Tanikawa R, Ishida S, Yoshida J, Mizuno T, Nakanishi H and Furuta N (2020) Elevated Expression of Vacuolar Nickel Transporter Gene IREG2 Is Associated With Reduced Root-to-Shoot Nickel Translocation in Noccaea japonica. Front. Plant Sci. 11:610. doi: 10.3389/fpls.2020.00610
Received: 06 February 2020; Accepted: 21 April 2020;
Published: 03 June 2020.
Edited by:
Tomoko Nozoye, Meiji Gakuin University, JapanCopyright © 2020 Nishida, Tanikawa, Ishida, Yoshida, Mizuno, Nakanishi and Furuta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sho Nishida, c2hvbkBjYy5zYWdhLXUuYWMuanA=; Naoki Furuta, bmZ1cnV0YUBjaGVtLmNodW8tdS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.