
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 May 2020
Sec. Plant Pathogen Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00605
This article is part of the Research Topic Plant-Arthropod Interactions: Effectors and Elicitors of Arthropods and Their Associated Microbes View all 10 articles
Cowpea, Vigna unguiculata, is a crop that is essential to semiarid areas of the world like Sub-Sahara Africa. Cowpea is highly susceptible to cowpea aphid, Aphis craccivora, infestation that can lead to major yield losses. Aphids feed on their host plant by inserting their hypodermal needlelike flexible stylets into the plant to reach the phloem sap. During feeding, aphids secrete saliva, containing effector proteins, into the plant to disrupt plant immune responses and alter the physiology of the plant to their own advantage. Liquid chromatography tandem mass spectrometry (LC-MS/MS) was used to identify the salivary proteome of the cowpea aphid. About 150 candidate proteins were identified including diacetyl/L-xylulose reductase (DCXR), a novel enzyme previously unidentified in aphid saliva. DCXR is a member of short-chain dehydrogenases/reductases with dual enzymatic functions in carbohydrate and dicarbonyl metabolism. To assess whether cowpea aphid DCXR (AcDCXR) has similar functions, recombinant AcDCXR was purified and assayed enzymatically. For carbohydrate metabolism, the oxidation of xylitol to xylulose was tested. The dicarbonyl reaction involved the reduction of methylglyoxal, an α-β-dicarbonyl ketoaldehyde, known as an abiotic and biotic stress response molecule causing cytotoxicity at high concentrations. To assess whether cowpea aphids induce methylglyoxal in plants, we measured methylglyoxal levels in both cowpea and pea (Pisum sativum) plants and found them elevated transiently after aphid infestation. Agrobacterium-mediated transient overexpression of AcDCXR in pea resulted in an increase of cowpea aphid fecundity. Taken together, our results indicate that AcDCXR is an effector with a putative ability to generate additional sources of energy to the aphid and to alter plant defense responses. In addition, this work identified methylglyoxal as a potential novel aphid defense metabolite adding to the known repertoire of plant defenses against aphid pests.
Cowpea (Vigna unguiculata) is one of the most important agronomic plant species grown in semiarid tropical regions of the world. Cowpea is well adapted to biotic and abiotic stresses and provides an excellent source of nutrition (Singh et al., 2002; Timko and Singh, 2008). However, a stress that is a limiting factor in cowpea production is infestation by the cowpea aphid, Aphis craccivora (Jackai and Daoust, 1986). Cowpea aphid infestation can cause devastating effects; it has been reported that young plants of highly susceptible cowpea cultivars were killed by an infestation of cowpea aphids initiated with fewer than ten aphids (Ofuya, 1995). Cowpea aphid feeding induced damage includes chlorosis, leaf curling, and stunted growth resulting in a decrease in yield (Blackman and Eastop, 2000; Kamphuis et al., 2012; Choudhary et al., 2017). In addition to cowpea aphid being a deadly pest, this aphid species is also known to vector over 50 plant viruses (Chan et al., 1991).
There are about 4500 species of aphids reported to date (Remaudiere and Remaudiere, 1997; Blackman and Eastop, 2000; Sorenson, 2009). Of these species, only 100 are considered to have an economic impact and 14 are considered to be serious pests, among which is the cowpea aphid (Sorenson, 2009). Aphids feed differently from chewing insects, which generate massive mechanical tissue damage. Aphids insert their specialized and flexible mouthparts, the stylets, through plant tissues to reach their source of food, the phloem sap, thus avoiding much of the mechanical tissue damage (Tjallingii and Esch, 1993; Tjallingii, 2006). En route to the phloem, aphids puncture cells and deposit saliva in the plant apoplast and the punctured cells to facilitate feeding and interfere with plant defenses (Miles, 1999; Will et al., 2007). Aphid feeding and colonization damage the plant, and aphids are categorized based on the type of damage they incur onto their hosts. Aphids that cause extensive direct damage are considered phytotoxic, whereas others that cause indirect damage – for example, by transmitting viruses – are considered non-phytotoxic (Nicholson et al., 2012). Phytotoxic aphids, such as the Russian wheat aphid (Diuraphis noxia) and greenbug (Schizaphis graminum), cause damage in low numbers and are believed to secrete salivary proteins into the plant that are responsible for the increased manifestation of the damage symptoms. In contrast, the non-phytotoxic aphids, like the pea aphid (Acyrthosiphon pisum) and potato aphid (Macrosiphum euphorbiae), do not cause damage at low numbers and secrete salivary proteins to enhance feeding and interfere with plant defenses (Nicholson et al., 2012; Nicholson and Puterka, 2014; Chaudhary et al., 2015).
Aphid saliva has been shown to contain effector proteins that are necessary for successful aphid colonization (Mutti et al., 2006, 2008; Bos et al., 2010; Atamian et al., 2013; Pitino and Hogenhout, 2013; Elzinga et al., 2014; Naessens et al., 2015; Wang et al., 2015; Will and Vilcinskas, 2015; Guy et al., 2016; Kaloshian and Walling, 2016). To characterize aphid salivary protein content, the saliva of several aphid species has been investigated with liquid chromatography tandem mass spectrometry (LC-MS/MS) (Harmel et al., 2008; Carolan et al., 2009; Cooper et al., 2010, 2011; Rao et al., 2013; Vandermoten et al., 2014; Chaudhary et al., 2015; Thorpe et al., 2016; Boulain et al., 2018; Loudit et al., 2018; Yang et al., 2018). These studies have identified numerous conserved salivary proteins common among the different aphid species as well as some that have only been identified in a single aphid species. The conserved proteins are presumed to be a core set of aphid effectors that are used by aphids to facilitate feeding or disrupt general plant defenses, while the unique proteins identified in only a single aphid species or biotype, act in a species-specific host-aphid interaction (Thorpe et al., 2016). This recent wealth of salivary protein identification stems from the release of additional aphid genomes and transcriptomes. Since the first aphid genome was released for the pea aphid, five additional aphid genomes are publicly available (International Aphid Genomics Consortium, 2010; Nicholson et al., 2015; Mathers et al., 2017; Wenger et al., 2017; Thorpe et al., 2018). Numerous aphid transcriptomes are also available including a transcriptome for the cowpea aphid (Agunbiade et al., 2013). Three main criteria have been used to identify putative aphid effectors: (1) expression of the candidate transcripts in aphid heads or salivary glands with prediction for secretion, (2) presence in saliva, and (3) sequence similarity to previously identified aphid effectors.
In general, microbial, nematode and pest effectors are diverse, lacking consensus sequences and features, making it difficult to predict effectors. This has led to reporting of mostly specific subclasses of effectors. For example, effectors from plant pathogenic fungi are small sized proteins with high cysteine content while those from Phytophthora contain a RXLR motif (Jiang et al., 2008; Stergiopoulos and de Wit, 2009; Petre and Kamoun, 2014; Sperschneider et al., 2015). To enhance plant fungal effector predictions, EffectorP was developed as a machine-learned predictor for fungal effectors that does not rely only on predetermined thresholds based on criteria including protein size and cysteine content (Sperschneider et al., 2016, 2018). It is therefore likely that the repertoire of aphid effectors can be enhanced with the development of machine learned effector identification programs.
Numerous studies have functionally characterized aphid effectors. These included overexpression of the candidate effector in planta or silencing it, through plant-mediated RNAi or injection with RNAi constructs, in the aphid and determining aphid performance on the plants. Of the effectors experimentally tested, about a dozen have shown altered aphid colonization phenotypes (Mutti et al., 2006, 2008; Bos et al., 2010; Atamian et al., 2013; Pitino and Hogenhout, 2013; Elzinga et al., 2014; Abdellatef et al., 2015; Naessens et al., 2015; Wang et al., 2015; Will and Vilcinskas, 2015; Guy et al., 2016; Kettles and Kaloshian, 2016). The altered survival/colonization phenotypes determined by some of these effectors act in species-specific and host-specific manner (Atamian et al., 2013; Pitino and Hogenhout, 2013; Elzinga et al., 2014; Rodriguez et al., 2017).
To date, the plant targets for only Mp1 and Me10 aphid effectors have been identified and the mechanism of effector function partially elucidated (Rodriguez et al., 2017; Chaudhary et al., 2019). The function of two additional aphid effectors MIF1 (Naessens et al., 2015) and Armet (Wang et al., 2015) have been predicted based on the function of homologous sequences from other organisms. Both MIF1 and Armet are highly conserved proteins in the animal kingdom. MIF1 encodes a macrophage migration inhibitory factor that is a cytokine deposited in aphid saliva during feeding (Calandra, 2003; Naessens et al., 2015). Armet in mammalian systems and in Drosophila has been reported in the cell as part of the unfolded protein response and extracellularly as a neurotrophic factor (Lindholm et al., 2007, 2008; Palgi et al., 2009, 2012). Both MIF1 and Armet are important for the pea aphid survival as knockdown of their expressions results in shortened lifespan (Naessens et al., 2015; Wang et al., 2015). The function of an additional effector, Me47 encoding a Glutathione S-transferase (GST), was shown based on its GST enzymatic activity and its ability to detoxify isothiocyanates that are implicated in herbivore defense (Kettles and Kaloshian, 2016).
Here we report the salivary proteome of a California population of the cowpea aphid using LC-MS/MS and publicly available aphid genomes and transcriptomes. We also characterize the function of a novel salivary protein, diacetyl/L-xylulose reductase (DCXR). DCXR is a member of short-chain dehydrogenases/reductases (Nakagawa et al., 2002). Mammalian orthologs of DCXR are involved in NADPH-dependent reduction of both carbohydrates and dicarbonyls (Nakagawa et al., 2002; Ishikura et al., 2003; Ebert et al., 2015). The reversible oxidative reduction of the carbohydrates xylitol and L-xylulose can lead to an additional energy source through the pentose phosphate pathway (Sochor et al., 1979; Nakagawa et al., 2002). The reduction of dicarbonyls detoxifies and prevents the formation of advanced glycation end-products (AGEs), also known as glycotoxins, associated with development of numerous degenerative human diseases (Chen et al., 2009; Gkogkolou and Bohm, 2012; Kizer et al., 2014). In plants, the build-up of dicarbonyls leads to oxidative stress and cell death resulting in stunted growth (Hoque et al., 2012; Ray et al., 2013; Sankaranarayanan et al., 2015; Li, 2016). One of these dicarbonyls, generated through multiple pathways in plants and animals, is methylglyoxal (Yadav et al., 2005a, b; Hoque et al., 2016; Mostofa et al., 2018). Depending on concentration, methylglyoxal can act as defense signaling molecule or as a cytotoxin during abiotic stress in plants (Li, 2016). Recently methylglyoxal has also been implicated in plant defense against biotic stresses (Melvin et al., 2017). Here we report the identification of DCXR in cowpea aphid saliva. We show that the recombinant cowpea aphid DCXR, AcDCXR, is able to catalyze the reversible xylitol to xylulose reaction as well as to utilize methylglyoxal as substrate. We also demonstrate that aphid feeding induced methylglyoxal accumulation and that expression of AcDCXR in planta enhanced aphid fecundity contributing to the success of the aphid as a pest.
Cowpea California blackeye cultivar 46 (CB46) and pea (Pisum sativum) cv ZP1130 were grown in UC Mix 3 soil1 in 32 oz plastifoam cups in a pesticide free room at 22–24°C with a 16:8 light:dark photoperiod. Plants were fertilized weekly with MiracleGro (18-18-21; Stern’s MiracleGro Products).
A colony of cowpea aphids, collected from a field in Riverside, California, in summer of 2016, was reared on cowpea cv CB46. A second colony, taken from the cowpea plants, was reared on pea cv ZP1130 for 3 months before use. The colonies was maintained separately in insect cages in growth chambers at 26–30°C with a 16:8 light:dark photoperiod. The colony on cowpea was used for aphid saliva collection and the colony on pea was used for aphid bioassays.
Cowpea aphid saliva was collected by feeding mixed developmental stages of the aphid on a water diet as previously described (Chaudhary et al., 2015). About 100–200 mixed stage aphids were loaded in a feeding chamber, consisting of a plastic cylinder with one end containing the diet inside a parafilm sachet, and the other end secured with a cheesecloth. Aphids were allowed to feed on the 200 μL of ultrapure autoclaved water for 16 h under yellow light. The components of the chamber were sterilized or treated with alcohol and all materials were handled in a laminar flow hood using aseptic conditions. After feeding the diet was collected aseptically using a pipet and stored at −80°C. A new cohort of aphids were used for each overnight collection and saliva was collected from an estimated 10,000 aphids over a three-month period.
Saliva was vacuum concentrated down to protein pellets and dissolved in 100 μL trypsin buffer (50 mM ammonium bicarbonate, pH 8.0, 10% v/v acetonitrile) containing 1 μg trypsin and treated overnight at 37°C. After trypsin digestion, the sample was centrifuged, the supernatant was collected, pelleted with a speedvac concentrator and suspended in 24 μL 0.1% formic acid for LC-MS/MS analysis.
A MudPIT approach was employed to analyze the trypsin-treated samples. A two-dimension nanoAcquity UPLC (Waters) and an Orbitrap Fusion MS (ThermoFisher Scientific) were configured together to perform online 2D-nano LC-MS/MS analysis. The 2D-nanoLC was operated with a 2D-dilution method that was configured with nanoAcquity UPLC. Two mobile phases for the first dimension LC fractionation were 20 mM ammonium formate (pH 10) and acetonitrile, respectively. Online fractionation was achieved by 5-min elution off a NanoEase trap column (PN# 186003682; Waters) using stepwise-increased concentration of acetonitrile. A total of five fractions were generated with 13, 18, 21.5, 27, and 50% of acetonitrile, respectively. A final flushing step used 80% acetonitrile to clean up the trap column. Each and every fraction was then analyzed online using a second dimension LC gradient.
For the second-dimension LC, a BEH130 C18 column (1.7 μm particle, 75 μm i.d., 20 cm long, PN# 186003544; Waters) was used for peptide separation. A Symmetry C18 (5 μm particle, 180 μm i.d., 20 mm long, PN# 186003514; Waters) served as a trap/guard column for desalting and pre-concentrating the peptides for each MudPIT fraction. The solvent components for peptide separation were as follows: mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in acetonitrile. The separation gradient was as follows: at 0 to 1 min, 3% B; at 2 min, 8% B; at 50 min, 45% B; at 52–55 min, 85% B; at 56–70 min, 3% B. The nano-flow rate was set at 0.3 μl/min without flow-splitting.
Spectra were obtained using Orbitrap Fusion MS (Thermo Fisher Scientific). The Orbitrap Fusion MS was in positive ion mode with an ion transfer tube temperature of 275°C. The isolation window used was 2 Da. Three different types of dissociation were used: Collision Induced Dissociation (CID), High-energy Collision Induced Dissociation (HCD), and Electron Transfer Dissociation (ETD). The energy for each of these was 30%. Three scan ranges were used (300–1800, 300–2000 400–1400 Da) with 30 s dynamic exclusion.
The MS/MS spectra were filtered for high confident peptides with strict FDR (1%), with enhanced peptide and protein annotations using the software Proteome Discoverer v2.3 (Thermo Fisher). Spectra with peptide sequences less than 6 residues were removed. The search parameters allowed for 0.5 Da mass tolerance and 2 missed cleavage sites. The following modifications were included: modification of Met Oxidation ± 15.99492 D, Lys Acetyl ± 42.01057 D, Ser, Thr, Tyr Phospho ± 79.966333 D, N-Terminus Formyl ± 27.99492 Da, Pyro-Glu ± 17.02655 Da, N-Terminus Acetyl ± 42.01057 Da. The identified peptides were then searched against an aphid proteome database compiled from every aphid genome available on NCBI and AphidBase (Pea aphid, Russian wheat aphid, soybean aphid (Aphis glycines), bird cherry-oat aphid (Rhopalosiphum padi), green peach aphid (Myzus persicae), and black cherry aphid (Myzus cerasi) and other aphid proteins deposited in NCBI in 2017. These other proteins included six-frame translations of a cowpea aphid transcriptome and the transcriptome of the potato aphid). The 13,330 PSMs identified corresponded to 2,119 proteins and were further filtered to 721 protein group hits. Only high confidence (99%) were considered further filtering the protein groups to 521 protein groups. Spectra that came up when filtering out possible contaminants with a FASTA file containing common contaminants. To accept proteins, they needed to have at least 3 peptides in at least 2 of the 3 replicates (CID, HCD, ETD). The raw peptide spectra were deposited in the Mass Spectrometry Interactive Virtual Environment (MassIVE) repository with the proteome ID: PXD017323.
The MS/MS identified proteins were annotated with BLASTP using OmicsBox (V 1.1.135 Hotfix) and the NCBI non-redundant protein database with the taxonomy filter for aphids, Aphidomorpha (3380) (e-value = 1e-3) (Gotz et al., 2008). The proteins were then subjected to BLASTP to the pea aphid annotation v2.1b proteins on Aphidbase to identify the corresponding ACYPI homologs (BIPAA, 2017). Gene ontology (GO) was determined for molecular function, biological process, and cellular component using InterProScan (v5.36-75.0) (Gotz et al., 2008; Jones et al., 2014). The identified proteins were screened with SignalP (V3.0 and V5.0) and SecretomeP 1.0 using eukaryote and mammalian filters, respectively, and by TMHMM V2.0 (Krogh et al., 2001; Bendtsen et al., 2004a, b; Armenteros et al., 2019). The proteins were further analyzed using EffectorP 2.0 (Sperschneider et al., 2018).
RNA was extracted from 10 mixed developmental stage aphids using Trizol (Invitrogen), and cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) according to manufactures instructions. Using AcDCXR (MN855408) gene-specific Gateway recombination primers (DCXRF- ACAAGTTTGTACAAAAAAGCAGGCTCCATGGAAGAATTC TTTGTCGGAAAAAAGTTCAT, DCXRR- GGGGACCACT TTGTACAAGAAAGCTGGGTCACTGGCCAAAAATCCACCA TC), the DCXR coding region, excluding the secretion signal peptide, was amplified using Q5® High-Fidelity DNA Polymerase (New England Biolabs) with the following conditions: an initial 98°C for 30 s, 98°C for 7 s, 54°C for 20 s, 72°C for 30 s, for 30 cycles and a final cycle of 72°C for 3 min. DCXR was purified using GeneJET PCR Purification Kit (Thermo Scientific) and recombined into vector pDONR207 (Invitrogen) using BP Clonase (Invitrogen). Following Sanger sequencing pDONR207-DCXR was recombined into the expression vectors pDEST17 (Invitrogen; pDEST17-DCXR), pEAQ-HT-DEST1 (Sainsbury et al., 2009; pEAQ-HT-DEST1-AcDCXR), or pCAMBIA1300-GW-mScarlet (pCAMBIA1300-AcDCXR-mScarlet). pCAMBIA1300-GW-mScarlet was developed by modifying pCAMBIA1300 using parts from pGWB614 and p#128060 by restriction digestion and ligations. After transformation into E. coli strain DH5α and the purified pDEST17-DCXR was transformed into E. coli strain ArcticExpress (Agilent) while pEAQ-HT-DEST1-AcDCXR and pCAMBIA1300-AcDCXR-mScarlet were transformed into Agrobacterium tumefaciens strains AGL01 and GV3101, respectively.
The pDEST17-AcDCXR was purified in a similar manner as previously described for the aphid effector Me47 (Kettles and Kaloshian, 2016). Briefly, pDEST17-AcDCXR (N-terminal 6xHis tag) in ArcticExpress was grown in LB media at 37°C to an OD600 of 0.8 and the expression induced by adding of 0.5 mM IPTG followed by incubation at 10°C for 16 h. After centrifugation (6,000 × g for 20 min) the cells were resuspended in chilled lysis buffer (300 mM NaCl, 50 mM NaH2PO4/Na2HPO4, pH 7.0). The cells were lysed using sonication (4 × 15 s pulses), the soluble protein fraction was separated by centrifugation (10,000 × g for 45 min) and incubated with Ni- NTA agarose beads (Qiagen) for 1 h at 4°C with gentle agitation. The column was washed with the lysis buffer containing 40 mM imidazole to remove non-specifically bound proteins. After four washes, DCXR was eluted with three washes of lysis buffer containing 150, 200, and 200 mM of imidazole, respectively. The eluted fractions were concentrated with VivaSpin 500 Centrifugal Concentrator PES (Sartorius, United Kingdom) and monitored using Bradford assay with BSA as the standard. The recombinant DCXR was analyzed on a 12% SDS–PAGE using Coomassie Brilliant Blue G-250 staining.
Oxidation of xylitol to xylulose by recombinant DCXR was measured through the reduction of NADP+ to NADPH as previously described (Yang et al., 2017) with minor modification. A 0.5 mL reaction mixture containing 10 μg AcDCXR 100 mM glycine buffer, pH 9.5, 3 mM MgCl2, NADP+, and 200 mM xylitol were used in 1 mL cuvettes and a Beckman Coulter Du® 730 Life Sciences spectrophotometer. Reactions began after the addition of AcDCXR, and changes in absorbance at 340 nm were monitored. The reaction rates were calculated based on the NADP+ concentrations.
Methylglyoxal reduction by recombinant DCXR was measured through the oxidation of NADPH to NADP+ using 1 mL cuvettes as previously described (Misra et al., 1996) and the Beckman Coulter Spectrophotometer. The 0.5 mL reaction was composed of 10 μg DCXR, 100 μM sodium phosphate buffer (NaH2PO4/Na2HPO4, pH 6.5), 200 μM NADPH and methylglyoxal. The reaction was initiated with the addition of NADPH and monitored by the decrease in absorbance at 340 nm. The reaction rates were calculated based on the methylglyoxal concentrations.
Agrobacterium tumefaciens strain AGL01, carrying either pEAQ-HT-GFP or pEAQ-HT-DEST1-AcDCXR, were used in transient expression of pea, Pisum sativum, cv. ZP1130 as described previously (Guy et al., 2016). Bacterial cells, grown up overnight in YEP media, were harvested, washed three times in infiltration buffer (10 mM MgCl2, 10 mM MES pH 5.6, and 150 μM acetosyringone) and resuspended at a final OD600 of 0.5. The youngest expanded leaf of a 2-week-old plant was infiltrated with a needleless syringe.
The duration of GFP expression in pEAQ-HT-GFP infiltrated leaves was monitored with Western blot analysis. Three 1 cm diameter leaf disks were cut from the same agroinfiltrated leaf using a cork borer after 2, 3, 5, 7, 8, 9, and 10 days post infiltration. Protein was extracted from the leaf disks by grinding in 200 μl lysis buffer (6 M Urea, 2 M Thiourea, 1% Protease inhibitor cocktail [Sigma P9599]). Samples were centrifuged at 14,000 × g for 5 min and the supernatant was resuspended in equal volume 2x loading buffer (100 mM Tris-HCl pH 6.8, 100 mM DTT, 10% glycerol, 2% SDS, 0.01% bromophenol blue). About 25 μg of protein were loaded per sample on 12% SDS–PAGE and transferred to a nitrocellulose membrane. The membrane was probed with mouse anti-GFP antibody (Sigma) and secondary antibody, goat anti-mouse HRP-conjugated (Santa Cruz Biotechnology). Primary antibody was used at 1:2000 and secondary antibody was used at 1:2000 dilution. Pierce ECL Western Blotting Substrate (Thermo Scientific) was used to detect the signal with autoradiography film (Denville Scientific Inc.).
Agrobacterium tumefaciens GV3101 carrying pCAMBIA1300-DCXR-mScarlet or pCAMBIA1300-GFP were grown and prepared as previously described for transient agroexpression. At an OD600 = 0.5 each, the constructs were co-infiltrated in Nicotiana benthamiana leaves. Three days post infiltration, leaf epidermal cells were analyzed using a Leica SP5 confocal microscope. GFP and mScarlet were excited by 488 nm and 543 nm filters, respectively, and images were collected through band emission filters at 498–520 nm and 553–650 nm, respectively.
A day after agroinfiltration, five adult cowpea aphids were caged onto the adaxial side of an agroinfiltrated leaf of 2-week-old pea plants. After 24 h (i.e., 2 days post infiltration; dpi), the adult aphids were removed, and 5 to 6 new-born nymphs were left on the leaf with both the adaxial and abaxial sides of the leaf accessible to the aphids. Eight days later (10 dpi), the surviving aphids were counted and transferred to a new infiltration site on a plant infiltrated 2 days earlier. The fecundity of these aphids was monitored two and five days later (i.e., when the aphids were 12 and 15 day-old). The nymphs were removed after each counting. This experiment was performed three times. Each experiment consisted of 13–15 plants per construct. All experiments were conducted at 22°C, 16:8 light:dark photoperiod.
Methylglyoxal levels were evaluated in 2-week-old cowpea and pea plants following the protocol by Borysiuk et al. (2018). Highly infested leaves were harvested at day 1, 2, and 3 after infestation. Briefly, samples were homogenized in 5% perchloric acid and centrifuged at 13,000 × g for 10 min at 4°C. The supernatant was decolorized with charcoal and neutralized with 1 M potassium carbonate. After centrifuging at 13,000 × g at 4°C the supernatant was used to estimate the methylglyoxal concentration in sodium dihydrogen phosphate buffer (pH 7.0). The absorbance was recorded after 10 min incubation with N-acetyl-L-cysteine to monitor the N-a-acetyl-S-(1-hydroxy-2-oxo-prop-1-yl) cysteine formation (Wild et al., 2012). Methylglyoxal concentration was determined using a standard curve of known methylglyoxal concentrations. The experiment with pea was performed once and with cowpea was performed twice.
We used generalized linear models (GLM) with a likelihood ratio and chi-square test to assess whether AcDCXR expression had an effect on aphid survival and fecundity. Data on aphid survival were analyzed with GLM following a binomial distribution and data on aphid fecundity were assumed to follow a Poisson distribution. The fit of all generalized linear models was checked by inspecting residuals and QQ plots. Methylglyoxal levels in plants were analyzed using a nested ANOVA (biological replicates treated as random factor) (package R: ‘nlme’). When a significant effect was detected, a pairwise comparison using multiple comparisons of the means (package R: ‘multcomp’) (Tukey contrasts, p-values adjustment with ‘fdr’ method) at the 0.05 significance level was used to test for differences between days after infestation. Statistical analyses were performed using the R software (version 3.6.0) (R Core Team, 2019).
To identify the protein composition of the cowpea aphid saliva, aphid saliva was collected in parafilm feeding pouches containing water. The contents of the pouches were concentrated and subjected to proteome analyses. The peptides identified by LC-MS/MS were searched against a custom aphid protein database. The database was composed of proteomes based on all aphid genomes available in the summer of 2017, as well as cowpea aphid-specific transcriptome and a transcriptome from the potato aphid, both with six-frame translations. Around 175 candidate proteins were identified with at least three peptides from at least two replicates and having at least one unique peptide (Supplementary Table 1). The identified proteins were then annotated using BLASTP with OmicsBox (TaxID: Aphididiae 27482). Among these annotated proteins, 18/175 (10.29%) were uncharacterized. In addition, functional redundancies were recorded among the proteins with annotations. To eliminate these redundancies, the proteins were subjected to BLASTP on AphidBase to identify their corresponding ACYPI homolog using the pea aphid protein database annotation v2.1b. Among these proteins, 47/175 (26.86%) shared one of 21 ACYPI top hits. Although these 47 proteins had at least one unique peptide, we grouped them as 21 proteins, resulting in a total of 149 salivary proteins (Supplementary Table 1).
Annotation of these proteins presented a wide range of functional attributes to the cowpea aphid salivary proteins. Among the 149 identified proteins, 33 proteins with similar functional annotations have been previously reported in the saliva of a cowpea aphid population from Gabon, Africa (Loudit et al., 2018). Among these 33 proteins are glucose dehydrogenases, carbonic anhydrases and a trehalase (Supplementary Table 1).
Of the 149 identified cowpea aphid proteins, gene ontology (GO) assigned 123 proteins with at least one GO term in the three most common ontological designations: molecular function, biological process and cellular component. The three most abundant biological process designations were carbohydrate metabolic process (19%), translation (16%) and catabolic process (11%) (Figure 1). The three most abundant molecular function designations were oxidoreductase activity (20%), structural constituent of ribosome (16%) and ATP binding (13%) (Figure 1). As for the most abundant cellular component designations, they were for protein-containing complexes (33%) and cytosol (29%) (Figure 1).
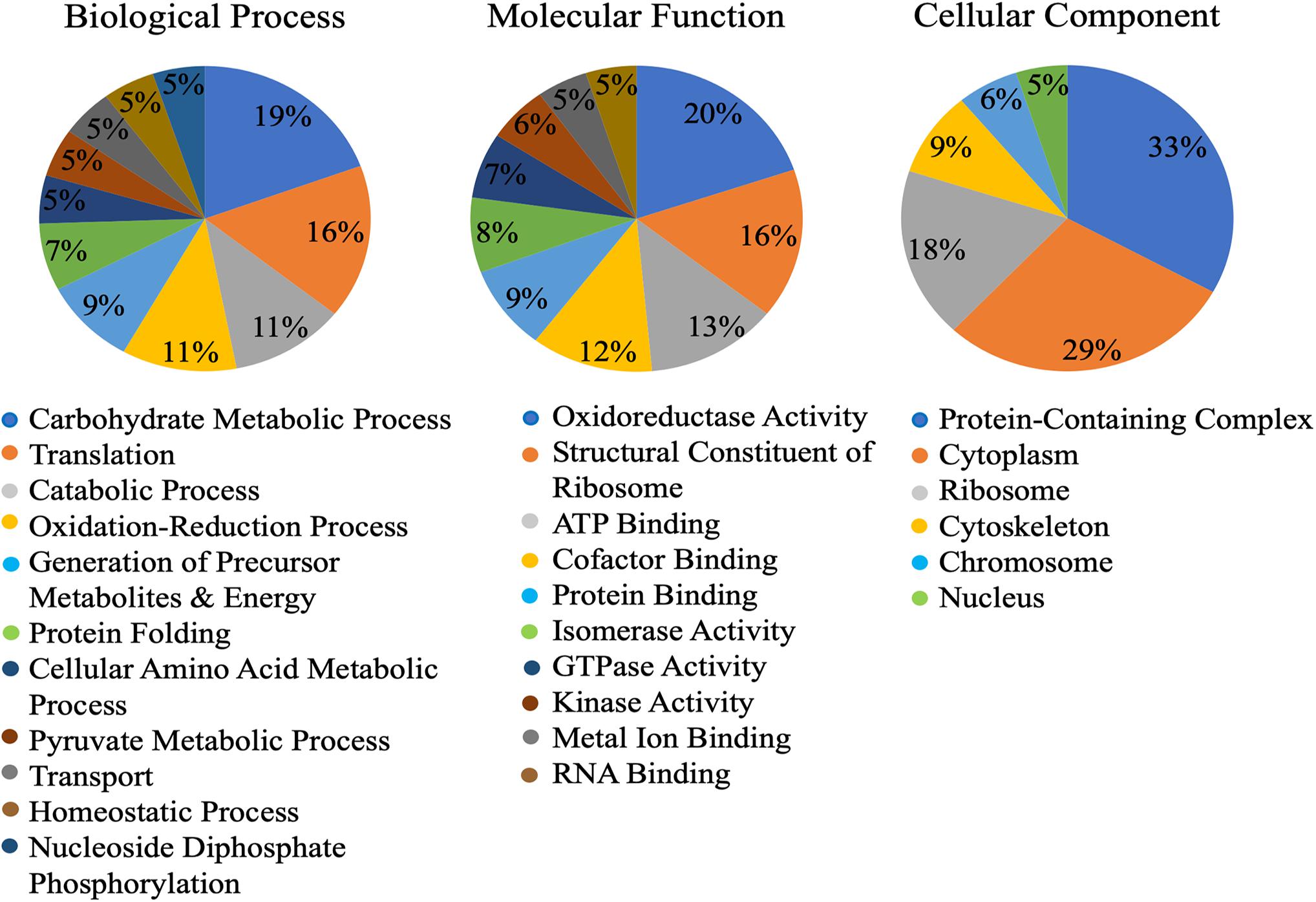
Figure 1. Gene ontology (GO) of the Cowpea aphid salivary proteins. Cowpea aphid salivary proteins were identified by LC-MS/MS and protein content were determined using a number of aphid genomes and the transcriptomes of cowpea aphid and potato aphid.
Since the cowpea aphid genome has not been sequenced, homologous proteins from the different aphid species or those based on cowpea aphid transcriptome, used in our custom database, were used for these analyses. Multiple bioinformatics tools were harnessed to screen the identified salivary proteins for putative effector function. First, the salivary proteins were evaluated for secretion using tools that predict classical and non-classical secretions, SignalP and SecretomeP (Bendtsen et al., 2004a, b; Armenteros et al., 2019), respectively. Using SignalP, a secretion signal was detected in 29 (19.46%) proteins, while SecretomeP predicted the secretion of an additional 23 (15.44%) of the 149 salivary proteins (Table 1). To eliminate proteins with transmembrane domains, presence of transmembrane helices was evaluated using TMHMM (Sonnhammer et al., 1998). Six of these predicted secreted proteins contained transmembrane helices.
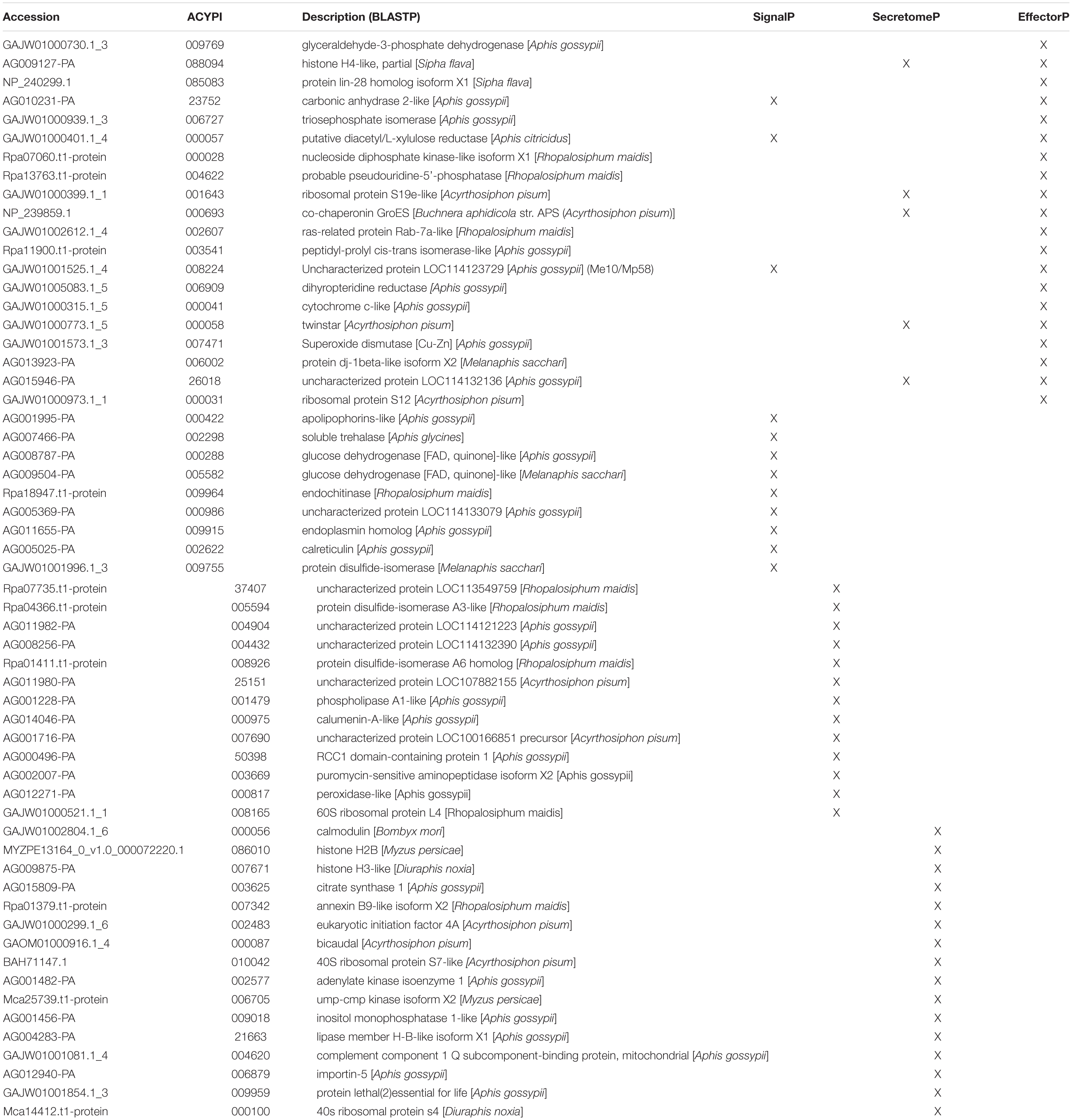
Table 1. Cowpea aphid salivary proteins identified for secretion or effector function using bioinformatic programs.
A machine learning approach was recently used to develop novel prediction program for fungal effectors (Sperschneider et al., 2016, 2018). We wondered whether this tool, EffectorP, could be used to predict aphid effectors. To test this, we first subjected known aphid effectors for EffectorP analysis. We tested the C002 effector, identified first in pea aphid (Mutti et al., 2008), and Me10, identified in potato aphid (Atamian et al., 2013). Both C002 and Me10 were identified as effectors by EffectorP indicating that EffectorP can be utilized as a tool to screen for aphid effectors. Using EffectorP, 20/149 (13.4%) of the cowpea aphid salivary proteins were identified as putative effectors (Table 1). Only eight of the 20 were identified for secretion by SignalP or SecretomeP. Taken together 58 proteins were predicted for secretion or for effector function encoding a wide range of functions with eight being unknowns (Figure 2 and Table 1)
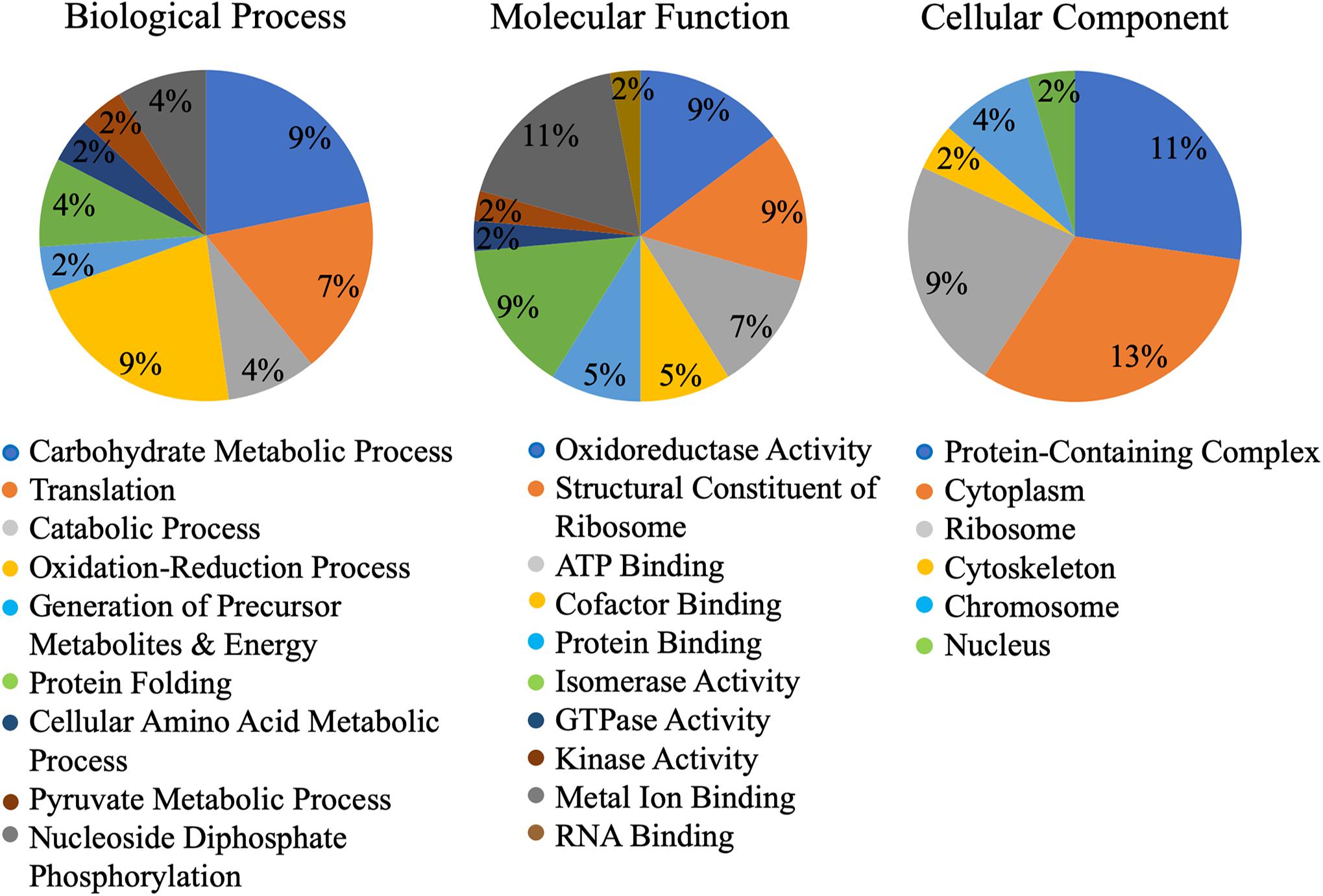
Figure 2. Gene ontology (GO) of putative Cowpea aphid effectors. Cowpea aphid putative effectors were identified by analyzing the salivary proteins with SignalP, SecretomeP, and EffectorP.
A set of criteria were applied to choose a putative effector protein identified by EffectorP for functional characterization. These included a previously unidentified effector predicted for secretion or with secretion signal peptide, a protein with predicted enzymatic activity, and high abundance in cowpea aphid saliva based on the SEQUEST score. Based on these criteria, DCXR was selected for further analysis.
Sequence prediction indicated that cowpea aphid DCXR (AcDCXR; GAJW01000401.1) consists of at least 263 amino acids, with the first 23 amino acids encoding a predicted signal peptide, and a conserved enzymatic domain for short-chain dehydrogenases/reductases (Supplementary Figure 1). Using AcDCXR in BLASTP searches identified DCXR homologs in seven aphid species. Interestingly, only the DCXR from cotton melon aphid (Aphis gossypii; XP_027848224.1) contains a secretion signal peptide (Supplementary Figure 1). Consistent with this information, DCXR has been reported previously from other aphid species but has not been previously identified in aphid saliva (Nguyen et al., 2008, 2009; Pinheiro et al., 2014).
Diacetyl/L-xylulose reductase is a multifunctional enzyme. Mammalian orthologs of DCXR have been shown to function in the glucuronic acid/uronate cycle, in a reversible reaction either oxidizing or reducing xylitol and xylulose, respectively (Sochor et al., 1979; Yang et al., 2017), as well as having α-β-dicarbonyl reductase activity to metabolize toxic carbonyls like methylglyoxal (Ebert et al., 2015). Direct comparison between AcDCXR and XP_027848224.1 showed 100% identity at the amino acid level with perfect conservation of the enzyme active site (Supplementary Figure 1). To test whether AcDCXR has similar functions as the mammalian orthologs, we expressed recombinant AcDCXR and performed enzymatic assays.
Aphid diacetyl/L-xylulose reductase, amplified from cDNAs developed from the whole bodies of mixed stages of the aphid, was cloned into the pDEST17 expression vector and expressed in E. coli strain ArcticExpress. Purified AcDCXR (Supplementary Figure 2) was used in two distinct enzymatic assays to check its functionality. To verify whether AcDCXR is able to oxidize xylitol to xylulose, AcDCXR was assayed using xylitol as the substrate and NADP+ as co-substrate. The reduction of NADP+ to NADPH was spectroscopically monitored by the increase of absorbance at 340 nm. AcDCXR was able to oxidize xylitol to xylulose in a NADP+ concentration-dependent manner (Figure 3A). Analysis of the Lineweaver-Burke plot data determined the enzymatic constants to be: kcat = 1.85 s–1, a Km = 0.56 mM and a Vmax = 79.4 μM/min (Figure 3B).
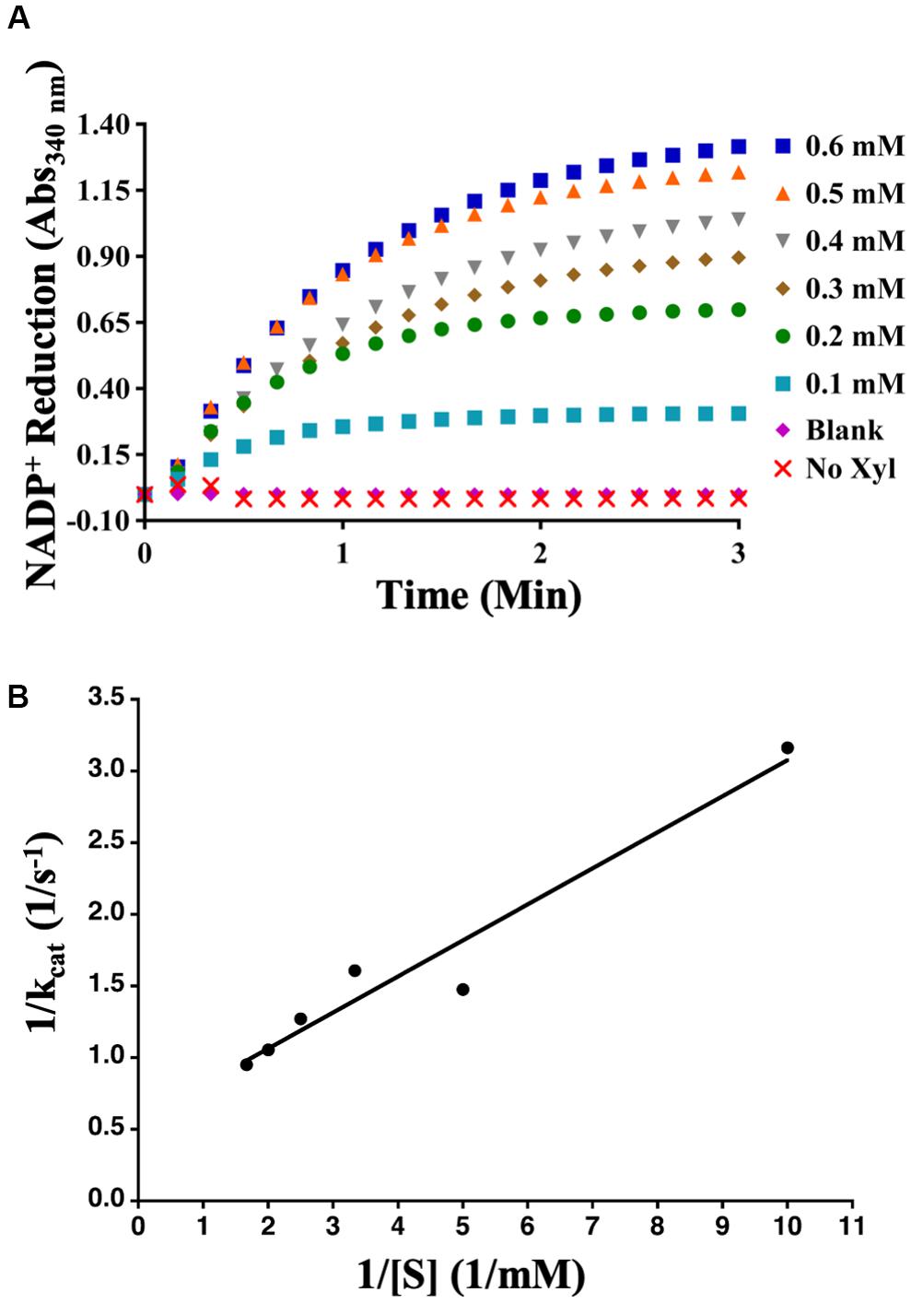
Figure 3. Recombinant AcDCXR oxidation activity. Xylitol oxidation by cowpea aphid recombinant AcDCXR. (A) Various concentrations of NADP+ were used to oxidize 200 mM xylitol in the presence of 10 μg of AcDCXR. Reactions containing no AcDCXR (Blank) or no xylitol (Xyl) were used as controls. (B) Lineweaver-Burk plot of xylitol oxidation. Data represent average of two technical replicates from a single experiment. The experiment was repeated once with similar results.
To determine whether AcDCXR was able to use methylglyoxal as a substrate, we tested the reduction of methylglyoxal by spectroscopically measuring the decrease in absorption of concomitant NADPH oxidation at 340 nm. We found that AcDCXR was able to reduce methylglyoxal in a concentration-dependent manner (Figure 4A). Analysis of the Lineweaver-Burke plot data determined the enzymatic constants to be: kcat = 0.23 s–1, a Km = 1.3 mM and a Vmax = 13.8 μM/min (Figure 4B). The control reactions, in the presence of AcDCXR and absence of a substrate, showed neither oxidation nor reduction (Figures 3A, 4A). Similarly, the control reactions in the absence of the enzyme showed neither oxidation nor reduction, indicating the AcDCXR’s presence was necessary to complete the reactions (Figures 3A, 4A). The kinetic constants in AcDCXR show that, in vitro, it was more efficient oxidizing xylitol with a kcat/Km of 3.32 mM–1 s–1 compared to reducing methylglyoxal that had only a kcat/Km of 0.174 mM–1 s–1, nearly a 20-fold difference in activity.
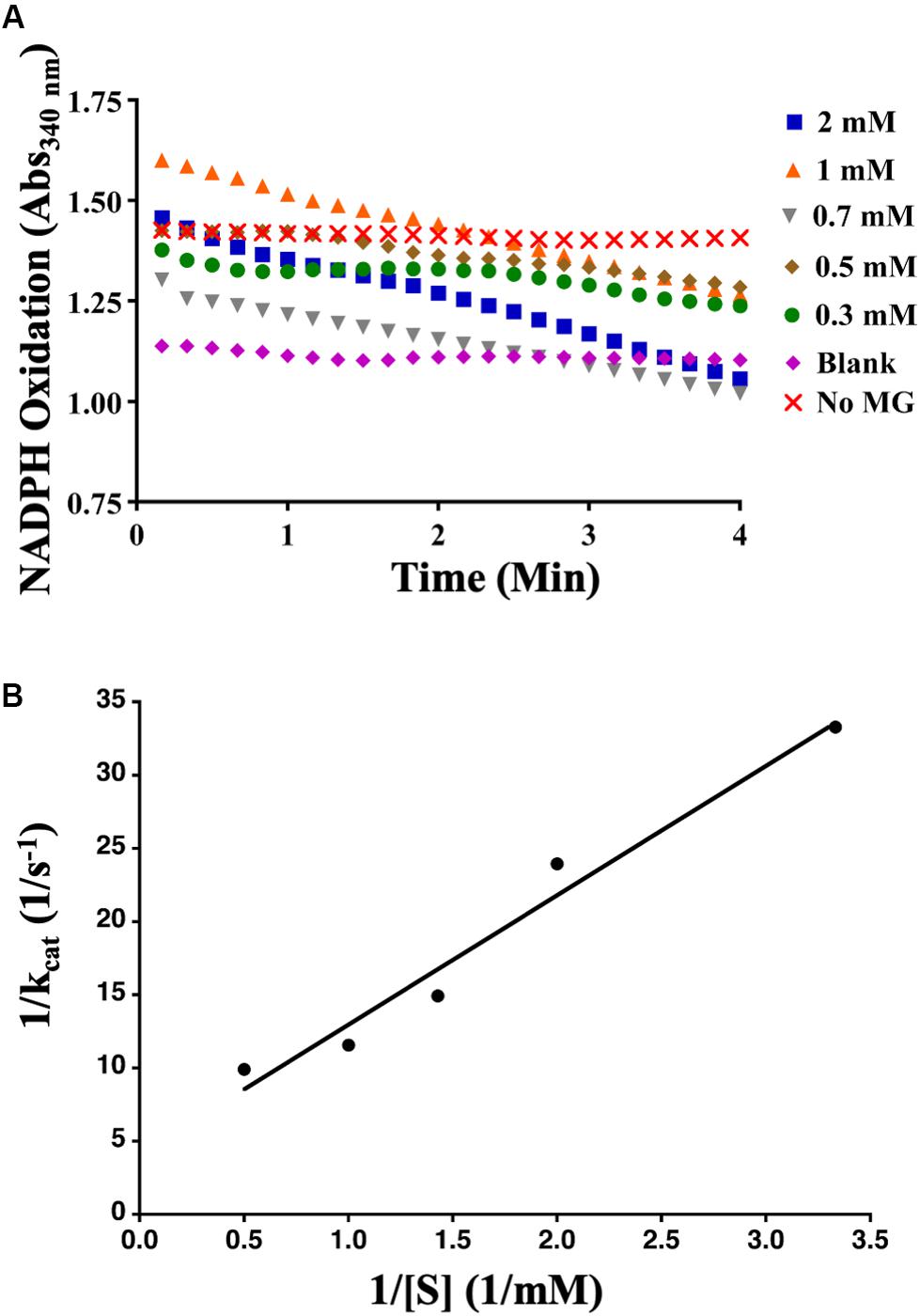
Figure 4. Recombinant AcDCXR reduction activity. Methylglyoxal reduction by cowpea aphid recombinant AcDCXR. (A) Various concentrations of methylglyoxal were reduced with 200 mM NADPH in the presence of 10 μg of AcDCXR. Reactions containing no AcDCXR (Blank) or no methylglyoxal (MG) were used as controls. (B) Lineweaver-Burk plot of methylglyoxal reduction. Data represent average of two technical replicates from a single experiment. The experiment was repeated once with similar results.
To functionally evaluate the role of AcDCXR on cowpea aphid colonization, AcDCXR was cloned into the binary vector pEAQ-DEST1 for Agrobacterium-mediated transient expression. Since Agrobacterium-mediated transient expression in cowpea has not yet been developed, pea plants were used for this experiment. Pea is a host for cowpea aphid and has been previously used successfully in transient expression experiments for evaluation of aphid effectors (Guy et al., 2016). Using the same cultivar of pea cv ZP1130, we first transiently expression GFP using A. tumefaciens strain AGL01. Monitoring GFP expression by western blot analysis, GFP was detected as early as 2 days after agroinfiltration and lasted at least for 10 days (Supplementary Figure 3). Based on the GFP expression in pea, a cowpea aphid bioassay was developed.
Aphid bioassays were performed to evaluate the effect of AcDCXR overexpression in pea plants on cowpea aphid. Plants were agroinfiltrated with AcDCXR or GFP control constructs as described earlier for the western blot analysis. A day post infiltration (dpi), adult cowpea aphids, maintained on pea cv ZP1130, were placed on a leaf, at the site of the infiltration, in a clip cage. After 24 h (2 dpi), all adult aphids were removed and six newborn nymphs were left at the infiltration site. At ten dpi, similar number of aphids were counted on GFP and AcDCXR infiltrated leaves indicating no effect on nymph survival rate (GLM, Chisq = 0.034, P = 0.854) (Figure 5A). To evaluate the fecundity of these aphids, one aphid per cage was transferred to a freshly agroinfiltrated (2 dpi) plant, with the same construct, and aphid survival and fecundity was monitored 4 and 7 days later. Sixteen days after initiation of the aphid bioassay, no difference in adult survival was detected between aphids feeding on AcDCXR compared to those feeding on the GFP infiltrated leaves (GLM, Chisq = 0.367, P = 0.544) (Figure 5B). However, a significant difference (GLM, Chisq = 16.901, P < 0.001) in aphid fecundity was observed between the aphids feeding on AcDCXR compared to those feeding on the GFP control indicating a role for AcDCXR in cowpea aphid colonization (Figure 5C). To determine the subcellular localization of AcDCXR in planta, AcDCXR was cloned into the binary vector pCAMBIA-1300-mScarlet and used in Agrobacterium-mediated transient expression in N. benthiamana. pCAMBIA-1300-AcDCXR-mScarlet was co-infiltrated with a GFP construct. As expected, GFP was detected throughout the cell including the nucleus, while AcDCXR-mScarlet was localized to the cytoplasm (Figure 6).
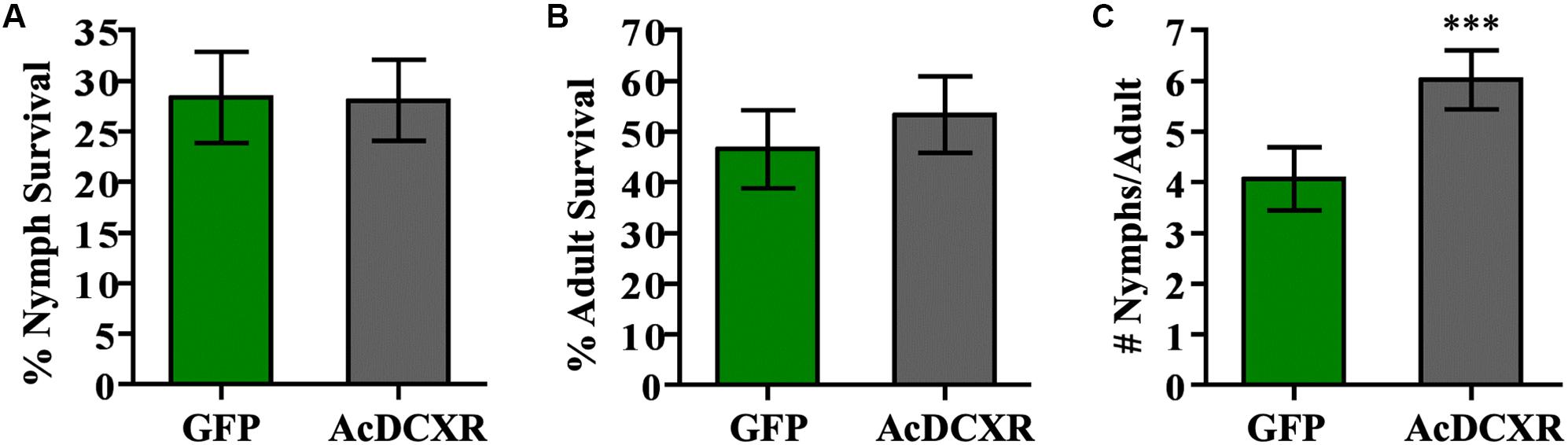
Figure 5. AcDCXR effect on aphid performance. Agrobacterium tumefaciens strain AGL01 was used to transiently express pEAQ-HT-DEST1-GFP and pEAQ-HT-DEST1-AcDCXR in Pisum sativum cv. ZP1130. Adult cowpea aphid adults were placed the infiltration site to lay nymphs and removed 24 h later. (A) The survival rate of the nymphs after 8 days on the site of infiltration. (B,C) A single adult was transferred to a new infiltration site of the same construct and the (B) survival of the adult and (C) fecundity were monitored. Graphs show the mean with error bars representing ± SE of the mean for n = 43 for GFP and n = 45 for AcDCXR from three independent experiments. ∗∗∗P < 0.001 as determined by generalized linear models (GLM).
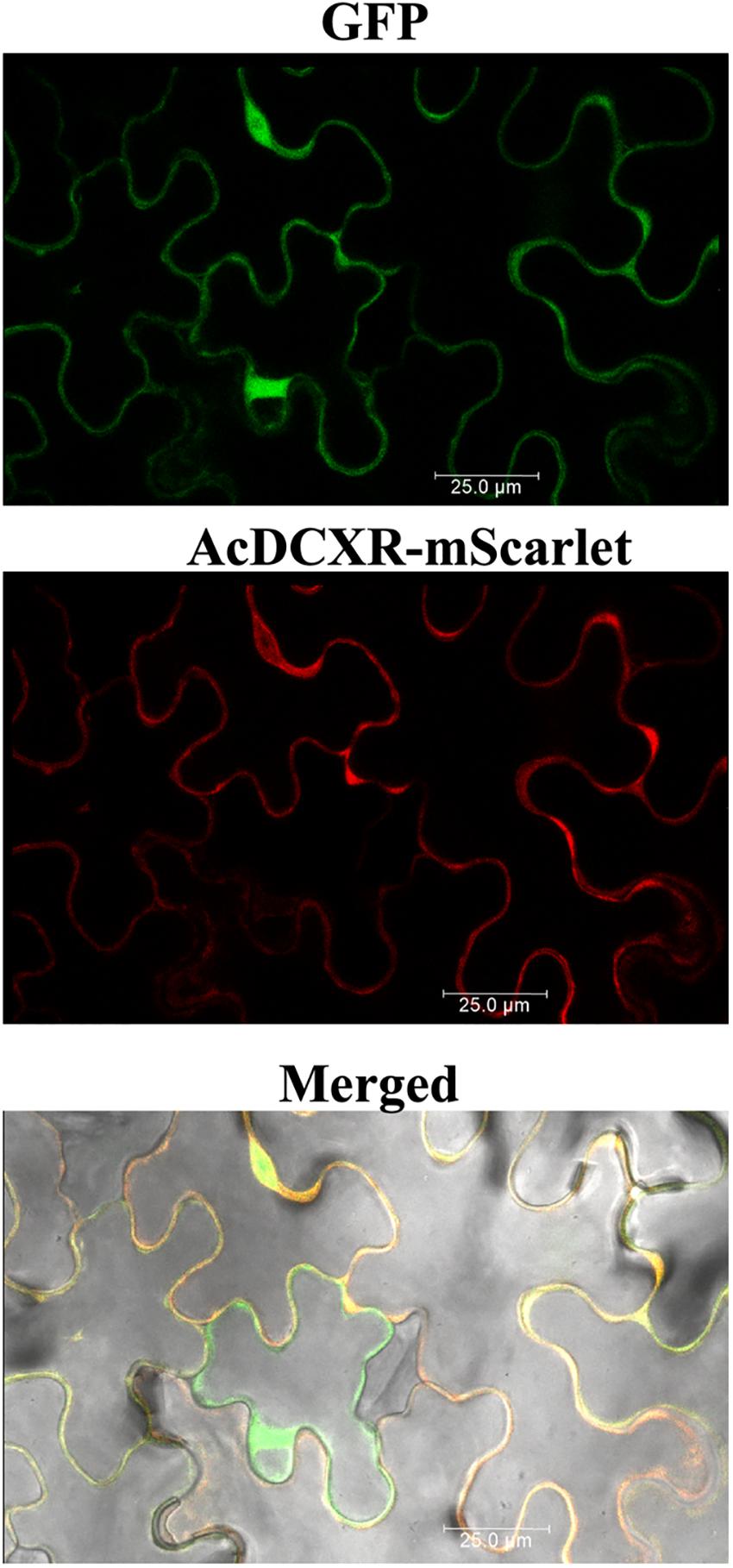
Figure 6. In planta subcellular localization of the recombinant AcDCXR. A. tumefaciens strain GV3101 containing pCAMBIA-1300-GFP or pCAMBIA-1300-AcDCXR-mScarlet were co-infiltrated into N. benthamiana leaves. Three days after agroinfiltration, leaf epidermal cells were used in confocal microscopy.
Methylglyoxal has been shown to accumulate in multiple plant species when exposed to abiotic stresses (Yadav et al., 2005a; Hossain et al., 2009; Mustafiz et al., 2014). Recently, it was also shown that methylglyoxal accumulates in plants exposed to biotic stresses (Melvin et al., 2017). To assess if methylglyoxal also accumulates by aphid infestation, methylglyoxal levels in cowpea and pea plants were monitored. A day after infestation of cowpea plants to cowpea aphids, a significantly higher (multiple comparisons, z = 2.812, P = 0.015) levels of methylglyoxal were detected in the infested leaves compared to the uninfested control leaves (Figure 7A). Methylglyoxal levels remained significantly higher (multiple comparisons, z = 3.832, P < 0.001) on day 2 but reduced to pre-infective levels on day 3 (multiple comparisons, z = 1.479, P = 0.208) (Figure 7A). A similar trend of methylglyoxal accumulation was detected in pea leaves exposed to cowpea aphids indicating that cowpea aphid feeding induces methylglyoxal levels irrespective of the host species (Figure 7B).
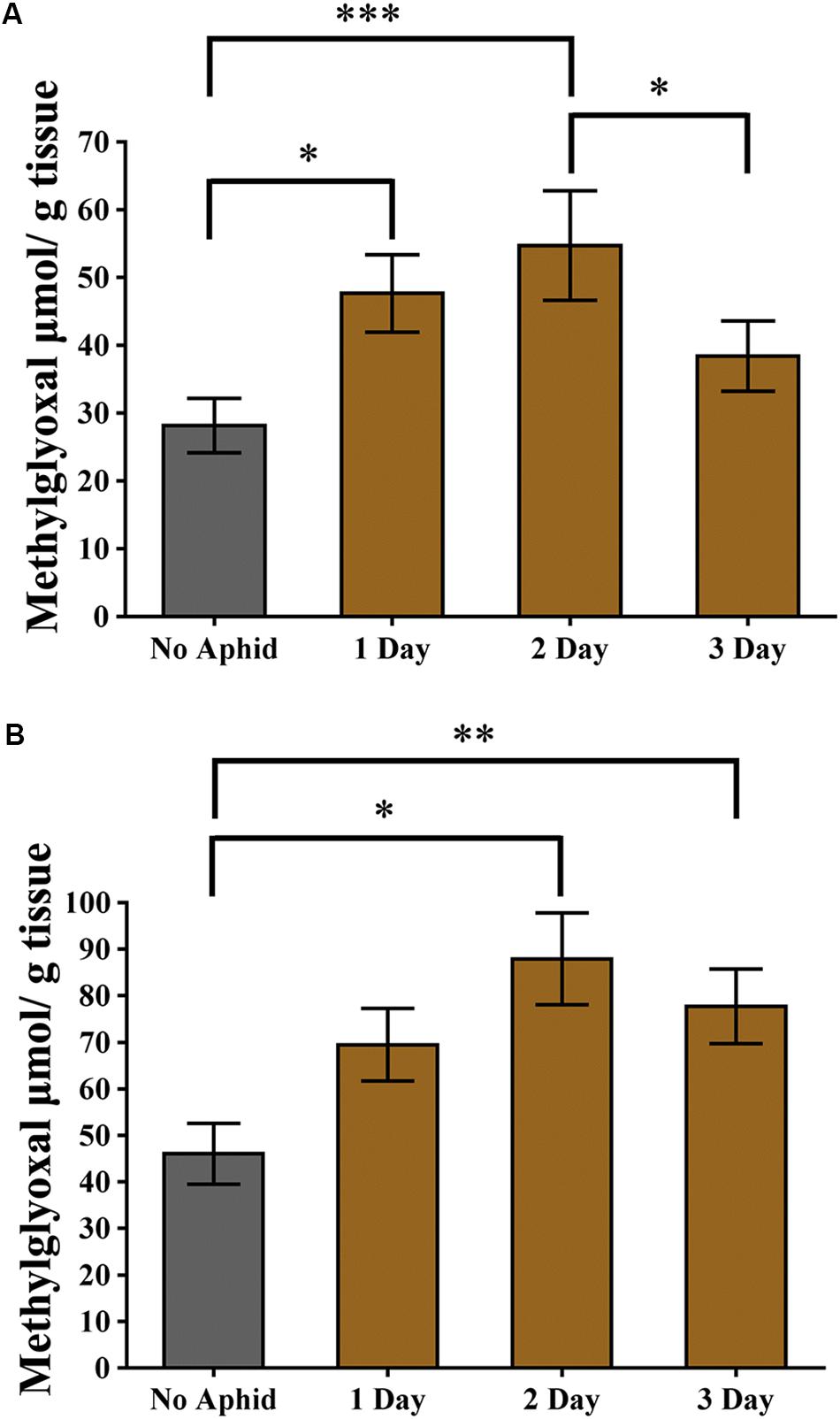
Figure 7. Methylglyoxal levels induced by aphid infestation. (A) Cowpea and (B) pea plants were exposed to a heavy infestation of cowpea aphids. Leaves were harvested at 1, 2, and 3 days post infestation. Uninfested plants of the same age were used as controls. Graphs show the mean with error bars representing ± SE of the mean of n = 6 for cowpea, from two independent experiments, and n = 3 for pea, from a single experiment, with two technical replicates each. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 as determined by nested ANOVA followed by multiple comparisons of means.
We carried out proteomics analysis to identify the salivary protein composition of a population of cowpea aphid from California. The identified proteins had a diverse range of functions including some that are uncharacterized. We were conservative in assessing the salivary proteome and used strict cut-off measures to identify the proteins. Nevertheless, we identified 149 non-redundant proteins. Previously, the salivary proteome from an African cowpea aphid population was reported (Loudit et al., 2018). The majority of the proteins identified in our study were not reported from this African population suggesting that our approach allowed us to identify higher numbers of proteins. While the cowpea aphid saliva in this work was collected in water, the African cowpea aphid saliva was collected in a sucrose-based diet and required clean up steps before undergoing mass spectrometry and that could have contributed to the low number of proteins identified in the saliva. Interestingly, both studies did not identify a set of functionally characterized aphid effectors such as Armet, Me23, Ap25, Mp2, Mp55 (Atamian et al., 2013; Pitino and Hogenhout, 2013; Elzinga et al., 2014; Wang et al., 2015; Guy et al., 2016). While in our study we identified Me10/Mp58 and SHP, the structural sheath protein, these two proteins were not identified in the African cowpea aphid saliva (Carolan et al., 2009; Chaudhary et al., 2015). The well characterized effector C002, was reported in the African population and not in this work (Mutti et al., 2006, 2008; Pitino and Hogenhout, 2013; Elzinga et al., 2014; Loudit et al., 2018). Although peptides for C002 and two additional effectors, Mp1 and MIF1, were detected in the saliva of the California cowpea aphids, this work, they did not fulfil the criteria used in our selection (Harmel et al., 2008; Naessens et al., 2015).
Unlike the salivary proteome of the African cowpea aphid, there were no proteins identified from secondary symbionts in the California cowpea aphid saliva (Loudit et al., 2018). The only bacterial proteins identified in the California cowpea aphid salivary proteome were from the primary endosymbiont Buchnera aphidicola, the chaperonin GroEL and GroES. GroEL has been previously identified in the saliva of several aphid species including the cowpea aphid (Chaudhary et al., 2014, 2015; Vandermoten et al., 2014; Loudit et al., 2018). GroEL is an aphid-associated molecular pattern triggering immune responses in plants (Chaudhary et al., 2014).
Our work was limited by the absence of a cowpea aphid genome and a gland/head specific transcriptome that could have been used for the peptide searches. In addition, homologous sequences from different aphid species were used in the secretion prediction analyses including some originating from transcriptomes that could have been truncated. Therefore, the number of proteins predicted for secretion, 46 out of 149 (30.9%), based on the bioinformatic programs SignalP and SecretomeP, are likely an underestimate (Table 1). Previous work describing salivary proteome from aphids with genome sequences and gland/head specific RNAseq generated sequences, also identified a large number of proteins from aphid saliva, collected in sugar and amino acid-based diets, with no prediction for secretion (Thorpe et al., 2016; Boulain et al., 2018). Boulain et al. (2018) reported 37/51 (72.5%) of the pea aphid salivary proteins with a secretion prediction. Thorpe et al. (2016), studying three different aphid species, green peach aphid, black cherry aphid, and bird cherry-oat aphid, reported only 61/204 (30%) secretion prediction of the identified salivary proteins. Taken together, this information indicates that the current bioinformatic prediction programs are likely limited in their ability to identify aphid secreted proteins.
Here we reported the use of a machine learning plant-pathogenic fungi effector prediction program, EffectorP, for prediction of aphid effectors (Sperschneider et al., 2016, 2018). We confirmed the use of EffectorP as a possible program for identifying aphid effector proteins by successfully subjecting the well-characterized aphid effectors C002 and Me10 to EffectorP analysis (Mutti et al., 2008; Atamian et al., 2013; Pitino and Hogenhout, 2013; Chaudhary et al., 2019). Interestingly, EffectorP predicted 20/149 of the cowpea aphid proteins as effectors. Among these 20 proteins, is the functionally characterized Me10 effector and three proteins which have been predicted for effector function (Atamian et al., 2013; Elzinga et al., 2014; Thorpe et al., 2016; Chaudhary et al., 2019). Orthologs of Me10 have been identified in multiple aphid species. Me10 has been detected in plant tissues fed on by aphids and expression of Me10 in plants has been shown to enhance the performance of potato aphid on tomato and green peach aphid on N. benthamiana (Atamian et al., 2013; Chaudhary et al., 2015, 2019). In addition, Me10 was shown to interact with the tomato scaffold protein Fourteen-Three-Three isoform 7 (TFT7) and predicted to interfere with a mitogen-activated protein kinase defense signaling pathway (Chaudhary et al., 2019).
The remaining three previously predicted putative effectors are carbonic anhydrase, superoxide dismutase, and peptidyl-prolyl cis-trans isomerase (PPIase). The latter two proteins were identified in the proteomes of the pea aphid salivary glands (Carolan et al., 2011). While carbonic anhydrases have been identified in aphid saliva, superoxide dismutase and PPIase have not been previously reported in aphid saliva (Rao et al., 2013; Nicholson and Puterka, 2014; Chaudhary et al., 2015; Loudit et al., 2018). A carbonic anhydrase and a superoxide dismutase have been shown to be under positive selection further implicating these proteins as effectors (Thorpe et al., 2016). While clear roles for carbonic anhydrases and PPIases have not been characterized in plant immune responses, superoxide dismutases are attributed to detoxify reactive oxygen species (ROS), the well-known defense signaling molecule.
Among the EffectorP identified putative effector proteins, that had not been previously identified in aphid saliva or as a putative effector, is AcDCXR (Table 1, Supplementary Table 1). DCXR has been identified in the pea aphid salivary gland but has not been reported in the saliva of this aphid species (Carolan et al., 2011; Boulain et al., 2018). Interestingly, pea aphid homolog of AcDCXR as well as homologs from five additional aphid species with genome sequences, do not have a secretion signal peptide. The homolog from the cotton melon aphid does have a secretion signal suggesting that DCXR is one of the differential pest arsenals utilized by a subset of aphid species. An increase in DCXR accumulation was reported in a virulent biotype of greenbug infesting resistant wheat (Pinheiro et al., 2014). Additionally, enhanced accumulation of DCXR in response to heat/UV stress as well as predation by parasitoids in the potato aphid were reported from whole insects (Nguyen et al., 2008, 2009). Taken together, these information suggest that aphids may have evolved different roles for DCXR to deal with stress conditions in the plant and within the aphid itself.
In mammals DCXRs are reported to be oxidoreductases for monosaccharides and dicarbonyls. Human DCXR was first discovered while investigating the disease pentosuria and found that an enzymatic defect in DCXR was the cause of the high excretion of L-xylulose. This lead to the conclusion that L-xylulose is a possible substrate of DCXR (Wang and Van Eys, 1970). DCXR has been shown also to catalyze reactions with other sugars. For example, xylitol is a sugar alcohol that is transported through the phloem as a carbon source (Lewis and Smith, 1967; Lemoine et al., 2013). Xylitol can be converted to xylulose and be used in the pentose phosphate pathway to generate glycolytic intermediates as a source of energy. Since the AcDCXR catalyzes the reversible reaction between xylulose and xylitol, the enzyme may provide the aphid an additional mode of generating energy.
Diacetyl/L-xylulose reductases also participates in the reductive metabolism of carbonyls. In this role, the enzyme is considered as a defense mechanism against harmful carbonyls (Nakagawa et al., 2002; Ebert et al., 2015; Yang et al., 2017). These molecules lead to formation of AGEs by reacting with lysine, cysteine and arginine, thus inactivating proteins (Thornalley, 2006; Ahmed and Thornalley, 2007). One of these harmful carbonyls is methylglyoxal which is reactive α-β-dicarbonyl ketoaldehyde. Interestingly, methylglyoxal has been shown to accumulate in a number of plant species under various abiotic stresses (Yadav et al., 2005a; Hossain et al., 2009; Mustafiz et al., 2014; Rahman et al., 2015; Borysiuk et al., 2018). Recently, methylglyoxal has also been implicated in biotic stresses. Increases in methylglyoxal levels were detected in tobacco plants exposed to the bacterium Pseudomonas syringae, or the Mungbean yellow mosaic virus, or to the fungus Alternaria alternata (Melvin et al., 2017). In addition, exogenous application of methylglyoxal in wheat and rice plants upregulated antioxidant and defense-related genes indicating a role for methylglyoxal in plant defense (Kaur et al., 2015; Li et al., 2017). In this work we showed that aphid feeding also enhanced accumulation of methylglyoxal in cowpea and pea, suggesting methylglyoxal also functions in aphid defense. Since methylglyoxal levels in aphid infested leaves were mostly transient, this suggests that aphids are able to counteract methylglyoxal accumulation possibly through AcDCXR activity.
Transient expression of AcDCXR indicates that this enzyme is localized in the plant cell cytoplasm. Likewise, both AcDCXR substrates tested in this study, methylglyoxal and xylitol/xylulose, are also located in the cell cytoplasm. In plants, the pentose phosphate pathway where xylitol/xylulose are used, takes place in both the cytoplasm and plastids. Methylglyoxal is generated in multiple pathways in the cytoplasm and in various organelles (Phillips and Thornalley, 1993; Dennis and Blakeley, 2000; Kruger and von Schaewen, 2003).
The transient expression of AcDCXR increased the fecundity of the cowpea aphid most likely due to its effect on one or both of these two substrates; either by increasing the obtained nutrient content and/or through diminishing defense responses. This increase in fecundity was seen despite no differences in the survival of both adult and nymphal stages of the aphid. Transient or stable overexpression of a number of aphid effectors in various plant species including, Arabidopsis, tomato, pea and N. benthamiana also yielded increases in aphid fecundity but no effect on aphid survival suggesting that overexpression of multiple effectors may be needed to observe a pronounced change in aphid survival.
In this work, using a classical and a novel bioinformatics programs, SignalP and EffectorP, respectively, we identified a novel aphid effector, AcDCXR. The functional annotation of DCXR and in vitro biochemical analysis of AcDCXR lead us to identify methylglyoxal as a potential novel metabolite involved in aphid defense. Therefore, identification of novel effectors may lead to the discovery of yet unknown defense pathways that may lead to novel approaches to engineer pest/pathogen resistance in crops.
The datasets generated for this study can be found in MassIVE, https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp, ID: PXD017323
IK conceived the project. IK, SD, and AS designed the experiments. JM conducted the experiments. IK, JM, SD, and QC analyzed the data. IK and JM wrote the manuscript with help from all other authors.
This research is partly funded by the USDA National Institute of Food and Agriculture Hatch project 1017522 to IK.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Monika Ostaszewska-Bugajska (University of Warsaw) for sharing methylglyoxal evaluation protocol, Dr. Philip Roberts (UC Riverside) for the cowpea aphids, and Dr. Jiangman He (UC Riverside) for help with confocal microscopy and constructing the pCAMBIA-1300-GW-mScarlet vector.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00605/full#supplementary-material
Abdellatef, E., Will, T., Koch, A., Imani, J., Vilcinskas, A., and Kogel, K. H. (2015). Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae. Plant Biotechnol. J. 13, 849–857. doi: 10.1111/pbi.12322
Agunbiade, T. A., Sun, W., Coates, B. S., Djouaka, R., Tamo, M., Ba, M. N., et al. (2013). Development of reference transcriptomes for the major field insect pests of cowpea: a toolbox for insect pest management approaches in west Africa. PLoS One 8:e79929. doi: 10.1371/journal.pone.0079929
Ahmed, N., and Thornalley, P. J. (2007). Advanced glycation endproducts: what is their relevance to diabetic complications? Diabetes Obes. Metab. 9, 233–245. doi: 10.1111/j.1463-1326.2006.00595.x
Armenteros, J. J. A., Tsirigos, K. D., Sonderby, C. K., Peterson, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Atamian, H. S., Chaudhary, R., Dal Cin, V., Bao, E., Girke, T., and Kaloshian, I. (2013). In planta expression or delivery of potato aphid Macrosiphum euphorbiae effectors Me10 and Me23 enhances aphid fecundity. Mol. Plant Microbe Interact. 26, 67–74. doi: 10.1094/Mpmi-06-12-0144-Fi
Bendtsen, J. D., Jensen, L. J., Blom, N., von Heijne, G., and Brunak, S. (2004a). Feature based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17, 349–356. doi: 10.1093/protein/gzh037
Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. (2004b). Improved prediction of signal peptides: signalp 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
BIPAA (2017). AphidBase. Available: https://bipaa.genouest.org/is/aphidbase/ (accessed June 27, 2017).
Blackman, R. L., and Eastop, V. F. (2000). Aphids on the World’s Crops. New York, NY: John Wiley & Sons Inc.
Borysiuk, K., Ostaszewska-Bugajska, M., Vaultier, M. N., Hasenfratz-Sauder, M. P., and Szal, B. (2018). Enhanced formation of methylglyoxal-derived advanced glycation end products in Arabidopsis under ammonium nutrition. Front. Plant Sci. 9:667. doi: 10.3389/fpls.2018.00667
Bos, J. I. B., Prince, D., Pitino, M., Maffei, M. E., Win, J., and Hogenhout, S. A. (2010). A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 6:e1001216. doi: 10.1371/journal.pgen.1001216
Boulain, H., Legeai, F., Guy, E., Morliere, S., Douglas, N. E., Oh, J., et al. (2018). Fast evolution and lineage-specific gene family expansions of aphid salivary effectors driven by interactions with host-plants. Genome Biol. Evol. 10, 1554–1572. doi: 10.1093/gbe/evy097
Calandra, T. (2003). Macrophage migration inhibitory factor and host innate immune responses to microbes. Scand. J. Infect. Dis. 35, 573–576. doi: 10.1080/00365540310016277
Carolan, J. C., Caragea, D., Reardon, K. T., Mutti, N. S., Dittmer, N., Pappan, K., et al. (2011). Predicted effector molecules in the salivary secretome of the pea aphid (Acyrthosiphon pisum): a dual transcriptomic/proteomic approach. J. Proteome Res. 10, 1505–1518. doi: 10.1021/pr100881q
Carolan, J. C., Fitzroy, C. I. J., Ashton, P. D., Douglas, A. E., and Wilkinson, T. L. (2009). The secreted salivary proteome of the pea aphid Acyrthosiphon pisum characterised by mass spectrometry. Proteomics 9, 2457–2467. doi: 10.1002/pmic.200800692
Chan, C. K., Forbes, A. R., and Raworth, D. A. (1991). Aphid-Transmitted Viruses and their Vectors of the World. Ottawa: Agriculture and Agri-Food Canada.
Chaudhary, R., Atamian, H. S., Shen, Z., Briggs, S. P., and Kaloshian, I. (2014). GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc. Natl. Acad. Sci. U.S.A. 111, 8919–8924. doi: 10.1073/pnas.1407687111
Chaudhary, R., Atamian, H. S., Shen, Z., Briggs, S. P., and Kaloshian, I. (2015). Potato aphid salivary proteome: enhanced salivation using resorcinol and identification of aphid phosphoproteins. J. Proteome Res. 14, 1762–1778. doi: 10.1021/pr501128k
Chaudhary, R., Peng, H. C., He, J. M., MacWilliams, J., Teixeira, M., Tsuchiya, T., et al. (2019). Aphid effector Me10 interacts with tomato TFT7, a 14-3-3 isoform involved in aphid resistance. New Phytol. 221, 1518–1528. doi: 10.1111/nph.15475
Chen, L., Wang, T., Wang, X., Sun, B. B., Li, J. Q., Liu, D. S., et al. (2009). Blockade of advanced glycation end product formation attenuates bleomycin-induced pulmonary fibrosis in rats. Respir. Res. 10:55. doi: 10.1186/1465-9921-10-55
Choudhary, A., Hussein, A., Samota, R. G., and Nehra, S. (2017). Effect of biotic and abiotic factors on the incidence of aphid, Aphis craccivora Koch on cowpea. J. Pharmacogn. Phytochem. 6, 1587–1590.
Cooper, W. R., Dillwith, J. W., and Puterka, G. J. (2010). Salivary proteins of Russian wheat aphid (Hemiptera: Aphididae). Environ. Entomol. 39, 223–231. doi: 10.1603/En09079
Cooper, W. R., Dillwith, J. W., and Puterka, G. J. (2011). Comparisons of salivary proteins from five aphid (Hemiptera: Aphididae) species. Environ. Entomol. 40, 151–156. doi: 10.1603/En10153
Dennis, D. T., and Blakeley, S. D. (2000). “Carbohydrate metabolism,” in Biochemistry & Molecular Biology of Plants, eds B. B. Buchanan, W. Gruissem, and R. L. Jones (Waldorf, MD: American Society of Plant Physiologists), 630–675.
Ebert, B., Kisiela, M., and Maser, E. (2015). Human DCXR - another ‘moonlighting protein’ involved in sugar metabolism, carbonyl detoxification, cell adhesion and male fertility? Biol. Rev. 90, 254–278. doi: 10.1111/brv.12108
Elzinga, D. A., De Vos, M., and Jander, G. (2014). Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol. Plant Microbe Interact. 27, 747–756. doi: 10.1094/Mpmi-01-14-0018-R
Gkogkolou, P., and Bohm, M. (2012). Advanced glycation end products: key players in skin aging? Dermatoendocrinol. 4, 259–270. doi: 10.4161/derm.22028
Gotz, S., Garcia-Gomez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Guy, E., Boulain, H., Aigu, Y., Le Pennec, C., Chawki, K., Morliere, S., et al. (2016). Optimization of agroinfiltration in Pisum sativum provides a new tool for studying the salivary protein functions in the pea aphid complex. Front. Plant Sci. 7:1171. doi: 10.3389/fpls.2016.01171
Harmel, N., Letocart, E., Cherqui, A., Giordanengo, P., Mazzucchelli, G., Guillonneau, F., et al. (2008). Identification of aphid salivary proteins: a proteomic investigation of Myzus persicae. Insect Mol. Biol. 17, 165–174. doi: 10.1111/j.1365-2583.2008.00790.x
Hoque, M. A., Uraji, M., Torii, A., Banu, M. N., Mori, I. C., Nakamura, Y., et al. (2012). Methylglyoxal inhibition of cytosolic ascorbate peroxidase from Nicotiana tabacum. J. Biochem. Mol. Toxicol. 26, 315–321. doi: 10.1002/jbt.21423
Hoque, T. S., Hossain, M. A., Mostofa, M. G., Burritt, D. J., Fujita, M., and Tran, L. S. (2016). Methylglyoxal: an emerging signaling molecule in plant abiotic stress responses and tolerance. Front. Plant Sci. 7:1341. doi: 10.3389/fpls.2016.01341
Hossain, M. A., Zakir, H. M., and Fujita, M. (2009). Stress-induced changes of methylglyoxal level and glyoxalase I activity in pumpkin seedlings and cDNA cloning of glyoxalase I gene. J. Crop Sci. 3:53.
International Aphid Genomics Consortium (2010). Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. doi: 10.1371/journal.pbio.1000313
Ishikura, S., Usami, N., El-Kabbani, O., and Hara, A. (2003). Structural determinant for cold inactivation of rodent L-xylulose reductase. Biochem. Biophys. Res. Comm. 308, 68–72. doi: 10.1016/s0006-291x(03)01336-6
Jackai, L., and Daoust, R. (1986). Insect pests of cowpeas. Annu. Rev. Entomol. 31, 95–119. doi: 10.1146/annurev.en.31.010186.000523
Jiang, R. H., Tripathy, S., Govers, F., and Tyler, B. M. (2008). RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. U.S.A. 105, 4874–4879. doi: 10.1073/pnas.0709303105
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kaloshian, I., and Walling, L. L. (2016). Hemipteran and dipteran pests: effectors and plant host immune regulators. J. Integr. Plant Biol. 58, 350–361. doi: 10.1111/jipb.12438
Kamphuis, L. G., Gao, L., and Singh, K. B. (2012). Identification and characterization of resistance to cowpea aphid (Aphis craccivora Koch) in Medicago truncatula. BMC Plant Biol. 12:101. doi: 10.1186/1471-2229-12-101
Kaur, C., Kushwaha, H. R., Mustafiz, A., Pareek, A., Sopory, S. K., and Singla-Pareek, S. L. (2015). Analysis of global gene expression profile of rice in response to methylglyoxal indicates its possible role as a stress signal molecule. Front. Plant Sci. 6:682. doi: 10.3389/fpls.2015.00682
Kettles, G. J., and Kaloshian, I. (2016). The potato aphid salivary effector Me47 is a glutathione-S-transferase involved in modifying plant responses to aphid infestation. Front. Plant Sci. 7:1142. doi: 10.3389/fpls.2016.01142
Kizer, J. R., Benkeser, D., Arnold, A. M., Ix, J. H., Mukamal, K. J., Djousse, L., et al. (2014). Advanced glycation/glycoxidation endproduct carboxymethyl-lysine and incidence of coronary heart disease and stroke in older adults. Atherosclerosis 235, 116–121. doi: 10.1016/j.atherosclerosis.2014.04.013
Krogh, A., Larsson, B., von Heijne, G., and Sonhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kruger, N. J., and von Schaewen, A. (2003). The oxidative pentose phosphate pathway: structure and organisation. Curr. Opin. Plant Biol. 6, 236–246. doi: 10.1016/s1369-5266(03)00039-6
Lemoine, R., La Camera, S., Atanassova, R., Dedaldechamp, F., Allario, T., Pourtau, N., et al. (2013). Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 4:272. doi: 10.3389/fpls.2013.00272
Lewis, D. H., and Smith, D. C. (1967). Sugar alcohols (polyols) in fungi and green plants. New Phytol. 66, 185–204. doi: 10.1111/j.1469-8137.1967.tb05998.x
Li, Z. G. (2016). Methylglyoxal and glyoxalase system in plants: old players, new concepts. Bot. Rev. 82, 183–203.
Li, Z. G., Duan, X. Q., Min, X., and Zhou, Z. H. (2017). Methylglyoxal as a novel signal molecule induces the salt tolerance of wheat by regulating the glyoxalase system, the antioxidant system and osmolytes. Protoplasma 254, 1995–2006. doi: 10.1007/s00709-017-1094-z
Lindholm, P., Peranen, J., Andressoo, J. O., Kalkkinen, N., Kokaia, Z., Lindvall, O., et al. (2008). MANF is widely expressed in mammalian tissues and differently regulated after ischemic and epileptic insults in rodent brain. Mol. Cell. Neurosci. 39, 356–371. doi: 10.1016/j.mcn.2008.07.016
Lindholm, P., Voutilainen, M. H., Lauren, J., Peranen, J., Leppanen, V. M., Andressoo, J. O., et al. (2007). Novel neurotrophic factor CDNF protects and rescues midbrain dopamine neurons in vivo. Nature 448, 73–77. doi: 10.1038/nature05957
Loudit, S. M. B., Bauwens, J., and Francis, F. (2018). Cowpea aphid-plant interactions: endosymbionts and related salivary protein patterns. Entomol. Exp. Appl. 166, 460–473. doi: 10.1111/eea.12687
Mathers, T. C., Chen, Y., Kaithakottil, G., Legeai, F., Mugford, S. T., Baa-Puyoulet, P., et al. (2017). Rapid transcriptional plasticity of duplicated gene clusters enables a clonally reproducing aphid to colonise diverse plant species. Genome Biol. 18:27. doi: 10.1186/s13059-016-1145-3
Melvin, P., Bankapalli, K., D’Silva, P., and Shivaprasad, P. V. (2017). Methylglyoxal detoxification by a DJ-1 family protein provides dual abiotic and biotic stress tolerance in transgenic plants. Plant Mol. Biol. 94, 381–397. doi: 10.1007/s11103-017-0613-9
Misra, K., Banerjee, A. B., Ray, S., and Ray, M. (1996). Reduction of methylglyoxal in Escherichia coli K12 by an aldehyde reductase and alcohol dehydrogenase. Mol. Cell. Biochem. 156, 117–124. doi: 10.1007/bf00426333
Mostofa, M. G., Ghosh, A., Li, Z. G., Siddiqui, M. N., Fujita, M., and Tran, L. P. (2018). Methylglyoxal - a signaling molecule in plant abiotic stress responses. Free Radic. Biol. Med. 122, 96–109. doi: 10.1016/j.freeradbiomed.2018.03.009
Mustafiz, A., Ghosh, A., Tripathi, A. K., Kaur, C., Ganguly, A. K., Bhavesh, N. S., et al. (2014). A unique Ni2+-dependent and methylglyoxal-inducible rice glyoxalase I possesses a single active site and functions in abiotic stress response. Plant J. 78, 951–963. doi: 10.1111/tpj.12521
Mutti, N. S., Louis, J., Pappan, L. K., Pappan, K., Begum, K., Chen, M. S., et al. (2008). A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. U.S.A. 105, 9965–9969. doi: 10.1073/pnas.0708958105
Mutti, N. S., Park, Y., Reese, J. C., and Reeck, G. R. (2006). RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. J. Insect Sci. 6:38. doi: 10.1673/031.006.3801
Naessens, E., Dubreuil, G., Giordanengo, P., Baron, O. L., Minet-Kebdani, N., Keller, H., et al. (2015). A secreted MIF cytokine enables aphid feeding and represses plant immune responses. Curr. Biol. 25, 1898–1903. doi: 10.1016/j.cub.2015.05.047
Nakagawa, J., Ishikura, S., Asami, J., Isaji, T., Usami, N., Hara, A., et al. (2002). Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney. J. Biol. Chem. 277, 17883–17891. doi: 10.1074/jbc.M110703200
Nguyen, T. T., Boudreault, S., Michaud, D., and Cloutier, C. (2008). Proteomes of the aphid Macrosiphum euphorbiae in its resistance and susceptibility responses to differently compatible parasitoids. Insect Biochem. Mol. Biol. 38, 730–739. doi: 10.1016/j.ibmb.2008.04.005
Nguyen, T. T., Michaud, D., and Cloutier, C. (2009). A proteomic analysis of the aphid Macrosiphum euphorbiae under heat and radiation stress. Insect Biochem. Mol. Biol. 39, 20–30. doi: 10.1016/j.ibmb.2008.09.014
Nicholson, S. J., Hartson, S. D., and Puterka, G. J. (2012). Proteomic analysis of secreted saliva from Russian Wheat Aphid (Diuraphis noxia Kurd.) biotypes that differ in virulence to wheat. J. Proteomics 75, 2252–2268. doi: 10.1016/j.jprot.2012.01.031
Nicholson, S. J., Nickerson, M. L., Dean, M., Song, Y., Hoyt, P. R., Rhee, H., et al. (2015). The genome of Diuraphis noxia, a global aphid pest of small grains. BMC Genomics 16:429. doi: 10.1186/s12864-015-1525-1
Nicholson, S. J., and Puterka, G. J. (2014). Variation in the salivary proteomes of differentially virulent greenbug (Schizaphis graminum Rondani) biotypes. J. Proteomics 105, 186–203. doi: 10.1016/j.jprot.2013.12.005
Ofuya, T. (1995). Studies on the capability of Cheilomenes lunata (Fabricius) (Coleoptera: Coccinellidae) to prey on the cowpea aphid, Aphis craccivora Koch (Homoptera: Aphididae) in Nigeria. Agric. Ecosyst. Environ. 52, 35–38. doi: 10.1016/0167-8809(94)09006-S
Palgi, M., Greco, D., Lindstrom, R., Auvinen, P., and Heino, T. I. (2012). Gene expression analysis of Drosophilaa Manf mutants reveals perturbations in membrane traffic and major metabolic changes. BMC Genomics 13:134. doi: 10.1186/1471-2164-13-134
Palgi, M., Lindstrom, R., Peranen, J., Piepponen, T. P., Saarma, M., and Heino, T. I. (2009). Evidence that DmMANF is an invertebrate neurotrophic factor supporting dopaminergic neurons. Proc. Natl. Acad. Sci. U.S.A. 106, 2429–2434. doi: 10.1073/pnas.0810996106
Petre, B., and Kamoun, S. (2014). How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 12:e1001801. doi: 10.1371/journal.pbio.1001801
Phillips, S. A., and Thornalley, P. J. (1993). Formation of methylglyoxal and D-lactate in human red blood cells in vitro. Biochem. Soc. Trans. 21:163S. doi: 10.1042/bst021163s
Pinheiro, P., Bereman, M. S., Burd, J., Pals, M., Armstrong, S., Howe, K. J., et al. (2014). Evidence of the biochemical basis of host virulence in the greenbug aphid, Schizaphis graminum (Homoptera: Aphididae). J. Proteome Res. 13, 2094–2108. doi: 10.1021/pr4012415
Pitino, M., and Hogenhout, S. A. (2013). Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol. Plant Microbe Interact. 26, 130–139. doi: 10.1094/Mpmi-07-12-0172-Fi
R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
Rahman, A., Mostofa, M. G., Alam, M. M., Nahar, K., Hasanuzzaman, M., and Fujita, M. (2015). Calcium mitigates arsenic toxicity in rice seedlings by reducing arsenic uptake and modulating the antioxidant defense and glyoxalase systems and stress markers. Biomed Res. Int. 2015:340812. doi: 10.1155/2015/340812
Rao, S. A. K., Carolan, J. C., and Wilkinson, T. L. (2013). Proteomic profiling of cereal aphid saliva reveals both ubiquitous and adaptive secreted proteins. PLoS ONE 8:e57413. doi: 10.1371/journal.pone.0057413
Ray, A., Ray, S., Mukhopadhyay, S., and Ray, M. (2013). Methylglyoxal with glycine or succinate enhances differentiation and shoot morphogenesis in Nicotiana tabacum callus. Biol. Plant. 57, 219–223.
Remaudiere, G., and Remaudiere, M. (1997). Catalogue des Aphididae du Monde (Homoptera Aphidoidea). Paris: Institut National de la Recherche Agronomique.
Rodriguez, P. A., Escudero-Martinez, C., and Bos, J. I. B. (2017). An aphid effector targets trafficking protein VPS52 in a host-specific manner to promote virulence. Plant Physiol. 173, 1892–1903. doi: 10.1104/pp.16.01458
Sainsbury, F., Thuenemann, E. C., and Lomonossoff, G. P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693. doi: 10.1111/j.1467-7652.2009.00434.x
Sankaranarayanan, S., Jamshed, M., and Samuel, M. A. (2015). Degradation of glyoxalase I in Brassica napus stigma leads to self-incompatibility response. Nat. Plants 1:15185. doi: 10.1038/nplants.2015.185
Singh, B. B., Ehlers, J. D., Sharma, B., and Freire, F. R. (2002). “Recent progress in cowpea breeding,” in Proceedings of the World Cowpea Conference III, eds C. A. Fatokun, B. B. Tarawali, B. B. Singh, P. M. Kormawa, and M. Tamo (Ibadan: IITA), 22–40.
Sochor, M., Baquer, N. Z., and McLean, P. (1979). Glucose overutilization in diabetes: evidence from studies on the changes in hexokinase, the pentose phosphate pathway and glucuronate-xylulose pathway in rat kidney cortex in diabetes. Biochem. Biophys. Res. Commun. 86, 32–39. doi: 10.1016/0006-291x(79)90378-4
Sonnhammer, E. L., von Heijne, G., and Krogh, A. (1998). “A hidden Markov model for predicting transmembrane helices in protein sequences,” in Proceedings of the 6th International Conference on Intelligent Systems for Molecular Biology, Vol. 6, Montreal, 175–182.
Sorenson, J. (2009). “Aphids,” in Encyclopedia of Insects, 2nd Edn, eds V. Resh and R. Carde (San Diego, CA: Academic Press), 27–30.
Sperschneider, J., Dodds, P., Gardiner, D., Singh, K. B., and Taylor, J. M. (2018). Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 19, 2094–2110. doi: 10.1111/mpp.12682
Sperschneider, J., Dodds, P. N., Gardiner, D. M., Manners, J. M., Singh, K. B., and Taylor, J. M. (2015). Advances and challenges in computational prediction of effectors from plant pathogenic fungi. PLoS Pathog. 11:e1004806. doi: 10.1371/journal.ppat.1004806
Sperschneider, J., Gardiner, D. M., Dodds, P. N., Tini, F., Covarelli, L., Singh, K. B., et al. (2016). EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol. 210, 743–761. doi: 10.1111/nph.13794
Stergiopoulos, I., and de Wit, P. J. (2009). Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. doi: 10.1146/annurev.phyto.112408.132637
Thornalley, P. J. (2006). Advanced glycation end products in renal failure. J. Ren. Nutr. 16, 178–184. doi: 10.1053/j.jrn.2006.04.012
Thorpe, P., Cock, P. J., and Bos, J. (2016). Comparative transcriptomics and proteomics of three different aphid species identifies core and diverse effector sets. BMC Genomics 17:172. doi: 10.1186/s12864-016-2496-6
Thorpe, P., Escudero-Martinez, C. M., Cock, P. J. A., Eves-van den Akker, S., and Bos, J. I. B. (2018). Shared transcriptional control and disparate gain and loss of aphid parasitism genes. Genome Biol. Evol. 10, 2716–2733. doi: 10.1093/gbe/evy183
Timko, M., and Singh, B. B. (2008). “Cowpea, a multifunctional legume,” in Genomics of Tropical Crop Plants, eds P. H. Moore and R. Ming (New York, NY: Springer), 227–258.
Tjallingii, W. F. (2006). Salivary secretions by aphids interacting with proteins of phloem wound responses. J. Exp. Bot. 57, 739–745. doi: 10.1093/jxb/erj088
Tjallingii, W. F., and Esch, T. H. (1993). Fine-structure of aphid stylet routes in plant-tissues in correlation with EPG signals. Physiol. Entomol. 18, 317–328. doi: 10.1111/j.1365-3032.1993.tb00604.x
Vandermoten, S., Harmel, N., Mazzucchelli, G., De Pauw, E., Haubruge, E., and Francis, F. (2014). Comparative analyses of salivary proteins from three aphid species. Insect Mol. Biol. 23, 67–77. doi: 10.1111/imb.12061
Wang, W., Dai, H. E., Zhang, Y., Chandrasekar, R., Luo, L., Hiromasa, Y., et al. (2015). Armet is an effector protein mediating aphid-plant interactions. FASEB J. 29, 2032–2045. doi: 10.1096/fj.14-266023
Wang, Y. M., and Van Eys, J. (1970). The enzymatic defect in essential pentosuria. N. Engl. J. Med. 282, 892–896. doi: 10.1056/NEJM197004162821604
Wenger, J. A., Cassone, B. J., Legeai, F., Johnston, J. S., Bansal, R., Yates, A. D., et al. (2017). Whole genome sequence of the soybean aphid, Aphis glycines. Insect Biochem. Mol. Biol. doi: 10.1016/j.ibmb.2017.01.005 [Epub ahead of print].
Wild, R., Ooi, L., Srikanth, V., and Munch, G. (2012). A quick, convenient and economical method for the reliable determination of methylglyoxal in millimolar concentrations: the N-acetyl-L-cysteine assay. Anal. Bioanal. Chem. 403, 2577–2581. doi: 10.1007/s00216-012-6086-4
Will, T., Tjallingii, W. F., Thonnessen, A., and van Bel, A. J. E. (2007). Molecular sabotage of plant defense by aphid saliva. Proc. Natl. Acad. Sci. U.S.A. 104, 10536–10541. doi: 10.1073/pnas.0703535104
Will, T., and Vilcinskas, A. (2015). The structural sheath protein of aphids is required for phloem feeding. Insect Biochem. Mol. Biol. 57, 34–40. doi: 10.1016/j.ibmb.2014.12.005
Yadav, S. K., Singla-Pareek, S. L., Ray, M., Reddy, M. K., and Sopory, S. K. (2005a). Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem. Biophys. Res. Commun. 337, 61–67. doi: 10.1016/j.bbrc.2005.08.263
Yadav, S. K., Singla-Pareek, S. L., Reddy, M. K., and Sopory, S. K. (2005b). Transgenic tobacco plants overexpressing glyoxalase enzymes resist an increase in methylglyoxal and maintain higher reduced glutathione levels under salinity stress. FEBS Lett. 579, 6265–6271. doi: 10.1016/j.febslet.2005.10.006
Yang, S., Jan, Y. H., Mishin, V., Heck, D. E., Laskin, D. L., and Laskin, J. D. (2017). Diacetyl/l-Xylulose reductase mediates chemical redox cycling in lung epithelial cells. Chem. Res. Toxicol. 30, 1406–1418. doi: 10.1021/acs.chemrestox.7b00052
Keywords: cowpea aphid, salivary proteins, effector, diacetyl/L-xylulose reductase, DCXR, methylglyoxal, host defense
Citation: MacWilliams JR, Dingwall S, Chesnais Q, Sugio A and Kaloshian I (2020) AcDCXR Is a Cowpea Aphid Effector With Putative Roles in Altering Host Immunity and Physiology. Front. Plant Sci. 11:605. doi: 10.3389/fpls.2020.00605
Received: 04 February 2020; Accepted: 21 April 2020;
Published: 15 May 2020.
Edited by:
Carolina Escobar, University of Castilla-La Mancha, SpainReviewed by:
Sam T. Mugford, John Innes Centre, United KingdomCopyright © 2020 MacWilliams, Dingwall, Chesnais, Sugio and Kaloshian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isgouhi Kaloshian, aXNnb3VoaS5rYWxvc2hpYW5AdWNyLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.