- 1Center for Plant Aging Research, Institute for Basic Science, Daegu, South Korea
- 2Department of New Biology, Daegu Gyeongbuk Institute of Science and Technology, Daegu, South Korea
Leaf senescence is a developmental process designed for nutrient recycling and relocation to maximize growth competence and reproductive capacity of plants. Thus, plants integrate developmental and environmental signals to precisely control senescence. To genetically dissect the complex regulatory mechanism underlying leaf senescence, we identified an early leaf senescence mutant, rse1. RSE1 encodes a putative glycosyltransferase. Loss-of-function mutations in RSE1 resulted in precocious leaf yellowing and up-regulation of senescence marker genes, indicating enhanced leaf senescence. Transcriptome analysis revealed that salicylic acid (SA) and defense signaling cascades were up-regulated in rse1 prior to the onset of leaf senescence. We found that SA accumulation was significantly increased in rse1. The rse1 phenotypes are dependent on SA-INDUCTION DEFICIENT 2 (SID2), supporting a role of SA in accelerated leaf senescence in rse1. Furthermore, RSE1 protein was localized to the cell wall, implying a possible link between the cell wall and RSE1 function. Together, we show that RSE1 negatively modulates leaf senescence through an SID2-dependent SA signaling pathway.
Introduction
Senescence is the last stage of plant development and is a genetically programmed and evolutionarily advantageous process. Plant senescence occurs at the cellular, tissue, organ, and organismal levels, and leaf senescence has been extensively studied in the past years. Leaf senescence is marked by an enhanced metabolic transition of macromolecules from biosynthesis to degradation, and the nutrients are recycled to sink organs such as young leaves, reproductive organs, and seeds. It has been shown that leaf senescence is modulated by multilayered, multidimensional regulatory mechanisms, including transcriptional and post-transcriptional regulation, protein modification, and hormone signaling (Woo et al., 2013; Schippers, 2015; Yolcu et al., 2018). The mechanism underlying the onset of leaf senescence is elusive, and NAC and WRKY family transcriptional factors have been identified as key regulators acting upstream of leaf senescence onset by turning on senescence-associated genes (SAGs) (Woo et al., 2013). Leaf senescence is also triggered by external stimuli for increasing plant fitness to environmental changes. For instance, hormones, light conditions, salt, and biotic stress-induced senescence involve differential regulation of age-dependent SAGs (Balazadeh et al., 2010; Al-Daoud and Cameron, 2011; Sakuraba et al., 2014; Fernandez-Calvino et al., 2016).
Plant hormones play a crucial role in the regulation of senescence by integrating internal and external signals (Khan et al., 2014). Salicylic acid (SA) is a well-known defense hormone, which plays a critical role in the plant immune response as well as in leaf senescence (Park and Paek, 2007; Vlot et al., 2009). The SA levels increase in senescing leaves, and the mutants for the SA biosynthesis or signaling genes, PHYTOALEXIN DEFICIENT 4 (PAD4) and NON-EXPRESSER OF PR GENES 1 (NPR1), as well as the NahG transgenic plants show delayed leaf senescence and reduced transcript levels of several SAGs (Morris et al., 2000). Furthermore, an increased SA accumulation results in early leaf senescence accompanying the up-regulation of age-dependent SAGs (Vogelmann et al., 2012; Lee et al., 2016; Li et al., 2016; Huang et al., 2018). Conversely, impaired senescence signaling pathways affect plant defense responses (Carviel et al., 2009; Zhu et al., 2011; Piisila et al., 2015), suggesting possible crosstalk between leaf senescence and the defense mechanisms. Interestingly, recent studies have revealed a link between SA-mediated leaf senescence and the modification of lipids and small molecules by glycosylation, S-palmitoylation, and mannosylation (von Saint Paul et al., 2011; Mortimer et al., 2013; Lai et al., 2015; Zhao et al., 2016; Huang et al., 2018). For example, loss-of-function mutations in UGT76B1 glycosyltransferase (GT) that facilitates the glycosylation of isoleucic acid result in early leaf senescence accompanied with increased levels in SA and an up-regulation of defense-related genes and SAGs (von Saint Paul et al., 2011). However, detailed underlying mechanisms remain unclear.
Glycosylation is involved in various cellular processes and plays a vital role in the glycodiversity of macromolecules and the signaling complexity in plants. GTs catalyze the transfer of carbohydrate moieties to a vast range of acceptor molecules including nucleic acids, saccharides, lipids, proteins, and other organic compounds. GTs exist in most living organisms including bacteria, virus, archaea, and eukaryotes, and are classified into over 100 distinct families (GT1–GT107)1 (Lombard et al., 2014). Particularly, plant genomes encode highly diverse GT enzymes (Coutinho et al., 2003), reflecting their functional divergence in cellular homeostasis and physiology. For example, the Arabidopsis genome encodes 565 GT genes, which consists over 40 families (see text footnote 1). Glycosylation by GTs plays a role in the regulation of plant development, defense, and adaptation (Wang and Hou, 2009).
One of the key functions of GTs is the biosynthesis of polysaccharides, a major component of the cell wall (Lampugnani et al., 2018). Broadly conserved GTs and plant-specific GTs including GT2, GT48, and GT14/14-like gene families have been suggested to partake in this process, and these enzymes are mainly localized in the endomembrane system (Zhou et al., 2009; Hansen et al., 2012; Lao et al., 2014; Culbertson and Zabotina, 2015). A subset of 152 cell wall GTs has been implicated in maintaining distinct cell wall properties of the shoot apical meristem (Yang et al., 2016). Glycosylation is a crucial biological reaction in living organisms, but the physiological role of the majority of GTs remains largely unknown. This may be due to the functional redundancy in the large gene families, substrate diversity, and substrate specificity.
Here, we show that RSE1, a GT-like protein, acts as a negative regulator of Arabidopsis leaf senescence in an SA-dependent manner. Transcriptome analysis revealed that genes involved in defense response and SA signaling are highly up-regulated in the rse1 mutant. Consistent with this, the early leaf senescence phenotype of rse1 was further enhanced upon external SA treatment, and SA levels were significantly increased in rse1. A null mutation in the SA biosynthesis gene SID2, but not in PAD4, restored the early leaf senescence of rse1, implying that RSE1 negatively regulates leaf senescence through the SID2-dependent SA biosynthesis pathway. The RSE1 protein is localized preferentially to the cell wall. Overall, our results provide an insight into leaf senescence modulated by the crosstalk between glycosylation and SA signaling.
Materials and Methods
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col) and Landsberg (Ler) were used for all experiments. Plants were grown on soil or 1/2 Murashige and Skoog (MS) salts at 22°C under long day condition (16 h light/8 h dark). The T-DNA insertion line in RSE1 (rse1-2; GK791C05) was obtained from the Nottingham Arabidopsis Stock Centre (NASC). sid2-1 and pad4-1 were previously described (Ng et al., 2011). At least five independent transgenic lines were used for each analysis.
Mapping of the rse1-1 mutation, whole genome sequencing library construction and analysis rse1-1 was crossed with Col-0 to generate mapping populations. Twenty-four F2 plants exhibiting the early leaf senescence phenotype were used for simple sequence length polymorphism mapping and the mutation was mapped to chromosome 5.
For whole genome sequencing, rse1-1 was backcrossed to Ler and 50 F2 plants showing the early leaf senescence phenotype were pooled to extract genomic DNA using the DNeasy plant mini kit (Qiagen). DNA library was prepared according to the illumina Truseq Nano DNA Library prep protocol. For sample library preparation, 0.1 ug of high molecular weight genomic DNA (for a 350 bp insert size) was randomly sheared to yield DNA fragments using the Covaris S2 system. The fragments were blunt ended, phosphorylated, and a single ‘A’ nucleotide was added to the 3′ ends of the fragments in preparation for ligation to an adapter with a single-base ‘T’ overhang. Adapter ligation at both ends of the genomic DNA fragment conferred different sequences at the 5′ and 3′ ends of each strand in the genomic fragment. The ligated DNA was PCR amplified to enrich for fragments with adapters on both ends. The quality of the amplified libraries was verified by capillary electrophoresis (Bioanalyzer, Agilent). The library was clustered on the Illumina cBOT station and sequenced paired end for 101 cycles on the HiSeq 2500 sequencer according to the Illumina cluster and sequencing protocols.
Sequence QC was done through FastqQC 0.11.52 and was mapped to customer genome assembly using bwa 0.7.12 (Li and Durbin, 2009). BAM files were realigned with the Genome Analysis Toolkit 3.5 (GATK) IndelRealigner (McKenna et al., 2010), and the base quality scores were recalibrated using the GATK base quality recalibration tool. Variants were called with GATK’s UnifiedGenotyper tool 3.5. The functional information of the variants was annotated using SnpEff 4.1 (Cingolani et al., 2012).
Plasmid Construction
In order to generate RSE1 complementary transgenic plants, the genomic DNA of RSE1 was amplified by PCR, cloned into pENTR/D-TOPO vector (Invitrogen), and subcloned into Gateway binary vectors by LR recombination (Karimi et al., 2005) (Invitrogen). To generate RSE1 promoter:GUS lines, an approximately 2.0 kb upstream region from the start codon was amplified by PCR and cloned into the pBI101.2 vector between the SalI and XmaI restriction sites. The constructs were then transformed into Arabidopsis plants by Agrobacterium-mediated transformation using the floral dipping method (Clough and Bent, 1998). The primers used for plasmid construction are listed in Supplementary Table S1.
RNA Extraction and Real-Time RT-PCR
For gene expression analysis, total RNA was extracted using plant RNA mini prep kit (ZYMO RESEARCH), and cDNA was synthesized using RNA to cDNA EcoDry Premix (Clontech) according to the manufacturer’s instructions. Quantitative real-time PCR was performed by using the iQTM SYBR Green Super mix (BIO-RAD) in three technical and biological replicates. Statistical analysis was performed using GraphPad Prism.
The gene-specific primers used for PCR are listed in Supplementary Table S1.
Assays for Measurement of Symptoms of Leaf Senescence
To analyze leaf senescence phenotypes, the third and fourth rosette leaves of each plant were detached and submerged in a lactophenol-trypan blue solution (0.03% trypan blue, 33% [w/v] lactic acid, 33% water-saturated phenol, and 33% glycerol). Samples were incubated at 99°C for 1 min and at RT for 24 h, and washed in a chloral hydrate solution (2.5 g mL–1) (Koch and Slusarenko, 1990). For detection of H2O2, the detached leaves were immersed in 1 mg/ml DAB solution followed by incubation for 4 h in the dark and then boiled in 100% ethanol for 10 min (Rea et al., 2004). For measurements of chlorophyll, the third and fourth rosette leaves of 20-day-old plants (approximately 150 mg) were collected and ground under liquid N2. For chlorophyll extraction, the powder was dissolved in 80% acetone (v/v) in the dark for 30 min, and centrifuged for 15 min at 4°C. The supernatant was collected, and the extraction procedure was repeated. The chlorophyll content was detected using a spectrophotometer at 663 and 645 nm, and the chlorophyll contents were calculated using the formula (8.02∗A663 + 20.21∗A645) × volume/weight (Lichtenthaler, 1987). The chlorophyll content in the Ler was defined as 100%. All experiments were repeated using at least three independent plants.
SA Measurement
The level of SA was measured as previously described (Segarra et al., 2006; Forcat et al., 2008). Briefly, seedling material (200 mg) was harvested into liquid nitrogen and ground under liquid N2. The powder was dissolved with 750 μL MeOH-H2O-HOAc (90:9:1, v/v/v) and centrifuged for 1 min at 10,000 rpm. The supernatant was collected, and the extraction procedure was repeated. Pooled supernatants were dried under N2 and resuspended in 200 μL 20% MeOH. Samples were analyzed using a liquid chromatography coupled to triple quadrupole mass spectrometry (TSQ Quantum ultra EMR, Thermo Fisher Scientific, San Jose, CA, United States) at the Korea Basic Science Institute (Seoul). The SA standard and samples were separated on the reverse-phase column (RocRM C18, 3.0 mm × 150 mm, 5 um, RESTECK). The mobile phases A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile) were used. The chromatographic run time was 20 min, and the gradient elution profile was 20% for 2 min, 20–100% for 10 min, 100% for 2 min (phase B), and equilibration for 6 min. The flow rate was 0.2 ml/min, and the sample injection volume was 10 ul. The ESI-MS was operated in the negative ion and selected reaction monitoring (SRM) mode. The mass parameters were a spray voltage of 3000 V, sheath gas pressure of 40 (arbitrary unit), aux gas pressure of 10 (arbitrary unit) and capillary temperature of 300°C. The SRM of SA and IS was acquired using an m/z 137.00 > 93.28, and 299.00 > 137.18 transition at a collision energy of 18%.
mRNA-Seq Library Construction and Data Analysis
RNA purity was determined by assaying 1 μL of total RNA extract on a NanoDrop8000 spectrophotometer (Thermo Scientific). Total RNA integrity was checked using an Agilent Technologies 2100 Bioanalyzer with an RNA integrity number value. mRNA sequencing libraries were prepared according to manufacturer’s instructions (Illumina Truseq stranded mRNA library prep kit). mRNA was purified and fragmented from total RNA (1 ug) using poly-T oligo-attached magnetic beads and two rounds of purification. Cleaved RNA fragments primed with random hexamers were reverse transcribed into first-strand cDNA using reverse transcriptase, random primers and dUTP in place of dTTP. (The incorporation of dUTP quenches the second strand during amplification because the polymerase does not incorporate past this nucleotide.) A single adenine base was added to these cDNA fragments and was followed by adapter ligation. The products were purified and enriched with PCR to create the final strand-specific cDNA library. The quality of the amplified libraries was verified by capillary electrophoresis (Bioanalyzer, Agilent). After qPCR using SYBR Green PCR Master Mix (Applied Biosystems), the index-tagged libraries were combined in equimolar amounts within a single pool. RNA sequencing was performed using an Illumina NextSeq 500 system following the provided protocols for 1 × 75 sequencing.
Data quality control and preprocessing were performed using FastQC. (v0.11.2)3. The Trimmomatic (v0.32.3) (Bolger et al., 2014) was used to trim the adapter sequences and low-quality sequences. Remaining reads were aligned to the Arabidopsis thaliana Ler-0 genome using TopHat (v2.0.9) (Trapnell et al., 2009). The Ler-0 genome was reconstructed using the Arabidopsis thaliana Col-0 genome based on the variant call format file for Ler-0 obtained from the 1001 Genomes Project website4. PCR duplicates were removed using Picard (v1.119)5, and ‘MarkDupicates’ and only uniquely mapped reads were counted using HTSeq (v0.6.1) (Anders et al., 2015). The differential expression analysis was performed using DESeq2 (Love et al., 2014) and the P-values were determined using the Benjamini–Hochberg method (Benjamini and Hochberg, 1995). Differentially expressed genes (DEGs) were identified as the ones that have the adjusted P-value < 0.05.
Gene Ontology Enrichment
Gene ontology (GO) enrichment analysis was performed using DAVID software6. The GO terms with Benjamini P-value < 0.05 were selected as the ones enriched by the DEGs used.
Subcellular Localization
For the analysis of subcellular localization of RSE1, RSE1 cDNA was subcloned into pB7m34GW destination vectors to generate C-terminal fusion with GFP. The expression constructs were co-transformed into Arabidopsis with mCherry-tagged markers for various organelles including the Golgi-localized Man49 (Saint-Jore-Dupas et al., 2006) and plasma membrane-localized PIP2 (Ivanov and Harrison, 2014). Endoplasmic reticulum (ER) localization constructs contained the ER retention sequences HDEL (Ivanov and Harrison, 2014). For the plasmolysis experiment, plants were treated with 0.8 M mannitol for 30 min (Truernit et al., 2012). Imaging was performed using an LSM 7 DUO (ZEISS) confocal laser microscopy. The primers used for plasmid construction are listed in Supplementary Table S1.
Results
rse1 Displays an Early Senescence and Cell Death Phenotypes
To identify genetic mutants with altered senescence phenotypes, we isolated a mutant, rapid senescence 1-1 (rse1-1) from a genetic screen of EMS-mutagenized plants. Compared with wild-type Ler plants that retain green and healthy rosette leaves, the cotyledons of rse1-1 were remarkably yellow, and rosette leaves rapidly turned yellow as they expanded (Figures 1A,B). In contrast to the rosette leaves of WT plants that just began yellowing 3 weeks after sowing, all rosette leaves of rse1-1 had already turned yellow (Supplementary Figure S1A), indicating enhanced leaf senescence in rse1-1. The rse1-1 mutant also showed other developmental defects, including dwarfism and severely reduced fertility (Supplementary Figure S1C).
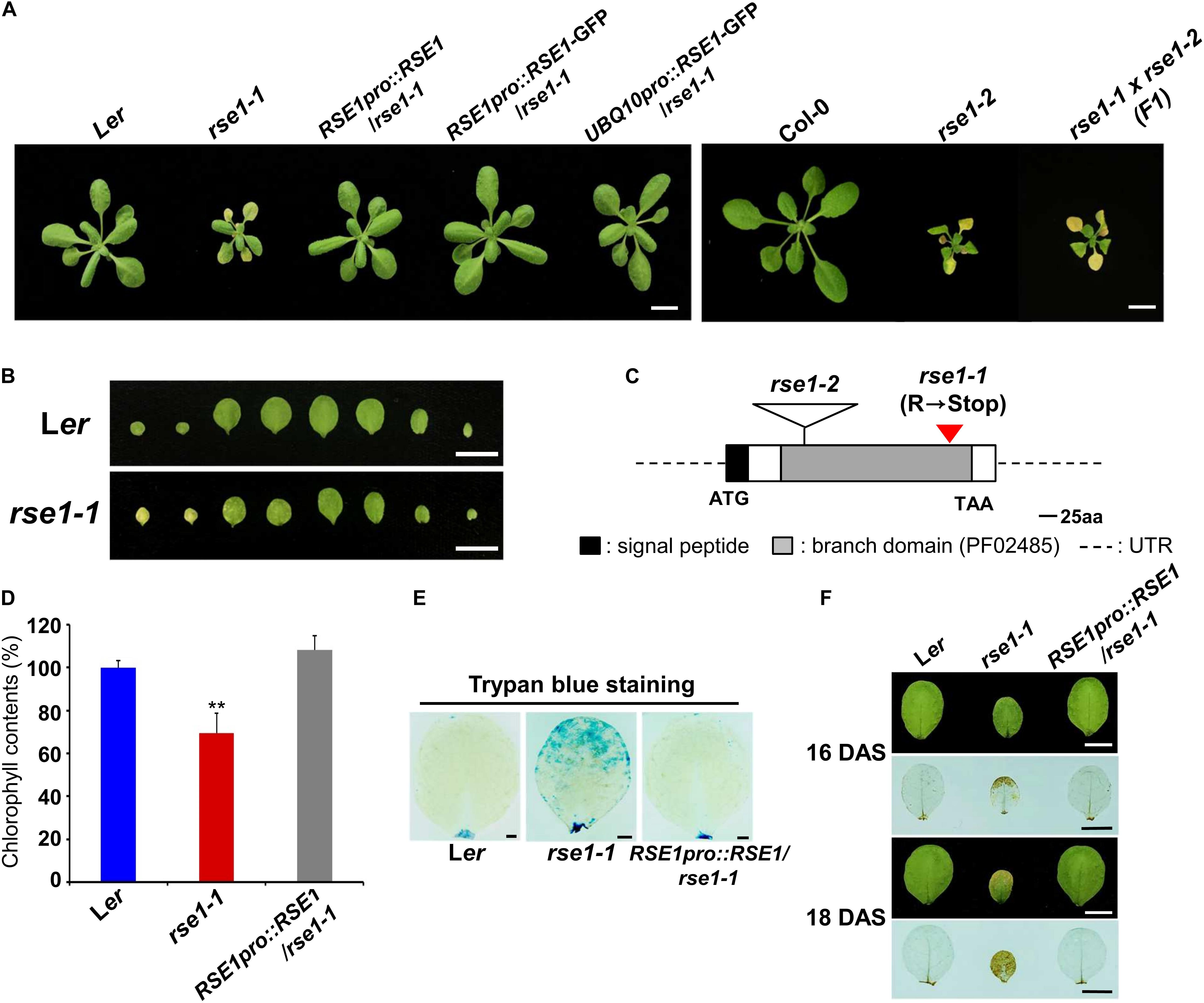
Figure 1. Loss-of-function mutations in RSE1 cause early senescence and cell death. (A) Senescence phenotypes of rse1-1, rse1-2, RSE1pro:RSE1/rse1-1, RSE1pro:RSE1/rse1-1, UBQ10pro:RSE1-GFP/rse1-1 at 21 days after sowing, and rse1-1 × rse1-2 (F1) plants at 22 days after sowing (DAS). Scale bar, 1 cm. Results are representative of at least five independent plants. (B) Detached leaves of Ler and rse1-1 at 16 DAS. Scale bar, 1 cm. Results are representative of more than 10 independent plants. (C) A schematic diagram of RSE1 encoding a glucosaminyl transferase (At5g22070) shows the position of the C- to-T mutation in rse1-1 resulting in a premature stop codon after 300 amino acid and the location of the T-DNA insert in rse1-2. (D) Relative chlorophyll contents in detached third and fourth leaves of WT, rse1-1, and RSE1pro:RSE1/rse1-1 plants at 20 DAS. Error bars indicate SD from three biological replicates. ∗∗p < 0.01. (E) Trypan blue staining of the third leaves of WT, rse1-1, and RSE1pro:RSE1/rse1-1 plants at 21 DAS. Scale bar, 0.5 cm. (F) 3.3′-Diaminobenzidine (DAB) staining of hydrogen peroxide in the third leaves of WT, rse1-1, and RSE1pro:RSE1/rse1-1 plants at 16 and 18 DAS. Leaf yellowing phenotype is observed from 18 DAS in the third leaves of rse1-1 mutants. Scale bar, 0.5 cm. (E,F) Results are representative of three independent plants.
To identify the mutated gene, we carried out map-based cloning and whole-genome sequencing in parallel and found a C-to-T mutation in a previously uncharacterized GT-like gene (At5g22070), which introduced a premature termination codon (Figure 1C). The gene product is a member of the GT14/GT14-like family (Ye et al., 2011) and consists of a putative transmembrane domain at the N terminus (1 to 29 amino acid residues) followed by a core-2/1-branching enzymatic domain (72 to 329 residues). The introduction of an RSE1 genomic fragment (RSE1pro:RSE1) and the native promoter/UBQ10 promoter-driven GFP-tagged RSE1 (RSE1pro:RSE1-GFP/UBQ10pro:RSE1-GFP) rescued the early leaf senescence phenotype of rse1-1 (Figure 1A). In addition, a T-DNA insertion mutant in the Col-0 background was obtained from the Nottingham Arabidopsis Stock Center and designated as rse1-2 (Figures 1A,C). In rse1-2 mutants, RSE1 expression was abolished (Supplementary Figure S1B), and leaf senescence was accelerated as in rse1-1 (Figure 1A). When rse1-1 was crossed to rse1-2, early leaf senescence phenotype was not recovered in F1 plants (Figure 1A), further supporting that functional impairment of RSE1 results in early leaf senescence.
A typical symptom of leaf senescence is the yellowing induced by chlorophyll degradation. We measured the chlorophyll content in rse1-1 leaves and found 30% reduction compared with WT and the complemented lines (Figure 1D). Next, we determined cell death and H2O2 levels using trypan blue and 3,3′-diaminobenzidine (DAB), respectively, because the production of reactive oxygen species (ROS) and the resulting cell death occur during leaf senescence (Koo et al., 2017). We found that both cell death and H2O2 levels were dramatically increased in rse1-1 mutants compared with WT and the complemented lines (Figures 1E,F). Furthermore, even when no visible symptoms of senescence were observed in WT, rse1-1, and the complemented lines, H2O2 was detected at the margin of rse1-1 leaves only (Figure 1F, upper panel). Hydrogen peroxide levels were further intensified and spread to most areas of the leaves in two additional days (Figure 1F, lower panel), suggesting that the enhanced cell death may contribute to leaf senescence in rse1-1 mutants. Together, these results imply that RSE1 negatively regulates leaf senescence.
SAGs Are Up-Regulated in rse1
Previous genetic and transcriptomics studies have shown roles of SAGs and the dynamics of genetic reprogramming in leaf senescence (Hinderhofer and Zentgraf, 2001; Guo and Gan, 2006; Kim et al., 2009; Woo et al., 2016). Transcriptional factors such as NACs and WRKYs also act at early stages of leaf senescence and turn on the downstream genes encoding catabolic enzymes for nutrient recycling (Schippers et al., 2015). We thus analyzed senescence marker genes in the rse1-1 mutant to examine whether the genetic program governing senescence was affected in rse1-1. qRT-PCR analyses showed that the early senescence markers and the genes encoding senescence-induced metabolic enzymes were strongly up-regulated in rse1-1 compared with WT and the complemented lines (Figure 2). The expression levels of SAG13 and SAG12 were increased by about 23,000- and 4,000-fold, respectively, in rse1-1. The expression of SIRK, which is implicated in immune response, cell death and senescence (Asai et al., 2002; Koo et al., 2017), was increased by 900-fold in rse1-1. Furthermore, the central regulator of leaf senescence ORE1 (Kim et al., 2009) was significantly (180-fold) up-regulated in rse1-1 compared with WT and the complemented lines (Figure 2). These data suggest that the rse1 mutation causes accelerated senescence.
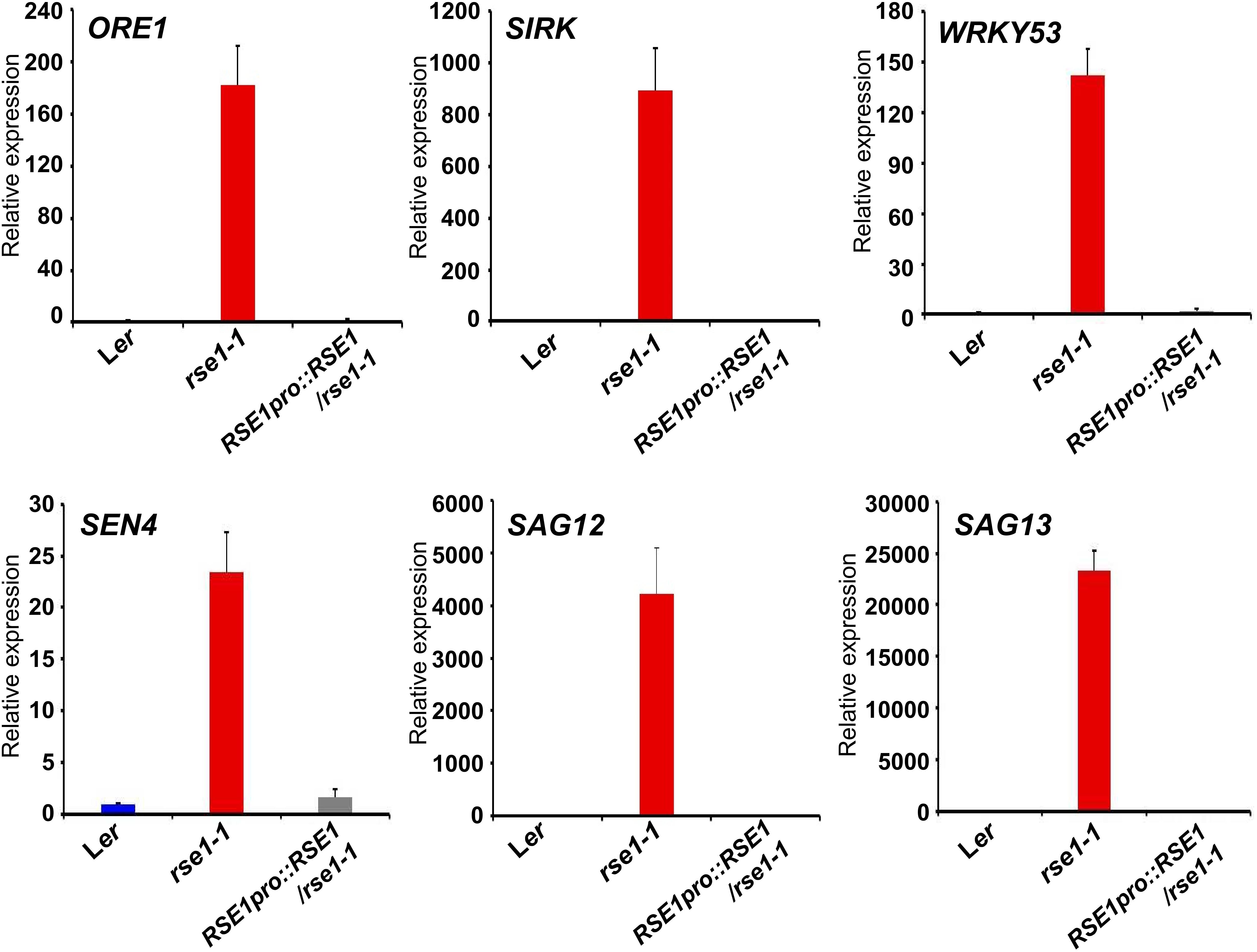
Figure 2. qRT-PCR analyses of senescence-associated genes in rse1-1. qRT-PCR indicates a strong up-regulation in expression of the senescence marker genes in rse1-1. Third and fourth leaves of WT, rse1-1, and RSE1pro:RSE1/rse1-1 plants at 20 DAS were used for RNA extraction and qRT-PCR. The expression levels were normalized to UBQ5 (At3g62250). The data have been obtained from three biological replicates, and error bars indicate SD.
Disruption of RSE1 Up-Regulates Defense-Related Gene Expression
Next, we conducted transcriptome analysis to further investigate the role of RSE1. To focus on the early molecular events that might have caused accelerated leaf senescence in rse1-1, leaves of rse1-1 and WT that showed no visible yellowing were used for analyses. The expression profiling identified 2,186 differentially expressed genes in rse1-1. Among these differentially expressed genes, 1,783 were up-regulated, and 403 were down-regulated (Supplementary Tables S2, S3). GO term analysis revealed that the loss-of-function mutation of RSE1 affected the expression of genes involved in various processes, including aging, stress signaling, and protein phosphorylation (Figure 3A and Supplementary Figure S2). Remarkably, genes involved in defense response were among the highly up-regulated genes in rse1-1 (Figure 3A). Moreover, genes involved in SA biosynthesis and signaling were also most highly up-regulated in rse1-1 compared with other hormones (Figure 3A).
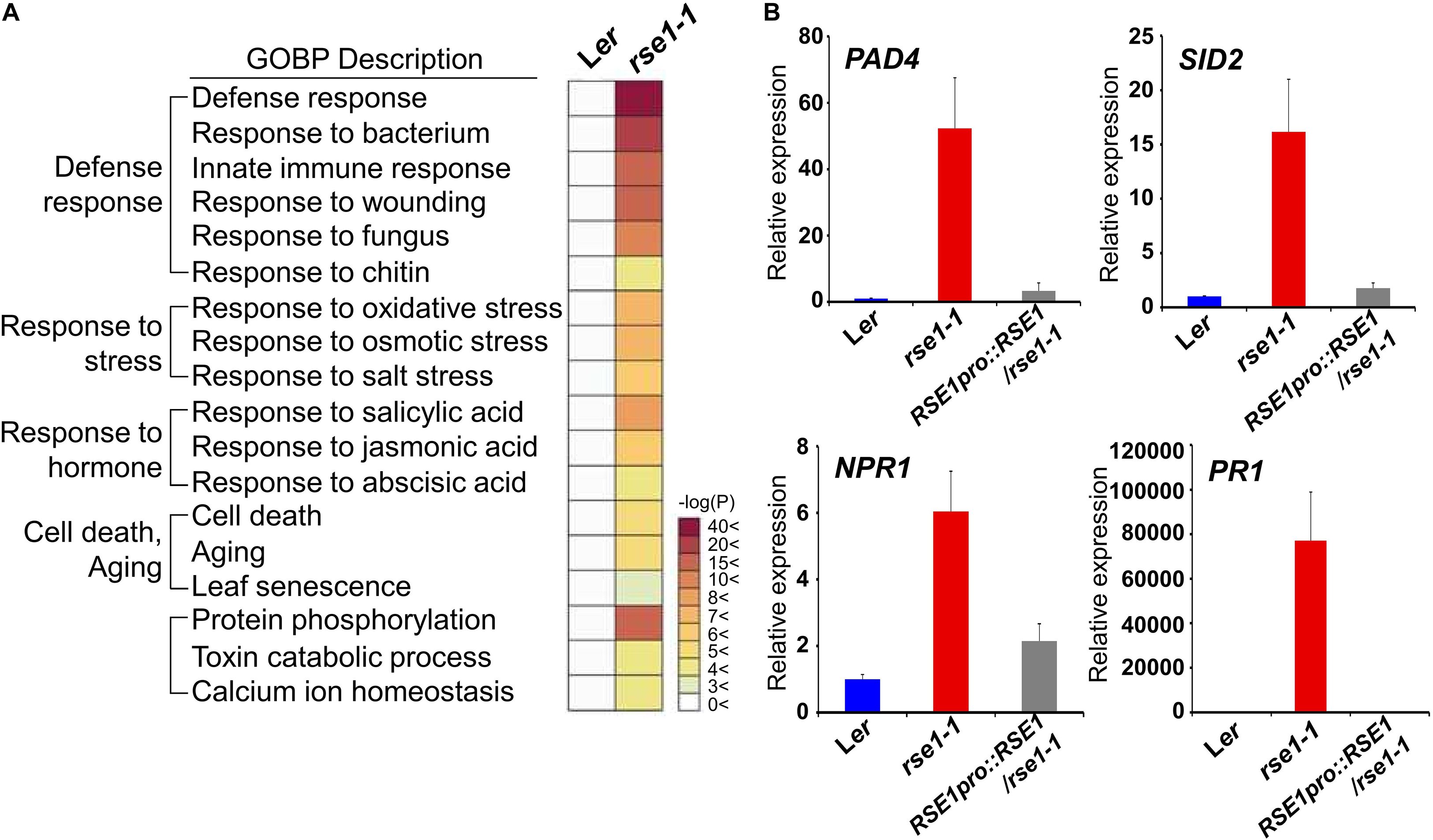
Figure 3. Genes related to defense response are highly up-regulated in rse1-1. (A) Gene Ontology Biological Processes (GOBP) analysis of differentially expressed genes shows an up-regulation of genes involved in the defense response in rse1-1. Third and fourth leaves of WT and rse1-1 plants at 16 DAS were used for mRNA-seq with three biological replicates. (B) qRT-PCR analysis of salicylic acid signaling genes, PAD4, SID2, NPR1, and PR1 in WT, rse1-1, and RSE1pro:RSE1/rse1-1 plants. Third and fourth leaves of WT, rse1-1, and RSE1pro:RSE1/rse1-1 plants at 16 DAS were used. The expression levels were normalized to UBQ5 (At3g62250). The data have been obtained from three biological replicates, and error bars indicate SD.
Salicylic acid plays an essential role in plant immunity. However, a constitutively high level of SA and activated defense response could be harmful to plants as the plants are forced on alert thereby resulting in stunted plant growth (Seyfferth and Tsuda, 2014). SALICYLIC ACID INDUCTION DEFICIENT 2 (SID2) and PAD4 regulate SA biosynthesis and accumulation, respectively (Vlot et al., 2009). NPR1 is an SA receptor, which initiates SA signaling (Wu et al., 2012). To validate the up-regulation of genes involved in SA and defense signaling in rse1-1, the expression levels of SID2, PAD4, and NPR1 were determined by qRT-PCR. All three genes were drastically up-regulated in rse1-1 compared with WT and the complemented lines (Figure 3B). Furthermore, the level of SA-responsive PATHOGENESIS-RELATED GENE 1 (PR1) was increased by about 80,000-fold (Figure 3B), implying that the disruption of RSE1 puts the plants on high alert against biotic stress. On the other hand, the expression levels of the early senescence markers such as ORE1 and WRKY53 were not changed at the time when defense-related genes were highly up-regulated in rse1-1 (Supplementary Tables S2, S3). Taken together, these results suggest that SA-mediated defense signaling is associated with enhanced leaf senescence in the rse1 mutant.
RSE1 Is Involved in SA Biosynthesis and Signaling Pathway
Since the genes involved in SA signaling were highly up-regulated in rse1-1 (Figure 3), we tested the involvement of RSE1 in SA-mediated responses. WT, rse1-1, and rse1-2 plants were grown in the absence and presence of SA to examine whether rse1 had an altered sensitivity to SA. In the absence of exogenously applied SA, no visible symptoms were observed in WT, rse1-1, and the complemented plants. However, rse1-1 showed retarded growth accompanied by early senescence compared to WT and the complemented plants in the presence of 25 μM SA (Figure 4A). Since rse1-2 is sterile, the progenies of rse1-2/+ heterozygous plants were grown in the absence and presence of SA. Twenty-four out of 141 plants displayed reduced growth and enhanced senescence in the presence of 25 μM SA but not in the absence of SA (Supplementary Figure S3). All the 24 plants turned out to be homozygous for the rse1-2 mutation, indicating that rse1 mutants are more sensitive to SA. To determine whether the retarded growth of rse1-1 was due to the altered level of SA or due to an enhanced sensitivity to SA, we measured contents of free and glucose-conjugated SA (SA-glc) in WT and rse1-1 mutant plants. It turned out that both free SA and SA-glc levels were increased 6-fold and 45-fold, respectively, in rse1-1 mutants compared with WT and the complemented lines (Figure 4B). The enhanced sensitivity of rse1 mutants to SA is probably due to the increased level of endogenous SA in rse1 mutants, resulting in the increased SA response. Together, these results indicate that RSE1 negatively regulates SA biosynthesis.
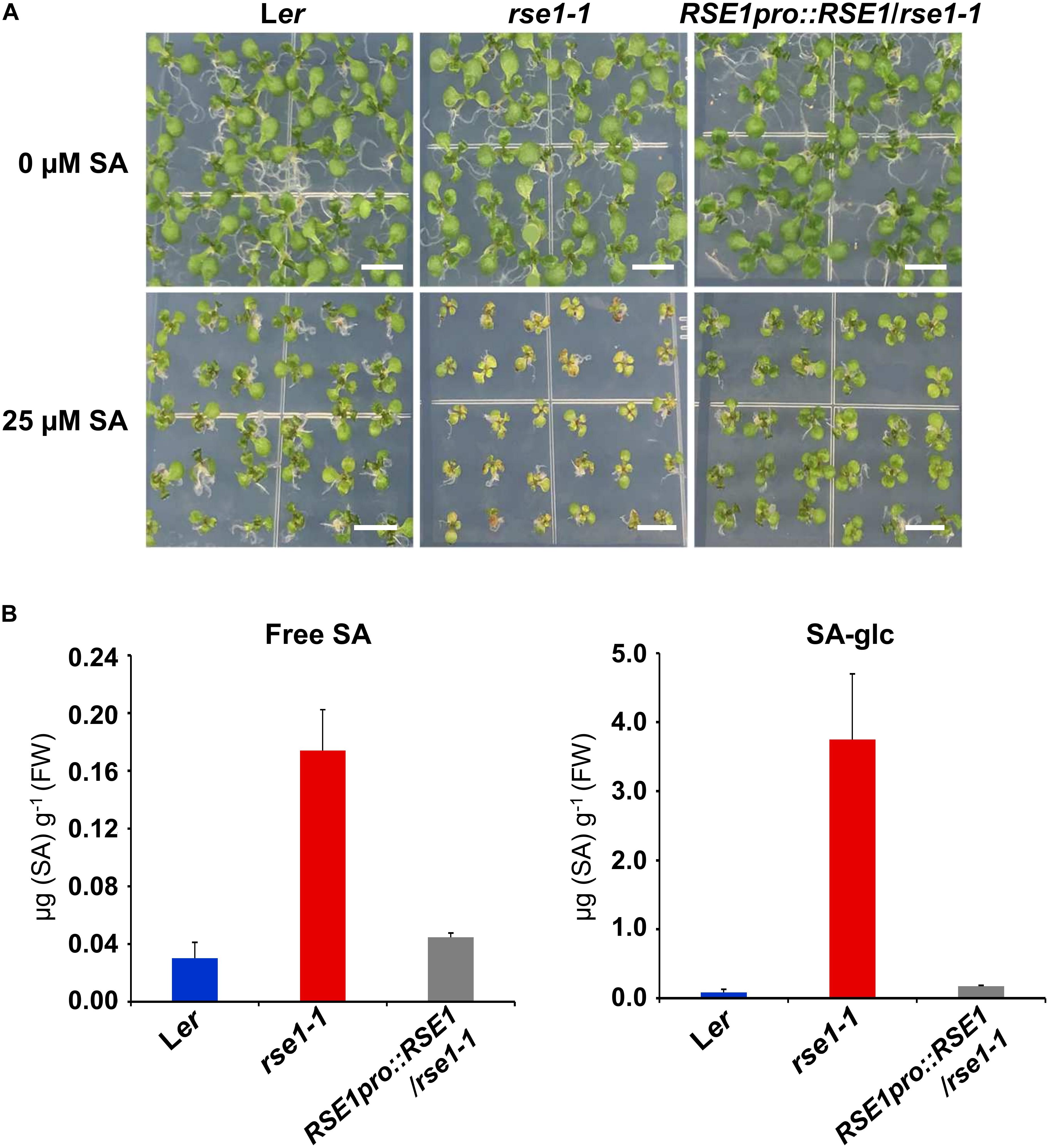
Figure 4. rse1 accumulates higher levels of SA and is sensitive to SA. (A) Exogenous SA inhibits the seedling growth of rse1-1. Seedlings were grown on MS media in the absence (0 μM) or presence (25 μM) of SA for 12 days. Results are representative of three independent experiments. Scale bar, 0.5 cm. (B) Measurements of free SA and glucose-conjugated SA (SA-glc) in 10-day-old seedlings of WT, rse1-1, and RSE1pro:RSE1/rse1-1 grown on 1/2 MS media. The data have been obtained from three biological replicates, and error bars indicate SD.
Early Leaf Senescence of rse1 Is in Part Attributable to SID2
SID2 and PAD4 are key enzymes in SA biosynthesis and accumulation, respectively (Zhou et al., 1998; Wildermuth et al., 2001). It has been suggested that SID2 and PAD4-dependent SA pathways play a role in leaf senescence (Nawrath and Metraux, 1999; Vogelmann et al., 2012; Guo et al., 2017). Our data suggested that the enhanced SA signaling in rse1-1 can be attributed to the increased levels of SA and associated with early leaf senescence. To test whether the RSE1 function in leaf senescence depends on the SA signaling pathway, rse1-2 was crossed to sid2-1 and pad4-1 mutants. Interestingly, rse1-2sid2-1 double mutants largely rescued the early leaf senescence phenotype of rse1-2 mutants (Figure 5A and Supplementary Figure S4). The size of the plant and developmental defects related to reproduction were also rescued (Supplementary Figure S4B). In contrast, rse1-2pad4-1 mutants were indistinguishable from the rse1-2 single mutants (Figure 5B). The phenotypes of the rse1-2sid2-1 double mutants were in agreement with the expression patterns of the senescence- and SA-related marker genes. The expression levels of ORE1, SAG12, and PR1 were reduced to the WT levels in the rse1-2sid2-1 double mutant (Figure 5C). These results suggest that SID2 contributes to the early leaf senescence of rse1. In contrast, the increased expression of SEN4 in rse1-2 was not rescued by sid2-1, suggesting that RSE1 may also regulate leaf senescence in a SID2-independent manner to a certain extent. The difference in the level of PR1 expression between rse1-1 and rse1-2 is possibly because the basal level of PR1 in Ler (rse1-1 background) at 16 days after sowing was much lower compared to that in Col-0 (rse1-2 background) at 20 days after sowing, resulting in the smaller fold-change in rse1-2. It could be also due to the ecotype difference.
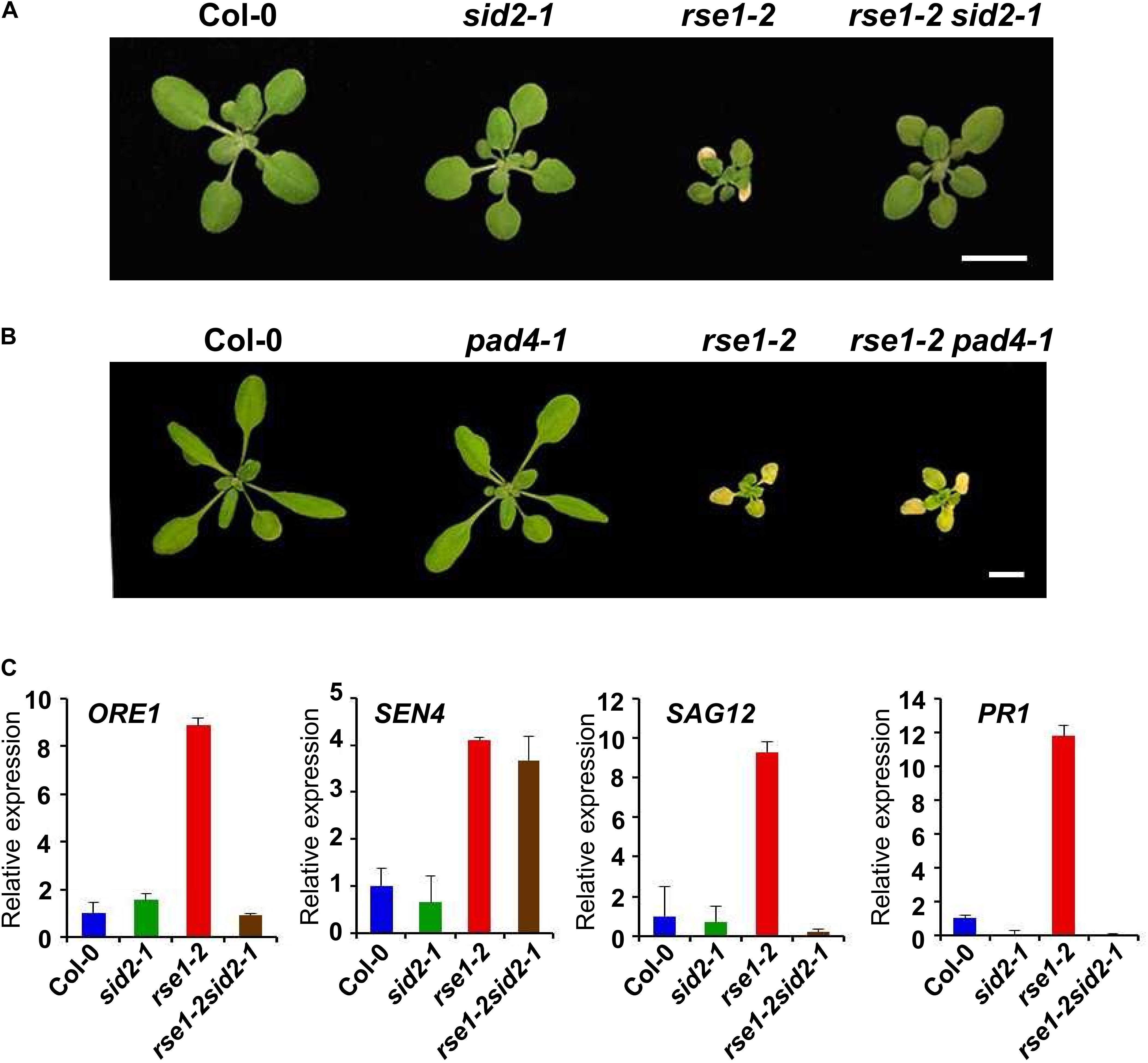
Figure 5. RSE1 modulates leaf senescence through SID2-dependent SA pathway. Phenotypes of WT, sid2-1, rse1-2, rse1-2sid2-1 double mutants (A) at 17 DAS and WT, pad4-1, rse1-2, rse1-2pad4-1 double mutants (B) at 22 DAS. Scale bar, 1 cm. Results are representative of at least three independent plants. (C) qRT-PCR analyses of senescence- and defense-related genes in WT, sid2-1, rse1-2 and rse1-2sid2-1 double mutants. Total RNA was extracted from the third and fourth leaves of plants at 20 DAS. The expression levels were normalized to UBQ5 (At3g62250). The data have been obtained from three biological replicates, and error bars indicate SD.
RSE1 Preferentially Localizes to the Cell Wall
We performed qPCR analysis using leaves, roots, siliques, and flowers to examine the spatial expression patterns of RSE1 and found that RSE1 was ubiquitously expressed in the tissues tested (Supplementary Figure S5A). RSE1 expression patterns were further determined by analyzing transgenic plants harboring an RSE1pro:GUS construct. Strong GUS expression was detected in the emerging young leaves, and the histochemical GUS staining decreased and was found only in trichomes as the leaves further developed (Supplementary Figure S5B). In roots, GUS expression was strongly detected in root hairs (Supplementary Figure S5B).
Plant GTs are predicted to mainly localize in the Golgi apparatus and the ER, which most post-translational modifications occur, including glycosylation (Ye et al., 2011; Lao et al., 2014). In order to obtain a clue regarding the function of RSE1, we examined the cellular localization of RSE1 protein by generating transgenic plants expressing a GFP-tagged RSE1 under the control of the native or UBQ10 promoter. Both the native and UBQ10 promoter-driven RSE1-GFP were able to rescue the early leaf senescence phenotype in rse1-1 (Figure 1A). We analyzed at least eight independent plants for each construct. Co-localization analyses with the Golgi, ER, and plasma membrane markers in UBQ10pro:RSE1-GFP plants showed that RSE1 is not localized to either the Golgi or ER (Figures 6A,B). In addition, DAPI staining revealed that RSE1-GFP is not localized in the nucleus (Supplementary Figure S6A). The RSE1-GFP pattern suggested its localization at the plasma membrane, but the RSE1-GFP signal did not overlap with the plasma membrane marker PIP2a-mCherry and was detected on the outside the plasma membrane (Figure 6C). Optical Z-sections of confocal microscopy show that RSE1-GFP is broadly detected in the cell (top sections) and becomes restricted to the outer layer of the cell (middle sections) (Supplementary Figure S6B). To determine RSE1 localization, we stained the plant cell walls with propidium iodide (PI) and found that RSE1-GFP signals were overlapped entirely with the PI staining (Figure 6D). To further prove RSE1 localization at the cell wall, plant cells expressing both RSE1-GFP driven by the native promoter and PIP2a-mCherry were treated with a high osmolarity solution causing plasmolysis. In contrast to the PIP2a-mCherry that shrank and dislocated, RSE1-GFP was again detected at the cell wall after plasmolysis (Figure 6E). This result implies that RSE1 preferentially localizes to the cell wall.
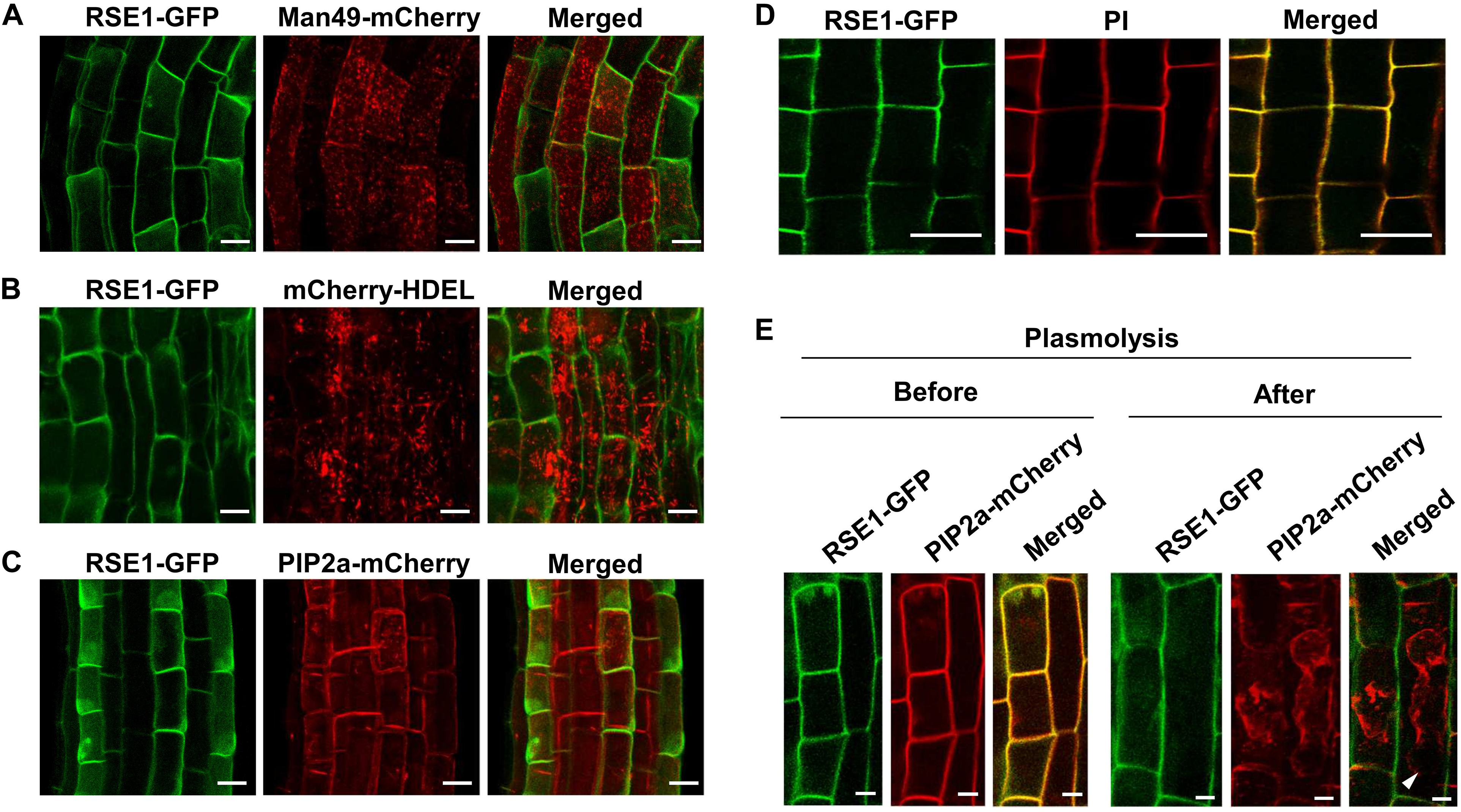
Figure 6. RSE1 is localized to the cell wall. (A–C) RSE1-GFP localization in combination with the Golgi localization marker MAN49-mCherry (A), the ER localization marker mCherry-HDEL (B), and the plasma membrane localization marker PIP2a-mCherry (C) in 7-day-old seedling roots of UBQ10pro:RSE1-GFP plants. Results are representative of at least eight independent transgenic lines. (D) UBQ10pro:RSE1-GFP transgenic lines were stained with propidium iodide (PI) for cell wall visualization. (E) Localization of RSE1-GFP and PIP2a-mCherry before (Mock) and after plasmolysis (0.8 M mannitol) in RSE1pro:RSE1-GFP plants. White arrows indicate the plasma membrane of shrunken cells. Scale bar, 20 μm. (D,E) Results are representative of at least three independent transgenic lines.
Discussion
In this study, we showed that RSE1 negatively regulates leaf senescence through an SA-dependent signaling pathway. Premature leaf senescence is enhanced in rse1 mutants, accompanying with the up-regulation of senescence marker genes. The phenotypes of the rse1 mutants, including up-regulation of pathogen-related genes, high accumulations of ROS and SA, and enhanced cell death, are typical defense responses (Jones and Dangl, 2006), which are also common molecular events during leaf senescence. Interestingly, defense signaling is activated prior to the onset of leaf senescence, and SA accumulation is increased in the rse1 mutant. In addition, early leaf senescence phenotype is rescued by sid2-1 defective in SA biosynthesis, suggesting that RSE1 may primarily be involved in the defense signaling. In addition to the early leaf senescence phenotype, the rse1 mutants display severely retarded growth (Supplementary Figures S1A,C). The high level of SA account for the phenotypes because a high dosage of exogenous and endogenous SA causes early leaf senescence and negatively modulates the vegetative growth of plants (Zhang et al., 2007, 2008; Kovacik et al., 2009; Miura et al., 2010; Vogelmann et al., 2012; Lee et al., 2016). Several studies have suggested cross-talk between defense responses and leaf senescence, in which mutants with constant activation of defense signaling display enhanced cell death and leaf senescence (Yoshida et al., 2002; Vogelmann et al., 2012; Lee et al., 2016; Zhang et al., 2017). Together, these findings suggest that RSE1 may primarily act in the SA-dependent defense signaling and cell death, consequently impacting leaf senescence.
RSE1 is previously thought to belong to the Domain of Unknown Function (DUF) 266 proteins. Bioinformatic analyses based on protein sequences and structures have revealed that DUF266 proteins are markedly close to the GT14 family of proteins in the GT superfamilies. Both DUF266 and GT14 proteins encode a branched domain that shows the highest similarity to the core-2/1-branching enzymatic function domain in animals, suggesting that the GT14 and DUF266 family of proteins are closely related to each other (Hansen et al., 2012). GT14 and DUF266 proteins are currently classified into the GT14/GT14-like family (Hansen et al., 2012). Unlike the GT14 proteins, there are high variations in the predicted localization of GT14-like proteins, including the Golgi apparatus, ER, the plasma membrane, extracellular space, and the peroxisomes (Ye et al., 2011). Indeed, the in vivo localization of GT14-like proteins remains elusive. For example, rice BC10 containing the DUF 266 domain localizes to the Golgi and regulates cell wall biosynthesis (Zhou et al., 2009). AtGnTL is a GT14-like family protein with the highest similarity to RSE1 among GT14-like proteins in Arabidopsis and localizes to the cell wall-associated plasmodesmata (Zalepa-King and Citovsky, 2013). Our data show that RSE1 is localized at the cell wall (Figure 6). This finding suggests that the GT14-like proteins might be involved in more diverse cellular processes due to their assorted localization compared with GTs in other families.
A recent study has revealed that the Premature Leaf Senescence (OsPLS) gene encoding a putative GT modulates leaf senescence in rice (Ke et al., 2019). OsPLS is a homolog of RSE1, which contains a putative transmembrane domain at the N-terminus and a predicted core-2/1-branching/DUF266 domain (Yang et al., 2017; Ke et al., 2019). Ospls mutants show premature leaf senescence accompanied by up-regulated SAGs, elevated ROS levels, and enhanced cell death (Yang et al., 2017; Ke et al., 2019), similar to the phenotypes observed in rse1. This indicates that both OsPLS and RSE1 are involved in leaf senescence and cell death. OsPLS regulates leaf senescence and cell death through protein O-glycosylation and ethylene-dependent signaling pathways (Ke et al., 2019). In contrast to Ospls in which O-GlcNAc transferase (OGT) genes SECRET AGENT1 (SEC1), SEC2 and SPINDLY (SPY) are significantly reduced (Ke et al., 2019), the expression level of AtSEC or AtSPY is slightly up-regulated or not altered in rse1 (Supplementary Tables S2, S3). Moreover, transcriptome analysis and GO term analysis revealed that the ethylene signaling pathway is not nearly affected in rse1 (Figure 3A; Supplementary Figure S2 and Supplementary Tables S2, S3). RSE1 appeared to modulate SA-mediated signaling thereby regulating leaf senescence and cell death. These studies suggest that molecular mechanisms involving RSE1 and OsPLS may be different, further supporting a functional diversity of the GT14-like genes.
To date, the biological function of GT14/GT14-like family proteins remained very elusive. A few studies have revealed a functional link between GT14/GT14-like proteins and cell wall regulation (Ye et al., 2011). Most of the cell wall biosynthesis-related GT proteins are membrane-associated and are typically localized to the Golgi or the plasma membrane (Ye et al., 2011). RSE1 was also predicted to localize to the Golgi and the plasma membrane. However, our data showed that RSE1 localized to the cell wall (Figure 6). Considering our results and the previous studies, it is plausible to speculate that RSE1 may contribute to the structure and/or function of the cell wall. Additionally, RSE1 contains the plant-specific DUF266 domain (Hansen et al., 2012), thereby raising a possibility that RSE1 may act in a plant-specific process. Furthermore, our transcriptome and Gene Ontology Biological Processes analyses showed that the genes regulating the organization or biogenesis of the cell wall are significantly reduced in rse1 (Supplementary Figure S2B). Over 130 cell wall-related genes encoding cell wall macromolecule modifying-, cell wall structure regulating-, and secondary cell wall biosynthesis-related proteins are differentially regulated in the rse1-1 mutants (Supplementary Tables S4, S5). This further implies a role of RSE1 in the regulation of the cell wall. Most sugar nucleotides, donor substrates for GTs, are synthesized in the cytosol and transported to the lumens of the Golgi apparatus and ER by nucleotide sugar transporters for the synthesis of non-cellulosic cell wall polysaccharides (Temple et al., 2016). Since no nucleotide sugar transporters responsible for delivering sugar nucleotides into the cell wall have been identified, we do not exclude the possibility that RSE1 may possess an additional enzymatic activity other than GT. Further biochemical characterization should be able to address RSE1’s enzymatic property.
It is intriguing that the loss-of-function mutation in RSE1 resulted in the activation of SA-dependent defense signaling. Various lines of emerging evidence have suggested a feedback link between the cell wall function and plant defense mechanisms (Nuhse, 2012). Since plant cell walls are a primary barrier against pathogen attacks, plants turn on defense signaling by monitoring the integrity and function of the cell wall (Nuhse, 2012). For instance, the mutants for POWDERY MILDEW RESISTANT 4 (PMR4) encoding a callose synthase and MURUS 3 (MUR3) encoding a xyloglucan galactosyltransferase display cell wall defects, consequently enhanced resistance to pathogens, and increased cell death due to the constitutive activation of SA-dependent defense signaling (Nishimura et al., 2003; Tedman-Jones et al., 2008; Vega-Sanchez et al., 2012). RSE1 localization and the altered gene expression related to the regulatory pathway of the cell wall function in rse1 indicates a possible role of RSE1 in the regulation of cell wall function. Moreover, defense signaling is highly activated in rse1, suggesting a potential link between RSE1-mediated cell wall regulation and defense mechanisms. RSE1, as a putative GT, might directly modify cell wall macromolecules or cell wall proteins that are required for cell wall function or structures. The defects in the cell wall integrity or function caused by the impaired function of RSE1 may be recognized as pathogen attacks, making the plant turn on the constant alert by activating SA-dependent defense signaling, which in turn promotes cell death and subsequent leaf senescence. A further detailed characterization of RSE1 including identification of its molecular targets is noteworthy of investigation.
Data Availability Statement
The datasets generated for this study can be found in the GenBank, accession no. GSE132740.
Author Contributions
SL, JK, and YK conceived the study and designed the experiments. SL, M-HK, and JJ performed the experiments. JL analyzed the RNA-seq data. SL, JK, and YK wrote the manuscript.
Funding
This work was supported by the Institute for Basic Science (IBSR013-G2 and in part by IBS-R013-D1) and a grant from National Research Foundation (2019R1A2C3007376), and in part by start-up funds from DGIST to JK.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kee Hoon Sohn (POSTECH) for providing sid2-1 and pad4-1 seeds and Sunghyun Hong (IBS) for helpful discussion.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00551/full#supplementary-material
Footnotes
- ^ http://www.cazy.org
- ^ http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- ^ http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- ^ https://1001genomes.org/
- ^ http://broadinstitute.github.io/picard/
- ^ https://david.ncifcrf.gov/
References
Al-Daoud, F., and Cameron, R. K. (2011). ANAC055 and ANAC092 contribute non-redundantly in an EIN2-dependent manner to age-related resistance in Arabidopsis. Physiol. Mol. Plant Pathol. 76, 212–222. doi: 10.1016/j.pmpp.2011.09.003
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Balazadeh, S., Siddiqui, H., Allu, A. D., Matallana-Ramirez, L. P., Caldana, C., Mehrnia, M., et al. (2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62, 250–264. doi: 10.1111/j.1365-313X.2010.04151.x
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57, 289–300.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Carviel, J. L., Al-Daoud, F., Neumann, M., Mohammad, A., Provart, N. J., Moeder, W., et al. (2009). Forward and reverse genetics to identify genes involved in the age-related resistance response in Arabidopsis thaliana. Mol. Plant Pathol. 10, 621–634. doi: 10.1111/j.1364-3703.2009.00557.x
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w(1118); iso-2; iso-3. Fly 6, 80–92. doi: 10.4161/fly.19695
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coutinho, P. M., Starn, M., Blanc, E., and Henrissat, B. (2003). Why are there so many carbohydrate-active enzyme-related genes in plants? Trends Plant Sci. 8, 563–565. doi: 10.1016/j.tplants.2003.10.002
Culbertson, A. T., and Zabotina, O. A. (2015). The Glycosyltransferases involved in synthesis of plant cell wall polysaccharides: present and future. JSM Enzymol. Protein Sci. 1:1004.
Fernandez-Calvino, L., Guzman-Benito, I., Del Toro, F. J., Donaire, L., Castro-Sanz, A. B., Ruiz-Ferrer, V., et al. (2016). Activation of senescence-associated Dark-inducible (DIN) genes during infection contributes to enhanced susceptibility to plant viruses. Mol. Plant Pathol. 17, 3–15. doi: 10.1111/mpp.12257
Forcat, S., Bennett, M. H., Mansfield, J. W., and Grant, M. R. (2008). A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods 4:16. doi: 10.1186/1746-4811-4-16
Guo, P. R., Li, Z. H., Huang, P. X., Li, B. S., Fang, S., Chu, J. F., et al. (2017). A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29, 2854–2870. doi: 10.1105/tpc.17.00438
Guo, Y. F., and Gan, S. S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612. doi: 10.1111/j.1365-313X.2006.02723.x
Hansen, S. F., Harholt, J., Oikawa, A., and Scheller, H. V. (2012). Plant glycosyltransferases beyond CAZy: a perspective on DUF families. Front. Plant Sci. 3:59. doi: 10.3389/Fpls.2012.00059
Hinderhofer, K., and Zentgraf, U. (2001). Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213, 469–473. doi: 10.1007/s004250000512
Huang, X. X., Zhu, G. Q., Liu, Q., Chen, L., Li, Y. J., and Hou, B. K. (2018). Modulation of plant salicylic acid-associated immune responses via glycosylation of dihydroxybenzoic acids. Plant Physiol. 176, 3103–3119. doi: 10.1104/pp.17.01530
Ivanov, S., and Harrison, M. J. (2014). A set of fluorescent protein-based markers expressed from constitutive and arbuscular mycorrhiza-inducible promoters to label organelles, membranes and cytoskeletal elements in Medicago truncatula. Plant J. 80, 1151–1163. doi: 10.1111/tpj.12706
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Karimi, M., De Meyer, B., and Hilson, P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10, 103–105. doi: 10.1016/j.tplants.2005.01.008
Ke, S., Liu, S., Luan, X., Xie, X. M., Hsieh, T. F., and Zhang, X. Q. (2019). Mutation in a putative glycosyltransferase-like gene causes programmed cell death and early leaf senescence in rice. Rice 12:7. doi: 10.1186/s12284-019-0266-1
Khan, M., Rozhon, W., and Poppenberger, B. (2014). The role of hormones in the aging of plants - a mini-review. Gerontology 60, 49–55. doi: 10.1159/000354334
Kim, J. H., Woo, H. R., Kim, J., Lim, P. O., Lee, I. C., Choi, S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057. doi: 10.1126/science.1166386
Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2, 437–445. doi: 10.1105/tpc.2.5.437
Koo, J. C., Lee, I. C., Dai, C., Lee, Y., Cho, H. K., Kim, Y., et al. (2017). The protein trio RPK1-CaM4-RbohF mediates transient superoxide production to trigger age-dependent cell death in Arabidopsis. Cell Rep. 21, 3373–3380. doi: 10.1016/j.celrep.2017.11.077
Kovacik, J., Gruz, J., Backor, M., Strnad, M., and Repcak, M. (2009). Salicylic acid-induced changes to growth and phenolic metabolism in Matricaria chamomilla plants. Plant Cell Rep. 28, 135–143. doi: 10.1007/s00299-008-0627-5
Lai, J., Yu, B., Cao, Z., Chen, Y., Wu, Q., Huang, J., et al. (2015). Two homologous protein S-acyltransferases, PAT13 and PAT14, cooperatively regulate leaf senescence in Arabidopsis. J. Exp. Bot. 66, 6345–6353. doi: 10.1093/jxb/erv347
Lampugnani, E. R., Khan, G. A., Somssich, M., and Persson, S. (2018). Building a plant cell wall at a glance. J. Cell Sci. 131, jcs207373. doi: 10.1242/jcs.207373
Lao, J. M., Oikawa, A., Bromley, J. R., McInerney, P., Suttangkakul, A., Smith-Moritz, A. M., et al. (2014). The plant glycosyltransferase clone collection for functional genomics. Plant J. 79, 517–529. doi: 10.1111/tpj.12577
Lee, I. H., Lee, I. C., Kim, J., Kim, J. H., Chung, E. H., Kim, H. J., et al. (2016). NORE1/SAUL1 integrates temperature-dependent defense programs involving SGT1b and PAD4 pathways and leaf senescence in Arabidopsis. Physiol. Plant 158, 180–199. doi: 10.1111/ppl.12434
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, Y., Chang, Y., Zhao, C., Yang, H., and Ren, D. (2016). Expression of the inactive ZmMEK1 induces salicylic acid accumulation and salicylic acid-dependent leaf senescence. J. Integr. Plant Biol. 58, 724–736. doi: 10.1111/jipb.12465
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382.
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Miura, K., Lee, J., Miura, T., and Hasegawa, P. M. (2010). SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 51, 103–113. doi: 10.1093/pcp/pcp171
Morris, K., MacKerness, S. A., Page, T., John, C. F., Murphy, A. M., Carr, J. P., et al. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23, 677–685. doi: 10.1046/j.1365-313x.2000.00836.x
Mortimer, J. C., Yu, X., Albrecht, S., Sicilia, F., Huichalaf, M., Ampuero, D., et al. (2013). Abnormal glycosphingolipid mannosylation triggers salicylic acid-mediated responses in Arabidopsis. Plant Cell 25, 1881–1894. doi: 10.1105/tpc.113.111500
Nawrath, C., and Metraux, J. P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. doi: 10.1105/tpc.11.8.1393
Ng, G., Seabolt, S., Zhang, C., Salimian, S., Watkins, T. A., and Lu, H. (2011). Genetic dissection of salicylic acid-mediated defense signaling networks in Arabidopsis. Genetics 189, 851–859. doi: 10.1534/genetics.111.132332
Nishimura, M. T., Stein, M., Hou, B. H., Vogel, J. P., Edwards, H., and Somerville, S. C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301, 969–972. doi: 10.1126/science.1086716
Nuhse, T. S. (2012). Cell wall integrity signaling and innate immunity in plants. Front. Plant Sci. 3:280. doi: 10.3389/Fpls.2012.00280
Park, J. M., and Paek, K. H. (2007). Recognition and response in plant-pathogen interactions. J. Plant Biol. 50, 132–138. doi: 10.1007/Bf03030621
Piisila, M., Keceli, M. A., Brader, G., Jakobson, L., Joesaar, I., Sipari, N., et al. (2015). The F-box protein MAX2 contributes to resistance to bacterial phytopathogens in Arabidopsis thaliana. BMC Plant Biol. 15:53. doi: 10.1186/s12870-015-0434-4
Rea, G., de Pinto, M. C., Tavazza, R., Biondi, S., Gobbi, V., Ferrante, P., et al. (2004). Ectopic expression of maize polyamine oxidase and pea copper amine oxidase in the cell wall of tobacco plants. Plant Physiol. 134, 1414–1426. doi: 10.1104/pp.103.036764
Saint-Jore-Dupas, C., Nebenfuhr, A., Boulaflous, A., Follet-Gueye, M. L., Plasson, C., Hawes, C., et al. (2006). Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18, 3182–3200. doi: 10.1105/tpc.105.036400
Sakuraba, Y., Jeong, J., Kang, M. Y., Kim, J., Paek, N. C., and Choi, G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5:4636. doi: 10.1038/ncomms5636
Schippers, J. H. (2015). Transcriptional networks in leaf senescence. Curr. Opin. Plant Biol. 27, 77–83. doi: 10.1016/j.pbi.2015.06.018
Schippers, J. H., Schmidt, R., Wagstaff, C., and Jing, H. C. (2015). Living to die and dying to live: the survival strategy behind leaf senescence. Plant Physiol. 169, 914–930. doi: 10.1104/pp.15.00498
Segarra, G., Jauregui, O., Casanova, E., and Trillas, I. (2006). Simultaneous quantitative LC-ESI-MS/MS analyses of salicylic acid and jasmonic acid in crude extracts of Cucumis sativus under biotic stress. Phytochemistry 67, 395–401. doi: 10.1016/j.phytochem.2005.11.017
Seyfferth, C., and Tsuda, K. (2014). Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Front. Plant Sci. 5:697. doi: 10.3389/fpls.2014.00697
Tedman-Jones, J. D., Lei, R., Jay, F., Fabro, G., Li, X. M., Reiter, W. D., et al. (2008). Characterization of Arabidopsis mur3 mutations that result in constitutive activation of defence in petioles, but not leaves. Plant J. 56, 691–703. doi: 10.1111/j.1365-313X.2008.03636.x
Temple, H., Saez-Aguayo, S., Reyes, F. C., and Orellana, A. (2016). The inside and outside: topological issues in plant cell wall biosynthesis and the roles of nucleotide sugar transporters. Glycobiology 26, 913–925. doi: 10.1093/glycob/cww054
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Truernit, E., Baub, H., Belcram, K., Barthelemy, J., and Palauqui, J. C. (2012). OCTOPUS, a polarly localised membrane-associated protein, regulates phloem differentiation entry in Arabidopsis thaliana. Development 139, 1306–1315. doi: 10.1242/dev.072629
Vega-Sanchez, M. E., Verhertbruggen, Y., Christensen, U., Chen, X. W., Sharma, V., Varanasi, P., et al. (2012). Loss of cellulose synthase-like f6 function affects mixed-linkage Glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 159, 56–69. doi: 10.1104/pp.112.195495
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Vogelmann, K., Drechsel, G., Bergler, J., Subert, C., Philippar, K., Soll, J., et al. (2012). Early senescence and cell death in Arabidopsis saul1 mutants involves the PAD4-dependent salicylic acid pathway. Plant Physiol. 159, 1477–1487. doi: 10.1104/pp.112.196220
von Saint Paul, V., Zhang, W., Kanawati, B., Geist, B., Faus-Kessler, T., Schmitt-Kopplin, P., et al. (2011). The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell 23, 4124–4145. doi: 10.1105/tpc.111.088443
Wang, J., and Hou, B. (2009). Glycosyltransferases: key players involved in the modification of plant secondary metabolites. Front. Biol. China 4, 39–46. doi: 10.1007/s11515-008-0111-1
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Woo, H. R., Kim, H. J., Nam, H. G., and Lim, P. O. (2013). Plant leaf senescence and death - regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 126(Pt 21), 4823–4833. doi: 10.1242/jcs.109116
Woo, H. R., Koo, H. J., Kim, J., Jeong, H., Yang, J. O., Lee, I. H., et al. (2016). Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol. 171, 452–467. doi: 10.1104/pp.15.01929
Wu, Y., Zhang, D., Chu, J. Y., Boyle, P., Wang, Y., Brindle, I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. doi: 10.1016/j.celrep.2012.05.008
Yang, W., Schuster, C., Beahan, C. T., Charoensawan, V., Peaucelle, A., Bacic, A., et al. (2016). Regulation of meristem morphogenesis by cell wall synthases in Arabidopsis. Curr. Biol. 26, 1404–1415. doi: 10.1016/j.cub.2016.04.026
Yang, Y., Yoo, C. G., Guo, H. B., Rottmann, W., Winkeler, K. A., Collins, C. M., et al. (2017). Overexpression of a Domain of Unknown Function 266-containing protein results in high cellulose content, reduced recalcitrance, and enhanced plant growth in the bioenergy crop Populus. Biotechnol. Biofuels 10:74. doi: 10.1186/s13068-017-0760-x
Ye, C. Y., Li, T., Tuskan, G. A., Tschaplinski, T. J., and Yang, X. (2011). Comparative analysis of GT14/GT14-like gene family in Arabidopsis, Oryza, Populus, Sorghum and Vitis. Plant Sci. 181, 688–695. doi: 10.1016/j.plantsci.2011.01.021
Yolcu, S., Li, X., Li, S., and Kim, Y. J. (2018). Beyond the genetic code in leaf senescence. J. Exp. Bot. 69, 801–810. doi: 10.1093/jxb/erx401
Yoshida, S., Ito, M., Nishida, I., and Watanabe, A. (2002). Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defence responses in Arabidopsis thaliana. Plant J. 29, 427–437. doi: 10.1046/j.0960-7412.2001.01228.x
Zalepa-King, L., and Citovsky, V. (2013). A plasmodesmal glycosyltransferase-like protein. PLoS One 8:e58025. doi: 10.1371/journal.pone.0058025
Zhang, S. C., Li, C., Wang, R., Chen, Y. X., Shu, S., Huang, R. H., et al. (2017). The arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol. 173, 2294–2307. doi: 10.1104/pp.16.00008
Zhang, Z., Lenk, A., Andersson, M. X., Gjetting, T., Pedersen, C., Nielsen, M. E., et al. (2008). A lesion-mimic syntaxin double mutant in Arabidopsis reveals novel complexity of pathogen defense signaling. Mol. Plant 1, 510–527. doi: 10.1093/mp/ssn011
Zhang, Z. G., Feechan, A., Pedersen, C., Newman, M. A., Qiu, J. L., Olesen, K. L., et al. (2007). A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J. 49, 302–312. doi: 10.1111/j.1365-313X.2006.02961.x
Zhao, X. Y., Wang, J. G., Song, S. J., Wang, Q., Kang, H., Zhang, Y., et al. (2016). Precocious leaf senescence by functional loss of PROTEIN S-ACYL TRANSFERASE14 involves the NPR1-dependent salicylic acid signaling. Sci. Rep. 6:20309. doi: 10.1038/srep20309
Zhou, N., Tootle, T. L., Tsui, F., Klessig, D. F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10, 1021–1030.
Zhou, Y. H., Li, S. B., Qian, Q., Zeng, D. L., Zhang, M., Guo, L. B., et al. (2009). BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.). Plant J. 57, 446–462. doi: 10.1111/j.1365-313X.2008.03703.x
Keywords: cell wall, glycosylation, glycosyltransferase, leaf senescence, salicylic acid
Citation: Lee S, Kim M-H, Lee JH, Jeon J, Kwak JM and Kim YJ (2020) Glycosyltransferase-Like RSE1 Negatively Regulates Leaf Senescence Through Salicylic Acid Signaling in Arabidopsis. Front. Plant Sci. 11:551. doi: 10.3389/fpls.2020.00551
Received: 04 October 2019; Accepted: 14 April 2020;
Published: 15 May 2020.
Edited by:
Tzung-Fu Hsieh, North Carolina State University, United StatesReviewed by:
Xin Li, University of British Columbia, CanadaLei Wang, Institute of Botany, Chinese Academy of Sciences, China
Copyright © 2020 Lee, Kim, Lee, Jeon, Kwak and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: June M. Kwak, amt3YWtAZGdpc3QuYWMua3I=; Yun Ju Kim, eWpraW03N0BpYnMucmUua3I=; eXVuanU3N0BnbWFpbC5jb20=
†These authors have contributed equally to this work
‡Present address: Jae Ho Lee, Korean Bioinformation Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, South Korea
§ORCID: June M. Kwak, orcid.org/0000-0001-6948-5155; Yun Ju Kim, orcid.org/0000-0003-4284-9144
 Seulbee Lee
Seulbee Lee Myung-Hee Kim
Myung-Hee Kim Jae Ho Lee
Jae Ho Lee Jieun Jeon1,2
Jieun Jeon1,2 June M. Kwak
June M. Kwak Yun Ju Kim
Yun Ju Kim