
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 May 2020
Sec. Plant Breeding
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00444
 Sylvester N. T. T. Addy1*
Sylvester N. T. T. Addy1* Karen A. Cichy2
Karen A. Cichy2 Hans Adu-Dapaah1
Hans Adu-Dapaah1 Isaac K. Asante3
Isaac K. Asante3 Afutu Emmanuel4
Afutu Emmanuel4 Samuel K. Offei3
Samuel K. Offei3Cowpeas provide food and income for many small-holder farmers in Africa. Cowpea grains contain substantial quantities of protein, carbohydrates, vitamins, and fiber. In areas where subsistence farming is practiced, cowpea’s protein is cheaper than that obtained from other sources such as fish, meat, poultry or dairy products and combines well with cereal grains in diets. However, long-cooking times, typical of many grain legumes, is a major limitation to the utilization of cowpeas especially among the low-income and growing middle-income population of Africa. Long periods of cooking cowpeas lead to loss of nutrients, loss of useful time and increased greenhouse gas emission through increased burning of firewood. Fast-cooking cowpeas has the potential to deliver highly nutritious food to the hungry within shorter periods, encourage less use of firewood, improve gender equity, increase the consumption of cowpeas, trigger an increase in demand for cowpeas and thus incentivize cowpea production by smallholder farmers in Sub-Saharan Africa. In this study, the inheritance of storage-induced cooking time in cowpeas was investigated. Two sets of bi-parental crosses were conducted involving three cowpea genotypes: CRI-11(1)-1, C9P(B) and TVu7687. Generation means from six generations were used to determine the phenotypic and genotypic variances and coefficients of variation. Broad and narrow sense heritabilities and genetic advance percentage of mean were estimated. Generation mean analysis showed that additive, dominant, additive–additive, additive–dominant, and dominant–dominant gene actions were significant (p < 0.001). Fast-cooking trait was dominant over the long-cooking trait. Broad sense heritability for crosses C9P(B) × CRI-11(1)-1 and TVu7687 × CRI-11(1)-1 were 0.94 and 0.99 respectively while narrow sense heritabilities were 0.84 and 0.88 respectively. Genetic advances were 27.09 and 40.40 respectively. High narrow-sense heritabilities and moderate genetic advance for the fast-cooking trait indicated the presence of additive genes in the trait and the possibility of introgressing the trait into farmer-preferred varieties using conventional selection methods. However, due to significant epistatic gene effects observed, effective selection for fast-cooking trait would be appropriate at advanced generations.
Cowpea [Vigna unguiculata (L.) Walp] is an edible legume belonging to the family Fabaceae. It is native to Africa and is widely cultivated and consumed in many tropical and subtropical areas of Africa, Latin America, Southeast Asia, and in Southern United States (Appiah et al., 2011). It is commonly known as Southern pea, China pea and Black-eye pea in parts of the world (Olalekan and Bosede, 2010). Cowpeas provide food for humans, livestock and serve as a valuable income-generating commodity for many farmers and grain traders in Ghana and West Africa (Langyintuo et al., 2003; Timko et al., 2008). Its richness in protein makes the crop a source of plant protein (Akpapunam and Sefa-Dedeh, 1997) to people who hardly can afford animal protein from meat, fish, milk, and eggs. As a result, cowpeas contribute immensely to addressing malnutrition among the rural poor in many communities in West Africa. In Ghana, a large section of consumers purchase cowpeas from markets although many cowpea farmers also store quantities for home consumption. Cowpeas are mainly boiled and consumed as whole grains in many parts of West Africa. In Ghana, cowpea is the second most important legume after groundnut. However, its per capita consumption is low (5 kg/year) (Ministry of Food and Agriculture, 2010) compared to common beans, a close relative of cowpeas, with a per capita consumption of up to 34 kg/year in many parts of eastern, central and southern Africa and parts of Latin America (Akibode and Maredia, 2011). One of the major limitations to the increased consumption of cowpeas is their longer cooking times. Long-cooking of cowpeas is therefore a disincentive to their cultivation and consumption. Long periods of cooking cowpeas lead to loss of nutrients, loss of useful time and increased greenhouse gas emission through increased burning of firewood. Fast-cooking cowpeas will help deliver highly nutritious food to the hungry within shorter periods, encourage less use of firewood, improve gender equity, increase the consumption of cowpeas, trigger an increase in demand for cowpeas and thus incentivize cowpea production by smallholder farmers in Sub-Saharan Africa (Addy et al., 2016).
Longer cooking periods render cowpea less economical and acceptable to consumers especially when compared to cereals such as rice which cook in shorter periods. Quaye et al. (2009, 2011) and Egbadzor et al. (2013) reported these conclusions from surveys in Ghana. For this very reason, in many parts of Northern Ghana, cowpeas are popular for this adage: “you don’t wait till you’re hungry before attempting to cook cowpeas.” Typically, cowpeas found on markets in Ghana would have been stored in ambient tropical conditions for between 3 and 6 months (Golob et al., 1998) until premium prices are attained. Storage of cowpeas in these tropical conditions of high temperatures and humidity trigger a grain hardening condition called the ‘hard-to-cook’ phenomenon which leads to increase in cooking duration. This phenomenon is typical of grain legumes (Liu, 1995; Coelho et al., 2007; Nasar-Abbas et al., 2008; Pirhayati et al., 2011). For instance, while common beans in their fresh or non-stored state would generally cook in 22 min, they would later cook in 51 and 140 min after storage in ambient tropical condition for 30 and 60 days respectively (Ribeiro et al., 2005; Coelho et al., 2007). The same phenomenon is experienced in cowpeas but with varying tendencies (Addy, 2016). Thus, increase in cooking time is an indication of the susceptibility of cowpeas to grain hardening. Various procedures including appropriate storage; controlled atmospheres; pretreatments (e.g., steaming, roasting, irradiation, solar drying, microwaving) can be used to prevent or mitigate the development and effect of the hard-to-cook (HTC) phenomenon. However, these procedures do not provide a holistic, economically viable and sustainable solution to this challenge especially in developing countries due to implications on the culture and food systems of its people (Michaels and Stanley, 1988; Reyes-Moreno et al., 1994). Breeding of cowpea varieties less prone to grain hardening or the hard-to-cook state for use in developing countries is by far a more sustainable solution (Reyes-Moreno et al., 1994). A number of studies have been conducted on fast-cooking trait in grain legumes. For instance, studies on cooking time have been conducted for lentils (Erskine et al., 1985); cowpeas (Nielsen et al., 1993) and beans (Elia et al., 1997; Jacinto-Hernandez et al., 2003; Cichy et al., 2015). However, these authors carried out determination of cooking time for grains in their fresh state and not after grains have been kept in storage under tropical conditions. Therefore, a study to determine the mode of inheritance and gene action of this trait would be useful for designing an appropriate breeding program to develop cowpeas possessing resistance to grain hardening or varieties that will remain fast-cooking even after storage.
Generation mean analysis is a simple technique useful for the estimation of gene effects of traits controlled by many genes (Kearsey and Pooni, 2004). The uniqueness of the generation mean model is founded on its ability to estimate additive × additive, dominance × dominance, and additive × dominance gene effects also known as epistatic gene effects (Singh and Singh, 1992). The objectives of this study were to determine the mode of inheritance and gene action for the fast-cooking trait and estimate its genetic advance.
Three cowpea genotypes: CRI-11(1)-1, C9P(B) and TVu7687 were used in this study (Table 1). Genotype CRI-11(1)-1 is an advanced breeding line developed by the Crops Research Institute, Ghana. Genotype C9P(B) is an advanced breeding line obtained from Cinzana Agronomic Research Station, Institut d’Economie Rurale (SRAC-IER), Mali. Seeds of this genotype have a creamy seed coat color, smooth seed coat texture, black helium color and an ovoid seed shape. Genotype TVu7687 is a brown-seeded advanced breeding line developed by the International Institute for Tropical Agriculture (IITA), Nigeria. It has a smooth seed coat texture and a globose seed shape. Genotype CRI-11(1)-1 possesses resistance to storage-induced cooking time while C9P(B) and TVu7687 shows susceptibility to the trait. Its seeds have a rough seed coat texture, a black helium color and a rhomboid seed shape.
Artificial hybridization was conducted using the crossing procedure by Rachie et al. (1975) involving two steps: emasculation and pollination. Genotype CRI-11(1)-1 was used as the fast-cooking male parent (P1) while C9P(B) and TVu7687 were used as female parents (P2 and P3) respectively. At podding stage the crossing block was covered with netting to prevent birds from picking the successful pods. Pods that developed from crossing were harvested at physiological maturity. These were dried, threshed, seeds labeled and stored in cold storage. For each cross, seeds for P1, P2, F1, F2, BCP1, and BCP2 generations were obtained. All precautions to ensuring purity and success of crosses were undertaken.
Six generations (P1, P2, F1, F2, BC1, and BC2) obtained for each cross were evaluated at Crops Research Institute, Fumesua in separate randomized complete block designs with three replicates. Experimental units comprised a row each for the non-segregating generations (P1, P2, F1), two rows each for (BC1 and BC2) and six rows each for the segregating generation (F2). Each replicate consisted of 20 plants in one row for each of the parents and F1. Thirty (30) plants in three rows each for the backcross generation and 60 plants in 6 rows for the F2 population. Two meters long rows with a plant spacing of 20 cm within row and 1.0 m space between rows were used. Two seeds were sown per hill and thinned to one plant per hill. All recommended cultural practices were performed during the growing season. Harvested pods were dried, threshed and kept for cooking quality evaluation.
Harvested seeds were manually sorted to remove extremely small grains, seeds with defective seed coat and extraneous materials such as unhealthy seed, insect infested seed, soil, dust and chaff. Seeds were not treated with storage chemical before or during storage. Sixty (60) seeds were randomly selected from each entry and soaked in 120 ml of 0.1 M acetate buffer (pH 4.0) at 37°C for 7 h and then dried at room temperature for 48 h (Paredes-López et al., 1991; Reyes-Moreno et al., 2001) before cooking time determinations were carried out. Seeds were soaked in (1:4) w/w distilled water for 12 h prior to cooking. Moisture content of seeds ranged from 10–14% as determined with a moisture meter. Data on cooking time were recorded on 10 individual plants from non-segregating populations (P1, P2, and F1) and 20 plants for BC1 and BC2 and 60 plants for F2 population for each replicate. The determination of cooking time was conducted using an experimental pin drop cooker (Wang and Daun, 2005).
Analyses of variance was done using SAS 9.3 Software (SAS Institute, 2004) to test for significance of difference among generations for cooking time. A significance in differences in generations would indicate the presence of genetic variation. Means were computed for each generation (P1, P2, F1, F2, BC1, and BC2) for each cross over their replications. XLSTAT 2020.1.1 (Addinsoft, 2020) was used to test for normality (Shapiro–Wilk test) for data obtained for F2, BC1, and BC2 generations.
Generation mean was expressed in terms of its genetic effects using the following equation:
Y = m + αa + βd + α2aa + 2αβad + β2dd, by Gamble (1962).
where genetic effects were:
Y = the observed mean for generation;
m = mean effect of the F2 (i.e., the base population);
a = average additive effects;
d = average dominance effects;
aa = average interactions between additive effects;
dd = average interactions between dominance effects;
ad = additive × dominance gene interaction effects;
α and β = are coefficients for a and d.
Generation mean analysis was performed using Hayman (1958) method. The means for the genetic effects were estimated using the means of each family or generation as:
m = mean of F2;
a = BCP1- BCP2;
d = F1-4F2-0.5P1 -0.5P2 + 2BCP1 + 2BCP2;
aa = 2BCP1 + 2BCP2 – 4F2;
ad = BCP1 -0.5P1- BCP2 + 0.5P2;
dd = P1 + P2 + 2F1 + 4F2 – 4BCP1 – 4BCP2.
Variances for the genetic effects were estimated using the variances for each family or generation as:
Vm = VF2;
Va = VBP1 + VBP2;
Vd = VF1 + 16VF2 + 0.25VP1 + 0.25VP2 + 4VBP1 + 4VBP2;
Vaa = 4VBP1 + 4VBP2 + 16VF2;
Vad = VBP1 + 0.25VP1 + VBP2 + 0.25VP2;
Vdd = VP1 + VP2 + 4VF1 + 16VF2 + 16VBP1 + 16VBP2.
Standard errors for each of the genetic effects were also estimated as the square root of their corresponding variances. Genetic parameters and their variances were tested for significance by calculating ‘t’ tests as ratios of genetic effects to their respective standard errors. Broad-sense (hb2) and narrow-sense heritabilities (hn2) were estimated using variances of F2 and backcross generations according to Warner (1952).
The degree of dominance (D) which is the deviation from the mid-parent value toward one of the parents was also calculated as the ratio of the dominance to the additive effect according to Falconer and Mackay (1960):
D (degree of dominance) = d/a;
Where
Means, variances, heritabilities, degree of dominance, genotypic and phenotypic coefficient of variation and genetic advance were computed using Microsoft Excel 2013. Genotypic and phenotypic variances and coefficients of variation were computed according to Burton and Devane (1953). Genetic advance to be expected for the selection of five percent of superior progenies was computed according to Robinson et al. (1951) as follows:
Phenotypic variance (δ2p) = Var F2
Environmental variance (δ2e) = Var F1
Genotypic variance (δ2g) = (δ2p – δ2e)
Genotypic coefficient of variance (GCV) =
Phenotypic coefficient of variance (GCV) =
Genetic advance (GA) = iδp
Where Var F2 = variance of F2 population;
Var F1 = variance of corresponding F1 population;
X = general mean of the character;
I = intensity of selection (5% with a value of K being 2.06);
δp = phenotypic standard deviation;
= broad sense heritability.
Results of the analysis of variance for the six basic generations (P1, P2, F1, F2, BC1, and BC2) for both crosses are presented (Table 2). The table shows highly significant (P < 0.001) differences were detected among generations for both crosses 1 and 2. Differences found among the six generations were observed in the results of population means distribution for cooking time for crosses 1 and 2 (Tables 3, 4). In cross 1, means of generations were in increasing order of P1 < BC1P1 < F1 < BC1P2 < F2, < P2 (Table 3). However, in cross 2, means of generations were in the increasing order of P1 < BC1P3 < F1 < F2 < BC2P3 < P3 (Table 4). In both crosses, the means of the F1 fell between cooking time values for both parents indicating some level of partial or incomplete dominance. In fact, means of all other generations were less than the higher cooking time parents P2 and P3. The population mean distributions obtained for both crosses showed that P1, BC1P1 and F1 were skewed toward the short cooking time parent. Furthermore, in cross 1, the coefficient of variation for F2 was high (1.9%), although lower than coefficients of variation for F1 and BC1P1 (Table 3). However, in Cross 2, the coefficient of variation of F2 was the highest (2.09%) among all generations, whiles coefficients of variation for P1, P2, and F1 were 1%, 0.03% and 1.27% respectively (Table 4). Test for normality for the F2 segregants and backcross populations did not show a normal distribution. A Shapiro–Wilk test yielded p-values (two-tailed) of 0.011 and 0.002 for an alpha value of 0.05 for crosses 1 and 2. The Jarque-Bera test for normality, however, gave a contrary result (0.442 and 0.314) for an alpha value of 0.05 for F2 populations of crosses 1 and 2 respectively. Assuming the absence of normality, clear phenotypic classes may be established. Using a cooking time classification of short (up to 39 min), medium-cooking (40–59 min), long or slow-cooking (60–79 min), very or extra-long or extra slow-cooking (80 min and beyond) three arbitrary phenotypic classes can be created for the F2 populations of crosses 1 and 2. These have ratios 1:3:1 and 1:2:7 for F2 populations obtained for crosses 1 and 2 respectively. However, these phenotypic ratios do not fit principles of qualitative inheritance.
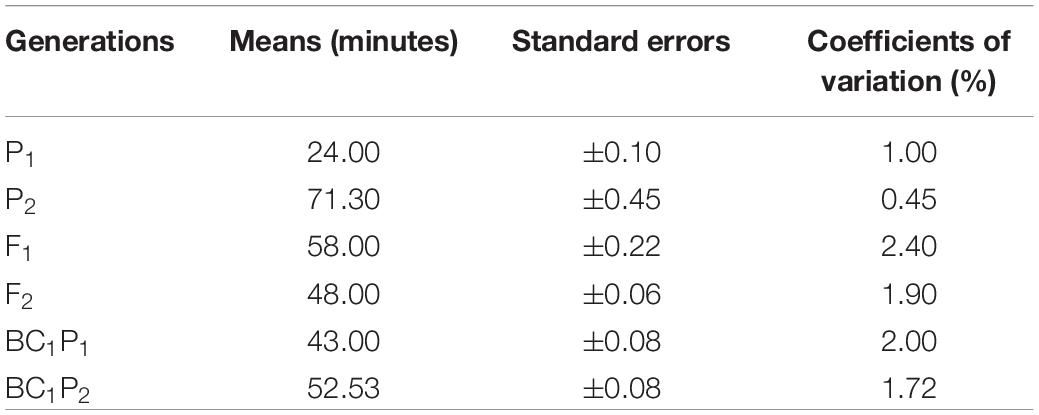
Table 3. Population mean distribution for cooking time among six basic generations derived from cross 1 [CRI-11(1)-1 × C9P(B)].
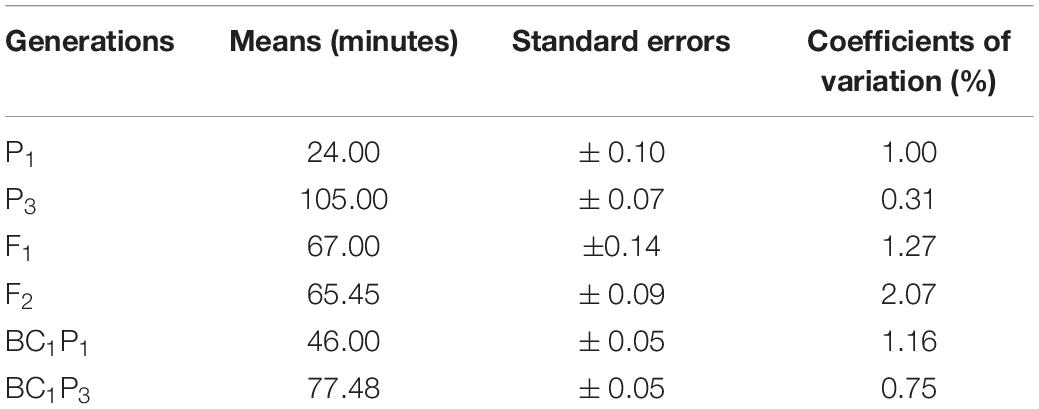
Table 4. Population mean distribution for cooking time among six basic generations derived from the cross 2 [CRI-11(1)-1 × TVu7687].
Mean values, variances and their standard errors for the analyzed traits of the two crosses are presented (Tables 5, 6). Additive and dominance gene effects were found to be significant at p = 0.001. In both crosses (1 and 2 respectively), means of dominant gene effect were higher (−103.00 and −67.65) than the additive dominant gene effect (−33.35 and −38.93). Furthermore, both dominance and additive gene effects were found to be negative.
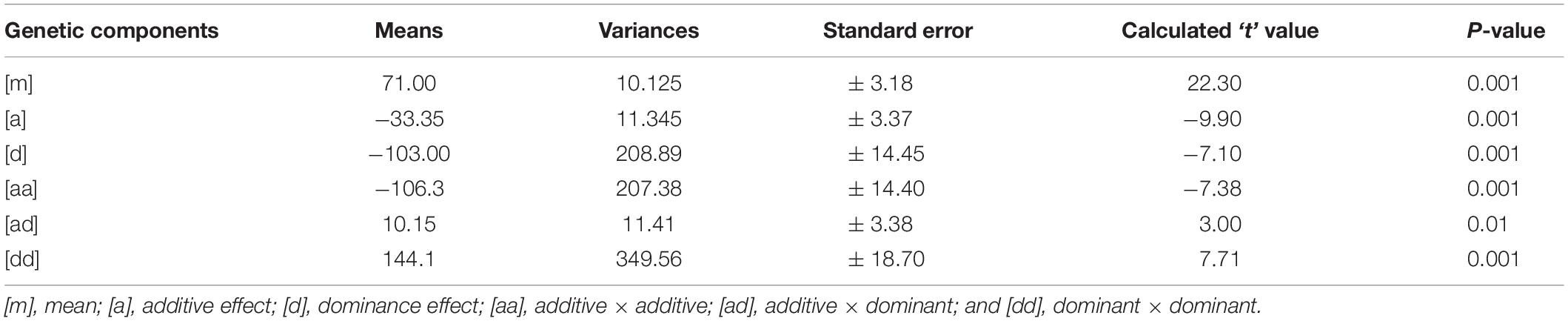
Table 5. Estimates of means, variances, and standard errors of gene effects for cross 1[CRI-11(1)-1 × C9P(B)].
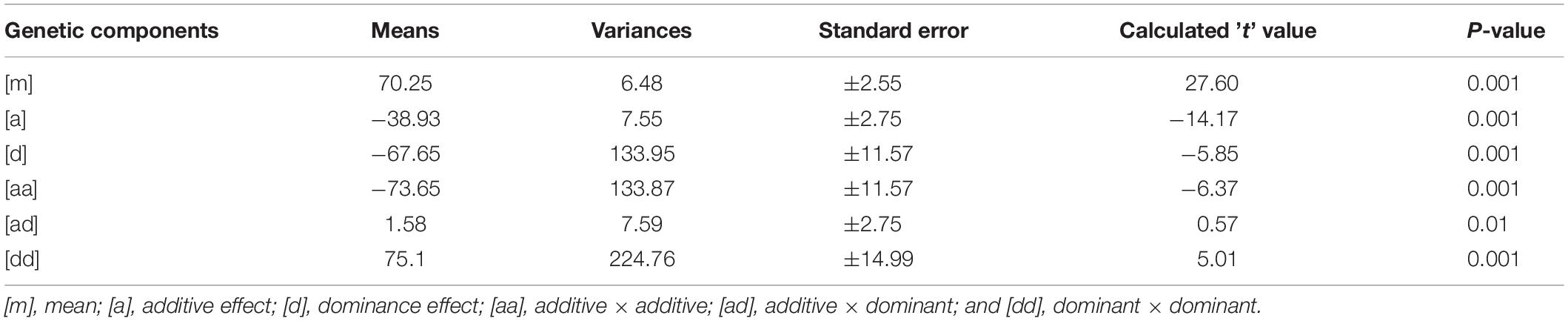
Table 6. Estimates of means, variances, and standard errors of gene effects for cross 2 [CRI-11(1)-1 × TVu7687].
Significant epistatic additive × additive, additive × dominant and dominant × dominant gene effects were detected for cooking time for cross 1 [CRI-11(1)-1 × C9P(B)]. Mean value for the epistatic dominant × dominant gene effect was the highest among the additive and dominant gene effects as well as among the epistatic gene effects. The mean value for the epistatic additive × additive gene effect was found to be the second highest among the epistatic gene effects and yet higher than the additive and dominant gene effects. Epistatic additive × dominant gene effect was found to have lowest mean value and was the only gene effect found to be significant at p = 0.01. Additive and dominant gene effects were both negative in direction. In addition, the additive × additive gene effect was the only epistatic gene effect found to be negative. Similar results were obtained for cross 2 [CRI-11(1)-1 × TVu7687] are presented in Table 6. Significant epistatic additive × additive and dominant × dominant gene effects were detected for cooking time for this cross. Epistatic additive × dominant gene effect was the only gene effect detected to be non-significant. All other gene effects were significant at p = 0.001. Similarly, dominant gene effect was higher than additive gene effect. In addition, epistatic dominant × dominant gene effect was also higher than the mean value for additive × additive gene effect. Similar to cross 1 [CRI-11(1)-1 × C9P(B)], the additive and dominant gene effects were found to be negative. Furthermore, the epistatic additive × additive gene effect was the only epistatic gene effect found to be negative.
All gene effects were found to be significant for both crosses except epistatic additive × dominant gene effect in cross 2 [CRI-11(1)-1 × TVu7687]. In addition, epistatic dominance × dominance, epistatic additive × dominance, dominance and additive gene effects were values in decreasing order of magnitude in both crosses. Furthermore, epistatic additive × dominance, dominance and additive gene effects were found to be negative. All remaining epistatic gene effects were found to be positive.
Broad sense and narrow sense heritabilities were computed for each cross and presented in Table 7. Broad sense heritabilities obtained were 0.94 and 0.99 for crosses 1 and 2 respectively. Broad sense heritabilities for the two crosses were generally high but marginally higher for cross 2 than for cross 1. Also, high narrow sense heritabilities of 0.88 and 0.84 were obtained for crosses 1 and 2 respectively, although the narrow sense heritability of cross 1 was marginally higher than cross 2. Generally, narrow sense heritabilities were lower than broad sense heritabilities for both crosses. Degree of dominance was higher (−0.15) in cross 2 than for cross 1 (0.08a). Thus, in both crosses, the degree of dominance was between zero and one (−1 < D < 0) for cooking time.
Variances, coefficient of variability and genetic advance of cooking time (after storage) in two crosses in cowpea are presented (Table 8). In both crosses, the differences between the genotypic and phenotypic coefficient of variation values were marginal. Genetic advance for cross 1 was 27.09 which was lower than the value obtained for cross 2 (40.4).

Table 8. Variances, coefficient of variability and genetic advance of cooking time (after storage) in two crosses in cowpea.
The differences among analyzed generations were sufficient to perform generation mean analysis for both crosses. In both crosses, the means of F1 fell between cooking time values for both parents indicating some level of partial or incomplete dominance. Observed variations among populations of both crosses indicate a high possibility of response to selection for cooking time trait. It also indicates that parents chosen for these bi-parental crosses, to a large extent, had the contrasting expressions for the cooking time trait. The extent of variation among populations is useful since the progress of any breeding program depends largely on the existence of genetic variability within the population available (Akhshi et al., 2014). Population mean values obtained for both crosses which showed that P1, BC1P1, and F1 were skewed toward the short cooking time parent indicate dominance over long-cooking time although other gene effects could play a role in the inheritance of cooking time. Elia (2003) attributed skewness of the distribution in cooking time in favor of short cooking parents to either maternal effects, or control by dominant genes.
Relatively higher coefficients of variations observed for especially the segregating generations such as F2 and the backcrosses, can be attributed to the combined action and interaction of various gene effects as well as the environment. Coefficients of variation indicate the level of dispersion around the mean. Consequently, the higher the coefficient of variation, the greater the level of dispersion around the mean. On the other hand, the relatively lower coefficients of variation for cooking time values of parents (P1, P2, and P3) is attributed mainly to effect of environmental factors (Acquaah, 2007). This is because parents are considered homozygous while F1 progenies are uniformly heterozygous. This phenomenon generally indicates that cooking time trait in cowpeas is quantitatively inherited. In this study, results of the Shapiro–Wilk test for normality suggest the occurrence of qualitative inheritance of the cooking time. However, ratios of computed phenotypic classes did not fit any expected Mendelian ratios for qualitative inheritance. Jarque–Bera test for normality for the F2 populations for both crosses however showed continuous distribution indicating the occurrence of quantitative inheritance. In an evaluation of cooking time on 140 F2:4 families segregating for cooking time, six QTL were identified for cooking time in common beans in a single environment (Garcia et al., 2012), In another study in common beans, Cichy et al. (2015) also found markers with small effects on cooking time. They found regions on chromosomes Pv02, Pv03, and Pv06 associated with cooking time.
Furthermore, ratios obtained for both crosses were not similar. This difference may be due to the brown-seeded parent (TVu7687) involved in the Cross 2. Generally, colored cowpeas are known to be long-cooking while white or cream-colored genotypes are fast-cooking (Addy et al., 2016). According to Cichy et al. (2015), three QTL on chromosome Pv06 which were associated with cooking time had locations also associated with three UDP-glucosyl transferase genes. This gene family is reported to be involved in flavonoid biosynthesis and the development of pigments (Bowles et al., 2005). Seed coat color due to the presence of anthocyanins was correlated with cooking time.
Significant but negative additive gene effects obtained for both crosses is an indication of assurance of gains with selection for fast-cooking trait in the long term (Acquaah, 2007). The negative value of the dominance gene effect, observed showed that the alleles responsible for less value of the trait (fast or fast-cooking) was dominant over the alleles controlling high value of the trait (long or slow-cooking). This result is in agreement with the work of Jacinto-Hernandez et al. (2003) who reported on the dominance of short cooking time in Phaseolus vulgaris L. in Mexico. Negative values for the additive gene effect indicates that alleles conferring short cooking time and not for the long-cooking time trait are responsible for parent-offspring resemblance. The existence of significant and negative additive gene effect would make selection for short cooking time at early stages of breeding effective. However, due to significant epistatic dominance × dominance, epistatic additive × dominance gene effects in cross [CRI-11(1)-1 × C9P(B)] and significant epistatic dominance × dominance gene effect observed in cross [CRI-11(1)-1 × TVu7687] selection for short cooking time trait at advanced generations would be the effective breeding decision to consider.
High estimates for broad and narrow sense heritabilities obtained for storage-induced cooking time are similar to values reported from other studies. Nielsen et al. (1993) reported broad sense heritability of 0.76 and a narrow sense heritability ranging from 58 to 85% for cooking time in cowpea. Investigations using lentils gave a broad sense heritability of 0.82 for cooking time (Erskine et al., 1985). Elia et al. (1997) also reported a high value of 0.9 narrow sense heritability for the same trait in Phaseolus vulgaris in Tanzania while Jacinto-Hernandez et al. (2003) reported a value of 0.74 narrow sense heritability for the same trait in Phaseolus vulgaris in Mexico. In a dry bean GxE study across nine locations in Africa, North America, and the Caribbean, the heritability of cooking time was 98% on presoaked beans and 60% on unsoaked beans (Cichy et al., 2019). Mashi (2006) also reported estimates of heritability in the broad sense ranged from 89 to 95% and a range of 58–85% for narrow sense heritability in cowpeas. Generally, both broad and narrow sense heritabilities reported are high for both cowpeas and beans. Similar range of heritability estimates between cowpeas and beans is probable because they are closely related species. These high estimates of both broad and narrow sense heritabilities obtained in this study agree with the generally high estimates reported for both cowpeas and beans. According to Stansfield (1988), traits with narrow sense heritability higher than 50% are considered to have a high heritability. High heritability estimates may be due to high additive genetic variance in relation to the total phenotypic variance for the trait. High narrow sense heritability indicates a high measure of the predictability of offspring trait values that is based on parental trait values. This means it is possible to make genetic advancements for the cooking time trait. Although, heritability estimates reported and obtained from this study are within similar ranges, there is need for some clarity in the type of cooking time examined by these authors. These authors may have worked on cowpeas or beans grains at different storage periods and conditions. This is due to the trigering and increasing effect of the ‘hard-to-cook’ phenomenon or grain hardening during storage. This is a phenomenon, typical of cowpeas and beans (Coelho et al., 2007), and occurs during storage in environmental conditions such as high temperatures (30–40°C) and high humidity (>75%) prevalent in the tropics and sub-tropics. A clear distinction, therefore, needs to be made for data of cooking time obtained in any study of this nature. This study specifically examined the heritability of storage-induced cooking time in cowpeas. It is likely that some or all studies on cooking time may have been carried out with seeds that had the hard-to-cook phenomenon already triggered.
The degree of dominance was between zero and one (−1 < D < 0) for cooking time indicating incomplete or partial dominance with marginal magnitude in the direction of the long cooking parent. In addition, mid-parent values were lower than F1 means for cooking time in the two crosses. It indicates that that the cooking time of a heterozygous genotype is distinct from and often intermediate to the phenotypes of the homozygous genotypes. However, the degree of partial dominance obtained for both crosses were skewed toward zero than −1. It follows also that for the two crosses studied the means of cooking time of the first filial generation were toward the fast-cooking trait than for the long-cooking trait.
Estimates of degree of dominance between zero and one indicate low effect of environment on the expression of cooking time in cowpeas. Low to moderate coefficients of variation of phenotype and genotype indicate low to moderate influence of environment on the trait thus making it a reliable one for selection. However, high genetic coefficient of variation recorded for cooking time alone is not sufficient for the determination of the extent of the advance to be expected by selection. Burton (1952) suggested that genetic coefficient of variation together with heritability estimates would give the best picture of the extent of the advance to be expected by selection. Considering heritability estimates in conjunction with the predicted genetic gain constitute a more reliable strategy for selection. Although heritability estimates are the true indicators of potential genetic ability of genotypes that can be used as a tool for selection, changes in the values of the heritability estimates due to changes in the environment suggest that total dependence on such estimates should be done with a lot of caution. Therefore, the high heritability estimates and moderate to high genetic advance for the fast-cooking trait in the present study indicate that it is controlled by additive gene action rendering phenotypic selection for cooking time an effective strategy (Tazeen et al., 2009). This means that the additive nature of genetic variation was transmitted from the parents to the progeny.
Genetic studies on storage-induced cooking time trait in cowpea have rarely been reported previously. This study showed that fast-cooking time was dominant over long-cooking time. High broad and narrow sense heritabilities with moderate genetic advances indicated the additive nature of inheritance. However, due to significant epistatic gene effects observed, selection for fast-cooking time trait at advanced generations would be the effective breeding decision to consider. This is so especially when cooking time evaluation is time consuming and somewhat laborious. Finally, since evaluation of cooking time in cowpea is expensive and time consuming, we recommend that additional studies be carried out to identify molecular markers for the fast-cooking trait as a means for indirect selection. The development of fast-cooking cowpea varieties acceptable to consumers have the potential to increase their per capita consumption and help reduce the emission of greenhouse gasses through prolonged burning of fuel during cooking.
The datasets generated for this study are available on request to the corresponding author.
SA initiated and designed the research, conducted the research, analyzed the data, and had primary responsibility for final content. SA and KC provided essential materials and workspace. SA, KC, HA-D, IA, SO, and AE wrote the manuscript. All authors have read and approved the final manuscript.
This work was supported in part by the CSIR-Crops Research Institute for the provision of land for field experiments. All other costs were borne by lead author.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to Edward Appiah of the CSIR-Crops Research Institute for his assistance with fieldwork and the Norman Borlaug Leadership Enhancement in Agriculture Program (LEAP) for facilitating collaboration with Dr. Karen Cichy of Michigan State University.
Acquaah, G. (2007). Principles of Plant Genetics and Breeding, 2nd Edn. Hoboken, NJ: Blackwell Publishing.
Addy, S. N. T. T. (2016). Breeding Fast-cooking and Iron Biofortified Cowpea in Ghana. Doctoral dissertation, University of Ghana, Ghana. 190.
Addy, S. N. T. T., Asante, I. K., Adu-Dapaah, H., Offei, S. K., Cichy, K. A., and Wiesinger, J. (2016). “Breeding Fast Cooking Cowpeas for Increased Utilization and Improved Nutrition in Sub-Saharan Africa,” in Proceedings of the Pan-African Grain Legume & World Cowpea Conference. 2016 International Year of Pulses, Livingstone.
Akhshi, N., Cheghamirza, K., Ahmadi, H., and Firouzabadi, N. F. (2014). Generation mean analysis to estimate genetic parameters for morphological traits in common bean (Phaseolus vulgaris L.). J. Biodiver. Environ. Sci. 4, 254–261.
Akibode, S., and Maredia, M. (2011). Global and Regional Trends in Production, Trade and Consumption of Food Legume Crops. Michigan: Michigan State University, 83.
Akpapunam, M. A., and Sefa-Dedeh, S. (1997). Jack bean (Canavalia ensiformis) nutrition related aspects and needed research. J. Plant Food Hum. Nutr. 10, 123–127.
Appiah, F., Asibuo, J. Y., and Kumah, P. (2011). Physicochemical and functional properties of bean flours of three cowpea (Vigna unguiculata (L.) Walp) varieties in Ghana. Afr. J. Food Sci. 5, 100–104.
Bowles, D., Isayenkova, J., Lim, E.-K., and Poppenberger, B. (2005). Glycosyltransferases: managers of small molecules. Curr. Opin. Plant Biol. 8, 254–263. doi: 10.1016/j.pbi.2005.03.007
Burton, G. W. (1952). Quantitative inheritance in grasses. Proc. Sixth Int. Grassland Congr. 1, 277–283.
Burton, G. W., and Devane, E. M. (1953). Estimating heritability in fall fescue (Festecd cirunclindcede) from replicated clonal material. Agron. J. 45, 478–481.
Cichy, K. A., Wiesinger, J. A., Matthew, B., Nchimbi-Msolla, S., Deidre, F., Porch, T. G., et al. (2019). The role of genotype and production environment in determining the cooking time of dry beans (Phaseolus vulgaris L.). Legume Sci. 1:e13.
Cichy, K. A., Wiesinger Jason, A., and Mendoza Fernando, A. (2015). Genetic diversity and genome-wide association analysis of cooking time in dry bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 128, 1555–1567. doi: 10.1007/s00122-015-2531-z
Coelho, C. M. M., de Mattos Bellato, C., Santos, J. C. P., Ortega, E. M. M., and Tsai, S. M. (2007). Effect of phytate and storage conditions on the development of the ‘hard-to-cook’ phenomenon in common beans. J. Sci. Food Agric. 87, 1237–1243. doi: 10.1002/jsfa.2822
Egbadzor, K. F., Yeboah, M., Offei, S. K., Ofori, K., and Danquah, E. Y. (2013). Farmers’ key production constraints and traits desired in cowpea in Ghana. J. Agricult. Extens. Rural Dev. 5:14. doi: 10.15414/raae.2019.22.01.14-23
Elia, F. M. (2003). Heritability of cooking time and water absorption traits in dry beans (Phaseolus vulgaris L.) using a North Carolina design II mating scheme. Tanzania J. Sci. 29, 25–35. doi: 10.4314/tjs.v29i1.18364
Elia, F. M., Hosfield, G. L., and Kelly, J. D. (1997). Genetic analysis and interrelationships between traits for cooking time, water absorption and protein and tannin content of Andean dry beans. J. Am. Soc. Horticult. Sci. 122, 512–518. doi: 10.21273/jashs.122.4.512
Erskine, W., Williams, P.C., and Nakkoul, H. (1985). Genetic and environmental variation in the seed size, protein, yield, and cooking quality of lentils. Field Crops Res. 12, 153–161. doi: 10.1016/0378-4290(85)90061-9
Falconer, D. S., and Mackay, T. F. C. (1960). Introduction to Quantitative Genetics, 4th Edn. New York, NY: Longman, 132–133.
Gamble, E. E. (1962). Gene effects in corn (Zea mays L.): I. Separation and relative importance of gene effects for yield. Can. J. Plant Sci. 42, 339–348. doi: 10.4141/cjps62-048
Garcia, R. A. V., Rangel, P. N., Bassinello, P. Z., Brondani, C., Melo, L. C., Sibov, S. T., et al. (2012). QTL mapping for the cooking time of common beans. Euphytica 186, 779–792. doi: 10.1007/s11032-018-0789-y
Golob, P., Andean, H. F., Atarigiya, J., and Ran, B. M. D. (1998). “On-farm storage losses of cowpea and bambara groundnut in Northern Ghana,” in Proceedings of the 7th International Working Conference on Stored-product Protection - Volume 2, Beijing.
Hayman, B. I. (1958). The separation of epistatic from additive and dominance variation in generation means. Heredity 12, 371–390. doi: 10.1038/hdy.1958.36
Jacinto-Hernandez, C., Azpiroz-Rivero, S., Acosta-Gallegos, J. A., Hernandez- Sanchez, H., and Bernal-Lugo, I. (2003). Genetic analysis and random amplified polymorphic DNA markers associated with cooking time in common bean. Crop Sci. Soc. Am. 43, 329–332.
Kearsey, M. J., and Pooni, H. S. (2004). The Genetical Analysis of Quantitative Traits. London: Chapman and Hall, 12–26.
Langyintuo, A. S., Lowenberg-DeBoer, J., Faye, M., Lambert, D., Ibro, G., Moussa, B., et al. (2003). Cowpea supply and demand in West and Central Africa. Field Crops Res. 82, 215–231. doi: 10.1016/s0378-4290(03)00039-x
Liu, K. (1995). Cellular, biological, and physicochemical basisdor the hard to cook defect in legume seeds. CRC Crt. Rev. Food Sci. Nutr. 35, 263–298. doi: 10.1080/10408399509527702
Mashi, D. S. (2006). Genetic Studies on Seed Coat Texture and Cooking Time in some varieties of Cowpea (Vigna unguiculata (L.) Walp. PhD thesis, Faculty of Natural Sciences, Slovakia, 131.
Michaels, T. M., and Stanley, D. W. (1988). “Is hard-to-cook defect in common beans under genetic control?,” in Beans Network, Proceedings of the 3rd Workshop, Termas de Panimavida, Chile, 25–28 November 1987, eds L. G. Elías and W. Edwardson (Ottawa: IDRC), 91–97.
Ministry of Food and Agriculture (2010). Statistics, Research and Information Directorate (SRID). Accra: Ministry of Food and Agriculture.
Nasar-Abbas, S. M., Plummer, J. A., Siddique, K. H. M., White, P., Harris, D., and Dods, K. (2008). Cooking quality of faba bean after storage at high temperature and the role of lignins and other phenolics in bean hardening. LWT Food Sci. Technol. 41, 1260–1267. doi: 10.1016/j.lwt.2007.07.017
Nielsen, S., Brandt, W., and Singh, B. (1993). Genetic variability for nutritional composition and cooking time of improved cowpea lines. Crop Sci. 33, 469–472. doi: 10.2135/cropsci1993.0011183x003300030010x
Olalekan, A. J., and Bosede, B. F. (2010). Comparative study on chemical composition and functional properties of three Nigerian legumes (Jack beans, pigeon pea and cowpea). J. Emerg. Trends Eng. Appl. Sci. 1, 89–95.
Paredes-López, O., Cárabez-Trejo, A., Palma-Tirado, L., and Reyes-Moreno, C. (1991). Influence of hardening procedure and soaking solution on cooking quality of common beans. Plant Foods Hum. Nutr. 41, 155–164. doi: 10.1007/bf02194084
Pirhayati, M., Soltanizadeh, N., and Kadivar, M. (2011). Chemical and microstructural evaluation of ‘hard-to-cook’ phenomenon in legumes (pinto bean and small-type lentil). Int. J. Food Sci. Technol. 46, 1884–1890. doi: 10.1111/j.1365-2621.2011.02697.x
Quaye, W., Adofo, K., Buckman, E. S., Frempong, G., Jongerden, J., and Guido, R. (2011). A socio-economic assessment of cowpea diversity on the Ghanaian market: implications for breeding. Int. J. Cons. Stud. 35, 679–687. doi: 10.1111/j.1470-6431.2010.00980.x
Quaye, W., Adofo, K., Madode, Y. E., and Abizari, A.-R. (2009). Exploratory and multidisciplinary survey of the cowpea network in Tolon-Kumbungu district of Ghana: a food sovereignty perspective. Afr. J. Agric. Res. 4, 311–320.
Rachie, K. O., Rawal, K. M., and Franckowiak, J. D. (1975). A Rapid Method of Handling Cowpeas. Ibadan: IITA.
Reyes-Moreno, C., Cárabez-Trejo, A., Paredes-López, O., and Ordorica-Falomir, C. (1994). Physicochemical and structural properties of two bean varieties which differ in cooking time and the HTC characteristic. Lebensm Wiss Technol. 23, 331–336. doi: 10.1006/fstl.1994.1068
Reyes-Moreno, C., Rouzaud-Sandez, O., Milán-Carrillo, J., Garzón-Tiznado, J. A., and Camacho-Hernández, L. (2001). Hard-to-cook tendency of chickpea (Cicer arietinum L) varieties. J. Sci. Food Agricult. 81, 1008–1012. doi: 10.1002/jsfa.872
Ribeiro, H. J. S. S., Prudencio-Ferreira, S. H., and Miyagui, D. T. (2005). Propriedades fisicas e quimicas de feijao comum preto, cultivar lapar 44 após envelhecimento acelerado. Ciênc. Tecnol. Alim. 25, 165–169. doi: 10.1590/s0101-20612005000100027
Robinson, H. E., Comstock, R. E., and Harvay, P. H. (1951). Genotypic and phenotypic correlations in corn and their implications in selection. Agron. J. 43, 282–287. doi: 10.2134/agronj1951.00021962004300060007x
Singh, R. P., and Singh, S. (1992). Estimation of genetic parameters through generation mean analysis in bread wheat. Indian J. Genet. 52, 369–375.
Tazeen, M., Nadia, K., and Farzana, N. N. (2009). Heritability, phenotypic correlation and path coefficient studies for some agronomic characters in synthetic elite lines of wheat. J. Food Agricult. Environ. 7, 278–282.
Timko, M. P., Rushton, P. J., Laudeman, T. W., Bokowiec, M. T., Chipumuro, E., Cheung, F., et al. (2008). Sequencing and analysis of the gene-rich space of cowpea. BMC Genome 9:103. doi: 10.1186/1471-2164-9-103
Wang, N., and Daun, J. K. (2005). Determination of cooking times of pulses using an automated Mattson cooker apparatus. J. Sci. Food Agricult. 85:1631e1635.
Keywords: heritability, cooking time, cowpea, hard-to-cook, gene action
Citation: Addy SNTT, Cichy KA, Adu-Dapaah H, Asante IK, Emmanuel A and Offei SK (2020) Genetic Studies on the Inheritance of Storage-Induced Cooking Time in Cowpeas [Vigna unguiculata (L.) Walp]. Front. Plant Sci. 11:444. doi: 10.3389/fpls.2020.00444
Received: 10 September 2019; Accepted: 25 March 2020;
Published: 05 May 2020.
Edited by:
Roberto Papa, Marche Polytechnic University, ItalyReviewed by:
Timothy Close, University of California, Riverside, United StatesCopyright © 2020 Addy, Cichy, Adu-Dapaah, Asante, Emmanuel and Offei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sylvester N. T. T. Addy, dmVzdGVyYTIwMDBAeWFob28uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.