- 1CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
- 2Center of Conservation Biology, Core Botanical Gardens, Chinese Academy of Sciences, Wuhan, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4Department of Botany and Plant Sciences, University of California, Riverside, Riverside, CA, United States
- 5School of Integrative Plant Science, Section of Plant Biology and the L.H. Bailey Hortorium, Cornell University, Ithaca, NY, United States
- 6Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
Dolomiaea is a medicinally important genus of Asteraceae endemic to alpine habitats of the Qinghai-Tibet Plateau (QTP) and adjacent areas. Despite significant medicinal value, genomic resources of Dolomiaea are still lacking, impeding our understanding of its evolutionary history. Here, we sequenced and annotated plastomes of four Dolomiaea species. All analyzed plastomes share the gene content and structure of most Asteraceae plastomes, indicating the conservation of plastome evolutionary history of Dolomiaea. Eight highly divergent regions (rps16-trnQ, trnC-petN, trnE-rpoB, trnT-trnL-trnF, psbE-petL, ndhF-rpl32-trnL, rps15-ycf1, and ycf1), along with a total of 51–61 simple sequence repeats (SSRs) were identified as valuable molecular markers for further species delimitation and population genetic studies. Phylogenetic analyses confirmed the evolutionary position of Dolomiaea as a clade within the subtribe Saussureinae, while revealing the discordance between the molecular phylogeny and morphological treatment. Our analysis also revealed that the plastid genes, rpoC2 and ycf1, which are rarely used in Asteraceae phylogenetic inference, exhibit great phylogenetic informativeness and promise in further phylogenetic studies of tribe Cardueae. Analysis for signatures of selection identified four genes that contain sites undergoing positive selection (atpA, ndhF, rbcL, and ycf4). These genes may play important roles in the adaptation of Dolomiaea to alpine environments. Our study constitutes the first investigation on the sequence and structural variation, phylogenetic utility and positive selection of plastomes of Dolomiaea, which will facilitate further studies of its taxonomy, evolution and conservation.
Introduction
Dolomiaea DC. is a medicinally important genus of the sunflower family (Asteraceae) with approximately 15 species endemic to alpine habitats between 2800 and 4800 m in the Qinghai-Tibet Plateau (QTP) and adjacent areas (Ling, 1965; Shih, 1986; Wang et al., 2007; Wang X. et al., 2013; Chen and von Raab-Straube, 2013). Morphologically, Dolomiaea is similar to Jurinea Cass. (Asteraceae), and Saussurea DC. (Asteraceae), but is distinguished from these related genera by the presence of scabrid pappus bristles and naked alveolate receptacles without scales (Ling, 1965; Shih, 1986; Wang et al., 2007; Wang X. et al., 2013; Chen and von Raab-Straube, 2013). Two sections were recognized by Shih (1986) within the genus, including the primary group sect. Dolomiaea DC. with short, compact and apex-rounded style branches, and sect. Vladimiria (Iljin) Shih with divergent style-branches. Molecular phylogenetic analysis of Dolomiaea and related genera (Wang et al., 2007), along with a comprehensive systematic revision of the tribe Cardueae (Susanna and Garcia-Jacas, 2007, 2009), suggested the status of Dolomiaea as an independent clade within subtribe Saussureinae (tribe Cardueae), albeit Dolomiaea was absent in a more recent phylogenomic study (Herrando-Moraira et al., 2019).
Dolomiaea is commonly known as “Chuan Mu Xiang” in traditional Chinese medicine (Wei et al., 2014; Huang et al., 2019). Species of Dolomiaea are used medicinally for their significant bioactivities such as antioxidant (Chen et al., 2013), anti-inflammatory (Chen et al., 2017; Shi et al., 2017) and antimicrobial (Chen et al., 2015) properties, and are also rich resources of multiple chemical components such as sesquiterpenes (Chen et al., 2015), triterpenes (Fan et al., 2016), and phenylpropanoids (Wei et al., 2014). Despite significant medicinal value, genomic resources of Dolomiaea are lacking. Hence, taxonomic delimitation and the understanding of the evolutionary history of Dolomiaea remain hindered by insufficient information provided by Sanger sequencing of DNA markers.
Plastid genomes (plastomes) of photosynthetic plants are highly conserved in terms of gene content and structure (Ruhlman and Jansen, 2014). Typical plastome architecture is comprised of a quadripartite structure with two copies of an inverted repeat (IR) separated by large and small single copy (SSC) regions (LSC and SSC, respectively) (Palmer, 1991). In photosynthetic angiosperms, most plastomes contain approximately 80 protein-coding genes (PCGs) including photosynthetic genes, transcription and translation related genes, as well as some proteins related to other metabolic and synthesis processes, along with 30 transfer RNA (tRNA) genes and four ribosomal RNA (rRNA) genes (Ruhlman and Jansen, 2014). Due to the lack of recombination, usually uniparental inheritance and high copy numbers per cells (Wicke et al., 2011; Ruhlman and Jansen, 2014), whole plastome sequences have been extensively used in reconstructing the plant Tree of Life (e.g., Jansen et al., 2007; Moore et al., 2007; Ruhfel et al., 2014; Gitzendanner et al., 2018; Li et al., 2019). Comparative plastome studies provide the opportunity to explore sequence variation and the molecular evolutionary patterns associated with genome rearrangements (e.g., Knox, 2014; Weng et al., 2014; Rabah et al., 2019; Shrestha et al., 2019) as well as gene loss, duplication, and transfer events (e.g., Downie and Jansen, 2015; Wu and Chaw, 2016; Sun et al., 2017), while also detecting signatures of positive selection in plastid genes facilitating our understanding of plants adapting to extreme environments (e.g., alpine areas) (Bock et al., 2014; Jiang et al., 2018; Liu et al., 2018). Highly divergent regions and simple sequence repeats (SSRs) obtained from whole plastome sequence hold promise as efficient molecular markers implemented in species delimitation and population genetics (Zhang et al., 2017; Cui et al., 2019). Use of these markers as DNA barcodes for herbal medicine is promising for the authentication and identification of specimens for quality assurance (Sgamma et al., 2017; Wang et al., 2017; Kreuzer et al., 2019). In addition, although studies of phylogenetic utility of plastid genes have been widely documented (e.g., Logacheva et al., 2007; Neubig et al., 2009; Granados-Mendoza et al., 2013; Goncalves et al., 2019; Walker et al., 2019), phylogenetic informativeness of plastid genes in inferring intergeneric and infrageneric relationships within Asteraceae remains poorly understood.
In the present study, we sequenced and annotated plastomes of four species in Dolomiaea: D. calophylla Y. Ling, D. denticulata (Y. Ling) C. Shih, Dolomiaea edulis (Franch.) C. Shih, and D. souliei (Franch.) C. Shih. Among them, D. calophylla belongs to sect. Dolomiaea, and the other three species are members of Sect. Vladimiria. We analyzed our data in a comparative genomic framework within Cardueae to detect structural variation of the plastomes in this diverse tribe (with ca. 2400 species in 73 genera) (Susanna and Garcia-Jacas, 2009), as well as to identify highly divergent hotspots as molecular markers. Furthermore, a phylogenomic backbone of Cardueae was inferred to investigate the systematic position of Dolomiaea and examine phylogenetic informativeness of plastid PCGs (Townsend, 2007) in resolving the Cardueae phylogeny. Nucleotide substitution rates of plastid genes were also characterized to uncover possible mechanisms associated with Dolomiaea adapting to alpine environments. Overall, the current study is the first to investigate the sequence and structural variation, phylogenetic utility and positive selection of plastomes of the medicinally important genus Dolomiaea, while incorporating a phylogenomic framework.
Materials and Methods
Taxon Sampling and Sequencing
Fresh leaves of four Dolomiaea species representing both traditionally recognized sections were collected from the QTP and adjacent regions. The circumscription and the taxonomy of Dolomiaea were determined following the Flora of China or the Plant List database1. Voucher specimens were deposited in the Herbarium of Kunming Institute of Botany (KUN), Chinese Academy of Sciences. The voucher information is provided in Supplementary Table S1. For all species, total genomic DNA was extracted following the procedure of Plant Genomic DNA Kit (DP305) from Tiangen Biotech Co., Ltd. (Beijing, China). Paired-end Illumina libraries were constructed with the NEBNext Ultra DNA Library Prep Kit (New England Biolabs, Ipswich, MA, United States) according to the manufacturer’s protocol. A 500-bp DNA TruSeq Illumina (Illumina Inc., San Diego, CA, United States) sequencing library was constructed using 2.5–5.0 ng sonicated DNA as input and final quantifications were done using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States) and real-time quantitative PCR. Libraries were multiplexed and sequenced using a 2 × 150 bp run on an Illumina HiSeq 2500 platform at Novogene Co., Ltd. in Kunming, Yunnan, China.
Plastome Assembly and Annotation
Raw sequence reads were cleaned using Trimmomatic v.0.36 (Bolger et al., 2014) by removing duplicate reads and trimming adapter-contaminated reads. Remaining high-quality reads were assembled de novo into contigs in NOVOPlasty v.2.7.2 (Dierckxsens et al., 2017) using a seed-and-extend algorithm with the plastome sequence of Saussurea japonica (Thunb.) DC. (GenBank accession: MK953481.1) as the seed input due to a close relationship of Dolomiaea with Saussurea (Wang et al., 2007). Other parameters were left at default settings (see NOVOPlasty manual). Assembled plastomes were annotated with the “Annotate from source” tool in Geneious v.9.0.5 (Kearse et al., 2012) using a BLAST-like algorithm to search for annotations in the specified “Source” folder (Kearse et al., 2012) which included all the publicly available plastomes of Cardueae (see Supplementary Table S1 for GenBank accession numbers). Start/stop codons and intron/exon boundaries were manually inspected, and the tRNA genes were identified with tRNAscan-SE (Lowe and Eddy, 1997) as implemented in GeSeq (Tillich et al., 2017). Graphical maps of the circular plastomes were visualized with OGDRAW (Lohse et al., 2013).
Comparative Plastome Analysis
To detect the presence of IR expansion or contraction among four plastomes of Dolomiaea, we used S. japonica as reference and visualized the borders of the large single-copy (LSC), small single-copy (SSC), and IR regions among the five species in Irscope (Amiryousefi et al., 2018). We downloaded the publicly available plastomes of eight species from different genera of Cardueae (i.e., Arctium lappa L., Atractylodes chinensis (Bunge) DC., Carthamus tinctorius L., Centaurea diffusa Lam., Cirsium arvense (L.) Scop., Cynara humilis L., S. japonica, and Silybum marianum (L.) Gaertn.; GenBank accession numbers are provided in Supplementary Table S1). The percentage of sequence identity was plotted with nine species representing the nine genera of Cardueae using the mVISTA program (Frazer et al., 2004) with LAGAN mode (Brudno et al., 2003).
Identification of Molecular Markers
Simple sequence repeats across the plastomes of four Dolomiaea species were detected using the MISA-web application (Beier et al., 2017)2. Thresholds for a minimum number of repeat units were established as follows: ten for mono-nucleotide, five for di-nucleotide, four for tri-nucleotide, and three for tetra-nucleotide, penta-nucleotide, or hexa-nucleotide SSR.
To observe the sequence divergence and determine highly divergent hotspots of Cardueae plastomes, 22 publicly available plastomes were downloaded from the National Center for Biotechnology Information (NCBI) database (Supplementary Table S1). All plastome sequences of Cardueae with only one IR region included were aligned using MAFFT v.7.22 (Katoh and Standley, 2013) under “–auto” strategy. Nucleotide diversity (π) was calculated by sliding window analysis conducted in DnaSP v.6.11.01 (Rozas et al., 2017). The step size was set to 200 bp, with a 600 bp window.
Phylogenetic Analyses
For phylogenetic analyses, 28 taxa were sampled (Supplementary Table S1), including 26 taxa of Cardueae (four newly sequenced and 22 downloaded from NCBI representing all available genera of Cardueae) and two outgroup taxa of tribe Cichorieae (Taraxacum officinale F. H.Wigg. and Lactuca sativa L.) based on the availability and previous study (Fu et al., 2016). All 79 PCGs were extracted in PhyloSuite v.1.1.16 (Zhang D. et al., 2019). Initial sequences were aligned using the codon-aware program MACSE v. 2.03 (Ranwez et al., 2018), which preserves reading frame and allows incorporation of sequencing errors or sequences with frameshifts. These aligned sequences were then concatenated into a supermatrix using PhyloSuite, and the number of parsimony informative sites was calculated using MEGA X (Kumar et al., 2018).
Both maximum-likelihood (ML) and Bayesian inference (BI) analyses were conducted. For the ML analysis, RAxML v.7.4.2 (Stamatakis, 2014) was used under the general time reversible model of nucleotide substitution with the gamma model of rate heterogeneity (GTR + G) as suggested (see RAxML manual). Twenty independent ML searches and 1000 rapid Bootstrap replicates were executed with “-f a -x 12345 -# 1000” option in RAxML analysis. Bayesian inference was conducted in MrBayes v.3.2.3 (Huelsenbeck and Ronquist, 2001) with the optimal model (GTR + G + I) calculated by jModelTest v.2.1.10 (Darriba et al., 2012) under the Bayesian information criterion (BIC). Two runs were conducted in parallel with four Markov chains (one cold and three heated), with each running for 2,000,000 generations from a random tree and sampled every 200 generations. Convergence was checked by examining the average standard deviation of split frequencies (ASDF). After ASDF reached < 0.01, the initial 25% of the sampled data were discarded as burn-in, and the remaining trees were used to construct a majority-rule consensus tree and calculate the posterior probability. We also conducted partitioned ML and BI analyses. PartitionFinder v.1.0.1 (Lanfear et al., 2012) was implemented to determine optimal partitioning scheme and evolutionary model selection under the BIC. The best-fit evolutionary models of partitioning subsets were used in the partitioned phylogenetic inferences. Other settings were consistent with the unpartitioned analyses. The final phylogenetic results were viewed using FigTree v.1.6.1 (Rambaut and Drummond, 2010).
Phylogenetic Informativeness of Plastid Genes
The PhyDesign web application3 was used to estimate the phylogenetic informativeness profiles for the 79 PCGs using the HyPhy substitution rates algorithm for DNA sequences with the default settings (Pond et al., 2005; Townsend, 2007; Lopez-Giraldez and Townsend, 2011). The inferred tree from the concatenated ML analysis in RAxML was used as an input tree to reconstruct a relative-time ultrametric tree in the dnamlk program in PHYLIP (Felsenstein, 1989). The converted relative-time ultrametric tree and alignment of 79 PCGs partitioned by genes were used as input files in PhyDesign to calculate phylogenetic informativeness.
Positive Selection on Plastid Genes
We used the branch-site model in codeml program of the package PAML v.4.9h (Yang, 2007) to identify plastid genes under positive selection in Dolomiaea. The 79 PCGs used in the phylogenetic analysis were tested. The ML tree generated via RAxML was used as a constraint topology, and the clade formed by Dolomiaea was set as a foreground branch. Likelihood ratio test (LRT) was conducted to compare a model allowing positive selection (the ratio of non-synonymous to synonymous substitutions: ω > 1) acting on a site in the foreground branch with a null model where the site may have undergone neutral evolution (ω = 1) or purifying selection (ω < 1). A Chi square p-value smaller than 0.05 were used as cutoff of significance. The Bayes Empirical Bayes (BEB) inference (Yang et al., 2005) was then implemented in site models M2a and M8 to estimate the posterior probabilities and positive selection pressures of the selected genes.
Results
Genome Assembly and Plastome Features
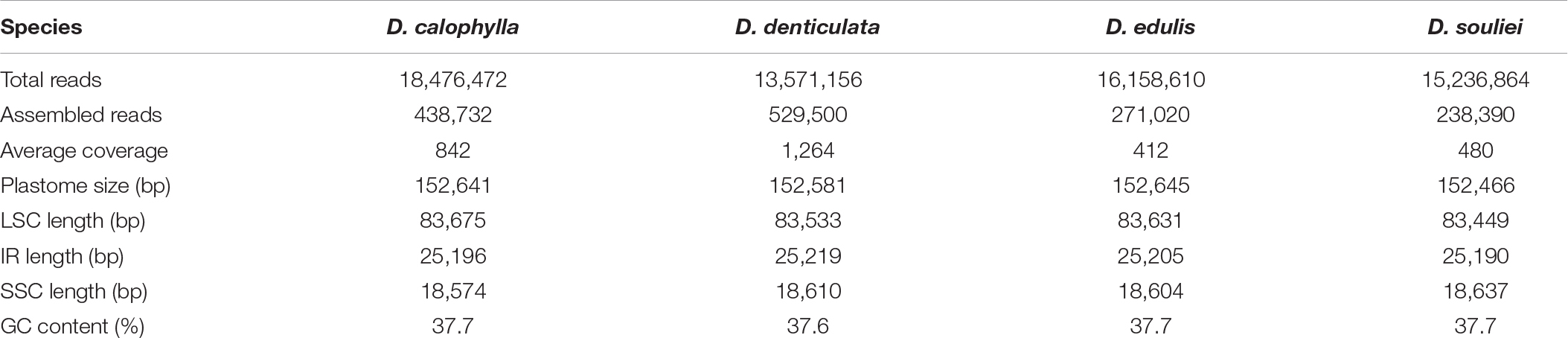
After Illumina sequencing, a total of 13,571,156–18,476,472 pair-end clean reads were obtained for each species (Table 1). The mean sequencing coverage ranged from 412 × (D. edulis) to 1,264 × (D. denticulata). All four plastomes displayed the typical quadripartite structure composed of a large single copy (LSC), a SSC, and two IRs (IRa and IRb). The length of the four plastomes ranged from 152,466 bp in D. souliei to 152,645 bp in D. edulis (Table 1). All the plastomes contain 79 PCGs, four rRNAs and 30 tRNAs arranged in the same gene order (Figure 1). A total of 18 genes (including 11 PCGs and seven tRNA genes) had introns, with 15 genes having one intron and three genes having two introns. The rps12 gene was found to be trans-spliced in all plastomes, with one of its exons located in the LSC region and the other duplicated in the IR (Figure 1).
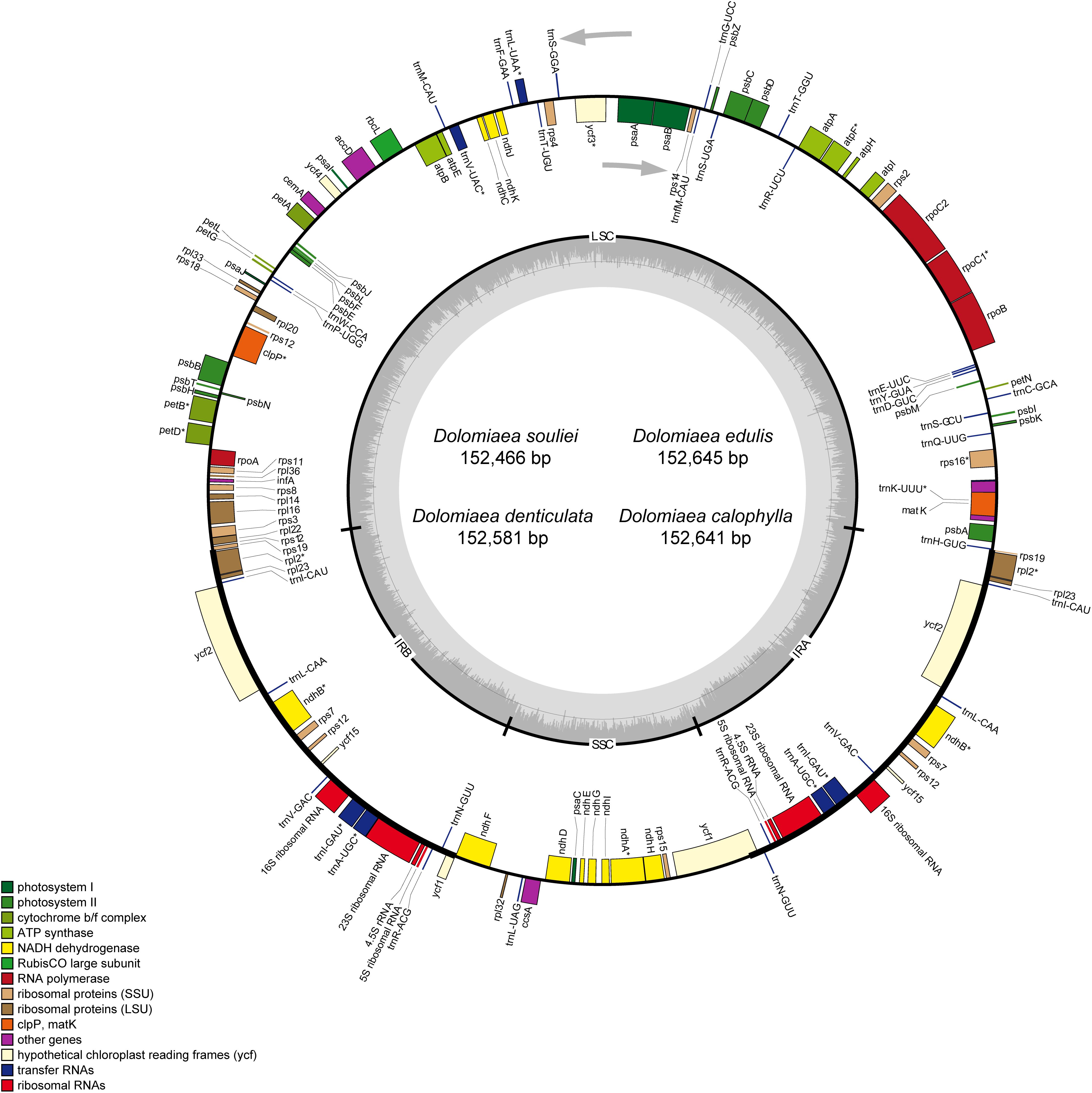
Figure 1. Map of four newly sequenced plastomes of Dolomiaea. Genes outside the circle are transcribed in a counter-clockwise direction, whereas those inside the circle are transcribed in a clockwise direction. The dark gray area in the inner circle indicates GC content and the thick line shows the extent of different regions. Different colors for genes indicate different functional groups. LSC: large single copy; SSC: small single copy; IR: inverted repeat.
The IR regions were highly consistent in plastomes of Dolomiaea, comprising a length of 25,190–25,219 base pairs and including 17 genes (six PCGs, seven tRNA genes, and four rRNA genes). Two pseudogenes, rps19 and ycf1, were identified. The IRb/LSC junction was located within the rps19 gene in all plastomes, resulting in the presence of a part of the rps19 gene in the IRa. Similarly, the IRa/SSC boundary positions in all species were located in the ycf1 gene, with part of this gene duplicated in the IRb (Figure 2).
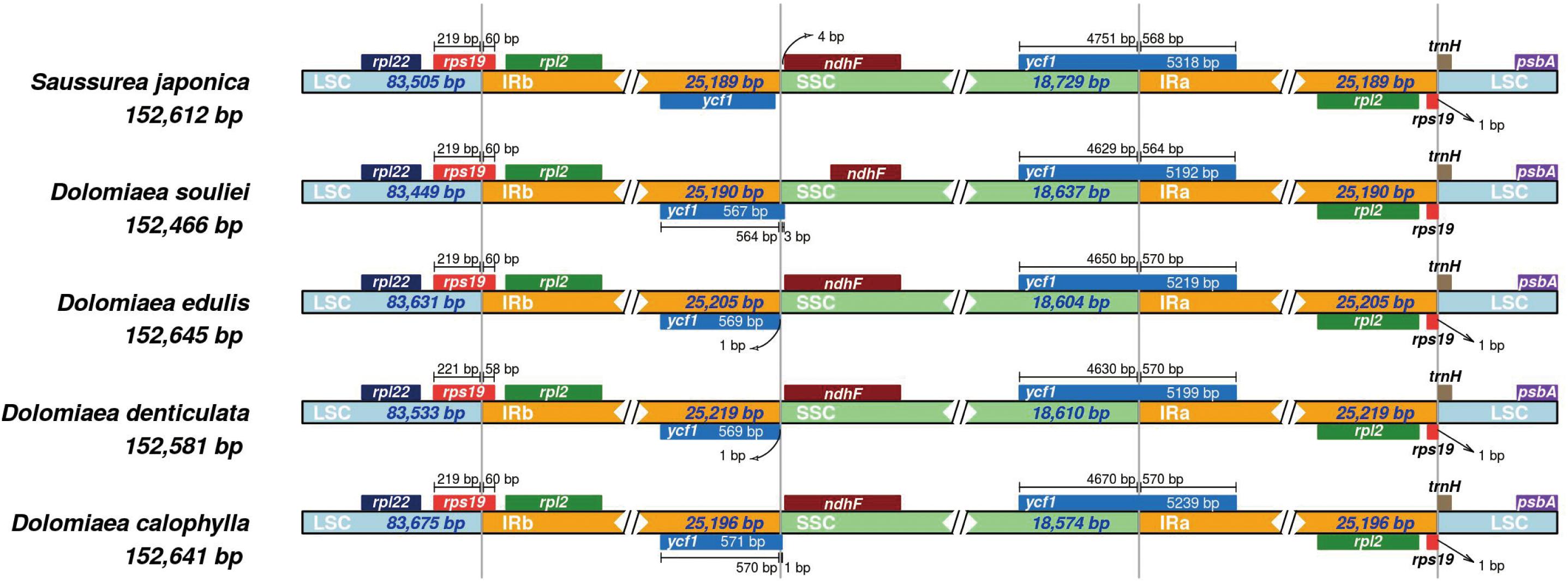
Figure 2. Comparison of IR-SC border positions across plastomes of four Dolomiaea species and Saussurea japonica. Genes are denoted by colored boxes. The gaps between the genes and the boundaries are indicated by the base lengths (bp).
To investigate the sequence divergence of plastomes, the percentage of sequence identity was plotted for nine representative species from different genera of Cardueae using the mVISTA program (Frazer et al., 2004) with A. lappa as the reference. High similarity was detected among those nine taxa, suggesting that plastome sequences are conserved in Cardueae. The IR regions were found to be more conserved than single-copy regions, and coding regions are more conserved than non-coding regions (Figure 3). All analyzed plastomes possessed the ∼20 and ∼3 kb inversions (Inv1 and Inv2), which have been widely detected in plastomes of Asteraceae (e.g., Kim et al., 2005; Liu et al., 2013; Walker et al., 2014; Zhang X. et al., 2019). The Inv2, located between the trnS-GCU and trnE-UUC genes, was nested within the Inv1, located between the trnG-UCC and trnS-GCU genes (Figure 3).
Figure 3. Visualization of the alignments of nine plastomes representing nine genera of Cardueae using mVISTA, with Arctium lappa as the reference. The gray arrows above the alignment indicate genes. Different colors represent different regions (coding and non-coding). The horizontal axis indicates the coordinates within the chloroplast genome. The vertical scale represents the percentage of identity, ranging from 50% to 100%.
Molecular Markers
The counts and types of SSRs were generally similar between the newly sequenced plastomes (Figure 4). The number of SSRs ranged from 51 (D. souliei) to 61 (D. denticulata) (Figure 4A). The types of SSRs in each plastome were quite similar (Figure 4A). In all four plastomes, the total number of SSRs for mono-nucleotide, di-nucleotide, tri-nucleotide, tetra-nucleotide, penta-nucleotide, and hexa-nucleotide were 153, 17, 12, 29, 1, and 4 (Figures 4B,C), respectively. Nucleotide diversity (π) was calculated by sliding window analysis to observe the sequence divergence and determine highly divergent hotspots. The single-copy regions have higher proportions of variable sites than in the IR regions (Figure 5). By scanning for variation throughout the whole plastome, we determined eight highly divergent regions, namely rps16-trnQ, trnC-petN, trnE-rpoB, trnT-trnL-trnF, psbE-petL, ndhF-rpl32-trnL, rps15-ycf1, and ycf1 (Figure 5).
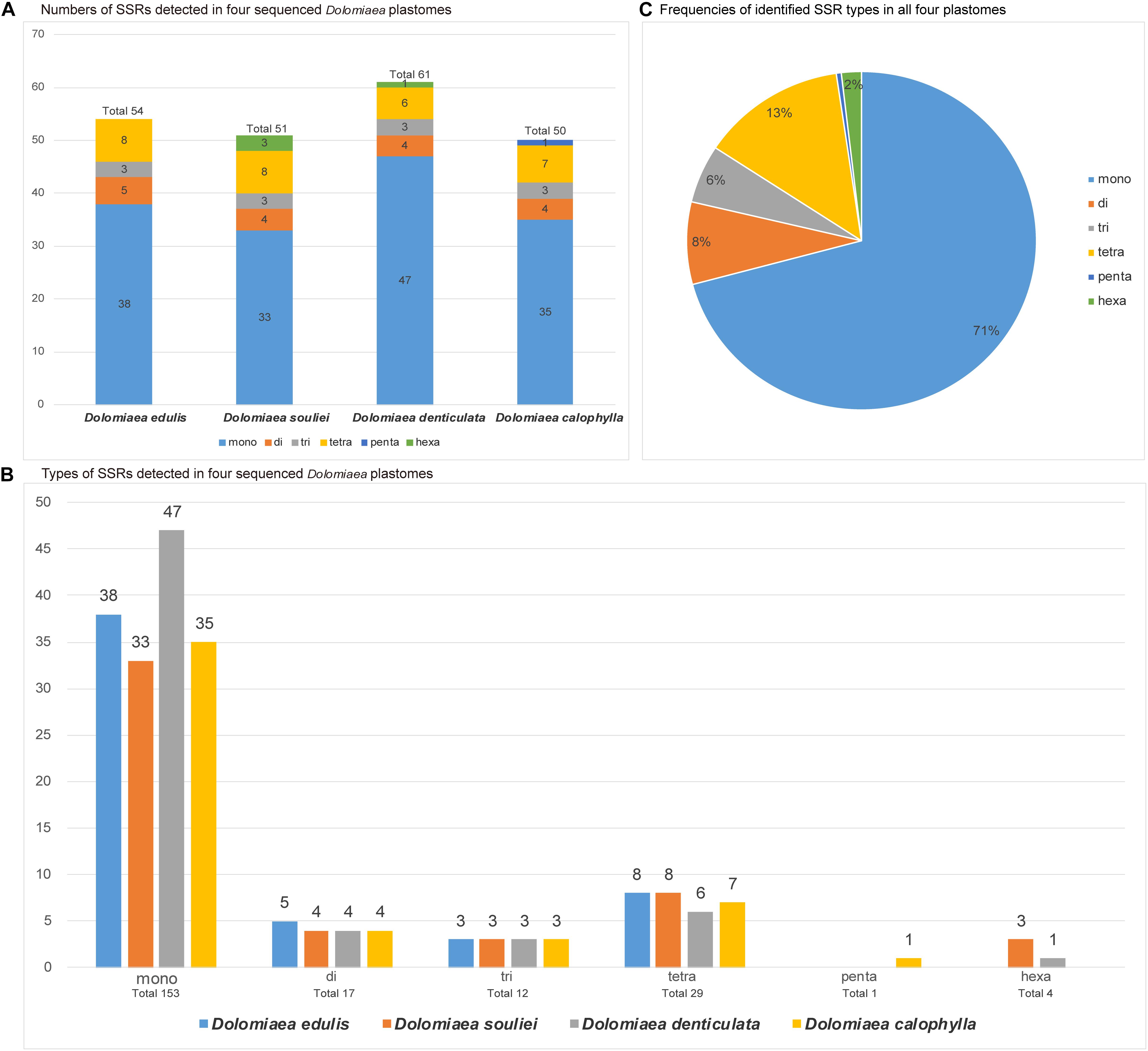
Figure 4. Comparison of simple sequence repeats (SSRs) among four plastomes. (A) Numbers of SSRs detected in the four newly sequenced Cardueae plastomes. (B) Types of SSR types detected in all four plastomes. (C) Frequencies of identified SSR types in all four plastomes.
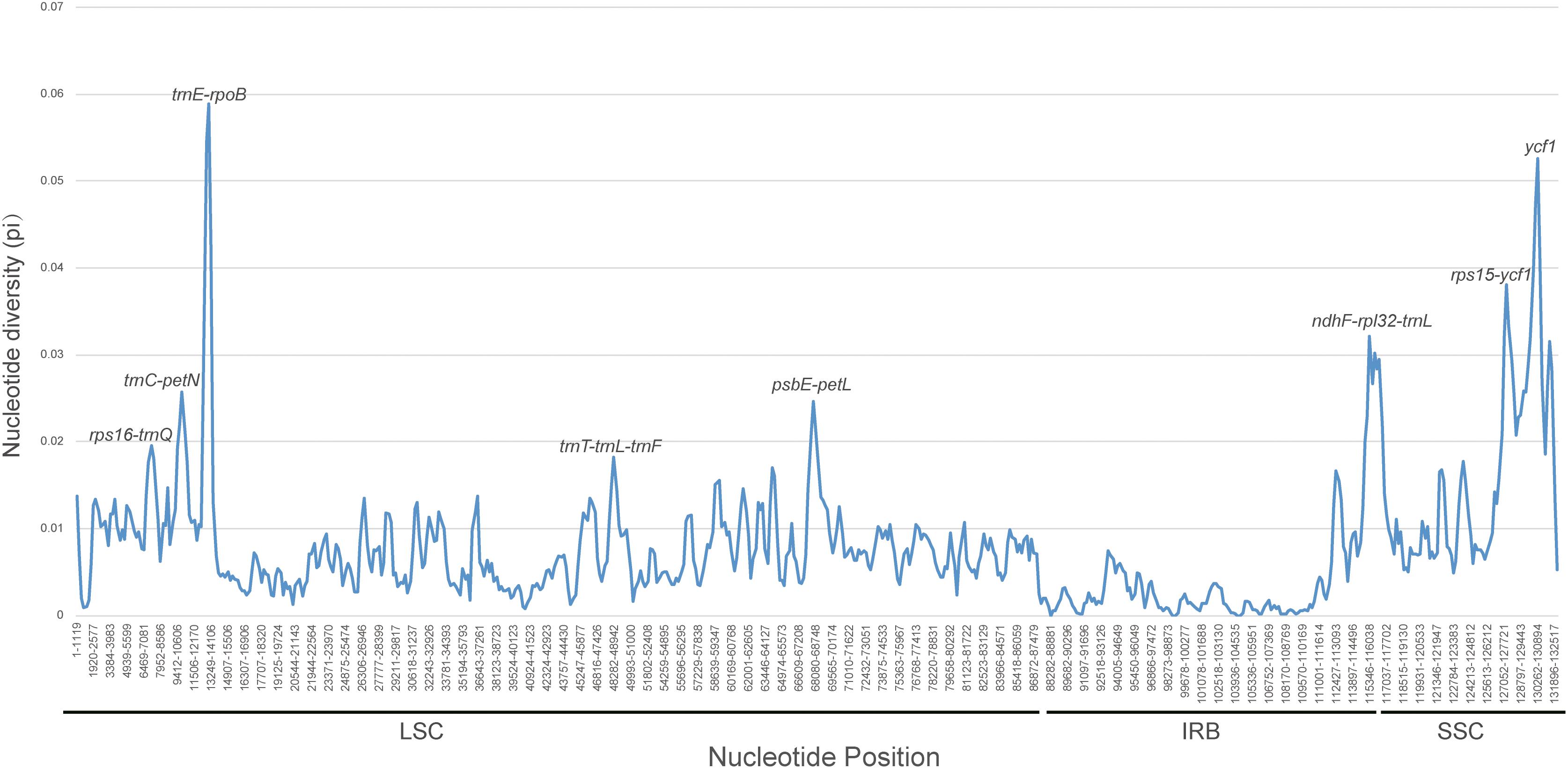
Figure 5. Sliding window analysis of nucleotide diversity (π) along the whole plastome for the 26 aligned plastomes of Cardueae with one IR copy included. Genes underlying peaks of nucleotide diversity are labeled.
Phylogenetic Relationships
The final alignment of the 79-PCGs, 28-species data set consisted of 69,546 bp containing 2,485 parsimony-informative sites. PartitionFinder identified 11 subsets, and the best-fit substitution models of partitioning subsets were provided in Supplementary Table S2. Both unpartitioned and partitioned ML and BI analyses yielded identical tree topology [Figure 6 and Supplementary Figure S1; bootstrap values (BS) and posterior probabilities (PP) are depicted in ML tree]. Four species of Dolomiaea formed a clade with 100% support (BS = 100, PP = 1.0). Dolomiaea souliei is the sister species to a clade formed by D. calophylla and D. edulis + D. demticulata. Our results do not support the two morphological sections, i.e., sect. Dolomiaea and sect. Vladimiria, identified by Shih (1986) via long and acute vs. short and round style, as D. calophylla is nested within the other Dolomiaea species. Dolomiaea was revealed as a sister to Saussurea albeit with low support (BS = 61, PP = 0.78). The two genera constitute the subtribe Saussureinae in our analysis and are sister to subtribe Arctiinae. In tribe Cardueae, four subtribes included (Carlininae, Caduinae, Centaureinae, and Saussureinae) were monophyletic. Carlininae was the earliest-diverging subtribe, Caduinae was next, and Centaureinae was sister to Arctiinae + Saussureinae. Most genera included were supported as monophyletic except Cirsium (Asteraceae), showing a nested position with Silybum (Asteraceae).
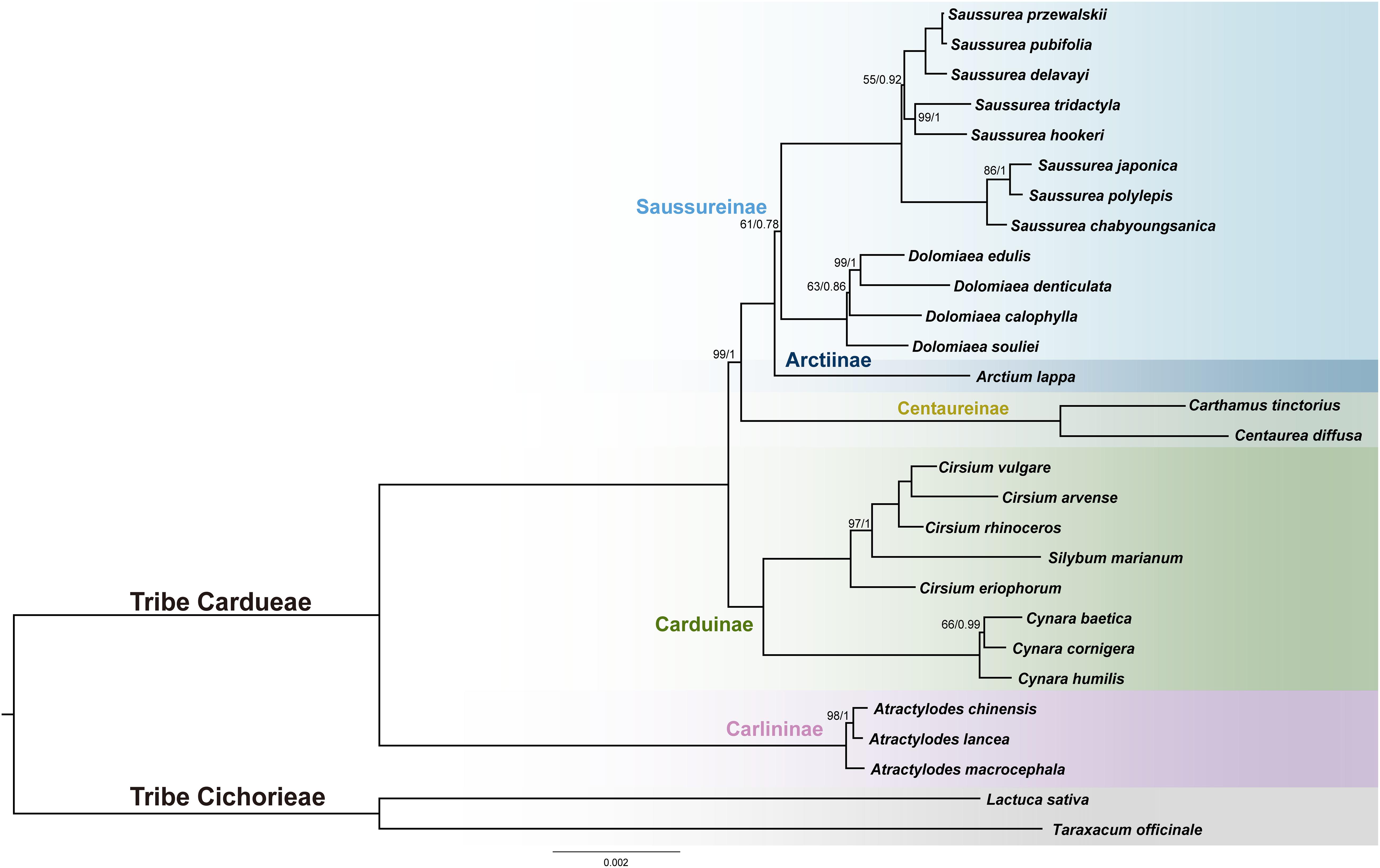
Figure 6. Phylogenetic reconstruction of Cardueae from maximum likelihood (ML) and Bayesian inference (BI) analyses using concatenated 79 protein-coding regions. The ML tree is shown. Maximum likelihood bootstrap values (BS) and posterior probabilities (PP) are shown at nodes. Branches with no values listed have 100% BS and PP of 1.00.
Phylogenetic Informativeness
The net and per-site phylogenetic informativeness (PI) profiles for the 79 PCGs were measured using PhyDesign (Figure 7 and Supplementary Table S3). The ycf1 gene had the highest net phylogenetic informativeness among all PCGs, followed by rpoC2, ndhF and ycf2. Genes with high net PI were also genes with longer length, suggesting a large contribution of gene length to phylogenetic informativeness. For the per-site PI, ycf1 also performed best among PCGs, followed by rps16 and ndhF (Supplementary Table S3). The long gene ycf2 had a comparatively low per-site PI possibly due to a low frequency of rapidly evolving sites (Shrestha et al., 2019). Relatively conservative genes with less net PI were primarily associated with photosynthesis and were shorter in length (<200 bp).
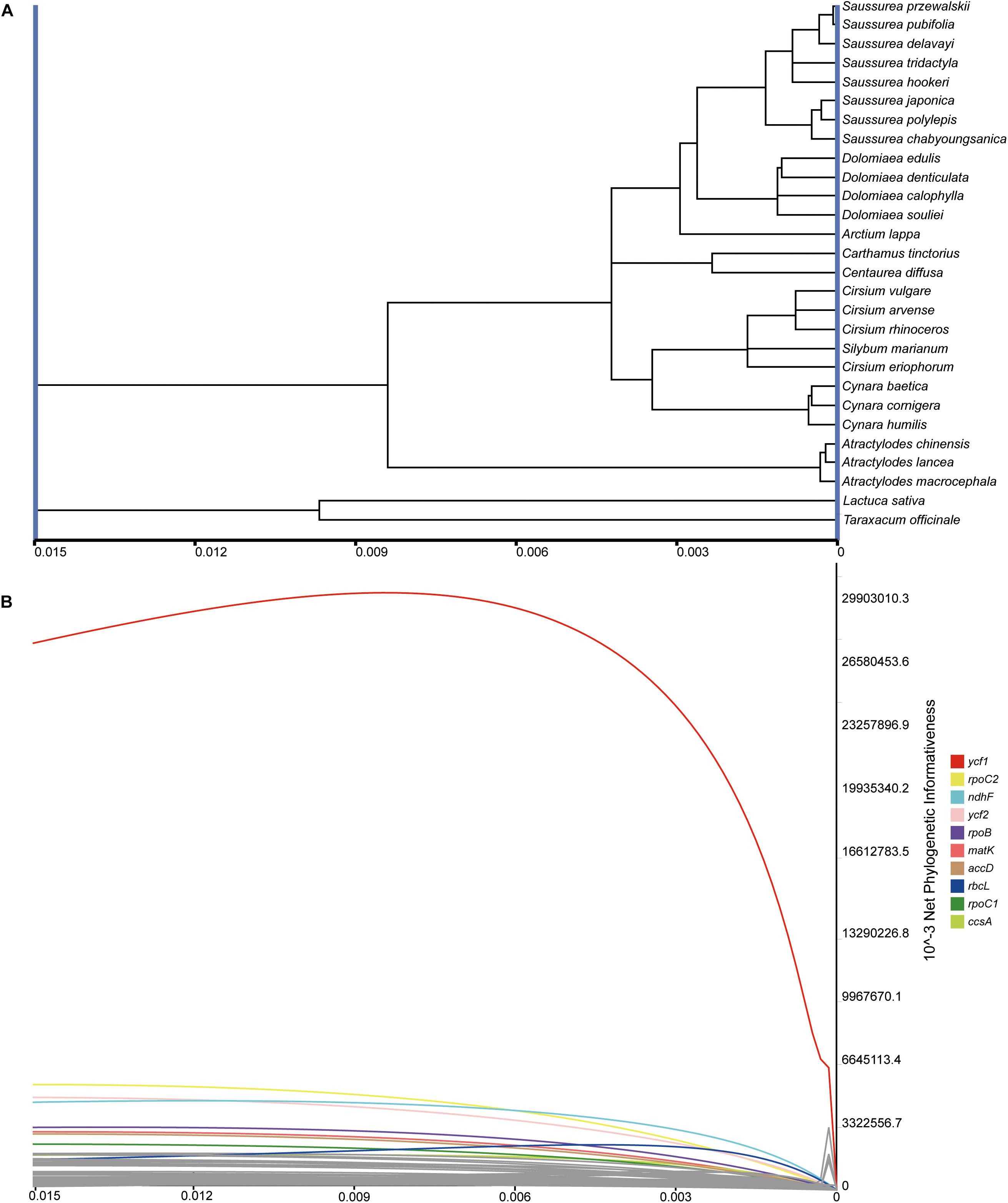
Figure 7. Phylogenetic net informativeness profiles of 79 PCGs of Cardueae estimated in PhyDesign (Lopez-Giraldez and Townsend, 2011). (A) The ultramteric tree of Cardueae converted in the dnamlk program in PHYLIP (Felsenstein, 1989). (B) Net phylogenetic informativeness profiles for 79 PCGs. Ten genes with the greatest informativeness are color-coded and indicated at the right. X- and Y-axes represent relative-time and net phylogenetic informativeness, respectively.
Positive Selection
Analyses of selection were conducted to identify signals of episodic selection occurring on PCGs along a specified branch (the clade formed by Dolomiaea in our analysis). Comparison against a branch−site model allowing positive selection along specified branches (Model A) with a null model (Model Anull) allowing neutral evolution and negative selection was employed (Yang et al., 2005). We observed signatures of positive selection in four PCGs, atpA (3 sites), ndhF (5 sites), rbcL (4 sites), and ycf4 (1 sites) at a significance level of 0.05 (Table 2).
Discussion
Plastome Features
In this study, we assembled and analyzed four complete plastomes of Dolomiaea species. All four plastomes share the gene content and structure of most Asteraceae plastomes, and are similar to other angiosperm plastomes. The expansion/contraction of IR regions has been demonstrated to be a significant contribution to substantial variation in plastome size (Ruhlman and Jansen, 2014). Occurrence of IR expansion/contraction is common in angiosperm plastomes and has been documented in several lineages, such as Pelargonium (Weng et al., 2017), Trochodendraceae (Sun et al., 2013), Plantago (Zhu et al., 2016), and Passiflora (Rabah et al., 2019). In the present study, however, no significant IR length variation was found among Dolomiaea plastomes and with plastomes of its sister genus (Saussurea) (Zhang X. et al., 2019), indicating the conservative nature of plastome evolutionary history of Dolomiaea.
The percentage of sequence identity show high similarity among nine plastomes of Cardueae, suggesting that plastome sequences are conserved in this tribe despite being one of the most diverse tribes in Asteraceae. The two large inversions (Inv1 and Inv2 in Figure 3) in LSC are detected in all analyzed plastomes. These two large inversions are commonly found in plastomes of Asteraceae, e.g., Lactuca (Kim et al., 2005), Artemisia (Liu et al., 2013), Lasthenia (Walker et al., 2014), Mikania (Su et al., 2018), and Saussurea (Zhang X. et al., 2019), implying that such genome organization may reflect common evolutionary history in the family. Although uncommon, the structural variation of inversions has been discovered in many other angiosperm lineages, such as the ∼49 kb inversion discovered in the early diverging eudicot family Circaeasteraceae (Sun et al., 2017); the ∼78 kb inversion present in the Fabaceae subtribe Phaseolinae (Bruneau et al., 1990), the ∼36 kb inversion shared by all core genistoid legumes (Martin et al., 2014); and ∼20 kb inversion occurring in Styracaceae of Ericales (Yan et al., 2017, 2018). These inversions are considered highly valuable in phylogenetics due to their rarity, easily determined homology and easily inferred state polarity (Jansen et al., 2008).
Molecular Markers
Species of Dolomiaea are commonly used in medicine for their significant bioactivities and multiple chemical components (Chen et al., 2015). However, the lack of genomic resources of Dolomiaea has hindered our deep exploration of its taxonomy, genetics and conservation. Assembling plastome sequences not only provides opportunity for illustrating the plastome evolution, but also generates valuable genetic resources such as SSRs and highly divergent regions helpful in further population genetics and taxonomic studies of Dolomiaea. In this study, the number of SSRs identified within Dolomiaea plastomes ranges from 51 to 61. The mononucleotide repeats with A and T repeat units are found to be the most abundant, which also were reported in other Asteraceae genera, e.g., Jacobaea (Doorduin et al., 2011), Artemisia (Shen et al., 2017), Dendrosenecio (Gichira et al., 2019) and Saussurea (Zhang X. et al., 2019), and other lineages such as Nymphoides and Menyanthes (Menyanthaceae) (Njuguna et al., 2019), Sinadoxa (Adoxaceae) (Wang et al., 2016), and Actinidia (Actinidiaceae) (Yao et al., 2015). Eight highly divergent regions (rps16-trnQ, trnC-petN, trnE-rpoB, trnT-trnL-trnF, psbE-petL, ndhF-rpl32-trnL, rps15-ycf1, and ycf1) are identified by scanning for variation throughout the whole plastomes. Most of the divergent regions are overlapping with those determined in plastomes of Saussurea (Zhang X. et al., 2019), reflecting their close affinity. Additionally, most of them, i.e., rps16-trnQ, trnL-trnF, psbE-petL, rpl32-trnL, and ycf1, have been previously reported as rapidly diverging loci and are broadly used for reconstructing phylogenies (Neubig et al., 2009; Prince, 2015; Gao et al., 2018; Su et al., 2018). For the herbal medicinal genus Dolomiaea, these genetic resources as molecular markers can serve as promising DNA barcoding in the authentication of specimens for medicinal quality assurance.
Phylogenetic Relationships and Phylogenetic Informativeness of Plastid Genes
Our phylogenetic analyses show a sister relationship between Saussureinae and Arctiinae, which is consistent with phylogenetic results of Herrando-Moraira et al. (2019) using nuclear data, but inconsistent with their plastome data results. We speculate this inconsistency likely results from the difference of sequencing strategy between the present study with the study of Herrando-Moraira et al. (2019), in which they recovered plastome sequences using a Hyb-Seq approach. Only targeted DNA regions are enriched in Hyb-Seq approaches using specific probes or “baits” (Herrando-Moraira et al., 2019), and thus plastome sequences generated from target enrichment are likely incomplete. Overall, the phylogenetic discordance from different data sets, along with weakly supported resolution of some relationships, suggests a possible complex evolutionary history involving hybridization and incomplete lineage sorting, and further investigation using more advanced methods, as well as a wider species sampling, are necessary.
The present phylogenomic framework confirms the systematic position of Dolomiaea as a monophyletic group within subtribe Saussureinae. Phylogenetic relationships within the Saussureinae on a large scale remain ambiguous, mainly because of the high number of small segregates separated from the large genera Saussurea or Jurinea using molecular data. As proposed, there may be three genera (Dolomiaea, Jurinea, and Saussurea) that constitute the Saussureinae (Susanna and Garcia-Jacas, 2009). Our analysis resolves Dolomiaea as a sister to Saussurea with weak support, indicating the relationship is not very robust, and future studies adding Jurinea species are needed. The early diverging position of D. edulis was consistent with the study of Wang et al. (2007) using nuclear rDNA (ITS) and plastid trnL-F + psbA-trnH regions. Furthermore, we reveal the discordance between the molecular phylogeny and morphological treatment of Dolomiaea, which may suggest the infrageneric classification of the circumscribed genus needs further revision.
As one of the largest tribes in Asteraceae, Cardueae is also one of the most complicated because of great morphological diversity and because it comprises some of the largest genera of the family, such as Cirsium Mill. (250 spp.), Centaurea L. (400 spp.), Cousinia Cass. (600 spp.), and Saussurea DC. (300 spp.) (Susanna and Garcia-Jacas, 2009). Although some progress in reconstructing phylogenetic relationships within Cardueae has been made using plastid markers, no study has yet to examine the phylogenetic utility of plastid genes in this tribe. A major bottleneck in molecular phylogenetic studies of recently diverged lineages is the insufficient resolution provided by loci with limited phylogenetic signal. Therefore, identifying plastid markers with high phylogenetic informativeness will aid efforts in resolving complex phylogenetic relationships at the species-level, as well as in plant barcoding. All three genes (matK, ndhF and rbcL) widely used in phylogenetic studies of Cardueae (Susanna et al., 2006; Barres et al., 2013; Wang Y.J. et al., 2013; Fu et al., 2016), showed a high value of phylogenetic informativeness. Nonetheless, ycf1 and rpoC2 exhibited high phylogenetic informativeness values and relatively long gene length, yet have not been employed in phylogenetic analyses of Cardueae. Previous studies have indicated the usefulness of ycf1 in plant phylogenetics, such as orchids (Neubig et al., 2009), Annonaceae (Neubig and Abbott, 2010), and Amaryllidaceae (García et al., 2014). A recent study of Walker et al. (2019) revealed the good performance of ycf1 and rpoC2 in the phylogenetic reconstruction of the angiosperm phylogeny, which is also supported by our current results. Hence, we advocate the usefulness of ycf1 and rpoC2 as promising plastid markers for inferring evolutionary relationships of Cardueae.
Given primer universality as an important criterion for an ideal DNA marker in phylogenetics and plant barcoding (Dong et al., 2015), primer design is a possible obstacle to the application of ycf1 and rpoC2 in Sanger sequencing. Our study presents a routine procedure for marker selection at the molecular level. Taking the availability of primers into account is essential for more practical evaluation of the phylogenetic informativeness of a marker in the further studies.
Signature of Positive Selection on Plastid Genes
Our study reveals that natural selection can target different functional groups of plastid genes and support the possible involvement of plastid genes in plant adaptation to alpine environments. Dolomiaea species are distributed mainly in high-altitude environments in the QTP, which pose a variety of stress factors including low temperature, strong UV radiation, low oxygen level, and capricious climate (Zhang T. et al., 2019). Genes under positive selection may play important roles in the adaptation of Dolomiaea to the harsh environments.
Among the 79 PCGs within Dolomiaea, four (atpA, ndhF, rbcL, and ycf4) show evidence of undergoing positive selection. The atpA gene, encoding the alpha subunit of ATP synthase, plays a vital role in energy-transduction during photosynthesis (Walker, 2013). Accelerated rates of evolution in atpA may promote the specialization of ATP synthases enhancing the efficiency of energy-transduction in photosynthesis at high-altitude environments with low CO2 concentration. The atpF gene encodes the NADH dehydrogenase unit which is involved in cyclic electron flow around photosystem I essential for photosynthesis (Munekage et al., 2004). Positive selection of ndhA is possibly related to the protection of Dolomiaea species from strong light damage in higher altitudes. Positive selection might be acting on the rbcL gene which encodes the large subunit of the photosynthetic enzyme Rubisco and is related to plants adaptation to low temperature, drought and carbon dioxide concentration. Selection on this gene has also been observed in various plant groups such as Panax (Araliaceae) (Jiang et al., 2018), Silene (Caryophyllaceae), and Potamogeton (Potamogetonaceae) (Iida et al., 2009), and is particularly prevalent following the evolution of C4 photosynthesis (Piot et al., 2018).
Conclusion
Previous phylogenetic analysis using fragment DNAs suggested Dolomiaea as a clade within subtribe Saussureinae (Wang et al., 2007), however the most recent phylogenomic study did not include any samples of Dolomiaea (Herrando-Moraira et al., 2019). Our phylogenomic framework based on whole plastomes confirm the systematic position of the genus, while revealing the discordance between the molecular phylogeny and morphological treatment suggesting the infrageneric classification of the circumscribed genus needs further revision. Our analysis also reveals that the rarely used plastid genes in Asteraceae phylogenetic inference, rpoC2 and ycf1, exhibit great phylogenetic informativeness and substantial promise in further phylogenetic studies of Cardueae. By comparative analyses, we detect the sequence and structural conservatism of plastomes indicating Dolomiaea may have a recently diverged history, and/or slow mutation rates of plastid genes. As a medicinally important genus endemic to the QTP, species identification and authentication, as well as resource conservation, are of importance for medicinal quality assurance. The highly divergent plastome regions determined in this study will be helpful in taxonomy and population genetics of Dolomiaea. Furthermore, given the lack of knowledge about the underlying mechanisms for plants adapting to alpine environments, the signature of positive selection on plastid genes identified here provides new insights into the role plastomes play in adaptive evolution.
Data Availability Statement
All newly sequenced plastome sequences have been deposited in NCBI. Accession numbers can be found in Supplementary Table S1.
Author Contributions
HW and HS conceived and designed the study. JS and XZ performed de novo assembly, genome annotation, phylogenetic, and other analyses. JS, XZ, JL, HW, and HS drafted the manuscript. XZ and TD collected the leaf materials. JS and HZ performed the experiments. All authors discussed the results and helped shape the research, analyses, and final manuscript.
Funding
This study was supported by the Key Projects of the Joint Fund of the National Natural Science Foundation of China (U1802232), the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0502), the National Key R&D Program of China (2017YFC0505200), the Strategic Priority Research Program of Chinese Academy of Sciences (XDA20050203), and the Major Program of the National Natural Science Foundation of China (31590823).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00376/full#supplementary-material
FIGURE S1 | Phylogenetic reconstruction of Cardueae from partitioned maximum likelihood (ML) and Bayesian inference (BI) analyses using concatenated 79 protein-coding regions. The ML tree is shown. Maximum likelihood bootstrap values (BS) and posterior probabilities (PP) are shown at nodes. Branches with no values listed have 100% BS and PP of 1.00.
TABLE S1 | Taxa included in the present study. NCBI accession numbers and voucher specimens’ information are provided for newly sequenced plastomes.
TABLE S2 | The best-fit evolutionary models of 11 partitioning subsets calculated by PartitionFinder. # Sites: the number of sites of the partitioning subsets.
TABLE S3 | Phylogenetic informativeness profiles for 79 plastid protein-coding genes. Included are number of sites for which substitution rates were calculated (#Rates), net phylogenetic informativeness at epochs: 0–0.015 (Net PI) and per-site phylogenetic informativeness (Per-site PI). Ten genes with the most net informativeness are colored in red.
Footnotes
- ^ http://www.theplantlist.org/
- ^ https://webblast.ipk-gatersleben.de/misa/
- ^ http://phydesign.townsend.yale.edu/
References
Amiryousefi, A., Hyvonen, J., and Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Barres, L., Sanmartin, I., Anderson, C. L., Susanna, A., Buerki, S., Galbany-Casals, M., et al. (2013). Reconstructing the evolution and biogeographic history of tribe Cardueae (Compositae). Am. J. Bot. 100, 867–882. doi: 10.3732/ajb.1200058
Beier, S., Thiel, T., Munch, T., Scholz, U., and Mascher, M. (2017). MISA-web: a web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi: 10.1093/bioinformatics/btx198
Bock, D. G., Andrew, R. L., and Rieseberg, L. H. (2014). On the adaptive value of cytoplasmic genomes in plants. Mol. Ecol. 23, 4899–4911. doi: 10.1111/mec.12920
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brudno, M., Do, C. B., Cooper, G. M., Kim, M. F., Davydov, E., Program, N. C. S., et al. (2003). LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13, 721–731. doi: 10.1101/gr.926603
Bruneau, A., Doyle, J. J., and Palmer, J. D. (1990). A Chloroplast DNA inversion as a subtribal character in the phaseoleae (Leguminosae). Syst. Bot. 15, 378–386.
Chen, J. J., Bai, W., Gobu, F. R., Wu, C. H., Zeng, J., Sun, M., et al. (2015). Sesquiterpenoids from the roots of Vladimiria muliensis. J. Asian Nat. Prod. Res. 17, 1188–1195. doi: 10.1080/10286020.2015.1119130
Chen, J. J., Wei, H. B., Xu, Y. Z., Zeng, J., and Gao, K. (2013). Antioxidant lignans from the roots of Vladimiria muliensis. Planta Med. 79, 1470–1473. doi: 10.1055/s-0033-1350801
Chen, L. P., Wu, G. Z., Zhang, J. P., Ye, J., Liu, Q. X., Shen, Y. H., et al. (2017). Vlasouliolides A-D, four rare C17/C15 sesquiterpene lactone dimers with potential anti-inflammatory activity from Vladimiria souliei. Sci. Rep. 7:43837. doi: 10.1038/srep43837
Chen, Y. S., and von Raab-Straube, E. (2013). A new record and new combination for Dolomiaea (Compositae, Cardueae) in China. Willdenowia 43, 287–291. doi: 10.3372/wi.43.43208
Cui, Y., Chen, X., Nie, L., Sun, W., Hu, H., Lin, Y., et al. (2019). Comparison and phylogenetic analysis of chloroplast genomes of three medicinal and edible Amomum species. Int. J. Mol. Sci. 20:4040. doi: 10.3390/ijms20164040
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Dierckxsens, N., Mardulyn, P., and Smits, G. (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. doi: 10.1093/nar/gkw955
Dong, W., Xu, C., Li, C., Sun, J., Zuo, Y., Shi, S., et al. (2015). ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 5:8348. doi: 10.1038/srep08348
Doorduin, L., Gravendeel, B., Lammers, Y., Ariyurek, Y., Chin-A-Woeng, T., and Vrieling, K. (2011). The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 18, 93–105. doi: 10.1093/dnares/dsr002
Downie, S. R., and Jansen, R. K. (2015). A Comparative analysis of whole plastid genomes from the apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot. 40, 336–351. doi: 10.1600/036364415x686620
Fan, G. X., Dong, L. L., Li, H. H., Li, Z. Y., Zhang, Z. X., and Fei, D. Q. (2016). Sesquiterpenoids and other chemical components from the roots of Dolomiaea souliei. Chem. Nat. Comp. 52, 754–757. doi: 10.1007/s10600-016-1766-5
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. doi: 10.1093/nar/gkh458
Fu, Z. X., Jiao, B. H., Nie, B., Zhang, G. J., and Gao, T. G. (2016). A comprehensive generic-level phylogeny of the sunflower family: implications for the systematics of Chinese Asteraceae. J. Syst. Evol. 54, 416–437. doi: 10.1111/jse.12216
Gao, X., Zhang, X., Meng, H., Li, J., Zhang, D., and Liu, C. (2018). Comparative chloroplast genomes of paris sect. marmorata: insights into repeat regions and evolutionary implications. BMC Genomics 19(Suppl. 10):878. doi: 10.1186/s12864-018-5281-x
García, N., Meerow, A. W., Soltis, D. E., and Soltis, P. S. (2014). Testing deep reticulate evolution in Amaryllidaceae tribe Hippeastreae (Asparagales) with ITS and chloroplast sequencedata. Syst. Bot. 39, 75–89. doi: 10.1600/036364414X678099
Gichira, A. W., Avoga, S., Li, Z., Hu, G., Wang, Q., and Chen, J. (2019). Comparative genomics of 11 complete chloroplast genomes of Senecioneae (Asteraceae) species: DNA barcodes and phylogenetics. Bot. Stud. 60:17. doi: 10.1186/s40529-019-0265-y
Gitzendanner, M. A., Soltis, P. S., Wong, G. K., Ruhfel, B. R., and Soltis, D. E. (2018). Plastid phylogenomic analysis of green plants: a billion years of evolutionary history. Am. J. Bot. 105, 291–301. doi: 10.1002/ajb2.1048
Goncalves, D. J. P., Simpson, B. B., Ortiz, E. M., Shimizu, G. H., and Jansen, R. K. (2019). Incongruence between gene trees and species trees and phylogenetic signal variation in plastid genes. Mol. Phylogenet. Evol. 138, 219–232. doi: 10.1016/j.ympev.2019.05.022
Granados-Mendoza, C., Wanke, S., Salomo, K., Goetghebeur, P., and Samain, M. S. (2013). Application of the phylogenetic informativeness method to chloroplast markers: atest case of closely related species in tribe Hydrangeeae (Hydrangeaceae). Mol. Phylogenet. Evol. 66, 233–242. doi: 10.1016/j.ympev.2012.09.029
Herrando-Moraira, S., Cardueae Radiations, G., Calleja, J. A., Galbany-Casals, M., Garcia-Jacas, N., Liu, J. Q., et al. (2019). Nuclear and plastid DNA phylogeny of tribe Cardueae (Compositae) with Hyb-Seq data: a new subtribal classification and a temporal diversification framework. Mol. Phylogenet. Evol. 137, 313–332. doi: 10.1016/j.ympev.2019.05.001
Huang, Y. Y., Qiu, X., Xie, Y. G., Wu, G. J., Zhu, S. L., Zhang, W. D., et al. (2019). New acetylenic compounds and lignans from Dolomiaea berardioidea (Franch.) Shih. Phytochem. Lett. 31, 125–130. doi: 10.1016/j.phytol.2019.03.012
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Iida, S., Miyagi, A., Aoki, S., Ito, M., Kadono, Y., and Kosuge, K. (2009). Molecular adaptation of rbcL in the heterophyllous aquatic plant Potamogeton. PLoS One 4:e4633. doi: 10.1371/journal.pone.0004633
Jansen, R. K., Cai, Z., Raubeson, L. A., Daniell, H., Depamphilis, C. W., Leebens-Mack, J., et al. (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U.S.A. 104, 19369–19374. doi: 10.1073/pnas.0709121104
Jansen, R. K., Wojciechowski, M. F., Sanniyasi, E., Lee, S. B., and Daniell, H. (2008). Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol. Phylogenet. Evol. 48, 1204–1217. doi: 10.1016/j.ympev.2008.06.013
Jiang, P., Shi, F. X., Li, M. R., Liu, B., Wen, J., Xiao, H. X., et al. (2018). Positive selection driving cytoplasmic genome evolution of the medicinally important ginseng plant genus Panax. Front. Plant Sci. 9:359. doi: 10.3389/fpls.2018.00359
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, K. J., Choi, K. S., and Jansen, R. K. (2005). Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol. Biol. Evol. 22, 1783–1792. doi: 10.1093/molbev/msi174
Knox, E. B. (2014). The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms. Proc. Natl. Acad. Sci. U.S.A. 111, 11097–11102. doi: 10.1073/pnas.1403363111
Kreuzer, M., Howard, C., Adhikari, B., Pendry, C. A., and Hawkins, J. A. (2019). Phylogenomic approaches to dna barcoding of herbal medicines: developing clade-specific diagnostic characters for berberis. Front. Plant Sci. 10:586. doi: 10.3389/fpls.2019.00586
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lanfear, R., Calcott, B., Ho, S. Y. W., and Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Li, H. T., Yi, T. S., Gao, L. M., Ma, P. F., Zhang, T., Yang, J. B., et al. (2019). Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 5, 461–470. doi: 10.1038/s41477-019-0421-0
Ling, Y. (1965). Genera nova vel minus cognita familiae compositarum, 1: vladimiria Ilj., diplazoptilon Ling et dolomiaea DC. Acta Phytotaxon. Sin. 10, 75–91.
Liu, M. L., Fan, W. B., Wang, N., Dong, P. B., Zhang, T. T., Yue, M., et al. (2018). Evolutionary analysis of plastid genomes of seven lonicera l. species: implications for sequence divergence and phylogenetic relationships. Int. J. Mol. Sci. 19:4039. doi: 10.3390/ijms19124039
Liu, Y., Huo, N., Dong, L., Wang, Y., Zhang, S., Young, H. A., et al. (2013). Complete chloroplast genome sequences of mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS One 8:e57533. doi: 10.1371/journal.pone.0057533
Logacheva, M. D., Penin, A. A., Samigullin, T. H., Vallejo-Roman, C. M., and Antonov, A. S. (2007). Phylogeny of flowering plants by the chloroplast genome sequences: in search of a lucky gene. Biochemistry 72, 1324–1330. doi: 10.1134/S0006297907120061
Lohse, M., Drechsel, O., Kahlau, S., and Bock, R. (2013). Organellargenomedraw–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41, W575–W581. doi: 10.1093/nar/gkt289
Lopez-Giraldez, F., and Townsend, J. P. (2011). PhyDesign: an online application for profiling phylogenetic informativeness. BMC Evol. Biol. 11:152. doi: 10.1186/1471-2148-11-152
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Martin, G. E., Rousseau-Gueutin, M., Cordonnier, S., Lima, O., Michon-Coudouel, S., Naquin, D., et al. (2014). The first complete chloroplast genome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann. Bot. 113, 1197–1210. doi: 10.1093/aob/mcu050
Moore, M. J., Bell, C. D., Soltis, P. S., and Soltis, D. E. (2007). Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. U.S.A. 104, 19363–19368. doi: 10.1073/pnas.0708072104
Munekage, Y., Hashimoto, M., Miyake, C., Tomizawa, K.-I., Endo, T., Tasaka, M., et al. (2004). Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429, 579–582. doi: 10.1038/nature02598
Neubig, K. M., and Abbott, J. R. (2010). Primer development for the plastid region ycf1 in Annonaceae and other magnoliids. Am. J. Bot. 97, e52–e55. doi: 10.3732/ajb.1000128
Neubig, K. M., Whitten, W. M., Carlsward, B. S., Blanco, M. A., Endara, L., Williams, N. H., et al. (2009). Phylogenetic utility of ycf1 in orchids: a plastid gene more variable than matK. Plant Syst. Evol. 277, 75–84. doi: 10.1007/s00606-008-0105-0
Njuguna, A. W., Li, Z. Z., Saina, J. K., Munywoki, J. M., Gichira, A. W., Gituru, R. W., et al. (2019). Comparative analyses of the complete chloroplast genomes of Nymphoides and Menyanthes species (Menyanthaceae). Aquat. Bot. 156, 73–81. doi: 10.1016/j.aquabot.2019.05.001
Palmer, J. D. (1991). “Plastid chromosomes: structure and evolution,” in The Molecular Biology of Plastids, eds L. Bogorad and I. K. Vasil (Cambridge, MA: Academic Press), 5–53. doi: 10.1016/b978-0-12-715007-9.50009-8
Piot, A., Hackel, J., Christin, P.-A., and Besnard, G. (2018). One-third of the plastid genes evolved under positive selection in PACMAD grasses. Planta 247, 255–266. doi: 10.1007/s00425-017-2781-x
Pond, S. L., Frost, S. D., and Muse, S. V. (2005). HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. doi: 10.1093/bioinformatics/bti079
Prince, L. M. (2015). Plastid primers for angiosperm phylogenetics and phylogeography. Appl. Plant Sci. 3:1400085. doi: 10.3732/apps.1400085
Rabah, S. O., Shrestha, B., Hajrah, N. H., Sabir, M. J., Alharby, H. F., Sabir, M. J., et al. (2019). Passiflora plastome sequencing reveals widespread genomic rearrangements. J. Syst. Evol. 57, 1–14. doi: 10.1111/jse.12425
Rambaut, A., and Drummond, A. (2010). TreeAnnotator Version 1.6. 1. Edinburgh: University of Edinburgh.
Ranwez, V., Douzery, E. J. P., Cambon, C., Chantret, N., and Delsuc, F. (2018). MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol. Biol. Evol. 35, 2582–2584. doi: 10.1093/molbev/msy159
Rozas, J., Ferrer-Mata, A., Sanchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Ruhfel, B. R., Gitzendanner, M. A., Soltis, P. S., Soltis, D. E., and Burleigh, J. G. (2014). From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol. 14:23. doi: 10.1186/1471-2148-14-23
Ruhlman, T. A., and Jansen, R. K. (2014). The plastid genomes of flowering plants. Chloropl. Biotechnol. 1132, 3–38. doi: 10.1007/978-1-62703-995-6_1
Sgamma, T., Lockie-Williams, C., Kreuzer, M., Williams, S., Scheyhing, U., Koch, E., et al. (2017). DNA barcoding for industrial quality assurance. Planta Med. 83, 1117–1129. doi: 10.1055/s-0043-113448
Shen, X., Wu, M., Liao, B., Liu, Z., Bai, R., Xiao, S., et al. (2017). Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules 22:1330. doi: 10.3390/molecules22081330
Shi, D., Song, X., Guo, Y., Xu, J., Liu, Y., Zhang, J., et al. (2017). Alismol, a sesquiterpenoid isolated from Vladimiria souliei, suppresses proinflammatory mediators in lipopolysaccharide-stimulated microglia. J. Mol. Neurosci. 62, 106–113. doi: 10.1007/s12031-017-0890-4
Shrestha, B., Weng, M. L., Theriot, E. C., Gilbert, L. E., Ruhlman, T. A., Krosnick, S. E., et al. (2019). Highly accelerated rates of genomic rearrangements and nucleotide substitutions in plastid genomes of Passiflora subgenus Decaloba. Mol. Phylogenet. Evol. 138, 53–64. doi: 10.1016/j.ympev.2019.05.030
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Su, Y., Huang, L., Wang, Z., and Wang, T. (2018). Comparative chloroplast genomics between the invasive weed Mikania micrantha and its indigenous congener Mikania cordata: structure variation, identification of highly divergent regions, divergence time estimation, and phylogenetic analysis. Mol. Phylogenet. Evol. 126, 181–195. doi: 10.1016/j.ympev.2018.04.015
Sun, Y., Moore, M. J., Lin, N., Adelalu, K. F., Meng, A., Jian, S., et al. (2017). Complete plastome sequencing of both living species of Circaeasteraceae (Ranunculales) reveals unusual rearrangements and the loss of the ndh gene family. BMC Genom. 18:592. doi: 10.1186/s12864-017-3956-3
Sun, Y. X., Moore, M. J., Meng, A. P., Soltis, P. S., Soltis, D. E., Li, J. Q., et al. (2013). Complete plastid genome sequencing of Trochodendraceae reveals a significant expansion of the inverted repeat and suggests a Paleogene divergence between the two extant species. PLoS One 8:e60429. doi: 10.1371/journal.pone.0060429
Susanna, A., and Garcia-Jacas, N. (2007). “Tribe cardueae cass,” in The Families And Genera of Vascular Plants, eds J. W. Kadereit and C. Jeffrey (Berlin: Springer), 123–146.
Susanna, A., and Garcia-Jacas, N. (2009). “Cardueae (Carduoideae),” in Systematics, Evolution, and Biogeography of Compositae, eds V. A. Funk, A. Susanna, T. F. Stuessy, and R. J. Bayer (Vienna: IAPT), 293–313.
Susanna, A., Garcia-Jacas, N., Hidalgo, O., Vilatersana, R., and Garnatje, T. (2006). The cardueae (Compositae) revisited: insights from its, trnL-trnF, and matK nuclear and chloroplast DNA analysis. Ann. Missouri Bot. Garden 93, 150–171. doi: 10.3417/0026-6493
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi: 10.1093/nar/gkx391
Townsend, J. P. (2007). Profiling phylogenetic informativeness. Syst. Biol. 56, 222–231. doi: 10.1080/10635150701311362
Walker, J. E. (2013). The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 41, 1–16. doi: 10.1042/BST20110773
Walker, J. F., Stull, G. W., Walker-Hale, N., Vargas, O. M., and Larson, D. A. (2019). Characterizing gene tree conflict in plastome-inferred phylogenies. PeerJ 7:e7747. doi: 10.1101/512079
Walker, J. F., Zanis, M. J., and Emery, N. C. (2014). Comparative analysis of complete chloroplast genome sequence and inversion variation in Lasthenia burkei (Madieae, Asteraceae). Am. J. Bot. 101, 722–729. doi: 10.3732/ajb.1400049
Wang, W., Yu, H., Wang, J., Lei, W., Gao, J., Qiu, X., et al. (2017). The complete chloroplast genome sequences of the medicinal plant Forsythia suspensa (Oleaceae). Int. J. Mol. Sci. 18:2288. doi: 10.3390/ijms18112288
Wang, X., Liu, B. B., Ma, Y. Z., Xie, P. H., He, X. Y., Shang, B. L., et al. (2013). Chromosomal studies on the alpine genus Dolomiaea (Asteraceae: Cardueae) from the qinghai-tibet plateau and adjacent regions. Caryologia 66, 186–193. doi: 10.1080/00087114.2013.823296
Wang, Y. J., von Raab-Straube, E., Susanna, A., and Liu, J. Q. (2013). Shangwua (Compositae), a new genus from the Qinghai-Tibetan plateau and Himalayas. Taxon 62, 984–996. doi: 10.12705/625.19
Wang, Y., Guo, X., Hao, G., Wang, T., and Wang, K. (2016). The complete chloroplast genome of Sinadoxa corydalifolia (Adoxaceae). Conserv. Genet. Resour. 8, 303–305. doi: 10.1007/s12686-016-0559-2
Wang, Y. J., Liu, J. Q., and Miehe, G. (2007). Phylogenetic origins of the Himalayan endemic Dolomiaea, Diplazoptilon and Xanthopappus (Asteraceae: Cardueae) based on three DNA regions. Ann. Bot. 99, 311–322. doi: 10.1093/aob/mcl259
Wei, H., Ma, G. X., Peng, Y., He, C. N., and Xiao, P. G. (2014). Chemical constituents of the roots of Dolomiaea souliei. Chem. Nat. Comp. 50, 455–457. doi: 10.1007/s10600-014-0985-x
Weng, M. L., Blazier, J. C., Govindu, M., and Jansen, R. K. (2014). Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 31, 645–659. doi: 10.1093/molbev/mst257
Weng, M. L., Ruhlman, T. A., and Jansen, R. K. (2017). Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol. 214, 842–851. doi: 10.1111/nph.14375
Wicke, S., Schneeweiss, G. M., dePamphilis, C. W., Muller, K. F., and Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Wu, C. S., and Chaw, S. M. (2016). Large-scale comparative analysis reveals the mechanisms driving plastomic compaction, reduction, and inversions in conifers II (Cupressophytes). Genome Biol. Evol. 8, 3740–3750. doi: 10.1093/gbe/evw278
Yan, M., Fritsch, P. W., Moore, M. J., Feng, T., Meng, A., Yang, J., et al. (2018). Plastid phylogenomics resolves infrafamilial relationships of the Styracaceae and sheds light on the backbone relationships of the Ericales. Mol. Phylogenet. Evol. 121, 198–211. doi: 10.1016/j.ympev.2018.01.004
Yan, M. H., Moore, M. J., Meng, A. P., Yao, X. H., and Wang, H. C. (2017). The first complete plastome sequence of the basal asterid family styracaceae (Ericales) reveals a large inversion. Plant Syst. Evol. 303, 61–70. doi: 10.1007/s00606-016-1352-0
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang, Z., Wong, W. S., and Nielsen, R. (2005). Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. doi: 10.1093/molbev/msi097
Yao, X. H., Tang, P., Li, Z. Z., Li, D. W., Liu, Y. F., and Huang, H. W. (2015). The first complete chloroplast genome sequences in Actinidiaceae: genome structure and comparative analysis. PLoS One 10:e0129347. doi: 10.1371/journal.pone.0129347
Zhang, D., Gao, F., Jakovlic, I., Zou, H., Zhang, J., Li, W. X., et al. (2019). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang, N., Erickson, D. L., Ramachandran, P., Ottesen, A. R., Timme, R. E., Funk, V. A., et al. (2017). An analysis of echinacea chloroplast genomes: implications for future botanical identification. Sci. Rep. 7:216. doi: 10.1038/s41598-017-00321-6
Zhang, T., Qiao, Q., Novikova, P. Y., Wang, Q., Yue, J., Guan, Y., et al. (2019). Genome of Crucihimalaya himalaica, a close relative of Arabidopsis, shows ecological adaptation to high altitude. Proc. Natl. Acad. Sci. U.S.A. 116, 7137. doi: 10.1073/pnas.1817580116
Zhang, X., Deng, T., Moore, M. J., Ji, Y., Lin, N., Zhang, H., et al. (2019). Plastome phylogenomics of Saussurea (Asteraceae: Cardueae). BMC Plant Biol. 19:290. doi: 10.1186/s12870-019-1896-6
Keywords: Dolomiaea, plastomes, alpine environments, phylogenomic framework, highly divergent regions, positive selection, adaptive evolution
Citation: Shen J, Zhang X, Landis JB, Zhang H, Deng T, Sun H and Wang H (2020) Plastome Evolution in Dolomiaea (Asteraceae, Cardueae) Using Phylogenomic and Comparative Analyses. Front. Plant Sci. 11:376. doi: 10.3389/fpls.2020.00376
Received: 12 January 2020; Accepted: 16 March 2020;
Published: 15 April 2020.
Edited by:
Angelica Cibrian-Jaramillo, Center for Research and Advanced Studies, National Polytechnic Institute, MexicoReviewed by:
Carolina Granados Mendoza, National Autonomous University of Mexico, MexicoItzi Fragoso-Martínez, National Autonomous University of Mexico, Mexico
Copyright © 2020 Shen, Zhang, Landis, Zhang, Deng, Sun and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hang Sun, c3VuaGFuZ0BtYWlsLmtpYi5hYy5jbg==; Hengchang Wang, aGN3YW5nQHdiZ2Nhcy5jbg==
†These authors have contributed equally to this work
 Jun Shen1,2,3†
Jun Shen1,2,3† Xu Zhang
Xu Zhang Jacob B. Landis
Jacob B. Landis Huajie Zhang
Huajie Zhang Tao Deng
Tao Deng Hang Sun
Hang Sun Hengchang Wang
Hengchang Wang