- 1College of Agriculture, Fujian Agricultural and Forestry University, Fuzhou, China
- 2Fujian Key Laboratory of Genetic Engineering for Agriculture, Biotechnology Research Institute, Fujian Academy of Agricultural Sciences, Fuzhou, China
- 3Marine and Agricultural Biotechnology Laboratory, Institute of Oceanography, Minjiang University, Fuzhou, China
Japonica rice has become increasingly popular in China owing to its superior grain quality. Over the past decades, “indica to japonica” projects have been proposed to promote cultivation of japonica rice in low latitudes in China. Traditionally, japonica varieties were planted mainly in mid latitudes in the northeast plain and Yangtze River region. The key obstacle for introducing elite mid-latitude japonica varieties to low latitudes is the severe shortening of growth period of the japonica varieties due to their sensitivity to low-latitude short photoperiod and high temperature. Here we report development of new japonica rice with prolonged basic vegetative growth (BVG) periods for low latitudes by targeted editing the Early heading date 1 (Ehd1) gene. Using CRISPR/Cas9 system, we generated both frame-shift and/or in-frame deletion mutants in four japonica varieties, Nipponbare, Longdao16, Longdao24, and Xiushui134. When planting at low-latitude stations, the frame-shift homozygous lines exhibited significantly longer BVG periods compared with wild-types. Interestingly, we observed that minor deletion of the first few residues within the receiver domain could quantitatively impair the function of Ehd1 on activation of Hd3a and RFT1, resulting in an intermediate-long BVG period phenotype in the homozygous in-frame deletion ehd1 lines. Field investigation further showed that, both the in-frame and frame-shift lines exhibited significantly improved yield potential compared with wild-types. Our study demonstrates an effective approach to rapid breeding of elite japonica varieties with intermediate-long and long BVG periods for flexible cropping systems in diverse areas or under different seasons in southern China, and other low-latitude regions.
Introduction
Rice (Oryza sativa L.) is one of the most important crops in China. Cultivated rice is comprised of japonica and indica subspecies. Compared to indica varieties, japonica varieties in general have more superior grain quality – higher head rice rates and gel consistency, lower chalky rice rates, chalkiness degree, and amylose contents (Feng et al., 2017). Over the past decades, japonica rice has become increasingly popular in China. To meet the demands of consumers, “indica to japonica” projects have been proposed to promote cultivation of japonica varieties in southern China (Zhang et al., 2013; Leng et al., 2019).
Traditionally, japonica varieties were planted mainly in mid latitudes in the northeast plain and Yangtze River region, and indica varieties were planted in low latitudes in southern China (Min et al., 2011; Wang et al., 2015). Over the past decades, many elite japonica varieties have been bred for mid latitudes in China. However, most of the mid-latitude japonica varieties were sensitive to photoperiod or temperature, and they were not suitable for cultivating in low-latitude regions. Under short photoperiod and high temperature environmental conditions in low latitudes, the mid-latitude japonica varieties commonly display early flowering, resulting in low grain yield (Saito et al., 2009; Wei et al., 2009; Yuan et al., 2009).
The rice Early heading date 1 (Ehd1) gene encodes a highly conserved B-type two component response regulator (RR), which acts as a key signal integrator in the networks regulating floral transition (Doi et al., 2004). Ehd1 promotes the expression of Heading date 3a (Hd3a) and Rice Flowering Locust1 (RFT1) that induce rice flowering under both short day (SD) and long day (LD) conditions (Doi et al., 2004; Komiya et al., 2008). The expression of Ehd1 is activated or repressed by multiple genes (Komiya et al., 2009; Shrestha et al., 2014). For example, LVP1/SDG724 (Sun et al., 2012), Ehd2/RID1/OsID1 (Matsubara et al., 2008; Park et al., 2008; Wu et al., 2008; Hu et al., 2013), Ehd3 (Matsubara et al., 2011), Ehd4 (Gao et al., 2013), OsMADS50/DTH3 (Lee et al., 2004; Bian et al., 2011), OsMADS51 (Kim et al., 2007), Hd-q/Ef7 (Yuan et al., 2009; Zhao et al., 2013) can positively regulate Ehd1 transcription. In contrast, Ehd1 expression is negatively regulated by HD1 (Yano et al., 2000; Nemoto et al., 2016), Ghd7 (Xue et al., 2008), DTH7/Ghd7.1/OsPRR37 (Koo et al., 2013; Yan et al., 2013; Gao et al., 2014), DTH8/EF8/Ghd8 (Wei et al., 2010; Yan et al., 2011; Feng et al., 2014) under LD, and negatively regulated by SE5 under both LD and SD (Andrés et al., 2009). Mutation in Ehd1 has a severe effect on the floral transition. A Gly-to-Arg point mutation in the GARP motif of Ehd1 leads to a reduced response to photoperiod in rice (Doi et al., 2004). This ehd1 mutation allele enabled japonica varieties from Taiwan region suitable for planting under low-latitude conditions. These japonica varieties exhibit long basic vegetative growth (BVG) periods and display elite agronomic traits in Taiwan region (Nishida et al., 2002; Saito et al., 2009; Wei et al., 2016). Overall, these previous studies demonstrated the potentiality of Ehd1 as a target for genome editing for regulating heading date in rice.
CRISPR/Cas9 technology has emerged as a powerful approach for rice research and breeding (Shan et al., 2014; Xie et al., 2015; Lu et al., 2017; Xu et al., 2017; Li et al., 2018). CRISPR/Cas9-mediated gene modification has been applied in rice for improvement of important agronomic traits, such as photo-sensitive and thermo-sensitive genic male sterility (Li et al., 2016; Zhou et al., 2016), herbicide-resistance (Sun et al., 2016), resistance to blast and tungro spherical virus (Wang et al., 2016; Macovei et al., 2018), low cadmium accumulation (Tang et al., 2017), grain yield (Miao et al., 2018), glutinous trait (Zhang J. et al., 2018), chilling tolerance (Mao et al., 2019), red pericarp (Zhu et al., 2019), and fixation of heterosis (Khanday et al., 2019; Wang et al., 2019). More recently, Cui et al. (2019) assessed the effect of ten heading time genes on reproductive transition and yield components in rice using a CRISPR/Cas9 system, revealing that heading time was often associated with yield-related traits. In the present study, we report development of new japonica rice exhibiting prolonged BVG periods for low latitudes by targeted editing the receiver domain region of Ehd1. Using CRISPR/Cas9 system, we generated both frame-shift and/or in-frame deletion mutants in four japonica varieties, Nipponbare, Longdao16, Longdao24, and Xiushui134. When planting under low-latitude conditions, the frame-shift mutants of Nipponbare, Longdao16, Longdao24, and Xiushui134 exhibited significantly longer BVG periods compared with their wild-type parents. Interestingly, the in-frame mutants of Longdao16, Longdao24, and Xiushui134 possessed intermediate-long BVG periods. Field investigation further showed that, both the in-frame and frame-shift mutant lines showed significantly improved yield potential compared with wild-type plants. Our study demonstrates an effective approach for breeding new japonica varieties with intermediate-long and more extremely long BVG periods for flexible cropping systems in southern China, as well as in other low-latitude regions.
Materials and Methods
Plant Materials
The japonica variety Nipponbare and three elite japonica varieties Longdao16, Longdao24 and Xiushui134 were used in this study.
Construction of sgRNA-Cas9 Plant Expression Vector and Rice Transformation
A single guide RNA (sgRNA) sequence targeted the receiver-domain region of the Ehd1 gene were designed according to Shan’s program (Shan et al., 2014). A sgRNA-Cas9 vector pVK005-01 (Viewsolid Biotech, Beijing, China) in which a codon-optimized Cas9 gene was under control of the maize ubiquitin promoter and the customized sgRNA was driven by the rice U6 promoter, was used for generation of the sgRNA-Cas9 construct for Ehd1. DNA oligos (Supplementary Table S1) corresponding to the designed sgRNA sequence were synthesized and the dimer was cloned into pVK005-01 according to the manufacturer’s instruction.
The Ehd1 sgRNA-Cas9 expression vector was introduced into Agrobacterium tumefaciens strain EHA105 by electroporation. Agrobacterium-mediated transformation of rice calli induced from seeds of Nipponbare, Longdao16, Longdao24, and Xiushui134 was carried out as described previously (Hiei et al., 1997).
Genotyping and Off-Target Analysis
Genomic DNA was extracted from rice leaves using the CTAB method. The T0 transgenic plants were determined by PCR amplification of the hygromycin phosphotransferase gene. To genotype mutations in the Ehd1 gene, PCR amplification was performed using primer pairs flanking the Ehd1 target site (Supplementary Table S1). The PCR products were sequenced by Sanger method. DNA sequences were aligned using Clustal Omega (Sievers et al., 2011).
Transgene-free homozygous mutant plants/lines were screened in T1 generation derived from self-pollination of T0 mutant plants, and were double confirmed in T2 generation. To identify transgene-free plants, PCR amplification was performed with DNA extracted from individual plants using specific primers (Supplementary Table S1) for the hygromycin phosphotransferase gene and the Cas9 gene.
Forty transgene-free T2 mutant plants in Nipponbare, Longdao16, Longdao24, and Xiushui134 backgrounds were selected for evaluation of the potential off-target effects. PCR amplification was performed using specific primers (Supplementary Table S1) for the potential off-target sites. The resulting PCR products were sequenced for mutations by Sanger method.
Plant Growth Conditions and Phenotype Analysis
T0 generation plants were grown in a standard greenhouse. Wild-type parents, T1 and T2 transgene-free homozygous mutant plants were investigated for agronomic traits. The plants were cultivated at Sanya experimental station, Hainan Province, under nature SD condition (December to next April, c. 11–12.5 h, 18°14′N, 109°31′E), and at Fuzhou experimental station, Fujian Province, under nature LD condition (mid-May to mid-September, c. 13.5 h, 26°08′N, 119°28′E). Longdao16 and Longdao24 were also cultivated at Haerbing experimental station, Heilongjiang Province, under nature LD condition (April to September, c. 13.5–15.7 h, 45°51′N, 126°50′E). About 100 seedlings of each line were transplanted in the field. Column space and row were both 20 cm. The field management was followed the normal agricultural practice. Head rice rate was determined by the percentage of the total head rice weight produced after milling. Grain chalkiness traits were determined using image analysis method (Fang et al., 2015), and grain amylose content was determined by iodine reaction (Juliano, 1971). Gel consistency and alkali spreading value were determined as described previously (Leng et al., 2013).
One T2 Nipponbare mutant line N-ehd1-#1, and two T2 Longdao16 mutant lines L16-end1-#2, L16-end1-#3 were used for analyzing gene expression of Hd3a and RFT1 at different developmental stages. The frame-shift mutant lines N-ehd1-#2, L16-end1-#1, and in-frame mutant line L16-end1-#6 were also analyzed for the mRNA expression levels of Ehd1, Hd3a, and RFT1 at 50d after sowing. All the lines were grown under LD condition (mid-May to mid-August, c. 13.5 h, before heading) in nature filed and SD condition (September to next January, c. 12.7–11.5 h) in a greenhouse at Fuzhou experimental station. Full expanded leaves were sampled 2 h after dawn. Three independent plants were used as biological replicates.
Gene Expression Analysis
Total RNAs were isolated from rice leaves using RNA isolater Total RNA Extraction Reagent (Vazyme Biotech Co., Ltd., Nanjing, China) according to the manufacturer’s instructions. The RNA samples were treated with Recombinant DNase I (TaKaRa, Shiga, Japan). First cDNA was synthesized with 2 μg of total RNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, UC, Lithuania). The synthesized cDNAs were used as templates for quantitative real-time RT-PCR with ChamQTM SYBR qPCR Master Mix (Vazyme Biotech, Nanjing, China) on a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific, Singapore). All experiments were conducted three times. The primers used for gene expression analysis are listed in Supplementary Table S1.
Results
Site-Specific Mutagenesis of Ehd1 Mediated by CRISPR/Cas9 System
To generate transgenic plants with targeted mutation in Ehd1, a sgRNA-Cas9 construct targeting the first exon of the Ehd1 gene (Figure 1A) was designed and used for rice transformation. A total of 30, 28, 25, and 20 T0 independent transgenic plants were generated from calli of japonica rice varieties Nipponbare, Longdao16, Longdao24, and Xiushui134, respectively (Table 1 and Supplementary Table S2). Genotyping of the transgenic plants identified 53 (14, 15, 13, and 11 in Nipponbare, Longdao16, Longdao24, and Xiushui134 backgrounds, respectively) with mutations in the predicted target site of Ehd1 (Figure 1, Table 1, and Supplementary Table S2). The mutations in T0 transgenic plants included homozygous, bi-allelic and heterozygous genotypes (Table 1 and Supplementary Table S2). Most of the mutations were frame-shift types in mutants in Nipponbare, Longdao16, Longdao24, and Xiushui134 backgrounds. In addition, there were two, three, and one mutants with in-frame deletion in the target site in Longdao16, Longdao24, and Xiushui134 backgrounds, respectively (Figure 1B and Supplementary Table S2). The mutants in Nipponbare, Longdao16, Longdao24, and Xiushui134 backgrounds were thus referred to as N-ehd1, L16-ehd1, L24-ehd1, and X-ehd1, respectively.
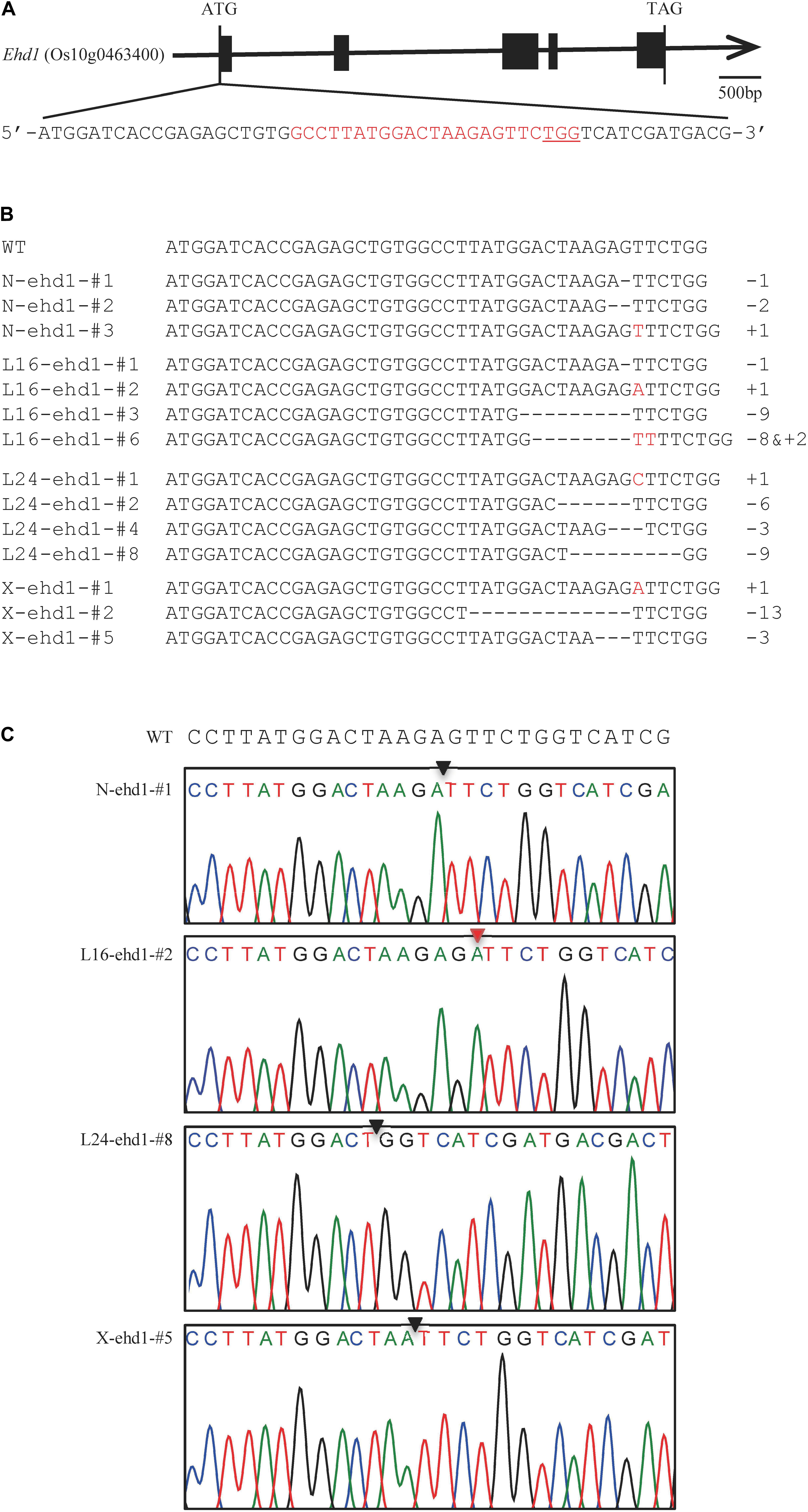
Figure 1. CRISPR/Cas9-mediated genome editing of the Ehd1 gene. (A) Schematic illustration of the Ehd1 gene structure. Black rectangles represent the 5 exons of Ehd1; Red characters indicate the sequence of the target site; PAM sequences are underlined. (B) Examples of mutations of Ehd1 in CRISPR/Cas9-mediated mutated plants. Deletions and insertions are indicated by dashes and red letters, respectively, Numbers on the right side indicate the numbers of nucleotides involved (+, insertion; −, deletion); WT indicates wild-type sequence; N-ehd1-#, L16-ehd1-#, L24-ehd1-#, and X-ehd1-# represent the mutant plants in Nipponbare, Longdao16, Longdao24, and Xiushui134 backgrounds, respectively. (C) Examples of sequencing chromatograms of an 1 bp deletion mutant (N-ehd1-#1), an 1 bp insertion mutant (L16-ehd1-#2), a 9 bp deletion mutant (L24-ehd1-#8), and a 3 bp deletion mutant (X-ehd1-#5), in Nipponbare, Longdao16, Longdao24, and Xiushui134 backgrounds, respectively. The deletion and insertion sites are indicated by the black triangle and red triangle, respectively.
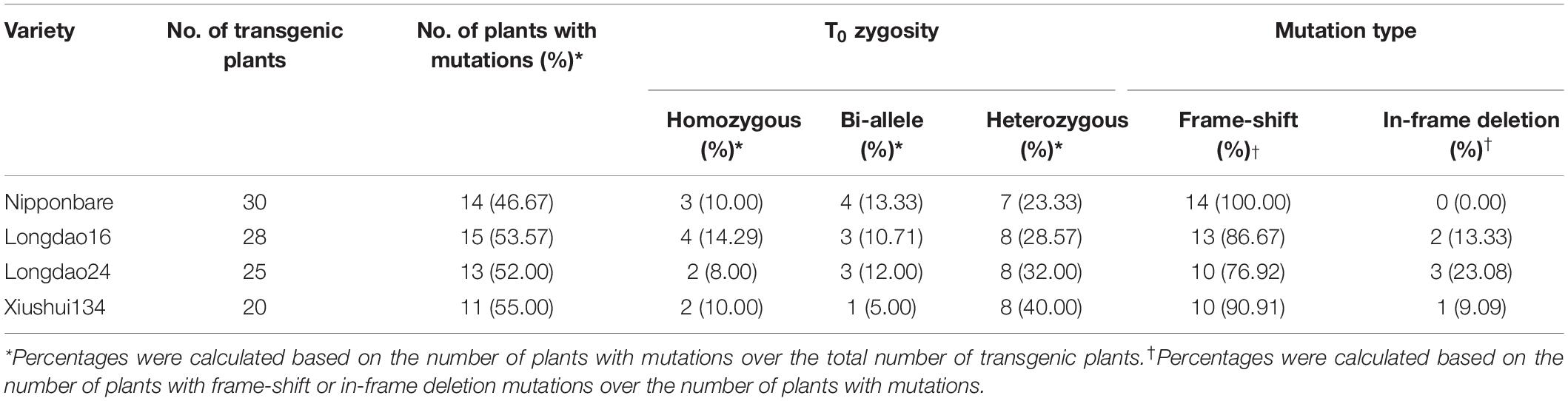
Table 1. Mutation analysis of T0 plants transformed with the sgRNA-Cas9 construct targeting the first exon of the Ehd1 gene.
Transgene-free T1 and T2 mutant lines were screened (Supplementary Figures S1, S2) and T2 mutant lines were examined for potential off-target mutations. Six potential off-target sites containing four or five mismatched bases were retrieved using the CRISPR-P 2.0 tool (Liu et al., 2017). PCR products amplified from 40 transgene-free T2 mutant plants (9, 11, 10, and 10 in Nipponbare, Longdao16, Longdao24, and Xiushui134 backgrounds, respectively) were sequenced. No mutations were found in all six potential sites (Supplementary Table S3), indicating the high specificity of the designed sgRNA in mediating mutagenesis of the predicted site.
Frame-Shift Mutations in Ehd1 Resulted in Long BVG Periods in Nipponbare, Longdao16, Longdao24, and Xiushui134
Transgene-free homozygous T1 and T2 mutant lines were investigated for heading date under nature SD (Sanya, December to next April, c. 11–12.5 h, 18°14′N, 109°31′E) and LD (Fuzhou, mid-May to mid-September, c. 13.5 h, 26°08′N, 119°28′E) environment conditions in low-latitude regions. As showed in Figure 2 and Supplementary Figure S3, the flowering times of the frame-shift mutants were significantly delayed compared with that of wild-type in Nipponbare, Longdao16, Longdao24, and Xiushui134 backgrounds. For example, the heading dates of N-ehd1-#1, an 1-bp deletion homozygous mutant line in Nipponbare background (Figure 2A), were 98.1 ± 3.1 days under SD and 92.3 ± 2.5 days under LD, respectively. In contrast, the heading dates of wild-type Nipponbare were about 57.7 ± 2.3 days and 61.1 ± 2.3 days under SD and LD, respectively (Figure 2A). The heading dates of L16-ehd1-#1, an 1-bp deletion homozygous mutant line in Longdao16 background, were 108.5 ± 2.4 days under SD and 106.4 ± 2.5 days under LD, which were about 40 days more than that of wild-type Longdao16 plants under SD (71.2 ± 2.2 days) and LD (64.9 ± 2.3 days), respectively (Figure 2B). Similar results were observed in other tested frame-shift mutant lines in all four backgrounds (Figure 2), indicating that frame-shift mutations in Ehd1 could result in long BVG periods in japonica varieties under low-latitude conditions.
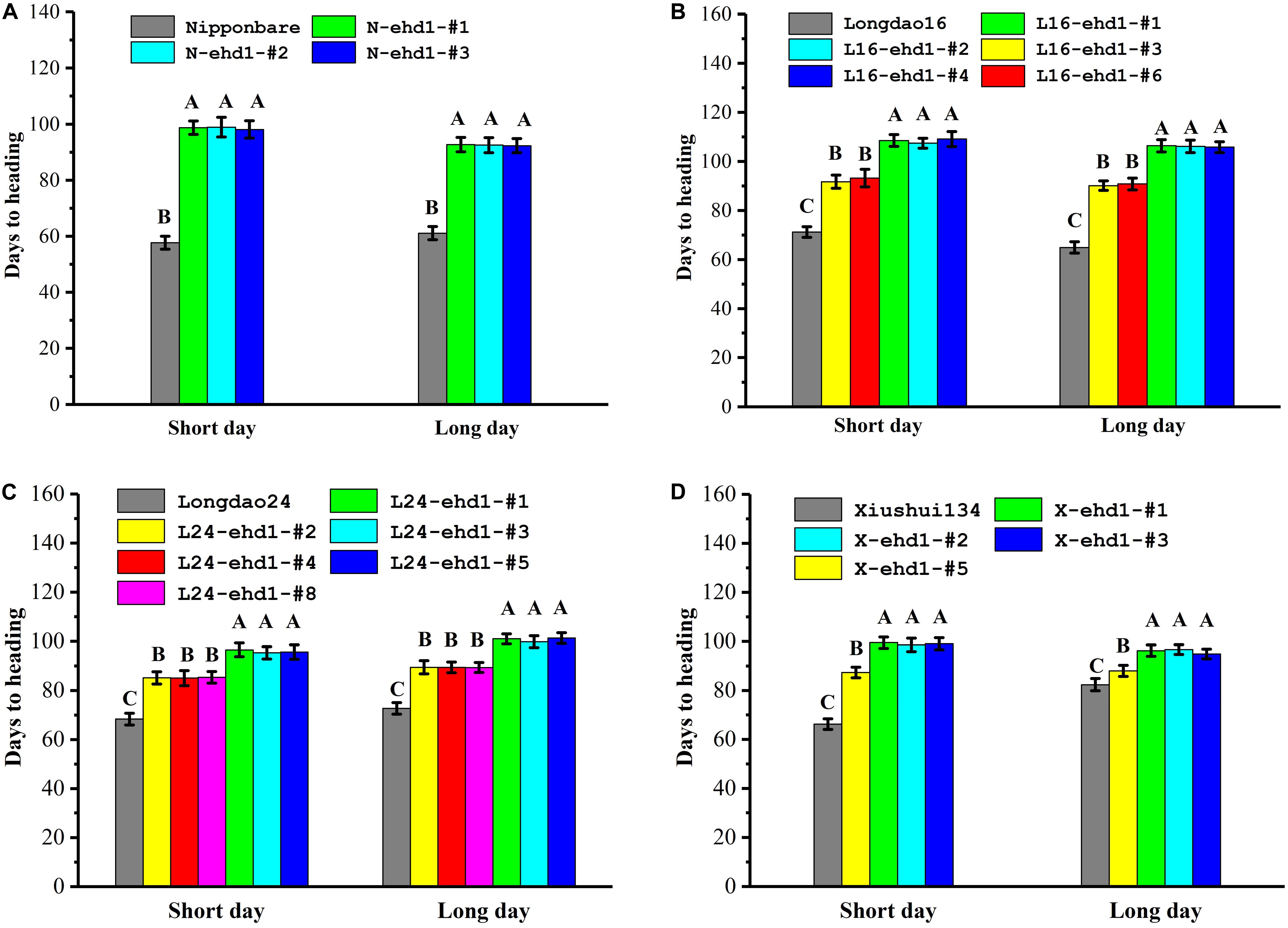
Figure 2. ehd1 mutant lines exhibited prolonged basic vegetative growth (BVG) periods under short day and long day conditions. Rice plants were cultivated at Sanya experimental station under nature short day condition (December to next April, c. 11–12.5 h, 18°14′ N, 109°31′E), and at Fuzhou experimental station under natural long day condition (mid-May to mid-September, c. 13.5 h, 26°08′N, 119°28′E). (A–D) Flowering times of homozygous mutant lines in Nipponbare (A), Longdao16 (B), Longdao24 (C) and Xiushui134 (D) backgrounds under both short day and long day conditions. Data were collected from 30 plants. The letters A, B, and C indicate significant differences according to LSD multiple range test at P ≤ 0.01.
In-Frame Deletion Mutations in the Receiver Domain Region of Ehd1 Resulted in Moderate Delays of Heading Date in Longdao16, Longdao24, and Xiushui134
Six in-frame deletion lines, L16-ehd1-#3, L16-ehd1-#6, L24-ehd1-#2, L24-ehd1-#4, L24-ehd1-#8, and X-ehd1-#5, were identified in Longdao16, Longdao24, or Xiushui134 backgrounds, respectively. Sequencing results of the mutated target site showed that the change numbers of nucleotides were multiples of three in the six lines (Figure 3 and Supplementary Figure S4). For example, L16-ehd1-#3 was a mutant with 9-bp deletion (5′-GACTAAGAG-3′) (Supplementary Figure S4) and L16-ehd1-#6 was a mutant with 8 bp deletion and 2 bp insertion (5′-ACTAAGAGTT-3′) (Supplementary Figure S4). To examine whether the mutated alleles had different splicing patterns of ehd1, RT-PCR was performed to amplify the open reading frame (ORF) of the mutated ehd1 in all six lines. Sequencing results revealed that the splicing pattern of these six novel alleles was unchanged (Supplementary Figure S4). Prediction from the mutated ehd1-ORF sequences revealed that there were minor deletions (two- to three-amino acid) in the receiver domain of ehd1 in L16-ehd1-#3, L24-ehd1-#2, and L24-ehd1-#8, and there were minor deletions (one- to two-amino acid) plus a Val-to-Phe, -Ser or -Ile substitution in L16-ehd1-#6, L24-ehd1-#4, and X-ehd1-#5 (Figure 3A).
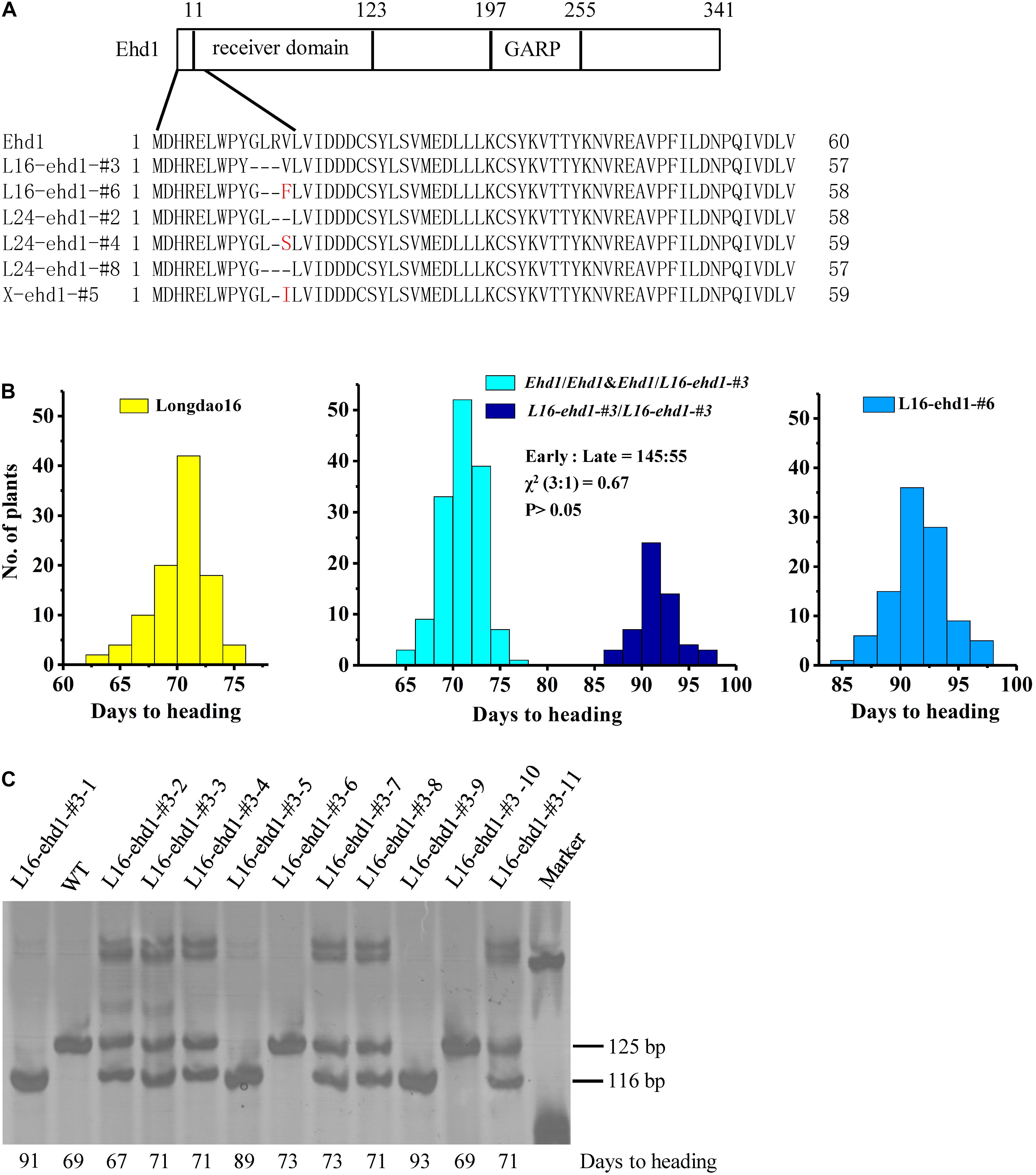
Figure 3. Minor in-frame deletion mutations in the receiver domain region of Ehd1 caused moderate delays of heading date in rice. (A) Schematic diagram of the domains within Ehd1 and comparison of deduced amino acid sequences of Ehd1 in the six in-frame deletion mutant lines L16-ehd1-#3, L16-ehd1-#6, L24-ehd1-#2, L24-ehd1-#4, L24-ehd1-#8, and X-ehd1-#5. Deletion and substitution are indicated by dash and red letter, respectively. (B) Frequency distributions of days to heading of Longdao16 (left), T1 segregating population of L16-ehd1-#3 (middle), and T1 homozygous line L16-ehd1-#6 (right). About 100 plants, 200 plants and 100 plants of Longdao16, L16-ehd1-#3, and L16-ehd1-#6, respectively, were investigated. (C) Examples of genotyping and phenotyping results showing co-segregation of the moderately delayed flowering phenotype with the homozygous L16-ehd1-#3 allele in T1 population of L16-ehd1-#3. Each individual plant was analyzed by PCR amplification using allele-specific primers. The 125 and 116 bp fragments represent wild-type Ehd1 and L16-ehd1-#3 alleles, respectively. Days to heading of each detected plant are shown at the bottom.
Under both SD and LD, the flowering times of the homozygous in-frame deletion plants in Longdao16 and Longdao24 backgrounds were delayed about 2–3 weeks compared with that of their wild-type parents, but were about 10 days to 2 weeks earlier than that of the frame-shift mutant lines in the same backgrounds (Figures 2B,C). Similar results were observed in the in-frame deletion line X-ehd1-#5 in Xiushui134 background, although the delay of flowering time under LD was not as great as that under SD (Figure 2D). Overall, the in-frame deletion mutant lines exhibited moderate delays of heading date. To further confirm the correlation between the in-frame deletion ehd1 alleles and the moderately-delayed flowering phenotype, a total of 200 segregating T1 progeny derived from T0 L16-ehd1-#3 were phenotyped and genotyped individually. The T1 L16-ehd1-#3 plants exhibited a segregation of 145 early flowering and 55 moderately-delayed flowering, fitting to a 3:1 ratio (χ2 = 0.67, P > 0.05) (Figure 3B). The genotyping result showed that the segregating ratios of wild-type (Ehd1/Ehd1): heterozygous type (Ehd1/L16-ehd1-#3): homozygous type (L16-ehd1-#3/L16-ehd1-#3) fitted the 1:2:1 ratio of segregation (42:103:55, χ2 = 0.87, P > 0.05). More importantly, the homozygous L16-ehd1-#3 allele was co-segregated with the moderately-delayed flowering phenotype (Figures 3B,C).
In-Frame Deletion ehd1 Allele Had a Moderate Effect on Activation of Hd3a and RFT1
The mRNA expression levels of Ehd1, Hd3a and RFT1 in homozygous T2 lines N-ehd1-#1, N-ehd1-#2, L16-end1-#1, L16-ehd1-#2, L16-ehd1-#3, and L16-end1-#6 under both SD and LD were detected. The results showed no significant difference in the mRNA levels of Ehd1 between Nipponbare and Longdao16 under SD. In contrast, Ehd1 mRNA level in Nipponbare was significantly lower than that in Longdao16 under LD (Supplementary Figure S5). In Nipponbare background, there was no significant difference in Ehd1 mRNA levels between N-ehd1-#1, N-ehd1-#2, and wild-type plants. In Longdao16 background, Ehd1 mRNA levels in the frame-shift mutant lines L16-end1-#1 and L16-ehd1-#2 were higher than that in wild-type plants or the in-frame deletion lines L16-ehd1-#3 and L16-end1-#6 (Supplementary Figure S5). These results suggested that mutagenesis mediated by CRISPR/Cas9 affected Ehd1 at the protein level but not the transcript level.
During the vegetative growth phrase, Hd3a and RFT1 were expressed at relatively high levels in wild-type varieties Nipponbare and Longdao16. In contrast, Hd3a and RFT1 were expressed at low levels in the frame-shift mutant lines N-ehd1-#1, N-ehd1-#2, L16-end1-#1, and L16-ehd1-#2 (Figure 4A and Supplementary Figure S6). The expression levels of Hd3a and RFT1 in the in-frame deletion lines L16-ehd1-#3 and L16-end1-#6 were lower than that in wild-type Longdao16, but were higher than that in L16-end1-#1 and L16-ehd1-#2 (Figure 4A and Supplementary Figure S6), although expression patterns were not exactly the same under SD and LD, respectively. We further performed the liner fitting analysis of the relationship between heading date and expression levels of Hd3a and RFT1 in L16-ehd1-#2, L16-ehd1-#3, and Longdao16. The analysis revealed that there was a significantly negative correlation between heading date and expression levels of Hd3a and RFT1 (Supplementary Figure S7). These results thus supported the conclusion that transcriptional activation of Hd3a and RFT1 mainly relied on the presence of functional Ehd1 (Doi et al., 2004; Zhao et al., 2015; Cho et al., 2016; Nemoto et al., 2016). Our results also indicated that minor deletions in the receiver domain could quantitatively impair the function of Ehd1 on activation of Hd3a and RFT1, resulting in a moderately-delayed flowering phenotype (Figure 4B).
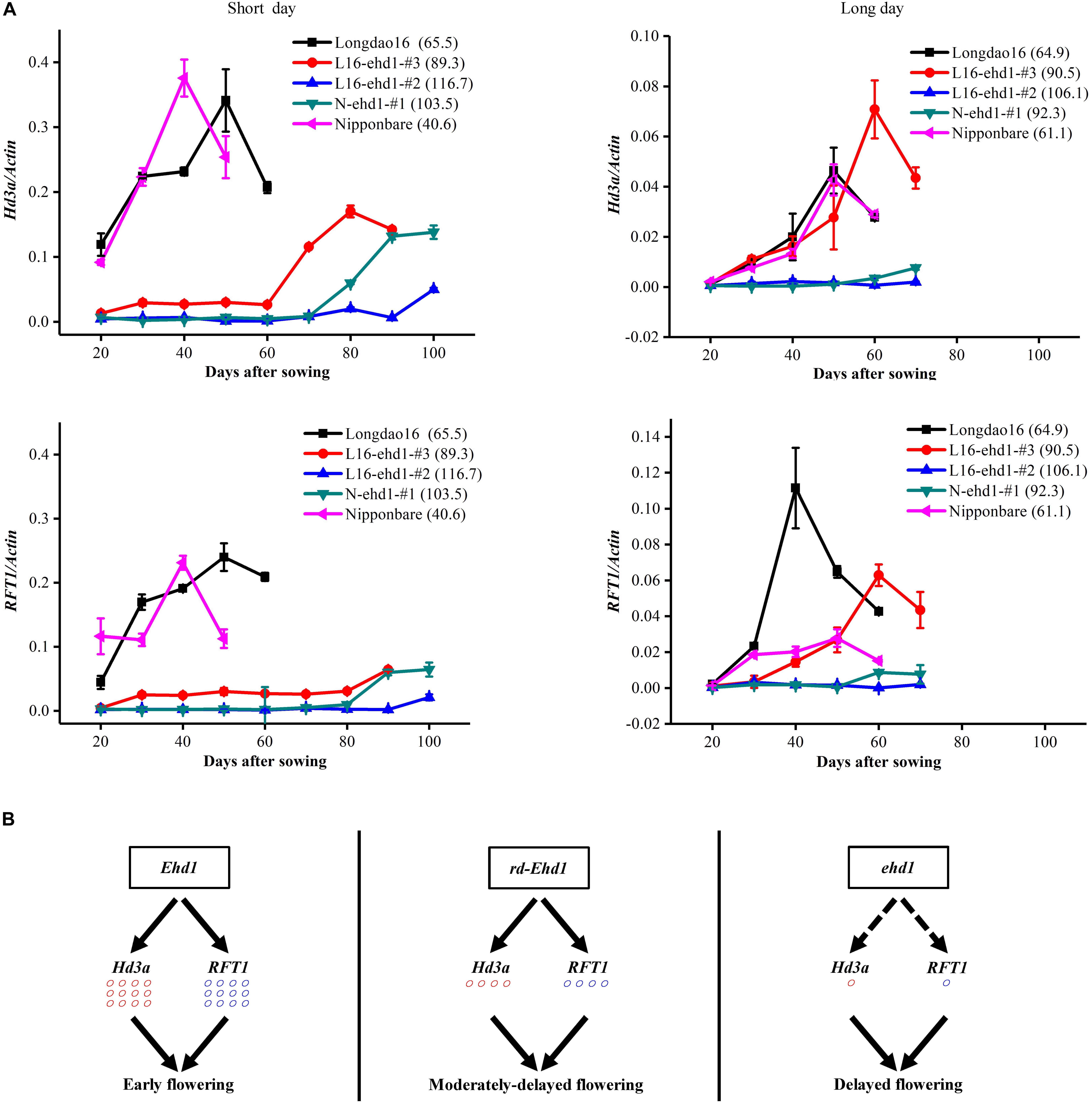
Figure 4. Minor in-frame deletion in the receiver domain partially impairs the function of Ehd1 on activation of Hd3a and RFT1. (A) Expression patterns of Hd3a and RFT1 in frame-shift mutant lines N-ehd1-#1 and L16-end1-#2, and in-frame mutant line L16-end1-#3. T2 homozygous lines were used for detection. Fully expanded leaves were sampled 2 h after dawn every 10 days (the first time to harvest samples was 20 days after sowing). Three different rice plants were used as biological replicates and the experiments were repeated three times with similar results. Gene expression values are shown as mean and standard deviation of the three biological replicates. Days to heading of each line are shown in parentheses. (B) Schematic model of the minor deletion-ehd1-Hd3a/RFT1 regulatory network. Functional Ehd1 promotes the expression of two florigen genes Hd3a and RFT1 (left panel). In ehd1 background, the expression of Hd3a and RFT1 was inhibited (right panel). In-frame deletion ehd1 partially activates the expression of Hd3a and RFT1 (middle panel), resulting in a moderately-delayed flowering phenotype. rd-Ehd1 represents in-frame deletion ehd1 alleles.
In-Frame and Fame-Shift Mutant Lines Exhibited High Grain Yield and Good Grain Quality in Low-Latitude Regions
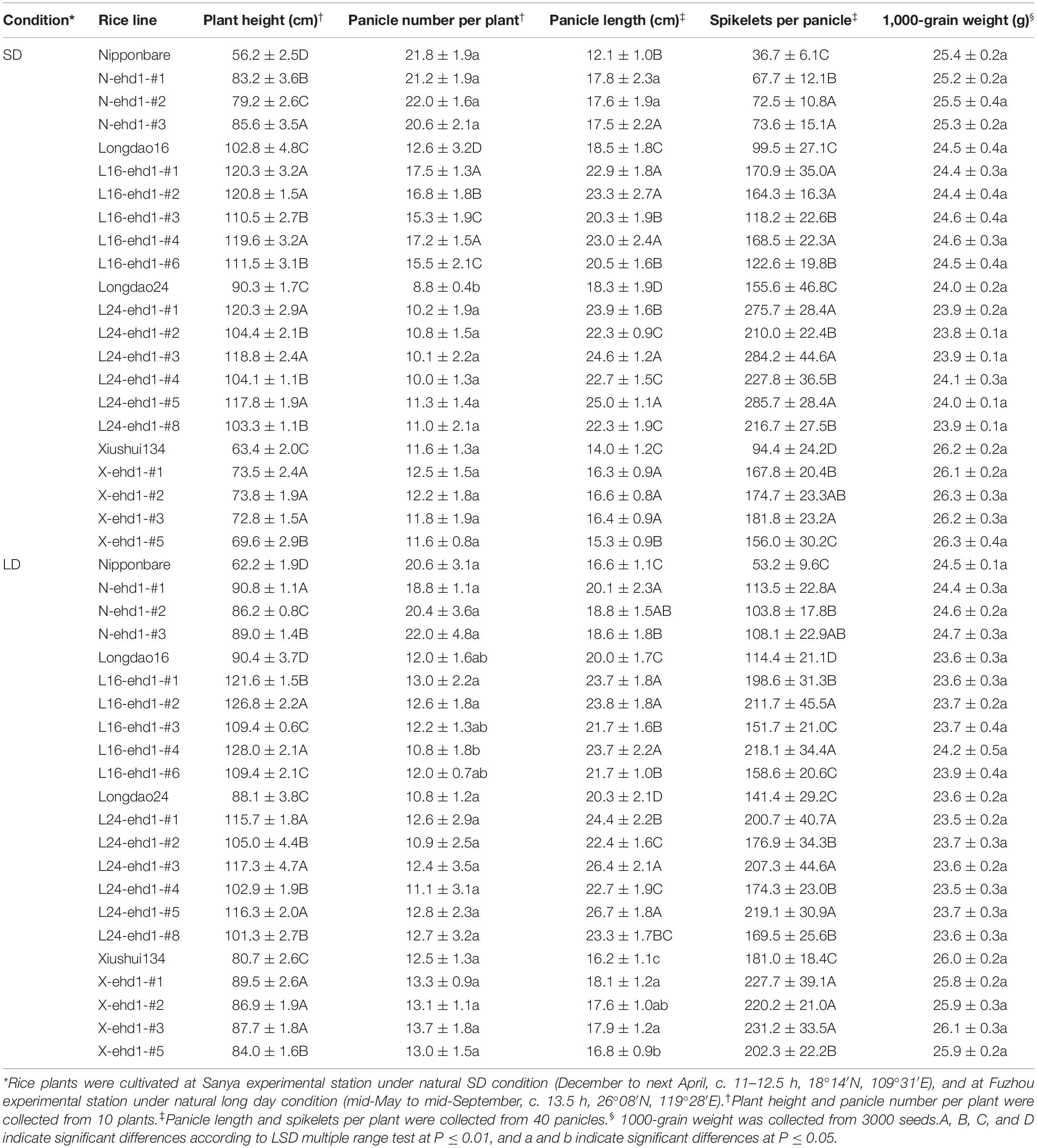
Homozygous in-frame and frame-shift mutant lines were investigated under both SD and LD in low-latitude regions for yield-associated traits, including plant height, panicle number per plant, panicle length, 1000-grains weight, spikelets per panicle, and grain yield per plant. Due to the prolongation of growth period, both the in-frame and frame-shift mutant lines exhibited significantly higher plant height, panicle length, especially spikelets per panicle, which in turn resulted in higher grain yield per plant compared with wild-types (Figure 5 and Table 2). For example, the grain yields per plant of the in-frame mutant lines L16-ehd1-#3 and L16-ehd1-#6 were about 1.37- to 1.43-fold, and the grain yields per plant of the frame-shift lines L16-ehd1-#1, L16-ehd1-#2, and L16-ehd1-#4 were about 1.90–2.28-fold compared with that in wild-type Longdao16 plants under SD or LD (Figure 5F). Similar results were observed in other homozygous frame-shift lines in Nipponbare, Longdao24, or Xiushui134 backgrounds (Figures 5E,G,H).
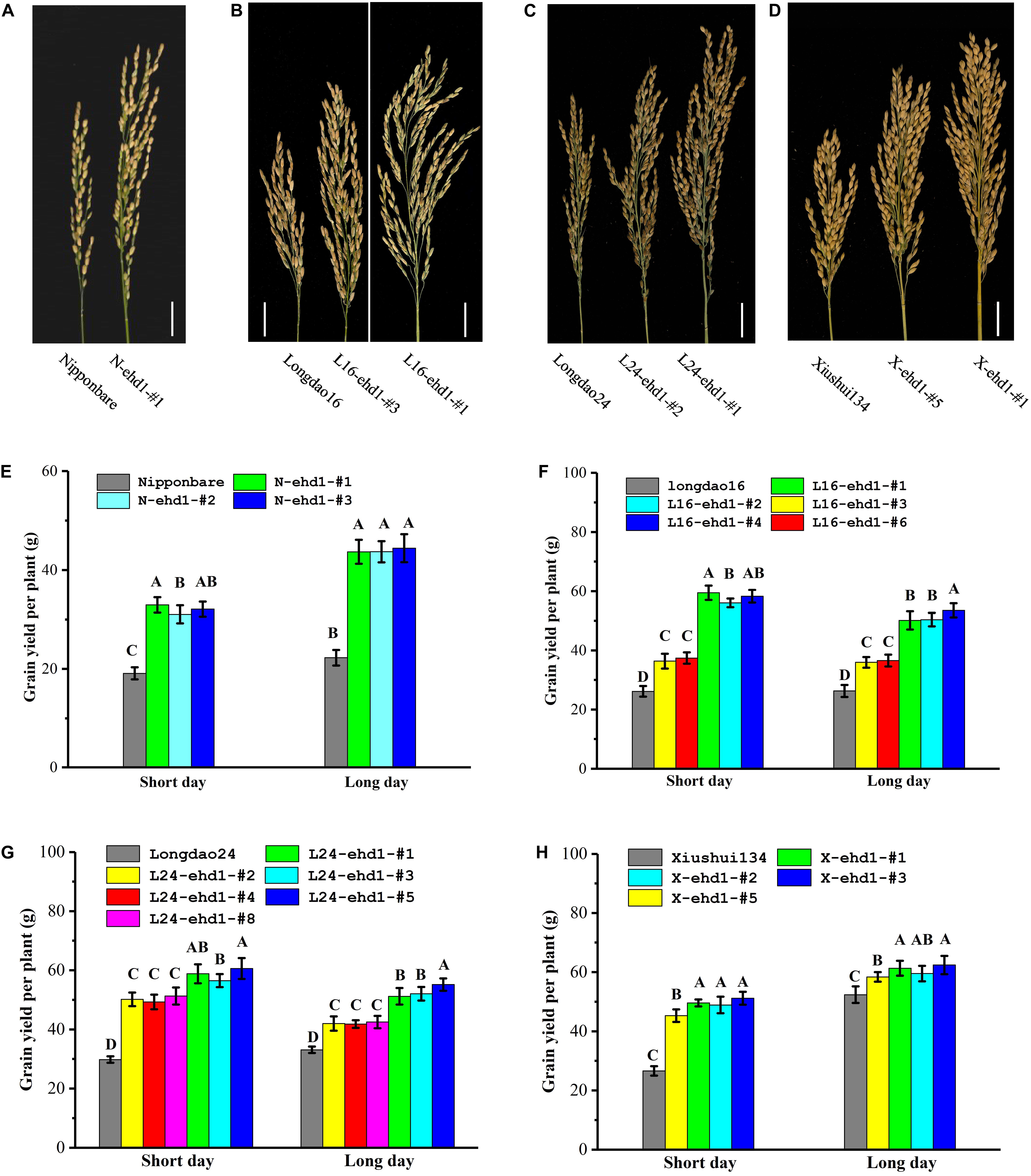
Figure 5. ehd1 mutant lines exhibited high grain yield under both LD and SD conditions. (A–D) ehd1 mutant lines exhibited significantly increased panicle length and grain number per panicle in Nipponbare (A), Longdao16 (B), Longdao24 (C), and Xiushui134 (D) backgrounds, respectively. Pictures were taken at Fuzhou experimental station under long day condition. Scale bar: 2 cm. (E–H) Statistical analysis for grain yield per plant of ehd1 mutant lines in Nipponbare (E), Longdao16 (F), Longdao24 (G), and Xiushui134 (H) backgrounds. Data were collected from 10 plants. The letters A, B, C, and D indicates the significant differences according to LSD multiple range test at P ≤ 0.01.
Rice grains harvested from wild-types and the ehd1 mutant lines cultivated in Fuzhou, and from the two Heilongjiang varieties Longdao16 and Longdao24 cultivated in Haerbing, Heilongjiang, were evaluated for quality characteristics, including head rice rate, chalkiness degree, chalky rice rate, amylose content, gel consistency, and alkali spreading value. The results showed that, Longdao16 and Longdao24 cultivated in Heilongjiang had higher head rice rate, lower chalky rice rate and chalkiness degree compared with Longdao16, Longdao24 and the ehd1 mutant lines cultivated in Fujian. There were no significant differences among the lines for other traits (Supplementary Figure S8). When compared with wild-types cultivated in Fuzhou, the in-frame and frame-shift ehd1 mutant lines showed higher head rice rate, lower chalky rice rate and chalkiness degree (Supplementary Figure S8), which may be due to that the ehd1 mutant lines were exposed to relatively lower temperature during grain-filling stage. There were no significant differences in amylose content, gel consistency and alkali spreading value between the ehd1 mutant lines and their corresponding wild-types (Supplementary Figure S8). Overall, the ehd1 lines exhibited high grain yield and good grain quality when cultivated in low-latitude regions.
Discussion
Rapid Breeding of New Elite Japonica Varieties With Prolonged BVG Periods and High Yield for Low-Latitude Regions Through Genome Editing of the Ehd1 Gene
Previous studies revealed that grain yield of rice is positively correlated with growth period. A longer BVG period could promote the accumulation and allocation of more resources to reproductive organs, resulting in higher yield (Xue et al., 2008; Wei et al., 2010; Hu et al., 2013; Yan et al., 2013; Zhang et al., 2015; Leng et al., 2019). Severe shortening of growth period is considered to be the main obstacle for introducing northern japonica varieties to southern areas in China. Therefore, breeding of japonica varieties with long BVG periods would be of great significance to promote “indica to japonica” projects in southern China (Leng et al., 2019).
A growing number of studies showed that enhanced expression of or mutations in flowering time genes could lead to heading date changes in rice (Xue et al., 2008; Wei et al., 2010; Hu et al., 2013; Koo et al., 2013; Yan et al., 2013; Zhang et al., 2015; Wu et al., 2017; Leng et al., 2019). For example, enhanced expression of Ghd7, an upstream negative regulator of Ehd1, could lead to delayed heading in rice under LD. In contrast, natural mutants with reduced function of Ghd7 exhibited early flowering (Xue et al., 2008). DTH7, DTH8 are two suppressors in the signal network of photoperiodic flowering. Expression of DTH7 or DTH8 led to delayed heading by down-regulating expression of Ehd1 and Hd3a in rice under LD (Wei et al., 2010; Yan et al., 2013; Gao et al., 2014). Natural mutation of DTH7 or DTH8 caused weak photoperiod sensitivity. More recently, a preponderant allele of Heading date 1 (Hd1) has been identified from indica cultivars, and backcrossing the preponderant Hd1 allele led to prolongation of growth period of a japonica variety Chunjiang06 (Leng et al., 2019). These studies also demonstrated that, due to the prolongation of growth period, rice lines with enhanced expression of Ghd7, DTH7, DTH8 or introgression of the preponderant Hd1 allele possessed significantly increased grain yield (Xue et al., 2008; Wei et al., 2010; Yan et al., 2013; Gao et al., 2014; Zhang et al., 2015; Leng et al., 2019).
In this study, we aimed to breed japonica rice with prolonged growth periods under both SD and LD. While many rice flowering time genes, such as Ghd7, DTH7 or DTH8, function only under LD, Ehd1 functions to promote rice flowering under both SD and LD (Doi et al., 2004; Komiya et al., 2008). More recently, Cui et al. (2019) demonstrated that a single−nucleotide insertion in the third exon of the Ehd1 gene resulted in delayed heading date of the japonica rice variety Sasanishiki. In the present study, we chose the first exon of Ehd1 as a target for CRISPR/Cas9-mediated genome editing for regulating heading date in rice. We generated both frame-shift and/or in-frame deletion mutants in four japonica varieties, Nipponbare, Longdao16, Longdao24, and Xiushui134. Similar to previous studies (Li et al., 2016; Zhou et al., 2016; Macovei et al., 2018; Zhang J. et al., 2018), we were able to obtain transgene-free mutant lines by segregating away the integrated T-DNA in T1 generations (Supplementary Figure S2). Field investigation further showed that, in all four japonica variety backgrounds, the flowering time of in-frame and frame-shift ehd1 mutant lines was significantly delayed compared with that of wild-type plants under both SD and LD. In the present study, we also observed that the delay of flowering times of the Xiushui134 mutant lines was not as great as that of the ehd1 mutant lines in Nipponbare, Longdao16, or Longdao24 backgrounds, suggesting that the effects of Ehd1 on heading date varied under different genetic backgrounds.
Accordingly, the ehd1 mutant lines displayed significantly improved yield potential (Figure 5). When planting as middle-season rice in mid-May at Fuzhou station, wild-type Nipponbare, Longdao16, Longdao24, and Xiushui134 flowered in late-July to early-August, and were exposed to high temperatures during grain-filling stage. In contrast, the in-frame and frame-shift ehd1 mutant lines started to flower in late-August to early-September. While high temperature during grain-filling stage has been shown to lead to higher chalk values (Yamakawa et al., 2007), the delay of flowering time allowed the ehd1 mutant lines to be exposed to relatively lower temperatures during grain-filling stage, resulting in higher head rice rate, lower chalky rice rate and chalkiness degree (Supplementary Figure S8). In addition, we did not observe obvious differences in stress tolerance, disease resistance or lodging resistance between the ehd1 mutant lines and their wild-type parents while they were cultivated in the field at Sanya station and Fuzhou station over two generations (data not shown). Taken together, our study demonstrates an effective approach to rapid breeding of elite japonica varieties with prolonged BVG periods and high yield for low-latitude regions through genome editing of Ehd1.
In-Frame Editing of Ehd1 for Developing Rice With Intermediate-Long BVG Periods
Japonica varieties with extremely long BVG periods are expected to be suitable for cultivating in low-latitude regions. In production practice, however, rice varieties with various growth periods are required for cultivation in diverse areas or under different cropping seasons. For example, low-altitude and single-season cropping areas are more likely to grow varieties with long growth periods. In contrast, varieties with intermediate-long growth periods would be more applicable to planting in relatively high altitude areas or under double-season cropping system. In the present study, we found it interesting that the in-frame deletion ehd1 lines in Longdao16, Longdao24, and Xiushui134 backgrounds, displayed a moderately-delayed flowering phenotype. The flowering time of the five in-frame deletion ehd1 lines in Longdao16 and Longdao24 backgrounds was delayed about 3 weeks compared with that of wild-type plants, but was about 2 weeks earlier than that of the frame-shift lines (Figure 5). Similar moderately-delayed flowering phenotypes were observed in the in-frame deletion Xiushui134 mutant line X-ehd1-#5, although the delay of flowering time was less extreme. Ehd1 has a receiver domain and a GARP DNA-binding domain (Doi et al., 2004; Cho et al., 2016). In the receiver domain, the conserved Asp-Asp-Lys (D17th -D63th -K117th) motif, is known to be important for Ehd1 function (Takahashi et al., 2009; Cho et al., 2016). For example, phosphorylation of the middle Asp is required for homodimerization of Ehd1, which is crucial for Ehd1 activity (Cho et al., 2016). In the in-frame deletion ehd1 lines, the editing caused minor deletion mutations of the first few residues of the Ehd1 receiver domain. These residues might not be critical for basic function of Ehd1. However, the moderately-delayed flowering phenotype of the six lines suggested that the minor in-frame mutations could quantitatively impair the biological function of Ehd1. Expression of Hd3a and RFT1 in L16-ehd1-#3 was lower than that in wild-type Longdao16, but higher than that in the frame-shift line L16-ehd1-#2 (Figure 4), further supporting that the in-frame deletion ehd1 allele had a moderate effect on activation of Hd3a and RFT1. Overall, our results showed that the in-frame deletion ehd1 alleles have common effect among different backgrounds. Therefore, the in-frame deletion ehd1 alleles could be valuable gene resources in developing rice varieties with intermediate-long BVG periods.
The majority of important agronomic traits in crops are quantitative traits. Thus, manipulation of quantitative traits has been extremely desired by breeders. Recently, CRISPR/Cas9-based cis-regulatory and upstream open reading frames (uORFs) mutagenesis approaches have been developed to create quantitative variations for crop breeding (Rodríguez-Leal et al., 2017; Zhang H. et al., 2018). Editing of the cis-regulatory elements or uORFs could cause transcriptional change of the targeted genes or the downstream primary ORFs (pORFs), leading to quantitative variation of the traits (Li et al., 2017; Zhang H. et al., 2018). Using the CRISPR/Cas9 system, Rodríguez-Leal et al. (2017) generated a continuum of variants of three major productivity traits (fruit size, inflorescence branching, and plant architecture) in tomato; Zhang H. et al. (2018) created mutant plant lines with varying amounts of mRNA translation in four pORFs. These results demonstrated the great feasibility of the two approaches for engineering quantitative traits for crop improvement. In the present study, we observed that minor in-frame deletion of non-critical residues of the Ehd1 receiver domain could result in quantitatively delayed flowering in rice. Our finding suggested that in-frame editing of non-critical residues could potentially be an alternative approach to creating quantitative variants of important traits. Unlike targeted editing in cis-regulatory regions that may change the expression patterns of the genes and cause unpredictable phenotypic variation (Rodríguez-Leal et al., 2017), in-frame editing does not affect expression pattern of the targeted genes, and may be applicable to traits controlled by specific expression patterns.
In summary, we report the development of japonica rice varieties with prolonged BVG periods and high yield potential by CRISPR/Cas9-mediated editing of Ehd1. Our results showed that, while frame-shift mutations in Ehd1 resulted in more extremely long BVG periods, minor in-frame deletion mutations in the receiver domain region of Ehd1 led to moderate delays of heading date in japonica rice. Field investigation further showed that, both the in-frame and frame-shift lines exhibited significantly improved yield potential compared with wild-types. Therefore, our study demonstrated an effective approach to rapid breeding of elite japonica varieties with intermediate-long and more extremely long BVG periods. The ehd1 mutant lines with the frame-shift or the novel in-frame deletion ehd1 alleles represent valuable resources to develop elite japonica rice varieties for flexible cropping systems in diverse areas or under different seasons in southern China, and other low-latitude regions. In addition, our study suggests that in-frame editing of non-critical residues could potentially be an effective approach for engineering quantitative traits for crop improvement.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
MW, SC, and FW conceived and designed the experiments. MW, HL, YL, JC, YF, JL, and ZZ performed the experiments. MW, KL, SC, and FW analyzed the data. MW, SC, and FW wrote the manuscript. All authors commented on the manuscript.
Funding
This work was supported by grants from the Crop Breeding Major Projects of Fujian Province (2015NZ0002-3), National Major Special Project on New Varieties Cultivation for Transgenic Organisms, China (2016ZX08001001-002), and Technological Innovation Projects of Fujian Academy of Agricultural Sciences (ZYTS2019014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Yin Meng at Tillage and Cultivation Researching Institute, Heilongjiang Academe of Agriculture Sciences for providing japonica varieties Longdao16 and Longdao24, and Prof. Haigen Yao at Jiaxing Academy of Agricultural Sciences for providing japonica variety Xiushui134.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00307/full#supplementary-material
References
Andrés, F., Galbraith, D. W., Talón, M., and Domingo, C. (2009). Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice. Plant Physiol. 151, 681–690. doi: 10.1104/pp.109.139097
Bian, X. F., Liu, X., Zhao, Z. G., Jiang, L., Gao, H., Zhang, Y. H., et al. (2011). Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. by down-regulating Ehd1. Plant Cell Rep. 30, 2243–2254. doi: 10.1007/s00299-011-1129-4
Cho, L. H., Yoon, J., Pasriga, R., and An, G. (2016). Homodimerization of Ehd1 is required to induce flowering in Rice. Plant Physiol. 170, 2159–2171. doi: 10.1104/pp.15.01723
Cui, Y., Zhu, M., Xu, Z., and Xu, Q. (2019). Assessment of the effect of ten heading time genes on reproductive transition and yield components in rice using a CRISPR/Cas9 system. Theor. Appl. Genet. 132, 1887–1896. doi: 10.1007/s00122-019-03324-1
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936. doi: 10.1101/gad.1189604
Fang, C., Hu, X., Sun, C., Duan, B., Xie, L., and Zhou, P. (2015). Simultaneous determination of multi rice quality parameters using image analysis method. Food Anal. Methods 8, 70–78. doi: 10.1007/s12161-014-9870-2
Feng, F., Li, Y., Qin, X., Liao, Y., and Siddique, K. H. M. (2017). Changes in rice grain quality of indica and japonica type varieties released in China from 2000 to 2014. Front. Plant Sci. 8:1863. doi: 10.3389/fpls.2017.01863
Feng, Z., Zhang, L., Yang, C., Wu, T., Lv, J., Chen, Y., et al. (2014). EF8 is involved in photoperiodic flowering pathway and chlorophyll biogenesis in rice. Plant Cell Rep. 33, 2003–2014. doi: 10.1007/s00299-014-1674-8
Gao, H., Jin, M., Zheng, X.-M., Chen, J., Yuan, D., Xin, Y., et al. (2014). Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 16337–16342. doi: 10.1073/pnas.1418204111
Gao, H., Zheng, X. M., Fei, G., Chen, J., Jin, M., Ren, Y., et al. (2013). Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genet. 9:e1003281. doi: 10.1371/journal.pgen.1003281
Hiei, Y., Komari, T., and Kubo, T. (1997). Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 35, 205–218.
Hu, S., Dong, G., Xu, J., Su, Y., Shi, Z., Ye, W., et al. (2013). A point mutation in the zinc finger motif of RID1/EHD2/OsID1 protein leads to outstanding yield-related traits in japonica rice variety Wuyunjing 7. Rice 6, 1–12. doi: 10.1186/1939-8433-6-24
Khanday, I., Skinner, D., Yang, B., Mercier, R., and Sundaresan, V. (2019). A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565, 91–95. doi: 10.1038/s41586-018-0785-8
Kim, S. L., Lee, S., Kim, H. J., Nam, H. G., and An, G. (2007). OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145, 1484–1494.
Komiya, R., Ikegami, A., Tamaki, S., Yokoi, S., and Shimamoto, K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774. doi: 10.1242/dev.008631
Komiya, R., Yokoi, S., and Shimamoto, K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136, 3443–3450. doi: 10.1242/dev.040170
Koo, B. H., Yoo, S. C., Park, J. W., Kwon, C. T., Lee, B. D., An, G., et al. (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 6, 1877–1888. doi: 10.1093/mp/sst088
Lee, S., Kim, J., Han, J. J., Han, M. J., and An, G. (2004). Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 38, 754–764. doi: 10.1111/j.1365-313x.2004.02082.x
Leng, Y., Gao, Y., Chen, L., Yang, Y., Huang, L., Dai, L., et al. (2019). Using Heading date 1 preponderant alleles from indica cultivars to breed high-yield, high-quality japonica rice varieties for cultivation in south China. Plant Biotechnol. J. 18, 119–128. doi: 10.1111/pbi.13177
Leng, Y., Xue, D., Yang, Y., Hu, S., Su, Y., Huang, L., et al. (2013). Mapping of QTLs for eating and cooking quality-related traits in rice (Oryza sativa L.). Euphytica 197, 99–108. doi: 10.1007/s10681-013-1055-3
Li, C., Zong, Y., Wang, Y., Jin, S., Zhang, D., Song, Q., et al. (2018). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19:59. doi: 10.1186/s13059-018-1443-z
Li, Q., Zhang, D., Chen, M., Liang, W., Wei, J., Qi, Y., et al. (2016). Development of japonica photo-sensitive genic male sterile rice lines by editing carbon starved anther using CRISPR/Cas9. J. Genet. Genomics 43, 415–419. doi: 10.1016/j.jgg.2016.04.011
Li, X., Xie, Y., Zhu, Q., and Liu, Y. G. (2017). Targeted genome editing in genes and cis-regulatory regions improves qualitative and quantitative traits in crops. Mol. Plant 10, 1368–1370. doi: 10.1016/j.molp.2017.10.009
Liu, H., Ding, Y., Zhou, Y., Jin, W., Xie, K., and Chen, L. L. (2017). CRISPR-P 2.0: an improved CRISPR/Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532. doi: 10.1016/j.molp.2017.01.003
Lu, Y., Ye, X., Guo, R., Huang, J., Wang, W., Tang, J., et al. (2017). Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant 10, 1242–1245. doi: 10.1016/j.molp.2017.06.007
Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Čermák, T., et al. (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927. doi: 10.1111/pbi.12927
Mao, D., Xin, Y., Tan, Y., Hu, X., Bai, J., Liu, Z. Y., et al. (2019). Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc. Natl. Acad. Sci. U.S.A. 116, 3494–3501. doi: 10.1073/pnas.1819769116
Matsubara, K., Yamanouchi, U., Nonoue, Y., Sugimoto, K., Wang, Z. X., Minobe, Y., et al. (2011). Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J. 66, 603–612. doi: 10.1111/j.1365-313X.2011.04517.x
Matsubara, K., Yamanouchi, U., Wang, Z.-X., Minobe, Y., Izawa, T., and Yano, M. (2008). Ehd2, a rice ortholog of the maize INDETERMINATE1 Gene, promotes flowering by up-regulating Ehd1. Plant Physiol. 148, 1425–1435. doi: 10.1104/pp.108.125542
Miao, C., Xiao, L., Hua, K., Zou, C., Zhao, Y., Bressan, R. A., et al. (2018). Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. U.S.A. 115, 6058–6063. doi: 10.1073/pnas.1804774115
Min, J., Zhu, Z., Jin, L., Xu, L., Zhang, L., and Tang, S. (2011). Analysis on grain quality of indica hybrid rice combinations bred during twenty-five years in China. Chin. J. Rice Sci. 25, 201–205.
Nemoto, Y., Nonoue, Y., Yano, M., and Izawa, T. (2016). Hd1,a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 86, 221–233. doi: 10.1111/tpj.13168
Nishida, H., Inoue, H., Okumoto, Y., and Tanisaka, T. (2002). A novel gene ef1-h conferring an extremely long basic vegetative growth period in rice. Crop Sci. 42, 348–354. doi: 10.2135/cropsci2002.3480
Park, S. J., Kim, S. L., Lee, S., Je, B. I., Piao, H. L., Park, S. H., et al. (2008). Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J. 56, 1018–1029. doi: 10.1111/j.1365-313X.2008.03667.x
Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., and Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480. doi: 10.1016/j.cell.2017.08.030
Saito, H., Yuan, Q., Okumoto, Y., Doi, K., Yoshimura, A., Inoue, H., et al. (2009). Multiple alleles at Early flowering 1 locus making variation in the basic vegetative growth period in rice (Oryza sativa L.). Theor. Appl. Genet. 119, 315–323. doi: 10.1007/s00122-009-1040-3
Shan, Q., Wang, Y., Li, J., and Gao, C. (2014). Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9, 2395–2410. doi: 10.1038/nprot.2014.157
Shrestha, R., Gómez-Ariza, J., Brambilla, V., and Fornara, F. (2014). Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann. Bot. 114, 1445–1458. doi: 10.1093/aob/mcu032
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Sun, C., Fang, J., Zhao, T., Xu, B., Zhang, F., Liu, L., et al. (2012). The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice. Plant Cell 24, 3235–3247. doi: 10.1105/tpc.112.101436
Sun, Y., Zhang, X., Wu, C., He, Y., Ma, Y., Hou, H., et al. (2016). Engineering herbicide-resistant rice plants through CRISPR/Cas9-Mediated homologous recombination of acetolactate synthase. Mol. Plant 9, 628–631. doi: 10.1016/j.molp.2016.01.001
Takahashi, Y., Teshima, K. M., Yokoi, S., Innan, H., and Shimamoto, K. (2009). Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl. Acad. Sci. U.S.A. 106, 4555–4560. doi: 10.1073/pnas.0812092106
Tang, L., Mao, B., Li, Y., Lv, Q., Zhang, L., Chen, C., et al. (2017). Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-14832-9
Wang, C., Liu, Q., Shen, Y., Hua, Y., Wang, J., Lin, J., et al. (2019). Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 37, 283–286. doi: 10.1038/s41587-018-0003-0
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One 11:e0154027. doi: 10.1371/journal.pone.0154027
Wang, Y., Wang, X., Li, Y., Xu, H., Wang, J., Zhao, M., et al. (2015). Analysis of yield and quality traits and their relationship in japonica rice in northern China. Acta Agron. Sin. 41, 84–92. doi: 10.1186/s12915-018-0572-x
Wei, F. J., Tsai, Y. C., Wu, H. P., Huang, L. T., Chen, Y. C., Chen, Y. F., et al. (2016). Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice. Plant Sci. 242, 187–194. doi: 10.1016/j.plantsci.2015.09.005
Wei, X., Jiang, L., Xu, J. F., Liu, X., Liu, S. J., Zhai, H. Q., et al. (2009). The distribution of japonica rice cultivars in the lower region of the yangtze river valley is determined by its photoperiod-sensitivity and heading date genotypes. J. Integr. Plant Biol. 51, 922–932. doi: 10.1111/j.1744-7909.2009.00866.x
Wei, X., Xu, J., Guo, H., Jiang, L., Chen, S., Yu, C., et al. (2010). DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153, 1747–1758. doi: 10.1104/pp.110.156943
Wu, C., You, C., Li, C., Long, T., Chen, G., Byrne, M. E., et al. (2008). RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc. Natl. Acad. Sci. U.S.A. 105, 12915–12920. doi: 10.1073/pnas.0806019105
Wu, W., Zheng, X. M., Chen, D., Zhang, Y., Ma, W., Zhang, H., et al. (2017). OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice. Plant Sci. 260, 60–69. doi: 10.1016/j.plantsci.2017.04.004
Xie, K., Minkenberg, B., and Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U.S.A. 112, 3570–3575. doi: 10.1073/pnas.1420294112
Xu, R., Wei, P., and Yang, J. (2017). Use of CRISPR/Cas genome editing technology for targeted mutagenesis in rice. Methods Mol. Biol. 1498, 33–40. doi: 10.1007/978-1-4939-6472-7_3
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yamakawa, H., Hirose, T., Kuroda, M., and Yamaguchi, T. (2007). Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 144, 258–277. doi: 10.1104/pp.107.098665
Yan, W., Liu, H., Zhou, X., Li, Q., Zhang, J., Lu, L., et al. (2013). Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 23, 969–971. doi: 10.1038/cr.2013.43
Yan, W., Wang, P., Chen, H., Zhou, H., Li, Q., Wang, C., et al. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4, 319–330. doi: 10.1093/mp/ssq070
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484.
Yuan, Q., Saito, H., Okumoto, Y., Inoue, H., Nishida, H., Tsukiyama, T., et al. (2009). Identification of a novel gene ef7 conferring an extremely long basic vegetative growth phase in rice. Theor. Appl. Genet. 119, 675–684. doi: 10.1007/s00122-009-1078-2
Zhang, H., Zhang, J., Gong, J., Chang, Y., Li, M., Gao, H., et al. (2013). The productive advantages and formation mechanisms of “indica rice to japonica rice”. Sci. Agric. Sin. 46, 686–704.
Zhang, J., Zhang, H., Botella, J. R., and Zhu, J. K. (2018). Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J. Integr. Plant Biol. 60, 369–375. doi: 10.1111/jipb.12620
Zhang, H., Si, X., Ji, X., Fan, R., Liu, J., Chen, K., et al. (2018). Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36, 894–898. doi: 10.1038/nbt.4202
Zhang, J., Zhou, X., Yan, W., Zhang, Z., Lu, L., Han, Z., et al. (2015). Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 208, 1056–1066. doi: 10.1111/nph.13538
Zhao, D., Zhang, C., Feng, G., Yang, Q., Gu, M., and Liu, Q. (2013). Hd-q, a novel allele of Ef7 from a Chinese rice landrace, confers weak photoperiod sensitivity and improves local adaptability and yield potential. Mol. Breed. 32, 651–662. doi: 10.1007/s11032-013-9898-9
Zhao, J., Chen, H., Ren, D., Tang, H., Qiu, R., Feng, J., et al. (2015). Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytol. 208, 936–948. doi: 10.1111/nph.13503
Zhou, H., He, M., Li, J., Chen, L., Huang, Z., Zheng, S., et al. (2016). Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci. Rep. 6, 1–12. doi: 10.1038/srep37395
Keywords: japonica rice, basic vegetative growth, heading date, CRISPR/Cas9, Ehd1, in-frame, frame-shift
Citation: Wu M, Liu H, Lin Y, Chen J, Fu Y, Luo J, Zhang Z, Liang K, Chen S and Wang F (2020) In-Frame and Frame-Shift Editing of the Ehd1 Gene to Develop Japonica Rice With Prolonged Basic Vegetative Growth Periods. Front. Plant Sci. 11:307. doi: 10.3389/fpls.2020.00307
Received: 29 September 2019; Accepted: 03 March 2020;
Published: 19 March 2020.
Edited by:
Goetz Hensel, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), GermanyReviewed by:
Yao-Guang Liu, South China Agricultural University, ChinaLing Jiang, Nanjing Agricultural University, China
Copyright © 2020 Wu, Liu, Lin, Chen, Fu, Luo, Zhang, Liang, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kangjing Liang, bGlhbmdral8yMDA1QDEyNi5jb20=; Songbiao Chen, c29uZ2JpYW9fY2hlbkBob3RtYWlsLmNvbQ==; Feng Wang, d2ZAZmphZ2Uub3Jn
 Mingji Wu1,2
Mingji Wu1,2 Huaqing Liu
Huaqing Liu Songbiao Chen
Songbiao Chen