- 1College of Life Sciences, Agriculture and Forestry, Qiqihar University, Qiqihar, China
- 2Heilongjiang Provincial Key Laboratory of Resistance Gene Engineering and Preservation of Biodiversity in Cold Areas, Qiqihar, China
- 3Qiqihar Horticultural Research Institute, Qiqihar, China
- 4College of Horticulture, Landscape Architecture, Northeast Agricultural University, Harbin, China
Cucurbits (Cucurbitaceae family) include many economically important fruit vegetable crops such as watermelon, pumpkin/squash, cucumber, and melon. Seed size (SS) is an important trait in cucurbits breeding, which is controlled by quantitative trait loci (QTL). Recent advances have deciphered several signaling pathways underlying seed size variation in model plants such as Arabidopsis and rice, but little is known on the genetic basis of SS variation in cucurbits. Here we conducted literature review on seed size QTL identified in watermelon, pumpkin/squash, cucumber and melon, and inferred 14, 9 and 13 consensus SS QTL based on their physical positions in respective draft genomes. Among them, four from watermelon (ClSS2.2, ClSS6.1, ClSS6.2, and ClSS8.2), two from cucumber (CsSS4.1 and CsSS5.1), and one from melon (CmSS11.1) were major-effect, stable QTL for seed size and weight. Whole genome sequence alignment revealed that these major-effect QTL were located in syntenic regions across different genomes suggesting possible structural and functional conservation of some important genes for seed size control in cucurbit crops. Annotation of genes in the four watermelon consensus SS QTL regions identified genes that are known to play important roles in seed size control including members of the zinc finger protein and the E3 ubiquitin-protein ligase families. The present work highlights the utility of comparative analysis in understanding the genetic basis of seed size variation, which may help future mapping and cloning of seed size QTL in cucurbits.
Introduction
Seed is the start and end points of plant life, and the important determinants of growth and development. Seed size is a key agronomic characteristic of evolutionary fitness in plants during domestication and breeding in many crops with seeds as the main product organ, and a key factor affecting seed yield, eating quality, tolerances to environmental stresses (Song et al., 2007; Bai et al., 2010; Gu et al., 2017; Smitchger and Weeden, 2018). In general, large seeds have more advantages than small ones in crop production because large seeds have bigger endosperm or cotyledons that can provide more nutrients for seedling establishment (Chen et al., 2006). Large seeds also have faster germination rate than small seeds, resulting in stronger seedlings that can better compete for light and nutrition, and stronger resistance or tolerance to adverse environmental conditions (Silvertown, 1981; Coomes and Grubb, 2003; Gómez, 2004). On the other hand, small seeds may have an advantage in seed transmission, thus affect the reproductive capacity of offspring (Wu et al., 2006).
For many crops, seed size is an important target of selection during domestication (Xia et al., 2018). The seeds of wild plants of modern crops are usually small and roundish in shape, while the domesticated ones in general are much larger. Changes in seed size are the results of natural selection and artificial selection in adaptation to different environments or human needs during domestication. Seed size varies greatly among crop plants (Stanton, 1984; Gómez, 2004; Moles et al., 2005). For crops with seeds as the final target of production (for example, rice, wheat and soybean), seed size is also the determinant of productivity and yield (Fan et al., 2006; Chettoor et al., 2016; Hu et al., 2018). As such, increase of seed size has been one of the primary goals in breeding in cereal crops (Zhang et al., 2016; Savadi, 2018).
Crops in the Cucurbitaceae family, which often referred to as ‘cucurbits,’ include several economically important fruit vegetables worldwide such as watermelon [Citrullus lanatus (Thunb.) Matsum. & Nakai], melon (Cucumis melo L.), cucumber (Cucumis sativus L.), and pumpkin/squash (Cucurbita spp.). Cucurbits are generally prized for their delicious fruits, and the seeds are also good sources of vegetable oil and proteins. For example, the watermelon seeds are rich in oil (Baboli and Kordi, 2010) and protein (Al-Khalifa, 1996; Baboli and Kordi, 2010). However, seed size and weight within cultivated cucurbits also vary considerably depending on their intended uses, which is especially true in watermelon and pumpkin whose seeds have dual uses and seed size is an important target in breeding (Silvertown, 1981; Xiang et al., 2002; Tian et al., 2012). On the other hand, seeds of melon and cucumber do not seem to be under selection in modern breeding, thus which do not seem show as much diversity in seed size and color as those in watermelon and pumpkin/squash (Figure 1). Therefore, the cucurbits may offer a good system to understand the differential roles of artificial selection for seed-related traits during cucurbits breeding.
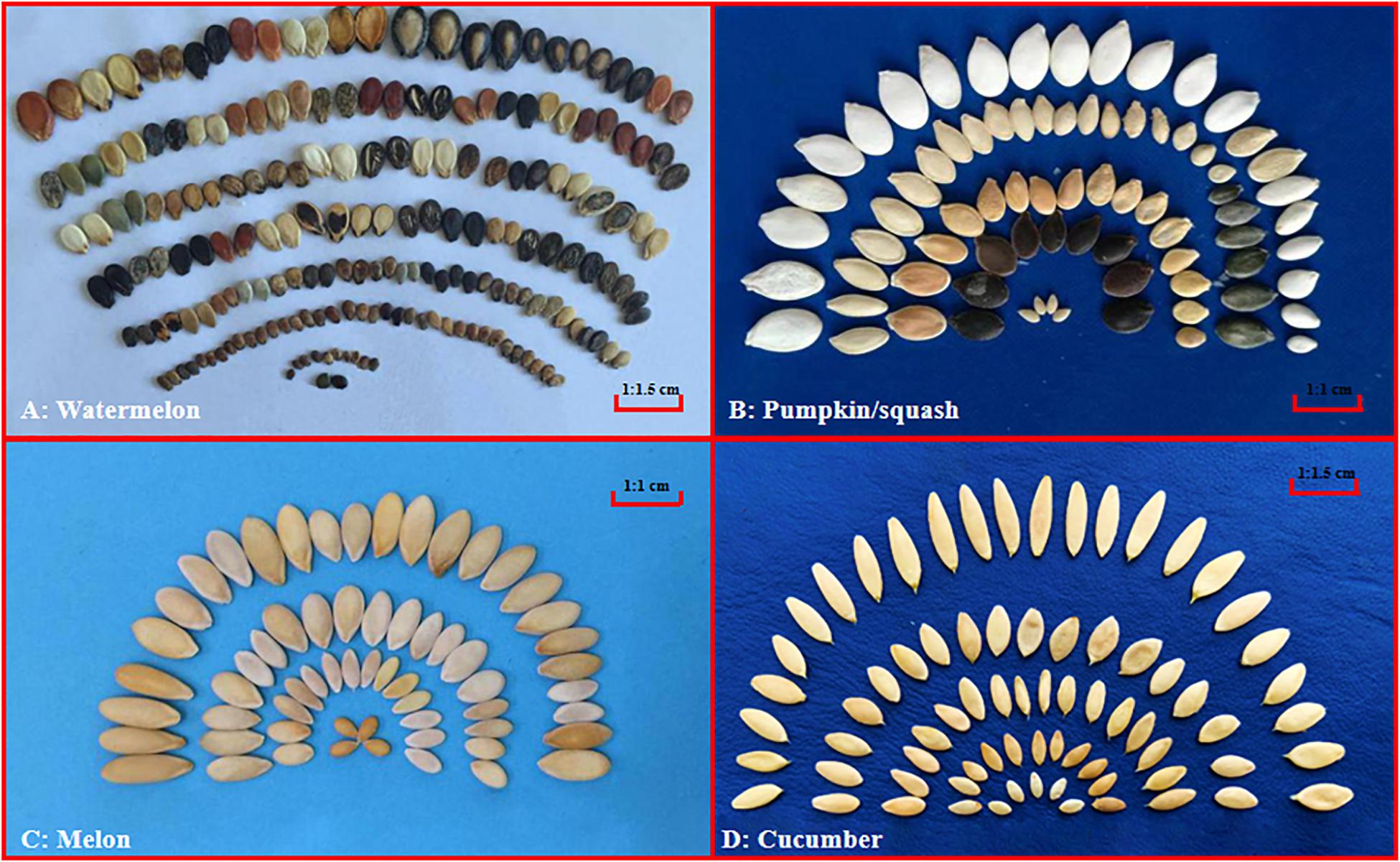
Figure 1. Seed size variation among cucurbit crops. Representative seeds of watermelon (Citrullus lanatus), pumpkin/squash (Cucurbita), melon (Cucumis melo) and cucumber (Cucumis sativus) are shown in (A–D), respectively. All images were taken by the authors.
In cucurbits, seed size and weight are quantitatively inherited in nature, and the genetic basis is still poorly understood. However, in recent years, rapid progress has been made in genome sequencing among cucurbits. So far draft genomes have been developed for cucumber (Huang et al., 2009; Yang L. M. et al., 2012; Li Q. et al., 2019), melon (Garcia-Mas et al., 2012; Ruggieri et al., 2018), watermelon (Guo et al., 2013), bottle gourd, Lagenaria siceraria (Wu et al., 2017), bitter gourd, Momordica charantia (Urasaki et al., 2017), pumpkin/squash (Sun et al., 2017; Montero-Pau et al., 2018), and wax gourd, Benincasa hispida (Thunb.) Cogn. (Xie et al., 2019). These advances have made it possible to understand the genetic architecture of seed size variation in cucurbits through QTL studies and genome-wide association analysis. Thus in this report, we attempt to summarize seed size QTL identified in four major cucurbits (watermelon, pumpkin/squash, cucumber, and melon). From these QTL, we inferred consensus seed size (SS) QTL across different cucurbits. We found structural and functional conservation of consensus SS QTL, as well as unique QTL in each crop. Through comparative analysis across four major cucurbits, we identified potential candidate genes in syntenic regions among several major-effect SS QTL.
Overall View of the Genes and Regulatory Networks Controlling Seed Size Variation in Plants
In angiosperms, a mature seed is composed of three parts: the embryo, the endosperm and the seed coat, all of which contribute to seed size. Seed development involves many complex processes and numerous genes (e.g., Chaudhury et al., 2001; Sun et al., 2010; Figueiredo and Kohler, 2014; Li and Li, 2016). Mutations in many of those genes may result in seed size change. Large amount of work has been done to understand the genetic control of seed size in primarily the model plants such as Arabidopsis thaliana, and rice (Oryza sativa L.). Major genes and regulatory pathways that play important roles in seed size control in plants are summarized in Supplementary Table S1. Major pathways include the ubiquitin pathway (Li et al., 2008; Xia et al., 2013; Li N. et al., 2019), the IKU pathway (Luo et al., 2005; Zhou et al., 2009; Wang et al., 2010), and hormone signaling pathways (Okushima et al., 2005; Du et al., 2017; Hu et al., 2018; Assefa et al., 2019), as well as other regulators (e.g., Huang et al., 2011; Wang S. et al., 2012; Wang et al., 2015; Si et al., 2016).
Several ubiquitin pathway genes have been shown in control of seed size (Li et al., 2008). The ubiquitination process involves ubiquitin activating enzymes (E1s), ubiquitin conjugating enzymes (E2s), and ubiquitin ligases (E3s) (Li et al., 2018b). In Arabidopsis, DA1 encodes a ubiquitin receptor and acts as a negative regulator in seed size and organ development (Li et al., 2008; Dong et al., 2017). DA2 encodes a RING-type E3 ligase with a similar function as DA1 (Xia et al., 2013). The rice GRAIN WIDTH AND WEIGHT2 (GW2) is a homolog of DA2; over-expression of GW2 and DA2 restricts seed/organ size (Song et al., 2007; Xia et al., 2013; Dong et al., 2017). The maize ZmGW2-CHR4 and wheat TaGW2 are homologs of rice GW2, both of which have also been shown to control grain size (Li et al., 2010; Su et al., 2011). In addition, the Brassica napus HECT E3 ligase gene BnaUPL3.C03 regulates seed weight; its down-regulation leads to higher seed weight per pod (Downes et al., 2003; Charlotte et al., 2019).
In Arabidopsis, three genes, HAIKU1 (IKU1), HAIKU2 (IKU2), and MINISEED3 (MINI3) in the same HAIKU (IKU) pathway coordinate to control seed size by influencing endosperm development during early seed growth (Li N. et al., 2019). IKU1 is a VQ motif-containing protein (Wang et al., 2010). IKU2 and MINI3 encode a leucine-rich repeat (LRR) kinase, and a WRKY transcription factor, respectively (Garcia et al., 2003; Luo et al., 2005). The IKU1, IKU2 and MINI3 mutants all show reduced endosperm size, and thus smaller seeds (Garcia et al., 2003; Luo et al., 2005; Wang et al., 2010). In addition, SHORT HYPOCOTYL UNDER BIUE1 (SHB1) is an upstream regulatory factor affecting the expression of MINI3 and IKU2 by interacting with other proteins in the early stage of seed development leading to enlarged seed cavity and improved endosperm growth (Zhou et al., 2009). Over-expression of IKU2 in canola (B. napus) increases seed size and seed yield (Xiao et al., 2016).
Phytohormones, such as brassinosteroids (BRs), gibberellin (GA), auxin and cytokinin (CK) play important roles in seed development which directly control seed size by enhancing cell proliferation, affecting embryo and endosperm development (Jiang et al., 2013; Du et al., 2017; Hu et al., 2018; Assefa et al., 2019; Li Y. D. et al., 2019). Many genes regulate seed development through integration into plant hormone metabolism. For example, ARF2 (AUXIN RESPONSE FACTOR 2) regulates seed size by limiting cell proliferation in the maternal integument (Okushima et al., 2005; Schruff et al., 2005). SRS5 (SMALL AND ROUND SEED), an α-tubulin protein, positively regulates grain length independent of the BR signaling pathway (Segami et al., 2012, 2017). In soybean, over-expression the P450 gene GmCYP78A5 results in increasing seed weight (Du et al., 2017). The rice gene qTGW3 encodes OsSK41 that interacts with OsARF4 (AUXIN RESPONSE FACTOR 4); loss-of-function mutant of OsSK41 has increased grain length and weight (Hu et al., 2018). The grape gene VvHB58 has a potential function in regulating seed number and development by impacting auxin, gibberellin, and ethylene signaling pathways (Li Y. D. et al., 2019). In soybean, Glyma.02g04660 encodes a histidine phosphotransfer protein ortholog that controls seed width through the cytokinin-mediated regulating pathway (Assefa et al., 2019). In B. nupus, BnaA9.CYP78A9 positively regulates seed length by affecting auxin metabolism (Shi et al., 2019).
Many transcription factors have been shown to play important roles in regulating seed development. For example, APETALA2 (AP2), a member of the AP2/EREBP transcription factor gene family, regulates cell expansion in the maternal integument of seed development (Jofuku et al., 2005; Zhang et al., 2016). AINTEGUMENTA (ANT) is another type of AP2 transcription factor affecting the seed size by regulating the cell division of integument during ovule development (Mizukami and Fischer, 2000). TRANSPARENT TESTA GLABRA2 (TTG2) encodes a WRKY transcription factor that is mainly expressed in the endosperm and integument with a similar function as AP2: it may also affect the seed size by directly regulating integument cell elongation (Johnson et al., 2002; Garcia et al., 2005). The MADS-box genes have been found to control seed size in rice. For example, OsMADS29 regulates development of the integument or seed coat, and OsMADS87 may affect seed size in endosperm cellularization period (Prasad et al., 2010; Yang X. et al., 2012; Chen et al., 2016). The WG7 (WIDE GRAIN 7) up-regulates OsMADS1 expression by directly binding to its promoter, and enhancing histone H3K4me3 enrichment in the promoter and ultimately increases grain width in rice (Huang et al., 2019). The SlPRE2 encodes a bHLH family transcription factor, which mediates plant response to gibberellin; silencing of SlPRE2 decreases tomato seed size by restricting cell expansion (Zhu et al., 2019).
Many other genes or QTL not belonging to above-mentioned categories have been cloned that affect seed/grain size. For example, the rice gene Os02g0192300 for a zinc finger protein seems to control grain weight (Huang et al., 2011). The rice grain width QTL GW8 encoding OsSPL16 increases grain width by promoting cell proliferation and grain filling (Wang S. et al., 2012). GLW7 (GRAIN LENGTH AND WEIGHT ON CHROMOSOME 7) also encoding a SPL protein (OsSPL13) positively regulates grain size by increasing spikelet hull cell expansion (Si et al., 2016). The rice GW7 (GRAIN WIDTH7) controls grain size and shape through modulating cell proliferation (Wang et al., 2015). The rice GS3 (GRAIN SIZE 3) also could regulate grain size and weight (Li et al., 2016; Zeng et al., 2020). In Arabidopsis, the cyclin gene CYCB1;4 controls the final seed size by regulating the cell cycles in maternal tissues and zygotic tissues (Ren et al., 2019).
QTL for Seed Size (SS) Variation in Major Cucurbit Crops
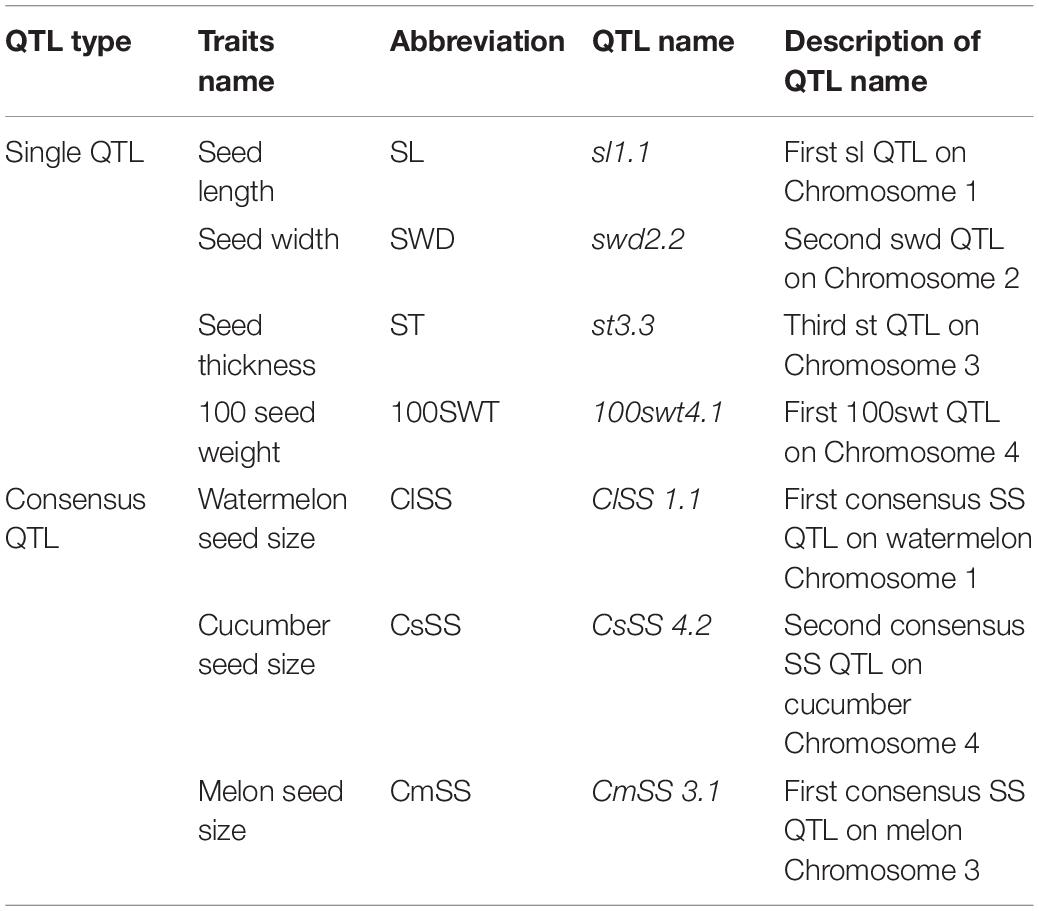
There is rich genetic diversity in seed size among major cucurbits (Figure 1), which was the result of selection during domestication or diversifying selection in breeding to adapt to crop production for various purposes (Tan et al., 2013; Wang et al., 2014; Li et al., 2018a; Zhang et al., 2018). In recent years, a number of seed size QTL mapping studies have been conducted in watermelon, pumpkin/squash, cucumber and melon QTL, which allows a comparative analysis of the genetic basis of seed size variation across different cucurbits. However, the names of the QTL are confusing in the literature. For convenience, following recommendations for QTL naming by Pan et al. (2020) and Wang et al. (2020), here we developed a QTL nomenclature for naming QTL for seed-related traits, which are presented in Table 1. In brief, SL, SWD, ST, and 100SWT stood for the seed length, seed width, seed thickness and 100-seed weight, respectively. The QTL was named according to Pan et al. (2020) which included information for its location and relative order on chromosome (e.g., sl1.1 presents the first QTL on chromosome 1). In each crop, the same QTL could be identified in different studies with different genetic backgrounds or environments. By examining their physical locations and LOD support intervals, it is possible to establish a consensus SS QTL for these related traits (Pan et al., 2020). Thus, a consensus SS QTL will be defined based on individual SL, SWD, ST, or 100SWT QTL.
Seed Size QTL in Watermelon
Watermelon could be divided into two types based on its uses: use of the edible flesh and use of seeds. For seed-use watermelons, large seeds are preferred because they are rich in oils and proteins, and easy to crack open (Al-Khalifa, 1996; Baboli and Kordi, 2010; Prothro et al., 2012b), while for edible flesh watermelons, small and few (or no) seeds could increase the pulp proportion and improve product quality. Thus, seed size is a target of selection in watermelon breeding, but the breeding direction of selection depends on its end use.
There is a wide range of variation in seed size among watermelon collections (e.g., Figure 1A). The watermelon seed could be classified into six representative types based on the size: giant, big, medium, small, tiny, and tomato seed (Yongjae et al., 2009). Poole et al. (1941) were pioneers who paid attention to the inheritance of seed size. They found that the length of large and medium-sized seeds was controlled by two genes (l and s), and that small seeds were dominant to medium seed size. Later, two gene, Ti for tiny seed and ts for tomato seed were also reported (Zhang et al., 1994; Tanaka et al., 1995; Zhang, 1996). Zhang and Zhang (2011) found that a pair of major genes and a pair of recessive genes determine seed size, but additional modifiers are possible. Kim et al. (2015) considered watermelon seed size as a quality trait that was controlled by a single dominant gene. Gao et al. (2016) carried out genetic analysis on seed size-related traits, and found that seed length, width and 1000-seed weight were quantitatively inherited that show continuous variation in segregating populations, and they may be controlled by major QTL. The quantitative nature of seed size variation in watermelon was further observed in other studies (e.g., Prothro et al., 2012a; Zhou et al., 2016). Prothro et al. (2012a) were probably the first to conduct QTL analysis for seed size in watermelon, but pre-draft genome studies were sporadic (Meru and McGregor, 2013). Using a recombinant inbred line (RIL) population from KBS (medium seed, cultivated) × NHM (medium seed, cultivated) and an F2 population from ZWRM (small seed, cultivated) × PI244019 (medium seed, Citroides), Prothro et al. (2012a) conducted QTL mapping on seed size related traits and identified 13 QTL on four linkage groups (LG2, LG4, LG9 and LG11). In both populations, three major QTL with phenotypic variance explained (PVE) being from 26.9 to 73.6% for SL, SWD and 100SWT, were identified at the same region on Chr6 (LG2). In addition, they also detected a major-effect QTL (PVE = 25.6%) for SWD on Chr2 (LG9). Using an F2 population from the normal type (PI 279261) × egusi type (PI 560023) cross, Meru and McGregor (2013) detected 3 major-effect QTL (PVE = 34.4–60.8%) on Chr6, which seem to be the same chromosomal region as identified by Prothro et al. (2012a).
The release of the watermelon draft genome (Guo et al., 2013) greatly facilitated QTL mapping studies. Ren et al. (2014) verified the major QTL on Chr2 and Chr6 reported by Prothro et al. (2012a). Several studies all detected SS-related QTL in the same Chr6 region (Sandlin et al., 2012; Prothro et al., 2012a, b; Ren et al., 2012). Meanwhile, using an F2 population developed from the cross between wild egusi type (PI 186490) and cultivated type (LSW-177) watermelons, Liu et al. (2014) identified 1 SL QTL (QSL) and 1 SWD QTL (QSWD1) on Chr6 (PVE = 18.8 and 15.7%), respectively. With the same population, Zhou et al. (2016) mapped more SS QTL (PVE = 3.2–28.9%) on Chr6 including 3 SL, 4 SWD, 1 ST, and 2 100SWT QTL. Kim et al. (2015) detected a 20-seed-weight QTL on Chr2 with an F2 population from the cross between the open-pollinated cultivar ‘Arka Manik’ and Jubilee-type inbred line ‘TS34.’ In our recent study, we identified a major-effect QTL, SS6.1 for seed size on Chr6 using an F2 population developed from the cross between a small size line and a medium size line. The PVE for QTL of SL, SWD, and 100SWT in this population was as high as 48.5, 42.2, and 45.3%, respectively. Interestingly, in an F2 population from the cross between two watermelon inbred lines with medium size seeds, we detected a major-effect QTL (PVE = 35.5–50.1%) for SL, SWD, and 20SWT (20 seed weight) on Chr2 (Gao et al. unpublished data).
Li et al. (2018a) conducted fine mapping of QTL for seed-related traits in watermelon, and identified a major-effect QTL qSS6 for seed size. The PVE of the QTL for SL, SWD, and 1000SWT (1000-seed weight) in this population were as high as 94.1, 95.3, and 93.0%, respectively. They further identified a region of qSS6 harboring three candidate genes including Cla009291, Cla009301, and Cla009310. Among them, Cla009291 encodes the MDR protein mdtK, which is differentially expressed at different seed developmental stages between large- and small-seeded lines. Cla009301 is a homolog of SRS3 (SMALL AND ROUND SEED) for a BY-kinesin-like protein 10 that is a seed size regulator in rice (Kanako et al., 2010). Cla009310 encodes an unknown protein that was proposed to be a candidate for qSS6 based on an SNP in the first exon between the two parental lines (Li et al., 2018a).
To summarize, so far 20 SL, 19 SWD, 3 ST, and 19 100SWT QTL (total 61) have been identified in watermelon. The details of these QTL are presented in Supplementary Table S2. Additional information for each QTL is also provided including flanking markers with sequences, their chromosomal positions, mapping populations and PVE are presented in Supplementary Table S2. Based on their chromosomal locations and LOD support intervals, 14 consensus SS QTL were inferred from different studies. The information of all 14 consensus SS QTL including independent QTL from each study is listed in Supplementary Table S3, their chromosomal positions are visually illustrated in Figure 2. The 14 consensus SS QTL were distributed on 7 of the 11 watermelon chromosomes. Of them, 4 QTL (ClSS2.2, ClSS6.1, ClSS6.2, and ClSS8.2) could be detected in at least two populations/studies. ClSS2.2 was only detected by the populations constructed from two cultivated lines, but ClSS6.1 and ClSS6.2 could be detected in populations derived from cultivated and wild watermelon lines. Especially, ClSS6.1 and ClSS6.2 were identified in most studies with F2 and RILs populations derived from different seed size materials, suggesting that this QTL may play the most important role in seed size/weight determination. Meanwhile, some ‘consensus’ QTL such as ClSS1.1 and ClSS11.1 were only detected in a single population/study (e.g., Zhou et al., 2016), which should be considered putative pending validation in future studies.
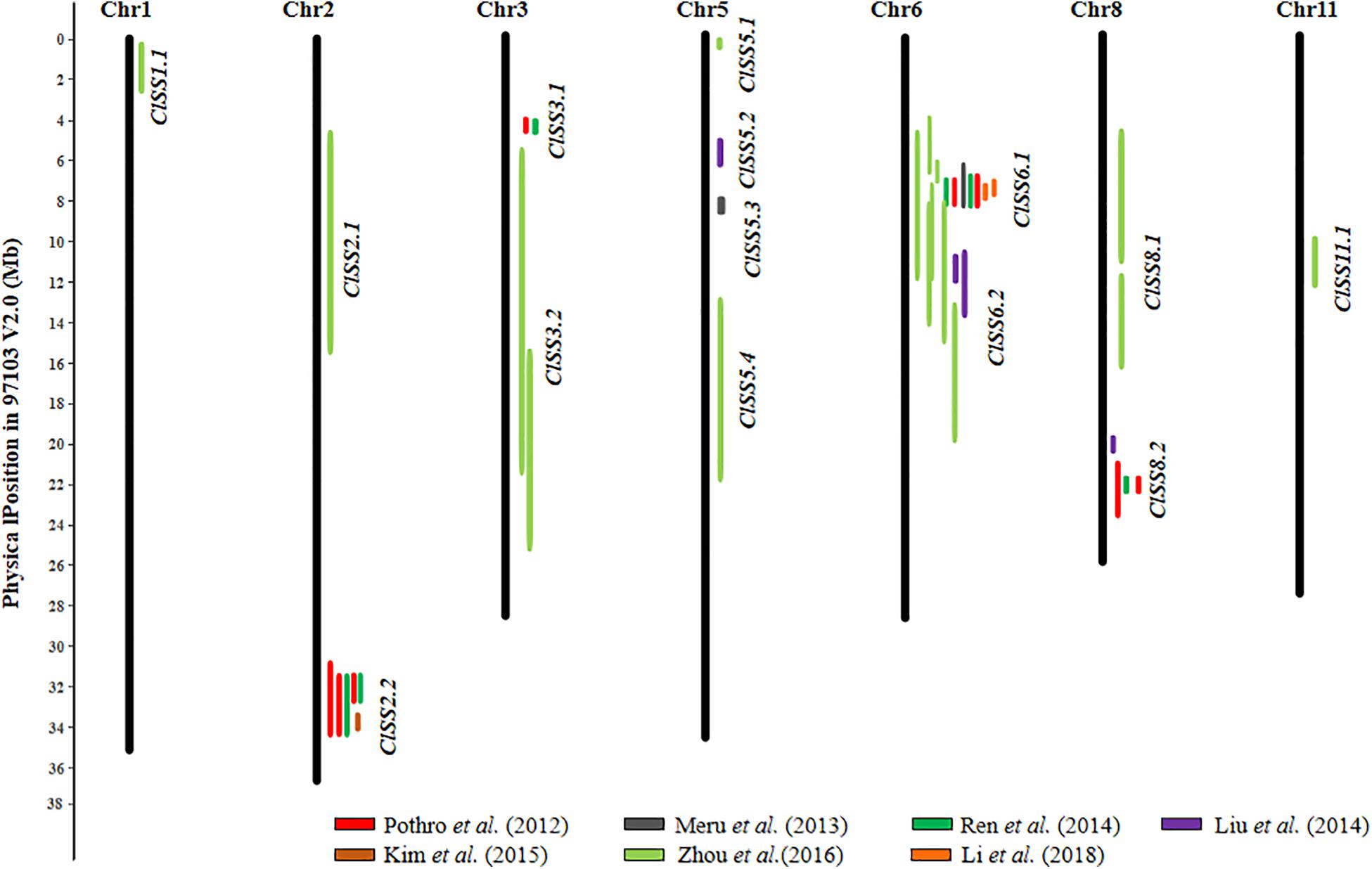
Figure 2. Distribution of 14 seed size (SS) consensus QTL in the watermelon (97103 V2.0) draft genome. Physical LOD intervals of QTL identified from different studies are presented to the right of each chromosome. QTL detected by the same study are showed with same color. Only chromosomes harboring seed size QTL are shown.
Seed Size QTL in Pumpkin/Squash
Pumpkin/squash (Cucurbit spp.) are important cucurbit crops cultivated worldwide. The seed size of seed-use pumpkin affects not only seed yield, but may also impact fruit quality. For example, Paris and Nerson (2003) found that pumpkin SL and SWD were positively correlated with the fruit size, while the seed shape was negatively correlated with the fruit shape. There was larger heterosis of SWD than that of SL (Xu et al., 2006). Seed number per fruit and 100SWT directly affect the yield of seed-use pumpkin (Wang P. et al., 2012). Meru et al. (2018) found that seed size was positively correlated with oil content, but negatively with seed protein in pumpkin. However, little is known about the genetic basis of seed size variation in pumpkin/squash. So far, relatively few QTL have been identified underlying seed size variation in pumpkin/squash. Tan et al. (2013) developed an F2 population using the Indian large-grain ‘0515-1’ and the small-grain ‘0460-1-1’ pumpkin lines; QTL analysis detected 4 major-effect QTL controlling SWD on LG2, LG3 and LG4 with PVE from 2.9 to 29.7%. In C. maxima, Wang et al. (2019) developed a high-density genetic map with the F2 population from the cross between two squash inbred lines (‘2013-12’ and ‘9-6’). In QTL mapping, they identified 10 QTL on six chromosomes (Chr) (Chr4, 5, 6, 8, 17, and 18), including 4 for SL, 4 for SW, and 2 for HSW (100-hundred-seed weight) with the PVE ranging from 7.0 to 38.6%. The major-effect QTL SL6-1 was located on Chr6, and explained 38.6% of observed phenotypic variance. Another major-effect QTL, SW6-1 was also detected on Chr6 with the PVE of 28.9%. The details of these QTL are presented in Supplementary Table S4. Since the limited number of seed size-related QTL were detected, no consensus QTL could be inferred in pumpkin/squash.
Seed Size QTL in Cucumber
A few studies have been conducted in cucumber for QTL mapping of seed size-related traits. Chen et al. (2012) suggested that SL in cucumber is a quantitative trait controlled by multiple genes. With a RIL population developed from the cross between two cultivated cucumber lines: the large seeded ‘D06157’ and small seeded ‘D0603,’ they detected 6 SL QTL. In the RIL population derived from PI 183967 (wild) × 931 (cultivated), Wang et al. (2014) detected 14 SS QTL on five chromosomes (Chr2, 3, 4, 5, and 6) with mainly additive effect including 6 for SL (PVE = 7.5–15.6%), 4 for SWD (PVE = 7.7–18.8%), and 4 for 100SWT (PVE = 11.0–28.3%). However, only SL5.1, SL6.1, and 100SWT6.1 could be detected in two seasons. In another study, using an F2:3 population derived from the cross between two US processing cucumber inbred lines ‘2A’ and ‘Gy8,’ Lietzow (2015) identified 8 QTL for SS and 50SWT (50 seed weight) with two major QTL (PVE = 11.9% for both) located on Chr4.
From the 28 QTL identified in cucumber (details in Supplementary Table S5), 9 consensus SS QTL could be established, which are listed in Supplementary Table S6. Their chromosomal locations are graphically presented in Figure 3. Five of the 9 consensus SS QTL, CsSS2.2, CsSS3.2, CsSS4.1, CsSS5.1, and CsSS6.1 were identified with populations derived from crosses between the wild and cultivated cucumbers, of which CsSS4.1 and CsSS5.1 were detected in all studies with segregating populations created by parental materials with different seed sizes and origins. Therefore, CsSS4.1 and CsSS5.1 may be the potential target regions for selection during both domestication and diversifying selection.
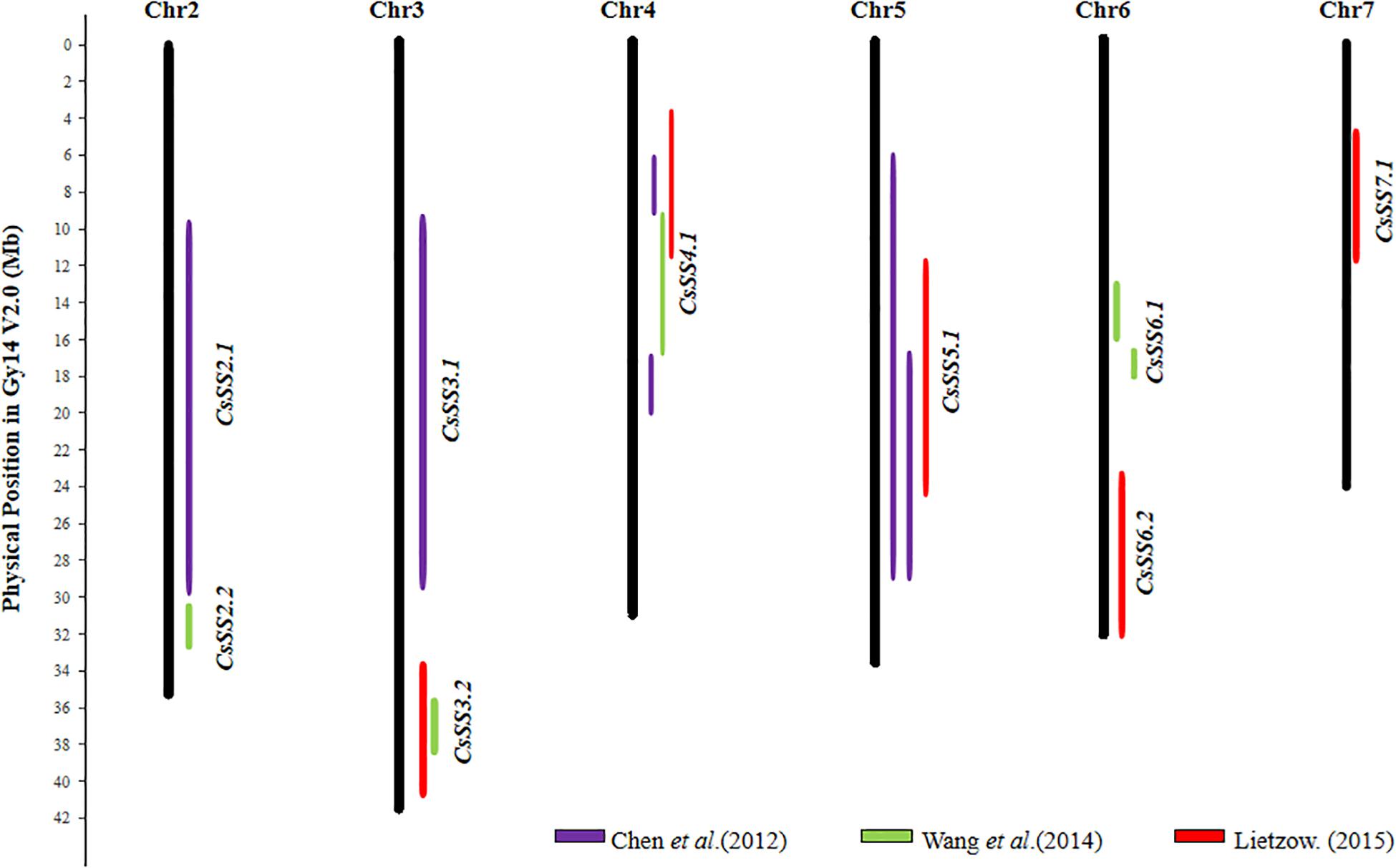
Figure 3. Distribution of 9 seed size (SS) consensus QTL in the cucumber (Gy14 V2.0) draft genome. Physical LOD intervals of QTL identified from different studies are presented to the right of each chromosome. QTL detected by the same study are showed with same color. Only chromosomes harboring seed size QTL are shown.
Seed Size QTL in Melon
Five studies conducted QTL mapping for seed size in melon. Jiao (2017) conducted QTL analysis for seed size with an F2:3 population derived from the cross of large seeded ‘ms-5’ and small seeded ‘HM-1.’ 7 QTL for SL, SWD, and 100SWT were identified on four chromosomes (Chr5, 6, 9, and 11). The 2 major-effect QTL, Sl11.1 and SW11.1 for SL and SWD were detected in the same region on Chr11 with PVE of 17.5 and 19.5%, respectively. With a BC1 population from ‘MR-1’ (long seed) × ‘M1-15’ (short/narrow seed), Ye et al. (2017) detected 4 QTL, including 2 for SL, 1 for SWD and 1 for 100SWT. The 2 QTL (SL5.1 and SD5.1) on Chr5 (PVE = 9.9–11.2%), and 2 QTL (SL1.1 and TGW1.2) on Chr1 were validated. In addition, using a BC1 population, Ning et al. (2018) also detected QTL for SL and 100SWT on Chr1 (LG1), but not in the same region reported by Ye et al. (2017). With a high-density genetic linkage map constructed by a RIL population, Pereira et al. (2018) identified 4 QTL for SWD with 1 major QTL (SWQU8.1) located on Chr8 with PVE of 19.9%. Zhang et al. (2018) mapped 6 QTL on SL, SWD and 100SWT in a F2:3 population from the cross between cantaloupe line ‘ms-5’ with large seed and muskmelon line ‘HM1-1’ with small seed, the major QTL (sl11.1 and sd2.1) for SL and SWD were identified on Chr11 and Chr2 (PVE = 17.5 and 18.1%), respectively. Three QTL (sl11.1, sl11.2, and 100swt11.1) for SL and 100SWT were detected in the same Chr11 region.
From 23 QTL detected in the six studies (Supplementary Table S7), 13 consensus SS QTL could be inferred, and the details are provided in Supplementary Table S8, and their chromosomal locations are illustrated in Figure 4. Both Jiao (2017) and Zhang et al. (2018) detected the QTL for SL, SWD, and 100SWT in the same region on Chr11, and other QTL were not detected reproducibly. Therefore, we speculated that the Cm11.1 may play a major role in regulating seed size/weight variation in melon.
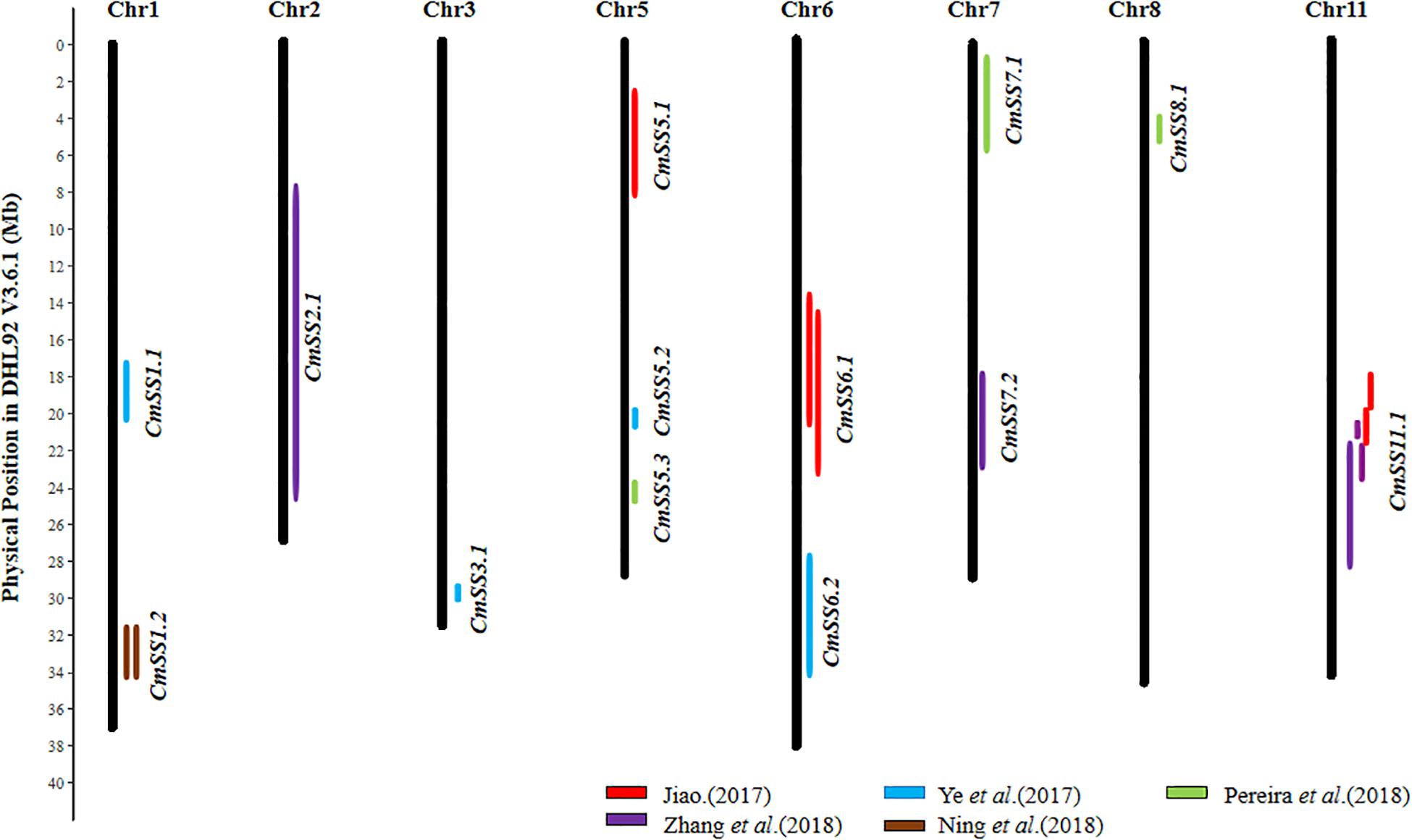
Figure 4. Distribution of 13 seed size (SS) consensus QTL in the melon (DHL92 V3.6.1) draft genome. Physical LOD intervals of QTL identified from different studies are presented to the right of each chromosome. QTL detected by the same study are showed with same color. Only chromosomes harboring seed size QTL are shown.
Structure and Function Conservation of SS QTL in Major Cucurbits
Previous studies have revealed high degree of sequence conservation and synteny across different cucurbit genomes, and the syntenic relationships among different chromosomal regions have also been well-established (e.g., Li et al., 2011; Garcia-Mas et al., 2012; Guo et al., 2013; Yang et al., 2014; Zhu et al., 2016; Pan et al., 2020). Comparison of consensus SS QTL locations identified from the present study may help reveal possible conservation of the genetic basis of seed size variation across major cucurbit crops. In total, 14, 9, and 13 consensus SS QTL were established based on their physical positions in the watermelon, cucumber, and melon draft genomes, respectively (Figures 2–4). Among the four major cucurbits, watermelon has the most extensive studies on QTL mapping of seed size related traits (Figure 2 and Supplementary Tables S2, S3). Four watermelon SS QTL have been repeatedly detected in multiple populations and environments, including ClSS2.2, ClSS6.1, ClSS6.2, and ClSS8.2 (Figure 2). To further investigate the syntenic relationships of these major-effect, highly stable SS consensus QTL with those in other cucurbits, we aligned watermelon chromosomes 2 (W2), 6 (W6) and 8 (W8) with cucumber, melon and pumpkin genomes. We looked into the syntenic relationships of the three watermelon chromosomes (W2, W6, and W8) in cucumber, melon and pumpkin/squash. We searched the syntenic blocks based on the physical positions of these consensus QTL in the cucurbits draft genomes, which could be conveniently conducted through the cucurbit genome database at http://cucurbitgenomics.org/. The syntenic relationships of the three watermelon chromosomes (W2, W6, and W8) in cucumber, melon and pumpkin/squash were calculated by the One Step MCScanx function, and the syntenic blocks were drawn by the visualization tools Dual Systeny Pltter function in TBtools software. The syntenic relationships of the three watermelon chromosomes with those in other three cucurbits are illustrated in Figure 5. The syntenic blocks harboring the four watermelon SS QTL and those in melon, cucumber, and pumpkin are highlighted in red. From Figure 5, it is easy to see that the chromosomal region harboring watermelon ClSS2.2 (W2) was syntenic to the regions where the cucumber CsSS4.1 (C4) and CsSS5.1 (C5), melon CmSS11.1 (M11) and pumpkin SL6-1, SW6-1 (P6), were respectively located. CmSS11.1 (M11) and CsSS4.1 (C4) were also in syntenic regions. Similarly, ClSS6.1 (W6) was syntenic to the regions CmSS11.1 (M11) in melon and pumpkin SL17-1, 100SWT17-1 (P17). CmSS1.2 (M1) and the regions harboring both ClSS6.1 (W6) and ClSS8.2 (W8) were syntenic. These observations suggested possible conservation of functions and structure genes/QTL for seed size control among cucurbit crops. Such conservation could also be possible for QTL in other regions. However, more QTL mapping work is needed to make such inferences.
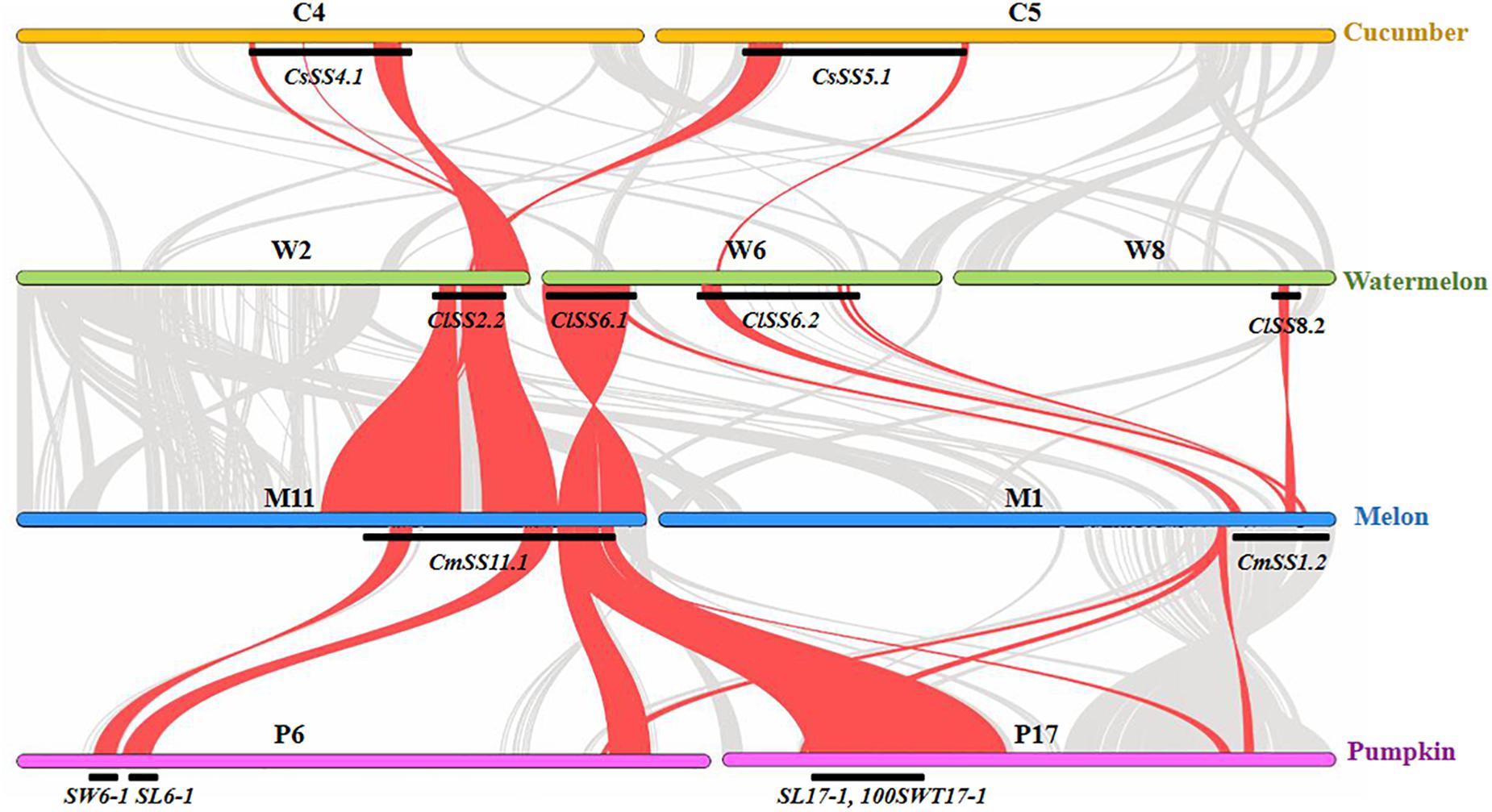
Figure 5. Syntenic relationships of watermelon chromosomes W2, W6, and W8 harboring major-effect consensus SS QTL with cucumber, melon and pumpkin chromosomes. The inference is based on watermelon “97103 V2.0,” cucumber “Gy14 V2.0,” melon “DHL92 V3.6.1,” and pumpkin “Rimu” draft genomes.
The molecular mechanisms of seed size regulation in cucurbits are unknown. The functions of many genes for organ development are high conserved (for example, fruit size and shape as reviewed in Pan et al., 2020). From QTL mapping studies, the four watermelon consensus SS QTL (ClSS2.2, ClSS6.1, ClSS6.2, and ClSS8.2) have been delimited to relatively small regions. We examined the annotated genes in the four regions from the watermelon draft genomes. In the watermelon 97103 V2.0 draft genome, 21, 11, 6, and 34 genes were predicted in the ClSS2.2, ClSS6.1, ClSS6.2 and ClSS8.2 consensus QTL regions, respectively, which are listed in Supplementary Table S9. Among those genes, several seem to be good candidates for these SS QTL. As discussed early (see section “Introduction,” Supplementary Table S1), the ubiquitination pathway may play important roles in control of seed size (e.g., Song et al., 2007; Li et al., 2010; Su et al., 2011; Xia et al., 2013; Wang et al., 2016; Dong et al., 2017). From our recent study in pumpkin, the E3 ubiquitin-protein ligase gene was located in the SL6-1 and SW6-1 major-effect QTL region (Wang et al., 2019). In rice, a gene encoding a zinc finger protein has been verified in control grain weight (Huang et al., 2011). In the watermelon SS QTL regions, there were six genes with predicted functions to encode zinc finger proteins including Cla97C02G045430, Cla97C02G045460 and Cla97C02G045500 in the ClSS2.2 region, Cla97C06G114460 in ClSS6.1 region, and Cla97C08G154340 and Cla97C08G154360 in ClSS8.2 region. Another gene, Cla97C08G154570 encoding E3 ubiquitin-protein ligase was located in ClSS8.2 region (Supplementary Table S9). These genes might be interesting candidates for the seed size QTL that merit consideration in future studies.
Conclusion Remarks
In this study, we summarized QTL for seed size-related traits in major cucurbits, and identified consensus SS QTL in watermelon, melon and cucumber. Many of these SS QTL were located in non-syntenic regions in the three cucurbit crops. In watermelon and melon, it seems that several stable consensus QTL are located in syntenic blocks in different cucurbit crops (Figure 5), which may suggest shared common QTL for seed size control in some genetic backgrounds. It should be noted that QTL mapping studies for seed size/weight are very limited in major cucurbit crops (none in minor cucurbits). Many of these consensus QTL were inferred from single study. The genomic regions harboring these QTL are still very large. The effects of these QTL need to be confirmed, and their locations need to be refined in future studies. Seed size/weight was the target of selection during domestication from wild ancestors, which usually have small seeds. In seed-use cucurbits like watermelon and pumpkin, seed size may be under further selection during breeding, while in melon and cucumber where flesh (endocarp) is consumed, seed size may not be a main target in breeding. Understanding the genetic basis and the roles of selection in diversifying and domestication shaping seed size variation might be some interesting topics in future studies. Such information is also important in marker-assisted selection in breeding for seed size in cucurbit crops.
Author Contributions
YGu and MG conducted literature review and wrote the manuscript. XXL, MX, and XSL performed the comparative analysis. FL and SQ helped review of watermelon, melon, and pumpkin data. YZ, JL, XJL, and YGa analyzed and reviewed cucumber and melon data. All authors read and approved the final submission.
Funding
This work was supported by the National Natural Science Foundation of China (Project ID: 31772334, 31972437, and 31401891), the Natural Science Foundation of Heilongjiang Province (Project ID: LC2018015), and the Fundamental Research Funds in Heilongjiang Provincial Universities (Project ID: 135209106) to MG.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00304/full#supplementary-material
References
Al-Khalifa, A. S. (1996). Physicochemical characteristics, fatty acid composition, and lipoxygenase activity of crude pumpkin and melon seed oils. J. Agric. Food Chem. 44, 964–966. doi: 10.1021/jf950519s
Assefa, T., Otyama, P. I., Brown, A. V., Kalberer, S. R., Kulkarni, R. S., and Cannon, S. B. (2019). Genome-wide associations and epistatic interactions for internode number, plant height, seed weight and seed yield in soybean. BMC Genomics 20:527. doi: 10.1186/s12864-019-5907-7
Baboli, Z. M., and Kordi, A. A. S. (2010). Characteristics and composition of watermelon seed oil and solvent extraction parameters effects. J. Am. Oil Chem. Soc. 87, 667–671. doi: 10.1007/s11746-010-1546-5
Bai, X., Luo, L., Yan, W., Kovi, M. R., and Xing, Y. (2010). Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet. 11:16. doi: 10.1186/1471-2156-11-16
Charlotte, M., Rachel, W., Neil, M., Martin, T., Joshua, B., Abdelhak, F., et al. (2019). Variation in expression of the HECT E3 ligase UPL3 modulates LEC2 levels, seed size, and crop yields in Brassica napus. Plant Cell 31, 2370–2385. doi: 10.1101/334581
Chaudhury, A. M., Koltunow, A., Payne, T., Luo, M., Tucker, M. R., Dennis, E. S., et al. (2001). Control of early seed development. Annu. Rev. Cell Dev. Biol. 17, 677–699. doi: 10.1146/annurev.cellbio.17.1.677
Chen, C., Begcy, K., Liu, K., Folsom, J. J., Wang, Z., Zhang, C., et al. (2016). Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity. Plant Physiol. 171, 606–622. doi: 10.1104/pp.15.01992
Chen, L. L., Qin, Z. W., Zhou, X. Y., Xin, M., and Wu, T. (2012). Molecular marker and genetic analysis of cucumber seed length. Chinese Sci. Bull. 28, 165–170.
Chen, N. L., Gao, Y. M., and Zhang, Y. X. (2006). Effects of seed size and light intensity on seedling growth of watermelon. China Cucurbits Vegetables 1–3. doi: 10.3969/j.issn.1673-2871.2006.03.001
Chettoor, A. M., Phillips, A. R., Coker, C. T., Dilkes, B., and Evans, M. M. S. (2016). Maternal gametophyte effects on seed development in maize. Genetics 204, 233–248. doi: 10.1534/genetics.116.191833
Coomes, D. A., and Grubb, P. J. (2003). Colonization, tolerance, competition and seed-size variation within functional groups. Trends Ecol. Evol. 18, 283–291. doi: 10.1016/S0169-5347(03)00072-7
Dong, H., Dumenil, J., Lu, F. H., Na, L., Vanhaeren, H., Naumann, C., et al. (2017). Ubiquitylation activates a peptidase that promotes cleavage and destabilization of its activating E3 ligases and diverse growth regulatory proteins to limit cell proliferation in Arabidopsis. Genes Dev. 31, 197–208. doi: 10.1101/gad.292235.116
Downes, B. P., Stupar, R. M., Gingerich, D. J., and Vierstra, R. D. (2003). The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 35, 729–742. doi: 10.1046/j.1365-313X.2003.01844.x
Du, J., Wang, S., He, C., Zhou, B., Ruan, Y. L., and Shou, H. (2017). Identification of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis. J. Exp. Bot. 68, 1955–1972. doi: 10.1093/jxb/erw460
Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Figueiredo, D., and Kohler, C. (2014). Signaling events regulating seed coat development. Biochem. Soc. Trans. 42, 358–363. doi: 10.1042/BST20130221
Gao, M. L., Zhao, F. F., Wang, Y. S., Yuan, C. Z., Wei, X. M., Li, J. Y., et al. (2016). Genetic analysis and correlation of seed traits in mini watermelon. Jiangsu Agric. Sci. 44, 259–261.
Garcia, D., Gerald, J. N. F., and Berger, F. (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17, 52–60. doi: 10.1105/tpc.104.027136
Garcia, D., Saingery, V., Chambrier, P., Mayer, U., Jürgens, G., and Berger, F. (2003). Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 131, 1661–1670. doi: 10.1104/pp.102.018762
Garcia-Mas, J., Benjak, A., Sanseverino, W., Bourgeois, M., Mir, G., Gonzalez, V. M., et al. (2012). The genome of melon (Cucumis melo L.). Proc. Natl Acad. Sci. U.S.A. 109, 11872–11877. doi: 10.1073/pnas.1205415109
Gómez, J. M. (2004). Bigger is not always better: conflicting selective pressure on seed size in Quercus ilex. Evolution 58, 71–80. doi: 10.1111/j.0014-3820.2004.tb01574.x
Gu, Y. Z., Li, W., Jiang, H. W., Wang, Y., Gao, H. H., Liu, M., et al. (2017). Differential expression of a WRKY gene between wild and cultivated soybeans correlates to seed size. J. Exp. Bot. 68, 2717–2729. doi: 10.1093/jxb/erx147
Guo, S. G., Zhang, J. G., Sun, H. H., Salse, J., Lucas, W., Zhang, H. Y., et al. (2013). The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 45, 51–58. doi: 10.1038/ng.2470
Hu, Z. J., Lu, S. J., Wang, M. J., He, H. H., Sun, L., Wang, H. R., et al. (2018). A novel QTL qTGW3 encodes the GSK3/SHAGGY-Like Kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 11, 736–749. doi: 10.1016/j.molp.2018.03.005
Huang, S. W., Li, R. Q., Zhang, Z. H., Li, L., Gu, X. F., Fan, W., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41, 1275–1281. doi: 10.1038/ng.475
Huang, X. H., Zhao, Y., Wei, X. H., Li, C. Y., Wang, A. H., Zhao, Q., et al. (2011). Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32–39. doi: 10.1038/ng.1018
Huang, Y., Bai, X. F., Cheng, N. N., Xiao, J. H., Li, X. H., and Xing, Y. Z. (2019). Wide Grain 7 increases grain width by enhancing H3K4me3 enrichment in the OsMADS1 promoter in rice (Oryza sativa L.). Plant J. [Epub ahead of print].
Jiang, W. B., Huang, H. Y., Hu, Y. W., Zhu, S. W., Wang, Z. Y., and Lin, W. H. (2013). Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 162, 1965–1977. doi: 10.1104/pp.113.217703
Jiao, S. Q. (2017). QTL Analysis of Fruit and Seed Related Traits in Melon. Harbin: Northeast Agricultural University, 4–25.
Jofuku, K. D., Omidyar, P. K., Gee, Z., and Okamuro, J. K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. U.S.A. 102, 3117–3122. doi: 10.1073/pnas.0409893102
Johnson, C. S., Kolevski, B., and Smyth, D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. doi: 10.1105/tpc.001404
Kanako, K., Shigeru, K., Katsuyuki, O., Yuki, A., Tsuyu, A., Izumi, K., et al. (2010). A novel kinesin 13 protein regulating rice seed length. Plant Cell Physiol. 51, 1315–1329. doi: 10.1093/pcp/pcq092
Kim, K. H., Hwang, J. H., Han, D. Y., Park, M., Kim, S., Choi, D., et al. (2015). Major quantitative trait loci and putative candidate genes for powdery mildew resistance and fruit-related traits revealed by an intraspecific genetic map for watermelon (Citrullus lanatus var. lanatus). PLoS One 10:e0145665. doi: 10.1371/journal.pone.0145665
Li, D. W., Cuevas, H. E., Yang, L., Li, Y. H., Garcia-Mas, J., Zalapa, J., et al. (2011). Syntenic relationships between cucumber (Cucumis sativus L.) and melon (C. melo L.) chromosomes as revealed by comparative genetic mapping. BMC Genomics 12:396. doi: 10.1186/1471-2164-12-396
Li, M. R., Li, X. X., Zhou, Z. J., Wu, P. Z., Fang, M. C., Pan, X. P., et al. (2016). Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 7:377. doi: 10.3389/fpls.2016.00377
Li, N., and Li, Y. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. doi: 10.1016/j.pbi.2016.05.008
Li, N., Shang, J. L., Wang, J. M., Zhou, D., Li, N. N., and Ma, S. W. (2018a). Fine mapping and discovery of candidate genes for seed size in watermelon by genome survey sequencing. Sci. Rep. 8, 843–854. doi: 10.1038/s41598-018-36104-w
Li, N., Xu, R., Duan, P. G., and Li, Y. H. (2018b). Control of grain size in rice. Plant Rep. 31, 237–251. doi: 10.1007/s00497-018-0333-6
Li, N., Xu, R., and Li, Y. H. (2019). Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 70, 435–463. doi: 10.1146/annurev-arplant-050718-095851
Li, Q., Li, H. B., Huang, W., Xu, Y. C., Zhou, Q., Wang, S. H., et al. (2019). A chromosome-scale genome assembly of cucumber (Cucumis sativus L.). Gigascience 8:giz072. doi: 10.1093/gigascience/giz072
Li, Q., Li, L., Yang, X., Warburton, M. L., Bai, G., Dai, J., et al. (2010). Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 10:143. doi: 10.1186/1471-2229-10-143
Li, Y. D., Zhang, S. L., Dong, R. Z., Wang, L., Yao, J., Van, N. S., et al. (2019). The grapevine homeobox gene VvHB58 influences seed and fruit development through multiple hormonal signaling pathways. BMC Plant Biol. 19:523. doi: 10.1186/s12870-019-2144-9
Li, Y. H., Zheng, L. Y., Corke, F., Smith, C., and Bevan, M. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336. doi: 10.1101/gad.463608
Lietzow, C. D. (2015). Characterization and QTL Mapping of Parthenocarpic Fruit Set in Processing Cucumber (Cucumis sativus L.). (Ann Arbor, MI: ProQuest LLC), 157–158.
Liu, C. Q., Gao, P., and Luan, F. S. (2014). Construction of a genetic linkage map and QTL analysis of fruit-associated traits in watermelon. Sci. Agric. Sin. 47, 2814–2829.
Luo, M., Dennis, E. S., Berger, F., Peacock, W. J., and Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 17531–17536. doi: 10.1073/pnas.0508418102
Meru, G., Fu, Y., Leyva, D., Sarnoski, P., and Yagiz, Y. (2018). Phenotypic relationships among oil, protein, fatty acid composition and seed size traits in Cucurbita pepo. Sci. Hortic. 233, 47–53. doi: 10.1016/j.scienta.2018.01.030
Meru, G., and McGregor, C. (2013). Genetic mapping of seed traits correlated with seed oil percentage in watermelon. Hortscience 48, 955–959. doi: 10.21273/HORTSCI.48.8.955
Mizukami, Y., and Fischer, R. L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. U.S.A. 97, 942–947. doi: 10.1073/pnas.97.2.942
Moles, A. T., Ackerly, D. D., Webb, C. O., Tweddle, J. C., Dickie, J. B., and Westoby, M. (2005). A brief history of seed size. Science 307, 576–580. doi: 10.1126/science.1104863
Montero-Pau, J., Blanca, J., Bombarely, A., Ziarsolo, P., Esteras, C., Martí-Gómez, C., et al. (2018). De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol. J. 16, 1161–1171. doi: 10.1111/pbi.12860
Ning, X. F., Wang, X. L., Yu, Z. J., Lu, S. M., and Li, G. (2018). Inheritance and molecular mapping of tight-placenta gene and seed traits in Chinese Hami Melon ‘Queen’. Hortscience 53, 788–794. doi: 10.21273/HORTSCI13046-18
Okushima, Y., Overvoorde, P. J., Arima, K., Alonso, J. M., Chan, A., Chang, C., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463. doi: 10.1105/tpc.104.028316
Pan, Y. P., Wang, Y. H., McGregor, C., Liu, S., Luan, F. S., Gao, M. L., et al. (2020). Genetic architecture of fruit size and shape variation in cucurbits: a comparative perspective. Theor. Appl. Genet. 133, 1–21. doi: 10.1007/s00122-019-03481-3
Paris, H. S., and Nerson, H. (2003). Seed dimensions in the subspecies and cultivar-groups of Cucurbita pepo. Gen. Resour. Crop Evol. 50, 615–625. doi: 10.1023/a:1024464831595
Pereira, L., Ruggieri, V., Pérez, S., Alexiou, K. G., Fernández, M., Jahrmann, T., et al. (2018). QTL mapping of melon fruit quality traits using a high-density GBS-based genetic map. BMC Plant Biol. 18:324. doi: 10.1186/s12870-018-1537-5
Poole, C. F., Grimball, P. C., and Porter, D. R. (1941). Inheritance of seed characters in watermelon. J. Agric. Res. 63, 433–456.
Prasad, K., Zhang, X., Tobón, E., and Ambrose, B. A. (2010). The Arabidopsis B-sister MADS-box protein. GORDITA, represses fruit growth and contributes to integument development. Plant J. 62, 203–214. doi: 10.4161/psb.5.7.12095
Prothro, J., Sandlin, K., Abdel-Haleem, H., Bachlava, E., and Mcgregor, C. (2012a). Main and epistatic quantitative trait loci associated with seed size in watermelon. J. Am. Soc. Hortic. Sci. 137, 452–457. doi: 10.21273/JASHS.137.6.452
Prothro, J., Sandlin, K., Gill, R., Bachlava, E., White, V., and Knapp, S. (2012b). Mapping of the Egusi seed trait locus (eg) and quantitative trait Loci associated with seed oil percentage in watermelon. J. Am. Soc. Hortic. Sci. 137, 311–315. doi: 10.21273/JASHS.137.5.311
Ren, D. Q., Wang, X. C., Yang, M., Yang, L., He, G. M., and Deng, X. W. (2019). A new regulator of seed size control in Arabidopsis identified by a genome-wide association study. New Phytol. 222, 895–906. doi: 10.1111/nph.15642
Ren, Y., Mcgregor, C., Zhang, Y., Gong, G., Zhang, H., Guo, S. G., et al. (2014). An integrated genetic map based on four mapping populations and quantitative trait loci associated with economically important traits in watermelon (Citrullus lanatus). BMC Plant Biol. 14:33. doi: 10.1186/1471-2229-14-33
Ren, Y., Zhao, H., Kou, Q. H., Jiang, J., Guo, S. G., Zhang, H. Y., et al. (2012). A high resolution genetic map anchoring scaffolds of the sequenced watermelon genome. PLoS One 7:e29453. doi: 10.1371/journal.pone.0029453
Ruggieri, V., Alexiou, K. G., Morata, J., Argyris, J., Pujol, M., Yano, R., et al. (2018). An improved assembly and annotation of the melon (Cucumis melo L.) reference genome. Sci. Rep. 8:8088. doi: 10.1038/s41598-018-26416-2
Sandlin, K., Prothro, J., Heesacker, A., Khalilian, N., Okashah, R., Xiang, W., et al. (2012). Comparative mapping in watermelon [Citrullus lanatus (Thunb.) Matsum. et Nakai]. Theor. Appl. Genet. 125, 1603–1618. doi: 10.1007/s00122-012-1938-z
Savadi, S. (2018). Molecular regulation of seed development and strategies for engineering seed size in crop plants. Plant Growth Regul. 84, 401–422. doi: 10.1007/s10725-017-0355-3
Schruff, M. C., Spielman, M., Tiwari, S., Adams, S., Fenby, N., and Scott, R. J. (2005). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signaling, cell division, and the size of seeds and other organs. Development 133, 251–261. doi: 10.1242/dev.02194
Segami, S., Kono, I., Ando, T., Yano, M., Kitano, H., Miura, K., et al. (2012). Small and round seed 5, gene encodes alpha-tubulin regulating seed cell elongation in rice. Rice 5:4. doi: 10.1186/1939-8433-5-4
Segami, S., Takehara, K., Yamamoto, T., Kido, S., and Miura, K. (2017). Overexpression of SRS5 improves grain size of brassinosteroid-related dwarf mutants in rice (Oryza sativa L.). Breed. Sci. 67, 393–397. doi: 10.1270/jsbbs.16198
Shi, L. L., Song, J. R., Guo, C. C., Wang, B., Guan, Z. L., Yang, P., et al. (2019). A CACTA-like transposable element in the upstream region of BnaA9.CYP78A9 acts as an enhancer to increase silique length and seed weight in rapeseed. Plant J. 98, 524–539. doi: 10.1111/tpj.14236
Si, L. Z., Chen, J. Y., Huang, X. H., Gong, H., Luo, J. H., Hou, Q. Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Silvertown, J. W. (1981). Seed size, life span, and germination date as coadapted features of plant life history. Am. Nat. 118, 860–864. doi: 10.2307/2460817
Smitchger, J., and Weeden, N. F. (2018). The ideotype for seed size: a model examining the relationship between seed size and actual yield in pea. Int. J. Agron. 2018:9658707. doi: 10.1155/2018/9658707
Song, X. J., Huang, W., Shi, M., Zhu, M. Z., and Lin, H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi: 10.1038/ng2014
Stanton, M. L. (1984). Seed variation in wild radish: effect of seed size on components of seedling and adult fitness. Ecology 65, 1105–1112. doi: 10.2307/1938318
Su, Z. Q., Hao, C. Y., Wang, L. F., Dong, Y. C., and Zhang, X. Y. (2011). Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 122, 211–223. doi: 10.1007/s00122-010-1437-z
Sun, H. H., Wu, S., Zhang, G. Y., Jiao, C., Guo, S. G., Ren, Y., et al. (2017). Karyotype stability and unbiased fractionation in the paleo-allotetraploid, cucurbita, genomes. Mol. Plant 10, 1293–1306. doi: 10.1016/j.molp.2017.09.003
Sun, X. D., Shantharaj, D., Kang, X. J., and Ni, M. (2010). Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr. Opin. Plant Biol. 13, 611–620. doi: 10.1016/j.pbi.2010.08.009
Tan, X. Z., Ge, Y., Xu, W. L., Cui, C. S., and Qu, S. P. (2013). Construction of genetic linkage map and QTL analysis for seed width in pumpkin (Cucurbita Maxima). Acta Bot. Borealioccidentalia Sin. 33, 697–702.
Tanaka, T., Wimol, S., and Mizutani, T. (1995). Inheritance of fruit shape and seed size of watermelon. J. Jpn. Soc. Hort. Sci. 64, 543–548. doi: 10.2503/jjshs.64.543
Tian, Y. F., Wei, Y. Y., Chang, X. K., Yang, M. K., Wu, W. J., and Wei, X. (2012). Effects of seed size on germinating period and seedling quality of cucumber, watermelon and squash. Shandong Agric. Sci. 44, 40–42. doi: 10.14083/j.issn.1001-4942.2012.08.039
Urasaki, N., Takagi, H., Natsume, S., Uemura, A., Taniai, N., Miyagi, N., et al. (2017). Draft genome sequence of bitter gourd (Momordica charantia), a vegetable and medicinal plant in tropical and subtropical regions. DNA Res. 24, 51–58. doi: 10.1093/dnares/dsw047
Wang, A. H., Garcia, D., Zhang, H. Y., Feng, K., Chaudhury, A., Berger, F., et al. (2010). The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 63, 670–679. doi: 10.1111/j.1365-313X.2010.04271.x
Wang, M., Miao, H., Zhang, S. P., Liu, S. L., Dong, S. Y., Wang, Y., et al. (2014). Inheritance analysis and QTL mapping of cucumber seed size. Acta Hortic. Sin. 41, 63–72. doi: 10.16420/j.issn.0513-353x.2014.01.010
Wang, P., Zhang, Y., Bai, L. H., Liu, J. C., and Yang, W. X. (2012). Correlation and path analysis about yield component factors of seed-used pumpkin (Cucurbita pepo L.). Northern Hortic. 36, 16–19.
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Wang, S. K., Li, S., Liu, Q., Wu, K., Zhang, J. Q., Wang, S. S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, Y. H., Bo, K. L., Gu, X. F., Pan, J. S., Li, Y. H., Chen, J. F., et al. (2020). Molecularly tagged genes and quantitative trait loci in cucumber with recommendations for QTL nomenclature. Hortic. Res. 7:3. doi: 10.1038/s41438-019-0226-3
Wang, Y. H., Wang, C. J., Han, H. Y., Luo, Y. S., Wang, Z. C., Yan, Y. L., et al. (2019). Construction of a high-density genetic map and analysis of seed-related traits using specific length amplified fragment sequencing for Cucurbita maxima. Front. Plant Sci. 10:1782. doi: 10.3389/fpls.2019.01782
Wang, Z., Li, N., Jiang, S., Gonzalez, N., Huang, X., Wang, Y., et al. (2016). SCFSAP controls organ size by targeting PPD proteins for degradation in Arabidopsis thaliana. Nat. Commun. 7:11192. doi: 10.1038/ncomms11192
Wu, G. L., Du, G. Z., and Shang, Z. H. (2006). Contribution of seed size and its fate to vegetation renewal: a review. Chin. J. Appl. Ecol. 17, 1969–1972. doi: 10.13287/j.1001-9332.2006.0387
Wu, S., Shamimuzzaman, M., Sun, H., Salse, J., Sui, X., Wilder, A., et al. (2017). The bottle gourd genome provides insights into Cucurbitaceae evolution and facilitates mapping of a Papaya ring-spot virus resistance locus. Plant J. 92, 963–975. doi: 10.1111/tpj.13722
Xia, D., Zhou, H., Liu, R., Dan, W., Li, P., Wu, B., et al. (2018). GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3, to form extra-long grains in rice. Mol. Plant 11, 754–756. doi: 10.1016/j.molp.2018.03.006
Xia, T., Li, N., Dumenil, J., Li, J., Kamenski, A., Bevan, M. W., et al. (2013). The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25, 3347–3359. doi: 10.1105/tpc.113.115063
Xiang, C. P., Shi, X. M., and Zhang, Y. (2002). Effect of seed size on seed quality and yield of bitter gourd. J. Changjiang Vegetables 35–36.
Xiao, Y. G., Sun, Q. B., Kang, X. J., Chen, C. B., and Ni, M. (2016). SHORT HYPOCOTYL UNDER BLUE1 or HAIKU2 mixepression alters canola and Arabidopsis seed development. New Phytol. 209, 636–649. doi: 10.1111/nph.13632
Xie, D. S., Xu, Y. C., Wang, J. P., Liu, W. R., Zhou, Q., Luo, S., et al. (2019). The wax gourd genomes offer insights into the genetic diversity and ancestral cucurbit karyotype. Nat Commun. 10:5158. doi: 10.1038/s41467-019-13185-3
Xu, D. H., Zhang, Y. W., Li, C. G., and Cui, C. S. (2006). Combining ability of economic characters in seed pumpkin (Cucurbita maxima Duch.). China Vegetables 01, 13–15.
Yang, L. M., Koo, D. H., Li, D. W., Zhang, T., Jiang, J. M., Luan, F. S., et al. (2014). Next-generation sequencing. FISH mapping and synteny-based modeling reveal mechanisms of decreasing dysploidy in Cucumis. Plant J. 77, 16–30. doi: 10.1111/tpj.12355
Yang, L. M., Koo, D. H., Li, Y. H., Zhang, X. J., Luan, F. S., Havey, M. J., et al. (2012). Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 71, 895–906. doi: 10.1111/j.1365-313x.2012.05017.x
Yang, X., Wu, F., Lin, X., Du, X., Chong, K., Gramzow, L., et al. (2012). Live and let die-the Bsister MADS-box gene OsMADS29 controls the degeneration of cells in maternal tissues during seed development of rice (Oryza sativa). PLoS One 7:e51435. doi: 10.1371/journal.pone.0051435
Ye, W. Z., Liu, S., Ma, H. Y., Liu, H. Y., Li, G. Y., and Luan, F. S. (2017). QTL analysis on related seed traits in melon based on CAPS markers. North. Hortic. 12, 119–128. doi: 10.11937/bfyy.201712028
Yongjae, K., Taejin, Y., Younghoon, P., Yongjik, L., Suncheol, K., Yongkwon, K., et al. (2009). Development of near isogenic lines with various seed sizes and study on seed size-related characteristics in watermelon. Korean J. Breed. Sci. 41, 403–411.
Zeng, Y. F., Wen, J. Y., Zhao, W. B., Wang, Q., and Huang, W. C. (2020). Rational improvement of rice yield and cold tolerance by editing the three genes OsPIN5b, GS3, and OsMYB30 with the CRISPR-Cas9 system. Front. Plant Sci. 10:1633. doi: 10.3389/fpls.2019.01663
Zhang, G. F., and Zhang, J. N. (2011). Inheritance of seed size in watermelon. Jiangsu Agric. Sci. 39, 216–217. doi: 10.15889/j.issn.1002-1302.2011.04.170
Zhang, J. (1996). Inheritance of seed size from diverse crosses in watermelon. Cucurbit Genet. Coop. Rep. 19, 67–69.
Zhang, K. X., Dai, D. Y., Wang, H. N., Yu, M. Y., and Sheng, Y. Y. (2018). Genetic and QTL analysis of seed traits in melon (Cucumis melon L.). Acta Agric. Zhejiang. 30, 1496–1503. doi: 10.1186/1471-2229-11-111
Zhang, X., Rhodes, B., and Wang, M. (1994). “Genes controlling watermelon seed size,” in Cucurbitaceae ’94: Evaluation and Enhancement of Cucurbit Germplasm, eds G. Lester and J. Dunlap, (Alexandria, VA: ASHS Press), 144–147.
Zhang, X. J., Jiang, W. B., and Pang, Y. Z. (2016). Advances in the regulation mechanism of plant seed size. Plant. Physiol. J. 998–1010. doi: 10.13592/j.cnki.ppj.2016.0159
Zhou, H. W., Lu, B. Y., Ma, H. Y., Gao, P., Luan, F. S., Gao, Q. F., et al. (2016). QTL mapping of watermelon seed traits. Acta Hortic. Sin. 43, 715–723. doi: 10.16420/j.issn.0513-353x.2015-0973
Zhou, Y., Zhang, X. J., Kang, X. J., Zhao, X. Y., Zhang, X. S., and Ni, M. (2009). SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 21, 106–117. doi: 10.2307/40537464
Zhu, H. Y., Song, P. Y., Koo, D. H., Guo, L. Q., Li, Y. M., Sun, S. R., et al. (2016). Genome wide characterization of simple sequence repeats in watermelon genome and their application in comparative mapping and genetic diversity analysis. BMC Genomics 17:557. doi: 10.1186/s12864-016-2870-4
Keywords: cucurbits, watermelon, pumpkin/squash, cucumber, melon, seed size, QTL, comparative analysis
Citation: Guo Y, Gao M, Liang X, Xu M, Liu X, Zhang Y, Liu X, Liu J, Gao Y, Qu S and Luan F (2020) Quantitative Trait Loci for Seed Size Variation in Cucurbits – A Review. Front. Plant Sci. 11:304. doi: 10.3389/fpls.2020.00304
Received: 30 August 2019; Accepted: 03 March 2020;
Published: 20 March 2020.
Edited by:
Alma Balestrazzi, University of Pavia, ItalyReviewed by:
Byoung-Cheorl Kang, Seoul National University, South KoreaYan Long, Institute of Biotechnology (CAAS), China
Fernando Juan Yuste-Lisbona, University of Almería, Spain
Copyright © 2020 Guo, Gao, Liang, Xu, Liu, Zhang, Liu, Liu, Gao, Qu and Luan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meiling Gao, Z2FvbWVpbGluZzA1MzlAMTYzLmNvbQ==
 Yu Guo
Yu Guo Meiling Gao
Meiling Gao Xiaoxue Liang
Xiaoxue Liang Ming Xu1
Ming Xu1 Shuping Qu
Shuping Qu Feishi Luan
Feishi Luan