
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 10 March 2020
Sec. Plant Pathogen Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00253
This article is part of the Research TopicAdvances and Challenges of RNAi Based Technologies for PlantsView all 11 articles
 Luisa Katharina Schaefer1
Luisa Katharina Schaefer1 Francis Parlange1
Francis Parlange1 Gabriele Buchmann1
Gabriele Buchmann1 Esther Jung1
Esther Jung1 Andreas Wehrli1
Andreas Wehrli1 Gerhard Herren1
Gerhard Herren1 Marion Claudia Müller1
Marion Claudia Müller1 Jonas Stehlin1
Jonas Stehlin1 Roman Schmid1
Roman Schmid1 Thomas Wicker1
Thomas Wicker1 Beat Keller1*
Beat Keller1* Salim Bourras1,2*
Salim Bourras1,2*Cross-kingdom RNA interference (RNAi) is a biological process allowing plants to transfer small regulatory RNAs to invading pathogens to trigger the silencing of target virulence genes. Transient assays in cereal powdery mildews suggest that silencing of one or two effectors could lead to near loss of virulence, but evidence from stable RNAi lines is lacking. We established transient host-induced gene silencing (HIGS) in wheat, and demonstrate that targeting an essential housekeeping gene in the wheat powdery mildew pathogen (Blumeria graminis f. sp. tritici) results in significant reduction of virulence at an early stage of infection. We generated stable transgenic RNAi wheat lines encoding a HIGS construct simultaneously silencing three B.g. tritici effectors including SvrPm3a1/f1, a virulence factor involved in the suppression of the Pm3 powdery mildew resistance gene. We show that all targeted effectors are effectively downregulated by HIGS, resulting in reduced fungal virulence on adult wheat plants. Our findings demonstrate that stable HIGS of effector genes can lead to quantitative gain of resistance without major pleiotropic effects in wheat.
RNA interference (RNAi) is a biological process in which small non-coding RNAs (sRNAs) are employed to selectively downregulate gene expression at the transcriptional or post-transcriptional level. Post-transcriptional gene silencing is a highly regulated mechanism relying on a cohort of proteins that direct gene silencing based on sequence complementarity between the sRNA and a target mRNA (Gheysen and Vanholme, 2007; Shabalina and Koonin, 2008). Plants encode functionally diverse populations of regulatory sRNAs which include microRNAs (miRNAs) and short interfering RNAs (siRNAs). While miRNAs correspond to 20–22-nt sequences typically derived from imperfect RNA hairpin structures, siRNAs refer to 20–24-nt sequences typically processed from long double-stranded RNA (dsRNA) precursors (Borges and Martienssen, 2015; D’Ario et al., 2017). Both siRNAs and miRNAs are processed from dsRNA precursors by the ribonuclease Dicer, and both can regulate gene expression. RNAi participates in the regulation of diverse biological processes including plant immunity (Brant and Budak, 2018; Deng et al., 2018), and several siRNAs and miRNAs have been described as important players in plant defense against viruses, bacteria, and fungi (Hua et al., 2018; Rosa et al., 2018; Muhammad et al., 2019).
Small non-coding RNAs can be expressed in a tissue-specific or stage-specific manner (Mohorianu et al., 2011; Mao et al., 2012) and correspond to highly mobile molecules that can travel from cell-to-cell (short-range) or systemically (long-range) in the plant (Dunoyer et al., 2013). There is increasing evidence demonstrating that sRNAs are also mobilized in bi-directional exchanges between plants and their parasites, thus providing a basis for cross-kingdom RNAi (ck-RNAi) as a plant defense mechanism (Knip et al., 2014; Hua et al., 2018). Prominent examples include miRNAs from the parasitic plant Cuscuta campestris, and siRNA Bc-siR37 from the fungal pathogen Botrytis cinerea, which act as virulence factors downregulating several Arabidopsis thaliana genes involved in immunity (Weiberg et al., 2013; Wang et al., 2017; Shahid et al., 2018). This phenomenon can be partially explained by the fact that the three proteins that are important for RNAi, an argonaute protein, a dicer-like (DCL) protein, and a RNA-dependent RNA polymerase, are commonly found across eukaryotes (Shabalina and Koonin, 2008). While experimental evidence for the transfer of sRNAs from fungi to their host has been provided recently (Weiberg et al., 2013; Wang et al., 2017), sRNA mobility from plant to fungi has been used for host-induced gene silencing (HIGS) for almost a decade (Nowara et al., 2010; Qi et al., 2019). In plant-pathogen interactions, HIGS can be based on the uptake of exosome-like vesicles, containing transgene-derived siRNAs from the host (Cai et al., 2018).
In the obligate biotrophic cereal powdery mildew formae speciales (Blumeria graminis ff. spp.), HIGS is an important tool for functional genomics (Nowara et al., 2010; Pliego et al., 2013). In this system, successful infection is typically characterized by the formation of a highly specialized feeding structure called the haustorium, shortly after an appressorium-mediated penetration of the plant cell wall (Praz et al., 2018). The haustorium is a poorly understood membrane invagination that develops inside the host epidermal cell, and serves as a basis for the emergence of a network of strictly epiphytic hyphae which can form secondary appressoria (Kwaaitaal et al., 2017). The latter will then infect the same cell or the surrounding cells and form additional haustoria. These invasive structures are the sites of molecular exchange between B. graminis and its hosts. In particular, it was demonstrated that haustoria are the main interface for the delivery of candidate small secreted effector proteins (CSEPs) which promote pathogen virulence (Bourras et al., 2018).
The B. graminis genomes encode one of the largest repertoires of effectors in fungi, consisting of over 800 annotated proteins (Bourras et al., 2018; Frantzeskakis et al., 2018; Müller et al., 2019). Transient HIGS has been successfully used to functionally validate such candidate virulence factors in barley powdery mildew (B. graminis f. sp. hordei), leading to the identification of 21 bona fide effectors involved in host penetration or promoting haustorium formation (Zhang et al., 2012; Pliego et al., 2013; Ahmed et al., 2015, 2016). Of these, HIGS of the B.g. hordei effectors BEC1054 and BEC1011 resulted in a 60–70% reduction of haustorium formation, suggesting that some B. graminis CSEPs are probably essential for virulence (Pliego et al., 2013). Further molecular and biochemical characterization of these HIGS-assayed effectors revealed that they are interacting with host proteins involved in plant immunity (reviewed in Bourras et al., 2018). Relevant examples include CSEP0055 which interacts with the barley pathogenesis-related protein PR17c involved in resistance to penetration (Zhang et al., 2012), and the ribonuclease-like CSEP0064/BEC1054 which targets several barley proteins implicated in defense responses (Pennington et al., 2016, 2019). Based on these results, it has been suggested that HIGS of a few essential B. graminis effectors via stable host transformation could lead to a significant to permanent loss of pathogen virulence.
In the recent decade, ck-RNAi has emerged as a possible approach to control diseases in crops (Koch and Kogel, 2014; Hua et al., 2018). Several studies have described the use of stable HIGS in plants to confer resistance to fungal pathogens (Nowara et al., 2010; Koch et al., 2013; Chen et al., 2016; Panwar et al., 2017; Guo et al., 2019). Based on experimental evidence from transient assays in barley, a preliminary study using stable transgenics showed that HIGS of the B.g. hordei 1,3-β-glucanosyl-transferase (GTF1) gene, resulted in a decrease of fungal virulence on barley T1 seedlings (Nowara et al., 2010). So far, fungal house-keeping genes and pathogenesis-related genes were the primary gene targets for stable HIGS. In B. graminis, the discovery of bona fide effectors such as BEC1054 and BEC1011 raised the question whether stable silencing of such CSEPs would be equivalent to silencing an essential gene. In the BEC1054 and BEC1011 HIGS assays, single barley epidermal cells were biolistically transformed with a HIGS construct encoding a long dsRNA sequence, perfectly complementary to a segment of the effector mRNA sequence (Pliego et al., 2013). B.g. hordei virulence was scored microscopically at the haustorial stage, based on the number of formed haustoria, normalized to the number of penetration attempts (Pliego et al., 2013). Because of the technical limitation of this strategy, it was not possible to quantify target gene expression or to assess macroscopic phenotypes. In this context, HIGS of effectors using stable transgenic lines would be an opportunity to understand the impact of ck-RNAi on pathogen virulence, based on accurate, whole tissue assessment of target gene silencing in relation to plant resistance.
Here, we demonstrate the applicability of HIGS in B.g. tritici based on the silencing of the β2-tubulin (β2-tub) housekeeping gene. We also present the generation, selection, and characterization of stable wheat lines expressing a HIGS construct targeting the suppressor of avirulence gene SvrPm3a1/f1. This wheat powdery mildew effector is involved in the suppression of several allelic Pm3 resistance gene variants in wheat (Bourras et al., 2015, 2019; Parlange et al., 2015). We also show that SvrPm3a1/f1 is expressed at significantly lower levels in B.g. tritici colonies growing on the HIGS lines as compared to the non-transgenic control. Consistent with in silico prediction of possible targets in the pathogen, two additional members of the SvrPm3a1/f1 effector gene family sharing sequence homology are also downregulated by HIGS. Finally, based on extensive phenotypic characterization of infected wheat material from laboratory and semi-field experiments, we show that HIGS of SvrPm3a1/f1 results in a quantitative gain of resistance against B.g. tritici.
Host-induced gene silencing of B. graminis genes was originally established in barley (Nowara et al., 2010). Here, we aimed at establishing HIGS in wheat for targeting B.g. tritici genes and at comparing HIGS efficacy in wheat and barley powdery mildew. The B. graminis β2-tub gene, an essential housekeeping gene and a canonical fungicide target (Zhou et al., 2016; Vela-Corcía et al., 2018), was selected as target gene for this assay. There is no β1-tub gene in B. graminis, indicating that transcriptional knockdown of the single-copy β2-tub gene is highly relevant for assessing the impact of HIGS on fungal virulence. Due to high sequence similarity between β2-tub genes across B. graminis ff. spp., the sequence from B.g. hordei was used as a template to design a HIGS construct capable of silencing this gene in B.g. tritici and B.g. hordei (Figure 1A). The silencing construct is based on a 140-nt segment of exon 6 from the B.g. hordei β2-tub gene (Supplementary File 1). The β2-tub-RNAi sequence used in this study was analyzed for possible off-targets in both formae speciales and their host species wheat and barley using the si-Fi software, which predicted no off-targets in either of the analyzed B. graminis and host genomes (Lück et al., 2019).
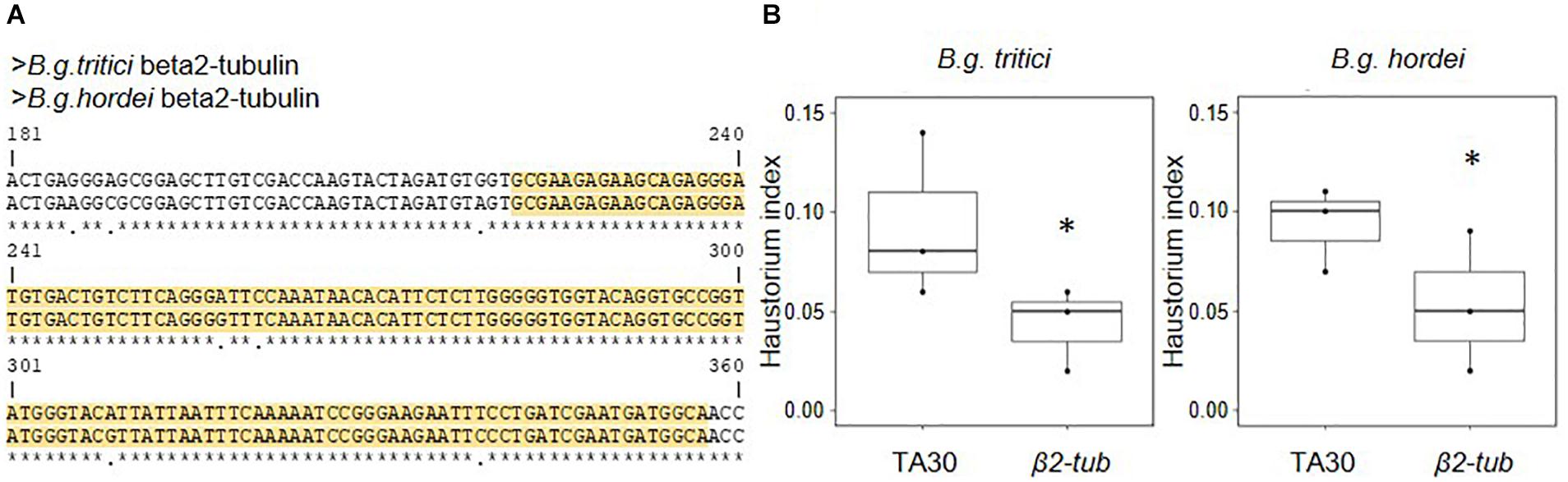
Figure 1. Transient HIGS of fungal β2-tubulin (β2-tub) reduces virulence in B.g. hordei and B.g. tritici. The β2-tub-RNAi construct was transiently expressed in barley and wheat epidermal cells, which were infected with B.g. hordei and B.g. tritici, respectively. (A) Alignment of the B.g. tritici and B.g. hordei β2-tub gene sequences. Only the fragment including the donor sequence for the β2-tub-RNAi construct (yellow) is depicted. The B.g. hordei sequence was used to construct the β2-tub-RNAi plasmid. (B) Effect of the β2-tub-RNAi construct on virulence in both formae speciales. The effect on fungal virulence was measured 2 days post infection, by scoring the ratio of successful over total infection attempts (haustorium index). Results were compared to the empty vector control pIPKTA30 (TA30) using a proportion test. ∗p < 0.05.
Wheat and barley leaves were biolistically co-transfected with a GUS reporter plasmid and the β2-tub-RNAi plasmid and infected with B.g. tritici and B.g. hordei, respectively. Fungal virulence was assessed microscopically 2 days post infection (dpi) based on successful haustoria formation (haustorium index, see section “Materials and Methods”). The empty pIPKTA30 vector was used as negative control. Results from three independent biological replicates showed statistically significant reduction of fungal virulence, corresponding to a decrease of the haustorium index by −42% in B.g. tritici and −43% in B.g. hordei as compared to the negative control (Figure 1B). We observed no effect on hyphae formation in the successful colonies. We conclude that transient HIGS of an essential housekeeping gene in B.g. tritici and B.g. hordei leads to very similar, quantitative loss of virulence in both formae speciales. Thus, HIGS studies are feasible in wheat using the same vector design and phenotyping strategy. We suggest that targeting of a housekeeping gene, such as β2-tub, can be used as a technical reference for assessing the relative efficiency of HIGS in future assays.
We designed a new HIGS construct targeting the B.g. tritici effector SvrPm3a1/f1. We selected a 500-nt fragment of the SvrPm3a1/f1 mRNA, which includes a segment of the 5′UTR and both exons excluding the last two codons (Figure 2A). Off-target analysis using the si-Fi software (Lück et al., 2019) predicted four putative siRNAs that map to a single 30-nt off-target locus in a putative pseudogene on the wheat chromosome 6A. However, these siRNAs are predicted to be inefficient in directing silencing (Supplementary Note 1 and Supplementary File 2). Off-target analysis using the B.g. tritici genome identified two additional target genes, Bgt_Bcg-6 and Bgt_Bcg-7 (Figure 2A and Supplementary Table 1). Consistent with the off-target predictions, both effector genes share nucleotide homology with SvrPm3a1/f1, with Bgt_Bcg-6 being identical in the 5′ first 120-nt of the HIGS construct (Supplementary Figure 1). Bgt_Bcg-6 and Bgt_Bcg-7 both belong to the SvrPm3a1/f1 effector gene family and are the closest homologs of the target effector (Parlange et al., 2015; Müller et al., 2019). Based on these results, we conclude that SvrPm3a1/f1, Bgt_Bcg-6, and Bgt_Bcg-7 are potent targets of the HIGS construct designed for the generation of stable transgenics.
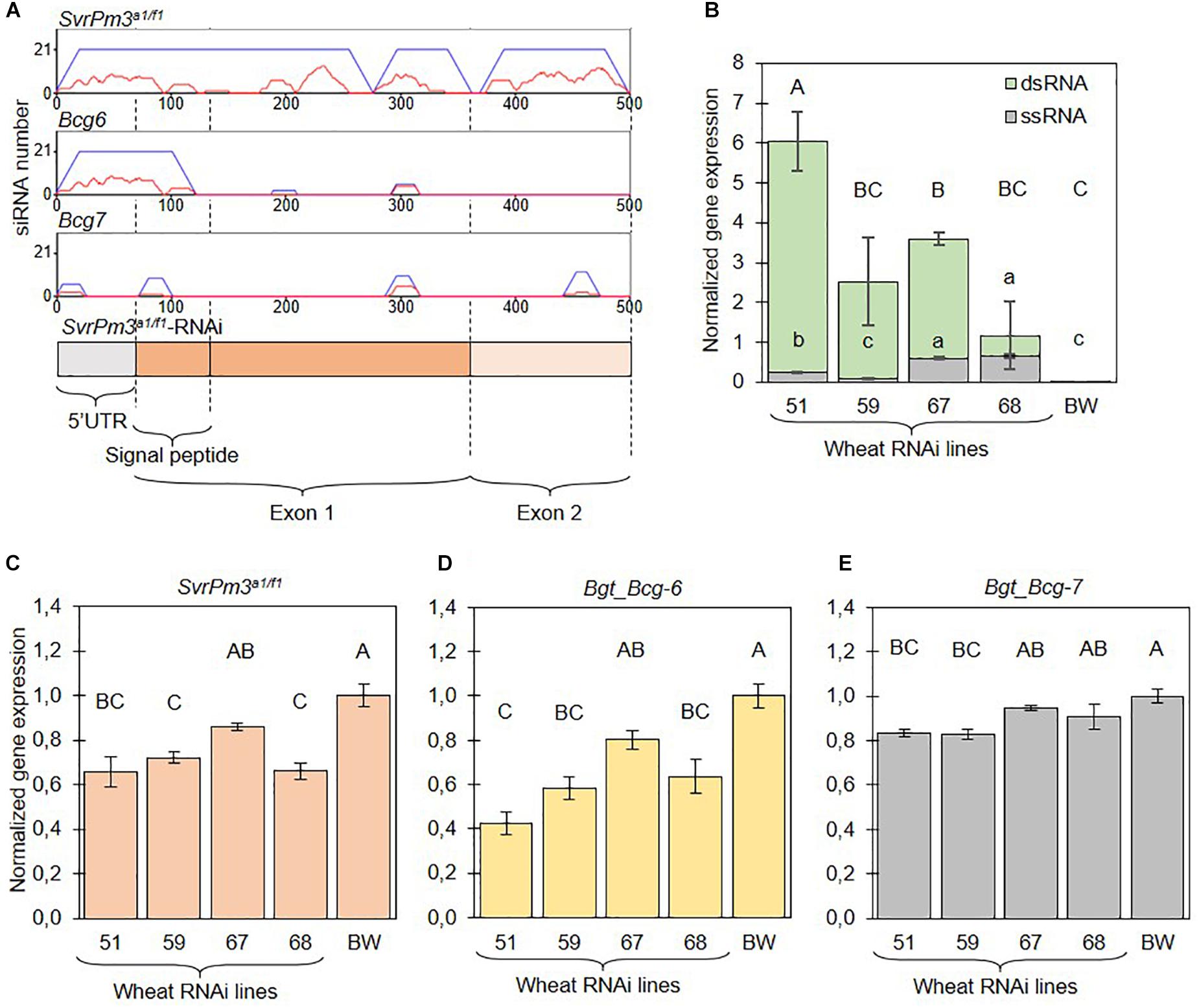
Figure 2. The SvrPm3a1/f1-RNAi transgene is constitutively expressed in transgenic wheat lines and silences three B.g. tritici effectors. (A) si-Fi software predictions of SvrPm3a1/f1-RNAi transgene-derived siRNAs that are efficient (red lines) or inefficient (blue lines) in directing silencing of the three putative target effectors SvrPm3a1/f1, Bgt_Bcg-6, and Bgt_Bcg-7. On the y-axis is the number of siRNAs that bind to a given locus on the x-axis. (B) Quantification of transgene-derived ssRNA and dsRNA transcripts from non-infected samples. (C–E) Quantification of target gene transcripts from infected samples. Samples were collected 3 days post infection. Error bars represent the standard error. Statistical difference was tested using ANOVA (B:dsRNA,D,E) or Welch ANOVA (B:ssRNA,C), according to the data distribution and variance. Different letters indicate statistical significance.
We cloned the selected SvrPm3a1/f1-RNAi sequence into the pIPKb007 vector, in which sense and antisense sequences are separated by a wheat RGA2 intron allowing hairpin formation (Himmelbach et al., 2007). Transgene expression is controlled by the maize ubiquitin promoter and the CaMV 35S terminator (Supplementary Figure 2). The spring wheat cultivar Bobwhite SH 98 26 (BW) was co-transformed with the linearized HIGS construct and the phosphomannose isomerase gene (Pmi) as a selection marker by particle bombardment. We obtained 138 independent T0 transformants (events), which were propagated to the T1 generation. Macroscopic scoring of leaf coverage by B.g. tritici colonies in T1 seedling leaves, did not allow us to discriminate between the segregating transgenic and non-transgenic T1 progeny. We hypothesized that our assay was not sensitive enough for scoring quantitative effects. We therefore aimed at selecting four events with the following criteria (i) 1–2 copies of the transgene can be detected by southern blot (Supplementary Figure 3), (ii) both the sense and antisense copy of the HIGS construct can be amplified by PCR indicating that the complete HIGS transgene is integrated into the genome, (iii) events are homozygous, (iv) transgene expression can be detected by RT-PCR, and (v) no major pleiotropic effects on plant development resulting from tissue culture can be observed. Based on this combination of criteria, we could confidently select four events, #51, #59, #67, and #68. All are single copy events, except event #67 which contains two genetically linked transgene copies.
We conclude that the generation of stable transgenic HIGS wheat plants can be restricted by the following challenges: First, T1 plants are difficult to phenotype for quantitative, small effects in which replication and scalability are key for inferring statistically significant differences. Second, the hairpin secondary structure inherent to the design of the HIGS construct complicates the molecular detection of transgene copies, both at the DNA and cDNA levels, using PCR-based methods. Considering these technical limitations and our selection process in which only events that pass all selection criteria are further characterized, we suggest that we are probably underestimating the proportion of successful transformation events.
In recent work by Wang et al. (2018), it was demonstrated that standard RT-qPCR assays are not suitable for the quantification of dsRNA transcripts produced by RNAi transgenes. Standard protocols for mRNA quantification by RT-qPCR primarily quantify aberrant single-stranded RNA (ssRNA) transcripts, which are inefficient in directing gene silencing (Wang et al., 2018). To further characterize the four selected events, we adapted the protocol described by Wang et al. (2018) and quantified dsRNA and ssRNA transcripts in non-infected seedlings. Consistent with previous findings by Wang et al. (2018), our assays show that expression of dsRNA and ssRNA transcripts are not correlated, thus further corroborating the importance of validating dsRNA expression from RNAi transgenes. But most importantly, we found that dsRNA transcripts are generally more abundant than ssRNA transcripts thus demonstrating that the HIGS transgene is giving rise to a proper template for siRNA biogenesis (Figure 2B). Together this data demonstrate that all four events contain a full, transcriptionally active copy of the SvrPm3a1/f1-RNAi transgene, which is constitutively expressed in absence of B.g. tritici infection.
We conclude that our selection procedure, based on careful molecular validation of transgene integrity at the DNA and RNA levels, led to the identification of relevant events, constitutively expressing the HIGS transgene. In this context, we propose that this material is ideal for assessing the effect of stable HIGS on mildew virulence at different stages of wheat development.
The B.g. tritici effectors SvrPm3a1/f1, Bgt_Bcg-6, and Bgt_Bcg-7 are equally valid potential targets of the SvrPm3a1/f1-RNAi transgene (Figure 2A). To test whether the three targets are effectively silenced, we quantified mRNA expression from B.g. tritici infected seedlings at three dpi, a time point corresponding to the haustorial stage. Previous studies have shown that gene expression levels of SvrPm3a1/f1 are the highest during haustorium formation (Bourras et al., 2015, 2019; Praz et al., 2018). Similarly, several members of the SvrPm3a1/f1 effector gene family, including Bgt_Bcg-6 and Bgt_Bcg-7, are induced at that same stage (Supplementary Figure 4) (Praz et al., 2018; Bourras et al., 2019), suggesting that this time point is particularly appropriate for the quantification of mRNA levels of all three effectors, and to assess HIGS efficiency. Transgenic events and wildtype BW, were infected with the B.g. tritici isolate Bgt_JIW2, which shows intermediate levels of expression of SvrPm3a1/f1 (Bourras et al., 2015, 2019). Primer design for specific amplification of the respective fungal SvrPm3a1/f1, Bgt_Bcg-6, and Bgt_Bcg-7 mRNAs was challenging, as the sequences of the three effectors are rather similar, and since the transgene corresponded to almost the complete SvrPm3a1/f1 coding sequence. Therefore, RT-qPCR primers could only be designed on the 3′UTR to discriminate between the transgene, SvrPm3a1/f1 and Bgt_Bcg-6 in particular. Specificity was verified by Sanger sequencing of amplicons from infected leaf material and non-infected controls.
The mRNA levels of the target genes SvrPm3a1/f1 and Bgt_Bcg-6 were significantly reduced as compared to the wildtype BW control in all events except #67 (Figures 2C,D). Interestingly, silencing of Bgt_Bcg-6 was consistently similar and sometimes even more effective than silencing of SvrPm3a1/f1 (Figures 2C,D). For Bgt_Bcg-7, we observed a less pronounced but yet significant reduction of mRNA levels in events #51 and #59, but not in event #68 (Figure 2E). Here, differences in target gene silencing are in agreement with the differences in sequence similarity between the three effectors. In particular, we propose that the 120-nt segment that is identical between SvrPm3a1/f1 and Bgt_Bcg-6 is possibly responsible for the silencing of both genes, while lower sequence identity between these two effectors and Bgt_Bcg-7 in that same segment is likely responsible for reduced HIGS efficiency on the latter. All effector targets considered, target gene downregulation ranged from −16% (Bgt_Bcg-7 on #51) to −58% (Bgt_Bcg-6 on #51), thus molecularly demonstrating that the HIGS transgene is functional in the events #51, #59 and #68, and does mediate ck-RNAi.
We conclude that stable wheat lines constitutively expressing a HIGS transgene can effectively mediate target gene silencing in B.g. tritici. We also conclude that the 5′ 120nt of the HIGS construct are probably sufficient for achieving high levels of target silencing, which could also be combined with other RNAi sequences for simultaneous silencing of entire gene families to potentially strengthen the impact of HIGS.
Based on the functional validation of target gene silencing, the impact of simultaneous silencing of three effectors, SvrPm3a1/f1, Bgt_Bcg-6, and Bgt_Bcg-7, on B.g. tritici virulence was assessed at different developmental stages (Figures 3A–D). First, we infected leaf segments from seedlings of the transgenic events and the wildtype control with the B.g. tritici isolate Bgt_JIW2 using a low-density inoculum, and quantitatively scored virulence based on the infected leaf area at six dpi. In this assay, we observed no reduction of virulence of B.g. tritici on seedlings (Supplementary Figure 5). We hypothesized that the effect of HIGS is best observed at the haustorial stage when the target effectors, in particular SvrPm3a1/f1 are transcriptionally active (Bourras et al., 2015, 2019; Praz et al., 2018). So, we prepared infected leaf segments for microscopy to score virulence at two dpi. Interestingly, we found a significant reduction in the haustorium index of −46% in event #68 compared to the wildtype (Figure 3E). The effect was stronger on the subgroup of immature haustoria, which are not yet showing the fully branched structure typical of mature haustoria (Figures 3A–C). For these, we found a significant reduction in the haustorium index of −41% in event #51 and −60% in event #68 compared to the wildtype (Figure 3F). The reduction of haustorium indices in the events #59 and #67 were not significant (Figures 3E,F). These results suggest that silencing of the three target effectors impairs B.g. tritici virulence at the haustorial stage in an event-specific, quantitative manner. Next, we microscopically assessed B.g. tritici development at four and six dpi on line #68, as it shows the strongest effect at the haustorial stage. Here, we scored B.g. tritici virulence by assessing the presence of haustoria, hyphae, and conidiophores. We found no significant differences at four dpi (Supplementary Figures 6C–H). However, we observed a significant reduction of the haustorium index of −30% in event #68 at six dpi compared to wildtype (Figure 3G), although no macroscopic differences in infected leaf area were observed in the initial seedling infection assays at this time point. Congruently, no additive effect was observed on hyphae and conidiophore formation (Figures 3D,H,I and Supplementary Figures 6A,B,I). Altogether, these results indicate that there is a consistent quantitative effect from HIGS of the three target effectors specifically impairing primary haustoria formation.
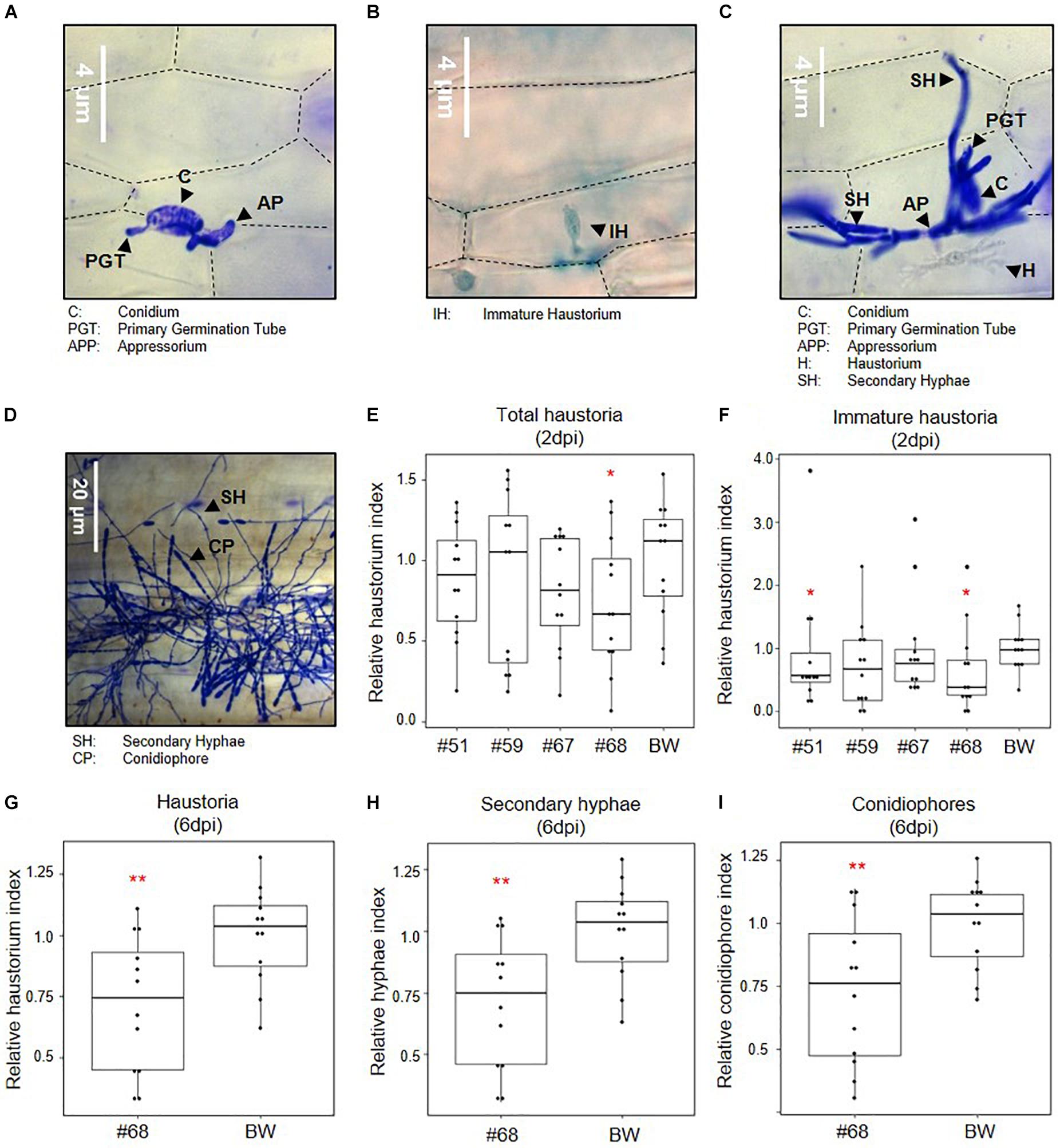
Figure 3. Effect of effector gene silencing on infection success. Seedlings of SvrPm3a1/f1-RNAi wheat lines were infected with B.g. tritici, and virulence was scored at two- and six-days post infection (dpi). All data is normalized to the wildtype control Bobwhite (BW). (A–D) Representative pictures of B.g. tritici developmental stages. (A) A spore has formed an appressorium, but fails at infection. (B) An immature haustorium. (D) A successful colonization at ca. two dpi with a mature haustorium and secondary hyphae. (D) A successful B.g. tritici colony at six dpi with mature conidiophores. (E) Total haustorium index at two dpi. (F) Immature haustorium index at two dpi. (G) Haustorium index at six dpi. (H) Hyphae index at six dpi. (I) Conidiophore index at six dpi. Pairwise comparison of transgenic events to the wildtype BW was carried out using a one-sided t-test (E,G–I) or a Wilcoxon rank sum test (F), according to the data distribution and variance. *p < 0.05; **p < 0.01.
We conclude that the observed effects of HIGS on B.g. tritici development on seedlings, is in agreement with previous evidence suggesting that the effectors SvrPm3a1/f1, Bgt_Bcg-6, and Bgt_Bcg-7 are important virulence factors particularly active at the haustorial stage. We also conclude that the observed effect is quantitative and event-specific, which could be due to genomic location of the transgene and subsequent tissue-specific variations of transgene expression.
In a final phenotypic analysis, we characterized all events for susceptibility to B.g. tritici at the adult stage in a semi-field setup. Here, the plants were exposed to local climate, and the local B.g. tritici population, We monitored B.g. tritici disease development over time according to Brunner et al. (2011) and calculated the area under the disease progression curve (AUDPC). We additionally collected representative samples for each test plot of flag leaves (F0), F-1 and F-2 leaves for precise quantification of infected leaf area. F0 leaves were sampled at the beginning of flag leaf infection at 70 days post sowing (dps), concurrent with spike emergence, and a second time at the end of the B.g. tritici disease progression at 92 dps, concurrent with seed ripening. F-1 and F-2 leaves were sampled at 78 dps.
Area under the disease progression curves based on all plant development stages did not indicate significant differences between the transgenic events and the wildtype control (Supplementary Figure 7A). However, AUDPCs based on flag leaves indicated significantly lower B.g. tritici infection than wildtype in the three events that also show target gene silencing (#51, #59, and #68) (Figure 4A). AUDPC is designed to score disease development on whole plants, but in our assay, it was only informative when considering the flag leaves. Therefore, we used image-based precise quantification of infected leaf area in the representative leaf samples to further support our results. In line with the AUDPC results, we found a significant reduction of infected leaf area in the flag leaf at 70 dps compared to wildtype in the same three events (−71% for #51, −50% for #59, and −70% for #68) (Figures 4B,D). Consistent with the results from seedling assays, this effect decreased over time to no significant difference at 92 dps with the exception of event #68, in which a significant reduction in infected leaf area (−69%) could still be observed (Figures 4C,E). The effect also decreased with the age of the leaves. While the F-1 leaves show a significant reduction in infected leaf area at 78 dps for events #59 (−36%) and #68 (−60%) (Figure 4F and Supplementary Figure 7B), there was no significant effect in the F-2 leaves (Supplementary Figures 7C,D).
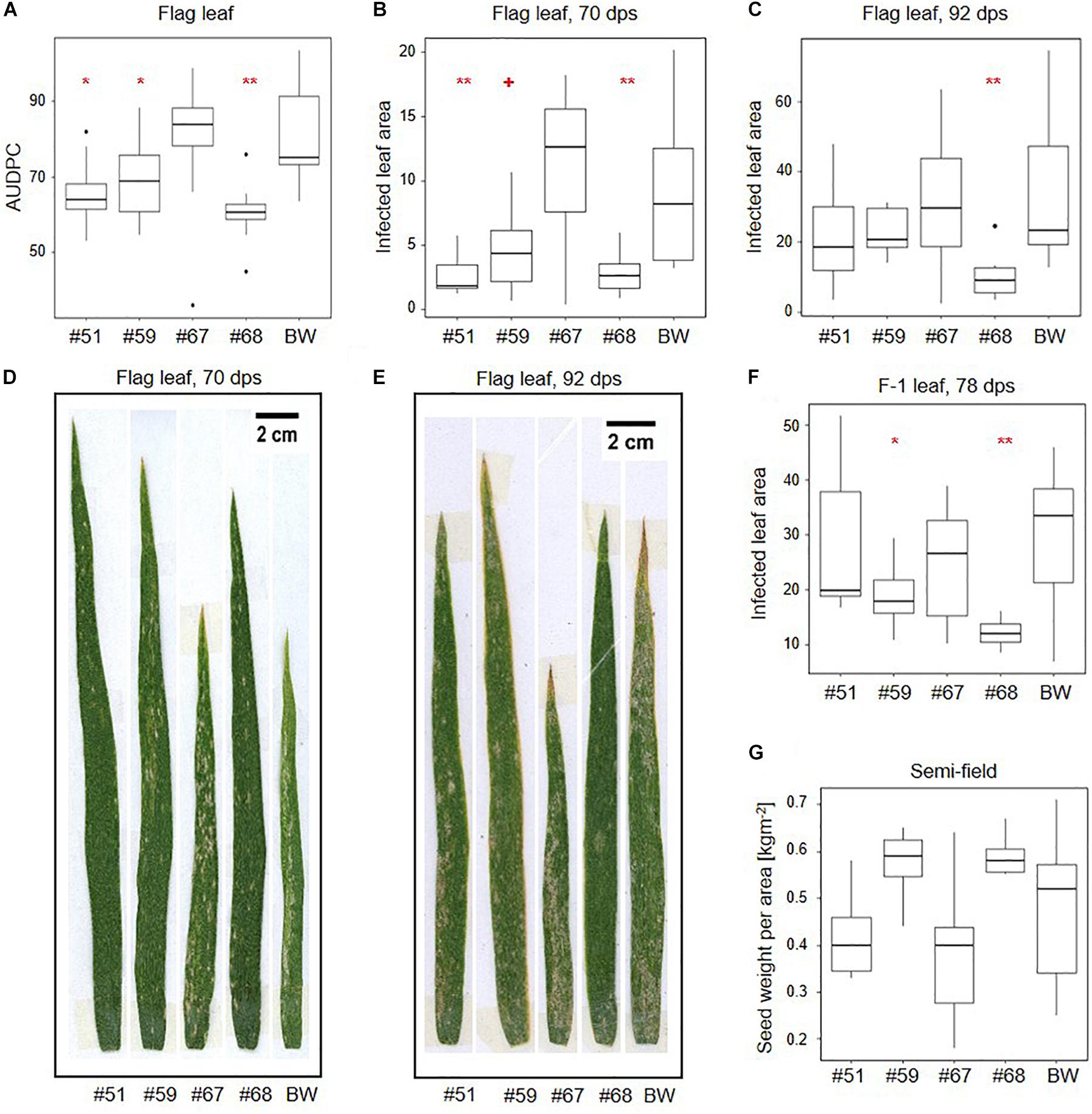
Figure 4. SvrPm3a1/f1-RNAi wheat lines show quantitative adult plant resistance in semi-field conditions. Transgenic events and wild-type Bobwhite (BW) were exposed to natural B.g. tritici infection in a semi-field trial. (A) Disease was monitored over time on the flag leaves and the area under the disease progression curve (AUDPC) was calculated. (B–F) Image-based quantification of the infected leaf area in flag and F-1 leaves. (B,D) At the beginning of flag leaf infection, 70 days post sowing (dps). (C,E) At the end of the B.g. tritici disease progression, 92 dps. (F) F-1 leaves at 78 dps. (G) Quantification of seed weight per area. Pairwise comparison of transgenic events with wildtype BW were carried out using one-sided t-test (A), Welch t-test (C,F,G) or Wilcoxon rank sum test (B) depending on the distribution and variance of the data. + < 0.06; *p < 0.05; **p < 0.01.
We also compared leaf area from adult leaves and took measurements of physiological traits including spike emergence and flowering time during the semi-field experiment. Additionally, we scored plant height, seed set, and seed weight per area both in the semi-field experiment and under controlled greenhouse conditions. While we acknowledge the small scale of these experiments, our results indicate that while there are no significant differences in seed weight per area in the semi-field, there is a trend toward higher seed weight per area in lines #59 and #68 which correlates with lower infected leaf area in F-1 leaves (Figure 4G). Under controlled greenhouse conditions, the seed weight per area was significantly higher in all three lines that showed target gene silencing and reduced infected leaf area in the semi-field (#51, #59, and #68) (Supplementary Figure 8A). Higher seed weight per area partially correlates with significantly higher spike number per plant in event #51 and #68 (Supplementary Figure 8B). We also observed that the pooled F0, F-1, and F-2 leaves of the three events showing increased adult plant resistance have significantly larger leaf area than wildtype, which could explain the increase in seed weight per area (Supplementary Figure 8C). We observed no meaningful differences in flowering time, plant height, or seed set neither in the semi-field nor in the greenhouse (Supplementary Figures 8D–H,J). But we observed a trend toward a 2- to 3-day delay in median spike emergence in all four transgenic lines, which was significant for events #51 and #67 (Supplementary Figure 8I). Overall, these results indicate that the virulence reducing effect of the SvrPm3a1/f1-RNAi transgene on the B.g. tritici pathogen is not accompanied by a major effect on host performance.
We conclude that stable HIGS of the three B.g. tritici effectors resulted in a quantitative gain of powdery mildew resistance in wheat. Resistant events can impair haustorium formation on seedlings, and restrict B.g. tritici growth on adult leaves in a quantitative manner.
Transient HIGS has been an important tool for functional genomics in B. graminis. In this study, we showed that HIGS can be used in stable transgenic wheat plants to effectively target three B.g. tritici effectors. Most importantly, we show that the use of this strategy to impair pathogen virulence results in a quantitative gain of resistance to wheat powdery mildew in adult plants.
The stable transgenic events generated in this study constitutively express the SvrPm3a1/f1-RNAi transgene. Therefore, we could confidently assess and correlate the observed phenotypes with the actual downregulation of the target genes, which is a major improvement to single cell transient HIGS assays. In this study, three effector genes were targeted (SvrPm3a1/f1, Bgt_Bcg-6, and Bgt_Bcg-7) and RT-qPCR assays showed that all three are downregulated when the pathogen is growing on transgenic HIGS events as compared to the non-transgenic control. The level of target gene silencing ranged from −16 to −58%, which is similar to the levels of HIGS reported in other fungal pathogens such as B. cinerea on Arabidopsis and Puccinia triticina on wheat (Wang et al., 2016; Panwar et al., 2017). In Fusarium graminearum, an even higher level of target gene silencing was reported (−77 to −92%) on Arabidopsis, suggesting that HIGS efficiency is intrinsically regulated by the two organisms involved in ck-RNAi (Koch et al., 2013).
The biology behind sRNA transport from the host to the fungal cell through the haustorial interface is not well understood. In principle, the transgene-derived hairpin transcripts are processed by host DCL enzymes into siRNAs. These siRNAs are then secreted in exosome-like vesicles that can be taken up by the fungus via the haustorial interface (Cai et al., 2018). Once the siRNAs are taken up, they guide the RNA-induced silencing complex (RISC) to cleave homologous, fungal mRNA transcripts. In plants and worms, mRNA-cleavage by a siRNA-primed RISC leads to the accumulation of secondary siRNAs through a specific RNA-dependent RNA polymerase (RdRP), which uses the diced mRNA molecules as a template to synthesize secondary dsRNA precursors or directly siRNAs. The accumulation of secondary siRNAs is known as transitivity, and is thought to result in a more persistent and stable target gene silencing (Calo et al., 2012).
In fungi, transitivity has been described in the zygomycete Mucor circinelloides and relies on its RdRP homolog McRdRP-2 (Calo et al., 2012). In ascomcyetes such as Neurospora crassa and Aspergillus nidulans, whole genome sequencing data indicates that the orthologs of the McRdRP-2 gene have been lost (Torres-Martínez and Ruiz-Vázquez, 2017). Instead, ascomycetes have orthologs of McRdRP-1, e.g., NcQde-1, which are necessary for sense but not for inverted repeat transgene-triggered RNAi. It is unclear whether the ascomycete orthologs of McRdRP-1 are capable of transitivity. In the case of the Blumeria clade, Kusch et al. (2018) found that this group of fungi has lost all RdRP paralogs except one single copy. It is unknown which specific RNA processing properties were retained by this single representative of the RdRP family in Blumeria, and how this protein is contributing to ck-RNAi. Based on our results, we hypothesize that ck-RNAi in Blumeria is possibly restricted by the absence of a transitivity-capable RdRP. This hypothesis provides an explanation for the relatively moderate silencing of target gene expression achieved by HIGS in this study. We propose that further characterization of the fungal RNAi machinery can provide meaningful information about the limitations and possible improvements of HIGS in Blumeria.
Efficient transgene-derived dsRNA accumulation in planta is thought to be decisive for efficient RNAi-directed control of insect pests (Wang et al., 2018). In fact, both dsRNA and ssRNA transcripts can be derived from a hairpin transgene, however, since only dsRNA is an efficient template for RNAi, it is anticipated that higher accumulation of dsRNA molecules should correlate with a higher accumulation of siRNAs, and thus higher target gene silencing efficiency. Here, we had a unique opportunity to test this hypothesis in a stable system in which we had clear evidence for HIGS. Interestingly, our results showed that the dsRNA proxy performed poorly in predicting the efficacy of ck-RNAi. While we could show that all events express a detectable dsRNA molecule, we found little correlation between ssRNA/dsRNA expression levels and target gene silencing. This could be explained by differences in transgene expression at the tissue level, which cannot be detected in our RT-qPCR assays based on RNA extraction from entire leaf segments. Here, higher levels of target gene silencing can possibly be associated with higher dsRNA expression in epidermal cells which are the only cells infected by mildew, and the primary target tissue of transient HIGS. It is also possible that dsRNA accumulation is not the only factor relevant for efficient ck-RNAi in Blumeria. In fungi, evidence suggests that dsRNA-derived siRNAs are the bioactive molecule driving HIGS (Cai et al., 2018), thus additional factors influencing dsRNA processing by the host DCL enzymes, siRNA packaging into exosome-like vesicles, and vesicle uptake by the fungus can have an impact on silencing efficiency, either acting as bottlenecks or enhancers. In humans, the discovery of sequence motifs that are over-represented in exosome-associated miRNAs led to the identification of specific proteins that control miRNA sorting into exosomes (Villarroya-Beltri et al., 2013). We therefore suggest that the identification of the genetic factors associated with RNA mobility from its hosts to Blumeria can provide an important leverage for improving HIGS efficiency. We speculate that favorable HIGS traits can be identified from the natural diversity of wheat cultivars, introduced through breeding or genome editing, and inform better HIGS transgene design.
Small secreted proteins with a predicted ribonuclease fold constitute the largest class of candidate effectors in the Blumeria genomes (Pedersen et al., 2012; Bourras et al., 2018). This RNase-like class of effectors also includes the SvrPm3a1/f1 family which encodes all three effectors targeted by HIGS in this study. Evidence from previous transient HIGS experiments indicated that this class of effectors is highly important for Blumeria virulence at the early stages of infection (Pliego et al., 2013). In this study, we extensively scored several attributes of B.g. tritici virulence on stable SvrPm3a1/f1-RNAi wheat lines at the macroscopic and microscopic level. We observed that HIGS of the three RNase-like effectors, SvrPm3a1/f1, Bgt_Bcg-6, and Bgt_Bcg-7, leads to a consistent loss of virulence at the haustorial stage, in the form of a reduced ability of the fungus to penetrate and form such feeding structures, and to a significant reduction in infected leaf area on adult leaves. Our results thereby further substantiate the importance of this class of proteins for Blumeria virulence. Previous transient HIGS experiments in B.g. hordei also described an effect primarily on haustorial development upon targeting of effectors (Nowara et al., 2010; Zhang et al., 2012; Pliego et al., 2013; Ahmed et al., 2015; Aguilar et al., 2016).
Several families of Blumeria effectors, including the effector targets in this study, are specifically induced at the haustorial stage (Praz et al., 2018), suggesting that temporal expression patterns of the target gene could explain the limitation of loss of virulence to this developmental stage. However, we observed the same limited impact on virulence to the haustorial stage when we used transient HIGS to target β2-tub. Thus, demonstrating that this stage of development is highly relevant to control the pathogen via HIGS, independently of temporal target expression patterns. Similarly, it was speculated previously that stable HIGS lines could cause complete loss of virulence, instead our results suggest that the effect of HIGS on fungal virulence is quantitative by nature.
The primary target of this study SvrPm3a1/f1 has first been characterized as a suppressor of the cell death triggered by the Pm3 wheat resistance gene alleles (Bourras et al., 2015, 2019). By interfering with the recognition of cognate AvrPm3 genes, SvrPm3a1/f1 reduces the resistance spectrum of Pm3 alleles. McNally et al. (2018) showed that the suppressor variant of SvrPm3a1/f1 is present in wheat powdery mildew populations globally. We therefore conclude that combining HIGS of SvrPm3a1/f1 with Pm3 resistance alleles has the potential to increase the resistance spectrum of such wheat lines.
To our knowledge, this is the first comprehensive study of ck-RNAi in Blumeria based on stable transgenic HIGS lines. We conclude that ck-RNAi has great potential to complement current pest control strategies by e.g. broadening the resistance spectra of host resistance genes. We suggest that using ck-RNAi to tap naturally occurring RNA exchanges can provide new routes for crop improvement through genetic engineering as well as classical breeding.
The United Kingdom B.g. tritici isolate Bgt_JIW2 (Wicker et al., 2013) and the Swiss B.g. hordei isolate K1 were maintained as described by Parlange et al. (2011) and Jordan et al. (2011), respectively. For high-density infection tests, inoculum for a 12 cm square plate was derived from three heavily infected 3 cm wheat leaves 10 dpi and applied using an infection tower of 25 cm height. Infected wheat leaves were scored macroscopically at 10 dpi. For low-density infection tests, inoculum was harvested at eight dpi from two mildly infected 2 cm wheat leaf segments, when the first mildew colonies start to sporulate, then dusted on leaf material in an infection tower. Infected wheat leaves were scanned six dpi using an HD-Scanner and “leaf area” and area covered by powdery mildew were estimated using Fiji1 based on color thresholding. The “infected leaf area” was calculated for each leaf by normalizing the infected area to the “leaf area”. The “relative infected leaf area” was further calculated by normalizing the “infected leaf area” to the mean infected leaf area of the non-transformed BW control from the same infection plate. For microscopy, high-density infection was used for two dpi samples and low-density infection for four and six dpi samples. Leave samples were destained (in 8% lactic acid, 16% glycerol, and 66% ethanol) and fungal structures were stained using coomassie blue. Aniline blue was used for papilla staining of a subset of two dpi samples, but since there were no observed effects on papilla formation it was omitted for the remaining samples. For each leaf, a minimum of 50 powdery mildew-wheat interactions, corresponding to an appressorium attacking an A- or B-type epidermal cell (Rubiales and Carver, 2000), were assessed for haustorium formation and shape, hyphae formation and number, and conidiophore formation and number.
The si-Fi software was used to select the donor sequences for the HIGS constructs (Lück et al., 2019). The candidate sequences were analyzed for putative off-targets in the respective donor genomes, B.g. tritici and B.g. hordei, and in the respective host genomes, wheat and barley (Mascher et al., 2017; Frantzeskakis et al., 2018; International Wheat Genome Sequencing Consortium, 2018; Müller et al., 2019). Additionally, CDS files of manually annotated SvrPm3a1/f1 family members were used for target prediction in the isolate Bgt_JIW2. The SvrPm3a1/f1-RNAi sequence was amplified from genomic DNA of the Swiss B.g. tritici isolate Bgt_96224 (Stirnweis et al., 2014) using the primers BgtSvr-RNAi-F/R (Supplementary Table 2), cloned in reverse into the pIPKTA38 entry vector according to Douchkov et al. (2005) and transferred via Gateway cloning to the pIPKb007-RNAi vector (Himmelbach et al., 2007). The β2-tub-RNAi sequence was cloned into the pIPKTA30-RNAi vector from genomic DNA of isolate K1 using the primers Bgtβ2tub-RNAi-F/R (Supplementary Table 2).
Transient HIGS was performed as described by Nowara et al. (2010). Here, the barley (Hordeum vulgare) cultivar “Golden Promise” and the wheat (Triticum aestivum) cultivar “Chancellor” were used. Three days after bombardment, leaves were infected with the barley powdery mildew isolate K1 and the wheat powdery mildew isolate Bgt_JIW2, respectively. The “haustorium index” represents the ratio of appressorium-forming spores that established a haustorium. Additionally, the presence of secondary hyphae was recorded. Results are derived from three independent experiments.
Wheat transformation and transformant selection is described in Brunner et al. (2011). The ubi:SvrPm3a1/f1-RNAi transgene was excised from the pIPKb007 plasmid using SfiI. SvrPm3a1/f1-RNAi transgene presence was confirmed by PCR using the primers BgtSvr-RNAi-F/R (Supplementary Table 2). The selected T0 seedlings were transferred to the greenhouse and allowed to self. To select transgenic events for further analysis presence of both repeat sequences, copy number and transgene expression was assessed. Genomic DNA extraction is described in Brunner et al. (2011). Antisense repeat-specific primers pIPKb007-4/BgtSvr-RNAi-F and the sense repeat-specific primers pIPKb007-5/BgtSvr-RNAi-R were used to confirm presence of both repeats by PCR (Supplementary Table 2). Copy number was assessed by southern blotting 20μg HindIII-digested genomic DNA as described in Graner et al. (1990). For the probe, the repeat sequence was amplified using the BgtSvr-RNAi-F/R primers. Transgene expression was assessed by RT-PCR on cDNA. RNA was isolated as described in Brunner et al. (2011) with the alteration that leaves were collected in tubes and ground using glass beads. cDNA was synthesized from 500 ng RNA using the iScriptTM Advanced cDNA Synthesis Kit. The qRT-BgtBcg1F primer from Bourras et al. (2015) and BgtSvr-RNAi-R were used for amplification.
Biological replicates consist of first leaves of three 10-days-old seedlings which were pooled, immediately frozen in liquid nitrogen and stored at −80°C. RNA was extracted as described above with the alteration that infected samples were ground using metal beads. For target gene expression analysis, cDNA was synthesized as described above. Primers were designed to be specific to the fungal SvrPm3a1/f1, Bgt_Bcg-6, and Bgt_Bcg-7 mRNAs (Supplementary Table 2). They were checked for specificity on plasmid DNA containing the transgene or either fungal target gene and on cDNA from infected non-transgenic and non-infected transgenic leaf material. Amplicons were verified by Sanger sequencing. B.g. tritici gapdh was used to normalize expression (Bourras et al., 2015). To further increase specificity, primer annealing was performed at 63°C and cDNA was quantified at 72°C. For consistency, these settings were used in all RT-qPCR analysis performed in this study. For transgene expression analysis, dsRNA and ssRNA were quantified according to Wang et al. (2018) with the following modifications: (1) RNA digested with RNase If was purified using the Promega SV Total RNA Isolation System, following the instructions in Promega Notes No. 86 2004 17, but using EconoSpin Micro Volume DNA/RNA Spin Columns. And (2) iScriptTM Advanced cDNA Synthesis Kit was used after hexamer incubation. qRT-BgtBcg1F/R primers from Bourras et al. (2015) were used to quantify transgene-derived transcripts. The wheat reference genes Ta2291, Ta.6863, and Ta.25640 were used to normalize transgene expression (Supplementary Table 2; Giménez et al., 2011; Hurni et al., 2013). Results represent 3–4 biological replicates. The Kapa Sybr Fast qPCR Kit and the CFX384 Real-Time PCR Detection System were used.
For the semi-field experiment we used the convertible glasshouse previously described in Romeis et al. (2007) and Boni et al. (2017) with a convertible roof programed to close in case of wind, rain, or cold. In this experimental setup the transgenic plants are retained from spreading to the environment in compliance with Swiss federal regulation. We planted a randomized setup of eight irrigated plots per event and wildtype BW, containing 10 plants each. Pre-germinated seeds were sown on March 29, 2018 into the 0.055 m2 central cylinders. More BW was sown as buffering plants around the central cylinders to reduce border effects. The soil was fertilized with 50 kg/ha P2O5, 85 kg/ha K2O, 40 kg/ha N, 11.7 kg/ha S, and 8.3 kg/ha Mg before sowing. At the start of stem elongation, test plants were reduced to 10 plants per cylinder and fertilized with 40 kg/ha N. The last fertilization with 40 kg/ha N was carried out before booting. Insecticides were applied as needed; weeding was done by hand. B.g. tritici infection was provided by natural inoculum. For AUDPC calculations, B.g. tritici disease was scored twice a week from beginning of May to end of June as described in Brunner et al. (2011). Representative leaves of adult plants were sampled per plot and infected leaf area was quantified as described above. Agronomical data was recorded per plot. 10 plants of each transgenic event and BW were also grown under greenhouse conditions. Data was recorded per plant. Plants were hand-harvested and threshed.
The statistical “exact and approximate test for proportions” in R was used to compare the β2-tub-RNAi construct results to the empty vector control. RT-qPCR experiment results were assessed for normal distribution and homogenous variance using the Shapiro–Wilk test and the Levene test, respectively. Based on this, one-way ANOVA or Welch ANOVA followed by the Tukey multiple pairwise comparison or the Kruskal–Wallis rank sum test followed by the Dunn’s post hoc test was used to test for statistical significance. Phenotyping results were assessed for normal distribution and homogenous variance using the Shapiro–Wilk test and the F-test, respectively. Based on this, the Wilcoxon rank sum, Welch t, or t-test was used to compare independent transgenic events to BW. All analyses were carried out using R software2.
All datasets generated for this study are included in the article/Supplementary Material.
LS, BK, and SB wrote the manuscript. LS, BK, SB, FP, and MM designed the experiments. LS, FP, GB, and GH performed the lab experiments. LS, EJ, JS, and RS performed the field experiments. LS, FP, TW, and SB performed the bioinformatics analyses. LS and AW performed the statistical analyses.
This project was supported by the Plant Science Center (PSC)-Syngenta Ph.D. fellowship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Susanne Brunner and her team from Agroscope in Zurich-Reckenholz for their crucial support of the semi-field trial. We also thank Dr. Teresa Koller, Dr. Javier Sanchez-Martin, and Dr. Rainer Böni for their support in the design of the semi-field trial.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00253/full#supplementary-material
Aguilar, G. B., Pedersen, C., and Thordal-Christensen, H. (2016). Identification of eight effector candidate genes involved in early aggressiveness of the barley powdery mildew fungus. Plant Pathol. 65, 953–958. doi: 10.1111/ppa.12476
Ahmed, A. A., Pedersen, C., Schultz-Larsen, T., Kwaaitaal, M., Jørgensen, H. J. L., and Thordal-Christensen, H. (2015). The barley powdery mildew candidate secreted effector protein CSEP0105 inhibits the chaperone activity of a small heat shock protein. Plant Physiol. 168, 321–333. doi: 10.1104/pp.15.00278
Ahmed, A. A., Pedersen, C., and Thordal-Christensen, H. (2016). The barley powdery mildew effector candidates CSEP0081 and CSEP0254 promote fungal infection success. PLoS One 11:e0157586. doi: 10.1371/journal.pone.0157586
Boni, R., Chauhan, H., Hensel, G., Roulin, A., Sucher, J., Kumlehn, J., et al. (2017). Pathogen-inducible Ta-Lr34res expression in heterologous barley confers disease resistance without negative pleiotropic effects. Plant Biotechnol. J. 16, 245–253. doi: 10.1111/pbi.12765
Borges, F., and Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Bourras, S., Kunz, L., Xue, M., Praz, C. R., Müller, M. C., Kälin, C., et al. (2019). The AvrPm3-Pm3 effector-NLR interactions control both race-specific resistance and host-specificity of cereal mildews on wheat. Nat. Commun. 10:2292. doi: 10.1038/s41467-019-10274-1
Bourras, S., McNally, K. E., Ben-David, R., Parlange, F., Roffler, S., Praz, C. R., et al. (2015). Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 27, 2991–3012. doi: 10.1105/tpc.15.00171
Bourras, S., Praz, C. R., Spanu, P. D., and Keller, B. (2018). Cereal powdery mildew effectors: a complex toolbox for an obligate pathogen. Curr. Opin. Microbiol. 46, 26–33. doi: 10.1016/J.MIB.2018.01.018
Brant, E. J., and Budak, H. (2018). Plant small non-coding RNAs and their roles in biotic stresses. Front. Plant Sci. 9:1038. doi: 10.3389/fpls.2018.01038
Brunner, S., Hurni, S., Herren, G., Kalinina, O., von Burg, S., Zeller, S. L., et al. (2011). Transgenic Pm3b wheat lines show resistance to powdery mildew in the field. Plant Biotechnol. J. 9, 897–910. doi: 10.1111/j.1467-7652.2011.00603.x
Cai, Q., Qiao, L., Wang, M., He, B., Lin, F., Palmquist, J., et al. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129. doi: 10.1126/science.aar4142
Calo, S., Nicolás, F. E., Vila, A., Torres-Martínez, S., and Ruiz-Vázquez, R. M. (2012). Two distinct RNA-dependent RNA polymerases are required for initiation and amplification of RNA silencing in the basal fungus Mucor circinelloides. Mol. Microbiol. 83, 379–394. doi: 10.1111/j.1365-2958.2011.07939.x
Chen, W., Kastner, C., Nowara, D., Oliveira-Garcia, E., Rutten, T., Zhao, Y., et al. (2016). Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J. Exp. Bot. 67, 4979–4991. doi: 10.1093/jxb/erw263
D’Ario, M., Griffiths-Jones, S., and Kim, M. (2017). Small RNAs: big impact on plant development. Trends Plant Sci. 22, 1056–1068. doi: 10.1016/J.TPLANTS.2017.09.009
Deng, Y., Wang, J., Tung, J., Liu, D., Zhou, Y., He, S., et al. (2018). A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 14:e1006756. doi: 10.1371/journal.ppat.1006756
Douchkov, D., Nowara, D., Zierold, U., and Schweizer, P. (2005). A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Mol. Plant Microbe Interact. 18, 755–761. doi: 10.1094/mpmi-18-0755
Dunoyer, P., Melnyk, C., Molnar, A., and Slotkin, R. K. (2013). Plant mobile small RNAs. Cold Spring Harb. Perspect. Biol. 5:a017897. doi: 10.1101/cshperspect.a017897
Frantzeskakis, L., Kracher, B., Kusch, S., Yoshikawa-Maekawa, M., Bauer, S., Pedersen, C., et al. (2018). Signatures of host specialization and a recent transposable element burst in the dynamic one-speed genome of the fungal barley powdery mildew pathogen. BMC Genomics 19:381. doi: 10.1186/s12864-018-4750-6
Gheysen, G., and Vanholme, B. (2007). RNAi from plants to nematodes. Trends Biotechnol. 25, 89–92. doi: 10.1016/J.TIBTECH.2007.01.007
Giménez, M. J., Pistón, F., and Atienza, S. G. (2011). Identification of suitable reference genes for normalization of qPCR data in comparative transcriptomics analyses in the Triticeae. Planta 233, 163–173. doi: 10.1007/s00425-010-1290-y
Graner, A., Siedler, H., Jahoor, A., Herrmann, R. G., and Wenzel, G. (1990). Assessment of the degree and the type of restriction fragment length polymorphism in barley (Hordeum vulgare). Theor. Appl. Genet. 80, 826–832. doi: 10.1007/BF00224200
Guo, X.-Y., Li, Y., Fan, J., Xiong, H., Xu, F.-X., Shi, J., et al. (2019). Host-induced gene silencing of MoAP1 confers broad-spectrum resistance to Magnaporthe oryzae. Front. Plant Sci. 10:433. doi: 10.3389/fpls.2019.00433
Himmelbach, A., Zierold, U., Hensel, G., Riechen, J., Douchkov, D., Schweizer, P., et al. (2007). A set of modular binary vectors for transformation of cereals. Plant Physiol. 145, 1192–1200. doi: 10.1104/pp.107.111575
Hua, C., Zhao, J.-H., and Guo, H.-S. (2018). Trans-kingdom RNA silencing in plant–fungal pathogen interactions. Mol. Plant 11, 235–244. doi: 10.1016/J.MOLP.2017.12.001
Hurni, S., Brunner, S., Buchmann, G., Herren, G., Jordan, T., Krukowski, P., et al. (2013). Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 76, 957–969. doi: 10.1111/tpj.12345
International Wheat Genome Sequencing Consortium (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/SCIENCE.AAR7191
Jordan, T., Seeholzer, S., Schwizer, S., Töller, A., Somssich, I. E., and Keller, B. (2011). The wheat Mla homologue TmMla1 exhibits an evolutionarily conserved function against powdery mildew in both wheat and barley. Plant J. 65, 610–621. doi: 10.1111/j.1365-313X.2010.04445.x
Knip, M., Constantin, M. E., and Thordal-Christensen, H. (2014). Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet. 10:e1004602. doi: 10.1371/journal.pgen.1004602
Koch, A., and Kogel, K. H. (2014). New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol. J. 12, 821–831. doi: 10.1111/pbi.12226
Koch, A., Kumar, N., Weber, L., Keller, H., Imani, J., and Kogel, K.-H. (2013). Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species. PNAS 110, 19324–19329. doi: 10.1073/pnas.1306373110
Kusch, S., Frantzeskakis, L., Thieron, H., and Panstruga, R. (2018). Small RNAs from cereal powdery mildew pathogens may target host plant genes. Fungal Biol. 122, 1050–1063. doi: 10.1016/j.funbio.2018.08.008
Kwaaitaal, M., Nielsen, M. E., Böhlenius, H., and Thordal-Christensen, H. (2017). The plant membrane surrounding powdery mildew haustoria shares properties with the endoplasmic reticulum membrane. J. Exp. Bot. 68, 5731–5743. doi: 10.1093/jxb/erx403
Lück, S., Kreszies, T., Strickert, M., Schweizer, P., Kuhlmann, M., and Douchkov, D. (2019). siRNA-Finder (si-Fi) software for RNAi-target design and off-target prediction. Front. Plant Sci. 10:1023. doi: 10.3389/fpls.2019.01023
Mao, W., Li, Z., Xia, X., Li, Y., and Yu, J. (2012). A combined approach of high-throughput sequencing and degradome analysis reveals tissue specific expression of microRNAs and their targets in cucumber. PLoS One 7:e33040. doi: 10.1371/journal.pone.0033040
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S. O., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. doi: 10.1038/nature22043
McNally, K. E., Menardo, F., Lüthi, L., Praz, C. R., Müller, M. C., Kunz, L., et al. (2018). Distinct domains of the AVRPM3A2/F2 avirulence protein from wheat powdery mildew are involved in immune receptor recognition and putative effector function. New Phytol. 218, 681–695. doi: 10.1111/nph.15026
Mohorianu, I., Schwach, F., Jing, R., Lopez-Gomollon, S., Moxon, S., Szittya, G., et al. (2011). Profiling of short RNAs during fleshy fruit development reveals stage-specific sRNAome expression patterns. Plant J. 67, 232–246. doi: 10.1111/j.1365-313X.2011.04586.x
Muhammad, T., Zhang, F., Zhang, Y., and Liang, Y. (2019). RNA interference: a natural immune system of plants to counteract biotic stressors. Cells 8:38. doi: 10.3390/cells8010038
Müller, M. C., Praz, C. R., Sotiropoulos, A. G., Menardo, F., Kunz, L., Schudel, S., et al. (2019). A chromosome−scale genome assembly reveals a highly dynamic effector repertoire of wheat powdery mildew. New Phytol. 221, 2176–2189. doi: 10.1111/nph.15529
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Panwar, V., Jordan, M., McCallum, B., and Bakkeren, G. (2017). Host-induced silencing of essential genes in Puccinia triticina through transgenic expression of RNAi sequences reduces severity of leaf rust infection in wheat. Plant Biotechnol. J. 16, 1013–1023. doi: 10.1111/pbi.12845
Parlange, F., Oberhaensli, S., Breen, J., Platzer, M., Taudien, S., Šimková, H., et al. (2011). A major invasion of transposable elements accounts for the large size of the Blumeria graminis f. sp. tritici genome. Funct. Integr. Genomics 11, 671–677. doi: 10.1007/s10142-011-0240-5
Parlange, F., Roffler, S., Menardo, F., Ben-David, R., Bourras, S., McNally, K. E., et al. (2015). Genetic and molecular characterization of a locus involved in avirulence of Blumeria graminis f. sp. tritici on wheat Pm3 resistance alleles. Fungal Genet. Biol. 82, 181–192. doi: 10.1016/j.fgb.2015.06.009
Pedersen, C., van Themaat, E. V. L., McGuffin, L. J., Abbott, J. C., Burgis, T. A., Barton, G., et al. (2012). Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 13:694. doi: 10.1186/1471-2164-13-694
Pennington, H. G., Gheorghe, D. M., Damerum, A., Pliego, C., Spanu, P. D., Cramer, R., et al. (2016). Interactions between the powdery mildew effector BEC1054 and barley proteins identify candidate host targets. J. Proteome Res. 15, 826–839. doi: 10.1021/acs.jproteome.5b00732
Pennington, H. G., Jones, R., Kwon, S., Bonciani, G., Thieron, H., Chandler, T., et al. (2019). The fungal ribonuclease-like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathog. 15:e1007620. doi: 10.1371/journal.ppat.1007620
Pliego, C., Nowara, D., Bonciani, G., Gheorghe, D. M., Xu, R., Surana, P., et al. (2013). Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. MPMI 26, 633–642. doi: 10.1094/MPMI-01-13-0005-R
Praz, C. R., Menardo, F., Robinson, M. D., Müller, M. C., Wicker, T., Bourras, S., et al. (2018). Non-parent of origin expression of numerous effector genes indicates a role of gene regulation in host adaption of the hybrid triticale powdery mildew pathogen. Front. Plant Sci. 9:49. doi: 10.3389/fpls.2018.00049
Qi, T., Guo, J., Peng, H., Liu, P., Kang, Z., and Guo, J. (2019). Host-induced gene silencing: a powerful strategy to control diseases of wheat and barley. Int. J. Mol. Sci 20:206. doi: 10.3390/ijms20010206
Romeis, J., Waldburger, M., Streckeisen, P., Hogervorst, P. A. M., Keller, B., Winzeler, M., et al. (2007). Performance of transgenic spring wheat plants and effects on non-target organisms under glasshouse and semi-field conditions. J. Appl. Entomol. 131, 593–602. doi: 10.1111/j.1439-0418.2007.01231.x
Rosa, C., Kuo, Y.-W., Wuriyanghan, H., and Falk, B. W. (2018). RNA interference mechanisms and applications in plant pathology. Annu. Rev. Phytopathol. 56, 581–610. doi: 10.1146/annurev-phyto-080417-050044
Rubiales, D., and Carver, T. L. (2000). Defence reactions of Hordeum chilense accessions to three formae speciales of cereal powdery mildew fungi. Can. J. Bot. 78, 1561–1570. doi: 10.1139/b00-132
Shabalina, S. A., and Koonin, E. V. (2008). Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 23, 578–587. doi: 10.1016/J.TREE.2008.06.005
Shahid, S., Kim, G., Johnson, N. R., Wafula, E., Wang, F., Coruh, C., et al. (2018). MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553, 82–85. doi: 10.1038/nature25027
Stirnweis, D., Milani, S. D., Jordan, T., Keller, B., and Brunner, S. (2014). Substitutions of two amino acids in the nucleotide-binding site domain of a resistance protein enhance the hypersensitive response and enlarge the PM3F resistance spectrum in wheat. MPMI 27, 265–276. doi: 10.1094/MPMI-10-13-0297-FI
Torres-Martínez, S., and Ruiz-Vázquez, R. M. (2017). The RNAi universe in fungi: a varied landscape of small RNAs and biological functions. Annu. Rev. Microbiol. 71, 371–391. doi: 10.1146/annurev-micro-090816-093352
Vela-Corcía, D., Romero, D., de Vicente, A., and Pérez-García, A. (2018). Analysis of β-tubulin-carbendazim interaction reveals that binding site for MBC fungicides does not include residues involved in fungicide resistance. Sci. Rep. 8:7161. doi: 10.1038/s41598-018-25336-5
Villarroya-Beltri, C., Gutiérrez-Vázquez, C., Sánchez-Cabo, F., Pérez-Hernández, D., Vázquez, J., Martin-Cofreces, N., et al. (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 4:2980. doi: 10.1038/ncomms3980
Wang, M., Weiberg, A., Dellota, E., Yamane, D., and Jin, H. (2017). Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 14, 421–428. doi: 10.1080/15476286.2017.1291112
Wang, M., Weiberg, A., Lin, F.-M., Thomma, B. P. H. J., Huang, H.-D., and Jin, H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2:16151. doi: 10.1038/nplants.2016.151
Wang, P.-H., Schulenberg, G., Whitlock, S., Worden, A., Zhou, N., Novak, S., et al. (2018). RNase If-treated quantitative PCR for dsRNA quantitation of RNAi trait in genetically modified crops. BMC Biotechnol. 18:3. doi: 10.1186/s12896-018-0413-6
Weiberg, A., Wang, M., Lin, F.-M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science (80-.). 342, 118–123. doi: 10.1126/science.1239705
Wicker, T., Oberhaensli, S., Parlange, F., Buchmann, J. P., Shatalina, M., Roffler, S., et al. (2013). The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat. Genet. 45, 1092–1096. doi: 10.1038/ng.2704
Zhang, W.-J., Pedersen, C., Kwaaitaal, M., Gregersen, P. L., Morch, S. M., Hanisch, S., et al. (2012). Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Mol. Plant Pathol. 13, 1110–1119. doi: 10.1111/j.1364-3703.2012.00820.x
Keywords: cross-kingdom RNAi, ck-RNAi, host-induced gene silencing, HIGS, ribonuclease-like effectors, effectors, Blumeria graminis, wheat
Citation: Schaefer LK, Parlange F, Buchmann G, Jung E, Wehrli A, Herren G, Müller MC, Stehlin J, Schmid R, Wicker T, Keller B and Bourras S (2020) Cross-Kingdom RNAi of Pathogen Effectors Leads to Quantitative Adult Plant Resistance in Wheat. Front. Plant Sci. 11:253. doi: 10.3389/fpls.2020.00253
Received: 15 November 2019; Accepted: 18 February 2020;
Published: 10 March 2020.
Edited by:
Bruno Mezzetti, Marche Polytechnic University, ItalyReviewed by:
Elena Baraldi, University of Bologna, ItalyCopyright © 2020 Schaefer, Parlange, Buchmann, Jung, Wehrli, Herren, Müller, Stehlin, Schmid, Wicker, Keller and Bourras. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beat Keller, YmtlbGxlckBib3RpbnN0LnV6aC5jaA==; Salim Bourras, c2FsaW0uYm91cnJhc0BzbHUuc2U=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.