
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 03 March 2020
Sec. Plant Biotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00231
This article is part of the Research Topic Microalgae Biology and Biotechnology View all 15 articles
 Young-Saeng Kim1
Young-Saeng Kim1 Seong-Im Park2,3
Seong-Im Park2,3 Jin-Ju Kim2,3
Jin-Ju Kim2,3 Joseph S. Boyd4
Joseph S. Boyd4 Joris Beld5
Joris Beld5 Arnaud Taton4
Arnaud Taton4 Kyoung-In Lee6
Kyoung-In Lee6 Il-Sup Kim7*
Il-Sup Kim7* James W. Golden4*
James W. Golden4* Ho-Sung Yoon2,3,7*
Ho-Sung Yoon2,3,7*An excess of reactive oxygen species (ROS) can cause severe oxidative damage to cellular components in photosynthetic cells. Antioxidant systems, such as the glutathione (GSH) pools, regulate redox status in cells to guard against such damage. Dehydroascorbate reductase (DHAR, EC 1.8.5.1) catalyzes the glutathione-dependent reduction of oxidized ascorbate (dehydroascorbate) and contains a redox active site and glutathione binding-site. The DHAR gene is important in biological and abiotic stress responses involving reduction of the oxidative damage caused by ROS. In this study, transgenic Synechococcus elongatus PCC 7942 (TA) was constructed by cloning the Oryza sativa L. japonica DHAR (OsDHAR) gene controlled by an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible promoter (Ptrc) into the cyanobacterium to study the functional activities of OsDHAR under oxidative stress caused by hydrogen peroxide exposure. OsDHAR expression increased the growth of S. elongatus PCC 7942 under oxidative stress by reducing the levels of hydroperoxides and malondialdehyde (MDA) and mitigating the loss of chlorophyll. DHAR and glutathione S-transferase activity were higher than in the wild-type S. elongatus PCC 7942 (WT). Additionally, overexpression of OsDHAR in S. elongatus PCC 7942 greatly increased the glutathione (GSH)/glutathione disulfide (GSSG) ratio in the presence or absence of hydrogen peroxide. These results strongly suggest that DHAR attenuates deleterious oxidative effects via the glutathione (GSH)-dependent antioxidant system in cyanobacterial cells. The expression of heterologous OsDHAR in S. elongatus PCC 7942 protected cells from oxidative damage through a GSH-dependent antioxidant system via GSH-dependent reactions at the redox active site and GSH binding site residues during oxidative stress.
The maintenance of cellular redox balance requires a steady-state equilibrium between oxidants and antioxidants. Under oxidative stress, loss of detoxification and accumulation of excess reactive oxygen species (ROS) leads to cell death through disruptions to redox balance, subsequent signaling, and cell rescue systems (Boyer, 1982; Owens, 2001; Rouhier et al., 2008). In aerobic cells, ROS degradation occurs through the GSH pools, which is catalyzed by a series of antioxidative enzymes localized primarily in intracellular and extracellular areas with high ROS production (Potters et al., 2002). Dehydroascorbate reductases (DHARs) are important to the GSH-dependent reduction of dehydroascorbate (DHA) to ascorbate, and their activities are shown to affect redox homeostasis in higher plants (Shimaoka et al., 2000; Urano et al., 2000; Dixon et al., 2002; Dixon and Edwards, 2010). The presence of a DHAR gene has yet to be conclusively established in the cyanobacterium Synechococcus elongatus PCC 7942.
In previous studies, we initially predicted that the GSH-bound Oryza sativa L. japonica DHAR (OsDHAR) structure might contain a GSH binding site (Do et al., 2016). A subsequent study reported the pronounced similarity of GSH binding sites in the conserved residues of glutathione S-transferases (GSTs), such as thioredoxin-like sites found in different species (Abdul Kayum et al., 2018). These residues indicate that the DHARs are GST-like proteins with GSH-dependent thiol transferase activity, with a cysteine at the active site instead of serine or tyrosine (Dixon et al., 2002). In DHAR activity, the first cysteine residue in the GSH binding site reacts with the sulfur atom in the GSH mixed disulfide bridges and promotes thiol transfer (Dixon et al., 2002; Dixon and Edwards, 2010). Mutation of the first cysteine residue induces significant losses of catalytic efficiency and GSH binding affinity at GSH binding site residues during abiotic stresses (Tang and Yang, 2013; Do et al., 2016).
There is also growing evidence that overexpression of DHAR enhances tolerance to many abiotic stresses. Overexpression of wheat DHAR conferred protection from ozone in transgenic tobacco (Chen and Gallie, 2005), and reinforcement of tobacco with the human DHAR gene increased tolerance to low temperatures and salt stress (Kwon et al., 2003). Overexpression of rice DHAR increased salt tolerance in Arabidopsis (Ushimaru et al., 2006) and resistance to oxidative stress in Escherichia coli (Shin et al., 2008). Overexpression of Arabidopsis cytosolic DHAR enhanced tolerance to drought and ozone stress in tobacco plants (Eltayeb et al., 2006), and overexpression of homologous OsDHAR containing a redox active site and GSH binding site residues in transgenic rice improved environmental adaptation and rice productivity in paddy fields (Kim et al., 2013). Although the relationship between DHAR expression and stress tolerance in plants is supported by a great deal of evidence, there is little information concerning the effect of DHAR overexpression in cyanobacteria. Cyanobacteria are believed to be the ancestors of photosynthetic eukaryotes as a result of an ancient endosymbiosis (Blank, 2013). The unicellular S. elongatus PCC 7942 has been used extensively as a cyanobacterial model for biochemical, physiological, and genetic studies of important cellular processes, such as prokaryotic nitrate reductases, cell division, responses to nutrient depletion, iron deprivation, and environmental abiotic stresses, such as ambient temperature and light intensity (Koksharova et al., 2006).
This study was undertaken to characterize biochemical and physiological features of OsDHAR-mediated stress tolerance in transgenic S. elongatus PCC 7942 under ROS-induced oxidative stress conditions. We found that the expression of heterologous OsDHAR in S. elongatus PCC 7942 conferred resistance to oxidative stress by helping to maintain cellular redox homeostasis through a GSH-dependent antioxidant system via GSH-dependent reactions. The multifaceted approach we employed provides new insights into the mechanisms of gene family expansion and functional evolution.
BLAST software1 was used to align OsDHAR with known DHAR sequences using the National Center for Biotechnology Information (NCBI) database. Amino acid sequences were as follows: O. sativa DHAR (OsDHAR; accession no. AAL71856.1), S. elongatus PCC 7942 GST (SeGST; accession no. WP_011243077.1), Synechocystis sp. PCC6803 GST (StGST; accession no. WP_010872521.1), Gloeobacter violaceus PCC 7421 GST (GvGST; accession no. WP_011143013.1), Prochlorococcus marinus MIT9313 GST (PcGST; accession no. WP_011124721.1), Acaryochloris marina MBIC11017 GST (AmGST; accession no. WP_012161393.1), and Cyanothece sp. ATCC 51142 GST (CtGST; accession no. WP_009546792.1). The GSTs from these cells are soluble monomeric enzymes that contain a GSH-binding redox active site and similarly functional conserved residues comprised of different amino acids in other active sites.
The DHAR gene from O. sativa L. japonica (accession no. AY074786.1; OsDHAR) coding region was amplified from cDNA using PCR with ExTaq polymerase (Takara Bio Inc., Shiga, Japan). PCR reaction conditions were as follows: initial denaturation at 95°C for 3 min, followed by 30 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 1 min, and a final extension for 5 min at 72°C. The OsDHAR gene was cloned by PCR using OsDHAR-F-SwaI and OsDHAR-R-SwaI as the sense and antisense primers, respectively (Supplementary Table S2). The insertion of heterologous OsDHAR in S. elongatus PCC 7942 was constructed using the pAM4957 (pCV0069) a conjugative plasmid vector for chromosomal integration into recombinant complementation constructs at neutral site II (NS2).
Competent E. coli DH5α cells that had been conjugated by the pAM4957:OsDHAR plasmid were selected using nourseothricin (cloNAT, 50 μg mL–1). Clones were confirmed by sequencing, then used to conjugate S. elongatus PCC 7942. The plasmids harboring the OsDHAR encoding gene were sequenced using neutral site II (NS2) primers (Supplementary Tables S1, S2) complementary to the NS2 region to confirm proper gene ligation and direction. Finally, the resulting plasmid was assembled using the GeneArt Seamless Cloning and Assembly Kit (Life Technologies, Carlsbad, CA, United States). Plasmids harboring the OsDHAR encoding gene were constructed by recombinant complementation, and chromosomal integration into S. elongatus PCC 7942 NS2 using published protocols (Golden et al., 1987). Integration of the pAM4957:OsDHAR transgene into genomic DNA of the S. elongatus PCC 7942 was verified by PCR using Ptrc-F and OsDHAR-R primers (Supplementary Table S2) and the PCR PreMix kit (Bioneer, Daejeon, South Korea) according to the manufacturer’s instructions.
Cyanobacteria were cultured in BG11 medium at 30°C in continuous light (80–120 μE m–2s–1) on a rotary shaker (120 rpm) for 7 days in Erlenmeyer flask. Then, the TA, EV, and WT S. elongatus PCC 7942 were cultured in liquid BG11 medium containing 2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 5 days before 2.5 mM H2O2 treatment at 7 days with shaking. The TA, and EV S. elongatus PCC 7942 were maintained in BG11 liquid or solid media supplemented with the appropriate antibiotics.
Initial absorbances at 730 nm (A730) were adjusted to 0.03. Aliquots of each sample were then measured to adjust them to 0.03 in BG11 medium after culturing for 18 days at 30°C with shaking in liquid microbial lab flasks (Supplementary Figure S2A). Responses to oxidative stress were evaluated by allowing cells to reach the exponential phase (A730 = 0.3–0.35) before exposing them to 2.5 mM H2O2 at 7 days with shaking, after which A730 values were monitored each day through the end of the experiment.
For the cell-spotting assay, the growth following the presence (10 days) or absence (7 days) of 2.5 mM H2O2 with 2 mM IPTG for 3 days were measured at the TA, EV, and WT S. elongatus PCC 7942 (A730 = 0.25) and were then serially diluted 10-fold with distilled water. The diluted samples were spotted on agar-solidified BG11 medium and incubated for 3 days at 30°C. They were then photographed or counted. All experiments were performed three or more times.
The cDNA was reverse transcribed from total RNA using the SuperScript III kit (Life Technologies). For semi-qRT-PCR, the OsDHAR-qRT-PCR-F and OsDHAR-qRT-PCR-R (Supplementary Table S2) primer sets were used to amplify OsDHAR. The reaction sequence consisted of one cycle at 95°C for 3 min followed by 26 cycles at 94°C for 30 s, 54°C for 30 s, and 72°C for 40 s, and then a final extension at 72°C for 5 min. The semi-qRT-PCR amplicons were resolved on a 1.0% agarose gel by electrophoresis in 0.5X Tris/borate/EDTA buffer. The rpoA gene primer sets were used as a housekeeping control and were also amplified (Supplementary Table S2).
Western blot analysis of the crude extracts was performed by obtaining the total protein from cyanobacteria (A730 = 0.3) with suspension in five volumes of cold extraction buffer containing 150 mM NaCl, 50 mM Tris-HCl (pH 7.3), 1 mM EDTA, 2% β-mercaptoethanol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride (PMSF), and an equal of volume of phenol saturated with Tris-HCl (pH 7.3). Protein was then extracted from the cells as previously described (Kim et al., 2018).
Briefly, crude protein extracts were prepared using glass beads. Cells grown for 7 days were exposed to 2.5 mM H2O2 for 24 h to induce cellular oxidative stress, followed by vigorous vortexing 10 times for 1 min each at 3-min intervals on ice. The protein extracts were cleared by centrifugation at 12,000 rpm for 20 min at 4°C. Finally, protein concentrations were determined using a Pierce bicinchoninic acid protein assay kit (Thermo Scientific, Waltham, MA, United States) (Komatsu et al., 2009; Peng et al., 2011).
Protein extracts (20 μg) were separated on a 10% SDS-PAGE gel at 100 V and transferred to polyvinylidene fluoride membranes (Bio-Rad, Hercules, CA, United States). Membranes were then incubated in blocking buffer consisting of 5% non-fat skim milk and 0.02% sodium azide in Tris-buffered saline plus Tween [TBST, 10% Tween-20, 20 mM Tris-HCl (pH 7.6), and 150 mM NaCl] for 1.5 h at room temperature. Antibodies to O. sativa DHAR (OsDHAR) were produced in rabbits hyperimmunized using proteins purified from E. coli (Younginfrontier, Seoul, South Korea). The blots were washed three times for 30 min with TBST, after which they were incubated with conjugated anti-rabbit secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA, United States) diluted with blocking buffer (without 0.02% sodium azide) for 4 h at room temperature. After washing with TBST, binding of antibody to protein immobilized on the blots was visualized using the SuperSignal West Femto substrate kit (Pierce, Rockford, IL, United States) and imaged using a MultiImage II Light Cabinet (DE-500; Alpha Innotech Corporation, San Leandro, CA, United States).
Chlorophyll content assays were conducted by harvesting the TA, EV, and WT S. elongatus PCC 7942 from a 100 mL culture at an A730 of 0.3 after a 2-day exposure to 2.5 mM H2O2 in liquid BG11 medium containing 2 mM IPTG. Chlorophyll content was analyzed by treating cultures (100 mL, A730 = 0.3) with 2.5 mM H2O2 for 2 days. The cultivated cells were harvested, and the cell pellets were suspended in 90% methanol. Chlorophyll was extracted as previously described (Kim et al., 2017, 2018) with some modifications. The chlorophyll content of the cells was measured by scanning them over 260 to 800 nm using an Infinite M200 Pro microplate reader (Tecan, Männedorf, Switzerland). All assays were performed in biologically least three independent experiments. The dry weights of cells at 9 days after 2-day exposure to H2O2 were determined gravimetrically as previously described (Gorain et al., 2013; Kim et al., 2017). Other dry weight samples were measured at 14 days after growth in the presence or absence of 2.5 mM H2O2 for 7 days (Supplementary Figure S2B).
The intracellular levels of H2O2 in cells exposed to H2O2 with IPTG (H2O2-IPTG) for 24 h were determined using the ferrous oxidation-xylenol orange (FOX) reagent (100 μM xylenol orange, 250 μM ammonium ferrous sulfate, 100 mM sorbitol, and 25 mM sulfuric acid) by ferrous ion oxidation in the presence of the ferric ion indicator xylenol orange (Kim et al., 2011, 2018).
Cellular ROS levels were measured in vivo by harvesting of the TA, EV, and WT S. elongatus PCC 7942 from 100 mL culture (A730 = 0.3) after exposure to 2.5 mM H2O2 -IPTG with 2 mM IPTG for 2 h; these were incubated for 20 min at 30°C with 10 μM DCFHDA (Invitrogen, Carlsbad, CA, United States) in the dark (Wang and Joseph, 1999; Kim et al., 2018). Cells were then collected and examined using a model LSM700 fluorospectrophotometer (Carl Zeiss, Jena, Germany) at an excitation wavelength of 488 nm and an emission wavelength of 535 nm. Images were also captured by laser scanning confocal microscopy at an excitation wavelength of 488 nm (Huang et al., 2012). Levels of lipid peroxidation were evaluated in cells exposed to H2O2-IPTG with 2 mM IPTG for 24 h. The extent of oxidative damage to the lipids was determined by measuring malondialdehyde (MDA) content; this was achieved adding 10% trichloroacetic acid containing 0.65% 2-thiobarbituric acid (TBA) and heating at 95°C for 25 min. Finally, the MDA concentration of the resulting supernatant was measured at 532 nm (based on 600 nm) and estimated using an absorbance coefficient of 1.56 × 105, as described by Jiang and Zhang (2001) and Kim et al. (2018).
We evaluated potential mechanisms by which the TA tolerates oxidative stress by measuring GSH/GSSG ratio as they related to total GSH content during a 24-h exposure to H2O2. The TA, EV, and WT S. elongatus PCC 7942 were harvested by centrifugation and then washed twice with cold phosphate-buffered saline (PBS). We then measured GST activity by adding the crude protein of samples exposed to the H2O2 stress for 24 h to a lysis buffer containing 50 mM sodium phosphate (pH 7.5), 3 mM MgCl2, 1 mM EDTA, 1 mM PMSF, and protease inhibitor cocktails. Total GSH content was measured as previously described (Jiang and Zhang, 2001). GST enzyme activity was detected as previously described using 1-chloro-2,4- dinitrobenzene as substrate (Dixit et al., 2015).
An RNA extraction of 3 different samples exposed to 2.5 mM H2O2 for 24 h for biologically least three independent experiments. Samples were frozen with liquid nitrogen and ground using a mortar and pestle. Total RNA was extracted using TRIzol reagent (Takara, Japan) according to the manufacturer’s instructions, and reverse transcription reactions were performed with 2 μg total RNA using the SuperScript III First-Strand Synthesis System. The qRT-PCR analysis was performed using the TB Green Premix Ex Taq II (Tli RNaseH Plus) Kit (Takara, Japan) and QuantStudio3 Real-Time PCR system (Applied Biosystems, Foster City, CA, United States). The 20 μL reactions contained 12.5 μL of TB Green Premix Ex Taq II (Tli RNaseH Plus), 0.4 μL of ROX Reference Dye II, 200 ng of cDNA template, 0.4 μL of each primer (10 μM), and 6.26 μL of PCR-grade water. All samples were analyzed in biological triplicates as follows: initial preheating at 95°C for 30 s, followed by 40 cycles of 95°C for 3 s and 55°C for 30 s. The rpoA gene primer sets were used as a positive control. Fold changes in mRNA were calculated relative to the calibrator. Expression levels of GSH-related genes in Figure 6 is presented relative to that in TA strains (which expression level was considered to be 1 of WT strains; described as relative values.) under stress condition (Supplementary Figure S3). Normalization and relative quantification were conducted using the 2–ΔΔCt method (Livak and Schmittgen, 2001). The primers used for qRT-PCR analysis are listed in the Supplementary Table S2.
All biochemical results from the TA were calculated relative to those of the EV and WT under normal conditions, which were defined as 100%. Significant differences among results for the TA, EV, and WT were determined by ANOVA. Comparisons between individual data points were performed using Student’s t-test, and a P-value of less than 0.05 was considered statistically significant. All experiments were carried out three or more times, and all results are expressed as mean ± SD.
The amino acid sequence motifs of OsDHAR were compared with the sequences of known GSTs to clarify the molecular properties of OsDHAR. Pairwise alignment of the sequences of OsDHAR and the other GSTs were conducted using BLAST software. OsDHAR displayed 24, 26, 25, 26, 20, and 23% similarity to SeGST, StGST, GvGST, PcGST, AmGST, and CtGST, respectively (Figure 1A, see section “Materials and Methods” for details of the GSTs). All contained a conserved site that bound GSH residues (Cys20, Pro61, and Ser74) (Figure 1A, red background) and similar functionally conserved residues (Pro21, Lys34, Lys47, Val60, and Asp73) (Figure 1A, blue background) that are likely involved in redox active site function. Other strictly conserved residues were not clearly evident (Figure 1A, green boxes). We used these results to construct a phylogenetic tree containing the S. elongatus PCC 7942 GST (SeGST; accession no. WP_011243077) that was 24% homologous to OsDHAR (accession no. AAL71856.1) (Figure 1B).
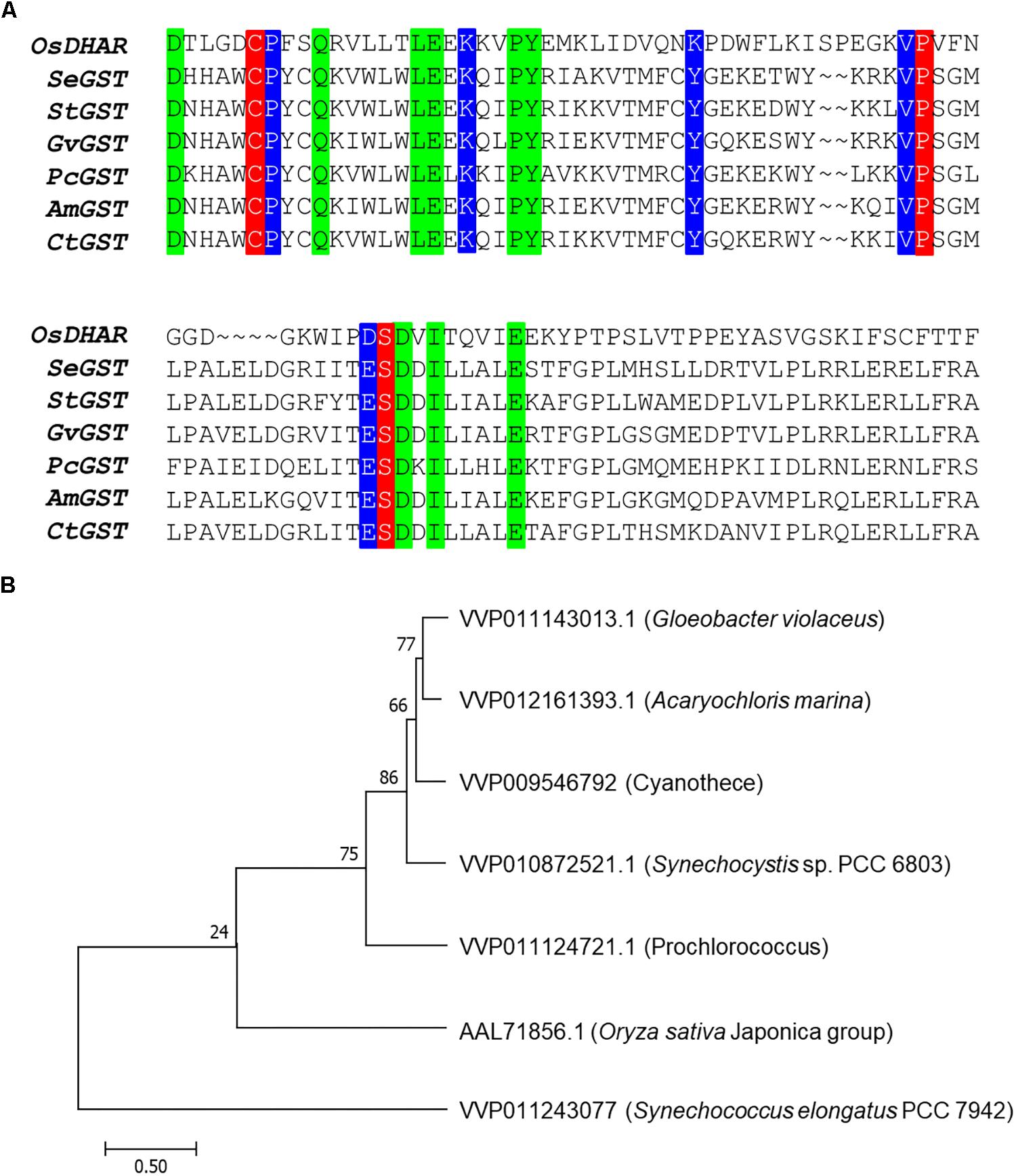
Figure 1. Amino acid alignment of OsDHAR and six glutathione S-transferase (GSTs). (A) Alignment showing GSH binding site and GSTs with similarly functional conserved residues. Colored boxes indicate identical residues across sequences. All contain a conserved site with GSH binding residues (red background), GSTs having similar functional conserved residues (blue background), and strictly conserved residues (green boxes). (B) The phylogenetic trees show the evolutionary relationship between rice DHAR and the amino acids of cyanobacterium GSTs. OsDHAR, Oryza sativa DHAR; SeGST, Synechococcus elongatus PCC 7942 GST; StGST, Synechocystis sp. PCC6803 GST; GvGST, Gloeobacter violaceus PCC 7421 GST; PcGST, Prochlorococcus marinus MIT9313 GST; AmGST, Acaryochloris marina MBIC11017 GST; CtGST, Cyanothece sp. ATCC 51142 GST.
The expression of heterologous OsDHAR in S. elongatus PCC 7942 was constructed to evaluate the effect on tolerance to hydrogen peroxide (H2O2)-induced oxidative stress. OsDHAR was cloned into the pAM4957 (pCV0069) vector for NS2 chromosomal integration under the control of the IPTG-inducible promoter (Ptrc) (Figure 2A). Resistance to oxidative stress was evaluated in cyanobacterial S. elongatus PCC 7942 conjugated with the pAM4957:OsDHAR plasmid containing the OsDHAR gene or the pAM4957 vector alone plasmid containing the empty neutral site vector (EV) (Supplementary Table S1). Genotypes were confirmed by PCR using primers external to the OsDHAR gene (Supplementary Table S2) that produced the expected fragment size of 690 bp (Figure 2B and Supplementary Table S1).
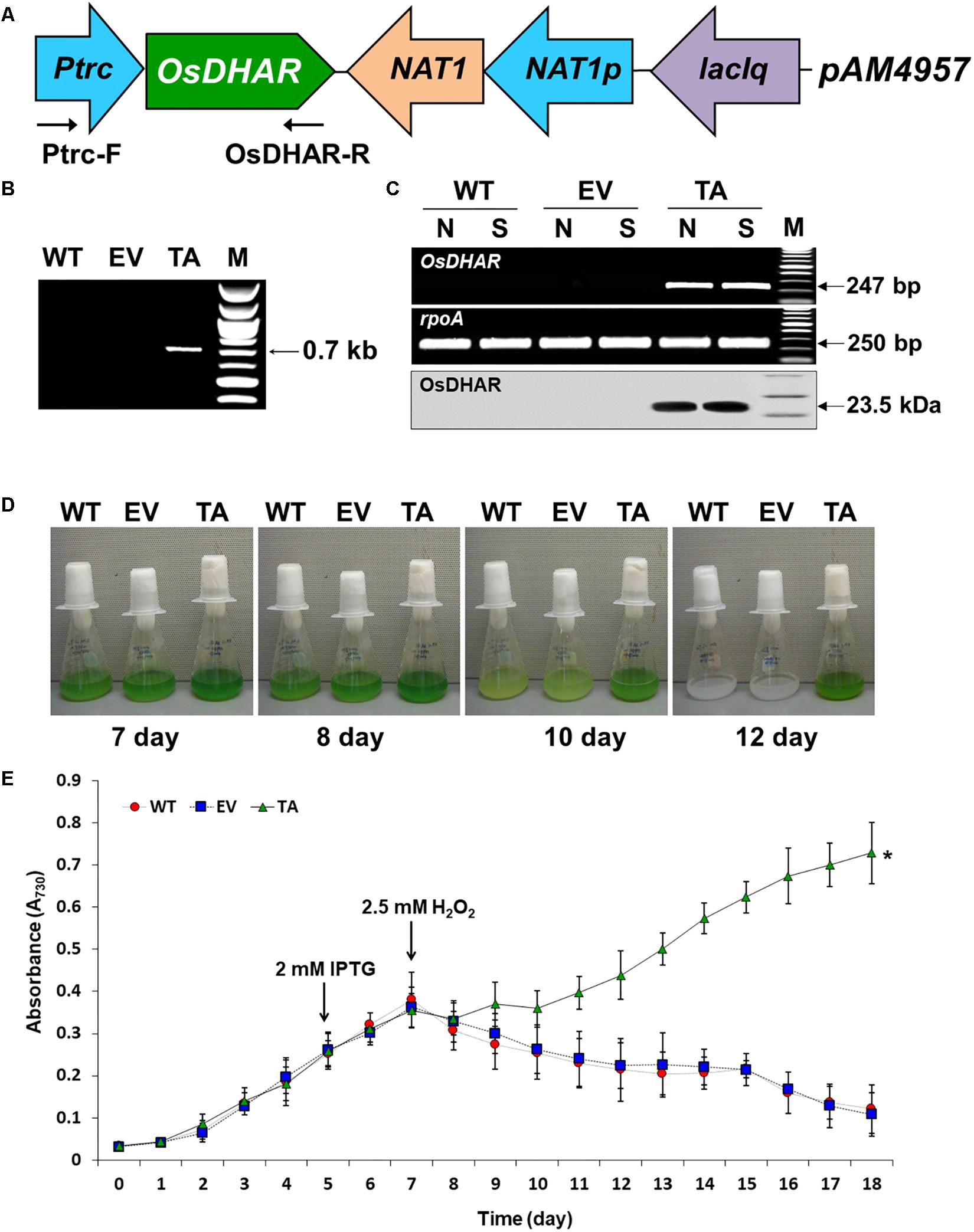
Figure 2. Expression of the OsDHAR gene in the S. elongatus PCC 7942. (A) Schematic showing expression of OsDHAR in of S. elongatus PCC 7942. Ptrc, IPTG-inducible promoter; OsDHAR, dehydroascorbate reductase gene of O. sativa; NAT, antibiotic nourseothricin selection marker (cloNAT); and LacIq, lactose repressor promoter. Arrows indicate the direction of each gene. (B,C) PCR results verifying OsDHAR gene integration in the genome of the transgenic TA. (B) PCR of OsDHAR gene in transgenic TA to confirm genotype (690 bp). (C) Expression of the OsDHAR gene was confirmed by semi-quantitative RT-PCR (247 bp) and Western blot (23.5 kDa). N, normal conditions; S, stress conditions. The rpoA transcript was used as a control standard in the analysis (250 bp). (D,E) Growth profiles of the TA, EV, and WT S. elongatus PCC 7942. Growth levels of the different S. elongatus PCC 7942 were compared to determine whether the TA grew better under H2O2 stress conditions. After 7 days of growth under standard conditions, the TA, EV, and WT S. elongatus PCC 7942 were adjusted to an absorbance at 730 nm of 0.3 to 0.35 and then grown in BG11 medium containing 2.5 mM H2O2, then cultured for 18 days at 30°C with shaking in liquid microbial lab flasks. WT, WT S. elongatus PCC 7942; EV, empty vector S. elongatus PCC 7942; TA, TA S. elongatus PCC 7942. Error bars indicate ± SD of biologically least three independent experiments. Asterisks indicate significant differences between treatments as estimated by Student’s t-test (P < 0.05).
We explored whether the OsDHAR gene was effectively expressed with a semi-quantitative RT-PCR analysis using primers internal to the OsDHAR gene (Supplementary Table S2). The OsDHAR gene was expressed in the TA, while no signal was detected in the empty vector (EV, pAM4957) or the wild-type (WT) S. elongatus PCC 7942 (Figure 2C and Supplementary Table S1). We also performed Western blot analyses to determine whether the OsDHAR gene was properly translated. The TA grown under H2O2 stress conditions presented with a single band in a Western blot analysis using the OsDHAR antibody. No signal was detected in the EV and WT under the same conditions (Figure 2C). These results indicated that the OsDHAR gene under the control of the Ptrc promoter was expressed in the TA during incubation in 2 mM IPTG at 30°C for 2 days before 2.5 mM H2O2 treatment.
Stress tolerance was measured based on growth kinetics, colony formation on plates, and chlorophyll content. The TA grew better than the EV and WT under H2O2 stress conditions (Figure 2E). The WT and control EV showed loss of pigment on days 7, 8, and 10 after exposure to H2O2 stress, and were completely bleached by day 12, whereas the TA showed loss of pigment on day 10 (Figure 2D).
Stress responses were also evaluated with a plate cell-spotting assay. Exponentially growing TA, EV, and WT (absorbance at 730 nm = 0.25) were exposed to 2.5 mM H2O2 with 2 mM IPTG for 3 days, and then serial dilutions were spotted on agar-solidified BG11 medium. Cell viability of the TA was better than that of the EV and WT following exposure to H2O2 stress. On the other hand, all individuals were similarly spotted under normal conditions (Figure 3A). Measurement of chlorophyll content after 2 days exposure to H2O2 revealed less loss of chlorophyll in the TA than in the EV and WT (Figure 3B). The total dry weight of the TA was twofold higher than that of the EA and WT under oxidative stress conditions (Figure 3C).
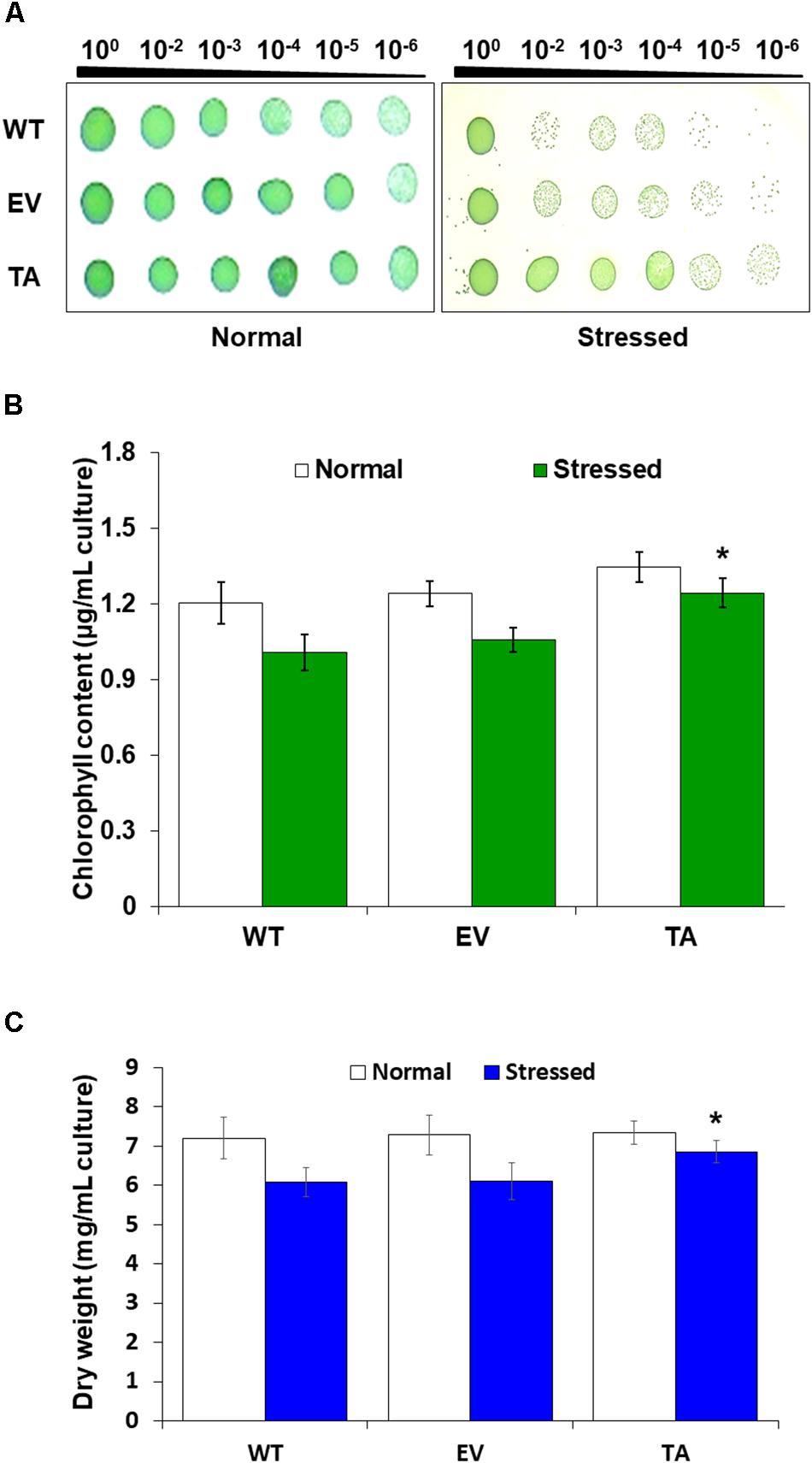
Figure 3. Stress sensitivity in the OsDHAR-expressing transgenic TA. (A) Cell viability was evaluated based on number of colony-forming units and spotting assay. (B) Chlorophyll contents of grown for 2 days in the presence of 2.5 mM H2O2 (A730 = 0.3). (C) Dry weights measured at 7 and 9 days after growth in the presence or absence of 2.5 mM H2O2 for 2 days. Error bars indicate ± SD of biologically least three independent experiments. Asterisks indicate significant differences between treatments as estimated by Student’s t-test (P < 0.05). WT, WT S. elongatus PCC 7942; EV, empty vector S. elongatus PCC 7942; TA, TA S. elongatus PCC 7942.
Redox states were determined based on cellular ROS levels assayed with the cytosolic oxidant-sensitive probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFHDA), which measures the oxidative conversion of DCFHDA to the highly fluorescent compound dichlorofluorescein (DCF). All three the TA, EV, and WT S. elongatus PCC 7942 showed an increase in DCF fluorescence following exposure to 2.5 mM H2O2 with 2 mM IPTG for 2 h, but the fluorescence intensity of the probe was more pronounced in the control EV and WT than in the TA (Figure 4A). When the TA, EV, and WT were subjected to oxidative stress, the TA displayed ROS-mediated cellular damage with or without ROS generation and contained moderate green fluorescence in the intracellular space. In contrast, EV and WT showed much more intense green fluorescence (Figure 4A). DCFHDA fluorescence was quantified by individually mixing the TA, EV, and WT with 10 μM DCFHDA followed by incubation for 20 min in the dark. Fluorescence intensity was 1.5-fold higher in the EV and WT than in the TA in the presence of 2.5 mM H2O2 with 2 mM IPTG (Figure 4D). In a biochemical assay using the FOX reagent, cellular ROS levels in the TA were determined to be approximately 20% lower than those in the EV and WT in the presence of H2O2 (Figure 4B). The lower ROS production in the TA resulted in reduced accumulation of MDA due to lipid peroxidation compared to the EV and WT. MDA accumulations were 1.2-fold higher in the EV and WT than in the TA (Figure 4C).
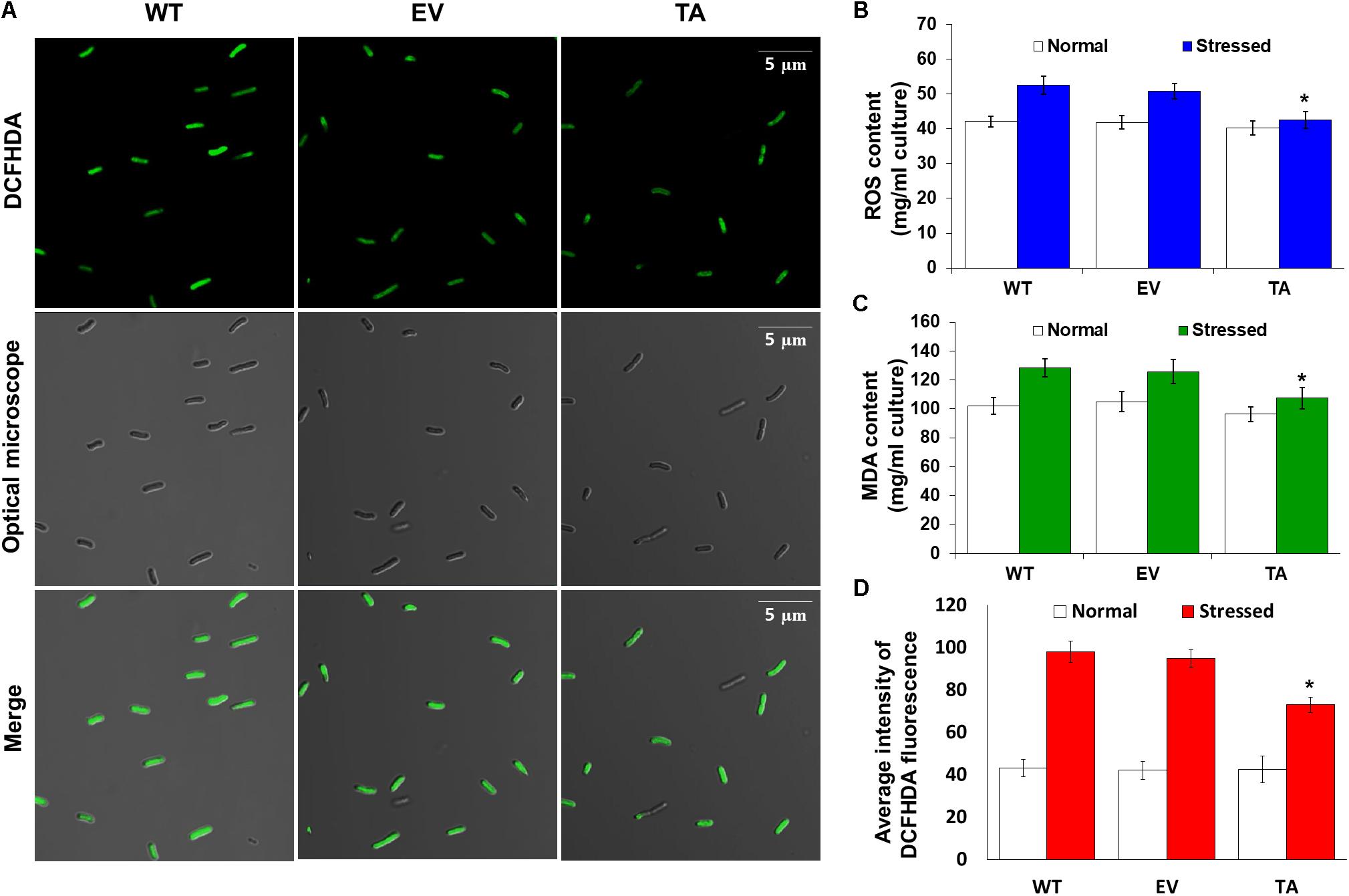
Figure 4. Expression of the OsDHAR gene in S. elongatus PCC 7942 improves intracellular redox homeostasis. (A) Confocal microscopy images of cells stained with DCFHDA to measure levels of ROS after treatment with 2.5 mM H2O2 for 2 h. (B) Intracellular ROS levels were determined using intracellular H2O2 after treatment with 2.5 mM H2O2 for 24 h. (C) Levels of lipid peroxidation were determined by TBA assay using MDA after treatment to 2.5 mM H2O2-IPTG with 2 mM IPTG for 24 h. (D) DCFHDA fluorescence they also measured ROS production via a microplate reader after treatment with 2.5 mM H2O2 for 2 h. Error bars indicate ± SD of biologically least three independent experiments. Asterisks indicate significant differences between treatments as estimated by Student’s t-test (P < 0.05).
We analyzed redox homeostasis in the OsDHAR-expressing transgenic by measuring the GSH pools to determine how OsDHAR expression affects total GSH content in cyanobacteria exposed to H2O2. In the TA, GSH redox status was expressed as the ratio of reduced GSH to oxidized GSSG (GSH/GSSG), was approximately 1.5-fold higher than those of the EV and WT (Figure 5B). The GSH/GSSG ratio in the TA were higher under oxidative stress than under normal conditions. Similarly, the GSH/GSSG ratio in the EV and WT were also moderately increased under oxidative stress, although overall values were lower than those of the TA (Figure 5B). Conversely, oxidized GSSG content in the EV and WT increased tremendously over baseline values compared to the TA under oxidative stress and normal conditions (Supplementary Figure S1).
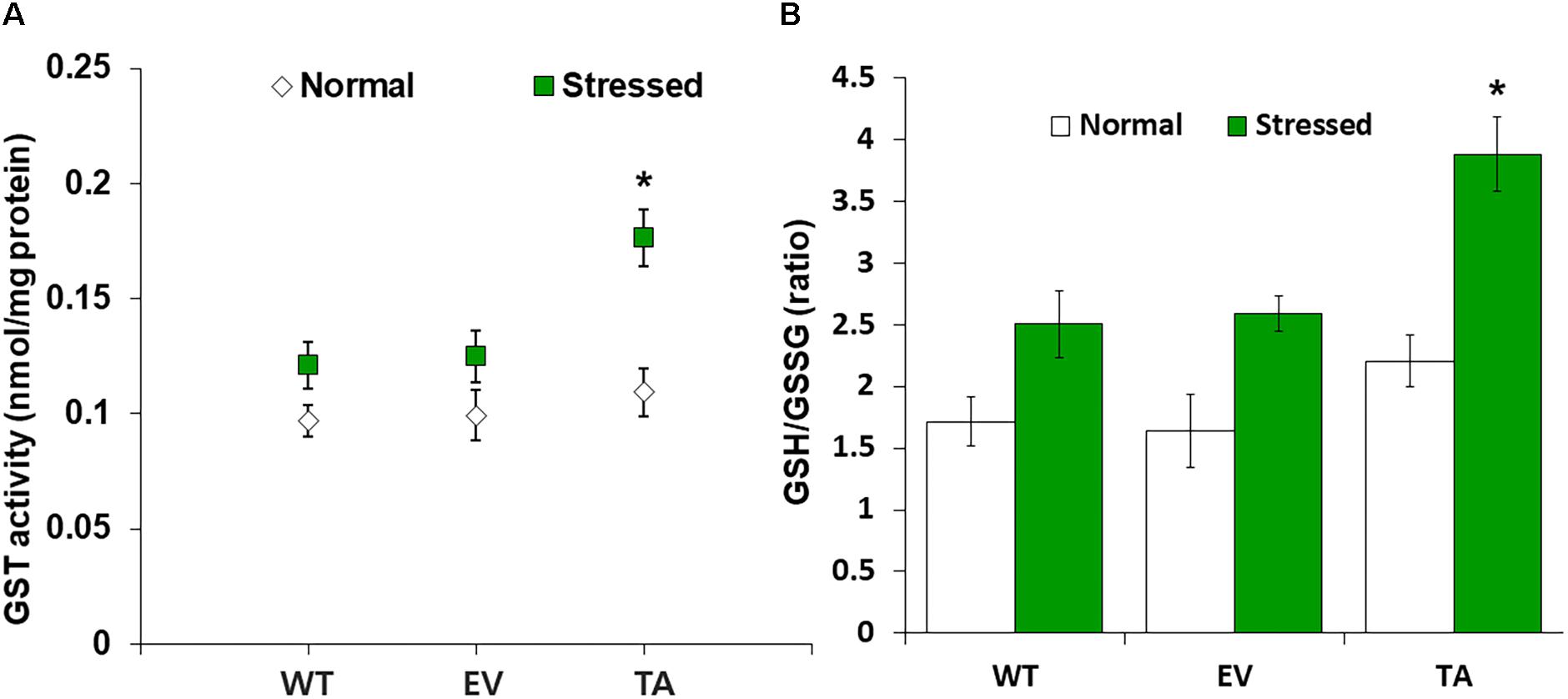
Figure 5. Analysis of enzyme activity and antioxidant redox status in TA after H2O2 treatment. (A) Activity of GST under normal and stress conditions. (B) Intracellular GSH/GSSG redox ratios in WT, EV, and TA after exposure to 2.5 mM H2O2 for 24 h. White bars correspond to conditions before H2O2 treatment (normal conditions), and the GSH/GSSG redox ratios (green bars) correspond to conditions after H2O2 treatment (stress conditions). Data are shown as mean values ± SD of biologically least three independent experiments. Asterisks indicate significant differences between treatments as estimated by Student’s t-test (P < 0.05). Measurements are an annotated in Supplementary Figure S1.
OsDHAR expression in the TA was also associated with increased GST activity under oxidative stress conditions (Figure 5A). These results indicate that enhanced resistance to the oxidative stress by maintaining of the cellular redox homeostasis during ROS-induced oxidative stress relates to the homology GST in cyanobacteria. For this reason, increased homeostasis correlated with total GSH content, and oxidized GSH and reduced GSH were observed in cyanobacteria harboring the DHAR gene. This result strongly suggests that the expression of DHAR (with its homology with GST) in S. elongatus PCC 7942 enhances tolerance of ROS-induced oxidative stress by maintaining cellular redox homeostasis using GSH alongside other antioxidant enzymes in cyanobacteria (Figure 6 and Supplementary Figure S3). Thus, heterologous OsDHAR expression in the TA responded to oxidative stress by increasing the number of redox active sites, such as GSH binding sites, and GSTs with similar functionally conserved residues under these conditions (Figures 1, 5). Altogether, our findings indicate that OsDHAR expression conferred tolerance to oxidative stress in S. elongatus PCC 7942 by interacting with GSH-dependent enzymes.
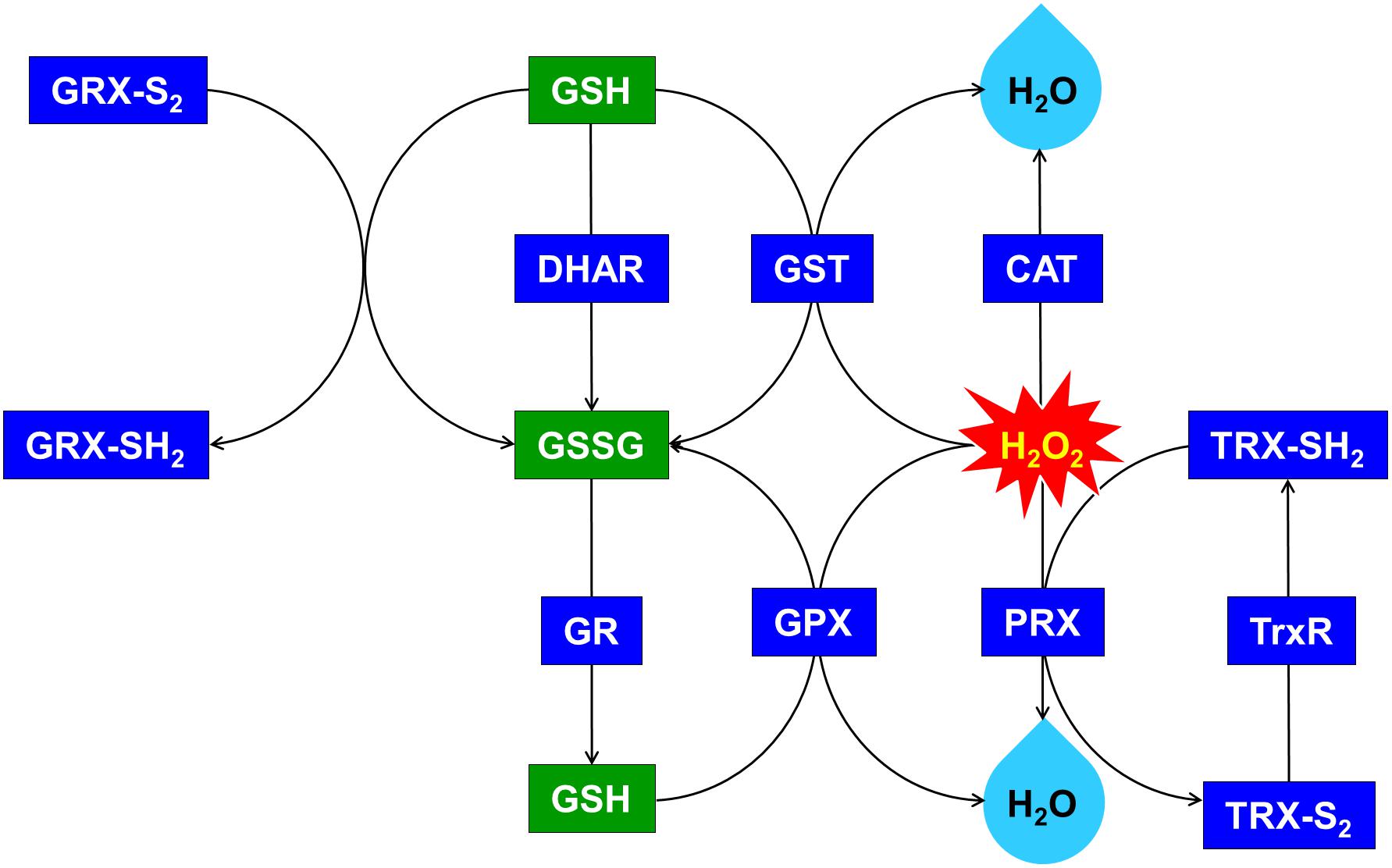
Figure 6. Upregulated glutathione (GSH)-dependent antioxidant system and maintenance of cellular redox status through the overexpression of OsDHAR in S. elongatus. GRX, glutaredoxin; GST, glutathione S-transferase; GR, glutathione reductase; GPX, glutathione peroxidase; CAT, catalase; PRX, peroxiredoxin; TRX, thioredoxin; TrxR, thioredoxin reductase. qRT-PCR identified GSH-related genes that were annotated in Supplementary Figure S3.
We explored this possibility further by conducting gene expression of the TA after exposure to H2O2-IPTG for 24 h. qRT-PCR identified GSH-dependent genes that were expressed in the TA after treatment with 2.5 mM H2O2-IPTG for 24 h in liquid BG11 medium containing 2 mM IPTG (Supplementary Figure S3). These genes included GST, thioredoxin reductase (TrxR), glutathione reductase (GR), catalase (CAT), glutathione peroxidase (GPX), 2-Cys peroxiredoxin (TRX), glutaredoxin (GRX), and peroxiredoxin (PRX) belonging to GSH-dependent reactions (Figure 6 and Supplementary Figure S3). Our results indicate that heterologous OsDHAR expression in the TA can enhance tolerance through the maintenance of cellular redox homeostasis via the GSH-dependent antioxidant system during ROS-induced oxidative stress.
Heterologous OsDHAR and the other GSTs demonstrated pairwise alignment with a conserved site that bound to GSH residues and other similarly functional conserved residues (Figure 1). OsDHAR also contains interacting residues of the GSH binding site. This indicates that OsDHAR can supply a redox active site during ROS-induced oxidative stress (Pulido et al., 2010; Do et al., 2016; Kim et al., 2017). The similarity of the aforementioned conserved DHAR sequences indicated that the OsDHAR redox active site possesses strictly conserved residues, including a GSH binding site and similar functional conserved residues from cyanobacteria GSTs. This GSH-dependent antioxidant system can facilitate GSH-dependent reactions during ROS-induced oxidative stress (Figure 6 and Supplementary Figure S3). As such, the redox active sites of OsDHAR participate in the maintenance of a stable cellular thiol/disulfide ratio, redox regulation, and refolding of oxidatively damaged proteins (Thomas et al., 1999; Hamnell-Pamment et al., 2005; Latifi et al., 2009; Anfelt et al., 2013; Kim et al., 2013; Abdul Kayum et al., 2018). In addition, we constructed a phylogenetic tree containing the S. elongatus PCC 7942 GST (SeGST; accession no. WP_011243077), which was 24% homologous to OsDHAR (accession no. AAL71856.1) (Figure 1B). The phylogenetic tree illustrated the evolutionary relationship between rice DHAR and cyanobacterial GST proteins.
We analyzed OsDHAR-mediated stress tolerance in transgenic S. elongatus PCC 7942 under H2O2 stress conditions because excess H2O2 is a major cause of oxidative stress in biological systems, and cyanobacteria are generally more sensitive to H2O2 than are other phototrophs (Drabkova et al., 2007; Banerjee et al., 2015). We investigated the mechanism by which OsDHAR gene overexpression increased H2O2 stress tolerance by examining the phenotypes and gene expression of the TA under H2O2 stress. There were no differences in phenotype among the TA, EV, and WT S. elongatus PCC 7942 grown under normal conditions (7 days; Figure 2D). Under H2O2 stress, however, most of the EV and WT had lost almost all their pigment after 10 days, while the TA lost considerably less pigment over this same period. The TA also recovered quickly from H2O2 damage (12 days; Figures 2D,E) and overall TA survival rates were higher than those of the EV and WT. Results also demonstrated that the TA, EV, and WT possessed clear phenotypic differences that correlated with their ability (or inability) to recover from H2O2 damage. Specifically, the TA recuperated quickly after H2O2 treatment, whereas the EV and WT did not (Figures 2D,E).
Stress responses were also evaluated by plate cell-spotting assay following H2O2 stress. Cell viability of the TA was better than the EV and WT following exposure to 2.5 mM H2O2 for 3 days (Figure 3A). Measurement of chlorophyll content after a 2-day exposure to 2.5 mM H2O2 revealed a more loss of 18% less chlorophyll in the EV and WT compared to the TA (Figure 3B). Based on these results, the total dry weight of the TA was twofold higher than the EA and WT to H2O2 stress (Figure 3C).
The TA, EV, and WT were additionally exposed to oxidative stress due to high salt concentrations (200 mM NaCl), low temperature (10°C), excess light (250 μE high-intensity light), and sodium dodecyl sulfate (SDS, 20%) (Figure 7). When cells are exposed to high salinities or low temperatures, major processes of photosynthesis, protein synthesis, and energy and lipid metabolism are slowed or halted (Demiral and Turkan, 2006). In addition, the stress of excess light can induce damage to the reaction center of photosystem II and may lead to the perturbation or inhibition of photosynthetic electron transport in cells (Fryer et al., 2003; Kim et al., 2018). Previous studies have shown that multiple ion channel pores in cell participate to stress responses in cyanobacterial cells (Los and Murata, 2004; Kim et al., 2018). SDS and cold stress were a surfactant that damage lipid membranes, thereby disrupting cellular redox homeostasis and leading to apoptosis (Kwon et al., 2003; Tahara et al., 2012; Shin et al., 2013).
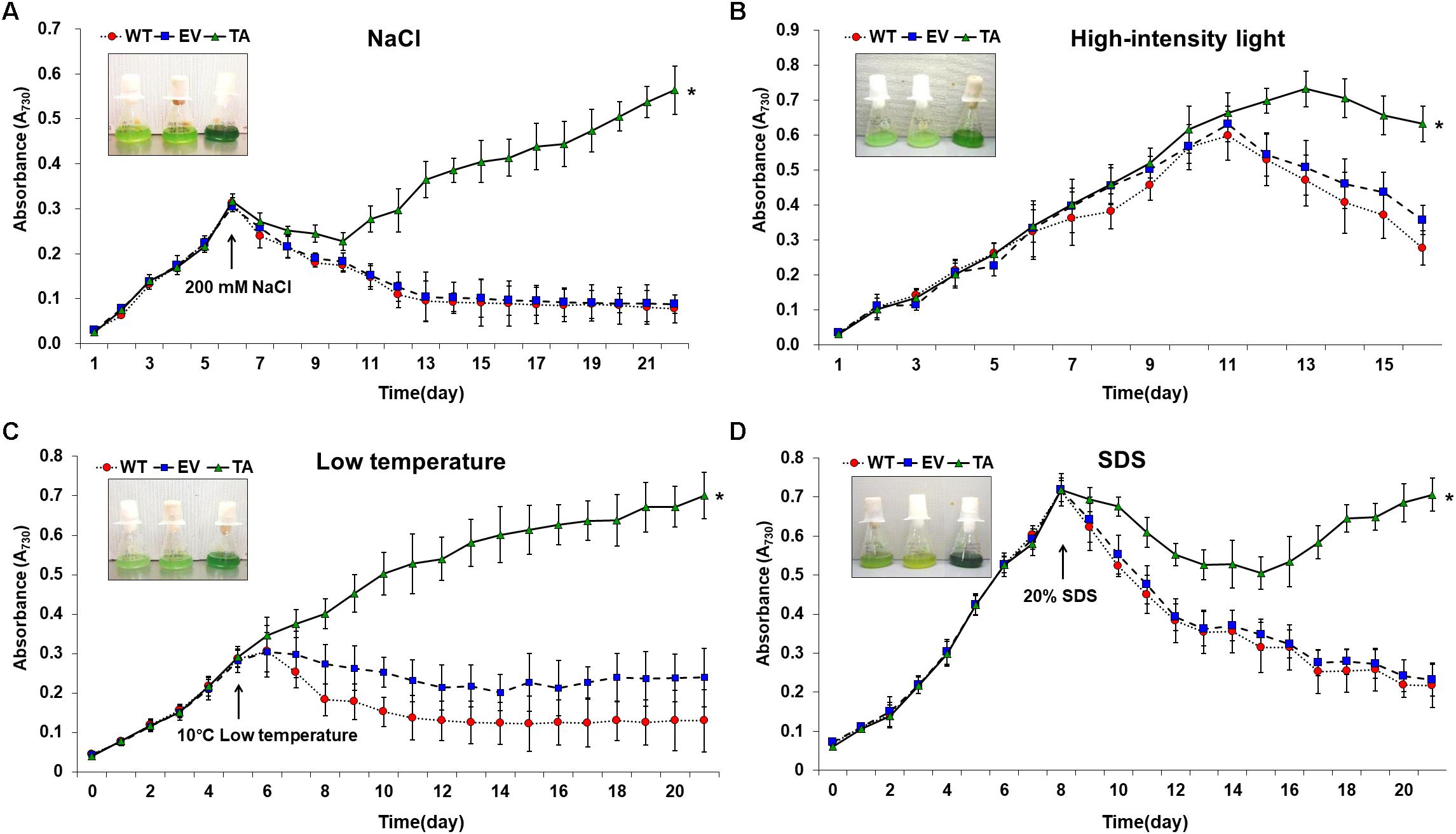
Figure 7. Growth profiles of WT, EV, and TA in the presence of oxidative stressors, including (A) 200 mM NaCl after incubation for 6 days, (B) 250 μE high-intensity light during continuous incubation, (C) low temperature (10°C) after incubation for 5 days, and (D) 20% SDS after incubation for 8 days. Photographs were taken approximately 3 days after stress treatments at which point the sensitive phenotype appeared. Error bars indicate ± SD of biologically least three independent experiments. Asterisks indicate significant differences between treatments as estimated by Student’s t-test (P < 0.05). WT, WT S. elongatus PCC 7942; EV, empty vector S. elongatus PCC 7942; TA, TA S. elongatus PCC 7942.
The TA displayed better growth than the control EV and WT in the presence of NaCl, high-intensity light, low temperature, and SDS (Figure 7). These results indicate that the expression of the OsDHAR gene in the TA conferred higher tolerance to oxidative stresses compared to that of the EV and WT. As already mentioned, DHAR participates in cellular detoxification, stress tolerance, and light signaling (Loyall et al., 2000; Thom et al., 2002; Benekos et al., 2010). Thus, DHAR as isozymes are critical to cellular detoxification of exogenous and endogenous harmful compounds. In aerobic cells, high DHAR activity enhances growth and development through increased chlorophyll fluorescence under oxidative stress, whereas it decreases chlorophyll in aging cells under the same conditions (Wang et al., 2010; Kim et al., 2013; Do et al., 2016). In cyanobacteria, H2O2-induced oxidative stress and the resulting excess ROS lead to cell death in several types of cyanobacteria (Drabkova et al., 2007; Qian et al., 2010; Banerjee et al., 2015). We investigated the effects of oxidative stress produced by H2O2-generated ROS by analyzing ROS levels in vitro and in vivo in the TA, EV, and WT. Cellular ROS levels were assayed to evaluate cell rescue redox states with the cytosolic oxidant-sensitive probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFHDA), which measures the oxidative conversion of DCFHDA to the highly fluorescent compound dichlorofluorescein (DCF). The TA displayed ROS-mediated cellular damage with and without the targeting signal, and contained weaker green fluorescence in the intracellular space than the EV and WT in the presence of 2.5 mM H2O2 (Figure 4A). DCFHDA fluorescence intensity was 1.5-fold higher in the EV and WT than the TA in the presence of 2.5 mM H2O2 (Figure 4D). Cellular ROS levels in the TA were found to be lower than those in the EV and WT through a biochemical assay using the FOX reagent in the presence of H2O2 (Figure 4B). Lower ROS production in the TA was associated with reduced accumulation of MDA due to lipid peroxidation compared to the EV and WT (Figure 4C). These results indicated that transgenic OsDHAR expression in the TA increased cell tolerance to oxidative stress by scavenging ROS and increasing chlorophyll content. These findings indicate that OsDHAR is key to oxidative tolerance through the maintenance of cellular redox homeostasis via a GSH-dependent antioxidant system.
Cells employ several mechanisms to combat toxic ROS generated by abiotic stresses like oxidants. The generated ROS can be scavenged by non-enzymatic antioxidants, such as GSH, or by enzymatic systems, such as the GSH pools involving DHAR (Noctor and Foyer, 1998; Kim et al., 2014). Different environmental stressors, including high salinity, low temperature, and drought, can activate DHAR expression, which goes on to regulate GSH pools (Urano et al., 2000; Jiang and Zhang, 2002; Kwon et al., 2003; Eltayeb et al., 2007; Gill and Tuteja, 2010). GSTs have been divided into eight classes: tau, phi, theta, zeta, lambda, EF1Bγ, DHAR, and tetrachlorohydroquinone dehalogenase (TCHQD) (Oakley, 2005; Lan et al., 2009; Dixon and Edwards, 2010; Jain et al., 2010; Liu et al., 2013).
Among these, DHAR and GST share considerable homology has been ubiquitously produced in eukaryotes and prokaryotes, such as plants, animals, fungi, and bacteria (Dixon et al., 2002; Liu et al., 2013; Rezaei et al., 2013; Ding et al., 2017). The homologous DHAR and GST have similar functions involving their redox active sites and GSH binding site residues (Dixon et al., 2002; Kim et al., 2013). Both DHAR and GST execute GSH-dependent thiol transferase activity to recycle antioxidants, including flavonoids, and quinones (Dixon et al., 2011). In this study, GST activity increased concomitantly with GSH via heterologous OsDHAR overexpression in cyanobacteria (Figure 5 and Supplementary Figure S1). Cellular oxidative stress depends on maintaining a balance between ROS content and the antioxidant system (Tang et al., 2007; Kim et al., 2017, 2018). Increased production of antioxidants (GSH) and antioxidant enzymes (GST) can ameliorate excess ROS during oxidative stress. In this study, these phenomena were associated with TA resistance to oxidative stress (Figures 2–7 and Supplementary Figure S1). We thereby conclude that the expression of heterologous OsDHAR in S. elongatus PCC 7942 enhanced tolerance to ROS-induced oxidative stress by maintaining cellular redox homeostasis through cooperation of the homologous GST in cyanobacterial cells.
Previous evidences have established a relationship between stress tolerance and the cellular redox homeostasis that is affected by DHAR overexpression in several cells (Horemans et al., 2000; Kwon et al., 2003; Ushimaru et al., 2006; Eltayeb et al., 2007; Qian et al., 2010; Wang et al., 2010). We confirmed that the TA was resistant to H2O2 stress and that this was related to an increased GSH/GSSG ratio (Figure 5B) and GST activity (Figure 5A). Our results indicated that heterologous OsDHAR expression in S. elongatus PCC 7942 confers tolerance to oxidative stress by interacting with GSH-dependent enzymes. Gene expression levels of the TA during exposure to H2O2 was also conducted to confirm this conclusion, and qRT-PCR identified GSH-related genes that were upregulated, including GST, TrxR, GR, CAT, GPX, TRX, GRX, and PRX, belonging to GSH-dependent reactions in the TA (Figure 6 and Supplementary Figure S3). GSH-dependent antioxidation involving GST, GR, CAT, GPX, GRX, TrxR, PRX, and TRX are all involved in protecting cells from oxidative stress and minimizing subsequent cellular damage (Kingston-Smith and Foyer, 2000). In particular, the GSH-dependent enzymes are among the most potent protein-based protection mechanisms and occur in nearly all cells, from bacteria like E. coli to higher plants and animals (Meyer et al., 2012). We surmise that oxidative stress tolerance in the TA would be increased through improved maintenance of redox homeostasis compared to the WT and EV (Figures 2–7 and Supplementary Figure S3).
Taken together, our findings indicate that the expression of OsDHAR in S. elongatus PCC 7942 maintained cellular redox homeostasis and protected cells from oxidative damage with the GSH-dependent antioxidant system via GSH-dependent reactions at the redox active site and GSH binding-site residues. However, genetic and molecular underpinnings of heterologous OsDHAR and homologous SeGST activity remain unclear. Cellular redox homeostasis and molecular processes involved in their response to various environmental conditions warrant further study.
We found that expression of the OsDHAR gene in the S. elongatus PCC 7942 increased tolerance to ROS-induced oxidative stress through the maintenance of cellular redox homeostasis with a GSH-dependent antioxidant system via GSH-dependent reactions. We anticipate that other antioxidant defense mechanisms might act synergistically to produce an effective defense response and protect cells during oxidative stress. The genetically modified cyanobacteria used in this study could help verify mutual functions between O. sativa DHAR and S. elongatus PCC 7942 GST genes. Further studies are needed to elucidate the mechanisms that underlie the increased oxidative stress tolerance observed in heterologous OsDHAR-expressing transgenic cyanobacterium.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Y-SK, I-SK, AT, JG, and H-SY designed the experiments. S-IP, J-JK, and K-IL performed the experiments. The article was written by Y-SK, and edited by JSB and JB. All authors read and approved the final manuscript.
This work was funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant no. 2016R1A6A1A05011910) and (grant no. 2018R1D1A3B07049385), Korea. We are grateful for the financial support of the Next-Generation BioGreen 21 Program (grant nos. PJ01366701 and PJ013240), Korea and the US NIH-NIGMS (grant no. R01GM118815), United States (JG).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Susan S. Golden (the University of California, San Diego and Center for Chronobiology) for supplying S. elongatus PCC 7942. We also thank Spencer Diamond, Benjamin Rubin, Ryan K. Shultzaberger, You Chen, Amy T. Ma, Mizuho Ota, Susan E. Cohen, Mark L. Paddock, Yong-Ick Kim, David G. Welkie, Roger Tseng, and Cigdem Sancar for helpful discussions and assistance with the section “Materials and Methods.”
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00231/full#supplementary-material
Abdul Kayum, M., Nath, U. K., Park, J. I, Biswas, M. K., Choi, E. K., Song, J. Y., et al. (2018). Genome-wide identification, characterization, and expression profiling of glutathione S-transferase (GST) family in pumpkin reveals likely role in cold-stress tolerance. Genes (Basel) 9:84. doi: 10.3390/genes9020084
Anfelt, J., Hallstrom, B., Nielsen, J., Uhlen, M., and Hudson, E. P. (2013). Using transcriptomics to improve butanol tolerance of Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 79, 7419–7427. doi: 10.1128/AEM.02694-13
Banerjee, M., Chakravarty, D., and Ballal, A. (2015). Redox-dependent chaperone/peroxidase function of 2-Cys-Prx from the cyanobacterium Anabaena PCC7120: role in oxidative stress tolerance. BMC Plant Biol. 15:60. doi: 10.1186/s12870-015-0444-2
Benekos, K., Kissoudis, C., Nianiou-Obeidat, I., Labrou, N., Madesis, P., Kalamaki, M., et al. (2010). Overexpression of a specific soybean GmGSTU4 isoenzyme improves diphenyl ether and chloroacetanilide herbicide tolerance of transgenic tobacco plants. J. Biotechnol. 150, 195–201. doi: 10.1016/j.jbiotec.2010.07.011
Blank, C. E. (2013). Origin and early evolution of photosynthetic eukaryotes in freshwater environments: reinterpreting proterozoic paleobiology and biogeochemical processes in light of trait evolution. J. Phycol. 49, 1040–1055. doi: 10.1111/jpy.12111
Boyer, J. S. (1982). Plant productivity and environment. Science 218, 443–448. doi: 10.1126/science.218.4571.443
Chen, Z., and Gallie, D. R. (2005). Increasing tolerance to ozone by elevating foliar ascorbic acid confers greater protection against ozone than increasing avoidance. Plant Physiol. 138, 1673–1689. doi: 10.1104/pp.105.062000
Demiral, T., and Turkan, I. (2006). Exogenous glycinebetaine affects growth and proline accumulation and retards senescence in two rice cultivars under NaCl stress. Environ. Exp. Bot. 56, 72–79. doi: 10.1016/j.envexpbot.2005.01.005
Ding, N., Wang, A., Zhang, X., Wu, Y., Wang, R., Cui, H., et al. (2017). Identification and analysis of glutathione S-transferase gene family in sweet potato reveal divergent GST-mediated networks in aboveground and underground tissues in response to abiotic stresses. BMC Plant Biol. 17:225. doi: 10.1186/s12870-017-1179-z
Dixit, G., Singh, A. P., Kumar, A., Singh, P. K., Kumar, S., Dwivedi, S., et al. (2015). Sulfur mediated reduction of arsenic toxicity involves efficient thiol metabolism and the antioxidant defense system in rice. J. Hazard. Mater. 298, 241–251. doi: 10.1016/j.jhazmat.2015.06.008
Dixon, D. P., Davis, B. G., and Edwards, R. (2002). Functional divergence in the glutathione transferase superfamily in plants. Identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J. Biol. Chem. 277, 30859–30869. doi: 10.1074/jbc.m202919200
Dixon, D. P., and Edwards, R. (2010). Roles for stress-inducible lambda glutathione transferases in flavonoid metabolism in plants as identified by ligand fishing. J. Biol. Chem. 285, 36322–36329. doi: 10.1074/jbc.M110.164806
Dixon, D. P., Steel, P. G., and Edwards, R. (2011). Roles for glutathione transferases in antioxidant recycling. Plant Signal. Behav. 6, 1223–1227. doi: 10.4161/psb.6.8.16253
Do, H., Kim, I. S., Jeon, B. W., Lee, C. W., Park, A. K., Wi, A. R., et al. (2016). Structural understanding of the recycling of oxidized ascorbate by dehydroascorbate reductase (OsDHAR) from Oryza sativa L. japonica. Sci. Rep. 6:19498. doi: 10.1038/srep19498
Drabkova, M., Matthijs, H. C. P., Admiraal, W., and Marsalek, B. (2007). Selective effects of H2O2 on cyanobacterial photosynthesis. Photosynthetica 45, 363–369. doi: 10.1007/s11099-007-0062-9
Eltayeb, A. E., Kawano, N., Badawi, G. H., Kaminaka, H., Sanekata, T., Morishima, I., et al. (2006). Enhanced tolerance to ozone and drought stresses in transgenic tobacco overexpressing dehydroascorbate reductase in cytosol. Physiol. Plantarum 127, 57–65. doi: 10.1111/j.1399-3054.2006.00624.x
Eltayeb, A. E., Kawano, N., Badawi, G. H., Kaminaka, H., Sanekata, T., Shibahara, T., et al. (2007). Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 225, 1255–1264. doi: 10.1007/s00425-006-0417-7
Fryer, M. J., Ball, L., Oxborough, K., Karpinski, S., Mullineaux, P. M., and Baker, N. R. (2003). Control of ascorbate peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J. 33, 691–705. doi: 10.1046/j.1365-313x.2003.01656.x
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Golden, S. S., Brusslan, J., and Haselkorn, R. (1987). Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 153, 215–231. doi: 10.1016/0076-6879(87)53055-5
Gorain, P. C., Bagchi, S. K., and Mallick, N. (2013). Effects of calcium, magnesium and sodium chloride in enhancing lipid accumulation in two green microalgae. Environ. Technol. 34, 1887–1894. doi: 10.1080/09593330.2013.812668
Hamnell-Pamment, Y., Lind, C., Palmberg, C., Bergman, T., and Cotgreave, I. A. (2005). Determination of site-specificity of S-glutathionylated cellular proteins. Biochem. Biophys. Res. Commun. 332, 362–369. doi: 10.1016/j.bbrc.2005.04.130
Horemans, N., Foyer, C. H., Potters, G., and Asard, H. (2000). Ascorbate function and associated transport systems in plants. Plant Physiol. Biochem. 38, 531–540. doi: 10.1016/s0981-9428(00)00782-8
Huang, T., Lin, J., Cao, J., Zhang, P., Bai, Y., Chen, G., et al. (2012). An exopolysaccharide from Trichoderma pseudokoningii and its apoptotic activity on human leukemia K562 cells. Carbohydr. Polym. 89, 701–708. doi: 10.1016/j.carbpol.2012.03.079
Jain, M., Ghanashyam, C., and Bhattacharjee, A. (2010). Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genomics 11:73. doi: 10.1186/1471-2164-11-73
Jiang, M., and Zhang, J. (2001). Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol. 42, 1265–1273. doi: 10.1093/pcp/pce162
Jiang, M. Y., and Zhang, J. H. (2002). Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 53, 2401–2410. doi: 10.1093/jxb/erf090
Kim, I. S., Jin, I., and Yoon, H. S. (2011). Decarbonylated cyclophilin A Cpr1 protein protects Saccharomyces cerevisiae KNU5377Y when exposed to stress induced by menadione. Cell Stress Chaperones 16, 1–14. doi: 10.1007/s12192-010-0215-9
Kim, Y. S., Kim, I. S., Bae, M. J., Choe, Y. H., Kim, Y. H., Park, H. M., et al. (2013). Homologous expression of cytosolic dehydroascorbate reductase increases grain yield and biomass under paddy field conditions in transgenic rice (Oryza sativa L. japonica). Planta 237, 1613–1625. doi: 10.1007/s00425-013-1862-8
Kim, Y. S., Kim, I. S., Boyd, J. S., Taton, A., Golden, J. W., and Yoon, H. S. (2017). Enhanced biomass and oxidative stress tolerance of Synechococcus elongatus PCC 7942 overexpressing the DHAR gene from Brassica juncea. Biotechnol. Lett. 39, 1499–1507. doi: 10.1007/s10529-017-2382-6
Kim, Y. S., Kim, I. S., Shin, S. Y., Park, T. H., Park, H. M., Kim, Y. H., et al. (2014). Overexpression of dehydroascorbate reductase confers enhanced tolerance to salt stress in rice plants (Oryza sativa L. japonica). J. Agron. Crop Sci. 200, 444–456. doi: 10.1111/jac.12078
Kim, Y. S., Kim, J. J., Park, S. I., Diamond, S., Boyd, J. S., Taton, A., et al. (2018). Expression of OsTPX gene improves cellular redox homeostasis and photosynthesis efficiency in Synechococcus elongatus PCC 7942. Front. Plant Sci. 9:1848. doi: 10.3389/fpls.2018.01848
Kingston-Smith, A. H., and Foyer, C. H. (2000). Overexpression of Mn-superoxide dismutase in maize leaves leads to increased monodehydroascorbate reductase, dehydroascorbate reductase and glutathione reductase activities. J. Exp. Bot. 51, 1867–1877. doi: 10.1093/jexbot/51.352.1867
Koksharova, O. A., Klint, I., and Rasmussen, U. (2006). The first protein map of Synechococcus sp. strain PCC 7942. Mikrobiologiia 75, 765–774.
Komatsu, S., Yamamoto, R., Nanjo, Y., Mikami, Y., Yunokawa, H., and Sakata, K. (2009). A comprehensive analysis of the soybean genes and proteins expressed under flooding stress using transcriptome and proteome techniques. J. Proteome Res. 8, 4766–4778. doi: 10.1021/pr900460x
Kwon, S. Y., Choi, S. M., Ahn, Y. O., Lee, H. S., Lee, H. B., Park, Y. M., et al. (2003). Enhanced stress-tolerance of transgenic tobacco plants expressing a human dehydroascorbate reductase gene. J. Plant Physiol. 160, 347–353. doi: 10.1078/0176-1617-00926
Lan, T., Yang, Z. L., Yang, X., Liu, Y. J., Wang, X. R., and Zeng, Q. Y. (2009). Extensive functional diversification of the Populus glutathione S-transferase supergene family. Plant Cell 21, 3749–3766. doi: 10.1105/tpc.109.070219
Latifi, A., Ruiz, M., and Zhang, C. C. (2009). Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 33, 258–278. doi: 10.1111/j.1574-6976.2008.00134.x
Liu, Y. J., Han, X. M., Ren, L. L., Yang, H. L., and Zeng, Q. Y. (2013). Functional divergence of the glutathione S-transferase supergene family in Physcomitrella patens reveals complex patterns of large gene family evolution in land plants. Plant Physiol. 161, 773–786. doi: 10.1104/pp.112.205815
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Los, D. A., and Murata, N. (2004). Membrane fluidity and its roles in the perception of environmental signals. Biochim. Biophys. Acta 1666, 142–157. doi: 10.1016/j.bbamem.2004.08.002
Loyall, L., Uchida, K., Braun, S., Furuya, M., and Frohnmeyer, H. (2000). Glutathione and a UV light-induced glutathione S-transferase are involved in signaling to chalcone synthase in cell cultures. Plant Cell 12, 1939–1950. doi: 10.1105/tpc.12.10.1939
Meyer, Y., Belin, C., Delorme-Hinoux, V., Reichheld, J. P., and Riondet, C. (2012). Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid. Redox Signal. 17, 1124–1160. doi: 10.1089/ars.2011.4327
Noctor, G., and Foyer, C. H. (1998). Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249–279. doi: 10.1146/annurev.arplant.49.1.249
Oakley, A. J. (2005). Glutathione transferases: new functions. Curr. Opin. Struct. Biol. 15, 716–723. doi: 10.1016/j.sbi.2005.10.005
Owens, S. (2001). Salt of the earth: genetic engineering may help to reclaim agricultural land lost due to salinisation. EMBO Rep. 2, 877–879. doi: 10.1093/embo-reports/kve219
Peng, L. F., Kapp, E. A., McLauchlan, D., and Jordan, T. W. (2011). Characterization of the asia oceania human proteome organisation membrane proteomics initiative standard using SDS-PAGE shotgun proteomics. Proteomics 11, 4376–4384. doi: 10.1002/pmic.201100169
Potters, G., De Gara, L., Asard, H., and Horemans, N. (2002). Ascorbate and glutathione: guardians of the cell cycle, partners in crime? Plant Physiol. Biochem. 40, 537–548. doi: 10.1016/s0981-9428(02)01414-6
Pulido, P., Spinola, M. C., Kirchsteiger, K., Guinea, M., Pascual, M. B., Sahrawy, M., et al. (2010). Functional analysis of the pathways for 2-Cys peroxiredoxin reduction in Arabidopsis thaliana chloroplasts. J. Exp. Bot. 61, 4043–4054. doi: 10.1093/jxb/erq218
Qian, H., Yu, S., Sun, Z., Xie, X., Liu, W., and Fu, Z. (2010). Effects of copper sulfate, hydrogen peroxide and N-phenyl-2-naphthylamine on oxidative stress and the expression of genes involved photosynthesis and microcystin disposition in Microcystis aeruginosa. Aquat. Toxicol. 99, 405–412. doi: 10.1016/j.aquatox.2010.05.018
Rezaei, M. K., Shobbar, Z. S., Shahbazi, M., Abedini, R., and Zare, S. (2013). Glutathione S-transferase (GST) family in barley: identification of members, enzyme activity, and gene expression pattern. J. Plant Physiol. 170, 1277–1284. doi: 10.1016/j.jplph.2013.04.005
Rouhier, N., Lemaire, S. D., and Jacquot, J. P. (2008). The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 59, 143–166. doi: 10.1146/annurev.arplant.59.032607.092811
Shimaoka, T., Yokota, A., and Miyake, C. (2000). Purification and characterization of chloroplast dehydroascorbate reductase from spinach leaves. Plant Cell Physiol. 41, 1110–1118. doi: 10.1093/pcp/pcd035
Shin, S. Y., Kim, I. S., Kim, Y. H., Park, H. M., Lee, J. Y., Kang, H. G., et al. (2008). Scavenging reactive oxygen species by rice dehydroascorbate reductase alleviates oxidative stresses in Escherichia coli. Mol. Cells 26, 616–620.
Shin, S. Y., Kim, M. H., Kim, Y. H., Park, H. M., and Yoon, H. S. (2013). Co-expression of monodehydroascorbate reductase and dehydroascorbate reductase from Brassica rapa effectively confers tolerance to freezing-induced oxidative stress. Mol. Cells 36, 304–315. doi: 10.1007/s10059-013-0071-4
Tahara, H., Uchiyama, J., Yoshihara, T., Matsumoto, K., and Ohta, H. (2012). Role of Slr1045 in environmental stress tolerance and lipid transport in the cyanobacterium Synechocystis sp. PCC6803. Biochim. Biophys. Acta 1817, 1360–1366. doi: 10.1016/j.bbabio.2012.02.035
Tang, D., Shi, S., Li, D., Hu, C., and Liu, Y. (2007). Physiological and biochemical responses of Scytonema javanicum (cyanobacterium) to salt stress. J. Arid Environ. 71, 312–322.
Tang, Z. X., and Yang, H. L. (2013). Functional divergence and catalytic properties of dehydroascorbate reductase family proteins from Populus tomentosa. Mol. Biol. Rep. 40, 5105–5114. doi: 10.1007/s11033-013-2612-5
Thom, R., Cummins, I., Dixon, D. P., Edwards, R., Cole, D. J., and Lapthorn, A. J. (2002). Structure of a tau class glutathione S-transferase from wheat active in herbicide detoxification. Biochemistry 41, 7008–7020. doi: 10.1021/bi015964x
Thomas, D. J., Thomas, J. B., Prier, S. D., Nasso, N. E., and Herbert, S. K. (1999). Iron superoxide dismutase protects against chilling damage in the cyanobacterium Synechococcus species PCC7942. Plant Physiol. 120, 275–282. doi: 10.1104/pp.120.1.275
Urano, J., Nakagawa, T., Maki, Y., Masumura, T., Tanaka, K., Murata, N., et al. (2000). Molecular cloning and characterization of a rice dehydroascorbate reductase. FEBS Lett. 466, 107–111. doi: 10.1016/s0014-5793(99)01768-8
Ushimaru, T., Nakagawa, T., Fujioka, Y., Daicho, K., Naito, M., Yamauchi, Y., et al. (2006). Transgenic Arabidopsis plants expressing the rice dehydroascorbate reductase gene are resistant to salt stress. J. Plant Physiol. 163, 1179–1184. doi: 10.1016/j.jplph.2005.10.002
Wang, H., and Joseph, J. A. (1999). Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic. Biol. Med. 27, 612–616. doi: 10.1016/s0891-5849(99)00107-0
Keywords: antioxidants, antioxidant-related enzymes, cyanobacterium, dehydroascorbate reductase, oxidative stress
Citation: Kim Y-S, Park S-I, Kim J-J, Boyd JS, Beld J, Taton A, Lee K-I, Kim I-S, Golden JW and Yoon H-S (2020) Expression of Heterologous OsDHAR Gene Improves Glutathione (GSH)-Dependent Antioxidant System and Maintenance of Cellular Redox Status in Synechococcus elongatus PCC 7942. Front. Plant Sci. 11:231. doi: 10.3389/fpls.2020.00231
Received: 24 June 2019; Accepted: 14 February 2020;
Published: 03 March 2020.
Edited by:
Dimitris Petroutsos, UMR 5168 Laboratoire de Physiologie Cellulaire Vegetale (LPCV), FranceReviewed by:
Corinne Cassier-Chauvat, UMR 9198 Institut de Biologie Intégrative de la Cellule (I2BC), FranceCopyright © 2020 Kim, Park, Kim, Boyd, Beld, Taton, Lee, Kim, Golden and Yoon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Il-Sup Kim, OTJraXNAaGFubWFpbC5uZXQ=; James W. Golden, andnb2xkZW5AdWNzZC5lZHU=; Ho-Sung Yoon, aHN5QGtudS5hYy5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.