- 1Hardwood Tree Improvement and Regeneration Center, Department of Forestry and Natural Resources, Purdue University, West Lafayette, IN, United States
- 2USDA Forest Service, Northern Research Station, Hardwood Tree Improvement and Regeneration Center, West Lafayette, IN, United States
Climate change may have unpredictable effects on the cold hardiness of woody species planted outside of their range of origin. Extreme undulations in temperatures may exacerbate susceptibility to cold stress, thereby interfering with productivity and ecosystem functioning. Juglans L. and their naturally occurring interspecific F1 hybrids, are distributed natively across many temperate regions, and J. regia has been extensively introduced. Cold hardiness, an environmental and genetic factor yet to be evaluated in many native and introduced Juglans species, may be a limiting factor under future climate change and following species introductions. We evaluated cold hardiness of native North American and Eastern Asian Juglans along with J. regia genotypes using field data from the Midwestern United States (Indiana), controlled freezing tests, and genome sequencing with close assessment of Juglans cold hardy genes. Many Juglans species previously screened for cold-hardiness were genotypes derived from the Midwest, California, and Europe. In 2014, despite general climate adaptation, Midwestern winter temperatures of −30°C killed J. regia originating from California; however, naturalized Midwestern J. regia survived and displayed low damage. Hybridization of J. regia with black walnut (J. nigra) and butternut (J. cinerea) produced F1s displaying greater cold tolerance than pure J. regia. Cold hardiness and growth are variable in Midwestern J. regia compared to native Juglans, East Asian Juglans, and F1 hybrids. Phylogeny analyses revealed that J. cinerea sorted with East Asian species using the nuclear genome but with North American species using the organellar genome. Investigation of selected cold hardy genes revealed that J. regia was distinct from other species and exhibited less genetic diversity than native Juglans species Average whole genome heterozygosity and Tajima’s D for cold hardy genes was low within J. regia samples and significantly higher for hybrid as well as J. nigra. We confirmed that molecular and morpho-physiological data were highly correlated and thus can be used effectively to characterize cold hardiness in Juglans species. We conclude that the genetic diversity within local J. regia populations is low and additional germplasm is needed for development of more regionally adapted J. regia varieties.
Introduction
Walnuts (Juglans spp.) occur around the world in a broad range of climates from Western and Eastern Asia to North, Central and South America (Ramos, 1997). Among Juglans species, Persian walnut (Juglans regia L.) is reported to have the most valuable edible nuts and high-quality timber. J. regia is found and grown worldwide, far from the Central Asian region where the species originated (Ramos, 1997; Guàrdia et al., 2013). In the Eastern and Midwestern United States, much of the introduced J. regia was derived from seeds collected in the Carpathian Mountain range during the mid-19th Century (Grimo, 1979). Seed collections from this eastern edge of Europe were designated “Carpathian walnut” and selected for tolerance to the cold temperatures found in Eastern and Midwestern North America (Ramos, 1997). Tracing the genetic origin of trees facilitates adaptive management for climate change (Aitken and Bemmels, 2016). Hamilton et al. (2013) evaluated the interaction of genotype with environment in spruce hybrids (Picea sitchensis × P. glauca) and noted that suitable genotypes could be identified for reforestation in areas where climatic conditions have changed or are predicted to change using growth and cold hardy traits.
Cold hardiness is a major limiting factor for J. regia cultivation and reforestation, particularly in the Midwest (Ebrahimi et al., 2017). J. regia has proven to be cold hardy but is not well adapted to the Eastern deciduous forests of the United States (Grimo, 1979; Pollegioni et al., 2017). Conditions such as winterkill, which destroys young branches, spring frost that damages flowers and new shoots, poor growth on acidic soils, and disease pressure given the higher humidity of North America relative to central Asia where the species originates are reoccurring issues (Manning, 1978; Grimo, 1979).
The greater environmental tolerance of interspecific hybrids, proven capacity to respond to variable climates, and quality timber production make them good candidates for reforestation efforts (Hamilton et al., 2013). Despite the known advantages of reforestation with adaptive hybrid species, the genetic background of interspecific F1 Juglans hybrids in the United States has not been determined. F1 hybrids of J. regia and J. nigra (J. intermedia) could be used for rootstocks to overcome the climate limitations and improve nut and timber production (McKay, 1965).
Cold hardiness is an important trait for abiotic stress adaptation but is difficult to track in the field, as the causative agent for injury or mortality can be difficult to pinpoint (Aitken and Bemmels, 2016). Native midwestern Juglans species (J. nigra and J. cinerea) can withstand cold temperatures to −40°C and -50°C, respectively (Weiser, 1970; Sakai and Weiser, 1973; George et al., 1977; Jacobs et al., 2008). Carpathian walnut is reportedly the most cold-hardy ecotype of J. regia, tolerating temperatures of −32 to −35°C without injury (Mitra et al., 1991). Researchers in France noted significant variability in cold hardiness among J. regia cultivars with a J. intermedia hybrid being the most tolerant (Poirier et al., 2004). Hardier than Carpathian walnut, J. intermedia endured −37°C temperatures with no damage (McKay, 1965; Grimo, 1979). Other factors associated with cold hardiness, such as carbohydrate content, also varied among species (Hyung et al., 2012).
Cold hardiness in woody plants may be evaluated in numerous ways, with visualization being the primary method for evaluating branches, buds, leaves, and twigs damaged by freezing temperatures (Sakai and Weiser, 1973; George et al., 1977). Cold hardiness can also be assessed using electrolyte leakage (McKay, 1992; Jacobs et al., 2008; Aslamarz et al., 2011), which occurs after cell death when loss of membrane integrity allows electrolytes, primarily potassium (K+), to cross the damaged membrane barrier unhindered. Quantification is obtained by measurement of increased electrolytic conductivity within the media containing dying cells (Lyons et al., 1979). Aslamarz et al. (2011) reported that levels of freezing damage for California J. regia cultivars were lower than Iranian cultivars. Sutinen et al. (1992) noted that visual and electrolyte leakage measurements of freezing damage in red pine (Pinus resinosa) were highly correlated. Patterson et al. (1976) suggested that measurement of electrolyte-leakage after chilling injury was much easier to evaluate than visible chilling injury.
Evaluation of cold hardy genes through qPCR is an inexpensive and reliable method for understanding gene expression in Juglans species. At present, no information regarding expression of cold hardy genes in Juglans species has been published. Numerous transcription factors have been found for evaluating freezing tolerance in woody plants (Benedict et al., 2006; Wisniewski et al., 2015). Dehydration-responsive element-binding proteins (DREBs) previously identified in Arabidopsis and woody plants were shown to play major roles in cold response (Gilmour et al., 1998; Wisniewski et al., 2015). Overexpression of a C-repeat binding factor (CBF) gene in apple (Malus domestica) resulted in delayed bud break in the spring and increased sensitivity to short photoperiods with respect to the start of dormancy (Wisniewski et al., 2015). Regulatory pathways in woody plants for cold hardy genes such as CBF are more complex than those of herbaceous plants as gene manipulation was shown to also affect growth and flowering (Benedict et al., 2006; Wisniewski et al., 2014).
Genome sequence technology can provide a greater depth of information for complex genes and pathways such as those involved in cold responses. A surge of interest in Juglans functional and comparative genomics was initiated after publication of a first draft of the J. regia genome in 2016 (Martínez-García et al., 2016). No genome sequence information linked to specific genes for cold hardiness and related traits has been published to date. However, chloroplast and genome-based data were used for phylogeny analyses of Juglans species (Hu et al., 2017; Stevens et al., 2018). Publication of a Juglans reference genome has provided researchers worldwide with the ability to make tremendous strides in a myriad of directions from additional phylogeny analyses and gene annotation to development of genetic linkage maps and gene filtering.
Statistical methods for analysis of nucleotide diversity and heterozygosity (i.e., Tajima’s D, π, FST), highlight the lower allelic diversity in domesticated verses wild populations (Thornton and Jensen, 2007; Stevens et al., 2018). Cultivation of J. regia began in Persia 2000–2500 years ago (Potts, 2018). Human selection and domestication of J. regia for nut production could result in lower levels of genetic diversity in J. regia compared with wild relatives. Natural selection of Juglans species before their introduction to the United States may have led to population or diversity bottlenecks for adaptation to colder climates, such as J. regia and J. ailantifolia. Limited natural regeneration combined with genetic introgression of local populations after introduction of nut producing genotypes resulted in decreased wild J. regia population sizes (Demesure, 1996). Research by Clair (2006) indicated fewer cold hardy traits were expressed in selectively bred Douglas-fir (Pseudotsuga menziesii) than wild or unselected populations. A recent study based on whole genome data reported heterozygosity and genetic diversity within J. regia populations is lower than J. nigra and Texas walnut (J. microcarpa) (Stevens et al., 2018). These authors found that the polyphenol oxidase (PPO) gene was the least diverse within J. regia compared to J. nigra and J. microcarpa. Similar work in chestnut (Castanea dentata) reported a lack of variation in genes for nut traits for domesticated populations compared to wild populations (LaBonte et al., 2018).
Some Juglans traits, such as cold hardiness, have evolved over time and can now be used in selection studies (Ramos, 1997). Prevalence of cold periods and undulations in temperature in response to climate change have negatively impacted exotic Juglans species in the Midwest U.S. (Ebrahimi et al., 2017). Despite the many research studies in the literature on Juglans, none have focused on genetic mechanisms for cold hardiness. This work seeks to: (i) compare and contrast cold hardiness traits found in J. regia with those of North American Juglans species, Eastern Asia, F1 hybrids (crosses of cold hardy by exotic non-cold hardy) using both visual approximations and controlled freezing experiments; (ii) determine levels of cold hardy gene expression using qPCR; (iii) sequence and assemble 12 Juglans samples with paired end read data combined with whole genome and chloroplast phylogeny analyses of cold hardy genes; and (iv) identify signatures of selection for the cold hardy genes from various Juglans pools.
Materials and Methods
Plant Material
The Hardwood Tree Improvement and Regeneration Center (HTIRC)1 at Purdue University in West Lafayette, Indiana, USA established a germplasm collection with seven Juglans species and eight F1 hybrids in 2003 (Table 1 and Supplementary Appendix A) for its walnut timber breeding program. Those samples originated from the Midwestern United States, California, and Europe (Table 1 and Supplementary Appendix A). Between 1 to 3 grafted ramets or 1 to 6 half-sib seedlings were planted for each accession. An additional 26 J. regia samples were obtained from the Indiana Nut and Fruit Growers Association (INFGA)2 (Supplementary Appendix A) for use in growth and winterkill data collection. The INFGA trees ranged from 15–95 years old (35-year-old average) and were relatively well adapted to the Midwestern U.S. climate.
Growth Data and Winterkill Ratings
The winter of 2014 was unusually cold in Indiana (Figures 1A,B and Supplementary Appendix B). Study trees were measured for height and diameter at breast height (dbh) (Figure 2 and Supplementary Appendix C) in January 2014. Extremely low temperatures (−30°C) were recorded on two consecutive days in February 2014. Winterkill damage was assessed in mid-June 2014 after mild weather to allow for full symptom development from cold injury. Damage was qualitatively rated from 1 to 5 (1 = no visible damage; 2 = minimal twig damage (5–20%); 3 = moderate twig damage (20–50%); 4 = severe damage, dieback to ground with resprouting (50–90%); 5 = mortal injury, dieback to ground without resprouting (90–100%). For each biological replicate, 5-10 twigs were assessed for winterkill ratings. Regional weather data were obtained from the Bureau of Reclamation airport weather station in West Lafayette, IN, United States (Figure 1A,B and Supplementary Appendix B).
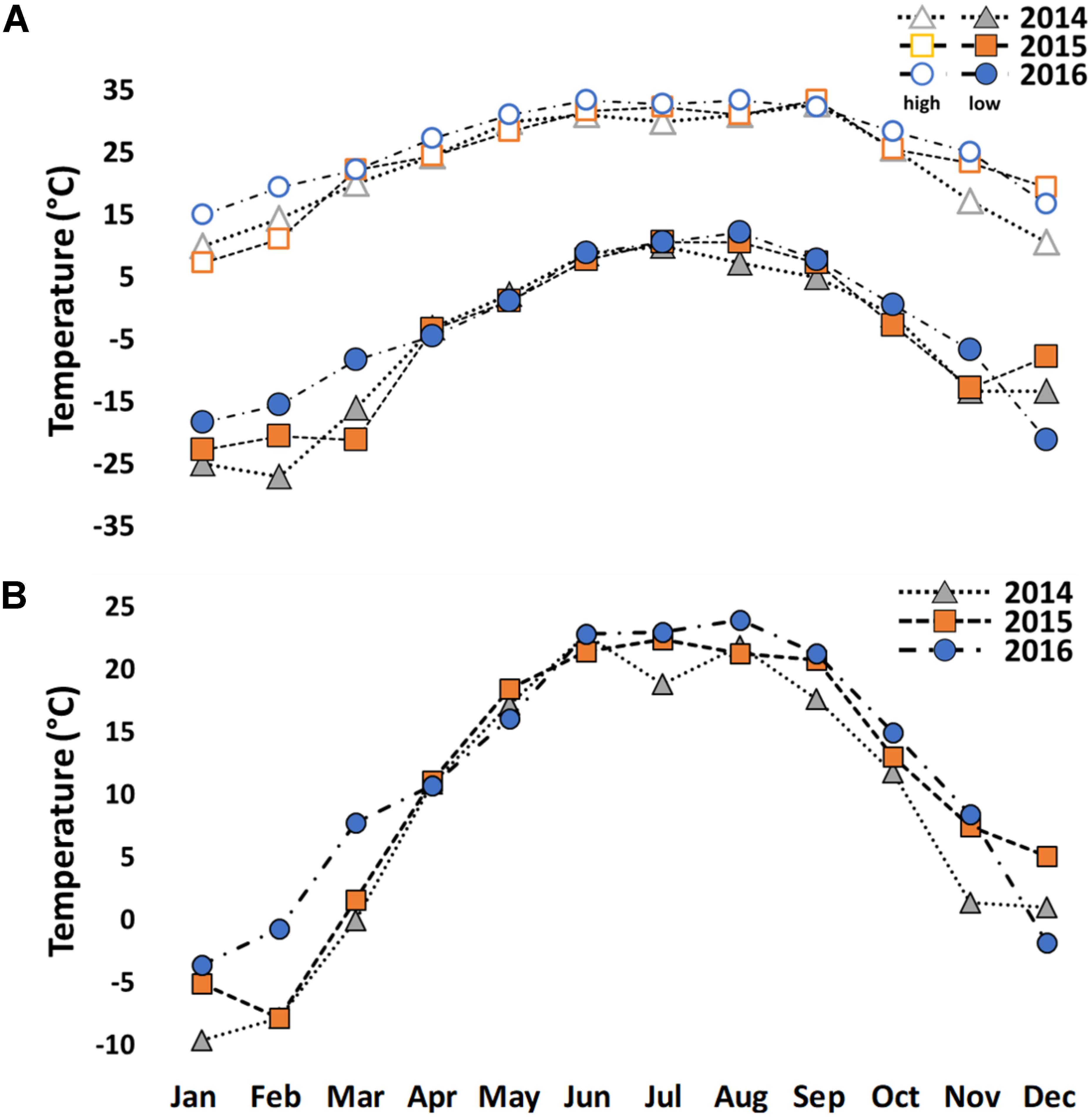
Figure 1. High and low temperatures (A) and average temperatures (B) from 2014 to 2016 in West Lafayette, IN, United States.
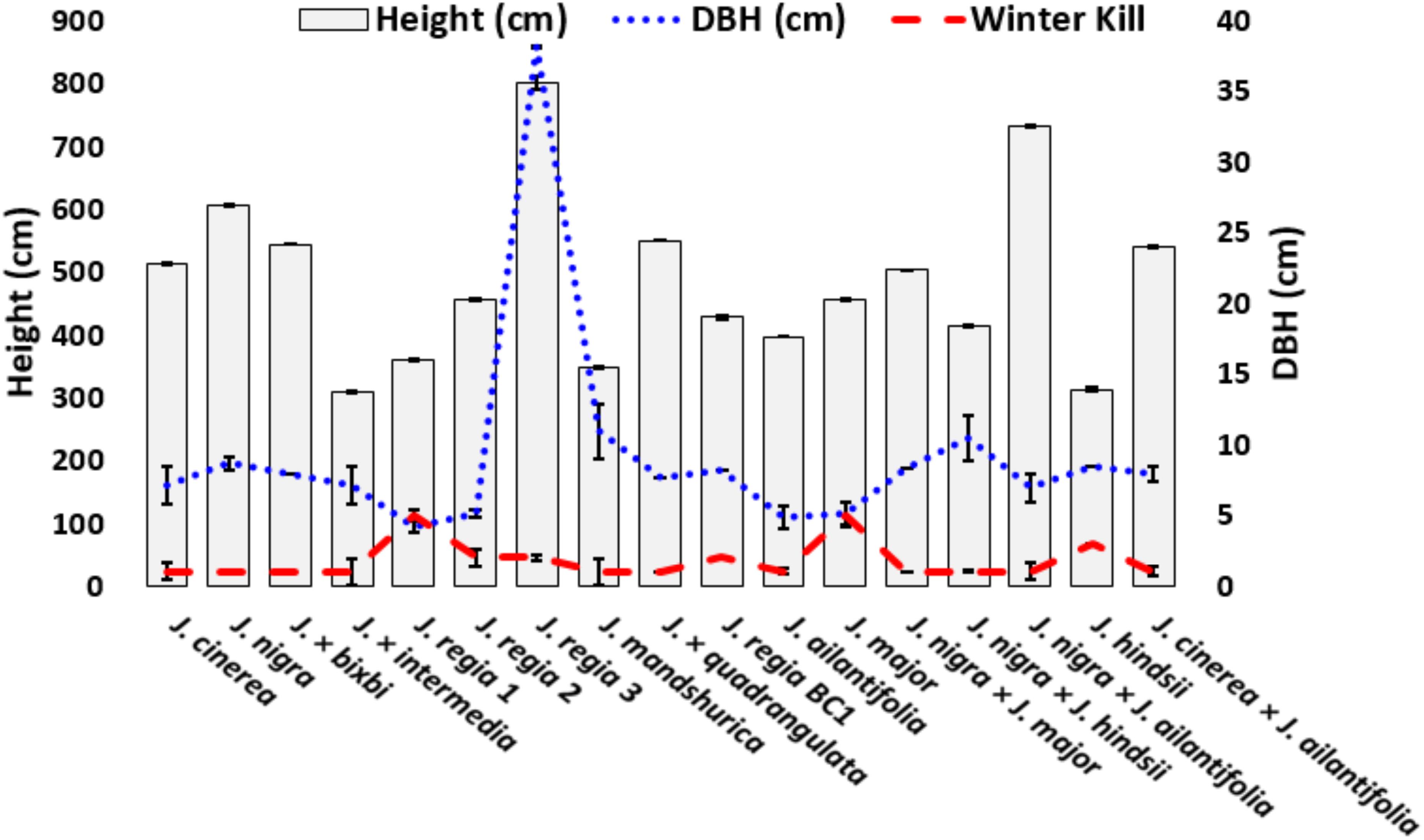
Figure 2. Mean (±SE) height and winterkill for studied Juglans species and interspecific hybrids. Heights were measured in winter 2014 and winterkill damage was measured in June 2014. J. regia 1 (California) and J. major (Arizona black walnut) were killed to the ground and did not re-sprout. Winterkill was rated from 1 to 5.
Electrolyte Leakage
Electrolyte leakage (EL) was measured based on 3 to 6 one-year-old twigs for each genotype (Figure 3 and Supplementary Appendix D). Twigs were collected in January 2016 at the maximum of frost damage hardiness (Charrier et al., 2011). Several slices from each twig sample were weighed and examined for frost damage to glean a whole twig cold hardiness value (Charrier et al., 2013). EL was evaluated for three temperatures in a programmable freezer at −10, −20, and −30°C. Twigs were transferred into 20-ml tubes before being placed in each pre-set freezer. The initial temperature of the programmable freezer was 10°C, and the temperature was decreased at 0.30°C min–1. Temperatures were maintained for 1.5 h before being returned to room temperature (RT). Initial electrical conductivity (ECi) (Mettler Toledo, United States) was measured 24 h after control samples (4°C) were transferred to test tubes with 20 mL of distilled water. After freezing treatments, twig samples were returned to room temperature for 24 h before of ECi was measured. All samples were autoclaved for 1 h at 100°C and final electrical conductivity (ECf) was assessed after the sample tubes cooled. Frost damage was approximated using relative frost index on (DI) values. Calculations of EL were based on the formula: DI = 100 (Rt – Ro)/(1 – Ro) where Rt = ECi/ECf of frozen samples and Ro = ECi/ECf of control samples (Earnshaw, 1993).
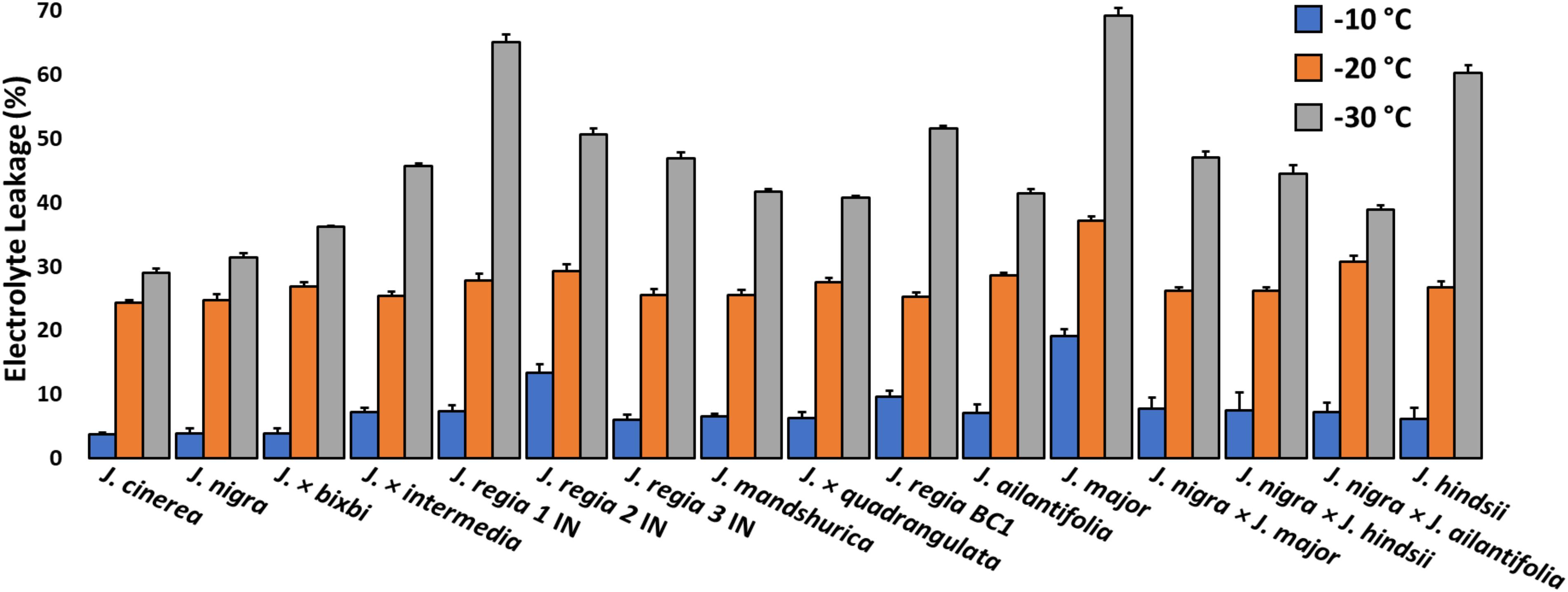
Figure 3. Index of freezing damage for 1-year-old twigs from Juglans species. and interspecific hybrids at –10, –20, and –30°C. Damage indices (Di) were obtained following protocols published by Earnshaw (1993). All J. regia used for electrolyte leakage analysis were obtained from Indiana and J. regia from California was excluded from this analysis. J. major was obtained from Indiana.
RNA Extraction and qPCR
Similar temperatures of EL test were used for gene expression analysis with the exception of −38°C that added to gene expression analysis. One of the subsamples was used to evaluate damage for EL, while the other was used for RNA analysis. Samples for RNA extraction were transferred to liquid nitrogen and stored at −80°C before isolation based on established protocols by Zarei et al. (2012). The qPCR template was generated from 1 μg total RNA used for First-strand cDNA synthesis with the Superscript III (Invitrogen, Carlsbad, CA, United States) kit. Primers for qPCR were designed with Primer33 using the J. regia genome sequence data available in the NCBI database4. Actin was used as an internal qPCR control to measure gene expression. qPCR was performed using the Bio-RAD Real-time System (CFX connect, United States) with a SYBR Premix ExTaq Kit (Takara Bio, United States). Fold change in expression for the target cold hardy genes was based on the delta-delta Ct method (ΔΔCT) described in Livak and Schmittgen (2001). Three biological replicates (three technical replicates) for each sample were used for gene expression (Figure 4 and Supplementary Appendices E, F).
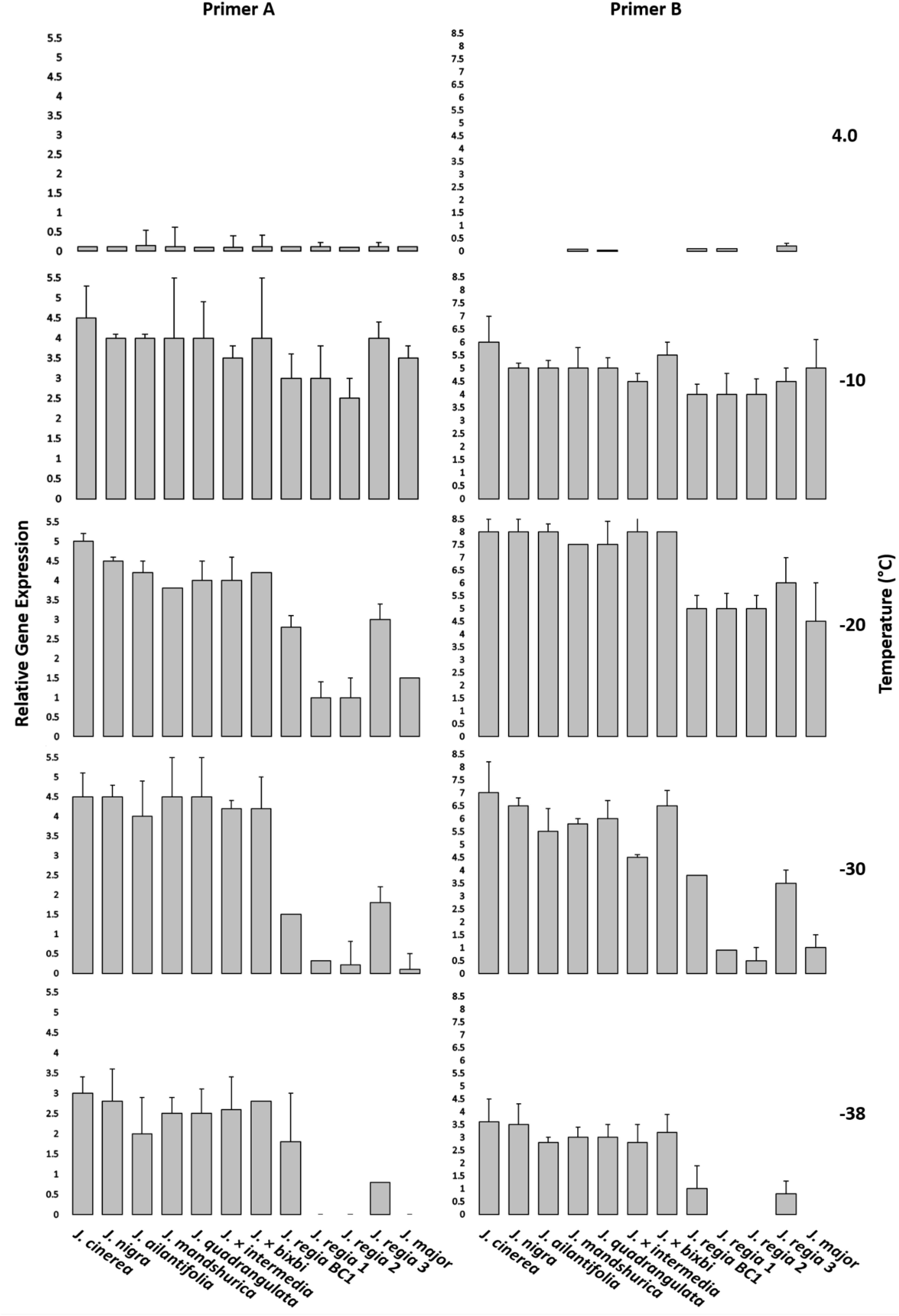
Figure 4. qPCR analysis. Two candidate primers for Juglans cold hardy genes were used to ascertain relative gene expression of twelve Juglans species and interspecific hybrid samples at 4, –10, –20, –30, and –38°C. Error bars represent standard error of the mean for three biological replicates (three technical replicates). All J. regia used for gene expression analysis were obtained from Indiana and J. regia from California was excluded from this analysis. J. major was obtained from Indiana.
Statistical Analyses
Field growth, electrolyte leakage, and gene expression data were analyzed using SAS software with PROC GLM mode (version 9.1; SAS Institute, Cary, NC, United States). When ANOVA was significant (P ≤ 0.05), protected Fisher’s least significant difference (LSD) test (Steel and Torrie, 1980) was applied to describe differences between different Juglans species. All the statistical output results are listed in Supplementary Appendices C–F. The number of genotypes varied across Juglans species (Table 1), but for those with single genotypes at least 3-6 replicates were used for the traits examined.
Genome Assembly and Sequencing
Analyses of field and electrolyte leakage data were made on 12 Juglans samples (Table 2). Genomic DNA was extracted from each sample following the protocol of Doyle and Doyle (1987). Samples were sequenced with 100 bp paired-end reads at Purdue Genomics Center5 using high throughput Illumina Hiseq 2500. Raw data were trimmed with Trimmomatic software (Bolger et al., 2014). Sequence fragments were assembled with SOAP-de novo software using trimmed reads using maximal read length = 100, average insert size = 400, and cutoff of pair number = 3 (Luo et al., 2012). Paired-end reads were mapped to the J. regia reference genome (Marrano et al., 2019) using the Burrow-Wheeler Aligner (BWA), Picard-tools (Langmead et al., 2009; McKenna et al., 2010; Wysoker et al., 2013) and SNP calling with the HaplotypeCaller tool from Genome Analysis Tool-kit (GATK) (McKenna et al., 2010). Consensus genome extracted with VCF tools and SAMtools. The assembled whole genome was used for whole gene prediction running AUGUSTUS (Stanke et al., 2006). A similar approach was used to assemble the chloroplast genomes of Juglans samples with J. regia chloroplasts used as reference (Martínez-García et al., 2016).
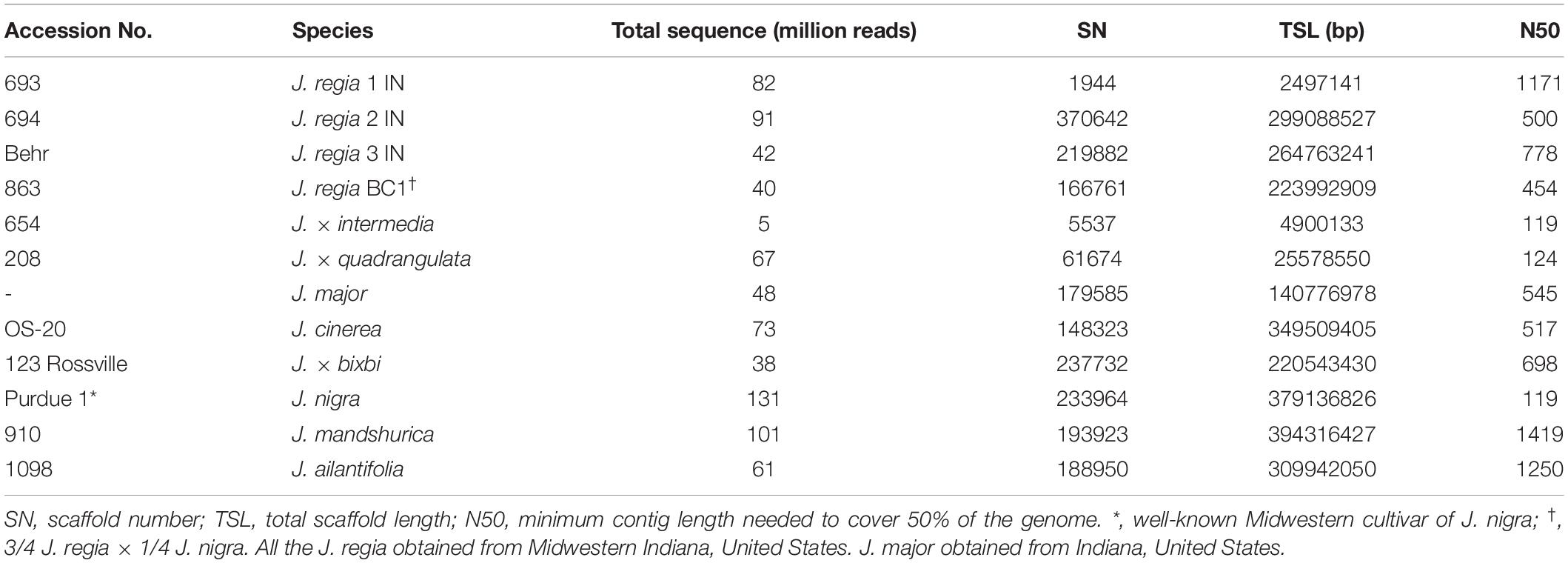
Table 2. Accession number, species, total sequence reads and assembly statistics for the 12 Juglans species used for evaluation of cold hardiness genes.
Phylogeny Analyses
Phylogenetic trees were created based on whole genome (paired-end reads) data using the Assembly and Alignment-Free (AAF)6 method described in Fan et al. (2015). The assembled chloroplast genomes were aligned with Molecular Evolutionary Genetic Analysis (MEGA) software (Tamura et al., 2007) to obtain a similarity/distance matrix for phylogeny analysis. After whole-genome annotation, cold hardiness-related proteins predicted by AUGUSTUS software in .gff (general feature format) format were converted to .fasta format with perl script. Each .fasta file of predicted proteins was subjected to Blastp analysis (Buchfink et al., 2015) that UniProt7 database used as a reference. Hits obtained from Blastp were averaged and the information used to create a similarity matrix. Genetic relatedness among Juglans samples was calculated by maximum likelihood following Tamura et al. (2007). A distance tree (Figure 5) was created with T-REX web server (Boc et al., 2012).
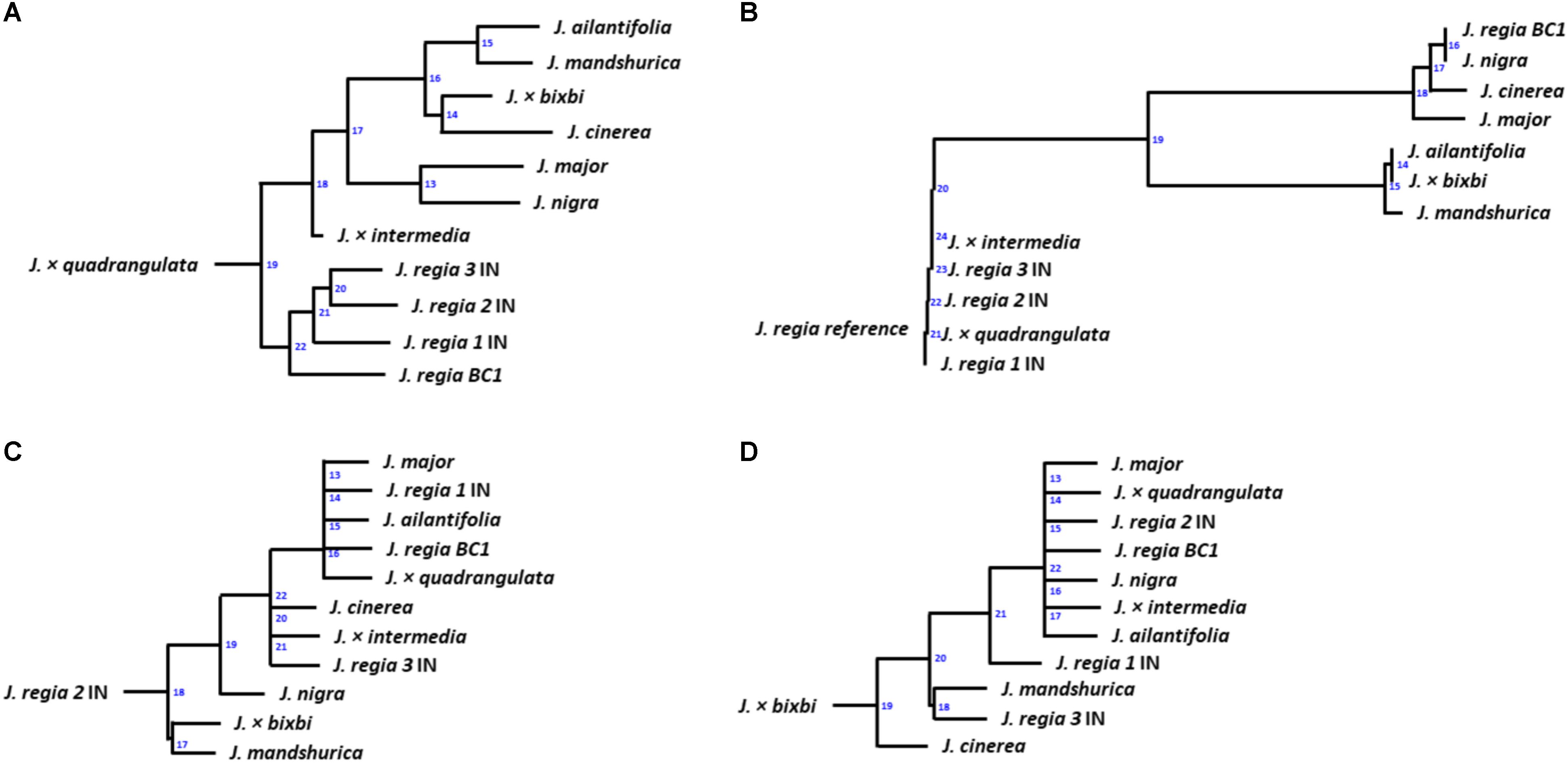
Figure 5. Dendrograms for samples of 12 Juglans: (A) based on whole nuclear genome; (B) based on chloroplast genome (C) based on whole proteins; (D) based on DREB-like genes filtered from the whole genome annotation. Numbers on transfers indicate the order of inference (Boc et al., 2012) for Juglans species.
Identification of Cold Hardy Genes
Evaluation of cold hardy genes predicted by AUGUSTUS was done following the protocol of LaBonte et al. (2018). The predicted genes were aligned to the UniProt7 database using DIAMOND sequence aligner on Blastp default mode using cold hardy gene keywords DREBs-like proteins (Buchfink et al., 2015). Genes identified after the initial blast search were again blasted against the UniProt protein database with DIAMOND sequence aligner on Blastp mode. Putative gene function was assigned based on of the top hits blasted.
Cold Hardy Genes Based on Signature of Selection
In addition to 12 Juglans samples described in Table 2, we downloaded 26 Juglans genomes previously deposited in the NCBI database (Bai et al., 2018; Stevens et al., 2018). Those samples contain five J. nigra, eight J. regia from Asia and United States, four J. mandshurica, four J. ailantifolia, four J. cinerea, two J. major. We used nine pools for calculation of population indices designated as Japanese walnut (J. ailantifolia); Black walnut (J. nigra); Butternut (J. cinerea); Hybrid (interspecific F1s); Arizona walnut (J. major); Manchurian walnut (J. mandshurica) and Persian walnut (J. regia from Asia and United States considered as a separate pools). Despite being composed of a variety of species, all hybrids (Table 2) were pooled together (J. × bixbi, J. × intermedia, J. × quadrangulata, and a BC1 [J. regia × [J. × intermedia]). Tajima’s D, heterozygosity (Het), SNP variation, nucleotide diversity (π), and FST for a nine pooled genome for cold hardy gene were calculated with VCF-tools8 and perl scripts obtained from LaBonte et al. (2018). A negative Tajima’s D value and low values for π and proportion of heterozygous loci for a predicted gene among Juglans samples was considered evidence for signature of selection (Table 3 and Supplementary Table 1). Additional information regarding selective sweep or bottleneck regions was obtained by calculating Tajima’s D, nucleotide diversity (π), FST and SNP variation for genes chosen at random across the whole genome for approving cold hardy gene (Supplementary Table 2).
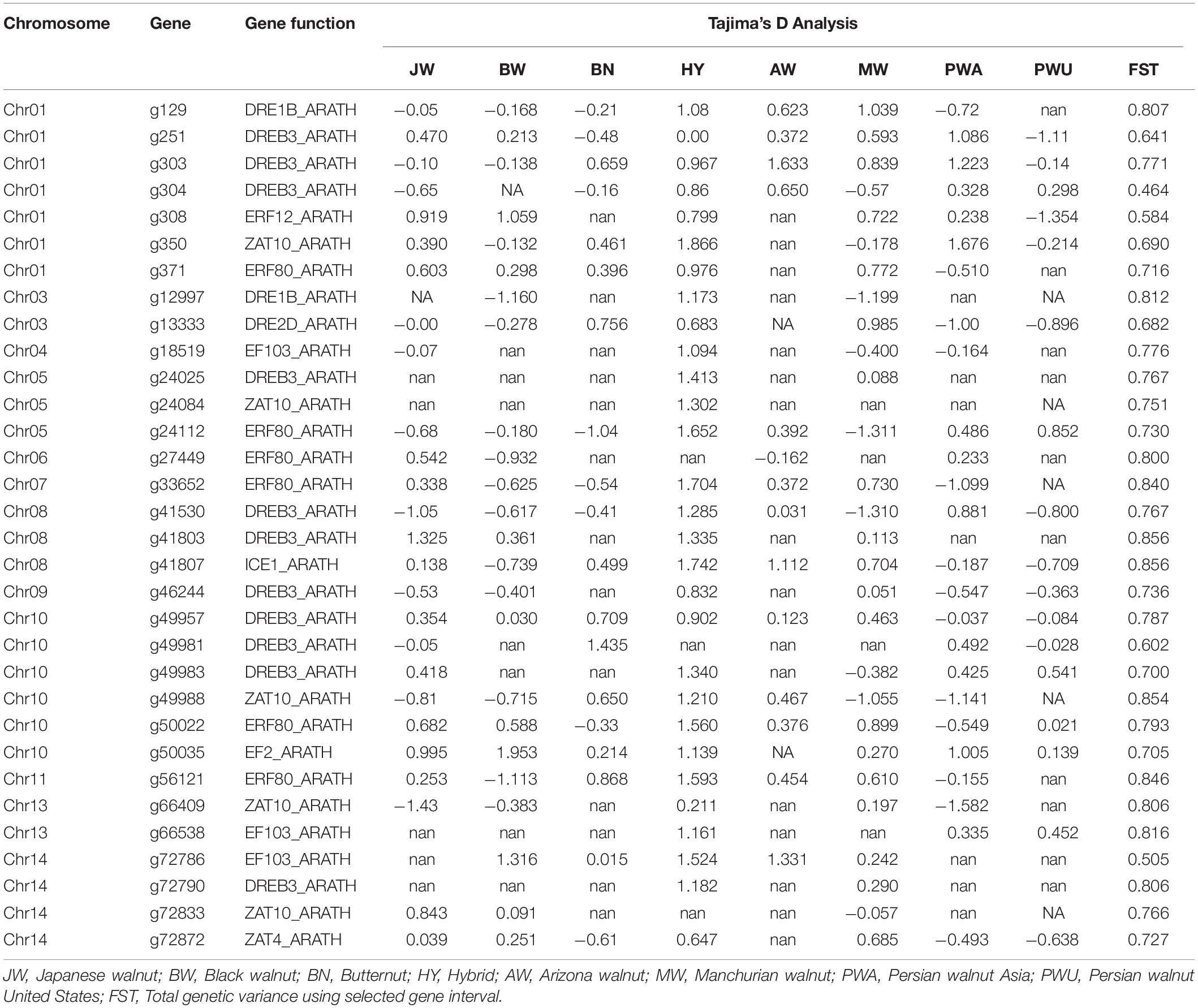
Table 3. Statistical data of population indices for cold hardy genes among the nine identified Juglans pools.
Results
Field Ratings
Juglans species exhibited a wide range of growth differences in response to cold temperatures (Figure 2 and Supplementary Appendix C). Timber stem quality of J. regia was average at best and most commonly appeared below average to poor. J. regia exhibited a reduction in growth compared to cold hardy J. cinerea and J. nigra (Figure 2 and Supplementary Appendix C). The J. regia BC1 ([J. nigra × J. regia] × J. regia)] grew comparable to J. cinerea and J. nigra as did the F1 hybrid J. × quadrangulata (J. regia × J. cinerea). Hybrids ‘Royal’ (J. hindsii × J. nigra) and J. × bixbyi (J. ailantifolia × J. cinerea) had greater growth than J. cinerea and J. nigra. Average dbh among Juglans samples varied from 4.2 cm (J. regia CA) to > 10.9 cm for Manchurian walnut (J. mandshurica) (Figure 2 and Supplementary Appendix A). The dbh values for native Juglans species (J. cinerea, J. nigra) and their F1 hybrids were all greater than that of J. regia originally from California and the Midwest U.S (Figure 2 and Supplementary Appendix C).
Very little cold damage was observed in the Juglans Germplasm block from 2003–2013. Damage occurred presumably in February 2014 after low temperatures dropped to a low of −30°C for about 4 h two consecutive days (Figures 1A,B). As a result, all J. regia had various levels of winterkill ranging from slight damage for Carpathian (J. regia IN 3) to 100% mortality for Mediterranean varieties (J. regia California) which were killed entirely to their graft unions (Figure 2 and Supplementary Appendix C). Arizona walnut (J. major), comprised of seven high-elevation half-sib families from Arizona U.S. displayed winterkill damage similar to J. regia. When cold-sensitive Juglans species. (J. hindsii, J. major, J. regia) were hybridized with cold hardy Juglans species. (J. cinerea, J. nigra), the resultant F1 hybrids exhibited tolerance akin to the pure cold hardy species (Figure 2 and Supplementary Appendix C). Like J. cinerea and J. nigra, Japanese walnut (J. ailantifolia) and Chinese walnut (J. mandshurica) showed little winterkill (Figure 2 and Supplementary Appendix C).
Cold-Hardiness Evaluation After Controlled Freezing
All of our J. regia from California perished in 2014 after exposure to unusually cold (−30°C) winter temperatures, and so this source was excluded from the controlled-freezing tests. A comparison of cold damage indices among the remaining Juglans species revealed that California black walnut (J. hindsii), J. major, and J. regia had significantly greater damage (p < 0.01) than other Juglans species (Figure 3 and Supplementary Appendix D). The two Juglans species that showed the least damage were J. cinerea and J. nigra. East Asian species (J. ailantifolia and J. mandshurica) had less damage than J. regia and F1 hybrids but more damage than J. cinerea and J. nigra. When less hardy Juglans species were crossed with hardier Juglans species, cold hardiness of the offspring significantly increased from previous levels. While cold damage was lower in F1 hybrids but not to the level of pure cold hardy species (Figure 3 and Supplementary Appendix D).
Cold Hardy Gene Expression
Two primers indicated significant variation in the freezing response for Juglans species. (Figure 4 and Supplementary Appendices E, F). J. cinerea and J. nigra had higher gene expression for all examined temperatures. Cold hardy genes were highly expressed in J. regia (a non-cold hardy species) at −10 and −20°C but levels decreased dramatically at lower temperatures (−30 and −38°C). Gene expression levels for F1 hybrids (J. regia × cold hardy species) were higher than for pure J. regia. East Asian species (J. ailantifolia and J. mandshurica) had lower gene expression compared with J. cinerea and J. nigra but displayed gene expression levels like F1 hybrids. J. regia had the lowest gene expression of the Juglans species sampled.
Phylogeny Analysis
Twelve Juglans samples were evaluated based on their chloroplast, whole-genome, protein, and DREB-like gene similarity (Table 2). Phylogeny analyses of whole genome data indicated that the Juglans samples sorted into three main groups based on their continent of origin. However, the North American J. cinerea sorted with East Asian species (Figure 5A). F1 hybrids sorted between the groups containing their parents (Figure 5A). Phylogeny analyses generated from the chloroplast data revealed a similar sorting pattern although J. cinerea now sorted with North American Juglans species and each hybrid sorted with their maternal parent (Figure 5B). Analyses of protein data revealed an inconsistency as the categorization of Juglans changed both within and among species (Figure 5C). For example, each of the three cold hardy J. regia samples from Indiana were placed into different groups. Phylogeny analyses created from DREB-like genes showed J. cinerea and J. × bixbi, the most cold hardy Juglans species sampled, sorted into separate groups but J. nigra, also cold hardy, sorted with the F1 hybrid samples (Figure 5D).
Juglans Genome Signature of Selection
Selective sweeps within cold hardy genes were evaluated within the nine Juglans pools based on heterozygosity and Tajima’s D analyses. Thirty-three cold hardy genes with various Tajima’s D in all Juglans pools were chosen for evidence of selective sweeps (Table 3 and Supplementary Table 1). Numerous intervals with elevated variation in Tajima’s D among different pools were selected for further gene annotation analysis and concomitantly, these regions with lower Tajima’s D also display lower genetic diversity. The average value of Tajima’s D for cold hardy genes among the Juglans species varied from -0.237 (J. regia from United States) to 1.146 (Hybrid) (Table 3 and Supplementary Table 1). Tajima’s D values for most of the cold hardy genes in hybrid samples were ≥1, a rare occurrence for other Juglans pools (Table 3 and Supplementary Table 1). Heterozygosity of cold hardy genes in the different Juglans species undulated from 0.0074 (Persian walnut United States) to 0.0264 (F1 hybrids) and 0.0216 (black walnut) (Table 3 and Supplementary Table 1). Analysis of the selected cold hardy genes revealed 1794 SNPs that varied across pools with Persian walnut from Asia (176, lowest) to Manchurian walnut from China (250, highest) (Supplementary Table 1). The cold hardy genes in this study showed unusually low heterozygosity and nucleotide diversity within the nine pools of Juglans species; however, average heterozygosity (Hetavg) was significantly lower for J. regia than for hybrids and black walnut. The whole genomes from each pool of Juglans species also were investigated for presence of signature of selection for random gene across chromosome one. Average random genome values for Tajima’s D were −0.0457 (Persian walnut, from Asia), to 0.782 (interspecific F1 hybrids), 0.0095 (J. cinerea, butternut), and −0.114 (J. nigra, black walnut) (Supplementary Table 2). Random genome heterozygosity varied from 0.01 in J. regia to 0.046 (F1 hybrids), 0.015 in butternut and 0.023 in black walnut (Supplementary Table 2). Assessment of genes chosen at random confirmed that J. regia exhibited the least diversity of all Juglans species studied (Supplementary Table 2), a result validated by a selective sweep of cold hardy genes.
Discussion
Field Ratings
Assessments of cold hardiness and growth for local and introduced Juglans species are vital for the success of Juglans plantations. In this study, native species (J. cinerea and J. nigra) and their F1 hybrids grew faster and were hardier than J. regia species. Growth of J. regia was poor compared to related F1 hybrids and native Juglans species (J. cinerea, J. nigra); however, growth of the J. regia BC1 (J. regia × [J. nigra × J. regia]) was comparable to pure J. cinerea and J. nigra. Overall, our data indicate a significant correlation between cold hardiness and growth and survival. Cold tolerant hybrids grew at rates similar to native cold hardy species and had improved timber form. Cold hardiness among J. regia selections varied by origin, with Carpathian varieties being much more cold hardy than Mediterranean varieties, from France and California (‘Franquette’, ‘Fernette’, and Fernor’) being not cold hardy (Figure 2 and Supplementary Appendix A). Severe cold temperatures of 2014 caused some damage to even our hardiest J. regia IN 3, ‘Behr,’ which was nearly 60 years old.
Guàrdia et al. (2013) evaluated cold hardiness in various Juglans species and rated J. nigra as most cold hardy and J. regia as least. In addition, they found that J. nigra acclimated quicker to cold than J. regia (Guàrdia et al., 2013). Cold hardiness of the F1 hybrid, J. × intermedia was intermediate to that of pure species (Mitra et al., 1991). Our findings confirmed that (cold-sensitive × cold-tolerant) hybrids inherit a combination of cold genes from each parent, and that cold hardiness is a genetic and highly heritable trait. Such interspecific hybrids show the potential for assisted migration of pure ecotypes where cold tolerant northern populations could be crossed with cold sensitive but heat tolerant populations (Hamilton et al., 2013). Natural hybrids can be genetically diverse and well suited to compete with conspecifics (Aitken and Bemmels, 2016).
Controlled Freezing
Electrolyte leakage (EL) results indicated that J. regia IN, J. major, and northern California black walnut (J. hindsii) had significantly greater damage indices (p < 0.01) than native species J. cinerea, J. nigra and F1 hybrids at −30°C (Figure 3 and Supplementary Appendix D). Research by Guàrdia et al. (2013) reported that J. nigra was hardier than J. intermedia and J. regia; and Aslamarz et al. (2011) found that California-based J. regia cultivars were hardier than Iranian cultivars. Patterson et al. (1976) examined different Passiflora spp. from diverse climates using EL and noted tropical lowland species were more-susceptible than those originating in colder climates. Many methods for evaluation of cold hardiness exist, but measurement of electrolyte leakage cell death is less difficult and more sensitive than visual observations of injury (Patterson et al., 1976). Laboratory estimates of cold tolerance and those from data collected in the field tend to be highly correlated. Laboratory estimates may be slightly higher compared to damage under field conditions (Schaberg and DeHayes, 2000), and our own observations. However, the ease and speed of screening material through EL is compelling for breeding and forest management.
Gene Expression Through qPCR
In addition to EL tests, gene expression of two putative cold hardy genes was investigated using qPCR (Figure 4 and Supplementary Appendices E, F). Close inspection of gene expression data indicated that cold hardy gene expression patterns mirrored results seen in the field as well as EL data. Cold-susceptible species such as J. regia showed little to no gene expression at −30 and −38°C, a plausible explanation for the increased mortality in winter 2014. Although one J. regia (J. regia 3 IN) had greater expression than the other J. regia, expression levels were much lower than for native Juglans species and F1 hybrids. Cold hardiness in relation to genetic composition of Juglans species was corroborated when F1 hybrids with J. regia parents were examined; F1 hybrids with a greater composition of J. regia had lower cold hardy gene expression. For example, J. × intermedia (50:50 J. regia: J. nigra) had lower gene expression than J. nigra and much more than pure J. regia. Our BC1 (75:25 J. regia: J. nigra) had lower gene expression than J. × intermedia but higher expression than pure J. regia as well. Some cold hardy gene expression studies reported that F1 hybrids of Ocimum kilimandscharicum (Camphor Basil) showed higher expression levels than pure species (Dhawan et al., 2016). Interspecific hybrid Pinus spp. with a genetic background composed of 75–87% cold hardy species exhibited greater levels of cold hardiness than others comprised of more cold-sensitive species (Lu et al., 2007). Assessment of cold hardiness using molecular markers represents a significant advancement over long-term (years to decades) field observations and even EL (Joosen et al., 2006). These DREB genes and their associated SNP’s that we have discovered can now serve as a marker assisted selection system for breeding cold hardiness into Juglans.
Phylogenetic Analyses
Whole genome data indicated that the most cold hardy species (J. cinerea) sorted with the East Asian species rather than other native Juglans species of United States (Figures 5A,B). The matK and ITS results from Stanford et al. (2000), also indicated that J. cinerea (Trachycaryon) sorted closely with Asian species (J. regia, Dioscaryon; J. mandshurica, J. ailantifolia; Cardiocaryon). Analyses of chloroplast data sorted J. cinerea with North American species (Figures 5A,B). These data lend support to the Stanford et al. (2000) hypothesis that J. cinerea moved from North America to Eurasia over geologic time following a warming trend during the mid-Oligocene.
Using DREB-like genes, cold hardy J. cinerea and J. × bixbi sorted separately. Other F1 hybrids sorted with J. nigra and J. major but could not be sorted according to geological history (Figures 5C,D). Protein analyses suggest that proteins coding for cold tolerance genes differed among Juglans species. Early research indicated that different proteins within the same species could evolve at dramatically different rates (Zuckerkandl and Pauling, 1965; Wang et al., 2004). This theory was supported by our results in which three J. regia IN all sorted to different groups according to DREB gene sequence. Interestingly, J. regia 3 IN, the hardiest Persian walnut variety in the study, sorted adjacent to the cold hardy J. mandshurica (Figure 5D). Stevens et al. (2018) reported that extra copies of a specific gene were due to assembly and demonstrated high heterozygosity, while Martínez-García et al. (2016) highlighted a PPO that may be differentially expressed in a wide range of tissues. Cold hardy genes may have numerous, slightly different copies throughout the genome that work as part of an uncharacterized network; however, further study is required to understand how these genes vary within species.
Juglans Genome Signature of Selection
The signature of selection reveals that cold hardy genes are under strong positive selection in interspecific F1 hybrids compared to J. regia from United States, which displayed negative Tajima’s D values. Positive selection shows genetic adaptation of species or hybrid in response to abiotic/biotic interactions (Bonhomme et al., 2015). Most hybrid Tajima’s D values were ≥1, a rarity unlike the other Juglans pools (Table 3 and Supplementary Table 1). J. regia was sought out for domestication thousands of years as a high-quality food source (Mercuri et al., 2013). This may explain much of the reduced genetic diversity in the species today. Screens of polymorphic SSR markers in J. regia cultivated in the Midwest showed evidence of a genetic bottleneck and severely reduced genetic diversity compared to European Carpathian samples (Ebrahimi et al., 2017). A longitudinal trend of genetic variability was reported in Eurasian J. regia populations Pollegioni et al. (2017). These authors highlighted the noticeable loss of allelic richness and heterozygosity from Eastern to Western Europe and the recent reduction in effective population size compared with Asian samples.
In our study, the amount of nucleotide diversity in J. regia pools was lower than other Juglans species based on cold hardiness genes (Table 3 and Supplementary Table 1) and whole genome analysis (Supplementary Table 2). New studies based on whole genome sequencing (Stevens et al., 2018) revealed that nucleotide diversity in J. regia (0.0056) is lower than J. nigra (0.0096) and J. microcarpa (0.0089), but higher than J. hindsii (0.0016). Other studies have noted that allelic numbers and heterozygosity in J. nigra were greater than wild J. regia populations (Robichaud et al., 2006; Pollegioni et al., 2017). Forward selected seed orchard parents of J. nigra displayed fewer alleles than wild selected parents but still maintained high heterozygosity values (Ebrahimi et al., 2018). A recent whole genome study with Chinese chestnut (Castanea mollissima) likewise showed that domesticated orchard cultivars were less diverse than wild trees (LaBonte et al., 2018). Studies with hybrid varieties are expected to reveal greater genetic diversity; however, heterozygosity values obtained in this work, although high, were only slightly higher than J. nigra, J. ailantifolia, and J. mandshurica populations.
Conclusion
Very little winterkill damage was observed among the Juglans species studied from 2003–2013. Most damage resulted from extremely cold temperatures in February 2014. We quantitatively illustrated the correlations between weather, physiological data, and expression of cold hardy genes using whole genome analyses to detect potential markers for assessing cold hardiness in Juglans species. Our results indicated that molecular and morpho-physiological data were highly correlated. In addition to morpho-physiology and molecular markers, we employed selective sweep to filter Juglans species based on genes related to cold hardiness into different whole genome pools. Should these genes be conserved in other species, these findings may be applicable to more tree genre and species that vary in cold hardiness. All analyses confirmed that cold hardy (J. nigra or J. cinerea) crossed with cold sensitive (J. regia and J. hindsii) interspecific F1 hybrids have higher cold hardy gene expression levels than all J. regia and the other cold sensitive species examined. As sequencing costs decline, utilization of RNA-sequencing and genome-wide association studies (GWAS) with these genes and their SNP’s should be useful for marker assisted selection of Juglans species for cold hardiness.
Data Availability Statement
The genome sequencing data associated with this study has been deposited in the Hardwood Genomic Project database, and can be accessed using the following links: https://www. hardwoodgenomics.org/bio_data/3641871, https://www.hardwo odgenomics.org/bio_data/3958579.
Author Contributions
AE designed the project, analyzed the data, and wrote the original draft manuscript. SL contributed to interpretation and visualization of the results and revised the manuscript. JM assisted with collecting field data and providing material for electrolyte leakage and gene expression analysis and revised the manuscript. DJ contributed to interpretation of the results and revised the manuscript. All authors have read and approved the submitted version.
Funding
This work was supported by Hardwood Tree Improvement and Regeneration Center (HTIRC) at Purdue University, the USDA Forest Service, and the Department of Forestry and Natural Resources at Purdue University, United States.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Indiana Nut and Fruit Growers Association for providing Julglans regia samples, Burchell Nursery (www.burchellnursery.com/; Oakdale, CA, United States) for providing the French cultivars ‘Fernette’ and ‘Fernor’ and the UC Davis Walnut Improvement Program for providing scion wood from California cultivars. Within the INFGA, Ken Dooley, Bill Heiman, Jerry Lehman, John Harrell, and Bill Deeter each provided scion wood and historical accounts of J. regia in Indiana. Additional materials were provided by Forest Glen Preserve (http://www.vccd.org/forest-glen-preserve/; Vermillion County, IL, United States) and Stark Brothers Nursery Inc. (https://www.starkbros.com/; Vera, MO, United States). We also thank Dr. Nicholas R. LaBonte, University of Illinois, Urbana-Champaign Samarth Mathur, Purdue University for providing bioinformatics scripts and consulting on evaluations of genomic data and Dr. Mercedes Uscola University of Alcalá, Spain for valuable assistance with this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00229/full#supplementary-material
Footnotes
- ^ https://htirc.org/
- ^ https://www.infga.org/
- ^ http://frodo.wi.mit.edu/primer3/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://www.purdue.edu/hla/sites/genomics/
- ^ https://omictools.com/aaf-tool
- ^ https://www.uniprot.org/blast/
- ^ http://vcftools.sourceforge.net/
References
Aitken, S. N., and Bemmels, J. B. (2016). Time to get moving: assisted gene flow of forest trees. Evol. Appl. 9, 271–290. doi: 10.1111/eva.12293
Aslamarz, A. A., Vahdati, K., Hassani, D., Rahemi, M., Mohammadi, N., and Leslie, C. (2011). Cold hardiness and its relationship with proline content in Persian walnut. Eur. J. Hort. Sci. 76, 84–90.
Bai, W. N., Yan, P. C., Zhang, B. W., Woeste, K. E., Lin, K., and Zhang, D. Y. (2018). Demographically idiosyncratic responses to climate change and rapid Pleistocene diversification of the walnut genus Juglans (Juglandaceae) revealed by whole−genome sequences. New Phytol. 217, 1726–1736. doi: 10.1111/nph.14917
Benedict, C. J., Skinner, S., Meng, R., Chang, Y., Bhalerao, R., Huner, N. P., et al. (2006). The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 29, 1259–1272. doi: 10.1111/j.1365-3040.2006.01505.x
Boc, A., Diallo, A. B., and Makarenkov, V. (2012). T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 40, 573–579. doi: 10.1093/nar/gks485
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bonhomme, M., Boitard, S., San Clemente, H., Dumas, B., Young, N., and Jacquet, C. (2015). Genomic signature of selective sweeps illuminates adaptation of Medicago truncatula to root-associated microorganisms. Mol. Biol. Evol. 32, 2097–2110. doi: 10.1093/molbev/msv092
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Charrier, G., Bonhomme, M., Lacointe, A., and Améglio, T. (2011). Are budburst dates, dormancy and cold acclimation in walnut trees (Juglans regia L.) under mainly genotypic or environmental control? Int. J. Biomet. 55, 763–774. doi: 10.1007/s00484-011-0470-1
Charrier, G., Poirier, M., Bonhomme, M., Lacointe, A., and Améglio, T. (2013). Frost hardiness in walnut trees (Juglans regia L.): how to link physiology and modeling. Tree Physiol. 33, 1229–1241. doi: 10.1093/treephys/tpt090
Clair, J. B. (2006). Genetic variation in fall cold hardiness in coastal Douglas-fir in western Oregon and Washington. Botany 84, 1110–1121. doi: 10.1139/b06-084
Demesure, B. (1996). “Conservation of genetic resources of noble hardwoods in France: overview,” in Noble Hardwoods Network. Report of the First Meeting, Escherode, Germany, eds J. Turok, G. Eriksson, J. Kleinschmit, and S. Canger (Rome: International Plant Genetic Resources Institute), 9–11.
Dhawan, S. S., Shukla, P., Gupta, P., and Lal, R. K. (2016). A cold-tolerant evergreen interspecific hybrid of Ocimum kilimandscharicum and Ocimum basilicum: analyzing trichomes and molecular variations. Protoplasma 253, 845–855. doi: 10.1007/s00709-015-0847-9
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Earnshaw, M. J. (1993). Stress Indicators: Electrolyte Leakage. Methods in Comparative Plant Ecology. London: Chapman & Hall, 152–154.
Ebrahimi, A., Lawson, S. S., Frank, G. S., Coggeshall, M. V., Woeste, K. E., and McKenna, J. R. (2018). Pollen flow and paternity in an isolated and non-isolated black walnut (Juglans nigra L.) timber seed orchard. PLoS One 13:e0207861. doi: 10.1371/journal.pone.0207861
Ebrahimi, A., Zarei, A., McKenna, J. R., Bujdoso, G., and Woeste, K. E. (2017). Genetic diversity of Persian walnut (Juglans regia) in the cold-temperate zone of the United States and Europe. Sci. Hortic. 220, 36–41. doi: 10.1016/j.scienta.2017.03.030
Fan, H., Ives, A. R., Surget-Groba, Y., and Cannon, C. H. (2015). An assembly and alignment-free method of phylogeny reconstruction from next-generation sequencing data. BMC Genom. 16:522. doi: 10.1186/s12864-015-1647-5
George, M. F., Hong, S. G., and Burke, M. J. (1977). Cold hardiness and deep supercooling of hardwoods: its occurrence in provenance collections of red oak, yellow birch, black walnut, and black cherry. Ecology 58, 674–680. doi: 10.2307/1939018
Gilmour, S. J., Zarka, D. G., Stockinger, E. J., Salazar, M. P., Houghton, J. M., and Thomashow, M. F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. doi: 10.1046/j.1365-313x.1998.00310.x
Grimo, E. (1979). “Carpathian (Persian) walnuts,” in Nut Tree Culture in North America, ed. R. A. Jaynes (Hamden, CT: Northern Nut Growers Association), 74–83.
Guàrdia, M., Díaz, R., Savé, R., and Aletà, N. (2013). Autumn frost resistance on several walnut species: methods comparison and impact of leaf fall. For. Sci. 59, 559–565. doi: 10.5849/forsci.12-094
Hamilton, J. A., Lexer, C., and Aitken, S. N. (2013). Genomic and phenotypic architecture of a spruce hybrid zone (Picea sitchensis× P. glauca). Mol. Ecol. 22, 827–841. doi: 10.1111/mec.12007
Hu, Y., Woeste, K. E., and Zhao, P. (2017). Completion of the chloroplast genomes of five Chinese Juglans and their contribution to chloroplast phylogeny. Front. Plant Sci. 7:1955. doi: 10.3389/fpls.2016.01955
Hyung, L. J., Yu, D. J., Kim, S. J., Choi, D., and Lee, H. J. (2012). Intraspecies differences in cold hardiness, carbohydrate content and β-amylase gene expression of Vaccinium corymbosum during cold acclimation and deacclimation. Tree Physiol. 32, 1533–1540. doi: 10.1093/treephys/tps102
Jacobs, D. F., Wilson, B. C., Ross-Davis, A. L., and Davis, A. S. (2008). Cold hardiness and transplant response of Juglans nigra seedlings subjected to alternative storage regimes. Ann. Forest. Sci. 65, 606–606. doi: 10.1051/forest:2008036
Joosen, R. V., Lammers, M., Balk, P. A., Brønnum, P., Konings, M. C., Perks, M., et al. (2006). Correlating gene expression to physiological parameters and environmental conditions during cold acclimation of Pinus sylvestris, identification of molecular markers using cDNA microarrays. Tree Physiol. 26, 1297–1313. doi: 10.1093/treephys/26.10.1297
LaBonte, N. R., Zhao, P., and Woeste, K. (2018). Signatures of selection in the genomes of Chinese chestnut (Castanea mollissima Blume): the roots of nut tree domestication. Front. Plant Sci. 9:810. doi: 10.3389/fpls.2018.00810
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 22DDCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, P., Colombo, S. J., and Sinclair, R. W. (2007). Cold hardiness of interspecific hybrids between Pinus strobus and P. wallichiana measured by post-freezing needle electrolyte leakage. Tree Physiol. 27, 243–250. doi: 10.1093/treephys/27.2.243
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18
Lyons, J. M., Graham, D., and Raison, J. K. (1979). “The plant membrane in response to low temperature: an overview,” in Low Temperature Stress in Crop Plant, eds J. M. LYONS, D. GRAHAM, and J. K. RAISON (New York, NY: Academic Press), 4–24.
Manning, W. E. (1978). The classification within the Juglandaceae. Ann. Mo. Bot. Gard. 65, 1058–1087.
Marrano, A., Britton, M., Zaini, P. A., Zimin, A., Workman, R., Puiu, D., et al. (2019). High-quality chromosome-scale assembly of the walnut (Juglans regia L) reference genome. bioRxiv [Preprint]. doi: 10.1101/809798x
Martínez-García, P. J., Crepeau, M. W., Puiu, D., Gonzalez−Ibeas, D., Whalen, J., Stevens, K. A., et al. (2016). The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non−structural polyphenols. Plant J. 87, 507–532. doi: 10.1111/tpj.13207
McKay, H. M. (1992). Electrolyte leakage from fine roots of conifer seedlings: a rapid index of plant vitality following cold storage. Can. J. For. Res. 22, 1371–1377. doi: 10.1139/x92-182
McKay, J. W. (1965). Progress in black x Persian walnut breeding. Annu. Rep. North Nut Growers Assoc. 56, 76–80.
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Mercuri, A. M., Bandini, M. M., Florenzano, A., Montecchi, M. C., and Rattighieri, E. O. (2013). Juglans and Castanea: the OJC group as pollen evidence of the development of human-induced environments in the Italian peninsula. Quat. Int. 303, 24–42. doi: 10.1016/j.quaint.2013.01.005
Mitra, S. K., Rathore, S. D., and Bose, T. K. (1991). Walnut. Temperate Fruits, Vol. 27. Kolkata: Horticulture and Allied Publishers, 377–414.
Patterson, B. D., Murata, T., and Graham, D. (1976). Electrolyte leakage induced by chilling in Passiflora species tolerant to different climates. Func. Plant Biol. 3, 435–442.
Poirier, M., Bodet, C., Ploquin, S., Saint-Joanis, B., Lacointe, A., and Améglio, T. (2004). “Walnut cultivar performance of cold resistance in south central France,” in Proceedings of the Fifth International Walnut Symposium, Sorrento, Vol. 705, 281–285. doi: 10.17660/actahortic.2005.705.35
Pollegioni, P., Woeste, K., Chiocchini, F., Del Lungo, S., Ciolfi, M., Olimpieri, I., et al. (2017). Rethinking the history of common walnut (Juglans regia L.) in Europe: its origins and human interactions. PLoS One 12:e0172541. doi: 10.1371/journal.pone.0172541
Potts, D. T. (2018). Arboriculture in ancient Iran: walnut (Juglans regia), plane (Platanus orientalis) and the “Radde dictum”. Dabir 6, 101–109.
Robichaud, R. L., Glaubitz, J. C., Rhodes, O. E., and Woeste, K. (2006). A robust set of black walnut microsatellites for parentage and clonal identification. New Forest. 32, 179–196. doi: 10.1007/s11056-005-5961-7
Sakai, A., and Weiser, C. J. (1973). Freezing resistance of trees in North America with reference to tree regions. Ecology 54, 118–126. doi: 10.2307/1934380
Schaberg, P. G., and DeHayes, D. H. (2000). “Physiological and environmental causes of freezing injury in red spruce,” in Responses of Northern US Forests to Environmental Change, eds M. A. Robert, B. A. Richard, and H. John (New York, NY: Springer), 181–227. doi: 10.1007/978-1-4612-1256-0_6
Stanford, A. M., Harden, R., and Parks, C. R. (2000). Phylogeny and biogeography of Juglans (Juglandaceae) based on matK and ITS sequence data. Am. J. Bot. 87, 872–882. doi: 10.2307/2656895
Stanke, M., Keller, O., Gunduz, I., Hayes, A., Waack, S., and Morgenstern, B. (2006). AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, 435–439.
Steel, R. G., and Torrie, J. H. (1980). Principles and Procedures of Statistics, a Biometrical Approach, 2nd Edn. Auckland: McGraw-Hill Kogakusha Ltd.
Stevens, K. A., Woeste, K., Chakraborty, S., Crepeau, M. W., Leslie, C. A., Martínez-García, P. J., et al. (2018). Genomic variation among and within six Juglans species. G3-Genes Genom Genet. 8, 2153–2165.
Sutinen, M. L., Palta, J. P., and Reich, P. B. (1992). Seasonal differences in freezing stress resistance of needles of Pinus nigra and Pinus resinosa: evaluation of the electrolyte leakage method. Tree Physiol. 11, 241–254. doi: 10.1093/treephys/11.3.241
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Boil. Evo. 24, 1596–1599. doi: 10.1093/molbev/msm092
Thornton, K. R., and Jensen, J. D. (2007). Controlling the false-positive rate in multilocus genome scans for selection. Genetics 175, 737–750. doi: 10.1534/genetics.106.064642
Wang, G., Kong, H., Sun, Y., Zhang, X., Zhang, W., Altman, N., et al. (2004). Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 135, 1084–1099. doi: 10.1104/pp.104.040436
Weiser, C. J. (1970). Cold resistance and injury in woody plants: knowledge of hardy plant adaptations to freezing stress may help us to reduce winter damage. Science 169, 1269–1278. doi: 10.1126/science.169.3952.1269
Wisniewski, M., Nassuth, A., Teulières, C., Marque, C., Rowland, J., Cao, P. B., et al. (2014). Genomics of cold hardiness in woody plants. Crit. Rev. Plant Sci. 33, 92–124.
Wisniewski, M., Norelli, J., and Artlip, T. (2015). Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 6:85. doi: 10.3389/fpls.2015.00085
Wysoker, A., Tibbetts, K., and Fennell, T. (2013). Picard Tools Version 1.90. Available online at: http://picard.sourceforge.net (accessesd April 22, 2013).
Zarei, A., Zamani, Z., Mousavi, A., Fatahi, R., Alavijeh, M. K., Dehsara, B., et al. (2012). An effective protocol for isolation of high-quality RNA from pomegranate seeds. Asian Aust. J. Plant Sci. Biotechnol. 6, 32–37.
Keywords: cold hardiness, interspecific F1 hybrids, Juglans, signature of selection, Carpathian walnut
Citation: Ebrahimi A, Lawson SS, McKenna JR and Jacobs DF (2020) Morpho-Physiological and Genomic Evaluation of Juglans Species Reveals Regional Maladaptation to Cold Stress. Front. Plant Sci. 11:229. doi: 10.3389/fpls.2020.00229
Received: 12 November 2019; Accepted: 14 February 2020;
Published: 10 March 2020.
Edited by:
Giorgio Gambino, Italian National Research Council, ItalyReviewed by:
Kim E. Hummer, National Clonal Germplasm Repository, United States Department of Agriculture, United StatesTingting Zhu, University of California, Davis, United States
Milic Curovic, University of Montenegro, Montenegro
Copyright © 2020 Ebrahimi, Lawson, McKenna and Jacobs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Douglass F. Jacobs, ZGphY29ic0BwdXJkdWUuZWR1
 Aziz Ebrahimi
Aziz Ebrahimi Shaneka S. Lawson2
Shaneka S. Lawson2 Douglass F. Jacobs
Douglass F. Jacobs