- 1College of Horticulture, Northwest A&F University, Yangling, China
- 2Tianjin Vegetable Research Center, Tianjin Academy of Agricultural Sciences, Tianjin, China
- 3Department of Horticulture, The University of Agriculture, Peshawar, Pakistan
Little information is available on the role of Squamosa promoter binding protein (SBP)-box genes in pepper plants. This family of genes is known to have transcription characteristics specific to plants and to regulate plant growth, development, stress responses, and signal transduction. To investigate their specific effects in pepper (Capsicum annuum), we screened pepper SBP-box family genes (CaSBP genes) for Phytophthora capsici (P. capsici) resistance genes using virus-induced gene silencing. CaSBP08, CaSBP11, CaSBP12, and CaSBP13, which are associated with plant defense responses against P. capsici, were obtained from among fifteen identified CaSBP genes. The function of CaSBP08 was identified in pepper defense response against P. capsici infection in particular. CaSBP08 protein was localized to the nucleus. Silencing of CaSBP08 enhanced resistance to P. capsici infection. Following P. capsici inoculation, the malondialdehyde content, peroxidase activity, and disease index percentage of the CaSBP08-silenced plants decreased compared to the control. Additionally, the expression levels of other defense-related genes, especially those of CaBPR1 and CaSAR8.2, were more strongly induced in CaSBP08-silenced plants than in the control. However, CaSBP08 overexpression in Nicotiana benthamiana enhanced susceptibility to P. capsici infection. This work provides a foundation for the further research on the role of CaSBP genes in plant defense responses against P. capsici infection.
Introduction
Pepper (Capsicum annuum L.) is a valuable solanaceous crop species with high economic as well as high nutritive value worldwide. However, it is prone to various diseases, especially Phytophthora blight, which is caused by Phytophthora capsici (Jin et al., 2016), a soil-borne pathogen that can infect various tissues of pepper plants, including roots, stems, leaves, flowers, and fruits (Wang et al., 2013a). Its host range is wide, including tomato, eggplant, cucumber, watermelon, pumpkin, snap peas, and lima beans (Granke et al., 2012). To combat pathogen infection, plants have developed a range of defense mechanisms including physiological, biochemical, molecular, and cellular processes and barriers along with inducible innate immunity (Mou et al., 2017; Hou et al., 2018). Inducible innate immunity is largely regulated at the transcriptional level by the action of many transcriptional factors. Transcription factors have important functions in combating the impact of pathogens through activating or repressing the expression of defense genes (Xu et al., 2011). For example, APETALA2/Ethylene Response Factor-domain transcription factor (AP2/ERF-ORA59) directly regulates the expression of PDF1.2 (PLANT DEFENSIN1.2) in the process of necrotrophic pathogen infection in Arabidopsis (Zarei et al., 2011). The basic leucine zipper (bZIP) of the Arabidopsis TGA6 transcription factor acts together specifically with the translocation of the NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1) ankyrin repeat protein, which activates the salicylic acid (SA) marker gene PATHOGENESIS-RELATED-1 (PR-1) in the Botrytis cinerea necrotrophic infection process (Zhang et al., 1999; Zander et al., 2010). JAMYC2 and JAMYC10 are two other MYC transcription factors that are jasmonic acid (JA)-regulated and involved in defense responses against pathogen infection through inducing PDF1.2 gene expression in tomato (Boter et al., 2004; Lorenzo and Solano, 2005).
Squamosa promoter binding protein (SBP)-box genes are a family of plant-specific transcription factors that contain a highly conserved SBP domain (Klein et al., 1996). The domain contains about 76 amino acids residues, including two zinc fingers and one nuclear localization signal (Yamasaki et al., 2004). Klein et al. (1996) first discovered Antirrhinum majus SBP-box genes, identified as AmSBP1 and AmSBP2, according to their capability to interact with the floral meristem identity gene SQUAMOSA promoter sequence. Subsequently, Lännenpää et al. (2004) identified SBP-box genes in another plant, Betula pendula (i.e., BpSPL1). Similarly, Hou et al. (2013) discovered VpSBP5 in Vitis vinifera, and Zhang B. et al. (2017) found two others in hexaploid wheat, i.e., TaSPL20 and TaSPL21. There are many reports of SBP-box gene involvement in the development and growth of plants. However, there are few reports about its involvement in responses to biotic and abiotic stresses, especially the former (Stone et al., 2005; Wang et al., 2016; Hou et al., 2018). For example, AtSPL14 is induced by the fungal toxin fumonisin B1, which induces programmed cell death in Arabidopsis (Stone et al., 2005). Transgenic Arabidopsis JASMONATE CARBOXYL METHYLTRANSFERASE (AtJMT) plants exhibit down-regulation of AtSPL2 (At5g43270), which has a role in the JA-mediated resistance pathway (Kim et al., 2007). Tobacco NbSPL6 is needed to develop N-mediated resistance for combating tobacco mosaic virus. Moreover, the ortholog AtSPL6 Arabidopsis gene is necessary for the Toll and Interleukin-1 Receptor Nucleotide Binding-Leucine Rich Repeat (TIR-NB-LRR) to function in mediating resistance against Pseudomonas syringae infection (Padmanabhan et al., 2013). Hou et al. (2013) reported that through the SA-induced systemic acquired resistance pathway, VpSBP5 participates in regulating resistance against Erysiphe necator and, in grapes, through the methyl jasmonate (MeJA)-induced wound signaling pathway. AtSPL9 interacts with jasmonate ZIM-domain (JAZ) proteins and negatively regulates the JA response as well as resistance to insects in Arabidopsis (Mao et al., 2017). SPL6 functions in the endoplasmic reticulum (ER) stress response to control ER stress signaling outputs and retain equilibrium between both adaptive and death signals to determine cell fates in rice during ER stress (Wang et al., 2018). However, compared with the widespread research on SBP-box genes in model species such as Arabidopsis, less information is available concerning pepper SBP-box genes, especially regarding their involvement in resistance against P. capsici.
In our previous research, we identified fifteen SBP-box genes in pepper (i.e., CaSBP01, Capana01g002647; CaSBP02, Capana01g002832; CaSBP03, Capana01g003073; CaSBP04, Capana01g003445; CaSBP05, Capana02g001917; CaSBP06, Capana05g002237; CaSBP07, Capana07g001731; CaSBP08, CA07g17550; CaSBP09, CA08g03640; CaSBP10, Capana10g000507; CaSBP11, Capana10g000709; CaSBP12, Capana10g000886; CaSBP13, Capana10g002379; CaSBP14, Capana11g002003; and CaSBP15, CA11g04690), and we named them according to their chromosomal order (Zhang et al., 2016). The CaSBP coding sequences ranged from 336 base pair (bp) to 3024 bp in length for CaSBP08 and CaSBP06, respectively, with a nuclear localization signal occurring for all gene family members except CaSBP08. Additionally, all the CaSBP genes encoded proteins with two zinc finger-like structures, i.e. C3H and C2HC, except for CaSBP09 and CaSBP15, which lack the C3H zinc finger-like structures (Zhang et al., 2016). All CaSBP genes are induced by compatible or incompatible strains of P. capsici, except for CaSBP15, whose expression is down-regulated during P. capsici infection. Some CaSBP genes (i.e., CaSBP11 and CaSBP12) may also be involved in SA and MeJA regulation mechanisms (Zhang et al., 2016). To further study the function of CaSBP genes in plant resistance against the pathogen P. capsici, CaSBP08, CaSBP11, CaSBP12, and CaSBP13, which are involved the plant defense response against P. capsica, were obtained from among the fifteen identified CaSBP genes using virus-induced gene silencing in this work. In addition, we further investigated the function of CaSBP08 and found it to be localized to the nucleus and to play a negative regulatory role in the plant defense response against P. capsici infection in pepper and transgenic Nicotiana benthamiana. This work provides a foundation for further research on the role of pepper SBP-box genes in plant defense responses against P. capsici infection.
Materials and Methods
Plant Material and Pathogen Preparation
Pepper cultivar AA3 and the P. capsici strain HX-9, provided by the Capsicum Research Group, College of Horticulture, Northwest A&F University, P. R. China, were tested. Plants were maintained in a growth chamber at 22°C/18°C (day/night temperatures) with a 16/8-hour photoperiod. The P. capsici strain HX-9 was cultured in darkness at 28°C with potato dextrose agar (PDA) medium. Sporulation induction and spore release were conducted using the method described by Wang et al. (2011) with modifications. Briefly, the HX-9 strain of P. capsici was first cultured on PDA in a Petri dish under darkness at 28°C for five days. Then, ten approximately 0.8-cm diameter discs were cut from PDA culture plates and grown in the dark for 3 days in 90-mm Petri dishes with 15–20 mL of 2% (w/v) cleared carrot broth at a constant temperature of 28°C. The cultures were then washed twice with sterile distilled water and covered with 15–20 mL of Petri broth [KH2PO4, 0.15 g; Ca(NO3)2, 0.4 g; CaCl2, 0.06 g; Mg(NO3)2, 0.15 g each per 1000 mL]. These cultures were further incubated at 28°C for five more days, before being chilled for 30 minutes at 4°C to induce zoospore release, followed by a 1-hour incubation at room temperature. A hemocytometer was used to measure zoospore concentration adjusted to 1 × 105 spores/mL following the method by Jin et al. (2016). Then, 5 mL of this zoospore culture was used to inoculate CaSBP08-silenced, control pepper plants, and transgenic N. benthamiana plants via the root-drench method as described by Wang (2013). A detached leaf inoculation assay was prepared as per the method described by Zhang et al. (2015) and maintained at 22°C/18°C (day/night temperature) under a 16/8-hour photoperiod and 60% relative humidity in a growth chamber.
Virus-Induced Gene Silencing of SBP-Box Family Genes in Pepper
The virus-induced gene silencing (VIGS) system based on tobacco rattle virus (TRV) was used to silence the SBP-box family pepper genes, as previously reported by Wang (2013). To generate the VIGS plasmid constructs of CaSBP genes, fifteen 200–500 bp fragments from the corresponding SBP-box genes were amplified using gene-specific primers. Then, their specificities were assessed using NCBI Primer BLAST (Table S1). Using double digestion, the acquired products were cloned into the TRV2 vectors with BamHI and KpnI restriction enzymes (Zhang et al., 2003). Then, they were sequenced by Sangon Biotech Company (Shanghai, China). The recombined vectors, i.e., TRV2:CaSBP01, TRV2:CaSBP02, TRV2:CaSBP03, TRV2:CaSBP04, TRV2:CaSBP05, TRV2:CaSBP06, TRV2:CaSBP07, TRV2:CaSBP08, TRV2:CaSBP09, TRV2:CaSBP10, TRV2:CaSBP11, TRV2:CaSBP12, TRV2:CaSBP13, TRV2:CaSBP14, TRV2 (negative control), TRV2:CaPDS (phytoene desaturase, positive control), and TRV1, were transformed into Agrobacterium tumefaciens strain GV3101 using freeze-thaw transformation. Pepper seedlings at the two true leaves stage were used for the procedures silencing the SBP-box genes family according to the method described by Zhang et al. (2013). All the injected plants were maintained in a growth chamber set at 18°C under darkness for two days and then moved to a growth chamber with 22°C/18°C (day/night temperatures), a 16/8-hour photoperiod, and 60% relative humidity. Forty-five days after infiltration, the silencing efficiency was measured from leaf samples collected from the silenced and control plants. Then, an assay of the detached leaves was conducted as described by Zhang et al. (2015). Five milliliters of 1 × 105 spores/mL zoospore culture of P. capsici strain HX-9 was used to inoculate the control and silenced plants by drenching roots. Lastly, the roots and leaves of control and silenced plants were collected and stored at −80°C.
Subcellular Localization of CaSBP08
The CaSBP08 coding region without a termination codon was amplified using sequence-specific primers (Table S1). The obtained product was cloned into the PMD-19 vector and then cloned into the pVBG2307:GFP vector between the XbaI and KpnI restriction sites to produce the final pVBG2307:CaSBP08:GFP plasmid. The recombinant fusion pVBG2307:CaSBP08:GFP plasmid was sequenced by Sangon Biotech Company and then transformed into A. tumefaciens strain GV3101 through freeze-thaw transformation. Then, the GV3101 cells carrying the pVBG2307:CaSBP08:GFP vector and pVBG2307:GFP vector (used as a control) were grown overnight in Luria-Bertani (LB) medium with the proper antibiotics. Then, infiltration buffer (10 mM MES, pH 5.7, 10 mM MgCl2, and 200 µM acetosyringone) was used for cell suspension, which was infiltrated into N. benthamiana leaves with a needleless syringe (Mou et al., 2017). After being injected, the plants were grown in a chamber set at 22°C/18°C (day/night) temperatures and a 16/8-hour photoperiod for two days and then assessed under a fluorescent confocal microscope (Olympus, Tokyo, Japan) with a 488 nm excitation wavelength.
N. benthamiana Transformation
The full encoding region of CaSBP08 (336 bp) was cloned into the pVBG2307:GFP vector between the XbaI and KpnI restriction sites to produce the final plasmid pVBG2307:CaSBP08:GFP for genetic transformation (Table S1). Overexpression lines of CaSBP08 were achieved through tobacco leaf disc transformation with Agrobacterium intervention (Oh et al., 2005). Eleven lines of transgenic N. benthamiana plants, each with resistance to kanamycin and having the pVBG2307:CaSBP08:GFP construct were obtained. Three transgenic lines of CaSBP08 (lines 2, 10, and 11) were randomly selected for further study. Transformation was confirmed using quantitative real-time PCR during the T2 generation (Table S2). T1 plants seeds were obtained from T0 regenerated plants, and T2 lines seedlings were generated on MS agar plates with 100 µg/mL kanamycin. For further analyses, T3 plants were used.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was isolated as per the procedures described by Guo et al. (2012). The first strand cDNA was synthesized using the PrimeScript Kit (Takara, Dalian, China) according to the manufacturer's instructions. The cDNA concentration was diluted to 50 ng/µL and used for quantitative real-time PCR (qRT-PCR). Then, qRT-PCR was performed in triplicate on an iCycler iQ™ Multicolor PCR Detection System (Bio-Rad, Hercules, CA, USA) with the following thermal cycling program: pre-denaturation at 95°C for 1 min followed by 40 cycles of denaturization at 95°C for 10 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s. All the primer specificities for qRT-PCR were assessed using NCBI Primer BLAST (Table S2). Gene expression was quantified and normalized to the expression level of actin (CaActin2, accession no. AY572427; Nbactin-97, accession No. LOC109206422) (Schmittgen and Livak, 2008; Du et al., 2015; Yin et al., 2015).
Malondialdehyde Measurement
Following inoculation with P. capsici, the malondialdehyde (MDA) content of the control plants and CaSBP08-silenced plants were measured using a colorimetric determination technique with thiobarbituric acid from Ma et al. (2013) with modifications. For this purpose, the crude enzyme used for MDA determination was extracted using 10% trichloroacetic acid (TCA). Then, 2 mL of crude enzyme extract was mixed with 5 mL of 0.5% thiobarbituric acid (TBA) reagent, boiled for 10 min, quickly cooled, and centrifuged at 5000 × g for 10 min. The control contained 2 mL of distilled water instead of the crude enzyme. Absorbance was measured at 600 nm, 532 nm, and 450 nm.
Peroxidase and Catalase Activity Measurements
After inoculation with P. capsici, the peroxidase (POD) and catalase (CAT) activities of the CaSBP08-silenced and control plants were measured using the guaiacol method and ultraviolet spectrophotometry, respectively (Hammerschmidt et al., 1982). The crude enzymes, used to determine POD and CAT activity, were extracted using 0.2 M phosphate buffer (pH 7.8) and 0.05M Tris-HCl buffer (pH7.0), respectively. The POD activity determination reaction included 0.1 mL of crude enzyme, 2 mL of 0.3% H2O2, and 0.9 mL of 0.2% guaiacol. The CAT activity determination reaction included 1.0 mL of Tris-HCl, 1.7 mL of distilled water, 0.5 mL of crude enzyme, and 0.1 mL of 100 µM H2O2.
Disease Index Percentage Statistics
Subsequent to P. capsici inoculation, the percent disease index values of plants were recorded following the procedure described by Zhang (2009). Sixteen days after P. capsici HX-9 strain inoculation, the CaSBP08-silenced and control plants infection symptoms were categorized into five levels: Level 0, no symptoms; Level 1, lower leaves of plants yellowing or wilting; Level 2, lower leaves of plants with obvious defoliation or whole plants wilting; Level 3, stem base black, except for new growth, with all leaves fallen; and Level 4, whole plant death. Thirteen days post-inoculation with P. capsici, the symptoms in the transgenic lines were also categorized into five levels: Level 0, no symptoms; Level 1, whole plant wilting, with no constriction between stems and leaves; Level 2, whole plant wilting, with death of lower leaves and constriction between stems and leaves; Level 3, all leaves dead, except those at the point of new growth, with constriction occurring between stems and leaves; Level 4: death of the whole plant. Disease index percentages were recorded based on the following formula:
Statistical Analysis
Least significant difference (LSD) values were calculated using Data Processing System 7.05 (DPS 7.05, China), a software package with comprehensive experimental design and statistical analysis functions. Significance was determined at P ≤ 0.05 or P ≤ 0.01 thresholds. All the experiments, with at least three biological replicates, were conducted and evaluated separately.
Results
Pepper Plant CaSBP08, CaSBP11, CaSBP12, and CaSBP13 Genes Are Involved in Resistance to P. capsici
Phenotypic Observation and Silencing Efficiency of CaSBP-Silenced Plants
To screen for genes that respond to P. capsici infection, fifteen CaSBP genes were silenced using the VIGS method. In this study, the pepper CaPDS gene (phytoene desaturase, GenBank accession number, X68058) was taken as a positive control, which induces a leaf photo-bleaching phenotype when silenced. For the negative control, an empty TRV2:00 vector was selected. Forty days after injection, the positive control (TRV2:CaPDS) plants showed photo-bleaching, while the TRV2:00 plants and plants with each of the fifteen CaSBP genes silenced exhibited no obvious phenotypic changes (Figure 1A). Subsequently, we detected the silencing efficiency of the fifteen CaSBP genes. As shown in Figure 1B, the fifteen CaSBP genes were silenced compared with the negative control, and the silencing efficiencies were between 50% and 90%. To ensure the silencing specificity of the target CaSBP genes, the expression of both the target genes as well as the genes with the highest homology to the target genes were also measured. As shown in Figure 1C, when the target gene was silenced, the expression levels of the CaSBP genes and their respective genes with the highest homology can be divided into three categories. The first category includes silenced target genes (i.e., CaSBP02 or CaSBP09) for which their corresponding gene of highest homology (i.e., CaSBP06 or CaSBP15) is also silenced, but with a silencing efficiency lower than that of the target gene. The second category included target genes that were silenced (i.e., CaSBP04, CaSBP11, CaSBP12, CaSBP14, or CaSBP15), but their corresponding highest homology genes (i.e., CaSBP12, CaSBP09, CaSBP04 or CaSBP09) had increased expression. The third category includes silenced target genes (i.e., CaSBP05 or CaSBP10) with corresponding highest homology genes (i.e. CaSBP10 or CaSBP05) having unchanged expression. The CaSBP genes with the highest homology with the targeted silenced genes are shown in Table S3.
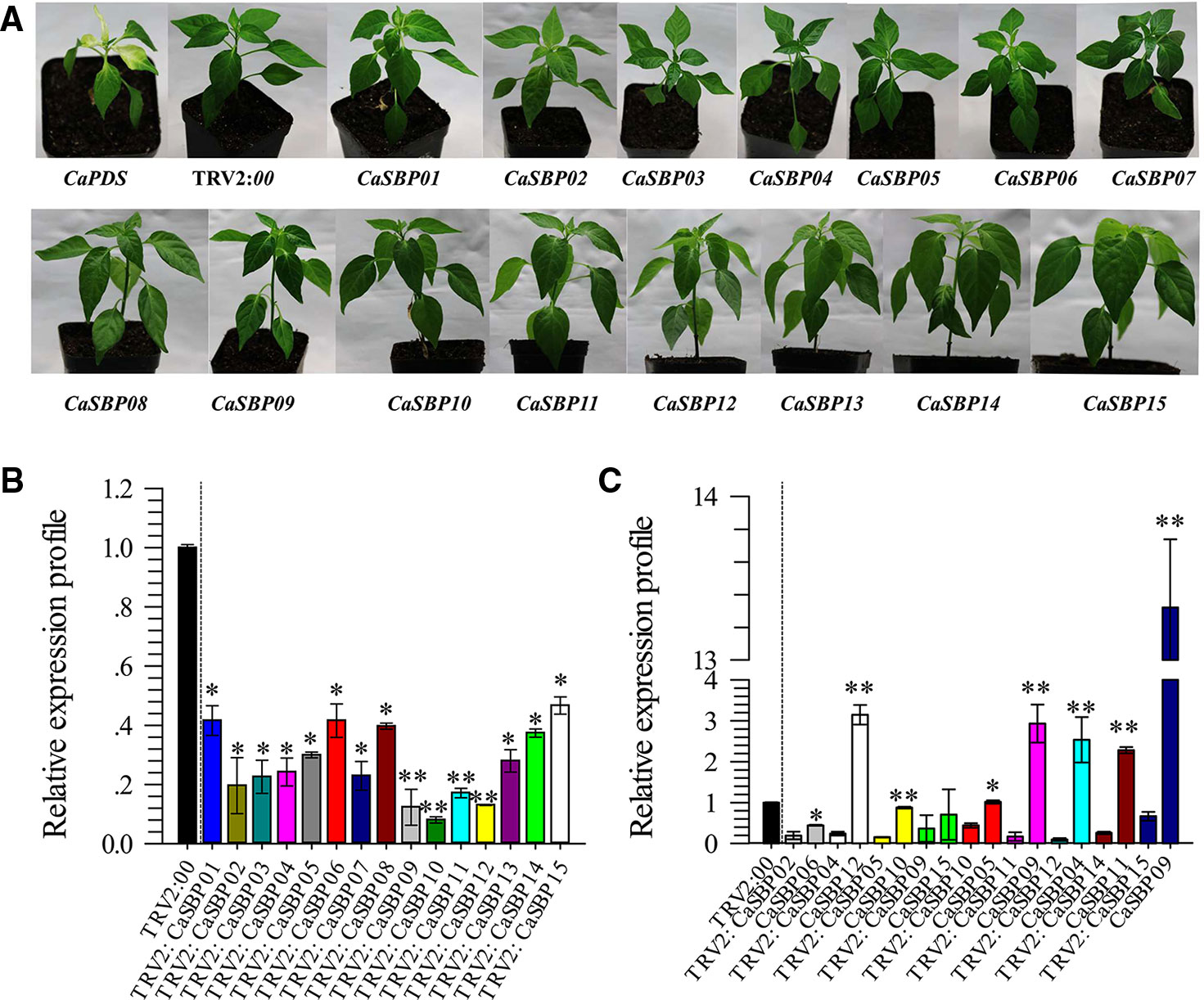
Figure 1 Phenotypes and silencing efficiency of the fifteen members of CaSBPs silenced plant. (A) Photographs were taken forty days after injection (the diameter of the pot is 7 cm). (B) Silencing efficiency of the fifteen members of CaSBPs in their corresponding silenced plants. (C) The expression level of the pepper SBP-box genes, which with the highest homology with their corresponding silenced gene. * and ** represent significant differences at P ≤ 0.05 and P ≤ 0.01 respectively. Mean values and SDs for three biological replicates are shown.
Identification of Resistance Associated With CaSBP Genes
Forty-five days after injection, detached leaves of plants silenced for each of the fifteen CaSBP genes and those of the negative control plants were inoculated with P. capsici strain HX-9. Three days post-inoculation with HX-9, the detached leaves of the negative control plants exhibited large hygrophanous lesions, which occupied almost 80% of the whole leaf area (Figure 2A). Additionally, the detached leaves of the plants silenced for CaSBP01, CaSBP02, CaSBP03, CaSBP04, CaSBP05, CaSBP06, CaSBP07, CaSBP09, CaSBP10, CaSBP14, and CaSBP15 also exhibited large hygrophanous lesions without any noticeable difference from the negative control (Figures 2A, B). However, the detached leaves of the CaSBP08-, CaSBP11-, CaSBP12-, and CaSBP13-silenced plants exhibited smaller hygrophanous lesions or lacked them altogether (Figure 2A). In addition, the percentages of lesion areas on the leaves of CaSBP08-, CaSBP11-, CaSBP12-, and CaSBP13-silenced plants and negative control plants also significantly differed (Figure 2B). Detailed statistics of the disease incidence of detached leaves from CaSBP-silenced and negative control plants are available in Table S4. These results indicated that four CaSBP genes (CaSBP08, CaSBP11, CaSBP12, and CaSBP13) responded to P. capsici infection in pepper.
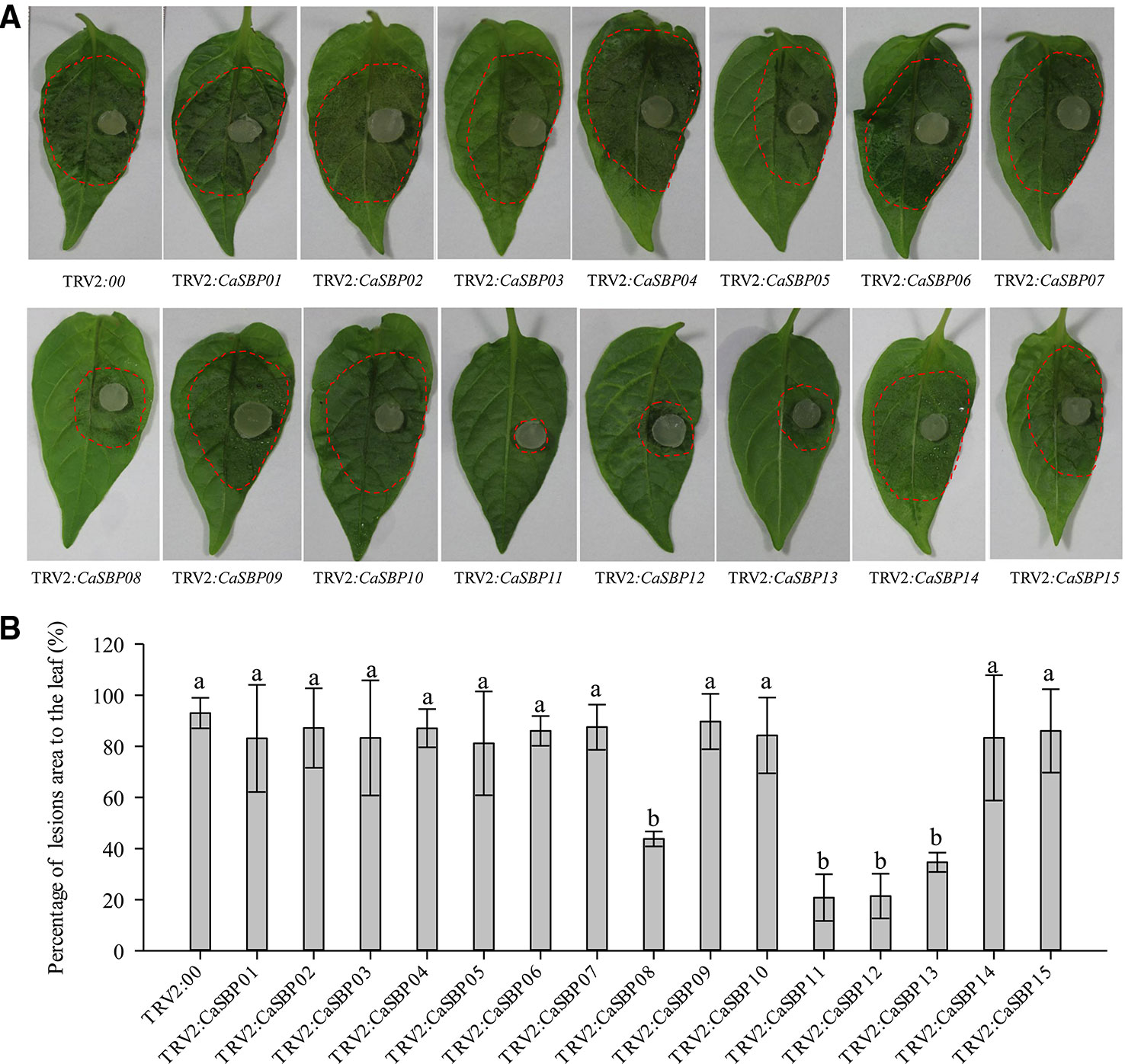
Figure 2 Phenotype and percentage of the lesion area of detached leaves of the fifteen members of CaSBPs, silenced and control plants after inoculation with P. capsici. (A) Phenotype of detached leaves of the fifteen members of CaSBPs, silenced and control plants after inoculation with P. capsici. Photographs were taken after inoculation with P. capsici three days. The diameter of the plug of P. capsici is 0.5cm. The red dotted line was used to label the lesion area in each leaf. (B) Percentage of the lesion area of the leaves three days after inoculation with P. capsici. Bars with different lower case letters indicate significant differences at P ≤ 0.05. Mean values and SDs for three biological replicates are shown.
CaSBP08 Protein Localization in the Nucleus
To determine the subcellular localization of CaSBP08 protein, the GV3101 strain of A. tumefaciens with pVBG2307:CaSBP08:GFP and pVBG2307:GFP (used as a control) vectors were rapidly expressed in the leaves of N. benthamiana plants. The results indicated that the control (pVBG2307:GFP) exhibited GFP signals in the whole cell, including the nucleus, cytoplasm, cell wall, and cell membrane, whereas, pVBG2307:CaSBP08:GFP only exhibited GFP signals in the nucleus (Figure 3). This indicated that CaSBP08 protein was localized in the nucleus.
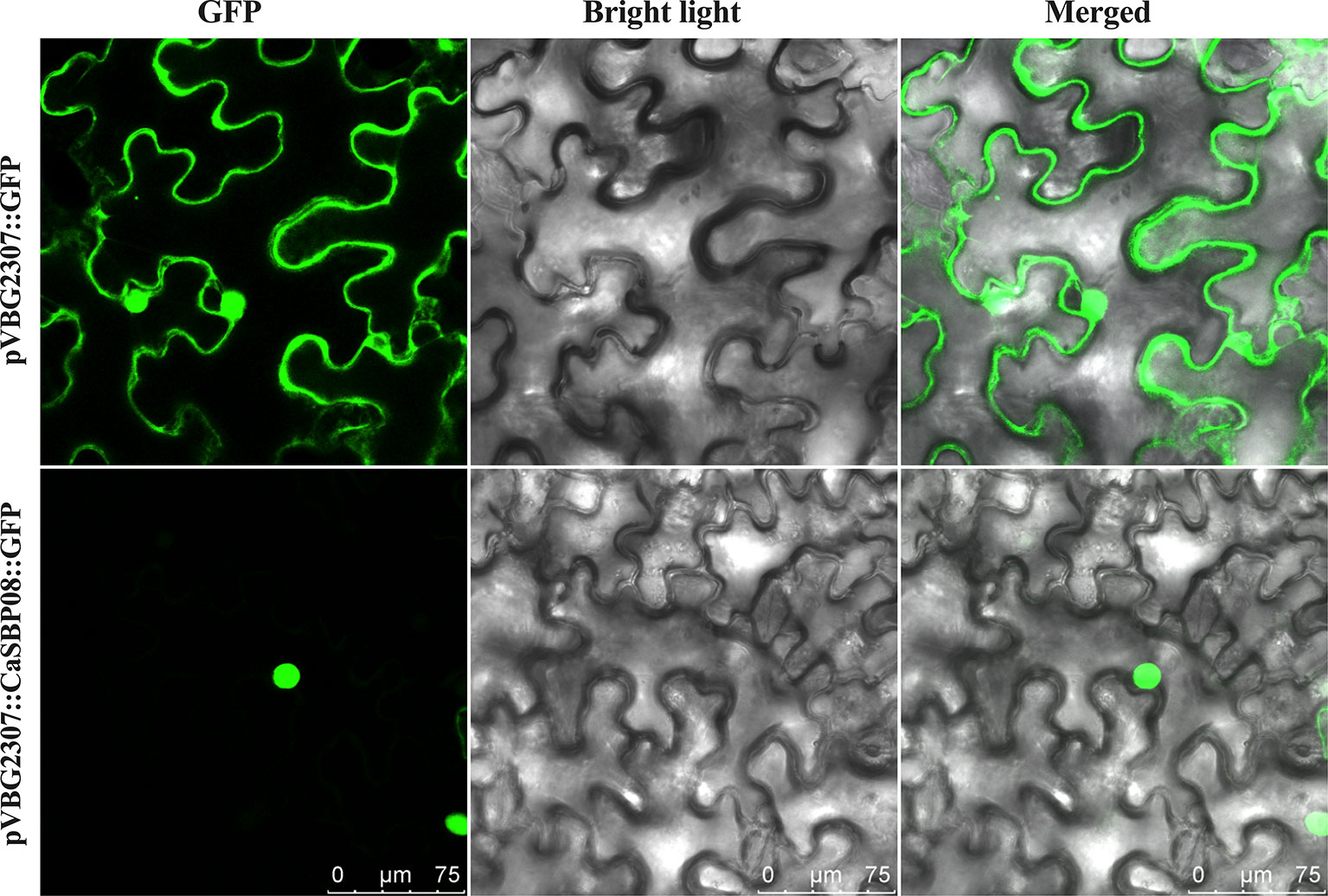
Figure 3 Subcellular localization of the CaSBP08 protein. Agrobacterium tumefaciens strain GV3101 with pVBG2307:CaSBP08:GFP and pVBG : GFP (used as a control) vectors were transiently expressed in N. benthamiana leaves. The fluorescence was visualized using a laser scanning confocal microscope under bright and fluorescent fields. The photographs were taken in a dark field for green fluorescence and under bright light for the morphology of the cell. Bars in this picture are 75μm.
Silencing of CaSBP08 Enhanced Pepper Resistance to P. capsici Infection
In order to confirm the CaSBP genes involved in resistance to P. capsici infection, we first selected CaSBP08 for further study using the virus-induced silencing procedure. As Figure S1 shows, the CaSBP08 gene was considerably silenced, and the silencing efficiency was 75%. Two days after inoculation with the P. capsici HX-9 strain, the detached leaves of negative control plants showed obvious hygrophanous lesions, while those of the CaSBP08-silenced plants displayed no or small hygrophanous lesions (Figure 4A). Additionally, the average diseased area of the leaves of CaSBP08-silenced plants was significantly smaller than that of the negative control plants (Figure 4B). Additionally, sixteen days after P. capsici infection, the damage to CaSBP08-silenced plants was less than that to control plants (Figure 4C). The disease index percentages of CaSBP08-silenced plants were substantially lower than those for the control plants (Figure 4D). Moreover, the MDA content in the CaSBP08-silenced plant was lower than that of the control plants (Figure 4E-1). The POD and CAT activities increased in the CaSBP08-silenced and negative control plants (Figures 4E-2, 4E-3). However, the POD and CAT activities in the CaSBP08-silenced plants were less than those in the negative control plants (Figures 4E-2, 4E-3). The CaSBP08 expression level increased in the beginning and then decreased, but the expression level in the silenced plants was considerably lesser than that in the negative control plants (Figure 5A). The expression of the defense genes (CaDEF1, AF442388; CaSAR8.2, AF112868; CaPO1, AF442386; CaBPR1, AF053343) increased in CaSBP08-silenced plants to different degrees, and the expression level was more than that in the negative control plants at day one (Figures 5B–E). However, the expression of the defense-related genes (i.e., CaPO1, CaDEF1, CaBPR1, and CaSAR8.2) in the CaSBP08-silenced and negative control plants decreased at day two (except for CaDEF1; Figures 5B–E). The results revealed that CaSBP08 played a negative role in the plant defense response against P. capsici infection.
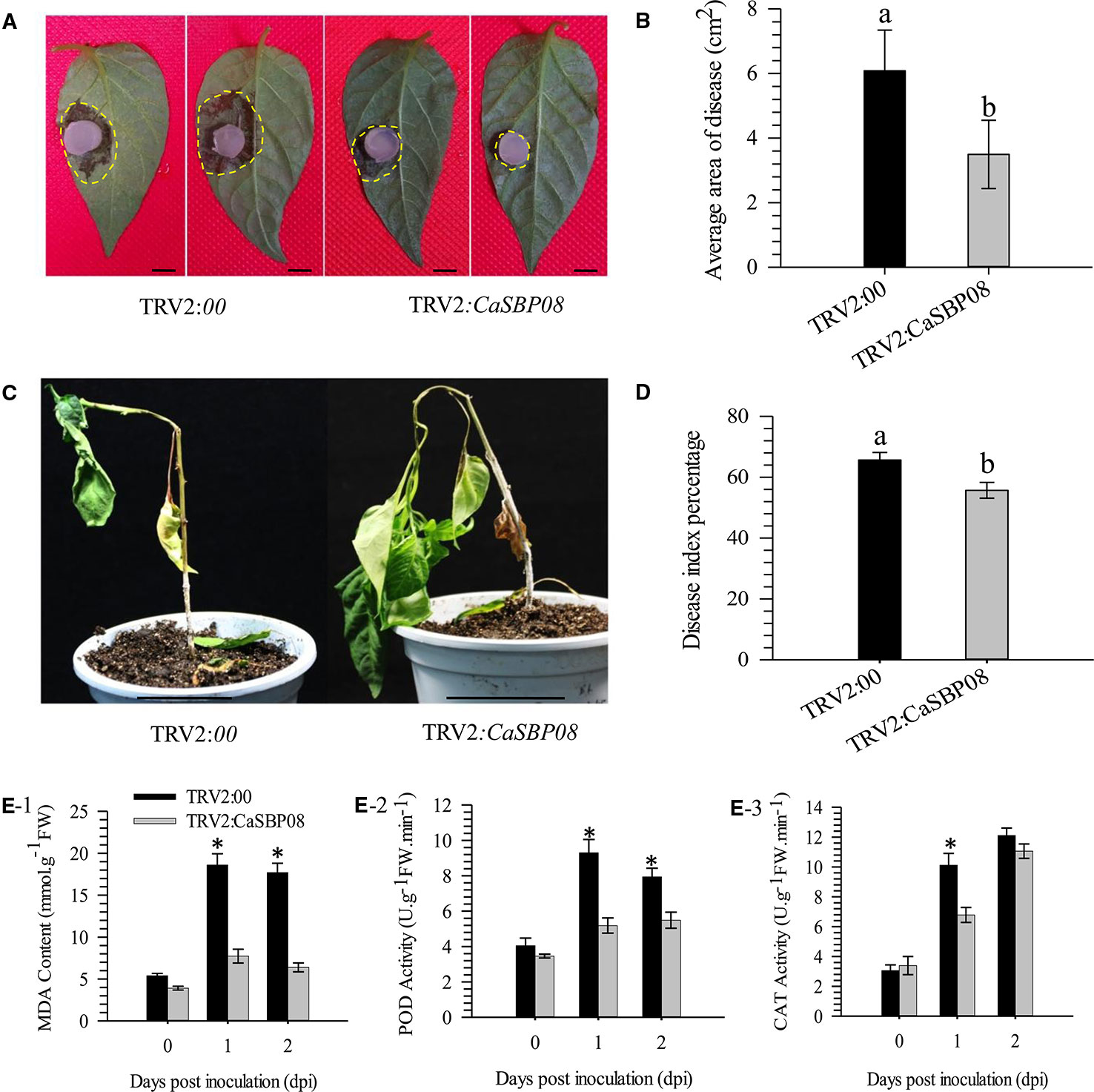
Figure 4 Loss function analysis of CaSBP08 in pepper plant defense response against P. capsici infection. (A) Phenotypes of detached leaves of CaSBP08 silenced and negative control plants after inoculation with P. capsici. Photographs were taken at two days after inoculation with P. capsici. The yellow dotted line was used to label the lesion area in each leaf. (B) The average diseased areas of the detached leaves of the CaSBP08 silenced and negative control plants. Data were collected, two days after inoculation with P. capsici. (C) Phenotypes of the CaSBP08 silenced and negative control plants after inoculation with P. capsici sixteen days. (D) The disease index percentage of the CaSBP08 silenced and negative control plants and data were collected sixteen days after inoculation with P. capsici. (E) Determination of MDA content (E-1), POD activity (E-2), and CAT activity (E-3) of CaSBP08 silenced and negative control plants after inoculating with P. capsici. Bars in Figure A are 0.5cm, and C are 4.5cm. Bars with different letters indicate significant differences at P ≤ 0.05. * Represent significant differences at P ≤ 0.05. Mean values and SDs for three biological replicates are shown.
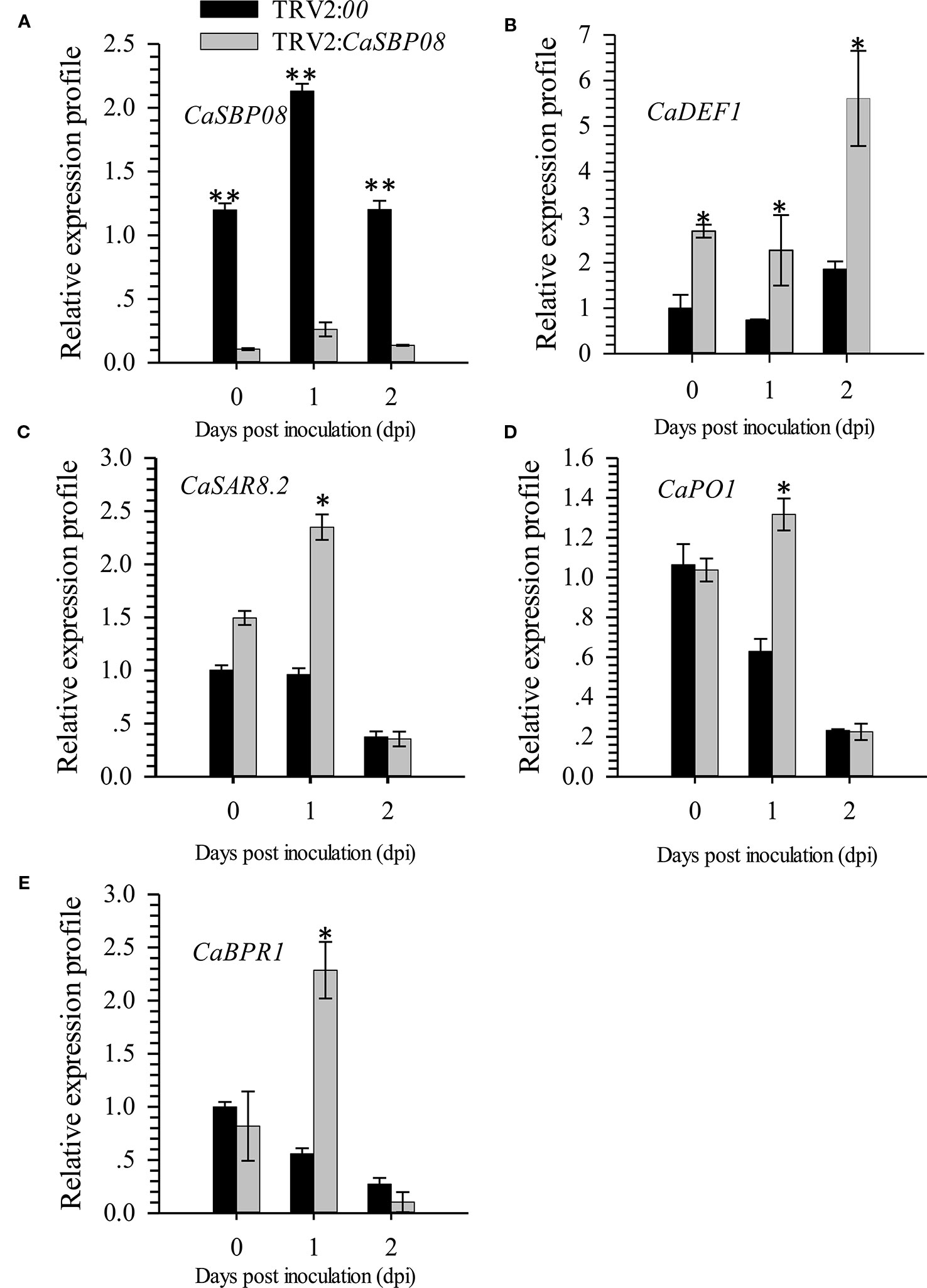
Figure 5 The expression of CaSBP08 (A), and defense-related genes, CaDEF1 (B), CaSAR8.2 (C), CaPO1 (D), and CaBPR1 (E) after inoculation with P. capsici in CaSBP08 silenced and negative control plants. * and ** represent significant differences at P ≤ 0.05 and P ≤ 0.01 respectively. Mean values and SDs for three biological replicates are shown.
Overexpression of CaSBP08 in N. benthamiana Increased Susceptibility to P. capsici Infection
In order to confirm CaSBP08 is involved in plant resistance to P. capsici infection, transgenic CaSBP08 lines were obtained by Agrobacterium-mediated tobacco leaf disc transformation, as the stable transformation of pepper plants remains challenging. Eleven transgenic lines were acquired, and there were no observable differences among their phenotypes. Then, three CaSBP08 transgenic lines (lines 2, 10, and 11) were randomly selected for disease resistance assays. Seedlings of forty-day-old plants were used for the following experiment. Two days after the P. capsici HX-9 strain inoculation, a small hygrophanous lesion area appeared on the detached leaves of wild-type (WT) plants, while the hygrophanous lesion area occupied almost half of the detached leaves from transgenic lines 2, 10, and 11 (Figure 6A). Additionally, the average areas of disease of transgenic lines 2, 10, and 11 were significantly higher than that of WT plants (Figure 6B). Three days after HX-9 strain inoculation, no disease symptoms were observed in WT plants, whereas in transgenic plants (lines 2, 10, and 11) wilting and constriction at the junction of the root and stem were observed (Figure 6C). In addition, the forty-five-day-old seedings were used for the disease index percentage statistics experiment. Thirteen days post-inoculation with P. capsici strain HX-9, the disease symptoms in WT and transgenic plants (lines 2, 10, and 11) were categorized into five levels (Figure 6D). The disease index percentages of transgenic lines 2, 4, and 11 were substantially higher than that of WT plants (Figure 6E). A detailed summary of disease index percentage data is provided in Table S5.
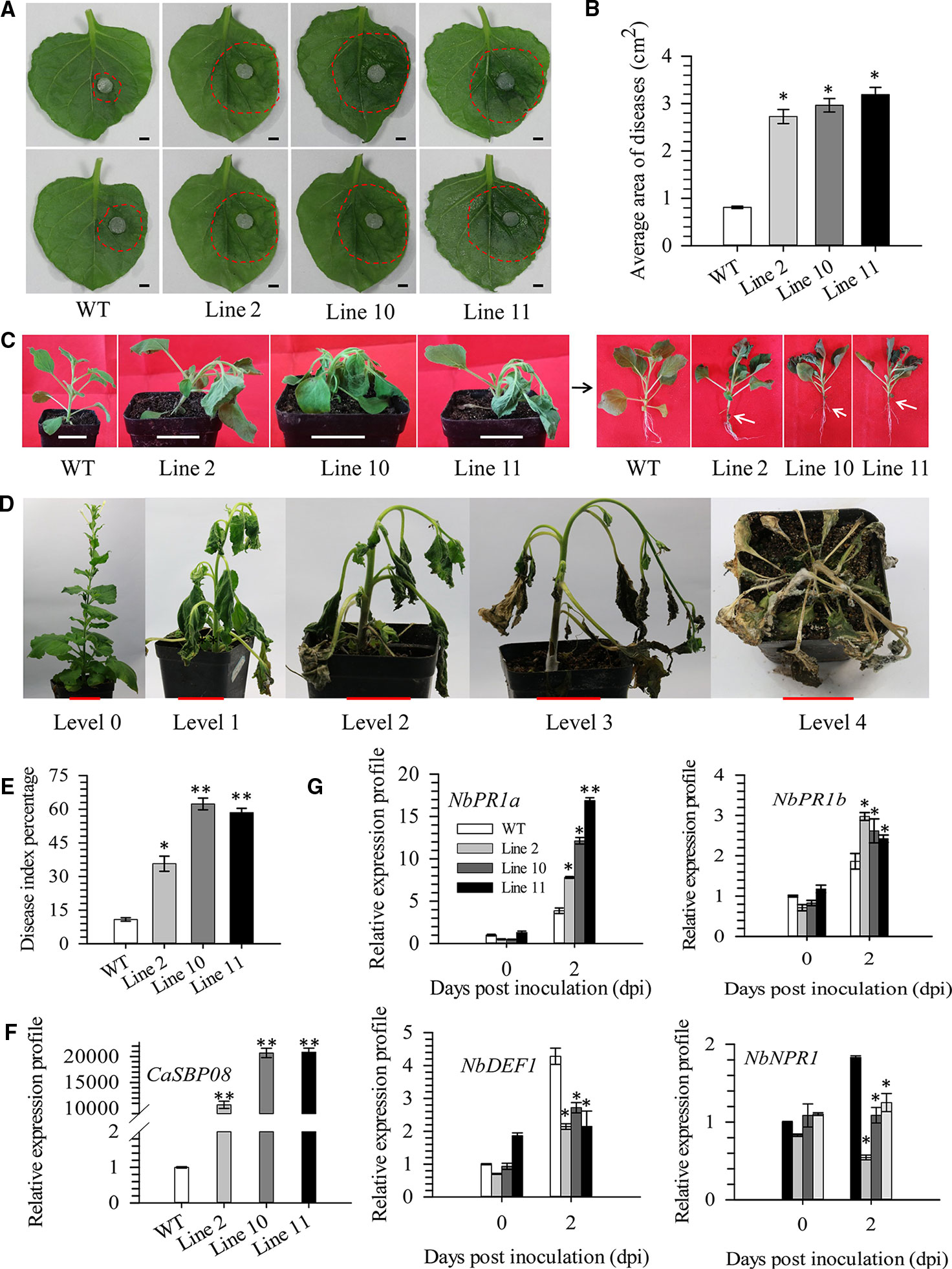
Figure 6 Function analysis of overexpression of CaSBP08 transgenic lines in defense response against P. capsici infection. (A) Phenotypes of the detached leaves of transgenic and wild-type plants after inoculation with P. capsici two days. The red dotted line was used to label the lesion area in each leaf. (B) The average diseased areas of the detached leaves of transgenic and wild-type plants. Data were collected, two days after inoculation with P. capsici. (C) Phenotypes of transgenic and wild-type plants after inoculation with P. capsici three days. The black arrow indicates the phenotype of the left transgenic and wild-type plants after removing the matrix. White arrows indicate the constricted area between root and stem. (D) Classification of disease index percentage of transgenic and wild-type plants after inoculation with P. capsici thirteen days. (E) Disease index percentage of transgenic and wild-type plants and data were collected thirteen days after inoculation with P. capsici. (F) The expression level of CaSBP08 in transgenic and wild-type plants. (G) The expression of defense-related genes in transgenic and wild-type plants after inoculation with P. capsici. Bars in Figure A are 0.4cm, C and D are 3.5cm. * and ** represent significant differences at P ≤ 0.05 and P ≤ 0.01 respectively. Mean values and SDs for three biological replicates are shown.
The CaSBP08 expression in transgenic lines 2, 10, and 11 differed from that of WT plants (Figure 6F). The expression of defense-related genes, i.e., NbPR1a (pathogenesis-related), NbPR1b (pathogenesis-related), NbDEF1 (defensin), and NbNPR1 (non-expressor pathogenesis-related), were measured. After P. capsici inoculation, the expression of NbPR1a (JN247448.1) and NbPR1b (XM_016587501.1) increased at day two, and its level in transgenic lines was more than that in the WT plants (Figure 6G). The expression levels of NbDEF1 (X99403) and NbNPR1 (AF480488) in WT plants increased at day two and were higher than that in the transgenic lines (Figure 6G). However, the expression levels of NbDEF1 and NbNPR1 in transgenic lines had no obvious changes between day one and day two. These results reveal that CaSBP08 has a negative role in the defense response of plants against P. capsici infection.
Discussion
The SBP-box gene family is comprised only of plant transcription factors. Gene family memebers participate in different pathways, including those related to plant morphogenesis, floral transition, male sterility, biosynthesis of gibberellic acid (GA), transition from the vegetative to reproductive stage, endoplasmic reticulum (ER) stress signaling, and environmental stress responses (Cardon et al., 1997; Zhang et al., 2007; Shikata et al., 2012; Ning et al., 2017; Zhang J. et al., 2017). These roles of SBP-box genes were investigated in other plants. However, the function of CaSBPs, especially in plant defense responses against P. capsici infection have not yet been studied.
We screened a gene (CaSBP05) identified as differentially expressed from our previously established transcriptome database of P. capsici across different affinity races. We analyzed the expression patterns of CaSBP genes under infections with P. capsici-compatible and -incompatible strains and hormonal treatment in our previous study (Zhang et al., 2016). We found that most CaSBP genes are induced by hormones and P. capsici infection, but there is no direct evidence proving that they are involved in pepper defence mechanisms against P. capsici infection (Zhang et al., 2016). Therefore, to screen P. capsici infection response genes from among CaSBP genes, fifteen CaSBP genes were silenced. The phenotypes of the plants with each of the fifteen silenced CaSBP genes exhibited no obvious differences compared with the control plants (Figure 1A). It is known that most of the genes in SBP-box family are related to plant growth, development, and morphogenesis. For example, overexpression of the small RNA molecule miR156/157, whose target is the protein-regulated SBP-box genes TfLFY and TfMIR172 in Torenia plants, can induce bushy plant architectures in Torenia fournieri (Shikata et al., 2012). Overexpression of tae-miR156, whose target is the squamosa promoter binding protein-like genes (TaSPL3/17) in the bread wheat cultivar ‘Kenong199,' leads to an increase in tiller number and serious flaws in spikelet development, and tae-miR156 mediated suppression of some squamosa promoter binding genes (Liu et al., 2017). Arabidopsis AtSPL14 mutants with a T-DNA insertion in their squamosa binding protein (SBP) domain exhibited altered architectures with petiole elongation and more serrated leaf margins (Stone et al., 2005). However, these phenomena were not observed in pepper plants with each of the fifteen CaSBP genes silenced. Moreover, in order to confirm the silencing specificity of each of the CaSBP genes, we also measured the relative expression of genes with the highest homology compared to each of the silent genes.
When the target pepper SBP-box genes were silenced, the expression levels of the pepper SBP-box genes and their corresponding genes with the highest homology can be divided into three categories (Figure 1C). In the first, the target gene was silenced (i.e., CaSBP02 or CaSBP09), and the genes with the highest homology (i.e., CaSBP06 or CaSBP15) were also silenced, but with a lower silencing efficiency. The second category included target genes that were silenced (i.e., CaSBP04, CaSBP11, CaSBP12, CaSBP14, or CaSBP15), while their corresponding genes with highest homology (i.e., CaSBP12, CaSBP09, CaSBP04, or CaSBP09) showed increased expression. The third category included target genes that were silenced (i.e., CaSBP05 or CaSBP10), with their genes of highest homology (i.e., CaSBP10 or CaSBP05) remaining unchanged (Figure 1C).
Among the fifteen CaSBP genes, two genes (CaSBP02 and CaSBP06) contained ankyrin repeats that are present in proteins with different biological roles and are involved in interactions between proteins (Zhang et al., 1999). Thus, there may be a functional relationship between the CaSBP genes. The detached leaves of the fifteen different CaSBP-silenced plants and the negative control plants were inoculated with the P. capsici HX-9 strain. After three days, the detached leaves of the CaSBP01-, CaSBP02-, CaSBP03-, CaSBP04-, CaSBP05-, CaSBP06-, CaSBP07-, CaSBP09-, CaSBP10-, CaSBP14-, and CaSBP15-silenced plants exhibited large hygrophanous lesions with no difference compared to the negative control (Figure 2). Additionally, the detached leaves of the CaSBP08-, CaSBP11-, CaSBP12-, and CaSBP13-silenced plants exhibited very small or no hygrophanous lesions, and the percentage of lesion area of the plants silenced for these genes exhibited a significant difference compared with the negative control (Figure 2). Therefore, we screened four CaSBP genes (CaSBP08, CaSBP11, CaSBP12, and CaSBP13) that are involved in plant defense responses to P. capsici infection. To further study the function of CaSBP genes in the process of P. capsici infection response, we chose one of our screened peppers SBP-box gene (CaSBP08) for further research.
CaSBP08 has a 336-bp open reading frame, encoding 111 amino acids (Zhang et al., 2016). The CaSBP08 protein was localized to the nucleus (Figure 3). Silencing of this gene enhanced resistance to P. capsici infection in pepper plants.
After P. capsici inoculation, the lesion areas of detached leaves of CaSBP08-silenced plants were smaller than the lesion areas of the negative control plants (Figures 4A, B). The disease index percentage of CaSBP08-silenced plants was also lower than that of the negative control plants (Figure 4D). Furthermore, after inoculation with P. capsici using the root-drench method, the MDA content as well as the POD and CAT activities of CaSBP08-silenced plants increased but were lower than those of the negative control treatment plants (Figure 4E). A plant under stress is closely related to membrane lipid peroxidation, which is induced by active oxygen accumulation. MDA is one of the most important products of membrane lipid peroxidation. Therefore, the degree of membrane lipid peroxidation can be determined by measuring MDA content. This can reflect the degree of damage to the membrane system and the resistance of plants (Ma et al., 2013). Moreover, it has been reported that during Phytophthora root rot development there is a relationship between the disease induced by P. capsici and the antioxidant system (Koç and Üstün, 2012). The protein encoded by the peroxidase CanPOD gene plays a positive role in plant defense responses to P. capsici infection in pepper plants (Wang et al., 2013b), as CanPOD is related to reactive oxygen species (ROS)-scavenging enzymes. CAT is also a major ROS-scavenging enzyme in plants (Mittler et al., 2004). POD and CAT activity levels were increased during P. capsici infection in pepper plants (Koç and Üstün, 2012). The high POD activity indicates that pepper plants are extra sensitive to infection by P. capsici as POD-mediated enzymatic reactions are enhanced in infected plants (Wu et al., 2016). POD activity increase is an essential factor in enhancing resistance to plant disease (Wu et al., 2016). The activity of POD increases during the first stage of Xanthomonas campestris pv. vesicatoria infection and then declines when the H2O2 accumulation reaches its maximum in pepper plants (Do et al., 2003). After P. capsici inoculation, the increased rate of POD activity in susceptible varieties was greater than that in resistant varieties of Kernel Pumpkin (Zhou et al., 2003). Moreover, catalase plays a key role in maintaining H2O2 homeostasis in cells and has been implicated in ROS signaling in response to pathogen attack (Magbanua et al., 2007). In addition, during infection, Phytophthora nicotianae increases its own peroxisomal catalase levels while concurrently down-regulating host catalase expression (Blackman and Hardham, 2008). The activity of CAT in cucumber varieties resistant to downy mildew was lower compared with susceptible varieties (Wang, 2001).
It has been reported that the strong suppression of pepper CaPO1 can cause dramatic H2O2 accumulation and a huge decrease in peroxidase activity during programmed cell death (Do et al., 2003). Therefore, we detected the expression level of CaPO1 subsequent to P. capsici inoculation into pepper plants. The level of CaPO1 expression was suppressed in the negative control plants and first increased and then decreased in the CaSBP08-silenced plants (Figure 5). Bae et al. (2006) reported that poplar PoPOD1 is suppressed under NaCl, methyl viologen, polyethylene glycol, gibberellic acid (GA3) and jasmonic acid (JA) treatments.
Furthermore, it has been reported that SA- and JA-mediated signal transduction pathways play a crucial role in plant resistance to diseases (Spoel et al., 2003). Most CaSBP genes can be induced under SA and MeJA treatment and inhibited during the early stage by SA synthesis inhibitor (paclobutrazol, PBZ) and MeJA (salicylhydroxamic acid, SHAM) synthesis inhibitor treatments (Zhang et al., 2016). To determine whether CaSBP08 is involved in the SA- and MeJA-mediated resistant pathways, we also studied and detected the expression of some defense-related genes. For example, the molecular marker CaSAR8.2 can be used for the detection of several pathogenic diseases that affect the SA-mediated signal transduction pathway (Lee and Hwang, 2003). CaBPR1 is involved in the hypersensitive response and was induced in an incompatible interaction of leaves with Xanthomonas campestris pv. vesicatoria (Kim and Hwang, 2000). CaDEF1 is involved in the MeJA-mediated signal transduction pathway, which has functions in microbial infection, as abiotic elicitors, and in response to some environmental stressors (Do et al., 2004). In this work, the CaSAR8.2 and CaBPR1 expression levels were induced at the first stage and then decreased in CaSBP08-silenced plants, while they were suppressed in the negative control plants (Figure 5). Furthermore, the expression level of CaDEF1 increased in negative control plants and CaSBP08-silenced plants and was higher in CaSBP08-silenced plants compared with negative control plants. The results demonstrate the involvement of CaSBP08 in the SA- and MeJA-mediated resistant pathways. Further research, however, is need in this regard.
Ectopic expression of BpSPL9 in the Betula platyphylla Suk. (birch) has been reported to enhance the ROS scavenging under drought and salt stress (Ning et al., 2017). SPL9 interacts with JA ZIM-domain (JAZ) proteins and negatively regulates JA response, though it promotes JAZ3 accumulation in Arabidopsis (Mao et al., 2017). Similarly, VpSBP5 likely participates in regulating resistance against Erysiphe necator though SA- and MeJA-mediated signal transduction pathway in grapes (Hou et al., 2013). Overexpression of CaSBP08 in transgenic N. benthamiana enhanced susceptibility to P. capsici infection, as demonstrated by the higher average disease area and disease index percentage compared to WT plants (Figures 6B, E). Besides, the NbPR1a and NbPR1b genes, which are involved in the SA-induced systemic acquired resistance pathway and JA-mediated disease resistance signaling pathway, respectively, were induced in the CaSBP08 transgenic and WT plants (Sohn et al., 2007; Cheol Song et al., 2016). PR1a and PR1b, two reported defense-related genes in tobacco, can be highly induced by tobacco mosaic virus (TMV) (van Huijsduijnen et al., 1985). The expression level of the SA signaling marker gene NbPR1a was associated with systemic acquired resistance (SAR) against Pseudomonas syringae pv. tabaci (Cheol Song et al., 2016). PR1b is a JA-responsive gene in tobacco (Sohn et al., 2007). Moreover, it has been reported that ectopic overexpression of CaC3H14 enhances resistance of tobacco to Ralstonia solanacearum infection, and the expression of PR1b was induced in transgenic and WT plants. However, the expression of PR1b in transgenic lines was significantly lower than that in WT plants (Qiu et al., 2018). The results of the present study suggest that CaSBP08 may be involved in resistance to P. capsici through regulating the expression of defense-related genes. However, further research is need to examine the regulatory mechanism. In N. benthamiana, SPL6 plays a positive regulatory role in nucleotide binding rich leucine repeat (N TIR-NB-LRR) receptor-mediated plant natural immunity (Padmanabhan et al., 2013). Overexpression of JcNAC1 in Jatropha curcas can enhance susceptibility of plants to Botrytis cinerea infection and inhibit the expression of some defense-related marker genes (Qin et al., 2014). Overexpression of ATAF2 in Arabidopsis can enhance the sensitivity of plants to Fusarium oxysporum infection and repress the expression of pathogenesis-related genes (Delessert et al., 2005).
Conclusions
In conclusion, we screened four genes (CaSBP08, CaSBP11, CaSBP12, and CaSBP13) out of the fifteen identified CaSBP genes, each of which responded to P. capsici infection. Additionally, we selected one of our screened pepper SBP-box genes (CaSBP08) for further research. CaSBP08 protein was thus observed to be localized to the nucleus. Silencing CaSBP08 enhanced resistance against P. capsici infection, such that the average disease area, the percent disease index, and the POD and CAT activities were lower in the CaSBP08-silenced plants compared with the negative control plants. Additionally, following inoculation with P. capsici, the defense genes CaPO1, CaDEF1, CaBPR1, and CaSAR8.2 were induced during the early stage of infection in CaSBP08-silenced plants, while CaPO1, CaBPR1, and CaSAR8.2 were suppressed in the negative control plants. In addition, overexpression of CaSBP08 in N. benthamiana enhanced susceptibility to P. capsici infection, as demonstrated by the average disease area and the percent disease index being greater than those of WT plants. Our work provides a basis for future research on the role of CaSBP genes in plant resistance to infections by P. capsici and similar pathogens.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
H-XZ and Z-HG perceived and designed research. H-XZ, X-HF, and J-HJ carried out the experiments. H-XZ analyzed data. H-XZ wrote the manuscript. MA and AK revised the manuscript. Z-HG and A-MW contributed reagents/materials/analysis tools. All authors read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Key R&D Program of China (No. 2016YFD0101900), National Natural Science Foundation of China (No.U1603102, No. 31772309).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00183/full#supplementary-material
References
Bae, E., Lee, H., Lee, J., Noh, E., Jo, J. (2006). Molecular cloning of a peroxidase gene from poplar and its expression in response to stress. Tree Physiol. 26, 1405–1412. doi: 10.1093/treephys/26.11.1405
Blackman, L. M., Hardham, A. R. (2008). Regulation of catalase activity and gene expression during Phytophthora nicotianae development and infection of tobacco. Mol. Plant Pathol. 9, 495–510. doi: 10.1111/J.1364-3703.2008.00478.X
Boter, M., Ruíz-Rivero, O., Abdeen, A., Prat, S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18, 1577–1591. doi: 10.1101/gad.297704
Cardon, G. H., Hohmann, S., Nettesheim, K., Saedler, H., Huijser, P. (1997). Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J. 12, 367–377. doi: 10.1046/j.1365-313X.1997.12020367.x
Cheol Song, G., Sim, H.-J., Kim, S.-G., Ryu, C.-M. (2016). Root-mediated signal transmission of systemic acquired resistance against above-ground and below-ground pathogens. Ann. Bot. 118, 821–831. doi: 10.1093/aob/mcw152
Delessert, C., Kazan, K., Wilson, I. W., Straeten, D., Van Der, M. J., Dennis, E. S., et al. (2005). The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 43, 745–757. doi: 10.1111/j.1365-313X.2005.02488.x
Do, H. M., Hong, J. K., Jung, H. W., Kim, S. H., Ham, J. H., Hwang, B. K. (2003). Expression of peroxidase-like genes, H2O2 production, and peroxidase activity during the hypersensitive response to Xanthomonas campestris pv. vesicatoria in Capsicum annuum. Mol. Plant-Microbe Interact. 16, 196–205. doi: 10.1094/Mpmi.2003.16.3.196
Do, H. M., Lee, S. C., Jung, H. W., Sohn, K. H., Hwang, B. K. (2004). Differential expression and in situ localization of a pepper defensin (CADEF1) gene in response to pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Plant Sci. 166, 1297–1305. doi: 10.1016/j.plantsci.2004.01.008
Du, Y., Berg, J., Govers, F. , Bouwmeester, K. (2015). Immune activation mediated by the late blight resistance protein R1 requires nuclear localization of R1 and the effector AVR1. New Phytol. 207, 735–747. doi: 10.1111/nph.13355
Granke, L. L., Quesada-Ocampo, L., Lamour, K., Hausbeck, M. K. (2012). Advances in research on Phytophthora capsici on vegetable crops in the united states. Plant Dis. 95, 1588–1600. doi: 10.1094/PDIS-02-12-0211-FE
Guo, W. L., Chen, R. G., Gong, Z. H., Yin, Y. X., Ahmed, S. S., He, Y. M.. (2012). Exogenous abscisic acid increases antioxidant enzymes and related gene expression in pepper (Capsicum annuum) leaves subjected to chilling stress. Genet. Mol. Res. 11, 4063–4080. doi: 10.4238/2012.September.10.5
Hammerschmidt, R., Nuckles, E. M., Kuć, J. (1982). Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrichum lagenarium. Physiol. Plant Pathol. 20, 73–82. doi: 10.1016/0048-4059(82)90025-X
Hou, H., Yan, Q., Wang, X., Xu, H. (2013). A SBP-Box gene VpSBP5 from Chinese wild Vitis species responds to erysiphe necator and defense signaling molecules. Plant Mol. Biol. Rep. 31, 1261–1270. doi: 10.1007/s11105-013-0591-2
Hou, H., Jia, H., Yan, Q., Wang, X. (2018). Overexpression of a SBP-box gene (VpSBP16) from Chinese wild Vitis species in Arabidopsis improves salinity and drought stress tolerance. Int. J. Mol. Sci. 19, 1–17. doi: 10.3390/ijms19040940
Jin, J.-H., Zhang, H.-X., Tan, J.-Y., Yan, M.-J., Li, D.-W., Khan, A., et al. (2016). A new ethylene-responsive factor CaPTI1 gene of pepper (Capsicum annuum L.) involved in the regulation of defense response to Phytophthora capsici. Front. Plant Sci. 6, 1–12. doi: 10.3389/fpls.2015.01217
Kim, Y. J., Hwang, B. K. (2000). Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol. Plant 108, 51–60. doi: 10.1034/j.1399-3054.2000.108001051.x
Kim, M., Do Choi, Y. (2007). Transcript profile of transgenic Arabidopsis constitutively producing methyl jasmonate. J. Plant Biol. 50, 12–17. doi: 10.1007/BF03030594
Klein, J., Saedler, H., Huijser, P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250, 7–16. doi: 10.1007/s004380050046
Koç, E., Üstün, A. S. (2012). Influence of Phytophthora capsici L. inoculation on disease severity, necrosis length, peroxidase and catalase activity, and phenolic content of resistant and susceptible pepper (Capsicum annuum L.) plants. Turk. J. Biol. 36, 357–371. doi: 10.3906/biy-1109-12
Lännenpää, M., Jänönen, I., Hölttä-Vuori, M., Gardemeister, M., Porali, I., Sopanen, T. (2004). A new SBP-box gene BpSPL1 in silver birch (Betula pendula). Physiol. Plant 120, 491–500. doi: 10.1111/j.0031-9317.2004.00254.x
Lee, S. C., Hwang, B. K. (2003). Identification of the pepper SAR8.2 gene as a molecular marker for pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Planta 216, 387–396. doi: 10.1007/s00425-002-0875-5
Liu, J., Cheng, X., Liu, P., Sun, J. (2017). miR156-targeted SBP-Box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol. 174, 1931–1948. doi: 10.1104/pp.17.00445
Lorenzo, O., Solano, R. (2005). Molecular players regulating the jasmonate signaling network. Curr. Opin. Plant Biol. 8, 532–540. doi: 10.1016/j.pbi.2005.07.003
Ma, N. N., Zuo, Y. Q., Liang, X. Q., Yin, B., Wang, G. D., Meng, Q. W. (2013). The multiple stress-responsive transcription factor SlNAC1 improves the chilling tolerance of tomato. Physiol. Plant 149, 474–486. doi: 10.1111/ppl.12049
Magbanua, Z. V., De Moraes, C. M., Brooks, T. D., Williams, W. P., Luthe, D. S. (2007). Is catalase activity one of the factors associated with maize resistance to Aspergillus flavus? Mol. Plant Microbe Interact. 20, 697–706. doi: 10.1094/MPMI-20-6-0697
Mao, Y. B., Liu, Y. Q., Chen, D. Y., Chen, F. Y., Fang, X., Hong, G. J., et al. (2017). Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 8, 1–13. doi: 10.1038/ncomms13925
Mittler, R., Vanderauwera, S., Gollery, M., Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Mou, S. L., Liu, Z. Q., Gao, F., Yang, S., Su, M. X., Shen, L., et al. (2017). CaHDZ27, a homeodomain-leucine zipper I protein, positively regulates the resistance to Ralstonia solanacearum infection in pepper. Mol. Plant-Microbe Interact. 30, 960–973. doi: 10.1094/MPMI-06-17-0130-R
Ning, K., Chen, S., Huang, H., Jiang, J., Yuan, H., Li, H. (2017). Molecular characterization and expression analysis of the SPL gene family with BpSPL9 transgenic lines found to confer tolerance to abiotic stress in Betula platyphylla Suk. Plant Cell Tissue Organ Cult. 130, 469–481. doi: 10.1007/s11240-017-1226-3
Oh, S. K., Jeong, M. P., Young, H. J., Lee, S., Chung, E., Kim, S. Y., et al. (2005). A plant EPF-type zinc-finger protein, CaPIF1, involved in defense against pathogens. Mol. Plant Pathol. 6, 269–285. doi: 10.1111/j.1364-3703.2005.00284.x
Padmanabhan, M. S., Ma, S., Burch-Smith, T. M., Czymmek, K., Huijser, P., Dinesh-Kumar, S. P. (2013). Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PloS Pathog. 9, e1003235. doi: 10.1371/journal.ppat.1003235
Qin, X., Zheng, X., Huang, X., Lii, Y., Shao, C., Xu, Y., et al. (2014). A novel transcription factor JcNAC1 response to stress in new model woody plant Jatropha curcas. Planta 239, 511–520. doi: 10.1007/s00425-013-1993-y
Qiu, A., Lei, Y., Yang, S., Wu, J., Li, J., Bao, B., et al. (2018). CaC3H14 encoding a tandem CCCH zinc finger protein is directly targeted by CaWRKY40 and positively regulates the response of pepper to inoculation by Ralstonia solanacearum. Mol. Plant Pathol. 19, 2221–2235. doi: 10.1111/mpp.12694
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shikata, M., Yamaguchi, H., Microrna, Á, Molecular, Á (2012). Overexpression of Arabidopsis miR157b induces bushy architecture and delayed phase transition in Torenia fournieri. Planta 236, 1027–1035. doi: 10.1007/s00425-012-1649-3
Sohn, S. I., Kim, Y. H., Kim, B. R., Lee, S. Y., Lim, C. K., Hur, J. H., et al. (2007). Transgenic tobacco expressing the hrpNEP gene from Erwinia pyrifoliae triggers defense responses against Botrytis cinerea. Mol. Cells 24, 232–239. doi: 10.1186/1476-511X-6-30
Spoel, S. H., Koornneef, A., Claessens, S. M. C., Korzelius, J. P., Pelt, J. A., Van, M. J., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15, 760–770. doi: 10.1105/tpc.009159.ceptibility
Stone, J. M., Liang, X., Nekl, E. R., Stiers, J. J. (2005). Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 41, 744–754. doi: 10.1111/j.1365-313X.2005.02334.x
van Huijsduijnen, R. A. M. H., Cornelissen, B. J. C., van Loon, L. C., van Boom, J. H., Tromp, M., Bol, J. F. (1985). Virus-induced synthesis of messenger RNAs for precursors of pathogenesis-related proteins in tobacco. EMBO J. 4, 2167–2171. doi: 10.1002/j.1460-2075.1985.tb03911.x
Wang, Y., Meng, Y., Zhang, M., Tong, X., Wang, Q., Sun, Y., et al. (2011). Infection of Arabidopsis thaliana by Phytophthora parasitica and identification of variation in host specificity. Mol. Plant Pathol. 12, 187–201. doi: 10.1111/J.1364-3703.2010.00659.X
Wang, J. E., Li, D. W., Zhang, Y. L., Zhao, Q., He, Y. M., Gong, Z. H. (2013a). Defence responses of pepper (Capsicum annuum L.) infected with incompatible and compatible strains of Phytophthora capsici. Eur. J. Plant Pathol. 136, 625–638. doi: 10.1007/s10658-013-0193-8
Wang, J. E., Liu, K. K., Li, D. W., Zhang, Y. L., Zhao, Q., He, Y. M., et al. (2013b). A novel peroxidase CanPOD gene of pepper is involved in defense responses to Phytophtora capsici infection as well as abiotic stress tolerance. Int. J. Mol. Sci. 14, 3158–3177. doi: 10.3390/ijms14023158
Wang, Z., Wang, Y., Kohalmi, S. E., Amyot, L., Hannoufa, A. (2016). SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 2 controls floral organ development and plant fertility by activating ASYMMETRIC LEAVES 2 in Arabidopsis thaliana. Plant Mol. Biol. 92, 661–674. doi: 10.1007/s11103-016-0536-x
Wang, Q., Sun, A.-Z., Chen, S.-T., Chen, L.-S., Guo, F.-Q. (2018). SPL6 represses signaling outputs of ER stress in control of panicle cell death in rice. Nat. Plants 4, 280–288. doi: 10.1038/s41477-018-0131-z
Wang, L. P. (2001). Studies on the relationships of some physilogical-chemical changes and disease resisantce in cucumber infected with Fusarium wilt disease (Taigu, China: College of Horticulture, Shanxi Agricultural University).
Wang, J. E. (2013). Expression analysis and functional identification of CaRGA1 and CaPOD genes induced by Phytophthora capsici in pepper (Yangling, China: Institute of Horticulture, Northwest A&F University).
Wu, L., Xian, W., Ma, Y., Guo, Q. (2016). Effects of Phytophthora capsici on activities of POD, SOD, MDA in leaves of grafted pepper. J. Qinghai Univ. 34, 2–5.
Xu, Z. S., Chen, M., Li, L. C., Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53, 570–585. doi: 10.1111/j.1744-7909.2011.01062.x
Yamasaki, K., Kigawa, T., Inoue, M., Tateno, M., Yamasaki, T., Yabuki, T., et al. (2004). A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 337, 49–63. doi: 10.1016/j.jmb.2004.01.015
Yin, Y. X., Wang, S. B., Zhang, H. X., Xiao, H. J., Jin, J. H., Ji, J. J., et al. (2015). Cloning and expression analysis of CaPIP1-1 gene in pepper (Capsicum annuum L.). Gene 563, 87–93. doi: 10.1016/j.gene.2015.03.012
Zander, M., La Camera, S., Lamotte, O., Métraux, J. P., Gatz, C. (2010). Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 61, 200–210. doi: 10.1111/j.1365-313X.2009.04044.x
Zarei, A., Körbes, A. P., Younessi, P., Montiel, G., Champion, A., Memelink, J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 75, 321–331. doi: 10.1007/s11103-010-9728-y
Zhang, Y., Fan, W., Kinkema, M., Li, X., Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. 96, 6523–6528. doi: 10.1073/pnas.96.11.6523
Zhang, C. L., Shi, D. J., Huang, J., Zhang, H. X., Peng, G. H. (2003). Clone of envelope protein VP28 gene of white spot syndrome virus (wssv) and expression vector construction for cyanobacteria. Mar. Sci. 27, 72–76. doi: 10.3969/j.issn.1000-3096.2003.02.021
Zhang, Y., Schwarz, S., Saedler, H., Huijser, P. (2007). SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 63, 429–439. doi: 10.1007/s11103-006-9099-6
Zhang, Y. L., Jia, Q. L., Li, D. W., Wang, J. E., Yin, Y. X., Gong, Z. H.. (2013). Characteristic of the pepper CaRGA2 gene in defense responses against Phytophthora capsici leonian. Int. J. Mol. Sci. 14, 8985–9004. doi: 10.3390/ijms14058985
Zhang, Z., Li, D.-W., Jin, J.-H., Yin, Y.-X., Zhang, H.-X., Chai, W.-G., et al. (2015). VIGS approach reveals the modulation of anthocyanin biosynthetic genes by CaMYB in chili pepper leaves. Front. Plant Sci. 6, 1–10. doi: 10.3389/fpls.2015.00500
Zhang, H.-X., Jin, J.-H., He, Y.-M., Lu, B.-Y., Li, D.-W., Chai, W.-G., et al. (2016). Genome-wide identification and analysis of the SBP-Box family genes under Phytophthora capsici stress in pepper (Capsicum annuum L.). Front. Plant Sci. 7, 1–14. doi: 10.3389/fpls.2016.00504
Zhang, B., Xu, W., Liu, X., Mao, X., Li, A., Wang, J., et al. (2017). Functional conservation and divergence among homoeologs of TaSPL20 and TaSPL21, two SBP-box genes governing yield-related traits in hexaploid wheat. Plant Physiol. 174, 1177–1191. doi: 10.1104/pp.17.00113
Zhang, J., Ping, A. M., Wang, X. T., Li, G. Z., Zhu, Z. J., Li, M. L., et al. (2017). Cloning and expression analysis of SPL8 homolog from pak choi (Brassica rapa subsp. chinensis). Biotechnol. Biotechnol. Equip. 31, 1132–1138. doi: 10.1080/13102818.2017.1382390
Zhang, Y. L. (2009). “Identification of physiological race of Phytophthora capsici and analysis of the efficiency of chemical control,” (Yangling, China: Institute of Horticulture, Northwest A&F University).
Keywords: pepper, Phytophthora capsici, SBP-box family genes, CaSBP08, defense genes, Nicotiana benthamiana
Citation: Zhang H-X, Feng X-H, Ali M, Jin J-H, Wei A-M, Khattak AM and Gong Z-H (2020) Identification of Pepper CaSBP08 Gene in Defense Response Against Phytophthora capsici Infection. Front. Plant Sci. 11:183. doi: 10.3389/fpls.2020.00183
Received: 02 December 2019; Accepted: 07 February 2020;
Published: 26 February 2020.
Edited by:
Brigitte Mauch-Mani, Université de Neuchâtel, SwitzerlandReviewed by:
Abid Khan, University of Haripur, PakistanDaolong Dou, Nanjing Agricultural University, China
Copyright © 2020 Zhang, Feng, Ali, Jin, Wei, Khattak and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen-Hui Gong, emhnb25nQG53c3VhZi5lZHUuY24=
 Huai-Xia Zhang1
Huai-Xia Zhang1 Muhammad Ali
Muhammad Ali Zhen-Hui Gong
Zhen-Hui Gong