Corrigendum: Expression of Pumpkin CmbHLH87 Gene Improves Powdery Mildew Resistance in Tobacco
- 1School of Horticulture Landscape Architecture, Henan Institute of Science and Technology, Xinxiang, China
- 2Henan Province Engineering Research Center of Horticultural Plant Resource Utilization and Germplasm Enhancement, Xinxiang, China
Powdery mildew (PM), caused by Podosphaera xanthii, is a major threat to the global cucurbit yield. The molecular mechanisms underlying the PM resistance of pumpkin (Cucurbita moschata Duch.) are largely unknown. A homolog of the basic helix-loop-helix (bHLH) transcription factor was previously identified through a transcriptomic analysis of a PM-resistant pumpkin. In this study, this bHLH homolog in pumpkin has been functionally characterized. CmbHLH87 is present in the nucleus. CmbHLH87 expression in the PM-resistant material was considerably downregulated by PM; and abscisic acid, methyl jasmonate, ethephon, and NaCl treatments induced CmbHLH87 expression. Ectopic expression of CmbHLH87 in tobacco plants alleviated the PM symptoms on the leaves, accelerated cell necrosis, and enhanced H2O2 accumulation. The expression levels of PR1a, PR5, and NPR1 were higher in the PM-infected transgenic plants than in PM-infected wild-type plants. Additionally, the chlorosis and yellowing of plant materials were less extensive and the concentration of bacteria at infection sites was lower in the transgenic tobacco plants than in the wild-type plants in response to bacterial wilt and scab pathogens. CmbHLH87 may be useful for genetic engineering of novel pumpkin cultivars in the future.
Introduction
Pumpkin (Cucurbita moschata Duch.) is an important vegetable crop and is widely cultivated in China, with total harvested area of 438,466 hectares in 2017 (i.e., 17.42% of the global area) (Food and Agriculture Organization, 2017). Powdery mildew (PM) is a fungal disease seriously affecting Cucurbitaceae crops yield including cucumber, melon, watermelon, pumpkin, and squash in the world. The disease is mainly caused by Podosphaera xanthii (formerly known as Sphaerotheca fuliginea), which is a biotrophic plant pathogen (Perez-Garcia et al., 2009; Fukino et al., 2013). Excessive fungicide application poorly control PM, because it not only increases selection pressure on PM pathogens to adapt increasing levels of fungicide resistance, but it also may be harmful for human health and the environment. Therefore, studying the moleculer mechanism of PM by exploiting the resistant genes to breed resistant varieties represents a favored strategy to control PM.
The basic helix-loop-helix (bHLH) transcription factors constitute one of the largest transcription factor families in plants, wherein they help regulate developmental processes and responses to environmental stresses. These proteins have a 60-amino-acid conserved domain, which contains the following two functionally distinct regions: an N-terminal basic region (13–17 amino acids) that functions as a DNA-binding domain and a C-terminal HLH region that contributes to the formation of homodimers or heterodimers (Heim et al., 2003; Toledo-Ortiz et al., 2003). Recent studies have indicated that a number of bHLH transcription factor genes are involved in responses to abiotic stresses including drought, salt, and cold. The overexpression of AtbHLH068 and OsbHLH148 in transgenic Arabidopsis thaliana and rice, respectively, reportedly induces drought stress tolerance via abscisic acid (ABA)– and jasmonic acid (JA)–mediated signaling pathways (Seo et al., 2011; Le Hir et al., 2017). In rice, OsbHLH062, OsJAZ9, and OsNINJA form a transcriptional regulatory complex that fine-tunes the expression of JA-responsive genes involved in salt stress tolerance (e.g., OsHAK21) (Wu et al., 2015). Additionally, VaICE1/VaICE2, ZmmICE1, and NtbHLH123 are key regulators in the C-repeat binding factor regulatory pathway controlling cold tolerance (Xu et al., 2014; Lu et al., 2017; Zhao et al., 2018). Moreover, bHLHs influence the adaptation and resistance of plants to pathogen stress. An earlier investigation revealed that OsDPF expression is induced in the leaves of blast-infected rice plants, and the overexpression and knockdown of DPF considerably increases and decreases the accumulation of momilactones and phytocassanes, respectively (Yamamura et al., 2015). The overexpression of wheat bHLH060 in transgenic A. thaliana negatively regulates the resistance to Pseudomonas syringae through JA and ethylene (ET) signaling pathways (Wang et al., 2015). A recent study on wheat indicated that the expression levels of 28 and 6 TabHLH genes are obviously downregulated and upregulated, respectively, in response to a PM infection (Wei and Chen, 2018). However, bHLH functions related to biotic stress resistance remain poorly characterized in plants.
In our previous study, the PM-resitant pumpkin resources were obtained in an 8-year outdoor field observation study (Zhou et al., 2010). However, the resistant mechanism underlying pumpkin response to biotic stress is not yet elucidated. Thus, a RNA sequencing analysis of PM-infected pumpkin identified 4,716 differentially expressed genes, including gene encoding bHLH transcription factor (bHLH87) (Guo et al., 2018). The expression of this bHLH87 homolog in response to PM, hydrogen peroxide (H2O2), salicylic acid (SA), ABA, methyl jasmonate (MeJA), ethephon (Eth), and NaCl treatments were analyzed through real-time quantitative PCR (RT-qPCR). To evaluate its function in disease resistance, we expressed CmbHLH87 in tobacco ectopically. Transgenic tobacco plants constitutively overexpressing CmbHLH87 were more resistant to PM, bacterial wilt, and scab than the control plants.
Results
Isolation of CmbHLH87 and Subcellular Localization
Pumpkin PM-related genes were identified in a transcriptome (Guo et al., 2018). One of these clones was 89% identical at the nucleotide level to C. melo bHLH87. Full length of this homolog was obtained by a homology-based candidate gene method (named CmbHLH87) and submitted to the GenBank database (accession number MH105822). The size of this gene was 1,222 bp, including a 1,068-bp open reading frame (355 amino acids). The predicted polypeptide was relatively acidic, with a pI of 5.96, and an Mw of 39.1 kDa. A phylogenetic tree was conducted between the overall amino acid sequences of CmbHLH87 and other known bHLH proteins. CmbHLH87 was clustered into the bHLH VIII subfamily (Heim et al., 2003) (Supplementary Figure S1). An alignment of the deduced CmbHLH87 amino acid sequence with homologous sequences is presented in Supplementary Figure S2. At the amino acid level, CmbHLH87 was highly similar to the bHLH87 transcription factors from C. moschata (99.1% identical), Cucurbita pepo (98.8% identical), and Cucurbita maxima (71.0% identical), but was relatively dissimilar to Nicotiana tabacum bHLH87 (38.3% identical) and Arabidopsis AtbHLH087 (41.7% identical). The predicted amino acid sequence contained a conserved bHLH domain (amino acids 256–305) and a helix-loop-helix structure at the C terminal.
The subcellular localization of CmbHLH87 was assessed with a CmbHLH87-GFP fusion protein into Arabidopsis protoplasts under the control of the 35S CaMV promoter. The GFP signal in protoplasts producing GFP alone was detected in the cytoplasm and nucleus (Figure 1) supported by Guo et al. (2019), whereas the signal from the CmbHLH87-GFP fusion protein was detected exclusively in the nucleus.
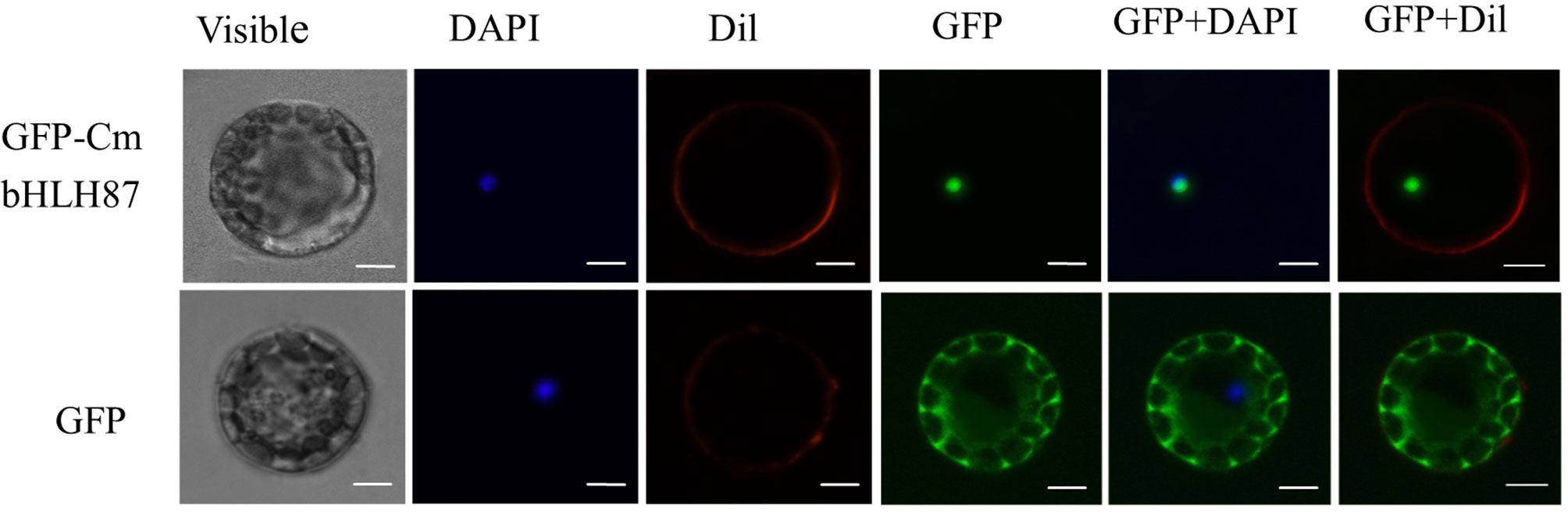
Figure 1 The subcellular localization of pumpkin CmbHLH87. The fused green fluorescent protein (GFP)–CmbHLH87 and alone GFP constructs were introduced into Arabidopsis protoplast by polyethylene glycol-mediated protoplast transformation. The fluorescent signals were detected using a confocal fluorescence microscope. Scale bars = 5 μm.
Expression Patterns of CmbHLH87
The CmbHLH87 expression patterns in both PM-resistant inbred line ‘112-2’ and PM-susceptible cultivar ‘JJJD’ were analyzed in response to PM and exogenous treatments (H2O2, SA, ABA, MeJA, Eth, or NaCl) (Figure 2). The CmbHLH87 expression level in the ‘112-2’ plants was significantly downregulated by PM (except at 0 hpi), but was significantly upregulated by H2O2 treatment (except at 6 hpi). In contrast, the CmbHLH87 expression level in ‘JJJD’ plants was not significantly altered by PM, but was downregulated by H2O2 treatments. Regarding the effects of SA, the CmbHLH87 expression level in ‘112-2’ plants decreased at 3 and 6 hpi, and was essentially unchanged thereafter. However, the CmbHLH87 expression level in ‘JJJD’ plants was significantly upregulated by the SA treatment. In response to ABA, MeJA, Eth, and NaCl treatments, CmbHLH87 expression was significantly higher in the ‘112-2’ and ‘JJJD’ seedlings than in the control (CK) seedlings. Specifically, the CmbHLH87 expression level in ‘112-2’ plants was upregulated by more than 40-fold by Eth and NaCl treatments over the duration of the study period. The results indicated that CmbHLH87 expression in PM-resistant inbred line ‘112-2’ was downregulated by PM and upregulated by H2O2, which differed from the results of the PM-sensitive cultivar ‘JJJD.’ Moreover, ABA, MeJA, Eth, and NaCl treatments obviously upregulated CmbHLH87 expression in both analyzed plant materials and these were not influenced by susceptibility or resistance status.
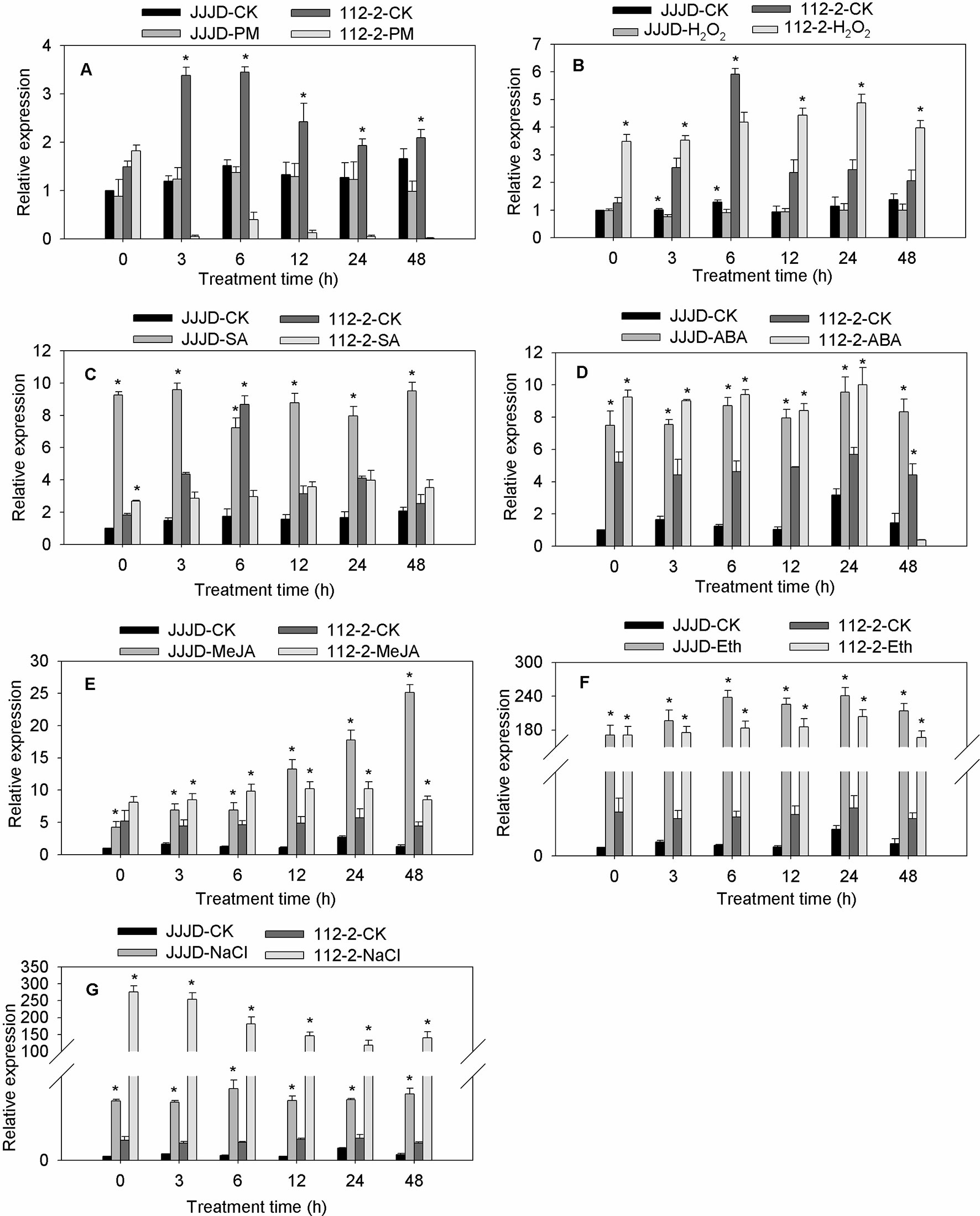
Figure 2 CmbHLH87 expression in response to powdery mildew and exogenous treatments. The pumpkin seedlings were sprayed with a spore suspension (A), exogenous hydrogen peroxide (H2O2) (B), salicylic acid (SA) (C), abscisic acid (ABA) (D), methyl jasmonate (MeJA) (E), ethephon (Eth) (F), and NaCl (G). The β-actin gene was used as an internal reference for qRT-PCR. The transcript level of CmbHLH87 in the cultivar ‘JJJD’ at 0 h was used as control (quantities of calibrator) and assumed as 1. The values are the means ± SEs of three biological replicates. Data between treatments (112-2-treatment vs. 112-2-CK and JJJD-treatment vs. JJJD-CK) were analyzed by one-way ANOVA, and *denotes statistical significance at p < 0.05.
Improved PM Resistance of CmbHLH87-Overexpressing Tobacco Plants
The transcript level of the transgenic plants CmbHLH87 under normal conditions was determined by qRT-PCR. The expression of CmbHLH87 in wild-type (WT) plants was not basically examined, whereas the transgenic plants CmbHLH87 expression was obviously upregulated (Figure 3), indicating that CmbHLH87 is overexpressed in the transgenic plants. The disease severity (DS) of the transgenic plants was 84% lower than that of WT plants at 10 days postinoculation (dpi) (Table 1). Furthermore, the visible symptoms of leaf damage in tobacco seedlings were observed to examine the resistance of CmbHLH87-expressing plants to PM. As shown in Figure 4A, powdery symptoms were observed in WT seedlings at 7 dpi, and expanded considerably at 28 dpi, while transgenic plants exhibited undetectable after 7 dpi and slight powdery pots at 28 dpi. Blue spots are actually trypan blue stained cell death. The blue spots on the transgenic leaves ocurred at 4 dpi, continued to expand at 5 and 7 dpi, and those blue spots were more and bigger than those on the WT leaves, implying that overexpression of CmbHLH87 in transgenic plants accelerated cell death following a PM infection (Figure 4B). Moreover, brown spots are H2O2 for DAB staining. Compared with WT leaves, there were more brown spots on the transgenic leaves at 1 dpi, larger and stained more intensely at 3 and 5 dpi. These results indicated that the overexpression of CmbHLH87 promoted the accumulation of H2O2 in transgenic plants infected with PM (Figure 4C).
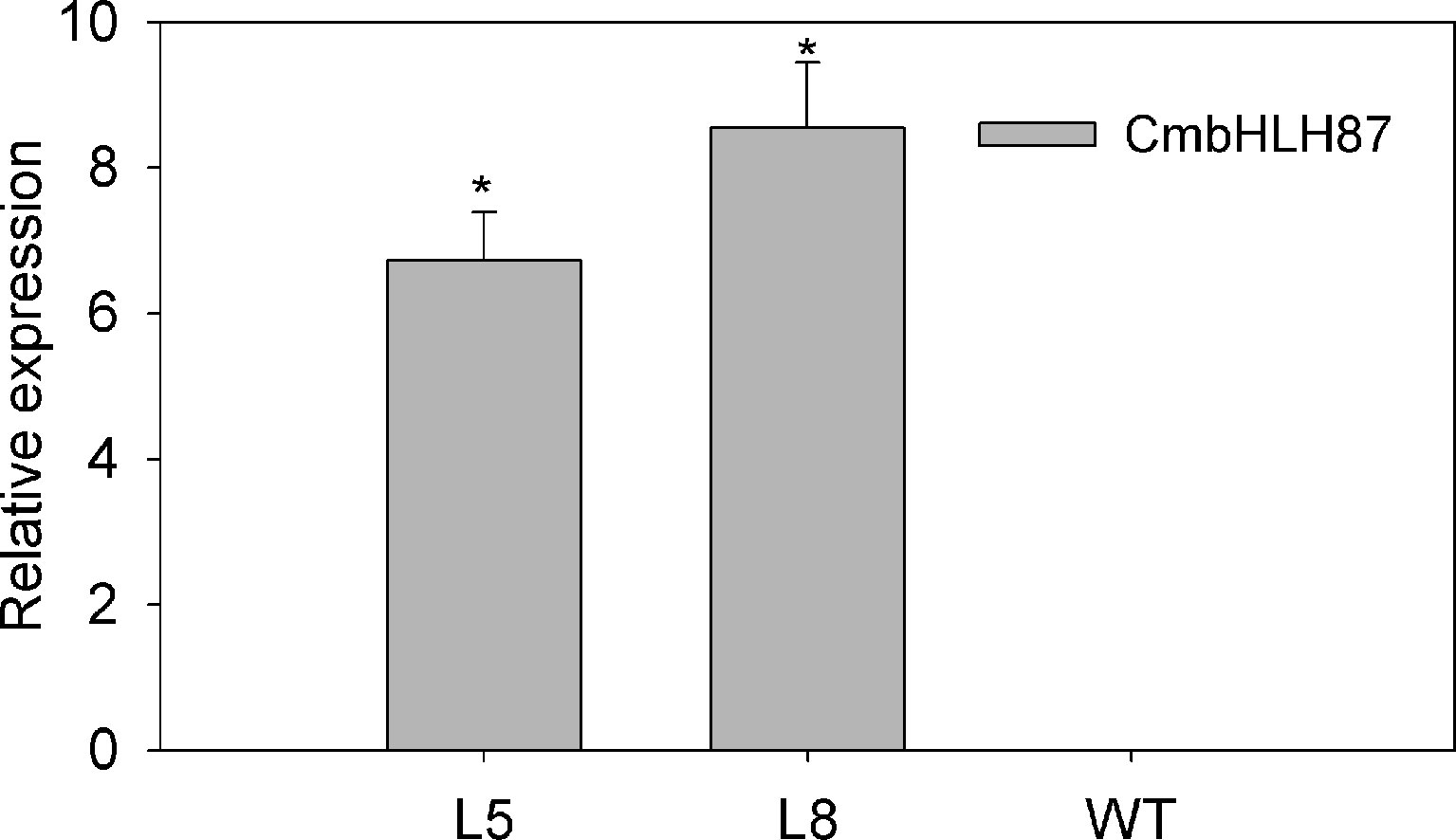
Figure 3 Relative expression of CmbHLH87 in transgenic or wild-type plants under normal growth conditions. L5 and L8 are two independent transgenic lines that overexpress CmbHLH87. Three biological triplicates were averaged and bars indicate standard error of the mean. *denotes statistical significance at p < 0.05 between both materials.

Figure 4 The pathogenic symptoms, trypan blue, and diaminobenzidine (DAB) staining of tobacco leaves infected with powdery mildew. The pathogenic symptoms of transgenic tobacco (L5) and wild type (WT) at 7 and 28 day post inoculation (A); trypan blue staining was performed to visualize cell death (B); DAB staining was performed to visualize hydrogen peroxide (H2O2) accumulation (C). Scale bars = 200 μm.
Expression of Signal-Related Genes
To investigate the signal transduction pathways affected by CmbHLH87 during plant defense responses to PM, the expression levels of five signaling-associated genes (NPR1, PR5, PR1a, PAL, and PDF1.2) in the SA, JA, and ET signal transduction pathways were analyzed by qRT-PCR (Figure 5). The NPR1, PR1a, and PR5 expression levels were higher in the transgenic plants infected with PM than in the transgenic plants-CK plants, implying that PM induced the expression of these genes. And the opposite pattern of these genes expression was observed in the WT plants infected with PM compared with WT-CK plants. There were no basically differences in the PAL and PDF1.2 (except at 12 hpi) expression levels of the transgenic plants between PM and CK treatments. In response to the PM infection, the PR1a, PR5, and NPR1 expression levels of the transgenic plants were higher than those of the WT plants (except at 0 hpi). The PAL expression level in the transgenic plants was significantly higher at 24 and 48 hpi, lower at 12, 72, and 120 hpi than that in the WT plants. Furthermore, the PDF1.2 expression level in the transgenic plants was slightly higher at 12 and 48 hpi, considerably lower at 24, 72, and 120 hpi than that in the WT plants. These results implied that CmbHLH87 overexpression increased the transcription of PR1a, PR5, and NPR1 and suppressed the expression of PDF1.2 in transgenic plants infect with PM. Furthermore, the increased PM resistance of the transgenic plants appeared to be related to the upregulated expression of these genes.

Figure 5 Expression of signal-related genes in transgenic and wild-type (WT) plants treated with powdery mildew. The samples of two genetically-modified tobacco lines (L5, L8) were used to analyze genes expression by qRT-PCR. A, NtNPR1; B, NtPAL; C, NtPR1a; D, NtPR5; E, NtPDF1.2. There were four treatments: L5/L8-CK represents transgenic lines under normal conditions; L5/L8-PM represents transgenic lines infected with powdery mildew; WT-CK represents WT plants under normal conditions; WT-PM represents WT plants infected with powdery mildew. Tobacco NtEF1-α gene (AF120093) was used as an internal reference. The expression levels of signal-related genes in WT plants at 0 h were used as control (quantities of calibrator) and assumed as 1. Three biological triplicates per line were averaged and Bars indicate standard error of the mean. Data between transgenic lines and WT plants (L5/L8-PM vs. WT-PM and L5/L8-CK vs. WT-CK) were analyzed by one-way ANOVA, and *denotes statistical significance at p < 0.05.
Improved Resistance of Transgenic Tobacco Plants to Bacterial Diseases
To analyze the effect of CmbHLH87 on other plant diseases, two common bacterial pathogens causing bacterial wilt (Ralstonia solanacearum) and scab (Xanthomonas euvesicatoria) were injected into tobacco plants (Figure 6). At 6 dpi, the chlorosis and yellowing of the 6th leaf at injection sites were less severe for the transgenic plants than for the WT plants. Additionally, the concentrations of bacterial wilt and scab bacteria in the transgenic plants were 0.14 and 0.10 times those of the WT plants, respectively. These observations suggested that overexpression of CmbHLH87 in tobacco increased the resistance to bacterial wilt and scab.

Figure 6 The resistance of CmbHLH87 in tobacco plants to bacterial wilt and scab. (A) Pathogens symptoms of the 6th-upper leaf injection sites was injected with bacterial wilt bacteria with a needle-removed syringe; (B) concentration bacteria of the 6th-upper leaf injection sites was injected with bacterial wilt bacteria with a needle-removed syringe; (C) pathogens symptoms of the 6th-upper leaf injection sites was injected with scab bacteria with a needle-removed syringe; (D) concentration bacteria of the 6th-upper leaf injection sites was injected with scab bacteria. Three biological triplicates were averaged and bars indicate standard error of the mean. **denotes significant differences between wild-type (WT) and transgenic plants at p < 0.01.
Discussion
In this study, we isolated a novel pumpkin bHLH87 gene and the predicted amino acid sequence included a conserved bHLH domain, suggesting that CmbHLH87 is a putative pumpkin bHLH transcription factor. Its predicted pI was 5.96, which is similar to a previous report that the isoelectric points of watermelon bHLH family proteins are mostly in the acidic range (He et al., 2016). The CmbHLH87 protein was localized to the nucleus in Arabidopsis protoplasts, which is consistent with the observations of earlier studies involving the bHLH transcription factors from other plant species (Wang et al., 2015).
The interplay among complex signaling networks, including various pathways regulated by phytohormones, such as SA, JA, ethylene (ET), and ABA, considerably influences plant resistance to diseases. An earlier investigation during an infection of wheat plants by the stripe rust pathogen infection, SA, MeJA, and particularly ET suppress the transcription of TabHLH060 (Wang et al., 2015). In the current study, ABA, MeJA, Eth, and NaCl treatments induced pumpkin CmbHLH87 expression, whereas PM infection had the opposite effect, suggesting that CmbHLH87 may have a regulatory role during responses to hormones, salt stress, and PM. Additionally, overexpression of the pumpkin CmbHLH87 gene in tobacco plants decreased the Disease severity by 84%, accelerated necrosis, and increased the accumulation of H2O2. These results indicated that the PM resistance of the transgenic tobacco plants was enhanced, likely because of the changes to the HR-related necrosis and H2O2 accumulation. Our findings are consistent with the results of an earlier investigation that H2O2 accumulation and subsequent cell death usually lead to the resistance to diseases caused by biotrophic pathogens (Li et al., 2011). The expression pattern of CmbHLH87 in response to PM is controversial to its transgenic phenotype in disease resistance. There were similar reports that pathogen-regulated ectopic expression of bHLH transcription factors was inconsistent with activation of pathogen resistance. Two Arabidopsis bHLH25 and bHLH27 positively influence cyst nematode parasitism (Heterodera schachtii). Transgenic Arabidopsis plants overexpressing either one or both of the bHLH genes exhibited an increased susceptibility to H. schachtii (Jin et al., 2011). Jin et al. document an example of pathogen-induced ectopic coexpression of two regulatory genes to enhance pathogen success. In our experiment, this is also an intriguing biological phenomenon that highlights the complexity of obligate biotrophic plant–pathogen interactions who are direct target of the CmbHLH87 protein and how to interact in response to pathogen resistance need to be further research. PM symptoms herein were observed in WT tobacco seedlings at 7 dpi of G. cichoracearum, and the phenotypes were not more robust than those of previous reports: PM disease occurs abundantly in tobacco plants at 12 dpi of the PM strain G. cichoracearum SICAU1 (Li et al., 2019). Different tobacco accessions may show various susceptible PM disease phenotypes to different PM isolates.
SA plays a central role in plant disease resistance against biotrophic pathogens, while JA is critical for activating plant defenses against necrotrophic pathogens (Wasternack and Hause, 2013). PDF1.2 is important for the JA/ET-dependent signaling pathway. At intermediate SA levels, NPR1 (nonexpresser of PR1) accumulates and interacts with the TGA transcription factor, functioning as a coactivator of SA-responsive genes, including PR genes (Caarls et al., 2015). The overexpression of wheat TabHLH060 gene in Arabidopsis increases the susceptibility to P. syringae by suppressing the transcription levels of PR1, PR2, and PR5, which are involved in the SA signaling pathway, and by upregulating the expression of PDF1.2 and ORA59, which are associated with the JA and ET signaling pathways (Wang et al., 2015). In bread wheat, TaJAZ1 overexpression increases PM resistance by promoting the accumulation of reactive oxygen species. The encoded TaJAZ1 directly interacts with TaMYC4 (JA-induced bHLH transcription factor) to repress its transcriptional activity (Jing et al., 2019). In the current study, following PM infection, the PR1a, PR5, and NPR1 expression levels in the transgenic plants were higher than those in the WT plants, whereas a different expression pattern was observed for the PDF1.2 expression level. These results suggest that in the SA pathway, the transactivation of PR1a, PR5, and NPR1 is dependent on CmbHLH87. Additionally, CmbHLH87 does not directly affect the JA/ET-dependent defense pathway to regulate PDF1.2 expression. We propose that CmbHLH87 activates stress-resistance mechanisms via SA-dependent defense pathways without upregulating PAL expression, but suppresses the activities of JA/ET-dependent defense pathways. Therefore, we speculate that CmbHLH87 positively regulates the H2O2 and SA pathways. Moreover, H2O2 might directly transfer the SA signal to regulate the expression of downstream response genes in the CmbHLH87-overexpressing transgenic plants infected with PM. In NPR1-dependent SA signal transduction pathways, the activation of PR gene expression requires an interaction between NPR1 and the TGA transcription factor that binds to the target promoters (Fan and Dong, 2002; Spoel and Dong, 2012). We speculated whether there is a relationship between the upregulation of NPR1 expression and SA-mediated transcriptional activation of PR genes. Notably, the phenotypes and genes influenced by CmbHLH87 overexpression in tobacco plants might not be the same as those regulated by CmbHLH87 expression in pumpkin in response to PM. Additional studies are necessary to reveal the biological functions of CmbHLH87 in pumpkin infected with PM.
Two globally important diseases that affect tobacco, bacterial wilt and scab, are caused by the necrotrophic R. solanacearum and the hemi-biotrophic X. euvesicatoria respectively. The overexpression of the bHLH transcription factor gene (HBI1) decreases the pathogen-associated defense responses and increases the susceptibility to bacteria P. syringae (Malinovsky et al., 2014). In the current study, the chlorosis and yellowing of the leaves near infection sites were less extensive in the transgenic plants than in the WT plants at 6 dpi. Moreover, the concentrations of bacterial wilt and scab bacteria were substantially lower in the transgenic plants than in the WT plants. These results imply overexpression of the pumpkin CmbHLH87 gene in tobacco enhances the resistance to bacterial wilt and scab, which is consistent with the effects of CmbHLH87 overexpression on the resistance to PM.
In conclusions, the results of this study indicate that overexpression of the pumpkin CmbHLH87 gene in tobacco may increase the resistance to PM, bacterial wilt, and scab. Additionally, CmbHLH87 overexpression may improve PM resistance by enhancing HR-related cell death and H2O2 accumulation and by upregulating the expression of SA-mediated defense genes. The data generated in this study may provide valuable genetic information for the breeding of new disease-resistant pumpkin varieties.
Materials and Methods
Plant Materials and Treatments
Pumpkin inbred line ‘112-2’ and cultivar ‘Jiujiangjiaoding’ (abbreviated ‘JJJD’), which are resistant and susceptible to PM, respectively, were provided by the Henan Institute of Science and Technology, Xinxiang, Henan, China (Zhou et al., 2010). Pumpkin seeds were germinated and the resulting seedlings were grown as previously described (Guo et al., 2018). Seedlings at the third-leaf stage were treated as previously described by Guo et al. (2019). Seedlings were sprayed with a freshly prepared spore suspension (106 spores/ml), exogenous 1.5 mM H2O2, 100 μM SA, 100 μM ABA, 100 μM MeJA, 0.5 g/L Eth and 0.4mM NaCl. Moreover, water alone was used for the control treatment (CK). The treated seedlings were maintained in a growth chamber with a 15-h light (28°C)/9-h dark (18°C) cycle (5,500 lux light intensity) and harvested after 0, 3, 6, 12, 24, and 48 h to examine the CmbHLH87 expression pattern. At each time point, two young leaves were collected from the upper parts of four seedlings (i.e., one sample), wrapped in foil, frozen in liquid nitrogen, and stored at −80°C. The treatments were arranged in a randomized complete block design, with three biological replicates.
Isolation of CmbHLH87 and
The bHLH homolog expressed sequence tag (GenBank accession number SRR5369792) was identified from a transcriptome of PM-resistant pumpkin seedling (Guo et al., 2018). Full length of this homolog was obtained using a homology-based candidate gene method (Guo et al., 2014). The theoretical molecular weight (Mw) and isoelectric point (pI) were calculated with the ExPASy Compute pI/Mw tool (Bjellqvist et al., 1993). Sequence data were analyzed with the ClustalW program (Thompson et al., 1994). The phylogenetic tree was constructed using Mega5.0 by the neighbor-joining method. The NCBI databases were screened for homologous sequences with the default parameters of the BLAST program (http://www.ncbi.nlm.nih.gov/blast) (Altschul et al., 1997).
Subcellular Localization Analysis
The CmbHLH87 ORF (without the termination codon) was ligated into the pBI221-GFP vector for the subsequent production of a green fluorescent protein (GFP)–tagged CmbHLH87 fusion protein. Polyethylene glycol was used during the transformation of Arabidopsis thaliana protoplasts with the recombinant plasmid (Lee et al., 2013). The subcellular localization of CmbHLH87 was determined based on the GFP signal, which was detected with the confocal fluorescence microscope (UltraVIEW VoX, Olympus, Japan) (Guo et al., 2019).
Generation of CmbHLH87 Transgenic Tobacco Plants
Pumpkin is known to be one of the plants most refractory for transformation. To date, only two reports on transformation in pumpkin existed using a combined method of vacuum infiltration and Agrobacterium infection (Nanasato et al., 2011; Nanasato and Tabei, 2015). So, we choosed an ectopic overexpression assay in tobacco instead of generating pumpkin transgenic plants. Forward and reverse primers with an added BamH I site and Kpn I site, respectively, were used to amplify CmbHLH87. The amplified sequence was then inserted into the pVBG2307 vector for the subsequent expression of CmbHLH87 under the control of the 35S cauliflower mosaic virus (CaMV) promoter. The recombinant plasmid was introduced into Agrobacterium tumefaciens GV3101 cells as previously described (Guo et al., 2014). The resulting A. tumefaciens cells were used to transform tobacco (Nicotiana tabacum L. cv. NC89) plants according to a previously described leaf disk method (Li et al., 2012). The transgenic plants were confirmed by examining the segregation ratio of the kanamycin selectable marker and by PCR analysis of NPTII and CmbHLH87, self-pollinated to obtain homozygous T2 offspring. T2 lines that produced 100% kanamycin-resistant plants in the T3 generation were considered as homozygous transformants. In each experiment, T2 generations of homozygous transgenic lines (L5, L8, and L12) were selected for further analysis. Similar phenotypes and results used for this study were observed in more than three independent lines of transgenic plants.
Primer Design
All primers designed and used in this study are provided in Supplementary Table S1.
Performance of Transgenic Lines Infected With PM, Bacterial Wilt and Scab
Conidia were collected from tobacco leaves naturally infected with Golovinomyces cichoracearum, which are main pathogen isolates of PM. The upper second leaf from the transgenic and WT seedlings at the fifth-leaf stage was sprayed with a spore suspension (106 spores/ml) and contine to in vitro grow for 10 d (Guo et al., 2019). At 10 dpi, mildew development on each leaf disk was recorded, using the following scale: 0 = no visible mildew development, 1 = 0 to 5%, 2 = 6 to 25%, 3 = 26 to 50%, 4 = 51 to 75%, and 5 = > 76% of disk surface covered with mildew, as described by Ishii et al. (2001). Disease severity was calculated as [(5A + 4B + 3C + 2D + E)/5F] × 100, where A, B, C, D, and E were the number of leaf disks corresponding to the scales, 5, 4, 3, 2, and 1, respectively, and F was the total number of leaf disks assessed. Additionally, the second leaf of the transgenic and WT plants was sprayed with the abovementioned spore suspension and sampled at 0, 12, 24, 48, 72, and 120 hpi for a subsequent extraction of total RNA. Furthermore, these leaves were harvested symmetrically along the sides of the main vein after 0, 1, 3, 4, 5, and 7 d to examine cell death and H2O2 accumulation. 3, 3′ -diaminobenzidine (DAB) and trypan blue staining were used to analyze H2O2 accumulation and cell death, respectively, as previously described (Choi et al., 2012). The treatments were arranged in a randomized complete block design with three replicates.
The upper sixth leaf from the transgenic and WT seedlings at the twelfth-leaf stage were inoculated with bacterial solutions (108 cfu/ml). The bacterial solutions were injected into the underside of leaves between the lateral veins with a syringe lacking a needle. After 6 d, the concentration of the bacteria at the injection sites was determined as previously described (Guo et al., 2019). The injection sites were sampled with a circular perforator (1cm diameter) and ground in aseptic water, then serially diluted to produce the 102-fold, 103-fold, and 104-fold diluents. Experiments were done in triplicate for each line.
qRT-PCR Analysis
The RNA extraction, first-strand cDNA synthesis and qRT-PCR were completed as described by Guo et al. (2015). Relative gene expression levels were determined with the 2-ΔΔCT method. Total RNA was extracted from the leaves of pumpkin seedlings treated with various stresses or distilled water for 0, 3, 6, 12, 24, or 48 h as described above. The β-actin gene was used as an internal reference for normalizing of gene expression levels in pumpkin (Wu and Cao, 2010).
Total RNA was extracted from CmbHLH87-overexpressing and WT tobacco seedlings to examine the expression of five hormone-related genes (NtNPR1, NtPR1a, NtPR5, NtPDF1.2, and NtPAL) at 0, 12, 24, 48, 72, and 120 hpi as described above. The tobacco NtEF1-α gene (AF120093) was used as an internal control in the assays.
Statistical Analyses
Values are herein provided as the mean ± standard error of three independent analyses. Data underwent a one-way ANOVA, and differences between WT and transgenic plants were evaluated with a post hoc comparison test (Student-Newman-Keuls method) at p < 0.05 with SPSS 19.0 for Windows (SPSS Inc, Chicago, IL).
Data Availability Statement
The datasets generated for this study can be found in the the GenBank database (accession number MH105822).
Author Contributions
W-LG and Y-YG conceived and designed the experiments. W-LG and B-HC performed the experiments. X-JC and Q-FL analyzed the data. H-LY, J-GZ, X-ZL, and W-LG contributed reagents/materials/analysis tools. W-LG wrote the paper. G-YW rectified the manuscript. All authors read and approved the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (Grant no. 31401876), Henan High-level Talent Introduction and Training Special Foundation (Grant no. 103020218003/008), the Henan Provincial commodity vegetable industry technology system (Grant no. S2010-03-G06).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Guizhou academy of tobacco sciences researcher Hancheng Wang for providing tobacco Golovinomyces cichoracearum. We thank Henan Institute of Science and Technology professor G-YW for interpretation of data in the revising manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00163/full#supplementary-material
Figure S1 | A Neighbor-joining phylogenetic tree of CmbHLH87 and bHLH proteins from different plant species. The CmbHLH87 protein is marked with arrow. The bHLH subgroup names are shown to the right of square.
Figure S2 | Amino acid sequences alignment of pumpkin CmbHLH87 with others. The conserved domains (bHLH) are shown by thin underlines. The genes included are bHLH87-like (C. moschata cv. ‘Rifu’, XP_022953326.1), bHLH87-like(C. pepo, XP_023547764.1), bHLH87-like (C. maxima, XP_022992322.1), bHLH87-like (C. melo, XP_008462383.1), bHLH87-like (C. sativus, XP_004141799.1), bHLH87-like (Nicotiana tabacum, XP_016504990.1), AtbHLH087 (Arabidopsis, AAM10960.1) and CmbHLH87 (C. moschata cv. ‘112-2’, MH105822).
Table S1 | Primers used in this investigation.
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Bjellqvist, B., Hughes, G. J., Pasquali, C., Paquet, N., Ravier, F., Sanchez, J. C., et al. (1993). The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14, 1023–1031. doi: 10.1002/elps.11501401163
Caarls, L., Pieterse, C. M. J., Wees, S. C. M. V. (2015). How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 6, 170. doi: 10.3389/fpls.2015.00170
Choi, D. S., Hwang, I. S., Hwang, B. K. (2012). Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24, 1675–1690. doi: 10.1105/tpc.112.095869
Fan, W., Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid–mediated gene activation in Arabidopsis. Plant Cell 14, 1377–1389. doi: 10.1105/tpc.001628
Fukino, N., Yoshioka, Y., Sugiyama, M., Sakata, Y., Matsumoto, S. (2013). Identification and validation of powdery mildew (Podosphaera xanthii)-resistant loci in recombinant inbred lines of cucumber (Cucumis sativus L.). Mol. Breed. 32 (2), 267–277. doi: 10.1007/s11032-013-9867-3
Guo, W. L., Chen, R. G., Du, X. H., Zhang, Z., Yin, Y. X., Gong, Z. H., et al. (2014). Reduced tolerance to abiotic stress in transgenic Arabidopsis overexpressing a Capsicum annuum multiprotein bridging factor1. BMC Plant Biol. 14, 138. doi: 10.1186/1471-2229-14-138
Guo, W. L., Wang, S. B., Chen, R. G., Chen, B. H., Du, X. H., Yin, Y. X., et al. (2015). Characterization and expression profile of CaNAC2 pepper gene. Front. Plant Sci. 6, 755. doi: 10.3389/fpls.2015.00755
Guo, W. L., Chen, B. H., Chen, X. J., Guo, Y. Y., Yang, H. L., Li, X. Z., et al. (2018). Transcriptome profiling of pumpkin (Cucurbita moschata Duch.) leaves infected with powdery mildew. PloS One 13(1), e0190175. doi: 10.1371/journal.pone.0190175
Guo, W. L., Chen, B. H., Guo, Y. Y., Yang, H. L., Mu, J. Y., Wang, Y. L., et al. (2019). Improved powdery mildew resistance of transgenic Nicotiana benthamiana overexpressing the Cucurbita moschata CmSGT1 Gene. Front. Plant Sci. 10, 955. doi: 10.3389/fpls.2019.00955
He, J., Gu, X. R., Wei, C. H., Yang, X. Z., Zhang, X. (2016). Identification and expression analysis under abiotic stresses of the bHLH transcription factor gene family in watermelon. Acta Hortic. Sin. 43 (2), 281–294. doi: 10.16420/j.issn.0513-353x.2015-0886
Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., Bailey, P. C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. doi: 10.1093/molbev/msg088
Ishii, H., Fraaije, B. A., Sugiyama, T., Noguchi, K., Nishimura, K., Takeda, T. (2001). Occurrence and molecular characterization of Strobilurin resistance in cucumber powdery mildew and downy mildew. Phytopathology 91, 1166–1171. doi: 10.1094/PHYTO.2001.91.12.1166
Jin, J., Hewezi, T., Baum, T. J. (2011). The Arabidopsis bHLH25 and bHLH27 transcription factors contribute to susceptibility to the cyst nematode Heterodera schachtii. Plant J. 65 (2), 319–328. doi: 10.1111/j.1365-313X.2010.04424.x
Jing, Y. X., Liu, J., Liu, P., Ming, D. F., Sun, J. Q. (2019). Overexpression of TaJAZ1 increases powdery mildew resistance through promoting reactive oxygen species accumulation in bread wheat. Sci. Rep. 9, 5691. doi: 10.1038/s41598-019-42177-y
Le Hir, R., Castelain, M., Chakraborti, D., Moritz, T., Dinant, S., Bellini, C. (2017). AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana. Physiol. Plant 160 (3), 312–327. doi: 10.1111/ppl.12549
Lee, M. H., Lee, Y., Hwang, I. (2013). “In Vivo Localization in Arabidopsis Protoplasts and Root Tissue,” in G Protein-Coupled Receptor Signaling in Plants. Methods in Molecular Biology (Methods and Protocols), vol. 1043. Ed. Running, M. (Totowa, NJ: Humana Press).
Li, A., Zhang, R., Pan, L., Tang, L., Zhao, G., Zhu, M., et al. (2011). Transcriptome analysis of H2O2-treated wheat seedlings reveals a H2O2-responsive ratty acid desaturase gene participating in powdery mildew resistance. PloS One 12, 1–16. doi: 10.1371/journal.pone.0028810
Li, Z., Wang, S., Tao, Q., Pan, J., Si, L., Gong, Z., et al. (2012). A putative positive feedback regulation mechanism in CsACS2 expression suggests a modified model for sex determination in cucumber (Cucumis sativus L.). J. Exp. Bot. 63, 4475–4484. doi: 10.1093/jxb/ers123
Li, R., Zhang, L. L., Yang, X. M., Cao, X. L., Wang, W. M. (2019). Transcriptome analysis reveals pathways facilitating the growth of tobacco powdery mildew in arabidopsis. Phytopathol. Res. 1, 7. doi: 10.1186/s42483-019-0012-z
Lu, X., Yang, L., Yu, M., Lai, J., Wang, C., McNeil, D., et al. (2017). A novel Zea mays ssp. mexicana L. MYC-type ICE-like transcription factor gene ZmmICE1, enhances freezing tolerance in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 113, 78–88. doi: 10.1016/j.plaphy.2017.02.002
Malinovsky, F. G., Batoux, M., Schwessinger, B., Youn, J. H., Stransfeld, L., Win, J., et al. (2014). Antagonistic regulation of growth and immunity by the arabidopsis basic helix-loop-helix transcription factor homolog of brassinosteroid enhanced expression2 interacting with increased leaf inclination1 binding bhlh1. Plant Physiol. 164 (3), 1443–1455. doi: 10.1104/pp.113.234625
Nanasato, Y., Tabei, Y. (2015). Cucumber (Cucumis sativus L.) and Kabocha Squash (Cucurbita moschata Duch). (eds) Agrobacterium Protocols. Methods In Mol. Biol. 1223, 299–310. doi: 10.1007/978-1-4939-1695-5_24
Nanasato, Y., Konagaya, K., Okuzaki, A., Tsuda, M., Tabei, Y. (2011). Agrobacterium-mediated transformation of kabocha squash (Cucurbita moschata Duch) induced by wounding with aluminum borate whiskers. Plant Cell Rep. 30 (8), 1455–1464. doi: 10.1007/s00299-011-1054-6
Perez-Garcia, A., Romero, D., Fernandez-Ortuno, D., Lopez-Ruiz, F., De-Vicente, A., Tores, J. A. (2009). The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mol. Plant Pathol. 10, 153–160. doi: 10.1111/j.1364-3703.2008.00527.x
Seo, J. S., Joo, J., Kim, M. J., Kim, Y. K., Nahm, B. H., Song, S. I., et al. (2011). OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65 (6), 907–921. doi: 10.1111/j.1365-313X.2010.04477.x
Spoel, S. H., Dong, X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. doi: 10.1038/nri3141
Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Toledo-Ortiz, G., Huq, E., Quail, P. H. (2003). The Arabidopsis basic/helixloop-helix transcription factor family. Plant Cell 15, 1749–1770. doi: 10.1105/tpc.013839
Wang, F., Lin, R., Feng, J., Qiu, D., Chen, W., Xu, S. (2015). Wheat bHLH transcription factor gene, TabHLH060, enhances susceptibility of transgenic Arabidopsis thaliana to Pseudomonas syringae. Physiol. Mol. Plant Pathol. 90, 123–130. doi: 10.1016/j.pmpp.2015.04.007
Wasternack, C., Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in annals of botany. Ann. Bot. 111 (6), 1021–1058. doi: 10.1093/aob/mct067
Wei, K. F., Chen, H. Q. (2018). Comparative functional genomics analysis of bHLH gene family in rice, maize and wheat. BMC Plant Biol. 18, 309. doi: 10.1186/s12870-018-1529-5
Wu, T., Cao, J. (2010). Molecular cloning and expression of a bush related CmV1, gene in tropical pumpkin. Mol. Biol. Rep. 37, 649–652. doi: 10.1007/s11033-009-9505-7
Wu, H., Ye, H., Yao, R., Zhang, T., Xiong, L. (2015). OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 232, 1–12. doi: 10.1016/j.plantsci.2014.12.010
Xu, W., Jiao, Y., Li, R., Zhang, N., Xiao, D., Ding, X., et al. (2014). Chinese wild-growing Vitis amurensis ICE1 and ICE2 encode MYC-type bHLH transcription activators that regulate cold tolerance in Arabidopsis. PloS One 9(7), e102303. doi: 10.1371/journal.pone.0102303
Yamamura, C., Mizutani, E., Okada, K., Nakagawa, H., Fukushima, S., Tanaka, A., et al. (2015). Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. Plant J. 84 (6), 1100–1113. doi: 10.1111/tpj.13065
Zhao, Q., Xiang, X., Liu, D., Yang, A., Wang, Y. (2018). Tobacco transcription factor NtbHLH123 confers tolerance to cold stress by regulating the NtCBF pathway and reactive oxygen species homeostasis. Front. Plant Sci. 9, 381. doi: 10.3389/fpls.2018.00381
Keywords: pumpkin, powdery mildew, CmbHLH87, functional analysis, tobacco
Citation: Guo W-L, Chen B-H, Guo Y-Y, Chen X-J, Li Q-F, Yang H-L, Li X-Z, Zhou J-G and Wang G-Y (2020) Expression of Pumpkin CmbHLH87 Gene Improves Powdery Mildew Resistance in Tobacco. Front. Plant Sci. 11:163. doi: 10.3389/fpls.2020.00163
Received: 27 September 2019; Accepted: 03 February 2020;
Published: 03 April 2020.
Edited by:
Giorgio Gambino, Italian National Research Council, ItalyReviewed by:
David Vela-Corcia, University of Málaga, SpainWen-Ming Wang, Sichuan Agricultural University, China
Zujun Yang, University of Electronic Science and Technology of China, China
Copyright © 2020 Guo, Chen, Guo, Chen, Li, Yang, Li, Zhou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Zheng Li, bHh6MjI4M0AxMjYuY29t
 Wei-Li Guo
Wei-Li Guo Bi-Hua Chen1,2
Bi-Hua Chen1,2