- 1Plant-Microbe Interactions Laboratory, School of Agriculture and Food Sciences, the University of Queensland, Brisbane, QLD, Australia
- 2Plant Protection Department, College of Agriculture, University of Baghdad, Baghdad, Iraq
Mediator subunits play key roles in numerous physiological pathways and developmental processes in plants. Arabidopsis Mediator subunits, MED18 and MED25, have previously been shown to modulate disease resistance against fungal and bacterial pathogens through their role in jasmonic acid (JA) signaling. In this study, Arabidopsis mutant plants of the two Mediator subunits, med18 and med25, were tested against three ssRNA viruses and one dsDNA virus belonging to four different families: Turnip mosaic virus (TuMV), Cauliflower mosaic virus (CaMV), Alternanthera mosaic virus (AltMV), and Cucumber mosaic virus (CMV). Although both subunits are utilized in JA signaling, they occupy different positions (Head and Tail domain, respectively) in the Mediator complex and their absence affected virus infection differently. Arabidopsis med18 plants displayed increased resistance to RNA viral infection and a trend against the DNA virus, while med25 mutants displayed increased susceptibility to all viruses tested at 2 and 14 days post inoculations. Defense marker gene expression profiling of mock- and virus-inoculated plants showed that med18 and med25 mutants exhibited an upregulated SA pathway upon virus infection at 2 dpi for all viruses tested. JA signaling was also suppressed in med18 plants after virus infection, independent of which virus infected the plants. The upregulation of SA signaling and suppression of JA signaling in med18 may have led to more targeted oxidative burst and programmed cell death to control viruses. However, the susceptibility exhibited by med25 mutants suggests that other factors, such as a weakened RNAi pathway, might play a role in the observed susceptibility. We conclude that MED18 and MED25 have clear and opposite effects on accumulation of plant viruses. MED18 is required for normal virus infection, while MED25 is important for defense against virus infection. Results from this study provide a better understanding of the role of Mediator subunits during plant-virus interactions, viral disease progression and strategies to develop virus resistant plants.
Introduction
Resistance to infection in plants is often due to plant defense that limit multiplication or spread of a pathogen. As discussed by Klessig et al. (2000), some plants have the potential to identify pathogens and then activate a defense reaction when the products of resistance (R) gene expression interact with products of the virulence (Avr) gene expression. According to Revers and Nicaise (2014), the R genes that exist in the majority of the plant-pathogen integrations are part of the nucleotide binding site-leucine-rich repeat (NB-LRR).
Some types of plants possess mechanisms to detect plant pathogens by Pathogen-Associated Molecular Patterns (PAMPs). The main role of PAMPs is to act as the elicitors that permit plants to identify pathogens to mount the mechanisms that are ideally suited for their effective defense (Nürnberger and Lipka, 2005). PAMPs are advanced by plants through a different process known as Pattern Recognitive Receptors (PRRs) (Chiriac, 2013). This procedure involves the receptors that are triggered once the effect of pathogen proteins or other gene products is perceived by the plant. However, just as pointed out by Zvereva and Pooggin (2012), since there is currently no proof that PAMPs are linked with viruses at the moment, the primary defense mechanism for plants entail RNA muzzling, while the proteins are related to inducible resistance. Despite the fact that different viruses cannot be detected by PAMPs, the viruses still establish Pattern-Triggered Immunity (PTI) that makes it possible for the resistance (R) genes to identify various nonviral effectors together with the viral virulence proteins that are capable of producing Effector-Triggered Immunity (ETI). The procedure is similar to PTI even though it tends to be more intensive, since this type of immunity has the potential to activate different signals that will result in a hypersensitive reaction together with automated cell death (Nürnberger and Brunner, 2002). This reaction is an effort to locally respond to the virus by limiting its spread in the plants and needs triggering of the salicylic acid (SA) pathway that can result in systematic acquired resistance (SAR).
Defense signaling encompasses a methodical resistance that involves triggering cellular and the molecular defenses (Pieterse et al., 2009; Dong et al., 2015). Some plant pathogens are necrotrophs that require dead tissues for nutrients, while others are biotrophs that required live cells as hosts for completing their life cycles. In other cases, pathogens use plants for nectrotrophic and biotrophic life styles, but usually at different phases. The SA defense pathway typically confers resistance to biotrophic and the jasmonic acid (JA) pathway is typically effective against necrotrophic pathogens, with some exceptions (Pieterse et al., 2009). In many cases, however, it appears that virulent pathogens have the ability to trick the plant with effector molecules that induce the wrong pathway and as a consequence damage plants more (Thatcher et al., 2009). As viruses are biotrophs, an upregulation of the SA pathway would be essential for resistance, as this can result in localized oxidative burst by reactive oxygen species (ROS) production, hypersensitive response (HR), and programmed cell death, thus limiting the spread of viruses. Bacteria and fungi can feature biotrophic and nectrotrophic pathogenesis, while in the case of viruses, they are completely biotrophic pathogens. It is however important to take note that viruses are only able to reproduce intracellularly and can migrate and spread from cell-to-cell through plasmodesmata as well as over long distances using the vascular system, resulting in overall infection of their vulnerable hosts (Ellis et al., 2006). However before this occurs, certain pathogen-linked genes can trigger SAR which then activates resistance in plants whose distal tissues have not been affected (Zvereva and Pooggin, 2012).
The Mediator complex provides the link between transcription factors (TFs) and RNA Polymerase II, required for transcription. It contains approximately 31 and 34 subunits in mammals and plants, respectively (Mathur et al., 2011; Allen and Taatjes, 2015; Samanta and Thakur, 2015). The Mediator complex comprises three domains which are Head, Middle, and Tail, that, when taken together, are referred to as the core Mediator. In addition, a fourth module comprises Cyclin-Dependent Kinase 8 (CDK8) that is reversibly linked with Mediator. For yeast and metazoa, CDK8 controls transcription through phosphorylation of TFs and induction of RNA Pol II-Mediator interactions (Parker et al., 1996). The Tail domain of the Mediator complex interacts with the DNA-bound TFs, while the Head domain interacts with RNA Pol II and might also be engaged in either basal or activator-free transcription. For the Middle domain, Karijolich and Hampsey (Karijolich et al., 2012) discussed that it offers the flexibility needed by the huge protein complex so that it can show the necessary conformational changes in its reaction to RNA Pol II binding. The Mediator is capable of modulating RNA Pol II-based transcription by impacting the makeup of the preinitiation complex, Pol II stopping, elongation, and reinitiation and chromatic buildup (Allen and Taatjes, 2015).
MED18 is part of the Head domain of the Mediator complex where it can bind to TFs (Kim et al., 2011; Lai et al, 2014; Fallath et al., 2017). MED18 plays a major role in plant growth, flowering and immunity, including the production of noncoding RNA and modulates crosstalk between JA- and SA-associated defense pathways. Moreover, its Arabidopsis mutant med18 plants show reduced miRNA levels and downregulated JA signaling and biosynthesis genes, while SA-associated PR and ROS producing genes are upregulated (Fallath et al., 2017). MED18 is believed to bind to MED20 in the Head domain of the Mediator complex and med18 has similar growth phenotypes to med20a with strong resistance against the hemibiotrophic fungus Fusarium oxysporum, while showing increased susceptibility to necrotrophic fungi Botrytis cinerea and Alternaria oxysporum.
Contrary to MED18, MED25 is located at the Tail domain of the Mediator complex that connects to TFs such as MYC2 (Kidd et al., 2011). MED25 comprises 836 amino acids and is mechanically segmented into three different domains that are preserved (Bäckström et al., 2007; Kidd et al., 2009; Chen et al., 2012). The amino terminal of MED25 comprises of the domain Von Willebrand factor type A that is located in a preserved section of the Mediator attachment region. The subsequent domain comprises an activator interaction domain (ACID), that is vital for the interactions with TFs. Finally, the carboxyl terminal of MED25 comprises a poly track that is rich in glutamine at its C terminus, and most likely engaged in transcriptional triggering (Cerdán and Chory, 2003). MED25 plays a critical role in the growth of plants as well as response to stress. MED25 is reported to add to shoot elongation as well as flowering, based on the amount of the light that is present (Bäckström et al., 2007). Moreover, MED25 controls the adaptive nature of the plants perceived to avoid shade and MED25 is also needed for uncompromised expression of JA-dependent defense genes (Kidd et al., 2009; Elfving et al., 2011).
Common for both, med18 and med25 mutants is that they were found to be insensitive or partially insensitive, respectively, to JA (Kidd et al., 2009; Fallath et al, 2017). This defect in JA signaling for both plants is believed to mediate resistance to the hemibiotrophic fungal pathogen F. oxysporum, while being more susceptible to necrotrophic fungi Botrytis cinerea and Alternaria brassicicola. In this study, plant defense responses and resistance Arabidopsis med18 and med25 were tested against four viruses belonging to different families and compared to those in wild-type Col-0 plants. It was aimed to establish how MED18 and MED25 influence viral pathogenesis outcomes and whether there are common defense responses to different viruses (as was previously observed for fungal pathogens). In preliminary experiments, we found that med18 mutants displayed resistance against Turnip mosaic virus (TuMV). Hence, we hypothesized that Mediator subunits may control common principles for broad resistance or susceptibility to different viruses, including TuMV, Cauliflower mosaic virus (CaMV), Alternanthera mosaic virus (AltMV), and Cucumber mosaic virus (CMV). All are ssRNA viruses apart from CaMV which is a dsDNA virus. This study can help to understand how plants interact with viruses and assist in establishing the pathways, including the roles of Mediator subunits MED18 and MED25, that were found to be required for normal virus infection and defense against viruses, respectively.
Materials and Methods
Cultivation of Arabidopsis Plants
Arabidopsis med18 and med25 homozygous mutant plants were previously characterized and maintained from our previous studies (Kidd et al., 2009; Fallath et al, 2017). The first stage included sowing wild-type Col-0, med18, and med25 Arabidopsis thaliana seeds and placing them at 4°C for 2 days. A growth chamber was used to cultivate seedlings under the following conditions: 8 h of 24°C during the day (160 μE m-2s-1) followed by 16 h at 21°C during the night. The last stage included transplanting the seedlings after 3 weeks (two plants per pot), and then inoculating them with viruses when they were 5 weeks old. All plants were grown in parallel [including mock-inoculated-/virus-infected plants for each genotype and both time points of sampling (2 and 14 days post inoculation; dpi)], but experiments for different viruses were carried out on separate occasions. At least 60 plants were grown for each genotype, treatment and time point. Each tray contained 60 plants (20 of each genotype) and was regularly repositioned within the growth cabinet to exclude positional effects on plant growth. Each treatment, genotype and time point had three biological replicates (total of 72 biological samples). Each biological replicate contained a pooled sample of 20 plants each.
Virus Inoculation
The TuMV-QLD1b isolate used in this study was a serially passaged isolate of an original sample (VIR-0745; TuMV-QLD1a) previously sourced from the Queensland Department of Agriculture and Fisheries (DAF) in 2007 (Pretorius et al., 2016). Similarly, CaMV- Dar78694 and the AltMV virus isolates were also supplied from the DAF collection (Pretorius et al., 2017). CMV isolate K was kindly supplied by John Randle in 2004 (Moyle et al., 2018). Nicotiana benthamiana plants were used to propagate TuMV and AltMV, while tomato wild type (Moneymaker) was used to propagate CMV. Brassica rapa subsp. chinensis leaves were inoculated with CaMV and used for propagation. All virus inoculations of the wild-type Arabidopsis plants were performed by fresh inoculums. A 100 mM sodium phosphate buffer, pH 7.4 containing 1 g/L sodium sulphite was used to suspend the leaves after crushing. Abrasion was done by using celite. The following steps included gently rubbing three leaves per plant and then leaving the inoculum on the leaves for about 5 min before washing it off. For controls, mock inoculations were performed using the same buffer and abrasive.
RNA Extraction and cDNA Synthesis
Two time points (2, 14 dpi) were used to collect the Arabidopsis plants. The foliar parts of the plants were used for analysis after cutting the plants at the base. Then, these parts were dropped into liquid nitrogen immediately including three replicates with 20 pooled plants per replicate (combined from two different trays each), for each genotype and time point. This was followed by grounding the tissue samples in liquid nitrogen. RNA was extracted using the Promega SV Total RNA Isolation System by following the manufacturer’s instructions. A Nanodrop spectrophotometer (Thermo Scientific, Australasia) was used to measure the concentrations of the purified RNA samples. Agarose gel electrophoresis was used to confirm the intact quality of the RNA samples. cDNA was generated by using components of the Tetro cDNA synthesis kit (BIOLINE, Australasia), including 1 µl reverse transcriptase for 2.5 µg of total RNA in 20 µl reactions containing 0.5 µl random hexamers, 0.5 µl oligo dT primers, 1 µl 10 mM dNTP mix, 4 µl 5x RT Buffer, and 1 µl RNase Inhibitor with the following thermocycling program 25°C for 10 min, 45°C for 30 min. The reaction was finished by incubating at 85°C for 5 min.
DNA Extraction
Extraction of plant DNA was performed by crushing plant material with a mortar and pestle in liquid nitrogen, then subjecting it to the CTAB method to extract DNA (Graham et al., 1994).
Real-Time Quantitative Reverse Transcriptase PCR
The ViiA 7 Real-Time PCR system with SYBR green was used to perform real-time quantitative PCR (qPCR) to measure the virus titers of CaMV or real-time quantitative reverse transcriptase PCR (qRT-PCR) to measure virus titers of TuMV, AltMV, CMV, or gene transcript abundances with three technical replicates per run. Primer sequences are shown in Supplementary Table 1. Primer Express software was used to quantify the abundance of virus sequences or gene transcripts relative to the housekeeping reference genes β-ACTIN2, β-ACTIN7, and β-ACTIN8. qRT-PCR assays were performed in a 10 µl reaction containing 5 µl SYBR Green Master MIX, 1 µl of primer mix (0.3 µM for each primer), and 4 µl of cDNA templates which were diluted 12.5 times prior to PCR reactions. The PCR conditions were as follows: 95°C for 10 min, 40 cycles for 15 s at 95°C, 60°C for 1 min. The melting curve conditions were 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. LinRegPCR™ software was used based on the following equation:
as previously described (Liu et al., 2016; Fallath et al., 2017).
Technical replicates where the higher value had more than 0.5 Ct difference to the other values were excluded from the analysis as this could be due to reaction inhibition. The change of the gene expression levels was determined by taking the average of the three biological replicates and comparing that to the control plants (either wild-type virus-inoculated or mock-inoculated plants). Statistically significant differences (P < 0.05) were determined using Student’s t-test for pairwise comparisons or ANOVA F test followed LSD analysis for multiple comparisons. Experiments for different viruses were carried out at different times. For this reason, all comparisons to different viruses are done on the basis on whether a gene was induced or repressed within the experiments (i.e., pairwise comparisons to mock-inoculated controls or to wild type), but not absolute levels of gene expression were used.
Results
Wild-type Col-0, med18, and med25 mutants of Arabidopsis thaliana were inoculated with TuMV, CaMV, AltMV, and CMV, to test for host defense marker gene expression and virus disease progression, and to compare these across different viruses. First the phenotypes of infected plants were determined, then the virus titers were quantified by qRT-PCR (TuMV, AltMV, CMV) or qPCR (CaMV) at both, 2 dpi and 14 dpi, followed by qRT-PCR analyses for host marker gene expression at 2 dpi to gain an understanding of early defense responses that may have influenced virus resistance.
med18 Mutants Display Broad RNA Virus Resistance While med25 Mutants are More Susceptible to RNA and DNA Viruses
Inoculation of wild-type med18 and med25 with TuMV, CaMV, AltMV, or CMV resulted in symptom development for all viruses tested (Supplementary Figure 1). However, it was noted that symptoms were generally less pronounced in med18 plants when compared to wild-type plants. On the contrary, med25 plants typically displayed more severe symptoms. Virus titer quantification by qRT-PCR confirmed that indeed, all med18 plants harbored significantly (P < 0.05) less RNA viruses than wild-type plants at an early stage (2 dpi) as well as at a more advanced state (14 dpi) of disease progression, and CaMV-infected plants also showed the same trend (Figure 1). However, med25 plants consistently contained significantly (P < 0.05) more viruses (RNA and DNA viruses) than wild-type plants at both time points. The following sections will present data for each virus and compare these with marker gene expression in all three genotypes tested.
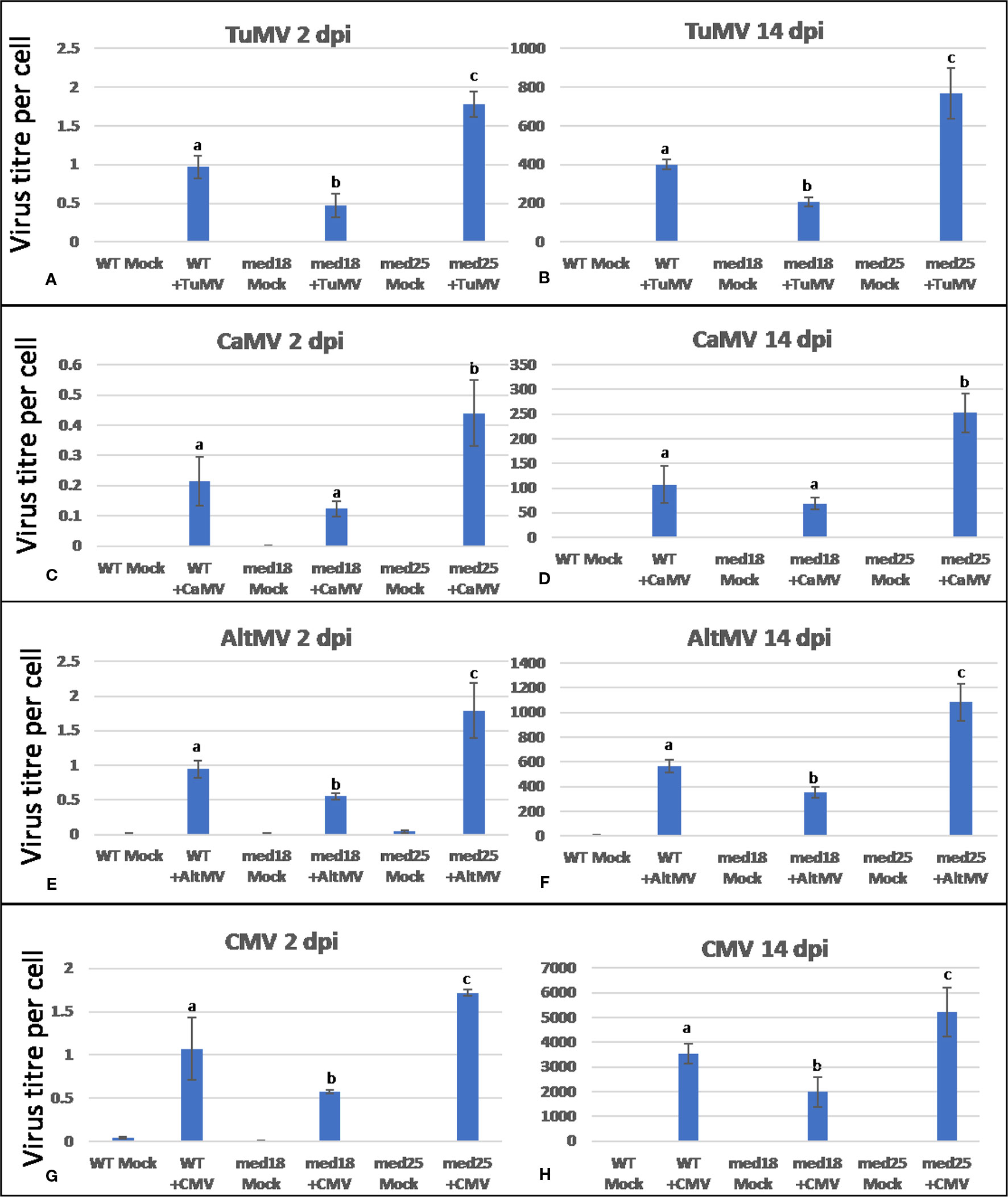
Figure 1 Virus titer per plant cell of Arabidopsis wild-type (Col-0), med18, and med25 mutant plants inoculated with Turnip mosaic virus (TuMV) (A, B), Cauliflower mosaic virus (CaMV) (C, D), Alternanthera mosaic virus (AltMV) (E, F), or Cucumber mosaic virus (CMV) (G, H) at 2 dpi and 14 dpi. Shown on the Y axes are quantitative reverse transcriptase PCR (qRT-PCR) or quantitative PCR (qPCR) mean values of virus abundance ± SE relative to plant cells (ACTIN). Different small letters indicate statistically significant (P < 0.05) differences between genotypes of infected plants for each time point and virus.
TuMV’s Suppression of SA Signaling and Induction of JA Signaling Is Reversed in med18 and med25 Mutant Plants
TuMV-inoculated plants showed clear growth reduction symptoms compared to mock-inoculated controls (Supplementary Figure 1A). Visual symptoms were strongest for med25 and weakest for med18. The virus titer of TuMV-inoculated wild-type Col-0 plants, as measured by qRT-PCR, increased from approx. 1 virus per ACTIN transcript at 2 dpi to 400 at 14 dpi (Figures 1A, B). qRT-PCR Col-0 gene expression data at 2 dpi showed that SA signaling marker gene expression of PR1 was reduced in the presence of TuMV, while marker genes for JA signaling (VSP2, PDF1.2) were upregulated (Figure 2). The virus titer of TuMV-inoculated med18 plants, as measured by qRT-PCR, increased from 0.48 viruses per ACTIN transcript at 2 dpi to 207 at 14 dpi (Figures 1A, B), suggesting that med18 plants are more resistant to TuMV than wild-type plants. The virus titer of TuMV-inoculated med25 plants, as measured by qRT-PCR, increased from 1.78 viruses per ACTIN transcript at 2 dpi to 769 at 14 dpi (Figures 1A, B), suggesting that med25 plants are more susceptible to TuMV than wild-type plants. In contrast to wild type, qRT-PCR gene expression data for both med18 and med25 plants, showed that marker genes for JA signaling (VSP2, PDF1.2) were downregulated, while SA marker gene expression of PR1 was significantly increased (and PR5 also in med18) in the presence of TuMV at 2 dpi (Figure 2). In med25, there was a trend to increased expression that was not significant with the number of experiments and replicates used.
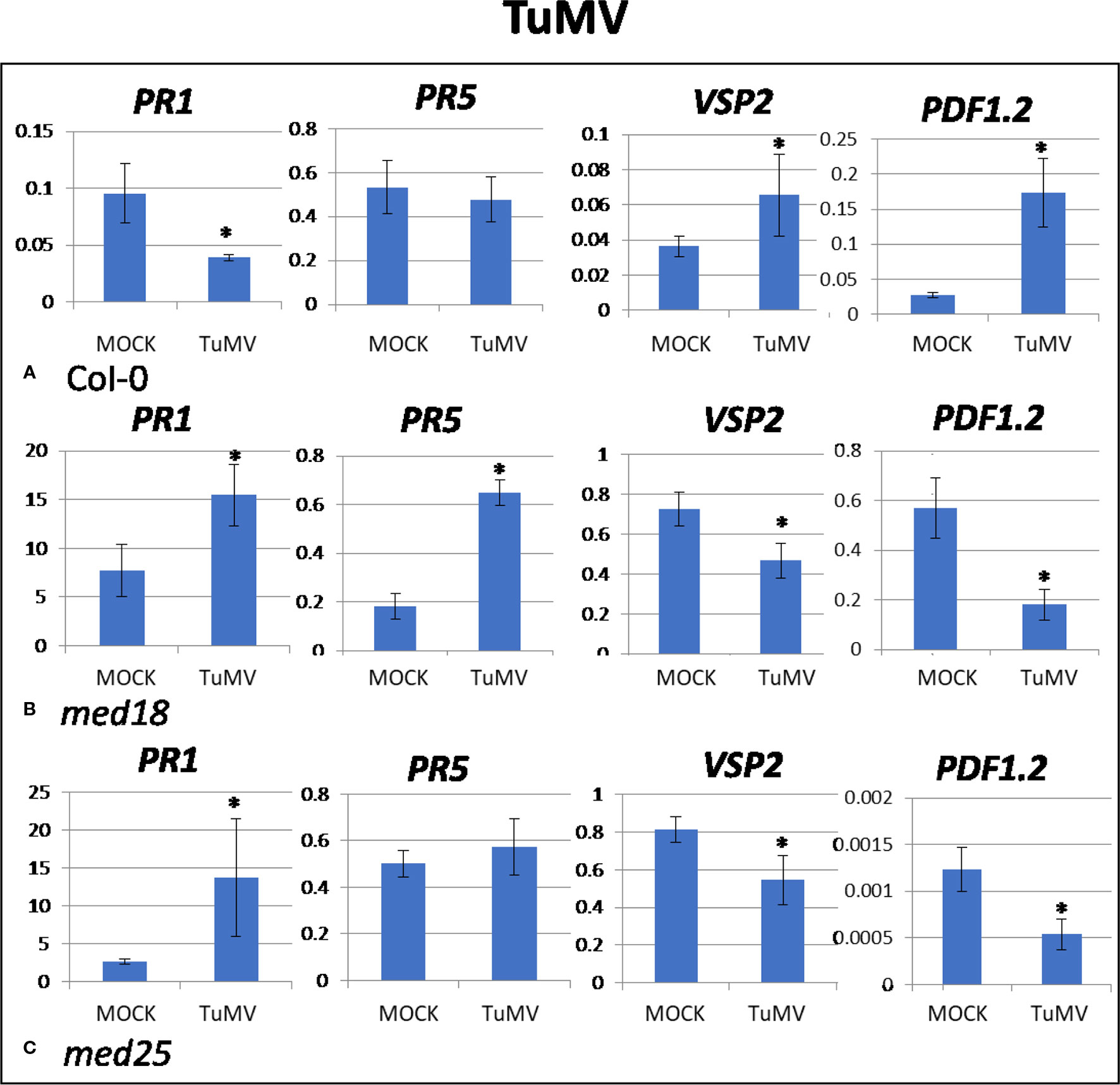
Figure 2 Quantitative reverse transcriptase PCR (qRT-PCR) assays with SA- (PR1, PR5) and JA- (PDF1.2, VSP2) signaling marker genes of Arabidopsis plants inoculated with TuMV. Shown on the Y axes are mean ± SE values of transcript abundances relative to housekeeping ACTIN genes for (A) wild-type Col-0, (B) med18, and (C) med25 plants at 2 days after inoculation. Asterisks indicate significant (P < 0.05) induction or repression of each gene by comparing the means of mock-inoculated to TuMV-infected samples.
CaMV Infection Induced SA Signaling, but JA Signaling Was Suppressed in med18 and Induced in med25 Plants
As shown in Supplementary Figure 1B, CaMV-inoculated plants showed clear growth reduction symptoms compared to mock-inoculated controls. Visual symptoms were strongest for med25 and weakest for med18. The virus titer of CaMV-inoculated wild-type plants, as measured by qPCR, increased from 0.21 viruses per cell at 2 dpi to 107 at 14 dpi (Figures 1C, D). qRT-PCR gene expression data showed that marker genes for JA signaling (VSP2, PDF1.2) were not differentially expressed, while SA marker gene expression (PR1) was induced in the presence of CaMV at 2 dpi (Figure 3). Base levels for PR1 differed for the different replicates and also across other virus experiments, but each replicate showed clear induction for PR1 following CaMV infection. To more broadly profile gene expression for SA signaling, but also for ABA signaling, PR2 (SA signaling), and RD22 (ABA signaling) were included in the analysis. Both of these genes also showed upregulation in the presence of CaMV at 2 dpi (Figure 3).
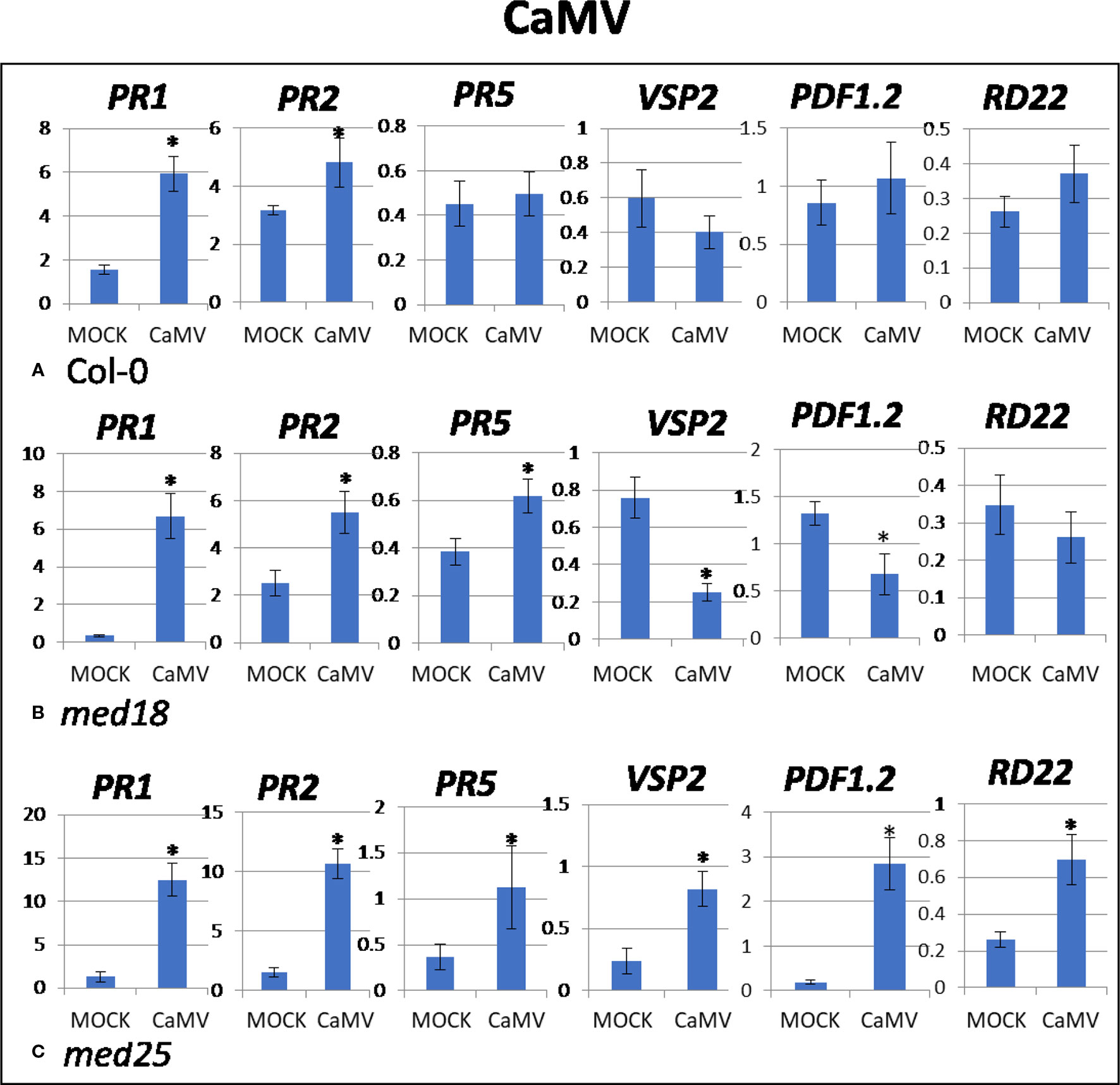
Figure 3 Quantitative reverse transcriptase PCR (qRT-PCR) assays with SA- (PR1, PR5), JA- (PDF1.2, VSP2) and ABA- (RD22) signaling marker genes of Arabidopsis plants inoculated with CaMV. Shown on the Y axes are mean ± SE values of transcript abundances relative to ACTIN housekeeping genes for (A) wild-type Col-0, (B) med18, and (C) med25 plants at 2 days after inoculation. Asterisks indicate significant (P < 0.05) induction or repression of each gene by comparing the means of mock-inoculated to CaMV-infected samples.
The virus titer of CaMV-inoculated med18 plants, as measured by qPCR, increased from 0.12 viruses per cell at 2 dpi to 69 at 14 dpi (Figures 1C, D). Although there was a trend for reduced CaMV accumulation in med18 plants, it was not significantly different from the wild-type plants at either 2 or 14 dpi. Similar to wild-type plants, qRT-PCR gene expression data from med18 plants in the presence of CaMV at 2 dpi showed that marker genes for SA signaling (PR1, PR2, and the trend for PR5) were upregulated, while JA signaling (VSP2, PDF1.2) was downregulated (Figure 3). The virus titer of CaMV-inoculated med25 plants, as measured by qPCR, increased from 0.44 viruses per cell at 2 dpi to 253 at 14 dpi (Figures 1C, D), indicating that med25 plants are more susceptible to CaMV compared to wild type. qRT-PCR gene expression data showed that marker genes for SA-, JA- and ABA- (RD22) signaling were all upregulated in the presence of CaMV at 2 dpi (Figure 3).
AltMV Infection Induced SA and JA Signaling, but JA Signaling Was Suppressed in med18 Plants
As shown in Supplementary Figure 1C, AltMV-inoculated plants showed clear growth reduction symptoms compared to mock-inoculated controls. Visual symptoms (based on biomass) across different genotypes were similar. However, the virus titer of AltMV-inoculated wild-type plants, as measured by qRT-PCR, increased from 0.94 viruses per ACTIN transcript at 2 dpi to 567 at 14 dpi (Figures 1E, F). qRT-PCR gene expression data showed that marker genes for JA signaling (PDF1.2) and SA signaling (PR1, PR2, PR5) were upregulated after infection with AltMV at 2 dpi, and a second JA signaling pathway gene (VSP2) showed the same trend (Figure 4).
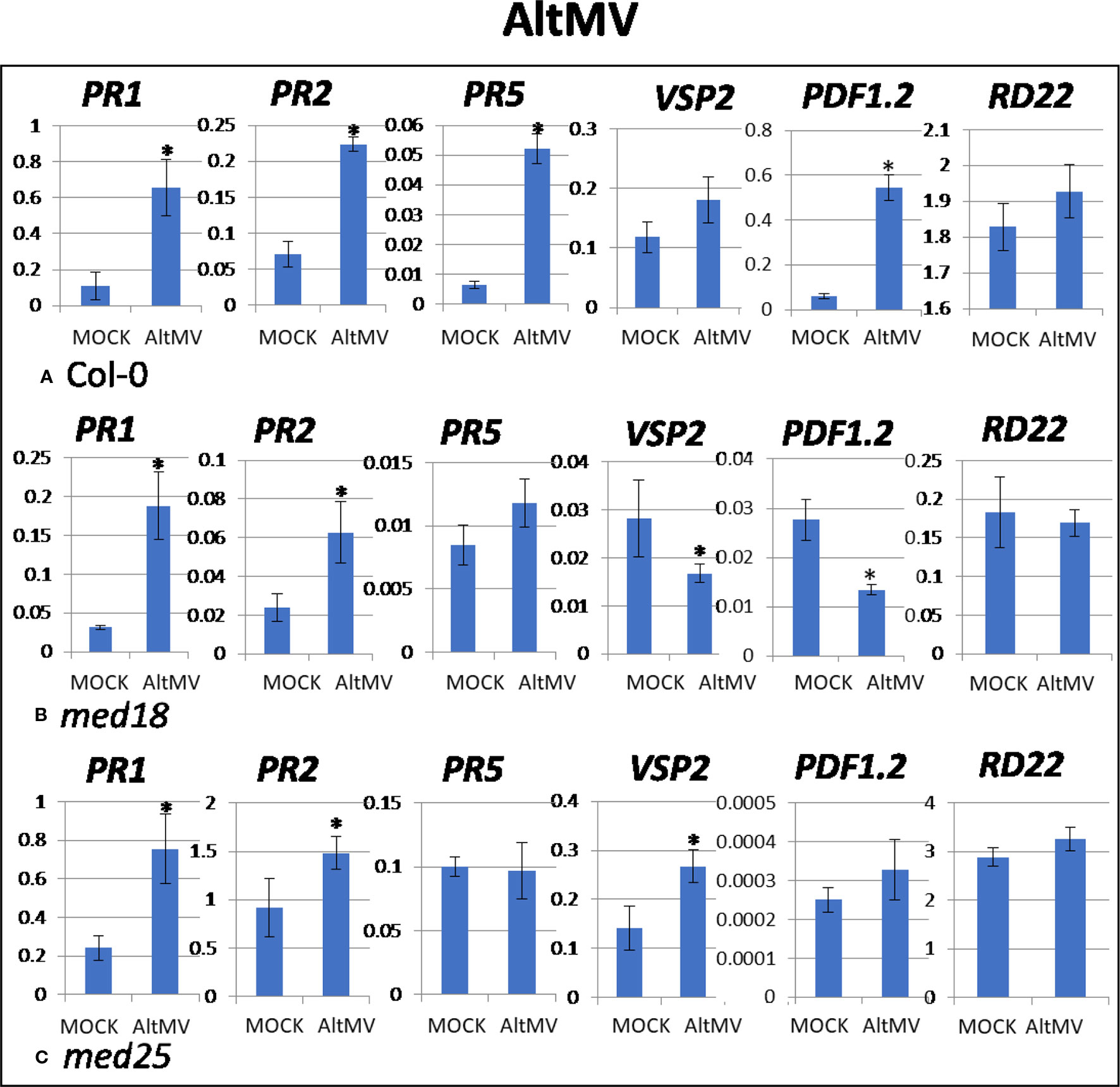
Figure 4 Quantitative reverse transcriptase PCR (qRT-PCR) assays with SA- (PR1, PR5), JA- (PDF1.2, VSP2), and ABA- (RD22) signaling marker genes of Arabidopsis plants inoculated with AltMV. Shown on the Y axes are mean ± SE values of transcript abundances relative to ACTIN housekeeping genes for (A) wild-type Col-0, (B) med18, and (C) med25 plants at 2 days after inoculation. Asterisks indicate significant (P < 0.05) induction or repression of each gene by comparing the means of mock-inoculated to Alternanthera mosaic virus (AltMV)–infected samples.
The virus titer of AltMV-inoculated med18 plants, as measured by qRT-PCR, increased from 0.55 viruses per ACTIN transcript at 2 dpi to 355 at 14 dpi (Figures 1E, F), indicating that med18 plants are more resistant compared to wild type. Similar to wild type, SA signaling (PR1, PR,2 and the trend for PR5) was upregulated in med18 after infection. Interestingly in contrast to wild-type and med25 plants, qRT-PCR gene expression data showed that marker genes for JA signaling (VSP2, PDF1.2) were downregulated in the presence of AltMV in med18 at 2 dpi (Figure 4).
For med25 AltMV-inoculated plants, the virus titer, as measured by qRT-PCR, increased from 1.79 viruses per ACTIN transcript at 2 dpi to 1,083 at 14 dpi (Figures 1E, F), indicating that med25 plants are more susceptible compared to wild type. qRT-PCR gene expression data showed that marker genes for SA signaling (PR1, PR2, but not PR5) and JA signaling (VSP2) were upregulated in med25 in the presence of AltMV at 2 dpi (Figure 4). ABA pathway marker gene RD22 was not significantly differentially expressed following AltMV infection for any time point or genotype.
CMV Infection Induced PR1 Involved in SA Signaling and JA Signaling was Further Suppressed in med18 and med25 Plants
As shown in Supplementary Figure 1D, CMV-inoculated plants showed clear growth reduction symptoms compared to mock-inoculated controls. Visual symptoms were strongest for med25 and weakest for med18. The virus titer of CMV-inoculated wild-type plants, as measured by qRT-PCR, increased from 1.07 viruses per ACTIN transcript at 2 dpi to 3,544 at 14 dpi (Figures 1G, H). qRT-PCR expression data showed that genes for SA signaling (PR1, PR5 but not PR2) were upregulated in the presence of CMV at 2 dpi (Figure 5). There was no significant change in JA-signaling markers on CMV infection of Col-0 plants.
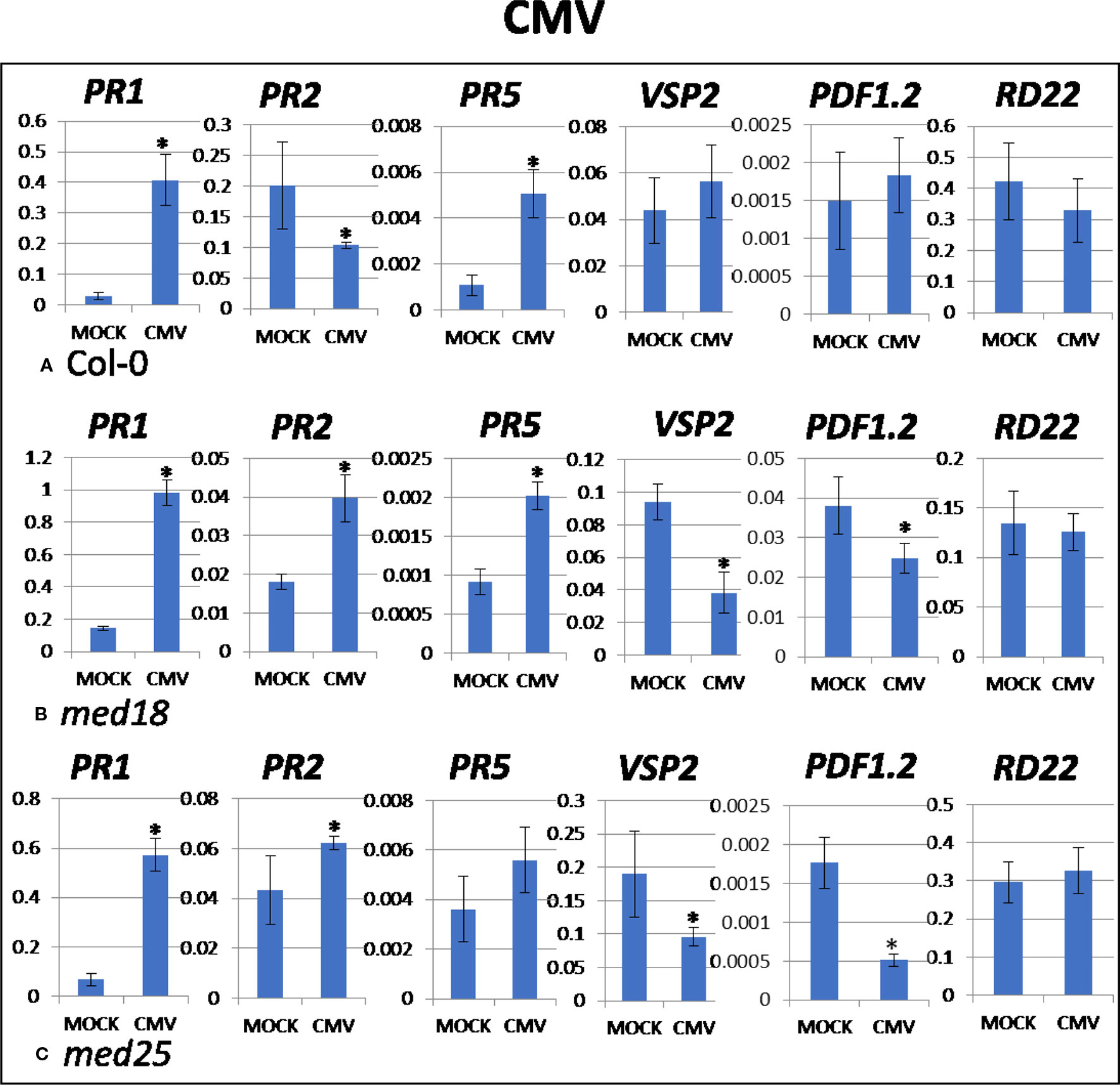
Figure 5 Quantitative reverse transcriptase PCR (qRT-PCR) assays with SA- (PR1, PR5), JA- (PDF1.2, VSP2), and ABA- (RD22) signaling marker genes of Arabidopsis plants inoculated with CMV. Shown on the Y axes are mean ± SE values of transcript abundances relative to ACTIN housekeeping genes for wild-type (A) Col-0, (B) med18, and (C) med25 plants at 2 days after inoculation. Asterisks indicate significant (P < 0.05) induction or repression of each gene by comparing the means of mock-inoculated to CMV-infected samples.
The virus titer of CMV-inoculated med18 plants, as measured by qRT-PCR, increased from 0.57 viruses per ACTIN transcript at 2 dpi to 1994 at 14 dpi (Figures 1G, H), indicating that med18 mutants are more resistant than wild-type plants. qRT-PCR expression data showed that SA signaling (PR1, PR2, PR5) was upregulated, while a gene for JA signaling (VSP2) was downregulated in the presence of CMV at 2 dpi (Figure 5). The virus titer of CMV-inoculated med25 plants increased from 1.72 viruses per ACTIN transcript at 2 dpi to 5218 at 14 dpi (Figures 1G, H), indicating that med25 mutants are more susceptible than wild-type plants. qRT-PCR expression data showed that SA signaling (PR1, PR2, but not PR5) was upregulated, while genes for JA signaling (VSP2, PDF1.2) were downregulated in the presence of CMV at 2 dpi (Figure 5). ABA pathway marker gene RD22 was not significantly differentially expressed following CMV infection for any time point or genotype.
Discussion
Studies on TuMV, CaMV, AltMV, and CMV are crucial in providing a better understanding of the impact that these viruses have on Arabidopsis. Many studies have attempted to elucidate the mechanism by which the virus affects plants leading to systemic infections (Zvereva and Pooggin, 2012; Revers and Nicaise, 2014; Roossinck, 2015). Results obtained in the present work suggest that the mechanisms vary widely for different viruses in Arabidopsis. For example, while SA signaling was activated upon CaMV, AltMV, and CMV inoculation of wild-type plants at 2 dpi (Figures 3A, 4A, and 5A), it was downregulated when plants were inoculated with TuMV (Figure 2A). JA signaling was also upregulated by TuMV and AltMV, but not CaMV and CMV. It should be emphasized that all viruses used in this study are compatible and are able to infect and proliferate in Arabidopsis plants (Figure 1; Supplementary Figure 1). Therefore, these viruses, and unlike their incompatible counterparts, have evolved to effectively evade the plant’s defense system. This can be achieved in different ways, but one of the strategies virulent plant pathogens can use, is to “hijack” a defense pathway that makes plants more susceptible. Wild-type plants infected with TuMV showed reduced SA and increased JA signaling (Figure 2), suggesting that TuMV may be able to hijack the JA pathway to make plants more susceptible (e.g. by counteracting SA signaling and oxidative burst). This phenomenon has been found for several plant pathogens, in particular F. oxysporum and P. syringae (Edgar et al., 2006; Kidd et al., 2009; Thatcher et al., 2009). Both are hemibiotrophic pathogens where early SA signaling/oxidative burst would be effective to fight the pathogens in their initial biotrophic phase. However, instead, JA signaling is upregulated which suppresses oxidative burst and SA signaling, presumably making plants more susceptible.
Many other mechanisms also play a role in viral disease progression, in particular the role of viral genes targeted to the nucleus and RNAi pathways should be considered. TuMV’s viral-genome-related protein of the nuclear inclusion protein interrelates with the potyviral VP-g interrelating protein that has a role in the transcriptional control and this interrelation might interfere with the host gene expression (Dunoyer et al., 2004). In addition, TuMV also has another nonstructural protein that tends to accrue in the nucleus and has been known to inhibit RNA silencing, assistance component proteinase (Mallory et al., 2002; Kasschau et al., 2003) that is comparable to CMV 2b that is described as a strong inhibitor of RNA silencing and RNA-directed DNA methylation (Wang et al., 2004). By comparison, AltMV protein gene block 1 is based in the nucleus and is also labeled as a suppressor of RNA silencing (Nam et al., 2013). Finally, CaMV even though a DNA virus, also duplicates in the cytoplasm through RNA intermediates. Moreover, its protein 6 is also known to accrue in the nucleus with one of its responsibility being to suppress RNA muzzling by interacting with the host nuclear protein DRB4 (Love et al., 2012). All viruses used in this research have the potential to interfere with the RNAi pathway and this should be the subject of further investigations.
Two Mediator subunits, MED18 and MED25, have been investigated in the present study with the aim to better understand their function in defense signaling and to establish their potential role in plant virus resistance. Both subunits play an established role in JA signaling, but are located at opposite ends of the Mediator complex (Kidd et al., 2009; Fallath et al., 2017). Mutants of both subunits show resistance against the root-infecting hemibiotrophic pathogen F. oxysporum, but are more susceptible to necrotrophic leaf pathogens B. cinerea and A. brassicicola (Kidd et al., 2009; Çevik et al., 2012; Fallath et al, 2017). This made these two subunits particularly interesting to study for virus resistance and defense against viruses that are obligate biotrophs and we hypothesized that the corresponding mutants may show an altered phenotype toward viral resistance. Indeed, med25 mutants were consistently more susceptible to all viruses tested, whereas med18 plants were consistently more resistant when compared to wild type. This was observed for both, 2 dpi and 14 dpi (Figure 1).
med18 plants reportedly show down-regulation of JA signaling defense and biosynthesis genes, while SA-associated PR-, ROS producing and scavenging genes are upregulated (Fallath et al, 2017). It is likely that the enhanced SA signaling capability in these plants led to a more pronounced HR resulting in faster oxidative burst and the production of ROS, leading to programmed cell death of virus-infected cells and therefore limiting spreading of viruses in the plants. Indeed, the gene expression analysis of virus-infected med18 plants consistently demonstrated that JA marker genes were downregulated, and SA genes were upregulated at 2 dpi, independent of which virus infected the plants (Figures 2–5). Similarly, JA signaling genes were suppressed under F. oxysporum infection in med18, while PR1 and PR5, as well as several genes linked to ROS production were upregulated (Fallath et al., 2017). med18 plants are more sensitive to SA signaling, but there were differences for different SA marker genes. After SA treatment, PR1 and to a lesser extend PR5 (but not PR2) expression was higher in med18 compared to wild type, indicating that MED18 differentially regulates various genes of the SA pathway in a different manner. The present study found that various viruses modulate SA signaling in a different manner in wild-type and med18 mutants. For example, PR1 was higher expressed in med18 upon TuMV and CMV infection but less expressed during AltMV infection, while PR5 was upregulated in AltMV- and CMV-infected med18 compared to wild type.
JA signaling and singlet oxygen stress signaling could be controlled through the Mediator complex via MED18, or otherwise, faulty JA signaling in med18 leads to increased ROS production and tolerance. No major lesions could be observed in virus-infected med18 plants upon microscopic observations, suggesting that the spread of viruses is similarly restricted as previously reported for F. oxysporum in med18 (Fallath et al., 2017). It was also shown that MED18 is recruited to the promoter of WRKY33 leading to increased WRKY33 expression (Liao et al., 2016). Similar to MED18, the inactivation of WRKY33 results in reduced JA and increased SA defense responses and higher susceptibility to B. cinerea (Liu et al., 2017) and therefore the mechanism of the MED18-controlled JA/SA crosstalk found in the present study could be based on MED18’s recruitment to the WRKY33 promoter. MED18 has also been reported to interact with TFs, YIN YANG1, ABA INSENSITIVE4 and SUPPRESSOR OF FRIGIDA4 (Lai et al., 2014), and future research could test whether these TFs also affect virus resistance.
med25 mutants, on the other hand, are only partially insensitive to JA but also show a reduction in JA-associated gene expression that was linked to the F. oxysporum resistance phenotype, although to a lesser extent than med18 plants (Kidd et al., 2009; Fallath et al., 2017). Interestingly, med25, showed increased susceptibility to all viruses tested (Figure 1), suggesting that this Tail-located subunit plays a role in antivirus defense signaling (rather than normal virus infection). This may include the RNAi pathway. As explained by Kim et al. (2011), MED25, MED18, and MED20a all play a role in the microRNA biogenesis pathway. Indeed, it was found that the subunits were engaged in encouraging the transcription of miRNA genes and were also in charge of recruiting RNA-directed RNA Polymerase 2 (RDR2) to certain promoters. In addition, the Mediator complex is involved when it comes to transcriptional gene silencing that is regulated by small interfering RNAs (Kim et al., 2011). The intricacy of the Mediator complex as well as the initiation of RDR2 transcription has been equated to an ancient type of cellular defense in averting DNA and RNA factors like transposons and viruses from attacking the host transcriptional machinery (Madhani, 2013). This is likely to expound the reason why Mediator subunits might be targeted by viruses as has previously been shown for the herpes simplex virus VP16 interaction domain (Bäckström et al., 2007; Aguilar et al, 2014). Plant viral interference with Mediator subunits would effectively modulate their roles in mRNA transcription, miRNA gene transcription, and transcriptional gene silencing.
Conclusion
The results obtained from this study provide a more comprehensive understanding of antiviral resistance in plants, progression of viral infections and the role of Mediator subunits in this process. The role of two Mediator subunits, MED18 and MED25, required for normal JA signaling, was tested, but that are located at the Head and Tail domain of the Mediator complex, respectively. Infection with all viruses exhibited clear growth reduction in plants, with visible symptoms being strongest in med25 mutants and weakest in med18 mutants. All wild-type plants showed an upregulation of SA signaling when infected with all viruses except TuMV which showed a downregulation in SA signaling and upregulation in JA signaling. This suggests that TuMV possesses a mechanism capable of increasing the plant’s susceptibility to infection. On infection with TuMV, CaMV, CMV, or AltMV, med25 plants were found to be susceptible to infection, while med18 plants were found to be resistant against all viruses tested. med18’s resistance could be explained by the finding that virus-infected med18 plants had JA marker genes downregulated, while SA genes were upregulated at 2 dpi, independent of which virus infected the plants. The results obtained from this study confirmed our hypothesis that med18 mutants would be resistant to viral infection based on their defective JA and increased SA signaling, therefore establishing a firm role of MED18 for normal virus infection. In contrast, med25 mutants were susceptible to all viral infections which suggest that MED25-mediated gene expression is required for plant defense signaling against viruses and further studies may reveal whether MED25 is also needed to interfere with viral mechanisms that inhibit RNA silencing.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
NH performed most experiments and contributed to manuscript writing and data interpretation. LS contributed to experimental design and data interpretation. EL performed experiments and interpreted data. JB performed experiments and interpreted data. EA performed experiments and interpreted data. SD contributed to manuscript writing. PS contributed to experimental design, data interpretation and manuscript writing.
Funding
We wish to thank the Iraqi Ministry of Higher Education and Iraqi Cultural Attaché for financial support and for kindly providing a scholarship to the first author Nasser Kadhum Hussein).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank the Iraqi government for financial support and for kindly providing a scholarship to NH. We also thank Dr. Lara-Simone Pretorius for performing preliminary experiments on TuMV during her PhD thesis and useful discussions and Drs. Andrew Geering and John Randle for kindly providing viral isolates used in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00162/full#supplementary-material
References
Aguilar, X., Blomberg, J., Brännström, C., Oloffson, A., Schleucher, J., Björklund, S. (2014). Interaction studies of the human and Arabidopsis thaliana MED25-ACID proteins with the herpes simplex virus VP16- and plant specific Dreb2a transcription factors. PLoS One 9 (5), e98575. doi: 10.1371/journal.pone.0098575
Allen, B. L., Taatjes, D. J. (2015). The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166. doi: 10.1038/nrm3951
Bäckström, S., Elfving, N., Nilsson, R., Wingsle, G., Björklund, S. (2007). Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26, 717–729. doi: 10.1016/j.molcel.2007.05.007
Cerdán, P. D., Chory, J. (2003). Regulation of flowering time by light quality. Nature 423, 881–885. doi: 10.1038/nature01636
Çevik, V., Kidd, B. N., Zhang, P., Hill, C., Kiddle, S., Denby, K. J., et al. (2012). MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 160 (1), 541–555. doi: 10.1104/pp.112.202697
Chen, R., Jiang, H., Li, L., Zhai, Q., Qi, L., Zhou, W., et al. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24, 2898–2916. doi: 10.1105/tpc.112.098277
Chiriac, C. (2013). Pattern recognition receptors and their role in plant immunity-a minireview. Extreme Life Biospeology Astrobiology 5 (2), 122–129.
Dong, S., Raffaele, S., Kamoun, S. (2015). The two-speed genomes of filamentous pathogens: waltz with plants. Curr. Opin. Genet. Dev. 35, 57–65. doi: 10.1016/j.gde.2015.09.001
Dunoyer, P., Thomas, C., Harrison, S., Revers, S., Maule, A. (2004). A cysteine-rich plant protein potentiates potyvirus movement through an interaction with the virus genome-linked protein VPg. J. Virol. 78 (5), 2301–2309. doi: 10.1128/JVI.78.5.2301-2309.2004
Edgar, C. I., McGrath, K. C., Dombrecht, B., Manners, J. M., Maclean, D. C., Schenk, P. M., et al. (2006). Salicylic acid mediates resistance to the vascular wilt pathogen Fusarium oxysporum in the model host Arabidopsis thaliana. Australas. Plant Pathol. 35 (6), 581–591. doi: 10.1071/AP06060
Elfving, N., Davoine, C., Benlloch, R., Blomberg, J., Brännström, K., Müller, D., et al. (2011). The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. U. S. A. 108, 8245–8250. doi: 10.1073/pnas.1002981108
Ellis, J., Catanzariti, A.-M., Dodds, P. (2006). The problem of how fungal and oomycete avirulence proteins enter plant cells. Trends Plant Sci. 11 (2), 61–63. doi: 10.1016/j.tplants.2005.12.008
Fallath, T., Kidd, B. N., Stiller, J., Davoine, C., Björklund, S., Manners, J. M., et al. (2017). MEDIATOR18 and MEDIATOR20 confer susceptibility to Fusarium oxysporum in Arabidopsis thaliana. PLoS One 12, e0176022. doi: 10.1371/journal.pone.0176022
Graham, G., Mayers, P., Henry, R. (1994). A simplified method for the preparation of fungal genomic DNA for PCR and RAPD analysis. BioTechniques 16, 47–50. doi: 10.1007/978-3-642-60441-6_5
Karijolich, J. J., Hampsey, M. (2012). The Mediator complex. Curr. Biol. 22, R1030–R1031. doi: 10.1016/j.cub.2012.11.011
Kidd, B. N., Edgar, C. I., Kumar, K. K., Aitken, E. A., Schenk, P. M., Manners, J. M., et al. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21, 2237–2252. doi: 10.1105/tpc.109.066910
Kidd, B. N., Cahill, D. M., Manners, J. M., Schenk, P. M., Kazan, K. (2011). Diverse roles of the Mediator complex in plants. Semin. Cell Dev. Biol. 22, 741–748. doi: 10.1016/j.semcdb.2011.07.012
Kim, Y. J., Zheng, B., Yu, Y., Won, S. Y., Mo, B., Chen, X. (2011). The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30 (5), 814–822. doi: 10.1038/emboj.2011.3
Klessig, D., Durner, J., Noad, R., Navarre, D., Wendehenne, D., Kumar, D., et al. (2000). Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. 97 (16), 8849–8855. doi: 10.1073/pnas.97.16.8849
Lai, Z., Schluttenhofer, C. M., Bhide, K., Shreve, J., Thimmapuram, J., Lee, S. Y., et al. (2014). MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 5 (1). doi: 10.1038/ncomms4064
Liao, C. J., Lai, Z., Lee, S., Yun, D. J., Mengiste, T. (2016). Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. Plant Cell. 28 (7), 1662–1681. doi: 10.1105/tpc.16.00105
Liu, H., Carvalhais, L. C., Kazan, K., Schenk, P. M. (2016). Development of marker genes for jasmonic acid signaling in shoots and roots of wheat. Plant Signaling Behav. 11 (5), e1176654. doi: 10.1080/15592324.2016.1176654
Liu, S., Ziegler, J., Zeier, J., Birkenbihl, B. P., Somssich, I. E. (2017). Botrytis cinerea B05.10 promotes disease development in Arabidopsis by suppressing WRKY33-mediated host immunity Plant Cell Environ. 40, 10, 2189–2206. doi: 10.1111/pce.13022
Love, A. J., Geri, C., Laird, J., Carr, C., Yun, B. W., Loake, G. J., et al. (2012). Cauliflower mosaic virus Protein P6 Inhibits signaling responses to salicylic acid and regulates innate immunity. PLoS One 7 (10), e47535. doi: 10.1371/journal.pone.0047535
Madhani, H. D. (2013). The frustrated gene: origins of eukaryotic gene expression. Cell 155 (4), 744–774. doi: 10.1016/j.cell.2013.10.003
Mallory, A. C., Reinhart, B. J., Bartel, D., Vance, V. B., Bowman, L. H. (2002). A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. 99 (23), 15228–15233. doi: 10.1073/pnas.232434999
Mathur, S., Vyas, S., Kapoor, S., Tyagi, A. K. (2011). The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol. 157, 1609–1627. doi: 10.1104/pp.111.188300
Moyle, R., Pretorius, L. S., Shuey, L. S., Nowak, E., Schenk, P. M. (2018). Analysis of the complete genome sequence of Cucumber mosaic virus strain K. Genome Announcements 6 (7), e00053–e00018. doi: 10.1128/genomeA.00053-18
Nürnberger, T., Brunner, F. (2002). Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5 (4), 318–324. doi: 10.1016/S1369-5266(02)00265-0
Nürnberger, T., Lipka, V. (2005). Non-host resistance in plants: new insights into an old phenomenon. Mol. Plant Pathology. 6 (3), 335–345. doi: 10.1111/j.1364-3703.2005.00279.x
Nam, J., Nam, M., Bae, H., Lee, C., Lee, B. C., Hammond, J., et al. (2013). AltMV TGB1 nucleolar localization requires homologous interaction and correlates with cell wall localization associated with cell-to-cell movement. Plant Pathol. J. 29 (4), 454–459. doi: 10.5423/PPJ.NT.04.2013.0045
Parker, J. E., Holub, E. B., Frost, L. N., Falk, A., Gunn, N. D., Daniels, M. J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell. 8, 2033–2204. doi: 10.2307/3870410
Pieterse, C. M., Leon-Reyes, A., Van der Ent, S., Van Wees, S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5 (5), 308. doi: 10.1038/nchembio.164
Pretorius, L., Moyle, R. L., Dalton-Morgan, J., Hussein, N., Schenk, P. M. (2016). Complete nucleotide sequence of an Australian isolate of Turnip mosaic virus before and after seven years of serial passaging. Genome Announcenments 4 (6), e01269–e01216. doi: 10.1128/genomeA.01269-16
Pretorius, L., Moyle, R. L., Dalton-Morgan, J., Schwinghamer, M. W., Crew, K., Schenk, P. M., et al. (2017). First fully sequenced genome of an Australian isolate of Cauliflower mosaic virus. Australas. Plant Pathol. 46 (6), 597–599. doi: 10.1007/s13313-017-0520-1
Revers, F., Nicaise, V. (2014). Plant resistance to infection by viruses. Ecol. Life Sci. doi: 10.1002/9780470015902.a0000757.pub3
Roossinck, M. (2015). Plants, viruses and the environment: ecology and mutualism. Virology, 479–480:271-277. doi: 10.1016/j.virol.2015.03.041
Samanta, S., Thakur, J. K. (2015). Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front. Plant Sci. 6, 757. doi: 10.3389/fpls.2015.00757
Thatcher, L. F., Manners, J. M., Kazan, K. (2009). Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. 58, 927–939. doi: 10.1111/j.1365-313X.2009.03831.x
Wang, M.-B., Bian, X. Y., Wu, L. M., Liu, L. X., Smith, N. A., Isenegger, D., et al. (2004). On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. U. S. A. 101 (9), 3275–3280. doi: 10.1073/pnas.0400104101
Keywords: Alternanthera mosaic virus, Arabidopsis thaliana, Cauliflower mosaic virus, Cucumber mosaic virus, mediator subunit, plant virus resistance, Turnip mosaic virus
Citation: Hussein NK, Sabr LJ, Lobo E, Booth J, Ariens E, Detchanamurthy S and Schenk PM (2020) Suppression of Arabidopsis Mediator Subunit-Encoding MED18 Confers Broad Resistance Against DNA and RNA Viruses While MED25 Is Required for Virus Defense. Front. Plant Sci. 11:162. doi: 10.3389/fpls.2020.00162
Received: 17 October 2019; Accepted: 03 February 2020;
Published: 04 March 2020.
Edited by:
Aardra Kachroo, University of Kentucky, United StatesReviewed by:
Rae-Dong Jeong, Chonnam National University, South KoreaRichard S. Nelson, Oklahoma State University, United States
Copyright © 2020 Hussein, Sabr, Lobo, Booth, Ariens, Detchanamurthy and Schenk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nasser K. Hussein, bi5odXNzZWluQHVxLmVkdS5hdQ==
 Nasser K. Hussein1,2*
Nasser K. Hussein1,2* Layla J. Sabr
Layla J. Sabr Swaminathan Detchanamurthy
Swaminathan Detchanamurthy Peer M. Schenk
Peer M. Schenk