- 1Cereal Pathology Lab, Agriculture and Agri-Food Canada, Lethbridge Research and Development Centre, Lethbridge, AB, Canada
- 2Department of Agricultural, Food and Nutritional Science, University of Alberta, Edmonton, AB, Canada
- 3The Sainsbury Laboratory, University of East Anglia, Norwich, United Kingdom
- 4Institute of Plant Science and Resources, Okayama University, Kurashiki, Japan
The fungus Pyrenophora tritici-repentis (Ptr) causes tan spot, a destructive foliar disease of wheat worldwide. The pathogen produces several necrotrophic effectors, which induce necrosis or chlorosis on susceptible wheat lines. Multiple races of Ptr have been identified, based on their ability to produce one or more of these effectors. Ptr has a wide host range of cereal and non-cereal grasses, but is known to cause damage only on wheat. Previously, we showed that Ptr can interact specifically with cultivated barley (Hordeum vulgare ssp. vulgare), and that the necrotrophic effector Ptr ToxB induces mild chlorosis in a highly selective manner when infiltrated into certain barley genotypes. In the present study, a barley doubled-haploid (DH) population was evaluated for reaction to Ptr race 5, a Ptr ToxB-producer. Then a comprehensive genetic map composed of 381 single nucleotide polymorphism (SNP) markers was used to map the locus conditioning this chlorosis. The F1 seedlings, and 92 DH lines derived from a cross between the resistant Japanese malting barley cultivar Haruna Nijo and the susceptible wild barley (H. vulgare ssp. spontaneum) OUH602 were inoculated with a conidial suspension of Ptr race 5 isolate at the two-leaf stage. The seedlings were monitored daily for symptoms and assessed for chlorosis development on the second leaf, 6 days after inoculation. All tested F1 seedlings exhibited chlorosis symptoms similar to the susceptible parent, and the DH lines segregated 1:1 for susceptible:resistant phenotypes, indicating the involvement of a single locus. Marker-trait linkage analysis based on interval mapping identified a single locus on the distal region of the short arm of chromosome 2H. We designate this locus Susceptibility to P. tritici-repentis1 (Spr1). The region encompassing this locus has 99 high confidence gene models, including membrane receptor-like kinases (RLKs), intracellular nucleotide-binding, leucine-rich repeat receptors (NLRs), and ankyrin-repeat proteins (ANKs). This shows the involvement of a dominant locus conferring susceptibility to Ptr in barley. Further work using high-resolution mapping and transgenic complementation will be required to identify the underlying gene.
Introduction
Pyrenophora tritici-repentis (Ptr), an ascomycete fungus, is a necrotrophic pathogen causing tan spot, an important foliar disease of wheat. Ptr infects its primary wheat host (Triticum aestivum L. and Triticum turgidum L.) worldwide, and has been isolated from numerous graminaceous species including rye, barley, oat, bromegrass, and several prairie grasses that may function as secondary hosts for the pathogen (Ciuffetti et al., 2014; De Wolf et al., 1998). Ptr was first isolated and characterized from the grass species Agropyron repens, almost a century before it was identified as a pathogen of wheat (De Wolf et al., 1998). Grasses were, for a long time, considered as the primary host for this fungus, then both A. repens and Triticum sp. were regarded as its main hosts, explaining why the fungus was given its hyphenated name P. tritici-repentis (De Wolf et al., 1998). Ptr has a wide host range of cereal and non-cereal grasses on which the fungus can survive (Ali and Langham, 2015). The vast majority of research on tan spot has focussed on understanding the interaction of Ptr with its primary wheat host (reviewed in Ciuffetti et al., 2014). Early research explored, albeit in a descriptive manner, the interaction between Ptr and other hosts by defining the severity of symptoms, or the ability of the fungus to reproduce, and evaluated the pathogenicity of Ptr isolates collected from grasses on wheat (reviewed in De Wolf et al., 1998).
Ptr was found to colonize barley (Hordeum vulgare ssp. vulgare) saprophytically (Summerell and Burgess, 1988), or to cause moderate to severe damage on this species (Morrall and Howard, 1975; Postnikova and Khasanov, 1997). It also was reported that Ptr produced a host-specific toxin of low molecular weight and an acidic nature that could cause moderate chlorosis on barley (Brown and Hunger, 1993); however, that toxin was not characterized further or identified in any subsequent studies. More recently, Ptr was found to interact specifically with barley, with the interaction mediated by the chlorosis-inducing necrotrophic effector Ptr ToxB (Aboukhaddour and Strelkov, 2016). While the symptoms induced by Ptr on barley were weaker than those on wheat, and a higher concentration of Ptr ToxB was needed to induce chlorosis on the barley (Aboukhaddour and Strelkov, 2016; See et al., 2019), the specificity between Ptr and barley was evident, since chlorosis developed on certain barley genotypes but not on others (Aboukhaddour and Strelkov, 2016). Furthermore, infiltration of Ptr ToxB by itself induced chlorosis on the same barley genotypes rated as susceptible to the producing fungal isolate, but not on genotypes rated as resistant. Thus, susceptibility to the pathogen and sensitivity to the effector appear to be associated (Aboukhaddour and Strelkov, 2016).
Despite the milder chlorosis that was developed on some barley genotypes, the pathogen was able to invade susceptible and resistant barley to the same extent, with no considerable difference in the cytology of infection, nor in the amount of fungal biomass detected in tissues after infection (Aboukhaddour and Strelkov, 2016). Ptr can infect barley and wheat in similar way, with few exceptions. On barley, Ptr invaded the vascular bundle without causing any wilting or yellowing of the vascular tissues, and on resistant barley, the fungus advanced in the mesophyll layer without causing any symptoms (Aboukhaddour and Strelkov, 2016). This may indicate a high adaptability of Ptr on barley and suggests that specificity and pathogenicity in Ptr are not under the same genetic control (Aboukhaddour and Strelkov, 2016). Variation in the genetic control of pathogenicity and specificity have been reported for several fungal pathogens (Freeman and Rodriguez, 1993; Ware, 2006; Stukenbrock and Mcdonald, 2008).
Ptr can induce chlorosis on 13.5% of 74 tested Canadian barley cultivars, representing over 100 years of breeding barley in Canada (Aboukhaddour and Strelkov, 2016), and a high concentration of Ptr ToxB caused symptoms on all five barley genotypes tested from Australia (See et al., 2019). Nonetheless, the genetic basis of the interaction of Ptr with barley or with other non-wheat hosts has not been investigated. These hosts may not exhibit as severe damage as wheat in response to Ptr, but they provide additional sources for pathogen inoculum and survival, and may impact pathogen genetic variability and therefore disease management. Ptr follows an inverse gene-for-gene interaction with its wheat host, meaning that specific recognition between a pathogen effector and the host leads to disease development (Lamari et al., 2003). So far, three different necrotrophic effectors have been identified in Ptr, the necrosis inducing effector, Ptr ToxA, and the two chlorosis inducing effectors Ptr ToxB and Ptr ToxC. Each effector interacts with a specific dominant sensitivity gene in the wheat host, and host sensitivity to each effector is associated with susceptibility to the producing fungal isolates [reviewed in (Faris et al., 2013)]. Here, we hypothesized that the Ptr-barley interaction is specific and likely follows a one-to-one relationship. Although this interaction is subtle, and slight changes in incubation temperature after inoculation can cause shifts in the barley reaction from susceptible to resistant (Aboukhaddour and Strelkov, 2016).
Although Canadian or Australian barley exhibits sensitivity to Ptr ToxB, this effector is absent from the pathogen population in Australia, and rarely reported in North America. In these regions, Ptr ToxA is the predominant effector (Aboukhaddour et al., 2013). Tsn1, encoding a serine/threonine protein kinase, nucleotide binding, leucine-rich repeat protein, is the sensitivity gene to PtrToxA in wheat (Faris et al., 2010; Faris et al., 2013). Ptr ToxA-Tsn1 interaction is the best characterized interaction for Ptr-wheat, and the remaining Ptr effector-wheat interactions await further characterization. Ptr ToxB-producing races of Ptr are common in the wheat centre of origin, and Ptr ToxB-producers were found mostly among isolates collected from durum wheat (Aboukhaddour et al., 2011). The aim of this study is to investigate the genetics of the Ptr-barley interaction to expand our understanding of the Ptr pathosystem in related species to wheat. quantitative trait locus (QTL) analysis for susceptibility to Ptr in barley was conducted using a doubled-haploid (DH) mapping population from a cross between a Japanese barley cultivar and wild barley.
Materials and Methods
Fungal Isolate and Inoculum Preparation
In this study, Ptr race 5 isolate Alg3-24 (Ptr-ToxB-producer) was used to inoculate barley genotypes. This is the same isolate that was used by Aboukhaddour and Strelkov (2016) to investigate the specificity of the Ptr-barley interaction. Alg3-24 was collected from durum wheat in eastern Algeria, and has been used as the standard Ptr race 5 isolate in several investigations on Ptr ToxB [reviewed in (Lamari and Strelkov, 2010)].
For inoculum preparation, a single-spore of Alg3-24 was recovered and grown on fresh V8-potato dextrose agar (V8-PDA) in a 9-cm diameter Petri plate (Lamari and Bernier, 1989b). Several mycelial plugs (0.5 cm in diameter) were then excised from the actively growing part (edge) of the colony, and transferred singly to 9-cm-diameter V8-PDA Petri plates. The fungal colonies were incubated in darkness for 5 days at room temperature, until the culture reached 4–5 cm in diameter, at which point sterile distilled water was added and the mycelium flattened with the bottom of flame-sterilized glass tube. The water was decanted and the plates were incubated under fluorescent light overnight at room temperature, following which they were transferred to the dark for 24 h at 15°C to induce sporulation. The sporulating cultures were then flooded with sterile distilled water and scraped gently with a sterilized wire loop to dislodge the conidia. The conidial suspensions were collected and the concentration of conidia was estimated with a Fuchs Rosenthal Counting Chamber (Hausser Scientific, Blue Bell, PA) and adjusted to 5,000 conidia ml−1. Two drops of Tween 20 (polyoxyethylene sorbitan monolaurate) were added per 100 ml of conidial suspension.
Plant Material and Inoculation
A DH barley population consisting of 92 lines previously derived from a cross of Haruna Nijo (H. vulgare ssp. vulgare) x OUH602 (H. vulgare ssp. spontaneum) at Okayama University, Japan (Sato and Takeda, 2009), was evaluated for its reaction to the Ptr race 5 isolate Alg3-24. F1 generated plants also were inoculated. The first parent, Haruna Nijo is a two-row malting cultivar grown in Japan, and was rated resistant to the Ptr isolate Alg3-24. The second parent, OUH602 is a wild barley (H. vulgare ssp. spontaneum) genotype, and was rated as susceptible to this isolate. The hexaploid wheat genotype 6B662 (sensitive to Ptr ToxB and susceptible to Ptr race 5), and the two barley lines, Rivers and Norbert, both of which are six-row barley, were included as controls. Rivers and Norbert were rated as susceptible and resistant to Ptr isolate Alg3-24, respectively (Aboukhaddour and Strelkov, 2016). These two genotypes also were evaluated for their reaction to infiltration with the purified Ptr ToxB, and Rivers was rated sensitive, while Norbert was insensitive (Aboukhaddour and Strelkov, 2016).
All plant genotypes were planted in 10 cm-diameter plastic pots filled with Sunshine Potting Mix (W.R. Grace and Co., Fogelsville, PA) at a rate of eight seeds per pot. Each genotype was seeded in two independent pots, and the bioassay was replicated three times independently. The seedlings were maintained in growth cabinets at 20/18°C (day/night) with a 16 h photoperiod (180 mmol m−2 s−1) until inoculation at the 2–3 leaf stage. Briefly, the seedlings were inoculated with the conidial suspension (5,000 conidia ml−1), prepared as described above, until runoff using a sprayer connected to an airline (Lamari and Bernier, 1989a). Immediately following inoculation, the seedlings were transferred to a humidity chamber (>95% relative humidity) for 24 h. The plants were then transferred to growth cabinets with a 16 h photoperiod (180 mmol m−2 s−1) at 20/18°C (day/night) and 60% relative humidity. The seedlings were monitored daily for symptom development and were rated for symptom development at 6 days post-inoculation (dpi).
Phenotypic Analysis
The 92 lines of the DH population, the parental genotypes Haruna Nijo and OUH602, and the Canadian control cultivars were screened in three experiments for symptom development at 6 dpi with Ptr race 5. Symptoms were rated on scale of 1 to 5 following Lamari and Bernier (1989b). In brief, infected plants were rated as follows: reactions 1 and 2 are resistant, and reaction 3 to 5 are susceptible. Reaction 1 (small dark spots without any surrounding chlorosis or necrosis); reaction 2 (small dark spots with a very small chlorotic halo at the site of infection); reaction 3 (small dark spots completely surrounded by a distinct chlorosis, with lesions not coalescing together); reaction 4 (small dark spots completely surrounded by a chlorotic zone with the lesions coalescing); and reaction 5 (small dark spots surrounded by a chlorotic zone, with almost all of the infected leaf chlorotic).
The phenotypic (disease severity) data were subjected to a χ2 test and ANOVA using the agricolae (v. 1.2–4) package of R (R v. 3.2.3) (R Core Team, 2018). For the ANOVA model, DH lines, parents, and controls were considered as fixed effects, while experiments were considered as random effects. The ANOVA was conducted across experiments. The least significance difference (LSD) and the coefficient of variation % (CV%) were calculated with agricolae.
Quantitative Trait Locus Analysis
A genetic map was previously constructed using an oligo-nucleotide pooled assay (OPA) for high-throughput single nucleotide polymorphism (SNP) genotyping, and 381 SNP markers were selected that were distributed across all seven barley chromosomes (Sato and Takeda, 2009; Muñoz-Amatriaín et al., 2011). Interval mapping was performed using scanone (R/qtl) with the expectation–maximization (EM) method and a 2.0 cM step size. Experimental-wide threshold was determined using 1,000 permutations and controlled at α = 0.05.
Comparison of Tsc2 and Spr1 Loci
Tsc2 is the dominant locus conditioning sensitivity to Ptr ToxB in wheat, and is located on the short arm of 2B chromosome (Friesen and Faris, 2004). Chromosome 2B was retrieved from URGI (Appels et al., 2018) and a 5.8 Mb interval representing the Tsc2 locus between markers XBE517745 and Xmag681 (Abeysekara et al., 2010) was extracted with Bedtools (Quinlan and Hall, 2010). Similarly, the barley genome was obtained from GeneBank (Mascher et al., 2017) and a 3.2 Mb interval representing the Spr1 locus on barley 2H chromosome between genes HORVU2Hr1G004230 and HORVU2Hr1G006010 was extracted with Bedtools. The two extracted loci were aligned and visualized by large-scale genome alignment tool progressiveMauve (v. 20150226 build 10) with the default settings (Darling et al., 2010). The predicted genes within the Spr1 locus were then compared in sequence identity to the Tsc2 locus. Protein and coding sequences for wheat were retrieved from the JGI Genome Portal (IWGSC, 2014). BLASTP and BLASTN searches (e−10) were performed using predicted gene sequences from high confidence gene models in barley (Mascher et al., 2017) and wheat (IWGSC, 2014; Appels et al., 2018). Orthologs in the wheat chromosome 2B Tsc2 region were identified when percent identity was greater than 50% over a region covering 50% of the BLASTP query length.
Results
Phenotypic Analysis
In all three phenotyping experiments, Haruna Nijo scored as highly resistant, and OUH602 was rated as susceptible to Ptr isolate Alg3-24 (Figure 1). The disease severity of the DH lines ranged from 1 to 4, with a mean of 2.144 (Table 1). Among the parents and the controls, the disease severity of the resistant parent Haruna Nijo and the resistant control Norbert were scored as 1, while the mean disease severity of the susceptible parent OUH602 and the susceptible control barley cultivars ranged from 3 to 4 (Table 1). In the first run of the experiment, 48 (52%) of the DH lines were rated as resistant and 44 (48%) were rated as susceptible. In the second experiment, 43 (47%) lines were rated as resistant and 49 (53%) were rated as susceptible, while in the third experiment, 39 (42%) lines were rated resistant and 53 (58%) were rated susceptible (Figure 2). In all experiments, the segregating ratio susceptible:resistant was not significantly different from the expected 1:1 ratio at the 0.05 level of probability (χ² = 0.93) (Table 2). F1 plants of Haruna Nijo x OUH602 exhibited a chlorotic reaction similar to the susceptible parent OUH602, indicating that susceptibility to Ptr isolate Alg3-24 in this cross is dominant.
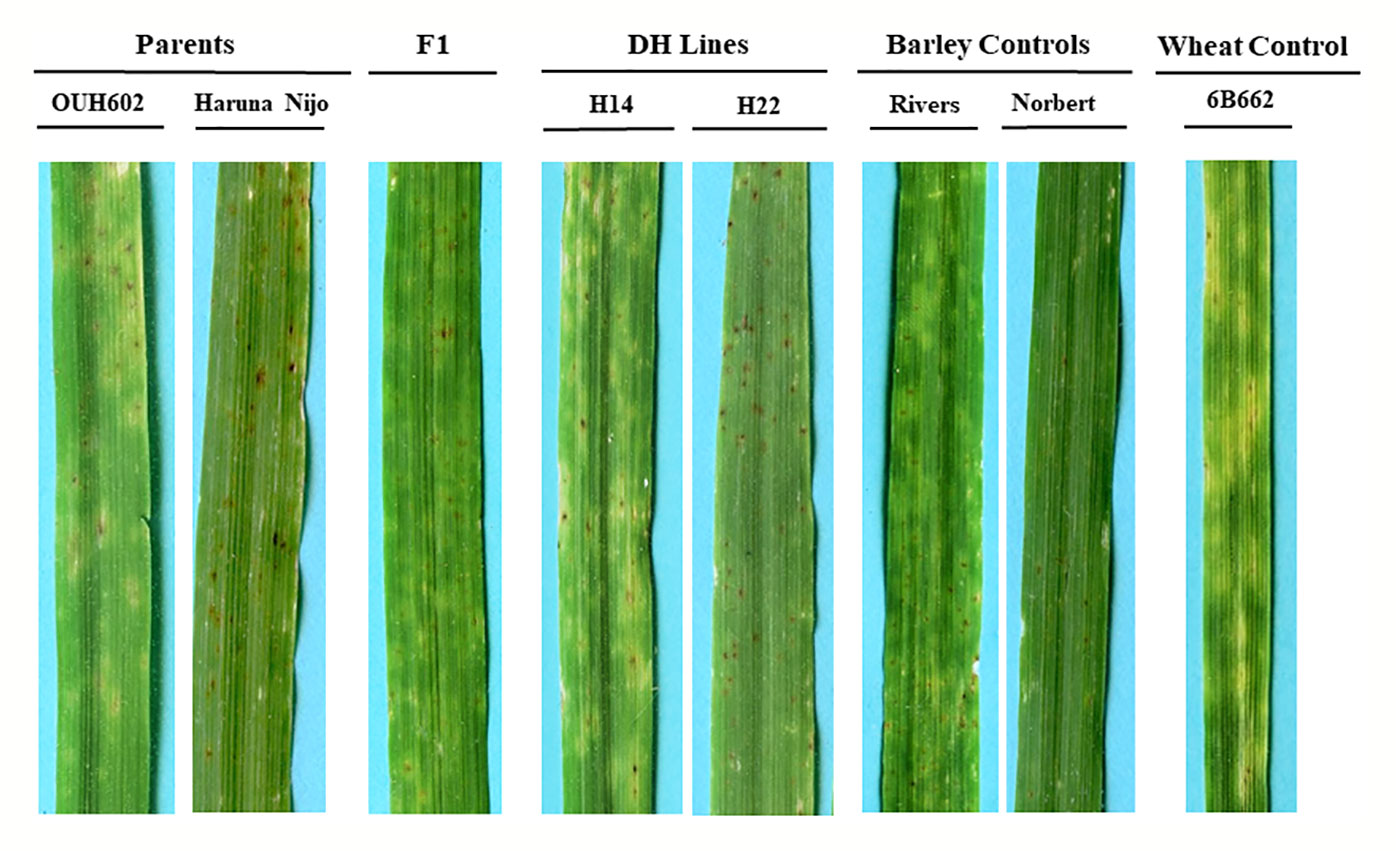
Figure 1 Reaction of barley to Ptr ToxB-producing race 5 isolate, Alg3-24. The two parents OUH602 and Haruna Nijo representing the susceptible and resistant reaction to race 5, respectively. The F1 plants exhibited the susceptible reaction. Two doubled haploid lines H24 and H22 representing a susceptible and resistant reaction, respectively. The barley controls, Rivers and Norbert were included as additional controls for susceptible and resistant reaction, respectively. The hexaploid wheat genotype 6B662 was also included as a susceptible wheat control.
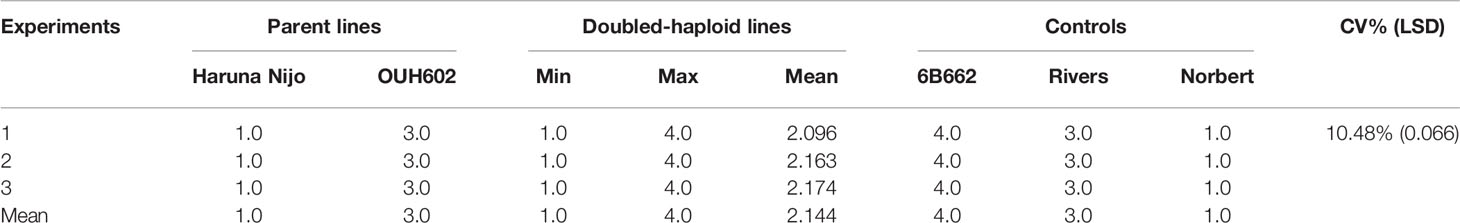
Table 1 Details of average and range of disease severity on 92 doubled-haploid lines, their parents, Haruna Nijo (Hordeum vulgare ssp. vulgare) and OUH602 (H. vulgare ssp. spontaneum), and control cultivars screened in three experiments after inoculation with Pyrenophora tritici-repentis race 5 isolate Alg3-24.
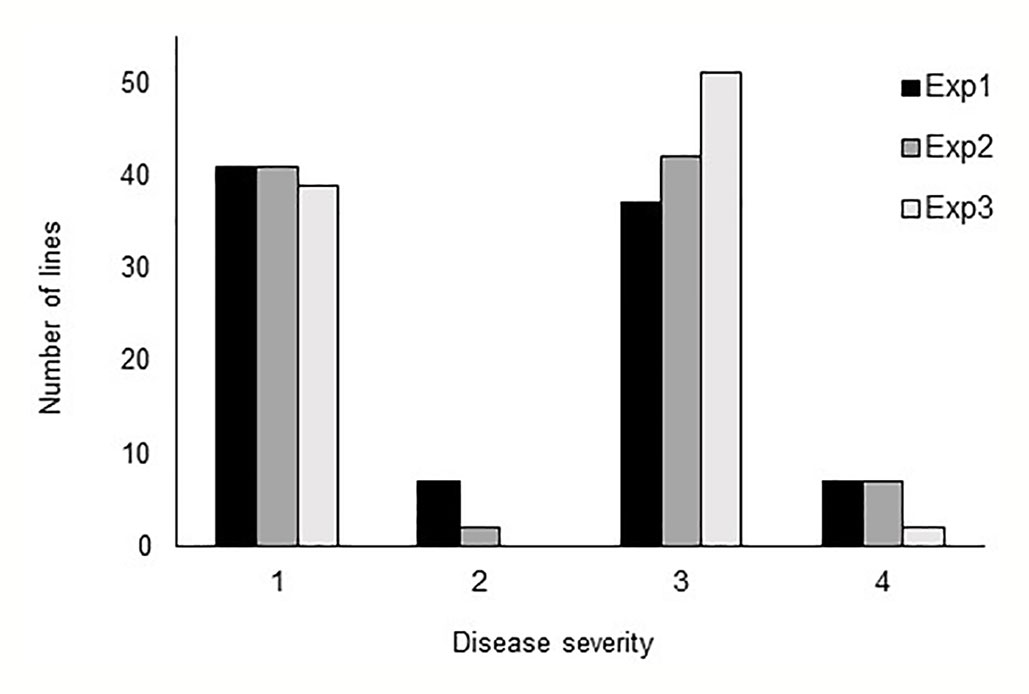
Figure 2 Barplot of the tan spot severity on barley DH lines. Disease severity was rated from 1 to 5, while 1 to 2 represent the resistance and 3 to 5 represent the susceptible reaction. The DH lines segregated in score 1 and score 3 into nearly 1:1 ratio.
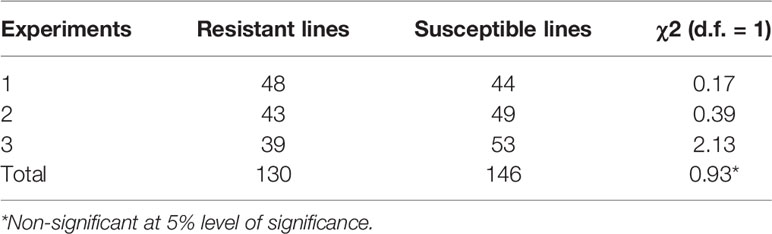
Table 2 Chi square table of doubled-haploid segregation in three experiments from a cross of Haruna Nijo and OUH602.
The LSD (p = 0.066), which was lower than the differences between parents, and the CV% (10.48%) (Table 2) showed that a large genetic effect contributed to disease resistance, and that the data were suitable for further analysis. The ANOVA (Table 3) indicated a highly significant genotype effect, as well as significant effect of experiments and experiments x genotype interactions, on tan spot disease severity.
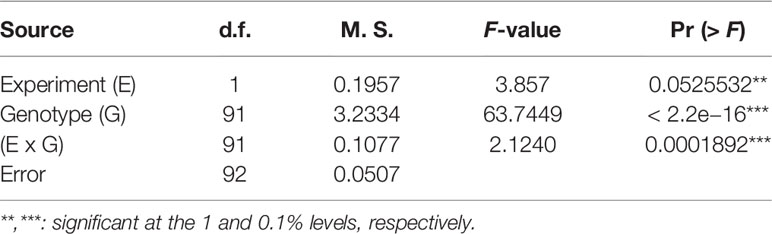
Table 3 ANOVA of doubled-haploid barley lines from a cross of Haruna Nijo and OUH602 under the experiment effect, genotype effect and their interactions.
Quantitative Trait Locus Analysis
Marker-trait linkage analysis based on interval mapping identified a single locus on the distal region of the short arm of chromosome 2H (Figures 3A, B). The QTL was flanked by SNP markers 1-1059 and 2-0562 in the genes HORVU2Hr1G004230 and HORVU2Hr1G006010, respectively (Figure 3B). LOD scores for this single QTL were 47.6 (experiment 1), 51.7 (experiment 2), and 92.8 (experiment 3) (Figure 3A). Susceptibility is the dominant trait, therefore we designate this locus Susceptibility to P. tritici-repentis1 (Spr1). On the barley physical map (Mascher et al., 2017), the interval ranges from 9.64 to 12.86 Mbp. The region encompassing the locus has 99 high confidence gene models, including membrane receptor-like kinases (RLKs), intracellular nucleotide-binding, leucine-rich repeat receptors (NLRs), and ankyrin-repeat proteins (Supplementary Table 1).
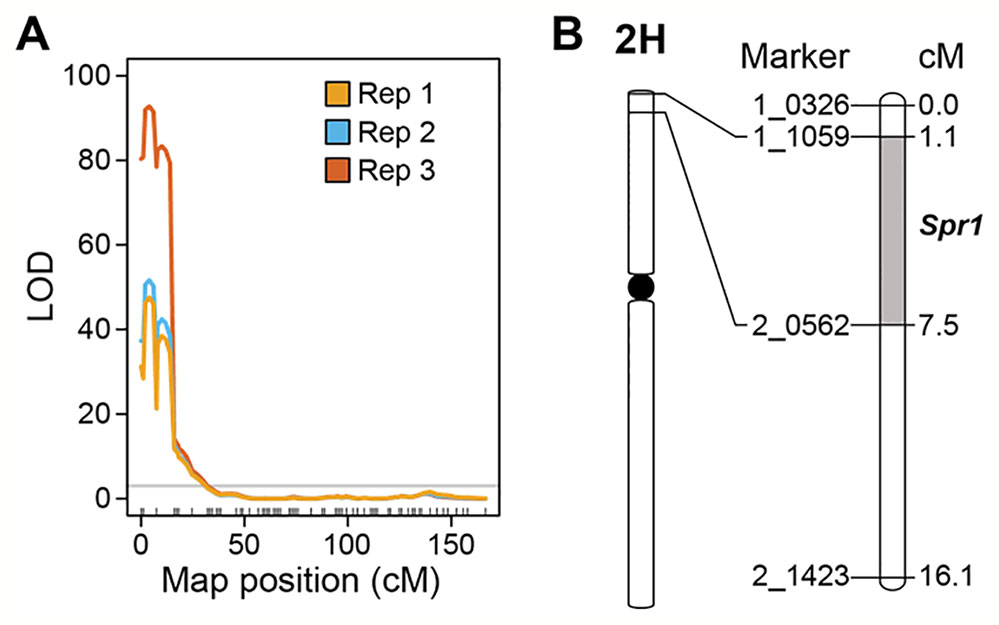
Figure 3 Mapping of Spr1. (A) Interval mapping based on replicated experiments. Permutation threshold of α = 0.05 is shown in grey. (B) Genetic interval encompassing the Spr1 locus on chromosome 2H of barley.
Comparison of Tsc2 and Spr1
The Spr1 region contained 99 high-confidence candidate gene models (Supplementary Table 1). Multiple genome and protein alignments of the genes in Spr1 and those in Tsc2 showed that 73 of those genes are present as homologs in the wheat Tsc2 locus based on BLASTP results (highlighted in the Supplementary Tables 1 and 2). An additional seven genes were identified as potential homologs based on BLASTN results (Supplementary Tables 1 and 3), these additional seven may be present only as non-coding sequences. Based on the 50-50 rule, 43 of the 73 homologous genes are present as orthologs in the Tsc2 locus. Many of the predicted protein coding genes are involved in biotic and abiotic stress tolerance.
Discussion
Genetic control of the Ptr-wheat interaction has been investigated for the last 50 years (reviewed in Faris et al., 2013). There is, however, no information on the genetics of the Ptr interaction with other hosts. Ptr is known to cause damage to wheat, but on other host species it is either non-pathogenic or causes moderate to severe symptoms (Morrall and Howard, 1975; Krupinsky, 1982; Postnikova and Khasanov, 1997; Ali and Francl, 2003). In one study, a number of Ptr isolates collected from 18 different grass species and cultivated barley were as aggressive on wheat cultivars as isolates recovered from wheat in the Northern Great Plains, and all the barley isolates tested were pathogenic on wheat (Krupinsky, 1992). Recently, an evidence of a specific interaction between Ptr and cultivated barley has been reported, and Ptr ToxB can act as a necrotrophic effector in barley as in wheat, albeit a higher concentration of this effector is needed to induce the chlorosis symptoms on barley (Aboukhaddour and Strelkov, 2016; See et al., 2019).
In this study, susceptibility to Ptr in barley was mapped to a single locus. The DH lines segregated in a 1:1 susceptible:resistant ratio following inoculation with Ptr race 5, and mapped to the short arm of chromosome 2H in barley. Moreover, F1 plants exhibited a chlorotic reaction similar to the susceptible parent OUH602, indicating that susceptibility to Ptr in this cross is dominant. Although the susceptible parent in this study was a wild barley, however, susceptibility in cultivated barley is also dominant. The F1 plants generated from a reciprocal crosses between two cultivated barley genotypes Rivers and Norbert, a susceptible and resistant lines, respectively, were susceptible to Ptr race 5 tested here (data not shown). This confirms that an inverse gene-for-gene model (Ellingboe, 1976), which mimics the Ptr-wheat interaction, is involved in the Ptr-barley interaction. This is the first genetic study on the interaction of Ptr with a secondary host, which will contribute to a greater understanding of the evolution of the Ptr pathosystem.
Ptr is not recognized as a barley pathogen and, indeed, in this study like in previous ones, we noted that the chlorosis on barley and wild barley was moderate and less intense than on susceptible wheat. Moreover, there was variation in the severity of the chlorosis that developed on the DH lines tested in this study (1 to 4 on a scale of 1 to 5), and on various barley genotypes in previous studies (Aboukhaddour and Strelkov, 2016; See et al., 2019). That may explain the wide range of symptoms described on barley in earlier studies by various groups (Morrall and Howard, 1975; Krupinsky, 1982; Summerell and Burgess, 1988; Brown and Hunger, 1993; Postnikova and Khasanov, 1997). This also suggests the presence of additional effectors produced by Ptr, which may contribute to the variation in symptom development on barley genotypes. Moreover, it was noted that the temperature after inoculation had a significant effect on symptom development, with declines or increases in the incubation temperature resulting in shifts in the host interaction from susceptible to resistant (Lamari and Bernier, 1994; Aboukhaddour and Strelkov, 2016). Therefore, establishing a consistent temperature for phenotype evaluation is critical.
The chromosomal region where the single locus was identified in this study encompasses 99 high confidence gene models, including genes from gene families known to be involved in plant immunity such as membrane receptor-like kinases (RLKs), intracellular nucleotide-binding, leucine-rich repeat receptors (NLRs), and ankyrin-repeat proteins (ANKs) (Supplementary Table 1). Multiple genome and protein alignments of the genes in Spr1 and those in Tsc2 showed the presence of 43 orthologous genes, and many of these genes have predicted function in abiotic and biotic stress tolerance. However, the exact identity and function of the gene mediating Ptr-barley interaction is unknown and cannot be predicted based on this information.
It is hypothesized that necrotrophic pathogens can utilize host resistance mechanisms for biotrophic fungi to their benefit, for example by proliferating in dead tissue resulting from the hypersensitive reaction and triggered by a host resistance gene (Shi et al., 2016). In wheat, Tsn1 confers sensitivity to Ptr and susceptibility to Ptr ToxA-producing isolates. Tsn1 is structurally related to plant disease resistance genes and includes serine/threonine protein kinase (S/TPK) and nucleotide-binding-leucine-rich repeat (NLR) domains (Faris et al., 2010). Interestingly, barley Rpg5 stem rust resistance gene encodes a NB-LRR-S/TPK (Brueggeman et al., 2008), although these two genes encode two unrelated proteins (Faris et al., 2010). The interaction between a necrotrophic effector and a dominant sensitivity gene that is structurally similar to a typical biotrophic pathogen resistance gene is not unique and has been reported in other pathosystems (Shi et al., 2016).
Ptr is considered as new pathogen of wheat (Friesen et al., 2006), and it was suggested to have evolved on wild grasses prior to a host jump onto wheat (Strelkov and Lamari, 2003). On grasses, the race structure of Ptr is different from that on wheat. For example, while the non-pathogenic Ptr race 4 appears to be predominant on grasses, it is almost absent on wheat (Ali and Francl, 2003). Nevertheless, Ptr race 4 does carry the toxb gene, which is a homolog of ToxB, the Ptr ToxB-coding gene (Strelkov et al., 2006). The sequences of ToxB and its homolog in Ptr race 4 exhibit 86% similarity over the length of the open-reading frame (ORF) (Martinez et al., 2004; Strelkov et al., 2006). ToxB-like sequences are also found in race 3 isolates of Ptr, other species of the genus Pyrenophora, and even other genera of the Pleosporacea (Martinez et al., 2004; Strelkov et al., 2006; Andrie et al., 2007). Isolates of Pyrenophora bromi, a sister species to Ptr causing brown spot of brome grass, has several ToxB-like sequences (termed Pb ToxB) with coding regions having 89% similarity to ToxB (Andrie et al., 2008; Andrie and Ciuffetti, 2011). However, none of the heterologously expressed Pb ToxB proteins induced symptoms on brome grass, while they did cause chlorosis on ToxB-sensitive wheat genotypes (Andrie and Ciuffetti, 2011).
Several leaf spot causing pathogens of cereals or grasses share the same necrotrophic effectors or homologous coding gene sequences. Similar to ToxB, a homolog of the ToxA gene, which encodes Ptr ToxA, is found in Bipolaris sorokiniana, a pathogen infecting both wheat and barley (Mcdonald et al., 2018). Another ToxA homolog is also present in the maize pathogen Cochliobolus heterostrophus (Lu et al., 2015), and an identical ToxA sequence is present in the wheat pathogen, Parastagonospora nodorum (Friesen et al., 2006). Parallel to the presence of one effector or its homologs in various necrotrophic pathogens, related or unrelated plant genes conditioning sensitivity to one effector can exist in various host species, and these genes may condition multiple interactions with various plant pathogens (Lorang et al., 2007). The LOV1 gene in Arabidopsis confers sensitivity to victorin, which is a secondary metabolite effector produced by the pathogen Bipolaris victoriae that devastated oat in the 1940s. LOV1 belongs to the NLR class of resistance genes (Lorang et al., 2007). Similarly, the Pc gene in sorghum, which confers susceptibility to Periconia circinata and its Pc-effector, encodes an NLR (Nagy and Bennetzen, 2008).
The presence of ToxB-like sequences and non-functional homologs of Ptr ToxB in several species within two fungal orders (Dothideomycetes and Sordariomycetes) (Ciuffetti et al., 2014) remains unexplained. Why do these species code for what appear to be non-functional proteins? Ptr ToxB, like the other Ptr-necrotrophic effectors, does not appear to control any essential biological function in the fungus (Strelkov and Lamari, 2003). On wheat, Ptr ToxB interacts with a dominant sensitivity gene Tsc2 on the short arm of the wheat chromosome 2B (Friesen and Faris, 2004). The exact mode of action to Ptr ToxB is not yet known, but treatment with this effector does cause chlorophyll photooxidation and an inhibition of photosynthesis (Lamari and Strelkov, 2010); this ultimately results in the development of chlorosis in wheat, similar to the symptoms observed here on cultivated and wild barley. On wheat, Ptr ToxB plays a considerable role in disease development, contributes to quantitative variation in the virulence of Ptr, and may influence development of fungal appressoria (Amaike et al., 2008; Aboukhaddour et al., 2012). Perhaps there are additional roles for Ptr ToxB and its various homologs that explain their presence in a wide range of fungal species, and which may also help to explain the interaction of Ptr with its secondary hosts.
Data Availability Statement
All datasets generated and analyzed for this study are included and cited in the article/Supplementary Material.
Author Contributions
BW performed most of the work in this manuscript and drafted the first version. KS developed the DH population and F1 plants, MM developed SNP markers and performed QTL mapping, RG performed sequence analysis of Tsc2 and Spr1 loci. SS, MM, and RA conceived the experiment and RA closely supervised the work. All authors reviewed, edited and contributed to this manuscript.
Funding
Funding from University of Alberta, Alberta Wheat Commission and Saskatchewan Wheat Development Commission to BW, Agriculture and Agri-Food Canada and Alberta Wheat Commission and Saskatchewan Wheat Development Commission to RA, and Biotechnology and Biological Sciences Research Council (BB/P012574/1) and Gatsby Foundation to MM. The funding bodies were not involved in the design of the experiments and collection, analysis and interpretation of data, nor in the writing of this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mrs. Therese Despins for her excellent technical assistance in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00158/full#supplementary-material
References
Abeysekara, N. S., Friesen, T. L., Liu, Z., Mcclean, P. E., Faris, J. D. (2010). Marker development and saturation mapping of the tan spot ptr toxb sensitivity locus Tsc2 in hexaploid wheat. Plant Genome J 3, 179–189. doi: 10.3835/plantgenome2010.07.0017
Aboukhaddour, R., Strelkov, S. E. (2016). Exploringde novospecificity: the Pyrenophora tritici-repentis-barley interaction. Plant Pathol. 65, 1347–1357. doi: 10.1111/ppa.12500
Aboukhaddour, R., Cloutier, S., Lamari, L., Strelkov, S. E. (2011). Simple sequence repeats and diversity of globally distributed populations of Pyrenophora tritici-repentis. Can. J. Plant Pathol. 33, 389–399. doi: 10.1080/07060661.2011.590821
Aboukhaddour, R., Kim, Y. M., Strelkov, S. E. (2012). RNA-mediated gene silencing of ToxB in Pyrenophora tritici-repentis. Mol. Plant Pathol. 13, 318–326. doi: 10.1111/J.1364-3703.2011.00748.x
Aboukhaddour, R., Turkington, T. K., Strelkov, S. E. (2013). Race structure of Pyrenophora tritici-repentis (tan spot of wheat) in Alberta, Canada. Can. J. Plant Pathol. 35, 256–268. doi: 10.1080/07060661.2013.782470
Ali, S., Francl, L. (2003). Population race structure of Pyrenophora tritici-repentis prevalent on wheat and noncereal grasses in the great plains. Plant Dis. 87, 418–422. doi: 10.1094/pdis.2003.87.4.418
Ali, S., Langham, M. A. (2015). Reaction of five non-cereal grasses to five races and two host selective toxins of Pyrenophora tritici-repentis. Plant Pathol. J. 31, 245–251. doi: 10.5423/PPJ.OA.03.2015.0028
Amaike, S., Ozga, J. A., Basu, U., Strelkov, S. E. (2008). Quantification of ToxB gene expression and formation of appressoria by isolates of Pyrenophora tritici-repentis differing in pathogenicity. Plant Pathol. 57, 623–633. doi: 10.1111/j.1365-3059.2007.01821.x
Andrie, R. M., Ciuffetti, L. M. (2011). Pyrenophora bromi, causal agent of brownspot of bromegrass, expresses a gene encoding a protein with homology and similar activity to Ptr ToxB, a host-selective toxin of wheat. Mol. Plant-Microbe Interact. 24, 359–367. doi: 10.1094/MPMI-06-10-0142
Andrie, R. M., Iovanna, P., Ciuffetti, L. M. (2007). A combination of phenotypic and genotypic characterization strengthens Pyrenophora tritici-repentis race identification. Phytopathology 97, 694–701. doi: 10.1094/PHYTO-97-6-0694
Andrie, R. M., Schoch, C. L., Hedges, R., Spatafora, J. W., Ciuffetti, L. M. (2008). Homologs of ToxB, a host-selective toxin gene from Pyrenophora tritici-repentis, are present in the genome of sister-species Pyrenophora bromi and other members of the Ascomycota. Fungal Genet. Biol. 45, 363–377. doi: 10.1016/j.fgb.2007.10.014
Appels, R., Eversole, K., Stein, N., Feuillet, C., Keller, B., Rogers, J., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Brown, D. A., Hunger, R. M. (1993). Production of a Chlorosis-inducing, host-specific, low-molecular weight toxin by isolates of Pyrenophora tritici-repentis. Cause of tan spot of wheat. Phytopathology 137, 221–232. doi: 10.1111/j.1439-0434.1993.tb01342.x
Brueggeman, R., Rostoks, N., Kudrna, D., Kilian, A., Han, F., Chen, J., et al. (2008). The barley stem rust-resistance gene Rpg1 is a novel. PNAS 99, 9328–9333. doi: 10.1073/pnas.142284999
Ciuffetti, L. M., Manning, V. A., Pandelova, I., Faris, J. D., Friesen, T. L., Strelkov, S. E., et al. (2014). “Pyrenophora tritici-repentis: A Plant Pathogenic Fungus with Global Impact,” in Genomics of Plant-Associated Fungi: Monocot Pathogens. Eds. Dean, R. A., Lichens-Park, A., Kole, C. (Berlin, Heidelberg: Springer Berlin Heidelberg), 1–39. doi: 10.1007/978-3-662-44053-7_1
Darling, A. E., Mau, B., Perna, N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS One 5, e11147. doi: 10.1371/journal.pone.0011147
De Wolf, E. D., Effertz, R. J., Ali, S., Francl, L. J. (1998). Vistas of tan spot research. Can. J. Plant Pathol. 20, 349–370. doi: 10.1080/07060669809500404
Ellingboe, A. H. (1976). “Genetics of Host-Parasite Interactions,” in Physiological Plant Pathology. Eds. Heitefuss, R., Williams, P. H. (Berlin, Heidelberg: Springer Berlin Heidelberg), 761–778. doi: 10.1007/978-3-642-66279-9_29
Faris, J. D., Zhang, Z., Lu, H., Lu, S., Reddy, L., Cloutier, S., et al. (2010). A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. U.S.A. 107, 13544–13549. doi: 10.1073/pnas.1004090107/-/DCSupplemental
Faris, J. D., Liu, Z., Xu, S. S. (2013). Genetics of tan spot resistance in wheat. Theor. Appl. Genet. 126, 2197–2217. doi: 10.1007/s00122-013-2157-y
Freeman, S., Rodriguez, R. J. (1993). Genetic conversion of a fungal plant pathogen to a nonpathogenic, endophytic mutualist. Science 260, 75. doi: 10.1126/science.260.5104.75
Friesen, T. L., Faris, J. D. (2004). Molecular mapping of resistance to Pyrenophora tritici-repentis race 5 and sensitivity to Ptr ToxB in wheat. Theor. Appl. Genet. 109, 464–471. doi: 10.1007/s00122-004-1678-9
Friesen, T. L., Stukenbrock, E. H., Liu, Z., Meinhardt, S., Ling, H., Faris, J. D., et al. (2006). Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956. doi: 10.1038/ng1839
International Wheat Genome Sequencing Consortium (IWGSC) (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345 (6194), 1251788. doi: 10.1126/science.1251788
Krupinsky, J. M. (1982). Observations on the host range of isolates of Pyrenophora trichostoma. Can. J. Plant Pathol. 4, 42–46. doi: 10.1080/07060668209501335
Krupinsky, J. M. (1992). Aggressiveness of Pyrenophora tritici-repentis isolated from grass and barley hosts. Plant Dis. 76, 783–789. doi: 10.1094/PD-76-0783
Lamari, L., Bernier, C. C. (1989a). Toxin of Pyrenophora tritici-repentis: host-specifity, significance in disease, and inhertance of host reaction. Phytopathology 79, 740–744. doi: 10.1080/07060668909501146
Lamari, L., Bernier, C. C. (1989b). Virulence of isolates of Pyrenophora tritici-repentis on 11 wheat cultivars and cytology of the differential host reactions. Can. J. Plant Pathol. 11, 284–290. doi: 10.1080/07060668909501114
Lamari, L., Bernier, C. C. (1994). Temperature-induced resistance to tan spot [Pyrenophora tritici-repentis] of wheat. Can. J. Plant Pathol. 16, 279–286. doi: 10.1080/07060669409500732
Lamari, L., Strelkov, S. E. (2010). Minireview/Minisynthèse the wheat/Pyrenophora tritici-repentis interaction: progress towards an understanding of tan spot disease†. Can. J. Plant Pathol. 32, 4–10. doi: 10.1080/07060661003594117
Lamari, L., Strelkov, S. E., Yahyaoui, L., Orabi, J., Smith, R. B. (2003). The identification of two new races of Pyrenophora tritici-repentis from the host center of diversity confirms a one-to-one relationship in tan spot of wheat. Phytopathology 93, 391–396. doi: 10.1094/phyto.2003.93.4.391
Lorang, J. M., Sweat, T. A., Wolpert, T. J. (2007). Plant disease susceptibility conferred by a “resistance” gene. PNAS 104, 14861–14866. doi: 10.1073/pnas.0702572104
Lu, S., Gillian, T. B., Edwards, M. C. (2015). A ToxA-like protein from Cochliobolus heterostrophus induces light-dependent leaf necrosis and acts as a virulence factor with host selectivity on maize. Fungal Genet. Biol. 81, 12–24. doi: 10.1016/j.fgb.2015.05.013
Martinez, J. P., Oesch, N. W., Ciuffetti, L. M. (2004). Characterization of the multiple copy host-selective toxin gene, ToxB, in pathogenic and nonpathogenic isolates of Pyrenophora tritici-repentis. MPMI 17, 467–474. doi: 10.1094/MPMI.2004.17.5.467
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S. O., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. doi: 10.1038/nature22043
Mcdonald, M. C., Ahren, D., Simpfendorfer, S., Milgate, A., Solomon, P. S. (2018). The discovery of the virulence gene ToxA in the wheat and barley pathogen Bipolaris sorokiniana. Mol. Plant Pathol. 19, 432–439. doi: 10.1111/mpp.12535
Morrall, R. A., Howard, R. J. (1975). The epidemiology of leaf spot disease in a native prairie. II. Airborne spore populations of Pyrenophora tritici-repentis. Can. J. Bontany 53, 2345–2353. doi: 10.1139/b75-260
Muñoz-Amatriaín, M., Moscou, M. J., Bhat, P. R., Svensson, J. T., Bartoš, J., Suchánková, P., et al. (2011). An improved consensus linkage map of barley based on flow-sorted chromosomes and single nucleotide polymorphism markers. Plant Genome J. 4, 238–249. doi: 10.3835/plantgenome2011.08.0023
Nagy, E. D., Bennetzen, J. L. (2008). Pathogen corruption and site-directed recombination at a plant disease resistance gene cluster. Genome Res. 18, 1918–1923. doi: 10.1101/gr.078766.108
Postnikova, E. N., Khasanov, B. A. (1997). “Tan Spot in Central Asia,” in Helminthosporium Blights of Wheat: Spot Blotch and Tan Spot (CIMMYT/UCL/BADC), 107–113.
Quinlan, A. R., Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. 10.1093/bioinformatics/btq033
R Core Team (2018). R: A Language and Environment for Statistical Computing (Vienna: R Foundation for Statistical Computing).
Sato, K., Takeda, K. (2009). An application of high-throughput SNP genotyping for barley genome mapping and characterization of recombinant chromosome substitution lines. Theor. Appl. Genet. 119, 613–619. doi: 10.1007/s00122-009-1071-9
See, P. T., Iagallo, E. M., Oliver, R. P., Moffat, C. S. (2019). Heterologous expression of the Pyrenophora tritici-repentis effector proteins ToxA and ToxB, and the prevalence of effector sensitivity in Australian cereal crops. Front. Microbiol. 10, 182. doi: 10.3389/fmicb.2019.00182
Shi, G., Zhang, Z., Friesen, T. L., Raats, D., Fahima, T., Brueggeman, R., et al. (2016). The hijacking of a receptor kinase–driven pathway by a wheat fungal pathogen leads to disease. Sci. Adv. 2, e1600822. doi: 10.1126/sciadv.1600822.
Strelkov, S. E., Lamari, L. (2003). Host-parasite interactions in tan spot [Pyrenophora tritici-repentis] of wheat. Can. J. Plant Pathol. 25, 339–349. doi: 10.1080/07060660309507089
Strelkov, S. E., Kowatsch, R. F., Ballance, G. M., Lamari, L. (2006). Characterization of the ToxB gene from North African and Canadian isolates of Pyrenophora tritici-repentis. Physiol. Mol. Plant Pathol. 67, 164–170. doi: 10.1016/j.pmpp.2005.12.004
Stukenbrock, E. H., Mcdonald, B. A. (2008). The origins of plant pathogens in agro-ecosystems. Annu. Rev. Phytopathol. 46, 75–100. doi: 10.1146/annurev.phyto.010708.154114
Summerell, B. A., Burgess, L. W. (1988). Saprophytic colonization of wheat and barley by Pyrenophora tritici-repentis in the field. Trans. Br. Mycol. Soc. 9, 551–556. doi: 10.1016/S0007-1536(88)80058-5
Keywords: tan spot, barley, race 5, quantitative trait locus mapping, Ptr ToxB, chlorosis
Citation: Wei B, Moscou MJ, Sato K, Gourlie R, Strelkov S and Aboukhaddour R (2020) Identification of a Locus Conferring Dominant Susceptibility to Pyrenophora tritici-repentis in Barley. Front. Plant Sci. 11:158. doi: 10.3389/fpls.2020.00158
Received: 12 November 2019; Accepted: 31 January 2020;
Published: 28 February 2020.
Edited by:
Morten Lillemo, Norwegian University of Life Sciences, NorwayReviewed by:
Justin Faris, Edward T. Schafer Agricultural Research Center (USDA-ARS), United StatesMarja Johaana Jalli, Natural Resources Institute Finland (Luke), Finland
Copyright © 2020 Wei, Moscou, Sato, Gourlie, Strelkov and Aboukhaddour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reem Aboukhaddour, cmVlbS5hYm91a2hhZGRvdXJAY2FuYWRhLmNh
 Bohan Wei
Bohan Wei Matthew J. Moscou
Matthew J. Moscou Kazuhiro Sato
Kazuhiro Sato Ryan Gourlie1
Ryan Gourlie1 Reem Aboukhaddour
Reem Aboukhaddour