
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 20 February 2020
Sec. Plant Abiotic Stress
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00115
A correction has been applied to this article in:
Corrigendum: C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants
Abiotic stresses such as drought and salinity are major environmental factors that limit crop yields. Unraveling the molecular mechanisms underlying abiotic stress resistance is crucial for improving crop performance and increasing productivity under adverse environmental conditions. Zinc finger proteins, comprising one of the largest transcription factor families, are known for their finger-like structure and their ability to bind Zn2+. Zinc finger proteins are categorized into nine subfamilies based on their conserved Cys and His motifs, including the Cys2/His2-type (C2H2), C3H, C3HC4, C2HC5, C4HC3, C2HC, C4, C6, and C8 subfamilies. Over the past two decades, much progress has been made in understanding the roles of C2H2 zinc finger proteins in plant growth, development, and stress signal transduction. In this review, we focus on recent progress in elucidating the structures, functions, and classifications of plant C2H2 zinc finger proteins and their roles in abiotic stress responses.
As environmental problems become more severe, abiotic stress factors such as high salinity, low temperature, and drought will increasingly limit plant growth and crop yields (Chen et al., 2018; Costa and Farrant, 2019). During the long process of evolution, plants have evolved protective mechanisms in response to stress involving morphological, physiological, and molecular adaptations (Meng et al., 2018; Bernstein, 2019; Kumari et al., 2019). When a plant is subjected to abiotic stress, a series of adaptive changes occur in the plant cells to maintain growth, including the upregulation or downregulation of various genes (Cui et al., 2018; Yang et al., 2018; Han et al., 2019b).
Regulatory proteins function in stress signal transduction by influencing the expression of downstream target genes (functional genes). These regulatory proteins include protein kinases [including mitogen activated protein kinases (MAPK), calcium dependent protein kinases (CDPK), receptor protein kinases, ribosomal protein kinases, and transcriptional regulatory protein kinases] (Tibbles and Woodgett, 1999; Wimalasekera and Scherer, 2018; Divya et al., 2019), protein phosphatases (Singh et al., 2010; Gong et al., 2018), transcription factors (TFs) (Han et al., 2014; Sun et al., 2018), and proteins involved in inorganic phosphate (Pi) turnover (Takahashi et al., 2001; Fabiańska et al., 2019). TFs bind to specific sequences in the promoters of their target genes, thereby regulating gene expression and affecting biological phenotypes (Han et al., 2019a). TFs are key regulatory components of abiotic stress signaling pathways (Schmidt et al., 2012; Castelán-Muñoz et al., 2019).
Zinc finger proteins form one of the largest TF families in plants and are categorized into subfamilies based on the order of the Cys and His residues in their secondary structures, such as Cys2/His2-type (C2H2), C3H, C3HC4, C2HC5, C4HC3, C2HC, C4, C6, and C8 (Miller et al., 1985; Klug, 2010; Han et al., 2014). Among these, C2H2-type zinc finger protein genes account for ~0.7% of Arabidopsis thaliana genes, 0.8% of yeast genes, and 3% of dipteran and mammalian genes (Böhm et al., 1997; Englbrecht et al., 2004; Ciftci-Yilmaz and Mittler, 2008).
The first C2H2-type zinc finger protein gene discovered in plants was EPF1 from Petunia, encoding a protein with two typical C2H2 zinc finger motifs. Fourteen EPF1-related genes are present in the Petunia genome (Takatsuji et al., 1992). Many C2H2-type zinc finger protein genes have since been cloned and studied in model plants such as Arabidopsis, wheat (Triticum aestivum), soybean (Glycine max), and rice (Oryza sativa) (Gao et al., 2011; Sun et al., 2012; Zhang et al., 2014; Hong et al., 2016). These genes encode proteins that play many roles in plant growth, development, and biotic stress resistance (Feurtado et al., 2011; An et al., 2012; Zhou et al., 2013; Yang et al., 2014; Weng et al., 2015; Cao et al., 2016).
In the past two decades, increasing evidence has indicated that C2H2-type zinc finger proteins also play important roles in abiotic stress resistance in plants. In this review, we describe the structures, classifications, and roles of plant C2H2-type zinc proteins in regulating abiotic stress responses in plants.
Zn2+ binds to several conserved amino acids in C2H2-type zinc finger proteins (generally Cys and His) to form a relatively independent region of the protein. Eukaryotic C2H2-type zinc finger proteins generally contain a specific conserved sequence consisting of 25 to 30 amino acids: C-X2~4-C-X3-P-X5-L-X2-H-X3-H (Shimeld, 2008; Finn et al., 2013). Two pairs of histidines at the C-terminus of the α-helix and the two cysteines at the end of the β-strand bind to Zn2+ to form a tetrahedral structure. Zn2+ is sandwiched between an α-helix and two antiparallel β-strands; the stability of the ββα structural system is maintained by the interlaced linkage provided by Zn2+ (Figure 1). The presence of Zn2+ ensures that the stability of the entire zinc finger structure and the normal helical structure are maintained (Wolfe et al., 2000; Alam et al., 2019).
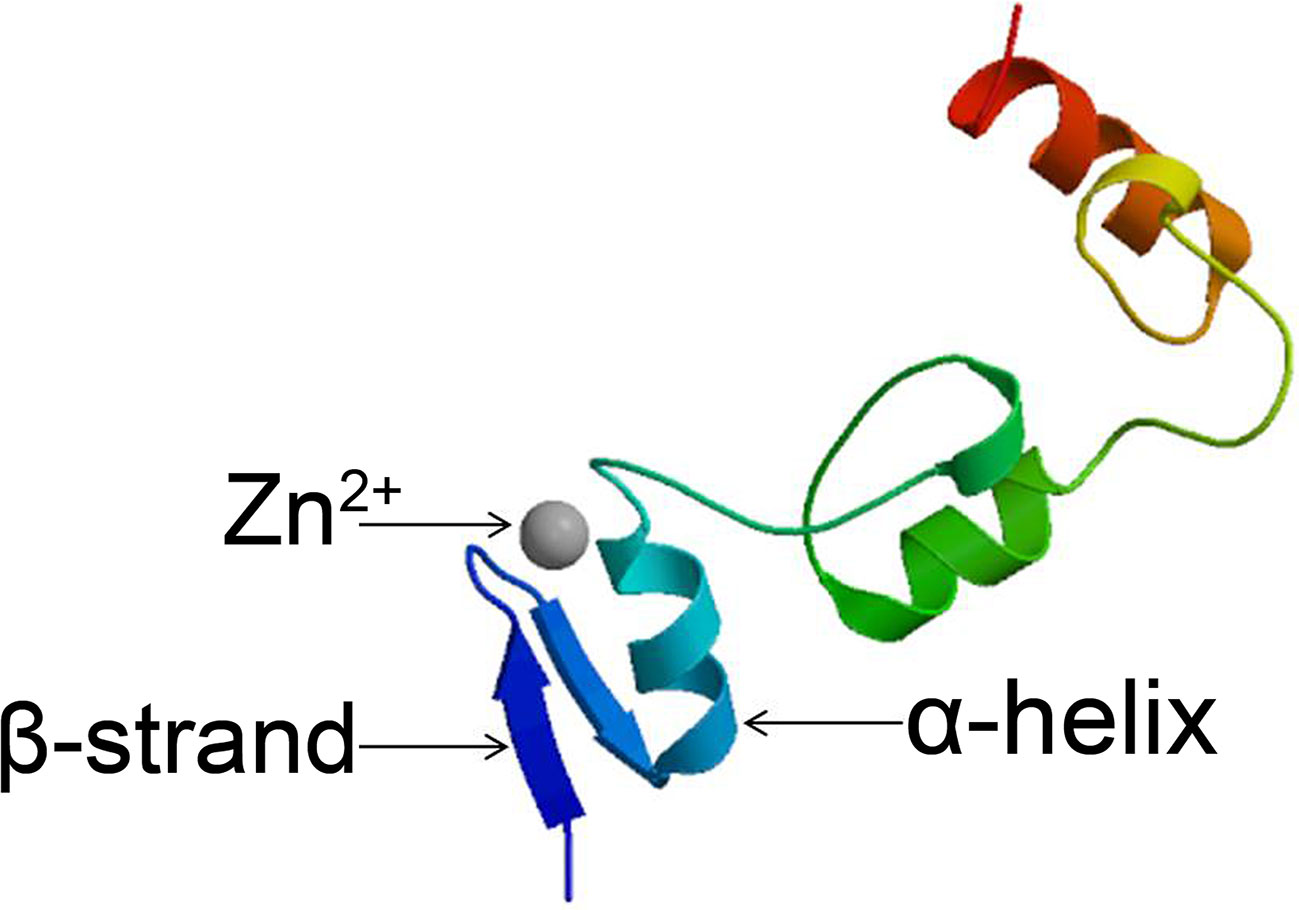
Figure 1 Structure of C2H2 zinc finger proteins. Structural model of the Arabidopsis C2H2 zinc finger protein STZ produced using the Protein Model Portal tool.
Most C2H2 zinc finger proteins in plants contain the highly conserved sequence QALGGH in their zinc finger domains: such proteins are referred to as Q-type zinc finger proteins. Unlike proteins containing the QALGGH sequence, a class of C2H2-type zinc finger proteins do not contain any conserved motifs in the zinc finger region are known as C-type proteins (Agarwal et al., 2007; Liu et al., 2015b).
According to the number and distribution of zinc fingers, C2H2-type zinc finger proteins in plants are divided into four categories: (1) proteins containing one C2H2 zinc finger (single-C2H2); (2) proteins containing three C2H2 zinc fingers (triple-C2H2 [tC2H2]); (3) proteins containing more than three adjacent C2H2 zinc fingers (multiple-adjacent-C2H2 [maC2H2]); and (4) proteins containing several C2H2 zinc finger pairs that are widely separated (separated-paired-C2H2 [spC2H2]) (Wang et al., 2019).
Although many C2H2 transcription factors show transcriptional activation activity, there is no general rule about particular domains that function as transcriptional activators among the various types of domains, such as proline-rich, serine-rich, glutamine-rich, and threonine-rich domains (De Pater et al., 1996; Kiełbowicz-Matuk, 2012; Tang et al., 2019). Many C2H2-type zinc finger proteins function as transcriptional activators during plant growth and abiotic stress, but the activation domain is poorly understood (Sakamoto et al., 2000; Xu et al., 2007; Zhou et al., 2011; Zhang et al., 2016a). Several stress-related C2H2-type zinc finger proteins contain a proline-rich region between the NLS and L-box motifs, which is considered to be the activation domain (Sakamoto et al., 2000).
The associated amphiphilic repression (EAR) motif (L/FDLN L/F(x)P) in plants is present in class II AP2/ERF proteins and in the C-termini of C2H2-type zinc finger proteins (TFIIIA class). The EAR motif reduces the underlying transcriptional level of the reporter gene, as well as the transcriptional activation activity of other TFs (Kazan, 2006). The conserved EAR motif in class II ERF TFs plays a key role in their inhibitory activity. For example, the C-terminal inhibitory domain of AtERF4 is DLDLNL; if a mutation occurs in this domain, the protein's inhibitory function is abolished (Ohta et al., 2001; Han et al., 2019a).
C2H2-type zinc finger proteins with EAR motifs play important roles in regulating abiotic-stress-related gene expression under conditions such as salt, drought, and low temperature (Han and Fu, 2019). The DLNL sequence (DLN-box) at the C-terminus of STZ, with trans-activation activity, functions in the transcriptional activation of DREB1A under salt stress (Sakamoto et al., 2004; Kazan, 2006; Zhang et al., 2016c; Li et al., 2018), while the DLN-box is not the only domain that determines the transcriptional inhibitory activity of C2H2 zinc finger proteins (Huang et al., 2009a).
Plant C2H2-type zinc finger proteins have unique structural features: the conserved, unique QALGGH (Q-type C2H2) motif and the long variable spacers between adjacent zinc finger domains (Klug and Schwabe, 1995; Takatsuji, 1999; Lyu and Cao, 2018). Both features are thought to be important for DNA-binding activity in plants (Kubo et al., 1998; Brayer et al., 2008). A mutation of any amino acid in QALGGH sequence except Q will cause the protein to completely lose its ability to bind to DNA, whereas a mutation in Q will greatly reduce its DNA binding ability (Kubo et al., 1998; Gourcilleau et al., 2011). Of course, characteristic region of a leucine-rich box (L-box) has also been identified to be related to protein interactions in these proteins (Sakamoto et al., 2000; Ciftci-Yilmaz et al., 2007). Bioinformatics tools such ZF Models (zinc finger specificity prediction based on the random forest model) (Gupta et al., 2014), ZifRC (zinc finger recognition code) (Najafabadi et al., 2015), and B1H screening of the C2H2-ZF domain (Persikov et al., 2015) have also been used for binding sequence prediction based on the C2H2-type zinc finger motif.
In addition to binding to DNA, C2H2-type zinc finger proteins can recognize RNA. Zinc fingers comprehensively recognize specific bases and specific folding backbones in diverse RNA structures (Brown, 2005; Lin and Lin, 2018). Contact with the phosphoric acid skeleton in RNA is essential for the recognition of this molecule. A phage display assay showed that the amino acids at positions −1 and +2 of the α-helix in C2H2 zinc finger proteins are important for RNA binding (Mcbryant et al., 1995).
Zinc finger proteins interact with other zinc finger proteins or other types of protein to regulate target gene expression (Brayer and Segal, 2008; Song et al., 2010). Zinc finger proteins interact with other zinc finger proteins to bind to other DNA sequences or to prevent binding to the corresponding DNA sequences, thereby regulating gene transcription and expression, the potential for mediating protein interactions is much larger compared to DNA binding (Shi and Berg, 1995; Mackay and Crossley, 1998; Gamsjaeger et al., 2007; Brayer et al., 2008; Yengo et al., 2018).
In vitro experiments such as gel-shift and yeast one-hybrid assays are widely used to study the binding activity of plant C2H2-type zinc finger proteins to cis-elements in the promoter regions of their downstream genes (Sakamoto et al., 2004). Several abiotic-stress-related C2H2-type zinc finger proteins, such as AZF1, AZF2, AZF3, and ZAT10/STZ, specifically bind to the A(G/C)T repeat sequences in their target promoters (Takatsuji et al., 1994; Sakamoto et al., 2004; Kodaira et al., 2011). The DNA sequence A[AG/CT]CNAC (Sakai et al., 1995; Kubo et al., 1998; Zhou et al., 2011; Han et al., 2019a), the TGCTANNATTG element (Huang et al., 2009b), and TACAAT motifs (Shi et al., 2014) are also possible binding domains of C2H2 TFs in plants. Under abiotic stress conditions, C2H2 zinc finger proteins directly target downstream stress-related genes to activate or inhibit their expression.
C2H2 zinc finger proteins directly target downstream ion balance-related genes to improve salt resistance in plants (Sakamoto et al., 2004; Huang et al., 2007; Ma et al., 2016). C2H2 zinc finger proteins directly target key genes involved in the biosynthesis of osmotic adjustment substances to improve osmotic stress resistance in plants (Xu et al., 2008; Sun et al., 2010; Huang et al., 2012; Yu et al., 2014; Wang et al., 2016; Han et al., 2019b). C2H2 zinc finger proteins also directly target antioxidant genes associated with reactive oxygen species (ROS) scavenging under abiotic stress conditions (Rizhsky et al., 2004; Davletova et al., 2005a; Huang et al., 2009a; Sun et al., 2010; Chu et al., 2016; Liu et al., 2017; Li et al., 2018). C2H2 zinc finger proteins directly target C-repeat/DRE-binding factor genes (CBFs) to improve cold resistance in plants (Vogel et al., 2005). These proteins enhance salt tolerance by interacting with the miRNA-transport-related proteins (Ciftci-Yilmaz et al., 2007). Finally, C2H2 zinc finger proteins directly target downstream genes involved in hormone signal transduction (Kodaira et al., 2011; Kim et al., 2016; Yin et al., 2017).
Salt stress is a serious problem that limits crop production worldwide (Munns and Tester, 2008; Chen et al., 2018). Many C2H2-type TFs act as transcriptional activators or repressors to regulate plant responses to salt stress (Han et al., 2014). Salt stress generates ionic, osmotic, and oxidative stress to plants (Song et al., 2011; Liu et al., 2018). C2H2-type zinc finger proteins participate in salt tolerance by influencing salt-regulated genes.
One way that C2H2-type zinc finger proteins can improve plant salt tolerance is by helping maintain ionic balance. STZ, the first salt-tolerance-related zinc finger protein identified, eliminates the Na+ sensitivity of calcineurin-deficient yeast. STZ appears to be partially dependent on ENA1/PMR2, a P-type ATPase required for Li+ and Na+ efflux in yeast. STZ might enhance salt tolerance in plants by regulating the expression of downstream ion-balance-related genes (Lippuner et al., 1996). Arabidopsis AZF1 and AZF3 respond rapidly to salt stress and may improve salt tolerance by regulating downstream ENA1-like genes (Sakamoto et al., 2000; Sakamoto et al., 2004). In rice, the ZFP182 promoter appears to respond only to monovalent cations such as Na+. Therefore, ZFP182 might regulate downstream ion-transport-related genes, such as Na+ transporter genes, in the salt regulatory signaling pathway (Huang et al., 2007). Finally, TaZNF improves salt tolerance in wheat by increasing the excretion of Na+ and reducing stomatal aperture (Ma et al., 2016).
C2H2-type zinc finger proteins also improve salt tolerance by increasing the concentrations of osmotic adjustment substances. ZFP252 (Xu et al., 2008) and ZFP179 (Sun et al., 2010) improve salt tolerance in rice by increasing the contents of free proline and soluble sugars and upregulating various genes involved in the biosynthesis of osmotic substances, such as OsDREB2A, OsP5CS, OsProT, and OsLea3. In addition, IbZFP1 in sweet potato (I. batatas (L.) Lam.) (Wang et al., 2016), ZFP3 (Zhang et al., 2016a), and AtSIZ1 (Han et al., 2019b) in Arabidopsis enhance plant salt tolerance by increasing proline biosynthesis and accumulation.
C2H2-type zinc finger proteins also improve plant salt tolerance by increasing the ability to scavenge ROS. Arabidopsis plants constitutively expressing ZAT10 show enhanced expression of the ROS scavenging enzymes ascorbate peroxidase1 (APX1), APX2, and Fe-superoxide dismutase1 (FSD1) (Mittler et al., 2006). Transgenic tomato and Arabidopsis plants overexpressing SlZF3 show significantly increased levels of ascorbic acid. Bimolecular fluorescence complementation (BiFC) and Co-Immunoprecipitation (Co-IP) experiments show that SlZF3 directly binds CSN5B, a key component of the COP9 signalosome; CSN5B can promote the degradation of AsA in Arabidopsis by binding to VTC1, so the interaction of SlZF3 and CSN5B inhibited the binding of CSN5B to VTC1, which contributes to AsA biosynthesis (Li et al., 2018).
Many C2H2-type zinc finger proteins enhance plant salt tolerance through abscisic acid (ABA)-mediated signaling pathways. In addition to high salt, AZF2 (Sakamoto et al., 2000; Sakamoto et al., 2004) and StZFP1 (Tian et al., 2010) respond rapidly to ABA, suggesting that they improve salt tolerance via an ABA-dependent pathway. In Thellungiella, ThZF1, like AZF2, regulates downstream gene expression under salt stress (Xu et al., 2007). Transgenic Arabidopsis plants harboring GhDi19-1 and GhDi19-2 from cotton were more sensitive to salt and ABA than the wild type, suggesting that these genes are involved in plant adaptation to salt stress via the ABA signaling pathway (Li et al., 2010).
The mitogen-activated protein kinase (MAPK) cascade is involved in various biotic and abiotic stress responses. ZAT6-overexpressing lines in Arabidopsis show improved seed germination under salt stress via ZAT6's effects on the MAPK cascade. ZAT6 interacts with the stress-responsive MAPK MPK6, and the phosphorylation of ZAT6 by MPK6 is required to improve salt tolerance during seed germination (Liu et al., 2013b). OsZFP213-overexpressing rice lines are more salt tolerant than wild-type and RNAi lines. Yeast two-hybrid, pull-down, and BiFC experiments demonstrate that OsZFP213 interacts with OsMAPK3 to regulate salt tolerance (Zhang et al., 2018).
C2H2-type zinc finger proteins can improve plant salt tolerance via several mechanisms simultaneously, such as promoting ionic balance, scavenging ROS, and increasing the levels of osmotic adjustment substances. GsZFP1 from soybean enhances salt tolerance in transgenic alfalfa by maintaining ionic balance, removing peroxides, and increasing the levels of osmotic adjustment substances (Tang et al., 2013). TaZFP1 overexpression lines show improved salt tolerance due to increased photosynthesis, osmolyte accumulation, and ROS scavenging. RNA-seq analysis show that TaZFP1 regulates photosynthesis, osmolyte metabolism, and ROS-homeostasis-related genes involved in salt stress (Sun et al., 2019a). AtRZFP enhances salt tolerance by increasing ROS scavenging and maintaining Na+ and K+ homeostasis (Zang et al., 2016).
C2H2-type zinc finger proteins can also improve salt tolerance via other mechanisms. ZAT7 may enhance salt tolerance by interacting with the miRNA-transport-related proteins WRKY70 and HASTY (Ciftci-Yilmaz et al., 2007). A DST loss-of-function rice mutant shows enhanced salt tolerance due to increased stomatal closure and reduced stomatal density (Huang et al., 2009b). ZjZFN1 from Zoysia japonica increases salt tolerance in transgenic Arabidopsis. RNA-seq analysis of these plants suggests that ZjZFN1 regulates phenylalanine metabolism, α-linolenic acid metabolism, and phenylpropanoid biosynthesis during salt stress (Teng et al., 2018).
Osmotic stress is induced by many abiotic stresses such as salinity, cold, and drought stress (Bashir et al., 2019). Osmotic stress causes physiological drought, ion imbalance, oxidative damage, and growth inhibition in plants (Yamaguchi-Shinozaki and Shinozaki, 2006). The ability of plants to resist osmotic stress is related to their ability to alter osmotic pressure and increase the biosynthesis of osmotic regulators (Zhu, 2016).
The C2H2 zinc finger proteins ZAT12 (Davletova et al., 2005b) and ZAT10 (Mittler et al., 2006) are involved in regulating the osmotic stress pathway in Arabidopsis. Both ZAT10-overexpressing lines and zat10 mutant showed enhanced tolerance to osmotic stress (Mittler et al., 2006). ZAT10 is phosphorylated by MPKs that are thought to function in abiotic stress tolerance (Nguyen et al., 2012). ZAT10 is a positive regulator of osmotic stress responses that is regulated by MAP kinases in Arabidopsis (Nguyen et al., 2016). In rice, RZF71 was strongly induced by 20% PEG6000 treatment, suggesting that RZF71 plays an important role in the response to osmotic stress (Guo et al., 2007). Moreover, six C2H2-type zinc finger proteins were reported to be involved in osmotic stress responses in poplar (Gourcilleau et al., 2011). Finally, heterologously expressing GmZAT4 from soybean enhanced the tolerance of Arabidopsis to 20% PEG treatment. Expression analysis of marker genes indicated that GmZAT4 enhances osmotic stress tolerance via an ABA pathway in both soybean and Arabidopsis (Sun et al., 2019b).
Cold snap often cause serious damage to plants and can kill plants when severe (Liu et al., 2016; Wang et al., 2017). A comprehensive understanding of the mechanism underlying cold damage in plants would help resolve the problem of cold stress injury (Yang et al., 2013; Cheng et al., 2014; Sui, 2015; Pareek et al., 2017).
C2H2 zinc finger proteins enhance cold resistance by directly regulating downstream cold-related genes in plants. For instance, ZAT12 regulates cold acclimation by controlling the expression of 15 cold-suppressed genes and 9 cold-inducible genes. ZAT12 also downregulates the expression of CBF genes, suggesting it plays a negative role in plant adaptation to cold stress (Vogel et al., 2005). Transgenic Arabidopsis and tobacco (Nicotiana tabacum) plants overexpressing SCOF-1 show increased expression of COR (cold-regulated) genes and enhanced cold tolerance. The SCOF-1 transgenic plants recover from chilling stress more rapidly than the control, and the T2 generation of SCOF-1 transgenic Arabidopsis still expresses cold regulatory genes, such as COR15a, COR47, and RD29B, at higher levels than the wild type at normal growth temperatures. SCOF-1 does not directly interact with the CTR/DRE, ABRE, or cis-acting elements in the promoter regions of COR genes in vitro, but yeast two-hybrid experiments demonstrated that SCOF-1 interacts with SGBF-1. Interestingly, SCOF-1 significantly enhances the activity of SGBF-1 bound to ABRE sequences in vitro, thereby promoting COR genes' expression and enhancing cold tolerance. The interaction of these two proteins is required for the participation of ABRE in the expression of cold-regulated genes to enhance cold tolerance. In addition, SCOF-1 increases cold tolerance in transgenic sweet potato (Kim et al., 2011) and transgenic potato (Kim et al., 2016). SlCZFP1 enhances cold tolerance in transgenic Arabidopsis and rice by inducing the constitutive expression of COR or cold-responsive genes (Zhang et al., 2011). The cold-stress-related gene COR6.6 was significantly upregulated in GmZF1 transgenic plants, suggesting that GmZF1 regulates cold stress resistance in transgenic Arabidopsis by binding to the COR6.6 promoter region (Yu et al., 2014). In banana, the overexpression of MaC2H2-2 and MaC2H2-3 significantly represses the transcription of MaICE1, a key component of the cold signaling pathway. Therefore, MaC2H2s might enhance cold resistance in banana by suppressing the transcription of MaICE1(Han and Fu, 2019).
C2H2 zinc finger proteins can enhance plant cold resistance by increasing the levels of osmotic substances. ZFP182 significantly enhances cold tolerance in rice overexpression lines by increasing the expression of OsP5CS and OsLEA3 and the accumulation of osmoprotectants (Huang et al., 2012). GmZF1 improved cold resistance in transgenic Arabidopsis by increasing proline and soluble contents and reducing membrane lipid peroxidation under cold stress (Yu et al., 2014).
Finally, several C2H2-type zinc finger proteins regulate low-temperature stress through the ABA signaling pathway. SCOF-1 enhances plant cold tolerance through ABA-dependent signaling pathways (Kim et al., 2001). GmZF1 is highly induced by ABA treatment, suggesting that GmZF1 might participate in an ABA-dependent signaling pathway (Yu et al., 2014).
Drought is an important factor limiting plant growth worldwide, inducing adverse reactions such as osmotic imbalance, membrane system damage, and decreased respiratory and photosynthetic rates. Drought not only hinders plant growth and metabolism at various stages but also affects crop quality and yields (Bartels and Sunkar, 2005; Liu et al., 2015a; Guo et al., 2018).
C2H2 zinc finger proteins enhance plant drought resistance by increasing the levels of osmotic adjustment substances. For example, rice plants overexpressing ZFP252 have a significantly higher survival rate than wild-type and antisense-ZFP252 plants under drought stress. The overexpression lines exhibit enhanced expression of stress-related genes, such as Oslea3, OsP5CS, and OsProT, which contribute to the accumulation of osmotic substances (Xu et al., 2008). Overexpressing OsMSR15 (Zhang et al., 2016c) and ZFP3 in transgenic Arabidopsis (Zhang et al., 2016a) confers increase in drought tolerance by maintaining higher proline contents, reducing electrolyte leakage, and increasing stress-responsive gene expression.
C2H2 zinc finger proteins also enhance drought resistance by improving the ability to scavenge ROS. ZFP245 improves drought resistance in rice by enhancing the activities of the ROS scavenging enzymes superoxide dismutase (SOD) and peroxide (POD) and increasing resistance to H2O2 (Huang et al., 2009a). When ZxZF from the highly drought-tolerant plant Zygophyllum xanthoxylum is expressed under the control of the drought-inducible promoter rd29A in Arabidopsis and poplar, the resulting transgenic plants have significantly increased photosynthetic efficiency, ROS, and scavenging ability (Chu et al., 2016). ZAT18 knock down mutant exhibits reduced drought tolerance, while the overexpression lines show higher leaf water content and higher antioxidant enzyme activity than wild-type plants under drought stress (Yin et al., 2017).
C2H2 zinc finger proteins also enhance drought resistance via the ABA and other signaling pathways. Under dehydration stress, AZF2 expression is lower in the aba1 and abi1 mutants than in the wild type, indicating that AZF2 confers drought stress tolerance through an ABA-dependent pathway (Sakamoto et al., 2004). GmZFP3 plays a negative role in plant tolerance to drought stress. Transgenic Arabidopsis plants of GmZFP3 show enhanced expression of ABA-related marker genes, including DREB2A, LHY1, MYB2, RCI3, PAD3, CCA1, and UGT71B6, suggesting that GmZFP3 regulates the drought stress response through an ABA-dependent signaling pathway (Zhang et al., 2016b). Transgenic plants overexpressing OsMSR15 show hypersensitivity to exogenous ABA. Transgenic plants also showed increased expressions of a number of stress-responsive genes, including RD29A and DREB1A under drought stress, which suggested that OsMSR15 may play important roles in response to drought stress both in ABA-dependent and -independent pathways (Zhang et al., 2016c). In the ZAT18 overexpression lines, RNA-seq analysis revealed the enrichment of pathways, such as hormone metabolism, stress response, and signaling pathways; stress-related genes, such as COR47, ERD7, LEA6, RAS1, and hormone-signaling-transduction-related genes, such as JAZ7 and PYL5, appear to be the downstream target genes of ZAT18 (Yin et al., 2017).
C2H2-type zinc finger proteins can also improve plant drought resistance through various mechanisms at the same time. Transformation with CgZFP1 (Gao et al., 2012) and DgZFP3 (Liu et al., 2013a) from chrysanthemum, BcZAT12 from tomato (Rai et al., 2013), and IbZFP1 from sweet potato (Wang et al., 2016) improve drought resistance in transgenic plants by increasing the levels of osmotic adjustment substances, improving ROS scavenging ability, and regulating downstream stress response genes. The Arabidopsis azf2 mutant expressing the Thellungiella halophila gene ThZF1 driven by the 35S promoter displays a flowering time phenotype similar to that of the wild type under drought stress. ThZF1 and AZF2 can both activate the transcription of AtEPSP (Xu et al., 2007). GsZFP1 overexpression enhances drought tolerance in transgenic alfalfa. The expression levels of stress-responsive marker genes, including MtCOR47, MtRAB18, MtP5CS, and MtRD2, were much higher in these plants than in the wild type under drought stress (Luo et al., 2012a).
Abiotic stress usually leads to the production of ROS. Low levels of ROS benefit plant growth and development by functioning as signals and might play other important roles as well (Choudhury et al., 2017), while the excessive accumulation of ROS causes secondary damage to plants. In the presence of excessive ROS, C2H2 zinc finger proteins are expressed at higher levels to help maintain stable ROS levels in plants (Gadjev et al., 2006).
The expression of antioxidase genes that are associated with ROS scavenging under abiotic stress can be regulated by C2H2 zinc finger proteins (Liu et al., 2017). Ascorbate peroxidase 1 (Apx1) is an important scavenger of H2O2 in plants (Rizhsky et al., 2004). The expression levels of TF genes ZAT12 and ZAT7 increase with increasing H2O2 levels in the loss-of-function mutant apx1. Plants lacking ZAT12 are more sensitive to high H2O2 contents than wild-type plants (Rizhsky et al., 2004; Davletova et al., 2005a). APX1, APX2, and FeSOD1 are significantly upregulated in ZAT10 and ZAT12 transgenic lines, but are significantly downregulated in zat10 loss-of-function mutants under high-light stress (Mittler et al., 2006). DST directly binds the promoter region of peroxidase24 precursor to regulate its expression and reduces the accumulation of H2O2 in plants (Huang et al., 2009b). SOD and POD activity is significantly enhanced in ZFP245 and ZFP179 overexpression rice plants, thereby increasing resistance to abiotic stress and ROS at the seedling stage (Huang et al., 2009a; Sun et al., 2010). Under water and oxidative stress, overexpressing ZFP36 reduces the damage from ROS by increasing SOD and APX activity, whereas ZFP36-RNAi lines exhibit severe ROS damage due to reduced SOD and APX activity (Zhang et al., 2014).
C2H2-type zinc finger proteins participate in antioxidative stress responses through ABA and MAPK signaling pathways. ZFP36 is significantly induced by ABA-triggered increases in H2O2 and OsMPK activity. ZFP36 also increases the expression levels of NADPH oxidase and MAPK genes in the ABA signaling pathway. These findings indicate that ZFP36 is involved in regulating the crosstalk among NADPH oxidase, H2O2, and MAPK in the ABA signaling pathway (Zhang et al., 2014).
Plants are highly sensitive to strong light. The ability of plants to withstand strong light depends on the duration of light exposure and light quality (Yang et al., 2019). Overexpressing the C2H2-type zinc finger protein gene RHL41, which is identical to the ZAT12 gene in Arabidopsis, significantly improved resistance to high-light conditions, as manifested by dramatic changes in plant morphology, such as increased production of thin-walled tissues of the barrier and increased anthocyanin and chlorophyll contents (Iida et al., 2000). The expression of ZAT12 increases in response to light stress (Iida et al., 2000; Davletova et al., 2005b). Perhaps ZAT12 is part of the active oxygen scavenging signal transduction pathway, which responds to H2O2 produced during light stress (Davletova et al., 2005b). ZAT10 expression is induced by light stress, and ZAT10-overexpressing transgenic lines showed enhanced tolerance to photo-inhibitory light stress (Mittler et al., 2006; Kilian et al., 2007). Under high-light stress, the levels of Apx1, Apx2, and iron superoxide dismutase 1 (FSD1) significantly increased in ZAT10-overexpressing transgenic plants, whereas suppressed expression was observed in ZAT10 knockout plants, suggesting that the increased tolerance of ZAT10-overexpressing plants to high light results from the specific expression of ROS clearance-related genes (Miller et al., 2008). However, to date, few studies have focused on the roles of zinc finger proteins in plant responses to high-light stress; more work needs to be done in this area.
As is well known, many plant hormones participate in physiological adaptations to abiotic stress, and many stress related plant hormones that play an important role in resistance to many stresses mediated by C2H2 type zinc finger proteins have been studied. Abscisic acid (ABA), salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) are important plant hormones in pathogens and abiotic stresses signaling pathways (Shi et al., 2014; Zhang et al., 2014; Ma et al., 2016; Zang et al., 2016).
Typically, ABA is responsible for plant defense against abiotic stresses such as salinity, cold, osmotic, drought, active oxygen, and heat (Lata and Prasad, 2011; Ryu and Cho, 2015; Wang et al., 2019). Many studies found that the application of exogenous ABA can induce the expression of many stress related marker genes (Zhu, 2002; Fujita et al., 2013; Nakashima and Yamaguchi-Shinozaki, 2013; Tang et al., 2013). There are at least two ABA-mediated signal pathways existing in plants to respond to abiotic stresses: ABA-dependent and ABA-independent pathways; the two pathways also have some degree of crosstalk in response to abiotic stresses (Zhu, 2002; Shinozaki et al., 2003; Yamaguchi-Shinozaki and Shinozaki, 2005; Yamaguchi-Shinozaki and Shinozaki, 2006). C2H2 zinc finger proteins can confer abiotic stress tolerance by increasing the contents of abscisic acid (ABA), proline, soluble sugars or chlorophyll, and by reducing the water loss rate (Luo et al., 2012b; Wang et al., 2016). Zinc finger proteins are also involved in high temperature (Kim et al., 2015) and H+ (Fan et al., 2015) tolerance mediated by ABA signaling pathways. C2H2 zinc finger protein genes improve plant tolerance to stresses by ABA-dependent pathway (Zhang et al., 2016b) or ABA-dependent pathway (Huang et al., 2007), whereas some genes improve plant tolerance to stresses through both ABA-dependent and ABA-independent pathways (Sun et al., 2010). Abiotic stresses, as well as ABA signaling pathways, together with C2H2 zinc finger proteins constitute a complex network and there is many overlaps between different signaling pathways (Knight and Knight, 2001; Kiełbowicz-Matuk, 2012; Alam et al., 2019).
Salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) play major roles in biotic stress responses, as their levels increase in response to pathogen infection and herbivorous insect attack (Bari and Jones, 2009; Gamalero and Glick, 2012; Wasternack and Hause, 2013). Of course, the stress response of hormones to stress is not completely strict. SA is a phenolic hormone that functions as an important signaling molecule in plant responses to biotic and abiotic stress. C2H2 TFs are involved in the early response to SA in Carica papaya (Jiang and Pan, 2012). AtZAT6 plays important roles in stress responses by activating the expression of SA-related genes in Arabidopsis (Shi et al., 2014). JAs are important class of lipid phytohormones. ZPT2-3 expression is induced by mechanical wounding and abiotic stress, a process mediated by a JA-dependent, ET-independent pathway (Sugano et al., 2003). AtZFP11-overexpressing lines exhibited changes in the expression of a number of genes involved in JA and stress responses (Dinkins et al., 2012). ETHYLENE RESPONSE FACTOR1 (ERF1), a downstream component of the ET signaling pathway, is involved in plant resistance to several necrotrophic fungi (Berrocal-Lobo et al., 2002). High salt levels and drought stress significantly induced the expression of ERF1 in Arabidopsis. ERF1 specifically regulates gene expression by integrating the JA, ET, and ABA signaling pathways and plays a positive regulatory role in plant resistance to salt, drought, and heat stress (Cheng et al., 2013).
It is now clear that C2H2-type zinc finger proteins play central roles in plant responses to high salt, cold, osmotic, drought, and oxidative stresses. Identifying abiotic stress sensors is crucial for understanding the molecular mechanisms underlying abiotic stress resistance in plants. For example, Glycosyl inositol phosphorylceramide (GIPC) was recently shown to be a salt receptor in plants (Jiang et al., 2019). REDUCED HYPEROSMOLALITY-INDUCED CALCIUM INCREASE 1 (OSCA1) is considered to be an osmotic stress sensor, but how it perceives osmotic stress is not clear (Yuan et al., 2014). Chilling-Tolerance Divergence 1 (COLD1), which regulates cold stress perception in rice, is another potential stress receptor (Ma et al., 2015; Shi and Yang, 2015). However, much more effort will be needed to identify abiotic stress sensors and to unravel the detailed roles of C2H2-type zinc finger proteins in abiotic stress resistance in plants.
After stress signals are perceived by the plant, second messengers (such as Ca2+ and ROS) are generated in the cytosol, where they mediate the phosphorylation of downstream proteins (by protein kinases such as MAPKs) as well as ABA-dependent or -independent pathways. Various cis-elements are present in the promoter regions of high salt-, drought-, and ABA-induced genes (Agarwal et al., 2006; Onishi et al., 2006). For example, C2H2-type zinc finger protein genes AZF2, AtSIZ1, and STZ contain cis-elements such as DRE (CCGAC), MYCRS (CANNTG), and MYBRS (RAACYR) elements in their promoter regions, conferring salt tolerance (Han et al., 2019). Protein kinases and ABA-dependent or -independent pathways (Zhang et al., 2014) directly or indirectly regulate C2H2 transcription factors, which in turn specifically activate or inhibit a group of target genes containing C2H2 zinc finger protein-binding sequences such as A [AG/CT]CNAC in their promoter regions, thereby enhancing plant stress tolerance (Figure 2).

Figure 2 Signaling pathways of C2H2-type zinc finger protein genes involved in abiotic stress responses (Xiong et al., 2002; Wang et al., 2019).
Although abiotic stress responses are regulated by multiple pathways controlled by C2H2 zinc finger proteins, there is a certain degree of overlap among these signaling pathways and regulatory networks (Kiełbowicz-Matuk, 2012). The roles of plant C2H2-type zinc finger proteins in different abiotic stress responses and signaling pathways are summarized in Supplementary Table 1. As shown in the table, the same C2H2 zinc finger protein gene can be induced by multiple abiotic stresses at the same time. In addition, different C2H2 zinc finger proteins increase plant stress resistance through similar regulatory mechanisms, and different C2H2 zinc finger proteins improve plant stress resistance through similar ABA or protein kinase signaling pathways. Therefore, the stress response network of plants is very complex, and the many unknown proteins and related pathways must be further elucidated.
C2H2-type zinc finger proteins play important roles in plant development and growth as well as abiotic stress resistance. In-depth research on these proteins is needed to better understand their functions and mechanisms.
The roles of C2H2 zinc finger proteins in plant stress-resistance mechanisms must be analyzed in more detail, such as how these transcription factors respond to various signaling molecules, how they regulate downstream gene expression, and how they interact with other transcription factors. Whether (and how) C2H2 zinc finger proteins mediate abiotic stress responses in plants by interacting with noncoding RNAs is currently unclear. The continuous development of biotechnology methods such as CRISPR-Cas should greatly facilitate the study of abiotic-stress-related C2H2-type zinc finger proteins in plants.
In addition to the model plant Arabidopsis, more studies should focus on the roles of C2H2-type zinc finger proteins in stress-resistant plants, such as halophytes (Yuan et al., 2015b) and xerophytes (Chu et al., 2016). During the course of evolution, stress-resistant plants have evolved various physiological and molecular mechanisms to adapt to stress (Sui et al., 2010; Sun et al., 2010; Feng et al., 2014). Halophytes can grow and finish their life cycles in saline soils with a salt concentration equivalent to ≥200mM NaCl (Santos et al., 2015; Yuan et al., 2015a; Zhou et al., 2016), and euhalophytes grow vigorously in sea water. C2H2 zinc finger proteins might be associated with these morphological features or play distinct roles in these plants. For instance, overexpressing the C2H2 zinc finger gene ZjZFN1 from the halophyte Z. japonica improved salt tolerance in Arabidopsis by altering phenylalanine metabolism, α-linolenic acid metabolism, and phenylpropanoid biosynthesis (Teng et al., 2018). The expression of a homolog of the C2H2 zinc finger gene AtSIZ1 from the halophyte Limonium bicolor improved salt tolerance in Arabidopsis by maintaining ionic homeostasis and osmotic balance (Han et al., 2019b).
These issues have been clarified in succession, which will be of great benefit to make full use of plant C2H2 zinc finger protein gene resources to improve crop varieties and increase plant stress resistance.
GH and BW conceived and designed this study. GH and BW wrote the manuscript. CL, JG, ZQ, NS, and NQ proposed related theories and assisted with the interpretation of some references. All authors have read, edited, and approved the current version of the manuscript.
This work was supported by the NSFC (National Natural Science Research Foundation of China, project No. 31570251; 31600200; 31770288), the Shandong Province Key Research and Development Plan (2017CXGC0313; 2016GNC113012), the Natural Science Research Foundation of Shandong Province (ZR2014CZ002; ZR2017MC003; ZR2019MC065), the Higher Educational Science and Technology Program of Shandong Province (J15LE08; J17KA136), and the Open Fund of Shandong Provincial Key Laboratory of Plant Stress (KLPS2018-01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00115/full#supplementary-material
Agarwal, M., Hao, Y., Kapoor, A., Dong, C.-H., Fujii, H., Zheng, X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281, 37636–37645. doi: 10.1074/jbc.M605895200
Agarwal, P., Arora, R., Ray, S., Singh, A. K., Singh, V. P., Takatsuji, H., et al. (2007). Genome-wide identification of C 2 H 2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 65, 467–485. doi: 10.1007/s11103-007-9199-y
Alam, I., Batool, K., Cui, D.-L., Yang, Y.-Q., Lu, Y.-H. (2019). Comprehensive genomic survey, structural classification and expression analysis of C2H2 zinc finger protein gene family in Brassica rapa L. PloS One 14, e0216071. doi: 10.1371/journal.pone.0216071
An, L., Zhou, Z., Sun, L., Yan, A., Xi, W., Yu, N., et al. (2012). A zinc finger protein gene ZFP5 integrates phytohormone signaling to control root hair development in Arabidopsis. Plant J. 72, 474–490. doi: 10.1111/j.1365-313X.2012.05094.x
Böhm, S., Frishman, D., Mewes, H. W. (1997). Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 25, 2464–2469. doi: 10.1093/nar/25.12.2464
Bari, R., Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Bartels, D., Sunkar, R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58. doi: 10.1080/07352680590910410
Bashir, K., Matsui, A., Rasheed, S., Seki, M. (2019). Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress. F1000Research 8, 658. doi: 10.12688/f1000research.18424.1
Bernstein, N. (2019). Plants and salt: Plant response and adaptations to salinity. In Model Ecosystems in Extreme Environments Elsevier, 101–112. doi: 10.1016/B978-0-12-812742-1.00005-2
Berrocal-Lobo, M., Molina, A., Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. doi: 10.1046/j.1365-313x.2002.01191.x
Brayer, K. J., Segal, D. J. (2008). Keep your fingers off my DNA: protein–protein interactions mediated by C2H2 zinc finger domains. Cell Biochem. Biophys. 50, 111–131. doi: 10.1007/s12013-008-9008-5
Brayer, K. J., Kulshreshtha, S., Segal, D. J. (2008). The protein-binding potential of C2H2 zinc finger domains. Cell Biochem. Biophys. 51, 9–19. doi: 10.1007/s12013-008-9007-6
Brown, R. S. (2005). Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struct. Biol. 15, 94–98. doi: 10.1016/j.sbi.2005.01.006
Cao, H., Huang, P., Zhang, L., Shi, Y., Sun, D., Yan, Y., et al. (2016). Characterization of 47 Cys2-His2 zinc finger proteins required for the development and pathogenicity of the rice blast fungus Magnaporthe oryzae. New Phytol. 211, 1035–1051. doi: 10.1111/nph.13948
Castelán-Muñoz, N., Herrera, J., Cajero-Sánchez, W., Arrizubieta, M., Trejo, C., Garcia-Ponce, B., et al. (2019). MADS-box genes are key components of genetic regulatory networks involved in abiotic stress and plastic developmental responses in plants. Front. Plant Sci. 10, 853. doi: 10.3389/fpls.2019.00853
Chen, M., Yang, Z., Liu, J., Zhu, T., Wei, X., Fan, H., et al. (2018). Adaptation mechanism of salt excluders under saline conditions and its applications. Int. J. Mol. Sci. 19, 3668. doi: 10.3390/ijms19113668
Cheng, M.-C., Liao, P.-M., Kuo, W.-W., Lin, T.-P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Cheng, S., Yang, Z., Wang, M., Song, J., Sui, N., Fan, H. (2014). Salinity improves chilling resistance in Suaeda salsa. Acta physiologiae plant. 36, 1823–1830. doi: 10.1007/s11738-014-1555-3
Choudhury, F. K., Rivero, R. M., Blumwald, E., Mittler, R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867. doi: 10.1111/tpj.13299
Chu, Y., Zhang, W., Wu, B., Huang, Q., Zhang, B., Su, X. (2016). Overexpression of the novel Zygophyllum xanthoxylum C2H2-type zinc finger gene ZxZF improves drought tolerance in transgenic Arabidopsis and poplar. Biologia 71, 769–776. doi: 10.1515/biolog-2016-0093
Ciftci-Yilmaz, S., Mittler, R. (2008). The zinc finger network of plants. Cell. Mol. Life Sci. 65, 1150–1160. doi: 10.1007/s00018-007-7473-4
Ciftci-Yilmaz, S., Morsy, M. R., Song, L., Coutu, A., Krizek, B. A., Lewis, M. W., et al. (2007). The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J. Biol. Chem. 282, 9260–9268. doi: 10.1074/jbc.M611093200
Costa, M.-C. D., Farrant, J. M. (2019). Plant Resistance to Abiotic Stresses. Plants 8 (12), 553. doi: 10.3390/plants8120553
Cui, F., Sui, N., Duan, G., Liu, Y., Han, Y., Liu, S., et al. (2018). Identification of metabolites and transcripts involved in salt stress and recovery in peanut. Front. Plant Sci. 9, 217. doi: 10.3389/fpls.2018.00217
Davletova, S., Rizhsky, L., Liang, H., Shengqiang, Z., Oliver, D. J., Coutu, J., et al. (2005a). Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17, 268–281. doi: 10.1105/tpc.104.026971
Davletova, S., Schlauch, K., Coutu, J., Mittler, R. (2005b). The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 139, 847–856. doi: 10.1104/pp.105.068254
De Pater, S., Greco, V., Pham, K., Memelink, J., Kijne, J. (1996). Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 24, 4624–4631. doi: 10.1093/nar/24.23.4624
Dinkins, R. D., Tavva, V. S., Palli, S. R., Collins, G. B. (2012). Mutant and overexpression analysis of a C2H2 single zinc finger gene of Arabidopsis. Plant Mol. Biol. Rep. 30, 99–110. doi: 10.1007/s11105-011-0320-7
Divya, K., Bhatnagar-Mathur, P., Sharma, K. K., Reddy, P. S. (2019). Heat Shock Proteins (Hsps) Mediated Signalling Pathways During Abiotic Stress Conditions. Plant Signaling Mol., 499–516. doi: 10.1016/B978-0-12-816451-8.00031-9
Englbrecht, C. C., Schoof, H., Böhm, S. (2004). Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5, 39. doi: 10.1186/1471-2164-5-39
Fabiańska, I., Bucher, M., Häusler, R. E. (2019). Intracellular phosphate homeostasis–A short way from metabolism to signaling. Plant Sci. 286, 57–67. doi: 10.1016/j.plantsci.2019.05.018
Fan, W., Lou, H. Q., Gong, Y. L., Liu, M. Y., Cao, M. J., Liu, Y., et al. (2015). Characterization of an inducible C2H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. New Phytol. 208, 456–468. doi: 10.1111/nph.13456
Feng, Z., Deng, Y., Fan, H., Sun, Q., Sui, N., Wang, B. (2014). Effects of NaCl stress on the growth and photosynthetic characteristics of Ulmus pumila L. seedlings in sand culture. Photosynthetica 52, 313–320. doi: 10.1007/s11099-014-0032-y
Feurtado, J. A., Huang, D., Wicki-Stordeur, L., Hemstock, L. E., Potentier, M. S., Tsang, E. W., et al. (2011). The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 23, 1772–1794. doi: 10.1105/tpc.111.085134
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2013). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Fujita, Y., Yoshida, T., Yamaguchi-Shinozaki, K. (2013). Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 147, 15–27. doi: 10.1111/j.1399-3054.2012.01635.x
Gadjev, I., Vanderauwera, S., Gechev, T. S., Laloi, C., Minkov, I. N., Shulaev, V., et al. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141, 436–445. doi: 10.1104/pp.106.078717
Gamalero, E., Glick, B. R. (2012). “Ethylene and abiotic stress tolerance in plants,” in Environmental adaptations and stress tolerance of plants in the era of climate change (Springer), 395–412. doi: 10.1007/978-1-4614-0815-4_18
Gamsjaeger, R., Liew, C. K., Loughlin, F. E., Crossley, M., Mackay, J. P. (2007). Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 32, 63–70. doi: 10.1016/j.tibs.2006.12.007
Gao, S.-Q., Chen, M., Xu, Z.-S., Zhao, C.-P., Li, L., Xu, H.-J., et al. (2011). The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol. Biol. 75, 537–553. doi: 10.1007/s11103-011-9738-4
Gao, H., Song, A., Zhu, X., Chen, F., Jiang, J., Chen, Y., et al. (2012). The heterologous expression in Arabidopsis of a chrysanthemum Cys2/His2 zinc finger protein gene confers salinity and drought tolerance. Planta 235, 979–993. doi: 10.1007/s00425-011-1558-x
Gong, Z., Chinnusamy, V., Zhu, J. K. (2018). The molecular networks of abiotic stress signaling. Annu. Plant Rev. Online, 33, 388–416. doi: 10.1002/9781119312994.apr0360
Gourcilleau, D., Lenne, C., Armenise, C., Moulia, B., Julien, J.-L., Bronner, G., et al. (2011). Phylogenetic study of plant Q-type C2H2 zinc finger proteins and expression analysis of poplar genes in response to osmotic, cold and mechanical stresses. DNA Res. 18, 77–92. doi: 10.1093/dnares/dsr001
Guo, S.-Q., Huang, J., Jiang, Y., Zhang, H. (2007). Cloning and characterization of RZF71 encoding a C2H2-type zinc finger protein from rice. Yi chuan= Hereditas 29, 607–613. doi: 10.1360/yc-007-0607
Guo, Y., Tian, S., Liu, S., Wang, W., Sui, N. (2018). Energy dissipation and antioxidant enzyme system protect photosystem II of sweet sorghum under drought stress. Photosynthetica, 56, 861–872. doi: 10.1007/s11099-017-0741-0
Gupta, A., Christensen, R. G., Bell, H. A., Goodwin, M., Patel, R. Y., Pandey, M., et al. (2014). An improved predictive recognition model for Cys2-His2 zinc finger proteins. Nucleic Acids Res. 42, 4800–4812. doi: 10.1093/nar/gku132
Han, Y.-C., Fu, C.-C. (2019). Cold-inducible MaC2H2s are associated with cold stress response of banana fruit via regulating MaICE1. Plant Cell Rep. 38, 673–680. doi: 10.1007/s00299-019-02399-w
Han, G., Wang, M., Yuan, F., Sui, N., Song, J., Wang, B. (2014). The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana. Plant Mol. Biol. 86, 237–253. doi: 10.1007/s11103-014-0226-5
Han, G.-L., Wei, X., Dong, X., Wang, C., Sui, N., Guo, J.-R., et al. (2019a). Arabidopsis ZINC FINGER PROTEIN1 Acts Downstream of GL2 To Repress Root Hair Initiation and Elongation by Directly Suppressing bHLH Genes. Plant Cell. 32, 206–225, on line. doi: 10.1105/tpc.19.00226
Han, G., Yuan, F., Guo, J., Zhang, Y., Sui, N., Wang, B. (2019b). AtSIZ1 improves salt tolerance by maintaining ionic homeostasis and osmotic balance in Arabidopsis. Plant Sci. 285, 55–67. doi: 10.1016/j.plantsci.2019.05.002
Hong, Y., Zhang, H., Huang, L., Li, D., Song, F. (2016). Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 7, 4. doi: 10.3389/fpls.2016.00004
Huang, J., Yang, X., Wang, M.-M., Tang, H.-J., Ding, L.-Y., Shen, Y., et al. (2007). A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochim. Biophys. Acta (BBA)-Gene Struct. Expression 1769, 220–227. doi: 10.1016/j.bbaexp.2007.02.006
Huang, J., Sun, S.-J., Xu, D.-Q., Yang, X., Bao, Y.-M., Wang, Z.-F., et al. (2009a). Increased tolerance of rice to cold, drought and oxidative stresses mediated by the overexpression of a gene that encodes the zinc finger protein ZFP245. Biochem. Biophys. Res. Commun. 389, 556–561. doi: 10.1016/j.bbrc.2009.09.032
Huang, X.-Y., Chao, D.-Y., Gao, J.-P., Zhu, M.-Z., Shi, M., Lin, H.-X. (2009b). A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23, 1805–1817. doi: 10.1101/gad.1812409
Huang, J., Sun, S., Xu, D., Lan, H., Sun, H., Wang, Z., et al. (2012). A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.). Plant Mol. Biol. 80, 337–350. doi: 10.1007/s11103-012-9955-5
Iida, A., Kazuoka, T., Torikai, S., Kikuchi, H., Oeda, K. (2000). A zinc finger protein RHL41 mediates the light acclimatization response in Arabidopsis. Plant J. 24, 191–203. doi: 10.1046/j.1365-313x.2000.00864.x
Jiang, L., Pan, L.-J. (2012). Identification and expression of C2H2 transcription factor genes in Carica papaya under abiotic and biotic stresses. Mol. Biol. Rep. 39, 7105–7115. doi: 10.1007/s11033-012-1542-y
Jiang, Z., Zhou, X., Tao, M., Yuan, F., Liu, L., Wu, F., et al. (2019). Plant cell-surface GIPC sphingolipids sense salt to trigger Ca 2+ influx. Nature 572, 341–346. doi: 10.1038/s41586-019-1449-z
Kazan, K. (2006). Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 11, 109–112. doi: 10.1016/j.tplants.2006.01.004
Kiełbowicz-Matuk, A. (2012). Involvement of plant C2H2-type zinc finger transcription factors in stress responses. Plant Sci. 185, 78–85. doi: 10.1016/j.plantsci.2011.11.015
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363. doi: 10.1111/j.1365-313X.2007.03052.x
Kim, J. C., Lee, S. H., Cheong, Y. H., Yoo, C. M., Lee, S. I., Chun, H. J., et al. (2001). A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J. 25, 247–259. doi: 10.1046/j.1365-313x.2001.00947.x
Kim, Y.-H., Kim, M. D., Park, S.-C., Yang, K.-S., Jeong, J. C., Lee, H.-S., et al. (2011). SCOF-1-expressing transgenic sweetpotato plants show enhanced tolerance to low-temperature stress. Plant Physiol. Biochem. 49, 1436–1441. doi: 10.1016/j.plaphy.2011.09.002
Kim, G.-D., Cho, Y.-H., Yoo, S.-D. (2015). Regulatory functions of evolutionarily conserved AN1/A20-like Zinc finger family proteins in Arabidopsis stress responses under high temperature. Biochem. Biophys. Res. Commun. 457, 213–220. doi: 10.1016/j.bbrc.2014.12.090
Kim, Y. H., Kim, M. D., Park, S. C., Jeong, J. C., Kwak, S. S., Lee, H. S. (2016). Transgenic potato plants expressing the cold-inducible transcription factor SCOF-1 display enhanced tolerance to freezing stress. Plant Breed. 135, 513–518. doi: 10.1111/pbr.12390
Klug, A., Schwabe, J. (1995). Protein motifs 5. Zinc fingers. FASEB J. 9, 597–604. doi: 10.1096/fasebj.9.8.7768350
Klug, A. (2010). The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 79, 213–231. doi: 10.1146/annurev-biochem-010909-095056
Knight, H., Knight, M. R. (2001). Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci. 6, 262–267. doi: 10.1016/S1360-1385(01)01946-X
Kodaira, K.-S., Qin, F., Tran, L.-S. P., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2011). Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 157, 742–756. doi: 10.1104/pp.111.182683
Kubo, K.-I., Sakamoto, A., Kobayashi, A., Rybka, Z., Kanno, Y., Nakagawa, H., et al. (1998). Cys2/His2 zinc-finger protein family of petunia: evolution and general mechanism of target-sequence recognition. Nucleic Acids Res. 26, 608–615. doi: 10.1093/nar/26.2.608
Kumari, P., Kumari, R., Sharma, B., Prasad, S. K., Singh, R. K. (2019). Abiotic Stress Response of Field Crops: Recent Approach. Int. J. Curr. Microbiol. App. Sci. 8, 1761–1769. doi: 10.20546/ijcmas.2019.804.205
Lata, C., Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Li, G., Tai, F.-J., Zheng, Y., Luo, J., Gong, S.-Y., Zhang, Z.-T., et al. (2010). Two cotton Cys2/His2-type zinc-finger proteins, GhDi19-1 and GhDi19-2, are involved in plant response to salt/drought stress and abscisic acid signaling. Plant Mol. Biol. 74, 437–452. doi: 10.1007/s11103-010-9684-6
Li, Y., Chu, Z., Luo, J., Zhou, Y., Cai, Y., Lu, Y., et al. (2018). The C2H2 zinc-finger protein Sl ZF 3 regulates AsA synthesis and salt tolerance by interacting with CSN 5B. Plant Biotechnol. J. 16, 1201–1213. doi: 10.1111/pbi.12863
Lin, C.-Y., Lin, L.-Y. (2018). The conserved basic residues and the charged amino acid residues at the α-helix of the zinc finger motif regulate the nuclear transport activity of triple C2H2 zinc finger proteins. PloS One 13, e0191971. doi: 10.1371/journal.pone.0191971
Lippuner, V., Cyert, M. S., Gasser, C. S. (1996). Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J. Biol. Chem. 271, 12859–12866. doi: 10.1074/jbc.271.22.12859
Liu, Q.-L., Xu, K.-D., Zhong, M., Pan, Y.-Z., Jiang, B.-B., Liu, G.-L., et al. (2013a). Overexpression of a novel chrysanthemum Cys2/His2-type zinc finger protein gene DgZFP3 confers drought tolerance in tobacco. Biotechnol. Lett. 35, 1953–1959. doi: 10.1007/s10529-013-1289-0
Liu, X.-M., Nguyen, X. C., Kim, K. E., Han, H. J., Yoo, J., Lee, K., et al. (2013b). Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochem. Biophys. Res. Commun. 430, 1054–1059. doi: 10.1016/j.bbrc.2012.12.039
Liu, M., Li, M., Liu, K., Sui, N. (2015a). Effects of drought stress on seed germination and seedling growth of different maize varieties. J. Agric. Sci. 7, 231. doi: 10.5539/jas.v7n5p231
Liu, Q., Wang, Z., Xu, X., Zhang, H., Li, C. (2015b). Genome-wide analysis of C2H2 zinc-finger family transcription factors and their responses to abiotic stresses in poplar (Populus trichocarpa). PloS One 10, e0134753. doi: 10.1371/journal.pone.0134753
Liu, X. X., Fu, C., Yang, W. W., Zhang, Q., Fan, H., Liu, J. (2016). The involvement of TsFtsH8 in Thellungiella salsuginea tolerance to cold and high light stresses. Acta physiologiae plant. 38, 62. doi: 10.1007/s11738-016-2080-3
Liu, D., Yang, L., Luo, M., Wu, Q., Liu, S., Liu, Y. (2017). Molecular cloning and characterization of PtrZPT2-1, a ZPT2 family gene encoding a Cys2/His2-type zinc finger protein from trifoliate orange (Poncirus trifoliata (L.) Raf.) that enhances plant tolerance to multiple abiotic stresses. Plant Sci. 263, 66–78. doi: 10.1016/j.plantsci.2017.07.012
Liu, Q., Liu, R., Ma, Y., Song, J. (2018). Physiological and molecular evidence for Na+ and Cl– exclusion in the roots of two Suaeda salsa populations. Aquat. Bot. 146, 1–7. doi: 10.1016/j.aquabot.2018.01.001
Luo, X., Bai, X., Zhu, D., Li, Y., Ji, W., Cai, H., et al. (2012a). GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress. Planta 235, 1141–1155. doi: 10.1007/s00425-011-1563-0
Luo, X., Cui, N., Zhu, Y., Cao, L., Zhai, H., Cai, H., et al. (2012b). Over-expression of GsZFP1, an ABA-responsive C2H2-type zinc finger protein lacking a QALGGH motif, reduces ABA sensitivity and decreases stomata size. J. Plant Physiol. 169, 1192–1202. doi: 10.1016/j.jplph.2012.03.019
Lyu, T., Cao, J. (2018). Cys2/His2 zinc-finger proteins in transcriptional regulation of flower development. Int. J. Mol. Sci. 19, 2589. doi: 10.3390/ijms19092589
Ma, Y., Dai, X., Xu, Y., Luo, W., Zheng, X., Zeng, D., et al. (2015). COLD1 confers chilling tolerance in rice. Cell 160, 1209–1221. doi: 10.1016/j.cell.2015.01.046
Ma, X., Liang, W., Gu, P., Huang, Z. (2016). Salt tolerance function of the novel C2H2-type zinc finger protein TaZNF in wheat. Plant Physiol. Biochem. 106, 129–140. doi: 10.1016/j.plaphy.2016.04.033
Mackay, J. P., Crossley, M. (1998). Zinc fingers are sticking together. Trends Biochem. Sci. 23, 1–4. doi: 10.1016/S0968-0004(97)01168-7
Mcbryant, S. J., Veldhoen, N., Gedulin, B., Leresche, A., Foster, M. P., Wright, P. E., et al. (1995). Interaction of the RNA binding Fingers ofXenopusTranscription Factor IIIA with Specific Regions of 5 S Ribosomal RNA. J. Mol. Biol. 248, 44–57. doi: 10.1006/jmbi.1995.0201
Meng, X., Zhou, J., Sui, N. (2018). Mechanisms of salt tolerance in halophytes: current understanding and recent advances. Open Life Sci. 13, 149–154. doi: 10.1515/biol-2018-0020
Miller, J., Mclachlan, A., Klug, A. (1985). Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4, 1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x
Miller, G., Shulaev, V., Mittler, R. (2008). Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133, 481–489. doi: 10.1111/j.1399-3054.2008.01090.x
Mittler, R., Kim, Y., Song, L., Coutu, J., Coutu, A., Ciftci-Yilmaz, S., et al. (2006). Gain-and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580, 6537–6542. doi: 10.1016/j.febslet.2006.11.002
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Najafabadi, H. S., Mnaimneh, S., Schmitges, F. W., Garton, M., Lam, K. N., Yang, A., et al. (2015). C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 33, 555. doi: 10.1038/nbt.3128
Nakashima, K., Yamaguchi-Shinozaki, K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Nguyen, X. C., Kim, S. H., Lee, K., Kim, K. E., Liu, X.-M., Han, H. J., et al. (2012). Identification of a C 2 H 2-type zinc finger transcription factor (ZAT10) from Arabidopsis as a substrate of MAP kinase. Plant Cell Rep. 31, 737–745. doi: 10.1007/s00299-011-1192-x
Nguyen, X. C., Kim, S. H., Hussain, S., An, J., Yoo, Y., Han, H. J., et al. (2016). A positive transcription factor in osmotic stress tolerance, ZAT10, is regulated by MAP kinases in Arabidopsis. J. Plant Biol. 59, 55–61. doi: 10.1007/s12374-016-0442-4
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. doi: 10.1105/TPC.010127
Onishi, M., Tachi, H., Kojima, T., Shiraiwa, M., Takahara, H. (2006). Molecular cloning and characterization of a novel salt-inducible gene encoding an acidic isoform of PR-5 protein in soybean (Glycine max [L.] Merr.). Plant Physiol. Biochem. 44, 574–580. doi: 10.1016/j.plaphy.2006.09.009
Pareek, A., Khurana, A., K Sharma, A., Kumar, R. (2017). An overview of signaling regulons during cold stress tolerance in plants. Curr. Genomics 18, 498–511. doi: 10.2174/1389202918666170228141345
Persikov, A. V., Wetzel, J. L., Rowland, E. F., Oakes, B. L., Xu, D. J., Singh, M., et al. (2015). A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic Acids Res. 43, 1965–1984. doi: 10.1093/nar/gku1395
Rai, A. C., Singh, M., Shah, K. (2013). Engineering drought tolerant tomato plants over-expressing BcZAT12 gene encoding a C2H2 zinc finger transcription factor. Phytochemistry 85, 44–50. doi: 10.1016/j.phytochem.2012.09.007
Rizhsky, L., Davletova, S., Liang, H., Mittler, R. (2004). The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 279, 11736–11743. doi: 10.1074/jbc.M313350200
Ryu, H., Cho, Y.-G. (2015). Plant hormones in salt stress tolerance. J. Plant Biol. 58, 147–155. doi: 10.1007/s12374-015-0103-z
Sakai, H., Medrano, L. J., Meyerowitz, E. M. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378, 199–203. doi: 10.1038/378199a0
Sakamoto, H., Araki, T., Meshi, T., Iwabuchi, M. (2000). Expression of a subset of the Arabidopsis Cys2/His2-type zinc-finger protein gene family under water stress. Gene 248, 23–32. doi: 10.1016/S0378-1119(00)00133-5
Sakamoto, H., Maruyama, K., Sakuma, Y., Meshi, T., Iwabuchi, M., Shinozaki, K., et al. (2004). Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136, 2734–2746. doi: 10.1104/pp.104.046599
Santos, J., Al-Azzawi, M., Aronson, J., Flowers, T. J. (2015). eHALOPH a database of salt-tolerant plants: helping put halophytes to work. Plant Cell Physiol. 57, e10–e10. doi: 10.1093/pcp/pcv155
Schmidt, R., Schippers, J. H., Welker, A., Mieulet, D., Guiderdoni, E., Mueller-Roeber, B. (2012). Transcription factor OsHsfC1b regulates salt tolerance and development in Oryza sativa ssp. japonica. AoB Plants 2012, pls011. doi: 10.1093/aobpla/pls011
Shi, Y., Berg, J. M. (1995). Specific DNA-RNA hybrid binding by zinc finger proteins. Science 268, 282–284. doi: 10.1126/science.7536342
Shi, Y., Yang, S. (2015). COLD1: a cold sensor in rice. Sci. China. Life Sci. 58, 409. doi: 10.1007/s11427-015-4831-6
Shi, H., Wang, X., Ye, T., Cheng, F., Deng, J., Yang, P., et al. (2014). The Cys2/His2-type zinc finger transcription factor ZAT6 modulates biotic and abiotic stress responses by activating salicylic acid-related genes and CBFs in Arabidopsis. Plant Physiol. pp, 114.242404. doi: 10.1104/pp.114.242404
Shimeld, S. M. (2008). C2H2 zinc finger genes of the Gli, Zic, KLF, SP, Wilms' tumour, Huckebein, Snail, Ovo, Spalt, Odd, Blimp-1, Fez and related gene families from Branchiostoma floridae. Dev. Genes Evol. 218, 639–649. doi: 10.1007/s00427-008-0248-6
Shinozaki, K., Yamaguchi-Shinozaki, K., Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. In Plant Biol. 6, 410–417. doi: 10.1016/S1369-5266(03)00092-X
Singh, A., Giri, J., Kapoor, S., Tyagi, A. K., Pandey, G. K. (2010). Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics 11, 435. doi: 10.1186/1471-2164-11-435
Song, B., Hong, Y., Wang, P., Wang, H., Fu, Y., Ding, X. (2010). Advances on plant C2H2-type zinc finger protein. Genomics Appl. Biol. 29, 1133–1141.
Song, J., Shi, G., Gao, B., Fan, H., Wang, B. (2011). Waterlogging and salinity effects on two Suaeda salsa populations. Physiol. Plant. 141, 343–351. doi: 10.1111/j.1399-3054.2011.01445.x
Sugano, S., Kaminaka, H., Rybka, Z., Catala, R., Salinas, J., Matsui, K., et al. (2003). Stress-responsive zinc finger gene ZPT2-3 plays a role in drought tolerance in petunia. Plant J. 36, 830–841. doi: 10.1046/j.1365-313X.2003.01924.x
Sui, N. (2015). Photoinhibition of Suaeda salsa to chilling stress is related to energy dissipation and water-water cycle. Photosynthetica 53, 207–212. doi: 10.1007/s11099-015-0080-y
Sui, N., Li, M., Li, K., Song, J., Wang, B.-S. (2010). Increase in unsaturated fatty acids in membrane lipids of Suaeda salsa L. enhances protection of photosystem II under high salinity. Photosynthetica 48, 623–629. doi: 10.1007/s11099-010-0080-x
Sun, S.-J., Guo, S.-Q., Yang, X., Bao, Y.-M., Tang, H.-J., Sun, H., et al. (2010). Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J. Exp. Bot. 61, 2807–2818. doi: 10.1093/jxb/erq120
Sun, Z.-H., Ding, C.-H., Li, X.-J., Kai, X. (2012). Molecular characterization and expression analysis of TaZFP15, a C2H2-type zinc finger transcription factor gene in wheat (Triticum aestivum L.). J. Integr. Agric. 11, 31–42. doi: 10.1016/S1671-2927(12)60780-9
Sun, X., Wang, Y., Sui, N. (2018). Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 503, 397–401. doi: 10.1016/j.bbrc.2018.07.123
Sun, B., Zhao, Y., Shi, S., Yang, M., Xiao, K. (2019a). TaZFP1, a C2H2 type-ZFP gene of T. aestivum, mediates salt stress tolerance of plants by modulating diverse stress-defensive physiological processes. Plant Physiol. Biochem. 136, 127–142. doi: 10.1016/j.plaphy.2019.01.014
Sun, Z., Liu, R., Guo, B., Huang, K., Wang, L., Han, Y., et al. (2019b). Ectopic expression of GmZAT4, a putative C2H2-type zinc finger protein, enhances PEG and NaCl stress tolerances in Arabidopsis thaliana. Biotech 9, 166. doi: 10.1007/s13205-019-1673-0
Takahashi, S., Katagiri, T., Hirayama, T., Yamaguchi-Shinozaki, K., Shinozaki, K. (2001). Hyperosmotic stress induces a rapid and transient increase in inositol 1, 4, 5-trisphosphate independent of abscisic acid in Arabidopsis cell culture. Plant Cell Physiol. 42, 214–222. doi: 10.1093/pcp/pce028
Takatsuji, H. (1999). Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39, 1073–1078. doi: 10.1023/A:1006184519697
Takatsuji, H., Mori, M., Benfey, P. N., Ren, L., Chua, N. H. (1992). Characterization of a zinc finger DNA-binding protein expressed specifically in Petunia petals and seedlings. EMBO J. 11, 241. doi: 10.1002/j.1460-2075.1992.tb05047.x
Takatsuji, H., Nakamura, N., Katsumoto, Y. (1994). A new family of zinc finger proteins in petunia: structure, DNA sequence recognition, and floral organ-specific expression. Plant Cell 6, 947–958. doi: 10.1105/tpc.6.7.947
Tang, L., Cai, H., Ji, W., Luo, X., Wang, Z., Wu, J., et al. (2013). Overexpression of GsZFP1 enhances salt and drought tolerance in transgenic alfalfa (Medicago sativa L.). Plant Physiol. Biochem. 71, 22–30. doi: 10.1016/j.plaphy.2013.06.024
Tang, T., Du, C., Song, H., Aziz, U., Wang, L., Zhao, C., et al. (2019). Genome-wide analysis reveals the evolution and structural features of WRINKLED1 in plants. Mol. Genet. Genomics 294, 329–341. doi: 10.1007/s00438-018-1512-8
Teng, K., Tan, P., Guo, W., Yue, Y., Fan, X., Wu, J. (2018). Heterologous expression of a novel Zoysia japonica C2H2 zinc finger gene, ZjZFN1, improved salt tolerance in Arabidopsis. Front. Plant Sci. 9, 1159. doi: 10.3389/fpls.2018.01159
Tian, Z. D., Zhang, Y., Liu, J., Xie, C. H. (2010). Novel potato C2H2-type zinc finger protein gene, StZFP1, which responds to biotic and abiotic stress, plays a role in salt tolerance. Plant Biol. 12, 689–697. doi: 10.1111/j.1438-8677.2009.00276.x
Tibbles, L., Woodgett, J. R. (1999). The stress-activated protein kinase pathways. Cell. Mol. Life Sci. CMLS 55, 1230–1254. doi: 10.1007/s000180050369
Vogel, J. T., Zarka, D. G., Van Buskirk, H. A., Fowler, S. G., Thomashow, M. F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41, 195–211. doi: 10.1111/j.1365-313X.2004.02288.x
Wang, F., Tong, W., Zhu, H., Kong, W., Peng, R., Liu, Q., et al. (2016). A novel Cys 2/His 2 zinc finger protein gene from sweetpotato, IbZFP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Planta 243, 783–797. doi: 10.1007/s00425-015-2443-9
Wang, J., Zhang, Q., Cui, F., Hou, L., Zhao, S., Xia, H., et al. (2017). Genome-wide analysis of gene expression provides new insights into cold responses in Thellungiella salsuginea. Front. In Plant Sci. 8, 713. doi: 10.3389/fpls.2017.00713
Wang, K., Ding, Y., Cai, C., Chen, Z., Zhu, C. (2019). The role of C2H2 zinc finger proteins in plant responses to abiotic stresses. Physiol. plant. 165, 690–700. doi: 10.1111/ppl.12728
Wasternack, C., Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Weng, L., Zhao, F., Li, R., Xu, C., Chen, K., Xiao, H. (2015). The zinc finger transcription factor SlZFP2 negatively regulates abscisic acid biosynthesis and fruit ripening in tomato. Plant Physiol. 167, 931–949. doi: 10.1104/pp.114.255174
Wimalasekera, R., Scherer, G. F. (2018). “Involvement of mitogen-activated protein kinases in abiotic stress responses in plants,” in Plant Metabolites and Regulation Under Environmental Stress (Elsevier), 389–395. doi: 10.1016/B978-0-12-812689-9
Wolfe, S. A., Nekludova, L., Pabo, C. O. (2000). DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. biophys. biomol. struct. 29, 183–212. doi: 10.1146/annurev.biophys.29.1.183
Xiong, L., Schumaker, K. S., Zhu, J.-K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14, S165–S183. doi: 10.1105/tpc.000596
Xu, S., Wang, X., Chen, J. (2007). Zinc finger protein 1 (ThZF1) from salt cress (Thellungiella halophila) is a Cys-2/His-2-type transcription factor involved in drought and salt stress. Plant Cell Rep. 26, 497–506. doi: 10.1007/s00299-006-0248-9
Xu, D.-Q., Huang, J., Guo, S.-Q., Yang, X., Bao, Y.-M., Tang, H.-J., et al. (2008). Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.). FEBS Lett. 582, 1037–1043. doi: 10.1016/j.febslet.2008.02.052
Yamaguchi-Shinozaki, K., Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yamaguchi-Shinozaki, K., Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yang, J. C., Li, M., Xie, X. Z., Han, G. L., Sui, N., Wang, B. S. (2013). Deficiency of phytochrome B alleviates chilling-induced photoinhibition in rice. Am. J. Bot. 100, 1860–1870. doi: 10.3732/ajb.1200574
Yang, Y., Ma, C., Xu, Y., Wei, Q., Imtiaz, M., Lan, H., et al. (2014). A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 26, 2038–2054. doi: 10.1105/tpc.114.124867
Yang, Z., Zheng, H., Wei, X., Song, J., Wang, B., Sui, N. (2018). Transcriptome analysis of sweet sorghum inbred lines differing in salt tolerance provides novel insights into salt exclusion by roots. Plant Soil 430, 423–439. doi: 10.1007/s11104-018-3736-0
Yang, B., Tang, J., Yu, Z., Khare, T., Srivastav, A., Datir, S., et al. (2019). Light Stress Responses and Prospects for Engineering Light Stress Tolerance in Crop Plants. J. Plant Growth Regul. 38, 1489–1506. doi: 10.1007/s00344-019-09951-8
Yengo, R. K., Nurmemmedov, E., Thunnissen, M. M. (2018). Structural view on the role of WT1's zinc finger 1 in DNA binding. BioRxiv, 284489. doi: 10.1101/284489
Yin, M., Wang, Y., Zhang, L., Li, J., Quan, W., Yang, L., et al. (2017). The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J. Exp. Bot. 68, 2991–3005. doi: 10.1093/jxb/erx157
Yu, G.-H., Jiang, L.-L., Ma, X.-F., Xu, Z.-S., Liu, M.-M., Shan, S.-G., et al. (2014). A soybean C2H2-type zinc finger gene GmZF1 enhanced cold tolerance in transgenic Arabidopsis. PloS One 9, e109399. doi: 10.1371/journal.pone.0109399
Yuan, F., Yang, H., Xue, Y., Kong, D., Ye, R., Li, C., et al. (2014). OSCA1 mediates osmotic-stress-evoked Ca 2+ increases vital for osmosensing in Arabidopsis. Nature 514, 367. doi: 10.1038/nature13593
Yuan, F., Chen, M., Yang, J., Song, J., Wang, B. (2015a). The optimal dosage of 60 co gamma irradiation for obtaining salt gland mutants of exo-recretohalophyte limonium bicolor (bunge) o. Kuntze. Pak J. Bot. 47, 71–76.
Yuan, F., Lyu, M. J., Leng, B. Y., Zheng, G. Y., Feng, Z. T., Li, P. H., et al. (2015b). Comparative Transcriptome Analysis of Developmental Stages of the Limonium bicolor Leaf Generates Insights into Salt Gland Differentiation. Plant Cell Environ. 38, 1637–1657. doi: 10.1111/pce.12514
Zang, D., Li, H., Xu, H., Zhang, W., Zhang, Y., Shi, X., et al. (2016). An Arabidopsis zinc finger protein increases abiotic stress tolerance by regulating sodium and potassium homeostasis, reactive oxygen species scavenging and osmotic potential. Front. Plant Sci. 7, 1272. doi: 10.3389/fpls.2016.01272
Zhang, X., Guo, X., Lei, C., Cheng, Z., Lin, Q., Wang, J., et al. (2011). Overexpression of SlCZFP1, a novel TFIIIA-type zinc finger protein from tomato, confers enhanced cold tolerance in transgenic Arabidopsis and rice. Plant Mol. Biol. Rep. 29, 185–196. doi: 10.1007/s11105-010-0223-z
Zhang, H., Liu, Y., Wen, F., Yao, D., Wang, L., Guo, J., et al. (2014). A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J. Exp. Bot. 65, 5795–5809. doi: 10.1093/jxb/eru313
Zhang, A., Liu, D., Hua, C., Yan, A., Liu, B., Wu, M., et al. (2016a). The Arabidopsis Gene zinc finger protein 3 (ZFP3) is involved in salt stress and osmotic stress response. PloS One 11, e0168367. doi: 10.1371/journal.pone.0168367
Zhang, D., Tong, J., Xu, Z., Wei, P., Xu, L., Wan, Q., et al. (2016b). Soybean C2H2-type zinc finger protein GmZFP3 with conserved QALGGH motif negatively regulates drought responses in transgenic Arabidopsis. Front. Plant Sci. 7, 325. doi: 10.3389/fpls.2016.00325
Zhang, X., Zhang, B., Li, M. J., Yin, X. M., Huang, L. F., Cui, Y. C., et al. (2016c). OsMSR15 encoding a rice C2H2-type zinc finger protein confers enhanced drought tolerance in transgenic Arabidopsis. J. Plant Biol. 59, 271–281. doi: 10.1007/s12374-016-0539-9
Zhang, Z., Liu, H., Sun, C., Ma, Q., Bu, H., Chong, K., et al. (2018). A C2H2 zinc-finger protein OsZFP213 interacts with OsMAPK3 to enhance salt tolerance in rice. J. Plant Physiol. 229, 100–110. doi: 10.1016/j.jplph.2018.07.003
Zhou, Z., An, L., Sun, L., Zhu, S., Xi, W., Broun, P., et al. (2011). Zinc finger protein5 is required for the control of trichome initiation by acting upstream of zinc finger protein8 in Arabidopsis. Plant Physiol. 157, 673–682. doi: 10.1104/pp.111.180281
Zhou, Z., Sun, L., Zhao, Y., An, L., Yan, A., Meng, X., et al. (2013). Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol. 198, 699–708. doi: 10.1111/nph.12211
Zhou, J., Fu, T., Sui, N., Guo, J., Feng, G., Fan, J., et al. (2016). The role of salinity in seed maturation of the euhalophyte Suaeda salsa. Plant Biosystems-An Int. J. Dealing all Aspects Plant Biol. 150, 83–90. doi: 10.1080/11263504.2014.976294
Zhu, J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247. doi: 10.1146/annurev.arplant.53.091401.143329
Keywords: abiotic stress, adaptation mechanism, C2H2 zinc finger proteins, plant, signaling pathways, stress response networks
Citation: Han G, Lu C, Guo J, Qiao Z, Sui N, Qiu N and Wang B (2020) C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 11:115. doi: 10.3389/fpls.2020.00115
Received: 03 October 2019; Accepted: 24 January 2020;
Published: 20 February 2020.
Edited by:
Vicent Arbona, University of Jaume I, SpainReviewed by:
Yong Hwa Cheong, Sunchon National University, South KoreaCopyright © 2020 Han, Lu, Guo, Qiao, Sui, Qiu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baoshan Wang, YnN3YW5nQHNkbnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.