
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 13 February 2020
Sec. Plant Biotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00056
This article is part of the Research Topic CRISPR-Cas in Agriculture: Opportunities and Challenges View all 12 articles
The terms genome engineering, genome editing, and gene editing, refer to modifications (insertions, deletions, substitutions) in the genome of a living organism. The most widely used approach to genome editing nowadays is based on Clustered Regularly Interspaced Short Palindromic Repeats and associated protein 9 (CRISPR-Cas9). In prokaryotes, CRISPR-Cas9 is an adaptive immune system that naturally protects cells from DNA virus infections. CRISPR-Cas9 has been modified to create a versatile genome editing technology that has a wide diversity of applications in medicine, agriculture, and basic studies of gene functions. CRISPR-Cas9 has been used in a growing number of monocot and dicot plant species to enhance yield, quality, and nutritional value, to introduce or enhance tolerance to biotic and abiotic stresses, among other applications. Although biosafety concerns remain, genome editing is a promising technology with potential to contribute to food production for the benefit of the growing human population. Here, we review the principles, current advances and applications of CRISPR-Cas9-based gene editing in crop improvement. We also address biosafety concerns and show that humans have been exposed to Cas9 protein homologues long before the use of CRISPR-Cas9 in genome editing.
The world population is predicted to reach 10 billion by 2050. While the available farm land and water are being reduced, the global demand for food will increase 25%–70% above current production levels (Hunter et al., 2017). Thus, feeding a rapidly growing population, particularly in the face of climate change, is a big challenge. There is, therefore, an urgent need to improve food production and accelerate sustainable agricultural development.
Long time before the field of genetics was established, humans genetically modified plants through breeding and selection. Without knowledge of genes, mutagenesis, or gene editing, our ancestors influenced the genetic make-up of plants and animals by selecting for traits conducive to food production (Wang et al., 1999; Clark et al., 2005; Li et al., 2013b). A prime example is maize (Zea mays subsp. mays L.), which is one of the most produced cereals worldwide. Molecular, cytological, and isozyme profiles have shown that maize is a descendant of an annual species of teosinte (Zea mays ssp. parviglumis) native to the Balsas River Valley on the Pacific slopes of the states of Michoacán and Guerrero, Mexico. The process started approximately 9,000 years ago. Teosinte has a popping ability that provided an incentive for its cultivation. Repetitive cycles of selection for traits conducive to kernel production led to the development of the maize plant as we know it (Doebley et al., 1990; Dorweiler et al., 1993; Piperno and Flannery, 2001).
To make plant breeding faster, more predictable, and amendable to a wide range of species, several techniques of plant genetic engineering have been developed. Genome editing through programmable endonucleases is the most recent approach to genetic engineering. Endonucleases are used to specifically induce double strand breaks in target genes of interest. The cellular DNA repair pathway then acts on the double strand break to restore the damage through non-homologous end joining (NHEJ) or homology-directed repair (HDR). In the process, insertions, deletions, substitutions, and DNA recombination may occur (Puchta et al., 1996; Puchta, 2005; Symington and Gautier, 2011).
Three kinds of programmable endonucleases are currently being used for plant genome editing. Zinc finger nucleases, transcription activator-like effector nucleases (TALENs), and CRISPR-Cas9 (Malzahn et al., 2017; Shah et al., 2018; Zhang et al., 2018a; Bao et al., 2019). Zinc finger nucleases are chimeric proteins composed of a synthetic zinc finger DNA binding domain and a DNA cleavage domain. The zinc finger DNA binding domain can be modified to specifically target any long stretch of double stranded DNA of interest (Kim et al., 1996; Cathomen and Joung, 2008). Zinc finger nucleases have been used to edit the genomes of several species, including maize, rice and Arabidopsis (Shukla et al., 2009; Osakabe et al., 2010; Ainley et al., 2013; Gallego-Bartolome et al., 2019).
TALENs are sequence-specific nucleases consisting of transcription activator-like effectors fused to the catalytic domain of the FokI endonuclease (Boch et al., 2009; Christian et al., 2010). The DNA-binding domain in TALE monomers in turn is comprised of a central repeat domain (CRD) that directs DNA binding and host specificity. The CRD is formed by tandem repeats of 34 amino acid residues, each binding to one nucleotide in the target nucleotide sequence which allows more flexible target design and increases the number of potential target sites relative to those that can be targeted by zinc finger nucleases (Moscou and Bogdanove, 2009). Genome editing by TALENs has been demonstrated in a wide variety of plants including Arabidopsis (Christian et al., 2013), barley (Budhagatapalli et al., 2015), Brachypodium (Shan et al., 2013), maize (Char et al., 2015), tobacco (Zhang et al., 2013), potato (Clasen et al., 2016; Nicolia et al., 2015), rice (Li et al., 2012; Shan et al., 2013; Shan et al., 2015), soybean (Du et al., 2016), sugarcane (Jung and Altpeter, 2016), tomato (Lor et al., 2014), and wheat (Liang et al., 2014).
The CRISPR-Cas9 system consists of a programmable Cas9 nuclease and a synthetic short guide RNA (sgRNA). DNA target specificity is provided by the guide RNA (Figure 1). Thus, the CRISPR-Cas9 system is much easier to be constructed than Zinc finger or TALENs, simple, efficient, has low cost and allows the targeting of multiple genes at once (Cong et al., 2013; Mali et al., 2013).
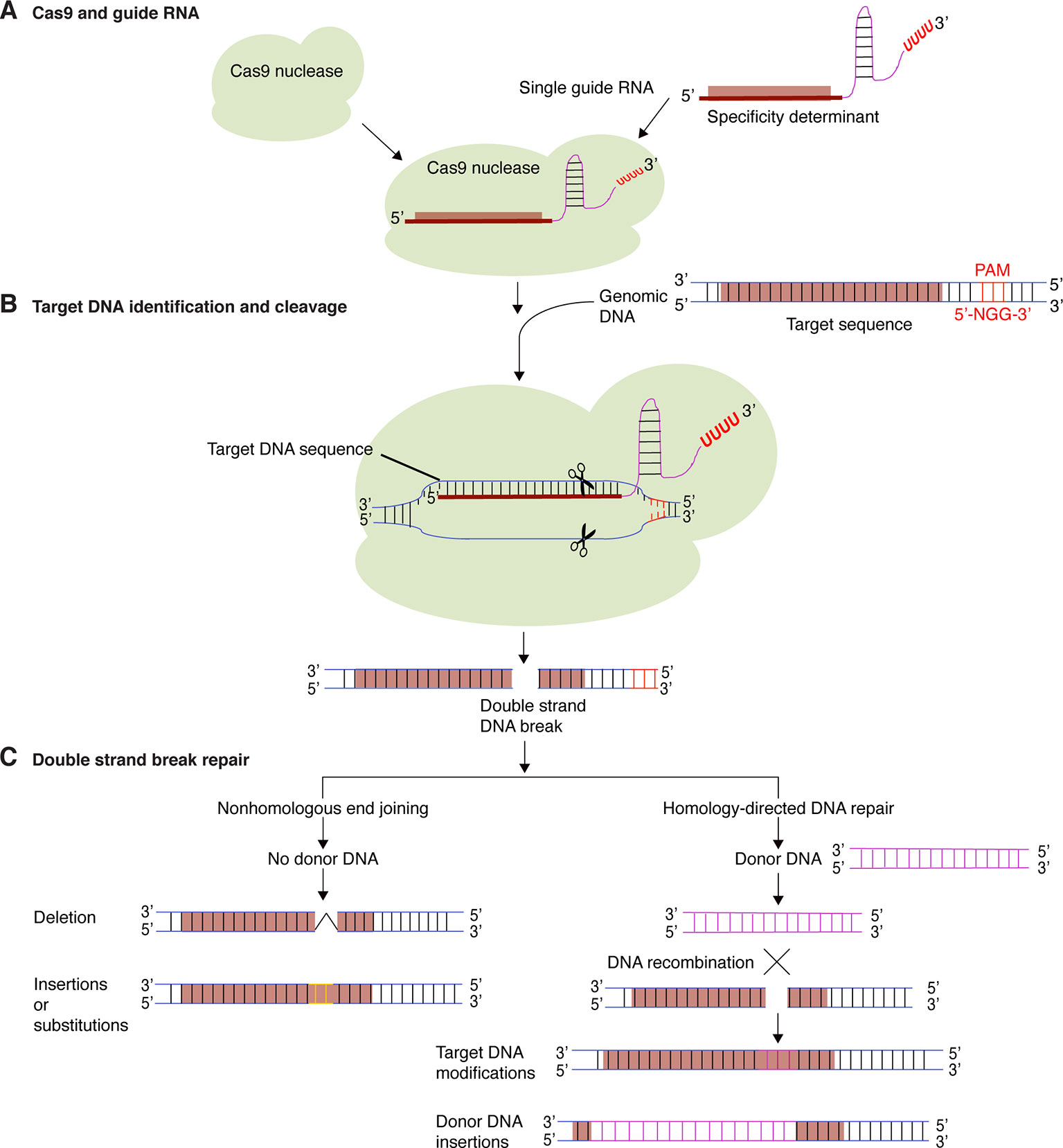
Figure 1 Targeted genome editing using CRISPR-Cas9. (A) The CRISPR-Cas9 system consists of a Cas9 protein and one or several guide RNA. Guide RNAs determine target DNA specificity by sequence complementarity. (B) Guide RNA and Cas9 protein form a binary complex that specifically cleaves target DNA creating a double-strand DNA break. (C) Cellular DNA repair mechanisms, non-homologous end joining (NHEJ) and homology-directed repair (HDR), repairs the double-strand DNA break. In the process, short insertions, deletions, nucleotide substitutions, or gene insertion may occur.
The CRISPR-Cas9 system has a wide diversity of applications. In medicine, it has been applied in research related to cancer, virus infections, genetic diseases and detection of pathogens. This system has been successfully used in mice to correct mutations in monogenic diseases (Schwank et al., 2013; Ye et al., 2014; Yin et al., 2014; Kang et al., 2015), including the one responsible for Duchenne muscular dystrophy (DMD) (Long et al., 2016; Nelson et al., 2016; Tabebordbar et al., 2016). CRISPR-Cas9 has also been used to disrupt HIV-1 provirus (Ebina et al., 2013), human papillomaviruses (Kennedy et al., 2014) and hepatitis B virus (Kennedy et al., 2015). Furthermore, CRISPR-Cas9 has also been used to target human hereditary liver diseases (Yang et al., 2016; Yin et al., 2016) and has shown great promise for the treatment of cancer (Chen et al., 2019) and Hutchinson–Gilford progeria syndrome (Beyret et al., 2019). In human cells, CRISPR-Cas9 has been used successfully to replace endogenously-encoded antibodies with antibodies protective against Respiratory Syncytial Virus (RSV), human immunodeficiency virus (HIV), influenza virus, and Epstein-Barr virus (EBV) (Moffett et al., 2019). This review is focused on applications of CRISPR-Cas9 in crop improvement.
Clustered regularly interspaced short palindromic repeats (CRISPR) are a family of DNA sequences found in the genomes of bacteria and archaea. CRISPRs were first discovered downstream of the alkaline phosphatase isozyme gene (iap) in Escherichia coli (Ishino et al., 1987). Palindromic repeats are separated by short (32 to 36 bp) sequences derived from the DNA of viruses that have previously infected the cell or its predecessors. These virus-derived sequences integrated into the bacterial genome provide a memory system of previous virus infection. Once integrated into the genome, CRISPRs are transcribed and the virus-derived sequences form short guide RNAs that are bound by CRISPR associated protein 9 (Cas9). Cas9 is a DNA endonuclease. In bacteria and archaea, the natural role of the CRISPR-Cas9 system is to provide adaptive antiviral immunity against DNA viruses. Binary complexes formed by guide RNA-Cas9 recognize and cleave DNA of incoming viruses with sequence similarity to the guide RNA (Garneau et al., 2010; Horvath and Barrangou, 2010; Jinek et al., 2012; Sternberg et al., 2014).
There are several CRISPR-Cas systems in bacteria and archaea. For a comprehensive review, see (Karginov and Hannon, 2010; Sorek et al., 2013). The CRISPR-Cas9 system most frequently used in plant genome editing is an adaptation of the type II CRISPR-Cas system of Streptococcus pyogenes (Garneau et al., 2010). S. pyogenes is a Gram-positive human-restricted pathogen that colonizes the pharynx and the skin causing an array of diseases ranging from mild sore throat and impetigo to invasive and life-threatening infections (Cunningham, 2000; Rosinski-Chupin et al., 2019). Type II CRISPR-Cas system consists of a Cas9 protein with DNA endonuclease activity and one CRISPR RNA transcript that is processed to form one or several short guide RNAs that direct Cas9 to the target DNA sequence (Figure 1) (Jinek et al., 2012; Lander, 2016; Jiang and Doudna, 2017). In the cell, Cas9 binds to the guide RNA and forms a binary complex that scans the genome for the DNA target for cleavage using Watson-Crick base pairing. The specificity is determined by the guide RNA. Cas9 also requires a specific proto-spacer adjacent motif (PAM) localized on the non-target DNA strand, directly downstream of the target DNA sequence (Figure 1B). Cas9 from S. pyogenes recognizes NGG as a PAM (Anders et al., 2014; Lander, 2016; Jiang and Doudna, 2017). Cas9 proteins have two signature nuclease domains: HNH and RuvC. The HNH-like nuclease domain cleaves the target DNA strand complementary to the guide RNA sequence. The RuvC-like nuclease domain cleaves the non-target strand (Jinek et al., 2012; Gao et al., 2017; Jiang and Doudna, 2017). This creates a DNA double strand break (DSB) at the target site, which can be subsequently used to introduce modifications by NHEJ or HDR (Figure 1C) (Symington and Gautier, 2011). In higher plants, NHEJ occurs most frequently than the more precise HDR, which requires a donor DNA template during homologous recombination to repair the dsDNA breaks. NHEJ does not require a homologous repair template (Puchta et al., 1996; Puchta, 2005). NHEJ has therefore become a popular way to disrupt genes by the creation of small base pair indels (insertions/deletions) at specific points in the target genes, while HDR can be used to precisely introduce specific point mutations and insert or replace desired sequences into the target DNA (Figure 1C) (Li et al., 2013a). Currently, it is also possible to precisely edit a single base pair in the genome without the introduction of DSBs by using engineered Cas9 base editors. The base editors consist of a dead Cas9 domain fused to a cytidine deaminase enzyme that can be programmed with a guide RNA and is able to convert G to A and C to T without inducing dsDNA breaks (Komor et al., 2016). A Cas9 fused with a transfer RNA adenosine deaminase that can mediate conversion of G to A and C to T was also created (Gaudelli et al., 2017). These base editors install point mutations without generating excess undesired editing byproducts. In plants, base editing has been used to efficiently generate point mutations in maize, rice and wheat (Rees and Liu, 2018). These base editors will allow more and better genome modifications and expand the type of cells that can be efficiently edited. In order to make CRISPR-Cas9 a successful genome editing technology in plants, several modifications have been implemented. These include codon modification of the protein Cas9 to ensure its stability in plants, the use of strong constitutive or inducible promotors and the development of versatile DNA cassettes to co-express guide RNAs and Cas9 in the same cells (Li et al., 2013a).
A fundamental part of the genome editing process is the identification of target genes that determine phenotypes of interest, such as susceptibility to viruses (Garcia-Ruiz, 2018), other pathogens, resistance to herbicides or adverse environmental factors (Table 1). Assessment of natural variation or systematic genome-wide screens are also powerful approaches to identify target genes (Kushner et al., 2003; Panavas et al., 2005; Pyott et al., 2016; Giner et al., 2017).
Guide RNAs are artificially designed to specifically direct Cas9 to the target gene to be edited. Bioinformatic programs that generate candidate guide RNAs while accounting for the possibility of off-targets are available (http://crispr.mit.edu/). Dynamic expression vectors have also been designed to clone and co-express guide RNAs and Cas9 (Li et al., 2013a; Fauser et al., 2014; Shimatani et al., 2017). Although variations have been developed recently (Toda et al., 2019), transformation of plant cells to express guide RNAs and Cas9 follows a process similar to these established for the generation of transgenic plants (Figure 2). The expression cassettes contain constitutive or inducible promoters, transcription terminators and antibiotic and/or herbicide resistance markers used for selection purposes (Li et al., 2013a; Fauser et al., 2014; Shimatani et al., 2017).
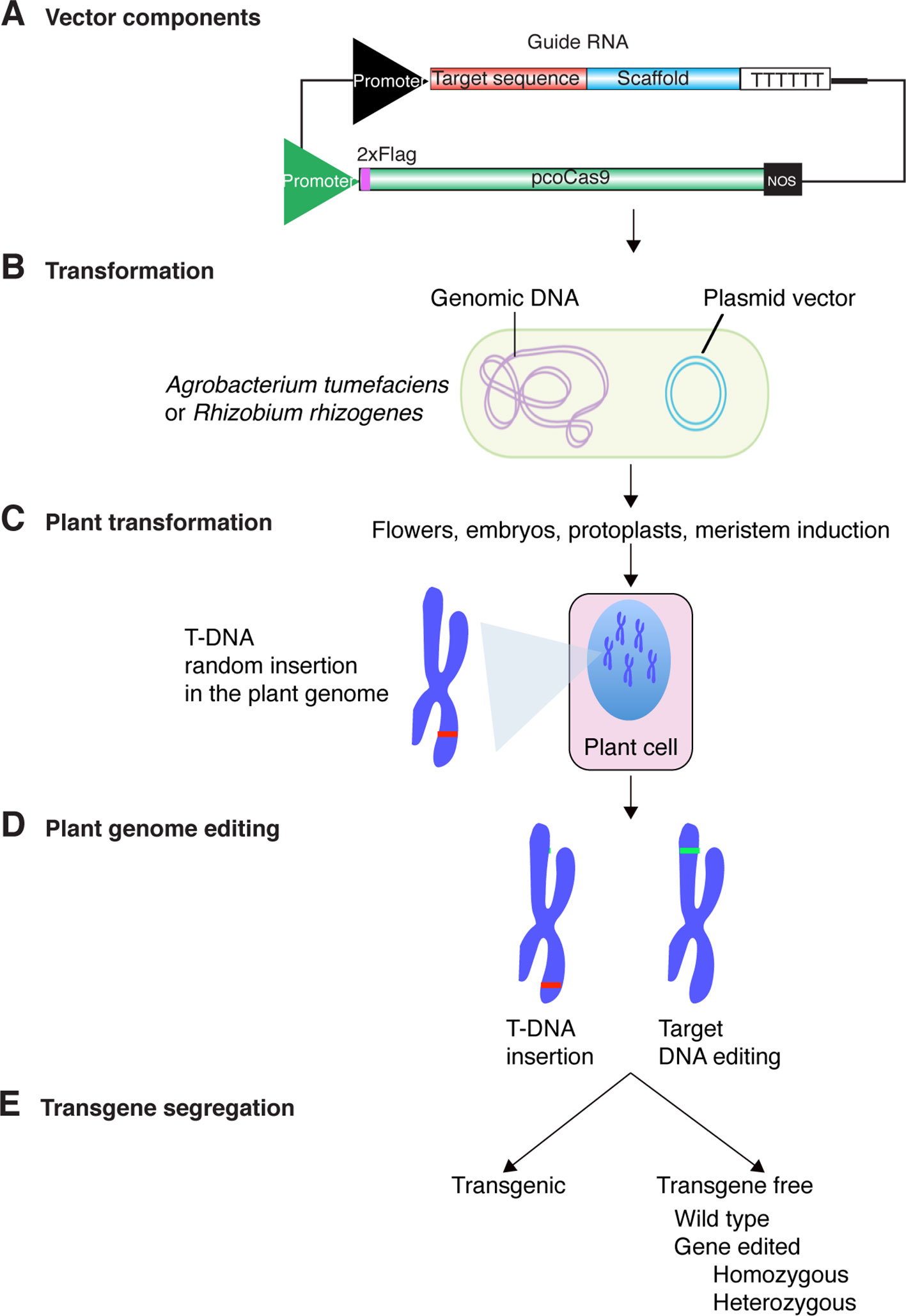
Figure 2 Genome editing process using CRISPR-Cas9 and Agrobacterium tumefaciens. (A) Cas9 protein and guide RNAs are cloned into the same plasmid vector containing transfer DNA (T-DNA) signals. Expression is driven by strong constitutive (U6, 35S, or other), inducible or tissue specific promoters. Transcription termination is programmed by addition of terminator such as the U6 or Nopaline synthase (NOS). For plant genome editing purposes, Cas9 has been codon-optimized and might contain an epitope tag to determine expression. (B) A. tumefaciens or R. rhizogens is transformed with the plasmid vector carrying the cassette for Cas9 protein and guide RNAs expression. (C) Bacteria is used to transform embryos, ovules in flowers, protoplasts, roots, or cells in leaves. Integration site of the T-DNA is random. (D) Expression of Cas9 protein and guide RNAs lead to editing of the target DNA. The T-DNA insertion site and the DNA target are likely not linked. (E) The T-DNA insertion and edited part of the genome can be separated by Mendelian segregation.
The vector carrying the Cas9 protein and the guide RNA is then introduced into Agrobacterium tumefaciens or Rhizobium rhizogenes (Figure 2B). Colonies containing the CRISPR-Cas9 construct are further used to transform plants by Agrobacterium-mediated transformation and first generation transgenic plants are identified by antibiotic or herbicide selection (Li et al., 2013a; Pyott et al., 2016; Veillet et al., 2019). Green fluorescent protein (GFP) has also been used to distinguish cells or calluses containing the CRISPR-Cas9 cassette (Doench et al., 2014). In all cases, sequencing the target gene is required in order to identify the mutations introduced by genome editing. The presence of the CRISPR-Cas9 cassette renders the plants transgenic and thus subject to the corresponding biosafety regulations (Callaway, 2018; Garcia Ruiz et al., 2018; Eckerstorfer et al., 2019). However, in sexually propagated plants, after identification of the genome edited plants, the CRISPR-Cas9 transgene can be eliminated by Mendelian segregation (Figures 2D–E) (Zhang et al., 2019a). This key part of the process removes the transgene in the third or subsequent generations resulting in the formation of genome-edited plants without a transgene (Pyott et al., 2016). Because of the absence of the transgene in these plants, they resemble those with mutations generated by natural means or chemical mutagenesis (Lellis et al., 2002; Pyott et al., 2016).
Because the introduction of the CRISPR-Cas9 cassette as a transgene might be controversial under certain regulations in some countries (Table 2), protocols have been developed to edit genomes without transgenes using guide RNA-Cas9 ribonucleoprotein complexes (Liang et al., 2017; Toda et al., 2019) or transient expression (Zhang et al., 2016).
Not all plant species are susceptible to A. tumefaciens. In species recalcitrant to Agrobacterium-mediated transformation, alternatives include Rhizobium rhizogenes-mediated or protoplast transformation. R. rhizogenes previously known as Agrobacterium rhizogenes, is a soil-borne gram-negative bacteria that causes hairy roots in plants. In most plant species, cells transformed with R. rhizogenes and its Ri plasmid differentiate into transformed roots, serving as a visual marker for marker-free screening and selection (Young et al., 2001; Bahramnejad et al., 2019). A comprehensive description of R. rhizogenes strains, binary vectors, and plants transformed using them is provided by (Bahramnejad et al., 2019). Examples of plants edited through CRISPR-Cas9 using R. rhizogenes include soybean (Du et al., 2016), tomato and rubber producing dandelion Taraxacum kok-saghyz (Iaffaldano et al., 2016).
For several plant species, including maize, soybean, wheat, rice, tomato, lettuce, arabidopsis, petunia, grapevine, apple, potato, and tobacco, protocols have been developed to isolate protoplast and transfect them with cassettes carrying CRISPR-Cas9 for genome editing purposes. Protoplast transfection has been used to rapidly optimize CRISPR-Ca9 parameters (Woo et al., 2015; Lin et al., 2018). However, isolation of single-protoplast has been used to regenerate stable transformants after transfection with cassettes carrying CRISPR-Cas9 or with ribonucleoprotein complexes assembled in vitro by synthesizing small guide RNAs and Cas9 protein. Gene editing using ribonucleoprotein complexes has the advantage of obtaining mutants without the presence of exogenous DNA. Preassembled Cas9-guide RNA ribonucleoproteins complexes can be delivered into protoplasts using polyethylene glycol-calcium-mediated transfection (Woo et al., 2015; Kim et al., 2017; Liang et al., 2017; Lin et al., 2018). To overcome the low efficiency of this approach, a protocol has been develop to transform plant zygotes by ribonucleoprotein complexes or by biolistic bombardment (Toda et al., 2019).
Delivering the CRISPR-Cas9 cassette into the germ line or protoplasts is technically challenging and inefficient. However, in dicotyledonous plants, those limitations might be eliminated through de novo induction of meristems. Developmental regulators and gene-editing components are delivered into somatic cells of whole plants. From treated tissue, shoots emerge that contain the targeted DNA modifications that are transmitted to the next generation (Maher et al., 2019).
Genome editing with CRISPR-Cas9 is amendable to edit any gene in any plant species. Because of its simplicity, efficiency, low cost, and the possibility to target multiple genes, it allows faster genetic modification than other techniques. It also can be used to genetically modify plants that were previously neglected. The potential that this represents for crop breeding and the development of sustainable agriculture is incommensurable (Cong et al., 2013; Mali et al., 2013; Zhang et al., 2017a; Toda et al., 2019; Wurtzel et al., 2019; Zhang et al., 2019b).
Impressive genetic modifications have been achieved with CRISPR-Cas9 to enhance metabolic pathways, tolerance to biotic (fungal, bacterial or viral pathogens), or abiotic stresses (cold, drought, salt), improve nutritional content, increase yield and grain quality, obtain haploid seeds, herbicide resistance, and others (Table 1). Notable cases include thermosensitive genic male sterility in maize (Li et al., 2017) and wheat (Okada et al., 2019), improved nutritional properties in sorghum and wheat (Li et al., 2018a; Zhang et al., 2018b), tolerance or resistance to pathogens (Zhang et al., 2017b; Pyott, 2016), and resistance to herbicides (Endo et al., 2016; Sun et al., 2016).
In potato CRISPR-Cas9 was used to knockout the gene encoding granule-bound starch synthase (GBSS) in one round of transfection resulting in the development of potato plants that produce amylopectin starch, a highly desirable commercial trait (Andersson et al., 2017). In cucumber CRISPR-Cas9 system was used to inactivate the eukaryotic translation initiation factor gene elF4E. The resulting non-transgenic homozygotic mutant plants were immune to Cucumber vein yellowing virus (Genus Ipomovirus) and resistant to the potyviruses Zucchini yellow mosaic virus and Papaya ring spot mosaic virus (Chandrasekaran et al., 2016). Engineering genetic resistance to viruses and other pathogens has immense potential to manage diseases for which no natural resistance has been detected, such as maize lethal necrosis disease and tomato brown rugose fruit virus (Luria et al., 2017; Garcia-Ruiz, 2018; Wamaitha et al., 2018).
In many bacteria and most archaea, CRISPR-Cas provides acquired immunity against viruses and plasmids by targeting nucleic acid in a sequence-specific manner (Horvath and Barrangou, 2010). Comparative genomic analyses revealed that CRISPR and genes coding for their associated proteins were present in diverse bacterial phylogenetic groups (Haft et al., 2005; Lillestol et al., 2006; Makarova et al., 2006). Since this adaptive immune system is useful for bacterial survival, it is likely to be present in all bacteria.
We compared the amino acid sequence of the Cas9 protein from S. pyogenes used in plant genome editing to proteins from bacteria to which humans are exposed through food consumption or in the environment. Results showed that Cas9 from S. pyogenes has 23% to 58% similarity to Cas9 protein from Streptococcus thermophilus, a bacterium widely used as a probiotic and in the production of cheese and yogurt (Figure 3). Additionally, Cas9 from S. pyogenes shares up to 35% similarity with Cas9 proteins from a wide range of bacteria used in food production such as Lactobacillus plantarum used to make cheese, yogurt, kefir and other fermented milk and meat products as well as fermented vegetables and beverages (Coloretti et al., 2007; Zago et al., 2011; Khemariya et al., 2016; Settachaimongkon et al., 2016; Sidira et al., 2017; Behera et al., 2018). L. plantarum is frequently encountered as a natural inhabitant of the human gastrointestinal tract, in which it is a transient guest acquirable through the diet (Vesa et al., 2000; De Vries et al., 2006). Additionally, L. plantarum is often used as a probiotic and can improve the balance of beneficial intestinal microflora (Nguyen et al., 2007; Nagpal et al., 2012; Kassayova et al., 2014).
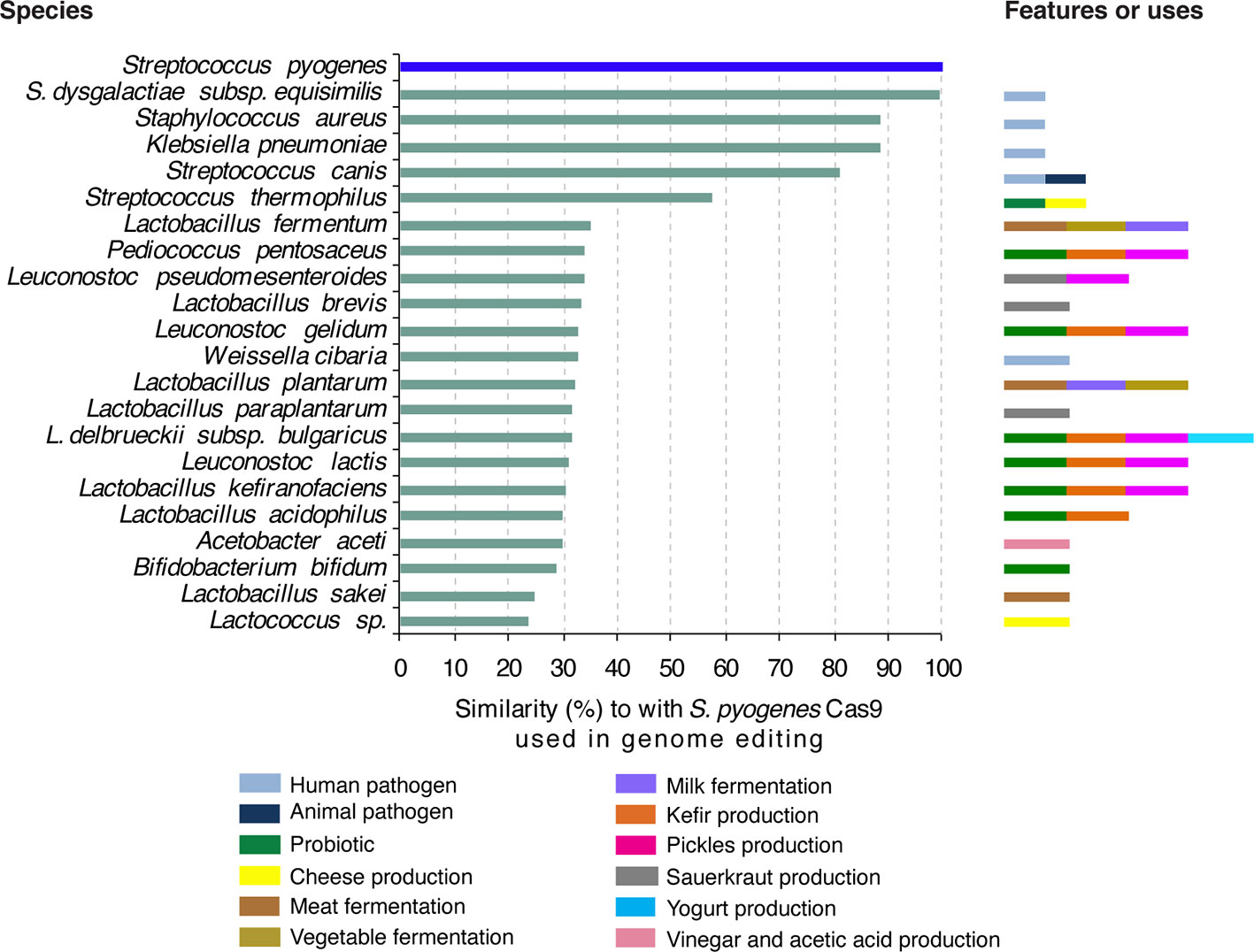
Figure 3 Bacteria frequently in contact with humans and similarity of their proteins to S. pyogenes Cas9 frequently used in plant genome editing. Amino acid sequence of S. pyogenes Cas9 was used to search for homologues proteins in GenBank. Proteins with more than 20% similarity are indicated. Features or anthropocentric use of each bacteria species are color-coded.
Furthermore, Cas9 from S. pyogenes has homologues in diverse Gram-positive and Gram-negative bacteria that occupy very diverse niches throughout the human body (Louwen et al., 2014). Some are commensals and others are pathogenic bacteria. More than 80% amino acid sequence similarity was detected between Cas9 from S. pyogenes and that from human commensal and pathogenic bacteria such as Streptococcus dysgalactiae subsp. equisimilis, Staphylococcus aureus, Klebsiella pneumonia and S. canis (Figure 3).
These observations show that humans have been exposed to Cas9 proteins in their food and environment long before the development of genome editing. The biosafety risk of human exposure to the Cas9 used for plant genome editing needs further assessment (Pineda et al., 2019) and our results do not mean that potential human exposure to Cas9 used in genome editing is irrelevant.
Methodological, biosafety and social concerns remain about the use of genome editing in plants. They mostly are related to target gene site selection, guide RNA design, off-target effects, and the delivery method. The major concern is the risk of generating unwanted genetic changes in plants due to off-target mutations (Liang et al., 2018; Pineda et al., 2019). Fragments of the CRISPR-Cas9 might be degraded into filler DNA and inserted into expected and/or unexpected genomic positions during the DNA repair process (Gorbunova and Levy, 1997; Zhang et al., 2016). However, transgene integration and the risk of off-target mutations can be prevented by delivering in vitro pre-assembled CRISPR-Cas9 ribonucleoproteins (Malnoy et al., 2016; Svitashev et al., 2016; Zhang et al., 2016; Liang et al., 2018). This technique has already been used in several crop species but there are still some drawbacks in its application such as low stability, high costs and high levels of technical requirements, which need to be improved (Malnoy et al., 2016; Subburaj et al., 2016; Murovec et al., 2018).
Substantial work has also been done to minimize off-target effects of Cas9 itself, including improving RNA guide–design strategies, ribonucleoprotein delivery, protein engineering, using spatiotemporally controlled Cas9, and/or gRNAs through a plethora of chemical or environmental inducers, or using synthetic genetic circuits that modulate CRISPR function according to predefined logic (Svitashev et al., 2016; Liang et al., 2018). Base editing is also being modified to improve the specificity of base editors by limiting deaminase activity outside of Cas9 binding through the use of different deaminase effectors or rationally engineering the deaminase to decrease its DNA binding ability (Shimatani et al., 2017).
Other concerns about CRISPR-Cas9 technology are related to the Cas9 protein itself as it was shown to induce an immune response when delivered by adeno-associated virus in mice, making immunogenic side effects a concern (Chew et al., 2016). There are also concerns about the specificity of Cas9 and the limited number of sites which can be targeted due to the requirement of the PAM (Spencer and Zhang, 2017). Protein engineering efforts led to the identification of mutations in Cas9 that alter its PAM recognition and enhance its fidelity and recognize other motifs (Kleinstiver et al., 2015; Kleinstiver et al., 2016; Leenay and Beisel, 2017). Further modifications to Cas9 and guide RNA design, such as FokI fusions, paired nicking, and the use of truncated guide RNAs, have provided additional improvements to specificity (Wyvekens et al., 2015). Furthermore, Cas9 variants, Cas9 homologs derived from other bacteria, or novel Cas proteins such as Cpf1 nucleases can be used (Nakade et al., 2017; Pineda et al., 2019).
The societal concerns about genome editing stem in part from the lack of information about its principles and applications. A fundamental feature here is the distinction between genetically modified plants, transgenic plants, and genome edited plants (Garcia Ruiz et al., 2018; Eckerstorfer et al., 2019). Genome edited plants may or may not be transgenic. As indicated above, the transgene carrying the CRISPR-Cas9 cassette might be removed by gene segregation (Figure 2). If this is done, a genome-edited plant might be classified as non-transgenic. Educating the public on the principles of genome editing has the potential to correct and prevent the spread of misconceptions (Garcia Ruiz et al., 2018; Eckerstorfer et al., 2019).
The term genetically modified refers to plants whose genome has been modified in a way that would not have been occurred naturally (Wang et al., 2016b; Duensing et al., 2018; Friedrichs et al., 2019).
In contrast, gene editing refers to DNA modifications similar to those potentially generated naturally (deletions, nt substitutions, insertions) of by conventional plant breeding (Nature Plants Editorial, 2018). The basis to regulate the release and international trade of living genetically modified organisms were established in the Cartagena Protocol on Biosafety. However, production, consumption, and regulation of genetically modified plants have followed contrasting patterns. While some countries reject consumption and ban production, others openly grow and consume them (Garcia Ruiz et al., 2018).
Regulation of genome-edited plants follows two frameworks. Some countries regulate the process, while others regulate characteristics of the final product (Eckerstorfer et al., 2019; Van Vu et al., 2019). While some countries have established biosafety regulations for genome edited plants, or declared their deregulation (Table 2), most countries have not yet established their position (Eckerstorfer et al., 2019). Challenges in regulating plant genome editing include market access, and addressing the societal concerns about its biological safety without limiting the development of the technology (Kupferschmidt, 2018; Eckerstorfer et al., 2019). Transgene-free, genome-edited plants are similar to varieties containing genetic variations created naturally (Figure 2). Therefore, commercialization of genome edited plants or their products might bypass the strict biosafety regulations required for transgenic plants (Tuteja et al., 2012; Van Vu et al., 2019).
The United States Department of Agriculture (USDA) declared in March 2018 that genome editing is the equivalent of conventional breeding in some instances and therefore does not require regulatory oversight within the American regulatory framework (Waltz, 2016a). A mushroom engineered to resist browning and a waxy corn engineered to contain starch composed exclusively of amylopectin are the first CRISPR edited crops to be approved for commercialization in the USA with no regulations (Waltz, 2016b). The decision not to regulate was based on the fact that no foreign DNA (transgene) was inserted during editing and that the resulting change did not involve resistance to pesticides or herbicides.
Canada, on the other hand, has remained committed to the scientific principles laid down in its domestic regulatory framework for plants with novel traits established 25 years ago. Canada's policy states that any gene editing technology that creates a novel product is subject to additional regulatory oversight on allergenicity, toxicity and impacts on non-target organisms (Smyth, 2017). Two products obtained by gene editing have been approved in Canada, non-browning apples and non-dark spots potatoes (Waltz, 2016b). The approval was granted after a lengthy evaluation process that determined that the changes made to the apples and the potatoes did not pose a greater risk to human health than apples and potatoes currently available on the Canadian market (Waltz, 2016b).
Argentina has developed a functional regulatory system for the approval of genome-edited products (Whelan and Lema, 2015). The regulatory system was developed to be consistent with the Cartagena Protocol on Biosafety and relies on case-by-case assessment. If a transgene technology was used in the development of a product, where the final product is free of the transgene, then this product can be classified as nontransgenic. Chile and Brazil followed Argentina's lead. Chile signed a normative resolution in 2017 while Brazil published a resolution in January 2018 (Duensing et al., 2018). Both regulate gene-edited products on a case-by-case basis and exempt them from regulation when there is no insertion of transgenes.
Meanwhile, European Union (EU) countries remain politically opposed to genetically modified crops (Waltz, 2016b). On July 2018, the Court of Justice of the European Union (ECJ) ruled that gene-edited crops should be subject to the same stringent regulations as conventional genetically modified (GM) organisms. In its ruling, the ECJ determined that only mutagenesis techniques that have conventionally been used in a number of applications and have a long safety record are exempt from this rule.
In Australia, the Gene Technology Act (GT Act), introduced in 2000, stipulates that a GMO is an organism produced by any technique that modifies genes or other genetic material. In 2001, the Gene Technology Regulations were introduced. Schedule 1 of these regulations, specifies that organisms resulting from an exchange of DNA in which the donor species is also the hosts species and the vector DNA does not contain heterogenous DNA as not GMOs. In October 2019, an amendment to schedule 1 came in effect. The amendment excludes organisms modified through CRISPR-Cas9 and other unguided repair of site-directed nuclease activity (SDN), from being regulated as GMOs. The amendment also indicates that organisms generated in the intermediated steps of the SDN method are deemed non GMOs if 1) no nucleic acid template is supplied to guide genome repair through homology-directed recombination, and 2) the organism has no other modifications as a result of the gene technology (Eckerstorfer et al., 2019).
In New Zealand, importation, development, field testing, and release of GMOs genetically modified are regulated by the Hazardous Substances and New Organisms Act 1996 (HSNO Act). The country has the most rigorous and comprehensive process for regulation of GMOs. As a result of that, no GMO commercial crops are grown in the country and no GM meat or fresh produce is sold in the country. Furthermore, processed food that contains imported GM ingredients is tested for safety and should be labeled as so. In 2016, the HSNO Act was amended with an article stating that plant breeding by genome editing is subject to the same regulations as the GMOs (Shimatani et al., 2017).
India's regulatory process for research, development and use of GMOs and their products, including new gene technologies was established in 1989. The Food Safety and Standards Authority of India define genetically engineered or modified food as “any food or food ingredient composed or containing genetically modified or engineered organisms obtained through modern biotechnology, or food and food ingredients produced from but not containing genetically modified or engineered organisms obtained through modern biotechnology”. Thus all new technologies including CRISPR-Cas9 gene technologies (including genome editing) are still regulated within the existing regulatory framework (Friedrichs et al., 2019).
Japan's Ministry of Health, Labor and Welfare (MHLW) has recently declared that foods derived from genome editing technologies which do not contain transgenic genes and/or fragments of transgenic genes are not considered GMOs and are not subject to regulations as long as the DNA double-strand break induced by the genetic engineering method is either a base-pair deletion, a naturally occurring gene deletion and/or a concomitant insertion of one to several base pairs. The new MHLW's policy also indicates that off-target mutations in GE foods should not be of concern as they can also be observed in multiple locations in the genome of crops produced by traditional breeding (South et al., 2019; Van Vu et al., 2019).
It seems that the decision to regulate or not regulate GE crops and foods depends mainly on the type of GMO regulatory system already in place in the country. Countries that have adopted a process-based GMO regulatory system and consider that products made using the regulated process are fundamentally different or more risky than similar products made using other methods will likely regulate GE crops and foods under the GMO laws. On the other hand, countries who follow a product-based regulatory system and regulate based on the characteristics of the final product rather than the process by which it was made might not regulate GE crops and foods under GMO laws. Countries, such as Malaysia and Thailand, who adopt a dual product and process approach will also likely to regulate GE crops and foods under GMO laws (Friedrichs et al., 2019).
Due to the many practical applications related to food production, genome editing can and will be used to solve agricultural issues that directly affect food security, such a citrus greening disease (Taylor et al., 2019), and the high yield losses in C3 plants, such as rice and barely, due to inefficient photorespiration in these crops. A recent report described the construction of three synthetic glycolate metabolic pathways in tobacco chloroplasts with the aim of improving the plant's photosynthetic efficiency. Flux through the synthetic pathways was maximized by inhibiting glycolate export from the chloroplast using RNA interference to down-regulate a native chloroplast glycolate transporter. In the transgenic tobacco plants, the photosynthetic yield increased by 20% while biomass productivity improved by more than 40% (South et al., 2019). While this study did not use genome editing technology and was carried out in Nicotiana tabacum, a model plant, the concept can be applied easily and successfully in staple crops using CRISPR-Cas9. Successful improvement in photorespiration efficiency in crops such as maize, rice and wheat, has the potential to substantially increase food production for the growing worldwide population while using the same cultivation areas and without having to destroy more forest areas for agricultural purposes. Producing crops with better quality food through genome editing will also help achieve food security (Li et al., 2018b; Narayanan et al., 2019).
Genome editing in general, and CRISPR-Cas9 in particular, is a revolutionary tool that can impact science, food production, and society. CRISPR-Cas9 has great potential for transforming agriculture by making plants tolerant to biotic and abiotic stresses and improving their nutritional value and yield. These attributes are necessary to meet the demand of an increasing world population. In order to be able to effectively and durably use this technology in crop improvement, the scientific community needs to address the various biosafety and societal concerns about it. There is also a need to re-evaluate the regulations of genome-edited plants and to educate the general public about their properties.
HG-R conceived the study. KE-M, MM-F, and HG-R performed the analysis. KE-M, MM-F, and HG-R wrote the paper.
This research was supported by NIH grant R01GM120108 to HG-R and by the Nebraska Agricultural Experiment Station with funding from the Hatch Act (Accession Number 1007272) through the USDA National Institute of Food and Agriculture. Open access costs were provided by the same grant. MM-F was the recipient of a graduate student fellowship from CONACYT, Mexico.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ainley, W. M., Sastry-Dent, L., Welter, M. E., Murray, M. G., Zeitler, B., Amora, R., et al. (2013). Trait stacking via targeted genome editing. Plant Biotechnol. J. 11, 1126–1134. doi: 10.1111/pbi.12107
Anders, C., Niewoehner, O., Duerst, A., Jinek, M. (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573. doi: 10.1038/nature13579
Andersson, M., Turesson, H., Nicolia, A., Falt, A. S., Samuelsson, M., Hofvander, P. (2017). Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 36, 117–128. doi: 10.1007/s00299-016-2062-3
Bahramnejad, B., Naji, M., Bose, R., Jha, S. (2019). A critical review on use of Agrobacterium rhizogenes and their associated binary vectors for plant transformation. Biotechnol. Adv. 37, 107405. doi: 10.1016/j.biotechadv.2019.06.004
Bao, A., Burritt, D. J., Chen, H., Zhou, X., Cao, D., Tran, L. P. (2019). The CRISPR/Cas9 system and its applications in crop genome editing. Crit. Rev. Biotechnol. 39, 1–16. doi: 10.1080/07388551.2018.1554621
Behera, S. S., Ray, R. C., Zdolec, N. (2018). Lactobacillus plantarum with functional properties: an approach to increase safety and shelf-life of fermented foods. BioMed. Res. Int. 2018, 9361614. doi: 10.1155/2018/9361614
Beyret, E., Liao, H. K., Yamamoto, M., Hernandez-Benitez, R., Fu, Y., Erikson, G., et al. (2019). Single-dose CRISPR-Cas9 therapy extends lifespan of mice with Hutchinson-Gilford progeria syndrome. Nat. Med. 25, 419–422. doi: 10.1038/s41591-019-0343-4
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512. doi: 10.1126/science.1178811
Budhagatapalli, N., Rutten, T., Gurushidze, M., Kumlehn, J., Hensel, G. (2015). Targeted Modification of Gene Function Exploiting Homology-Directed Repair of TALEN-Mediated Double-Strand Breaks in Barley. G3. (Bethesda). 5, 1857–1863.
Cai, Y., Chen, L., Liu, X., Guo, C., Sun, S., Wu, C., et al. (2018). CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 16, 176–185. doi: 10.1111/pbi.12758
Callaway, E. (2018). CRISPR plants now subject to tough GM laws in European Union. Nature 560, 16. doi: 10.1038/d41586-018-05814-6
Cathomen, T., Joung, J. K. (2008). Zinc-finger nucleases: the next generation emerges. Mol. Ther. 16, 1200–1207. doi: 10.1038/mt.2008.114
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Char, S. N., Unger-Wallace, E., Frame, B., Brigss, S. A., Main, M., Spalding, M. H., et al. (2015). Heritable site-specific mutagenesis using TALENs in maize. Plant Biotechnol. J. 13, 1002–1010.
Chen, M., Mao, A., Xu, M., Weng, Q., Mao, J., Ji, J. (2019). CRISPR-Cas9 for cancer therapy: opportunities and challenges. Cancer Lett. 447, 48–55. doi: 10.1016/j.canlet.2019.01.017
Chew, W. L., Tabebordbar, M., Cheng, J. K., Mali, P., Wu, E. Y., Ng, A. H., et al. (2016). A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 13, 868–874. doi: 10.1038/nmeth.3993
Christian, M., Cermak, T., Doyle, E. L., Schmidt, C., Zhang, F., Hummel, A., et al. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761. doi: 10.1534/genetics.110.120717
Christian, M., Qi, Y., Zhang, Y., Voytas, D. F. (2013). Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3. (Bethesda). 3, 1697–1705.
Clark, R. M., Tavare, S., Doebley, J. (2005). Estimating a nucleotide substitution rate for maize from polymorphism at a major domestication locus. Mol. Biol. Evol. 22, 2304–2312. doi: 10.1093/molbev/msi228
Clasen, B. M., Stoddard, T. J., Luo, S., Demorest, Z. L., Li, J., Cedrone, F., et al. (2016). Improving cold storage and processing traits in potato through targeted gene knockout. Plant. Biotechnol. J. 14, 169–176.
Coloretti, F., Carri, S., Armaforte, E., Chiavari, C., Grazia, L., Zambonelli, C. (2007). Antifungal activity of lactobacilli isolated from salami. FEMS Microbiol. Lett. 271, 245–250. doi: 10.1111/j.1574-6968.2007.00723.x
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Cunningham, M. W. (2000). Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13, 470–511. doi: 10.1128/CMR.13.3.470
D'ambrosio, C., Stigliani, A. L., Giorio, G. (2018). CRISPR/Cas9 editing of carotenoid genes in tomato. Transgenic Res. 27, 367–378. doi: 10.1007/s11248-018-0079-9
De Vries, M. C., Vaughan, E. E., Kleerebezem, M., De Vos, W. M. (2006). Lactobacillus plantarum—survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 16, 1018–1028. doi: 10.1016/j.idairyj.2005.09.003
Doebley, J., Stec, A., Wendel, J., Edwards, M. (1990). Genetic and morphological analysis of a maize-teosinte F2 population: implications for the origin of maize. Proc. Natl. Acad. Sci. U. S. A. 87, 9888–9892. doi: 10.1073/pnas.87.24.9888
Doench, J. G., Hartenian, E., Graham, D. B., Tothova, Z., Hegde, M., Smith, I., et al. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267. doi: 10.1038/nbt.3026
Dorweiler, J., Stec, A., Kermicle, J., Doebley, J. (1993). Teosinte glume architecture 1: a genetic locus controlling a key step in maize evolution. Science 262, 233–235. doi: 10.1126/science.262.5131.233
Du, H., Zeng, X., Zhao, M., Cui, X., Wang, Q., Yang, H., et al. (2016). Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 217, 90–97. doi: 10.1016/j.jbiotec.2015.11.005
Duensing, N., Sprink, T., Parrott, W. A., Fedorova, M., Lema, M. A., Wolt, J. D., et al. (2018). Novel features and considerations for ERA and regulation of crops produced by Genome editing. Front. Bioeng. Biotechnol. 6, 79. doi: 10.3389/fbioe.2018.00079
Ebina, H., Misawa, N., Kanemura, Y., Koyanagi, Y. (2013). Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 3, 2510. doi: 10.1038/srep02510
Eckerstorfer, M. F., Engelhard, M., Heissenberger, A., Simon, S., Teichmann, H. (2019). Plants developed by new genetic modification techniques—comparison of existing regulatory frameworks in the EU and non-EU countries. Front. In Bioeng. Biotechnol. 7, 26. doi: 10.3389/fbioe.2019.00026
Endo, M., Mikami, M., Toki, S. (2016). Biallelic gene targeting in rice. Plant Physiol. 170, 667–677. doi: 10.1104/pp.15.01663
Fauser, F., Schiml, S., Puchta, H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359. doi: 10.1111/tpj.12554
Friedrichs, S., Takasu, Y., Kearns, P., Dagallier, B., Oshima, R., Schofield, J., et al. (2019). Meeting report of the OECD conference on “genome editing: applications in agriculture—implications for health, environment and regulation”. Transgenic Res. 28, 419–463. doi: 10.1007/s11248-019-00154-1
Gallego-Bartolome, J., Liu, W., Kuo, P. H., Feng, S., Ghoshal, B., Gardiner, J., et al. (2019). Co-targeting RNA polymerases IV and V promotes efficient de novo DNA methylation in Arabidopsis. Cell. 176, 1068–1082. doi: 10.1016/j.cell.2019.01.029
Gao, L., Cox, D. B. T., Yan, W. X., Manteiga, J. C., Schneider, M. W., Yamano, T., et al. (2017). Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 35, 789–792. doi: 10.1038/nbt.3900
Garcia Ruiz, M. T., Knapp, A. N., Garcia-Ruiz, H. (2018). Profile of genetically modified plants authorized in Mexico. GM Crops Food 9, 152–168. doi: 10.1080/21645698.2018.1507601
Garcia-Ruiz, H. (2018). Susceptibility genes to plant viruses. Viruses 10, 484. doi: 10.3390/v10090484
Garneau, J. E., Dupuis, M. E., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. doi: 10.1038/nature09523
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471.
Giner, A., Pascual, L., Bourgeois, M., Gyetvai, G., Rios, P., Picó, B., et al. (2017). A mutation in the melon vacuolar protein sorting 41prevents systemic infection of Cucumber mosaic virus. Sci. Rep. 7, 10471. doi: 10.1038/s41598-017-10783-3
Gorbunova, V., Levy, A. A. (1997). Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 25, 4650–4657. doi: 10.1093/nar/25.22.4650
Haft, D. H., Selengut, J., Mongodin, E. F., Nelson, K. E. (2005). A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PloS Comput. Biol. 1, e60. doi: 10.1371/journal.pcbi.0010060
Horvath, P., Barrangou, R. (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170. doi: 10.1126/science.1179555
Hunter, M. C., Smith, R. G., Schipanski, M. E., Atwood, L. W., Mortensen, D. A. (2017). Agriculture in 2050: recalibrating targets for sustainable intensification. Bioscience 67, 385–390. doi: 10.1093/biosci/bix010
Iaffaldano, B., Zhang, Y., Cornish, K. (2016). CRISPR/Cas9 genome editing of rubber producing dandelion Taraxacum kok-saghyz using Agrobacterium rhizogenes without selection. Ind. Crops Prod. 89, 356–362. doi: 10.1016/j.indcrop.2016.05.029
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169, 5429–5433. doi: 10.1128/JB.169.12.5429-5433.1987
Jia, H., Orbovic, V., Jones, J. B., Wang, N. (2016a). Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccDeltapthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 14, 1291–1301. doi: 10.1111/pbi.12495
Jia, Y., Ding, Y., Shi, Y., Zhang, X., Gong, Z., Yang, S. (2016b). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212, 345–353. doi: 10.1111/nph.14088
Jia, H., Zhang, Y., Orbovic, V., Xu, J., White, F. F., Jones, J. B., et al. (2017). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15, 817–823. doi: 10.1111/pbi.12677
Jiang, F., Doudna, J. A. (2017). CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529. doi: 10.1146/annurev-biophys-062215-010822
Jiang, W. Z., Henry, I. M., Lynagh, P. G., Comai, L., Cahoon, E. B., Weeks, D. P. (2017). Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 15, 648–657. doi: 10.1111/pbi.12663
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jung, J. H., Altpeter, F. (2016). TALEN mediated targeted mutagenesis of the caffeic acid O-methyltransferase in highly polyploid sugarcane improves cell wall composition for production of bioethanol. Plant. Mol. Biol. 92, 131–142
Kang, H., Minder, P., Park, M. A., Mesquitta, W. T., Torbett, B. E., Slukvin, II (2015). CCR5 disruption in induced Pluripotent stem cells using CRISPR/Cas9 provides selective resistance of immune cells to CCR5-tropic HIV-1 virus. Mol. Ther. Nucleic Acids 4, e268. doi: 10.1038/mtna.2015.42
Karginov, F. V., Hannon, G. J. (2010). The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell 37, 7–19. doi: 10.1016/j.molcel.2009.12.033
Kassayova, M., Bobrov, N., Strojny, L., Kiskova, T., Mikes, J., Demeckova, V., et al. (2014). Preventive effects of probiotic bacteria Lactobacillus plantarum and dietary fiber in chemically-induced mammary carcinogenesis. Anticancer Res. 34, 4969–4975.
Kennedy, E. M., Kornepati, A. V., Goldstein, M., Bogerd, H. P., Poling, B. C., Whisnant, A. W., et al. (2014). Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J. Virol. 88, 11965–11972. doi: 10.1128/JVI.01879-14
Kennedy, E. M., Bassit, L. C., Mueller, H., Kornepati, A. V. R., Bogerd, H. P., Nie, T., et al. (2015). Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology 476, 196–205. doi: 10.1016/j.virol.2014.12.001
Khemariya, P., Singh, S., Jaiswal, N., Chaurasia, S. N. S. (2016). Isolation and identification of lactobacillus plantarum from vegetable samples. Food Biotechnol. 30, 49–62. doi: 10.1080/08905436.2015.1132428
Kim, Y. G., Cha, J., Chandrasegaran, S. (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U. S. A 93, 1156–1160. doi: 10.1073/pnas.93.3.1156
Kim, H., Kim, S.-T., Ryu, J., Kang, B.-C., Kim, J.-S., Kim, S.-G. (2017). CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8, 14406. doi: 10.1038/ncomms14406
Klap, C., Yeshayahou, E., Bolger, A. M., Arazi, T., Gupta, S. K., Shabtai, S., et al. (2017). Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 15, 634–647. doi: 10.1111/pbi.12662
Kleinstiver, B. P., Prew, M. S., Tsai, S. Q., Topkar, V. V., Nguyen, N. T., Zheng, Z., et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485. doi: 10.1038/nature14592
Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z., et al. (2016). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. doi: 10.1038/nature16526
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424.
Kupferschmidt, K. (2018). EU verdict on CRISPR crops dismays scientists. Science 361, 435–436. doi: 10.1126/science.361.6401.435
Kushner, D. B., Lindenbach, B. D., Grdzelishvili, V. Z., Noueiry, A. O., Paul, S. M., Ahlquist, P. (2003). Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. U. S. A. 100, 15764–15769. doi: 10.1073/pnas.2536857100
Leenay, R. T., Beisel, C. L. (2017). Deciphering, communicating, and engineering the CRISPR PAM. J. Mol. Biol. 429, 177–191. doi: 10.1016/j.jmb.2016.11.024
Lellis, A. D., Kasschau, K. D., Whitham, S. A., Carrington, J. C. (2002). Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12, 1046–1051. doi: 10.1016/S0960-9822(02)00898-9
Li, J. F., Norville, J. E., Aach, J., Mccormack, M., Zhang, D., Bush, J., et al. (2013a). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. doi: 10.1038/nbt.2654
Li, Y., Vonholdt, B. M., Reynolds, A., Boyko, A. R., Wayne, R. K., Wu, D. D., et al. (2013b). Artificial selection on brain-expressed genes during the domestication of dog. Mol. Biol. Evol. 30, 1867–1876. doi: 10.1093/molbev/mst088
Li, M., Li, X., Zhou, Z., Wu, P., Fang, M., Pan, X., et al. (2016). Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 System. Front. Plant Sci. 7, 377. doi: 10.3389/fpls.2016.00377
Li, J., Zhang, H., Si, X., Tian, Y., Chen, K., Liu, J., et al. (2017). Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J. Genet. Genomics 44, 465–468. doi: 10.1016/j.jgg.2017.02.002
Li, A., Jia, S., Yobi, A., Ge, Z., Sato, S. J., Zhang, C., et al. (2018a). Editing of an Alpha-Kafirin gene family increases, digestibility and protein quality in Sorghum. Plant Physiol. 177, 1425–1438. doi: 10.1104/pp.18.00200
Li, T., Yang, X., Yu, Y., Si, X., Zhai, X., Zhang, H., et al. (2018b). Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163. doi: 10.1038/nbt.4273
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392
Liang, Z., Zhang, K., Chen, K., Gao, C. (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 41, 63–68
Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., et al. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8, 14261. doi: 10.1038/ncomms14261
Liang, Z., Chen, K., Zhang, Y., Liu, J., Yin, K., Qiu, J. L., et al. (2018). Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 13, 413–430. doi: 10.1038/nprot.2017.145
Lillestol, R. K., Redder, P., Garrett, R. A., Brugger, K. (2006). A putative viral defence mechanism in archaeal cells. Archaea 2, 59–72. doi: 10.1155/2006/542818
Lin, C.-S., Hsu, C.-T., Yang, L.-H., Lee, L.-Y., Fu, J.-Y., Cheng, Q.-W., et al. (2018). Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 16, 1295–1310. doi: 10.1111/pbi.12870
Long, C., Amoasii, L., Mireault, A. A., Mcanally, J. R., Li, H., Sanchez-Ortiz, E., et al. (2016). Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403. doi: 10.1126/science.aad5725
Lor, V. S., Starker, C. G., Voytas, D. F., Weiss, D., Olszewski, N. E. (2014). Targeted Mutagenesis of the Tomato PROCERA; Gene Using Transcription Activator-Like Effector Nucleases. Plant Physiology 166, 1228. doi: 10.1126/science.aad5725
Louwen, R., Staals, R. H., Endtz, H. P., Van Baarlen, P., Van Der Oost, J. (2014). The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol. Mol. Biol. Rev. 78, 74–88. doi: 10.1128/MMBR.00039-13
Luria, N., Smith, E., Reingold, V., Bekelman, I., Lapidot, M., Levin, I., et al. (2017). A new Israeli tobamovirus isolate infects tomato plants harboring Tm-22 resistance genes. PloS One 12, e0170429. doi: 10.1371/journal.pone.0170429
Maher, M. F., Nasti, R. A., Vollbrecht, M., Starker, C. G., Clark, M. D., Voytas, D. F. (2019). Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38, 84–89. doi: 10.1038/s41587-019-0337-2
Makarova, K. S., Grishin, N. V., Shabalina, S. A., Wolf, Y. I., Koonin, E. V. (2006). A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1, 7. doi: 10.1186/1745-6150-1-7
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., Dicarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 7, 1904. doi: 10.3389/fpls.2016.01904
Malzahn, A., Lowder, L., Qi, Y. (2017). Plant genome editing with TALEN and CRISPR. Cell Biosci. 7, 21. doi: 10.1186/s13578-017-0148-4
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Moffett, H. F., Harms, C. K., Fitzpatrick, K. S., Tooley, M. R., Boonyaratankornkit, J., Taylor, J. J. (2019). B cells engineered to express pathogen-specific antibodies protect against infection. Sci. Immunol. 4, eaax0644.
Moscou, M. J., Bogdanove, A. J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501. doi: 10.1126/science.1178817
Murovec, J., Gucek, K., Bohanec, B., Avbelj, M., Jerala, R. (2018). DNA-free genome editing of Brassica oleracea and B. rapa Protoplasts using CRISPR-Cas9 Ribonucleoprotein complexes. Front. Plant Sci. 9, 1594. doi: 10.3389/fpls.2018.01594
Nagpal, R., Kumar, A., Kumar, M., Behare, P. V., Jain, S., Yadav, H. (2012). Probiotics, their health benefits and applications for developing healthier foods: a review. FEMS Microbiol. Lett. 334, 1–15. doi: 10.1111/j.1574-6968.2012.02593.x
Naim, F., Dugdale, B., Kleidon, J., Brinin, A., Shand, K., Waterhouse, P., et al. (2018). Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 27, 451–460. doi: 10.1007/s11248-018-0083-0
Nakade, S., Yamamoto, T., Sakuma, T. (2017). Cas9, Cpf1 and C2c1/2/3-What's next? Bioengineered 8, 265–273. doi: 10.1080/21655979.2017.1282018
Narayanan, N., Beyene, G., Chauhan, R. D., Gaitan-Solis, E., Gehan, J., Butts, P., et al. (2019). Biofortification of field-grown cassava by engineering expression of an iron transporter and ferritin. Nat. Biotechnol. 37, 144–151. doi: 10.1038/s41587-018-0002-1
Nature Plants Editorial. (2018). A CRISPR definition of genetic modification. Nat. Plants 4, 233. doi: 10.1038/s41477-018-0158-1
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 482. doi: 10.1038/s41598-017-00578-x
Nelson, C. E., Hakim, C. H., Ousterout, D. G., Thakore, P. I., Moreb, E. A., Castellanos Rivera, R. M., et al. (2016). In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407. doi: 10.1126/science.aad5143
Nguyen, T. D., Kang, J. H., Lee, M. S. (2007). Characterization of Lactobacillus plantarum PH04, a potential probiotic bacterium with cholesterol-lowering effects. Int. J. Food Microbiol. 113, 358–361. doi: 10.1016/j.ijfoodmicro.2006.08.015
Nicolia, A., Proux-Wera, E., Ahman, I., Onkokesung, N., Andersson, M., Andreasson, E., et al. (2015). Targeted gene mutation in tetraploid potato through transient TALEN expression in protoplasts. J. Biotechnol. 204, 17–24
Okada, A., Arndell, T., Borisjuk, N., Sharma, N., Watson-Haigh, N. S., Tucker, E. J., et al. (2019). CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 17, 1905–1913. doi: 10.1111/pbi.13106
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., Solano, R. (2018). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 17, 665–673. doi: 10.1111/pbi.13006
Osakabe, K., Osakabe, Y., Toki, S. (2010). Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Natl. Acad. Sci. U. S. A 107, 12034–12039. doi: 10.1073/pnas.1000234107
Panavas, T., Serviene, E., Brasher, J., Nagy, P. D. (2005). Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 102, 7326–7331. doi: 10.1073/pnas.0502604102
Papikian, A., Liu, W., Gallego-Bartolome, J., Jacobsen, S. E. (2019). Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 10, 729. doi: 10.1038/s41467-019-08736-7
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Pineda, M., Lear, A., Collins, J. P., Kiani, S. (2019). Safe CRISPR: challenges and possible solutions. Trends Biotechnol. 37, 389–401. doi: 10.1016/j.tibtech.2018.09.010
Piperno, D. R., Flannery, K. V. (2001). The earliest archaeological maize (Zea mays L.) from highland Mexico: new accelerator mass spectrometry dates and their implications. Proc. Natl. Acad. Sci. U. S. A 98, 2101–2103. doi: 10.1073/pnas.98.4.2101
Puchta, H., Dujon, B., Hohn, B. (1996). Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. U. S. A 93, 5055–5060. doi: 10.1073/pnas.93.10.5055
Puchta, H. (2005). The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56, 1–14. doi: 10.1093/jxb/eri025
Pyott, D. E., Sheehan, E., Molnar, A. (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 17, 1276–1288. doi: 10.1111/mpp.12417
Rees, H. A., Liu, D. R. (2018). Base editing: precision chemistry on the genome and transcriptome of living cells. Nature Reviews Genetics. 19, 770–788.
Rosinski-Chupin, I., Sauvage, E., Fouet, A., Poyart, C., Glaser, P. (2019). Conserved and specific features of Streptococcus pyogenes and Streptococcus agalactiae transcriptional landscapes. BMC Genomics 20, 236. doi: 10.1186/s12864-019-5613-5
Schwank, G., Koo, B. K., Sasselli, V., Dekkers, J. F., Heo, I., Demircan, T., et al. (2013). Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13, 653–658. doi: 10.1016/j.stem.2013.11.002
Settachaimongkon, S., Van Valenberg, H. J., Gazi, I., Nout, M. J., Van Hooijdonk, T. C., Zwietering, M. H., et al. (2016). Influence of Lactobacillus plantarum WCFS1 on post-acidification, metabolite formation and survival of starter bacteria in set-yoghurt. Food Microbiol. 59, 14–22. doi: 10.1016/j.fm.2016.04.008
Shah, T., Andleeb, T., Lateef, S., Noor, M. A. (2018). Genome editing in plants: advancing crop transformation and overview of tools. Plant Physiol. Biochem. 131, 12–21. doi: 10.1016/j.plaphy.2018.05.009
Shan, Q., Wang, Y., Chen, K., Liang, Z., Li, J., Zhang, Y., et al. (2013). Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant 6, 1365–1368.
Shan, Q., Zhang, Y., Chen, K., Zhang, K., Gao, C. (2015). Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol. J. 13, 791–800.
Shimatani, Z., Kashojiya, S., Takayama, M., Terada, R., Arazoe, T., Ishii, H., et al. (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441. doi: 10.1038/nbt.3833
Shukla, V. K., Doyon, Y., Miller, J. C., Dekelver, R. C., Moehle, E. A., Worden, S. E., et al. (2009). Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459, 437–441. doi: 10.1038/nature07992
Sidira, M., Santarmaki, V., Kiourtzidis, M., Argyri, A. A., Papadopoulou, O. S., Chorianopoulos, N., et al. (2017). Evaluation of immobilized Lactobacillus plantarum 2035 on whey protein as adjunct probiotic culture in yoghurt production. LWT 75, 137–146. doi: 10.1016/j.lwt.2016.08.026
Smyth, S. J. (2017). Canadian regulatory perspectives on genome engineered crops. GM Crops Food 8, 35–43. doi: 10.1080/21645698.2016.1257468
Sorek, R., Lawrence, C. M., Wiedenheft, B. (2013). CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82, 237–266. doi: 10.1146/annurev-biochem-072911-172315
South, P. F., Cavanagh, A. P., Liu, H. W., Ort, D. R. (2019). Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363, eaat9077. doi: 10.1126/science.aat9077
Soyk, S., Muller, N. A., Park, S. J., Schmalenbach, I., Jiang, K., Hayama, R., et al. (2017). Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 49, 162–168. doi: 10.1038/ng.3733
Spencer, J. M., Zhang, X. (2017). Deep mutational scanning of S. pyogenes Cas9 reveals important functional domains. Sci. Rep. 7, 16836. doi: 10.1038/s41598-017-17081-y
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C., Doudna, J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67. doi: 10.1038/nature13011
Subburaj, S., Chung, S. J., Lee, C., Ryu, S. M., Kim, D. H., Kim, J. S., et al. (2016). Site-directed mutagenesis in Petunia x hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 35, 1535–1544. doi: 10.1007/s00299-016-1937-7
Sun, Y., Zhang, X., Wu, C., He, Y., Ma, Y., Hou, H., et al. (2016). Engineering Herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of Acetolactate synthase. Mol. Plant 9, 628–631. doi: 10.1016/j.molp.2016.01.001
Sun, Y., Jiao, G., Liu, Z., Zhang, X., Li, J., Guo, X., et al. (2017). Generation of high-Amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 8, 298. doi: 10.3389/fpls.2017.00298
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., Mark Cigan, A. (2016). Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274. doi: 10.1038/ncomms13274
Symington, L. S., Gautier, J. (2011). Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271. doi: 10.1146/annurev-genet-110410-132435
Tabebordbar, M., Zhu, K., Cheng, J. K. W., Chew, W. L., Widrick, J. J., Yan, W. X., et al. (2016). In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351, 407–411. doi: 10.1126/science.aad5177
Taylor, R. A., Ryan, S. J., Lippi, C. A., Hall, D. G., Narouei-Khandan, H. A., Rohr, J. R., et al. (2019). Predicting the fundamental thermal niche of crop pests and diseases in a changing world: a case study on citrus greening. J. Appl. Ecol. 56, 2057–2068. doi: 10.1111/1365-2664.13455
Toda, E., Koiso, N., Takebayashi, A., Ichikawa, M., Kiba, T., Osakabe, K., et al. (2019). An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat. Plants 5, 363–368. doi: 10.1038/s41477-019-0386-z
Tripathi, J. N., Ntui, V. O., Ron, M., Muiruri, S. K., Britt, A., Tripathi, L. (2019). CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2, 46. doi: 10.1038/s42003-019-0288-7
Tuteja, N., Verma, S., Sahoo, R. K., Raveendar, S., Reddy, I. N. (2012). Recent advances in development of marker-free transgenic plants: regulation and biosafety concern. J. Biosci. 37, 167–197. doi: 10.1007/s12038-012-9187-5
Ueta, R., Abe, C., Watanabe, T., Sugano, S. S., Ishihara, R., Ezura, H., et al. (2017). Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 7, 507. doi: 10.1038/s41598-017-00501-4
Van Vu, T., Sung, Y. W., Kim, J., Doan, D. T. H., Tran, M. T., Kim, J.-Y. (2019). Challenges and perspectives in homology-directed gene targeting in monocot plants. Rice 12, 95. doi: 10.1186/s12284-019-0355-1
Veillet, F., Perrot, L., Chauvin, L., Kermarrec, M. P., Guyon-Debast, A., Chauvin, J. E., et al. (2019). Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 Cytidine base editor. Int. J. Mol. Sci. 20, 402–452. doi: 10.3390/ijms20020402
Vesa, T., Pochart, P., Marteau, P. (2000). Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment. Pharmacol. Ther. 14, 823–828. doi: 10.1046/j.1365-2036.2000.00763.x
Waltz, E. (2016a). CRISPR-edited crops free to enter market, skip regulation. Nat. Biotechnol. 34, 582. doi: 10.1038/nbt0616-582
Waltz, E. (2016b). Gene-edited CRISPR mushroom escapes US regulation. Nature 532, 293. doi: 10.1038/nature.2016.19754
Wamaitha, M. J., Nigam, D., Maina, S., Stomeo, F., Wangai, A., Njuguna, J. N., et al. (2018). Metagenomic analysis of viruses associated with maize lethal necrosis in Kenya. Virol. J. 15, 90. doi: 10.1186/s12985-018-0999-2
Wang, R. L., Stec, A., Hey, J., Lukens, L., Doebley, J. (1999). The limits of selection during maize domestication. Nature 398, 236–239. doi: 10.1038/18435
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016a). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PloS One 11, e0154027. doi: 10.1371/journal.pone.0154027
Wang, G. P., Yu, X. D., Sun, Y. W., Jones, H. D., Xia, L. Q. (2016b). Generation of marker- and/or backbone-free transgenic wheat plants via Agrobacterium-mediated transformation. Front. Plant Sci. 7, 1324. doi: 10.3389/fpls.2016.01324
Wang, L., Chen, L., Li, R., Zhao, R., Yang, M., Sheng, J., et al. (2017). Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 65, 8674–8682. doi: 10.1021/acs.jafc.7b02745
Wang, X., Tu, M., Wang, D., Liu, J., Li, Y., Li, Z., et al. (2018). CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 16, 844–855. doi: 10.1111/pbi.12832
Whelan, A. I., Lema, M. A. (2015). Regulatory framework for gene editing and other new breeding techniques (NBTs) in Argentina. GM Crops Food 6, 253–265. doi: 10.1080/21645698.2015.1114698
Woo, J. W., Kim, J., Kwon, S. I., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Wurtzel, E. T., Vickers, C. E., Hanson, A. D., Millar, A. H., Cooper, M., Voss-Fels, K. P., et al. (2019). Revolutionizing agriculture with synthetic biology. Nat. Plants. 5, 1207–1210. doi: 10.1038/s41477-019-0539-0
Wyvekens, N., Topkar, V. V., Khayter, C., Joung, J. K., Tsai, S. Q. (2015). Dimeric CRISPR RNA-Guided FokI-dCas9 nucleases directed by truncated gRNAs for highly specific genome editing. Hum. Gene Ther. 26, 425–431. doi: 10.1089/hum.2015.084
Yang, Y., Wang, L., Bell, P., Mcmenamin, D., He, Z., White, J., et al. (2016). A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 34, 334–338. doi: 10.1038/nbt.3469
Yao, L., Zhang, Y., Liu, C., Liu, Y., Wang, Y., Liang, D., et al. (2018). OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants 4, 530–533. doi: 10.1038/s41477-018-0193-y
Ye, L., Wang, J., Beyer, A. I., Teque, F., Cradick, T. J., Qi, Z., et al. (2014). Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. U. S. A 111, 9591–9596. doi: 10.1073/pnas.1407473111
Yin, H., Xue, W., Chen, S., Bogorad, R. L., Benedetti, E., Grompe, M., et al. (2014). Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 32, 551–553. doi: 10.1038/nbt.2884
Yin, H., Song, C. Q., Dorkin, J. R., Zhu, L. J., Li, Y., Wu, Q., et al. (2016). Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 34, 328–333. doi: 10.1038/nbt.3471
Young, J. M., Kuykendall, L. D., Martínez-Romero, E., Kerr, A., Sawada, H. (2001). A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al., 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola, and R. vitis. Int. J. Syst. Evol. Microbiol. 51, 89–103. doi: 10.1099/00207713-51-1-89
Zago, M., Fornasari, M. E., Carminati, D., Burns, P., Suarez, V., Vinderola, G., et al. (2011). Characterization and probiotic potential of Lactobacillus plantarum strains isolated from cheeses. Food Microbiol. 28, 1033–1040. doi: 10.1016/j.fm.2011.02.009
Zhang, Y., Zhang, F., Li, X., Baller, J. A., Qi, Y., Starker, C. G., et al. (2013). Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 161, 20–27.
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. doi: 10.1038/ncomms12617
Zhang, H., Zhang, J. S., Lang, Z. B., Botella, J. R., Zhu, J. K. (2017a). Genome editing-principles and applications for functional genomics research and crop improvement. Crit. Rev. Plant Sci. 36, 291–309. doi: 10.1080/07352689.2017.1402989
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017b). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91, 714–724. doi: 10.1111/tpj.13599
Zhang, Y., Li, D., Zhang, D., Zhao, X., Cao, X., Dong, L., et al. (2018a). Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. Plant J. 94, 857–866. doi: 10.1111/tpj.13903
Zhang, Y., Massel, K., Godwin, I. D., Gao, C. (2018b). Applications and potential of genome editing in crop improvement. Genome Biol. 19, 210. doi: 10.1186/s13059-018-1586-y
Zhang, A., Liu, Y., Wang, F., Li, T., Chen, Z., Kong, D., et al. (2019a). Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 39, 47. doi: 10.1007/s11032-019-0954-y
Zhang, Y., Malzahn, A. A., Sretenovic, S., Qi, Y. (2019b). The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 5, 778–794. doi: 10.1038/s41477-019-0461-5
Zhou, J., Peng, Z., Long, J., Sosso, D., Liu, B., Eom, J. S., et al. (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643. doi: 10.1111/tpj.12838
Zhou, H., He, M., Li, J., Chen, L., Huang, Z., Zheng, S., et al. (2016). Development of commercial thermo-sensitive Genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci. Rep. 6, 37395. doi: 10.1038/srep37395
Keywords: genome editing, CRISPR-Cas9, Cas9 human exposure, plant breeding, biosafety regulations
Citation: El-Mounadi K, Morales-Floriano ML and Garcia-Ruiz H (2020) Principles, Applications, and Biosafety of Plant Genome Editing Using CRISPR-Cas9. Front. Plant Sci. 11:56. doi: 10.3389/fpls.2020.00056
Received: 16 September 2019; Accepted: 15 January 2020;
Published: 13 February 2020.
Edited by:
Hiroshi Ezura, University of Tsukuba, JapanReviewed by:
David Gagneul, Lille University of Science and Technology, FranceCopyright © 2020 El-Mounadi, Morales-Floriano and Garcia-Ruiz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hernan Garcia-Ruiz, aGdhcmNpYXJ1aXoyQHVubC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.