
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 21 February 2020
Sec. Plant Systematics and Evolution
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00022
This article is part of the Research Topic Orchid Genomics and Developmental Biology View all 13 articles
A correction has been applied to this article in:
Corrigendum: Plastome Evolution and Phylogeny of Orchidaceae, With 24 New Sequences
In order to understand the evolution of the orchid plastome, we annotated and compared 124 complete plastomes of Orchidaceae representing all the major lineages in their structures, gene contents, gene rearrangements, and IR contractions/expansions. Forty-two of these plastomes were generated from the corresponding author's laboratory, and 24 plastomes—including nine genera (Amitostigma, Bulbophyllum, Dactylorhiza, Dipodium, Galearis, Gymnadenia, Hetaeria, Oreorchis, and Sedirea)—are new in this study. All orchid plastomes, except Aphyllorchis montana, Epipogium aphyllum, and Gastrodia elata, have a quadripartite structure consisting of a large single copy (LSC), two inverted repeats (IRs), and a small single copy (SSC) region. The IR region was completely lost in the A. montana and G. elata plastomes. The SSC is lost in the E. aphyllum plastome. The smallest plastome size was 19,047 bp, in E. roseum, and the largest plastome size was 178,131 bp, in Cypripedium formosanum. The small plastome sizes are primarily the result of gene losses associated with mycoheterotrophic habitats, while the large plastome sizes are due to the expansion of noncoding regions. The minimal number of common genes among orchid plastomes to maintain minimal plastome activity was 15, including the three subunits of rpl (14, 16, and 36), seven subunits of rps (2, 3, 4, 7, 8, 11, and 14), three subunits of rrn (5, 16, and 23), trnC-GCA, and clpP genes. Three stages of gene loss were observed among the orchid plastomes. The first was ndh gene loss, which is widespread in Apostasioideae, Vanilloideae, Cypripedioideae, and Epidendroideae, but rare in the Orchidoideae. The second stage was the loss of photosynthetic genes (atp, pet, psa, and psb) and rpo gene subunits, which are restricted to Aphyllorchis, Hetaeria, Hexalectris, and some species of Corallorhiza and Neottia. The third stage was gene loss related to prokaryotic gene expression (rpl, rps, trn, and others), which was observed in Epipogium, Gastrodia, Lecanorchis, and Rhizanthella. In addition, an intermediate stage between the second and third stage was observed in Cyrtosia (Vanilloideae). The majority of intron losses are associated with the loss of their corresponding genes. In some orchid taxa, however, introns have been lost in rpl16, rps16, and clpP(2) without their corresponding gene being lost. A total of 104 gene rearrangements were counted when comparing 116 orchid plastomes. Among them, many were concentrated near the IRa/b-SSC junction area. The plastome phylogeny of 124 orchid species confirmed the relationship of {Apostasioideae [Vanilloideae (Cypripedioideae (Orchidoideae, Epidendroideae))]} at the subfamily level and the phylogenetic relationships of 17 tribes were also established. Molecular clock analysis based on the whole plastome sequences suggested that Orchidaceae diverged from its sister family 99.2 mya, and the estimated divergence times of five subfamilies are as follows: Apostasioideae (79.91 mya), Vanilloideae (69.84 mya), Cypripedioideae (64.97 mya), Orchidoideae (59.16 mya), and Epidendroideae (59.16 mya). We also released the first nuclear ribosomal (nr) DNA unit (18S-ITS1-5.8S-ITS2-28S-NTS-ETS) sequences for the 42 species of Orchidaceae. Finally, the phylogenetic tree based on the nrDNA unit sequences is compared to the tree based on the 42 identical plastome sequences, and the differences between the two datasets are discussed in this paper.
Orchidaceae is one of the most flourishing flowering plants and contains about 736 known genera and 28,000 species worldwide (Christenhusz and Byng, 2016). Recent studies recognize five subfamilies within Orchidaceae (Apostasioideae, Vanilloideae, Cypripedioideae, Orchidoideae, and Epidendroideae) as a monophyletic group (Chase et al., 2015). The most recently differentiated subfamily, Epidendroideae, includes about 505 genera and 20,600 species, and accounts for most of Orchidaceae (Chase et al., 2015). Orchidaceae is widely distributed throughout the world, and most members in temperate regions have terrestrial life forms, but orchids in tropical rainforests are known to have mainly epiphyte life forms (Givnish et al., 2015). Non-photosynthetic mycoheterotrophic orchids are found in a total of 43 genera and belong to three subfamilies: Vanilloideae, Orchidoideae, and Epidendroideae (Merckx et al., 2013).
Complete Orchidaceae plastomes have been reported in 38 genera and 118 species (NCBI GenBank, June 30, 2019). There are only five genera (Corallorhiza, 10 spp.; Cymbidium, 9 spp.; Dendrobium, 40 spp.; Holcoglossum, 11 spp.; and Neottia, 7 spp.) in which at least five plastomes per genus have been decoded (Logacheva et al., 2011; Barrett and Davis, 2012; Yang et al., 2013; Barrett et al., 2014; Feng et al., 2016; Niu et al., 2017b; Barrett et al., 2018; Kim et al., 2018). Among them, Corallorhiza and Neottia have been subjected to extensive evolutionary studies of their plastomes because both photosynthetic and non-photosynthetic species occur in a congeneric group (Barrett and Davis, 2012; Barrett et al., 2014; Feng et al., 2016; Barrett et al., 2018). In addition to the two genera, evolutionary studies have been carried out on the plastomes of some species of other orchid genera. For examples, extensive gene losses have been reported in several independent mycoheterotrophic orchid lineages—i.e., Aphyllorchis (Feng et al., 2016), Cyrtosia (Kim et al., 2019), Epipogium (Schelkunov et al., 2015), Gastrodia (Yuan et al., 2018), Hexalectris (Barrett and Kennedy, 2018) and Rhizanthella (Delannoy et al., 2011). In addition, ndh deletion and pseudogenization are assumed to be phenomena that occur independently in many orchid lineages such as Apostasia (Lin et al., 2017; Niu et al., 2017a), Calypso (Barrett et al., 2018), Cattleya (da Rocha Perini et al., 2016), Cephalanthera (Feng et al., 2016), Cremastra (Dong et al., 2018), Cymbidium (Yang et al., 2013; Kim et al., 2018; Wang et al., 2018), Dendrobium (Niu et al., 2017b), Epipactis (Dong et al., 2018), Eulophia (Huo et al., 2017), Holcoglossum (Li et al., 2019), Limodorum (Lallemand et al., 2019), Liparis (Krawczyk et al., 2018), Neuwiedia (Niu et al., 2017a), Oncidium (Wu et al., 2010; Kim et al., 2015a), Paphiopedilum (Niu et al., 2017b; Hou et al., 2018), Phalaenopsis (Chang et al., 2006), Phragmipedium (Kim et al., 2015a), Platanthera (Dong et al., 2018), Vanilla (Lin et al., 2015), and Vanda (Li et al., 2019). On the other hand, full ndh genes have been reported in members of Anoectochilus (Yu et al., 2016), Calanthe (Dong et al., 2018), Cypripedium (Kim et al., 2015b; Lin et al., 2015), Habenaria (Lin et al., 2015; Kim et al., 2017b), Masdevallia (Kim et al., 2015a), Ophrys (Roma et al., 2018), Pleione (Shi et al., 2018), and Sobralia (Kim et al., 2015a). Gene relocations within a plastome often occur in the reduced plastome of orchids such as Cyrtosia (Kim et al., 2019), Hexalectris (Barrett and Kennedy, 2018), and Rhizanthella (Delannoy et al., 2011). Similar gene rearrangement events within a plastome have been reported in Campanulaceae (Cosner et al., 1997), conifers (Hirao et al., 2008), Fabaceae (Cai et al., 2008), Geraniaceae (Chumley et al., 2006), and Oleaceae (Lee et al., 2007). Contractions of the small single copy (SSC) region similar to that of Geraniaceae were also reported in Paphiopedilum (Kim et al., 2015a; Niu et al., 2017b; Hou et al., 2018) and Vanilla (Lin et al., 2015; Amiryousefi et al., 2017).
Phylogenetic studies of Orchidaceae using entire plastomes are in a relatively early stage because of limited available plastome sequences (Givnish et al., 2015; Kim et al., 2015a; Lin et al., 2015; Niu et al., 2017b; Dong et al., 2018). But the relationships among major orchid lineages determined using whole plastomes agree well to the large-scale phylogenetic studies of Orchidaceae using two or three genes (Górniak et al., 2010; Freudenstein and Chase, 2015). Therefore, several outstanding phylogenetic problems in Orchidaceae will be resolved if more plastome sequences are accumulated.
In this study we first completely decoded the plastomes of 24 taxonomic groups of Orchidaceae, including nine genera (Amitostigma, Bulbophyllum, Dactylorhiza, Dipodium, Galearis, Gymnadenia, Hetaeria, Oreorchis, and Sedirea) for which the plastomes had not yet been decoded previously. Second, this study re-annotated and compared the entire plastome sequences of 129 taxa comprising 124 Orchidaceae and five outgroups to investigate evolutionary directions in orchid plastomes, such as sizes, gene contents, gene losses, gene rearrangements, and inverted repeat (IR) expansions/contractions. Third, three stages of gene loss patterns were inferred from mycoheterotrophic orchids and the minimum genes required for plastid maintenance were inferred. Fourth, the phylogenetic trees of Orchidaceae were constructed using whole plastome sequences, and the times at which each taxonomic group differentiated were inferred. Among the 124 orchid plastomes completely decoded, 42 were produced by the NGS method in the laboratory of the corresponding author, and the nuclear ribosomal RNA gene unit (18S-ITS1-5.8S-ITS2-28S-NTS-ETS) was also annotated at the same time. Finally, the common features and differences among the 42 species were compared and explained by comparing their plastome trees and nrDNA gene unit trees.
Plant leaf materials used in this study and their voucher information are given in Table 1. Fresh leaf samples were ground into fine powder with liquid nitrogen in a mortar. Ground samples were used to extract genomic DNA using a G-spin™ II Genomic DNA Extraction Kit (Intron, Seoul, Korea). The quality of DNA was checked by a UVVIS spectrophotometer. Extracted DNAs were deposited into the Plant DNA Bank in Korea (PDBK) and voucher specimens were deposited into the Korea University (KUS) herbarium and National Institute of Biological Resources (NIBR) herbarium.
Four samples—Calanthe bicolor, Dendrobium moniliforme, D. moniliforme “Royal Dream,” and D. moniliforme “Sangeum”—were sequenced by Illumina HiSeq 2000. Twenty other samples were sequenced by Illumina MiSeq. The resulting raw reads from HiSeq were trimmed by Geneious 6.1.8 (Kearse et al., 2012) with a 0.05 error probability limitation. Dendrobium officinale (NC024019) was used as a reference sequence to perform reference-guided assembly using a Geneious assembler. Contigs from the reference-guided assembly were used as reference sequences to perform reference-guided assembly repeatedly until complete plastome sequences were obtained.
The resulting raw reads from MiSeq were trimmed by BBDuk 37.64, implemented in Geneious 11.1.5 (length: 27 kmer). BBNorm 37.64 was used to normalize trimmed reads (target coverage level: 30; minimum depth: 12). Normalized reads were used to perform de-novo assembly by Geneious assembler. Resulting de-novo assembly contigs were used as reference sequences to perform reference-guided assembly to obtain complete plastome sequences when the de-novo assembly did not produce complete plastome contigs. All complete plastome sequences were used as reference sequences to gather chloroplast reads from trimmed read sets. Gathered reads were reused to perform de-novo assembly and validate the level of completeness. Due to its unusual plastome structure, Gastrodia elata was assembled again by SPAdes 3.10.0 (Bankevich et al., 2012) using an error correction tool and assembly module. NGS results are given in Table 1.
Complete plastome sequences were annotated with BLASTn, tRNAscan-SE 2.0 (Lowe and Chan, 2016), ORF finder, and find annotation function in Geneious 11.1.5 (Habenaria radiata plastome sequences, NC035834, used as a reference). Alternative start codons—such as ACG and TTG—were also included in the ORF finder. Genes with many stop codons in the middle of sequences, uncorrectable frame shift mutations, and several large abnormal indels, were judged to have a pseudogene. Gene sequences with complete CDS or few (five or fewer) internal stop codons and for which RNA editing was possible were analyzed to have a gene. These criteria were based on a character reconstructions study of the gene status of ndh in Orchidaceae (Kim et al., 2015a). ORFs from the plastome sequence of four mixotrophic orchids (Corallorhiza maculata var. maculata, Dipodium roseum, Gastrodia elata, and Hetaeria shikokiana) were translated into protein sequences. The translated sequences were exported in the fasta format files to perform psi-blast (Altschul et al., 1997) based on several Orchidaceae plastome databases. Circular plastome maps were constructed using the OGdraw web server (Lohse et al., 2007). All downloaded NCBI plastome data used in our study were re-annotated because the published data contained many annotation errors. Finally, we manually edited some ambiguous annotation area using our comparative alignments of orchid sequences.
Ninety-nine plastome sequences were downloaded from the NCBI to perform phylogenetic analysis (Table 2; 94 Orchidaceae sequences, four Asparagales sequences, and one Liliales sequences). Eight to 79 CDS and two to four rRNA genes were extracted from plastome sequences. Each extracted region was aligned by MAFFT (Katoh et al., 2002) and all alignments were checked manually. Each aligned gene sequences were subjected to jModeltest (Darriba et al., 2012) in CIPRES Science Gateway (Miller et al., 2010) to obtain the best model. GTR-I-G-X or GTR-G-X were the best-fit models for all genes except rrn5 (short sequence), for which SYM-G was the best-fit model. Eighty-three alignments were concatenated to a length of 87,399 bp. Concatenated alignments were used to perform jModeltest (Darriba et al., 2012) in CIPRES Science Gateway (Miller et al., 2010) to obtain the best model. GTR+G+I was the best-fit model for concatenated data. Lost genes were treated as missing data because they do not affect the phylogenetic signals in the remaining genes (Lam et al., 2016). A maximum likelihood (ML) tree was constructed using RaxML-HPC2 on XSEDE in CIPRES Science Gateway with a GTR+G+I model and 100 bootstrap replicates (−672774.637133) of ML optimization likelihood. Concatenated alignment was also used to construct a Bayesian inference (BI) tree. MrBayes_CIPRES api (Miller et al., 2015) was used to construct a BI tree with a Markov chain Monte Carlo (MCMC) chain length of 1,000,000 and GTR+G+I model. The trees obtained were treated graphically using Treegraph2 (Stöver and Müller, 2010).
IRScope (Amiryousefi et al., 2018) was used to describe SC-IR junction regions among 41 plastome sequences generated in the laboratory (Gastrodia elata was excluded due to its lack of IR). The length information of 24 new plastome sequences was visualized by ggplot2 package in R (Wickham et al., 2016). Gene contents of 129 plastome sequences were displayed as a heatmap. The ProgressiveMauve algorithm (Darling et al., 2010) was used to check plastome rearrangements among 116 Orchidaceae plastome sequences, excluding non-photosynthetic orchids with an extremely short plastome (Cyrtosia, Epipogium, Gastrodia, Lecanorchis, and Rhizanthella). The resulting LCBs (locally collinear blocks) from ProgressiveMauve were extracted and numbered to visualize their features (Tables S1 and S2).
The nuclear rDNA region—which contains 18S rRNA, internal transcribed spacers, 5.8S rRNA, 28S rRNA, NTS, and ETS—was generated in 42 Orchidaceae species (Table 2). Forty-two rDNA sequences were aligned by MAFFT (6,043 bp long). The alignment was tested with jModeltest in CIPRES Science Gateway. RaxML-HPC2 on XSEDE in CIPRES Science Gateway was used to construct an ML tree with a GTR+G+I model and 100 bootstrap replicates. The plastome sequence gene data of 42 species were also used to construct an ML tree. Eighty-three CDS and rRNA genes were extracted and aligned by MAFFT. Eighty-three alignments were concatenated into one (80,798 bp long). Concatenated alignments were used to perform jModeltest. An ML tree was constructed by RaxML-HPC2 on XSEDE with a GTR+G+I model with 100 bootstrap replicates. The trees obtained were treated by Treegraph2.
The alignments used in the ML tree construction were also used to estimate divergence time. The GTR estimated model was selected to build a time divergence tree, following the results of PartifionFinder v2.1.1 (Lanfear et al., 2012). An XML file was prepared by BEAUti 2.5.2 (Bouckaert et al., 2019). The XML file was submitted to the CIPRES Science Gateway to perform BEAST2-XSEDE (Bouckaert et al., 2019). A relaxed clock log normal model (Drummond et al., 2006) and Yule model were chosen to perform MCMC with a chain length of 300,000,000. Logs and trees were collected every 5,000 generations, and three independent runs were performed. Three fossil data (Asparagales, normal distribution, sigma 8.0, mean 105.3; Dendrobium, log-normal distribution, sigma 2.0, offset 23.2; and Goodyera, log-normal distribution, sigma 2.0, offset 15.0) were used to calibrate nodes (Ramírez et al., 2007; Conran et al., 2009; Gustafsson et al., 2010; Iles et al., 2015).
Three log and tree files were concatenated by Logcombiner v2.5.2 (Rambaut and Drummond, 2014) by discarding 20% of files. The concatenated log files were checked by Tracer v1.6 (Rambaut et al., 2014) to validate the effective sample size (ESS). Major parameters—including posterior, likelihood, and the prior—exceeded an ESS of 100, and all other parameters exceeded an ESS of 50. The concatenated tree files were treated by Treeannotator (Rambaut and Drummond, 2007) in CIPRES Science Gateway with an option of 0.95 posterior probability. The concatenated maximum clade credibility tree generated by Treeannotator was treated by FigTree v1.4 (Rambaut, 2012) and “phytools” and “ape” packages in R.
The taxonomic positions, NGS methods, raw read numbers, trimmed read numbers, plastome lengths, coverage depth, voucher information, etc. of the 24 new plastomes are summarized in Table 1. The coverage depths ranged from 220x (Amitostigma gracile) to 1,734x (Dactylorhiza viridis var. coreana), so that each plastome sequence was sequenced at least several hundred times.
Among the plastomes of the 24 newly decoded species, the length of the plastome of Gastrodia elata, a non-photosynthetic species, was the shortest at 35,056 bp and that of Cremastra unguiculata, a photosynthetic species, was the longest at 159,341 bp. Gastrodia elata is unique in that it only has a single copy of the plastome because the IR was lost. The plastomes of the remaining 23 species have quadripartite structures consisting of an large single copy (LSC), an SSC, and two IR regions. The total lengths of the plastomes and the relative lengths of the IR, SSC, and LSC of the 24 newly decoded species were compared, as shown in Figure 1.
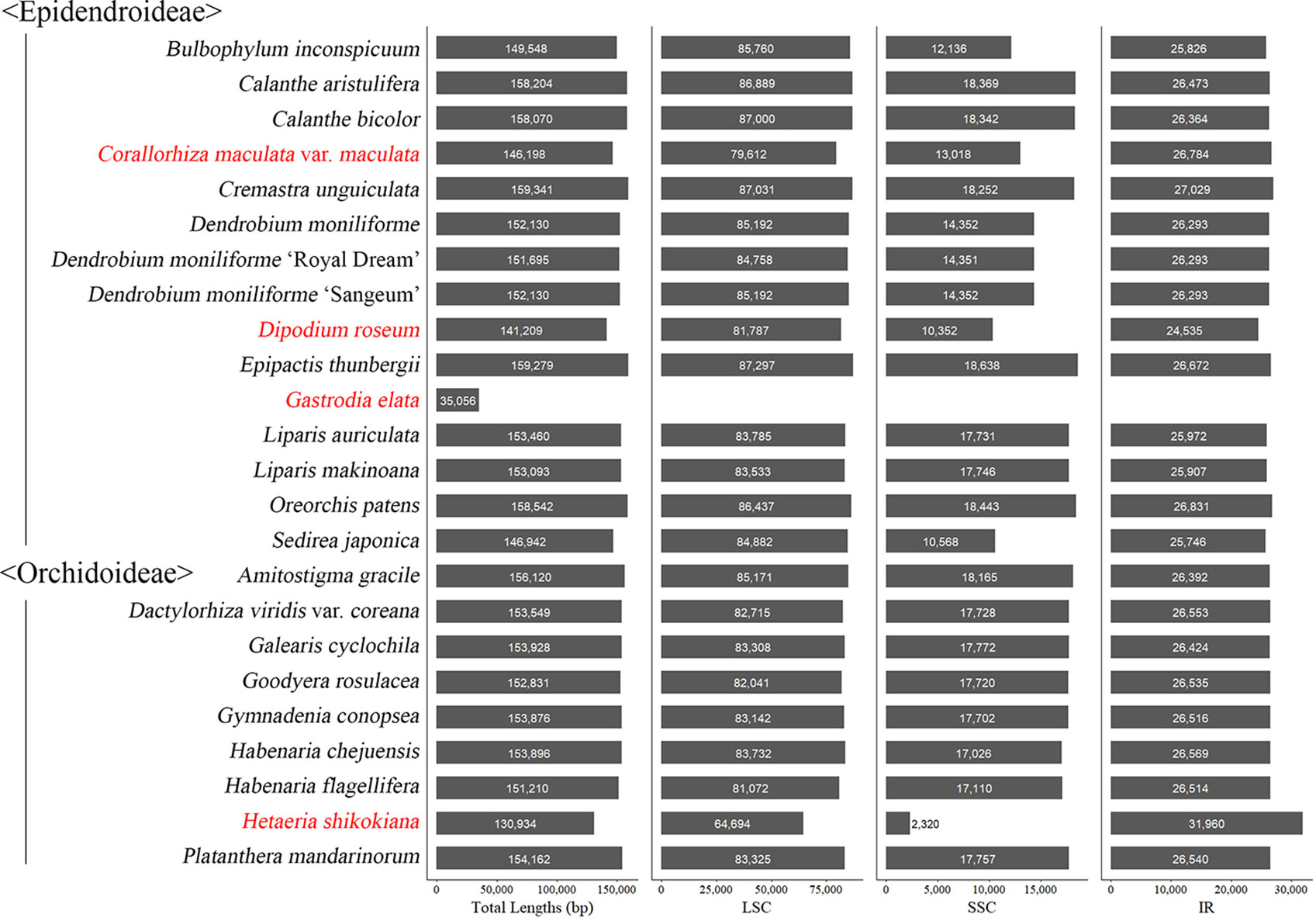
Figure 1 Plastome length variation in 24 newly sequenced orchid species. The plastome of Gastrodia elata is 35,056 bp long and consists of only one single copy region. The species names in red indicate mycoheterotrophic species.
Among the 24 species, the plastomes of 13 (Amitostigma gracile, Calanthe aristulifera, C. bicolor, Dactylorhiza viridis var. coreana, Epipactis thunbergii, Galearis cyclochila, Goodyera rosulacea, Habenaria chejuensis, H. flagellifera, Liparis auriculata, L. makinoana, Oreorchis patens, and Platanthera mandarinorum) had all the same genes as typical plant plastids. Various subunits of the ndh gene class were found to be pseudogenized or lost in eight species (Bulbophyllum inconspicuum, Cremastra unguiculata, Dendrobium moniliforme, D. moniliforme “Sangeum”, D. moniliforme “Royal Dream”, Dipodium roseum, Gymnadenia conopsea, and Sedirea japonica). In addition, petL was lost in Cremastra unguiculata, psbD was pseudogenized in Dipodium roseum, and trnG-UCC was lost in Gymnadenia conopsea. In the case of the non-photosynthetic species Corallorhiza maculata var. maculata, ccsA, cemA, ndhB, ndhC, ndhD, ndhG, ndhH, ndhI, ndhJ, ndhK, petA, petD, petG, psaA, psaB, psbA, psbC, rbcL, rpoA, rpoB, rpoC1, and ycf1 existed as pseudogenes and ndhA, ndhE, ndhF, psbB, psbJ, psbL, psbM, and rpoC2 were lost. In the case of Hetaeria shikokiana, another non-photosynthetic species, many genes were lost or pseudogenized. Among the genes, cemA, ndhA, ndhB, ndhC, ndhH, petA, petD, petG, petN, psaB, psaI, psbC, psbF, psbN, rpoA, rpoC1, rpoC2, ycf3, and ycf4 were pseudogenized, and atpA, atpB, ccsA, ndhD, ndhE, ndhF, ndhG, ndhI, ndhJ, petB, psaA, psaC, psbA, psbB, psbJ, psbL, psbM, rbcL, rpl23, and rpoB were lost. Gastrodia elata, which has the shortest plastome of the 24 species examined, only had the following genes: accD, clpP, matK, rpl2, rpl14, rpl16, rpl20, rpl36, rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps14, rps18, rps19, rrn5, rrn16, rrn23, ycf1, and ycf2. Two pseudogenes (psaI and psbK) remained, and all the other genes were lost.
The Orchidaceae plastomes that have been completely decoded and can be used in comparative studies comprise 60 genera, 142 species, and 146 accessions, including the 24 new plastomes (NCBI database, June 30, 2019). Since the main purpose of this study was to identify the evolutionary trends in plastomes of the entire Orchidaceae, there were three genera—Cymbidium (9 spp.), Dendrobium (40 spp.), and Holcoglossum (11 spp.)—for which the plastomes of many species were decoded, but only three species each were included in the comparative study. In the case of Corallorhiza (10 spp.) and Neottia (7 spp.), the plastomes of all species were included in the comparative study, even though the plastomes of many species were decoded, because these genera include both photosynthetic and non-photosynthetic species. In the case of the remaining genera, all available plastomes were included in the comparative study except for one species of Cypripedium, in which many problems were found in the re-annotation process. Therefore, five subfamilies, 17 tribes, 60 genera, 118 species, and 124 accessions were used in the comparative study in this paper. In addition, five outgroup plastomes were also used for comparison. Therefore, the GenBank accession numbers, taxonomic classification, habitats, plastome sizes, LSC lengths, IR lengths, SSC lengths, GC contents, and numbers of genes and pseudogenes of the 129 plastomes used in this study, along with whether or not their corresponding species is photosynthetic, are listed in Table 2. The plastomes of land plants usually have an AT-biased base composition. Furthermore, highly reduced plastomes have more AT-biased substitutions than typical plastomes because they experience relaxed selection. The average GC content of orchid plastomes in this study was 36.40 ± 1.71%. The three highly reduced orchid plastomes—Epipogium aphyllum, E. roseum, and Rhizanthella—had 32.8%, 30.6%, and 34.2% GC contents, respectively. The CDS of E. aphyllum., E. roseum, and Rhizanthella had 38.4%, 34.3%, and 38.1% GC contents, respectively. They showed 2–6% more AT-biased substitutions than general orchid plastomes. Extreme AT bias was reported in the plastome of Rhopalocnemis phalloides, which had a GC content of only 13.2% (Schelkunov et al., 2019). Among the 124 Orchidaceae plastomes used in the comparative study, 42 were decoded using the NGS method in the laboratory of the corresponding author. In addition to the plastomes, the base sequences of nuclear ribosomal RNA repeating unit DNA (nrDNA, 18S-ITS1-5.8S-ITS2-28S-NTS-ETS), which are present as tandem repeats, could also be assembled in these 42 species (Figure 2). Therefore, the GenBank accession numbers of these species are also published in Table 2 for the first time. Consequently, these 42 species were used to compare nrDNA repeating unit-based trees and plastome-based trees.

Figure 2 Diagram of nuclear ribosomal (nr) DNA repeat units consisting of 18S-ITS1-5.8S-ITS2-28S-NTS-ETS. The total length of the unit is approximately 10 kb, and it is arranged as a tandem repeat. The nrDNA repeat sequences of 42 orchid species were first reported in this paper.
Among the 124 Orchidaceae plastomes, 121 had an IR region. In the case of Aphyllorchis montana, the IR region was lost and the plastome existed only as a single copy (Table 2), similarly to Gastrodia elata 1 and 2. The plastome of Aphyllorchis montana is small at 94,559 bp. Another example of the SSC region being lost and the plastome consisting of the LSC and two IRs was reported in Epipogium aphyllum (Figure 3). These three species are examples of the quadripartite structure of a plastome not being maintained. The quadripartite structure was maintained in all the remaining species. Epipogium aphyllum is the only example in which the IRs appear consecutively without any SSC. However, in the case of Epipogium roseum, which is a related species, an SSC region, although short, existed at 890 bp, and the IR region was shortened to 261 bp (Table 2, Figure 3).
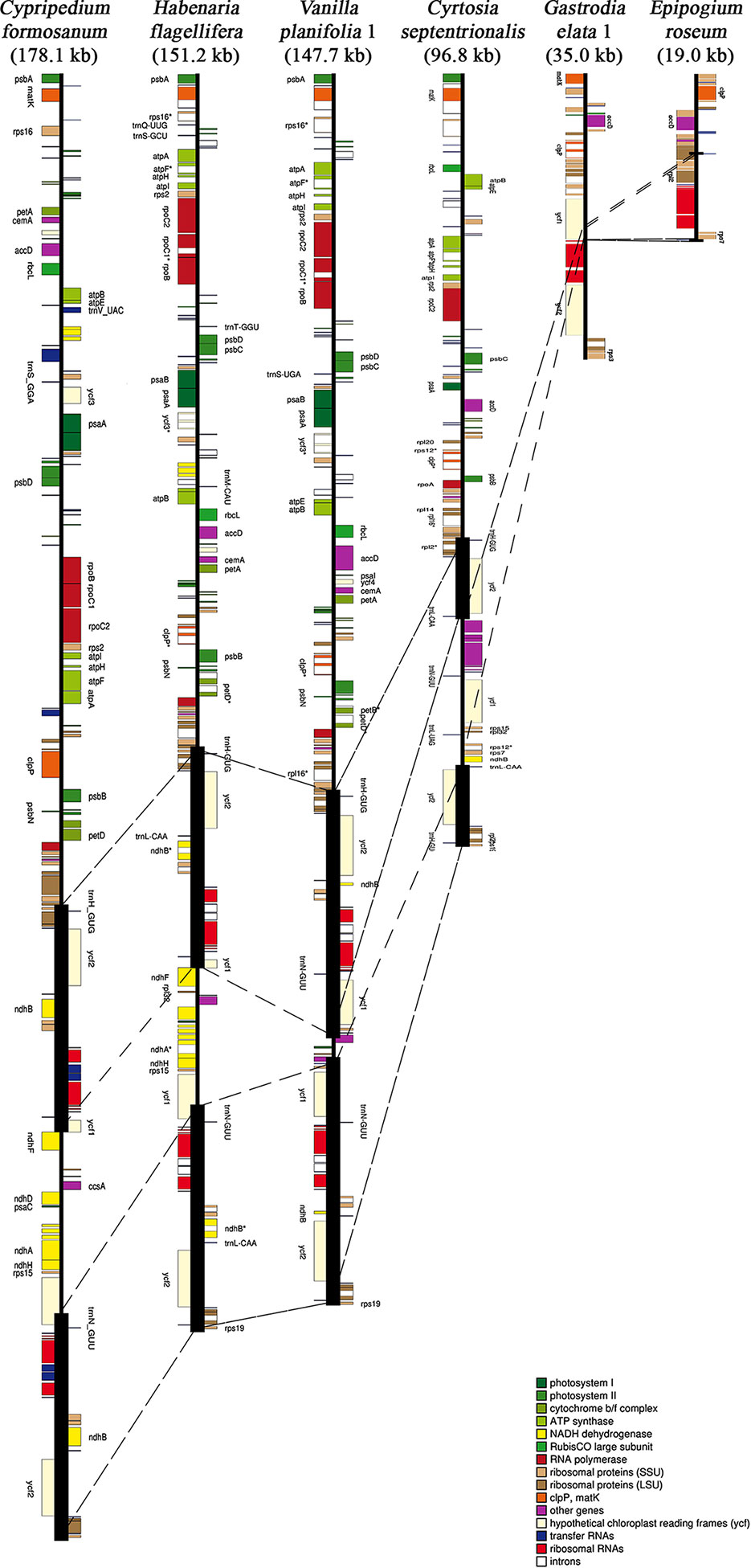
Figure 3 Six representative plastomes of Orchidaceae. Cypripedium formosanum has the longest plastome (178.1 kb), while Epipogium roseum has the shortest. Gastrodia elata and Epipogium roseum hold 27 genes in their plastome, even though they have substantially different plastome sizes. The heavy vertical bars indicate inverted repeat (IR) regions and the broken lines among plastomes indicate the boundaries of IR and single copy regions.
The largest plastome in Orchidaceae was 178,131 bp, in Cypripedium formosanum, and the smallest was 19,047 bp, in Epipogium roseum (Figure 3). The plastome sizes of all photosynthetic orchids were at least 140 kb. On the other hand, the plastome sizes of non-photosynthetic mycoheterotrophic orchids were mostly smaller than 150 kb and showed high positive correlations with gene numbers (Figure 4A). In addition, unlike the plastome sizes of epiphytic orchids, which were at least 140 kb, those of terrestrial orchids varied greatly because they include mycoheterotrophic species (Figure 4B). Although the plastome sizes of photosynthetic orchids mostly ranged from 145–160 kb, those of Cypripedium had a much larger derange; this was highly correlated with the expansion of the LSC region, and not correlated much with the expansion of IRs (Figures 4C, D). On the other hand, in the case of mycoheterotrophic species, plastome sizes showed high correlations with the size of both the LSC and IR.
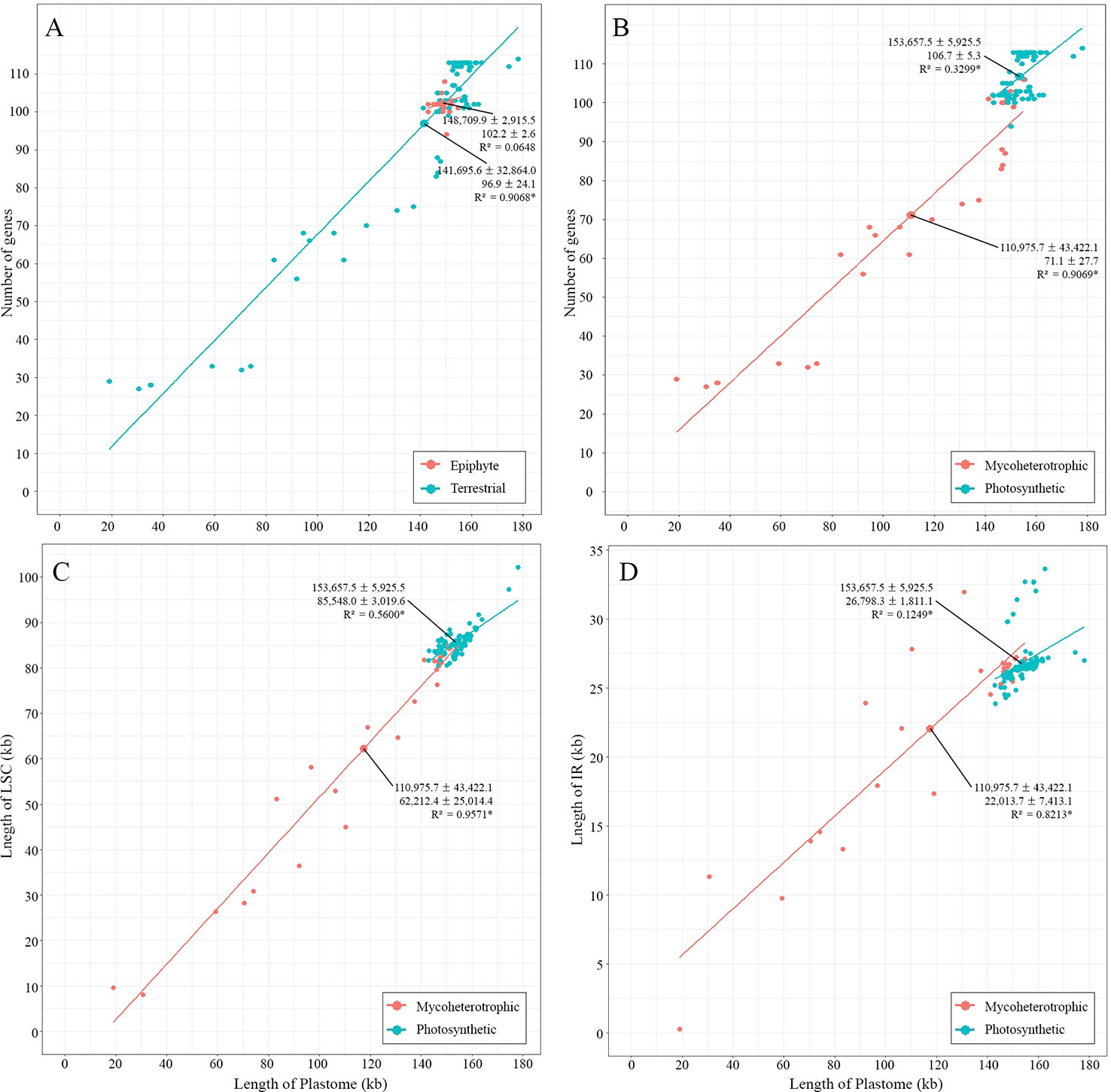
Figure 4 Relationships between plastome lengths and gene numbers. (A): Terrestrial orchids show a wider range of variation than epiphytic orchids. (B): Mycoheterotrophic orchids show a wider range of variation than photosynthetic orchids. (C, D): Plastome lengths are more strongly correlated with LSC lengths than IR lengths in both mycoheterotrophic and photosynthetic orchids.
The gene contents of the 124 orchid species showed high variation (Figure 5). A plastome usually contains a total of 113 genes, comprising 6 atp, 11 ndh, 6 pet, 9 rpl, 4 rpo, 12 rps, 4 rrn, 5 psa, 15 psb, 30 trn, and 11 ungrouped genes. Among the 124 species of Orchidaceae, 27 (22%) have all 113 genes existing in an active status. In eight species, one to three genes were pseudogenized or lost. These 35 species fall into the category of having plastomes with almost all their plastid genes, and they make up 28% of all 124 species in this study. There are 69 species where four to 11 of the 11 genes in the ndh gene class or all 11 ndh genes plus one or two other gene(s) do not function, indicating that the plastid ndh gene class does not function in 56% of all 124 species. Furthermore, ndh genes, photosynthesis light reaction genes (pet, psa, psb), and rpo gene were shown to be lost in 13 species. Such cases are also accompanied by the loss of rpo genes. Finally, in the case of Lecanorchis (two species), Epipogium (two species), Gastrodia (one species, two accs.), and Rhizanthella (one species), many housekeeping genes such as rpl, rps, and trn were also lost.
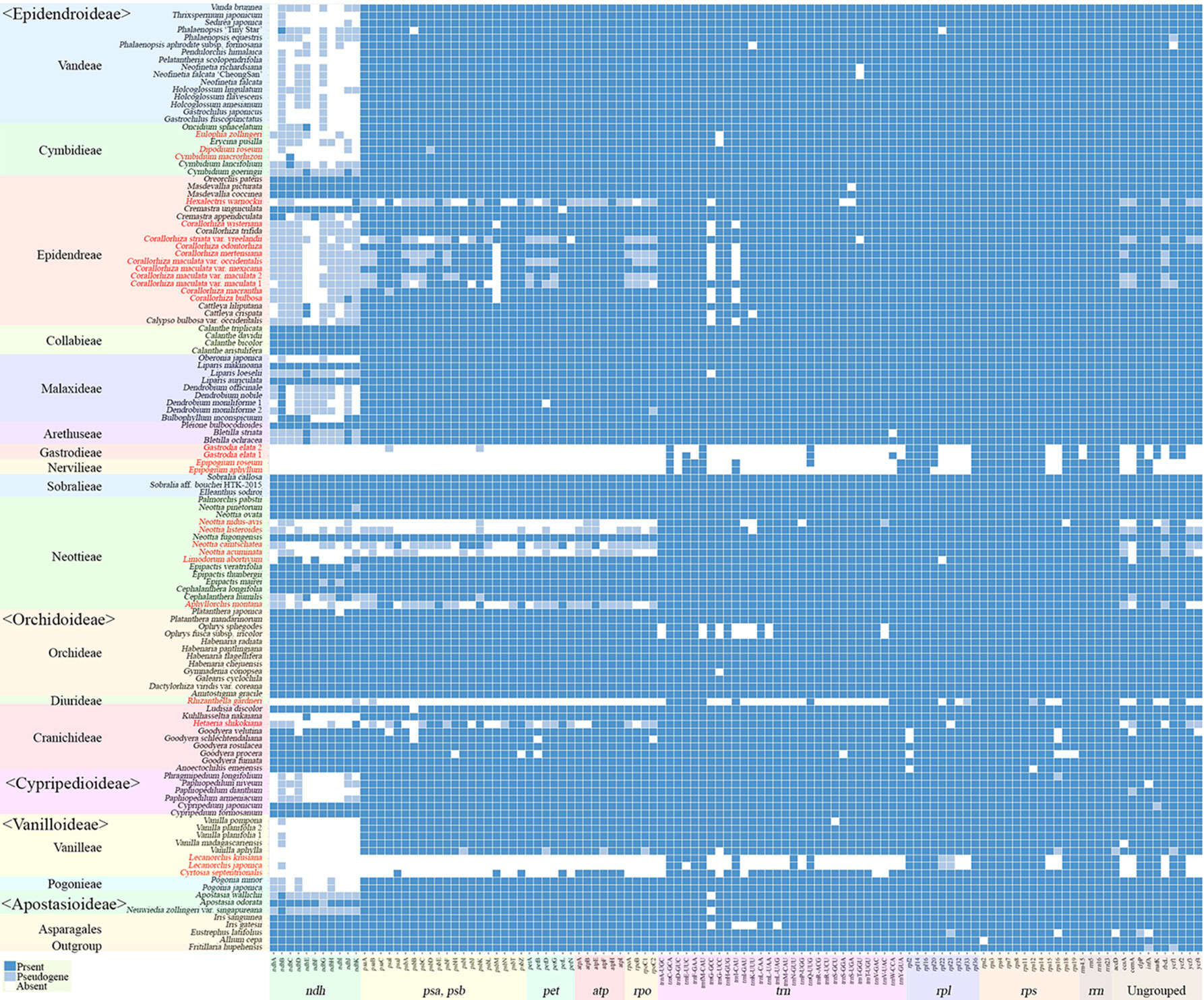
Figure 5 Distribution patterns of gene loss in Orchidaceae. The dark blue, light blue, and white blocks indicate presence, pseudogene, and absence of each gene, respectively. The non-functionalization of 11 ndh genes are distributed widely across all taxonomic groups of Orchidaceae. This frequently occurs in Epidendroideae and Vanilloideae. The non-functionalization of psa, psb, pet, and rpo gene classes are confined to mycoheterotrophic lineages. In addition, the loss of housekeeping genes such as the rps, rpl, or trn gene classes occur independently in four genera, Epipogium, Gastrodia, Lecanorchis, and Rhizanthella. The species names in red indicate mycoheterotrophic orchids.
The presence of genes and pseudogenization in the 124 Orchidaceae and five outgroup species are set forth in Figure 5 by subfamily, tribe, species, and gene class. In the case of photosynthetic orchids, up to 13 genes were lost. In most of these cases, 11 ndh genes and one or two other genes were lost. In an exceptional case with photosynthetic orchids, 19 genes were lost in Vanilla aphylla with epiphytic habitats. The lost genes seem to consist of 11 ndh genes and eight other pseudogenized genes. However, given that only 11 to 12 genes were lost from the plastids in other Vanilla species, it is inferred that the plastid gene annotation of this species is problematic. On the other hand, among the species known to be mycoheterotrophic species, the fewest genes were lost in Limodorum abortivum, where the loss of seven genes comprising cemA, five ndh genes, and rpl22 was observed. Limodorum abortivum was followed by Cymbidium macrorhizon, in which 10 ndh genes were lost, Eulophia zollingeri, where 11 ndh genes and trnG-UCC were lost, and Dipodium roseum, where 11 ndh genes and a psbD gene were lost. However, although these four species have the mycoheterotrophic nutritional mode, they also have chlorophyll, and are reported or observed to be orchids that carry out low levels of photosynthesis. In the case of Corallorhiza, both mycoheterotrophic and photosynthetic species exist, and the levels of photosynthesis vary according to variety or habitat, even in the same species. When seen based on the degree of gene loss, species in which 11 ndh genes and one to three other genes were lost are very likely to preserve photosynthetic activity. When the gene contents of the 124 orchid plastomes were compared, a large gap was found in the area where 14–25 genes were lost. Therefore, whether photosynthetic ability was lost or not can be divided around this range (Figures 4 and 5).
Among the plastomes that are classified into completely non-photosynthetic orchids because at least 25 genes were lost, two groups are recognized based on the numbers of lost genes and preserved genes. That is, they are a group of plastomes in which the number of preserved genes is similar to or larger than the number of lost genes (13 plastomes), and a group of plastomes in which the number of lost genes is overwhelmingly larger (seven plastomes). Among the 124 plastomes, the Orchidaceae species that belong to the latter are Epipogium aphyllum (86 lost, 27 preserved), Epipogium roseum (84 lost, 29 preserved), Gastrodia elata 1 and 2 (85 lost, 28 preserved), Lecanorchis kiusiana (80 lost, 33 preserved), Lecanorchis japonica (81 lost, 32 preserved), and Rhizanthella gardneri (80 lost, 33 preserved). These seven plastomes have 27–33 genes preserved, and among them, 20 genes are common (Table 3). These 20 genes are inferred to be the minimum common plastid genes necessary to maintain non-photosynthetic plastids. However, among these 20 plastid genes, five are missing in any of the remaining 117 Orchidaceae plastomes. Therefore, 15 genes are common to all orchid plastids. The 15 genes comprise 14 housekeeping genes (rpl14, rpl16, rpl26, rps2, rps3, rps4, rps7, rps8, rps11, rps14, rrn5, rrn16, rrn23, and trnC-GGA) and clpP.
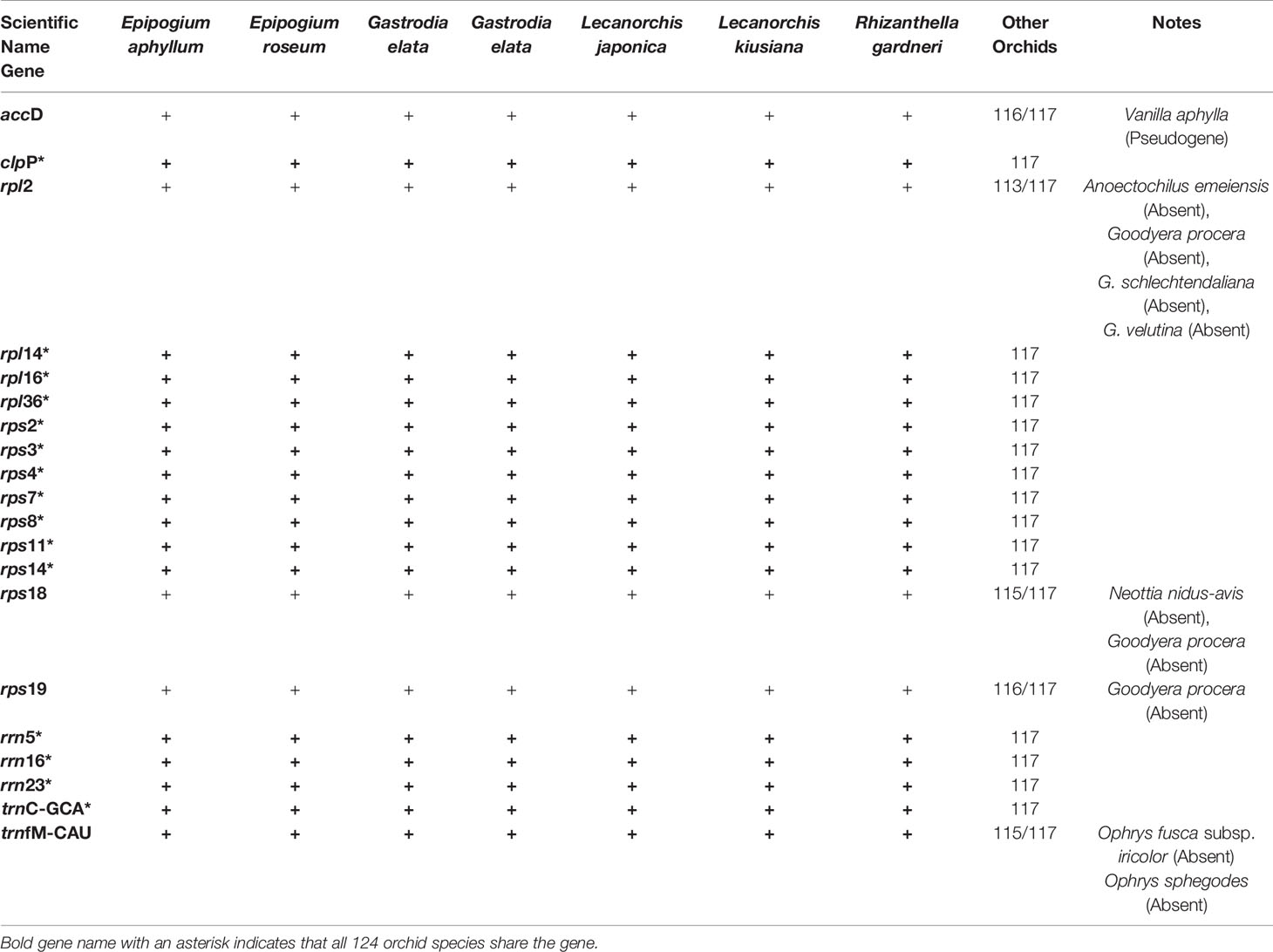
Table 3 Minimum genes required for seven orchid species with extremely degraded plastomes (Epipogium aphyllum, E. roseum, two Gastrodia elata, Lecanorchis japonica, L. kiusiana, and Rhizanthella gardneri).
To determine the patterns of plastome gene loss over Orchidaceae evolution, we plotted the gene loss patterns on the ML phylogenetic trees using Bayesian estimation approaches (Figure 6). Apostasioideae, Vanilloideae, Cypripedioideae, and Orchidoideae are shown in Figure 6A, and Epidendroideae is shown in Figure 6B. In the case of the ndh gene class, gene loss or pseudogenization occurred independently in almost all lineages of Epidendroideae. On the other hand, in the case of the four remaining subfamilies, the loss of ndh genes, although relatively rare, was observed independently in at least five lineages. The loss of genes directly involved in the photosynthetic light reaction—such as psa, psb, and pet—was observed in six lineages of Epidendroideae, two lineages of Orchidoideae, and one lineage of Vanilloideae. The loss of housekeeping genes such as rpl, rps, and trn occurred independently in three lineages: Gastrodia-Epipogium of Epidendroideae, Lecanorchis of Vanilloideae, and Rhizanthella of Orchidoideae.
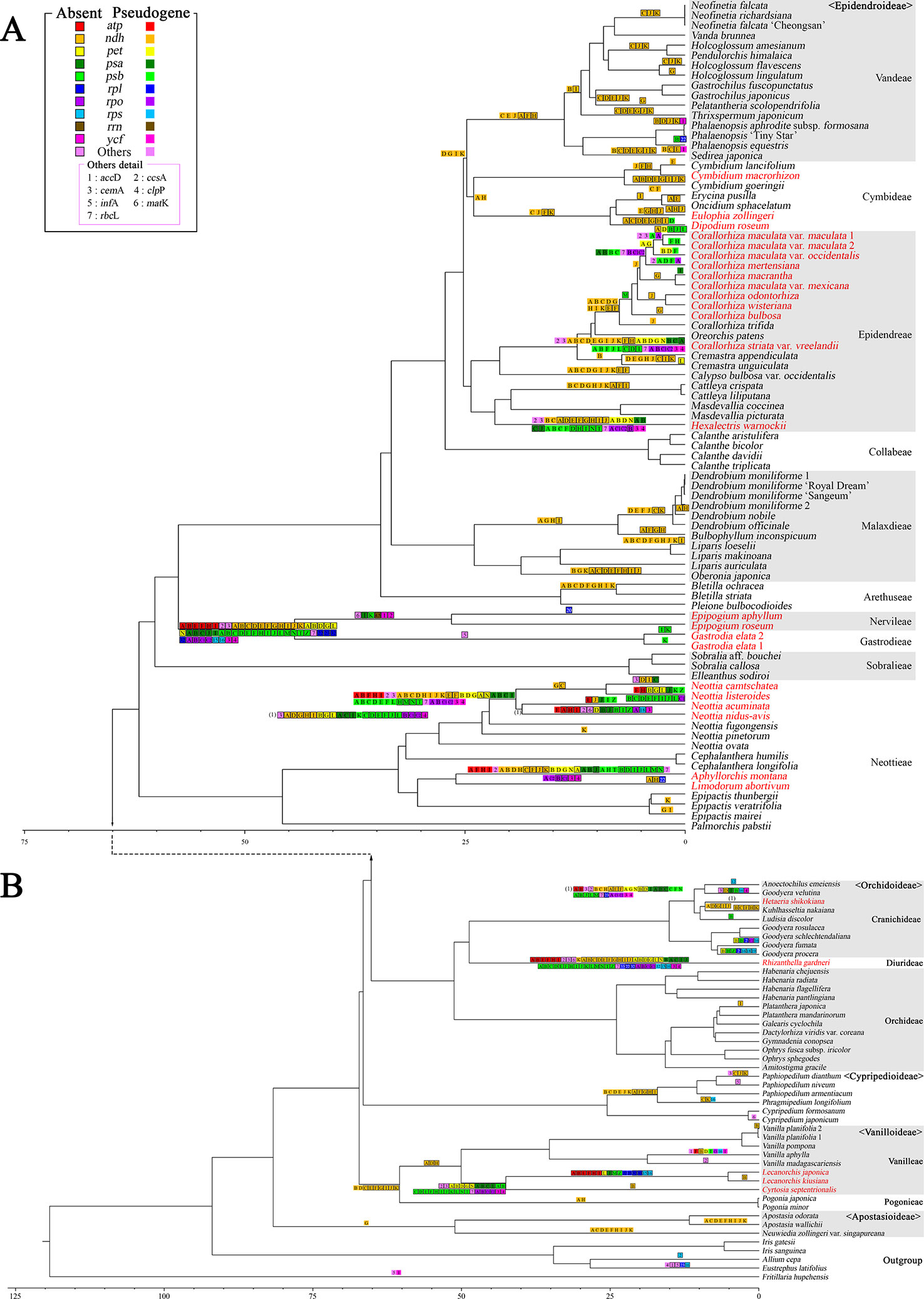
Figure 6 Evolution of gene losses in the phylogenetic tree of Orchidaceae. A total of 129 taxa—124 Orchidaceae and five outgroup taxa—were the subject of tree reconstruction. The sequences of 83 protein coding genes were concatenated to a length of 87,399 bp. A maximum likelihood (ML) tree was constructed using RaxML-HPC2 with a GTR+G+I model (ML = −672774.637133 of ML optimization likelihood). All the genes were then plotted on the tree node using parsimony criteria under the condition of no parallel gains of the same gene. The species names in red indicate mycoheterotrophic orchids. (A): The basal portion of the tree showing the subfamilies Apostasioideae, Vanilloideae, Cypripedioideae, and Orchidoideae. (B): The upper portions of the tree showing the subfamily Epidendroideae.
Sixteen genes in the typical land plant plastomes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) generally have one intron, and two genes (clpP and ycf3) have two introns. The absence or presence of these 20 introns are summarized (Figure 7). The majority of intron losses are associated with the loss of their corresponding gene; the exceptions are rpl16, rps16, and clpP(2), in which the introns were lost but the corresponding genes were not in some terminal clade of the tree.
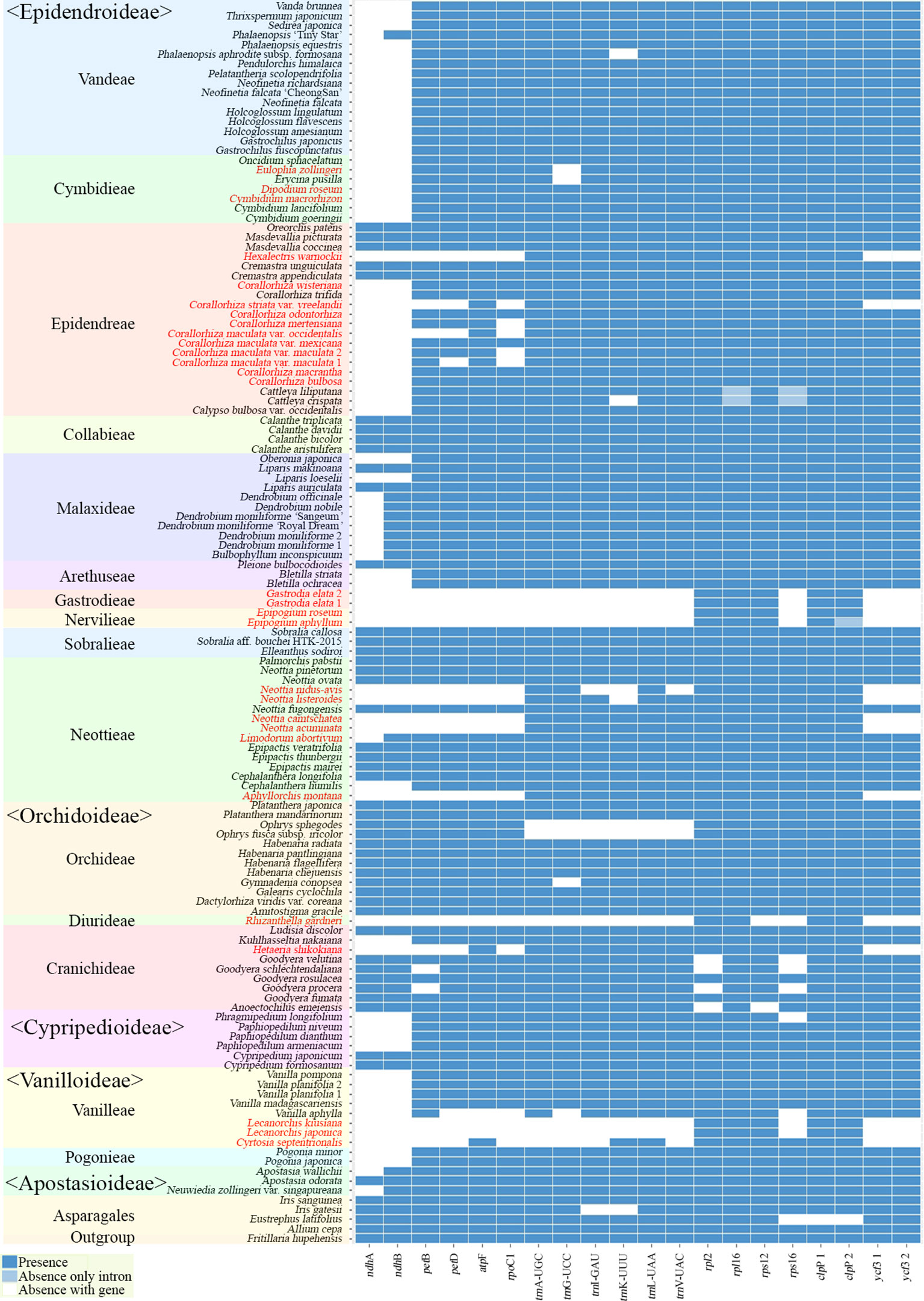
Figure 7 Distribution patterns of the loss of 20 introns in Orchidaceae. The dark blue, light blue, and white blocks indicate intron presence, absence without corresponding gene loss, and absence with corresponding gene loss, respectively.
To identify gene rearrangements among Orchidaceae plastomes, gene block analysis was carried out using MAUVE. Among the 124 Orchidaceae plastomes, only 116 taxonomic groups were analyzed; eight were excluded—Cyrtosia septentrionalis, Epipogium roseum, Epipogium aphyllum, Gastrodia elata 1, G. elata 2, Lecanorchis kiusiana, L. japonica, and Rhizanthella gardneri—because they had severe gene losses and were therefore meaningless in gene block analysis. A total of 134 gene blocks were identified. The aligned lengths (kb), gene regions, forward/reverse orientations, and names of taxa distributed in individual gene blocks are listed in Tables S1 and S2. In addition, the orientations of the 134 gene block and distribution patterns by taxonomic group (Figure 8A), and the lengths of individual blocks and the number of shared taxonomic groups (Figure 8B), were set forth. Among the 134 blocks, 104 were identified as genome rearrangement blocks because they showed variations, and 30 were identified as constant blocks. In terms of plastome regions, 48 blocks were found in the LSC region and among them, 34 were identified as rearrangement blocks and 14 were identified as constant blocks. Only three blocks were found in the IR region, and they were rearranged blocks. Eighty-three blocks (~60%) were found in the SSC region and 24 small blocks were identified in the ycf1 gene region existing at the IR/SSC boundary.
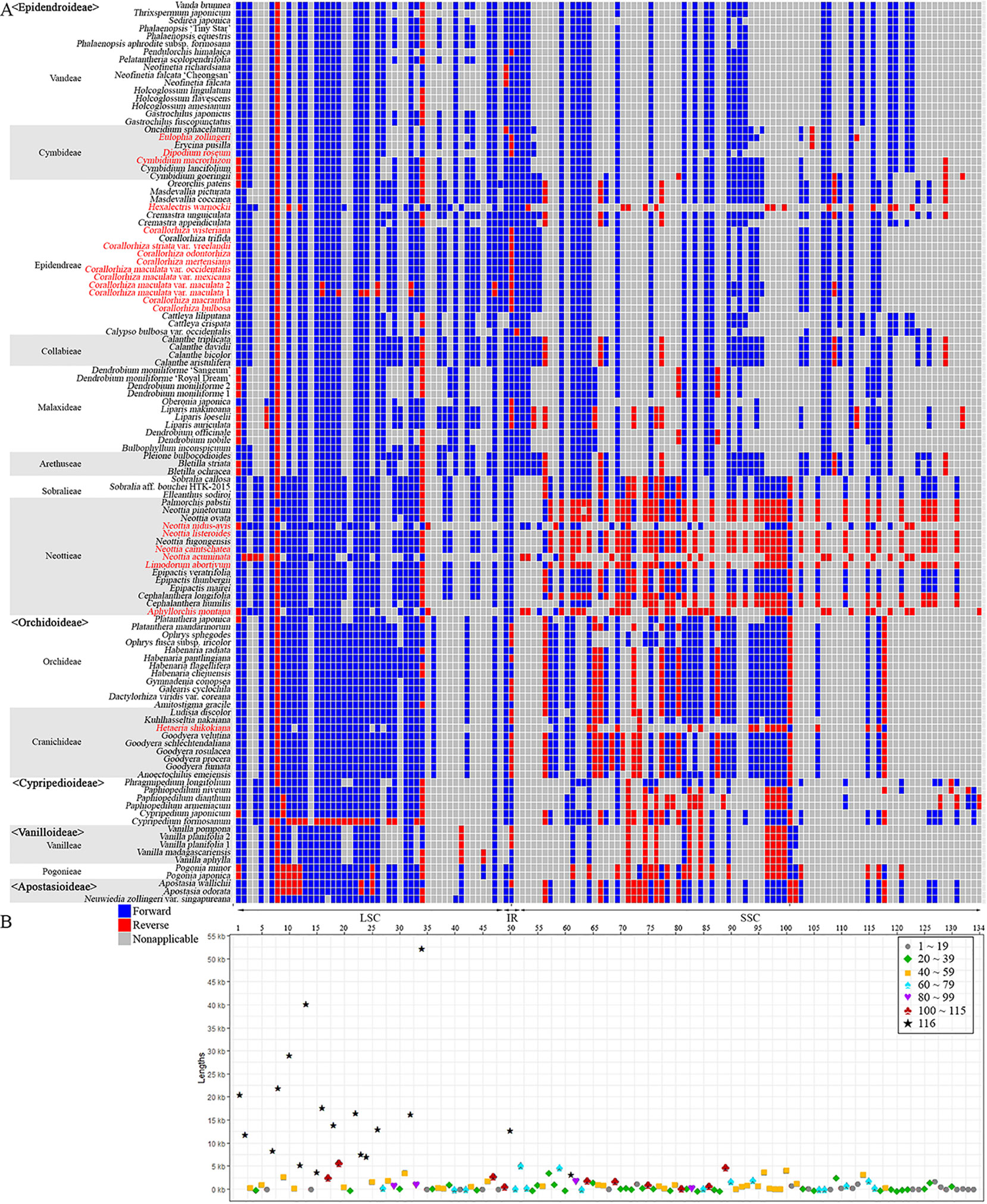
Figure 8 Distribution of gene blocks. A total of 134 gene blocks were recognized in the 124 orchid plastomes. Thirty were constant blocks. 104 blocks show both forward (dark blue) and reverse (red) orientations and confirmed the block rearrangements. Blocks that are non-applicable because of gene losses or shifts are indicated in gray. All large rearrangements longer than 15 kb are located on the LSC. The species names in red indicate mycoheterotrophic orchids. (A): Forward and reverse orientations of each gene block in the orchid species. (B): Locations, lengths, and frequencies of 134 gene blocks.
Based on block length, 81 blocks shorter than 1 kb, 35 blocks 1 to 5 kb long, six blocks 5 to 10 kb long, and 12 blocks longer than 10 kb were found. The longest gene block is 52.63 kb in length, located between petD-rps12 in the LSC region that has rearranged. This block was followed by trnE-UUC-ycf3 (40.6 kb, rearrangement), rpoC2-rpoB (29.49 kb, rearrangement), trnG-UUC-rpoC2 (22.42 kb, rearrangement), psbA-trnK-UUU (21.00 kb, rearrangement), cemA-trnP-UGG (18.15 kb, rearrangement), trnV-UAC-atpB (16.92 kb, rearrangement), psbB-petD (16.64 kb, rearrangement), trnL-UAA-ndhJ (14.42 kb rearrangement), psaJ-clpP (14.42 kb, rearrangement), rrn16-rrn5 (13.19 kb, rearrangement), and rps16 (12.32 kb, rearrangement). Among the 12 gene blocks longer than 10 kb long, 10 are distributed in the LSC region and two in the IR region (Figure 8B).
In the gene block analysis, to compare structures in the vicinity of the IR/SSC junctions where many small rearrangements are concentrated, the SC-IR junctions of 42 species of Orchidaceae (including 24 new plastome sequences) decoded in this laboratory were schematized by taxonomic group and expressed as shown in Figure S2. Here, the IR of Gastrodia elata was deleted. All the LSC-IRb junctions of 19 taxonomic groups, not including non-photosynthetic taxonomic groups (Cyrtosia, Hetaeria, Lecanorchis), were formed in the vicinity of rpl22. The LSC-IRb junction of Cyrtosia septentrionalis was found in the vicinity of rps19, that of Lecanorchis was found in the vicinity of rpl2, and that of Hetaeria shikokiana was found in the vicinity of rpl2. As with LSC-IRb junctions, LSC-IRa junctions were identified to have been formed in the IGS before psbA in all taxonomic groups except for three non-photosynthetic taxonomic groups (Hetaeria shikokiana, Lecanorchis kiusiana, and L. japonica). The IRa-SSC and IRb-SSC junctions of the remaining taxonomic groups—except non-photosynthetic taxonomic groups, seven species of Vanilloideae (in Cyrtosia, Lecanorchis, Pogonia, and Vanilla) and one species of Orchidoideae (Hetaeria)—were located on ndhF and ycf1, respectively. Seven taxonomic groups belonging to Vanilloideae commonly showed a phenomenon of a shorter SSC and among them, in the case of non-photosynthetic taxonomic groups Cyrtosia and Lecanorchis, the rRNA genes generally present in the IR were relocated to the SSC. Although Hetaeria shikokiana belongs to Orchidoideae, it has a shortened SSC (2,320 bp), similar to the taxonomic groups of Vanilloideae (Figure 1).
The phylogenetic relationships connected to Apostasioideae {Vanilloideae [Cypripedioideae (Orchidoideae, Epidendroideae)]} were identified based on a ML tree made using 83 genes (Figure S3). Although bootstrap values were high at most nodes, those of the nodes of Anoectochilus emeiensis and Goodyera velutina, and G. procera and G. fumata of Cranichideae, were shown to be relatively low at 69.4% and 77.6%, respectively, as was the bootstrap value of the node dividing Neottia camtschatea, N. listeroides, and N. nidus-avis at 63.3%. The results of a BI tree made using the same data matrix are shown in Figure S4. The BI value of Cranichideae, which had low bootstrap values in the ML tree, showed a high degree of support at 1.0, but the BI value of Corallorhiza's branches showed a degree of support of 0.5. Despite the bootstrap and BI support problems in several sub-taxonomic groups, the relationships among the tribes (Pogonieae, Vanilleae, Cranichideae, Diurideae, Orchideae, Neottieae, Sobralieae, Nervilieae, Gastrodieae, Arethuseae, Malaxideae, Collabieae, Epidendreae, Cymbidieae, and Vandeae) represented by the data matrix used were identified to be strongly supported in both the ML and BI trees. In addition, both ML and BI trees had very long branch lengths leading to the taxa with highly reduced plastomes, such as Epipogium, Gastrodia, and Rhizanthella (Figures S3 and S4).
Among the 42 Orchidaceae species decoded in this laboratory, both plastome sequences and nrDNA units (18S-ITS1-5.8S-ITS2-28S) about 10 kb long exist (Table 2). Therefore, phylogenetic trees using the nrDNA regions and phylogenetic trees using the base sequences of the 83 plastid genes were made using the same method and compared (Figure 9). The best nrDNA tree was obtained at ML = −35673.363572 and the best plastid tree was obtained at ML = −332414.394814. These phylogenetic trees included only three subfamilies—Vanillioideae, Orchidoideae, and Epidendroideae—and the mutual relationships among the three subfamilies and the tree topologies in Vanilloideae and Orchidoideae were identical. However, differences between the two trees were found in the tree topologies in Epidendroideae. In particular, the phylogenetic positions of Gastrodia elata, Bulbophyllum inconspicuum, Calanthe bicolor, C. aristulifera, Cremastra unguiculata, Corallorhiza maculata var. maculata, Oreorchis patens, Sedirea japonica, and Thrixspermum japonicum differed between the two trees (Figure 9).
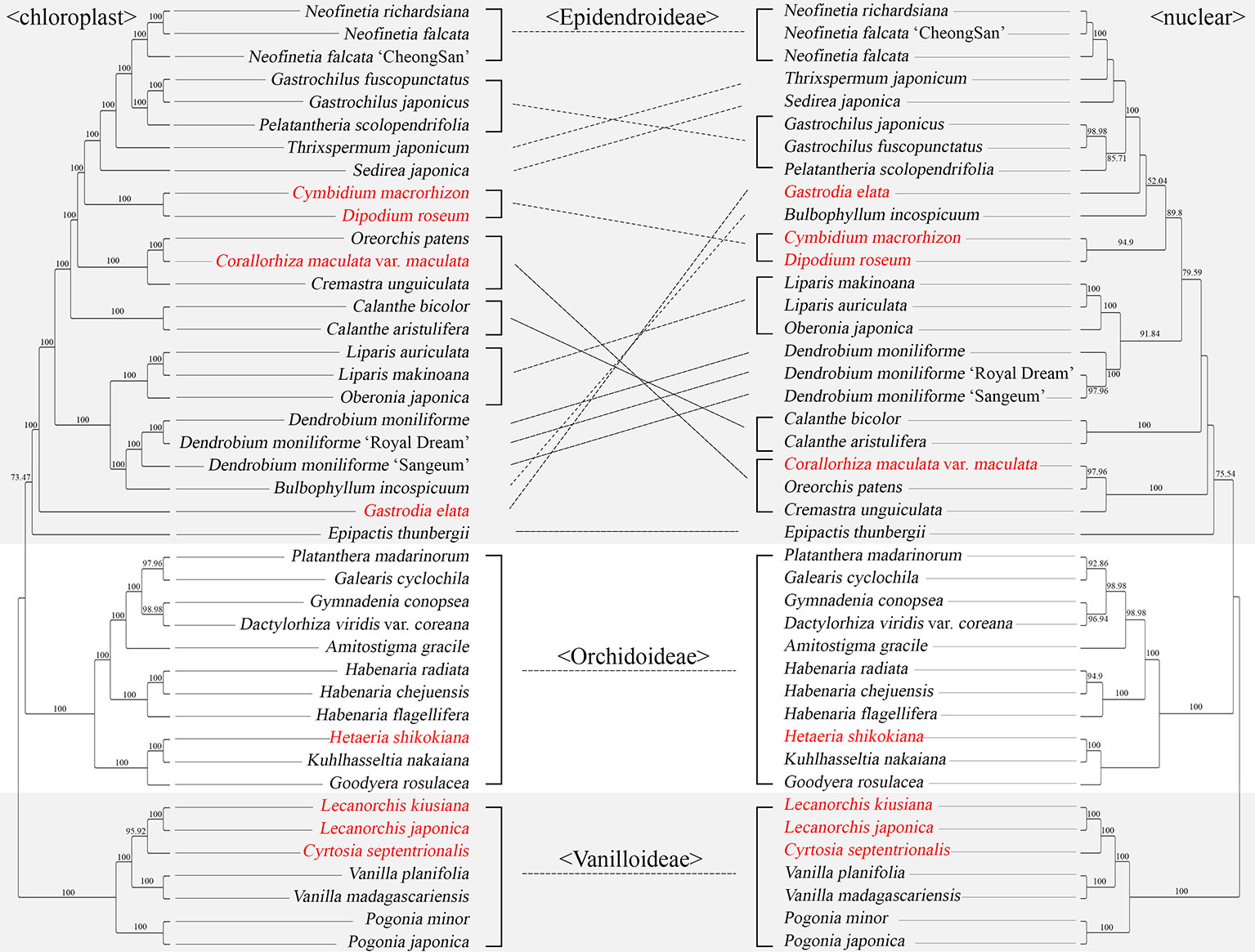
Figure 9 Comparison of a plastid tree and nrDNA tree for the same 42 orchid species. The 83 aligned protein coding genes were 80,798 bp long. A maximum likelihood (ML) tree was constructed using RaxML-HPC2 with a GTR+G+I model with 100 bootstrap replicates. The best plastid tree was obtained with ML = −332414.394814. The nrDNA unit (18S-ITS1-5.8S-ITS2-28S) was approximately 10 kb long. The tree reconstruction methods for nrDNA were identical to those of the plastid tree. The species names in red indicate mycoheterotrophic orchids. The lines between the two trees indicate the topological differences between them.
A phylogenetic tree was constructed using 83 plastid genes in 124 Orchidaceae species and five outgroup species, and the divergence times of individual tree nodes were estimated using BEAST2. The results are shown in Figure 10. The inferred branching times of 21 main clades are set forth in Table 4. For easy comparison with existing studies, literature materials are also presented in Table 4. In this study, Orchidaceae was inferred to have diverged from other Asparagales at 99.20 (83.78–114.92) mya; Apostasioideae, which is a basal subfamily, was inferred to have diverged from other subfamilies at 79.91 (59.65–99.00) mya; and Vanilloideae was inferred to have diverged at 69.84 (52.59–89.63) mya. It was inferred that Pogonieae and Vanilleae of Vanilloideae diverged at 57.52 (37.88–77.68) mya, Cypripedioideae diverged at 64.97 (48.54–84.93) mya, and Orchidoideae diverged from Epidendroideae at 59.16 (43.99–78.66) mya. Cranichideae of Orchidoideae diverged from Orchideae at 46.64 (29.49–66.26) mya and Diurideae diverged from Cranichideae at 37.59 (23.13–57.30) mya. Neottieae diverged at 55.06 (40.55–73.82) mya and other taxonomic groups of Epidendroideae and Sobralieae diverged at 51.46 (37.65–69.44) mya. In addition, Gastrodieae and Nervilleae diverged from other Epidendroideae at 49.21 (35.89–66.60) mya, and the two tribes diverged from each other at 38.27 (27.59–51.84) mya. Arethuseae diverged at 39.57 (27.20–54.68) mya, Malaxideae at 34.95 (24.93–47.40) mya, Collabieae at 32.10 (23.20–44.64) mya, Epidendreae at 29.96 (21.36–41.98) mya, and Cymbideae and Vandeae at 28.13 (18.92–39.72) mya (Figure 10, Table 4).
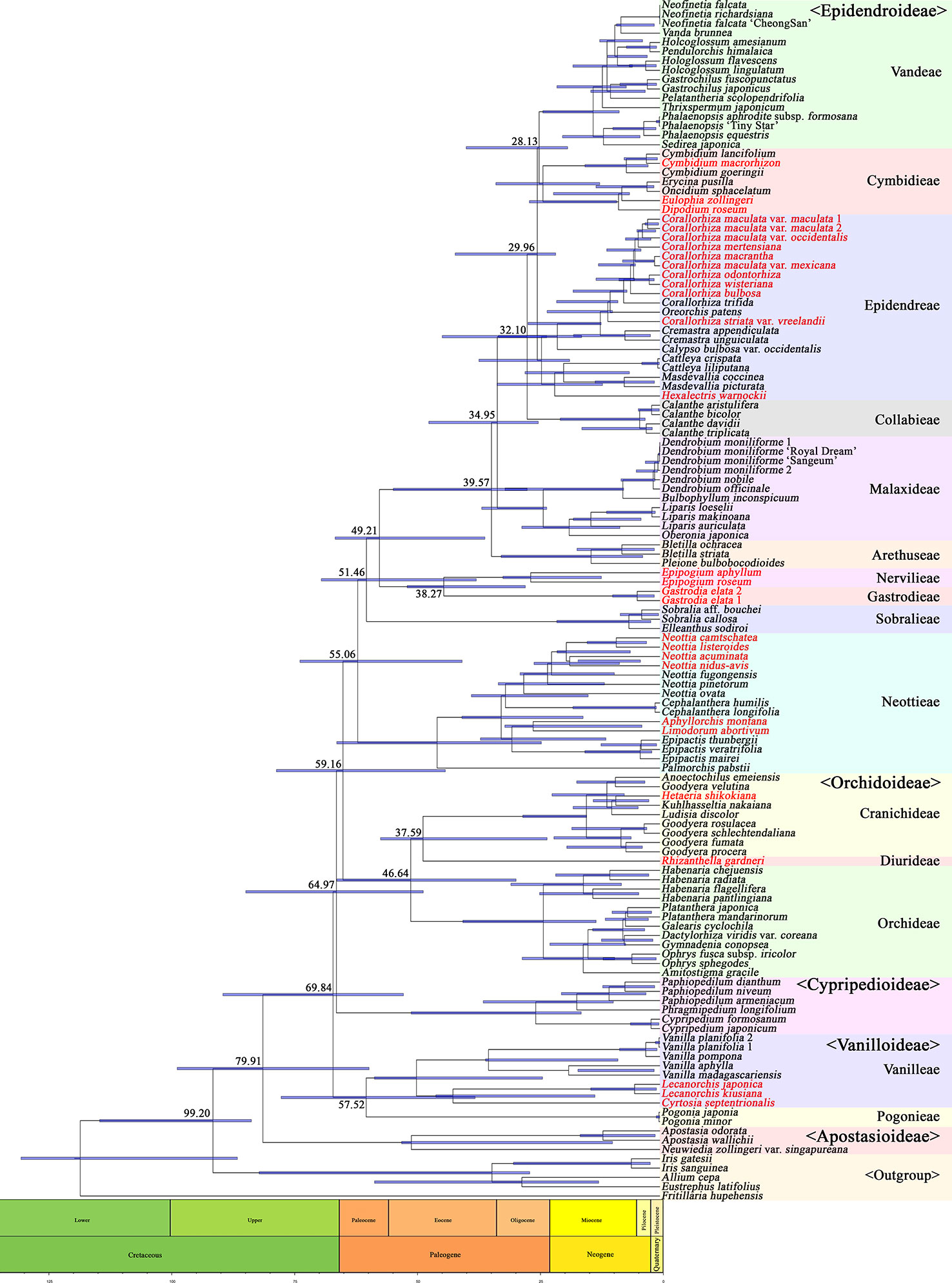
Figure 10 Fossil data showing the estimated divergence time of each node. Three fossil data were used to calibrate nodes (Asparagales—mean 105.3 mya, Dendrobium—23.2 mya, and Goodyera—15.0 mya). Orchidaceae diverged from its sister family at 99.20 mya, and then five subfamilies subsequently diverged in the order of Apostasioideae (79.91 mya), Vanilloideae (69.84 mya), Cypripedioideae (64.97 mya), Orchidoideae (59.16 mya), and Epidendroideae (59.16 mya). However, several specious subtribes within the Epidendroideae diverged in relatively short time periods (39.57–28.13 mya). The species names in red indicate mycoheterotrophic orchids.
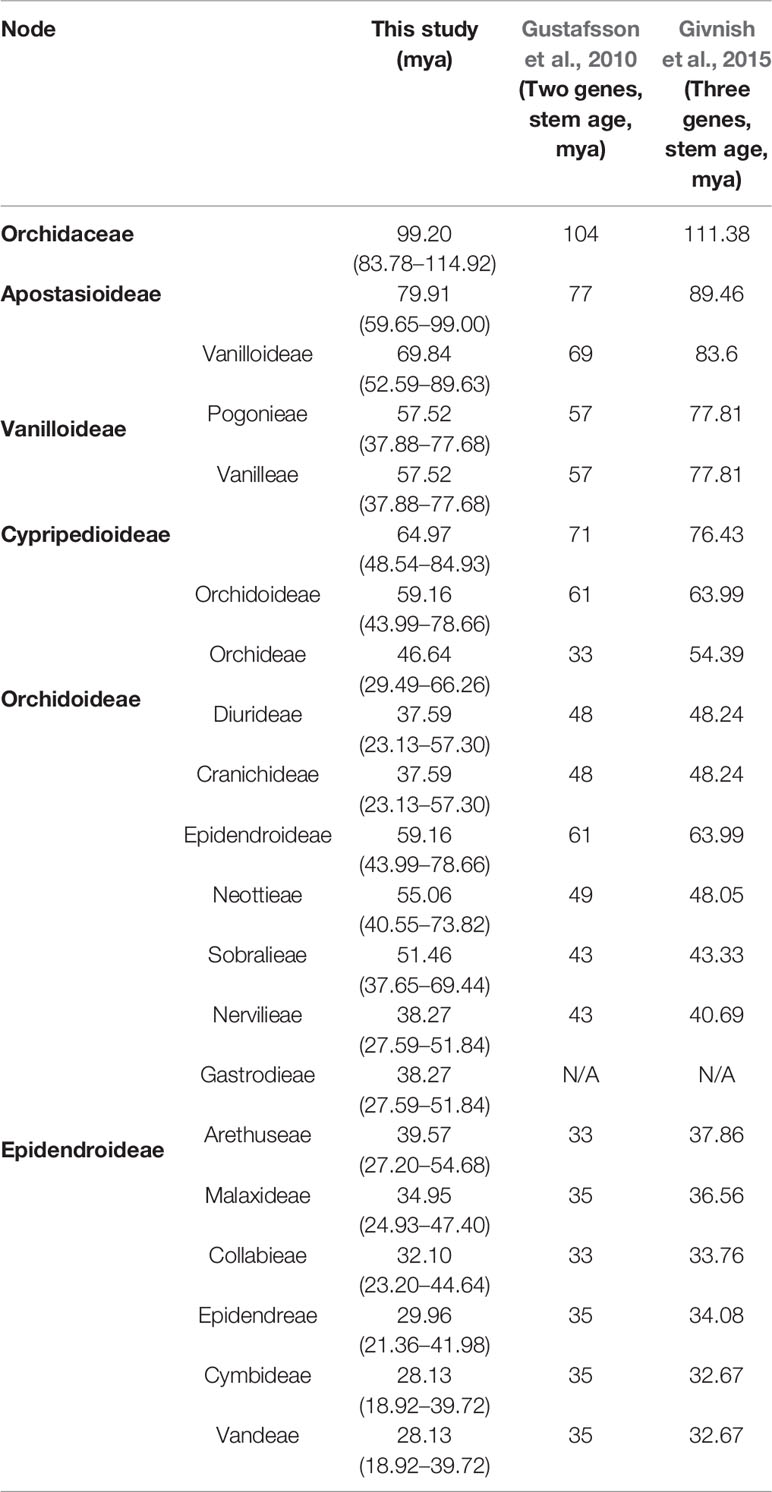
Table 4 Comparing time estimation results with two previous studies that used two and three genes for analysis.
Most plastomes of flowering plants have quadripartite structures consisting of LSC and SSC regions with two IR regions between the two SCs (Bock, 2007). The IR regions are generally known to play a role in the structural stability of plastomes (Palmer and Thompson, 1982), and Orchidaceae plastomes are no exception. When the 124 Orchidaceae plastomes were compared, all but Aphyllorchis montana, Epipogium aphyllum, and Gastrodia elata (2 accessions) had quadripartite structures (Table 2). Gastrodia elata (Epidendroideae-Gastrodieae) is a special case in which the plastome exists only as one single copy because the IR region was lost when the plastome contracted to about 35.1 kb (Figure 1). In the case of Aphyllorchis montana (Epidendroideae-Neottieae), the plastome also exists as only one single copy region because the IR region was lost when the plastome size decreased to 94.6 kb (Feng et al., 2016). In the case of Epipogium aphyllum (Epidendroideae-Nervilieae), the plastome has a tripartite structure consists of the SC and IR because the two IR regions were combined to head-to-head formation when the SSC region disappeared and the plastome contracted to 30.6 kb (Schelkunov et al., 2015). However, in the case of Epipogium roseum in the same genus, the SSC region exists at 890 bp (shorter than average) and the IR decreased to 261 bp (Figure 3). Although these structural variations are related to gene losses associated with mycoheterotrophic habitats, the phenomenon of broken quadripartite structures appears only in the three species mentioned above of the 28 mycoheterotrophic species compared. Plastomes exist as only SC regions due to the loss of IR in several gymnosperms (Wu et al., 2011), Trifolium (Fabaceae), Medicago (Fabaceae), and Cicer (Fabaceae) (Cai et al., 2008). However, all these are photosynthetic species, which are different from the Orchidaceae mentioned above.
Another feature that can be found frequently in Orchidaceae plastomes is IR expansion/shift toward the SSC region. Therefore, the SSC size has been greatly reduced. Due to this feature, which can be found in Pogonia and Vanilla of Vanilloideae, the lengths of IR became 29,808–32,683 bp because both IRa and IRb expanded toward the SSC (Lin et al., 2015; Amiryousefi et al., 2017). In contrast, the lengths of the SSC were shortened by as much as 1,254–5,387 bp. The similar SSC contractions were also found in three species of Cypripedioideae: Paphiopedilum armeniacum, P. dianthum, and P. niveum (Table 2). The same phenomenon was observed in the newly decoded plastome of Hetaeria shikokiana, and the length of the IR and the SSC were found to be 31,960 bp and 2,320 bp, respectively (Figure 1). Among the four genera in which SSC contraction occurred, Paphiopedilum, Pogonia, and Vanilla carry out photosynthesis, while Hetaeria does not. As for life forms, three genera (Hetaeria, Paphiopedilum, and Pogonia) have terrestrial life forms, while one genus (Vanilla) has an epiphytic life form. In all four genera, ndh genes were deleted in most cases. This is thought to be a widely occurring phenomenon in orchids due to parallel evolution and is not considered attributable to the direct effect of SSC contraction. Since ndh genes exist in the vicinity of IR-SSC—such as ndhA, ndhB, and ndhH in Pogonia and Hetaeria, which are pseudogenes—ndh genes are not a direct cause of SSC contraction. However, many small sequence block rearrangements appear in many Orchidaceae lineages near the IR-SSC boundary, indicating that the ndhF and ycf1 genes at the IR-SSC boundary may affected the stability of Orchidaceae plastomes. This is a common phenomenon, especially in species that have lost their ndh gene class or the capacity for photosynthesis.
On the other hand, no IR shift toward the IR-LSC is observed in the Orchidaceae plastomes (Figure S2). The largest plastome size in Orchidaceae was 178,131 bp in Cypripedium formosanum, but there was no IR expansion toward the LSC in this case either. The increase in plastome sizes in photosynthetic plant species is mainly related to IR expansion (Chumley et al., 2006; Weng et al., 2014; Blazier et al., 2016). However, in the case of Cypripedium, the genome size increased due to scattered AT-rich repeats among IGS in the LSC region without IR expansion (Figure S2) (Kim et al., 2015b). Mycoheterotrophic orchids have plastome sizes below 150 kb and show high correlations between length of the LSC region and the IR length (R2 = 0.9571 and R2 = 0.8213, respectively). On the other hand, the plastome sizes of all photosynthetic orchids were at least 140 kb and showed a weak correlation with the length of the LSC region (R2 = 0.5606), but little correlation with the IR length (R2 = 0.1251) (Figures 4C, D). This means that, in mycoheterotrophic orchids, genome contraction occurs regardless of region, but genome size expansion is affected more by the expansion of the LSC region than the expansion of the IR region.
Gene block analysis was conducted using MAUVE to identify gene rearrangements among Orchidaceae plastomes and, according to the results, 104 out of 134 blocks had rearrangements in at least one taxonomic group (Figure 8, Tables S1 and S2). Of the 104 rearrangements, 34 were distributed in the LSC region, three were distributed in the IR region, and the remaining 67 were concentrated on the SSC region. Among the LSC rearrangements, 11 were at least 10 kb long. On the other hand, the SSC rearrangements were mostly concentrated on ndhF and ycf1 in the IR/SSC boundary region, and most were short rearrangements that did not exceed 1 kb (Figure 8, Table S2). Many of the IRa-SSC junction rearrangements are assumed to be attributable to the unequal crossing over among repeats during the processes of pseudogenization and gene loss of ndhF and ycf1. In the IR region, IR shift occurred as the genes in the SSC region were lost and became smaller, and the region containing the rrn4.5-rrn23 gene moved to the SSC region. Their position changes have been reported in mycoheterotrophs with large gene losses, such as Aphyllorchis montana, Cyrtosia septentrionalis, Hexalectris warnockii, Lecanorchis japonica, L. kiusiana, and Rhizanthella gardneri (Delannoy et al., 2011; Feng et al., 2016; Barrett and Kennedy, 2018; Kim et al., 2019). Given that the relevant shift did not occur in other mycoheterotrophs (Corallorhiza, Dipodium, Hetaeria, or Neottia), this is considered to be an independent evolutionary phenomenon associated with gene loss.
Based on taxonomic groupings, although some species in the genera that include both photosynthetic and non-photosynthetic nutritional modes such as Cephalanthera, Cymbidium, Epipactis, and Platanthera (Merckx et al., 2013) shared rearrangements, species of other genera in the same category (e.g., Cremastra) did not share any rearrangements, indicating that the gene relocation in the plastome occurred after development of non-photosynthetic mycoheterotrophy. Therefore, the plastid gene rearrangements are judged to have developed independently in many orchid lineages, and this is supported by the fact that each of photosynthetic groups (Apostasia, Bulbophyllum, Dendrobium, Elleanthus, Oberonia, Ophrys, Palmorchis, Pleione, Pogonia, Sobralia, and Vanilla) has multiple rearrangements. Furthermore, since a number of rearrangements have been reported in Aphyllorchis montana, Cyrtosia septentrionalis, Hexalectris warnockii, Lecanorchis japonica, L. kiusiana, and Rhizanthella gardneri, which are mycoheterotrophs that reached stage 2 of gene loss, it is assumed that rearrangements played some role in gene loss. However, given that there is no gene rearrangement in mycoheterotrophs (Corallorhiza, Hetaeria, and Neottia) (Delannoy et al., 2011; Feng et al., 2016; Barrett and Kennedy, 2018; Kim et al., 2019), whose gene losses reached stage 1 or 2, further comparative studies are necessary to determine the relationships between gene relocation and gene loss.
Almost all orchid species are initially mycoheterotrophs at an early stage of development, then subsequently develop into full autotrophs. Some orchids exhibit both autotrophic and mycoheterotrophic (mixotrophic) nutritional modes, even at adult stages. Finally, several orchid species maintain an obligate (full) mycoheterotrophic (non-photosynthetic) nutritional mode throughout their life cycle. Forty-three genera in Orchidaceae are known to include non-photosynthetic mycoheterotrophs (Merckx et al., 2013). Among them, 17 were used in this study (Aphyllorchis, Cephalanthera, Corallorhiza, Cremastra, Cymbidium, Cyrtosia, Dipodium, Epipogium, Eulophia, Gastrodia, Hetaeria, Hexalectris, Lecanorchis, Limodorum, Neottia, Platanthera, and Rhizanthella), and only photosynthetic species were analyzed in the case of three of these genera (Cephalanthera, Cremastra, and Platanthera). However, when the plastomes were checked, it was found that Cymbidium macrorhizon, Eulophia zollingeri, Dipodium roseum, and Limodorum abortivum only lost the ndh gene (Figure 5, Figure S1). In addition, after many literature reviews and firsthand observation by these researchers, it was found that, regarding these four species, chlorophylls exist in stems and flowers and low-level photosynthesis is carried out during a certain period in the life cycle (Blumenfeld, 1935; Girlanda et al., 2006; Kim et al., 2017a; Suetsugu et al., 2018). Therefore, it is reasonable to treat these four species as mixotrophs rather than obligate mycoheterotrophs. Among the species thought to be non-photosynthetic, Corallorhiza, which was studied the most extensively, is very likely to have photosynthetic capability if only ndh genes had been lost. In addition, cases have been reported in which members of the same species of Corallorhiza had different, mixotrophic nutritional modes depending on living environments (Barrett and Davis, 2012; Barrett et al., 2014; Barrett et al., 2018). The estimated divergence times of these mixotrophs are 7.44 (2.41–15.37) mya (Cymbidium), 17.34 (8.87–26.76) mya (Dipodium and Eulophia), and 17.77 (3.68–31.79) mya (Limodorum), and the evolution of these genera are thought to be relatively recent events. Furthermore, the divergence times of the obligate mycoheterotrophic groups were also estimated as follows: 23.61 (16.05–33.12) mya (Corallorhiza and Cremastra), 21.56 (11.77–33.40) mya (Hexalectris), 38.27 (27.60–51.84) mya (Gastrodia and Epipogium), 28.11 (15.75–40.61) mya (Aphyllorchis, Cephalanthera, Epipactis, and Neottia), 5.32 (1.71–9.80) mya (Platanthera), 37.59 (23.13–57.30) mya (Rhizanthella), 7.48 (2.26–13.66) mya (Hetaeria), and 27.82 (13.31–45.95) mya (Cyrtosia and Lecanorchis) (Figure 10). It can be seen that, except for Gastrodieae (Gastrodia), Nervilleae (Epipogium), and Diurideae (Rhizanthella), all these species diverged more recently than 30 mya (Figure 10). When estimated based on the foregoing, there are two possibilities: the obligate mycoheterotrophic taxa are relatively recently evolved taxonomic groups within Orchidaceae, and information about the older obligate mycoheterotrophic taxonomic groups is not yet available. To test these, further studies should be conducted on other species in Gastrodieae and Nervilleae, whom are estimated to have diverged for long times ago.
While epiphytic orchids consisted only of photosynthetic species and all had genomes at least 140 kb long, terrestrial orchids showed a large variation in plastome sizes and had much different numbers of functional genes because they included non-photosynthetic species (Figure 4A). In addition, the correlation between genome size and the number of functional genes was high (R2 = 0.9069) in mycoheterotrophs, and relatively low (R2 = 0.3320) in photosynthetic species (Figure 4B). Also, mycoheterotrophs are distributed into three clusters: one with about 100 functional genes, one with 55–90 functional genes, and one with 25–30 functional genes (Figures 4B and 5). This means that in the evolutionary process of mycoheterotrophic species, gene loss occurred step by step in three stages rather than on a continuous spectrum.
The first stage of gene loss was the pseudogenization or loss of the ndh gene class. There are 11 ndh gene subunits in each plastome. ndh gene losses are observed in all five subfamilies of Orchidaceae, and appear at especially high frequencies in Cypripedioideae, Epidendroideae, and Vanilloideae. On the other hand, the frequencies of non-functionalization are low in Apostasioideae and Orchidoideae. The ndh genes of the epiphytic orchids (Bulbophyllum, Cattleya, Dendrobium, Erycina, Gastrochilus, Holcoglossum, Neofinetia, Oberonia, Phalaenopsis, Pelatantheria, Pendulorchis, Sedirea, Thrixspermum, Vanda, and Vanilla) used in the analysis were lost or pseudogenized (Figures 5 and 6). Although the loss or pseudogenization of ndh genes was also observed in terrestrial taxonomic groups (Apostasia, Calypso, Cephalanthera, Cremastra, Cymbidium, Epipactis, Goodyera, Kuhlhasseltia, Limodorum, Liparis, Neuwiedia, Oncidium, Paphiopedilum, Phragmipedium, Platanthera, and Pogonia), the frequencies are low compared to epiphytes. Although the loss or pseudogenization of ccsA, cemA, and infA is often observed in addition to ndh gene non-functionalization, but no clear trend has been found (Barrett et al., 2014; Kim et al., 2015a; Feng et al., 2016; Niu et al., 2017b; Hou et al., 2018). In the phylogenetic tree, the non-functionalization of ndh appears to occur gradually and independently in the process of pseudogenization in many independent lineages (Figure 6). However, given that most orchid species in which ndh gene non-functionalization occurred retain photosynthetic capacity, it is inferred that the function of this gene class may be affected by the nuclear or mitochondrial genomes. Of course, the gene function may be maintained by RNA editing after pseudogenization, but the possibility is low because most pseudogenization entail not only base changes but also indels. However, given that ndh genes were lost in all non-photosynthetic species, the loss of the ndh gene class is considered to be a precondition for the development of mycoheterotrophs.
The second stage of gene loss is the loss of functions of genes involved in the photosynthetic light reaction, such as pet (six genes), psa (five genes), and psb (15 genes). This includes the rpo (four genes) gene class, which includes housekeeping genes. Gene losses at this stage are shown to have progressed independently in at least 10 clades including six independent lineages of Epidendroideae (Aphyllorchis, Corallorhiza, Epipogium, Gastrodia, Hexalectris, and Neottia), two Orchidoideae clades (Hetaeria and Rhizanthella), and one Vanilloideae clade (Cyrtosia and Lecanorchis) (Figure 6). In particular, in the case of Corallorhiza and Neottia, gene losses progressed independently depending on species, even within the same genus, indicating that these are important taxonomic groups for understanding second stage gene losses. In addition, the plastomes of Cyrtosia and Lecanorchis of Vanilloideae degraded significantly so that ccsA, cemA, rbcL, ycf3, and ycf4 were commonly deleted in addition to ndh, pet, psa, psb, and rpo, and it was identified that some subunits of the housekeeping genes, such as rpl and rps, were also deleted from Lecanorchis (Figure 6A). The degradation of Cyrtosia and Lecanorchis of Vanilloideae is thought to be part of the transition process from plastome degradation stage 2–4 (Wicke et al., 2016). On the other hand, Hetaeria shikokiana of Orchidoideae and Corallorhiza maculata var. maculata of Epidendroideae correspond to plastome degradation stage 2 because some of their ndh, psa, psb, pet, and rpo genes were deleted or pseudogenized. However, since plastome degradation stages 2 and 3 (Wicke et al., 2013; Wicke et al., 2016) appeared to occur simultaneously in Orchidaceae, this stage was defined as the second stage of gene loss.
The third stage of gene loss in Orchidaceae includes cases where only 27 to 33 out of 113 unique plastome genes remain, meaning that further gene loss has occurred than in stage 2 cases. Since at least 55 genes are preserved in stage 2 and fewer than 33 genes are preserved in stage 3, a large gap exists between the two stages (Figure 4B). In stage 3, all the photosynthesis-related gene functions were lost and most housekeeping genes—such as trn, rpl, and rps—were also lost. In addition, the fact that pseudogenes do not exist or are limited to three or fewer is also a characteristic of stage 3. This stage is observed in a total of four lineages: two lineages of Epidendroideae (Epipogium, 27 to 29 conserved genes; Gastrodia, 28 conserved genes), one of Orchidoideae (Rhizanthella, 33 conserved genes), and one of Vanilloideae (Lecanorchis, 32–33 conserved genes) (Figures 5 and 6, Table 2). Regarding the number of genes, the 27 in Epipogium aphyllum are the minimum number of genes found in Orchidaceae plastomes. Furthermore, 20 genes that commonly exist in the four lineages of seven plastomes, which are in gene loss stage 3, were identified, comprising rpl (12, 14, 16, 36), rps (2, 3, 4, 7, 8, 11, 14, 18, 19), rrn (5, 16, 23), trnC-GCA, trnfM-CAU, accD, and clpP (Table 3). The fact that the same 20 genes have been preserved even though the extremely contracted orchid plastomes evolved from four independent mycoheterotrophic lineages means that those genes selectively remained because they perform the minimum functions necessary to maintain the plastomes. Since five (rpl12, rps18, rps19, trnfM-CAU, and accD) of these 20 genes were lost in one to four other orchid species (Table 3), it was concluded that there are 15 common orchid plastome genes.
Thus far, phylogenetic studies using a portion of the plastome genes have been common (Cameron et al., 1999; Givnish et al., 2015), and phylogenetic studies using the entire plastome genes have also progressed thanks to the development of NGS technology (Yang et al., 2013; Kim et al., 2015a; Feng et al., 2016). The use of chloroplast genes in phylogenetic studies has many advantages such as ease of use, but this has been pointed out to be vulnerable to lineage sorting and problems such as plastome capture due to hybridization because it only tracks maternal lineages. Although NGS technology and transcriptome analysis are developing, there are still cost limitations to using the entire nuclear genome for phylogenetic studies. Therefore, nuclear ribosomal ITS regions have been extensively used in studies of the relationships between species or allied genera. An advantage of the genome skimming NGS is that, for plastome sequences with high depths of coverage (see Table 1), it can decode about 10 kb of regions that exist as tandem repeats (18S-ITS1-5.8S-ITS2-28S-NTS-ETS) (Figure 2). Although it has difficulties in recovering entire NTS-ETS when the plastome coverage depth is below then 500x, genome skimming for NGS can easily recover the 6 kb region of the 18S-ITS1-5.8S-ITS2-28S region. Therefore, in this study, an attempt was made to compare phylogenetic trees constructed using all the same plastome genes and phylogenetic trees constructed using the nrDNA unit sequences. Orchid plastomes from 42 species, representing three subfamilies of Orchidaceae, were produced in the laboratory of the corresponding author and directly compared (Figure 9).
According to the results, the relationships among the three subfamilies were the same as (Vanilloideae (Orchidoideae, Epidendroideae)). Furthermore, the associations among the tribes and species in Vanilloideae and Orchidoideae were completely identical. However, the topologies of the two trees were different for the relationships among Epidendroideae taxa. In the Epidendroideae, there was no difference in generic or tribal grouping, but the positions of Gastrodia, in which the plastome reduced greatly, and Bulbophyllum, which is a photosynthetic species, were very different. In addition, clades with bootstrap support values of less than 50% had different topologies. However, it is not clear from this comparative analysis alone whether these differences are due to lineage sorting, ancient plastome capture, other evolutionary histories, or sampling issues. Although plastome data for diverse orchid species are currently known, the sequence data for nrDNA repeats for the 42 species in this study are presented for the first time. Although some nodes in Epidendroideae show discrepancies, major parts of the trees are generally identical to each other. In addition, these data are thought to be easily obtainable because nrDNA repeats can be recovered by simply carrying out additional data mining while conducting plastome sequencing using the genome skimming NGS method. If more data are accumulated for the same sample, then the evolution of orchids can be more easily understood.
The results of estimating the divergence times of major clades of Orchidaceae using all 83 genes in the plastome were similar to the findings of previous studies conducted using two or three genes at some points, and different at others (Figure 10, Table 4). First, in this study, the time of origin of orchids was estimated to be 99.20 (83.78–114.92) mya, which is similar to the previous estimate of 104 mya using two genes, and not significantly different from the 111.38 mya estimated using three genes (Gustafsson et al., 2010; Givnish et al., 2015). However, the finding in this study shows the divergence time of the basal taxonomic groups ranging from Orchidoideae and Cypripedioideae to Epidendroideae to be a little earlier than the previous finding estimated using two genes (1.84–6.03 mya differences), and a little more recent than the result using three genes. That is, the times estimated in the two previous studies are very different and the one in this study fell between the other two. However, the origin time of terminal tribes in Epidendroideae was inferred to be more recent by this study than the previous studies, which only used two or three genes.
Orchidoideae is estimated to consist of 198 genera and 4,931 species, and the Epidendroideae is estimated to consist of 505 genera and 20,606 species (Chase et al., 2015; Christenhusz and Byng, 2016). These two clades account for most of Orchidaceae and diverged 71.85 mya (Figure 10, Table 4). The divergence times of Arethuseae (25 genera, 723 species), Collabieae (20 genera, 453 species), Cymbideae (165 genera, 3,997 species), Epidendreae (99 genera, 6,935 species), Malaxideae (16 genera, 4,631 species), Podochileae (27 genera, 1,292 species), and Vandeae (136 genera, 2,340 species)—tribes of Epidendroideae consisting of many species—are later than 39.57 (27.20–54.68) mya, which is relatively recent. These data mean that many species differentiated in a short time. In studies that analyzed diversification rates, the relevant tribes had higher rates compared to the basal lineage in most cases, thereby supporting the hypothesis that many species underwent evolutionary radiation in a short time (Givnish et al., 2015).
Sixty genera, 140 species, 146 accessions of Orchidaceae plastomes have been completed decoded and can be used in comparative studies of plastomes, including the 24 plastomes newly reported in this study (NCBI database, June 30, 2019). In the present study, the evolutionary trends of plastomes were compared and analyzed to understand the evolutionary processes of orchid plastomes and mycoheterotrophic orchids. Only three different types of species were selected in the case of Cymbidium (9 spp.), Dendrobium (40 spp.), and Holcoglossum (11 spp.) because many species in these genera have been studied. Therefore, the plastomes of 60 genera, 118 species, and 124 accessions were analyzed. In addition, for the 42 species decoded in the corresponding author's laboratory, a plastome tree and nrDNA tandem repeat tree were compared to discuss evolution. Orchidaceae is known to include about 736 genera and 28,000 species. Therefore, those genera for which their plastomes have been decoded thus far make up approximately 8% of all the known genera of the Orchidaceae, and those species whose plastomes have been decoded thus far comprise less than 1% of all the known species of the Orchidaceae. There are 43 genera known to include non-photosynthetic mycoheterotrophs in Orchidaceae. In this study, plastome gene loss patterns of 17 genera that correspond to 40% of the foregoing genera were compared and analyzed to derive third-stage gene losses and 15 common genes. However, if further studies are conducted with more genera and species, Orchidaceae plastome gene losses can be understood better. In particular, genera such as Corallorhiza and Neottia, in which photosynthetic species coexist with non-photosynthetic mycoheterotrophs, should be intensively studied with all species, including several populations per species. Studies of the taxonomic groups in which many species were recently differentiated should be continuously conducted to elucidate the mechanism of evolutionary radiation. In this regard, the use of both plastid and nuclear genomes using NGS technology will increase for orchid evolution studies. If high-quality NGS data are accumulated by 50% at the genus level and 10% at the species level, then the evolution of orchids can be better understood.
The datasets generated for this study can be found in the National Center for Biotechnology Information (NCBI), please find accession numbers in Table 2.
K-JK and MK designed research. Y-KK, SJ, S-HC, J-RH, and MK collected the research materials. Y-KK and SJ performed research. Y-KK, SJ S-HC, J-RH, and M-JJ analyzed data and deposited the data to data libraries. Y-KK and K-JK wrote the manuscript. MK and K-JK secured the research funds.
This work was supported by the National Research Foundation of Korea (NRF) under grant no. NRF-2015M3A9B8030588 to K-JK and by the National Institute of Biological Resources (NIBR) under the genetic evaluation of vascular plants IV-1 (2018, grant no. NIBR201803102) to MK and K-JK.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank to two anonymous reviewers for their helpful comments to improving the manuscript. We would like to thank Noah Last of Third Draft Editing for his English language editing. We also thank to Dr. Sanghun Oh of Daejeon University for the leaf materials of Calanthe aristulifera and Dr. Kyeongwon Kang of Babo Orchid Farm for the materials of Habenaria chejuensis and H. flagellifera. We thank the curator of the Korea University Herbarium (KUS) for voucher specimen preparation. The genomic and chloroplast DNAs are deposited in the Plant DNA Bank in Korea (PDBK).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00022/full#supplementary-material
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Amiryousefi, A., Hyvönen, J., Poczai, P. (2017). The plastid genome of Vanillon (Vanilla pompona, Orchidaceae). Mitochondrial DNA Part B 2, 689–691. doi: 10.1080/23802359.2017.1383201
Amiryousefi, A., Hyvo, J., Poczai, P. (2018). Genome analysis IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barrett, C. F., Davis, J. I. (2012). The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am. J. Bot. 99, 1513–1523. doi: 10.3732/ajb.1200256
Barrett, C. F., Kennedy, A. H. (2018). Plastid genome degradation in the endangered, mycoheterotrophic, North American orchid Hexalectris warnockii. Genome Biol. Evol. 10, 1657–1662. doi: 10.1093/gbe/evy107
Barrett, C. F., Freudenstein, J. V., Li, J., Mayfield-Jones, D. R., Perez, L., Pires, J. C., et al. (2014). Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol. 31, 3095–3112. doi: 10.1093/molbev/msu252
Barrett, C. F., Wicke, S., Sass, C. (2018). Dense infraspecific sampling reveals rapid and independent trajectories of plastome degradation in a heterotrophic orchid complex. New Phytol. 218, 1192–1204. doi: 10.1111/nph.15072
Blazier, J. C., Jansen, R. K., Mower, J. P., Govindu, M., Zhang, J., Weng, M. L., et al. (2016). Variable presence of the inverted repeat and plastome stability in Erodium. Ann. Bot. 117, 1209–1220. doi: 10.1093/aob/mcw065
Blumenfeld, H. (1935). Beitrage zur Physiologie des Wurzepilzes von Limodorum abortivum (L.) Sw. Universitat Basel.
Bock, R. (2007). “Structure, function and inheritance of plastid genomes,” in Cell and molecular biology of plastids. Ed. Bock, R. (Berlin: Springer), 29–63. doi: 10.1007/978-3-642-12422-8_3
Bouckaert, R., Vaughan, T. G., Barido-Sottani, J., Duchêne, S., Fourment, M., Gavryushkina, A., et al. (2019). BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PloS Comput. Biol. 15, e1006650. doi: 10.1371/journal.pcbi.1006650
Cai, Z., Guisinger, M., Kim, H. G., Ruck, E., Blazier, J. C., McMurtry, V., et al. (2008). Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J. Mol. Evol. 67, 696–704. doi: 10.1007/s00239-008-9180-7
Cameron, K. M., Chase, M. W., Whitten, W. M., Kores, P. J., Jarrell, D. C., Albert, V. A., et al. (1999). A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. Am. J. Bot. 86, 208–224. doi: 10.2307/2656938
Chang, C. C., Lin, H. C., Lin, I. P., Chow, T. Y., Chen, H. H., Chen, W. H., et al. (2006). The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 23, 279–291. doi: 10.1093/molbev/msj029
Chase, M. W., Cameron, K. M., Freudenstein, J. V., Pridgeon, A. M., Salazar, G., van den Berg, C., et al. (2015). An updated classification of Orchidaceae. Bot. J. Linn. Soc 177, 151–174. doi: 10.1111/boj.12234
Christenhusz, M. J. M., Byng, J. W. (2016). The number of known plants species in the world and its annual increase. Phytotaxa 261, 201–217. doi: 10.11646/phytotaxa.261.3.1
Chumley, T. W., Palmer, J. D., Mower, J. P., Fourcade, H. M., Calie, P. J., Boore, J. L., et al. (2006). The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23, 2175–2190. doi: 10.1093/molbev/msl089
Conran, J. G., Bannister, J. M., Lee, D. E. (2009). Earliest orchid macrofossils: early miocene dendrobium and earina (Orchidaceae: Epidendroideae) from New Zealand. Am. J. Bot. 96, 466–474. doi: 10.3732/ajb.0800269
Cosner, M. E., Jansen, R. K., Palmer, J. D., Downie, S. R. (1997). The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr. Genet. 31, 419–429. doi: 10.1007/s002940050225
da Rocha Perini, V., Leles, B., Furtado, C., Prosdocimi, F. (2016). Complete chloroplast genome of the orchid Cattleya crispata (Orchidaceae: Laeliinae), a neotropical rupiculous species. Mitochondrial DNA Part A. Anal. 27, 4075–4077. doi: 10.3109/19401736.2014.1003850
Darling, A. E., Mau, B., Perna, N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS One 5, e11147. doi: 10.1371/journal.pone.0011147
Darriba, D., Taboada, G. L., Doallo, R., Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2106
Delannoy, E., Fujii, S., Colas Des Francs-Small, C., Brundrett, M., Small, I. (2011). Rampant gene loss in the underground orchid Rhizanthella Gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 28, 2077–2086. doi: 10.1093/molbev/msr028
Dong, W. L., Wang, R. N., Zhang, N. Y., Fan, W. B., Fang, M. F., Li, Z. H. (2018). Molecular evolution of chloroplast genomes of orchid species: insights into phylogenetic relationship and adaptive evolution. Int. J. Mol. Sci. 19. doi: 10.3390/ijms19030716
Drummond, A. J., Ho, S. Y. W., Phillips, M. J., Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PloS Biol. 4, e88. doi: 10.1371/journal.pbio.0040088
Feng, Y. L., Wicke, S., Li, J. W., Han, Y., Lin, C. S., Li, D. Z., et al. (2016). Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol. Evol. 8, 2164–2175. doi: 10.1093/gbe/evw144
Freudenstein, J. V., Chase, M. W. (2015). Phylogenetic relationships in epidendroideae (Orchidaceae), one of the great flowering plant radiations: progressive specialization and diversification. Ann. Bot. 115, 665–681. doi: 10.1093/aob/mcu253
Górniak, M., Paun, O., Chase, M. W. (2010). Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, xdh: congruence with organellar and nuclear ribosomal DNA results. Mol. Phylogenet. Evol. 56, 784–795. doi: 10.1016/j.ympev.2010.03.003
Girlanda, M., Selosse, M. A., Cafasso, D., Brilli, F., Delfine, S., Fabbian, R., et al. (2006). Inefficient photosynthesis in the Mediterranean orchid Limodorum abortivum is mirrored by specific association to ectomycorrhizal Russulaceae. Mol. Ecol. 15, 491–504. doi: 10.1111/j.1365-294X.2005.02770.x
Givnish, T. J., Spalink, D., Ames, M., Lyon, S. P., Hunter, S. J., Zuluaga, A., et al. (2015). Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B Biol. Sci. 282, 20151553. doi: 10.1098/rspb.2015.1553
Gustafsson, A. L. S., Verola, C. F., Antonelli, A. (2010). Reassessing the temporal evolution of orchids with new fossils and a Bayesian relaxed clock, with implications for the diversification of the rare South American genus Hoffmannseggella (Orchidaceae: Epidendroideae). BMC Evol. Biol. 10. doi: 10.1186/1471-2148-10-177
Hirao, T., Watanabe, A., Kurita, M., Kondo, T., Takata, K. (2008). Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol. 8, 1–20. doi: 10.1186/1471-2229-8-70
Hou, N., Wang, G., Zhu, Y., Wang, L., Xu, J. (2018). The complete chloroplast genome of the rare and endangered herb Paphiopedilum dianthum (Asparagales: Orchidaceae). Conserv. Genet. Resour. 10, 709–712. doi: 10.1007/s12686-017-0907-x
Huo, X., Zhao, Y., Qian, Z., Liu, M. (2017). Characterization of the complete chloroplast genome of Eulophia zollingeri, an endangered orchid in China. Conserv. Genet. Resour. 10, 817–819. doi: 10.1007/s12686-017-0938-3
Iles, W. J. D., Smith, S. Y., Gandolfo, M. A., Graham, S. W. (2015). Monocot fossils suitable for molecular dating analyses. Bot. J. Linn. Soc. 178, 346–374. doi: 10.1111/boj.12233
Katoh, K., Misawa, K., Kuma, K., Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, H. T., Kim, J. S., Moore, M. J., Neubig, K. M., Williams, N. H., Whitten, W. M., et al. (2015a). Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PloS One 10. doi: 10.1371/journal.pone.0142215
Kim, J. S., Kim, H. T., Kim, J.-H. (2015b). The largest plastid genome of monocots: a novel genome type containing AT residue repeats in the slipper orchid Cypripedium japonicum. Plant Mol. Biol. Rep. 33, 1210–1220. doi: 10.1007/s11105-014-0833-y
Kim, Y. K., Kwak, M. H., Chung, M. G., Kim, H. W., Jo, S., Sohn, J. Y., et al. (2017a). The complete plastome sequence of the endangered orchid Cymbidium macrorhizon (Orchidaceae). Mitochondrial DNA Part B Resour. 2, 725–727. doi: 10.1080/23802359.2017.1390411
Kim, Y. K., Kwak, M. H., Hong, J. R., Kim, H. W., Jo, S., Sohn, J. Y., et al. (2017b). The complete plastome sequence of the endangered orchid Habenaria radiata (Orchidaceae). Mitochondrial DNA Part B Resour. 2, 704–706. doi: 10.1080/23802359.2017.1390410
Kim, H. T., Shin, C. H., Sun, H., Kim, J. H. (2018). Sequencing of the plastome in the leafless green mycoheterotroph Cymbidium macrorhizon helps us to understand an early stage of fully mycoheterotrophic plastome structure. Plant Syst. Evol. 304, 245–258. doi: 10.1007/s00606-017-1472-1
Kim, Y. K., Jo, S., Cheon, S. H., Joo, M. J., Hong, J. R., Kwak, M. H., et al. (2019). Extensive losses of photosynthesis genes in the plastome of a mycoheterotrophic orchid, Cyrtosia septentrionalis (Vanilloideae: Orchidaceae). Genome Biol. Evol. 11, 565–571. doi: 10.1093/gbe/evz024
Krawczyk, K., Wiland-Szymańska, J., Buczkowska-Chmielewska, K., Drapikowska, M., Maślak, M., Myszczyński, K., et al. (2018). The complete chloroplast genome of a rare orchid species Liparis loeselii (L.). Conserv. Genet. Resour. 10, 305–308. doi: 10.1007/s12686-017-0809-y
Lallemand, F., Logacheva, M., Clainche, I. L., Bérard, A., Zheleznaia, E., May, M., et al. (2019). Thirteen new plastid genomes from mixotrophic and autotrophic species provide insights into heterotrophy evolution in Neottieae orchids. Genome Biol. Evol. 11, 2457–2467. doi: 10.1093/gbe/evz170
Lam, V. K. Y., Merckx, V. S. F. T., Graham., S. W. (2016). A few-gene plastid phylogenetic framework for mycoheterotrophic monocots. Am. J. Bot. 103, 692–708. doi: 10.3732/ajb.1500412
Lanfear, R., Calcott, B., Ho, S. Y. W., Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Lee, H. L., Jansen, R. K., Chumley, T. W., Kim, K. J. (2007). Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 24, 1161–1180. doi: 10.1093/molbev/msm036
Li, Z. H., Ma, X., Wang, D. Y., Li, Y. X., Wang, C. W., Jin, X. H. (2019). Evolution of plastid genomes of Holcoglossum (Orchidaceae) with recent radiation. BMC Evol. Biol. 19, 1–10. doi: 10.1186/s12862-019-1384-5
Lin, C. S., Chen, J. J. W., Huang, Y. T., Chan, M. T., Daniell, H., Chang, W. J., et al. (2015). The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 5, 1–10. doi: 10.1038/srep09040
Lin, C. S., Chen, J. J. W., Chiu, C. C., Hsiao, H. C. W., Yang, C. J., Jin, X. H., et al. (2017). Concomitant loss of ndh complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 90, 994–1006. doi: 10.1111/tpj.13525
Logacheva, M. D., Schelkunov, M. I., Penin, A. A. (2011). Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol. Evol. 3, 1296–1303. doi: 10.1093/gbe/evr102
Lohse, M., Drechsel, O., Bock, R. (2007). OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 52, 267–274. doi: 10.1007/s00294-007-0161-y
Lowe, T. M., Chan, P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Merckx, V. S. F. T., Freudenstein, J. V., Kissling, J., Christenhusz, M. J. M., Stotler, R. E., Crandall-Stotler, B., et al. (2013). Taxonomy and Classification BT - Mycoheterotrophy: The Biology of Plants Living on Fungi. Ed. Merckx, V. (New York, NY: Springer New York), 19–101. doi: 10.1007/978-1-4614-5209-6_2
Miller, M. A., Pfeiffer, W., Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees”, in 2010 gateway computing environments workshop (GCE). (IEEE), 1–8.
Miller, M. A., Schwartz, T., Pickett, B. E., He, S., Klem, E. B., Scheuermann, R. H., et al. (2015). A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol. Bioinforma. 11, EBO–S21501. doi: 10.4137/EBO.S21501
Niu, Z., Pan, J., Zhu, S., Li, L., Xue, Q., Liu, W., et al. (2017a). Comparative analysis of the complete plastomes of Apostasia wallichii and Neuwiedia singapureana (Apostasioideae) reveals different evolutionary dynamics of IR/SSC boundary among photosynthetic orchids. Front. Plant Sci. 8, 1–11. doi: 10.3389/fpls.2017.01713
Niu, Z., Xue, Q., Zhu, S., Sun, J., Liu, W., Ding, X. (2017b). The complete plastome sequences of four orchid species: insights into the evolution of the Orchidaceae and the utility of plastomic mutational hotspots. Front. Plant Sci. 8, 1–11. doi: 10.3389/fpls.2017.00715
Palmer, J. D., Thompson, W. F. (1982). Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29, 537–550. doi: 10.1016/0092-8674(82)90170-2
Ramírez, S. R., Gravendeel, B., Singer, R. B., Marshall, C. R., Pierce, N. E. (2007). Dating the origin of the Orchidaceae from a fossil orchid with its pollinator. Nature 448, 1042–1045. doi: 10.1038/nature06039
Rambaut, A., Drummond, A. J. (2007). TreeAnnotator in Computer Program and Documentation Distributed by the Author. Available online at: https://www.beast2.org/
Rambaut, A., Drummond, A. J. (2014). LogCombiner v2.1.3 in Computer Program and Documentation Distributed by the Author. Available online at: https://www.beast2.org/
Rambaut, A., Suchard, M. A., Xie, D., Drummond, A. J. (2014). Tracer 1.6 in Computer Program and Documentation Distributed by the Author. Available online at: https://www.beast2.org/tracer-2/
Rambaut, A. (2012). FigTree v1. 4. in Computer Program and Documentation Distributed by the Author. Available online at: http://tree.bio.ed.ac.uk/software/figtree/
Roma, L., Cozzolino, S., Schlüter, P. M., Scopece, G., Cafasso, D. (2018). The complete plastid genomes of Ophrys iricolor and O. Sphegodes (Orchidaceae) and comparative analyses with other orchids. PloS One 13, 1–15. doi: 10.1371/journal.pone.0204174
Schelkunov, M. I., Shtratnikova, V. Y., Nuraliev, M. S., Selosse, M. A., Penin, A. A., Logacheva, M. D. (2015). Exploring the limits for reduction of plastid genomes: a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol. Evol. 7, 1179–1191. doi: 10.1093/gbe/evv019
Schelkunov, M. I., Nuraliev, M. S., Logacheva, M. D. (2019). Rhopalocnemis phalloides has one of the most reduced and mutated plastid genomes known. PeerJ 7, e7500. doi: 10.7717/peerj.7500
Shi, Y., Yang, L., Yang, Z., Ji, Y. (2018). The complete chloroplast genome of Pleione bulbocodioides (Orchidaceae). Conserv. Genet. Resour. 10, 21–25. doi: 10.1007/s12686-017-0753-x
Stöver, B. C., Müller, K. F. (2010). TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinf. 11, 1–9. doi: 10.1186/1471-2105-11-7
Suetsugu, K., Ohta, T., Tayasu, I. (2018). Partial mycoheterotrophy in the leafless orchid Cymbidium macrorhizon. Am. J. Bot. 105, 1595–1600. doi: 10.1002/ajb2.1142
Wang, H., Park, S. Y., Lee, A. R., Jang, S. G., Im, D. E., Jun, T. H., et al. (2018). Next-generation sequencing yields the complete chloroplast genome of C. goeringii acc. smg222 and phylogenetic analysis. Mitochondrial DNA Part B Resour. 3, 215–216. doi: 10.1080/23802359.2018.1437812
Weng, M. L., Blazier, J. C., Govindu, M., Jansen, R. K. (2014). Reconstruction of the ancestral plastid genome in geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 31, 645–659. doi: 10.1093/molbev/mst257
Wicke, S., Muller, K. F., de Pamphilis, C. W., Quandt, D., Wickett, N. J., Zhang, Y., et al. (2013). Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25, 3711–3725. doi: 10.1105/tpc.113.113373
Wicke, S., Müller, K. F., dePamphilis, C. W., Quandt, D., Bellot, S., Schneeweiss, G. M. (2016). Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc. Natl. Acad. Sci. 113, 9045–9050. doi: 10.1073/pnas.1607576113
Wickham, H., Chang, W., Wickham, M. H. (2016), Package ‘ggplot2.' Creat. Elegant Data Vis. Using Gramm. Graph. Version 2. 1–189.
Wu, F.-H., Chan, M.-T., Liao, D.-C., Hsu, C.-T., Lee, Y.-W., Daniell, H., et al. (2010). Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 10, 68. doi: 10.1186/1471-2229-10-68
Wu, C. S., Lin, C. P., Hsu, C. Y., Wang, R. J., Chaw, S. M. (2011). Comparative chloroplast genomes of pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol. Evol. 3, 309–319. doi: 10.1093/gbe/evr026
Yang, J.-B., Tang, M., Li, H.-T., Zhang, Z.-R., Li, D.-Z. (2013). Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 13, 84. doi: 10.1186/1471-2148-13-84
Yu, C. W., Lian, Q., Wu, K. C., Yu, S. H., Xie, L. Y., Wu, Z. J. (2016). The complete chloroplast genome sequence of Anoectochilus roxburghii. Mitochondrial DNA 27, 2477–2478. doi: 10.3109/19401736.2015.1033706
Keywords: Orchidaceae, plastome evolution, gene loss, IR contraction/expansion, genome rearrangement
Citation: Kim Y-K, Jo S, Cheon S-H, Joo M-J, Hong J-R, Kwak M and Kim K-J (2020) Plastome Evolution and Phylogeny of Orchidaceae, With 24 New Sequences. Front. Plant Sci. 11:22. doi: 10.3389/fpls.2020.00022
Received: 24 September 2019; Accepted: 10 January 2020;
Published: 21 February 2020.
Edited by:
Jen-Tsung Chen, National University of Kaohsiung, TaiwanReviewed by:
Aleksey Penin, Lomonosov Moscow State University, RussiaCopyright © 2020 Kim, Jo, Cheon, Joo, Hong, Kwak and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ki-Joong Kim, a2lta2pAa29yZWEuYWMua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.