
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 29 January 2020
Sec. Plant Abiotic Stress
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01746
 Meng-qi Wang1
Meng-qi Wang1 Qiu-xiang Huang1
Qiu-xiang Huang1 Ping Lin1
Ping Lin1 Qin-han Zeng1
Qin-han Zeng1 Yan Li2*
Yan Li2* Qing-lin Liu1*
Qing-lin Liu1* Lei Zhang1
Lei Zhang1 Yuan-zhi Pan1
Yuan-zhi Pan1 Bei-bei Jiang1
Bei-bei Jiang1 Fan Zhang1
Fan Zhang1Cold stress poses a serious threat to the survival and bloom of Verbena bonariensis. The enhancement of the cold tolerance of V. bonariensis is the central concern of our research. The WRKY transcription factor (TF) family was paid great attention to in the field of abiotic stress. The VbWRKY32 gene was obtained from V. bonariensis. The VbWRKY32 predicted protein contained two typical WRKY domains and two C2H2 zinc-finger motifs. Under cold stress, VbWRKY32 in leaves was more greatly induced than that in stems and roots. The overexpression (OE) in V. bonariensis increased cold tolerance compared with wild-type (WT). Under cold stress, the OE lines possessed showed greater recovery after cold-treatment restoration ratios, proline content, soluble sugar content, and activities of antioxidant enzymes than WT; the relative electrolyte conductivity (EL), the accumulation of malondialdehyde (MDA), hydrogen peroxide (H2O2), and superoxide anion (O2−) are lower in OE lines than that in WT. In addition, a series of cold-response genes of OE lines were compared with WT. The results revealed that VbWRKY32 worked as a positive regulator by up-regulating transcription levels of cold-responsive genes. The genes above can contribute to the elevation of antioxidant activities, maintain the membrane stability, and raise osmotic regulation ability, leading to the enhancement of the survival capacity under cold stress. According to this work, VbWRKY32 could serve as an essential gene to confer enhanced cold tolerance in plants.
Plants are often subjected to various stresses which affects their growth and development. Some genes, including WRKY transcription factors (TFs), are induced to help plants adapt to stresses by changing physiology and morphology. WRKY TFs were a valuable family which resisted abiotic stress, such as cold, NaCl, drought, salicylic acid, ethylene, abscisic acid, methyl jasmonate, and hydrogen peroxide (H2O2) (Wang et al., 2013; Zhou et al., 2015; Xiu et al., 2016). The WRKY family comprised one or two DNA-binding domains consisted of 60 amino acid regions and the highly conservative sequence WRKYGQK at its N-terminus and a zinc-finger motif (Cx4−5 Cx22−23HxH or Cx7Cx23HxC) at C-terminus (Eulgem et al., 2000; Rushton et al., 2010). In terms of the structure of WRKY protein, it could be divided into three groups: WRKY group I (contained two WRKY domains with Cx4−5Cx22−23HxH zinc-finger motifs), group II (one WRKY domain with a Cx4−5Cx22−23HxH motif), and group III (one WRKY domain with a C-X7-C-X23-H-X-C motif).
Several WRKY-overexpressed plants had successfully enhanced the resistance to various abiotic stresses. For example, overexpression of IIWRKY2 (WRKY group II) gene elevated salt tolerance in overexpression (OE) lines of Iris lactea var. chinensis (Tang et al., 2018). OsWRKY11 (WRKY group II) functioned as a positive regulator in tolerance to heat and salt stress of the transgenic rice seedlings (Wu et al., 2009). OsWRKY45 promoted the resistance to disease and drought in Arabidopsis (Qiu and Yu, 2009). Compared with wild type (WT), the CsWRKY46 (WRKY group II)-overexpressed cucumber via regulating a series of regulated cold-responsive genes raised cold tolerance (Ying et al., 2016). OsWRKY71 (WRKY group II) has a positive function in cold tolerance by regulating downstream target genes in rice (Kim et al., 2016). Overexpressed FcWRKY70 (WRKY group III) in tobacco and lemon conferred enhanced tolerance to drought stresses (Gong et al., 2015). Overexpression of GhWRKY25 (WRKY group I) in Nicotiana benthamiana enhanced plant tolerance to salt stress (Liu et al., 2016). TFs are significant for cold signaling and tolerance by modulating the expression of related functional genes (Nair et al., 2009). WRKY are vital regulators in certain development processes. Overexpressed OsWRKY30 (WRKY group I) enhanced rice resistance to disease by the salicylic acid (SA) signaling pathway (Ryu et al., 2006). The WRKY34 (WRKY group I) TF negatively regulated cold sensitivity of mature Arabidopsis pollen and might be involved in the C-repeat binding factor (CBF) signal cascade in mature pollen (Zou et al., 2010). However, WRKY group I and III members have been rarely reported compared with group II members, in particular in response to stress. Therefore, combined with the analysis of transcriptomic data, WRKY group I will be the focus of our attention.
Verbena bonariensis is a perennial herb native to South America (Brazil, Argentina, etc.). With its high ornamental value and supreme drought resistance, V. bonariensis is widely used in flower border and sightseeing farms. However, V. bonariensis owns inferior resistance of low temperature, which causes damage during flowering and impacts yield. When below 0°C or worse, chilling injury would result in destruction or death in production. In China, the studies on V. bonariensis are now mainly focused on seedling breeding, garden application, and salt tolerance. The low temperature molecular research in V. bonariensis has not been reported. Thus, it is urgent to determine and improve the cold resistance in V. bonariensis. The transcriptomic data of V. bonariensis in cold stress displayed that WRKY TFs worked as a vital role in helping plants cope with low temperatures stress.
In this study, we isolated and cloned VbWRKY32 gene from V. bonariensis. The analysis of VbWRKY32 expression responding to cold stress was elaborated from multiple angles. Overexpressed VbWRKY32 in V. bonariensis elevated tolerance of cold stress, compared with WT. The statistics indicated that VbWRKY32 could serve as a new candidate gene to accomplish the cultivation of cold-tolerant plants and contribute more detailed information to WRKY family.
V. bonariensis, c.v. Finesse was selected as research materials. The VbWRKY32 gene was cloned from tissue-cultured seedlings which were grown on Murashige and Skoog (MS) medium. V. bonariensis were cultivated in the greenhouse of Sichuan Agricultural University. The two OE lines (OE-1 and OE-5) and WT of V. bonariensis were tested in our study.
Total RNA extraction of tissue-cultured V. bonariensis leaves was achieved by TRIzol Reagent (MyLab, Beijing, China). The synthesis of complementary DNA (cDNA) was accomplished by Takara® PrimeScript™ RT Reagent Kit with gDNA Eraser (Perfect Real Time) (TAKARABIOIN, Beijing, China). The full-length cDNA of VbWRKY32 was acquired by PCR. The VbWRKY32 gene was cloned using specific primers (F: CGTAAAGAAAAGAAAAAGCTTTTAT; R: CGCTACCACTACAATCAACCTATAT). PCR was performed in 25-μl reaction volume containing 1 μl cDNA, 0.5 μl each primer, 10.5 μl double-distilled H2O (ddH2O), and 12.4 μl Taq Mix. The program employed was 30 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min; finally stored at 4°C. The cloned sequence was inserted into pCAMBIA 2300 with the control of cauliflower mosaic virus (CaMV) 35S promoter to obtain PVbWRKY32. The enzyme restriction sites were BamH I and Kpn I.
Phylogenetic analysis was conducted by MEGA version 4.0 using 34 VbWRKY32 homologs selected from multiple sequence alignment. Sequence alignment was performed using DNAMAN.
The regenerated explants were selected from upper middle leaves of V. bonariensis. The PVbWRKY32 construct was introduced into Agrobacterium tumefaciens strain GV3101 and transformed into leaves of V. bonariensis. The mediums used for transformation were showed in Table 1. The explants were cultured in the M1 medium for 2 days at 35°C and under the 16/8 h light/dark photoperiod with 3,000 lx intensity of illumination. The samples were infected with bacterial solution of A. tumefaciens (OD600 = 0.4) for 10 min and placed on the M1 medium. After 2-day dark culture, materials were transferred to the M2 medium in culture bottles. These explants were transferred to fresh M2 medium every 2 weeks to maintain appropriate selective pressure and at the same time to prevent the growth of Agrobacteria. After 35 days, the adventitious buds were cut off, transplanting to M3 medium. After rooting and elongation, the plantlets were identified by PCR and adapted to growth in the soil in the soil. The DNA secure Plant Kit (TIANGEN®DNAsecure) was used for the PCR procedure for the transgenic identification.
Three-month old seedlings of V. bonariensis were transferred from the greenhouse (16 h photoperiod, 28°C/20°C day/night temperature) to the thermoregulating incubator. The seedlings were treated at the following temperature (this cooling process was continuous.): 4°C for 24 h (T2), followed by −4°C for 4 h (T3) and 6 h (T4). The upper leaves were harvested at the time points of the control (T1), T2, T3, and T4 for physiological experiments and histochemical detection of ROS, which were frozen in liquid nitrogen instantly and stored at −80°C. Roots, stems, and leaves of the same untreated seedlings were collected for the analysis of tissue-specific expression.
The materials were treated under following conditions: 4°C for 24 h, followed by −4°C for 4 h. The quantitative real-time polymerase chain reaction (qRT-PCR) was performed in the SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA, United States) and Bio-Rad CFX96TM detection system. The Actin-11 gene served as a quantitative control to detect expression level of VbWRKY32 and of nine cold-related genes, including VbCor413im1, VbCor413pm2, VbPOD, VbCAT, VbSOD, VbAPX6, VbP5CS, VbAMY3, and VbBAM1. The 20 μl qRT-PCR reaction mixture was incubated at 95°C for 30 s, followed by 40 cycles at 95°C for 15 s, at 60°C for 30 s, then by a final single melt cycle from 65 to 95°C. Each reaction was carried out for three biological repetitions. Relative expression levels were calculated by the 2−ΔΔCT method. All correlative primers of qRT-PCR were exhibited in Table 2.
Three-month-old seedlings (WT, OE-1, and OE-5) were selected to detect freezing recovery ratios. The materials were treated under following conditions: 4°C for 24 h, followed by −4°C for 4 h (wilting symptoms began to appear), finally allowed to recover for 10 days at 28°C. The freezing recovery ratios were measured. It was recorded that less than 25% of withered leaves in seedlings were regarded as recovery.
Malondialdehyde (MDA) content and electrolyte conductivity (EL) were measured according to Lei et al. (2009) and Wang et al. (2017b), respectively. Activities of superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and ascorbate peroxidase (APX) were measured by the methods of Beauchamp and Fridovich (1971), Ranieri et al. (2000), and Zhang et al. (2011), respectively. The content of soluble protein, soluble sugar, and proline was measured following Wang et al. (2013) and Irigoyen et al. (2010).
The accumulation of H2O2 and superoxide anion (O2−) in leaves was detected by the method of histochemical staining using 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT), respectively (Wang et al., 2017a). Finally, the stained leaves were photographed. H2O2 and O2− content were measured following An (2012) and Yamamoto et al. (2001).
All experiments were performed for three biological repeats. The data were analyzed via one-way analysis of variance using SPSS version 19.0, and statistically significant differences were calculated with p < 0.05 as the thresholds for significance.
To obtain valuable candidate genes from V. bonariensis and to facilitate the molecular culture of cold-resistant varieties, next generation sequencing technique were applied to construct a cDNA library out of V. bonariensis leaves (Wang et al., 2019). The RNASEQ data it was deposited in National Center for Biotechnology Information (NCBI) (NCBI GEO series GSE112477, NR ID: XP_011083976.1). The WRKY32 TF (Cluster-14918.129050) was selected from differentially expressed genes (DEGs) with functional annotations, and named VbWRKY32. Its log2 (fold change) was 5.45 and q value was less than 0.01.
The full-length VbWRKY32 gene was 1,689 bp, of these, the open reading frame (ORF) was 1,500 bp and encode 499 amino acids with predicted protein molecular weight of 55.138 kDa. It contained two WRKY domains of WRKYGQK and two C2H2 zinc-finger motifs (Cx4Cx22HxH and Cx4Cx23HxH) (Figure 1). Comparisons of the amino acid sequences between VbWRKY32 and other WRKY proteins in plants showed that WRKYGQK and two C2H2 zinc-finger motifs were highly conserved in four plant species (Figure 2). The phylogenetic tree was constructed with full-length amino acid sequences, the result demonstrated that VbWRKY32 belonged to group I of the WRKY family (Figure 3).

Figure 1 Nucleotide and deduced amino acid sequences of VbWRKY32. The WRKY domain is underlined. The two cysteines and two histidines in the zinc-finger motifs are framed.
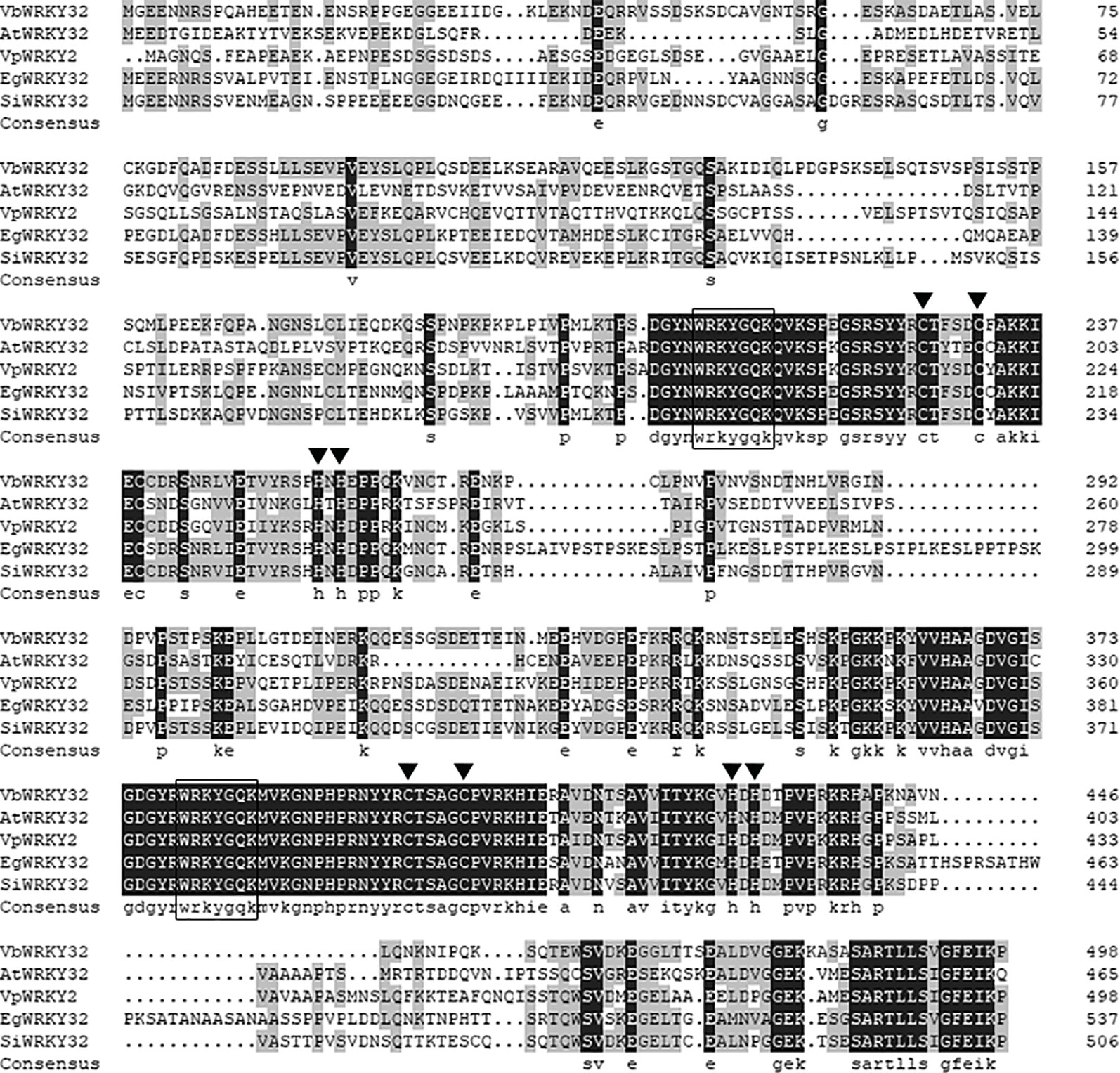
Figure 2 Comparison between the amino acid sequences deduced by VbWRKY32 gene and the multisequences of WRKY protein from other plants. Identical amino acid residues are shaded in black boxes, others meet two identical residues in gray boxes. The completely conserved WRKYGQK amino acids are enclosed by oblong. The cysteine (C) and histidine (H) in zinc-finger motifs are pointed out by black trigon. AtWRKY2 (NP_200438.1) from Arabidopsis thaliana; VpWRKY2 (GU565706) from Vitis pseudoreticulata; EgWRKY32 (XP_012841117.1) from Erythranthe guttata; SiWRKY32 (XP_011083976.1) from Sesamum indicum.
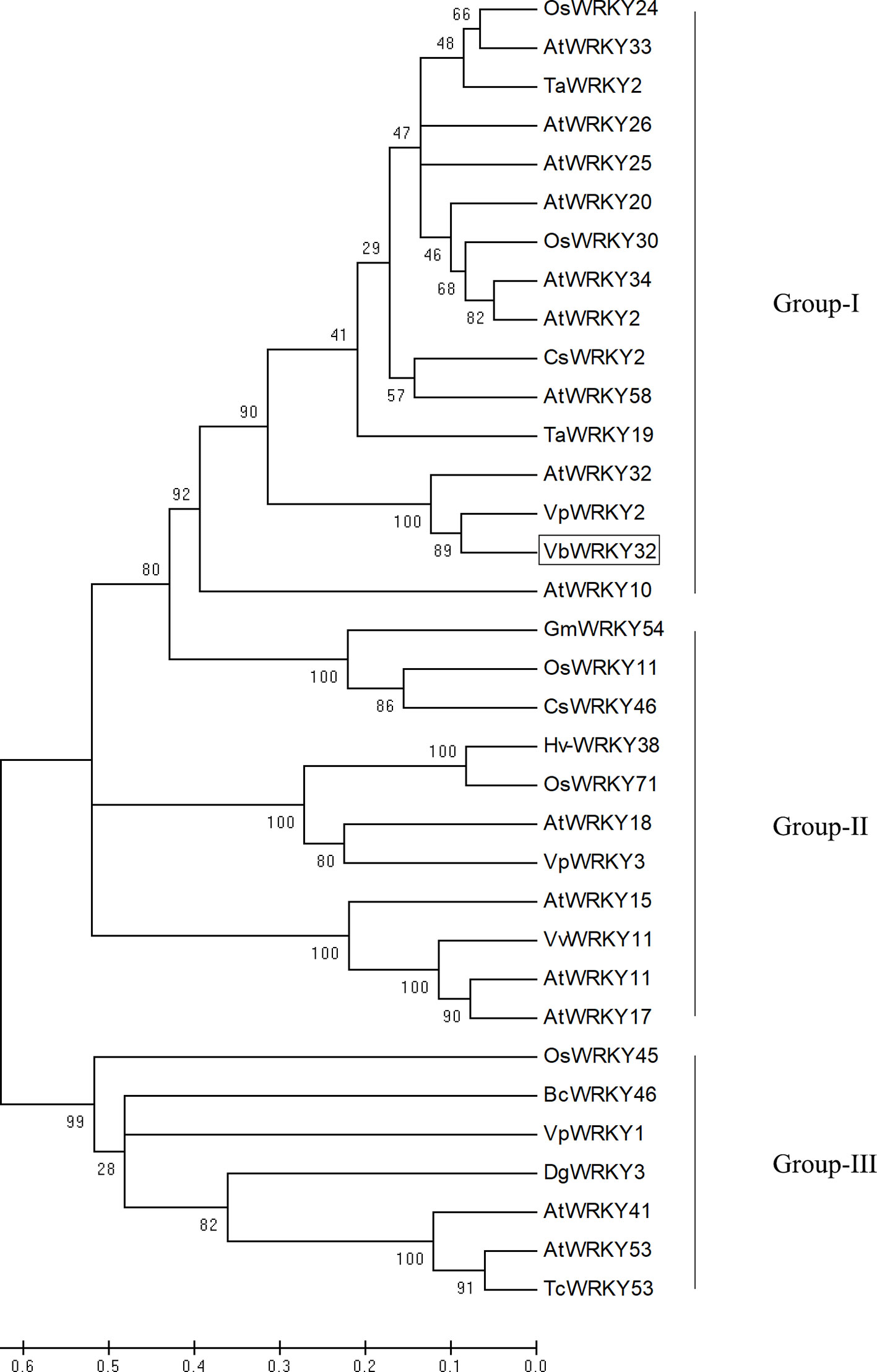
Figure 3 Phylogenetic tree analysis of VbWRKY32 and WRKY proteins from different species. VbWRKY32 is boxed. The plant WRKY proteins used for the phylogenetic tree are as follows: OsWRKY11 (AK108745), OsWRKY24 (NC_029256), OsWRKY30 (NP_001062148), OsWRKY45 (AY870611), and OsWRKY71 (NC_029257) from Oryza sativa; AtWRKY2, AtWRKY10, AtWRKY11 (NP_849559), AtWRKY15 (NP_179913.1), AtWRKY17 (NP_565574.1), AtWRKY18 (NP_567882), AtWRKY20, AtWRKY25 (NP_180584), AtWRKY26 (AAK28309), AtWRKY32, AtWRKY34, AtWRKY41, AtWRKY53 (NP_194112), and AtWRKY58 from Arabidopsis thaliana; TaWRKY2 (EU665425), TaWRKY19 (EU665430) from Triticicum aestivum; CsWRKY2 (AFJ54352), CsWRKY46 from Camellia sinensis; VpWRKY2 (GU565706) and VpWRKY3 from Vitis pseudoreticulata; GmWRKY54 (DQ322698) from Glycine max; Hv-WRKY38 from barley; VvWRKY11 (EC935078) from Vitis vinifera; BcWRKY46 (HM585284) from Brassica campestris; BcWRKY46 (HM585284) from B. campestris.
To determine the involvement of VbWRKY32 under cold stresses, the expression of VbWRKY32 in different tissues of V. bonariensis was measured by RT-qPCR. The results were showed in Figure 4A. The transcript level of VbWRKY32 was higher in leaves than that in stems and roots. The change of VbWRKY32 expression was not significant during the course of the experiment under normal condition (28°C). The expression of VbWRKY32 was increased during 4°C and reached a peak at 4 h during −4°C, then decreased at 5 h (Figure 4B). The result indicated that VbWRKY32 was involved in the cold response.
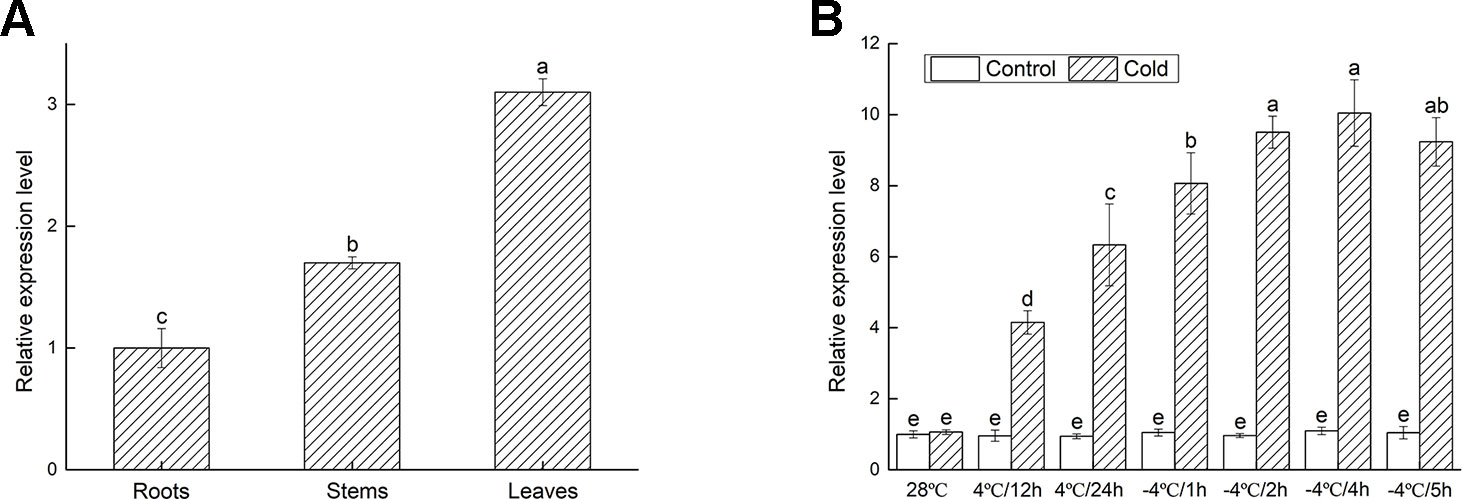
Figure 4 The expression analysis of VbWRKY32 in leaves of Verbena bonariensis. (A) Relative expression level of VbWRKY32 in roots, stems, and leaves of WT lines at 28°C. (B) Cold treatment. Control stands for non-stress treatment and at 28°C. The process of temperature treatment was continuous. The temperature was at 28°C, then drop to 4°C for 12 h, 24 h, and then to −4°C for 1, 2, 4, 5 h. Data indicate means ± standard errors (SE) of three biological replicates. The different letters above the columns represent significant differences (P < 0.05) on the basis of Duncan’s multiple range test.
To further identify the function of VbWRKY32, V. bonariensis transgenic plants overexpressing VbWRKY32 were obtained. The expression level of VbWRKY32 in leaves was measured through RT-qPCR. The results of RT-qPCR showed the VbWRKY32 transcript abundance of lines OE-1 and OE-5 was evidently (P < 0.05) higher than that of other OE and WT lines under 28°C (Figure 5). Therefore, OE-1 and OE-5 lines were selected for cold stress experiment. The plants were treated under following conditions: 4°C for 24 h, followed by −4°C for 4 h, finally allowed to recover for 10 days at 28°C (Figure 6). Under 28°C for 7 days, there were no obvious phenotypic difference between OE and WT lines. Under 4°C for 24 h, leaves of WT plants began to wilt, while transgenic plants’ leaves remained unchanged. The WT and OE plants were then treated at −4°C for 4 h, WT plants obviously fell over and even dead, leaves of OE lines partially wilted. The plants recovered at 28°C for 10 days, most of leaves of WT turned yellow and withered. The leaves of OE lines were not affected much and only a few of leaves turned yellow. The transgenic V. bonariensis showed better recovery than WT. The recovery ratio of OE-1 and OE-5 were 84.45 and 86.67%, respectively, whereas WT plants was 44.44% (Table 3).
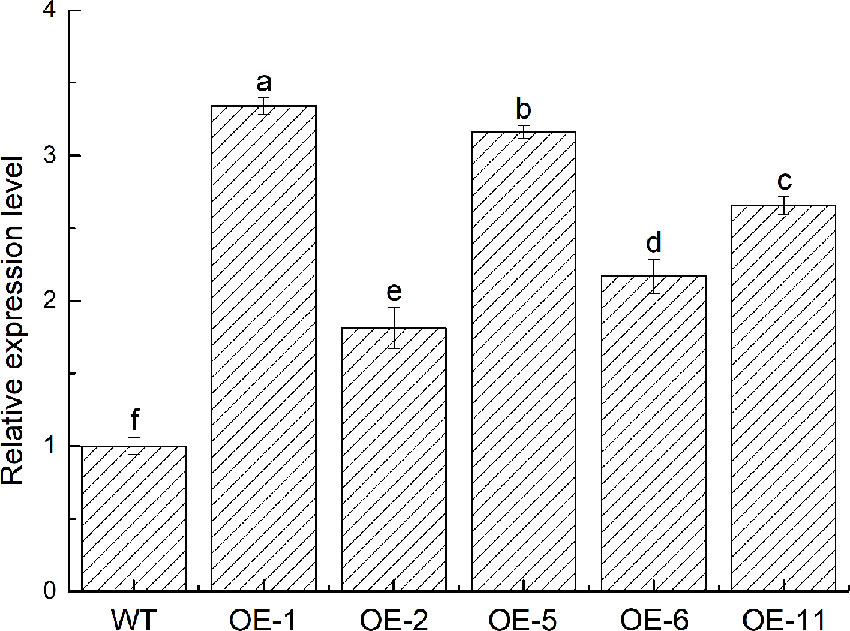
Figure 5 Acquisition of transgenic VbWRKY32 Verbena bonariensis. Expression level of VbWRKY32 in wild-type (WT) and transgenic V. bonariensis. Actin-11 served as the internal reference gene. The different letters above the columns represent significant differences (P < 0.05) on the basis of Duncans multiple range test.
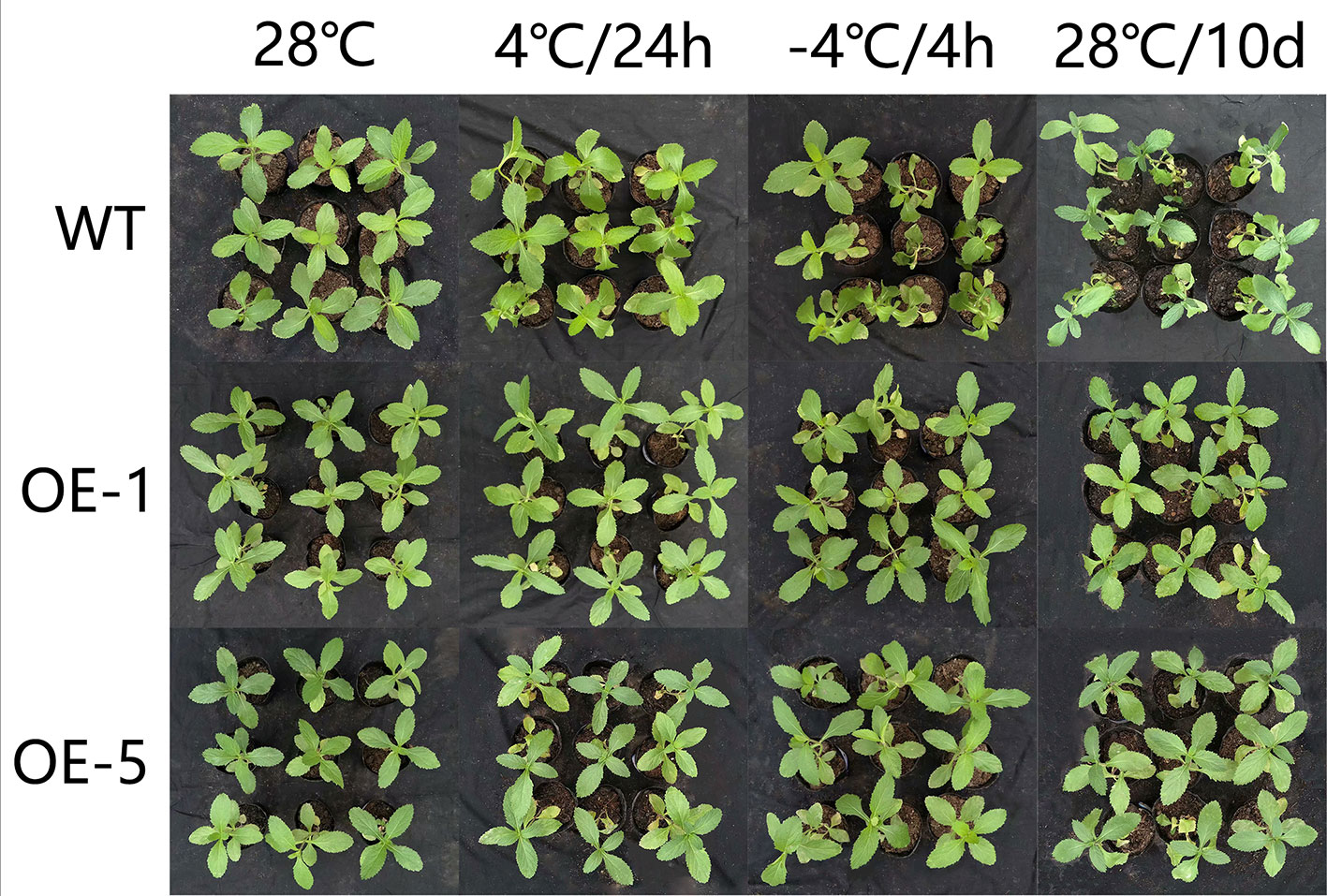
Figure 6 Phenotypic comparison of wild-type (WT) and VbWRKY32 overexpressed lines (OE-1 and OE-5) under cold stress.
In the process of aerobic metabolism of plants, ROS such as H2O2 and O2− was accumulated under cold stress, harming the membrane and related biological macromolecules. To visualize H2O2 and O2− produced in V. bonariensis leaves under cold stress, the leaves were stained by using DAB and NBT chemical. Histochemical staining showed that less brown or blue precipitations were observed in overexpressed lines (OE-1 and OE-5) than that in WT (Figures 7A, B). In addition, quantitative analysis also exhibited that the accumulated level of H2O2 and O2− in leaves of all lines were increased when exposed to cold condition, WT significantly (P < 0.05) produced more H2O2 and O2− than OE lines (Figures 7C, D).
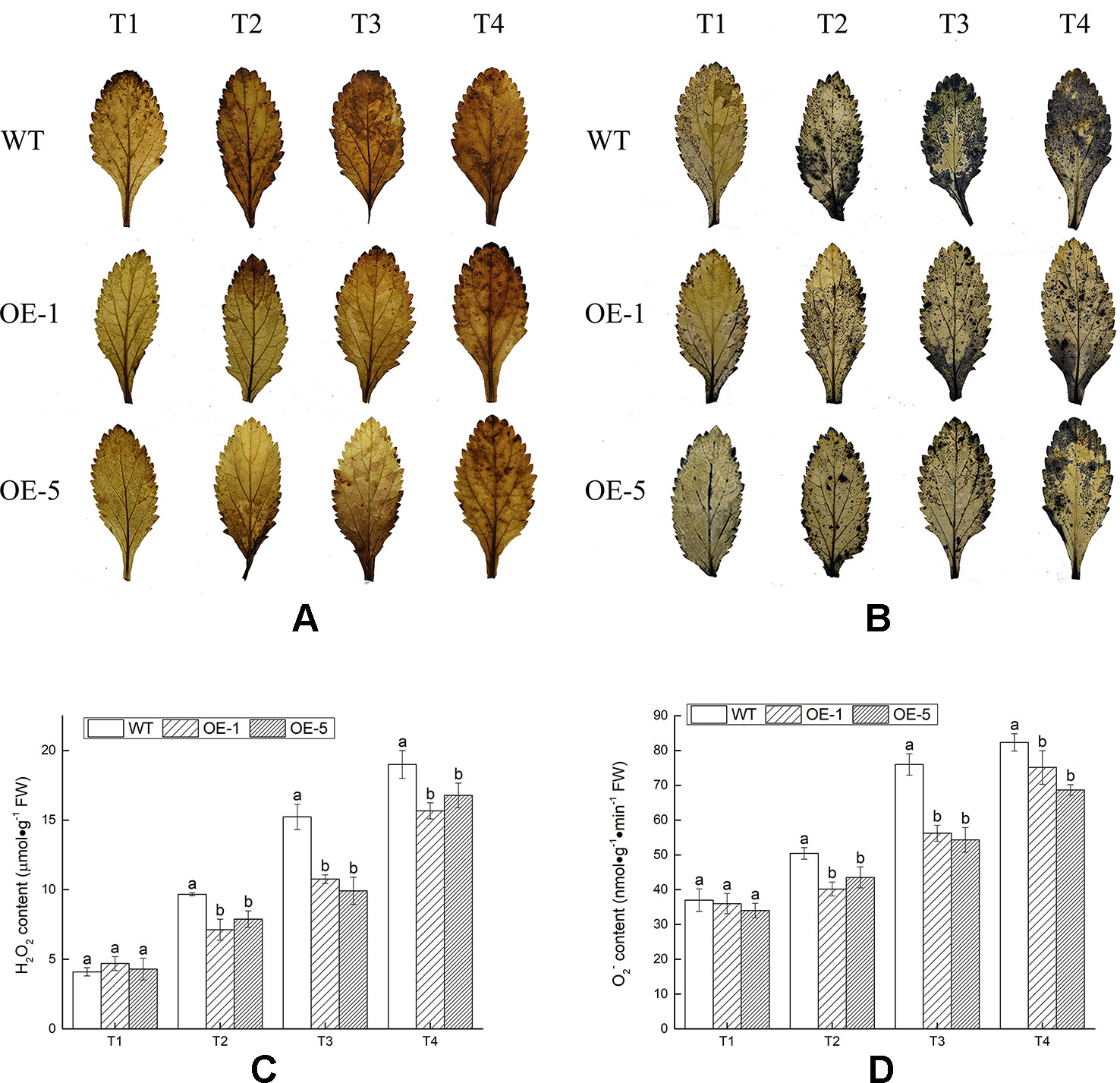
Figure 7 Analysis of the reactive oxygen species (ROS) accumulation levels in wild-type (WT) and VbWRKY32 overexpression (OE)-1 and OE-5 lines of Verbena bonariensis (OE-1 and OE-5) under cold stress. (A, B) Histochemical staining with 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) for observing the accumulation situation of H2O2 and O2−. (C, D) The quantitative measurement of H2O2 and O2−. The temperature kept at 28°C (T1), then drop to 4°C for 24 h (T2), then to −4°C for 4 h (T3), 6 h (T4). Data indicate means ± standard errors (SE) of three biological replicates. The different letters above the columns represent significant differences (P < 0.05) on the basis of Duncan’s multiple range test.
MDA and EL, the two important indexes of cell damage, could reflect the extent of membrane injury (Huang et al., 2013). The MDA content and EL were prominently (P < 0.05) lower in transgenic V. bonariensis than that in WT (Figure 8). Under T4 treatment, the MDA content of OE-1, OE-5 lines increased to 2.78- and 2.76-fold, while WT increased to 3.65-fold; The EL of WT, OE-1, and OE-5 raised to 2.18-, 2.07-, and 1.87-fold of that before stress.
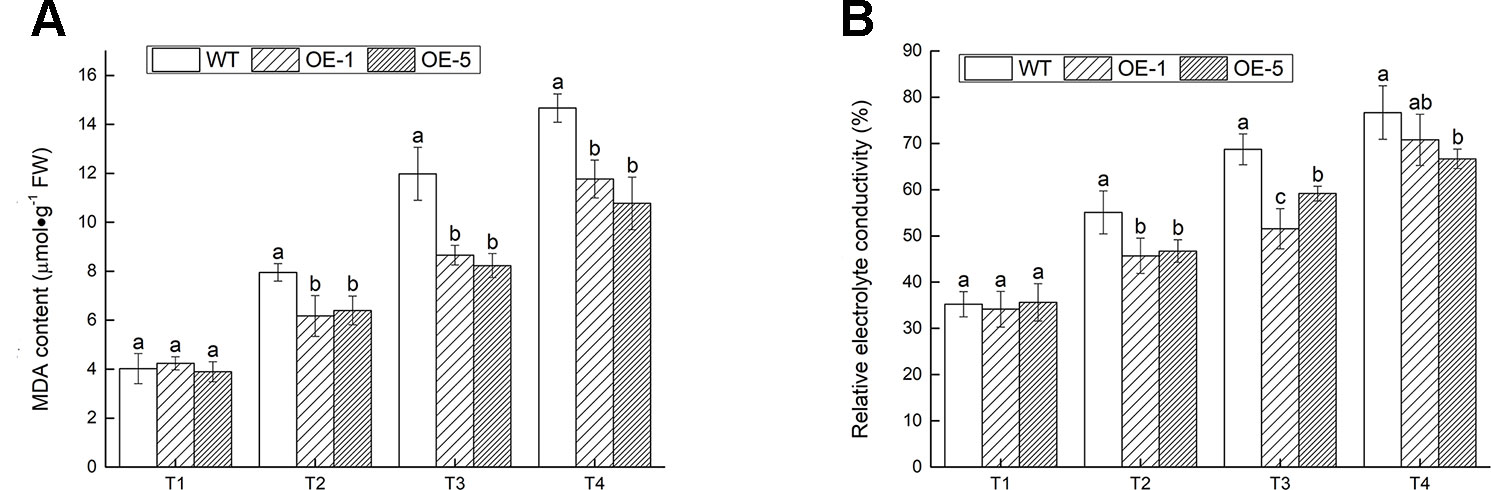
Figure 8 The extent of membrane injury. (A) Malondialdehyde (MDA) content. (B) Relative electrolyte conductivity. Data indicate means ± standard errors (SE) of three biological replicates. The different letters above the columns represent significant differences (P < 0.05) on the basis of Duncan’s multiple range test.
The conclusion above indicated that the degree of cell damage in WT plant was more severe than the transgenic lines under cold stress.ss
The existence of antioxidant enzymes was essential for scavenging ROS and alleviating cell injury. Hence the activities of antioxidant enzymes (SOD, POD, APX, and CAT) at various time and temperature points were monitored (Figures 9A–D). Under 28°C condition (T1), these enzymes activities revealed no remarkable difference in between WT and OE lines. After chilling treatment of −4°C for 4 h (T3), there was an evident increase in all lines; exposure to −4°C treatment for 6 h (T4), the activities of SOD, POD, and APX decreased in OE and WT lines. The contents of proline, soluble sugar, and soluble protein were measured to investigate the regulated capable of osmotic mechanism in VbWRKY32 OE lines of V. bonariensis under cold stress. Compared with WT, OE lines accumulated distinctly (P < 0.05) higher content of proline, soluble sugar, and soluble protein under cold condition (Figures 9E–G). In addition, SOD, POD, APX, and CAT were normalized by protein data (Figure S1). As a whole, the value of SOD, POD, APX, and CAT after normalized treatment were lowered in OE lines than that in WT during cold condition.
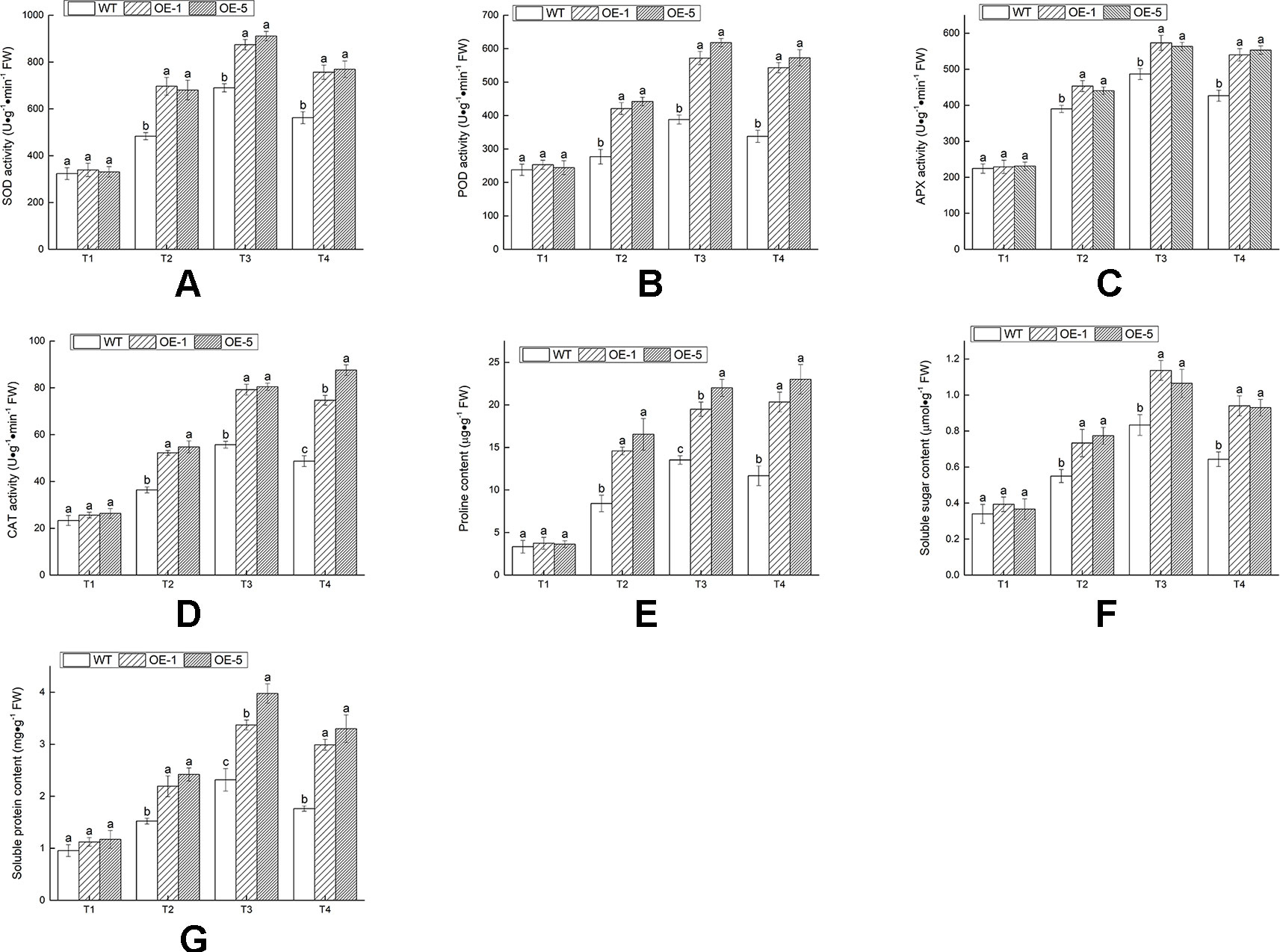
Figure 9 The changes of physiological indexes on wild type (WT) and VbWRKY32 overexpression (OE) lines leaves of Verbena bonariensis (OE-1 and OE-5) under cold stress. (A) Superoxide dismutase (SOD) activity. (B) Peroxidase (POD) activity. (C) Ascorbate peroxidase (APX) activity. (D) Catalase (CAT) activity. (E) Proline content. (F) Soluble sugar content. (G) Soluble protein content. Data indicate means ± standard errors (SE) of three biological replicates. The different letters above the columns represent significant differences (P < 0.05) on the basis of Duncan’s multiple range test.
These results indicated that overexpression of VbWRKY32 could raise the ability of plants against ROS persecution and alleviate cell injury under cold stress.
Based on the above physiological data of V. bonariensis under low temperature stress, it was found that the antioxidant enzyme activity and osmotic adjustment substance content of plants reached a high level under −4°C for 4 h. Considering that the physiological changes depended on the change of molecular mechanism, it was speculated that the gene expression should be at a high level at this time. Therefore, in order to understand the effect of VbWRKY32 overexpression on the cold resistance at gene level, expressions of nine cold-related genes were detected by qRT-PCR at T1 (28°C), T2 (4°C), and T3 (−4°C).
These genes selected were related to antioxidant enzyme, cold regulated protein, and osmotic adjustment substances. The antioxidant enzyme activities and the content of osmotic adjustment substances were increased, which helped clear excess ROS, maintain the balance of cell osmotic pressure, and stabilize the cell structure. Under normal environment, there was no remarkable difference in transcript accumulation between WT and OE lines. Under chilling stress (4°C), the gene expression levels of OE lines were specifically up-regulated than that of WT. Compared with the control (28°C), VbSOD in the WT, OE-1, and OE-5 lines was remarkably (P < 0.05) increased by 1.80, 2.10, and 2.69 times under freezing stress (−4°C) (Figure 10A), respectively. The gene expression level of VbPOD and VbCAT matched similar pattern with VbSOD, which was 3.01 and 3.35 times in OE-5 lines, respectively, higher than that of the control (Figures 10B, C). VbAPX6 in the WT, OE-1, and OE-5 lines was increased to 3.14, 4.93, and 4.44 times under −4°C than that of the control (Figure 10D). The transcription levels of VbCor413im1 in OE lines were over 7.56-fold at 4°C and 9.13-fold at −4°C, greater than that in control, respectively; the expression level of VbCor413pm2 in OE lines was more than 6.30 times at 4°C and 8.11 times at −4°C higher than that of WT, respectively (Figures 10E, F). The expression level of VbAMY3, VbBAM1, and VbP5CS were significantly (P < 0.05) increased by over 3.49, 3.64, and 5.99 times in OE lines under −4°C stress than that of control, a maximum reached (Figures 10G–I). The results indicated that VbWRKY32 transcription factor could increase the expression level of cold-related genes and the cold tolerance of transgenic plants.
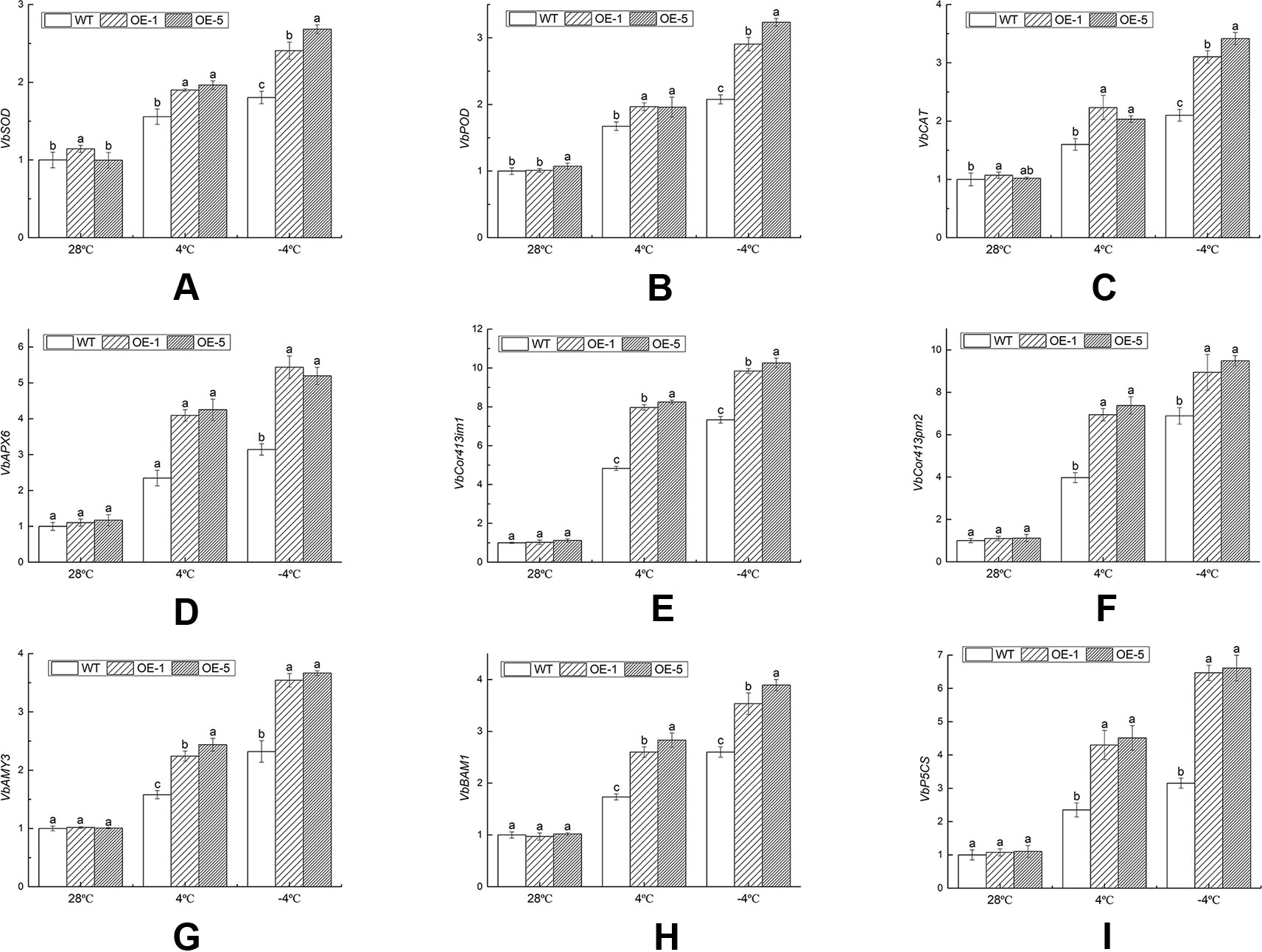
Figure 10 The expression analysis of transcripts of stress-response genes in wild-type (WT) and overexpression (OE) lines (OE-1 and OE-5) at various time and temperature points. (A) VbSOD. (B) VbPOD. (C) VbCAT. (D) VbAPX6. (E) VbCor413im1. (F) VbCor413pm2. (G) VbAMY3. (H) VbBMY1. (G) VbP5CS. Actin-11 was amplified as a control. Data indicate means ± standard errors (SE) of three biological replicates. The different letters above the columns represent significant differences (P < 0.05) on the basis of Duncan’s multiple range test. (I) The seedlings were treated at the following temperature (this cooling process was continuous.): 28°C (the control), 4°C for 24 h, followed by −4°C for 4 h.
Despite low-cost cultivation and high-ornamental value, the study on V. bonariensis has generally been slower compared with many landscape plants, especially in terms of the abiotic stress response. Cold stress decrease production and ornamental value of V. bonariensis. The WRKY TF family was paid great attention to in the field of abiotic stress. It represented as one of major plant-specific transcriptional regulators. Many WRKY TFs were up-regulated under drought, heat, or salt stresses (Wang et al., 2018b; Xu et al., 2018). However, few researches reported the function of WRKY TF under cold stress. In our experiment, VbWRKY32 was separated from V. bonariensis on its differential expression in response to freezing stress.
The results of multiple sequence alignment and phylogenetic tree analyses showed that the VbWRKY32 gene belonged to the group I of WRKY family. The transgenic plants of VpWRKY2, which are in the same branch as VbWRKY32, possessed high cold resistance (Li et al., 2010). Overexpression of TaWRKY19 enhanced the ability of responding salt, drought, and freezing stresses in transgenic plants (Niu et al., 2012). In addition, WRKY32 was actively upregulated in kenaf responding to drought and salinity stresses (Niu et al., 2015). Moreover, the expression level of VbWRKY32 in leaves was remarkably raised under cold stress. The above results showed that VbWRKY32 could participate in the cold resistance process in V. bonariensis.
The expression level of VbWRKY32 in leaves is higher than in stems and roots and the transcript peak was after 4 h at −4°C. The leaves reflected the chilling and freezing injury. We speculated that the leaves activated the expression of VbWRKY32 more quickly than other tissues exposed directly to cold air. CsWRKY2 expression was highly promoted in leaves than in other organs exposed to coldness, drought, and exogenous abscisic acid (Wang et al., 2016). The previous result may be consistent with our speculation. The VbWRKY32 gene in WT could increase expression under cold stress. The elevated expression of VbWRKY32 transgenic lines under normal circumstances could account for its rapid response to cold. The damage of WT plants could have occurred in the early stages responding to cold stress in the process of resistance.
In the experiment for exploring the cold tolerance of V. bonariensis, the OE lines were promoted in response to chill compared with WT. The results were revealed by observing phenotypic changes and measuring recovery ratios, MDA, EL, antioxidant enzyme activities, and osmotic regulating substance contents. VbWRKY32 overexpression caused the elevated expression of down-stream genes and the changes of their related substances in plants.
The MDA content and EL could be used to test the degree of lipid peroxidation and the change of membrane permeability, respectively. Due to cold stress, the high concentration of ROS which was reactive and toxic caused the enhanced production of MDA. The accumulation of MDA under abiotic stress, which caused membrane lipid peroxidation, injury of plant cells, lead to the death of plants ultimately (Mittler et al., 2004; Wang et al., 2018a). Under the chilly condition, the cell membrane of plant transformed from liquid crystalline phase to gel phase with the deviation of selective permeability, which resulted in the cellular electrolyte exosmosis. Therefore, the damage of plants under low temperature stress could be measured by MDA and EL. The production of MDA and the EL of WT were greater compared with VbWRKY32 OE lines. The results illustrated that VbWRKY32 might reduce the accumulation of MDA and stable cell membrane structure.
The antioxidant enzyme system, including POD, SOD, CAT, and APX, could scavenge excessive ROS and improve plant resistance under various types of abiotic stresses (Yu et al., 2014; Singh et al., 2017; Xu et al., 2017; Ma et al., 2018). Compared with the OE lines, the WT plants showed deeper intense histochemical staining by cold treatment. The appearance suggested that less ROS was accumulated in OE lines than the WT. To scavenge ROS, the expression level of ROS-scavenging related genes (e.g., VbSOD, VbPOD, VbCAT, and VbAPX6) had a significant increase in OE lines than that in WT. The transcript levels of these genes were in line with the antioxidant enzyme (SOD, POD, CAT, and APX) activities. The antioxidant enzymes converted toxic superoxide radicals to harmless ion and eliminated hydrogen peroxide in plants (Sarvajeet Singh and Narendra, 2010). The activities of SOD, POD, CAT, and APX in OE lines were remarkably (P < 0.05) increased than that in WT. However, intrinsic antioxidant systems of plants only eliminate a certain amount of ROS and excessive ROS could destroy antioxidant systems.
During cold treatment, we found the enzymes activities of SOD, POD, APX, and CAT were higher in OE lines than that in WT. On the one hand, this may be due to an increase in the level of transcription, which in turn leaded to an increase in protein activities. Further studies need to determine the relation of the transcription of SOD, POD, APX, and CAT to protein activities. On the other hand, the protein normalization treatment of SOD, POD, APX, and CAT was performed, the results have showed that the concentration of enzymes (including protein per FW) in OE lines were lower than that in WT (Figure S1). While the decreases in MDA, H2O2, O2− content (expressed per g FW) in the WRKY OE lines might be explained by an increased retention of water in the OE lines. However, SOD, POD, APX, and CAT (expressed per g FW) in the OE lines increased and they are no significantly different when normalized for protein (expressed per mg protein, Figure S1), it was speculated that increased water loss of in OE lines. These two phenomena were contradictory and not easy to explain. Therefore, the relation of the loss water of plants under cold stress to the protein activities needed further study.
The osmotic adjustment substances such as soluble sugar, soluble protein, and free proline could keep the stability of cellular structure and cell osmotic pressure. It was proved that they aided to maintain proteins and cell structures, particularly under severe or prolonged stress (Hoekstra et al., 2001). Under freezing stress, the transcription levels of VbAMY3, VbBAM1, and VbP5CS had been greatly improved, the expression levels of these gene in OE lines were evidently higher than WT. The expression levels of AMY3 increased significantly under cold conditions in Arabidopsis (Thalmann et al., 2016). By degrading starch into maltose under stress, BAM1 sustained the biosynthesis of proline and soluble sugars, alleviating the oxidative stress (Kyonoshin et al., 2009). The lack of AMY3 and BAM1 prevented plants from mobilizing starch in leaves in face of stress. Carbon exported to the root was reduced, which ultimately affected osmolyte accumulation for water, nutrient intake, and root growth (Zanella et al., 2016). The expression level of VbP5CS which functioned in osmotic adjustment was up-regulated in OE lines. The expression of the P5CS gene induced by environmental stress could promote proline synthesis and increase proline content in plants (He et al., 2018). Therefore, the proline, soluble protein, and soluble sugar in OE lines was of improved content compared with WT under freezing stress.
Massive studies had revealed that expression of COR genes was positively related to cold tolerance in plants (Liang et al., 2011; Wathugala et al., 2011; Wan et al., 2014). The Cor413 family were divided into Cor413-inner membrane (Cor413im) and Cor413-plasma membrane (Cor413pm) proteins (Ghislain et al., 2003). In this study, the expression level of VbCor413im1 and VbCor413pm2 were up-regulated in OE lines compared with WT. Arabidopsis AtCor413im mapped in the inner membrane of chloroplasts might stabilize the chloroplast membrane under cold stress (Okawa et al., 2010; Okawa et al., 2014). Overexpression of PsCor413im1 isolated from Phlox subulata improved cold resistance of Arabidopsis plants (Zhou et al., 2018b). PsCor413pm2, a plasma membrane protein, enhanced cold tolerance of transgenic Arabidopsis. The plasma membrane suffered most from cold injury (Zhou et al., 2018a).
The TFs played an important role in activating multiple biological processes to insulate plant cells from cold. They functioned as a pivotal regulator for adaption of the plant through the binding of TFs to cognate cis-acting elements present in the promoter region of their target genes (Zou et al., 2010; Yang et al., 2012; Peng et al., 2013). The downstream-related genes were regulated by TFs to adapt to external environmental changes. The cold tolerance in plants was enhanced by regulating various series of cold-responsive genes (Thomashow, 1999).
Therefore, the data indicated VbWRKY32 improve the activities of antioxidant enzymes and the content of osmotic adjustment substances by influencing downstream genes, and then alleviate the oxidative damage and stabilize the plasma membrane of V. bonariensis under cold stress.
It is worth mentioning that in addition to affecting the expression of downstream genes and physiological changes, WRKY may also have an impact on water retention, and thus increase the cold resistance of plants. In Arabidopsis, the WRKY54-WRKY70 double mutants exhibited clearly enhanced tolerance to osmotic stress. The enhanced tolerance was correlated with improved water retention and enhanced stomatal closure (Li et al., 2013). The water retention also may be related to EL, enzyme activities, MDA, H2O2, O2− and proline. In addition, the VbWRKY32 over-expression transgenic plants were validated by qRT-PCR in our study, the VbWRKY32 gene played a vital role in improving cold stress tolerance. Whether there will be changes in protein levels in the transgenic plants in cold stress, further studies need to determine the relation of the VbWRKY32 gene to protein level. These issues will be focused in the future.
Taken together, the overexpression of VbWRKY32 in the seedling stage of V. bonariensis resulted in elevated cold tolerance without abnormal growth by phenotypic observation. The expression level of down-stream genes was remarkably promoted, which accounted for the improvements in antioxidant enzyme activities and the contents of osmotic adjustment substance. The VbWRKY32 gene alleviated the damage of membrane lipid peroxidation and relieved the electrolyte exosmosis of the cell. These performances proved the positive function of VbWRKY32 in V. bonariensis under cold stress. Currently, this cold resistance experiment was provisionally limited to the laboratory. Further exploration and verification would be required on whether the transgenic V. bonariensis plants’ safely survival and winter duration in the field environment below 4°C.
The raw sequencing data of the RNASEQ experiment have been submitted to the NCBI Sequence Read Archive database with accession number GSE112477.
M-qW, YL, and Q-lL conceived and designed the experiments. M-qW, Q-xH, PL, and Q-hZ performed the experiments. M-qW, LZ, Y-zP, B-bJ, and FZ analyzed the data. M-qW wrote the paper. All authors read and approved the final manuscript.
This research was supported by National Natural Science Foundation of China (31971707), Sichuan Science and Technology Program (2019YJ0512), Natural Science Foundation of Guizhou (20181044), the introduction of talent project of Guizhou University (201756), and Guizhou University cultivation project (20175788-33).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01746/full#supplementary-material
Figure S1 | The protein normalization treatment of SOD, POD, APX and CAT. (A) SOD. (B) POD. (C) APX. (D) CAT.
An, D. (2012). Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress. BMC Genomics 13, 64. doi: 10.1186/1471-2164-13-64
Beauchamp, C., Fridovich, I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287. doi: 10.1016/0003-2697(71)90370-8
Caterina, M., Mazzucotelli, E., Crosatti, C., Francia, E., Stanca, M. A., Cattivelli, L. (2004). Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol. Biol. 55 (3), 399–416. doi: 10.1007/s11103-004-0906-7
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Ghislain, B., Jean, D., Jean-Benoit Frenette, C., Fathey, S. (2003). Expression profiling and bioinformatic analyses of a novel stress-regulated multispanning transmembrane protein family from cereals and Arabidopsis. Plant Physiol. 132, 64–74. doi: 10.1104/pp.102.015255
Gong, X., Zhang, J., Hu, J., Wang, W., Wu, H., Zhang, Q., et al. (2015). FcWRKY70, a WRKY protein of Fortunella crassifolia, functions in drought tolerance and modulates putrescine synthesis by regulating arginine decarboxylase gene. Plant Cell Environ. 38 (11), 2248–2262. doi: 10.1111/pce.12539
He, L., Wu, Y. H., Zhao, Q., Wang, B., Liu, Q. L., Zhang, L. (2018). ChrysanthemumDgWRKY2Gene enhances tolerance to salt stress in transgenic chrysanthemum. Int. J. Mol. Sci. 19, 2062. doi: 10.3390/ijms19072062
Hoekstra, F. A., Golovina, E. A., Buitink, J. (2001). Mechanisms of plant desiccation tolerance. Trends Plant Sci. 6, 431–438. doi: 10.1016/s1360-1385(01)02052-0
Huang, X. S., Wang, W., Zhang, Q., Liu, J. H. (2013). A basic helix-loop-helix transcription factor, PtrbHLH, of poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol. 162, 1178–1194. doi: 10.1104/pp.112.210740
Irigoyen, J. J., Emerich, D. W., Sanchez-Diaz, M. (2010). Water stress induced changes in concentrations of proline and total soluble sugars in nodulated alfalfa (Medicago sativa) plants. Physiol. Plant 84, 55–60. doi: 10.1111/j.1399-3054.1992.tb08764.x
Kim, C. Y., Vo, K. T. X., Cong, D. N., Jeong, D. H., Lee, S. K., Kumar, M., et al. (2016). Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71. Plant Biotechnol. Rep. 10, 13–23. doi: 10.1007/s11816-015-0383-2
Kyonoshin, M., Migiwa, T., Satoshi, K., Kohji, Y., Yoh, S., Kaoru, U., et al. (2009). Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150, 1972–1980. doi: 10.1104/pp.109.135327
Lei, Z., Li-Hong, T., Jun-Feng, Z., Yun, S., Cui-Jun, Z., Yi, G. (2009). Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 149, 916–928. doi: 10.1104/pp.108.131144
Li, H., Yan, X., Yu, X., Zhu, Z., Xie, X., Zhao, H., et al. (2010). Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from chinese wild vitis pseudoreticulata. Planta 232, 1325–1337. doi: 10.1007/s00425-010-1258-y
Li, J., Besseau, S., Toronen, P., Sipari, N., Kollist, H., Holm, L., et al. (2013). Defense-related transcription factors wrky70 and wrky54 modulate osmotic stress tolerance by regulating stomatal aperture in arabidopsis. New Phytol. 200 (2), 457–472. doi: 10.1111/nph.12378
Liang, C., Hui, Z., Feng, R., Qian-Qian, G., Xu-Peng, H., Xue-Bao, L. (2011). A novel cold-regulated gene, COR25, of Brassica napus is involved in plant response and tolerance to cold stress. Plant Cell Rep. 30, 463–471. doi: 10.1007/s00299-010-0952-3
Liu, X., Song, Y., Xing, F., Wang, N., Wen, F., Zhu, C. (2016). GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenicnicotiana benthamiana. Protoplasma 253 (5), 1265–1281. doi: 10.1007/s00709-015-0885-3
Ma, N. L., Che, L. W., Abd, K. N., Mustaqim, M., Rahmat, Z., Ahmad, A., et al. (2018). Susceptibility and tolerance of rice crop to salt threat: physiological and metabolic inspections. PloS One 13, e0192732. doi: 10.1371/journal.pone.0192732
Mittler, R., Vanderauwera, S., Gollery, M., Breusegem, F. V. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Nair, D., Maria, T. S. W., Luiz, A. S. D. (2009). Chromosome numbers of Jatropha curcas L.: an important agrofuel plant. Crop Breed. Appl. Biotechnol. 9, 386–389. doi: 10.12702/1984-7033.v09n04a14
Niu, C. F., Wei, W., Zhou, Q. Y., Tian, A. G., Hao, Y. J., Zhang, W. K., et al. (2012). Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Env. 35, 1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x
Niu, X., Qi, J., Chen, M., Zhang, G., Su, J. (2015). Reference genes selection for transcript normalization in kenaf (hibiscus cannabinus l.) under salinity and drought stress. PeerJ 3 (3), e1347. doi: 10.7717/peerj.1347
Okawa, K., Nakayama K.K., T., Yamashita, T., Inaba, T. (2010). Identification and characterization of Cor413im proteins as novel components of the chloroplast inner envelope. Plant Cell Env. 31, 1470–1483. doi: 10.1111/j.1365-3040.2008.01854.x
Okawa, K., Inoue, H., Adachi, F., Nakayama, K., Ito-Inaba, Y., Schnell, D. J., et al. (2014). Targeting of a polytopic membrane protein to the inner envelope membrane of chloroplasts in vivo involves multiple transmembrane segments. J. Exp. Bot. 65, 5257–5265. doi: 10.1093/jxb/eru290
Peng, X., Zhang, L., Zhang, L., Liu, Z., Cheng, L., Yang, Y., et al. (2013). The transcriptional factor LcDREB2 cooperates with LcSAMDC2 to contribute to salt tolerance in Leymus chinensis. Plant Cell Tissue Organ Cult. 113, 245–256. doi: 10.1007/s11240-012-0264-0
Qiu, Y., Yu, D. (2009). Over-expression of the stress-induced oswrky45 enhances disease resistance and drought tolerance in arabidopsis. Environ. Exp. Bot. 65 (1), 35–47. doi: 10.1016/j.envexpbot.2008.07.002
Ranieri, A., Petacco, F., Castagna, A., Soldatini, G. F. (2000). Redox state and peroxidase system in sunflower plants exposed to ozone. Plant Sci. 159, 159–167. doi: 10.1016/s0168-9452(00)00352-6
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15 (5), 247–258. doi: 10.1016/j.tplants.2010.02.006
Ryu, H. S., Han, M., Lee, S. K., Cho, J. I., Ryoo, N., Heu, S., et al. (2006). A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 25, 836–847. doi: 10.1007/s00299-006-0138-1
Sarvajeet Singh, G., Narendra, T. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Singh, D., Singh, C. K., Taunk, J., Tomar, R. S. S., Chaturvedi, A. K., Gaikwad, K., et al. (2017). Transcriptome analysis of lentil (Lens culinaris Medikus) in response to seedling drought stress. BMC Genomics 18, 206. doi: 10.1186/s12864-017-3596-7
Tang, J., Liu, Q., Yuan, H., Zhang, Y., Wang, W., Huang, S. (2018). Molecular cloning and characterization of a novel salt-specific responsive WRKY transcription factor gene IlWRKY2 from the halophyte Iris lactea var. chinensis. Genes Genomics 40, 893–903. doi: 10.1007/s13258-018-0698-9
Thalmann, M., Pazmino, D., Seung, D., Horrer, D., Nigro, A., Meier, T., et al. (2016). Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell. 28, 1860. doi: 10.1105/tpc.16.00143
Thomashow, M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Wan, F. X., Pan, Y., Li, J. H., Chen, X. F., Pan, Y. L., Wang, Y. Q., et al. (2014). Heterologous expression of Arabidopsis C-repeat binding factor 3 (AtCBF3) and cold-regulated 15A (AtCOR15A) enhanced chilling tolerance in transgenic eggplant (Solanum melongena L.). Plant Cell Rep. 33, 1951–1961. doi: 10.1007/s00299-014-1670-z
Wang, C., Deng, P., Wang, X., Hu, W., Li, K., Chang, J., et al. (2013). A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PloS One 8 (6), e65120. doi: 10.1371/journal.pone.0065120
Wang, Y., Shu, Z., Wang, W., Jiang, X., Li, D., Pan, J., et al. (2016). CsWRKY2, a novel WRKY gene from Camellia sinensis, is involved in cold and drought stress responses. Biol. Plant. 60, 1–9. doi: 10.1007/s10535-016-0618-2
Wang, K., Wu, Y. H., Tian, X. Q., Bai, Z. Y., Liang, Q. Y., Liu, Q. L., et al. (2017a). Overexpression of DgWRKY4 enhances salt tolerance in chrysanthemum seedlings. Front. Plant Sci. 8, 1592. doi: 10.3389/fpls.2017.01592
Wang, K., Zhong, M., Wu, Y. H., Bai, Z. Y., Liang, Q. Y., Liu, Q. L., et al. (2017b). Overexpression of a chrysanthemum transcription factor gene DgNAC1 improves the salinity tolerance in chrysanthemum. Plant Cell Rep. 36, 571–581. doi: 10.1007/s00299-017-2103-6
Wang, Y., Jiang, L., Chen, J., Tao, L., An, Y., Cai, H., et al. (2018a). Overexpression of the alfalfa WRKY11 gene enhances salt tolerance in soybean. PloS One 13, e0192382. doi: 10.1371/journal.pone.0192382
Wang, C. T., Ru, J. N., Liu, Y. W., Li, M., Zhao, D., Yang, J. F., et al. (2018b). Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. Mol. Sci. 19 (10), E3046. doi: 10.3390/ijms19103046
Wang, M. Q., Zeng, Q. H., Huang, Q. X., Lin, P., Li, Y., Liu, Q. L., et al. (2019). Transcriptomic Analysis of Verbena bonariensis Leaves Under Low-Temperature Stress. DNA Cell Biol. 38 (11), 1–16. doi: 10.1089/dna.2019.4707
Wathugala, D. L., Richards, S. A., Heather, K., Knight, M. R. (2011). OsSFR6 is a functional rice orthologue of SENSITIVE TO FREEZING-6 and can act as a regulator of COR gene expression, osmotic stress and freezing tolerance in Arabidopsis. New Phytol. 191, 984–995. doi: 10.1111/j.1469-8137.2011.03759.x
Wu, X. L., Yoko, S., Sachie, K., Yukihiro, I., Kinya, T. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xiu, H., Nuruzzaman, M., Guo, X., Cao, H., Huang, J., Chen, X., et al. (2016). Molecular cloning and expression analysis of eight PgWRKY genes in panax ginseng responsive to salt and hormones. Int. J. Mol. Sci. 17, 319. doi: 10.3390/ijms17030319
Xu, B., Wang, Y., Zhang, S., Guo, Q., Jin, Y., Chen, J., et al. (2017). Transcriptomic and physiological analyses of Medicago sativa L. roots in response to lead stress. PloS One 12, e0175307. doi: 10.1371/journal.pone.0175307
Xu, Z., Raza, Q., Xu, L., He, X., Huang, Y. X., Yi, J., et al. (2018). GmWRKY49, a Salt-responsive nuclear protein, improved root length and governed better salinity tolerance in transgenic Arabidopsis. Front. Plant Sci. 9, 809. doi: 10.3389/fpls.2018.00809
Yamamoto, Y., Kobayashi, Y., Matsumoto, H. (2001). Lipid peroxidation is an early symptom triggered by aluminum, but not the primary cause of elongation inhibition in pea roots. Plant Physiol. 125, 199–208. doi: 10.1104/pp.125.1.199
Yang, A., Dai, X., Zhang, W. H. (2012). A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63, 2541–2556. doi: 10.1093/jxb/err431
Ying, Z., Yu, H., Yang, X., Qiang, L., Jian, L., Hong, W., et al. (2016). CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol. Biochem. Ppb. 108, 478–487. doi: 10.1016/j.plaphy.2016.08.013
Yu, C., Jiafu, J., Qingshan, C., Chunsun, G., Aiping, S., Sumei, C., et al. (2014). Cold acclimation induces freezing tolerance via antioxidative enzymes, proline metabolism and gene expression changes in two chrysanthemum species. Mol. Biol. Rep. 41, 815–822. doi: 10.1007/s11033-013-2921-8
Zanella, M., Borghi, G. L., Pirone, C., Thalmann, M., Pazmino, D., Costa, A., et al. (2016). β-amylase 1 (BAM1) degrades transitory starch to sustain proline biosynthesis during drought stress. J. Exp. Bot. 67, 1819–1826. doi: 10.1093/jxb/erv572
Zhang, L., Xi, D., Luo, L., Meng, F., Li, Y., Wu, C. A., et al. (2011). Cotton GhMPK2 is involved in multiple signaling pathways and mediates defense responses to pathogen infection and oxidative stress. FEBS J. 278, 1367–1378. doi: 10.1111/j.1742-4658.2011.08056.x
Zhou, L., Wang, N. N., Gong, S. Y., Lu, R., Li, Y., Li, X. B. (2015). Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiol. Biochem. 96, 311–320. doi: 10.1016/j.plaphy.2015.08.016
Zhou, A., Liu, E., Li, H., Li, Y., Feng, S., Gong, S., et al. (2018a). PsCor413pm2, a plasma membrane-localized, cold-regulated protein from phlox subulata, confers low temperature tolerance in Arabidopsis. Int. J. Mol. Sci. 30, 19 (9), E2579. doi: 10.3390/ijms19092579
Zhou, A., Sun, H., Feng, S., Zhou, M., Gong, S., Wang, J., et al. (2018b). A novel cold-regulated gene from Phlox subulata, PsCor413im1, enhances low temperature tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 495, 1688–1694. doi: 10.1016/j.bbrc.2017.12.042
Keywords: Verbena bonariensis, cold stress, VbWRKY32, antioxidant enzymes, osmotic adjustment, down-stream genes
Citation: Wang M-q, Huang Q-x, Lin P, Zeng Q-h, Li Y, Liu Q-l, Zhang L, Pan Y-z, Jiang B-b and Zhang F (2020) The Overexpression of a Transcription Factor Gene VbWRKY32 Enhances the Cold Tolerance in Verbena bonariensis. Front. Plant Sci. 10:1746. doi: 10.3389/fpls.2019.01746
Received: 05 July 2019; Accepted: 12 December 2019;
Published: 29 January 2020.
Edited by:
Eric Ruelland, UMR7618 Institut d’écologie et des sciences de l’environnement de Paris (IEES), FranceReviewed by:
Stephen K. Randall, Purdue University Indianapolis, United StatesCopyright © 2020 Wang, Huang, Lin, Zeng, Li, Liu, Zhang, Pan, Jiang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Li, eWxpQGd6dS5lZHUuY24=; Qing-lin Liu, cWluZ2xpbmxpdUAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.