- 1State Key Laboratory of Crop Stress Biology for Arid Areas and College of Plant Protection, Northwest A&F University, Yangling, China
- 2Agricultural Research Service, United States Department of Agriculture and Department of Plant Pathology, Washington State University, Pullman, WA, United States
- 3Pest & Pathogen Ecology, NIAB East Malling Research, Kent, United Kingdom
RPM1 is a CC-NBS-LRR protein that was first shown to be required for resistance to Pseudomonas syringae pv. maculicola in Arabidopsis thaliana. Our previous study showed that TaRPM1 gene in wheat was upregulated about six times following infection by Puccinia striiformis f. sp. tritici (Pst) under high temperature, compared with normal temperature. To study the function of TaRPM1 in wheat high-temperature seedling-plant (HTSP) resistance to Pst, the full length of TaRPM1 was cloned, with three copies each located on chromosomes 1A, 1B, and 1D. Transient expression of the TaRPM1-GFP fusion protein in Nicotiana benthamiana indicated that TaRPM1 localizes in the cytoplasm and nucleus. Profiling TaRPM1 expression indicated that TaRPM1 transcription was rapidly upregulated upon Pst inoculation under high temperature. In addition, TaRPM1 was induced by exogenous salicylic acid hormone application. Silencing TaRPM1 in wheat cultivar Xiaoyan 6 (XY 6) resulted in reduced HTSP resistance to Pst in terms of reduced number of necrotic cells and increased uredinial length, whereas no obvious phenotypic changes were observed in TaRPM1-silenced leaves under normal temperature. Related defense genes TaPR1 and TaPR2 were downregulated in TaRPM1-silenced plants under high temperature. We conclude that TaRPM1 is involved in HTSP resistance to Pst in XY 6.
Introduction
Wheat (Triticum aestivum) yield can be greatly reduced by stripe rust caused by obligate biotrophic pathogen Puccinia striiformis f. sp. tritici (Pst) (Wellings, 2011; Chen, 2014). Currently, stripe rust control is primarily achieved through the use of resistant cultivars (Chen, 2014) and fungicide sprays (Chen et al., 2013). However, the loss of resistance in wheat cultivars has been happening frequently due to rapid emergence of new virulent races in Pst (Chen et al., 2009; Hu et al., 2014).
Wheat high-temperature (HT) resistance to Pst, a non-race-specific and durable resistance, can be classified into two types: high-temperature adult-plant (HTAP) and high-temperature seedling-plant (HTSP) resistance (Chen, 2013). HTAP resistance expresses or increases when plants are in the adult stage and weather becomes warm, whereas HTSP resistance expresses when wheat seedlings are temporarily exposed to 20°C for only 24 h at the initial stage of Pst incubation (Wang et al., 2017a; Wang et al., 2019). Xiaoyan 6 (XY 6) is a typical example of wheat cultivars with HTSP resistance to Pst.
Plants have evolved various defense mechanisms against biotic (e.g., bacterial and fungal pathogens) and abiotic stresses (e.g., high temperature, drought, salt, and heavy metals) (Jones and Dangl, 2006; Bhattarai et al., 2016; Jiang et al., 2018). One efficient mechanism, effector-triggered immunity (ETI), is activated upon recognition of a pathogen avirulence (Avr) gene by a resistance (R) protein, leading to an array of defense responses, including hypersensitive response (HR) (Cui et al., 2015), reactive oxygen species (ROS) bursts, and induction of defense-related genes (Kandoth and Mitchum, 2013; Liang et al., 2014; Gao et al., 2017).
To date, more than 100 R genes against 122 different pathogens have been cloned from numerous plant species such as Arabidopsis, tomato, potato, barely, rice, and wheat (Hinsch and Staskawicz, 1996; Anderson et al., 1997; Ellis et al., 1999; Feuillet et al., 2003; Tör et al., 2004; Fu et al., 2009; Zhou et al., 2014; Li N. Y. et al., 2017; Qian et al., 2017; He et al., 2018; Klymiuk et al., 2018). R proteins can be divided into several super-families based on their specific conserved motifs, including nucleotide-binding sites (NBS), leucine-rich repeats (LRR), toll-interleukin-1 receptors (TIR), coiled-coils (CC), transmembrane motifs (TM), and protein kinases (PK) (Li X. et al., 2017). The NBS-LRR resistance genes represent the largest R-gene family, and can be further subdivided into two major subclasses: those having a putative CC domain (CC-NB-LRR) and those having a TIR domain (TIR-NB-LRR) (Ellis and Jones, 1998; Dangl and Jones, 2001). The CC-NB-LRR subclass includes genes such as RPM1 and RPS2 of Arabidopsis, which could recognize specifically avrB and avrRpt2 effectors, respectively (Gopalan et al., 1996; Warren et al., 1998; Xu et al., 2018). TIR-NB-LRR subclass includes rust R gene L6 in flax (Lawrence et al., 1995) and downy mildew R-genes RPP5 (Parker et al., 1997) and RPP1 (Botella et al., 1998) in Arabidopsis. The Arabidopsis RPM1 gene confers resistance against bacterium Pseudomonas syringae expressing either of the Type III effectors AvrRpm1 or AvrB (Mackey et al., 2002). RIN4 (RPM1-interacting protein 4) has been identified as a membrane protein for resistance against P. syringae via its interaction with RPM1. AvrB and AvrRpm1, secreted into plant cells by the Type III protein secretion system, induce phosphorylation of RIN4, which is perceived by RPM1 and then serves to activate host resistance responses (Gururani et al., 2012). Therefore, RPM1 “guards” the plant against P. syringae by perceiving the Avr-dependent modifications of RIN4 (Dangl and Jones, 2001). We showed that TaRPM1 gene in XY 6 is upregulated rapidly following infection by Pst under high temperature, compared with normal temperature (Tao et al., 2018). Thus, TaRPM1 is associated with HTSP; however, the precise roles played by TaRPM1 in the HTSP resistance to Pst has not been elucidated.
Temperature sensitivity of R genes has been widely reported in numerous plants. For example, tobacco N-mediated HR against tobacco mosaic virus is activated at 22°C but not at 30°C (Wang et al., 2009). The tomato Mi-1 gene against root-knot nematodes is inactive above 28°C (Hwang et al., 2000; Jablonska et al., 2007). The Arabidopsis RPW8 gene, conferring resistance to powdery mildew, is suppressed above 30°C (Xiao et al., 2003). The defense responses conferred by Arabidopsis NB-LRR receptor gene SNC1 is activated at 22°C, but not at 28°C (Yang and Hua, 2004). Yr36, an R gene involved in HTAP, confers resistance to Pst at relatively high temperatures (25°C to 35°C) but not at low temperatures (e.g., 15°C) (Fu et al., 2009). Previously, we showed transcriptional factors TaWRKY70 (Wang et al., 2017a), TaWRKY62 (Wang et al., 2017b), and receptor like kinase TaXa21 (Wang et al., 2019) positively regulate HTSP resistance to Pst.
In the present study, we identified and cloned a highly upregulated NBS-LRR gene TaRPM1 from XY 6 infected with Pst and subsequently exposed to high temperature for 24 h. Silencing TaRPM1 in XY 6 impaired HTSP resistance to Pst with reduced host defense responses, increased Pst growth, and decreased the expression levels of TaPR1 and TaPR2. We thus conclude that TaRPM1 positively regulates the HTSP resistance to Pst through the salicylic acid (SA) signaling pathway.
Materials And Methods
Identification and Characterization of TaRPM1
To clone the TaRPM1 gene, full-length primers based on the XY 6 transcriptome sequences (Tao et al., 2018) were designed using Primer 5.0 software (Table S1). The PCR products were purified, and cloned into the PMD18-T vector (TaKaRa, Tokyo, Japan) for sequencing. A phylogenetic tree of TaRPM1 and RPM1 members in other species were generated by the neighbor-joining method (1,000 bootstrap replicates) using MEGA6.0 software. For confirming the copy number of TaRPM1 in the wheat genome, nucleotide sequence of TaRPM1 was aligned with the sequence from the wheat genome database (http://www.wheatgenome.org/). Multiple sequence alignment was performed using DNAMAN6.0 software.
Plant and Fungal Materials, Inoculations, and Treatments
Wheat cultivar XY 6 and Pst race CYR32 were used in this study. The methods of growing wheat seedlings, inoculation, and temperature treatment regimes were the same as those described by Wang et al. (2017a). To analyze the expression of TaRPM1 under different treatments, leaves were sampled at 0, 48, 96, 192, 194, 198, 204, 216, 240, 264, and 312 hpi with Pst. At 192 hpi, some plants were exposed to the HT (20°C) treatment for 24 h, whereas the others remained at the NT (15°C) treatment. Control wheat seedlings were inoculated with sterile distilled water. All samples were immediately flash-frozen in liquid nitrogen and stored at −80°C. In all experiments, there were three biological replicates for each treatment and sampling time combination, and three technical replicates for each sample were conduced to qRT-PCR analysis.
Hormone Treatments and Tissue-Specific Expression Analysis
For hormone treatments, leaves of 2-week-old wheat seedlings were sprayed with one of the following hormones: 100 µM SA, 100 µM MeJA, 100 µM ET, and 100 µM ABA. Each of the hormones was dissolved in 0.1% (v/v) ethanol (Wang et al., 2017a). The mock wheat seedlings were sprayed with 0.1% (v/v) ethanol. Leaves were sampled at 0, 0.5, 2, 6, 12, and 24 h post-treatment. For tissue-specific expression analysis of TaRPM1, roots, stems, and leaves were sampled from 2-week-old wheat seedlings under the NT conditions. Three independent biological replicates were used for each treatment and sampling time combination, and three technical replicates were performed for each sample to qRT-PCR analysis.
RNA Extraction and qRT-PCR Analysis
Total RNA of sampled leaves was extracted using the SV Total RNA Isolation System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. First-strand cDNA was synthesized using the PrimeScript RT Reaction System (TaKaRa, Tokyo, Japan) according to the manufacturer’s instruction. qRT-PCR was performed using UltraSYBR Mixture (Kangwei, Beijing, China) to quantify TaRPM1 expression. Based on our previous study (Wang et al., 2014), the wheat Ta26s gene (ATP dependent 26s proteasome regulatory subunit) expressed stably among different treatments; therefore, Ta26s was used as a reference gene for analyses. Relative expression of TaRPM1 was analyzed using the comparative 2–∆∆Ct method. In all the experiments, three independent biological replicates and three technical replicates of each biological replicate for each sample were analyzed to ensure reproducibility and reliability.
BSMV-Mediated TaRPM1 Gene Silencing
To generate the BSMV: TaRPM1-1as and BSMV: TaRPM1-2as recombined plasmids, two specific cDNA fragments of TaRPM1 with the NotI and PacI restriction sites were inserted into the BSMV:r vector, respectively. The wheat phytoene desaturase (TaPDS) gene was inserted into the BSMV:r vector as a positive control. Two-week-old wheat seedlings were inoculated with each of the four viruses: BSMV:r, BSMV: TaPDS, BSMV: TaRPM1-1as, and BSMV: TaRPM1-2as following a previously published method (Wang et al., 2017a). Wheat seedlings treated with 1x FES buffer were used as a negative control. After incubation for 24 h in the dark, all wheat plants were placed in a growth chamber at 25 ± 1°C. Once photobleaching was observed in the BSMV: PDS infected leaves, the fourth leaves were inoculated with Pst race CYR32 and then maintained at 15 ± 1°C. For estimating the silencing efficiency of TaRPM1, leaves infected with Pst were sampled at 0, 24, 48, and 120 hpi for qRT-PCR. To confirm the TaRPM1 silencing efficiency and expression level of PR genes, leaves were harvested at 0, 12, 24, 48, 72, and 120 hptt for RNA extraction and qRT-PCR (HT was applied at 0 hptt). Three independent biological replicates were performed for each treatment and sampling time combination, and three technical replicates for each sample were conducted to qRT-PCR analysis.
Histological Observations
The sampled wheat leaves were decolorized and stained as previously described (Wang et al., 2007). The stained leaf segments were observed under a microscope for hyphal length, colony linear length, number of haustoria, and uredinial length using DP-BSW software (Olympus, Corp., Tokyo, Japan). Autofluorescence of wheat necrotic cells was observed through epifluorescence microscopy. About 30–50 infection sites were examined from 8–10 randomly selected leaf segments for each treatment at each sampling time point. Ten leaves were selected randomly for each treatment to assess pustules number/leaves area. There were three biological replicates for each treatment at each sampling time point.
Subcellular Localization Analysis
The pCambia-TaRPM1-GFP fusion protein and pCambia1302-GFP control vector were separately introduced into Agrobacterium tumefaciens strain GV3101 through electroporation. Five-week-old tobacco plants were transiently transformed with A. tumefaciens GV3101 containing pCambia-TaRPM1-GFP or pCambia1302-GFP constructs and then assessed under a fluorescent microscope. An H2B-mcherry recombination plasmid was used as a nuclear location marker.
For Western blotting, the total protein was extracted from 500 mg of N. benthamiana leaves carrying pCambia-TaRPM1-GFP and was separated by SDS-PAGE gel. GFP protein was detected with the anti-GFP antibody (Sigma-Aldrich, Shanghai, China).
Statistical Analysis
Analysis of variance (ANOVA) was conducted using SAS software (SAS Institute Inc., Cary, NC, USA). Individual mean comparisons were based on the least significant difference (LSD) test.
Results
Cloning and Characterization of TaRPM1
We used the 5’ and 3’ rapid amplification of cDNA ends (RACE) method to clone a 3,325bp cDNA fragment from XY 6, which was selected due to its high expression level of the HTSPresistance after inoculation with Pst race CYR32 and subsequently exposed to high temperature for 24 h. The cDNA nucleotide sequence contains an open reading frame (ORF) of 2,754 bp encoding a predicted protein of 917 amino acids with an estimated molecular weight of 103.31 kDa and an isoelectric point value of 6.84. The predicted amino acid sequence of this protein was identical to that of AetRPM1-like of Aegilops tauschii; thus, this gene is named as TaRPM1 (GenBank accession number MN647923). Phylogenetic analysis of TaRPM1 and other RPM1 proteins showed that TaRPM1 is most similar to rice OsRPM1 in addition to AetRPM1-like (Figure 1). A BlastN search at the International Wheat Genomic Sequence Consortium (IWGSC) revealed that TaRPM1 shares 94.71% nucleotides with TaRPM1-1AL (Figure S1).
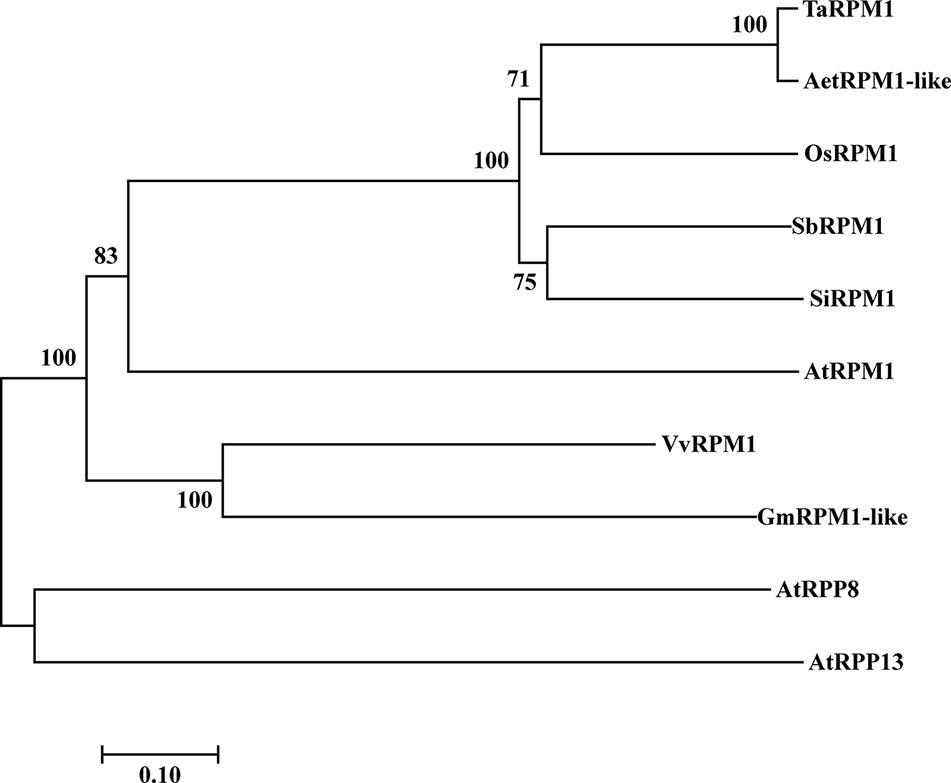
Figure 1 A phylogenetic tree of RPM1 amino acid sequences based on multiple alignments. The GenBank accession numbers of RPM1 protein sequences are as follows: AetRPM1-like (XP_020156667.1), OsRPM1 (XP_015616849.1), SbRPM1 (XP_002449338.1), SiRPM1 (XP_004979032.1), AtRPM1 (AGC12588.1), VvRPM1 (XP_002265617.2), GmRPM1 (XP_003551698.1), AtRPP8 (AAL32592.1), AtRPP13 (AAF42832.1). Aet, Aegilops tauschii; Os, Oryza sativa; Sb, Sorghum bicolor; Si, Setaria italic; At, Arabidopsis thaliana; Vv, Vitis vinifera; Gm, Glycine max.
TaRPM1Transcript Levels in Different Tissues and Its Response to Hormones
The relative expression levels of TaRPM1 in different wheat tissues were determined using quantitative reverse transcription PCR (qRT-PCR). The TaRPM1 gene was mainly expressed in leaves and, to a less extent, in roots and stems (Figure 2A). To study the expression level of TaRPM1 in response to plant hormones, wheat seedlings were treated with SA, ethylene (ET), methyl jasmonate (MeJA), and abscisic acid (ABA). TaRPM1 transcription was significantly (P < 0.05) increased only upon the SA treatment for 0.5 h. In contrast, when treated with ABA, the expression level of TaRPM1 was significantly reduced during 2–24 h (Figure 2B).
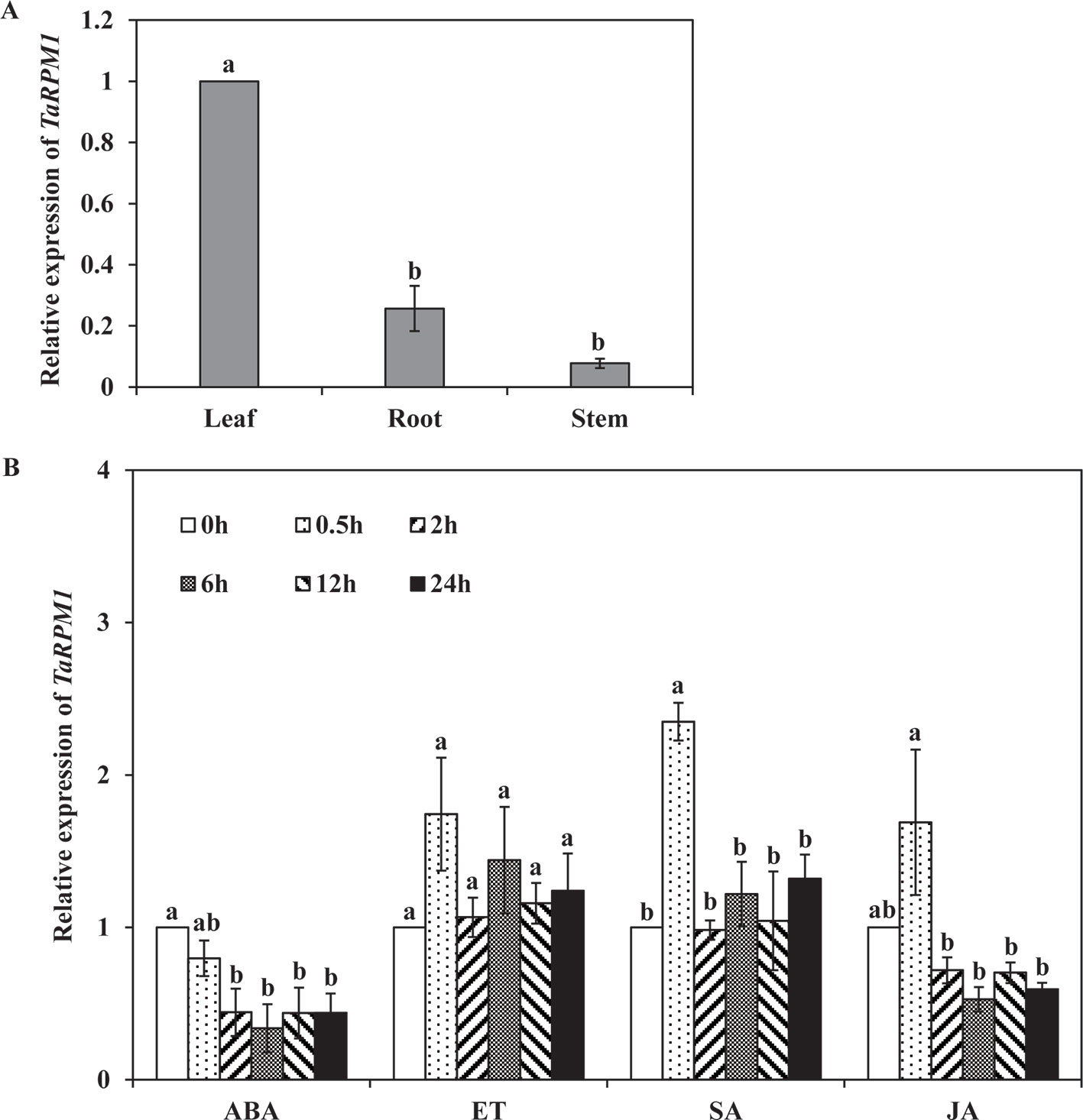
Figure 2 The relative transcript levels of TaRPM1 in different wheat tissues and in response to different hormones. (A) Tissue-specific expression level of TaRPM1. (B) Responses to hormones: ABA, abscisic acid; ET, ethylene; SA, salicylic acid; MeJA, methyl jasmonate. The mock control was treated with 0.1% (v/v) ethanol. Wheat leaves were sampled at 0, 0.5, 2, 6, 12, and 24 h post–hormone treatment. There were three biological replicates for each treatment and each sampling time point. Three technical replicates for each sample were conducted. Relative transcript levels of TaRPM1 were calculated using the comparative threshold (2−ΔΔCT) method, relative to the mock control at every sampling point. The expression level was standardized as 1 at 0 h. Duncan’s multiple comparison test was conducted to compare between time points for each hormone treatment. The TaRPM1 expression levels do not differ significantly if they contain at least one common lowercase letter among time points for each hormone treatment.
TaRPM1 Transcript Level in HTSP Resistance to Pst
To investigate the expression profile of TaRPM1 in the HTSP resistance to Pst, both inoculated and non-inoculated wheat seedlings were subjected to two temperature regimes [high temperature (HT): 20°C for 24 h to induce HTSP, or normal temperature (NT): 15°C] at 192 h post–Pst inoculation (hpi), and the leaves were sampled at several time points up to 312 hpi. Compared with the mRNA levels in the mock leaves (non-inoculated leaves), the TaRPM1 transcript level increased in response to Pst inoculation. TaRPM1 expression from plants exposed to 20°C for 24 h was higher (P < 0.05) than that under 15°C at 204, 216, and 264 hpi, and peaked at 204 hpi (Figure 3).
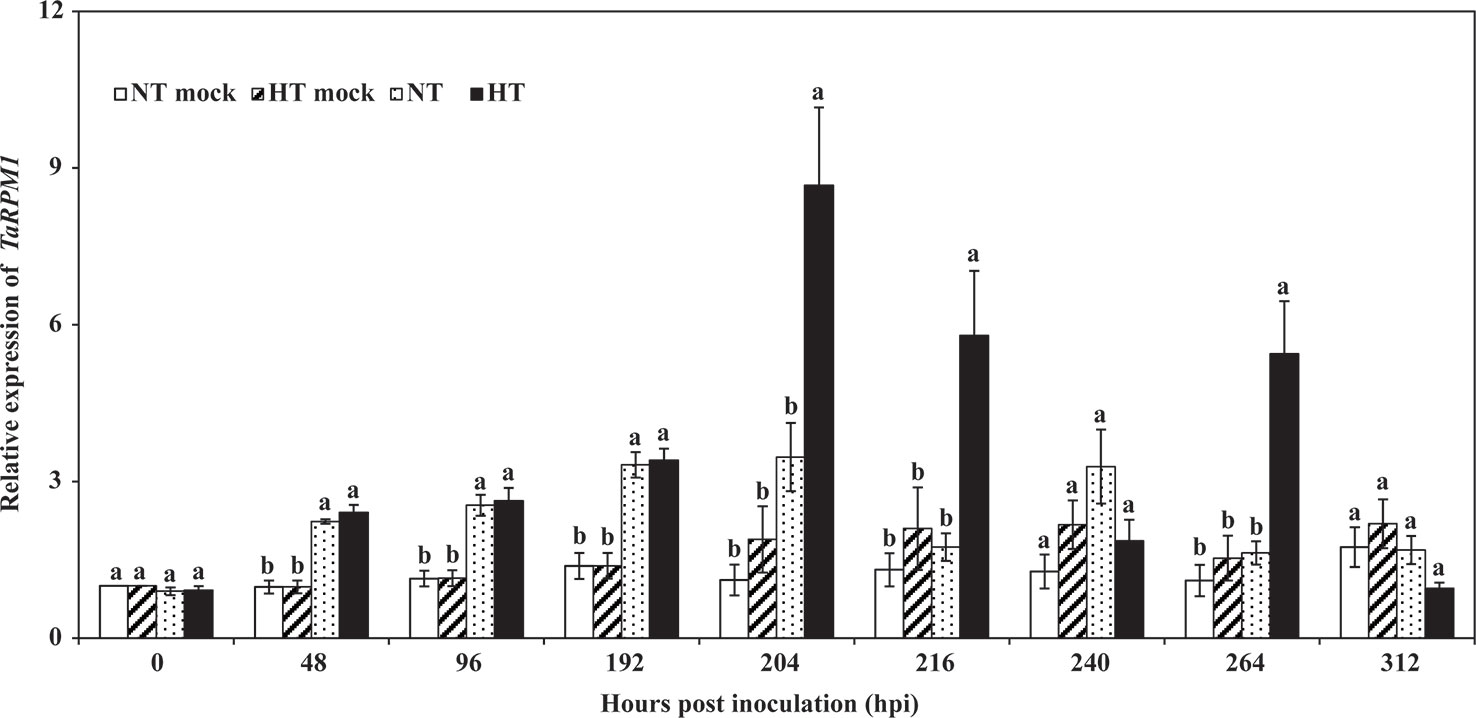
Figure 3 The expression profiles of TaRPM1 in high-temperature seedling-plant (HTSP) resistance to Puccinia striiformis f. sp. tritici (Pst). NT, wheat leaves were maintained under normal temperature (15°C) after inoculated with Pst. HT, wheat leaves were inoculated with Pst and transferred to high temperature (20°C) for 24 h at 192 h post-inoculation (hpi). NT mock, non-inoculated wheat leaves were exposed to 15°C. HT mock, non-inoculated wheat leaves were exposed to 20°C for 24 h at 192 hpi. Three independent biological replicates were performed for each treatment and sampling time combination, and three technical replicates for each sample were conducted. The TaRPM1 expression level in the NT mock leaves at 0 hpi was standardized as 1. Duncan’s multiple comparison test was conducted at the same time point within four different treatments. There are no significant differences at the same time point among the treatments if they share at least one lowercase letter.
TaRPM1 Knockdown Compromised Wheat HTSP Resistance to Pst
To determine the contribution of TaRPM1 to HTSP in XY 6, we performed barley stripe mosaic virus (BSMV)–based virus-induced gene silencing (VIGS). Two TaRPM1-specifc fragments were integrated separately into the BSMV:γ vector to generate BSMV: TaRPM1-1as and BSMV: TaRPM1-2as silencing plants. All of the BSMV-inoculated plants showed stripe mosaic symptoms 10 days after virus inoculation, and the plants inoculated with BSMV: PDS (phytoene desaturase) displayed a photobleaching phenotype 15 days after BSMV inoculation, suggesting the induction of BSMV-mediated silencing (Figure 4A). qRT-PCR results indicated that TaRPM1 silencing efficiency was in the range of 70%–80% (Figure 4E). Next, the fourth leaves of XY 6 plants were inoculated with Pst race CYR32. Fifteen days after Pst inoculation, XY 6 showed a susceptible response under the NT treatment with numerous uredinia on the mock and BSMV:00 leaves (Figures 4B, D). The non-silenced leaves with a higher expression level of TaRPM1 showed a resistant response to Pst under the HT treatment; and in contrast, the TaRPM1-silenced leaves had greater fungal growth than with the non-silenced leaves under HT (Figures 4C, D, F).
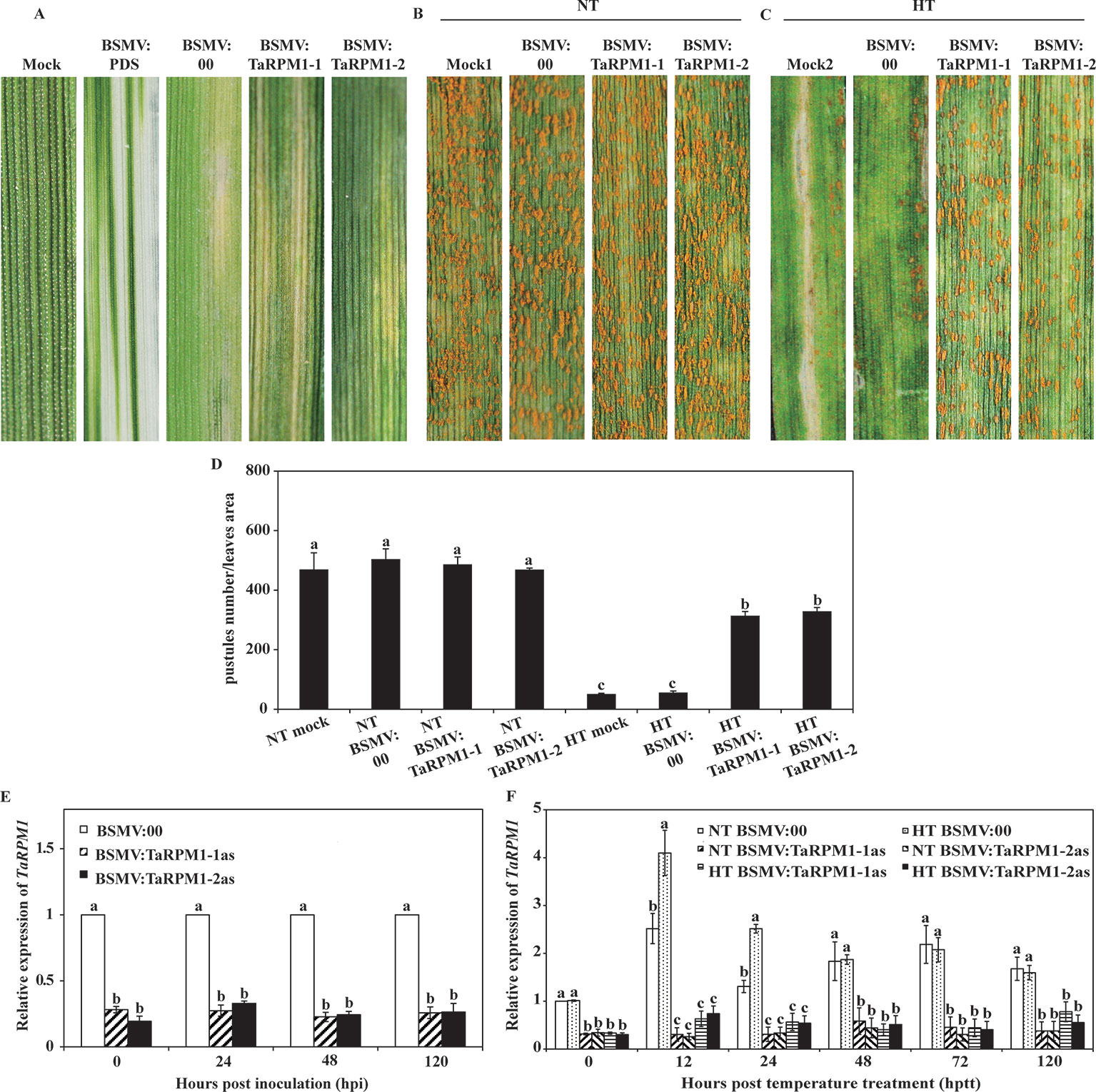
Figure 4 Functional analysis of TaRPM1 in HTSP resistance to Pst using virus-induced gene silencing assay. (A) Phenotypic observation on the second leaves inoculated with FES buffer (mock), BSMV: TaPDS, BSMV:00, BSMV: TaRPM1-1as, and BSMV: TaRPM1-2as. The mock leaves and leaves pre-inoculated with BSMV virus were all challenged with Pst race CYR32, followed by NT (B) and HT (C) treatment at 192 hpi. (D) Quantification of pustules number in a certain area on different treatment leaves. (E) The relative expression levels of TaRPM1 in silenced and non-silenced leaves infected with CYR32. Leaves were collected at 0, 24, 48, and 120 hpi with CYR32 for RNA extraction and quantitative reverse transcription PCR (qRT-PCR analysis). The TaRPM1 transcript level in non-silenced leaves at every sampled time point was standardized as 1. (F) The relative expression levels of TaRPM1 in the silenced and non-silenced leaves inoculated with CYR32 under NT and HT treatment. RNA samples were isolated from the leaves first infected with barley stripe mosaic virus (BSMV), and then inoculated with CYR32 at 0, 12, 24, 48, 72, and 120 h post-temperature treatment (hptt) (0 hptt: HT was applied, namely 192 hpi). Three independent biological replicates were performed for each treatment and sampling time combination, and three technical replicates for each sample were conducted. The TaRPM1 transcript level in NT-treated non-silenced leaves at 0 hptt was standardized as 1. If the treatments share at least one common lowercase letter at the same time point, the TaRPM1 expression levels do not differ significantly among the these treatments.
To confirm the host response in TaRPM1-silenced plants upon Pst infection and the HT treatment, we measured the transcript levels of two pathogenesis-related (PR) protein genes in the SA-mediated signaling pathway. TaPR1 (Figure 5A) and TaPR2 (Figure 5B) were both significantly decreased (P < 0.05) in the two HT-treated TaRPM1-silenced leaves compared with the HT-treated but non-silenced leaves.
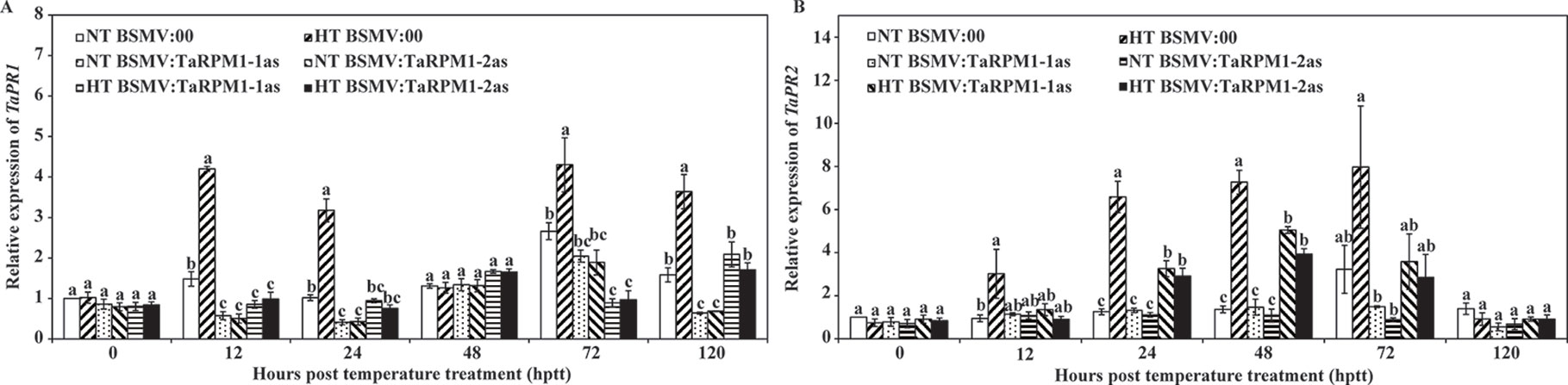
Figure 5 The relative transcript levels of TaPR1 (A) and TaPR2 (B) in TaRPM1-silenced plants infected with race CYR32 of P. striiformis f. sp. tritici exposed to different temperature treatments. Three independent biological replicates were performed for each treatment and sampling time combination, and three technical replicates for each sample were conducted. The expression levels of TaPR1 and TaPR2 in the NT-treated leaves pre-inoculated with BSMV:00 at 0 hptt was standardized as 1. There are no significant differences at the same time point among treatments if they share at least one common lowercase letter.
Histological Observation of Pst Growth and Host Response
We examined leaves microscopically to determine histological changes in TaRPM1-silenced leaves at the initial stage of Pst development (Figures 6A–F). At 48 and 120 hpi, hyphal length and number of haustorial mother cells did not differ significantly between BSMV:00 control and TaRPM1-silenced plants (Figures 6G, H).
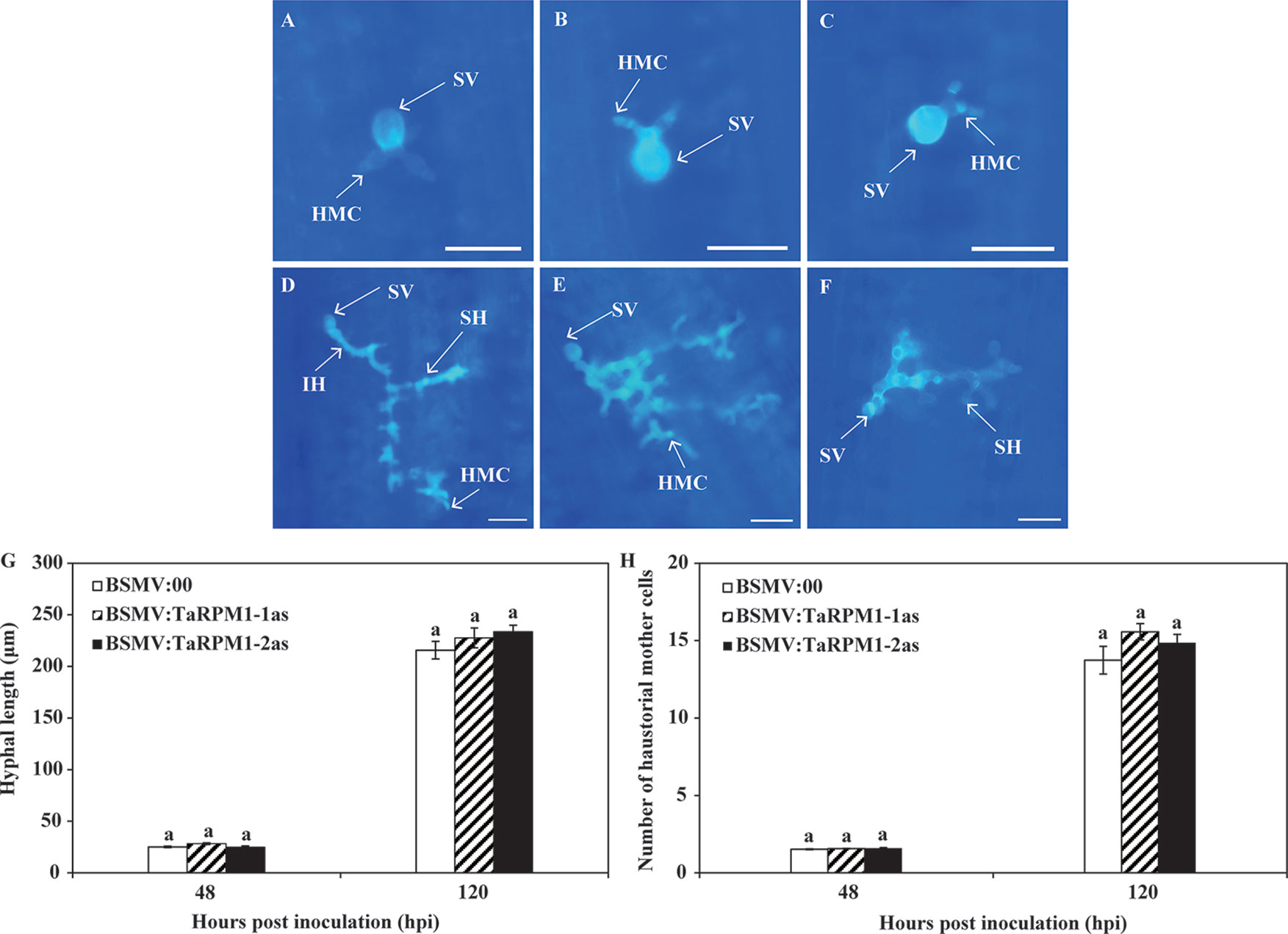
Figure 6 Histological observations of Pst development and host responses in TaRPM1-silenced leaves under normal temperature. The images of the fourth leaves inoculated with BSMV:00 (A, D), BSMV: TaRPM1-1as (B, E), and BSMV: TaRPM1-2as (C, F) were taken under a fluorescence microscope at 48 hpi (A, B, C, bar, 50 μm) and 120 hpi (D, E, F, bar, 100 μm). SV, substomatal vesicle; HMC, haustorial mother cell; IH, infection hypha; SH, secondary hypha. Hyphal length (G) and the number of haustorial mother cells (H) were assessed at 48 and 120 hpi using the DP-BSW software after Pst were stained. There are no significant differences at the same time point among treatments if they share at least one common lowercase letter. Three independent biological replicates were performed for each treatment and sampling time combination.
At 192 hpi, plants inoculated with Pst were subjected to one of the two temperature regimes (HT and NT). For the NT treatment, fusion of urediniospores was observed (Figures 7A–C), and there were no apparent differences in the uredinium length, the linear length of colony per infection site, and the number of necrotic cells between NT-treated BSMV:00 and NT-treated TaRPM1-silenced plants. In contrast, uredinia were sparsely distributed in the HT-treated leaves (Figures 7D–F), the colony length in the HT-treated TaRPM1-silenced leaves was grater (P < 0.05) than that the HT-treated but non-silenced leaves at 24 h post-temperature treatment (hptt) (Figure 7G), and similarly the uredinium length in the HT-treated TaRPM1-silenced leaves was greater (P < 0.05) than that in the HT-treated non-silenced leaves 312 hpi, i.e., 120 h hptt (Figure 7H). There were fewer (P < 0.05) necrotic cells in the HT-treated TaRPM1-silenced leaves than in the HT-treated non-silenced leaves from 24 to 120 hptt (Figure 7I).
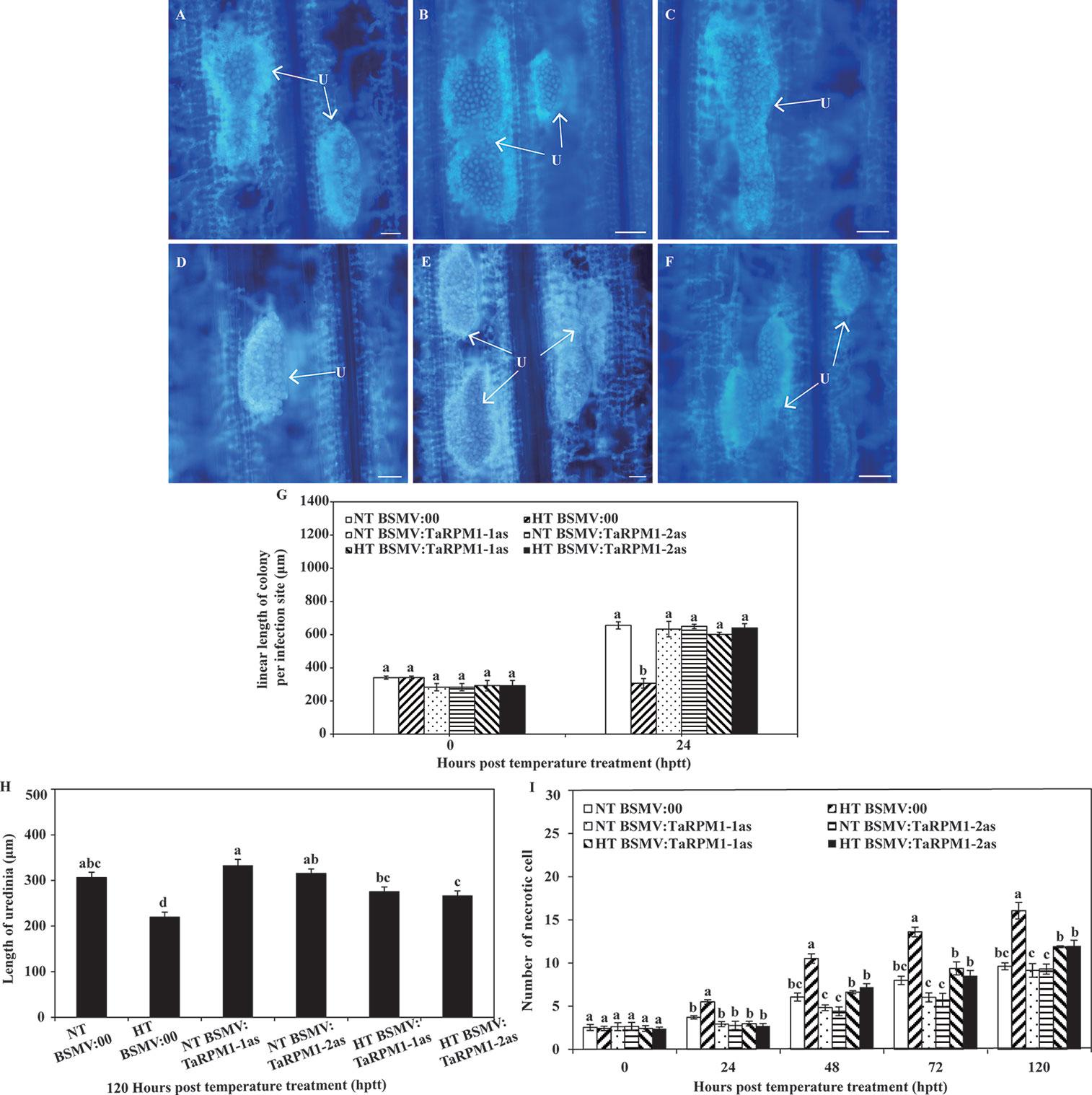
Figure 7 Silencing TaRPM1 in wheat leaves induces growth of the Pst under high temperature. The urediniospores in non-silenced leaves (A, D, bar 100 μm) and TaRPM1-silenced (B, C, E, F, bar, 100 μm) under NT (A, B, C) and HT (D, E, F) were observed and photographed at 24 hptt. The colony length (G) uredinium lengths (H) and numbers of necrotic cells (I) from 30–50 randomly selected infection sites were calculated. Three independent biological replicates were performed for each treatment and sampling time combination. There are no significant differences at the same time point among treatments if they share at least one common lowercase letter.
TaRPM1 Localization
Prediction of subcellular localization using WoLF PSORT indicated that TaRPM1 was most likely located in the cytoplasm (kNN value, cyto: 4, plas: 4, chlo: 2, E.R.: 2, nucl: 1). To verify this prediction, we produced the fusion construct 35S-TaRPM1-GFP and the control vector 35S-GFP and transformed them into tobacco (Nicotiana benthamiana) cells. Fluorescence microscopy showed that the TaRPM1-GFP fusion protein was expressed in both the cytoplasm and the nucleus, similarly to the control vector 35S-GFP (Figure 8A). To confirm these results, we further conducted Western blot to analyze the stability of the TaRPM1-GFP fusion protein and found both GFP and the TaRPM1-GFP fusion protein expressed successfully (Figure 8B).
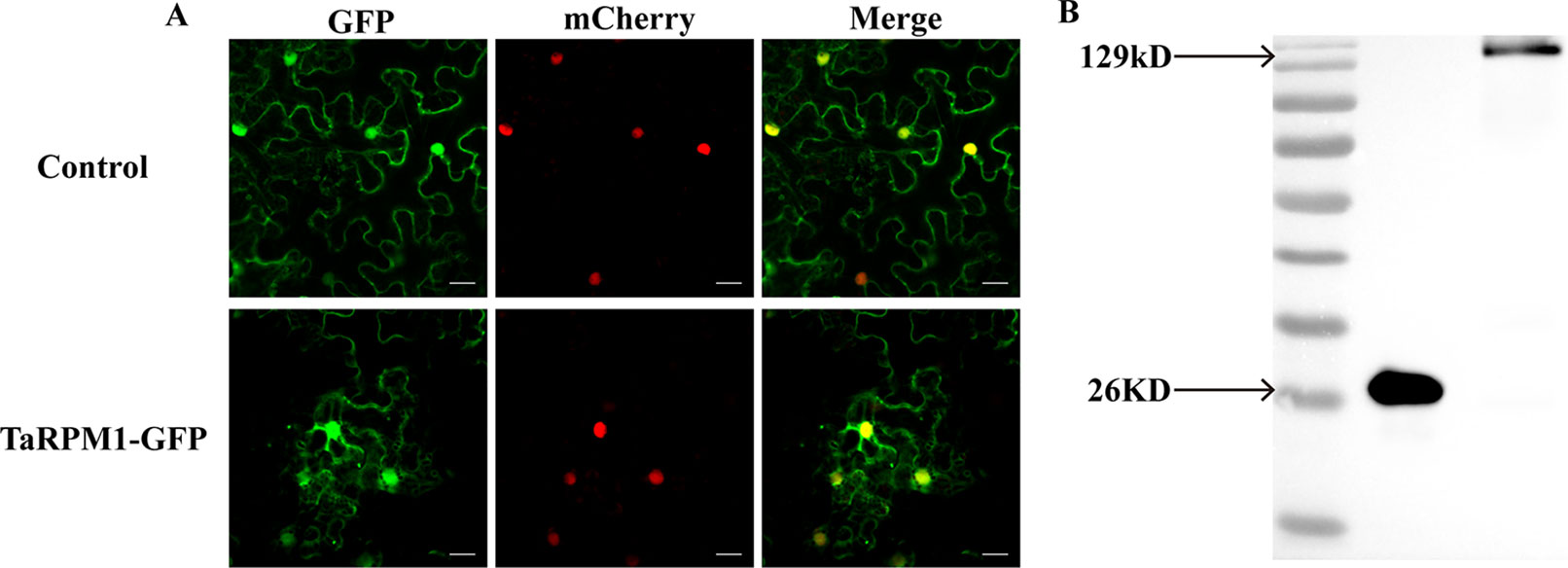
Figure 8 Subcellular localization of protein TaRPM1. (A) The TaRPM1-GFP fusion protein and green fluorescent protein (GFP) were separately expressed in Nicotiana benthamiana. The histone protein, AtH2B, was used as a nuclear location marker gene. Bar, 20 μm. (B) Western blot analysis of GFP and TaRPM1-GFP fusion protein.
Discussion
NBS-LRR genes, the largest class of R genes, have been reported to play important roles in initiating plant pathogen-triggered immunity (Tameling and Takken, 2008; Gao et al., 2010). As intracellular receptors, NBS-LRR proteins could bind pathogen effectors directly or indirectly through perceiving effector-induced modifications to other proteins, inducing host defense responses (Eitas and Dangl, 2010; Li et al., 2015). To date, numerous NBS-LRR genes have been reported to be involved in resistance to pathogens (Axtell and Staskawicz, 2003; Mackey et al., 2003; Shao et al., 2003; Cherkis et al., 2012; Jun et al., 2016; Macqueen et al., 2016; Zhu et al., 2017). However, functions of NBS-LRR genes in the wheat HTSP to Pst remain unknown. In the present study, we demonstrated that TaRPM1 is a candidate gene required for HTSP resistance to Pst. Furthermore, silencing TaRPM1 also affected Pst colony development as well as uredinium length. It may be, therefore, concluded that TaRPM1 positively contributes to the HTSP resistance to Pst in wheat cultivar XY 6.
There are differences in effects of temperature on various R gene–mediated resistance. Many defense responses activated by R genes can be suppressed at higher temperature. For instance, SNC1 (Yang and Hua, 2004), N (Wang et al., 2009), and RPS4-mediated resistance are inhibited at 28°C (Gassmann et al., 1999), whereas Rx gene against Potato virus X is inactive at 30°C (Bendahmane and Baulcombe, 1999). In contrast, Yr36 could induce HTAP resistance to Pst when wheat were grown above 25°C (Fu et al., 2009). Different from these R genes, a short period of exposure 20°C in the early stage of Pst development can activate TaRPM1-meditated resistance to Pst, but not at 15°C. These results showed differences in the temperature sensitivity of R protein activation. Plants with temperature-sensitive R genes may have evolved to balance the energies required for growth and defense when perception of a temperature change (Alcázar and Parker, 2011). For example, the temperature change could result in (i) reorganization of energy resources, which may be followed by reducing available nutrient to rust (Viola and Davies, 1994; Grof et al., 2010), or (ii) production of metabolites that may inhibit the fungal growth (Berger et al., 2007). Thus, we speculate that resistance changes induced by temperature may be through dynamic interactions between plants and the fungal pathogen.
P. syringae effectors AvrRpm1 and AvrB induce RIN4 phosphorylation, which is required for activation of RPM1-mediated plant defense responses (Mackey et al., 2002). In contrast to AtRPM1, the interaction between TaRPM1 and TaRIN4 was not detected using the yeast two-hybrid system in the present study (data not shown), indicating that the molecular mechanism of TaRPM1 involved in the HTSP resistance to Pst differs from AtRPM1-mediated resistance against P. syringae. Further investigation will shed light on whether TaRPM1 recognizes the effector(s) directly or indirectly with help from other proteins.
NBS-LRR proteins that are involved in the host defense responses may differ in their subcellular locations. RRS1-R (Bernoux et al., 2008), RPS4 (Wirthmueller et al., 2007), MLA10 (Shen et al., 2007), N (Caplan et al., 2008), and Rx1 (Slootweg et al., 2010) are located in the nucleus; host defense responses are critically dependent on the nuclear localization of these NBS-LRR proteins. In addition, plasma-membrane localization of Arabidopsis thaliana RPS5 and RPM1 (Boyes et al., 1998; Gao et al., 2011; Qi et al., 2012) is required for their functions. The present study showed that TaRPM1 is located in both nucleus and cytoplasm, differing from A. thaliana RPM1. Further research is needed to assess whether TaRPM1 nuclear and/or cytoplasm distribution is of critical importance for its function in the HTSP resistance to Pst.
Various host defense responses, including ROS production and programmed cell death, are activated upon perception of pathogens through NBS-LRR proteins (Andersson et al., 2006; Gao et al., 2013; Qi and Innes, 2013). Arabidopsis plants with the GhDSC1-overexpressing heterologously display strong resistance to Verticillium, with ROS accumulation abundantly and cell death (Li et al., 2019). In our study, silencing TaRPM1 has no effects on the number of necrotic cells for the NT treatment. In contrast, the number of necrotic cells in the HT-treated TaRPM1-silenced leaves significantly decreased compared with the HT-treated non-silenced leaves, indicating cell death induced by TaRPM1 was only promoted for the HT treatment. Moreover, number of necrotic cells in the HT-treated TaRPM1-silenced leaves was still more than that in the NT-treated TaRPM1-silenced leaves at 48, 72, and 120 hptt, suggesting that genes other than TaRPM1 might also be involved in regulating the HTSP to Pst.
Several studies have shown that hormones are involved in the NBS-LRR protein-mediated defense responses (Qi and Innes, 2013; Roberts et al., 2013). SA, an important plant defense signaling molecule against biotrophic pathogens, increases transcription of many PR proteins and systemic acquired resistance (SAR) (Liu et al., 2018). Numerous R gene–mediated resistance responses are related to the SA signaling pathway (Shirano et al., 2002). Treatment with SA in Arabidopsis leads to increased expression of R genes RPW8.1 and RPW8.2, and resistance to powdery mildew (Xiao et al., 2003). NBS-LRR R gene TaRGA is induced significantly by exogenous applications of SA, and silencing TaRGA leads to compromised resistance to wheat powdery mildew cause by Blumeria graminis f. sp. tritici and reduced expression of PR1 (Wang et al., 2016). In the present study, the highest induction of TaRPM1 occurred after the SA treatment for 0.5 h, and TaPR1 and TaPR2 (two marker genes of the SA pathway) had significantly lower expressions in the HT-treated TaRPM1-silenced plants than in non-silenced HT-treated plants, indicating that TaRPM1 plays a positive role in the HTSP resistance to Pst through the SA signaling pathway.
In summary, we demonstrated that NBS-LRR gene TaRPM1 positively contributes to the HTSP resistance to Pst in wheat cultivar XY 6. In addition, the present results suggested that TaRPM1 confers HTSP resistance to Pst though the SA signaling pathway. However, further research is needed to investigate whether TaRPM1 could recognize effector(s) and to ascertain the exact molecular mechanisms in the TaRPM1-mediated HTSP resistance to Pst.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
XH, HS, XC, and XX planned and designed the research. JiaW, WT, FT, and JinW performed the experiments. XH, JiaW, XC, and XX wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Xiaojie Wang (Northwest A&F University) for providing the BSMV vectors. This work was supported by grants from the National Key Research and Development Program of China (no. 2018YFD0200402, 2016YFD0300702) and the National Natural Science Foundation of China (no. 31271985 and 31972219).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01679/full#supplementary-material
Figure S1 | Sequence alignment of TaRPM1 and three copies in the wheat genomes. The blue and yellow boxes represent the primer locations for the VIGS1 and VIGS2 fragments of TaRPM1, respectively and the red box represents the primer locations for qRT-PCR of TaRPM1.
Table S1 | Primers used in this study.
References
Alcázar, R., Parker, J. E. (2011). The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 16, 666–675. doi: 10.1016/j.tplants.2011.09.001
Anderson, P. A., Lawrence, G. J., Morrish, B. C., Ayliffe, M. A., Finnegan, E. J., Ellis, J. G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell. 9, 641–651. doi: 10.2307/3870513
Andersson, M. X., Kourtchenko, O., Dangl, J. L., Mackey, D., Ellerström, M. (2006). Phospholipase-dependent signalling during the AvrRpm1- and AvrRpt2-induced disease resistance responses in Arabidopsis thaliana. Plant J. 47, 947–959. doi: 10.1111/j.1365-313X.2006.02844.x
Axtell, M. J., Staskawicz, B. J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. doi: 10.1016/s0092-8674(03)00036-9
Bendahmane, A., Baulcombe, K. D. C. (1999). The Rxhttps://www.frontiersin.org/Registration/Register.aspx gene from potato controls separate virus resistance and cell death responses. Plant Cell. 11, 781–791. doi: 10.1105/tpc.11.5.781
Berger, S., Sinha, A. K., Roitsch, T. (2007). Plant physiology meets phytopathology: plant primary metabolism and plant-pathogen interactions. J. Exp. Bot. 58, 4019–4026. doi: 10.1093/jxb/erm298
Bernoux, M., Timmers, T., Jauneau, A., Brière, C., Wit, P.J.G.M.d., Marco, Y., et al. (2008). RD19, an Arabidopsis cysteine protease required for RRS1-R: mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell. 20, 2252–2264. doi: 10.2307/25224327
Bhattarai, K., Louws, F. J., Williamson, J. D., Panthee, D. R. (2016). Differential response of tomato genotypes to Xanthomonas-specific pathogen-associated molecular patterns and correlation with bacterial spot (Xanthomonas perforans) resistance. Hortic. Res. 3, 16035. doi: 10.1038/hortres.2016.35
Botella, M. A., Parker, J. E., Frost, L. N., Bittner-Eddy, P. D., Beynon, J. L., Daniels, M. J., et al. (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell. 10, 1847–1860. doi: 10.2307/3870908
Boyes, D. C., Nam, J., Dangl, J. L. (1998). The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Plant Biol. 95, 15849–15854. doi: 10.2307/46468
Caplan, J., Padmanabhan, M., Dinesh-Kumar, S. P. (2008). Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host. Microbe 3, 126–135. doi: 10.1016/j.chom.2008.02.010
Chen, W. Q., Wu, L. R., Liu, T. G., Xu, S. C., Jin, S. L., Peng, Y. L., et al. (2009). Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust in China from 2003 to 2007. Plant Dis. 93, 1093–1101. doi: 10.1094/PDIS-93-11-1093
Chen, W. Q., Kang, Z. S., Ma, Z. H., Xu, S. C., Jin, S. L., Jiang, Y. Y. (2013). Integrated management of wheat stripe rust caused by Puccinia striiformis f. sp. tritici in China. Sci. Agr. Sin. 46, 4254–4262. doi: 10.3864/j.issn.0578-1752.2013.20.008
Chen, X. M. (2013). Review article: high-temperature adult-plant resistance, key for sustainable control of stripe rust. Am. J. Plant Sci. 4, 608–627. doi: 10.4236/ajps.2013.43080
Chen, X. M. (2014). Integration of cultivar resistance and fungicide application for control of wheat stripe rust. Can. J. Plant Pathol. 36, 16. doi: 10.1080/07060661.2014.924560
Cherkis, K. A., Temple, B. R. S., Eui-Hwan, C., John, S., Dangl, J. L. (2012). AvrRpm1 missense mutations weakly activate RPS2-mediated immune response in Arabidopsis thaliana. PloS One 7, e42633. doi: 10.1371/journal.pone.0042633
Cui, H., Tsuda, K., Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487. doi: 10.1146/annurev-arplant-050213-040012
Dangl, J. L., Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Eitas, T. K., Dangl, J. L. (2010). NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 13, 472–477. doi: 10.1016/j.pbi.2010.04.007
Ellis, J., Jones, D. (1998). Structure and function of proteins controlling strain-specific pathogen resistance in plants. Curr. Opin. Plant Biol. 1, 288–293. doi: 10.1016/1369-5266(88)80048-7
Ellis, J. G., Lawrence, G. J., Luck, J. E., Dodds, P. N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell. 11, 495–506. doi: 10.2307/3870876
Feuillet, C., Travella, S., Stein, N., Albar, L., Nublat, A., Keller, B. (2003). Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. P. Natl. Acad. Sci. U. S. A. 100, 15253–15258. doi: 10.1073/pnas.2435133100
Fu, D., Uauy, C., Distelfeld, A., Blechl, A., Epstein, L., Chen, X., et al. (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323, 1357–1360. doi: 10.1126/science.1166289
Gao, Y., Xu, Z., Jiao, F., Yu, H., Xiao, B., Li, Y., et al. (2010). Cloning, structural features, and expression analysis of resistance gene analogs in tobacco. Mol. Biol. Rep. 37, 345–354. doi: 10.1007/s11033-009-9749-2
Gao, X., Britt, R. C., Jr., Shan, L., He, P. (2011). Agrobacterium-mediated virus-induced gene silencing assay in cotton. J. Vis. Exp. 54, 2938. doi: 10.3791/2938
Gao, X., Chen, X., Lin, W., Chen, S., Lu, D., Niu, Y., et al. (2013). Bifurcation of Arabidopsis NLR immune signaling via Ca²⁺-dependent protein kinases. PloS Pathog. 9, e1003127. doi: 10.1371/journal.ppat.1003127
Gao, Y., Wu, Y., Du, J., Zhan, Y., Sun, D., Zhao, J., et al. (2017). Both light-induced SA accumulation and ETI mediators contribute to the cell death regulated by BAK1 and BKK1. Front. Plant Sci. 8, 622. doi: 10.3389/fpls.2017.00622
Gassmann, W., Hinsch, M. E., Staskawicz, B. J. (1999). The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 20, 265–277. doi: 10.1046/j.1365-313X.1999.00600.x
Gopalan, S., Bauer, D. W., Alfano, J. R., Loniello, A. O., He, S. Y., Collmer, A. (1996). Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell. 8, 1095–1105. doi: 10.1385/ABAB:105:1-3:53
Grof, C. P. L., Campbell, J. A., Kravchuk, O., Lambrides, C. J., Albertson, P. L. (2010). Temperature effect on carbon partitioning in two commercial cultivars of sugarcane. Funct. Plant Biol. 37, 334–341. doi: 10.1071/FP09216
Gururani, M. A., Venkatesh, J., Upadhyaya, C. P., Nookaraju, A., Pandey, S. K., Park, S. W. (2012). Plant disease resistance genes: current status and future directions. Physiol. Mol. Plant Pathol. 78, 51–65. doi: 10.1016/j.pmpp.2012.01.002
He, H. G., Zhu, S. Y., Zhao, R. H., Jiang, Z. N., Ji, Y. Y., Ji, J., et al. (2018). Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant 11, 879–882. doi: 10.1016/j.molp.2018.03.004
Hinsch, M., Staskawicz, B. (1996). Identification of a new Arabidopsis disease resistance locus, RPs4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol. Plant Microbe Interact. 9, 55–61. doi: 10.1094/MPMI-9-0055
Hu, X. P., Wang, B. T., Kang, Z. S. (2014). Research progress on virulence variation of Puccinia striiformis f. sp. tritici in China. J. Triticeae Crops. 35, 709–716. doi: 10.2478/s13545-014-0139-6
Hwang, C. F., Bhakta, A. V., Truesdell, G. M., Pudlo, W. M., Williamson, V. M. (2000). Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell. 12, 1319–1329. doi: 10.2307/3871132
Jablonska, B., Ammiraju, J. S. S., Bhattarai, K. K., Mantelin, S., Ilarduya, O.M.d., Roberts, P. A., et al. (2007). The Mi-9 Gene from Solanum arcanum conferring heat-stable resistance to root-knot nematodes is a homolog of Mi-1. Plant Physiol. 143, 1044–1054. doi: 10.2307/40065453
Jiang, N., Cui, J., Meng, J., Luan, Y. (2018). A tomato NBS-LRR gene is positively involved in plant resistance to Phytophthora infestans. Phytopathology 8, 108. doi: 10.1094/PHYTO-12-17-0389-R
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jun, Y., Qing, M., Yan, Z., Xingfen, W., Guiyin, Z., Zhiying, M. (2015). Molecular cloning and functional analysis of GbRVd, a gene in Gossypium barbadense that plays an important role in conferring resistance to Verticillium wilt. Gene 575, 687–694. doi: 10.1016/j.gene.2015.09.046
Kandoth, P. K., Mitchum, M. G. (2013). War of the worms: how plants fight underground attacks. Curr. Opin. Plant Biol. 16, 457–463. doi: 10.1016/j.pbi.2013.07.001
Klymiuk, V., Yaniv, E., Huang, L., Raats, D., Fatiukha, A., Chen, S., et al. (2018). Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat. Commun. 9, 3735. doi: 10.1038/s41467-018-06138-9
Lawrence, G. J., Finnegan, E. J., Ayliffe, M. A., Ellis, J. G. (1995). The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell. 7, 1195. doi: 10.2307/3870095
Li, X., Kapos, P., Zhang, Y. (2015). NLRs in plants. Curr. Opin. Immunol. 32, 114–121. doi: 10.1016/j.coi.2015.01.014
Li, N. Y., Ma, X. F., Short, D., Li, T. G., Zhou, L., Gui, Y. J., et al. (2017). The island cotton NBS-LRR gene GbaNA1 confers resistance to the non-race 1 Verticillium dahliae isolate Vd991. Mol. Plant Pathol. 19, 1466–1479. doi: 10.1111/mpp.12630
Li, X., Zhang, Y., Yin, L., Lu, J. (2017). Overexpression of pathogen-induced grapevine TIR-NB-LRR gene VaRGA1 enhances disease resistance and drought and salt tolerance in Nicotiana benthamian. Protoplasma 254, 957–969. doi: 10.1007/s00709-016-1005-8
Li, T. G., Wang, B. L., Yin, C. M., Zhang, D. D., Wang, D., Song, J., et al. (2019). The Gossypium hirsutum TIR-NBS-LRR gene GhDSC1 mediates resistance against Verticillium wilt. Mol. Plant Pathol. 20, 857–876. doi: 10.1111/mpp.12797
Liang, W., Chen, H., Curtis, C., Zheng, Q. F. (2014). Go in for the kill: How plants deploy effector-triggered immunity to combat pathogens. Virulence 5, 12. doi: 10.4161/viru.29755
Liu, P., Yinghui, D., Cong, L., Qinghe, X., Jia, G., Tuo, Q., et al. (2018). The calcium sensor TaCBL4 and its interacting protein TaCIPK5 are required for wheat resistance to stripe rust fungus. J. Exp. Bot. 12, 69. doi: 10.1093/jxb/ery307
Mackey, D., Holf, B.F.III., Wiig, A., Dangl, J. L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 0–754. doi: 10.1016/s0092-8674(02)00661-x
Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J. R., Dangl, J. L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector avrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. doi: 10.1016/s0092-8674(03)00040-0
Macqueen, A., Sun, X., Bergelson, J. (2016). Genetic architecture and pleiotropy shape costs of Rps2-mediated resistance in Arabidopsis thaliana. Nat. Plants 2, 16110–16110. doi: 10.1038/nplants.2016.110
Parker, J. E., Coleman, M. J., Szabò, V., Frost, L. N., Schmidt, R., Biezen, E. A., et al. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell. 9, 879–894. doi: 10.1105/tpc.9.6.879
Qi, D., Innes, R. W. (2013). Recent advances in plant NLR structure, function, localization, and signaling. Front. Immuno. 4, 348. doi: 10.3389/fimmu.2013.00348
Qi, D., Deyoung, B. J., Innes, R. W. (2012). Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol. 158, 1819–1832. doi: 10.2307/41496322
Qian, L. H., Zhou, G. C., Sun, X. Q., Lei, Z., Zhang, Y. M., Xue, J. Y., et al. (2017). Distinct patterns of gene gain and loss: diverse evolutionary modes of NBS-encoding genes in three Solanaceae crop species. G3. (Bethesda) 7, 1577–1585. doi: 10.1534/g3.117.040485
Roberts, M., Tang, S., Stallmann, A., Dangl, J. L., Bonardi, V. (2013). Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PloS Genet. 9, e1003465. doi: 10.1371/journal.pgen.1003465
Shao, F., Catherine, G., Jules, A., Mark, S., Jack, E. D., Roger, W. I. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301, 1230–1233. doi: 10.1126/science.1085671
Shen, Q. H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., et al. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103. doi: 10.1126/science.1136372
Shirano, Y., Kachroo, P., Shah, J., Klessig, D. F. (2002). A gain-of-function mutation in an Arabidopsis toll interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell. 14, 3149–3162. doi: 10.1105/tpc.005348
Slootweg, E., Roosien, J., Spiridon, L. N., Petrescu, A. J., Tameling, W., Joosten, M., et al. (2010). Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell. 22, 4195–4215. doi: 10.2307/41059420
Tör, M., Brown, D., Cooper, A., Woods-Tör, A., Sjölander, K., Jones, J. D. G., et al. (2004). Arabidopsis downy mildew resistance gene RPP27 encodes a receptor-like protein similar to CLAVATA2 and tomato Cf-9. Plant Physiol. 135, 1100–1112. doi: 10.2307/4281828
Tameling, W. I. L., Takken, F. L. W. (2008). Resistance proteins: scouts of the plant innate immune system. Eur. J. Plant Pathol. 121, 243–255. doi: 10.1007/s10658-007-9187-8
Tao, F., Wang, J. J., Guo, Z. F., Hu, J. J., Xu, X. M., Yang, J. R., et al. (2018). Transcriptomic analysis reveal the molecular mechanisms of wheat higher-temperature seedling-plant resistance to Puccinia striiformis f. sp. tritici. Front. Plant Sci. 9, 240. doi: 10.3389/fpls.2018.00240
Viola, R., Davies, H. V. (1994). Effect of temperature on pathways of carbohydrate metabolism in tubers of potato (Solanum-tuberosum L). Plant Sci. 103, 135–143. doi: 10.1016/0168-9452(94)90201-1
Wang, C. F., Huang, L. L., Buchenauer, H., Han, Q. M., Zhang, H. C., Kang, Z. S. (2007). Histochemical studies on the accumulation of reactive oxygen species (O2- and H2O2) in the incompatible and compatible interaction of wheat-Puccinia striiformis f. sp. tritici. Physiol. Mol. Plant Pathol. 71, 230–239. doi: 10.1016/j.pmpp.2008.02.006
Wang, Y., Bao, Z., Zhu, Y., Hua, J. (2009). Analysis of temperature modulation of plant defense against biotrophic microbes. Mol. Plant Microbe Interact. 22, 498–506. doi: 10.1094/MPMI-22-5-0498
Wang, J. J., Tao, F., An, F., Xu, X. M., Hu, X. P. (2014). Selection of reference genes in wheat stressed by temperature and Puccinia striiformis f. sp. tritici. Acta Phytopathol. Sin. 44, 497–503. doi: 10.13926/j.cnki.apps.2014.05.007
Wang, D., Wang, X., Mei, Y., Dong, H. (2016). The wheat homolog of putative nucleotide-binding site–leucine-rich repeat resistance gene TaRGA contributes to resistance against powdery mildew. Funct. Integr. Genomics 16, 1–12. doi: 10.1007/s10142-015-0471-y
Wang, J. J., Tao, F., An, F., Zou, Y. P., Tian, W., Chen, X. M., et al. (2017a). Wheat transcription factor TaWRKY70 is positively involved in high-temperature seedling plant resistance to Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 18, 649–661. doi: 10.1111/mpp.12425
Wang, J. J., Tao, F., Tian, W., Guo, Z. F., Chen, X. M., Xu, X. M., et al. (2017b). The wheat WRKY transcription factors TaWRKY49 and TaWRKY62 confer differential high-temperature seedling-plant resistance to Puccinia striiformis f. sp. tritici. PloS One 12, e0181963. doi: 10.1371/journal.pone.0181963
Wang, J. H., Wang, J. J., Shang, H. S., Chen, X. M., Xu, X. M., Hu, X. P. (2019). TaXa21, a LRR-rich receptor like kinase associated with TaWRKY76 and TaWRKY62, plays positive roles in wheat high-temperature seedling plant resistance to Puccinia striiformis f. sp. tritici. Mol. Plant Microbe In. 32, 1526–1535. doi: 10.1094/MPMI-05-19-0137-R
Warren, R. F., Henk, A., Mowery, P., Holub, E., Innes, R. W. (1998). A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell. 10, 1439–1452. doi: 10.1105/tpc.10.9.1439
Wellings, C. R. (2011). Global status of stripe rust: a review of historical and current threats. Euphytica 179, 129–141. doi: 10.1007/s10681-011-0360-y
Wirthmueller, L., Zhang, Y., Jones, J. D., Parker, J. E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17, 2023–2029. doi: 10.1016/j.cub.2007.10.042
Xiao, S., Brown, S., Patrick, E., Brearley, C., Turner, J. G. (2003). Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell. 15, 33–45. doi: 10.1105/tpc.006940
Xu, Y., Liu, F., Zhu, S., Li, X. (2018). The maize NBS-LRR gene ZmNBS25 enhances disease resistance in rice and Arabidopsis. Front. Plant Sci. 9, 1033. doi: 10.3389/fpls.2018.01033
Yang, S., Hua, J. (2004). A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell. 16, 1060–1071. doi: 10.1105/tpc.020479
Zhou, X. L., Wang, M. N., Chen, X. M., Lu, Y., Kang, Z. S., Jing, J. X. (2014). Identification of Yr59 conferring high-temperature adult-plant resistance to stripe rust in wheat germplasm PI 178759. Theor. Appl. Genet. 127, 935–945. doi: 10.1007/s00122-014-2269-z
Keywords: wheat stripe rust, high-temperature seeding plant resistance (HTSP), non-species-specific, virus-induced gene silencing (VIGS), NBS-LRR
Citation: Wang J, Tian W, Tao F, Wang J, Shang H, Chen X, Xu X and Hu X (2020) TaRPM1 Positively Regulates Wheat High-Temperature Seedling-Plant Resistance to Puccinia striiformis f. sp. tritici. Front. Plant Sci. 10:1679. doi: 10.3389/fpls.2019.01679
Received: 15 September 2019; Accepted: 28 November 2019;
Published: 15 January 2020.
Edited by:
Sébastien Duplessis, INRA Centre Nancy-Lorraine, FranceReviewed by:
Clemence Marchal, John Innes Centre (JIC), United KingdomWolfgang Moeder, University of Toronto, Canada
Copyright © 2020 Wang, Tian, Tao, Wang, Shang, Chen, Xu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoping Hu, eHBodUBud3N1YWYuZWR1LmNu
 Jiahui Wang1
Jiahui Wang1 Fei Tao
Fei Tao Jingjing Wang
Jingjing Wang Xianming Chen
Xianming Chen Xiangming Xu
Xiangming Xu Xiaoping Hu
Xiaoping Hu