
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 17 December 2019
Sec. Plant Development and EvoDevo
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01625
Flowering is a critical stage of plant development and is closely correlated with seed production and crop yield. Flowering transition is regulated by complex genetic networks in response to endogenous and environmental signals. FLOWERING LOCUS C (FLC) is a central repressor in the flowering transition of Arabidopsis thaliana. The regulation of FLC expression is well studied at transcriptional and post-transcriptional levels. A subset of antisense transcripts from FLC locus, collectively termed cold-induced long antisense intragenic RNAs (COOLAIR), repress FLC expression under cold exposure. Recent studies have provided important insights into the alternative splicing of COOLAIR and FLC sense transcripts in response to developmental and environmental cues. Herein, at the 20th anniversary of FLC functional identification, we summarise new research advances in the alternative splicing of FLC sense and antisense transcripts that regulates flowering.
FLOWERING LOCUS C (FLC) is a key repressor in flowering transition. The alternative splicing of FLC sense and antisense transcripts regulated by external and internal cues modulates flowering transition.
RNA splicing is a critical step in the post-transcriptional regulation of gene expression. This process occurs by removing intronic sequences and joining exons by spliceosome and numerous splicing factors (Jurica and Moore, 2003; Wahl et al., 2009; Matera and Wang, 2014; Vosseberg and Snel, 2017). Spliceosome is a highly dynamic ribonucleoprotein complex that catalyses RNA splicing and is composed of five small nuclear ribonucleoprotein (snRNP) particles (U1, U2, U4/U6 and U5) (Will and Lührmann, 2011; Fica et al., 2019). Splicing factors are one of the key determinants as accessory non-snRNP proteins regulating RNA splicing (Cho et al., 2011; Liu et al., 2016; Long et al., 2019; Wang et al., 2019; Xiong et al., 2019a; Xiong et al., 2019b). A pre-mRNA may undergo different splicing patterns, creating various mature transcripts that encode distinct functional proteins (Samach et al., 2011; Wang et al., 2015; Zhu et al., 2017; Lockhart, 2018; Okumoto et al., 2018). This phenomenon is called alternative splicing (AS).
AS fulfils important biological functions in plants, such as flowering transition and flower development (Wang et al., 2014; Melzer, 2017; Rodríguez-Cazorla et al., 2018; Park et al., 2019a; Park et al., 2019b; Wang et al., 2019). AS is also implicated in plant response to circadian rhythm regulation (Filichkin and Mockler, 2012), phytohormone (Hrtyan et al., 2015; Wang et al., 2015; Zhu et al., 2017; Xiong et al., 2019a), ambient temperature (Verhage et al., 2017) and abiotic and biotic stresses (Lyons and Kazan, 2016; Huang et al., 2017; Mei et al., 2017; Shang et al., 2017; Laloum et al., 2018). These cues are all important for flowering transition; therefore, AS plays multiple roles in flowering by integrating endogenous developmental and exogenous environmental signals. The flowering inhibitor gene flowering locus c (FLC) encodes a MADS-box transcription factor and is a key regulator of vernalisation and autonomous pathways in Arabidopsis and related species (Michaels and Amasino, 1999; Sheldon et al., 1999; Sheldon et al., 2000; Chen et al., 2019; Coupland, 2019). FLC inhibits flowering by repressing the expression of a subset of key genes in promoting flowering, such as flowering locus t (FT), suppressor of overexpression of constans 1 (SOC1) and target of flc and svp1 (TFS1) (Helliwell et al., 2006; Searle et al., 2006; Luo et al., 2019; Richter et al., 2019). Thus, FLC regulation is central for flowering at the transcriptional, post-transcriptional and post-translational levels (Michaels et al., 2003; Lempe et al., 2005; Li et al., 2016; Kwak et al., 2017; Whittaker and Dean, 2017; Xiong et al., 2019a). The AS of FLC sense and antisense transcripts is required for flowering transition in Arabidopsis and other dicots (Michaels and Amasino, 1999; Sheldon et al., 1999; Helliwell et al., 2006; Yuan et al., 2009; Wu et al., 2012).
Twenty years ago, the works of Michaels and Amasino (1999), and Sheldon et al. (1999) provided a first glimpse of the central functions of FLC in flowering and the molecular basis of FLC in vernalisation (Coupland, 2019). Subsequent research works have demonstrated the regulation of FLC at the transcriptional and post-transcriptional levels, especially the epigenetic silencing of FLC by histone methylation in vernalisation, and the splicing regulation of FLC sense and antisense transcripts by splice factors (Bastow et al., 2004; Liu et al., 2007; Liu et al., 2010; Marquardt et al., 2014). In this review, we describe the current understanding of the AS of FLC sense and antisense transcripts in modulating flowering time and the splice factors involved in these processes.
Arabidopsis accessions exhibit markedly different flowering behaviour from different environments. The AS of FLC sense transcripts in these accessions generate multiple splice isoforms (Figure 1A). The naturally occurring splice variants of FLC are related to different vernalisation responses of various Arabidopsis accessions (Bloomer and Dean, 2017). Arabidopsis ecotype Bur-0 is late flowering and vernalisation insensitive. FLC cDNA from Bur-0 contains 64 bp of intron sequence immediately upstream of exon 7, causing a mutation at the final position of intron 6. The 64 bp intron retention causes a frame shift and a premature stop codon in FLC cDNA. Thus, the encoded FLC lacking the C-terminal 33 amino acid residues is a null function protein in Bur-0 (Werner et al., 2005). Similarly, in variations of Cen-0 and Cal-0, alternative splice acceptor sites in the last exon and last intron are used, respectively. These aberrant splicing forms all lead to a frame shift of cDNA sequences and severely compromise protein function (Lempe et al., 2005). In Col-0 Arabidopsis, there are several additional splicing variants from FLC locus besides the canonical transcript (Severing et al., 2012; Park et al., 2019a); however, their roles in flowering transition remains to be investigated. An additional shorter transcript from FLC locus is induced after vernalisation treatment for 15 days in Est-0 and Le-0 ecotypes, which is not observed at normal temperature (Caicedo et al., 2004). Whether this short transcript is involved in vernalisation-induced flowering in Arabidopsis is unknown.
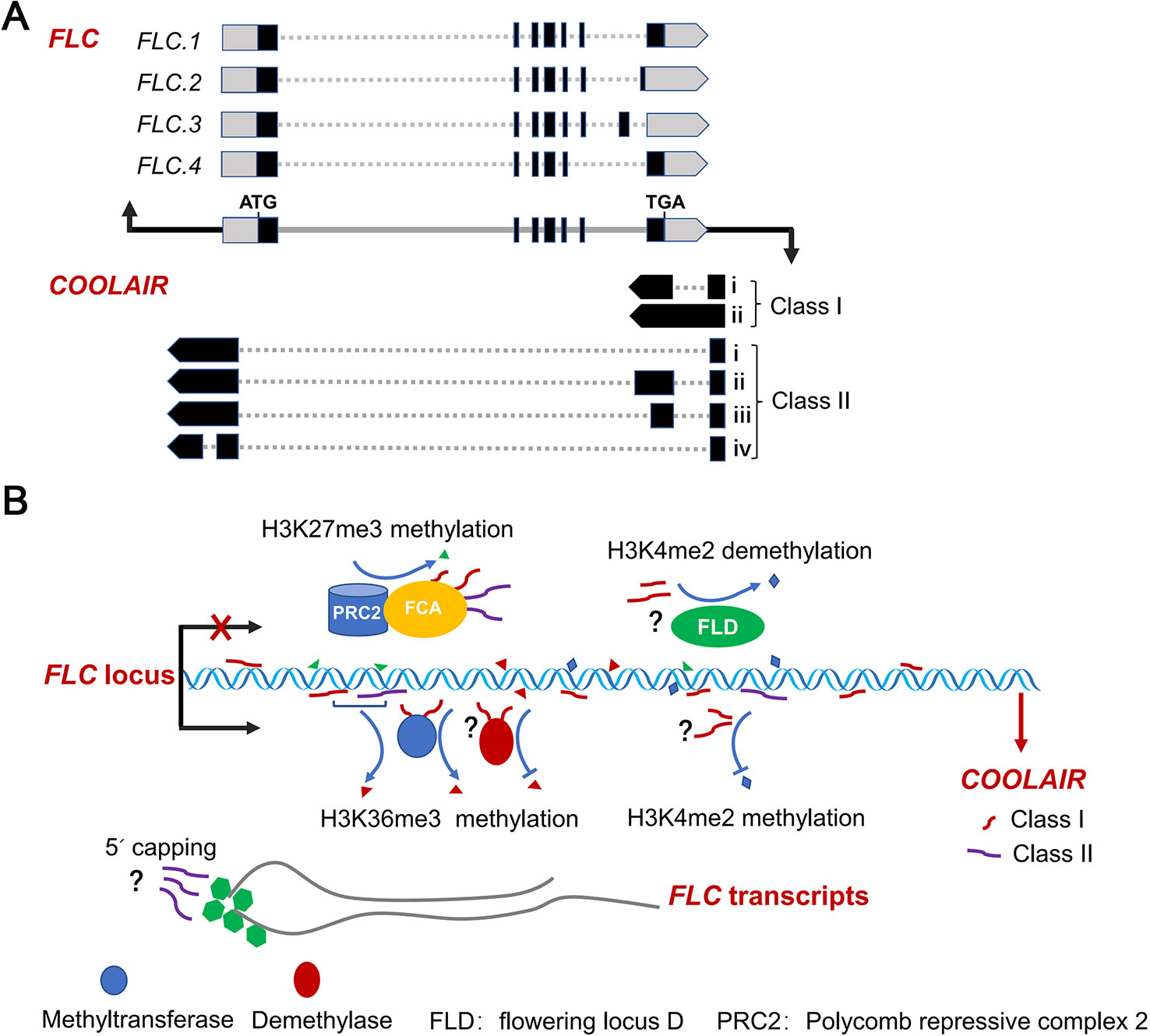
Figure 1 Splicing patterns of FLC sense and antisense transcripts, and AS of FLC antisense transcripts regulating FLC expression. (A) Schematic diagram of the splicing patterns of FLC sense and antisense transcripts at FLC locus in Arabidopsis. Black rectangles, gray dash lines and boxes indicate exons, introns and non-translated regions of FLC RNAs, respectively. Gray and black lines represent the genes and promoter regions of FLC and COOLAIR loci, respectively. (B) AS variants of COOLAIR transcripts effect FLC expression. Nascent COOLAIR RNAs are physically associated with the FLC locus to regulate the switching of chromatin states (Csorba et al., 2014; Li et al., 2015; Rosa et al., 2016). Two classes of COOLAIR transcripts bind to FCA and recruit PRC2 complex to FLC locus (Tian et al., 2019). The class I variants with proximal polyadenylation and the proximal–distal ratio of COOLAIR transcripts effect the histone methylation of H3K4me2, H3K36me3 and H3K27me3 (Marquardt et al., 2014; Berry and Dean, 2015; Xiao et al., 2015; Rosa et al., 2016). The class II COOLAIR transcripts with distal polyadenylation influence the degree of capping of the FLC nascent transcripts (Li et al., 2015).
In other plant species, the naturally occurring splice variants of the FLC locus also reveal that FLC AS is important for the control of flowering time. For example, four different transcripts of the FLC homologs in Brassica rapa BrFLC1 and BrFLC2 have been identified from naturally occurring splicing mutations (Yuan et al., 2009; Zhao et al., 2010; Wu et al., 2012). These different splice types of BrFLCs are significantly associated with a natural variation of flowering time in different germplasms of B. rapa. Additionally, differential splicing variants of BnFLC.A3b, a FLC homolog in B. napus, have been observed in leaves at the seedling stage between winter cultivar Tapidor and semi-winter cultivar Ningyou7. The transcripts from Tapidor are usually spliced canonically, but numerous incompletely spliced transcripts have been identified in Ningyou7, causing decreased functional transcripts (Zou et al., 2012). In tetraploid Capsella bursa-pastoris, splice site polymorphisms in the FLC loci create different transcripts which are nonfunctional (Slotte et al., 2009). These findings partially explain the differential flowering times of C. bursa-pastoris from different districts. A natural splicing site mutation in the BrpFLC1 gene causes early flowering in the cultivated variety of purple flowering stalk (Bras. campestris L. ssp. chinensis var. purpurea) compared with that of pakchoi (Bras. campestris ssp. chinensis var. communis) (Hu et al., 2011). Additionally, in an early flowering trifoliate orange (Poncirus trifoliata) mutant, five alternatively spliced transcripts of PtFLC have been identified; furthermore, their abundances are variable at the juvenile and adult stages, suggesting that the AS of PtFLC is related to flowering time (Zhang et al., 2009). Therefore, the AS of FLC is a target of natural selection for flowering regulation under natural conditions.
COOLAIR is a set of long noncoding RNAs expressed at the FLC locus in the antisense direction that was first identified in Arabidopsis (Liu et al., 2007; Swiezewski et al., 2007; Swiezewski et al., 2009; Ietswaart et al., 2012). COOLAIR RNAs functionally repress FLC sense expression at an early stage of cold exposure (vernalisation) via different ways, such as by directly associating with FLC chromatin or affecting H3K36me3 and H3K27me3 levels (Csorba et al., 2014; Berry and Dean, 2015; Rosa et al., 2016; Pajoro et al., 2017; Whittaker and Dean, 2017; Tian et al., 2019) (Figure 1B). The AS of antisense transcripts generated from the FLC locus produces two main classes of COOLAIR isoforms, terminating at proximal (sense intron 6, class I) and distal (sense promoter, class II) sites of FLC locus (Liu et al., 2007; Swiezewski et al., 2009; Marquardt et al., 2014). Each class contains several subclasses produced by AS associated with the usage of alternative 3′polyadenylation in spliced variants (Figure 1A). These multiple spliced variants with alternative polyadenylation are linked to different FLC expression states. Class I COOLAIR RNAs with proximal polyadenylation are associated with FLC repression (Liu et al., 2007; Swiezewski et al., 2009; Liu et al., 2010; Hornyik et al., 2010; Csorba et al., 2014). By contrast, class II COOLAIR transcripts are associated with high FLC expression levels. These COOLAIR RNAs with distal polyadenylation affect the capping of FLC nascent transcripts (Li et al., 2015).
The efficient splicing of class I intron promotes proximal polyadenylation in the antisense transcripts of FLC. As a result, this proximal polyadenylation inhibits the transcription of FLC through triggering H3K4me2 demethylation in the FLC locus (Marquardt et al., 2014). A few 3′-end processing factors containing RNA-recognition motifs, such as flowering control locus a (FCA) and FPA, glycine-rich rna-binding protein7 (GRP7), cleavage stimulation factor 64 (CstF64), CstF77, and nuclear speckle RNA binding protein a (NSRa), affect the splice site selection and polyadenylation site usage of COOLAIR transcripts, leading to an altered ratio of proximal–distal spliced variants (Liu et al., 2007; Streitner et al., 2008; Liu et al., 2010; Hornyik et al., 2010; Streitner et al., 2012; Xiao et al., 2015; Bazin et al., 2018). Loss-of-function mutations in these factors decrease or increase the usage of COOLAIR proximal polyadenylation site, leading to upregulated or downregulated FLC transcription.
The AS of COOLAIR is altered by environmental conditions and natural intronic polymorphisms. Cold exposure influences COOLAIR splicing (Swiezewski et al., 2009). For example, class I COOLAIR RNAs increase more rapidly than do class II ones during vernalisation (Csorba et al., 2014; Eom et al., 2018). Arabidopsis accessions show variable COOLAIR splicing patterns that affect FLC expression and flowering time. A single nucleotide polymorphism (SNP) specifically regulates COOLAIR AS (Li et al., 2015). This SNP is located next to the acceptor splice site of the intron of class IIi COOLAIR. In later flowering accessions, such as Var2-6 and Eden-1, this SNP reduces use of the splice acceptor site of the class IIi COOLAIR intron, leading to a shift to a downstream distal splice acceptor site and the inclusion of an internal exon. This change of splicing site produces isoforms with altered secondary structure and upregulates FLC expression (Li et al., 2015; Hawkes et al., 2016).
Splice sites, including 5′ donor splice site, branch point site, polypyrimidine tract and 3′ acceptor splice site, in pre-mRNA introns are precisely recognized by some splicing factors. Numerous splicing factors, such as BRR2, SC35 and SC35-like (SCL), RZ-1B and RZ-1C, are involved in the splicing of FLC in Arabidopsis flowering transition (Table 1). BRR2, an ATP-dependent RNA helicase, is an integral component of the U5 snRNP that is required for the activation of the spliceosome complex (Raghunathan and Guthrie, 1998). A missense mutation in Arabidopsis BRR2a results in defective FLC splicing and reduced FLC transcript levels (Mahrez et al., 2016). SC35 is a serine/arginine-rich (SR) protein that functions in the selection of splice sites (Valcárcel and Green, 1996). SC35 and SC35-like (SCL) proteins in Arabidopsis simultaneously modulate the splicing and transcription of FLC (Yan et al., 2017). Arabidopsis RZ-1B and RZ-1C, two heterogeneous nuclear ribonucleoproteins (hnRNPs), regulate FLC splicing and transcription by directly interacting with the SR protein (Wu et al., 2016). Interestingly, the retention of FLC introns 1, 5 and 6 in the brr2a mutant increases and the splicing efficiency of FLC intron 1 decreases in rz-1b rz-1c double mutants. These findings suggest that BRR2a, RZ-1B and RZ-1C promote the splicing of FLC introns. By contrast, the splicing efficiency of intron 1 in FLC increases compared with that in wild-type seedlings in quintuple mutants of SC35 and SCL genes, indicating that SC35 and SCL proteins inhibit the splicing of the first intron of FLC. The splicing efficiency of FLC introns is also inhibited by KHZ1 and KHZ2, two RNA-binding proteins containing CCCH zinc-finger and K homology (KH) domain (Yan et al., 2019).
U2 auxiliary factor (U2AF) regulates flowering via modulating FLC splicing. U2AF65, a large subunit of U2AF in mammalians, recognises and binds to the 3′ polypyrimidine tract of introns (Wang et al., 2008; Shao et al., 2014). The binding site of U2AF65 with RNA is regulated and shifted in noncanonical introns (Shao et al., 2014; Howardet al., 2018). The genes AtU2AF65a and AtU2AF65b encode the U2AF large subunit in Arabidopsis (Jang et al., 2014). AtU2AF65b plays roles in regulating flowering transition by splicing the introns 1 and 6 of FLC (Xiong et al., 2019a) (Table 1). AtU2AF65b expression is responsive to ABA, by which AtU2AF65b is involved in ABA-regulated flowering. AtU2AF65a is also implicated in FLC splicing (Park et al., 2019a). Strikingly, the loss-of-function mutants of AtU2AF65a and AtU2AF65b display opposite flowering phenotypes. Mutations in atu2af65a cause late flowering, whereas AtU2AF65b mutants exhibit early flowering (Park et al., 2019a; Xiong et al., 2019a). The differences in the noncanonical splicing variants between atu2af65a and atu2af65b null mutants (Park et al., 2019a) indicate that AtU2AF65a and AtU2AF65b might recognise different FLC introns.
In addition, some proteins play roles in RNA splicing by interaction with splicing factors. For example, the mRNA cap-binding complex (CBC) is involved in modulating pre-mRNA splicing activities via interaction with the splicing factors that recognise the 5′ splice site of the cap proximal intron (Izaurralde et al., 1994; Lewis et al., 1996). The CBP80/ABA Hypersensitive 1 (ABH1) and CBP20 are the large and small subunits of CBC protein complex in Arabidopsis, respectively (Hugouvieux et al., 2001). Knockout mutants of ABH1/CBP80 and CBP20 showing early-flowering phenotypes result from the defective splicing of FLC introns, especially the large first intron (Kuhn et al., 2007) (Table 1). In the abh1 knockout mutant, the most prominent products are the splice intermediates containing the first intron, causing the downregulation of FLC transcript and the early-flowering phenotypes (Kuhn et al., 2007). Similarly, CBP20 null mutation also results in increased unspliced–spliced ratio of FLC introns and low FLC mRNA levels (Geraldo et al., 2009).
To date, little is known about the splicing regulation of COOLAIR transcripts mediated by splicing factors. Only one splice factor, PRP8, a core spliceosome component, is found to function in modulating COOLAIR RNAs splicing (Marquardt et al., 2014) (Table 1). PRP8 is specifically required for the splicing of antisense transcripts COOLAIR but not for that of FLC sense transcripts. Single-base mutations of PRP8 reduce the splicing efficiency of COOLAIR introns, especially class Ii introns. The decreased splicing efficiency of COOLAIR class Ii reduces proximal poly(A) site usage, leading to increased H3K4me2 and the transcriptional upregulation of FLC expression (Liu et al., 2010; Marquardt et al., 2014). NSRa, as a nuclear speckle RNA binding protein, modules flowering time through regulation of the COOLAIR AS (Table 1). Only distal variants are decrease in the nsra mutant. This change in relative variant usage of proximal-distal RNAs leads to a down-regulation of FLC mRNA and an early flowering phenotype of nsra mutant (Bazin et al., 2018).
FLC is a key inhibitor in flowering transition in Arabidopsis and other dicots (Michaels and Amasino, 1999; Sheldon et al., 1999; Helliwell et al., 2006; Yuan et al., 2009; Wu et al., 2012). Therefore, the regulation of FLC is central to the transition to flowering in these plants. The AS of FLC sense and antisense transcripts is a critical step for FLC expression regulation. In Arabidopsis, multiple spliced variants of FLC sense and antisense transcripts have been determined (Kuhn et al., 2007; Severing et al., 2012; Li et al., 2015; Hawkes et al., 2016; Park et al., 2019a); however, we know little about functions in vivo of these spliced isoforms, especially, the COOLAIR-mediated regulation of switching of chromatin states at FLC and processing of FLC sense transcripts (Figure 1B). The further investigation will focus on how regulation of COOLAIR AS is linked to FLC chromatin modifications in response to external and internal influences.
Alternative splicing is regulated by splicing factors; however, only few splicing factors have been identified to be involved in the AS of FLC, especially those in the AS of COOLAIR transcripts. The identification of splicing factors and their functions is important for understanding the AS regulation of FLC sense and antisense transcripts in flowering transition. Additionally, the patterns of FLC AS is altered in response to environmental and signal stimuli (Swiezewski et al., 2009; Hornyik et al., 2010; Liu et al., 2010; Xiong et al., 2019a). Thus, the mechanisms by which the activities of splicing factors are regulated in response to external and internal cues must be investigated to study the AS of FLC sense and antisense transcripts in flowering regulation.
In agriculture, flowering is a prerequisite for crop production. Changes in the splicing patterns of FLC sense and antisense transcripts have enabled adaptation in response to changing environment for Arabidopsis accessions. Moreover, cold-induced sense and antisense RNAs of FLC are evolutionarily conserved in Arabidopsis perennial relatives and sugar beet (Reeves et al., 2007; Castaings et al., 2014; Li et al., 2015; Hawkes et al., 2016). Therefore, the molecular dissection of the diversity in splicing patterns of FLC across natural populations of Arabidopsis provides an important insight into how splicing regulation influences the switch from vegetative to reproductive growth. These findings propose a possible application for cultivating new varieties and augmenting the control of flowering time to adapt the environmental changes via modulating FLC expression in some crops, such as Brassicaceae.
X-LW designed the concept, organized and drafted the text. H-DQ performed the meta-analysis, drafted text, and prepared figures. H-DQ and YL edited the manuscript with help of Q-PR, Y-YW and FX.
This work was supported by National Natural Science Foundation of China (31970330) and Major Basic Research Projects of Shandong Natural Science Foundation (ZR2018ZC08N3).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 (6970), 164–167. doi: 10.1038/nature02269
Bazin, J., Romero, N., Rigo, R., Charon, C., Blein, T., Ariel, F., et al. (2018). Nuclear speckle RNA binding proteins remodel alternative splicing and the non-coding Arabidopsis transcriptome to regulate a cross-talk between auxin and immune responses. Front. Plant Sci. 9 (1209). doi: 10.3389/fpls.2018.01209
Berry, S., Dean, C. (2015). Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J. 83 (1), 133–148. doi: 10.1111/tpj.12869
Bloomer, R., Dean, C. (2017). Fine-tuning timing: natural variation informs the mechanistic basis of the switch to flowering in Arabidopsis thaliana. J. Exp. Bot. 68 (20), 5439–5452. doi: 10.1093/jxb/erx270
Caicedo, A. L., Stinchcombe, J. R., Olsen, K. M., Schmitt, J., Purugganan, M. D. (2004). Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. U.S.A. 101 (44), 15670–15675. doi: 10.1073/pnas.0406232101
Castaings, L., Bergonzi, S., Albani, M. C., Kemi, U., Savolainen, O., Coupland, G. (2014). Evolutionary conservation of cold-induced antisense RNAs of FLOWERING LOCUS C in Arabidopsis thaliana perennial relatives. Nat. Commun. 5, 4457. doi: 10.1038/ncomms5457
Chen, Y., Shen, Q., Lyu, P., Lin, R., Sun, C. (2019). Identification and expression profiling of selected MADS-box family genes in Dendrobium officinale. Genetica 147 (3-4), 303–313. doi: 10.1007/s10709-019-00071-5
Cho, S., Hoang, A., Sinha, R., Zhong, X. Y., Fu, X. D., Krainer, A. R., et al. (2011). Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. U.S.A. 108 (20), 8233–8238. doi: 10.1073/pnas.1017700108
Coupland, G. (2019). FLOWERING LOCUS C isolation and characterization: two articles that opened many doors. Plant Cell 31 (6), 1190–1191. doi: 10.1105/tpc.19.00325
Csorba, T., Questa, J. I., Sun, Q., Dean, C. (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 11; 111 (45), 16160–16165. doi: 10.1073/pnas.1419030111
Eom, H., Park, S. J., Kim, M. K., Kim, H., Kang, H., Lee, I. (2018). TAF15b, involved in the autonomous pathway for flowering, represses transcription of FLOWERING LOCUS C. Plant J. 93 (1), 79–91. doi: 10.1111/tpj.13758
Fica, S. M., Oubridge, C., Wilkinson, M. E., Newman, A. J., Nagai, K. (2019). A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science 363 (6428), 710–714. doi: 10.1126/science.aaw5569
Filichkin, S. A., Mockler, T. C. (2012). Unproductive alternative splicing and nonsense mRNAs: A widespread phenomenon among plant circadian clock genes. Bio. Direct. 7, 20. doi: 10.1186/1745-6150-7-20
Geraldo, N., Bäurle, I., Kidou, S., Hu, X., Dean, C. (2009). FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol. 150 (3), 1611–1618. doi: 10.1104/pp.109.137448
Hawkes, E. J., Hennelly, S. P., Novikova, I. V., Irwin, J. A., Dean, C., Sanbonmatsu, K. Y. (2016). COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep. 16 (12), 3087–3096. doi: 10.1016/j.celrep.2016.08.045
Helliwell, C. A., Wood, C. C., Robertson, M., James Peacock, W., Dennis, E. S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46 (2), 183–192. doi: 10.1111/j.1365-313X.2006.02686.x
Hornyik, C., Terzi, L. C., Simpson, G. G. (2010). The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev. Cell 18 (2), 203–213. doi: 10.1016/j.devcel.2009.12.009
Howard, J. M., Lin, H., Wallace, A. J., Kim, G., Draper, J. M., Haeussler, M., et al. (2018). HNRNPA1 promotes recognition of splice site decoys by U2AF2 in vivo. Genome Res. 28 (5), 689–698. doi: 10.1101/gr.229062.117
Hrtyan, M., Slikova, E., Hejatko, J., Ruzicka, K. (2015). RNA processing in auxin and cytokinin pathways. J. Exp. Bot. 66 (16), 4897–4912. doi: 10.1093/jxb/erv189
Hu, G., Hu, Z., Li, Y., Gu, F., Zhao, Z., Chen, G. (2011). A splicing site mutation in BrpFLC1 and repressed expression of BrpFLC genes are associated with the early flowering of purple flowering stalk. Russ. J. Plant Physiol. 58 (3), 431–438.
Huang, J., Gu, L., Zhang, Y., Yan, T., Kong, G., Kong, L., et al. (2017). An oomycete plant pathogen reprograms host pre-mRNA splicing to subvert immunity. Nat. Commun. 8 (1), 2051. doi: 10.1038/s41467-017-02233-5
Hugouvieux, V., Kwak, J. M., Schroeder, J. I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106 (4), 477–487. doi: 10.1016/s0092-8674(01)00460-3
Ietswaart, R., Wu, Z., Dean, C. (2012). Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 28 (9), 445–453. doi: 10.1016/j.tig.2012.06.002
Izaurralde, E., Lewis, J., McGuigan, C., Jankowska, M., Darzynkiewicz, E., Mattaj, I. W. (1994). A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78 (4), 657–668. doi: 10.1016/0092-8674(94)90530-4
Jang, Y. H., Park, H. Y., Lee, K. C., Thu, M. P., Kim, S. K., Suh, M. C. (2014). A homolog of splicing factor SF1 is essential for development and is involved in the alternative splicing of pre-mRNA in Arabidopsis thaliana. Plant J. 78 (4), 591–603. doi: 10.1111/tpj.12491
Jurica, M. S., Moore, M. J. (2003). Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 12 (1), 5–14. doi: 10.1016/s1097-2765(03)00270-3
Kuhn, J. M., Breton, G., Schroeder, J. I. (2007). mRNA metabolism of flowering-time regulators in wild-type Arabidopsis revealed by a nuclear cap binding protein mutant, abh1. Plant J. 50 (6), 1049–1062. doi: 10.1111/j.1365-313X.2007.03110.x
Kwak, J. S., Son, G. H., Song, J. T., Seo, H. S. (2017). Post-translational modifications of FLOWERING LOCUS C modulate its activity. J. Exp. Bot. 68 (3), 383–389. doi: 10.1093/jxb/erw431
Laloum, T., Martin, G., Duque, P. (2018). Alternative splicing control of abiotic stress responses. Trends Plant Sci. 23 (2), 140–150. doi: 10.1016/j.tplants.2017.09.019
Laubinger, S., Sachsenberg, T., Zeller, G., Busch, W., Lohmann, J. U., Rätsch, G., et al. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105 (25), 8795–8800. doi: 10.1073/pnas.0802493105
Lempe, J., Balasubramanian, S., Sureshkumar, S., Singh, A., Schmid, M., Weigel, D. (2005). Diversity of flowering responses in wild Arabidopsis thaliana strains. PloS Genet. 1 (1), 109–118. doi: 10.1371/journal.pgen.0010006
Lewis, J. D., Izaurralde, E., Jarmolowski, A., McGuigan, C., Mattaj, I. W. (1996). A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5' splice site. Genes Dev. 10 (13), 1683–1698. doi: 10.1101/gad.10.13.1683
Li, P., Tao, Z., Dean, C. (2015). Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev. 29 (7), 696–701. doi: 10.1101/gad.258814.115
Li, S., Yamada, M., Han, X., Ohler, U., Benfey, P. N. (2016). High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincrna regulation. Dev. Cell 39 (4), 508–522. doi: 10.1016/j.devcel.2016.10.012
Liu, F., Quesada, V., Crevillen, P., Baurle, I., Swiezewski, S., et al. (2007). The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell 28 (3), 398–407. doi: 10.1016/j.molcel.2007.10.018
Liu, F., Marquardt, S., Lister, C., Swiezewski, S., Dean, C. (2010). Targeted 3' processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327 (5961), 94–97. doi: 10.1126/science.1180278
Liu, L., Wu, F., Zhang, W. (2016). A conserved interaction between SKIP and SMP1/2 aids in recruiting the second-step splicing factors to the spliceosome in Arabidopsis. Mol. Plant 9 (12), 1660–1663. doi: 10.1016/j.molp.2016.09.007
Lockhart, J. (2018). Fresh as an Exitron: a flower-specific splice variant of AUXIN RESPONSE FACTOR8 helps shape the stamen. Plant Cell 30 (3), 518–519. doi: 10.1105/tpc.18.00208
Long, Y., Sou, W. H., Yung, K. W. Y., Liu, H., Wan, S. W. C., et al. (2019). Distinct mechanisms govern the phosphorylation of different SR protein splicing factors. J. Biol. Chem. 294 (4), 1312–1327. doi: 10.1074/jbc.RA118.003392
Luo, X., Chen, T., Zeng, X., He, D., He, Y. (2019). Feedback regulation of FLC by FLOWERING LOCUS T (FT) and FD through a 5' promoter region in Arabidopsis. Mol. Plant 12 (3), 285–288. doi: 10.1016/j.molp.2019.01.013
Lyons, R., Kazan, K. (2016). The link between flowering time and stress tolerance. J. Exp. Bot. 67 (1), 47–60. doi: 10.1093/jxb/erv441
Mahrez, W., Shin, J., Muñoz-Viana, R., Figueiredo, D. D., Trejo-Arellano, M. S., Exner, V., et al. (2016). BRR2a affects flowering time via FLC splicing. PloS Genet. 12 (4), e1005924. doi: 10.1371/journal.pgen.1005924
Marquardt, S., Raitskin, O., Wu, Z., Liu, F., Sun, Q. (2014). Dean, CFunctional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 54 (1), 156–165. doi: 10.1016/j.molcel.2014.03.026
Matera, A. G., Wang, Z. (2014). A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15 (2), 108–121. doi: 10.1038/nrm3742
Mei, W., Liu, S., Schnable, J. C., Yeh, C. T., Springer, N. M., Schnable, P. S., et al. (2017). A comprehensive analysis of alternative splicing in paleopolyploid maize. Front. Plant Sci. 8, 694. doi: 10.3389/fpls.2017.00694
Melzer, R. (2017). Regulation of flowering time: a splicy business. J. Exp. Bot. 68 (18), 5017–5020. doi: 10.1093/jxb/erx353
Michaels, S. D., Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 (5), 949–956. doi: 10.1105/tpc.11.5.949
Michaels, S. D., He, Y., Scortecci, K. C., Amasino, R. M. (2003). Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100 (17), 10102–10107. doi: 10.1073/pnas.1531467100
Okumoto, K., Ono, T., Toyama, R., Shimomura, A., Nagata, A., Fujiki, Y. (2018). New splicing variants of mitochondrial Rho GTPase-1 (Miro1) transport peroxisomes. J. Cell Biol. 217 (2), 619–633. doi: 10.1083/jcb.201708122
Pajoro, A., Severing, E., Angenent, G., Immink, R. (2017). Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Biol. 18 (1), 102. doi: 10.1186/s13059-017-1235-x
Park, H. Y., Lee, H. T., Lee, J. H., Kim, J. K. (2019a). Arabidopsis U2AF65 regulates flowering time and the growth of pollen tubes. Front. Plant Sci. 10, 569. doi: 10.3389/fpls.2019.00569
Park, Y. J., Lee, J. H., Kim, J. Y., Park, C. M. (2019b). Alternative RNA splicing expands the developmental plasticity of flowering transition. Front. Plant Sci. 10, 606. doi: 10.3389/fpls.2019.00606
Raghunathan, P. L., Guthrie, C. (1998). RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2 . Curr. Biol. 8 (15), 847–855. doi: 10.1016/s0960-9822(07)00345-4
Reeves, P. A., He, Y., Schmitz, R. J., Amasino, R. M., Panella, L. W., Richards, C. M. (2007). Evolutionary conservation of the FLOWERING LOCUS C-mediated vernalization response: evidence from the sugar beet (Beta vulgaris). Genetics 176 (1), 295–307. doi: 10.1534/genetics.106.069336
Richter, R., Kinoshita, A., Vincent, C., Martinez-Gallegos, R., Gao, H., van Driel, A. D., et al. (2019). Floral regulators FLC and SOC1 directly regulate expression of the B3-type transcription factor TARGET OF FLC AND SVP 1 at the Arabidopsis shoot apex via antagonistic chromatin modifications. PloS Genet. 15 (4), e1008065. doi: 10.1371/journal.pgen.1008065
Rodríguez-Cazorla, E., Ortuño-Miquel, S., Candela, H., Bailey-Steinitz, L. J., Yanofsky, M. F., Martínez-Laborda, A., et al. (2018). Ovule identity mediated by pre-mRNA processing in Arabidopsis. PloS Genet. 14 (1), e1007182. doi: 10.1371/journal.pgen.1007182
Rosa, S., Duncan, S., Dean, C. (2016). Mutually exclusive sense-antisense transcription at FLC facilitates environmentally induced generepression. Nat. Commun. 7, 13031. doi: 10.1038/ncomms13031
Samach, A., Melamed-Bessudo, C., Avivi-Ragolski, N., Pietrokovski, S., Levy, A. A. (2011). Identification of plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. Plant Cell 23 (12), 4266–4279. doi: 10.1105/tpc.111.091744
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Kröber, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20 (7), 898–912. doi: 10.1101/gad.373506
Severing, E. I., van Dijk, A. D., Morabito, G., Busscher-Lange, J., Immink, R. G., van Ham, R. C. (2012). Predicting the impact of alternative splicing on plant MADS domain protein function. PloS One 7 (1), e30524. doi: 10.1371/journal.pone.0030524
Shang, X., Cao, Y., Ma, L. (2017). Alternative splicing in plant genes: a means of regulating the environmental fitness of plants. Int. J. Mol. Sci. 18 (2). doi: 10.3390/ijms18020432
Shao, C., Yang, B., Wu, T., Huang, J., Tang, P., Zhou, Y., et al. (2014). Mechanisms for U2AF to define 3' splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 21 (11), 997–1005. doi: 10.1038/nsmb2906
Sheldon, C. C., Burn, J. E., Perez, P. P., Metzger, J., Edwards, J. A., Peacock, W. J., et al. (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 (3), 445–458. doi: 10.1105/tpc.11.3.445
Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J., Dennis, E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. 97 (7), 3753–3758. doi: 10.1073/pnas.060023597
Slotte, T., Huang, H.-R., Holm, K., Ceplitis, A., Onge, K. S., Chen, J., et al. (2009). Splicing variation at a FLOWERING LOCUS C homeolog is associated with flowering time variation in the tetraploid Capsella bursa-pastoris. Genetics 183 (1), 337–345. doi: 10.1534/genetics.109.103705
Streitner, C., Danisman, S., Wehrle, F., Schöning, J. C., Alfano, J. R., Staiger, D. (2008). The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. Plant J. 56 (2), 239–250. doi: 10.1111/j.1365-313X.2008.03591.x
Streitner, C., Köster, T., Simpson, C. G., Shaw, P., Danisman, S., Brown, J. (2012). WAn hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana. Nucleic. Acids Res. 40 (22), 11240–11255. doi: 10.1093/nar/gks873
Swiezewski, S., Crevillen, P., Liu, F., Ecker, J. R., Jerzmanowski, A., Dean, C. (2007). Small RNA-mediated chromatin silencing directed to the 3' region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc. Natl. Acad. Sci. U.S.A. 104 (9), 3633–3638. doi: 10.1073/pnas.0611459104
Swiezewski, S., Liu, F., Magusin, A., Dean, C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462 (7274), 799–802. doi: 10.1038/nature08618
Tian, Y., Zheng, H., Zhang, F., Wang, S., Ji, X., Xu, C., et al. (2019). PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci. Adv. 5 (4), eaau7246. doi: 10.1126/sciadv.aau7246
Valcárcel, J., Green, M. R. (1996). The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci. 21 (8), 296–301. doi: 10.1016/S0968-0004(96)10039-6
Verhage, L., Severing, E., Bucher, J., Lammers, M., Busscher-Lange, J., Bonnema, G., et al. (2017). Splicing-related genes are alternatively spliced upon changes in ambient temperatures in plants. PLoS One 12 (3), e0172950. doi: 10.1371/journal.pone.0172950
Vosseberg, J., Snel, B. (2017). Domestication of self-splicing introns during eukaryogenesis: the rise of the complexspliceosomal machinery. Biol. Direct. 12 (1), 30. doi: 10.1186/s13062-017-0201-6
Wahl, M. C., Will, C. L., Lührmann, R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136 (4), 701–718. doi: 10.1016/j.cell.2009.02.009
Wang, H., You, C., Chang, F., Wang, Y., Wang, L., Qi, J., et al. (2014). Alternative splicing during Arabidopsis flower development results in constitutive and stage-regulated isoforms. Front. Genet. 5, 25. doi: 10.3389/fgene.2014.00025
Wang, Q., Zhang, L., Lynn, B., Rymond, B. C. (2008). A BBP-Mud2p heterodimer mediates branchpoint recognition and influences splicing substrate abundance in budding yeast. Nucleic Acids Res. 36 (8), 2787–2798. doi: 10.1093/nar/gkn144
Wang, Z., Ji, H., Yuan, B., Wang, S., Su, C., Yao, B., et al. (2015). ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat. Commun. 6, 8138. doi: 10.1038/ncomms9138
Wang, Y. Y., Xiong, F., Ren, Q. P., Wang, X. L. (2019). Regulation of flowering transition by alternative splicing: the role of the U2 auxiliary factors. J. Exp. Bot. doi: 10.1093/jxb/erz416
Werner, J. D., Borevitz, J. O., Uhlenhaut, N. H., Ecker, J. R., Chory, J., Weigel, D. (2005). FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170 (3), 1197–1207. doi: 10.1534/genetics.104.036533
Whittaker, C., Dean, C. (2017). The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell. Dev. Biol. 33, 555–575. doi: 10.1146/annurev-cellbio-100616-060546
Will, C. L., Lührmann, R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. doi: 10.1101/cshperspect.a003707
Wu, J., Wei, K., Cheng, F., Li, S., Wang, Q., Zhao, J., et al. (2012). A naturally occurring InDel variation in BraA.FLC.b (BrFLC2) associated with flowering time variation in Brassica rapa. BMC Plant Biol. 12, 151. doi: 10.1186/1471-2229-12-151
Wu, Z., Zhu, D., Lin, X., Miao, J., Gu, L., Deng, X., et al. (2016). RNA binding proteins RZ-1B and RZ-1C play critical roles in regulating pre-mRNA splicing and gene expression during development in Arabidopsis. Plant Cell 28 (1), 55–73. doi: 10.1105/tpc.15.00949
Xiao, J., Li, C., Xu, S., Xing, L., Xu, Y., Chong, K. (2015). JACALIN-LECTIN LIKE1 Regulates the Nuclear Accumulation of GLYCINE-RICH RNA-BINDING PROTEIN7, influencing the RNA processing of FLOWERING LOCUS C antisense transcripts and flowering time in Arabidopsis. Plant Physiol. 169 (3), 2102–2117. doi: 10.1104/pp.15.00801
Xiong, F., Ren, J. J., Yu, Q., Wang, Y. Y., Lu, C. C., Kong, L. J., et al. (2019a). AtU2AF65b functions in ABA-mediated flowering via regulating the pre-mRNA splicing of ABI5 and FLC in Arabidopsis. New Phytol. 223 (1), 277–292. doi: 10.1111/nph.15756
Xiong, F., Ren, J. J., Yu, Q., Wang, Y. Y., Kong, L. J., Otegui, M. S., et al. (2019b). AtBUD13 affects pre-mRNA splicing and is essential for embryo development in Arabidopsis. Plant J. 98 (4), 714–726. doi: 10.1111/tpj.14268
Yan, Q., Xia, X., Sun, Z., Fang, Y. (2017). Depletion of Arabidopsis SC35 and SC35-like serine/arginine-rich proteins affects the transcription and splicing of a subset of genes. PloS Genet. 13 (3), e1006663. doi: 10.1371/journal.pgen.1006663
Yan, Z., Shi, H., Liu, Y., Jing, M., Han, Y. (2019). KHZ1 and KHZ2, novel members of the autonomous pathway, repress the splicing efficiency of FLC pre-mRNA in Arabidopsis. J. Exp. Bot. erz499. doi: 10.1093/jxb/erz499
Yuan, Y. X., Wu, J., Sun, R. F., Zhang, X. W., Xu, D. H., Bonnema, G., et al. (2009). A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J. Exp. Bot. 60 (4), 1299–1308. doi: 10.1093/jxb/erp010
Zhang, J. Z., Li, Z. M., Mei, L., Yao, J. L., Hu, C. G. (2009). PtFLC homolog from trifoliate orange (Poncirus trifoliata) is regulated by alternative splicing and experiences seasonal fluctuation in expression level. Planta. 229 (4), 847–859. doi: 10.1007/s00425-008-0885-z
Zhao, J., Kulkarni, V., Liu, N., Del Carpio, D. P., Bucher, J., Bonnema, G. (2010). BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J. Exp. Bot. 61 (6), 1817–1825. doi: 10.1093/jxb/erq048
Zhu, F. Y., Fan, T., Liu, Y. G., Zhu, F. Y., Chen, M. X., Shi, L., et al (2017). Proteogenomic analysis reveals alternative splicing and translation as part of the abscisic acid response in Arabidopsis seedlings. Plant J. 91 (3), 518–533. doi: 10.1111/tpj.13571
Keywords: alternative splicing, FLOWERING LOCUS C, COOLAIR, flowering transition, splicing factor, vernalization
Citation: Qi H-D, Lin Y, Ren Q-P, Wang Y-Y, Xiong F and Wang X-L (2019) RNA Splicing of FLC Modulates the Transition to Flowering. Front. Plant Sci. 10:1625. doi: 10.3389/fpls.2019.01625
Received: 24 September 2019; Accepted: 19 November 2019;
Published: 17 December 2019.
Edited by:
Stefan de Folter, Center for Research and Advanced Studies (CINVESTAV), MexicoReviewed by:
Lee Jeong Hwan, Chonbuk National University, South KoreaCopyright © 2019 Qi, Lin, Ren, Wang, Xiong and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiu-Ling Wang, eGx3YW5nQHNkYXUuZWR1LmNu
†ORCID: Xiu-Ling Wang, orcid.org/0000-0003-4430-4674
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.