
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 29 November 2019
Sec. Plant Development and EvoDevo
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01594
This article is part of the Research TopicOrchid Genomics and Developmental BiologyView all 13 articles
Somatic embryogenesis is commonly used for clonal propagation of a wide variety of plant species. Induction of protocorm-like-bodies (PLBs), which are capable of developing into individual plants, is a routine tissue culture-based practice for micropropagation of orchid plants. Even though PLBs are often regarded as somatic embryos, our recent study provides molecular evidence to argue that PLBs are not derived from somatic embryogenesis. Here, we report and characterize the somatic embryonic tissues induced by Phalaenopsis aphrodite LEAFY COTYLEDON1 (PaLEC1) in Phalaenopsis equestris. We found that PaLEC1-induced somatic tissues are morphologically different from PLBs, supporting our molecular study that PLBs are not of somatic embryonic origin. The embryonic identity of PaLEC1-induced embryonic tissues was confirmed by expression of the embryonic-specific transcription factors FUSCA3 (FUS3) and ABSCISIC ACID INSENSITIVE3 (ABI3), and seed storage proteins 7S GLOBULIN and OLEOSIN. Moreover, PaLEC1-GFP protein was found to be associated with the Pa7S-1 and PaFUS3 promoters containing the CCAAT element, supporting that PaLEC1 directly regulates embryo-specific processes to activate the somatic embryonic program in P. equestris. Despite diverse embryonic structures, PaLEC1-GFP-induced embryonic structures are pluripotent and capable of generating new shoots. Our study resolves the long-term debate on the developmental identity of PLB and suggests that somatic embryogenesis may be a useful approach to clonally propagate orchid seedlings.
Plant totipotency provides a means of clonally propagating a wide variety of plant species (Ikeuchi et al., 2013; Kareem et al., 2016; Radhakrishnan et al., 2017). In vitro regeneration bypasses the embryogenic program and directly generates seedlings with identical genetic makeup. One of the most common in vitro regeneration methods is somatic embryogenesis (Zimmerman, 1993; Pulianmackal et al., 2014). Somatic embryogenesis is crucial for establishing genetic transformation platforms for many non-model plant species and for clonal propagation of numerous high-value plants. For example, somatic embryos are used as transformation materials for alfalfa, American chestnut, cassava, cotton, grapevine, maize, mango, melon, Norway spruce, papaya, rose, tea tree, and walnut (Umbeck et al., 1987; Mcgranahan et al., 1988; Robertson et al., 1992; Fitch et al., 1993; Li et al., 1996; Brettschneider et al., 1997; Trinh et al., 1998; Mondal et al., 2001; Akasaka-Kennedy et al., 2004; Chavarri et al., 2004; Li et al., 2006; Polin et al., 2006; Vergne et al., 2010). In addition, the regeneration capacity of somatic embryos has made somatic embryogenesis a common method through which to clonally propagate economically important trees or herbal plants (Joshee et al., 2007; Nordine et al., 2014; Guan et al., 2016; Kim et al., 2019).
Embryogenesis is a defined developmental program during which the zygote grows and develops into a mature embryo. Somatic embryogenesis, on the other hand, activates the embryogenesis program in the absence of gamete fusion (von Arnold et al., 2002; Braybrook and Harada, 2008; Yang and Zhang, 2010; Feher, 2015). Zygotic embryogenesis and somatic embryogenesis programs not only share similar morphogenesis and maturation phases, they also share similar if not completely identical genetic and molecular networks (Zimmerman, 1993; Mordhorst et al., 2002; Gaj et al., 2005). Moreover, ectopic expression of several key embryo-associated transcription factors (TFs) is capable of inducing the embryogenesis program in somatic tissues (Lotan et al., 1998; Hecht et al., 2001; Stone et al., 2001; Boutilier et al., 2002; Zuo et al., 2002; Harding et al., 2003; Kwong et al., 2003; Gaj et al., 2005; Wang et al., 2009), demonstrating the developmental plasticity of plant tissues.
Orchids evolve specialized developmental programs including the co-evolution of diverse floral structures and pollinators (Waterman and Bidartondo, 2008), formation of pollen dispersal units (pollinia) (Pacini and Hesse, 2002), lack of cotyledon organogenesis during embryogenesis (Kull and Arditti, 2002; Yeung, 2017), and mycorrhizal fungi-assisted seed germination (Rasmussen, 2002), and all of these developmental processes contribute to their distinct morphology and physiological characteristics. These unique developmental strategies have not only fascinated many evolutionary and plant biologists; the beauty of the resulting floral structures is also enthusiastically admired by the general public. Much effort has been put into tissue culture-based clonal propagation of elite orchids over the past decades and this technology has transformed the orchid business into a multimillion-dollar orchid biotechnology industry (Winkelmann et al., 2006; Liao et al., 2011; Hossain et al., 2013).
Generally, embryogenesis of angiosperm plants starts from morphogenesis with continuous changes in embryo morphology and establishment of shoot-root polarity followed by maturation and desiccation processes (Bentsink and Koornneef, 2008; Braybrook and Harada, 2008). One of the characteristic features that defines the somatic embryo is the formation of the embryonic cotyledons. Even though orchid embryos go through a maturation and desiccation process, they lack characteristic cotyledons (organogenesis) and fail to establish a shoot-root axis during embryogenesis (Arditti, 1992; Dressler, 1993; Burger, 1998). Instead, a tubular embryo structure with an anterior meristem is formed. Upon germination, a tubular embryo emerges as a protocorm and new leaves and roots are generated from the anterior meristem of the protocorm (Nishimura, 1981).
Protocorm-like body (PLB)-based regeneration is commonly used to produce huge amounts of orchid seedlings of elite cultivars (Arditti and Krikorian, 1996; Chen et al., 2002; Arditti, 2009; Chugh et al., 2009; Yam and Arditti, 2009; Paek et al., 2011; Yam and Arditti, 2017). For years, much effort has been devoted to develop in vitro protocols to induce PLB and somatic embryo development either directly or indirectly (via the callus tissue) from explants to improve micropropagation in Phalaenopsis orchids (Tokuhara and Mii, 2001; Tokuhara and Mii, 2003; Kuo et al., 2005; Chen and Chang, 2006; Gow et al., 2009; Gow et al., 2010; Pramanik et al., 2016). PLBs are often induced from somatic tissues such as protocorms, floral stalk internodes, leaves, and root tips (Chen et al., 2002; Park et al., 2002; Chen and Chang, 2004; Chen and Chang, 2006; Teixeira da Silva et al., 2006; Zhao et al., 2008; Guo et al., 2010; Paek et al., 2011). Because embryogenesis produces tubular embryos, which, upon germination, develop into protocorms, and PLBs resemble protocorms morphologically, initiation, and development of PLBs is often regarded as somatic embryogenesis (Jones and Tisserat, 1990; Begum et al., 1994; Ishii et al., 1998). However, our recent study provides molecular evidence to argue that PLBs are not somatic embryos. Instead, PLB development seems to adopt a shoot pole-related organogenesis program (Fang et al., 2016). So if PLBs are not somatic embryos, what does the somatic embryo of orchids look like? Are somatic embryos of orchid species also prolific in propagating clonal plants? To address these questions and to explore the potential of using orchid somatic embryos for clonal propagation, we generated transgenic plants overexpressing Phalaenopsis aphrodite LEAFY COTYLEDON1 (PaLEC1) gene in Phalaenopsis equestris. LEC1 encodes a HAP3 subunit of CCAAT-binding TF required for embryo development (Lotan et al., 1998). Moreover, ectopic expression of LEC1 isolated from Arabidopsis and other plant species has been demonstrated to be a potent inducer of the somatic embryogenesis program (Lotan et al., 1998; Yazawa et al., 2004; Garcês et al., 2014; Zhu et al., 2014; Min et al., 2015; Fang et al., 2016). Consistently, we have shown that PaLEC1 is capable of inducing somatic embryogenesis in Arabidopsis (Fang et al., 2016). Here, we report PaLEC1-induced embryonic tissues in P. equestris and demonstrate their embryonic identity by validation of expression of the embryonic-specific genes. To the best of our knowledge, this is the first time that somatic embryonic tissues was shown to be induced by the embryonic factor, PaLEC1, in the transgenic orchid plants. Our studies reveal that PaLEC1 may be a useful tool for large scale propagation of clonal plants in orchid biotechnology.
P. equestris plants were grown and maintained in a growth chamber with alternating 12 h light (23°C)/12 h dark (18°C) cycles. Hand pollination was conducted for each flower, as described previously (Chen and Fang, 2016).
Seeds were allowed to germinate in germination medium (0.1% tryptone, 1.8% sucrose, 2% potato, 0.75% agar, 1.08 g/l MS salt, 1X MS vitamin pH 5.7). For regeneration assay, the protocorm segments (explants) were transferred to T2 medium containing 0.1% (w/v) tryptone, 2% (w/v) sucrose, 2% (w/v) potato homogenate, 2.5% (w/v) banana homogenate, 0.01% (v/v) citric acid, 0.1% (w/v) charcoal, and 1% (w/v) agar adjusted to pH 5.5 (Chen et al., 2009) to induce PLBs.
Protocorms or PLBs of P. equestris were used for transformation. Agrobacterium-based transformation was conducted as described previously with slight modification (Liau et al., 2003). Briefly, protocorms or PLBs were co-cultured with Agrobacterium tumefaciens strain EHA105 culture (OD600 = ~ 0.8) containing the 35S:PaLEC1-GFP or pH7FWG2 (vector only control) construct in the presence of 100 μM acetosyringone overnight (for protocorms) or 3 days (for PLBs) in the dark. After removing Agrobacterium inoculum, the infected tissues were washed four to six times with distilled water containing 40 mg/L meropenem (China Chemical & Pharmaceutical). The infected tissues were allowed to recover in New Dogashima medium (NDM) (Belarmino and Mii, 2000) containing 10 g/L maltose, 0.1 mg/L naphthaleneacetic acid (NAA), and 0.4 mg/L benzyladenine (BA) and supplemented with 40 mg/L meropenem for 1 week in the dark. All the NDM media used in this study contained 10 g/L maltose, 0.1 mg/L NAA, and 0.4 mg/L BA. The recovered tissues were then selected on NDM agar plates supplemented with 20 mg/L hygromycin and 40 mg/L meropenem at 23°C in the dark. The infected tissues were selected continuously by sub-culturing to fresh NDM plates supplemented with 20 mg/L hygromycin and 40 mg/L meropenem at 2- to 4-week intervals until new leaves emerged. The hygromycin-resistant transgenic plants were then transferred to T2 (Chen et al., 2009) medium supplemented with 20 mg/L hygromycin and 40 mg/L meropenem to encourage growth.
Genomic DNA from young seedlings (1 to 1.5 cm in size) of T1 transgenic plants and wild-type plants was isolated by cetyltrimethylammonium bromide (CTAB)-based method (Murray and Thompson, 1980; Saghai-Maroof et al., 1984) with some modifications. Briefly, freeze-dried tissue was ground in the presence of liquid nitrogen, resuspended in 1 g/10 ml of the CTAB extraction buffer [100 mM Tris, pH 8.0, 1.4 M NaCl, 20 mM EDTA, 2% CTAB, 0.2% (w/v) polyvinylpyrrolidone 40, and 0.2% β-mercaptoethanol], and incubated at 65oC for 60 min with occasional gentle vortexing. Equal volume of chloroform:isoamylalcohol (24:1) was then added and mixed by inversion for 15 min. Plant extract was then centrifuged at 3220 × g for 15 min at 4oC. The aqueous phase was extracted with equal volume of chloroform:isoamylalcohol (24:1) one more time. The aqueous phase was transferred to a new tube and equal volume of isopropanol was added, mixed, and incubated at ‑20oC overnight. DNA was precipitated by centrifugation at 3,220 × g at 4oC for 10 min. DNA was washed once with 75% ethanol followed by 99% ethanol. DNA was then resuspended in H2O supplemented with 10 µg/ml RNase A and incubated at 37oC overnight to remove RNA. This method yielded approximately 150‑350 µg per g of orchid tissue. Approximately 15 µg DNA was digested and Southern blotting and hybridization were carried out as described previously (Fang et al., 2006).
To generate polyclonal antibodies specific for PaLEC1 protein, PaLEC1 peptide YLHRYRELEGDHRGSIRG (LTK BioLaboratories, Taiwan) was synthesized to generate rabbit polyclonal antisera. Polyclonal antibodies raised against PaLEC1 were affinity purified for western blotting analysis.
To prepare total protein extract, tissues were ground to a fine powder in the presence of liquid nitrogen using a pestle and mortar. One gram of ground tissues was homogenized in 1 ml 1X Urea-based protein extraction buffer (250 mM Tris-HCl, pH 6.8, 3.5% sodium dodecyl sulfate (SDS), 10% glycerol, 1 M urea) supplemented with 1X protease inhibitor cocktail (Sigma-Aldrich), 1 mM phenylmethylsulfonyl fluoride by vortexing. Cell debris was removed by centrifugation at 16,100 × g for 10 min at 4°C. Concentration of protein lysates was determined by Bio-Rad DC Protein Assay Kit (Bio-Rad).
Protein lysates were resolved by a standard 10% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to Immobilon-P PVDF membrane (Merck Millipore). Blots were blocked in 1 × TBST (20 mM Tris base, 150 mM NaCl, 0.1% Tween 20) supplemented with 5% non-fat milk at room temperature (RT) for 1 h and incubated with anti-GFP antibodies (Roche) diluted 1:2,000 in 1 × TBST supplemented with 5% non-fat milk at 4°C overnight. For detection of PaLEC1 and PaRbCL proteins, blots were incubated with PaLEC1 (1:2,000) and RbCL (1:5,000, Agrisera) antibodies diluted in 1 × TBST with 5% non-fat milk for 1 h at RT. Blots were washed three times for 15 min each time and then incubated with horseradish peroxidase (HRP) conjugated goat-anti-mouse-IgG (1:20,000, Jackson ImmunoResearch Laboratories), or goat-anti-rabbit-IgG (1:20,000, PerkinElmer) for 1 h at RT. After washing in 1 × TBST for 15 min three times, the blots were processed for chemiluminescence detection using Amersham ECL select Western Blotting detection reagent. The images were acquired by a ChemiDoc XRS+ imager (Bio-Rad).
Plant tissues (young seedlings or callus tissues) were homogenized by pestle and mortar in the presence of liquid nitrogen. RNA was isolated using RNA extraction reagent (3-Zol, MDBio) according to the manufacturer’s instructions. Total RNA was treated with RNase-free DNase (Qiagen) to remove DNA followed by RNeasy column purification (Qiagen) according to the manufacturer’s instructions.
Approximately 5 μg DNA-free RNA was reverse transcribed in the presence of a mixture of oligo dT and random primers (9:1 ratio) using the GoScript Reverse Transcription System (Promega) according to the manufacturer’s instructions. Ten microliters of quantitative RT-PCR reaction was set up as follows: 2.5 μl of 1/20 diluted complementary DNA, 0.2 μM of primers, and 5 μl of 2X KAPA SYBR FAST master mix (KAPA Biosystems). The PCR program was used for DNA amplification: 95°C for 1 min, 40 cycles of 95°C for 5 s, and 58°C, 60°C, or 62°C for 20 s. PCR was performed in triplicate. Standard error was calculated from three technical replicates. Fold change in expression was calculated as 2 ‑△△CT. A melting curve of each PCR was examined to ensure no spurious products were present. Primer pairs used for quantitative PCR are listed in Supplementary Table S1. A ubiquitin gene, PaUBI (Lin et al., 2014), was used as an internal control.
Tissues were fixed in paraformaldehyde fixative and sectioned to 10 μm in thickness using the hybrid-cut method as previously described (Chen et al., 2016). Tissues sections were then stained with hematoxylin solution and photographed on a Zeiss Axio Scope A1 microscope equipped with an Axio-Cam HRc camera (Zeiss, Germany).
Fresh tissues of transgenic plants were hand sectioned and fluorescence images were photographed on a LSM 710 Confocal Microscope (Zeiss).
Chromatin immunoprecipitation was conducted as described previously (Villar and Köhler, 2010) with slight modification. One gram of young seedling (1 to 1.5 cm in size, pooled from approximately 10 T1 seedlings) tissue was fixed in 37 ml of 1% formaldehyde and vacuum-infiltrated for 20 min. Two and half milliliters of 2 M glycine was added to stop crosslinking. Fixed tissues were then homogenized by pestle and mortar and chromatin was extracted and filtered by a 70 μM cell strainer (Biologix Group Limited) as described (Villar and Köhler, 2010). Chromatin was resuspended in 200 μl of nuclei lysis buffer (50 mM Tris-HCl, pH 8.0; 10 mM EDTA, 1% SDS, and 1X protease inhibitor cocktail) before sonication. Chromatin was then transferred to a micro TUBE-500 AFA Fiber Screw-Cap tube and sonicated for 12 min or 20 min under the condition: peak incident power 150 S; duty factor 2%, cycles per burst 200, in a Covaris S220 system (Covaris). The sheered chromatin was precleared by incubating with 40 μl protein A Magnetic Sepharose beads (Merck) at 4°C for 1.5 h. In the meantime, 5 μg PaLEC1 polyclonal antibody or histone H3 antibody (Agrisera) was incubated with protein A beads at 4°C for 1.5 h. After preclearing, protein A/antibody mix was added to each sheered chromatin sample and incubated at 4°C overnight. Magnetic beads were then washed as described (Villar and Köhler, 2010) and eluted by adding 150 μl of elution buffer (1% SDS and 0.1% NaHCO3) and incubating at 65°C for 15 min. Elution was repeated one more time and elutes were pooled into a single tube. Twelve microliters of 5 M NaCl was added into 200 μl elutes and incubated at 65°C overnight. Proteins were removed by incubating with 6 μl of 2 mg/ml proteinase K, 6 μl of 0.5 M EDTA, and 12 μl of 1 M This-HCl (pH 6.5) at 45°C for 2 h. Immunoprecipated DNA was then purified by QIAquick PCR Purification Kit (Qiagen) following the manufacturer’s instructions.
Twenty microliters of PCR reaction was set up as follows: 2 or 3 μl of chromatin immunoprecipitation (ChIP) DNA, 0.5 μM of primers, 2 μl of 10X PCR buffer, 3% dimethyl sulfoxide, and 1.25 unit Power Taq polymerase (Genomics, Taiwan). The PCR program was used for amplification of immunoprecipitated DNA: 95°C for 2 min, 42 cycles of 94°C for 5 s, 58–60°C for 20 s, and 72°C for 20 s. Primers used for ChIP-PCR are listed in Supplementary Table S1. ChIP-PCR was repeated three times using three independent biological samples.
To explore the possibility of inducing a somatic embryonic program in Phalaenopsis orchids and to investigate whether somatic embryonic potential can be utilized for micropropagation, transgenic P. equestris plants overexpressing the embryonic maker PaLEC1 that has been shown to induce somatic embryogenesis in Arabidopsis (Fang et al., 2016) were generated. After transformation, we successfully obtained multiple independent transgenic lines carrying the 35S:PaLEC1-GFP transgene with the hygromycin phosphotransferase (hpt) marker (Supplementary Figure S1A). Expression of PaLEC1-GFP was also confirmed in four out of seven independent transgenic lines by western blotting using the GFP antibody (Supplementary Figure S1B). However, multiple transformation attempts failed to generate transgenic plants carrying the empty pH7FWG2 vector. It is not clear why only 35S:PaLEC1-GFP transgenic plants were obtained. Overexpression of the embryonic genes BABYBOOM has been reported to enhance embryogenic callus proliferation, overall transformation efficiency, and regeneration capacity in several monocot plants recalcitrant for transformation (Lowe et al., 2016). Hence, it is possible that ectopic PaLEC1 enhances transformation efficiency by increasing embryogenic callus formation and regeneration ability.
The 35S:PaLEC1-GFP transgenic plants were allowed to grow in growth chamber for approximately 5 years before reaching maturity for flowering. 35S:PaLEC1-GFP transgenic lines #1, #2, and #3 started to flower in March of 2018 and were therefore used in this study. The flowers were hand-pollinated and T1 seeds were allowed to produce in developing capsules. Seeds were sown and allowed to germinate on medium containing hygromycin.
As expected, seeds harvested from wild-type control plants failed to grow and were arrested on hygromycin-containing plates 5 months after sowing (MAS, Figure 1A). T1 seeds from 35S:PaLEC1-GFP transgenic lines #1 and #2, on the other hand, started to expand and turned green at 2 MAS (Figure 1B). The germinated protocorms continued to grow and produce new shoots on hygromycin-containing plates at 5 MAS (Figure 1B). Transgenic 35S:PaLEC1-GFP line #3 failed to produce viable seeds and was therefore omitted from further study. Southern blotting was used to confirm independent integration of the transgene in 35S:PaLEC1-GFP #1 and 35S:PaLEC1-GFP #2 lines (Supplementary Figure S1C).
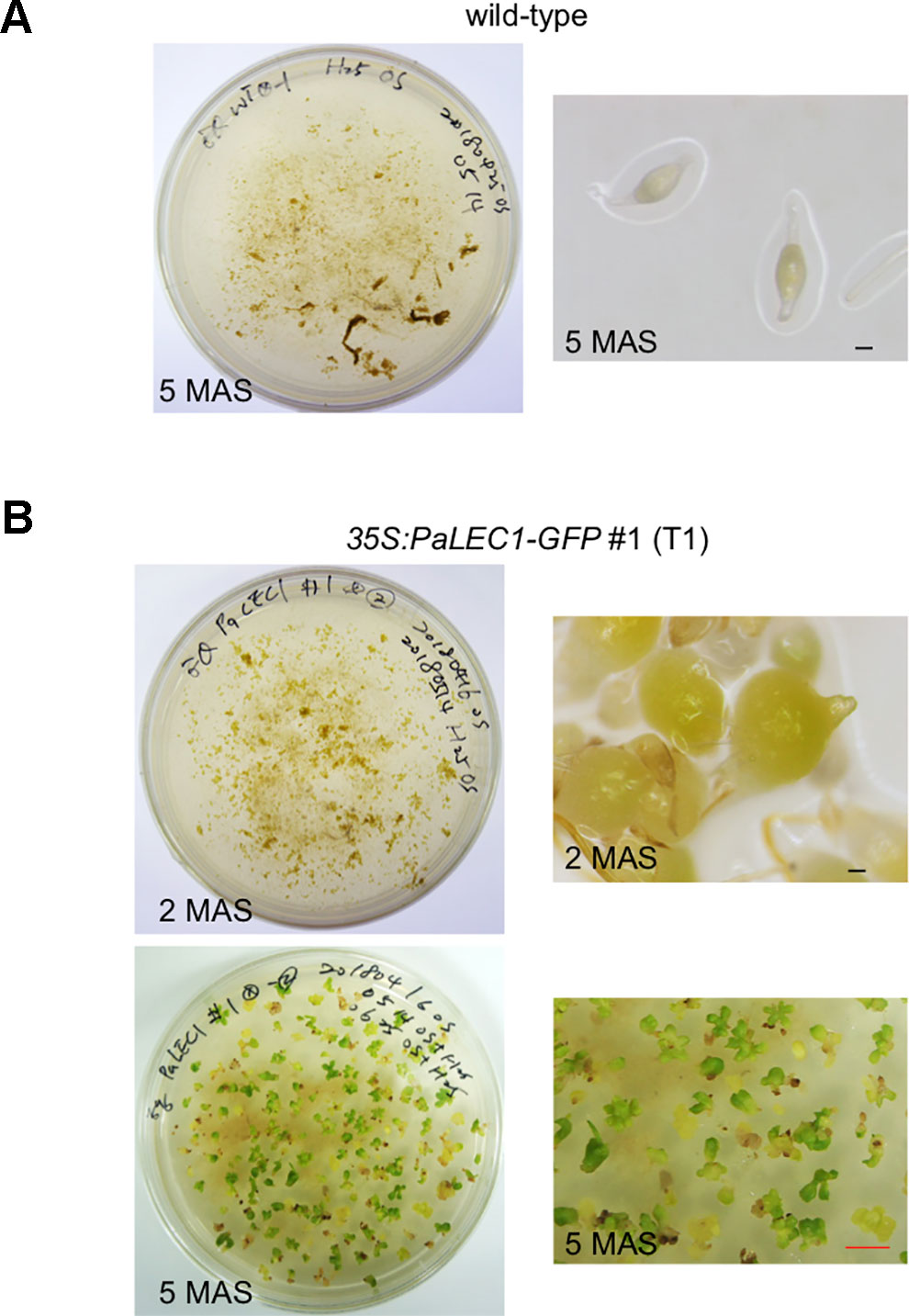
Figure 1 Selection of T1 35:PaLEC1-GFP #1 transgenic plants on hygromycin plates. (A) Wild-type protocorms failed to develop on hygromycin-containing medium. (B) 35:PaLEC1-GFP #1 protocorms continued to grow and developed into seedlings 5 months after sowing. MAS, month after sowing. Black scale bar = 100 μm. Red scale bar = 0.5 cm.
Protocorms from wild-type plants continued to develop and grow into young seedlings at approximately 80–90 days after sowing (DAS, Figures 2A, B). Instead of growing into individual seedlings, protocorms of the transgenic 35S:PaLEC1-GFP #1 plants developed into various structures (Figure 2A) that did not completely resemble seedlings of wild-type seedlings at 80–90 DAS (Table 1). For example, approximately 25% germinated protocorms underwent uncontrolled cell division and formed unorganized callus-like structures (callus type, Figure 2C), which is similar to somatic embryonic calli commonly observed in other plant species (Zimmerman, 1993; Yang and Zhang, 2010; Ikeuchi et al., 2013). Around 48% of protocorms, on the other hand, developed into a fan-like structure with multiple meristems forming on the surface (multiple-meristem type, Figure 2C). Around 27% protocorms formed a dorsal-fin-like structure with newly emerging leaves growing from it (dorsal-fin type, Figure 2C). The callus-like structure was capable of acquiring meristematic fate and developed into shoots (Supplementary Figure S2). Even though only one shoot was developed from the dorsal-fin type structure, new shoots could occasionally emerge from the base of the dorsal-fin type structure.
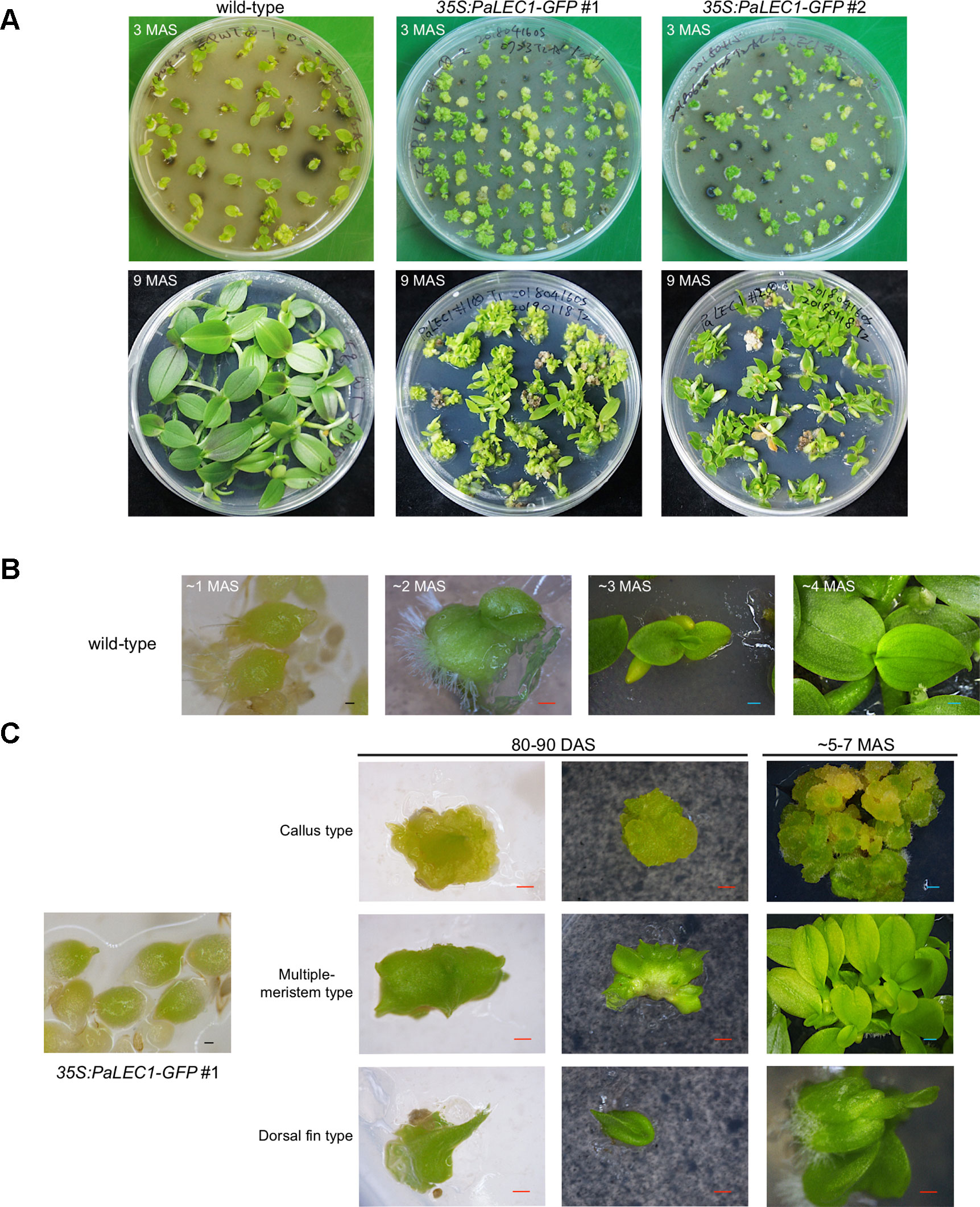
Figure 2 Developmental processes of T1 35:PaLEC1-GFP #1 transgenic plants. (A) Developing seedlings of wild-type plant, 35:PaLEC1-GFP #1, and 35:PaLEC1-GFP #2 transgenic plants at 3 and 9 months after germination. (B) Germinated protocorms of wild-type plants developed into young seedlings at 1, 2, 3, and 4 months after germination. (C) Germinated protocorms of 35:PaLEC1-GFP #1 transgenic plants developed into callus-type, multiple-meristem type, and dorsal fin-like structures. MAS, month after sowing; DAS, day after sowing; Red scale bar = 500 μm. Blue scale bar = 1 mm.
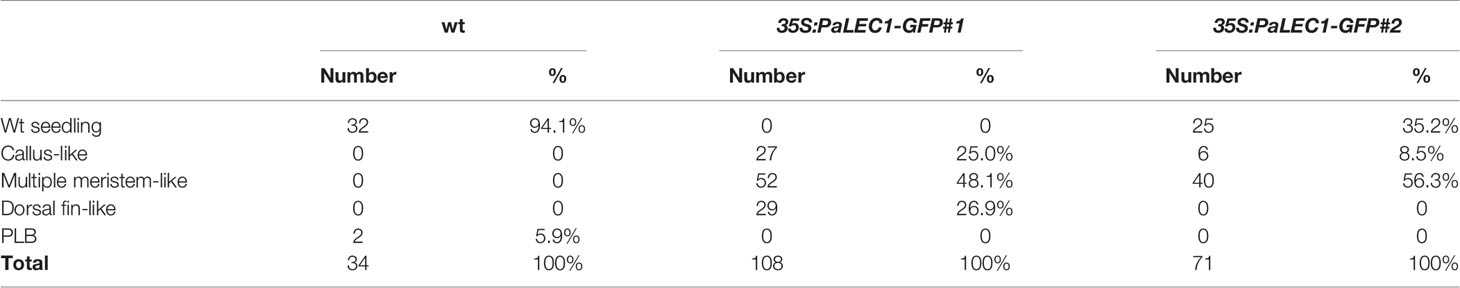
Table 1 Different types of structures were formed from germinating protocorms of 35S:PaLEC1-GFP transgenic lines. Number of the represented type (number) and percentage of the represented type (%) of structures are shown.
Even though PaLEC1-GFP protein could not be detected in T0 35S:PaLEC1-GFP #2 transgenic plants, some of the germinated T1 seeds also developed into structures that were similar to those observed in 35S:PaLEC1-GFP #1 line 80–90 DAS (Supplementary Figure S3). Approximately 35.2% of protocorms grew and became normal seedlings as seen for wild-type protocorms (Table 1). Around 56.3% protocorms developed into multiple-meristem type structures and 8.5% protocorms developed into callus-like structures. No dorsal-fin type structure was observed in the T1 35S:PaLEC1-GFP #1 line. Quantitative RT-PCR and western blotting were conducted to validate and re-evaluate the expression of PaLEC1-GFP mRNA and its encoded protein in 35S:PaLEC1-GFP #1 and 35S:PaLEC1-GFP #2. As compared to 35S:PaLEC1-GFP #1 seedlings, 35S:PaLEC1-GFP #2 had much less PaLEC1-GFP mRNA than 35S:PaLEC1-GFP #1 line (Figure 3). Because GFP monoclonal antibody may be less sensitive than polyclonal antibodies in detecting PaLEC1-GFP protein, PaLEC1 polyclonal antibodies were generated. Similar to hygromycin-resistant T0 35S:PaLEC1-GFP #1 parent, PaLEC1-GFP protein was expressed in high abundance in callus-like, multiple-meristem, and dorsal-fin types of structures germinated from T1 seeds (Figure 3). Using the PaLEC1 polyclonal antibodies, PaLEC1-GFP protein was also detected in callus-like tissues but was barely detectable in multiple-meristem-like structures and wild-type-like seedlings of 35S:PaLEC1-GFP #2 transgenic line (Figure 3), indicating PaLEC1-GFP protein was expressed but accumulated in very low abundance. We noticed that PaLEC1-induced callus tissues had a reduced amount of ribulose bisphosphate carboxylase large subunit (RbcL) protein (Figure 3), that is consistent with the pale-yellow appearance of PaLEC1-induced callus tissues.
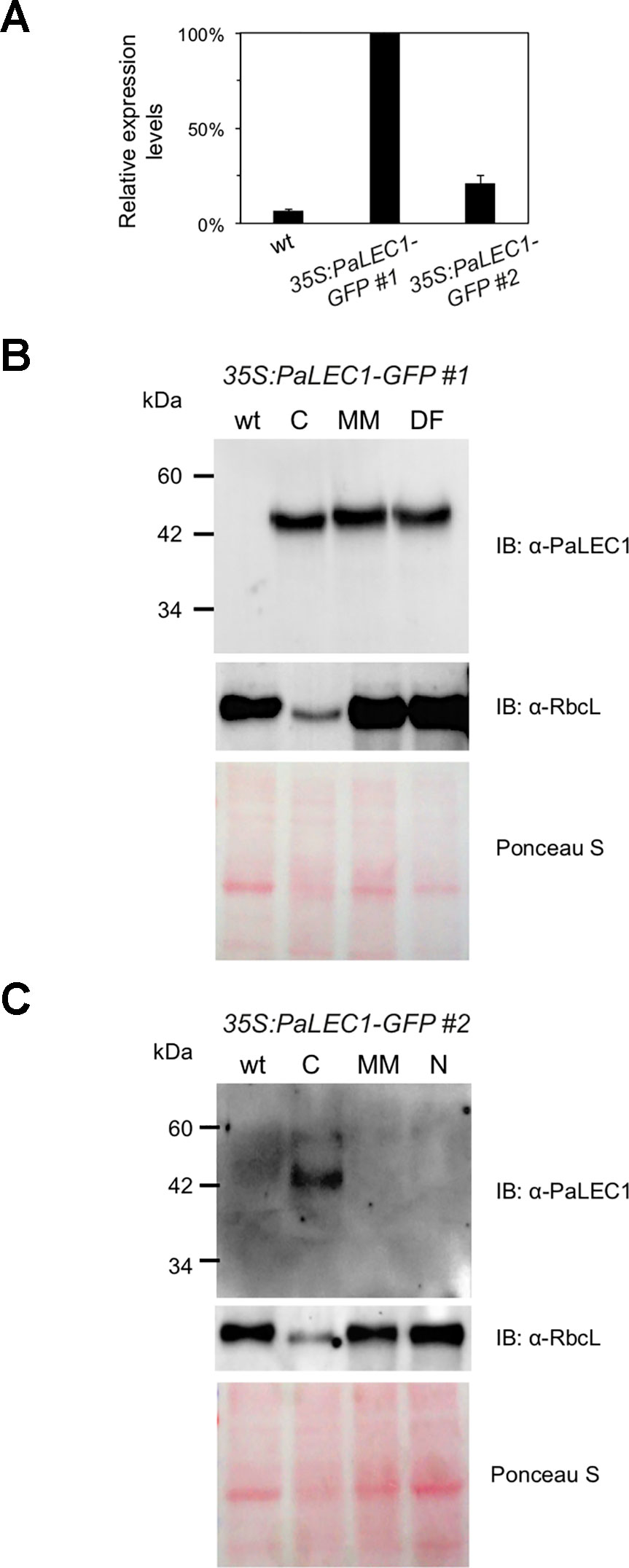
Figure 3 Expression of PaLEC1-GFP protein in 35:PaLEC1-GFP transgenic lines. (A) Quantitative RT-PCR analysis of expression of PaLEC1-GFP in wild-type (wt) and two 35:PaLEC1-GFP transgenic lines. (B) Western blotting showing expression of PaLEC1-GFP and PaRbcL proteins in callus-type, multiple-meristem type, and dorsal fin-like embryonic tissues of 35:PaLEC1-GFP#1 plants. (C) Western blotting showing expression of PaLEC1-GFP and PaRbcL proteins in callus-type, embryonic tissues of 35:PaLEC1-GFP#2 plants. Blots stained with Ponceau S staining were used to visualize total loaded protein. wt, wild-type plants; C, callus-like tissues; MM, multiple meristem-like structures; DF, dorsal fin-like structures; N, normal wt-like tissues.
To gain further structural details of the PaLEC1-induced structures, histological section and histochemical staining were conducted. As described previously (Yeung, 2017), shoot apical meristem was initiated from the apical end of a protocorm in the wild-type plants (Figure 4). In the 35S:PaLEC1-GFP transgenic plants, active cell division appeared to take place on the outer cell layer (epidermal cells) of developing protocorms (early stage, Figure 4). Protocorms then grew into friable and unstructured cellular masses with small cells showing dense cytoplasm on the surface (late stage, Figure 4). As for multiple meristem-type structures, the dividing cells on the surface of developing protocorms gradually organized into the shoot apical meristems at the early stage (Figure 4). As development proceeded, leaf primordia started to emerge from the organized shoot apical meristems (late stage, Figure 4) and multiple shoots were formed.
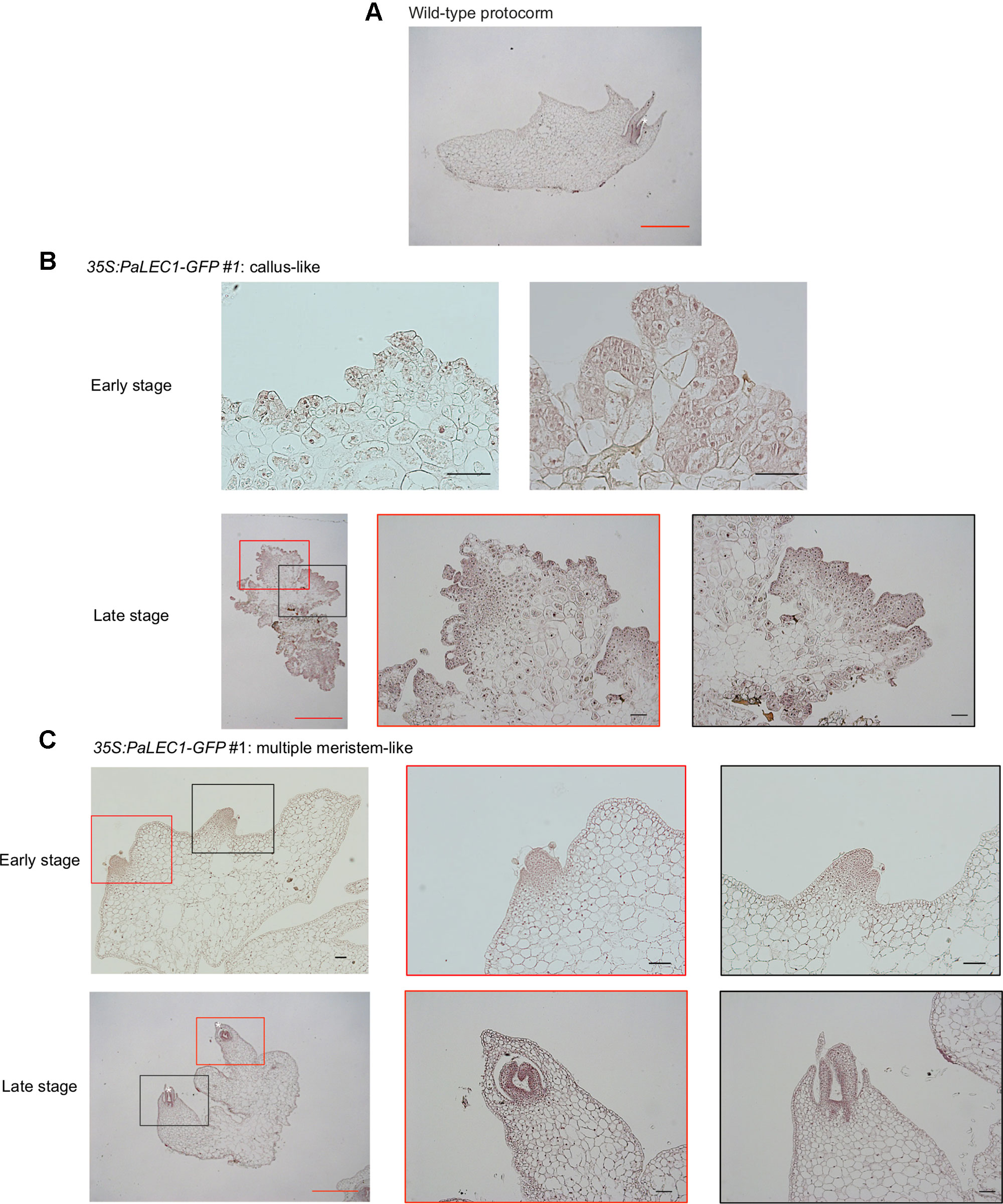
Figure 4 Histological examination of embryonic tissues of 35:PaLEC1-GFP#1 transgenic plants. (A) Light micrograph of 10 μm longitudinal section of developing wild-type protocorm stained with hematoxylin. (B) Light micrograph of a 10 μm section of the early and late stages of callus-like tissues derived from protocorms of the T1 35:PaLEC1-GFP#1 seeds. (C) Light micrograph of a 10 μm section of the early and late stages of multiple meristem-like tissues derived from protocorms of the T1 35:PaLEC1-GFP#1 seeds. Shoot apical meristem (SAM) with the emerging leaf primordia are marked by a white asterisk. For panel (B and C), low and high magnification of structures marked with red and black rectangles are shown. Red scale bar = 1 mm. Black scale bar = 100 μm.
Overexpression of LEC1 gene is known to be a potent inducer of somatic embryos (Lotan et al., 1998; Mu et al., 2008; Horstman et al., 2017a). We were therefore interested in asking what types of somatic embryonic structures could be induced by PaLEC1 in P. equestris. To do so, protocorms were used as the explants for induction of somatic embryonic tissues. As described previously (Zhao et al., 2008; Mahendran and Bai, 2012; Fang et al., 2016), cutting protocorms of wild-type plants induced green PLBs (Figure 5). Cutting protocorms of T1 35S:PaLEC1-GFP #1 transgenic line, on the other hand, induced callus-like tissues and pale yellow-green protrusions (Figure 5). The appearance of pale yellow-green color is consistent with a previous report stating that overexpressing LEC1 downregulates light responsive genes in Arabidopsis (Junker et al., 2012). Protrusions and callus-like tissues continued to grow and develop into unorganized thick and flat tissues, which then eventually grew into multiple-meristem type structures that were capable of producing multiple shoots (Figure 5). Using protocorms of T1 35S:PaLEC1-GFP #2 line as explants also induced callus-like and multiple-meristem type structures (Figure 5). Taken together, we conclude that PaLEC1-induced tissues have the ability to establish new shoot apical meristems for shoot regeneration.

Figure 5 Clonal propagation from protocorm-based explants of 35:PaLEC1-GFP#1 transgenic plants. (A) PLBs generated from cutting of wild-type protocorms. (B) Callus-like tissues produced by cutting of 35:PaLEC1-GFP#1 protocorms. (C) Callus-like tissues generated from 35:PaLEC1-GFP#1 grew into multiple-shoot like structures. (D) Callus-like and multiple shoot-like tissues produced by cutting of 35:PaLEC1-GFP#2 protocorms. For each panel, two independent clusters of tissues were photographed and showed. Red scale bar = 500 μm. Blue scale bar = 1 mm.
To evaluate whether PaLEC1-GFP protein was regulated post-transcriptionally as somatic embryonic calli programmed to initiate shoot formation, PaLEC1-GFP protein was monitored by western blotting. As shown in Supplementary Figure S4, PaLEC1-GFP protein remained stable throughout the process.
To evaluate the embryonic identity of PaLEC1-GFP overexpressing somatic tissues, expression of previously identified embryonic-specific genes (Fang et al., 2016) were examined in T1 35S:PaLEC1-GFP #1 protocorm-derived young seedling tissues. Expression of the embryonic TFs ABSCISIC ACID INSENSITITVE3-like1 (PeABI3L1), PeABI3L2, BABY BOOM (PeBBM), FUSCA3 (PeFUS3), and WRINKLED1 (PeWRI1) and seed storage proteins, 7S GLOBULIN1 (Pe7S-1), Pe7S-2, OLEOCIN1 (PeOLE1), and PeOLE2 genes were confirmed to be preferentially expressed in ovary tissues collected at 130 days after pollination (DAP) of P. equestris (Figure 6). As expected, overexpression of PaLEC1-GFP caused significant induction of embryonic TFs PeFUS3 and PeABI3L1, and seed storage proteins, Pe7S-1, Pe7S-2, and PeOLE2 genes (Figure 6). Overexpression of PaLEC1-GFP protein only led to mild elevation of PeABI3L2 mRNA and did not affect expression of PeBBM, PeWRI1, and PeOLE1 genes (Figure 6), suggesting that PaLEC1 activates only part of the embryonic program. Similar expression patterns were also confirmed in independently collected young seedlings and callus tissues except PeABI3L2 whose expression was not induced in PaLEC1-induced callus tissues (Supplementary Figures S5A, B). It is likely that the moderate level of PaLEC1-GFP (Supplementary Figure S5B) in callus was not enough to induce PeABI3L2. Activation of PeFUS3 was also confirmed in leaf tissues of five independent T0 35S:PaLEC1-GFP transgenic plants (Supplementary Figure S6).
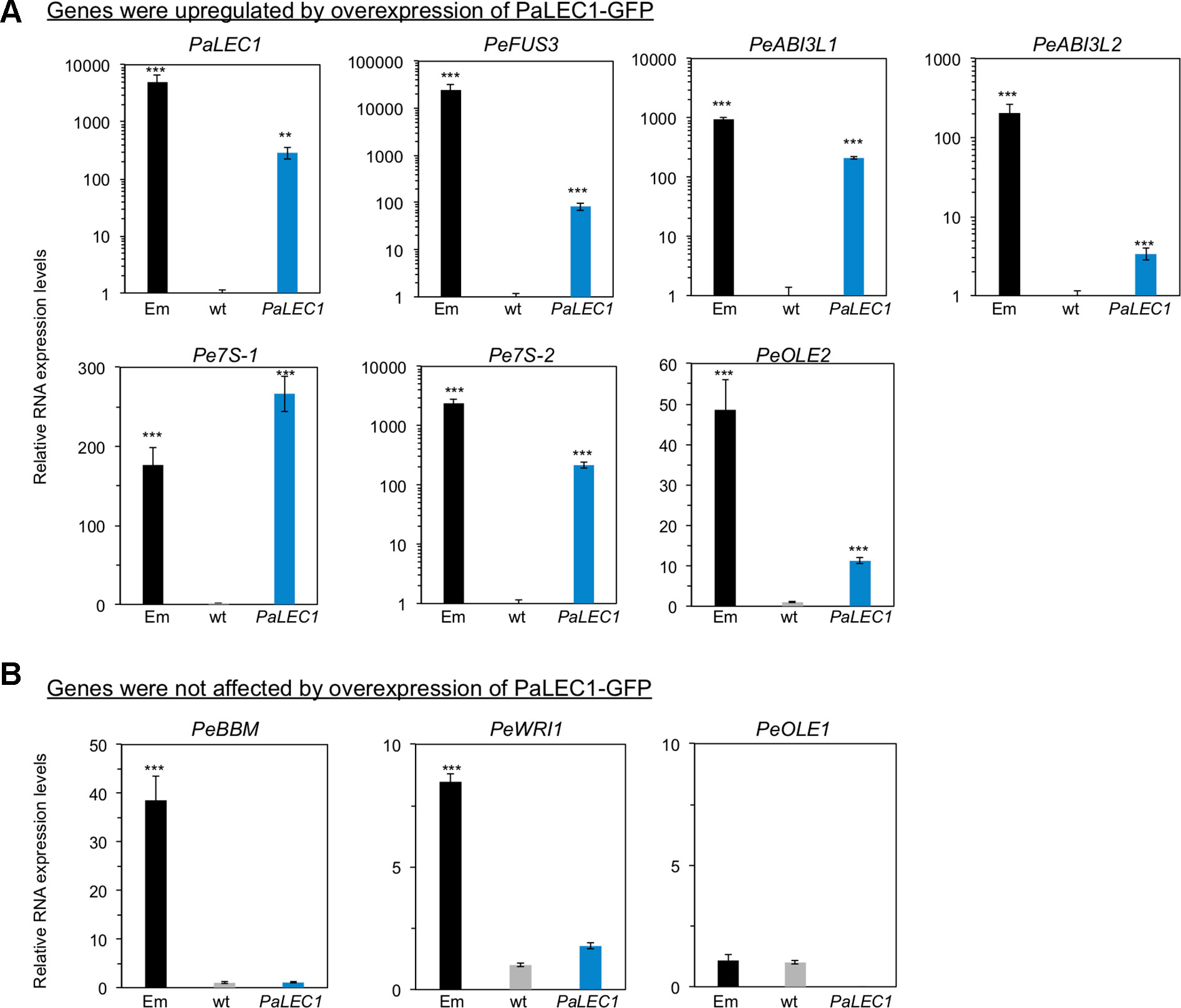
Figure 6 Expression of embryonic genes in 35:PaLEC1-GFP#1 transgenic plants. (A) Expression of indicated embryonic genes was induced in 35:PaLEC1-GFP#1 transgenic plants. (B) Expression of indicated embryonic genes was not induced significantly by overexpression of PaLEC1-GFP protein. Em, zygotic embryonic tissues collected at 130 day after pollination. wt, wild-type plant. PaLEC1, 35:PaLEC1-GFP#1 transgenic plants. ***, P < 0.001; **, P < 0.01. Three independent biological replicates were performed.
Nuclear Transcription Factor-Y (NF-Y) is an evolutionarily conserved, DNA-binding, trimeric protein complex composed of NF-YA, NF-YB, and NF-YC subunits (Dolfini et al., 2012). LEC1 encodes an atypical subunit of the (NF-YB) CCAAT-binding TF. To validate subcellular localization of PaLEC1, confocal microscopy was used to examine the localization of PaLEC1-GFP protein in transgenic plants. As predicted, PaLEC1-GFP was predominantly localized in the nuclei in the cells of the 35S:PaLEC1-GFP#1 line (Figure 7). The nuclear localized GFP signal was detected in all of the cells investigated in the 35S:PaLEC1-GFP#1 line. PaLEC1-GFP protein, however, was only detected in a few cells in callus tissues and was therefore much less abundant in 35S:PaLEC1-GFP#2 transgenic line (Figure 7), supporting the low expression level shown by western blot. Hence, PaLEC1 protein is a nuclear protein.
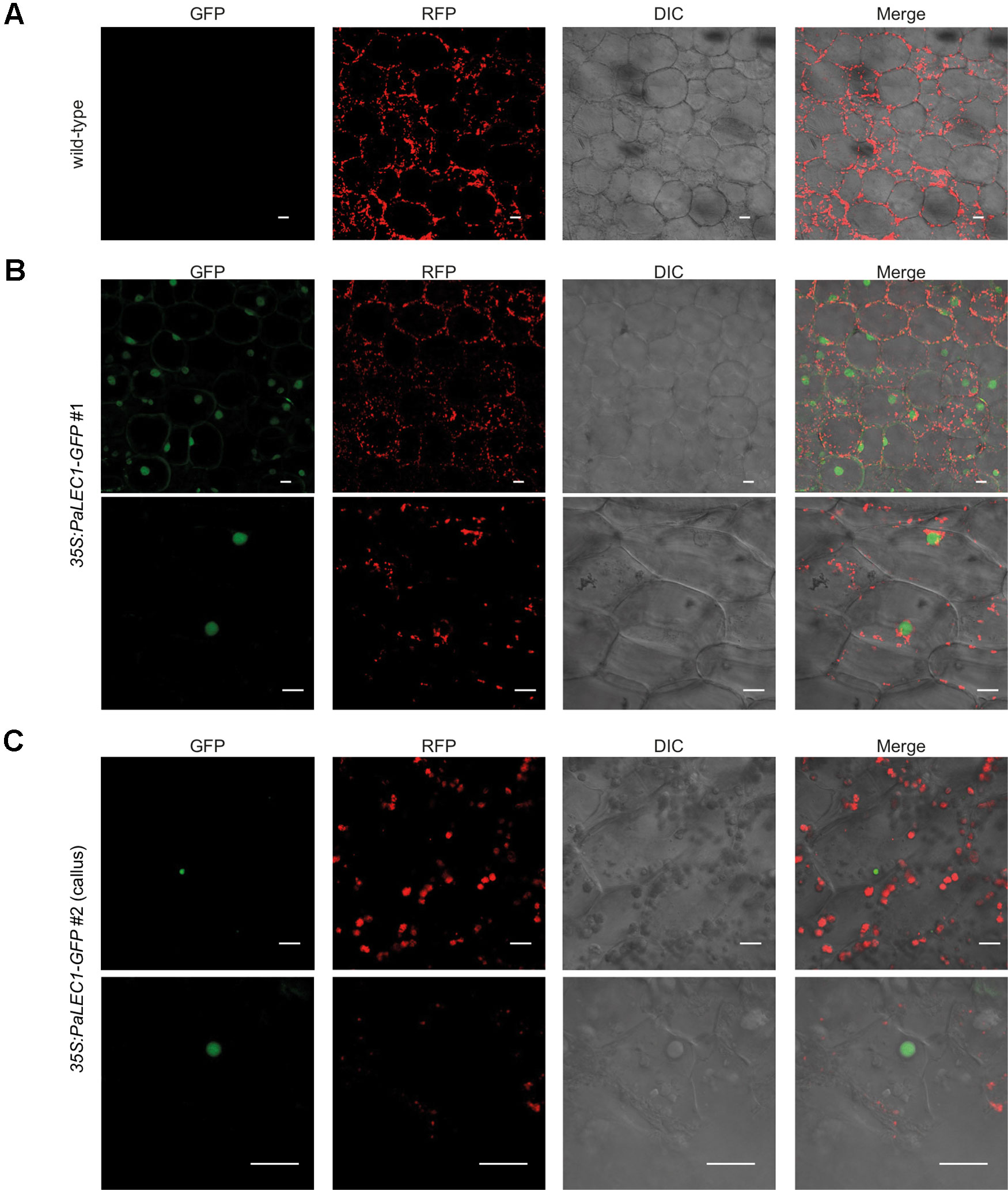
Figure 7 Nuclear localization of PaLEC1-GFP. (A) No GFP signal was detected in wild-type plants. (B) PaLEC1-GFP was detected in nuclei of cells in roots of 35:PaLEC1-GFP#1 transgenic line. Nuclear localization of PaLEC1-GFP was confirmed in multiple (more than three) independent seedlings. (C) PaLEC1-GFP was detected in very few nuclei within callus cells of 35:PaLEC1-GFP#2 transgenic line. This sparsely distribution of nuclear PaLEC1-GFP signal was confirmed in multiple (more than three) protocorm-derived callus tissues. Photos with lower magnification are shown in the upper panel in panels (B and C). Photos with higher magnification are shown in the bottom panel in panels (B and C). GFP, photographs taken in the GFP channel. RFP, photographs taken in the RFP channel. DIC, differential interference contrast images. Merge, differential interference contrast images of cells superimposed with PaLEC1 (GFP) and chloroplast (RFP) channels. Scale bar = 20 μm.
It has been reported that LEC1 regulates many embryonic-specific TFs and storage proteins by directly binding to their promoters through the CCAAT DNA element (Lee et al., 2003; Gnesutta et al., 2017; Pelletier et al., 2017). To investigate whether PaLEC1 activates its targets in the same manner, we retrieved the 1 kb DNA sequences upstream of the start codon of PeFUS3, PeABI3L1, PeABI3L2, PeBBM, PeWRI1, Pe7S-1, Pe7S-2, PeOLE1, and PeOEL2 (Cai et al., 2015) and surveyed the presence of the CCAAT element. Because of a large gap within the promoter regions of PeABI3L2, it was omitted from further study. Intriguingly, two to five CCAAT DNA elements were found within the 1 kb promoter sequences of PeFUS3, PeABI3L1, Pe7S-1, Pe7S-2, and PeOLE2 (Supplementary Figure S7) whose expression was induced by overexpression of PaLEC1 (Figure 6). None or only one CCAAT element was present in promoters of PeBBM, PeWRI1, and PeOLE1 (Supplementary Figure S7) whose expression was not induced in the PaLEC1 overexpressor (Figure 6). The correlation between the number of CCAAT elements and activation capability of PaLEC1-GFP suggests that PaLEC1-GFP protein may activate its targets in a CCAAT DNA element dosage-dependent manner.
To test whether PaLEC1-GFP binds the CCAAT element directly, ChIP using PaLEC1 antibody followed by PCR analysis was conducted. Because PeFUS3 and Pe7S-2 mRNAs were significantly upregulated by PaLEC1 and tandem CCAAT elements are present in their promoters (Figure 8 and Supplementary Figure S8), PeFUS3 and Pe7S-2 were chosen for further ChIP analysis. Histone H3 antibody was used as a positive control and no antibody was included as the negative control. We found that PaLEC1-GFP protein interacted with the PeFUS3 promoter region containing the CCAAT element but did not interact with PeBBM promoter that lacks the CCAAT element (Figure 8). Similarly, PaLEC1-GFP was found to interact with the Pe7S-2 promoter carrying the CCAAT element but not to the PeBBM promoter lacking the element (Figure 8). These results were confirmed in three independent biological samples. Taken together, it is very likely that PaLEC1 activates PeFUS3 and Pe7S-2 through binding to the CCAAT element-containing promoters.
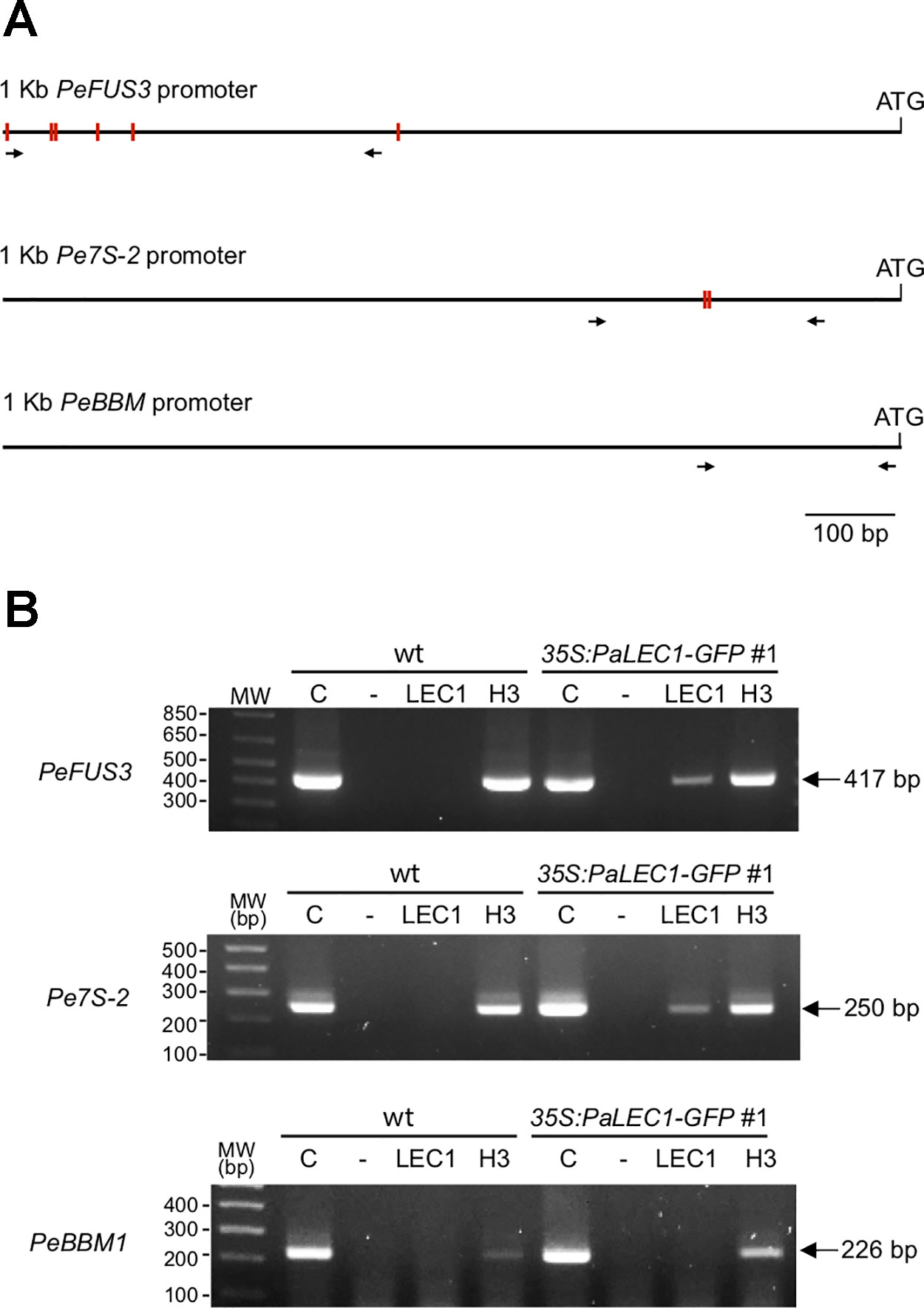
Figure 8 Interaction between PaLEC1-GFP protein and the CCAAT element containing promoters. (A) Schematic representation of distribution of the CCAAT elements (red rectangles) in 1 kb upstream of the start codon of PeFUS3 and Pe7S-2 genomic DNAs. No CCAAT element was found in 1 Kb promoter region of PeBBM. Primers used for PCR reactions were shown as black arrows. (B) PCR of chromatin immunoprecipitation isolated DNA fragments to confirm interaction between PaLEC1-GFP protein and the CCAAT-containing promoters of the indicated genes. C, total cell lysate; “-”, no antibody control; PaLEC1, PaLEC1 antibody precipitated chromatin; H3, histone H3 antibody precipitated chromatin. The amplified PCR fragments were verified by DNA sequencing. The same result was validated in two other independent biological samples (Supplementary Figure S8).
Our recent comparative transcriptome study provided evidence that PLB regeneration does not follow the somatic embryogenesis program (Fang et al., 2016) and raises a question about the morphology and anatomy of orchid somatic embryos. In this study, overexpression of PaLEC1 was utilized to induce the somatic embryonic program in P. equestris. Similar to PLB induction, active cell division is a prerequisite for PaLEC1-induced somatic embryonic structures (Figures 4B, C). For PLB development, the polarized growth of proliferating cells gives rise to small protuberances with a distinct gradient of cell size, with the smaller cells occupying the future shoot pole and larger and vacuolated cells forming the base of the structure (Lee et al., 2013). For PaLEC1-induced somatic embryonic tissues, on the other hand, diverse types of somatic embryonic structures were formed from protocorms or explants of the two 35S:PaLEC1-GFP transgenic lines regardless of the large difference in the amounts of PaLEC1-GFP protein accumulated. The most common type (~50% in both transgenic lines) was tissue clusters containing multiple shoot apical meristems (multiple-meristem type). For this type of regeneration, multiple meristems were initiated directly from developing protocorms and establishment of unorganized callus tissue did not seem to be a prerequisite. It is important to note that this type of regeneration is very different from direct shoot regeneration during which leaf primordia are generated directly (Košir et al., 2004; Wu and Chen, 2008; Yam and Arditti, 2017). In addition to producing multiple meristems for shoot regeneration, new shoots could also be produced indirectly from callus-type tissue (Supplementary Figure S2). The frequency of obtaining the callus-type tissue was higher in the strong PaLEC1-GFP overexpressors than in the weak ones (Table 1), indicating formation of the embryonic callus may be PaLEC1 dosage-dependent. Dosage-dependent induction of somatic embryogenesis has also been reported for Arabidopsis BBM and PLT2 (Horstman et al., 2017b). Intriguingly, the fin-like structure was only observed in the strong overexpressor (PaLEC1-GFP #1) and normal protocorm-derived seedlings were only observed in the weak overexpressor (PaLEC1-GFP #2). Taken together these results indicate that PaLEC1-induced embryonic tissues were morphologically distinct from PLBs. Regardless of the type of structures induced by PaLEC1, all embryonic structures documented here are totipotent and capable of generating clonal orchid seedlings.
The embryonic identity of PaLEC1-induced embryonic tissues was verified by expression of embryonic TFs, PeFUS3, PeABI3L1, PeABI3L2, as well as seed storage proteins Pe7S-1, and Pe7S-2. In Arabidopsis, FUS3, ABI3, and WRI1 are reported to be the direct targets of LEC1 (Pelletier et al., 2017). However, overexpression of PaLEC1 failed to induce PeWRI1. LEC1 is reported to directly activate WRI1 or indirectly acts through upregulation of FUS3 and LEC2 (Baud et al., 2007; Mu et al., 2008; Shen et al., 2010; Wang and Perry, 2013; Pelletier et al., 2017). Despite a high level of PaLEC1 protein and elevation of PeFUS3 mRNA in 35S:PaLEC1-GFP#1 transgenic line, we did not observe induction of PeWRI1, suggesting a PaLEC1-independent pathway is required to induce PeWRI1 in Phalaenopsis orchids. Similar to Arabidopsis on the other hand, PaLEC1 cannot activate PeBBM and only induces a subset of storage proteins (West et al., 1994; Pelletier et al., 2017). In Arabidopsis, BBM acts upstream of the LEC1-FUS3-ABI3-LEC2 network to induce the somatic embryogenesis program (Horstman et al., 2017b). Whether a similar regulatory module exists in Phalaenopsis orchids remains to be tested. Taken together, despite being a potent inducer of somatic embryogenesis in Phalaenopsis orchids, the PaLEC1-dependent regulatory network is similar but not identical to that reported in Arabidopsis.
LEC1 belongs to a distinct subgroup of the NF-YB family that is diverged from non-LEC1-type NF-YB genes in non-seed vascular plants (Xie et al., 2008). It has been proposed that LEC1-type NF-YB is recruited into a seed-specific regulatory network during evolution. Similar to Arabidopsis LEC1-type NF-YB TFs (Calvenzani et al., 2012; Laloum et al., 2013), PaLEC1 also binds to the CCAAT-containing promoters of its downstream genes, PeFUS3 and Pe7S-2. Therefore, PaLEC1 is very likely to regulate its targets by directly acting through binding of the CCAAT DNA element. Even though one CCAAT element was present in the 1 kb promoters of PeWRI1 and PeOLE1 genes (Supplementary Figure S4), PeWRI1 and PeOLE1 mRNAs were not activated by overexpression of PaLEC1-GFP protein, indicating the CCAAT DNA element may not be sufficient for PaLEC1 binding. This notion is supported by studies of Arabidopsis and soybean that the G-box (CACGTG), abscisic acid response element (ABRE)-like (C/G/T)ACGTG(G/T)(A/C), and RY (CATGCA) motifs are significantly overrepresented in LEC1 target genes (Pelletier et al., 2017). The involvement of G-box, ABRE-like, and RY motifs in PaLEC1 functions remain to be determined.
In conclusion, our study of PaLEC1-induced somatic embryogenesis has demonstrated that somatic tissues of Phalaenopsis orchids do not resemble PLB, a result that is consistent with our previous comparative transcriptome studies. In addition to confirming the role of PaLEC1 in activating the somatic embryonic program, we obtained evidence supporting a direct role for PaLEC1 in controlling its targets and revealed the potential of PaLEC1-induce somatic embryonic tissues for orchid propagation. Conservation of the developmental processes and gene regulatory networks controlled by PaLEC1 is consistent with the notion that LEC1 is a major regulator of embryogenesis.
All datasets generated for this study are included in the article/Supplementary Material.
S-CF designed the experiments, coordinated the studies, and wrote the manuscript. J-CC maintained and characterized transgenic orchid plants, and developed the ChIP protocol. J-CC and H-YL carried out the molecular biology experiments. C-GT developed and coordinated the orchid transformation process. All the authors have read and approved the final manuscript.
This work was supported by the Innovative Translational Agricultural Research Grants (to S-CF); and in part by a grant (to S-CF) from the Biotechnology Center in Southern Taiwan, Academia Sinica.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Yao-Cheng Lin for his assistance with bioinformatics, Ms. Miao-Ju Wei for her technical support during the initial work on transgenic orchid plants, the Confocal Microscopy Core, and Greenhouse Core Facilities for their supporting services, and Ms. Miranda Loney for English editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01594/full#supplementary-material
Akasaka-Kennedy, Y., Tomita, K. O., Ezura, H. (2004). Efficient plant regeneration and Agrobacterium-mediated transformation via somatic embryogenesis in melon (Cucumis melo L.). Plant Sci. 166 (3), 763–769. doi: 10.1016/j.plantsci.2003.11.020
Arditti, J., Krikorian, A. D. (1996). Orchid micropropagation: the path from laboratory to commercialization and an account of several unappreciated investigators. Bot. J. Linean Soc. 122 (3), 183–241. doi: 10.1111/j.1095-8339.1996.tb02073.x
Baud, S., Mendoza, M. S., To, A., Harscoet, E., Lepiniec, L., Dubreucq, B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50 (5), 825–838. doi: 10.1111/j.1365-313X.2007.03092.x
Begum, A. A., Tamaki, M., Tahara, M., Kako, S. (1994). Somatic embryogenesis in Cymbidium through in vitro culture of inner tissue of protocorm-like bodies. J. Jpn Soc. Hortic. Sci. 63 (2), 419–427. doi: 10.2503/jjshs.63.419
Belarmino, M. M., Mii, M. (2000). Agrobacterium-mediated genetic transformation of a Phalaenopsis orchid. Plant Cell Rep. 19 (5), 435–442. doi: 10.1007/s002990050752
Bentsink, L., Koornneef, M. (2008). Seed dormancy and germination. Arabidopsis Book 6, e0119. doi: 10.1199/tab.0119
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L., et al. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14 (8), 1737–1749. doi: 10.1105/tpc.001941
Braybrook, S. A., Harada, J. J. (2008). LECs go crazy in embryo development. Trends In Plant Sci. 13 (12), 624–630. doi: 10.1016/j.tplants.2008.09.008
Brettschneider, R., Becker, D., Lorz, H. (1997). Efficient transformation of scutellar tissue of immature maize embryos. Theor. Appl. Genet. 94 (6-7), 737–748. doi: 10.1007/s001220050473
Burger, W. C. (1998). The question of cotyledon homology in angiosperms. Bot. Rev. 64 (4), 356–371. doi: 10.1007/BF02857623
Cai, J., Liu, X., Vanneste, K., Proost, S., Tsai, W. C., Liu, K. W., et al. (2015). The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 47 (1), 65–72. doi: 10.1038/ng.3149
Calvenzani, V., Testoni, B., Gusmaroli, G., Lorenzo, M., Gnesutta, N., Petroni, K., et al. (2012). Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PloS One 7 (8), e42902. doi: 10.1371/journal.pone.0042902
Chavarri, M., Vegas, A., Zambrano, A. Y., Demey, J. R. (2004). Transformation of mango somatic embryos by biobalistics. Interciencia 29 (5), 261–266.
Chen, J. T., Chang, W. C. (2004). Induction of repetitive embryogenesis from seed-derived protocorms of Phalaenopsis amabilis var. formosa Shimadzu. In Vitro Cell Dev. Biol. Plant 40 (3), 290–293. doi: 10.1079/IVP2003527
Chen, J., Chang, W. (2006). Direct somatic embryogenesis and plant regeneration from leaf explants of Phalaenopsis amabilis. Biol. Plantarum 50 (2), 169–173. doi: 10.1007/s10535-006-0002-8
Chen, J. C., Fang, S. C. (2016). The long pollen tube journey and in vitro pollen germination of Phalaenopsis orchids. Plant Reprod. 29 (1-2), 179–188. doi: 10.1007/s00497-016-0280-z
Chen, L. R., Chen, J. T., Chang, W. C. (2002). Efficient production of protocorm-like bodies and plant regeneration from flower stalk explants of the sympodial orchid Epidendrum radicans. In Vitro Cell Dev. Biol. Plant 38 (5), 441–445. doi: 10.1079/IVP2002315
Chen, W.-H., Kao, Y.-L., Tang, C.-Y. (2009). Method for producing polyploid plants of orchids (USA patent application).
Chen, T. K., Yang, H. T., Fang, S. C., Lien, Y. C., Yang, T. T., Ko, S. S. (2016). Hybrid-Cut: an improved sectioning method for recalcitrant plant tissue samples. J. Vis Exp. (117), e54754. doi: 10.3791/54754
Chugh, S., Guha, S., Rao, I. U. (2009). Micropropagation of orchids: a review on the potential of different explants. Sci. Hortic. 122 (4), 507–520. doi: 10.1016/j.scienta.2009.07.016
Dolfini, D., Gatta, R., Mantovani, R. (2012). NF-Y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Biol. 47 (1), 29–49. doi: 10.3109/10409238.2011.628970
Dressler, R. L. (1993). Phylogeny and classification of the orchid family (Portland, Oregon: Dioscorides Press).
Fang, S. C., de los Reyes, C., Umen, J. G. (2006). Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PloS Genet. 2 (10), e167. doi: 10.1371/journal.pgen.0020167
Fang, S. C., Chen, J. C., Wei, M. J. (2016). Protocorms and protocorm-like bodies are molecularly distinct from zygotic embryonic tissues in Phalaenopsis aphrodite. Plant Physiol. 171 (4), 2682–2700. doi: 10.1104/pp.16.00841
Feher, A. (2015). Somatic embryogenesis - Stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta 1849 (4), 385–402. doi: 10.1016/j.bbagrm.2014.07.005
Fitch, M. M., Manshardt, R. M., Gonsalves, D., Slightom, J. L. (1993). Transgenic papaya plants from Agrobacterium-mediated transformation of somatic embryos. Plant Cell Rep. 12 (5), 245–249. doi: 10.1007/BF00237128
Gaj, M. D., Zhang, S., Harada, J. J., Lemaux, P. G. (2005). Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222 (6), 977–988. doi: 10.1007/s00425-005-0041-y
Garcês, H. M., Koenig, D., Townsley, B. T., Kim, M., Sinha, N. R. (2014). Truncation of LEAFY COTYLEDON1 protein is required for asexual reproduction in Kalanchoë daigremontiana. Plant Physiol. 165 (1), 196–206. doi: 10.1104/pp.114.237222
Gnesutta, N., Saad, D., Chaves-Sanjuan, A., Mantovani, R., Nardini, M. (2017). Crystal structure of the Arabidopsis thaliana L1L/NF-YC3 histone-fold dimer reveals specificities of the LEC1 Family of NF-Y subunits in plants. Mol. Plant 10 (4), 645–648. doi: 10.1016/j.molp.2016.11.006
Gow, W. P., Chen, J. T., Chang, W. C. (2009). Effects of genotype, light regime, explant position and orientation on direct somatic embryogenesis from leaf explants of Phalaenopsis orchids. Acta Physiol. Plant 31 (2), 363–369. doi: 10.1007/s11738-008-0243-6
Gow, W. P., Chen, J. T., Chang, W. C. (2010). Enhancement of direct somatic embryogenesis and plantlet growth from leaf explants of Phalaenopsis by adjusting culture period and explant length. Acta Physiol. Plant 32 (4), 621–627. doi: 10.1007/s11738-009-0438-5
Guan, Y., Li, S. G., Fan, X. F., Su, Z. H. (2016). Application of somatic embryogenesis in woody plants. Front. Plant Sci. 7, 938. doi: 10.3389/fpls.2016.00938
Guo, W. L., Chang, Y. C. A., Kao, C. Y. (2010). Protocorm-like Bodies initiation from root tips of Cyrtopodium paranaense (Orchidaceae). HortScience 45 (9), 1365–1368. doi: 10.21273/HORTSCI.45.9.1365
Harding, E. W., Tang, W., Nichols, K. W., Fernandez, D. E., Perry, S. E. (2003). Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol. 133 (2), 653–663. doi: 10.1104/pp.103.023499
Hecht, V., Vielle-Calzada, J. P., Hartog, M. V., Schmidt, E. D., Boutilier, K., Grossniklaus, U., et al. (2001). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127 (3), 803–816. doi: 10.1104/pp.010324
Horstman, A., Bemer, M., Boutilier, K. (2017a). A transcriptional view on somatic embryogenesis. Regenerat. (Oxf.) 4 (4), 201–216. doi: 10.1002/reg2.91
Horstman, A., Li, M., Heidmann, I., Weemen, M., Chen, B., Muino, J. M., et al. (2017b). The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 175 (2), 848–857. doi: 10.1104/pp.17.00232
Hossain, M. M., Kant, R., Van, P. T., Winarto, B., Zeng, S. J., da Silva, J. A. T. (2013). The application of biotechnology to orchids. Crit. Rev. Plant Sci. 32 (2), 69–139. doi: 10.1080/07352689.2012.715984
Ikeuchi, M., Sugimoto, K., Iwase, A. (2013). Plant callus: mechanisms of induction and repression. Plant Cell 25 (9), 3159–3173. doi: 10.1105/tpc.113.116053
Ishii, Y., Takamura, T., Goi, M., Tanaka, M. (1998). Callus induction and somatic embryogenesis of Phalaenopsis. Plant Cell Rep. 17 (6), 446–450. doi: 10.1007/s002990050423
Jones, D., Tisserat, B. (1990). Clonal propagation of orchids. Methods Mol. Biol. 6, 181–191. doi: 10.1385/0-89603-161-6:181
Joshee, N., Biswas, B. K., Yadav, A. K. (2007). Somatic embryogenesis and plant development in Centella asiatica L., a highly prized medicinal plant of the tropics. HortScience 42 (3), 633–637. doi: 10.21273/HORTSCI.42.3.633
Junker, A., Monke, G., Rutten, T., Keilwagen, J., Seifert, M., Thi, T. M., et al. (2012). Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 71 (3), 427–442. doi: 10.1111/j.1365-313X.2012.04999.x
Kareem, A., Radhakrishnan, D., Sondhi, Y., Aiyaz, M., Roy, M. V., Sugimoto, K., et al. (2016). De novo assembly of plant body plan: a step ahead of Deadpool. Regenerat. (Oxf.) 3 (4), 182–197. doi: 10.1002/reg2.68
Kim, J. Y., Adhikari, P. B., Ahn, C. H., Kim, D. H., Kim, Y. C., Han, J. Y., et al. (2019). High frequency somatic embryogenesis and plant regeneration of interspecific ginseng hybrid between Panax ginseng and Panax quinquefolius. J. Ginseng Res. 43 (1), 38–48. doi: 10.1016/j.jgr.2017.08.002
Košir, D., Škof, S., Luthar, Z. (2004). Direct shoot regeneration from nodes of Phalaenopsis orchids. Acta Agriu. Slovenica 83, 233–242.
Kull, T., Arditti, J. (2002). Orchid biology reviews and perspectives (Dordrecht; London: Kluwer Academic Publishers). doi: 10.1007/978-94-017-2500-2
Kuo, H. L., Chen, J. T., Chang, W. C. (2005). Efficient plant regeneration through direct somatic embryogenesis from Leaf explants of Phalaenopsis ‘Little Steve’. In Vitro Cell Devl. Biol. Plant 41 (4), 453–456. doi: 10.1079/IVP2005644
Kwong, R. W., Bui, A. Q., Lee, H., Kwong, L. W., Fischer, R. L., Goldberg, R. B., et al. (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15 (1), 5–18. doi: 10.1105/tpc.006973
Laloum, T., De Mita, S., Gamas, P., Baudin, M., Niebel, A. (2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18 (3), 157–166. doi: 10.1016/j.tplants.2012.07.004
Lee, H., Fischer, R. L., Goldberg, R. B., Harada, J. J. (2003). Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 100 (4), 2152–2156. doi: 10.1073/pnas.0437909100
Lee, Y. I., Hsu, S. T., Yeung, E. C. (2013). Orchid protocorm-like bodies are somatic embryos. Am. J. Bot. 100 (11), 2121–2131. doi: 10.3732/ajb.1300193
Li, H. Q., Sautter, C., Potrykus, I., PuontiKaerlas, J. (1996). Genetic transformation of cassava (Manihot esculenta Crantz). Nat. Biotech. 14 (6), 736–740. doi: 10.1038/nbt0696-736
Li, Z. T., Dhekney, S., Dutt, M., Van Aman, M., Tattersall, J., Kelley, K. T., et al. (2006). Optimizing Agrobacterium-mediated transformation of grapevine. In Vitro Cell Dev. Biol. Plant 42 (3), 220–227. doi: 10.1079/IVP2006770
Liao, Y. J., Tsai, Y. C., Sun, Y. W., Lin, R. S., Wu, F. S. (2011). In vitro shoot induction and plant regeneration from flower buds in Paphiopedilum orchids. In Vitro Cell Dev. Biol. Plant 47 (6), 702–709. doi: 10.1007/s11627-011-9370-7
Liau, C. H., You, S. J., Prasad, V., Hsiao, H. H., Lu, J. C., Yang, N. S., et al. (2003). Agrobacterium tumefaciens-mediated transformation of an Oncidium orchid. Plant Cell Rep. 21 (10), 993–998. doi: 10.1007/s00299-003-0614-9
Lin, H. Y., Chen, J. C., Wei, M. J., Lien, Y. C., Li, H. H., Ko, S. S., et al. (2014). Genome-wide annotation, expression profiling, and protein interaction studies of the core cell-cycle genes in Phalaenopsis aphrodite. Plant Mol. Biol. 84 (1-2), 203–226. doi: 10.1007/s11103-013-0128-y
Lotan, T., Ohto, M., Yee, K. M., West, M. A., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 (7), 1195–1205. doi: 10.1016/S0092-8674(00)81463-4
Lowe, K., Wu, E., Wang, N., Hoerster, G., Hastings, C., Cho, M. J., et al. (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell. 28 (9), 1998–2015. doi: 10.1105/tpc.16.00124
Mahendran, G., Bai, V. N. (2012). Direct somatic embryogenesis and plant regeneration from seed derived protocorms of Cymbidium bicolor Lindl. Sci. Hortic. 135, 40–44. doi: 10.1016/j.scienta.2011.12.003
Mcgranahan, G. H., Leslie, C. A., Uratsu, S. L., Martin, L. A., Dandekar, A. M. (1988). Agrobacterium-mediated transformation of salnut somatic embryos and regeneration of transgenic plants. Nat. Biotech. 6 (7), 800–804. doi: 10.1038/nbt0788-800
Min, L., Hu, Q., Li, Y., Xu, J., Ma, Y., Zhu, L., et al. (2015). LEAFY COTYLEDON1-CASEIN KINASE I-TCP15-PHYTOCHROME INTERACTING FACTOR4 network regulates somatic embryogenesis by regulating auxin homeostasis. Plant Physiol. 169 (4), 2805–2821. doi: 10.1104/pp.15.01480
Mondal, T. K., Bhattacharya, A., Ahuja, P. S., Chang, P. K. (2001). Transgenic tea [Camellia sinensis (L.) O. Kuntze cv. Kangra Jat] plants obtained by Agrobacterium-mediated transformation of somatic embryos. Plant Cell Rep. 20 (8), 712–720. doi: 10.1007/s002990100382
Mordhorst, A. P., Hartog, M. V., El Tamer, M. K., Laux, T., de Vries, S. C. (2002). Somatic embryogenesis from Arabidopsis shoot apical meristem mutants. Planta 214 (6), 829–836. doi: 10.1007/s00425-001-0700-6
Mu, J., Tan, H., Zheng, Q., Fu, F., Liang, Y., Zhang, J., et al. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148 (2), 1042–1054. doi: 10.1104/pp.108.126342
Murray, M. G., Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8 (19), 4321–4325. doi: 10.1093/nar/8.19.4321
Nishimura, G. (1981). Comparative morphology of Cattleya and Phalaenopsis (Orchidaceae) seedlings. Bot. Gazette 142 (3), 360–365. doi: 10.1086/337235
Nordine, A., Tlemcani, C. R., El Meskaoui, A. (2014). Regeneration of plants through somatic embryogenesis in Thymus hyemalis Lange, a potential medicinal and aromatic plant. In Vitro Cell Dev. Biol. Plant 50 (1), 19–25. doi: 10.1007/s11627-013-9577-x
Pacini, E., Hesse, M. (2002). Types of pollen dispersal units in orchids, and their consequences for germination and fertilization. Ann. Bot. 89 (6), 653–664. doi: 10.1093/aob/mcf138
Paek, K. Y., Hahn, E. J., Park, S. Y. (2011). Micropropagation of Phalaenopsis orchids via protocorms and protocorm-like bodies. Methods Mol. Biol. 710, 293–306. doi: 10.1007/978-1-61737-988-8_20
Park, S. Y., Yeung, E. C., Chakrabarty, D., Paek, K. Y. (2002). An efficient direct induction of protocorm-like bodies from leaf subepidermal cells of Doritaenopsis hybrid using thin-section culture. Plant Cell Rep. 21 (1), 46–51. doi: 10.1007/s00299-002-0480-x
Pelletier, J. M., Kwong, R. W., Park, S., Le, B. H., Baden, R., Cagliari, A., et al. (2017). LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. U.S.A. 114 (32), E6710–E6719. doi: 10.1073/pnas.1707957114
Polin, L. D., Liang, H. Y., Rothrock, R. E., Nishii, M., Diehl, D. L., Newhouse, A. E., et al. (2006). Agrobacterium-mediated transformation of American chestnut (Castanea dentata (Marsh.) Borkh.) somatic embryos. Plant Cell Tiss. Org. Cult. 84 (1), 69–78. doi: 10.1007/s11240-005-9002-1
Pramanik, D., Rachmawati, F., Winarto, B. (2016). Propagation of Phalaenopsis ‘Puspa Tiara Kencan’?™ via somatic embryogenesis. Acta Hortic. (1113), 21–26. doi: 10.17660/ActaHortic.2016.1113.3
Pulianmackal, A. J., Kareem, A. V., Durgaprasad, K., Trivedi, Z. B., Prasad, K. (2014). Competence and regulatory interactions during regeneration in plants. Front. Plant Sci. 5, 142. doi: 10.3389/fpls.2014.00142
Radhakrishnan, D., Kareem, A., Durgaprasad, K., Sreeraj, E., Sugimoto, K., Prasad, K. (2017). Shoot regeneration: a journey from acquisition of competence to completion. Curr. Opin. Plant Biol. 41, 23–31. doi: 10.1016/j.pbi.2017.08.001
Rasmussen, H. N. (2002). Recent developments in the study of orchid mycorrhiza. Plant Soil 244 (1-2), 149–163. doi: 10.1023/A:1020246715436
Robertson, D., Weissinger, A. K., Ackley, R., Glover, S., Sederoff, R. R. (1992). Genetic transformation of Norway Spruce (Picea abies (L) Karst) using somatic embryo explants by microprojectile bombardment. Plant Mol. Biol. 19 (6), 925–935. doi: 10.1007/BF00040525
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. U.S.A. 81 (24), 8014–8018. doi: 10.1073/pnas.81.24.8014
Shen, B., Allen, W. B., Zheng, P., Li, C., Glassman, K., Ranch, J., et al. (2010). Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 153 (3), 980–987. doi: 10.1104/pp.110.157537
Stone, S. L., Kwong, L. W., Yee, K. M., Pelletier, J., Lepiniec, L., Fischer, R. L., et al. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A. 98 (20), 11806–11811. doi: 10.1073/pnas.201413498
Teixeira da Silva, J. A., Singh, N., Tanaka, M. (2006). Priming biotic factors for optimal protocorm-like body and callus induction in hybrid Cymbidium (Orchidaceae), and assessment of cytogenetic stability in regenerated plantlets. Plant Cell Tissue Organ Cult. 84 (2), 135–144. doi: 10.1007/s11240-005-9003-0
Tokuhara, K., Mii, M. (2001). Induction of embryogenic callus and cell suspension culture from shoot tips excised from flower stalk buds of Phalaenopsis (Orchidaceae). In Vitro Cell Dev. Biol. Plant 37 (4), 457–461. doi: 10.1007/s11627-001-0080-4
Tokuhara, K., Mii, M. (2003). Highly-efficient somatic embryogenesis from cell suspension cultures of Phalaenopsis orchids by adjusting carbohydrate sources. In Vitro Cell Dev. Biol. Plant 39 (6), 635–639. doi: 10.1079/IVP2003466
Trinh, T. H., Ratet, P., Kondorosi, E., Durand, P., Kamate, K., Bauer, P., et al. (1998). Rapid and efficient transformation of diploid Medicago truncatula and Medicago sativa ssp falcata lines improved in somatic embryogenesis. Plant Cell Rep. 17 (5), 345–355. doi: 10.1007/s002990050405
Umbeck, P., Johnson, G., Barton, K., Swain, W. (1987). Genetically transformed cotton (Gossypium Hirsutum-L) Plants. Nat. Biotech. 5 (3), 263–266. doi: 10.1038/nbt0387-263
Vergne, P., Maene, M., Gabant, G., Chauvet, A., Debener, T., Bendahmane, M. (2010). Somatic embryogenesis and transformation of the diploid Rosa chinensis cv Old Blush. Plant Cell Tiss. Org. Cult. 100 (1), 73–81. doi: 10.1007/s11240-009-9621-z
Villar, C. B., Köhler, C. (2010). Plant chromatin immunoprecipitation. Methods Mol. Biol. 655, 401–411. doi: 10.1007/978-1-60761-765-5_27
von Arnold, S., Sabala, I., Bozhkov, P., Dyachok, J., Filonova, L. (2002). Developmental pathways of somatic embryogenesis. Plant Cell Tiss. Org. Cult. 69 (3), 233–249. doi: 10.1023/A:1015673200621
Wang, F., Perry, S. E. (2013). Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161 (3), 1251–1264. doi: 10.1104/pp.112.212282
Wang, X., Niu, Q. W., Teng, C., Li, C., Mu, J., Chua, N. H., et al. (2009). Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 19 (2), 224–235. doi: 10.1038/cr.2008.276
Waterman, R. J., Bidartondo, M. I. (2008). Deception above, deception below: linking pollination and mycorrhizal biology of orchids. J. Exp. Bot. 59 (5), 1085–1096. doi: 10.1093/jxb/erm366
West, M., Yee, K. M., Danao, J., Zimmerman, J. L., Fischer, R. L., Goldberg, R. B., et al. (1994). LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6 (12), 1731–1745. doi: 10.1105/tpc.6.12.1731
Winkelmann, T., Geier, T., Preil, W. (2006). Commercial in vitro plant production in Germany in 1985-2004. Plant Cell Tiss. Org. Cult. 86 (3), 319–327. doi: 10.1007/s11240-006-9125-z
Wu, H.-H., Chen, F.-C. (2008). Effect of plant growth regulators on shoot multiplication from flower stalk nodal buds of Phalaenopsis and Doritaenopsis. J. Taiwan Soc Hortic. Sci. 54 (2), 151–159.
Xie, Z., Li, X., Glover, B. J., Bai, S., Rao, G. Y., Luo, J., et al. (2008). Duplication and functional diversification of HAP3 genes leading to the origin of the seed-developmental regulatory gene, LEAFY COTYLEDON1 (LEC1), in nonseed plant genomes. Mol. Biol. Evol. 25 (8), 1581–1592. doi: 10.1093/molbev/msn105
Yam, T. W., Arditti, J. (2009). History of orchid propagation: a mirror of the history of biotechnology. Plant Biotechnol. Rep. 3 (1), 1–56. doi: 10.1007/s11816-008-0066-3
Yam, T. W., Arditti, J. (2017). Micropropagation of Orchids, Third Edition (Chichester, West Sussex, UK: John Wiley & Sons Ltd.). doi: 10.1002/9781119187080
Yang, X. Y., Zhang, X. L. (2010). Regulation of somatic embryogenesis in higher plants. Crit. Rev. Plant Sci. 29 (1), 36–57. doi: 10.1080/07352680903436291
Yazawa, K., Takahata, K., Kamada, H. (2004). Isolation of the gene encoding carrot LEAFY COTYLEDON1 and expression analysis during somatic and zygotic embryogenesis. Plant Physiol. Biochem. 42 (3), 215–223. doi: 10.1016/j.plaphy.2003.12.003
Yeung, E. C. (2017). A perspective on orchid seed and protocorm development. Bot. Stud. 58 (1), 33. doi: 10.1186/s40529-017-0188-4
Zhao, P., Wu, F., Feng, F.-S., Wang, W.-J. (2008). Protocorm-like body (PLB) formation and plant regeneration from the callus culture of Dendrobium candidum Wall ex Lindl. In Vitro Cell Dev. Biol. Plant 44 (3), 178–185. doi: 10.1007/s11627-007-9101-2
Zhu, S. P., Wang, J., Ye, J. L., Zhu, A. D., Guo, W. W., Deng, X. X. (2014). Isolation and characterization of LEAFY COTYLEDON 1-LIKE gene related to embryogenic competence in Citrus sinensis. Plant Cell Tiss. Org. Cult. 119 (1), 1–13. doi: 10.1007/s11240-014-0509-1
Zimmerman, J. L. (1993). Somatic embryogenesis: a model for early development in higher plants. Plant Cell 5 (10), 1411–1423. doi: 10.1105/tpc.5.10.1411
Keywords: somatic embryogenesis, Phalaenopsis orchids, LEAFY-COTYLEDON1, protocorm-like-body, Phalaenopsis equestris
Citation: Chen J-C, Tong C-G, Lin H-Y and Fang S-C (2019) Phalaenopsis LEAFY COTYLEDON1-Induced Somatic Embryonic Structures Are Morphologically Distinct From Protocorm-Like Bodies. Front. Plant Sci. 10:1594. doi: 10.3389/fpls.2019.01594
Received: 01 July 2019; Accepted: 13 November 2019;
Published: 29 November 2019.
Edited by:
Jen-Tsung Chen, National University of Kaohsiung, TaiwanReviewed by:
Sonia Gazzarrini, University of Toronto, CanadaCopyright © 2019 Chen, Tong, Lin and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Su-Chiung Fang c2NmYW5nQGdhdGUuc2luaWNhLmVkdS50dw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.