- 1Laboratory of Cotton Genetics, Genomics and Breeding/Key Laboratory of Crop Heterosis and Utilization of Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement, College of Agronomy and Biotechnology, China Agricultural University, Beijing, China
- 2Institute of Economic Crops, Xinjiang Academy of Agricultural Sciences, Urumqi, China
Genetic improvement in fiber quality is one of the main challenges for cotton breeders. Fiber quality traits are controlled by multiple genes and are classified as complex quantitative traits, with a negative relationship with yield potential, so the genetic gain is low in traditional genetic improvement by phenotypic selection. The availability of Gossypium genomic sequences facilitates the development of high-throughput molecular markers, quantitative trait loci (QTL) fine mapping and gene identification, which helps us to validate candidate genes and to use marker assisted selection (MAS) on fiber quality in breeding programs. Based on developments of high density linkage maps, QTLs fine mapping, marker selection and omics, we have performed trait dissection on fiber quality traits in diverse populations of upland cotton. QTL mapping combined with multi-omics approaches such as, RNA sequencing datasets to identify differentially expressed genes have benefited the improvement of fiber quality. In this review, we discuss the application of molecular markers, QTL mapping and MAS for fiber quality improvement in upland cotton.
Introduction
Among 53 Gossypium species, there are four cultivated species, including Gossypium Hirsutum, G. Barbadense, G. Arboreum, and G. Herbaceum (Fryxell, 1992; Wendel and Grover, 2015; Gallagher et al., 2017; Wang et al., 2018). Upland Cotton (G. Hirsutum L.) is extensively cultivated due to its wide adaptability to the environment, high production, and better yield potential, which fulfils over 95% of the output of global cotton yield (Chen et al., 2007). Sea Island cotton (G. Barbadense) is known for excellent fiber quality with long, strong, and fine fibers (Avci et al., 2013). To meet the demands of modern textile industry, genetic improvements in cotton have been performed in fiber quality and yield traits in different populations along with cotton production. Therefore, it is a great challenge to improve the fiber quality and to increase the yield potential simultaneously in diverse areas and planting systems in cotton production.
Fiber quality traits generally include fiber length, fiber uniformity, fiber strength, fiber elongation, and micronaire value. Fiber strength and fiber length are considered as the most important properties affecting yarn quality (Yang, et al., 2016). Fiber strength is very important for advanced spinning technologies in the textile industry (Felker, 2001). The micronaire value, a measure of fiber fineness and fiber maturity, influences the fiber processing and dyeing consistency (Rodgers et al., 2017).
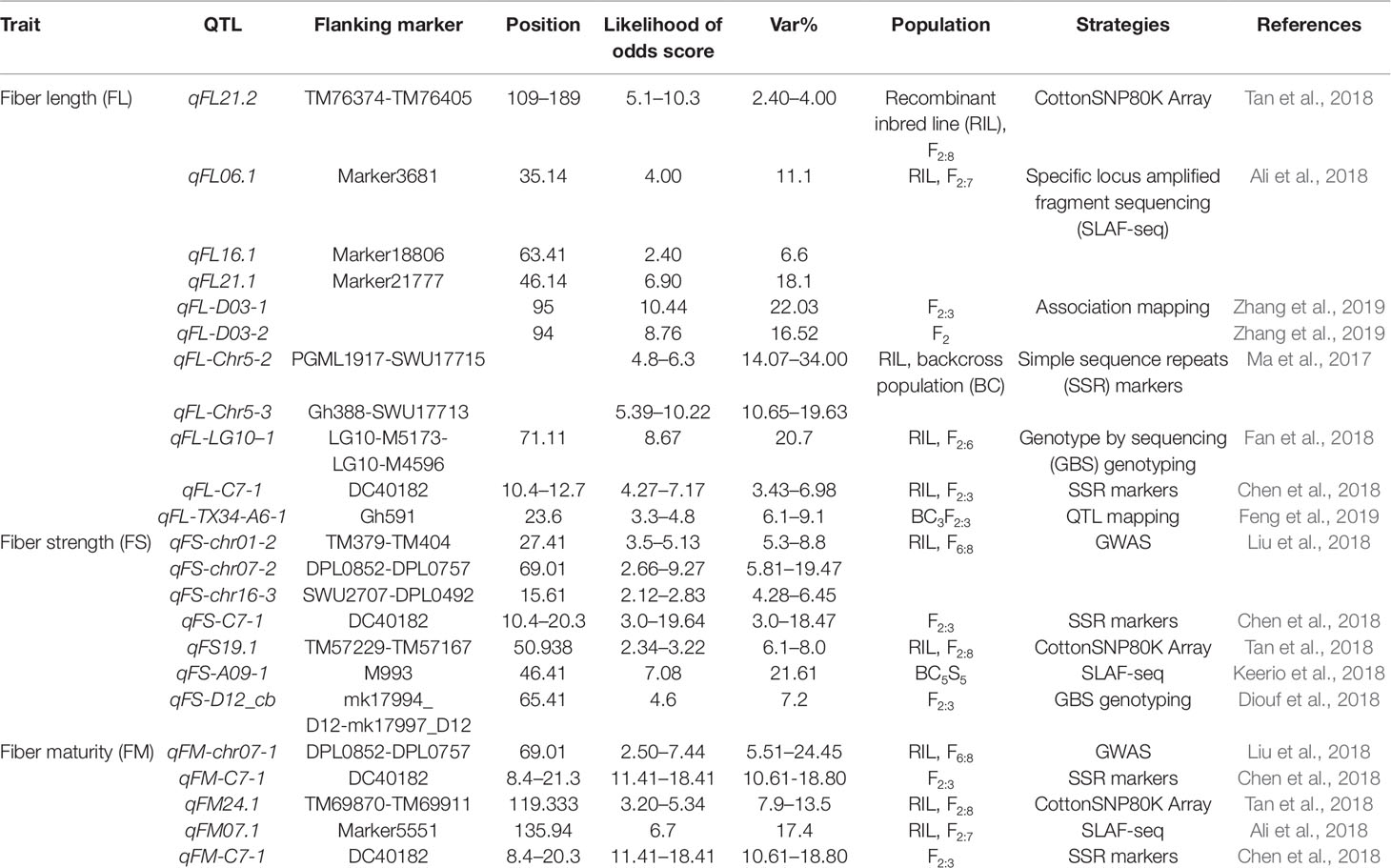
Although studies in fiber development have been engaged for many years, the molecular mechanism is still not well explained (Zhang et al., 2013). Quantitative trait loci (QTL) mapping with molecular markers provides a powerful approach to dissect the molecular mechanism underlying complex fiber quality traits. At least 1075 QTLs from 58 studies and 1059 QTLs within 30 studies have been reported with intraspecific and interspecific crosses (Said et al., 2015b), which provide a platform for follow-up QTL validation and gene cloning in dissecting the mechanism involving complex traits. Functional genomic studies provide new insights to better understand fiber development mechanisms in Gossypium species (Figure 1). Here, we endeavor to provide comprehensive information on QTL mapping and utilization in cotton.
Figure 1 Overview of the strategies for marker-assisted selection (MAS) to improve fiber quality in Gossypium hirsutum.
Overview of Fiber Development Mechanism in Cotton
Cotton fiber is a seed-borne epidermal trichome composed of a highly elongated and thickened single cell. Cotton fiber achieves the expansion through water influx with changes of turgor pressure in a cell (Cosgrove, 1997). Generally, the fiber cell develops into a trichome in four stages: a) fiber initiation, b) cell elongation [primary cell wall (PCW) synthesis], c) cell wall biosynthesis [secondary cell wall (SCW) deposition], and d) fiber maturation (Kim and Triplet, 2001; Ruan, 2005; Pang et al., 2010; Zhang et al., 2013; Zhang and Liu, 2013). The fiber initiation starts from 3 days before anthesis to 3 days post anthesis (DPA) with trichome protrusion and enlargement of epidermal cells (Qin and Zhu, 2011). In some G. hirsutum cultivars, lint percentage and lint index are positively correlated with the density of 0 DPA (fiber protrusion) and 1 DPA fiber initials (fiber elongation; Li et al., 2009).
Fiber elongation starts from 2 DPA to 20 DPA (typically) after the initiation period, these elongating fibers twist together to develop into bundles of fiber (Singh et al., 2009; Haigler et al., 2012). The closure of plasmodesmata is important in fiber cell elongation with the increase of cellulose synthesis at 10-16 DPA (Ruan et al., 2004). At a cellular level, several mechanisms facilitate the rapid cotton fiber elongation; as an example, GhVIN1 expression increased in G. barbadense at a higher level until 10 DPA during fiber elongation (Wang et al., 2010). Aquaporin proteins also facilitate cotton fiber elongation and the corresponding genes, including GhγTIP1 and GhTIP1-2 in the tonoplast and plasma membrane, respectively (Liu et al., 2008). The expression of pectin biosynthesis genes promotes fiber elongation by ethylene signaling pathways (Qin et al., 2007; Pang et al., 2010). Expansins are likely to play a role in regulating fiber elongation (Li et al., 2016). Dynamic DNA methylation has been reported during cotton fiber differentiation, and additionally, lncRNAs are associated with chromatin modifications (Wang et al., 2015a; Wang et al., 2016a).
During the transition phase of cell elongation to cell wall biosynthesis, fiber elongation stops gradually and SCW is thickened with the biosynthesis of SCW genes up-regulated (Hinchliffe et al., 2010). SCW thickening depends upon the increased rate of cellulose synthesis, orientation of cellulose fibrils, and greater length of individual cellulose chains (Haigler et al., 2009; Betancur et al., 2010). Cellulose fibrils in primary and secondary walls are synthesized by cellulose synthases (CesAs) and cellulose synthase-like enzymes, which initiates cell wall matrix polymers (Taylor et al., 2003). Several flavonoid and lignin biosynthesis pathways regulate the start of SCW synthesis in sclerenchyma cells (Ehlting et al., 2005; Zhao and Dixon, 2011). Single-cell expression systems will provide a better understanding for the molecular control of cotton fiber elongation dynamics and cell wall biosynthesis in the future. Genes controlling cellulose synthesis of cotton fiber are orthologs of other vascular plants (Haigler et al., 2005; Haigler et al., 2009). The PCW features of Arabidopsis leaf trichomes are integrated with SCW synthesis of cotton the fiber during cell wall thickening (Betancur et al., 2010), which offers a strategy and technology roadmap to validate functions of related genes.
The Underlying Genomic and Proteomic Modifications in Control of Fiber Development
Overview of the Genes Involved in Fiber Development
Transcriptome profiles reveal novel information in different stages of fiber development. A number of transcription factor (TF) families, for example, MYB, WRKY, AP2/EREBP, C2H2, and bHLH, were involved in fiber initiation (Table 1; Yang et al., 2006). The diverse TF genes in different Gossypium species (GbML1, GaMYB2, GhMYB109, and GaHOX1) are expressed differentially during fiber initiation and the early stage of fiber development (Wang et al., 2004; Guan et al., 2008; Pu et al., 2008; Zhang et al., 2010).
The transcriptional expression of GhHD1, GhMYB-25, GhSusA1, GhFLA1, GbPDF1, and GhVIN1 genes contributes to the initial stage of fiber development (Machado et al., 2009; Walford et al., 2011; Deng et al., 2012; Jiang et al., 2012; Huang et al., 2013; Wang et al., 2014), in which GhFLA1, an arabinogalactan protein gene, enhanced fiber initiation and fiber elongation with the alteration of PCW integrity (Huang et al., 2013). The Myb2 TF triggered the expression of R22-like (RDL) gene in the stage of fiber initiation (Suo et al., 2003; Wang et al., 2004). The silencing of the cotton GbPDF1 (PROTODERMAL FACTOR1) gene retarded fiber initiation and produced shorter fibers compared with wild type (Deng et al., 2012).
Cellulose synthase catalytic subunits (CesAs) regulated the cellulose synthesis pathways by catalytic sites within a multi-subunit complex, at least 15, 14, and 29 CesA genes have been reported in G. raimondii, G. arboreum, and G. barbadense, respectively (Paterson et al., 2012). Moreover, the up-regulation of CESa genes (GhCES5–GhCESA10 in G. hirsutum; CesA4, CesA7 and CesA8 in G. barbadense) were involved in primary and SCW biosynthesis (Li et al., 2013; Liu, et al., 2015; Nawaz et al., 2017), post-translational modifications, and trafficking (Polko and Kieber 2019).
Plant hormones control fiber cells development by regulating gene expression at different growth levels. The roles of brassinosteroid (BR), jasmonic acid (JA), auxin, and arabinogalactan protein (AGP) biosynthetic pathways have been revealed to be involved in up-regulating or suppressing the expression of genes in both fiber initiation and fiber elongation (Yang et al., 2014; Zhang et al., 2016; Qin et al., 2017). GhPIN-mediated auxin transport was responsible for ovule-specific suppression of the GhPIN gene, demonstrating its roles in fiber initiation via the auxin accumulation (Zhang et al., 2016). DELLA-like proteins acted as repressors in gibberellic acid (GA) signaling pathway, for example, four genes (GhGAI3a, GhGAI3b, GhGAI4a, and GhGAI4B) containing DELLA domains expressed during fiber initiation and elongation (Wen et al., 2012). GhCaM7 played an important part in the initiation and elongation stages of fiber cells by controlling the production of reactive oxygen species and H2O2 which regulated Ca2+ influx into the fiber (Tang et al., 2014).
ACTIN1, GhPIP2, GhCaM7, WLIM1a, GhPAG1, and GhHOX3 played important roles in the fiber elongation process (Li et al., 2005; Han et al., 2013; Li et al., 2013; Shan et al., 2014; Tang et al., 2014; Yang et al., 2014). The GhBZR1 protein regulated fiber elongation by binding to the promoters of genes GhXTH1 and GhEXP (Zhou et al., 2015). Expansin proteins play critical roles in cell wall loosening and fiber elongation by producing polysaccharide complexes including xyloglucan and pectin, with the 3-year field performance of GhEXPA8 transgenic plants showed longer fiber and better micronaire (Bajwa et al., 2015). PAG1 gene controlled endogenous BRs level by encoding a cytochrome P450 and played an important role in regulating fiber elongation (Yang et al., 2014). GhMYB212 regulates the expression of GhSWEET12 during cell expansion at developing phases of fiber elongation (Sun et al., 2019). Further studies involved in gene function elucidation will enhance the understanding in the mechanism underlying fiber development.
The Major Proteins Involved in Cotton Fiber Development
The availability of cotton genome sequences makes post-genomic proteomic studies possible, which further helps to provide new understandings into cotton fiber initiation and elongation mechanisms. All the genes function by accomplishing protein modifications in different development stages of cotton fiber. Many proteins, acting as enzymes, make contributions in the basic energy metabolism pathways, such as polysaccharide biosynthesis, glycolytic pathway, the pentose-phosphate, and the tricarboxylic acid cycle (Yang et al., 2008; Pang et al., 2010; Liu et al., 2012; Du et al., 2013; Li et al., 2013a; Zhang and Liu, 2013). Some proteins are also involved in hormone signaling pathways, such as GA, jasmonate (JA), BR, ethylene (ET) and abscisic acid (ABA) (Pang et al., 2010; Liu et al., 2012; Du et al., 2013; Zhang et al., 2013).
Proteomic analyses showed the involvement of redox homeostasis-related proteins, including dehydroascorbate reductase (DHAR), ascorbate peroxidase (APX), catalase (CAT), nicotineamide adenine dinucleotide phosphate-isocitrate dehydrogenase (NADP-ICDH), and phospholipase D alpha (PLDα), in cotton fiber elongation (Yang et al., 2008). H2O2 acted as a signaling element involved in controlling the growth and development of plants, that is, the lower the level of H2O2 was associated with the higher levels of DHAR, CAT at 5 DPA and 10 DPA, respectively, showing the H2O2 producing and scavenging systems in elongating fiber cells (Mittler et al., 2004; Du et al., 2013).
Cytosolic pyruvate kinase involved in catalyzing the glycolytic pathway was highly expressed at 20 DPA, suggesting its role in the process of fiber elongation to secondary wall thickness (Duggleby and Dennis, 1973; Zhang et al., 2015). Some enzymes (such as transketolase) catalyze several reactions in glycolysis and were up-regulated in cotton fiber elongation, implying the key role of carbohydrate metabolism in fiber development at transcriptional level (Zhang et al., 2015). Similarly, the expression of the Sus gene was higher at early stages and lower at later stages of fiber elongation because sucrose was converted into fructose and UDP-glucose in cell expansion and cell wall biosynthesis (Delmer, 1987; Ruan et al., 2003). Furthermore, the transition phase from PCW to SCW synthesis was accompanied with the biosynthesis of guanosine diphosphate fucose, suggesting the roles of more fucosylation (Freshour et al., 2003). The proteomic profiles of fuzzless-lintless (fl) mutant and wild-type upland cotton fiber revealed that pectin synthesis was necessary for fiber elongation (Pang et al., 2010), supporting the hypothesis that fiber elongation was regulated through redox homeostasis by the evidence that proteins involved in the cellular redox homeostasis were expressed differentially in fl mutant (Liu et al., 2012).
Proteomic approaches in cotton fiber domestication and evolution provide databases for functional genomics analysis in cotton fiber. In total, 190 differentially expressed proteins were identified between wild and cultivated accessions using iTRAQ technology (Hu et al., 2013). Proteome comparison between G. hirsutum and G. barbadense with their diploid progenitors revealed that the two allopolyploid species have attained superficially ‘modern’ fiber phenotypes during evolution (Hu et al., 2015). However, the early events in fiber elongation have been neglected so far and could be characterized at the proteome level.
Molecular Markers and Linkage Maps for Fiber Quality Traits
The applications of molecular markers allow the development of high density genetic linkage map and germplasm evaluation, as well as phylogenetic and evolutionary analysis. Desired alleles and high density linkage maps will be helpful for mapping of stable QTLs and efficient gene identification controlling good quality fiber. Molecular markers including random amplified polymorphic DNA, restriction fragment length polymorphism, simple sequence repeats (SSRs), inter simple sequence repeats, and single nucleotide polymorphisms (SNPs) (Tatineni et al., 1996; Liu et al., 2000; Abdalla et al., 2001; Lu and Myers 2002; Reddy et al., 2002; Zhu et al., 2003; Alvarez and Wendel, 2006; Li et al., 2015) have been used for QTL identification for fiber quality traits (Said et al., 2015b). Genetic maps have also been proven to be useful to uncover the molecular bases of multi-genic traits such as fiber quality. Mapping populations between diverse sets of cultivars from distinct geographic origins were used to determine the positive associations between genetic markers and fiber traits. Several major QTLs that are persistent among the populations show high phenotypic variation (PV) and likelihood of odds score. The first genetic map of cotton that covered a length of 4675 cM with 705 restriction fragment length polymorphism markers was developed by Reinisch et al. (1994). Afterward, different genetic maps have been developed and identify 14, 26 and 13 QTLs for fiber quality (Jiang et al., 1998; Ulloa et al., 2000; Kohel et al., 2001), 28 QTLs for fiber length, 9 QTLs for fiber length uniformity, and 8 QTLs for short fiber index (Chee et al., 2005).
SSR and SNP Markers for Fiber Quality Traits
With the availability of cotton genome sequences and the development of bioinformatics tools, the rapid increase of SSR markers and SNPs has become more effective for genotyping and QTL mapping in cotton. SSRs have been widely utilized for genetic dissection of QTL mapping in different wild and domesticated cotton species. There are a number of studies conducted to identify fiber quality related QTLs using SSR genetic maps with the use of intraspecific populations and under multiple environments and generations (Zhang et al., 2003; Lacape et al., 2005; Zhang et al., 2005; Wang et al., 2006; Wang et al., 2007; Zhang et al., 2009; Sun et al., 2012; Liang et al., 2013; Fang et al., 2014; Zhiyuan et al., 2014; Shao et al., 2014;Ma et al., 2019). Shen et al. (2005) performed extensive SSR genotyping using 1378 markers in an F2 population and detected 39 QTLs linked with fiber quality traits among them, 11 QTLs for fiber length, 10 for fiber strength, 9 for micronaire, and 9 for fiber elongation.
A significant association had been found between SSRs and fiber quality traits in Gossypium species (Zeng et al., 2009). Association mapping of fiber quality traits linked with SSR markers identified 52.86% (70 stable loci) for target traits including 30 for fiber length, 27 for fiber strength, and 13 for fiber fineness (Cai et al., 2014). Wang et al. (2015) constructed a genetic linkage map containing 644 polymorphic loci and identified a total of 64 QTLs (10 QTLs for fiber length, 18 for fiber strength, 13 for micronaire, 16 for fiber elongation, and 7 for fiber length uniformity ratio) associated with fiber quality in seven environments. Another linkage map comprising of 579 markers was developed using SSRs, conserved intron scanning primers, and transcript-derived fragment amplifications, with an average distance of 7.19 cM between markers, in a backcross population (BC) derived from a cross among G. hirsutum and G. barbadense (Yang et al., 2015). Nie et al. (2016) used 494 genome-wide SSR markers and 503 indigenous upland cotton lines to map 216 marker loci linked with fiber quality traits (61 loci with fiber strength, 46 for fiber upper half mean length, 25 for fiber elongation, 23 for micronaire value, 19 for fiber uniformity, and 42 for short fiber) and some novel alleles that could be considered as selection tags for breeding programs.
Shang et al. (2016) explored fiber quality in two recombinant inbred line (RIL) populations along-with their corresponding BC populations and identified 62 common QTLs in multiple environments and populations; 286 QTLs were also detected by the digenic interactions and their environmental interactions (QEs) analyses under multiple environments; such results concluded that single-locus and epistasis along with minor measureable major effects showed an important role in regulating fiber quality in upland cotton. Ma et al. (2017) developed a BC population from a RIL population and compared the genetic mapping effects on RIL and BC populations. The 26 and 37 QTLs were detected for fiber quality in RIL and BC populations, respectively, yet there was no significant role of heterosis observed in fiber quality traits (Ma et al., 2017). Li et al. (2018) utilized immortalized F2 population with two reciprocal BC populations and identified total 167 QTLs, whereas, multi-environment analysis offered 104 main-QTLs and 114 epistasis-QTLs in both populations referred over-dominance and epistatic factors are important to fiber quality heterosis in cotton. In total, 134 SSR marker-trait associations were identified for four fiber quality traits; of these, 15 significant associations explained 12%–16% of total phenotypic variance for the markers NAU5120, DPL0378, and JESPR101 (Adhikari et al., 2017). A total of 68 QTLs including 11 stable QTLs for five fiber quality traits were detected in RIL by SSR map containing 2051 loci (Liu et al., 2017). Huang et al. (2018) applied 284 SSR markers to identified 14 stable QTLs linked with fiber quality traits in a multi-parent advanced generation inter-cross population with 960 lines developed from eight parents. Similarly, three sets of introgression line (ILs) populations developed from G. hirsutum race stocks (TX-34, TX-48, and TX-114) were genotyped with 452 SSR markers, and identified 38 new QTLs related to fiber quality traits, among them 17 favorable QTLs demonstrate the importance of race stock germplasm in cotton fiber improvement (Feng et al., 2019).
High-Throughput Sequencing Techniques for Fiber Quality Traits
The availability of genomic sequences of the G. raimondii (D5), G. arboreum (A2), G. hirsutum (AD1), and G. barbadense (AD2) provides more opportunities to improve the density of intraspecific genetic maps (Paterson et al., 2012; Wang et al., 2012; Li et al., 2014; Li et al., 2015; Liu et al., 2015; Yuan et al., 2015; Zhang et al., 2015). Soon after the genome sequences in Gossypium became available, molecular markers were used to develop high precision QTL linkage maps. High-throughput genotyping offers a deep insight into genome wide genetic maps for a wide range of applications in QTL mapping and marker assisted selection. For example, the development of CottonSNP63K (Hulse-Kemp et al., 2015) and CottonSNP80K (Cai et al., 2017) arrays has allowed genetic mapping, genomic selection, genome-wide association studies, and an understanding of genomic diversity among Gossypium breeding populations. Common QTLs related to fiber quality in different environments identified through SNP markers can be deployed as a priority for fine mapping and candidate genes identification (Li et al., 2016; Su et al., 2016; Tan et al., 2018).
The advance techniques in next generation sequencing have boosted the genetic maps quality and validity with SNP polymorphisms to increase marker coverage across the genome. These technologies include restriction-site associated DNA (Wang et al., 2015; Jia et al., 2016), specific locus amplified fragment sequencing (SLAF-seq) (Zhang et al., 2016), and genotyping by sequencing (Qi et al., 2017). Zhang et al. (2019) performed GWAS analysis with the combined association and linkage mapping approach and identified two potential QTLs on chromosome D03 and D08 (qFL-D03-1/2 and qFL-D08-1/2) in multiple environments. Further identification of candidate genes reveals 26 genes on chromosome D03 and among them, the Gh_D03G1338 was potentially involved in the fiber development stages (Zhang et al., 2019). Another study, performed multi-locus GWAS and genotyping was done by the SLAF-seq approach, identifying several quantitative traits nucleotides (QTNs), D11_21619830, A05_28352019, and D03_34920546 for fiber quality traits and detected six genes highly expressed in cotton fibers (Su et al., 2018). Li et al. (2018) conducted both single locus and multi-locus GWAS and identified collectively 342 QTNs for fiber quality traits, and nine QTNs with maximum PV. A multi-parent advanced generation inter-cross population with 550 individuals were utilized for GWAS analysis combined with RNA-seq of lines with extreme fiber characteristics, identified highly significant QTL on D11 which further dissected into 12 genes and an auxin-responsive GH3 gene (Gh_D11G1989) as the candidate gene for fiber length (Naoumkina et al., 2019). Fan et al. (2018) constructed another high-density map with a G. barbadense (5917 and American Pima S-7) RIL population locating 42 QTLs for fiber quality traits, distributed among 14 linkage groups, with the number of QTLs located in A subgenome was 28 whereas only 14 QTLs were associated with D subgenome. An intraspecific population of G. hirsutum was utilized for genotyping by sequencing mapping and 30 consistent QTLs were identified from 110 common QTLs, divided into two main clusters on chromosome D03 and D12 (Diouf et al., 2018). Five candidate genes (Gh_D03G0889, Gh_D12G0093, Gh_D12G0410, Gh_D12G0435, and Gh_D12G0969) were associated with QTLs (qFM-D03_cb, qFS-D12_cb, and qFY-D12_cb), functions in protein kinases and phosphorylation, respectively (Diouf et al., 2018).
Distribution and Resources of QTL Mapping for Fiber Quality Traits
A number of QTLs associated with fiber quality traits have been identified in different mapping populations such as RILs, bi-parental segregating populations, and BC populations within intraspecific populations among G. hirsutum and interspecific crosses of G. hirsutum and G. barbadense. In particular, nearly 80% of fiber quality QTLs were identified from the interspecific populations were due to the wide genetic base (https://www.cottongen.org/; Yu et al., 2014). To date, more than 1500 QTLs for fiber traits have been mapped (Said et al. 2015a;Yang et al., 2015), providing the potential to be manipulated by MAS for the improvement of cotton fiber length and strength. Overall, for six fiber quality traits (FL, FS, FE, FM, FU, and Mic), At sub-genome linked with more number of QTLs than Dt sub-genome (Figure 2A). The number of QTLs for different fiber quality traits also varies over the chromosomes, with the maximum number of 359 QTLs distributed across the genome for fiber strength followed by fiber length with 318 QTLs spread over 26 chromosomes (Figure 2B). Whereas, the fiber maturity had minimum number of 62 QTLs in cotton genome. According to Said et al. (2015b), majority of the QTLs were located around the first 0–20 cM of each chromosome with few exceptions, and this seems to be evolutionarily conserved.
The number of fiber quality trait QTLs over the chromosomes of the cotton genome is not identical, some chromosomes containing more QTLs than the others. QTLs associated with cotton fiber quality obtained from CottonQTLdb database (http://www.cottonqtldb.org) were distributed unevenly across the 26 chromosomes of the cotton genome (Figure 2C; Said et al., 2015a). In total, 132 QTLs were linked within chromosome 25, while 15 mapped in chromosome 22 with the least number of QTLs. Chromosome 25 had been dissected for stable QTLs associated with fiber strength and fiber length (Sun et al., 2012; Jamshed et al., 2016).
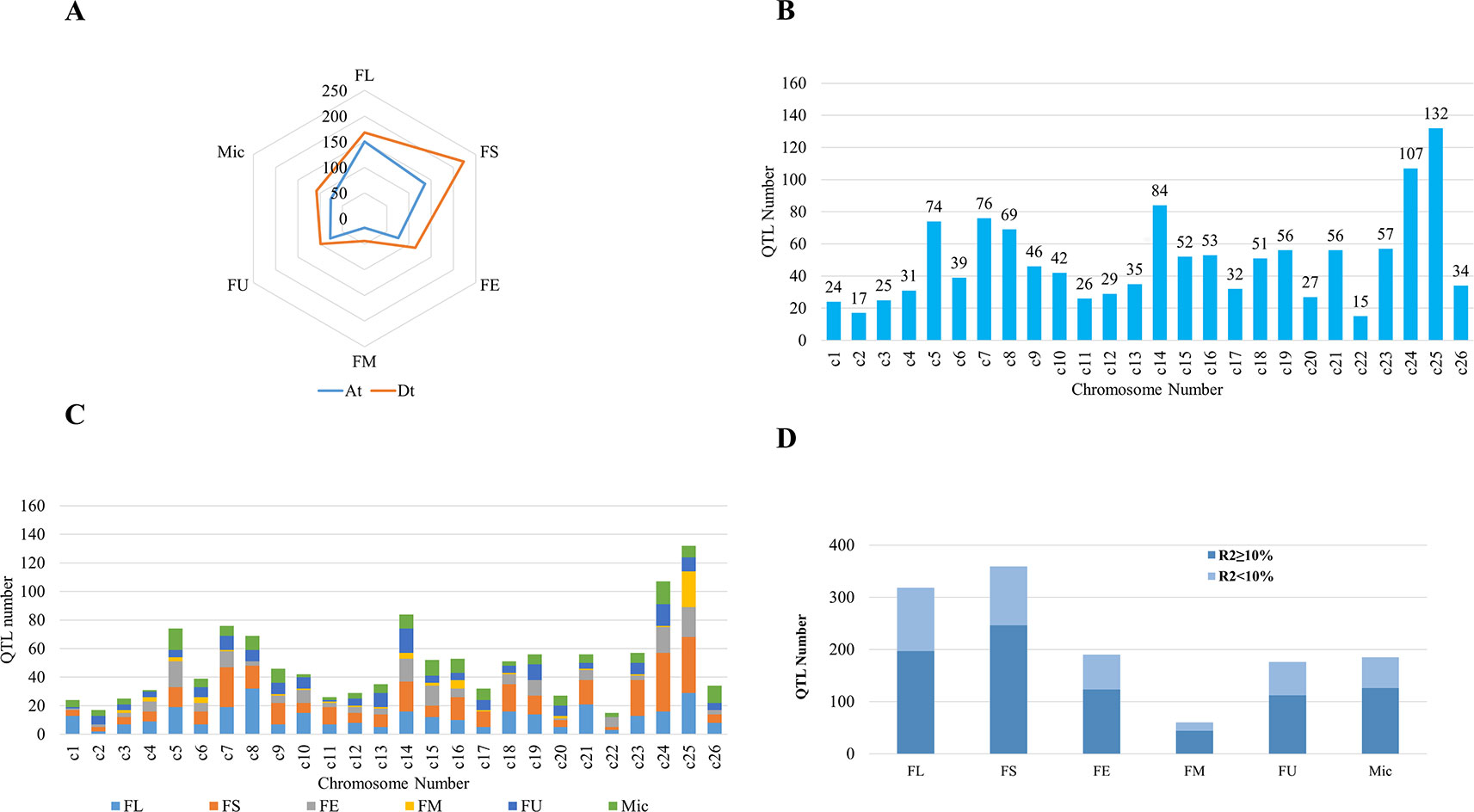
Figure 2 Quantitative trait loci (QTLs) distributed across each chromosome in cotton genome. (A) QTLs of fiber quality traits distributed across At and Dt sub-genomes. (B) QTLs mapped in each chromosome. c1–c26 represents chromosome 1 to chromosome 26, respectively. (C) QTLs for different fiber quality traits. FL, fiber length; FS, fiber strength; FE, fiber elongation; FM, fiber maturity; FU, fiber uniformity; Mic, micronaire. (D) The variance (R2) explained by a single QTL related to fiber quality traits. FL, FS, FE, FM, FU, Mic. The same as used above.
More than 50% of QTLs for each fiber quality traits showed R2 ≥ 10% (Figure 2D), implying that genome-wide PV explained by single QTL was comparatively high. These results suggested different mechanisms are involved in the fiber quality traits and showed complex genetic architecture.
Potential QTLs Mapped for Key Fiber Quality Traits
QTL studies across multiple generations and environments allow the identification of common QTLs for MAS. As mentioned above, fiber length and fiber strength are important economic indicators for fiber quality in cotton. Tan et al. (2015) identified several QTLs for fiber length (qFL07.1), fiber strength (qFS07.1), fiber micronaire (qFM07.1), and fiber elongation (qFE07.1) by SSR markers (DPL0757 and BNL1604) in different environments. Wang et al. (2016) utilized immortalized BC populations and identified 22 stable QTLs in many environments and found a potential stable QTL (qFL-c10-1) in RIL and BCF1populations in three environments. Ali et al. (2018) identified stable QTLs for fiber length and micronaire (qFL06.1, qFL16.1, qFL21.1, and qFM07.1), which could be further verified by fine mapping and used for candidate gene identification (Ali et al., 2018).
For fiber strength, Zhang et al. (2015) dissected chromosome 25 and identified a comparatively stable QTL for fiber strength (qFS-chr25-4) explaining PV of 6.53%–11.83% identified in seven environments. Another, stable QTL (qFS-C9-1) on chromosome 09 was mapped within the marker interval of NAU2395 and NAU1092 using an interspecific cross between G. barbadense and G. hirsutum in different environments and populations (Yang et al., 2016). They focused on fine mapping and cloning genes underlying qFS-C9-1, which could provide valuable resources to study the mechanisms for fiber strength. Jamshed et al. (2016) also verified QTL clusters for fiber quality traits on chromosomes c4, c7, c14, and c25 as common QTLs in different environments and populations. Besides, 65 QTLs for fiber length and 15 QTLs for fiber strength were identified by association mapping (Wang et al., 2017). Liu et al. (2018) constructed a high-density genetic map with GWAS analysis of 4,729 SNPs and 122 SSR markers with an average interval of 0.51 cM and harboring a total of 134 QTLs for fiber quality traits in nine environments. A chromosome segment substitution line population developed by crossing between two tetraploid genome species (G. hirsutum and G. barbadense) detected 103 QTLs associated with fiber quality traits (Li et al., 2019). The introgression of superior alleles of fiber quality from G. barbadense contributed to G. hirsutum have been linked in five QTL clusters for fiber quality which further associated with significant candidate genes (Gh_A07G1752, Gh_A07G1753, Gh_A07G1754, Gh_A07G1755, and Gh_A07G1756) in fiber development (Chen et al., 2018).
The non-domesticated allotetraploid species (G. tomentosum, G. mustelinum, and G. darwinii) have also been widely utilized in genetic mapping programs for the improvement of fiber quality traits (Keerio et al., 2018). The interspecific population between G. hirsutum and G. mustelinum (AD4) demonstrated potential value in cotton breeding programs for the improvement of fiber quality (Wang et al., 2017). The ILs obtained from crosses between G. hirsutum × G. darwinii detected 278 polymorphic loci and 13 loci associated with fiber strength among 105 ILs (Wang et al., 2012). Several QTLs were detected across two environments in a RIL population by a genetic map harboring 2618 SNP markers for fiber quality traits and identified 12 QTLs for fiber length and 8 QTLs for fiber strength (Li et al., 2016). A SLAF-seq genetic mapping study conducted with 107 ILs from G. tomentosum, identified 30 QTLs linked with fiber quality traits (Keerio et al., 2018). The CottonSNP80K array was utilized for a GWAS study combined with RNA-seq analysis of fiber development stages to predict several candidate genes with differential expression patterns (Dong et al., 2019). Some studies including stable QTLs for fiber quality traits obtained from different mapping techniques have been described in (Table 2). The introgression of these stable QTLs into an elite cultivar by marker-assisted BCing will improve fiber traits in cotton.
Strategy for Fine Mapping of Fiber Quality Traits QTLs
Cotton fiber quality traits are controlled by multiple genes with minor effects and are affected by environmental factors, which take a long period to improve fiber quality through conventional genetic improvement techniques. The DNA markers provide promising means to dissect the mechanisms underlying the traits with complex nature, which allows the breeders to select the region with desirable effects after cycles of MAS (Young and Tanksley 1989). Several studies have mapped fiber QTLs in past decades (Said et al., 2013; Fang et al., 2014; Shang et al., 2015; Tang et al., 2015; Tan et al., 2015; Ma et al., 2017; Ma et al., 2018); these QTLs mapped in large genomic regions that may include hundreds or thousands of genes. Therefore, the target genes with less ‘linkage drag’ can be identified by fine-mapping of QTLs.
Studies Involved Fine Mapping of Fiber Quality Traits
Fine mapping of QTLs related to fiber quality traits in marker-assisted breeding relies on narrowing down the genetic basis by producing near isogenic lines (NILs) and ILs. NILs have been constructed with the selection of only one locus from diverse species (Fridman et al., 2000; Brouwer and Clair 2004; Thomson et al., 2006; Szalma et al., 2007; Zhang et al., 2009; Ding et al., 2011; Chen et al., 2012). ILs have been utilized in reducing the QTLs regions from 20 to 3 cM by fine mapping for important traits in tomato, rice, and rape (Paterson et al., 1990). Later, stable QTL for fiber bundle strength was mapped by utilizing two NILs MD52ne and MD90ne, which provided the prospects to study the genetic components involved in cotton fiber strength (Islam et al., 2014).
Su et al. (2013) developed ILs using a marker-assisted breeding approach with TM-1 and fine-mapped fiber strength QTL (QFS-D11-1) that exhibited 35.8% PV, into a marker interval of 0.6 cM within flanking markers (NAU2110 and NAU2950). Later, a G. barbadense introgressed line, developed by Cao et al. (2015), led to a fine-mapped QTL at same position between the two SSR markers, NAU3735 and NAU845. The intervals of putative QTLs, qFL-chr.7 for fiber length and qFS-chr.7 for fiber strength, were decreased to 0.36 and 0.44 cM, respectively (Cao et al., 2015). Because the fiber length and fiber strength are usually positively correlated, there might be evidence of pleiotropy. Furthermore, the integration of Mapping-By-Sequencing technique facilitated the identification of candidate genes in mapped QTLs for fiber bundle strength, short fiber index, and fiber length. The outcome of Mapping-By-Sequencing strategy validated the QTLs and suggested the role of the receptor-like kinases pathway genes in fiber strength (Islam et al., 2016).
RNA-Seq Facilitates Fine-Mapping in Identifying Fiber Quality Related Genes
The choice of fine mapping, enhanced by development of transcriptomic datasets and candidate genes identification was supported by the integration of quantitative genetics and transcriptomics. The combination of fine-mapping of QTLs and RNA-seq is a potential strategy for the identification of candidate genes for several complex fiber quality traits. Liu et al. (2016) established a combine approach of fine-mapping and RNA-seq to elucidate the molecular basis of fiber quality by identifying candidate genes underlying a previous QTL near T1 locus on chromosome 6. Recently, one QTL was mapped to an interval of 0.28 cM with the flanking markers (HAU2119 and SWU2302), whereas RNA-seq results of ovules at 0 DPA and 5 DPA fibers showed that four genes in this QTL region were expressed differentially and the expression of three genes encoding XTH, ALDH and GPI-anchored protein were significantly linked to fiber elongation (Liu et al., 2016). A potential candidate gene (RLK family protein) for QTL qFS07.1 was revealed by a comparative approach of RT-qPCR and fine-mapping (Fang et al., 2017). A fiber length QTL (qFL-chr1) on chromosome 1 was fine-mapped to a 0.9 cM interval and the candidate genes (GOBAR07705 and GOBAR25992) were associated with fiber length in a BC4F2 population (Xu et al., 2017). The ILs between G. hirsutum and G. barbadense were dissected for fiber quality QTL analysis and 13 QTLs were integrated with 235 highly expressed genes during fiber development, and the transcriptome analysis of 10 DPA of these lines led to the identification of eQTLs (expression QTLs) for 125 of these 235 genes (Wang et al., 2019). Among these 235 genes, the Ghir_D09G014120 and Ghir_D09G014460 that respectively encoded ubiquitin extension protein and a microtubule-associated protein could be possible candidate genes for fiber strength (Wang et al., 2019). The fuzzless gene (GaFzl) was fine mapped to a 70-kb region linked with seven genes in the region, further RNA-seq and re-sequencing analysis helped to narrow down the region up to two candidate genes (Cotton_A_11941 and Cotton_A_11942) with a single-base mutation in the promoter region in a NIL (DPL972) of G. arboreum (Feng et al., 2019).
Fiber Quality Improvement by Mas
The QTLs that have been identified in different populations and in different environments provide predominant evidence of putative loci related with fiber quality traits for MAS. Therefore, high density genetic mapping is a pre-requisite for marker assisted breeding. Once a stable QTL is found to be linked with the desired trait in a certain population, a composite fine-mapping approach will help to dissect the markers close to the target genes. Generally, MAS will be less effective for traits that are governed by many genes such as fiber quality traits. The breeding design and implementation scheme also depend on the efficiency of MAS. Therefore, QTLs that were consistently detected across multi-environments and retained the maximum proportion of PV should be selected for MAS (Flint-Garcia et al., 2003).
Fiber length and strength are important fiber quality traits and plenty of associated QTLs have been identified in previous decades. During the last decade, SSRs have been widely used for MAS for fiber quality in many interspecific population combinations. The stable QTLs for fiber length and strength (qFL-C7-1, qFL-chr.7, and QFS-D11-1) can be utilized in MAS programs to enhance fiber length and fiber strength of commercial cultivars in different populations. QTLs (qFL19.1, qFS03.1, and qFM19.1) identified under multi-environments and different populations have more chance to be a successful MAS program. Furthermore, markers flanking the QTL interval help to identify QTLs with different populations in different environments. Flanking markers of QTLs for fiber length (NAU1085-BNL1694, NAU1085-TMB1618, and NAU3298b-CM067) and fiber strength (BNL2572-BNL1440) could be the best targets to find novel QTLs controlling fiber quality traits in mapping populations (Lacape et al., 2005; Sun et al., 2012; Zhang et al., 2015). Fine-mapping of putative QTLs can also enhance the reliability of MAS and be helpful in developing the best fiber quality cotton cultivars.
Recently, GWAS analyses have been used to identify the key genes that were altered during the cotton fiber evolution. Two genomic loci for LINT YIELD INCREASING (GhLYI-A02 and GhLYI-D08) were identified with the mix of GWAS and gene-based association with functional annotation of identified orthologs in Arabidopsis, providing the quick method to identify the candidate genes associated with fiber quality (Fang et al., 2017a). Ma et al. (2018) characterized genes related to fiber length (GhFL1 and GhFL2) and fiber strength (Gh_A07G1769), giving targets for genetic improvement to fulfill the demand for fiber quality (Ma et al. 2018b). Cotton fiber quality improvement has occurred under domestication during evolution process and is certainly associated with fiber strength and fiber micronaire genes (MYB, ADF, and CAD) (Wang et al., 2017b). Genome wide analyses of introgression populations by genome re-sequencing and the expression of homologous genes related to fiber quality provide references to produce superior fiber (Fang et al., 2017b).
Efficiency and Targets for Future Fiber Quality Improvement
Markers assisted selection is one of the widely used breeding strategies to improve traits including cotton fiber quality traits with a multifaceted genetic basis. Several genes associated with fiber development have been reported to function at different stages of fiber development in many articles. In past decades, thousands of QTLs in diploid and tetraploid cotton species have been identified in populations developed by interspecific crosses between parents differing in fiber characteristics. The genomic sequences of cotton provide precious resources to develop high-density SSR or SNP based genetic maps. Establishing linkage between phenotype and genotypic interactions, the identification of stable QTLs lays a basis for fine mapping to dig out the related genes.
With genomic and bioinformatics approaches, it is more feasible to retrieve target QTL regions. The integration of transcriptomic analysis and QTL mapping can reveal more concrete information about fiber development mechanism. These studies should be improved in corresponding cotton genomes and among the populations developed by interspecific crosses. Cotton genomes, DNA markers and transcriptomic studies can play a major role in dissecting the mechanisms underlying fiber development to cultivate superior varieties with improved cotton fiber quality.
Author Contributions
BI wrote the manuscript. NZ participated in the discussion and the manuscript revision. JK participated in data collection and discussion. JH proposed the idea, instructed the research, revised the manuscript, and provided the work platform. All authors approved the final manuscript.
Funding
This research was supported by a grant from National Key R & D Program for Crop Breeding (2016YFD0100305) to JH, and a part from Key Cultivating Projects of Scientific and Technological Innovation for Xinjiang Academy of Agricultural Sciences (xjkcpy-001) to JK.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to Dr. Lingling Ma for helpful discussion.
Abbreviations
QTL, quantitative trait loci; MAS, marker assisted selection; GS, genomic selection; PCW, primary cell wall; SCW, secondary cell wall; TF, transcription factors; DPA, days post anthesis; RAPD, random amplified polymorphic DNA; RFLP, restriction fragment length polymorphism; SSR, simple sequence repeats; ISSR, inter simple sequence repeats; SNP, single nucleotide polymorphism; PV, phenotypic variation; LOD, likelihood of odds score; RIL, recombinant inbred line; ILs, introgression lines; NIL, near isogenic lines; IF2, immortalized F2; MAGIC, multi-parent advanced generation inter-cross; FBS, fiber bundle strength; CISPs, conserved intron scanning primers; TDFs, transcript-derived fragments.
References
Abdalla, A. M., Reddy, O. U. K., El-Zik, K. M., Pepper, A. E. (2001). Genetic diversity and relationships of diploid and tetraploid cottons revealed using AFLP. Theor. Appl. Genet. 102, 222–229. doi: 10.1007/s001220051639
Adhikari, J., Das, S., Wang, Z., Khanal, S., Chandnani, R., Patel, J. D., et al. (2017). Targeted identification of association between cotton fiber quality traits and microsatellite markers. Euphytica 213, 65. doi: 10.1007/s10681-017-1853-0
Ali, I., Teng, Z., Bai, Y., Yang, Q., Hao, Y., Hou, J., et al. (2018). A high density SLAF-SNP genetic map and QTL detection for fibre quality traits in Gossypium hirsutum. BMC Genomics 19, 879. doi: 10.1186/s12864-018-5294-5
Alvarez, I., Wendel, J. F. (2006). Cryptic interspecific introgression and genetic differentiation within Gossypium aridum (Malvaceae) and its relatives. Evol. Int. J. Org. Evol. 60, 505–517. doi: 10.1111/j.0014-3820.2006.tb01132.x
Avci, U., Pattathil, S., Singh, B., Brown, V. L., Hahn, M. G., Haigler, C. H. (2013). Cotton fiber cell walls of Gossypium hirsutum and Gossypium barbadense have differences related to Loosely-Bound xyloglucan. PloS One 8, e56315. doi: 10.1371/journal.pone.0056315
Bajwa, K. S., Shahid, A. A., Rao, A. Q., Bashir, A., Aftab, A., Husnain, T. (2015). Stable transformation and expression of GhEXPA8 fiber expansin gene to improve fiber length and micronaire value in cotton. Front. Plant Sci. 6, 838. doi: 10.3389/fpls.2015.00838
Betancur, L., Singh, B., Rapp, R. A., Wendel, J. F., Marks, M. D., Roberts, A. W., et al. (2010). Phylogenetically distinct cellulose synthase genes support secondary wall thickening in Arabidopsis shoot trichomes and cotton fiber. J. Integ. Plant Biol. 52, 205–220. doi: 10.1111/j.1744-7909.2010.00934.x
Bai, W., Xiao, Y., Zhao, J., Song, S., Hu, L., Zeng, J., et al. (2014). Gibberellin overproduction promotes sucrose synthase expression and secondary cell wall deposition in cotton fibers. PLoS One 9, e96537. doi: 10.1371/journal.pone.0096537
Brouwer, D. J., Clair, D. S. (2004). Fine mapping of three quantitative trait loci for late blight resistance in tomato using near isogenic lines (NILs) and sub-NILs. Theor. Appl. Genet. 108, 628–638. doi: 10.1007/s00122-003-1469-8
Cai, C., Ye, W., Zhang, T., Guo, W. (2014). Association analysis of fiber quality traits and exploration of elite alleles in Upland cotton cultivars/accessions (Gossypium hirsutum L.). J. Integ. Plant Biol. 56, 51–62. doi: 10.1111/jipb.12124
Cai, C., Zhu, G., Zhang, T., Guo, W. (2017). High-density 80 K SNP array is a powerful tool for genotyping G. hirsutum accessions and genome analysis. BMC Genomics 18, 654. doi: 10.1186/s12864-017-4062-2
Cao, Z. B., Zhu, X. F., Chen, H., Zhang, T. Z. (2015). Fine mapping of clustered quantitative trait loci for fiber quality on chromosome 7 using a Gossypium barbadense introgressed line. Mol. Breed. 35, 215. doi: 10.1007/s11032-015-0393-3
Chee, P. W., Draye, X., Jiang, C. X., Decanini, L., Delmonte, T. A., Bredhauer, R., et al. (2005). Molecular dissection of phenotypic variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: III Fiber length. Theor. Appl. Genet. 111, 772–781. doi: 10.1007/s00122-005-2062-0
Chen, J., Burke, J. J. (2015). Developing fiber specific promoter-reporter transgenic lines to study the effect of abiotic stresses on fiber development in cotton. PloS One 10, e0129870. doi: 10.1371/journal.pone.0129870
Chen, Z. J., Scheffler, B. E., Dennis, E., Triplett, B. A., Zhang, T., Guo, W., et al. (2007). Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 145, 1303–1310. doi: 10.1104/pp.107.107672
Chen, G. D., Li, H. B., Zheng, Z., Wei, Y. M., Zheng, Y. L., McIntyre, C. L., et al. (2012). Characterization of a QTL affecting spike morphology on the long arm of chromosome 3H in barley (Hordeum vulgare L.) based on near isogenic lines and a NIL-derived population. Theor. Appl. Genet. 125, 1385–1392. doi: 10.1007/s00122-012-1918-3
Chen, Y., Liu, G., Ma, H., Song, Z., Zhang, C., Zhang, J., et al. (2018). Identification of introgressed alleles conferring high fiber quality derived from GOSSYPIUM BARBADENSE L. @ in secondary mapping populations of G. hirsutum L. Front. Plant Sci. 9, 1023. doi: 10.3389/fpls.2018.01023
Cheng, Y., Lu, L., Yang, Z., Wu, Z., Qin, W., Yu, D., et al. (2016). GhCaM7-like, a calcium sensor gene, influences cotton fiber elongation and biomass production. Plant Physiol. Biochem. 109, 128–136. doi: 10.1016/j.plaphy.2016.09.009
Cosgrove, D. J. (1997). Assembly and enlargement of the primary cell wall in plants. Ann. Rev. Cell Dev. Biol. 13, 171–201. doi: 10.1146/annurev.cellbio.13.1.171
Delmer, D. P. (1987). Cellulose biosynthesis. Ann. Rev. Plant Physiol. 38, 259–290. doi: 10.1146/annurev.pp.38.060187.001355
Deng, F., Tu, L., Tan, J., Li, Y., Nie, Y., Zhang, X. (2012). GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol. 158, 890–904. doi: 10.1104/pp.111.186742
Ding, X., Li, X., Xiong, L. (2011). Evaluation of near-isogenic lines for drought resistance QTL and fine mapping of a locus affecting flag leaf width, spikelet number, and root volume in rice. Theor. Appl. Genet. 123, 815–826. doi: 10.1007/s00122-011-1629-1
Diouf, L., Magwanga, R. O., Gong, W., He, S., Pan, Z., et al. (2018). QTL mapping of fiber quality and yield-related traits in an intra-specific upland cotton using genotype by sequencing (GBS). Int. J. Mol. Sci. 19, 441. doi: 10.3390/ijms19020441
Dong, C., Wang, J., Yu, Y., Ju, L., Zhou, X., Ma, X., et al. (2019). Identifying functional genes influencing Gossypium hirsutum fiber quality. Front. Plant Sci. 9, 1968. doi: 10.3389/fpls.2018.01968
Du, S. J., Dong, C. J., Zhang, B., Lai, T. F., Du, X. M., Liu, J. Y. (2013). Comparative proteomic analysis reveals differentially expressed proteins correlated with fuzz fiber initiation in diploid cotton (Gossypium arboreum L.). J. Proteom. 82, 113–129. doi: 10.1016/j.jprot.2013.02.020
Duggleby, R. G., Dennis, D. T. (1973). Pyruvate kinase, a possible regulatory enzyme in higher plants. Plant Physiol. 52, 312–317. doi: 10.1104/pp.52.4.312
Ehlting, J., Mattheus, N., Aeschliman, D. S., Li, E., Hamberger, B., Cullis, I. F., et al. (2005). Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 42, 618–640. doi: 10.1111/j.1365-313X.2005.02403.x
Fan, L., Wang, L., Wang, X., Zhang, H., Zhu, Y., Guo, J., et al. (2018). A high-density genetic map of extra-long staple cotton (Gossypium barbadense) constructed using genotyping-by-sequencing based single nucleotide polymorphic markers and identification of fiber traits-related QTL in a recombinant inbred line population. BMC Genomics 19, 489. doi: 10.1186/s12864-018-4890-8
Fang, D. D., Jenkins, J. N., Deng, D. D., McCarty, J. C., Li, P., Wu, J. (2014). Quantitative trait loci analysis of fiber quality traits using a random-mated recombinant inbred population in Upland cotton (Gossypium hirsutum L.). BMC Genomics 15, 397. doi: 10.1186/1471-2164-15-397
Fang, X., Liu, X., Wang, X., Wang, W., Liu, D., Zhang, J., et al. (2017). Fine-mapping qFS07.1 controlling fiber strength in upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 130, 795–806. doi: 10.1007/s00122-017-2852-1
Fang, L., Gong, H., Hu, Y., Liu, C., Zhou, B., Huang, T., et al. (2017a). Genomic insights into divergence and dual domestication of cultivated allotetraploid cottons. Genome Biol. 18, 33. doi: 10.1186/s13059-017-1167-5
Fang, L., Wang, Q., Hu, Y., Jia, Y., Chen, J., Liu, B., et al. (2017b). Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat. Genet. 49, 1089–1098. doi: 10.1038/ng.3887
Felker, G. S. (2001). “Fiber quality and new spinning technologies,” in Beltwide cotton conferences. National Cotton Council of America. Eds. Dugger, P., Richter, D. C. (Anaheim, U.S.A.), 5–7.
Feng, L., Zhang, S., Xing, L., Yang, B., Gao, X., Xie, X., et al. (2019). QTL analysis for yield and fibre quality traits using three sets of introgression lines developed from three Gossypium hirsutum race stocks. Mol. Genet. Genomics 294, 789. doi: 10.1007/s00438-019-01548-w
Flint-Garcia, S. A., Darrah, L. L., McMullen, M. D., Hibbard, B. E. (2003). Phenotypic versus marker-assisted selection for stalk strength and second-generation European corn borer resistance in maize. Theor. Appl. Genet. 107, 1331–1336. doi: 10.1007/s00122-003-1387-9
Freshour, G., Bonin, C. P., Reiter, W. D., Albersheim, P., Darvill, A. G., Hahn, M. G. (2003). Distribution of fucose-containing xyloglucans in cell walls of the mur1 mutant of Arabidopsis. Plant Physiol. 131, 1602–1612. doi: 10.1104/pp.102.016444
Fridman, E., Pleban, T., Zamir, D. (2000). A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl. Acad. Sci. U.S.A. 97, 4718–4723. doi: 10.1073/pnas.97.9.4718
Fryxell, P. A. (1992). A revised taxonomic interpretation of Gossypium L. (Malvaceae). Rheedea 2, 108–116.
Gallagher, J. P., Grover, C. E., Rex, K., Moran, M., Wendel, J. F. (2017). A new species of cotton from Wake Atoll, Gossypium stephensii (Malvaceae). Syst. Bot. 42, 115–123. doi: 10.1600/036364417X694593
Gao, P., Zhao, P. M., Wang, J., Wang, H. Y., Wu, X. M., Xia, G. X. (2007). Identification of genes preferentially expressed in cotton fibers: a possible role of calcium signaling in cotton fiber elongation. Plant Sci. 173, 61–69. doi: 10.1016/j.plantsci.2007.04.008
Guan, X. Y., Li, Q. J., Shan, C. M., Wang, S., Mao, Y. B., et al. (2008). The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiol. Plant. 134, 174–182. doi: 10.1111/j.1399-3054.2008.01115.x
Haigler, C. H., Zhang, D., Wilkerson, C. G. (2005). Biotechnological improvement of cotton fibre maturity. Physiol. Plant. 124, 285–294. doi: 10.1111/j.1399-3054.2005.00480.x
Haigler, C. H., Singh, B., Zhang, D., Hwang, S., Wu, C., Cai, W. X., et al. (2007). Transgenic cotton over-producing spinach sucrose phosphate synthase showed enhanced leaf sucrose synthesis and improved fiber quality under controlled environmental conditions. Plant Mol. Biol. 63, 815–832. doi: 10.1007/s11103-006-9127-6
Haigler, C. H., Singh, B., Wang, G., Zhang, D. (2009). “Genomics of cotton fiber secondary wall deposition and cellulose biogenesis,” in Genetics and Genomics of Cotton. Ed. Paterson, A. H. (New York: Springer), 385–417.
Haigler, C. H., Betancur, L., Stiff, M. R., Tuttle, J. R. (2012). Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front. Plant Sci. 3, 104. doi: 10.3389/fpls.2012.00104
Han, L. B., Li, Y. B., Wang, H. Y., Wu, X. M., Li, C. L., Luo, M., et al. (2013). The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell 25, 4421–4438. doi: 10.1105/tpc.113.116970
Hinchliffe, D. J., Meredith, W. R., Yeater, K. M., Kim, H. J., Woodward, A. W., et al. (2010). Near-isogenic cotton germplasm lines that differ in fiber-bundle strength have temporal differences in fiber gene expression patterns as revealed by comparative high-throughput profiling. Theor. Appl. Genet. 120, 1347–1366. doi: 10.1007/s00122-010-1260-6
Hu, G., Koh, J., Yoo, M. J., Grupp, K., Chen, S., Wendel, J. F. (2013). Proteomic profiling of developing cotton fibers from wild and domesticated Gossypium barbadense. New Phytol. 200, 570–582. doi: 10.1111/nph.12381
Hu, G., Koh, J., Yoo, M. J., Chen, S., Wendel, J. F. (2015). Gene-expression novelty in allopolyploid cotton: a proteomic perspective. Genetics 200, 91–104. doi: 10.1534/genetics.115.174367
Huang, G. Q., Gong, S. Y., Xu, W. L., Li, W., Li, P, Zhang, C. J., et al. (2013). A fasciclin-like arabinogalactan protein, GhFLA1, is involved in fiber initiation and elongation of cotton. Plant Physiol. 161, 1278–1290. doi: 10.1104/pp.112.203760
Huang, C., Shen, C., Wen, T., Gao, B., Zhu, D., Li, X., et al. (2018). SSR-based association mapping of fiber quality in upland cotton using an eight-way MAGIC population. Mol. Genet. Genomics 293, 793. doi: 10.1007/s00438-018-1419-4
Hulse-Kemp, A. M., Lemm, J., Plieske, J., Ashrafi, H., Buyyarapu, R., Fang, D. D., et al. (2015). Development of a 63K SNP array for cotton and high-density mapping of intraspecific and interspecific populations of Gossypium spp. G3. Genes Genomes Genet. 5, 1187–1209. doi: 10.1534/g3.115.018416
Islam, M. S., Zeng, L., Delhom, C. D., Song, X., Kim, H. J., Li, P., et al. (2014). Identification of cotton fiber quality quantitative trait loci using intraspecific crosses derived from two near-isogenic lines differing in fiber bundle strength. Mol. Breed. 34, 373–384. doi: 10.1007/s00122-016-2684-4
Islam, M. S., Zeng, L., Thyssen, G. N., Delhom, C. D., Kim, H. J., Li, P., et al. (2016). Mapping by sequencing in cotton (Gossypium hirsutum) line MD52ne identified candidate genes for fiber strength and its related quality attributes. Theor. Appl. Genet. 129, 1071–1086. . doi: 1007/s00122-016-2684-4
Jamshed, M., Jia, F., Gong, J., Palanga, K. K., Shi, Y., et al. (2016). Identification of stable quantitative trait loci (QTLs) for fiber quality traits across multiple environments in Gossypium hirsutum recombinant inbred line population. BMC Genomics 17, 197. doi: 10.1186/s12864-016-2560-2
Jia, X., Pang, C., Wei, H., Wang, H., Ma, Q., Yang, J., et al. (2016). High density linkage map construction and QTL analysis for earliness-related traits in Gossypium hirsutum L. BMC Genomics 17, 909. doi: 10.1186/s12864-016-3269-y
Jiang, Y, Guo, W, Zhu, H, Ruan, YL, Zhang, T. (2012). Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality. Plant Biotechnol. J. 10, 301–12. doi: 10.1111/j.1467-7652.2011.00662.x
Jiang, C. X., Wright, R. J., El-Zik, K. M., Paterson, A. H. (1998). Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc. Natl. Acad. Sci. U. S. A. 95, 4419–4424. doi: 10.1073/pnas.95.8.4419
Jiang, Y., Guo, W., Zhu, H., Ruan, Y. L., Zhang, T. (2012). Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality. Plant Biotechol. J. 10, 301–312. doi: 10.1073/pnas.95.8.4419
Kawai, M., Aotsuka, S., Uchimiya, H. (1998). Isolation of a cotton cap gene: a homologue of adenylyl cyclase-associated protein highly expressed during fiber elongation. Plant Cell Physiol. 39, 1380–1383. doi: 10.1093/oxfordjournals.pcp.a029346
Keerio, A. A., Shen, C., Nie, Y., Ahmed, M. M., Zhang, X., Lin, Z. (2018). QTL Mapping for Fiber Quality and Yield Traits Based on Introgression Lines Derived from Gossypium hirsutum G. tomentosum. Int. J. Mol. Sci. 19, 243. doi: 10.3390/ijms19010243
Kim, H. J., Triplett, B. A. (2001). Cotton fiber growth in planta and in vitro. Models Plant Cell elongation Cell wall biogenesis. Plant Physiol. 127, 1361–1366. doi: 10.1104/pp.010724
Kohel, R. J., Yu, J., Park, Y. H., Lazo, G. R. (2001). Molecular mapping and characterization of traits controlling fiber quality in cotton. Euphytica 121, 163–172. doi: 10.1023/A:1012263413418
Lacape, J. M., Nguyen, T. B., Courtois, B., Belot, J. L., Giband, M., Gourlot, J. P., et al. (2005). QTL analysis of cotton fiber quality using multiple Gossypium hirsutum × Gossypium barbadense backcross generations. Crop Sci. 45, 123–140. doi: 10.2135/cropsci2005.0123a
Lee, J. J., Woodward, A. W., Chen, Z. J. (2007). Gene expression changes and early events in cotton fibre development. Ann. Bot. 100, 1391–1401. doi: 10.1093/aob/mcm232
Li, X. B., Fan, X. P., Wang, X. L., Cai, L., Yang, W. C. (2005). The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17, 859–875. doi: 10.1105/tpc.104.029629
Li, X. B., Xiao, Y. H., Luo, M., Hou, L., Li, D. M., Luo, X. Y., et al. (2005a). Cloning and expression analysis of two Rac genes from cotton (Gossypium hirsutum L.). Acta Genetica Sin. 32, 72–78.
Li, C., Guo, W., Zhang, T. (2009). Fiber initiation development in Upland cotton (Gossypium hirsutum L.) cultivars varying in lint percentage. Euphytica 165, 223. doi: 10.1007/s10681-008-9740-3
Li, A., Xia, T., Xu, W., Chen, T., Li, X., Fan, J., et al. (2013). An integrative analysis of four CESA isoforms specific for fiber cellulose production between Gossypium hirsutum and Gossypium barbadense. Planta 237, 1585–1597. doi: 10.1007/s00425-013-1868-2
Li, D. D., Ruan, X. M., Zhang, J., Wu, Y. J., Wang, X. L., Li, X. B. (2013). Cotton plasma membrane intrinsic protein 2s (PIP2s) selectively interact to regulate their water channel activities and are required for fibre development. New Phytol. 199, 695–670. doi: 10.1111/nph.12309
Li, Y. J., Zhang, X. Y., Wang, F. X., Yang, C. L., Liu, F., Xia, G. X., et al. (2013). A comparative proteomic analysis provides insights into pigment biosynthesis in brown color fiber. J. Proteom. 78, 374–388. doi: 10.1016/j.jprot.2012.10.005
Li, Y. J., Wang, F. X., Wang, Y. Q., Liu, Y. C., Zhang, X. Y., Sun, Y. Q., et al. (2013a). Identification of the proteins in green cotton fiber using a proteomics-based approach. Biotechnol. Lett. 35, 1519–1523. doi: 10.1007/s10529-013-1227-1
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Li, C., Dong, Y., Zhao, T., Li, L., Yu, E., Mei, L., et al. (2016). Genome-wide SNP linkage mapping and QTL analysis for fiber quality and yield traits in the upland cotton recombinant inbred lines population. Front. Plant Sci. 7, 1356. doi: 10.3389/fpls.2016.01356
Li, C., Fu, Y., Sun, R., Wang, Y., Wang, Q. (2018). Single-locus and multi-locus genome-wide association studies in the genetic dissection of fiber quality traits in upland cotton (Gossypium hirsutum L.). Front. Plant Sci. 9, 1083. doi: 10.3389/fpls.2018.01083
Li, C., Yu, H., Li, C., Zhao, T., Dong, Y., et al. (2018). QTL mapping and heterosis analysis for fiber quality traits across multiple genetic populations and environments in upland cotton. Front. Plant Sci. 9, 1364. doi: 10.3389/fpls.2018.01364
Li, S. Q., Liu, A. Y., Kong, L. L., Gong, J. W., Li, J. W., Gong, W. K., et al. (2019). QTL mapping and genetic effect of chromosome segment substitution lines with excellent fiber quality from Gossypium hirsutum Gossypium barbadense. Mol. Genet. Genom. 294, 1123-1136. doi: 10.1007/s00438-019-01566-8
Liang, Q., Hu, C., Hua, H., Li, Z., Hua, J. (2013). Construction of a linkage map and QTL mapping for fiber quality traits in upland cotton (Gossypium hirsutum L.). Chin. Sci. Bull. 58, 3233–3243. doi: 10.1007/s11434-013-5807-1
Liu, H. C., Creech, R. G., Jenkins, J. N., Ma, D. P. (2000). Cloning and promoter analysis of the cotton lipid transfer protein gene Ltp3. Bioch. Biophys. Acta 1487, 106–111. doi: 10.1016/S1388-1981(00)00072-X
Liu, D., Tu, L., Wang, L., Li, Y., Zhu, L., Zhang, X. (2008). Characterization and expression of plasma and tonoplast membrane aquaporins in elongating cotton fibers. Plant Cell Rep. 27, 1385–1394. doi: 10.1007/s00299-008-0545-6
Liu, K., Han, M., Zhang, C., Yao, L., Sun, J., Zhang, T. (2012). Comparative proteomic analysis reveals the mechanisms governing cotton fiber differentiation and initiation. J. Proteom. 75, 845–856. doi: 10.1016/j.jprot.2011.09.025
Liu, X., Zhao, B., Zheng, H. J., Hu, Y., Lu, G., Yang, C. Q., et al. (2015). Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci. Rep. 5, 14139. doi: 10.1038/srep14139
Liu, D., Zhang, J., Liu, X., Wang, W., Liu, D., Teng, Z., et al. (2016). Fine mapping and RNA-Seq unravels candidate genes for a major QTL controlling multiple fiber quality traits at the T1 region in upland cotton. BMC Genomics 17, 295. doi: 10.1186/s12864-016-2605-6
Liu, X., Teng, Z., Wang, J., Wu, T., Zhang, Z., Deng, X., et al. (2017). Enriching an intraspecific genetic map and identifying QTL for fiber quality and yield component traits across multiple environments in Upland cotton (Gossypium hirsutum L.). Mol. Genet. Genom. 292, 1281. doi: 10.1007/s00438-017-1347-8
Liu, R., Gong, J., Xiao, X., Zhang, Z., Li, J., Liu, A., et al. (2018). GWAS analysis and QTL identification of fiber quality traits and yield components in upland cotton using enriched high-density SNP markers. Front. Plant Sci. 9, 1067. doi: 10.3389/fpls.2018.01067
Lu, H., Myers, G. (2002). Genetic relationships and discrimination of ten influential upland cotton varieties using RAPD markers. Theor. Appl. Genet. 105, 325–331. doi: 10.1007/s00122-002-0947-8
Ma, G. J., Zhang, T. Z., Guo, W. Z. (2006). Cloning and characterization of cotton GhBG gene encoding β- glucosidase. DNA Seq. 17, 355–62.
Ma, L., Zhao, Y., Wang, Y., Shang, L., Hua, J. (2017). QTLs analysis and validation for fiber quality traits using maternal backcross population in Upland cotton. Front. Plant Sci. 8, 2168. doi: 10.3389/fpls.2017.02168
Ma, Z., He, S., Wang, X., Sun, J., Zhang, Y., et al. (2018). Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 50, 803–813. doi: 10.1038/s41588-018-0119-7
Ma, L., Wang, Y., Ijaz, B., Hua, J. (2019). Cumulative and different genetic effects contributed to yield heterosis using maternal and paternal testcross populations in Upland cotton. Sci. Rep. 9, 3984. doi: 10.1038/s41598-019-40611-9
Machado, A., Wu, Y., Yang, Y., Llewellyn, D. J., Dennis, E. S. (2009). The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 59, 52–62. doi: 10.1111/j.1365-313X.2009.03847.x
Mittler, R., Vanderauwera, S., Gollery, M., Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Morgante, M., Hanafey, M., Powell, W. (2002). Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 30, 194. doi: 10.1038/ng822
Naoumkina, M., Thyssen, G. N., Fang, D. D., Jenkins, J. N., McCarty, J. C., Florane, C. B. (2019). Genetic and transcriptomic dissection of the fiber length trait from a cotton (Gossypium hirsutum L.) MAGIC population. BMC Genomics 20, 112. doi: 10.1186/s12864-019
Nawaz, M. A., Rehman, H. M., Baloch., F. S., Ijaz, B., Ali, M. A., Khan, I. A., et al. (2017). Genome and transcriptome-wide analyses of cellulose synthase gene superfamily in soybean. J. Plant Physiol. 215, 163–175. doi: 10.1016/j.jplph.2017.04.009
Nie, X., Huang, C., You, C., Li, W., Zhao, W., Shen, C., et al. (2016). Genome-wide SSR-based association mapping for fiber quality in nation-wide upland cotton inbreed cultivars in China. BMC Genomics 17, 352. doi: 10.1186/s12864-016-2662-x
Pang, C. Y., Wang, H., Pang, Y., Xu, C., Jiao, Y., Qin, Y. M., et al. (2010). Comparative proteomics indicates that biosynthesis of pectic precursors is important for cotton fiber and Arabidopsis root hair elongation. Mol. Cel. Proteom. 9, 2019–2033. doi: 10.1074/mcp.M110.000349
Paterson, A. H., DeVerna, J. W., Lanini, B., Tanksley, S. D. (1990). Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in an interspecies cross of tomato. Genetics 124, 735–742. doi: 10.1038/nature11798
Paterson., A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423.
Pear, J. R., Kawagoe, Y., Schreckengost, W. E., Delmer, D. P., Stalker, D. M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. U. S. A. 93, 12637–12642. doi: 10.1073/pnas.93.22.12637
Polko, J. K., Kieber, J. J. (2019). The regulation of cellulose biosynthesis in plants. Plant Cell. 31, 282–296. doi: 10.1105/tpc.18.00760
Powell, W., Machray, G. C., Provan, J. (1996). Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1, 215–222. doi: 10.1016/1360-1385(96)86898-1
Pu, L., Li, Q., Fan, X., Yang, W., Xue, Y. (2008). The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180, 811–820. doi: 10.1534/genetics.108.093070
Qi, H., Wang, N., Qiao, W., Xu, Q., Zhou, H., Shi, J., et al. (2017). Construction of a high-density genetic map using genotyping by sequencing (GBS) for quantitative trait loci (QTL) analysis of three plant morphological traits in upland cotton (Gossypium hirsutum L.). Euphytica 213, 83. doi: 10.1007/s10681-017-1867-7
Qin, Y. M., Zhu, Y. X. (2011). How cotton fibers elongate: a tale of linear cell-growth mode. Curr. Opin. Plant Biol. 14, 106–111. doi: 10.1016/j.pbi.2010.09.010
Qin, Y. M., Hu, C. Y., Pang, Y., Kastaniotis, A. J., Hiltunen, J. K., Zhu, Y. X. (2007). Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 19, 3692–3704. doi: 10.1105/tpc.107.054437
Qin, L. X., Chen, Y., Zeng, W., Li, Y., Gao, L., Li, D. D., et al. (2017). The cotton β-galactosyltransferase 1 (GalT1) that galactosylates arabinogalactan proteins participates in controlling fiber development. Plant J. 89, 957–971. doi: 10.1111/tpj.13434
Reddy, M. P., Sarla, N., Siddiq, E. A. (2002). Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 128, 9–17. doi: 10.1023/A:1020691618797
Reinisch, A. J., Dong, J. M., Brubaker, C. L., Stelly, D. M., Wendel, J. F., Paterson, A. H. (1994). A detailed RFLP map of cotton, Gossypium hirsutum x Gossypium barbadense: chromosome organization and evolution in a disomic polyploid genome. Genetics 138, 829–847.
Ribaut, J. M., Hoisington, D. (1998). Marker-assisted selection: New tools and strategies. Trends Plant Sci. 3, 236–239. doi: 10.1016/S1360-1385(98)01240-0
Rodgers, J., Zumba, J., Fortier, C. (2017). Measurement comparison of cotton fiber micronaire and its components by portable near infrared spectroscopy instruments. Text. Res. J. 87, 57–69. doi: 10.1177/0040517515622153
Ruan, Y. L., Llewellyn, D. J., Furbank, R. T. (2003). Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15, 952–964. doi: 10.1105/tpc.010108
Ruan, Y. L., Xu, S. M., White, R., Furbank, R. T. (2004). Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fiber elongation mediated by callose turnover. Plant Physiol. 136, 4104–4113. doi: 10.1104/pp.104.051540
Ruan, Y. L. (2005). Recent advances in understanding cotton fibre and seed development. Seed Sci. Res. 15, 269–280. doi: 10.1079/SSR2005217
Said, J. I., Lin, Z., Zhang, X., Song, M., Zhang, J. (2013). A comprehensive meta QTL analysis for fiber quality, yield, yield related and morphological traits, drought tolerance, and disease resistance in tetraploid cotton. BMC Genomics 14, 776. doi: 10.1186/1471-2164-14-776
Said, J. I., Knapka, J. A., Song, M. Z., Zhang, J. F. (2015a). Cotton QTLdb: a cotton QTL database for QTL analysis, visualization, and comparison between Gossypium hirsutum and G. hirsutum × G. barbadense populations. Mol. Genet. Genom. 290, 1615–1625. doi: 10.1007/s00438-015-1021-y
Said, J. I., Song, M., Wang, H., Lin, Z., Zhang, X., Fang, D., et al. (2015b). A comparative meta-analysis of QTL between intraspecific Gossypium hirsutum and interspecific G. hirsutum × G. barbadense populations. Mol. Genet. Genomics 290, 1003. doi: 10.1007/s00438-014-0963-9
Shan, C. M., Shangguan, X. X., Zhao, B., Zhang, X. F., Chao, L. M., et al. (2014). Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat. Commun. 5, 5519. doi: 10.1038/ncomms6519
Shang, L., Liang, Q., Wang, Y., Wang, X., Wang, K., Abduweli, A., et al. (2015). Identification of stable QTLs controlling fiber traits properties in multi-environment using recombinant inbred lines in Upland cotton (Gossypium hirsutum L.). Euphytica 205, 877–888. doi: 10.1007/s10681-015-1434-z
Shang, L., Wang, Y., Wang, X., Liu, F., Abduweli, A., Cai, S., et al. (2016). Genetic analysis and QTL detection on fiber traits using two recombinant inbred lines and their backcross populations in upland cotton. G3. Gene Genom. Genet. 6, 2717–2724. doi: 10.1534/g3.116.031302
Shao, Q., Zhang, F., Tang, S., Liu, Y., Fang, X., Liu, D., et al. (2014). Identifying QTL for fiber quality traits with three upland cotton (Gossypium hirsutum L.) populations. Euphytica 198, 43–58. doi: 10.1007/s10681-014-1082-8
Shappley, Z. W., Jenkins, J. N., Zhu, J., McCarty, J. C. (1998). Quantitative trait loci associated with agronomic and fiber traits of Upland Cotton. J. Cotton Sci. 4, 153–163.
Shi, Y. H., Zhu, S. W., Mao, X. Z., Feng, J. X., Qin, Y. M., Zhang, L., et al. (2006). Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell. 18, 651–664. doi: 10.1105/tpc.105.040303
Shen, X., Guo, W., Zhu, X., Yuan, Y., John, Z. Y., Kohel, R. J., et al. (2005). Molecular mapping of QTLs for fiber qualities in three diverse lines in Upland cotton using SSR markers. Mol. Breed. 15, 169–181. doi: 10.1007/s11032-004-4731-0
Shi, H., Wang, X., Li, D., Tang, W., Wang, H., Xu, W., et al. (2007). Molecular characterization of Cotton 14-3-3L gene preferentially expressed during fiber elongation. J. Genet. Genomics 34, 151–159. doi: 10.1016/s1673-8527(07)60016-2
Shi, Y., Li, W., Li, A., Ge, R., Zhang, B., Li, J., et al. (2015). Constructing a high-density linkage map for Gossypium hirsutum × Gossypium barbadense and identifying QTLs for lint percentage. J. Integ. Plant Biol. 57, 450–467. doi: 10.1111/jipb.12288
Singh, B., Avci, U., Inwood, S. E. E., Grimson, M. J., Landgraf, J., Mohnen, D., et al. (2009). A specialized outer layer of the primary cell wall joins elongating cotton fibers into tissue-like bundles. Plant Physiol. 150, 684–699. doi: 10.1104/pp.109.135459
Su, C., Wang, W., Qiu, X., Yang, L., Li, S., Wang, M., et al. (2013). Fine-mapping a fibre strength QTL QFS-D11-1 on cotton chromosome 21 using introgressed lines. Plant Breed. 132, 725–730. doi: 10.1007/s00122-017-2852-1
Su, J., Li, L., Pang, C., Wei, H., Wang, C., Song, M., et al. (2016). Two genomic regions associated with fiber quality traits in Chinese upland cotton under apparent breeding selection. Sci. Rep. 6, 38496. doi: 10.1038/srep38496
Su, J., Ma, Q., Li, M., Hao, F., Wang, C. (2018). Multi-locus genome-wide association studies of fiber-quality related traits in Chinese early-maturity upland cotton. Front. Plant Sci. 9, 1169. doi: 10.3389/fpls.2018.01169
Sun, F. D., Zhang, J. H., Wang, S. F., Gong, W. K., Shi, Y. Z., Liu, A. Y., et al. (2012). QTL mapping for fiber quality traits across multiple generations and environments in upland cotton. Mol. Breed. 30, 569–582. doi: 10.1007/s11032-011-9645-z
Sun, W., Gao, Z., Wang, J., Huang, Y., Chen, Y., Li, J., et al. (2019). Cotton fiber elongation requires the transcription factor GhMYB212 to regulate sucrose transportation into expanding fibers. New Phytol. 222, 864–881. doi: 10.1111/nph.15620
Suo, J., Liang, X., Pu, L., Zhang, Y., Xue, Y. (2003). Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Bioch. Biophys. Acta 1630, 25–34. doi: 10.1016/j.bbaexp.2003.08.009
Szalma, S. J., Hostert, B. M., LeDeaux, J. R., Stuber, C. W., Holland, J. B. (2007). QTL mapping with near-isogenic lines in maize. Theor. Appl. Genet. 114, 1211–1228. doi: 10.1007/s00122-007-0512-6
Tan, Z. Y., Fang, X. M., Tang, S. Y., Zhang, J., Liu, D. J., Teng, Z. H., et al. (2015). Genetic map and QTL controlling fiber quality traits in Upland cotton (Gossypium hirsutum L.). Euphytica 203, 615–628. doi: 10.1007/s10681-014-1288-9
Tan, Z., Zhang, Z., Sun, X., Li, Q., Sun, Y., Yang, P., et al. (2018). Genetic map construction and fiber quality QTL mapping using the CottonSNP80K array in upland cotton. Front. Plant Sci. 9, 225. doi: 10.3389/fpls.2018.00225
Tang, W., Tu, L., Yang, X., Tan, J., Deng, F., Hao, J., et al. (2014). The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol. 202, 509–520. doi: 10.1111/nph.12676
Tang, S. Y., Teng, Z. H., Zhai, T. F., Fang, X. M., Liu, F., Liu, D. J., et al. (2015). Construction of genetic map and QTL analysis of fiber quality traits for Upland cotton (Gossypium hirsutum L.). Euphytica 201, 195–213. doi: 10.1007/s10681-014-1189-y
Tatineni, V., Cantrell, R. G., Davis, D. D. (1996). Genetic diversity in elite cotton germplasm determined by morphological characteristics and RAPDs. Crop Sci. 36, 186–192. doi: 10.2135/cropsci1996.0011183X003600010033x
Taylor, N. G., Howells, R. M., Huttly, A. K., Vickers, K., Turner, S. R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 1450–1455. doi: 10.1073/pnas.0337628100
Thomson, M. J., Edwards, J. D., Septiningsih, E. M., Harrington, S. E., McCouch, S. R. (2006). Substitution mapping of dth1.1, a flowering-time quantitative trait locus (QTL) associated with transgressive variation in rice, reveals multiple sub-QTL. Genetics 172, 2501–2514. doi: 10.1534/genetics.105.050500
Ulloa, M., Cantrell, R. G., Percy, R. G., Zeiger, E., Lu, Z. (2000). QTL analysis of stomatal conductance and relationship to lint yield in an interspecific cotton. J. Cotton Sci. 4, 10–18.
Walford, S. A., Wu, Y., Llewellyn, D. J., Dennis, E. S. (2011). GhMYB25-like: a key factor in early cotton fibre development. Plant J. 65, 785–797. doi: 10.1111/j.1365-313X.2010.04464.x
Wang, S., Wang, J. W., Yu, N., Li, C. H., Luo, B., Gou, J. Y., et al. (2004). Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16, 2323–2334. doi: 10.1105/tpc.104.024844
Wang, B., Guo, W., Zhu, X., Wu, Y., Huang, N., Zhang, T. (2006). QTL mapping of fiber quality in an elite hybrid derived-RIL population of upland cotton. Euphytica 152, 367–378. doi: 10.1007/s10681-006-9224-2
Wang, B., Wu, Y., Guo, W., Zhu, X., Huang, N., Zhang, T. (2007). QTL analysis and epistasis effects dissection of fiber qualities in an elite cotton hybrid grown in second generation. Crop Sci. 47, 1384–1392. doi: 10.2135/cropsci2006.10.0647
Wang, L., Li, X. R., Lian, H., Ni, D. A., He, Y. K., et al. (2010). Evidence that high activity of vacuolar invertase is required for cotton fiber andArabidopsis root elongation through osmotic dependent and independent pathways, respectively. Plant Physiol. 154, 744–756. doi: 10.1104/pp.110.162487
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103. doi: 10.1038/ng.2371
Wang, L., Cook, A., Patrick, J. W., Chen, X. Y., Ruan, Y. L. (2014). Silencing the vacuolar invertase gene GhVIN1 blocks cotton fiber initiation from the ovule epidermis, probably by suppressing a cohort of regulatory genes via sugar signaling. Plant J. 78, 686–696. doi: 10.1111/tpj.12512
Wang, H., Huang, C., Guo, H., Li, X., Zhao, W., Dai, B., et al. (2015). QTL mapping for fiber and yield traits in upland cotton under multiple environments. PloS One 10,pe0130742. doi: 10.1371/journal.pone.0130742
Wang, H., Jin, X., Zhang, B., Shen, C., Lin, Z. (2015). Enrichment of an intraspecific genetic map of upland cotton by developing markers using parental RAD sequencing. DNA Res. 22, 147–160. doi: 10.1093/dnares/dsu047
Wang, M., Yuan, D., Tu, L., Gao, W., He, Y., Hu, H., et al. (2015a). Long noncoding RNAs and their proposed functions in fibre development of cotton (Gossypium spp.). New Phytol. 207, 1181–1197. doi: 10.1111/nph.13429
Wang, M., Wang, P., Tu, L., Zhu, S., Zhang, L., Li, Z., et al. (2016). Multi-omics maps of cotton fibre reveal epigenetic basis for staged single-cell differentiation. Nucleic Acids Res. 44, 4067–4079. doi: 10.1093/nar/gkw238
Wang, H., Huang, C., Zhao, W., Dai, B., Shen, C., Zhang, B., et al. (2016a). Identification of QTL for fiber quality and yield traits using two immortalized backcross populations in Upland Cotton. PloS One 11,pe0166970. doi: 10.1371/journal.pone.0166970
Wang, B., Draye, X., Zhuang, Z., Zhang, Z., Liu, M., Lubbers, E. L., et al. (2017). QTL analysis of cotton fiber length in advanced backcross populations derived from a cross between Gossypium hirsutum and G. mustelinum. Theor. Appl. Genet. 6, 1297–1308. doi: 10.1007/s00122-017-2889-1
Wang, B., Zhuang, Z., Zhang, Z., Draye, X., Shuang, L.-S., Shehzad, T., et al. (2017a). Advanced Backcross QTL Analysis of Fiber Strength and Fineness in a Cross between Gossypium hirsutum and G. mustelinum. Front. Plant Sci. 8, 1848. doi: 10.3389/fpls.2017.01848
Wang, M., Tu, L., Lin, M., Lin, Z., Wang, P., Yang, Q., et al. (2017b). Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat Genet. 49, 579–587. doi: 10.1038/ng.3807
Wang, K., Wendel, J. F., Hua, J. (2018). Designations for individual genomes and chromosomes in Gossypium. J. Cotton Res. 1, 3. doi: s42397-018-0002-1
Wang, M., Tu, L., Yuan, D., Zhu, D., Shen, C., Li, J., et al. (2019). Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 51, 224–229. doi: 10.1038/s41588-018-0282-x
Wen, W., Cui, B., Yu, X., Chen, Q., Zheng, Y., Xia, Y., et al. (2012). Functional analysis of cotton DELLA-Like genes that are differentially regulated during fiber development. Plant Mol. Biol. Rep. 30, 1014–1024. doi: 10.1007/s11105-012-0412-z
Wendel, J. F., Grover, C. E. (2015). Taxonomy and evolution of the cotton genus. Eds. Fang, D., Percy, R. (Madison, WI: Cotton American Society of Agronomy, Inc, Crop Science Society of America, Inc, and Soil Science Society of America, Inc), 25–44.
Wu, J. X., Gutierrez, O. A., Jenkins, J. N., McCarty, J. C., Zhu, J. (2009). Quantitative analysis and QTL mapping for agronomic and fiber traits in an RI population of Upland cotton. Euphytica 165, 231–245. doi: 10.1007/s10681-008-9748-8
Xu, P., Gao, J., Cao, Z., Chee, P. W., Guo, Q., Xu, Z., et al. (2017). Fine mapping and candidate gene analysis of qFL-chr1, a fiber length QTL in cotton. Theor. Appl. Genet. 130, 1309–1319. doi: 10.1007/s00122-017-2890-8
Xu, Y. (2010). “Marker-assisted Selection,” in Theory. Molecular Plant Breeding (Wallingford, UK: CABI), 249–285.
Yang, S. S., Cheung, F., Lee, J. J., et al. (2006). Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 47, 761–775. doi: 10.1111/j.1365-313X.2006.02829.x
Yang, Y. W., Bian, S. M., Yao, Y., Liu, J. Y. (2008). Comparative proteomic analysis provides new insights into the fiber elongating process in cotton. J. Prot. Res. 7, 4623–4637. doi: 10.1021/pr800550q
Yang, Z., Zhang, C., Yang, X., Liu, K., Wu, Z., Zhang, X., et al. (2014). PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 203, 437–448. doi: 10.1111/nph.12824
Yang, X., Zhou, X., Wang, X., Li, Z., Zhang, Y., Liu, H., et al. (2015). Mapping QTL for cotton fiber quality traits using simple sequence repeat markers, conserved intron-scanning primers, and transcript-derived fragments. Euphytica 201, 215–230. doi: 10.1007/s10681-014-1194-1
Yang, X., Wang, Y., Zhang, G., Wang, X., Wu, L., et al. (2016). Detection and validation of one stable fiber strength QTL on c9 in tetraploid cotton. Mol. Genet. Genom. 291, 1625–1638. doi: 10.1007/s00438-016-1206-z
Young, N. D., Tanksley, S. D. (1989). RFLP analysis of the size of chromosomal segments retained around the Tm-2 locus of tomato during backcross breeding. Theor. Appl. Genet. 77, 353–359. doi: 10.1007/BF00305828
Yu, J., Jung, S., Cheng, C. H., Ficklin, S. P., Lee, T., Zheng, P., et al. (2014). CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42, 1229–1236. doi: 10.1093/nar/gkt1064
Yuan, D., Tang, Z., Wang, M., Gao, W., Tu, L., Jin, X., et al. (2015). The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 4, 17662. doi: 10.1038/srep17662
Zeng, L., Meredith, W. R., Gutierrez, O. A., Boykin, D. L. (2009). Identification of associations between SSR markers and fibre traits in an exotic germplasm derived from multiple cross among Gossypium tetraploid species. Theor. Appl. Genet. 119, 93–103. doi: 10.1007/s00122-009-1020-7
Zhang, B., Liu, J. Y. (2013). Mass spectrometric identification of in vivo phosphorylation sites of differentially expressed proteins in elongating cotton fiber cells. PloS One 8, e58758. doi: 10.1371/journal.pone.0058758
Zhang, T., Yuan, Y., Yu, J., Guo, W., Kohel, R. J. (2003). Molecular tagging of a major QTL for fiber strength in Upland cotton and its marker-assisted selection. Theor. Appl. Genet. 106, 262–268. doi: 10.1007/s00122-002-1101-3
Zhang, Z. S., Xiao, Y. H., Luo, M., Li, X. B., Luo, X. Y., Hou, L., et al. (2005). Construction of a genetic linkage map and QTL analysis of fiber-related traits in upland cotton (Gossypium hirsutum L.). Euphytica 144, 91–99. doi: 10.1007/s10681-005-4629-x
Zhang, Z. S., Hu, M. C., Zhang, J., Liu, D. J., Zheng, J., Zhang, K., et al. (2009). Construction of a comprehensive PCR-based marker linkage map and QTL mapping for fiber quality traits in upland cotton (Gossypium hirsutum L). Mol. Breed. 24, 49–61. doi: 10.1007/s11032-009-9271-1
Zhang, F., Zuo, K., Zhang, J., Liu, X., Zhang, L., Sun, X., et al. (2010). An L1 box binding protein, GbML1, interacts with GbMYB25 to control cotton fibre development. J. Exp. Bot. 61, 3599–3613. doi: 10.1093/jxb/erq173
Zhang, M., Zhang, Y., Huang, J. J., Zhang, X., Lee, M.-K., et al. (2012). Genome physical mapping of polyploids: a BIBAC physical map of cultivated tetraploid cotton, Gossypium hirsutum L. PloS One 7, e33644. doi: 10.1371/journal.pone.0033644
Zhang, B., Yang, Y. W., Zhang, Y., Liu, J. Y. (2013). A high-confidence reference dataset of differentially expressed proteins in elongating cotton fiber cells. Proteomics 13, 1159–1163. doi: 10.1002/pmic.201200176
Zhang, T., Qian, N., Zhu, X., Chen, H., Wang, S., Mei, H., et al. (2013). Variations and transmission of QTL alleles for yield and fiber qualities in upland cotton cultivars developed in China. PloS One 8,pe57220. doi: 10.1371/journal.pone.0057220
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhang, Z., Li, J., Muhammad, J., Cai, J., Jia, F., Shi, Y., et al. (2015). High resolution consensus mapping of quantitative trait loci for fiber strength, length and micronaire on chromosome 25 of the upland cotton (Gossypium hirsutum L.). PloS One 10,pe0135430. doi: 10.1371/journal.pone.0135430
Zhang, M., Zeng, J. Y., Long, H., Xiao, Y. H., Yan, X. Y., Pei, Y. (2016). Auxin regulates cotton fiber initiation via GhPIN-mediated auxin transport. Plant Cell Physiol. 58, 85–397. doi: 10.1093/pcp/pcw203
Zhang, Z., Shang, H., Shi, Y., Huang, L., Li, J., Ge, Q., et al. (2016). Construction of a high-density genetic map by specific locus amplified fragment sequencing (SLAF-seq) and its application to Quantitative Trait Loci (QTL) analysis for boll weight in upland cotton (Gossypium hirsutum). BMC Plant Biol. 16, 79. doi: 10.1186/s12870-016-0741-4
Zhang, C., Li, L., Liu, Q., Gu, L., Huang, J., et al. (2019). Identification of loci and candidate genes responsible for fiber length in upland cotton (Gossypium hirsutum L.) via association mapping and linkage analyses. Front. Plant Sci. 10, 53. doi: 10.3389/fpls.2019.00053
Zhao, Q., Dixon, R. A. (2011). Transcriptional networks for lignin biosynthesis: more complex than we thought. Trends Plant Sci. 16, 227–233. doi: 10.1016/j.tplants.2010.12.005
Zhiyuan, N., Chen, H., Mei, H., Zhang, T. (2014). Molecular tagging of QTLs for fiber quality and yield in the upland cotton cultivar Acala-Prema. Euphytica 195, 143–156. doi: 10.1007/s10681-013-0990-3
Zhong, M., McCarty, J. C., Jenkins, J. N., Saha, S. (2002). Assessment of day-neutral backcross populations of cotton using AFLP markers. J. Cotton Sci. 6, 97–103.
Zhou, Y., Zhang, Z. T., Li., M., Wei, X. Z., Li, X. J., Li, B. Y., et al. (2015). Cotton (Gossypium hirsutum) 14-3-3 proteins participate in regulation of fibre initiation and elongation by modulating brassinosteroid signaling. Plant Biotechnol. J. 13, 269–280. doi: 10.1111/pbi.12275
Zhu, H., Han, X., Lv, J., et al. (2011). Structure, expression differentiation and evolution of duplicated fiber developmental genes in Gossypium barbadense and G. hirsutum. BMC Plant Biol 11, 40. doi: 10.1186/1471-2229-11-40
Zhu, H., Lv, J., Zhao, L., Tong, X., Zhou, B., Zhang, T., et al. (2012). Molecular evolution and phylogenetic analysis of genes related to cotton fibers development from wild and domesticated cotton species in Gossypium. Molecular Phylogenetics Evol. 63, 589-597. doi: 10.1016/j.ympev.2012.01.025
Keywords: fiber quality, genetic mechanism, molecular markers, QTL mapping, fine mapping, Gossypium species
Citation: Ijaz B, Zhao N, Kong J and Hua J (2019) Fiber Quality Improvement in Upland Cotton (Gossypium hirsutum L.): Quantitative Trait Loci Mapping and Marker Assisted Selection Application. Front. Plant Sci. 10:1585. doi: 10.3389/fpls.2019.01585
Received: 26 August 2019; Accepted: 12 November 2019;
Published: 11 December 2019.
Edited by:
Sean Mayes, University of Nottingham, United KingdomReviewed by:
Kai Wang, Fujian Agriculture and Forestry University, ChinaYongzhong Xing, Huazhong Agricultural University, China
Copyright © 2019 Ijaz, Zhao, Kong and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinping Hua, jinping_hua@cau.edu.cn
 Babar Ijaz
Babar Ijaz Nan Zhao
Nan Zhao Jie Kong
Jie Kong Jinping Hua
Jinping Hua