- 1Beijing Vegetable Research Center (BVRC), Beijing Academy of Agricultural and Forestry Sciences, National Engineering Research Center for Vegetables, Beijing, China
- 2Beijing Key Laboratory of Vegetable Germplasms Improvement, Beijing, China
Pericarp wax of cucumber is an important economic trait, determining sales and marketing. Grafting of cucumber onto pumpkin rootstock (Cucurbita moschata) is an effective way to produce glossy cucumber fruits. However, the molecular regulation mechanism of this phenomenon remains largely unknown. In the present study, transcriptome analyses, genome-wide DNA methylation sequencing, and wax metabolite analysis were performed on the pericarp of self-rooted versus grafted cucumber. We identified the AP2/ERF-type transcription factor CsWIN1 as methylated and significantly upregulated in grafted cucumber compared to self-rooted cucumber. The increased expression of CsWIN1 was also positively correlated with several key wax biosynthesis genes, including CsCER1, CsCER1-1, CsCER4, CsKCS1, and the wax transporter gene CsABC. The transcriptome expression level of these genes was validated through qRT-PCR profiles. Furthermore, wax metabolite analysis showed that more wax ester (C20 fatty acid composition), but fewer alkanes (C29 and C31) were deposited in grafted cucumber pericarp. The higher expression of CsWIN1 and wax biosynthesis genes was reflected in the glossier appearance of grafted pericarp, possibly the result of higher wax ester content and higher integration of small trichomes in the pericarp. This study demonstrates that grafting can affect the content and composition of pericarp wax in cucumber grafted on pumpkin, and a unique regulation model of CsWIN1 for wax biosynthesis may exist in cucumber.
Introduction
Cucumber is one of the major vegetables cultivated worldwide (Huang et al., 2009). China is the largest producer and consumer of cucumber with about 77.4% of the total cucumber yield in the world (Year 2017, www.fao.org/faostat/en/).
Glossy cucumber is a dominant player in the Chinese market. The pericarp appearance of cucumber is an important economical factor, appealing to customers and plays an important role in sales and marketing of the fruit worldwide. The pericarp wax is mainly composed of wax and cutin derived from the cuticle and is a critical trait affecting the appearance and quality of cucumber (Wang et al., 2015b; Li et al., 2019). The waxy cucumber has a natural white powder-like substance on the pericarp surface, which is less desirable than cucumber varieties with a glossy pericarp.
Tian et al., (2015) identified five QTL (WP1.1, WP3.1, WP5.1, WP6.1 and WP6.2) promoting pericarp wax accumulation and two QTL (WP5.1 and WP6.2) with moderate effect. Other researchers revealed that the pericarp wax in cucumber is affected by both potential biosynthesis genes (CsCER1, CsWAX2) and environmental factors, such as light intensity, temperature, moisture, and grafting (Liu et al., 2014; Wang et al., 2015b; Wang et al., 2015c). However, there has been little research focusing on the genetic mapping of pericarp wax biosynthesis genes, which are critical to the discovery of the pericarp wax biosynthesis pathway in cucumber. Grafting onto pumpkin rootstock (Cucurbita moschata) proved to be an effective way to brighten the pericarp of cucumber (Samuels et al., 1993; Buschhaus et al., 2015; Shen et al., 2015). Still, the molecular mechanisms in regulating pericarp wax biosynthesis in cucumber grafted on pumpkin is unknown. There is a need to elucidate the biosynthesis pathway of pericarp wax in cucumber and identify the gene loci that confer glossy appearance in cucumber.
The cuticle layer covers the epidermal cells of leaves and other plant parts. It protects the plant from various environmental stresses, including fungal pathogens, UV radiation, water loss and non-stomatal transpiration, and dust deposits (Kunst and Samuels, 2009; Buschhaus et al., 2015; Blanc et al., 2018).The chemical components of cuticular wax has been well studied in Arabidopsis and other crops. Several functional genes involved in wax biosynthesis (CER1, WAX2, and FAC4) and transportation (ABC/LTP transporters) were identified (Broun et al., 2004; Suh et al., 2005; Bernard et al., 2012; Park et al., 2016). Cuticular wax was shown to be biosynthesized from C16 and C18-CoAsin plastids and then elongated into a long chain fatty acids (VLCFAs, chain length >20 carbons) in the endoplasmic reticulum membrane (Suh et al., 2005; Kunst and Samuels, 2009). Cuticular wax covers the outer surface of the cutin layer and presents in the form of crystals, a complex mixture that contains various primary and secondary alcohols, alkenes, ketones, and esters derived from VLCFAs through alkane- and alcohol-forming pathways (Lee et al., 2009; Gan et al., 2016). However, the chemical composition and content of cuticular wax are different among species and even tissues (Kunst and Samuels, 2009). Moreover, several studies have found that cuticular wax biosynthesis is affected by environmental factors, such as water deficit, lower temperature, and high levels of UV light (Kunst and Samuels, 2009; Lee and Mi, 2015). AP2-ERF (APETALA2/ethylene) transcription factors (TFs) were involved in the regulation of ET-response genes and had proved to possess a variety of functions in plants (Gutterson and Reuber, 2004; Feng et al., 2005; Li et al., 2011; Huang et al., 2016). Among these, WIN1 was first identified in a mutant in Arabidopsis thaliana; the overexpression of WIN1 caused a glossy phenotype, indicating that WIN1 is a regulator of wax biosynthesis, and it was reported to activate the expression of wax biosynthesis genes such as CER1, CER2, and KCS1(Broun et al., 2004). The other AP2/ERF gene DEWAX was found to negatively regulate cuticular wax biosynthesis in A. thaliana (Go et al., 2014). Furthermore, a positive regulator of wax biosynthesis “WRI4”, a member of the ERF family was identified in Arabidopsis stems (Park et al., 2016). There is a need to determine the role of each of the AP2/ERF transcription factors in regulating wax biosynthesis in cucumber.
Glossy cucumber is a dominant player in the Chinese market. Grafting is used for defending cucumber plants against soil-borne diseases. At the same time, it is an important method for production of glossy cucumbers. In this study, the cucumber variety “Jingyan 118” was grafted onto the pumpkin rootstock variety “Jingxinzhen 6,” an elite line used in grafting to brighten the scion cucumber pericarp. We conducted associated transcriptome and genome-wide methylation analyses in conjunction with changes in the cucumber pericarp wax in response to grafting. We determined that the CsWIN1 gene of cucumber (homologous to WIN1 in Arabidopsis) is methylated and upregulated and at the same time transcriptionally regulates several wax biosynthesis genes, including CsCER1 and CsCER4 in the pericarp of grafted cucumber.
Materials and Methods
Plant Materials and Grafting Experiment
The experiment was conducted in the Beijing Vegetable Research Center (BVRC) from 26 March to 10 July, 2017. Seeds of the cucumber variety “Jingyan 118” with high pericarp wax and a pumpkin rootstock “Jingxinzhen 6” were sown in a standard potting mix (peat: sand: pumice, 1:1:1, V/V/V). The cuttage grafting system was applied when the scions were growing at the one true leaf and the cotyledon of the rootstock was in the expansion stage. To enhance the survival rate, grafted seedlings were kept in the shade (24–28°C, 80–90% RH) for 3 days. Two weeks later, self-rooted and grafted seedlings were transplanted into soil in a greenhouse. Fertilization and cultivation management methods were as commonly recommended in cucumber production. The pericarp of self-rooted and grafted cucumber at commodity maturity were extracted for the following experiments below.
Pericarp Wax Observation and Chemical Component Analysis
Using a sharp thin blade, a 1 cm2 pericarp was carefully cut off from cucumber fruits at marketable mature stage. Images of the cuticular wax crystals were visualized at 200× magnification using a scanning electron microscope (Semagn et al., 2014) (S4700, Hitachi, Japan). Cellular morphology under the microscope was also observed using cryosection techniques. Long alkanes analysis of wax by gas chromatography was performed, following a method described by Park (Park et al., 2016). Wax esters containing saturated and unsaturated fatty acids from five biological replicates were detected by using specific multiple-reaction monitoring (MRM) scanning (Lam et al., 2013).
RNA Isolation and Library Construction
Total RNA of the pericarp from self-rooted, grafted, and failed grafted cucumber (cucumber scion which developed roots connected with the stock pumpkin and/or raised new roots into the soil) with three biological replicates were extracted using a DNeasy Kit and miRNeasy Kit, respectively (QIAGEN, USA). The concentration and quality of DNA and RNA were evaluated by a NanoDrop 2000C Spectrophotometer and an Agilent 2100 Bioanalyzer. Pyro-sequencing assays were designed and performed by BIOMARKER Company with both programs and assay result data supplied. mRNA was isolated by Oligo-dT magnetic beads from RNA, then the cDNA was synthesized using a QiaQuick PCR Extraction Kit (QIAGEN, USA). The cDNA library was constructed and sequenced by Illumina Hiseq 2500.
Differential Expressed Genes Analysis
Raw sequencing reads containing adaptors and low-quality (Q30 < 85%) were filtered. Then the remained reads were aligned to the genome of cucumber (9930 Version2) with TopHat2 (Kim et al., 2013), which mismatch was set as 2 and other parameters as the default value. FPKM (Fragments per Kilobase of transcript per Million fragments mapped) was used to detect the transcript abundance of each gene and estimate the expression values in all samples (Trapnell et al., 2010). Differentially expressed genes (DEGs) were identified according to the following criteria of |log2(fold change)| > 1 and false discovery rate (FDR) < 0.01 by DESeq2 (Love et al., 2014).
Genomic Methylation Analysis of Pericarp in Grafted Cucumber
In this study, the genome-wide methylation sequencing method was used according to the protocol described by Wang et al. (2015a), which was proved as a simple and scalable method. Total DNA of pericarp from self-rooted, grafted, and failed grafted cucumber with three biological replicates was extracted using a DNeasy Kit. According to the AFSM methylation technology, two restriction enzyme pairs of EcoRI-MspI and EcoRI-HpaII were used in the treatment of cucumber pericarp DNA samples in this study (Xia et al., 2014). The isoschizomers EcoRI was used as a frequent cutter while MspI and HpaII were used as rare cutters, and the methylation-susceptible sequences 59-CCGG and their methylation statuses were assessed by the AFSM method (Xia et al., 2014). The results of AFSM methylation were based on comparisons of the EcoRI-MspI and EcoRI-HpaII assembled sequences at the 59-CCGG sites using custom Perl scripts (http://afsmseq.sourceforge.net/) for each DNA sample analyzed by the China Golden Marker (Beijing) biotechnology company.
Expression Validation of Differentially Expressed and Methylated Genes
Quantitative real-time RT-PCR (qRT-PCR) technology was employed to verify the expression level of some candidate genes from RNA-seq results. The first-strand cDNA was amplified using a SYBR Green PCR Master Mix Kit (TaKaRa, Japan). Primer sequences for RT-PCR were designed on the “Quant Prime” website (http://quantprime.mpimp-golm.mpg.de/) (Table S1). qRT-PCR was conducted on an Applied Biosystems 7500 RT-PCR system (Thermo Fisher, USA) according to the manufacturer’s instruction. The relative transcript levels of candidate genes and TUA (internal control) were calculated using the 2−ΔΔCt method. Three biological and technical replicates were performed; the differential gene expression level was determined by t-test (p < 0.01).
Transcriptional Regulation Validation of CsWIN1 in Pericarp Wax Biosynthesis
To investigate the transcriptional regulation of CsWIN1, cDNA of CsWIN1 was amplified using specific primers (Table S1) and constructed into the vector pGADT7-Rec. The promoter sequences of four targeted genes (CsCER1, CsCER1-1, CsCER4, and CsKCS1) were amplified from genomic DNA and transformed into the pAbAi vector. Then the linearized library vectors pGADT7-Rec and pAbAi-HYT were co-transformed into the yeast strain Y1HGold. The transformed yeast cell with an empty pGADT7 vector was set as a negative control. The cultivation and transformation of yeast were carried out as described in the manufacturer’s protocol. All primers used in this study are list in Table S1.
Results
Observation of Pericarp Wax in Grafted Cucumber
The grafted cucumber exhibited a glossy pericarp with a light green color, in contrast to the self-rooted and failed grafted cucumber (Figure 1A). SEM (scanning electron microscope) observation revealed that the content and form of the wax was significantly different in grafted cucumber versus self-rooted cucumber. Fewer wax crystals were detected on the fruit surface, and the form of the wax was smaller and unbroken in grafted cucumber (Figure 1B). Wax crystals were more abundant in self-rooted and failed grafted cucumber and the wax exhibited a broken form, expressed in rods and tubes (Figure 1B). Cryosectioning that was conducted in both self-rooted and grafted cucumber showed that a thicker cuticle (15mm × 15mm) was clearly observed in self-rooted cucumber (Figure 1C).
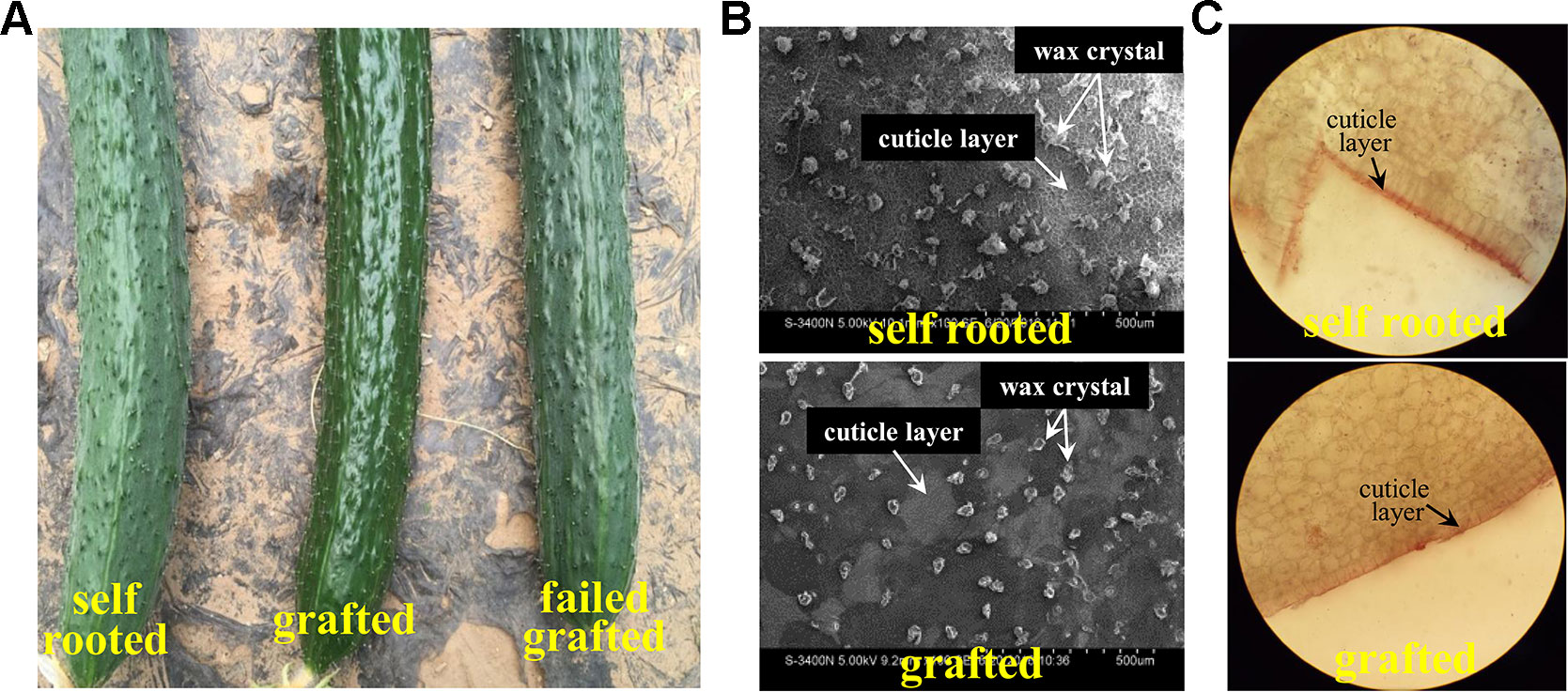
Figure 1 Observation of pericarp appearance in grafted cucumber and in self-rooted cucumber. Figure 1A shows the pericarp of self-rooted, grafted, and failed grafted cucumber. The self-rooted and failed grafted pericarp samples were waxy while the grafted cucumber pericarp was glossy. Figure 1B displays the SEM observation of self-rooted and grafted cucumber pericarp. The small trichomes of self-rooted pericarp were broken, while those in the grafted pericarp were integrated. Figure 1C presents the frozen pathological examination. The cutin in self-rooted pericarp was found to be much thicker than in grafted cucumber.
Components Analysis of Pericarp Wax in Grafted Cucumber
The significantly reduced formation of wax powder in grafted cucumber prompted us to investigate the changes in chemical composition of cuticular wax using GC-mass spectrometry at the commercial maturity stage. We detected a significant decrease in the content of alkanes with length of 29 and 31 carbons (which are important components of wax) in grafted cucumber pericarp compared with the self-rooted and failed grafted cucumber (Figure 2A). The levels of these two alkanes components were decreased in the grafted versus self-rooted cucumber by approximately 50 and 30%, respectively. Grafted cucumber contained increased amounts of wax esters as compared to self-rooted cucumber (p < 0.01), especially these wax esters derived from C20 fatty acids, such wax ester C20:C22, wax ester C20:C24, wax ester C20:C26, wax ester C20:C28, and wax ester C20:29 (Figure 2B). Total wax ester levels were increased by approximately 15% in grafted cucumber compared to self-rooted cucumber. No noticeable changes in other alkanes and wax esters were detected in the pericarp of grafted cucumber versus self-rooted cucumber.
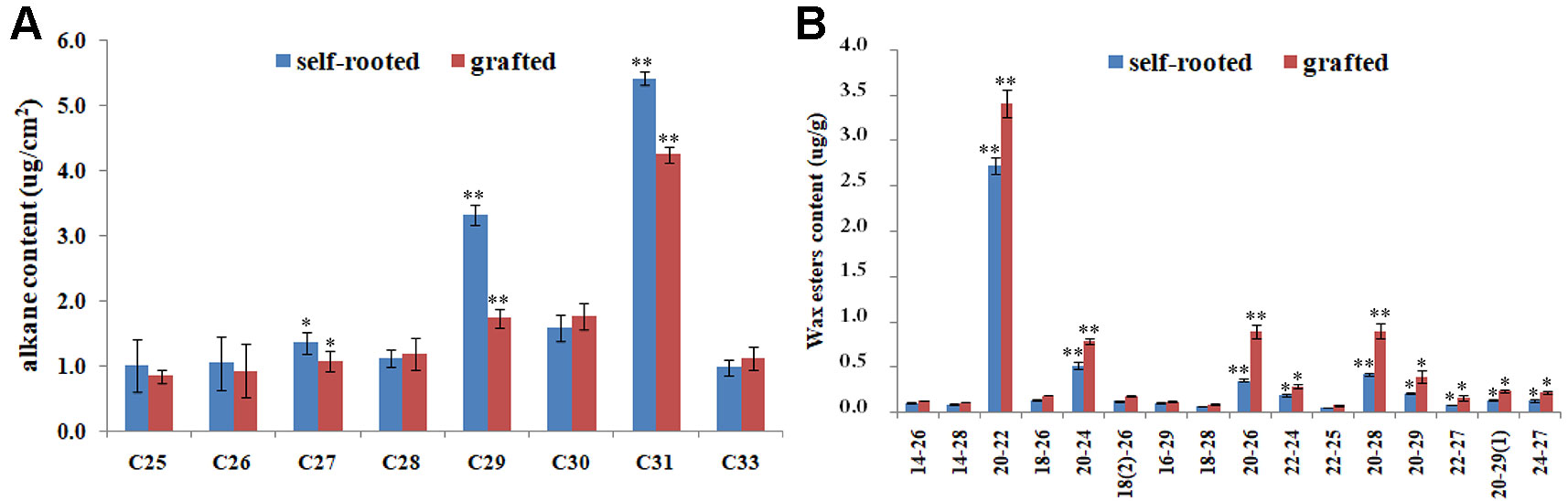
Figure 2 Comparison of the composition of pericarp wax between self-rooted and grafted cucumber, analyzed by GC-MS. The major alkanes in Figure 2A were designated by carbon chain length in x axis and the amount of alkanes were expressed as μg/cm2 pericarp area in y axis. The numbers in Figure 2B showed the carbon chain length of fatty acid and fatty alcohol in x axis, the corresponding wax esters amount were expressed as μg/g in y axis. Numbers in brackets indicated the amount of double bond. Error bars indicate standard errors. * and ** indicated significant differences of t test at p < 0.05 and p < 0.01, respectively.
Transcriptome Analysis of Pericarp Wax Biosynthesis After Grafting
Nine cucumber cDNA libraries from self-rooted, grafted, and failed grafted cucumber were sequenced on the Illumina HiSeq2500 platform; and 33.87 Gb clean reads were generated in total. Each library generated about 4.84 Gb clean reads; all Q30 reached more than 94.69% (Table S2). All of the clean sequencing data used in the present study are deposited in the NCBI Sequence Read Archive database (SRA accession: SRR10113591, SRR10113592, SRR10113593, SRR10113594, SRR 10113569, and SRR10113570). The sequencing reads from nine libraries showed a significant positive correlation (0.98) among the three replications, indicating the reliable phenotype variations and high-quality sequencing data (Figure S1). All clean reads were aligned to Chinese long cucumber reference genome (9930 V2) and the mapping rate varied from 87.91 to 89.51% (Table S2). Then, all mapped reads were aligned with the databases of Swiss-Prot, KEGG, and GO using BLAST. Four hundred and twenty-four new genes were identified, 347 of which were annotated depended on the test for functional annotation.
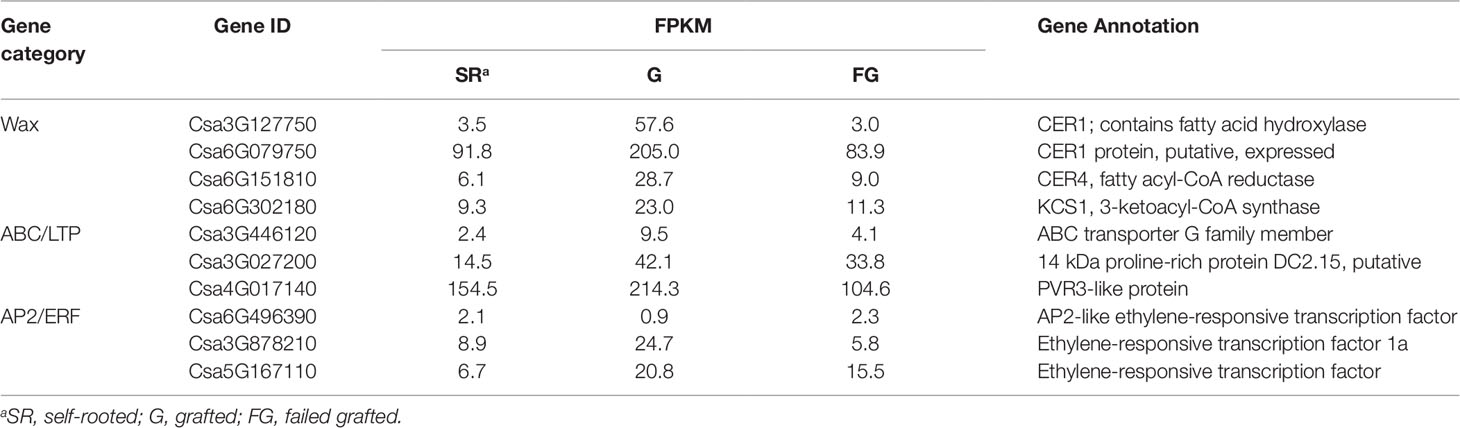
Differentially expressed genes (DEGs) were identified by calculating the FPKM values among self-rooted, grafted, and failed grafted cucumber with the following criteria (p < 0.05, FDR < 0.01). Finally, a total of 384 DEGs were identified in this study (Figure 3A). Compared with self-rooted cucumber, 111 genes were upregulated while 82 genes were downregulated in grafted cucumber. In contrast to failed grafted cucumber, 118 genes were upregulated and 111 genes were downregulated in grafted cucumber. Above all, 68 genes were found to have significant differential expression levels in grafted cucumber compared with both self-rooted and failed grafted cucumber (Table S3), indicating that these genes could play a role in the pericarp wax biosynthesis of grafted cucumber. Based on the gene functions involved in wax biosynthesis and highest fold changes in grafted cucumber, 10 of the 68 DEGs were considered to be important candidate genes affecting pericarp wax formation (Table 1). These 10 DEGs had different functions associated with wax metabolism, including four DEGs involved in wax biosynthesis, three involved in wax transportation, and three AP2/ERF transcriptional factors that may be involved in the regulation of the gene expression of wax biosynthesis.
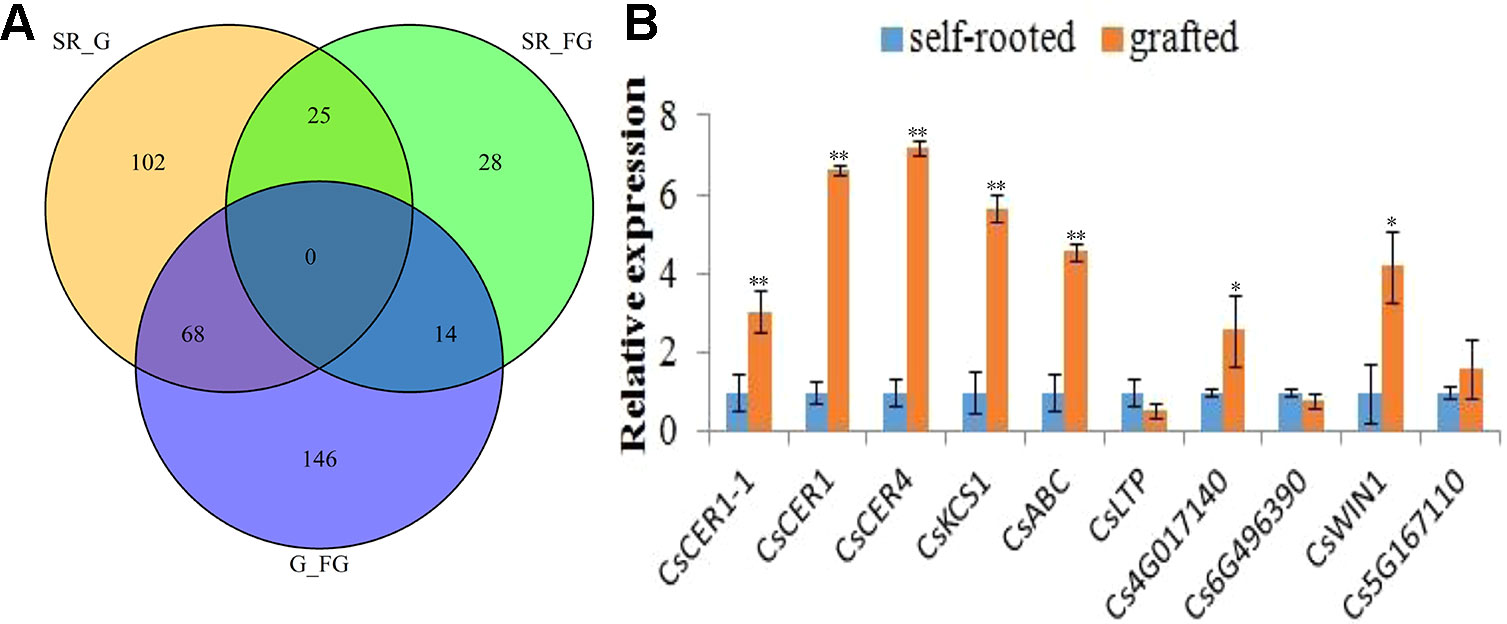
Figure 3 Transcriptome dataset and expression validation of differentially expressed genes affected by grafting at commercial mature stage. Figure 3A presents a Venn diagram of differentially expressed genes (DEGs) obtained from the self-rooted, grafted, and failed grafted cucumber pericarp. Figure 3B shows the qRT-PCR validation (three technical replicates per biological replicate) of the 10 differentially expressed genes involved pericarp wax biosynthesis. Error bars indicate the standard deviations. * and ** indicated significant differences of t test at p < 0.05 and p < 0.01, respectively.
Genomic Methylation Analysis of Pericarp Wax Synthesis After Grafting
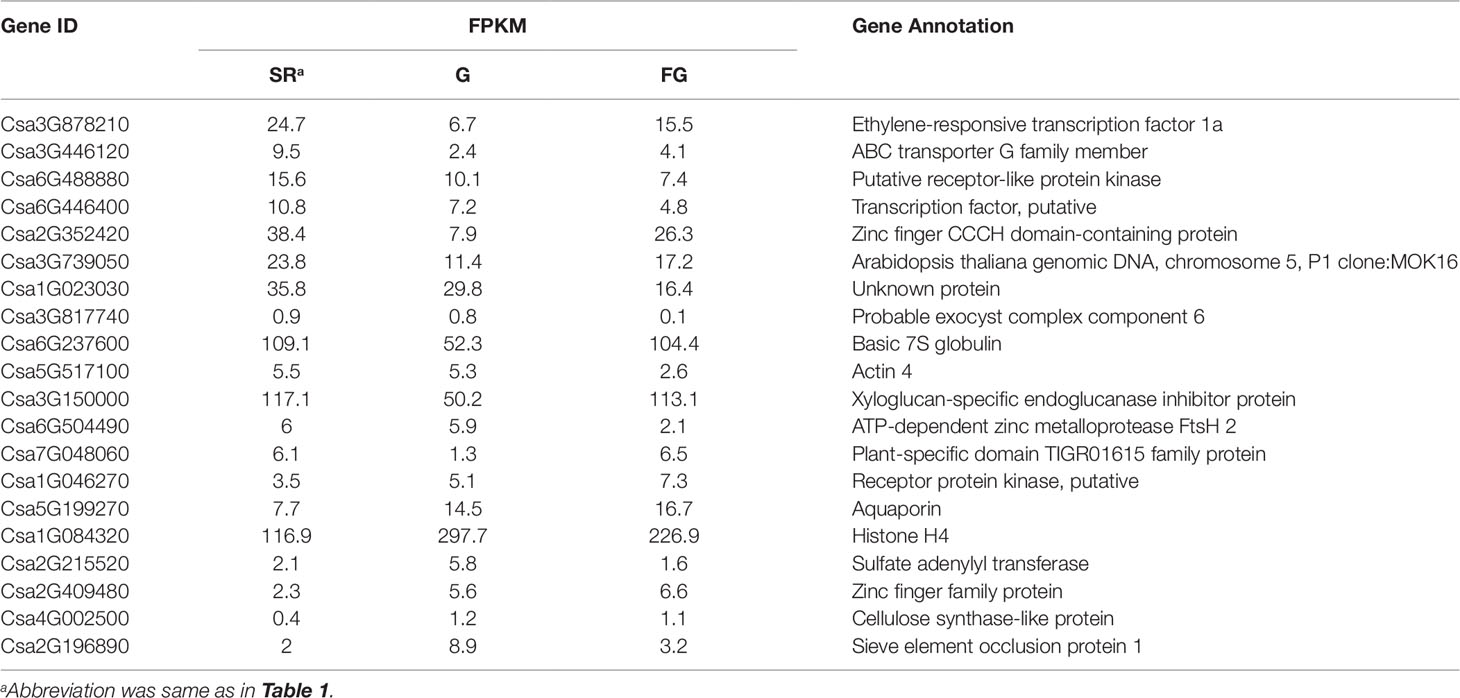
A total of 4,247 methylated genes were detected in this study, 1,184 of which were specifically observed in grafted cucumber (as compared to self-rooted and failed grafted cucumber). Among these genes, the DNA methylated regions in the UTR, exon, intron, and promoter regions were distributed as 4, 48.8, 32.7, and 14.2% in the grafting-induced methylated genes, respectively. Twenty grafting-induced methylated genes were observed in the profile of differentially expressed genes in the grafted, but not in the un-grafted cucumber pericarp. These were identified as critical candidate genes affecting the changes in appearance of the scion pericarp in cucumber (Table 2). In contrast, we observed 2,521 methylated genes in the negative control but not in grafted treatment, these genes were assumed as the demethylated genes after graft, which included 43 DEGs based on transcriptome analysis (Table S4).
The methylation sequencing results indicated that the intron region of Csa3G878210 possesses a CCGG type methylation in grafted cucumber which was not detected in both negative control treatments. Furthermore, the expression of Csa3G878210 was up-regulated in grafted cucumber compared with the self-rooted and failed grafted cucumber treatment. This gene is an AP2/ERF transcriptional factor and the homologous gene of the wax biosynthesis regulator gene WIN1 (AP2/ERF gene) in Arabidopsis. Hence, this grafting-induced methylated gene (Csa3G878210) was named CsWIN1, which may play a critical role in regulation of wax biosynthesis in cucumber, especially in brightening the appearance of grafted pericarp, resulting in glossy cucumbers.
Expression Validation of Wax Biosynthesis-Related Genes Affected by Grafting
In this study, the 10 obtained wax biosynthesis genes in transcriptome profiles showed considerable differences in gene expression levels between self-rooted and grafted cucumber pericarp. To confirm this results, qRT-PCR evaluation was performed in the self-rooted and grafted cucumber pericarp. There were eight genes (CsCER1, CsCER1-1, CsCER4, CsKCS1, CsABC, Csa4G017140, Csa6G496390, and CsWIN1) that were significantly (p < 0.01) upregulated in grafted cucumber, while one ERF gene (Cs6G496390) was downregulated. Thus, these results validated the transcriptome data (Figure 3B). However, one gene (CsLTP) showed a difference in expression level without significance. This validation result was consistent with the transcriptome profiles. Most of the wax biosynthesis-related genes were upregulated in the grafted cucumber pericarp (Table 1).
Transcription Regulation Validation of CsWIN1 in Pericarp Wax Biosynthesis
In this study, five well-investigated AP2/ERF TFs in the regulation of wax biosynthesis in Arabidopsis were analyzed in the cucumber genome, and three homologue genes were obtained among the 68 DEGs identified in the transcriptome data, including the key CsWIN1 gene, which was specifically methylated by grafting in the cucumber pericarp, as well as two ERF genes. A phylogenetic tree aligned using the neighbor-joining (NJ) method through MEGA7 software were constructed andCsWIN1 was confirmed to be closely related to the key wax biosynthesis regulatorWIN1in Arabidopsis (Figure 4A) (Shi et al., 2011). In this study, the transcription regulation validation of CsWIN1 was examined in the regulation of wax biosynthesis genes by a yeast one-hybrid system (Y1H). The promoter sequences of two targeted genes (CsCER1and CsCER4) including the element “GCCGGC” were amplified from genomic DNA and transformed into the pAbAi vector. The yeast cells that grew on the Y1H Gold-carrying PGADT7-CsWIN1 vectors and the pBait-AbAi vectors could be grown in SD/-Leu/AbA 100 medium, and the positive clones increased gradually with the increase of the dilution concentration. This result indicated that pGADT7-CsWIN1 harbors a transcriptional activation domain preferentially binding to the “GCCGGC” box of the CsCER1 and CsCER4 promoter (Figure 4B). The key grafting-induced regulator CsWIN1 transcriptionally activates the expression of target wax biosynthesis genes, thus regulating the content and composition of pericarp wax in cucumber, resulting in the brightening and increased glossiness of the pericarp.
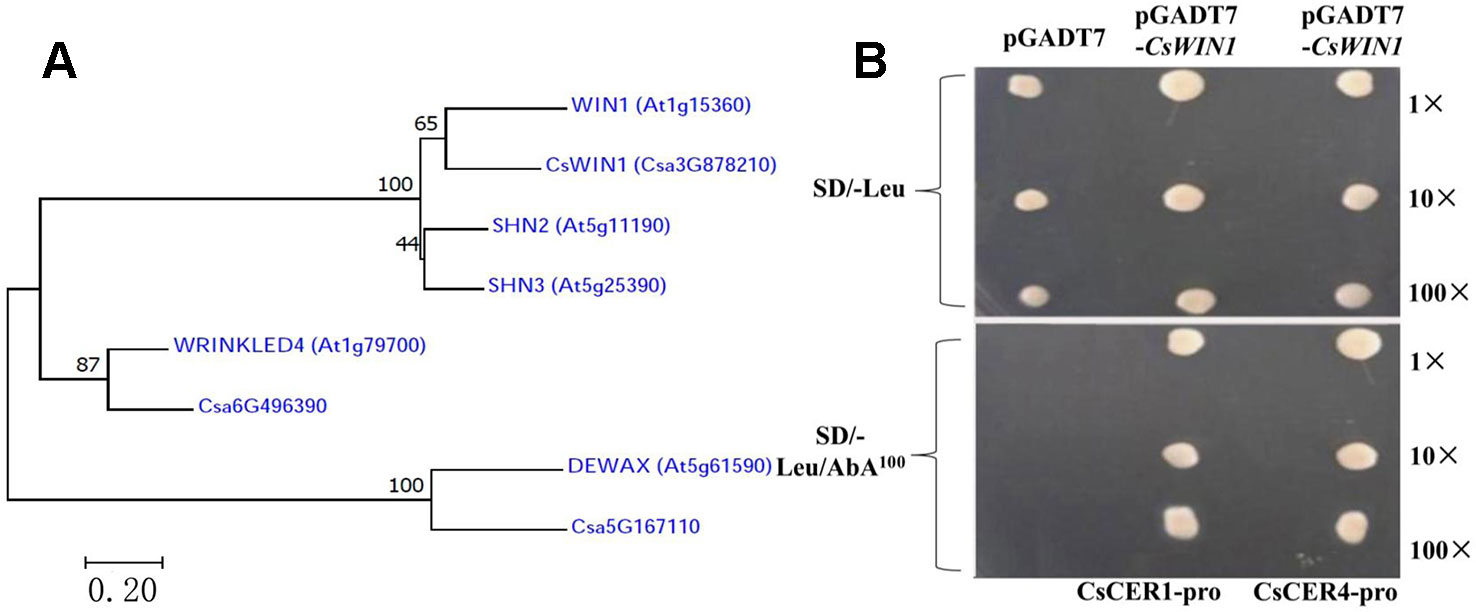
Figure 4 Phylogenetic analysis and Y1H transcriptional test of CsWIN1 in the regulation of wax biosynthesis. Figure 4A displays the key wax regulator genes in Arabidopsis and their homologues in cucumber. Figure 4B shows the transcriptional binding test of CsWIN1 with CsCER1 and CsCER4 promoters by Y1H.
Discussion
The Unique Character of Cucumber Pericarp, as Well as Appearance After Grafting
In Arabidopsis, the wax appearance was crystal-like in the stem and leaves as well as in siliques (Haslam and Kunst, 2012). The chemical composition and content of cuticular wax is different among species and tissues (Kunst and Samuels, 2009).The cuticle wax responds to external environment stresses and is a physical and chemical barrier on the outer surfaces of terrestrial plants (Yeats and Rose, 2013; Ziv et al., 2018). The wax of cucumber fruit was observed as small balls or trichomes, quite different from that in Arabidopsis and other crops like wheat (Hen-Avivi et al., 2016). The small balls or trichomes in the glossy cucumber are integrated, while as those in waxy cucumbers appear broken and exist as fragmentary forms that are uniformly distributed on the surface of the cucumber fruit (Figure 1B). Previous studies also found that the chemical components of wax was different between Arabidopsis and cucumber, which phenols and alkenes were uniquely detected, while the ketones was unable to identified in cucumber (Wang et al., 2015b; Wang et al., 2015c).
In this study, we observed that after grafting with the rootstock of “Jingxinzhen 6”, the cucumber pericarp on the scion became glossy, as compared with the waxy appearance of the self-rooted cucumber (Figure 1A). The metabolite analysis indicates that there is a lower alkane content, but more wax esters were accumulated in the grafted cucumber.
Several studies have shown that the surface of cucumber fruit is more affected by silica, which is a natural powder covering on the surface of cucumber fruits, rather than the cuticular wax (Mitani et al., 2011). This finding was also investigated in the grafting system by comparing the effects of different rootstocks—C. moschata and Cucurbita ficifolia. The former pumpkin rootstock enhanced the glossy appearance of cucumber fruit, whereas the latter rootstock did not (Seki and Hotta, 1997). Another study showed that one amino acid mutation in the silicon influx transporter gene (AQP family genes) could lead to less silicon uptake in the soil, resulting in a glossy cucumber fruit when grafted onto C. moschata (Mitani et al., 2011). Here, we observed that four AQP family genes were all downregulated by grafting. These results are consistent with these previous studies (Table S3). Combined with the fact that the cuticular wax composition was significantly changed after grafting in this study, we hypothesized that there were two regulation pathways (wax biosynthesis and Si absorption) through which the rootstock affects the scion in grafting systems, though further research is required to validate this.
Regulated Wax Biosynthesis Genes in the Pericarp After Grafting in Cucumber
Grafting is an important way to improve plant growth, stress tolerance, and fruit quality, and has been widely used in commercial horticultural crop production (Zahaf et al., 2012; Liu et al., 2016). The molecular mechanisms regulating plant growth via grafting have been investigated by several groups (Kyriacou et al., 2017), for example using high-throughput sequencing in watermelon (Liu et al., 2013), tomato (Estan et al., 2005), apple (An et al., 2018), and grapevine (Pagliarani et al., 2017). These studies showed that grafting affects differential expression of genes in grafted plants. miRNAs are also exchanged between the scion and rootstock in grafted watermelon, which may regulate the growth and development of the scion (Cookson and Ollat, 2013; Liu et al., 2013). A genomic DNA methylation analysis was performed in cucumber and melon scions by a grafting system using cucurbitaceous inter-grafting (Avramidou et al., 2015). Recently, some research identified many DEGs involved in various metabolic processes in grafted cucumber and verified by qRT-PCR (Miao et al., 2019a; Miao et al., 2019b). In this study, we observed that pericarp phenotype of cucumber grafted on pumpkin rootstock is distinct from these of the failed grafted cucumber and self-rooted cucumber. A previous study (Jun et al., 2016) also reported that the pericarp wax powder content in the self-grafted cucumber had no difference with self-rooted cucumber.
The present study identified 68 significantly DEGs in grafted cucumber and 10 of them were annotated in the wax biosynthesis pathway (Table 1). These 10 DEGs associated with wax biosynthesis were validated by qRT-PCR (Figure 3B). These DEGs may also function in VLCFAs biosynthesis and wax ester biosynthesis, as well as in the transportation of cuticular wax inside or outside of the cell in cucumber scion (McFarlane et al., 2010; Kim et al., 2012).
This study also examined the genomic methylation of cucumber scion grafted in pumpkin rootstock based on a genome-wide methylation sequencing method (Wang et al., 2015a), which was proved as a simple and scalable method. Although it may not detect all methylated genes in the genome because of limited sites by enzymes and the low coverage sequencing, it was a cost-effectively method in identifying candidate genes and had been used in many researches (Mastan et al., 2012; Rathore et al., 2014; Tang et al., 2014; Xia et al., 2014). Here, 20 key genes were methylated and the association analysis between the transcriptome and methylation identified one key wax biosynthesis regulator—CsWIN1, homologous to the ERF family members WIN, WR4, and DEWAX in Arabidopsis. The close phylogenetic relationship between CsWIN1 and WIN1 provides confirmation evidence that the ERF gene CsWIN1 is an important regulator in wax biosynthesis. Moreover, this gene also affects expression of other genes (CsCER1 and CsCER4) in the wax biosynthesis pathway, in addition to the downstream transporter gene CsABC in the cucumber scion.
Potential Regulation Model of CsWIN1 Affecting Pericarp Wax Biosynthesis
The key CsWIN1 gene was observed in the transcriptome data and in the genomic DNA methylation profile. Furthermore, it is homologous to a key wax biosynthesis regulator in DEWAX in the model plant Arabidopsis (Table S2; Figure 4A). The expression of CsWIN1 is positively correlated with four cuticular wax biosynthesis genes (CsCER1, CsCER1-1, CsCER4, and CsKCS1) that were upregulated after grafting on the pumpkin rootstock (Figure 3). In addition, the potential wax transporter genes (ABC/LTP transporter) were upregulated and co-expressed with the key regulator CsWIN1 in the graft treatment (Table S2). Based on these results, we hypothesized that the key regulator CsWIN1 affected by grafting could be a master switch that transcriptionally regulates the expression of the wax biosynthesis pathway genes in response to grafting, specifically activating CsCER1, CsCER1-1, CsCER4, and CsKCS1 as well as transportation members of the ABC/LTP transporter family in the scion. TheY1H experiment validated that the key regulator could bind to the promoter region of CsCER1 and CsCER4, illustrating the feasibility of transcriptional regulation (Figure 4B). Moreover, the metabolite dataset showed that the content composition of cuticular wax the cucumber scion was significantly affected by grafting on pumpkin.
Following grafting, fewer alkanes and more wax esters were accumulated on the pericarp and the wax balls were not broken. Therefore, we propose a possible model of CsWIN1 in the regulation of wax biosynthesis in grafted cucumber (Figure 5). CsWIN1 was methylated and upregulated in grafted cucumber, and then transcriptionally activated the expression of wax biosynthesis genes CsCER1 and CsCER4, and may regulate the expression of transporter gene CsABC, resulting in the biosynthesis and transportation of more wax esters into the pericarp. This process makes the small trichomes less prone to breaking in the grafted cucumber, as opposed to being easily broken in the self-rooted cucumber. This model shows a molecular regulation pathway of wax biosynthesis in cucumber and could provide a reference to other crops. However, the in vitro function of CsWIN1 still needs to be validated by transformation experiments. The pumpkin’s genetic factors (genes, RNAs and proteins) that affect the cucumber pericarp are unknown and could be identified in future studies.
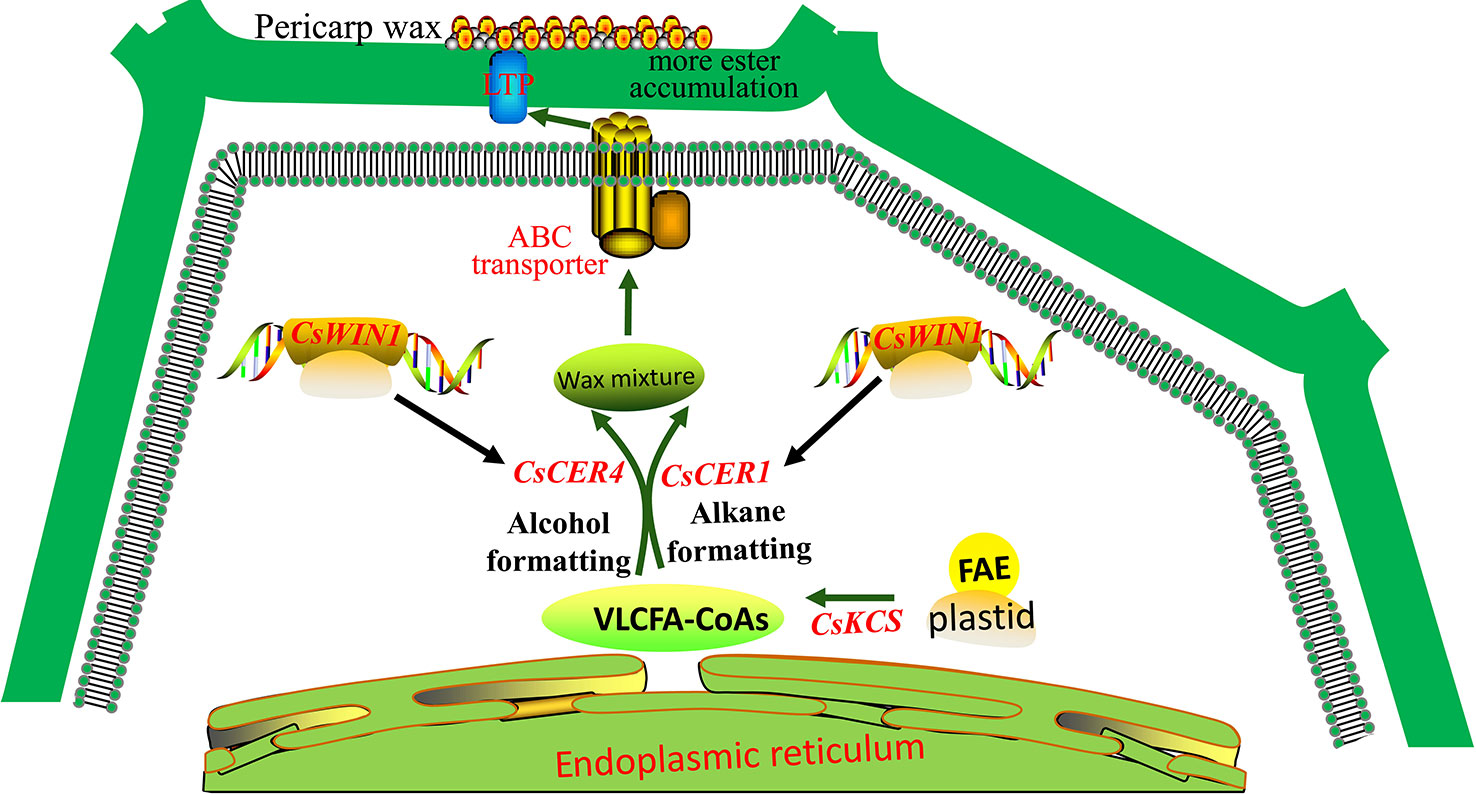
Figure 5 The potential regulation model of CsWIN1 in wax biosynthesis. CsWIN1 was methylated and upregulated by a grafting test. It then transcriptionally activated the expression of wax biosynthesis genes CsCER1 and CsCER4, and may regulate the expression of transporter gene CsABC, resulted in the biosynthesis and transportation of more wax esters into the pericarp. This make the small trichomes not so fragile, so they do not break up in the grafted cucumber, as opposed to being easily broken in the self-rooted cucumber.
Data Availability Statement
All datasets for this study are included in the article/Supplementary Material.
Author Contributions
CW and YX designed this research. JZ and YY performed the grafting experiment, JY and JZ analyzed the bioinformatics data, and JL, JZ, and XZ performed the validation test. JZ, CW, and YX wrote the manuscript. All authors have read and approved the final manuscript.
Funding
This research was financially supported by National Natural Science Foundation of China (No.31801887), China Postdoctoral Science Foundation (2018M631381), Beijing Postdoctoral Science Foundation (ZZ2019-49), Beijing Academy of Agricultural and Forestry Sciences (KJCX20170402/2018-ZZ-006/QNJJ201810), Beijing Nova Program (Z181100006218060), Beijing Municipal Department of Organization (2016000021223ZK22), and Beijing Youth Talent Promotion Project (2018).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01564/full#supplementary-material
Figure S1 | Heat map of all DEGs in self-rooted (SR), grafted (G), and failed grafted (FG) cucumber in three biological replications.
Table S1 | All primer sequences used in this study.
Table S2 | The basic analysis of RNA-seq reads.
Table S3 | FPKM values of differentially expressed genes affected by grafting.
Table S4 | List of 43 DEGs with DNA demethylation in grafted cucumber.
References
An, N., Fan, S., Yang, Y., Chen, X., Dong, F., Wang, Y., et al. (2018). Identification and Characterization of miRNAs in Self-Rooted and Grafted Malus Reveals Critical Networks Associated with Flowering. Int. J. Mol. Sci. 19, 2384. doi: 10.3390/ijms19082384
Avramidou, E., Kapazoglou, A., Aravanopoulos, F. A., Xanthopoulou, A., Ganopoulos, I., Tsaballa, A., et al. (2015). Global DNA methylation changes in Cucurbitaceae inter-species grafting. Crop Breed. Appl. Biotechnol. 15, 112–116. doi: 10.1590/1984-70332015v15n2n20
Bernard, A., Domergue, F., Pascal, S., Jetter, R., Renne, C., Faure, J. D., et al. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24, 3106–3118. doi: 10.1105/tpc.112.099796
Blanc, C., Coluccia, F., L’Haridon, F., Torres, M., Ortiz-Berrocal, M., Stahl, E., et al. (2018). The Cuticle Mutant eca2 Modifies Plant Defense Responses to Biotrophic and Necrotrophic Pathogens and Herbivory Insects. Mol. Plant-Microbe Interact. : MPMI 31, 344–355. doi: 10.1094/MPMI-07-17-0181-R
Broun, P., Poindexter, P., Osborne, E., Jiang, C. Z., Riechmann, J. L., Chrispeels, M. J. (2004). WIN1, a Transcriptional Activator of Epidermal Wax Accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 101, 4706–4711. doi: 10.1073/pnas.0305574101
Buschhaus, C., Hager, D., Jetter, R. (2015). Wax Layers on Cosmos bipinnatus Petals Contribute Unequally to Total Petal Water Resistance. Plant Physiol. 167, 80. doi: 10.1104/pp.114.249235
Cookson, S. J., Ollat, N. (2013). Grafting with rootstocks induces extensive transcriptional re-programming in the shoot apical meristem of grapevine. BMC Plant Biol. 13, 147. doi: 10.1186/1471-2229-13-147
Estan, M. T., Martinez-Rodriguez, M. M., Perez-Alfocea, F., Flowers, T. J., Bolarin, M. C. (2005). Grafting raises the salt tolerance of tomato through limiting the transport of sodium and chloride to the shoot. J. Exp. Bot. 56, 703–712. doi: 10.1093/jxb/eri027
Feng, J. X., Liu, D., Pan, Y., Gong, W., Ma, L. G., Luo, J. C., et al. (2005). An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol. 59, 853–868. doi: 10.1007/s11103-005-1511-0
Gan, L., Wang, X., Cheng, Z., Liu, L., Wang, J., Zhang, Z., et al. (2016). Wax crystal-sparse leaf 3 encoding a β-ketoacyl-CoA reductase is involved in cuticular wax biosynthesis in rice. Plant Cell Rep. 35, 1687–1698. doi: 10.1007/s00299-016-1983-1
Go, Y. S., Kim, H., Kim, H. J., Mi, C. S. (2014). Arabidopsis Cuticular Wax Biosynthesis Is Negatively Regulated by the DEWAX Gene Encoding an AP2/ERF-Type Transcription Factor. Plant Cell 26, 1666–1680. doi: 10.1105/tpc.114.123307
Gutterson, N., Reuber, T. L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7, 465–471. doi: 10.1016/j.pbi.2004.04.007
Haslam, T. M., Kunst, L. (2012). Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol. 160, 1164. doi: 10.1104/pp.112.201640
Hen-Avivi, S., Savin, O., Racovita, R. C., Lee, W.-S., Adamski, N. M., Malitsky, S., et al. (2016). A metabolic gene cluster in the wheat W1 and the barley Cer-cqu loci determines β-diketone biosynthesis and glaucousness. Plant Cell 28, 1440–1460. doi: 10.1105/tpc.16.00197
Huang, S., Ruiqiang, L., Zhonghua, Z., Li, L., Xingfang, G., Wei, F., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41, 1275. doi: 10.1038/ng.475
Huang, P. Y., Catinot, J., Zimmerli, L. (2016). Ethylene response factors in Arabidopsis immunity. J. Exp. Bot. 67, 1231–1241. doi: 10.1093/jxb/erv518
Jun, H. U., Xiang, C., Wang, C., Duan, Y., Chen, H. (2016). Effect of Rootstock on Silicon and Fruit Surface Wax Powder Content in Grafted Cucumber (Northern Horticulture).
Kim, H., Lee, S. B., Kim, H. J., Min, M. K., Hwang, I., Suh, M. C. (2012). Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 53, 1391–1403. doi: 10.1093/pcp/pcs083
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. doi: 10.1186/gb-2013-14-4-r36
Kunst, L., Samuels, L. (2009). Plant cuticles shine: advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 12, 721–727. doi: 10.1016/j.pbi.2009.09.009
Kyriacou, M., Rouphael, Y., Colla, G., Zrenner, R., Schwarz, D. (2017). Vegetable Grafting: The Implications of a Growing Agronomic Imperative for Vegetable Fruit Quality and Nutritive Value. Front. In Plant Sci. 8, 741. doi: 10.3389/fpls.2017.00741
Lam, S. M., Tong, L., Reux, B., Lear, M. J., Wenk, M. R., Shui, G. (2013). Rapid and sensitive profiling of tear wax ester species using high performance liquid chromatography coupled with tandem mass spectrometry. J. Chromatogr. A 1308, 166–171. doi: 10.1016/j.chroma.2013.08.016
Lee, S. B., Mi, C. S. (2015). Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 56, 48. doi: 10.1093/pcp/pcu142
Lee, S. B., Jung, S. J., Go, Y. S., Kim, H. U., Kim, J. K., Cho, H. J., et al. (2009). Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. : Cell Mol. Biol. 60, 462–475. doi: 10.1111/j.1365-313X.2009.03973.x
Li, C. W., Su, R. C., Cheng, C. P., Sanjaya, You, S. J., Hsieh, T. H., et al. (2011). Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol. 156, 213–227. doi: 10.1104/pp.111.174268
Li, L., Du, Y., He, C., Dietrich, C. R., Li, J., Ma, X., et al. (2019). Maize glossy6 is involved in cuticular wax deposition and drought tolerance. J. Exp. Bot. 70 (12), 3089–3099. doi: 10.1093/jxb/erz131
Liu, N., Yang, J., Guo, S., Xu, Y., Zhang, M. (2013). Genome-wide identification and comparative analysis of conserved and novel microRNAs in grafted watermelon by high-throughput sequencing. PloS One 8, e57359. doi: 10.1371/journal.pone.0057359
Liu, J., Li, J., Su, X., Xia, Z. (2014). Grafting improves drought tolerance by regulating antioxidant enzyme activities and stress-responsive gene expression in tobacco. Environ. Exp. Bot. 107, 173–179. doi: 10.1016/j.envexpbot.2014.06.012
Liu, N., Yang, J., Fu, X., Zhang, L., Tang, K., Guy, K. M., et al. (2016). Genome-wide identification and comparative analysis of grafting-responsive mRNA in watermelon grafted onto bottle gourd and squash rootstocks by high-throughput sequencing. Mol. Genet. Genomics 291, 621–633. doi: 10.1007/s00438-015-1132-5
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Mastan, S. G., Rathore, M. S., Bhatt, V. D., Yadav, P., Chikara, J. (2012). Assessment of changes in DNA methylation by methylation-sensitive amplification polymorphism in Jatropha curcas L. Subjected Salinity Stress Gene 508, 125–129. doi: 10.1016/j.gene.2012.07.063
McFarlane, H. E., Shin, J. J., Bird, D. A., Samuels, A. L. (2010). Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 22, 3066–3075. doi: 10.1105/tpc.110.077974
Miao, L., Di, Q., Sun, T., Li, Y., Duan, Y., Wang, J., et al. (2019a). Integrated Metabolome and Transcriptome Analysis Provide Insights into the Effects of Grafting on Fruit Flavor of Cucumber with Different Rootstocks. Int. J. Mol. Sci. 20 (14), 3592. doi: 10.3390/ijms20143592
Miao, L., Qin, X., Gao, L., Li, Q., Li, S., He, C., et al. (2019b). Selection of reference genes for quantitative real-time PCR analysis in cucumber (Cucumis sativus L.), pumpkin (Cucurbita moschata Duch.) and cucumber-pumpkin grafted plants. PeerJ 7, e6536. doi: 10.7717/peerj.6536
Mitani, N., Yamaji, N., Ago, Y., Iwasaki, K., Ma, J. F. (2011). Isolation and functional characterization of an influx silicon transporter in two pumpkin cultivars contrasting in silicon accumulation. Plant J. Cell Mol. Biol. 66, 231–240. doi: 10.1111/j.1365-313X.2011.04483.x
Pagliarani, C., Vitali, M., Ferrero, M., Vitulo, N., Incarbone, M., Lovisolo, C., et al. (2017). Accumulation of MicroRNAs Differentially Modulated by Drought Is Affected by Grafting in Grapevine. Plant Physiol. 173, pp.01119–02016. doi: 10.1104/pp.16.01119
Park, C. S., Go, Y. S., Suh, M. C. (2016). Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 88, 257–270. doi: 10.1111/tpj.13248
Rathore, M. S., Mastan, S. G., Agarwal, P. K. (2014). Evaluation of DNA methylation using methylation-sensitive amplification polymorphism in plant tissues grown in vivo and in vitro. Plant Growth Regul. 75, 11–19. doi: 10.1007/s10725-014-9926-8
Samuels, A. L., Glass, A. D. M., Ehret, D. L., Menzies, J. G. (1993). The Effects of Silicon Supplementation on Cucumber Fruit: Changes in Surface Characteristics. Ann. Bot. 72, 433–440. doi: 10.1006/anbo.1993.1129
Seki, M., Hotta, Y. (1997). Effect of bloomless stock cultivar on the growth and mineral uptake of cucumber [Cucumis sativus] plants ((Japan): Research Bulletin of the Aichi-ken Agricultural Research Cente).
Semagn, K., Babu, R., Hearne, S., Olsen, M. (2014). Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol. Breed. 33, 1–14. doi: 10.1007/s11032-013-9917-x
Shen, Q. H., Ren, Z. H., Wei, M., Li, Y., Wang, X. F., Cui, J. (2015). Effects of silicon supply on silicon absorption and bloom formation on fruit surface of cucumber grafted on different rootstocks. Acta Hortic. 1086, 231–236. doi: 10.17660/ActaHortic.2015.1086.29
Shi, J. X., Malitsky, S., De, O. S., Branigan, C., Franke, R. B., Schreiber, L., et al. (2011). SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PloS Genet. 7, e1001388. doi: 10.1371/journal.pgen.1001388
Suh, M. C., Samuels, A. L., Jetter, R., Kunst, L., Pollard, M., Ohlrogge, J., et al. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139, 1649–1665. doi: 10.1104/pp.105.070805
Tang, X.-M., Tao, X., Wang, Y., Ma, D.-W., Li, D., Yang, H., et al. (2014). Analysis of DNA methylation of perennial ryegrass under drought using the methylation-sensitive amplification polymorphism (MSAP) technique. Mol. Genet. Genomics 289, 1075–1084. doi: 10.1007/s00438-014-0869-6
Tian, G.-L., Zhang, S.-P., Song, Z.-C., Zhang, S., Cui, J.-Y., Miao, H., et al. (2015). Genetic Analysis and QTL Mapping of Wax Powder on the Surface of Cucumber Fruit. Sci. Agric. Sin. 2015, 10.
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Wang, S., Lv, J., Zhang, L., Dou, J., Sun, Y., Li, X., et al. (2015a). MethylRAD: a simple and scalable method for genome-wide DNA methylation profiling using methylation-dependent restriction enzymes. Open Biol. 5, 150130. doi: 10.1098/rsob.150130
Wang, W., Liu, X., Gai, X., Ren, J., Liu, X., Cai, Y., et al. (2015b). Cucumis sativus L. WAX2 Plays a Pivotal Role in Wax Biosynthesis, Influencing Pollen Fertility and Plant Biotic and Abiotic Stress Responses. Plant Cell Physiol. 56, 1339–1354. doi: 10.1093/pcp/pcv052
Wang, W., Zhang, Y., Xu, C., Ren, J., Liu, X., Black, K., et al. (2015c). Cucumber ECERIFERUM1 (CsCER1), which influences the cuticle properties and drought tolerance of cucumber, plays a key role in VLC alkanes biosynthesis. Plant Mol. Biol. 87, 219–233. doi: 10.1007/s11103-014-0271-0
Xia, Z., Zou, M., Zhang, S., Feng, B., Wang, W. (2014). AFSM sequencing approach: a simple and rapid method for genome-wide SNP and methylation site discovery and genetic mapping. Sci. Rep. 4, 7300. doi: 10.1038/srep07300
Yeats, T. H., Rose, J. K. C. (2013). The Formation and Function of Plant Cuticles. Plant Physiol. 163, 5–20. doi: 10.1104/pp.113.222737
Zahaf, O., Blanchet, S., De Zélicourt, A., Alunni, B., Plet, J., Laffont, C., et al. (2012). Comparative transcriptomic analysis of salt adaptation in roots of contrasting Medicago truncatula genotypes. Mol. Plant 5, 1068–1081. doi: 10.1093/mp/sss009
Keywords: pericarp wax, transcriptome, methylation, CsWIN1, grafted cucumber
Citation: Zhang J, Yang J, Yang Y, Luo J, Zheng X, Wen C and Xu Y (2019) Transcription Factor CsWIN1 Regulates Pericarp Wax Biosynthesis in Cucumber Grafted on Pumpkin. Front. Plant Sci. 10:1564. doi: 10.3389/fpls.2019.01564
Received: 15 June 2019; Accepted: 07 November 2019;
Published: 29 November 2019.
Edited by:
Amnon Levi, United States Department of Agriculture, United StatesReviewed by:
Francisco Perez-Alfocea, Spanish National Research Council, SpainJinghua Yang, Zhejiang University, China
Copyright © 2019 Zhang, Yang, Yang, Luo, Zheng, Wen and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changlong Wen, d2VuY2hhbmdsb25nQG5lcmN2Lm9yZw==; Yong Xu, eHV5b25nQG5lcmN2Lm9yZw==
 Jian Zhang
Jian Zhang Jingjing Yang
Jingjing Yang Yang Yang1,2
Yang Yang1,2 Changlong Wen
Changlong Wen Yong Xu
Yong Xu