- 1Laboratory of Medicinal Plant, Institute of Basic Medical Sciences, School of Basic Medicine, Biomedical Research Institute, Hubei Key Laboratory of Wudang Local Chinese Medicine Research, Hubei Key Laboratory of Embryonic Stem Cell Research, Hubei University of Medicine, Shiyan, China
- 2Center for Multi-omics Research, Key Laboratory of Plant Stress Biology, School of Life Sciences, Henan University, Kaifeng, China
- 3College of Life Sciences, Nanjing Agricultural University, Nanjing, China
- 4National Engineering Laboratory for Resource Development of Endangered Crude Drugs in Northwest China, Key Laboratory of Medicinal Resources and Natural Pharmaceutical Chemistry, Ministry of Education, College of Life Sciences, Shaanxi Normal University, Xi'an, China
- 5Department of Biological Science, Florida State University, Tallahassee, FL, United States
- 6Jiangsu Key Laboratory of Marine Biological Resources and Environment, Jiangsu Key Laboratory of Marine Pharmaceutical Compound Screening, Co-Innovation Center of Jiangsu Marine Bio-industry Technology, Jiangsu Ocean University, Lianyungang, China
- 7Key Laboratory of Three Gorges Regional Plant Genetics & Germplasm Enhancement (CTGU)/Biotechnology Research Center, China Three Gorges University, Yichang, China
Light is essential for plant organogenesis and development. Light-regulated shoot morphogenesis has been extensively studied; however, the mechanisms by which plant roots perceive and respond to aboveground light are largely unknown, particularly because the roots of most terrestrial plants are usually located underground in darkness. To mimic natural root growth conditions, we developed a root-covered system (RCS) in which the shoots were illuminated and the plant roots could be either exposed to light or cultivated in darkness. Using the RCS, we observed that root growth of wild-type plants was significantly promoted when the roots were in darkness, whereas it was inhibited by direct light exposure. This growth change seems to be regulated by ELONGATED HYPOCOTYL 5 (HY5), a master regulator of photomorphogenesis. Light was found to regulate HY5 expression in the roots, while a HY5 deficiency partially abolished the inhibition of growth in roots directly exposed to light, suggesting that HY5 expression is induced by direct light exposure and inhibits root growth. However, no differences in HY5 expression were observed between illuminated and dark-grown cop1 roots, indicating that HY5 may be regulated by COP1-mediated proteasome degradation. We confirmed the crucial role of HY5 in regulating root development in response to light under soil-grown conditions. A transcriptomic analysis revealed that light controls the expression of numerous genes involved in phytohormone signaling, stress adaptation, and metabolic processes in a HY5-dependent manner. In combination with the results of the flavonol quantification and exogenous quercetin application, these findings suggested that HY5 regulates the root response to light through a complex network that integrates flavonol biosynthesis and reactive oxygen species signaling. Collectively, our results indicate that HY5 is a master regulator of root photomorphogenesis.
Introduction
Root system architecture (RSA) is vital for plant fitness, crop performance, and plant productivity, and its development is regulated by both genetic components and environmental factors (Jansen et al., 2013; Rogers and Benfey, 2015; Li et al., 2018a; Xiao et al., 2018). Light has largely been neglected as an environmental factor controlling RSA since the root systems of most terrestrial plants remain underground in darkness (Osmont et al., 2007; Bellini et al., 2014). Intriguingly, the genes encoding photoreceptors are expressed in the roots, enabling them to detect ambient light and trigger downstream responses that mediate root tropism and elongation in the soil (Galen et al., 2007; Salisbury et al., 2007; Mo et al., 2015; Van Gelderen et al., 2018b). For instance, red light was found to activate the phytochrome A (phyA)- and phyB-mediated signal transduction pathways in the root and dramatically inhibit root elongation in etiolated seedlings (Correll and Kiss, 2005). Roots exposed to direct illumination immediately generate a strong burst of reactive oxygen species (ROS), leading to the redistribution of auxin (Yokawa et al., 2011); however, the mechanisms by which light regulates RSA development have not been fully elucidated.
Light-mediated shoot development (i.e., shoot photomorphogenesis) has been much more extensively studied than light-mediated root development (root photomorphogenesis) (Briggs and Lin, 2012; Gommers and Monte, 2018; Van Gelderen et al., 2018b). Numerous downstream components of the photoreceptor signaling pathways have been characterized (Arsovski et al., 2012; Li et al., 2012). The RING E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), a major integrator of light responses, ubiquitinates two downstream transcription factors, ELONGATED HYPOCOTYL 5 (HY5) and HY5 HOMOLOG (HYH), to mediate their degradation (Ang et al., 1998; Osterlund et al., 2000; Holm et al., 2002). HY5 is a positive regulator of photomorphogenesis (Lee et al., 2007; Li et al., 2010; Wang et al., 2019). The loss-of-function hy5 mutant produces elongated hypocotyls when grown under constant light, while the hyh mutant exhibits no obvious aberrant phenotype (Oyama et al., 1997; Holm et al., 2002; Sibout et al., 2006). The expression levels of HY5 and HYH are upregulated by light in the seedlings (Osterlund et al., 2000; Holm et al., 2002; Singh et al., 2012).
Generally, root growth and development in response to light are controlled by both shoot-derived and root-autonomous signals. Among the shoot-derived signals, phytohormones and peptides are regarded as important players (Bhalerao et al., 2002; Li et al., 2018b; Qu et al., 2019); for example, regardless of whether roots were exposed to light or darkness, the illumination of shoots efficiently promoted root growth by inducing the expression of DWF4, which encodes a brassinosteroid (BR) biosynthesis enzyme, leading to an increased accumulation of BRs in the roots (Sakaguchi and Watanabe, 2017). The COP1-mediated shoot-to-root transport of auxin is also involved in adapting root growth to the ambient light conditions (Sassi et al., 2012), with the downstream transcription factor HY5 regulating auxin accumulation in the roots by modulating the intracellular distribution of the auxin transporter PIN-FORMED2 (PIN2) (Laxmi et al., 2008).
Besides phytohormones, HY5 acts as a mobile shoot-to-root signal that promotes nitrate uptake in roots in response to light (Chen et al., 2016). In addition, aboveground light is directly transmitted through the stem to activate phytochrome B in the roots, thereby triggering HY5-mediated light responses (Lee et al., 2016). Root-autonomous signals might also be partially generated through metabolic processes; for example, Silva-Navas et al. (2016) reported that the accumulation of flavonols in the root transition zone mediates the root response to light and therefore proposed that flavonols function as local positional signals (Silva-Navas et al., 2016).
In the laboratory, seedlings are often grown on transparent Petri dishes, exposing the roots to light, despite roots naturally growing in the soil and avoiding light. To achieve a better understanding of RSA development under natural light conditions, researchers have developed different strategies to culture shoots and roots in separate light conditions (Xu et al., 2013; Yokawa et al., 2014; Silva-Navas et al., 2015; Sakaguchi and Watanabe, 2017). In this study, we developed a root-covered system (RCS) to investigate the mechanism by which light regulates root development. We found that the RSA is enhanced in plants grown in darkness compared to roots exposed to light, which was partially dependent on HY5. Furthermore, a transcriptomic analysis revealed that light-mediated regulation of root development might be based on a complex network involving phytohormones, stress signaling, and biosynthetic and metabolic processes and that some of these regulatory pathways are dependent on HY5.
Materials and Methods
Plant Materials and Growth Conditions
All Arabidopsis thaliana mutants used in this study were originally generated in the Columbia-0 (Col-0) background. The hy5 (SALK_056405), hyh (WISCDSLOX253D10), hy5 hyh, and cop1-6 mutants were described previously (Sassi et al., 2012; Zhang et al., 2017). The seeds were germinated on half-strength Murashige and Skoog (1/2 MS) agar plates or on soil (covered with a thin layer of sand to shield the roots from light) and were incubated at 22°C under continuous white light (120 µmol m−2 s−1) or in darkness (covered with foil) in a plant growth chamber for 7 days, or as indicated.
To accomplish the covered-root culture condition, two pinholes were made on the top edge of a square plastic Petri dish (9 × 9 cm) containing 1/2 MS medium. Two 3-day-old etiolated seedlings were transferred into the corresponding pinholes using fine forceps, ensuring that the roots were placed on the medium inside the dish with the hypocotyls passing through the pinholes, leaving the cotyledons outside the dish. The Petri dishes were subsequently either covered with double layers of foil to conceal the root but not the shoot (roots are covered) or sealed without foil (roots are illuminated). The dishes were then placed into larger dishes (13 × 13 cm) to create a relatively sterile environment and the seedlings were grown for 12 additional days, or as indicated (Figure S1 and Figure 1).
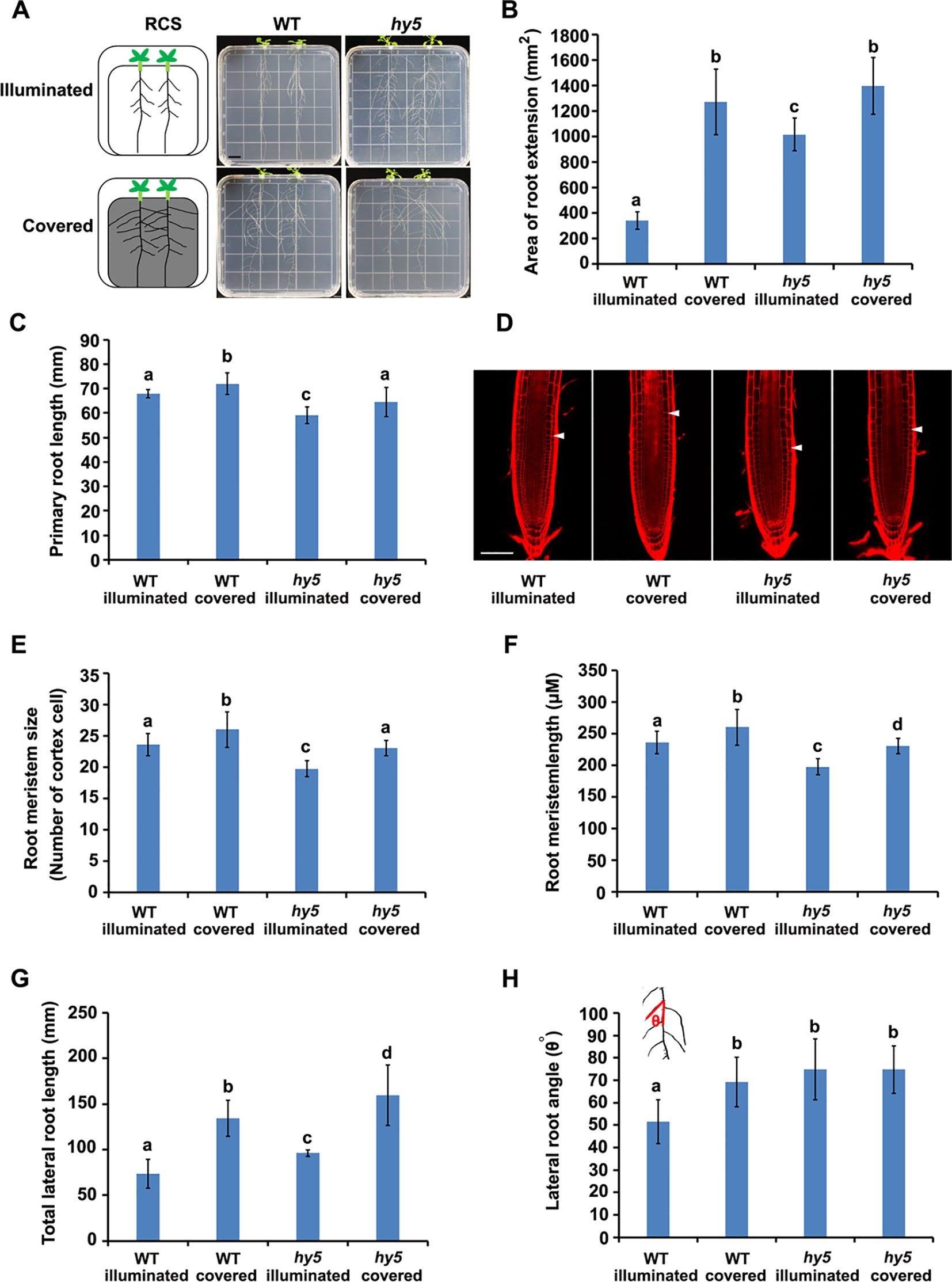
Figure 1 Phenotypes of root system architectures (RSA) of wild-type (WT) plants and hy5 mutants under root-covered system (RCS). (A) Representative pictures of the RSA phenotypes of 15-day-old WT and hy5 plants. Scale bar represents 10 mm. (B) Quantification of the root extension area (in square millimeters) of WT-illuminated, WT-covered, hy5-illuminated, and hy5-covered. (C) Quantification of the primary root lengths of 9-day-old WT and hy5 plants. (D–F) Root meristem phenotypes of 9-day-old WT and hy5 plants presented as representative confocal images (D), meristem sizes (measured as the number of cortex cells) (E), and the length of root meristems (F). Scale bar in (D) represents 50 μm. (G) Total lateral root lengths of 9-day-old WT and hy5 plants. (H) Average lateral root angles of 9-day-old WT and hy5 plants. The inset on the upper left denotes how the angles were measured. Error bars represent the standard deviation (SD) of biological triplicates (n = 6). Different letters indicate significantly different values at P < 0.05 (pairwise Student's t test).
For the light–dark transition experiments, the seedlings were grown in the soil under continuous light or darkness for 5 days, after which they were transferred to the opposite condition for 24 h. At least three biological replicates were performed.
For quercetin treatment, 3-day-old etiolated seedlings were transferred onto a solid 1/2 MS medium containing either a mock control or 50 µM quercetin for the indicated number of days.
Expression and Phenotypic Analyses
β-glucuronidase (GUS) staining was performed as described previously (Zhang et al., 2017; Liu et al., 2019a; Zhang et al., 2019). Briefly, 7-day-old seedlings were incubated in the assay buffer [50 mM sodium phosphate (pH 7.0), 10 mM EDTA, 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide, 0.1% Triton X-100, 20% methanol, and 0.5 mg ml–1 X-Gluc A] at 37°C until sufficient staining was observed. The GUS activity was analyzed using a Nikon 80i microscope with Nomarski interference contrast optics.
The confocal imaging was performed using a Leica TCS SP2 microscope. Propidium iodide (PI; Sigma-Aldrich; 10 µg ml–1 dissolved in water) was used to counterstain the root cells. The fluorescence signals of green fluorescent protein (GFP) and PI were excited using an argon ion laser. The excitation and emission wavelengths for GFP were 488 and 510 nm, respectively. The excitation and emission wavelengths for PI were 561 and 630 nm, respectively.
To analyze the root phenotypes, the seedlings were imaged with a Canon 40D camera. The areas of root extension were indicated by tracking all the lateral roots (Ristova et al., 2013) and measuring their area using ImageJ with the SmartRoot plug-in (Lobet et al., 2011). Quantitative analyses of the root apical meristem (RAM) and lateral root angles were performed as previously described (Li et al., 2017a; Zhang et al., 2017). Statistical analyses (pairwise two-tailed Student's t tests) were performed using Microsoft Excel. Each experiment was performed in three biological replicates.
RNA Extraction and Quantitative Reverse Transcription PCR
Total RNA was isolated from the roots using Tranzol (Transgene), in accordance with the manufacturer's protocol. The RNA purity was checked using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific). Only samples with absorbance ratios of OD260/280 ≥ 1.8 and OD260/230 ≥ 1.8 were selected for further use. The cDNA was prepared using a PrimeScript RT Reagent Kit (RR047A; Takara Bio). The relative expression levels were determined by performing a quantitative reverse transcription PCR (qRT-PCR) analysis using an ABI 7500 Real-Time PCR System (Thermo Fisher Scientific) or a ViiA 7 Real-Time PCR System (Life Technologies). The PCR thermocycling protocol was as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. After the amplification cycles, dissociation analyses were performed to confirm the validity of the target (Guo et al., 2018; Jiao et al., 2019; Li et al., 2019c) Three biological replicates and three qRT-PCR technical replicates were performed for each sample. EF1α (AT5G60390) was used as the reference gene for normalization. The primer sequences used for the qRT-PCR analyses are listed in Table S1.
RNA Sequencing and Data Analyses
The roots of 15-day-old wild-type (WT) and hy5 seedlings grown in illuminated or covered conditions were collected, immediately frozen in liquid nitrogen, and stored at −80°C before an RNA extraction. Three independent biological replicates were collected for each genotype and growth condition, and the total RNA was extracted as described above. After purification, the integrity of the RNA was verified using the RNA 6000 Nano Labchip Kit on an Agilent 2100 Bioanalyzer (Agilent Technologies), following the manufacturer's protocol. The enrichment of the mRNA from the total RNA, the cDNA synthesis, and the construction of the cDNA library were performed by the Beijing Genome Institute (BGI, China). An Illumina HiSeq 2000 sequencing system was used to sequence the libraries. After removing the sequences containing more than 5% unknown bases or more than 30% nucleotides with a sequence quality value below 10, the clean reads were aligned to the Arabidopsis genome using HISAT (Kim et al., 2015). The expression level of each gene was normalized as the number of clean reads per kilobase of exon per million mapped reads (RPKM) (Mortazavi et al., 2008). The method DESeq2 was used to select differentially expressed transcripts, while a false discovery rate (FDR) < 0.05 was further used to estimate the correction for false positive and false negative errors (Love et al., 2014). A heat map of differential expression was generated using custom R scripts based on Bioconductor packages (Gentleman et al., 2004), and the Venn diagrams were produced using the online software jvenn (Bardou et al., 2014). Gene Ontology (GO) annotations and GO singular enrichment analysis (SEA) were performed using goseq R Bioconductor package (Young et al., 2010). An FDR cutoff of 0.05 was used to determine the enriched GO pathways.
ROS Level Determination
Fluorescein diacetate (FDA) was used to detect the ROS level in the roots, following the method described previously by Silva-Navas et al. (2016), with a slight modification. Briefly, the roots were submerged in a solution containing 0.01% Triton X-100 and 0.83 mg ml–1 fluorescein diacetate for 10 min. After being washed in deionized water for 2 min, the root tips were mounted on slides and imaged using a Nikon/A1+N-SIM confocal laser scanning microscope. The excitation and emission wavelengths for FDA were 488 and 520 nm, respectively. Three biological replicates were performed.
Extraction and Determination of Flavonols
The root samples were homogenized in liquid nitrogen, and the flavonols were extracted in 80% acetone. The simultaneous rapid separation of the flavonoids was performed using high-performance liquid chromatography (HPLC) with 5-µm (150 × 4.6 mm) Bridge C18 columns. The linear gradient elution was performed using a mobile phase (0.1% formic acid and acetonitrile acidified with 0.1% formic acid) starting with 95% formic acid and ending with 5% formic acid over a total run time of 35 min. The flow rate of the mobile phase was 1 ml min–1. A 2998 PDA detector was used.
Results
HY5 Regulates RSA Development, as Revealed Using the RCS
In nature, plant roots grow in the soil and perceive little to no light (Galen et al., 2007; Sassi et al., 2012). For a long time, plant biologists have been growing Arabidopsis seedlings on transparent Petri dishes with the roots exposed to light, an approach we designated as a traditional culture system (TCS). Using a TCS, plant roots that would naturally grow in darkness were instead exposed to light (Figures S1A, B), which is controversial in the field of root research (Xu et al., 2013; Yokawa et al., 2013; Mo et al., 2015). To better understand the natural RSA of a plant, various approaches have been developed to mimic natural conditions (Xu et al., 2013; Yokawa et al., 2013; Silva-Navas et al., 2015; Lee et al., 2016; Sakaguchi and Watanabe, 2017). In this study, we developed a laboratory culture system called a root-covered system (RCS). Under the RCS, the plant shoots were exposed to light, while the exposure of the roots to light could be manually controlled by the presence or absence of a foil cover (Figure S1 and Figure 1).
We examined the RSA of plants grown using the RCS. In WT seedlings, we observed a dramatic increase in the area of root extension of the seedlings grown with the foil cover (WT-covered) compared to those grown without the foil cover over the roots (WT-illuminated) (Figures 1A, B), demonstrating the regulatory role of light on root growth.
HY5 has been characterized as a key regulator of photomorphogenesis. To investigate whether HY5 is involved in the light-mediated regulation of RSA development, we grew the hy5 loss-of-function mutant in the RCS under illuminated and covered conditions. Intriguingly, the area of root extension of the hy5 mutants grown in the illuminated condition was dramatically higher than that of the WT illuminated seedlings (Figures 1A, B). Furthermore, the increase in root growth observed in the WT seedlings grown with the foil cover (WT-covered) relative to the illuminated condition was greater than that of the hy5 seedlings (Figures 1A, B), with a 4.0-fold increase in the WT and a 1.3-fold increase in hy5. Thus, HY5 plays a role in regulating RSA in response to light.
A further analysis showed that both darkness and the hy5 mutation increased the total lateral root length, lateral root angle, primary root length, and root hair lengths of the plants (Figures 1C–E and Figure S2). As root length is tightly correlated with root meristem activity (Sassi et al., 2012), we also analyzed the meristem size of the primary roots, revealing that darkness increased the size and length of the root meristem in both the WT and hy5 plants (Figures 1F–H).
Auxin is the main regulator of lateral root development (Audenaert et al., 2013). To test whether the local accumulation of auxin was altered in the hy5 mutants, we introduced the auxin-responsive marker, DR5::GUS (Ulmasov et al., 1997), into hy5 mutants. DR5::GUS expression was strongly enhanced in the lateral roots of the hy5 mutant compared to the WT plants (Figure S3), indicating that the loss of HY5 function may alter the local accumulation of auxin to accelerate lateral root growth, thereby promoting RSA development.
Expression Dynamics of HY5 in Response to Light
Lateral roots are a major component of the RSA (Smith and De Smet, 2012). As HY5 regulates RSA, we first examined the expression pattern of HY5 during lateral root development using a HY5 promoter reporter, HY5::GUS. This experiment revealed that HY5 was abundantly expressed from the early stages of lateral root primordia development to the maturation of the lateral roots (Figure 2). HY5::GUS expression was predominately detected at the meristem and in the meristem-elongation transition zone of the mature lateral roots. In contrast, HYH, the HY5 homolog, was only expressed in the vascular tissues of mature lateral roots. These data suggest that HY5 may be a crucial factor controlling RSA.
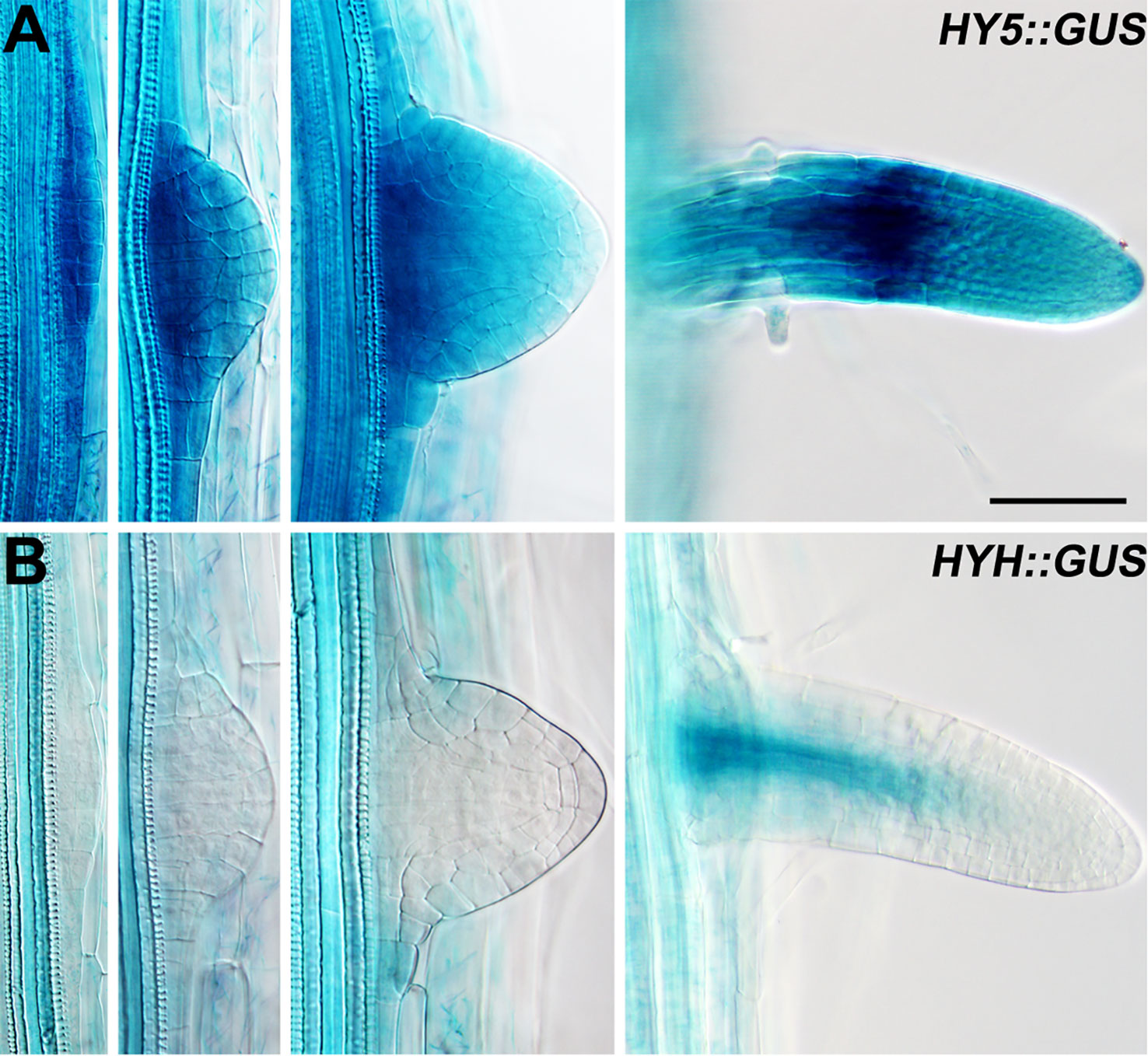
Figure 2 The expression patterns of HY5 and HYH during lateral root development. (A) HY5::GUS roots. (B) HYH::GUS roots. The seedlings were grown in constant light for 8 days before the GUS staining. Scale bar represents 20 μm.
Subsequently, we determined the expression of HY5 and HYH in the roots in response to light. The transcript levels of HY5 and HYH were measured in the roots of the illuminated and covered WT and hy5 seedlings using qRT-PCR. As expected, the expression levels of HY5 and HYH were significantly decreased in the foil-covered WT seedlings (WT-covered) compared to those exposed to light (WT-illuminated) (Figure S4). Consistently, the GFP signals in the HY5 and HYH transcriptional GFP reporter lines expressing HY5::ERGFP or HYH::ERGFP, respectively, were dramatically enhanced by exposure to light (Figures 3A–D). The light-induced expression of HYH was abolished in the hy5 mutant (Figures 3C–F and Figure S4), demonstrating that the transcriptional regulation of HYH is governed by HY5 in response to light.
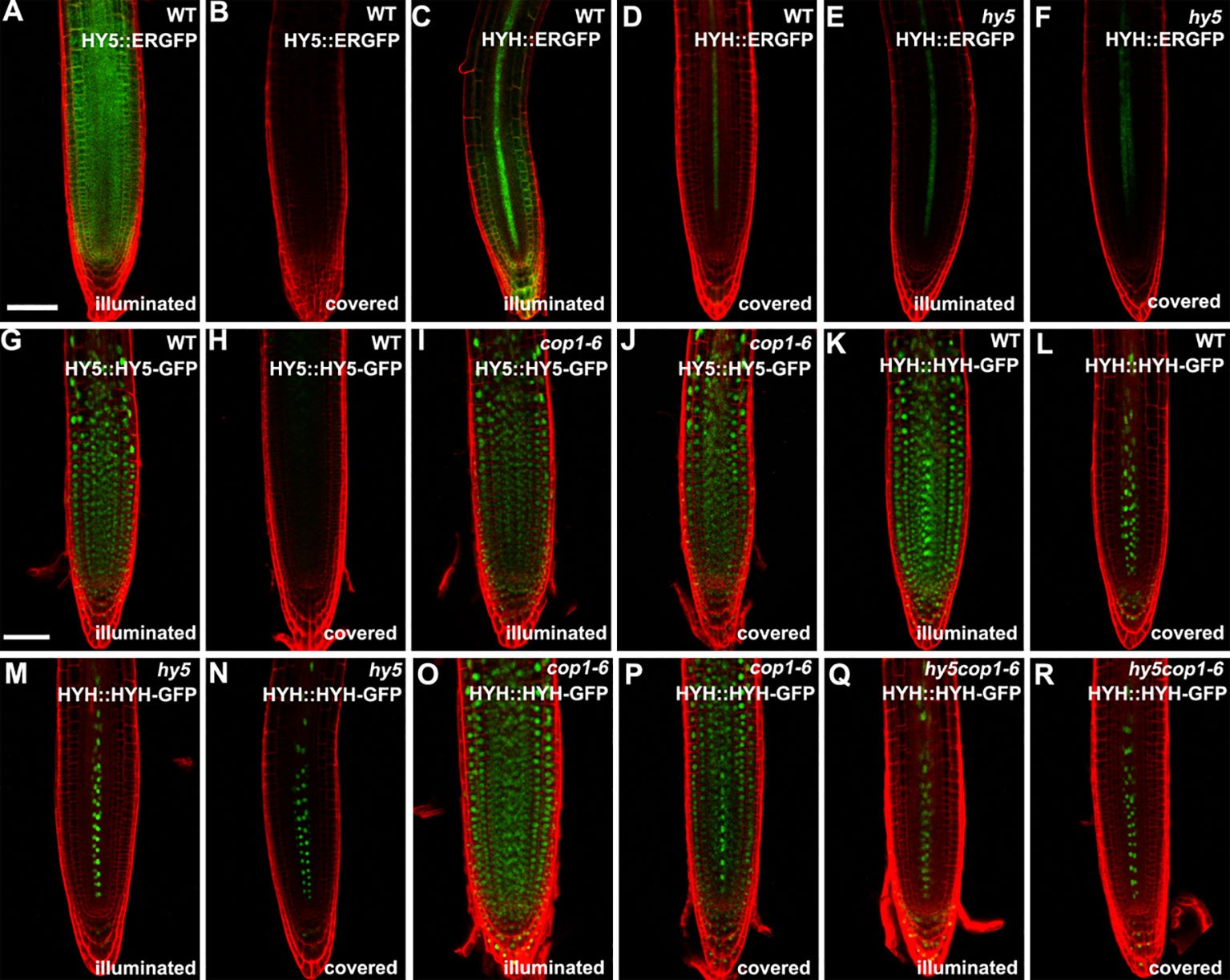
Figure 3 The expression patterns of HY5 and HYH in response to darkness and light in the roots of seedlings grown in the root-covered system (RCS). (A, B, G, H) Expression of HY5 promoter fusion (A, B) and protein fusion (G, H) in roots of wild-type (WT) seedlings in either illuminated or covered conditions as indicated. (C, D, K, L) Expression of the HYH promoter fusion (C, D) and protein fusion (K, L) in the roots of WT seedlings grown in either illuminated or covered conditions as indicated. (E, F, M, N) Expression of the HYH promoter fusion (E, F) and protein fusion (M, N) in the roots of hy5 mutant grown in either illuminated or covered conditions as indicated. (I, J) Expression of the HY5 protein fusion lines in roots of cop1-6 mutant in either illuminated or covered conditions as indicated. (O–R) Expression of the HYH protein fusion lines in the roots of cop1-6 mutant (O, P) and hy5 cop1-6 mutants (Q, R) grown in either illuminated or covered conditions as indicated. Scale bar represents 20 μm.
COP1 Mediates the Accumulation of HY5 Protein in Roots Directly Exposed to Light
The COP1-mediated degradation of proteins is known to be crucial for light-mediated growth and development (Osterlund et al., 2000; Holm et al., 2002). To determine whether COP1 mediates the accumulation of HY5 and HYH proteins in response to light in the roots, we next employed translational GFP reporters to investigate the levels of HY5 and HYH in plants of various genotypes with dark-grown or illuminated roots. The GFP signals of the WT plants expressing HY5::HY5-GFP or HYH::HYH-GFP were strongly decreased in plants with covered roots compared to the illuminated controls (Figures 3G, H, K, L), demonstrating that the expression of HY5 and HYH in response to light occurs in a root-autonomous manner. Light did not appear to alter the GFP signals of the cop1-6 mutant plants expressing HY5::HY5-GFP or HYH::HYH-GFP (Figures 3I, J, O, P). The HYH::HYH-GFP expression was found to be constantly inhibited in the hy5 and hy5 cop1-6 double mutants grown under the RCS (Figures 3M–R), indicating that the COP1-mediated proteasome degradation of HYH is dependent on HY5. Collectively, our data indicate that HY5 is a master coordinator of light-mediated root development.
Expression Dynamics of HY5 in Response to Light Under the Soil-Grown Condition
Arabidopsis roots naturally grow in darkness in soil. To investigate whether HY5 plays a crucial role in soil-grown root development, we investigated the expression of HY5::HY5-GFP and HYH::HYH-GFP in plants whose shoots were subjected to light–dark transitions in soil-grown conditions. The seedlings were germinated on soil covered with a thin layer of sand to shield the roots from light. The shoot was either exposed to light or kept in darkness by covering with foil (Figure 4A). A qRT-PCR analysis revealed that the level of HYH expression was similar in the roots of soil-grown hy5 and WT plants with light-exposed shoots (Figure 4B). Consistently, HY5::HY5-GFP accumulated at a relatively low level in the roots of soil-grown WT plants under shoot-illuminated and shoot-covered conditions (Figures 4C, D). HYH::HYH-GFP expression was much greater in the root vascular tissues of soil-grown seedlings when their shoots were exposed to light rather than darkness (Figure 4E, F) and in those transferred from darkness to light (Figure 4H). In contrast, almost no HYH::HYH-GFP expression was observed in the roots of plants whose shoots were either grown in darkness or transferred from light to dark (Figures 4E–H). No significant differences were observed in the HYH::HYH-GFP expression levels of the hy5 and WT plants with shoots exposed to different light conditions (Figures 5E–L).
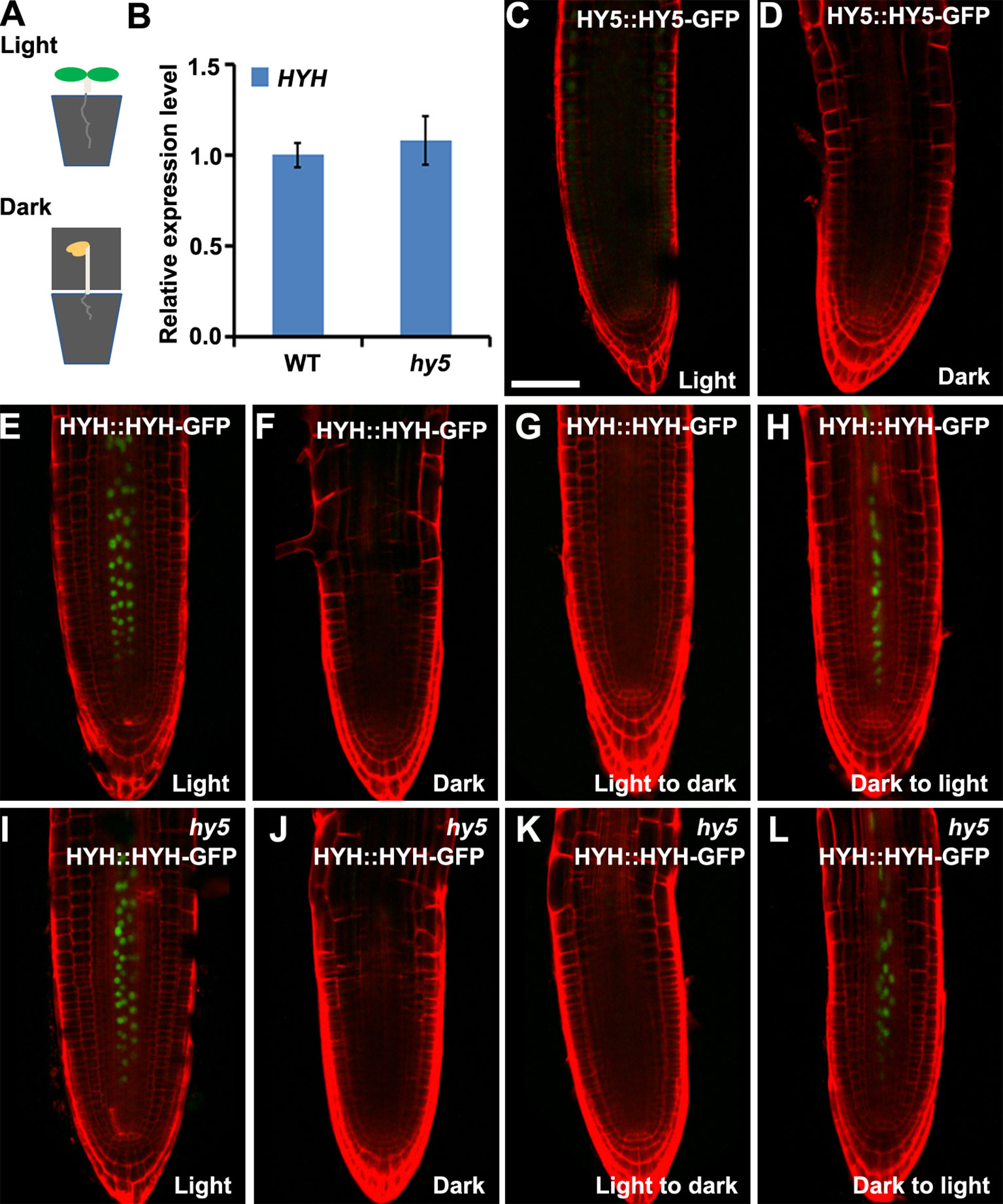
Figure 4 The expressions of HY5 and HYH in response to light in the soil. The expression levels of HY5 and HYH in soil-grown seedlings in response to illumination of the shoot. (A) Illustration of the soil growing system used to perform the light treatments on the Arabidopsis seedlings. (B) qRT-PCR quantification of HYH transcripts in soil-grown wild-type (WT) and hy5 seedlings when the shoots were exposed to light. Error bars represent the standard deviation (SD) of biological triplicates. HYH transcripts were normalized to the EF1α gene. (C, D) Expression of HY5 protein fusion lines in the roots of soil-grown WT plants whose shoots were exposed to light (C) or darkness (D). (E–H) Expression of HYH protein fusion lines in the roots of soil-grown WT plants whose shoots were subjected to light (E), darkness (F), or a light-to-dark (G) or dark-to-light (H) transition. (I–L) Expression of the HYH protein fusion lines in the roots of soil-grown hy5 plants whose shoots were subjected to light (I), darkness (J), or a light-to-dark (K) or dark-to-light (L) transition. Scale bar represents 20 μm.
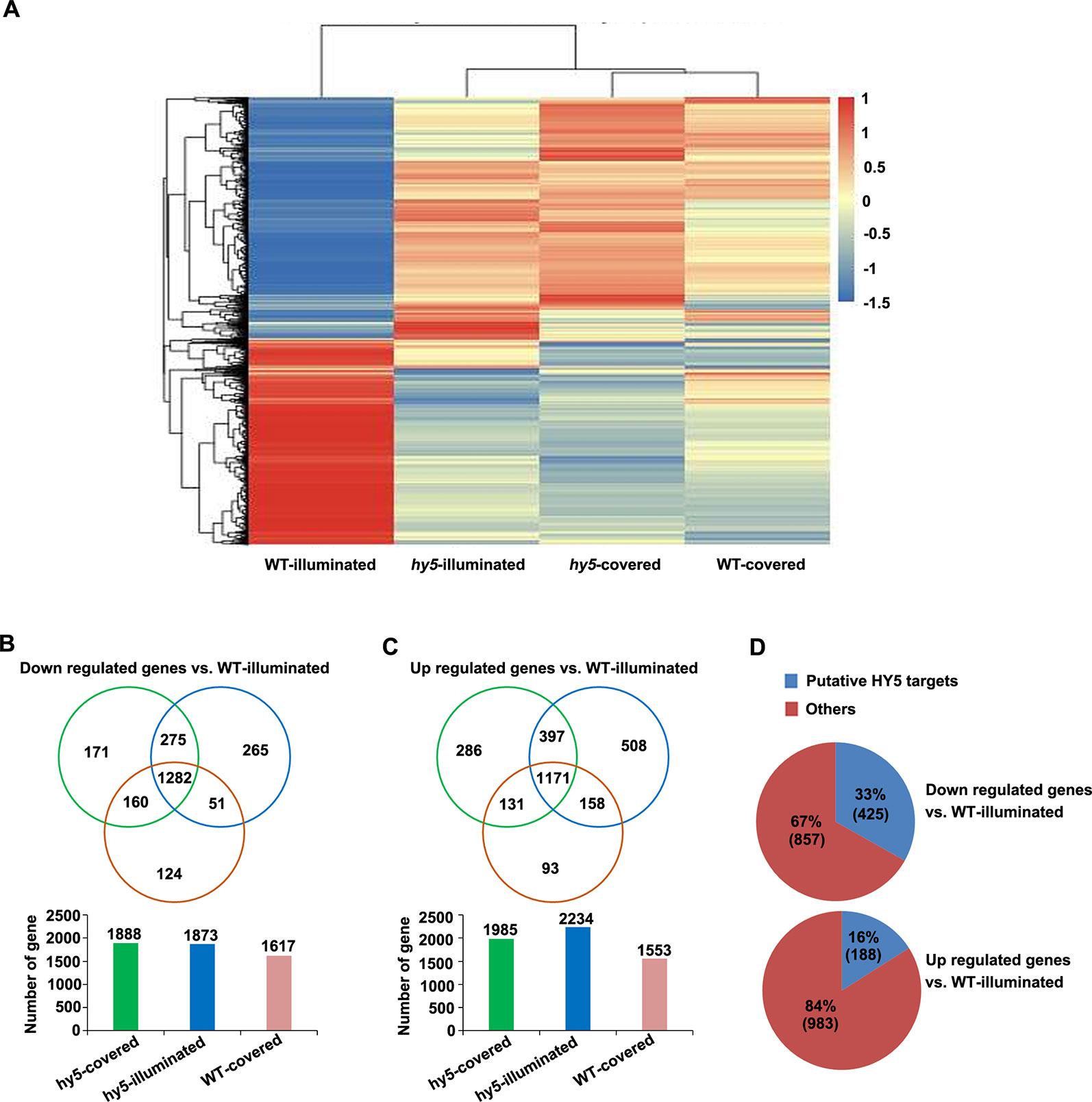
Figure 5 Analysis of the differentially expressed genes (DEGs) identified in the comparisons between the transcriptome of hy5-illuminated, hy5-covered, and wild-type (WT)-covered samples with those of WT-illuminated. (A) Overall clustering analysis and heat map of the four groups of root samples (WT-illuminated, hy5-illuminated, hy5-covered, and WT-covered). The gene expression values of each sample were averaged and z-transformed, resulting in scaled expression values between −1.5 and +1.5 for each gene. Hierarchical clustering was calculated using the hclust() function in R. (B, C) Venn diagram illustrating the number of unique (non-overlapping circles) and common (overlapping circles) upregulated and downregulated transcripts identified in hy5-illuminated, hy5-covered, and WT-covered compared to WT-illuminated. (D) Proportion of upregulated and downregulated transcripts that overlap with the list of HY5 targets.
Transcriptomic Analysis of HY5-Regulated Genes in Plants Grown Under RCS
To obtain new insights into the light-mediated regulation of RSA development by HY5 at a genome-wide level, we performed an RNA sequencing (RNA-Seq) study to compare the transcriptomic profiles of the light-exposed or dark-grown roots of 15-day-old WT and hy5 seedlings. Differentially expressed genes (DEGs) were determined by comparing the transcriptomes of the illuminated or covered roots in hy5 plants and by comparing the transcriptomes of the roots of WT plants grown in the two conditions. The clustered patterns of all DEGs (5,066 unique genes) were created based on their relative expression level values in all four samples (Figure 5). The results indicate that the gene expression patterns of the WT roots grown in darkness and the hy5 roots grown in both illuminated and covered conditions contrasted with the expression pattern of the WT roots exposed to light.
Compared to the illuminated WT root sample (WT-illuminated), 1,985 genes, 2,234 genes, and 1,553 genes were upregulated in the hy5 plants with covered roots (hy5-covered), the hy5 roots exposed to light (hy5-illuminated), and the WT dark-grown roots (WT-covered), respectively, while 1,888 genes, 1,873 genes, and 1,617 genes were downregulated in these lines, respectively (Figures 5B, C and Table S1). We next investigated the overlapping DEGs in clusters of hy5-illuminated, hy5-covered, and WT-covered. One thousand two hundred eighty-two genes were downregulated and 1,171 genes were upregulated relative to their expression in WT plants with illuminated roots (WT-illuminated) (Figures 5B, C and Table S1).
Previous studies documented that the ACE element (ACGT) and G-box motif are the putative binding sites of HY5 (Lee et al., 2007). A comparative analysis of our data with the previous genome-wide microarray data of putative HY5 binding targets revealed that 425 of the 1,282 downregulated genes (33%) and 188 of the 1,171 upregulated genes (16%) are putative HY5 targets (Figure 5D and Table S2). In a previous summary of light signaling genes (Shikata et al., 2014), 29 of the downregulated genes identified here were shown to be involved in categories of COP/DET complex, such as phyA-105 (SPA1)-related 3 (SPA3) and SPA4; PHY and Phot signaling, namely, ROOT PHOTOTROPISM 2 and RPT3/NON-PHOTOTROPIC HYPOCOTYL3 (NPH3); and COP1 substrates encoding genes including HY5 and BZS1/B-BOX DOMAIN PROTEIN 20 (BBX20). Among these 29 genes, 16 were putative HY5 targets (Table S3). Taken together, the HY5-targeted genes are enriched in light growth responses, which make a major contribution to root photomorphogenesis when grown under the RCS.
Phytochrome signaling induces genome-wide alternative splicing (AS) during de-etiolation to mediate light responses in Arabidopsis seedlings (Shikata et al., 2014). To reveal whether HY5-regulated RSA development is associated with AS changes, we explored four major AS events—exon skipping, intron retention, alternative 5′ splice sites, and alternative 3′ splice sites—in the transcriptome profiles of WT-illuminated, hy5-illuminated, hy5-covered, and WT-covered. Intron retention was found to be the most abundant AS event in each group, while exon skipping was the least common (Figure S5 and Table S4). There were no differences among the frequency of these four AS events in either the individual samples or the overlapping clusters (Figure S5). Moreover, a low proportion of these AS events were accompanied by altered transcriptional expression levels (Table S4). Collectively, these data indicated that light-regulated AS might not be involved in the HY5-mediated regulationof RSA development in response to light.
GO Functional Enrichments of HY5-Regulated Genes in Plants Grown Under RCS
To investigate the biological processes regulated by HY5, we categorized the common DEGs found in clusters of hy5-illuminated, hy5-covered, and WT-covered into GO terms. The processes were first grouped according to ontologies reflecting cellular components (CC) (Figure 6A and Table S5). The downregulated DEGs were mostly enriched in the chloroplast, organelle membrane, and ribosome categories, while the upregulated DEGs were enriched in the category of the plasma membrane (Figure 6A and Table S5).
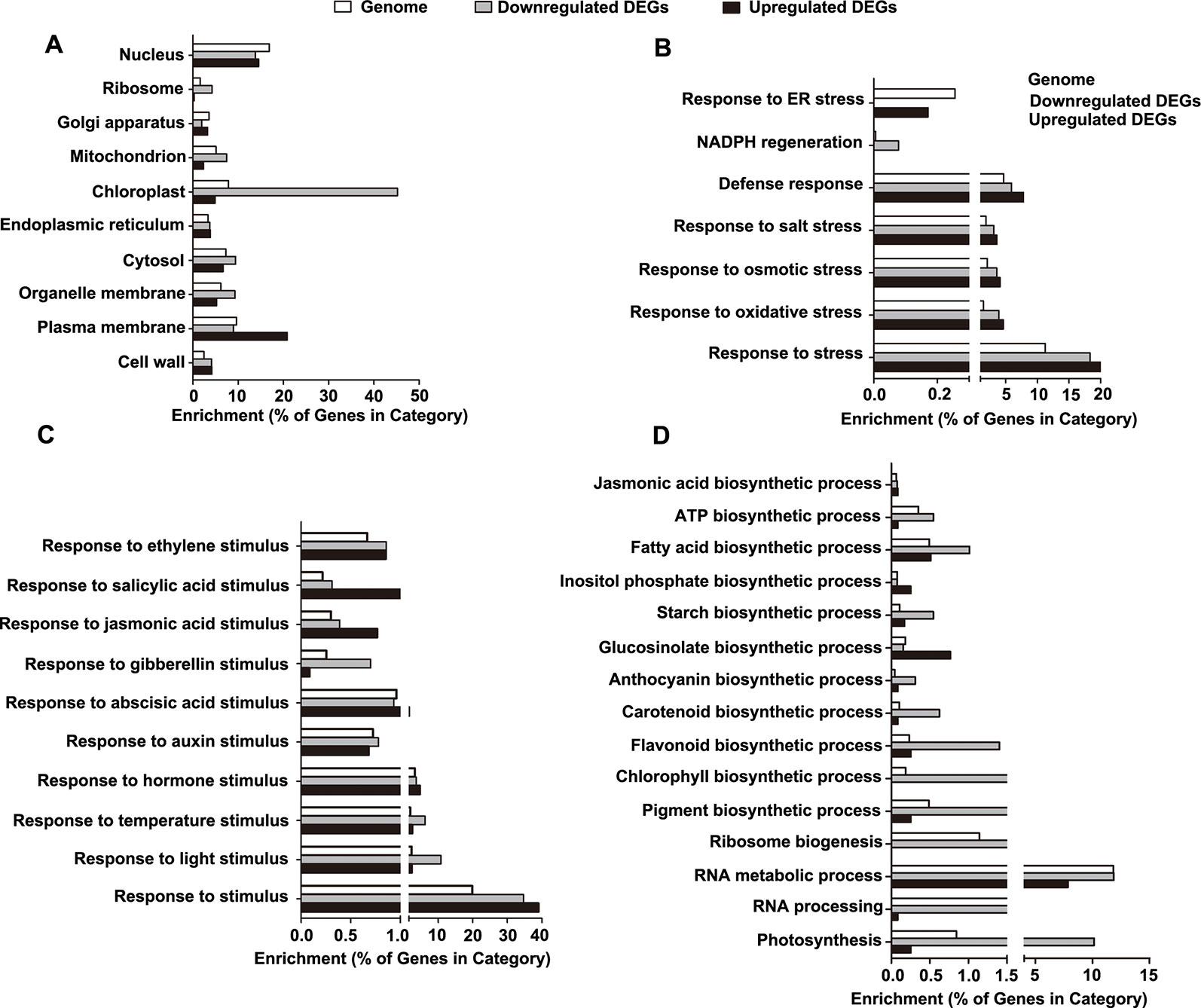
Figure 6 Gene ontology (GO) enrichment analysis of the differentially expressed genes (DEGs). (A) Categories of cellular component-related GO terms. (B) Categories of stress-related GO terms. (C) Categories of stimulus-related GO terms. (D) Categories of biosynthesis process-related GO terms.
Next, we determined the enrichment of the GO terms reflecting biological processes (BP), particularly those related to stress, stimuli, and biosynthetic processes. For stress-related GO terms, the categories of defense, salt stress, osmotic stress, NADPH regeneration, and oxidative stress were significantly overrepresented in the downregulated DEGs, while the category of endoplasmic reticulum (ER) stress was exclusively enriched in the upregulated DEGs (Figure 6B and Table S5). For stimulus-related GO terms, the categories of both environmental stimuli (light and temperature) and hormonal stimuli (auxin, abscisic acid, gibberellins, jasmonic acid, salicylic acid, and ethylene) were enriched in the downregulated DEGs (Figure 6C and Table S5). For biosynthetic process-related GO terms, the downregulated DEGs were enriched in the categories involved in the processes of ribosome biogenesis, RNA metabolic process, RNA processing, photosynthesis, and biosynthesis of jasmonic acid, ATP, fatty acids, inositol phosphate, starch, glucosinolate, anthocyanin, carotenoid, flavonoid, chlorophyll, and pigment (Figure 6D and Table S5).
Transcriptomic Comparison of Plants Grown in the RCS With Those Grown in the D-Root System
Using a novel D-root system, in which the roots of the cultivated plants were grown in darkness while the shoot was in the light, Silva-Navas and colleagues revealed that light illumination inhibits root elongation (Silva-Navas et al., 2015; Silva-Navas et al., 2016). In a transcriptomic analysis, they identified 1,553 upregulated and 1,617 downregulated genes in the roots of seedlings with illuminated roots compared to the dark-grown roots (Silva-Navas et al., 2016). To compare the transcriptomes of plants grown in the RCS with those grown in the D-root system, we first compared the DEGs of the WT plants grown in the RCS with dark-grown or light-exposed roots to the DEGs identified for plants grown in the D-root system. In both systems, 575 genes were differentially regulated, among which, 114 common genes were upregulated and 316 common genes were downregulated in the roots grown in darkness compared to those under illumination (Figure S6 and Table S6). A GO analysis suggested the response to salicylic acid, particularly the terpene biosynthetic and metabolic process, was significantly overrepresented in the upregulated transcripts, while in the downregulated genes, the significantly overrepresented categories were related to the response to abiotic stimulus, response to light stimulus, and photosynthesis (Table S7).
To further identify candidate HY5-related components involved in the responses of the root to its surrounding light and dark conditions, we further overlapped the DEGs of WT-covered, hy5-illuminated, and hy5-covered compared to WT-illuminated under RCS with that under D-roots (Silva-Navas et al., 2016). The DEGs between the roots of the dark-grown WT roots and the illuminated WT roots were compared to those of the hy5 mutant under these two conditions. The expression patterns of 11 upregulated genes and eight downregulated genes in the WT comparison were abolished or partially compromised in the hy5 mutant background (Figure S6 and Table S6). This suggests that these genes might contribute to the HY5-mediated regulation of the root response to light. Accordingly, four of these 19 genes harbor putative HY5 binding sites in their promoter regions. It will be of interest to dissect the functions of these genes in follow-up work.
HY5 Regulates the Root Response to Light by Mediating the Flavonol Content in the Roots
Silva-Navas et al. (2016) reported that the accumulation of flavonols in the root transition zone mediates the root response to light and therefore proposed that flavonols function as local positional signals. Interestingly, four of the flavonoid biosynthetic genes (4CL3, TT7/F3'H, FLS, and CHS), which are putative HY5 targets, were downregulated in the DEG clusters of hy5-illuminated, hy5-covered, and WT-covered in comparison to that of WT-illuminated (Table S1), indicating that flavonoid biosynthesis might be involved in the HY5 mediation regulation of RSA development in response to light. To test this hypothesis, the flavonol contents in the illuminated and dark-grown roots of the WT and hy5 plants were determined using HPLC. The roots of the WT plants grown under light accumulated significantly higher levels of kaempferol and quercetin, the main flavonol derivatives, than WT-covered, hy5-illuminated, and hy5-covered (Figure S7), suggesting that illumination induces the accumulation of flavonols in a HY5-dependent manner. Next, we investigated whether the application of exogenous quercetin could revert the RSA phenotypes of the hy5 mutant. The quercetin treatment decreased the primary root elongation, area of root extension, and number of lateral roots in both the WT and hy5 roots, regardless of whether they were exposed to light or darkness (Figures S8A–D).
Flavonols regulate root growth in response to light by decreasing the accumulation of ROS in the root meristem (Silva-Navas et al., 2016). Since HY5 is indispensable for the accumulation of flavonols in response to light, we monitored the ROS levels in the roots of the hy5 mutant. We found that the ROS levels were much higher in the roots of the dark-grown and illuminated hy5 roots than those of the dark-grown and illuminated WT roots (Figure S9). Taken together, these results suggest that HY5 might regulate the root response to light by mediating the accumulation of flavonol in the roots.
Discussion
The RCS Is a Reliable Culture System That Can Mimic Soil Conditions
Traditionally, Arabidopsis seedlings were cultured in transparent Petri dishes in the laboratory, where both the shoots and roots are exposed to light. In nature, the roots of most terrestrial plants grow underground in darkness and grow away from light in a process known as negative phototropism. It has been reported that all photoreceptors are expressed in the roots to sense external light changes (Yokawa et al., 2014; Mo et al., 2015); therefore, many researchers have argued that investigating root development in transparent Petri dishes is inappropriate (Han et al., 2015; Mo et al., 2015). Recently, several improved cultural systems have been developed to study root development. In these systems, the shoots and roots are grown in separate light environments, which can be altered to explore the effects of light and darkness on the roots (Xu et al., 2013; Yokawa et al., 2013; Silva-Navas et al., 2015; Sakaguchi and Watanabe, 2017). Here, we developed an RCS, in which the roots can be grown in darkness by covering the Petri dish with foil. Our comparative analysis indicated that the RCS provides growth conditions similar to the soil environment; for example, the expression levels of HY5 and HYH were similar between plants grown in the RCS and in the soil (Figures 3 and 4). Compared to the microtomography method, which uses X-ray CT scans to image the RSA (Lucas et al., 2011; Rellán-Álvarez et al., 2015), our RCS is more conventional and effective for use in the laboratory.
Using the RCS system, we revealed that the RSA was dramatically enhanced when the roots were covered with foil. Similar RSA phenotypes were observed in two recent reports (Silva-Navas et al., 2015; Van Gelderen et al., 2018a). This darkness-promoted root growth has been observed in other species; for example, Weller et al. reported that soil-grown pea (Pisum sativum) seedlings produced more lateral roots than seedlings grown in transparent plants with roots directly exposed to light (Weller et al., 2009). These data clearly show that our RCS is a useful and reliable culture approach for mimicking soil growth conditions.
HY5 Is a Major Regulator of RSA Development
HY5 plays multifaceted roles during plant growth and development (Gangappa and Botto, 2016). Here, we demonstrated that the avoidance of direct light exposure could enhance the RSA in a HY5-dependent manner. The inhibition of RSA development by direct light exposure was not observed in the hy5 mutants (Figure 1). Direct light exposure induced the transcriptional upregulation and protein accumulation of HY5 in the WT, which were decreased by darkness (Figures 3 and 4). This might explain the observation that the RSA of the hy5 roots exposed to light was similar to that of the dark-grown WT plants (Figure 1). The inhibition of growth by HY5 was also previously observed for the hypocotyl and lateral roots (Oyama et al., 1997; Sibout et al., 2006). Moreover, the phytochrome-dependent accumulation of HY5 was found to regulate lateral root development when the shoots were exposed to far-red light (Van Gelderen et al., 2018a). In addition, light-grown pea seedlings deficient in HY5 activity showed a phenotype similar to that of the dark-grown WT roots, although RSA development was significantly inhibited when the WT roots were exposed to light, suggesting that the mechanism by which HY5 inhibits growth might be conserved among species (Weller et al., 2009). A dramatic accumulation of HY5 occurred in the dark-grown cop1-6 roots (Figure 3), suggesting that the accumulation of HY5 in dark-grown roots may be negatively regulated by COP1-mediated proteasome degradation (Osterlund et al., 2000). HY5 stability in early seedlings is also affected by the phosphorylation status in its COP1 binding domain (Hardtke et al., 2000; Osterlund et al., 2000). These evidences suggest that plants might utilize a similar process for the regulation of HY5 activity in different tissues.
Possible Mechanisms by Which HY5 Regulates RSA
Our transcriptomic analysis revealed that over 2,000 genes were differentially expressed between roots grown in darkness and those exposed to light (Figure 5). Around 42% of DEGs (848 of the 2,021 DEGs) contained putative HY5 binding targets, highlighting the central role that HY5 may play in regulating RSA in response to light. These HY5-dependent genes were involved in various biological processes (Figure 6), suggesting that the mechanisms by which HY5 regulates RSA are complex.
Phytohormones are known to be important for root development (Zhao et al., 2015; Li et al., 2019b; Liu et al., 2019b); therefore, the modulation of hormone signaling may be one mechanism by which HY5 mediates RSA development. The stimulus categories containing the responses to plant hormones, including auxin, abscisic acid, gibberellins, jasmonic acid, salicylic acid, and ethylene, were all overrepresented in the HY5-dependent DEGs. All of these hormones, particularly auxin, have been well known in regulating root developments (Jiao et al., 2013; Pacifici et al., 2015; Li et al., 2017b; Lv et al., 2018). We also found that the loss of HY5 function increased the local auxin accumulation in the lateral roots (Figure S3).
HY5-mediated stress responses may also contribute to the regulation of RSA development. Genes involved in responses to stresses such as salt, osmosis, and oxidation were downregulated in roots grown in darkness compared to those exposed to light (Figure 6), suggesting that the defense response may be more active in the light-grown roots (Cui et al., 2019). A higher defense capacity is predicted to consume more energy, thereby reducing the energy available for growth (Bhosale et al., 2018; Li et al., 2019b). Our data suggest that HY5 might control the activation of the defense systems, which is consistent with the previous report that HY5 negatively regulates the plant responses to ER stress (Nawkar et al., 2017). Increasing light intensities evoke ER stress by affecting the protein-folding capacity of this organelle. Consequently, the unfolded protein response (UPR) in the ER is negatively controlled by HY5, which transcriptionally represses the UPR marker gene BINDING PROTEIN3 (BIP3) by directly binding to its promoter (Nawkar et al., 2017). It was reasoned that the increased tolerance of the hy5 mutant to the ER stress conditions was caused by the induction of the UPR marker genes (Nawkar et al., 2017). Although the upregulated genes were exclusively enriched in the response to ER stress category in our dataset, BIP3 was only found to be upregulated in the illuminated hy5 roots, with no difference in expression between the light-exposed WT roots compared to the dark-grown WT or hy5 roots. We cannot rule out the involvement of the ER stress response in the RSA response to root illumination, but it may not be a primary contributor to the HY5-mediated regulation of root growth in response to light exposure.
As a central regulator in the response to light, HY5-regulated biosynthetic processes might also affect RSA development. The downregulated DEGs detected in the various comparisons with the illuminated WT roots are enriched in the categories related to the chloroplasts and photosynthesis. Root greening did not occur under our 15-day illumination; however, a previous study revealed that a long-term exposure to light (30 days) dramatically promotes chloroplast development and results in root greening in the WT, but not in the hy5 mutant (Oyama et al., 1997). This demonstrates that cellular and subcellular changes occur as a result of the altered biosynthetic processes in light-exposed roots.
Further study is still needed to clarify which biological process is disturbed by light to cause the inhibition of RSA. The direct illumination of the root increases the expression of the flavonoid biosynthesis genes in a HY5-dependent manner (Figure 6D and Table S1 and 5), suggesting that the carotenoid and flavonoid biosynthesis pathways might contribute to the inhibition of RSA development. An uncharacterized carotenoid-derived molecule, apocarotenoid, was reported to function in a non-cell-autonomous manner in the establishment of the lateral root pattern and the regulation of lateral root formation (Van Norman et al., 2014). Flavonoids are ubiquitous plant secondary metabolites, which are involved in the regulation of basipetal auxin transport, thereby regulating root development (Winkel-Shirley, 2002; Buer et al., 2010; Li et al., 2019a). Defects in flavonoid biosynthesis result in a conditional increase in lateral root density (Buer et al., 2010). Until recently, flavonols were proposed to function as local signals, integrating the hormonal and ROS pathways to regulate root growth in response to light (Silva-Navas et al., 2016). In this study, we found that both darkness and the loss of HY5 function decreased the flavonol content in the roots (Figure S7), while an exogenous supply of the major active flavonol intermediate quercetin restored the RSA phenotype of the hy5 mutant regardless of the exposure of its root to illumination or darkness (Figure S8). This suggests that HY5 might regulate RSA in response to light by mediating flavonol production in the roots.
Lastly, ROS signaling might also be involved in the HY5-mediated regulation of RSA in response to light. Our transcriptome data showed that the genes involved in NADPH regeneration were enriched in the downregulated genes (Figure 6B; Table S5). NADPH is particularly important for ROS generation and metabolism (Noctor, 2006). Previous reports have also revealed that ROS plays a key role in regulating root growth and development (Jiao et al., 2013; Bai et al., 2014; Li et al., 2015). Additionally, a higher accumulation of ROS was previously observed in roots grown in darkness using a D-root system (Silva-Navas et al., 2015). We observed much higher ROS levels in the roots of the hy5 mutant than in the WT root, whether they were grown in the darkness or exposed to light (Figure S9). These findings suggest that HY5 regulates RSA development in response to light by integrating flavonol production and ROS signaling.
Collectively, our results indicate that HY5 regulates RSA development through a complex network. Although we identified many putative HY5 targets that might influence RSA (Table S2 and 6), future studies should verify these targets and further dissect the mechanisms by which HY5 regulates RSA.
Data Availability Statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center (Wang et al., 2017; Members, 2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers CRA001994 that are publicly accessible at https://bigd.big.ac.cn/gsa.
Author Contributions
YHZ, CL, GW, and LZ: Conception and design, execution of experiment, analysis and interpretation of the data, and drafing of the article. HX, XS, JH, WZ, YZ, PH, K-XZ, XX, CW, XH, and CZ: Execution of experiment, analysis and interpretation of the data. ZH and WZ: Interpretation of the data and drafing of the article. XW, ZH, CW, HX, and WZ: Critical revision of the article for important intellectual content.
Funding
This work was funded by the National Natural Science Foundation of China (31801210 to L.Z., 31701294 to C.L., and 31601191 to K.Z.), the Start-up Foundation of Hubei University of Medicine (2017QDJZR26, 2016QDJZR11, and 2016QDJZR14), the Open Research Fund of Key Laboratory of Medicinal Resources and Natural Pharmaceutical Chemistry Ministry of Education (2019005), the Foundation of Health Commission of Hubei Province (ZY2019Q004), the Fund for Key Laboratory Construction of Hubei Province (Grant No. WLSP201905), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (16KJB180002 to K.Z., in part), and the Priority Academic Program Development of Jiangsu Higher Education Institutions of China (in part).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Jian Xu for initiating this project and generous helpful guidance. We are grateful to Dr. Ronald Pierik for helpful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01490/full#supplementary-material
Figure S1 | Illustration of a traditional culture system and the root-covered system. Representative photographs and corresponding schematic diagrams showing Arabidopsis seedlings grown on Petri dishes under TCS (A, B) and RCS (C). Scale bar represents 10 mm.
Figure S2 | Root hair phenotypes of WT and hy5 roots in illuminated and dark-grown conditions grown using in the RCS. Representative images (A) and the quantification of root hair length (B) of WT and hy5 seedlings grown in illuminated and covered conditions under the RCS. Scale bar represents 1 mm. Error bars represent the SD of biological triplicates (n ≥ 20). Different letters indicate significantly different values at P < 0.05 (pairwise Student’s t-test).
Figure S3 | DR5::GUS expression in the lateral roots of WT and hy5 seedlings. DR5::GUS expression in the early stage of lateral root growth (A and B) and in the mature lateral root (C and D). Scale bar represents 20 μm.
Figure S4 | HY5 and HYH expression levels in the roots of WT and hy5 plants grown in illuminated and covered conditions in the RCS. qRT-PCR quantification of HY5 and HYH expression. Error bars represent the standard deviation (SD) of biological triplicates. HY5 and HYH transcripts were normalized to the EF1α gene. **P < 0.01, pairwise Student’s t-test.
Figure S5 | Analysis of alternative splicing events in differentially expressed genes. Four types of splicing events were investigated from the transcripts of WT-illuminated, WT-covered, hy5-illuminated, and hy5- covered, respectively.
Figure S6 | Transcriptomic comparison of plants grown in the RCS with those grown in the D-root system. (A) Venn diagram illustrating the overall overlaps of the DEGs between the illuminated and the dark-grown WT roots grown in the RCS (this study) and the same comparison in the D-root system (Silva‐Navas et at., 2015). (B and C) Venn diagram illustrating the overlap of upregulated DEGs (B) and downregulated DEGs (C) in the dark-grown versus illuminated WT roots. (D) Venn diagram illustrating the overlap of light-repressed genes (upregulated genes compared to the illuminated WT roots) in different clusters. (E) Venn diagram illustrating the overlap of light-induced genes (downregulated genes compared with illuminated WT roots) in different clusters.
Figure S7 | The determination of flavonols in the roots of the WT and hy5 roots grown in illuminated and dark conditions. (A-B) An HPLC chromatogram of the flavonols in WT covered (A) and hy5 illuminated (B) compared to WT illuminated. (C-D) An HPLC chromatogram of the flavonols in hy5 covered compared to WT covered (C) and hy5 illuminated (D), respectively.
Figure S8 | Flavonols regulate the root system architecture of plants grown under the RCS. (A) Representative pictures of the RSA phenotypes of 12-d-old WT and hy5 plants grown in 1/2 MS medium with a mock control or 50 μM quercetin. Scale bar represents 10 mm. (B-E) Quantification of the RSA phenotypes of 12-d-old WT and hy5 plants grown in 1/2 MS medium with a mock control or 50 μM quercetin under the indicated light conditions, including the primary root lengths (B), root extension area (C), lateral root number (D), and lateral root angle (E). Error bars represent the SD of biological triplicates (n ≥ 6). Different letters indicate significantly different values at P < 0.05 (pairwise Student’s t-test).
Figure S9 | The ROS levels in the roots of WT and hy5 plants grown in illuminated or covered conditions. Fluorescein diacetate staining was used to visualize ROS. Scale bar represents 50 μm.
Table S1 | Upregulated and downregulated genes in dark-grown hy5 and WT roots and illuminated hy5 roots compared to the illuminated WT roots.
Table S2 | Putative HY5 target genes identified in the clusters of upregulated and downregulated differentially expressed genes (DEGs), respectively.
Table S3 | Genes found to overlap in a comparison with a previous summary of light signaling genes reported by Shikata et al. (2014).
Table S4 | Alternative splicing (AS) events analysis of DEGs.
Table S5 | Gene Ontology (GO) enrichments of DEGs.
Table S6 | Overlap of genes in the DEGs of dark-grown WT roots grown under the RCS and those grown in the D-root system.
Table S7 | GO enrichment analysis in common regulated genes related to HY5.
References
Ang, L. H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., et al. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. doi: 10.1016/S1097-2765(00)80022-2
Arsovski, A. A., Galstyan, A., Guseman, J. M., Nemhauser, J. L. (2012). Photomorphogenesis. Arabidopsis Book, e0147. doi: 10.1199/tab.0147
Audenaert, D., De Rybel, B., Nguyen, L., Beeckman, T. (2013). Small-molecule screens to study lateral root development in Plant Organogenesis (Springer). Methods Mol. Biol. 189–195. doi: 10.1007/978-1-62703-221-6_12
Bai, L., Ma, X. N., Zhang, G. Z., Song, S. F., Zhou, Y., Gao, L. J., et al. (2014). A receptor-like kinase mediates ammonium homeostasis and is important for the polar growth of root hairs in Arabidopsis. Plant Cell 26, 1497–1511. doi: 10.1105/tpc.114.124586
Bardou, P., Mariette, J., Escudié, F., Djemiel, C., Klopp, C. (2014). jvenn: an interactive Venn diagram viewer. BMC Bioinf. 15, 293. doi: 10.1186/1471-2105-15-293
Bellini, C., Pacurar, D. I., Perrone, I. (2014). Adventitious roots and lateral roots: similarities and differences. Annu. Rev. Plant Biol. 65, 639–666. doi: 10.1146/annurev-arplant-050213-035645
Bhalerao, R. P., Eklof, J., Ljung, K., Marchant, A., Bennett, M., Sandberg, G. (2002). Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 29, 325–332. doi: 10.1046/j.0960-7412.2001.01217.x
Bhosale, R., Boudolf, V., Cuevas, F., Lu, R., Eekhout, T., Hu, Z. B., et al. (2018). A spatiotemporal dna endoploidy map of the Arabidopsis root reveals roles for the endocycle in root development and stress adaptation. Plant Cell 30, 2330–2351. doi: 10.1105/tpc.17.00983
Briggs, W. R., Lin, C. T. (2012). Photomorphogenesis-from one photoreceptor to 14: 40 years of progress. Mol. Plant 5, 531–532. doi: 10.1093/mp/sss059
Buer, C. S., Imin, N., Djordjevic, M. A. (2010). Flavonoids: new roles for old molecules. J. Integr. Plant Biol. 52, 98–111. doi: 10.1111/j.1744-7909.2010.00905.x
Chen, X., Yao, Q., Gao, X., Jiang, C., Harberd, N. P., Fu, X. (2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26, 640–646. doi: 10.1016/j.cub.2015.12.066
Correll, M. J., Kiss, J. Z. (2005). The roles of phytochromes in elongation and gravitropism of roots. Plant Cell Physiol. 46, 317–323. doi: 10.1093/pcp/pci038
Cui, F., Wu, W., Wang, K., Zhang, Y., Hu, Z., Brosché, M., et al. (2019). Cell death regulation but not abscisic acid signaling is required for the enhanced Botrytis immunity in cuticle permeable mutants. J. Exp. Bot. 70, 5971–5984.doi: 10.1093/jxb/erz345
Galen, C., Rabenold, J. J., Liscum, E. (2007). Light-sensing in roots. Plant Signal Behav. 2, 106–108. doi: 10.4161/psb.2.2.3638
Gangappa, S. N., Botto, J. F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9, 1353–1365. doi: 10.1016/j.molp.2016.07.002
Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M., Dudoit, S., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. doi: 10.1186/gb-2004-5-10-r80
Gommers, C. M., Monte, E. (2018). Seedling establishment: a dimmer switch-regulated process between dark and light signaling. Plant Physiol. 176, 1061–1074. doi: 10.1104/pp.17.01460
Guo, S. Y., Dai, S. J., Singh, P. K., Wang, H. Y., Wang, Y. N., Tan, J. L. H., et al. (2018). A membrane-bound nac-like transcription factor OsNTL5 Represses the flowering in Oryza sativa. Front. Plant Sci. 9, 9. doi: 10.3389/fpls.2018.00555
Han, S. A., Fang, L., Ren, X. J., Wang, W. L., Jiang, J. (2015). MPK6 controls H2O2-induced root elongation by mediating Ca2+ influx across the plasma membrane of root cells in Arabidopsis seedlings. New Phytol. 205, 695–706. doi: 10.1111/nph.12990
Hardtke, C. S., Gohda, K., Osterlund, M. T., Oyama, T., Okada, K., Deng, X. W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19, 4997–5006. doi: 10.1093/emboj/19.18.4997
Holm, M., Ma, L. G., Qu, L. J., Deng, X. W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259. doi: 10.1101/gad.969702
Jansen, L., Hollunder, J., Roberts, I., Forestan, C., Fonteyne, P., Quickenborne, C., et al. (2013). Comparative transcriptomics as a tool for the identification of root branching genes in maize. Plant Biotechnol. J. 11, 1092–1102. doi: 10.1111/pbi.12104
Jiao, Y. H., Sun, L. R., Song, Y. L., Wang, L. M., Liu, L. P., Zhang, L. Y., et al. (2013). AtrbohD and AtrbohF positively regulate abscisic acid-inhibited primary root growth by affecting Ca-2 signalling and auxin response of roots in Arabidopsis. J. Exp. Bot. 64, 4183–4192. doi: 10.1093/jxb/ert228
Jiao, K., Li, X., Guo, Y., Guan, Y., Guo, W., Luo, D., et al. (2019). Regulation of compound leaf development in mungbean (Vigna radiata L.) by CUP-SHAPED COTYLEDON/NO APICAL MERISTEM (CUC/NAM) gene. Planta 249, 765–774. doi: 10.1007/s00425-018-3038-z
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357. doi: 10.1038/nmeth.3317
Laxmi, A., Pan, J., Morsy, M., Chen, R. (2008). Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PloS One 3, e1510. doi: 10.1371/journal.pone.0001510
Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., et al. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749. doi: 10.1105/tpc.106.047688
Lee, H. J., Ha, J. H., Kim, S. G., Choi, H. K., Kim, Z. H., Han, Y. J., et al. (2016). Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots. Sci. Signal 9, ra106. doi: 10.1126/scisignal.aaf6530
Li, J. G., Li, G., Gao, S. M., Martinez, C., He, G. M., Zhou, Z. Z., et al. (2010). Arabidopsis transcription factor elongated HYPOCOTYL5 plays a role in the feedback regulation of phytochrome a signaling. Plant Cell 22, 3634–3649. doi: 10.1105/tpc.110.075788
Li, J., Terzaghi, W., Deng, X. W. (2012). Genomic basis for light control of plant development. Protein Cell 3, 106–116. doi: 10.1007/s13238-012-2016-7
Li, N., Sun, L. R., Zhang, L. Y., Song, Y. L., Hu, P. P., Li, C., et al. (2015). AtrbohD and AtrbohF negatively regulate lateral root development by changing the localized accumulation of superoxide in primary roots of Arabidopsis. Planta 241, 591–602. doi: 10.1007/s00425-014-2204-1
Li, C., Zheng, L., Zhang, J., Lv, Y., Liu, J., Wang, X., et al. (2017a). Characterization and functional analysis of four HYH splicing variants in Arabidopsis hypocotyl elongation. Gene 619, 44–49. doi: 10.1016/j.gene.2017.04.001
Li, K., Yang, F. B., Zhang, G. Z., Song, S. F., Li, Y., Ren, D. T., et al. (2017b). AIK1, A mitogen-activated protein kinase, modulates abscisic acid responses through the MKK5-MPK6 Kinase Cascade. Plant Physiol. 173, 1391–1408. doi: 10.1104/pp.16.01386
Li, L., Hou, M. J., Cao, L., Xia, Y., Shen, Z. G., Hu, Z. B. (2018a). Glutathione S-transferases modulate Cu tolerance in Oryza sativa. Environ. Exp. Bot. 155, 313–320. doi: 10.1016/j.envexpbot.2018.07.007
Li, W. R., De Ollas, C., Dodd, I. C. (2018b). Long-distance ABA transport can mediate distal tissue responses by affecting local ABA concentrations. J. Integr. Plant Biol. 60, 16–33. doi: 10.1111/jipb.12605
Li, B., Fan, R., Guo, S., Wang, P., Zhu, X., Fan, Y., et al. (2019a). The Arabidopsis MYB transcription factor, MYB111 modulates salt responses by regulating flavonoid biosynthesis. Environ. Exp. Bot. 166, 103807. doi: 10.1016/j.envexpbot.2019.103807
Li, C., Zheng, L., Wang, X., Hu, Z., Zheng, Y., Chen, Q., et al. (2019b). Comprehensive expression analysis of Arabidopsis GA2-oxidase genes and their functional insights. Plant Sci. 285, 1–13. doi: 10.1016/j.plantsci.2019.04.023
Li, X., Liu, W., Zhuang, L., Zhu, Y., Wang, F., Chen, T., et al. (2019c). Bigger organs and elephant ear-likE LEAF1 control organ size and floral organ internal asymmetry in pea. J. Exp. Bot. 70, 179–191. doi: 10.1093/jxb/ery352
Liu, C., Cui, D., Zhao, J., Liu, N., Wang, B., Liu, J., et al. (2019a). Two Arabidopsis receptor-like cytoplasmic kinases SZE1 and SZE2 associate with the ZAR1-ZED1 complex and are required for effector-triggered immunity. Mol. Plant 12, 967–983. doi: 10.1016/j.molp.2019.03.012
Liu, H., Guo, S., Lu, M., Zhang, Y., Li, J., Wang, W., et al. (2019b). Biosynthesis of DHGA 12 and its roles in Arabidopsis seedling establishment. Nat. Commun. 10, 1768. doi: 10.1038/s41467-019-09467-5
Lobet, G., Pagès, L., Draye, X. (2011). A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol. 157, 29–39. doi: 10.1104/pp.111.179895
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Lucas, M., Swarup, R., Paponov, I. A., Swarup, K., Casimiro, I., Lake, D., et al. (2011). Short-root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol. 155, 384–398. doi: 10.1104/pp.110.165126
Lv, S. F., Yu, D. Y., Sun, Q. Q., Jiang, J. (2018). Activation of gibberellin 20-oxidase 2 undermines auxin-dependent root and root hair growth in NaCl-stressed Arabidopsis seedlings. Plant Growth Regul. 84, 225–236. doi: 10.1007/s10725-017-0333-9
Members, B.I.G.D.C. (2019). Database resources of the big data center in 2019. Nucleic Acids Res. 47, D8–D14. doi: 10.1093/nar/gky993
Mo, M., Yokawa, K., Wan, Y., Baluška, F. (2015). How and why do root apices sense light under the soil surface?. Front. Plant Sci. 6, 775. doi: 10.3389/fpls.2015.00930
Mortazavi, A., Williams, B. A., Mccue, K., Schaeffer, L., Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621. doi: 10.1038/nmeth.1226
Nawkar, G. M., Kang, C. H., Maibam, P., Park, J. H., Jung, Y. J., Chae, H. B., et al. (2017). HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, 2084–2089. doi: 10.1073/pnas.1609844114
Noctor, G. (2006). Metabolic signalling in defence and stress: the central roles of soluble redox couples. Plant Cell Environ. 29, 409–425. doi: 10.1111/j.1365-3040.2005.01476.x
Osmont, K. S., Sibout, R., Hardtke, C. S. (2007). Hidden branches: developments in root system architecture. Annu. Rev. Plant Biol. 58, 93–113. doi: 10.1146/annurev.arplant.58.032806.104006
Osterlund, M. T., Hardtke, C. S., Wei, N., Deng, X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. doi: 10.1038/35013076
Oyama, T., Shimura, Y., Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. doi: 10.1101/gad.11.22.2983
Pacifici, E., Polverari, L., Sabatini, S. (2015). Plant hormone cross-talk: the pivot of root growth. J. Exp. Bot. 66, 1113–1121. doi: 10.1093/jxb/eru534
Qu, X., Cao, B., Kang, J., Wang, X., Han, X., Jiang, W., et al. (2019). Fine-tuning stomatal movement through small signaling peptides. Front. Plant Sci. 10, 69. doi: 10.3389/fpls.2019.00069
Rellán- lvarez, R., Lobet, G., Lindner, H., Pradier, P.-L., Sebastian, J., Yee, M.-C., et al. (2015). GLO-Roots: an imaging platform enabling multidimensional characterization of soil-grown root systems. Elife 4, e07597. doi: 10.7554/eLife.07597
Ristova, D., Rosas, U., Krouk, G., Ruffel, S., Birnbaum, K. D., Coruzzi, G. M. (2013). RootScape: a landmark-based system for rapid screening of root architecture in Arabidopsis. Plant Physiol. 161, 1086–1096. doi: 10.1104/pp.112.210872
Rogers, E. D., Benfey, P. N. (2015). Regulation of plant root system architecture: implications for crop advancement. Curr. Opin. Biotechnol. 32, 93–98. doi: 10.1016/j.copbio.2014.11.015
Sakaguchi, J., Watanabe, Y. (2017). Light perception in aerial tissues enhances DWF4 accumulation in root tips and induces root growth. Sci. Rep. 7, 1808. doi: 10.1038/s41598-017-01872-4
Salisbury, F. J., Hall, A., Grierson, C. S., Halliday, K. J. (2007). Phytochrome coordinates Arabidopsis shoot and root development. Plant J. 50, 429–438. doi: 10.1111/j.1365-313X.2007.03059.x
Sassi, M., Lu, Y., Zhang, Y., Wang, J., Dhonukshe, P., Blilou, I., et al. (2012). COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development 139, 3402–3412. doi: 10.1242/dev.078212
Shikata, H., Hanada, K., Ushijima, T., Nakashima, M., Suzuki, Y., Matsushita, T. (2014). Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 18781–18786. doi: 10.1073/pnas.1407147112
Sibout, R., Sukumar, P., Hettiarachchi, C., Holm, M., Muday, G. K., Hardtke, C. S. (2006). Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PloS Genet. 2, e202. doi: 10.1371/journal.pgen.0020202
Silva-Navas, J., Moreno-Risueno, M. A., Manzano, C., Pallero-Baena, M., Navarro-Neila, S., Téllez-Robledo, B., et al. (2015). D-Root: a system for cultivating plants with the roots in darkness or under different light conditions. Plant J. 84, 244–255. doi: 10.1111/tpj.12998
Silva-Navas, J., Moreno-Risueno, M. A., Manzano, C., Tellez-Robledo, B., Navarro-Neila, S., Carrasco, V., et al. (2016). Flavonols mediate root phototropism and growth through regulation of proliferation-to-differentiation transition. Plant Cell 28, 1372–1387. doi: 10.1105/tpc.15.00857
Singh, A., Ram, H., Abbas, N., Chattopadhyay, S. (2012). Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabidopsis thaliana. J. Biol. Chem. 287, 25995–26009. doi: 10.1074/jbc.M111.333906
Smith, S., De Smet, I. (2012). Root system architecture: insights from Arabidopsis and cereal crops. Philos. Trans. R Soc. Lond B Biol. Sci. 367, 1441–1452. doi: 10.1098/rstb.2011.0234
Ulmasov, T., Murfett, J., Hagen, G., Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. doi: 10.1105/tpc.9.11.1963
Van Gelderen, K., Kang, C., Paalman, R., Keuskamp, D. H., Hayes, S., Pierik, R. (2018a). Far-red light detection in the shoot regulates lateral root development through the HY5 transcription factor. Plant Cell 30, 101–116. doi: 10.1105/tpc.17.00771
Van Gelderen, K., Kang, C., Pierik, R. (2018b). Light signaling, root development, and plasticity. Plant Physiol. 176, 1049–1060. doi: 10.1104/pp.17.01079
Van Norman, J. M., Zhang, J., Cazzonelli, C. I., Pogson, B. J., Harrison, P. J., Bugg, T. D., et al. (2014). Periodic root branching in Arabidopsis requires synthesis of an uncharacterized carotenoid derivative. Proc. Natl. Acad. Sci. U. S. A. 111, E1300–E1309. doi: 10.1073/pnas.1403016111
Wang, Y., Song, F., Zhu, J., Zhang, S., Yang, Y., Chen, T., et al. (2017). GSA: genome sequence archive. Genomics Proteomics Bioinf. 15, 14–18. doi: 10.1016/j.gpb.2017.01.001
Wang, W., Chen, Q., Botella, J. R., Guo, S. (2019). Beyond light: insights into the role of constitutively photomorphogenic1 in plant hormonal signaling. Front. Plant Sci. 10, 557. doi: 10.3389/fpls.2019.00557
Weller, J. L., Hecht, V., Schoor, J. K. V., Davidson, S. E., Ross, J. J. (2009). Light regulation of gibberellin biosynthesis in pea is mediated through the COP1/HY5 pathway. Plant Cell 21, 800–813. doi: 10.1105/tpc.108.063628
Winkel-Shirley, B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5, 218–223. doi: 10.1016/S1369-5266(02)00256-X
Xiao, T. W., Mi, M. M., Wang, C. Y., Qian, M., Chen, Y. H., Zheng, L. Q., et al. (2018). A methionine-R-sulfoxide reductase, OsMSRB5, is required for rice defense against copper toxicity. Environ. Exp. Bot. 153, 45–53. doi: 10.1016/j.envexpbot.2018.04.006
Xu, W., Ding, G., Yokawa, K., Baluska, F., Li, Q. F., Liu, Y., et al. (2013). An improved agar-plate method for studying root growth and response of Arabidopsis thaliana. Sci. Rep. 3, 1273. doi: 10.1038/srep01273
Yokawa, K., Kagenishi, T., Kawano, T., Mancuso, S., Baluška, F. (2011). Illumination of Arabidopsis roots induces immediate burst of ROS production. Plant Signal Behav. 6, 1460–1464. doi: 10.4161/psb.6.10.18165
Yokawa, K., Kagenishi, T., Baluška, F. (2013). Root photomorphogenesis in laboratory-maintained Arabidopsis seedlings. Trends Plant Sci. 18, 117–119. doi: 10.1016/j.tplants.2013.01.002
Yokawa, K., Fasano, R., Kagenishi, T., Baluška, F. (2014). Light as stress factor to plant roots - case of root halotropism. Front. Plant Sci. 5, 718. doi: 10.3389/fpls.2014.00718
Young, M. D., Wakefield, M. J., Smyth, G. K., Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Zhang, Y., Li, C., Zhang, J., Wang, J., Yang, J., Lv, Y., et al. (2017). Dissection of HY5/HYH expression in Arabidopsis reveals a root-autonomous HY5-mediated photomorphogenic pathway. PloS One 12, e0180449. doi: 10.1371/journal.pone.0180449
Zhang, L., Shi, X., Zhang, Y., Wang, J., Yang, J., Ishida, T., et al. (2019). CLE9 peptide-induced stomatal closure is mediated by abscisic acid, hydrogen peroxide, and nitric oxide in Arabidopsis thaliana. Plant Cell Environ. 42, 1033–1044. doi: 10.1111/pce.13475
Keywords: Arabidopsis, HY5, light, root photomorphogenesis, root system architecture, soil, transcriptome
Citation: Zhang Y, Wang C, Xu H, Shi X, Zhen W, Hu Z, Huang J, Zheng Y, Huang P, Zhang K-X, Xiao X, Hao X, Wang X, Zhou C, Wang G, Li C and Zheng L (2019) HY5 Contributes to Light-Regulated Root System Architecture Under a Root-Covered Culture System. Front. Plant Sci. 10:1490. doi: 10.3389/fpls.2019.01490
Received: 19 February 2019; Accepted: 28 October 2019;
Published: 28 November 2019.
Edited by:
Stephan Pollmann, National Institute of Agricultural and Food Research and Technology, SpainReviewed by:
Gabriela Toledo-Ortiz, Lancaster University, United KingdomJuan Carlos Del Pozo, National Institute of Agricultural and Food Research and Technology, Spain
Copyright © 2019 Zhang, Wang, Xu, Shi, Zhen, Hu, Huang, Zheng, Huang, Zhang, Xiao, Hao, Wang, Zhou, Wang, Li and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guodong Wang, guodong_wang@snnu.edu.cn; Chen Li, lee.childe@gmail.com; Lanlan Zheng, zhenglanlan0622@163.com
†These authors have contributed equally to this work
 Yonghong Zhang
Yonghong Zhang Chunfei Wang2†
Chunfei Wang2† Zhubing Hu
Zhubing Hu Xuanbin Wang
Xuanbin Wang Guodong Wang
Guodong Wang Chen Li
Chen Li Lanlan Zheng
Lanlan Zheng