
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 13 November 2019
Sec. Plant Breeding
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01452
Aphids are phloem sap-feeding insects common as pests in various crops. Here we review 62 omics studies of aphid/plant interactions to search for indications of how aphids may manipulate the plants to make them more suitable as hosts, i.e. more susceptible. Our aim is to try to reveal host plant susceptibility (S) genes, knowledge which can be exploited for making a plant more resistant to its pest by using new plant breeding techniques to knock out or down such S genes. S genes may be of two types, those that are involved in reducing functional plant defense and those involved in further increasing plant factors that are positive to the aphid, such as facilitated access to food or improved nutritional quality. Approximately 40% of the omics studies we have reviewed indicate how aphids may modify their host to their advantage. To exploit knowledge obtained so far, we suggest knocking out/down candidate aphid S genes using CRISPR/Cas9 or RNAi techniques in crops to evaluate if this will be sufficient to keep the aphid pest at economically viable levels without severe pleiotropic effects. As a complement, we also propose functional studies of recessively inherited resistance previously discovered in some aphid–crop combinations, to potentially identify new types of S genes that later could be knocked out or down also in other crops to improve their resistance to aphids.
Aphids are phloem sap-feeding insects and most cultivated plant species are hosts of one or more aphid species. Aphids damage plants mainly by plant nutrient withdrawal, by virus transmission, and via indirect reduction of photosynthesis due to sooty mould growing in their excreta. Some species also change form and color of the plant tissues locally. Aphids can have complicated life cycles alternating between primary and secondary hosts, reproduce sexually or asexually, and be winged (alate) or without wings (apterous). Crops are invaded by alate aphids which give rise to successive apterous generations parthenogenetically (Blackman and Eastop, 1984). Aphids in these clonal colonies are sedentary if weather conditions are favorable, if host plant quality is high and if there is minimal disturbance by natural enemies. Insects with this life style have evolved so that they can modify their host plants locally to their own benefit (Walling, 2008; Giordanengo et al., 2010; Zust and Agrawal, 2016). Here we review such evidence of host manipulation and suggest how this knowledge can be used to breed plants which are less suitable as aphid hosts.
The number of aphid species in the family Aphididae is approximately 4 700 and circa 10% of those occur on crops (Blackman and Eastop, 2007). Most aphid species are specialized in their host range. It is common for aphids to only use plant species within a plant family as hosts, or species from two distinct families if aphids are host alternating (Blackman and Eastop 1984). This high degree of specialization requires that aphids have the ability to distinguish hosts from non-hosts using their sensory organs (Pettersson et al., 2007). Despite this host specialisation aphids have certain feeding characteristics in common. After alighting on a plant, aphids start probing with their stylet-shaped mouthparts, on the plant surface and in the plant tissue. The stylet is predominantly pushed in between plant cells, but during its way to the phloem most plant cells along the route are briefly punctured (Pettersson et al., 2007). This is believed partly to take place for the aphids to localize the stylet in relation to phloem tissue and to assess host status, but also to inject watery saliva to improve host quality. Watery saliva is also injected into the phloem, before the phloem sap is taken up. During ingestion, watery saliva is produced as well, transported with the phloem sap into the food canal (Pettersson et al., 2007). Over the whole plant probing period, gelling saliva is also produced. This type of saliva forms a protective sheet around the stylet and seals punctured plant cells. The chemical content of gelling saliva is more conserved between aphid species than is the content of the watery saliva. The main compound in gelling saliva is a sheath-forming protein, and probably there are also cell wall-degrading proteins and oxidases (van Bel and Will, 2016). The watery saliva contains various compounds like carbohydrates, phospholipids and proteins, of which many are in the categories of Ca2+-binding, proteolytic and oxidising proteins and effectors (Thompson and Goggin, 2006; Giordanengo et al., 2010). Effectors are compounds that interact with the host plant and thereby change plant function and structures in various ways (Rodriguez and Bos, 2013). Several proteins from aphid watery saliva have been shown to promote or reduce aphid performance in a species-specific way. In compatible plant interactions, aphid performance may be promoted by effector-driven downregulation of functional plant defense or by regulation of plant factors that are positive to the aphid (Giordanengo et al., 2010; van Bel and Will, 2016; Mondal, 2017).
The knowledge about aphid-induced plant responses has increased dramatically during recent years. There are now many pairs of aphid species/plant species combinations where “global” plant gene regulations have been studied, mainly at the transcriptional level, but to some extent also at the protein and metabolite levels. Aphid-induced plant genes may change the way the plant functions as an aphid host, to the better or to the worse. Approximately half of the 62 studies presented in Table 1 have included both susceptible and resistant plant genotypes, with the aim to distinguish differences between these two categories of plants in order to better understand mechanisms of plant resistance. This might in turn be used for more precise selection of resistant plants in plant breeding programs. In the present review we instead aim at emphasizing similarities in gene regulation between susceptible plants subjected to aphid feeding, keeping in mind that there might also be aphid/plant-specific interactions. This knowledge may be applied in breeding programs with the aim to reduce susceptibility rather than to increase resistance to aphids.
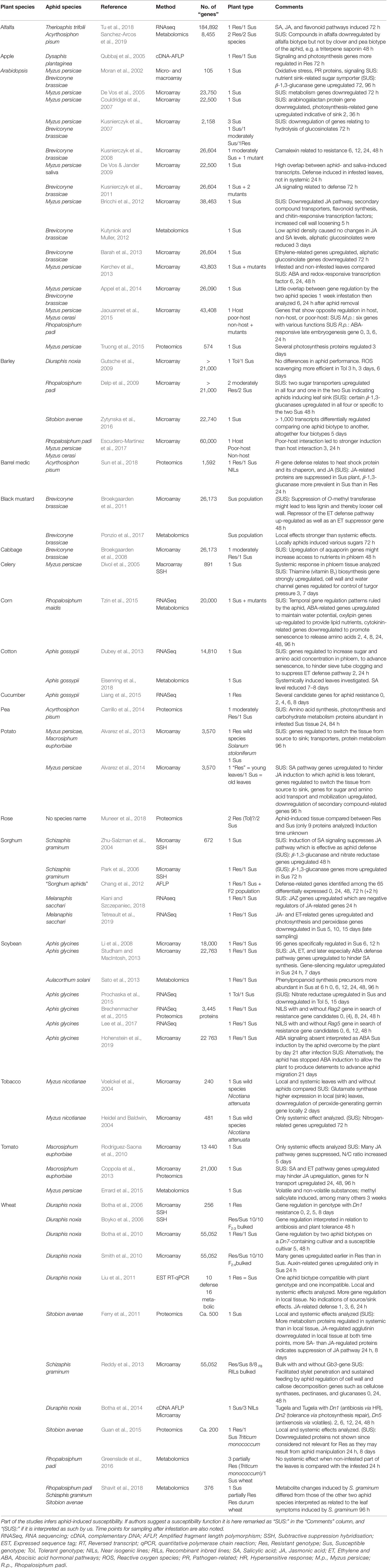
Table 1 Global studies of plant genes as DNA contigs, RNA sequences, probes, proteins, or metabolites regulated by aphid feeding.
In recent years two new plant breeding approaches offer possibilities to exploit such knowledge. RNA interference (RNAi) may knock down expression of susceptibility (S) genes and site-directed mutagenesis techniques such as CRISPR/Cas9 may knock out such genes. Since these new breeding techniques are best suited to reduce gene expression levels, we have focussed on candidate S genes that are upregulated by aphids, even though also plant genes that are downregulated by aphids may facilitate aphid host acceptance and performance. Furthermore, there must be numerous plant genes that through their constitutive expression favor the aphid. However, it is very difficult to distinguish which of these are the most important S genes and most of them are likely to be essential also for plant fitness. Apart from using omics information to try to discern S genes, there is also the possibility to investigate the molecular mechanisms behind previously detected resistance to aphids that is inherited recessively. Potentially such resistance may be due to S genes that have been spontaneously mutated in the host.
Candidate genes for induced susceptibility are little addressed in the studies of global gene expressions in the aphid host plants (Table 1). On the contrary, genes are often referred to as defense genes even in studies of aphids on susceptible hosts. One of the difficulties to infer a gene function in induced susceptibility is that susceptibility is often a quantitative trait. An induced reduction in functional plant defense makes the plant somewhat more susceptible and vice versa. Another difficulty is that both up and down gene regulation may lead to increased host susceptibility depending on the function of the gene product. Yet another difficulty is that RNA transcript abundances are rarely linearly related to the quantities of the metabolic products with potential effects on aphid performance. Partly, this discrepancy is due to timing since gene regulation and expression precede metabolism. This dynamic nature of the induction requires sampling and analyzing at many time points, which is costly. Other reasons for discrepancies between transcripts and metabolites are the complicated interactions between gene products and limitations in access to substrates in the metabolic pathways. Despite difficulties like this, in ca. 40% of the global expression studies listed in Table 1, the authors infer certain gene classes, proteins or metabolites to have a susceptibility function. Here we summarize the results from these studies. Moreover, we include also those global expression studies where susceptibility is not discussed, for providing information regarding other potential target genes for future studies. In the following, the susceptibility concept is discussed in terms of how it relates to aphid-induced changes in plant defense pathways (see Plant Hormonal Induction in Relation to Functional Plant Defense), local and systemic induction patterns (Local and Systemic Induction Patterns Influencing Aphid Performance), and improvements of food accessibility (Aphid Modification of Food Accessibility) and quality (Aphid Modification of Food Quality), mainly with examples from the studies listed in Table 1.
Plant receptors can detect invaders such as herbivorous insects and induce defense signaling pathways, mainly involving the salicylate (SA) and the jasmonate (JA) pathways, modified primarily by the ethylene (ET), abscisic acid (ABA), and gibberellic acid (GA) related ones. Whereas chewing insects predominantly induce the JA pathway genes, aphids commonly induce genes related to the SA pathway. Since aphids, in general, have been shown to be more sensitive to plant defense involving the JA signaling pathway, SA induction is considered to be a way for aphids to deceive the plant because cross-talk between hormonal pathways hinders the SA-induced plant to fully induce the JA and ET pathways (Thompson and Goggin 2006; De Vos et al., 2007; Goggin, 2007; Walling, 2008). Indications of this SA–JA antagonism have been found in the global expression studies of Acyrthosiphon pisum Harris on susceptible barrel medic (Sun et al., 2018), Myzus persicae Sulzer on potato (Alvarez et al., 2014), Schizaphis graminum Rondani on susceptible sorghum (Zhu-Salzman et al., 2004), Melanaphis sacchari Zehntner on sorghum (Kiani and Szczepaniec, 2018), Macrosiphum euphorbiae Thomas on susceptible tomato (Coppola et al., 2013), and Sitobion avenae Fabricius on susceptible wheat (Ferry et al., 2011). However, in the study by Bricchi et al. (2012), there was no indication of SA-dependant crosstalk and yet many genes in the JA pathway were downregulated by M. persicae on susceptible Arabidopsis at the early sampling of 5 h after aphid infestation. A similar result was found with M. euphorbiae in susceptible tomato by Rodriguez-Saona et al. (2010). Perhaps contrary to expectations, JA-defense type oxylipins induced by M. persicae in Arabidopsis produced in roots facilitated leaf infestation by the aphid (Nalam et al., 2012).
There are thus reasons to believe that the explanation for aphid-induced plant susceptibility via SA/JA hormonal competition is somewhat simplified. Also ABA-signaling is inferred to be manipulated by aphids to negatively regulate plant defense; in the non-host relationship of Rhopalosiphum padi L. on Arabidopsis (Jaouannet et al., 2015) as well as in the host relationship of M. persicae on Arabidopsis (Kerchev et al., 2013; Hillwig et al., 2016) and by Aphis glycines Matsumura on susceptible soybean (Studham and MacIntosh, 2013). In addition, the authors of the soybean study speculated that also ET-related gene induction might be a way of blocking the JA induction, since A. glycines also upregulated ET-related genes but the aphid’s performance was not affected by artificial ET induction. The finding by Mantelin et al. (2009) that impaired ET signaling or biosynthesis decreases tomato susceptibility to M. euphorbiae might support this. However, there are also studies that do not lend support to this aphid-induced plant hormone regulatory role of ET. Brevicoryne brassicae L. upregulates a negative regulator gene for ET signaling and an ET suppressor gene whereas SA- and JA-related genes are unaffected in its host black mustard (Broekgaarden et al., 2011). ET-related genes are suppressed also by Aphis gossypii Glover in susceptible cotton (Dubey et al., 2013). Furthermore, Zhu et al. (2018) found that M. persicae upregulated the transcription factor MYB102 in Arabidopsis which in turn upregulated ET biosynthesis. This led to increased host susceptibility but with no evidence for other hormones to be involved. Thus, the relations between plant hormonal regulation by aphids and aphid performance are complicated and perhaps aphid/plant specific.
The SA defense pathway is typically induced by biotrophic pathogens and may lead to local cell death hindering the pathogen to infest and proliferate in the plant. However, such a hypersensitive response is not always seen when aphids upregulate the SA pathway in resistant plants (Thompson and Goggin, 2006). There are so far only three aphid resistance genes cloned and sequenced. Mi-1 in tomato confers resistance to certain biotypes of M. euphorbiae. Vat confers resistance to A. gossypii in melon (Dogimont et al., 2010) and SLI1 to M. persicae in Arabidopsis (Kloth et al., 2017). Mi-1 and Vat are of the type nucleotide-binding-site leucine-rich repeat (NBS-LLR) known to be involved in recognition of specific effectors secreted by plant-associated organisms and commonly denoted as R genes. Upon recognition they trigger plant hormonal defense signaling, which in turn activates plant defense. Interestingly, Mi-1 mostly relies upon the SA signaling (Dogimont et al., 2010), challenging the ideas that induction of the SA signaling pathway will hinder the plant to embark supposedly efficient JA-dependant plant defense against aphids and that SA defense is not functional against aphids. In line with the Mi-1-based resistance, SA application increases the effect of R-gene-based resistance in soybean to A. glycines whereas SA application to a susceptible soybean has no influence on aphid performance (Studham and MacIntosh, 2013). Vat gene function involves the JA, ET, and auxin signaling pathways but with a peroxidase burst soon after aphid attack. Upon aphid recognition by Vat in melon plants, peroxidase activity, and callose and lignin lining of the cell walls increase along the stylet path. When the aphids probe the plants they may be deterred by the oxidative burst in the plant cells or hindered to probe and feed efficiently by the cell wall thickening (Boissot et al., 2016). Certain short RNAs (microRNAs) with phytohormonal-related gene transcripts as targets have a role in this resistance reaction, for example by inactivating auxin receptors and signaling (Sattar and Thompson, 2016). The third aphid resistance gene sequenced so far, SLI1, codes for a sieve element-lining protein associated with organelles and proteins in the thin layer of cytoplasm along the phloem sieve tube plasma membrane, potentially making it more difficult for aphids to penetrate and reach the sieve element lumen, alternatively to cause aphid food canal occlusion (Kloth et al., 2017).
It is common for aphids to induce plant resistance to subsequent aphid colonization at the systemic level (i.e. in leaves or tissue away from the infested site) whereas direction of local effects of previous infestation is generally the opposite, i.e. induced susceptibility, according to Zust and Agrawal (2016). This is in line with our expectations that aphids modify their feeding site to their advantage. Some of the studies in Table 1 analyzed both types of tissues, in pairs of aphids and hosts representing compatible interactions. Not unexpectedly, gene regulation and metabolites in the infested tissue were more different from control than systemic tissue; as shown in Arabidopsis exposed to M. persicae (Kerchev et al., 2013), in black mustard infested by B. brassicae (Ponzio et al., 2017) as well as in wheat infested by Diuraphis noxia Kurdjumov (Liu et al., 2011), S. avenae (Guan et al., 2015), or R. padi (Greenslade et al., 2016). However, certain gene classes are sometimes found more activated in systemic than in infested tissue. In a wheat study with S. avenae, relatively more of proteins involved in metabolic processes were regulated in systemic than in infested tissue of a susceptible cultivar (Ferry et al., 2011). This may fit with the sink-source theory; that the aphids’ feeding site is a sink tissue that withdraws nutrients produced in non-infested plant tissue, as has been shown for nitrogen in alfalfa infested by A. pisum (Girousse et al., 2005).
Apart from nutritional aspects, defense-related aphid-induced plant moderations support the resistance/susceptibility pattern in systemic/infested leaves as suggested by Zust and Agrawal (2016). In wheat susceptible to S. avenae, a JA-regulated agglutinin was downregulated initially both in the infested and in the systemic tissue but after 8 days only in the infested tissue (Ferry et al., 2011). Several agglutinins, also called lectins, of plant origin have given negative effects on aphid performance when tested in aphid diets or transgenic plants (Vandenborre et al., 2011). Furthermore, Myzus nicotianae Blackman downregulated a germin gene in infested but not in systemic leaves of wild tobacco (Voelckel et al., 2004). Germin is an oxalate oxidase involved in release of H202- and Ca2+-mediated signaling, and in cross-linking of cell wall polymers (Lane et al., 1993). In systemic leaves of Arabidopsis, JA-induced anti-insect storage protein genes were upregulated as well as genes for cell-wall strengthening and synthesis of plant secondary compounds whereas in infested leaves, ABA and a redox-responsive transcription factor for ABA signaling were suggested to favor M. persicae reproduction (Kerchev et al., 2013) (Table 1).
However, there are studies of aphids on host plants where pre-infestation by conspecifics causes subsequent aphids to avoid or perform less well on the same leaves, possibly due to high infection pressure at pre-infestation (De Vos and Jander, 2009; Zust and Agrawal, 2016). The effects of induction by pre-infestation also vary depending on if plants are susceptible or resistant to the aphid. R-gene based resistance in barrel medic against the aphid Acyrthosiphon kondoi Shinji was induced by previous feeding by conspecifics, whereas no evidence of induced susceptibility was found in a susceptible host (Klingler et al., 2005). In peach susceptible to M. persicae, aphid performance was improved by pre-infestation whereas in various resistant peach genotypes, M. persicae pre-infestation either led to reduced, improved, or no change in aphid performance (Sauge et al., 2006). The pre-infestation effect is also dependant on aphid biotype; e.g. in barley with monogenic resistance to S. graminum, pre-infestation with a non-compatible aphid biotype led to reduced feeding by a compatible biotype, indicating defense induction by the former (Hays et al., 1999). Similarly, Zytynska et al. (2016) found biotype-dependant induction of numerous transcripts and reduction of aphid population growth of S. avenae in barley. In neither of these two barley studies were there aphid biotypes that induced susceptibility to another biotype though.
To successfully access and feed on phloem sap, aphids need to penetrate the plant tissue with their mouthparts, prevent sieve tube clogging as a result of the mechanical damage they cause, and maintain leaf water potential in the vascular bundles.
In order for aphids to reach the phloem, their stylets first penetrate the epidermis in the cell wall between two cells and then proceed deeper between other cells in the plant tissues. Aphids also penetrate the cell walls when they probe cells along the pathway to the vascular bundles, as well as when they feed from phloem or drink from xylem (Pettersson et al., 2007). Thus there is reason to believe that the structure of the cell walls may influence the ease with which the phloem and xylem can be reached, especially by the small, newborn nymphs in a colony. The main component of the cell walls is cellulose which is combined with other polysaccharides; various hemicelluloses, pectin (galacturons) and callose (β-1,3-glucans), and glycoproteins (arabinogalactan proteins and extensins). The primary cell wall is flexible whereas the secondary cell wall that is deposited after cessation of cell expansion is rigid, strengthened by the polyphenol lignin (Zhong et al., 2019). Some of the studies listed in Table 1 infer cell wall manipulations by aphids. Reddy et al. (2013) found some pectinase genes upregulated by D. noxia in susceptible compared to resistant wheat, indicating aphid modification of cell walls to its favor. Furthermore, an arabinogalactan protein gene was downregulated by M. persicae in Arabidopsis and the authors suggest that this may facilitate cell wall penetration by the aphid (Couldridge et al., 2007). Many expansin genes were found to be upregulated in another study of the same aphid/host combination (Bricchi et al., 2012). Expansins are proteins involved in cell wall loosening. In black mustard, the O-methyltransferase gene was downregulated by B. brassicae which might in turn lead to less lignin and loosened secondary cell walls (Broekgaarden et al., 2011) (Table 1).
Certain β-1,3-glucanases are among the pathogenesis-related (PR) proteins, induced via the SA defense pathways by various pathogens (van Loon and van Strien, 1999). β-1,3-Glucanase genes have been found upregulated by aphids as well. However, rather than contributing to plant defense, these enzymes may facilitate aphid feeding since they might reduce callose sealing of phloem tubes through breaking down callose. Pores in the phloem sieve plates are lined with callose collars that may regulate phloem sap flow from one cell to another. In undamaged phloem tissues there is a balance between build-up of callose by callose synthases and break-down by β-1,3-glucanases. When a sieve element is damaged, there is a rapid plugging of the sieve plate pores, within seconds to minutes. However, there are several examples of compatible aphid/host relationships where β-1,3-glucanases in the plants are upregulated by aphid infestation indicating that this is a way for these aphids to reduce or hinder phloem plugging (van Bel and Will, 2016), which otherwise might be permanent (Pirselova and Matusikova, 2013). Both M. persicae and B. brassicae upregulated β-1,3-glucanase gene expression in the susceptible host Arabidopsis (Moran et al., 2002). Zhu-Salzman et al. (2004) found the same in sorghum susceptible to S. graminum. Comparing two R. padi-susceptible and two partially resistant barley lines, Delp et al. (2009) found one glucanase gene upregulated in all four, one upregulated specifically in the two susceptible and one constitutively expressed only in the two susceptible lines with some upregulation by R. padi. Reddy et al. (2013) compared bulks of susceptible and resistant wheat lines and found that three β-1,3-glucanase genes were more upregulated by S. graminum in the susceptible than in the resistant bulk. Sun et al. (2018) found the same relationship between the A. pisum infestation and the glucanase regulation when comparing one partially resistant and one susceptible near-isogenic barrel medic line, so did Park et al. (2006) who compared sorghum genotypes resistant and susceptible to S. graminum (Table 1).
High leaf water potential facilitates xylem feeding which is a way for aphids to compensate for the high osmolarity of phloem sap (Sun et al., 2015). ABA signaling controls stomatal closure and leaf transpiration and aphid upregulation of this pathway might serve to maintain leaf water potential, as suggested in studies of corn infested by Rhopalosiphum maidis Fitch (Tzin et al., 2015) and of barrel medic by A. pisum (Sun et al., 2015). Aquaporins are proteins that facilitate water and other small molecules movement across cell membranes in either direction (Kapilan et al., 2018). In susceptible and moderately resistant cabbage, aquaporin genes were upregulated by B. brassicae (Broekgaarden et al., 2008) and in celery by M. persicae (Divol et al., 2005), something which in turn might increase water availability for the xylem and allocation of phloem sap.
Plant tissue infested by aphids functions as a sink to which photo-assimilates are directed (Hawkins et al., 1987). In many plant species the disaccharide sucrose is the predominant sugar transported in the phloem. Sucrose is produced in the green tissues during photosynthesis and its apoplastic cell-to-cell movement to the phloem is mediated by transporter and facilitator proteins. Transporters also help sugar uptake in sink tissue cells, in the form of sucrose or the monosaccharides glucose and fructose (Lemoine et al., 2013). A monosaccharide symporter gene was upregulated in Arabidopsis by M. persicae as well as by B. brassicae, suggesting that infested tissues function as a nutrient sink favorable to the aphids (Moran et al., 2002). The sink theory was also referred to by Delp et al. (2009) since two sugar transporter genes were upregulated locally by R. padi in both susceptible and partially resistant barley lines and another only in the susceptible lines. In cotton A. gossypii strongly induced a phosphate translocator gene that potentially increases sugar content in phloem sap as well as an asparaginase gene that increases the access to nitrogen (Dubey et al., 2013) (Table 1). Aphid control of plant genes related to sink effects deserves more attention in future studies.
Phloem sap is imbalanced as aphid food since it is rich in sugars and relatively low in amino acids (Dinant et al., 2010). Genes for glutamate and glutamine synthases and a nitrate transporter were found to be upregulated by M. nicotianae in a wild tobacco host, more so in infested than in systemic leaves (Heidel and Baldwin, 2004; Voelckel et al., 2004). In tomato, M. euphorbiae upregulated nitrogen transporters genes (Coppola et al., 2013) and increased the N/C ratio (Rodriguez-Saona et al., 2010). A gene for nitrate reductase was upregulated in soybean susceptible to A. glycines (Prochaska et al., 2015) and in sorghum susceptible to S. graminum (Zhu-Salzman et al., 2004), but was downregulated in an aphid-resistant soybean (Prochaska et al., 2015). Nitrate reductase is required for nitrogen assimilation into amino acids. Furthermore, in addition to being low in amino acids in general, the amino acid composition of phloem sap is imbalanced for certain essential amino acids. Nutrient-focused studies have shown that D. noxia and S. graminum increased phloem concentrations of essential amino acids in susceptible barley and wheat (Telang et al., 1999; Sandström et al., 2000), even though the aphids’ obligate endosymbionts ensure that aphids are not dependant on the plant for provision of such amino acids (Baumann, 2005). Both of these aphid species cause typical leaf symptoms on susceptible hosts whereas R. padi infesting the same hosts does not, neither does it change the host amino acid profile. Among the non-essential amino acids, glutamine was significantly increased by D. noxia and S. graminum. This amino acid has been associated with phloem unloading sites but also senescing tissues (Sandström et al., 2000). In line with this, Dubey et al. (2013) found a senescence-related gene upregulated by A. gossypii in susceptible cotton and Tzin et al. (2015) showed cytokinin-related genes downregulated by R. maidis in susceptible corn resulting in lowered cytokinin concentration and thereby earlier senescence. However, comparing one susceptible and one moderately resistant pea genotype infested by A. pisum, Carrillo et al. (2014) found a relative decrease in a putative senescence-related protein in the susceptible genotype along with increase in proteins involved in photosynthesis, carbohydrate metabolism, and amino acid synthesis (Table 1).
Apart from nutrients, there is a large array of secondary compounds in the phloem sap, which might affect feeding aphids negatively depending on aphid host adaptation (Dinant et al., 2010; Foyer et al., 2015). M. persicae downregulated many genes related to flavonoid biosynthesis as well as transporter genes related to secondary metabolites in Arabidopsis (Bricchi et al., 2012) and potato (Alvarez et al., 2014), while S. avenae downregulated a lectin-like protein in wheat, previously inferred to have a role in resistance to Hessian fly (Ferry et al., 2011). Sanchez-Arcos et al. (2019) compared inductions by A. pisum biotypes adapted to alfalfa, clover, and pea and found that certain compounds, e.g. a triterpene saponin, were downregulated in alfalfa by the adapted biotype but not by the other two (Table 1).
Thus aphids have the capacity to modify the composition of phloem sap at their feeding site to their advantage.
Several aphid effectors have now been shown to contribute to aphid virulence; C002 and Armet from A. pisum and M. persicae, Me10 and Me23 from M. euphorbiae, and Mp1, Mp2, and Mp55 from M. persicae; but their molecular interactions with the plants have just begun to be revealed (Rodriguez and Bos, 2013; Elzinga et al., 2014; Jaouannet et al., 2014; Rodriguez et al., 2017; Cui et al., 2019). Armet from A. pisum and M. persicae promotes SA accumulation (Cui et al., 2019). Mp1 from M. persicae hampers the Vacuolar Protein Sorting Associated Protein52 (VPS52) in the hosts Arabidopsis and potato. VPS52 vesicle trafficking is suggested to either contribute to plant defense or to reduce nutrient availability (Rodriguez et al., 2017). Mp55 when expressed in Arabidopsis, reduces callose deposition, hydrogen peroxide accumulation and glucosinolate conversion and increases host acceptance to M. persicae (Elzinga et al., 2014).
Other components of watery saliva may have a more direct influence on host plant suitability. Ca2+-binding proteins play a role in preventing sieve plate occlusion by protein and callose plugs and sieve element protein-lining. Proteases in aphid saliva may break down protein plugs in sieve plates and proteins in cytoplasm lining the sieve tube walls as well as degrade free proteins in the phloem sap, among them plant defense proteins. Proteases thereby also increase amino acid availability in phloem. Polyphenoloxidases, peroxidases, and oxidoreductases may reduce phenols and reactive oxygen species involved in plant defense (van Bel and Will, 2016; Kloth et al., 2017).
Plant genes that facilitate aphid host acceptance and performance can be called S genes, in line with the nomenclature for the corresponding genes in pathogen/plant interactions (van Schie and Takken, 2014). If an S gene is mutated so that it no longer functions favorably to the aphid the plant will be more resistant. Recessively inherited resistance genes may be such mutated susceptibility genes. Although most of the resistance genes characterized in aphid resistant germplasm are dominantly inherited there are a number of documented recessive genes; one in peanut associated with resistance to Aphis craccivora Koch, two in soybean against A. glycines, two in corn associated with resistance to R. maidis, and two introgressed in wheat; of which one causes resistance to D. noxia and the other to S. graminum (Dogimont et al., 2010). However, as far as we know none of these genes have been cloned and tested for their functions in aphid resistance. One well-known example of plant S genes in relation to pathogens is the recessive resistance to powdery mildew by mlo alleles which cause resistance to all races of that pathogen in barley. The exact function of the MLO protein is not known but it is involved in negative regulation of plant defense (Pavan et al., 2010; Kusch and Panstruga, 2017). Resistance breeding via mutagenesis or RNAi of S genes has been suggested for increasing resistance to plant pathogens and nematodes (De Almeida Engler et al., 2005; Pavan et al., 2010). Site-directed mutagenesis has been successful in introducing mlo resistance to powdery mildew in wheat, race-specific resistance to bacterial blight in rice and canker resistance in citrus for example. In the latter two examples, the promoter regions of the susceptibility genes were mutated. In yet another example, a gene for an ET responsive element that is a negative regulator of resistance to rice blast was mutated. Also resistance to several viral diseases has been achieved by plant-mediated gene mutations in the viral genome, or by mutations of S genes in the plant genome (Weeks et al., 2015; Mushtaq et al., 2019; Sedeek et al., 2019). In the following section, we will discuss the possibility to exploit knowledge about S genes for breeding resistance to aphids, namely to modify S gene function by site-directed mutagenesis or via RNAi.
Mutation techniques are now available that allow mutations specifically in target genes as opposed to the conventional chemical or radiation mutation methods, which generate mutations in non-targeted genes in addition to the targeted ones. The most recent and versatile technique by which to make such mutations is CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeat/CRISPR-associated protein9), a technique that mimics a microbe defense mechanism. There are different approaches for applying this technique, while the most common one is to knock out the target gene function. With this approach, a guide RNA sequence binds to a matching DNA sequence in the target gene and the Cas9 enzyme makes a double DNA strand break. The break is then repaired by the cell machinery and normally results in short insertion or deletion mutations which may result in knock-out of the gene function. Moreover, this technique is also possible to use for a specific single base pair editing, or overexpression or downregulation of target genes although these approaches are not yet as well developed and applied as the knock out approach (http://www.addgene.org/crispr/guide/; accessed on 3 July 2019). Alternatively, it is possible to use the RNAi technique to downregulate the gene expression. This method builds upon transformation of DNA constructs that give rise to short double-stranded RNA that after plant processing changes the plant transcriptome (Younis et al., 2014).
There are numerous examples of mutants produced with the older mutation techniques (i.e. by chemicals, radiation or T-DNA insertion) used in studies of gene function, often in model species such as Arabidopsis (De Vos et al., 2007; Louis and Shah, 2013). However, to be applied in aphid resistance breeding of crops it is necessary to test the effects in the target crop and with the target pest. Furthermore, it is essential to make sure that no additional mutations interfere. Existing mutants tested in earlier studies are, however, useful in providing indications of potential susceptibility mechanisms to exploit and also which general pleiotropic plant effects can be expected for a given gene knock-out (Thompson and Goggin, 2006; Huckelhoven et al., 2013; van Schie and Takken, 2014).
Since many studies have shown that the JA-related plant defense is functional for reducing host plant susceptibility to aphids and that aphids can counteract the induction of this pathway via upregulation of the SA-based defense, it is possible to exploit this knowledge for aphid resistance breeding by mutating genes in the SA defense signaling pathway so that the SA–JA cross-talk is disturbed and JA signaling can take place. However, this strategy may be risky because it will probably make the plant more susceptible to biotrophic pathogens, against which SA-induced defense is important. Besides, as described above, there is probably cross-talk also between other hormonal signaling pathways, which might be important for aphid-induced susceptibility, but at the same time important for other plant functions like tolerance to abiotic stresses.
Ideally the S gene candidate for mutation should be more specific to the aphid/plant interaction and have functions that are not absolutely vital for plant growth and development. This is possible if there are homologues of the gene differing in whether they are regulated by aphids or not. An example of this is β-1,3-glucanase genes, of which only some are upregulated by aphids, more so in the susceptible hosts (Park et al., 2006; Delp et al., 2009, Reddy et al., 2013; Mehrabi et al., 2016; Sun et al., 2018). Recently, Shoala et al. (2018) tested Arabidopsis β-1,3-glucanase mutants and proposed that one out of the four targeted β-1,3-glucanases is a susceptibility factor for M. persicae. We are currently testing barley with one or two of such genes mutated for potentially increased resistance to R. padi. The rationale for this attempt is that we expect less of glucanases and therefore more callose accumulating in the phloem in response to aphid infestation, which in turn may reduce aphid access to the phloem sap.
Another possibility to reduce susceptibility to aphids might be to hinder aphid-induced loosening of plant cell walls by knocking out or down genes associated with cell wall structure. Previous global gene expression studies have indicated that certain cell wall-related genes, which showed increased expression levels upon aphid infestation, such as pectinase (Reddy et al., 2013) and expansin genes (Bricchi et al., 2012), might be potential S genes to target by CRISPR/Cas9 or RNAi. One type of cell wall-related candidate S genes has already been confirmed via transient RNAi, i.e. (1,3;1,4)-β-glucanases that can degrade plant cell wall cellulose. Corresponding RNA sequences were shown to be induced to higher levels in susceptible than in resistant wheat lines 5 h after infestation by D. noxia. Knock-down of plant (1,3;1,4)-β-glucanase sequences increased plant resistance to D. noxia, suggesting that they are aphid-induced susceptibility factors (Anderson et al., 2014). Moreover, instead of directly targeting the S gene, a transcription factor gene with a regulatory role for the target gene might be knocked out or down. The transcription factor WRKY22 that enhances the susceptibility to M. persicae in Arabidopsis can serve as an example. Aphids took longer time to penetrate the epidermis and mesophyll and reach the vascular bundles of the wrky22 mutants, possibly due to the downregulation of the cell-wall loosening genes for pectin lyases and expansins (Kloth et al., 2016).
Genes involved in nutrient transport such as certain sugar and nitrogen transporters may be another type of genes to target. Like with some aphid examples mentioned above (Table 1), sugar transporters have been found to be regulated also by pathogens and disease resistance has been confirmed in the mutants with edited promoter regions or in genotypes with natural mutations in coding regions of sugar transporter genes (Julius et al., 2017;Sedeek et al., 2019). Based on the imbalanced N/C ratio of phloem sap, nitrogen-related genes are possibly more important to target for reducing aphid performance than carbohydrate-related genes. Genes for turgor-regulating proteins, aquaporins, offer another possibility for gene editing in order to indirectly reduce the aphid access to nutrients in the phloem.
Although several omics studies have been carried out in aphid-infested plants, the molecular mechanisms explaining aphid-induced plant susceptibility are still largely unknown. The fact that the global expression studies performed until now commonly include a single susceptible host genotype or compare solely one resistant and one susceptible genotype makes it difficult to draw conclusions about potential susceptibility roles of plant genes in relation to aphid performance. Therefore several representatives of resistant and susceptible plant genotypes should be compared in future studies in order to unravel more general patterns. Thus, there are so far only few candidate genes for induced plant susceptibility to aphid attack which might be exploited for improving crop resistance to aphids. We expect this field of research and plant resistance breeding to expand in the coming years.
Based on the knowledge we have so far, we have suggested certain types of potential susceptibility-associated genes for improving crop resistance to aphids. We also propose functional studies of recessively inherited resistance discovered in some aphid–crop combinations which might reveal currently unknown types of susceptibility genes that could be knocked out or down in other crops to improve their resistance to aphids. For application in commercial plant breeding, it will be necessary to find susceptibility genes that do not compromise the crop yield, or are at least so effective in controlling a severe pest that some crop loss can be accepted. Reluctance to accept crops developed by gene technology as for instance in EU is currently an obstacle for efficient reduction of host susceptibility as a resistance breeding approach.
IÅ wrote the main part of the manuscript. L-HZ suggested manuscript improvements and added suggestions for candidate gene types to target, along with S-YK.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Swedish Research Council FORMAS is thanked for providing financial support via the grant 220-2014-1330.
Alvarez, A. E., D’Amato, A. M. A., Tjallingii, W. F., Dicke, M., Vosman, B. (2014). Response of Solanum tuberosum to Myzus persicae infestation at different stages of foliage maturity. Insect Sci. 21, 727–740. doi: 10.1111/1744-7917.12072
Alvarez, A. E., Broglia, V. G., D’Amato, A. M. A., Wouters, D., van der Vossen, E., Garzo, E., et al. (2013). Comparative analysis of Solanum stoloniferum responses to probing by the green peach aphid Myzus persicae and the potato aphid Macrosiphum euphorbiae. Insect Sci. 20, 207–227. doi: 10.1111/j.1744-7917.2012.01505.x
Anderson, V. A., Haley, S. D., Peairs, F. B., van Eck, L., Leach, J. E., Lapitan, N. L. V. (2014). Virus-induced gene silencing suggests (1,3;1,4)- β -glucanase is a susceptibility factor in the compatible Russian wheat aphid-wheat interaction. MPMI 27, 913–922. doi: 10.1094/MPMI-05-13-0141-R
Appel, H. M., Fescemyer, H., Ehlting, J., Weston, D., Rehrig, E., Joshi, T., et al. (2014). Transcriptional responses of Arabidopsis thaliana to chewing and sucking insect herbivores. Front. Plant Sci. 5, 565. doi: 10.3389/fpls.2014.00565
Barah, P., Winge, P., Kusnierczyk, A., Tran, D. H., Bones, A. M. (2013). Molecular signatures in Arabidopsis thaliana in response to insect attack and bacterial infection. PloS One 8, e58987. doi: 10.1371/journal.pone.0058987
Baumann, P. (2005). Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59, 155–189. doi: 10.1146/annurev.micro.59.030804.121041
Blackman, R. L., Eastop, V. F. (1984). Aphids on the World’s Crops – an Identification Guide. Avon: John Wiley and Sons.
Blackman, R. L., Eastop, V. F. (2007). “Taxonomic issues,” in Aphids as Crop Pests. Eds. van Emden, H. F., Harrington, R. (Trowbridge, UK: CABI), 1–29.
Boissot, N., Schoeny, A., Vanlerberghe-Masutti, F. (2016). Vat, an amazing gene conferring resistance to aphids and viruses they carry: From molecular structure to field effects. Front. Plant Sci. 7, 1420. doi: 10.3389/fpls.2016.01420
Botha, A.-M., Lacock, L., van Niekerk, C., Matsioloko, M. T., du Preez, F. B., Loots, S., et al. (2006). Is photosynthetic transcriptional regulation in Triticum aestivum L. cv. ‘TugelaDN’ a contributing factor for tolerance to Diuraphis noxia (Homoptera: Aphididae)? Plant Cell Rep. 25, 41–54. doi: 10.1007/s00299-005-0001-9
Botha, A.-M., Swanewelder, Z. H., Lapitan, N. L. V. (2010). Transcript profiling of wheat genes expressed during feeding by two different biotypes of Diuraphis noxia. Environ. Entomol. 39, 1206–1231. doi: 10.1603/EN09248
Botha, A.-M., van Eck, L., Burger, N. F. V., Swanevelder, Z. H. (2014). Near-isogenic lines of Triticum aestivum with distinct modes of resistance exhibit dissimilar transcriptional regulation during Diuraphis noxia feeding. Biol. Open 3, 1116–1126. doi: 10.1242/bio.201410280
Boyko, E. V., Smith, C. M., Thara, V. K., Bruno, J. M., Deng, Y., Starkey, S. R., et al. (2006). Molecular basis of plant gene expression during aphid invasion: Wheat Pto- and Pti-like sequences are involved in interactions between wheat and Russian wheat aphid (Homoptera: Aphididae). J. Econ. Entomol. 99, 1430–1445. doi: 10.1603/0022-0493-99.4.1430
Brechenmacher, L., Nguyen, T. H. N., Zhang, N., Jun, T.-H., Xu, D., Mian, M. A. R., et al. (2015). Identification of soybean proteins and genes differentially regulated in near isogenic lines differing in resistance to aphid infestation. J. Proteome Res. 14, 4137–4146. doi: 10.1021/acs.jproteome.5b00146
Bricchi, I., Bertea, C. M., Occhipinti, A., Paponov, I. A., Maffei, M. E. (2012). Dynamics of membrane potential variation and gene expression induced by Spodoptera littoralis, Myzus persicae, and Pseudomonas syringae in Arabidopsis. PloS One 7, e46673. doi: 10.1371/journal.pone.0046673
Broekgaarden, C., Poelman, E. H., Steenhuis, G., Vorrips, R. E., Dicke, M., Vosman, B. (2008). Responses of Brassica oleracea cultivars to infestation by the aphid Brevicoryne brassicae: an ecological and molecular approach. Plant Cell Environ. 31, 1592–1605. doi: 10.1111/j.1365-3040.2008.01871.x
Broekgaarden, C., Voorrips, R. E., Dicke, M., Vosman, B. (2011). Transcriptional responses of Brassica nigra to feeding by specialist insects of different feeding guilds. Insect Sci. 18, 259–272. doi: 10.1111/j.1744-7917.2010.01368.x
Carrillo, E., Rubiales, D., Castillejo, M. A. (2014). Proteomic analysis of pea (Pisum sativum L.) response during compatible and incompatible interactions with the pea aphid (Acyrthosiphon pisum H.). Plant Mol. Biol. Rep. 32, 697–718. doi: 10.1007/s11105-013-0677-x
Chang, J., Duan, X., Cui, J., Xue, W., Zhang, Q. (2012). Differential molecular responses of aphid-sensitive and aphid-resistant sorghum lines to aphid infestation. Arthropod-Plant Interact. 6, 113–120. doi: 10.1007/s11829-011-9159-y
Coppola, V., Coppola, M., Rocco, M., Digilio, M. C., D’Ambrosio, C., Renzone, G., et al. (2013). Transcriptomic and proteomic analysis of a compatible tomato-aphid interaction reveals a predominant salicylic acid-dependent plant response. BMC Genomics 14, 515. doi: 10.1186/1471-2164-14-515
Couldridge, C., Newbury, H. J., Ford-Lloyd, B., Bale, J., Pritchard, J. (2007). Exploring plant responses to aphid feeding using a full Arabidopsis microarray reveals a small number of genes with significantly altered expression. Bull. Entomol. Res. 97, 523–532. doi: 10.1017/S0007485307005160
Cui, N., Lu, H., Wang, T., Zhang, W., Kang, L., Cui, F. (2019). Armet, an aphid effector protein, induces pathogen resistance in plants by promoting the accumulation of salicylic acid. Phil. Trans. R. Soc B 374, 20180314. doi: 10.1098/rstb.2018.0314
De Almeida Engler, J., Favery, B., Engler, G., Abad, P. (2005). Loss of susceptibility as an alternative for nematode resistance. Curr. Opin. Biotechnol. 16, 112–117. doi: 10.1016/j.copbio.2005.01.009
De Vos, M., Jander, G. (2009). Myzus persicae (green peach aphid) salivary components induce defence responses in Arabidopsis thaliana. Plant Cell Environ. 32, 1548–1560. doi: 10.1111/j.1365-3040.2009.02019.x
De Vos, M., Kim, J. H., Jander, G. (2007). Biochemistry and molecular biology of Arabidopsis-aphid interactions. BioEssays 29, 871–883. doi: 10.1002/bies.20624
De Vos, M., van Oosten, V. R., van Poecke, R. M. P., van Pelt, J. A., Pozo, M. J., Mueller, M. J., et al. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. MPMI 18, 923–937. doi: 10.1094/MPMI-18-0923
Delp, G., Gradin, T., Åhman, I., Jonsson, L. M. V. (2009). Microarray analysis of the interaction between the aphid Rhopalosiphum padi and host plants reveals both differences and similarities between susceptible and partially resistant barley lines. Mol. Genet. Genomics 281, 233–248. doi: 10.1007/s00438-008-0409-3
Dinant, S., Bonnemain, J.-L., Girousse, C., Kehr, J. (2010). Phloem sap intricacy and interplay with aphid feeding. Comptes Rendus Biologies 333, 504–515. doi: 10.1016/j.crvi.2010.03.008
Divol, F., Vilaine, F., Thibivilliers, S., Amselem, J., Palauqui, J.-C., Kusiak, C., et al. (2005). Systemic response to aphid infestation by Myzus persicae in the phloem of Apium graveolens. Plant Mol. Biol. 57, 517–540. doi: 10.1007/s11103-005-0338-z
Dogimont, C., Bendahmane, A., Chovelon, V., Boissot, N. (2010). Host plant resistance to aphids in cultivated crops: Genetic and molecular bases, and interactions with aphid populations. Comptes Rendus Biologies 333, 566–573. doi: 10.1016/j.crvi.2010.04.003
Dubey, N. K., Goel, R., Ranjan, A., Idris, A., Singh, S. K., Bag, S. K., et al. (2013). Comparative transcriptome analysis of Gossypium hirsutum L. in response to sap sucking insects: aphid and whitefly. BMC Genomics 14, 241. doi: 10.1186/1471-2164-14-241
Eisenring, M., Glauser, G., Meissle, M., Romeis, J. (2018). Differential impact of herbivores from three feeding guilds on systemic secondary metabolite induction, phytohormone levels and plant-mediated herbivore interactions. J. Chem. Ecol. 44, 1178–1189. doi: 10.1007/s10886-018-1015-4
Elzinga, D. A., de Vos, M., Jander, G. (2014). Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. MPMI 27, 747–756. doi: 10.1094/MPMI-01-14-0018-R
Errard, A., Ulrichs, C., Kuhne, S., Mewis, I., Drungowski, M., Schreiner, M., et al. (2015). Single- versus multiple-pest infestation affects differently the biochemistry of tomato (Solanum lycopersicum‘Ailsa Craig’). J. Agric. Food Chem. 63, 10103–10111. doi: 10.1021/acs.jafc.5b03884
Escudero-Martinez, C. M., Morris, J. A., Hedley, P. E., Bos, J. I. B. (2017). Barley transcriptome analyses upon interaction with different aphid species identify thionins contributing to resistance. Plant Cell Environ. 40, 2628–2643. doi: 10.1111/pce.12979
Ferry, N., Stavroulakis, S., Guan, W., Davison, G. M., Bell, H. A., Weaver, R. J., et al. (2011). Molecular interactions between wheat and cereal aphid (Sitobion avenae): Analysis of changes to the wheat proteome. Proteomics 11, 1985–2002. doi: 10.1002/pmic.200900801
Foyer, C. H., Verrall, S. R., Hancock, R. D. (2015). Systematic analysis of phloem-feeding insect-induced transcriptional reprogramming in Arabidopsis highlights common features and reveals distinct responses to specialist and generalist insects. J. Exp. Bot. 66, 495–512. doi: 10.1093/jxb/eru491
Giordanengo, P., Brunissen, L., Rusterucci, C., Vincent, C., van Bel, A., Dinant, S., et al. (2010). Compatible plant-aphid interactions: How aphids manipulate plant responses. Comptes Rendus Biologies 333, 516–523. doi: 10.1016/j.crvi.2010.03.007
Girousse, C., Moulia, B., Silk, W., Bonnemain, J.-L. (2005). Aphid infestation causes different changes in carbon and nitrogen allocation in alfalfa stems as well as different inhibitions of longitudinal and radial expansion. Plant Physiol. 137, 1474–1484. doi: 10.1104/pp.104.057430
Goggin, G. L. (2007). Plant-aphid interactions: molecular and ecological perspectives. Curr. Opin. Plant Biol. 10, 399–408. doi: 10.1016/j.pbi.2007.06.004
Greenslade, A. F. C., Ward, J. L., Martin, J. L., Corol, D. I., Clark, S. J., Smart, L. E., et al. (2016). Triticum monococcum lines with distinct metabolic phenotypes and phloem-based partial resistance to the bird cherry-oat aphid Rhopalosiphum padi. Ann. Appl. Biol. 168, 435–449. doi: 10.1111/aab.12274
Guan, W., Ferry, N., Edwards, M. G., Bell, H. A., Othman, H., Gatehouse, J. A., et al. (2015). Proteomic analysis shows that stress response proteins are significantly up-regulated in resistant diploid wheat (Tritcum monococcum) in response to attack by the grain aphid (Sitobion avenae). Mol. Breed. 35, 57. doi: 10.1007/s11032-015-0220-x
Gutsche, A., Heng-Moss, T., Sarath, G., Twigg, P., Xia, Y., Lu, G., et al. (2009). Gene expression profiling of tolerant barley in response to Diuraphis noxia (Hemiptera: Aphididae) feeding. Bull. Entomol. Res. 99, 163–173. doi: 10.1017/S0007485308006184
Hawkins, C. D. B., Aston, M. J., Whitecross, M. I. (1987). Short-term effects of aphid feeding on photosynthetic CO2 exchange and dark respiration in legume leaves. Physiologia Plantarum 71, 379–383. doi: 10.1111/j.1399-3054.1987.tb04359.x
Hays, D. B., Porter, D. R., Webster, J. A., Carver, B. F. (1999). Feeding behavior of biotypes E and H greenbug (Homoptera: Aphididae) on previously infested near-isolines of barley. J. Econ. Entomol. 92, 1223–1229. doi: 10.1093/jee/92.5.1223
Heidel, A. J., Baldwin, I. T. (2004). Microarray analysis of salicylic acid- and jasmonic acid-signalling in responses of Nicotiana attenuata to attack by insects from multiple feeding guilds. Plant Cell Environ. 27, 1362–1373. doi: 10.1111/j.1365-3040.2004.01228.x
Hillwig, M. S., Chiozza, M., Casteel, C. L., Lau, S. T., Hohenstein, J., Hernandez, E., et al. (2016). Abscicic acid deficiency increases defence responses against Myzus persicae in Arabidopsis. Mol. Plant Pathol. 17, 225–235. doi: 10.1111/mpp.12274
Hohenstein, J. D., Studham, M. E., Klein, A., Kovinich, N., Barry, K., Lee, Y.-J., et al. (2019). Transcriptional and chemical changes in soybean leaves in response to long-term aphid colonization. Front. Plant Sci. 10, 310. doi: 10.3389/fpls.2019.00310
Huckelhoven, R., Eichmann, R., Weis, C., Hoefle, C., Proels, R. K. (2013). Genetic loss of susceptibility: a costly route to disease resistance? Plant Pathol. 62 (Suppl. 1), 52–62. doi: 10.1111/ppa.12103
Jaouannet, M., Morris, J. A., Hedley, P. E., Bos, J. I. B. (2015). Characterization of Arabidopsis transcriptional responses to different aphid species reveals genes that contribute to host susceptibility and non-host resistance. PloS Pathog. 11, e1004918. doi: 10.1371/journal.ppat.1004918
Jaouannet, M., Rodriguez, P. A., Thorpe, P., Lenoir, C. J. G., MacLeod, R., Escudero-Martinez, C., et al. (2014). Plant immunity in plant-aphid interactions. Front. Plant Sci. 5, 663. doi: 10.3389/fpls.2014.00663
Julius, B. T., Leach, K. A., Tran, T. M., Mertz, R. A., Braun, D. M. (2017). Sugar transporters in plants: new insights and discoveries. Plant Cell Physiol. 58, 1442–1460. doi: 10.1093/pcp/pcx090
Kapilan, R., Vaziri, M., Zwiazek, J. J. (2018). Regulation of aquaporins in plants under stress. Biol. Res. 51, 4. doi: 10.1186/s40659-018-0152-0
Kerchev, P. I., Karpinska, B., Morris, J. A., Hussain, A., Verrall, S. R., Hedley, P. E., et al. (2013). Vitamin C and the abscisic acid-insensitive 4 transcription factor are important determinants of aphid resistance in Arabidopsis. Antioxidants Redox Signaling 18, 2091–2105. doi: 10.1089/ars.2012.5097
Kiani, M., Szczepaniec, A. (2018). Effects of sugarcane aphid herbivory on transcriptional responses of resistant and susceptible sorghum. BMC Genomics 19, 774. doi: 10.1186/s12864-018-5095-x
Klingler, J., Creasy, R., Gao, L., Nair, R. M., Calix, A. S., Jacob, H. S., et al. (2005). Aphid resistance in Medicago truncatula involves antixenosis and phloem-specific, inducible antibiosis, and maps to a single locus flanked by NBS-LRR resistance gene analogs. Plant Physiol. 137, 1445–1455. doi: 10.1104/pp.104.051243
Kloth, K. J., Busscher-Lange, J., Wiegers, G. L., Kruijer, W., Buijs, G., Meyer, R. C., et al. (2017). Sieve element-lining chaperone1 restricts aphid feeding on Arabidopsis during heat stress. Plant Cell 29, 2450–2464. doi: 10.1105/tpc.16.00424
Kloth, K. J., Wiegers, G. L., Busscher-Lange, J., van Haarst, J. C., Kruijer, W., Bouwmeester, H. J., et al. (2016). AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. J. Exp. Bot. 67, 3383–3396. doi: 10.1093/jxb/erw159
Kusch, S., Panstruga, R. (2017). mlo-based resistance: an apparently universal “weapon” to defeat powdery mildew disease. MPMI 30, 179–189. doi: 10.1094/MPMI-12-16-0255-CR
Kusnierczyk, A., Tran, D. H. T., Winge, P., Jörstad, T. S., Reese, J. C., Troczynska, J., et al. (2011). Testing the importance of jasmonate signalling in induction of plant defences upon cabbage aphid (Brevicoryne brassicae) attack. BMC Genomics 12, 423. doi: 10.1186/1471-2164-12-423
Kusnierczyk, A., Winge, P., Jörstad, T. S., Troczynska, J., Rossiter, J. T., Bones, A. M. (2008). Towards global understanding of plant defence against aphids – timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell Environ. 31, 1097–1115. doi: 10.1111/j.1365-3040.2008.01823.x
Kusnierczyk, A., Winge, P., Midelfart, H., Armbruster, W. S., Rossiter, J. T., Bones, A. M. (2007). Transcriptional responses of Arabidopsis thaliana ecotypes with different glucosinolate profiles after attack by polyphagous Myzus persicae and oligophagous Brevicoryne brassicae. J. Exp. Bot. 58, 2537–2552. doi: 10.1093/jxb/erm043
Kutyniok, M., Muller, C. (2012). Crosstalk between above– and belowground herbivores is mediated by minute metabolic responses of the host Arabidopsis thaliana. J. Exp. Bot. 63, 6199–6210. doi: 10.1093/jxb/ers274
Lane, B. G., Dunwell, J. M., Ray, J. A., Schmitt, M. R., Cuming, A. C. (1993). Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 268, 12239–12242.
Lee, S., Cassone, B. J., Wijeratne, A., Jun, T.-H., Michel, A. P., Mian, M. A. R. (2017). Transcriptomic dynamics in soybean near-isogenic lines differing in alleles for an aphid resistance gene, following infestation by soybean aphid biotype 2 . BMC Genomics 18, 472. doi: 10.1186/s12864-017-3829-9
Lemoine, R., La Camera, S., Atanassova, R., Dédaldéchamp, F., Allario, T., Pourtau, N., et al. (2013). Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 4, 272. doi: 10.3389/fpls.2013.00272
Li, Y., Zou, J., Li, M., Bilgin, D. D., Vodkin, L. O., Hartman, G. L., et al. (2008). Soybean defense responses to the soybean aphid. New Phytol. 179, 185–195. doi: 10.1111/j.1469-8137.2008.02443.x
Liang, D., Liu, M., Hu, Q., He, M., Qi, X., Xu, Q., et al. (2015). Identification of differentially expressed genes related to aphid resistance in cucumber (Cucumis sativus L.). Sci. Rep. 5, 9645. doi: 10.1038/srep09645
Liu, X., Meng, J., Starkey, S., Smith, C. M. (2011). Wheat gene expression is differentially affected by a virulent Russian wheat aphid biotype. J. Chem. Ecol. 37, 472–482. doi: 10.1007/s10886-011-9949-9
Louis, J., Shah, J. (2013). Arabidopsis thaliana– Myzus persicae interaction: shaping the understanding of plant defense against phloem-feeding aphids. Front. Plant Sci. 4, 213. doi: 10.3389/fpls.2013.00213
Mantelin, S., Bhattarai, K. K., Kaloshian, I. (2009). Ethylene contributes to potato aphid susceptibility in a compatible tomato host. New Phytol. 183, 444–456. doi: 10.1111/j.1469-8137.2009.02870.x
Mehrabi, S., Åhman, I., Jonsson, L. M. V. (2016). The constitutive expression and induction of three β-1,3-glucanases by bird-cherry oat aphid in relation to aphid resistance in 15 barley breeding lines. Arthropod-Plant Interact. 10, 101–111. doi: 10.1007/s11829-016-9415-2
Mondal, H. A. (2017). Shaping the understanding of saliva-derived effectors towards aphid colony proliferation in host plant. J. Plant Biol. 60, 103–115. doi: 10.1007/s12374-016-0465-x
Moran, P. J., Cheng, Y., Cassell, J. L., Thompson, G. A. (2002). Gene expression profiling of Arabidopsis thaliana in compatible plant-aphid interactions. Arch. Insect Biochem. Physiol. 51, 182–203. doi: 10.1002/arch.10064
Muneer, S., Jeong, H. K., Park, Y. G., Jeong, B. R. (2018). Proteomic analysis of aphid-resistant and –sensitive rose (Rosa hybrida) cultivars at two developmental stages. Proteomes 6, 25. doi: 10.3390/proteomes6020025
Mushtaq, M., Sakina, A., Wani, S. H., Shikari, A. B., Tripathi, P., Zaid, A., et al. (2019). Harnessing genome editing techniques to engineer disease resistance in plants. Front. Plant Sci. 10, 550. doi: 10.3389/fpls.2019.00550
Nalam, V. J., Keeretaweep, J., Sarowar, S., Shah, J. (2012). Root-derived oxylipins promote green peach aphid performance on Arabidopsis foliage. Plant Cell 24, 1643–1653. doi: 10.1105/tpc.111.094110
Park, S.-J., Huang, Y., Ayoubi, P. (2006). Identification of expression profiles of sorghum genes in response to greenbug phloem-feeding using cDNA subtraction and microarray analysis. Planta 223, 932–947. doi: 10.1007/s00425-005-0148-1
Pavan, S., Jacobsen, E., Visser, R. G. F., Bai, Y. (2010). Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 25, 1–12. doi: 10.1007/s11032-009-9323-6
Pettersson, J., Tjallingii, W. F., Hardie, J. (2007). “Host-plant selection and feeding,” in Aphids as Crop Pests. Eds. van Emden, H. F., Harrington, R. (Trowbridge, UK: CABI), 87–113.
Pirselova, B., Matusikova, I. (2013). Callose: the plant cell wall polysaccharide with multiple biological functions. Acta Physiol. Plant 35, 635–644. doi: 10.1007/s11738-012-1103-y
Ponzio, C., Papazian, S., Albrectsen, B. R., Dicke, M., Gols, R. (2017). Dual herbivore attack and herbivore density affect metabolic profiles of Brassica nigra leaves. Plant Cell Environ. 40, 1356–1367. doi: 10.1111/pce.12926
Prochaska, T. J., Donze-Reiner, T., Marchi-Werle, L., Palmer, N. A., Hunt, T. E., Sarath, G., et al. (2015). Transcriptional responses of tolerant and susceptible soybeans to soybean aphid (Aphis glycines Matsumura) herbivory. Arthropod-Plant Interact. 9, 347–359. doi: 10.1007/s11829-015-9371-2
Qubbaj, T., Reineke, A., Zebitz, C. P. W. (2005). Molecular interactions between rosy apple aphids, Dysaphis plantaginea, and resistant and susceptible cultivars of its primary host Malus domestica. Entomol. Exp. Appl. 115, 145–152. doi: 10.1111/j.1570-7458.2005.00255.x
Reddy, S. K., Weng, Y., Rudd, J. C., Akhunova, A., Liu, S. (2013). Transcriptomics of induced defense responses to greenbug aphid feeding in near isogenic wheat lines. Plant Sci. 212, 26–36. doi: 10.1016/j.plantsci.2013.08.002
Rodriguez, P. A., Bos, J. I. B. (2013). Toward understanding the role of aphid effectors in plant infestation. MPMI 26, 25–30. doi: 10.1094/MPMI-05-12-0119-FI
Rodriguez, P. A., Escudero-Martinez, C., Bos, J. I. B. (2017). An aphid effector targets trafficking protein VPS52 in a host-specific manner to promote virulence. Plant Physiol. 173, 1892–1903. doi: 10.1104/pp.16.01458
Rodriguez-Saona, C. R., Musser, R. O., Vogel, H., Hum-Musser, S. M., Thaler, J. S. (2010). Molecular, biochemical, and organismal analyses of tomato plants simultaneously attacked by herbivores from two feeding guilds. J. Chem. Ecol. 36, 1043–1057. doi: 10.1007/s10886-010-9854-7
Sanchez-Arcos, C., Kai, M., Svatos, A., Gershenzon, J., Kunert, G. (2019). Untargeted metabolomics approach reveals differences in host plant chemistry before and after infestation with different pea aphid host races. Front. Plant Sci. 10, 188. doi: 10.3389/fpls.2019.00188
Sandström, J., Telang, A., Moran, N. A. (2000). Nutritional enhancement of host plants by aphids – a comparison of three aphid species on grasses. J. Insect Physiol. 46, 33–40. doi: 10.1016/S0022-1910(99)00098-0
Sato, D., Akashi, H., Sugimoto, M., Tomita, M., Soga, T. (2013). Metabolomic profiling of the response of susceptible and resistant soybean strains to foxglove aphid, Aulacorthum solani Kaltenbach. J. Chromatography B 925, 95–103. doi: 10.1016/j.jchromb.2013.02.036
Sattar, S., Thompson, G. A. (2016). Small RNA regulators of plant-hemipteran interactions: micromanagers with versatile roles. Front. Plant Sci. 7, 1241. doi: 10.3389/fpls.2016.01241
Sauge, M.-H., Mus, F., Lacroze, J.-P., Pascal, T., Kervella, J., Poessel, J.-L. (2006). Genotypic variation in induced resistance and induced susceptibility in the peach-Myzus persicae aphid system. OIKOS 113, 305–313. doi: 10.1111/j.2006.0030-1299.14250.x
Sedeek, K. E. M., Mahas, A., Mahfouz, M. (2019). Plant genome engineering for targeted improvement of crop traits. Front. Plant Sci. 10, 114. doi: 10.3389/fpls.2019.00114
Shavit, R., Batyrshina, Z. S., Dotan, N., Tzin, V. (2018). Cereal aphids differently affect benzoxazinoid levels in durum wheat. PloS One 13, e0208103. doi: 10.1371/journal.pone.0208103
Shoala, T., Edwards, M. G., Knight, M. R., Gatehouse, A. M. R. (2018). OXI1 kinase plays a key role in resistance of Arabidopsis towards aphids (Myzus persicae). Transgenic Res. 27, 355–366. doi: 10.1007/s11248-018-0078-x
Smith, C. M., Liu, X., Wang, L. J., Liu, X., Chen, M.-S., Starkey, S., et al. (2010). Aphid feeding activates expression of a transcriptome of oxylipin-based defense signals in wheat involved in resistance to herbivory. J. Chem. Ecol. 36, 260–276. doi: 10.1007/s10886-010-9756-8
Studham, M. E., MacIntosh, G. C. (2013). Multiple phytohormone signals control the transcriptional response to soybean aphid infestation in susceptible and resistant soybean plants. MPMI 26, 116–129. doi: 10.1094/MPMI-05-12-0124-FI
Sun, Y., Guo, H., Yuan, E., Ge, F. (2018). Elevated CO2 increases R gene-dependent resistance of Medicago truncatula against the pea aphid by up-regulating a heat shock gene. New Phytol. 217, 1696–1711. doi: 10.1111/nph.14892
Sun, Y., Guo, H., Yuan, L., Wei, J., Zhang, W., Ge, F. (2015). Plant stomatal closure improves aphid feeding under elevated CO2. Global Change Biol. 21, 2739–2748. doi: 10.1111/gcb.12858
Telang, A., Sandström, J., Dyreson, E., Moran, N. A. (1999). Feeding damage by Diuraphis noxia results in a nutritionally enhanced phloem diet. Entomol. Exp. Appl. 91, 403–412. doi: 10.1046/j.1570-7458.1999.00508.x
Tetreault, H. M., Grover, S., Scully, E. D., Gries, T., Palmer, N. A., Sarath, G., et al. (2019). Global responses of resistant and susceptible sorghum (Sorghum bicolor) to sugarcane aphid (Melanaphis sacchari). Front. Plant Sci. 10, 145. doi: 10.3389/fpls.2019.00145
Thompson, G. A., Goggin, F. L. (2006). Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J. Exp. Bot. 57, 755–766. doi: 10.1093/jxb/erj135
Truong, D.-H., Bauwens, J., Delaplace, P., Mazzuccelli, G., Lognay, G., Francis, F. (2015). Proteomic analysis of Arabidopsis thaliana (L.) Heynh responses to a generalist sucking pest (Myzus persicae Sulzer). Plant Biol. 17, 1210–1217. doi: 10.1111/plb.12363
Tu, X.-B., Zhao, H.-L., Zhang, Z.-H. (2018). Transcriptome approach to understand the potential mechanisms of resistant and susceptible alfalfa (Medicago sativa L.) cultivars in response to aphid feeding. J. Integr. Agric. 17, 60345–60347. doi: 10.1016/S2095-3119(17)61843-4
Tzin, V., Fernandez-Pozo, N., Richter, A., Schmelz, E. A., Schoettner, M., Schäfer, M., et al. (2015). Dynamic maize responses to aphid feeding are revealed by a time series of transcriptomic and metabolomic assays. Plant Physiol. 169, 1727–1743. doi: 10.1104/pp.15.01039
van Bel, A. J. E., Will, T. (2016). Functional evaluation of proteins in watery and gel saliva of aphids. Front. Plant Sci. 7, 1840. doi: 10.3389/fpls.2016.01840
Vandenborre, G., Smagghe, G., van Damme, E. J. M. (2011). Plant lectins as defense proteins against phytophagous insects. Phytochem. 72, 1538–1550. doi: 10.1016/j.phytochem.2011.02.024
van Loon, L. C., van Strien, E. A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. doi: 10.1006/pmpp.1999.0213
van Schie, C. C. N., Takken, F. L. W. (2014). Susceptibility genes 101: How to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-045854
Voelckel, C., Weisser, W. W., Baldwin, I. T. (2004). An analysis of plant-aphid interactions by different microarray hybridization strategies. Mol. Ecol. 13, 3187–3195. doi: 10.1111/j.1365-294X.2004.02297.x
Walling, L. L. (2008). Avoiding effective defenses: Strategies employed by phloem-feeding insects. Plant Physiol. 146, 859–866. doi: 10.1104/pp.107.113142
Weeks, D. P., Spalding, M. H., Yang, B. (2015). Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol. J. 14 483–495. doi: 10.1111/pbi.12448
Younis, A., Siddique, M. I., Kim, C.-K., Lim, K.-B. (2014). RNA interference (RNAi) induced gene silencing: a promising approach of hi-tech plant breeding. Int. J. Biol. Sci. 10, 1150–1158. doi: 10.7150/ijbs.10452
Zhong, R., Cui, D., Ye, Z.-H. (2019). Secondary cell wall biosynthesis. New Phytol. 221, 1703–1723. doi: 10.1111/nph.15537
Zhu, L., Guo, J., Ma, Z., Wang, J., Zhou, C. (2018). Arabidopsis transcription factor MYB102 increases plant susceptibility to aphids by substantial activation of ethylene biosynthesis. Biomolecules 8, 39. doi: 10.3390/biom8020039
Zhu-Salzman, K., Salzman, R. A., Ahn, J.-E., Koiwa, H. (2004). Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol. 134, 420–431. doi: 10.1104/pp.103.028324
Zust, T., Agrawal, A. A. (2016). Mechanisms and evolution of plant resistance to aphids. Nat. Plants 2, 16206. doi: 10.1038/NPLANTS.2015.206
Keywords: aphid, susceptibility gene, omics, plant resistance, plant defense, gene editing, CRISPR/Cas9, RNAi
Citation: Åhman I, Kim S-Y and Zhu L-H (2019) Plant Genes Benefitting Aphids—Potential for Exploitation in Resistance Breeding. Front. Plant Sci. 10:1452. doi: 10.3389/fpls.2019.01452
Received: 05 July 2019; Accepted: 18 October 2019;
Published: 13 November 2019.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Isabel Diaz, Polytechnic University of Madrid, SpainCopyright © 2019 Åhman, Kim and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Inger Åhman, aW5nZXIuYWhtYW5Ac2x1LnNl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.