- 1Normandie Université, UNICAEN, SF ICORE 4206, Caen, France
- 2INRA Unité Expérimentale Fourrages Environnement Ruminants (FERLUS) et Système d’Observation et d’Expérimentation pour la Recherche en Environnement (SOERE), Les Verrines CS 80006, Lusignan, France
- 3INRA—Agrocampus Ouest—Université de Rennes 1, UMR 1349 Institut de Génétique, Environnement et Protection des Plantes (IGEPP) Université de Rennes 1, Rennes, France
- 4NIAB EMR, Crop Science and Production Systems New Road, East Malling, United Kingdom
Background and Aims: Although AVG (aminoethoxyvinylglycine) is intensely used to decipher signaling in ethylene/indol-3-acetic acid (IAA) interactions on root morphogenesis, AVG is not a specific inhibitor of aminocyclopropane-1-carboxylate synthase (ACS) and tryptophan aminotransferase (TAA) and tryptophan aminotransferase related (TAR) activities since it is able to inhibit several aminotransferases involved in N metabolism. Indeed, 1 mM glutamate (Glu) supply to the roots in plants treated with 10 μM AVG partially restores the root growth. Here, we highlight the changes induced by AVG and AVG + Glu treatments on the N metabolism impairment and root morphogenetic program.
Methods: Root nitrate uptake induced by AVG and AVG + Glu treatments was measured by a differential labeling with 15NO3- and 15Nglutamate. In parallel a profiling of amino acids (AA) was performed to decipher the impairment of AA metabolism.
Key Results: 10 μM AVG treatment increases K15NO3 uptake and 15N translocation during root growth inhibition whereas 10 μM AVG + 1 mM 15Nglutamate treatment inhibits K15NO3 uptake and increases 15Nglutamate uptake during partial root growth restoration. This is explained by a nitrogen (N) limitation condition induced by AVG treatment and a N excess condition induced by AVG + Glu treatment. AA levels were mainly impaired by AVG treatment in roots, where levels of Ser, Thr, α-Ala, β-Ala, Val, Asn and His were significantly increased. His was the only amino acid for which no restoration was observed in roots and shoots after glutamate treatment suggesting important control of His homeostasis on aminotransferase network. Results were discussed in light of recent findings on the interconnection between His homeostasis and the general amino acid control system (GAAC) in eukaryotes.
Conclusions: These results demonstrate that AVG concentration above 5 μM is a powerful pharmacological tool for unraveling the involvement of GAAC system or new N sensory system in morphological and metabolic changes of the roots in leguminous and non-leguminous plants.
Introduction
Inhibitors of aminotransferases such as aminoethoxyvinyl-glycine (AVG), Rhizobitoxine and aminooxyacetic acid (AOA) have recently been demonstrated to be non-specific, possessing a broad inhibitory spectrum (Berkowitz et al., 2006; Soeno et al., 2010; Le Deunff and Lecourt, 2016; Le Deunff, 2018). This offers the possibility to highlight the effects of nitrogen metabolism impairment on the root and shoot morphogenetic program and root nitrate uptake (Le Deunff and Lecourt, 2016; Le Deunff et al., 2016). Indeed, nitrogen nutrition and plant development depend not only on nitrate uptake and reduction, but also on the assimilation of nitrogen into amino acids, nucleic acid bases, proteins, polyamines, chlorophylls and hormones due to the action of many aminotransferases downstream the GS/GOGAT cycle (Stitt et al., 2002; Lea and Azevedo, 2006). Aminotransferases are the key enzymes involved in nitrogen (N), sulphur (S) and carbon (C) shuttling and distribution in plants through the interconversions between organic acids and amino acids (Christen and Mehta, 2001; Liepman and Olsen, 2004; Le Deunff, 2018). They catalyze amino group transfers from amino donor to amino acceptor. For their catalytic activity, they require pyridoxal-5’-phosphate (PLP) a phosphorylated form of vitamin B6 that is a versatile coenzyme. PLP biosynthesis depends on the glutamine produced by the GS/GOGAT cycle, glyceraldehyde 3-phosphaste (G3P) and ribose 5-phosphate (5RP) provided by the glycolytic and pentose phosphate pathways, respectively (Tambasco-Studart et al., 2007; Colinas et al., 2016). Aminotransferases have been classified in four families: alpha (α), beta (β), D-alanine aminotransferase and alanine racemase (Mehta et al., 1993; Christen and Mehta, 2001). The aminotransferases belonging to the α family have been further subdivided into four evolutionary subgroups (I–IV) based on their protein sequences and catalytic site structure (Mehta et al., 1993; Christen and Mehta, 2001). The subgroup I contains major enzymes involved in N, S and C assimilation and shuttling such as aspartate, alanine, tyrosine, histidine-phosphate and phenylalanine aminotransferases (Christen and Mehta, 2001). Interestingly, key enzymes involved in auxins (IAA and PAA) and ethylene biosynthesis such as tryptophan aminotransferase of Arabidopsis (TAA1), tryptophan aminotranferase related (TARs) and 1-aminocyclopropane-1-carboxylate synthase (ACS), respectively, belong also to the subgroup I of aminotransferases (Christen and Mehta, 2001; Stepanova et al., 2008; Tao et al., 2008; Sugawara et al., 2015). For example, X-ray structure analysis of ACS catalytic site showed that AA surrounding the active site share the same function as aspartate aminotransferase (AAT) counterpart (Tarun and Theologis, 1998; Capitani et al., 1999).
Despite the essential role played by TAA, TARs and ACS aminotransferases in the root morphogenetic program (Rahman et al., 2000; Stepanova et al., 2008; Tao et al., 2008; Okamoto et al., 2008; Yamada et al., 2009; Ma et al., 2014; Sugawara et al., 2015), the involvement of other aminotransferases within subgroup I of the α family on root development has been overlooked (Le Deunff et al., 2016; Le Deunff, 2018). Arabidopsis mutants of aminotransferases of N metabolism such as aspartate aminotransferase (AAT), alanine aminotransferase (AlaAT) and histidinol phosphate aminotransferase (HPA) displayed root developmental defects (Schultz et al., 1998; Miesak and Coruzzi, 2002; McAllister and Good, 2015; Mo et al., 2006). In Arabidopsis a comparison of the three aat2-1, att2-2 and att2-3 mutants deficient in AAT2 cytosolic isoform, found that only the aat2-2 mutant showed a dramatic reduction in root length that was exaggerated after an addition of 20 mM of aspartate in the medium (Schultz et al., 1998; Miesak and Coruzzi, 2002). Alanine aminotransferase catalyzes the reversible conversion of alanine and 2-oxoglutarate into glutamate and pyruvate. In Arabidopsis, the alaat1;2 double knockout mutants showed a significant reduction in the tap root length and primary lateral roots, whereas over-expression of AlaAT variants from diverse plant sources and different subcellular locations significantly increased root development at non-limiting and limiting external nitrate concentrations (McAllister and Good, 2015). The mutant hpa1 carries a mutation in one of the two plastidial histidinol phosphate aminotransferases (HPA) that converts imidazoleacetol-phosphate and glutamate to histidinol-phosphate and α-ketoglutarate. The mutant presents a 30% reduction in root His content and displays defect in root meristems maintenance and reduction in the primary root and lateral roots development (Mo et al., 2006).
Furthermore, the use of non-specific inhibitors of aminotransferases such as AVG at a concentration greater than 5 μM has revealed that the impairment of N metabolism affects the root morphogenetic program and N uptake (Leblanc et al., 2008; Lemaire et al., 2013). Thus, in Brassica napus, the reduction of root elongation and root hairs formation by AVG treatment were counteracted by a significant increase of nitrate uptake and BnNrt2.1 nitrate transporter gene expression (Leblanc et al., 2008). In comparison to controls, AVG-treated plantlets showed significant increases in the root levels of primary AA such as glutamate, glutamine, aspartate and asparagine (Beauclair et al., 2009; Lemaire et al., 2013). Moreover, root elongation was almost completely restored with 1 mM glutamate treatment in 10 μM AVG-treated plantlets suggesting that major aminotransferases of N metabolism were strongly impaired (Leblanc et al., 2008). Taken together, these results confirm that AVG is a non-specific suicide inhibitor of many PLP-dependent enzymes that belongs to the subgroup I of aminotransferases (Werck-Reichhart et al., 1988; Berkowitz et al., 2006; Soeno et al., 2010; Lin et al., 2010; Le Deunff et al., 2016). Owing to its broad inhibitory spectrum, there is a need to analyse responses of 10 μM AVG treatment on the root growth and development (Soeno et al., 2010). For example, it is well known that treatment with ACC (aminocyclopropane-1-carboxylate), the ethylene precursor, in AVG-treated plantlets does not restore the AVG-induced effect on root hair and primary root elongation. This suggests that other aminotransferases, in addition to ACS and TAA, are also targets for AVG and potentially involved in inhibition of the root morphogenetic program through N sensing systems such as TOR and GCN2 (General Control Non-derepressible-2) kinases (Leblanc et al., 2008; Soeno et al., 2010; Le Deunff et al., 2016; Le Deunff, 2018).
Otherwise, AVG and rhizobitoxine belong to the enol ether family and are a subclass of naturally occurring α-vinylic amino acids (Berkowitz et al., 2006; Boibessot et al., 2016) that are secreted by symbiotic rhizobacteria and pathogenic bacteria (Pruess et al., 1974; Owens et al., 1968; Mitchell et al., 1986; Yasuta et al., 2001). If these two natural compounds at low concentrations (< 5 μM) are known as inhibitors of two enzymes of methionine pathway involved in ethylene biosynthesis: cystathionine β-lyase and ACC synthase (Clausen et al., 1997; Yasuta et al., 1999; Rahman et al., 2000; Berkowitz et al., 2006), the role of their wide inhibitory spectrum on aminotransferases of N metabolism has been neglected in establishing symbiosis and increasing the nodule number (Peters and Crist-Estes, 1989; Xia et al., 2017).
Therefore, the non-specificity of AVG enzymatic inhibition allows us to explore the effects of N metabolism impairment downstream GS/GOGAT cycle assimilation in order to investigate the fundamental role of aminotransferases network on the root morphogenetic program and root nutrient ion uptake through the AA content imbalance and structure-function analyses (Le Deunff and Lecourt, 2016; Le Deunff et al., 2016). This study investigated to what extent root architecture modifications induced by high concentrations of AVG treatments (>5 μM) are solely attributed to ethylene and IAA biosynthesis inhibition. It examined whether the elongation changes of exploratory root systems induced by AVG affect nitrate uptake and N metabolism. Changes in 15N absorption and allocation in the plantlets were achieved using differential labeling of K15NO3 and 15Nglutamate. Quantitative analyses of AA profiles using Ultra Performance Liquid Chromatography (UPLC) reveal significant changes in N metabolism induced by AVG treatment and the restoration effect caused by AVG + Glu treatment. Overall, the results demonstrate that changes in the root morphogenetic program induced by AVG treatment are associated with an alteration of N status and histidine homeostasis and a remodelling of N and C metabolism through root aminotransferases network.
Materials And Methods
Plant Material and Growth Conditions
The oil seed rape (Brassica napus L.) seeds used in this study were the winter oil seed rape cultivar Capitol. The seeds were treated with 70% alcohol for 3 min, followed by three rinses in sterile water and placed on imbibed filter paper (Watmann 3M) saturated with sterile water within Petri dishes (12 × 12 cm) for 48 h in the dark at room temperature. The germinated seeds were then selected according to their radicle length (5–6 mm) and 4 plantlets were transferred onto new Petri dishes (12 × 12 cm), filled with 50 ml of solidified agar culture medium with different chemical treatments. Basic medium contained 0.4 mM KH2PO4, 0.15 mM K2HPO4, 1 mM K2S04, 0.5 mM MgSO4, 3 mM CaCl2, 0.2 mM Fe-Na EDTA, 14 μM H3B03, 5 μM MnSO4, 3μM ZnSO4, 0.7 μM CuSO4, 0.7 μM (NH4)6 Mo7O24 and 0.1 μM CoCl2 and solidified with 0.8% (W/V) agar (Sigma-Aldrich, A-7002), pH 6.75. This basic medium was supplemented with 1 mM KNO3 as a sole nitrogen source for all the treatments. Ethylene biosynthesis inhibitor AVG (aminoethoxyvinylglycine, sigma-aldrich, A 6685) was dissolved in sterile water to final stock concentration of 10 mM. Adequate volumes of chemicals were added to 50 ml autoclaved cultured medium with or without 1 mM Glutamate in a falcon tube, mixed and then poured into Petri dishes under laminar sterile air flux. After addition of two-day-old germinated plantlets, Petri dishes were half sealed with adhesive tape. The dishes were placed vertically in a growth chamber at 22°C under 16/8 light/dark regime with a light intensity of 200 μmol. m-2 s-1 and 70% relative humidity.
Morphometric Analyses of Root System Length and Cotyledon Surface Area
Different concentrations of AVG (0.5; 1; 5; 10; 20 μM) or treatments with 10 μM AVG and 10 μM AVG + 1 mM glutamate in the presence of 1 mM KNO3 were applied to the seedlings during 5 d as described by Leblanc et al., 2008.
Effects of treatments on the exploratory root system (primary and lateral roots) and shoot area were measured every day during a kinetic of 5 days or after 120 h of treatment. For each treatment four repeats corresponding to four different agar plates comprising 4 plantlets were harvested. The root and shoot parts of the plantlets were excised. Plantlet roots were washed in 1 mM CaSO4 solution for 1 min at room temperature before to be placed in demineralized water solution and analyzed by WinRHIZO scan system. Shoots were placed on transparent slide and photocopied (copier Kyocera Mita, KM-2030) to further quantify the total leaf area by WinRHIZO scan system (Regent Instruments inc., Quebec, Canada). After these treatments, the roots and the shoots were dried on tissue paper and placed into 2 ml Eppendorf tubes. The tubes were weighed to get root and shoot fresh weight. Then, tubes were dried in oven for 72 h at 60°C and root and shoot parts were weighed to get their dry weight. Before isotope analyses, the shoots and roots were ground separately for 2 min to fine powder with inox beads of 0.4 mm diameter in oscillating grinder (Retsch mixer mill, MM301).
Net K15NO3- Uptake and Isotope Analysis
Net uptake of NO3- and glutamate was obtained by 15N labeling. For each point of the time-course study (24, 48, 72, 96 and 120 h) the medium was supplemented with K15NO3 or K 15NGlutamate (atom% 15N: 1%). The total 15N amount was determined for roots and shoots. Then, the 15N absorption and translocation were calculated as 15N μg .cm-1 root length and 15N μg shoot/(15Nμg shoot + 15Nμg root)*100, respectively. The 15N analyses were performed using an analyser (EA 3000, Eurovector, Milan Italy) coupled with a isotopic mass spectrometer (isoprime X, GV instrument, Manchester, UK).
Amino Acids Profiling
Amino acid profiling was performed on shoot and root material using the ACQUITY Ultra Performance LC (UPLC) separation system (Waters corp., Milford, USA). Plant tissues were collected, freeze-dried and homogenized with a 4-mm steel ball for 1 min at 30/s frequency. Mehtanol-chloroform-water-based extractions were made on 10 mg of the resulting dry powder. The powder was suspended in 400 µl of Mehtanol containing 200 µM DL-3-aminobutyric acid (BABA) as an internal standard and agitated at 1,500 rpm for 15 min at room temperature. 200 µl of chloroform was then added and samples were agitated at 1,500 rpm for 5 min at room temperature. Finally, 400 µl of ultra-pure water was added and samples were vigorously mixed and then centrifuged at 13,000 g for 5 min at 4°C. The upper phase containing AA was transferred to a clean microtube, dried under vacuum and the dry residue was resuspended in 600 µl of ultra-pure water. A 5 µl aliquot of the resulting extract was used for derivatization according to the AccQ•Tag Ultra Derivatization Kit protocol (Waters corp., Milford, USA) and then derivatized AA were analysed using an ACQUITY UPLC® system (Waters corp., Milford, USA). One µl of the reaction mix was injected onto an ACQUITY UPLC BEH C18 1.7 µm 2.1 × 100 mm column heated at 55°C. Elution of AA was performed with a mix of 10-fold diluted AccQ•Tag Ultra Eluent (A) and acetonitrile (B) at 0.7 ml.min-1 flow according to the following gradient: initial, 99.9% A; 0.54 min, 99.9% A; 6.50 min, 90.9% A, curve 7; 8.50 min, 78.8% A, curve 6; 8.90 min, 40.4% A, curve 6; 9.50 min, 40.4% A, curve 6; 9.60 min, 99.9% A, curve 6; 10.10 min, 99.9% A. AA were detected at 260 nm using a photo diode array detector and were subsequently identified and quantified with the individual external standard calibration curves.
Results
Chronic Treatments With Increasing AVG Concentrations Reduce the Root Elongation Without Altering the Shoot Growth
Changes in the exploratory root system elongation (primary and lateral roots) and shoot surface area expansion in B. napus plantlets were examined on vertical agar plates under a homogeneous supply of 1 mM nitrate with different concentrations of AVG (0.5 to 20 μM) after 120 h of treatment (Figure 1). AVG-treated plantlets exhibited a biphasic dose-response curve with an increase in root elongation below 1 μM followed by a sharp reduction in exploratory root system development compared to control plantlets AVG-untreated (Figure 1A). Roots grown with 10 and 20 μM AVG reduced roughly 18.6% and 59.7% their root elongation compared to roots grown without AVG. The dose-dependent reduction in the elongation of exploratory root system was accompanied by a progressive and significant reduction in root fresh and dry weights (f.wt and d.wt) as indicated by high linear correlations obtained between root length and root f.wt and d.wt (Figures 1B, C). Although AVG concentrations did not significantly affect shoot surface expansion, AVG induced a significant increase in leaf biomass resulting in an increase in leaf specific mass (weight per cm2, data not shown) as previously observed (Leblanc et al., 2008). Therefore, the differential morphological effects measured between shoot and root raised the question: does AVG treatment induce a profound remodeling of N and C metabolism in the roots and shoots irrespective of the inhibition of ethylene and IAA biosynthesis?
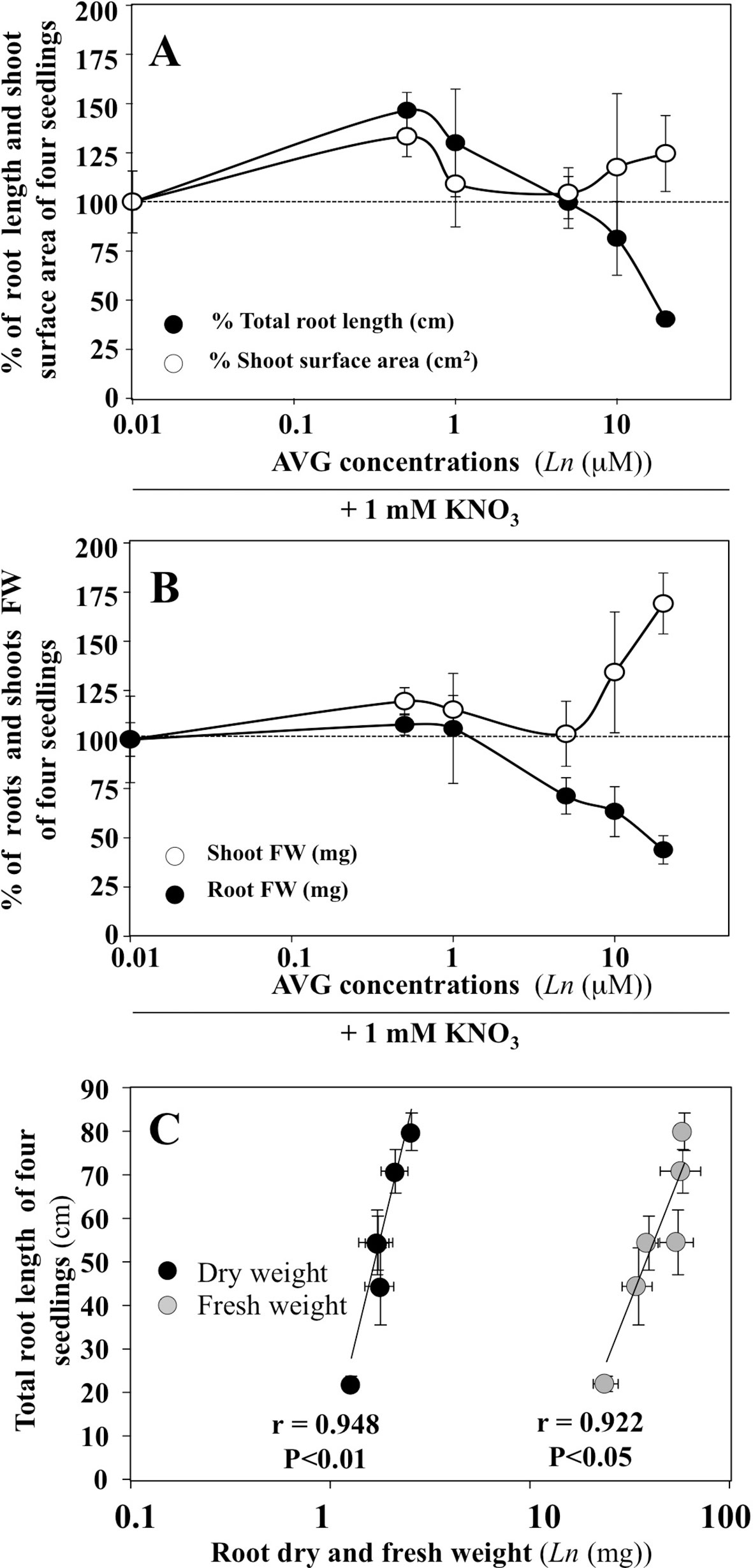
Figure 1 Impacts of AVG treatments on growth of Brassica napus seedlings treated over 5 days on agar plates under homogeneous feeding of 1 mM KNO3. (A) Dose-response curves of AVG treatments on % of total root length and shoot surface area compared to control seedlings. (B) Dose-response curves of AVG treatments on % of shoot and root FW compared to control seedlings. (C) Correlation between total root length and root dry and fresh weight. Values are the average ( ± SE) of 4 agar plates (N 4) of four seedlings each. Significant differences between control (1mM KNO3) and treatments are given for *p < 0.05; ***p < 0.005; (t-test).
Root Elongation Is Significantly Inhibited by AVG Treatment But Partially Restored by 1 Mm L-Glutamate Treatment
Dynamics of the inhibition of the exploratory root system elongation by 10 μM AVG treatment were followed during the time course of experiment (Figure 2A). Root growth inhibition by AVG became significant only after 72 h of treatment. Until 72 h, the plantlets may have heterotrophic growth and develop only a primary root from their cotyledon lipid reserves. Then, the lateral roots (LRs) appear with the greening of cotyledons and the beginning of autotrophic growth of the plantlets (Figure 2A; Marek and Stewart, 1992). Co-treatment of the plantlets with 10 µM AVG + 1 mM Glu, a major amino acid in N assimilation, induced a restoration of the length and biomass of exploratory root system after 72 h of treatment (Figures 2A, B). Glutamate was chosen because of its central position in N and AA metabolism in plant after N assimilation by the GS/GOGAT cycle. This partial morphological recovery indicates that the AVG inhibition spectrum is likely to extend beyond enzymes involved in biosynthesis of ethylene and IAA such as ACS and TAA (Figures 2A, B), while other PLP-dependent enzymes of α family involved in N metabolism are also the potential targets of AVG inhibition (Berkowitz et al., 2006).
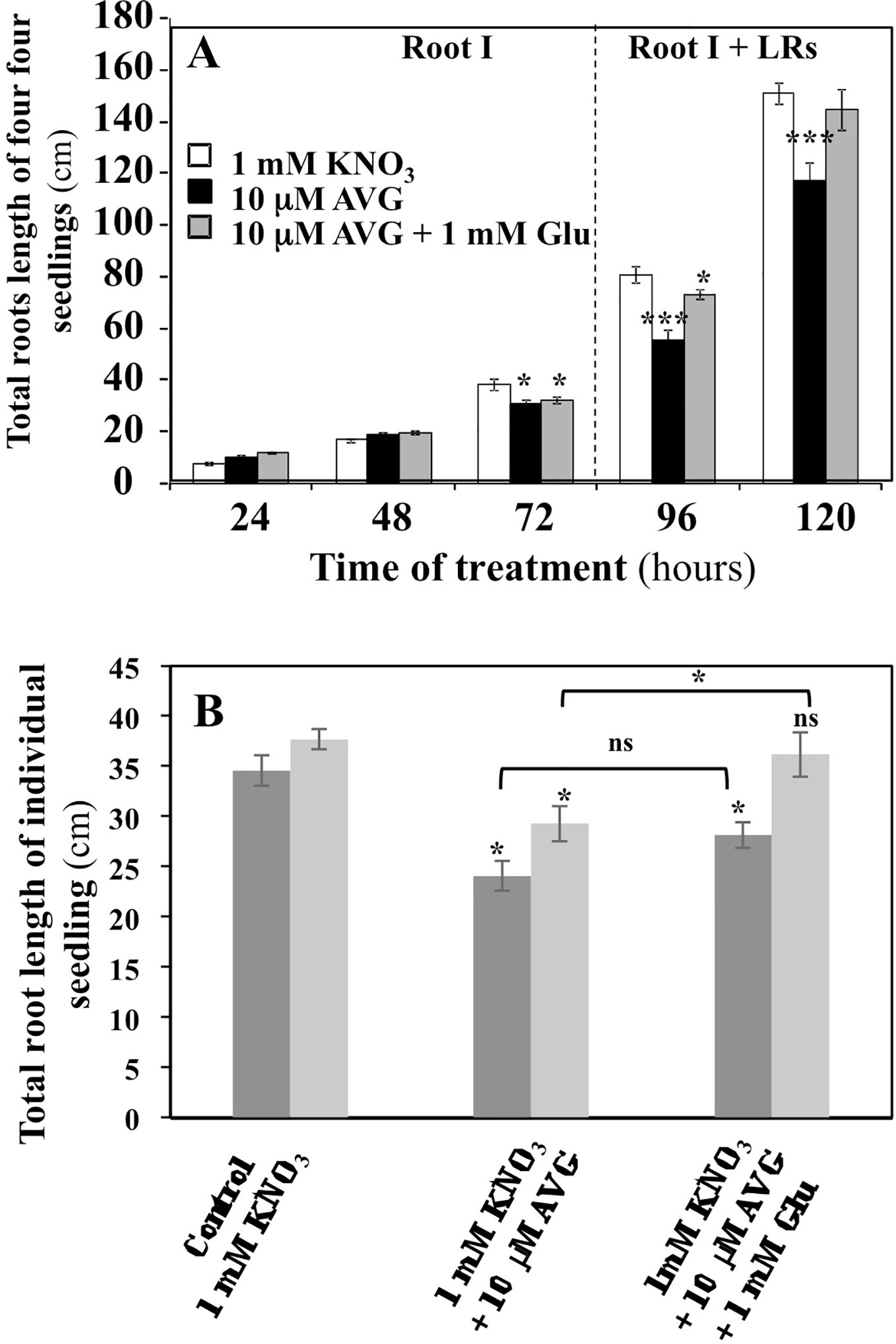
Figure 2 Effects of chronic treatment by L-glutamate on the restoration of root length in AVG-treated seedlings growing over 5 days on agar plates under homogeneous feeding of 1 mM KNO3. (A) Restoration of root length induced by 1 mM glutamate treatment in AVG-treated seedlings during time course of experiment. Values are the average ( ± SE) of 4 agar plates (n = 4) of four seedlings each. (B) Partial restoration of seedlings root length induced by 1 mM glutamate treatment on 10 μM AVG-treated seedlings after 120 h of treatment between two different and independent experiments. Values are the average ( ± SE) of 11–16 plantlets. Significant differences between control (1 mM KNO3) and treatments are given for *p < 0.05 (t-test).
AVG Treatment Increases K15NO3 Uptake Whereas AVG + K15Nglutamate Treatment Inhibits K15NO3 Uptake and Increases 15Nglutamate Uptake
In order to better characterize the root morphological inhibition and restoration effects induced by AVG and AVG + Glu treatments in relation with N uptake and allocation, a differential 15N labeling of nitrate (K15NO3) and glutamate (K15NGlu) was used to examine 15N absorption and allocation issue from nitrate or glutamate in the plantlets after 120 h of treatment (Figure 3). 15NO3- accumulation revealed that AVG treatment induces a significant increase in 15NO3 uptake and 15N translocation to the shoots (Figure 3B). Accordingly, compared to control (KNO3) in KNO3 + AVG treatment the shoot/root ratios of N and C contents were significantly increased (Figure 3A). By contrast, in co-treated plantlets with 1 mM K15NO3 + 10 μM AVG + 1 mM Glu, 15NO3 uptake and 15N translocation were significantly decreased compared to control whereas in 1 mM KNO3 + 10 μM AVG + 1 mM K15NGlu, 15NGlu uptake and 15N translocation were significantly increased (Figure 3B). The high 15N accumulation in the shoots (90%) in plantlets treated with AVG + 1 mM K15NGlu indicated that the 15NGlu taken up by the roots was rapidly translocated to the shoots for assimilation and/or storage. This result is consistent with the compensation mechanism between KNO3 and KNO3 + Glu uptake observed in a previous work (Leblanc et al., 2013). Indeed, when Glu availability increases, 15NGlu is strongly taken up and translocated to the shoots whereas nitrate uptake is progressively inhibited (Leblanc et al., 2013). This ensures the maintenance of a high N status of the plant under a mineral or organic N supply. Since Glu treatment also inhibits nitrogen assimilation in roots and shoots (Leblanc et al., 2013), this suggests that a qualitative reprogramming of N metabolism is induced during an organic N supply like Glu. To highlight the effects of AVG and AVG + Glu treatment on changes in the N metabolism and root and shoot growth, changes in total free AA contents were determined after 5 days of treatment by ultra high-performance liquid chromatography (UPLC) analysis.
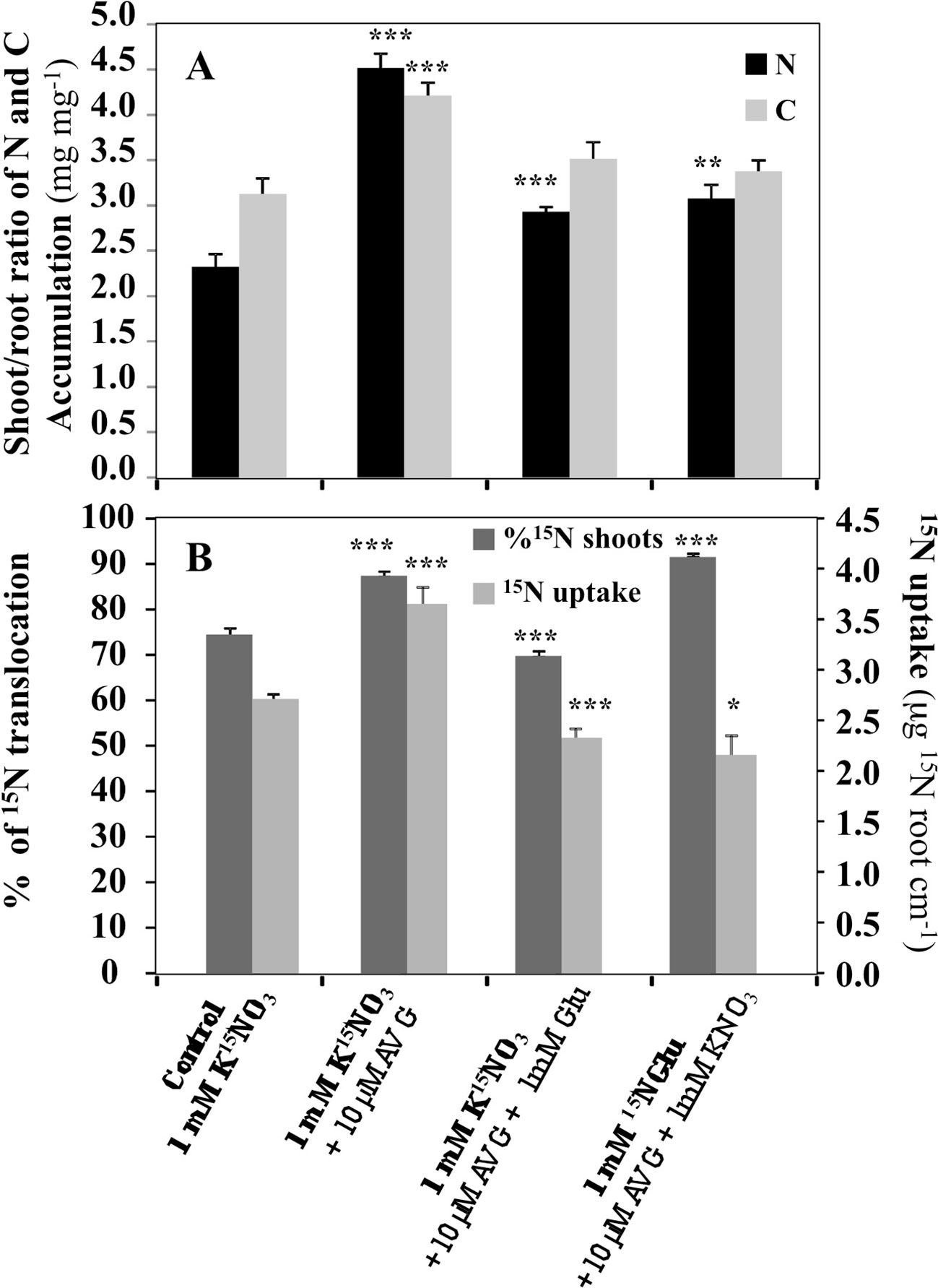
Figure 3 Comparison of AVG and AVG + Glu treatments on the shoot/root ratio of total amounts of nitrogen and carbon (A) and 15N uptake and translocation (B) in Brassica napus seedlings treated over 5 days on agar plate with 1 mM KNO3 (control). 15N uptake was measured either by treating the seedlings with K15NO3 or 15NGlu. Values are the average ( ± SE) of 4 agar plates (n = 4) of four seedlings each. Significant differences between control and treatments are given for *p < 0.05; **p < 0.01; ***p < 0.005; (t-test).
Inhibition of the Aminotransferase Network by AVG Treatment Creates a Physiological Situation of N Limitation
The effect of a high range of external concentrations in nitrate on the content of total free amino acids in the roots and shoots of B. napus seedlings growing in agar plates has been previously determined (Le Ny et al., 2013). Amino acids are the first products of nitrate assimilation and varied in the roots and shoots depending on nitrate supply and the plant N status (Figure 4A). Low N availability, decreased the translocation of 15N to the shoot compartment and was associated with a depletion of AA levels in shoot tissues, while AA levels were maintained or increased in the root. Conversely, when increasing nitrate availability, the restoration of high 15N translocation to the shoot was associated with a two-fold increase in AA levels in the shoot and a decrease in the root (Le Ny et al., 2013). So, we used the root/shoot ratio of total free amino acids (Figure 4B) to define a synthetic parameter of global N status of B. napus seedlings in a steady-state growth under homogeneous feeding of nitrate. This parameter allows characterising the effect of AVG and AVG + Glu treatments on the plant N status through aminotransferase network activities in relation with nitrate availability (Figure 4C). Thus, high ratio values indicate a physiological situation corresponding to a N limitation with an increase in total AA in the root and a decrease in the shoot (Figures 4A, B). As shown in Figure 4C, treatment with AVG induces a near-deprivation nitrogen limitation condition equivalent to 0.2 mM KNO3 availability. This corresponds to a 5-fold reduction in the nitrate concentration perceived by the roots (Figure 4C) while AVG + Glu treatment induced a near-excess nitrogen condition equivalent to 4.8 mM KNO3 availability (Figure 4C). To highlight the qualitative and quantitative changes of N metabolism induced by AVG and AVG + Glu treatment, the individual free AA contents of the roots and shoots were then analysed.
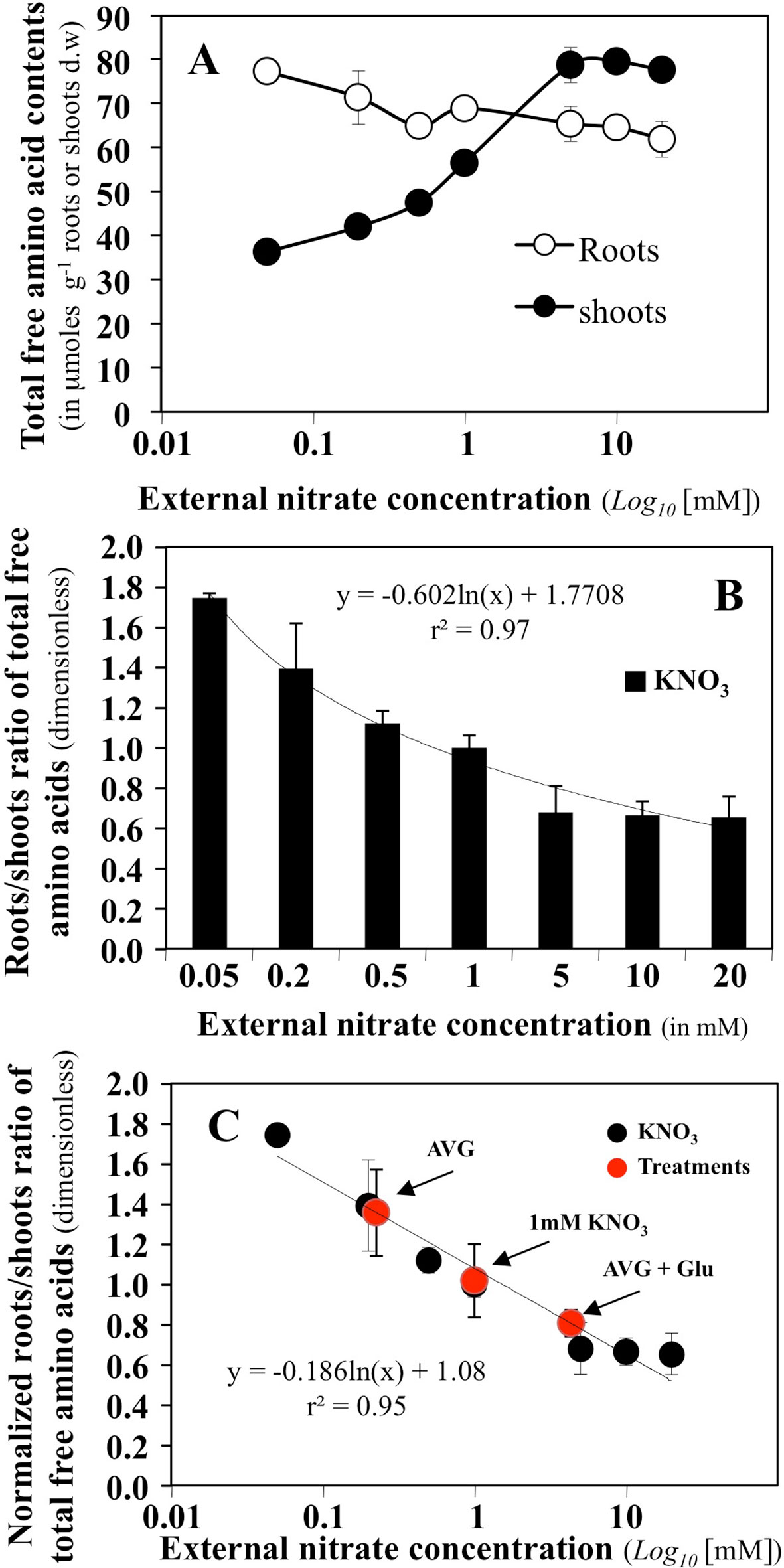
Figure 4 Determination of a synthetic N status parameter from variations of total amino acid contents in the roots and shoots of Brassica napus seedlings growing over 5 days on agar plates under homogeneous feeding of nitrate. (A) Changes in total free amino acid contents in the roots and shoots of B. napus seedlings treated with different external nitrate concentrations. (B) Roots/shoots ratio of total free amino acids defined as a synthetic parameter of N status of B. napus seedlings. (C) Normalized N status parameter from 1 mM KNO3 treatment and estimation of the values of this parameter for seedlings treated with 10 μM AVG + 1 mM KNO3 and 10 μM AVG + 1mM Glu + 1 μM KNO3.Values are the average ( ± SE) of 3 agar plates (n = 3) of four seedlings each.
The Nitrogen Assimilation Pathway is More Impaired by AVG Treatment in the Root Than in the Shoot
The effect of AVG treatment on the content of individual free AA was more severe in roots than in shoots (Figure 5). In roots, AVG treatment induced a significant increase of the three major AA: glutamine, aspartate and glutamate (Figure 5A). Since glutamine synthase (GS) and glutamate synthase (GOGAT) are not PLP-dependent enzymes, the increase in glutamine, glutamate and NH4+ contents in root tissues must be caused by inhibition in the protein synthesis or non-specific inhibition of some aminotransferases belonging to the aminotransferases network downstream GS/GOGAT cycle. Likewise, the restoration of glutamine and NH4+ concentrations to control levels following glutamate treatment can be explained by the reduction of nitrate uptake rate that in turn induces a decrease in NH4+ and glutamine production by GS/GOGAT cycle (Figures 3B, 5B). In addition, the accumulation of significantly higher levels of glutamate is caused to higher absorption of K15NGlu during AVG + Glu treatment (Figures 3B, 5A). The observed increase in aspartate levels could result from a differential inhibition of both types of AAT by AVG (Maeda et al., 2011; Maeda and Dudareva, 2012; de la Torre et al., 2014). Indeed, the plastidic (p) Eukaryotic Type (ET-pAAT) and Prokaryotic Type (PT-pAAT) of AAT belong to the subfamily Iα and Iβ respectively and present only 15% identity in the protein sequence between their members (Jensen and Gu, 1996). Alternatively, inhibition by AVG of the cystathionine β-lyase (CbL) and ACS activities may explain decrease in the use of Asp and increase in the root Asp levels (Clausen et al., 1997; Ravanel et al., 1998). Likewise, cellular compartmentation of AAT isoenzymes localised within cytosolic, plastidial, peroxisomal and mitochondrial compartments would explain aspartate accumulation in AVG-treated plantlets and the lack of recovery after AVG + Glu treatment (Figure 5A).
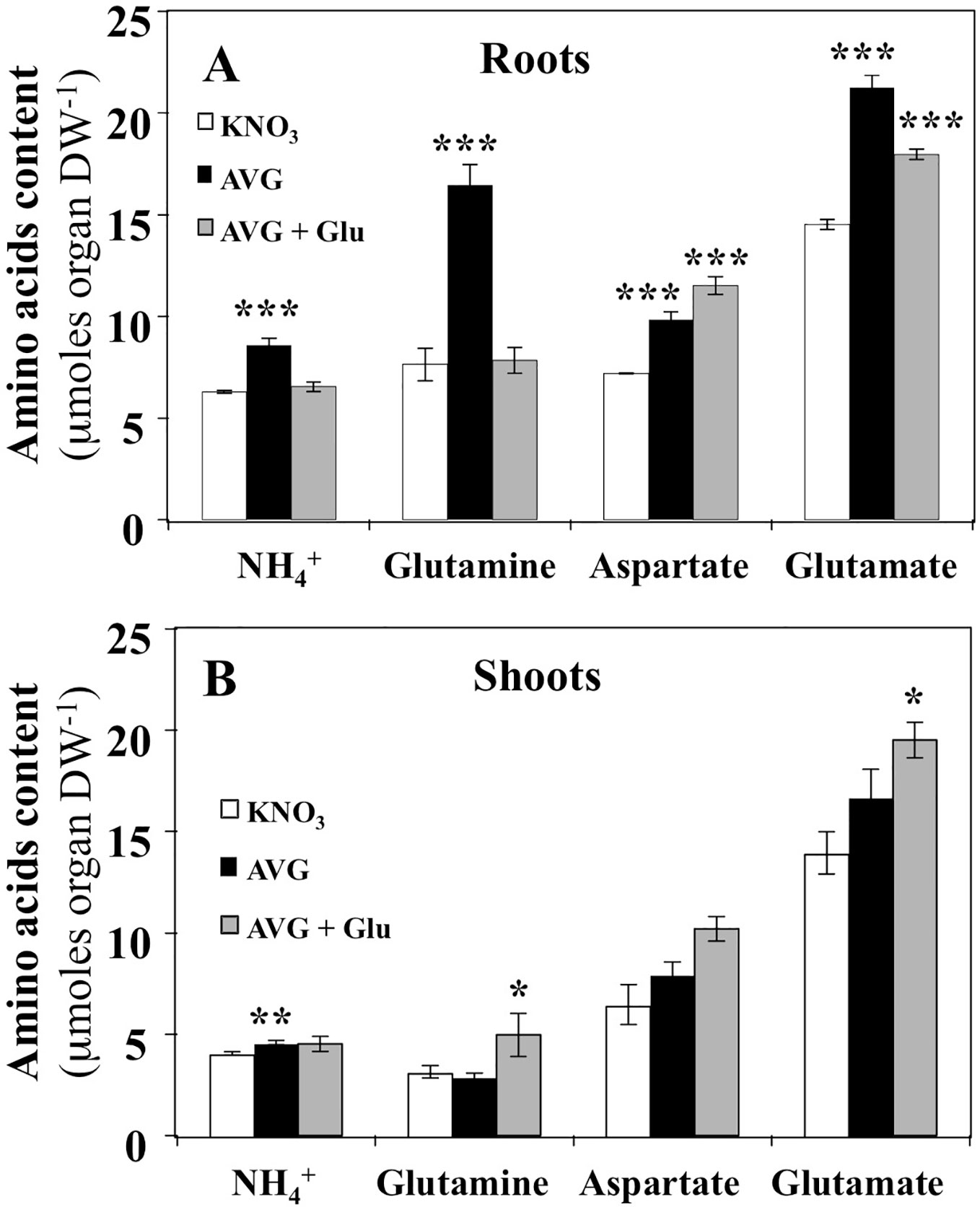
Figure 5 Changes in NH4+ and free amino acids contents induced by 10 μM AVG and 10 μM AVG + 1 mM Glu treatments in the root and shoot tissues of Brassica napus seedlings treated over 5 days on agar plates under homogeneous feeding of 1 mM KNO3. (A) Changes of NH4+, glutamine, aspartate and glutamate contents in the root tissues. (B) Changes of NH4+, glutamine, aspartate and glutamate contents in the shoot tissues. Values are the average ( ± SE) of 3 agar plates (n = 3) of four seedlings each. Significant differences between control (1mM KNO3) and treatments are given for *p < 0.05; **p < 0.01; ***p < 0.001; (t-test).
Variations Induced by AVG and AVG + Glu Treatments on the Precursor Levels of Ethylene and Auxins
Since ethylene and auxins (IAA and PAA) play a central role in root growth and development (Muday et al., 2012; Sugawara et al., 2015; Le Deunff, 2018), we checked whereas significant changes in contents of their precursors methionine, tryptophan and phenylalanine respectively, could explain the observed differences in root elongation between treatments (Figure 6). Indeed, ACS and TAA1/TARs are cytosolic aminotransferases targeted by AVG, Rhizobitoxine and AOA inhibitors (Lieberman et al., 1974; Hanson and Kende, 1976; Berkowitz et al., 2006; Soeno et al., 2010). ACS is a PLP-dependent enzyme that catalyzes the irreversible conversion of S-adenosyl-methionine into 1-aminocyclopropane-1-carboxylic acid (ACC, ethylene precursor) and 5-methylthio-adenosine. TAA1/TARs are PLP-dependent enzymes essential in indol-3-pyruvic acid (IPA) branch of the auxins biosynthetic pathway (Stepanova et al., 2008; Tao et al., 2008). They convert tryptophan and phenylalanine with α-ketoglutarate or pyruvate to indol-3-pyruvic acid and phenylpyruvic acid corresponding respectively to the precursors of IAA and PAA (Sugawara et al., 2015). In roots of AVG-treated plantlets, methionine and phenylalanine contents were significantly increased (Figure 6A), while in shoots only methionine showed slight and non-significant accumulation (Figure 6B). A recovery of methionine and a significant decrease in phenylalanine contents in roots was induced by AVG + Glu treatment (Figure 6A). However, tryptophan concentrations were unchanged in the roots and shoots regardless of the treatment used (Figures 6A, B). To highlight the quantitative changes in the levels of methionine, tryptophan and phenylalanine with respect to the nitrogen metabolism and shoot and root morphological changes, the AA levels induced by the treatments were repositioned on the responses curves obtained across a whole range of external nitrate concentrations.
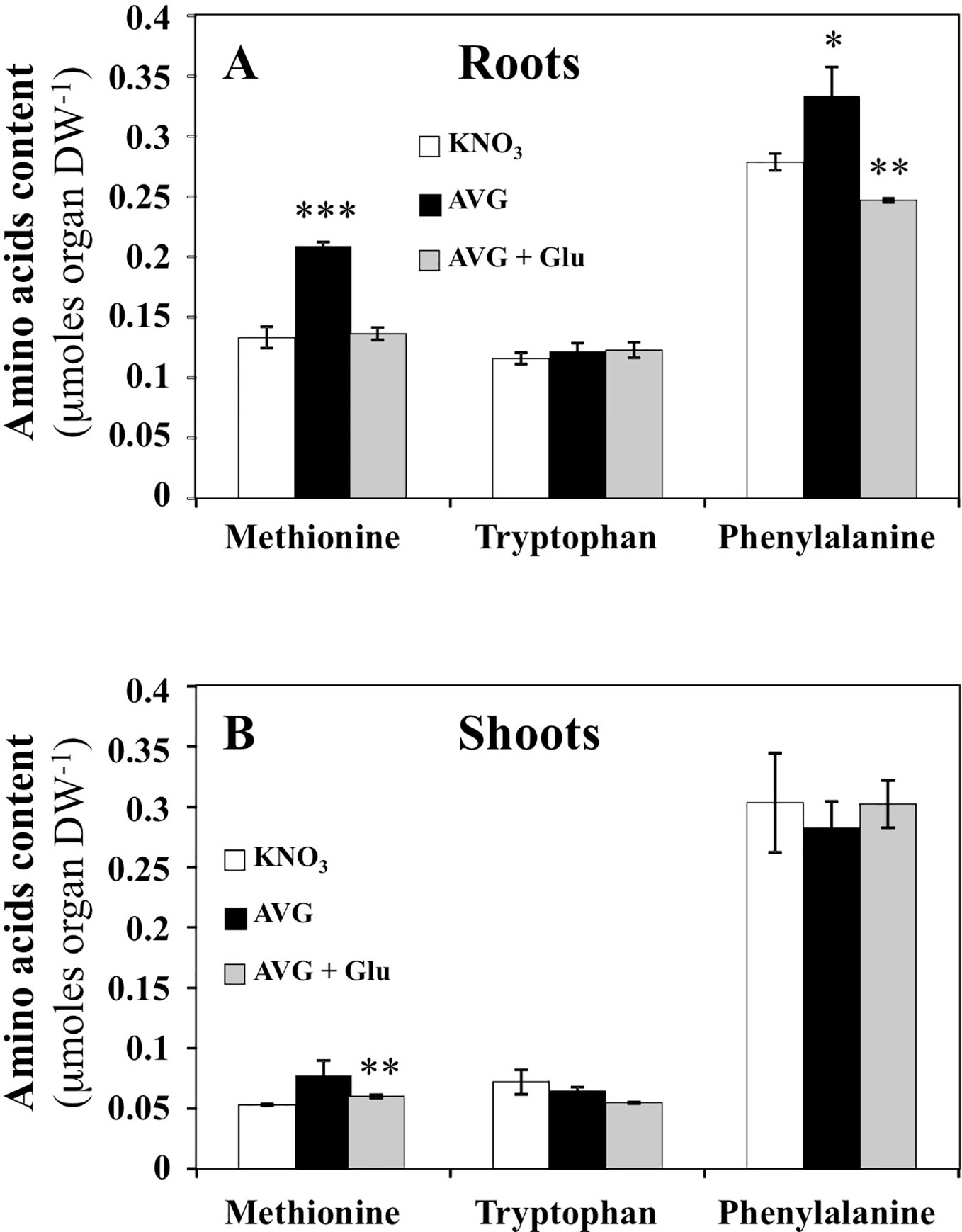
Figure 6 Changes in free amino acids contents induced by 10 μM AVG and 10 μM AVG + 1 mM Glu treatments in the root and shoot tissues of Brassica napus seedlings treated over 5 days on agar plates under homogeneous feeding of 1 mM KNO3. (A) Changes of methionine, tryptophan and phenylalanine contents in the root tissues. (B) Changes of methionine, tryptophan and phenylalanine contents in the shoot tissues. Values are the average ( ± SE) of 3 agar plates (n = 3) of four seedlings each. Significant differences between control (1 mM KNO3) and treatments are given for *p < 0.09; **p< 0.01; ***p < 0.001; (t-test).
Changes in Trp Levels Are significantly Correlated With Morphological Changes and Nitrogen or Carbon Accumulation in the Shoots and Roots
Relationships between changes in Met, Trp and Phe levels with common indicators of root and shoot growth, such as the N translocation and accumulation, shoot surface area, root length, fresh and dry weights in response to a whole range of external nitrate concentrations have been systematically sought from a previous study (Le Ny et al., 2013). Significant correlations with shoot and root growth changes were only found for changes in Trp levels (Figures 7 and S1). Figure 7 showed that changes in Trp contents in the shoots are significantly correlated with 15N translocation (Figures 7A, B) and 15N accumulation (Figures S1). The same is not true for the roots where changes in Trp content are mainly related to variations in root DW and length (Figures 7C, D and S1). After normalization of all the graphs relative to 1 mM KNO3 treatment, the values obtained after treatments with 10 μM AVG + 1 mM KNO3 and 10 μM AVG + 1 mM Glu + 1 mM KNO3 were positioned on the N response curves.
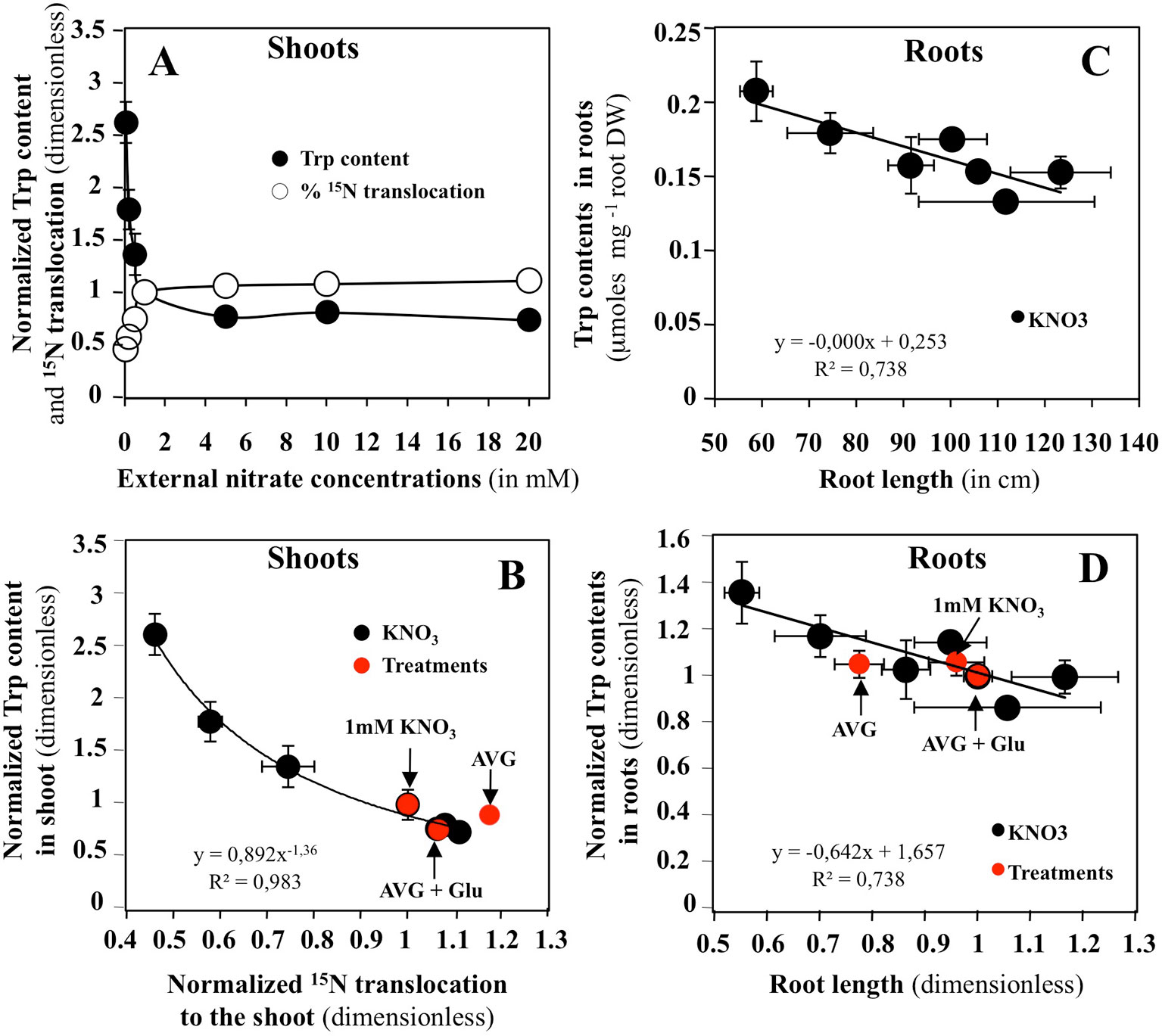
Figure 7 Effects of nitrate on different roots and shoots bichemical and morphological variables in Brassica napus seedlings treated with varying external nitrate concentrations for 5 days on agar plates. (A) Responses of Trp content and % of 15N translocation in shoots to increasing nitrate concentrations. (B) Relationship between the shoots Trp content and % of 15N translocation after normalization of the values relative to 1 mM KNO3 (control). (C) Relationship between the roots Trp content and the root length. (D) Same relationship as in C but the values have been normalized relative to 1 mM KNO3 (control). Values are the average ( ± SE) of four repeats of four seedlings each. Red points correspond to estimation of the values of same variables in another experiment for seedlings treated with 1 mM KNO3, 10 μM AVG + 1 mM KNO3 and 10 μM AVG + 1mM Glu + 1 mM KNO3.
In shoots, the results confirm AVG treatment is associated with an increase in nitrate uptake and translocation rates (Figure 7B). Intriguingly, AVG induced a translocation rate of N greater than 87% (Figure 7B). Such a translocation rate is only found in seedlings treated with more than 20 mM nitrate. Indeed, the control seedlings treated with 1 mM KNO3 have a translocation rate of 75%. Even AVG + Glu treatment fails to induce such a level of translocation rate (87%). AVG treatment leads to a decrease in Trp content of the shoots up to a 2 mM KNO3 treatment, whereas the Trp level induced by AVG + Glu treatment is equivalent to approximately 6 mM of treatment with KNO3 (Figure S1B). Similarly, AVG + Glu treatment increases the amount of 15N uptake and translocation provided by nitrate and Glu and provoked a greater decrease in Trp levels in the shoots. Because shoot growth is not impaired by the treatments, the results suggest that in shoots, the alternative IAA biosynthesis pathways are used from Trp to produce IAA necessary for growth. In roots, Trp levels were mainly correlated with root DW and root length (Figures 7C, D and S1). The results showed that AVG treatment induced a reduction in root length and DW associated with an accumulation of Trp, whereas AVG + Glu treatment resulted in an increased length of the root associated with a decrease in Trp level (Figures 7C, D). This leads to the conclusion that the alternative biosynthesis pathways of IAA are not or little used in roots to compensate for AVG inhibition of TAA catalytic functions. In order to find other specific determinants induced by AVG in inversion of shoot and root growth compared to treatment with increasing nitrate concentrations, changes in individual levels of the other AAs in roots and shoots have also been analyzed.
AVG Treatment Impairs the Content in Most Amino Acids in the Roots More Sharply Than in the Shoots
Contents of the other AA showed that roots are more responsive than shoots to AVG treatment (Figure 8). In addition, in most cases glutamate treatment induced a recovery of AA in the roots to the control levels (Figure 8). In control plantlets (1 mM KNO3), total AA content in root tissues was about 1.8-fold higher than total AA content in the shoots (Figure 8). This result was in agreement with previous AA profiling obtained with the same B. napus line grown in similar conditions (Figure 4A, Le Ny et al., 2013). However, relative to external nitrate concentrations, the 1 mM KNO3 + 10 μM AVG treatment provoked a quantitative effect on the N metabolism balance between the shoots and roots as previously exemplified in Figures 4C. In the root tissues, significant increases in levels of several AA such as serine (Ser), threonine (Thr), α-alanine (Ala), β-alanine (β-Ala), valine (Val), asparagine (Asn) and histidine (His) were observed in AVG-treated plantlets relative to controls (Figure 8A). In plants, AA can be grouped depending on whether their carbon skeleton originates from glycolytic, TCA cycle or the oxidative pentose phosphate pathway (Figure S2, Coruzzi 2003; Stitt et al., 2002). As most of these AA are the end products of several metabolic pathways (Figure S1), this leads to the conclusion that PLP-dependent aminotransferases involved in their biosynthesis were strongly inhibited by root AVG treatment (Figure 8A). As the contents of almost all AA increased with AVG treatment, several hypotheses non-mutually exclusive may be proposed for AVG effects: i) reduction of protein synthesis, ii) induction of proteolysis, and iii) inhibition of catabolism of AA in the roots.
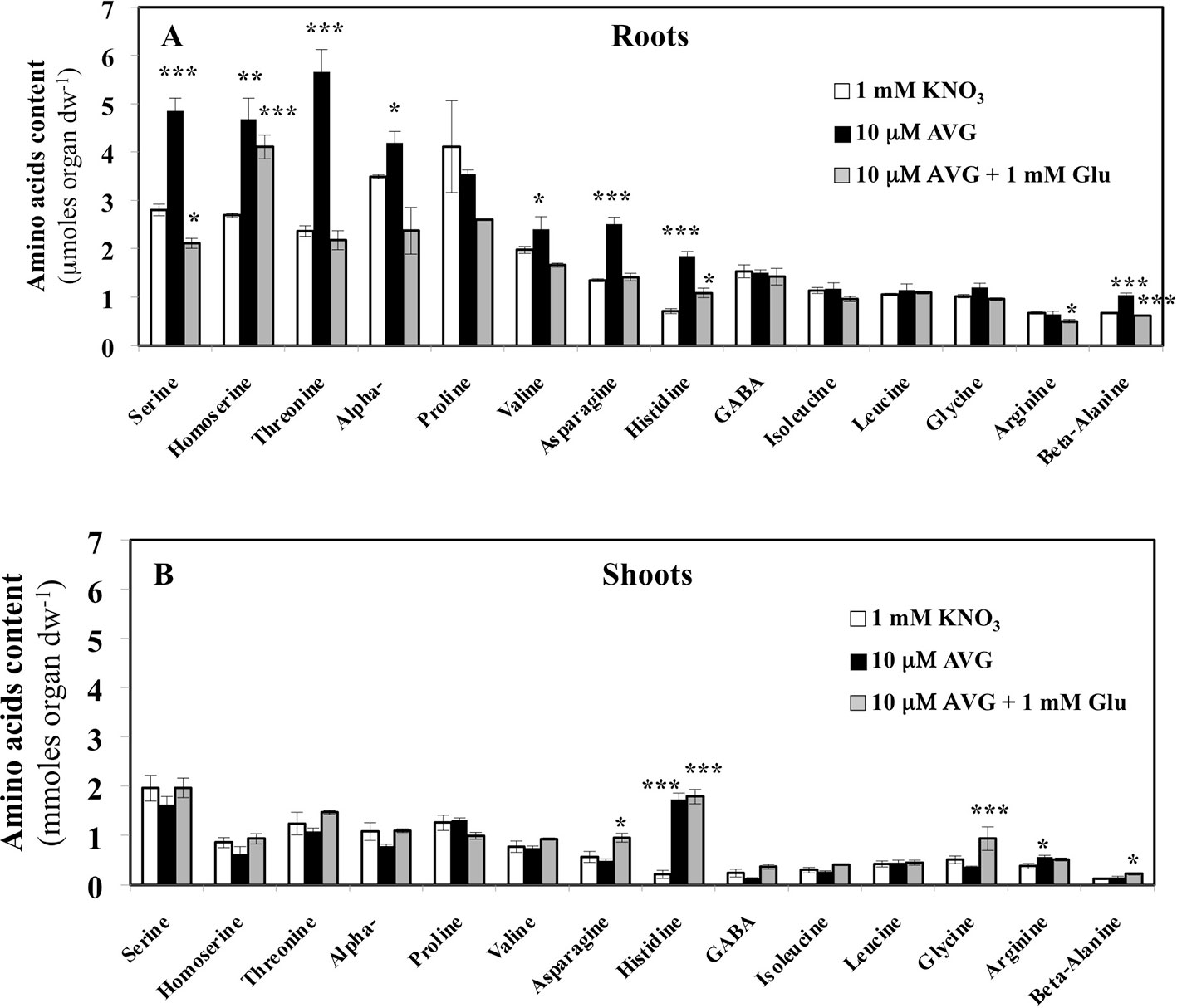
Figure 8 Changes in free amino acid contents induced by 10 μM AVG and 10 μM AVG + 1 mM Glu treatments in the root (A) and shoot tissues (B) of Brassica napus seedlings treated over 5 days on agar plates under homogeneous feeding of 1 mM KNO3. Values are the average ( ± SE) of 3 agar plates (n = 3) of four seedlings each. Significant differences between control (1 mM KNO3) and treatments are given for *p < 0.05; **p< 0.01; ***p < 0.005; (t-test).
In the shoot tissues, the free AA pools were less affected by AVG and AVG + Glu treatments (Figure 8B). This can be explained by the lowest AA content in shoots compared to the roots at 1 mM KNO3 external concentration (Figure 4A), itself further reduced by the AVG-induced N limitation (Figure 4C). The overall trend for the three treatments was opposite to the AA patterns found in the root tissues (Figures 8A, B). In particular, AA showing a slight decrease in the shoot content induced by AVG treatment were strongly and significantly increased in the roots suggesting a tight coordination of AA levels and recycling between shoots and roots by a N sensing mechanism of N status that remains to be determined. However, AVG and AVG + Glu treatments had no effect on AA levels, except for some AAs such as His, Asp, β-Ala and Gly (Figures 8B, S4). Amongst amino acid, only His showed a significant increase by 8-fold after AVG treatment and its control level was not restored by glutamate (Figure 8B). Further, relative changes in AA levels belonging to the same metabolic pathways compared to control plantlets confirmed conclusions from changes of individuals AA in the roots (Figure 9A) and shoots (Figure 9B). Repetition of the experiment gave similar results regarding changes in AA contents taken individually (Figures S3, S4) or grouped by family (Figure S5). Only levels of pentose-P family (His) in shoots were lower in this second experiment for both treatments but still significantly different (Figure S5).
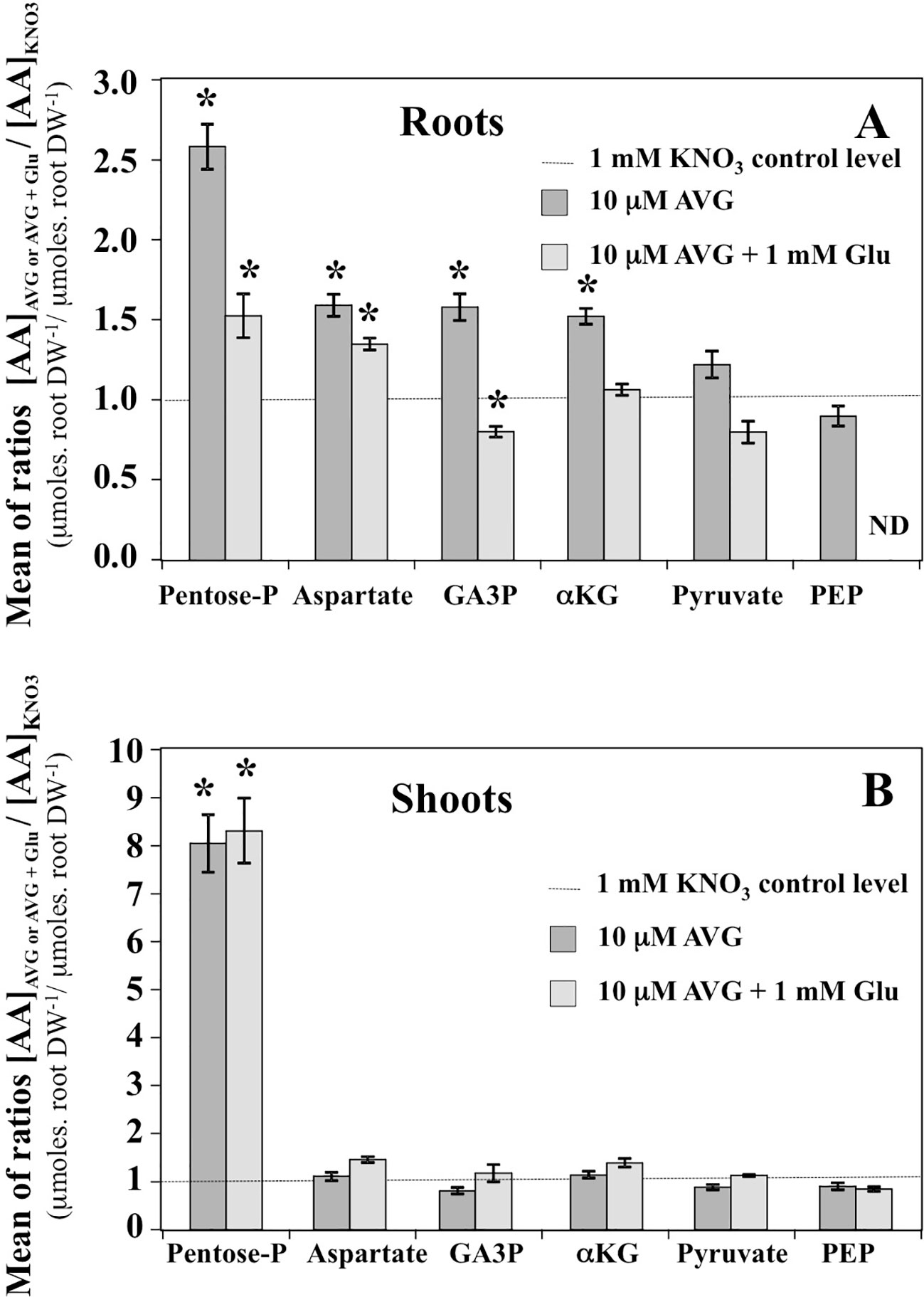
Figure 9 Mean contents of amino acid families in the roots (A) and shoots (B) tissues of Brassica napus seedlings treated with 10 μM AVG or 10 μM AVG + 1 mM Glu relative to those in KNO3 treated seedlings (control). Seedlings were growing on agar plates under homogeneous feeding of 1 mM KNO3 whatever the treatment. Values are the average ( ± SE) of 3 agar plates (n = 3) of four seedlings each. A significant difference in amino acid ratio is indicated by *p < 0.05 (Benjamini and Hochberg correction for multiple t-test analyses with a false discovery rate of 0.05 between the treatments and KNO3 control plants have been done). The composition of the amino acid families is as follows: Pentose-P (His), α-KG (Glu, Gln, Arg, GABA and Pro), Aspartate (Asn, Asp, Thr, Met, Ile and Lys), PEP (Trp, Phe, Tyr), Pyruvate (a-Ala; b-Ala, Val and Leu) and GA3P (Ser and Gly).
Discussion
Although the use of AVG inhibitor is generally associated to ethylene and IAA biosynthesis inhibition in relation with root development (Rahman et al, 2000; Le et al., 2001; Stepanova et al., 2007; Swarup et al., 2007; Růžička et al., 2007), the results demonstrate that the non-specificity of AVG inhibitor creates a physiological situation of N limitation associated with a profound remodelling of AA metabolism that impairs regulation of nitrate uptake and translocation, His homeostasis and root and shoot growth.
AVG and AVG + Glu Treatments Impair N Metabolism and Challenges the Canonical Regulation of Nitrate Uptake Rate by NH4+ and Glutamine
Despite an average 22.5% decrease in the exploratory root system, 10 μM AVG-treated plantlets accumulated as much as 15N as KNO3 control plantlets after 120 h of treatment. This finding confirms earlier data that seedlings treated with 10 μM AVG, which only have three-quarter to half of their exploratory root system and few root hairs, accumulated as much as 15N as the control seedlings (Leblanc et al., 2008; Le Deunff et al., 2016). Accordingly, relative to the total root length, nitrate uptake rate was significantly increased in AVG-treated plantlets. As previously demonstrated, the increase in nitrate uptake rate was accompanied by an over-expression of BnNRT2.1 nitrate transporter gene, the transcription level of which adapts to changes in the roots’ absorbing surface (Leblanc et al., 2008; Le Ny et al., 2013; Lemaire et al., 2013). The up-regulation of nitrate uptake can be explained by AVG induction of a physiological condition similar to a N limitation (low N status) as indicated by a high value of root/shoot ratio of total free amino acids. Indeed, N deprivation conditions is known to induce an up-regulation of NRT2.1 nitrate transporter expression (Lejay et al., 1999; Beuve et al., 2004). Interestingly, the synthetic N status parameter used allows to demonstrate that AA are not directly involved in feed-back regulation of nitrate uptake and nitrate transporters expression as previously supposed (Cooper and Clarkson, 1989; Muller and Touraine, 1992; Krapp et al., 1998; Tillard et al., 1998; Vidmar et al., 2000; Gansel et al., 2001; Beuve et al., 2004). Indeed, it is generally accepted that increase contents of NH4+ and glutamine down regulated 15NO3- influx rate and NRT2.1 expression (Vidmar et al., 2000; Nazoa et al., 2003; Beuve et al., 2004). In AVG-treated plantlets, the endogenous contents of NH4+, glutamine and glutamate were 2.6-, 1.6- and 1.5-fold higher than the control values, respectively. Likewise, restoration of NH4+ and glutamine to low levels in roots after AVG + Glu treatment induced a significant decrease rather than an increase in nitrate uptake rate.
By contrast, AVG + Glu treatment was associated with a decrease in root/shoot ratio of total AA corresponding to a theoretical supply of 4.8 mM KNO3. This glutamate-induced high N status is likely due to the N form used for N uptake and assimilation (e.g. organic versus mineral) rather than qualitative changes in individual free amino acid contents per se. In this regard, these data confirm a previous work (Leblanc et al., 2013) and indicate that a high glutamate supply inhibits nitrate uptake and increases 15NGlu uptake and translocation. Taken together, the results also suggest that Glu induces a general switch in the seedling N status regulating nitrate uptake and AA biosynthesis independently or in parallel to AVG effects.
Plants Treated With AVG Mimic Some Root Morphological and Biochemical Traits of Mine/Pdx3 Mutants
Because AVG is a strong inhibitor of TAA1/TAR activity and roots elongation in dose-dependent manner (Leblanc et al., 2008; Soeno et al., 2010), the lack of tryptophan (Trp) accumulation after AVG treatment is very intriguing (Figure 1). This could explain either from ethylene-directed IAA biosynthesis (He et al., 2011; Kim et al., 2018) and/or diversion of Trp flow to other Trp-dependent IAA biosynthetic pathways (Wang et al., 2015). Indeed, transcription factor EIN3, an ethylene-signaling component, is a major inducer of local IAA biosynthesis pathway via the transcription of α- and β-subunits of anthranilate synthase (AnS) and TAA1/TARs genes (He et al., 2011). Therefore, AVG inhibition of ethylene biosynthesis could block the transcription of EIN3, which in turn may impede the Trp biosynthesis pathway and IAA production via AnS. Similarly, ethylene-directed IAA biosynthesis is also dependant of pyridoxine 5’-phosphate/pyridoxamine 5’-phosphate (PNP/PMP) oxidase (PDX3), an enzyme involved in pyridoxal-5’-phosphate (PLP) salvage pathway and PLP homeostasis (Colinas et al., 2016; Kim et al., 2018). Indeed, in dark-grown condition, root IAA treatment restores the ethylene insensitivity of mine/pdx3 mutants suggesting that lack of PLP or increase imbalance in the ratio of PMP/PLP contents in the mutants impedes activity of TAA1/TAR necessary to produce IAA in roots (Kim et al., 2018).
Intriguingly, mine/pdx3 mutants that develop under normal conditions show strong inhibition of root growth but not shoot growth (Kim et al., 2018). This result is very similar to data obtained with AVG treatment. In fact, the differential inhibition of AVG can be explained by a diversion of Trp flow to other Trp-dependent IAA biosynthetic pathways in shoots to produce IAA required for growth. Because in the roots, these alternative pathways could be less or not used, this would lead to over accumulation of Trp. This explanation is reinforced by the significant relationships found between Trp contents with the N translocation and accumulation (Figure 7B) and morphological variables in shoots (Figure S1B). Such relationships have never been found with roots. This also supports the fact that in different species such as corn, wheat, rice and Arabidopsis TAA/TARs mutants induced pleiotropic auxin-related processes associated with shoots and roots growth (Philipps et al., 2011; Ma et al., 2014; Yoshikawa et al., 2014; Shao et al., 2017). For example, in maize, TAA (vt2) mutants are more affected in their reproductive phase than their vegetative phase (Philipps et al., 2011) and in Arabidopsis, the root growth of tar2 mutants is more altered than the shoots growth (Ma et al., 2014). Furthermore, in mine/pdx3 mutants, AA were over accumulated in rosette leaves (Colinas et al., 2016) as in N deprivation condition (Figure 4A; Le Ny et al., 2013) or after a treatment with AVG (this study). Taken together these data suggest that catalytic inhibition of PLP-dependent aminotransferases by AVG strongly mimics the AA metabolism impairment observed in mine/pdx3 mutants.
How to Explain the High Accumulation of His Contents After AVG Treatment
In both sets of experiments (Figures 8A, B and S5), His contents showed the highest increase compared to other AA in the shoots and roots (by 2.5- and 8-fold, respectively). Because histidinol-phosphate aminotransferase (HPA), the last enzyme of His biosynthesis, is a potential target of AVG (Figure S2), the increased levels of His were unexpected. Two non-mutually exclusive biochemical hypotheses could explain this result: (i) a diversion of 5’-phosphoribosyl-1-pyrophosphate (PRPP) for His biosynthesis and (ii) an inhibition of His catabolism pathway via the involvement of TAA1/TAR aminotransferases (Figure 10). The first hypothesis could be explained by a cross-regulation mechanism in the metabolic pathway of His and Trp (Stepansky and Leustek, 2006). Indeed, His is metabolically the only other amino acid with Trp (Figure S2) that derives a part of its skeleton from PRPP. PRPP is required in tryptophan biosynthetic pathway for the conversion of anthranilate to N-(5’-phosphoribosyl)-anthranilate via phosphoribosyl-anthranilate transferase (Pendyala and Wellman, 1975; Last et al., 1991). Therefore, strong increase in His levels of the roots and shoots in response to 10 μM AVG would be caused by a diversion of PRPP required for His biosynthesis, nucleotides production and biosynthesis of NAD and NADP cofactors instead of Trp biosynthesis (Figure S1 and Figure 10). This cross-pathway between His and Trp metabolism was confirmed in Arabidopsis by inhibition of imidazole glycerol-phosphate dehydratase (IGPD) catalyzing the sixth step of His biosynthesis (Guyer et al., 1995) and in Nicotiana by the over-expression of PRPP synthase (Koslowsky et al., 2008).
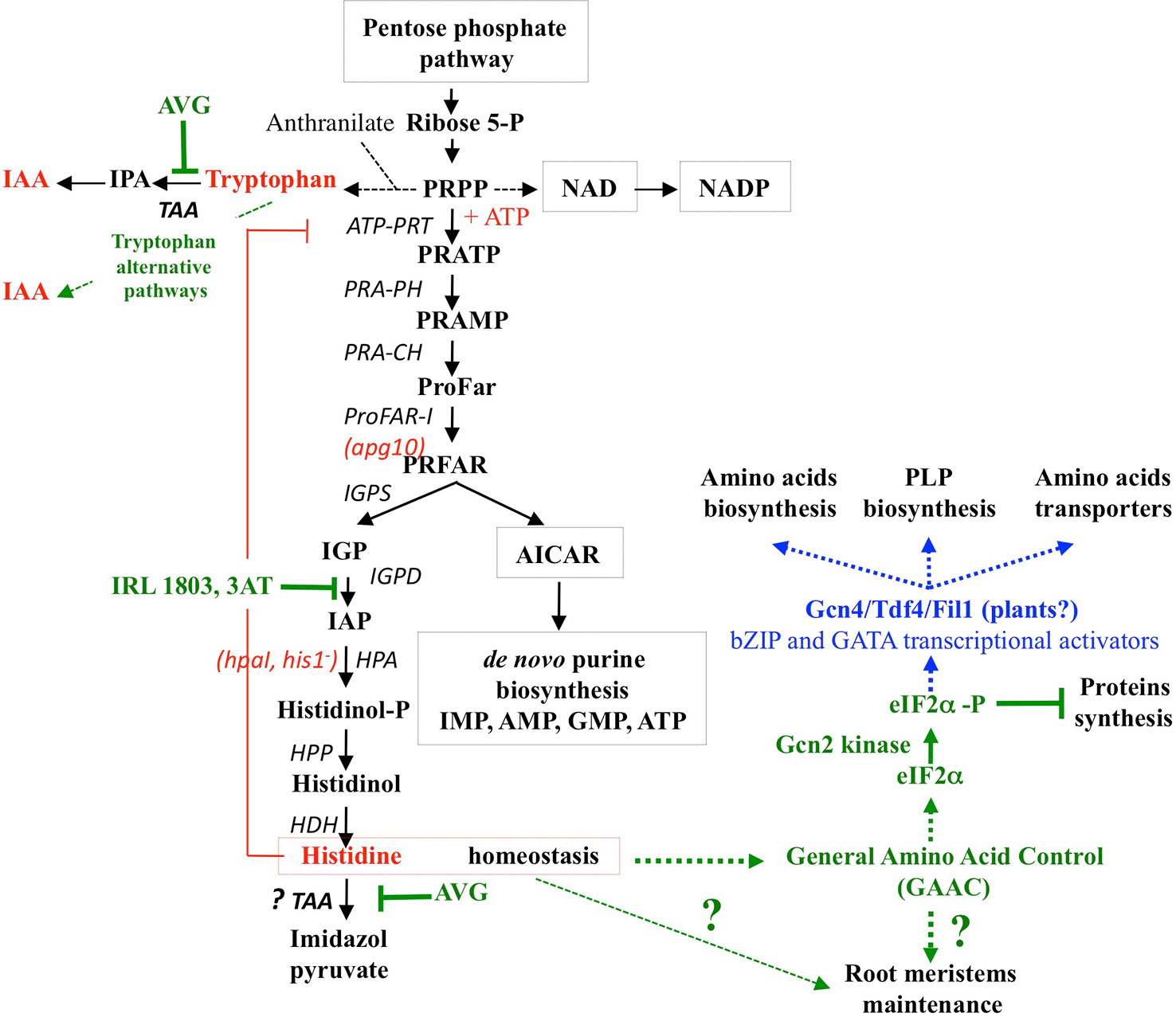
Figure 10 Inhibitors and mutants of the histidine biosynthesis pathway. Enzyme names are shown in black letters with the mutant name in red letters in parentheses. Inhibitors of Histidine biosynthesis and catabolism are designated in green letters and their effects on other metabolic or signalling pathways are indicated by green dotted arrows. 3AT: 3-aminotriazole; ATP-PRT: ATP-phosphoribosyl-transferase, PRA-PH: Phosphoribosyl-ATP Pyrophosphohydrolase, PRA-PH Phosphoribosyl-AMP Cyclohydrolase, ProFAR-I: N´-[(5´-phosphoribosyl)formimino]-5-aminoimidazole-4-carboxamide ribonucleotide isomerase, IGPS: Imidazole glycerol-phosphate synthase, IGPD: Imidazole glycerol-phosphate dehydratase, HPA: Histidinol-phosphate aminotransferase. HPP: Histidinol-phosphate phosphatase, HDH: Histidinol dehydrogenase, TAA: Tryptophan aminotransferase. Missing actors in Gcn2 kinase signaling in plants are indicated in blue. (adapted from Ingle, 2011 and Immanuel et al., 2012).
The second hypothesis could be provided by the promiscuity in TAA activity. In other words, the ability of TAA to catalyse a fortuitous side reaction such as His catabolism in addition to its main reaction. This hypothesis is also supported by the fact that the pathway of His catabolism has not yet been elucidated in plants (Ingle, 2011; Hildebrandt et al., 2015). Thus, in C. glabrata and S. cerevisiae, Brunke et al. (2014) have recently demonstrated that TAA1/TARs aminotransferases also catalyse the conversion of L-histidine and α–ketoglutarate into imidazol-5yl-pyruvate and glutamate (Figures 10, S2). Since the enzyme activity of cytosolic TAA1/TARs is strongly inhibited by AVG (Soeno et al., 2010), this second hypothesis would explain the high accumulation of His. This explanation is also corroborated by high levels of His accumulation in roots and shoots, whereas Trp-dependent pathways other than IPA pathway are only used in shoots to maintain growth. Furthermore, this hypothesis would reinforce the cross-pathway regulation of enzymes in the biosynthesis of Trp and His already observed in plants and fungi (Carsiotis and Jones, 1974; Carsiotis et al., 1974; Niederberger et al., 1981; Hinnebusch, 2005; Stepansky and Leustek, 2006).
Are AVG and Rhizobitoxine Secreted by Symbiotic and Pathogen Bacteria Only Involved in the Inhibition of Aminocyclopropane-1-Carboxylate Synthase?
Ethylene biosynthesis inhibition by AVG and Rhizobitoxine plays a major role in establishing symbiosis and increasing the nodule number between rhizobacteria and their host legumes (Berkowitz et al., 2006; Boibessot et al., 2016). However, N deprivation or low nitrate concentrations (<50 mg/L = 0.5 mM) are commonly used with AVG treatments to induce nodulation (Xia et al., 2017). Indeed, reduction of nitrogen supply such as nitrate significantly increases the number and weight of nodules (Gibson and Harper, 1985; Macduff et al., 1996; Gan et al, 2004). The nitrogen limitation of AVG-treated seedlings from 1 mM nitrate to 0.2 mM (expressed as N status) indicates that the increase in nodule numbers induced by 10 μM AVG could be caused by both deprivation of N and inhibition of ethylene biosynthesis (Peters and Crist-Estes, 1989). This suggests that the major effects of AVG and Rhizobitoxine secreted by rhizobacteria and pathogenic bacteria on the N metabolism of their host legumes have been overlooked. In this respect, Lotus NIN (Nodule INception) transcription factors have been shown to be involved in early-steps of N-regulated symbiosis between rhizobia and legumes roots (Schauser et al., 1999; Liu et al., 2019). In Arabidopsis, NIN homologous protein: NLP7 (Nin Like Protein 7) is involved in the control of nitrate and N starvation response (Castaings et al., 2009).
Is AVG-Induced Remodeling of Plant Nitrogen Status due to Inhibition of a Major N-Sensory System?
Induction of AA imbalance associated with nitrate starvation response suggest that the suicide inhibition by AVG of PLP-dependent enzymes probably mimics or impairs a major plant N sensory system. Since Histidine biosynthesis has the highest energy cost among AA and His contents exhibit the greatest variations in plantlets treated with AVG, it is tempting to assign a regulatory role of His homeostasis in these responses. Indeed, previous studies with mutant of different enzymes of His biosynthesis have demonstrated that His homeostasis is crucial for root meristem maintenance, root development and plant growth (El Malki and Jacobs, 2001; Noutoshi et al., 2005; Mo et al., 2006). Similarly, inhibition by the IRL1803 herbicide of IGPD catalysing the sixth step of His biosynthesis (Figure 10) increased the transcription of eight genes involved in the synthesis of aromatic AA, His, Lys and purines (Guyer et al., 1995). This suggests a cross-pathway regulation of AA biosynthesis genes via a general amino acid control system (GAAC) as in yeast treated by another IGPD inhibitor: 3-aminotriazole (3-AT) (Hinnebusch, 1984; Natarajan et al., 2001). Although most of the components of GAAC system mediated by the Gcn2 kinase signaling (General Control Non-derepressible-2) are conserved in all eukaryotes (Zhang et al., 2003; Zhang et al., 2008; Lageix et al., 2008; Casthilo et al., 2014), the bZIP transcription factor Gnc4/Atf4 is always the missing component of this N sensory system in plants (Lam et al., 2006; Hey et al., 2010; Immanuel et al., 2012). Recently, Fil1 a transcription factor belonging to GATA family and non-orthologous to GCN4/ATF4 bZIP family has been discovered in Schizosaccharomyces pombe (Duncan et al., 2018). In plants, two other transcription factors of GATA family such as NTL1 (NiT-2-Like) and GNC (GATA, Nitrate-inducible, Carbon metabolism-involved) are already involved in nitrate assimilation and glucose responses (Reyes et al., 2004; Bi et al., 2005).
In conclusion, AVG is not a sufficiently reliable inhibitor to validate the genetic processes controlling the effects of ethylene and ethylene/auxin crosstalk on the root and shoot growth when its concentrations exceed 5 μM (Rahman et al, 2000; Okamoto et al., 2008; Leblanc et al., 2008; Le Deunff and Lecourt, 2016; Le Deunff 2018). As shown in this study, 10 μM AVG treatment induces a N limitation condition close to deprivation associated with an increase of nitrate uptake and translocation, imbalance in AA metabolism and a deregulation of His homeostasis. These effects appear to be associated with differential use of Trp in roots and shoots to produce IAA through other biosynthetic pathways that do not utilize PLP-dependent enzymes. Therefore, the use of high AVG concentrations range could be an original and valuable tool to highlight, by using RNAseq and metabolic profiling, the major N sensory system that is involved in reprogramming aminotranferases network associated with the root and shoot morphological changes.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
EL and JL designed the research. EL, JL, PB, and CD performed the experiments. CD performed UPLC analyses. EL and CD analyzed the data. EL, JL, and CD wrote the paper.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01387/full#supplementary-material
References
Beauclair, P., Leblanc, A., Lemauviel-Lavenant, S., Deleu, C., Le Deunff, E. (2009). Ethylene modifies architecture of root system in response to stomatal opening and water allocation change between root and shoot. Plant Signal. Behav. 4 (1), 44–46. doi: 10.4161/psb.4.1.7268
Berkowitz, D. B., Charrette, B. D., Karukurichi, K. R., McFadden, J. M. (2006). α-Vinylic amino acids: occurrence, asymmetric synthesis, and biochemical mechanisms. Tetrahedron-Asymmetr. 17, 869–882. doi: 10.1016/j.tetasy.2006.02.026
Beuve, N., Rispail, N., Lainé, P., Ourry, A., Le Deunff, E. (2004). Putative role of γ-aminobutyric acid (GABA) as long distance signal in up-regulation of nitrate uptake in Brassica napus L. Plant Cell Environ. 27, 1035–1046. doi: 10.1111/j.1365-3040.2004.01208.x
Bi, Y. M., Zhang, Y., Signorelli, T., Zhao, R., Zhu, T., Rothstein, S. (2005). Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J 44 (4), 680–692. doi: 10.1111/j.1365-313X.2005.02568.x
Boibessot, T., Bénimèlis, D., Jean, M., Benfoda, Z., Meffre, P. (2016). Synthesis of a novel Rhizobitoxine-like triazol-containing amino acid. Synlett 27, A–D. doi: 10.1055/s-0036-1588300
Brunke, S., Seider, K., Richter, M. E., Bremer-Streck, S., Ramachandra, S., Kiehntopf, M., et al. (2014). Histidine degradation via an aminotransferase increases the nutritional flexibility of Candida glabrata. Eukaryot Cell 13 (6), 758–765. doi: 10.1128/EC.00072-14
Capitani, G., Hohenester, E., Feng, L., Storici, P., Kirsch, J. F., Jansonius, J. N. (1999). Structure of 1-aminocyclopropane-1- carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene. J. Mol. Biol. 294, 745–756. doi: 10.1006/jmbi.1999.3255
Carsiotis, M., Jones, R. F. (1974). Cross-pathway regulation: tryptophan-mediated control of histidine and arginine biosynthetic enzymes in Neurospora crassa. J. Bacteriol. 119 (3), 889–892.
Carsiotis, M., Jones, R. F., Wesseling, A. C. (1974). Cross-pathway regulation: histidine-mediated control of histidine, tryptophan, and arginine biosynthetic enzymes in Neurospora crassa. J bacteriol. 119 (3), 893–898.
Castaings, L., Camargo, A., Pocholle, D., Gaudon, V., Texier, Y., Boutet-Mercey, S., et al. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57, 426–435. doi: 10.1111/j.1365-313X.2008.03695.x
Casthilo, B. A., Shanmugam, R., Silva, R. C., Ramesh, R., Himme, B. M., Sattleger, E. (2014). Keeping the eIF2 alpha kinase Gcn2 in check. Biochem. Phys. Acta. 1843, 1948–1968. doi: 10.1016/j.bbamcr.2014.04.006
Christen, P., Mehta, P. K. (2001). From cofactor to enzymes. The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Chem. Rec. 1, 436–447. doi: 10.1002/tcr.10005
Clausen, T., Huber, R., Messerschmidt, A., Pohlenz, H. D., Laber, B. (1997). Slow-binding inhibition of Escherichia coli cystathionine β-lyase by L-aminoethoxyvinylglycine: a kinetic and X-ray study. Biochemistry 36, 12633–12643. doi: 10.1021/bi970630m
Colinas, M., Esenhut, M., Tohge, T., Pesquera, M., Fernie, A. R., Weber, A. P. M., et al. (2016). Balancing of B6 vitaminers is essential for plant development and metabolism in Arabidopsis. Plant Cell 28, 439–453. doi: 10.1105/tpc.15.01033
Cooper, H.D., Clarkson, D.T. (1989) Cycling of amino nitrogen and other nutrients between shoots and roots in cereals: a possible mechanism integrating shoot and root in the regulation of nutrient uptake. J. Exp. Bot. 40, 753–762. doi: 10.1093/jxb/40.7.753
Coruzzi, G. M. (2003). “Primary N-assimilation into amino acids in Arabidopsis,” in The Arabidopsis book (Rockville, Maryland: American Society of Plant Biologists), 2. doi: 10.1199/tab.0010
de la Torre, F., Cañas, R. A., Pascual, M. B., Avila, C., Cánovas, F. M. (2014). Plastidic aspartate aminotransferases and the biosynthesis of essential amino acids in plants. J. Exp. Bot. 65, 5527–5534. doi: 10.1093/jxb/eru240
Duncan, C. D. S., Rodríguez-López, M., Ruis, P., Bähler, J., Mata, J. (2018). General amino acid control in fission yeast is regulated by a nonconserved transcription factor, with functions analogous to Gcn4/Atf4. Proc. Natl. Acad. Sci. USA 115 (8), 1829–1838. doi: 10.1073/pnas.1713991115
El Malki, F., Jacobs, M. (2001). Molecular characterization and expression study of a histidine auxotrophic mutant (his1-) of Nicotiana plumbaginifolia. Plant Mol. Biol. 45, 191–199. doi: 10.1023/A:1006493021557
Gan, Y. B., Stulen, I., Keuelen van, H., Kuiper, P. J. C. (2004). Low concentration of nitrate and ammonium stimulate nodulation and N2 fixation while inhibiting specific nodulation (nodule DW g-1 root dry weight) and specific N2 fixation (N2 fixed g-1 root dry weight) in soybean. Plant Soil. 258, 281–292. doi: 10.1023/B:PLSO.0000016558.32575.17
Gansel, X., Muños, S., Tillard, P., Gojon, A. (2001) Differential regulation of the NO3– and NH4+-transporter genes AtNrt2.1 and AtAmt1.1. Arabidopsis: relation with long-distance and local controls by N status of the plant. Plant J. 26, 143–155. doi: 10.1046/j.1365-313x.2001.01016.x
Gibson, A. H., Harper, J. E. (1985). Nitrate effect on nodulation of soybean by Bradyrhizobium japonicum. Crop Sci. 25, 497–501. doi: 10.2135/cropsci1985.0011183X002500030015x
Guyer, D., Patton, D., Ward, E. (1995). Evidence for cross-pathway regulation of metabolic gene expression in plants. Proc. Natl. Acad. Sci. USA 92, 4997–5000. doi: 10.1073/pnas.92.11.4997
Hanson, A., Kende, H. (1976). Biosynthesis of wound ethylene in morning-glory fl ower tissue. Plant Physiol. 57, 538–541. doi: 10.1104/pp.57.4.538
He, W., Brumos, J., Li, H., Ji, Y., Ke, M., Gong, X., et al (2011). A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23, 3944–3960. doi: 10.1105/tpc.111.089029
Hey, S. J., Byrne, E., Halford, N. G. (2010). The interface between metabolic and stress signalling. Ann. Bot-London. 105, 197–203. doi: 10.1093/aob/mcp285
Hildebrandt, T. M., Nunes-Nesi, A., Araújo, W. L., Braun, H.-P. (2015). Amino acid catabolism in plants. Mol. Plant 8, 1563–1579. doi: 10.1016/j.molp.2015.09.005
Hinnebusch, A. G. (1984). Evidence for translational regulation of the activator of general amino acid control in yeast. Proc. Natl. Acad. Sci. USA 81, 6442–6446. doi: 10.1073/pnas.81.20.6442
Hinnebusch, A. G. (2005). Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450. doi: 10.1146/annurev.micro.59.031805.133833
Immanuel, T. R., Greenwood, D. R., MacDiarmid, R. M. (2012). A critical review of translation initiation factor eIF2α kinases in plants-regulating protein synthesis during stress. Funct. Plant Biol. 39, 717–735. doi: 10.1071/FP12116.
Ingle, R. A. (2011). Histidine biosynthesis. Arabidopsis book. Am. Soc. Plant Biol. 9, e0141. doi: 10.1199/tab.0141
Jensen, R. A., Gu, W. (1996). Evolutionary recruitment of biochemically specialized subdivisions of family I within the protein superfamily of aminotransferases. J. Bacteriol. 178, 2161–2171. doi: 10.1128/jb.178.8.2161-2171.1996
Kim, G., Jang, S., Yoon, E. K., Lee, S. A., Dhar, S., Kim, J., et al. (2018). Involvement of pyridoxine/pyridoxamine 5'-phosphate oxidase (PDX3) in ethylene-induced auxin biosynthesis in the Arabidopsis root. Mol. Cells 41 (12), 1033–1044. doi: 10.14348/molcells.2018.0363
Koslowsky, S., Riegler, H., Bergmüller, E., Zrenner, R. (2008). Higher biomass accumulation by increasing phosphoribosylpyrophosphate synthetase activity in Arabidopsis thaliana and Nicotiana tabacum. Plant Biotechnol. J. 6, 281–294. doi: 10.1111/j.1467-7652.2007.00314.x
Krapp, A., Fraisier, V., Scheible, W. R., Quesada, A., Gojon, A., Stitt, M., et al. (1998). Expression studies of NRT2.1Np, a putative high affinity nitrate transporter: evidence for its role in nitrate uptake. Plant J. 14, 723–731. doi: 10.1046/j.1365-313x.1998.00181.x
Lageix, S., Lanet, E., Pouch-Pélissier, M.-N., Espagnol, M.-C., Robaglia, C., Deragon, J.-M., et al. (2008). Arabidopsis eIF2A kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC plant Biol. 8, 134. doi: 10.1186/1471-2229-8-134
Lam, H.-M., Chiao, Y. A., Li, M.-W., Yung, Y.-K., Sang, J. (2006). Putative nitrogen sensing systems in plants. J. Integr. Plant Biol. 43, 873–888. doi: 10.1111/j.1744-7909.2006.00313.x
Last, R. L., Bissinger, P. H., Mahoney, D. J., Radwanski, E. R., Fink, G. R. (1991). Tryptophan mutants in Arabidopsis: the consequences of duplicate tryptophan synthase β genes. Plant Cell 3, 345–358. doi: 10.1105/tpc.3.4.345
Le Deunff, E. (2018). From aspartate to ethylene: central role of N, C, and S shuttles by aminotransferases during biosynthesis of a major plant growth hormone. Progress in Bot. 80, 253–293. doi: 10.1007/124_2018_17
Le Deunff, E., Lecourt, J. (2016). Non-specificity of ethylene inhibitors: ‘double edged’ tools to find out new targets involved in the root morphogenetic programme. Plant Biol. 18, 353–361. doi: 10.1111/plb.12405
Le Deunff, E., Lecourt, J., Malagoli, P. (2016). Fine-tuning of root elongation by ethylene: a tool to study dynamic structure-function relationships between root architecture and nitrate absorption. Ann. Bot. 118 (4), 607–620. doi: 10.1093/aob/mcw123.
Le Ny, F., Leblanc, A., Beauclair, P., Deleu, C., Le Deunff, E. (2013). In low transpiring conditions, nitrate and water fluxes for growth of B. napus plantlets correlate with changes in BnNrt2.1 and BnNrt1.1 nitrate transporters expression. Plant Signal. Behav. 8, e22902. doi: 10.4161/psb.22902
Le, J., Vandenbussche, F., Van Der Straeten, D., Verbelen, J.-P. (2001). In the early response of Arabidopsis roots to ethylene, cell elongation is up-and down-regulated and uncoupled from differentiation. Plant Physiol. 125, 519–522. doi: 10.1104/pp.125.2.519
Lea, P. J., Azevedo, R. A. (2006). Nitrogen use efficiency. 1. Uptake of nitrogen from the soil. Ann. Appl. Biol. 149 (3), 243–247. doi: 10.1111/j.1744-7348.2006.00101.x
Leblanc, A., Renault, H., Lecourt, J., Etienne, P., Deleu, C., Le Deunff, E. (2008). Elongation changes of exploratory and root hair systems induced by AVG and ACC affect nitrate uptake and BnNrt2.1 and BnNrt1.1 gene expression in oil seed Rape. Plant Physiol. 146, 1028–1040. doi: 10.1104/pp.107.109363
Leblanc, A., Segura, R., Deleu, C., Le Deunff, E. (2013) In low transpiring conditions, uncoupling the BnNrt2.1 and BnNrt1.1 NO3- transporters by glutamate treatment reveals the essential role of BnNrt2.1 for nitrate uptake and the nitrate-signaling cascade during growth. Plant Signal. Behav. 8 (2), e22904. doi: 10.4161/psb.22904
Lejay, L., Tillard, P., Lepetit, M., Olive, F.D., Filleur, S., Daniel-Vedele, F., et al. (1999). Molecular and functional regulation of two NO3- uptake systems by N- and C-status of Arabidopsis plants. Plant J. 18, 509–519. doi: 10.1046/j.1365-313X.1999.00480.x
Lemaire, L., Deleu, C., Le Deunff, E. (2013). Modulation of ethylene biosynthesis by ACC and AIB reveals a structural and functional relationship between the K15NO3 uptake rate and root absorbing surfaces. J. Exp. Bot. 64, 2725–2737. doi: 10.1093/jxb/ert124
Lieberman, M., Kunishi, A., Owens, L. (1974). Specific inhibitors of ethylene production as retardants of the ripening process in fruits. In, Facteurs et Régulation de la Maturation des Fruits. Colloques Internationaux CNRS No. 238, 161–170
Liepman, A. H., Olsen, L. J. (2004). Genomic analysis of aminotransferases in Arabidopsis thaliana. Crit. Rev. Plant Sci. 23, 73–89. doi: 10.1080/07352680490273419
Lin, L.-C., Hsu, J.-H., Wang, L.-C. (2010). Identification of novel inhibitors of 1-aminocyclopropane-1-carboxylic acid synthase by chemical screening in Arabidopsis thaliana. J. Biol. Chem. 285, 33445–33456. doi: 10.1074/jbc.M110.132498
Liu, C.-W., Breakspear, A., Guan, D., Cerri, M. R., Jackson, K., Robson, F., et al. (2019). NIN acts as a nework hub controlling a growth module required for rhizobial infection. Plant Physiol. 179, 1704–1722. doi: 10.1104/pp.18.01572.
Ma, W., Li, J., Qu, B., He, X., Zhao, X., Li, B., et al. (2014). Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. 78, 70–79. doi: 10.1111/tpj.12448
Macduff, J. H., Jarvis, S. C., Davidson, I. A. (1996). Inhibition of N2 fixation by white clover (Trifolium repens L.) at low concentrations of NO3- in flowing solution culture. Plant Soil. 180, 287–295. doi: 10.1007/BF00015312
Maeda, H., Dudareva, N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant. Biol. 63, 73–105. doi: 10.1146/annurev-arplant-042811-105439
Maeda, H., Yoo, H., Dudareva, N. (2011). Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nat. Chem. Biol. 7 (1), 19–21. doi: 10.1038/nchembio.485
Marek, L. F., Stewart, C. R. (1992). Photosynthesis and photorespiration in presenescent, senescent, and rejuvenated soybean cotyledons. Plant Physiol. 98, 694–699. doi: 10.1104/pp.98.2.694
McAllister, C. H., Good, A. G. (2015). Alanine aminotransferase variants conferring diverse NUE phenotypes in Arabidopsis thaliana. PLoS One 10, e0121830. doi: 10.1371/journal.pone.0121830
Mehta, P. K., Hale, T. I., Christen, P. (1993). Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214, 549–561. doi: 10.1111/j.1432-1033.1993.tb17953.x
Miesak, B., Coruzzi, G. M. (2002). Molecular and physiological analysis of Arabidopsis mutants defective in cytosolic or chloroplastic aspartate aminotransferase. Plant Physiol. 129, 650–660. doi: 10.1104/pp.005090
Mitchell, R. E., Frey, E. J., Benn, M. H. (1986). Rhizobitoxine and L-threohydroxythreonine production by the plant pathogen Pseudomonas andropogonis. Phytochemistry 25, 2711–2715.
Mo, X., Zhu, Q., Li, X., Li, J., Zeng, Q., Rong, H., et al. (2006). The hpa1 mutant of Arabidopsis reveals a crucial role of histidinehomeostasis in root meristem maintenance. Plant Physiol. 141, 1425–1435. doi: 10.1104/pp.106.084178
Muday, G. K., Rahman, A., Binder, B. M. (2012). Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 17, 181–195. doi: 10.1016/j.tplants.2012.02.001
Muller, B., Touraine, B. (1992) Inhibition of NO3– uptake by various phloem-translocated amino acids in soybean seedlings. J. Exp. Bot. 43, 617–623. doi: 10.1093/jxb/43.5.617
Natarajan, K., Meyer, M. R., Jackson, B. M., Slade, D., Roberts, C., Hinnebusch, A. G., et al. (2001). Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 13, 4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001
Nazoa, P., Vidmar, J. J., Tranbarger, T. J., Mouline, K., Damiani, I., Tillard, P., et al (2003). Regulation of the nitrate transporter gene AtNRT2. 1 in Arabidopsis thaliana: responses to nitrate, amino acids and developmental stage. Plant Mol. Biol. 52, 689–703. doi: 10.1023/A:1024899808018
Niederberger, P., Miozzari, G., Hutter, R. (1981). Biological role of the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1, 584–593. doi: 10.1128/MCB.1.7.584
Noutoshi, Y., Ito, T., Shinozaki. (2005). ALBINO AND PALE GREEN 10 encodes BBMII isomerase involved in histidine biosynthesis in Arabidopsis thaliana. Plant Cell Physiol. 46 (7), 1165–1172. doi: 10.1093/pcp/pci119
Okamoto, T., Tsurumi, S., Shibasaki, K., Obana, Y., Tajaji, H., Oono, Y., et al. (2008). Genetic dissection of hormonal responses in the roots of Arabidopsis grown under continuous mechanical impedance. Plant Physiol 146, 1651–1662. doi: 10.1104/pp.107.115519
Owens, L. D., Guggenheim, S., Hilton, J. L. (1968). Rhizobium-synthetized phytotoxin: an inhibitor of β-cystathionase in Samonella typhimurium. Biochim. Biophys. Acta, Gen. Subj. 158, 219–225. doi: 10.1016/0304-4165(68)90134-7
Pendyala, L., Wellman, A. (1975). Effect of histidine on purine nucleotide synthesis and utilization in Neurospora crassa. J. Bacteriol. 124, 78–85.
Peters, N. K., Crist-Estes, D. K. (1989). Nodule formation is stimulated by the ethylene inhibitor aminoethoxyvinylglycine. Plant Physiol. 91, 690–693. doi: 10.1104/pp.91.2.690
Philipps, K., Skripan, A. L., Liu, X., Christensen, A., Slewinsky, T. L., Hudson, C., et al. (2011). Vanishing tassel2 encodes a grass-specific tryptophan aminoitransferase required for vegetative and reproductive development in maize. Plant Cell 23, 550–560. doi: 10.1105/tpc.110.075267
Pruess, D. L., Scannell, J. P., Kellett, M., AX, H. A., Janecek, J., William, T. H., et al. (1974). Antimetabolites produced by microorganisms. X, L-2-amino4-(2-aminoethoxy)-trans-3-butenoic acid. J. Antibiot. 27, 229–233. doi: 10.7164/antibiotics.27.229
Rahman, A., Tsurumi, S., Amakawa, T., Soga, K., Hoson, T., Goto, N., et al. (2000). Involvement of ethylene and gibberellin signalings in chromosaponin I-induced cell division and cell elongation in the roots of Arabidopsis seedlings. Plant Cell Physiol 41 (1), 1–9. doi: 10.1093/pcp/41.1.1
Ravanel, S., Gakière, B., Job, D., Douce, R. (1998). The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. USA 95, 7805– 7812. doi: 10.1073/pnas.95.13.7805
Reyes, J. C., Muro-Pastor, M. I., Florencio, F.-J. (2004). The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 134 (4), 1718–1732. doi: 10.1104/pp.103.037788
Růžička, K., Ljung, K., Vanneste, S., Podhorská, R., Beeckman, T., Friml, J., et al. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19, 2197–2212. doi: 10.1105/tpc.107.052126
Schauser, L., Roussis, A., Stiller, J., Stougaard, J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402, 191–195. doi: 10.1038/46058
Schultz, C. J., Hsu, M., Miesak, B., Coruzzi, G. M. (1998). Arapidopsis mutants define an in vivo role for isoenzymes of aspartate aminotransferase in plant nitrogen assimilation. Genetics 149, 491–499
Shao, A., Ma, W., Zhao, X., Hu, M., He, X., teng, W., et al . (2017). The auxin biosynthetic Tryptophan Aminotransferase Related TaTAR2.1-3A increases grain yield of wheat. Plant Physiol 174 (4), 2274–2288. doi: 10.1104/pp.17.00094
Soeno, K., Goda, H., Ishii, T., Ogura, T., Tachikawa, T., Sasaki, E., et al. (2010). Auxin biosynthesis inhibitors, identified by a genomics-based approach, provide insights into auxin biosynthesis. Plant Cell Physiol. 51, 524–536. doi: 10.1093/pcp/pcq032
Stepanova, A. N., Hoyt, J. M., Hamilton, A. A., Alonso, J. M. (2005). A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17, 2230–2242. doi: 10.1105/tpc.105.033365
Stepanova, A. N., Robertson-Hoyt, J., Yun, J., Benavente, L.M., Xie, D-Y., Dolezal, K., et al. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191. doi: 10.1016/j.cell.2008.01.047
Stepanova, A. N., Yun, J., Likhacheva, A. V., Alonso, J. M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19, 2169–2185. doi: 10.1105/tpc.107.052068
Stepansky, A., Leustek, T. (2006). Histidine biosynthesis in plants. Amino Acids 30, 127–142. doi: 10.1007/s00726-005-0247-0
Stitt, M., Muller, C., Matt, P., Gibon, Y., Carillo, P., Morcuende, R., et al. (2002). Steps towards an integrated view of nitrogen metabolism. J. Exp. Bot. 53, 959–970. doi: 10.1093/jexbot/53.370.959.
Sugawara, S., Mashiguchi, K., Tanaka, K., Hishiyama, S., Sakai, T., Hanada, K., et al. (2015). Distinct characteristics of indole-3-acetic acid and phenylacetic acid, two common auxins in plants. Plant Cell Physiol. 56 (8), 1641–1654. doi: 10.1093/pcp/pcv088
Swarup, R., Perry, P., Hagenbeek, D., Van Der Straeten, D., Beemster, G.T.S., Sandberg, G., et al. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis plantlets to enhance inhibition of root cell elongation. Plant Cell 19, 2186–2196. doi: 10.1105/tpc.107.052100
Tambasco-Studart, M., Tews, I., Amrhein, N., Fitspatrick, T. B. (2007). Functional analysis of PDX2 from Arabidopsis, a glutaminase involved in vitamin B6 biosynthesis. Plant Physiol. 114, 915–925. doi: 10.1104/pp.107.096784
Tao, Y., Ferrer, J.-L., Ljung, K., Van Der Straeten, D., Beemster, G.T.S., Sandberg, G., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. doi: 10.1016/j.cell.2008.01.049.
Tarun, A. S., Theologis, A. (1998). Complementation analysis of mutants of 1-aminocyclopropane 1-carboxylate synthase reveals the enzyme is a dimer with shared active sites. J. Biol. Chem. 273, 12509–12514. doi: 10.1074/jbc.273.20.12509
Tillard, P., Passama, L., Gojon, A. (1998) Are phloem amino acids involved in the shoot to root control of NO3– uptake in Ricinus communis plants. J. Exp. Bot. 49, 1371–1379. doi: 10.1093/jxb/49.325.1371
Vidmar, J. J., Zhuo, D., Siddiqui, M. Y., Schjoerring, J. K., Touraine, B., Glass, A. D. M. (2000). Regulation of high-affinity transporter genes and high-affinity nitrate influx by nitrogen pools in roots of barley. Plant Physiol. 123, 307–318. doi: 10.1104/pp.123.1.307
Wang, B., Chu, J., Yu, T., Xu, Q., Sun, X., Yuan, J., et al. (2015). Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 112 (15), 4821–4826. doi: 10.1073/pnas.1503998112
Werck-Reichhart, D., Jones, O. T. G., Durst, F. (1988). Haem synthesis during cytochrome P-450 induction in higher plants. Biochem. J. 249, 473–480. doi: 10.1042/bj2490473
Xia, X., Ma, C., Dong, S., Xu, Y., Gong, Z. (2017). Effects of nitrogen concentrations on nodulation and nitrogenase activity in dual root systems of soybean plants. Soil Sci and Plant nut. 63 (5), 470–482. doi: 10.1080/00380768.2017.1370960
Yamada, M., Greenham, K., Prigge, M. J., Jensen, P. J., Estelle, M. (2009). The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 151, 168–179. doi: 10.1104/pp.109.138859
Yasuta, T., Satoh, S., Minamisawa, K. (1999) New assay for Rhizobitoxine based on inhibition of 1-Aminocyclopropane-1-Carboxylate synthase. Appl. Environ. Microbiol. 65 (2), 849–852.
Yasuta, T., Okazaki, S., Mitsui, H., Yuhashi, K.-I., Ezura, H., Minamisawa, K. (2001). DNA sequence and mutational analysis of Rhizobitoxine biosynthesis genes in Bradyrhizobium elkanii. Appl. Environ. Microbiol. 67 (11), 4999–5009. doi: 10.1128/AEM.67.11.4999-5009.2001
Yoshikawa, T., Ito, M., Sumikura, T., Nakayama, A., Nishimura, T., Kitano, H., et al. (2014). The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant Journal 78, 927–936. doi: 10.1111/tpj.12517
Zhang, Y., Dickinson, J. R. K., Paul, M. J., Halford, N. G. (2003). Molecular cloning of an Arabidopsis homologue of GCN2, a protein kinase involved in co-ordinated response to amino acid starvation. Planta 217, 668–675. doi: 10.1007/s00425-003-1025-4
Keywords: Aminoethoxyvinylglycine, tryptophan aminotransferase, histidine catabolism, aminocyclopropane-1-carboxylate synthase, Brassica napus, nitrate uptake, root morphogenesis, general amino acid control
Citation: Le Deunff E, Beauclair P, Deleu C and Lecourt J (2019) Inhibition of Aminotransferases by Aminoethoxyvinylglycine Triggers a Nitrogen Limitation Condition and Deregulation of Histidine Homeostasis That Impact Root and Shoot Development and Nitrate Uptake. Front. Plant Sci. 10:1387. doi: 10.3389/fpls.2019.01387
Received: 06 June 2019; Accepted: 08 October 2019;
Published: 07 November 2019.
Edited by:
Michael J. Haydon, The University of Melbourne, AustraliaReviewed by:
Abidur Rahman, Iwate University, JapanMary L. TIerney, University of Vermont, United States
Copyright © 2019 Le Deunff, Beauclair, Deleu and Lecourt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erwan Le Deunff, erwan.ledeunff@unicaen.fr
 Erwan Le Deunff
Erwan Le Deunff Patrick Beauclair2
Patrick Beauclair2 Carole Deleu
Carole Deleu