- 1Chinese Academy of Agricultural Science (ICR, CAAS) /State Key Laboratory of Cotton Biology, Institute of Cotton Research, Anyang, China
- 2School of Biological and Physical Sciences (SBPS), Jaramogi Oginga Odinga University of Science and Technology (JOOUST), Bondo, Kenya
We found 33, 17, and 20 Alba genes in Gossypium hirsutum, Gossypium arboretum, and Gossypium raimondii, respectively. The Alba protein lengths ranged from 62 to 312 aa, the molecular weight (MW) from 7.003 to 34.55 kDa, grand average hydropathy values of −1.012 to 0.609 and isoelectric (pI) values of −3 to 11. Moreover, miRNAs such as gra-miR8770 targeted four genes, gra-miR8752 and gra-miR8666 targeted three genes, and each and gra-miR8657 a, b, c, d, e targeted 10 genes each, while the rests targeted 1 to 2 genes each. Similarly, various cis-regulatory elements were detected with significant roles in enhancing abiotic stress tolerance, such as CBFHV (RYCGAC) with a role in cold stress acclimation among others. Two genes, Gh_D01G0884 and Gh_D01G0922, were found to be highly induced under water deficit and salt stress conditions. Through virus-induced gene silencing (VIGS), the VIGS cotton plants were found to be highly susceptible to both water deficit and salt stresses; the VIGS plants exhibited a significant reduction in root growth, low cell membrane stability (CMS), saturated leaf weight (SLW), chlorophyll content levels, and higher excised leaf water loss (ELWL). Furthermore, the stress-responsive genes and ROS scavenging enzymes were significantly reduced in the VIGS plants compared to either the wild type (WT) and or the positively controlled plants. The VIGS plants registered higher concentration levels of hydrogen peroxide and malondialdehyde, with significantly lower levels of the various antioxidants evaluated an indication that the VIGS plants were highly affected by salt and drought stresses. This result provides a key foundation for future exploration of the Alba proteins in relation to abiotic stress.
Introduction
Alba family proteins are mainly referred to as basic, small, and dimeric nucleic acid-binding proteins and are mainly distributed in a number of eukaryotes and the archaeal organisms (Subota et al., 2011). The Alba protein family is integral in the organization and regulation of the euryarchea genome possessing histone and as well as the crenarchea, with no histone (Reeve, 2003). The Alba proteins have a distinctive property in the regulation and organization of the organism’s genomes through acetylation and deacetylation (Goyal et al., 2012). The Alba protein binding has a very high affinity towards double-strand deoxyribonucleic acid (DNA), thus named as acetylation lower binding affinity (Alba) (Crnigoj et al., 2011). In Sulfolobus solfatataricus, a species of thermophilic archaeon, Alba proteins have been found to reversibly acetylated at lysine 6 (Lys16) by a homologous protein acetyltransferase (Pat) and deacetylated by a sirtuin family deacetylase (Sir2) (Starai and Escalante-Semerena, 2004). Apart from the acetylation at the N-terminal of the lysine residues, the Alba proteins also contain arginine–glycine–glycine (RGG) repeat at the C-terminal, which are important mediators of protein:RNA, and protein: protein interactions resulting in the formation of the membraneless ribonucleoprotein granules (Chong et al., 2018). The RGG domain, are closely spaced arginine–glycine–glycine repeats, it is a DNA and RNA-binding domain in various nucleic acid-binding proteins (Arribas-Layton et al., 2016). The RGG repeats affinity for the RNA is regulated by the methylation of the arginine various RGG-box proteins (Aravind et al., 2003). Structural analysis of the Alba proteins reveals the homodimer (dimeric) nature of the proteins (Bell et al., 2002).
Alba family proteins have been found to bind the DNA with no sequence specificity (Aravind et al., 2003; Kumar Verma et al., 2018). Moreover, the Alba proteins also do interact with diverse kinds of ribonucleic acid (RNA) and in addition to a number of ribonucleo-protein complexes, such as ribosomal ribonucleic acid (rRNA) and messenger ribonucleic acid (mRNA) (Xue et al., 2000). The Alba proteins have also been found to associate with both DNA and RNA (Liu et al., 2012). The Alba protein, SshAlba or Ssh10b isolated from Sulfolobus shibatae, has been found to bind well with double-stranded deoxyribonucleic acid and even the single-stranded deoxyribonucleic acid but do prefer the ribonucleic acids as the physiological substrates and RNA (Guo et al., 2003). In the recent past, studies have shown that SshAlba interacts with double-stranded ribonucleic acid (dsRNA), leading to the destabilization of the secondary structure of the RNA (Guo et al., 2014). Plants being sessile have evolved a number of survival strategies, one of which is the evolution of a diverse number of the stress-responsive genes (Nakashima and Yamaguchi-Shinozaki, 2006). The plant’s response to abiotic stress factors through molecular mechanism has been considered as the most complex mechanism, being based on the inductions and regulation of transcriptional activity of stress-related genes (Shinozaki and Yamaguchi-Shinozaki, 2007). Plants acquire tolerance to various abiotic stress factors through metabolism reprogramming and gene expression, and in turn gaining a balance among all the plant’s faculties which are necessary for plant performance (Muñoz et al., 2016). Several plants transcription factors have been found to be integral in enhancing tolerance in plants to various environmental stress factors, but it has been shown that despite the overexpression of these genes, their overexpression is not sufficient to boost plants tolerance levels toward various abiotic factors, being additional post-translational modifications may be needed—for instance, the dehydration-responsive element-binding protein 2 (DREB2) (Thirumalaikumar et al., 2018). Phosphorylation is a vital process in protein post-translational modifications, which affects the protein conformation, stability, and localization (Kemp, 2018). Phosphorylation functions in a number of biological processes; it translates external stimuli which in turn illicit specific response by the cell (Boudsocq, 2005). Similarly, protein degradation has also been found to play a significant role in enhancing plant stress tolerance; this occurs through ubiquitination, which refers to the covalent addition of the small protein ubiquitin to selected target proteins (Weissman, 2001).
Alba proteins are predominantly abundant in eukaryotes, more specifically in plants and protozoan (Aravind et al., 2003). Functional analysis of the Alba proteins has been done in rice (Kumar Verma et al., 2018) and other protozoans such as Trypanosoma brucei, Leishmania infantum, Toxoplasma gondii, and Plasmodiumfalciparum (Wardleworth et al., 2002). OsAlba1, an Alba gene, isolated from Oryza sativa, of indica species, was up-regulated by water deficit condition; it has also been found to complement yeast VIGSs, lacking the Pop6 gene, thereby enhancing their tolerance to dehydration (Verma et al., 2014). Rice is a water plant, upregulation of the dehydration-responsive nuclear protein.
OsAlba1 under water deficit condition indicated the integral role played by the Alba proteins in plants in enhancing their tolerance to various environmental stress factors (Choudhary et al., 2009). In addition, two Alba protein domains, LiAlba1 and LiAlba3, with a molecular weight of 13 kDa and 30 kDa, respectively, to interact with each other, thus do associate with RNA-binding proteins, ribosomal units, and translation factors (Dupé et al., 2015). Two Alba homologs, TgAlba1 and TgAlba2 isolated from T. gondii, are dual in relation to subcellular localization, found both in the nucleus and cytoplasm, but are predominantly cytoplasmic proteins, and their presence in the two cellular structures shows their integral role in both nucleus and cytoplasm (Olguin-Lamas et al., 2011). The gene silencing of TgAlbas revealed in regulating response to stress and differentiation, TgAlbas 1 and TgAlbas 2, are associated with a high number of proteins, such as the RNA-binding proteins (Gissot et al., 2013).
Being an important commercial crop, cotton production is threatened by various abiotic stress factors such as drought, salinity, and cold among others. No studies have been done elucidating the role of the Alba proteins in relation to their stress response, despite the significant contribution of the Alba proteins in eukaryotic organisms. The completion and sequencing of the three cotton species, Gossypium arboreum (Li et al., 2014), Gossypium raimondii (Wang et al., 2012), and Gossypium hirsutum (Li et al., 2015), have provided the needed materials for molecular studies in cotton. Therefore, in this research work, we carried out genome-wide identification of the Alba proteins in cotton, by determining the number, their distribution, gene structure, phylogenetic relationship, their expression levels, and further carried the functional analysis of two key Alba genes through virus-induced gene silencing (VIGS). These results will provide new insights into the biological relevance of the proteins encoded by the Alba genes in plants, and their future use in developing a more water-deficit and salt stress–resilient cotton genotypes.
Materials and Methods
Alba Protein Identification, Sequence Analysis, Phylogenetic Tree Analysis, and Subcellular Localization Prediction
The whole sequences for the Alba proteins in G. hirsutum, G. arboreum, and G. raimondii were retrieved from cotton research institute (http://mascotton.njau.edu.cn), Beijing genome database (https://www.bgi.com/),and phytozome 12 (http://www.phytozome.net/), respectively. The conserved domain of Alba proteins, PF01918, was downloaded from Pfam protein families’ database (http://pfam.xfam.org). The HMM profile of the Alba proteins were submitted to HMMER search (http://hmmer.janelia.org/) against G. hirsutum, G. raimondii, and G. arboreum protein sequences. The amino acid sequences were analyzed in order to determine the Alba domain using online tools: the NCBI Conserved Domain Database (Marchler-Bauer et al., 2017), the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/), and the ScanProsite tool (http://prosite.expasy.org/scanprosite/). The Alba proteins for G. hirsutum AD genome, G. arboreum of A genome, and G. raimondii of the D genome [together with the whole sequences for Arabidopsis thaliana obtained from TAIR (http://www.arabidopsis.org/)], O. sativa obtained from http://rice.plantbiology.msu.edu/index.shtml , Theobroma cacao, Sorghum bicolor, Glycine max, and Populus trichocarpa were all downloaded from Phytozome v12.0 (https://phytozome.jgi.doe.gov/pz/portal.html) and were used to investigate the evolutionary relationships of the Alba proteins in plants (Table S1). The multiple sequence alignments of all the Alba proteins were carried out by Clustal Omega, MEGA 7.0 software, using an algorithm with 1,000 bootstraps, using complete deletion of site coverage for gaps and missing data as previously outlined in the analysis of the cotton LEA proteins (Magwanga et al., 2018). An online program, ExPASy Server tool (http://www.web.xpasy.org/compute_pi/), was applied in the investigation of the physiochemical properties of all the Alba proteins obtained for the three cotton species. Finally, the subcellular localization predictions were carried out for all the three cotton species Alba proteins, through an online tool, WoLF PSORT (https://www.wolfpsort.hgc.jp/), and the results obtained were later validated by other two online tools, the Protein Prowler Subcellular Localisation Predictor version 1.2 (http://www.bioinf.scmb.uq.edu.au/pprowler_webapp_1-2/) and TargetP1.1 server (http://www.cbs.dtu.dk/services/TargetP/).
Chromosome Location, Sequence Analysis, and Structural Analysis of the Alba Genes in Cotton
The information for the Alba protein sequences, genomic sequences, cDNA sequences, and chromosomal positions was retrieved from phytozome (www.phytozome.net) for G. raimondii and cotton functional database (https://cottonfgd.org) for G. hirsutum and G. arboreum. The genomic sequences, the coding sequences (CDS), and Newick structure for each of the cotton species protein sequence analyses were submitted to an online tool, Gene Structure Displayer Server (http://gsds.cbi.pku.edu.cn/), to analyze their respective gene structures in relation to intron-exon ratio. The gene structures were combined by the various Alba proteins distinctive motifs; the Alba protein motifs were determined by analyzing their respective protein sequences, through an online tool MEME, with default parameters set at 50 for maximum motif length and a minimum of 6, with the largest number of 15 (Brown et al., 2013). All the Alba genes were mapped into their respective chromosomes using the mapping tool, MapChart (Voorrips, 2002).
Prediction of miRNA Targets and Cis-Regulatory Element Analysis in Cotton Alba Genes
In promoter sequences, 1,500 bp DNA sequence of each the Alba gene for the diploid cotton, G. raimondii was obtained from phytozome (www.phytozome.net), while for G. hirsutum then tetraploid cotton and that of G. arboreum, the diploid cotton of the A genome was obtained from the cotton functional database (https://cottonfgd.org). The Alba genes cis-regulatory elements were predicted by use of an online tool, the PLACE database (http://www.dna.affrc.go.jp/PLACE/signalscan.html), while the Alba genes targeted by miRNAs were predicted by using the online tool, the psRNATarget server with default parameters (http://plantgrn.noble.org/psRNATarget/).
Plant Materials and Abiotic Stress Exposure
The seeds of the three cotton species, G. hirsutum (AD1), G. raimondii (D5), and G. arboreum (A2), were used. The three cotton germplasms are regularly maintained by our Institute of Cotton Research, Chinese Academy of Agricultural Sciences, CRI-CAAS. G. hirsutum, coded as CRI, was developed by our research institute and currently is the most preferred upland cotton grown in China; it covers more than 90% of the cotton growing regions. The seeds were delinted and then pre-treated before being germinated. Upon germination for 3 days, the seedlings were then transferred to a Hoagland solution (Hoagland and Arnon, 1950) in a hydroponic set up in the greenhouse, with temperature set at 28°C day/25°C night, 14 h photoperiod, and 60 to 70% relative humidity, a condition suitable for cotton growth. At three-leaf stages, the seedlings were exposed to water deficit and salinity stress, in which water deficit was initiated by transferring the seedlings into Hoagland nutrient solution supplemented with 15% of PEG-600 and samples collected at 0, 3, 6, 12, and 24 h, while salinity stress was imposed by supplementing the Hoagland solution with 250 mM of NaCl and samples collected at 0, 3, 6, 12, and 24 h. The tissues collected for both RNA extraction and expression analysis were root, leaf, and stem tissues.
RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis
Total RNA was extracted from the two organs: leaf and root tissues of both treated and control plants under the two forms of abiotic stress condition, water deficit, and salt stress by using TRI reagent (Sigma Life Science, St. Louis, MO, USA). The quality and quantity of the RNA samples extracted were evaluated through NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, DE, USA) and agarose gel electrophoresis. The cDNAs were synthesized using RNA samples with quality and quantity value of 260/280 ratio between 1.8 and 2.1, and 260/230 ratio between 2.0 and 2.5; all the RNA samples which felt out of the range were discarded and not used due to protein contamination. The Alba gene-specific primers were designed by Primer Premier 5 with melting temperatures of 55–60°C, primer lengths of 18–25 bp, and amplicon lengths of 101–221 bp (Table S2). Three biological replicates from each of the treatment, under drought, and salt stress, which comprised of three technical replicates, were analyzed. The transcript analysis was conducted by RT-qPCR using an ABI Prism 7500 Detection System (Applied Biosystems, Foster City, CA, USA). The cotton GhActin gene was used as the internal reference gene. The RT-qPCR reaction mixtures were carried out in a volume of 20 μl, containing 10 μl of SYBR Green Master Mix (Takara, Beijing, China), 2 μl of cDNA template, 6 μl of ddH2O, and 2 μl of each primer to make a final concentration of 10 μM. Reaction conditions were carried out with 95°C for 10 min, followed by 40 cycles of 95°C for 5 s, 59°C for 15 s, and 72°C for 30 s. The relative gene expression levels were calculated using the 2− ΔΔCt methods (Rao et al., 2013).
Generation of Transiently Transformed G. hirsutum Plants With Repression of Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5) Genes
In this study, two types of transiently transformed G. hirsutum plants were generated in order to verify the function of Alba genes. For the VIG silencing of Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5), the tobacco rattle virus based VIGS technique was employed (Mu et al., 2016). The CDS of the two genes where used in designing the primers, 402 and 373 bp gene-specific fragments from Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5), respectively, were amplified by PCR using the vGh_A01G0884 and vGh_D01G0922 gene-specific primers (Table S2). The PCR products were cloned into the pTRV2 vector to produce TRV: Gh_A01G0884 (ghAlba_4) and TRV: Gh_D01G0922 (ghAlba_5) constructs. The Agrobacterium tumefaciens, strain LBA4404, was used in transforming the recombinant plasmids pTRV1, pTRV2, and the two TRV: Alba constructs, the process followed as outlined by Gao et al. (Gao et al., 2011b). After 2 weeks, samples were collected from the wild types, the positive control, and the VIGS plants. At three-leaf stage, water deficit and salt stress treatments were initiated. For water deficit conditions, irrigation was totally withdrawn, then after 6 days, various morphological, physiological, and biochemical parameters were determined. Moreover, for salt stress, the VIGS plant seedlings were irrigated with water supplemented with 250 mM of NaCl solution for a period of 4 days; thereafter, evaluation was carried out.
Evaluation of Physiological Traits in the Two Transformed G. hirsutum Seedlings Under Stress Conditions
The cell membrane stability (CMS), excised leaf water loss (ELWL), and relative leaf water content (RLWC) were evaluated. The three physiological traits evaluated have been explored widely in determining the tolerance levels of plants to various abiotic stress—for instance, RLWC helps in evaluating plant water status (Tanentzap et al., 2015) and water stress and increased high levels of electrolytes affected the integrity of the cell membrane (Petrov et al., 2018). The CMS was determined as outlined by Fokar (Fokar et al., 1998). The RLWC determination was carried out as outlined by Barrs (Barrs and Weatherley, 1962) and, lastly, ELWL (McCaig and Romagosa, 1989). All the measurements were carried out in three biological repeats.
Evaluation of Chlorophyll Content, Oxidants, and Antioxidant in the Transformed and Non-Transformed G. hirsutum Cotton Plant Under Stress Conditions
In order to determine the role of the Alba genes in cotton, we examined the chlorophyll content, oxidants, and antioxidant levels within the transformed seedlings and the non-transformed ones under salt, and water deficit conditions. Just as described in section 2.8, after 24 h of stress exposure, we measured the leaf chlorophyll content using MINOLTA SPAD, a non-destructive method (do et al., 2008). The oxidants and antioxidants were determined as described by Lu et al. (Lu et al., 2018b), in their analysis of GPCR genes role under salt stress condition in transgenic Arabidopsis plants. We evaluated three antioxidants, SOD, CAT, and POD. The CAT antioxidant enzyme activity was evaluated by determining the reducing level of H2O2, as outlined by Cakmak and Marschner (Cakmak and Marschner, 1992); SOD activity was evaluated by the determination of the inhibition of the photochemical reduction of nitro blue tetrazoliumas described by Giannopolitis and Ries (Giannopolitis and Ries, 1977), and finally the POD evaluated as described by Van Assche (Van Assche et al., 1988). The MDA and H2O2 were evaluated between the two plants, non-transformed, and the transformed under the three stress levels. MDA was determined a measure of lipid peroxidation (Cakmak and Horst, 1991), and H2O2 concentration was measured as described by Loreto and Velikova (Loreto and Velikova, 2001).
Root Assays of the Two Cotton Plants, the VIGS, and Wild Type (WT) Under Water Deficit and Salt Stress Conditions
In order to evaluate the effect of knockdown of the two Alba genes on the plant root growth, vermiculite and sand were used as the rooting medium. The roots were evaluated after 14 days of stress exposure, the roots were carefully removed from the rooting medium and washed, and various parameters were measured. The root evaluation was carried out by determining the root length, by the use of root scanner, WinRHIZO Epson V700 Photo Scanner JZZIA (model number expression 11000XL), obtained from Seiko Epson Corp. Japan. The dry weights were determined after oven drying at 80°C.
Statistical Analysis
All the experimental data derived from this research were computed from the mean values of three biological replicates, and statistical analysis was carried out using IBM SPSS Statistics 20. The variations between the VIGS cotton and the control plants under water deficit and salt stress treatment were evaluated by one way ANOVA.
Results
Identification of the Alba Proteins in Cotton Species
A total of 60 proteins encoded by the cotton Alba genes were determined and found to be distributed across the three cotton genomes, with 33, 20, and 17 Alba proteins in G. hirsutum (AD1), G. raimondii (D5), and G. arboreum (A2), respectively. The Alba proteins in the three cotton species were confirmed both through SMART and Pfam scan for all the sequences obtained from the HMM profile and BLASTP search. The number of Alba proteins in G. hirsutum was higher than in either of the two diploid cotton species, though an element of gene loss was detected, being G. hirsutum evolved through whole genome duplication (WGD) between A and D genomes (Li et al., 2016). The cotton Alba protein lengths ranged from 62 to 312 aa, in which the highest protein lengths were detected in G. hirsutum and G. raimondii with both having the highest protein lengths of 312 aa, for Gh_A04G1077 (RPP25L) and Gorai.012G160400 (RPP25L) in G. hirsutum and G. raimondii, respectively. The two Alba proteins were all members of ribonuclease P protein subunit p25-like protein (RPP25L), being in agreement with the previous results in rice with the highest and lowest protein lengths of 320 and 132 aa, respectively (Kumar Verma et al., 2018).
The other physiochemical properties for the cotton Alba proteins were varied across the three cotton species. In G. hirsutum, the molecular weight (MW) ranged from 7.003 to 34.55 kDa, the electric charge from −3 to 21.5, isoelectric (pI) values from 4.761 to 10.91 and the grand average hydropathy (GRAVY) values from −1.012 to 0.311. In G. arboreum, their protein lengths ranged from 122 to 284 aa, MW from 13 to 32 kDa, a charge of −3 to 19, pI values from 5 to 11, and the GRAVY values of −0.972 to −0.183. Finally, in G. raimondii, Alba protein lengths ranged from 90 to 312 aa, MW from 10 to 34 kDa, pI values from 5 to 11, and the GRAVY values of −0.985 to 0.609 (Table 1). Over 99% of the cotton Alba proteins had GRAVY values below zero, which showed that they are hydrophilic, a property shared among the proteins encoded by the various stress-responsive genes such as the LEA (Magwanga et al., 2018), GPCR (Lu et al., 2018b), and MATE (Lu et al., 2018a).
Phylogenetic Tree Analysis, Chromosomal Mapping, and Subcellular Localization Prediction of the Cotton Alba Proteins
By integrating all the three cotton Alba protein sequences together with O. sativa, T. cacao, A. thaliana, S. bicolor, Populus trichocarpa, and G. max were aligned them through Clustal and constructed phylogenetic tree by MEGA 7. The Alba proteins from cotton and the other plants were clustered into four distinct groups, designated as clade 1 to clade 4. Clade 4 was the largest, then closely followed by clade1, while clades 2 and 3 had a fewer number of Alba proteins (Figure S1). In all the clades, cotton proteins encoded by the Alba genes were found to have an orthologous gene pair with other plants, though the majority of the ortholog genes were formed between the three proteins encoded by Alba genes derived from the three cotton species. In clade 1, the orthologous gene pairs between the cotton Alba genes were Gorai.002G121600 and AT1G76010 and Glyma.06G148800 and Gh_A12G0762, and the third orthologous pair were Thecc1EG006429 and Gorai.012G160400.
In clades 2 and 3, no ortholog gene pairs were formed between the cotton Alba genes with any other plant used in the phylogenetic tree analysis. In clade 4, Thecc1EG038396 and Gh_D06G0537, Cotton_A_30556, and Thecc1EG026310 were the only ortholog gene pairs between the cotton and the other plants; the rest of the ortholog gene pairs were between the cotton proteins encoded by Alba genes. The detection of the ortholog gene pairs between the cotton proteins encoded by the Alba genes showed that these proteins might have evolved from a common origin. T. cacao and Gossypium species shared a common evolutionary origin. The results obtained in the analysis of the phylogeny of the cotton Alba genes are in agreement with previous reports in rice with similar domain compositions clustered in the same clade (Kumar Verma et al., 2018).
The distribution of the Alba genes across the 26 chromosomes of the tetraploid cotton was uneven, only 19 chromosomes out of the 26 chromosomes in tetraploid cotton; G. hirsutum were found to harbor the Alba genes. The highest loci density among the mapped chromosomes in the tetraploid cotton was noted in chromosomes Ah05, Ah11, Dh01, and Dh11 with three genes each. The following chromosomes harbored no Alba genes: chrAh03, chrAh07, chrAh09, chrAh10, chrDh07, chrDh09, and chrDh10. The most interesting is that three sets of chromosomes, chrAh07, chrAh09, and chrAh10 and their corresponding homologs harbored no genes. The uneven distribution of the Alba genes could be attributed to their low numbers, only 33 genes in a relatively large genome of the tetraploid cotton, G. hirsutum. In the diploid cotton, 10 and 11 chromosomes were found to harbor the Alba genes in G. raimondii and G. arboreum, respectively. In G. raimondii, the highest gene loci were observed in chrD502, with four genes; chrD507, chrD509, and chrD510, with three genes each; and chrD503, chrD505, chrD508, chrD510, and chrD512, with a single gene each. No genes were detected to be mapped in chrD501, chrD506, and chrD511. In the diploid cotton of the A genome, the highest gene loci were detected in chrA202, chrA203, chrA206, chr A208, and chrA212 with two genes in each; the rest of the mapped chromosomes, chr A204, chrA205, chrA209, chrA210, and chrA213, had a gene in each. The chromosomes were named as described by Wang et al. (Kunbo et al., 2018).
In the analysis of subcellular predictions, a higher percentage of the proteins encoded by the Alba genes was found to be located within the nucleus, which was evident across the three cotton species. Among the proteins encoded by the Alba genes, obtained for the tetraploid cotton, out of the 33 proteins, 16 were found to be located within the nucleus, 6 endoplasmic reticulum (E.R), 4 cytoplasm, 3 in the extracellular structures (Extr), 3 within the mitochondrion, and 1 within the plasma membrane. In the diploid cotton of D genome, G. raimondii, 13 proteins encoded by the Alba genes were predicted to be located within the nucleus, 3 in E.R, and 2 in the cytoplasm, 1 in extracellular structures, and 1 in the mitochondrion. Finally, in G. arboreum, nine proteins encoded by the Alba genes were predicted to be localized within the nucleus; four in the E.R, two in the cytoplasm, and one each were found to be embedded in the extracellular structures and mitochondrion (Table 2). The subcellular localization predictions of the cotton Alba proteins encoded by the Alba genes showed that the majority of the cotton alba proteins are located within the nucleus, 48.5, 65, and 52% of all the Alba proteins encoded by the Alba genes in G. hirsutum, G. raimondii, and G. arboreum, respectively. The high number of the proteins encoded by the alba genes embedded within the nucleus could possibly mean that these proteins could be playing an integral role within the nucleus, in relation to gene expression regulation, more so stress-responsive genes (Verma et al., 2014).
Gene Structure and Motif Identification
For the analysis of the Alba gene structures in the three cotton species, all the Alba genes were found to be disrupted, except two genes, Gh_D01G0359 in the tetraploid cotton and Gorai.002G047400 in diploid cotton of the D genome, which were intronless and members of the ribonuclease P protein subunit p25-like protein (Rpp25l). Among the Alba genes in the tetraploid cotton, the lowest intron disruption of only one was observed in two genes, Gh_D03G1718 and Gh_Sca129121G01, while the highest intron disruption of eight was detected in Gh_A12G0762 (Figure S2A). In G. raimondii, the highest level of intron disruption was detected in Gorai.003G023400 with nine disruptions, and the least intron disruption was observed for Gorai.002G140000, with two introns (Figure S2B). Similarly, in G. arboreum, the least intron disruption was one, while the highest intron disruption was eight also, as observed in Cotton_A_14221 and Cotton_A_03232, respectively (Figure S2C). These results were in agreement with the previous reports in rice, sAlba1, which was found to be interrupted by four introns (Verma et al., 2014). Cotton Alba genes had distinctive motifs. In the tetraploid cotton among 33 Alba genes, the following motifs were common: motif 1, motif 2, and motif 6. In G. arboreum, motif 1 and motif 2 were common among its all Alba genes, while in G. raimondii, motif 1 motif 2 and motif 5 were common among its all Alba genes. In combined analysis of all the three cotton Alba genes, very specific distinctive motifs were identified, which can be used for the identification and characterization of the Alba genes in cotton. The common motifs identified were motif 1 motif 2 and motif 3.
miRNA Target and Cis-Regulatory Element Analysis of the Cotton Alba Genes
The plants small/micro ribonucleic acids (miRNAs) have emerged as a significant player in translational, transcriptional, and post-transcriptional regulation of plant genes, which are vital plant responsiveness to various kinds of abiotic and biotic stress factors (Kumar, 2014). In the analysis of the possible miRNAs targets to various cotton Alba genes, no miRNAs were detected to target any of the Alba genes obtained from G. arboreum; however, in the G. raimondii, a diploid cotton of the D genome, high level of miRNAs target, was observed; 52 miRNAs were found to target all the 20 Alba genes in G. raimondii. The miRNAs with the highest gene targets were gra-miR8770 that targeted 4 genes, gra-miR8752 and gra-miR8666 that targeted 3 genes each, and gra-miR8657a, b, c, d, and e that targeted 10 genes, while the rest targeted either 1 to a maximum of 2 genes each. Some of the Alba genes were found to be targeted by more than 5 miRNAs—for instance, Gorai.002G206900 was targeted by 8 miRNAs, Gorai.004G274000 was targeted by 6 miRNAs, Gorai.007G063100 and Gorai.012G160400 were targeted by 11 miRNAs, and Gorai.013G105600 was targeted by 7 miRNAs. Low level of miRNAs targets was observed among the Alba genes obtained for the tetraploid cotton, G. hirsutum; only 16 miRNAs were found to target 17 genes; the genes with the highest miRNA target were Gh_A04G1077, Gh_A05G1575, Gh_A11G2262, Gh_D04G2019, and Gh_D05G1753 with three miRNA each; and the rest of the genes were either target by 1 to 2 miRNAs. Only one miRNA, ghr-miR7498, was found to target four genes, such as Gh_A05G3960, Gh_A11G2262, Gh_D05G0083, and Gh_D11G2569 (Table S3). One of the most significant miRNA detected was ghr-miR394a; the same miRNA has been found to be highly upregulated in Arabidopsis under water deficit condition (Liu et al., 2009).
Plant responses and acclimations under various environmental stress factors require differential gene expression, which is modulated by a given plant transcription factor (TF) (Saibo et al., 2008; Banerjee and Roychoudhury, 2017; Hernandez and Sanan-Mishra, 2017). The TFs are proteins with a DNA domain that binds to the cis-regulatory element found within the promoter regions of the targeted gene (Song et al., 2001). Several plant TFs have been identified—for instance, abscisic acid (ABA) responsive element (ABRE), CBF/DREB, myeloblastosis (MYB/MYC), AP2/ERF, and the NAM, ATAF1/2, and CUC2 (NAC) domain, which are the major plant-specific families of the TFs with significant role in the regulation of the abiotic stress-induced multiple gene expression in an ABA-dependent or independent processes (Qin et al., 2011). The most abundant forms of the cis-regulatory elements detected across the three cotton species were CAATBOX1 (disease resistance/putative functions in response to environmental stresses), GATABOX (required for high level, light-regulated, and tissue-specific expression), MYCCONSENSUSAT (MYC recognition site found in the promoters of the dehydration-responsive gene rd22), GT1CONSENSUS (light regulation), WRKY71OS (positive and negative regulators of ABA signaling), MYBCORE (dehydration/ water stress), and ABRELATERD1 (function in induction by dehydration stress and dark-induced senescence) (Figure S3 and Table S4). The detection of these cis-regulatory elements showed that the cotton Alba genes indicated their broader functions in enhancing abiotic and biotic stress tolerances.
RNA Sequence Data Analysis and RT-qPCR Validation of the Various Alba Genes Under Salt and Water Deficit Conditions
The RNA sequence profiling of leaf, root, and stem under water deficit and salt stress conditions showed that the upland cotton, G. hirsutum Alba genes, were grouped into three groups as per their expression pattern. In both salt and water deficit conditions, group 1 gene exhibited significant upregulations across all the tissues tested and in different time points of stress exposure. Group 2 showed differential expression, though in most of the time points, more were up-regulated. In group 3, they were either down-regulated or showed no expressions at all in all the tissues tested. Some of the genes were found to have significant upregulation under salt and water deficit, such as Gh_D05G0083 (DNA-/RNA-binding protein), Gh_A08G2091 (DNA-/RNA-binding protein), Gh_D01G0922 (ribonuclease P protein), Gh_A01G0884 (ribonuclease P protein), Gh_D13G2120 (DNA-/RNA-binding protein), and Gh_A13G1770 (DNA-/RNA-binding protein) (Figure S4).
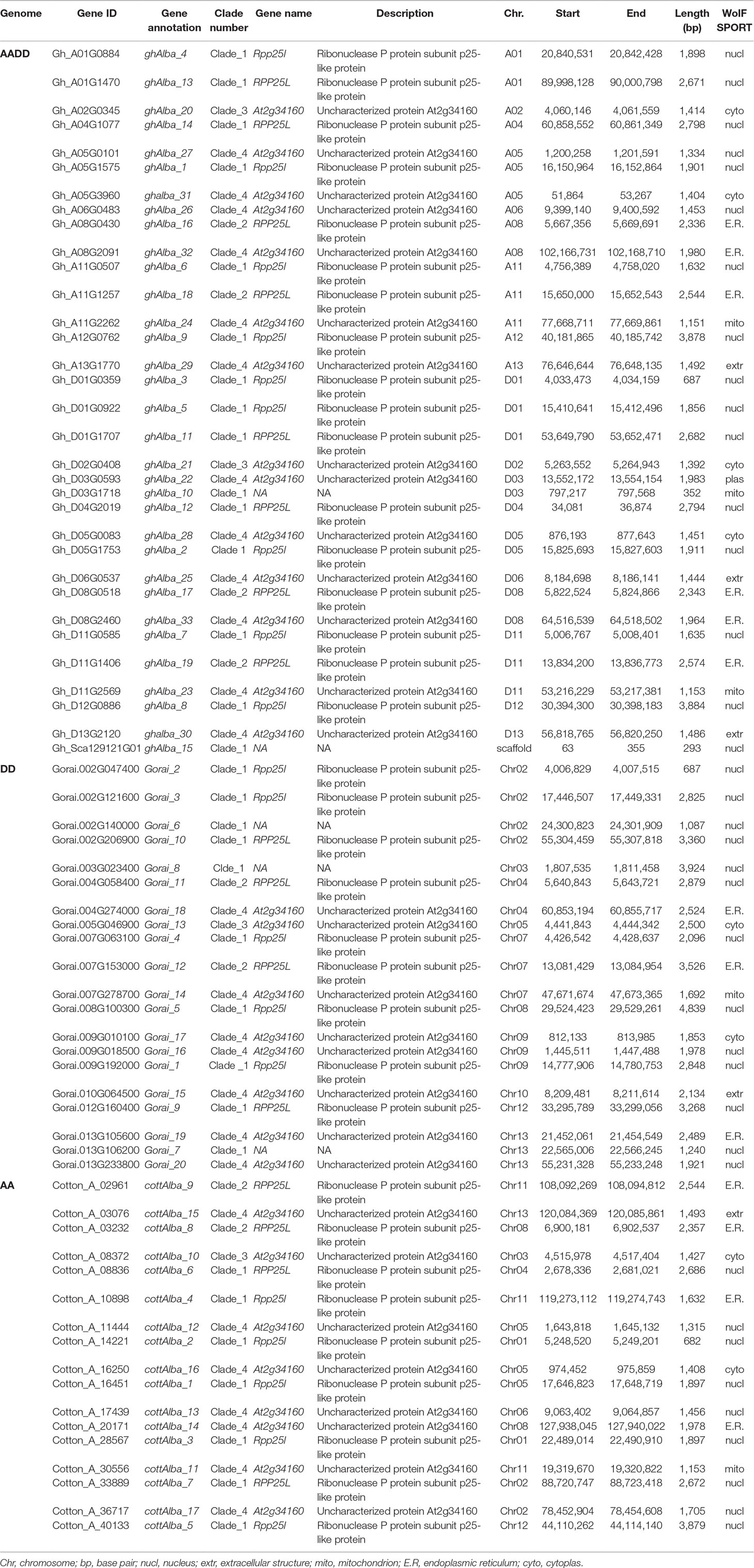
For RT-qPCR validation, 20 (61%), 13 (65%), and 10 (59%) genes were used for G. hirsutum, G. raimondii, and G. arboreum, respectively. The genes were chosen as per the results obtained from the phylogenetic tree analysis, RNA expression, and gene structure analysis. Based on phylogenetic tree analysis, the proteins encoded by the cotton Alba genes were grouped into four clades, but clade 3 members were least, which were majorly composed of Alba proteins from other plants used for the tree analysis; so, clades 1, 2, and 4 were considered for the gene selections. In relation to gene structure, we analyzed to take into consideration the nature of intron disruption; the selected genes shared a common gene structure attribute, with intron number ranging from five to nine. Finally, the secondary RNA sequenced data was obtained from the cotton genome database (https://cottonfgd.org/analyze/); only those genes which showed significance upregulation was finally chosen for further analysis through RT-qPCR validation, just as it has been previously described by Magwanga et al. (Magwanga et al., 2018), in the analysis of the LEA genes in upland cotton (Table S5). Only two tissues root and leaf were investigated under water deficit and salt stress conditions. The Alba genes across the three cotton species exhibited a similar expression pattern, in which more genes were found to be significantly up-regulated in the root tissues but not in the leaf (Table S6). In G. hirsutum, two genes were found to be significantly up-regulated in the leaf under water deficit and salt stress conditions; the same expression pattern was replicated in the roots under similar stress conditions, Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5) (Figure 1A). A unique expression pattern was noted among the highly upregulated genes; the significantly up-regulated genes were observed both in root and leaves under water deficit and salt stress conditions—for instance, Gorai.002G121600, Gorai.009G018500, and Gorai.010G064500 were highly upregulated in root and leaf tissues (Figure 1B); similarly, so were Cotton_A_28567, Cotton_A_20171, Cotton_A_08836, and Cotton_A_03076 (Figure 1C).
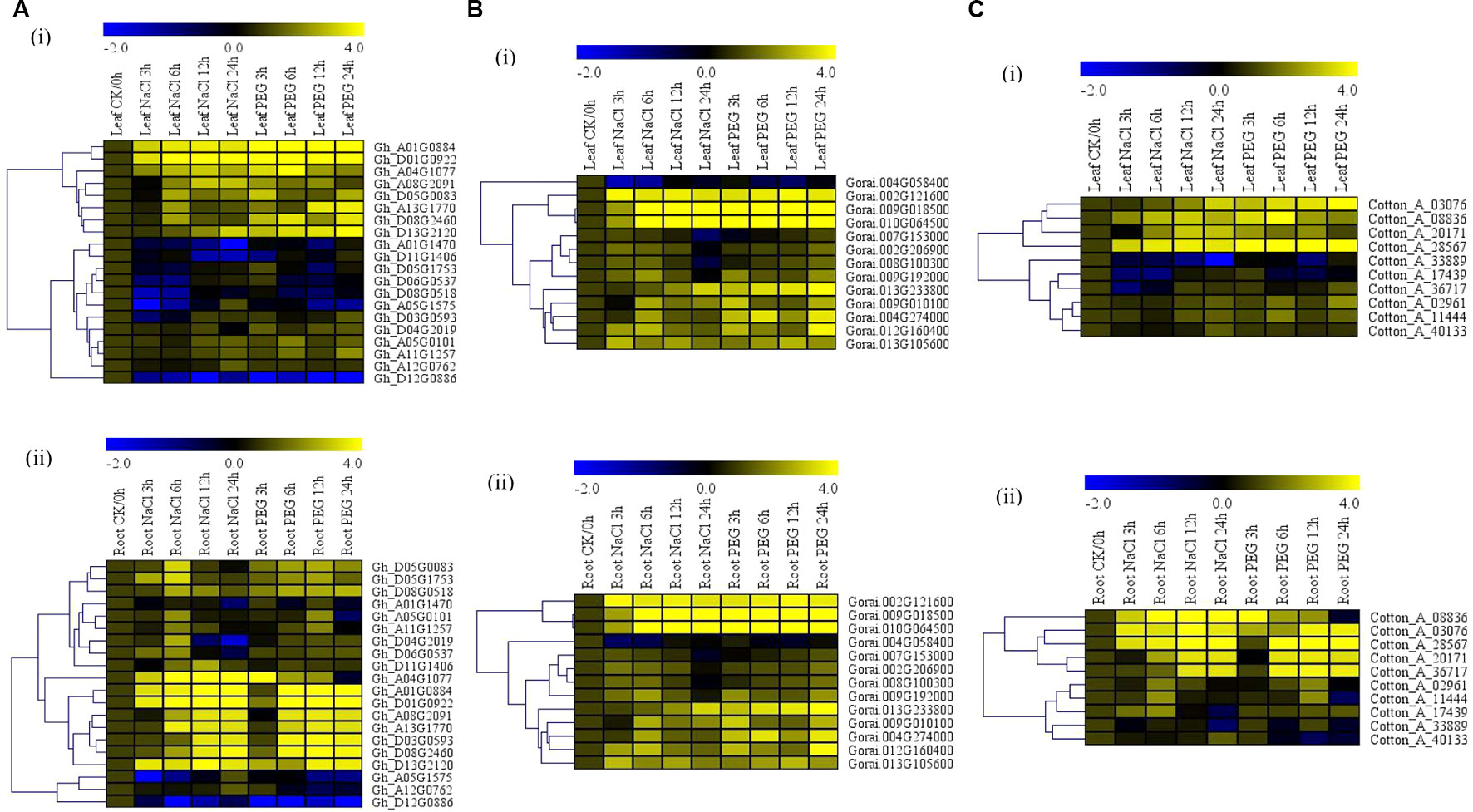
Figure 1 RT-qPCR analysis of the selected upland cotton Alba genes under water deficit and salt stress conditions. (A–C) Expression level of the selected cotton, G. hirsutum, G. raimondii and G. arboreum Alba genes under drought and salt stress conditions. The heatmap was visualized using MeV.exe program (showed by log 2 values). In control, and in treated samples, 1, 3, 6, and 12h poststress treatment. (i) Yellow—up-regulated, blue—down-regulated, and black—no expression.
The Efficiency of Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5) Gene Silencing in Cotton
The albino trait was observed among the plants infused with the phytoene desaturase gene (TRV-PDS) after 12 days of post-inoculation (dpi). The leaves and the stem region above the cotyledon became chlorotic thus exhibited the albino type of characteristics while the VIGS, wild types, and the positively controlled plants showed normal leaf color (Figure 2A). Moreover, the knockdown of the two Alba genes was further confirmed through carrying out a half RT-qPCR from the RNAs extracted from the PDS infused plants wild type, the positively controlled plants, and the two VIGS-plants using their specific primers. The TRV1 and 2 bands were never detected on the WT plants, but bands were formed in TRV:00, PDS, TRV: Alba_4, and TRV:Alba_5 infused plants; similarly, the two knocked genes bands were amplified in PDS, WT, and TRV:00 but showed thin bands on either of their extracted RNAs (Figure 2B). The bands were checked with an internal control gene, GhActin. These results showed that the targeted genes were effectively knocked down in the cotton plants. The efficiency of the VIGS on the plants is monitored phenotypically by the albino-like appearance on the leaves (Gao et al., 2011b). To further determine the efficiency level of the gene knockdown through VIGS, RT-qPCR assay was carried out on the leaf, stem, and root tissues collected from the TRV:Gh_A01G0884 (ghAlba_4) and TRV:Gh_D01G0922 (ghAlba_5) constructs, wild type, and the positively controlled plants. The transcript expression levels of the knocked genes, Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5), were significantly reduced in the Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants compared with their expression levels in the wild type and the positive control plants; though in the VIGS-plants, the expression levels of the knocked genes were relatively higher in the leaves compared to other tissues, such as the stem and the roots (Figures 2C–D).
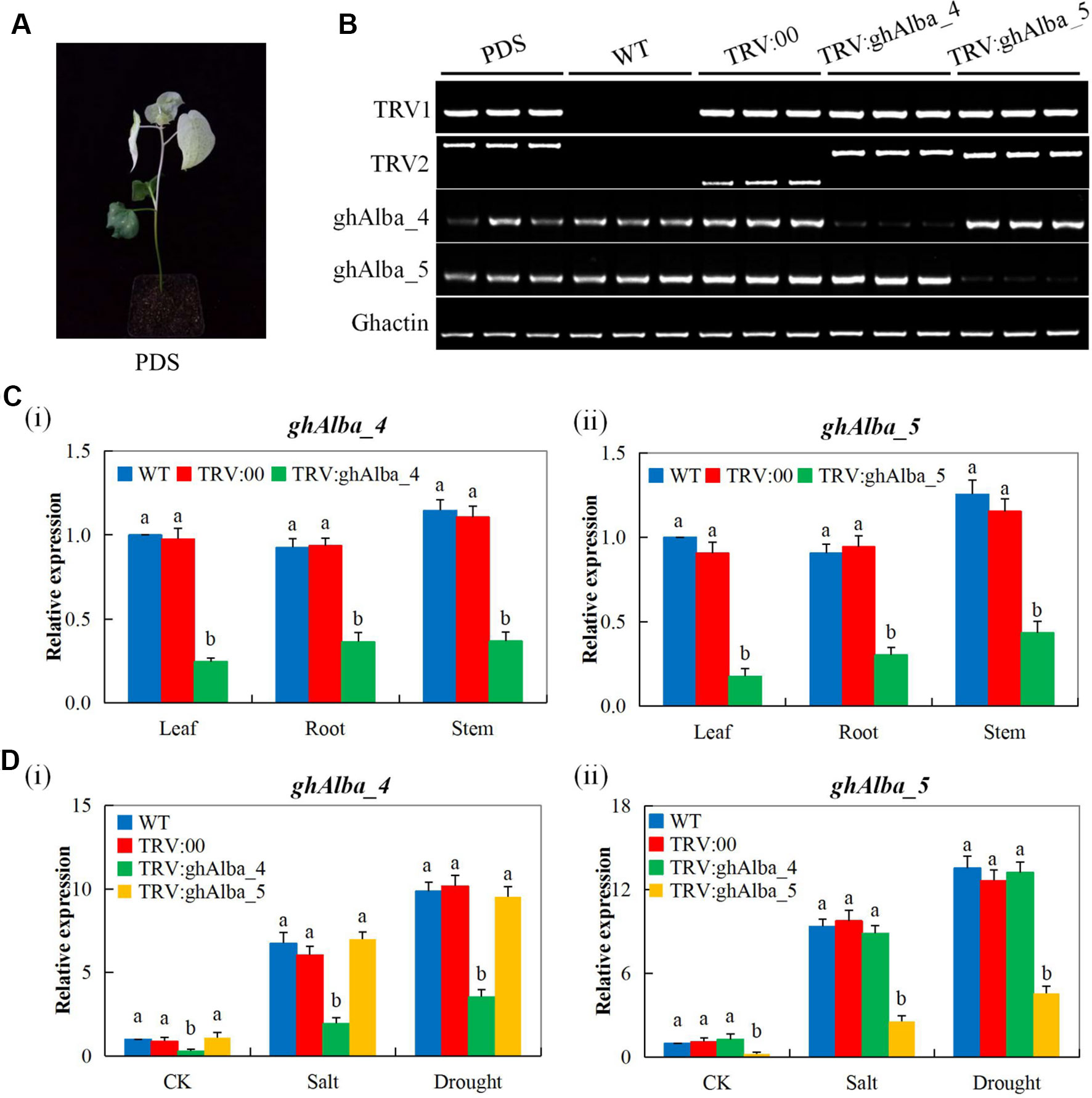
Figure 2 Phenotype observed in the silenced plants with the TRV2:00 empty vector, wild type plants, and Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants. (A) Albino appearance on the leaves of the Phytoene desaturase (PDS) infused plants. (B) Gel electrophoresis in determining the effectiveness of gene silencing by the vector. (C) RT-qPCR analysis of the change in the expression level of Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5) genes in cotton plants treated with VIGS. "Gh_A01G0884 (ghAlba_4) and TRV2:Gh_D01G0922 (ghAlba_5)” represent the Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5)-silenced plants. (D) Gel electrophoresis in determining the effectiveness of gene silencing by the vector. The letters a/b indicate statistically significant differences (two-tailed, p < 0.05) between the samples in each treatment. Error bars of the Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5) gene expression levels represent the standard deviation of three biological replicates.
Physiological Traits Evaluation and Root Assays of the VIGS Plants and the Wild Types Under Water Deficit and Salt Stress Conditions
Evaluation of the physiological traits, the Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants showed a significant reduction in CMS, ELWL, chlorophyll content, and RLWC compared with the wild types and the positive control plants (Figures 3A–D). The reduction in CMS as evident by high ion leakage showed that Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants suffered more of oxidative stress, and their membrane integrity was highly affected; the results were coherent with previous findings in which the knockdown of trihelix (TH), the plant TFs, affected the CMS and, in turn, increased the level of ion leakage (Magwanga et al., 2019a). Under environmental stress condition, the plant’s inability to assimilate sufficient amount of carbon (IV) oxide lead to increased photorespiration, thus higher production of hydrogen peroxide (Choudhury et al., 2017). Excess accumulation of reactive oxygen species (ROS) does cause cellular damage, which eventually leads to plant death (Mirza et al., 2013).
Figure 3 Evaluation of physiological traits in the VIGS-treated plants. (A) Chlorophyll content in the TRV2 empty vector-carrying plants and Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants. (B) The excised leaf water loss (ELWL) level in the TRV2 empty vector-carrying plants and Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants. (C) The relative leaf water content (RLWC) in the TRV2 empty vector-carrying plants and Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants. (D) The cell membrane stability (CMS) evaluated through ion leakage in the TRV2 empty vector-carrying plants and Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants. The letters a/b indicated statistically significant differences (two-tailed, p < 0.05) between the samples in each treatment. Error bars represent the standard deviation of three biological replicates.
The two cotton plants showed significant variation in root lengths and biomass accumulation. The Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants under treatment and control conditions compared with the pTRV2 (empty vector) were infused, and wild types had reduced root growth with relatively low emergence of lateral roots (Figures 4A–C). The results showed that the silencing of the Alba genes had a negative effect on root growth. Genes have been found to have a dominant effect on plant root growth—for instance, overexpression of water deficit inducible OsERF48 gene has been found to regulate rice calmodulin-like protein (OsCML16) gene, which promotes plant root growth and in turn enhance water deficit tolerance (Jung et al., 2017). The root is an important organ; it contributes directly to crop performance (Rogers and Benfey, 2015) and is the primary organ for the uptake of water and nutrients, which are the raw materials for photosynthesis in plants (Yamauchi et al., 2018). We hypothesize that the downregulation of the Alba genes could have an effect on the quiescent center on the root primordial region, thereby lowering the rate of cell division, enlargement, and elongation, which are the main cellular processes contributing to root growth.
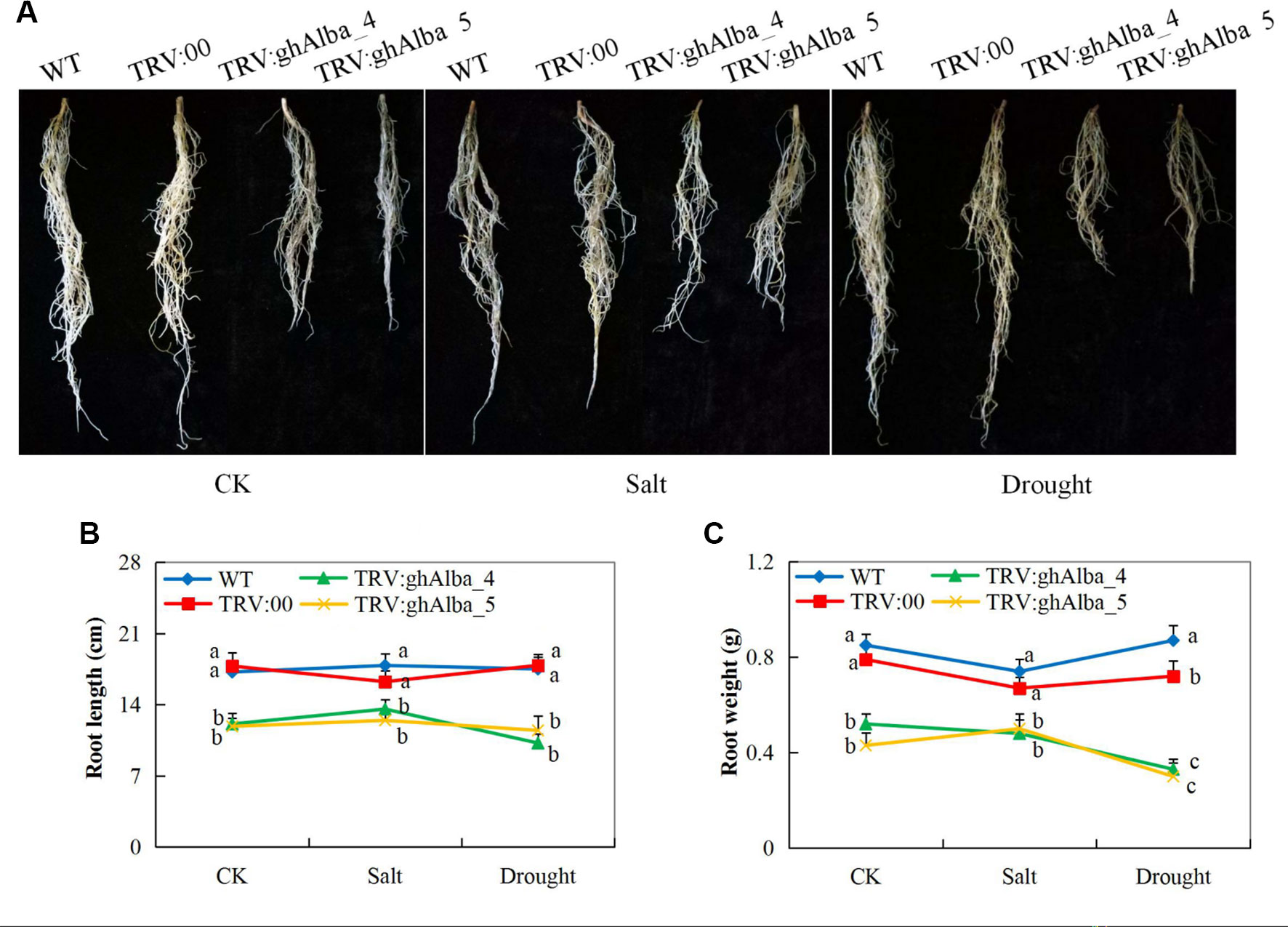
Figure 4 Root evaluation in the VIGS-treated plants (A) Root length in the TRV2 empty vector-carrying plants and Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5)-silenced plants, photographs taken after 8 days of stress exposure (B) Statistical determination of the root length in the TRV2 empty vector-carrying plants and Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5)-silenced plants (C) Root weight measurements in the TRV2 empty vector-carrying plants and Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5)-silenced plants. The letters a,b,c indicated statistically significant differences (two-tailed, p < 0.05) between the samples in each treatment. Error bars represent the standard deviation of three biological replicates.
The Oxidant, Antioxidant, and Abiotic Stress-Responsive Evaluation Between the VIGS and the Wild Cotton Varieties Under Water Deficit and Salt Stress Conditions
Evaluation of the oxidants and antioxidant enzymes showed that Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants were highly affected under water deficit and salt stress compared with the wild types and the control plants. The VIGS plants showed drought and salt stress symptoms on their leaves compared to the wild types (Figure 5), but when the positively controlled, the wild types, and the VIGS plants under drought stress were re-watered for a period of 3 days, the positively controlled and the wild type plants showed a significantly higher level of recovery compared to the VIGS plants (Figure S5A). The survival rate of the wilt type, the positively controlled, and the VIGS plants were 55% (11 of 20 plants), 50% (10 of 20 plants), and 20% (5 of 20 plants), respectively (Figure S5B). The results were in agreement to the finding obtained when the SpMPK1, SpMPK2, and SpMPK3 were knocked down in tomato; the survival rate of the VIGS plants was significantly lower than the wild types (Li et al., 2013). Moreover, evaluation of the oxidant and antioxidant enzymes such as POD, SOD, CAT, MDA, and H2O2 revealed that the VIGS-plants were significantly affected under drought and salt stress conditions compared to the wild types. The VIGS-plants and the wild types exhibited no significance difference under controlled conditions in all the biochemical parameters evaluated; however, under drought and salt stress conditions, MDA and H2O2 were significantly higher in concentration in the leaves of the VIGS plants but lower in the wild types (Figures 5B–C). Furthermore, three antioxidant enzymes were assayed; POD, SOD, and CAT all registered significant reduction on the leaves of VIGS plants under drought and salt stress, while there were no significant differences observed in their levels on the VIGS and wild types under controlled conditions (Figures 5D–F). The results obtained were in agreement to previous findings in which plants which are susceptible to any form of abiotic stress factor do register higher levels of oxidant enzymes as opposed to antioxidant under stress (Lu et al., 2018b).When plants are exposed to either abiotic or biotic stress conditions, the normal balance between ROS production and elimination shifts, leading to excessive accumulation of ROS and, in turn, resulting in massive oxidative damage, causing extensive cellular damage and inhibition of photosynthesis which limit the plant productivity. The excess ROS is then catalyzed into non-destructive form by antioxidant enzymes, such as catalase (CAT), peroxidase (POD) and superoxide dismutase (SOD), ascorbate peroxidase (APX), and polyphenoloxidase (PPO), among others (Wang et al., 2017a). The SOD is the first enzyme involved in the detoxification of ROS and converts superoxide (O2−) radicals to H2O2 (Kuo et al., 2013). The significant reduction in the concentration levels of the various antioxidant enzymes evaluated showed that the Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants were highly susceptible to drought and salt stresses compared with to the control and wild types, an indication showing that the Alba genes are integral in enhancing abiotic stress tolerance in plants.
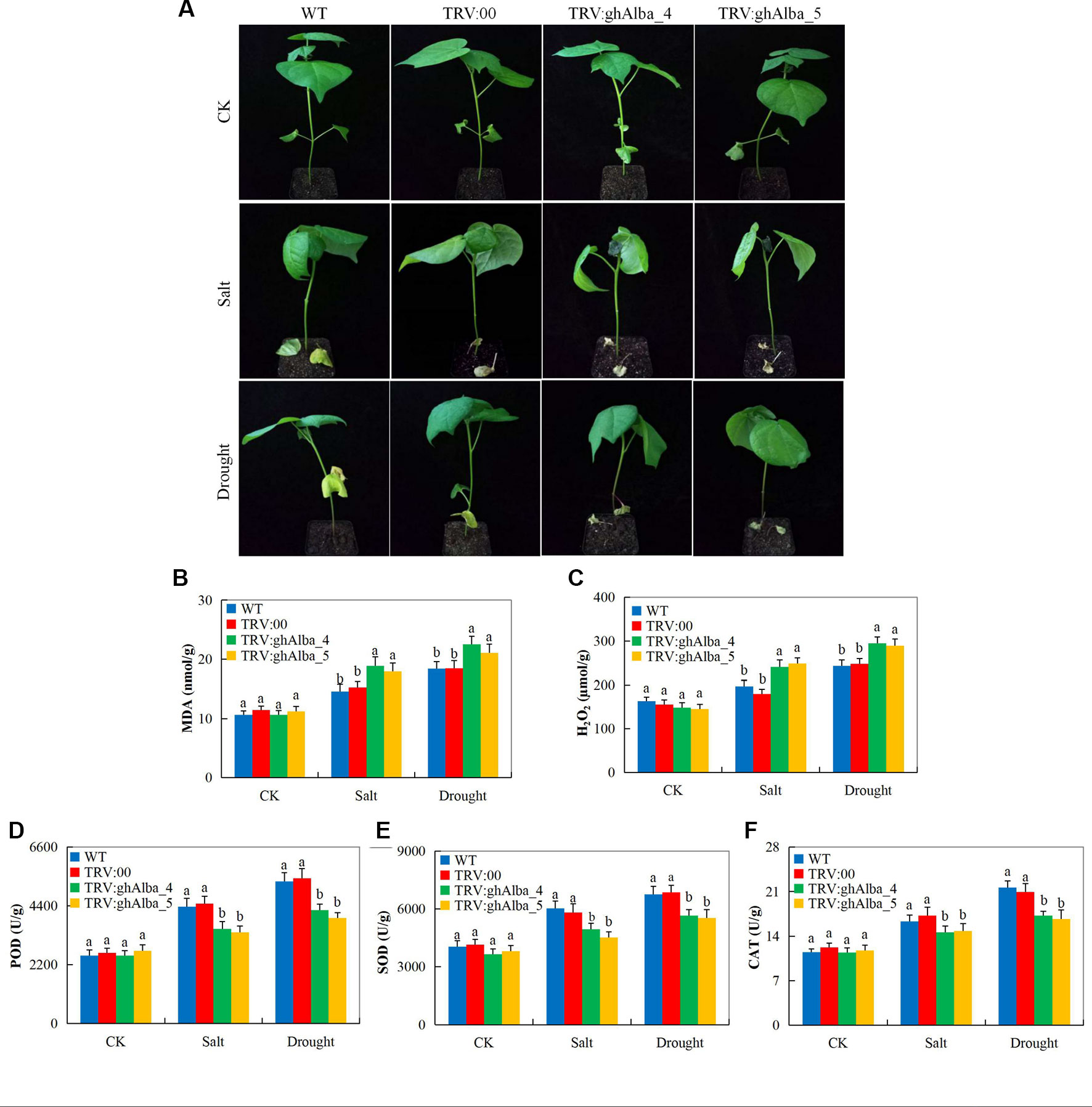
Figure 5 Phenotypic observation and the determination levels of the accumulation of oxidants and antioxidant enzymes in the VIGS-treated plants. (A) The phenotypic observation on the VIGS and wild types after drought and salt stress treatments, drought imposed by withdrawal of watering for 6 days while salt stress was initiated by irrigating the plants with 250 mM of NaCl solution for a period of 4 days. (B–F) The H2O2, POD, SOD, and CAT contents in the TRV2 empty vector-carrying plants and Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants. The letters a/b indicated statistically significant differences (two-tailed, p < 0.05) between the samples in each treatment set. Error bars of the H2O2, POD, SOD, and CAT contents represent the standard deviation of three biological replicates.
Transcription Analysis of Abiotic Stress-Responsive Genes on the Tissues of VIGS-Plants and Wild Types Under Drought and Salt Stress Conditions
The ability of the plants to induct stress-responsive genes indicates their ability to tolerate the stress levels (Jorge et al., 2017). In the evaluation of three abiotic stress-responsive genes, cotton superoxide dismutase (GhSOD), cotton myeloblastosis (GhMYB), and cotton delta-1-pyrroline-5-carboxylate synthetase (GhP5CS) showed the knockdown of the two Alba genes, ghAlba_4 and ghAlba_5, significantly affected the ability of the VIGS plants to induce more stress-responsive genes in order to improve their ability to tolerate the effects caused by drought and salt stresses. The expression levels of all the genes showed significant downregulation in the VIGS plants compared to their wild types under drought and salt stress conditions; however, under normal condition, no significant variation was observed among the VIGS and the wild types, an indication that the stress-responsive genes are only induced by the plants under stress conditions (Figure 6). The results obtained were in agreement to the previous finding in which the knockdown of cotton CYP450 genes significantly affected the ability of the plants to tolerate drought and or salt stress, and thus GhSOD, GhP5CS, and GhMYB genes were all downregulated in the VIGS plants (Magwanga et al., 2019b). The first line of defense by plants against the deleterious effects of ROS due to abiotic stresses is the SODs which convert O2− into H2O2; this is because of its presence in all the cellular compartments (Balamurugan et al., 2018). Furthermore, the MYB is among the top-ranked stress-responsive plant’s TFs together with NAC family members, and thus a number of investigations have revealed the key regulatory roles played by the MYBs in plant growth, development, and abiotic stress response (Tang et al., 2019). Moreover, delta-1-pyrroline-5-carboxylate synthase genes have been demonstrated to be vital in the proline biosynthesis pathways and are significantly induced by drought stress (Xia et al., 2017). The ability of the plants to induct the Alba proteins encoded by the Alba genes enables the plants to maintain the photosynthetic process and other drought and or salinity stress related tolerant mechanisms thus enhances the plants survival under drought and salt stress conditions (Figure S6). Thus, the downregulation of these stress-responsive genes showed that the knockdown of the Alba genes significantly reduced the tolerance levels of the cotton plants to drought and salt stresses.
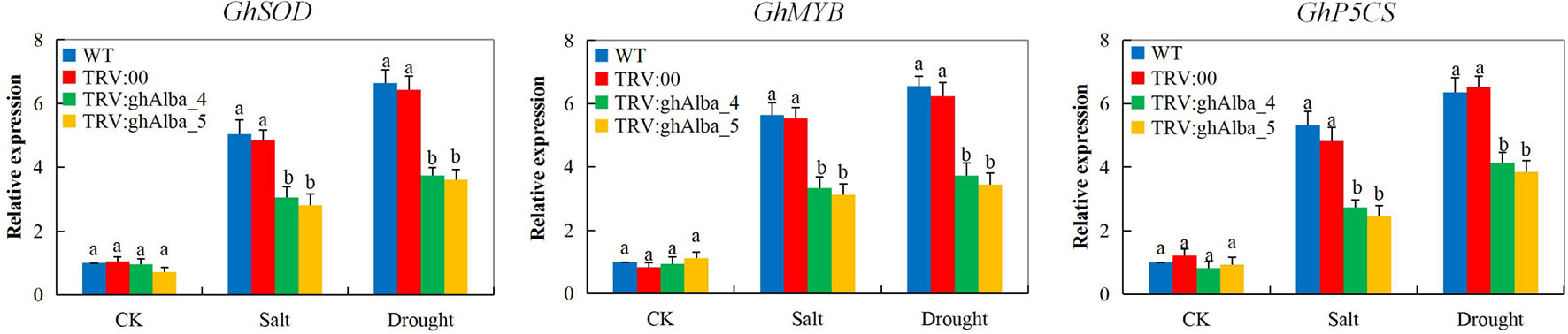
Figure 6 RT-qPCR analysis of the change in the expression levels of the GhP5CS, GhMYB, and GhSOD stress response genes in cotton plants treated with VIGS. “TRV2:00” represents the plants carrying control the TRV2 empty vector; “TRV2:Gh_A01G0884 (ghAlba_4) and TRV2:Gh_D01G0922 (ghAlba_5)” represent the Gh_A01G0884 (ghAlba_4)- and Gh_D01G0922 (ghAlba_5)-silenced plants. The letters a/b indicate statistically significant differences (two-tailed, p < 0.05) between the samples in each treatment. Error bars of the gene expression levels represent the standard deviation of three biological replicates.
Discussion
Unfavorable environmental changes have become reality, and this is projected to worsen if the rate of environmental degradation is not abated (Yadav et al., 2018). Plants being poikilothermic and sessile in nature, erratic environmental changes, do result in major losses in both yield and quality of the products (Halford et al., 2015). Non-edible plant with bushy architecture is a challenge to be grown under greenhouse conditions; thus, a number of crops are mainly grown in the fields, more so cotton, due to mechanization requirement and long growth periods. Several studies have been carried out in order to investigate the effects of abiotic stress on cotton production, and it has been shown that drought, salt, and extreme temperature stress are the major factors hindering full potential in cotton production (Isoda, 2010; Nachimuthu and Webb, 2017). Due to extreme growing conditions, plants have evolved various adaptive strategies at morphological, physiological, and molecular levels in order to reduce the effects of different abiotic stress factors (Rasool et al., 2019). At the molecular level, several stress-responsive genes and plant TFs have undergone tremendous transitions. Plants are capable to induct more of the genes and TFs to enable themselves to tolerate various stress factors. One of the gene families highly associated with stress tolerance is Alba genes (Kumar Verma et al., 2018). In this study, we identified various Alba genes in the three cotton species, G. hirsutum, G. arboreum, and G. raimondii, and found 33, 17, and 20 genes, respectively. The number of genes detected is in line with the nature of the three cotton genomes; G. hirsutum is a tetraploid cotton (AADD), having emerged through WGD of both the G. arboreum (AA) and G. raimondii (DD) (Yu-xiang et al., 2013); thus, the high number of genes in G. hirsutum affirms this evolution process. Some level of gene loss or duplication was detected; the numbers of genes in tetraploid cotton were less than the exact the number of proteins encoded by the Alba genes in the two diploid cotton parents. If the principle of WGD was to hold, it therefore means that either the tetraploid Alba genes lost some function or either of the two diploid Alba genes underwent duplication over time in the course of their evolution; though, this needs further investigation.
RNA expression profiling and RT-qPCR validation of the genes under water deficit and salt stresses showed that major genes were highly induced in the root tissues compared to other organs. The roots are the primary tissues which bear the full effect of water deficit and or salt stress being into contact with dry soil in case of water deficit or saline soil for the salt stress condition (Kunert et al., 2016). The highly upregulated genes in the root tissues were also found to be targeted by specific miRNAs—for instance, Gh_D01G0922 was target by ghr-miR396a and ghr-miR396b; the same miRNA has been found to be highly expressed in Arabidopsis (Liu et al., 2008), Zea mays (Ding et al., 2009), O. sativa (Gao et al., 2011a), and G. max (Li et al., 2011) under salt and water deficit conditions. In addition, miR396b has been found to exhibit at least two-fold changes under water deficit only in CB46, a drought-sensitive cowpea genotype (Barrera-Figueroa et al., 2011). Moreover, the highly up-regulated genes within the root tissues under water deficit and salt stresses were also found to be associated with some unique cis-regulatory elements—for instance, ABRELATERD1 (ACGTG )with a role in early responsive to dehydration, ARFAT (TGTCTC) in dehydration-responsiveness, CBFHV (RYCGAC) for dehydration-responsive element (DRE)/low temperature, LTRE1HVBLT49 (CCGAAA) as low-temperature-responsive element, MYBCORE (CNGTTR) in dehydration/water stress, and MYCCONSENSUSAT (CANNTG) as dehydration-responsive gene/cold stress. Similar cis-regulatory elements have been found to regulate some of the stress-responsive genes, such as the LEA genes (Magwanga et al., 2018).
In the functional characterization of the two highly upregulated Alba genes under water deficit and salt stress conditions, we carried out VIGS of the two genes, Gh_A01G0884 (ghAlba_4) and Gh_D01G0922 (ghAlba_5); in upland cotton, the VIGS and the wild type were exposed to salt and water deficit conditions. The VIGS cotton genotype was highly affected by water deficit and salt stress compared to the wild type. Chlorophyll content, CMS, saturated leaf weight (SLW), ELWL, and root traits showed negative deviation compared to the wild type, indicating that the wild type had a higher capacity to tolerate the effects caused by water deficit and salt stresses. Similarly, analysis of the reactive oxygen scavenging species, the antioxidant enzymes, POD, SOD, and CAT was significantly reduced in the leaves of the VIGS cotton. Moreover, the evaluation of the oxidants showed that H2O2 and MDA concentrations were significantly higher in the leaves of the VIGS than the wild type. When plants are exposed to any stress, the equilibrium between ROS release and detoxification becomes altered, thus leading to higher accumulation of ROS. Excess ROS results in oxidative injuries, which eventually lead to plant death. The low ROS scavenging enzymes in the leaves of the VIGS exhibited higher oxidative injuries compared to the wild type.
The SOD does constitute the first line of the plant’s defense against the deleterious effects of the ROS when plants are exposed to any form of abiotic stress; the ROS production increases leading to excessive accumulation (Sharma et al., 2012). In plants, O2− is produced at any cellular sites as long as the electron transport chain is involved (Grene, 2002); thus, O2 activation is likely to occur in plant cellular structures such as the mitochondria microsomes, glyoxysomes, peroxisomes, chloroplasts, cytosol, and the apoplasts (Elstner and Osswald, 1994). Thus, the level of ROS is checked by the activation of the antioxidant enzymes such as the SOD; thus, the lower concentration of this protein encoded by the GhSOD genes within the leaves of the VIGS cotton showed that the plants ability to regulate the amount of ROS was highly affected and thus were subjected to oxidative damage as a result of salt and water deficit exposure. In addition, the expression of the GhMYB gene was significantly downregulated in the VIGS cotton compared to the wild types. The plant exposure to various stress factors triggers a well-coordinated changes in gene expression (Virlouvet et al., 2018); the MYBs are among the top-ranked plants TFs highly associated with significant roles in promoting plants tolerance to various abiotic stress factors (Wang et al., 2017b), but it is worth noting that the genes work in a synchronized manner, the downregulation of the two Alba genes, affected the expression levels of the MYBs. Moreover, the pyrroline-5-carboxylate synthase (P5CS) enzyme is critical in catalyzing the various reaction leading to proline biosynthesis, and proline has been found to have a protective role against environmental and non-environmental stress effects in plants (Rai and Penna, 2013).
The proteins encoded by the Alba genes have an integral role in the genome construction of an organism and in turn control the expression dynamics of a number of genes in the organisms (Kumar Verma et al., 2018). Moreover, characterization of ArabidopsisAlba genes, AtALBA1 and AtALBA2, revealed that, despite their differences in nucleic acid binding properties, they are located within the localized within the nucleus and mainly form a heterodimers in the nucleus and do bind the R-loop structures, and their depletion results in hypersensitivity of the plants to DNA damaging agents as a result of abiotic stress factors (Yuan et al., 2019). The heterodimers are vital in immune response—for instance, in rice, OsCERK1 forms a heterodimer complex with OsCEBiP, which is a LysM-containing receptor-like protein and directly binds chitin, to induce immune responses (Kouzai et al., 2014). The downregulation of the cotton GhP5CS gene in the tissues of the VIGS cotton indicated that the proline biosynthesis cycle is altered, and plant ability to tolerate salt and water deficit was highly compromised. Moreover, the increased levels of the oxidant enzymes, such as the MDA and H2O2, showed that the knockdown of ghAlba_4 and ghAlba_5 significantly affected the ability of the plants to tolerate the effects of drought and salt stresses and a thus higher level of oxidative.
Conclusion
In conclusion, the identification and functional characterization of the Alba proteins in upland cotton provide fundamental information for future exploration of this diverse and yet underexplored plant protein family. This study gives the very first insight evaluation of the proteins encoded by the Alba genes in cotton. A total of 33, 20, and 17 proteins encoded by the Alba genes were identified in G. hirsutum, G. raimondii, and G. arboreum, respectively. The total number of the Alba proteins in the two diploid cottons, G. raimondii of the D genome and G. arboreum of the A genome, is less than the number of Alba proteins obtained for the tetraploid cotton G. hirsutum, even though the tetraploid cotton emerged as a result of WGD of the A and D (Lee and Fang, 2015), The low number could be attributed to gene loss after the emergence of the tetraploid cotton. The virus gene silencing (VIGS) of the two novel Alba genes in cotton revealed that the proteins encoded by the Alba genes are critical in enhancing root growth; primary growth is an important trait in xerophytic plants; and long and widely extended roots increase the rate of water absorption, thus improving the drought responsible mechanism among the xerophytic plants (Moriuchi and Winn, 2005). Furthermore, the VIGS plants when subjected to osmotic and salt stresses were found to have higher levels of the oxidant and significant reduction in antioxidant enzymes such as CAT, POD, and SOD, an indication that the seedlings were under intense oxidative stress compared to their wild types under similar conditions. Moreover, known stress-responsive genes such as GhSOD, GhMYB, and GhP5CS were all downregulated in the tissues of the VIGS cotton but were significantly upregulated in wild types under water deficit and salt stress conditions, which further augmented our results, in the validation of the possible roles of the proteins encoded by the Alba genes in enhancing water deficit and salt stress tolerance in cotton. We hereby propose further research to explore the exact role of the proteins encoded by the Alba genes at the cellular level.
Data Availability Statement
All datasets generated for this study are included in the manuscript/Supplementary Files.
Author Contributions
RM and FL designed the experiment, RM, PL and JK implemented and collected the data. RM analyzed the results and prepared the manuscript. RM, JK, PL, SA, FL, XW, XC, ZZ, YX, YH, KW and FL revised the manuscript. All authors reviewed and approved the final manuscript.
Funding
The research work was funded by the National key research and development plan (2016YFD0100306) and the National Natural Science Foundation of China (31671745, 31530053).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are deeply indebted to our supervisors, more specifically Prof Wang Kunbo and Prof Liu Fang for their valuable advice, supervision and guidance in the course of the research work. To all the teachers and the students, we are grateful for your support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01292/full#supplementary-material
Figure S1 | Phylogenetic tree analysis (B): protein sequence alignment of the Alba Genes in G. hirsutum, G. raimondii, G. arboreum, Oryza sativa, Theobroma cacao, Arabidopsis thaliana, Sorghum bicolor, Populus trichocarpa and Glycine max. The red enclosure indicates the cloned genes.
Figure S2 | Cotton Alba gene structure and motif analysis (A) Phylogenetic tree, gene structure, and motif of all the G. hirsutum Alba genes. (B): Phylogenetic tree analysis, gene structure, and motif identification of the proteins encoded by the Alba genes for G. raimondii. (C): Phylogenetic tree, gene structure, and motif identification of the proteins encoded by the Alba genes for G. arboreum. Red: exons, blue: up/downstream, and the grey lines: the introns.
Figure S3 | Average number of the cis-regulatory elements in the promoter region of the three cotton species Alba genes. The cis-regulatory elements were analyzed in the 1 kb up/ down stream promoter region of the translation start site using the online tool, PLACE database.
Figure S4 | RNA sequence analysis of the Alba genes under water deficit and salt stress conditions. The heat map was visualized using the MeV4.9.0 program. Yellow indicates upregulation; blue indicates downregulation and black indicates no expression. (A) Heat map showing gene expression under salt stress conditions. (B) Heat map showing gene expression under water deficit condition. The abbreviations Rt: root; St: stem; Lf: leaf; CK: control condition; PEG: Polyethylene glycol for initiating osmotic stress. The RNA sequence analysis was profiled at 0h, 1h, 3h, 6h and 12h of stress exposure under salt and osmotic stress conditions.
Figure S5 | Phenotype.
Table S1 | Protein sequences for the Alba proteins used in the phylogenetic tree analysis. Gh: G. hirsutum; Gorai: G. raimondii, Cotton_A: G. arboreum; Thecca: Theobroma cacao, AT: Arabidopsis thaliana, Sobic: S. bicolor, Glyma: G. max, Potri: P. trichorcarpa and LOC_Os: Oryza sativa.
Table S2 | Details of the primers used for RT-qPCR analysis.
Table S3 | The predicted miRNAs found to target various cotton Alba genes cis-Alba.
Table S4 | Detailed information on the various cis-regulatory elements found to be associated with Alba genes in the three sequenced cotton species.
Table S5 | RNA sequence data for the upland cotton Alba genes profiled under drought and salt stress conditions.
Table S6 | The RT-qPCR results for the expression profiling of the selected Alba genes for G. hirsutum, G. raimondii and G. arboreum. A: The leaf tissues and B: The root tissues. The 2-△△CT values were transformed into log 2. CK: control, NaCl: sodium chloride (salt stress), PEG: polyethylene glycol (drought stress); the leaf and root tissues were collected at 0h, 3h, 6h, 12h and 24h of stress exposure.
References
Aravind, L., Iyer, L. M., Anantharaman, V. (2003). The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 4, R64. doi: 10.1186/gb-2003-4-10-r64
Arribas-Layton, M., Dennis, J., Bennett, E. J., Damgaard, C. K., Lykke-Andersen, J. (2016). The C-terminal RGG domain of human Lsm4 promotes processing body formation stimulated by arginine dimethylation. Mol. Cell. Biol. 3, 2226–2235. doi: 10.1128/MCB.01102-15
Balamurugan, M., Santharaman, P., Madasamy, T., Rajesh, S., Sethy, N. K., Bhargava, K., et al. (2018). Recent trends in electrochemical biosensors of superoxide dismutases. Biosens. Bioelectron. 116, 89–99. doi: 10.1016/j.bios.2018.05.040
Banerjee, A., Roychoudhury, A. (2017). Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254, 3–16. doi: 10.1007/s00709-015-0920-4
Barrera-Figueroa, B. E., Gao, L., Diop, N. N., Wu, Z., Ehlers, J. D., Roberts, P. A., et al. (2011). Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biol. 11, 127. doi: 10.1186/1471-2229-11-127
Barrs, H., Weatherley, P. (1962). A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust. J. Biol. Sci. 15, 413. doi: 10.1071/BI9620413
Bell, S. D., Botting, C. H., Wardleworth, B. N., Jackson, S. P., White, M. F. (2002). The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 80-(296) 148–151. doi: 10.1126/science.1070506
Boudsocq, M. (2005). Osmotic signaling in plants. Multiple pathways mediated by emerging kinase families. Plant Physiol. 138, 1185–1194. doi: 10.1104/pp.105.061275
Brown, P., Baxter, L., Hickman, R., Beynon, J., Moore, J. D., Ott, S. (2013). MEME-LaB: motif analysis in clusters. Bioinformatics (29), 1696–1697. doi: 10.1093/bioinformatics/btt248
Cakmak, I., Horst, W. J. (1991). Effect of aluminium on lipid peroxidation, siiperoxide dismutase, catalase, and peroxidase activities in root tips of soybean (Glycine max). Physiol. Plant. 83, 463–468. doi: 10.1111/j.1399-3054.1991.tb00121.x
Cakmak, I., Marschner, H. (1992). Magnesium deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase, and glutathione reductase in bean leaves. Plant Physiol. 98, 1222–1227. doi: 10.1104/pp.98.4.1222
Chong, P. A., Vernon, R. M., Forman-Kay, J. D. (2018). RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 430, 4650–4665. doi: 10.1016/j.jmb.2018.06.014
Choudhary, M. K., Basu, D., Datta, A., Chakraborty, N., Chakraborty, S. (2009). Dehydration-responsive nuclear proteome of rice (Oryza sativa L.) illustrates protein network, novel regulators of cellular adaptation, and evolutionary perspective. Mol. Cell. Proteomics 8, 1579–1598. doi: 10.1074/mcp.M800601-MCP200
Choudhury, F. K., Rivero, R. M., Blumwald, E., Mittler, R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867. doi: 10.1111/tpj.13299
Crnigoj, M., Hanzlowsky, A., Vilfan, T., Ulrih, N. P. (2011). Heterologous expression of the Alba protein from the hyperthermophilic Archaeon Aeropyrum Pernix. Croat. Chem. Acta 84, 499–504. doi: 10.5562/cca1772
Ding, D., Zhang, L., Wang, H., Liu, Z., Zhang, Z., Zheng, Y. (2009). Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot. 103, 29–38. doi: 10.1093/aob/mcn205
do, A. C., Bisognin, D., Steffens, C. (2008). Non-destructive quantification of chlorophylls in leaves by means of a colorimetric method. Hortic. Bras. 26, 471–475. doi: 10.1590/S0102-05362008000400009
Dupé, A., Dumas, C., Papadopoulou, B. (2015). Differential subcellular localization of leishmania alba-domain proteins throughout the parasite development. PLoS One (9), 10. doi: 10.1371/journal.pone.0137243
Elstner, E. F., Osswald, W. (1994). Mechanisms of oxygen activation during pl ant stress. Proc. R. Soc. Edinburgh. Sect. B. Biol. Sci. 102, 131–154. doi: 10.1017/S0269727000014068
Fang, D. D., Percy, R. G. (2015). Cotton. Agron. Monogr. 57. ASA, CSSA, and SSSA, Madison, WI. doi: 10.2134/agronmonogr57.frontmatter
Fokar, M., Nguyen, H. T., Blum, A. (1998). Heat tolerance in spring wheat. I. Estimating cellular thermotolerance and its heritability. Euphytica 104, 1–8. doi: 10.1023/A:1018346901363
Gao, P., Bai, X., Yang, L., Lv, D., Pan, X., Li, Y., et al. (2011a). Osa-MIR393: a salinity- and alkaline stress-related microRNA gene. Mol. Biol. Rep. 38, 237–242. doi: 10.1007/s11033-010-0100-8
Gao, X., Britt, R. C., Jr., Shan, L., He, P. (2011b). Agrobacterium-mediated virus-induced gene silencing assay in cotton. J. Vis. Exp. 20 (54), 2938. doi: 10.3791/2938
Giannopolitis, C. N., Ries, S. K. (1977). Superoxide dismutases: II. Purification and quantitative relationship with water-soluble protein in seedlings. Plant Physiol. 59, 315–318. doi: 10.1104/pp.59.2.315
Gissot, M., Walker, R., Delhaye, S., Alayi, T. D., Huot, L., Hot, D., et al. (2013). Toxoplasma gondii alba proteins are involved in translational control of gene expression. J. Mol. Biol. 425, 1287–1301. doi: 10.1016/j.jmb.2013.01.039
Goyal, M., Alam, A., Iqbal, M. S., Dey, S., Bindu, S., Pal, C., et al. (2012). Identification and molecular characterization of an Alba-family protein from human malaria parasite Plasmodium falciparum. Nucleic Acids Res. 40, 1174–1190. doi: 10.1093/nar/gkr821
Grene, R. (2002). Oxidative stress and acclimation mechanisms in plants. Arabidopsis Book 1, e0036. doi: 10.1199/tab.0036.1
Guo, L., Ding, J., Guo, R., Hou, Y., Wang, D. C., Huang, L. (2014). Biochemical and structural insights into RNA binding by Ssh10b, a member of the highly conserved Sac10b protein family in archaea. J. Biol. Chem. 289, 1478–1490. doi: 10.1074/jbc.M113.521351
Guo, R., Xue, H., Huang, L. (2003). Ssh10b, a conserved thermophilic archaeal protein, binds RNA in vivo. Mol. Microbiol. 50, 1605–1615. doi: 10.1046/j.1365-2958.2003.03793.x
Halford, N. G., Curtis, T. Y., Chen, Z., Huang, J. (2015). Effects of abiotic stress and crop management on cereal grain composition: Implications for food quality and safety. J. Exp. Bot. 66, 1145–1156. doi: 10.1093/jxb/eru473
Hernandez, Y., Sanan-Mishra, N. (2017). miRNA mediated regulation of NAC transcription factors in plant development and environment stress response. Plant Gene. (11), 190–198. doi: 10.1016/j.plgene.2017.05.013
Hoagland, D. R., Arnon, D. I. (1950). The water-culture method for growing plants without soil. 1884-1949. Berkeley, Calif.: University of California, College of Agriculture, Agricultural Experiment Station, 1938.
Isoda, A. (2010). Effects of water stress on leaf temperature and chlorophyll fluorescence parameters in cotton and peanut. Plant Prod. Sci. 13, 269–278. doi: 10.1626/pps.13.269
Jorge, T. F., Duro, N., da Costa, M., Florian, A., Ramalho, J. C., Ribeiro-Barros, A. I., et al. (2017). GC-TOF-MS analysis reveals salt stress-responsive primary metabolites in Casuarina glauca tissues. Metabolomics 13, 95. doi: 10.1007/s11306-017-1234-7
Jung, H., Chung, P. J., Park, S. H., Redillas, M. C. F. R., Kim, Y. S., Suh, J. W., et al. (2017). Overexpression of OsERF48 causes regulation of OsCML16, a calmodulin-like protein gene that enhances root growth and drought tolerance. Plant Biotechnol. J. 15, 1295–1308. doi: 10.1111/pbi.12716
Kouzai, Y., Mochizuki, S., Nakajima, K., Desaki, Y., Hayafune, M., Miyazaki, H., et al. (2014). Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Mol. Plant-Microbe Interact. 27, 975–982. doi: 10.1094/MPMI-03-14-0068-R
Kumar, R. (2014). Role of microRNAs in biotic and abiotic stress responses in crop plants. Appl. Biochem. Biotechnol. 174, 93–115. doi: 10.1007/s12010-014-0914-2
Kumar Verma, J., Wardhan, V., Singh, D., Chakraborty, S., Chakraborty, N. (2018). Genome-wide identification of the Alba gene family in plants and stress-responsive expression of the rice alba genes. Genes (Basel). 9, 1–23. doi: 10.3390/genes9040183
Kunbo, W., Wendel, J. F., Hua, J. (2018). Designations for individual genomes and chromosomes in Gossypium. (1), 3–7. doi: 10.1186/s42397-018-0002-1
Kunert, K., Vorster, B. J., Fenta, B. A., Kibido, T., Dionisio, G., Foyer, C. H. (2016). Drought stress responses in soybean roots and nodules. Front. Plant Sci. 7, 1015. doi: 10.3389/fpls.2016.01015
Kuo, W.-Y., Huang, C.-H., Shih, C., Jinn, T.-L. (2013). Cellular extract preparation for superoxide dismutase (SOD) activity assay. Bio-Protocol 3, 3–5. doi: 10.21769/BioProtoc.811
Lee, J. A., Fang, D. D. (2015). “Cotton as a world crop: origin, history, and current status,” in Cotton. Eds. Fang, D. D., Percy, R. G.. Madison, WI: Cotton. Agron. Monogr. 57. ASA, CSSA, and SSSA. doi: 10.2134/agronmonogr57.frontmatter
Li, C., Yan, J. M., Li, Y. Z., Zhang, Z. C., Wang, Q. L., Liang, Y. (2013). Silencing the SpMPK1, SpMPK2, and SpMPK3 genes in tomato reduces abscisic acid-mediated drought tolerance. Int. J. Mol. Sci. 14, 21983–21996. doi: 10.3390/ijms141121983
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboretum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
Li, H., Dong, Y., Yin, H., Wang, N., Yang, J., Liu, X., et al. (2011). Characterization of the stress associated microRNAs in Glycine max by deep sequencing. BMC Plant Biol. 11, 170. doi: 10.1186/1471-2229-11-170
Li, X., Jin, X., Wang, H., Zhang, X., Lin, Z. (2016). Structure, evolution, and comparative genomics of tetraploid cotton based on a high-density genetic linkage map. DNA Res. 23, 283–293. doi: 10.1093/dnares/dsw016
Liu, H. H., Tian, X., Li, Y. J., Wu, C. A., Zheng, C. C. (2008). Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14, 836–843. doi: 10.1261/rna.895308
Liu, Q., Zhang, Y. C., Wang, C. Y., Luo, Y. C., Huang, Q. J., Chen, S. Y., et al. (2009). Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett. 583, 723–728. doi: 10.1016/j.febslet.2009.01.020
Liu, Y.-F., Zhang, N., Liu, X., Wang, X., Wang, Z.-X., Chen, Y., et al. (2012). Molecular mechanism underlying the interaction of typical Sac10b family proteins with DNA. PLoS One 7, e34986. doi: 10.1371/journal.pone.0034986
Loreto, F., Velikova, V. (2001). Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quenches ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol. 127, 1781–1787. doi: 10.1104/pp.010497
Lu, P., Magwanga, R. O., Guo, X., Kirungu, J. N., Lu, H., Cai, X. (2018a). Genome-wide analysis of Multidrug and toxic compound extrusion (MATE) family in diploid cotton, G. raimondii and G. arboreum and its expression analysis under salt. G3 (Bethesda) 8 (7), 2483–2500. doi: 10.1534/g3.118.200232
Lu, P., Magwanga, R. O., Lu, H., Kirungu, J. N., Wei, Y., Dong, Q., et al. (2018b). A novel G-protein-coupled receptors gene from upland cotton enhances salt stress tolerance in transgenic Arabidopsis. Genes (Basel). 9 (4). doi: 10.3390/genes9040209
Magwanga, R. O., Kirungu, J. N., Pu, L., Xiu, Y., Dong, Q., Cai, X., et al. (2019a). Genome wide identification of the trihelix transcription factors and overexpression of Gh_A05G2067 (GT-2), a novel gene contributing to increased drought and salt stresses tolerance in cotton. Physiol. Plant. 2067. doi: 10.1111/ppl.12920
Magwanga, R. O., Lu, P., Kirungu, J. N., Dong, Q., Cai, X., Zhou, Z., et al. (2019b). Knockdown of Cytochrome P450 Genes Gh_D07G1197 and Gh_A13G2057 on chromosomes D07 and A13 reveals their putative role in enhancing drought and salt stress tolerance in Gossypium hirsutum. Genes (Basel). 10, 226. doi: 10.3390/genes10030226
Magwanga, R. O., Lu, P., Kirungu, J. N., Lu, H., Wang, X., Cai, X., et al. (2018). Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 19 (1), 6. doi: 10.1186/s12863-017-0596-1
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
McCaig, T. N., Romagosa, I. (1989). Measurement and use of excised-leaf water status in wheat. Crop Sci. 29, 1140–1145. doi: 10.2135/cropsci1989.0011183X002900050008x
Mirza, H., Kamrun, N., Singh, G. S., Masayuki, F. (2013). Drought stress responses in plants, oxidative stress, and antioxidant defense. Clim. Chang. Plant Abiotic Stress Toler. 209–250 .doi: 10.1002/9783527675265.ch09
Moriuchi, K. S., Winn, A. A. (2005). Relationships among growth, development and plastic response to environment quality in a perennial plant. New Phytol. 166, 149–158. doi: 10.1111/j.1469-8137.2005.01346.x
Mu, C., Zhou, L., Li, M.-Y., Du, M.-W., Zhang, M.-C., Tian, X.-L., et al. (2016). Establishment and optimisation of virus-induced gene silencing in system hydroponic cotton. Acta Agron. Sin. 42, 844. doi: 10.3724/SP.J.1006.2016.00844
Muñoz, N., Li, M. W., Ngai, S. M., Lam, H. M. (2016). “Use of proteomics to evaluate soybean response under abiotic stresses,” in Abiotic and Biotic Stresses in Soybean Production. Academic Press (1), 80–105. doi: 10.1016/B978-0-12-801536-0.00004-9
Nachimuthu, G., Webb, A. A. (2017). Closing the biotic and abiotic stress-mediated yield gap in cotton by improving soil management and agronomic practices. In: Plant tolerance to individual and concurrent stresses. Ed. Senthil-Kumar, M. (New Delhi: Springer). doi: 10.1007/978-81-322-3706-8_2
Nakashima, K., Yamaguchi-Shinozaki, K. (2006). Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol. Plant. 126, 62–71. doi: 10.1111/j.1399-3054.2005.00592.x
Olguin-Lamas, A., Madec, E., Hovasse, A., Werkmeister, E., Callebaut, I., Slomianny, C., et al. (2011). A novel Toxoplasma gondii nuclear factor TgNF3 is a dynamic chromatin-associated component, modulator of nucleolar architecture and parasite virulence. PLoS Pathog. 7 (3), e1001328. doi: 10.1371/journal.ppat.1001328
Petrov, P., Petrova, A., Dimitrov, I., Tashev, T., Olsovska, K., Brestic, M., et al. (2018). Relationships between leaf morpho-anatomy, water status and cell membrane stability in leaves of wheat seedlings subjected to severe soil drought. J. Agron. Crop Sci. 204, 219–227. doi: 10.1111/jac.12255
Qin, F., Shinozaki, K., Yamaguchi-Shinozaki, K. (2011). Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 52, 1569–1582. doi: 10.1093/pcp/pcr106
Rai, A. N., Penna, S. (2013). Molecular evolution of plant P5CS gene involved in proline biosynthesis. Mol. Biol. Rep. 40, 6429–6435. doi: 10.1007/s11033-013-2757-2
Rao, X., Huang, X., Zhou, Z., Lin, X. (2013). An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 3, 71–85. doi: 10.1016/j.biotechadv.2011.08.021
Rasool, S., Mir, B. A., Rehman, M. U., Amin, I., Mir, M. U. R., Ahmad, S. B. (2019). “Abiotic stress and plant senescence,” in Senescence Signalling and Control in Plants, (Academic Press) 15–27. doi: 10.1016/B978-0-12-813187-9.00002-0
Reeve, J. N. (2003). Archaeal chromatin and transcription. Mol. Microbiol. 48, 587–598. doi: 10.1046/j.1365-2958.2003.03439.x
Rogers, E. D., Benfey, P. N. (2015). Regulation of plant root system architecture: implications for crop advancement. Curr. Opin. Biotechnol. 32, 93–98. doi: 10.1016/j.copbio.2014.11.015
Saibo, N. J. M., Lourenco, T., Oliveira, M. M. (2008). Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann. Bot. 103, 609–623. doi: 10.1093/aob/mcn227
Sharma, P., Jha, A. B., Dubey, R. S., Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 1–26. doi: 10.1155/2012/217037
Shinozaki, K., Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. (58), 221–227. doi: 10.1093/jxb/erl164
Song, S. U., Oh, I.-S., Lee, B., Suh, J.-K., Kim, J.-H., Cha, Y.-D., et al. (2001). Identification of a negative Cis-regulatory element and multiple DNA binding proteins that inhibit transcription of the transforming growth factor-β type II receptor gene. Gene 262 (1–2), 179–87. doi: 10.1016/S0378-1119(00)00534-5
Starai, V. J., Escalante-Semerena, J. C. (2004). Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340, 1005–1012. doi: 10.1016/j.jmb.2004.05.010
Subota, I., Rotureau, B., Blisnick, T., Ngwabyt, S., Durand-Dubief, M., Engstler, M., et al. (2011). ALBA proteins are stage regulated during trypanosome development in the tsetse fly and participate in differentiation. Mol. Biol. Cell 22, 4205–4219. doi: 10.1091/mbc.e11-06-0511
Tanentzap, F. M., Stempel, A., Ryser, P. (2015). Reliability of leaf relative water content (RWC) measurements after storage: consequences for in situ measurements. Botany 93, 535–541. doi: 10.1139/cjb-2015-0065
Tang, Y., Bao, X., Zhi, Y., Wu, Q., Guo, Y., Yin, X., et al. (2019). Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice. Front. Plant Sci. 10, 168. doi: 10.3389/fpls.2019.00168
Thirumalaikumar, V. P., Devkar, V., Mehterov, N., Ali, S., Ozgur, R., Turkan, I., et al. (2018). NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 16, 354–366. doi: 10.1111/pbi.12776
Van Assche, F., Cardinaels, C., Clijsters, H. (1988). Induction of enzyme capacity in plants as a result of heavy metal toxicity: dose-response relations in Phaseolus vulgaris L., treated with zinc and cadmium. Environ. Pollut. 52, 103–115. doi: 10.1016/0269-7491(88)90084-X
Verma, J. K., Gayali, S., Dass, S., Kumar, A., Parveen, S., Chakraborty, S., et al. (2014). OsAlba1, a dehydration-responsive nuclear protein of rice (Oryza sativa L. ssp. indica), participates in stress adaptation. Phytochemistry 100, 16–25. doi: 10.1016/j.phytochem.2014.01.015
Virlouvet, L., Avenson, T. J., Du, Q., Zhang, C., Liu, N., Fromm, M., et al. (2018). Dehydration stress memory: gene networks linked to physiological responses during repeated stresses of Zea mays. Front. Plant Sci. 9, 1058. doi: 10.3389/fpls.2018.01058
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103. doi: 10.1038/ng.2371
Wang, N., Zhang, W., Qin, M., Li, S., Qiao, M., Liu, Z., et al. (2017a). Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol. 58, 1764–1776. doi: 10.1093/pcp/pcx111
Wang, Y., Wang, Q., Liu, M. L., Bo, C., Wang, X., Ma, Q., et al. (2017b). Overexpression of a maize MYB48 gene confers drought tolerance in transgenic arabidopsis plants. J. Plant Biol. 60, 612–621. doi: 10.1007/s12374-017-0273-y
Wardleworth, B. N., Russell, R. J. M., Bell, S. D., Taylor, G. L., White, M. F. (2002). Structure of Alba : an archaeal chromatin protein modulated by acetylation. EMBO J. 21, 4654–4662. doi: 10.1093/emboj/cdf465
Weissman, A. M. (2001). Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2, 169–178. doi: 10.1038/35056563
Xia, Y., Li, R., Bai, G., Siddique, K. H. M., Varshney, R. K., Baum, M., et al. (2017). Genetic variations of HvP5CS1 and their association with drought tolerance related traits in barley (Hordeum vulgare L.). Sci. Rep. 7 (1), 7870. doi: 10.1038/s41598-017-08393-0
Xue, H., Guo, R., Wen, Y., Liu, D., Huang, L. (2000). An abundant DNA binding protein from the hyperthermophilic archaeon Sulfolobus shibatae affects DNA supercoiling in a temperature-dependent fashion. J. Bacteriol. 182, 3929–3933. doi: 10.1128/JB.182.14.3929-3933.2000
Yadav, S. S., Redden, R. J., Hatfield, J. L., Ebert, A. W., Hunter, D., Dass, S., et al (2018). “Crop production management to climate change,” in Food Security and Climate Change, 251–287. doi: 10.1002/9781119180661.ch12
Yamauchi, T., Colmer, T. D., Pedersen, O., Nakazono, M. (2018). Regulation of root traits for internal aeration and tolerance to soil waterlogging-flooding stress. Plant Physiol. (176), 1118–1130. doi: 10.1104/pp.17.01157
Yu-xiang, W., Jin-hong, C., Qiu-ling, H., Shui-jin, Z. (2013). Parental origin and genomic evolution of tetraploid Gossypium species by molecular marker and GISH analyses. Caryologia 66, 368–374. doi: 10.1080/00087114.2013.857830
Keywords: cotton Alba genes, miRNAs, cis-regulatory elements, VIGS and wild cotton, virus-induced gene silencing (VIGS), water deficit and salt stresses, oxidant and antioxidant enzymes
Citation: Magwanga RO, Kirungu JN, Lu P, Cai X, Xu Y, Wang X, Zhou Z, Hou Y, Agong SG, Wang K and Liu F (2019) Knockdown of ghAlba_4 and ghAlba_5 Proteins in Cotton Inhibits Root Growth and Increases Sensitivity to Drought and Salt Stresses. Front. Plant Sci. 10:1292. doi: 10.3389/fpls.2019.01292
Received: 09 April 2019; Accepted: 17 September 2019;
Published: 15 October 2019.
Edited by:
Goetz Hensel, Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK), GermanyReviewed by:
Liang Chen, University of Chinese Academy of Sciences, ChinaGregory Thyssen, Southern Regional Research Center (USDA-ARS), United States
Zhen Su, China Agricultural, University (CAU), China
Copyright © 2019 Magwanga, Kirungu, Lu, Cai, Xu, Wang, Zhou, Hou, Agong, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Liu, bGl1ZmNyaUBjYWFzLmNvbQ==
 Richard Odongo Magwanga
Richard Odongo Magwanga Joy Nyangasi Kirungu
Joy Nyangasi Kirungu Pu Lu1
Pu Lu1