- 1Newe Ya’ar Research Center, Agricultural Research Organization, Ramat Yishay, Israel
- 2Germplasm Resource Unit, John Innes Center, Norwich, United Kingdom
- 3Agricultural Research Organization, Volcani Center, Rishon LeZion, Israel
- 4Robert W. Holley Center for Agriculture and Health, USDA-ARS, Cornell University, Ithaca, NY, United States
Carotenoids have various roles in plant physiology. Plant carotenoids are synthesized in plastids and are highly abundant in the chromoplasts of ripening fleshy fruits. Considerable research efforts have been devoted to elucidating mechanisms that regulate carotenoid biosynthesis, yet, little is known about the mechanism that triggers storage capacity, mainly through chromoplast differentiation. The Orange gene (OR) product stabilizes phytoene synthase protein (PSY) and triggers chromoplast differentiation. OR underlies carotenoid accumulation in orange cauliflower and melon. The OR’s ‘golden SNP’, found in melon, alters the highly evolutionary conserved Arginine108 to Histidine and controls β-carotene accumulation in melon fruit, in a mechanism yet to be elucidated. We have recently shown that similar carotenogenic metabolic flux is active in non-orange and orange melon fruit. This flux probably leads to carotenoid turnover but known carotenoid turnover products are not detected in non-orange fruit. Arrest of this metabolic flux, using chemical inhibitors or mutations, induces carotenoid accumulation and biogenesis of chromoplasts, regardless of the allelic state of OR. We suggest that the ‘golden SNP’ induces β-carotene accumulation probably by negatively affecting the capacity to synthesize downstream compounds. The accumulation of carotenoids induces chromoplast biogenesis through a metabolite-induced mechanism. Carotenogenic turnover flux can occur in non-photosynthetic tissues, which do not accumulate carotenoids. Arrest of this flux by the ‘golden SNP’ or other flux-arrest mutations is a potential tool for the biofortification of agricultural products with carotenoids.
Introduction
Carotenoids exhibit diverse roles during the plant life cycle, are essential components of the photosynthetic apparatus, color agents, and precursors of hormones and aroma compounds, and are also signaling molecules involved in various developmental and environmental signaling pathways (Nisar et al., 2015; Rodriguez-Concepcion et al., 2018a). Carotenoids are synthesized in plastids from the methylerythritol phosphate (MEP) pathway, followed by the biosynthesis of geranylgeranyl diphosphate (GGPP), a precursor of several essential isoprenoid pathways, including carotenoids (Ruiz-Sola et al., 2016). The first committed step in carotenogenesis requires the condensation of two GGPP molecules into phytoene, in an enzymatic reaction catalyzed by phytoene synthase (PSY), who’s activity divergence exists among its isozymes (Cao et al., 2019). Phytoene biosynthesis is followed by four desaturation reactions catalyzed by phytoene desaturase (PDS) and ζ-carotene desaturase (ZDS), to introduce cis double bonds, and isomerization to trans-configuration through ζ-carotene isomerase (ZISO) and carotene isomerase (CRTISO), producing all-trans-lycopene. Lycopene undergoes two cyclization steps during biosynthesis of either β-carotene (two β rings) or α-carotene (ε and β rings), converted by lycopene β-cyclase (CRTL-B) and lycopene ε-cyclase (CRTL-E) (Hirschberg, 2001; Alagoz et al., 2018). Carotenes undergo hydroxylation and epoxidation to form xanthophylls. Apocarotenoids, carotenoid-derived compounds, consist of volatile phytohormones ABA and strigolactones. Due to their critical role during the plant life cycle, carotenoids are subjected to multiple layers of regulation, affecting their biosynthesis, turnover, channeling and sequestration, including regulation of plastid development, transcriptional regulation of carotenogenic, as well as non-carotenogenic gene expression and post-transcriptional regulation, all of which determine the final composition and abundance of carotenoids.
Carotenoids are highly abundant in the chromoplasts and accumulate mainly in flowers, fruits, and vegetables (Yuan et al., 2015b; Sun et al., 2018). Chromoplasts can differentiate from different plastids, and exhibit considerable variance in structure and biochemical composition, all of which are highly enriched in sequestration substructures, enabling high rates of biosynthesis and stable storage capacities (Sun et al., 2018). As such, chromoplast formation during fruit ripening is one of the main factors governing fruit pigmentation, nutritional value and taste. Chromoplast biogenesis involves a matrix of interactions, including hormonal signaling, gene expression and post-transcriptional modifications (Harris and Spurr, 1969; Egea et al., 2010; Li and Yuan, 2013).
Melon’s, Cucumis melo L. (Cucurbitaceae), fruit flesh color appears either as white, green or orange (Burger et al., 2009). Green- and white-flesh melon fruit contain negligible amounts of carotenoids, whilst orange-flesh melon accumulates massive amount of β-carotene. Carotenoid accumulation in melon fruit is conferred by the dominant allele at the gf (green flesh) locus (Hughes, 1948; Clayberg, 1992). The melon’s ORANGE gene (CmOR) resides at the gf locus and its ‘golden’ SNP (single nucleotide polymorphism), replacing the evolutionary conserved Arginine108 (Arg108) with Histidine (His), controls chromoplast differentiation and carotenoid accumulation during melon fruit development (Tzuri et al., 2015). Except for this single amino-acid substitution, not much is known about the initial molecular signaling pathway triggering chromoplast differentiation. In this mini-review, we emphasize the role of the carotenogenic metabolic flux, its association with the ‘golden SNP’ and its arrest during the process of fruit ripening, which we suggest initiates chromoplast differentiation and carotenoid accumulation.
Regulation of Carotenoid Biosynthesis
Regulation of carotenoid biosynthesis and storage in non-photosynthetic tissues has drawn much attention in recent decades (Giuliano, 2017). Most of our knowledge about the regulation of the carotenoid pathway during fruit development stems from tomato, Solanum lycopersicum L. (Solanaceae), which serves as a model plant for the study of climacteric fleshy fruit development. The main carotenoid accumulated during tomato fruit ripening is lycopene. This accumulation, one of many ripening-related modifications, is governed by a transcriptional network, which regulates the ethylene burst, results in chromatin modification, and underlies the massive transcriptional modifications during fruit ripening (Oeller et al., 1991; Alba et al., 2005; Zhong et al., 2013; Giovannoni et al., 2017; Li et al., 2019). At the carotenogenic transcriptional level, lycopene upstream coding genes, including PSY1, PDS, and CRTISO, are transcriptionally upregulated, while the lycopene cyclases, CRTL-B and CRTL-E, are downregulated, enabling lycopene accumulation (Ronen et al., 1999; Isaacson et al., 2002). Among these processes, the transcriptional activation of PSY1 is one of the major flux-controlling factors during carotenoid accumulation in ripening fruit (Fraser et al., 1994; Fraser et al., 2002). In contrast to tomato, carotenoid accumulation during melon fruit ripening does not depend on the ethylene climacteric burst. Moreover, inhibition of ethylene biosynthesis does not affect fruit carotenoid accumulation in ‘Védrantais’ melon fruit (Ayub et al., 1996; Pech et al., 2008). These observations indicate two divergent regulating mechanisms of carotenoid accumulation in ripening tomato and melon fruits, consistent with recent findings indicating that different transcriptional networks regulate ethylene signaling in these species (Lu et al., 2018). As in tomatoes, carotenoid accumulation during melon fruit ripening is associated with the transcriptional upregulation of β-carotene upstream carotenogenic structural genes (PSY1, PDS, and ZDS), along with an increase in PSY1 protein abundance (Chayut et al., 2015). However, these transcriptional changes are not sufficient to induce carotenoid accumulation in melon, as similar patterns of carotenogenic gene expression along with PSY1 protein abundance have been found in both β-carotene-accumulating and non-accumulating melon accessions, similar to that found in cauliflower (Brassica oleracea) (Li et al., 2006; Chayut et al., 2015; Chayut et al., 2017). Enzymatic carotenoid’s turn over to apocarotenoids is regulated by carotenoid cleavage dioxygenases (CCDs). A comparison between melon fruit flesh of carotenoid accumulating versus non-accumulating accessions correlates apocarotenoid with carotenoid accumulation, while both orange and non-orange fruit show elevated CmCCD1 transcription during fruit ripening, suggesting substrate availability to be the limiting factor in apocarotenoid production in melon (Ibdah et al., 2006).
Several genes have been reported as plastid size and development regulators, involving different signaling pathways (Levin et al., 2006; Galpaz et al., 2008) but, to date, the only gene which has been found to directly regulate chromoplast biogenesis is the OR gene. OR belongs to a DnaJ-like family termed DnaJ-E, a non-canonical DnaJ-related protein coding gene family, coding for cysteine-rich zinc finger domain proteins, which have undergone vast duplication in plants during evolution. OR and several other members of the family possess plastid-related, protein-assembly factor activity (Pulido and Leister, 2018). OR was first described in cauliflower, as a single semi-dominant gene mutation conferring orange-colored curds (Crisp et al., 1975), it triggers chromoplast differentiation associated with high levels of β-carotene accumulation (Li et al., 2001; Paolillo et al., 2004). Positional cloning identified the causative mutation as an insertion of a copia element in the protein coding region of the BoOR gene, which leads to various altered transcripts and to the orange curd phenotype. The OR gene is highly conserved in the plant kingdom, and also found in algae, but not in photosynthetic bacteria or non-photosynthetic species (Lu et al., 2006). Ectopic overexpression (OE) of the single-cell green alga Chlamydomonas reinhardtiiOR gene enhances carotenoid content (Morikawa et al., 2018).
The Arabidopsis genome harbors two homologs of the OR gene, termed AtOR and AtOR-like (Zhou et al., 2015). Both interact with PSY in Nicotiana benthamiana leaf chloroplasts, are physically associated with PSY in-vitro, and are needed to stabilize PSY protein levels, activity, and carotenoid biosynthesis, as observed in AtOR/AtOR-like double knockout. The OR protein consists of two domains: the N-terminus binds PSY, protecting it from Clp-mediated proteolysis (Zhou et al., 2015; D’Andrea et al., 2018; Rodriguez-Concepcion et al., 2018b; Welsch et al., 2018). These findings, indicate that the DnaJ-like proteins have maintained the original function of canonical DnaJ proteins, acting as chaperones, synergistically determining protein turnover and quality along with proteases (Bukau et al., 2006). The C-terminus contains two putative transmembrane domains, along with the cysteine-rich ZF domain, which has been found to mediate OR protein dimerization (Zhou et al., 2015).
In melon, the CmOR gene was found to reside at the gf (green flesh) locus, regulating β-carotene accumulation in the fruit mesocarp. CmOR exhibits two major haplotypes differentially associated with β-carotene accumulation within a population of 49 melon accessions. A single SNP, termed the ‘golden SNP’, replaces a conserved amino acid Arg108 with His, in OR, underlies the trait (Tzuri et al., 2015). Recently, genome-wide, linkage-disequilibrium mapping of 177 melon accessions re-confirmed the causality of the ‘golden SNP’ (Gur et al., 2017). In the mesocarp of the CmOR knockout mutant, small amounts of carotenoids are accumulated in late fruit developmental stages associated with an increase in PSY abundance, which is related to an increase in transcriptional abundance of CmOR-like (Chayut et al., 2017).
Overexpression of AtORHis significantly increases carotenoid accumulation in dark-grown, seed-derived Arabidopsis calli, as compared with AtORArg (Tzuri et al., 2015; Yuan et al., 2015a). Similar to BoOrMut, AtORHis triggers membranous chromoplast biogenesis, while AtORArg does not (Paolillo et al., 2004; Yuan et al., 2015a). Both alleles of CmOR and AtOR (His and Arg) are equally capable of binding and stabilizing PSY protein levels, a characteristic governed by the N-terminus of the OR protein. The ‘golden SNP’ governs an additional role of OR gene, regulating carotenoid accumulation and plastid formation in melon fruit. This function in melon, can be introduced to other plant systems (Yuan et al., 2015a; Chayut et al., 2017; Yazdani et al., 2019). Ectopic overexpression of AtOR and AtOrHis in tomatoes has been shown to have a minimal effect on photosynthetic tissues, similar to OE in Arabidopsis plants or the effect of natural variation in OR ‘golden SNP’ on melon plant performance. However, tomato flowers and fruit are dramatically affected by the ‘golden SNP’; carotenoid abundance in flower petals increases by more than two-fold in AtORHis OE lines, mainly due to an increase in β-carotene, but it is not affected by AtOR OE (Yazdani et al., 2019). During early fruit developmental stages, AtORHis shows a significant increase in carotenoid accumulation, dominated by elevation of β-carotene and associated with chromoplast differentiation, while AtOR only has a minor effect on carotenoid content and composition. During fruit ripening, the climacteric ethylene induces lycopene accumulation in AtOR and AtORHis OE, as well as in the control, an M82-type processing tomato cultivar. Nevertheless, the effect of AtORHis OE on carotenoid content is much more dramatic; a 250% increase in lycopene and 300% in β-carotene compared to the control, with AtOR OE exhibiting only a 25% increase in lycopene content (Yazdani et al., 2019). The effect of AtORHis OE during the pre-climacteric green stages of tomato fruit development, i.e., ethylene-independent β-carotene accumulation associated with chromoplast differentiation, exhibits a striking similarity to the effect of CmORHis during melon fruit development.
The increase in carotenoid accumulation during ripening stages of the OE lines suggests that OR also serves as a limiting factor in carotenoid accumulation during tomato fruit ripening; this is further supported by the transcriptional upregulation of both SlOR and SlOR-like during fruit ripening (Tomato Expression Atlas database: http://tea.solgenomics.net) (Fernandez-Pozo et al., 2017). These studies strongly suggest a conserved function of the OR and OR-like genes.
More insight into the relationship between OR/OR-like and chromoplast development can be gained from a phylogenetic analysis (Figure 1). The hierarchical clustering of published OR and OR-like sequences indicates that lower plant evolution is generally associated with duplication of the OR gene, while clear differentiation into two OR and OR-like distinct clades is highly conserved from gymnosperms onwards, indicating a different unique function of each protein, in addition to their common PSY binding activity (Zhou et al., 2015). Although chromoplasts are mainly associated with angiosperm flowers and fruit, examples of similar tubular structures that are frequently present in fruit can be found in ripe red seeds of cycads (Whatley, 1985). This associates OR/OR-like evolutionary specialization with the early evolution of seed-producing plants, designating the vast majority of agricultural crops as candidates for biofortification through OR-based modification.
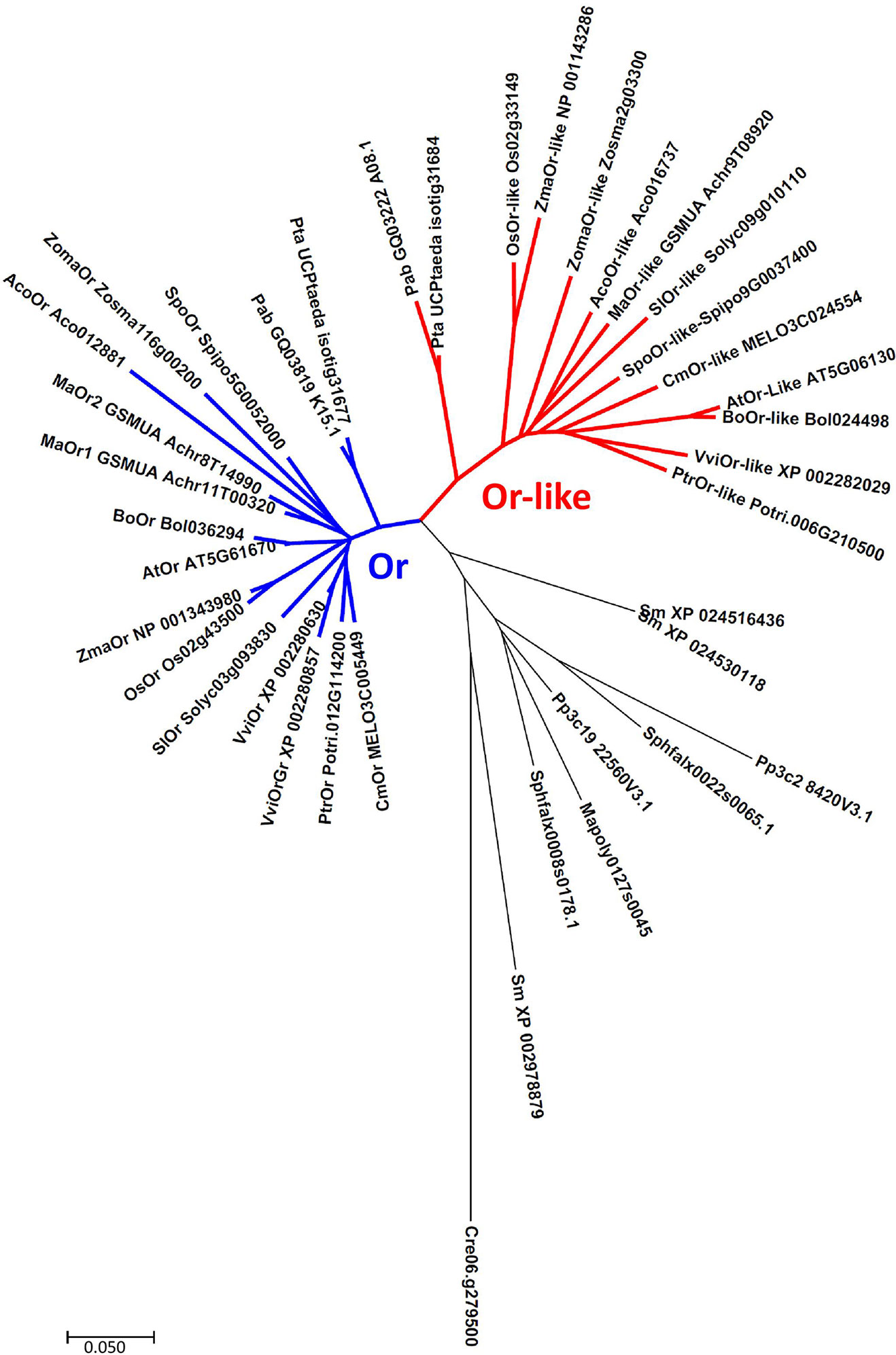
Figure 1 Neighbor-joining phylogenetic tree of OR and OR-like proteins. Cluster analysis associate’s gene duplication with bryophytes and exhibits two distinctively separate clades, conserved from gymnosperms onwards. Abbreviations- Algae: Cre (Chlamydomonas reinhardtii). Liverworts: Mapoly (Marchantia polymorpha). Mosses: Pp (Physcomitrella patens), Sphfalx (Sphagnum fallax). Lycophytes: Sm (Selaginella moellendorffii). Gymnosperms: Pte (Pinus taeda), Pab (Picea abies). Angiosperms: Aco (Ananas comosus), Ma (Musa acuminata), Spo (Spirodela polyrhiza), Zoma (Zostera marina), Os (Oryza sativa), Zma (Zea mays), Sl (Solanum lycopersicum), Vvi (Vitis vinifera), Ptr (Populus trichocarpa), At (Arabidopsis thaliana), Bo (Brassica oleracea), Cm (Cucumis melo). Sequences were downloaded from Phytozome (http://www.phytozome.net), National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), TAIR (https://www.arabidopsis.org), Sol Genomics Network (https://solgenomics.net), CuGenDB (http://cucurbitgenomics.org), and ConGenIE (http://congenie.org). Created with MEGA 7 (Kumar et al., 2016).
The difference between climacteric and ORHis-dependent, non-climacteric fruit carotenoid accumulation, demonstrated here by tomato and melon, suggest distinct regulating mechanisms of fruit-carotenoid accumulation, which can stand alone or complement each other. The major factor in climacteric-regulated carotenoid accumulation is the developmentally associated transcriptional changes in carotenogenic gene expression regulated by ethylene (Alba et al., 2005). The ORHis-dependent carotenoid accumulation mechanism is probably associated with post-translational regulation of the carotenogenic metabolic flux, which will be discussed in the next section.
Controlling the Carotenoid Metabolic Flux and Accumulation
The increase in PSY1 activity during fruit ripening is a major factor that limits lycopene accumulation in tomatoes (Cao et al., 2019). This increased activity, serves as an example of a direct relationship between carotenogenic gene activity and carotenoid accumulation. However, an immature green tomato does not accumulate significant carotenoid levels, even though it exhibits higher enzymatic activities of PSY, PDS and CRTL-B compared to ripening fruit, irrespective of the dramatic increase in β-carotene and lycopene abundance during fruit ripening (Fraser et al., 1994). The increase in carotenogenic activity during immature fruit stages is attributed to actively photosynthetic tissue resulting in a high rate of carotenoid turnover (Lytovchenko et al., 2011; Brazel and O’Maoileidigh, 2019). This indicates that intensive carotenogenic flux during fruit development does not necessarily relate to carotenoid accumulation, raising questions about the mechanism mediating ORHis-dependent carotenoid accumulation and chromoplast biogenesis. Additionally, this indicates that turnover of carotenoids creates a metabolic flux that could be utilized for carotenoid accumulation if combined with ORHis (Yazdani et al., 2019).
Several reports have documented that changes in the carotenoid biochemical flux by itself, without any additional developmental program, could be sufficient to trigger carotenoid accumulation. Over-expression of PSY1 in tomato results in chromoplast-like plastids in premature fruit, independent of the ripening program, and exhibit a metabolite-induced plastid transition (Fraser et al., 2007). Over-expression of PSY1 in Arabidopsis increases the PSY protein level, leading to increased carotenoid (β-carotene) levels in dark-grown, seed-derived calli but not in leaves (Maass et al., 2009). In addition, overexpression of crtB, a bacterial PSY, in white carrots, result in a similar increase in carotenoids deposited as crystals, indicating that sequestration into crystals can be achieved through an increase of the pathway flux (Maass et al., 2009).
Cauliflower calli containing either BoORWT or BoORMut produce a similar pattern of phytoene accumulation following treatment with norflurazon (NF, a PDS inhibitor), indicating that the carotenoid metabolism flux is similarly active in both calli, even though only BoORMut induces carotenoid accumulation (Li et al., 2006). In melon, the natural CmOR genetic variation is based on the ‘golden SNP’ (Tzuri et al., 2015). Carotenogenic gene expression and the carotenoid metabolic flux are similarly active in orange and non-orange melons (Chayut et al., 2017). Ethyl methanesulfonate (EMS)-induced mutagenesis of CEZ, an orange-flesh Charentais-type melon, generated an additional allele of CmOR, a nonsense allele called ‘low-β’. The CmOR ‘low-β’ allele completely lacks the CmOR protein, resulting in a low PSY1 protein level and low carotenoid biosynthesis metabolic flux, as revealed by PSY1 western blot analysis and the reduced accumulation of phytoene after treatment with NF (Chayut et al., 2017).
An additional mutant isolated from the CEZ mutation library, called “Yellow-Orange Flesh I’ (yofi), exhibits a yellow-orange flesh color due to a nonsense mutation in CRTISO, stopping the carotenogenic metabolic flux in prolycopene (tetra-cis-lycopene), since the isomerized product, all-trans-lycopene, is a necessary precursor of the β-cyclase enzymes (Galpaz et al., 2013). Yofi was a parental line of two segregating F2 populations. The first population was generated from a cross between yofi and the green-fleshed inbred line ‘Noy Yizre’el’, harboring the CmORArg allele, generating a population that segregates to CmORArg/CmORHis and CRTISO/crtiso, wild type and nonsense (yofi) alleles. The second population originated from a cross between yofi and ‘low-β’, which segregated to CmOrHis/’low β’ in addition to CRTISO/crtiso alleles. High Performance Liquid Chromatography (HPLC) carotenoid analysis of F2 segregants selected for defined genotypes revealed that, as expected, CmORHis segregants accumulate β-carotene in the CRTISO background and pro-lycopene in the yofi background. Interestingly, CmORArg and ‘low-β’ segregants, which harbor the yofi allele, accumulate prolycopene, similar to CmORHis/yofi segregants. This indicates that arrest of the carotenogenic metabolic flux can induce carotenoid accumulation and chromoplast formation regardless of CmOR allelic variation. In addition, it indicates that a low metabolic flux, as observed in ‘low-β’, is sufficient to induce carotenoid accumulation and chromoplast formation, similar to a high metabolic flux, as observed in the CmOR natural variation, when the flux is blocked. This suggests that the ‘golden SNP’ stabilizes β-carotene by inhibiting beta-ring hydroxylase activity (Chayut et al., 2017). Moreover, these results prompted us to suggest an arrest of the carotenogenic metabolic flux, by mutations or by genome editing, as an optional strategy to induce carotenoid accumulation in non-carotenoid accumulating tissues.
Conclusions and Future Perspectives
This mini-review summarizes studies that indicate the role of the OR gene in stabilizing the carotenogenic metabolic flux for chromoplast differentiation and carotenoid accumulation.
Selected studies, presented here, suggest that the OR ‘golden SNP’ mediates storage capacity in melon fruit, and in other non-photosynthetic tissues, by arresting the carotenogenic metabolic flux downstream to β-carotene, leading to chromoplast biogenesis and carotenoid accumulation, mainly β-carotene. This suggests an arrest of the carotenogenic metabolic flux in plant tissues, which normally do not accumulate carotenoids, as a potential biofortification tool. In the age of CRISPR-mediated genome editing, introgression of the ‘golden SNP’ in horticultural crops should prove an efficient tool for the development of novel products of high nutritional value, and for further elucidating the mechanism of chromoplast biogenesis.
The close relationship between OR and OR-like, and hints to a partially complementary function of OR-like, should be looked at more closely to gain further insight into the biogenesis and evolution of chromoplasts.
Carotenoid accumulation in tomatoes and in other fruit is regulated by a climacteric ethylene burst. In melon, fruit-flesh carotenoid accumulation depends on the ‘golden SNP’, independent of ethylene regulation. Combining these polyphyletic regulation processes may well enhance attempts to break the limits of carotenoid biofortification of agricultural products.
Author Contributions
All authors contributed to the OR gene work in melon, Arabidopsis and tomato. All authors contributed to the writing of this mini review.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was partially supported by BARD, the United States Israel Binational Agricultural Research and Development Fund, Award No. US-4918-16CR and by the ARO’s Center for the Genetic Enhancement of Cucurbit Fruit Quality. We thank Carly Golodets and Harry S. Paris for English language editing.
References
Alagoz, Y., Nayak, P., Dhami, N., Cazzonelli, C. I. (2018). cis-carotene biosynthesis, evolution and regulation in plants: the emergence of novel signaling metabolites. Arch. Biochem. Biophys. 654, 172–184. doi: 10.1016/j.abb.2018.07.014
Alba, R., Payton, P., Fei, Z., McQuinn, R., Debbie, P., Martin, G. B., et al. (2005). Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17 (11), 2954–2965. doi: 10.1105/tpc.105.036053
Ayub, R., Guis, M., Amor, M. B., Gillot, L., Roustan, J.-P., Latché, A., et al. (1996). Expression of ACC oxidase antisense gene inhibits ripening of cantaloupe melon fruits. Nat. Biotechnol. 14 (7), 862–866. doi: 10.1038/nbt0796-862
Brazel, A. J., O’Maoileidigh, D. S. (2019). Photosynthetic activity of reproductive organs. J. Exp. Bot. 70 (6), 1737–1754. doi: 10.1093/jxb/erz033
Bukau, B., Weissman, J., Horwich, A. (2006). Molecular chaperones and protein quality control. Cell 125 (3), 443–451. doi: 10.1016/j.cell.2006.04.014
Burger, Y., Paris, H., Cohen, R., Katzir, N., Tadmor, Y., Lewinsohn, E., et al. (2009). Genetic diversity of Cucumis melo. Hortic. Rev. 36, 165–198. doi: 10.1002/9780470527238.ch3
Cao, H., Luo, H., Yuan, H., Eissa, M., Thannhauser, T., Welsch, R., et al. (2019). A neighboring aromatic-aromatic amino acid combination governs activity divergence between tomato phytoene synthases. Plant Physiol. 180 (4), 1988–2003. doi: 10.1104/pp.19.00384
Chayut, N., Yuan, H., Ohali, S., Meir, A., Sa’ar, U., Tzuri, G., et al. (2017). Distinct mechanisms of the ORANGE protein in controlling Carotenoid flux. Plant Physiol. 173 (1), 376–389. doi: 10.1104/pp.16.01256
Chayut, N., Yuan, H., Ohali, S., Meir, A., Yeselson, Y., Portnoy, V., et al. (2015). A bulk segregant transcriptome analysis reveals metabolic and cellular processes associated with orange allelic variation and fruit beta-carotene accumulation in melon fruit. BMC Plant Biol. 15, 274. doi: 10.1186/s12870-015-0661-8
Clayberg, C. (1992). Interaction and linkage test of flesh color genes in Cucumis melo L. Rep. Cucurbit Genet. Coop. 15, 53.
Crisp, P., Walkey, D. G. A., Bellman, E., Roberts, E. (1975). A mutation affecting curd colour in cauli¯ower (Brassica oleracea L. var. botrytis DC). Euphytica 24, 173–176. doi: 10.1007/BF00147182
D’Andrea, L., Simon-Moya, M., Llorente, B., Llamas, E., Marro, M., Loza Alvarez, P., et al. (2018). Interference with Clp protease impairs carotenoid accumulation during tomato fruit ripening. J. Exp. Bot. 69 (7), 1557–1568. doi: 10.1093/jxb/erx491
Egea, I., Barsan, C., Bian, W., Purgatto, E., Latche, A., Chervin, C., et al. (2010). Chromoplast differentiation: current status and perspectives. Plant Cell Physiol. 51 (10), 1601–1611. doi: 10.1093/pcp/pcq136
Fernandez-Pozo, N., Zheng, Y., Snyder, S. I., Nicolas, P., Shinozaki, Y., Fei, Z., et al. (2017). The tomato expression atlas. Bioinformatics 33 (15), 2397–2398. doi: 10.1093/bioinformatics/btx190
Fraser, P. D., Enfissi, E. M., Halket, J. M., Truesdale, M. R., Yu, D., Gerrish, C., et al. (2007). Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell 19 (10), 3194–3211. doi: 10.1105/tpc.106.049817
Fraser, P. D., Romer, S., Shipton, C. A., Mills, P. B., Kiano, J. W., Misawa, N., et al. (2002). Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc. Natl. Acad. Sci. U. S. A. 99 (2), 1092–1097. doi: 10.1073/pnas.241374598
Fraser, P. D., Truesdale, M. R., Bird, C. R., Schuch, W., Bramley, P. M. (1994). Carotenoid biosynthesis during tomato fruit development (Evidence for tissue-specific gene expression). Plant Physiol. 105, 405–413. doi: 10.1104/pp.105.1.405
Galpaz, N., Burger, Y., Lavee, T., Tzuri, G., Sherman, A., Melamed, T., et al. (2013). Genetic and chemical characterization of an EMS induced mutation in Cucumis melo CRTISO gene. Arch. Biochem. Biophys. 539 (2), 117–125. doi: 10.1016/j.abb.2013.08.006
Galpaz, N., Wang, Q., Menda, N., Zamir, D., Hirschberg, J. (2008). Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J. 53 (5), 717–730. doi: 10.1111/j.1365-313X.2007.03362.x
Giovannoni, J., Nguyen, C., Ampofo, B., Zhong, S., Fei, Z. (2017). The epigenome and transcriptional dynamics of fruit ripening. Annu. Rev. Plant Biol. 68, 61–84. doi: 10.1146/annurev-arplant-042916-040906
Giuliano, G. (2017). Provitamin A biofortification of crop plants: a gold rush with many miners. Curr. Opin. Biotechnol. 44, 169–180. doi: 10.1016/j.copbio.2017.02.001
Gur, A., Tzuri, G., Meir, A., Sa’ar, U., Portnoy, V., Katzir, N., et al. (2017). Genome-wide linkage-disequilibrium mapping to the candidate gene level in melon (Cucumis melo). Sci. Rep. 7 (1), 9770. doi: 10.1038/s41598-017-09987-4
Harris, W. M., Spurr, A. R. (1969). Chromoplasts of tomato fruits. I. Ultrastructure of low-pigment and high- beta mutants. Am. J. Bot. 56, 369–379. doi: 10.1002/j.1537-2197.1969.tb07546.x
Hirschberg, J. (2001). Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 4 (3), 210–218. doi: 10.1016/S1369-5266(00)00163-1
Hughes, M. (1948). The inheritance of two characters of Cucumis melo and their interrelationship. Proc. Am. Soc. Hortic. Sci. 59, 399–402.
Ibdah, M., Azulay, Y., Portnoy, V., Wasserman, B., Bar, E., Meir, A., et al. (2006). Functional characterization of CmCCD1, a carotenoid cleavage dioxygenase from melon. Phytochemistry 67 (15), 1579–1589. doi: 10.1016/j.phytochem.2006.02.009
Isaacson, T., Ronen, G., Zamir, D., Hirschberg, J. (2002). Cloning of tangerine from tomato reveals a Carotenoid isomerase essential for the production of β-Carotene and Xanthophylls in plants. Plant Cell 14 (2), 333–342. doi: 10.1105/tpc.010303
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Levin, I., de Vos, R. C., Tadmor, Y., Bovy, A., Lieberman, M., Oren-Shamir, M., et al. (2006). High pigment tomato mutants—more than just lycopene (a review). Isr. J. Plant Sci. 54 (3), 179–190. doi: 10.1560/IJPS_54_3_179
Li, L., Lu, S., Cosman, K. M., Earle, E. D., Garvin, D. F., O’Neill, J. (2006). beta-Carotene accumulation induced by the cauliflower Or gene is not due to an increased capacity of biosynthesis. Phytochemistry 67 (12), 1177–1184. doi: 10.1016/j.phytochem.2006.05.013
Li, L., Paolillo, D. J., M.V., P., E.M., D., D.F., G. (2001). A novel gene mutation that confers abnormal patterns of β-carotene accumulation in cauliflower (Brassica oleracea var. botrytis). Plant J. 26 (1), 59–67. doi: 10.1046/j.1365-313x.2001.01008.x
Li, S., Chen, K., Grierson, D. (2019). A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol. 221 (4), 1724–1741. doi: 10.1111/nph.15545
Li, L., Yuan, H. (2013). Chromoplast biogenesis and carotenoid accumulation. Arch. Biochem. Biophys. 539 (2), 102–109. doi: 10.1016/j.abb.2013.07.002
Lu, P., Yu, S., Zhu, N., Chen, Y. R., Zhou, B., Pan, Y., et al. (2018). Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat. Plants 4 (10), 784–791. doi: 10.1038/s41477-018-0249-z
Lu, S., Van Eck, J., Zhou, X., Lopez, A. B., O’Halloran, D. M., Cosman, K. M., et al. (2006). The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell 18 (12), 3594–3605. doi: 10.1105/tpc.106.046417
Lytovchenko, A., Eickmeier, I., Pons, C., Osorio, S., Szecowka, M., Lehmberg, K., et al. (2011). Tomato fruit photosynthesis is seemingly unimportant in primary metabolism and ripening but plays a considerable role in seed development. Plant Physiol. 157 (4), 1650–1663. doi: 10.1104/pp.111.186874
Maass, D., Arango, J., Wust, F., Beyer, P., Welsch, R. (2009). Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS One 4 (7), e6373. doi: 10.1371/journal.pone.0006373
Morikawa, T., Uraguchi, Y., Sanda, S., Nakagawa, S., Sawayama, S. (2018). Overexpression of DnaJ-like chaperone enhances Carotenoid synthesis in Chlamydomonas reinhardtii. Appl. Biochem. Biotechnol. 184 (1), 80–91. doi: 10.1007/s12010-017-2521-5
Nisar, N., Li, L., Lu, S., Khin, N. C., Pogson, B. J. (2015). Carotenoid metabolism in plants. Mol. Plant 8 (1), 68–82. doi: 10.1016/j.molp.2014.12.007
Oeller, P. W., Lu, M. W., Taylor, L. P., Pike, D. A., Theologis, A. (1991). Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254, 437–439. doi: 10.1126/science.1925603
Paolillo, D. J., Jr., Garvin, D. F., Parthasarathy, M. V. (2004). The chromoplasts of Or mutants of cauliflower (Brassica oleracea L. var. botrytis). Protoplasma 224 (3-4), 245–253. doi: 10.1007/s00709-004-0059-1
Pech, J. C., Bouzayen, M., Latché, A. (2008). Climacteric fruit ripening: Ethylene-dependent and independent regulation of ripening pathways in melon fruit. Plant Sci. 175 (1-2), 114–120. doi: 10.1016/j.plantsci.2008.01.003
Pulido, P., Leister, D. (2018). Novel DNAJ-related proteins in Arabidopsis thaliana. New Phytol. 217, 480–490. doi: 10.1111/nph.14827
Rodriguez-Concepcion, M., Avalos, J., Bonet, M. L., Boronat, A., Gomez-Gomez, L., Hornero-Mendez, D., et al. (2018a). A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Prog. Lipid Res. 70, 62–93. doi: 10.1016/j.plipres.2018.04.004
Rodriguez-Concepcion, M., D’Andrea, L., Pulido, P. (2018b). Control of plastidial metabolism by the Clp protease complex. J. Exp. Bot. 70 (7), 2049–2058. doi: 10.1093/jxb/ery441
Ronen, G., Cohen, M., Zamir, D., Hirschberg, J. (1999). Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J. 17 (4), 341–351. doi: 10.1046/j.1365-313X.1999.00381.x
Ruiz-Sola, M. A., Coman, D., Beck, G., Barja, M. V., Colinas, M., Graf, A., et al. (2016). Arabidopsis GERANYLGERANYL DIPHOSPHATE SYNTHASE 11 is a hub isozyme required for the production of most photosynthesis-related isoprenoids. New Phytol. 209 (1), 252–264. doi: 10.1111/nph.13580
Sun, T., Yuan, H., Cao, H., Yazdani, M., Tadmor, Y., Li, L. (2018). Carotenoid metabolism in plants: the role of plastids. Mol. Plant 11 (1), 58–74. doi: 10.1016/j.molp.2017.09.010
Tzuri, G., Zhou, X., Chayut, N., Yuan, H., Portnoy, V., Meir, A., et al. (2015). A ‘golden’ SNP in CmOr governs the fruit flesh color of melon (Cucumis melo). Plant J. 82 (2), 267–279. doi: 10.1111/tpj.12814
Welsch, R., Zhou, X., Yuan, H., Alvarez, D., Sun, T., Schlossarek, D., et al. (2018). Clp protease and OR directly control the proteostasis of PHYTOENE synthase, the crucial enzyme for carotenoid biosynthesis in Arabidopsis. Mol. Plant 11 (1), 149–162. doi: 10.1016/j.molp.2017.11.003
Whatley, J. M. (1985). Chromoplasts in some Cycads. New Phytol. 101, 595–604. doi: 10.1111/j.1469-8137.1985.tb02865.x
Yazdani, M., Sun, Z., Yuan, H., Zeng, S., Thannhauser, T. W., Vrebalov, J., et al. (2019). Ectopic expression of ORANGE promotes carotenoid accumulation and fruit development in tomato. Plant Biotechnol. J. 17 (1), 33–49. doi: 10.1111/pbi.12945
Yuan, H., Owsiany, K., Sheeja, T. E., Zhou, X., Rodriguez, C., Li, Y., et al. (2015a). A single amino acid substitution in an ORANGE protein promotes Carotenoid overaccumulation in Arabidopsis. Plant Physiol. 169 (1), 421–431. doi: 10.1104/pp.15.00971
Yuan, H., Zhang, J., Nageswaran, D., Li, L. (2015b). Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2, 15036. doi: 10.1038/hortres.2015.36
Zhong, S., Fei, Z., Chen, Y. R., Zheng, Y., Huang, M., Vrebalov, J., et al. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31 (2), 154–159. doi: 10.1038/nbt.2462
Keywords: carotenoids accumulation, OR genes, melon (Cucumis melo L.), tomato (Solanum lycopersicum), metabolic flux
Citation: Feder A, Chayut N, Gur A, Freiman Z, Tzuri G, Meir A, Saar U, Ohali S, Baumkoler F, Gal-On A, Shnaider Y, Wolf D, Katzir N, Schaffer A, Burger J, Li L and Tadmor Y (2019) The Role of Carotenogenic Metabolic Flux in Carotenoid Accumulation and Chromoplast Differentiation: Lessons From the Melon Fruit. Front. Plant Sci. 10:1250. doi: 10.3389/fpls.2019.01250
Received: 16 May 2019; Accepted: 09 September 2019;
Published: 30 October 2019.
Edited by:
Aleš Svatoš, Max Planck Institute for Chemical Ecology, GermanyCopyright © 2019 Feder, Chayut, Gur, Freiman, Tzuri, Meir, Saar, Ohali, Baumkoler, Gal-On, Shnaider, Wolf, Katzir, Schaffer, Burger, Li and Tadmor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaakov Tadmor, dGFkbW9yeUBhZ3JpLmdvdi5pbA==.
 Ari Feder
Ari Feder Noam Chayut
Noam Chayut Amit Gur1
Amit Gur1 Amit Gal-On
Amit Gal-On Li Li
Li Li Yaakov Tadmor
Yaakov Tadmor