- 1Computational Structural Biology Lab, Department of Biotechnology, Indian Institute of Technology Kharagpur, Kharagpur, India
- 2Laboratory of Plant Stress Biology, Department of Biotechnology, Visva-Bharati, University Santiniketan, India
Plants being sessile are always exposed to various stresses including biotic and abiotic stresses. Some of these stresses are genotoxic to cells causing DNA damage by forming lesions which include altered bases, cross-links, and breaking of DNA strands, which in turn hamper the genomic integrity. In order to survive through all these adverse conditions, plants have evolved different DNA repair mechanisms. As seen from the mammalian system and different human diseases, various microRNAs (miRNAs) can target the 3′-untranslated region of mRNAs that code for the proteins involved in DNA repair pathways. Since miRNAs play an important role in plant cells by regulating various metabolic pathways, it can also be possible that miRNAs play an important role in DNA repair pathways too. However, till date, only a handful of plant miRNAs have been identified to play important role in combating genotoxic stresses in plants. Limitation of information regarding involvement of miRNAs in DNA repair as well as in ROS scavenging prompted us to gather information about plant miRNAs specific for these tasks. This mini-review aims to present pertinent literature dealing with different genotoxic stresses that cause genome instability as well as plant specific responses to survive the damage. This is intertwined with the involvement of miRNAs in genotoxic stress in plants, challenges of applying miRNAs as a tool to combat DNA damage along with ways to overcome these challenges, and finally, the future prospective of these understudied aspects.
Introduction
Plants are always subjected to various environmental stresses which cause severe DNA damage along with genotoxic stress, which in turn may reduce the development, genome stability, and crop productivity. Drought, extreme temperature stress, salt stress, oxidative stress, and damage due to UV irradiation are the abiotic stresses encountered by the plants on daily basis (Tuteja et al., 2011). Plants are also exposed to several biotic stresses through infection by bacteria, virus, pathogens, fungi, and insects (Huang et al., 2016). These genotoxic stresses cause serious damages to plant genome and put the genome integrity at risk (Tuteja et al., 2009).
There are numerous DNA-damaging agents including but not limited to bromouracil, nitrous acid, ethyl methane sulfonate, ethidium bromide like chemical mutagens, or ROS molecules, such as hydrogen peroxide (H2O2), superoxide [], hydroxyl radical (.OH), and even different types of radiations (UV rays, gamma rays, X-rays) (Tuteja et al., 2001). Chemical mutagens damage DNA by altering DNA structure, base pairing, and base structure along with frameshift mutation. UV radiation from sunlight, consisting of UV-A, UV-B, and UV-C types of radiation, causes DNA damage by producing pyrimidine photodimers including cyclobutane pyrimidine dimers (CPD) (Hu and Adar, 2017). These pyrimidine dimers inhibit transcription and replication and induce oxidative stress. ROS overproduction is toxic to plant cells causing damage to DNA, lipids, cell membranes, and proteins (Caverzan et al., 2019).
Plants have evolved strategies to withstand continuous DNA damage and to maintain genome stability. Photoreactivation is the major DNA repair pathway in which the lesions induced by UV radiation are directly reversed back to its normal form (Friedberg, 2015). Single-strand breaks, deaminated, oxidized, or alkylated bases, are repaired by base excision repair (BER) while nucleotide excision repair (NER) works to repair CPDs, although with low capacity (Ries et al., 2000). Homologous recombination (HR) and nonhomologous end-joining (NHEJ)—mediated pathways help in repairing double-strand breaks (Spampinato, 2017). Plants have also evolved mechanisms to scavenge oxidative stress-generated free radicals through different enzymatic processes involving catalases (CATs), peroxidases (POXs), and superoxide dismutases (SODs), as well as non-enzymatic processes involving ascorbic acid and secondary metabolites (Das and Roychoudhury, 2014).
microRNAs (miRNAs) are 20–24 nucleotides long, small non-coding ribonucleic acids, involved in the regulation of gene expression by interfering with various post-transcriptional processes (Yu et al., 2017). Plant miRNAs play vital roles in growth and development as well as in tolerating several types of biotic and abiotic stresses like extreme temperatures, nutrient deprivation, and salinity (Li et al., 2016). Plant miRNAs control gene expression either by cleavage of the target mRNA or through translational inhibition (Xie et al., 2015). miRNAs specifically identify targets by base complementarity and, in turn cleave, translationally repress or destabilize the target mRNAs (Moro et al., 2018). Perfect base pairing of miRNA with target mRNA leads to the cleavage of the targets, whereas the imperfect binding results in translational repression of the target mRNAs (Djami-Tchatchou et al., 2017).
A highly controlled regulation is required to maintain the DNA damage response (DDR) network and ROS scavenging mechanisms to combat genotoxic stresses in plant cells. It is yet to explore whether plant miRNAs play substantial roles in regulating the expression of the genes that are directly or indirectly involved in genotoxic stresses. The fact that only a handful of studies have considered the involvement of miRNAs in DDR and ROS scavenging prompted us to gather information about plant miRNAs specific for these functions. However, we have faced several hurdles in this task due to the unavailability of miRNAs and direct involvement of their corresponding targets within the DDR network. Based on the available information, we have discussed about different genotoxic stresses in plants, role of plant miRNAs in combating genotoxic stresses, hurdles in applying miRNAs as a tool to combat genotoxic stresses in plants, and ways to overcome these problems. The gathered information will be helpful for future practical application of miRNAs as potential tools to secure and stabilize crop yield in view of the continuous climatic changes.
Genotoxic Stresses Leading to Instability in Plant Genome
There are numerous DNA-damaging processes continuously threatening the integrity of the plant genome, including various chemical mutagens and UV radiation, the latest being amongst the most hazardous. These types of stresses generate various DNA lesions that includes altered, missing, and mismatched bases; single- or double-strand breaks; insertion or deletion of bases; pyrimidine dimers; and cross-linked DNA strands, which are genotoxic to plant cells (Tuteja et al., 2001). These damages in turn inhibit transcriptional and translational processes which ultimately affect plant growth and crop yield.
DNA Damage Due to the Production of Free Radicals
Plant cells get damaged by the excess production of ROS which includes free radicals like H2O2, [], and.OH (Inzé and Montagu, 1995; Sharma et al., 2012). Although [] and H2O2 can damage the DNA, these two radicals are very unstable and can easily be removed from the system in the absence of metal catalysts (Tuteja et al., 2009). Conversion of [] and H2O2 to.OH is catalyzed by metals, and.OH is the major source of toxicity in the cells as it reacts with almost all the cellular macromolecules including DNA (Sharma et al., 2012). Lipid peroxidation can also induce the production of ROS which leads to cross-linking of DNA and proteins; hence, they are toxic and mutagenic for cells (Gaschler and Stockwell, 2017). The reactive electrophiles are responsible for the production of various DNA adducts, namely, propano adducts, adducts of acrolein, and crotonaldehyde. The 4-hydroxynonenal compound is the most genotoxic whereas malondialdehyde (MDA) is considered the most mutagenic products of lipid peroxidation (Łuczaj and Skrzydlewska, 2003).
DNA Damage Induced by UV Radiation
UV radiation plays an important role in damaging plant genome stability by producing pyrimidine hydrates as a result of oxidative damage and cross-links between DNA and/or protein, and in turn, inhibits plant growth and development (Gill et al., 2015). UV-B, being the most harmful form of UV radiation, is responsible for the production of DNA lesions like CPD and pyrimidine (6–4) pyrimidinone adducts (6–4 PPs) (Ries et al., 2000; Law et al., 2013). CPDs are found to block transcribing complexes, which in turn is responsible for the alteration of gene expression patterns. In addition to CPD-mediated damage, UV-B also induces delay of G1-to-S phase transition within the plant cell cycle (Jiang et al., 2011). UV-C induces both single-stranded breaks and double-stranded breaks in Arabidopsis (Abas et al., 2007). Oxidative DNA damages are also found to be responsible for UV-associated mutagenicity and instability of plant genome (Manova and Gruszka, 2015).
Plant Cellular Responses to DNA Damage
Plants respond to DNA damage by activating a complex DDR network consisting of mechanisms like DNA repair, cell cycle arrest, and apoptosis (Yoshiyama et al., 2013). Different types of DNA repair mechanisms like photoreactivation, BER, NER, mismatch repair (MMR), and double-strand break (DSB) repair gets activated in plants in response to DNA damage (Kimura and Sakaguchi, 2006). It is found that large numbers of protein components are involved in these repair mechanisms, a handful of which being potential target of miRNAs. The involvements of some of these protein components in the repair mechanisms are briefly discussed.
In the photoreactivation-mediated DNA repair mechanism, thymine dimer structures are found to be cleaved by CPD lyase or (6–4) photolyase (Waterworth et al., 2002). Photolyases bind specifically to the damaged site of double-stranded DNA in a light independent manner, although it gets activated through UV-A for correction of the lesions. This is followed by the splitting of the covalent bonds of the dimers in an error-free manner (Manova and Gruszka, 2015).
Within BER, apurinic/apyrimidinic sites are found to be recognized by lesion-specific DNA glycosylases which cleave the N-glycosidic bond following the removal of the affected base and the generation of abasic sites in plants (Manova and Gruszka, 2015). In Arabidopsis, carrot, and rice, several DNA glycosylases were identified—as a couple of examples, 3-methyladenine-DNA glycosylase (MAG), formamido-pyrimidine-DNA glycosylase (FPG) 8-oxoG DNA-glycosylases (OGG), uracil-DNA glycosylase (UNG), and DNA glycosylase/lyase DNG701 (Santerre and Britt, 1994; Dany and Tissier, 2001; Talpaert-Borlé and Liuzzi, 2005; La et al., 2011). NER recognizes and repairs several types of DNA lesions induced by UV-rays and other mutagens, and the process was extensively studied in Arabidopsis thaliana. Xeroderma pigmentosum complementation of group C (XP-C)/AtRAD4 recognizes DNA damage. This is followed by the unwinding of DNA containing damaged portion by the transcription factor IIH (TFIIH), including AtXP-D. Damaged oligonucleotides get excised by AtXP-F or other Arabidopsis homologs (e.g., AtERCC1). The excised gap is filled through the activity of proliferating cell nuclear antigen (PCNA) and replication factor C (RFC)–mediated DNA synthesis. Finally, DNA ligase I joins the DNA strands (Xu et al., 1998; Ishibashi et al., 2003; Kimura and Sakaguchi, 2006; Molinier et al., 2008; Liu et al., 2012).
Within MMR, homologs of MutS (MSH) recognize the mismatch generated by the incorporation of incorrect bases by DNA polymerase. This is followed by generation of nicks through the activity of MutL (MLH) homologues and the successive steps of the repair system (Dion et al., 2007; Lario et al., 2015).
DSBs are repaired in plants by HR- and NHEJ-mediated pathways (Puchta, 2005). In Arabidopsis and rice, many components of DSB repair mechanisms have been identified—for example, AtRad51, AtRadA, AtRad50, OsRadA, AtMre11, AtKu70, AtKu80, Arabidopsis DNA ligase IV, AtXRCC4, AtXP-F, and AtERCC1 (Manova and Gruszka, 2015). Additionally, it has been discovered that Ataxia telangiectasia–mutated (ATM) and ATM-Rad3-related (ATR) proteins play important roles in DNA repair. Checkpoint kinases (CHK), including Chk1 and Chk2, work downstream of ATM and ATR proteins, where activated ATR initiates G-2 phase arrest by phosphorylating CHK1. This in turn is responsible for DDR-induced transcriptional repression (Culligan et al., 2004).
Involvement of Plant MiRNAs in Genotoxic Stress Tolerance
Till date, a handful of plant miRNAs with active role in combating genotoxic stresses have been identified in plants. Although the number is considerably low, we can divide these miRNAs into two categories. One set of miRNAs is involved in tolerating oxidative stress, while the other type of miRNAs may play an active part in DNA repair. A large set of enzymes acting as the key regulators of both ROS scavenging and DNA repair mechanism may be targeted by both these types of miRNAs. We have tabulated plant miRNAs found to target various enzymes involved in ROS scavenging and DDR network in Table 1. A schematic depiction of the involvement of miRNAs in these two processes is also shown in Figure 1.
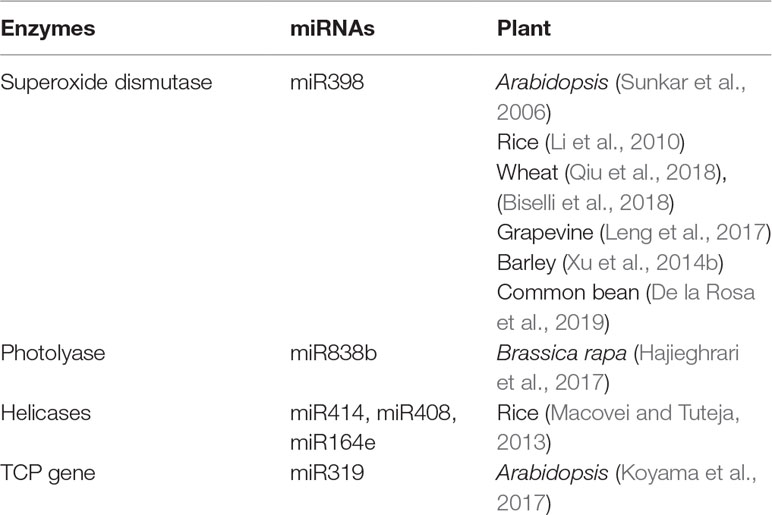
Table 1 Involvement of miRNAs in targeting various enzymes involved in ROS scavenging as well as DDR network in plants.
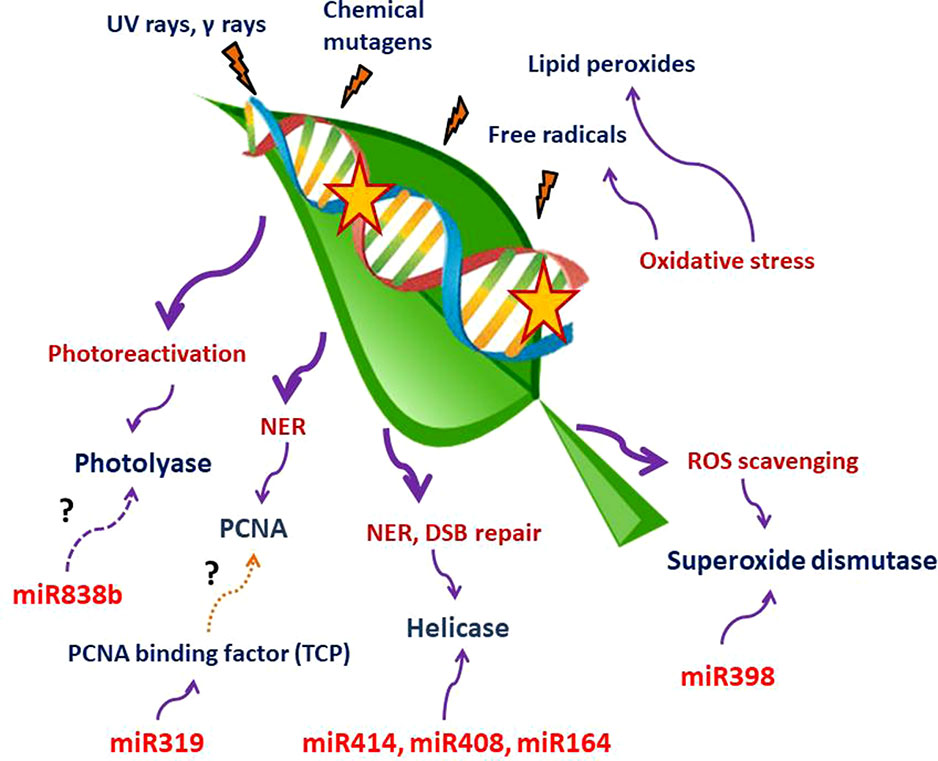
Figure 1 Schematic representation of miRNAs involvement in ROS scavenging and DNA repair mechanisms. UV and γ-rays, chemical mutagens, free radicals, and lipid peroxidation produced due to oxidative stress are among the most important DNA damage–causing agents. miR398, miR319, miR838b, miR414, miR408, and miR164 have been shown to target components involved in the abovementioned processes. The question mark indicates that more studies are required to establish the link with the adjacent pathways. There is no direct evidence of miR319 targeting TCPs that bind to the PCNA promoter (brown dotted line). Targeting of photolyase by miR838b through computational prediction needs experimental confirmation (magenta dashed line).
miRNA Involvement in ROS Scavenging
SOD, a metal containing enzyme, is one of the most important enzymes for the removal of ROS produced by oxidative stress (Das and Roychoudhury, 2014). There are different types of SODs, including mitochondrial MnSOD, cytosolic and chloroplastic Cu/ZnSOD, and chloroplastic FeSOD (Van Camp et al., 1990; Bowler et al., 1994; Fukai and Ushio-Fukai, 2011). The complete regulatory mechanism of CSD1 and CSD2 is unknown, but recent studies have proved that miRNAs are having a role in this. Sunkar et al. (2006) have shown that the Arabidopsis ath-miR398 has a role in the regulation of CSD1 and CSD2. They found that downregulation of miR398 expression is important for the accumulation of CSD1/2 mRNA. While experimenting with different tissues, the same group of scientists have found that tissues like cauline leaves, stems, and roots (having high levels of miR398) showed lower expression of CSD1/2 mRNAs, whereas tissues like old rosette, leaves, and inflorescence (having low level of the same miRNAs) showed higher levels of CSD1/2 expression. Additionally, in rice, it was evidenced that miR159 targets Cu/ZnSOD (At5g18100-CSD3) (Li et al., 2010). Another group of scientists has shown that CSD is a potential targets of miR398 in wheat seedlings exposed to drought (Qiu et al., 2018). Cytosolic CSD1/2 was found to be targeted by miR398 in wheat in response to the fungal attack by Fusarium graminearum which causes Fusarium head blight disease (Biselli et al., 2018). Cytosolic CSD1 and chloroplastic CSD2 are potential targets of Vv-miR398 in grapevines (Leng et al., 2017). The Vv-miR398 family is highly conserved, and three loci, namely, miR398a (located on the chromosome no. 1), miR398b, and miR398c (located on the chromosome no. 6) encode the MIR gene in grapevine. In barley, hvu-miR398 was found to be negatively regulated by Mildew resistance locus a (Mla) and Mla resistance1 (rom1), and the overexpression of miR398 is responsible for the reduction in CSD1 (Xu et al., 2014b). In another experiment carried out in Phaseolus vulgaris, it has been shown that repression of miR398 leads to upregulation of CSD1 expression in case of water deficit (De la Rosa et al., 2019).
miRNA Involvement in DNA Repair Mechanism
When considering the direct implication of miRNAs in plant DNA repair mechanisms, some helicases, important enzymes involved in both NER and DSB repairs, have been shown to be targeted by miR164, miR408, and miR414 in rice (Macovei and Tuteja, 2013). Several studies have addressed putative miRNA targeting mRNAs of genes involved in DDR by in silico analyses. For example, using psRNATarget server, a computational tool to predict miRNA targets, it was found that Brassica rapa miR838b putatively targets the photolyase mRNA (Hajieghrari et al., 2017). However, this prediction remains to be confirmed experimentally.
As an example of indirect involvement of miRNAs in DDR downstream processes, several studies established the contribution of miR319 to the regulation of TCP (teosinte-branched1/Cincinnata/proliferating cell factor) transcription factors, playing direct roles in leaf development (Danisman, 2016; Koyama et al., 2017; Bresso et al., 2018). It has been shown that two transcription factors, namely, PCF1 and PCF2, containing the non-canonical basic helix–loop–helix motif TCP domain, can regulate the transcription of PCNA (proliferating cell nuclear antigen) gene, an important component of the DDR network (Danisman, 2016; Nicolas and Cubas, 2016). Among the 24 members of Arabidopsis TCP family, miR319 targets the transcripts of TCP2, TCP3, TCP4, TCP10, and TCP24 genes (Koyama et al., 2017). However, till date, there is no direct evidence showing that miR319 targeting TCPs can directly bind to the PCNA promoter.
Challenges Related to MiRNA Applications in Combating Genotoxic Stresses in Plants
Hurdle I: Although many miRNAs have been found to be involved in the DDR network in human cells, very few miRNAs are found to be involved in DNA repair in plants. DNA repair processes are well characterized in mammalian systems; in contrast, very few studies are done in plants, and these are mainly limited to Arabidopsis and rice (Ueda and Nakamura, 2011; Macovei and Tuteja, 2013; Manova and Gruszka, 2015).
Hurdle II: There is absence of focused research on targeting genes involved in DNA repair even when considering miRNAs that are extensively studied in plants in relation to stress response; however, information about targets of miRNAs that are specifically involved in combating genotoxic stresses in plants is much more limited. Differently, many studies are being performed to find the targets of miRNAs in DDR in human diseases, including cancer studies (Hu and Gatti, 2011; He et al., 2016).
How to Overcome the Challenges?
Identification of more plant miRNAs involved in DNA repair and ROS scavenging is one of the ways to overcome the challenge. In this regard, RNA sequencing of plants exposed to genotoxic stresses will lead to identification of new miRNAs specifically associated with genotoxic stresses. Once a substantial number of miRNAs are being identified in plants, their targets need to be validated. For target identification, bioinformatics approaches, along with experimental validation of the same by 5’ rapid amplification of cDNA ends (RACE), parallel analysis of RNA ends (PARE), degradome-seq, or genome-wide mapping of uncapped transcripts, can be of great help. References from miRNAs involved in DDR in human diseases can be taken, and extensive studies must be performed to find out the role of plant miRNAs in targeting the homolog proteins involved in DNA repair in plant species.
Several human diseases are found to be associated with miRNA-dependent regulation of DNA repair pathways. For example, RAD23 and CDK7 are two important enzymes in NER pathway. miR-494 was found to target RAD23 homolog B (Comegna et al., 2014), while CDK7 is targeted by miR-210 (Abdullah et al., 2016). Human miR-103a-2-5p and miR-585-5p target poly ADP-ribose polymerase (PARP), an important BER enzyme (Dluzen et al., 2017). Human miR-422a base pairs with MLH1 3′-untranslated region and suppresses the expression of the same which in turn downregulate MutLα, a key protein of the MMR (Mao et al., 2012). In colorectal cancer cells, miR-7 targets XRCC2, a core protein involved in HR (Xu et al., 2014a). In cancer cell lines, RAD51 and BRCA1/2 (breast/ovarian cancer susceptibility gene products) are key proteins responsible for catalyzing HR, and both the proteins are potential target of miR-103 and miR-107 (Huang et al., 2013). BRCA1/2 is also found to be targeted by miR-15/107/182 in breast cancer (Petrovic et al., 2017). MSH2, another essential MMR component, is downregulated by miR-21 in human (Valeri et al., 2010). Homologs of all the human RAD23, CDK7, PARP, MSH, MLH, XRCC2, RAD51, and BRCA1/2 are present in rice. However, there is no information available about potential miRNAs that target these mRNAs. While studying NHEJ repair in lung-cancer cell line, Yan et al. (2010) reported that miR-101 targets 3′- UTR of DNA-PKcs, a core component of NHEJ. In the same study, it has been proved that ATM, another key protein of HR-mediated repair, is a target of miR-101. Even tough plant homologues of ATM are reported in Arabidopsis and rice, no miRNA associated to their sequences was identified. Hence, it is noteworthy to mention that, even though it is well-established, several human genes involved in DNA repair are targeted by miRNAs, and some of their homologs are also reported in plants. No information is available about miRNAs targeting these genes in plants. With the advancement of genome annotation techniques and the availability of published and draft genomes, miRNAs can be searched firstly through bioinformatics approach. Once detected, these miRNAs can be experimentally verified for differential regulation of the target miRNAs associated with genotoxic stresses.
Conclusion and Future Perspectives
DNA repair is a very important mechanism that allows plant cells to overcome genotoxic stresses and to maintain genome integrity. Impaired DNA repair mechanisms are the reason for plant slow growth and development, which in turn causes the reduction in crop production. In recent years, miRNAs have been identified as potentially novel and vital regulators of biological processes, including developmental processes and diseases. Considering their importance, it is essential to know more about miRNAs and their targets associated with DNA repair mechanism in plants. Once we find out the specific role of miRNAs and their targets in DNA repair and ROS scavenging, we could engineer them with genome-editing technologies like CRISPR-Cas, and hence aiming to combat a great number of genotoxic stresses.
Author Contributions
MC wrote the manuscript. JB conceived the study and participated in its coordination.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abas, Y., Touil, N., Kirsch-Volders, M., Angenon, G., Jacobs, M., Famelaer, I. D. H. (2007). Evaluation of UV damage at DNA level in Nicotiana plumbaginifolia protoplasts using single cell gel electrophoresis. Plant Cell Tissue Organ Cult. 91, 145–154. doi: 10.1007/s11240-007-9257-9
Abdullah, A. I., Zhang, H., Nie, Y., Tang, W., Sun, T. (2016). CDK7 and miR-210 co-regulate cell-cycle progression of neural progenitors in the developing neocortex. Stem Cell Rep. 7, 69–79. doi: 10.1016/j.stemcr.2016.06.005
Biselli, C., Bagnaresi, P., Faccioli, P., Hu, X., Balcerzak, M., Mattera, M. G., et al. (2018). Comparative Transcriptome profiles of near-isogenic hexaploid wheat lines differing for effective alleles at the 2DL FHB resistance QTL. Front. Plant Sci. 9, 37. doi: 10.3389/fpls.2018.00037
Bowler, C., Camp, W. V., Montagu, M. V., Inzé, D., Asada, P. K. (1994). Superoxide dismutase in plants. Crit. Rev. Plant Sci. 13, 199–218. doi: 10.1080/07352689409701914
Bresso, E. G., Chorostecki, U., Rodriguez, R. E., Palatnik, J. F., Schommer, C. (2018). Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol. 176, 1694–1708. doi: 10.1104/pp.17.00823
Caverzan, A., Piasecki, C., Chavarria, G., Stewart, C. N., Vargas, L. (2019). Defenses against ROS in crops and weeds: the effects of interference and herbicides. Int. J. Mol. Sci. 20, 1086. doi: 10.3390/ijms20051086
Comegna, M., Succoio, M., Napolitano, M., Vitale, M., D’Ambrosio, C., Scaloni, A., et al. (2014). Identification of miR-494 direct targets involved in senescence of human diploid fibroblasts. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 28, 3720–3733. doi: 10.1096/fj.13-239129
Culligan, K., Tissier, A., Britt, A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16, 1091–1104. doi: 10.1105/tpc.018903
Danisman, S. (2016). TCP transcription factors at the interface between environmental challenges and the plant’s growth responses. Front. Plant Sci. 7, 1930. doi: 10.3389/fpls.2016.01930
Dany, A. L., Tissier, A. (2001). A functional OGG1 homologue from Arabidopsis thaliana. Mol. Genet. Genomics MGG 265, 293–301. doi: 10.1007/s004380000414
Das, K., Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2, 53. doi: 10.3389/fenvs.2014.00053
De la Rosa, C., Covarrubias, A. A., Reyes, J. L. (2019). A dicistronic precursor encoding miR398 and the legume-specific miR2119 coregulates CSD1 and ADH1 mRNAs in response to water deficit. Plant Cell Environ. 42, 133–144. doi: 10.1111/pce.13209
Dion, E., Li, L., Jean, M., Belzile, F. (2007). An Arabidopsis MLH1 mutant exhibits reproductive defects and reveals a dual role for this gene in mitotic recombination. Plant J. Cell Mol. Biol. 51, 431–440. doi: 10.1111/j.1365-313X.2007.03145.x
Djami-Tchatchou, A. T., Sanan-Mishra, N., Ntushelo, K., Dubery, I. A. (2017). Functional roles of microRNAs in agronomically Important plants—potential as targets for crop improvement and protection. Front. Plant Sci. 8, 378. doi: 10.3389/fpls.2017.00378
Dluzen, D. F., Kim, Y., Bastian, P., Zhang, Y., Lehrmann, E., Becker, K. G., et al. (2017). MicroRNAs modulate oxidative stress in hypertension through PARP-1 regulation. Oxid. Med. Cell. Longev. 2017, 3984280. doi: 10.1155/2017/3984280
Friedberg, E. C. (2015). A history of the DNA repair and mutagenesis field: I. The discovery of enzymatic photoreactivation. DNA Repair 33, 35–42. doi: 10.1016/j.dnarep.2015.06.007
Fukai, T., Ushio-Fukai, M. (2011). Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal. 15, 1583–1606. doi: 10.1089/ars.2011.3999
Gaschler, M. M., Stockwell, B. R. (2017). Lipid peroxidation in cell death. Biochem. Biophys. Res. Commun. 482, 419–425. doi: 10.1016/j.bbrc.2016.10.086
Gill, S. S., Anjum, N. A., Gill, R., Jha, M., Tuteja, N. (2015). DNA damage and repair in plants under ultraviolet and ionizing radiations. Sci. World J. 2015, 250158. doi: 10.1155/2015/250158
Hajieghrari, B., Farrokhi, N., Goliaei, B., Kavousi, K. (2017). Computational identification of MicroRNAs and their transcript target(s) in field mustard (Brassica rapa L.). Iran. J. Biotechnol. 15, 22–32. doi: 10.15171/ijb.1390
He, M., Zhou, W., Li, C., Guo, M. (2016). MicroRNAs, DNA damage response, and cancer treatment. Int. J. Mol. Sci. 17, 2087. doi: 10.3390/ijms17122087
Hu, H., Gatti, R. A. (2011). MicroRNAs: new players in the DNA damage response. J. Mol. Cell Biol. 3, 151–158. doi: 10.1093/jmcb/mjq042
Hu, J., Adar, S. (2017). The cartography of UV-induced DNA damage formation and DNA repair. Photochem. Photobiol. 93, 199–206. doi: 10.1111/php.12668
Huang, J., Yang, M., Zhang, X. (2016). The function of small RNAs in plant biotic stress response. J. Integr. Plant Biol. 58, 312–327. doi: 10.1111/jipb.12463
Huang, J.-W., Wang, Y., Dhillon, K. K., Calses, P., Villegas, E., Mitchell, P. S., et al. (2013). Systematic screen identifies miRNAs that target RAD51 and RAD51D to enhance chemosensitivity. Mol. Cancer Res. MCR 11, 1564–1573. doi: 10.1158/1541-7786.MCR-13-0292
Inzé, D., Montagu, M. V. (1995). Oxidative stress in plants. Curr. Opin. Biotechnol. 6, 153–158. doi: 10.1016/0958-1669(95)80024-7
Ishibashi, T., Kimura, S., Yamamoto, T., Furukawa, T., Takata, K., Uchiyama, Y., et al. (2003). Rice UV-damaged DNA binding protein homologues are most abundant in proliferating tissues. Gene 308, 79–87. doi: 10.1016/S0378-1119(03)00447-5
Jiang, L., Wang, Y., Björn, L. O., Li, S. (2011). UV-B-induced DNA damage mediates expression changes of cell cycle regulatory genes in Arabidopsis root tips. Planta 233, 831–841. doi: 10.1007/s00425-010-1340-5
Kimura, S., Sakaguchi, K. (2006). DNA repair in plants. Chem. Rev. 106, 753–766. doi: 10.1021/cr040482n
Koyama, T., Sato, F., Ohme-Takagi, M. (2017). Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol. 175, 874–885. doi: 10.1104/pp.17.00732
La, H., Ding, B., Mishra, G. P., Zhou, B., Yang, H., Bellizzi, M., et al. (2011). A 5-methylcytosine DNA glycosylase/lyase demethylates the retrotransposon Tos17 and promotes its transposition in ricet. Proc. Natl. Acad. Sci. U.S.A. 108, 15498–15503. doi: 10.1073/pnas.1112704108
Lario, L. D., Botta, P., Casati, P., Spampinato, C. P. (2015). Role of AtMSH7 in UV-B-induced DNA damage recognition and recombination. J. Exp. Bot. 66, 3019–3026. doi: 10.1093/jxb/eru464
Law, Y. K., Forties, R. A., Liu, X., Poirier, M. G., Kohler, B. (2013). Sequence-dependent thymine dimer formation and photoreversal rates in double-stranded DNA. Photochem. Photobiol. Sci. Off. J. Eur. Photochem. Assoc. Eur. Soc. Photobiol. 12, 1431–1439. doi: 10.1039/c3pp50078k
Leng, X., Wang, P., Zhu, X., Li, X., Zheng, T., Shangguan, L., et al. (2017). Ectopic expression of CSD1 and CSD2 targeting genes of miR398 in grapevine is associated with oxidative stress tolerance. Funct. Integr. Genomics 17, 697–710. doi: 10.1007/s10142-017-0565-9
Li, H., Wang, Y., Wang, Z., Guo, X., Wang, F., Xia, X.-J., et al. (2016). Microarray and genetic analysis reveals that csa-miR159b plays a critical role in abscisic acid-mediated heat tolerance in grafted cucumber plants. Plant Cell Environ. 39, 1790–1804. doi: 10.1111/pce.12745
Li, Y.-F., Zheng, Y., Addo-Quaye, C., Zhang, L., Saini, A., Jagadeeswaran, G., et al. (2010). Transcriptome-wide identification of microRNA targets in rice. Plant J. Cell Mol. Biol. 62, 742–759. doi: 10.1111/j.1365-313X.2010.04187.x
Liu, J., Tang, X., Gao, L., Gao, Y., Li, Y., Huang, S., et al. (2012). A role of tomato UV-damaged dna binding protein 1 (DDB1) in organ size control via an epigenetic manner. PLoS One 7, e42621. doi: 10.1371/journal.pone.0042621
Łuczaj, W., Skrzydlewska, E. (2003). DNA damage caused by lipid peroxidation products. Cell. Mol. Biol. Lett. 8, 391–413.
Macovei, A., Tuteja, N. (2013). Different expression of miRNAs targeting helicases in rice in response to low and high dose rate γ-ray treatments. Plant Signal. Behav. 8, e25128. doi: 10.4161/psb.25128
Manova, V., Gruszka, D. (2015). DNA damage and repair in plants—from models to crops. Front. Plant Sci. 6, 885. doi: 10.3389/fpls.2015.00885
Mao, G., Lee, S., Ortega, J., Gu, L., Li, G.-M. (2012). Modulation of microRNA processing by mismatch repair protein MutLα. Cell Res. 22, 973–985. doi: 10.1038/cr.2012.18
Molinier, J., Lechner, E., Dumbliauskas, E., Genschik, P. (2008). Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress. PLoS Genet. 4, e1000093. doi: 10.1371/journal.pgen.1000093
Moro, B., Chorostecki, U., Arikit, S., Suarez, I. P., Höbartner, C., Rasia, R. M., et al. (2018). Efficiency and precision of microRNA biogenesis modes in plants. Nucleic Acids Res. 46, 10709–10723. doi: 10.1093/nar/gky853
Nicolas, M., Cubas, P. (2016). TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 33, 33–41. doi: 10.1016/j.pbi.2016.05.006
Petrovic, N., Davidovic, R., Bajic, V., Obradovic, M., Isenovic, R. E. (2017). MicroRNA in breast cancer: The association with BRCA1/2. Cancer Biomark. Sect. Dis. Markers 19, 119–128. doi: 10.3233/CBM-160319
Puchta, H. (2005). The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56, 1–14. doi: 10.1093/jxb/eri025
Qiu, Z., He, Y., Zhang, Y., Guo, J., Wang, L. (2018). Characterization of miRNAs and their target genes in He-Ne laser pretreated wheat seedlings exposed to drought stress. Ecotoxicol. Environ. Saf. 164, 611–617. doi: 10.1016/j.ecoenv.2018.08.077
Ries, G., Buchholz, G., Frohnmeyer, H., Hohn, B. (2000). UV-damage-mediated induction of homologous recombination in Arabidopsis is dependent on photosynthetically active radiation. Proc. Natl. Acad. Sci. 97, 13425–13429. doi: 10.1073/pnas.230251897
Santerre, A., Britt, A. B. (1994). Cloning of a 3-methyladenine-DNA glycosylase from Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 91, 2240–2244. doi: 10.1073/pnas.91.6.2240
Sharma, P., Jha, A. B., Dubey, R. S., Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 1–26. doi: 10.1155/2012/217037
Spampinato, C. P. (2017). Protecting DNA from errors and damage: an overview of DNA repair mechanisms in plants compared to mammals. Cell. Mol. Life Sci. CMLS 74, 1693–1709. doi: 10.1007/s00018-016-2436-2
Sunkar, R., Kapoor, A., Zhu, J.-K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18, 2051–2065. doi: 10.1105/tpc.106.041673
Talpaert-Borlé, M., Liuzzi, M. (2005). Base-excision repair in carrot cells: partial purification and characterization of uracil-DNA glycosylase and apurinic/apyrimidinic endodeox yribonuclease. Eur. J. Biochem. 124, 435–440. doi: 10.1111/j.1432-1033.1982.tb06611.x
Tuteja, N., Ahmad, P., Panda, B. B., Tuteja, R. (2009). Genotoxic stress in plants: shedding light on DNA damage, repair and DNA repair helicases. Mutat. Res. Mutat. Res. 681, 134–149. doi: 10.1016/j.mrrev.2008.06.004
Tuteja, N., Gill, S., Tuteja, R. (2011). “Plant responses to abiotic stresses: shedding light on salt, drought, cold and heavy metal stress,” in Omics and Plant Abiotic Stress Tolerance. (Sharjah: Bentham Science Publisher), 3, 39–64. doi: 10.2174/97816080505811110101
Tuteja, N., Singh, M. B., Misra, M. K., Bhalla, P. L., Tuteja, R. (2001). Molecular mechanisms of DNA damage and repair: progress in plants. Crit. Rev. Biochem. Mol. Biol. 36, 337–397. doi: 10.1080/20014091074219
Ueda, T., Nakamura, C. (2011). Ultraviolet-defense mechanisms in higher plants. Biotechnol. Biotechnol. Equip. 25, 2177–2182. doi: 10.5504/BBEQ.2011.0001
Valeri, N., Gasparini, P., Braconi, C., Paone, A., Lovat, F., Fabbri, M., et al. (2010). MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc. Natl. Acad. Sci. U.S.A. 107, 21098–21103. doi: 10.1073/pnas.1015541107
Van Camp, W., Bowler, C., Villarroel, R., Tsang, E. W., Van Montagu, M., Inzé, D. (1990). Characterization of iron superoxide dismutase cDNAs from plants obtained by genetic complementation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 87, 9903–9907. doi: 10.1073/pnas.87.24.9903
Waterworth, W. M., Jiang, Q., West, C. E., Nikaido, M., Bray, C. M. (2002). Characterization of Arabidopsis photolyase enzymes and analysis of their role in protection from ultraviolet-B radiation. J. Exp. Bot. 53, 1005–1015. doi: 10.1093/jexbot/53.371.1005
Xie, M., Zhang, S., Yu, B. (2015). microRNA biogenesis, degradation and activity in plants. Cell. Mol. Life Sci. CMLS 72, 87–99. doi: 10.1007/s00018-014-1728-7
Xu, H., Swoboda, I., Bhalla, P. L., Sijbers, A. M., Zhao, C., Ong, E. K., et al. (1998). Plant homologue of human excision repair gene ERCC1 points to conservation of DNA repair mechanisms. Plant J. Cell Mol. Biol. 13, 823–829. doi: 10.1046/j.1365-313X.1998.00081.x
Xu, K., Chen, Z., Qin, C., Song, X. (2014a). miR-7 inhibits colorectal cancer cell proliferation and induces apoptosis by targeting XRCC2. OncoTargets Ther. 7, 325–332. doi: 10.2147/OTT.S59364
Xu, W., Meng, Y., Wise, R. P. (2014b). Mla- and Rom1-mediated control of microRNA398 and chloroplast copper/zinc superoxide dismutase regulates cell death in response to the barley powdery mildew fungus. New Phytol. 201, 1396–1412. doi: 10.1111/nph.12598
Yan, D., Ng, W. L., Zhang, X., Wang, P., Zhang, Z., Mo, Y.-Y., et al. (2010). Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One 5, e11397. doi: 10.1371/journal.pone.0011397
Yoshiyama, K. O., Sakaguchi, K., Kimura, S. (2013). DNA damage response in plants: conserved and variable response compared to animals. Biology 2, 1338–1356. doi: 10.3390/biology2041338
Keywords: UV radiation, genotoxic stress, microRNAs, DNA lesions, DNA damage response network
Citation: Roy Chowdhury M and Basak J (2019) Tiny Yet Indispensable Plant MicroRNAs Are Worth to Explore as Key Components for Combating Genotoxic Stresses. Front. Plant Sci. 10:1197. doi: 10.3389/fpls.2019.01197
Received: 19 April 2019; Accepted: 30 August 2019;
Published: 04 October 2019.
Edited by:
Anca Macovei, University of Pavia, ItalyReviewed by:
Vinay Kumar, Pune University, IndiaJose Luis Reyes, National Autonomous University of Mexico, Mexico
Copyright © 2019 Roy Chowdhury and Basak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jolly Basak, am9sbHkuYmFzYWtAdmlzdmEtYmhhcmF0aS5hYy5pbg==
 Moumita Roy Chowdhury
Moumita Roy Chowdhury Jolly Basak
Jolly Basak