
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 09 October 2019
Sec. Plant Systematics and Evolution
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01196
 Qiu-jie Li1,2
Qiu-jie Li1,2 Xi Wang1,2
Xi Wang1,2 Jun-ru Wang1,2
Jun-ru Wang1,2 Na Su1,2
Na Su1,2 Ling Zhang3
Ling Zhang3 Yue-ping Ma4
Yue-ping Ma4 Zhao-yang Chang1,2
Zhao-yang Chang1,2 Liang Zhao1,2*
Liang Zhao1,2* Daniel Potter5
Daniel Potter5Pulsatilla (Ranunculaceae) comprises about 40 species, many of which have horticultural and/or medicinal importance. However, the recognition and identification of wild Pulsatilla species is difficult due to the presence of complex morphological characters. DNA barcoding is a powerful molecular tool capable of rapidly and accurately distinguishing between species. Here, we assessed the effectiveness of four commonly used DNA barcoding loci—rbcL (R), trnH-psbA ( T ), matK (M), and ITS (I)—to identify species of Pulsatilla from a comprehensive sampling group. Among the four barcoding single loci, the nuclear ITS marker showed the highest interspecific distances and the highest rate of correct identification. Among the eleven combinations, the chloroplast multi-locus R+T and R+M+T combinations were found to have the best species discrimination rate, followed by R+M. Overall, we propose that the R+M+T combination and the ITS marker on its own are, respectively, the best multi- and single-locus barcodes for discriminating among species of Pulsatilla. The phylogenetic analysis was able to distinguish species of Pulsatilla to the subgenus level, but the analysis also showed relatively low species resolution. This may be caused by incomplete lineage sorting and/or hybridization events in the evolutionary history of the genus, or by the resolution limit of the candidate barcodes. We also investigated the leaf epidermis of eight representative species using scanning electronic microscopy. The resulting micro-morphological characters were valuable for identification of related species. Using additional genome fragments, or even whole chloroplast genomes combined with micro-morphological data may permit even higher resolution of species in Pulsatilla.
The Ranunculaceae is a large and complex plant family, including approximately 59 genera and 2,500 species (Tamura, 1995). Pulsatilla Miller, first described in 1753, consists of about 40 species that are restricted to temperate subarctic and mountainous areas in the Northern Hemisphere (Tamura, 1995). Plants of Pulsatilla species are often covered with long, soft hairs. Their flowers are solitary and bisexual, with three bracts forming a bell-shaped involucre. The tepal number is always six, and stamens are generally numerous, with the outermost ones resembling degenerated petals (although Pulsatilla kostyczewii is a notable exception to this tendency) (Figure 1; Wang et al., 2001; Ren et al., 2011; Ren et al., 2015).

Figure 1 Representatives of species illustrating the morphological variation and similarities in Pulsatilla. (A–F) plants in flower; (G–L) anthetic flower. (A) P. chinensis; (B) P. cernua; (C) P. patens; (D) P. camoanella; (E) P. ambigua; (F) P. dahurica; (G) P. chinensis; (H) P.cernua; (I) P. patens; (J) P. camoanella; (K) P. ambigua; (L) P. dahurica; (M–P) style strongly elongate and plumose in fruit; (Q) lateral view of flower showing the retarded stamen in outermost; (R) sepal; (S) stamens and pistil.
Most authors have treated Pulsatilla as a subgenus or section of the genus Anemone s.l. (Linnaeus, 1753; Endlicher, 1839; Tamura, 1967; Tamura, 1995; Hoot et al., 1994). However, Miller (1754), Adanson (1763), and Wang et al. (2001) have supported a model that separates Pulsatilla from Anemone as an independent genus. Recent phylogenetic studies have shown that all species within Pulsatilla are clustered in a monophyletic group, which is nested within Anemone (Hoot et al., 1994; Jiang et al., 2017). Morphologically, Pulsatilla can easily be distinguished from Anemone s.s., since species of the former have a long, plumose beak on the achenes formed by the persistent style and stamens (Tamura, 1995; Wang et al., 2001) whereas species of the latter do not. Because the primary goal of the present study is to test the use of DNA barcodes for species in the Pulsatilla clade, we here follow the treatment of Wang et al. (2001) and Grey-Wilson (2014), regarding Pulsatilla as a distinct genus.
There are eleven species of Pulsatilla found in China, most of which are found primarily in the northern part of the country (Grey-Wilson, 2014). Some species of Pulsatilla have been used in traditional Chinese medicine for many years for “blood-cooling” or “detoxification” (Pharmacopoeia, 2015). In particular, the root of Pulsatilla chinensis (Bunge) Regel is a well-known ingredient included in the Chinese Pharmacopoeia (2015). Many species (e.g. P. ambigua, P. campanella, P. cernua, P. chinensis, P. dahurica and P. turczaninovii) used in folk medicine have been found to contain pharmacologically useful chemical components, including those with anti-cancer and anti-inflammatory activities (Xu et al., 2012; Wang et al., 2016; Suh and An 2017). The contents of these components differ in various species, resulting in different clinical pharmacological effects. Thus, in cases where target species can be easily confused with their close relatives, undesired species can be inadvertently collected, resulting in negative effects on drug efficacy and patient safety, as has been shown in other plant groups of medicinal importance in China (Zhang et al., 2015a; Zhang et al., 2015b).
Pulsatilla is an especially challenging, complex group. In all treatments published to date, the genus has been treated as comprising two to four subgenera: subgenus Miyakea, which contains only one species, P. integrifolia; subgenus Kostyczewianae, which has only one species, located in Central Asia and northwestern China; subgenus Preonanthus, which includes six species; and the largest subgenus Pulsatilla, which comprises 29 species (Tamura, 1995; Wang et al., 2001; Grey-Wilson, 2014; Sramkó et al., 2019). However, Pulsatilla shows a frustratingly complicated pattern of intrageneric morphological variability (Grey-Wilson, 2014). The recognition and identification of wild Pulsatilla species based on traditional approaches is difficult due to transitional intraspecific morphological characteristics in many Pulsatilla species. For instance, P. turczaninovii and P. tenuiloba were considered to be two separate species that could be told apart by the number of pairs of lateral leaflets (i.e. a leaf blade with 3 or 4 pairs of lateral leaflets vs. a leaf blade with 5 or 6 pairs of lateral leaflets) (Wang et al., 2001). After carefully checking specimens and population investigation, we found that the leaflet numbers of P. turczaninovii and P. tenuiloba are overlapping, and some individuals have both 4 and 5 (even 6) pairs of lateral leaflets. Flowers nodding before anthesis is recorded as a diagnostic character of P. campanella, but this character was also found in P. ambigua, P. cernua and P. dahurica, and their flower colors show continuous transitional shades of blue (Wang et al., 2001). Thus, these characters are not reliable and make Pulsatilla difficult to identify.
DNA barcoding aims to achieve rapid and accurate species recognition by sequencing short DNA sequences or a few small DNA regions (Fay et al., 1997; Hebert et al., 2003a; Hebert and Gregory, 2005; Kress et al., 2005; China Plant BOL Group, 2011; Kress, 2017). This technology was first developed to identify animal species; for example, Hebert et al. (2003b) argued that “the mitochondrial gene cytochrome c oxidase I (COI), can serve as the core of a global bio-identification system for animals”. Studies have continued to demonstrate that the COI gene fragment efficiently discriminates among animal species, including amphibians (Vences et al., 2005), birds (Hebert et al., 2004; Yoo et al., 2006; Kerr et al., 2007; Tavares and Baker, 2008), fish (Yancy et al., 2008; Ward et al., 2009), and insects (Hajibabaei et al., 2006; Pons et al., 2006). In plants, however, frequent recombination and low mutation rates restrict the utility of mitochondrial barcode markers (Cho et al., 1998; Cho et al., 2004). The search for suitable candidates has therefore focused on chloroplast and nuclear DNA markers (De Salle et al., 2005; Kress et al., 2005; CBOL Plant Working Group, 2009; China Plant BOL Group, 2011; Yan et al., 2015), although such markers are not always easy to amplify and sequence in all plant taxa using universal primers. Numerous studies have suggested that four standard barcodes — three from the chloroplast genome [the ribulose-bisphosphate/carboxylase Large-subunit gene (rbcL), the maturase-K gene (matK), and the trnH-psbA intergenic spacer and the nuclear ribosomal internal transcribed spacers (nrITS)] — should be used as core barcode markers for the molecular identification of plants (China Plant BOL Group, 2011; Xiang et al., 2011; Zhang et al., 2012; Li et al., 2015).
Significant progress has been made in DNA barcoding in plants (Zhang et al., 2015b; Han et al., 2016; Li et al., 2016). However, the discrimination of closely related species using only molecular data is still a major challenge in some genera (Piredda et al., 2011; Zhang et al., 2012; Chen et al., 2015; Guo et al., 2015). Morphological characters, including the shape of nutritive and reproductive organs, remain highly valuable for plant identification and studies of plant evolution (Ronse De Craene, 2010). Micromorphological characters have been shown to have great value for species identification and systematics (i.e., Kong, 2001; Ren et al., 2003), and these have rarely been considered by previous barcode studies. However, the combination of morphological data and DNA barcodes may be essential for species discrimination, especially in closely related species (Zhang et al., 2012; Li et al., 2016).
Previous molecular phylogenetic studies have included few species from the genus Pulsatilla (Hoot et al., 1994; Hoot, 1995; Ehrendorfer and Samuel, 2001; Schuettpelz et al., 2002; Ehrendorfer et al., 2009; Meyer et al., 2010; Kylem et al., 2010; Hoot et al., 2012; Mlinarec et al., 2012a; Mlinarec et al., 2012b; Jiang et al., 2017). In a recent phylogenetic study of Pulsatilla, few species were from Asia and few individuals were collected for one species (Sramkó et al., 2019). Obtaining DNA barcode data from a dataset created by comprehensive sampling of a taxonomically difficult genus such as Pulsatilla should contribute to understanding the discriminatory potential of barcodes in morphologically complex clades. The establishment of an available barcoding system for Pulsatilla may also facilitate further utilization of these taxa, as well as further research into their taxonomy.
In this study, four DNA barcode regions (rbcL, matK, trnH-psbA, and ITS) were assessed in 19 species (representing three subgenera) of Pulsatilla. Approximately 50% of the accepted species of Pulsatilla found in Europe and the Americas were included, as were 90% of the species found in China (Tamura, 1995; Grey-Wilson, 2014; Wang et al., 2001). Our objectives were to: test the effectiveness of common core DNA barcodes (rbcL+matK) in Pulsatilla, evaluate the resolution of these four barcodes, and use 2- to 4-region combinations to correctly identify individuals. We also aimed to develop a protocol that could effectively discriminate among closely related species, primarily for species discrimination of medicinal plants. In addition, we added micro-morphological analyses of leaf tissue obtained using scanning electronic microscopy (SEM) to reveal the taxonomic relationships among Pulsatilla.
In total, 52 accessions representing 19 Pulsatilla species (including widely used medicinal species) were involved in this study (Table 1). This sample covered each of the three subgenera from Asia, Europe, and America. Nine samples were sourced from herbarium specimens, while 43 samples were newly collected. All samples were taxonomically identified using published floras, monographs, and references. In total, one to five individuals per species were sampled from different populations in the wild. Fresh leaves were dried in silica gel upon collection and the longitude, latitude, and altitude of each collection site (population) were recorded using a GPS unit (Table 1). Voucher specimens were stored in the Herbarium of Northwest A&F University (WUK) and the US National Herbarium (US). Singleton species (species represented by one individual) were only used as potential causes of failed discrimination, and were not included in the calculation of the identification success rate. Three members of Anemone, two of Clematis, one of Anemoclema, and one of Hepatica were selected as outgroups for tree-based analyses.
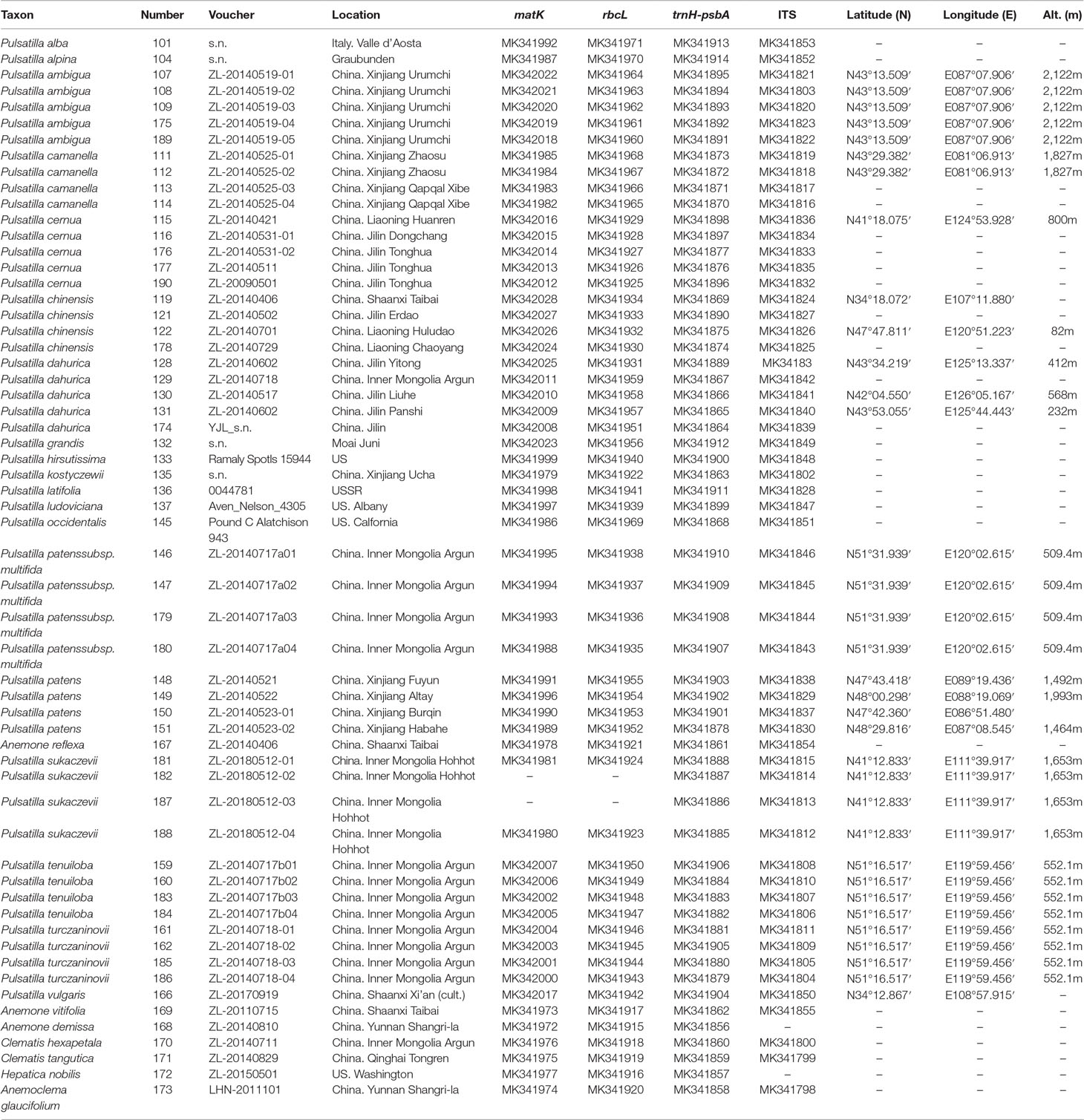
Table 1 Voucher information and GenBank accession numbers for Pulsatilla and outgroups sampled in this study. Classification follows Grey-Wilson (2014) and Tamura (1995). All voucher specimens are deposited in the Northwest A&F University Herbarium (WUK) and the US National Herbarium (US).
Total genomic DNA from freshly collected samples was extracted from approximately 20 mg of silica-dried leaves using a modified cetyltrimethylammonium bromide (CTAB) protocol (Ma and Dai, 2009). For herbarium specimens, we extracted DNA using DNeasy Plant mini kits (QIAGEN, Guangzhou, China). Amplification of DNA regions was performed by standard polymerase chain reaction (PCR). The primer sequences and thermocycling conditions for PCR amplification, are listed in Table 2. PCR reactions were conducted in 25 µl reaction volumes containing 12.5 µl 2 × Taq PCR mix (CWBIO, Xi’an, China), 1.0 µl of each primer (10 µmol/µl), 10.5 µl ddH2O, and 1.0 µl template DNA (30–50 ng). PCR products were run on 1% agarose gels to check whether PCR products showed a clear single band. For those PCR products that did not show a clear single band, the corresponding template DNA was amplified again with two/one pair segmented primers (Table 2, S1, S2). High-quality PCR products wereSequencing was performed from both directions to reduce sequencing error.
All sequence assemblies and adjustments were performed using Geneious v.9.0 (Kearse et al., 2012). Sequences were aligned with MUSCLE (Edgar, 2004). In particular, the number of indel and variable sites events for each dataset was inferred by deletion/insertion polymorphism (DIP) and polymorphic site analyses performed by DnaSP v5 (Librado and Rozas, 2009). To assess the barcoding resolution for all barcodes (rbcL, matK, trnH-psbA, ITS, and combinations of these), three analytical methods were employed. These included the pair-wise genetic distance method (PWG-distance), the sequence similarity method (TAXONDNA), and phylogenetic-based methods (NJ, BI, and ML). Each of these analyses is described in detail below.
For the pair-wise genetic distance-based method, five parameters — i.e. average distance, average interspecific distance, average intraspecific distance, smallest interspecific distance, and largest intraspecific distance — were calculated in MEGA 7 using the Kimura two-parameter distance model (K2P), to explore intra- and inter-species variation (Kumar et al., 2016).
Furthermore, to assess the candidate barcodes for the PWG-distance method, two analytical computations were made; i.e. the barcoding gap between interspecific and intraspecific distances and the local barcoding gap for species resolution. The barcoding gap was used to test for appropriate barcode markers, which show high interspecific but low intraspecific genetic divergence (Han et al., 2016). We graphed the distribution of intra- and inter-specific divergence of each candidate barcode with their combinations to show the barcoding gap. Next, we graphed the local barcoding gap to reveal the species resolution power of candidate barcodes. We considered discrimination to be successful if the smallest interspecific distance, involving more than one individual for one species, was larger than its largest intraspecific distance.
We used the proportion of correct identifications to assess the potential of all markers for accurate species identification with TAXONDNA (Species Identifier 1.8 program). The “Best Match” (which assigns queries to species with the best-matching sequences, regardless of their similarity), and “Best Close Match” (which assigns queries to species if a threshold similarity is met) tests in TAXONDNA were run for all species that were represented by more than one individual (Meier et al., 2006).
To evaluate the species discrimination power of the four single barcoding markers, three different tree-building analyses—i.e. the Neighbor Joining (NJ) tree, Bayesian inference (BI) tree, and Maximum likelihood (ML) analyses—were used. The NJ and ML analyses of all markers were conducted by K2P model using MEGA7. For the BI analysis, best substitution models were selected according to the Akaike information criterion (AIC) by jModeltest version 2.1.7 (Posada and Buckley 2004; Posada, 2008; Darriba et al., 2012). BI trees were conducted in MrBayes v 3.1 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003; Kumar et al., 2016). The Markov chain Monte Carlo (MCMC) analysis was run for 10,000,000 generations. The first 25% of the generations were discarded as burn-in after checking for stationarity and convergence of the chains, and a consensus tree was constructed using the remaining trees. Generally, species forming separate clusters in the tree with bootstrap support >50% were considered to be distinct.
Leaf collection information for Pulsatilla species is shown in Table 1. Leaves from 8 species, which were difficult to identify by barcode, were fixed in FAA (Formalin: acetic acid: ethanol: water = 10:5:50:35). The materials were first dissected and dehydrated in an ethanol and iso-amyl acetate series. Next, they were subjected to critical-point drying in CO2, sputter-coating with gold, and imaged using a HITACHI S-3500 scanning electron microscope (SEM). The backgrounds of SEM images were edited and details were colored using Adobe Photoshop. Photographs of mature leaves were taken with a Nikon D7100 digital camera against a black background. Descriptions of leaf morphology were based on 30 mature leaves.
The characteristics of the four DNA barcoding regions are shown in Table 3. For each of the four DNA barcoding regions (rbcL, matK, trnH-psbA, and ITS), PCR amplification and sequencing using a universal primer pair had a high success rate — i.e. 96.15, 96.15, 100, and 100%, for rbcL, matK, trnH-psbA, and ITS, respectively. It was difficult to obtain target barcodes for herbarium samples and the success rates from these samples were low. We could not get target barcodes for the 80% of the herbarium samples even when segmented primer pairs were used. A total of 232 new sequences were obtained from 52 accessions, which included 57 sequences for each of rbcL and matK, and 59 sequences for each of trnH-psbA and ITS. All sequences were submitted to the NCBI (Table 1).
The aligned lengths of the rbcL, matK, trnH-psbA, and ITS barcode sequences in the dataset were 1,207, 835, 379, and 589 bp. Compared to the other markers, rbcL and matK were the most highly conserved, with lower percentages of variable sites and fewer indels. The lengths of the rbcL and matK sequences were always uniform. However, length variation existed for both the ITS (587–589 bp) and trnH-psbA (299–379 bp) markers. The number of variable sites was the highest for ITS markers (16.98%), followed by trnH-psbA (4.22%), matK (3.71%), and rbcL (1.99%). Eleven indels (1–25bp) were found at the trnH-psbA locus and five were found for ITS (1–2bp), while none were found for matK or rbcL.
rbcL showed the lowest intraspecific and interspecific divergence as measured by the PWG-distance method, while ITS showed the highest interspecific divergence (0.1161), followed by trnH-psbA (0.0381) and matK (0.0207) (Table 3).
We found overlaps for both single markers and combinations of the candidate loci, but we found no distinct barcoding gaps (Figure 2, S1). Among single barcodes, the rbcL marker showed the highest species resolution (48.78%), followed by ITS (44.19%), with matK and trnH-psbA showing the lower species resolution (14.63% and 9.30%, respectively). Of the eleven combinations, rbcL+matK+trnH-psbA (R+M+T) from the chloroplast genome exhibited the best species resolution (70.73%), followed by rbcL+matK (R+M) (68.29%), while trnH-psbA+ITS (T+I) had the lowest species resolution (44.19%) (Figure 3).
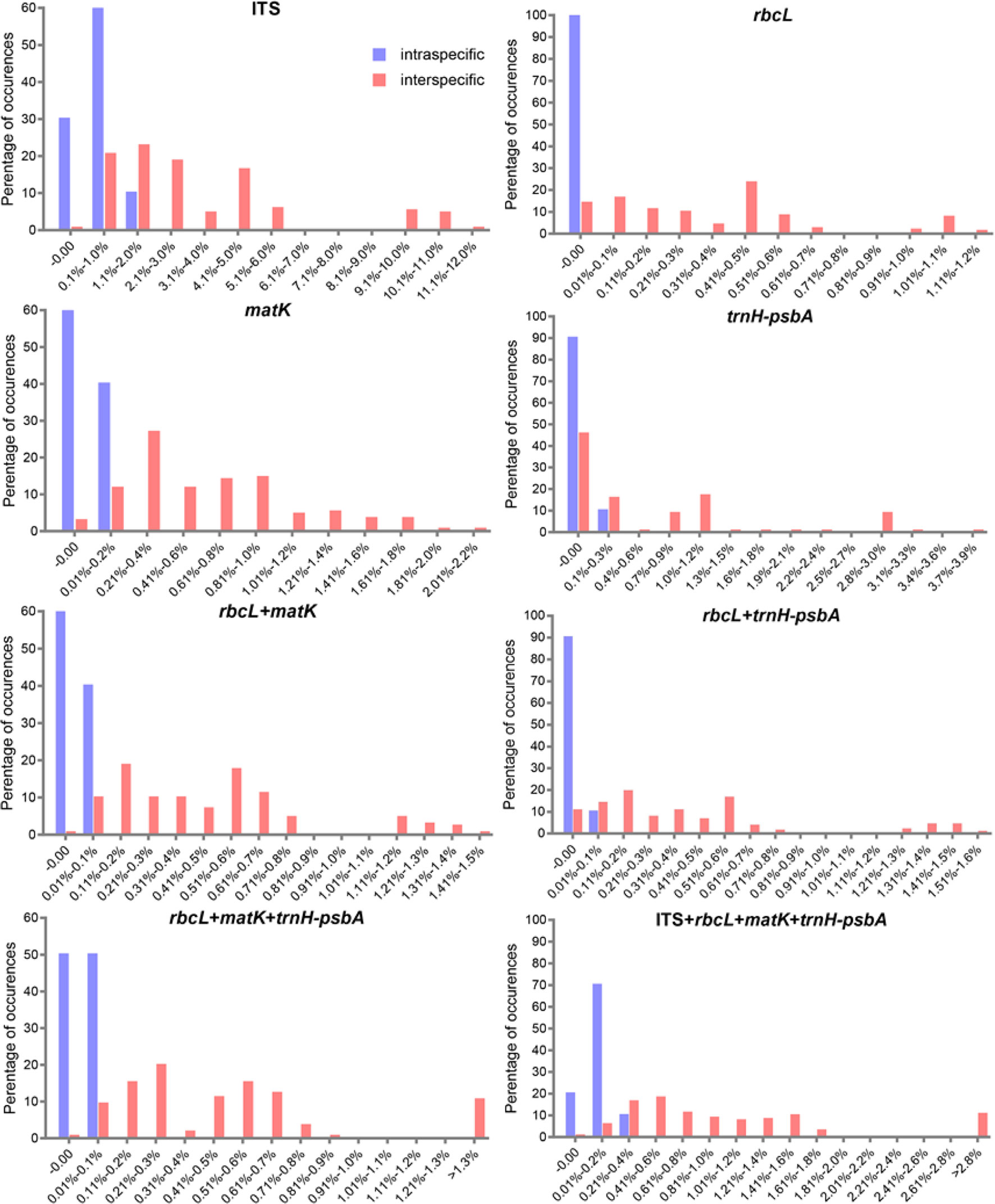
Figure 2 Histograms of the frequencies (y-axes) of pair wise intraspecific (blue bars) and interspecific (red bars) divergences based on the K2P distance (x-axes) for individual rbcL matK, trnH-psbA, and ITS markers and combined markers.
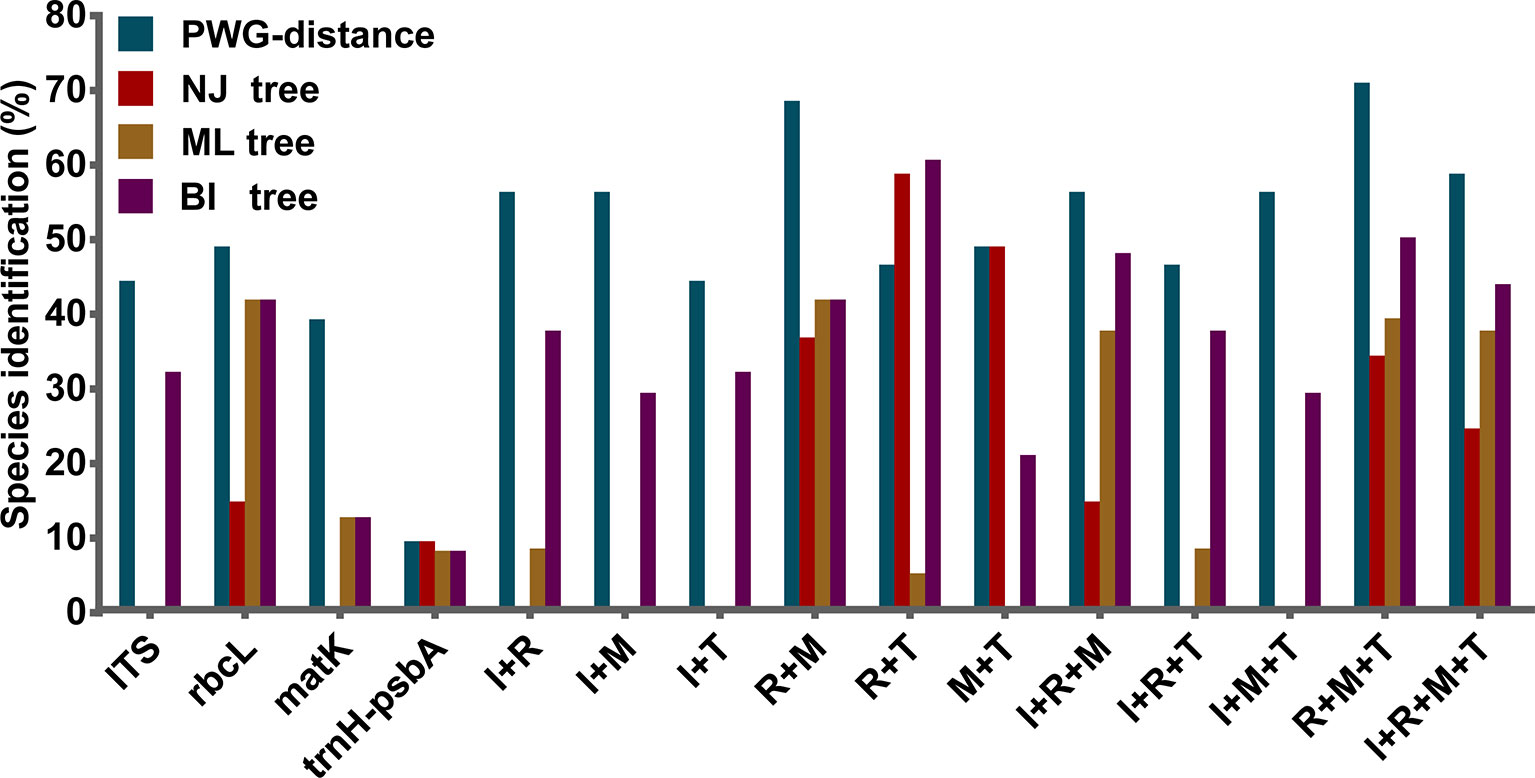
Figure 3 Species discrimination rates of all tested single- and multi-locus barcodes in Pulsatilla, rbcL (R), matK (M), trnH-psbA (T), and ITS (I).
TAXONDNA analyses using the “Best Match” and “Best Close Match” methods exhibited similar discrimination successes (Table 4). Among the four single barcodes, the ITS and trnH-psbA markers had the highest success rate (79.06%) for the correct identification of species, followed by matK (58.53%) and rbcL (48.78%), respectively. Among the eleven combinations, matK+ITS performed the best (83.72%). The rbcL+matK+ITS and rbcL+matK+trnH-psbA+ITS combinations had a success rate that was similar to matK+ITS.
The discrimination power for all markers and their combinations are shown in Table 5. Among the four single barcodes, the rbcL demonstrated the best discrimination power (NJ tree and BI tree: 48.78%), followed by ITS (BI tree: 39.02%), while matK and trnH-psbA both had lower discrimination power. When barcoding loci were combined, the rbcL+ITS combination had the highest resolution power (BI tree: 70.73%, NJ tree and ML tree: 58.54%), followed by rbcL+matK+trnH-psbA(BI tree: 58.54%) and rbcL+matK+ITS (BI tree: 56.10%). The core rbcL+matK combination recommended by COBL had relatively low resolution (NJ tree: 36.59%, ML tree and BI tree: 48.78%), and the rbcL+ITS combination was clearly better than all combinations (i.e. rbcL+matK+trnH-psbA+ITS; NJ tree: 24.39%, ML tree: 49.30%, BI tree:51.22%).
The BI phylogenetic trees based on ITS (left) and chloroplast marker data (right) are shown in Figure 4, respectively. In all phylogenetic analyses, Pulsatilla formed a monophyletic clade with high bootstrap support (PP = 0.97/1.00), and all barcodes could discriminate between subgenera of Pulsatilla.
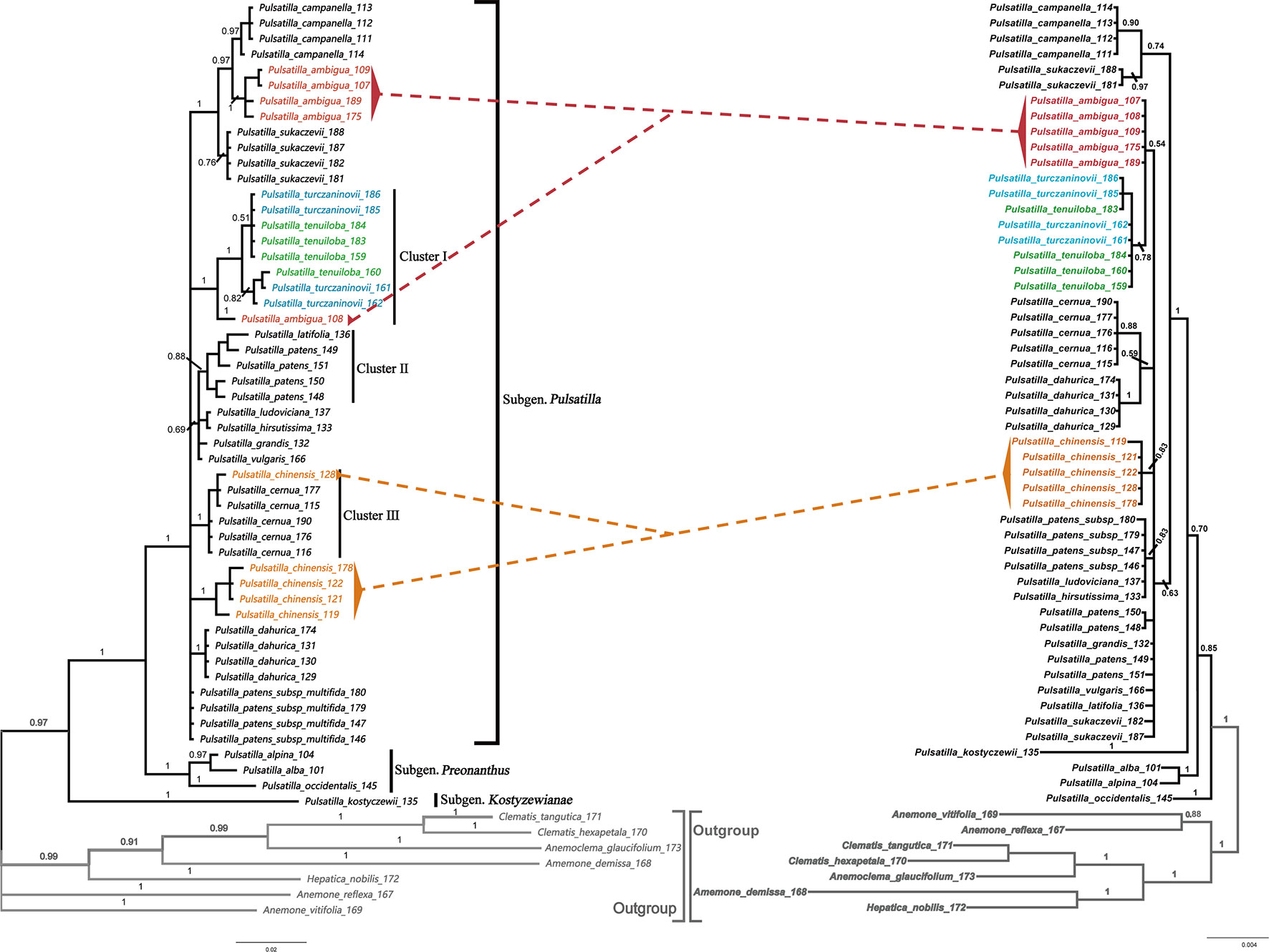
Figure 4 Bayesian inference (BI) trees based on ITS (left) and the combination of rbcL+matK+trnH-psbA sequences (right) in Pulsatilla. Bayesian posterior probabilities are given above branches.
All samples in this study formed three subclades, corresponding to the three subgenera. In subgenus Pulsatilla, some species represented by two or more individuals formed monophyletic groups, e.g. P. camoanella and P. dahurica. However, several groups were mixed with individuals of other species, e.g. P. turczaninovii continually clustered with P. tenuiloba, and they are mixed with one individual from P. ambigua (cluster I); P. latifolia_136 was clustered with samples of P. patens (cluster II); P. chinese_128 was mixed with individuals of P. cernua (Figure 4, cluster III). We also found that P. ludoviciana and P. hirsutissima were mixed with individuals of P. patens subsp. multifida.
We used scanning electronic microscopy to examine the leaf epidermis of eight species: P. ambigua, P. camoanella, P. turczaninovii, P. tenuiloba, P. vulgaris (Figure 5), P. chinensis, P. patens, and P. patens subsp. multifida (Figure 6). All observations were performed by imaging the back of the leaf. We found that the leaf epidermises of P. ambigua (Figures 5A, B), P. camanella (Figures 5E, F), P. chinensis (Figures 6A, B), and P. patens (Figures 6E, F) were pilose, and their trichomes were dense and long. The order of the density and length of trichomes were as follows: P. ambigua > P. patens > P. chinensis > P. camanella. In contrast, the leaf epidermises of P. tenuiloba (Figures 5M, N) was glabrous and those of P. turczaninovii (Figures 5I, J), P. vulgaris (Figures 5Q, R), and P. patens subsp. multifida (Figures 6I, J) showed sparsely short trichomes. The order of the density and length of trichomes were as follows: P. patens subsp. multifida > P. turczaninovii > P. vulgaris > P. tenuiloba.
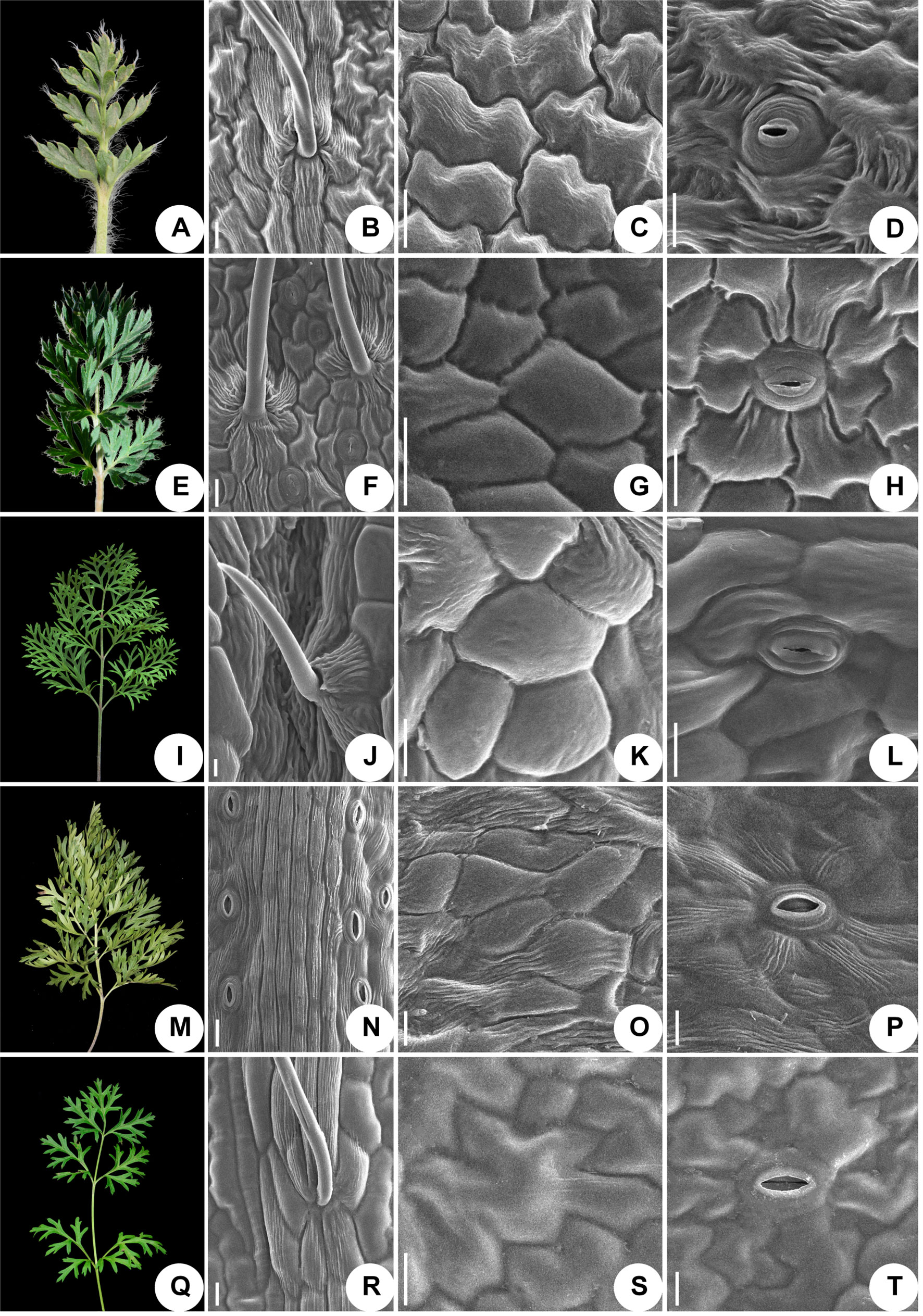
Figure 5 Micromorphological variation of leaf surfaces of Pulsatilla (A–D) P. ambigua; (E–H) P. camoanella; (I–L) P. turczaninovii; (M–P) P. tenuiloba; (Q–T) P. vulgaris. Scale bars: B–D, F–H, J–L, N–P, R–T, 200 µm.
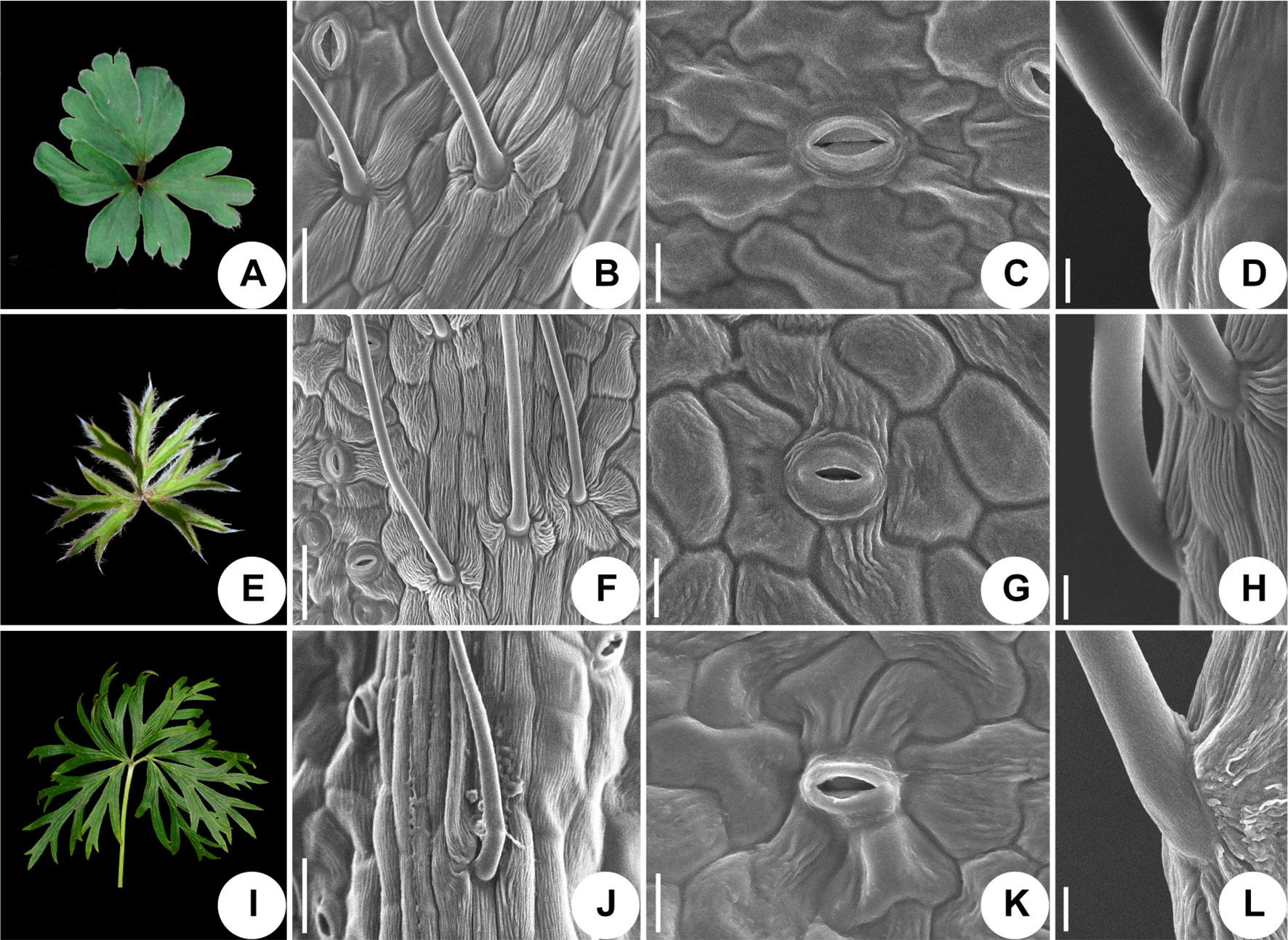
Figure 6 Micromorphological variation of leaf surfaces of Pulsatilla. (A–D) P. chinensis; (E–H) P. patens; (I–L) P. patens subsp. multifida; Scale bars: B, F, J, 0.5 mm; C, G, K, 200 µm; D, H, L, 100 µm.
The epidermal cells in all species were found to have a smooth polygonal shape on the epidermal walls, with the notable exceptions of P. ambigua (Figure 5C) and P. vulgaris (Figure 5S), which were found to have an irregular shape with sinuate anticlinal striation. Different species showed differences in stomatal organ type and cell shape. We found anomocytic (P. ambigua, Figure 5D; P. vulgaris, Figure 5T; P. patens, Figure 6G), actinocytic (P. camanella, Figure 5H; P. tenuiloba, Figure 5P; P. chinensis, Figure 6C; P. patens subsp. multifida, Figure 6K), and diacytic (P. turczaninovii, Figure 5L) stomata respectively.
In this study, the short DNA sequences ITS and trnH-psbA had the best performance in PCR amplification and sequencing among the four barcode markers (quantified by amplification success). Moreover, successful sequencing rates for sequences ITS and trnH-psbA were over 90% for silica-dried samples but lower for herbarium specimens. These findings are consistent with many previous studies (Xu et al., 2015; Yan et al., 2015; Han et al., 2016). In addition, the varying lengths of insertions/deletions (indels) found at the trnH-psbA loci for different species provide important phylogenetic information and species discrimination power (Liu et al., 2011). Thus, sequence alignments of this region must be performed with great care to avoid overestimating substitution events.
The rbcL and matK genes are approximately 1,428 bp and 1,570 bp in length, respectively (Jing et al., 2011; Dong et al., 2012; Dong et al., 2014). The greatest problem with rbcL and matK was that it was difficult to amplify them from the degraded DNA isolated from old herbarium specimens, since the short lengths of remaining fragments hampered the extension phase of the PCR for these longer genes. Although some problems may be alleviated by using additional pairs of primers, the amplification and sequencing success rate of the old herbarium samples remained poor. Thus, we were not able to obtain all sequences for all herbarium samples.
An ideal DNA barcode should be universal, reliable, cost effective, and show considerable discriminatory power. Because none of the proposed single-locus barcodes perfectly meets all these criteria. It is generally necessary to use multi-locus barcodes for land plants (Kress and Erickson, 2007; Fazekas et al., 2008). Multi-locus barcodes can often improve the resolution rate of species identification (CBOL Plant Working Group, 2009;China Plant BOL Group, 2011).
In the present study, when evaluated alone, the species resolutions based on tree-building for the three chloroplast regions rbcL, matK, and trnH-psbA were 48.78, 14.63, and 9.30%, respectively. Low resolution phylogenetic trees made using the chloroplast regions mentioned above have been reported for other taxa, including Curcuma (2.3–7.9%) (Chen et al., 2015) and Sisyrinchium (5.11%–20.41%) (Alves et al., 2014). The inadequate resolution may be due to the lower substitution rates and lack of variation found in single plastid regions. Thus, we do not recommend single plastid regions as DNA barcodes for this the genus.
Among the candidate barcode genes, the Consortium for the Barcode of Life (CBOL) Plant Working Group suggested that rbcL, matK, and the rbcL+matK combination should be sufficient for a plant barcode, and that this combination should be supplemented with additional markers as required (Clegg, 1993; CBOL Plant Working Group, 2009; Hollingsworth et al., 2011; Dong et al., 2014). In addition, Kress et al. (2005) and Chase et al. (2007) proposed that trnH-psbA can be used in two-locus or three-locus barcode systems to improve resolution. For instance, two of the three combinations of the three chloroplast loci tested in this study, rbcL+trnH-psbA (BI tree: 70.73%, PWG: 46.34%) and rbcL+matK+trnH-psbA (PWG: 70.73%, BI tree: 58.54) exhibited higher discriminatory performance than any single marker. Consequently, this highlights the need to use chloroplast multi-locus barcodes (rbcL+matK+trnH-psbA) to improve the resolution of species identification in Pulsatilla.
The nuclear ITS region provided the highest inter-and intraspecific divergences (0.1161 and 0.0116, respectively) and had a higher success rate for the correct identification of species in TAXONDNA (Best match: 79.06%). However, as for the tree-building method, the discriminatory performance of ITS is not satisfactory, as its highest resolution is 39.02% (BI).
As evidenced by previous studies, the multi-locus barcode (rbcL+matK+trnH-psbA+ITS) is one of the combinations that demonstrated the highest species resolution rate, e.g., Aceraceae (90.5%) (Han et al., 2016), Lysimachia (95.5%) (Zhang et al., 2012), Oberonia (62.99%) (Li et al., 2016), Rhodiola (73.01%) (Zhang et al., 2015b) and Schisandraceae (75%) (Zhang et al., 2015a). However, in this study, addition of ITS to different kinds of combinations of chloroplast markers did not increase the resolution rate obviously (Figure 4). The resolution rate based on tree-building analyses was 51.22% for BI and 58.54% for PWG. In addition, we found no distinct barcoding gap. This phenomenon may be due to the one or more of several reasons. First, incomplete lineage sorting and non-homogeneous concerted evolution are likely to occur at the ITS locus (Liu et al., 2011; Liu et al., 2018; Wang et al., 2011; Wirta et al., 2016). Second, the three chloroplast regions (rbcL, matK, and trnH-psbA) cannot compensate for the drawbacks of ITS because they are sourced from a different genome. Although the nuclear genome (and the ITS region) is inherited biparentally, the chloroplast genome is inherited uniparentally. Thus, the chloroplast genome experiences more complete lineage sorting than the ITS locus does. Third, hybridizations may cause conflicts between ITS and chloroplast loci, as well as problematic results in ITS phylogeny due to the possibility of homogenization to paternal copies in some lineages and maternal copies in others.
A combination of DNA markers from different genomes—which have different modes of inheritance and conflicting phylogenies—can hinder our understanding of species delimitation and the evolutionary processes of speciation. Because of its myriad variable sites that can reliably distinguish species, resulting from a high mutation rate and rapid concerted evolution, we recommend ITS as a good single barcode for the genus Pulsatilla.
Phylogenetic identification and species recognition are foundationally important for biology (Moritz, 1994; Wirta et al., 2016). The results of the phylogenetic analyses performed in this study may shed some light on the identification and taxonomy of the genus Pulsatilla (Figure 3). Here, we found that Pulsatilla formed a monophyletic group with high support. Moreover, the three recognized subgenera — i.e. subg. Pulsatilla, subg. Kostyczewianae, and subg. Preonanthus (Grey-Wilson, 2014) — were resolved as distinct monophyletic groups, which is consistent with the recent phylogenetic result (Sramkó et al., 2019).
Within subgenus Pulsatilla, our analyses found that P. camanella and P. ambigua were resolved as sister to one another with high support. These two species share many common morphological characters, such as almost fully expanded leaves at anthesis, and dense, long trichomes. The flowers of both species nod before anthesis (Wang et al., 2001). However, during anthesis, the sepals of P. camanella can easily be distinguished from those of P. ambigua by color (blue-violet vs. dark violet). At the same time, the micro-morphological characters of the leaves are also different (smooth polygonal shape epidermal walls vs. irregular shape with sinuate anticlinal striation). Actinocytic and anomocytic stomata exist in both species, but most stomata in P. camanella are actinocytic, whereas most are anomocytic in P. ambigua. Thus, molecular data as well as micro-morphological characters can distinguish between these two species relatively well. Both types of evidence may be helpful to accurately identify specimens that are damaged or lack sufficient diagnostic characters.
In addition to its use in identifying specimens, DNA barcoding is also useful for resolving taxonomic uncertainty (Burns et al., 2008; Li et al., 2016). Our phylogenetic trees showed that P. turczaninovii always clustered with P. tenuiloba. They did not have distinct barcodes. The micro-morphological characters were also found to be the same, since both plants showed polygonal epidermal cells with striation, a dense distribution of stomata, and glabrous or sparsely short trichomes. In addition, the geographical distribution of these two species overlaps in Inner Mongolia. Taken together, these distinct lines of evidence collectively suggest that P. turczaninovii and P. tenuiloba are the same species.
The discovery of hybridization, introgression, and/or incomplete lineage sorting among species is another useful application of DNA barcoding (Wang et al., 1999, Xiao and Zhu, 2009). The chloroplast region is inherited maternally, but the nuclear genome, including the ITS region, is inherited biparentally (Alvarez and Wendel, 2003). Thus, if there are different results in different phylogenetic analyses from chloroplast and nuclear data, we speculate that these differences may be caused by hybridization and/or introgression among species, which could result in a non-monophyletic clade. In subg. Pulsatilla, we found several complex groups. The samples of P. chinensis and P. cernua in cluster III, were indistinguishable. In the Bayesian inference (BI) tree based on ITS sequences (Figure 3), the samples of P. cernua clustered in a clade along with sample P. chinensis128. However, in the Bayesian inference (BI) tree based on the combination of chloroplast sequences (Figure 3), sample P. chinensis128 clustered in a clade with all other samples of P. chinensis. P. chinensisis a widespread species and has a geographical range that covers that of P. cernua; in addition, sample P. chinensis128 was collected near populations of P. cernua in Jilin Province, China. Hybridization or introgression might have occurred during the speciation of P. chinensisis and P. cernua. A similar situation was also found for sample P. ambigua108 and cluster I (Figure 3), suggesting hybridization may have occurred between P. ambigua and P. tenuiloba/P. turczaninovii.
The resolution of the present study is relatively low compared to the 70% resolution reported by Fazekas et al. (2009) or other plant groups (Zhang et al., 2012; Zhang et al., 2015a; Zhang et al., 2015b; Han et al., 2016). Factors specific to the evolution of the Pulsatilla and/or the sampling strategy of this study may affect the ability to discriminate between species. Such factors include incomplete lineage sorting and hybridization, the rapid radiation of Pulsatilla species, the variation present at the barcode loci, and the sampling density used in this study.
Unlike animal species, many plant species have paraphyletic or polyphyletic origins due to the higher frequency of reticulate evolution, which is facilitated by hybridization and polyploidization (Rieseberg and Brouillet, 1994; Tate and Simpson 2003). Given that this is the case, barcoding based solely on plastid markers may not reliably distinguish species. For example, in our study, some species are resolved to paraphyletic groups, such as P. patens. In these cases, the use of nuclear DNA sequences (e.g. ITS markers) may improve the resolution among plant species because nuclear loci have higher overall synonymous substitution rates, thus making nuclear markers such as ITS more sensitive. In our study, P. patens samples 148, 149, 150, and 151 formed a monophyletic group with sample P. latifolia 136. However, these samples were not clustered together by chloroplast marker data (Figure 3).
DNA barcoding promotes the development of high-resolution phylogenies (Guo et al., 2015). In this study, we selected nuclear ITS and three chloroplast barcodes to evaluate their suitability for use in classifying a comprehensive array of Pulsatilla samples. We found that ITS was the most efficient single-locus barcode, as marker data from this locus was able to identify accurately more than half of all Pulsatilla species. We also found that the combination of rbcL+matK+trnH-psbA was the most efficient multi-locus barcode. However, there is an upper limit to the information provided by the barcodes tested here and adding more fragments may not increase the discrimination power of the DNA barcoding process.
Due to hybridization and/or introgression into the genus Pulsatilla, supplementary use of other identification methods may assist DNA barcoding methods and permit more precise identification (Geißler et al., 2017; Valentini et al., 2017; Liu et al., 2018; Soares et al., 2018). In cases where rbcL and matK are difficult to amplify and/or perform unsatisfactorily, using the whole chloroplast genome as a marker represents a useful alternative to circumvent possible issues with gene deletion and low PCR efficiency (Huang et al., 2005). The idea of using whole chloroplast genomes, termed “super-barcoding”, to identify plant species, was proposed by Kane and Cronk (2008) and encouraged by Li et al. (2015). Recently, this approach has been used in practice in the feather grass genus Hippophae (Krawczyk et al., 2018). Future studies should aim to explore super-barcoding using next generation sequencing; such an approach may offer even more efficient discrimination of closely related species in the genus Pulsatilla. In addition, micro-morphological characters — e.g., pollen and seed grain, leaf and/or stem epidermis — may also provide useful supplementary data for plant identification. Combining molecular and micro-morphological data may also be an advisable strategy in the future.
All the sequences were deposited as GenBank: Accessions MK341798–MK342029.
LiaZ planned and designed the research. Q-JL, XW and J-RW performed experiments and analyzed data, Q-JL analyzed data, and Q-JL, XW, J-RW, NS, LinZ, Y-PM, Z-YC, LiaZ and DP wrote the manuscript.
This project was supported by the Fundamental Research Funds for the Central Universities (No. 2452019068) and the National Nature Science Foundation of China (No. 31770200, 31300158, 31872710).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are very grateful to Dr. Fuzhen Guo, Ningjuan Fan, Guoyun Zhang, and Xiaohua He of Northwest A&F University for assistance with SEM. We sincerely thank Prof. Zhong-hu Li and Prof. Jian-qiang Zhang for help with data analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01196/full#supplementary-material
Figure S1 | Histograms of the frequencies (y-axes) of pair wise intraspecific (blue bars) and interspecific (red bars) divergences based on the K2P distance (x-axes) for combined markers of rbcL matK, trnH-psbA, and ITS.
Alvarez, I., Wendel, J. F. (2003). Ribosomal its sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29, 417–434. doi: 10.1016/s1055-7903(03)00208-2
Alves, T. L. S., Chauveau, O., Eggers, L., de Souza-Chies, T. T. (2014). Species discrimination in Sisyrinchium (Iridaceae): assessment of DNA barcodes in a taxonomically challenging genus. Mol. Ecol. Resour. 14, 324–335. doi: 10.1111/1755-0998.12182
Asmussen, C. B., Chase, M. W. (2001). Coding and noncoding plastid DNA in palm systematics. Amer. J. Bot. 88, 1103–1117. doi: 10.2307/2657094
Burns, J. M., Janzen, D. H., Hajibabaei, M., Hallwachs, W., Hebert, P. D. N. (2008). DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservación Guanacaste, Costa Rica. Proc. Natl. Acad. Sci. U.S.A. 105, 6350–6355. doi: 10.1073/pnas.0712181105
China Plant BOL Group(2011). Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. U.S.A. 108, 19641–19646. doi: 10.1073/pnas.1104551108
CBOL Plant Working Group (2009). A DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A. 106, 12794–12797. doi: 10.1073/pnas.0905845106
Chase, M. W., Cowan, R. S., Hollingsworth, P. M., Berg, C. V. D., Madriñán, S., et al. (2007). A proposal for a standardised protocol to barcode all land plants. Taxon. 56, 295–299. doi: 10.1002/tax.562004
Chen, J., Zhao, J. T., Erickson, D. L., Xia, N. H., Kress, W. J. (2015). Testing DNA barcodes in closely related species of Curcuma (Zingiberaceae) from Myanmar and China. Mol. Ecol. Resour. 15, 337–348. doi: 10.1111/1755-0998.12319
Cho, Y., Mower, J. P., Qiu, Y. L., Palmer, J. D. (2004). Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc. Natl. Acad. Sci. U.S.A. 101, 17741–17746. doi: 10.1073/pnas.0408302101
Cho, Y., Qiu, Y. L., Kuhlman, P., Palmer, J. D. (1998). Explosive invasion of plant mitochondria by a group I intron. Proc. Natl. Acad. Sci. U.S.A. 95, 14244–14249. doi: 10.1073/pnas.95.24.14244
Clegg, M. T. (1993). Chloroplast gene sequences and the study of plant evolution. Proc. Natl. Acad. Sci. U.S.A. 90, 363–367. doi: 10.1073/pnas.90.2.363
Cuénoud, P., Savolainen, V., Chatrou, L. W., Powell, M., Grayer, R. J., et al. (2002). Molecular phylogenetics of caryophyllales based on nuclear 18s rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 89, 132–144. doi: 10.3732/ajb.89.1.132
Darriba, D., Taboada, G. L., Doallo, R., Posada, D. (2012). jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2109
De Salle, R., Egan, M. G., Siddall, M. (2005). The unholy trinity: taxonomy, species delimitation and DNA barcoding. Philos. Trans. R. Soc. Lond. B 360, 1905–1916. doi: 10.1098/rstb.2005.1722
Dong, W. P., Cheng, T., Li, C. H., Xu, C., Long, P., et al. (2014). Discriminating plants using the DNA barcode rbcL b: an appraisal based on a large data set. Mol. Ecol. Resour. 14, 336–343. doi: 10.1111/1755-0998.12185
Dong, W. P., Liu, J., Yu, J., Wang, L., Zhou, S. L. (2012). Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One 7, e35071. doi: 10.1371/journal.pone.0035071
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ehrendorfer, F., Samuel, R. (2001). Contributions to a molecular phylogeny and systematics of Anemone and related genera (Ranunculaceae-Anemoninae). Acta Phytotax. Sin. 39, 293–307
Ehrendorfer, F., Ziman, S. N., König, C., Keener, C. S., Dutton, B. E., et al. (2009). Taxonomic revision, phylogenetics and transcontinental distribution of Anemone section Anemone (Ranunculaceae). Bot. J. Linn. Soc. 160, 312–354. doi: 10.1111/j.1095-8339.2009.00861.x
Fay, M. F., Swensen, S. M., Chase, M. W. (1997). Taxonomic affinities of Medusagyne oppositifolia (Medusagynaceae). Kew Bull. 52, 111–120. doi: 10.2307/4117844
Fazekas, A. J., Burgess, K. S., Kesanakurti, P. R., Graham, S. W., Newmaster, S. G., et al. (2008). Multiple multi locus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS One 3, e2802. doi: 10.1371/journal.pone.0002802
Fazekas, A. J., Kesanakurti, P. R., Burgess, K. S., Percy, D. M., Graham, S. W., et al. (2009). Are plant species inherently harder to discriminate than animal species using DNA barcoding markers? Mol. Ecol. Resour. 9, 130–139. doi: 10.1111/j.1755-0998.2009.02652.x
Geißler, K., Greule, M., Schäfer, U., Hans, J., Geißler, T., et al. (2017). Vanilla authenticity control by DNA barcoding and isotope data aggregation. Flavour Fragr. J. 32, 228–237. doi: 10.1002/ffj.3379
Grey-Wilson, C. (2014). Pasque-flowers. The genus Pulsatilla. Kenning hall: The Charlotte Louise Press.
Guo, Y. Y., Huang, L. Q., Liu, Z. J., Wang, X. Q. (2015). Promise and challenge of DNA barcoding in Venus slipper (Paphiopedilum). PLoS One, 11, e0146880. doi: 10.1371/journal.pone.0146880
Han, Y. W., Duan, D., Ma, X. F., Jia, Y., Liu, Z. L., et al. (2016). Efficient identification of the forest tree species in Aceraceae using DNA barcodes. Front. Plant Sci. 7, 1707. doi: 10.3389/fpls.2016.01707
Hajibabaei, M., Janzen, D. H., Burns, J. M., Hallwachs, W., Hebert, P. D. N. (2006). DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl. Acad. Sci. U.S.A. 103, 968–971. doi: 10.1073/pnas.0510466103
Hebert, P. D. N., Cywinska, A., Ball, S. L., Waard, J. R. (2003a). Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 270, 313–321. doi: 10.1098/rspb.2002.2218
Hebert, P. D. N., Ratnasingham, S., Waard, J. R. D. (2003b). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B 270 (Suppl 1), S96. doi: 10.1098/rsbl.2003.0025
Hebert, P. D. N., Gregory, T. R. (2005). The promise of DNA barcoding for taxonomy. Syst. Biol. 54, 852–859. doi: 10.1080/10635150500354886
Hebert, P. D. N., Stoeckle, M. Y., Zemlak, T. S., Francis, C. M. (2004). Identification of birds through DNA barcodes. PLoS Biol. 2, e312. doi: 10.1371/journal.pbio.0020312
Hollingsworth, P. M., Graham, S. W., Little, D. P. (2011). Choosing and using a plant DNA barcode. PLoS One 6, e19254. doi: 10.1371/journal.pone.0019254
Hoot, S. B., Reznicek, A. A., Palmer, J. D. (1994). Phylogenetic relationshipin Anemone (Ranunculaceae) based on morphology and chloroplast DNA. Syst. Bot. 19, 169–200. doi: 10.2307/2419720
Hoot, S. B. (1995). “Phylogenetic relationships in Anemone (Ranunculaceae) based on DNA restriction site variation and morphology,” in Systematics and evolution of the Ranunculiflorae. Eds. Jensen, U., Kadereit, J. W.. (Vienn: Springer), 295–300. doi: 10.1007/978-3-7091-6612-3_30
Hoot, S. B., Meyer, K. M., Manning, J. C. (2012). Phylogeny and reclassification of Anemone (Ranunculaceae), with an emphasis on austral species. Syst. Bot. 37, 139–152. doi: 10.1600/036364412X616729
Huang, C. Y., Günheit, N., Ahmadinejad, N., Timmis, J. N., Martin, W. (2005). Mutational decay and age of chloroplast and mitochondrial genomes transferred recently to angiosperm nuclear chromosomes. Plant Physiol. 138, 1723–1733. doi: 10.1104/pp.105.060327
Huelsenbeck, J. P., Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jiang, N., Zhou, Z., Yang, J. B., Zhang, S. D., Guan, K. Y., et al. (2017). Phylogenetic reassessment of tribe Anemoneae (Ranunculaceae): non-monophyly of Anemone s.l. revealed by plastid datasets. PLoS One 12, e0174792. doi: 10.1371/journal.pone.0174792
Jing, Y. U., Xue, J. H., Zhou, S. L. (2011). New universal matK, primers for DNA barcoding angiosperms. J. Syst. Evol. 49, 176–181. doi: 10.1111/j.1759-6831.2011.00134.x
Kane, N. C., Cronk, Q. (2008). Botany without borders: barcoding in focus. Mol. Ecol. 17, 5175–5176. doi: 10.1111/j.1365-294x.2008.03972.x
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kerr, K. C. R., Stoeckle, M. Y., Dove, C. J., Weigt, L. A., Francis, C. M., et al. (2007). Comprehensive DNA barcode coverage of North American birds. Mol. Ecol. Notes 7, 535–543. doi: 10.1111/j.1471-8286.2007.01670.x
Kong, H. Z. (2001). Comparative morphology of leaf epidermis in the Chloranthaceae. Bot. J. Linn. Soc. 136, 279–294. doi: 10.1111/j.1095-8339.2001.tb00573.x
Krawczyk, K., Nobis, M., Myszczyński, K., Klichowska, E., Sawicki, J. (2018). Plastid super-barcodes as a tool for species discrimination in feather grasses (Poaceae: Stipa). Sci. Rep. 1924. doi: 10.1038/s41598-018-20399-w
Kress, W. J., Wurdack, K. J., Zimmer, E. A., Weigt, L. A., Janzen, D. H. (2005). Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. U.S.A. 102, 8369–8374. doi: 10.1073/pnas.0503123102
Kress, W. J., Erickson, D. L. (2007). A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One 2, e508. doi: 10.1371/journal.pone.0000508
Kress, W. J. (2017). Plant DNA barcodes: Applications today and in the future. Plant Syst. Evol. 55, 291–307. doi: 10.1111/jse.12254
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kylem, M., Sarab, H., Maryt, A. (2010). Phylogenetic affinities of South American Anemone (Ranunculaceae), including the endemic segregate genera, Barneoudia and Oreithales. Int. J. Plant Sci. 171, 323–331. doi: 10.1086/650153
Li, X. W., Yang, Y., Henry, R. J., Rossetto, M., Wang, Y. T., et al. (2015). Plant DNA barcoding: from gene to genome. Biol. Rev. 90, 157–166. doi: 10.1111/brv.12104
Li, Y. L., Tong, Y., Xing, F. W. (2016). DNA barcoding evaluation and its taxonomic implications in the recently evolved genus Oberonia Lindl. (Orchidaceae) in China. Front. Plant Sci. 7, 1791. doi: 10.3389/fpls.2016.01791
Liu, Y., Xiang, L., Zhang, Y., Lai, X. R., Xiong, C., et al. (2018). DNA barcoding based identification of Hippophae species and authentication of commercial products by high resolution melting analysis. Food Chem. 242, 62–67. doi: 10.1016/j.foodchem.2017.09.040
Liu, J., Möller, M., Gao, L. M., Zhang, D. Q., Li, D. Z. (2011). DNA barcoding for the discrimination of Eurasian yews (Taxus L., Taxaceae) and the discovery of cryptic species. Mol. Ecol. Resour. 11, 89–100. doi: 10.1111/j.1755-0998.2010.02907.x
Librado, P., Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Ma, Y. P., Dai, S. L. (2009). Genomic DNA extraction from chrysanthemum using high salt CTAB method. Biotechnol. Bull. 7, 90–93.
Meier, R., Shiyang, K., Vaidya, G., Ng, P. K. (2006). DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identifications success. Syst. Biol. 55, 715–728. doi: 10.1080/10635150600969864
Meyer, K. M., Hoot, S. B., Arroyo, M. T. K. (2010). Phylogenetic affinities of south American Anemone (Ranunculaceae), including the endemic segregate genera, Barneoudia and Oreithales. Int. J. Plant Sci. 171, 323–331. doi: 10.1086/650153
Mlinarec, J., Šatović, Z., Malenica, N., Ivančićbaće, I., Besendorfer, V. (2012a). Evolution of the tetraploid anemone multifida (2n = 32) and hexaploid A. baldensis (2n = 48) (Ranunculaceae) was accompanied by rDNA loci loss and intergenomic translocation: evidence for their common genome origin. Ann. Bot. 110, 703–712. doi: 10.1093/aob/mcs128
Mlinarec, J., Šatović, Z., Mihelj, D., Malenica, N., Besendorfer, V. (2012b). Cytogenetic and phylogenetic studies of diploid and polyploid members of tribe Anemoninae (Ranunculaceae). Plant Biol. 14, 525–536. doi: 10.1111/j.1438-8677.2011.00519.x
Moritz, C. (1994). Defining ‘Evolutionarily-Significant-Units’ for conservation. Trends Ecol. Evol. 9, 373–375. doi: 10.1016/0169-5347(94)90057-4
Pharmacopoeia (2015). Pharmacopoeia of the People’s Republic of China. Beijing: Chemical Industry Press.
Piredda, R., Simeone, M. C., Attimonelli, M., Bellarosa, R., Schirone, B. (2011). Prospects of barcoding the Italian wild dendroflora: oaks reveal severe limitations to tracking species identity. Mol. Ecol. Resour. 11, 72–83. doi: 10.1111/j.1755-0998.2010.02900.x
Pons, J., Barraclough, T. G., Gomez-Zurita, J., Cardoso, A., Duran, D. P., et al. (2006). Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55, 595–609. doi: 10.1080/10635150600852011
Posada, D., Buckley, T. R. (2004). Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53, 793–808. doi: 10.1080/10635150490522304
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Ren, H., Pan, K. Y., Chen, Z. D., Wang, R. Q. (2003). Structural characters of leaf epidermis and their systematic significance in Vitaceae. Acta Phytotax. Sin. 41, 531–544.
Ren, Y., Chang, H. L., Endress, P. K. (2015). Floral development in Anemoneae (Ranunculaceae). Bot. J. Linn. Soc. 162, 77–100. doi: 10.1111/j.1095-8339.2009.01017.x
Ren, Y., Gu, T. Q., Chang, H. L. (2011). Floral development of Dichocarpum, Thalictrum, and Aquilegia (Thalictroideae, Ranunculaceae). Plant Syst. Evol. 292, 203–213. doi: 10.1007/s00606-010-0399-6
Rieseberg, L. H., Brouillet, L. (1994). Are many plant species paraphyletic? Taxon. 43, 21–32. doi: 10.2307/1223457
Ronquist, F., Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Ronse De Craene, L. P. (2010). Floral diagrams: an aid to understanding flower morphology and evolution. New York: Cambridge University Press. doi: 10.1017/CBO9780511806711
Sang, T., Crawford, D. J., Stuessy, T. F. (1997). Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 84, 1120–1136. doi: 10.2307/2446155
Saitoh, T., Sugita, N., Someya, S., Iwami, Y., Kobayashi, S., et al. (2015). DNA barcoding reveals 24 distinct lineages as cryptic bird species candidates in and around the Japanese Archipelago. Mol. Ecol. Resour. 15, 177–186. doi: 10.1111/1755-0998.12282
Schuettpelz, E., Hoot, S. B., Samuel, R., Ehrendorfer, F. (2002). Multiple origins of southern hemisphere Anemone (Ranunculaceae) based on plastid and nuclear sequence data. Plant Syst. Evol. 231, 143–151. doi: 10.1007/s006060200016
Soares, S., Grazina, L., Costa, J., Amaral, J. S., Oliveira, M. B. P. P., et al. (2018). Botanical authentication of lavender (Lavandula, spp.) honey by a novel DNA-barcoding approach coupled to high resolution melting analysis. Food Control 86, 367–373. doi: 10.1016/j.foodcont.2017.11.046
Sramkó, G., Laczkó, L., Volkova, P. A., Bateman, R. M., Mlinarec, J. (2019). Evolutionary history of the Pasque-flowers (Pulsatilla, Ranunculaceae): Molecular phylogenetics, systematics and rDNA evolution. Mol. Phylogenet. Evol. 135, 45–61. doi: 10.1016/j.ympev.2019.02.015
Stanford, A. M., Harden, R., Parks, C. R. (2000). Phylogeny and biogeography of Juglans (Juglandaceae) based on matK and its sequence data. Am. J. Bot. 87, 872–882. doi: 10.2307/2656895
Suh, S. Y., An, W. G. (2017). Systems pharmacological approach of Pulsatillae radix on treating crohn’s disease. Evid. Based Complement. Alternat. Med. 2017, 1–21. doi: 10.1155/2017/4198035
Tamura, M. (1967). Morphology, ecology and phylogeny of the Ranunculaceae VII. Sci. Rep. Osaka Univ. 16, 21–43.
Tamura, M. (1995). “Ranunculaceae,” in Die natürlichen pflanzenfamilien. Ed. Hiepko, P.. (Berlin: Duncker und Humblot), 17a IV.
Tate, J. A., Simpson, B. B. (2003). Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst. Bot. 28, 723–737. doi: 10.1043/02-64.1
Tavares, E. S., Baker, A. J. (2008). Single mitochondrial gene barcodes reliably identify sister-species in diverse clades of birds. BMC Evol. Biol. 8, 1–14. doi: 10.1186/1471-2148-8-81
Valentini, P., Galimberti, A., Mezzasalma, V., De Mattia, F., Casiraghi, M., et al. (2017). DNA barcoding meets nanotechnology: development of a universal colorimetric test for food authentication. Angew. Chem. 129, 8206–8210. doi: 10.1002/ange.201702120
Vences, M., Thomas, M., Bonett, R. M., Vieites, D. R. (2005). Deciphering amphibian diversity through DNA barcoding: chances and challenges. Philos. Trans. R. Soc. Lond. B 360, 1859–1868. doi: 10.1098/rstb.2005.1717
Wang, J. B., Zhang, W. B., Chen, J. K. (1999). Application of ITS sequences of nuclear rDNA in phylogenetic and evolutionary studies of angiosperms. Acta Phytotax. Sin. 37, 407–416.
Wang, Q., Yu, Q. S., Liu, J. Q. (2011). Are nuclear loci ideal for barcoding plants? A case study of genetic delimitation of two sister species using multiple loci and multiple intraspecific individuals. J. Syst. Evol. 49, 182–188. doi: 10.1111/j.1759-6831.2011.00135.x
Wang, W. C., Fu, D. Z., Li, L. Q., Bruce, B., Anthony, R. B., et al. (2001). Flora of China. Vol. 6. Beijing: Science Press; St. Louis, Mo.: Missouri Botanical Garden Press, 133–438.
Wang, X., Fan, F., Cao, Q. (2016). Modified Pulsatilla decoction attenuates oxazolone-induced colitis in mice through suppression of inflammation and epithelial barrier disruption. Mol. Med. Rep. 14, 1173–1179. doi: 10.3892/mmr.2016.5358
Ward, R. D., Hanner, R., Hebert, P. D. N. (2009). The campaign to DNA barcode all fishes, FISH-BOL. J. Fish Biol. 74, 329–356. doi: 10.1111/j.1095-8649.2008.02080.x
Wirta, H., Várkonyi, G., Rasmussen, C., Kaartinen, R., Schmidt, N. M., et al. (2016). Establishing a community-wide DNA barcode library as a new tool for arctic research. Mol. Ecol. Resour. 16, 809–822. doi: 10.1111/1755-0998.12489
Xiang, X. G., Hu, H., Wang, W., Jin, X. H. (2011). DNA barcoding of the recently evolved genus Holcoglossum (Orchidaceae: Aeridinae): a test of DNA barcode candidates. Mol. Ecol. Resour. 11, 1012–1021. doi: 10.1111/j.1755-0998.2011.03044.x
Xiao, L. Q., Zhu, H. (2009). Incomplete nrDNA ITS concerted evolution in plants and its evolutionary implications. Acta Bot. Boreal. Occident. Sin. 29, 1708–1713.
Xu, Q. M., Shu, Z., He, W. J., Chen, L. Y., Yang, S. L., et al. (2012). Antitumor activity of Pulsatilla chinensis (Bunge) regel saponins in human liver tumor 7402 cells in vitro and in vivo. Phytomedicine 19, 293–300. doi: 10.1016/j.phymed.2011.08.066
Xu, S. Z., Li, D. Z., Li, J. W., Xiang, X. G., Jin, W. T., et al. (2015). Evaluation of the DNA barcodes in Dendrobium (Orchidaceae) from Mainland Asia. PLoS One 10, e0115168. doi: 10.1371/journal.pone.0115168
Yan, L. J., Liu, J., Möller, M., Zhang, L., Zhang, X. M., et al. (2015). DNA barcoding of Rhododendron (Ericaceae), the largest Chinese plant genus in biodiversity hotspots of the Himalaya-Hengduan Mountains. Mol. Ecol. Resour. 15, 932–944. doi: 10.1111/1755-0998.12353
Yancy, H. F., Zemlark, T. S., Mason, J. A., Washington, J. D., Tenge, B. J., et al. (2008). Potential use of DNA barcodes in regulatory science: applications of the regulatory fish encyclopedia. J. Food Prot. 71, 210–217. doi: 10.4315/0362-028X-71.1.210
Yoo, H. S., Eah, J. Y., Kim, J. S., Kim, Y. J., Min, M. S., et al. (2006). DNA barcoding Korean birds. Mol. Cells. 22, 323–327. doi: 10.1007/s10059-013-3151-6
Zhang, C. Y., Wang, F. Y., Yan, H. F., Hao, G., Hu, C. M., et al. (2012). Testing DNA barcoding in closely related groups of Lysimachia L. (Myrsinaceae). Mol. Ecol. Resour. 12, 98–108. doi: 10.1111/j.1755-0998.2011.03076.x
Zhang, J., Chen, M., Dong, X. Y., Lin, R. Z., Fan, J. H., et al. (2015a). Evaluation of four commonly used DNA barcoding loci for Chinese medicinal plants of the family Schisandraceae. PLoS One 10, e0125574. doi: 10.1371/journal.pone.0125574
Keywords: barcoding markers, ITS, pulsatilla, ranunculaceae, species identification
Citation: Li Q-j, Wang X, Wang J-r, Su N, Zhang L, Ma Y-p, Chang Z-y, Zhao L and Potter D (2019) Efficient Identification of Pulsatilla (Ranunculaceae) Using DNA Barcodes and Micro-Morphological Characters. Front. Plant Sci. 10:1196. doi: 10.3389/fpls.2019.01196
Received: 06 July 2019; Accepted: 30 August 2019;
Published: 09 October 2019.
Edited by:
Nina Rønsted, National Tropical Botanical Garden, United StatesReviewed by:
Eduardo Cires, Universidad de Oviedo, SpainCopyright © 2019 Li, Wang, Wang, Su, Zhang, Ma, Chang, Zhao and Potter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Zhao, YmlvbG9neV96aGFvbGlhbmdAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.