- 1The Flowering Lab, School of Biological Sciences, University of Auckland, Auckland, New Zealand
- 2Noble Research Institute, Ardmore, OK, United States
Optimizing flowering time is crucial for maximizing crop productivity, but gaps remain in the knowledge of the mechanisms underpinning temperate legume flowering. Medicago, like winter annual Arabidopsis, accelerates flowering after exposure to extended cold (vernalization, V) followed by long-day (LD) photoperiods. In Arabidopsis, photoperiodic flowering is triggered through CO, a photoperiodic switch that directly activates the FT gene encoding a mobile florigen and potent activator of flowering. In Arabidopsis, several CYCLING DOF FACTORs (CDFs), including AtCDF1, act redundantly to repress CO and thus FT expression, until their removal in LD by a blue-light-induced F-BOX1/GIGANTEA (FKF1/GI) complex. Medicago possesses a homolog of FT, MtFTa1, which acts as a strong activator of flowering. However, the regulation of MtFTa1 does not appear to involve a CO-like gene. Nevertheless, work in pea suggests that CDFs may still regulate flowering time in temperate legumes. Here, we analyze the function of Medicago MtCDF genes with a focus on MtCDFd1_1 in flowering time and development. MtCDFd1_1 causes strong delays to flowering when overexpressed in Arabidopsis and shows a cyclical diurnal expression in Medicago with peak expression at dawn, consistent with AtCDF genes like AtCDF1. However, MtCDFd1_1 lacks predicted GI or FKF1 binding domains, indicating possible differences in its regulation from AtCDF1. In Arabidopsis, CDFs act in a redundant manner, and the same is likely true of temperate legumes as no flowering time phenotypes were observed when MtCDFd1_1 or other MtCDFs were knocked out in Medicago Tnt1 lines. Nevertheless, overexpression of MtCDFd1_1 in Medicago plants resulted in late flowering relative to wild type in inductive vernalized long-day (VLD) conditions, but not in vernalized short days (VSDs), rendering them day neutral. Expression of MtCO-like genes was not affected in the transgenic lines, but LD-induced genes MtFTa1, MtFTb1, MtFTb2, and MtSOC1a showed reduced expression. Plants carrying both the Mtfta1 mutation and 35S:MtCDFd1_1 flowered no later than the Mtfta1 plants. This indicates that 35S:MtCDFd1_1 likely influences flowering in VLD via repressive effects on MtFTa1 expression. Overall, our study implicates MtCDF genes in photoperiodic regulation in Medicago by working redundantly to repress FT-like genes, particularly MtFTa1, but in a CO-independent manner, indicating differences from the Arabidopsis model.
Introduction
Plants integrate several molecular pathways to control when they flower to maximize reproductive fitness and successful development of seeds and fruit (Fornara et al., 2010; Srikanth and Schmid, 2011; Andrés and Coupland, 2012). One of these pathways involves the responsiveness to changes in day length (photoperiod), which plays a vital role in the plant’s ability to synchronize flowering time with favorable seasonal conditions (Putterill et al., 2004). For example, in temperate plants such as winter annual Arabidopsis thaliana (Arabidopsis) and the legume Medicago truncatula (Medicago), extended winter cold (vernalization, V) followed by exposure to long-day (LD) photoperiods—a feature of spring and early summer—promotes flowering.
The well-characterized Arabidopsis LD pathway promotes flowering via the accumulation of CONSTANS (CO) protein in the leaves, which directly activates the expression of the potent floral activator FLOWERING LOCUS T (FT) in the late afternoon of LD, but not in short days (SDs). FT encodes a mobile florigen that moves to the shoot apical meristem and initiates the transition to flowering via activation of genes such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1; Andrés and Coupland, 2012). Several factors converge to facilitate the accumulation of CO protein in LD including releasing the CO gene from transcriptional repression by CYCLING DOF FACTOR (CDF) transcription factors. This occurs via the light-induced formation of the FLAVIN-BINDING, KELCH REPEAT, F-BOX1/GIGANTEA (FKF1/GI) complex which targets the CDFs for degradation via the proteasome, which in turn enables the transcription of CO (Imaizumi et al., 2005; Sawa et al., 2007; Fornara et al., 2009; Song et al., 2012; Goralogia et al., 2017). In addition, there is direct regulation of FT by AtCDF1 (Song et al., 2012).
The acceleration of flowering by FT-like genes is conserved in a diverse range of species (Wickland and Hanzawa, 2015; Putterill and Varkonyi-Gasic, 2016) including the FTa1 gene in the temperate legumes Pisum sativum (pea) and Medicago (Hecht et al., 2011; Laurie et al., 2011). Temperate legumes are of particular interest as many serve as important agricultural crops with flowering time playing a significant role in annual production yields (Graham and Vance, 2003; Weller and Ortega, 2015).
However, increasing evidence suggests that temperate legume species operate with a CO-independent mechanism for the regulation of FT-like genes and thus flowering (Putterill et al., 2013; Weller and Ortega, 2015). Analysis of Medicago CO-like (COL) genes revealed that they were unable to complement the Arabidopsis co null mutant and did not promote flowering when overexpressed (Wong et al., 2014). Medicago col null mutant lines did not have a flowering phenotype under LD and therefore were unlikely to be involved in the Medicago photoperiodic response (Wong et al., 2014). An additional difference is that there are three LD-induced FT genes in Medicago, but none have the same diurnal pattern of expression as Arabidopsis FT, suggesting a different regulatory mechanism (Laurie et al., 2011). Thus, there is a substantial knowledge gap in our understanding of photoperiodic flowering in these species (Hecht et al., 2005, Hecht et al., 2011; Laurie et al., 2011; Putterill et al., 2013; Weller and Ortega, 2015; Ridge et al., 2016).
Despite the apparent lack of a functional CO in temperate legumes, legume CDFs appear to still participate in photoperiodic flowering. Specifically in garden pea, the dominant late-flowering LATE2 mutant was recently mapped to a CDF homolog, PsCDFc1. Yeast two-hybrid assays indicate that the mutation disrupts the binding of PsFKF1 to PsCDFc1, indicating that increased PsCDFc1 protein stability may be the basis of the dominant phenotype (Ridge et al., 2016). Plants carrying the late2/Pscdfc1 mutation have reduced expression of LD-induced FT-like genes, but not PsCOL genes. This indicates that CDFs participate in the photoperiodic regulation of flowering in pea but that the mechanism differs to that of Arabidopsis (Ridge et al., 2016).
CDFs were first characterized in Arabidopsis and are a subset of the plant-specific DNA-binding One Zinc Finger (DOF) gene family of transcription factors (Yanagisawa, 2002; Noguero et al., 2013). They are distinguished by their cyclical diurnal transcript levels, with the majority of genes showing peak transcript accumulation early in the day. In Arabidopsis, CDFs have an overlapping role in photoperiodic flowering control as single AtCDF mutants have either no or only weak flowering time phenotypes, but a quadruple Atcdf1–3,5 mutant has day-neutral early flowering (Imaizumi et al., 2005; Fornara et al., 2009).
In Medicago, phylogenetic analysis has revealed a total of 42 Medicago DOF proteins clustered into four phylogenetic clades (Shu et al., 2015). One of these clades, MCOGD, contains all of the 13 MtCDF-like proteins, which in turn group into several subclades (Shu et al., 2015; Ridge et al., 2016). These are expressed predominantly in leaf blades, nodules, and buds (Shu et al., 2015), with expression in leaves consistent with a role in photoperiodic flowering (Turck et al., 2008).
Here, we analyze the function of MtCDF genes in the regulation of Medicago flowering, with a focus on MtCDFd1_1. We analyzed the gene expression patterns of MtCDFs in VLD and VSD RNA-Seq morning time courses and surveyed plants carrying transposon insertions in MtCDF genes. While flowering time phenotypes were not observed in individual Medicago mutants, overexpressing the genes in Arabidopsis identified five genes, including MtCDFd1_1, which caused strong delays to flowering. We then examined the effect of overexpressing MtCDFd1_1 in Medicago on plant development, flowering time, and the expression of known flowering time genes. Collectively, our results implicate MtCDF genes as regulators of photoperiodic flowering and plant architecture via the repression of FT-like genes, such as MtFTa1.
Materials And Methods
Bioinformatics
Legume and other plant CDF protein sequences were obtained from the literature (Shu et al., 2015; Ridge et al., 2016) and by BLASTP searches with AtCDF1 of the J. Craig Venter Institute (JCVI) Medicago genome (Mt4.0 http://www.jcvi.org/medicago/) and National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/). The Medicago MtCDF gene identifiers and names are listed in Table S1. The phylogenetic tree of CDF-like proteins from Medicago, other legumes, tomato, potato, and Arabidopsis was constructed by aligning full-length amino acid sequences using MUSCLE (version 3.8.425; Edgar, 2004) as implemented in (version 11.1.5) and using the neighbor-joining algorithm implemented in PAUP* (version 4.0; Swofford, 2003). An existing RNA-Seq dataset (Thomson et al., 2019) comprising three biological replicates was consulted to obtain the mean abundance of MtCDF-like gene transcripts in leaf tissue at 0, 2, and 4 h after dawn in transcripts per million (TPMs) in SD and LD photoperiods. Medicago Tnt1 retroelement insertion lines were identified by screening the FST database (https://medicago-mutant.noble.org/mutant/blast/blast.php) and are listed in Table S1.
Plant Materials and Growth Conditions
Medicago truncatula (Medicago) wild type Jester (Hill, 2000) and R1 08-1_C3 (R108; Trinh et al., 1998) used in this study belong to Medicago truncatula Gaertn (barrel medic), ssp. truncatula and ssp. tricycla, respectively. All Tnt1 insertion mutants in the R108 background listed in Table S1 were obtained from the Noble Research Institute, LLC (Ardmore, OK, USA). Arabidopsis thaliana (Arabidopsis) wild type Columbia was used. The Mtfta1 mutant utilized was NF1634 (Jaudal et al., 2019).
Medicago and Arabidopsis plants were grown in controlled environments under ∼200 μM m−2 s−1 cool white fluorescent light at 22°C or 24°C and under ∼140 μM m−2 s−1 at 22°C, respectively, in LDs (16 h light/8 h dark) or SDs (8 h light/16 h dark), with or without prior vernalization of germinated seeds at 4°C for 21 days, as previously described (Laurie et al., 2011; Yeoh et al., 2013; Jaudal et al., 2015). Medicago flowering time was measured in days to when the first floral bud was observed by eye and the number of nodes on the primary axis at flowering. Arabidopsis flowering time was measured in days to when the first floral buds were observed by eye and the total number of rosette and cauline leaves at flowering.
CaMV 35S overexpression constructs were made by inserting complementary DNA (cDNA) sequences into vector pB2GW7 (Karimi et al., 2007) using Gateway® Technology (Invitrogen®, CA, USA). Forward and reverse primers used for Gateway cloning are shown in Table S2. Transgenic Arabidopsis plants overexpressing MtCDF genes were generated using Agrobacterium tumefaciens GV3101 containing overexpression constructs via floral dipping and Basta selection of the T1 population as previously described (Martinez-Trujillo et al., 2004; Jaudal et al., 2015).
Transgenic R108 Medicago plants overexpressing MtCDFd1_1 were generated using A. tumefaciens EHA105 with the 35S:MtCDFd1_1 construct via somatic embryogenesis and subsequent BASTA selection in soil as previously described (Cosson et al., 2006; Laurie et al., 2011).
35S:MtCDFd1_1 plants and Mtfta1 heterozygous plants were crossed together (Chabaud et al., 2006) and then bred and genotyped to identify F235S:MtCDFd1_1/Mtfta1 homozygous mutant plants.
Quantitative Reverse Transcriptase PCR (qRT-PCR) for Gene Expression
RNA extraction and cDNA synthesis using an oligo dT primer was carried out as previously described (Laurie et al., 2011; Yeoh et al., 2013; Jaudal et al., 2015). qRT-PCR was performed using SYBR® green chemistry on Applied Biosystems® 7900HT Sequence Detection System (Applied Biosystems®, CA, USA) or QuantStudio™ 5 Real-Time PCR System (Applied Biosystems®, CA, USA). Each data point is derived from three biological replicates harvested in parallel. Each replicate consisted of a pool of leaf tissue from either two or three independent plants. Primer sequences used for qRT-PCR are listed in Table S2. Gene expression was calculated using the comparative Ct method (Livak and Schmittgen, 2001) with modifications (Bookout and Mangelsdorf, 2003). Samples were normalized to PROTEIN PHOSPHATASE 2A (PP2A; Medtr6g084690).
The statistical testing for the gene expression data presented in Figures 4 and 6 was performed using a one-way analysis of variance (ANOVA) test between the means (α = 0.05). The Shapiro–Wilk normality assumption test was performed on all data presented. Multiple pairwise comparisons adjusted for false discovery rate (FDR) were utilized to highlight statistically significant differences in the data presented.
Yeast Two-Hybrid Assays
Full-length coding sequences of MtCDFd1_1, MtCDFc1, and AtCDF1 and the KELCH-repeat region of AtFKF1 (amino acids 284 to 619; Imaizumi et al., 2005; Ridge et al., 2016) were used for the yeast two-hybrid assay. Gene fragments were cloned into Invitrogen destination vectors pDEST22 (AD, prey) and pDEST32 (BD, bait). The prey and bait constructs were transformed into the haploid yeast strains PJ69-4A and PJ69-4α (James et al., 1996), respectively, and selected on synthetic defined (SD) medium lacking tryptophan (Trp; prey) or leucine (Leu; bait). PJ69-4A and PJ69-4α strains were then mated, and diploid clones with both constructs were selected on medium lacking Trp and Leu (SD −Trp −Leu). Haploids containing empty pDEST22 and pDEST32 were also included to test autoactivation. Two independent diploid clones from each mating were diluted in 100 µl of water and plated on nonselective medium (SD −Trp −Leu) and selective medium [SD −Trp −Leu −histidine (His)] with different 3-amino-1,2,4-triazol (3-AT) concentrations (0, 1, 2, 10, 25, 50, and 100 mM). Colonies developed over 11 days at 28°C. Photos were taken on days 4, 7, 9, and 11. Similar results were obtained for each of the two independent clones. The positive control interactors were AtCDF1 and AtFKF1.
Results
Initial Characterization of 14 MtCDF Genes
To investigate the role of MtCDF genes in Medicago flowering time, we selected 14 MtCDF genes for initial analysis. These were 13 MtCDFs identified previously (Shu et al., 2015; Ridge et al., 2016) and a 14th related gene (MtCDF1) that we previously observed to have cyclical diurnal expression with an afternoon peak (Thomson et al., 2019). Table S1 lists the MtCDF gene identifiers (JCVI Medicago genome Mt4.0) and corresponding gene names following the nomenclature in Ridge et al. (2016). The phylogenetic groupings of the predicted proteins along with AtCDFs are shown in Figure 1A, with a more comprehensive phylogenetic tree containing additional CDF proteins from legumes, tomato, and potato shown in Figure S1.
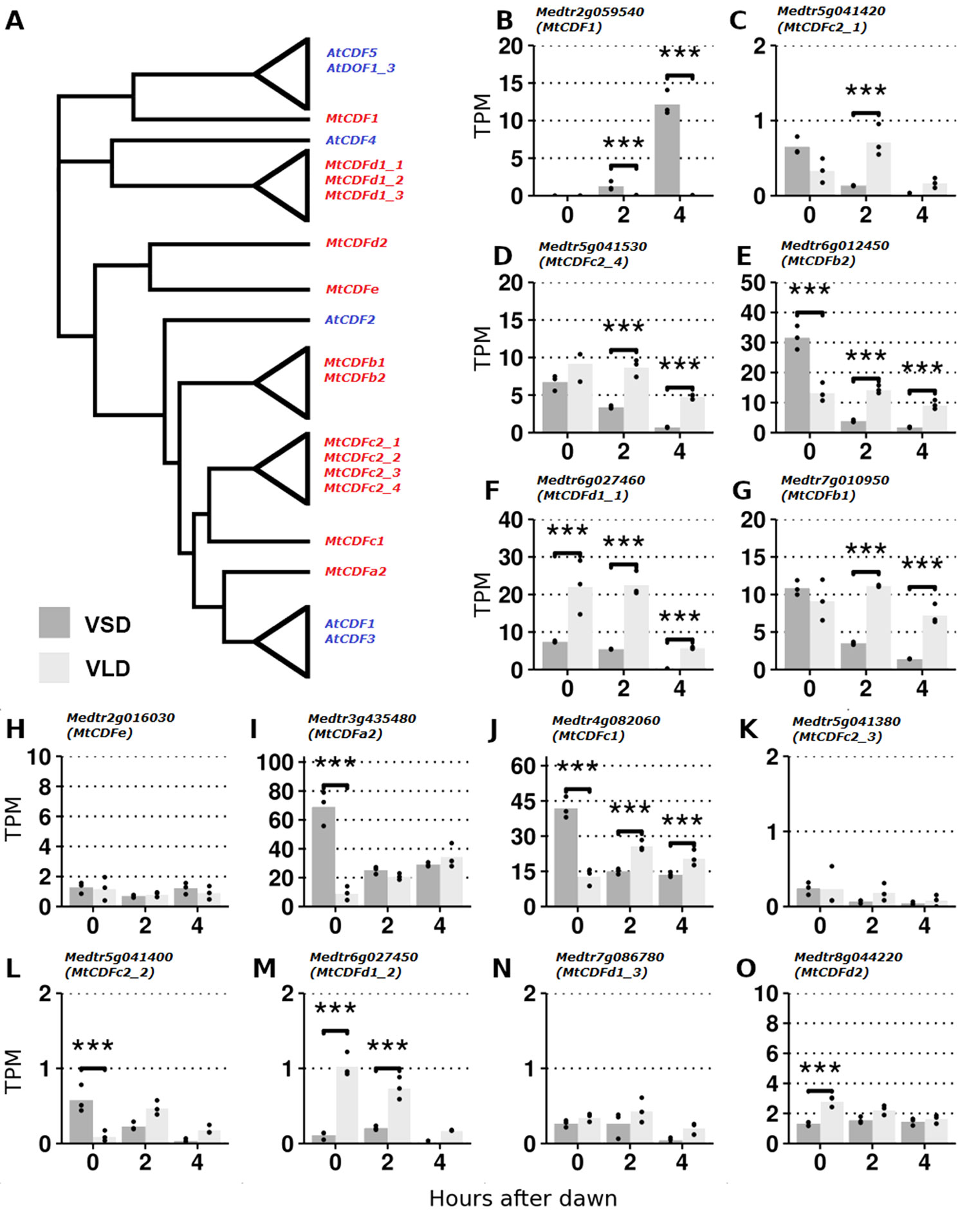
Figure 1 RNA-Seq analysis of MtCDF gene expression in Medicago Jester leaf tissue under vernalized short-day (VSD) and vernalized long-day (VLD) photoperiods. (A) Neighbor joining tree diagram of CDF-like proteins in Arabidopsis and Medicago using their full-length amino acid sequences. Clades of similar proteins were collapsed. See Figure S1 for a more comprehensive tree. (B–O) Derived from RNA-Seq data (Thomson et al., 2019); the mean abundance of MtCDF gene transcripts in leaf tissue at 0, 2, and 4 h after dawn in transcripts per million (TPMs) in VSD and VLD. Abundances for the three biological replicates are plotted as points, and asterisks indicate significant differential expression (Wald significance tests; α = 0.05).
Protein sequence alignments of Medicago and Arabidopsis CDFs (Figure S2) highlighted the highly conserved DOF domain in all the MtCDF proteins and MtCDF1. However, five proteins (MtCDF1, MtCDFd1_1, MtCDFd1_2, MtCDFd1_3, and MtCDFe) and two Arabidopsis CDFs (AtCOG1 and AtCDF4) lacked the two C-terminal regions that in Arabidopsis function as FKF1- and GI-binding domains. Two MtCDFs (MtCDF1 and MtCDFe) also lacked the predicted N-terminal TOPLESS (TPL)-binding domain. Recently, CDFs in Arabidopsis have been shown to form a complex with TPL (Goralogia et al., 2017); hence, the lack of TPL domains in these MtCDFs may indicate a functional divergence.
We analyzed expression of the 14 MtCDF genes (Figure 1) in an RNA-Seq dataset (Thomson et al., 2019) derived from leaves of plants grown in LD and SD after vernalization (V) and harvested at three time points: dawn and 2 and 4 h after dawn. We detected reads mapping to all 14 MtCDF genes, confirming that they are expressed in leaves as previously observed (Shu et al., 2015) and consistent with a potential role in photoperiodic flowering.
Transcript abundance varied >70-fold between the genes (Figures 1B–O). The four most abundant were MtCDFa2, MtCDFc1, MtCDFb2, and MtCDFd1_1. Most genes (11/14; MtCDF1, MtCDFa2, MtCDFb1, MtCDFb2, MtCDFc1, MtCDFc2_1, MtCDFc2_2, MtCDFc2_4, MtCDFd1_1, MtCDFd1_2, and MtCDFd2) were significantly differentially expressed between the two photoperiods. These included three genes, MtCDFd1_1, MtCDFb2, and MtCDFc1, that were differentially expressed between the photoperiods at all three time points.
Further analysis of MtCDFd1_1 by qRT-PCR over a full day (Figure S3), indicated that the transcript of this gene has a diurnal cycle that is modulated by LD and SD photoperiods similar to the Arabidopsis CDFs (AtCDF1-3,5; Imaizumi et al., 2005; Fornara et al., 2009).
No Altered Flowering Time Phenotypes Were Observed in Medicago MtCDF Tnt1 Insertion Lines
To investigate the function of the MtCDF genes, we screened the Medicago Tnt1 flanking sequence database for candidate mutant Medicago plant lines with knockout Tnt1 retroelement insertions in MtCDF genes. The results are summarized in Table S1. Lines homozygous for Tnt1 insertions in 13 out of the 14 genes (the exception was MtCDFd2) were found.
In total, we identified 27 candidate plant lines, genotyped them for the presence of the Tnt1 insertion, examined their gene expression, and scored their flowering time in VLD, LD, and VSD. Knockout, or knockdown, of gene expression was confirmed by qRT-PCR in 11/13 homozygous lines, except MtCDFb1 and MtCDFb2, where the insertions were located in introns. However, no altered flowering time phenotypes were observed in any single mutant, which may be attributable to functional redundancy between some of the genes, as observed in Arabidopsis (Fornara et al., 2009).
Overexpression of MtCDFd1_1 and Four Other MtCDF Genes Causes Delayed Flowering in Arabidopsis
In previous work, overexpression of AtCDF genes, including AtCDF1, caused delayed Arabidopsis flowering (Imaizumi et al., 2005; Fornara et al., 2009). On the other hand, overexpression of wild-type pea PsCDFc1 in Arabidopsis did not give late-flowering transgenic plants (Ridge et al., 2016). Only overexpression of the mutant version of PsCDFc1 from the late2 mutant resulted in late-flowering Arabidopsis plants (Ridge et al., 2016).
Here, having not observed mutant phenotypes in Medicago MtCDF knockout lines (Table S1), we turned to Arabidopsis to use as a rapid heterologous system for testing if any of the MtCDFs might regulate Arabidopsis flowering time. If such MtCDF genes were to be identified in this screen, then one would be selected for the overexpression functional analysis in Medicago.
We constitutively expressed 11 genes (MtCDF1, MtCDFa2, MtCDFb1, MtCDFb2, MtCDFc1, MtCDFc2_1, MtCDFc2_4, MtCDFd1_1, MtCDFd1_2, MtCDFd1_3, and MtCDFe) from across different subclades in wild-type Arabidopsis and measured flowering time (Figure 1A and Figure S1). Expression constructs were made by fusing the MtCDFs to the 35S promoter and then introduced into wild-type Columbia plants with the flowering time of T1Arabidopsis transformants and photographs of selected T2 and T3 progeny presented in Figure 2.
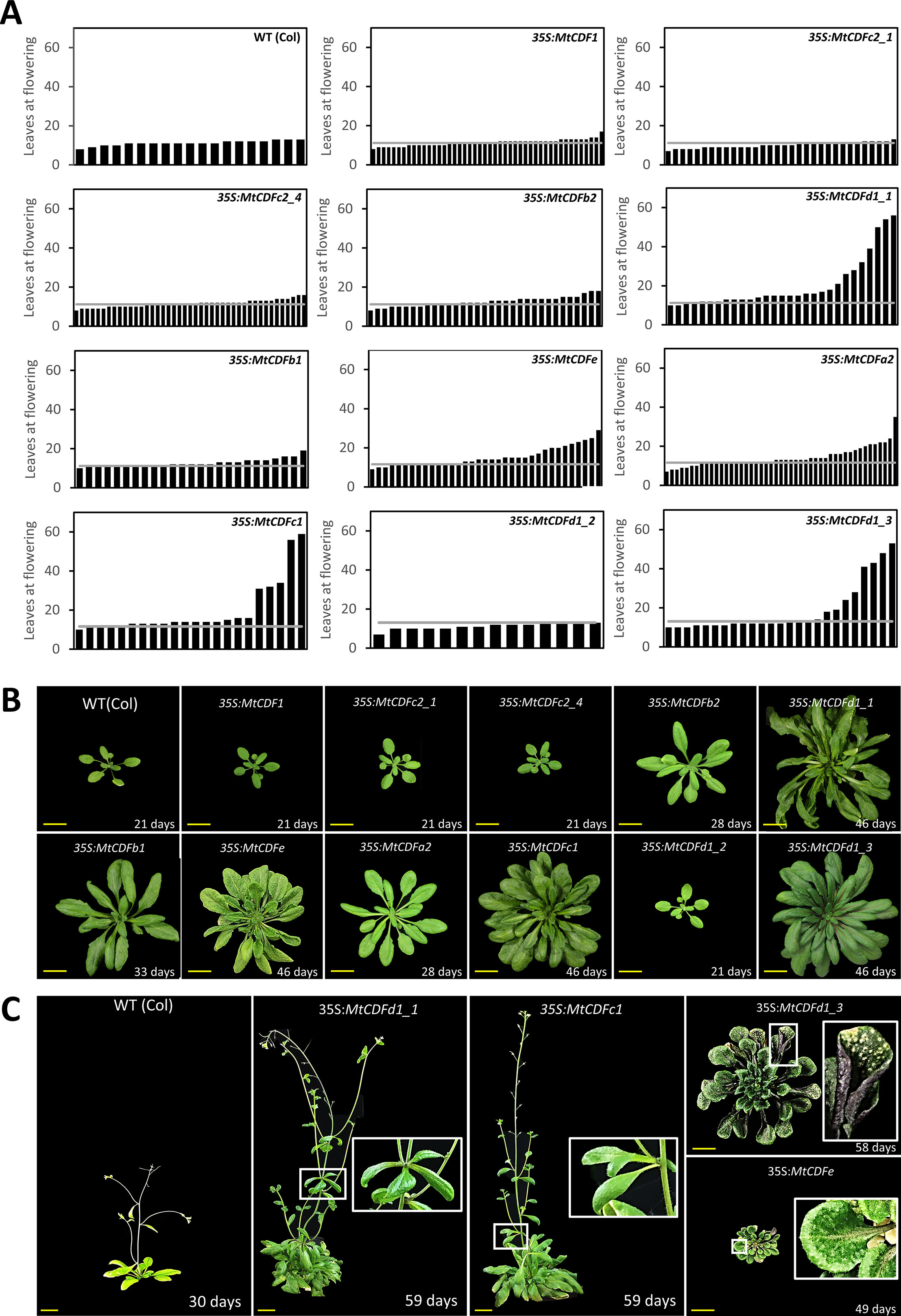
Figure 2 Overexpression of Medicago CDF genes in Arabidopsis can result in late flowering. (A) Flowering time of independent T1 transgenic plants (n ≥ 14) derived from 11 35S:MtCDF expression vectors and Columbia wild-type Arabidopsis in LD conditions. The gray line represents the average leaves at flowering for Columbia; 11.2 ± 0.63 leaves (t.SE 0.05; n = 19). (B) Photographs of selected T2- and T3-generation 35S:MtCDF plants at the time of flowering. (C) Several 35S:MtCDFd1_1 and 35S:MtCDFc1 transgenic plants displayed aerial rosette phenotypes (white boxes) and poor fertility. Multiple 35S:MtCDFd1_3 and 35S:MtCDFe transgenic plants had an upright rosette leaf stature with rigid long-handle spoon-shaped leaves. Additionally, these plants were darker in color with purple abaxial surfaces but had light-colored spots on the older leaves (white boxes in the last panel) and had poor fertility. Age of the plants indicated in days. Yellow scale bars = 2 cm.
Overexpression of five of the genes tested (MtCDFa2, MtCDFc1, MtCDFd1_1, MtCDFd1_3, and MtCDFe) resulted in strong delays to flowering in multiple independent T1 lines in LD, compared to Columbia (Figures 2A, B). Interestingly, these genes arise from different MtCDF subclades (Figure 1A and Figure S1). Overexpression of two other genes (MtCDFb1 and MtCDFb2) produced several transgenic plants that showed a slight delay in flowering time, while overexpression of four genes (MtCDF1, MtCDFc2_1, MtCDFc2_4, and MtCDFd1_2) had little to no effect on Columbia flowering time (Figures 2A, B).
Apart from being late flowering, unusual aerial architectural phenotypes were seen compared to Arabidopsis Columbia plants (Figure 2C). Specifically, an abnormal late-flowering phenotype characterized by aerial rosettes and poor fertility was observed in several independent transgenic lines carrying either of two transgenes, 35S:MtCDFd1_1 or 35S:MtCDFc1. The aerial rosette phenotype is a feature also seen in some Arabidopsis plants where the floral transition is delayed including resulting from disruptions in the floral transition genes including SOC1, AGAMOUS-like 42 (AGL42), AGL71, AGL72 (Dorca-Fornell et al., 2011), FT, TWIN SISTER OF FT (TSF; Hiraoka et al., 2013), FLOWERING LOCUS C (FLC), FRIGIDA (FRI), and AERIAL ROSETTE 1 (ART1; Poduska et al., 2003).
In addition, multiple independent lines carrying either of two transgenes, 35S:MtCDFe or 35S:MtCDFd1_3, displayed an upright rosette leaf stature with rigid, long-handled spoon-shaped leaves (Figure 2C). These plants also were smaller than wild type, infertile with a lack of primary inflorescence bolting, and darker in color. In addition, in some 35S:MtCDFd1_3 lines, the older leaves of some plants developed spotty lesions (Figure 2C).
In summary, among the MtCDFs, MtCDFa2, MtCDFc1, MtCDFd1_1, MtCDFd1_3, and MtCDFe were able to cause strong delays to flowering in multiple transgenic lines when overexpressed in wild-type Arabidopsis. The remaining MtCDF genes we tested did not appear to have much effect on flowering time in Arabidopsis in our experiments, but this may be due to factors such as transgene expression level.
Constitutive Expression of MtCDFd1_1 in Medicago Causes Late Flowering in VLD
We selected MtCDFd1_1 for further functional analysis by overexpression in Medicago. This was because its transcript was relatively abundant in Medicago leaves and exhibited diurnal cycling in VLD and VSD similar to AtCDFs that regulate flowering time redundantly in Arabidopsis. Additionally, it caused a strong delay to flowering in multiple independent lines when overexpressed in Arabidopsis. However, it was interesting also because its predicted protein sequence differs from these AtCDF proteins and from PsCDFc1/LATE2, which has already been characterized in pea (Ridge et al., 2016), falling into a different subclade (d1, Figure S1). It lacks the predicted GI- and FKF1-binding domains (Figure S2) and appears not to interact with AtFKF1 in yeast two-hybrid assays (Figure S4).
We overexpressed MtCDFd1_1 in Medicago to assay the effect this would have on flowering time. After co-cultivation of Medicago wild-type R108 leaf disks with Agrobacterium carrying the 35S:MtCDFd1_1 transgene, we selected six independent T0 transformants. T1 or T2 progeny was scored for flowering time in two photoperiodic conditions, with and without prior vernalization (VLD, LD, and VSD; Figure 3A).
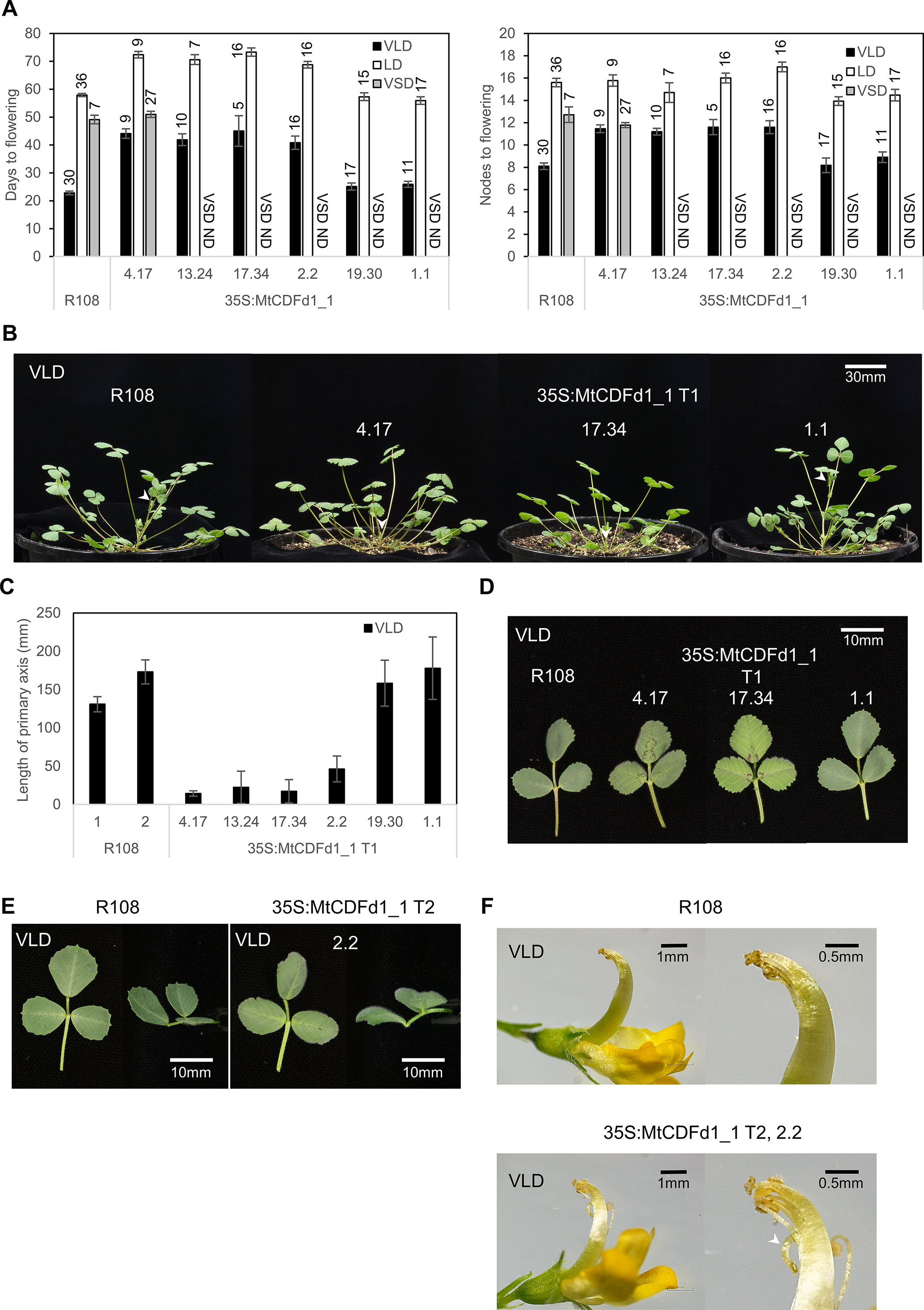
Figure 3 Overexpression of MtCDFd1_1 in Medicago results in late flowering and reduced primary axis elongation. (A) Flowering time of six independent 35S:MtCDFd1_1 Medicago R108 transgenic lines and R108 wild type in different conditions [vernalized long day (VLD), long day (LD), and vernalized short day (VSD)]. Either T1 or T2 generation plants were scored; data from different generations were not combined. Sample sizes are indicated above each bar. Flowering time was presented as the mean number of days, or the number of nodes on the primary axis when the first floral bud was observed ( ± t.SE 0.05) for each of the six independent transgenic lines and R108 control. ND meant that flowering time was not done under VSD. (B) Photographs of T135S:MtCDFd1_1 plants on day 29 under VLD. White arrows indicate the tip of the primary axis. (C) Mean length of the primary axis of the six independent T1 generation lines (41–50 days old) in VLD. The average primary axis length of each line was presented as ± t.SE (0.05), n = 5–10. The control line, R108-1, was planted and measured at the same time as lines 4.17, 13.24, 17.34, 19.30, and 1.1, while R108-2 was planted alongside line 2.2. (D) Photographs of 63-day-old fully expanded trifoliate leaves from different T1 plants and R108 in VLD. Trifoliate leaves photographed, from the top and from the side (E), and flower (F) comparisons between R108 and 35S:MtCDFd1_1 line 2.2. Photographs were taken when VLD R108 and 35S:MtCDFd1_1 plants were 71 and 86 days old, respectively. The white arrow indicates the abnormal curled-down pistil in the 35S:MtCDFd1_1 line compared to wild-type plants.
As expected, VLD most strongly accelerated the flowering of R108 wild-type plants, out of the three conditions tested (Figure 3A). In contrast, most of the transgenic lines (four of six lines: 4.17, 13.24, 17.34, and 2.2) showed delayed flowering in VLD, in both days and nodes at flowering (Figure 3A). In LD, the same four lines showed later flowering than R108 in days to flowering. However, only line 2.2 flowered marginally later in nodes, indicating overall a much weaker flowering time phenotype in LD.
Line 4.17 was then chosen as the representative transgenic line to test in VSD conditions. It had previously shown no phenotypic differences in VLD conditions from three other independent transgenic lines (13.24, 17.34, and 2.2) that also strongly overexpressed MtCDFd1_1 (Figure 4A). Line 4.17 flowering time was not statistically significantly different to R108 in VSD, indicating that 35S:MtCDFd1_1 did not confer late flowering relative to wild type in VSD conditions in this line. Additionally, we observed that line 4.17 flowered at a similar time in VSD and VLD. In summary, while 35S:MtCDFd1_1 caused late flowering in VLD compared to wild type, it had no significant effect in VSD in line 4.17, resulting in day-neutral flowering. Thus, flowering time analysis in VSD was not pursued further.
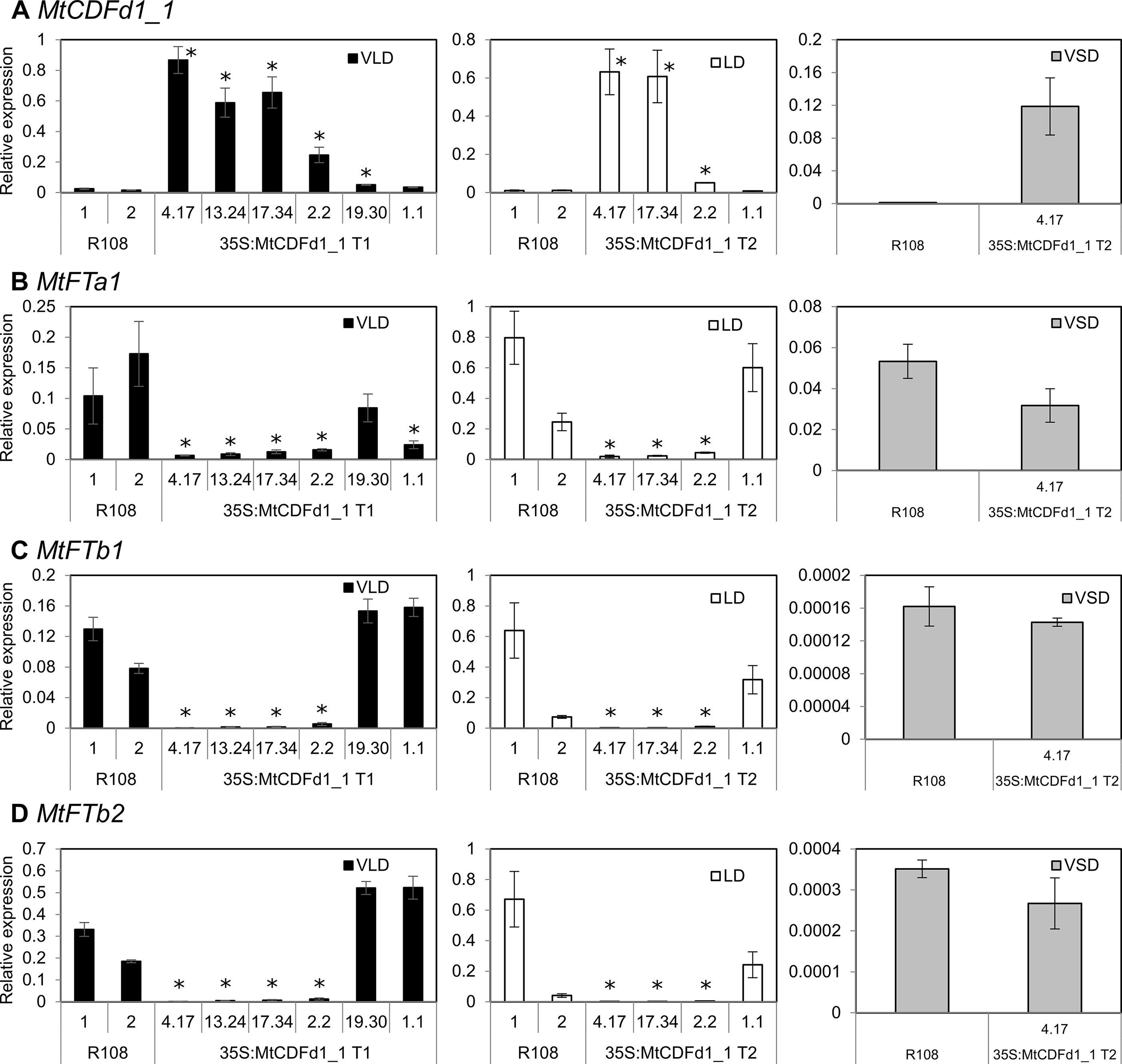
Figure 4 MtCDFd1_1 overexpression in Medicago reduces MtFTa1, MtFTb1, and MtFTb2 transcript levels. (A–D) Expression of MtCDFd1_1, MtFTa1, MtFTb1, and MtFTb2 in the 35S:MtCDFd1_1 Medicago R108 transgenic lines in vernalized long day (VLD), long day (LD), and vernalized short day (VSD). Data were derived from fully expanded trifoliate leaves harvested on days 14 and 15 (VLD), day 46 (LD), and day 43 (VSD) at ZT4. Gene expression levels are means ± SE of three biological replicates, normalized to PP2A. Data were presented relative to the highest value of a gene across the three growth conditions. In VLD, R108-1 was grown at the same time as lines 4.17, 13.24, 17.34, 19.30, and 1.1, while R108-2 was grown with line 2.2. In LD, R108-1 was grown at the same time as lines 17.34 and 1.1, while R108-2 with lines 4.17 and 2.2. All plants grown in VLD were T1 generation, while T2 populations were grown in LD and VSD. Asterisks indicate transgenic lines with significantly different expression from R108 [multiple pairwise comparisons adjusted for false discovery rate (FDR); α = 0.05].
Wild-type R108 plants grown in VLD conditions also typically show elongation of the primary shoot axis at the time of flowering. Therefore, as might be expected from their late-flowering phenotype, the four late-flowering transgenic lines had a shorter primary axis in VLD compared to R108. This was observed at the flowering of R108 and the 35S:MtCDFd1_1 plants (Figures 3B, C).
In addition, the leaves of later-flowering transgenic plants were sometimes paler in color than R108 and the transgenic plants that did not flower late (Figure 3D). In the later stage of plant growth, they had trifoliate leaves that curved down (epinastic) while R108 leaves curved upwards (Figure 3E). Some late-flowering transgenic plants displayed sterility. This was likely because the top of the pistil was curled down, causing the stigma to be away from anthers, leading to failure in pollination (Figure 3F).
In summary, four of the six independent lines carrying the 35S:MtCDFd1_1 transgene showed delayed flowering and changes to architecture including shorter primary stems, leaf curling, and infertility in VLD conditions.
MtCDFd1_1 Overexpression Is Negatively Correlated With Transcript Levels of MtFT-Like Genes but Not MtCOL Genes
To investigate the basis of the late-flowering phenotypes observed in the 35S:MtCDFd1_1 transgenic lines (Figure 3A), we analyzed gene expression by qRT-PCR (Figure 4). The genes assayed were MtCDFd1_1 and the three LD-induced MtFT genes, which are expressed at higher levels in VLD than in VSD: MtFTa1, MtFTb1, and MtFTb2 (Laurie et al., 2011). MtFTa1 has been shown to accelerate flowering when overexpressed in Medicago, while loss-of-function mutants show late flowering compared to wild type, particularly in VLD conditions (Laurie et al., 2011; Jaudal et al., 2019).
In VLD, 35S:MtCDFd1_1 transcript levels in the four late-flowering lines (4.17, 13.24, 17.34, and 2.2) were significantly higher compared to those in R108 controls (Figure 4A). However, MtCDFd1_1 expression in the fifth line was only very weakly elevated, while the sixth line, 1.1, was not significantly different from R108. These latter two lines, 19.30 and 1.1, also flowered at a similar time to R108 (Figure 3A).
The increased expression of MtCDFd1_1 in VLD observed in the four transgenic lines 4.17, 13.24, 17.34, and 2.2 (Figure 4A) correlated with significantly lower abundance of MtFTa1, MtFTb1, and MtFTb2 transcripts (Figures 4B–D) and late flowering (Figure 3A) in those lines compared to wild-type R108 plants.
In contrast, qRT-PCR analysis of five MtCOL genes (MtCOLa–MtCOLd and MtCOLh; Figure 5) indicates that there is no consistent change to the expression of these genes in the four Mt MtCDFd1_1 overexpression lines (4.17, 13.24, 17.34, and 2.2) compared to R108 and the two remaining transgenic lines that do not overexpress MtCDFd1_1 (19.30 and 1.1).
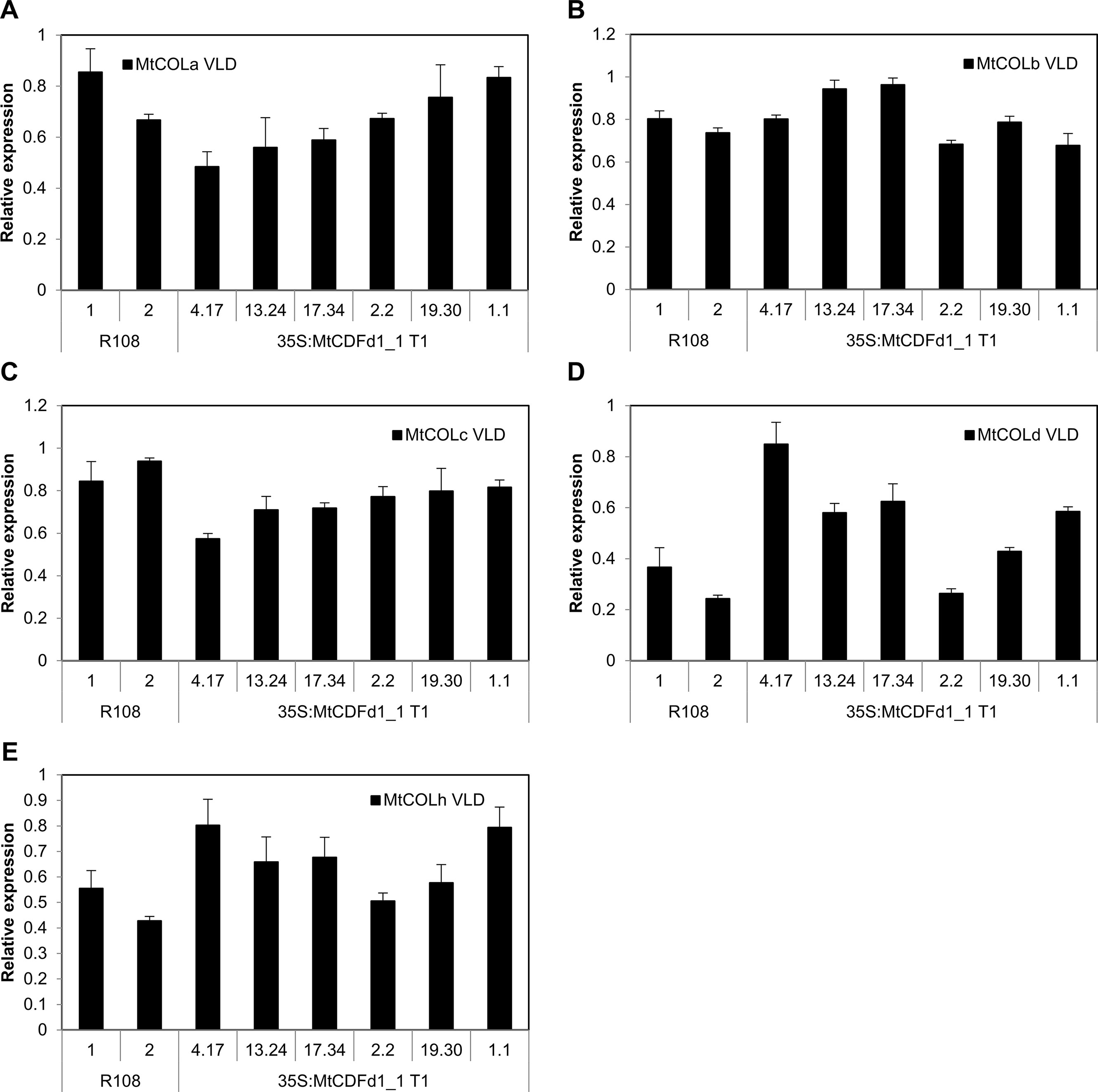
Figure 5 Overexpression of MtCDFd1_1 in Medicago does not reduce COL gene expression. (A–E) Relative gene expression of five Medicago COL genes in vernalized long day (VLD) in 35S:MtCDFd1_1 lines. Data were derived from fully expanded trifoliate leaves harvested from T1 plants on days 14 and 15 at ZT4. Gene expression levels were means of three biological replicates ± SE, normalized to PP2A. Data were presented relative to the sample with the highest expression. R108-1 was grown at the same time as lines 4.17, 13.24, 17.34, 19.30, and 1.1, while R108-2 was grown with line 2.2.
In LD, a subset of lines was tested for gene expression. Like in VLD, overexpression of MtCDFd1_1 correlated with significantly reduced expression of MtFTa1, MtFTb1, and MtFTb2 (Figure 4).
In VSD, no significant difference could be seen in the expression of MtFTa1 in representative MtCDFd1_1 overexpressing line 4.17 relative to R108 (Figure 4B). This is consistent with the absence of a flowering time phenotype in this transgenic line relative to R108 in VSD. MtFTb1 and MtFTb2 transcript levels were barely detectable in VSD in the transgenic line or R108 (Figures 4C, D) as expected (Laurie et al., 2011). Thus, gene expression analysis in VSD was not pursued further.
Flowering Time and Gene Expression in 35S:MtCDFd1_1/Mtfta1 Homozygous Lines
MtFTa1 is a strong promoter of Medicago flowering, particularly in VLD conditions (Laurie et al., 2011). This suggests that the delayed flowering in the 35S:MtCDFd1_1 plants in VLD might be due to the reduced average MtFTa1 expression we observed. Therefore, to analyze the interaction between 35S:MtCDFd1_1 and MtFTa1, two late-flowering 35S:MtCDFd1_1 lines, 4.17 and 2.2, were crossed with the late-flowering Mtfta1 mutant and the resulting F2 populations scored in VLD (Figure 6A).
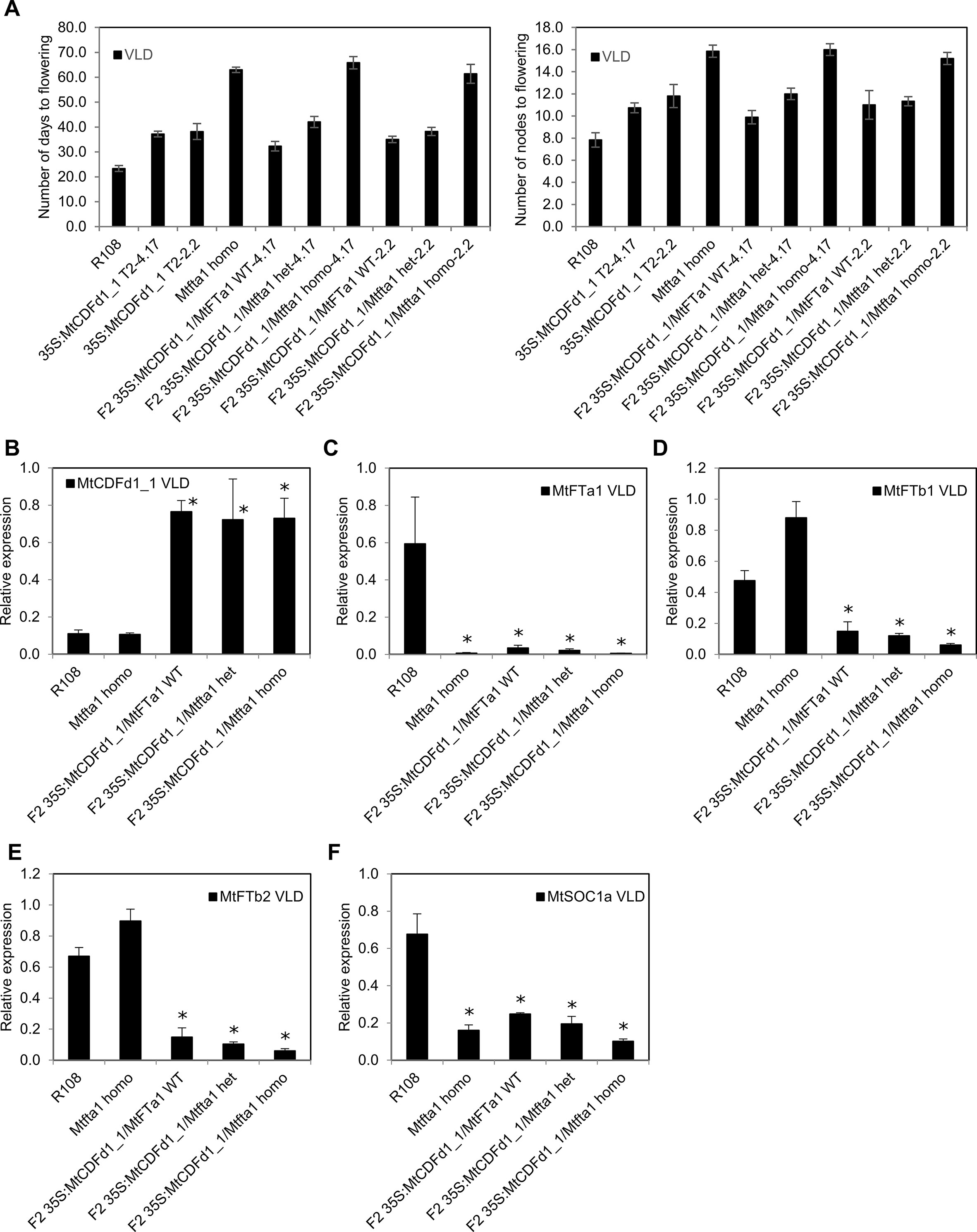
Figure 6 Analysis of an F2 population from a cross between late-flowering Medicago plants overexpressing MtCDFd1_1 and the late-flowering Mtfta1 R108 mutant. (A) Flowering time for each genotype in a segregating F2 population (n = 72) derived from a cross between the Mtfta1 mutant and transgenic plants overexpressing MtCDFd1_1 is presented as either the mean number of days or the number of nodes on the primary axis when the first floral bud was observed ± t.SE (0.05). Homo is homozygous for the Mtfta1 mutation, and het is heterozygous for the Mtfta1 mutation. One wild-type F2 segregant plant (without the 35S:MtCDFd1_1 transgene and wild type for MtFTa1) was obtained. It flowered at 26 days and eight nodes, similar to wild-type R108. (B–F) Relative gene expression in 35S:MtCDFd1_1 F2 plants, with or without the Mtfta1 mutation of MtCDFd1_1, MtFTa1, MtFTb1, MtFTb2 and MtSOC1a in vernalized long day (VLD). Data were derived from fully expanded trifoliate leaves harvested on day 23 at ZT4. Three biological samples each consisting of leaves from three plants were harvested per genotype. Gene expression levels were means of the three biological replicates ± SE, normalized to PP2A. Data were presented relative to the highest value of that specific gene. Asterisks indicate genotypes with significantly different expression from R108 [multiple pairwise comparisons adjusted for false discovery rate (FDR); α = 0.05].
35S:MtCDFd1_1/Mtfta1 homozygous F2 plants flowered ∼1 month later than 35S:MtCDFd1_1 lines homozygous for wild-type MtFTa1, but at a similar time to Mtfta1 homozygous mutant plants. Thus, no additive effect was observed in 35S:MtCDFd1_1 on the late flowering already conferred by the Mtfta1 homozygous mutation in VLD.
As previously observed in the four late-flowering 35S:MtCDFd1_1 transgenic plants (lines 4.17, 13.24, 17.34, and 2.2, Figure 4), the presence of the 35S:MtCDFd1_1 transgene correlated with significantly lower transcript levels of MtFTa1, MtFTb1, and MtFTb2 compared to R108 (Figures 6B–E).
We also analyzed the expression of MtSOC1a (Figure 6F), a SOC1-like gene which promotes flowering and primary stem growth and whose expression is partly dependent on MtFTa1 (Fudge et al., 2018; Jaudal et al., 2018). Plants with the 35S:MtCDFd1_1 transgene and wild type for MtFTa1 showed a statistically significant, moderate decrease (∼2.7-fold) in average MtSOC1a transcript levels compared to wild-type R108 plants.
Discussion
While the photoperiodic pathways in Medicago and pea promote flowering through LD-induced FT genes such as FTa1, in contrast to Arabidopsis, they appear to act in a CO-independent manner. To test whether MtCDF genes regulate Medicago photoperiodic flowering time, we analyzed the expression and function of members of the MtCDF clade with a focus on MtCDFd1_1. Our work on the MtCDFs has revealed similarities and differences between Medicago and the well-characterized Arabidopsis system and indicates how MtCDFs may contribute to Medicago flowering time control.
MtCDF genes, MtCDFd1_1 (here) and MtCDFc2-1 and MtCDFb2 (Thomson et al., 2019), showed a diurnal cycle of expression, with peak transcript levels at or near dawn, which was similar to the best characterized AtCDFs that regulate flowering time (AtCDF1-3,5). We also observed that overexpression of MtCDFd1_1 in Medicago caused VLD plants to flower late, as if they had been grown in VSD, rendering the transgenic plants day neutral. These results are similar to those reported for the dominant pea mutation late2/Pscdfc1 (Ridge et al., 2016) and for overexpression of AtCDFs in Arabidopsis. Thus, MtCDFs may normally function in wild-type plants predominantly to delay flowering in VSD.
35S:MtCDFd1_1 appears to regulate flowering in Medicago via repressing MtFTa1, a known strong promoter of flowering in VLD (Laurie et al., 2011), but not via MtCOL genes. The transcript levels of the LD-induced genes MtFTa1, MtFTb1, MtFTb2, and MtSOC1a were significantly reduced in the 35S:MtCDFd1_1 transgenic plants, while five MtCOL genes were unaffected. Genetic analysis showed that 35S:MtCDFd1_1/Mtfta1 plants flowered no later than the later-flowering parent, Mtfta1. Thus, in VLD, 35S:MtCDFd1_1 influenced flowering in the same pathway as MtFTa1, and the late flowering of 35S:MtCDFd1_1 plants in VLD likely results from reduced MtFTa1 gene expression. The short primary stem phenotype observed is also consistent with the repressive effect of 35S:MtCDFd1_1 on expression of MtFTa1 and MtSOC1a, previously indicated to be important for stem elongation in VLD and LD conditions (Laurie et al., 2011; Jaudal et al., 2018).
What might be the role of the other two MtFT-like genes, MtFTb1 and MtFTb2, whose expression is also strongly reduced by 35S:MtCDFd1_1? The 35S:MtCDFd1_1/Mtfta1 plants show no additional delay to flowering time, beyond that conferred by the Mtfta1 mutation in VLD conditions, and as previously reported (Laurie et al., 2011), MtFTb1 and MtFTb2 expression is not affected by the single Mtfta1 mutation. Overall, these results indicate that neither MtFTb1 nor MtFTb2 has non-redundant roles in Medicago flowering time in VLD. It is possible they may affect flowering via regulating MtFTa1, but testing this awaits the identification of single and double MtFTb1/2 mutant plants.
While no MtCDF Tnt1 insertion mutant plants had a flowering time phenotype, this is overall consistent with Arabidopsis CDF single mutants and is likely due to redundancy in function between the genes (Fornara et al., 2009). On the other hand, five genes (MtCDFd1_1, MtCDFa2, MtCDFc1, MtCDFd1_3, and MtCDFe), out of the 11 tested, stood out for their ability to cause late flowering when overexpressed in Arabidopsis. It is possible that sequence variation within key MtCDF functional domains, or their absence, could affect the other MtCDFs’ ability to interact with potential binding partners or target genes and regulate flowering time. For example, differential susceptibility to the Arabidopsis FKF1/GI protein degradation system may affect MtCDFs’ ability to repress flowering and could help explain some of the variation in flowering times observed between the different genes (Kloosterman et al., 2013; Ridge et al., 2016). On the other hand, it is possible that the inability of the other MtCDF genes tested to affect Arabidopsis flowering time was due to the differences in transgene expression levels.
In our case, 35S:MtCDFd1_1 strongly represses flowering, and its predicted protein lacks the predicted GI- and FKF1-binding domains. This provides some indication that MtCDFd1_1 protein may not be targeted for degradation by the endogenous FKF1/GI system in Arabidopsis or Medicago, suggesting an alternative method of regulation of its activity in LD from the AtCDF system. On the other hand, MtCDFc1 and its predicted pea ortholog PsCDFc1 (Ridge et al., 2016) do interact with AtFKF1 in yeast two-hybrid assays but have different effects on flowering in Arabidopsis. In our experiments, 35S:MtCDFc1 strongly delayed Arabidopsis flowering, while 35S:PsCDFc1 was reported not to (Ridge et al., 2016). This indicates that other differences in sequence may be important, or perhaps differences in cultivation or levels of expression in the transgenic plants may be responsible.
Apart from a delayed transition to flowering, other phenotypes were seen in multiple 35S:MtCDF transgenic Arabidopsis plants implicating MtCDFs in a variety of plant processes that extend beyond involvement in photoperiodic regulation (Corrales et al., 2014, Corrales et al., 2017). In plants such as Arabidopsis and tomato, CDF genes also modulate other processes such as abiotic stress tolerance (Corrales et al., 2014, Corrales et al., 2017). In addition, a different photoperiodic process, namely, SD-induced tuber development, is regulated by StCDF1 in Solanum tuberosum L. (potato; Kloosterman et al., 2013). The abnormal phenotypes we observed included an upright rosette leaf stature with rigid long-handle spoon-shaped curved leaves, which may indicate effects on hormone homeostasis (e.g. Sun et al., 2010). Interestingly, in addition to late flowering in some independent 35S:MtCDFd1_3 lines, the older leaves of some of the plants developed spotty lesions, perhaps indicative of effects on senescence or cell death and/or disease resistance processes (Lorrain et al., 2003).
Overall, our results expand the understanding of the features and functions of members of the MtCDF clade. MtCDF genes are implicated as regulators of the Medicago photoperiodic pathway, where they are likely to have overlapping functions in wild-type plants probably by repressing flowering in VSD conditions. In terms of mechanism, the absence of an effect of overexpression of MtCDFd1_1 in transgenic lines (4.17, 13.24, 17.34, and 2.2) on the expression of five MtCOL genes (Figure 5), but strong repression of LD-induced MtFT genes compared to R108 (Figure 4), adds further support to the idea that MtCDFs may function in a photoperiod pathway that is independent of CO. This is consistent with work in pea (Ridge et al., 2016). Future work to determine the function of MtCDFs and to overcome the challenges of functional redundancy will focus on generating plants carrying mutations in multiple MtCDF genes using the CRISPR–Cas9 system in Medicago (Meng et al., 2017; Curtin et al., 2018). In addition, since there is direct regulation of FT by AtCDF1 (Song et al., 2012), direct interactions of MtCDFs with the LD-induced MtFT-like genes could be tested to examine if this is conserved in legumes.
Data Availability
All datasets generated for this study are included in the manuscript and the supplementary files.
Author Contributions
LZ, AJ, MK-P, GT, TK, CP, and MJ performed the experiments, LZ, AJ, GT, and JP prepared the figures. JW and KM provided the Medicago Tnt1 insertion lines. JP conceived the project and wrote the article with contributions from AJ, LZ, MJ, and GT.
Funding
The research was funded by the New Zealand Foundation for Research Science and Technology (www.msi.govt.nz/) contract number C10X0816 MeriNET and the New Zealand Marsden Fund (www.royalsociety.org.nz/programmes/funds/marsden/) contract 14-UOA-125. The development of the Medicago Tnt1 insertion lines and reverse genetics screenings were supported by the National Science Foundation, USA (DBI 0703285 and IOS-1127155), and Noble Research Institute, LLC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Davide Zazarro and Nathan Deed for glasshouse support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01148/full#supplementary-material
References
Andrés, F., Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Bookout, A. L., Mangelsdorf, D. J. (2003). Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl. Recept. Signal. 1, e012. doi: 10.1621/nrs.01012
Chabaud, M., Lichtenzveig, J., Ellwood, J., Pfaff, T., Journet, E-P. (2006) Vernalization, crossings and testing for pollen viability. In: Mathesius, U, Journet, E, Sumner, L, editors. The Medicago truncatula handbook. Available: http://www.noble.org/MedicagoHandbook/.
Corrales, A.-R., Nebauer, S. G., Carrillo, L., Fernández-Nohales, P., Marqués, J., Renau-Morata, B., et al. (2014). Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 65, 995–1012. doi: 10.1093/jxb/ert451
Corrales, A. R., Carrillo, L., Lasierra, P., Nebauer, S. G., Dominguez-Figueroa, J., Renau-Morata, B., et al. (2017). Multifaceted role of Cycling DOF Factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 40, 748–764. doi: 10.1111/pce.12894
Cosson, V., Durand, P., Erfurth, I., Kondorosi, A., Ratet, P. (2006). Medicago truncatula transformation using leaf explants. Methods Mol. Biol. 343, 115–127. doi: 10.1385/1-59745-130-4:115
Curtin, S. J., Xiong, Y., Michno, J. M., Campbell, B. W., Stec, A. O., Čermák, T., et al. (2018). CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotechnol. J. 16, 1125–1137. doi: 10.1111/pbi.12857
Dorca-Fornell, C., Gregis, V., Grandi, V., Coupland, G., Colombo, L., Kater, M. M. (2011). The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J. 67, 1006–1017. doi: 10.1111/j.1365-313X.2011.04653.x
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fornara, F., de Montaigu, A., Coupland, G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141, 550. doi: 10.1016/j.cell.2010.04.024
Fornara, F., Panigrahi, K. C. S., Gissot, L., Sauerbrunn, N., Rühl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. doi: 10.1016/j.devcel.2009.06.015
Fudge, J. B., Lee, R. H., Laurie, R. E., Mysore, K. S., Wen, J., Weller, J. L., et al. (2018). Medicago truncatula SOC1 genes are up-regulated by environmental cues that promote flowering. Front. Plant Sci. 9, 496. doi: 10.3389/fpls.2018.00496
Geneious 11.1.5 from Biomatters. Available at: https://www.geneious.com.
Goralogia, G. S., Liu, T. K., Zhao, L., Panipinto, P. M., Groover, E. D., Bains, Y. S., et al. (2017). CYCLING DOF FACTOR 1 represses transcription through the TOPLESS co-repressor to control photoperiodic flowering in Arabidopsis. Plant J. 92, 244–262. doi: 10.1111/tpj.13649
Graham, P. H., Vance, C. P. (2003). Legumes: importance and constraints to greater use. Plant Physiol. 131, 872–877. doi: 10.1104/pp.017004
Hecht, V., Foucher, F., Ferrándiz, C., Macknight, R., Navarro, C., Morin, J., et al. (2005). Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137, 1420–1434. doi: 10.1104/pp.104.057018
Hecht, V., Laurie, R. E., Vander Schoor, J. K., Ridge, S., Knowles, C. L., Liew, L. C., et al. (2011). The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23, 147–161. doi: 10.1105/tpc.110.081042
Hiraoka, K., Yamaguchi, A., Abe, M., Araki, T. (2013). The florigen genes FT and TSF modulate lateral shoot outgrowth in Arabidopsis thaliana. Plant Cell Physiol. 54, 352–368. doi: 10.1093/pcp/pcs168
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., Kay, S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. doi: 10.1126/science.1110586
James, P., Halladay, J., Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436.
Jaudal, M., Thomson, G., Zhang, L., Che, C. E., Wen, J., Mysore, K., et al. (2019). “Forward and reverse screens to identify genes that control vernalisation and flowering time in Medicago,” in The model legume Medicago truncatula. Ed. de Bruijn, F. (Chichester, UK: Wiley-Blackwell).
Jaudal, M., Zhang, L., Che, C., Li, G., Tang, Y., Wen, J., et al. (2018). A SOC1-like gene MtSOC1a promotes flowering and primary stem elongation in Medicago. J. Exp. Bot. 69, 4867–4880. doi: 10.1093/jxb/ery284
Jaudal, M., Zhang, L., Che, C., Putterill, J. (2015). Three Medicago MtFUL genes have distinct and overlapping expression patterns during vegetative and reproductive development and 35S:MtFULb accelerates flowering and causes a terminal flower phenotype in Arabidopsis. Front. Genet. 5. doi: 10.3389/fgene.2015.00050
Karimi, M., Depicker, A., Hilson, P. (2007). Recombinational cloning with plant Gateway vectors. Plant Physiol. 145, 1144–1154. doi: 10.1104/pp.107.106989
Kloosterman, B., Abelenda, J. A., Gomez, M., del, M. C., Oortwijn, M., de Boer, J. M., et al. (2013). Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495, 246–250. doi: 10.1038/nature11912
Laurie, R. E., Diwadkar, P., Jaudal, M., Zhang, L., Hecht, V., Wen, J., et al. (2011). The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 156, 2207–2224. doi: 10.1104/pp.111.180182
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(–delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lorrain, S., Vailleau, F., Balagué, C., Roby, D. (2003). Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8, 263–271. doi: 10.1016/S1360-1385(03)00108-0
Martinez-Trujillo, M., Limones-Briones, V., Cabrera-Ponce, J. L., Herrera-Estrella, L. (2004). Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Mol. Biol Report. 22, 63–70. doi: 10.1007/BF02773350
Meng, Y., Hou, Y., Wang, H., Ji, R., Liu, B. (2017). Targeted mutagenesis by CRISPR/Cas9 system in the model legume Medicago truncatula.. Plant Cell Rep. 36, 371–374. doi: 10.1007/s00299-016-2069-9
Noguero, M., Atif, R. M., Ochatt, S., Thompson, R. D. (2013). The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 209, 32–45. doi: 10.1016/j.plantsci.2013.03.016
Poduska, B., Humphrey, T., Redweik, A., Grbić, V. (2003). The synergistic activation of FLOWERING LOCUS C by FRIGIDA and a new flowering gene AERIAL ROSETTE 1 underlies a novel morphology in Arabidopsis.. Genetics 163, 1457–1465.
Putterill, J., Laurie, R., Macknight, R. (2004). It’s time to flower: the genetic control of flowering time. BioEssays 26, 363–373. doi: 10.1002/bies.20021
Putterill, J., Varkonyi-Gasic, E. (2016). FT and florigen long-distance flowering control in plants. Curr. Opin. Plant Biol. 33, 77–82. doi: 10.1016/j.pbi.2016.06.008
Putterill, J., Zhang, L., Yeoh, C. C., Balcerowicz, M., Jaudal, M., Varkonyi-Gasic, E. (2013). FT genes and regulation of flowering in the legume Medicago truncatula. Funct. Plant Biol. 40, 1199–1207. doi: 10.1071/FP13087
Ridge, S., Sussmilch, F. C., Hecht, V., Schoor, K., Vander Schoor, J.K., Lee, R., et al. (2016). Identification of LATE BLOOMER2 as a CYCLING DOF FACTOR homolog reveals conserved and divergent features of the flowering response to photoperiod in pea. Plant Cell 28, 2545–2559. doi: 10.1105/tpc.15.01011
Sawa, M., Nusinow, D. A., Kay, S. A., Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Shu, Y. J., Song, L. L., Zhang, J., Liu, Y., Guo, C. H. (2015). Genome-wide identification and characterization of the Dof gene family in Medicago truncatula. Genet. Mol. Res. 14, 10645–10657. doi: 10.4238/2015.September.9.5
Song, Y. H., Smith, R. W., To, B. J., Millar, A. J., Imaizumi, T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049. doi: 10.1126/science.1219644
Srikanth, A., Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Sun, Y., Yang, Y., Yuan, Z., Muller, J. L., Yu, C., Xu, Y., et al. (2010). Overexpression of the Arabidopsis gene UPRIGHT ROSETTE reveals a homeostatic control for indole-3-acetic acid. Plant Physiol. 153, 1311–1320. doi: 10.1104/pp.110.154021
Swofford, D. L. (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, Massachusetts, Sinauer Associates. Available: http://paup.phylosolutions.com/
Thomson, G., Taylor, J., Putterill, J. (2019). The transcriptomic response to a short day to long day shift in leaves of the reference legume Medicago truncatula. PeerJ 7, e6626. doi: 10.7717/peerj.6626
Trinh, T. H., Ratet, P., Kondorosi, E., Durand, P., Kamaté, K., Bauer, P., et al. (1998). Rapid and efficient transformation of diploid Medicago truncatula and Medicago sativa ssp. falcata lines improved in somatic embryogenesis. Plant Cell Rep. 17, 345–355. doi: 10.1007/s002990050405
Turck, F., Fornara, F., Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Weller, J. L., Ortega, R. (2015). Genetic control of flowering time in legumes. Front. Plant Sci. 6, 207. doi: 10.3389/fpls.2015.00207
Wickland, D. P., Hanzawa, Y. (2015). The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol. Plant 8, 983–997. doi: 10.1016/j.molp.2015.01.007
Wong, A. C. S., Hecht, V. F. G., Picard, K., Diwadkar, P., Laurie, R. E., Wen, J., et al. (2014). Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 5, 486. doi: 10.3389/fpls.2014.00486
Yanagisawa, S. (2002). The Dof family of plant transcription factors. Trends Plant Sci. 7, 555–560. doi: 10.1016/S1360-1385(02)02362-2
Keywords: CYCLING DOF FACTOR, MtCDFd1_1, MtFTa1, MtFTb, CO, Medicago, flowering time, primary axis elongation
Citation: Zhang L, Jiang A, Thomson G, Kerr-Phillips M, Phan C, Krueger T, Jaudal M, Wen J, Mysore KS and Putterill J (2019) Overexpression of Medicago MtCDFd1_1 Causes Delayed Flowering in Medicago via Repression of MtFTa1 but Not MtCO-Like Genes. Front. Plant Sci. 10:1148. doi: 10.3389/fpls.2019.01148
Received: 08 April 2019; Accepted: 22 August 2019;
Published: 19 September 2019.
Edited by:
Matthew Nicholas Nelson, Commonwealth Scientific and Industrial Research Organisation, AustraliaReviewed by:
Jim Weller, University of Tasmania, AustraliaTakato Imaizumi, University of Washington, United States
Copyright © 2019 Zhang, Jiang, Thomson, Kerr-Phillips, Phan, Krueger, Jaudal, Wen, Mysore and Putterill. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanna Putterill, ai5wdXR0ZXJpbGxAYXVja2xhbmQuYWMubno=
 Lulu Zhang1
Lulu Zhang1 Andrew Jiang
Andrew Jiang Geoffrey Thomson
Geoffrey Thomson Chau Phan
Chau Phan Thorben Krueger
Thorben Krueger Mauren Jaudal
Mauren Jaudal Jiangqi Wen
Jiangqi Wen Kirankumar S. Mysore
Kirankumar S. Mysore Joanna Putterill
Joanna Putterill