- 1Key Laboratory of Sweetpotato Biology and Biotechnology, Ministry of Agriculture/Beijing Key Laboratory of Crop Genetic Improvement/Laboratory of Crop Heterosis and Utilization, Ministry of Education, College of Agronomy & Biotechnology, China Agricultural University, Beijing, China
- 2College of Agronomy, Qingdao Agricultural University, Qingdao, China
Several members of the MYB transcription factor family have been found to regulate growth, developmental processes, metabolism, and biotic and abiotic stress responses in plants. However, the role of MYB116 in plants is still unclear. In this study, a MYB transcription factor gene IbMYB116 was cloned and characterized from the sweetpotato [Ipomoea batatas (L.) Lam.] line Xushu55-2, a line that is considered to be drought resistant. We show here that IbMYB116 is a nuclear protein and that it possesses a transactivation domain at the C terminus. This gene exhibited a high expression level in the leaf tissues of Xushu55-2 and was strongly induced by PEG6000 and methyl-jasmonate (MeJA). The IbMYB116-overexpressing Arabidopsis plants showed significantly enhanced drought tolerance, increased MeJA content, and a decreased H2O2 level under drought stress. The overexpression of IbMYB116 in Arabidopsis systematically upregulated jasmonic acid (JA) biosynthesis genes and activated the JA signaling pathway as well as reactive oxygen species (ROS)-scavenging system genes under drought stress conditions. The overall results suggest that the IbMYB116 gene might enhance drought tolerance by activating a ROS-scavenging system through the JA signaling pathway in transgenic Arabidopsis. These findings reveal, for the first time, the crucial role of IbMYB116 in the drought tolerance of plants.
Introduction
Drought seriously affects the productivity of agricultural crops in the world (Yang et al., 2010; Boyer et al., 2013). Improving the drought tolerance of crops has become important for food security (Tester and Langridge, 2010; Zhu, 2016). Plants adapt to drought stress by developing a variety of mechanisms, including growth and development regulation, osmotic adjustment, ion homeostasis, and detoxification (Bohnert et al., 1995; Zhu, 2002). In response to water deficits, plants increase water uptake by forming long roots to promote their survival (Yu et al., 2008; Wang et al., 2016a). At the phytohormone level, jasmonic acid (JA), abscisic acid (ABA), salicylic acid (SA), and ethylene (ETH) play important roles in plant growth and development as well as protective responses against biotic and abiotic stresses (Fujita et al., 2005; Dong et al., 2014; Vargas et al., 2014; Zhang et al., 2015a; Jin et al., 2016; Xu et al., 2017).
Jasmonates (JAs), including JA, methyl-jasmonate (MeJA), and JA-isoleucine (JA-Ile), are important regulators of plant responses to environmental stresses, such as drought, salt, and ozone exposure (Overmyer et al., 2000; Turner et al., 2002; Lorenzo et al., 2003; Rojo et al., 2003; Wasternack et al., 2006; Dhakarey et al., 2017; Zhang et al., 2017). In JA biosynthesis, plastidial 13-lipoxygenase (LOX), allene oxide synthase (AOS), and allene oxide cyclase (AOC) catalyze linolenic acid to cis-12-oxophytodienoic acid (OPDA); OPDA reductase (OPR) further reduces OPDA to 3-oxo-2(2’(Z)-pentenyl)-cyclopentane-1-octanoic acid (OPC-8:0). Subsequently, the carboxyl chain of OPC-8:0 is shortened by three β-oxidation steps, which form JA with the key enzymes CoA ligase 1 (OPCL1), acyl-CoA oxidase (ACOX1, ACOX3), enoyl-CoA hydratase (MFP2), acetyl-CoA acyltransferase (fadA), and acetyl-CoA acyltransferase 1 (ACAA1) (Mueller, 1997; Berger, 2002; Wasternack and Hause, 2013). Furthermore, coronatine-insensitive 1 (COI1), jasmonate ZIM-domain (JAZ) and myelocytomatosis (MYC) proteins are the components of the core JA signaling pathway (Chini et al., 2007; Wasternack and Hause, 2013; Liu and Avramova, 2016). The JA signaling pathway plays a critical role in regulating the response to drought stress in plants (Liu et al., 2015; Ahmad et al., 2016).
In plants, several types of transcription factors (TFs), including MYB/MYC, NAC, bZIP, AP2/ERF, PHD, and WRKY, control biological processes by regulating target genes in response to stresses (Shiriga et al., 2014; Zong et al., 2016; Wang et al., 2017a; Wang et al., 2017b). MYB is a large transcription factor family in higher plants (Riechmann et al., 2000). Based on the number of repeats of the MYB domain, this family is divided into four major classes: 1R-MYB, R2R3-MYB, 3R-MYB, and 4R-MYB. ZmC1 from Zea mays was the first plant MYB gene (Paz-Ares et al., 1987). To date, more than 200 MYB TFs have been identified in several plant species, such as Arabidopsis, rice, and soybean (Dubos et al., 2010; Aoyagi et al., 2014; Smita et al., 2015). R2R3-MYB TFs have been found to play important roles in plant responses to biotic and abiotic stresses (Dubos et al., 2010). Accumulated evidence has indicated that R2R3-MYB TFs mediate stress-signaling pathways, such as salinity, drought, and extreme temperature (Hajiebrahimi et al., 2017). In Arabidopsis, several R2R3-MYB TFs, including AtMYB2, AtMYB15, AtMYB44, AtMYB60, AtMYB61, AtMYB96, and AtMYB102, were involved in drought stress responses (Denekamp and Smeekens, 2003; Abe et al., 2003; Liang et al., 2005; Chen et al., 2006; Jung et al., 2008; Rusconi et al., 2013; Lee et al., 2014). The overexpression of GmMYB84 and GmMYB174 in soybean, GaMYB85, GmMYBJ1, and GmMYBJ2 in Arabidopsis, OsMYB2 in rice, MdSIMYB1 in tobacco and apple, and SbMYB8 in tobacco improved drought tolerance in transgenic plants (Yang et al., 2012; Su et al., 2014; Wang et al., 2014; Su et al., 2015; Wang et al., 2015; Yuan et al., 2015; Butt et al., 2017; Wang et al., 2017b). Until now, the role of MYB116 in plants has not been reported.
The productivity of sweetpotato, Ipomoea batatas (L.) Lam., as an important food crop is seriously affected by drought stress. It has been shown that drought tolerance of sweetpotato could be improved through gene engineering (Park et al., 2011; Wang et al., 2016b; Fan et al., 2017; Kang et al., 2018a, Kang et al., 2018b; Kim et al., 2018). However, MYB TFs from sweetpotato have not been functionally characterized to date. In this study, we cloned a novel MYB TF gene named IbMYB116 from a drought-tolerant sweetpotato line Xushu55-2 and found its overexpression in Arabidopsis enhanced drought tolerance through the JA signaling pathway.
Materials and Methods
Plant Materials
The drought-tolerant sweetpotato line Xushu55-2 was applied to isolate the IbMYB116 gene. The expressed sequence tag (EST) was screened from the transcriptome sequencing data of Xushu55-2 (Zhu et al., 2018). The function of IbMYB116 was identified using Arabidopsis thaliana Columbia-0.
Cloning and Sequence Analysis of IbMYB116 and Its Promoter
The total RNA from in vitro-grown plants of Xushu55-2 was transcribed into the first-strand cDNA (Kang et al., 2018a). The rapid amplification of cDNA ends (RACE) procedure was used to amplify the full-length cDNA of IbMYB116. The genomic sequence of IbMYB116 was amplified from the genomic DNA of in vitro-grown Xushu55-2. Its promoter was obtained from the genomic DNA of in vitro-grown Xushu55-2 by homology cloning strategy. All of the specific primers are listed in Supplementary Table S1. IbMYB116 was analyzed with online BLAST, ORF Finder, DNAMAN software, Splign tool, and MEGA 7.0, and the cis-acting regulatory elements in its promoter region were defined online (Kang et al., 2018b).
Subcellular Localization of IbMYB116
For subcellular localization, the coding sequence of IbMYB116 was inserted into the pMDC83 vector. The fusion construct (35S::IbMYB116::GFP) and the empty vector (35S::GFP) were separately transformed into the EHA105 strain of Agrobacterium tumefaciens and further infiltrated the leaves of Nicotiana benthamiana (Strasser et al., 2007; Li et al., 2017). After 48 h of growth in a greenhouse, agroinfiltrated leaf sections were imaged at room temperature using a laser scanning confocal microscope with an Argon laser (LSM710, Zeiss, Germany). GFP was excited at 488 nm, and the emitted light was captured at 505–555 nm.
Transactivation Activity Assay of IbMYB116
The transactivation activity of IbMYB116 was tested in yeast (Saccharomyces cerevisiae) (Jiang et al., 2014). The full-length (construct 1) and a series of deletion mutations of IbMYB116 (constructs 2–9) were fused to the yeast expression vector pGBKT7 (pBD). The expression vector pBD-IbMYB116, pGAL4 (positive control), and pBD (negative control) were transferred into the host strain AH109. After the cells were cultured on the SD/-Trp medium, the yeast was further cultured on medium with X-α-Gal (SD/-Trp/-His/X).
Expression Analysis of IbMYB116 in Sweetpotato
The transcript levels of IbMYB116 in the roots, stems, and leaves of the in vitro-grown Xushu55-2 plants were analyzed using quantitative real-time PCR (qRT-PCR), the expression levels were normalized to Ibactin (AY905538), and the relative expression levels in different tissues were calibrated using the roots (Schmittgen and Livak, 2008; Liu et al., 2014). Furthermore, in vitro-grown Xushu55-2 plants were treated in Hoagland solution with H2O, 30% PEG6000, and 100 μM MeJA; the plants were sampled 0, 1, 3, 6, 12, and 24 h after treatments and then analyzed for the expression of IbMYB116. The expression levels were normalized to Ibactin (AY905538), and the relative expression levels in different treatments were calibrated using the plant sampled 0 h after treatment. The specific primers were designed in the nonconserved domain (Figure S1 and Supplementary Table S1).
Production of the Transgenic Arabidopsis Plants
The overexpression vector pC3301-35S-IbMYB116 was generated by inserting the expression cassette 35S-IbMYB116 into pCAMBIA3301, and then, it was transferred into the A. tumefaciens strain GV3101. The transgenic Arabidopsis plants were produced and further grown in pots to obtain T3 seeds as described by Kang et al. (2018b). The transcript levels of IbMYB116 in wild-type and transgenic lines were analyzed using qRT-PCR, and the expression levels were normalized to Atactin (NM112764) (Supplementary Table S1). The relative expression levels in transgenic lines were calibrated using the transgenic line with the lowest IbMYB116 expression.
Assay for Drought Tolerance
For the germination rate assay, T3 and wild-type (WT) seeds sterilized with 2% NaClO for 5 min were sown on 1/2 MS medium with mannitol (0, 100, 200, and 300 mM). Approximately 50 seeds per line were sown for each experiment, and their germination rates were investigated after 3 days. For the root length assay, 5-day-old seedlings of T3 and WT formed on 1/2 MS basal medium were vertically cultured on 1/2 MS medium with 0 (as a control) or 300 mM mannitol, and their roots were took out, and the primary root lengths were measured with a ruler after 3 weeks.
Furthermore, the 7-day-old seedlings obtained on 1/2 MS basal medium were transplanted in pots; after 2 weeks, they were stressed by a 4-week drought followed by 2-day rewatering for investigating their phenotypes. The 6-week normal treatment with water was used as a control.
Expression Analysis of the Related Genes and Measurements of MeJA and H2O2 Contents
T3 and WT plants were pot grown for 2 weeks under drought stress conditions after 2 weeks of normal treatment with water, and they were employed to analyze the expression of the genes involved in JA biosynthesis and signaling pathways as well as the reactive oxygen species (ROS)-scavenging system with qRT-PCR and Atactin (NM112764) as an internal control (Supplementary Table S1), and the relative expression levels in T3 and WT plants were calibrated using WT. Their MeJA and H2O2 contents were determined using high-performance liquid chromatography (HPLC) and an H2O2 Assay Kit (Comin Biotechnology Co., Ltd. Suzhou, China), respectively. The 4-week normal treatment with water was used as a control.
Statistical Analysis
Three biological replicates were performed for all experiments. Difference analysis of data presented as the mean ± SE was done with Student’s t-test (two-tailed analysis) using SPSS 20.0 Statistic Program. Significance levels at P < 0.05, P < 0.01, and P < 0.001 were indicated with ∗, ∗∗, and ∗∗∗, respectively.
Results
Cloning and Sequence Analysis of IbMYB116 and Its Promoter
The IbMYB116 full-length cDNA was 958 bp in length and contained an 849-bp ORF that encoded a polypeptide of 282 residues with a predicted molecular weight of 31.998 kDa and a pI of 6.79. The IbMYB116 protein had two conserved MYB domains that belonged to R2R3-MYB TFs and shared a high-sequence identity with MYB TFs from Ipomoea nil (InMYB21, XP_019189643.1, 91.23%), Glycine max (GmMYB184, NP_001235837.1, 49.12% and GmMYB84, NP_001235789.1, 43.83%), and Arabidopsis thaliana (AtMYB116, AT1G25340.1, 43.24%) (Figure 1A). The protein was characterized with a highly conserved DNA-binding sequence at the N-terminus (Figure 1A). Phylogenetic analysis along with 125 Arabidopsis R2R3-MYB TFs belonging to 24 groups (Dubos et al., 2010) revealed that IbMYB116 belonged to group S20 of the MYB family and had a close relationship with AtMYB116 (At1G25340.1) (Figure 1B). The genomic DNA of IbMYB116 was 1,498 bp with three exons and two introns (Figure 1C). Its promoter region (∼1,455 bp) contained the cis-acting regulatory elements associated with stresses and phytohormones, including LTR, HSE, MBS, GARE-motif, CGTCA-motif, TGACG-motif, P-box, and SARE (Figure S2).
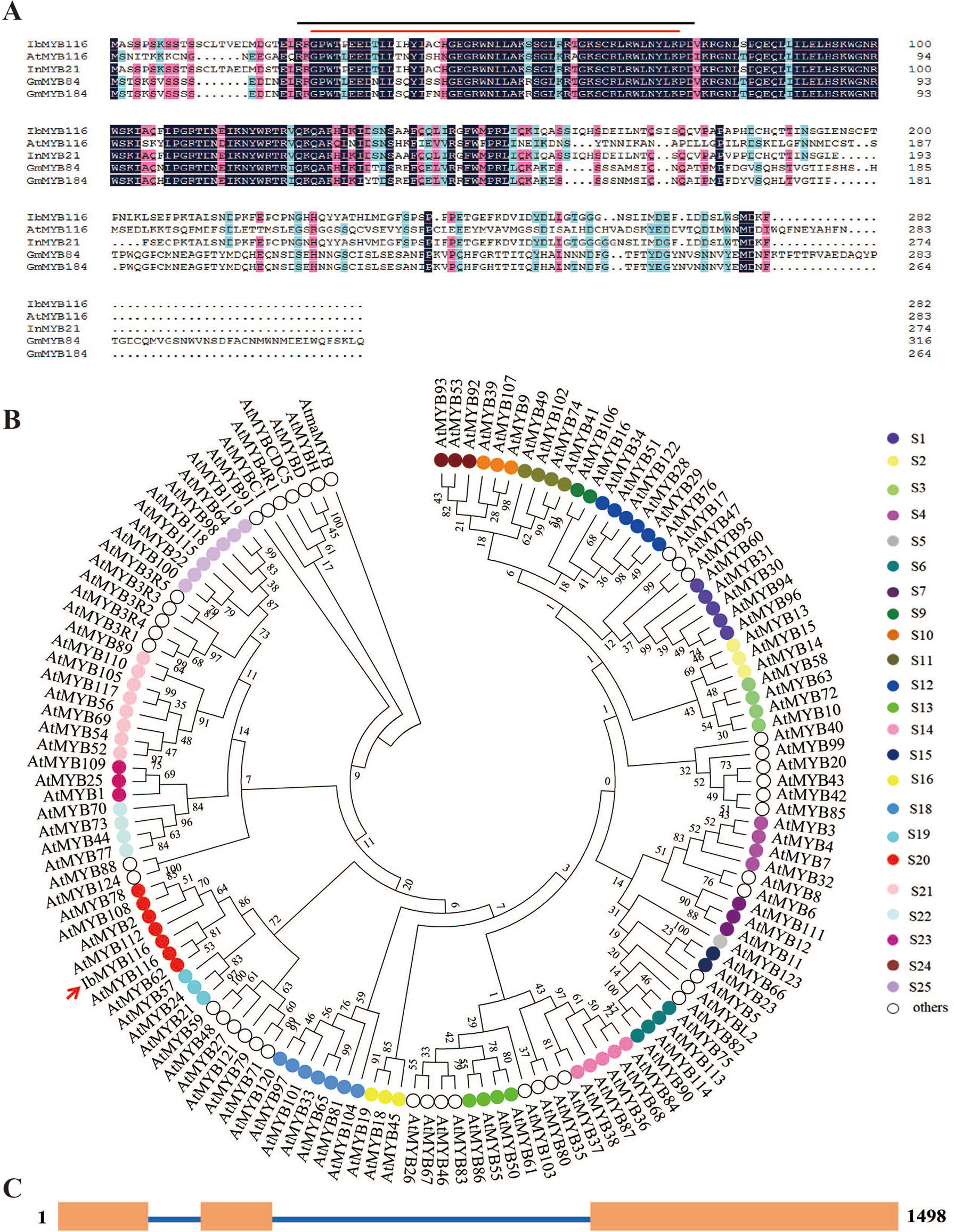
Figure 1 Sequence analysis of IbMYB116. (A) Sequence alignment of IbMYB116 with its homologues from other plants. Conserved amino acids are indicated by a dark background. Characteristic regions of MYB are indicated above the IbMYB116 sequence. —, R2 SANT domain; —, R3 SANT domain. (B) Phylogenetic analysis of IbMYB116 and MYB transcription factors from Arabidopsis thaliana. (C) Exon and intron constituents of IbMYB116. Exons are represented by boxes and introns by lines.
Subcellular Location of IbMYB116
IbMYB116-GFP showed fluorescence in the nuclei of Nicotiana benthamiana leaf hypodermal cells, while the fluorescence of GFP was distributed in the entire cell (Figure 2). These results clearly indicated that IbMYB116 is a nuclear protein.
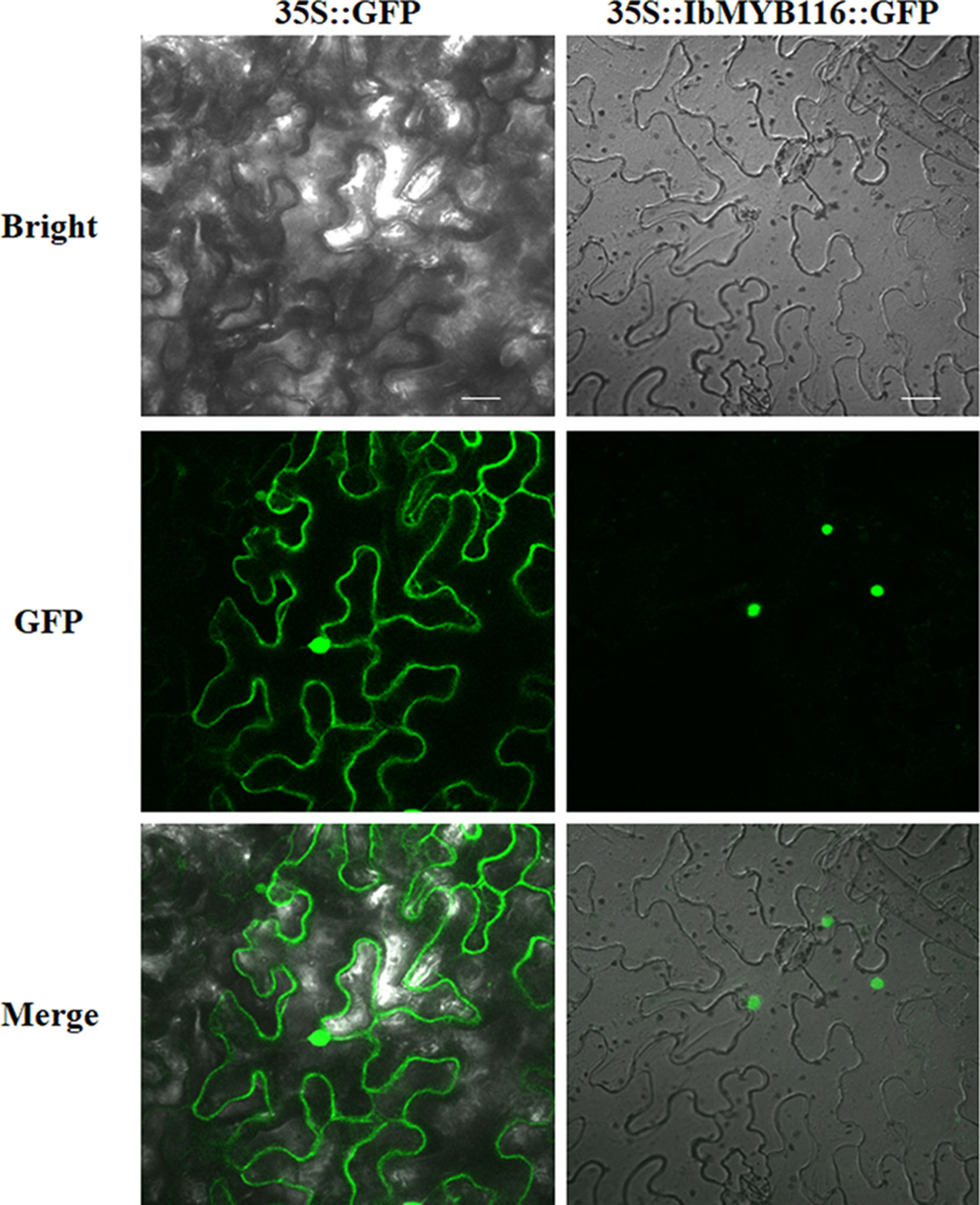
Figure 2 Subcellular localization of IbMYB116 in tobacco leaf hypodermal cells. Confocal scanning microscopic images show localization of IbMYB116-GFP fusion proteins to nuclei in the right column vs. GFP as the control in the left column. Bars = 20 µm.
Transactivation Activity of IbMYB116 in Yeast
All of the transformed yeast cells grew well on the SD/-Trp medium, which indicated that expression vectors (pBD-IbMYB116-1, -2, -3, -4, -5, -6, -7, -8, -9; Figure 3A) had been successfully transferred into yeast cells (Figure 3B). The yeast cells harboring pBD-IbMYB116-1, -4, -5, -6, -7, and pGAL4 (positive control) grew well and turned blue, but the cells with pBD-IbMYB116-2, -3, -8, -9, and pBD (negative control) failed to grow on medium with X-α-Gal (SD/-Trp/-His/X) (Figure 3C). These results demonstrated that IbMYB116 contained the transactivation activity domain at the region of amino acids 215–282.
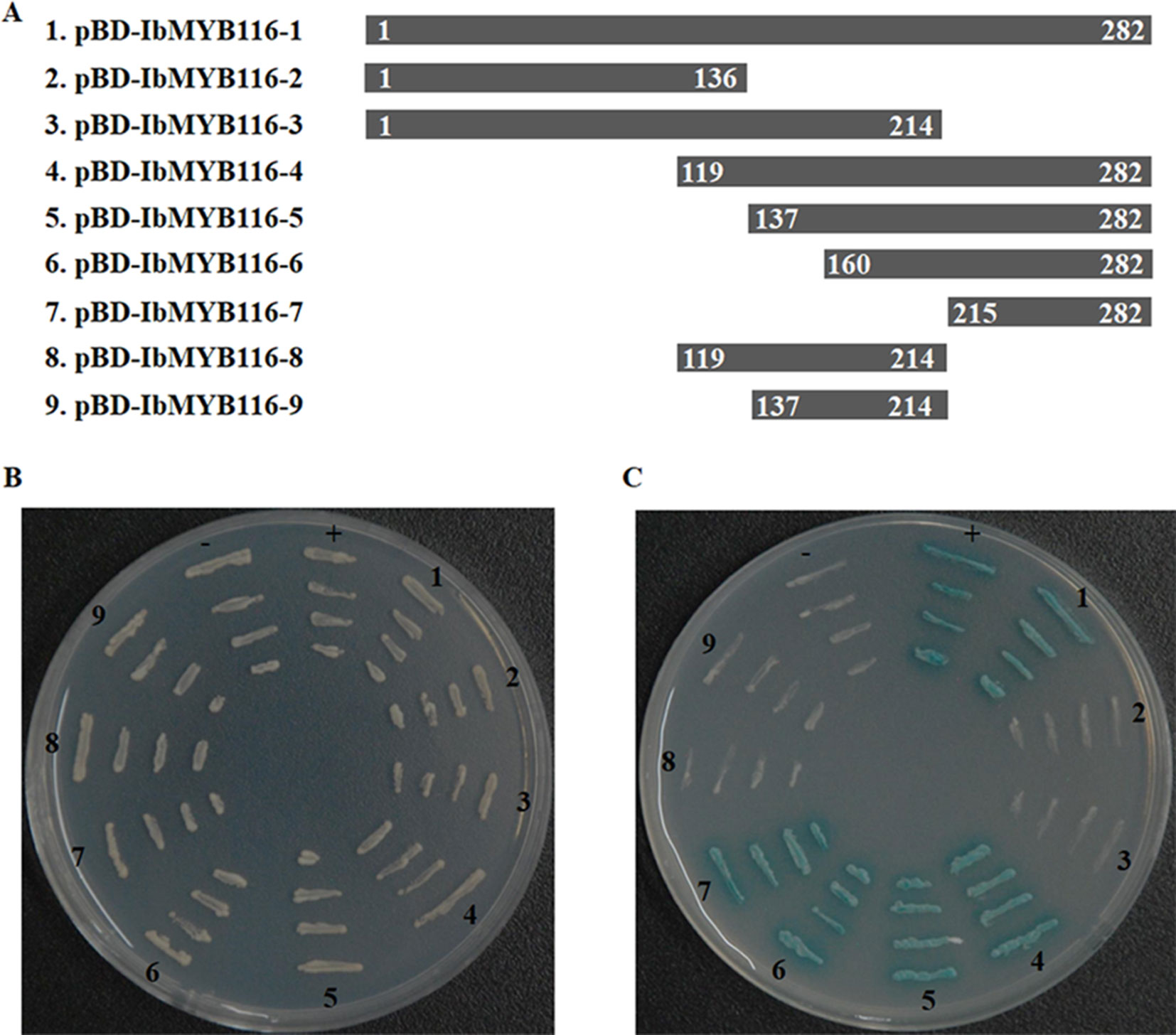
Figure 3 Transactivation activity assay of IbMYB116 in yeast. (A) Diagram showing a full-length construct (construct 1) and a series of deletion mutations of IbMYB116 (constructs 2–9). (B) The transformed yeast cells harboring different expression vectors (constructs 1–9) were drawn onto SD/-Trp medium. pBD (–) and pGAL4 (+) were used as negative and positive controls, respectively. (C) The transformed yeast cells harboring different expression vectors (constructs 1–9) were drawn onto SD/-Trp/-His medium supplemented with X-α-Gal. pBD (−) and pGAL4 (+) were used as negative and positive controls, respectively.
Expression of IbMYB116 in Sweetpotato
A significantly higher expression level of IbMYB116 was found in the leaves of Xushu55-2 than that in the roots and stems (Figure S3). Its expression in Xushu55-2 was strongly induced by 30% PEG6000 and 100 μM MeJA, and it reached the highest level at 12 h (4.78-fold) and 24 h (4.15-fold), respectively (Figure 4).
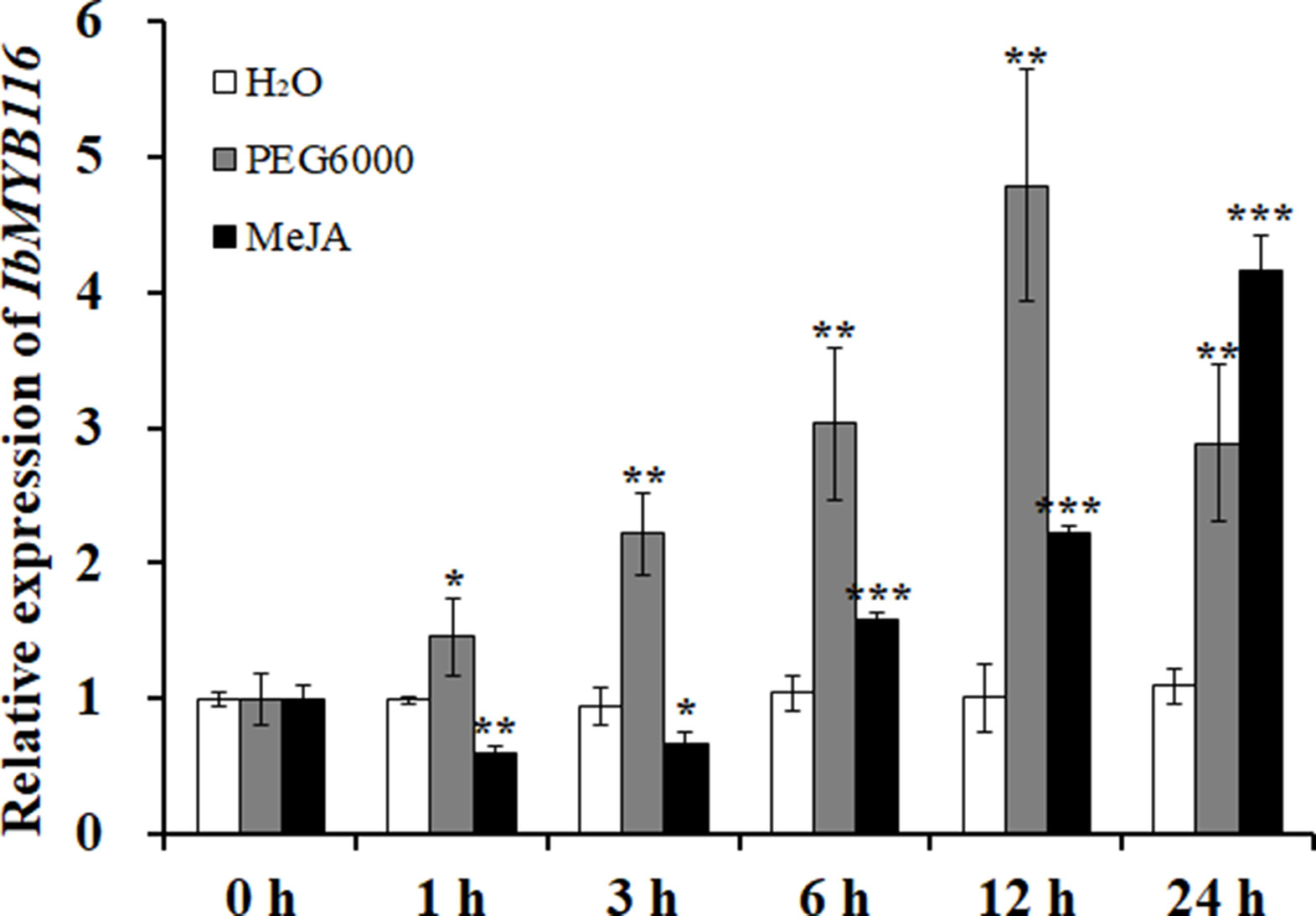
Figure 4 Expression analysis of IbMYB116 in the in vitro-grown plants of Xushu55-2 after different times (h) in response to H2O, 30% PEG6000, and 100 µM MeJA, respectively. Data are presented as the means ± SE (n = 3). *, ** and *** indicate significant differences from WT at P <0.05, P <0.01, and P <0.001, respectively, according to the Student’s t-test.
Production of the Transgenic Arabidopsis Plants
The transgenic Arabidopsis plants were obtained using the protocols of Kang et al. (2018b). Eight transgenic plants named L1, L2, …, L8 were randomly selected to analyze the expression level of IbMYB116 with qRT-PCR (Figure S1). The results showed that all of the transgenic plants exhibited significantly higher expression levels of IbMYB116 than the WT (Figure S4).
Enhanced Drought Tolerance
Three transgenic Arabidopsis plants (L1, L2, and L3) with high expression levels of IbMYB116 were selected for investigating their drought tolerance. The transgenic and WT plants showed no differences in germination rates on 1/2 MS media with 0 (control) and 100 mM mannitol (Figure 5). However, the transgenic plants provided significantly higher germination rates than the WT under 200 and 300 mM mannitol stress conditions (Figure 5). The transgenic and WT plants showed similar growth on 1/2 MS media without mannitol, but the transgenic plants formed significantly longer roots than the WT at the level of 300 mM mannitol (Figure 6).
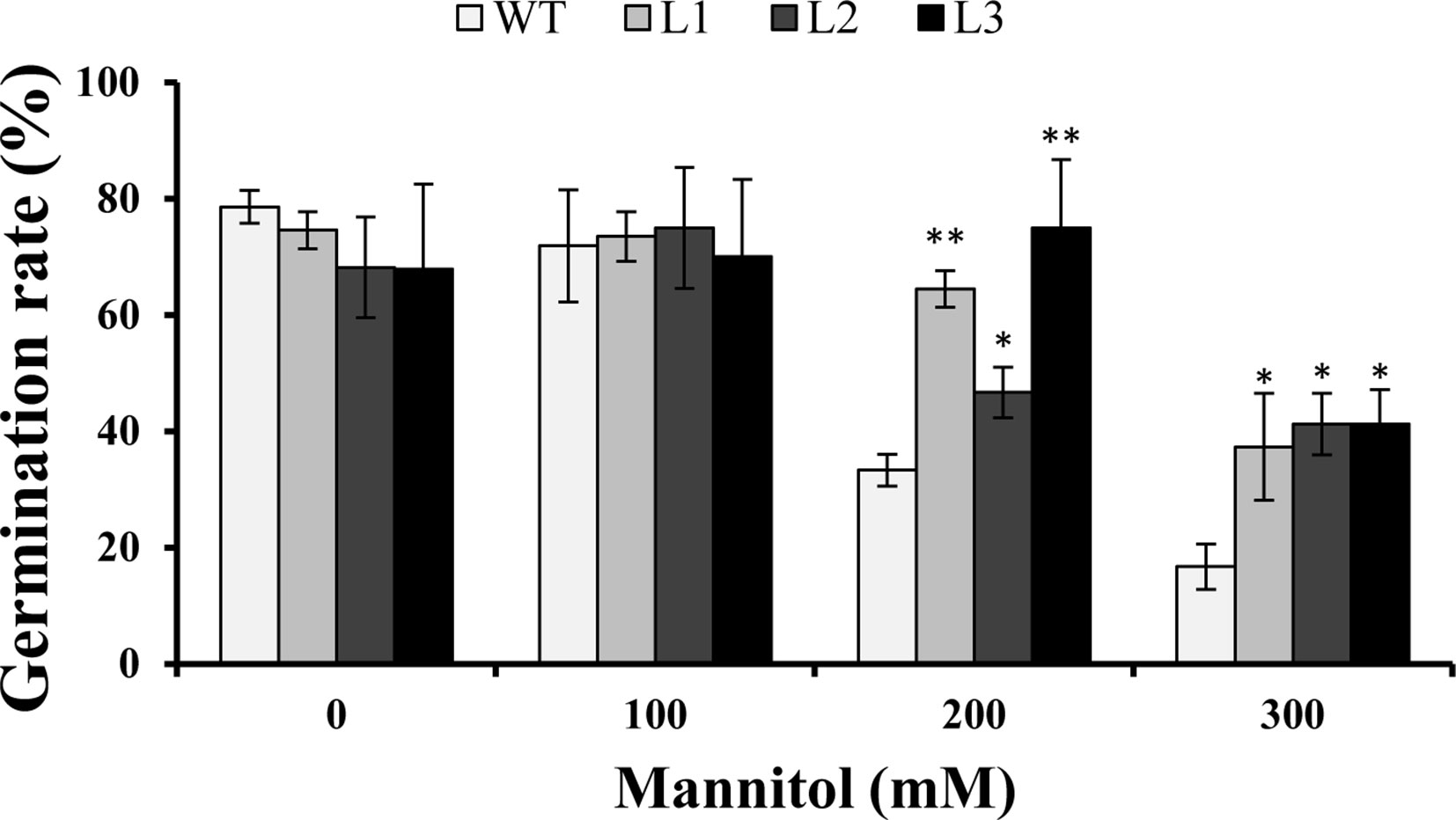
Figure 5 Germination assay of the transgenic Arabidopsis and WT seeds sown for 3 days on 1/2 MS medium with 0, 100, 200, and 300 mM mannitol, respectively. * and ** indicate significant differences from WT at P < 0.05 and P < 0.01, respectively, according to the Student’s t-test.
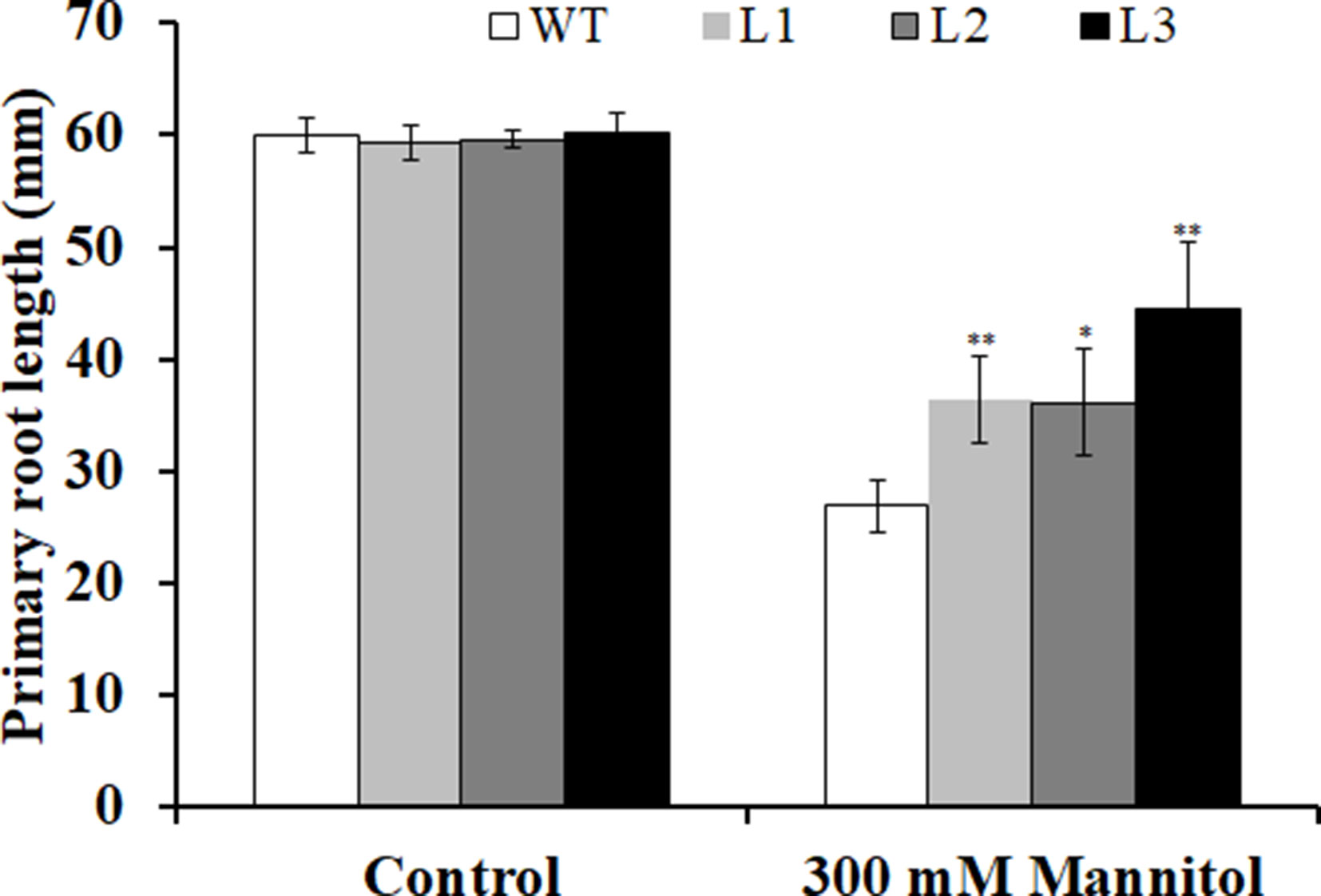
Figure 6 Primary root lengths of the transgenic Arabidopsis and WT seedlings cultured for 3 weeks on 1/2 MS medium without (control) and with 300 mM mannitol, respectively. All the plants were grown in a standard square plate of 12 × 12 cm. Data are presented as the mean ± SE (n = 3). * and ** indicate significant differences from WT at P < 0.05 and P < 0.01, respectively, according to the Student’s t-test.
Furthermore, the transgenic and WT plants grown in pots had no differences in growth without drought stress (Figure 7A). However, after being stressed by drought, the transgenic plants exhibited better growth, increased MeJA content, and decreased H2O2 content compared with the WT (Figures 7A, B). These results indicated that the transgenic Arabidopsis plants had significantly enhanced drought tolerance compared with the WT.
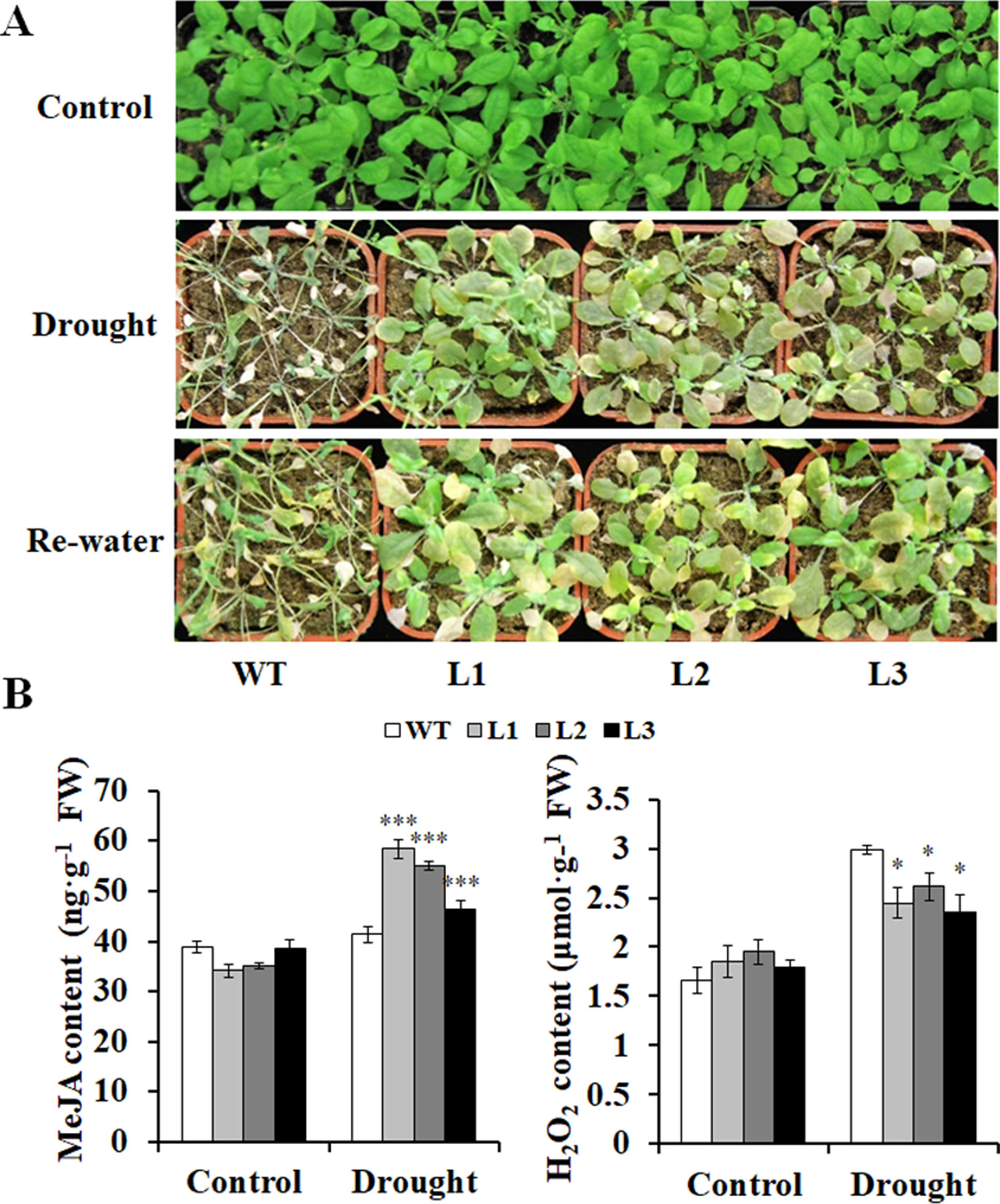
Figure 7 Responses of the transgenic Arabidopsis plants and WT to drought stress. (A) Phenotypes of the transgenic plants vs. WT stressed by a 4-week drought followed by 2 days rewatering after 2 weeks of normal treatment with water. The 6-week normal treatment was used as a control. (B) Contents of MeJA and H2O2 in the leaves of the transgenic and WT plants grown for 4 weeks under normal treatment (control) and 2 weeks under drought stress after 2 weeks of normal treatment, respectively. *, **, and *** indicate significant differences from WT at P < 0.05, P < 0.01, and P < 0.001, respectively, according to the Student’s t-test.
Expression of the Related Genes
Systematic upregulation of the JA biosynthesis key enzyme genes AtLOX, AtAOS, AtAOC, AtOPR, AtOPCL, AtACOX3, AtfadA, and AtACAA1, except for AtACOX1 (no change) and AtMFP2 (downregulated), was found in the transgenic Arabidopsis plants under drought stress conditions (Figure 8A). In the JA signaling pathway, AtCOI1 and AtJAZ were downregulated, but AtMYC2 was upregulated under drought stress conditions (Figure 8B). The genes encoding ROS scavenging enzymes superoxide dismutase (SOD), glutathione peroxidase (GPX), and peroxidase (POD) were also upregulated, and both AtCAT and AtDHAR apparently had not changed (Figure 8C).
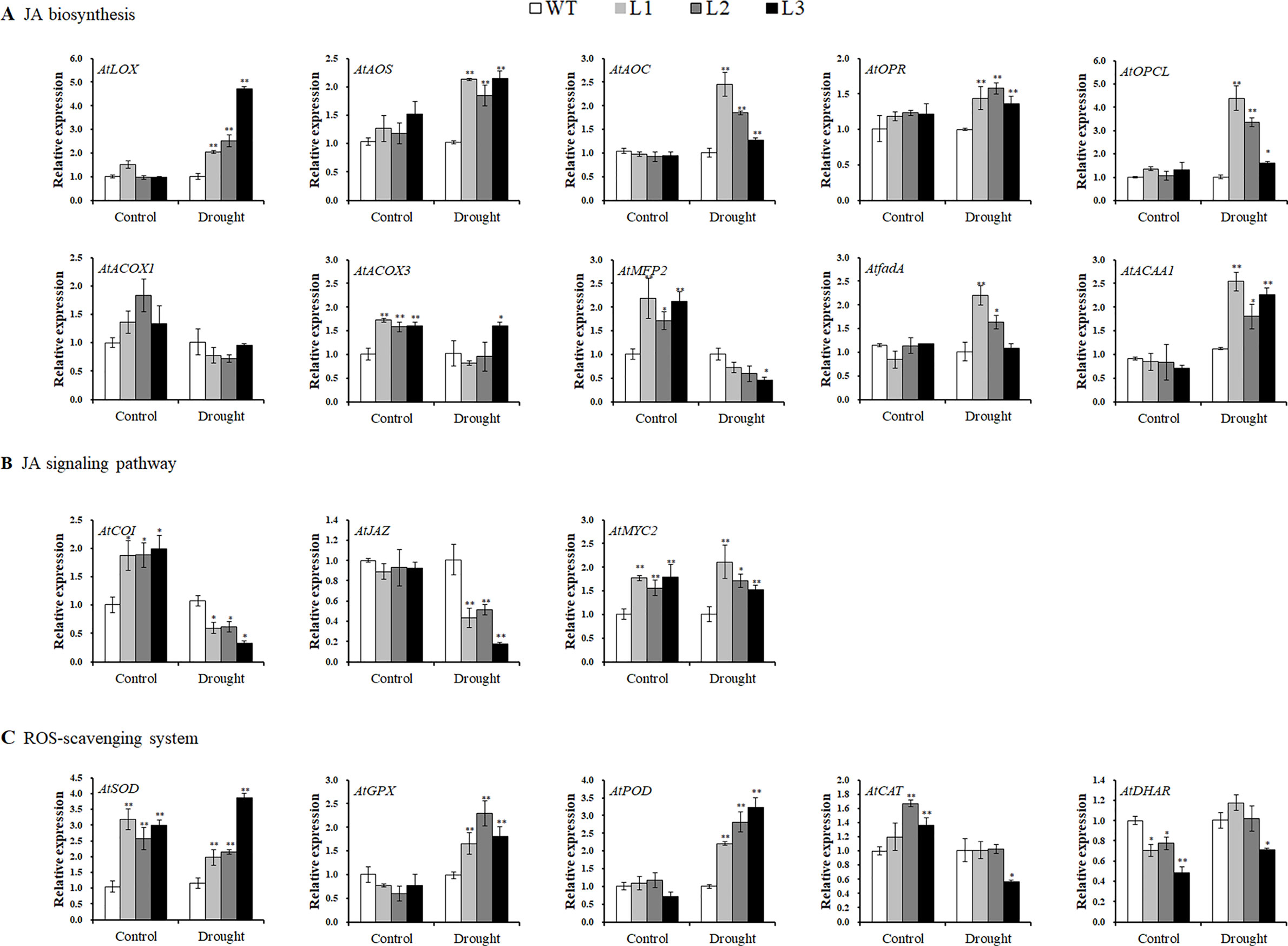
Figure 8 Transcript levels of the related genes in the leaves of the transgenic Arabidopsis and WT plants pot grown for 4 weeks under normal treatment with water (control) and 2 weeks under drought stress after 2 weeks of normal treatment, respectively. (A) Transcript levels of JA biosynthesis genes. (B) Transcript levels of JA signaling pathway genes. (C) Transcript levels of ROS-scavenging system genes. * and ** indicate significant differences from WT at P < 0.05 and P < 0.01, respectively, according to the Student’s t-test.
Discussion
IbMYB116 Enhances Drought Tolerance
It has been shown that several MYB TFs play crucial roles in the response to drought stress as positive stress response TFs (Rusconi et al., 2013; Lee et al., 2014; Yuan et al., 2015; Butt et al., 2017; Wang et al., 2017b). To date, the role of MYB116 in plants is still unknown. In this study, we cloned the IbMYB116 gene from the drought-tolerant sweetpotato line Xushu55-2 and found that the IbMYB116 protein belonged to R2R3-MYB TFs (Figure 1) (Jia et al., 2004; He et al., 2016). This gene was strongly induced by PEG6000 and MeJA, and its overexpression significantly improved the drought tolerance of the transgenic Arabidopsis plants (Figures 4–7). This study reveals, for the first time, the crucial role of IbMYB116 in the drought tolerance of plants.
IbMYB116 Upregulates JA Signaling Pathway Genes
JAs are naturally occurring signaling compounds that regulate plant responses to abiotic stresses, such as drought, salt, and ozone exposure (Rojo et al., 2003; Ahmad et al., 2016; Dhakarey et al., 2017). Rohwer and Erwin (2008) found that JA and JA-induced protein levels were increased in sorbitol-treated barley leaves, which confirmed the role of JAs in osmotic stress. MeJA enhanced drought tolerance by improving the water status of wheat plants (Ma et al., 2004) or increasing antioxidant activities in soybean plants (Anjum et al., 2011). In Arabidopsis, four 13-LOX forms (AtLOX2, 3, 4, and 6) contributed to JA formation in the stress response (Caldelari et al., 2011; Chauvin et al., 2013). The upregulation of OsLOX, OsAOS2, and OsOPR7 led to massive accumulation of endogenous JA in the rice rim1 mutant (Yoshii et al., 2010). The tolerance to drought as well as osmotic and salinity changes were increased in the Arabidopsis plants overexpressing CaLOX1 from pepper (Lim et al., 2015). The TaAOC1-overexpressing wheat and Arabidopsis plants accumulated more JA and exhibited enhanced tolerance to changes in salinity, which indicated that JA is related to the plant salinity response (Zhao et al., 2014). The expression of OsOPR7 was induced by drought stress and wounding, and the increase in OsOPR7 expression led to an elevated endogenous JA level (Tani et al., 2008).
The JA signaling pathway plays an important role in regulating the plant response to drought stress (Liu et al., 2015; Ahmad et al., 2016). As components of the core JA signaling pathway, COI1 participates in removing repressors of JA transduction (Xie et al., 1998; Ahmad et al., 2016); JAZs are transcription repressors of JA-responsive genes (Chini et al., 2007), and MYC2 acts as a positive regulator of JA signals (Zhang et al., 2015b). The overexpression of AtMYC2 in Arabidopsis improved osmotic stress tolerance (Abe et al., 2003). The OsMYC2-overexpressing rice exhibited increased resistance against Xoo (Uji et al., 2016).
Our results showed that overexpression of IbMYB116 in Arabidopsis systematically upregulated the JA biosynthesis genes, including AtLOX, AtAOS, AtAOC, AtOPR, AtOPCL, AtCOX3, AtfadA, and AtACAA1, and the MeJA content increased under drought stress conditions (Figures 7, 8). AtCOI1 and AtJAZ were downregulated, and AtMYC2 was upregulated in the transgenic Arabidopsis plants under drought stress conditions (Figure 8). These findings suggest that IbMYB116 enhances drought tolerance through the JA signaling pathway in transgenic Arabidopsis (Figure 9).
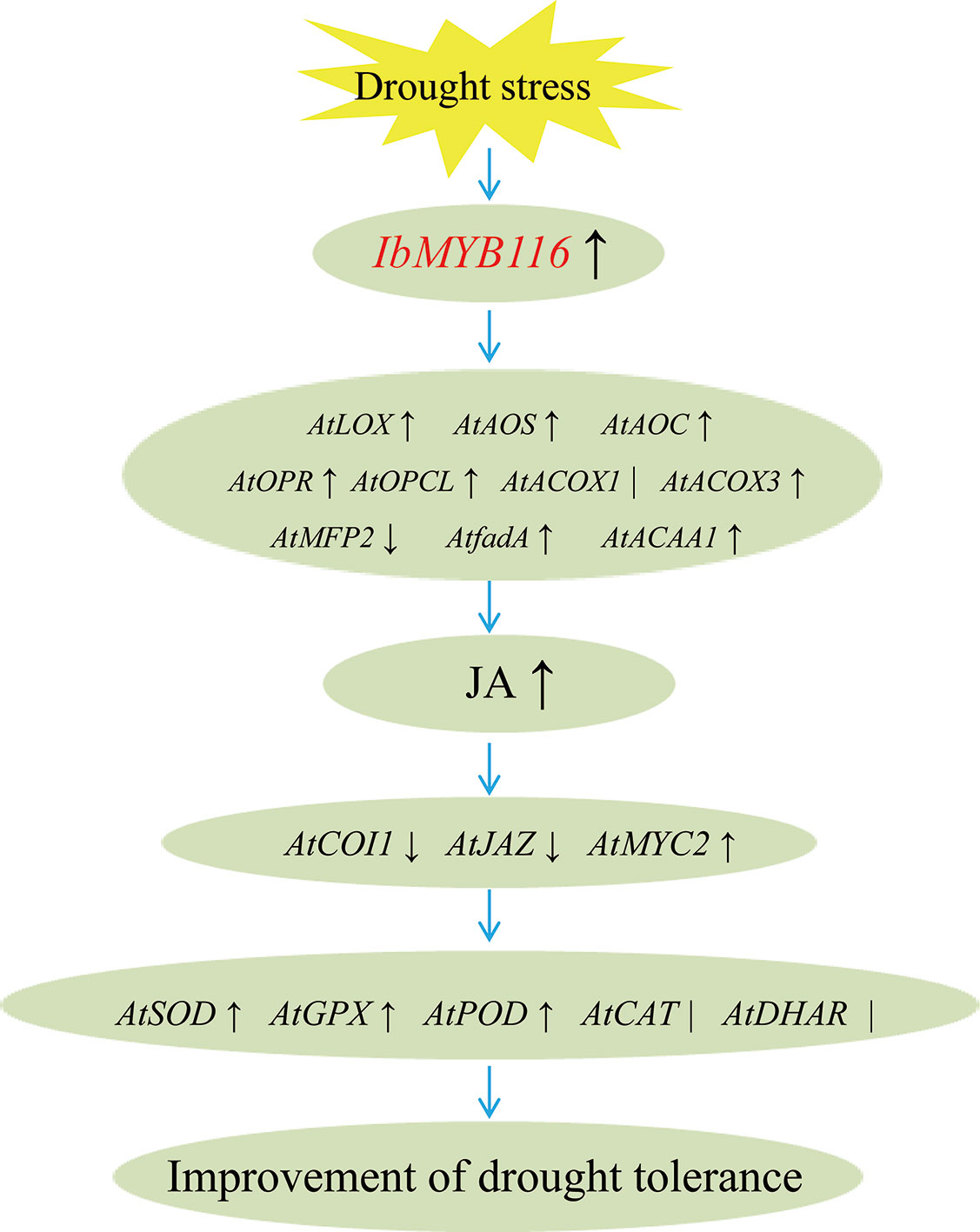
Figure 9 Diagram showing a proposed model for the regulation of IbMYB116 in drought tolerance for transgenic Arabidopsis. ↑, ↓and ∣ indicate upregulation, downregulation, and no obvious change of expression for genes coding these enzymes, respectively.
IbMYB116 Activates the ROS-Scavenging System
In plants, ROS cause damage to the structure and function of biomolecules, which leads to oxidative stress. H2O2, as one of the prominent ROS, is considered to be an important signaling molecule in plant cells (Wang et al., 2017a). The ROS-scavenging system can detoxify ROS to reduce oxidative damage in plant cells (Gill and Tuteja, 2010; Liu et al., 2014). It has been shown that JA can activate the ROS-scavenging system in plants. The overexpression of CaLOX1 in Arabidopsis systematically upregulated the ROS-scavenging system genes, which resulted in the reduced H2O2 level under drought and salt stress conditions (Lim et al., 2015). The JA-deficient tomato mutant def-1 showed higher ROS levels than the WT under salt stress conditions (Abouelsaad and Renault, 2018). JA influenced oxidative stress through its direct effect on the activities of ROS-scavenging enzymes in raspberry and strawberry (Wang, 1999; Ghasemnezhad and Javaherdashti, 2008). Reduced levels of JA led to more GLN18:3-induced ROS production in Arabidopsis and tomato (Block et al., 2018).
The results of this study demonstrated that ROS-scavenging system genes, including AtSOD, AtGPX, and AtPOD, were upregulated and that the H2O2 level was decreased in the IbMYB116-overexpressing Arabidopsis plants under drought stress conditions (Figures 7, 8). These results suggest that IbMYB116 might enhance drought tolerance by activating the ROS-scavenging system through the JA signaling pathway in transgenic Arabidopsis (Figure 9).
Conclusion
We found a crucial role for IbMYB116 in the drought tolerance of plants. The results suggest that the overexpression of IbMYB116 might enhance drought tolerance by activating the ROS-scavenging system through the JA signaling pathway in transgenic Arabidopsis. This gene has the potential to be used for improving the drought tolerance of sweetpotato and other plants.
Author Contributions
QL and YZ conceived and designed the experiments. YZ and HZu performed the experiments. YZ, SH, and HZa analyzed the data. QL, NZ, SX, and ZW contributed reagents, materials, and analysis tools. QL and YZ wrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31461143017) and the China Agriculture Research System (CARS-10, Sweetpotato).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01025/full#supplementary-material
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Abouelsaad, I., Renault, S. (2018). Enhanced oxidative stress in the jasmonic acid-deficient tomato mutant def-1 exposed to NaCl stress. J. Plant Physiol. 226, 136–144. doi: 10.1016/j.jplph.2018.04.009
Ahmad, P., Rasool, S., Gul, A., Sheikh, S. A., Akram, N. A., Ashraf, M., et al. (2016). Jasmonates: multifunctional roles in stress tolerance. Front. Plant Sci. 7, 813–827. doi: 10.3389/fpls.2016.00813
Anjum, S., Xie, X. Y., Wang, L. C., Saleem, M. F., Man, C., Wang, L. (2011). Morphological, physiological and biochemical responses of plants to drought stress. Afr. J. Agri. Res. 6, 2026–2032. doi: 10.5897/AJAR10.027
Aoyagi, L. N., Lopes-Caitar, V. S., de Carvalho, M., Darben, L. M., Polizel-Podanosqui, A., Kuwahara, M. K., et al. (2014). Genomic and transcriptomic characterization of the transcription factor family R2R3-MYB in soybean and its involvement in the resistance responses to Phakopsora pachyrhizi. Plant Sci. 229, 32–42. doi: 10.1016/j.plantsci.2014.08.005
Berger, S. (2002). Jasmonate-related mutants of Arabidopsis as tools for studying stress signaling. Planta 214, 497–504. doi: 10.1007/s00425-001-0688-y
Block, A., Christensen, S. A., Hunter, C. T., Alborn, H. T. (2018). Herbivore-derived fatty-acid amides elicit reactive oxygen species burst in plants. J. Exp. Bot. 69, 1235–1245. doi: 10.1093/jxb/erx449
Bohnert, H. J., Nelson, D. E., Jensen, R. G. (1995). Adaptations to environmental stresses. Plant Cell 7, 1099–1111. doi: 10.1105/tpc.7.7.1099
Boyer, J. S., Byrne, P., Cassman, K. G., Cooper, M., Delmer, D., Greene, T., et al. (2013). The US drought of 2012 in perspective: a call to action. Glob. Food Secur-Agr. 2, 139–143. doi: 10.1016/j.gfs.2013.08.002
Butt, H. I., Yang, Z., Gong, Q., Chen, E., Wang, X., Zhao, G., et al. (2017). GaMYB85, an R2R3 MYB gene, in transgenic Arabidopsis plays an important role in drought tolerance. BMC Plant Biol. 17, 142–158. doi: 10.1186/s12870-017-1078-3
Caldelari, D., Wang, G., Farmer, E. E., Dong, X. (2011). Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 75, 25–33. doi: 10.1007/s11103-010-9701-9
Chauvin, A., Caldelari, D., Wolfender, J. L., Farmer, E. E. (2013). Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: a role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 197, 566–575. doi: 10.1111/nph.12029
Chen, Y. H., Zhang, X. B., Wu, W., Chen, Z. L., Gu, H. Y., Qu, L. J., et al. (2006). Overexpression of the wounding-responsive gene AtMYB15 activates the shikimate pathway in Arabidopsis. J. Integr Plant Biol. 48, 1084–1095. doi: 10.1111/j.1744-7909.2006.00311.x
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Denekamp, M., Smeekens, S. C. (2003). Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol. 132, 1415–1423. doi: 10.1104/pp.102.019273
Dhakarey, R., Raorane, M. L., Treumann, A., Peethambaran, P. K., Schendel, R. R., Sahi, V. P., et al. (2017). Physiological and proteomic analysis of the rice mutant cpm2 suggests a negative regulatory role of jasmonic acid in drought tolerance. Front. Plant Sci. 8, 1903–1919. doi: 10.3389/fpls.2017.01903
Dong, Y. P., Fan, G. Q., Zhao, Z. L., Deng, M. J. (2014). Compatible solute, transporter protein, transcription factor, and hormone-related gene expression provides an indicator of drought stress in Paulownia fortunei. Funct. Integr. Genomic 14, 479–491. doi: 10.1007/s10142-014-0373-4
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Fan, W. J., Wang, H. X., Wu, Y. L., Yang, N., Yang, J., Zhang, P. (2017). H (+) -pyrophosphatase IbVP1 promotes efficient iron use in sweetpotato [Ipomoea batatas (L.) Lam.]. Plant Biotechnol. J. 15, 698–712. doi: 10.1111/pbi.12667
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488. doi: 10.1105/tpc.105.035659
Ghasemnezhad, M., Javaherdashti, M. (2008). Effect of methyl jasmonate treatment on antioxidant capacity internal quality and postharvest life of raspberry on antioxidant capacity internal quality and postharvest life of raspberry fruit. Caspian J. Environ. Sci. 6, 73–78.
Gill, S. S., Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Hajiebrahimi, A., Owji, H., Hemmati, S. (2017). Genome-wide identification, functional prediction, and evolutionary analysis of the R2R3-MYB superfamily in Brassica napus. Genome 60, 797–814. doi: 10.1139/gen-2017-0059
He, Q., Jones, D. C., Li, W., Xie, F., Ma, J., Sun, R., et al. (2016). Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii. Sci. Rep. 6, 22980–22993. doi: 10.1038/srep22980
Jia, L., Clegg, M. T., Jiang, T. (2004). Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 134, 575–585. doi: 10.1104/pp.103.027201
Jiang, X. Q., Zhang, C. Q., Lu, P. T., Jiang, G. M., Liu, X. W., Dai, F. W., et al. (2014). RhNAC3, a stress-associated NAC transcription factor, has a role in dehydration tolerance through regulating osmotic stress-related genes in rose petals. Plant Biotechnol. J. 12, 38–48. doi: 10.1111/pbi.12114
Jin, Y., Zheng, H. Y., Jin, X. J., Zhang, Y. X., Ren, C. Y. (2016). Effect of ABA, SA and JA on soybean growth under drought stress and re-watering. Soybean Sci. s565, 1. doi: 10.11861/j.issn.1000-9841
Jung, C., Seo, J. S., Han, S. W., Koo, Y. J., Kim, C. H., Song, S. I., et al. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146, 623–635. doi: 10.1104/pp.107.110981
Kang, C., He, S. Z., Zhai, H., Li, R. J., Zhao, N., Liu, Q. C. (2018b). A sweetpotato auxin response factor gene (IbARF5) is involved in carotenoid biosynthesis and salt and drought tolerance in transgenic Arabidopsis. Front. Plant Sci. 9, 1307. doi: 10.3389/fpls.2018.01307
Kang, C., Zhai, H., Xue, L. Y., Zhao, N., He, S. Z., Liu, Q. C. (2018a). A lycopene β-cyclase gene, IbLCYB2, enhances carotenoid contents and abiotic stress tolerance in transgenic sweetpotato. Plant Sci. 272, 243–254. doi: 10.1016/j.plantsci.2018.05.005
Kim, H. S., Ji, C. Y., Lee, C. J., Kim, S. E., Park, S. C., Kwak, S. S. (2018). Orange: a target gene for regulating carotenoid homeostasis and increasing plant tolerance to environmental stress in marginal lands. J. Exp. Bot. 69, 3393–3400. doi: 10.1093/jxb/ery023
Lee, S. B., Kim, H., Kim, R. J., Suh, M. C. (2014). Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Rep. 33, 1535–1546. doi: 10.1007/s00299-014-1636-1
Li, Y., Wang, Y. N., Zhang, H., Zhang, Q., Zhai, H., Liu, Q. C., et al. (2017). The plasma membrane-localized sucrose transporter IbSWEET10 contributes to the resistance of sweet potato to Fusarium oxysporum. Front. Plant Sci. 8, 197–211. doi: 10.3389/fpls.2017.00197
Liang, Y. K., Dubos, C., Dodd, I. C., Holroyd, G. H., Hetherington, A. M., Campbell, M. M. (2005). AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr. Biol. 15, 1201–1206. doi: 10.1016/j.cub.2005.06.041
Lim, C. W., Han, S. W., Hwang, I. S., Kim, D. S., Hwang, B. K., Lee, S. C. (2015). The pepper lipoxygenase CaLOX1 plays a role in osmotic, drought and high salinity stress response. Plant Cell Physiol. 56, 930–942. doi: 10.1093/pcp/pcv020
Liu, D. G., He, S. Z., Zhai, H., Wang, L. J., Zhao, Y., Wang, B., et al. (2014). Overexpression of IbP5CR enhances salt tolerance in transgenic sweetpotato. Plant Cell Tiss. Org. Cult. 117, 1–16. doi: 10.1007/s11240-013-0415-y
Liu, N., Avramova, Z. (2016). Molecular mechanism of the priming by jasmonic acid of specific dehydration stress response genes in Arabidopsis. Epigenet. Chromatin 9, 8–30. doi: 10.1186/s13072-016-0057-5
Liu, Z., Zhang, S. M., Sun, N., Liu, H. Y., Zhao, Y. H., Liang, Y. L., et al. (2015). Functional diversity of jasmonates in rice. Rice (N Y) 8, 42–54. doi: 10.1186/s12284-015-0042-9
Lorenzo, O., Piqueras, R., Sanchez-Serrano, J. J., Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Ma, C., Wang, Z. Q., Zhang, L. T., Sun, M. M., Lin, T. B. (2004). Photosynthetic responses of wheat (Triticum aestivum L.) to combined effects of drought and exogenous methyl jasmonate. Photosynthetica 3, 377–385. doi: 10.1007/s11099-014-0041-x
Mueller, M. J. (1997). Enzymes involved in jasmonic acid biosynthesis. Physiol. Plant 100:, 653–663. doi: 10.1111/j.1399-3054.1997.tb03072.x
Overmyer, K., Tuominen, H., Kettunen, R., Betz, C., Langebartels, C., Sandermann, H. J., et al. (2000). Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12, 1849–1862. doi: 10.1105/tpc.12.10.1849
Park, S. C., Kim, Y. H., Jeong, J. C., Kim, C. Y., Lee, H. S., Bang, J. W., et al. (2011). Sweetpotato late embryogenesis abundant 14 (IbLEA14) gene influences lignification and increases osmotic- and salt stress-tolerance of transgenic calli. Planta 233, 621–634. doi: 10.1007/s00425-010-1326-3
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P. A., Saedler, H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6, 3558–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C. Z., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Rohwer, C. L., Erwin, J. E. (2008). Horticultural applications of jasmonates. J. Hortic. Sci. Biotechnol. 83, 283–304. doi: 10.1016/j.biotechadv.2013.09.009
Rojo, E., Solano, R., Sanchez-Serrano, J. J. (2003). Interactions between signaling compounds involved in plant defense. J. Plant Growth Reg. 22, 82–98. doi: 10.1007/s00344-003-0027-6
Rusconi, F., Simeoni, F., Francia, P., Cominelli, E., Conti, L., Riboni, M., et al. (2013). The Arabidopsis thaliana MYB60 promoter provides a tool for the spatio-temporal control of gene expression in stomatal guard cells. J. Exp. Bot. 64, 3361–3371. doi: 10.1093/jxb/ert180
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shiriga, K., Sharma, R., Kumar, K., Yadav, S. K., Hossain, F., Thirunavukkarasu, N. (2014). Genome-wide identification and expression pattern of drought-responsive members of the NAC family in Maize. Meta Gene. 2, 407–417. doi: 10.1016/j.mgene.2014.05.001
Smita, S., Katiyar, A., Chinnusamy, V., Pandey, D. M., Bansal, K. C. (2015). Transcriptional regulatory network analysis of MYB transcription factor family genes in rice. Front. Plant Sci. 6, 1157–1175. doi: 10.3389/fpls.2015.01157
Strasser, R., Singh, J., Bondili, J. S., Schoberer, J., Svoboda, B., Liebminger, E., et al. (2007). Enzymatic properties and subcellular localization of Arabidopsisβ-N-acetylhexosaminidases. Plant Physiol. 145, 5–16. doi: 10.1104/pp.107.101162
Su, L. T., Li, J. W., Liu, D. Q., Zhai, Y., Zhang, H. J., Li, X. W., et al. (2014). A novel MYB transcription factor, GmMYBJ1, from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene 538, 46–55. doi: 10.1016/j.gene.2014.01.024
Su, L. T., Wang, Y., Liu, D. Q., Li, X. W., Zhai, Y., Sun, X., et al. (2015). The soybean gene, GmMYBJ2, encodes a R2R3-type transcription factor involved in drought stress tolerance in Arabidopsis thaliana. Acta Physiol Plant. 37, 155–157. doi: 10.1007/s00192-015-2823-5
Tani, T., Sobajima, H., Okada, K., Chujo, T., Arimura, S., Tsutsumi, N., et al. (2008). Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta 227, 517–526. doi: 10.1007/s00425-007-0635-7
Tester, M., Langridge, P. (2010). Breeding technologies to increase crop production in a changing world. Science 327, 818–822. doi: 10.1126/science.1183700
Turner, J. G., Ellis, C., Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14 Suppl, S153–S164. doi: 10.1105/tpc.000679
Uji, Y., Taniguchi, S., Tamaoki, D., Shishido, H., Akimitsu, K., Gomi, K. (2016). Overexpression of OsMYC2 results in the up-regulation of early JA-responsive genes and bacterial blight resistance in rice. Plant Cell Physiol. 57, 1814–1827. doi: 10.1093/pcp/pcw101
Vargas, L., Santa, B. A., Mota, F. J., de Carvalho, T. G., Rojas, C. A., Vaneechoutte, D., et al. (2014). Drought tolerance conferred to sugarcane by association with Gluconacetobacter diazotrophicus: a transcriptomic view of hormone pathways. PLoS One 9, e114744. doi: 10.1371/journal.pone.0114744
Wang, F. B., Tong, W. J., Zhu, H., Kong, W. L., Peng, R. H., Liu, Q. C., et al. (2016b). A novel Cys2/His2 zinc finger protein gene from sweetpotato, IbZFP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Planta 243, 783–797. doi: 10.1007/s00425-015-2443-9
Wang, F., Chen, H. W., Li, Q. T., Wei, W., Li, W., Zhang, W. K., et al. (2015). GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 83, 224–236. doi: 10.1111/tpj.12879
Wang, L. Q., Li, Z., Lu, M. Z., Wang, Y. C. (2017a). ThNAC13, a NAC transcription factor from Tamarix hispida, confers salt and osmotic stress tolerance to transgenic Tamarix and Arabidopsis. Front. Plant Sci. 8, 635–647. doi: 10.3389/fpls.2017.00635
Wang, N., Zhang, W. X., Qin, M. Y., Li, S., Qiao, M., Liu, Z. H., et al. (2017b). Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol. 58, 1764–1776. doi: 10.1093/pcp/pcx111
Wang, R. K., Cao, Z. H., Hao, Y. J. (2014). Overexpression of a R2R3 MYB gene MdSIMYB1 increases tolerance to multiple stresses in transgenic tobacco and apples. Physiol. Plant 150, 76–87. doi: 10.1111/ppl.12069
Wang, S. Y. (1999). Methyl jasmonate reduces water stress in strawberry. J. Plant Growth Reg. 18, 127–134. doi: 10.1007/PL00007060
Wang, Z., Wang, F., Hong, Y., Huang, J., Shi, H., Zhu, J. K. (2016a). Two chloroplast proteins suppress drought resistance by affecting ROS production in guard cells. Plant Physiol. 172, 2491–2503. doi: 10.1104/pp.16.00889
Wasternack, C., Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Wasternack, C., Stenzel, I., Hause, B., Hause, G., Kutter, C., Maucher, H., et al. (2006). The wound response in tomato-role of jasmonic acid. J. Plant Physiol. 163, 297–306. doi: 10.1016/j.jplph.2005.10.014
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Xu, Y., Burgess, P., Huang, B. (2017). Transcriptional regulation of hormone-synthesis and signaling pathways by overexpressing cytokinin-synthesis contributes to improved drought tolerance in creeping bentgrass. Physiol. Plant 161, 235–256. doi: 10.1111/ppl.12588
Yang, A., Dai, X., Zhang, W. H. (2012). A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63, 2541–2556. doi: 10.1093/jxb/err431
Yang, S. J., Vanderbeld, B., Wan, J. X., Huang, Y. F. (2010). Narrowing down the targets: towards successful genetic engineering of drought-tolerant crop. Mol. Plant. 3, 469–490. doi: 10.1093/mp/ssq016
Yoshii, M., Yamazaki, M., Rakwal, R., Kishi-Kaboshi, M., Miyao, A., Hirochika, H. (2010). The NAC transcription factor RIM1 of rice is a new regulator of jasmonate signaling. Plant J. 61, 804–815. doi: 10.1111/j.1365-313X.2009.04107.x
Yu, H., Chen, X., Hong, Y. Y., Wang, Y., Xu, P., Ke, S. D., et al. (2008). Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20, 1134–1151. doi: 10.1105/tpc.108.058263
Yuan, Y., Qi, L. J., Yang, J., Wu, C., Liu, Y. J., Huang, L. Q. (2015). A Scutellaria baicalensis R2R3-MYB gene, SbMYB8, regulates flavonoid biosynthesis and improves drought stress tolerance in transgenic tobacco. Plant Cell Tiss. Org. Cult. 120, 973–973. doi: 10.1007/s11240-014-0650-x
Zhang, C., Zhang, L., Zhang, S., Zhu, S., Wu, P., Chen, Y., et al. (2015a). Global analysis of gene expression profiles in physic nut (Jatropha curcas L.) seedlings exposed to drought stress. BMC Plant Biol. 15, 17–30. doi: 10.1186/s12870-014-0397-x
Zhang, F., Yao, J., Ke, J. Y., Zhang, L., Lam, V. Q., Xin, X. F., et al. (2015b). Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525, 269–273. doi: 10.1038/nature14661
Zhang, H., Zhang, Q., Zhai, H., Li, Y., Wang, X. F., Liu, Q. C., et al. (2017). Transcript profile analysis reveals important roles of jasmonic acid signalling pathway in the response of sweet potato to salt stress. Sci. Rep. 7, 40819–40830. doi: 10.1038/srep40819
Zhao, Y., Dong, W., Zhang, N. B., Ai, X. H., Wang, M. C., Huang, Z. G., et al. (2014). A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 164, 1068–1076. doi: 10.1104/pp.113.227595
Zhu, H., Zhou, Y. Y., Zhai, H., He, S. Z., Zhao, N., Liu, Q. C. (2018). Transcriptome profiling reveals insights into the molecular mechanism of drought tolerance in sweetpotato. J. Inter. Agri. 17, 60345–60347. doi: 10.1016/S2095-3119(18)61934-3
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant. Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Keywords: sweetpotato, IbMYB116, Arabidopsis, drought tolerance, JA signaling pathway
Citation: Zhou Y, Zhu H, He S, Zhai H, Zhao N, Xing S, Wei Z and Liu Q (2019) A Novel Sweetpotato Transcription Factor Gene IbMYB116 Enhances Drought Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 10:1025. doi: 10.3389/fpls.2019.01025
Received: 10 December 2018; Accepted: 22 July 2019;
Published: 15 August 2019.
Edited by:
Susanne Berger, Julius Maximilian University of Würzburg, GermanyReviewed by:
Ratna Karan, University of Florida, United StatesEric Ruelland, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2019 Zhou, Zhu, He, Zhai, Zhao, Xing, Wei and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingchang Liu, bGl1cWNAY2F1LmVkdS5jbg==
 Yuanyuan Zhou1
Yuanyuan Zhou1 Shaozhen He
Shaozhen He Hong Zhai
Hong Zhai Qingchang Liu
Qingchang Liu