- 1School of Life Sciences and Biotechnology, The Joint International Research Laboratory of Metabolic and Developmental Sciences, Shanghai Jiao Tong University, Shanghai, China
- 2School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
- 3National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China
Brassinosteroid (BR) is a family of bioactive steroid hormones that plays vital roles in plant growth and development. The BR-mediated regulation of plant growth and architecture has been well studied. However, relatively few studies have investigated the BR-related regulation of reproductive development because of the difficulties in excluding non-specific regulation and secondary responses from severe vegetative phenotypes and poor nutritional status. Furthermore, differentially regulating the BR signal in vegetative and reproductive organs is problematic. Thus, establishing a method for modulating the BR signal only in reproductive organs or during reproductive developmental stages will be beneficial. Additionally, the utility of BR applications for crop production is limited because of deleterious side-effects, including the associated decrease in the planting density and lodging resistance. Moreover, enhancing the BR signal may lead to feedback inhibition. In this study, we developed a transformation system for modulating the BR signal differentially during reproductive and vegetative developmental stages. This system involves transformations with different combinations of a reproductive tissue-specific promoter, coding sequences that increase or decrease the BR signal, and various genotypic backgrounds with enhanced or decreased BR signals. The enhanced BR signal generated in transformants was targeted to reproductive organs without affecting vegetative organs. This system may be useful for studying the BR-specific regulation of plant reproductive development and shows promise for optimizing seed yield.
Introduction
Phytohormones play essential roles in the regulation of seed development, seed size, seed weight, and crop yield. Brassinosteroid (BR) is plant steroid hormones that affect cell elongation and division, tissue differentiation, organogenesis, reproductive development, photomorphogenesis, and immunity (Morinaka et al., 2006; Sakamoto et al., 2006). In rice, BR can also decrease the amount of residues of common organophosphorus, organochlorine, and carbamate pesticides (Zhou et al., 2015) and induce crop tolerance to these pesticides (Ahammed et al., 2012a,b).
The BR signal transduction pathway has been well studied. The BR receptor BRASSINOSTEROID INSENSITIVE1 (BRI1, a cell-surface receptor kinase) (Li et al., 2001a) and the BR-induced transcription factors BRASSINAZOLE RESISTANT1 (BZR1) (Wang et al., 2002) and BRI1-EMS-SUPPRESSOR1 (BES1) (Yin et al., 2002) are key components for activating the BR signal. BRASSINOSTEROID INSENSITIVE2 (BIN2), a GSK3-like kinase, is the main repressor of the BR signal (Li et al., 2001b; He et al., 2002). The strong alleles of BRI1, such as bri1-116, show severe phenotypes of strongly dwarf, very small and round leaves, and almost complete sterility (Li and Chory, 1997). In contrast, the weak alleles of BRI1, such as bri1-5, exhibit semi-dwarfism, with small and round leaves as well as decreased fertility (Li et al., 2001a). A gain-of-function mutant of BIN2, bin2-1, that exhibits phenotypes associated with decreased BR signal levels, similar to the bri1 mutant (Li et al., 2001b). A gain-of-function mutant of BZR1, bzr1-1D, which phenotype is associated with enhanced BR signaling (Wang et al., 2002; He et al., 2005). Compared with wild-type plants, this mutant produces longer and curved petioles, longer and kinked inflorescence stems, larger floral organs, and more seeds. The increased BZR1 dephosphorylation in the bzr1-1D mutant is indicative of high BZR1 activity and higher BR signals.
Brassinosteroid positively regulates seed development and seed size/weight by modulating the transcription of SHORT HYPOCOTYL UNDER BLUE 1 (SHB1)–MINISEED 3 (MINI3)–HAIKU2 (IKU2), APETALA 2 (AP2), and AUXIN RESPONSE FACTOR 2 (ARF2) (Schruff et al., 2006; Ohto et al., 2009; Zhou et al., 2009; Jiang et al., 2013). Additionally, BR positively regulates ovule development and seed production (Huang et al., 2013), implying BR enhances seed yield. Brassinosteroid is well known to regulate rice architecture and grain yield (Zhang et al., 2018), and are important for regulating rice plant height (Fujioka and Yokota, 2003). The D11 gene encoding a cytochrome P450 (CYP724B1), functions in BR biosynthesis and affects rice plant height (Tanabe et al., 2005). The BR-deficient mutant d2 has a smaller leaf angle (Hong et al., 2003), implying that BR has a significant impact on leaf bending. In another BR-deficient mutant, brd1, the internodes essentially do not elongate (Hong et al., 2002). Increased BR contents can lead to the production of more tillers, larger spikes, and more grains per spike (Wu et al., 2008). Brassinosteroid also positively regulates rice seed/grain size/weight (Hong et al., 2005; Tanabe et al., 2005; Morinaka et al., 2006; Sahni et al., 2016). Although a number of specific regulators and different regulatory mechanisms have been identified in rice, including ENHANCED LEAF INCLINATION AND TILLER NUMBER1 (ELT1, Yang et al., 2017), RICE LEAF AND ILLER ANGLE INCREASED CONTROLLER1 (OsLIC1, Wang et al., 2008), and DWARF AND TILLERING (DLT, Tong et al., 2009), BR signal transduction and BR-regulated growth and development are essentially conserved in Arabidopsis, rice, soybean, and other plant species (Nakamura et al., 2006; Sakamoto et al., 2006; Bai et al., 2007; Tanaka et al., 2009; Zhang et al., 2016). Arabidopsis BR-related mutants are useful for identifying the BR signal regulators in other species.
Brassinosteroid also regulates the development of pollen grains (Ye et al., 2010), ovules (Huang et al., 2013), the embryo sac (Pérez-España et al., 2011), and seeds (Jiang et al., 2013). However, investigating the BR-specific regulation of plant reproductive development is difficult because severe vegetative phenotypes and poor nutrient accumulation in strong alleles of BR-deficient or insensitive mutants would affect reproductive development indirectly. Therefore, it is difficult to exclude the possibility of a secondary response or non-specific regulation. Establishing a transformation system that enables the differential modulation of the BR signal in various organs or developmental stages may be much helpful. Such a system may be relevant for increasing crop production, especially rice.
Increasing the total yield is an important aim of crop breeding. The total yield depends on the yield of individual plants and the planting density. The grain yield of individual rice plants may be improved by enhancing the BR signal, but the growing area of an individual plant would also increase, thereby leading to decreased planting density. Furthermore, plant height would increase and lodging resistance would decrease, which would negatively affect grain yield. Previous studies revealed that enhancing the BR signal contributes to improved nutritional status, greater efficiency of carbohydrate transport from the source to the sink, and increased grain yield. However, an overall increase in BR content also results in larger laminar joints and increased height, leading to decreased lodging resistance and planting density (Choe et al., 2001; Wu et al., 2008). These negative side-effects may limit the utility of BR applications for agricultural production. A system that specifically modulates BR signals in reproductive organs may be highly beneficial for increasing the total rice yield. In this study, we established a transformation system for modulating the BR signal during reproductive development. This system could be used to investigate the BR-specific regulation of plant reproductive development and optimize seed yield.
Materials and Methods
Plant Materials, Growth Conditions, and Transformation Procedure
The pSTK::GUS (pBI101.3) vector was transformed into wild-type Arabidopsis plants (Columbia ecotype; Col) to examine STK promoter activity. The pSTK::bzr1-1D-GUS (pBI101.3) and pSTK::bzr1-1D-GFP (pCAMBIA1302) vectors were transformed into bri1-5 plants (Wassilewskija ecotype) to enhance the BR signal in reproductive organs. Similarly, the pSTK::bzr1-1D-GFP vector was transformed into bin2-1 ± plants (Col ecotype) to enhance the BR signal in reproductive organs. The pSTK::bin2-1-GUS vector was transformed into bzr1-1D and DWF4-OX (Col ecotype) plants to repress the BR signal in reproductive organs. Additionally, the pSTK::bin2-1-GFP vector was transformed into bzr1-1D, DWF4-OX, and wild-type (Col ecotype) plants to repress the BR signal in reproductive organs.
All plants were grown in a greenhouse maintained at 22°C under a 16-h light/8-h dark photoperiod. Agrobacterium-mediated transformation was performed using the floral dip method (Clough and Bent, 1998). The positive clones were cultured for 10 h at 28°C in YEP medium (10 g/L peptone, 10 g/L yeast extract, and 5 g/L NaCl, pH 7.2) supplemented with 50 mg/L rifampicin and 50 mg/L kanamycin. The Agrobacterium cells were collected and diluted with 5% sucrose solution containing 0.02% (v/v) Silwet L-77 (SL77080596, GE) to OD600 0.8–1.0. The Arabidopsis inflorescence apices were dipped in the mixture buffer, sealed, and kept in the dark overnight.
Construction of Vectors
The 2,008 bp promoter and 1,085 bp coding sequence (gDNA) of STK was amplified by PCR with KOD-FX DNA polymerase and primers STK-F and STK-R, and cloned into the pBI101.3 vector (digested with BamHI) to generate the pSTK::GUS vector.
To obtain the bzr1-1D coding sequence, the former 1,055 bp and the latter 321 bp gDNA of BZR1 were amplified with KOD-FX DNA polymerase. The specific primer pairs containing a point mutation were BZR1-F and bzr1-1D-R, and bzr1-1D-F and BZR1-R, respectively. We then amplified 1,355 bp gDNA of bzr1-1D with the specific primers BZR1-F and BZR1-R by fusion PCR.
To insert pSTK::bzr1-1D in the pBI101.3 vector, pSTK was amplified with the primer pair STK-F and STK-R1, and bzr1-1D was amplified with the primer pair bzr1-1D-F1 and bzr1-1D-R1. STK-bzr1-1D was amplified with the primer pair STK-F and bzr1-1D-R1 by fusion PCR, after which the amplicon was cloned into pBI101.3 (digested with BamHI) to generate the pSTK::bzr1-1D-GUS vector.
To generate the pSTK::bzr1-1D-GFP construct, STK-bzr1-1D was amplified with the primer pair STK-F1 and bzr1-1D-R2 using the pSTK::bzr1-1D-GUS vector as the template, and then was cloned into pCAMBIA1302 (digested with EcoRI and NocI) to generate the pSTK::bzr1-1D-GFP vector.
Similar to bzr1-1D, we amplified bin2-1 sequence by fusion PCR, and then constructed the pCAMBIA1302 pSTK::bin2-1-GFP and pBI101.3 pSTK::bin2-1-GUS vectors. All primer sequences are shown in Supplementary Table S1.
qRT-PCR Assay
The quantitative real-time PCR (qRT-PCR) procedure was carried out as described previously (Zhang et al., 2016). Total RNA was extracted from 15-day-old rosette leaves and the apex of 40-day-old inflorescences. CPD sequence was amplified with CPD RT-F and CPD RT-R. DWF4 sequence was amplified with DWF4 RT-F and DWF4 RT-R. bzr1-1D-GFP sequence was amplified with bzr1-1D RT-F and GFP RT-R. The ACTIN gene was amplified as an internal reference using the primers ACTIN RT-F and ACTIN RT-R. The procedure was 40 cycles of 94°C for 10 s, 60°C for 15 s, and 72°C for 20 s. Primer sequences are shown in Supplementary Table S2. The experiments were biologically repeated for 3 times and data are shown as means ± SD (n = 3).
β-Glucuronidase (GUS) Staining Assay
The inflorescence apex of pSTK::GUS lines was dipped in GUS staining solution and vacuum-infiltrated for 30 min, maintained in the GUS staining solution at 37°C overnight, then chlorophyll was eluted with 75% alcohol twice. The stained tissues were observed with a Leica S8APO stereomicroscope and photographed with a Leica DFC450 digital camera.
Identification and Characterization of Transgenic Lines
The pSTK::GUS, pSTK::bzr1-1D-GUS, and pSTK::bin2-1-GUS transgenic lines were verified with primers 77-R and GUS. The pSTK::bzr1-1D-GFP and pSTK::bin2-1-GFP transgenic lines were verified with the primer pairs 77-R and GFP. Primer sequences are shown in Supplementary Table S3.
The phenotype of 21-day-old plants and the leaf shape of transgenic lines were photographed using a Canon EOS60D digital camera. The siliques from different lines were observed with a Leica S8APO stereomicroscope and photographed with a Leica DFC450 digital camera and the experiments were biologically repeated for 3 times and data are shown as means ± SD (n = 15). The seed yield (seed weight per plant) was measured by an analytical balance and the experiments were biologically repeated for 3 times and data are shown as means ± SD (n = 5).
Results
Designing Transgenic Plants to Modulate the BR Signal in Reproductive Organs
The normal BR signal positively regulates plant growth and development in both vegetative and reproductive organs (Figure 1A, R1V1 and R0V0). The reproductive development of plants with the strong alleles of BR-deficient/insensitive mutants was severely impacted. However, we could not exclude the possibility that the phenotypes were the result of the non-specific regulation of poor vegetative growth and poor nutrient accumulation. To investigate the BR-specific regulation of plant reproductive development, we generated transgenic plants in which the BR signal was specifically modulated in the reproductive organs during the reproductive developmental stages (Figure 1A, R1V0 and R2V0). To examine the relevant regulatory processes and to distinguish BR signals in various organs, we also generated transgenic lines with the opposite modification to the BR signal (Figure 1A, R1V2 and R2V1).
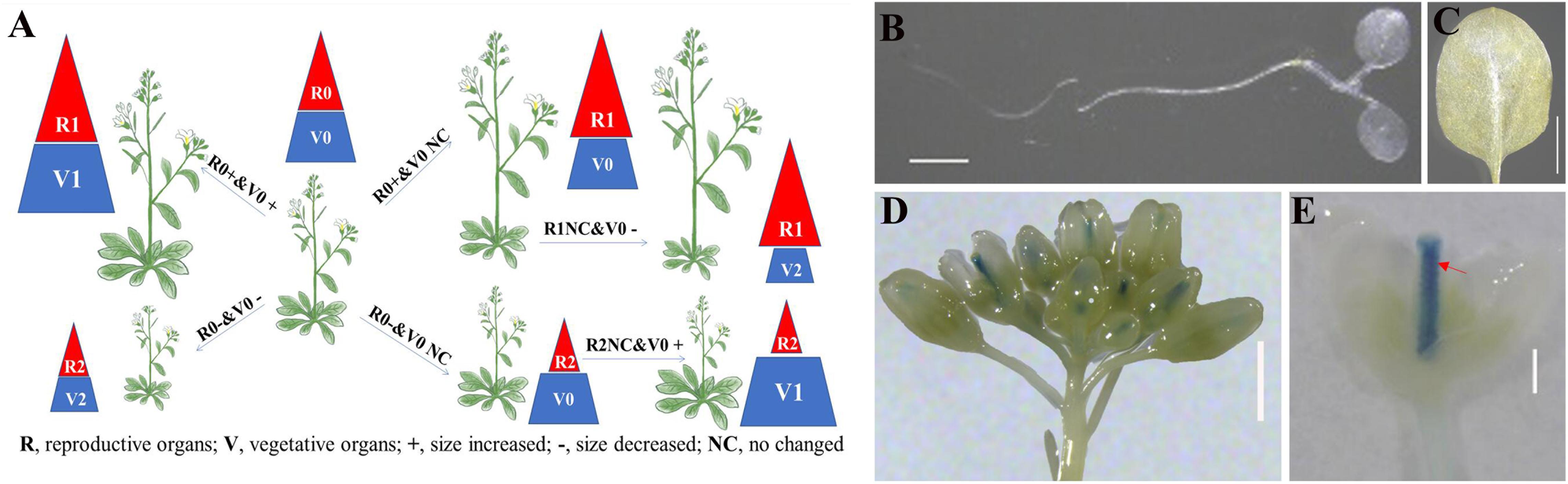
Figure 1. Design of transformation system for differential manipulation of brassinosteroid (BR) signal in reproductive and vegetative organs. (A) 0: Original BR signal and normal organ size; 1: increased BR signal and organ size; 2: decreased BR signal and organ size, R: reproductive organs, V: vegetative organs, +: size increased, -: size increased, NC: no changed (B–E) β-Glucuronidase (GUS) staining assay of pSTK::GUS (pBI101.3) transgenic lines. The GUS signal was detected in the cotyledons (B), rosette leaves (C), inflorescence apex (D), and pistil and ovules (E), bar = 1 cm. Red arrow indicates ovules.
The transformation system developed in this study may be useful for improving crop production. Specifically, R1V0 enhanced reproductive development without influencing vegetative growth, thereby avoiding side-effects. Additionally, R1V2 was the optimal combination, which resulted in increased reproductive organ size and appropriately decreased vegetative organ size. Developing the transformation system involved the following three steps. The first step was identifying specific promoters that are active in reproductive tissues. The second step was constructing the coding sequences for the efficient modulation of the BR signal. The final step was selecting plants in which the BR signal was modulated for transformation.
Selection of Tissue-Specific Promoters
Selecting tissue-specific promoters that are sufficiently active is an important step. Previous studies indicated that MADS-box genes, such as SEPALLATA 1 (SEP1), SEP2, SEP3, AGAMOUS (AG), SHATTERPROOF 1 (SHP1), SHP2, and SEEDSTICK (STK), are expressed in reproductive organs and control flower and ovule identity (Bowman et al., 1989; Liljegren et al., 2000; Pelaz et al., 2000; Favaro et al., 2003; Pinyopich et al., 2003). For example, STK was predicted to be highly transcribed (Supplementary Figure S1), mainly in the pistil and ovule. The results of GUS staining assays (Pinyopich et al., 2003; Kooiker et al., 2005; Nain et al., 2008) revealed that STK is only expressed in the septum and ovules. Consequently, we cloned the longest STK promoter into the pBI101.3 vector with a portion of genomic DNA (from -2,008 bp to 1,085 bp) to ensure accurate expression. We introduced the pSTK::GUS vector (pBI101.3) into wild-type Arabidopsis (Col) plants and assessed the transcription activity in a GUS staining assay. The results illustrated that the STK promoter was active in the septum and ovules, but not in the cotyledons and rosette leaves (Figures 1B–E), which was consistent with previous findings (pSTK::GUS in the pCAMBIA1300-H vector; Kooiker et al., 2005) (Supplementary Figure S2). Thus, the STK promoter would be useful for enhancing the BR signal in the septum and ovule. Additionally, the STK promoter-controlled vector appears to be a suitable tool for studying the BR-specific regulation of septum and ovule development.
Identification of Appropriate Coding Sequences to Directly Regulate the BR Signal
We selected specific coding sequences to efficiently modulate BR signaling and responses. The pBI101.3 and modified pCAMBIA1302 (35S promoter removed) vectors were used to harbor the coding sequences driven by the STK promoter. The activity of the BR-induced transcription factor BZR1 directly influences BR signaling and responses, and consequently seed production (Huang et al., 2013; Jiang et al., 2013; Zhang et al., 2016). Thus, we constructed a coding sequence (P234L) to mimic the gain-of-function mutant of BZR1, bzr1-1D. Finally, we inserted the pSTK::bzr1-1D construct (Supplementary Figure S3) into the pBI101.3 (pSTK::bzr1-1D-GUS) and pCAMBIA1302 (pSTK::bzr-1D-GFP) vectors.
Using a similar method, we inserted the pSTK::bin2-1 construct (Supplementary Figure S3) into the pBI101.3 (pSTK::bin2-1-GUS) and pCAMBIA1302 (pSTK::bin2-1-GFP) vectors to mimic the bin2-1 mutant and decrease the BR signal in reproductive organs.
Selection of Plants in Which the BR Signal Is Modulated for Transformation
We introduced the prepared vectors into wild-type Arabidopsis (Col) plants and observed the specifically enhanced or decreased BR signal in the reproductive organs of the transgenic lines. To investigate whether the BR signal in vegetative organs influences the BR-mediated regulation of reproductive development, we also transformed BR-insensitive mutants (bri1-5 and bin2-1) (Li et al., 2001a,b), a BR-signal-enhanced line (bzr1-1D), and the transgenic line DWF4-OX, which overexpresses the BR biosynthesis gene DWF4 (Choe et al., 2001).
Specific Enhancement of the BR Signal in Reproductive Organs
We transformed pSTK::bzr1-1D-GUS and pSTK::bzr1-1D-GFP into the bri1-5 mutant plants to increase the BR signal in reproductive organs. Given that bri1-5 is a weak allele that leads to smaller vegetative organs and not bad nutrient accumulation compared with wild-type plants, these transformations were expected to be appropriate for investigating whether the BR signal separately regulates vegetative and reproductive development. Lines 2, 3, and 5 were transgenic lines harboring pSTK::bzr1-1D-GUS (Figure 2). The shape and size of the rosette leaves in the transgenic lines were not significantly different from those of the bri1-5 mutant (Figure 2A), indicating that pSTK::bzr1-1D did not distinctly modify vegetative growth. As expected, the floral organs in the transgenic plants were visibly larger than those of the bri1-5 mutant (Figure 2B) and the siliques of the transgenic plants were 30% longer than those of the bri1-5 mutant (Figures 2C,E). Lines 2, 3, and 5 had an average of 42.7, 39, and 43.3 seeds per silique, respectively, whereas the bri1-5 mutant had 32 seeds per silique (Figure 2F). The seed weight per plant was higher for lines 2, 3, and 5 than for the bri1-5 mutant (Figure 2I). The qRT-PCR results revealed that CPD and DWF4 transcription levels decreased in reproductive organs, implying that the BR signal was enhanced in reproductive organs (Figure 2G). Moreover, there were no significant changes to the transcription of CPD and DWF4 in vegetative organs (Figure 2H), suggesting that the transformation did not increase the BR signal in vegetative organs (i.e., the BR signal was regulated separately in the reproductive and vegetative organs).
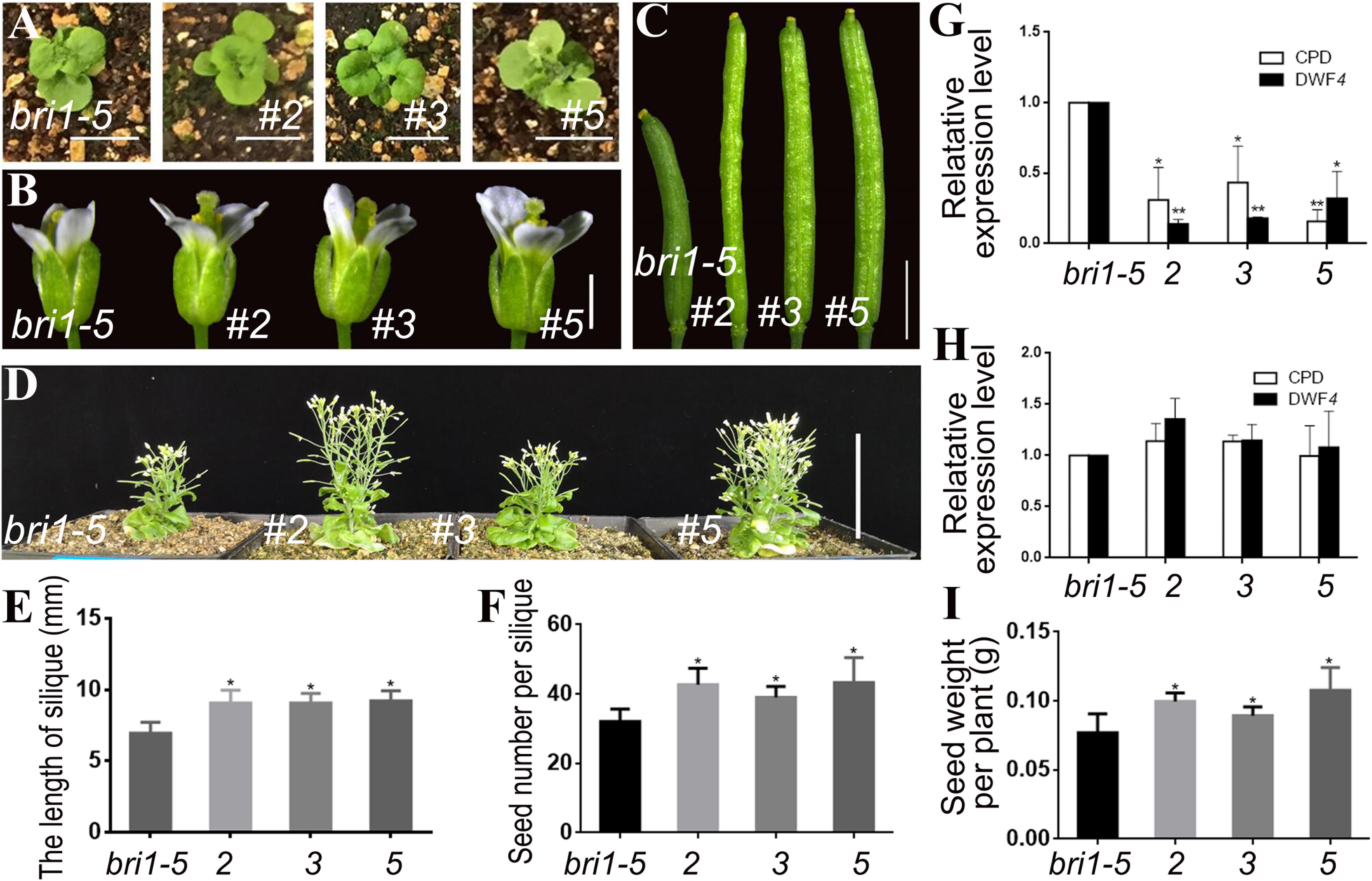
Figure 2. Phenotypic analysis of pSTK::bzr1-1D-GUS lines in bri1-5 background. (A) Shape and size of rosette leaves, bar = 1 cm. (B) Floral organs, bar = 2 mm. (C) Siliques, bar = 2 mm. (D) Adult plants, bar = 5 cm. (E,F) Silique length and seed number. Bars represent the means ± SD of three biological replicates (n = 15). (G,H) Relative expression level of CPD and DWF4 in inflorescence apex and rosette leaves. Values correspond to the arithmetic means ± SD of three biological replicates (n = 3). (I) Seed weight per plant. Bars represent the means ± SD of three biological replicates (n = 5). Asterisks represent significant differences (Student’s t-test, *P < 0.05, ∗∗P < 0.01).
Transgenic lines 86, 90, and 106 harboring pSTK::bzr1-1D-GFP were generated to assess the phosphorylation of the BZR1 protein and to verify the observed phenotypes. A western blot analysis involving GFP antibodies revealed more dephosphorylated BZR1 in the reproductive organs than in the vegetative organs of the transgenic lines, which indicated that BR was more active in reproductive organs than in vegetative organs (Figure 3J). Similar to lines 2, 3, and 5, the shape and size of the rosette leaves in lines 86, 90, and 106 were not significantly different from those of the bri1-5 mutant (Figures 3A,B). The transgenic lines produced enlarged flowers and siliques (Figures 3C,D,F) and more seeds than the bri1-5 mutant (Figure 3G). Additionally, lines 86, 90, and 106 were considerably taller than lines 2, 3, and 5 (i.e., longer inflorescence stems) (Figures 2D, 3E), which suggested that the enhanced BR signal spread from the expected organs to the inflorescence stem. The qRT-PCR results also revealed that CPD and DWF4 transcription levels decreased in the vegetative and reproductive organs of the pSTK::bzr1-1D-GFP lines (Figures 3H,I), indicating that the BR signal was enhanced in vegetative and reproductive organs. These observations were in contrast to the response of pSTK::bzr1-1D-GUS transgenic lines, suggesting the more specific modulation of BR signal by pBI101.3 vector. Accompanied by the varied plant height, silique size and seeds number, the qRT-PCR data illustrated that the varied transcription of the introduced bzr1-1D-GFP in pSTK::bzr1-1D-GFP lines (Figures 3F,G,K,L).
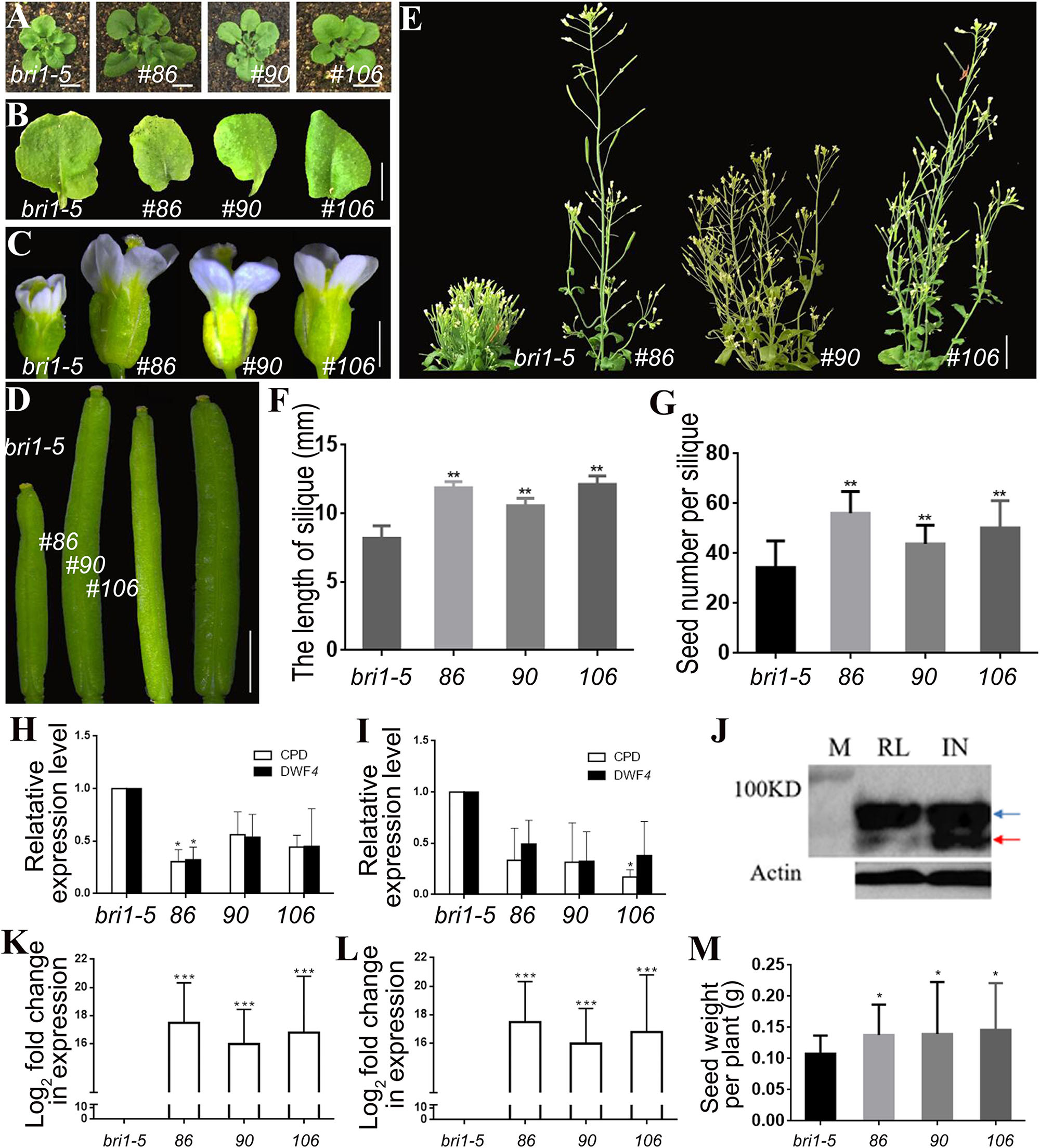
Figure 3. Phenotypic analysis of pSTK::bzr1-1D-GFP lines in bri1-5 background. (A,B) Shape and size of rosette leaves, bar = 1 cm. (C) Floral organs, bar = 2 mm. (D) Siliques, bar = 2 mm. (E) Adult plants, bar = 2 cm. (F,G) Silique length and seed number. Bars represent the means ± SD of three biological replicates (n = 15). (H,I) Relative expression level of CPD and DWF4 in inflorescence apex and rosette leaves. Values correspond to the arithmetic means ± SD of three biological replicates (n = 3). (J) Western-blot showing BZR1 (red arrow) and pBZR1 (blue arrow) in inflorescences (IN) and rosette leaves (RL). The protein marker (M) bands are 100 kD. Actin was the loading control. (K,L) Relative expression level of bzr1-1D-GFP in inflorescence apex and rosette leaves. Values correspond to the arithmetic means ± SD of three biological replicates (n = 3). (M) Seed weight per plant. Bars represent the means ± SD of three biological replicates (n = 5). Asterisks represent significant differences (Student’s t-test, *P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
Regarding the study objective, pSTK::bzr1-1D-GUS lines were suitable for the efficient investigation of the BR-specific regulation of reproductive development, but the seed yield of the pSTK::bzr1-1D-GFP lines increased to a greater extent (Figure 3M). The enhanced BR signal in reproductive organs and decreased BR signal in vegetative organs increased the seed yield of individual plants and enhanced the planting density, which considerably increased the total yield of pSTK::bzr1-1D lines, especially the pSTK::bzr1-1D-GFP lines.
To further verify that the transgenic lines could differentially regulate the BR signal in various organs and enhance the seed yield, we introduced the pSTK::bzr1-1D-GFP construct into the bin2-1 mutant. The shape and size of the rosette leaves of three independent transgenic lines (134, 217, and 221) did not change significantly (Figures 4A,B). However, these plants produced larger flowers and siliques (Figures 4C,D,F), grew taller (Figure 4E), and produced more seeds (Figures 4G,H) than the bin2-1 mutant. A western blot analysis revealed that BZR1 dephosphorylation and the BR signal was greater in reproductive organs than in vegetative organs (Figure 4I), implying that pSTK::bzr1-1D enhanced the BR signal and seed yield without changing the vegetative organ size of the bin2-1 mutant.
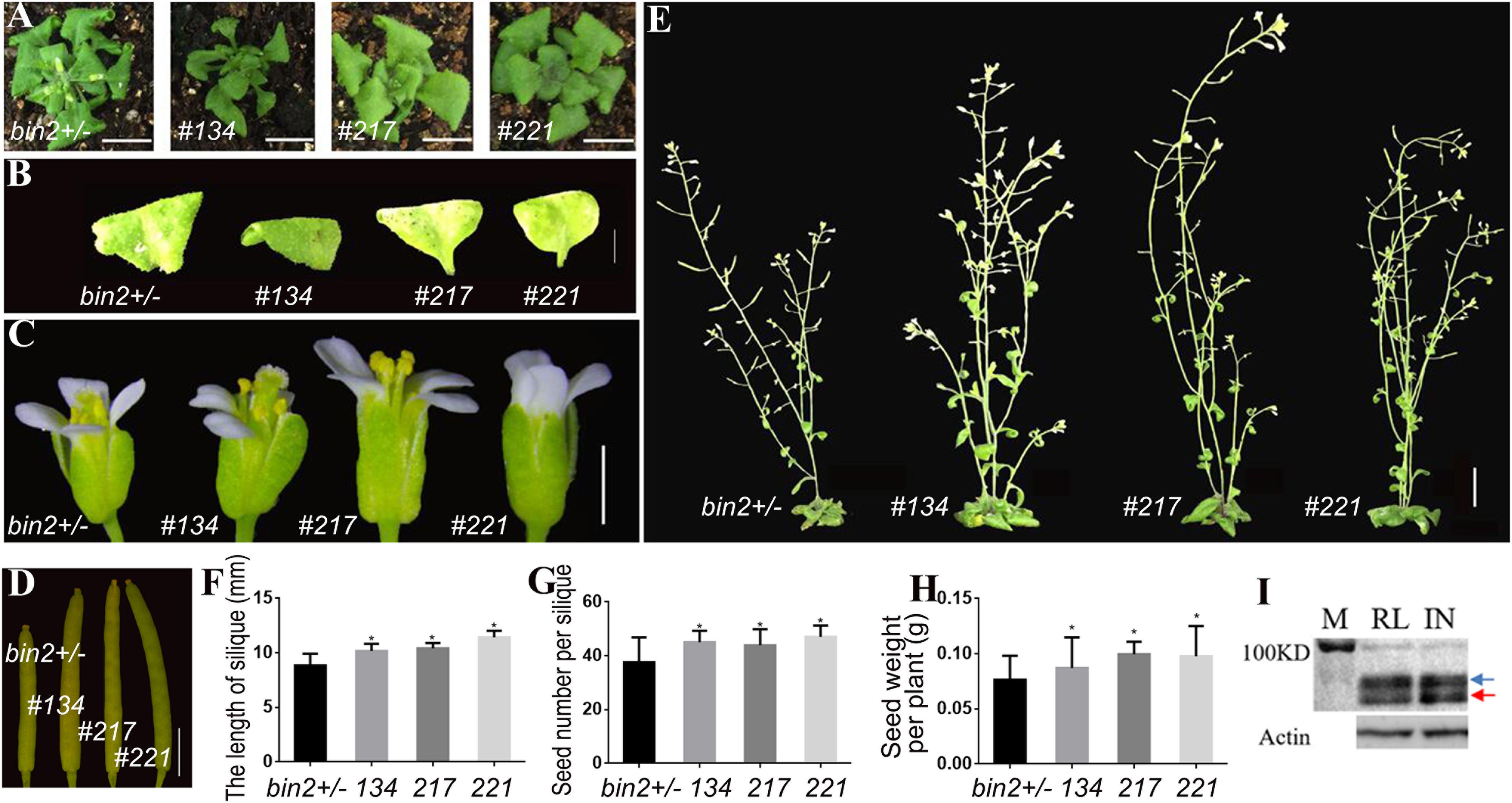
Figure 4. Phenotypic analysis of pSTK::bzr1-1D-GFP lines in bin2-1 background. (A,B) Shape and size of rosette leaves, bar = 1 cm. (C) Floral organs, bar = 2 mm. (D) Siliques, bar = 2 mm. (E) Adult plants, bar = 1 cm. (F,G) Silique length and seed number. Bars represent the means ± SD of three biological replicates (n = 15). (H) Seed weight per plant. Bars represent the means ± SD of three biological replicates (n = 5). (I) Western-blot showing BZR1 (red arrow) and pBZR1 (blue arrow) in inflorescences (IN) and rosette leaves (RL). The protein marker (M) bands are 100 kD. Actin was the loading control. Asterisks indicate a significant difference (Student’s t-test, *P < 0.05).
Specific Depression of the BR Signal in Reproductive Organs
Using a similar method, we incorporated the pSTK::bin2-1-GUS vector into the bzr1-1D mutant and DWF4-OX plants. Lines 6, 7, and 9 were transgenic lines in the bzr1-1D background (Figures 5A–F), whereas lines 2, 3, and 5 were transgenic lines in the DWF4-OX background (Figures 5G–K). The transgenic lines produced similar rosette leaves (Figures 5A,G), but smaller floral organs and shorter inflorescence stems (Figures 5B,H), than the control plants. Additionally, the siliques of the transgenic lines were very short and sterile (Figures 5C,I). The height of transgenic lines decreased compared with control plant (Figure 5D). The qRT-PCR results indicated that CPD and DWF4 expression levels in the inflorescence apex were higher in transgenic lines than in the bzr1-1D mutant and DWF4-OX plants (Figures 5E,J), suggesting that the BR signal was relatively weak in reproductive organs. The transcription of CPD and DWF4 in the rosette leaves was almost unchanged (Figures 5F,K), which indicated that the BR signal was unaffected in vegetative organs. These results implied that we decreased the BR signal specifically in reproductive organs.
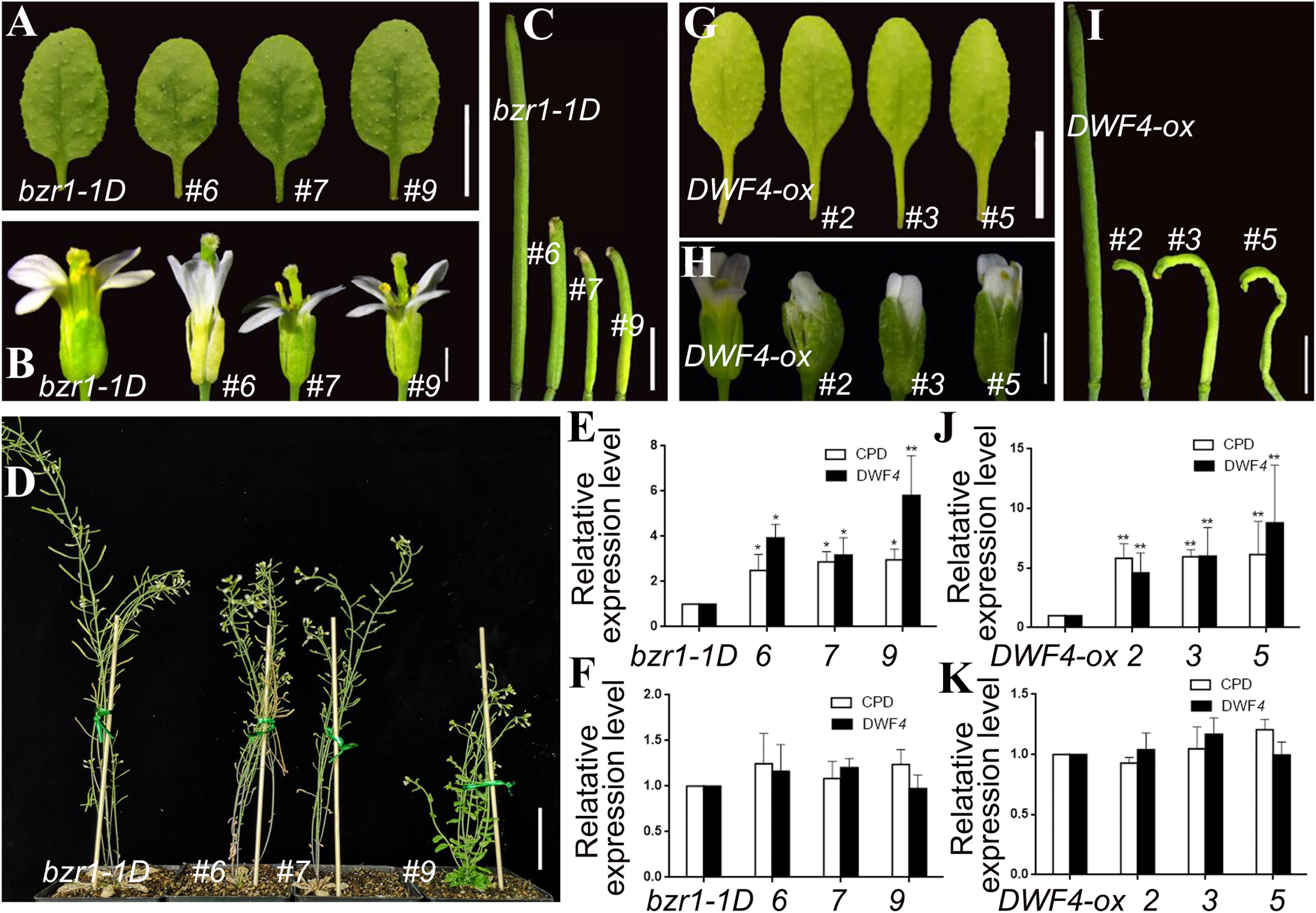
Figure 5. Phenotypic analysis of pSTK::bin2-1-GUS lines in bzr1-1D (A–F) and DWF4-OX (G–K) background. (A,G) Shape and size of rosette leaves, bar = 1 cm. (B,H) Floral organs, bar = 2 mm (C,I) Siliques, bar = 2 mm. (D) Adult plants, bar = 5 cm. (E,J,F,K) Relative expression level of CPD and DWF4 in inflorescence apex and rosette leaves. Values correspond to the arithmetic means ± SD of three biological replicates (n = 3). Asterisks represent significant differences (Student’s t-test, *P < 0.05, ∗∗P < 0.01).
Considering the effect of pSTK::bin2-1-GUS was excessive and led to sterility, we transformed the pSTK::bin2-1-GFP vector into the bzr1-1D mutant and DWF4-OX plants. However, no significant phenotypic changes were observed in the transgenic lines (Supplementary Figure S4). We then introduced the pSTK::bin2-1-GFP vector into wild-type Arabidopsis (Col) plants. Transgenic lines 31, 32, and 34 exhibited inhibited reproductive development (Figure 6). The shape and size of the rosette leaves did not change significantly (Figures 6A,B), whereas the flower size, silique length, plant height, and number of seeds decreased (Figures 6C–G) relative to the corresponding values for the wild-type control. The qRT-PCR results revealed that the CPD and DWF4 transcript levels in the inflorescence apex were higher in the transgenic lines than that in Col (Figure 6H). The data also suggested that the BR signal was weaker in reproductive organs than in vegetative organs. The CPD and DWF4 transcript levels in rosette leaves were higher in the transgenic lines than in the control plants (Figure 6I), indicating that the BR signal also changed in the rosette leaves, but not as markedly as in the inflorescence apex. These results also confirmed that we specifically decreased the BR signal in reproductive organs.
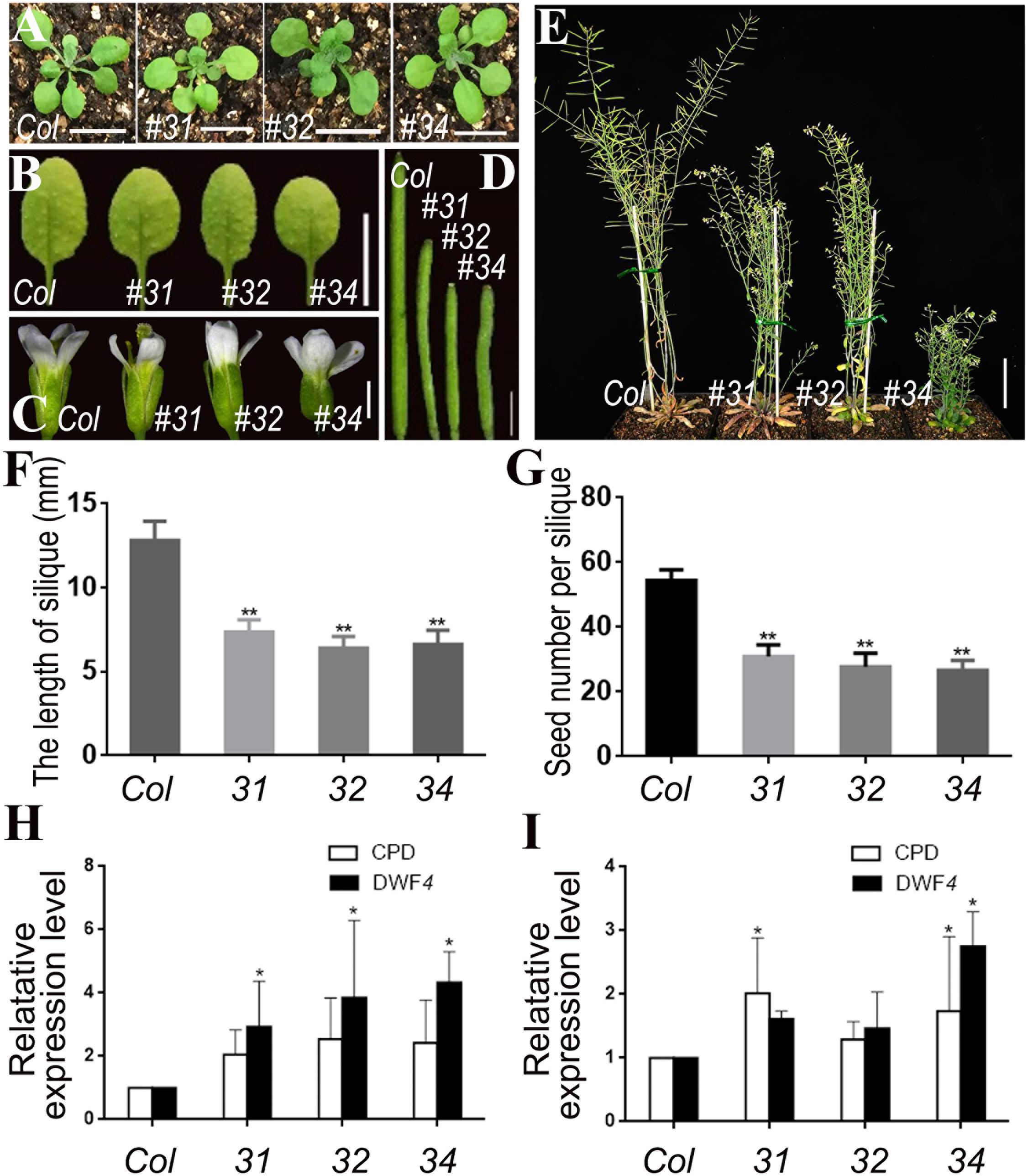
Figure 6. Phenotypic analysis of pSTK::bin2-1-GFP lines in Col background. (A,B) Shape and size of rosette leaves, bar = 1 cm. (C) Floral organs, bar = 2 mm. (D) Siliques, bar = 2 mm. (E) Adult plants, bar = 2 cm. (F,G) Silique length and seed number. Bars represent the means ± SD of three biological replicates (n = 15). (H,I) Relative expression level of CPD and DWF4 in inflorescence apex and rosette leaves. Values correspond to the arithmetic means ± SD of three biological replicates (n = 3). Asterisks represent significant differences (Student’s t-test, *P < 0.05, ∗∗P < 0.01).
Discussion
The regulation of BR is difficult to manipulate because of the complexity of the BR signal. A previous study (Wang et al., 2008) suggested that BR can enhance rice yield by upregulating the expression of BR biosynthesis genes driven by an artificial promoter with an enhancer. The promoter used in this earlier study was not a reproductive tissue-specific promoter, but was active in vegetative organs. The authors proposed that the BR signal stimulated the flow of assimilates from the source to the sink, ultimately resulting in enhanced rice yield. Moreover, they predicted that the expression of the same gene driven by an embryo-specific promoter in transgenic lines would not affect rice yield. The modulation of BR biosynthesis gene expression with a tissue-specific promoter does not appear to alter the BR signal in specific organs. In our study, the coding sequences directly regulated by the BR signal (bzr1-1D and bin2-1) and driven by tissue-specific promoters enhanced the BR signal only in the reproductive organs of transgenic lines, indicating the BR signal could be accurately manipulated in specific organs. The ideal combination (Figure 1A, R1V2) for increasing seed yield was determined, and the transgenic systems functioned as predicted.
The STK promoter is predicted to be highly active in various organs, including vegetative and reproductive organs, but especially in the pistil and ovule (Supplementary Figure S1). However, previously reported GUS assay results (Kooiker et al., 2005) and our results suggest that STK transcription is limited to the septum and ovule. The expression of our coding sequences driven by the STK promoter enhanced the BR signal in reproductive organs. Additionally, the two vectors used in this study differed regarding efficiency. For example, the enhanced BR signal was limited to flowers and siliques in the pSTK::bzr1-1D-GUS lines, and the adult plants were slightly taller. In contrast, the enhanced BR signal spread to additional organs in the pSTK::bzr1-1D-GFP lines, and the adult plants were considerably taller, with the architecture of some lines resembling that of wild-type plants. It seemed that the STK promoter was highly and more widely active in GFP vector than GUS vector, which was consistent with the predicted transcription based on electric northern (Supplementary Figure S4). The pSTK::bzr1-1D-GFP lines exhibited a greater expansion of the BR signal (i.e., in reproductive and vegetative organs) for unreported reasons. Fortunately, the BR signal was preferentially enhanced in the reproductive organs of pSTK::bzr1-1D-GFP lines, which indicated that the transformation system functioned as intended. Additionally, the seed yield of pSTK::bzr1-1D-GFP lines was considerably higher than that of pSTK::bzr1-1D-GUS lines.
Moreover, the efficiency of the pSTK::bin2-1 vectors varied. Inflorescence growth was suppressed in the transgenic lines of the bzr1-1D mutant and DWF4-OX plants harboring pBI101.3 vectors. But the transgenic lines are sterile. In the opposite way, there were no significant phenotypic changes in the transgenic lines harboring pSTK::bin2-1-GFP in DWF4-OX and bzr1-1D backgrounds. The BR signal may have been too strong to be suppressed in the DWF4-OX and bzr1-1D backgrounds. However, pSTK::bin2-1-GFP was effective in the Col background, suggesting that it could be used to study the BR-specific regulation of reproductive development. Theoretically, this system may be exploited to shorten inflorescences and enhance lodging resistance via appropriate modifications.
Fortunately, the pSTK::bin2-1-GFP lines in the Col background suppressed the reproductive development and produced less seed. The pBI101.3 vector was associated with specific STK promoter activity, resulting in the highly efficient modulation of the BR signal in the expected organs. In contrast, the pCAMBIA1302 vector was associated with expanded STK promoter activity and the relatively less-specific regulation of the BR signal. Considering this system may be used for future investigations and crop improvement, the combinations appropriate for use will depend on the specific aims.
In this study, we used a reproductive tissue-specific promoter (pSTK), coding sequences for modulating the BR signal (bzr1-1D and bin2-1), and vectors differing in efficiency (pBI101.3 and pCAMBIA1302) to preferentially modulate the BR signal in reproductive organs. The transgenic constructs enabled the differential regulation of the BR signal in vegetative and reproductive organs, which could be useful for studying the BR-specific regulation of reproductive development and for improving crop production. We increased flower/fruit size, seed production, and yield without affecting the vegetative organ size and planting density. On the basis of our results, the ideal procedure involves the introduction of pSTK::bzr1-1D-GFP into the weak allele of BR-deficient/insensitive plants to increase reproductive organ size and decrease vegetative organ size, and consequently enhance total yield by increasing the seed yield of individual plants and increasing planting density (Figure 1A, R1V2). Additional combinations of promoters, coding sequences, vectors, and genotypic backgrounds can be optimized in the future to study the BR-specific regulation of various developmental stages and potentially improve crop yield.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
W-HL designed the study, wrote and modified the manuscript, and acquired funding. S-HZ performed the experiments, organized the figures, and wrote the manuscript. Y-TJ, L-QH, and J-HC completed the experiments. Y-JZ helped to modify the figures. H-WX helped to organize the results. All authors agreed to be accountable for the contents of this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31771591 and 31761163003), and the Opening Research Projects of the National Key Laboratory of Plant Molecular Genetics.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Lucia Colombo (University of Milan) and Prof. Zhi-Yong Wang (Carnegie Institution for Science) for kindly providing the pSTK::GUS (pCAMBIA 1300-H) transgenic plant and DWF4-OX seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00980/full#supplementary-material
References
Ahammed, G. J., Gao, C. J., Ogweno, J. O., Zhou, Y. H., Xia, X. J., Mao, W. H., et al. (2012a). Brassinosteroids induce plant tolerance against phenanthrene by enhancing degradation and detoxification in Solanum lycopersicum L. Ecotoxicol. Environ. Saf. 80, 28–36. doi: 10.1016/j.ecoenv.2012.02.004
Ahammed, G. J., Yuan, H. L., Ogweno, J. O., Zhou, Y. H., Xia, X. J., Mao, W. H., et al. (2012b). Brassinosteroid alleviates phenanthrene and pyrene phytotoxicity by increasing detoxification activity and photosynthesis in tomato. Chemosphere 86, 546–555. doi: 10.1016/j.chemosphere.2011.10.038
Bai, M. Y., Zhang, L. Y., Gampala, S. S., Zhu, S. W., Song, W. Y., Chong, K., et al. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. U.S.A. 104, 13839–13844. doi: 10.1073/pnas.0706386104
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. doi: 10.1105/tpc.1.1.37
Choe, S., Fujioka, S., Noguchi, T., Takatsuto, S., Yoshida, S., and Feldmann, K. A. (2001). Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26, 573–582. doi: 10.1046/j.1365-313x.2001.01055.x
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., et al. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15, 2603–2611. doi: 10.1105/tpc.015123
Fujioka, S., and Yokota, T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54, 137–164. doi: 10.1021/bk-1991-0474.ch008
He, J. X., Gendron, J. M., Sun, Y., Gampala, S. S., Gendron, N., and Sun, C. Q. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638. doi: 10.1126/science.1107580
He, J. X., Gendron, J. M., Yang, Y., Li, J., and Wang, Z. Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99, 10185–10190. doi: 10.1073/pnas.152342599
Hong, Z., Ueguchi-Tanaka, M., Fujioka, S., Takatsuto, S., Yoshida, S., Hasegawa, Y., et al. (2005). The Rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17, 2243–2254. doi: 10.1105/tpc.105.030973
Hong, Z., Ueguchi-Tanaka, M., Shimizu-Sato, S., Inukai, Y., Fujioka, S., Shimada, Y., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32, 495–508. doi: 10.1046/j.1365-313X.2002.01438.x
Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., et al. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15, 2900–2910. doi: 10.1105/tpc.014712
Huang, H. Y., Jiang, W. B., Hu, Y. W., Wu, P., Zhu, J. Y., Liang, W. Q., et al. (2013). BR signal influences Arabidopsis ovule and seed number through regulating related genes expression by BZR1. Mol. Plant 6, 456–469. doi: 10.1093/mp/sss070
Jiang, W. B., Huang, H. Y., Hu, Y. W., Zhu, S. W., Wang, Z. Y., and Lin, W. H. (2013). Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 162, 1965–1977. doi: 10.1104/pp.113.217703
Kooiker, M., Airoldi, C. A., Losa, A., Manzotti, P. S., Finzi, L., Kater, M. M., et al. (2005). BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant Cell 17, 722–729. doi: 10.1105/tpc.104.030130
Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. doi: 10.1016/S0092-8674(00)80357-8
Li, J., Lease, K. A., Tax, F. E., and Walker, J. C. (2001a). BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 5916–5921. doi: 10.1073/pnas.091065998
Li, J., Nam, K. H., Vafeados, D., and Chory, J. (2001b). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127, 14–22. doi: 10.1104/pp.127.1.14
Liljegren, S. J., Ditta, G. S., Eshed, Y., Savidge, B., Bowman, J. L., and Yanofsky, M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. doi: 10.1038/35008089
Morinaka, Y., Sakamoto, T., Inukai, Y., Agetsuma, M., Kitano, H., Ashikari, M., et al. (2006). Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 141, 924–931. doi: 10.1104/pp.106.077081
Nain, V., Verma, A., Kumar, N., Sharma, P., Ramesh, B., and Kumar, P. A. (2008). Cloning of an ovule specific promoter from Arabidopsis thaliana and expression of beta-glucuronidase. Indian J. Exp. Biol. 46, 207–211.
Nakamura, A., Fujioka, S., Sunohara, H., Kamiya, N., Hong, Z., Inukai, Y., et al. (2006). The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 140, 580–590. doi: 10.1104/pp.105.072330
Ohto, M. A., Floyd, S. K., Fischer, R. L., Goldberg, R. B., and Harada, J. J. (2009). Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex Plant Reprod. 22, 277–289. doi: 10.1007/s00497-009-0116-1
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Pérez-España, V. H., Sánchez-León, N., and Vielle-Calzada, J. P. (2011). CYP85A1 is required for the initiation of female gametogenesis in Arabidopsis thaliana. Plant Signal. Behav. 6, 321–326. doi: 10.4161/psb.6.3.13206
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Sahni, S., Prasad, B. D., Liu, Q., Grbic, V., Sharpe, A., Singh, S. P., et al. (2016). Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci. Rep. 6:28298. doi: 10.1038/srep28298
Sakamoto, T., Morinaka, Y., Ohnishi, T., Sunohara, H., Fujioka, S., Ueguchi-Tanaka, M., et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24, 105–109. doi: 10.1038/nbt1173
Schruff, M. C., Spielman, M., Tiwari, S., Adams, S., Fenby, N., and Scott, R. J. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251–261. doi: 10.1242/dev.02194
Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S., Yano, M., et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17, 776–790. doi: 10.1105/tpc.104.024950
Tanaka, A., Nakagawa, H., Tomita, C., Shimatani, Z., Ohtake, M., Nomura, T., et al. (2009). BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 151, 669–680. doi: 10.1104/pp.109.140806
Tong, H., Jin, Y., Liu, W., Li, F., Fang, J., Yin, Y., et al. (2009). DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58, 803–816. doi: 10.1111/j.1365-313X.2009.03825.x
Wang, L., Xu, Y., Zhang, C., Ma, Q., Joo, S. H., Kim, S. K., et al. (2008). OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS One 3:e3521. doi: 10.1371/journal.pone.0003521
Wang, Z. Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513. doi: 10.1016/S1534-5807(02)00153-3
Wu, C. Y., Trieu, A., Radhakrishnan, P., Kwok, S. F., Harris, S., Zhang, K., et al. (2008). Brassinosteroids regulate grain filling in rice. Plant Cell 20, 2130–2145. doi: 10.1105/tpc.107.055087
Yang, B. J., Lin, W. H., Fu, F. F., Xu, Z. H., and Xue, H. W. (2017). Receptor-like protein ELT1 promotes brassinosteroid signaling through interacting with and suppressing the endocytosis-mediated degradation of receptor BRI1. Cell Res. 27, 1182–1185. doi: 10.1038/cr.2017.69
Ye, Q., Zhu, W., Li, L., Zhang, S., Yin, Y., Ma, H., et al. (2010). Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. U.S.A. 107, 6100–6105.
Yin, Y., Wang, Z. Y., Mora-Garcia, S., Li, J., Yoshida, S., Asami, T., et al. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. doi: 10.1016/s0092-8674(02)00721-3
Zhang, Y., Zhang, Y. J., Yang, B. J., Yu, X. X., Wang, D., Zu, S. H., et al. (2016). Functional characterization of GmBZL2 (AtBZR1 like gene) reveals the conserved BR signaling regulation in Glycine max. Sci. Rep. 6:31134. doi: 10.1038/srep31134
Zhang, Y. J., Zhang, Y., Zhang, L. L., Huang, H. Y., Yang, B. J., Luan, S., et al. (2018). OsGATA7 modulates brassinosteroids-mediated growth regulation and influences architecture and grain shape. Plant Biotechnol. J. 16, 1261–1264. doi: 10.1111/pbi.12887
Zhou, Y., Xia, X., Yu, G., Wang, J., Wu, J., Wang, M., et al. (2015). Brassinosteroids play a critical role in the regulation of pesticide metabolism in crop plants. Sci. Rep. 5:9018. doi: 10.1038/srep09018
Keywords: brassinosteroid, architecture, organ size, planting density, yield
Citation: Zu S-H, Jiang Y-T, Hu L-Q, Zhang Y-J, Chang J-H, Xue H-W and Lin W-H (2019) Effective Modulating Brassinosteroids Signal to Study Their Specific Regulation of Reproductive Development and Enhance Yield. Front. Plant Sci. 10:980. doi: 10.3389/fpls.2019.00980
Received: 03 April 2019; Accepted: 11 July 2019;
Published: 26 July 2019.
Edited by:
Zuhua He, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Jianming Li, University of Michigan, United StatesAndrzej Bajguz, University of Białystok, Poland
Copyright © 2019 Zu, Jiang, Hu, Zhang, Chang, Xue and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Hui Lin, d2hsaW5Ac2p0dS5lZHUuY24=
 Song-Hao Zu
Song-Hao Zu Yu-Tong Jiang
Yu-Tong Jiang Li-Qin Hu
Li-Qin Hu Yan-Jie Zhang
Yan-Jie Zhang Jin-Hui Chang
Jin-Hui Chang Hong-Wei Xue
Hong-Wei Xue Wen-Hui Lin
Wen-Hui Lin