- 1Plant Biotechnology Research Center, Fudan-SJTU-Nottingham Plant Biotechnology R&D Center, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
- 2Instrumental Analysis Center, Shanghai Jiao Tong University, Shanghai, China
Catharanthus roseus contains a variety of monoterpenoid indole alkaloids (MIAs), among which bisindole alkaloids vinblastine and vincristine are well-known to have antitumor effects and widely used in clinical treatment. However, their contents in C. roseus is extremely low and difficult to meet market demands. Therefore, it is of great significance to study the transcriptional regulation mechanism of MIAs biosynthesis for high yielding of bisindole alkaloids in C. roseus. Studies have shown that MIAs biosynthesis in C. roseus has complex temporal and spacial specificity and is under tight transcriptional regulation, especially bisindole alkaloids. In this study, an AP2/ERF transcription factor CrERF5 was selected by RNA-seq of C. roseus organs, and its full-length sequence was cloned and characterized. CrERF5 responds to both ethylene and JA signals and is localized in the nucleus. CrERF5 could activate the transcriptional activity of the TDC promoter. Transient overexpressing CrERF5 in C. roseus petals caused a significant increase of the expression levels of key genes in both the upstream and downstream pathways of MIAs biosynthesis while silencing CrERF5 resulted in a decrease of them. Accordingly, the contents of bisindole alkaloids anhydrovinblastine and vinblastine, monoindole alkaloids ajmalicine, vindoline, and catharanthine were strongly enhanced in CrERF5-overexpressing petals while their contents decreased in CrERF5-silenced plants. These results suggested that CrERF5 is a novel positive ethylene-JA-inducible AP2/ERF transcription factor upregulating the MIAs biosynthetic pathway leading to the bisindole alkaloids accumulation.
Introduction
Catharanthus roseus produces a wide range of bioactive monoterpenoid indole alkaloids (MIAs), which could be divided into two categories: monoindole alkaloids and bisindole alkaloids (Pan et al., 2016a). Both have high economic values due to their wide spectrum of pharmaceutical applications, among which bisindole alkaloids vinblastine and vincristine are the best-known antitumor compounds of significant clinical medical values. The first MIA strictosidine is formed by the coupling of tryptamine and secologanin, products of the indole and iridoid pathways, respectively. In recent years, key biosynthetic enzymes in the steps from strictosidine to ajmalicine, vindoline, and catharanthine have been identified and characterized. Strictosidine is deglucosylated by Strictosidine β-glucosidase (SGD) to form a highly reactive aglycone, which can be converted to ajmalicine by Heteroyohimbine Synthase (HYS), or to stemmadenine by Geissoschizine Synthase 1 and 2 (GS1 and 2) in conjunction with Geissoschizine Oxidase (GO) and the recently published Redox1 and Redox2 (Tatsis et al., 2017; Qu et al., 2018). Next, stemmadenine is acetylated by Stemmadenine-O-Acetyltransferase (SAT) to form O-acetylstemmadenine, which is further converted to catharanthine by α/β Hydrolase 1 (HL1) or to tabersonine by α/β Hydrolase 2 (HL2) (Qu et al., 2018). And tabersonine is converted into vindoline by the action of seven enzymes (Yang et al., 2015). Finally, the monoindole alkaloids vindoline and catharanthine are coupled by CrPRX1 to produce bisindole alkaloids anhydrovinblastine, the direct precursor of vinblastine and vincristine (Verpoorte et al., 1997; Costa et al., 2008).
Monoterpenoid indole alkaloid biosynthesis in C. roseus is an organ and cell-type specific pathway under tight transcriptional regulation (Courdavault et al., 2014; Thomas et al., 2015). Up to now, six types of transcription factor (TF) genes involved in the MIA pathways have been cloned and identified. AP2/ERF TFs, ORCAs (octadecanoid-responsive Catharanthus AP2-domain proteins), are firstly identified as the major regulators of several MIA pathway genes in C. roseus (Menke et al., 1999; Van der Fits and Memelink, 2000). ORCA3 has been reported to have the ability to upregulate the expression of several MIAs biosynthetic genes, like anthranilate synthase (AS), tryptophan decarboxylase (TDC), D-1-deoxyxylulose 5-phosphate synthase (DXS), cytochrome P450 reductase (CPR), strictosidine synthase (STR),and desacetoxyvindoline 4-hydroxylase (D4H) but not SGD, deacetylvindoline 4-O-acetyltransferase (DAT), CrPRX1 and the iridoid pathway genes (Van der Fits and Memelink, 2000). ORCA4 and 5 form a physical cluster and functionally overlap with ORCA3 (Paul et al., 2017). CrMYC2 in bHLH (basic helix-loop-helix) family could regulate the expression of ORCAs and also had a strong effect on the accumulation of catharanthine and tabersonine (Montiel et al., 2011; Zhang et al., 2011). CrMYC2 and ORCA3 have both been shown to be capable of transactivating the TDC promoter in tobacco protoplasts (Paul et al., 2017), but no report of transactivation on the downstream genes promoters of MIA pathway. Another two bHLH TFs, BIS1 and BIS2 act in a complementary manner to ORCAs and transactivated the expression of all genes in the iridoid pathway (Van Moerkercke et al., 2015). CrWRKY1 possesses overlapping functions with the ORCA TFs. Similar to ORCA3, CrWRKY1 activates pathway enzymes including AS, DXS, SLS1, SGD, and TDC (Suttipanta et al., 2011). Negative regulators, like CR1 from AP2/ERF family, RMT1 from bHLH family, zinc finger-binding proteins (ZCTs), CrGBF1, and CrGBF2 from bZIP family, are characterized to repress the transcription of CrMYC2, ORCAs, STR, and TDC (Memelink et al., 2001; Sibéril et al., 2001; Chatel et al., 2003; Pauw et al., 2004; Zhang et al., 2011; Patra et al., 2013, 2017; Thamm et al., 2016; Zhou and Memelink, 2016; Liu et al., 2017). Besides, MYB-like transcription-factor CrBPF1 was found binding to an enhancer domain of the STR promoter (Van der Fits et al., 2000).
Positive and negative regulatory loops consisting of these activating and repressing TFs operate in the MIA pathway. Upon JA induction, active CrMYC2 released by JAZ proteins induces expression of BIS and ORCA TFs, forming positive auto-or cross-amplification loops for the MIA biosynthesis genes. In parallel, active CrMYC2 is supposed to induce transcription of JAZrepressors, leading to a negative regulatory loop involving BIS1 and RMT1, which maintains a balance of MIA precursor flux and also limits the increase of MIAs accumulation (Patra et al., 2017; Paul et al., 2017; Schweizer et al., 2018). What’s more, the primary regulatory loop of CrMYC2, BIS1/2, and ORCAs has limited effect on the accumulation of bisindole alkaloids (Schweizer et al., 2018). Overexpressing either ORCA2 or ORCA3 in C. roseus suspension cells or hairy roots enhanced the contents of tryptamine, catharanthine, serpentine, and ajmalicine (Van der Fits and Memelink, 2000; Peebles et al., 2009; Li et al., 2013; Sun and Peebles, 2016). Overexpressing ORCA4 in hairy roots increased the accumulation of tabersonine, catharanthine, and ajmalicine (Paul et al., 2017). Bisindole alkaloids (like anhydrovinblastine, vinblastine, and vincristine) were below the detection levels even when overexpressing ORCAs in C. roseus cell cultures and hairy roots (Wang et al., 2010; Li et al., 2013). In C. roseus plants ORCA3 overexpression significantly increased the accumulation of strictosidine, vindoline, catharanthine, and ajmalicine but failed to affect the anhydrovinblastine and vinblastine levels (Pan et al., 2012). Negative AP2/ERF TF CR1 only involved in the accumulation of vindoline and serpentine in C. roseus (Liu et al., 2017). It was reported that an AP2/ERF TF might interact with CrPRX1 to play a key role in the vinblastine content (Wang X. et al., 2016). But no further investigation was performed. These researches strongly suggest that there should be undiscovered AP2/ERF TFs participating in the regulation of bisindole alkaloids.
In our study, combined with transcriptome sequencing data, a newly AP2/ERF TF, CrERF5, was selected as a potential regulator on the MIA pathway leading to the accumulation of binsindole alkaloids. Petal transient expression system and virus-induced gene silencing (VIGS), two efficient and effective screening platforms for gene discovery and assessment, were used to test the gene function of CrERF5, which provided experimental evidence that CrERF5 plays a positive role in regulating the expression of several key genes in the upstream and downstream pathways of MIA biosynthesis and boosts the bisindole alkaloids accumulation.
Materials and Methods
Plant Growth and Treatments
Seeds of C. roseus (cv. Vitae Rose Red) were purchased from Floranova Ltd. (Dereham Norfolk NR20 4SS United Kingdom). The seeds were surface sterilized in 75% (v/v) ethanol for 1 min and 10% (v/v) NaClO2 for another 10 min. Subsequently, seeds were washed 10 times with sterile distilled water and germinated on Petri dishes containing MS basal medium. Cultures were grown under 16 h light and 8 h dark photoperiod at 25 ± 2°C. After germination for 2 weeks, seedlings were transplanted into soil and grown in the greenhouse at 25 ± 2°C. Flowers, leaves, stems, and roots of at least three biological replicates were collected in September 2018 during the blooming season of C. roseus. Two types of leaf samples were collected according to their position in the plants: younger leaves from the top four upper layers and older leaves from the bottom four lower layers.
MeJA and ethephon (ETH) were purchased from Sigma-Aldrich. The stocks were prepared by dissolving them in ethanol to make a concentration of 1 M. The working solution of MeJA and ETH was 0.1 mM (diluted by water) and used for the treatments. The sterile water was used as mock treatment for the controls. After spraying the working solutions, C. roseus leaves were collected at 0, 3, 6, 12, and 24 h. The experiments were conducted in randomized block design (RBD) with three replications.
Analysis of the RNA-Seq Data
The transcriptome sequence of C. roseus retrieved from the NCBI was used as the reference database (Góngora-Castillo et al., 2012). The RNA-seq data of different tissues (SRR122239 for flowers; SRR122251 for leaves; SRR122253 for stems; SRR122254 for roots) were downloaded from the NCBI. Transcript abundant estimation and expression level in fragment per kilobase exon per million mapped fragment (FPKM) were performed as described by Wang A. et al., 2016 (Supplementary Table S2). AP2/ERF TFs were identified in the C. roseus transcriptome sequences using BLASTP queries with the known Arabidopsis AP2/ERFs. Hierarchical cluster (HCL) analysis was performed using a Multi Experiment Viewer (MEV, v.4.9.0) (Howe et al., 2010). Sample clustering was carried out using the HCL method and the evolutionary distances were computed with the Pearson correction and average linkage clustering (default parameters). The maximum FPKM in the color scale of the HCL heatmap was set up as 100. The putative AP2/ERF transcription factor sequences of C. roseus and five other plant species were aligned by CLUSTAL X (Trinity College, Dublin, Ireland). The phylogenetic tree was constructed by MEGA 5 software (Kumar et al., 2016). Protein sequences of AP2/ERF transcription factors (TFs) from other species were also gathered using the NCBI.
Cloning of CrERF5 Gene
The full-length ORF of CrERF5 was PCR-amplified from the leaf cDNA of C. roseus (primers shown in Supplementary Table S1). The PCR product was gel-purified (Axygen, United States), and cloned into the pLB-Simple vector (MIAngen, Shanghai, China) for sequencing (Sangon Biotech, Shanghai, China) and expression constructs generation.
Subcellular Localization
To analyze the subcellular localization, the full-length ORF of CrERF5 was cloned into BamHI and XbaI sites of the pHB-YFP expression vector (Zhong et al., 2018). The construct was introduced into Agrobacterium tumefaciens strain GV3101, which was transiently transformed into 5-week-old Nicotiana benthamiana leaves following the published protocol (Voinnet et al., 2003; Yan et al., 2016). The YFP signal was observed after 48–72 h cultivation in low-light conditions using confocal microscopy (Leica Microsystems, Wetzlar, Germany), with argon laser excitation at 514 nm and a 505–550 nm emission filter set.
Dual-Luciferase (Dual-LUC) Assay
The assay was performed as previously reported (Shan et al., 2014). P2300-CrERF5 was transformed into A. tumefaciens strain GV3101 to act as effectors. PGREEN-TDC-pro, PGREEN-SGD-pro, PGREEN-DXS1-pro, and PGREEN-CrPRX1-pro were separately fused to the luciferase and subsequently co-transformed with the helper plasmid pSoup19 into GV3101 to act as the reporter. The P2300 empty vector (EV) was used as a negative control. The 35S promoter-driven Renilla was used as the internal reference. After 48 h cultivation in low-light conditions, the leaf samples were collected for Dual-LUC assay using commercial Dual-LUC reaction reagents (Promega). Six biological repeats were measured for each sample.
Virus-Induced Gene Silencing
Seedlings of C. roseus with two to three pairs of true leaves were used for VIGS. The genomic DNA of C. roseus was used as template to amplify the partial fragment of CrERF5 (386 bp) with specific primers (Supplementary Table S1). The PCR-generated fragment was then introduced into the terminal vector pTRV2 by Gateway LR ClonaseTM II (Invitrogen, Thermo Fisher Scientific, United States). Catharanthus Protoporphyrin IX magnesium chelatase subunit H (ChlH) was selected as a useful marker for determining the best time to collect samples for testing (Liscombe and O’Connor, 2011). The purified plasmid pTRV2-CrERF5, pTRV2-EV, pTRV2-ChlH were respectively transformed into A. tumefaciens strain GV3101. Three kinds of strains and GV3101 harboring pTRV1 were cultured overnight at 28°C in 30 mL Luria-Bertani medium containing 10 Mm MES, 20 μM acetosyringone, and 50 μg/mL kanamycin. These bacterial pellets were collected and then resuspended in 10 mL infiltration buffer (10 mM MES, 200 μM acetosyringone, and 1 mM MgCl2), and further incubated at 28°C for 3 h with shaking. The suspension of strain harboring pTRV1 was mixed with pTRV2-CrERF5, pTRV2, or pTRV2-ChlH in equal volume just before the infection. Young plants were wounded using toothpicks through the stem just below the apical meristem, and infiltrated with sterile water (mock inoculation), with the mixture of A. tumefaciens cultures harboring pTRV2-EV/pTRV1, pTRV2-CrERF5/pTRV1, and pTRV2-ChlH/pTRV1, respectively. Each treatment was injected into nine plants. After injection, the plants were cultured in a constant temperature incubator. The ChlH phenotype was observed 3 weeks after inoculation, and the other three sample plants were harvested at this stage. Leaves from three different plants per sample were cut in two. One sample was used for RNA extraction for gene expression analysis and the other sample was used for metabolites analysis. After recording the fresh weights of harvested materials, the samples were frozen in liquid nitrogen. Three replicates were made for each sample.
Transient Transformation in Catharanthus Roseus Petals
The p2300-CrERF5 construct was made for Agrobacterium-mediated transient transformation in C. roseus petals. Flower petals of 1-year-old C. roseus plants (grown under greenhouse conditions) were infiltrated with A. tumefaciens GV3101 harboring the respective constructs p2300-pCaMV35S:CrERF5 and p2300-pCaMV35S:GUS (as a control vector) for overexpression as previously described (Schweizer et al., 2018). After 48 h of incubation with A. tumefaciens solution, petals from five different flowers per sample were cut in two and immediately flash-frozen in liquid nitrogen. One sample was used for RNA extraction for gene expression analysis and the other sample was used for metabolite profiling. Three replicates were made for each sample.
Relative Expression Analysis via Quantitative RT-PCR
For quantitative RT-PCR (qRT-PCR), total RNA was isolated from the leaves stored at −20°C. DNA contamination was removed using DNase I following the protocol provided by the manufacturer (TaKaRa, Japan). The cDNAs were synthesized from the RNA samples using Prime ScriptTM Reverse Transcriptase Reagent according to the manufacturer’s instructions, using oligo (dT) as primer. The qRT-PCR analysis was performed in Peltier Thermal Cycler PTC200 (Bio-Rad), using the cDNAs as templates and gene-specific primers (in Supplementary Table S1) for the gene analysis. The primers for the MIA biosynthetic genes are listed in Supplementary Table S1 (Schweizer et al., 2018). SYBR Green (SYBR Premix Ex Taq; TaKaRa) was used in the PCR reactions to quantify the amount of dsDNA. The relative Ct (threshold cycle value) method (User Bulletin 2, ABIPRISM700 Sequence Detection System, update 2001; PerkinElmer/Applied Biosystems) was used to estimate the initial amount of template present in the reactions.
Metabolite Analysis by UPLC-Q/TOF MS
UPLC-Q/TOF MS analyses were performed using a Primer UPLC-Q-TOF mass spectrometer (Waters Corp., Milford, MA, United States) equipped with an electrospray ionization source. Data acquisition, handling and instrument control were performed using MassLynx 4.1 software. Mass range, m/z 50–1000 in positive mode; capillary, 3.0 kV; sample cone voltage, 35 V; extraction cone, 3.0; ion guide, 3.0; source temperature, 115°C; desolvation gas temperature, 300°C; flow rate of desolvation gas, 700 L h–1.
To ensure accuracy and reproducibility, all analyses were conducted using an independent reference spray via the Lock Spray interface; Tyr–Gly–Gly–Phe–Leu (leucine-encephalin, 200 pg μL–1) was used as a lock mass (m/z 556.2771) under positive-ion conditions for real-time calibration (flow rate of 30 μL min–1). Before the experiment, a single-point calibration was performed against the lock mass compound (leucine-encephalin). A multiple-point calibration was then performed over the range m/z 50–1000 using sodium formate solution (prepared from 10% formic acid/0.1 mol L–1 sodium hydroxide solution/acetonitrile, 10 mL/10 mL/80 mL). All points fell within 1 ppm during calibration. The resolving power of the instrument was 8000.
UPLC conditions: a BEH C18 (100 mm × 2.1 mm, 1.7 μm) column (Waters) was used; the column temperature was maintained at 40°C. Mobile phases A (water, 0.1% formic acid) and B (acetonitrile, 0.1% formic acid) were used; the gradient program was as follows: 0–4 min 5–25% B, 9–12 min 45–85% B, 14 min 100% B; 14.5–16 min 5% B, flow rate 0.35 mL min–1; injection volume 2 μL. Acquity PDA detector wavelength was fixed at 210, 254, and 278 nm.
A mixture of reference standards of MIAs and precursors (secologanin, ajmalicine, catharanthine, and vinblastine were purchased from Sigma-Aldrich, St. Louis, MO, United States; vindoline and anhydrovinblastine were purchased from Shanghai R&D Center for Standardization of Chinese Medicines, China) were detected and identified based on MS/MS spectra (Supplementary Figures S1, S2). Samples were applied in triplicate for quantification using calibration curves of the standards.
Data Statistical Analysis
All experiments were conducted with three replicates. Statistical analysis was performed using the student’s t-test by SPSS (version 14.0, Chicago, IL, United States). The values are mean ± SD for replicates in each group. p values ≤ 0.05 were considered as significant.
Results
An AP2/ERF TF Was Selected by RNA-Seq
To investigate more MIA biosynthesis-related AP2/ERF TFs, we screened the transcriptome database of C. roseus tissues and identified 69 AP2/ERF proteins by performing a BLASTP analysis with Arabidopsis AP2/ERF proteins. Hierarchical cluster analysis of 69 C. roseus AP2/ERF TFs and key regulatory and biosynthetic genes in the MIAs pathway revealed one AP2/ERF TF gene (ID: cra_locus_1827_iso=2_len=1147_ver=1) that was found to be clustered with SGD, ORCA3, and D4H in the heatmap (Figure 1A).
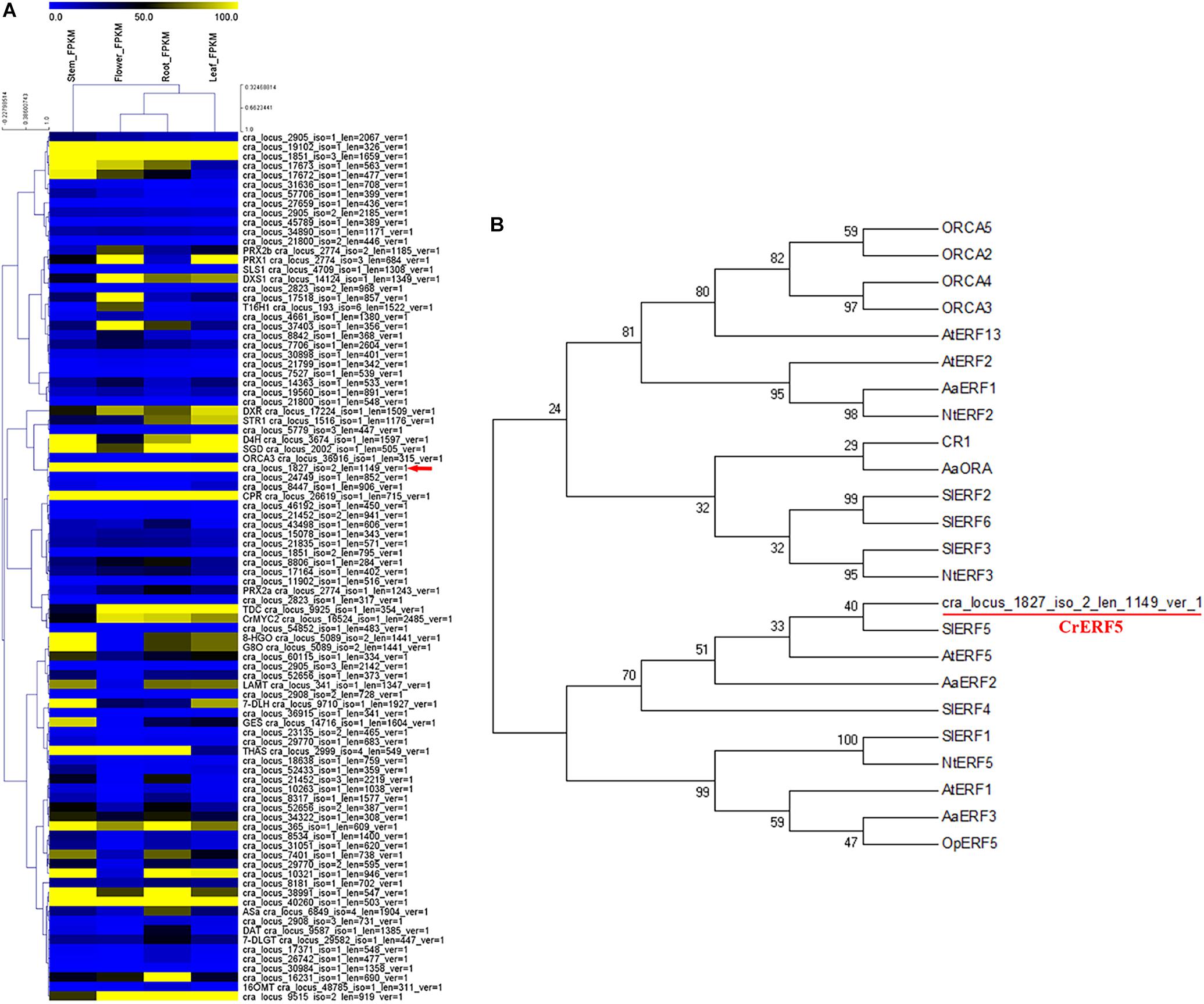
Figure 1. Identification and sequence analysis of CrERF5. (A) Hierarchical cluster analysis of AP2/ERF transcription factors in C. roseus flower, leaf, stem, and root. The average linkage hierarchical clustering with Pearson correlation was used. The color scale at the top represents the value of transformed reads per kilobase per million mapped reads. (B) Phylogenetic analysis of CrERF5 and other known AP2/ERF TFs from five plant species. Arabidopsis (At), Solanum lycopersicum (Sl), Artemisia annua (Aa), Nicotiana tabacum (Nt), and Ophiorrhiza pumila (Op). The tree presented here is a neighbor-joining tree based on amino acid sequence alignment.
Phylogenetic analysis was performed to investigate the evolutionary relationships between the putative AP2/ERF TF and 22 AP2/ERF proteins from five other plant species (Arabidopsis, Solanum lycopersicum, Artemisia annua, Nicotiana tabacum, Ophiorrhiza pumila). The putative AP2/ERF TF was classified to the same group with SlERF5, AtERF5, NtERF5, SlERF4, and AaERF2, and showed the closest relationship with SlERF5 (Figure 1B). Therefore, the candidate AP2/ERF TF was designated as CrERF5 for further functional study. CrERF5 encoded 342 amino acids and its predicted protein consisted of an AP2/ERF domain with 60 amino acids (177–227) in the central region (Supplementary Figure S3).
Subcellular Localization and Expression Pattern of CrERF5 Protein
To investigate the subcellular localization of CrERF5, laser scanning confocal microscopy was used to detect the YFP fluorescence signal of the fusion protein CrERF5-YFP expressed in N. benthamiana leaves. As illustrated in Figure 2A and Supplementary Figure S4, the YFP signal is specifically localized in the nucleus, consistent with CrERF5-YFP functioning as a transcription factor.
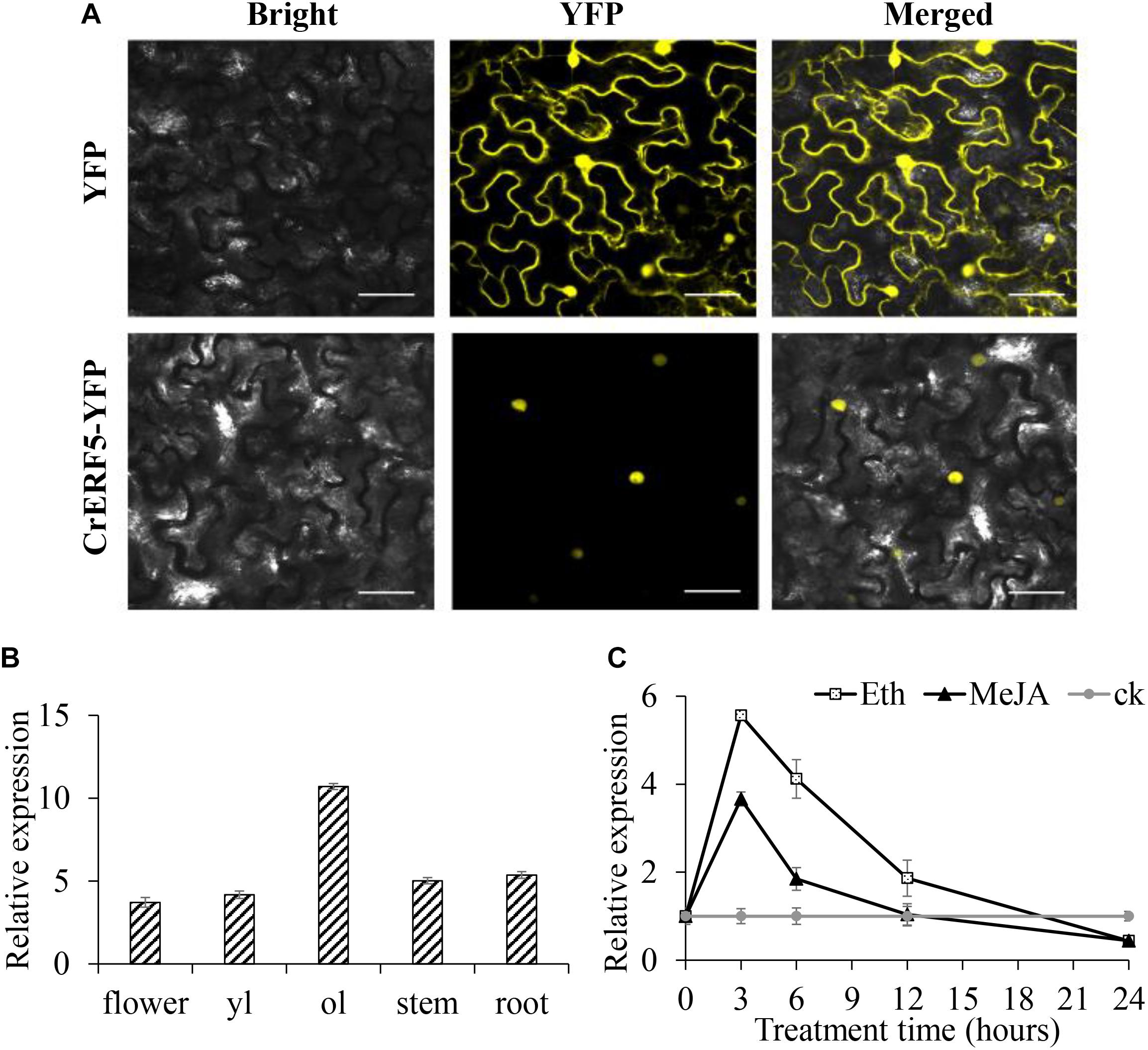
Figure 2. Subcellular localization and expression of CrERF5. (A) Subcellular localization of CrERF5 in tobacco leaves. Bars = 30 μm. (B) Relative expression of CrERF5 in flower, younger leaf (yl), older leaf (ol), stem, and root of C. roseus. (C) The expression pattern of CrERF5 in C. roseus leaves after treatment with 0.1 mM MeJA, 0.1 mM ETH, and sterile water (ck). The values of hormone treated samples were normalized to the ck samples at each time point, and the ck samples were normalized to themselves. The C. roseus N2227 gene was used as an internal control. The error bars represent the means ± SD (standard deviation) from three replicates.
The expression pattern of CrERF5 in the various tissues of C. roseus was analyzed through qRT-PCR. The results revealed that CrERF5 showed the highest expression level in the older leaves, and had a higher expression level in the flowers and stems than in the younger leaves and roots (Figure 2B). To further investigate the expression characteristics of CrERF5, we examined the time-course expression of CrERF5 under MeJA and ETH hormone treatments. As shown in Figure 2C, CrERF5 expression increased 5.6-fold over the control at 3 h and then back to the normal level during 12–24 h after ETH treatment. Under MeJA treatment CrERF5 showed a similar expression pattern with an increase of 3.7-fold over the control at 3 h. The results suggested that CrERF5 is a regulator that responds to both ethylene and JA signal.
Transactivation Activity of CrERF5 on MIA Biosynthetic Genes
In order to experimentally verify the involvement of CrERF5 in MIA pathway, Dual-LUC assay was used to determine the transactivation ability of CrERF5. Given the co-expression analysis, the SGD, DXS1, TDC, and CrPRX1 promoters were employed in the Dual-LUC assay (Figure 3). The results showed that CrERF5 significantly activated the TDC promoter by 2.7-fold compared to the controls, but slightly activate the CrPRX1 promoter and did not activate the SGD promoter. Furthermore, CrERF5 significantly repressed the transactivation of DXS1 promoter, which was not co-expressed with CrERF5.
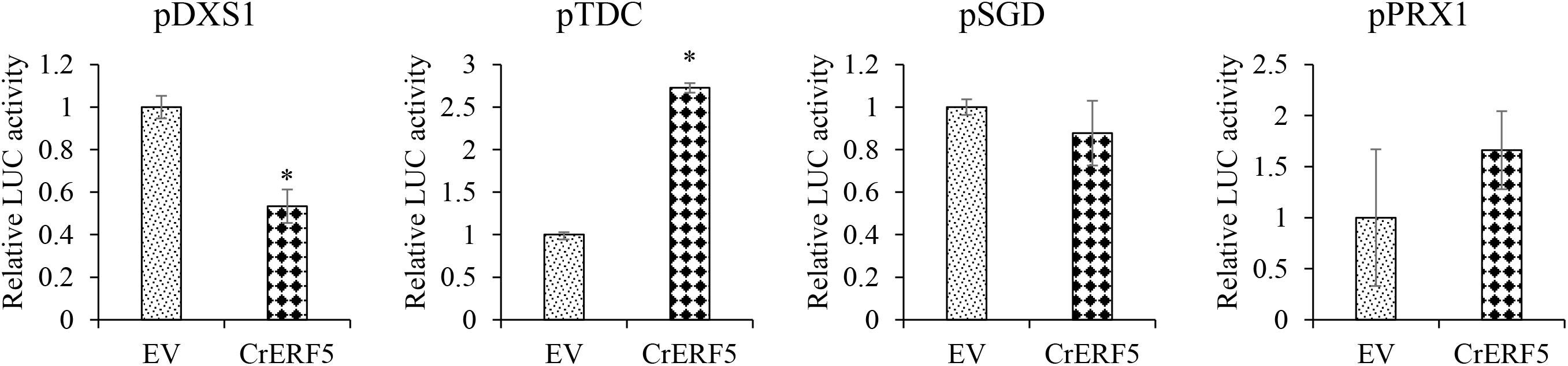
Figure 3. Transient Dual-luciferase assay of CrERF5 with MIA pathway genes (TDC, DXS1, SGD, and CrPRX1) promoters in tobacco. The relative LUC activity was normalized to the reference renilla (REN) luciferase. The error bars represent the means ± SD from three biological replicates, and asterisks indicate statistically significant differences compared with the controls. *P < 0.05.
Virus-Induced Gene Silence of CrERF5 Repressed the MIA Biosynthetic Pathway
In order to identify the gene function of CrERF5 in MIA pathway, VIGS was used to silence the expression of CrERF5 in C. roseus leaves. Transcripts analysis of CrERF5 and other 25 genes in the indole pathway, iridoid pathway, vindoline pathway and MIA biosynthetic pathway were monitored in C. roseus leaves by qRT-PCR. CrERF5 expression was suppressed by approximately 86% in CrERF5-silenced plants compared with the controls. In the CrERF5-silenced plants, the expression levels of two iridoid pathway genes (GES and SLS1), vindoline pathway gene T16H1, indole pathway gene TDC, and downstream MIA pathway genes (STR, SGD, Redox1, SAT, and PRX1) were significantly downregulated compared to the controls. But the expression levels of other genes did not significantly differ between the CrERF5-silenced plants and the controls (Figures 4A–C).
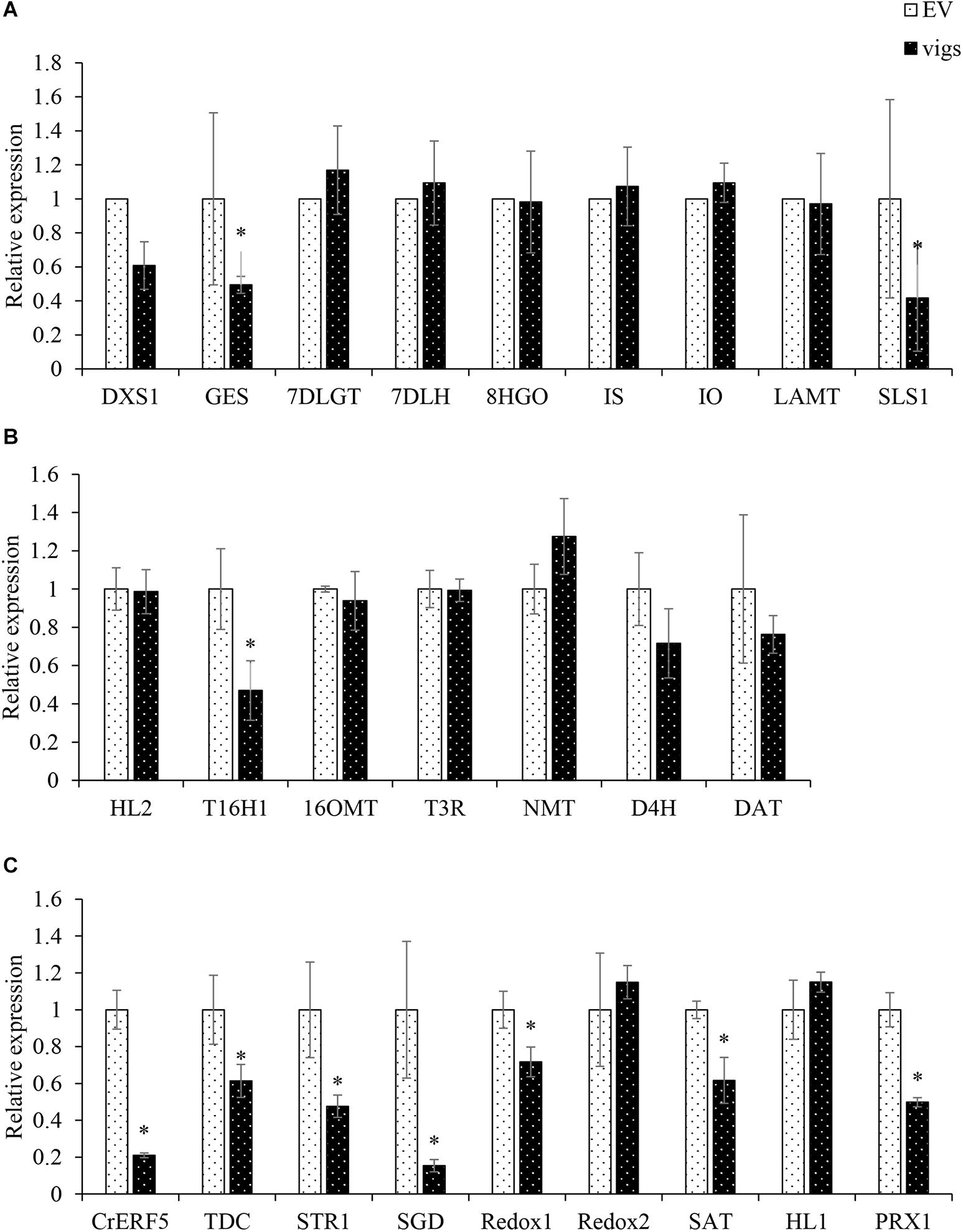
Figure 4. Expression of CrERF5 and MIA pathway genes in CrERF5-silencing C. roseus lines. (A) Iridoid pathway genes; (B) Vindoline pathway genes; (C) Indole and MIA pathway genes. The error bars represent the means ± SD from three biological replicates, and asterisks indicate statistically significant differences compared with the controls. *P < 0.05.
Subsequently, the contents of five MIAs (catharanthine, vindoline, ajmalicine, anhydrovinblastine, and vinblastine) and one precursor secologanin were measured by UPLC-Q/TOF MS (Figure 5). The accumulation of secologanin, catharanthine, ajmalicine, anhydrovinblastine and vinblastine in the leaves of CrERF5-silenced plants significantly downregulated with a decrease of 39.4, 28.9, 56.1, 20.9, and 54.4%, respectively, compared to the controls. The levels of catharanthine and vindoline showed a slight decrease. These results are consistent with the changes of the expression levels of MIAs biosynthetic genes in CrERF5-silenced plants, which indicated that CrERF5 might function as a positive regulator to regulate MIAs biosynthesis.
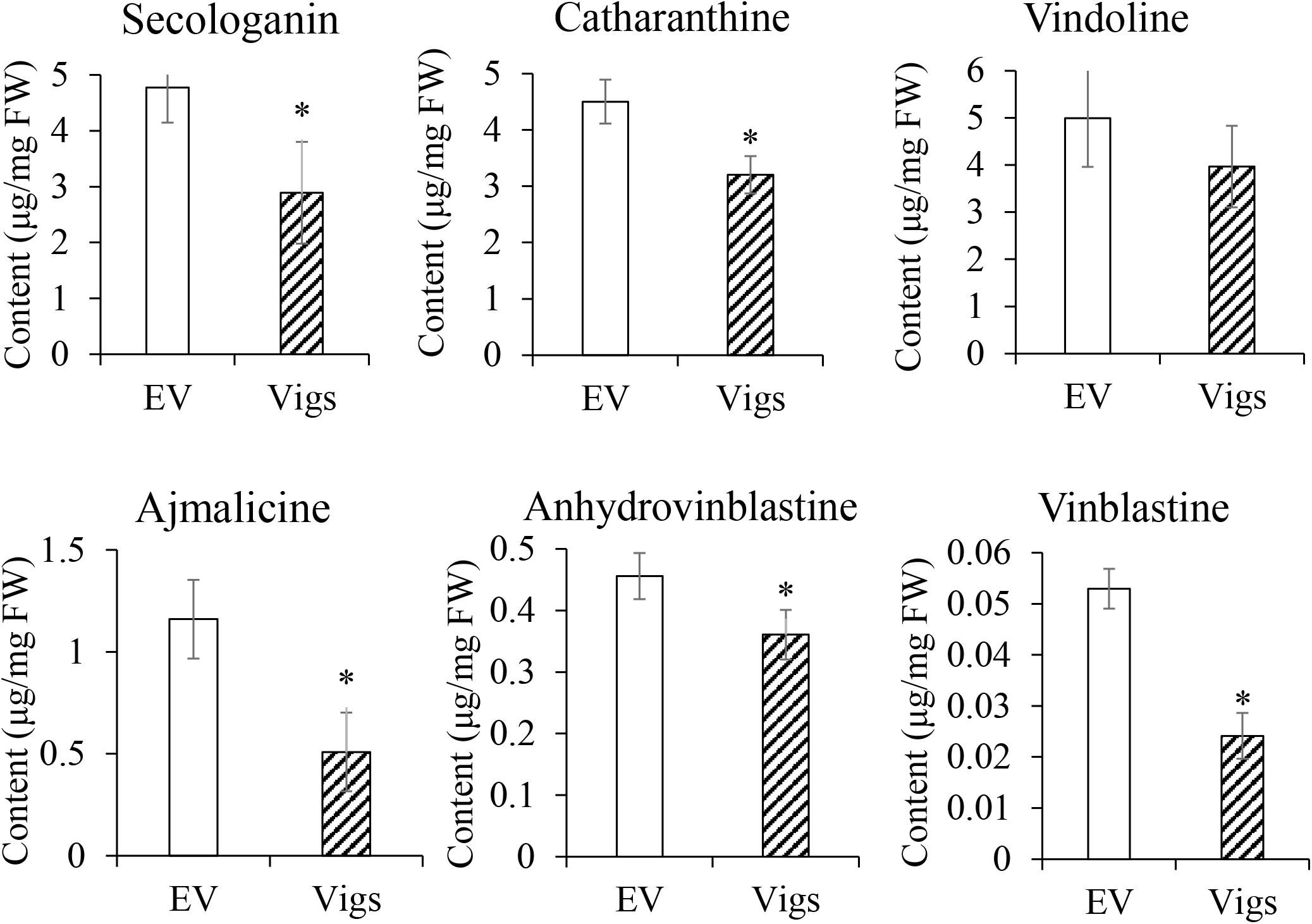
Figure 5. Contents of secologanin, ajmalicine, catharanthine, vindoline, anhydrovinblastine, and vinblastine in CrERF5-silencing C. roseus lines. The error bars represent the means ± SD from three biological replicates, and asterisks indicate statistically significant differences compared with the controls. *P < 0.05.
Transient Expression of CrERF5 Induces Expression of MIA Pathway Genes in C. roseus Flower Petals and Boosts MIAs Accumulation
To further confirm the function of CrERF5 in MIA biosynthesis, CrERF5 was transiently overexpressed in C. roseus flower petals. The results showed that overexpressing CrERF5 strongly activated the expression of MIA biosynthetic genes in both upstream and downstream pathways (Figures 6A–C). The significant induction of iridoid pathway genes GES and SLS1, indole pathway gene TDC, MIA biosynthetic genes STR1, Redox1, SAT, HL1, and CrPRX1, and vindoline pathway genes T16H1 and D4H was observed by transient CrERF5 overexpression. Next, the contents of five MIAs and one precursor were detected and analyzed as the previous part. Levels of the detectable MIAs and precursor were all significantly enhanced (Figure 7 and Supplementary Figure S5). Compared to the controls, the content of secologanin increased 34.9%, catharanthine increased 42.1%, anhydrovinblastine increased 71.9%, and vinblastine increased 50.7%. The results confirmed that CrERF5 plays a positive role in the regulation of the MIA pathway.
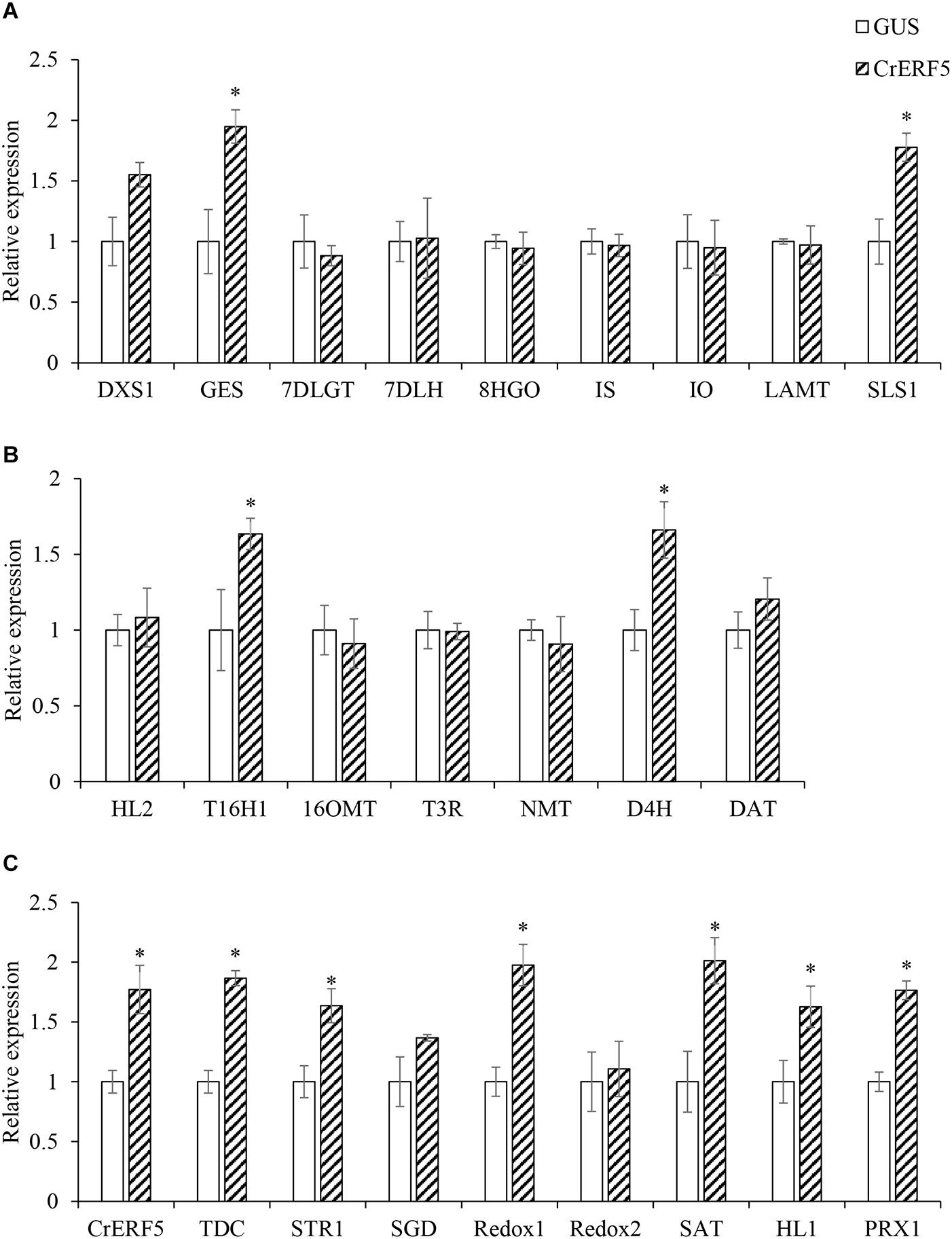
Figure 6. Expression of CrERF5 and MIA pathway genes in C. roseus flower petals transiently overexpressing CrERF5. (A) Iridoid pathway genes; (B) Vindoline pathway genes; (C) Indole and MIA pathway genes. The error bars represent the means ± SD from three biological replicates, and asterisks indicate statistically significant differences compared with the controls. *P < 0.05.
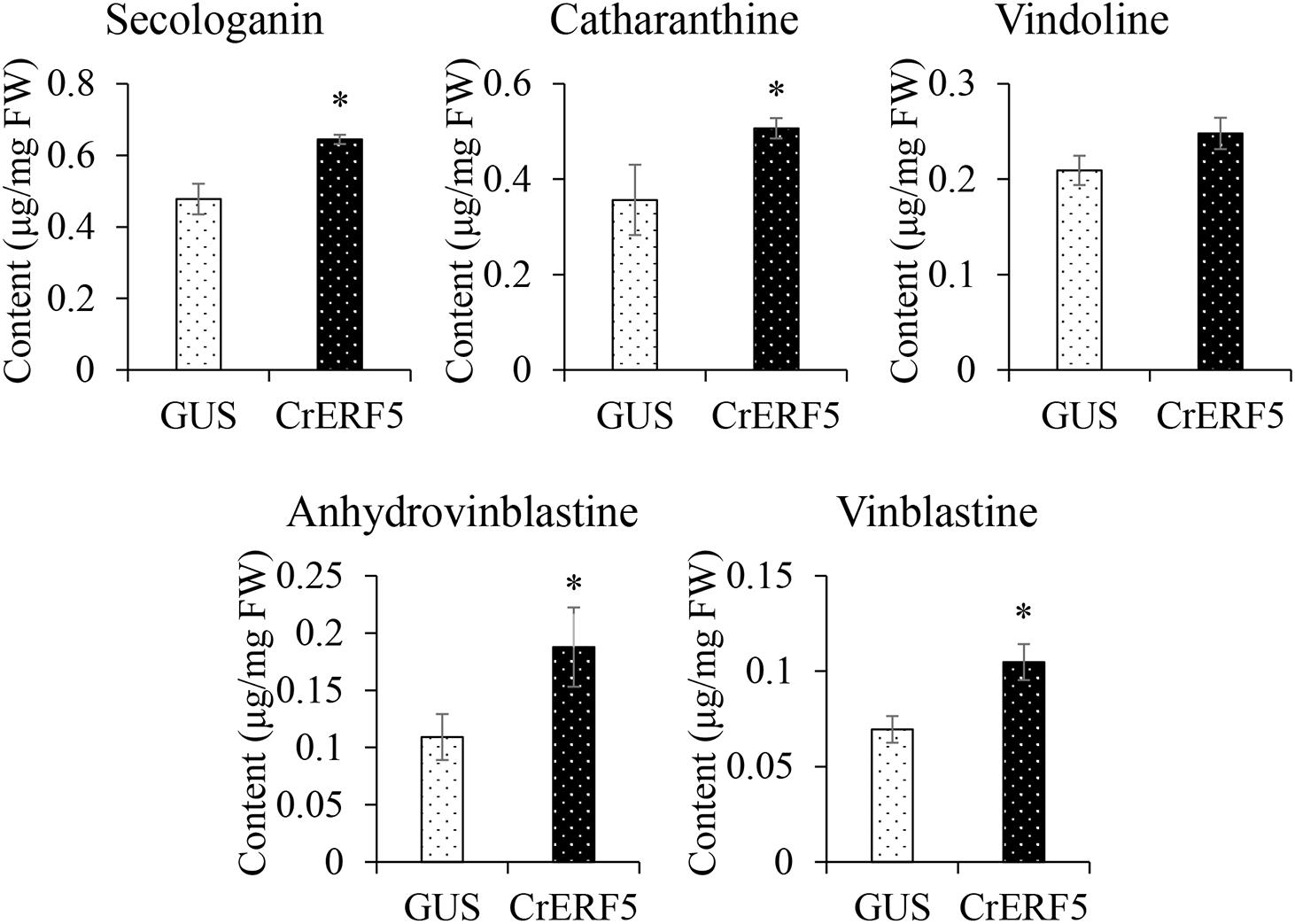
Figure 7. Contents of secologanin, ajmalicine, catharanthine, vindoline, anhydrovinblastine, and vinblastine in C. roseus flower petals transiently overexpressing CrERF5. The error bars represent the means ± SD from three biological replicates, and asterisks indicate statistically significant differences compared with the controls. *P < 0.05.
Discussion
In C. roseus, the reported AP2/ERF transcription factors, ORCAs and CR1, are mainly induced by the plant hormone jasmonate (JA) to regulate the MIA biosynthesis (Van der Fits and Memelink, 2000, 2001). The CrMYC2 JA-signaling module upregulates ORCAs expression, in turn activates several genes (TDC, CPR, and STR) in the upstream MIA pathway, steering the steps leading to strictosidine. Although more TFs are reported to be involved in JA-mediated regulation of MIA biosynthesis, such as BIS1/2, RMT1, CrWRKY1, CrGBF1/2, and the ZCTs, the regulatory network of MIA biosynthesis in C. roseus organs remains not well understood yet. For example, expression of the vinblastine, catharanthine and vindoline pathway genes is not regulated by the JA-responsive CrMYC2/BIS/ORCA module (Schweizer et al., 2018). All above indicated that besides JA-inducible TFs, many other hormone-inducible regulatory TFs need to be discovered still. Here, we have demonstrated that CrERF5 acts as a new ethylene-JA-inducible transcription factor upregulating the MIA pathway leading to the bisindole alkaloids accumulation.
In Arabidopsis, AtERF5 functioned as an activator of GCC box-dependent transcription and upregulated the expression of peroxisome-associated gene PEX1 (Salt-induced) (Fujimoto et al., 2000; Charlton et al., 2005; Mito et al., 2011). SlERF5 is a transcription activator for genes involved in the responses to ABA, biotic and abiotic stress (Chuang et al., 2010). NtERF5-Overexpressing plants show enhanced resistance to TMV with reference to reduced size of local hypersensitive-response lesions and impaired systemic spread of the virus (Fischer and Dröge-Laser, 2004). In our transcriptome analysis of C. roseus tissues (flower, leaf, stem, and root), CrERF5 was co-expressed with SGD, ORCA3, and D4H, and the activation of TDC promoter by CrERF5 verified its involvement in MIA biosynthesis.
The expression pattern of CrERF5 in organs indicated that CrERF5 transcripts increased with leaf age. As reported, the anhydrovinblastine level increases with leaf age, in parallel with an increase of CrPrx1 expression level (Costa et al., 2008; Pan et al., 2016b). CrERF5 expression pattern consisted with the accumulation pattern of bisindole alkaloids, which give an explanation why CrERF5 had a positive effect on the anhydrovinblastine and vinblastine contents. And CrERF5 expression was induced by ethylene or JA treatment. Moreover, ethylene induction had a stronger effect on CrERF5 expression than JA induction, not like other reported AP2/ERF TFs that were mainly induced by JA signaling. Previous studies have showed that exogenous ethylene not only elevate the Cd resistance of C. roseus but also effectively influence MIAs biosynthesis in C. roseus (Chen et al., 2017). Significant accumulation of vinblastine was found in response to high concentration of ethylene and Cu, in which ERF and MPK formed a positive feedback loop connecting two pathways actively involved in C. roseus (Pan et al., 2014). Comprehensive profiling of the vinblastine biosynthesis in response to ethephon suggested that ERF could regulate and interact with CrPRX1 to play a key role in vinblastine accumulation and peroxidase activity (Wang X. et al., 2016). These previous researches supported our results that CrERF5, responding to both ethylene and JA signals, might be involved in the bisindole alkaloids accumulation.
Further investigation via VIGS and transient petal expression showed that CrERF5 had a positive regulation on both upstream and downstream MIA pathways (Figure 8). The expression of indole pathway gene TDC was induced and activated by CrERF5, in turns which facilitated the boost of MIAs in the downstream pathway branches. CrERF5 upregulated the expression of GES and SLS and secologanin accumulation. Except GES and SLS, other genes in the iridoid pathway, like 7-DLGT, 7-DGT, IO, IS, and LAMT, were not under the control of CrERF5. Moreover, DXS1, the first step enzyme gene of MEP pathway, was repressed by CrERF5 in Dual-LUC assay. Given that GES and SLS are the enzymes in the closer catalyzing steps to form secologanin than DXS1, CrERF5 repression on DXS1 might have little effect on the secologanin biosynthesis. Vindoline pathway genes were partly regulated by CrERF5. But both overexpressing and silencing CrERF5 had limited effect on vindoline accumulation, indicating that CrERF5 might not control vindoline pathway. It has been reported that light-induced vindoline biosynthesis is regulated by GATA and PIF transcription factors in C. roseus (Liu et al., 2019). But the genes in the steps from strictosidine to vinblastine, like STR1, Redox1, SAT, and CrPRX1, were upregulated in CrERF5-overexpressing petals and downregulated in CrERF5-silencing plants, leading to a significant increase of the catharanthine, anhydrovinblastine and vinblastine accumulation. These results inferred that CrERF5 played a multiple positive role in regulating the MIA pathways, especially leading to the bisindole alkaloids biosynthesis.
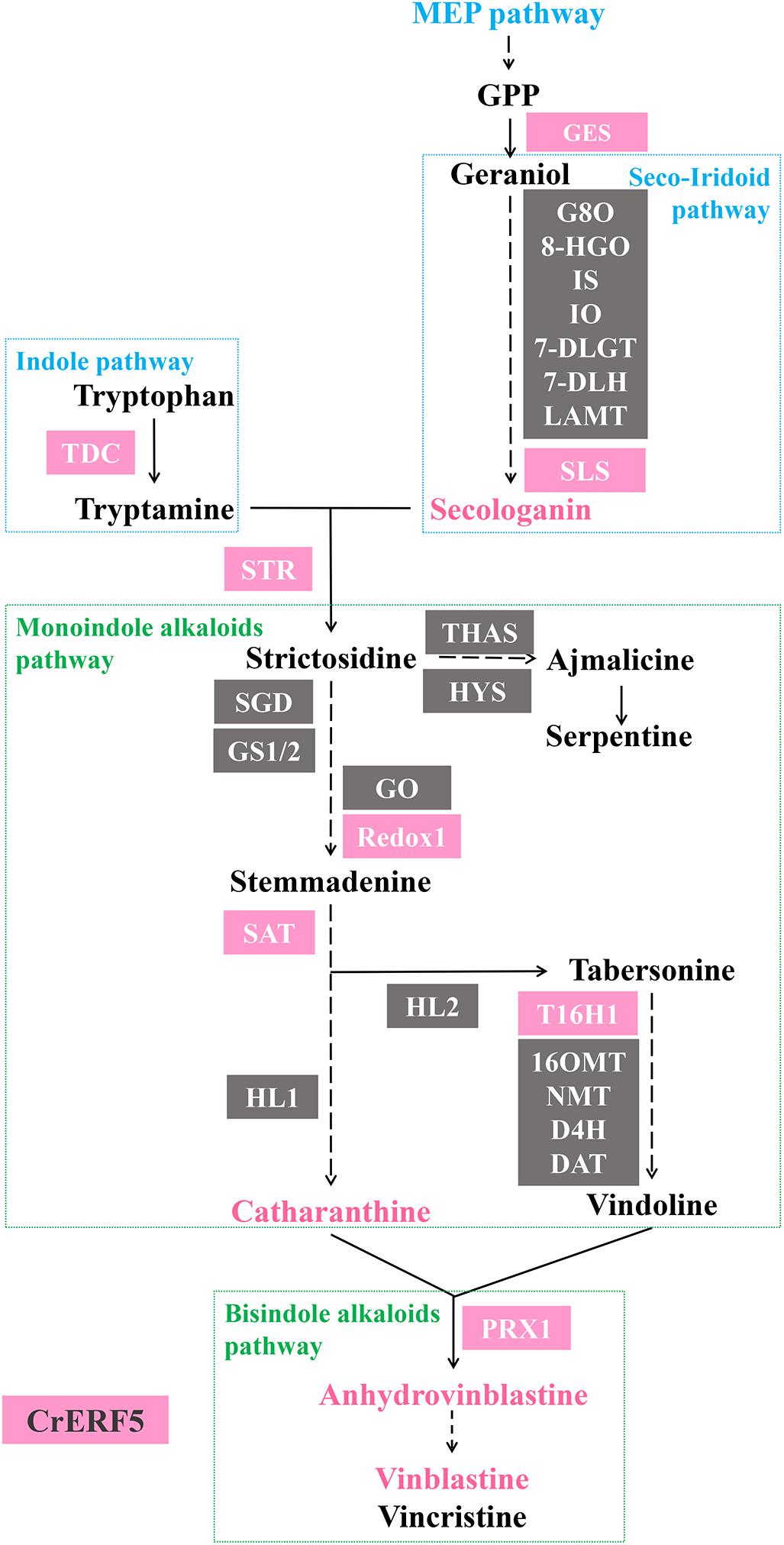
Figure 8. A scheme of CrERF5 regulation on MIA biosynthetic pathway. Genes regulated by CrERF5 are boxed in pink, respectively. Genes not regulated by CrERF5 are boxed in gray. MIAs upregulated by CrERF5 are in pink.
Conclusion
Our study found an important ethylene-JA-induced regulator CrERF5 that upregulates both the upstream and downstream MIA pathway branches, which could be used as a potent powerful tool to boost the high-value bisindole alkaloids production in the future. Combined with all these results, it indicated that in C. roseus ethylene signaling pathway might play an important role in transcriptional regulation on MIAs biosynthesis, especially bisindole alkaloids, which offered a new view to understand the complex molecular mechanism of MIAs biosynthesis.
Accession Numbers
Gene sequences have been deposited in the GeneBank database under the accession numbers CrERF5 (MK862158), ORCA2 (CAB93940.1), ORCA3 (ABW77571.1), ORCA4 (KR703577), ORCA5 (KR703578), AtERF1 (O80337), AtERF2 (O80338), AtERF5 (O80341), AtERF13 (NM_130048.3), AaERF1 (AEQ93554.1), AaERF2 (AEQ93555.1), AaERF3 (JN695782.1), AaORA (JQ797708), NtERF2 (NP_001311965.1), NtERF3 (NM_001325724.1), NtERF5 (AY655738.1), SlERF1 (NM_001247912.2), SlERF2 (NM_001247379.2), SlERF3 (NM_001246867), SlERF4 (NM_001247384.2), SlERF5 (AAS72389), SlERF6 (JN616265.1), and OpERF5 (XP_009764685.1).
Data Availability
Publicly available datasets were analyzed in this study. This data can be accessed from: https://www.ncbi.nlm.nih.gov.
Author Contributions
QP made her contributions to the concept or the design of the work, data analysis, and manuscript writing. CW made her contributions in CrERF5 gene cloning and VIGS experiments. ZX contributed to the Dual Luciferase assays and the results analysis. HW contributed to the metabolite analysis by UPLC-Q/TOF MS. QS contributed to the analysis of RNA-seq data. XF contributed to the subcellular localization of CrERF5. BP contributed to the expression profile of CrERF5 in C. roseus tissues and under hormone treatments. YM contributed to the transient overexpression of CrERF5 in C. roseus flowers. XS assisted to the data collection and analysis. KT contributed to the experiment design and manuscript correction.
Funding
This work was financially supported by the National Natural Science Foundation of China (31500240).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00931/full#supplementary-material
FIGURE S1 | Monoterpenoid indole alkaloid (MIA) component analysis in C. roseus by UPLC-Q/TOF MS. (A) Chromatogram spectra of MIAs. (B) Mass spectra of MIAs and precursors.
FIGURE S2 | MS/MS spectra of MIAs and precursor in petals and leaves. (A) MS/MS spectra of ajmalicine in flower, leaf, and standard; (B) MS/MS spectra of secologanin in flower, leaf, and standard; (C) MS/MS spectra of vindoline in flower, leaf, and standard; (D) MS/MS spectra of catharanthine in flower, leaf, and standard; (E) MS/MS spectra of anhydrovinblastine in flower, leaf, and standard; (F) MS/MS spectra of vinblastine in flower, leaf, and standard.
FIGURE S3 | Amino acid sequence alignments of the CrERF5 protein with ERF5 from other species. Identical amino acid residues are highlighted in black, and the upper line indicates the AP2/ERF domain.
FIGURE S4 | Subcellular localization of 35S:CrERF5-YFP (top) and 35S:YFP (below) in Nicotiana benthamiana leaf epidermal cells. Yellow, yellow fluorescent protein (YFP); blue, DAPI. Bars, 25 μm.
FIGURE S5 | The contents of ajmalicine/geissoschizine in C. roseus flower petals transiently overexpressing CrERF5. The error bars represent the means ± SD from three biological replicates, and asterisks indicate statistically significant differences compared with the controls. *P < 0.05.
TABLE S1 | Polymerase chain reaction (PCR) primers used in this study.
TABLE S2 | Fragments per kilobase of exon per million fragments mapped (FPKM) values of C. roseus AP2/ERF TFs, MIA pathway genes in flower, leaf, stem, and root. To tackle multiple contigs for certain genes, the contig with the highest FPKM was selected for the HCL analysis.
References
Charlton, W. L., Matsui, K., Johnson, B., Graham, I. A., Ohme-Takagi, M., and Baker, A. (2005). Salt-induced expression of peroxisome-associated genes requires components of the ethylene, jasmonate and abscisic acid signalling pathways. Plant Cell Environ. 28, 513–524. doi: 10.1111/j.1365-3040.2004.01293.x
Chatel, G., Montiel, G., Pré, M., Memelink, J., Thiersault, M., Saint-Pierre, B., et al. (2003). CrMYC1, a Catharanthus roseus elicitor-and jasmonate-responsive bHLH transcription factor that binds the G-box element of the strictosidine synthase gene promoter. J. Exp. Bot 54, 2587–2588. doi: 10.1093/jxb/erg275
Chen, Q., Wu, K., Tang, Z., Guo, Q. X., Guo, X., and Wang, H. (2017). Exogenous ethylene enhanced the cadmium resistance and changed the alkaloid biosynthesis in Catharanthus roseus seedlings. Acta Physiol. Plant 39:267.
Chuang, H. W., Harnrak, A., Chen, Y. C., and Hsu, C. M. (2010). A harpin-induced ethylene-responsive factor regulates plant growth and responses to biotic and abiotic stresses. Biochem. Biophys. Res. Commun. 402, 414–420. doi: 10.1016/j.bbrc.2010.10.047
Costa, M. M. R., Hilliou, F., Duarte, P., Pereira, L. G., Almeida, I., Leech, M., et al. (2008). Molecular cloning and characterization of a vacuolar class III peroxidase involved in the metabolism of anticancer alkaloids in Catharanthus roseus. Plant Physiol. 146, 403–417. doi: 10.1104/pp.107.107060
Courdavault, V., Papon, N., Clastre, M., Giglioli-Guivarc’h, N., St-Pierre, B., and Burlat, V. (2014). A look inside an alkaloid multisite plant: the Catharanthus logistics. Curr. Opin. Plant Biol. 19, 43–50. doi: 10.1016/j.pbi.2014.03.010
Fischer, U., and Dröge-Laser, W. (2004). Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Mol. Plant Microbe Interact. 17, 1162–1171. doi: 10.1094/mpmi.2004.17.10.1162
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Góngora-Castillo, E., Childs, K. L., Fedewa, G., Hamilton, J. P., Liscombe, D. K., Magallanes-Lundback, M., et al. (2012). Development of transcriptomic resources for interrogating the biosynthesis of monoterpene indole alkaloids in medicinal plant species. PLoS One 7:e52506. doi: 10.1371/journal.pone.0052506
Howe, E., Holton, K., Nair, S., Schlauch, D., Sinha, R., and Quackenbush, J. (2010). “MeV: multiexperiment viewer,” in Biomedical Informatics for Cancer Research, eds M. Ochs, J. Casagrande, and R. Davuluri (Berlin: SPRINGER), 267–277. doi: 10.1007/978-1-4419-5714-6_15
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, C. Y., Leopold, A. L., Sander, G. W., Shanks, J. V., Zhao, L., and Gibson, S. I. (2013). The ORCA2 transcription factor plays a key role in regulation of the terpenoid indole alkaloid pathway. BMC Plant Biol. 13:155. doi: 10.1186/1471-2229-13-155
Liscombe, D. K., and O’Connor, S. E. (2011). A virus-induced gene silencing approach to understanding alkaloid metabolism in Catharanthus roseus. Phytochemistry 72, 1969–1977. doi: 10.1016/j.phytochem.2011.07.001
Liu, J., Gao, F., Ren, J., Lu, X., Ren, G., and Wang, R. (2017). A novel AP2/ERF transcription factor CR1 regulates the accumulation of vindoline and serpentine in Catharanthus roseus. Front. Plant Sci. 8:2082. doi: 10.3389/fpls.2017.02082
Liu, Y., Patra, B., Pattanaik, S., Wang, Y., and Yuan, L. (2019). GATA and PIF transcription factors regulate light-induced vindoline biosynthesis in Catharanthus roseus. Plant Physiol. 180, 1316–1350. doi: 10.1104/pp.19.00489
Memelink, J., Verpoorte, R., and Kijne, J. (2001). ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism. Trends Plant Sci. 6, 212–219. doi: 10.1016/s1360-1385(01)01924-0
Menke, F. L. H., Champion, A., Kijne, J. W., and Memelink, J. (1999). A novel jasmonate-and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate-and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 18, 4455–4463. doi: 10.1093/emboj/18.16.4455
Mito, T., Seki, M., Shinozaki, K., Ohme-Takagi, M., and Matsui, K. (2011). Generation of chimeric repressors that confer salt tolerance in Arabidopsis and rice. Plant Biotechnol. J. 9, 736–746. doi: 10.1111/j.1467-7652.2010.00578.x
Montiel, G., Zarei, A., Körbes, A. P., and Memelink, J. (2011). The jasmonate-responsive element from the ORCA3 promoter from Catharanthus roseus is active in Arabidopsis and is controlled by the transcription factor AtMYC2. Plant Cell Physiol. 52, 578–587. doi: 10.1093/pcp/pcr016
Pan, Q., Mustafa, N. R., Tang, K., Choi, Y. H., and Verpoorte, R. (2016a). Monoterpenoid indole alkaloids biosynthesis and its regulation in Catharanthus roseus: a literature review from genes to metabolites. Phytochem. Rev. 15, 221–250. doi: 10.1007/s11101-015-9406-4
Pan, Q., Saiman, M. Z., Mustafa, N. R., Verpoorte, R., and Tang, K. (2016b). A simple and rapid HPLC-DAD method for simultaneously monitoring the accumulation of alkaloids and precursors in different parts and different developmental stages of Catharanthus roseus plants. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 1014, 10–16. doi: 10.1016/j.jchromb.2016.01.034
Pan, Q., Wang, Q., Yuan, F., Xing, S., Zhao, J., Choi, Y. H., et al. (2012). Overexpression of ORCA3 and G10H in Catharanthus roseus plants regulated alkaloid biosynthesis and metabolism revealed by NMR-metabolomics. PLoS One 7:e43038. doi: 10.1371/journal.pone.0043038
Pan, Y.-J., Liu, J., Guo, X.-R., Zu, Y.-G., and Tang, Z.-H. (2014). Gene transcript profiles of the TIA biosynthetic pathway in response to ethylene and copper reveal their interactive role in modulating TIA biosynthesis in Catharanthus roseus. Protoplasma 252, 813–824. doi: 10.1007/s00709-014-0718-9
Patra, B., Pattanaik, S., Schluttenhofer, C., and Yuan, L. (2017). A network of jasmonate-responsive bHLH factors modulate monoterpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol. 217, 1556–1581. doi: 10.1111/nph.14910
Patra, B., Schluttenhofer, C., Wu, Y., Pattanaik, S., and Yuan, L. (2013). Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta 1829, 1236–1247. doi: 10.1016/j.bbagrm.2013.09.006
Paul, P., Singh, S. K., Patra, B., Sui, X., Pattanaik, S., and Yuan, L. (2017). A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol. 213, 1107–1123. doi: 10.1111/nph.14252
Pauw, B., Hilliou, F. A. O., Martin, V. S., Chatel, G., de Wolf, C. J. F., Champion, A., et al. (2004). Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus. J. Biol. Chem. 279, 52940–52948. doi: 10.1074/jbc.m404391200
Peebles, C. A. M., Hughes, E. H., Shanks, J. V., and San, K. Y. (2009). Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with jasmonic acid elicitation of Catharanthus roseus hairy roots over time. Metab. Eng. 11, 76–86. doi: 10.1016/j.ymben.2008.09.002
Qu, Y., Easson, M. E. A. M., Simionescu, R., Hajicek, J., Thamm, A. M. K., Salim, V., et al. (2018). Solution of the multistep pathway for assembly of corynanthean, strychnos, iboga, and aspidosperma monoterpenoid indole alkaloids from 19E-geissoschizine. Proc. Natl. Acad. Sci. U.S.A. 115, 3180–3185. doi: 10.1073/pnas.1719979115
Schweizer, F., Colinas, M., Pollier, J., Moerkercke, A. V., Bossche, R. V., Clercq, R. D., et al. (2018). An engineered combinatorial module of transcription factors boosts production of monoterpenoid indole alkaloids in Catharanthus roseus. Metab. Eng. 48, 150–162. doi: 10.1016/j.ymben.2018.05.016
Shan, C. M., Shangguan, X. X., Zhao, B., Zhang, X. F., Chao, L. M., Yang, C. Q., et al. (2014). Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat. Commun. 5:5519. doi: 10.1038/ncomms6519
Sibéril, Y., Benhamron, S., Memelink, J., Giglioli-Guivarc’h, N., Thiersault, M., Boisson, B., et al. (2001). Catharanthus roseus G-box binding factors 1 and 2 act as repressors of strictosidine synthase gene expression in cell cultures. Plant Mol. Biol. 45, 477–488.
Sun, J., and Peebles, C. A. M. (2016). Engineering overexpression of ORCA3 and strictosidine glucosidase in Catharanthus roseus hairy roots increases alkaloid production. Protoplasma 253, 1255–1264. doi: 10.1007/s00709-015-0881-7
Suttipanta, N., Pattanaik, S., Kulshrestha, M., Patra, B., Singh, S. K., and Yuan, L. (2011). The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 157, 2081–2093. doi: 10.1104/pp.111.181834
Tatsis, E. C., Carqueijeiro, I., Bernonville, T. D. D., Franke, J., Dang, T. T. T., Oudin, A., et al. (2017). A three enzyme system to generate the Strychnos alkaloid scaffold from a central biosynthetic intermediate. Nat. Commun. 8:316. doi: 10.1038/s41467-017-00154-x
Thamm, A. M. K., Qu, Y., and Luca, V. D. (2016). Discovery and metabolic engineering of iridoid/secoiridoid and monoterpenoid indole alkaloid biosynthesis. Phytochemistry Rev. 15, 1–23.
Thomas, D. D. B., Marc, C., Sébastien, B., Audrey, O., Vincent, B., Gaëlle, G., et al. (2015). Phytochemical genomics of the Madagascar periwinkle: unravelling the last twists of the alkaloid engine. Phytochemistry 113, 9–23. doi: 10.1016/j.phytochem.2014.07.023
Van der Fits, L., and Memelink, J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289, 295–297. doi: 10.1126/science.289.5477.295
Van der Fits, L., and Memelink, J. (2001). The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 25, 43–53. doi: 10.1111/j.1365-313x.2001.00932.x
Van der Fits, L., Zhang, H., Menke, F. L. H., Deneka, M., and Memelink, J. (2000). A Catharanthus roseus BPF-1 homologue interacts with an elicitor-responsive region of the secondary metabolite biosynthetic gene Str and is induced by elicitor via a JA-independent signal transduction pathway. Plant Mol. Biol. 44, 675–685.
Van Moerkercke, A., Steensma, P., Schweizer, F., Pollier, J., Gariboldi, I., Payne, R., et al. (2015). The bHLH transcription factor BIS1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus. Proc. Natl. Acad. Sci. U.S.A. 112, 8130–8135. doi: 10.1073/pnas.1504951112
Verpoorte, R., van der Heijden, R., and Moreno, P. R. (1997). Biosynthesis of terpenoid indole alkaloids in Catharanthus roseus Cells. Alkaloids Chem. Pharmacol. 49, 221–299. doi: 10.1016/s0099-9598(08)60017-6
Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956. doi: 10.1046/j.1365-313x.2003.01676.x
Wang, A., Hu, J., Huang, X., Xia, L., Zhou, G., and Yan, Z. (2016). Comparative transcriptome analysis reveals heat-responsive genes in Chinese cabbage (Brassica rapassp. chinensis). Front. Plant Sci. 7:939. doi: 10.3389/fpls.2016.00939
Wang, C. T., Liu, H., Gao, X. S., and Zhang, H. X. (2010). Overexpression of G10H and ORCA3 in the hairy roots of Catharanthus roseus improves catharanthine production. Plant Cell Rep. 29, 887–894. doi: 10.1007/s00299-010-0874-0
Wang, X., Pan, Y. J., Chang, B. W., Hu, Y. B., Guo, X. R., and Tang, Z. H. (2016). Ethylene-induced vinblastine accumulation is related to activated expression of downstream TIA pathway genes in Catharanthus roseus. Biomed. Res. Int. 2016:3708187. doi: 10.1155/2016/3708187
Yan, T., Chen, M., Shen, Q., Li, L., Fu, X., Pan, Q., et al. (2016). Homeodomain Protein 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 213, 1145–1155. doi: 10.1111/nph.14205
Yang, Q., Easson, M. L. A. E., Jordan, F., Razvan, S., Tomas, H., and Vincenzo, D. L. (2015). Completion of the seven-step pathway from tabersonine to the anticancer drug precursor vindoline and its assembly in yeast. Proc. Natl. Acad. Sci. U.S.A. 112, 6224–6229. doi: 10.1073/pnas.1501821112
Zhang, H., Hedhili, S., Montiel, G., Zhang, Y., Chatel, G., Pre, M., et al. (2011). The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 67, 61–71. doi: 10.1111/j.1365-313X.2011.04575.x
Zhong, Y., Li, L., Hao, X., Fu, X., and Tang, K. (2018). AaABF3, an abscisic acid–responsive transcription factor, positively regulates artemisinin biosynthesis in Artemisia annua. Front. Plant Sci. 9:1777. doi: 10.3389/fpls.2018.01777
Keywords: Catharanthus roseus, AP2/ERF transcription factor, bisindole alkaloids, transcriptional regulation, MIAs biosynthesis
Citation: Pan Q, Wang C, Xiong Z, Wang H, Fu X, Shen Q, Peng B, Ma Y, Sun X and Tang K (2019) CrERF5, an AP2/ERF Transcription Factor, Positively Regulates the Biosynthesis of Bisindole Alkaloids and Their Precursors in Catharanthus roseus. Front. Plant Sci. 10:931. doi: 10.3389/fpls.2019.00931
Received: 16 February 2019; Accepted: 03 July 2019;
Published: 18 July 2019.
Edited by:
Danièle Werck, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Johan Memelink, Leiden University, NetherlandsVincent Courdavault, Université de Tours, France
Jacob Pollier, Flanders Institute for Biotechnology, Belgium
Copyright © 2019 Pan, Wang, Xiong, Wang, Fu, Shen, Peng, Ma, Sun and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qifang Pan, cGFucWZAc2p0dS5lZHUuY24=
 Qifang Pan
Qifang Pan Chenyi Wang
Chenyi Wang Zhiwei Xiong
Zhiwei Xiong Hang Wang2
Hang Wang2 Qian Shen
Qian Shen Bowen Peng
Bowen Peng Yanan Ma
Yanan Ma Kexuan Tang
Kexuan Tang