- 1Laboratoire de Génétique Moléculaire, Immunologie et Biotechnologie, Faculté des Sciences de Tunis, Université de Tunis El Manar, Tunis, Tunisia
- 2Unidad Técnica del IPNA-CSIC, Laboratorio de Agrobiología Juan José Bravo Rodríguez (Cabildo Insular de La Palma), Santa Cruz de La Palma, Spain
- 3Instituto de Hortofruticultura Subtropical y Mediterránea La Mayora (IHSM La Mayora -UMA-CSIC), Algarrobo, Spain
During the last decade, S-genotyping has been extensively investigated in fruit tree crops such as those belonging to the Prunus genus, including plums. In plums, S-allele typing has been largely studied in diploid species but works are scarcer in polyploid species due to the complexity of the polyploid genome. This study was conducted in order to analyze the S-genotypes of 30 diploid P. salicina, 17 of them reported here for the first time, and 29 hexaploid plums (24 of P. domestica and 5 of P. insititia). PCR analysis allowed identifying nine S-alleles in the P. salicina samples allocating the 30 accessions in 16 incompatibility groups, two of them identified here for the first time. In addition, pollen tube growth was studied in self-pollinated flowers of 17 Tunisian P. salicina under the microscope. In 16 samples, including one carrying the Se allele, which has been correlated with self-compatibility, the pollen tubes were arrested in the style. Only in one cultivar (“Bedri”), the pollen tubes reached the base of the style. Twelve S-alleles were identified in the 24 P. domestica and 5 P. insititia accessions, assigning accessions in 16 S-genotypes. S-genotyping results were combined with nine SSR loci to analyze genetic diversity. Results showed a close genetic relationship between P. domestica and P. salicina and between P. domestica and P. insititia corroborating that S-locus genotyping is suitable for molecular fingerprinting in diploid and polyploid Prunus species.
Introduction
Plums, belonging to the Prunus genus (family Rosaceae, subfamily Prunoideae, sub-genus Prunophora), have been cultivated for at least 2000–4000 years, being among the first fruit species to attract human interest and appear to have been domesticated very early (Gautier, 1977). In fact, one of the reasons for the frequent domestication of Prunus species might have been the coincidence between the location of the center of diversity of Prunus and the first ancient high civilizations of human history (OCDE, 2002). Within Prunophora, the section Euprunus contains the Asian (such as P. salicina Lindl. and P. simonii Carr.) and European (such as P. domestica L., P. cerasifera Ehrh., P. insititia L. and P. spinosa L.) plums, whereas the section Prunocerasus contains the North American species (such as P. americana Marsh., P. angustifolia Marsh., P. hortulana Bailey, P. munsoniana Wight & Hedr., P. maritima Marsh. and P. subcordata Benth).
The basic somatic chromosome number for Prunus is x = 8. However, natural interspecific hybridization is responsible for several cases of polyploidy in this genus (Das et al., 2011). Thus, chromosome numbers of different plum species varies from diploid to hexaploid: P. salicina, P. cerasifera, P. americana and P. simonii are diploid with 2n = 16, P. spinosa is tetraploid with 2n = 32 whereas P. domestica and P. insititia are hexaploid with 2n = 48. Polyploidy is of widespread occurrence in plants and a major mechanism of adaptation and speciation. It is thus recognized as a major force in evolution (Van de Peer et al., 2017). Polyploidy is important, too, from a practical point of view, since polyploid plants are often more vigorous and may be more resistant to frost and the attacks of parasitic fungi than their diploid counterparts (OCDE, 2002). In contrast, the complexity of the polyploid genome makes molecular studies in these species a complex and elusive task.
Plums are mainly self-incompatible species and show, like other Prunus species, a Gametophytic Self-Incompatibility system (GSI). Within Rosaceae, the self-incompatibility mechanism (SI) is a widespread genetic system that promotes outcrossing by enabling hermaphrodite plants to avoid self- and cross-fertilization with close relatives. This system comprises recognition of self-related pollen, by cells of the pistil, followed by rejection of the incompatible pollen. In GSI, pollen tube growth is aborted during pollen tube growth in the style (De Nettancourt, 1977). GSI is the most common self-incompatibility system in angiosperms, and has been reported in more than 60 families of flowering plants (Kao and McCubbin, 1996). The recognition in GSI is genetically determined by a polymorphic locus (S), encoding at least two linked genes that determine the pistil and pollen phenotypes (Kao and Tsukamoto, 2004). In Prunus species, the S-locus is less than 70 kb (Ushijima et al., 2001) and contains two linked genes; one, S-RNase, is expressed in the pistil while the second, SFB (S-haplotype-specific F-box), is expressed in the pollen (Ushijima et al., 2001; Entani et al., 2003). Genotypes sharing the same S-genotypes are genetically inter-incompatible and included in the same Incompatibility Group (IG), while cultivars having different S-genotypes are inter-compatible and are allocated to different IG. As consequence, assignment of each cultivar to its corresponding IG is essential for orchard planning and appropriate crosses in breeding programs (Guerra and Rodrigo, 2015; Halász et al., 2017).
In the last two decades, cloning and characterization of the S-locus genes in species of the Rosaceae have allowed the development of fast and accurate PCR methods for S-allele typing based on the polymorphism of amplified fragment length. This strategy does not require controlled pollinations to evaluate the percentage of fruit set in orchard conditions or microscopic observations of pollen tube growth in flowers of controlled crosses (Guerra et al., 2009; Guerra and Rodrigo, 2015; Matsumoto and Tao, 2016). Several PCR S-allele typing methods (Beppu et al., 2002, 2003) were widely and successfully developed in many Prunus species such as Japanese plum (Yamane et al., 1999), sweet cherry (Tao et al., 1999), almond (Tamura et al., 2000), European apricot (Romero et al., 2004) and Japanese apricot (Yaegaki et al., 2001).
In Japanese plum, “Sordum” (SaSb) was the first S-genotyped cultivar by cloning the cDNA of the S-RNases in the style (Yamane et al., 1999). Since then, numerous S-genotyping works have been conducted using PCR and analytic and/or capillary electrophoresis to detect intron length polymorphisms of both S-RNase and SFB genes (Tao et al., 1999; Yamane et al., 2001; Beppu et al., 2002, 2003, 2012a; Sonneveld et al., 2003; Sapir et al., 2004, 2007; Vaughan et al., 2006; Halász et al., 2007; Zhang et al., 2007, 2008; Guerra et al., 2009, 2012; Guerra and Rodrigo, 2015). Until 2015 (Guerra and Rodrigo, 2015). A total of 42 S-RNase alleles and 15 SFB alleles have been identified in Japanese plum, allocating cultivars in 26 incompatibility groups (IG).
Publications regarding SI in polyploid plums are scarcer. Most of these studies were conducted to test whether polyploidy influences self-compatibility (SC). The limited works of S-genotyping in polyploid plums might be explained by the fact that SI occurs less frequently in polyploid than in diploid species (Halász et al., 2014). The first study regarding the molecular genetics of SI in P. domestica (2n = 48) was published by Sutherland et al. (2004). Later, six S-locus-specific markers previously used for other Prunus species were successfully tested on 33 domestic plum cultivars (P. domestica) by Kota-Dombrovska and Lācis (2013). Preliminary information regarding the S-genotypes of 16 P. domestica cultivars grown in Hungary was conducted by Halász et al. (2014). More recently, Halász et al. (2017) provided an S-allele profiling of 17 SI polyploid Hungarian plums (10 P. spinosa L., 4 P. insititia L. and 3 P. spinosa X P. domestica) and identified the sizes of 23 S-RNase alleles using PaConsII consensus primers spanning the second intron of the S-RNase gene.
Due to the economic importance, risk of vulnerability and the high diversity of Tunisian plum germplasm, the first objective of this study was to establish the S-genotypes, the incompatibility groups and the self-(in)compatibility phenotypes of 17 Tunisian P. salicina plums with unknown S-genotypes by combining PCR analysis and observation of pollen tube growth in self-pollinated flowers. The second objective was to fingerprint the S-alleles in hexaploid plums. Thus, 29 polyploid plums (5 P. insititia from Tunisia and 24 P. domestica accessions from Spain) with unknown S-genotypes were S-genotyped for the first time. The third objective was to elucidate the pattern of genetic diversity based on nine SSR loci. Hence, the results of S-locus and those of SSR markers were combined to investigate the genetic relationships between the studied Prunus species.
Materials and Methods
Plant Material
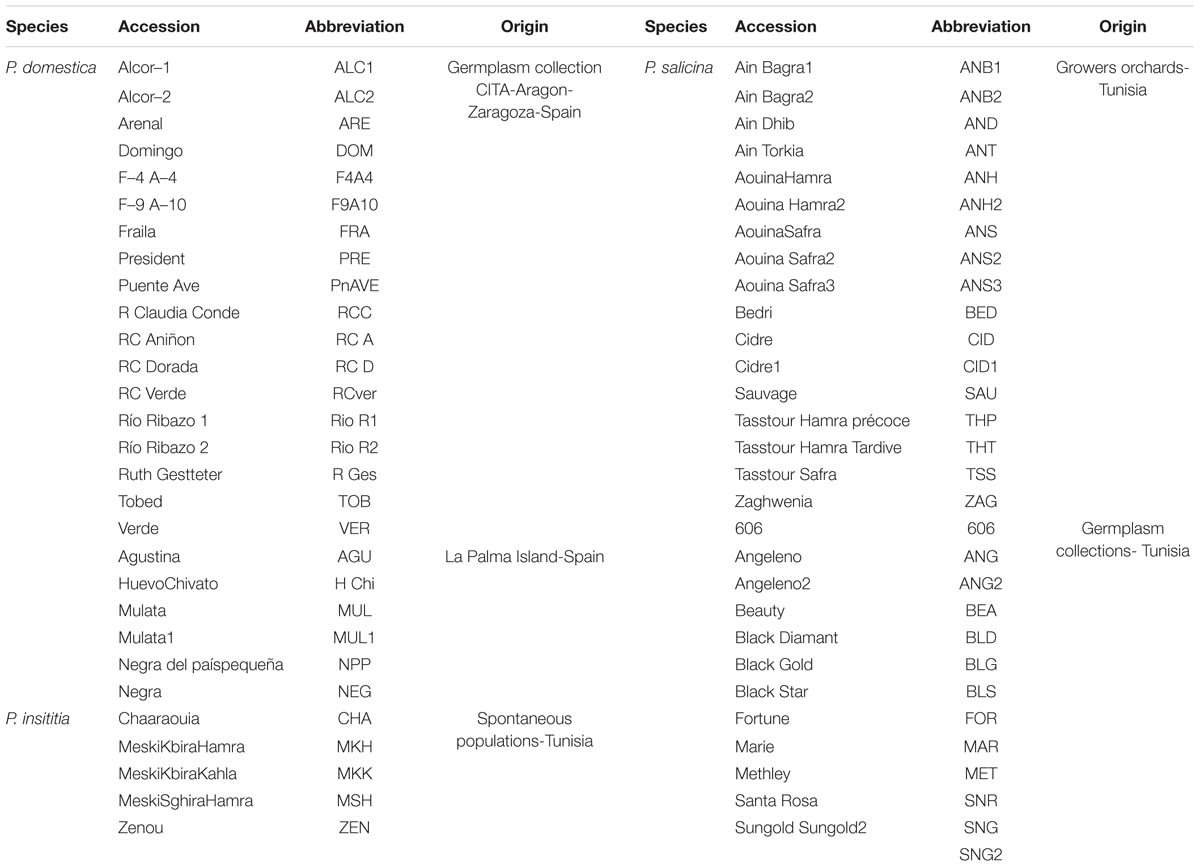
A total of 59 plum accessions were used in this study. Twenty four accessions belonged to hexaploid P. domestica (2n = 2x = 48), eighteen of them conserved at the CITA-Aragon germplasm collection in Zaragoza (Spain) and six collected in La Palma Island (Spain). Additionally, five accessions belonging to hexaploid P. insititia (2n = 2x = 48) were collected from non-cultivated populations in Tunisia. Thirty accessions belonging to diploid P. salicina (2n = 2x = 16) were collected from Tunisia, thirteen of them from two collections (Sodon and Sadira) and represent commercial varieties, and seventeen from growers orchards. Information related to plant material is summarized in Table 1.
S-Locus Analysis
Young leaves from each of these accessions were used as the starting material to extract DNA using the cetyltrimethyl ammonium bromide (CTAB) following the protocol described by Doyle and Doyle (1987). DNA was quantified using a Nanodop 1000 and diluted to 10 ng/μL.
S-genotyping of plum cultivars was performed using three pairs of consensus primers. The second intron of the S-RNase gene was amplified using the primer pair PruC2/PCER (Tao et al., 1999; Yamane et al., 2001). The primer pairs PaCons1F/PaCons1R2 (Sonneveld et al., 2006) and Fbox5’F/Fbox–IntronR (Vaughan et al., 2006) were used to amplify the first intron of the S-RNase gene and the SFB gene intron, respectively.
PCRs were performed in a volume of 15 μl, with 20 mM Tris–HCl, pH 8.4, 50 mM KCl, 2 mM MgCl2, 0.1 mM of each dNTP, 0.2 μM of each primer, 40 ng of genomic DNA and 0.45 U of BioTaqTM DNA polymerase (Bioline, London, United Kingdom). All PCR reactions were carried out in an I-cycler (Bio-Rad Laboratories, Hercules, CA, United States) thermocycler using the following temperature profile: an initial denaturation step at 94°C for 1 min, 35 cycles of 94°C for 3 min, 56°C for 1 min, and 72°C for 3 min. A final extension was programmed at 72°C for 7 min.
Fragments amplified with the PruC2/PCER primer combination, with sizes were greater than 500 bp, were separated using a 2% agarose gel electrophoresis, stained with SYBRGreen and visualized with UV light. Fragment size estimation was done using a size standard (1 kb DNA Ladder; Invitrogen, Carlsbad, CA, United States). Three P. salicina cultivars (“Fortune,” “Santa Rosa,” and “Beauty” with known S-genotypes (SbSc, ScSe, and SbSh, respectively) were used as references.
Fragments amplified with PaCons1F/PaCons1R2 andFbox5’F/Fbox–IntronR primers with sizes smaller than 500 bp, were analyzed by capillary electrophoresis. Forward primers were labeled with a fluorescent dye on the 5-end and PCR products were detected and sized with a Beckman Coulter Genome Lab GeXPTM capillary DNA analysis system. Samples were denaturalized at 90°C for 120 s, injected at 2.0 kV for 30 s, and separated at 6.0 kV for 35 min.
Pollination Tests
To establish self-(in)compatibility in the Tunisian diploid cultivars, 17 genotypes were self-pollinated in the laboratory and pollen tube growth was observed under the microscope.
Flowers of each genotype were collected at the balloon stage and pollen was obtained by manually removing and drying the anthers at room temperature during 24 h. The pollen was then sieved through a fine mesh and stored at −20°C until required (Guerra et al., 2009; Guerra and Rodrigo, 2015).
To obtain the pistils, flowers were collected at balloon stage 24 h before anthesis. These flowers were emasculated by removing petals, sepals and stamens with fingernails, then the pistils were maintained on wet florist foam at laboratory temperature (Rodrigo and Herrero, 1996). On the following day, a group of 25–45 flowers was hand-pollinated using a fine paintbrush. Three days later, the pollinated pistils were fixed in FAA [70% ethanol: acetic acid: formaldehyde (18 : 1 : 1, v/v/v)] (Guerra et al., 2009).
For microscope preparations, the fixed pistils were washed three times for 1 h with distilled water and left in 5% sodium sulphite at 4°C. To soften the tissues, the pistils were autoclaved at 1 kg/cm2 during 10 min in sodium sulphite (Jefferies and Belcher, 1974), and stained with 0.1% (v/v) aniline blue in 0.1 N K3PO4 to stain callose (Linskens and Esser, 1957). Pollen tube growth in the style was observed under an Olympus BH2 microscope with UV.
SSR Analysis
The 59 plum genotypes were also analyzed using nine SSR primer pairs previously developed in Prunus (Supplementary Table S1). PCR reactions were carried out in a volume of 20 μl, with 20 mM Tris–HCl, pH 8.4, 50 mM KCl, 4 mM MgCl2, 0.1 mM of each dNTP, 0.2 μM of each primer, 40 ng of genomic DNA and 0.45 U of BioTaqTM DNA polymerase (Bioline, London, United Kingdom). PCR reactions were run in an I-cycler (Bio-Rad Laboratories, Hercules, CA, United States) thermocycler using the following temperature cycles: an initial step of 2 min at 94°C, 35 cycles of 45 s at 94°C, 45 s at 57°C, 1 min at 72°C, and a final step of 5 min at 72°C. The obtained fragments were analyzed by capillary electrophoresis. Forward primers were labeled with a fluorescent dye on the 5-end and PCR products were detected and sized with a Beckman Coulter GenomeLabGeXPTM capillary DNA analysis system. Samples were denaturalized at 90°C for 120 s, injected at 2.0 kV for 30 s, and separated at 6.0 kV for 35 min.
Diversity Analyses
To explore the genetic diversity of the studied cultivars, different diversity parameters of the S-locus and SSR markers were calculated with Arlequin ver. 3.5 (Excoffier et al., 2005) for each plum species. Detected alleles for both SSR and S-locus markers were scored as present (1) or absent (0). Genetic distances were calculated with Maximum Composite Likelihood (Tamura et al., 2011) in Popgene ver. 1.31 software, and then imported to Mega5 to construct a UPGMA tree. Numbers on major branches represent bootstrap support from 2000 replicates. For MDS analysis, genetic distances were calculated with the Euclidean index (Taguchi and Oono, 2005) with three-dimensional coordinate system using the PAST software (Ver. 3.21) (Hammer et al., 2001).
Results
S-Locus Analysis
S-Allele Typing in Diploid P. salicina
PruC2/PCER markers, spanning the second intron of the S-RNase gene, showed a high degree of length polymorphism. Hence, nine fragments were amplified with sizes ranging from 400 to 1600 bp (Figure 1A). The sizes of these bands were consistent with the S-alleles previously identified in P. salicina. As band sizes were larger than 500 bp, results were analyzed by agarose gel electrophoresis which make difficult to distinguish differences between some alleles with similar sizes such Sg-Sq (1250 and 1270 bp, respectively) and Sa-Sh (470 and 500 bp, respectively).
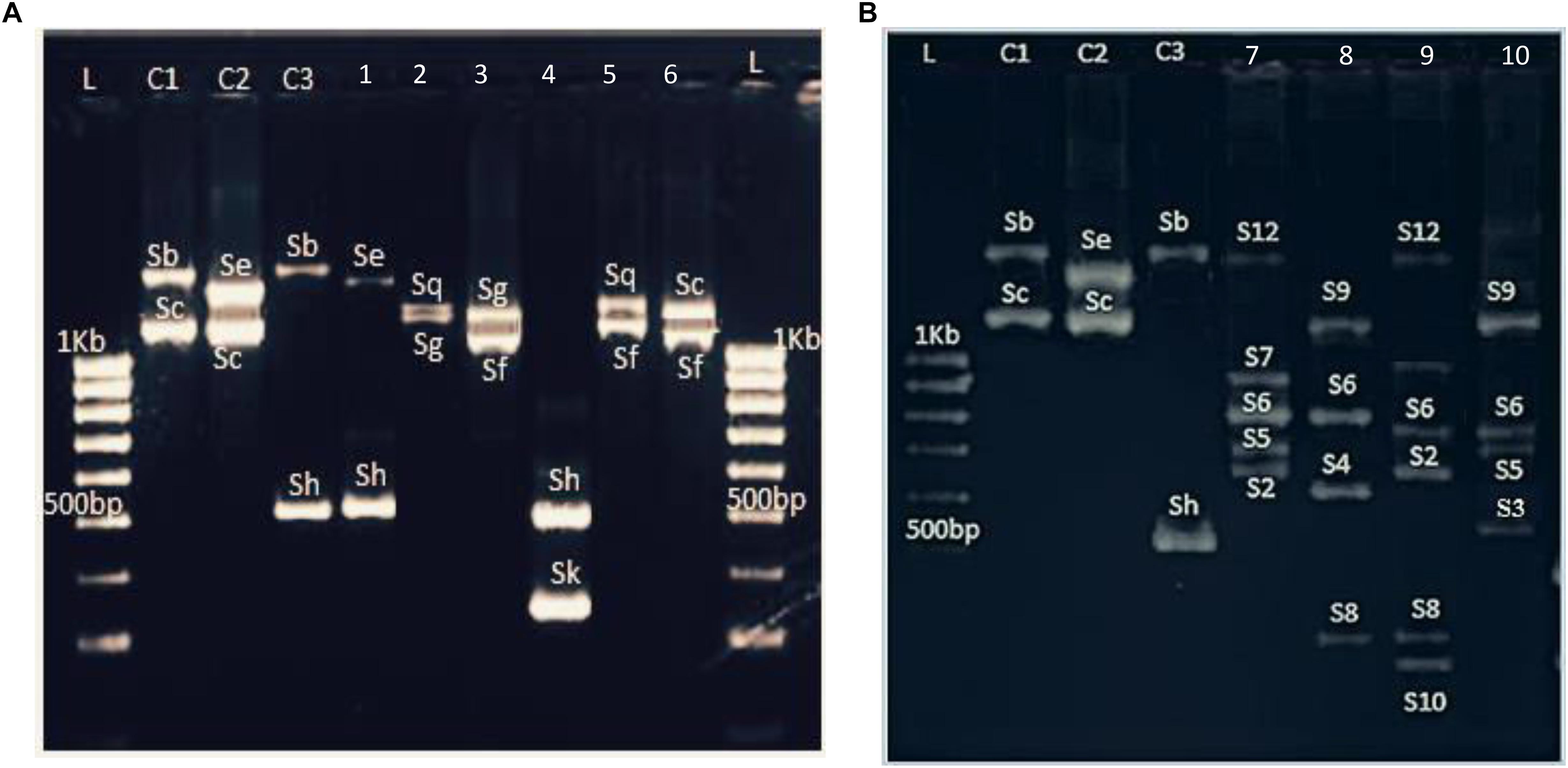
Figure 1. Example of S-allele identification in (A) six P. salicina samples and (B) six hexaploid plums amplified with PruC2-PCER. L: 1KB ladder. C1, C2, and C3: Varieties used as control with previously known S-genotypes. C1, Fortune; C2, Santa Rosa, C3, Beauty; 1, Bedri 1; 2, Aouina Hamra; 3, Tasstour Hamra précoce; 4, Sungold; 5, Ain Dhib; 6, Zaghwenia; 7, Mulata; 8, Negra; 9, Fraila; 10, Verde.
To confirm these results, a second round of amplification was conducted with the PaCons1F/PaCons1R2 primer pair, spanning the first intron of the S-RNase gene. Eight fragments were amplified with sizes ranged from 208 to 389 bp. Although the differentiation between Sg and Sq alleles was successful (Supplementary Table S2), it was not possible to distinguish between the Sa and Sh alleles since they showed similar sizes (388 and 389 bp).
Fbox5’F/Fbox–IntronR primer pair was additionally used to amplify the SFB gene intron. Capillary electrophoresis analysis showed nine fragments whose sizes ranged from 170 to 190 bp. Despite the reduced level of length polymorphism between bands (ranging between 1 and 2 bp), it was possible to differentiate the Sa (174 bp) and Sh (187 bp) alleles (Supplementary Table S2).
Hence, after combining the results of the three primer pairs used, nine S-alleles were identified in 17 Tunisian P. salicina genotypes of unknown S-allele composition (Supplementary Table S2). Additionally, the S-alleles of 13 P. salicina cultivars with previously reported S-genotypes were amplified in order to confirm the size of the S-alleles.
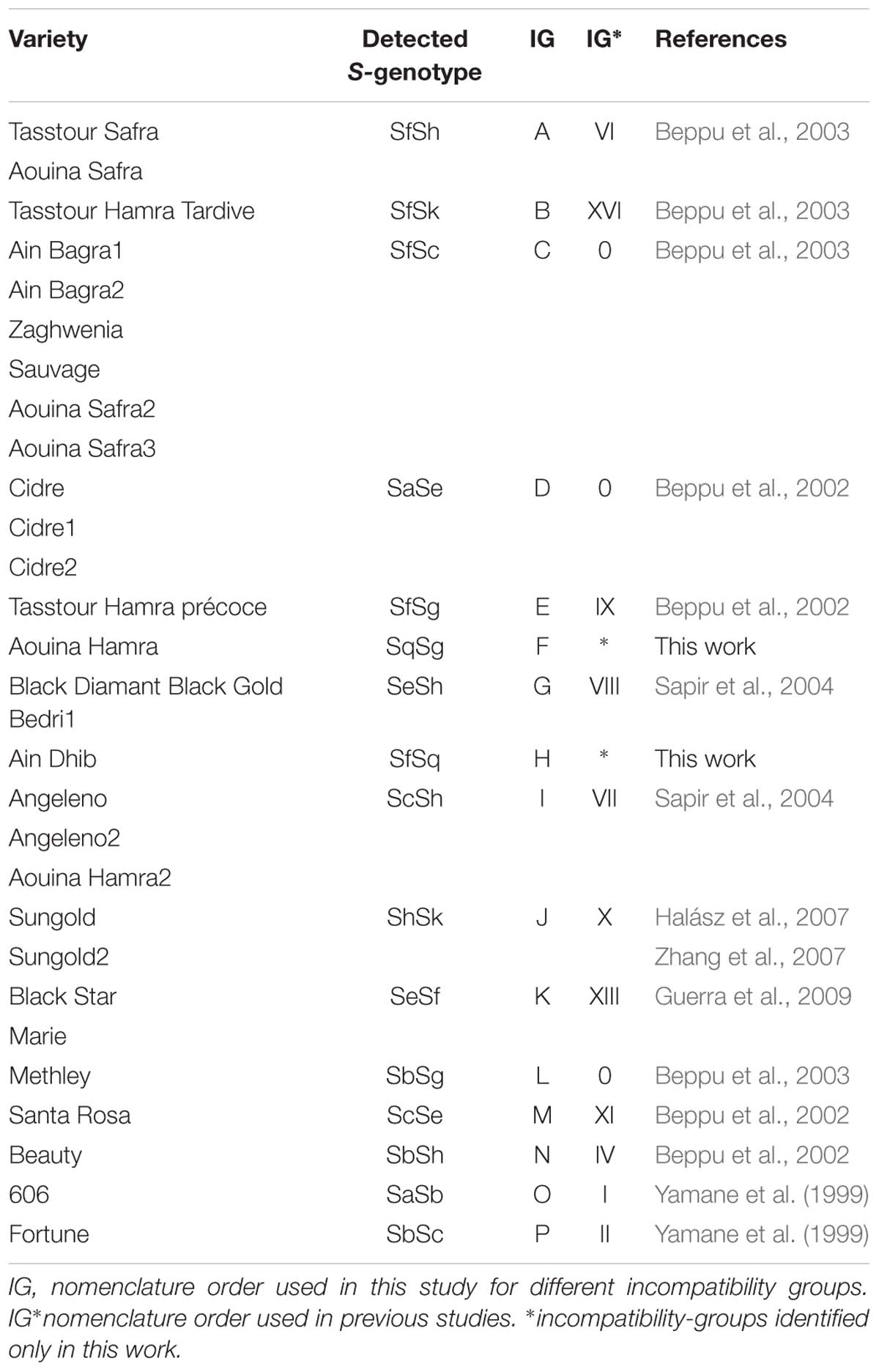
In all, 30 S-genotypes were reported, 17 of them for the first time. These cultivars have been assigned to 16 incompatibility groups (IG), three of them allocated to group 0 identified as rare genotypes and two new IG reported here for the first time (IG-F, IG-H) (Table 2).
Pollination Experiments in Diploid P. salicina
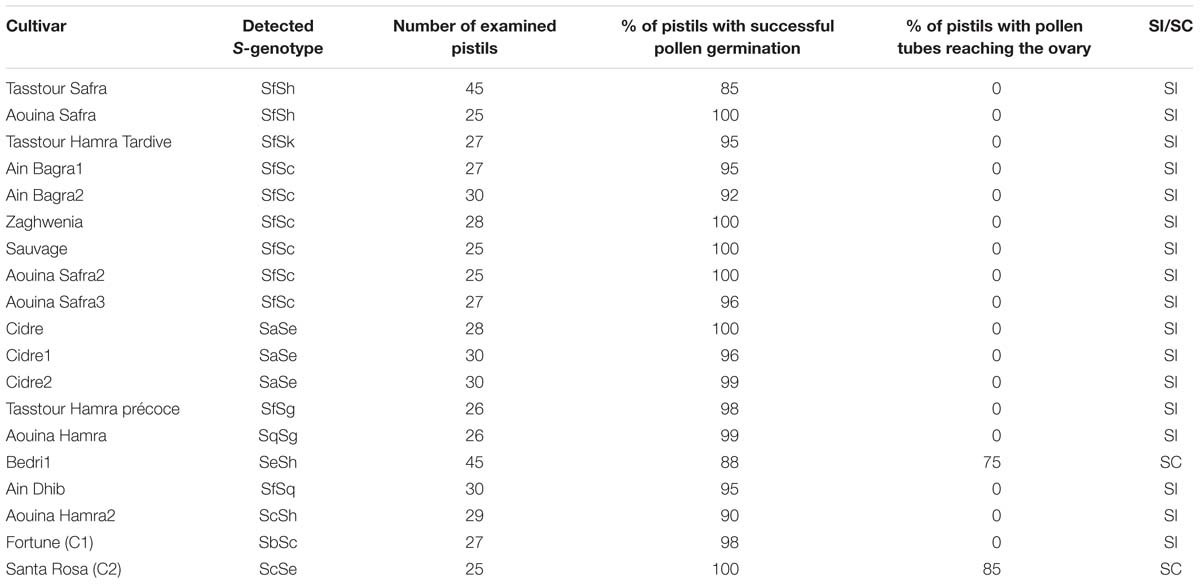
To explore self-(in)compatibility of the 17 P. salicina accessions analyzed, self-pollinations were carried out in the laboratory. For each cultivar, 25 to 45 flowers were collected at balloon stage (Figures 2A.a,b), emasculated (Figures 2A.c,d) and hand pollinated with the help of a paintbrush. Then, pollen tube growth in the style was observed in self-pollinated flowers under the microscope. As control, self-pollinations were tested in two cultivars: “Santa Rosa” (ScSe), known to be self-compatible, and “Fortune” (SbSc), considered as self-incompatible.

Figure 2. Pollen germination and pollen tube growth in flowers of Tunisian Japanese plums. (A) Flowers. (a) Balloon stage. (b) Full bloom. (c) Reproductive organs. (d) The pistil. (B) Pollen germination. (a) Germination at the stigma surface. (b) Pollen tube arrested in the style in an incompatible reaction. (c) The pollen tube tip. (d) Pollen tube growing in the base of the style in a compatible reaction. Scale bars = 50 μm. Sta, stamens; Stg, stigma; Sty, style; B.Sty, the base of the style; Ova, ovary; P, Pollen tube.
Pollen germination on the stigma was successful in the 17 cultivars tested (Figure 2B.a). In fifteen cultivars (IG-A-B-C-E-F-H-I), pollen tube growth was arrested at halfway the length of the style (Figures 2B.b,c). These self-pollination experiments confirmed the SI phenotypes of these plum accessions revealed by PCR (Table 3).
As expected, in the cultivar “Bedri” (IG-G), which carries the Se allele, previously known as related to the SC phenotype (Beppu et al., 2005), pollen tubes reached the base of the style in 75% of self-pollinated flowers confirming the SC phenotype in this cultivar (Figure 2B.d). However, an unexpected result was observed in the three accessions of “Cidre” (IG-D). Pollen tube growth was arrested at halfway the length of the styles despite the identification of the Se allele by PCR, which suggested that “Cidre” is rather a self-incompatible cultivar (Table 3).
S-Genotyping in Hexaploid P. insititia and P. domestica
The three tested primer pairs exhibited successful PCR amplifications for the 29 hexaploid plum accessions (Figure 1B). Furthermore, the same fragments were amplified in both P. insititia and P. domestica.
PCR amplifications performed with PruC2/PCER showed a multiallelic profile with genotypes carrying up to six fragments (Figure 1B). The S-typing revealed 12 alleles with sizes ranging between 300 and 1600 bp. These alleles were named with numbers to allow distinguishing them from the alleles of diploid plums, which are named with letters (Table 4). Three alleles showed the same sizes than those reported in diploid plums: S12–Sb (1580 bp), S9–Sf (1090 bp) and S1–Sa (470 bp).

Table 4. Sizes of S-alleles amplified in hexaploid P. domestica and P. insititia with three primers pairs.
To confirm these results, PCR amplifications were conducted using PaCons1F/PaCons1R2 primers. Capillary electrophoresis revealed 11 fragments with sizes ranging from 228 to 413 bp (Table 4). The use of the PaCons1F/PaCons1R2 primer pair allowed amplification of all S-alleles previously detected with PruC2/PCER, except the S4 allele for which no amplification with PaCons1F/PaCons1R2 was obtained. Moreover, the amplified alleles showed similar sizes between S12–Sb (368 bp) and S9–Sf (326 bp) alleles, but a difference of 15 bp was revealed between S1 and Sa (374 and 389 bp, respectively).
A third round of amplification was conducted using the Fbox5’F/Fbox–IntronR primer pair. Amplified bands showed small differences in length (in order of 1 or 2 bp) which required several repetitions to ensure the exact size of each band. All the S-alleles previously defined were detected with the Fbox5’F/Fbox–IntronR primer pair with sizes ranging between 172 and 208 bp (Table 4), except the three S-alleles S2, S8 and S10. In addition, the alleles S12 and S9 showed the same lengths than Sb (185 bp) and Sf (176 bp), respectively.
Overall, twelve S-alleles were detected in 29 hexaploid plums (Table 4) assigned to sixteen S-genotypes (Table 5).

Table 5. Identified S-genotypes and incompatibility groups for hexaploid Prunus domestica and Prunus insititia.
Genetic Diversity at the S Locus
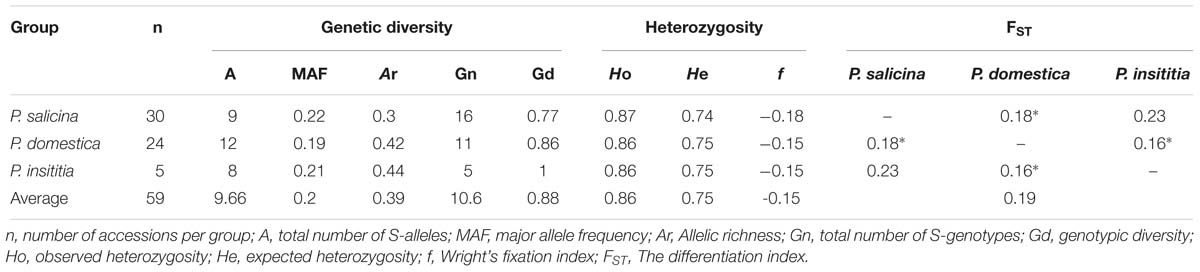
Using the three S-markers, nine S-alleles have been identified in 30 diploid P. salicina cultivars; among them, the Sf allele was the most frequent (0.22). In hexaploid plums, among 12 S-alleles, S1 (0.19) and S10 (0.21) were the most frequent in P. domestica and P. insititia, respectively (Table 5). In P. salicina, allelic richness (Ar) ranged between 0.11 and 0.48 with an average of 0.3. In P. domestica, Ar ranged between 0.35 and 0.49 with an average of 0.42, while in P. insititia it ranged between 0.37 and 0.51 with an average of 0.44 (Table 6).
A total of 32 S-genotypes were identified, 16 of them belonging to P. salicina, 11 to P. domestica and 5 to P. insititia. Genotypic diversity (Gd) showed high values and ranged between 0.54 and 1 in P. salicina (an average of 0.77), 0.72 and 1 in P. domestica (an average of 0.86) and 1in P. insititia (Table 6). This Gd depends on allele frequency in each genotype (whether the allele is frequent or rare) and on the number of accessions sharing the same genotype.
These high GD reflected an important frequency of the heterozygous fraction. The observed heterozygosity (Ho) ranged from 0.8 to 0.94 in diploid plums with an average of 0.87. In hexaploid plums, Ho ranged between 0.83 and 0.91 with an average of 0.86 for both P. domestica and P. insititia. These values were higher than the expected heterozygosities (He), which ranged between 0.71 and 0.78 in diploid plums with an average of 0.74 and between 0.74 and 0.77 in hexaploid plums with an average of 0.75. Consequently, negative values of Wright’s fixation index were observed in P. salicina (f = −0.18) as well as in hexaploid plums (f = −0.15) (Table 6).
The differentiation index (FST) between the three groups was 0.19. FST values were statistically significant between P. domestica and P. salicina and between P. insititia and P. domestica (0.18, 0.16, respectively with P < 0.05); however, FST values were not significant between P. salicina and P. insititia (0.23, 0.05 < P < 0.1) (Table 6). In fact, a value of FST ranging between 0.15 and 0.25 indicated that the analyzed populations are genetically different, independently from their geographic origins (Weir and Cockerham, 1984).
SSR Analysis
The nine SSR markers proved to be highly polymorphic in the three species tested. The BPPCT012 primer pairs amplified the highest number of alleles while the UDP96-008 amplified the lowest number of alleles in the different species.
In diploid P. salicina, 94 alleles (Ao) were amplified in the 30 accessions resulting in a value of 10.5 alleles per locus (Al) (Table 7). A total of 33.1 of them represent effective alleles (Ae) indicating that an important number of alleles appeared at low frequency (less than 5%) which explains the high allelic richness observed (Ar = 0.78). The loci analyzed allowed identifying 24 genotypes (Table 7).
In hexaploid P. domestica, the number of amplified alleles per genotype ranged from 1 to 6. The used SSR markers amplified 195 alleles (Ao) giving a value of 21.6 alleles per locus (Al) with 50.4 effective alleles (Ae) leading to an important allelic richness (Ar = 0.82). With the whole set of nine SSRs used it was possible to identify 20 different genotypes among the 24 cultivars studied (Table 7).
In hexaploid P. insititia, 73 alleles were amplified (Ao) in the 5 tested cultivars with an average of 8.1 alleles per locus (Al). A total of 29.3 were defined as effective alleles (Ae) reflecting the high allelic richness (Ar = 0.73). The five accessions had different genotypes using the whole set of SSR markers (Gn = 5) (Table 7).
In the three tested species, observed heterozygosity values were higher than those of expected heterozygosity leading to negative values of f (−0.06). In P. salicina, the observed heterozygosity ranged between 0.65 and 0.78 with an average of 0.71, whilst the expected heterozygosity ranged between 0.61 and 0.73 with an average of 0.67. In P. domestica, the observed heterozygosity varied from 0.71 to 0.76 with an average of 0.73, while the frequency of expected heterozygosity ranged between 0.66 and 0.73 with an average of 0.69. In P. insititia, the average of observed heterozygosity was 0.74 and the expected heterozygosity was 0.7 (Table 7).
Genetic Relationships Among the Accessions
Based on the defined S-alleles, the constructed UPGMA tree divided the cultivars into two heterogeneous groups (Figure 3A.a). The first group (G1) clustered together Japanese plums (P. salicina) with some European plums (P. domestica). This result suggests a close genetic relationship between P. salicina and P. domestica. The second group (G2) grouped the remaining P. domestica accessions together with P. insititia plums.
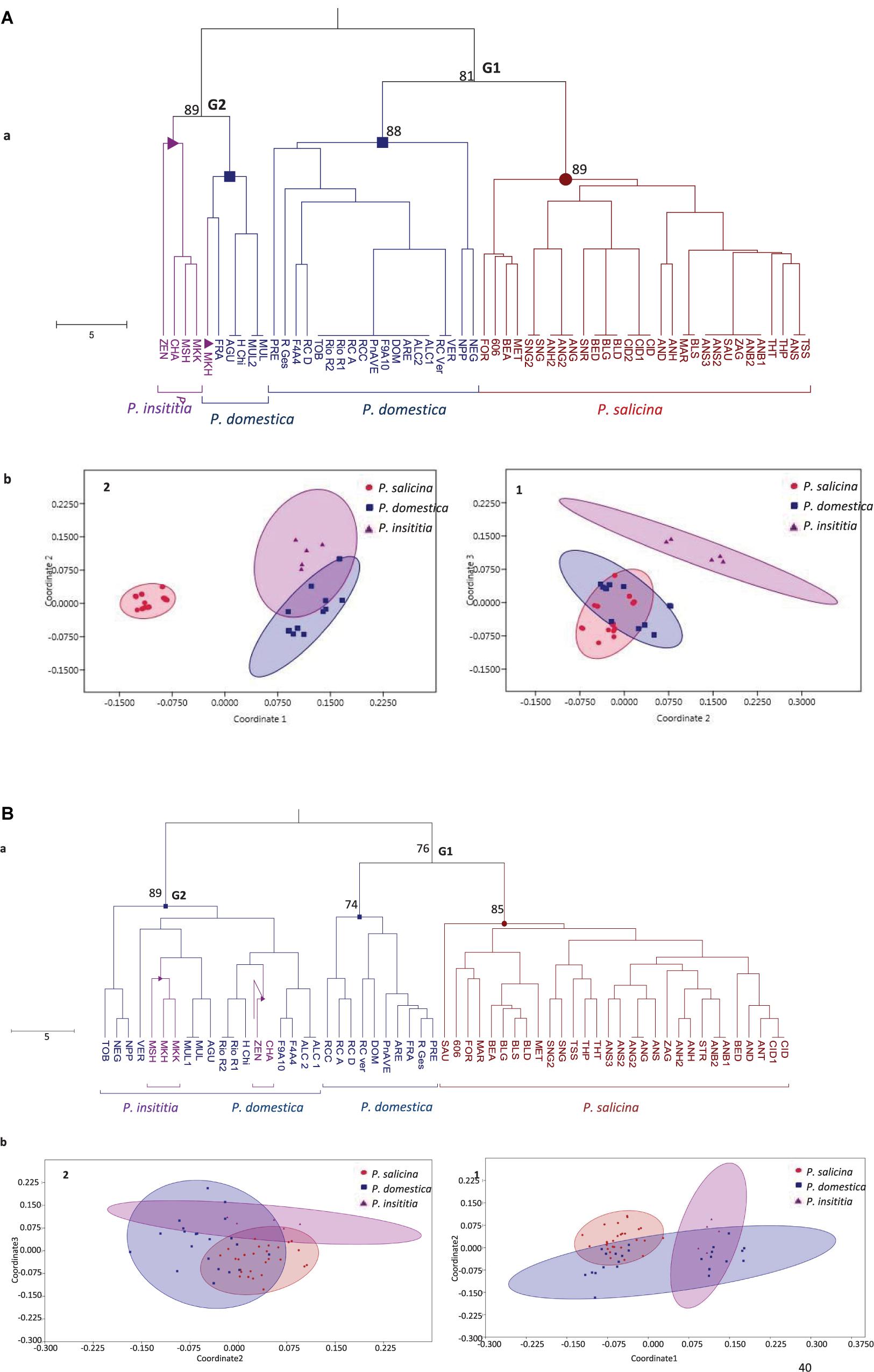
Figure 3. Genetic relationships between the 59 tested plums based on (A) S-locus data and (B) SSR data. (a) UPGMA tree. Numbers on major branches represent bootstrap support from 2000 replicates. (b) Non-metric multidimensional scaling MDS plots: (1) using dimensions 2–3. (2) using dimensions 1–2.
Multidimensional scatter plots (MDS) yielded similar results than the UPGMA tree (Figure 3A.b). Using dimensions 2 and 3, the first MDS plot (1) clearly separated P. insititia accessions from P. domestica and P. salicina, and confirmed a close genetic relationship between P. domestica and P. salicina. In contrast, using dimensions 1 and 2, the second MDS plot (2) showed a close relationship between European plums (P. domestica and P. insititia), while Japanese plums (P. salicina) were relatively separated.
Based on SSR loci, the UPGMA dendrogram (Figure 3B.a) showed the same topology than that constructed with S-markers and clearly divided the tested accessions into two main groups: G1 and G2. The first group (G1) gathered P. salicina and P. domestica accessions. The first sub-group in that main group clustered the diploid P. salicina accessions and the second sub-group gathered 10 P. domestica accessions. The remaining P. domestica accessions and the five P. insititia cultivars fell into the second group (G2).
In addition, MDS plots were drawn based on these SSR markers. Using dimensions 1 and 2, the first MDS plot (1) confirmed the close genetic relationship between P. salicina and P. domestica and between P. domestica and P. insititia. Using dimensions 2 and 3, the MDS plot (2) showed an overlap between the three species (Figure 3B.b).
Discussion
This study was conducted in order to report the S-genotypes of 59 plum accessions covering two levels of ploidy: diploid P. salicina (2n = 16) and hexaploid P. domestica and P. insititia (2n = 46). Among them, thirteen P. salicina accessions were previously S-genotyped and 46 accessions were analyzed for the first time. In addition, S-locus and SSR data were combined to investigate the genetic relationships between the species studied.
S-Locus Analysis
Applicability of Intron Length Polymorphism Technique (ILP) in Diploid and Hexaploid Plums
The ILP technique, the most common method for S-allele profiling in Prunus species (Sonneveld et al., 2003), was used with three S-locus specific consensus primer pairs. Two primer pairs amplified the first and second introns of the S-RNase coding region and one primer pair amplified the SFB intron. These markers were developed and used earlier to study SI in several Prunus species (Ushijima et al., 1998; Tao et al., 1999; Sutherland et al., 2004; Sonneveld et al., 2006; Vaughan et al., 2006; Guerra et al., 2009; Herrera et al., 2018) and showed good applicability for plum germplasm characterization and cultivar discrimination which confirmed the high conservation of S-genes among Prunus species and primer pair transferability among species (Kota-Dombrovska and Lācis, 2013).
PruC2/PCE-R, spanning the second intron of the S-RNase gene, amplified up to six fragments in P. salicina, P. insititia and P. domestica. Although it has been suggested that primers specific to the second intron of S-RNase gene were not suitable for S-genotyping (Sutherland et al., 2007), this primer pair was sufficient for successful genotyping in the species tested.
Primer pairs PaCons1F/PaCons1R2, specific to the first intron of the S-RNase gene, and Fbox5’F/Fbox–IntronR, spanning the SFB gene intron, showed limited polymorphism and amplified alleles with similar sizes, which make the amplified fragments hardly distinguishable on agarose gel electrophoresis. However, these markers should be employed to ensure a more accurate characterization. When amplification results are analyzed by capillary electrophoresis, these primers can be used as supplementary markers in the characterization of plum genetic resources.
Self-Compatibility in Japanese Plums
Pollination experiments provided complementary information to the PCR amplification in order to establish the self-(in)compatibility phenotypes of Tunisian diploid plums. Although Japanese plum is known to be SI, the SC reversion is not excluded in this species as in other Prunus species. The SC in species of this genus has been associated to particular S-alleles; examples include apricot (Vilanova et al., 2006; Halász et al., 2014; Herrera et al., 2018), almond (Fernandez i Marti et al., 2014; Kodad et al., 2015), peach (Tao et al., 2007; Hanada et al., 2014), sour cherry (Yamane et al., 2003) and sweet cherry (Wünsch and Hormaza, 2004; Cachi and Wünsch, 2014). In P. salicina, the SC was firstly related to the Se haplotype expression (Beppu et al., 2005, 2010; Guerra et al., 2009; Guerra and Rodrigo, 2015), for which an accumulation of insufficient levels of Se-RNase due to very low transcription levels in the pistil has been described (Watari et al., 2007; Guerra et al., 2009). After that, other S-haplotypes have been identified as SC related, such as Sb (Guerra et al., 2009; Beppu et al., 2010), St (Beppu et al., 2012a) and Sg (Beppu et al., 2012b).
Pollen tube growth and PCR analysis showed that “Bedri,” carrying the Se allele, is a SC cultivar, which may confirm the implication of this allele in SC. In contrast, three accessions of “Cidre,” also carrying the Se allele, were revealed as SI. Similar results were observed in previous studies where some Japanese plum cultivars, described as SI, which carried one of the alleles related to the SC in their S-genotypes (Beppu et al., 2002, 2003; Guerra et al., 2009; Guerra and Rodrigo, 2015). Thus, SC in Japanese plum could be also related to a double phenotypic expression of the same allele, as it has been reported in almond (Fernandez i Marti et al., 2010, 2014), than associated uniquely with specific S-alleles (Guerra and Rodrigo, 2015).
S-Genotyping in Hexaploid Plums
Sixteen S-genotypes were assigned for polyploid individuals. Only one genotype carrying six alleles (IG-3) was defined, while the remaining S-genotypes carried four or five alleles. Similar results were observed using different molecular approaches (S-genotyping and SSR markers) in polyploid Prunus species such P. domestica, P. spinosa, and P. insititia (Kota-Dombrovska and Lācis, 2013; Halász et al., 2014, 2017; Urrestarazu et al., 2018; Baraket et al., 2019). These incomplete S-genotypes in some accessions can be the consequence of the multiple appearance of a given allele (Walsh et al., 1992; Kota-Dombrovska and Lācis, 2013). This homozygosity leads to the question of whether polyploidy can cause SC in these species. Hence, direct association between polyploidy and SC has not been observed in some polyploid Prunus species. In the allotetraploid P. cerasus, the heteroallelic pollen retained its SI phenotype and SC was related to the co-expression of two non-functional S-haplotypes in the same genotype (Hauck et al., 2006). Additionally, polyploidy seemed not to cause SC in P. spinosa and P. insititia (Nunes et al., 2006). In contrast, polyploidy influences SC in Solanaceae and Scrophulariaceae (Mable, 2004; Nunes et al., 2006; Vieira et al., 2008).
In this work, an average of 0.41 alleles per polyploid accession was reported using three S-markers. This number is relatively low for species whose allopolyploid genome structure tends to increase the number of alleles and promote the emergence of new alleles (Ainouche and Wendel, 2014; Halász et al., 2017). Additionally, 23 S-alleles were assigned in 17 polyploid plums in Halász et al. (2017). This result may be explained by the low heterogeneity of our tested samples. Halász et al. (2017) used the PaConsII primer pair, spanning the second intron of the S-RNase gene. Compared to the result of PruC2/PCE-R specific to the same region, four S-alleles (S2, S4, S6, and S11) showed sizes similar with SR, SZ, SS, and SJ, respectively (Halász et al., 2017). In addition, four alleles (S1, S3, S5, and S9) showed differences less than 20 bp with SO, SB, SD, and SM, respectively, and two alleles (S7 and S12) showed differences less than 80 bp with SI and SV, respectively (Halász et al., 2017).
SSR Polymorphism
The level of SSR polymorphism observed in 30 P. salicina (10.5) is comparable to that revealed in 29 P. salicina accessions using eight SSR loci (12) by Carrasco et al. (2012). This variability is higher than that reported for P. salicina in other works, i.e., Mnejja et al. (2004) and Klabunde et al. (2014). It is also higher than that observed in other Prunus species such as almond (Fathi et al., 2008), cherry (Wünsch and Hormaza, 2002; Baris et al., 2017) and apricot (Gürcan et al., 2015).
Due to their polyploid genomes, few studies have focused on evaluating the genetic diversity of hexaploid plum species (Horvath et al., 2011; Sehic et al., 2015; Halász et al., 2017; Urrestarazu et al., 2018). A high degree of polymorphism has been revealed in hexaploid plums in previous works (Urrestarazu et al., 2018).
In general, among the Prunus genus, plums are considered as one of the most polymorphic species (with almond) (Salazar et al., 2014) due to their SI system. In SI populations, the fact that self-fertilization is avoided decreases the probability of obtaining homozygote offspring (Balloux, 2004; Stoeckel et al., 2006) and tends to increase the genetic diversity as well as heterozygosity levels elsewhere in the genome (Hartl and Clark, 1997; Stoeckel et al., 2006).
Genetic Richness and Allelic Singularity of Tunisian Plums
The characterization of 17 Tunisian P. salicina plums has allowed assigning them into nine IG (IG-A→IG-I). Three S-genotypes, previously defined as rare genotypes and included under IG-0 (Beppu et al., 2003; Guerra et al., 2009; Guerra and Rodrigo, 2015) were amplified in ten accessions and assigned to IG C, D, and L. Additionally, two new genotypes (IG F and H) were reported here for the first time. SSR results showed a significant genetic diversity in the P. salicina tested genotypes. These results highlight the allelic singularity and the rich variability characterizing the Tunisian plum germplasm as an unexplored source of genetic variation. In fact, several Tunisian Prunus varieties have disappeared during the last years due to the lack of programs addressed to preserve these local germplasm (Nabli, 2011; Abdallah et al., 2016; Baraket et al., 2019), which require the establishment of appropriate germplasm collections. The results obtained in this study could complement previous work of Baraket et al. (2019) in which 23 Tunisian plums accessions were phenotypically and genetically characterized.
Genetic Relationships in Prunus Species
Phylogenetic tree and Multidimensional scatter plots (MDS) for both S-locus and SSR loci underlined genetic relationships between the three plum species studied, especially between P. domestica and P. salicina and between P. domestica and P. insititia. Similar results were reported in previous works such in Hegedus and Halasz (2006) where the same alleles were reported in P. domestica, P. domestica × P. cerasifera hybrids, P. spinosa and P. salicina.
The close genetic relationship found between P. domestica and P. insititia is expected. In fact, P. insititia has been reported in wild form in three continents (southern Europe, Western Asia and northern Africa) (Malcolm, 2006). Although the ancestral form of this species remains still unknown, P. insititia has often been proposed to be a subspecies of P. domestica and chloroplast DNA analysis seems to corroborate this hypothesis (Reales et al., 2010).
The origin of P. domestica has also been debated for over a century (Crane and Lawrence, 1929; Bajashvili, 1991; Eryomine, 1991; Zohary, 1992; Hegedus and Halasz, 2006; Liu et al., 2007; Zhebentyayeva et al., 2019). The most accepted hypothesis is that of Crane and Lawrence (1929) in which P. domestica would be an allopolyploid hybrid species between tetraploid P. spinosa (2n = 2x = 32) and diploid P. cerasifera (2n = 2x = 16). Thus, P. domestica is believed to have arisen in the Caucasian region where these species are originated (Crane and Lawrence, 1929) via either chromosome doubling of the hybrid triploid or as a product of unreduced gametes from both parents (Liu et al., 2007). Bajashvili (1991) suggested that European plum could have also been originated directly from hexaploid P. divaricata (=P. cerasifera) or from P. spinosa by alloploidization. Moreover, Zohary (1992) reported that comparative morphology and available cytogenetic evidence do not support the assumption that P. spinosa contributed its two genomes (or even a single genome) to hexaploid P. domestica and suggested that P. domestica would be an autopolyploid P. cerasifera (Zohary, 1992). Eryomine (1991) proposed that P. domestica originated by artificial selection by early Eurasian societies and that several species participate in the origin of P. domestica probably explaining why P. domestica has never been found in the wild. He suggested that hybrids between (P. cerasifera × P. salicina) × P. spinosa and (P. cerasifera subsp. macrocarpa) presented the highest morphological similarities with P. domestica. In addition, P. cerasifera (CC) × P. salicina (SaSa) hybrids developed unreduced gametes (Eryomine, 1991). Hence, the genotypic formula suggested by Eryomine (1991) for P. domestica is SSCCCSa, where part S represents the genome of P. spinosa, part C represents the genome of diploid P. cerasifera and part Sa represents the genome of diploid P. salicina inherited into domestic plum. Recent results seem to support the hypothesis that P. domestica could be a hybrid of interspecific hybrids including P. cerasifera, P. spinosa, and other Eurasian plum species (Zhebentyayeva et al., 2019). Additionally, the close genetic relationship between P. domestica and P. salicina obtained by S-locus and SSR markers may represent an argument in favor of Eryomine proposal and may confirm that diploid P. salicina contributed to P. domestica formation.
Conclusion
The main objective of this study was to analyze the S-genotypes of 30 diploid P. salicina and 29 hexaploid plums (24 of P. domestica and 5 of P. insititia). The results obtained are relevant at three levels: (i) The identification of new S-alleles and new S-genotypes in the Prunus species analyzed should facilitate future work focused on exploring and understanding the SI system in diploid and, especially, in polyploid Prunus species. (ii) The identification of S-genotypes and the genetic characterization of Tunisian local plums showed that the Tunisian germoplasm could represent a new source of variability useful in future breeding programs. In addition, such results could be used for defining a Tunisian core collection aiming to preserve the extant diversity in local Tunisian plums. (iii) Combined with SSR markers, the information obtained from the S-locus may supply additional information regarding the genetic relationships among Prunus species.
Author Contributions
DA conducted the experiments and statistical analyses, developed the genetic analyses, and wrote the manuscript. GB provided some plant material, and discussed and corrected the content. VP provided some plant material and discussed the results. SBM conducted the pollination experiments and the microscope observations. AS-H and JH provided experimental instructions, supervised the work, and assisted in writing the manuscript.
Funding
This research was supported by the Tunisian “Ministère de l’Enseignement Supérieur et de la Recherche Scientifique” and “Ministerio de Economía y Competitividad – European Regional Development Fund, European Union (AGL2016-77267-R).”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to gratefully thank “Sodon collection” (Bou-Selem, Tunisia), Tunisian farmers and the CITA of Zaragoza (Spain) for kindly providing plant material.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00896/full#supplementary-material
References
Abdallah, D., Baraket, G., Ben Tamarzizt, H., Ben Mustapha, S., and Salhi-Hannachi, A. (2016). Identification, evolutionary patterns and intragenic recombination of the gametophytic self incompatibility pollen gene (SFB) in Tunisian Prunus Species (Rosaceae). Plant Mol. Biol. Rep. 34, 339–352. doi: 10.1007/s11105-015-0922-6
Ainouche, M. L., and Wendel, J. F. (2014). “Polyploid speciation and genome evolution: lessons from recent allopolyploids,” in Evolutionary Biology: Genome Evolution, Speciation, Coevolution and Origin of Life, ed. P. Pontarotti (Berlin: Springer International Publishing), 87–113. doi: 10.1007/978-3-319-07623-2_5
Bajashvili, E. I. (1991). Studies of some species of Prunus Mill. genus. Acta Hortic. 283, 31–34. doi: 10.1094/PDIS.2001.85.5.558B
Balloux, F. (2004). Heterozygote excess in small populations and the heterozygote-excess effective population size. Evolution 58, 1891–1900. doi: 10.1111/j.0014-3820.2004.tb00477.x
Baraket, G., Abdallah, D., Ben Mustapha, S., Ben Tamarzizt, H., and Salhi-Hannachi, A. (2019). Combination of simple sequence repeat, S-locus polymorphism and phenotypic data for identification of Tunisian plum species (Prunus spp.). Biochem. Genet. doi: 10.1007/s10528-019-09922-4 [Epub ahead of print].
Baris, E., Bedő, J., Edosa, O. S., Kiss, E., and Veres, A. (2017). Preliminary results of SSR based characterization of sour (Prunus cerasus L.) and sweet cherry (Prunus avium L.) genotypes cultivated in Hungary. Collumella 4,31–45.
Beppu, K., Komatsu, N., Yamane, H., Yaegaki, H., Yamaguchi, M., Tao, R., et al. (2005). Se-haplotype confers self-compatibility in Japanese plum (Prunus salicina Lindl.). J. Hortic. Sci. Biotechnol. 80, 760–764. doi: 10.1080/14620316.2005.11512011
Beppu, K., Konishi, K., and Kataoka, I. (2012a). S-haplotypes and self-compatibility of the Japanese plum cultivar ‘Karari’. Acta Hortic. 929, 261–266. doi: 10.17660/actahortic.2012.929.38
Beppu, K., Kumai, M., Yamane, H., Tao, R., and Kataoka, I. (2012b). Molecular and genetic analyses of the S-haplotype of the self-compatible Japanese plum (Prunus salicina Lindl.) ‘Methley’. J. Hortic. Sci. Biotechnol. 87, 493–498. doi: 10.1080/14620316.2012.11512900
Beppu, K., Syogase, K., Yamane, H., Tao, R., and Kataoka, I. (2010). Inheritance of self-compatibility conferred by the Se-haplotype of Japanese plum and development of Se-RNase gene-specific PCR primers. J. Hortic. Sci. Biotechnol. 85, 215–218. doi: 10.1080/14620316.2010.11512657
Beppu, K., Takemoto, Y., Yamane, H., Yaegaki, H., Yamaguchi, M., Kataoka, I., et al. (2003). Determination of S-haplotypes of Japanese plum (Prunus salicina Lindl.) cultivars by PCR and cross-pollination tests. J. Hortic. Sci. Biotechnol. 78, 315–318. doi: 10.1080/14620316.2003.11511624
Beppu, K., Yamane, H., Yaegaki, H., Yamaguchi, M., Kataoka, I., and Tao, R. (2002). Diversity of S-RNase genes and S-haplotypes in Japanese plum (Prunus salicina Lindl.). J. Hortic. Sci. Biotechnol. 77, 658–664.
Cachi, A. M., and Wünsch, A. (2014). Characterization of self-compatibility in sweet cherry varieties by crossing experiments and molecular genetic analysis. Tree Genet. Genomes 10, 1205–1212. doi: 10.1007/s11295-014-0754-9
Carrasco, B., Díaz, C., Moya, M., Gebauer, M., and García-González, R. (2012). Genetic characterization of Japanese plum cultivars (Prunus salicina) using SSR and ISSR molecular markers. Cien. Inv. Agric. 39, 533–543. doi: 10.4067/s0718-16202012000300012
Cipriani, G., Lot, G., Huang, W. G., Marrazzo, M. T., Peterlunger, E., and Testolin, R. (1999). AC/GT and AG/CT microsatellite repeats in peach [Prunus persica (L) Batsch]: isolation, characterisation and cross-species amplification in Prunus. Theor. Appl. Genet. 99, 65–72. doi: 10.1007/s001220051209
Crane, M. B., and Lawrence, J. C. (1929). Genetical and cytological aspects of incompatibility and sterility in cultivated fruits. J. Pomol. Hortic. Sci. 7, 276–301. doi: 10.1080/03683621.1928.11513345
Das, B., Ahmed, N., and Singh, P. (2011). Prunus diversity–early and present development: a review. Int. J. Biodiv. Conserv. 14, 721–734.
Dirlewanger, E., Cosson, P., Tavaud, M., Aranzana, M. J., Poizat, C., Zanetto, A., et al. (2002). Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor. Appl. Genet. 105, 127–138. doi: 10.1007/s00122-002-0867-7
Entani, T., Iwano, M., Shiba, H., Che, F. S., Isogai, A., and Takayama, S. (2003). Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed-box gene with allelic diversity. Genes Cells 8, 203–213. doi: 10.1046/j.1365-2443.2003.00626.x
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinf. Online 1, 47–50. doi: 10.1177/117693430500100003
Fathi, A., Ghareyazi, B., Haghnazari, A., Ghaffari, M. R., Pirseyedi, S. M., Kadkhodaei, S., et al. (2008). Assessment of the genetic diversity of almond (Prunus dulcis) using microsatellite markers and morphological traits. Iran. J. Biotechnol. 6, 98–106.
Fernandez i Marti, A., Gradziel, T. M., and Socias i Company R. (2014). Methylation of the Sf locus in almond is associated with S-RNase loss of function. Plant Mol. Biol. 86, 681–689. doi: 10.1007/s11103-014-0258-x
Fernandez i Marti, A., Hanada, T., Alonso, J. M., Yamane, H., Tao, R., and Sociasi Company, R. (2010). The almond Sf haplotype shows a double expression despite its comprehensive genetic identity. Sci. Hortic. 125, 685–691. doi: 10.1016/j.scienta.2010.05.024
Guerra, M. E., Lopez-Corrales, M., and Wünsch, A. (2012). Improved S genotyping and new incompatibility groups in Japanese plum. Euphytica 186, 445–452. doi: 10.1007/s10681-012-0636-x
Guerra, M. E., and Rodrigo, J. (2015). Japanese plum pollination: a review. Sci. Hortic. 197, 674–686. doi: 10.1016/j.scienta.2015.10.032
Guerra, M. E., Rodrigo, J., López-Corrales, M., and Wünsch, A. (2009). S-RNase genotyping and incompatibility group assignment by PCR and pollination experiments in Japanese plum. Plant Breed. 128, 304–311. doi: 10.1111/j.1439-0523.2008.01552.x
Gürcan, K., Öcal, N., Yılmaz, K. U., Ullah, S., Erdoðan, A., and Zengin, Y. (2015). Evaluation of Turkish apricot germplasm using SSR markers: genetic diversity assessment and search for plum pox virus resistance alleles. Sci. Hortic. 193, 155–164. doi: 10.1016/j.scienta.2015.07.012
Halász, J., Hegedus, A., Szabo, Z., Nyéki, J., and Pedryc, A. (2007). DNA-based S-genotyping of Japanese plum and pluot cultivars to clarify incompatibility relationships. Hortic. Sci. 42, 46–50. doi: 10.21273/hortsci.42.1.46
Halász, J., Kurilla, A., and Hegedûs, A. (2014). Preliminary characterization of the self-incompatibility genotypes of European plum (Prunus domestica L.) cultivars. Int. J. Hortic. Sci. 20, 23–26.
Halász, J., Makovics-Zsohàr, N., Szöke, F., Ercisli, S., and Hegedüs, A. (2017). Simple sequence repeat and S-locus genotyping to explore genetic variability in polyploid Prunus spinosa and P. insititia. Biochem. Genet. 55, 22–33. doi: 10.1007/s10528-016-9768-3
Hammer, O., Harper, D. A. T., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:9.
Hanada, T., Watari, A., Kibe, T., Yamane, H., Wünsch, A., Gradziel, T. M., et al. (2014). Two novel self-compatible S haplotypes in peach (Prunus persica). J. Jpn. Soc. Hortic. Sci. 83, 203–213. doi: 10.2503/jjshs1.ch-099
Hartl, D., and Clark, A. (1997). Principles of Population Genetics, 3rd Edn. Sunderland, MA: Sinauer Associates.
Hauck, N. R., Yamane, H., Tao, R., and Iezzoni, A. F. (2006). Accumulation of non-functional S-haplotypes results in the breakdown of gametophytic self-incompatibility in tetraploid Prunus. Genetics 172, 1191–1198. doi: 10.1534/genetics.105.049395
Hegedus, A., and Halasz, J. (2006). Self-incompatibility in plums (Prunus salicina Lindl., Prunus cerasifera Ehrh. and Prunus domestica L.). A minireview. J. Hortic. Sci. 12, 137–140.
Herrera, S., Lora, J., Hormaza, J. I., Herrero, M., and Rodrigo, J. (2018). Optimizing production in the new generation of apricot cultivars: self-incompatibility, S-RNase allele identification, and incompatibility group assignment. Front. Plant Sci. 9:527. doi: 10.3389/fpls.2018.00527
Horvath, A., Balsemin, E., Barbot, J.-C., Christmann, H., Manzano, G., Reynet, P., et al. (2011). Phenotypic variability and genetic structure in plum (Prunus domestica L.), cherry plum (P. cerasifera Ehrh.) and sloe (P. spinosa L.). Sci. Hort. 129, 283–293. doi: 10.1016/j.scienta.2011.03.049
Jefferies, C. J., and Belcher, A. R. (1974). A fluorescent brightener used for pollen tube identification in vivo. Stain Technol. 49, 199–202. doi: 10.3109/10520297409116977
Kao, T. H., and McCubbin, A. G. (1996). How flowering plants discriminate between self and non-self pollen to prevent inbreeding. PNAS 93, 12059–12065. doi: 10.1073/pnas.93.22.12059
Kao, T. H., and Tsukamoto, T. (2004). The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 16, 72–83.
Klabunde, G. H. F., Dalbó, M. A., and Nodari, R. O. (2014). DNA fingerprinting of Japanese plum (Prunus salicina) cultivars based on microsatellite markers. Crop Breed. Appl. Biotechnol. 14, 139–145. doi: 10.1590/1984-70332014v14n3a21
Kodad, O., Company, R. S. I., and Alonso, J. M. (2015). Unilateral recognition of the Sf allele in almond. Sci. Hortic. 185, 29–33. doi: 10.1016/j.scienta.2015.01.016
Kota-Dombrovska, I., and Lācis, G. (2013). Evaluation of self-incompatibility locus diversity of domestic plum (Prunus domestica L.) using DNA-based S-genotyping. Proc. Latv. Acad. Sci. B Nat. Exact Appl. Sci. 67, 109–115. doi: 10.2478/prolas-2013-0017
Linskens, H. F., and Esser, K. (1957). Uber eine spezifische anfarbung der pollen schlauche im griffel unddie zahl der kallosepfropfen nach selbstung und fremdung. Naturwiss 44, 16–17.
Liu, Z. Q., Xu, G. H., and Zhang, S. L. (2007). Pyrus pyrifolia stylar S-RNase induces alterations in the actin cytoskeleton in self-pollen and tubes in vitro. Protoplasma 232, 61–67. doi: 10.1007/s00709-007-0269-4
Mable, B. K. (2004). Polyploidy and self-compatibility: is there an association? New Phytol. 162, 803–811. doi: 10.1111/j.1469-8137.2004.01055.x
Malcolm, P. (2006). History of Plum Trees and Their Hybrids. Available at: http://thephantomwriters.com/free_content/db/m/history-plum-trees-hybrids.shtml (accessed June 30, 2019).
Matsumoto, D., and Tao, R. (2016). Distinct self-recognition in the Prunus S-RNase-based gametophytic self-incompatibility system. Hortic. J. 85, 289–305. doi: 10.2503/hortj.mi-ir06
Mnejja, M., Garcia-Mas, J., Howad, W., Badenes, M. L., and Arús, P. (2004). Simple-sequence repeat (SSR) markers of Japanese plum (Prunus salicina Lindl.) are highly polymorphic and transferable to peach and almond. Mol. Ecol. 4, 163–166. doi: 10.1111/j.1471-8286.2004.00603.x
Nunes, M. D. S., Santos, R. A. M., Ferreira, S. M., Vieira, J., and Vieira, C. P. (2006). Variability patterns and positively selected sites at the gametophytic self-incompatibility pollen SFB gene in a wild self-incompatible Prunus spinosa (Rosaceae) population. New Phytol. 172, 577–587. doi: 10.1111/j.1469-8137.2006.01838.x
OCDE (2002). Consensus Document on the Biology of Prunus spp. (Stone fruits), Environment Directorate. Paris: OECD Environment Directorate, 1–42.
Reales, A., Sargent, D. J., Tobutt, K. R., and Rivera, D. (2010). Phylogenetics of Eurasian plums, Prunus L. section Prunus (Rosaceae), according to coding and non-coding chloroplast DNA sequences. Tree Genet. Genomes 6, 37–45. doi: 10.1007/s11295-009-0226-9
Rodrigo, J., and Herrero, M. (1996). Evaluation of pollination as the cause of erratic fruit set in apricot Moniqui. J. Hortic. Sci. 71, 801–805. doi: 10.1080/14620316.1996.11515461
Romero, C., Vilanova, S., Burgos, L., Martínez-Calvo, J., Vicente, M., Llácer, G., et al. (2004). Analysis of the S-locus structure in Prunus armeniaca L. Identification of S-haplotype specific S-RNase and F-box genes. Plant Mol. Biol. 56, 145–157.
Salazar, J. A., Ruiz, D., Campoy, J. A., Sánchez-Pérez, R., Crisosto, C. H., Martínez-García, P. J., et al. (2014). Quantitative Trait Loci (QTL) and Mendelian Trait Loci (MTL) Analysis in Prunus: a breeding perspective and beyond. Plant Mol. Biol. Rep. 32, 1–18. doi: 10.1007/s11105-013-0643-7
Sapir, G., Stern, R. A., Eisikowitch, D., and Goldway, M. (2004). Cloning of four new Japanese plum S-alleles and determination ofthe compatibility between cultivars by PCR analysis. J. Hortic. Sci. Biotechnol. 79, 223–227. doi: 10.1080/14620316.2004.11511752
Sapir, G., Stern, R. A., Goldway, M., and Shafir, S. (2007). SFBs of Japanese plum (Prunus salicina): cloning seven alleles and determining their linkage to the S-RNase gene. Hort. Sci. 42, 1509–1512. doi: 10.21273/hortsci.42.7.1509
Sehic, J., Nybom, H., Hjeltnes, S.-H., and Gasi, F. (2015). DNA marker-assisted identification of Prunus accessions. Acta Hortic. 1101, 153–158. doi: 10.17660/ActaHortic.2015.1101.23
Sonneveld, T., Robbins, T. P., and Tobutt, K. R. (2006). Improved discrimination of self-incompatibility S-RNase alleles in cherry and high throughput genotyping by automated sizing of first intron polymerase chain reaction products. Plant Breed. 125, 305–307. doi: 10.1111/j.1439-0523.2006.01205.x
Sonneveld, T., Tobutt, K. R., and Robbins, T. P. (2003). Allele-specific PCR detection of sweet cherry self-incompatibility (S) alleles S1 to S16 using consensus and allele-specific primers. Theor. Appl. Genet. 107, 1059–1070. doi: 10.1007/s00122-003-1274-4
Stoeckel, S., Grange, J., Fernández-Manjarres, J. F., Bilger, I., Frascaria-Lacoste, N., and Mariette, S. (2006). Heterozygote excess in a self-incompatible and partially clonal forest tree species Prunus avium L. Mol. Ecol. 15, 2109–2118. doi: 10.1111/j.1365-294x.2006.02926.x
Struss, D. A. R., Southwick, S. M., and Boritzki, M. (2003). Analysis of sweet cherry (Prunus avium L.) cultivars using SSR and AFLP markers. J. Am. Soc. Hortic. Sci. 128, 904–909. doi: 10.21273/jashs.128.6.0904
Sutherland, B. G., Robbins, T. P., and Tobutt, K. R. (2004). Primers amplifying a range of Prunus S-alleles. Plant Breed. 123, 582–584. doi: 10.1111/j.1439-0523.2004.01016.x
Sutherland, B. G., Tobutt, K. R., and Robbins, T. P. (2007). Molecular genotyping of self-incompatible plum cultivars. Acta Hortic. 734, 47–51. doi: 10.17660/actahortic.2007.734.3
Taguchi, Y. H., and Oono, Y. (2005). Relational patterns of gene expression via non-metric multidimensional scaling analysis. Bioinformatics 21, 730–740. doi: 10.1093/bioinformatics/bti067
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tamura, M., Ushijima, K., Sassa, H., Hirano, H., Tao, R., Gradziel, T. M., et al. (2000). Identification of self-incompatibility genotypes of almond by allele-specific PCR analysis. Theor. Appl. Genet. 101, 344–349. doi: 10.1007/s001220051489
Tao, R., Watari, A., Hanada, T., Habu, T., Yaegaki, H., Yamaguchi, M., et al. (2007). Self compatible peach (Prunus persica) has mutant versions of the Shaplotypes found in self-incompatible Prunus species. Plant Mol. Biol. 63, 109–123. doi: 10.1007/s11103-006-9076-0
Tao, R., Yamane, H., Sugiura, A., Murayama, H., Sassa, H., and Mori, H. (1999). Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNases in sweet cherry. J. Am. Soc. Hort. Sci. 124, 224–233. doi: 10.21273/jashs.124.3.224
Testolin, R., Marrazzo, T., Cipriani, G., Quarta, R., Verde, I., Dettori, M. T., et al. (2000). Microsatellite DNA in peach (Prunus persica L. Batsch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome 43, 512–520. doi: 10.1139/gen-43-3-512
Urrestarazu, J., Errea, P., Miranda, C., Santesteban, L. G., and Pina, A. (2018). Genetic diversity of Spanish Prunus domestica L. germplasm reveals a complex genetic structure underlying. PLoS One 13:e0195591. doi: 10.1371/journal.pone.0195591
Ushijima, K., Sassa, H., Kusaba, M., Tao, R., Tamura, M., Gradziel, T. M., et al. (2001). Characterization of the S-locus region of almond (Prunus dulcis): analysis of a somaclonal mutant and a cosmid contig for an S-haplotype. Genetics 158, 379–386.
Ushijima, K., Sassa, H., Tao, R., Yamane, H., Dandekar, A. M., Gradziel, T. M., et al. (1998). Cloning and characterization of cDNAs encoding S-RNases from almond (Prunus dulcis): primary structural features and sequence diversity of the S-RNases in Rosaceae. Mol. Genet. Genome 260, 261–268. doi: 10.1007/s004380050894
Van de Peer, Y., Mizrachi, E., and Marchal, K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424. doi: 10.1038/nrg.2017.26
Vaughan, S. P., Russell, K., Sargent, D. J., and Tobutt, K. R. (2006). Isolation of S-locus F-box alleles in Prunus avium and their application in a novel method to determine self-incompatibility genotype. Theor. Appl. Genet. 112, 856–866. doi: 10.1007/s00122-005-0187-9
Vieira, J., Santos, R. A., Ferreira, S. M., and Vieira, C. P. (2008). Inferences on the number and frequency of S-pollen gene (SFB) specificities in the polyploidy Prunus spinosa. Heredity 101, 351–358. doi: 10.1038/hdy.2008.60
Vilanova, S., Badenes, M. L., Burgos, L., Martínez-Calvo, J., Llácer, G., and Romero, C. (2006). Self-compatibility of two apricot selections is associated with two pollen-part mutations of different nature. Plant Physiol. 142, 629–641. doi: 10.1104/pp.106.083865
Walsh, P. S., Erlich, H. A., and Higuchi, R. (1992). Preferential PCR amplification of alleles: mechanisms and solutions. Genome Res. 1, 241–250. doi: 10.1101/gr.1.4.241
Watari, A., Hanada, T., Yamane, H., Esumi, T., Tao, R., Yaegaki, H., et al. (2007). A low transcriptional level of Se-RNase in the Se-haplotype confers self-compatibility in Japanese plum. J. Am. Soc. Hort. Sci. 132, 396–406. doi: 10.21273/jashs.132.3.396
Weir, B. S., and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x
Wünsch, A., and Hormaza, J. I. (2002). Molecular characterization of sweet cherry (Prunus avium L.) genotypes using peach [Prunus persica (L.) Batsch] SSR sequences. Heredity 89, 56–63. doi: 10.1038/sj.hdy.6800101
Wünsch, A., and Hormaza, J. I. (2004). S-allele identification by PCR analysis in sweet cherry cultivars. Plant Breed. 123, 327–331. doi: 10.1111/j.1439-0523.2004.00984.x
Yaegaki, H., Shimada, T., Moriguchi, T., Hayama, H., Haji, T., and Yamaguchi, M. (2001). Molecular characterization of S-RNase genes and S- genotypes in the Japanese apricot (Prunus mume Sieb. Et Zucc.). Sex. Plant Reprod. 13, 251–257. doi: 10.1007/s004970100064
Yamane, H., Ikeda, K., Ushijima, K., Sassa, H., and Tao, R. (2003). A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol. 44, 764–769. doi: 10.1093/pcp/pcg088
Yamane, H., Tao, R., and Sugiura, A. (1999). Identification and cDNA cloning for S-RNases in self-incompatible Japanese plum (Prunus salicina Lindl. cv. Sordum). Plant Biotechnol. 16, 389–396. doi: 10.5511/plantbiotechnology.16.389
Yamane, H., Tao, R., Sugiura, A., Hauck, N. R., and Iezzoni, A. F. (2001). Identification and characterization of S-RNases in tetraploid sour cherry (Prunus cerasusL.). J. Am. Soc. Hortic. Sci. 126, 661–667. doi: 10.21273/jashs.126.6.661
Zhang, L., Chen, X., Chen, X., Zhang, C., Liu, X., Ci, Z., et al. (2008). Identification of self-incompatibility (S-) genotypes of Chinese apricot cultivars. Euphytica 160, 241–248. doi: 10.1007/s10681-007-9544-x
Zhang, S. L., Huang, S. X., Kitashiba, H., and Nishio, T. (2007). Identification of S-haplotype F-box gene in Japanese plum (Prunus salicina Lindl.). Sex Plant Reprod. 20, 1–8. doi: 10.1007/s00497-006-0037-1
Zhebentyayeva, T., Shankar, V., Scorza, R., Callahan, A., Ravelonandro, M., Castro, S., et al. (2019). Genetic characterization of worldwide Prunus domestica (plum) germplasm using sequence-based genotyping. Hortic. Res. 6:12. doi: 10.1038/s41438-018-0090-6
Keywords: plums, polyploidy, S-genotyping, self-(in)compatibility, pollination, pollen tube
Citation: Abdallah D, Baraket G, Perez V, Ben Mustapha S, Salhi-Hannachi A and Hormaza JI (2019) Analysis of Self-Incompatibility and Genetic Diversity in Diploid and Hexaploid Plum Genotypes. Front. Plant Sci. 10:896. doi: 10.3389/fpls.2019.00896
Received: 29 January 2019; Accepted: 24 June 2019;
Published: 12 July 2019.
Edited by:
Petr Smýkal, Palacký University Olomouc, CzechiaReviewed by:
Josef Patzak, Hop Research Institute Co., Ltd., CzechiaPedro Martinez-Gomez, Center for Edaphology and Applied Biology of Segura, Spanish National Research Council (CSIC), Spain
Copyright © 2019 Abdallah, Baraket, Perez, Ben Mustapha, Salhi-Hannachi and Hormaza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donia Abdallah, YWJkYWxsYWhkb25pYUB5YWhvby5mcg==
 Donia Abdallah
Donia Abdallah Ghada Baraket1
Ghada Baraket1 Veronica Perez
Veronica Perez Amel Salhi-Hannachi
Amel Salhi-Hannachi Jose I. Hormaza
Jose I. Hormaza