
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 July 2019
Sec. Plant Pathogen Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00886
 Amornrat Changkwian1
Amornrat Changkwian1 Jelli Venkatesh1
Jelli Venkatesh1 Joung-Ho Lee1
Joung-Ho Lee1 Ji-Woong Han1
Ji-Woong Han1 Jin-Kyung Kwon1
Jin-Kyung Kwon1 Muhammad Irfan Siddique1
Muhammad Irfan Siddique1 Abate Mekonnen Solomon1
Abate Mekonnen Solomon1 Gyung-Ja Choi2
Gyung-Ja Choi2 Eunji Kim3
Eunji Kim3 Yunhee Seo3
Yunhee Seo3 Young-Ho Kim3
Young-Ho Kim3 Byoung-Cheorl Kang1*
Byoung-Cheorl Kang1*The root-knot nematode (RKN) Meloidogyne incognita severely reduces yields of pepper (Capsicum annuum) worldwide. A single dominant locus, Me7, conferring RKN resistance was previously mapped on the long arm of pepper chromosome P9. In the present study, the Me7 locus was fine mapped using an F2 population of 714 plants derived from a cross between the RKN-susceptible parent C. annuum ECW30R and the RKN-resistant parent C. annuum CM334. CM334 exhibits suppressed RKN juvenile movement, suppressed feeding site enlargement and significant reduction in gall formation compared with ECW30R. RKN resistance screening in the F2 population identified 558 resistant and 156 susceptible plants, which fit a 3:1 ratio confirming that this RKN resistance was controlled by a single dominant gene. Using the C. annuum CM334 reference genome and BAC library sequencing, fine mapping of Me7 markers was performed. The Me7 locus was delimited between two markers G21U3 and G43U3 covering a physical interval of approximately 394.7 kb on the CM334 chromosome P9. Nine markers co-segregated with the Me7 gene. A cluster of 25 putative nucleotide-binding site and leucine-rich repeat (NBS-LRR)-type disease resistance genes were predicted in the delimited Me7 region. We propose that RKN resistance in CM334 is mediated by one or more of these NBS-LRR class R genes. The Me7-linked markers identified here will facilitate marker-assisted selection (MAS) for RKN resistance in pepper breeding programs, as well as functional analysis of Me7 candidate genes in C. annuum.
Root-knot nematodes (RKN; Meloidogyne spp.) are obligate phytopathogens causing significant economic losses in several crops including Solanaceae species (Eisenback and Triantaphyllou, 1991; Bernard et al., 2017). There are more than 98 Meloidogyne species (Jones et al., 2013). Four of these, Meloidogyne arenaria, M. incognita, M. hapla, and M. javanica, are widely distributed with diverse host ranges and cause considerable yield losses in many crops (Jones et al., 2013; Noling, 2014; Wu et al., 2018). Strategies to mitigate the RKN threat include biological and chemical control measures as well as the use of resistant cultivars/rootstocks. Due to the inefficiency of chemical applications, as well as increasing awareness of food safety and environmental concerns, the application of nematicides has been restricted. Since nematode-resistant cultivars provide an efficient and environmentally safe alternative to chemical measures, much effort has been devoted to identifying host resistance against RKN in cultivated or in wild species (Taylor and Sasser, 1978; Pegard et al., 2005).
Several dominant nematode resistance genes including Mi1.1-2 and Mi-9 have been reported in tomato (Solanum lycopersicum; Nombela et al., 2003; Seah et al., 2004; van der Vossen et al., 2005; Jablonska et al., 2007; Sanchez-Puerta and Masuelli, 2011; de Carvalho et al., 2015). In potato (Solanum tuberosum), the genes Hero and Gpa2 confer resistance to Globodera pallida (white potato cyst nematode, van der Voort et al., 1999; van der Vossen et al., 2000; Ernst et al., 2002) and Gro1-4 confers resistance to Globodera rostochiensis (yellow potato cyst nematode; Paal et al., 2004: Williamson and Kumar, 2006). In pepper, at least 10 dominant genes, Me1 to Me7, Mech1, Mech2 and N have been reported to confer resistance to Meloidogyne spp. (Wang and Bosland, 2006; Djian-Caporalino et al., 2007; Wang et al., 2009; Bucki et al., 2017). Me1, Me3, Me7, and N confer resistance to a wide range of RKNs, including M. arenaria, M. javanica, and M. incognita. Mech1 and Mech2 confer resistance to M. chitwoodi. These R genes originated from Capsicum annuum accessions PI 201234 (Me1 and Mech2), PI 322719 (Me3 and Me4), Criollo de Morelos 334 (CM334) (Me7 and Mech1), and “Missisippi Nemaheart” (N) (Hare, 1957). Each of these genes have been deployed in pepper breeding programs (Fery et al., 1998; Djian-Caporalino et al., 2007; Wang et al., 2009, 2018; Fazari et al., 2012; Celik et al., 2016).
These RKN resistance genes, including Me1, Me3, Me4, Me7, Mech1, Mech2, and N have been mapped on the P9 chromosome and are clustered in a 28 cM genetic interval (Djian-Caporalino et al., 2001, 2007; Fazari et al., 2012; Uncu et al., 2015). N is linked to the Me1 and Me3 loci, but is not allelic to them. Me3 and Me7 were originally mapped to different loci 12.1 cM apart (Djian-Caporalino et al., 2007) on chromosome P9, but were later found to be allelic (Thies and Ariss, 2009; Djian-Caporalino et al., 2011; Fazari et al., 2012). Several molecular markers linked to the Me1, Me3/Me7, and N loci have been developed for marker-assisted selection (MAS) and are used in breeding programs (Wang et al., 2009, 2018; Fazari et al., 2012; Gisbert et al., 2013; Uncu et al., 2015; Guo et al., 2016). Despite extensive mapping studies, the molecular aspects of pepper RKN resistance genes remain largely unexplored.
Plant defense responses against RKN are often associated with gene-for-gene resistance (effector-triggered immunity). For instance, the well-known dominant R gene, Mi-1 from Solanum peruvianum (Peruvian tomato), confers gene for gene resistance to some RKN Meloidogyne spp. Mi-1 belongs to an R gene family that shares several canonical sequences and structural motifs of nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains (Milligan et al., 1998; Chen et al., 2007; Mao et al., 2015). The LRR domain induces hypersensitive response (HR)-like localized cell death (Hwang et al., 2000). Two RKN effectors, MAP-1 from M. incognita and Cg1 from M. javanica, have been suggested to be the cognate avirulence (Avr) genes of Mi-1 (Gleason et al., 2008; Castagnone-Sereno et al., 2009). However, the interactions of these effectors with Mi-1, and Mi-1-mediated resistance, are not well understood. The potato cyst nematode RBP-1 and VAP1 effectors have been demonstrated to physically interact with their canonical R protein such as potato Gpa2 and a tomato Rcr3pim/Cf-2 and induce a foliar HR upon the transient co-expression (Sacco et al., 2009; Lozano-Torres et al., 2012).
CM334 is used by breeders as a source of resistance to a range of pathogens including several viruses, Phytophthora capsaici (chili pepper blight) and RKN (Palloix et al., 1990; Dogimont et al., 1996; Djian-Caporalino et al., 2007). Molecular markers have been developed to map the Me7 locus (Djian-Caporalino et al., 2007; Fazari et al., 2012), with the closest markers, HM2, SCAR_PM6a, and SSCP_PM5 delimiting it to a 3.8 cM genetic interval (Fazari et al., 2012). However, the exact location of the Me7 locus and the molecular basis of RKN resistance are poorly understood. To identify and clone the Me7 gene, it is essential to construct a fine genetic map of the Me7 locus and develop more closely linked markers.
Our aims were to construct a high-resolution map of the Me7 locus and predict candidate genes for RKN resistance. A total of 28 markers, including high-resolution melting (HRM) and cleaved amplified polymorphic sequences (CAPS) markers, linked to RKN resistance were developed and used for fine mapping of the Me7 locus using the F2 population derived from a cross between Early Calwonder 30R (ECW30R) and CM334. The tightly linked Me7 markers we identified will be useful for marker-assisted selection for RKN resistance in pepper breeding programs.
An F2 mapping population consisting of 714 plants, derived from a cross between M. incognita RKN-resistant CM334 and RKN-susceptible ECW30R lines from Horticultural Crops Breeding and Genetics Lab, Seoul National University, was used to fine-map the Me7 locus. Phenotype screening of 504 of these F2 plants was performed at the Research Center for Biobased Chemistry, Korea Research Institute of Chemical Technology (KRICT), Daejoen, in 2014–2015 and phenotype screening of the other 210 F2 plants was performed at Seoul National University, Seoul, Korea, in 2015.
Meloidogyne incognita race 1 was kindly provided by Prof. Y-HK (Clinical Plant Pathology and Nematology Laboratory, Seoul National University). M. incognita was propagated using susceptible tomato (S. lycopersicum cv. Micro-Tom). One-month old Micro-Tom plants were cultivated in pots with fresh commercial potting mixture (Hanarum, Minong Fertilizer, Korea) and sand in a 2:1 ratio were inoculated with 1,000 juvenile stage 2 (J2) of M. incognita. The inoculated plants were kept in the glasshouse condition with an average temperature of 26 ± 2°C. The nematode egg masses were collected from approximately 55-day-old infected susceptible tomato roots. Briefly, infected roots were cleaned with water and cut into 1 cm pieces, and then stirred in 1% NaOCl solution for 5 min (Coyne and Ross, 2014). The suspension was passed sequentially through a stack of sieves of 250, 180 and 25 μm, under running water, to remove any NaOCl residue. The eggs and J2 were captured on the 25 μm sieve with distilled water. The suspension with egg masses and J2 were filtered through a tissue paper that was placed on a Baermann funnel glass at room temperature (25°C) for three to four days. Thus, the J2 collected were counted under a light microscope. Four-leaf stage plants of testing populations were inoculated with 1,000 freshly hatched J2 M. incognita as mentioned above. Inoculated plants were kept in the glasshouse maintained at 26 ± 2°C.
The resistance phenotype was evaluated at 45 days after inoculation (dai). For egg mass production and viability, four roots from each of the parental lines were uprooted and cleaned with tap water. Egg masses were handpicked from root system, and incubated in distilled water at room temperature for 2 h before observing under the magnifying microscope (Carl Zeiss, Thornwood, NY, United States). The root systems of each plant were examined, and the RKN resistance phenotype was scored using a root galling index (GI), rated on 0–4 scales, where 0 = 0–25%, 1 = 26–50%, 2 = 51–75%, 3 = 76-99%, and 4 = ≥ 100% (Baker, 1985). The percentage was calculated by dividing the total number of galls from each root system by the mean of the number of galls from susceptible ECW30R. The plants were classified as resistant when GI = 0 and as susceptible when GI ≥1, a classification modified from Seo et al. (2014).
To study the histological response in the parental lines, infected roots as prepared above were harvested at 5, 10, and 15 dai for microscopic observation according to the procedure described by Seo et al. (2014). The specimens were sectioned by ultramicrotome (MT-X, RMC, Tucson, AZ, United States) to a thickness of 700 nm. The slides were stained with 1% toluidine blue O in 2% sodium tetraborate. The root systems of both parents were observed for HR by staining with fuchsin-acetic acid solution (Bybd et al, 1983). Specimens were analyzed using a Carl Zeiss microscope (Carl Zeiss, Thornwood, NY, United States).
Total genomic DNA was extracted from young leaf tissues using the cetyltrimethylammonium bromide (CTAB) protocol (Kang et al., 2010). DNA concentration and quality were analyzed using a NanoDrop spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, United States).
Previously reported PCR-based markers including sequence characterized amplified region (SCAR), simple sequences repeats (SSR) and CAPS linked to the Me1, Me3, Me7, and N loci (Djian-Caporalino et al., 2007; Fazari et al., 2012; Guo et al., 2016) were amplified from the ECW30R and CM334 parents and sequenced at the National Instrumentation Center for Environmental Management (NICEM, Seoul National University, Korea). We analyzed these marker sequences for polymorphism and used BLAST to identify their physical positions in the CM334 genome version v.1.5 (scaffold) and v.1.55 (chromosome) (Kim et al., 2014). The SSR marker, CASSR37, linked to the N locus (Celik et al., 2016) and BAC-end markers PE43N9R, PE242G21R, PE11F6F, and PE25F15F, linked to the Me3 locus derived from a double haploid pepper DH149 (Guo et al., 2016), were also used for marker development. The Me-loci and N linked markers are listed in Supplementary Table S1.
To develop additional markers for fine mapping Me7, PCR primers were designed from the v.1.55 chromosome and v.1.5 scaffold versions of the C. annuum “CM334” genome (Kim et al., 2014). Amplified PCR products were sequenced at NICEM and Macrogen1 to detect polymorphisms. Identified SNPs were converted into HRM or CAPS markers listed in Supplementary Table S2.
The HRM assays were performed as described (Liu et al., 2016) with slight modifications in the PCR conditions as follows: 95°C for 4 min, followed by 50 cycles of denaturing at 95°C for 20 s, touchdown annealing from 60 to 53°C for 30 s, 72°C extension for 25 s, and then 25°C for 30 s. The HRM used 0.1°C increments between 65 and 90°C, with a Rotor-GeneTM 6000 thermocycler (Corbett Research, Sydney, Australia). The HRM curve profiles of homozygous parents and heterozygous F1 plants were used to assign the genotypes (Supplementary Figure S1). For the CAPS marker (18660) assay, PCR was performed as described by Jo et al. (2016). The digestion was performed in a reaction mixture containing 10 μl of PCR product, 2 μl of digestion buffer, 0.5 μl of MspI and sterile distilled water in a total volume of 20 μl. Reactions were incubated for 3 h at 37°C. Digested PCR products were viewed using agarose gel electrophoresis (Supplementary Figure S2).
To expand the distal ends of scaffolds 1640 and 1578, markers from these scaffolds were used to screen a BAC library (Yoo et al., 2003). A three-step BAC screening procedure was described by Jo et al. (2016). BAC ends of 10 positive BAC clones were sequenced using the universal primers T7 and SP6. The BAC end sequences were then BLAST searched against the C. annuum genome databases2. The selected BAC611K18 clones were PacBio Pacific BioSciences) sequenced at Macrogen (see text footnote 1). The BAC sequence-derived SNPs and InDels were used for developing additional markers. The dot plot analysis of repeat sequences in the scaffold and BAC clones was performed using the NCBI server3.
To identify the most suitable sequence information to delimit the Me7 locus, the total genome sequences from C. annuum “CM334” version v.1.55, v.1.6 (Kim et al., 2014, 2017) and UCD10X v1.0 (Hulse-Kemp et al., 2018) were used. CLC Main Workbench 8.1 (QIAGEN, Aarhus, Denmark) was used to obtain the physical position of the Me7 linked markers, where map reads with ≥98% nucleotide similarity to the genomic sequence on chromosome P9 were used for comparative mapping.
Genetic linkage analysis was performed using Carthagene ActiveTcl 8.4 (de Givry et al., 2005). The mapping distance was calculated using Kosambi’s mapping function with the LOD threshold set at 3.0 and distance threshold at 0.5. The genetic linkage map was drawn using the MapChart 2.2 tool (Voorrips, 2002). The Me7 locus was initially mapped using 192 F2 plants and fine-mapped with 714 F2 plants. The physical map data for the Me7 locus were retrieved from the pepper genome database version v.1.64. The coding sequences (Annuum.v.2.0.CDS) from the genomic interval of the Me7 locus were BLAST searched at the NCBI server (see text footnote 3) for identification of conserved domains and functional annotation.
To know the resistance response to RKN in our experimental conditions, M. incognita gall formation and egg mass production were observed in resistant C. annuum “CM334” and susceptible C. annuum “ECW30R” at 45 dai (Figure 1A and Supplementary Figure S3). Viable egg masses with J2 were observed in both CM334 and ECW30R root systems (Supplementary Figures S3B,D,E), however, the root systems of CM334 formed only a few, small galls, while ECW30R roots had many, large galls (Figures 1A,B). We performed histological studies to investigate the intercellular resistance responses in the root systems. At 10 dai, no cell necrosis was noticed in ECW30R, whereas necrosis was observed in CM334 cells, as a result of a HR (Figure 1C), indicating that CM334 is highly resistant to M. incognita.
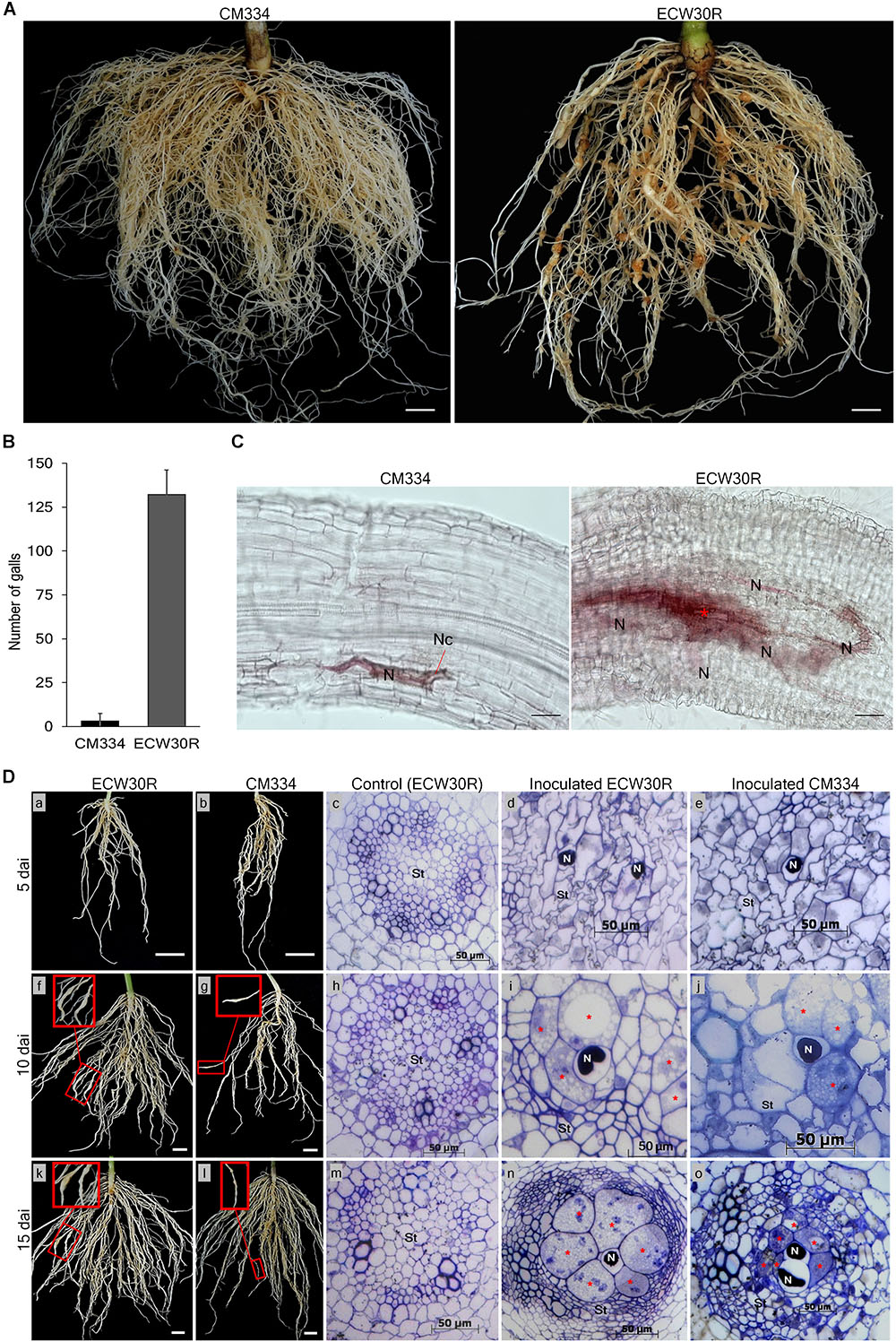
Figure 1. Comparison of resistant responses in the root systems of the parental lines. (A) A resistant CM334 and a susceptible ECW30R observed at 45 dai infected with Meloidogyne incognita J2. Bar = 1 cm. (B) Average number of root galls observed at 45 dai from 18 plants of each parent. Bar = standard deviation. (C) Resistance responses at 10 dai in CM334 and ECW30R root systems during juvenile penetration. Cell necrosis (Nc), nematode (N) and initiation of giant cell formation (red ∗). Bar = 100 μm. (D) Root systems and cross-sections of susceptible ECW30R and resistant CM334. The inoculated roots were harvested at 5, 10, and 15 dai. Pictures in red boxes pointing to galls development on the infected roots in close view (f, g, k, l), Bar = 1 cm. The root systems of ECW30R and CM334 and gall formation were observed at 5 (a–e), 10 (f–j), and 15 (k–o) dai. Cross-sections of inoculated ECW30R, CM334 and non-inoculated ECW30R (control) at 5 (c–e), 10 (h–j), and 15 (m–o) dai. Nematode (N), enlarged multinucleate giant cells (red ∗) and stele (St). Bar = 50 μm.
Cross-sections of the root system of RKN-infected plants revealed no morphological differences between susceptible ECW30R and resistant CM334 at five dai; however, juveniles of RKN were detected in both infected parents with no sign of feeding site formation (Figure 1D; a–e). At 10 dai, although giant cell development was initiated in both lines, the stele cells were slightly narrower and denser with enlarged feeding sites in ECW30R compared to CM334 (Figure 1D; f–j). At 15 dai, the infected cells in ECW30R grew much larger than in CM334 and caused increased compaction of cell layers in vascular tissues (Figure 1D; k–o). Overall, these results demonstrate that the RKN resistance of CM334 involves cell necrosis as well as the suppression of establishment and/or enlargement of feeding sites.
The RKN resistance was evaluated at 45 dai. All tested CM334 and F1 plants were resistant to RKN, while all ECW30R plants were susceptible. Out of 714 F2 individuals, 558 plants showed resistance and 156 F2 plants showed susceptible phenotypes, which fit a segregation ratio of 3 resistant:1 susceptible, as expected for the inheritance of a single dominant gene (X2 = 3.782, P = 0.0518) (Table 1). These results are consistent with previous phenotyping studies (Djian-Caporalino et al., 2007; Fazari et al., 2012).
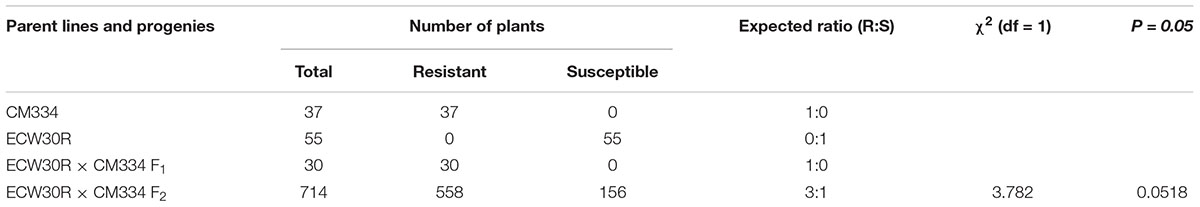
Table 1. Segregation analysis of RKN resistance in CM334, ECW30R, F1 and F2 mapping populations at 45 dai.
A high-density map for the Me7 locus was developed using the Me loci and N locus-linked markers from previous studies (Supplementary Table S1), and new markers developed using genomic information from CM334 (Supplementary Table S2). Among the previously reported markers, only SCAR_PM6a, SCAR_PM6b (linked to Me3, Me7, and N loci) and CASSR37 (linked to N locus) were found to be polymorphic and therefore used for mapping Me7 in this study. No recombinant were found for SCAR_PM6a and SCAR_PM6b, whereas three recombinants were detected for CASSR37 in a subset of the F2 population containing 192 individuals. These results indicate that the N and Me7 loci are not allelic, which concurs with previous reports (Fazari et al., 2012; Celik et al., 2016). Four Me3-linked markers (PE43N9R, PE242G21R, PE11F6F, and PE25F15F) (Guo et al., 2016) were also tested; however, these markers were not polymorphic in our mapping population (Supplementary Table S1).
To develop additional markers, we BLAST searched these Me3, Me7, and N linked marker sequences against the C. annuum CM334 genome (v.1.55) and scaffold (v.1.5) (Kim et al., 2014), and identified their genomic positions on chromosome P9 and unassigned scaffold sequences. Using the genomic information of CM334 and scaffolds (1640, 1646, 1677, and 1578) containing SCAR_PM6a, SCAR_PM6b, CASSR37, and the four Me3-linked markers, we developed 21 HRM markers and one CAPS marker (Figure 2 and Supplementary Table S2). In 192 F2 individuals, four newly developed markers and two previous markers (SCAR_PM6a and SCAR_PM6b) co-segregated with the Me7 locus (Figure 2) with a genetic interval of 0.9 cM on P9 (v.1.6) between G43U3, G79U3, and CA63a, which are located on scaffolds (1578, 1640, and 1677) (Figure 2 and Supplementary Table S2).
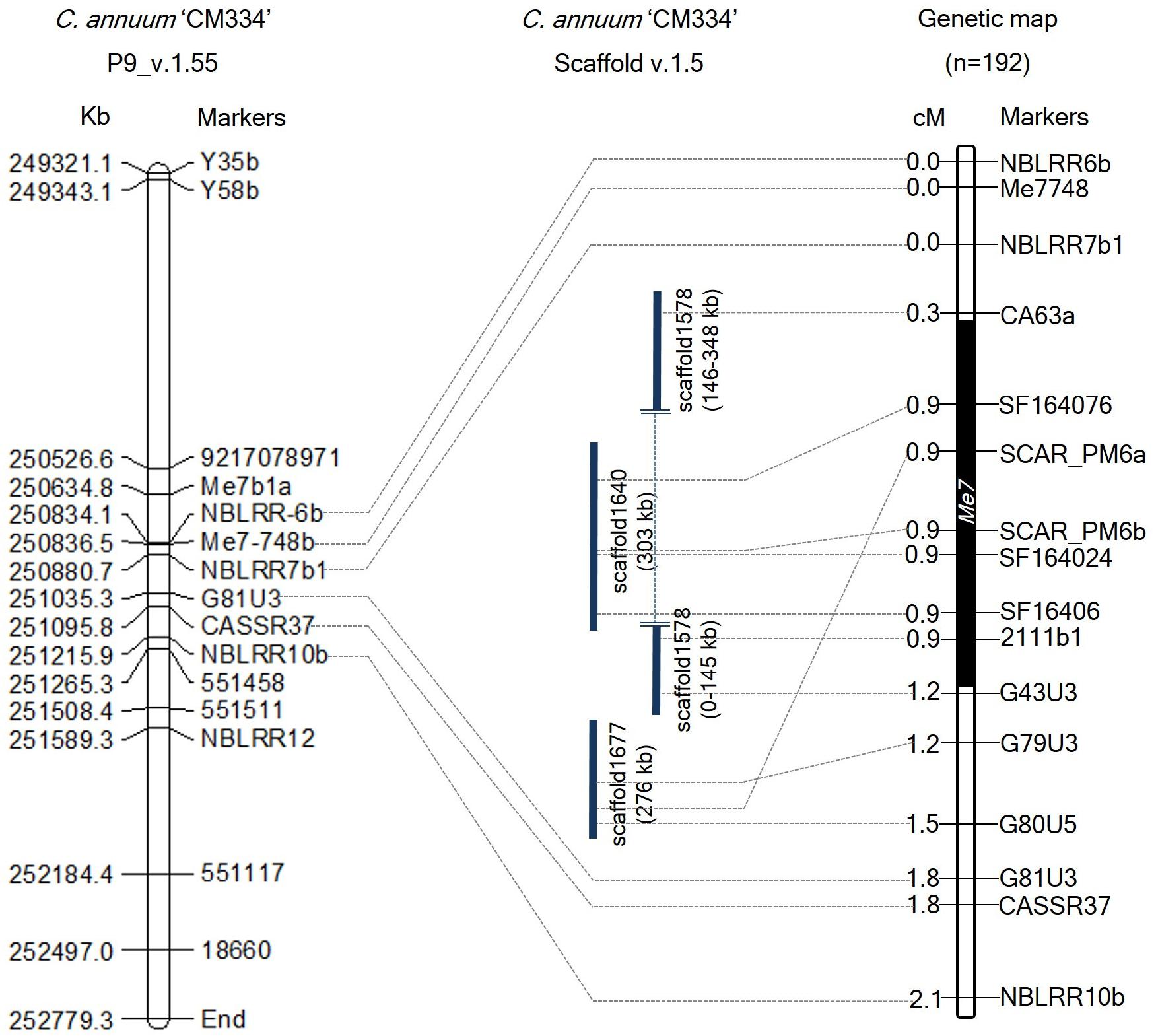
Figure 2. Comparative physical and genetic maps of the Me7 locus developed from 192 F2 plants. The left panel indicates the physical map of the Me7 linked markers on the “CM334” genome reference P9 (v.1.55). The right panel indicates the genetic map of the Me7 locus. Six markers co-segregating with the Me7 locus were detected on scaffolds (v.1.5). The Me7 locus was delimited to a 0.9 cM region between the G43U3/G79U3 and CA63a markers. Physical and genetic distances between markers are indicated on the left side of the chromosome P9.
To extend the distal ends of scaffold 1640 and to identify the regions overlapping with scaffolds 1578, we performed BAC library screening using three markers: the CA63a marker from scaffold 1578 and the SF164076 and SF164024 markers from scaffold1640 (Figure 3A). BAC library screening with the marker CA63a detected the 760A3 clone, and both its BAC ends aligned to scaffold 1578 (Figure 3A). BAC screening with the marker SF164024 identified five clones, 399A15, 765B13, 564D22, 694N7, and 714B12. Of these, only 765B13 and 714B12 clones overlapped with 611K18 and 514E15 (Figure 3A). BAC library screening with the SF164076 marker detected three BAC clones, 711J20, 514E15, and 611K18. BAC end sequences of the 611K18 clone aligned to the sequence of the scaffolds 1640 and 1578 (Figure 3A). Therefore, we selected the 611K18 clone for full-length sequencing and developed five markers (Supplementary Table S2). Further BAC screening with the 6119403 marker developed from the 611K18 BAC clone detected a single clone, 742O5, which bridged the gap between the 760A3 and 611K18 clones (Figure 3A). Furthermore, the comparative dot plot analysis revealed an association between scaffolds 1640 and 1578 bridging and overlapping the BAC clones (Figure 3B). A total of five additional HRM markers (Supplementary Table S2) were produced using genomic information of BAC clone 611K18 and a flanking marker indicated by G21U3 for fine mapping of the Me7 locus.
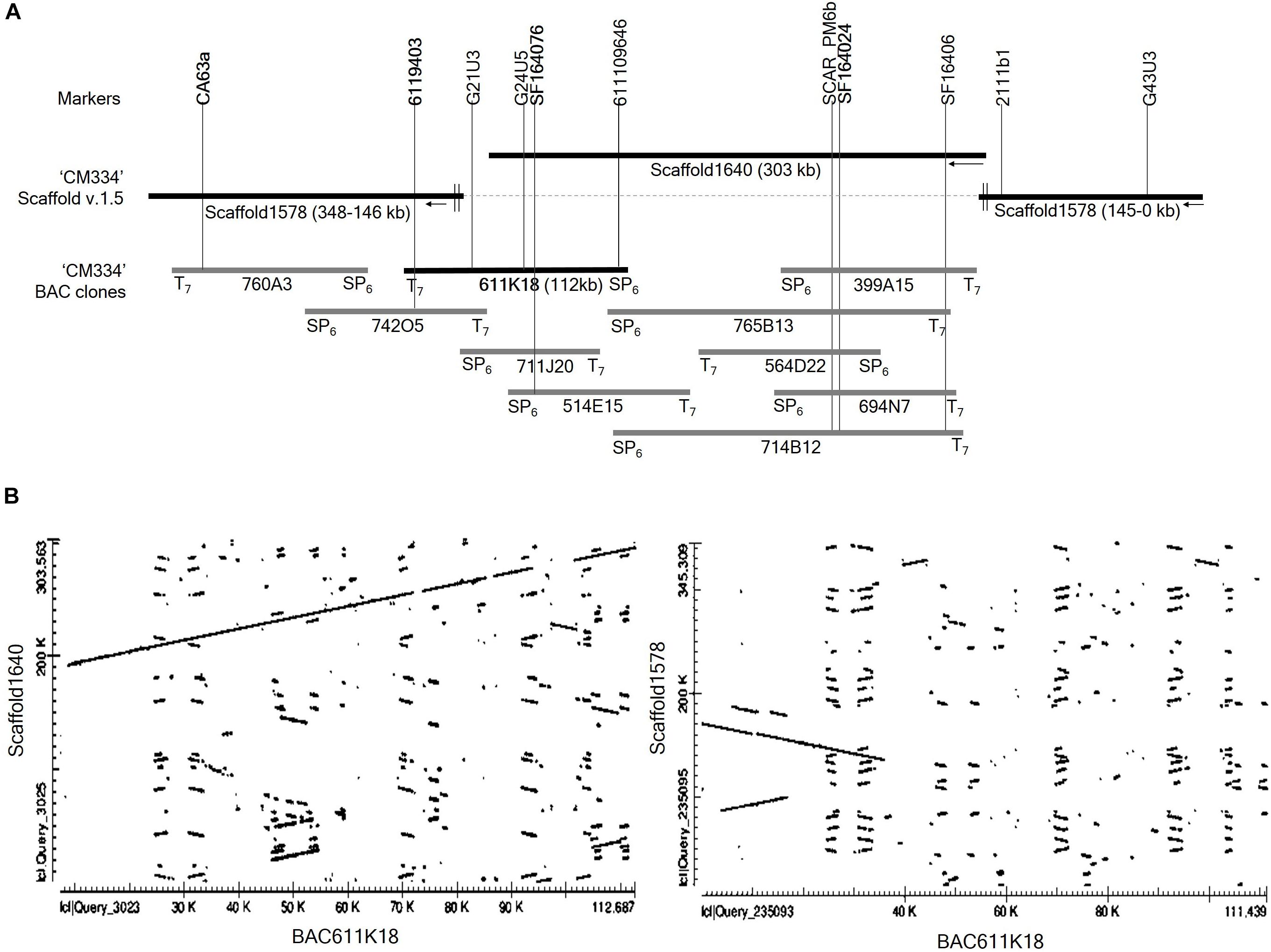
Figure 3. BAC library screening and alignment analysis. (A) Four markers (CA63a, 6119403, SF164076, and SF164024) derived from scaffold 1578, scaffold 1640 and BAC611K18 were used to screen the Capsicum annuum “CM334” BAC library (Yoo et al., 2003). BAC screening detected ten positive clones which were aligned on scaffold 1640 and 1578. (B) Dot plot analysis of full-length sequence of the BAC clone 611K18 revealed that 611K18 overlapped and bridged the gap between scaffold 1640 and 1578.
Initial mapping of the Me7 locus in an F2 population consisting of 192 individuals, placed SCAR_PM6a and SCAR_PM6b 0 cM from the Me7 locus. We used 31 markers, including 28 newly developed ones, to narrow the target region and we increased the size of the mapping population to 714 plants (Figure 4A and Supplementary Tables S1, S2). This fine mapping analysis produced seven recombinant plants (Figure 4A; 38, 161, 175, 209, 300, 450, and 578); one recombinant for each of G21U3, G43U3, and G79U3, two recombinants for 6119403 and three recombinants for CA63a (Figure 4A). Three markers (CA63a, 6119403, and G21U3) were placed on the one side of the Me7 locus, whereas two markers (G43U3 and G79U3) were placed on the other side (Figure 4A). Nine markers including seven newly developed markers and two reference markers (SCAR_PM6a and SCAR_PM6b) co-segregated with the Me7 locus. The Me7 locus was thus delimited to a 0.28 cM region with an interval of approximately 394.7 kb between G21U3 and G43U3 covered by BAC clone 611K18 and scaffolds 1640 and 1578 (Figures 4A,B).
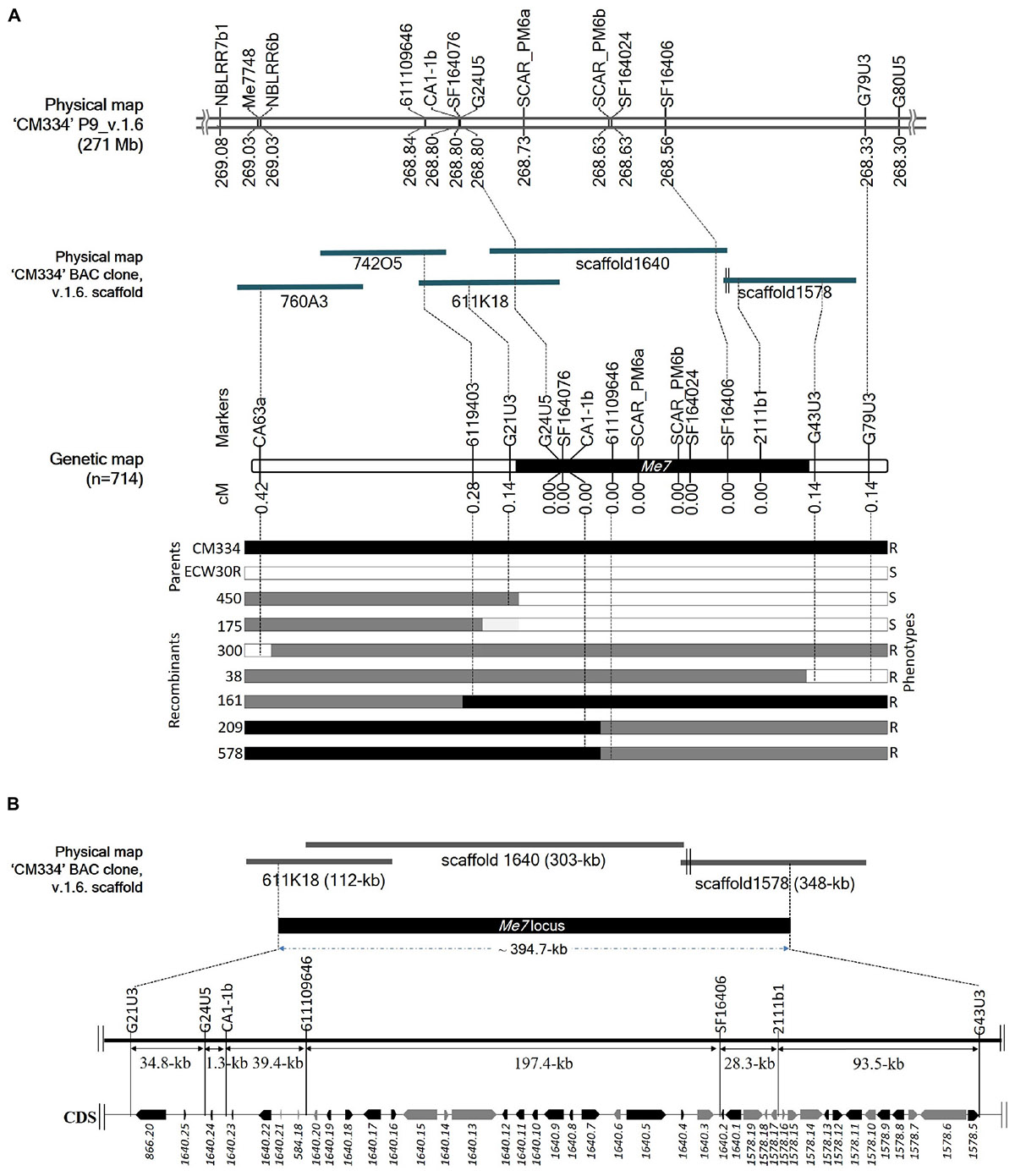
Figure 4. Fine mapping of the Me7 locus. (A) The physical position of known and newly developed linked markers to the Me7 locus on “CM334” chromosome P9, scaffold v.1.6 and BAC clones is shown. Fine mapping of the Me7 locus showing the genetic map constructed from 714 F2 population and the introgression patterns. (B) The Me7 locus was delimited to a ∼394.7 kb region (black area) between the G21U3 and G43U5 markers covered by the Capsicum annuum “CM334” BAC contig 611K18, scaffold 1640 and 1578. Gene prediction and BLAST search analysis against Annuum.v.2.0.CDS showed 42 genes in the target region. The genes colored in black are NBS-LRR type and genes in gray indicate non- NBS-LRR genes.
A total of 42 CDS were identified from the ∼394.7 kb target region (Figure 4B) covered by scaffold 1640, scaffold 1578, and BAC611K18. BLAST analysis of the corresponding predicted proteins revealed that 25 belonged to the NBS-LRR family and the other 17 were unknown hypothetical proteins (Table 2). The NBS-LRR candidates contained four different types of other conserved domains, including the potato virus X resistance protein (RX), ATPases associated with a variety of cellular activities (AAA), reverse transcriptase (RNA-dependent DNA polymerase) group 2 (RVT_2), or reverse transcriptase (RT) (Table 2). Among the 25 predicted NBS-LRR family genes, many of them are truncated, lack both Toll/interleukin-1 receptor (TIR) and coiled-coil (CC) motifs. Only six NBS-LRR family genes (1578.8, 1578.12, 1640.1, 1640.7, 1640.17, and 1640.22) were found to be related to CC-NBS-LRR (CNL) type R gene class. Furthermore, these NBS-LRR proteins shared sequence similarity with putative late blight resistance proteins including R1A-3, R1A-4, R1A-10, R1B-8, R1B-14, R1B-16, and R1B17 (Table 2). We thus consider these tightly linked NBS-LRR class resistance (R) genes as strong candidates to underly the RKN resistance conferred by the Me7 locus.
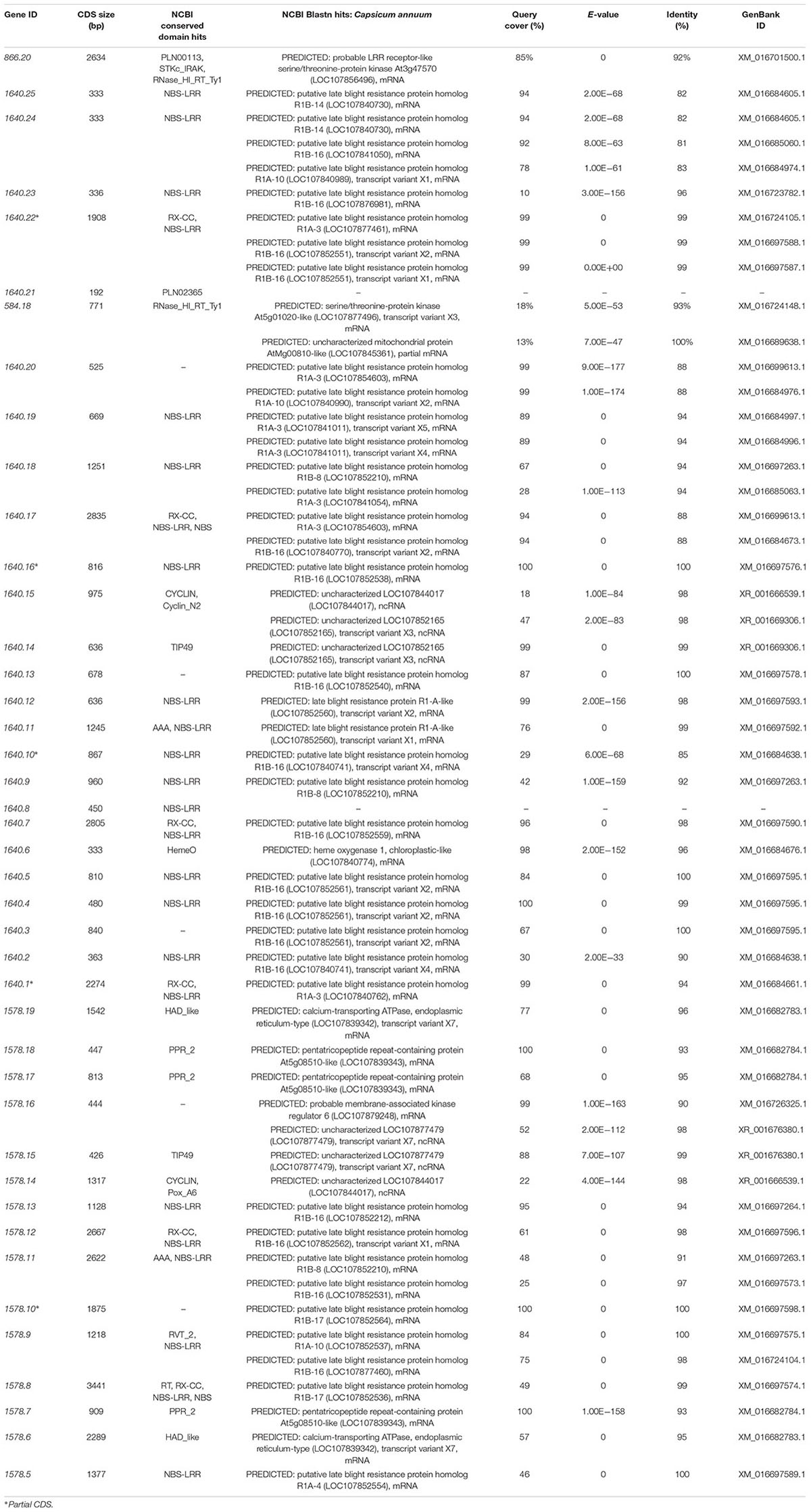
Table 2. Predicted genes from the ∼394.7 kb Me7 locus by BLAST alignment from the predicted CDS Annuum.v.2.0.CDS.
Understanding the molecular and genetic mechanisms of RKN resistance is of utmost importance for the development of RKN-resistant pepper lines. Although the RKN resistance gene Me7 from CM334 was previously mapped to a 3.8 cM genetic interval, its identity remained unknown. The primary aim of current study was to develop a high-resolution map of the Me7 locus and predict candidate genes. We were able to narrow down the Me7 locus to an approximately 394.7 kb region covered by the BAC clone 611K18 and unassigned scaffolds 1640 and 1578 (Kim et al., 2014) containing 42 predicted genes. Among these, 25 genes were predicted to belong to the NBS-LRR family of disease resistance genes. The Me1 gene (CA09g16830), which is a homolog of the resistance protein R1A-3 (Wang et al., 2018) belong to the CNL type R gene was found to be located approximately 3.3 and 1.1 Mb from the Me7 locus based on the CM334 v.1.6 (Kim et al., 2017) and UCDv1.0 (Hulse-Kemp et al., 2018) reference genomes, respectively (Supplementary Figure S4).
During phenotype screening, we found galls and egg masses of M. incognita in the root system of the resistant CM334 variety, in contrast to the previous studies of Pegard et al. (2005), who did not observe egg masses at 6 weeks after inoculation. In the present study, inoculations were done with 1000 J2 instead of 300 to 600 J2 used in the previous works (Djian-Caporalino et al., 1999, 2001, 2007; Pegard et al., 2005), which may explain the development of egg masses. Similarly, occasional egg masses were also reported to be produced on HDA149 pepper line, carrying Me3 (an allele of Me7) after inoculation with >1,000 J2 or ≥3,000 eggs (Castagnone-Sereno et al., 1996; Thies and Ariss, 2009). Thus, evidence from this study and previous research suggest that high inoculum densities (J2 ≥ 1,000 or eggs ≥3,000), has potential for resistant breaking in populations carrying Me (s) genes, such as Me3 and Me7 (Castagnone-Sereno et al., 1996; Thies and Ariss, 2009; Djian-Caporalino et al., 2011).
The ability of the nematode to overcome the plants early HR is highly correlates with RKN virulence levels. For instance, in Me3-resistant PM687 and HDA149 peppers formation of giant cells are no longer be prevented if cell necrosis in the outer barriers (epidermis and root cortex) has been overcome (Castagnone-Sereno et al., 1996). By contrast, in Me1 carrying peppers, PM217 and HDA330 defense reactions could takes place even later infection stages, at near or inside the vascular tissues and thus blocks nematode development (Castagnone-Sereno et al., 1996). In the present study, CM334 root tissue cross-sections suggested that it has at least two kinds of resistance to Meloidogyne spp.: firstly, resistance that supresses penetration by J2 and secondly, resistance that blocks development after penetration (Pegard et al., 2005). Furthermore, in our histological study, we also observed inhibition of feeding site enlargement and minimal vascular tissue damage in CM334 resistant line, suggesting an extra layer of resistance in CM334 that suppressed feeding site enlargement.
To fine-map the Me7 locus in pepper, we utilized information from previous mapping studies (Fazari et al., 2012; Celik et al., 2016) in combination with comparative mapping results (Supplementary Figure S4) from different versions of pepper genome databases (Kim et al., 2014, 2017; Hulse-Kemp et al., 2018). However, gaps in the genome sequence data and assembly errors due to multiple homologs with high nucleotide similarity hindered marker development and subsequent mapping of the Me7 locus. BAC library screening has previously been integrated to assist marker development in complicated repetitive genomic regions or unreliable genome sequence assemblies (Jo et al., 2016). Here, by screening a BAC library, we were able to fill gaps between scaffold sequences (1640 and 1578) with a bridge from the BAC611K18 clone, which aided the development of tightly linked Me7 markers. Furthermore, the release of the latest pepper genome update (version v.1.6, Kim et al., 2017) also partially allowed the integration of clone 611K18 and unassigned scaffolds 1640 and 1677 into chromosome P9.
A number of genes conferring resistance to parasitic plant nematodes encode NBS-LRR domains. Notably, the cyst nematode-resistance gene Hero from tomato (Ernst et al., 2002), Gro1-4 (Williamson and Kumar, 2006) and Gpa2 (van der Voort et al., 1999; Sacco et al., 2009) from potato and the RKN-resistance genes, Mi-1, 2, and 9 from tomato (Milligan et al., 1998; Seah et al., 2004; Jablonska et al., 2007), pepper RKN-resistance genes, CaMi which is homologous to Mi1.2 (Chen et al., 2007) and Me1, the homolog of putative late blight resistance protein R1A-3 gene (Wang et al., 2018) and Ma from Prunus spp. (Claverie et al., 2011) are reported to encode NBS-LRR proteins (Milligan et al., 1998; van der Vossen et al., 2000; Paal et al., 2004; Claverie et al., 2011). Similarly, we identified a cluster of 25 genes that were predicted to belong to the NBS-LRR R gene family, and which are strong candidates to be the Me7 gene. Several plant NBS-LRR class R genes cluster at specific genomic locations due to tandem and/or segmental duplications in the course of evolution (Hulbert et al., 2001; Leister, 2004; Huang et al., 2005; McDowell and Simon, 2006). For instance, dominant R genes such as Ph-3 from tomato, R1 from potato, and RpsUN1, RpsUN2, and Rpg1-b from soybean (Ballvora et al., 2002; Ashfield et al., 2003; Zhang et al., 2014; Li et al., 2016) belong to NBS-LRR domain-encoding complex R gene clusters. R gene clusters could be sources of novel R genes, as such clusters enhance the possibility of structural and copy number variation arising through various molecular mechanisms (Michelmore and Meyers, 1998; Torii, 2004; Nagy and Bennetzen, 2008). The RKN resistance-related R gene clusters identified in this study could be associated with RKN resistance in CM334. However, further studies are required to reveal the specific NBS-LRR gene encoding the Me7 gene.
Cloning of the R genes would be a key step toward unraveling the molecular mechanisms involved in disease resistance. However, the complexity of a large genome size as well as assembly errors at the Me7 locus hinders identification of the Me7 gene (Jupe et al., 2012). Recent developments such as resistance gene enrichment sequencing (RenSeq) and Single-Molecule Real Time (SMART) RenSeq techniques hold great promise for yielding higher sequencing read depth for individual genes, and aid in accurate identification of sequence variations in plants with large complex genomes (Jupe et al., 2012, 2013; Witek et al., 2016). RenSeq allows genome complexity reduction and enrichment of NBS-LRR type plant disease resistance genes in fully or partially assembled genomes (Jupe et al., 2012, 2013; Witek et al., 2016) and has been utilized for reannotation and identification of NBS-LRR R genes of Solanaceae plants such as tomato, potato, and tobacco. For instance, in potato, the number of NB-LRR identified increased from 438 to 755 through the RenSeq approach (Jupe et al., 2013). In pepper, studies to accurately determine the pepper R genes are now underway (we are currently undertaking RenSeq studies), which will further aid in the accurate prediction and functional characterization of the NB-LRR R genes identified in the present study.
In conclusion, we successfully delimited the Me7 locus to a genomic region of approximately 394.7 kb. We developed a total of 28 markers linked to the Me7 locus and identified nine markers co-segregating with RKN resistance. Notably, the newly delimited Me7 target region included a cluster of 25 NBS-LRR class candidate R genes. The tightly linked Me7 markers will facilitate MAS in pepper breeding programs as well as assisting with mapping of other nematode-resistant genes in C. annuum.
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
AC led the design of this study, including marker development, genetic and physical mapping, and BAC library screening. AC and G-JC developed mapping populations. J-WH and MS conducted DNA extraction. J-HL and AS were involved in the bioinformatics analysis. Y-HK provided M. incognita race 1. AC, J-KK, EK, and YS conducted nematode extraction and histological studies. AC and JV analyzed histological images and drafted and revised the manuscript. B-CK participated in the conception of this study, in discussions, and in the revision of the manuscript. All the authors have read and approved the final version of this manuscript.
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Agriculture, Food and Rural Affairs Research Center Support Program (Vegetable Breeding Research Center, 710011-03), funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) and Rural Affairs and Golden Seed Project (213006-05-3-SB540), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA), and Korea Forest Service (KFS), Republic of Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00886/full#supplementary-material
FIGURE S1 | Example of HRM analysis of the Me7 locus-linked markers. The analysis curves included HRM curves and melt curves. Blue, resistant (homozygous); red, susceptible; and green, resistant (heterozygous).
FIGURE S2 | Restriction patterns of the CAPS marker, 18660. MspI digested PCR product from susceptible and resistant lines show a clear bands of 560 and 808 bp, respectively. Heterozygous resistant lines show both 560 and 808 bp bands. L indicate 1 kb DNA molecular size marker.
FIGURE S3 | The RKN egg masses produced in CM334 and ECW30R roots, imaged at 45 dai. Eggs are exuded egg mass on the outside of the female RKN on roots of CM334 (A,B) and ECW30R (C,D). The egg mass from both CM334 (B) and ECW30R (D). Different developmental stages of RKN, from egg to hatching of J2 are indicated (E). e, egg; em, egg mass; r, root; rg, root gall; J0-1, egg developing undergo to J1. Bar = 0.5 mm.
FIGURE S4 | Comparative analysis of the Me7 linked markers on P9 from three genome references. The physical position of the Me7 linked markers on “CM334” P9 version v.1.55, v.1.6 and UCD10X v1.0 (Kim et al., 2014, 2017; Hulse-Kemp et al., 2018) is shown. BLAST alignment was performed using CLC Main Workbench 8.1 (QIAGEN, Aarhus, Denmark), where map reads with ≥98% nucleotide similarity are shown. Markers that co-segregated with the Me7 locus in the screening with 714 F2 individuals are represented in green.
TABLE S1 | Me loci and N linked markers from previous studies.
TABLE S2 | Newly developed markers used in the Me7 locus mapping.
Ashfield, T., Bocian, A., Held, D., Henk, A. D., Marek, L. F., Danesh, D., et al. (2003). Genetic and physical localization of the soybean Rpg1-b disease resistance gene reveals a complex locus containing several tightly linked families of NBS-LRR genes. Mol. Plant Microb. Interact. 16, 817–826. doi: 10.1094/MPMI.2003.16.9.817
Baker, K. R. (1985). “Nematode extractions and bioassays,” in An Advanced Treatis on Meloidogyne. Methodology, Vol. II, eds K. R. Baker, C. C. Carter, and J. N. Sasser (Raleigh, NC: North Carolina State University), 19–35.
Ballvora, A., Ercolano, M. R., Weiß, J., Meksem, K., Bormann, C. A., Oberhagemann, P., et al. (2002). The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 30, 361–371. doi: 10.1046/j.1365-313X.2001.01292.x
Bernard, G. C., Marceline, E. M., and Bonsi, C. (2017). “The impact of plant-parasitic nematodes on agriculture and methods of control,” in Nematology - Concepts, Diagnosis and Control, eds M. M. Shah and M. Mahamood (London: Intech).
Bucki, P., Paran, I., Ozalvo, R., Iberkleid, I., Ganot, L., and Miyara, S. B. (2017). Pathogenic variability of Meloidogyne incognita populations occurring in pepper-production greenhouses in Israel toward Me1, Me3 and N pepper resistance genes. Plant Dis. 101, 1391–1401. doi: 10.1094/PDIS-11-16-1667-RE
Bybd, D. W. Jr., Kirkpatrick, T., and Barker, K. R. (1983). An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 15, 142–143.
Castagnone-Sereno, P., Bongiovanni, M., Palloix, A., and Dalmasso, A. (1996). Selection for Meloidogyne incognita virulence against resistance genes from tomato and pepper and specificity of the virulence/resistance determinants. Eur. J. Plant Pathol. 102, 585–590. doi: 10.1007/BF01877026
Castagnone-Sereno, P., Semblat, J. P., and Castagnone, C. (2009). Modular architecture and evolution of the map-1 gene family in the root-knot nematode. Mol. Genet. Genomics 282:547. doi: 10.1007/s00438-009-0487-x
Celik, I., Sogut, M. A., Doganlar, S., and Frary, A. (2016). Physical mapping of NBS-coding resistance genes to the Me-gene cluster on chromosome P9 reveals markers tightly linked to the N gene for root-knot nematode resistance in pepper. Mol. Breed. 36:137. doi: 10.1007/s11032-016-0560-1
Chen, R., Li, H., Zhang, L., Zhang, J., Xiao, J., and Ye, Z. (2007). CaMi, a root-knot nematode resistance gene from hot pepper (Capsium annuum L.) confers nematode resistance in tomato. Plant Cell Rep. 26, 895–905. doi: 10.1007/s00299-007-0304-0
Claverie, M., Dirlewanger, E., Bosselut, N., van Ghelder, C., Voisin, R., Kleinhentz, M., et al. (2011). The Ma gene for complete-spectrum resistance to Meloidogyne species in Prunus is a TNL with a huge repeated C-terminal post-LRR region. Plant Physiol. 156, 779–792. doi: 10.1104/pp.111.176230
Coyne, D., and Ross, J. L. (2014). Protocol for Nematode Resistance Screening: Root Knot Nematodes, Meloidogyne spp. Ibadan: IITA.
de Carvalho, L. M., Benda, N. D., Vaughan, M. M., Cabrera, A. R., Hung, K., Cox, T., et al. (2015). Mi-1-mediated nematode resistance in tomatoes is broken by short-term heat stress but recovers over time. J. Nematol. 47:133.
de Givry, S., Bouchez, M., Chabrier, P., Milan, D., and Schiex, T. (2005). CARTHA GENE: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 21, 1703–1704. doi: 10.1093/bioinformatics/bti222
Djian-Caporalino, C., Fazari, A., Arguel, M. J., Vernie, T., VandeCasteele, C., Faure, I., et al. (2007). Root-knot nematode (Meloidogyne spp.) Me resistance genes in pepper (Capsicum annuum L.) are clustered on the P9 chromosome. Theor. Appl. Genet. 114, 473–486. doi: 10.1007/s00122-006-0447-3
Djian-Caporalino, C., Molinari, S., Palloix, A., Ciancio, A., Fazari, A., Marteu, N., et al. (2011). The reproductive potential of the root-knot nematode Meloidogyne incognita is affected by selection for virulence against major resistance genes from tomato and pepper. Eur. J. Plant Pathol. 131, 431–440. doi: 10.1007/s10658-011-9820-4
Djian-Caporalino, C., Pijarowski, L., Fazari, A., Samson, M., Gaveau, L., O’byrne, C., et al. (2001). High-resolution genetic mapping of the pepper (Capsicum annuum L.) resistance loci Me3 and Me4 conferring heat-stable resistance to root-knot nematodes (Meloidogyne spp.). Theor. Appl. Genet. 103, 592–600. doi: 10.1007/PL00002914
Djian-Caporalino, C., Pijarowski, L., Januel, A., Lefebvre, V., Daubèze, A., Palloix, A., et al. (1999). Spectrum of resistance to root-knot nematodes and inheritance of heat-stable resistance in in pepper (Capsicum annuum L.). Theor. Appl. Genet. 99, 496–502. doi: 10.1007/s001220051262
Dogimont, C., Palloix, A., Daubze, A. M., Marchoux, G., Selassie, K. G., and Pochard, E. (1996). Genetic analysis of broad spectrum resistance to potyviruses using doubled haploid lines of pepper (Capsicum annuum L.). Euphytica 88, 231–239. doi: 10.1007/BF00023895
Eisenback, J. D., and Triantaphyllou, H. H. (1991). “Root-knot nematodes: Meloidogyne species and races,” in Manual of Agricultural Nematology, ed. W. R. Nickle (New York, NY: Marcel Dekker, Inc), 191–274.
Ernst, K., Kumar, A., Kriseleit, D., Kloos, D. U., Phillips, M. S., and Ganal, M. W. (2002). The broad-spectrum potato cyst nematode resistance gene (Hero) from tomato is the only member of a large gene family of NBS-LRR genes with an unusual amino acid repeat in the LRR region. Plant J. 31, 127–136. doi: 10.1046/j.1365-313X.2002.01341.x
Fazari, A., Palloix, A., Wang, L., Yan, H. M., Sage-Palloix, A. M., Zhang, B. X., et al. (2012). The root-knot nematode resistance N-gene co-localizes in the Me-genes cluster on the pepper (Capsicum annuum L.) P9 chromosome. Plant Breed. 131, 665–673. doi: 10.1111/j.1439-0523.2012.01994.x
Fery, R. L., Dukes, P. D., and Thies, J. A. (1998). ‘Caroline Wonder’ and ‘Charleston Belle’: New Southern root-knot nematode resistant bell peppers. HortScience 33, 900–902. doi: 10.21273/hortsci.33.5.900
Gisbert, C., Trujillo-Moya, C., Sánchez-Torres, P., Sifres, A., Sánchez-Castro, E., and Nuez, F. (2013). Resistance of pepper germplasm to Meloidogyne incognita. Ann. Appl. Biol. 162, 110–118. doi: 10.1111/aab.12006
Gleason, C. A., Liu, Q. L., and Williamson, V. M. (2008). Silencing a candidate nematode effector gene corresponding to the tomato resistance gene Mi-1 leads to acquisition of virulence. Mol. Plant Microbe Interact. 21, 576–585. doi: 10.1094/MPMI-21-5-0576
Guo, X., Yang, X. H., Yang, Y., Mao, Z. C., Liu, F., Ma, W. Q., et al. (2016). Bacterial artificial chromosome library construction of root-knot nematode resistant pepper genotype HDA149 and identification of clones linked to Me3 resistant locus. J. Integr. Agric. 16, 57–64. doi: 10.1016/S2095-3119(16)61446-6
Hare, W. W. (1957). Inheritance of resistance to root-knot nematodes in pepper. Phytopathology 47, 455–459.
Huang, S., van der Vossen, E. A., Kuang, H., Vleeshouwers, V. G., Zhang, N., Borm, T. J., et al. (2005). Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 42, 251–261. doi: 10.1111/j.1365-313X.2005.02365.x
Hulbert, S. H., Webb, C. A., Smith, S. M., and Sun, Q. (2001). Resistance gene complexes: evolution and utilization. Annu. Rev. Phytopathol. 39, 285–312. doi: 10.1146/annurev.phyto.39.1.285
Hulse-Kemp, A. M., Maheshwari, S., Stoffel, K., Hill, T. A., Jaffe, D., Williams, S. R., et al. (2018). Reference quality assembly of the 3.5-Gb genome of Capsicum annuum from a single linked-read library. Hortic. Res. 5:4. doi: 10.1038/s41438-017-0011-0
Hwang, C. F., Bhakta, A. V., Truesdell, G. M., Pudlo, W. M., and Williamson, V. M. (2000). Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 12, 1319–1329. doi: 10.1105/tpc.12.8.1319
Jablonska, B., Ammiraju, J. S. S., Bhattarai, K. K., Mantelin, S., de Ilarduya, O. M., Roberts, P. A., et al. (2007). The Mi-9 gene from Solanum arcanum conferring heat-stable resistance to root-knot nematodes is a homolog of Mi-1. Plant Physiol. 143, 1044–1054. doi: 10.1104/pp.106.089615
Jo, Y. D., Ha, Y., Lee, J. H., Park, M., Bergsma, A. C., Choi, H. I., et al. (2016). Fine mapping of restorer-of-fertility in pepper (Capsicum annuum L.) identified a candidate gene encoding a pentatricopeptide repeat (PPR)-containing protein. Theor. Appl. Genet. 129, 2003–2017. doi: 10.1007/s00122-016-2755-6
Jones, J. T., Haegeman, A., Danchin, E. G. J., Gaur, H. S., Helder, J., Jones, M. G. K., et al. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14, 946–961. doi: 10.1111/mpp.12057
Jupe, F., Pritchard, L., Etherington, G. J., MacKenzie, K., Cock, P. J. A., Wright, F., et al. (2012). Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genomics 13:75. doi: 10.1186/1471-2164-13-75
Jupe, F., Witek, K., Verweij, W., Sliwka, J., Pritchard, L., Etherington, G. J., et al. (2013). Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 76, 530–544. doi: 10.1111/tpj.12307
Kang, W. H., Hoang, N. H., Yang, H. B., Kwon, J. K., Jo, S. H., Seo, J. K., et al. (2010). Molecular mapping and characterization of a single dominant gene controlling CMV resistance in peppers (Capsicum annuum L.). Theor. Appl. Genet. 120, 1587–1596. doi: 10.1007/s00122-010-1278-9
Kim, S., Park, J., Yeom, S. I., Kim, Y. M., Seo, E., Kim, K. T., et al. (2017). New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 18:210. doi: 10.1186/s13059-017-1341-9
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Leister, D. (2004). Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 20, 116–122. doi: 10.1016/j.tig.2004.01.007
Li, L., Lin, F., Wang, W., Ping, J., Fitzgerald, J. C., Zhao, M., et al. (2016). Fine mapping and candidate gene analysis of two loci conferring resistance to Phytophthora sojae in soybean. Theor. Appl. Genet. 129, 2379–2386. doi: 10.1007/s00122-016-2777-0
Liu, L., Venkatesh, J., Jo, Y. D., Koeda, S., Hosokawa, M., Kang, J. H., et al. (2016). Fine mapping and identification of candidate genes for the sy-2 locus in a temperature-sensitive chili pepper (Capsicum chinense). Theor. Appl. Genet. 129, 1541–1556. doi: 10.1007/s00122-016-2723-1
Lozano-Torres, J. L., Wilbers, R. H. P., Gawronski, P., Boshoven, J. C., Finkers-Tomczak, A., Cordewener, J. H. G., et al. (2012). Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. U.S.A. 109, 10119–10124. doi: 10.1073/pnas.1202867109
Mao, Z., Zhu, P., Liu, F., Huang, Y., Ling, J., Chen, G., et al. (2015). Cloning and functional analyses of pepper CaRKNR involved in Meloidogyne incognita resistance. Euphytica 205, 903–913. doi: 10.1007/s10681-015-1438-8
McDowell, J. M., and Simon, S. A. (2006). Recent insights into R gene evolution. Mol. Plant Pathol. 7, 437–448. doi: 10.1111/j.1364-3703.2006.00342.x
Michelmore, R. W., and Meyers, B. C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130. doi: 10.1101/gr.8.11.1113
Milligan, S. B., Bodeau, J., Yaghoobi, J., Kaloshian, I., Zabel, P., and Williamson, V. M. (1998). The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10, 1307–1319. doi: 10.1105/tpc.10.8.1307
Nagy, E. D., and Bennetzen, J. L. (2008). Pathogen corruption and site-directed recombination at a plant disease resistance gene cluster. Genome Res. 18, 1918–1923. doi: 10.1101/gr.078766.108
Noling, J. W. (2014). Nematode Management in Tomatoes, Peppers, and Eggplant. IFAS Extension, University of Flolida. Available at: https://edis.ifas.ufl.edu/ng032
Nombela, G., Williamson, V. M., and Muñiz, M. (2003). The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol. Plant Microb. Interact. 16, 645–649. doi: 10.1094/MPMI.2003.16.7.645
Paal, J., Henselewski, H., Muth, J., Meksem, K., Menéndez, C. M., Salamini, F., et al. (2004). Molecular cloning of the potato Gro1-4 gene conferring resistance to pathotype Ro1 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant J. 38, 285–297. doi: 10.1111/j.1365-313X.2004.02047.x
Palloix, A., Daubèze, A. M., Phaly, T., and Pochard, E. (1990). Breeding transgressive lines of pepper for resistance to Phytophthora capsici in a recurrent selection system. Euphytica 51, 141–150.
Pegard, A., Brizzard, G., Fazari, A., Soucaze, O., Abad, P., and Djian-Caporalino, C. (2005). Histological characterization of resistance to different root-knot nematode species related to phenolics accumulation in Capsicum annuum. Phytopathology 95, 158–165. doi: 10.1094/PHYTO-95-0158
Sacco, M. A., Koropacka, K., Grenier, E., Jaubert, M. J., Blanchard, A., Goverse, A., et al. (2009). The cyst nematode SPRYSEC protein RBP-1 elicits Gpa2- and RanGAP2-dependent plant cell death. PLoS Pathog. 5:e1000564. doi: 10.1371/journal.ppat.1000564
Sanchez-Puerta, M. V., and Masuelli, R. W. (2011). Evolution of nematode-resistant Mi-1 gene homologs in three species of Solanum. Mol. Genet. Genomics 285, 207–218. doi: 10.1007/s00438-010-0596-6
Seah, S., Yaghoobi, J., Rossi, M., Gleason, C. A., and Williamson, V. M. (2004). The nematode-resistance gene, Mi-1, is associated with an inverted chromosomal segment in susceptible compared to resistant tomato. Theor. Appl. Genet. 108, 1635–1642. doi: 10.1007/s00122-004-1594-z
Seo, Y., Park, J., Kim, Y. S., Park, Y., and Kim, Y. H. (2014). Screening and histopathological characterization of Korean carrot lines for resistance to the root-knot nematode Meloidogyne incognita. Plant Pathol. J. 30:75. doi: 10.5423/PPJ.OA.08.2013.0082
Taylor, A. L., and Sasser, J. N. (1978). Biology, Identification and Control of Root-Knot Nematodes (Meloidogyne Species). Raleigh: North Carolina State University Graphics.
Thies, J. A., and Ariss, J. J. (2009). Comparison between the N and Me3 genes conferring resistance to the root-knot nematode (Meloidogyne incognita) in genetically different pepper lines (Capsicum annuum). Eur. J. Plant Pathol. 125:545. doi: 10.1007/s10658-009-9502-7
Torii, K. U. (2004). Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int. Rev. Cytol. 234, 1–46. doi: 10.1016/S0074-7696(04)34001-5
Uncu, A. T., Celik, I., Devran, Z., Ozkaynak, E., Frary, A., Frary, A., et al. (2015). Development of a SNP-based CAPS assay for the Me1 gene conferring resistance to root knot nematode in pepper. Euphytica 206, 393–399. doi: 10.1007/s10681-015-1489-x
van der Voort, J. R., Kanyuka, K., van der Vossen, E., Bendahmane, A., Mooijman, P., Klein-Lankhorst, R., et al. (1999). Tight physical linkage of the nematode resistance gene Gpa2 and the virus resistance gene Rx on a single segment introgressed from the wild species Solanum tuberosum subsp. andigena CPC 1673 into cultivated potato. Mol. Plant Microbe Interact. 12, 197–206. doi: 10.1094/MPMI.1999.12.3.197
van der Vossen, E. A., Gros, J., Sikkema, A., Muskens, M., Wouters, D., Wolters, P., et al. (2005). The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 44, 208–222. doi: 10.1111/j.1365-313X.2005.02527.x
van der Vossen, E. A. G., van der Voort, J., Rouppe, N. A. M., Kanyuka, K., Bendahmane, A., Sandbrink, H., et al. (2000). Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: a virus and a nematode. Plant J. 23, 567–576. doi: 10.1046/j.1365-313X.2000.00814.x
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, D., and Bosland, P. W. (2006). The genes of Capsicum. HortScience 41, 1169–1187. doi: 10.21273/hortsci.41.5.1169
Wang, L. H., Gu, X. H., Hua, M. Y., Mao, S. L., Zhang, Z. H., Peng, D. L., et al. (2009). A SCAR marker linked to the N gene for resistance to root knot nematodes (Meloidogyne spp.) in pepper (Capsicum annuum L.). Sci. Hortic. 122, 318–322. doi: 10.1016/j.scienta.2009.04.011
Wang, X., Fazari, A., Cao, Y., Zhang, Z., Palloix, A., Mao, S., et al. (2018). Fine mapping of the root-knot nematode resistance gene Me1 in pepper (Capsicum annuum L.) and development of markers tightly linked to Me1. Mol. Breed. 38:39. doi: 10.1007/s11032-018-0793-2
Williamson, V. M., and Kumar, A. (2006). Nematode resistance in plants: the battle underground. Trends Genet. 22, 396–403. doi: 10.1016/j.tig.2006.05.003
Witek, K., Jupe, F., Witek, A. I., Baker, D., Clark, M. D., and Jones, J. D. (2016). Accelerated cloning of a potato late blight–resistance gene using RenSeq and SMRT sequencing. Nat. Biotechnol. 34:656. doi: 10.1038/nbt.3540
Wu, X., Zhu, X., Wang, Y., Liu, X., Chen, L., and Duan, Y. (2018). The cold tolerance of the northern root-knot nematode, Meloidogyne hapla. PLoS One 13:e0190531. doi: 10.1371/journal.pone.0190531
Yoo, E. Y., Kim, S., Kim, Y. H., Lee, C. J., and Kim, B. D. (2003). Construction of a deep coverage BAC library from Capsicum annuum, “CM334”. Theor. Appl. Genet. 107, 540–543. doi: 10.1007/s00122-003-1279-z
Keywords: co-segregating markers, dominant locus, fine mapping, Me7, NBS-LRR, resistance gene, root-knot nematode
Citation: Changkwian A, Venkatesh J, Lee J-H, Han J-W, Kwon J-K, Siddique MI, Solomon AM, Choi G-J, Kim E, Seo Y, Kim Y-H and Kang B-C (2019) Physical Localization of the Root-Knot Nematode (Meloidogyne incognita) Resistance Locus Me7 in Pepper (Capsicum annuum). Front. Plant Sci. 10:886. doi: 10.3389/fpls.2019.00886
Received: 02 April 2019; Accepted: 21 June 2019;
Published: 09 July 2019.
Edited by:
Bruno Favery, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Caroline Djian-Caporalino, Institut National de la Recherche Agronomique (INRA), FranceCopyright © 2019 Changkwian, Venkatesh, Lee, Han, Kwon, Siddique, Solomon, Choi, Kim, Seo, Kim and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byoung-Cheorl Kang, Yms1NEBzbnUuYWMua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.