
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 27 June 2019
Sec. Plant Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00848
 Shengjie Liu1,2†
Shengjie Liu1,2† Hua Fu3†
Hua Fu3† Jieming Jiang2
Jieming Jiang2 Zhongjian Chen1
Zhongjian Chen1 Jiadong Gao1
Jiadong Gao1 Haoran Shu2
Haoran Shu2 Sheng Zhang4
Sheng Zhang4 Chengwei Yang2*
Chengwei Yang2* Jun Liu1*
Jun Liu1*Glutaredoxins (Grxs) are a ubiquitous group of oxidoreductase enzymes that are important in plant growth and development; however, the functions of rice Grxs have not been fully elucidated. In this paper, we showed that one of the Grxs, encoded by OsGrxC2.2, exhibited Grx activity. Furthermore, we demonstrated that OsGrxC2.2 was able to regulate embryo development during embryogenesis. Transgenic rice lines overexpressing OsGrxC2.2 unexpectedly exhibited degenerate embryos as well as embryoless seeds. Our data indicated that the embryonic abnormalities occurred at an early stage during embryogenesis. We found that the expression of several endodermal layer marker genes for embryo development, such as OSH1 (apical region marker), OsSCR (L2 ground tissue marker), and OsPNH1 (L3 vascular tissue marker), were significantly decreased in the OsGrxC2.2-overexpressed transgenic rice lines. In contrast, the transcript levels of the majority of protodermal layer markers, including HAZ1, ROC2, ROC3, and RAmy1A, and the shoot apical meristem marker HB, showed little change between the wild-type (WT) and OsGrxC2.2-overexpressing embryos. Surprisingly, the seed weight of the overexpressed transgenic rice was remarkably increased in comparison to that of the WT. These results indicate that the overexpression of OsGrxC2.2 interferes with the normal embryogenesis of rice embryos and leads to increased grain weight. To the best of our knowledge, this is the first report that OsGrxC2.2 is a rice embryo development-associated gene.
Rice (Oryza sativa L.) is the predominant staple food for more than half of the global human population. Although only accounting for 2–3% of the seed weight, the rice embryo is crucial for plant growth. Advancing our understanding of the molecular mechanisms of rice embryogenesis is important and could be used to improve grain weight and seed quality. The genetic control of embryogenesis has been studied via analyses of embryonic mutants and marker gene expression in rice (Hong et al., 1995; Kamiya et al., 2003; Yi et al., 2016). Rice mutants with an embryonic lethal phenotype have been identified and characterized, and several genes have been used as molecular markers for rice embryogenesis studies (Hong et al., 1995; Di Laurenzio et al., 1996; Kamiya et al., 2003; Ito et al., 2004). Despite these advancements in phenotypic identification, the molecular mechanisms of rice embryogenesis remain largely unknown.
Glutaredoxins (Grxs) are a ubiquitous group of oxidoreductases in both prokaryotes and eukaryotes (Holmgren, 1989; Holmgren and Aslund, 1995; Lillig et al., 2008). They are small glutathione-dependent oxidoreductases that belong to the thioredoxin (TRX) superfamily (El-Kereamy et al., 2015). Their predominant function is believed to be the reduction of disulfide bridges, and some members have recently also been shown to interact with iron-sulfur clusters (Riondet et al., 2012). Grxs can be divided into three major classes based on the predicted amino acid sequences and arrangement of the cysteine residues in the active-site motifs: CPYC-type, CGFS-type, and CC-type. The CC-type has only been identified in land plants, whereas the other two types have been documented in a range of organisms from prokaryotes to eukaryotes (Rouhier et al., 2004, 2006; Garg et al., 2010).
As a large gene family, the Grx family contains members with diverse roles. One of the most well-documented functions of Grxs in plants is their involvement in the oxidative stress response. They are implicated in various processes, such as in directly reducing peroxides or dehydroascorbate (DHA), reducing peroxiredoxins (Prxs), and also protecting the thiol groups on other enzymes via glutathionylation/deglutathionylation mechanisms (Rouhier et al., 2006). Both AtGRXcp (also termed AtGRXS14) and AtGRX4 (also termed AtGRXS15) play a pivotal role in protecting cells against oxidative stress (Cheng et al., 2006; Cheng, 2008). OsGRX8 in rice has also been implicated in the plant response to auxin, salinity, osmotic stress, and oxidative stress (Sharma et al., 2013).
Grxs can also regulate plant development by participating in DNA synthesis, signal transduction and stress responses, and [Fe–S] cluster aggregation (Rouhier et al., 2004, 2007, 2015). Recently, a few studies elucidated the roles of class I and class II GRXs in plant development. For instance, a CC-type glutaredoxin, OsGRX6, affects hormone and nitrogen status in rice plants, with its overexpression leading to a semi-dwarf phenotype (El-Kereamy et al., 2015). As for CPYC-type Grxs, AtGrxC1 and its homolog AtGrxC2 were found to be functionally redundant in dicotyledonous plants (Riondet et al., 2012). Knock-out mutants in grxc1 or grxc2 are aphenotypic, but the double mutant produces a lethal phenotype at an early stage after pollination, implying that GRXC1 and GRXC2 share redundant and vital functions (Riondet et al., 2012). In addition, the CGFS-type Grx AtGRXS17 (At4g04950) is critical in redox homeostasis and hormone perception where it mediates temperature-dependent postembryonic growth, with grxs17 mutant plants showing abnormal apical meristems, elongated leaves, and impaired flowering under long photoperiods or at high temperature (Cheng et al., 2011; Knuesting et al., 2015).
The involvement of class III GRXs in the development of floral organs largely originates from genetic studies performed in Arabidopsis thaliana and O. sativa. In A. thaliana, the roxy1 mutant exhibits a reduced number of petal primordia and abnormalities in petal morphogenesis (Xing et al., 2005), while the roxy1 roxy2 double mutant is sterile and does not produce pollen (Xing and Zachgo, 2008). AtROXY4 was suggested to participate in gibberellin signaling and floral organ development. Indeed, plants overexpressing AtROXY4 displayed undeveloped petals and stamens and male sterility due to non-dehiscent anthers (Hou et al., 2008). In rice, the mil1 mutant does not produce microspores in the anthers and is thus male-sterile, but female-fertile. The role of class III GRXs in the control of floral development notably relies on the redox regulation of TGA transcription factor activity, as shown primarily in Arabidopsis (Lee et al., 2006), and rice (Hong et al., 2012).
Although Grxs in rice have been known for decades, only limited information on their functions is available. Our previous study showed that OsGrx may protect embryo proteins from oxidation and thus might be correlated with seed storability (Gao et al., 2016). Another study demonstrated that OsGrxC2.2, a class I Grx isoform, is abundantly expressed in the aleurone layers in developing seeds and participates in oxidative stress defense in rice (Morita et al., 2015). We thus cloned OsGrxC2.2 with the CPFC-type active site, a typical dithiol isoform, to investigate the specific roles of OsGrxC2.2 in seed storage and protection from aging. We show that OsGrxC2.2 overexpression in rice impairs embryo development and leads to increased grain weight.
Wild-type rice Zhonghua11 (O. sativa ssp. japonica, WT) and transgenic lines seeds were cultivated in a growth chamber with a 28°C/26°C light/dark cycle of 14 h/10 h. The seedlings were transplanted under natural growing seasons into an experimental field of South China Normal University (Guangzhou, China).
The DNA fragments for fusing the promoter of OsGrxC2.2 to β-glucuronidase (GUS) were constructed using a polymerase chain reaction (PCR)-mediated fusion strategy. The ∼2 kb promoter sequence of OsGrxC2.2 was amplified using the primers listed in Supplementary Table S1. The PCR products were digested with SacI and SalI and inserted into the pCambia1301 vector to generate the OsGrxC2.2promoter:GUS. Fifteen independent transgenic lines were selected for GUS activity analysis. To generate OsGrxC2.2 overexpression lines, the full-length OsGrxC2.2 cDNA from the WT was inserted into the plant binary vector pCambia1390 using PCR-mediated gene fusion, as described above. Both constructs were transferred into the callus of the WT, using Agrobacterium tumefaciens strain EHA105. The resultant 12 independent rice lines were labeled OE1–OE12, respectively, and 4 of them (OE2, OE6, OE11, OE12) were used for the subsequent experiments.
For GUS staining, various tissues including four-leaf-stage root; tillering stage stem and leaf; heading stage flag leaf; a hull before flowering; flower; grain-filling-stage developing seeds (3–5, 10–12, and 15–17 days after flowering); and germinating seeds at 24 h, 36 h, 2 days, 3 days, 4 days, 5 days, and 6 days from the pOsGrxC2.2:GUS transgenic plants were incubated in a solution containing 50 mM sodium phosphate (NaPO4) buffer (pH 7.0), 5 mM potassium ferricyanide (K3Fe(CN)6), 5 mM potassium ferrocyanide (K4Fe(CN)6), 0.1% Triton X-100, 20% (v/v) methanol (MeOH), and 1 mM X-Gluc and stained at 37°C for 6 h and destained twice with 75% ethanol. They were then observed under a stereoscopic microscope SteREO Lumar V12 (Carl Zeiss, Heidenheim, Germany).
The TTC assay was performed as described by Wei et al. (2015), with some modifications. Hulled seeds were imbibed in distilled water for 12 h and then cut longitudinally into two identical halves. The cut seeds were incubated in freshly prepared 1× phosphate-buffered solution (PBS) containing 0.1% (w/v) TTC (Sigma-Aldrich, St. Louis, MO, United States) for 30 min at 37°C in a darkened incubator. Following incubation, the seeds were immediately rinsed with distilled water and washed twice with 75% (v/v) ethanol. The stained seeds were air-dried on filter paper and observed using a stereoscopic microscope.
Total RNA was isolated from various tissues of the WT plants, including the root, stem, leaf, flag leaf, tiller, and panicle, and WT developing seeds (3, 6, 9, 12, 15, 18, and 21 days after pollination), and WT embryo and endosperm of the dry seeds, and 6 days embryos after pollination of the WT and transgenic rice lines using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. Developing seeds were collected at 6 days after pollination (DAP), and the embryos were isolated under a microscope using a sterile blade and immediately placed in liquid nitrogen, following which they were stored at −80°C until analysis. The RNA samples were reverse-transcribed into first-strand cDNA using a PrimeScript RT Reagent Kit (Takara, Japan) and qRT-PCR reactions were carried out on a Bio-Rad Laboratories CFX ConnectTM Real-Time PCR Detection System with SYBR Premix Ex Taq (Takara, Japan). All experiments were conducted with at least three biological replicates. The qRT-PCR reactions were normalized based on the rice Actin1 gene (OsActin1), which was used as an internal control using the 2–ΔΔCt method (Livak and Schmittgen, 2001). The primers designed by Primer Premier 5.0 software for the analysis of transcript levels are listed in Supplementary Table S1.
To verify the subcellular localization of OsGrxC2.2, the yellow fluorescent protein (YFP) gene was fused in-frame to the C-terminal of OsGrxC2.2 into the pSAT6-EYFP vector to generate 35S:OsGrxC2.2-YFP, where the fusion gene was expressed under the control of the cauliflower mosaic virus 35S promoter.
The plasmid was transformed into rice protoplasts prepared from the leaf sheaths of 10-days-old seedlings. Rice protoplast transformation was performed according to previously reported methods (Yoo et al., 2007; Zhang et al., 2011). The YFP signal of the OsGrxC2.2-YFP fusion protein was visualized under a confocal microscope (LSM710, Carl Zeiss, Heidenheim, Germany).
To express the OsGrxC2.2 protein in prokaryotic cells, OsGrxC2.2 was cloned into the expression vector PET-28a to construct the OsGrxC2.2-PET-28a recombinant plasmid, which was transformed into the BL21 (DE3) Escherichia coli strain. Expression and purification of recombinant proteins were performed as previously reported (Chi et al., 2012), with minor modifications. The recombinant OsGrxC2.2 was expressed as a His6-tagged fusion protein and purified using affinity chromatography with nickel-chelating Sepharose (Sigma-Aldrich). Fifteen microliters of each fraction was loaded into each lane of a 15% SDS-PAGE gel, followed by Coomassie brilliant blue R-250 staining.
The purified OsGrxC2.2 activity was determined using the β-hydroxyethyl disulfide (HED) assay (Chi et al., 2012; Riondet et al., 2012). The reaction mixture consisted of 100 mM Tris-Cl (pH 7.4), 3 μg glutathione reductase (GR; Sigma-Aldrich), 0.75 mM HED, 0.2 mM nicotinamide adenine dinucleotide phosphate (NADPH; Sigma-Aldrich), and 1 mM glutathione (GSH, Sigma-Aldrich) in a final volume of 100 μL. A mixed disulfide between HED and GSH was formed within 2 min, and the reaction was initiated by the addition of 0.06–0.40 μg OsGrxC2.2 and was followed by a decrease in absorbance at A340 due to the oxidation of NADPH. The control reaction included all the reagents except OsGrxC2.2.
The developing seeds at various stages after pollination, including 3, 5, 7, and 10 days, were fixed with 3% (w/v) paraformaldehyde and dehydrated in a graded ethanol with series of 75, 85, 90, 95, and 100%. The samples were embedded in paraplast and then sectioned into 8 μm-thick sections, stained with 0.1% toluidine blue, and observed with a Zeiss Axio Scope A1 microscope (Carl Zeiss, Heidenheim, Germany).
The grain weight was estimated using a 1000 seeds of the transgenic lines and WT in three biological replicates; the seed weight was estimated using a hundred dehulled seeds of the transgenic lines and WT in three biological replicates. The data were presented as mean ± standard error (SE).
Three-sample t-tests or one-way analysis of variance (ANOVA), in conjunction with LSD tests, were used to identify significant differences at p < 0.05 or p < 0.01.
To determine the OsGrxC2.2 expression pattern, total RNA from various tissues and organs was extracted. RT-PCR using OsActin1 as an internal control, was performed as described in the Materials and Methods. In all the examined tissues and organs, OsGrxC2.2 was found to be highly expressed in the leaf, flag leaf, and developing seeds at different stages compared to the root, stem, tiller, and panicle (Figure 1A). Interestingly, the expression of OsGrxC2.2 was even higher in the developing seeds, particularly in the late stages (18 and 21 DAP). The expression was also much higher in the embryo than in the endosperm (Figure 1B), suggesting that the OsGrxC2.2 enzyme may be involved in the developing seeds and embryos.
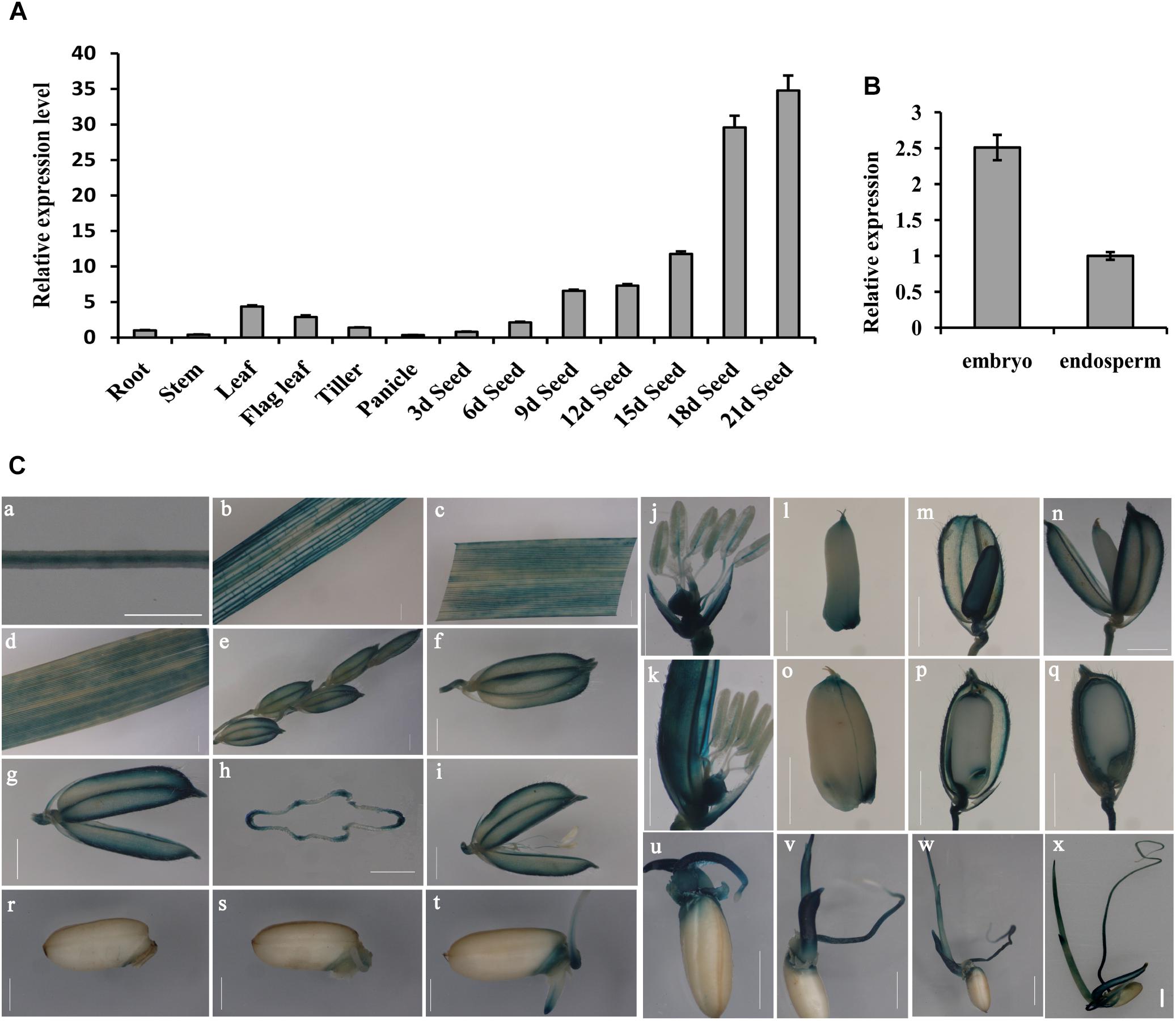
Figure 1. Expression pattern analysis of OsGrxC2.2. (A) Measurement of OsGrxC2.2 gene expression in various rice tissues. (B) Comparison of OsGrxC2.2 gene expression in the embryo and endosperm from dry seeds. The rice Actin1 gene (OsActin1) was used as an internal control. (C) Histochemical analysis of GUS activity in pOsGrxC2.2:GUS plants. Four-leaf-stage root (a); tillering stage stem (b), leaf (c); heading stage flag leaf (d), a hull before flowering (e–h), flower (i–k); grain-filling stage developing seeds of 3–5 days (l–n), 10–12 days (o, p), and 15–17 days (q) after flowering; and germinating seeds of 24 h (r), 36 h (s), 2 days (t), 3 days (u), 4 days (v), 5 days (w), and 6 days (x); (h) is the cross section of (g). Bar = 0.5 cm.
The YFP signal of the OsGrxC2.2-YFP fusion protein was visualized under a confocal microscope, which demonstrated that the OsGrxC2.2 protein was expressed and localized in the cytoplasm (Supplementary Figure S1). To elucidate the spatiotemporal expression profiles of OsGrxC2.2 in rice, the OsGrxC2.2 promoter was fused to the GUS reporter gene and transformed into WT plants. The GUS activity was detected in most of the examined tissues and organs, including the roots, culms, leaves, flowers, young spikelets, and developing caryopses at various stages. High GUS activity was measured in the vascular bundles of the roots, stems, leaf blades, hulls, caryopses, and filaments (Figures 1C,a–k). Additionally, high activity was also observed in various regions of the developing seeds, particularly in the embryos and outer layers of the endosperm, including the aleurone and subaleurone tissues (Figures 1C,l–q). GUS staining analysis of the germinating seeds (1–6 days) indicated that the activity of the promoter of OsGrxC2.2 was stronger in the embryo, coleorhizae, coleoptiles, and young leaf blades (Figures 1C,r–x). As shown in Figure 1C, relatively weak GUS activity was noted in the young leaf blades and blade sheaths.
In order to evaluate the enzymatic characteristics of OsGrxC2.2, the coding region of OsGrxC2.2 (384 bp) was amplified by PCR and subcloned in an expression vector, pET-28a (+), as described in the Materials and Methods, and overexpressed in E. coli. An SDS-PAGE image for the E. coli lysate showed an over-expressed 14.5 kD protein (Figure 2A). As expected, the corresponding gel band was identified and confirmed to be OsGrxC2.2 by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. An HED assay was then used to examine the Grx activity of OsGrxC2.2. The results show that the purified OsGrxC2.2 enabled the reduction of the disulfide bond of HED with specific activity at 54.1 (μmol/min/mg) (Figures 2B,C). Although weak NADPH oxidation activity was also observed in elution buffer (EB), it was negligible in comparison to that of OsGrxC2.2 (Figure 2D).
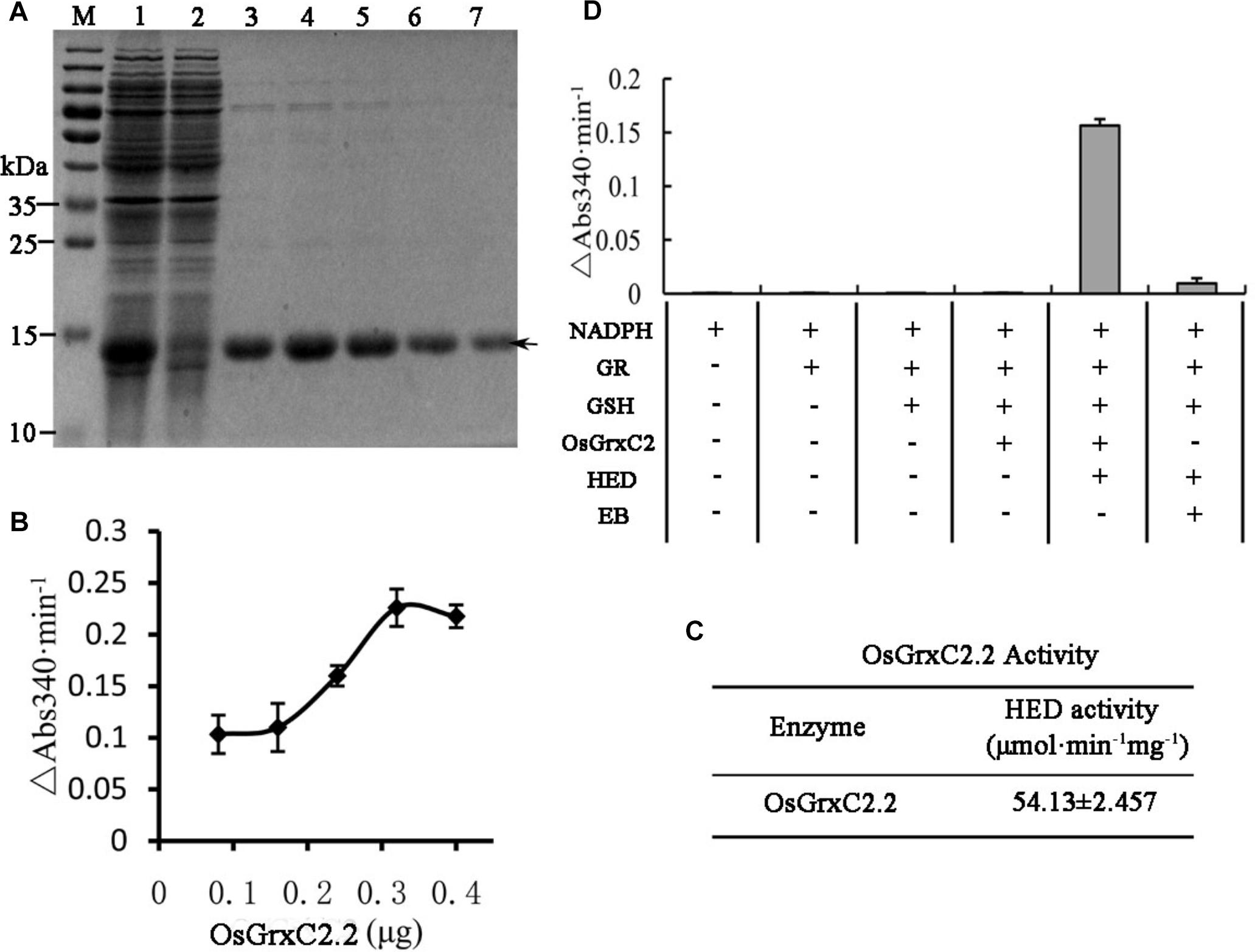
Figure 2. Expression, purification, and enzymatic characterization of OsGrxC2.2. (A) Expression and purification of recombinant OsGrxC2.2 in E. coli. Fifteen microliters of each fraction was loaded into each lane of a 15% SDS-PAGE gel, followed by Coomassie brilliant blue R-250 staining. Lane 1, crude extracts from E. coli expressing OsGrxC2.2; lane 2, flow-through proteins from the Ni-NTA column; lanes 3–7, OsGrxC2.2 eluted from the Ni-NTA column. Molecular masses (in kDa) of standards are shown on the left. The arrow indicates the target protein. (B) Determination of OsGrxC2.2 protein enzymatic activity followed by NADPH oxidation rate. NADPH oxidation was monitored at 340 nm in a 100 μL reaction mixture containing 0.1 M Tris-HCl (pH 7.4), 0.6 μg GR, 0.75 mM HED, 0.2 mM NADPH, and OsGrxC2.2 at concentrations varying from 0.06 to 0.3 μg. The rate of NADPH oxidation is shown by △Abs340min–1. (C) Enzymatic activity of OsGrxC2.2. (D) Effects of various combinations of GR, GSH, HED, EB (elution buffer), and OsGrxC2.2 on the initial rates of NADPH oxidation.
To further characterize the potential influence of OsGrxC2.2 on rice growth and development, transgenic rice lines overexpressing OsGrxC2.2 under the Ubi promoter (pUbi::OsGrxC2.2) were constructed. As indicated in Figure 3A, there were four representative overexpression lines demonstrating significant up-regulation of OsGrxC2.2 compared to the WT rice. Furthermore, the seeds obtained from the four homozygous transgenic lines revealed degraded and/or aborted embryos (Figure 3B). The overexpression lines OE11 and OE12 exhibited the most significant OsGrxC2.2 up-regulation compared with the OE2 and OE6 lines (Figure 3A). Accordingly, lines OE11 and OE12 possessed considerably more abnormal embryos, such as organless and radicleless embryos and displayed a more obvious embryoless seed phenotype than observed in OE2 and OE6 (Figure 3C). As indicated in Figure 3D, there was an average of approximately 60% abnormal embryo seeds in lines OE11 and OE12 (39% embryoless seeds plus 21% other abnormal seeds), while this value was 31% in lines OE2 and OE6 (1% embryoless seeds plus 30% other abnormal seeds).
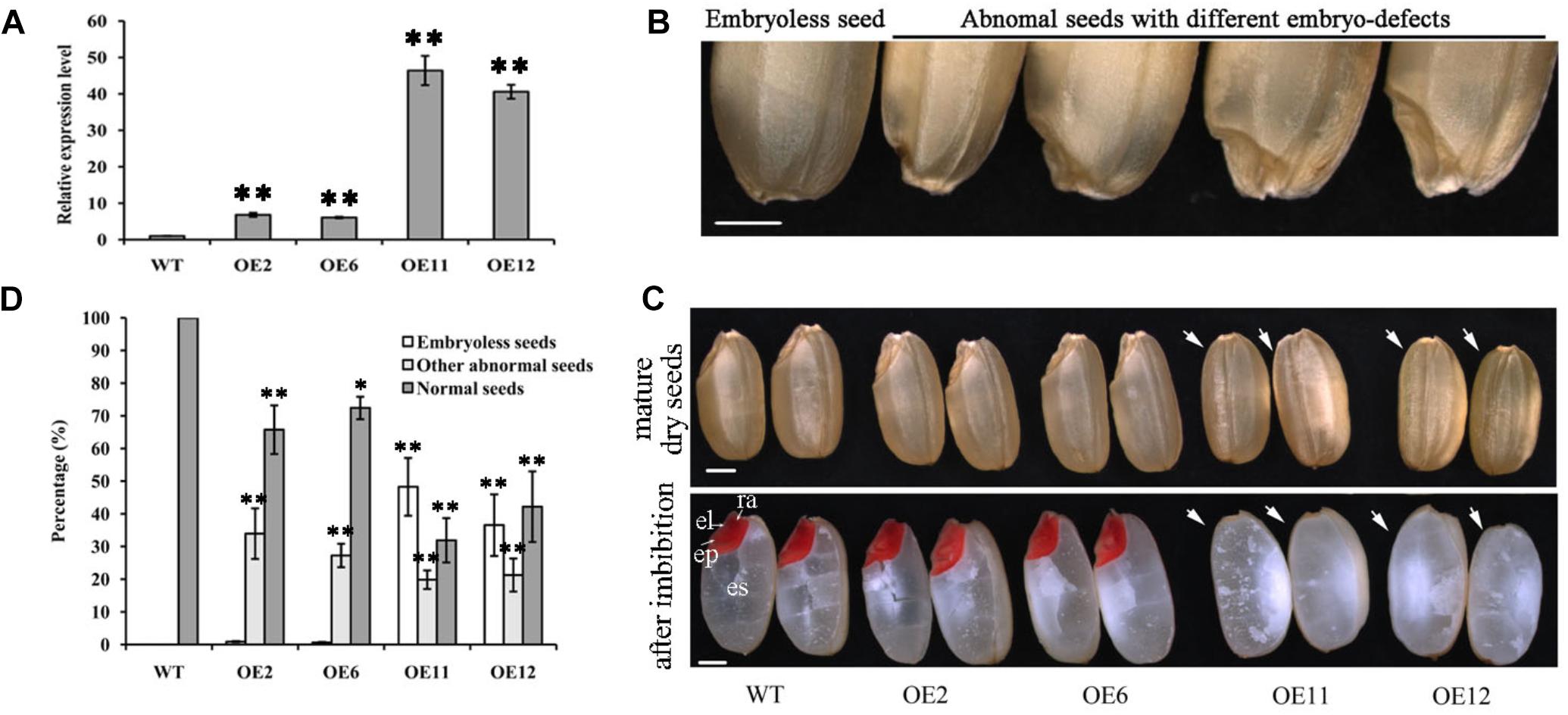
Figure 3. Phenotypic analysis of seeds overexpressing OsGrxC2.2. (A) qRT-PCR analysis of OsGrxC2.2 expression in the four independent transgenic lines OE2, OE6, OE11, and OE12. (B) Seeds with different types of embryo. (C) Embryo phenotype of the WT and OsGrxC2.2 overexpression lines. Upper panel, mature embryos from dry seeds; lower panel, TTC staining of rice seeds with imbibition at 12 h. cp, coleoptiles; el, embryonic leaves; ra, radical; arrow, embryos. Bars = 1 mm. (D) Percentage of different types of embryos in the WT seeds and OsGrxC2.2-overexpression line seeds. Error bars show the standard deviation. Statistical significance is indicated by ∗∗p < 0.01 and *p < 0.05.
To determine when the embryonic defects occurred in the overexpression lines, the developing embryos of the transgenic lines were subjected to histochemical analysis. At 3 DAP, a similar globular embryo was formed in the zygote in both the WT and overexpression lines (Supplementary Figure S2). At 5 DAP, the coleoptile primordium began to differentiate in the WT, and the shoot apical meristem (SAM) was recognizable as a bulging protrusion at the base (Figure 4A). However, some embryos in the overexpression lines differed from those of the WT at this stage and appeared to form abnormal embryos, including an elliptic curved embryo, spindle embryo, minor embryo, and other embryo types (Figures 4B–E). Consequently, some embryos failed to develop organs, including the coleoptile primordium and the SAM. At 7 DAP, the primordium of the first leaf in the WT emerged below the shoot apex, the radicle was formed, and the coleoptile and SAM were elongated (Figure 4F). In contrast, some abnormal embryos in the overexpression lines showed little differentiation from the radicleless embryos, sickle-shape embryos, “T”-shaped embryos, and the oval germinal embryos (Figures 4G–J). By 10 DAP, all organs had developed in the WT and embryogenesis was complete (Figure 4K), with the seeds continuing to develop until maturation. On the contrary, neither growth nor differentiation was noted in the abnormal embryos of the overexpression lines (Figures 4M–O), which constituted abnormal seeds or embryoless seeds that failed to germinate.
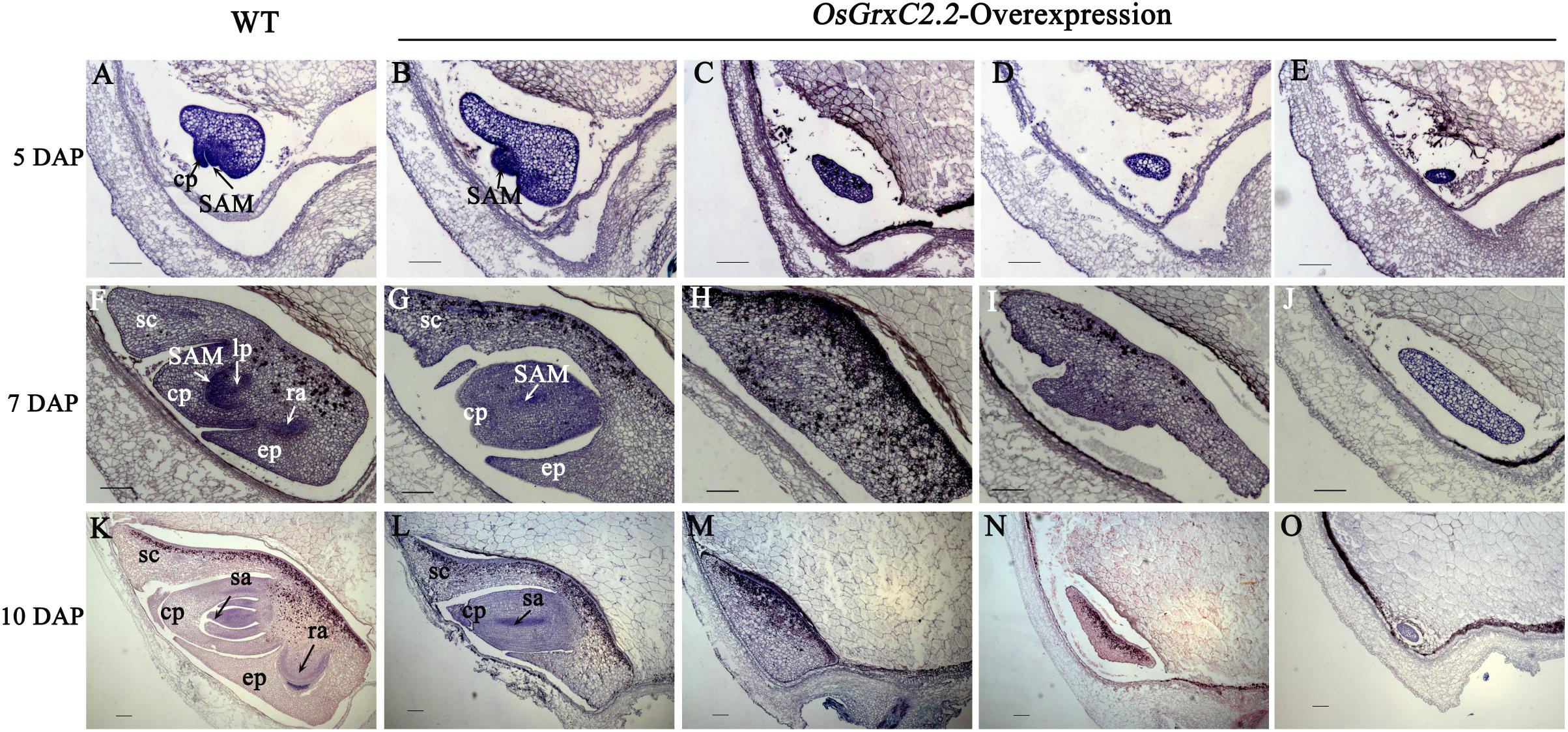
Figure 4. Median longitudinal sections of WT and OsGrxC2.2-overexpression lines embryos at various developmental stages. Developing embryos of WT (A,F,K) and representative overexpression line OE11 (B–E,G–J,L–O) at 5 DAP (A–E), 7 DAP (F–J), and 10 DAP (K–O). cp, coleoptiles; ep, epiblast; lp, leaf primordium; ra, radical apex; sa, shoot apex; SAM, shoot apical meristem; sc, scutellum are as indicated. Bars = 100 μm.
To investigate the molecular mechanisms underlying the abnormal embryos, we isolated embryos from the overexpression lines and WT at 6 DAP and determined the transcript level of OsGrxC2.2 and several representative embryogenesis-related genes.
Among the three ROCs, which are markers for the L1 layer, only ROC1 showed relatively lower expression levels in the overexpression lines embryos, while the expression of ROC2 and ROC3 was the same as that in the WT (Figures 5A–C). The transcript level of the outer layer gene HAZ1 was similar between the overexpression lines and the WT (Figure 5D). Furthermore, no impact on the expression of the L1 epithelium layer marker RAmy1A and the SAM markers HB1 and HB2 was observed. HB3 was decreased in the overexpression line embryos (Figures 5E–H).
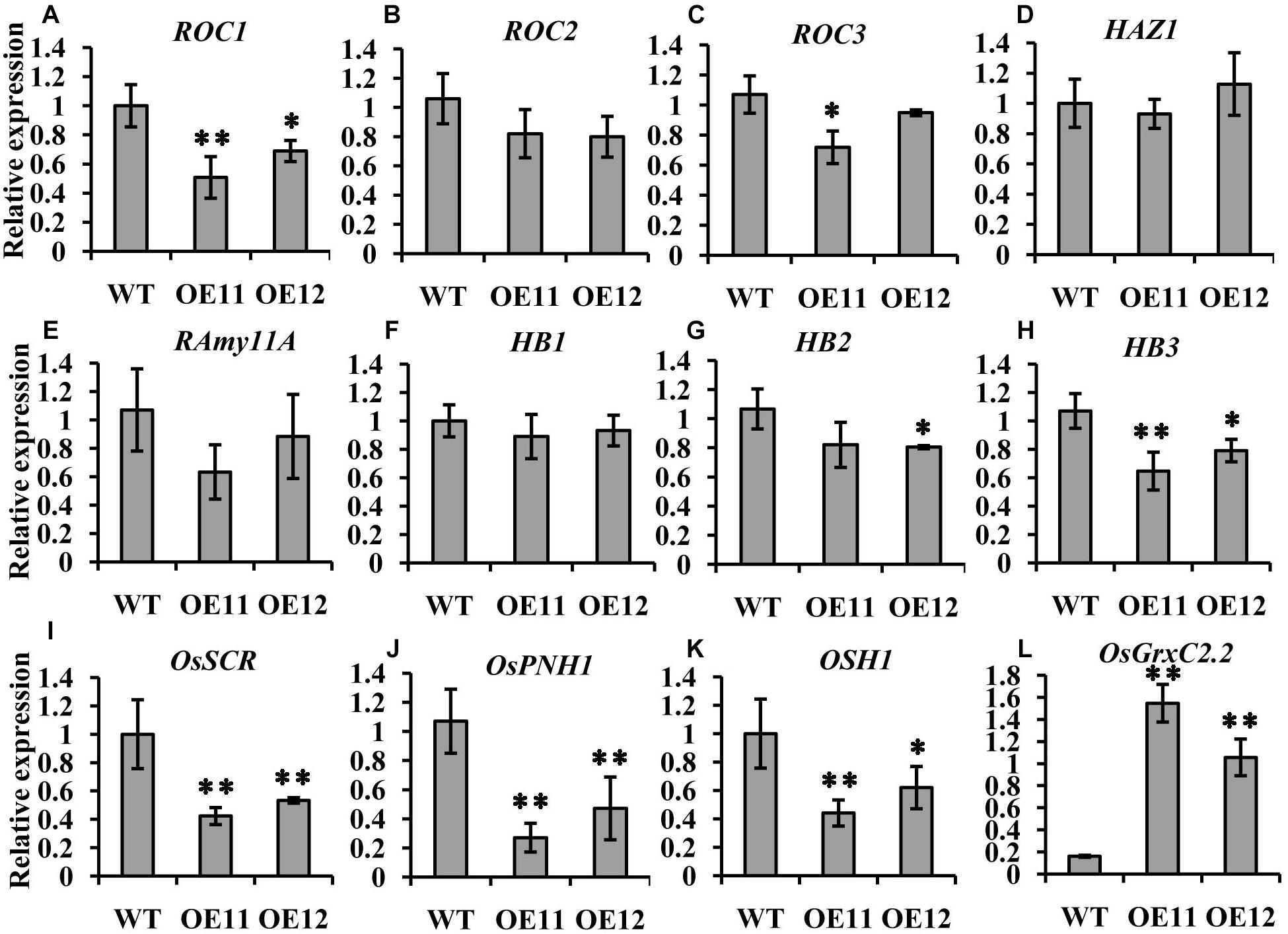
Figure 5. Expression comparison of molecular marker genes between the OsGrxC2.2-overexpression lines (OE11 and OE12) and the WT control. qRT-PCR of ROC1 (A), ROC2 (B), ROC3 (C), HAZ1 (D), RAmy11A (E), HB1 (F), HB2 (G), HB3 (H), OsSCR (I), OsPNH1 (J), OSH1 (K), and OsGrxC2.2 (L). The y-axis represents the gene expression relative to the OsActin1 transcript level. Results are the averages of three independent experiments. Error bars show the standard deviation. Statistical significance is indicated by ∗∗p < 0.01 and *p < 0.05.
Interestingly, two other markers, OsSCR at the L2 ground tissue and OsPNH1 at the L3 vascular tissue were significantly changed. The transcript levels of OsSCR and OsPNH1 were dramatically decreased in the overexpression line embryos (Figures 5I,J). The expression of OSH1, a marker of the apical region, was also considerably decreased in the overexpression lines embryos (Figure 5K). However, the transcript level of OsGrxC2.2 was significantly higher in the overexpression lines than in the WT (Figure 5L). These results are consistent with the abnormal embryo phenotypes observed by histochemical analysis (Figure 4), which demonstrated that OsGrxC2.2 is involved in negatively regulating the differentiation of the embryonic organs and the basic axis during embryogenesis.
Although embryo development was affected by OsGrxC2.2, the seed length and hull color of the seeds in the overexpression lines did not differ from that of the WT (Supplementary Figure S3). To further investigate whether OsGrxC2.2 affects endosperm development, we analyzed the transcripts of the genes involved in starch synthesis in the caryopses at 10 DAP, the time point when starch is being actively accumulated (Yamakawa et al., 2007). We found that the expression levels of starch biosynthesis genes encoding ADP-glucose pyrophosphorylase large subunit 1 (AGPL1), Starch synthase IIIa (SSIIIa), and Starch branching enzyme I (BEI) did not differ between the WT and transgenic lines (Supplementary Figure S4). The expression of a starch debranching enzyme gene, pullulanase, in the overexpression lines was slightly higher than that in the WT seeds (Supplementary Figure S4), which corroborates previous studies (Ohdan et al., 2005; Lee et al., 2015).
Since seed weight is an important indicator of crop yield, the seed weights from the overexpression lines and the WT plants were determined and compared. Statistical analysis indicated that the seed weights of the overexpression lines OE11 and OE12 were very significantly greater than those of the OE2, OE6 and WT (p < 0.01) (Figure 6A). The 1000-grain weight of lines OE11 and OE12 were 26.15 and 25.58 g, respectively, while the 1000-grain weight of the WT was 23.7 g. However, the 1000-grain weights of lines OE2 and OE6 were 24.37 and 24.18 g, respectively, which is higher but not significantly different from that of the WT. A similar trend is presented in Figure 6B about the dehulled seed weight. As the overexpression lines OE11 and OE12 possessed many more embryoless seed (39%) (Figure 6C) than in OE2 and OE6 (1%) (Figure 3D), the results suggest that the contribution to the increased seed weight mainly comes from the embryoless seeds.
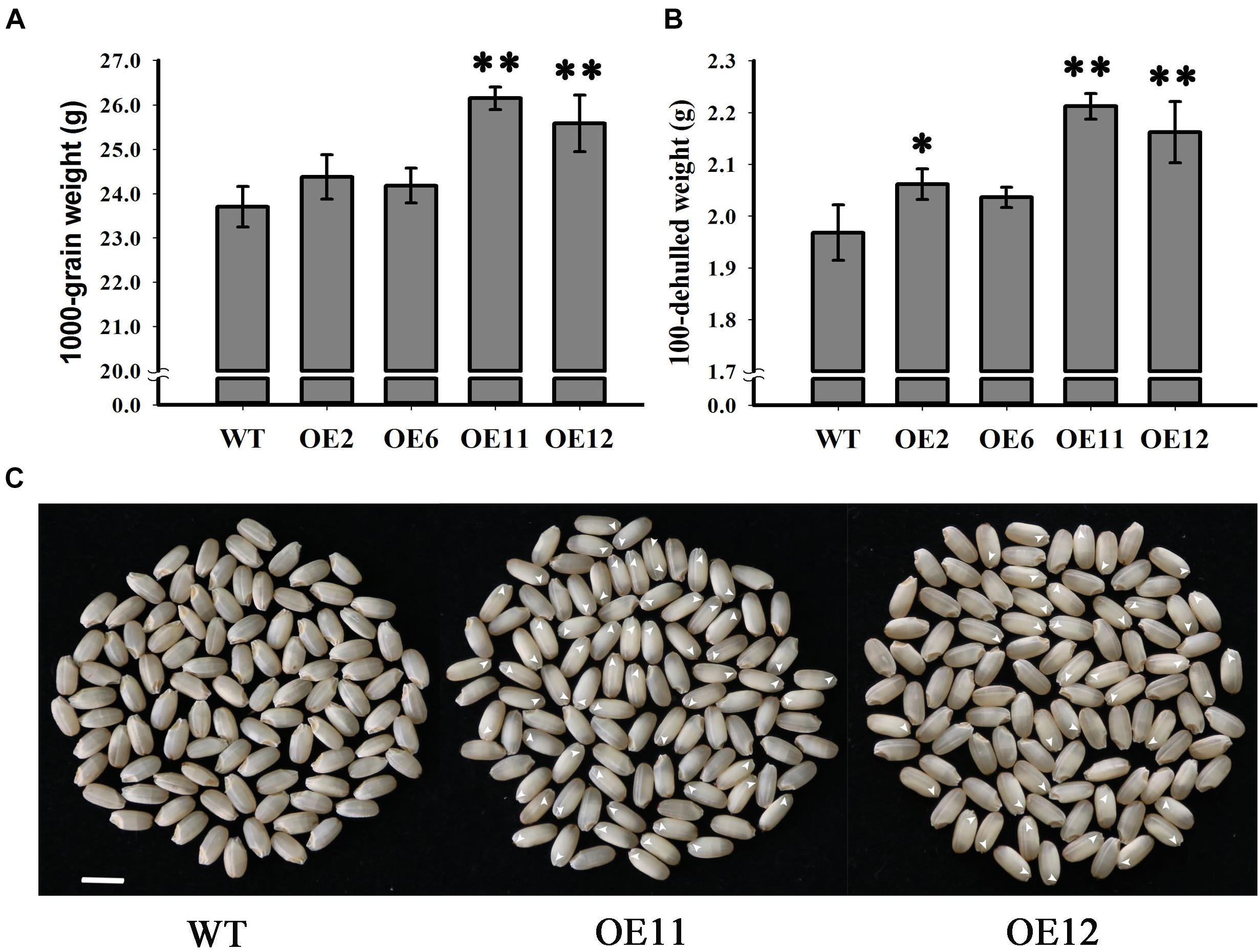
Figure 6. Analysis of grain weight in WT and OsGrxC2.2-Overexpression lines. (A) 1000-grain and (B) 100-seed weight of WT and OsGrxC2.2 overexpression line (OE2, OE6, OE11, and OE12) seeds. (C) Phenotypical comparison of seeds from WT and the OsGrxC2.2 overexpression plants. Bar = 0.5 cm. Arrows: embryoless seeds. Statistical significance is indicated by ∗∗p < 0.01 and *p < 0.05.
Although plant genomes contain many Grxs (Lemaire, 2004; Rouhier et al., 2006), only a few have been characterized (Rouhier et al., 2004). Garg et al. (2010) identified 48 genes encoding Grx proteins in the rice genome and suggested diverse roles for these Grx genes during growth and development. Some of the Grx genes were only expressed in specific organs/developmental stages, and the expression of many rice Grx genes was modulated by various phytohormones and abiotic and biotic stress conditions, suggesting an important role for Grx proteins in response to these stimuli (Garg et al., 2010). For instance, OsGRX8 in rice has been implicated in the plant response to stress (Sharma et al., 2013). However, the functions of most of these Grx genes are unclear.
In our previous study, several redox regulation proteins, mainly glutathione-related proteins including one Grx protein (CAA54397.1), exhibited various degrees of change in protein abundance during storage between hybrid rice seeds with different storability, and thus might play an important role in protecting embryo proteins from oxidation (Gao et al., 2016). Another study reported that the OsGrxC2.2 protein accumulates abundantly in the embryo and aleurone layers, and participates in oxidative stress defense in developing and mature seeds (Morita et al., 2015).
Given these findings, our original intention was to explore the function of OsGrxC2.2 in seed stress resistance. We thus cloned the Class I Grx isoform OsGrxC2.2 (Os04g42930) with a CPFC active site, which is a typical dithiol isoform (Supplementary Figure S5). However, we unexpectedly discovered that OsGrxC2.2 overexpression induces embryo malformation or abortion (Figure 3B). This finding motivated us to redirect our research to investigate the roles of the OsGrxC2.2 protein in embryonic development and explore the possible associated mechanisms.
Grxs have recently been found to play an important role in plant developmental processes (Rouhier et al., 2015). The overexpression of OsGRX6 led to a semi-dwarf phenotype (El-Kereamy et al., 2015), while OsMIL1 is involved in the floral organ development (Hong et al., 2012). A previous study reported the OsGrxC2.2 homologous gene AtGrxC2 in A. thaliana, found that the double grxc1 grxc2 mutant is lethal. The most likely hypothesis is that a very early stage of seed development is affected in the double grxc1 grxc2 mutant (Riondet et al., 2012), but no such reports on the equivalent OsGrxC2.2 were found in rice. Since rice is a crop plant, the unique role of OsGrxC2.2 in rice embryonic development could have significant economic implications for grain production. We thus speculated that OsGrxC2.2 might be crucial in seed maturation and particularly involved in embryo development.
Our histochemical assay revealed a higher level of GUS activity (representing OsGrxC2.2 localization as a fusion protein) in the vascular tissues (including the root, stem, leaf, and filament), embryo, and aleurone layer (Figure 1B). This corroborates a previous study by Garg et al. (2010), which demonstrated that OsGrxC2.2 mRNA was expressed at a high level in various tissues, including the leaves, roots, and developing seeds in comparison to other CPFC-type Grxs in rice. However, our findings differ from those of Morita et al. (2015) in that Grx expression was not detected in the leaf in their study. This inconsistency might be related to post-transcriptional regulation, where a number of genes are transcribed but cannot be translated into proteins in certain tissues due to their modification.
In addition, the qRT-PCR data did not well match with those from the GUS reporter system, for instance in the leaf. Studies have shown that sequences in the intron and/or 3’UTR can regulate gene transcription. We speculate that additional regulatory regions might exist in the intron and/or 3’UTR of OsGrxC2.2. Alternatively, we also cannot exclude the possibility that the promoter region we used was not long enough, thus resulting in some regulatory sequences being missed. Future studies are required to test these possibilities.
During the early stages of embryogenesis, a rice embryo first establishes three axes: apical–basal, radial, and dorsal–ventral (Sato, 2008; Yi et al., 2016). Along the apical–basal axis, the SAM, cotyledon, hypocotyls, and root apical meristem (RAM) form, while the radial axis consists of the epidermis (L1 layer), ground tissue (L2 layer), and central vascular cylinder (L3 layer) form perpendicular to the apical–basal axis, and develop from the outside in Mayer et al. (1991), Kamiya et al. (2003), and Yi et al. (2016).
To identify the point at which the morphological distinction appeared during embryogenesis, we conducted a precise histological analysis of the overexpression lines and WT embryos. Rice embryos complete their morphogenic events within 9–10 days under normal conditions, and the globular stage, which does not exhibit any morphological organ differentiation, continues until 3 DAP (Sato et al., 1996; Ito et al., 2002). No significant difference was found between the overexpression lines and WT embryos at the globular stage. However, from 5 DAP, the overexpression lines embryos began to differ from those of the WT, in that some did not undergo differentiation of the stem-tip tissues (Figures 4C–E), or otherwise displayed abnormal embryos, including organless embryos (Figures 4H–J,M–O) and radicleless embryos (Figures 4G,L).
Following this, we measured the expression of embryo development-related marker genes in the OsGrxC2.2-overexpressed rice lines. Several genes that are expressed in specific cell types and regions have been used as molecular markers in rice embryogenesis studies. For example, the rice SCARECROW (OsSCR) is specifically localized in the cortex/endodermis cell layer that corresponds to the L2 layer (Di Laurenzio et al., 1996); the rice PINHEAD/ZWILLE gene (OsPNH1) is expressed in the vascular regions of the leaf primordia and is used as an L3 vascular tissue marker (Nishimura et al., 2002; Kamiya et al., 2003); and OSH1 is expressed in the indeterminate cells around the SAM and is used as a marker for the apical region in rice embryos (Kamiya et al., 2003; Sato, 2008). Other developed markers include Rice outermost cell-specific gene (ROC, marker for the L1 layer) (Ito et al., 2002), Rice α-amylase1 (RAmy1A) (marker for the L1 layer of the epithelium) (Sugimoto et al., 1998), and OSH1 (marker for the apical region) (Sato et al., 1996).
The expression levels of most of the outer layers were similar between the overexpression lines and the WT (Figure 5).
However, the expression of OSH1 was significantly decreased in the overexpression line embryos (Figure 5K). Furthermore, the transcript levels of OsSCR and OsPNH1 were also decreased in the overexpression line embryos (Figures 5I,J). The significant decrease in the transcript levels of OsSCR, OsPNH1, and OSH1 in the central region indicated that differentiation in the ground tissue and vascular primordium had been affected. Importantly, the overexpression lines were partly defective in the establishment of the L2 and L3 layers from the inner cell mass.
Matching conversely with these marker genes in the central region, the expression of OsGrxC2.2 was significantly higher in the overexpression lines (Figure 5L). OsGrxC2.2 overexpression appears to be involved in rice embryogenesis, and negatively affects embryo development, resulting in embryo degeneration or abortion, or even the formation of embryoless seeds (Figure 3) by reducing the expression of several L2 and L3 marker genes associated with embryo development. However, it is unclear whether the overexpression of OsGrxC2.2 directly reduces the expression of several L2 and L3 marker genes, or whether another yet unknown redox-based process leads to defects in embryo L2 and L3 differentiation and a reduction of marker genes expression. Further research is required to clarify these possibilities.
In the present study, we demonstrated that the overexpression of OsGrxC2.2 in rice impacts embryo development, resulting in seeds without embryos or with a smaller embryo, with the seed embryos being completely or partially replaced by endosperms. The increased weight of the OsGrxC2.2-overexpressing seeds can probably be attributed to the large endosperm, which may occupy the entire space in some instances (Figure 6B). This mechanism differs from that of OsGRX6, which affects hormone signaling and nitrogen status in rice plants leading to a semi-dwarf phenotype and delayed chlorophyll degradation (El-Kereamy et al., 2015).
Grxs possess peroxidase activity and the capability to reduce DHA and type II Prxs, regenerate methionine sulfoxide reductase, and interact with thioredoxins. Therefore, they play important roles in various cellular processes, including the maintenance and regulation of cellular redox state, iron homeostasis, and redox-dependent signaling pathways, which eventually contribute to the removal of reactive oxygen species (ROS) and the repair of lipid and protein oxidative damage (Wells et al., 1990; Sha et al., 1997; Rouhier et al., 2001). Furthermore, there is growing evidence suggesting that ROS-mediated redox signals (redox homeostasis) function during plant development and adaptation to stress conditions, such as those occurring during seed germination, root hair development, stomatal closure, and root gravitropic responses (Joo et al., 2001; Foreman et al., 2003; Gapper and Dolan, 2006; Kwak et al., 2006; Achard et al., 2008). Therefore, we speculate that OsGrxC2 might be crucial in seed embryo development, and alters normal embryogenesis through interfering with redox homeostasis in the early rice embryo, thus leading to increased grain weight. We believe that our observation could be further explored in future studies to improve grain weight and yield using biotechnology.
In higher plants, embryo and endosperm originate from double fertilization. Rice embryos contain grass-specific coleoptiles, scutella, and epiblasts, suggesting that there are more complicated molecular mechanisms controlling rice embryo development. As the rice endosperm grows continuously and occupies most of the space in the mature seed, the developmental coordination of the embryo and endosperm is of particular importance for the proper development of rice seeds. However, the associated molecular pathways remain poorly understood. It has been suggested that the endosperm and embryo in rice modulate one another’s growth due to space limitations (Hong et al., 1996). Indeed, temperature-sensitive embryoless1 mutants produce embryoless seeds with large endosperm at high temperature, but display large embryos with small or even no endosperm at low temperature (Hong et al., 1995). The down-regulation of Orysa;CycB1;1 resulted in severe endosperm defects, but also resulted in the production of very large embryos (Guo et al., 2010). In our study, the overexpression lines OE11 and OE12 exhibited significantly higher OsGrxC2.2 level than the OE2 and OE6 lines (Figure 3A), and possessed much more embryoless seeds than those observed in OE2 and OE6 (Figure 3C). Consequently, both OE11 and OE12 display a significantly higher weight than that of the OE2 and OE6, and WT. This result suggests that OsGrxC2.2 overexpression impairs embryo development, and subsequently leads to increased endosperm size and seed weight.
In summary, we found that transgenic rice overexpressing OsGrxC2.2 resulted in a novel phenotype with degenerate embryos and even embryoless seeds. The embryonic abnormalities began at the late globular stage during embryogenesis. In addition, the grain weight of the overexpression lines was significantly increased in comparison to the WT. These results indicate that overexpression of OsGrxC2.2 interferes with the normal embryogenesis and leads to increased grain weight.
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
SL, CY, and JL conceived the project and designed the research. SL, HF, JJ, HS, and JG performed the research. SL, HF, ZC, SZ, CY, and JL analyzed the data. SL and JL wrote the manuscript. All authors read and approved the final version of the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (31371715, 31640059, and 31871716), the Natural Science Foundation of Guangdong Province (S2013010015346), and the Science and Technology Planning Project of Guangdong Province (2014A020208049, 2016B030303007, and 2018B020202004).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank professors Chenlong Li and Yuhai Cui for helpful discussions and comments on the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00848/full#supplementary-material
FIGURE S1 | Subcellular localization of the OsGrxC2.2 protein.
FIGURE S2 | Median longitudinal sections of the WT and OsGrxC2.2-over expression lines embryos at 3 DAP. Developing embryos of WT (A) and representative overexpression line OE11 (B).
FIGURE S3 | Phenotypic analysis of embryoless and WT seeds. (A) Hulled seeds. (B) Shelled embryoless and WT seeds.
FIGURE S4 | Analysis of endosperm development maker genes in WT and OsGrxC2.2-overexpression lines. Expression levels of genes involved in starch biosynthesis obtained via qRT-PCR analyses of AGPL1, OsSSIIa, BE1, and PUL from 10 DAP seeds of WT and OsGrxC2.2-overexpression lines. The y-axis represents the gene expression relative to the OsActin1 transcript level. Results are the averages of three independent experiments.
FIGURE S5 | Amino acid sequence alignment between OsGrxC2.2 and other class I Grx sequences in rice using ClustalW software.
TABLE S1 | Primer sequences used in this study.
Achard, P., Renou, J.-P., Berthomé, R., Harberd, N. P., and Genschik, P. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18, 656–660. doi: 10.1016/j.cub.2008.04.034
Cheng, N.-H. (2008). AtGRX4, an Arabidopsis chloroplastic monothiol glutaredoxin, is able to suppress yeast grx5 mutant phenotypes and respond to oxidative stress. FEBS Lett. 582, 848–854. doi: 10.1016/j.febslet.2008.02.006
Cheng, N.-H., Liu, J.-Z., Brock, A. J., Nelson, R. S., and Hirschi, K. D. (2006). AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J. Biol. Chem. 281, 26280–26288. doi: 10.1074/jbc.M601354200
Cheng, N.-H., Liu, J.-Z., Liu, X., Wu, Q., Thompson, S. M., Lin, J., et al. (2011). Arabidopsis monothiol glutaredoxin, AtGRXS17, is critical for temperature-dependent postembryonic growth and development via modulating auxin response. J. Biol. Chem. 286, 20398–20406. doi: 10.1074/jbc.M110.201707
Chi, X. W., Lin, C. T., Jiang, Y. C., and Wen, L. (2012). A dithiol glutaredoxin cDNA from sweet potato (Ipomoea batatas [L.] lam): enzyme properties and kinetic studies. Plant Biol. 14, 659–665. doi: 10.1111/j.1438-8677.2011.00544.x
Di Laurenzio, L., Wysocka-Diller, J., Malamy, J. E., Pysh, L., Helariutta, Y., Freshour, G., et al. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433. doi: 10.1016/s0092-8674(00)80115-4
El-Kereamy, A., Bi, Y. M., Mahmood, K., Ranathunge, K., Yaish, M. W., Nambara, E., et al. (2015). Overexpression of the CC-type glutaredoxin, OsGRX6 affects hormone and nitrogen status in rice plants. Front. Plant Sci. 6:934. doi: 10.3389/fpls.2015.00934
Foreman, J., Demidchik, V., Bothwell, J. H. F., Mylona, P., Miedema, H., Torres, M. A., et al. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. doi: 10.1038/nature01485
Gao, J., Fu, H., Zhou, X., Chen, Z., Luo, Y., Cui, B., et al. (2016). Comparative proteomic analysis of seed embryo proteins associated with seed storability in rice (Oryza sativa L) during natural aging. Plant Physiol. Biochem. 103, 31–44. doi: 10.1016/j.plaphy.2016.02.026
Gapper, C., and Dolan, L. (2006). Control of plant development by reactive oxygen species. Plant Physiol. 141, 341–345. doi: 10.1104/pp.106.079079
Garg, R., Jhanwar, S., Tyagi, A. K., and Jain, M. (2010). Genome-wide survey and expression analysis suggest diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Res. 17, 353–367. doi: 10.1093/dnares/dsq023
Guo, J., Wang, F., Song, J., Sun, W., and Zhang, X. S. (2010). The expression of Orysa;CycB1;1 is essential for endosperm formation and causes embryo enlargement in rice. Planta 231, 293–303. doi: 10.1007/s00425-009-1051-y
Hong, L., Tang, D., Zhu, K., Wang, K., Li, M., and Cheng, Z. (2012). Somatic and reproductive cell development in rice anther is regulated by a putative glutaredoxin. Plant Cell 24, 577–588. doi: 10.1105/tpc.111.093740
Hong, S. K., Aoki, T., Kitano, H., Satoh, H., and Nagato, Y. (1995). Phenotypic diversity of 188 rice embryo mutants. Dev. Genet. 16, 298–310. doi: 10.1002/dvg.1020160403
Hong, S. K., Kitano, H., Satoh, H., and Nagato, Y. (1996). How is embryo size genetically regulated in rice? Development 122, 2051–2058.
Hou, X., Hu, W. W., Shen, L., Lee, L. Y., Tao, Z., Han, J. H., et al. (2008). Global identification of DELLA target genes during Arabidopsis flower development. Plant Physiol. 147, 577–588. doi: 10.1104/pp.108.121301
Ito, M., Sentoku, N., Nishimura, A., Hong, S. K., Sato, Y., and Matsuoka, M. (2002). Position dependent expression of GL2-type homeobox gene, Roc1: significance for protoderm differentiation and radial pattern formation in early rice embryogenesis. Plant J. 29, 497–507. doi: 10.1046/j.1365-313x.2002.01234.x
Ito, Y., Chujo, A., Eiguchi, M., and Kurata, N. (2004). Radial axis differentiation in a globular embryo is marked by HAZ1, a PHD-finger homeobox gene of rice. Gene 331, 9–15. doi: 10.1016/j.gene.2004.02.040
Joo, J. H., Bae, Y. S., and Lee, J. S. (2001). Role of auxin-induced reactive oxygen species in root gravitropism. Plant Physiol. 126, 1055–1060. doi: 10.1104/pp.126.3.1055
Kamiya, N., Nishimura, A., Sentoku, N., Takabe, E., Nagato, Y., Kitano, H., et al. (2003). Rice globular embryo 4 (gle4) mutant is defective in radial pattern formation during embryogenesis. Plant Cell Physiol. 44, 875–883. doi: 10.1093/pcp/pcg112
Knuesting, J., Riondet, C., Maria, C., Kruse, I., Bécuwe, N., König, N., et al. (2015). Arabidopsis glutaredoxin S17 and its partner, the nuclear factor Y Subunit C11/Negative cofactor 2a, contribute to maintenance of the shoot apical meristem under. Long Day Photoperiod. Plant Physiol. 167, 1643–1658. doi: 10.1104/pp.15.00049
Kwak, J. M., Nguyen, V., and Schroeder, J. I. (2006). The role of reactive oxygen species in hormonal responses. Plant Physiol. 141, 323–329. doi: 10.1104/pp.106.079004
Lee, D. W., Lee, S. K., Phee, B. K., and Jeon, J. S. (2015). Proteomic analysis of the rice endosperm starch-deficient mutants osagps2 and osagpl2. J. Plant Biol. 58, 252–258. doi: 10.1007/s12374-015-0160-3
Lee, M. Y., Shin, K. H., Kim, Y. K., Suh, J. Y., Gu, Y. Y., Kim, M. R., et al. (2006). Induction of thioredoxin is required for nodule development to reduce reactive oxygen species levels in soybean roots. Plant Physiol. 139, 1881–1889. doi: 10.1104/pp.105.067884
Lemaire, S. D. (2004). The glutaredoxin family in oxygenic photosynthetic organisms. Photosynth. Res. 79, 305–318. doi: 10.1023/b:pres.0000017174.60951.74
Lillig, C. H., Berndt, C., and Holmgren, A. (2008). Glutaredoxin systems. Biochim. Biophys. Acta 1780, 1304–1317. doi: 10.1016/j.bbagen.2008.06.003
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mayer, U., Ruiz, R. A. T., Berleth, T., Miséra, S., and Jürgens, G. (1991). Mutations affecting body organization in the Arabidopsis embryo. Nature 353, 402–407. doi: 10.1038/353402a0
Morita, S., Yamashita, Y., Fujiki, M., Todaka, R., Nishikawa, Y., Hosoki, A., et al. (2015). Expression of a rice glutaredoxin in aleurone layers of developing and mature seeds: subcellular localization and possible functions in antioxidant defense. Planta 242, 1195–1206. doi: 10.1007/s00425-015-2354-9
Nishimura, A., Ito, M., Kamiya, N., Sato, Y., and Matsuoka, M. (2002). OsPNH1 regulates leaf development and maintenance of the shoot apical meristem in rice. Plant J. 30, 189–201. doi: 10.1046/j.1365-313x.2002.01279.x
Ohdan, T., Francisco, P. B. Jr., Sawada, T., Hirose, T., Terao, T., Satoh, H., et al. (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 56, 3229–3244. doi: 10.1093/jxb/eri292
Riondet, C., Desouris, J. P., Montoya, J. G., Chartier, Y., Meyer, Y., and Reichheld, J. P. (2012). A dicotyledon-specific glutaredoxin GRXC1 family with dimer-dependent redox regulation is functionally redundant with GRXC2. Plant Cell Environ. 35, 360–373. doi: 10.1111/j.1365-3040.2011.02355.x
Rouhier, N., Cerveau, D., Couturier, J., Reichheld, J. P., and Rey, P. (2015). Involvement of thiol-based mechanisms in plant development. Biochim. Biophys. Acta 1850, 1479–1496. doi: 10.1016/j.bbagen.2015.01.023
Rouhier, N., Couturier, J., and Jacquot, J. P. (2006). Genome-wide analysis of plant glutaredoxin systems. J. Exp. Bot. 57, 1685–1696. doi: 10.1093/jxb/erl001
Rouhier, N., Gelhaye, E., and Jacquot, J. P. (2004). Plant glutaredoxins: still mysterious reducing systems. Cell Mol. Life Sci. 61, 1266–1277. doi: 10.1007/s00018-004-3410-y
Rouhier, N., Gelhaye, E., Sautiere, P. E., Brun, A., Laurent, P., Tagu, D., et al. (2001). Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiol. 127, 1299–1309. doi: 10.1104/pp.127.3.1299
Rouhier, N., Unno, H., Bandyopadhyay, S., Masip, L., Kim, S. K., Hirasawa, M., et al. (2007). Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc. Natl. Acad. Sci. U.S.A. 104, 7379–7384. doi: 10.1073/pnas.0702268104
Sato, Y., Hong, S. K., Tagiri, A., Kitano, H., Yamamoto, N., Nagato, Y., et al. (1996). A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 93, 8117–8122. doi: 10.1073/pnas.93.15.8117
Sha, S., Minakuchi, K., Higaki, N., Sato, K., Ohtsuki, K., Kurata, A., et al. (1997). Purification and characterization of glutaredoxin (thioltransferase) from rice (Oryza sativa L.). J. Biochem. 121, 842–848. doi: 10.1093/oxfordjournals.jbchem.a021663
Sharma, R., Priya, P., and Jain, M. (2013). Modified expression of an auxin-responsive rice CC-type glutaredoxin gene affects multiple abiotic stress responses. Planta 238, 871–884. doi: 10.1007/s00425-013-1940-y
Sugimoto, N., Takeda, G., Nagato, Y., and Yamaguchi, J. (1998). Temporal and spatial expression of the α-amylase gene during seed germination in rice and barley. Plant Cell Physiol. 39, 323–333. doi: 10.1093/oxfordjournals.pcp.a029373
Wei, Y., Xu, H., Diao, L., Zhu, Y., Xie, H., Cai, Q., et al. (2015). Protein repair L-isoaspartyl methyltransferase 1 (PIMT1) in rice improves seed longevity by preserving embryo vigor and viability. Plant Mol. Biol. 89, 475–492. doi: 10.1007/s11103-015-0383-1
Wells, W. W., Xu, D. P., Yang, Y. F., and Rocque, P. A. (1990). Mammalian thioltransferase (glutaredoxin) and protein disulfide isomerase have dehydroascorbate reductase activity. J. Biol. Chem. 265, 15361–15364.
Xing, S., Rosso, M. G., and Zachgo, S. (2005). ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132, 1555–1565. doi: 10.1242/dev.01725
Xing, S., and Zachgo, S. (2008). ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 53, 790–801. doi: 10.1111/j.1365-313x.2007.03375.x
Yamakawa, H., Hirose, T., Kuroda, M., and Yamaguchi, T. (2007). Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 144, 258–277. doi: 10.1104/pp.107.098665
Yi, J., Lee, Y. S., Lee, D. Y., Cho, M. H., Jeon, J. S., and An, G. (2016). OsMPK6 plays a critical role in cell differentiation during early embryogenesis in Oryza sativa. J. Exp. Bot. 67, 2425–2437. doi: 10.1093/jxb/erw052
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Keywords: embryogenesis, glutaredoxin (GrxC2.2), rice (Oryza sativa L.), seed, grain weight
Citation: Liu S, Fu H, Jiang J, Chen Z, Gao J, Shu H, Zhang S, Yang C and Liu J (2019) Overexpression of a CPYC-Type Glutaredoxin, OsGrxC2.2, Causes Abnormal Embryos and an Increased Grain Weight in Rice. Front. Plant Sci. 10:848. doi: 10.3389/fpls.2019.00848
Received: 18 February 2019; Accepted: 13 June 2019;
Published: 27 June 2019.
Edited by:
Nicolas Rouhier, Université de Lorraine, FranceReviewed by:
Pascal Rey, Commissariat à l’Énergie Atomique et aux Énergies Alternatives (CEA), FranceCopyright © 2019 Liu, Fu, Jiang, Chen, Gao, Shu, Zhang, Yang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengwei Yang, WWFuZ2Nod0BzY251LmVkdS5jbg==; Jun Liu, bGl1anVuQGdkYWFzLmNu; bGl1anVuMTM5QDEzOS5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.