
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 27 June 2019
Sec. Plant Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00847
This article is part of the Research TopicNAD Metabolism and Signaling in PlantsView all 7 articles
Unicellular cyanobacteria are thought to be the evolutionary ancestors of plant chloroplasts and are widely used both for chemical production and as model organisms in studies of photosynthesis. Although most research focused on increasing reducing power (that is, NADPH) as target of metabolic engineering, the physiological roles of NAD(P)(H) in cyanobacteria poorly understood. In cyanobacteria such as the model species Synechocystis sp. PCC 6803, most metabolic pathways share a single compartment. This complex metabolism raises the question of how cyanobacteria control the amounts of the redox pairs NADH/NAD+ and NADPH/NADP+ in the cyanobacterial metabolic pathways. For example, photosynthetic and respiratory electron transport chains share several redox components in the thylakoid lumen, including plastoquinone, cytochrome b6f (cyt b6f), and the redox carriers plastocyanin and cytochrome c6. In the case of photosynthesis, NADP+ acts as an important electron mediator on the acceptor-side of photosystem I (PSI) in the linear electron chain as well as in the plant chloroplast. Meanwhile, in respiration, most electrons derived from NADPH and NADH are transferred by NAD(P)H dehydrogenases. Therefore, it is expected that Synechocystis employs unique NAD(P)(H) -pool control mechanisms to regulate the mixed metabolic systems involved in photosynthesis and respiration. This review article summarizes the current state of knowledge of NAD(P)(H) metabolism in Synechocystis. In particular, we focus on the physiological function in Synechocystis of NAD kinase, the enzyme that phosphorylates NAD+ to NADP+.
NAD(P)(H) are important electron carriers, employed by all living cells in energy conversion. NAD+ and NADP+ (oxidized forms), or NADH and NADPH (reduced forms) act as oxidizing agents or reducing agents in electron-transfer steps in several metabolic pathways. It is thought that the presence of the non-phosphorylated forms (NAD+ and NADH) and phosphorylated forms (NADP+ and NADPH) make it possible for multiple redox reactions to coexist simultaneously within the cell (Alberts et al., 2002). In plants, intensive studies have been performed to clarify the importance of the individual NAD(P)(H) species in growth and stress responses (Dutilleul et al., 2005; Schippers et al., 2008; Takahashi et al., 2009; Sienkiewicz-Porzucek et al., 2010; Takahara et al., 2010; Bernhardt et al., 2012; Pétriacq et al., 2012, 2016; Maruta et al., 2016). The influence of quantitative changes in the NAD(P)(H) balance on plant growth has been elucidated by reverse genetics using knockout or overexpression strains. The Arabidopsis old5 mutant harbors a mutation in the nuclear locus encoding the chloroplast-localized quinolinate synthase; the mutant exhibits increased NAD+ levels and early senescence phenotypes (Schippers et al., 2008). In contrast, overexpression in Arabidopsis of the cytoplasmic NAD synthase yields decreases NAD+ levels and accelerated senescence (Hashida et al., 2016). The Arabidopsis nudx19 mutant harbors a loss-of-function in the nuclear locus encoding a NADPH pyrophosphohydrolase that localizes to the chloroplast and peroxisome; this mutant shows increased NADPH contents in whole cell extracts, but not increased NADP levels. In contrast, plants overexpressing the Arabidopsis chloroplast-localized NAD kinase (NADK2) show an increased NADP(H) pool (i.e., elevations in both NADP+ and NADPH concentrations). Interestingly, both plants (Arabidopsis nudx19 mutant and NADK2-overexpressed transgenic plant) have been reported to exhibit enhanced carbon assimilation and tolerance to photooxidative stresses (Takahara et al., 2010; Maruta et al., 2016). The relationship between balancing of NAD(P)(H) and the physiological roles of these molecules in plants is complicated. This lack of clarity reflects the fact that plants have compartmentalized cell areas (chloroplast, mitochondria, vacuole, nucleus, etc.) within the cell. At present, experimental techniques do not permit measurement of the NAD(P)(H) contents of individual compartments in plant cells. As a result, there is no alternative to using quantitative results for the whole cell extracts.
On the other hand, cyanobacteria, which have comparatively simple cell structures, perform most metabolic reactions in a shared space (Figure 1A). Therefore, we expect that cyanobacteria are better suited to examining the direct effects of changes in NAD(P)(H) contents or balance. However, in cyanobacteria, there were (until recently) few reports on the use of the genetics of NAD(P)(H) metabolism for understanding the physiological roles of NAD(P)(H) species.
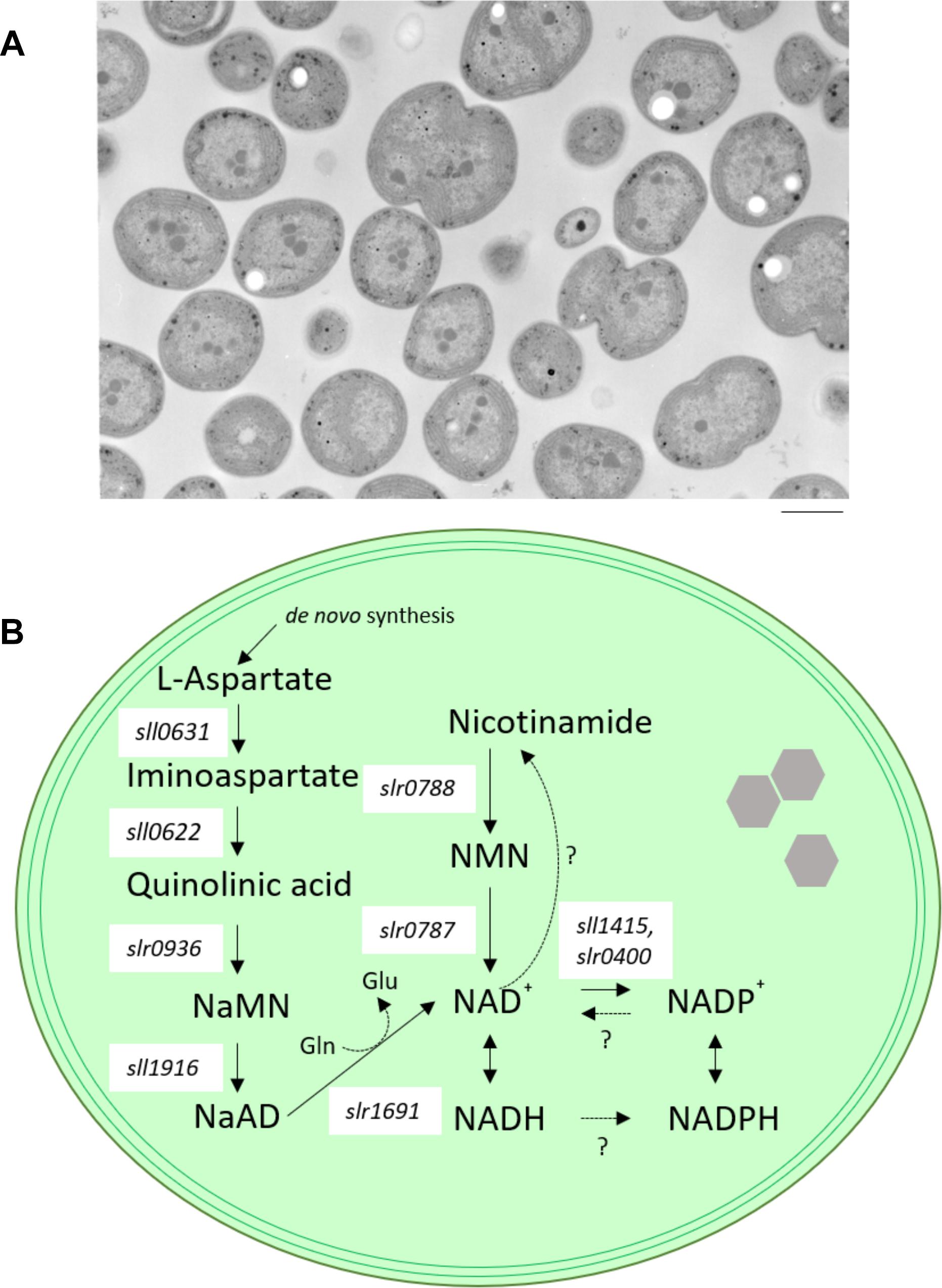
Figure 1. Model cyanobacterium Synechocystis sp. PCC 6803. (A) transmission electron microscope (TEM) images of wild-type cells of Synechocystis sp. PCC 6803. Bar indicates 1 μm. (B) Schematic diagram illustrating NAD+ biosynthesis in Synechocystis sp. PCC 6803. Enzymes contributing to NAD synthesis are the products of individual genes. Only NAD kinase (NADK), which catalyzes the phosphorylation of NAD+ to NADP+, is encoded by two separate genes (sll1415 and slr0400) in Synechocystis sp. PCC 6803. NaMN, nicotinate mononucleotide; NaAD, nicotinate adenine dinucleotide; NMN, nicotinamide mononucleotide; Gln, glutamine; Glu, glutamic acid; sll0631, L-aspartate oxidase-encoding gene; sll0622, quinolinate synthetase-encoding gene; slr0936, quinolinate phosphoribosyl transferase-encoding gene; sll1916, nicotinate nucleotide adenyl transferase-encoding gene; slr1691, NAD synthase-encoding gene; slr0787, nicotinamide nucleotide adenyl transferase-encoding gene; slr0788, nicotinamide phosphoribosyl transferase-encoding gene.
Cyanobacteria are the only prokaryotes capable of using light energy to perform plant-like oxygenic photosynthesis; these organisms often are used as production platforms in synthetic biology. For example, in recent years, research on biofuel and chemical production using cyanobacteria has been conducted. To enhance the productivity of these bacteria and the economic feasibility of bioproduction, most research in cyanobacteria has focused on increasing reducing power (that is, NADH or NADPH) by genetic improvement of cofactor metabolism (Hauf et al., 2013; Shabestary and Hudson, 2016; Wang et al., 2016; Zhou et al., 2016). The NADPH is involved in some cellular metabolism, e.g., the Calvin cycle, hydrogen biosynthesis, lipid biogenesis, amino acid biosynthesis and chlorophyll synthesis. Therefore, the molecular basis of NAD(P)(H) metabolism is crucial for planning a strategy to raise the yield of valuable substances.
In cyanobacteria, photosynthesis and respiration share some components of electron transport chain. So far, electron transport pathways in thylakoid membrane has been determined (summarized in Lea-Smith et al., 2016). In the case of photosynthesis, excitation energy trapping by PS II results in water splitting, and transport of electrons to the primary and secondary electron accepting quinone molecules QA and QB, respectively (Barthel et al., 2013). Following double reduction and protonation, electron is released from QB into the plastoquinone (PQ) pool and delivers electrons to the cytb6f complex (Kurisu et al., 2003). Cytb6f transfers one electron to either plastocyanin or cytochrome, and these small soluble electron carriers donate electrons to P700+ (Duran et al., 2004). In addition to respiration, it is now reported that many respiratory components may participate in PQ pool reduction including NAD(P)H dehydrogenase-like complex type I (NDH-1), succinate dehydrogenase (SDH) and one to three NDH-2s (Mi et al., 1992; Howitt et al., 1999; Cooley et al., 2000; Ohkawa et al., 2000). Furthermore, the cytochrome bd quinol oxidase, encoded by cydAB, has been identified in thylakoid of Synechocystis. Cytochrome bd quinol reduces oxygen with electrons presumably taken directly from the PQ pool (Berry et al., 2002; Ermakova et al., 2016). Finally, the aa3-type cytochrome oxidase complex, encoded by coxABC, can accept electrons from plastcyanin/cytochrome c6 to produce the H+ gradient (Howitt and Vermass, 1998). Plants perform above mentioned electron transports (photosynthesis and respiration) in chloroplast and mitochondria. While, in cyanobacteria, these two redox pathways occur in a single cell compartment (Figure 1A), and share with a several components such as PQ, cytb6f and plastcyanin/cytochrome c6. Currently, a central question has never addressed in cyanobacteria: how photosynthesis and respiration are regulated in a same membrane? In addition, most primary metabolic pathways, such as the Calvin-Benson cycle, the tricarboxylic acid (TCA) cycle, and glycolysis, also share the same cytosolic space (Knoop et al., 2010). Additionally, Synechocystis sp. PCC 6803 employ metabolic adaptative systems to provide alternative supplies of required substances. For example, under photoheterotrophic conditions (e.g., in medium containing glucose and 3-(3,4-dichlorophenyl)-1,1-demethylurea (DCMU), a compound that inhibits photosynthesis), and under low-light conditions (where illumination is too weak for photosynthesis to occur) in the presence of glucose, cyanobacteria utilize medium-supplied glucose as both an energy and carbon source, and generate NADPH through the oxidative pentose phosphate pathway to compensate for photosynthesis-derived NADPH (Narainsamy et al., 2013; Nakajima et al., 2014; Ueda et al., 2018). Based on these unique metabolic characteristics, cyanobacteria provide a unique tool for examining several relevant topics, including: (1) How do cyanobacteria control the amount of the redox pairs NADH/NAD+ and NADPH/NADP+ in mixed metabolism in the single compartment? (2) Which metabolic pathway is controlled by NAD(P)(H) under the various growth conditions in cyanobacteria?
This review focuses on the current state of knowledge of NAD(P)(H) metabolism in cyanobacteria. Notably, the physiological functions of NADKs, the enzymes that catalyze the synthesis of NADP+, are discussed.
Several genes involved in NAD+ biogenesis have been identified by comparative genomics in a model cyanobacterium, Synechocystis sp. PCC 6803 (Figure 1B; Gerdes et al., 2006). Additionally, four of the NAD biosynthetic enzymes, sll1916 (NaMNAT), slr1691 (NADS/GAT), slr0787 (NMPRT), slr0788 (NMNAT) were biochemically characterized (Raffaelli et al., 1999; Gerdes et al., 2006). The state of knowledge of the process of cyanobacterial NAD+ biogenesis can be summarized as follows. Firstly, aspartate serves as a precursor of the de novo pathway. The conversion of aspartate to nicotinate mononucleotide (NaMN) is implemented via three consecutive reactions catalyzed by L-aspartate oxidase (nadB), product of the Synechocystis sp. PCC 6803 sll0631 gene), quinolinate synthase (nadA), product of Synechocystis sp. PCC 6803 sll0622), and quinolinate phosphoribosyl transferase (nadC); product of Synechocystis sp. PCC 6803 slr0936). Next, NaMN is converted to nicotinate adenine dinucleotide (NaAD) by nicotinate-nucleotide adenylyl transferase (NaMNAT; product of the Synechocystis sp. PCC 6803 sll1916 gene). Subsequently, NaAD is converted to NAD+ by amidation, followed by phosphorylation of NAD+ to NADP+. In this final step of NAD+ biogenesis in cyanobacterium, NAD synthase (NADS/GAT; product of the Synechocystis sp. PCC 6803 slr1961 gene) requires glutamine as a source of an amide group for the ATP-mediated amidation reaction (Raffaelli et al., 1999). Such NAD+ synthesis pathway is consistent with all analyzed cyanobacteria (Gerdes et al., 2006) and Arabidopsis thaliana. Arabidopsis thaliana use a salvage pathway in which NaMN is synthesized from nicotinamide (Katoh et al., 2006; Wang and Pichersky, 2007). On the other hand, the routes for salvage or recycling of NAD+ vary significantly between cyanobacterial species (Gerdes et al., 2006). Moreover, Synechocystis sp. PCC 6803 possesses an alternative nicotinamide mononucleotide (NMN)-specific adenylyltransferase (encoded by slr0787), a novel bifunctional enzyme. Specifically, the N-terminal domain of the Slr0787 protein catalyzes NAD synthesis from NMN and ATP, while the C-terminal region of Slr0787 possesses Nudix hydrolase activity. Nudix hydrolases cleave an X-linked nucleoside diphosphate (where X is phosphate, sugar, nucleoside mono/diphosphate, etc.), an activity that is involved in the control of the cellular levels of metabolites and toxic compounds (Bessman et al., 1996). However, under standard growth conditions, the slr0787-deficient mutant shows no phenotype compared to wild-type cells (Gerdes et al., 2006). The presence of the bifunctional Slr0787 protein indicates that cyanobacteria have a special system for controlling NAD(P)(H) metabolism.
Furthermore, Synechocystis sp. PCC 6803 possesses a conversion nicotinamide phosphoribosyl transferase (NMPRT, the product of slr0788), which catalyzes nicotinamide to NMN. This observation indicates that Synechocystis sp. PCC 6803 can synthesize NAD+ from nicotinamide via slr0787 or slr0788. In contrast, the Synechocystis sp. PCC 6803 pathways for recycling and consuming NAD+ and NADP+ remain to be clarified. Additionally, Gerdes et al. (2006) showed that active uptake of nicotinamide is absent in Synechocystis sp. PCC 6803. Therefore, we believe that it will be necessary to study the contribution of the NAD-consuming pathway (including NAD consumption by ADP-ribosylating enzymes (Silman et al., 1995), NADP+ phosphatase, and Nudix hydrolases) in order to fully understand the homeostasis of NAD(P)(H) in Synechocystis sp. PCC 6803.
NADP+, the phosphorylated form of nicotinamide adenine dinucleotide (NAD), plays an essential role in many cellular processes. Notably, NADP+ is an electron mediator in the linear electron transfer chain of photosynthesis, where the NADP+ serves as a final acceptor of electrons in photosynthetic organisms (Hurley et al., 2002). NADP+ also is involved in primary metabolic reactions, such as those of the oxidative pentose phosphate pathway for the synthesis of NADPH, and those of the TCA cycle. Notably, in primary metabolism, NADP+ is an essential co-factor for the enzyme activities of glucose-6-phosphate dehydrogenase (G6PDH), 6-phosphogluconate dehydrogenase (6PGDH), and isocitrate dehydrogenase (ICD) in Synechocystis sp. PCC 6803 (Muro-Pastor and Florencio, 1992). The Synechocystis zwf mutant, in which G6PDH is inactive, becomes bleached under photoheterotrophic growth conditions and under light-activated heterotrophic conditions (Jansen et al., 2010). Deficiency for the icd gene is known to be lethal in Synechocystis sp. PCC 6803 (Muro-Pastor et al., 1996). Thus, NADP+-related enzymes have fundamental, essential functions required for this cyanobacterium to maintain carbon metabolism.
PntAB (pyridine nucleotide transdehydrogenase) is yet another important factor of NAD(P): NAD(P)H redox homeostasis maintenance which is located in the thylakoid membrane in Synechocystis (Kamarainen et al., 2017). PntAB is an integral membrane protein complex coupling the oxidation of NADH and concurrent reduction of NADP+ to proton translocation across the membrane. PntAB-deficient mutant exhibits growth defects under low-light and day-night rhythm in the presence of glucose. On the other hands, there was no apparent difference between the pntAB-deficient mutant and WT under the mixotrophic condition (20 μE/m2/s). Interestingly, sll1415-deficient mutant, one of the NADK-deficient mutants in Synechocystis, and pntAB-deficient mutant showed the similar phenotype under the day-night rhythm in the presence of glucose and the mixotrophic condition (20 μE/m2/s) (Ishikawa et al., 2016, 2019; Kamarainen et al., 2017). Therefore, pntAB may maintain the NADPH pool with sll1415 by unknown coordination system.
NAD kinase (NADK) phosphorylates NAD+ to NADP+,and is an activity that is present in all living organisms. In Arabidopsis thaliana, AtNADK2 produces NADP+ from NAD+ in the chloroplast. The nadk2 knockout mutant exhibits a pale-green phenotype and is hypersensitive to environmental stresses (Chai et al., 2005; Takahashi et al., 2009). The primary sequence of AtNADK2 includes a long N-terminal extension (compared to cyanobacterial homologs) that includes a chloroplast-targeting sequence as well as a calmodulin-binding site (Turner et al., 2004; Figure 2), and its additional N-terminal sequence exists in most plant chloroplast-localized species (Li et al., 2014). It is assumed that the NADP+ dynamics of the chloroplast are regulated by the enzyme activity of NADK2. On the other hand, AtNADK3, which localizes to the peroxisomal matrix (Waller et al., 2010), exhibits a different primary structure (Figure 2). Notably, NADK3 harbors a C-terminal extension (compared to homologs). To our knowledge, there have been no reports (to date) regarding the role of the NADK3 C-terminal region.
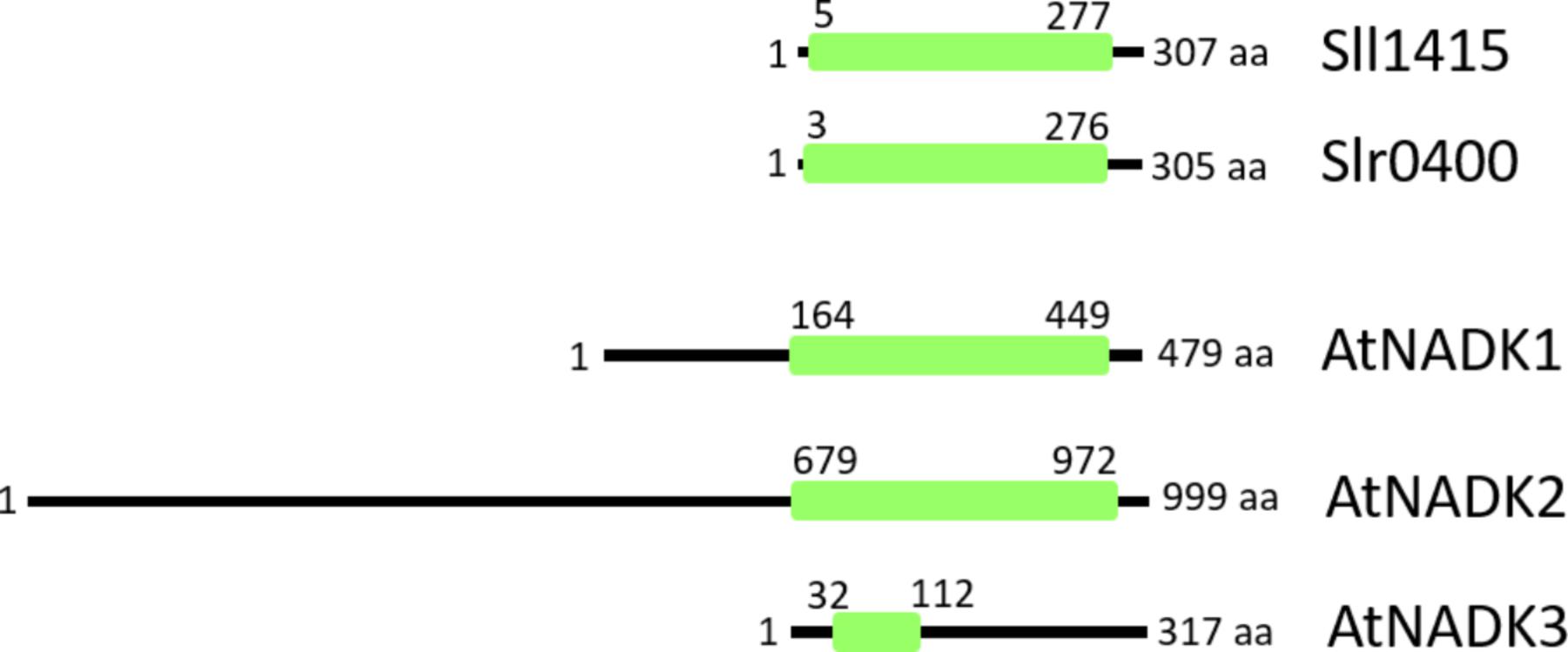
Figure 2. Comparison of primary structures of NADKs in Synechocystis sp. PCC 6803 (Sll1415 and Slr0400) and Arabidopsis (AtNADK1, AtNADK2, and AtNADK3). Conserved NADK regions (green box) span the entire length of the NADKs in Synechocystis sp. PCC 6803. In Arabidopsis, AtNADK1 and AtNADK2 possess N-terminal extensions, while, AtNADK3 possesses a C-terminal extension, compared to the various homologs. AtNADK1, At3G21070; AtNADK2, At1G21640; AtNADK3, At1G78590.
Based on BLAST searches of the cyanobacterial genomes available in the NCBI GenBank database, it was reported that almost all cyanobacteria possess two types of NADKs, despite the apparent lack of subcellular compartments within these cells (Gao and Xu, 2012). Consistent with these BLAST analyses, Synechocystis sp. PCC 6803 also harbors two NADK-encoding genes (sll1415 and slr0400). Both proteins include conserved amino acid sequence motifs for NAD binding (GGDG) (Raffaelli et al., 2004; Mori et al., 2005) and ATP binding (NE/D) (Liu et al., 2005). Moreover, the Sll1415 and Slr0400 proteins do not harbor N- and C-terminal extensions like those found in Arabidopsis (Figure 2). Therefore, in cyanobacteria, two short-type NADKs appear to suffice for controlling the NADP+/NAD+ ratio in cells.
When the proteomes of the 72 species registered in cyanobase1 were searched for NADK homologs, 69 species had two typical NADKs, such that one of could be classified as being of the Slr0400 type (Group 1) or the Sll1415 type (Group 2). On the other hand, 2 species (Atelocyanobacterium thalassa and Epithemia turgida) harbored only one NADK, which belonged to the Sll1415 type (Group 2 in Figure 3 and Table 1). Interestingly, in the case of E. turgida spheroid body (EtSB), EtSB has lost photosynthetic ability and is metabolically dependent on its host cell (Nakayama et al., 2014). Additionally, Atelocyanobacterium thalassa was found to lack the oxygen-evolving photosystem II, RuBisCo (ribulose-1,5-bisphophate carboxylase-oxygenase that fixes CO2) and the tricarboxylic acid cycle (Zehr et al., 2008; Tripp et al., 2010). Additionally, Atelocyanobacterium thalassa has a symbiotic association with a unicellular prymnesiophyte (Thompson et al., 2012), prymnesiophyte receives fixed N in exchange for transferring fixed carbon to Atelocyanobacterium thalassa. Thus, we speculate that slr0400 seems to be one of the key metabolic factors to understand the relationship between lost-photosynthesis, symbiotic and metabolically regulation in cyanobacteria. However, to our knowledge, there have been no reports (to date) regarding the role of the slr0400. Therefore, it remains why Atelocyanobacterium thalassa does not have NADK that group with Slr0400 in Figure 3. Based on our previous study, we speculate that slr0400 may have key role in photosynthesis and suppression of the heterotrophic metabolism in Synechocystis sp. PCC 6803. Interestingly, Synechococcus sp. UTEX 2973 encodes two NADKs, both of which can be classified into Group 2 based on protein phylogeny (Figure 3). Synechococcus sp. UTEX 2973 is a fast-growing cyanobacterium that has been proposed as a candidate biomass-producing platform (Yu et al., 2015). Moreover, the markers associated with fast-growth in this species were determined (Ungerer et al., 2018); that analysis revealed that a NADK-encoding locus is one of the genetic factors that contributes to fast-growth.

Figure 3. Phylogenetic tree of cyanobacterial NADKs. Most cyanobacteria possess two NADKs, representing one each of NADKs that group with Sll1415 (Group 2) or Slr0400 (Group 1). Bootstrap resampling analyses (100 times) with maximum likelihood was performed with PhyML (Guindon et al., 2010).
These genetic results raise several questions, including the following: (1) How do the typical types of NADKs (Group 1 and Group 2 in Figure 3) contribute to NAD(P)(H) homeostasis in cyanobacteria? (2) How is the NAD(P)(H) balance regulated in cyanobacteria possessing unusual NADKs (notably including Atelocyanobacterium thalassa, E. turgida and Synechococcus sp. UTEX 2973)?
Based on the phylogenetic tree (Figure 3), we hypothesized that the two types of cyanobacterial NADKs (Group 1 and Group 2) possess distinct roles in NAD(P)(H) metabolism. Therefore, we conducted metabolic analyses of Synechocystis sp. PCC 6803 harboring single mutations in either of the two NADK-encoding genes (Δ1415 or Δ0400). Notably, the wild-type (WT) strain and the single-mutant strains exhibited similar growth properties when cultured under photoautotrophic conditions (Ishikawa et al., 2016). However, Δ0400 (but not Δ1415) possessed a metabolic profile that differed from that of WT (Ishikawa et al., 2016). Specifically, Δ0400 showed metabolic changes in the levels of sugar phosphates (glucose-6-phosphate, ribulose-5-phosphate, 6-phosphogluconate, and fructose-6-phosphate), implying that the upper parts of the glycolytic pathway, the oxidative pentose phosphate pathway, and the Calvin cycle were affected by the disruption of slr0400 (Ishikawa et al., 2016).
The unicellular cyanobacterium Synechocystis sp. PCC 6803 is a model organism that can grow under a wide range of conditions, including photoautotrophy, photoheterotrophy in the presence of glucose (that is, the ability to grow under illumination even in the absence of photosynthesis), and photomixotrophy in the presence of glucose. Gao and Xu (2012) showed that a sll1415-deficient mutant exhibits growth deficiency under photoheterotrophic conditions. Those authors reported that a sll1415 null mutation affected the strain’s NADK activity and NADP(H) levels more severely than did a slr0400 null mutation. Furthermore, we reported that the sll1415-deficient mutant showed a growth-impaired phenotype under growth conditions that would typically yield photomixotrophy (with 12-h light/12-h dark cycling), and under light-activated heterotrophic condition (LAHG) conditions (in which cells are maintained in darkness but exposed to light for 15 min every 24 h) (Ishikawa et al., 2019). Moreover, we showed that WT cells exhibited approximately 2.2- and 1.8-fold increases in the levels of NADP+ and NADPH (respectively; compared to 0 h) at 96 h after transfer from autotrophic to photoheterotrophic conditions. Furthermore, analysis of sll1415 and slr0400 mRNA levels in WT during the acclimation to photoheterotrophic conditions suggested dynamic changes in NADP+ biosynthesis. Based on detailed molecular analysis, we showed that Sll1415 appears to have a specific role in the oxidative pentose phosphate pathway, sustaining cell growth under photoheterotrophic conditions (Ishikawa et al., 2019).
The NADK-encoding gene slr0400 also is an important factor in NAD(P)(H) homeostasis in cyanobacteria. When the slr0400 mutant was grown under photoautotrophic conditions, the amount of glycogen was decreased compared to that in WT. Additionally, the levels of the agp transcript (encoding ADP-glucose pyrophosphorylase) decreased in Δ0400 grown under photoautotrophic conditions (Ishikawa et al., 2019). Furthermore, the metabolomic profile of Δ0400 was different from those of WT and Δ1415 when grown under photoheterotrophic conditions (Ishikawa et al., 2016; Ishikawa et al., 2019). Notably, the intracellular levels of major metabolites (such as fructose-6-phosphate and 6-phosphogluconate) were decreased in Δ0400 (Ishikawa et al., 2016). Interestingly, Δ0400 exhibited an increased NAD+/NADH ratio and a decreased NADPH level (compared to WT and Δ1415) under photoautotrophic conditions (Ishikawa et al., 2019). Based on these data, we hypothesize that Δ0400 cells are forced to remain in the heterotrophic mode even in the absence of glucose.
NAD(P)(H) play an essential role in the metabolism of all organisms. However, it remains unclear how organisms maintain NAD(P)(H) homeostasis, and the physiological roles of NAD(P)(H) remain poorly characterized. Most cyanobacteria possess two structurally distinct NADKs, so it is assumed that these two protein types represent paralogs, that is, the two have distinct roles in cyanobacterial metabolism. As reviewed in this paper, one of the cyanobacterial NADKs (corresponding to Sll1415 of Synechocystis sp. PCC 6803) is critical for photoheterotrophic growth. However, it is not apparent why cyanobacteria, which have simple (prokaryotic) cell structures, have multiple NADKs, and the role of the separate NADK paralogs remain unclear. We believe that functional differences in the two typical NADKs must be crucial for cyanobacterial metabolic adaptation through control of NAD(P)(H) homeostasis. To elucidate the significance of NADKs in cyanobacteria, further characterization of the biochemical properties of NADKs, subcellular localizations of NADKs, and membrane permeability of NAD(P)(H) will need to perform.
YI wrote this review. Both authors approved the manuscript for publication.
This work was supported by the KAKENHI Grant Numbers 17H05714, 26292190, and T18H02165 to MK-Y, and KAKENHI Grant Number 18J11305 to YI.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Kaneko, Ms. Tanaka, and Ms. Tsuji (Saitama University, Saitama) for technical advises on transmission electron microscopy.
Alberts, B., Johnson, A., and Lewis, J. (2002). Catalysis and the Use of Energy by Cells, Molecular Biology of the Cell, 4th Edn. New York, NY: Garland Science.
Barthel, S., Bernat, G., Seidel, T., Rupprecht, E., Kahmann, U., and Schneider, D. (2013). Thylakoid membrane maturation and PSII activation are linked in greening Synechocystis sp. PCC 6803 cells. Plant Phys. 163, 1037–1046. doi: 10.1104/pp.113.224428
Bernhardt, K., Wilkinson, S., Weber, A. P., and Linka, N. (2012). Peroxisomal carrier delivers NAD + and contributes to optimal fatty acid degradation during storage oil mobilization. Plant J. 69, 1–13. doi: 10.1111/j.1365-313x.2011.04775.x
Berry, S., Schneider, D., Vermass, W. F., and Ronger, M. (2002). Electron transport routes in whole cells of Synechocystis sp. strain PCC 6803: the role of the cytochrome bd-type oxidase. Biochemistry 41, 3422–3429. doi: 10.1021/bi011683d
Bessman, M. J., Frick, D. N., and O’Handley, S. F. (1996). The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271, 25059–25062. doi: 10.1074/jbc.271.41.25059
Chai, M. F., Chen, Q. J., An, R., Chen, Y. M., Chen, J., and Wang, X. C. (2005). NADK2, an Arabidopsis chloroplastic NAD kinase, plays a vital role in both chlorophyll synthesis and chloroplast protection. Plant Mol. Biol. 59, 553–564. doi: 10.1007/s11103-005-6802-y
Cooley, J. W., Howitt, C. A., and Vermass, W. F. (2000). Succinate:quinol oxidoreductases in the cyanobacterium Synechocystis sp. PCC 6803: presence and function in metabolism and electrontransport. J. Bacteriol. 182, 714–722. doi: 10.1128/jb.182.3.714-722.2000
Duran, R. V., Hervas, M., De La Rosa, M. A., and Navarro, J. A. (2004). The efficient functioning of photosynthesis and respiration in Synechocystis sp. PCC 6803 strictly requires the presence of either cytochrome c6 or plastocyanin. J. Biol. Chem. 279, 7229–7233. doi: 10.1074/jbc.m311565200
Dutilleul, C., Lelarge, C., Prioul, J. L., De Paepe, R., Foyer, C. H., and Noctor, G. (2005). Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrarte assimilation and the integration of carbon and nitrogen metabolism. Plant Phys. 139, 64–78. doi: 10.1104/pp.105.066399
Ermakova, M., Huokko, T., Richaud, P., Bersanini, L., Howe, C. J., Lea-smith, D. J., et al. (2016). Distinguishing the roles of thylakoid respiratory terminal oxidases in the cyanobacterium Synechocystis sp. PCC 6806. Plant Phys. 171, 1307–1319. doi: 10.1104/pp.16.00479
Gao, H., and Xu, X. (2012). The cyanobacterial NAD kinase gene sll1415 is required for photoheterotrophic growth and cellular redox homeostasis in Synechocystis sp. strain PCC 6803. J. Bacteriol. 194, 218–224. doi: 10.1128/JB.05873-11
Gerdes, S. Y., Kurnasov, O. V., Shatalin, K., Polanuyer, B., Sloutsky, R., Vonstein, V., et al. (2006). Comparative genomics of NAD biosynthesis in cyanobacteria. J. Bacteriol. 188, 3012–3023. doi: 10.1128/jb.188.8.3012-3023.2006
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Mol. Biol. Evol. 34, 307–321. doi: 10.1093/sysbio/syq010
Hashida, S. N., Itami, T., Takahara, K., Hirabayashi, T., Uchimiya, H., and Kawai-Yamada, M. (2016). Increased rate of NAD metabolism shortens plant longevity by accelerating developmental senescence in Arabidopsis. Plant Cell Physiol. 57, 2427–2439. doi: 10.1093/pcp/pcw155
Hauf, W., Schlebusch, M., Huge, J., Kopka, J., Hagemann, M., and Forchhammer, K. (2013). Metabolic changes in Synechocystis PCC6803 upon nitrogen-starvation: excess NADPH sustains polyhydroxybutyrate accumulation. Metabolites 3, 101–118. doi: 10.3390/metabo3010101
Howitt, C. A., Udall, P. K., and Vermass, W. F. (1999). Type 2 NADH dehydrogenases in the cyanobacterium Synechocystis sp. PCC 6803 are involved in regulation rather than respiration. J. Bacteriol. 181, 3994–4003.
Howitt, C. A., and Vermass, W. F. (1998). Quinol and cytochrome oxidases in the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry 37, 17944–17951. doi: 10.1021/bi981486n
Hurley, J. K., Morales, R., Martinez-Julez, M., Brodie, T. B., Medina, M., Gomez-Moreno, C., et al. (2002). Structure-function relationships in Anabaena ferredoxin/ferredoxin: NADP (+) reductase electron transfer: insights from site-directed mutagenesis, transient absorption spectroscopy and X-ray crystallography. Biochem. Biophys. Acta 1554, 5–21. doi: 10.1016/s0005-2728(02)00188-3
Ishikawa, Y., Miyagi, A., Haishima, Y., Ishikawa, T., Nagano, M., Yamaguchi, M., et al. (2016). Metabolomic analysis of NAD kinase-deficient mutants of the cyanobacterium Synechocystis sp. PCC 6803. J. Plant Physiol. 205, 105–112. doi: 10.1016/j.jplph.2016.09.002
Ishikawa, Y., Miyagi, A., Ishikawa, T., Nagano, M., Yamaguchi, M., Hihara, Y., et al. (2019). One of the NAD kinases, sll1415, is required for the glucose metabolism of Synechocystis sp. PCC 6803. Plant J. 98, 654–666. doi: 10.1111/tpj.14262
Jansen, T., Kurian, D., Raksajit, W., York, S., Summers, M. L., and Maenpaa, P. (2010). Characterization of trophic changes and a functional oxidative pentose phosphate pathway in Synechocystis sp. PCC 6803. Acta Physiol. Plant 32, 511–518. doi: 10.1007/s11738-009-0428-7
Kamarainen, J., Huokko, T., Kreula, S., Jones, P. R., Aro, E. M., and Kallio, P. (2017). Pyridine nucleotide transhydrogenase PntAB is essential for optimal growth and photosynthetic integrity under low-light mixotrophic conditions in Synechocystis sp. PCC 6803. New Phytol. 214, 194–204. doi: 10.1111/nph.14353
Katoh, A., Uenohara, K., Akita, M., and Hashimoto, T. (2006). Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Phys. 141, 851–857. doi: 10.1104/pp.106.081091
Knoop, H., Zilliges, Y., Lockau, W., and Steuer, R. (2010). The metabolic network of Synechocystis sp. PCC 6803: systemic properties of autotrophic growth. Plant Phys. 154, 410–422. doi: 10.1104/pp.110.157198
Kurisu, G., Zhang, J. L., and Smith, W. A. (2003). Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science 302, 1009–1014. doi: 10.1126/science.1090165
Lea-Smith, D. J., Bombelli, P., Vasudevan, R., and Howe, C. J. (2016). Photosynthetic, respiratory and extracellular electron transport pathways in cyanobacteria. Biochim. Biophys. Acta Bioenerg. 1857, 247–255. doi: 10.1016/j.bbabio.2015.10.007
Li, W. Y., Wang, X., Li, R., Li, W. Q., and Chen, K. M. (2014). Genome-wide analysis of the NADK gene family in plants. PLoS One 9:e101051. doi: 10.1371/journal.pone.0101051
Liu, J., Lou, Y., Yokota, H., Adams, P. D., Kim, R., and Kim, S. H. (2005). Crystal structures of an NAD kinase from Archaeoglobus fulgidus in complex with ATP, NAD, or NADP. J. Mol. Biol. 354, 289–303. doi: 10.1016/j.jmb.2005.09.026
Maruta, T., Ogawa, T., Tsujimura, M., Ikemoto, K., Yoshida, T., Takahashi, H., et al. (2016). Loss-of-function of an Arabidopsis NADPH pyrophosphohydrolase, AtNUDX19, impacts on the pyridine nucleotides status and confers photooxidative stress tolerance. Sci. Rep. 6, 37432. doi: 10.1038/srep37432
Mi, H., Endo, T., Schreiber, U., Ogawa, T., and Asada, K. (1992). Electron donation from cyclic, and respiratory flows to the photosynthetic intersystem chain is medeated by pyridine nucleotide dehydrogenase in the cyanobacterium Synechocystis PCC. Plant Cell Physiol. 33, 1233–1237.
Mori, S., Yamasaki, M., Maruyama, Y., Momma, K., Kawai, S., Hashimoto, W., et al. (2005). NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex. Biochem. Biophys. Res. Commun. 327, 500–508. doi: 10.1016/j.bbrc.2004.11.163
Muro-Pastor, M., Reyes, J., and Florencio, F. (1996). The NADP+-isocitrate dehydrogenase gene (icd) is nitrogen regulated in cyanobacteria. J. Bacteriol. 178, 4070–4076. doi: 10.1128/jb.178.14.4070-4076.1996
Muro-Pastor, M. I., and Florencio, F. J. (1992). Purification and properties of NADP+-isocitrate dehydrogenase from the unicellular cyanobacterium Synechocystis sp. PCC 6803. Eur. J. Biochem. 203, 99–105. doi: 10.1111/j.1432-1033.1992.tb19833.x
Nakajima, T., Kajihata, S., Yoshikawa, K., Matsuda, F., Furusawa, C., Hirasawa, T., et al. (2014). Integrated metabolic flux and omics analysis of Synechocystis sp. PCC 6803 under mixotrophic and photoheterotrophic conditions. Plant Cell Physiol. 55, 1605–1612. doi: 10.1093/pcp/pcu091
Nakayama, T., Kamikawa, R., Tanifuji, G., Kashiyama, Y., Ohkouchi, N., Archibald, J. M., et al. (2014). Complete genome of a nonphotosynthetic cyanobacterium in a diatom reveals recent adaptations to an intracellular lifestyle. Proc. Natl. Acad. Sci. U.S.A. 111, 11407–11412. doi: 10.1073/pnas.1405222111
Narainsamy, K., Cassier-Chauvat, C., Junot, C., and Chauvat, F. (2013). High performance analysis of the cyanobacterial metabolism via liquid chromatography coupled to a LTQ-Orbitrap mass spectrometer: evidence that glucose reprograms the whole carbon metabolism and triggers oxidative stress. Metabolomics 9, 21–32. doi: 10.1007/s11306-011-0382-4
Ohkawa, H., Pakrasi, H. B., and Ogawa, T. (2000). Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain. PCC 6803. J. Biol. Chem. 275, 31630–31634. doi: 10.1074/jbc.m003706200
Pétriacq, P., de Bont, L., Hager, J., Didierlaurent, L., Mauve, C., Guérard, F., et al. (2012). Inducible NAD overproduction in Arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to Pst-AvrRpm1. Plant J. 70, 650–665. doi: 10.1111/j.1365-313X.2012.04920.x
Pétriacq, P., Ton, J., Patrit, O., Tcherkez, G., and Gakière, B. (2016). NAD acts as an integral regulator of multiple defense layers., NAD acts as an integral regulator of multiple defense layers. Plant Phys. 172, 1465–1479. doi: 10.1104/pp.16.00780
Raffaelli, N., Finaurini, L., Mazzola, F., Pucci, L., Sorci, L., Amici, A., et al. (2004). Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by site-directed mutagenesis. Biochemistry 43, 7610–7617. doi: 10.1021/bi049650w
Raffaelli, N., Lorenzi, T., Amici, A., Emanuelli, M., Ruggieri, S., and Magni, G. (1999). Synechocystis sp. slr0787 protein is a novel bifunctional enzyme endowed with both nicotinamide mononucleotide adenylyltransferase and ‘Nudix’ hydrolase activities. FEBS Lett. 444, 222–226. doi: 10.1016/s0014-5793(99)00068-x
Schippers, J. H., Nunes-Nesi, A., Apetrei, R., Hille, J., Fernie, A. R., and Dijkwel, P. P. (2008). The Arabidopsis onset of leaf death5 mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. Plant Cell 20, 2909–2925. doi: 10.1105/tpc.107.056341
Shabestary, K., and Hudson, E. P. (2016). Computational metabolic engineering strategies for growth-coupled biofuel production by Synechocystis. Metab. Eng. Commun. 3, 216–226. doi: 10.1016/j.meteno.2016.07.003
Sienkiewicz-Porzucek, A., Sulpice, R., Osorio, S., Krahnert, I., Leisse, A., Urbanczyk-Wochniak, E., et al. (2010). Mild reductions in mitochondrial NAD-dependent isocitrate dehydrogenase activity result in altered nitrate assimilation and pigmentation but do not impact growth. Mol. Plant 3, 156–173. doi: 10.1093/mp/ssp101
Silman, N. J., Carr, N. G., and Mann, N. H. (1995). ADP-ribosylation of glutamine synthetase in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 177, 3527–3533. doi: 10.1128/jb.177.12.3527-3533.1995
Takahara, K., Kasajima, I., Takahashi, H., Hashida, S. N., Itami, T., Onodera, H., et al. (2010). Metabolome and photochemical analysis of rice plants overexpressing Arabidopsis NAD kinase gene. Plant Phys. 152, 1863–1873. doi: 10.1104/pp.110.153098
Takahashi, H., Takahara, K., Hashida, S. N., Hirabayashi, T., Fujimori, T., Kawai-Yamada, M., et al. (2009). Pleiotropic modulation of carbon and nitrogen metabolism in Arabidopsis plants overexpressing the NAD kinase2 gene. Plant Phys. 151, 100–113. doi: 10.1104/pp.109.140665
Thompson, A. W., Foster, R. A., Krupke, A., Carter, B. J., Musat, N., Vaulot, D., et al. (2012). Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337, 1546–1550. doi: 10.1126/science.1222700
Tripp, H. J., Bench, S. R., Turk, K. A., Foster, R. A., Desany, B. A., Niazi, F., et al. (2010). Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature 464, 90–94. doi: 10.1038/nature08786
Turner, W. L., Waller, J. C., Vanderbeld, B., and Snedden, W. A. (2004). Cloning and characterization of two NAD kinases from Arabidopsis identification of a calmodulin binding isoform. Plant Phys. 135, 1243–1255. doi: 10.1104/pp.104.040428
Ueda, K., Nakajima, T., Yoshikawa, K., Toya, Y., Matsuda, F., and Shimizu, H. (2018). Metabolic flux of the oxidative pentose phosphate pathway under low light conditions in Synechocystis sp. PCC 6803. J. Biosci. Bioeng. 126, 38–43. doi: 10.1016/j.jbiosc.2018.01.020
Ungerer, J., Wendt, K. E., Hendry, J. I., Maranas, C. D., and Pakrasi, H. B. (2018). Comparative genomics reveals the molecular determinants of rapid growth of the cyanobacterium Synechococcus elongatus UTEX 2973. Proc. Natl. Acad. Sci. U.S.A. 115, E11761–E11770. doi: 10.1073/pnas.1814912115
Waller, J. C., Dhanoa, P. K., Schumann, U., Mullen, R. T., and Snedden, W. A. (2010). Subcellular and tissue localization of NAD kinases from Arabidopsis: compartmentalization of de novo NADP biosynthesis. Planta 231, 305–317. doi: 10.1007/s00425-009-1047-7
Wang, G., and Pichersky, E. (2007). Nicotinamidase participates in the salvage pathway of NAD biosynthesis in Arabidopsis. Plant J. 49, 1020–1029. doi: 10.1111/j.1365-313x.2006.03013.x
Wang, X., Liu, W., Xin, C., Zheng, Y., Cheng, Y., Sun, S., et al. (2016). Enhanced limonene production in cyanobacteria reveals photosynthesis limitations. Proc. Natl. Acad. Sci. U.S.A. 113, 14225–14230. doi: 10.1073/pnas.1613340113
Yu, J., Liberton, M., Cliften, P. F., Head, R. D., Jacobs, J. M., Smith, R. D., et al. (2015). Synechococcus elongatus UTEX 2973, a fast-growing cyanobacterial chassis for biosynthesis using light and CO2. Sci Rep. 5:8132. doi: 10.1038/srep08132
Zehr, J. P., Bench, S. R., Carter, B. J., Hewson, I., Niazi, F., Shi, T., et al. (2008). Globally distributed uncultivated oceanic N2-fixing cyanobacteria lack oxygenic photosystem II. Science 322, 1110–1112. doi: 10.1126/science.1165340
Keywords: cyanobacteria, Synechocystis, NAD, NAD kinase, metabolism
Citation: Ishikawa Y and Kawai-Yamada M (2019) Physiological Significance of NAD Kinases in Cyanobacteria. Front. Plant Sci. 10:847. doi: 10.3389/fpls.2019.00847
Received: 20 March 2019; Accepted: 13 June 2019;
Published: 27 June 2019.
Edited by:
Alisdair Fernie, Max Planck Institute of Molecular Plant Physiology, GermanyReviewed by:
David John Lea-Smith, University of East Anglia, United KingdomCopyright © 2019 Ishikawa and Kawai-Yamada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maki Kawai-Yamada, bWthd2FpQG1haWwuc2FpdGFtYS11LmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.