
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 19 June 2019
Sec. Crop and Product Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00792
This article is part of the Research Topic Bioactive Compounds Biosynthesis and Metabolism in Fruit and Vegetables View all 12 articles
Tomato fruit ripening is a complex process, which determines the formation of fruit quality. Many factors affect fruit ripening, including environmental conditions and genetic factors. Transcription factors (TFs) play key roles in regulating fruit ripening and quality formation. Current studies have found that the TDR4 gene is an important TF for tomato fruit ripening, but its effects on fruit metabolism and quality are less well studied. In this study, suppression of TDR4 gene expression obtained through virus-induced gene silencing (VIGS) technology resulted in an orange pericarp phenotype. Transcriptomic analysis of TDR4-silenced fruit showed changes in the expression of genes involved in various metabolic pathways, including amino acid and flavonoid biosynthesis pathways. Metabolomic analysis showed that levels of several amino acids including phenylalanine and tyrosine, and organic acids were reduced in TDR4-silenced fruit, while α-tomatine accumulated in TDR4-silenced fruit. Taken together, our RNA-seq and metabolomics analyses of TDR4-silenced fruit showed that TDR4 is involved in ripening and nutrient synthesis in tomato fruit, and is therefore an important regulator of fruit quality.
Fruit is an important source of human healthy diet which can provide vitamins, minerals, and a wide range of bioactive compounds, including antioxidant carotenoids and various polyphenols (Seymour et al., 2013). The quality and nutrition of fresh fruits are gradually formed during ripening. Studying the molecular mechanism of fruit ripening is an important way to understand the formation of fruit quality. Fruit ripening is a complex biological process to form delicious and nutritious fruits for attracting animals to eat and spread seeds (Martel et al., 2011). Some general ripening-associated changes take place among some fruit species, including the cell wall degradation for fruit softening, alteration of the composition and levels of secondary metabolites, such as pigments, flavors, and aromas during fruit ripening (Martel et al., 2011). These changes are influenced by multiple genetic and biochemical pathways that are regulated by several critical transcription factors (TFs) (Giovannoni, 2007).
The tomato (Solanum lycopersicum) is the main horticultural crops and it is hot popular food for consumers. Tomato is considered as an ideal model material for studying fleshy fruit ripening (Klee and Giovannoni, 2011; Karlova et al., 2014). In climacteric fruits, including tomatoes, increased ethylene production is required for the onset of ripening (Barry and Giovannoni, 2007). During fruit development and ripening, the biosynthesis and signal transduction of ethylene are both regulated by several TFs, including RIPENING INHIBITOR (MADS-RIN), COLORLESS NON-RIPENING (CNR), NON-RIPENING (NOR), TOMATO AGAMOUS-LIKE1 (TAGL1), NOR-like1, and APETALA2a (AP2a) (Vrebalov et al., 2002, 2009; Manning et al., 2006; Giovannoni, 2007; Karlova et al., 2011; Gao et al., 2018b, 2019). TDR4/FUL1 and its homolog MBP7/FUL2 are MADS-box family TFs with high sequence similarity to Arabidopsis FRUITFULL. In contrast to the above-mentioned TFs, TDR4/FUL1 and MBP7/FUL2 do not regulate ethylene biosynthesis but affect fruit ripening in an ethylene-independent manner (Bemer et al., 2012). A previous study revealed that TDR4/FUL1 mRNA and protein accumulate during ripening in tomato fruit, while MBP7/FUL2 mRNA and protein accumulate during the pre-ripening stage and throughout ripening process (Shima et al., 2013). RNAi-silencing of each of the FUL homologs independently results in very mild changes to tomato fruit pigmentation, while the silencing of both genes results in an orange ripe fruit with highly reduced levels of lycopene, suggesting that FUL1/TDR4 and FUL2/MBP7 possess redundant functions in fruit ripening (Bemer et al., 2012). The expression of genes involved in cell wall modification, cuticle production, volatile production, and glutamate accumulation was also altered in TDR4 silencing tomato fruit (Bemer et al., 2012). Chromatin immunoprecipitation coupled with microarray analysis (ChIP-chip) revealed that FUL homologs take part in many biological processes through the regulation of ripening-related gene expression, both in cooperation with and independent of RIN (Fujisawa et al., 2014).
In order to further study the effect of TDR4 on tomato quality metabolism, we utilized virus-induced gene silencing (VIGS) to silence TDR4 in tomato fruit. Analysis of transcripts and metabolites of TDR4-silened fruit indicated that it was involved in the metabolism of several amino acids and biosynthesis of secondary metabolites, altering fruit nutrient levels and flavor. The result shows that TDR4 regulates the nutrient levels and quality of tomato fruit.
Tomato plants (S. lycopersicum “Ailsa Craig”) were planted in commercial tomato-cultivated soil and grown under standard glasshouse conditions of 16-h day length and 25°C, with a night temperature of 18°C with 75% relative humidity. Flowers were tagged at 1 day post-anthesis (DPA). Ten plants are for control and 10 plants were used to silence TDR4 gene; each plant was no less than 15 fruits.
The tobacco rattle virus (TRV)-based vectors pTRV1 and pTRV2 were used for VIGS. To construct a pTRV2-TDR4 recombinant, a 360-bp EcoRI/BamHI-containing DNA fragment of the TDR4 gene, corresponding to nucleotides 323–682 (NM_001247244.2), was amplified from tomato fruit complementary DNA (cDNA) using primers TDR4-VIGS-For and TDR4-VIGS-Rev (Supplementary Table S1). The resulting products and pTRV2 vector were digested with EcoRI/BamHI and ligated by T4 ligase.
The VIGS assay was carried out as previously described (Fu et al., 2005) with slight modification. All plant inoculations were performed using a 1:1 (v/v) mixture of two Agrobacterium tumefaciens GV3101 cultures, one containing the pTRV1 vector and the other containing the pTRV2 or pTRV2-derived vector. Bacterial clones were grown overnight at 28°C in Luria-Bertani medium containing 10 mM MES and 20 mM acetosyringone with kanamycin, gentamycin, and rifampicin antibiotics. They were then harvested and transferred to the infiltration medium [10 mM MgCl2, 10 mM MES (pH 5.6), 200 mM acetosyringone] to a final OD600 of 6.0. For co-infiltration studies, 1:1 mixtures of pTRV1, pTRV2-00, or pTRV2-TDR4 were used. The Agrobacterium mixture was injected into the carpopodium of the tomato fruit at 7–10 DPA after pollination using a 1-ml syringe with a syringe needle. The control fruits were infected by A. tumefaciens containing a pTRV2 empty vector, and the TDR4-silenced fruits were infected by A. tumefaciens containing a pTRV2-TDR4 vector. Each infected fruit was not less than 100 from 10 different plants.
Total RNA was extracted from the fruit pericarp of TRV2-00 infected control fruits and TRV2-TDR4 silenced fruits (three biological replicates in which each sample was collected from six different fruits) using a RNeasy MiniKit (Qiagen, Hilden, Germany) (Wang et al., 2016). RNA integrity was evaluated on 1% agarose gels stained with ethidium bromide (EB). RNA concentrations were measured using a Nano Photometer® spectrophotometer (Implen, CA, United States). cDNA libraries were generated using the NEBNext® Ultra RNA Library Prep Kit for Illumina® (New England Biolabs, Ipswich, MA, United States) following the manufacturer’s instructions. Briefly, mRNA was enriched using oligo (dT)-attached magnetic beads. Fragmentation was performed by divalent cations in NEBNext First Strand Synthesis Reaction Buffer. These fragments were used to synthesize first-strand cDNA using random hexamer primers and M-MuLV Reverse Transcriptase. Then, second-strand cDNA synthesis was achieved using DNA Polymerase I and RNase H. Exonuclease/polymerase activities were used to convert overhangs into blunt ends. In order to select cDNA fragments of the appropriate size, library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, MA, United States). USER Enzyme (New England Biolabs) was subsequently used with size-selected, adaptor-ligated cDNA. Then, PCR was carried out with Phusion High-Fidelity DNA polymerase, universal PCR primers, and Index (X) Primer. Finally, PCR products were purified, and library quality was evaluated on the Agilent Bioanalyzer 2100 system (Palo Alto, CA, United States). Clustering of the index-coded samples was performed on a cBot Cluster Generation System using the HiSeq 4000 PE Cluster Kit (Illumina, San Diego, CA, United States) according to the manufacturer’s instructions. Then, library preparations were sequenced on an Illumina HiSeq 4000 platform, and 150-bp paired-end reads were generated.
We used cutadapt1 and the FASTX-Toolkit2 to trim raw reads in order to remove barcode and adaptor sequences, and the resulting clean reads were checked for quality using a threshold of Q < 20. Clean reads from each library were aligned to the tomato reference genome (SGN release version SL2.503) using TopHat4. We used Cufflinks5 to assemble reads with fewer than two mismatches. Differentially expressed genes (DEGs) between pTRV2-TDR4 andpTRV2-00 were identified with the following criteria: fold-change ≥ 2 and Q-value < 0.05. Clean reads of RNA-seq were deposited in the National Center for Biotechnology Information Sequence Read Archive6 under accession number SRP201254.
To assess the distribution of DEG functions, Gene Ontology (GO) enrichment analysis was performed using WEGO7. FASTA format files containing DEG cDNA sequences were obtained using Perl scripts, and then pathway analysis was conducted with the Kyoto Encyclopedia of Genes and Genomes (KEGG) in KOBAS8, based on native BLAST tools and organism annotation libraries.
Total RNA (2 μg) from three biological replicates was purified and reverse-transcribed into cDNA using TranScript One-step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) with oligo(dT). Then, qRT-PCR was performed with SYBR Green PCR SuperMix (TransGen Biotech) on a Bio-Rad Real-Time PCR System CFX96 (Bio-Rad, Hercules, CA, United States), using a tomato actin gene as a reference gene. All primer sequences are listed in Supplementary Table S1. The reaction proceeded as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. The fluorescence signal was monitored automatically at each cycle. Relative expression levels of specific mRNAs were measured using the 2-ΔΔCt method. Standard errors were calculated based on a minimum of three biological replicates.
Frozen TRV2-00 and TRV2-TDR4 tomato pericarp samples (six biological replicates) were milled into powder using a mortar and pestle, and the powder from each sample was weighed. Next, 100 mg was suspended in either 1.0 ml pure methanol or 1.0 ml 75% aqueous methanol for the extraction of lipid-soluble and water-soluble metabolites, respectively. Both types of methanol contained 20 mg l-1 lidocaine and 20 g l-1 CHAPS. The suspensions were vortexed and then extracted at 4°C overnight. Following centrifugation at 12,000 × g for 10 min, the supernatants of both the lipid-soluble and water-soluble metabolites were collected and mixed in a ratio of 1:1 before being filtered through a 0.45-μm membrane and subjected to LC-MS analysis.
A high performance liquid chromatography unit, equipped with a photodiode array detector (HPLC-20A, Shimadzu, Japan), was used to analyze the metabolites in the tomato extract. The separation of metabolites was carried out under the following conditions: column: Eclipse XDB-C18 (3.0 mm × 50 mm); solvent A: water with 0.2% formic acid; solvent B: acetonitrile; gradient program: 95:5 A:B (v/v) at 0 min, 5:95 A:B at 12 min, 5:95 A:B at 15 min, 95:5 A:B at 15.1 min, 95:5 A:B at 22 min; flow rate: 0.2 ml min-1; temperature: 45°C; injection volume: 2 μl. The masses of the eluted compounds ranging from 50 to 1500 m z-1 were monitored with a Triple Quad LC/MS equipped with an electrospray ionization (ESI) source.
Quantitative detection was performed using an UHPLC-ESI-QQQ-MS (Agilent 1290 and 6460 triple quadrupole mass spectrometry series). An ESI source working either in positive or negative ion mode was used for all MS analyses, with nitrogen as the drying agent. The MS conditions in positive mode were as follows: HV voltage: 4000 kV; capillary: 7 μg; nozzle voltage: 500 V; delta EMV: 300 V; gas flow: 5 l min-1; gas temperature: 400°C; sheath gas flow: 11 l min-1. Collision energy was optimized based on the standards. Helium was used as the collision gas for collision-induced dissociation (CID). Quantification was performed using the multiple reaction monitoring (MRM) mode under unit mass-resolution conditions. The data were processed with MassHunter software.
To silence TDR4 gene and analyze its effect on tomato fruit metabolism, a mixture of Agrobacterium cultures containing pTRV-TDR4 and pTRV1 was injected into the carpopodium of the tomato fruit at 7–10 days after pollination using a 1-ml syringe with a needle. Around 35 days post-Agro-injection, approximately 35 tomato fruits injected with pTRV-TDR4 failed to turn red and developed an orange phenotype at the red ripening (RR) stage. All control fruit injected with A. tumefaciens containing pTRV1 and pTRV2-00 turned red normally, like the wild-type fruit (Figure 1A). Quantitative real-time PCR (qRT-PCR) was performed to confirm TDR4 silencing. The primers that anneal to the TDR4 gene outside the region targeted for silencing were used (Figure 1B). In pTRV-TDR4 injected fruits, the TDR4 message was reduced by more than 80% compared with the TRV injected controls (Figure 1C). The level of actin gene RNA was similar in TRV-TDR4 and TRV alone injected tissue and served as an internal control for RNA quality and RT-qPCR. Based on the above results, we can conclude that the TDR4 gene has been successfully silenced in tomato fruit and TDR4-silenced fruits can be used for subsequent studies on the effects of TDR4 gene on tomato fruit metabolism.
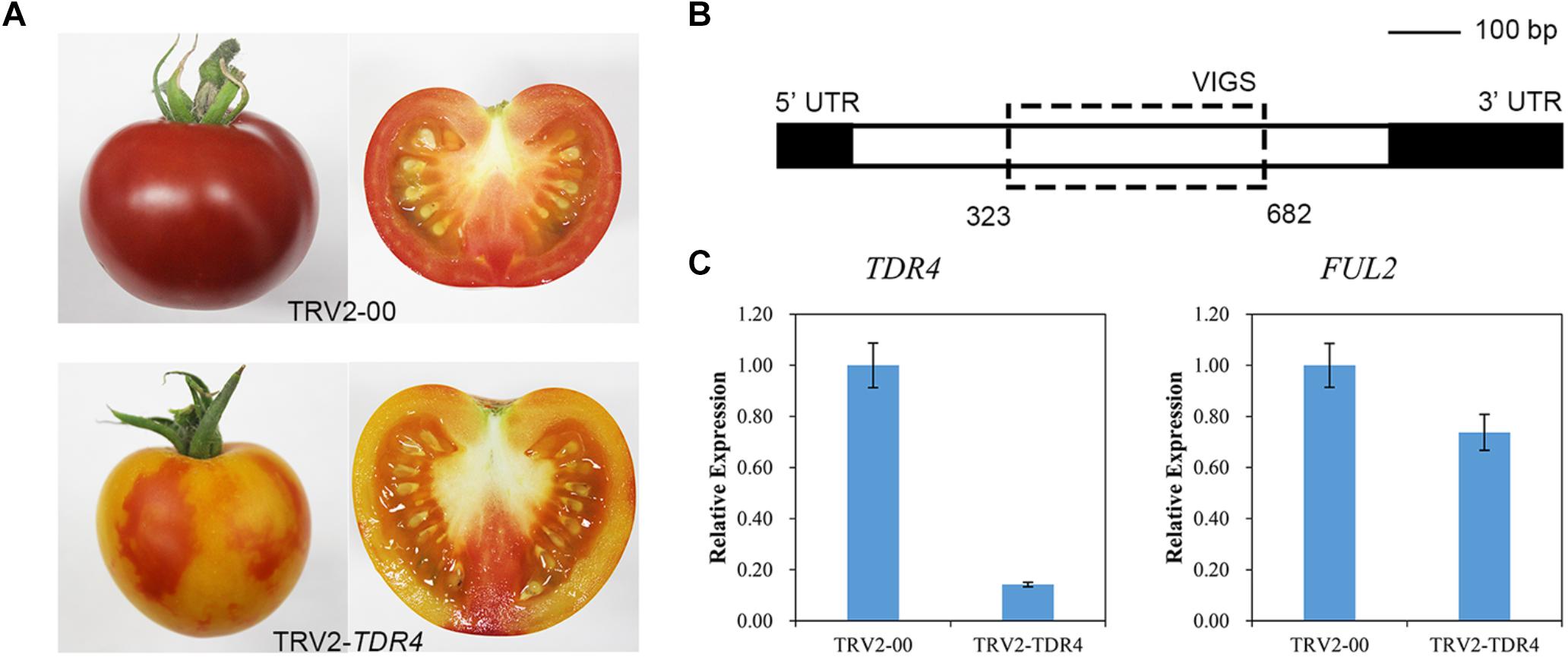
Figure 1. VIGS in tomato fruit. (A) Phenotype of TDR4-silenced tomato fruit. TRV2-00 was used as a control. (B) Schematic representation of the structure of the TDR4 coding sequence and the positions of the fragments used for VIGS. (C) Silencing efficiency of TDR4 and FUL2 genes.
To test the molecular consequences of silencing the TDR4 gene in TDR4-silenced fruit, we compared the gene expression levels in the pericarp of TDR4-silenced fruit with that in control pericarp at red stage using strand-specific mRNA sequencing. The result showed that all clean reads were mapped and aligned against the tomato reference genome (ITAG2.4). Within each file, 79.1 ± 2.1% of the reads were uniquely aligned, suggesting that the sequencing results were effective and reliable (Table 1).
Using cutoff criteria with an expression ratio of ≥2 and P < 0.05 between TDR4-silencd and control tissues, analysis of DEGs revealed that 1245 genes were upregulated while 639 genes were downregulated in the TDR4-silenced fruits compared with that in control fruit (Figure 2). To provide an overview of the role of TDR4, we evaluated the DEGs using GO and KEGG pathway enrichment analyses. GO analysis indicated that TDR4 silencing affected multiple metabolic pathways including 9 cellular component GO terms, with cell and cell part being the most enriched terms; 10 molecular function terms, with binding and catalytic being the most enriched; and 11 biological process terms, with metabolic process being the most enriched (Figure 3). KEGG pathway enrichment analysis showed that TDR4 was involved in photosynthesis and the biosynthesis of secondary metabolites (Figure 4). Eleven genes related to fruit ripening and nutrient metabolism were selected for qRT-PCR validation of the RNA-seq data. The RT-qPCR results were consistent with the sequencing data, indicating the reliability of the sequencing results (Figure 5).
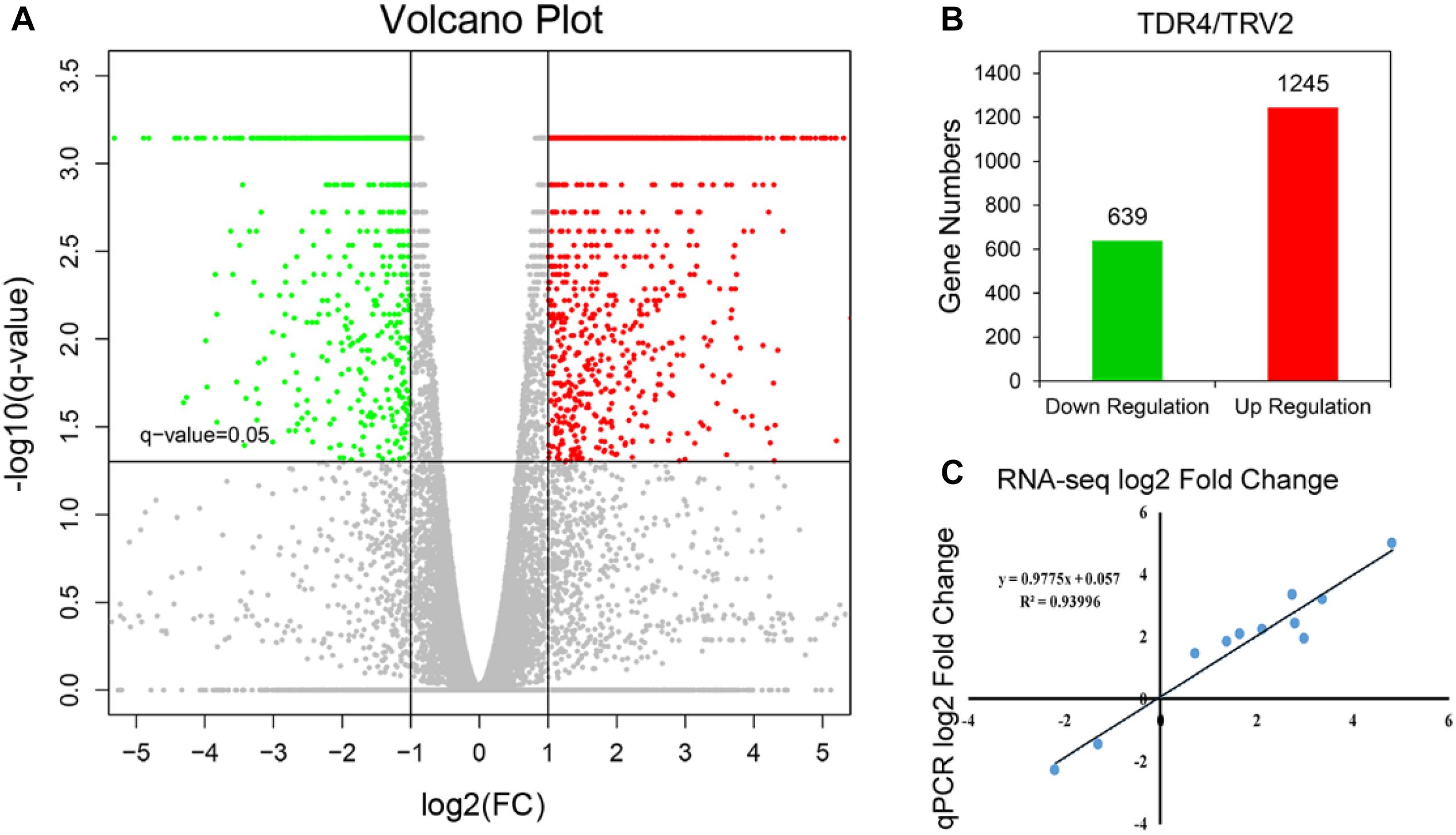
Figure 2. Global overview of DEGs between TDR4-silenced and control tomato fruit. (A) Volcano plot of DEGs. Spots above the threshold line (Q-value = 0.05) indicate significant DEGs. Genes for which expression in TDR4-silenced fruit was less than half that in control fruit, with a Q-value < 0.05, are shown in green, while those for which expression in TRD4-silenced fruit was more than two fold that in the control group are shown in red. Genes in gray were neither up- nor downregulated. (B) Number of down- (639) and upregulated (1245) genes. (C) Expression levels of 11 genes as determined by qRT-PCR are closely correlated with those according to RNA-seq.
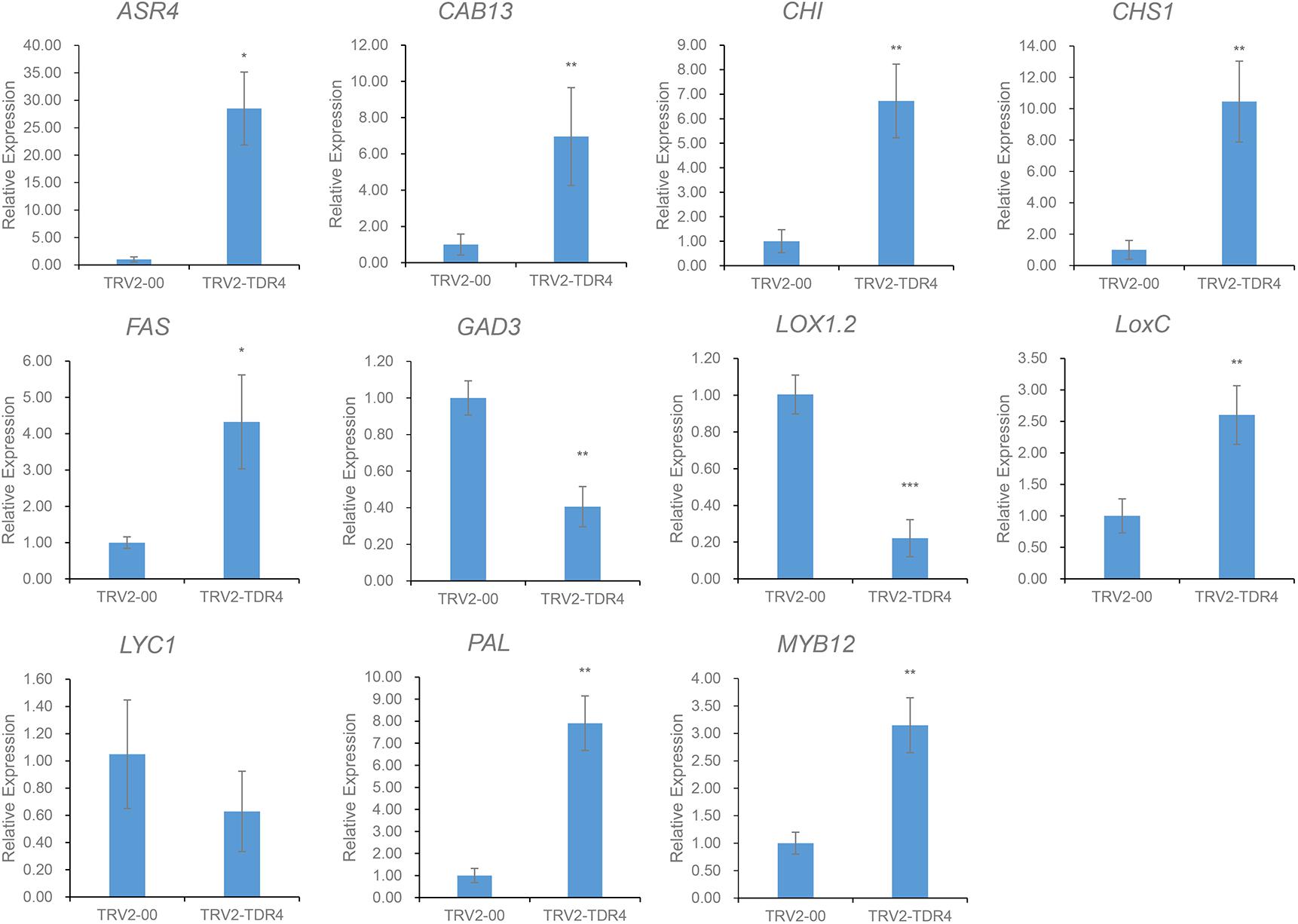
Figure 5. Expression levels of 11 genes according to qRT-PCR. Error bars indicate the standard deviation of three biological replicates. Asterisks indicate significant differences as determined by Student’s t-tests (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001).
To examine metabolic changes in the TDR4 silenced fruit, LC-MS/MS metabolite analysis was performed in TDR4-silenced and control tomato fruits. The result indicates that 50 metabolites were identified in TDR4-silenced tomato fruit. According to a two-way analysis of variance (ANOVA), 17 of these metabolites significantly differed in abundance between control (TRV2-00) and TDR4 silenced (TRV2-TDR4) fruit tissues (Table 2). All differential metabolites were further classified into four groups: amino acids, organic acids, phenolics, and solanum alkaloids. Possible pathways for each metabolite were determined by searching the KEGG database (Figure 6). These results suggest that the silencing of TDR4 altered the tomato fruit metabolism.
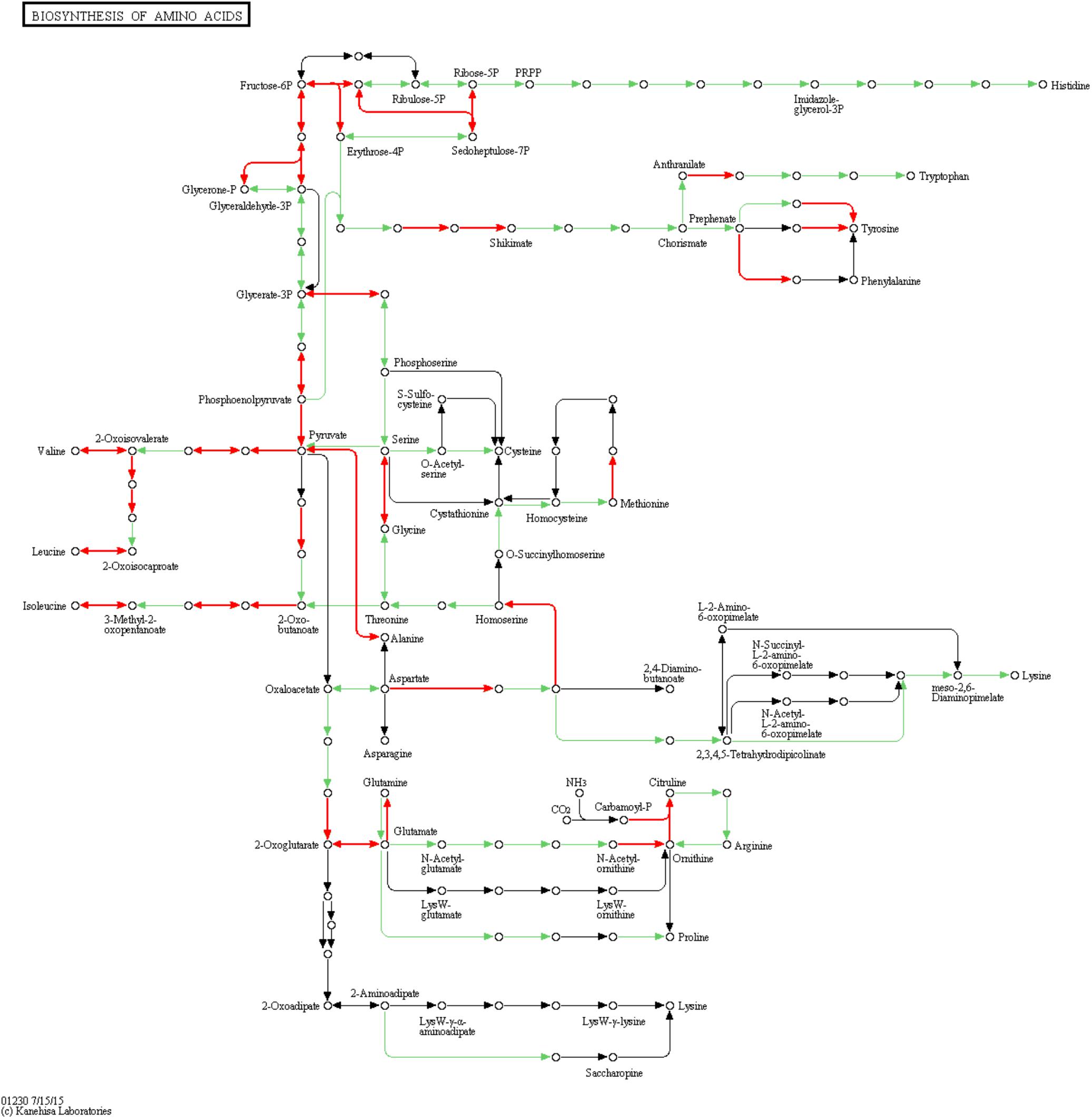
Figure 6. Schematic diagram of amino acid biosynthesis pathway. Diagram constructed using KEGG pathway analysis. Circles represent metabolites, and arrows indicate steps. Red arrows indicate steps catalyzed by DEGs.
Amino acids are primary metabolites that contribute to the flavor and nutritional value of tomato fruits. In TDR4-silenced fruit, levels of three amino acids were significantly reduced, including L-tyrosine (–65%), L-phenylalanine (–75%), and L-glutamic acid (–47%) (Table 2). KEGG pathway analysis revealed that the metabolic pathway of aromatic amino acids, such as phenylalanine and tyrosine, biosynthesis, and glutamate metabolism were also altered in TDR4-silenced fruits compared to control fruit (Figure 4), indicating that TDR4 gene plays a role in the accumulation of certain amino acids during tomato fruit ripening, which contributes to flavor formation of tomato fruit. Tomato fruit synthesizes flavor and nutrients during its ripening, and ripening process is regulated by ripening-related TFs, such as AP2a and Rin TFs, so these TFs may also participate in the regulation of flavor synthesis. The majority of the amino acids (15 out of 22) were present at significantly lower levels in the AP2i fruits than in the wild type, and the most dramatic reductions were in β-Ala, Ile, Met, Phe, and Trp (Karlova et al., 2011). Rin protein can target the promoter of TomloxC and ADH2 genes, which encode lipoxygenase (LOX) and alcohol dehydrogenase, respectively, and are critical for the production of characteristic tomato aromas derived from LOX pathway (Qin et al., 2012).
Phenylalanine is an important precursor of many aroma volatiles and flavonoids. For example, 2-phenylacetaldehyde and 2-phenylethanol are derived from phenylalanine (Tieman et al., 2006); both of these have pleasant fruity, floral odors, and important biological functions in plants (Knudsen et al., 1993). They attract mammals and other seed dispersers and exhibit antimicrobial properties (Goff and Klee, 2006).
RNA-seq result show that two previously unreported genes (Solyc11g066890 and Solyc06g050630), that encode prephenate dehydratase proteins, were significantly upregulated in TDR4-silenced fruit (Supplementary Table S3). These are probably involved in the first step of the sub-pathway that synthesizes L-phenylalanine or L-tyrosine, respectively, from L-arogenate. A gene (Solyc10g038080) encoding a shikimate dehydrogenase appears to be downregulated in TDR4-silenced fruits (Supplementary Table S2), which may contribute to a reduction in shikimate, an important precursor of L-phenylalanine and L-tyrosine.
Benzoic acid, synthesized from trans-cinnamate, is hypothesized to be the functional group in salicylic acid, and its derivatives are assumed to be involved in inducing stress tolerance in plants (Senaratna et al., 2003). In our study, the content of benzoic acid increased (3.89-fold) (Table 2) in TDR4-silenced fruit, along with the expression of two phenylalanine ammonia-lyase (PAL) genes. PALs are key enzymes in plant metabolism, catalyzing the first step of the sub-pathway that synthesizes trans-cinnamate from L-phenylalanine. One of the two upregulated genes is PAL5 (Supplementary Table S2), which is strongly expressed in old leaves and flowers and may function in response to biotic and abiotic stresses (Guo and Wang, 2009; Puthoff et al., 2010). Our findings suggest that the TDR4 gene negatively regulates the expression of PALs to inhibit the synthesis of benzoic acid in tomato fruit.
Glutamic acid is the most abundant amino acid in the diet, and a high level of free glutamate in some foods results in an umami taste (e.g., tomatoes, mushrooms, cheeses) (Bellisle, 1999). During tomato ripening, the glutamic acid content rises dramatically (Bemer et al., 2012). In our study, the expression levels of five related genes were significantly different in TDR4-silenced and control tomato fruit, and four of these were upregulated, including glutamate dehydrogenase (GDH1) (1.11-fold), glutamine synthetase (GS) (6.04-fold), glutamate decarboxylase 1 (GAD3) (2.20-fold), and a gene encoding isocitrate dehydrogenase (1.99-fold) (Supplementary Table S2). GDH1, which acts in the mitochondria, catalyzes the reversible amination of 2-oxoglutarate to L-glutamic acid (Ferraro et al., 2015). GS is a chloroplast glutamine synthetase that assimilates ammonia into glutamine (Perez-Rodriguez and Valpuesta, 1996), which is a metabolic intermediate in the synthesis of other nitrogen-containing compounds in plants (Perez-Rodriguez and Valpuesta, 1996). GAD3 converts L-glutamic acid to γ-aminobutanoic acid (GABA). The increased transcript abundances of these genes indicate the acceleration of glutamate metabolism in TDR4-silenced fruit.
Glutathione (GSH) is a tripeptide (γ-glutamylcysteinylglycine) that exists in a broad range of organisms, from bacteria to humans (Frendo et al., 2013). In humans, GSH plays an important role in the metabolism and detoxification of cytotoxic and carcinogenic compounds and reactive oxygen species (ROS) (Knapen et al., 1999). In plants, GSH is crucial for plant development and the plant response to the abiotic and biotic environment, and it is also involved in the detoxification of xenobiotics (Frendo et al., 2013; Yu et al., 2013). In our analysis of tomato fruit, TDR4 silencing resulted in a significant reduction in glutathione (–47%) (Table 2). KEGG pathway analysis showed that the GST gene, encoding glutathione S-transferase, was upregulated, which would promote the conversion of glutathione to glutamate.
Eriodictyol chalcone is a type of flavonoid. It has been reported that eriodictyol chalcone accumulates predominantly in the tomato peel and exhibits the highest accumulation at the breaker stage, gradually decreasing during ripening (Iijima et al., 2008). Tomato SlAN11 regulates flavonoid biosynthesis (Gao et al., 2018a). In our study, the content of eriodictyol chalcone was significantly increased in TDR4-silenced fruit (3.06-fold) (Table 2), compared to that in controls. KEGG pathway analysis showed that flavonoid biosynthesis was altered in TDR4-silenced fruit, which was consistent with the observation that chalcone synthase 1 (CHS1) and chalcone synthase 2 (CHS2) were upregulated (Supplementary Table S2).
5-Caffeoylquinic acid (chlorogenic acid) is one of the most abundant and widespread soluble phenolics among vascular plants (Mahesh et al., 2007). Evidence suggests that it can protect plant cells against oxidative stress, and plays a role in resistance to phytopathogens (Mahesh et al., 2007). The existence of another route involving the direct 3′-hydroxylation of p-coumaryol quinic acid was first suggested in carrot cell cultures, and studies of the impact of the expression of the hydroxycinnamoyl quinic acid gene in tobacco and tomato plants demonstrated that this route may be predominant in the Solanaceae family (Niggeweg et al., 2004). Analysis of TDR4-silenced tomato fruit revealed a significant reduction in 5-caffeoylquinic acid levels (–61%) (Table 2).
α-Tomatine is an anti-nutritional factor for humans (Friedman, 2013). In tomato, α-tomatine is present at high concentrations during the mature green (MG) stage and dramatically decreases during fruit ripening (Mintz-oron et al., 2008). In the present study, levels of α-tomatine were significantly elevated in TDR4-silenced fruit (7.69-fold) (Table 2). At the same time, the expression level of GAME11 was increased in TDR4-silenced fruit (Supplementary Table S3). It was reported that using VIGS technology to silence GAME11a putative dioxygenase in the cluster resulted in a significant reduction in α-tomatine levels and accumulation of several cholestanol-type steroidal saponins in tomato leaves (Itkin et al., 2013), which was consistent with our results. Putatively, GAME11 catalyzes the closure of the E-ring of 22,26-dihydroxycholesterol to form the furostanol-type aglycone (Itkin et al., 2013). In summary, the silencing of TDR4 promotes α-tomatine biosynthesis by enhancing the expression of GAME11 and TDR4 is a negative regulator of GAM11 gene.
The silencing of TDR4 using VIGS resulted in a non-ripening phenotype with an orange pericarp. RNA-seq analysis of TDR4-silenced fruit showed the altered expression of genes involved in various metabolic pathways. Analysis of metabolites by LC-MS/MS showed reductions in several amino acids as well as the accumulation of α-tomatine in TDR4-silenced fruit. These results suggest that TDR4 regulates the accumulation of nutrients and flavor in tomato fruit via the transcriptional regulation of target genes.
XZ designed the experiments and wrote the manuscript. XY and SC did the experiments. D-QF analyzed the data. C-ZJ revised the manuscript.
This work was supported by the National Natural Science Foundation of China, 31601518.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Dr. S. P. Dinesh-Kumar (University of California, Davis) for kindly providing pTRV1 and pTRV2 vectors.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00792/full#supplementary-material
TABLE S1 | Oligonucleotide primers used in the study.
TABLE S2 | Representative DEGs in TDR4-silenced tomato fruits.
TABLE S3 | The DEGs in TDR4-silenced tomato fruits.
Barry, C. S., and Giovannoni, J. J. (2007). Ethylene and fruit ripening. J. Plant Growth Regul. 26, 143–159.
Bellisle, F. (1999). Glutamate and the UMAMI taste: sensory, metabolic, nutritional and behavioural considerations. A review of the literature published in the last 10 years. Neurosci. Biobehav. Rev. 23, 423–438. doi: 10.1016/s0149-7634(98)00043-8
Bemer, M., Karlova, R., Ballester, A. R., Tikunov, Y. M., Bovy, A. G., Wolters-Arts, M., et al. (2012). The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24, 4437–4451. doi: 10.1105/tpc.112.103283
Ferraro, G., D’Angelo, M., Sulpice, R., Stitt, M., and Valle, E. M. (2015). Reduced levels of NADH-dependent glutamate dehydrogenase decrease the glutamate content of ripe tomato fruit but have no effect on green fruit or leaves. J. Exp. Bot. 66, 3381–3389. doi: 10.1093/jxb/erv150
Frendo, P., Baldacci-Cresp, F., Benyamina, S. M., and Puppo, A. (2013). Glutathione and plant response to the biotic environment. Free Radic. Biol. Med. 65, 724–730. doi: 10.1016/j.freeradbiomed.2013.07.035
Friedman, M. (2013). Anticarcinogenic, cardioprotective, and other health benefits of tomato compounds lycopene,à-tomatine, and tomatidine in pure form and in fresh and processed tomatoes. J. Agric. Food Chem. 61, 9534–9550. doi: 10.1021/jf402654e
Fu, D. Q., Zhu, B. Z., Zhu, H. L., Jiang, W. B., and Luo, Y. B. (2005). Virus-induced gene silencing in tomato fruit. Plant J. 43, 299–308. doi: 10.1111/j.1365-313x.2005.02441.x
Fujisawa, M., Shima, Y., Nakagawa, H., Kitagawa, M., Kimbara, J., and Ito, Y. (2014). Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26, 89–101. doi: 10.1105/tpc.113.119453
Gao, Y., Liu, J., Chen, Y., Tang, H., Wang, Y., He, Y., et al. (2018a). Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Hortic. Res. 5:27. doi: 10.1038/s41438-018-0032-3
Gao, Y., Wei, W., Zhao, X., Tan, X., Fan, Z., Zhang, Y., et al. (2018b). A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic. Res. 5:75. doi: 10.1038/s41438-018-0111-5
Gao, Y., Zhu, N., Zhu, X., Wu, M., Jiang, C. Z., Grierson, D., et al. (2019). Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas9 CNR and NOR mutants. Hortic Res. 6:39. doi: 10.1038/s41438-019-0122-x
Giovannoni, J. J. (2007). Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 10, 283–289. doi: 10.1016/j.pbi.2007.04.008
Goff, S. A., and Klee, H. J. (2006). Plant volatile compounds: sensory cues for health and nutritional value? Science 311, 815–819. doi: 10.1126/science.1112614
Guo, J., and Wang, M. H. (2009). Characterization of the phenylalanine ammonia-lyase gene (SlPAL5) from tomato (Solanum lycopersicum L.). Mol. Biol. Rep. 36, 1579–1585. doi: 10.1007/s11033-008-9354-9
Iijima, Y., Suda, K., Suzuki, T., Aoki, K., and Shibata, D. (2008). Metabolite profiling of chalcones and flavanones in tomato fruit. J. Japan. Soc. Hort. Sci. 77, 94–102. doi: 10.2503/jjshs1.77.94
Itkin, M., Heinig, U., Tzfadia, O., Bhide, A. J., Shinde, B., Cardenas, P. D., et al. (2013). Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341, 175–179. doi: 10.1126/science.1240230
Karlova, R., Chapman, N., David, K., Angenent, G. C., Seymour, G. B., and De Maagd, R. A. (2014). Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 65, 4527–4541. doi: 10.1093/jxb/eru316
Karlova, R., Rosin, F. M., Busscher-Lange, J., Parapunova, V., Do, P. T., Fernie, A. R., et al. (2011). Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23, 923–941. doi: 10.1105/tpc.110.081273
Klee, H. J., and Giovannoni, J. J. (2011). Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45, 41–59. doi: 10.1146/annurev-genet-110410-132507
Knapen, M. F. C., Zusterzeel, P. L., Peters, W. H., and Steegers, E. A. (1999). Glutathione and glutathione-related enzymes in reproduction: a review. Eur. J. Obstet. Gynecol. Reprod. Biol. 82, 171–184.
Knudsen, J. T., Tollsten, L., and Bergström, L. G. (1993). Floral scents-a checklist of volatile compounds isolated by head-space techniques. Phytochemistry 33, 253–280. doi: 10.1016/0031-9422(93)85502-i
Mahesh, V., Million-Rousseau, R., Ullmann, P., Chabrillange, N., Bustamante, J., Mondolot, L., et al. (2007). Functional characterization of two p-coumaroyl ester 3’-hydroxylase genes from coffee tree: evidence of a candidate for chlorogenic acid biosynthesis. Plant Mol. Biol. 64, 145–159. doi: 10.1007/s11103-007-9141-3
Manning, K., Tör, M., Poole, M., Hong, Y., Thompson, A. J., King, G. J., et al. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38, 948–952. doi: 10.1038/ng1841
Martel, C., Vrebalov, J., Tafelmeyer, P., and Giovannoni, J. J. (2011). The tomato MADS-Box transcription factor ripening inhibitor interacts with promoters involved in numerous ripening processes in a colorless nonripening-dependent manner. Plant Physiol. 157, 1568–1579. doi: 10.1104/pp.111.181107
Mintz-oron, S., Mandel, T., Rogachev, I., Feldberg, L., Lotan, O., Yativ, M., et al. (2008). Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol. 147, 823–851. doi: 10.1104/pp.108.116004
Niggeweg, R., Michael, A. J., and Martin, C. (2004). Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol. 22, 746–754. doi: 10.1038/nbt966
Perez-Rodriguez, J., and Valpuesta, V. (1996). Expression of glutamine synthetase genes during natural senescence of tomato leaves. Physiol. Plant 97, 576–582. doi: 10.1034/j.1399-3054.1996.970321.x
Puthoff, D. P., Holzer, F. M., Perring, T. M., and Walling, L. L. (2010). Tomato pathogenesis-related protein genes are expressed in response to trialeurodes vaporariorum and bemisia tabaci biotype B feeding. J. Chem. Ecol. 36, 1271–1285. doi: 10.1007/s10886-010-9868-1
Qin, G., Wang, Y., Cao, B., Wang, W., and Tian, S. (2012). Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 70, 243–255. doi: 10.1111/j.1365-313X.2011.04861.x
Senaratna, T., Merritt, D., Dixon, K., Bunn, E., Touchell, D., and Sivasithamparam, K. (2003). Benzoic acid may act as the functional group in salicylic acid and derivatives in the induction of multiple stress tolerance in plants. Plant Growth Regul. 39, 77–81.
Seymour, G. B., Chapman, N. H., Chew, B. L., and Rose, J. K. C. (2013). Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnol. J. 11, 269–278. doi: 10.1111/j.1467-7652.2012.00738.x
Shima, Y., Kitagawa, M., and Fujisawa, M. (2013). Tomato FRUITFULL homologues act in fruit ripening via forming MADS-box transcription factor complexes with RIN. Plant Mol. Biol. 82, 427–438. doi: 10.1007/s11103-013-0071-y
Tieman, D., Taylor, M., Schauer, N., Fernie, A. R., Hanson, A. D., and Klee, H. J. (2006). Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl. Acad. Sci. U.S.A. 103, 8287–8292. doi: 10.1073/pnas.0602469103
Vrebalov, J., Pan, I. L., Javier, A., Arroyo, M., Mcquinn, R., Chung, M., et al. (2009). Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21, 3041–3062. doi: 10.1105/tpc.109.066936
Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., et al. (2002). A MADS-Box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science 296, 343–346. doi: 10.1126/science.1068181
Wang, R. H., Yuan, X. Y., Meng, L. H., Zhu, B. Z., Zhu, H. L., Luo, Y. B., et al. (2016). Transcriptome analysis provides a preliminary regulation route of the ethylene signal transduction component, SlEIN2, during tomato ripening. PLoS One 11:e168287. doi: 10.1371/journal.pone.0168287
Keywords: transcription factors, virus-induced gene silencing, TDR4, fruit quality, metabolomic, transcriptomic
Citation: Zhao X, Yuan X, Chen S, Fu D-Q and Jiang C-Z (2019) Metabolomic and Transcriptomic Analyses Reveal That a MADS-Box Transcription Factor TDR4 Regulates Tomato Fruit Quality. Front. Plant Sci. 10:792. doi: 10.3389/fpls.2019.00792
Received: 17 December 2018; Accepted: 31 May 2019;
Published: 19 June 2019.
Edited by:
Antonio Ferrante, University of Milan, ItalyReviewed by:
Daniel Alexandre Neuwald, Competence Centre for Fruit Growing - Lake Constance, GermanyCopyright © 2019 Zhao, Yuan, Chen, Fu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodan Zhao, ZG9jZGFuQDEyNi5jb20=; Cai-Zhong Jiang, Y2ppYW5nQHVjZGF2aXMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.