- 1Bioproducts Research Unit, Western Regional Research Center, Agricultural Research Service, United States Department of Agriculture, Albany, CA, United States
- 2Guayule Research Farm, Section Manager Agricultural Operations, Bridgestone Americas, Inc., Eloy, AZ, United States
- 3Department of Biochemistry, University of Nebraska–Lincoln, Lincoln, NE, United States
- 4Monsanto Company, St. Louis, MO, United States
We report functional genomics studies of a CYP74 rubber particle protein from Parthenium argentatum, commonly called guayule. Previously identified as an allene oxide synthase (AOS), this CYP74 constitutes the most abundant protein found in guayule rubber particles. Transgenic guayule lines with AOS gene expression down-regulated by RNAi (AOSi) exhibited strong phenotypes that included agricultural traits conducive to enhancing rubber yield. AOSi lines had higher leaf and stem biomass, thicker stembark tissues, increased stem branching and improved net photosynthetic rate. Importantly, the rubber content was significantly increased in AOSi lines compared to the wild-type (WT), vector control and AOS overexpressing (AOSoe) lines, when grown in controlled environments both in tissue-culture media and in greenhouse/growth chambers. Rubber particles from AOSi plants consistently had less AOS particle-associated protein, and lower activity (for conversion of 13-HPOT to allene oxide). Yet plants with downregulated AOS showed higher rubber transferase enzyme activity. The increase in biomass in AOSi lines was associated with not only increases in the rate of photosynthesis and non-photochemical quenching (NPQ), in the cold, but also in the content of the phytohormone SA, along with a decrease in JA, GAs, and ABA. The increase in biosynthetic activity and rubber content could further result from the negative regulation of AOS expression by high levels of salicylic acid in AOSi lines and when introduced exogenously. It is apparent that AOS in guayule plays a pivotal role in rubber production and plant growth.
Introduction
Natural rubber is a secondary metabolite synthesized by more than 2,500 plant species (Lewinsohn, 1991; Cornish, 2001; Konno, 2011), yet its function in plant physiology remains unclear. It has been proposed that natural rubber defends against pathogen or insect infestation, repairs tissue damage caused by mechanical wounding, and/or protects cell damage induced by environmental stress (Tangpakdee and Tanaka, 1998; Kim et al., 2003; Konno, 2011). Regardless of its biological role, natural rubber remains one of the most economically valuable agricultural products worldwide, and is indispensable for modern transportation, medicine, and national defense. Hevea brasiliensis (rubber tree) is the main source of natural rubber, but is limited geographically, susceptible to diseases such as leaf blight (Radziah and Chee, 1989; Nyaka-Ngobisa et al., 2013), and its latex causes allergic reactions in specific groups of individuals (Siler and Cornish, 1994). Alternative sources of natural rubber would assure adequate supplies, protect national security, and safeguard human health.
One plant known to be a promising source of natural rubber is guayule (Parthenium argentatum A. Gray), a desert shrub native to the southwestern United States and northern Mexico (Mooibroek and Cornish, 2000; van Beilen and Poirier, 2007). Most rubber synthesis in guayule occurs during the cold season when the shrub is semi-dormant (Downes and Tonnet, 1985; Gilliland and van Staden, 1986; Madhavan et al., 1989; Ji et al., 1993). Guayule synthesizes rubber within subcellular organelles (Archer and Audley, 1987) known as rubber particles (RPs), located in the stembark tissues (Gilliland et al., 1984; Macrae et al., 1986; Kajiura et al., 2018). These particles consist mostly of hydrophobic cis-1,4 polyisoprene (i.e., natural rubber) molecules partitioned inside a phospholipid and protein-containing membrane (Nawamawat et al., 2011; Sansatsadeekul et al., 2011). In guayule, the most abundant rubber particle protein is allene oxide synthase (AOS) (Backhaus et al., 1991; Whalen et al., 2013). AOS is well-known as the first committed enzyme in the jasmonic acid (JA) biosynthetic pathway (Harms et al., 1995; Wang et al., 1999; Wasternack and Song, 2017), but the function of high levels of AOS protein in rubber particles is unknown. Guayule AOS has been shown to be a monooxygenase, cytochrome P450, heme-binding protein (Backhaus et al., 1991; Pan et al., 1995; Brash, 2009) and a distinct member of the CYP74A enzyme family in that it does not require oxygen or NADPH, nor does it contain a chloroplast targeting sequence (Song and Brash, 1991; Song et al., 1993; Pan et al., 1995). Recent genome analysis estimates there are three AOS genes in diploid guayule (Valdes Franco et al., 2018). To date, however, only one guayule AOS gene has been cloned and characterized (AOS1). While in most plants AOS is present at relatively low levels, AOS CYP74A P450 in flax (Linum usitatissimum) accumulates relatively large quantities in seeds. The function of the high levels of AOS in flax seeds is likewise unknown.
Overexpression of flax seed AOS led to higher levels of JA in potato (Solanum tuberosum; Harms et al., 1995) and tobacco (Nicotiana tabacum; Wang et al., 1999). Similarly, increases in the JA content was observed when Arabidopsis AOS was overexpressed both in Arabidopsis thaliana and N. tabacum (Laudert et al., 2000) and rice AOS in Oryza sativa (Mei et al., 2006). Constitutive overexpression of AOS in the mutant cea62 in rice also showed a dramatic increase in production of JA (Liu et al., 2012). In most cases wounding stress was a requirement for increased in JA level and substrate availability (Stenzel et al., 2003). On the other hand, down-regulation of AOS reduced or eliminated JA accumulation following wounding stress in Arabidopsis (Park et al., 2002) and further induced male sterility (Park et al., 2002; von Malek et al., 2002; Bae et al., 2010). These experiments are among those that documented AOS as the major control point in JA biosynthesis.
In this study, for the first time, a functional genomics approach was used to explore the role of AOS in guayule. We investigated the impact of AOS gene expression on guayule plant growth and development, phytohormone levels, and most importantly natural rubber production in plants grown in laboratory and greenhouse environments.
Materials and Methods
Plasmid Construction and Plant Transformation
The down-regulated and overexpressed guayule AOS gene in this study is the same gene reported in Backhaus et al. (1991), Pan et al. (1995, 1998), Chang et al. (2008), Li et al. (2008). The pND6 transformation vector was constructed from pPZP200 (Hajdukiewicz et al., 1994) with the potato Ubiquitin 409 promoter (Rockhold et al., 2008) and the Ubiquitin 1 sequence (Garbarino and Belknap, 1994) controlling β-glucuronidase (GUSplus) expression with the potato 409T terminator on the 3′ end. The pPZP200 Nos promoter driving the nptII (Herrera-Estrella et al., 1983) gene for conferring kanamycin resistance with Nos terminator on the 3′ end was retained. The guayule AOS gene was amplified by PCR using cDNA as a template. Gene specific primers for amplification of AOS comprised the following sequences: 5′-cttaagaggtggtATGGACCCATCGTCTAAACCC-3′ and 5′-ggtccTCATATACTAGCTCTCTTCAGGG-3′. The resulting PCR products were subcloned into pGEM T Easy vector (Promega Corp., Madison, WI, United States) and sequenced. Subsequently, the AOS gene was excised using the AflII and BamHI restriction enzymes and inserted into pND6. Plasmid pND6::AOSoe (Supplementary Figure S1) was generated by replacing the GUSplus gene with full-length guayule AOS gene. Plasmid pND6::AOSi was constructed by replacing the GUSplus gene with an inverted AOS sequence repeat (Supplementary Figure S1). The repeat is a reverse complementary 313 bp sequence in its 5′ end and the forward 313 bp sequence in its 3′ end. A BAR gene (Christensen and Quail, 1996) of 552 bp was inserted into the middle of this inverted repeat. The plasmids pND6, pND6::AOSoe and pND6::AOSi were used to transform Agrobacterium EHA101 competent cells (Hood et al., 1986). The transformed Agrobacterium EHA101 strains were used to transform guayule cultivar G7-11 following Dong et al. (2013). The expression of the GUSplus reporter gene in the pND6 transformed guayule plants was confirmed by histochemical staining (Dong et al., 2006).
Plant Germplasm and Growth Conditions
Wild-type (G7-11) and transgenic guayule lines were maintained in tissue culture as described in Dong et al. (2013). G7-11 is the source for publicly released USDA germplasm line AZ-2 (Dierig et al., 1989; Ray et al., 1999). It is apomictic, a tetraploid and a putative interspecific hybrid (Illut et al., 2017). This germplasm line is the most utilized line by industry as commercialization develops because of its vigor and rapid biomass accumulation. Our experiments included one WT, three pND6 vector controls, four AOSi and four AOSoe independent lines. These lines are all considered highly genetically similar except for targeted genes and expression of affected genes since polyploid guayule reproduces apomictically. For daytime simulation, all plants were grown at a temperature of 25–27°C for 16 h; for night time simulation, cold-treated plants were grown at 10°C for 8 h in the dark, and room temperature (RT) plants remained at 25–27°C for 8 h in the dark. For phenotypic analysis (plant architecture measurements, gas exchange, chlorophyll fluorescence and rubber content), plantlets were carefully uprooted from the tissue culture medium and transplanted into a pot (6″ diameter × 4.25″ depth; ITML Horticulture, Canby, OR) with SS#1 F1P RSI 3.8 CFC CDN planting mix (Sun Gro Horticulture, McClellan Park, CA, United States). Plants were grown in the greenhouse for 1 month then transferred into Conviron E8 growth chambers (Conviron, Temecula, CA, United States) at 40–45% relative humidity, light intensity ∼150–200 μmol m−2 s−2 (Spectrum Technologies, Inc., Aurora, IL, United States) and subjected to cold treatment as described above for 2 more months, after which the plants were 5-months-old. At this point the growth chamber cold treatment showed increased rubber production for WT plants, as is well documented in field conditions. One month and 3 months cold treatment were tested but did not show significant differences in the natural rubber content. Cold treatment experiments were performed three separate times from August 2015 to March 2016. The design included 3–4 replications per line, placed randomly inside the growth chambers. Plants were watered and fertilized as needed. Gas exchange, chlorophyll fluorescence, plant height, number of stems, stem diameter and biomass were measured at the end of the 2-month exposure to cold stress. For rubber particle extractions, plants were grown in the greenhouse for 1 year, watered and fertilized as needed. The greenhouse conditions were the following: temperature, 25–27°C; relative humidity, 50–65%; and light intensity, ∼1100–1300 μmol⋅m−2⋅s−2 (Spectrum Technologies, Inc.).
Genomic DNA Extraction and PCR Analysis
DNA was extracted using a Sigma GenElute Plant Genomic DNA Miniprep Kit (Sigma-Aldrich, Carlsbad, CA, United States). Approximately 150 mg leaf tissue was cut from the plants grown in tissue-culture, placed into 2 mL tubes and snap-frozen in liquid nitrogen. A bead was added to pulverize the tissues into a fine powder at a frequency of 30/s for 1 min using the mixer mill MM 400 tissue lyser (Verder Scientific, Inc., Newtown, PA, United States). PCR was carried out in 50 μL of a mixture containing GoTaq Green Master Mix (Promega Corp., Fitchburg, WI, United States), 200 ng guayule genomic DNA or 20 pg plasmid DNA, and 1 μM of vector and AOS specific primers 5′-CTTAAGAGGTGGTATGGACC-3′ and 5′-GGTTTCTTCCGGGTTCGAG-3′ for the AOSoe lines and 5′-ATGAGCCCAGAACGACGCCCGGCC-3′ and 5′-GATCTCGGTGACGGGCAGGACCGG-3′ for the AOSi lines. After heating the samples to 95°C for 2 min, the reaction proceeded with 35 cycles of 95°C for 30 s, 56°C to amplify the product ∼1.4 kb in the AOSoe lines (or 71°C for the product ∼0.5 kb in the AOSi lines) for 30 s and 72°C for 1 min. A final elongation step was carried out at 72°C for 5 min. PCR products were separated by electrophoresis on a 1% (w/v) agarose gel.
RNA Extraction, cDNA Synthesis and qPCR
Leaf, stembark and root tissues were collected and snap-frozen in liquid nitrogen for total RNA extraction. RNA was extracted from ∼100 mg tissues using the TRIzol method (Ambion, Pittsburg, PA, United States), cleaned with RNeasy MiniElute kit (Qiagen Inc., Valencia, CA, United States) and DNase1 treated (Qiagen Inc., Valencia, CA, United States). RNA concentration was quantified with NanoDrop ND1000 (ThermoScientific, Wilmington, DE, United States).
The iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, United States) was used to synthesize complementary DNA (cDNA) for real-time qPCR. Four μL of 5× iScript reaction mix, 1 μL of iScript reverse transcriptase, 1 μg of total RNA and nuclease-free water were used in a 20 μL reaction volume. The reaction mix was incubated in Eppendorf thermal cycler (Hauppauge, NY) with the following set-up: 5 min at 25°C for priming, 20 min at 46°C for reverse transcription, 1 min at 95°C for enzyme inactivation. For qPCR, 100 nM of forward and reverse primers, 7.5 μL of iQ SYBR Green Supermix Bio-Rad, Hercules, CA, United States), 2 μL of diluted cDNA (1:20) and nuclease free water were used in a 15 μL reaction volume. The 7500 Fast Real-Time PCR system (Applied Biosystem, Foster City, CA, United States) was use with the following thermal cycle: 95°C pre-incubation for 3 min; amplification for 40 cycles at 95°C for 15 s and 60°C for 30 s; the dissociation stage for 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. Each qPCR run was performed with three independent tissue samples, each sample having two technical replicates. The 18S gene was used as an internal control with the following respective forward and reverse primer pair: 5′-CAACAAACCCCGACTTCTGG-3′ and 5′-CACCCGTCACCACCATAGTA-3′. The AOS gene specific forward and reverse primers used were: 5′-AACCCGGAAGAAACCAAACT-3′ and 5′-CGCAACCGACTGGAAATAAT-3′, respectively. These primers were designed such that the product to be amplified is external to the sequences used in down-regulating the AOS with the RNAi technology. The putative hydroperoxide lyase (HPL) gene specific forward and reverse primers used were: 5′-GGTTTCAAGGCTCAGAGACG-3′ and 5′-AACGCGCCTATCTCCAGTAA-3′, respectively. The putative divinyl ether synthase (DES) gene specific forward and reverse primers used were: 5′-GACTCTGGTCCGGTCAGCTA-3′ and 5′- CGGTTATCGGGACAATGGGT-3′, respectively. The melting curve data were collected for all samples and genes to ensure a single peak, indicating amplification of a specific region by a pair of primers. The relative expression values were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Expression of AOS gene was normalized to the expression of the endogenous reference 18S gene and then to the expression of the WT plant.
Gas Exchange and Chlorophyll Fluorescence Measurements
Simultaneous measurements of gas exchange and chlorophyll fluorescence were taken on leaf tissues using LI-COR 6400xt (LI-COR Biosciences, Lincoln, NE, United States). Measurements were conducted between 0900 and 1200 h. Plants were dark adapted 24 h prior to measuring chlorophyll fluorescence. The healthy and fully expanded middle leaf position was chosen because this position showed significant differences based on chlorophyll meter measurements (Spectrum Technologies Inc.; Supplementary Figure S4). The concentration of CO2 coming in from the CO2 mixer was held fixed at 400 μmol⋅s−1 with a constant flow rate of 500 μmol⋅s−1. The leaf (6 cm2) was clamped on the Li-COR cuvette under a leaf temperature of approximately 23°C, relative humidity between 60–65% and photosynthetic photon flux density (PPFD) artificially supplied by the red-blue light source at 1,000 μmol⋅m−1⋅s−1. Measurements were made once the leaf reached the stable net CO2 fixation rate. Three leaves were measured per plant and three to four plants per genotype were sampled.
Natural Rubber Content
Fresh tissue from growth chamber plants (stems) and tissue-cultured plants (stems and leaves from 8 biological clones) was used for natural rubber content determinations. Tissues were harvested and lyophilized for 48 h. Tissues were then frozen in liquid nitrogen, ground to a fine power (30 Hz, 1 min) using a mixer mill MM 400 (Retsch, Haan, Germany), and stored at −80°C until analyzed. Approximately 300 mg of ground tissue was placed into an 11 mL ASE extraction cell (Dionex, Sunnyvale, CA, United States) with Ottawa sand as dispersant (Fisher Scientific, United States). A three-solvent sequential extraction method was applied (Pearson et al., 2013). Resin was extracted with acetone at room temperature, followed by a methanol extraction at room temperature, then rubber extracted with cyclohexane at 100°C (Fisher Sci., United States). Solvent containing analytes were evaporated using a TurboVap LV (Biotage, Charlotte, NC, United States) evaporator at 50°C with 15 psi N2. Rubber and resin dry weights were determined gravimetrically (w/w%).
Rubber Molecular Weight
Gel permeation chromatography (GPC) was used to determine the natural rubber molecular weight. For tissue cultured plants, cyclohexane extractables collected from ASE were re-suspended in approximately 3 mL of tetrahydrofuran (THF; Fisher Sci., United States) with gentle, overnight shaking (Multi-Purpose Rotator, Thermo Sci., United States). For greenhouse plants, washed rubber particles were solubilized directly in THF. Solutions were syringe-filtered through a 1.6 μm glass microfiber Whatman grade GF/A filter (GE Healthcare Life Sciences, United States), then 50 μL injected onto a Hewlett Packard 1100 series HPLC and size exclusion separated (THF continuous phase, 1.0 mL/min) by two Agilent PL gel 10 μm Mixed-B columns in series at 35°C. The first peak elutes at ∼12 mins by refractive index (RID; Agilent 1260 Infinity, dn/dc = 0.129) and light scattering (DAWN Heleos-II, Wyatt Technology, Santa Barbara, CA, United States) detectors and represents the high molecular weight natural rubber. Measurements were repeated at least three times with three technical replicates performed for each genotype.
Washed Rubber Particle Isolation
Rubber particles were extracted from WT, vector controls, and transgenic 1-year-old, greenhouse plants according to Cornish and Backhaus (1990). Briefly, ∼100 g of dirt-free stembark tissues were peeled off from each plant, ground twice, 1 min each, in a 4L blender containing 1L ice-cold extraction buffer. The resulting homogenate was filtered through eight layers of cheesecloth and distributed evenly in 250 mL centrifuge bottles. Centrifugation was performed in a swinging bucket rotor Sorvall RC5C 4°C centrifuge (DuPont, Wilmington, DE, United States) in a series of 2,000, 2,500, 3,500, 4,500, 5,500 and 7,000 rpm for 10 min. After each spin, the floating rubber particles were scooped with a stainless-steel spatula and transferred into 50 mL centrifuge tubes containing ∼25 mL ice-cold wash buffer and kept on ice. Purification by the aforementioned series of centrifugation speeds and subsequent collection into fresh, ice-cold wash buffer was performed two additional times. The rubber concentration (in μg⋅μL−1) was determined gravimetrically from three 50 μL aliquots of the final washed particles. Glycerol to a final concentration of 10% was added to the 3× washed rubber particles (WRPs) which were subsequently stored under liquid nitrogen until used.
Rubber Particle Protein Extraction and Analysis
WRP proteins were extracted by adding 500 μL of WRPs into 500 μL extraction buffer (7 M urea, 2 M thiourea, 6.5 mM CHAPS, 1% Nonidet-40, 60 mM DTT, 5 mM EDTA). Samples were sonicated three times in 1 min intervals, resting on ice for 1 min after each sonication. To separate the solubilized proteins from rubber, samples were centrifuged at 14,000 x g at 4°C for 5 min, and aqueous fraction filtered with a Millex GV 0.22 μm filter (MilliporeSigma, Burlington, MA, United States). Proteins were further concentrated with Microcon-10 kDa centrifugal filter unit (MilliporeSigma, Burlington, MA, United States). Total protein concentration was estimated with Quick StartTM Bradford Protein Assay (Bio-Rad, Hercules, CA, United States). After normalizing protein concentrations, the protein extracts were run on a 4–12% NuPAGE® Bis-Tris pre-cast polyacrylamide gel under reducing conditions (Life Technologies, Carlsbad, CA, United States) and detected with Bio-SafeTM Coomassie stain (Bio-Rad, Hercules, CA, United States). For western blot, proteins from the PAGE gel were transferred to a PVDF membrane, incubated for 45 min with 10 ng/mL anti-AOS-biotin conjugated monoclonal antibodies (Antibody Solution, Sunnyvale, CA, United States), following 30 min incubation with 2 ng/mL Streptavidin-Horse Radish Peroxidase conjugate (EMD Millipore, Temecula, CA, United States), and final chemiluminescent detection with SuperSignal® Femto Maximum Sensitivity Substrate (Thermo Scientific, Waltham, MA, United States). Membrane was wrapped with plastic wrap and exposed to CL-XPosureTM film (Thermo Scientific, Waltham, MA, United States) for 10–30 s. Recombinant AOS (Sigma-Aldrich, Saint Louis, MO, United States) was used as positive control. Protein molecular weight markers are PageRulerTM Plus Prestained Protein Ladder (Thermo Scientific, Rockford, IL, United States).
In vitro Assay of Rubber Synthesis
Enzymatically active WRPs were assayed for in vitro [1-14C]IPP incorporation in a Ultrafree® MC-VV centrifugal filter unit (MilliporeSigma), in a 50 μL reaction volume containing ∼1 mg of RP, 20 μM farnesyl pyrophosphate (FPP) initiator, 1 mM unlabeled isopentenyl pyrophosphate (IPP), and 0.9 nmol (55 mCi⋅mmol−1) [1−14C]IPP in buffer (100 mM Tris–HCl, pH 7.5; 1.25 mM MgSO4, 5 mM dithiothreitol). The reactions, performed in triplicates, were incubated at 16°C for 3–4 h, stopped with addition of 40 mM ethylene diaminetetraacetic acid (EDTA), pH 8.0, and washed by centrifugation at 14,000 x g three times with water. The filter unit was inserted in a vial containing 2 mL ScintiVerse BD Cocktail (Fisher Sci., United States) and the amount of [14C]-IPP in each individual filter was quantified with LS 6500 scintillation counting instrument (Beckman Coulter, Brea, CA, United States).
Rubber Particle Size
The size of extracted WRPs was determined using the Nano ZS ZEN 3600 (Malvern Instruments Ltd., United Kingdom) dynamic light scattering instrument. Briefly, the 3× WRPs extracted from 1-year-old greenhouse wild-type and transgenic guayule lines were filtered with cheesecloth. The sample volume (pH = 7.5) was adjusted to 1 mL with double distilled water. The liquid was transferred into the cuvette and inserted into the zetasizer instrument. The input refractive index was 1.51. The analyses were performed in triplicate. Data were analyzed using the average and standard deviation by student t-test. Significant differences were assessed relative to the wild-type and/or pND6 controls, where values of p < 0.05 were considered statistically significant. Measurements were repeated at least three times with three technical replicates were performed for each genotype.
Hormone Quantification
To determine the endogenous plant hormone content in the control and transgenic AOS plants using ESI-MS/MS, we followed the protocol by Pan et al. (2010) with modifications. Three biological plants with three technical replicates for each plant were used in this experiment. Briefly, leaves and stems were snapped-frozen and ground to powder with mortar and pestle. Solvent extraction solution and internal standards were added to ∼50 mg of pre-weighed tissues. ESI-MS/MS was performed as previously described (Luttgeharm et al., 2015) using a Shimadzu Prominence UPLC system, with ion detection by a QTRAP 4000 triple quadrupole mass spectrometer (Applied Biosystems) operated in negative mode and using previously published MRMs (Pan et al., 2010). Quantitation based on comparison of analyte to standard peak area was done using Multiquant 2.1 software (ABSciEX, Redwood City, CA, United States).
Exogenous Salicylic Acid and Hydrogen Peroxide Treatment
Wild type G7-11 plants were grown on half-strength MS medium (PhytoTechnology Laboratories, Lenexa, KS, United States) in Magenta boxes (Caisson Labs, Smithfield, UT, United States) for 8 weeks with/out the addition of salicylic acid (SA) at the following concentration: 0, 10, and 100 μM diluted in 65% ethanol. For hydrogen peroxide, the following concentrations were tested: 0 nM, 0.1 nm and 1 nM diluted in distilled water. Leaf and stembark tissues were collected and snap-frozen in liquid nitrogen for RNA extraction and expression analysis by qPCR. For SA and hydrogen peroxide treatment of potting mix plants growing in growth chamber, 1-month old G7-11 plants were water with 50 mL solution containing 10 μM of SA and/or 0.1 nM of H2O2 directly into the potting mix three times per week for 8 weeks. Both SA and hydrogen peroxide treatment experiments were repeated at least three separate times with three biological replicates.
AOS Enzyme Activity
Total proteins from 1 mL of WRPs extracted from stembark tissues of 1-year old greenhouse plants were solubilized in 0.5% CHAPS solution and sonicated three times at 1 min interval. Centrifugation at 14,000 x g at 4°C for 5 min followed thereafter. The clear solution (bottom layer) was aspirated from the rubber layer (top) into 1 mL syringe (Fisher Sci., United States) and passed through a Millex GV 0.22 μm filter (MilliporeSigma, Burlington, MA, United States). Total protein concentration was determined with Quick StartTM Bradford Protein Assay (Bio-Rad, Hercules, CA, United States). For AOS activity, 400 ng of WRP total protein and 300 μM ethanolic solution of 13S-hydroperoxy-9Z,11E,15Z-octadecatrienoic acid (13S-HpOTrE; Cayman Chemical Co.; Ann Arbor, MI, United States) was added into 50 mM potassium phosphate buffer (pH = 7.0) to make a total volume of 200 μL. The 13S-HpOTrE was chosen because it is a precursor of JA (Song et al., 1993; Hughes et al., 2009). Reaction was carried out in specialized 96-well quartz microplate (Molecular Devices, San Jose, CA, United States) with absorbance measured at 235 nm in 3 s intervals over a 10-min period using SpectraMax M3 Multi-Mode Microplate Reader (Molecular Devices, San Jose, CA, United States). Vmax was determined based on the slope of optical density (OD) versus time in min, which was then normalized against WT to calculate the relative AOS activity. AOS activity was assayed in three biological replicates of each genotype.
Hydrogen Peroxide Quantification
Hydrogen peroxide content was determined with optimized colorimetric determination using potassium iodide (Junglee et al., 2014). Eight-weeks-old guayule shoot (leaf and stem) were snap-frozen and ground to powder with mortar and pestle. One hundred fifty milligrams were homogenized in 1 mL of 0.25 mL of trichloroacetic acid (0.1% TCA; w/v), 0.5 mL potassium iodide (1 M) and 0.25 mL of potassium phosphate buffer (0.01 M; pH = 7) on ice for 10 mins with gentle shaking (Fisher Sci., United States). Homogenates were centrifuged in an Eppendorf 5403 at 15,000 x g for 15 min at 4°C. From each supernatant, an aliquot of 0.2 mL was aliquoted in a 96-well UV-microplate flat-round bottom (Greiner Bio-One Cat. #655801, Fisher Sci., United States) and incubated at RT (20–22°C) for 20 mins. Absorbance was measured for 1 min at 390 nm. The blank probe consisted of 0.1% TCA in the absence of the shoot extracts. H2O2 was quantified using a calibration curve of known H2O2 concentrations. Results are expressed as μmol H2O2 per gram of fresh weight.
Peroxidase Activity Assay
The activity of peroxidase was assayed following the method described by Putter (1974) with a few minor alterations. Prior to sample collection, tissue-cultured WT and transgenic lines were treated at RT (25–27°C) and cold temperature (10°C) for 2 h in growth chamber with relative humidity of 50–65% and light intensity of ∼1100–1300 μmol⋅m−2⋅s−2 (Spectrum Technologies, Inc., United Kingdom). Approximately 0.5 g of shoot tissue was homogenized with 5 mL of phosphate buffer (0.1 M; pH 6.0) with mortar and pestle. Homogenate was transferred into a 15 mL tube followed by centrifugation in a Sorvall Instrument RC5C (DuPont) at 4,000 x g for 10 mins. A 50 μL aliquot was aspirated from the supernatant into a 96-flat bottom well (Greiner Bio-one Cat # 655101, Fisher Sci., United States). A 150 μL of reaction mixture consisting of phosphate buffer (1 M), guaiacol (0.23 M) and hydrogen peroxide (0.018 M) was added into the same 96 well plate. Absorbance was read at 470 nm in 15 s intervals for 2 mins using SpectraMax M3 Multi-Mode Microplate Reader (Molecular Devices, San Jose, CA, United States). Experiments were repeated at least three times with three technical replicates performed for each genotype. Activity per unit was calculated by 6 times the slope of the linear fit of A470 vs. time (min).
Lipid Peroxidation Assay
Lipid peroxidation in tissue-cultured plants was determined by thiobarbituric acid (TBA) test (Heath and Parker, 1968). On ice, 0.1 mg of powdered shoot sample was added to 1 mL of trichloroacetic acid (TCA, 0.1%) in a 2 mL Eppendorf tube and mixed by vortexing. Samples were centrifuged for 10 mins at 13,000 rpm at 4°C. A 400 μL sample aliquot was mixed with 1 mL TBA (0.5%) and TCA (20%) mixture solution in a syringe-needle punctured cap 2 mL Eppendorf tube and incubated for 30 mins at 80°C. Tubes were immediately placed on ice for 5 mins followed by centrifugation at 13,500 rpm at 4°C for 5 mins. A 0.2 mL of the supernatant was aliquoted into the 96-well flat bottom Greiner plate (Greiner Bio-one Cat # 655101, Fisher Sci., United States) and absorption at 532 nm and 600 nm. The malondialdehyde (MDA) concentration was calculated using the following formula:
nmol MDA/ g FW = ; ΔAcorrected = A532–A600 corrected with ΔA of the blank; b = light path length (0.56 cm for 200 μl); ε = millimolar extinction coefficient (155 mM−1cm−1); 3.5 (dilution factor from 400 μl extract + 1 ml TBA/TCA solution); X (ml) TCA 0.1% used for extraction (1 ml); y (g) FW used for extraction; 1000 = conversion factor (nmol to μmol). Experiments were repeated at least three times with three technical replicates performed for each genotype.
Statistical Analysis
Student’s t-test from MS Excel was used for all statistical tests.
Accession Numbers
Sequence data from this article can be found in the GenBank data libraries under the following accession numbers: GUSplus (AF354045.1), potato 409T terminator (HK352815.1) and guayule AOS (X78166.2).
Results
Generation and Confirmation of AOS Transgenic Lines
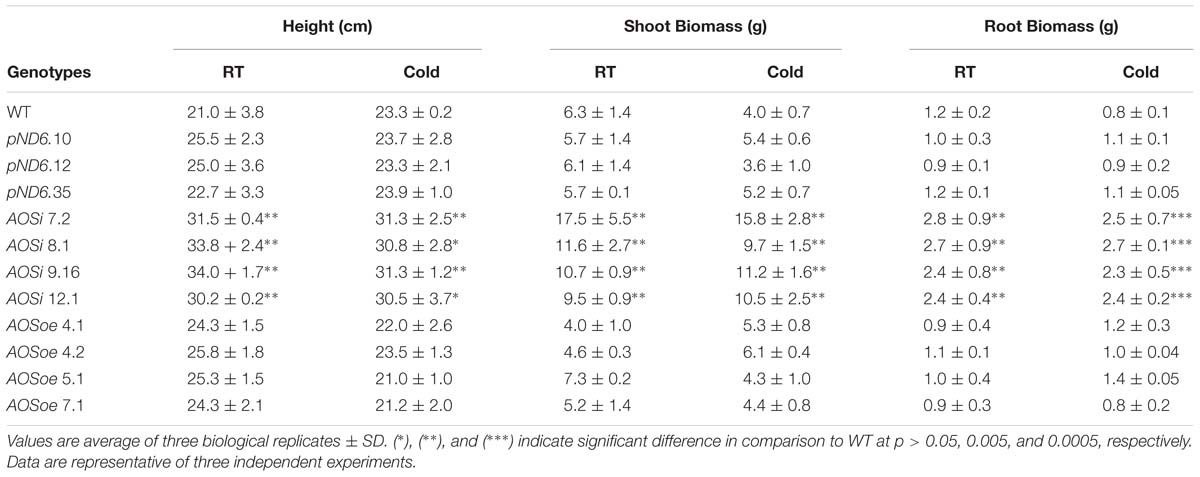
While highly abundant in rubber particles, AOS can be detected in all tissues of guayule. The expression pattern of AOS in 8-week-old tissue-cultured and 2-month-old greenhouse grown plants revealed the highest level of AOS expression in stems and roots, and stembark tissues (Figure 1). Interestingly, natural rubber is also mainly found in the stembark tissues of mature plants and some in the roots and leaves (Curtis, 1947; Kuruvadi et al., 1997).
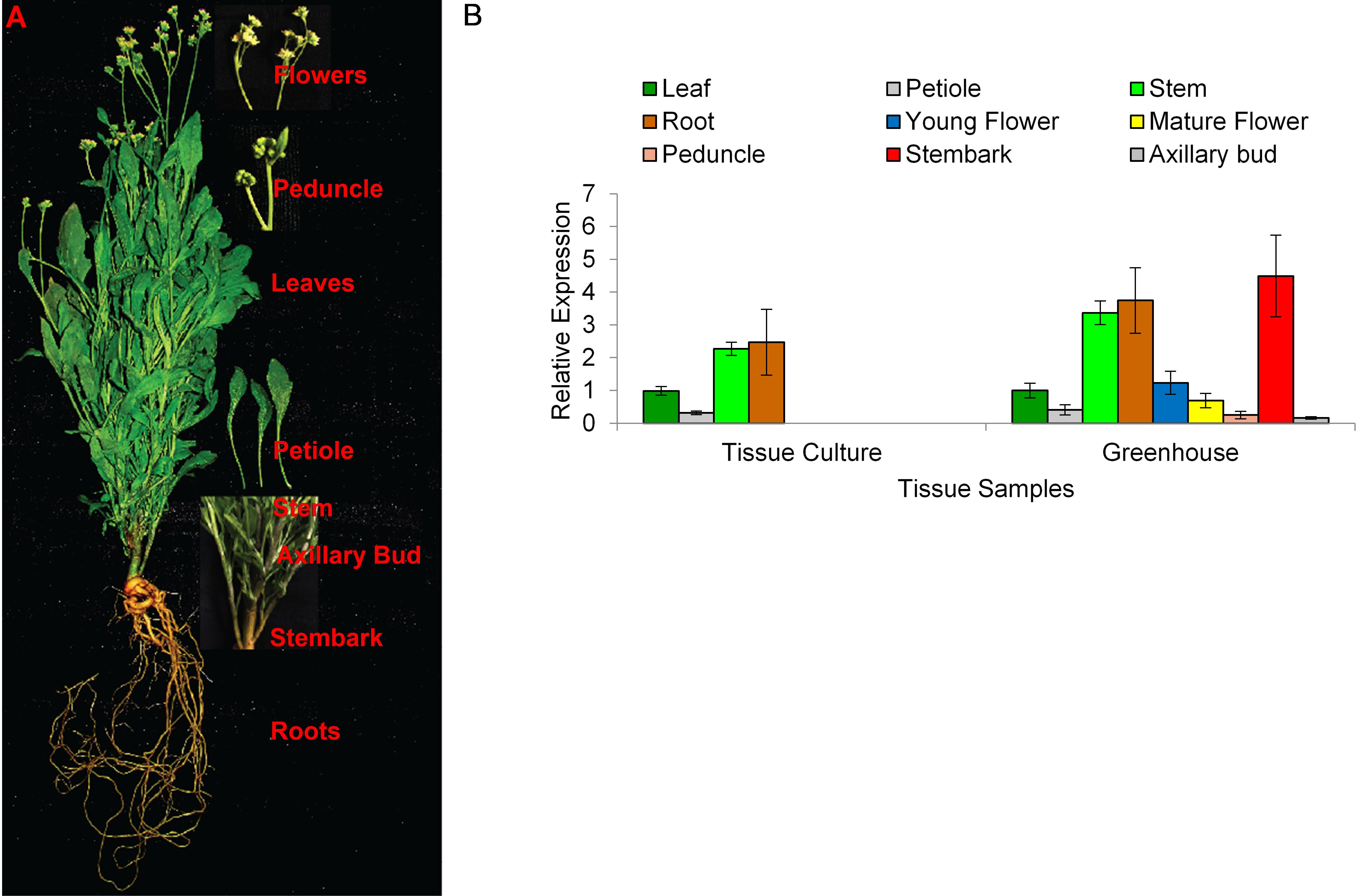
Figure 1. Endogenous AOS expression in guayule. (A) Guayule tissues analyzed. (B) AOS relative expression in WT G7-11 tissue-culture and greenhouse grown plant tissues. Expression levels for each tissue were compared to that of the corresponding leaf tissue, whether from tissue cultured or greenhouse grown plants, and normalized to the 18S reference gene. Error bars represent SD from the average of three biological replicates.
To better understand the role of AOS in guayule, we generated transgenic plants in which AOS was overexpressed (AOSoe) or silenced (AOSi) by RNAi (Supplementary Figure S1). Four plants harboring the vector control (plant transformation plasmid with GUSPlus gene, pND6) were confirmed by GUS staining (Supplementary Figure S2). To confirm the presence of the AOS transgenes, genomic DNA from AOS transgenic lines was screened with PCR (Figures 2A,B). Ten each AOSi and AOSoe lines were found to be positively transformed. The expected impact of transgenes on AOS expression was confirmed with quantitative RT-PCR (qPCR) on total RNA from stembark tissues. AOSi lines had significantly lower levels of AOS expression and AOSoe lines had significantly higher levels of expression (Figure 3). Denaturing protein gel analysis (PAGE) of washed rubber particle total proteins from transgenic lines indicated reduced levels of AOS protein in AOSi lines (Figure 4A), confirmed by Western blot analysis (Figure 4B). Despite higher levels of AOS gene expression, AOSoe lines appear to not contain higher levels of AOS protein (Figures 4A,B). Moreover, relative AOS enzyme activity (13-HPOT conversion to allene oxide) was reduced as expected for the AOSi lines but not increased for the AOSoe lines compared to the pND6 empty vector control (Figure 5). The discrepancy in the AOS transcript levels and protein levels and activity in the AOSoe lines indicates the absence of simple control of AOS protein levels.
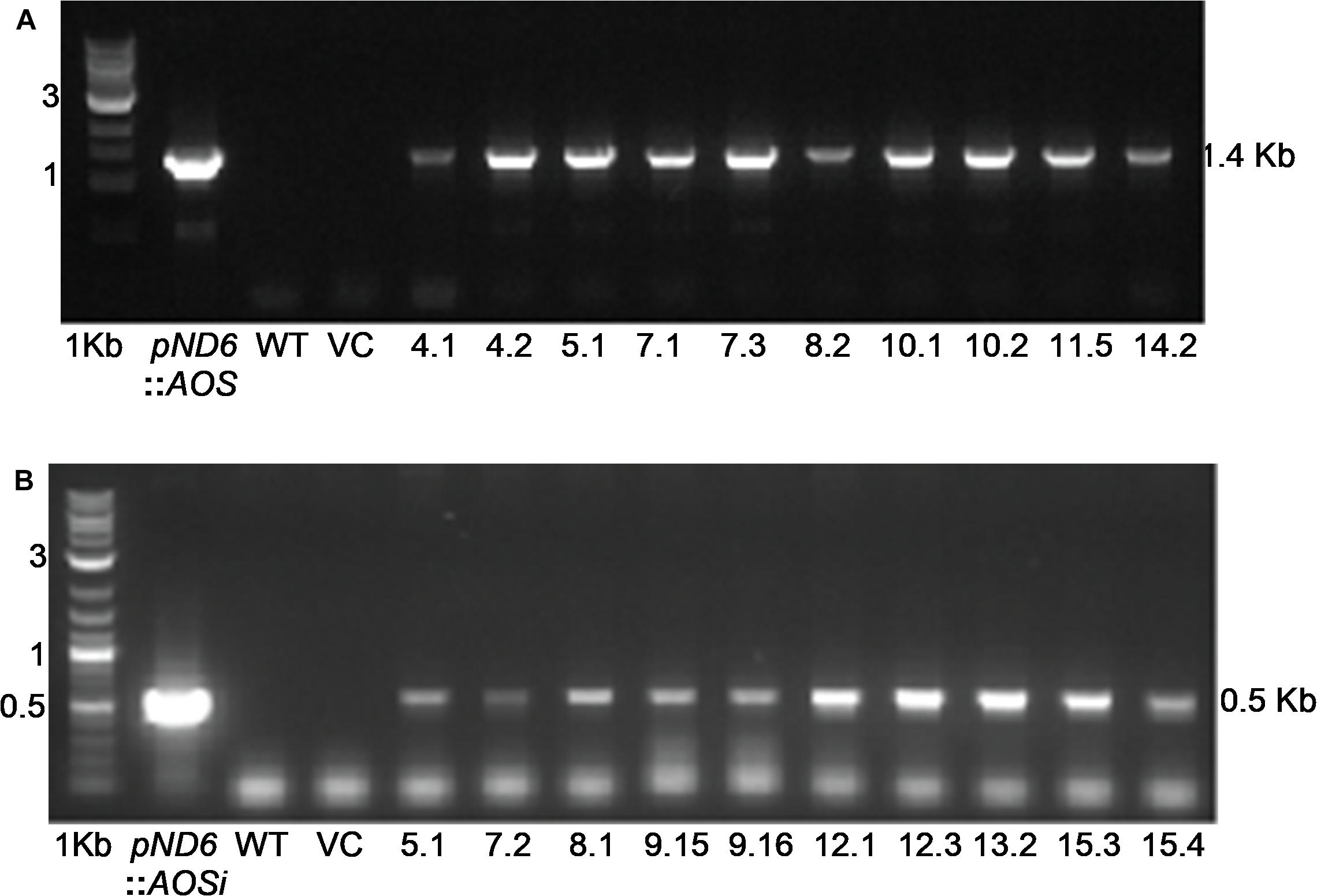
Figure 2. Genomic DNA PCR screening of (A) Overexpressing (AOSoe) and (B) downregulated (AOSi) transgenic guayule lines confirmed the presence of their corresponding transgenes. pND6::AOSoe and pND6::AOSi, transformation plasmid controls; WT, G7-11 wild-type control; VC, pND6 vector control.
Figure 3. Expression analysis of AOS in guayule. WT, wild-type G7-11; pND6, vector control; AOSi, downregulated AOS; AOSoe, overexpressed AOS. Asterisks (∗) indicates significant difference in comparison to WT at p > 0.05. Total RNA from stembark of 5-month-old growth chamber-grown control and transgenic lines were template for qPCR. Expression levels were compared to WT and normalized to the 18S reference gene. Average expression is from three biological replicates + SD.
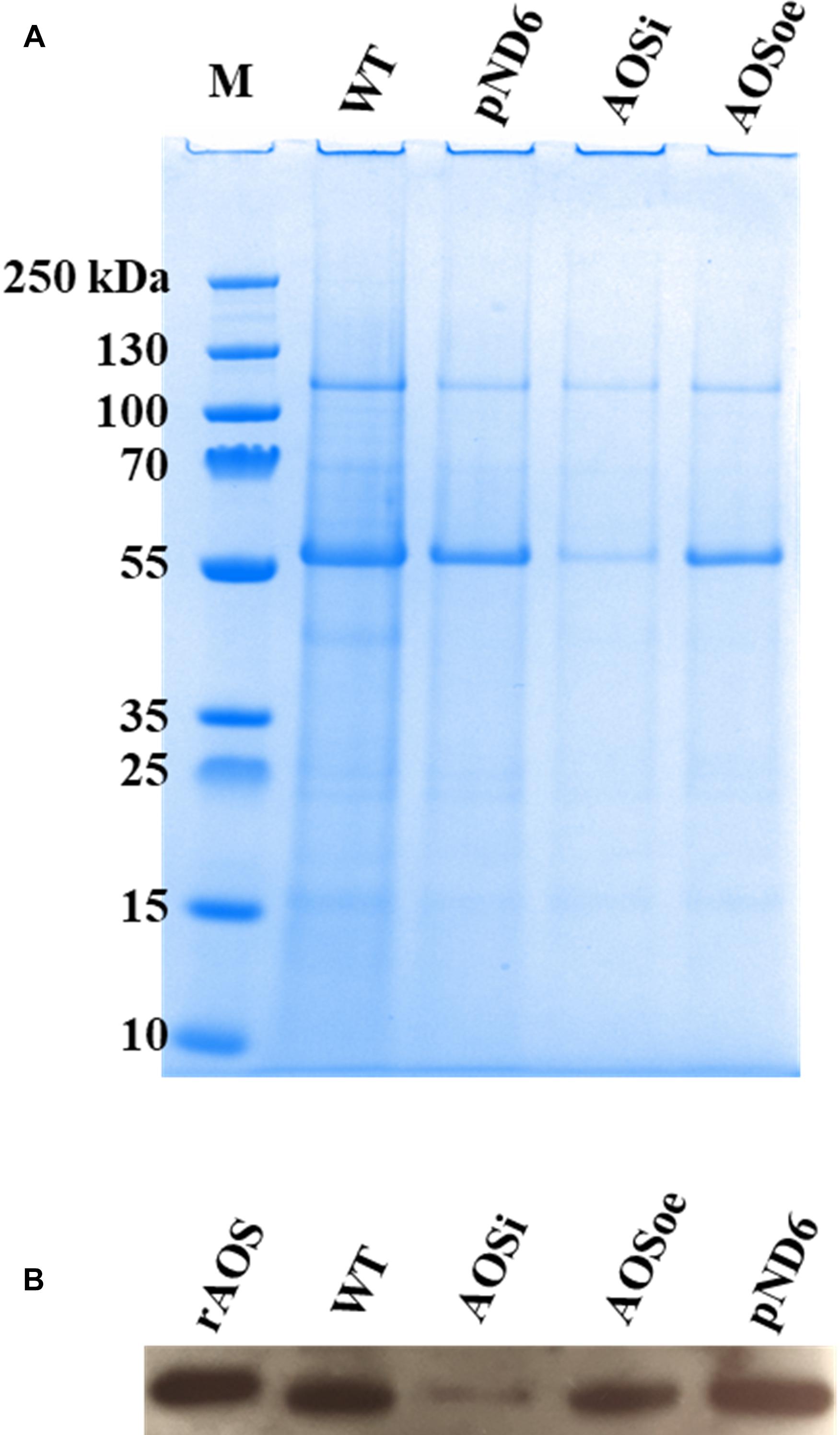
Figure 4. Washed rubber particle (WRP) proteins from guayule control plant and transgenic lines. Total proteins (3 μg) extracted from WRPs were separated by denaturing PAGE. (A) gel stained with Bio-SafeTM Coomassie, and (B) western blot. WT, wild-type; pND6, vector control; AOSi, downregulated AOS; AOSoe, overexpressed AOS; rAOS, recombinant AOS protein (225 ng).
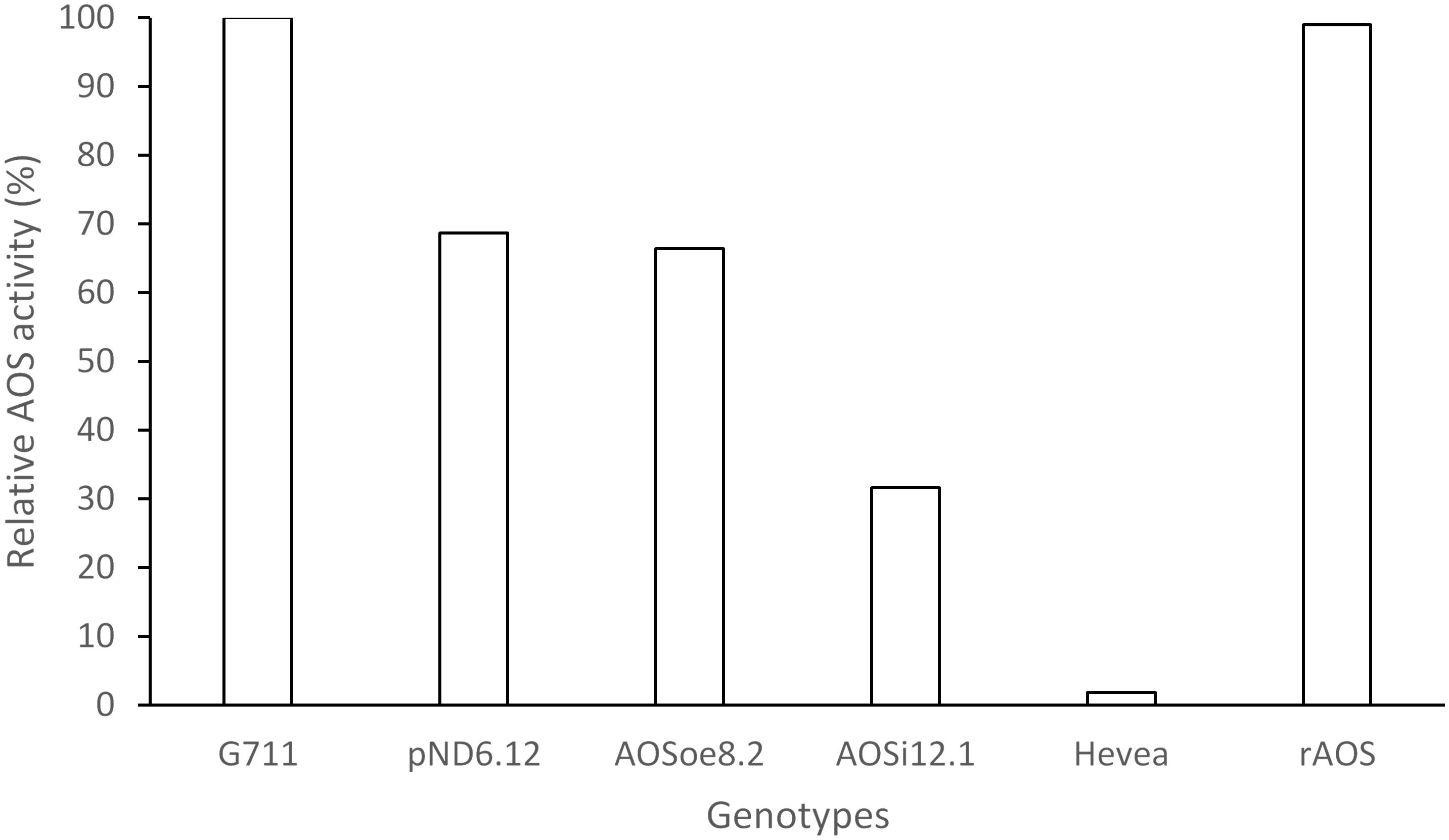
Figure 5. Relative AOS enzyme activity assay. WRP total protein extracts (400 ng) were assayed for AOS activity with 300 μM 13S-HpOTrE at 235 nm. Relative AOS activity was calculated by normalizing Vmax values against the WT. G7-11, wild-type control; pND6, vector control; AOSi, downregulated AOS; AOSoe, overexpressed AOS; Hevea, negative control; rAOS, commercial AOS recombinant protein, positive control.
Since rubber biosynthesis in guayule is highest in the cold (Goss et al., 1984; Ponciano et al., 2012), a series of experiments were conducted to evaluate the phenotype response of cold treatment on guayule plants varying in AOS expression. All plants were grown for 2 months at 25–27°C for 16 h in the light; RT plants were maintained at the same temperature for 8 h in the dark; cold-treated plants were grown at 10°C for 8 h in the dark.
Downregulation of AOS Resulted in Larger Plants With Enhanced Photosynthesis
All AOSi lines were significantly taller (20 to 30%) and wider (28 to 30%) and had higher shoot (leaves and stems; 34 to 57%) and root biomass (50 to 61%; Table 1; Supplementary Figure S3) than WT, pND6 and AOSoe. All AOSi lines also had more stems per plant than WT and pND6 vector controls (Table 2). Additionally, the mature stembark tissues in AOSi lines had a thicker diameter (35 to 54%) in comparison to WT, pND6 and AOSoe (Table 2). For all these phenotypes, cold treatment generally amplified the differences between AOSi lines and control lines.
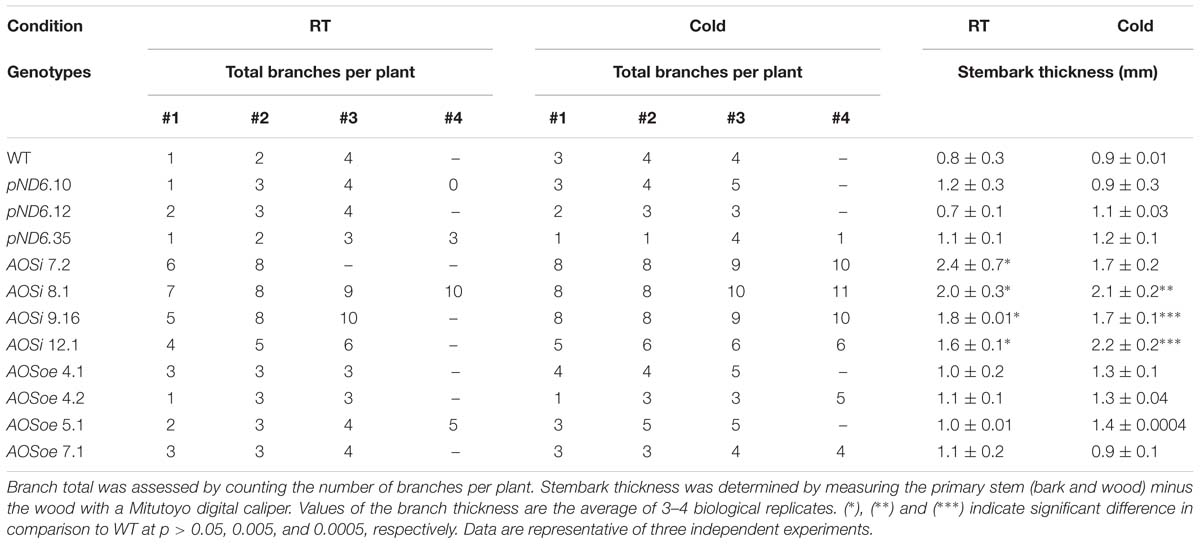
Table 2. Total branches and thickness of the primary stem of 5-month-old guayule control and AOS-transgenic lines.
The AOSi lines grown initially in the greenhouse and later in growth chambers were larger and had darker green leaves (Supplementary Figure S3) and higher chlorophyll levels (Supplementary Figure S4) than WT, pND6 and AOSoe. Gas exchange rate measurements showed a higher (23 to 31%) photosynthetic rate (Pn) in AOSi lines compared to WT, pND6 and AOSoe lines (Table 3). In addition, the AOSi lines showed higher stomatal conductance (g) and transpiration rate (€) as compared to all other genotypes (Table 3). Moreover, chlorophyll fluorescence measurements clearly showed efficiency of photosystem II (PSII) and electron transport rate (ETR) parameters are significantly higher in AOSi lines (Table 3) at both room and cold temperatures. Higher PSII and ETR would indicate more efficient light energy absorption and carbon assimilation in AOSi lines for growth and development as well as rubber synthesis compared to WT, pND6, and AOSoe genotypes. The non-photochemical quenching (NPQ) for AOSi lines was lower than controls under room temperature. However, under cold treatment, NPQ was higher in AOSi lines in comparison with WT, pND6 and AOSoe genotypes, suggesting that AOSi lines have a more effective mechanism of protecting the PSII apparatus under cold stress.
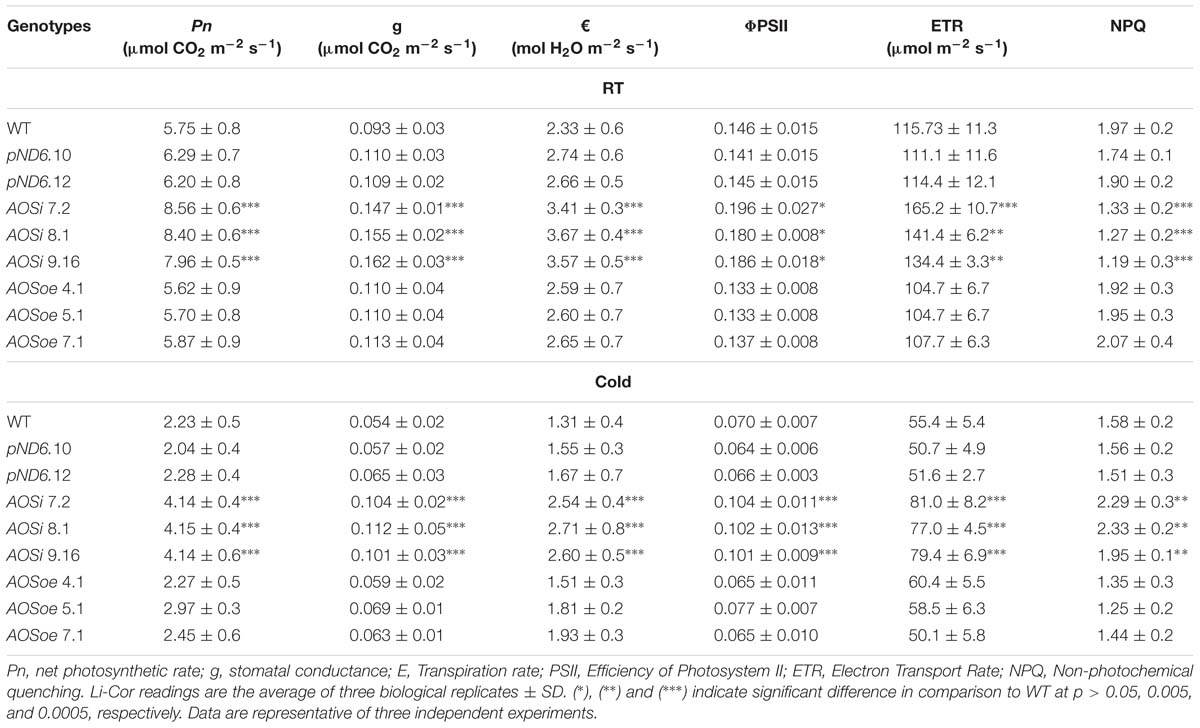
Table 3. Gas exchange rate and chlorophyll fluorescence measured with Li-Cor of 5-month-old guayule control and AOS-transgenic lines.
Natural Rubber Biosynthesis Rate and Rubber Content Increased When AOS Was Downregulated
The natural rubber content in shoot tissues (stems and leaves) was increased in AOSi lines for both tissue cultured plants (in media) and growth-chamber grown plants (in potting mix). The AOSi lines produced more rubber (30 to 54%) than WT, pND6 and AOSoe lines in the tissue-culture environment (Supplementary Table S1). Under cold treatment in growth chamber conditions, representing a microcosm of what guayule plants experience in the field, AOSi lines also exhibited higher rubber content in the shoots, with 16–21% more rubber in comparison with the WT (Table 4). Rubber also accumulated in the root tissues especially under cold treatment, where AOSi lines stored 18–38% higher rubber content compared with WT, pND6 and AOSoe lines. In all cases AOSoe lines showed the same rubber content as control lines. Also, in all cases, terpene resins content showed no differences (data not shown).
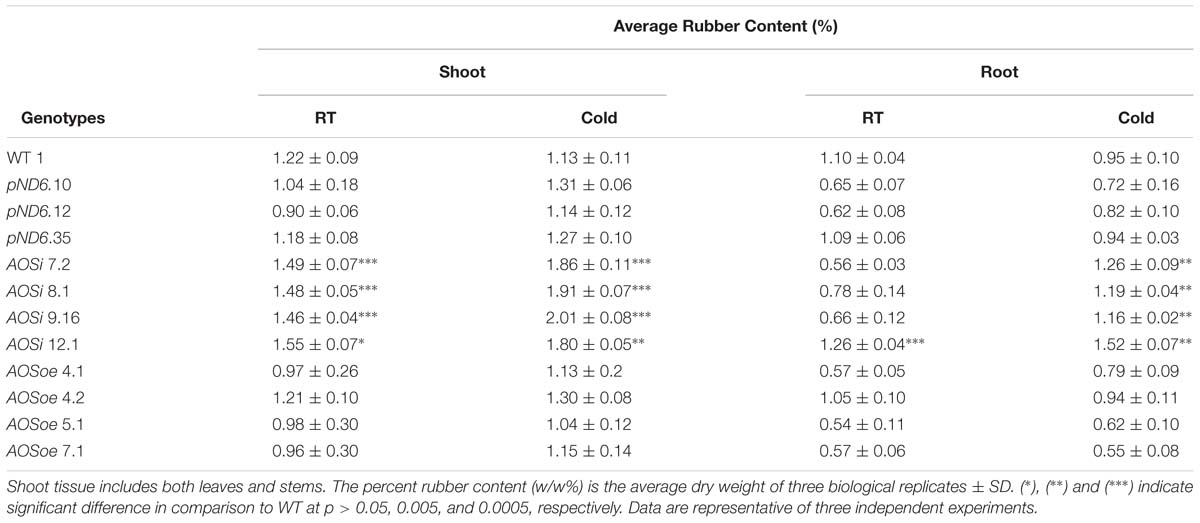
Table 4. Rubber content of growth chamber-grown 5-month-old guayule control and AOS-transgenic lines by Accelerated Solvent Extraction (ASE).
The molecular weight of the extracted rubber was analyzed to assess its quality. Under the tissue culture growing environment, when plants are very young (8 weeks old), AOSi lines showed significantly higher molecular weight rubber compared to all other genotypes (Supplementary Figure S5A). Natural rubber from all 1-year old plants in the greenhouse, however, displayed no differences in molecular weight (Supplementary Figure S5B).
Washed rubber particles isolated from stembark tissues of greenhouse plants were analyzed by light scattering to assess particle size. WRPs from the AOSi lines had the smallest diameters compared to all other genotypes (Figure 6A). Importantly, the rubber transferase activity, quantified by 14C IPP incorporation into new polymer, was significantly enhanced in the AOSi WRPs (Figure 6B). Again, the AOSoe lines were equivalent to WT and pND6 controls.
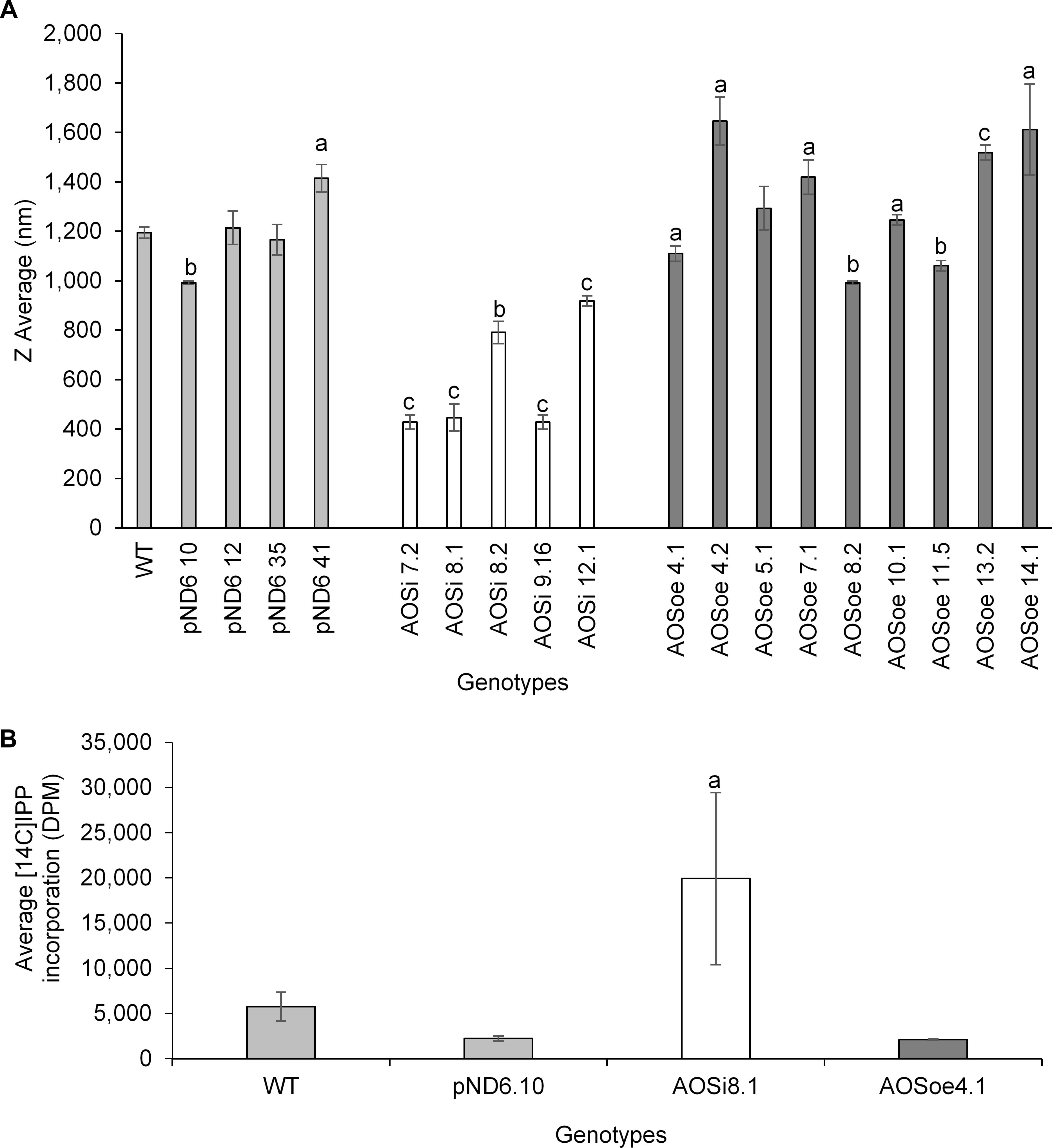
Figure 6. (A) WRP size determination by dynamic light scattering from 1-year old greenhouse-grown WT control and transgenic lines. Statistical significance analysis of the differences was performed with t-test with a, p < 0.05; b, p < 0.005; c, p < 0.0005 relative to the WT. (B) In vitro [14C]IPP incorporation assay. One mg of WRPs from 1-year old WT control and transgenic lines greenhouse plants was assayed in a final volume of 50 μL. Incorporation of labeled IPP was determined using a scintillation counter. Error bars represent SD from the average of three technical replicates. Data are representative of three independent experiments.
AOS Downregulation Resulted in Less Jasmonic Acid, Abscisic Acid, Gibberellic Acid, 12-Oxo-Phytodienoic Acid and More Salicylic Acid
AOS is the first enzyme committed to the production of JA, which was reduced as expected in AOSi lines (Table 5). Hormone profile analysis of tissue-culture grown lines revealed the content (ng/gfw) of JA and its precursor, 12-oxo-phytodienoic acid (OPDA), were significantly reduced to half (or less, down 10-fold) of that found in control lines. Abscisic acid (ABA) and gibberellic acid (GA), were diminished in the AOSi lines compared to WT, pND6 and AOSoe lines, to a lesser but still significant extent (Table 5). Conversely, the SA content was higher in the AOSi lines than in the rest of the lines by about 2-fold. Reducing expression of AOSi in guayule was associated with reduced levels of JA and other hormones as well, but higher levels of SA.
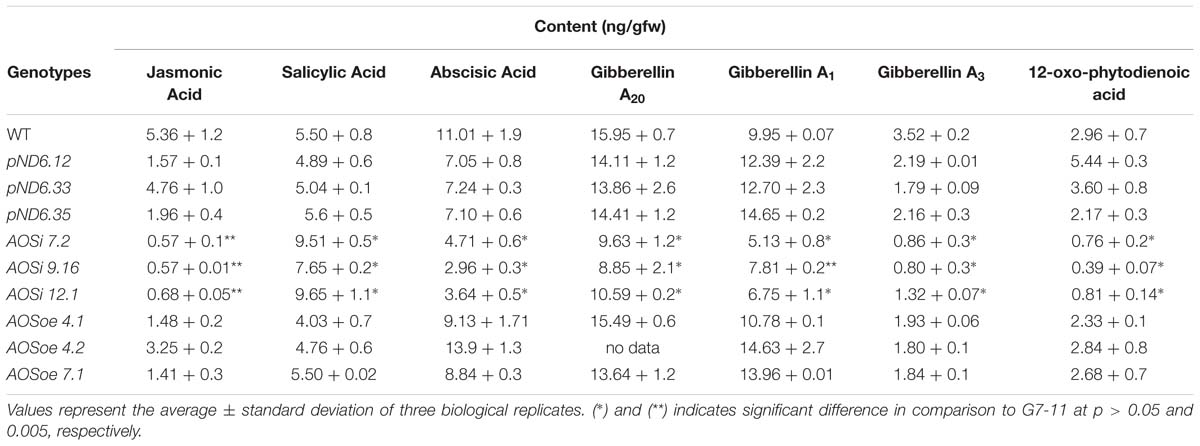
Table 5. Liquid chromatography-tandem spectrometry hormone analysis of stem and leaf tissue from 8-week-old tissue-cultured guayule control and AOS-transgenic lines.
Exogenous Treatment With Salicylic Acid Enhanced Rubber Content
SA (or its analogs) is known to have an inhibitory effect on AOS transcription (Harms et al., 1995) and activity (Pan et al., 1998; Norton et al., 2007). When SA was applied to guayule WT plants grown in tissue culture, the expression of AOS decreased, and the amount of rubber increased, in a dose-dependent manner (Supplementary Figures S6A,B). Under growth chamber conditions, SA treatment in the potting mix reduced AOS expression in guayule stems (Table 6). Rubber content increased, and plant architecture was impacted (increased height and stem diameter; Table 6) with SA potting mix treatment relative to mock or water treated WT plants. We generally observed that, in guayule, exogenous SA treatment resulted in similar phenotypes to AOSi downregulation.
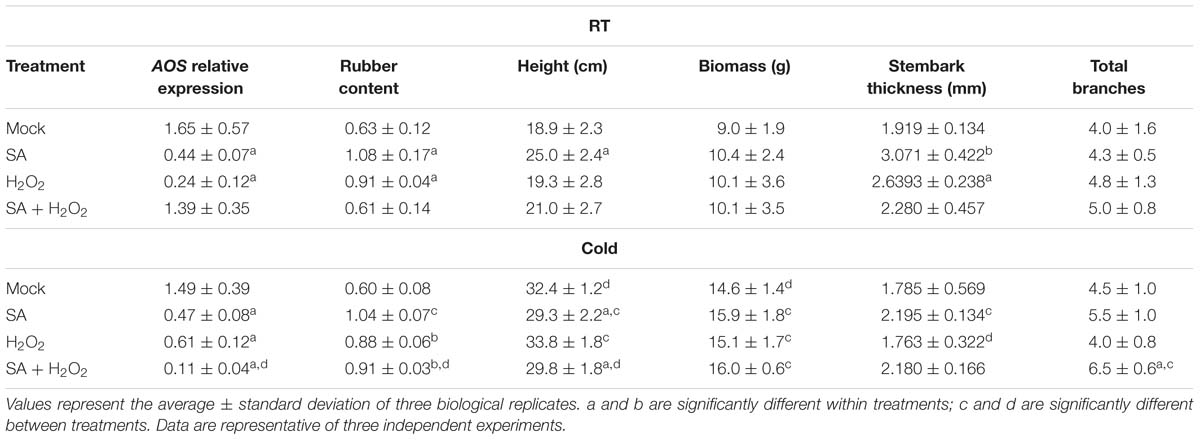
Table 6. Exogenous treatments with SA and H2O2. AOS relative expression in the stem, rubber content and phenotypes of 5-month-old WT guayule plants.
Hydrogen Peroxide Levels and Peroxidase Activity Are Impacted by AOS Downregulation
SA is known to influence hydrogen peroxide (H2O2), in either a self-amplifying or antagonistic manner (Quan et al., 2008). We tested the impact of AOS downregulation on peroxide level and peroxidase activity in tissues. Hydrogen peroxide levels measured in planta were higher for AOSi lines than for controls, under unstressed conditions (Figure 7A), suggesting enhancement between H2O2 and SA (and antagonistic to JA). Extracts from the AOSi lines had significantly lower peroxidase activity than WT, pND6 and AOSoe lines in a RT growth environment, consistent with the higher H2O2 level (Figure 7A). When plants were cold-treated, the WT, pND6 and AOSoe genotypes showed lower peroxidase activity compared to the RT condition. However, the AOSi plants showed no change in peroxidase activity with cold treatment (Figure 7A).
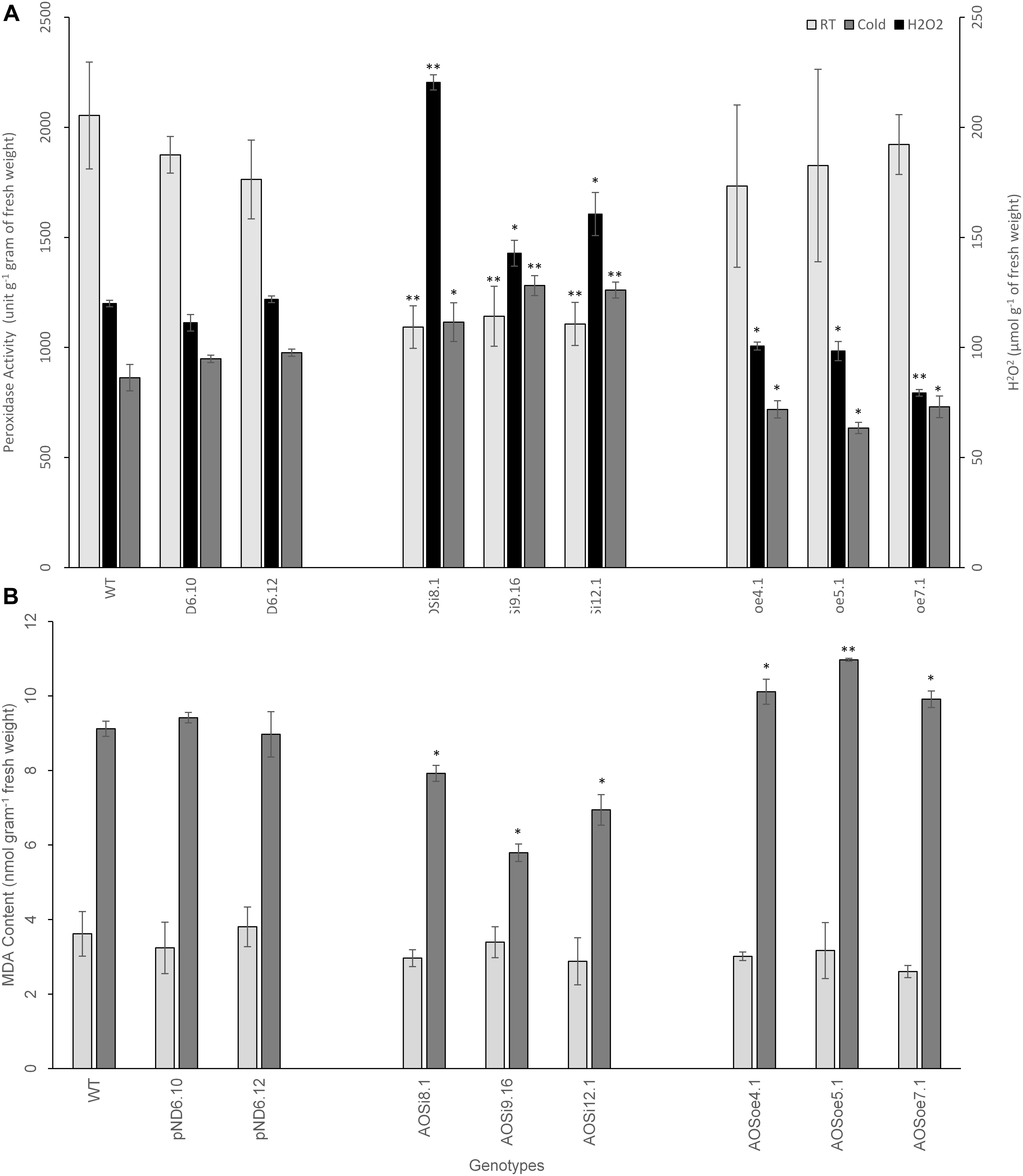
Figure 7. Spectrophotometric measurement from 8-week-old tissue culture-grown guayule control and transgenic lines for (A) H2O2 level and peroxidase activity and (B) lipid peroxidation as measured by MDA content in planta. Average values are from three biological replicates ± SD. ∗ and ∗∗ indicate significant difference at p > 0.05 and p > 0.005, respectively, in comparison to the WT. Data are representative of three independent experiments.
Exogenous Treatment With Hydrogen Peroxide Also Enhanced Rubber Content
Exogenous treatment of WT plants with H2O2 reduced AOS expression (Table 6), and increased rubber content relative to untreated plants both in RT and cold treatments (Table 6). Under room temperature, thickness of the primary stembark was significantly larger. However, thickness of the primary stembark was considerably smaller in cold treated plants (Table 6). Cold significantly increased the height and biomass of all plants compared to RT (Table 6). Other plant growth parameters did not show differences. The addition of both SA and H2O2 together did not enhance the rubber content under room temperature; AOS expression was reduced only under cold treatment with a concomitant increase in rubber content (Table 6).
Lipid Peroxidation Is Reduced Under Cold Stress for Downregulated Lines
Previous studies have suggested that plants produce natural rubber in order to maintain the integrity of the cellular membrane and keep protein-membrane interaction intact upon and/or during stress (Tangpakdee and Tanaka, 1998; Kim et al., 2003). We tested whether the differences in peroxidase activity may have resulted in cellular damage from oxidation. A lipid peroxidation assay was used to estimate the integrity of the cellular membrane for the tissue culture-grown WT and transgenic lines. This method measured the amount of the product of lipid peroxidation, malondialdehyde (MDA). High MDA levels generally indicate internal damage to the cells due to reactive oxygen species such as H2O2. No differences were observed in MDA content between genotypes for guayule plant shoot tissues grown in RT (Figure 7B). However, when cold treatment was imposed, the MDA level increased significantly, suggesting stress-induced cellular damage. Further, the MDA level was slightly lower for the AOSi lines and higher for the AOSoe lines in comparison to the WT and pND6 controls. Therefore, under cold treatment, the higher peroxidase activity in the AOSi lines (Figure 7A) may have mitigated oxidative stress thereby reducing cell wall damage in AOSi lines (Figure 7B). Sundar et al. (2004) reported that protective antioxidant metabolism, including peroxidase activity, increased to mitigate such oxidative stress in guayule exposed to low temperatures.
Discussion
A functional genomics approach was used to dissect the role of AOS in guayule. AOS is a member of the CYP74A enzyme family that metabolizes 13-hydroperoxylinolenic acid (13-HPOT) into allene oxide leading to JA synthesis (Song et al., 1993; Hughes et al., 2009). Levels of phytohormones and plant phenotypes associated with rubber production and growth were investigated in transgenic lines in which AOS was downregulated (AOSi) or overexpressed (AOSoe). Surprisingly, given its prevalence in the rubber-producing sub-cellular rubber particles, down-regulation of AOS in guayule (AOSi) resulted in higher natural rubber content (weight/weight %) compared to the WT, pND6 controls and overexpressed AOS-transformed plants. Interestingly, the physically larger AOSi guayule lines maintained a higher rate of photosynthesis conducive to increased carbon flux and rubber biosynthesis. Salvucci et al. (2010) linked guayule’s carbon fixation rate to higher rubber content (but not higher biomass) in greenhouse plants grown in winter-like conditions. Here, increased photosynthetic activity led to both higher biomass and higher rubber content in AOSi lines. Moreover, AOSi lines were associated with significant architectural changes which could contribute to enhanced natural rubber yield. Hormonal balances were disrupted in the AOSi lines, with changes that ultimately led to increased branching and stem thickness, for instance. Phytohormone interaction networks are complex, and it is clear that additional studies would be required to directly tease out cause and effect of manipulating AOS expression in guayule. Importantly, there were no discernible phenotypes associated with AOS overexpression, regardless of the scale used. The lack of an AOSoe phenotype suggests active control of protein levels, for example at the translational level or through a degradation mechanism. Evidence for transcriptional and post-transcriptional regulation in the JA signaling pathway has been previously reported in Hevea (Pirrello et al., 2014).
While the evolutionary function of rubber in plants is unknown, it has been suggested that natural rubber production serves as a protective mechanism against abiotic and biotic stresses (Tangpakdee and Tanaka, 1998; Kim et al., 2003). Our studies suggest that JA and SA levels may play a role in translation of stress signals relevant to natural rubber biosynthesis in guayule. Young guayule plants with lower endogenous JA due to down-regulation of AOS had consistently higher rubber content. In Hevea, exogenous application of JA (and its precursor, linolenic acid) increased the number of primary laticifers in young Hevea plants (Hao and Wu, 2000), which is related to higher rubber yield (Yu et al., 2008; Tan et al., 2017).
This generally contrasting result could be explained by a multitude of possibilities. It is not unexpected that there would be many physiological differences between the cells in stembark tissues (Kajiura et al., 2018) where rubber is synthesized in guayule, and laticifer cells, where it is synthesized in Hevea and Taraxacum kok-saghz (another rubber producing plant). In T. kok-saghz, treatment with methyl JA (MeJA) upregulated the expression of AOS and other JA associated genes (Cao et al., 2017). These results indicate a feedback loop mechanism exists connecting JA and AOS in T. koksaghyz. Conversely, in Hevea, treatment with JA had no effect on AOS enzyme activity (Norton et al., 2007).
AOS is well-known for its role in the biosynthesis of JA, but may have other functions as well (Pan et al., 1998). Furthermore, we cannot rule out unintentional silencing of other CYP74 family members such as three putative AOS-like sequences identified in a guayule stem RNASeq database (Supplementary Figure S7). Two of those sequences are likely isoforms of AOS as they share 87 and 79% amino acid identity with AOS (AOSL2 and AOSL3, respectively, in Supplementary Figure S7. This is not surprising as the recent guayule draft genome predicts three AOS genes (Valdes Franco et al., 2018). Although the RNAi construct and the potential target region of AOSL2 and -3 are not perfectly matched, it is possible they could be silenced. Silencing of AOSL2 and -3 could certainly contribute to some of the phenotypes observed in the AOSi lines. However, since none of the deduced proteins of AOSL2 and -3 appear to associate with the RP (Kajiura et al., 2018), silencing of these sequences would not directly affect the rubber biosynthetic complex. The third sequence (AOSL4) encodes a putative protein of 147 amino acids (a little over half the size of AOS) with 99% identity to AOS C-terminus. The RNAi construct matches this novel protein 100% and thus is likely silenced. Rubber particle proteomic analysis, however, did not identify this novel protein (Kajiura et al., 2018) and therefore its impact on rubber synthesis if any, remains unknown.
Other potential RNAi off-targets are DES and HPL, both CYP74 enzymes. Like AOS, DES, and HPL are key enzymes of plant oxylipin metabolism (Hughes et al., 2009) and their silencing would disrupt oxylipin signaling other than JA, with potential consequences in plant development. For example, colnelenic acid, a product of DES, affected root development in A. thaliana seedlings (Hughes et al., 2009). Putative guayule DES and HPL transcript sequences were cloned by PCR and sequenced. Expression analysis of these two putative genes showed down-regulation of DES but not HPL (Supplementary Figure S7) in the AOSi lines. It is possible therefore that some of the phenotypes observed in the AOSi lines are in part due to down-regulation of DES.
Down-regulating AOS in guayule not only decreased the amount of JA in planta, but also impacted other endogenous plant hormones suggesting a broad disruption in signaling. Importantly, SA levels were higher in the AOSi lines compared to WT, pND6 and AOSoe genotypes, suggesting that AOS directly or indirectly plays a role in suppressing SA production. The indirect route may be mediated by JA, as the two hormones are known to have an antagonistic relationship. On the other side of the interaction, SA treatment of guayule resulted in downregulation of AOS expression (Table 6). Most notably, the AOS expression, physical phenotypes and rubber content (Figure 3 and Tables 1, 2, 4) of the SA-treated plants (Table 6) showed similar phenotypes to the AOSi lines.
Salicylic acid is known to affect plant growth and development, photosynthesis, and stress response. Since JA is conversely known to inhibit photosynthesis (Qi et al., 2015; Zhu et al., 2015), the enhanced photosynthetic rate could be due to lower JA, higher SA, or both. In AOSi lines; higher levels of SA not only lower barriers to photosynthesis but could also enhance the efficiency of anti-oxidative response during cold stress. Further evidence was observed under cold treatment when NPQ was higher in AOSi lines in comparison with WT, pND6 and AOSoe suggesting that AOSi lines have a better PSII system protection in response to stressful environment. This result conforms with that of Turan et al. (2014), where guayule plants’ NPQ increased systematically with light exposure. They suggested that the higher NPQ served to alleviate the occurrence of photo-oxidative damage and thereby acclimate the plants to high light stress. Therefore, increases in SA, by enhancing photosynthesis, along with a more efficient cold stress response (i.e., higher NPQ) in guayule AOSi lines, result in more carbon supply for rubber synthesis and growth. These observations corroborate earlier reports by Allen et al. (1987) and Sundar and Reddy (2000) regarding the contribution of higher photosynthetic rates to biomass and rubber formation in guayule under cold conditions.
SA’s protective capacity in response to environmental stress in plants, sometimes functions antagonistically to JA (Pieterse et al., 2012), thus allowing the plant to fine tune its stress response and efficiently utilize its resources (Walters and Heil, 2007). The observed higher SA levels in AOSi guayule plants may have been a consequence of the lowered JA synthesis, and induced downstream stress responses. Rubber biosynthesis, usually initiated by cold stress, may instead respond to SA signaling in AOSi plants. Moreover, exogenous addition of SA or H2O2 increased rubber content in guayule, possibly as an induced stress response. Guayule AOS is competitively inhibited by SA (Pan et al., 1998) so it is not surprising that addition of SA and downregulation of AOS produced similar phenotypes. It is tempting to speculate that SA used as a foliar spray on guayule would circumvent the need for seasonal cold periods to maximize rubber production.
Downregulation of AOS reduced the plant tissues’ content of JA, it’s precursor, OPDA, several GAs, and ABA compared to the controls and overexpressed lines, probably due to hormonal crosstalk (Hou et al., 2013; Wasternack and Hause, 2013; Song et al., 2014). Stem morphology phenotypes have been associated with lower concentration of GA, using mutants or inhibitors, resulting in cortical cell expansion in pea plants (Pisum sativum; Tanimoto, 2012) and production of thicker stems in tobacco (Biemelt et al., 2004), consistent with thicker stems found for guayule with lower GA. The guayule AOSi lines also exhibited more branching, along with lower ABA content compared to the controls and AOSoe lines. Disruption in ABA signaling has been shown to negatively regulate auxillary bud growth (Yao and Finlayson, 2015), including in transgenic poplar (Populus × canescens), where reduced sensitivity or lower detectability toward ABA resulted in more lateral bud growth (Arend et al., 2009).
Lastly, OPDA, precursor to JA, has been reported to have a role, independent of JA, in stomatal closure in response to drought in Arabidopsis, tomato and Brassica napus (Savchenko et al., 2014). The Savchenko et al. (2014) study correlated higher OPDA, independent of and in concert with ABA, with reduced stomatal aperture. Therefore our observed physiological phenotypes showing low OPDA, higher stomatal conductance, and improved photosynthetic rate in the AOSi lines is a pertinent and significant finding. Overall, by disrupting hormonal balances in the AOSi lines, a guayule plant was developed that exhibited higher biomass accumulation from higher shoot and root biomass, increased stem branching, thicker stembark tissues and enhanced photosynthetic rate, all advantageous traits for increased natural rubber production.
A remaining question is why do guayule plants maintain high levels of AOS on the surface of rubber particles? The significant decrease in peroxidase activity found in the AOSi lines provides evidence that guayule rubber particle AOS may have an alternative or additional function in addition to or instead of JA biosynthesis. Pan et al. (1998) found recombinant guayule AOS to have a higher affinity for 15S-HPETE (an animal lipid derivative) than for 13S-HPODE or 13S-HPOTE (the usual plant substrates). Another AOS from flaxseed (Song et al., 1993) that is highly homologous to AOS from guayule, except for the chloroplast localization signal sequence (Pan et al., 1998), can also metabolize 15S-HPETE, suggesting alternative, but still unknown, functions in the two plant species.
The AOSi lines’ rubber particles showed reduced particle size as well as higher rubber transferase activity. In Hevea, smaller rubber particles (SRP) have higher transferase activity compared to the larger rubber particles (LRP; Ohya et al., 2000; Rojruthai et al., 2010; Schmidt et al., 2010; Yamashita et al., 2017); and our results provide the first evidence that this may be the case in guayule. Dai et al. (2017) propose a role for the two most abundant proteins in Hevea rubber particles (rubber elongation factor, REF, and small rubber particle protein, SRPP): as rubber is synthesized and the rubber particles grow in size, REF and SRPP associate to maintain the needed structure to stabilize the particle as the volume increases. It is possible then that in guayule less AOS on the particle surface may prevent enlargement and consequently retaining a higher rubber transferase activity in AOSi lines, leading to a more active form of the transferase complex. Hughes et al. (2006) proposed that differences in Arabidopsis CYP74 enzyme activity and substrate specificity are related to the extent of membrane association, even to the point of solubilization. They further pointed out that during pest or pathogen attack, AOS may be released from its membrane-bound state. The possibility that release of AOS from guayule rubber particles, for example, under cold stress, contributes to increased rubber biosynthesis, suggests a new view of the role of the abundant CYP74 rubber particle protein in guayule. This opens several possible avenues to pursue further, including assessment of conformational changes of the rubber transferase complex, and dissecting the role of SA in transducing the cold-induced increase in rubber production and enhancement in biomass. In conclusion, downregulation of AOS in guayule resulted in a plant with disruption of endogenous hormones, and higher photosynthetic rate, biomass, and natural rubber content. Furthermore, SA was found to be a mediating factor in the regulation of AOS expression.
Data Availability
The datasets generated for this study can be found in NCBI, Sequence data from this article can be found in the GenBank data libraries under the following accession numbers: GUSplus (AF354045.1), potato 409T terminator (HK352815.1) and guayule AOS (X78166.2).
Author Contributions
DP, ND, MW, and CM conceived and performed the original research plans and supervised the experiments. DP, GP, MW, and CM wrote the manuscript. GP performed rubber transferase assay, PAGE and western blot analysis. DP and CD performed the AOS enzyme activity and hydrogen peroxide assays, rubber and latex extractions and washed rubber particle analysis. VC and DD provided technical assistance and complemented the writing. RC performed the hormone analysis. ND prepared transformation vectors. ND, B-gK, and TH performed the guayule plant transformation and maintenance of the tissue-cultured plants.
Funding
The work was supported by a Cooperative Research and Development Agreement (58-3k95-3-1653) between Bridgestone Americas, Incorporated and USDA-ARS Western Regional Research Laboratory.
Disclaimer
Mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Conflict of Interest Statement
B-gK is currently employed by Monsanto. VC and DD are employed by Bridgestone Americas.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The work was supported by a Cooperative Research and Development Agreement (58-3k95-3-1653) between Bridgestone Americas, Incorporated and USDA-ARS Western Regional Research Laboratory. The funder played a role in the following: the study design, data analysis, and preparation of the manuscript.
Acknowledgments
We would like to thank Bridgestone Americas, Inc., and the USDA-ARS for the financial support. We sincerely appreciate the assistance of Dr. William Belknap (USDA-ARS, retired) for the guayule genome discussion; Dr. Roger Thilmony (USDA-ARS) for helpful discussions; Dr. Edgar Cahoon and the Lipidomics Core Facility at University of Nebraska, Lincoln for the hormone analysis; Dr. Grace Chen (USDA-ARS), Dr. Charles Lee (USDA-ARS), Dr. Kurt Wagschal (USDA-ARS), Dr. Kent McCue (USDA-ARS) and Dr. Tara McHugh (USDA-ARS) for instrument and equipment access; and Dr. Gary Wall and Ms. Laura Olivieri (USDA-ARS-ALARC) for use of the Li-Cor instrument.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00760/full#supplementary-material
References
Allen, S. G., Nakayama, F. S., Dierig, D. A., and Rasnick, B. A. (1987). Plant water relations, photosynthesis, and rubber content of young guayule plants during water stress. Agron. J. 79, 1030–1035.
Archer, B. L., and Audley, B. G. (1987). New aspects of rubber biosynthesis. Bot. J. Linn. Soc. 94, 181–196. doi: 10.1111/j.1095-8339.1987.tb01045.x
Arend, M., Schnitzler, J. P., Ehlting, B., Hansch, R., Lange, T., Rennenberg, H., et al. (2009). Expression of the Arabidopsis mutant abi1 gene alters abscisic acid sensitivity, stomatal development and growth morphology in gray poplars. Plant Physiol. 151, 2110–2119. doi: 10.1104/pp.109.144956
Backhaus, R. A., Cornish, K., Chen, S. F., Huang, D. S., and Bess, V. H. (1991). Purification and characterization of an abundant rubber particle protein from guayule. Phytochemistry 30, 2493–2497. doi: 10.1016/0031-9422(91)85088-h
Bae, H. K., Kang, H. G., Kim, G. J., Eu, H. J., Oh, S. A., Song, J. T., et al. (2010). Transgenic rice plants carrying RNA interference constructs of AOS (allene oxide synthase) genes show severe male sterility. Plant Breed. 129, 647–651. doi: 10.1111/j.1439-0523.2010.01784.x
Biemelt, S., Tschiersch, H., and Sonnewald, U. (2004). Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis and photosynthesis in transgenic tobacco plants. Plant Physiol. 135, 254–265. doi: 10.1104/pp.103.036988
Brash, A. R. (2009). Mechanistic aspects of CYP74 allene oxide synthases and related cytochrome P450 enzymes. Phytochemistry 70, 1522–1531. doi: 10.1016/j.phytochem.2009.08.005
Cao, X., Yan, J., Lei, J., Li, J., Zhu, J., and Zhang, H. (2017). De novo transcriptome sequencing of MeJA-induced Taraxacum kok-saghyz Rodin to identify genes related to rubber formation. Sci. Rep. 7, 1–13. doi: 10.1038/s41598-017-14890-z
Chang, Z., Li, L., Pan, Z., and Wang, X. (2008). Crystallization and preliminary X-ray analysis of allene oxide synthase, cytochrome P450 CYP74A2, from Parthenium argentatum. Acta Crystallogr. Sec. F Struct. Biol. Cryst. Commun. 64, 668–670. doi: 10.1107/S1744309108017545
Christensen, A. H., and Quail, P. H. (1996). Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5, 213–218. doi: 10.1007/bf01969712
Cornish, K. (2001). Similarities and differences in rubber biochemistry among plant species. Phytochemistry 57, 1123–1134. doi: 10.1016/s0031-9422(01)00097-8
Cornish, K., and Backhaus, R. A. (1990). Rubber transferase activity in rubber particles of guayule. Phytochemistry 29, 3809–3813. doi: 10.1016/0031-9422(90)85337-f
Curtis, O. F. Jr. (1947). Distribution of rubber and resins in guayule. Plant Physiol. 22, 333–359. doi: 10.1104/pp.22.4.333
Dai, L., Nie, Z., Kang, G., Li, Y., and Zeng, R. (2017). Identification and subcellular localization analysis of two rubber elongation factor isoforms on Hevea brasiliensis rubber particles. Plant Physiol. Biochem. 111, 97–106. doi: 10.1016/j.plaphy.2016.11.006
Dierig, D. A., Ray, D. T., and Thompson, A. E. (1989). Variation of agronomic characters among and between guayle lines. Euphytica 44, 265–271. doi: 10.1007/bf00037534
Dong, N., Montanez, B., Creelman, R. A., and Cornish, K. (2006). Low light and low ammonium are key factors for guayule leaf tissue shoot organogenesis and transformation. Plant Cell Rep. 25, 26–34. doi: 10.1007/s00299-005-0024-2
Dong, N., Ponciano, G., McMahan, C. M., Coffelt, T. A., Johnson, L., Creelman, R., et al. (2013). Overexpression of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Parthenium argentatum (guayule). Ind. Crops Prod. 46, 15–24. doi: 10.1016/j.indcrop.2012.12.044
Downes, E. W., and Tonnet, M. (1985). Effect of environmental conditions on growth and rubber production of guayule (Parthenium argentatum). Aust. J. Agric. Res. 36, 285–294.
Garbarino, J. E., and Belknap, W. R. (1994). Isolation of a ubiquitin-ribosomal protein gene (ubi3) from potato and expression of its promoter in transgenic plants. Plant Mol. Biol. 24, 119–127. doi: 10.1007/bf00040579
Gilliland, M. G., and van Staden, J. (1986). Cyclic patterns of growth and rubber deposition in guayule Parthenium argentatum. Suggestions for a management programme. S. Afr. J. Plant Soil 3, 21–26. doi: 10.1080/02571862.1986.10634180
Gilliland, M. G., van Staden, J., and Bruton, A. G. (1984). Studies on the translocation system of guayule (Parthenium argentatum Gray). Protoplasma 122, 169–177. doi: 10.1007/bf01281694
Goss, R. A., Benedict, C. R., Keithly, J. H., Nessler, C. L., and Stipanovic, R. D. (1984). Cis- polyisoprene synthesis in guayule plants (Parthenium argentatum) exposed to low nonfreezing temperatures. Plant Physiol. 74, 534–537. doi: 10.1104/pp.74.3.534
Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. doi: 10.1007/bf00014672
Hao, B. Z., and Wu, J. L. (2000). Laticifer differentiation in Hevea brasiliensis: induction by exogenous jasmonic acid and linolenic acid. Ann. Bot. 85, 37–43. doi: 10.1006/anbo.1999.0995
Harms, K., Atzorn, R., Brash, A., Kühn, H., Wasternack, C., Willmitzer, L., et al. (1995). Expression of a flax allene oxide synthase cDNA leads to increased endogenous jasmonic acid (JA) levels in transgenic potato plants but not to a corresponding activation of JA-responding genes. Plant Cell 7, 1645–1654. doi: 10.1105/tpc.7.10.1645
Heath, R. L., and Parker, L. (1968). Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. doi: 10.1016/0003-9861(68)90654-1
Herrera-Estrella, L., DeBlock, M., Messens, E., Hernalsteens, J. P., Van Montagu, M., and Schell, J. (1983). Chimeric genes as dominant selectable markers in plant cells. EMBO J. 2, 987–996.
Hood, E. E., Helmer, G. L., Fraley, R. T., and Chilton, M. (1986). The hypervirulence of Agrobacterium tumefaciens A281 is encoded a region of pTBo542 outside of T-DNA. J. Bacteriol. 168, 1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986
Hou, X., Ding, L., and Yu, H. (2013). Crosstalk between GA and JA signaling mediates plant growth and defense. 32, 1067-1074. Plant Cell Rep. 32, 1067–1074. doi: 10.1007/s00299-013-1423-4
Hughes, R. K., Belfield, E. J., Ashton, R., Fairhurst, S. A., Göbel, C., Stumpe, M., et al. (2006). Allene oxide synthase from Arabidopsis thaliana (CYP74A1) exhibits dual specificity that is regulated by monomer-micelle association. FEBS Lett. 580, 4188–4194. doi: 10.1016/j.febslet.2006.06.075
Hughes, R. K., De Domenico, S., and Santino, A. (2009). Plant cytochrome CYP74 family, Biochemical features, endocellular localization, activation mechanism in plant defense and improvements for industrial applications. Chembiochem 10, 1122–1133. doi: 10.1002/cbic.200800633
Illut, D. C., Sanchez, P. L., Coffelt, T. A., Dyer, J. M., Jenks, M. A., and Gore, M. A. (2017). A century of guayule, comprehensive genetic characterization of the guayule (Parthenium argentatum A. Gray) USDA germplasm collection. Ind. Crops Prod. 109, 300–309. doi: 10.1016/j.indcrop.2017.08.029
Ji, W., Benedict, C. R., and Foster, M. A. (1993). Seasonal variations in rubber biosynthesis, 3- hydroxy-3-methylglutaryl-coenzyme A reductase, and rubber transferase activities in Parthenium argentatum in the Chihuahuan desert. Plant Physiol. 103, 535–542. doi: 10.1104/pp.103.2.535
Junglee, S., Urban, L., Sallanon, H., and Lopez-Lauri, F. (2014). Optimized assay for hydrogen peroxide determination in plant tissue using potassium iodide. Am. J. Analyt. Chem. 5, 730–736. doi: 10.4236/ajac.2014.511081
Kajiura, H., Suzuki, N., Mouri, H., Watanabe, N., and Nakazawa, Y. (2018). Elucidation of rubber biosynthesis and accumulation in the rubber producing shrub, guayule (Parthenium argentatum Gray). Planta 247, 513–526. doi: 10.1007/s00425-017-2804-7
Kim, J. S., Kim, Y. O., Ryu, H. J., Kwak, Y. S., Lee, J. Y., and Kang, H. (2003). Isolation of stress-related genes or rubber particles and latex in Fig Tree (Ficus carica) and their expressions by abiotic stress or plant hormone. Plant Cell Physiol. 44, 412–414. doi: 10.1093/pcp/pcg058
Konno, K. (2011). Plant latex and other exudates as plant defense systems, roles of various defense chemical and proteins contained therein. Phytochemistry 52, 1510–1530. doi: 10.1016/j.phytochem.2011.02.016
Kuruvadi, S., Canto, D. J., and Angulo-Sanchez, J. L. (1997). Rubber content in different plant parts and tissues of Mexican guayule shrubs. Ind. Crops Prod. 7, 19–25. doi: 10.1016/s0926-6690(97)00033-2
Laudert, D., Schaller, F., and Weiler, E. W. (2000). Transgenic Nicotiana tabacum and Arabidopsis thaliana plants overexpressing allene oxide synthase. Planta 211, 163–167.
Lewinsohn, T. M. (1991). The geographical distribution of plant latex. Chemoecology 2, 64–68. doi: 10.1007/bf01240668
Li, L., Chang, Z., Pan, Z., Fu, Z.-Q., and Wang, X. (2008). Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 105, 13883–13888. doi: 10.1073/pnas.0804099105
Liu, X., Li, F., Tang, J., Wang, W., Zhang, F., Wang, G., et al. (2012). Activation of the jasmonic acid pathway by depletion of the hydroperoxide lyase OsHPL3 reveals crosstalk between the HPL and AOS branches of the oxylipin pathway in rice. PLoS One 7:e50089. doi: 10.1371/journal.pone.0050089
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real- time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luttgeharm, K. D., Chen, M., Mehra, A., Cahoon, R. E., Markham, J. E., and Cahoon, E. B. (2015). Overexpression of Arabidopsis ceramide synthases differentially affects growth, sphingolipid metabolism, programmed cell death and mycotoxin resistance. Plant Physiol. 169, 1108–1117. doi: 10.1104/pp.15.00987
Macrae, S., Gilliland, M. G., and Van Staden, J. (1986). Rubber production in guayule, determination of rubber producing potential. Plant Physiol. 81, 1027–1032. doi: 10.1104/pp.81.4.1027
Madhavan, S., Greenblatt, G. A., Foster, M. A., and Benedict, C. R. (1989). Stimulation of isopentenyl pyrophosphate incorporation into polyisoprene in extracts from guayule plants (Parthenium argentatum Gray) by low temperature and 2- (3,4- dichloro- phenoxy)triethylamine. Plant Physiol. 89, 506–511. doi: 10.1104/pp.89.2.506
Mei, C., Qi, M., Sheng, G., and Yang, Y. (2006). Inducible overexpression of a rice oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 10, 1127–1137. doi: 10.1094/mpmi-19-1127
Mooibroek, H., and Cornish, K. (2000). Alternative sources of natural rubber. Appl. Microbiol. Biotechnol. 53, 355–365. doi: 10.1007/s002530051627
Nawamawat, K., Sakdapipanich, J. T., Ho, C. C., Ma, Y., Song, J., and Vancso, J. G. (2011). Surface nanostructure of Hevea brasiliensis natural rubber latex particles. Colloids Surf. A 390, 157–166. doi: 10.1016/j.saa.2011.07.024
Norton, G., Pappusamy, A., Yusof, F., Pujade-Renaud, V., Perkins, M., Griffiths, D., et al. (2007). Characterization of recombinant Hevea brasiliensis allene oxide synthase, Effects of cyclooxygenase inhibitors, lipoxygenase inhibitors and salicylates on enzyme activity. Plant Physiol. Biochem. 45, 129–138. doi: 10.1016/j.plaphy.2007.01.003
Nyaka-Ngobisa, A. I. C., Zainal-Abidin, M. A., Wong, M. Y., and Wan-Noordin, M. W. D. (2013). Neofusicoccum ribis associated with leaf blight on rubber (Hevea brasiliensis) in Peninsular Malaysia. Plant Pathol. J. 29, 10–16. doi: 10.5423/PPJ.OA.07.2012.0110
Ohya, N., Tanaka, Y., Wititsuwannakul, R., and Koyama, T. (2000). Activity of rubber transferase and rubber particle size in Hevea latex. J. Rubb. Res. 3, 214–221.
Pan, X., Welti, R., and Wang, X. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 5, 986–992. doi: 10.1038/nprot.2010.37
Pan, Z., Camara, B., Gardner, H. W., and Backhaus, R. A. (1998). Aspirin inhibition and acetylation of the plant cytochrome P450 allene oxide synthase, resembles that of animal prostaglandin endoperoxide H synthase. J. Biol. Chem. 273, 18139–18145. doi: 10.1074/jbc.273.29.18139
Pan, Z., Durst, F., Werck-Reichhart, D., Gardner, H. W., Camara, B., Cornish, K., et al. (1995). The major protein of guayule rubber particles is a cytochrome P450. Characterization based on cDNA cloning and spectroscopic analysis of the solubilized enzyme and its reaction products. J. Biol. Chem. 270, 8487–8494. doi: 10.1074/jbc.270.15.8487
Park, J. H., Halitschke, R., Kim, H. B., Baldwin, I. T., Feldman, K. A., and Feyereisen, R. (2002). A knock- out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31, 1–12. doi: 10.1046/j.1365-313x.2002.01328.x
Pearson, C. H., Cornish, K., and Rath, D. J. (2013). Extraction of natural rubber and resin from guayule using an accelerated solvent extractor. Ind. Crops Prod. 43, 506–510. doi: 10.1016/j.indcrop.2012.06.052
Pieterse, C. M. J., Van der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S. C. M. (2012). Hormonal modulation of plant immunity. Ann. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Pirrello, J., Leclercq, J., Dessailly, F., Rio, M., Piyatrakul, P., Kuswanhadi, K., et al. (2014). Transcriptional and post-transcriptional regulation of the jasmonate signaling pathway in response to abiotic and harvesting stress in Hevea brasiliensis. BMC Plant Biol. 14:341. doi: 10.1186/s12870-014-0341-0
Ponciano, G., McMahan, C. M., Xie, W., Lazo, G. R., Coffelt, T. A., Collins-Silva, J., et al. (2012). Transcriptome and gene expression analysis in cold-acclimated guayule (Parthenium argentatum) rubber- producing tissue. Phytochemistry 79, 57–66. doi: 10.1016/j.phytochem.2012.04.007
Putter, J. (1974). “Peroxidases,” in Methods of Enzymatic Analysis, Vol. 2, ed. H. U. Bergmeyer (Cambridge, MA: Academic Press), 685–690.
Qi, T., Wang, J., Huang, H., Liu, B., Gao, H., Liu, Y., et al. (2015). Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27, 1634–1649. doi: 10.1105/tpc.15.00110
Quan, L. J., Zhang, B., Shi, W. W., and Li, H. Y. (2008). Hydrogen peroxide, a versatile molecule of the reactive oxygen species network. J. Integr. Plant Biol. 50, 2–18. doi: 10.1111/j.1744-7909.2007.00599.x
Radziah, N. Z., and Chee, K. H. (1989). A new foliar disease of rubber. Plant Pathol. 38, 293–296. doi: 10.1007/s00484-018-1598-z
Ray, D. T., Dierig, D. A., Thompson, A. E., and Coffelt, T. A. (1999). Registration of six guayule germplasms with high yielding ability. Crop. Sci. 39:300. doi: 10.2135/cropsci1999.0011183x003900010073x
Rockhold, D. R., Chang, S., Taylor, N., Allen, P. V., McCue, K. F., and Belknap, W. R. (2008). Structure of two Solanum bulbocastanum polyubiquitin genes and expression of their promoters. Am. J. Pot. Res. 85, 219–226. doi: 10.1007/s12230-008-9015-5
Rojruthai, P., Sakdapipanich, J. T., Takahashi, S., Hyegin, L., Noike, M., Koyama, T., et al. (2010). In vitro synthesis of high molecular weight rubber by Hevea small rubber particles. J. Biosci. Bioeng. 109, 107–114. doi: 10.1016/j.jbiosc.2009.08.009
Salvucci, M. E., Barta, C., Byers, J. A., and Canarini, A. (2010). Photosynthesis and assimilate partitioning between carbohydrates and isoprenoid products in vegetatively active and dormant guayule, physiological and environmental constraints on rubber accumulation in a semiarid shrub. Physiol. Plant. 140, 368–379. doi: 10.1111/j.1399-3054.2010.01409.x
Sansatsadeekul, J., Sakdapipanich, J., and Rojruthai, P. (2011). Characterization of assocated proteins and phospholipids in natural rubber latex. J. Biosci. Bioeng. 111, 628–634. doi: 10.1016/j.jbiosc.2011.01.013
Savchenko, T., Kolla, V. A., Wang, C.-Q., Nasafi, Z., Hicks, D. R., Phadungchod, B., et al. (2014). Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol. 164, 1151–1160. doi: 10.1104/pp.113.234310
Schmidt, T., Lenders, M., Hillebrand, A., van Deenen, N., Munt, O., Reichelt, R., et al. (2010). Characterization of rubber particles and rubber chain elongation in Taraxacum kok-saghyz. BMC Biochem. 11:11. doi: 10.1186/1471-2091-11-11
Siler, D. J., and Cornish, K. (1994). Hypoallergenicity of guayule rubber particle proteins compared to Hevea latex proteins. Ind. Crops Prod. 2, 307–313. doi: 10.1016/0926-6690(94)90122-8
Song, S., Qi, T., Wasternack, C., and Xie, D. (2014). Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 21, 112–119. doi: 10.1016/j.pbi.2014.07.005
Song, W. C., and Brash, A. R. (1991). Purification and characterization of an allene oxide synthase and identification of the enzyme as a cytochrome P450. Science 253, 781–784. doi: 10.1126/science.1876834
Song, W. C., Funk, C. D., and Brash, A. R. (1993). Molecular cloning of an allene oxide synthase, a cytochrome P450 specialized for the metabolism of fatty acid hydroperoxides. Proc. Natl. Acad. Sci. U.S.A. 90, 8519–8523. doi: 10.1073/pnas.90.18.8519
Stenzel, I., Hause, B., Maucher, H., Pitzschke, A., Miersch, O., Ziegler, J., et al. (2003). Allene oxide cyclase dependence of the wound response and vascular bundle-specific generatin of jasmonates in tomato-amplication in wound signalling. Plant J. 33, 577–589. doi: 10.1046/j.1365-313x.2003.01647.x
Sundar, D., Chaitanya, K. V., Jutur, P. P., and Reddy, R. A. (2004). Low temperature-induced changes in antioxidative metabolism in rubber-producing shrub, guayule (Parthenium argentatum Gray). Plant Growth Regul. 44, 175–181. doi: 10.1023/b
Sundar, D., and Reddy, R. (2000). Low night temperature-induced changes in photosynthesis and rubber accumulation in guayule (Parthenium argentatum Gray). Photosynthetica 3, 421–427.
Tan, D., Hu, X., Fu, L., Kumpeangkeaw, A., Ding, Z., Sun, X., et al. (2017). Comparative morphology and transcriptome analysis reveals distinct functions of the primary and secondary laticifer cells in the rubber tree. Sci. Rep. 7, 1–17. doi: 10.1038/s41598-017-03083-3
Tangpakdee, J. A., and Tanaka, Y. (1998). Why rubber trees produce polysioprene-a possible role of natural rubber in the Hevea Tree. J. Rubb. Res. 1, 77–83.
Tanimoto, E. (2012). Tall or short? Slender or thick? A plant strategy for regulating elongation growth of roots by low concentrations of gibberellin. Ann. Bot. 110, 373–381. doi: 10.1093/aob/mcs049
Turan, S., Kumar, S., and Cornish, K. (2014). Photosynthetic response of in vitro guayule plants in low and high lights and the role of non-photochemical quenching in plant acclimation. Ind. Crops Prod. 54, 266–271. doi: 10.1016/j.indcrop.2014.01.022
Valdes Franco, J. A., Wang, Y., Huo, N., Ponciano, G., Colvin, H. A., McMahan, C. M., et al. (2018). Modular assembly of transposable element arrays by microsatellite targeting in the guayule and rice genomes. BMC Genomics 19:271. doi: 10.1186/s12864-018-4653-6
van Beilen, J. B., and Poirier, Y. (2007). Establishment of new crops for the production of natural rubber. Trends Biotechnol. 25, 522–529. doi: 10.1016/j.tibtech.2007.08.009
von Malek, B., van der Graaff, E., Schneitz, K., and Keller, B. (2002). The Arabidopsis male- sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216, 187–192. doi: 10.1007/s00425-002-0906-2
Walters, D., and Heil, M. (2007). Costs and trade-offs associated with induced resistance. Physiol. Mol. Plant Pathol. 71, 3–17. doi: 10.1016/j.pmpp.2007.09.008
Wang, C., Avdiushko, S., and Hildebrand, D. F. (1999). Overexpression of a cytoplasm- localized allene oxide synthase promotes the wound-induced accumulation of jasmoinic acid in transgenic tobacco. Plant Mol. Biol. 40, 783–793.
Wasternack, C., and Hause, B. (2013). Jasmonates, biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Wasternack, C., and Song, S. (2017). Jasmonates, biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68, 1303–1321. doi: 10.1093/jxb/erw443
Whalen, M., McMahan, C., and Shintani, D. (2013). “Development of crops to produce industrially useful natural rubber,” in Isoprenoid Synthesis in Plants and Microorganisms, New Concepts and Experimental Approaches, eds T. Bach and M. Rohmer (New York, NY: Springer), 329–345. doi: 10.1007/978-1-4614-4063-5_23
Yamashita, S., Mizuno, M., Hayashi, H., Yamaguchi, H., Miyagi-Inoue, Y., Fushihara, K., et al. (2017). Purification and characterization of small and large rubber particles from Hevea brasiliensis. Biosci. Biotechnol. Biochem. 82, 1011–1020. doi: 10.1080/09168451.2017.1401913
Yao, C., and Finlayson, S. A. (2015). Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol. 169, 611–626. doi: 10.1104/pp.15.00682
Yu, J. H., Zeng, X., Yang, S. G., Huang, H. S., and Tian, W. M. (2008). Relationship between rate of laticifer differentiation, number of laticifer rows and rubber yield among 1981 IRRDB Germplasm collections of Hevea brasiliensis. J. Rubb. Res. 11, 43–51.
Keywords: allene oxide synthase, guayule, natural rubber, RNAi, jasmonic acid, salicylic acid
Citation: Placido DF, Dong N, Dong C, Cruz VMV, Dierig DA, Cahoon RE, Kang B-g, Huynh T, Whalen M, Ponciano G and McMahan C (2019) Downregulation of a CYP74 Rubber Particle Protein Increases Natural Rubber Production in Parthenium argentatum. Front. Plant Sci. 10:760. doi: 10.3389/fpls.2019.00760
Received: 29 March 2019; Accepted: 24 May 2019;
Published: 26 June 2019.
Edited by:
Goetz Hensel, Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK), GermanyReviewed by:
Guangyuan He, Huazhong University of Science and Technology, ChinaThomas J. Bach, Université de Strasbourg, France
Copyright © 2019 Placido, Dong, Dong, Cruz, Dierig, Cahoon, Kang, Huynh, Whalen, Ponciano and McMahan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Grisel Ponciano, Z3Jpc2VsLnBvbmNpYW5vQGFycy51c2RhLmdvdg==; Colleen McMahan, Y29sbGVlbi5tY21haGFuQGFycy51c2RhLmdvdg==
 Dante F. Placido
Dante F. Placido Niu Dong
Niu Dong Chen Dong
Chen Dong Von Mark V. Cruz
Von Mark V. Cruz David A. Dierig
David A. Dierig Rebecca E. Cahoon3
Rebecca E. Cahoon3 Byung-guk Kang
Byung-guk Kang Grisel Ponciano
Grisel Ponciano Colleen McMahan
Colleen McMahan