- Key Laboratory of Plant Hormones and Development Regulation of Chongqing, School of Life Sciences, Chongqing University, Chongqing, China
Environmental stresses, such as temperature, heavy metals, drought, cold, and microbial infections adversely damage various aspects of plant growth and development. Salinity and drought are among major hazardous factors, which adversity affects plant growth and productivity. Transcription factors, such as basic helix-loop-helix play critical roles in regulating plant physiological processes under abiotic stresses. In this study, we presented the characterization of a tomato SlbHLH22 gene under abiotic stresses such as drought and salinity. Plants overexpressing SlbHLH22 showed short height with small leaves and enhanced flavonoid accumulation. In wild type (WT) plant, the elevated levels of SlbHLH22 were detected under salt and D-mannitol stresses. Subcellular localization analysis revealed that SlbHLH22 protein was targeted to the nucleus in onion epidermal cells. Transactivation assay in yeast demonstrated that SlbHLH22 had transcriptional activation ability. The transgenic plants overexpressing SlbHLH22 displayed enhanced vigor and more tolerant to drought and salinity than WT. Overexpression of SlbHLH22 significantly peaked the activities of catalase (CAT), superoxide dismutase (SOD), and peroxidase (POD) to minimize the impacts of reactive oxygen species such as H2O2, which was reduced significantly in transgenic plants along with Malondialdehyde (MDA). Moreover, the expression levels of ROS defense genes (SlPOD, SlCAT, SlSOD), ABA biosynthesis genes, proline biosynthesis, and flavonoids synthesis genes were also activated under salinity and drought. Taken together, our study implies that the overexpression of SlbHLH22 improved tomato plant stress resistance by improving ROS scavenging system, increasing osmotic potential and enhanced accumulation of secondary metabolites in tomato plants.
Introduction
Environmental stresses, such as salinity, drought, temperature, and pathogen invasion can adversely affect plant growth, development, and subsequently impacting its productivity. Plants have evolved various strategies to cope with such stresses, including biochemical, physiological, cellular, and molecular strategies (Thomashow, 1999; Nakashima et al., 2009). In general, plant responses to abiotic stresses are under the transcriptional control of various stress-induced genes and their activation or suppression results in a response to stimuli (Pang et al., 2011; Han et al., 2014; Sun et al., 2015).
The basic helix-loop-helix (bHLH) is one of the largest gene family in plants and has DNA binding and dimerization capabilities as bHLH domain existed. bHLH domain contains approximately 60 amino acid with a basic region and 2 functionally distinct regions of the HLH region in their protein sequences (Peng et al., 2013). bHLH proteins identified in various plant species, such as Arabidopsis, poplar, rice (Carretero-Paulet et al., 2010), maize (Zhang T. et al., 2018), grapes (Wang et al., 2018), and peaches (Zhang C. et al., 2018). However, this large number of bHLH genes in plants may leads to demonstrations that these plant bHLH proteins may severe as a key regulatory components in transcriptional activation or suppression of wide range of plant development, metabolic processes, and responses related to abiotic stresses (Peng et al., 2013).
The members of bHLH gene family have been reported to be related to responses to abiotic stresses, such as salinity, drought, and cold. RsICE1 from Raphanus sativus, a stress-responsive bHLH TF enhances cold tolerance in rice through interacting with CBF/DREB1 (Man et al., 2017). ICE1 of Pyrus ussuriensis play pivotal roles in improving cold tolerance by increasing the transcriptional regulation of PuDREB via interaction with PubHLH1 (Huang et al., 2015). Rice OsbHLH148 could functions in JA-mediated drought tolerance (Seo et al., 2011), while Arabidopsis bHLH122 acts as a transcriptional activator provides drought and osmotic resistance through enhanced proline accumulation and by activating ROS scavenging system (Liu et al., 2014, 2015). Vitis vinifera VvbHLH1 has potential to improve tolerance to drought and salinity by regulating the accumulation of flavonoids and acts as regulator of ABA signaling (Wang et al., 2016). In addition to these diverse biological functions, the bHLH proteins were also reported to involve in various biosynthetic pathways such as anthocyanins and flavanols (Winkel-Shirley, 2001; Baudry et al., 2004).
Solanum lycopersicum L. is an ideal model plant for fruit development and its productivity is adversely affected by various abiotic stresses, such as salinity, drought, and temperature. The establishment of stress tolerant crop is key challenge in genetic engineering and biotechnology. We have previously shown that overexpression of SlbHLH22 promotes flowering and fruit ripening and enhanced sensitivity to phytohormones with decreased fruit shelf life in tomato (Waseem and Li, 2019; Waseem et al., 2019). To investigate, whether overexpression of SlbHLH22 improves plants tolerance to salinity and drought. The transgenic plants subjected to salinity and drought stresses. In this study, we found that SlbHLH22 was peaked significantly under salt stress. Our studies demonstrated that the overexpression of SlbHLH22 confers abiotic stress tolerance by regulating the expression of stress-inducible genes that are involved in physiological changes, including reactive oxygen species (ROS) scavenging system, abscisic acid (ABA) signaling, flavonoid biosynthesis pathway, and proline biosynthesis. We believe, our study might provide a new insight into the functional characterization of the bHLH gene family members during stress tolerance in tomato and other pant species.
Materials and Methods
Plant Growth Conditions and Collection
The surface sterilized seeds of Solanum lycopersicum L. cv. Micro-Tom wildtype (WT) and transgenic lines and empty vector (VC) were grown in green house under following conditions: 16 h/8 h light/dark cycle, 25°C/18°C day/night temperature, light intensity 250 μ mol m−2s−1, and 80% relative humidity. For gene expression analysis, plant parts, such as root, leaves, stem, flowers (in bud and fully opened), and flower parts (sepal, petal, carpel, and stamens) were harvested from 4-week-old plants (Waseem et al., 2018). For each sample, each plant part was collected from 10 plants were mixed and frozen in liquid nitrogen.
Plasmid Construction, Transformation, and Generation of Transgenic Plants
For overexpression, the K303 expression vector (Gateway technology) under CaMV 35S promoter was constructed as described in Waseem et al. (2019). For RNAi a 217 bp long SlbHLH22 fragment for sense and antisense silencing in pCAMBIA 2301 vector. The specific primers used for RNAi are listed in Supplementary Table S1. The transgenic line plants were generated by agrobacterium-mediated (Agrobacterium tumefaciens strain, GV310) transformation in WT tomato plants as described by Xian et al. (2014). The transgenic plants were screened on MS media supplemented with kanamycin (100 mg L−1). The generated kanamycin resistant seedlings were transferred to green house for further growth under control conditions and then verified with successful qRT-PCR. The homozygous T3 lines were used for further analysis.
Subcellular Localization and Transcriptional Activity of SlbHLH22
The SlbHLH22 ORF without the stop codon was amplified and cloned into the pGreen0029 vector. The recombinant plasmid containing the SlbHLH22-GFP fusion gene and the control plasmid with GFP alone were transformed into onion epidermal cells using Agrobacterium-mediated transformation as described by Sun et al. (2007). For trans-activation assays, to produce pBD- SlbHLH22 the coding sequence of SlbHLH22 was amplified and ligated into the yeast expression vector pGBKT7 (Clontech, United States). pBD-SlbHLH22, pGBKT7 (plasmid for negative control), and pGBKT7-53+pGADT7-T (plasmid combination for positive control) were transformed separately into the yeast strain AH109 according to the manufacturer’s protocol. Transformants were selected on SD/-Trp or SD/-Ade/-His/-Trp drop-out medium (Clontech, United States). After colony formation, the trans-activation activity of each protein was examined by comparing growth on permissive and selective medium and the activity of X-gal (40 μg mL−1, 5-bromo-4-chloro-3-indoxyl-α-D-galactopyranoside).
Determination of Total Flavonoids
Total flavonoid contents were determined by the AlCl3 method as described by Koolen et al. (2013) with slight modifications. 2.5 g of tomato leaves (WT, 35S:SlbHLH22 and RNAi lines) were ground in liquid nitrogen and dissolved in 70% (by vol.) ethanol solution and incubated at room temperature for 24 h. 1 mL of ethanol extract was diluted with 1 mL of AlCl3 (5%, w/v) and incubated for 1 h at room temperature. The mixture was centrifuged at 10,000 rpm for 10 min and supernatant was collected in a new tube. One volume of chloroform was added to remove chlorophylls. The mixture was centrifuged at 8000 rpm for 5 min and supernatant was used to measure absorbance at 430 nm. Total flavonoids were expressed in mg quercetin equivalent/g dry weight (Koolen et al., 2013).
Salt and Drought Stress Assay
For salt, osmotic, and oxidative stresses, the leaves from 5-week-old WT seedling were sprayed with 200 mM NaCl (Waseem and Ahmad, 2019), 100 mM mannitol, and 100 mM hydrogen peroxide, respectively. Leaves were harvested at 0, 3, 6, 9, 12, and 24 h. Leaves harvested at 0 h were used as control. For each sample, leaves were collected from 10 plants, mixed and all the experiments were performed in triplicate.
For salinity and drought stresses, each 15 plants of WT and transgenic lines were placed in big pot, watered twice in week to make sure water was uniform in all pots and were grown under same light and temperature conditions. For salinity stress, 6-week-old plants were irrigated with 200 mM (200 mL per pot, 9 cm) NaCl for every 48 h in the following 18 days. However, for drought stress, withhold water for up to 30 days followed by rehydrated for 10 days. The control plants were watered normally. During treatment, the relative water content (RWC, %) and total chlorophyll content was assessed (Pan et al., 2012). The leaves at same development stage were harvested and store immediately at −80°C till further analysis.
Assessment of Antioxidant Enzyme Activity and ABA Content Measurement
During salinity and drought treatment, leaves at same development stages were harvested from plants for antioxidant enzyme activity such as catalase (CAT. EC 1.11.1.6) superoxide dismutase (SOD, EC 1.15.1.1), peroxidase (POD, EC 1.11.1.7), H2O2 content, Malondialdehyde (MDA), soluble sugar content, and proline content assessment. MDA following method by Heath and Packer (1968). Briefly, about 0.5 g of tomato leaves were ground in 2 mL of the chilled reagent [0.25% (w/v) thio-barbituric acid in 10% (w/v) trichloroacetic acid]. The extracts were incubated at 100°C for 30 min, cooled to room temperature. The extracts were centrifuged at 12,000 rpm for 15 min and absorbance of the supernatant was measured at 450, 532, and 600 nm. The MDA content was calculated based on the following equation: 6.45 × (OD532–OD600) −0.559 × OD450.
Soluble sugar content was measured according to method described by Fukao et al. (2006). Proline contents were determined following Bates et al. (1973). About 0.5 g of tomato leaves were ground into powder with liquid nitrogen and extracted in 3% sulfosalicylic acid. After centrifuging at 12,000 rpm for 10 min, the supernatant (2 mL) was mixed with equal volume of ninhydrin reagent [2.5% (w/v) ninhydrin, 60% (v/v) glacial acetic acid, and 40% 6 M phosphoric acid] and of glacial acetic acid, incubated at 100°C for 40 min. The reaction was terminated in an ice bath. Then, the reaction mixture was extracted with 4 mL of toluene. The absorbance was measured at 520 nm with a UV-5200 spectrophotometer.
For SOD activity, 1 g of frozen leaves tomato leaves were homogenized in 5 ml of cold 20 mM HEPES buffer (pH 7.2, 1 mM EGTA, 210 mM mannitol, 70 mM sucrose) then centrifuged at 2,500 rpm for 5 min at 4°C. The enzyme activity SOD were measured following Mittova et al. (2000). Total protein from tomato leaves was extracted with 0.05 M potassium phosphate buffer (pH 7.0). After centrifuging at 12,000 rpm for 15 min at 4°C, the supernatant was used for the measurement of POD and CAT activities. POD activity was determined using the previously described method by Morohashi (2002) and Morohashi et al. (2003). The 5 mL reaction mixture contained 0.1 mL of the supernatant, 1 mL of 0.5% (v/v) H2O2, 2.9 mL of 0.05 M potassium phosphate buffer (pH 5.5), and 1 mL of 0.05 M guaiacol as substrates. The oxidation of guaiacol was monitored by the absorbance measured at 470 nm every 10 s. CAT activity was confirmed using a Catalase Assay Kit (Jiancheng Bioengineering Company, Nanjing, China) according to the manufacturer’s instructions. H2O2 content was determined according to instruction available in commercial kit from Jiancheng Bioengineering Company (Nanjing, China). For ABA quantification, ABA extracted from 1 g of leaves (WT, transgenic lines under stress, and mock) as described by Jia et al. (2011).
RNA Extraction, cDNA Preparation, and qRT-PCR Analysis
Total RNA was extracted from all harvested samples using InvitrogenTM TRIzol® reagent (Thermo Fisher Scientific, New York, NY, United States) according to the manufacturer’s instruction. The RNA concentration was determined using NanoDrop Lite UV-Vis spectrophotometer (Thermo Fisher ScientificTM). The cDNA was synthesized with 2 μg of total RNA using PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRA, Japan). All the primers (Supplementary Table S1) used in this study were designed in primer premier 5 (PREMIER Biosoft International, Palo Alto CA, United States). The real-time PCR was performed using SYBR® Premix Ex TaqTM II (TliRNaseH Plus) (Clontech, TaKaRa, Shiga, Japan) in 96 well plate, Bio-Rad CFX system (Bio-Rad, United States). The relative changes in gene expression was calculated by adopting 2−Δ(ΔCT) method (Livak and Schmittgen, 2001) using SlUBI3 (Solyc01g056940) as an internal control. All the experiments were performed in triplicate.
Statistical Analysis
All the experiments were performed in triplicate, reproducible and were presented as means ± standard error (SE). Statistical analysis of data was performed using Sigmaplot 12.1. (SYSTAT and MYSTAT Products, United States, and Canada) and two-tailed Student’s t-tests for salinity and drought comparison or Dunnett’s tests were used to compare between WT, empty vector plants, and each overexpression line to determine significant differences. The significance values of p ≤ 0.01∗∗/0.05∗ were considered.
Results
SlbHLH22 Expression in Tomato WT Plants
To gain insight into the roles of SlbHLH22 in plant growth and development, we analyzed the expression profile of SlbHLH22 in various plant parts using qRT-PCR. The results suggested that SlbHLH22 expressed in all the tested tissues, but high expressions were found in leaves and flowers compared to root (Figure 1A). We further examined the expression of SlbHLH22 response to osmotic and oxidative stresses. WT plants were treated with NaCl, H2O2, and D-mannitol. The results indicated that the transcript of SlbHLH22 was upregulated after salt treatment across all time points (Figure 1B). For D-mannitol, SlbHLH22 was upregulated after exposure, but was peaked at 24 h time point (Figure 1C). For H2O2 was only peaked at 3 h interval and was downregulated in the remaining time points (Figure 1D). The observation of expression profile for SlbHLH22 indicated that SlbHLH22 gene might be very important for plant resistance against abiotic stress.
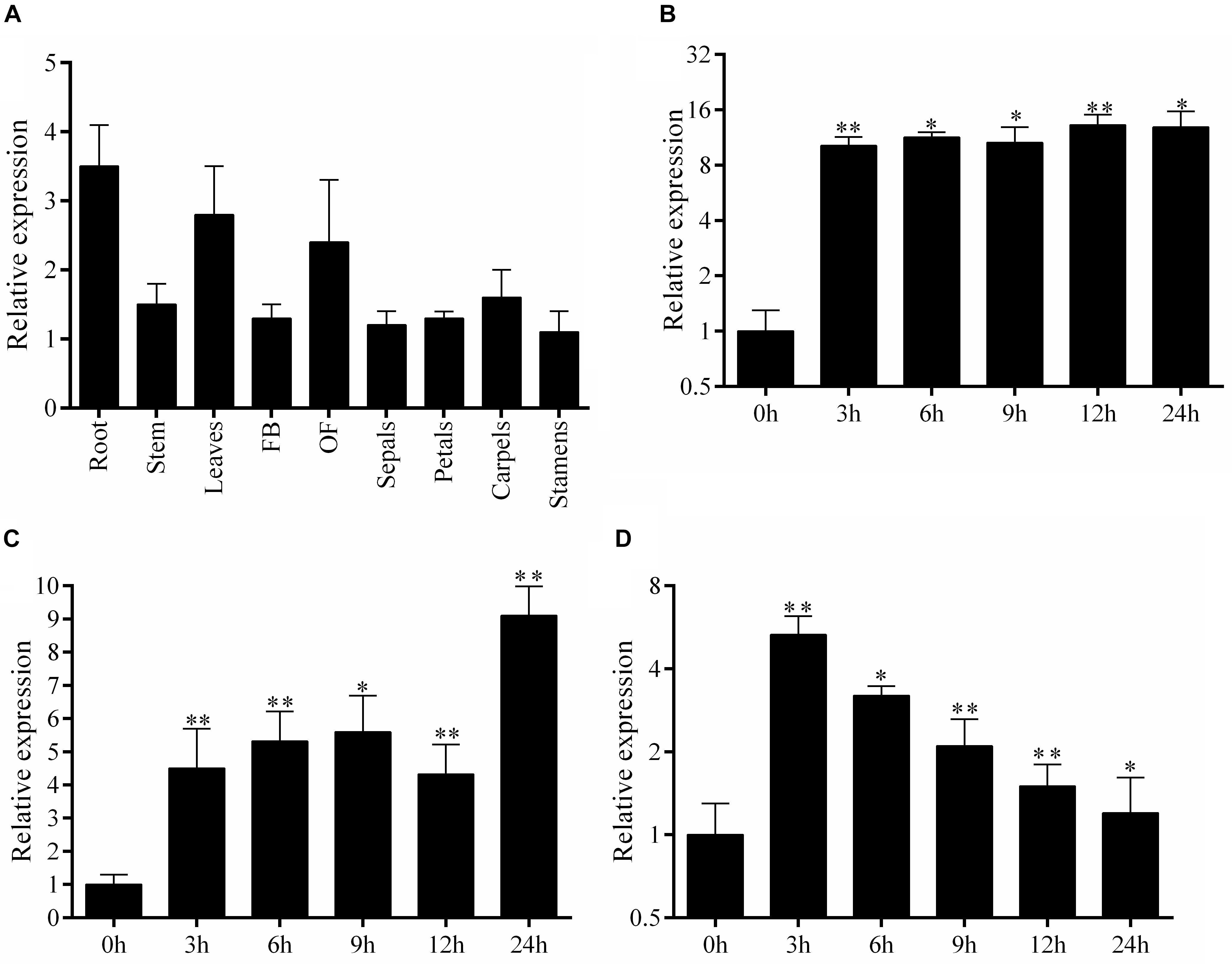
Figure 1. Endogenous expression profile of SlbHLH22 and expression levels under abiotic stress. (A) Expression profile of SlbHLH22 in different parts of WT plants, including root; stem; leaves; FB, flower bud; OF, opened flower; petals; sepals; stamens; and carpels. Expression analysis of SlbHLH22 from leaves of WT spray with (B) 200 mM NaCl, (C) 100 mM D-mannitol, (D) 100 mM H2O2 over 0 to 24 h time intervals. Student’s t-tests were used in comparison. Data represent mean of ± SE for three independent biological replicates (n = 3). ∗ and ∗∗ represent the significant difference at P-value ≤ 0.01∗∗/0.05∗.
SlbHLH22 Encodes a TF Targeted to the Nucleus
To determine the subcellular localisation of SlbHLH22 protein, the vector 35S-SlbHLH22-GFP was transiently expressed in living onion epidermal cells. Confocal imaging of protein fluorescence showed that the cells transformed with the vector containing GFP alone displayed fluorescence throughout the cells, whereas the green fluorescence signal of 35S-SlbHLH22-GFP was exclusively detected in the nucleus (Figure 2A). A Y2H experiment was used to examine the transcriptional activity of SlbHLH22. A GAL4 DNA-binding domain SlbHLH22 fusion protein was expressed in yeast cells, which were then assayed for their ability to activate transcription from the GAL4 sequence. SlbHLH22 promoted yeast growth in the absence of histidine and adenine, and showed X-α-gal activity, whereas the control vector pGBKT7 did not (Figure 2B). Moreover, string database was used to predict interaction network of SlbHLH22 with other proteins. It was found that tomato SlbHLH22 may interact with genes involved in flavonoid and ABA biosynthesis pathway include; CHS, CHI, PAL, PSY1, FLS, AAO, NCED, and ZEP (Supplementary Figure S1). Taken together, our results suggested that SlbHLH22 has transcriptional activity and is targeted to the nucleus in plant cells.
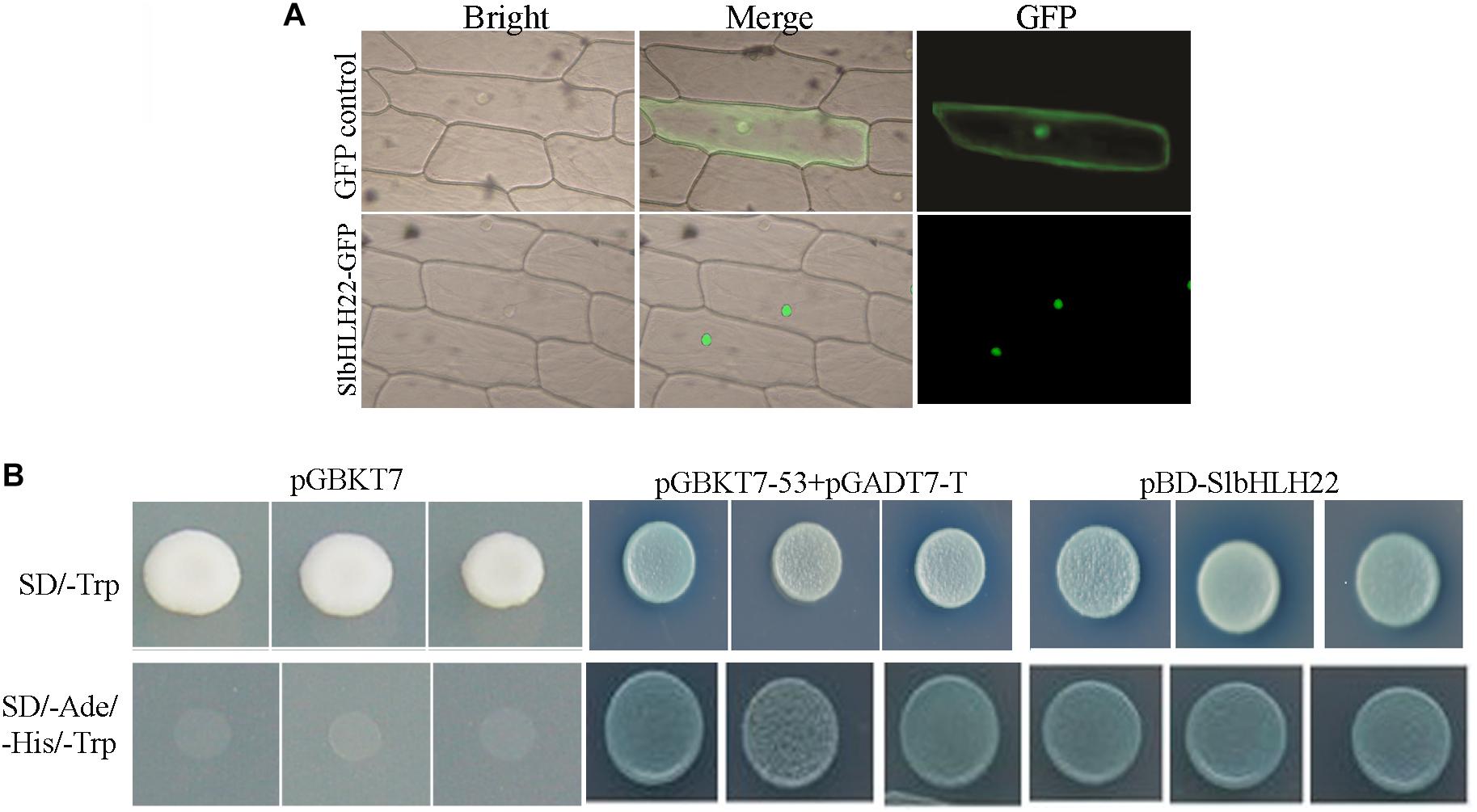
Figure 2. Subcellular localization and transcription activity of SlbHLH22. (A) The images were taken under bright light, merged, and in the dark field for the GFP-derived green fluorescence, respectively. (B) Analysis of the transactivation activity of SlbHLH22. Up, SD/-Trp-drop medium, below, SD/-Ade/-His/-Trp-drop medium both supplemented with X-gal for assaying yeast reporter (MEL1) gene.
Phenotypic Characterization of Transgenic Tomato With SlbHLH22
To further study the function of SlbHLH22 gene, the transgenic plant lines were generated by overexpressing the ORF and RNAi silencing by agrobacterium-mediated transformation. Three independent transgenic lines for overexpression (L18, L20, L23) and two RNAi lines (L14i and L19i) were detected exhibiting significant changes in expression fold (Figure 3A). Two transgenic plant lines L18 and L23 for overexpression and two lines for RNAi L14i and L19i were selected for further characterization. The transgenic plants showed pleiotropic phenotypes, such as plant height and leaf size (Figures 3B,C). Our results displayed that the tomato SlbHLH22 have remarkable effects on development of tomato plant.
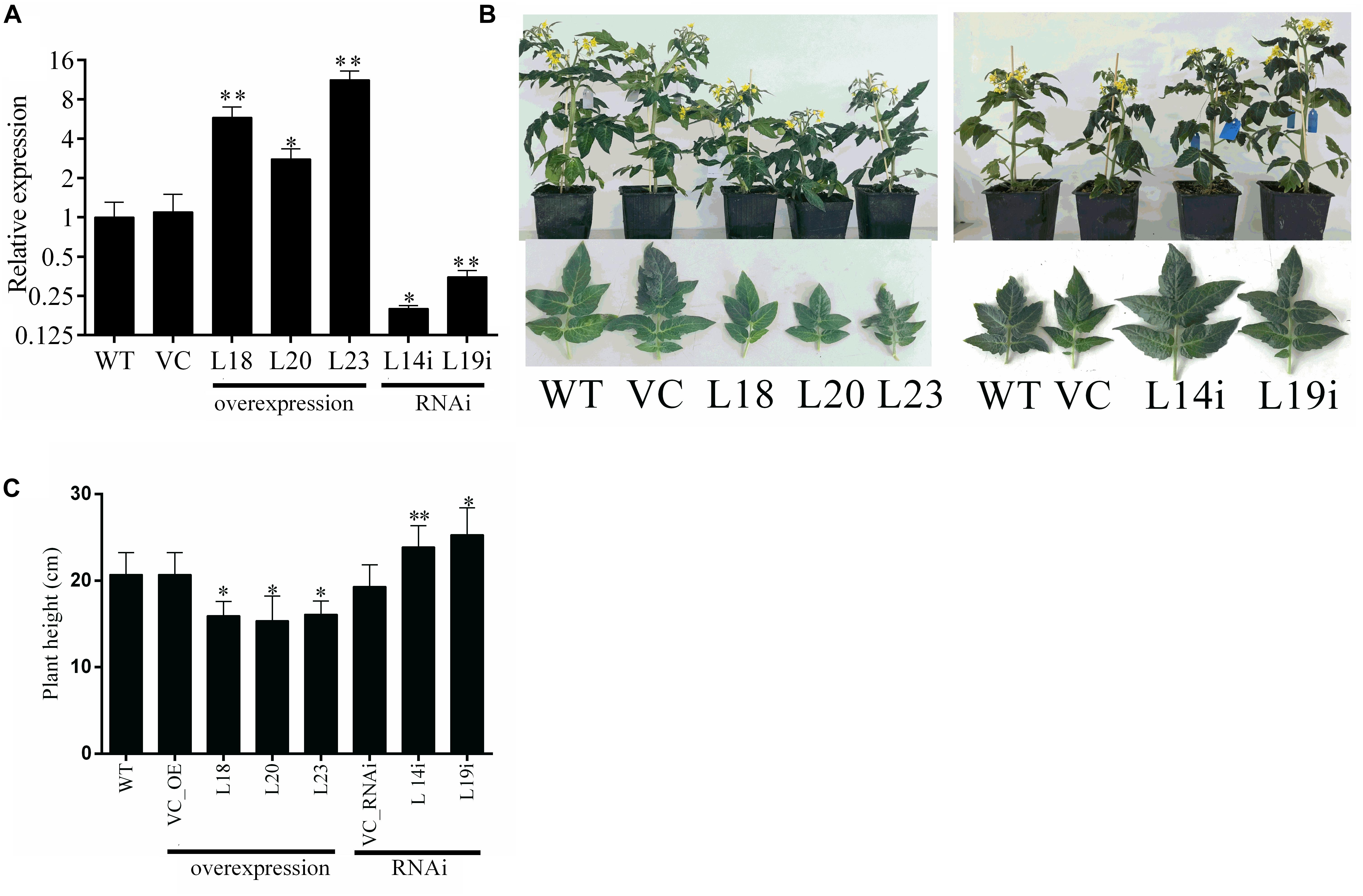
Figure 3. Transgenic line generation and phenotypic characterization of WT, empty vector, and transgenic plants. (A) Expression levels of SlbHLH22 in WT, empty vector (VC), and transgenic lines (L18, L20, L23) overexpressing and RNAi-silencing (L14i and L19i) of SlbHLH22 in leaves. Dunnett’s tests was used in the comparison between WT, VC, and SlbHLH22 overexpression lines (L18, L20, L23) (B) 6-week-old WT, VC, and three independent transgenic lines (L18, L20, L23, L14i, and L19i) plants. Phenotype of fifth leaves in WT, empty vector (VC) plants, and in SlbHLH22 transgenic line plants (C) Height of plants WT, VC, and transgenic lines. Data represent mean of ± SE for three independent biological replicates (n = 3) and each replicate contained 15 plants. ∗ and ∗∗ represent the significant difference as determined by t-test (P-value ≤ 0.01∗∗/0.05∗).
SlbHLH22 Enhances Transgenic Tomato Plant Resistance to Salt and Drought
As it was observed, SlbHLH22 expressions was activated by salinity. Thus, we hypothesized that SlbHLH22 might increase the resistance of transgenic plants to salinity and drought. To prove it, we explore the role the performance of transgenic and WT plants treated with salt solution for 18 days and deprived of water for 30 days. As shown in Figure 4, significant morphological changes were observed between transgenic plant lines with SlbHLH22 and WT plants after stresses. However, after stresses, the overexpression plant lines showed slight changes in their physical appearance, but RNAi lines and WT plants showed typical severe desiccation symptoms (Figure 4). In comparison, WT and RANi lines plants under drought showed severe damages than under salt treatment. However, upon exposure to normal conditions for 10 days, the overexpression plants recover very fast, but RNAi and WT plants under drought stress was unable to recover (Figure 4). The relative water content (RWC %) and total chlorophyll content was decreased in WT and transgenic lines during treatment, but relatively higher in the overexpression plant lines (Supplementary Figures S2A,B).
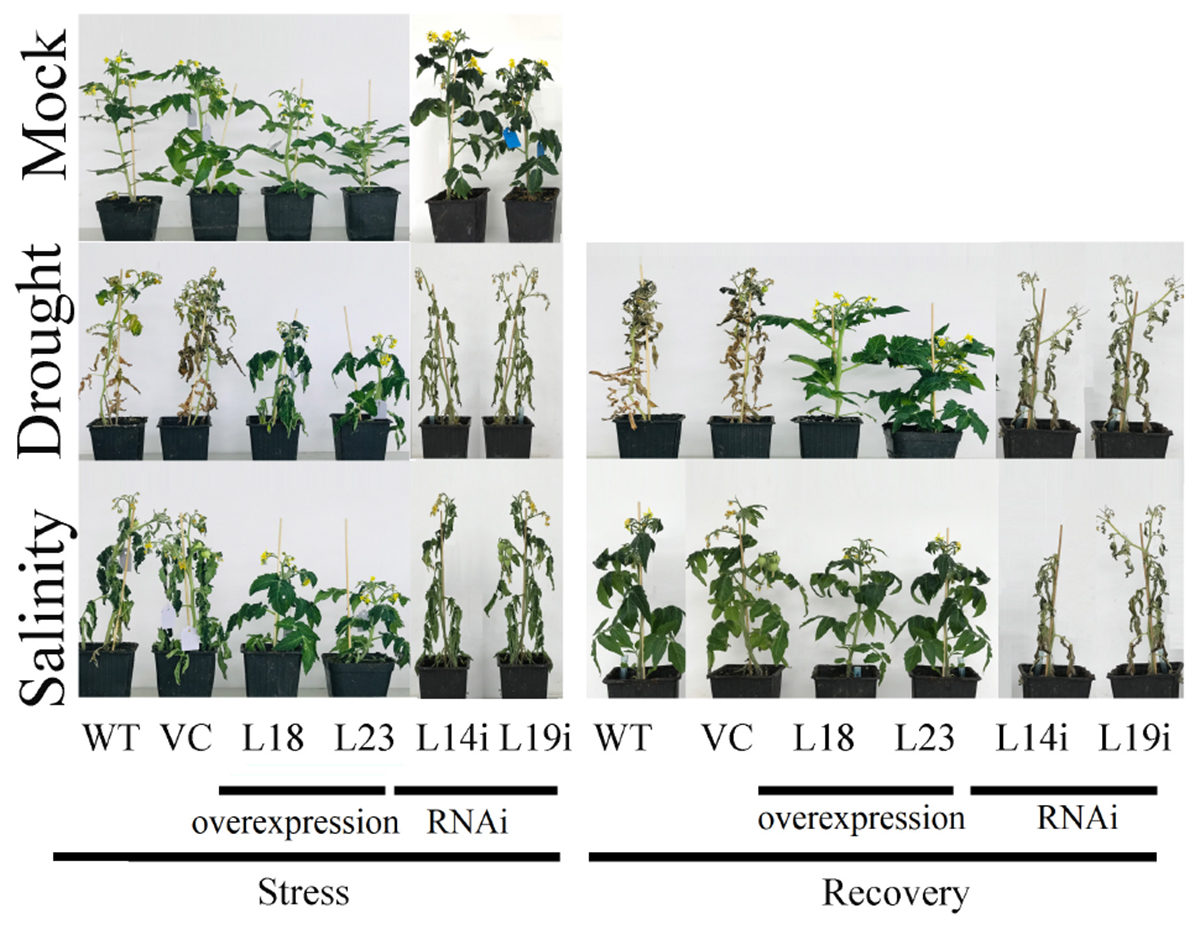
Figure 4. Drought and salinity assay. Phenotypes obtained after 30 days drought and 18 days salinity stress and recovery from drought and salinity for 10 days.
Upregulation of Flavonoid Accumulation Under Salt and Drought Stress
The total flavonoid contents were assessed in WT and transgenic lines under normal conditions and under salinity and drought stresses. It was found that the total flavonoid contents in overexpression plant lines were induced and suppressed significantly in RNAi lines compared to WT (Supplementary Figure S3). The expression levels of genes in the flavonoid biosynthesis pathway were further analyzed at molecular level in the overexpression and RNAi lines. The results indicated that the transcript levels of the flavonoid biosynthesis genes, such as SlCHS, SlCHI, SlF3’H, SlF3H, SlFLS, and SlPAL were peaked in overexpression lines than in WT. In SlbHLH22-RNAi lines the expression of all genes significantly downregulated than in WT plants (Supplementary Figure S3). These results demonstrating that SlbHLH22 affect flavonoid accumulation by modulating flavonoid biosynthesis pathway.
Improved Antioxidant Activity in Transgenic Plants
Salinity and drought lead to production of reactive oxygen species (ROS), that cause damages to membrane structure (Zhai et al., 2016). We investigated the changes in the accumulation of H2O2 in transgenic lines and WT plant under salinity and drought stresses. It was found that more H2O2 accumulated in WT plant than in transgenic lines (Figure 5). To explore the possible physiological mechanism responsible for the increased drought and salt tolerance, we compared the changes in contents of proline, MDA concentration, and total soluble sugars in the leaves from transgenic and WT plants grown under normal and stress conditions. MDA content was significantly peaked in RNAi-lines and WT plants under drought and salt stresses compared to overexpression lines (Figure 5). It was examined that the soluble sugars were accumulated more in overexpression lines than in RNAi-lines and WT plant, after salt and drought stresses (Figure 5). Furthermore, we examined the enzyme activities of antioxidants enzymes such as SOD, POD, and CAT of the leaves from transgenic plant lines and WT plant under stresses and normal growth. The enzymatic activities of SOD, POD, and CAT in transgenic lines and WT were almost same under normal conditions. For drought stress, the activities of CAT and SOD were significantly upregulated in overexpression lines as compared with WT and SlbHLH22-RNAi lines. However, POD activity was 10 points more in SlbHLH22 overexpression lines than in the WT and RNAi (Figure 5). The activities of SOD, POD, and CAT were significantly upregulated in overexpression lines under salt stress in compassion with the mocked corresponding transgenic lines, RNAi lines, and WT plants. The proline contents were increased in overexpression plant lines under drought and salt stress in mocked transgenic lines but peaked in transgenic lines under salt stress. However, in RNAi lines proline content was significantly downregulated than WT plants. It was found that the overexpression plant accumulated more prolines contents under salinity stress than under drought (Figure 5). Collectively, our results show that SlbHLH22 in tomato can help improve the resistance of transgenic plant to salinity and drought stresses.
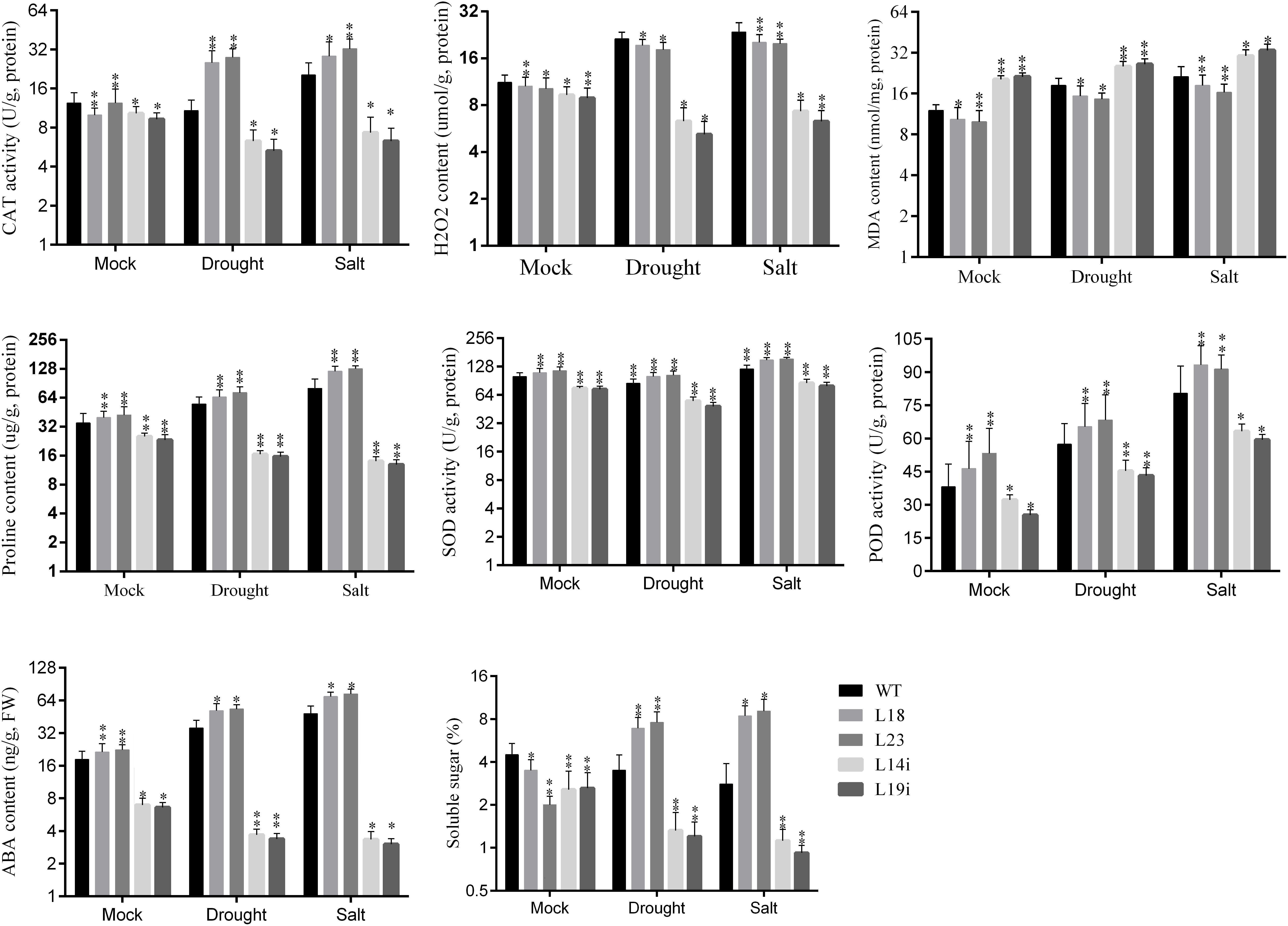
Figure 5. ABA content, soluble sugar, Malondialdehyde (MDA), hydrogen peroxide (H2O2) and antioxidant enzymes [catalase (CAT), superoxide dismutase (SOD), and peroxidase (POD)] activities in WT and transgenic lines (L18 and L23) under drought and salinity. Data represent mean of ± SE for three independent biological replicates (n = 3) and each replicate contained 6 plants. ∗ and ∗∗ represent the significant difference as determined by t-test (P-value ≤ 0.01∗∗/0.05∗).
Up-Regulation of ABA and Stress Related Pathways in Tomato
The overexpression of SlbHLH22 improved plant lines resistance against salinity and drought leads us to examine whether the overexpression of SlbHLH22 affects the endogenous level of ABA and its biosynthesis genes in transgenic tomato. We measured the ABA levels in leaves of transgenic lines overexpressing SlbHLH22 and WT plants under normal growth conditions and under stresses. The results showed a significant difference in ABA levels in WT and transgenic lines under normal conditions. However, the endogenous ABA contents was significantly higher in transgenic lines than that in the WT under salt and drought stresses (Figure 5). To further insight into the role of SlbHLH22 in ABA biosynthesis, we examined ABA levels by generating RNAi lines. It was observed that ABA level in RNAi is downregulated than WT under mock and salinity and drought stress (Figure 5). To ascertain the molecular mechanism involved in ABA biosynthesis, we analyzed the expression level of ABA biosynthesis genes between transgenic lines and WT plants. The results indicated that the expression of genes involved in ABA biosynthesis, such as SlAAO, SlABA2, SlNCED, and SlZEP were upregulated in transgenic lines under salt and drought than in WT and RNAi lines (Figure 6). Meanwhile, we investigated the expression profiles of genes involved in ROS scavenging system, including SlCAT, SlPOD, SlSOD (Supplementary Figure S4), and proline biosynthesis such as SlP5CS and SlP5CR (Supplementary Figure S4) in WT and transgenic lines under normal and stress conditions. The results suggested minor changes in the expression levels of SlCAT, SlPOD, and SlSOD in WT plants under normal and stress conditions but significantly downregulated in SlbHLH22-RNAi lines. Moreover, the transcripts of SlCAT, SlPOD, and SlSOD accumulated more in overexpression lines under stresses than WT and mocked corresponding overexpression plant lines grown under normal conditions (Supplementary Figure S4). SlP5CS and SlP5CR are two key genes in proline biosynthesis. The expression levels of SlP5CS and SlP5CR were upregulated under stress in WT plants, but more high transcript levels were detected in overexpressing plant lines grown under stress treatments (Supplementary Figure S4). These observations suggested that tomato SlbHLH22 may enhance tomato resistance to salt and drought stresses through ABA and/or other pathways.
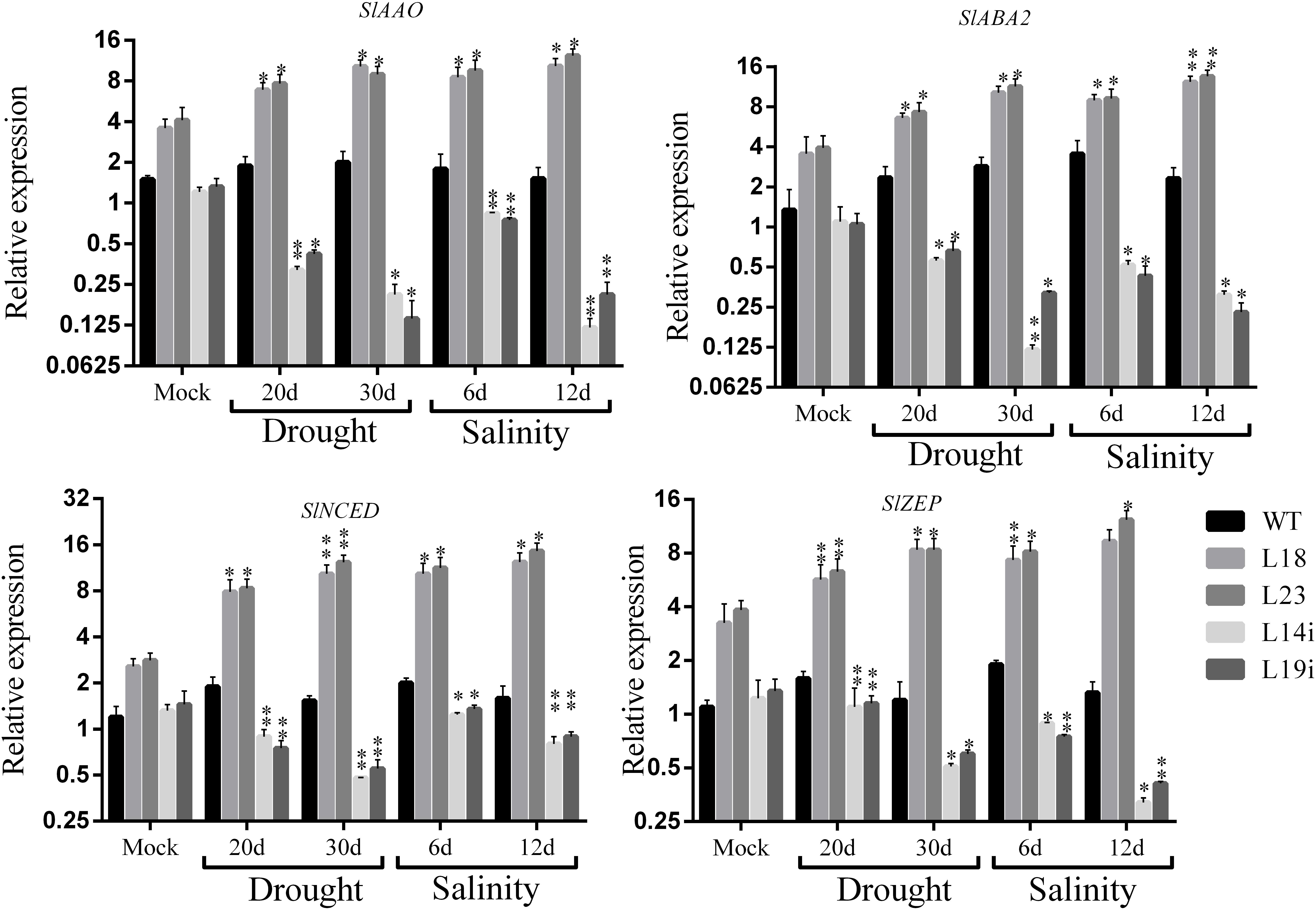
Figure 6. Expression profile of ABA biosynthesis related genes in WT and transgenic lines (L18 and L23) under drought and salinity. Data represent mean of ± SE for three independent biological replicates (n = 3). ∗ and ∗∗ represent the significant difference as determined by t-test (P-value ≤ 0.01∗∗/0.05∗).
Discussion
Plants, being sessile organisms encounter a range of biotic and abiotic stresses, such as pathogen invasion, temperature, light, drought, salinity, and so on. These stresses negatively affect growth, yield, and survival rate of plants (Sharma et al., 2013). Plants develop various mechanisms to counter the effects of these stresses, which enables them to adopt under such conditions (Gerszberg and Hnatuszko-Konka, 2017). Molecular biology and biotechnology approaches enables us to find out large number of genes, including transcription factors (TFs) that play critical roles in spatial and temporal genes expression during stress resistance which could be induced by one or more abiotic and biotic stresses (Ashrafi-Dehkordi et al., 2018). Moreover, the functions of most TFs are still unknown. The current study is important in elucidating the roles of a basic helix-loop-helix transcription factor gene, SlbHLH22, in tomato in response to salinity and drought stresses.
The results of subcellular localization of SlbHLH22-GFP show that the GFP signal was in the nucleus (Figure 2A), which suggests that SlbHLH22 might function in nucleus. Transcriptional activation assay suggested that SlbHLH22 showed transcriptional activation ability (Figure 2B). Further investigation should be performed to examine whether SlbHLH22 protein can function as activators or repressors of transcription in plants. In this study, phenotype analysis showed that the transgenic plant lines displayed changes in plant height and in leaves (Figure 3B) and altered flavonoid accumulation (Supplementary Figure S3). The plants overexpressing SlbHLH22 showed delay symptoms of necrosis, wilting, and leaf senescence than in SlbHLH22-RNAi and WT plants grown under salinity and drought stresses (Figure 4). The total chlorophyll and relative water content in leaves remain high in overexpressing lines (Supplementary Figures S2A,B). These findings suggested that the overexpression of SlbHLH22 enhances the tomato plant tolerance to salinity and drought. Flavonoids are a diverse group of naturally occurring secondary metabolites in plants, have strong antioxidant capacity (Verhoeyen et al., 2002). Flavonoids can enhance plant tolerance to drought and salinity stresses due to their ability to remove superoxide, peroxides, and free radicals produced during stress (Balasundram et al., 2006; Gao et al., 2011). For an instance, in Vitis vinifera, the overexpression of VvbHLH01 in Arabidopsis increased resistance to drought and salinity through improves flavonoids accumulation (Wang et al., 2016). The flavonoid biosynthesis genes such as CHS, F3H, FDR were also upregulated in transgenic Arabidopsis plants overexpressing AtbHLH8 (Shin et al., 2007). Thus, we hypothesized that the accumulation of flavonoids can enhance tolerance to osmotic and oxidative stresses. In order to validate it, the tomato transgenic plant lines were subjected to these stresses. It was observed that plants overexpressing SlbHLH22 accumulate more flavonoids and the expression levels of flavonoid biosynthesis genes were upregulated in transgenic plant lines grown under salinity and drought stress than in WT and silencing lines plant (Supplementary Figure S3). This implying that higher the flavonoid accumulation more will be the tolerance to the oxidative stresses.
Abscisic acid is a prime mediator known to regulate various plant physiological processes in adaptive responses to abiotic and biotics stresses (Umezawa et al., 2010). ABA regulates the expression of various stress-induced genes involved in proline, carbohydrate, and LEA biosynthesis that help plant to maintain the cellular water content and protect cellular proteins or enzymes (Verslues et al., 2006; Sreenivasulu et al., 2012). In our study, the ABA significantly peak under saline and drought stresses (Figure 6) and genes involved in ABA biosynthesis, such as SlAAO, SlABA2, SlZEP, and SlNCED were also upregulated (Figure 6). Similarly, this net upregulation of ABA can promote the expression of proline biosynthesis genes such as SlP5CS and SlP5CR, the key genes in proline biosynthesis (Supplementary Figure S5) and encourage soluble sugar content accumulation (Figure 5). We supposed that the overexpression of SlbHLH22 enhances the plant ability to scavenge reactive oxygen species (ROS). In plants, ROS scavenging enzymes such as POD, SOD, and CAT helps to minimize osmotic and oxidative damages to plasma membrane integrity, proteins, and cellular enzymes (Zhang et al., 2012; Zhai et al., 2016). It was found that H2O2 accumulated more in WT plants then the transgenic plants grown under salinity and drought, (Figure 5). This subsequently leads to systematic upregulation of ROS scavenging genes, such as SlSOD, SlPOD, and SlCAT in WT plants (Supplementary Figure S4). Moreover, the elevated levels of MDA damages the integrity of phospholipid bilayer membranes, which reduces plant tolerance to salt and drought (Zou et al., 2012). The MDA levels were decreased in tomato transgenic plant lines overexpressing SlbHLH22 (Figure 5). Thus, our results supported the fact that the overexpression of SlbHLH22 enhances tomato tolerance to drought and salinity due to elevated levels of ABA. The upregulation of ABA, proline biosynthesis genes, and genes involved in ROS scavenging system lead to enhance ability of transgenic tomato plants to cope with applied stress (Figure 7).
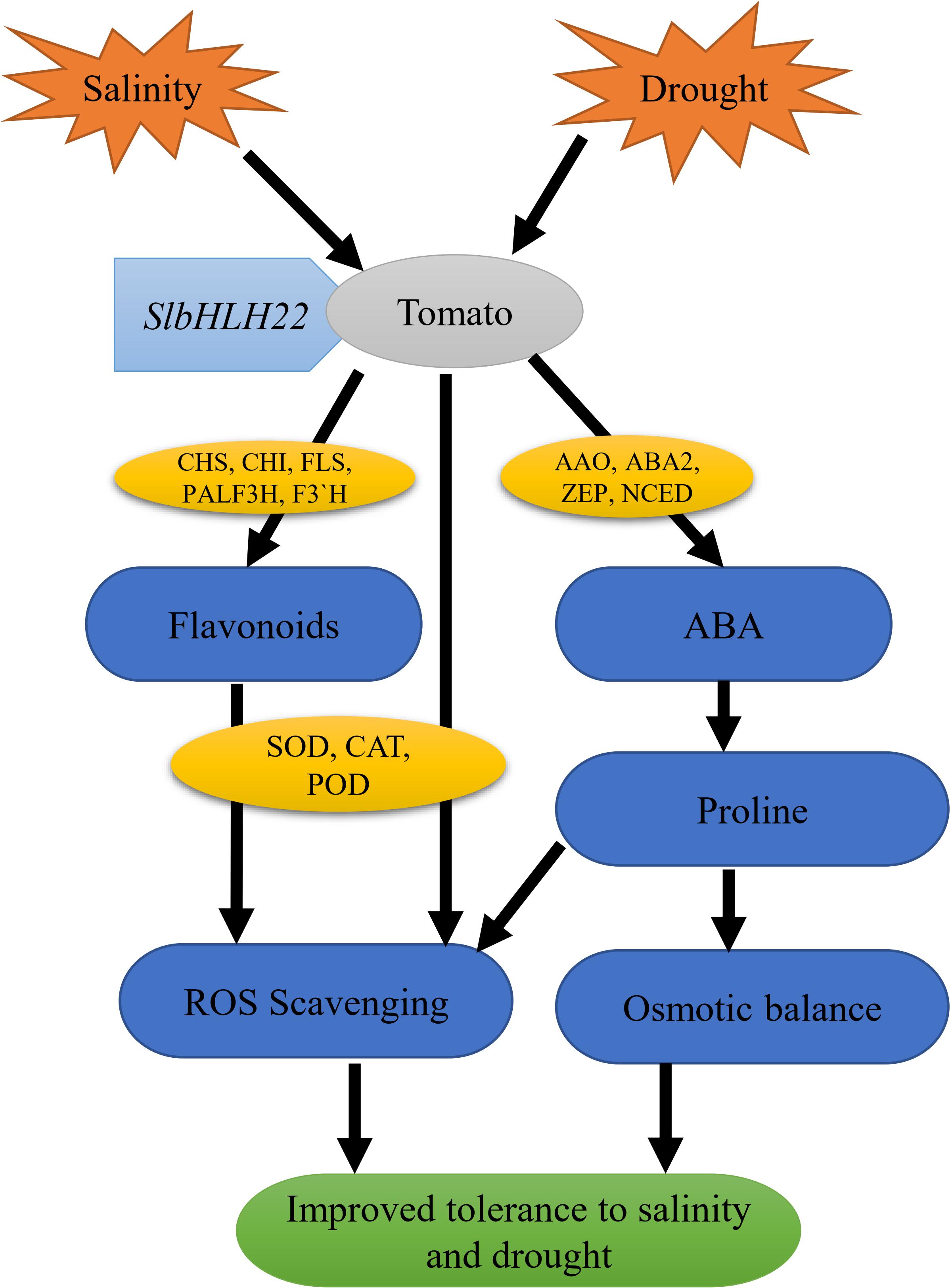
Figure 7. Hypothetical model of SlbHLH22 gene involved in improving drought and salinity tolerance in tomato. Overexpression of SlbHLH22 induced the expression of genes involved in flavonoid biosynthesis, ABA biosynthesis pathway and ROS scavenging genes. This subsequently resulted in altered physiology such as increased ABA accumulation, proline content, and enhanced CAT, POD, SOD activities with reduced ROS accumulation. This led to improve tolerance under abiotic stress.
Conclusion
In summary, the overexpression of tomato bHLH TF gene, SlbHLH22, enhances resistance to drought and salinity by increased in flavonoids accumulation, ABA accumulation and ABA-induced-stress related pathways. All these physiological changes lead to improve plant ability to survive under abiotic stress conditions. This study not only provides the evidences of bHLH roles in resisting abiotic stresses but, also helps to improve our understanding about their role in abiotic stresses.
Data Availability
No datasets were generated or analyzed for this study.
Author Contributions
MW designed and performed all the experiments and data analysis. XR performed the data analysis. MW drafted the manuscript. ZL revised the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Key Research and Development Program (2016YFD0400101) and the National Natural Science Foundation of China (Nos. 31772370 and 31572175).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00734/full#supplementary-material
FIGURE S1 | In silico bioinformatics analysis of SlbHLH22 protein interaction network predicted using STRING program.
FIGURE S2 | Relative water and chlorophyll content of WT and transgenic line plants under salinity and drought stresses. Relative water content (%), total chlorophyll content during various time points under (A) salinity and (B) drought stresses, respectively. Data represent mean of ± SE for three independent biological replicates (n = 3) and each replicate contained 15 plants. ∗ and ∗∗ represent the significant difference as determined by t-test (P-value ≤ 0.01∗∗/0.05∗).
FIGURE S3 | Flavonoid content and expression profile of flavonoid biosynthesis genes in WT and transgenic lines under drought and salinity. Data represent mean of ± SE for three independent biological replicates (n = 3). ∗ and ∗∗ represent the significant difference as determined by t-test (P-value ≤ 0.01∗∗/0.05∗). 1Expressed in mg quercetin equivalent/g dry weight.
FIGURE S4 | Expression profile of ROS scavenging related genes catalase (SlCAT) peroxidase (SlPOD), superoxide dismutase (SlSOD) genes in WT and transgenic lines under drought and salinity. Data represent mean of ± SE for three independent biological replicates (n = 3). ∗ and ∗∗ represent the significant difference as determined by t-test (P-value ≤ 0.01∗∗/0.05∗).
FIGURE S5 | Expression profile of proline biosynthesis genes in WT and transgenic lines under drought and salinity. Data represent mean of ± SE for three independent biological replicates (n = 3). ∗ and ∗∗ represent the significant difference as determined by t-test (P-value 0.01∗∗/0.05∗).
TABLE S1 | List of primer used in cloning and qRT-PCR for the expression analysis in this study.
Abbreviations
ABA, abscisic acid; bHLH, basic helix-loop-helix; CAT, catalase; CBF, C-repeat binding factor; DREB, dehydration responsive element-binding protein; H2O2, hydrogen peroxide; MDA, Malondialdehyde; POD, peroxide dismutase; ROS, reactive oxygen scavenging system; SOD, superoxide dismutase; TFs, transcription factors.
References
Ashrafi-Dehkordi, E., Alemzadeh, A., Tanaka, N., and Razi, H. (2018). Meta-analysis of transcriptomic responses to biotic and abiotic stress in tomato. Peer J. 6, e4631. doi: 10.7717/peerj.4631
Balasundram, N., Sundram, K., and Samman, S. (2006). Phenolic compounds in plants and agri-industrial by-products: antioxidant activity, occurrence, and potential uses. Food Chem. 99, 191–203. doi: 10.1016/j.foodchem.2005.07.042
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1016/j.dental.2010.07.006
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313x.2004.02138.x
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martínez-García, J. F., Bilbao-Castro, J. R., and Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Fukao, T., Xu, K., Ronald, P. C., and Bailey-Serres, J. (2006). A variable cluster of ethylene response factor–like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18:2021. doi: 10.1105/tpc.106.043000
Gao, J.-J., Zhang, Z., Peng, R.-H., Xiong, A.-S., Xu, J., Zhu, B., et al. (2011). Forced expression of Mdmyb10, a myb transcription factor gene from apple, enhances tolerance to osmotic stress in transgenic Arabidopsis. Mol. Biol. Rep. 38, 205–211. doi: 10.1007/s11033-010-0096-0
Gerszberg, A., and Hnatuszko-Konka, K. (2017). Tomato tolerance to abiotic stress: a review of most often engineered target sequences. Plant Growth Regul. 83, 175–198. doi: 10.1007/s10725-017-0251-x
Han, G., Wang, M., Yuan, F., Sui, N., Song, J., and Wang, B. (2014). The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana. Plant Mol. Biol. 86, 237–253. doi: 10.1007/s11103-014-0226-5
Heath, R. L., and Packer, L. (1968). Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. doi: 10.1016/0003-9861(68)90654-1
Huang, X., Li, K., Jin, C., and Zhang, S. (2015). ICE1 of Pyrus ussuriensis functions in cold tolerance by enhancing PuDREBa transcriptional levels through interacting with PuHHP1. Sci. Rep. 5:17620. doi: 10.1038/srep17620
Jia, H.-F., Chai, Y.-M., Li, C.-L., Lu, D., Luo, J.-J., Qin, L., et al. (2011). Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 157, 188–199. doi: 10.1104/pp.111.177311
Koolen, H. H. F., da Silva, F. M. A., Gozzo, F. C., de Souza, A. Q. L., and de Souza, A. D. L. (2013). Antioxidant, antimicrobial activities and characterization of phenolic compounds from buriti (Mauritia flexuosa L. f.) by UPLC–ESI-MS/MS. Food Res. Int. 51, 467–473. doi: 10.1016/j.foodres.2013.01.039
Liu, W., Tai, H., Li, S., Gao, W., Zhao, M., Xie, C., et al. (2014). bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 201, 1192–1204. doi: 10.1111/nph.12607
Liu, Y., Ji, X., Nie, X., Qu, M., Zheng, L., Tan, Z., et al. (2015). Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 207, 692–709. doi: 10.1111/nph.13387
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Man, L., Xiang, D., Wang, L., Zhang, W., Wang, X., and Qi, G. (2017). Stress-responsive gene RsICE1 from Raphanus sativus increases cold tolerance in rice. Protoplasma 254, 945–956. doi: 10.1007/s00709-016-1004-9
Mittova, V., Volokita, M., Guy, M., and Tal, M. (2000). Activities of SOD and the ascorbate-glutathione cycle enzymes in subcellular compartments in leaves and roots of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii. Physiol. Plant. 110, 42–51. doi: 10.1034/j.1399-3054.2000.110106.x
Morohashi, K., Minami, M., Takase, H., Hotta, Y., and Hiratsuka, K. (2003). Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression. J. Biol. Chem. 278, 20865–20873. doi: 10.1074/jbc.m301712200
Morohashi, Y. (2002). Peroxidase activity develops in the micropylar endosperm of tomato seeds prior to radicle protrusion. J. Exp. Bot. 53, 1643–1650. doi: 10.1093/jxb/erf012
Nakashima, K., Ito, Y., and Yamaguchi-Shinozaki, K. (2009). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. doi: 10.1104/pp.108.129791
Pan, Y., Seymour, G. B., Lu, C., Hu, Z., Chen, X., and Chen, G. (2012). An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 31, 349–360. doi: 10.1007/s00299-011-1170-3
Pang, C. H., Li, K., and Wang, B. (2011). Overexpression of SsCHLAPXs confers protection against oxidative stress induced by high light in transgenic Arabidopsis thaliana. Physiol. Plant 143, 355–366. doi: 10.1111/j.1399-3054.2011.01515.x
Peng, H.-H., Shan, W., Kuang, J.-F., Lu, W.-J., and Chen, J.-Y. (2013). Molecular characterization of cold-responsive basic helix-loop-helix transcription factors MabHLHs that interact with MaICE1 in banana fruit. Planta 238, 937–953. doi: 10.1007/s00425-013-1944-7
Seo, J. S., Joo, J., Kim, M. J., Kim, Y. K., Nahm, B. H., Song, S. I., et al. (2011). OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65, 907–921. doi: 10.1111/j.1365-313X.2010.04477.x
Sharma, R., De Vleesschauwer, D., Sharma, M. K., and Ronald, P. C. (2013). Recent advances in dissecting stress-regulatory crosstalk in rice. Mol. Plant 6, 250–260. doi: 10.1093/mp/sss147
Shin, J., Park, E., and Choi, G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49, 981–994. doi: 10.1111/j.1365-313x.2006.03021.x
Sreenivasulu, N., Harshavardhan, V. T., Govind, G., Seiler, C., and Kohli, A. (2012). Contrapuntal role of ABA: does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 506, 265–273. doi: 10.1016/j.gene.2012.06.076
Sun, H., Fan, H. J., and Ling, H. Q. (2015). Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics 16:9. doi: 10.1186/s12864-014-1209-2
Sun, W., Cao, Z., Li, Y., Yanxiu, Z., and Hui, Z. (2007). A simple and effective method for protein subcellular localization using Agrobacterium-mediated transformation of onion epidermal cells. Biologia 62, 529–532. doi: 10.2478/s11756-007-0104-6
Thomashow, M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Umezawa, T., Nakashima, K., Miyakawa, T., Kuromori, T., Tanokura, M., Shinozaki, K., et al. (2010). Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839. doi: 10.1093/pcp/pcq156
Verhoeyen, M. E., Bovy, A., Collins, G., Muir, S., Robinson, S., de Vos, C. H. R., et al. (2002). Increasing antioxidant levels in tomatoes through modification of the flavonoid biosynthetic pathway. J. Exp. Bot. 53, 2099–2106. doi: 10.1093/jxb/erf044
Verslues, P. E., Agarwal, M., Katiyar-Agarwal, S., Zhu, J., and Zhu, J. K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45, 523–539. doi: 10.1111/j.1365-313x.2005.02593.x
Wang, F., Zhu, H., Chen, D., Li, Z., Peng, R., and Yao, Q. (2016). A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 125, 387–398. doi: 10.1007/s11240-016-0953-1
Wang, P., Su, L., Gao, H., Jiang, X., Wu, X., Li, Y., et al. (2018). Genome-wide characterization of bHLH genes in grape and analysis of their potential relevance to abiotic stress tolerance and secondary metabolite biosynthesis. Front. Plant Sci. 9:64. doi: 10.3389/fpls.2018.00064
Waseem, M., and Ahmad, F. (2019). The phosphoenolpyruvate carboxylase gene family identification and expression analysis under abiotic and phytohormone stresses in Solanum lycopersicum L. Gene 690, 11–20. doi: 10.1016/j.gene.2018.12.033
Waseem, M., Ahmad, F., Habib, S., Gao, Y., and Li, Z. (2018). Genome-wide identification of FK506-binding domain protein gene family, its characterization, and expression analysis in tomato (Solanum lycopersicum L.). Gene 678, 143–154. doi: 10.1016/j.gene.2018.08.021
Waseem, M., Li, N., Su, D., Chen, J., and Li, Z. (2019). Overexpression of a basic helix-loop-helix transcription factor gene, SlbHLH22,. (promotes)early flowering and accelerates fruit ripening in tomato (Solanum lycopersicum L.). Planta [Epub ahead of print].
Waseem, M., and Li, Z. (2019). Overexpression of tomato SlbHLH22 transcription factor gene enhances fruit sensitivity to exogenous phytohormones, and shortens fruit shelf-life. J. Biotechnol. 299, 50–56. doi: 10.1016/j.jbiotec.2019.04.012
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126:485. doi: 10.1104/pp.126.2.485
Xian, Z., Huang, W., Yang, Y., Tang, N., Zhang, C., Ren, M., et al. (2014). miR168 influences phase transition, leaf epinasty, and fruit development via SlAGO1s in tomato. J. Exp. Bot. 65, 6655–6666. doi: 10.1093/jxb/eru387
Zhai, H., Wang, F., Si, Z., Huo, J., Xing, L., An, Y., et al. (2016). A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol. J. 14, 592–602. doi: 10.1111/pbi.12402
Zhang, C., Feng, R., Ma, R., Shen, Z., Cai, Z., Song, Z., et al. (2018). Genome-wide analysis of basic helix-loop-helix superfamily members in peach. PLoS One 13:e0195974. doi: 10.1371/journal.pone.0195974
Zhang, H., Han, B., Wang, T., Chen, S., Li, H., Zhang, Y., et al. (2012). Mechanisms of plant salt response: insights from proteomics. J. Proteome Res. 11, 49–67. doi: 10.1021/pr200861w
Zhang, T., Lv, W., Zhang, H., Ma, L., Li, P., Ge, L., et al. (2018). Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) transcription factor family in maize. BMC Plant Biol. 18:235. doi: 10.1186/s12870-018-1441-z
Keywords: tomato, ROS scavenging system, proline, flavonoids, tolerance, drought, salinity
Citation: Waseem M, Rong X and Li Z (2019) Dissecting the Role of a Basic Helix-Loop-Helix Transcription Factor, SlbHLH22, Under Salt and Drought Stresses in Transgenic Solanum lycopersicum L. Front. Plant Sci. 10:734. doi: 10.3389/fpls.2019.00734
Received: 18 February 2019; Accepted: 16 May 2019;
Published: 04 June 2019.
Edited by:
Paula Casati, National Council for Scientific and Technical Research (CONICET), ArgentinaReviewed by:
Guangyuan He, Huazhong University of Science and Technology, ChinaHaitao Shi, Hainan University, China
Copyright © 2019 Waseem, Rong and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhengguo Li, emhlbmdndW9saUBjcXUuZWR1LmNu orcid.org/0000-0002-4643-9540
 Muhammad Waseem
Muhammad Waseem Xiangyi Rong
Xiangyi Rong Zhengguo Li
Zhengguo Li